Abstract
The intermediate chains (ICs) are the subunits of the cytoplasmic dynein that provide binding of the complex to cargo organelles through interaction of their N termini with dynactin. We present evidence that in Drosophila, the IC subunits are represented by at least 10 structural isoforms, created by the alternative splicing of transcripts from a unique Cdic gene. The splicing pattern is tissue specific. A constitutive set of four IC isoforms is expressed in all tissues tested; in addition, tissue-specific isoforms are found in the ovaries and nervous tissue. The structural variations between isoforms are limited to the N terminus of the IC molecule, where the interaction with dynactin takes place. This suggests differences in the dynactin-mediated organelle binding by IC isoforms. Accordingly, when transiently expressed in Drosophila Schneider-3 cells, the IC isoforms differ in their intracellular targeting properties from each other. A mechanism is proposed for the regulation of dynein binding to organelles through the changes in the content of the IC isoform pool.
Cytoplasmic dynein is a multisubunit complex composed of two heavy chains, three intermediate chains (ICs), several light ICs, and one light chain (11, 20). It acts as a minus end-directed microtubule motor, participating in a number of events including anterograde organelle movement (1, 6, 22), mitosis (25), nuclear migration (28), slow axonal transport in nervous tissue (7), and transport from nurse cell cytoplasm to oocytes in Drosophila ovaries (12). These events call for binding of the dynein complex to multiple target organelles in the cell. Regulation of this binding is also required to enable relocation of the dynein between targets during the cell cycle and development of the organism. One of the proposed mechanisms implemented in the cell cycle-dependent regulation of dynein binding is through the phosphorylation of the subunits of the dynein complex (17).
Although the heavy chain comprises the catalytic dynein subunit and is capable by itself of the ATP-dependent moving force production on the microtubules (14), the presence of other subunits is apparently required for dynein function in vivo. For one class of these so-called accessory subunits of cytoplasmic dynein, the IC subunits, a key role in linking cytoplasmic dynein to the intracellular targets was suggested and then proved (20, 23). In particular, the N-terminal part of IC is directly involved in binding to the organelles (23) through the interaction with p150/Glued, the major component of the dynactin complex (26). Dynactin, also a multisubunit complex, is an activator of dynein in vitro (9) and is required for dynein function in vivo (4, 15, 16). Dynein and dynactin are colocalized in the cell, and overexpression of components of the dynactin complex disrupts dynein binding to organelles (5, 8).
Considering dynactin as a dynein “receptor” or at least a modulator of dynein binding, the interaction of dynein ICs with dynactin is likely to be the point where the regulation of dynein binding takes place. A number of IC isoforms were detected, and the content of IC isoform pool is highly regulated (21). The complexity of IC isoforms is due to the expression of a family of structurally different polypeptides, some of which are further modified by phosphorylation (21, 26). The structural differences are limited to the N-terminal part of the ICs, in the region essential for dynactin binding (26). The phosphorylation is strongly suggested to occur in the same region which contains the serine-rich domain. Thus, the observed complexity of ICs presumably provides a diversity in dynactin-mediated dynein binding to organelles. This means that changing the content of the IC isoform pool would result in relevant changes in dynein targeting.
The mechanism for generating the structural complexity of ICs has been unclear. Alternative splicing of a limited number of transcripts was suggested (26) but never shown directly. In this paper, we demonstrate that in Drosophila, the structural IC isoforms are created by the alternative splicing of transcripts from a single-copy Cdic gene. The isoforms differ in the polymorphic region located near the N terminus of IC. The exact positions of the polymorphic regions differ in ICs from Drosophila and rats, suggesting independent evolution of IC isoform complexity in the ancestry of distant orders.
The splicing pattern of Cdic and therefore the content of the IC isoform pool appear to be tissue specific. In addition to the constitutive set, tissue-specific IC isoforms are present in ovaries and neural tissue, where tissue-specific kinds of dynein-dependent transport take place. The IC isoforms differ in their intracellular targeting properties, thus providing the mechanism for developmental regulation of dynein binding to organelles by changing the content of the IC isoform pool.
MATERIALS AND METHODS
RNA isolation and Northern analysis.
Total RNA was isolated from various developmental stages and from adult body parts of Drosophila melanogaster and from adults of D. simulans with Trizol (Gibco-BRL). Poly(A)+ RNA was purified from the total RNA preparations with Poly(A)-Tract magnetic particles (Promega Corp).
For Northern analysis, 10 μg of total RNA or 2 μg of poly(A)+ RNA was electrophoresed through a 1% agarose–formaldehyde gel and transferred onto a Hybond N membrane in 10× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]). Single-stranded 32P-labeled RNA probes were generated by T7 RNA polymerase from the pTZ19R-based plasmid containing the sequence of exon 6 (probe A in Fig. 2). Random priming with the Prime-It system (Stratagene) was used to generate 32P-labeled DNA probes from the same fragment A or from the fragment representing the first 680 bp of Cdic cDNA (probe B in Fig. 2). Hybridization procedures were as described previously (19).
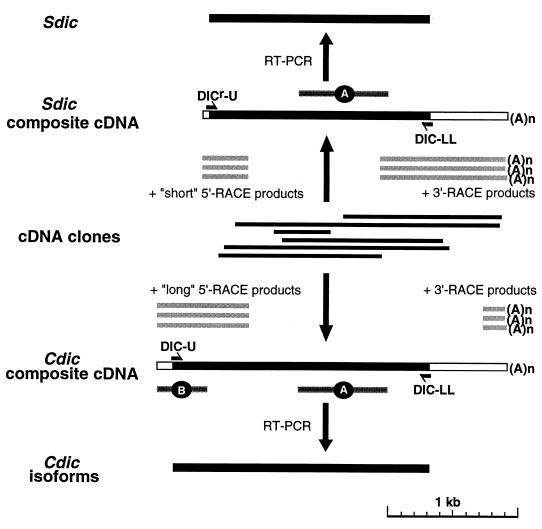
FIG. 2.
