Abstract
The mouse homologue of the human receptor-interacting protein 140 (RIP140) was isolated from a mouse embryonic cDNA library in yeast two-hybrid screening experiments by using the ligand binding domain (LBD) of nuclear orphan receptor TR2 as the bait. The receptor-interacting domains of mouse RIP140 were mapped to the regions containing the LXXLL motif (where L is leucine and X is any amino acid), and the RIP140-interacting domain of TR2 was mapped to its C-terminal 10- to 20-amino-acid sequence, a putative activation function 2 (AF-2) region. In a GAL4 reporter system and a reporter driven by the proximal region of the TR2 promoter, RIP140 functioned as a corepressor for both a GAL4 DNA binding domain (BD)–TR2 fusion and the wild-type receptor. When tethered to the BD of GAL4, RIP140 exerted a trans-repressive effect on the GAL4 reporter. In addition, RIP140 suppressed the retinoic acid (RA) receptor-mediated RA induction in a dose-dependent manner. Finally, it was demonstrated that in the presence of RIP140, a cytosolic, green fluorescent protein-tagged TR2 LBD translocated into the nucleus, and TR2 and RIP140 were coimmunoprecipitated from the cell extract, indicating that the interaction between RIP140 and the LBD of TR2 occurred in vivo. The potential biological role of RIP140 in TR2-modulated transcriptional activity is discussed.
Nuclear receptors regulate target gene expression by binding to their cognate DNA response elements and recruiting associate proteins to the transcription machinery (19, 45). Recently, different coactivators and corepressors for several nuclear receptors have been identified (16). For instance, the nuclear receptor corepressor (N-CoR) and the silencing mediator for retinoid and thyroid hormone receptors (SMRT) have been shown to function as corepressors for retinoic acid (RA) receptor α (RARα), thyroid hormone receptors (T3Rs), and orphan receptor COUP-TFI (4, 15, 36). The newly identified nuclear protein Nab1 is a corepressor for the orphan receptor NGFI-A family (38), and SUN-CoR is able to potentiate transcriptional repression by T3Rs and RevErb (44). In the coactivator category, the transcriptional mediator/intermediary factor 2 and steroid receptor coactivator 1 (SRC-1) are known to mediate transcriptional activation of RAR, estrogen receptor (ER), retinoid X receptor (RXR), and T3Rs (8, 14, 33, 39, 42). With respect to the working mechanism of corepressors and coactivators, it has been demonstrated that their actions involve the alteration of chromatin structure, such as the acetylation status of histone proteins (13, 34, 37).
The orphan receptor TR2 belongs to the superfamily of nuclear receptors (21, 31). This receptor gene is expressed most abundantly in the testes of adult animals, particularly the developing germ cell populations (22, 24, 25). During embryonic stages, TR2 expression is highest between embryonic day 8 (E8) and E12 (22). It is also known that this gene encodes two isoform receptors, one retaining the entire ligand binding domain (LBD) and the other truncated at the LBD, which exhibit differential expression patterns in developing testes (24). The biological function of the full-length TR2 has been examined in a variety of systems, such as the cellular RA binding protein I promoter containing a direct repeat 4 (DR4) element (6), a RAR-responsive element (RARE) of the DR5 type from RARβ (24, 30), a DR2 from the simian virus 40 (SV40) promoter (26), and a DR2 from the erythropoietin gene promoter (27). In all tested systems, the full-length TR2 consistently represses reporter gene expression in cell cultures supplemented with either regular serum or charcoal-depleted serum. In contrast, no specific biological activity has been detected for the LBD-truncated TR2 isoform in these reporter gene systems.
To understand the molecular mechanisms of TR2 functions and identify the associate proteins for TR2, we performed yeast two-hybrid screening experiments using the LBD of TR2 as the bait. Based on the expression pattern of TR2, we constructed two cDNA libraries for the screening, one prepared from poly(A) RNA of mouse embryos at E11.5 and E12.5 and another from poly(A) RNA of adult mouse testes. By screening these libraries, we isolated and characterized one strongly interacting clone which appeared to encode the mouse homologue of the human receptor-interacting protein 140 (hRIP140), a coactivator for ligand-activated receptors such as RAR, ER, vitamin D receptor, T3Rβ1, and androgen receptor (3, 8, 18, 20, 29, 32). However, the cloned mouse RIP140 (mRIP140) functioned as a corepressor for mouse TR2 and exerted a trans-repressive activity when it was tethered to the DNA binding domain (BD) of GAL4. In this study, we report the cloning and characterization of mRIP140, demonstrate its interaction with TR2 both in vitro and in vivo, and provide evidence for a corepressor activity of RIP140 for orphan receptor TR2.
MATERIALS AND METHODS
Construction of expression vectors.
To construct the bait for the yeast two-hybrid screening experiments as well as the expression vector for deletion studies, the hinge and LBD domain (residues 166 to 590) of TR2 (22) (TR2DEF) was cloned into pBD-GAL4 Cam and pAD-GAL4 vectors (Stratagene, La Jolla, Calif.) at EcoRI and SmaI sites, resulting in the constructs pBD-TR2DEF and pAD-mTR2 (166–590), respectively. The pBD-TR2DEF vector was used as the bait to screen the libraries. Various C-terminal fragments of TR2 were generated by PCR, flanked by HindIII and SmaI sites, and used to replace the HindIII/SmaI fragment of the pAD-mTR2 (166–590) vector. For interaction tests, the C-terminal portion of mouse N-CoR (residues 1843 to 2453) (15), the TR2, the mouse RARα, and the mRIP140 cDNAs were each cloned into the bait or prey vector (pBD-GAL4 Cam or pAD-GAL4) at EcoRI and SalI sites. Various RIP140 deletions were generated by restriction enzyme digestion (restriction enzyme sites are shown in Fig. 3A) and cloned into the pAD-GAL4 vector. For the mammalian two-hybrid interaction tests, the same TR2DEF, C-terminal deletions, and full-length TR2 and RARα cDNAs, as well as different partial RIP140 fragments, were cloned into the mammalian version of the bait and prey vectors, pM for GAL4 BD fusion and pVP16 for VP16 fusion (Clontech, Palo Alto, Calif.), respectively.
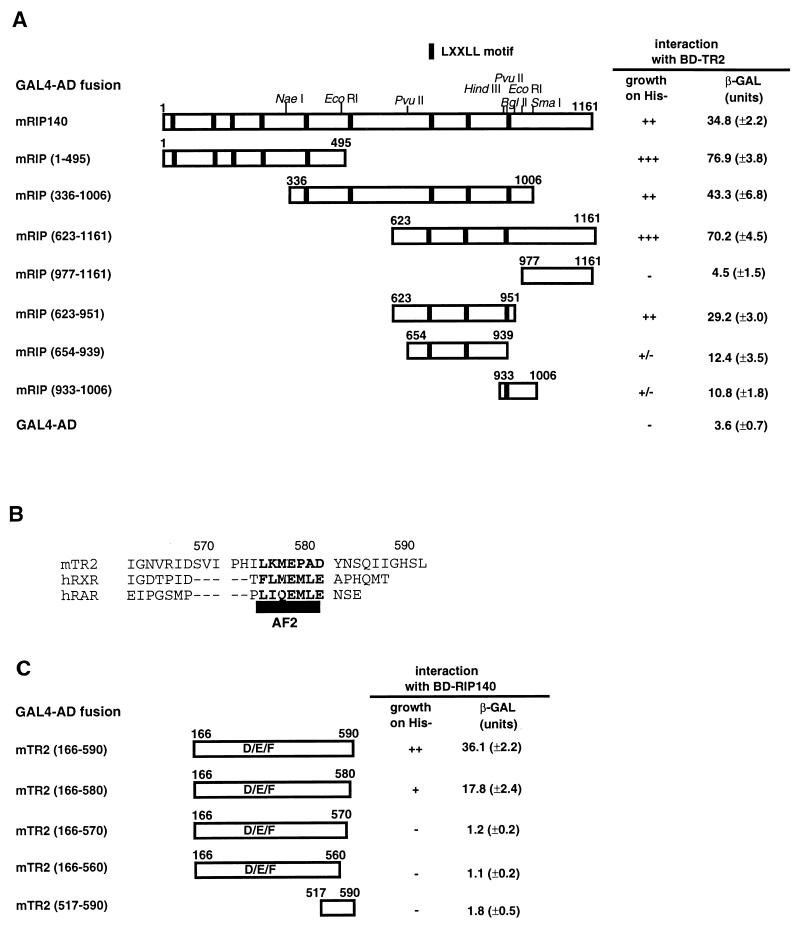
FIG. 3.