Cloning and characterization of Cdic and Sdic cDNAs. λZAP clones are indicated by thin black bars, and RACE products are indicated by shaded bars. In the composite cDNAs, coding regions are black. The positions for the DIC-U and DIC-LL primers, used to amplify cDNAs for Cdic isoforms, and for the DICr-U and DIC-LL primers, used for Sdic, are shown. A and B are the Cdic/Sdic-specific and Cdic-specific probes, respectively.
Southern hybridization with oligonucleotides.
Reverse transcription-PCR (RT-PCR) products corresponding to IC isoforms were separated in a 3% agarose–Tris-borate-EDTA (TBE) gel and transferred to a Nybond N membrane in 0.5 M NaOH–1 M NaCl by capillary blotting. The membrane was neutralized in 1 M ammonium acetate, air dried, and baked for 90 min at 80°C. The following primers, covering specific variable exon junctions, were synthesized (see Fig. 7): "iso2" (5′-TTATTATGATGAATAC-3′), covering the v2/v3 junction specific for Cdic2 and Cdic5; "iso2‴ (5′-CGGCGATGCTCATGCT-3′), covering the 4/v2 junction specific for Cdic1, Cdic2, and Cdic5; "iso3" (5′-CGGCGATGATGAATAC-3′), covering the 4/v3 junction specific for Cdic3; "iso4" (5′-CGGCGATGTGCTTGCA-3′), covering the 4/v4 junction specific for Cdic4; and "iso5" (TATATGGAGGACTGGT-3′), representing exon v1, specific for Cdic5. The primers were labeled with 32P by T4 DNA kinase and hybridized with the membrane in 4× SSPE–1% Sarkosyl, 0.1% each Ficoll-400, polyvinylpirrolidone, and sodium pyrophosphate for 2 to 14 h at the following temperatures: "iso2" at 42°C; "iso2‴ and "iso4" at 55°C; and "iso3" and "iso5" at 50°C. The membrane was washed in 100 mM sodium phosphate (pH 8.0)–1% Sarkosyl–1 mM EDTA three times for 15 min at room temperature and then once in 4× SSPE–1% Sarkosyl for 20 min at hybridization temperature.
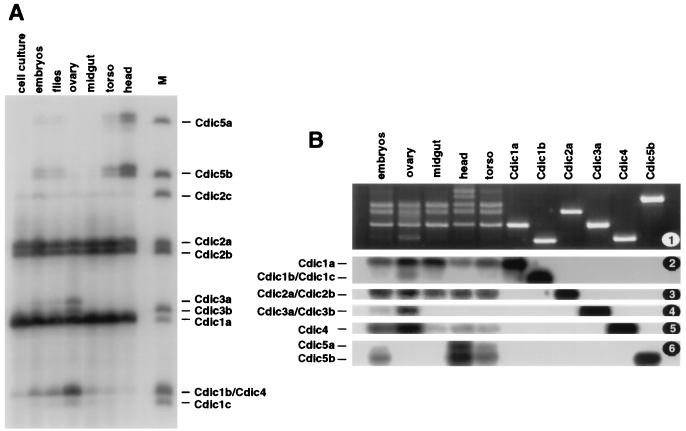
FIG. 7.
Tissue specificity of Cdic isoforms. RT-PCR fragments were amplified across the variable region of Cdic transcripts. (A) PCR fragments were labeled at one DNA strand with 32P and separated in a 5% acrylamide sequencing gel. The source of the RNA is indicated at the top. Lane M contains marker fragments generated from the of cDNA clones representing Cdic isoforms. (B) PCR fragments were separated in a 3% agarose gel (1) and, after Southern transfer, hybridized with oligonucleotides "iso2" (2), "iso2‴ (3), "iso3" (4), "iso4" (5), and "iso5" (6). The source of the RNA is indicated at the top; individual cDNA clones Cdic1a, Cdic1b, Cdic2a, Cdic3a, Cdic4, and Cdic5b were used to generate the marker fragments in the six right-hand lanes.
DNA cloning and sequencing.
cDNA clones were obtained by screening a λZAP cDNA library made from poly(A)+ RNA from D. melanogaster ovaries (supplied by Stratagene Corp.). Individual lambda clones were converted into the plasmid form by in vivo excision, and the inserts were transferred into the vector pSP72 and sequenced with an ABI 373A automated DNA sequencer after saturation with gamma-delta transposon insertions (24). Sequence data were analyzed with Sequencher software (GeneCodes Corp.).
The 5′- and 3′-RACE (rapid amplification of cDNA ends) PCR products were generated with the Marathon system (Clontech), using female poly(A)+ RNA as a template, and were sequenced after T-A cloning in the pCRII vector (Invitrogen).
A D. melanogaster P1 genomic library was screened by a PCR-based assay as described previously (10). The P1 clone containing the cytoplasmic dynein IC genes was subcloned in the λSCAN vector (18), and the subclone of interest was transferred into the vector pSP72 and sequenced as described above.
Plasmid constructs.
The green fluorescence protein (GFP) fusion expression plasmids were made by inserting the Cdic open reading frames (ORFs) upstream of the GFP ORF in pGreenLantern (Gibco-BRL). ORFs containing the specific Cdic isoforms were amplified by PCR with the corresponding cDNA clones as the templates and with the primers DIC-F (5′-GGTACCAGCTAATCGCCCCGAGAAATGGAT-3′) and DIC-LL (5′-GGCGGCCGCGTTCATCTTGATCTCGCTAAG-3′). The N-terminal domains were amplified with the primers DIC-F and DIC-R (5′-GCGGCCGCACGCACCACGAACCGCTGGAAG-3′). PCR products were cloned in the vector pCRII and transferred into the pGreenLantern as KpnI-NotI fragments. Partial digestions with NotI were used when necessary.
All the PCR fragments used for plasmid construction were generated with a polymerase mixture possessing proofreading activity (Elongase; Gibco-BRL), and their sequence was checked after cloning in the pCRII vector.
Cell culture transfections.