Mapping of the domains contributing to the interaction between TR2 and RIP140 in yeast two-hybrid interaction tests. (A) Different portions of RIP140 were cloned into the yeast expression vector pAD-GAL4 to test their abilities to interact with BD-TR2 (full-length TR2 fused to GAL4 BD). The activation domain (AD) fusion constructs and BD-TR2 were introduced into yeast strain YRG-2, and the criteria for positive interaction were based on their growth on histidine-deficient medium and LacZ activities as shown on the right. The LXXLL motif is depicted with small solid bars. The numbers for each construct correspond to amino acid residues, and the restriction sites shown on the top indicate cloning sites. (B) Sequence comparison of the putative AF-2 domain of TR2 (bold letters) with RARα and RXRα (2, 35). (C) Interactions between the AD fusions of TR2 C-terminal deletion mutants and BD-RIP140 (full-length RIP140 fused to GAL4 BD).
Green fluorescent protein (GFP) fusion proteins were constructed by placing the cDNAs of the full-length TR2, RIP140, and TR2DEF downstream of the GFP gene individually, at BglII and SalI (for TR2 and RIP140) or SmaI (for TR2DEF) sites of the pEGFP-C1 vector (Clontech). Glutathione S-transferase (GST) fusion proteins were constructed by inserting full-length TR2 and TR2DEF, individually, at the BamHI site of pGEX-2T vector (Pharmacia, Piscataway, N.J.), resulting in GST-TR2 and GST-TR2DEF constructs, respectively.
The reporter construct for the mammalian two-hybrid system was made by placing five copies of the GAL4 binding site (5′CGGAGGACAGTACTCCG3′) upstream of the thymidine kinase-luciferase (TK-Luc) reporter (41). The reporter for TR2 autoregulation was constructed by fusing the 5′ untranscribed sequence, exon 1, intron 1, and exon 2 containing the Kozak sequence and ATG codon of TR2 in frame to the β-galactosidase (β-Gal) gene (lacZ) (see Fig. 6C). By leaving its splicing junctions unchanged, a large portion of the intron 1 sequence was deleted from this construct due to its size (11.5 kb). A hemagglutinin (HA)-tagged TR2 expression vector under the control of a human Ha-ras promoter was constructed as described elsewhere (43). All other expression vectors for transfection experiments in COS-1 cells were under the control of the cytomegalovirus promoter. Full-length TR2 and RIP140 were cloned into pSG5 vector at BglII and XhoI sites for in vitro transcription-translation (TNT) reactions.
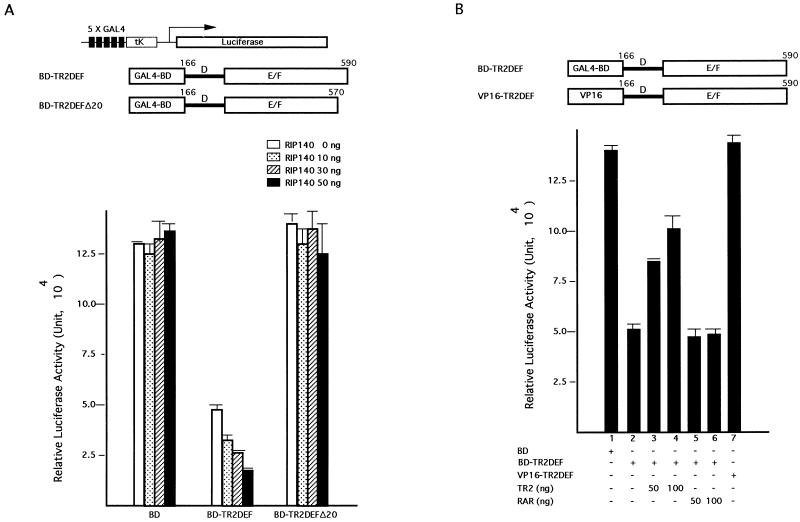
FIG. 6.
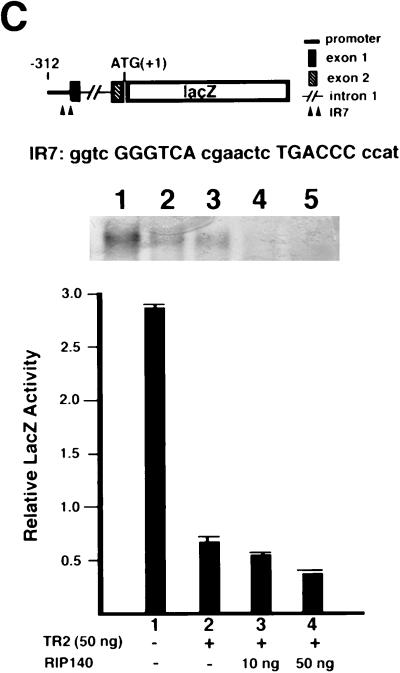
Corepressive activity of RIP140 for TR2. (A) Corepressive activity of RIP140 for the LBD of TR2 fused to the BD of GAL4. Cells were cotransfected with GAL4 BD or a BD fusion to TR2DEF (BD-TR2DEF) or the C-terminal 20-amino-acid deletion of TR2DEF (BD-TR2DEFΔ20) (20 ng) with different amounts of RIP140 expression vector (0 to 50 ng), along with the luciferase reporter (400 ng) and the lacZ internal control (30 ng). An equal amount of the transfected DNA was maintained for each experiment by supplementation with the corresponding control vector. Activity in RLU was measured as described for Fig. 5. (B) Sequestration of the trans-repressive activity of BD-TR2DEF by TR2 but not RAR. COS-1 cells were transfected with 20 ng of BD-TR2DEF and an increasing amount of either TR2 or RARα expression vector (50 to 100 ng), along with 400 ng of the luciferase reporter and 30 ng of the lacZ internal control. GAL4 BD and VP16-TR2DEF were included for control reactions. (C) RIP140 enhances the trans-repressive activity of TR2 on a natural TR2 promoter containing an IR7 element. The diagram at the top shows a lacZ reporter driven by the proximal region of TR2 promoter, the IR7 sequence (shown in capitals), and the gel mobility shift experiment. For the gel shift, the TR2-IR7 complex is shown in lane 1; lanes 2 to 5 represent competition experiments in which 2-, 10-, 50-, and 100-fold-excess amounts of unlabeled IR7 DNA fragment were included. For the transfection assay, COS-1 cells were transfected with the TR2-lacZ reporter (400 ng) and a TK-Luc internal control (30 ng), with or without TR2 (50 ng) and RIP140 (10 to 50 ng) expression vectors.