The D. melanogaster Schneider-3 cell culture was maintained in Drosophila Shields and Sang M3 medium (Sigma) supplemented with 10% insect medium supplement (Sigma) at room temperature. For transfection, the cells were plated on chamber slides (Falcon) at a density of 5 × 105 to 7 × 105 cells/ml and the next day were transfected with the Lipofectin reagent (Gibco-BRL). A 1.5-μg portion of DNA and 9 μl of Lipofectin were used per 5 × 105 to 7 × 105 cells. After 6 to 12 h, the medium was changed to M3 supplemented with 10% insect medium supplement; the cells were then allowed to grow for another 1 or 2 days and fixed for 10 min with 3.7% paraformaldehyde at room temperature. The cellular content was stained with propidium iodide. Alternatively, for Golgi-specific staining, the cells were permeabilized with 0.2% Triton X-100 for 15 min at room temperature and incubated with 20 μg of rhodamine-labeled Lens culinaris lectin per ml (Sigma). Lysosome-specific staining was obtained by in vivo incubation of cells with 50 nM LysoTracker DND-99 (Molecular Probes) for 2 h before fixation. The cells were mounted in Permount medium and imaged in a laser scanning microscope (Axiovert 100TV; Zeiss). The images were processed with Adobe Photoshop software.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank and are available under accession no. AF070687 to AF070699.
RESULTS
Multiple RNAs code for the cytoplasmic dynein IC proteins.
In a previous study, we characterized a tandem repeat in cytological region 19F of D. melanogaster (2). The unit of this repeat is 7.2 kb long and contains a fragment of the annexin X gene. It also contains a long ORF coding for a polypeptide with high similarity to the dynein IC proteins. Using a fragment of this ORF as a probe (probe A in Fig. 2), we were able to detect two bands on a Northern blot, one of 2.4 kb and one of 2.8 kb (Fig. 1).
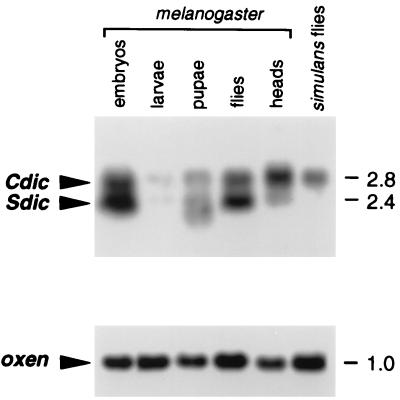
FIG. 1.
A. Dynein IC transcripts in Drosophila. Samples (10 μg) of total RNA isolated from various developmental stages of D. melanogaster, as indicated at the top, along with RNA samples from the heads of D. melanogaster adults or D. simulans adults, were separated in a 1% agarose–formaldehyde gel. Hybridization with probe A (Fig. 2) revealed two major bands, corresponding to the Cdic and Sdic transcripts. Only the Cdic transcripts were detected in D. simulans. (B) Control hybridization with the probe for the constitutively expressed gene oxen (1a) shows sufficient RNA loading on all lanes. The numbers on the right indicate the sizes of transcripts in kilobases.
Using the same probe, we isolated numerous cDNA clones from a D. melanogaster λZAP library. Six overlapping clones were sequenced, and the sequences were aligned, resulting in a composite cDNA sequence possessing an ORF for dynein IC polypeptide (Fig. 2). This composite sequence, however, obviously lacked both the 3′ and 5′ ends of the transcript, since neither a poly(A) tail nor a methionine initiation codon was detected. The 3′ end of the RNA was unambiguously mapped by performing 3′-RACE and sequencing several cloned PCR products. Mapping the 5′ end by 5′-RACE led to the description of two major classes of RNAs suggested from Northern analysis. The sequences of the 5′-RACE products could easily be sorted in two subsets, the long and short subsets. Aligning the short 5′ ends with the composite cDNA resulted in a 2.4-kb sequence, apparently representing a 2.4-kb RNA. The same alignment with the long 5′ ends produced a 2.8-kb sequence corresponding to the larger RNA.
The 70-kDa polypeptides encoded by the long, 2.8-kb mRNAs have extensive homology to the cytoplasmic dynein ICs from rats and Dictyostellium discoideum. The major stretch of homology covers more than 400 amino acids in the C-terminal part of molecule, which is 61% identical (79% similar) to the rat homolog and 50% identical (69% similar) to the protein from Dictyostellium discoideum. Included in the C-terminal region are four WD-40 repeats (see Fig. 6; a fifth repeat, described in reference 27, is very degenerate and is not shown here). This set of repeats is extremely strongly conserved among all dynein ICs and probably accounts for the interaction with other dynein subunits.
FIG. 6.
Sequence alignment of coding regions of the Cdic gene with Cdic cDNAs. The genomic sequence is at the top, with the gaps introduced in place of introns. cDNA sequences are below, shown as a single line in “constitutive” regions where they are identical and shown individually in the variable region. A conceptual protein sequence is shown at bottom; for the isoform Cdic5a, a frameshifted translation of exon v1 is included (peptide 5a). Coiled-coil donains, outlined as black boxes, were predicted with the PAIRCOILS (3) and COILS (13) algorithms. In the N-terminal region, serine-rich domain and the PPE/TQT conserved region are boxed. In the C-terminal region, four of five WD-40 repeats (27) are underlined.
An additional feature characteristic for the cytoplasmic dynein ICs is the presence of a coiled-coil domain at the N terminus followed by a serine-rich domain. Both these domains were detected in the 70-kDa polypeptide. Analysis of the alignment of this polypeptide with other cytoplasmic dynein ICs revealed another conserved stretch of amino acids in the N-terminal region, called PPE/TQT (Fig. 3).
FIG. 3.
Comparison of the sequence of the N-terminal regions of Cdic and other known cytoplasmic ICs from rats (GenBank accession no. U39046) and Dictyostellium (accession no. U25116). Conserved amino acids are outlined and presented in the consensus line. Coiled-coil domains, shown by solid boxes, were predicted with the PAIRCOILS (3) and COILS (13) algorithms. The serine-rich domain and PPE/TQT conserved block are outlined by boxes. The positions of variable regions in the rat IC (var-1 and var-2) and the beginning of the variable region in Cdic (var) are indicated. Cdic isoform shown is Cdic5b.