Yeast two-hybrid screening and interaction assay.
Yeast two-hybrid screening (HybriZAP two-hybrid system; Stratagene) was conducted according to the manufacturer’s instructions. Briefly, the phagemid library, from mRNA of either mouse embryos or adult testes, was prepared in the HybriZAP two-hybrid lambda vector. The primary HybriZAP lambda library, containing a total of 5 × 107 individual clones, was amplified and converted to the pAD-GAL4 plasmid library by in vivo mass excision. A portion of the amplified library (109 PFU) was transformed into Escherichia coli XLOLR cells to obtain the plasmid DNA representing the target cDNA library. To screen the libraries, the pBD-TR2DEF plasmid DNA and the library DNA (10 mg each) were introduced together into Saccharomyces cerevisiae YGR-2 cells containing a his3 marker and a LacZ reporter. Approximately 5 × 106 transformants were plated on selection medium lacking leucine, tryptophan, and histidine, and the plates were incubated at 30°C for 5 days. The positive clones were confirmed by a LacZ filter lift assay (11).
For the interaction assay, different combinations of the baits and the preys were cotransformed into S. cerevisiae YGR-2 cells and plated on the triple-selection medium. The liquid LacZ assay was performed as described previously (9), and 1 U of LacZ activity was defined as the amount that hydrolyzes 1 μmol of o-nitrophenyl-β-d-thiogalactopyranoside (ONPG) to o-nitrophenol and d-galactose per min. Luciferase activity was determined as relative luciferase units (RLU).
To obtain the full-length RIP140 clone, the EcoRI/BglII fragment from the original clone was used to screen the embryonic phagemid library. The cDNA library was screened under high-stringency conditions (42°C and 50% formamide) as described previously (22). DNA sequencing was conducted by the dideoxy-chain termination method, and sequence comparison was performed by using the Pro-Dom program (1).
GST pull-down assay.
GST fusion protein was purified as instructed by the manufacturer (Pharmacia). For in vitro interaction, 5 μg each of GST-TR2, GST-TR2DEF, and GST control, made in E. coli BL21, was bound to a glutathione-Sepharose column and incubated with 35S-labeled RIP140 protein prepared in TNT reactions (Promega, Madison Wis.) in binding buffer (20 mM HEPES [pH 7.4], 150 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.1% Nonidet P-40, 5 mg of bovine serum albumin [BSA] per ml, 10% glycerol, protease inhibitor cocktail) for 60 min at 4°C. Unbound proteins were removed by five washes with binding buffer without BSA and protease inhibitors. Subsequently, the specifically bound protein was eluted with a solution containing 50 mM reduced glutathione in 50 mM Tris (pH 8.0), resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% gel) and visualized by autoradiography.
Cell culture techniques and Northern blot analyses.
The technique for culturing COS-1 cells, transfection experiments, and luciferase and LacZ assays were as described previously (23). All cultures were maintained in Dulbecco modified Eagle medium containing dextran charcoal-treated serum. For RA induction experiments, all-trans RA was added at a final concentration of 5 × 10−7 M for 24 h. Each experiment was carried out in triplicate cultures. At least three independent experiments were conducted to obtain the means and standard errors of the means.
For GFP fusion protein expression, COS-1 cells were plated on a coverglass in 3-cm-diameter dishes. Forty-eight hours after transfection, the cells were fixed in a 4% formaldehyde solution and visualized by microscopy.
The methods for total RNA isolation and Northern blot hybridization were as described previously (22). The RIP140 probe was derived from the EcoRI/BglII fragment of the original clone.
Electrophoretic mobility shift assay.
The gel shift assay was conducted as described previously (23). Briefly, in vitro-translated TR2 protein was incubated with 1 ng of probe in a 20-μl reaction solution containing 20 mM HEPES (pH 7.4), 50 mM KCl, 1 mM β-mercaptoethanol, 10% glycerol, 1 μg of poly(dI-dC), and 5 mg of BSA per ml at 4°C for 60 min. The protein-DNA complex was analyzed on a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. The probes were prepared by annealing oligonucleotides containing an inverted repeat 7 (IR7)-type element derived from TR2 promoter (5′GGATCCAAGCCGAGGGTGGGGTCACGAACTCTGACCCCCATCCCCAAAACACAAACTCGAG3′) and labeled with [α-32P]dCTP, using Klenow enzyme. For competition experiments, 2 to 100 ng of unlabeled IR7 fragment was included in the reaction.
Immunoprecipitations and Western blot analyses.
HA-tagged TR2 was cotransfected with either GFP-tagged RIP140 or GFP expression vector in COS-1 cells. Forty-eight hours after transfection, the cells were harvested and resuspended in 200 μl of lysis buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40, 1 mM dithiothreitol, 2 μM phenylmethylsulfonyl fluoride, 10% glycerol, and protease inhibitor cocktail. After incubation on ice for 10 min, the cells were subjected to sonication and centrifugation. Fifteen microliters of the cell lysate was used for Western blot analysis to compare protein expression levels. For immunoprecipitation, 100 μl of the cell lysate was incubated with a mouse anti-HA monoclonal antibody (for HA-TR2) (Boehringer Mannheim, Indianapolis, Ind.) at 4°C for 2 h, and 15 μl of protein A-Sepharose CL4B resin (Sigma, St. Louis, Mo.) was added to the reaction for another 2 h of incubation. The resin was then washed three times with lysis buffer and resuspended in SDS-PAGE loading buffer for immunoblot analysis.
For Western blot analysis, the polyacrylamide gel was transferred to a polyvinylidene membrane (Millipore, Bedford, Mass.). Duplicate blots were made from the same set of immunoprecipitation experiments. One blot was probed with rabbit anti-GFP antibody (Clontech) to detect GFP-RIP140 in the HA-TR2–GFP-RIP140 immunocomplex, and another was probed with a rabbit anti-TR2 antibody (22) to monitor the amount of HA-TR2 protein that had been immunoprecipitated by the anti-HA antibody in each reaction. The blots were subsequently probed with a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) antibody (Gibco BRL, Gaithersburg, Md.) and detected with an enhanced chemiluminescence system (Amersham, Arlington Heights, Ill.).
Nucleotide sequence accession numbers.
The 3-kb 5′ untranscribed region, exon 1, exon 2, and partial intron 1 sequences of the TR2 gene have been deposited in GenBank under accession no. U96095 and U28269 (unpublished data). The nucleotide sequences containing the partial 5′ untranslated region, the coding sequence, and the entire 3′ untranslated region of RIP140 (Fig. 1) have been submitted to GenBank under accession no. AF053062.
FIG. 1.
Complete coding region of mRIP140 aligned to that of hRIP140. Colons show conserved residues, and dashes represent deleted residues. The mRIP140 contains 1161 amino acids, of which 970 are identical to the human clone. The sequences for the receptor-interacting signature motif LXXLL, where L is leucine and X is any amino acid, are underlined.
RESULTS
Cloning of mouse RIP140.
Two cDNA libraries, one of mixed mouse embryos at E11.5 and E12.5 and another of adult mouse testes, were constructed in the pAD-GAL4 prey vector. The bait was prepared by fusing the entire TR2 DEF in the pBD-GAL4 vector. S. cerevisiae YRG-2 was cotransformed with the bait and prey libraries. Positive clones were selected on selection medium and identified based on positive LacZ activity on the filters. The positive clones were further confirmed by a liquid LacZ assay, and their inserts were characterized by DNA sequencing. None of the total of 60 positive clones represented any known nuclear receptor corepressor. Interestingly, one strongly positive clone appeared to be homologous to RIP140, a coactivator of RAR, ER, vitamin D receptor, and T3Rβ1. By rescreening the original embryonic cDNA library with the partial mRIP140 cDNA, the full-length mRIP140 cDNA was isolated and completely sequenced. Figure 1 shows the alignment of mRIP140 and hRIP140, which exhibits 83% (970/1,161) amino acid identity between the two sequences. Like RIP140, the mouse protein contains nine copies of the LXXLL signature motif scattered within the molecule (12) (Fig. 1, underlined).