Based both on sequence homology and structural similarity to the cytoplasmic dynein ICs from rats and Dictyostelium discoideum, the 70-kDa polypeptide encoded by the long mRNAs was defined as the D. melanogaster cytoplasmic dynein IC (Cdic). In contrast, the 60.4-kDa polypeptide encoded by the short 2.4-kb mRNA lacks the N-terminal coiled-coil and serine-rich domains characteristic of ICs of cytoplasmic dyneins, although it shares most of its sequence with the Cdic polypeptide. It was shown to represent a novel sperm-specific IC subunit of axonemal dynein and was called Sdic (17a).
Amplification and sequencing of full-length Cdic cDNAs revealed multiple transcripts that differ by small insertions and deletions in the N-terminal part of the ORF and apparently code for the Cdic isoforms.
2.8-kb Cdic RNAs are transcribed from the single-copy cytoplasmic dynein IC (Cdic) gene.
Although all dynein IC cDNAs were isolated with a fragment of the 7.2-kb annexin-dynein repeat, the very 5′ end of the Cdic transcripts is not homologous to the repeated unit. This sequence, represented by probe B specific for Cdic transcripts (Fig. 2), was found to be unique in the genome on the basis of Southern analysis and mapped by in situ hybridization in the site 19E, i.e., in close vicinity to the annexin-dynein repeat. Northern analysis demonstrated that, as expected, the same Cdic-specific probe B hybridized with only the 2.8-kb Cdic mRNAs and not with the 2.4-kb Sdic mRNAs (data not shown).
When a P1 phage genomic library was screened for the Cdic-specific sequence, three clones that also contained the annexin-dynein repeat were obtained. None of these three clones, however, contained the complete annexin X gene located at the end of annexin-dynein tandem cluster (2). Considering the length of the tandem repeat (about 10 copies at 7.2 kb each) and the average length of a P1 clone (80 kb), these data suggest that the Cdic-specific sequence is located in the vicinity of the tandem cluster of annexin-dynein repeats, at the opposite end from the annexin X gene.
Cloning and sequencing of the corresponding genomic region revealed the structure of the gene encoding the Cdic transcripts. This Cdic gene is located at the 5′ end of the tandem cluster, and its 3′ end is directly fused to the initial 7.2-kb annexin-dynein repeated unit. The exon-intron structure of the 8.3-kb Cdic transcription unit was determined by aligning the Cdic cDNA sequences to the genomic sequence. A perfect match was obtained between Cdic cDNAs and the exons of the Cdic gene. A number of significant differences, however, were detected between the Cdic genomic sequence and the 2.4-kb Sdic cDNA, indicating that, unlike 2.8-kb Cdic cDNAs, this one does not represent the transcript from the Cdic gene. The true origin of Sdic transcripts was exposed, since a perfect match was achieved between the sequences of the Sdic cDNA and the annexin-dynein repeat (Fig. 4). The identity of 2.4-kb Sdic mRNAs as the transcripts from the annexin-dynein repeat was further supported by the fact that in D. simulans, a close relative of D. melanogaster that does not have any repeated structure analogous to the annexin-dynein repeat, no 2.4-kb Sdic transcripts were found: the only class of dynein mRNAs detected corresponds to the 2.8-kb Cdic mRNAs (Fig. 1).
FIG. 4.
Sequence comparison shows that Cdic cDNA represents the transcripts from the Cdic gene and that Sdic cDNA corresponds to the transcripts from the annexin-dynein repeat. The entire Cdic gene sequence is presented; for cDNAs and the annexin-dynein repeat, only the differences are shown. Gaps introduced in the sequences are marked with dots.
Transcription of Cdic changes throughout the development of D. melanogaster (Fig. 1). The transcripts are abundant in embryos and adult flies and apparently are up-regulated in the heads of adult flies, but they are hardly detectable in larvae and pupae.
Multiple Cdic isoforms are generated by alternative splicing.
Previous data state that all Cdic mRNAs are transcribed from the unique Cdic gene, even though multiple variants of Cdic transcripts were detected. Analysis of the exon-intron structure of the Cdic gene demonstrated that these variants, coding for the Cdic isoforms, are created by alternative splicing.
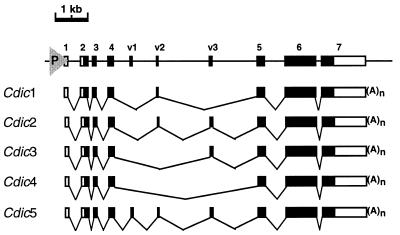
The transcription unit is 8.3 kb long and consists of 10 exons. The first four exons (1 to 4 in Fig. 5) are separated by relatively small introns, as are the last three exons (5 to 7 in Fig. 5). These two groups of exons are separated by 4.2-kb “spacer” region containing three “variable” exons, v1, v2, and v3. Alternative splicing of transcripts leads to the skipping of the variable exons, providing shortened versions of mRNAs apparently carrying the corresponding deletions in the dynein IC ORF (Fig. 5 and 6). Additional polymorphism of the mRNAs is created by using two alternative splice acceptor sites preceding exon v1 and three alternative splice acceptor sites of the intron preceding exon d5, which also results in insertions/deletions in the same region of the dynein IC ORF (Fig. 6). Use of one of the acceptor sites preceding exon v1 leads to the frameshift and premature termination of translation (isoform Cdic5a in Fig. 6). Except for this one, as many as 10 full-sized Cdic isoforms are generated by alternative splicing of Cdic transcripts.
FIG. 5.
Exon-intron structure of the Cdic gene. The genomic sequence is shown at the top, with exons indicated by boxes. Coding sequences are shown as solid boxes. The promoter is shown as triangle. 1 to 7, constitutive exons present in all Cdic mRNAs. v1 to v3, variable exons. Five classes of Cdic transcripts are shown below the sequence.
The content of the Cdic isoform pool is tissue specific.