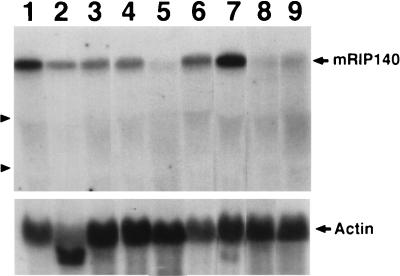
Since no expression data were provided for hRIP140, we then performed a Northern blot analysis to determine its tissue distribution pattern (Fig. 2). Total RNAs isolated from different adult mouse tissues, as well as E12.5 placenta and embryos, were examined on the Northern blot by hybridizing to an RIP140-specific probe followed by an actin-specific probe. mRIP140 mRNA has a size of approximately 8 kb and is detected in all samples examined. The strongest expression is observed in the testis (lane 7) and brain (lane 1), a relatively constant but weaker expression is observed in the heart (lane 2), lung (lane 3), stomach (lane 4), and kidney (lane 6), and the spleen appears to express RIP140 at the lowest level (lane 5) among all tissues examined. In the embryonic stages, this message is also weakly expressed in E12.5 embryos (lane 8) and placenta (lane 9).
FIG. 2.
Northern blot analysis of RIP140 expression. Total RNAs (30 μg) from various adult mouse tissues and E12.5 embryos and placenta were loaded onto a denaturing agarose gel, transferred to a nylon membrane, and hybridized with a RIP140-specific probe. An actin probe was included as a quantitative control. Arrowheads on the left indicate positions of 28S and 18S rRNAs. Lane 1, brain; 2, heart; 3, lung; 4, stomach; 5, spleen; 6, kidney; 7, testis; 8, E12.5 embryo; 9, E12.5 placenta.
TR2-RIP140 interaction.
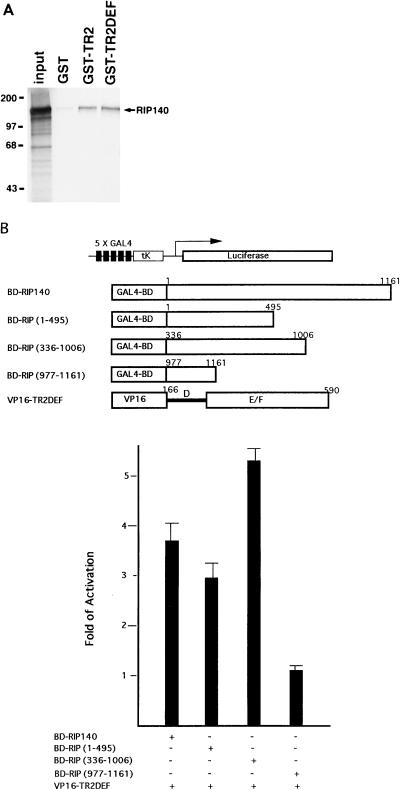
To dissect the interaction domains, serial deletions of RIP140 and TR2 were each constructed in the yeast two-hybrid expression vectors. In the first series of experiments, the full-length TR2 was cloned into pBD-GAL4 (BD-TR2), and different portions of RIP140 were cloned into pAD-GAL4; the interaction test was conducted in yeast (Fig. 3A). It appears that all RIP140 constructs that contain LXXLL clusters interact strongly with TR2, including N-terminal mRIP (1–495), central mRIP (336–1006), and C-terminal mRIP (623–1161). The very C-terminal part of this molecule, mRIP (977–1161), which contains no LXXLL signature motif, fails to interact with TR2. Consistent with this result, construct mRIP (623–951), in which this very C-terminal sequence was deleted from mRIP (623–1161), interacts well with TR2, although with lower efficiency. mRIP (623–951) was further dissected to determine whether a single motif is sufficient for the interaction. Intriguingly, both mRIP (654–939) and mRIP (933–1006), which contain two motifs and one motif, respectively, interact poorly with TR2, as demonstrated by slow growth of yeast cells and low β-Gal activity. In summary, these data suggest that the receptor-interacting domains of mRIP140 are scattered within the molecule, a finding which correlates with the presence of the LXXLL clusters. Although a deletion mutant containing one LXXLL is sufficient for a weak interaction, multiple LXXLLs are required for efficient interaction with TR2. In addition, since yeast culture contains no animal sera, it is suggested that RIP140 interacts with TR2, presumably the apo-form receptor.
To define the RIP140-interacting domains of TR2, we conducted the second series of experiments by deleting various segments from the C terminus of TR2 LBD in the pAD-GAL4 vectors and tested them against the full-length RIP140 cloned in the pBD-GAL4 vector (BD-RIP140) as shown in Fig. 3B. In contrast to a previous report (30), BD-RIP140 alone did not induce any β-Gal reporter activity. It appears that construct mTR2 (166–580), with 10 amino acids deleted from the C terminus, is capable of interacting with RIP140, albeit at an approximately 50% efficiency compared to the wild type [construct mTR2 (166–590)]. However, deletion of 20 amino acids of TR2 LBD (spanning the putative AF-2 region) or more [constructs mTR2 (166–570) and (166–560)] completely abolishes its interaction with RIP140. Intriguingly, although the C-terminal 20-amino-acid sequence appears to be important for this interaction (from comparison of the results for 10-, 20-, and 30-amino-acid-deletion mutants), the C-terminal 73-amino-acid sequence alone [construct mTR2 (517–590)] does not interact with RIP140.
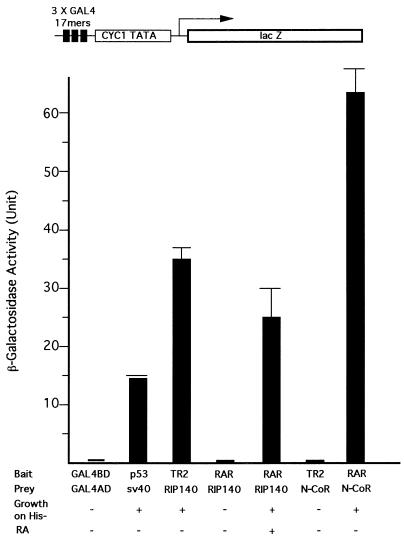
It is known that corepressors N-CoR and SMRT are involved in silencing activity of RAR, T3R, and orphan receptor COUP-TFI. To determine whether apo-TR2 also interacts with the common corepressor N-CoR and to compare the apo-TR2–RIP140 interaction with that of holo-RAR and RIP140, two-hybrid interaction tests for different combinations of receptors and coregulators were performed (Fig. 4). The positive control in this system (the p53-SV40 pair) results in moderate LacZ activity, whereas TR2-RIP140 interaction results in a stronger reporter activity that is comparable to or even stronger than that for RIP140 interaction with the holo-RAR. As expected, the apo-RAR does not interact with RIP140. The lack of TR2 interaction with N-CoR is demonstrated by comparison of the basal reporter activity in the TR2–N-CoR pair to that in the positive control (the apo-RAR–N-CoR pair), which induces a strong reporter activity. Therefore, we conclude that RIP140 interacts with TR2 in the absence of putative ligands and with RAR in a ligand-dependent manner. Furthermore, TR2 does not interact with N-CoR, which is consistent with our screening results.
FIG. 4.
RIP140 interaction with apo-TR2. Full-length mouse TR2 and RARα were each fused to pBD-GAL4 to examine their interaction with pAD-GAL4 fusions of the full-length RIP140 as well as the common receptor-interacting region of N-CoR (residues 1843 to 2453). The bait and prey were introduced into yeast cultures, and RA (10−6 M) was added to the medium for the RAR-RIP140 pair. Addition of RA at this concentration did not cause trans activation of lacZ and HIS3 reporters by BD-RAR in this system. The lacZ reporter with three GAL4 binding sites is shown on the top. The pairs GAL4 BD/GAL4 AD and p53-SV40 are negative and positive controls, respectively.
TR2 interaction with RIP140 in solution and in mammalian cells.
A GST pull-down assay was performed to examine TR2 interaction with RIP140 in solution. Full-length TR2 and TR2DEF were each fused to the GST vector. The GST control or a fusion protein was applied to a glutathione column and incubated with 35S-labeled RIP140 prepared in in vitro TNT reactions. After extensive washing, RIP140 was eluted and analyzed on a polyacrylamide gel as shown in Fig. 5A. Compared to the control (GST alone), both GST-TR2 and GST-TR2DEF columns retained a significant amount of the labeled RIP140. This result indicates that RIP140 interacts with the LBD of apo-TR2 in solution.
FIG. 5.
RIP140-TR2 interaction in solution and in mammalian cells. (A) GST pull-down assay for specific interaction between TR2 and RIP140. Purified GST or GST fusion to full-length TR2 (GST-TR2) or TR2DEF (GST-TR2DEF) was bound to glutathione-Sepharose beads and incubated with 35S-labeled RIP140. After extensive washes, specific interacting protein was eluted and analyzed by SDS-PAGE and autoradiography. Positions of the protein ladder (in kilodaltons) and RIP140 are shown on the left and right, respectively. (B) RIP140 interaction with TR2 in mammalian two-hybrid interaction tests. The diagram at the top shows a luciferase reporter containing five copies of GAL4 binding sites, the BD-GAL4 fusion constructs, and the VP16 fusion construct. The reporter (400 ng) and different combinations of GAL4 BD and VP16 fusion vectors (50 ng of each) were cotransfected, along with SV40-LacZ (30 ng) as an internal control, into COS-1 cells. Forty-eight hours after transfection, cells were harvested, and luciferase and LacZ activities were determined. Activity in RLU was calculated by normalizing the specific luciferase activity to that of β-Gal. Fold activation was determined by comparing the RLU of each GAL4BD-VP16 fusion pair to the RLU of the corresponding GAL4 BD-VP16 empty vector.
To conduct mammalian two-hybrid tests, various portions of RIP140 were each fused to the pBD-GAL4 mammalian expression vector, and the LBD of TR2 was fused to the mammalian pVP16 expression vector. COS-1 cells were cotransfected with one of the BD-RIP140 vectors and VP16-TR2DEF, together with the GAL4-TK-Luc reporter and an internal control lacZ vector. As shown in Fig. 5B, all three RIP constructs that contain the LXXLL motif are able to interact with TR2, whereas the C-terminal RIP140 segment which contains no LXXLL motif [construct BD-RIP (977–11610)] remains negative in this mammalian two-hybrid interaction test. This finding is consistent with results of the yeast experiments. Therefore, it is concluded that TR2 interacts with RIP140 in both yeast and mammalian cells.
Corepressor function of RIP140 in TR2 repressive activity.
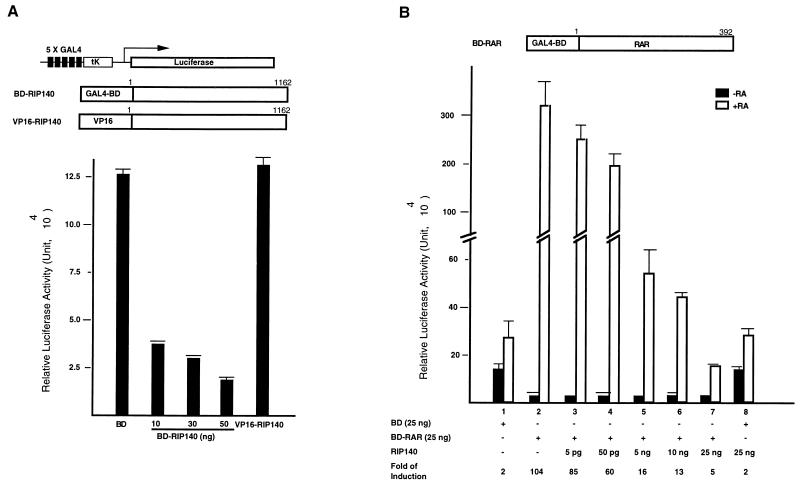
We have previously observed a trans-repressive activity of the LBD of TR2 fused to the GAL4 BD (5, 23). To verify whether the association of RIP140 contributes to this trans-repressive activity of TR2, we perform two series of experiments in the BD-TR2DEF fusion system. In the first series of experiments, COS-1 cells were cotransfected with the BD-TR2DEF and RIP140 expression vectors together with the GAL4-TK-Luc reporter and the internal control lacZ reporter. As shown in Fig. 6A, BD-TR2DEF exerted a trans-repressive activity that was enhanced by the addition of RIP140 in a dose-dependent manner, indicating a corepressive activity of RIP140 for BD-TR2DEF. As a control, cotransfection of RIP140 with the BD vector had no effect on the reporter activity. Furthermore, the C-terminal TR2 deletion mutant (BD-TR2DEFΔ20) lost its trans-repressive activity, which could not be rescued by the addition of RIP140, suggesting that the interaction between TR2 and RIP140 is required for this trans-repression function. This result strongly supports a corepressive activity of RIP140 for TR2, which is mediated by a specific interaction of RIP140 with the C terminus of TR2.
In the second series of experiments, we examined whether the corepressive activity of RIP140 in the BD-TR2DEF system could be sequestered by providing the cells with extra amounts of free receptors. COS-1 cells were transfected with BD-TR2DEF and the reporter, with the addition of different amounts of TR2 and RAR expression vectors. As shown in Fig. 6B, BD-TR2DEF represses reporter activity (column 2) compared to the control using the empty BD vector (column 1), and the addition of 50 and 100 ng of TR2 (columns 3 and 4) is able to rescue the GAL4 reporter gene activity from the repression by BD-TR2DEF, presumably due to the sequestration of the endogenous RIP140 by extra TR2 molecules. In contrast, the addition of apo-RAR (columns 5 and 6) fails to do so, indicating that apo-RAR cannot sequester the endogenous RIP140 because of the lack of interaction between RIP140 and apo-RAR. For a control of this trans-repressive activity of TR2, TR2DEF was fused to the VP16 vector and tested. As expected, VP16-TR2DEF had no effect on the reporter activity due to the lack of specific DNA binding (column 7).