The representation of the Cdic isoforms in the total Cdic pool was determined by two methods. First, RT-PCR was performed on the mRNA isolated from the embryos and from the adult flies by using the primers flanking the Cdic ORF, as shown in Fig. 2. PCR products were cloned, and a number of randomly picked clones were sequenced, resulting in the data presented in Table 1. Second, RT-PCR was carried out on the RNA isolated from embryos and adult females and also from the dissected female parts (ovaries, midguts, heads, and torsos) by using the primers var-U (5′-GCAGGCTACGAACATTCCA-3′) and var-L (5′-GCAGGCCGTGGGTGAGATA-3′), positioned close to the variable region of the ORF. PCR products were purified and subjected to 10 cycles of mock sequencing with the Taq polymerase in the presence of deoxynucleoside triphosphates and primer var-L labeled with 32P by T4 DNA kinase. The reaction products were separated in the sequencing gel and detected by autoradiography (Fig. 7A). The gel image was analyzed with BioMax 1D software (Kodak), producing the quantitative data in Table 2. Females were used here to avoid interference with the testis-specific expression of the Sdic. In males, similar representation of isoforms was detected in the midgut, torso, and head.
TABLE 1.
Representation of isoforms in Cdic cDNA clonesa
| Cdic isoform | % of Cdic cDNAs (no. of clones) inb:
|
|
|---|---|---|
| Embryos | Flies | |
| 1a | 38 (19) | 45 (27) |
| 2a | 14 (7) | 27 (16) |
| 2b | 20 (10) | 8 (5) |
| 2c | 2 (1) | 0 (0) |
| 1b | 8 (4) | 8 (5) |
| 4 | 8 (4) | 3 (2) |
| 1c | 0 (0) | 2 (1) |
| 3a | 6 (3) | 2 (1) |
| 3b | 2 (1) | 0 (0) |
| 5a | 0 (0) | 2 (1) |
| 5b | 2 (1) | 3 (2) |
| Total | 50 | 60 |
cDNAs for Cdic isoforms were amplified as shown in Fig. 2, and PCR fragments were cloned. A total of 50 randomly chosen clones were sequenced for cDNA from embryos, and 60 clones were sequenced for flies.
For each isoform, the percent representation in the cDNA pool is shown, as calculated from the number of relevant clones detected (in parentheses).
TABLE 2.
Representation of isoforms in Cdic mRNAs from different sourcesa
| Cdic isoform | % of Cdic mRNA inb:
|
|||||
|---|---|---|---|---|---|---|
| Embryo | Fly | Ovary | Midgut | Head | Torso | |
| Constitutive | ||||||
| 1a | 41 ± 3 | 54 ± 4 | 43 ± 1 | 51 ± 1 | 36 ± 6 | 49 ± 5 |
| 2a | 27 ± 4 | 28 ± 5 | 22 ± 1 | 24 ± 5 | 21 ± 1 | 27 ± 4 |
| 2b | 15 ± 2 | 20 ± 4 | 11 ± 2 | 16 ± 3 | 11 ± 1 | 17 ± 2 |
| 2c | 4 ± 1 | 4 ± 1 | 6 ± 3 | 4 ± 1 | 3 ± 2 | 2 ± 2 |
| Ovary-specific | ||||||
| 1b+4 | 10 ± 4 | 8 ± 2 | 17 ± 2 | 2 ± 1 | ND | ND |
| 1c | 3 ± 1 | 1 ± 1 | 3 ± 2 | ND | ND | ND |
| 3a | 4 ± 1 | 2 ± 1 | 8 ± 2 | ND | ND | ND |
| Head-specific | ||||||
| 5a | NDc | 1 ± 1 | ND | ND | 11 ± 4 | 2 ± 1 |
| 5b | 3 ± 2 | 3 ± 1 | ND | ND | 23 ± 3 | 6 ± 1 |
Cdic RT-PCR products amplified from the RNA extracted from indicated sources were end-labeled with 32P and separated in a sequencing gel, as shown in Fig. 7. The gel was exposed to Biomax-MR film (Kodak), the film was scanned, and the image was analyzed with Biomax-1D software (Kodak) to quantitate the intensity of the bands.
Representation of the isoform in the total mRNA pool was calculated by normalizing the intensity of relevant band by the total intensity of all specific bands on the lane. The numbers indicate the mean and standard error of results obtained in three experiments.
ND, nondetectable.
The data presented in both tables are in good agreement, indicating that the bulk of Cdic mRNAs is represented by three isoforms, Cdic1a, Cdic2a, and Cdic2b. They usually make up over 70% of Cdic mRNAs and are constitutively expressed in flies and embryos and in all tested body parts. The other eight isoforms usually make up to less than 30% of the pool. Only one of these “minor” isoforms, Cdic2c, is constitutively expressed at low levels. All the others demonstrate tissue specificity, being differentially expressed in the fly body. In particular, it appears that the isoforms Cdic1b and/or Cdic4, Cdic1c, and Cdic3a are up-regulated in the ovaries and Cdic5a and Cdic5b are overexpressed in the head and, to a lesser extent, in the torso. In these cases, the representation of some of the tissue-specific isoforms in the organ (17% of Cdic1b/Cdic4 in the ovary and 23% of Cdic5b in the head) is at the same level as that of major constitutively expressed isoforms.
Since isoforms Cdic1b and Cdic4 have exactly the same length of variable region, they could not be resolved by gel electrophoresis. To discriminate between these two isoforms and to confirm the identity of the RT-PCR products seen in the sequencing gel, they were separated in a 3% agarose gel, transferred to the nylon membrane, and hybridized to the labeled oligonucleotides specific to the particular isoforms. As seen in Fig. 7B, Cdic4, as well as Cdic3 and Cdic1b/Cdic1c, is abundant in ovaries, while Cdic1a and Cdic2a/Cdic2b are constitutively expressed. As expected, Cdic5a and Cdic5b are overexpressed in head and torso.
Cdic isoforms differ in intracellular distribution.