Our recent unpublished data suggested an autoregulatory mechanism for TR2 expression. This negative feedback control is mediated by an IR7 element that serves as the binding site for TR2. This IR7 element, which is located in the proximal region of the TR2 promoter, is highly conserved in all species examined, including human, zebrafish, Xenopus, and mouse (28). To investigate whether RIP140 could also function as a corepressor for TR2 in the context of this natural promoter, we used a lacZ reporter containing the TR2 promoter. The diagram at the top of Fig. 6C shows the TR2 promoter-lacZ reporter, IR7 sequence, and gel shift result demonstrating the specific binding of TR2 to the IR7 element (lane 1). This specific binding can be completely competed out by a 50-fold-excess amount of the unlabeled DNA molecule (lanes 2 to 5). The lacZ reporter, TR2 and RIP140 expression vectors, and an internal control TK-Luc construct were cotransfected into COS-1 cells. As shown at the bottom of Fig. 6C, TR2 represses the TR2-lacZ reporter activity (column 2), and the repression is enhanced by the addition of RIP140 (columns 3 and 4). This result provides evidence that in a reporter system that contains a natural TR2 binding site, RIP140 potentiates the trans repression by TR2. Collectively, our data suggest that RIP140 functions as a corepressor for TR2.
trans-repressive activity of RIP140.
From the above experiments, we conclude that RIP140 itself, when present in the free form, has no effect on GAL4 reporter activity. We then tested whether RIP140, if itself rendered DNA bound, could exert any repressive effect on this reporter. The entire RIP140 was fused to pBD-GAL4 (BD-RIP140) and tested in the GAL4-TK-Luc reporter system as shown in Fig. 7A. Compared to the control pBD-GAL4 vector (column 1), RIP140 tethered to the GAL4 BD represses the GAL4 reporter activity, and the repression is dose dependent (columns 2 to 4). For a control, RIP140 fused to the VP16 vector (VP16-RIP140) has no effect on this reporter (column 5), again suggesting that the corepressive activity of RIP140 is mediated by its recruitment to DNA. Therefore, it is concluded that RIP140 encodes a transferable, repressive activity when tethered to gene promoters.
FIG. 7.
RIP140 encodes an active repressive activity and suppresses RA induction. (A) Transcriptional repressive activity of RIP140 when tethered to the BD of GAL4. The full-length RIP140 was fused to GAL4 BD or VP16 (top panel), and effects on the GAL4 reporter were examined in COS-1 cells. BD, VP16-RIP140 (50 ng), or an increasing amount of BD-RIP140 (10 to 50 ng) was introduced into cells together with the reporter (400 ng) containing the GAL4 binding sites and an internal control lacZ vector (30 ng). The total amount of transfected DNA for each experiment was kept constant by supplementation with the corresponding control vector. (B) RIP140 suppresses RA induction mediated by BD-RAR fusion. Full-length RARα was fused to GAL4 BD and designated BD-RAR. The GAL4 reporter (400 ng), BD-RAR (25 ng), lacZ internal control vector, and increasing amounts of wild-type RIP140 (0 to 25 ng) were cointroduced into COS-1 cells, and all-trans RA was added at a final concentration of 5 × 10−7 M. Lanes 1 and 8 are control experiments in which GAL4 BD control vector was used. A nonspecific twofold increase in reporter activity was observed in the control (column 1). This increased activity was not affected by the addition of RIP140 (column 8).
A recent study showed that at low doses, RIP140 weakly (less than twofold) enhanced trans activation by ER whereas it strongly suppressed ER trans activation when the amount of RIP140 increased (3). Since the interaction of RAR and RIP140 is also ligand dependent, we used the GAL4 BD-RAR and GAL4 reporter system to study the function of RIP140 in the ligand-induced trans activation by nuclear receptor. The full-length RAR was fused to the BD of GAL4 (BD-RAR). As shown in Fig. 7B, unliganded BD-RAR represses GAL4 reporter activity as found previously (columns 1 and 2, −RA). In the presence of RA, the activity of this reporter can be induced up to 100-fold (column 2). However, the RA induction of the GAL4 reporter mediated by BD-RAR is strongly suppressed by RIP140 (columns 3 to 7). The suppression of RA induction was also observed in a system utilizing wild-type RARs and a DR5-type RARE-containing reporter (data not shown). Although the interaction of RAR and RIP140 is ligand dependent, our data do not support a coactivator role for RIP140 in RAR-mediated gene activation.
Demonstration of in vivo interaction between TR2 and RIP140.
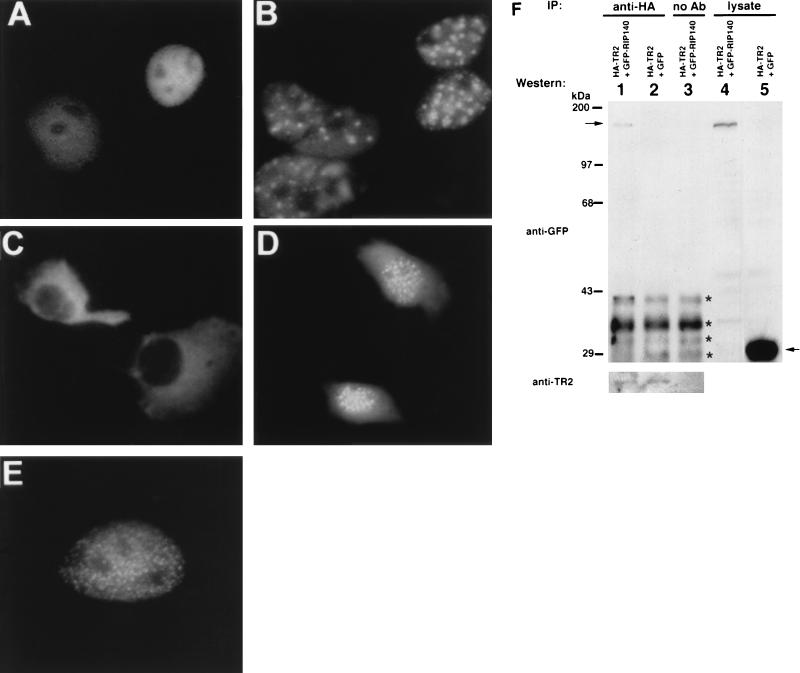
To provide evidence for TR2 interaction with RIP140 in vivo, GFP was used to tag these molecules for tracing their intracellular distribution and translocation. TR2 tagged with GFP exhibits a homogeneous, nuclear distribution (Fig. 8A), whereas RIP140 tagged with GFP exhibits a very different intranuclear distribution pattern, predominantly in many localized foci (Fig. 8E). In the presence of RIP140, the GFP-TR2 fusion exhibits a pattern (Fig. 8B) mimicking that of GFP-RIP140 fusion (Fig. 8E). Upon deletion of the nuclear localization signal of TR2 (43), the GFP-tagged TR2-DEF becomes cytosolic (Fig. 8C). Interestingly, in the presence of untagged RIP140, this otherwise cytosolic TR2-DEF mutant is retained in the nucleus (Fig. 8D), strongly supporting the notion that the interaction of TR2 with RIP140 occurs in vivo.
FIG. 8.
In vivo interaction of RIP140 with TR2. (A) GFP-tagged TR2; (B) GFP-TR2 plus untagged RIP140; (C) GFP-tagged TR2-DEF; (D) GFP-tagged TR2-DEF plus untagged RIP140; (E) GFP-tagged RIP140. COS-1 cells were transiently transfected with different combinations of expression vectors; 48 h after transfection, cells were fixed and observed under a microscope. Magnifications: A, B, and E, ×400; C and D, ×200. (F) Immunoprecipitation (IP) experiment showing the association of RIP140 and TR2. COS-1 cells were cotransfected with HA-tagged TR2 and either GFP-tagged RIP140 or GFP control vector. The cell lysate was incubated with a mouse anti-HA monoclonal antibody in the presence of protein A-Sepharose beads. The immunocomplex was then analyzed on a Western blot, which was probed first with a rabbit anti-GFP polyclonal antibody and then with a horseradish peroxidase-conjugated goat anti-rabbit IgG antibody and detected with an enhanced chemiluminescence system (top, lanes 1 to 3). Lanes 1 and 2 are from cells expressing HA-TR2–GFP-RIP140 and HA-TR2–GFP, respectively. Lane 3 is a control for lane 1 with no antibody (Ab) added. One-sixth of the total cell lysate used in immunoprecipitation was loaded onto lanes 4 and 5 as an indication of the relative expression level for GFP-RIP140 and GFP protein. The positions of GFP-RIP140 (∼170 kDa) and GFP (∼30 kDa) are indicated with arrows on the left and right, respectively. The nonspecific bands present in all the reactions (recognized by anti-IgG antibody) are labeled with asterisks. A duplicate blot was also probed with a rabbit anti-TR2 polycolonal antibody to monitor the amount of TR2 protein precipitated in each reaction (bottom).
The physical association of TR2 and RIP140 in vivo was further confirmed by coimmunoprecipitation assay. HA-tagged TR2 was cotransfected with GFP or GFP-tagged RIP140 into COS-1 cells. The cell lysate was then incubated with or without a mouse anti-HA monoclonal antibody in the presence of protein A-Sepharose beads. The immunocomplex was analyzed on a Western blot and detected with a rabbit anti-GFP antibody. It appears that GFP-RIP140, but not GFP, can be coprecipitated with HA-TR2 by the anti-HA antibody (Fig. 8F, lanes 1 and 2). The reaction for lane 3, a negative control, is similar to that for lane 1 except that the antibody was omitted. Lanes 4 and 5 are total cell lysates from GFP-RIP and GFP-transfected cells, showing the relative amounts of GFP-RIP and GFP expression, respectively. At the bottom of Fig. 8F is a duplicate blot probed with anti-TR2 antibody, confirming that equal amounts of HA-TR2 were immunoprecipitated in all reactions. These data clearly indicate that TR2 and RIP140 physically interact with each other in vivo.
DISCUSSION
In this study, we report the cloning of mRIP140 by using the LBD of orphan receptor TR2 as the bait in yeast two-hybrid screening experiments. We also characterize the receptor-interacting domains of the mRIP140, which correlate with the presence of multiple LXXLL signature motifs scattering within this molecule. The RIP-interacting domain of TR2 is mapped to the C-terminal 10- to 20-amino-acid sequence of TR2, but the C terminus of TR2 is not sufficient for this interaction. In addition, we provide the evidence for a corepressor function of RIP140 in the GAL4 BD-TR2 fusion system as well as in the natural TR2 promoter. A transferable repressive activity of RIP140 is also demonstrated in the GAL4 BD-RIP140 fusion system. The presence of RIP140 suppresses RA induction by GAL4 BD-RAR on a GAL4 reporter, even though the interaction of RAR with RIP140 is ligand dependent. Finally, the interaction of TR2 and RIP140 in vivo is demonstrated in the GFP-tagged protein translocation and coimmunoprecipitation studies.
As reported for hRIP140, SRC-1, and CREB-binding protein (CBP) (12), mRIP140 employs an LXXLL signature motif for receptor interaction, further supporting the notion that this sequence motif is conserved for interaction with nuclear receptors. In contrast to a recent study showing that a short peptide, derived from RIP140 containing a single LXXLL sequence, when fused to GAL4 BD interacted strongly with liganded GAL4 AD-ER in yeast (12), our data suggest that although a deletion mutant containing one LXXLL can interact weakly with TR2, a cluster of three LXXLLs, in the case of the C-terminus region of RIP140, is required for an efficient association between TR2 and RIP140 (Fig. 3A). However, it is also possible that sequences adjacent to the LXXLL motif that are important for maintaining the conformation for interaction are interrupted in the GAL4 activation domain fusion construct. Detailed point mutation studies will answer this question in the future. On the other hand, like all nuclear receptors that are capable of interacting with RIP140, the RIP140-interacting domain of TR2 is located at its C terminus, a presumptive AF-2 domain. A recent structural study has revealed that the ligand-induced interaction between the nuclear receptor and LXXLL motif of the coactivator involved helices 3, 5, 6, and 12 (AF-2) of the LBD of the receptor (10). This may explain why the C-terminal 73-amino-acid sequence of TR2 alone does not interact with RIP140. As expected, mRIP140 interacts with the holo-RAR but not the apo-RAR. The ligand-independent association of peroxisome proliferator-activated receptor (PPAR) with RIP140 was observed in the yeast system but not pull-down assays (40). However, for TR2, RIP140 appears to interact with its apo form, since their interaction occurs in yeast and mammalian cultures supplemented with serum depleted with charcoal as well as in the GST pull-down assay. It cannot be ruled out that ligands for TR2 exist in charcoal-depleted serum or buffer solutions. From the structural study, the hydrophobic cleft of the LBD for LXXLL binding comprises two parts. Helices 3, 5, and 6 form the constitutive part; helix 12, which contains the AF-2 domain and responds to active hormone, forms the second part. As no ligands have been identified for TR2, it remains to be determined how conformational changes of TR2 (i.e., by second messenger) may affect its interaction with RIP140. We are testing the hypothesis that mutations that alter the TR2 LBD conformation will affect this receptor’s ability to interact with RIP140 and its repressive activity.
The consequence of RIP140 interaction with liganded nuclear receptors remains controversial. hRIP140 was shown to function as a coactivator for the androgen receptor in mammalian cells (18) and for RAR and ER only in the yeast system (20, 29). Although it was originally identified as an ER-associating protein, RIP140 strongly suppressed the trans activation of ER at a higher dose (3). It was recently suggested that RIP140 suppressed PPAR/RXR trans activation by a possible mechanism that involves competition with SRC-1 for receptor binding (40). We (Fig. 3B) and others (40) were not able to detect coactivator activity for RIP140 in yeast. Our data also suggested that RIP140 not only suppresses RAR-mediated RA induction of the reporter gene but also serves as a corepressor for TR2. Moreover, once tethered to the GAL4 BD, RIP140 is found to possess a transferable repressive activity in the GAL4 system (Fig. 7A). This result is in contrast to that for hRIP140 tethered to the BD of the B-cell-specific activator protein in the pBS4 reporter system (29). This may be due to the difference in the BD and the promoter systems used in these studies; the present study used the standard GAL4 system, whereas in the other study a transcription factor system specific to B cells was used. A recent report has also shown that the function of RIP140 depends on the promoter context (7). It is possible that RIP140 can function as a coactivator or a corepressor, depending on the cell environment, the interacting nuclear receptor, and the context of target promoter.
Regardless of the transcription outcome in the different promoter systems, the high homology between human and mouse RIP140 suggests that this molecule has an essential function(s) that is conserved during evolution. It is tempting to speculate that functional constraints have been placed on the RIP140 structure and that this molecule may interact with many proteins that constitute a functional complex. According to their nuclear distribution patterns, RIP140 appears in localized foci, whereas TR2 is homogeneous. In the presence of exogenous RIP140, the distribution of TR2 changes to a pattern resembling that of the RIP140, providing evidence for their interaction inside living cells. Their interaction results in changing of intranuclear positioning of TR2, which is probably required for specific events in the processes of gene expression. The fact that RIP140 is able to render the translocation of the otherwise cytosolic TR2 LBD into the nucleus suggests that their in vivo interaction is fairly strong and requires only the LBD of TR2. The ability of TR2 to interact with RIP140, which also interacts with many ligand-bound receptors, suggests that TR2 may be able to sequester the endogenous RIP140, thereby modulating other hormonal signaling pathways.