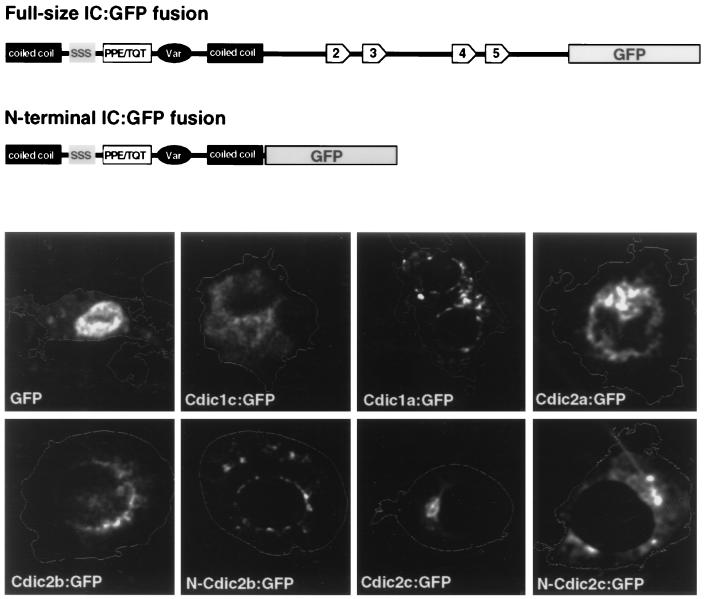
Structural variations between Cdic isoforms are limited to the N-terminal region involved in the interaction with dynactin and binding of the dynein complex to the organelles. We suggested that these variations may impel the differences in the targeting properties of Cdic isoforms. To check whether this may be true, Cdic isoforms were expressed in Drosophila Schneider-3 cell culture, and their intracellular distribution was analyzed. Since Schneider-3 cells possess only the constitutive set of endogenous Cdic isoforms (Fig. 7A), we excluded the tissue-specific isoforms from this analysis. To visualize the molecules in the cell, the polypeptides were fused at the carboxyl terminus to the green fluorescent protein (GFP).
When overexpressed at high levels, fusion proteins uniformly stained the cytoplasm. At low levels of expression, however, pronounced differences in the intracellular distribution of the fusion proteins were observed (Fig. 8). One of the ovary-specific isoforms, Cdic1c, was still distributed more or less diffusely throughout the cytoplasm, apparently reflecting the lack of ovary-specific target in the cultured cells. This isoform was chosen as a control, in contrast to the constitutive isoforms, which should have the relevant targets in the cultured cells, and clearly demonstrated different intracellular localizations.
FIG. 8.
Localization of the Cdic-GFP fusion proteins in cultured Schneider-3 cells. A schematic representation of the full-size fusion and N-terminal fusion proteins is shown at the top. Cells were transfected with plasmids expressing fusion proteins under the control of the cytomegalovirus promoter and stained with propidium iodide. Staining of the cellular content with propidium iodide was detected in the rhodamine channel, and the image was converted to the contour of the cell. GFP fluorescence was detected in the fluorescein isothiocyanate channel and pseudocolored in white. IC isoforms are indicated; for example, Cdic1a:GFP is a full-size fusion of Cdic1a isoform with GFP, and N-Cdic2b and N-Cdic2c are the N-terminal fusions. The localization of the GFP expressed alone is shown for comparison.
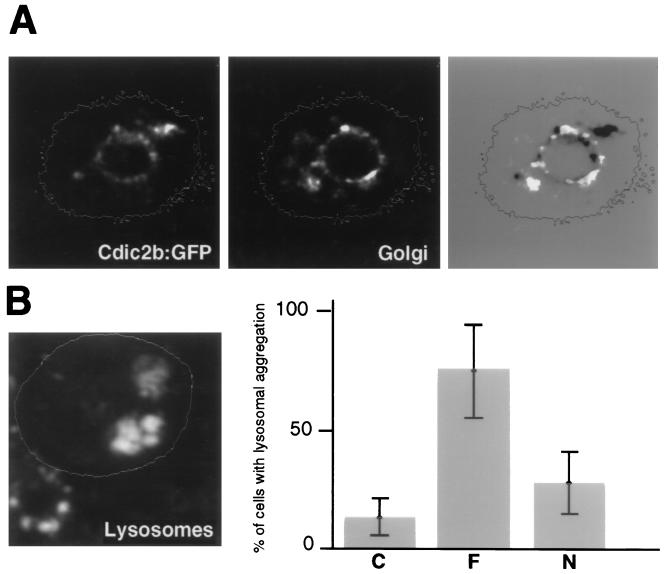
Fusion proteins corresponding to the constitutively expressed isoforms Cdic2a and Cdic2b possessed perinuclear localization, which is tighter for Cdic2b. The aggregation of Cdic2b fusion is more proximal than the perinuclear structures stained for Golgi (Fig. 9A) and most likely represents binding to the nuclear envelope. For the isoform Cdic1a, numerous small aggregations were detected and were distributed in the cytoplasm and around the nucleus. In the case of the isoform Cdic2c, the protein accumulated in one local area, as in Fig. 8, or in several patches often clustered in a sector of cytoplasm. In all cases, distribution of the fusion proteins was strictly different from the strong nuclear localization of GFP tag expressed alone (Fig. 8).
FIG. 9.
(A) Tight association of the Cdic2b fusion protein with the nucleus. In this case, Golgi staining mostly reveals the perinuclear elements. On the right, the GFP fluorescence image was converted to the negative and overlaid on the Golgi staining image. Note that the GFP fusion protein is located more proximal than the Golgi elements. (B) Redistribution of the lysosomes in the cells overexpressing the Cdic2c:GFP fusion protein. On the left, two transfected cells are contoured. Staining with Lysotracker DND-99 revealed aggregation of the lysosomes in transfected cells, in contrast to the random and mostly juxtranuclear distribution in nontransfected cell in the left lower corner. On the right, the frequency of the lysosomal aggregation in cells expressing Cdic2c:GFP (F) or N-Cdic2c:GFP (N) versus nontransfected cells (C) is shown. Error bars represent 95% confidence intervals.