Based on the criteria of interaction with nuclear receptors, the recently identified receptor-interacting protein NSD1 was reported to exhibit characteristics of both corepressors and coactivators (10). Two distinct domains of this protein are responsible for its interaction with liganded RAR, T3R, RXR, and ER and with unliganded RAR and T3R. It is not clear whether different clusters of the LXXLL motifs of RIP140 preferentially interact with apo or holo forms of receptors. However, from both interaction and functional studies, RIP140 may fit into this new category of bifunctional transcriptional coregulator. In summary, the studies reported here provide the evidence that RIP140 can function as a corepressor for the orphan receptor TR2. The corepressor activity of RIP140 depends on its interaction with the LBD of TR2 and is mediated by a transferable repressive activity of RIP140. The biochemical nature of RIP140 awaits further examination.
ACKNOWLEDGMENTS
This work was supported by NIH grants DK46866 and DA11190, a grant-in-aid from the graduate school of the University of Minnesota, a Leukemia Research Fund, and grant SMF2005-98 from the Minnesota Medical Foundation to L.-N.W. We thank the Core B of a PPG (DA08131) for help in oligonucleotide synthesis.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 3.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 5.Chinpaisal C, Lee C-H, Wei L-N. Mechanisms of the mouse orphan receptor TR2-11 mediated gene suppression. J Biol Chem. 1998;273:18077–18085. doi: 10.1074/jbc.273.29.18077. [DOI] [PubMed] [Google Scholar]
- 6.Chinpaisal C, Chang L, Hu X, Lee C-H, Wen W-N, Wei L-N. The orphan receptor TR2 suppresses a DR4 hormone response element of the mouse CRABP-I gene promoter. Biochemistry. 1997;36:14088–14095. doi: 10.1021/bi971598z. [DOI] [PubMed] [Google Scholar]
- 7.Chuang F M, West B L, Baxter J D, Schaufele F. Activities in Pit-1 determine whether receptor interacting protein 140 activates or inhibits Pit-1/nuclear receptor transcriptional synergy. Mol Endocrinol. 1997;11:1332–1341. doi: 10.1210/mend.11.9.9978. [DOI] [PubMed] [Google Scholar]
- 8.Collingwood T N, Rajanayagam O, Adams M, Wangner R, Cavailles V, Kalkhoven E, Matthews C, Nystrom E, Stenlof K, Lindstedt G, Tisell L, Fletterick R J, Parker M G, Chatterjee V K K. A natural transactivation mutation in the thyroid hormone β receptor: impaired interaction with putative transcriptional mediators. Proc Natl Acad Sci USA. 1997;94:248–253. doi: 10.1073/pnas.94.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagan R, Flint K J, Jones N. Phosphorylation of E2F-1 modulates its interaction with the retinoblastoma gene product and the adenoviral E4 19 kDa protein. Cell. 1994;78:799–811. doi: 10.1016/s0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 10.Feng W, Ribeiro R C J, Wanger R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 11.Hannon G J, Demetrick D, Beach D. Isolation of the RB-related p 130 through its interaction with CDK2 and cyclins. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 12.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 13.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 14.Henttu P M A, Kalkhoven E, Parker M G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamel Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature (London) 1995;377:397–403. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 17.Huang N, van Baur E, Garnier J-M, Lerouge T, Vonesch J-L, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikonen T, Palvimo J J, Janne O A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 19.Jenster G, Spencer T E, Burcin M N, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor induction of gene transcription—a two-step model. Proc Natl Acad Sci USA. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyeux A, Cavailles V, Balaguer P, Nicolas J C. RIP 140 enhances nuclear receptor-dependent transcription in vivo in yeast. Mol Endocrinol. 1997;11:193–202. doi: 10.1210/mend.11.2.9884. [DOI] [PubMed] [Google Scholar]
- 21.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 22.Lee C-H, Chang L, Wei L-N. Molecular cloning and characterization of a mouse nuclear orphan receptor expressed in embryos and testes. Mol Reprod Dev. 1996;44:305–314. doi: 10.1002/(SICI)1098-2795(199607)44:3<305::AID-MRD4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 23.Lee, C.-H., C. Chinpaisal, and L.-N. Wei. A novel nuclear receptor heterodimerization pathway mediated by orphan receptors TR2 and TR4. J. Biol. Chem., in press. [DOI] [PubMed]
- 24.Lee C-H, Chang L, Wei L-N. Distinct expression patterns and biological activities of two isoforms of the mouse orphan receptor TR2. J Endocrinol. 1997;152:245–255. doi: 10.1677/joe.0.1520245. [DOI] [PubMed] [Google Scholar]
- 25.Lee C-H, Copeland N G, Gilbert D J, Jenkins N A, Wei L-N. Genomic structure, promoter identification, and chromosomal mapping of a mouse nuclear orphan receptor expressed in embryos and adult testes. Genomics. 1995;30:46–52. doi: 10.1006/geno.1995.0007. [DOI] [PubMed] [Google Scholar]
- 26.Lee H-J, Chang C. Identification of human TR2 orphan receptor response element in the transcriptional initiation site of the simian virus 40 major late promoter. J Biol Chem. 1995;270:5434–5440. doi: 10.1074/jbc.270.10.5434. [DOI] [PubMed] [Google Scholar]
- 27.Lee H-J, Young W-J, Shih C C-Y, Chang C. Suppression of the human erythropoietin gene expression by the TR2 orphan receptor, a member of the steroid receptor superfamily. J Biol Chem. 1996;271:10405–10412. doi: 10.1074/jbc.271.17.10405. [DOI] [PubMed] [Google Scholar]
- 28.Le Jossic C, Michel D. Striking evolutionary conservation of a cis-element related to nuclear receptor target sites and present in TR2 orphan receptor genes. Biochem Biophys Res Commun. 1998;245:64–69. doi: 10.1006/bbrc.1998.8376. [DOI] [PubMed] [Google Scholar]
- 29.L’Horset F, Dauvois S, Heery D M, Cavailles V, Parker M G. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol Cell Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin T-M, Young W-J, Chang C. Multiple functions of the TR2-11 orphan receptor in modulating activation of two key cis-acting elements involved in retinoic acid signal transduction system. J Biol Chem. 1995;270:30121–30128. [PubMed] [Google Scholar]
- 31.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 32.Masuyama H, Brownfield C M, St-Arnaud R, MacDonald P N. Evidence for ligand-dependent intramolecular folding of the AF-2 domain in vitamin D receptor-activated transcription and coactivator interaction. Mol Endocrinol. 1997;11:1507–1517. doi: 10.1210/mend.11.10.9990. [DOI] [PubMed] [Google Scholar]
- 33.McInerney E M, Tsai M-J, O’Mally B M, Katzenellenbogen B S. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 35.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 36.Shibata H, Nawaz Z, Tsai S Y, O’Malley B W, Tsai M-J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 37.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 38.Swirnoff A H, Apel E D, Svaren J, Sevetson B R, Zimonjic D B, Popescu N C, Milbrandt J. Nab1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Mol Cell Biol. 1998;18:512–524. doi: 10.1128/mcb.18.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeshita A, Yen P M, Misiti S, Cardona G R, Liu Y, Chin W W. Molecular cloning and properties of a full-length putative thyroid hormone receptor coactivator. Endocrinology. 1996;137:3594–3597. doi: 10.1210/endo.137.8.8754792. [DOI] [PubMed] [Google Scholar]
- 40.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson J-A. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 41.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, Z., C.-H. Lee, C. Chinpaisal, and L.-N. Wei. A constitutive nuclear localization signal from the second zinc-finger of orphan nuclear receptor TR2. J. Endocrinol., in press. [DOI] [PubMed]
- 44.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]