In the cells overexpressing Cdic fusion proteins at a high level, morphological changes which would result from competitive inhibition of dynein binding to organelles may be expected. In particular, the distribution of lysosomes in the cytoplasm was checked for all constitutive isoforms, and only in the case of the Cdic2c fusion was redistribution of lysosomes observed. This effect has been proved to be indicative of disruption of the dynein IC-dynactin interaction and, therefore, of inhibition of dynein binding (5). In 65% of transfected cells, lysosomes were aggregated in one local area, as seen in Fig. 9B, or along the periphery of one side of the cell. This was found in only 12% of the untransfected cells, in which much more random and mostly juxtranuclear distribution was usually observed.
Since interaction with dynactin seems to be limited to the N-terminal portion of dynein ICs (26), we tested whether fusion proteins containing only this N-terminal part of ICs can possess isoform-specific intracellular distribution. For N-Cdic2b, perinuclear localization was still observed, but it was not as specific as that with the complete DIC2b fusion (Fig. 8). Overexpression of the N-Cdic2c protein caused lysosomal aggregation in 30% of transfected cells—somewhat greater than the 12% in nontransfected cells, but far less than the 65% in cells expressing the complete Cdic2c fusion (Fig. 9). This may explain the differences in intracellular localization between Cdic2c and N-Cdic2c; while the first usually accumulated in a local area of cytoplasm, presumably binding to the lysosomal aggregates, the localization of the other suggests binding to the normally distributed lysosomes (Fig. 8).
DISCUSSION
Alternative splicing of Cdic transcripts generates the diversity of cytoplasmic dynein ICs.
Although the isoform complexity of dynein ICs has been studied for a long time (21, 26), the molecular basis for it has been unknown, since no gene has been cloned to date. However, the suggestion has been presented that in the rat, the structural isoforms are produced by alternative splicing of transcripts from two highly homologous genes (26).
Our data directly demonstrate that in the D. melanogaster species group, the Cdic diversity is created by alternative splicing of transcripts from a unique gene. Alternative splicing of exons and multiple splice acceptor sites are used, resulting in both length and amino acid sequence variability in the short region near the N terminus of the polypeptide. As many as 10 isoforms were detected and characterized.
Comparison of the N-terminal part of Drosophila Cdic polypeptides with the cytoplasmic dynein ICs from rat revealed that the presence of the variable region created by the alternative splicing is conserved. This does not apply, however, to the location of this region: there are two variable regions in the rat polypeptide, flanking the serine-rich domain, and only one variable region in the Drosophila polypeptide (Fig. 3). Moreover, the position of the single variable region in the Drosophila protein does not correspond to the position of any of the two regions in rats, since these two are located on the different side of a well-conserved PPE/TQT block of amino acids. Therefore, it may be suggested that the mechanism of alternative splicing, generating the diversity of the N termini of dynein ICs, evolved independently in Drosophila and rats. This finding emphasizes the importance of IC heterogeneity for dynein functions and raises further questions about the implications of this heterogeneity.
The tissue specificity of splicing alters the content of Cdic isoform pool.
Although at least 10 isoforms are created by alternative splicing, only 4 are represented in consistent amounts in all tested tissues. These are referred to as constitutive, and in some tissues, for example in the midgut, or in cultured cells, they make up all the content of isoform pool. This means that these four Cdic isoforms provide at least the requisite set of the dynein-mediated activities. It still is not apparently enough in the other cases, requiring more specialized actions, since up-regulation of other isoforms was observed in nervous tissue (in particular, in the head) and ovaries. This up-regulation clearly reflects changes in the splicing pathways and leads to a lowered percent representation of constitutive isoforms, since the tissue-specific isoforms contribute a significant proportion (up to a quarter) of the total pool. This, however, does not seem to cause depletion of the constitutive isoforms, since at least in the head it is compensated for by the overall increase in expression of Cdic (Fig. 1).
In neurons, a specific and very active process of axonal transport takes place, and it has been shown to be cytoplasmic dynein dependent (7). In the ovaries, extensive transport takes place from the nurse cell cytoplasm to the oocyte, and this transport has also been shown to depend on cytoplasmic dynein (12). It seems than wherever specific transport takes place, specific Cdic isoforms are added to the basic set. Also, since the specificity of these kinds of transport mainly concerns the type of cargo organelle to be moved, it seems very likely that the specific Cdic isoforms provide targeting of dynein complex to these peculiar cargoes.
Cdic isoforms differ in intracellular targeting.
Considering the multiplicity of intracellular targets for dynein binding and the diversity of IC isoforms, it seems very likely that different IC isoforms provide targeting of the dynein complex to distinct organelles. If this were the case, the constitutive IC isoforms would furnish the targeting to organelles regularly bound to dynein, for example, lysosomes, Golgi complex, nuclei, and mitotic chromosomes. Tissue-specific IC isoforms, in turn, would provide dynein binding to cargo organelles that are specific to the tissue and are not regularly present or are not bound with dynein in other tissues.
When IC isoforms fused to the GFP at the C terminus were expressed in the cultured cells, they possessed different patterns of intracellular distribution. Although it is not clear whether these patterns reflect the distribution of endogenous ICs, it is certain that they are due to the variable N termini of ICs, since all other parts of the expressed fusion proteins are identical.
For the fusion proteins corresponding to the tissue-specific Cdic1c, a uniform diffuse distribution in the cytoplasm or aggregation around small, unidentified structures was observed (data not shown). This pattern, which may reflect the absence of tissue-specific targets for these ICs in the cytoplasm of cultured cells, is clearly different from that observed for the constitutive ICs. An apparent binding to the nuclear envelope was detected for Cdic2b. Overexpression of the Cdic2c fusion protein caused relocation of the lysosomes. Considering that this effect was not detected for other isoforms, Cdic2c could be called the lysosome-specific IC isoform.
It has been demonstrated that targeting of the organelles by ICs requires the N-terminal portion of the molecule (23). We checked whether this N-terminal portion of IC, fused to GFP, is capable of the specific targeting observed for the full-size IC fusion. This was somewhat true for isoform Cdic2b, although the pattern was not as well defined as with the full-size fusion protein. Overexpression of the N-terminal fusion for isoform Cdic2c had little effect on the lysosome relocation, compared to that of the full-size protein. This implies that the C-terminal part of the IC molecule, which has been considered to be a domain responsible for interaction with other components of the dynein complex, is also important for the organelle targeting. It is conceivable that the incorporation of the ICs in the dynein complex plays a substantial role. Resolution of this question requires additional experiments that include localization of endogenous IC isoforms in fly tissues. This would also provide additional data on the specificity of organelle targeting by isoforms.
The data presented here support a model in which alternative splicing generates a number of cytoplasmic dynein IC isoforms that differ in their targeting properties. These IC subunits mediate dynein binding to organelles, enabling various types of minus-end directed transport along microtubules. The default splicing pattern provides isoforms necessary for dynein-mediated “housekeeping” activities necessary for maintaining any cell or tissue, including anterograde movement and positioning of lysosomes and Golgi complex, mitosis, and nuclear migration. Tissue-specific alteration in the splicing supplies additional IC isoforms, capable of targeting cargo organelles that are subject to unusual tissue-specific transport. This mechanism provides a long-term modulation of the dynein binding to organelles in the development of organism, in addition to more swift regulation by phosphorylation of dynein subunits described previously (17).
ACKNOWLEDGMENTS
Y. Y. Shevelyov and V. A. Gvozdev were supported by grants 96-04-49015 and 96-15-98072 from the Russian Foundation for Basic Research.
REFERENCES
- 1.Aniento F, Emans N, Griffiths G, Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Benevolenskaya, E. Unpublished data.
- 2.Benevolenskaya E V, Nurminsky D I, Gvozdev V A. Structure of the Drosophila melanogaster annexin X gene. DNA Cell Biol. 1994;14:349–357. doi: 10.1089/dna.1995.14.349. [DOI] [PubMed] [Google Scholar]
- 3.Berger B, Wilson D B, Wolf E, Tonchev T, Milla M, Kim P S. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruno K S, Tinsley J H, Minke P F, Plamann M. Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc Natl Acad Sci USA. 1996;93:4775–4780. doi: 10.1073/pnas.93.10.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burkhardt J K, Echeverri C J, Nilsson T, Vallee R B. Overexpression of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corthesy-Theulaz I, Pauloin A, Rfeffer S R. Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dillman J F, Dabney L P, Pfister K K. Cytoplasmic dynein is associated with slow axonal transport. Proc Natl Acad Sci USA. 1996;93:141–144. doi: 10.1073/pnas.93.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echeverri C J, Paschal B M, Vaughan K T, Vallee R B. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill S R, Schroer T A, Szilak I, Steuer E R, Sheetz M P, Cleveland D W. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartl D L, Nurminsky D I, Jones R W, Lozovskaya E R. Genome structure and evolution in Drosophila: application of the framework P1 map. Proc Natl Acad Sci USA. 1994;91:6824–6829. doi: 10.1073/pnas.91.15.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King S M, Barbarese E, Dillman J F, Patel-King R S, Carson J H, Pfister K K. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996;271:19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- 12.Li M, McGrail M, Serr M, Hays T S. Drosophila cytoplasmic dynein, a microtubule motor that is asymmetrically localized in the oocyte. J Cell Biol. 1994;126:1475–1494. doi: 10.1083/jcb.126.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 14.Mazumdar M, Mikami A, Gee M A, Vallee R B. In vitro motility from recombinant dynein heavy chain. Proc Natl Acad Sci USA. 1996;93:6552–6556. doi: 10.1073/pnas.93.13.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGrail M, Gepner J, Silvanovich A, Ludmann S, Serr M, Hays T S. Regulation of cytoplasmic dynein function in vivo by the Drosophila Glued complex. J Cell Biol. 1995;131:411–425. doi: 10.1083/jcb.131.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muhua L, Karpova T S, Cooper J A. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- 17.Niclas J, Allan V J, Vale R D. Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J Cell Biol. 1996;133:585–593. doi: 10.1083/jcb.133.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Nurminsky, D. Unpublished data.
- 18.Nurminsky D I, Hartl D L. Sequence scanning: a method for rapid sequence acquisition from large-fragment DNA clones. Proc Natl Acad Sci USA. 1996;93:1694–1698. doi: 10.1073/pnas.93.4.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurminsky D I, Moriyama E N, Lozovskaya E R, Hartl D L. Molecular phylogeny and genome evolution in the Drosophila virilis species group: duplications of the alcohol dehydrogenase gene. Mol Biol Evol. 1996;13:132–149. doi: 10.1093/oxfordjournals.molbev.a025551. [DOI] [PubMed] [Google Scholar]
- 20.Paschal B M, Mikami A, Pfister K K, Vallee R B. Homology of the 74-kD cytoplasmic dynein subunit with a flagellar dynein polypeptide suggests an intracellular targeting function. J Cell Biol. 1992;118:1133–1143. doi: 10.1083/jcb.118.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfister K K, Salata M W, Dillman J F, Torre E, Lye R J. Identification and developmental regulation of a neuron-specific subunit of cytoplasmic dynein. Mol Biol Cell. 1996;7:331–343. doi: 10.1091/mbc.7.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroer T A, Steuer E R, Sheetz M P. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 23.Steffen W, Karki S, Vaughan K T, Vallee R B, Holzbaur E L F, Weiss D G, Kuznetsov S A. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol Biol Cell. 1997;8:2077–2088. doi: 10.1091/mbc.8.10.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strathmann M, Hamilton B A, Mayeda C A, Simon M I, Meyerowitz E M, Palazzolo M J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci USA. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaisberg E A, Koonce M P, McIntosh J R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaughan K T, Vallee R B. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkerson C G, King S M, Koutoulis A, Pazour G J, Witman G B. The 78,000 M(r) intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang X, Beckwith S M, Morris N R. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]