Significance
Bacteroidota are key members of the human gut microbiota that can modulate the enteric environment by producing vesicles. Bacterial vesiculation is a poorly understood process. Here, we developed a high-throughput screen to identify genes required for vesiculation in Bacteroides thetaiotaomicron (Bt). Our screen identified a family of Dual membrane-spanning anti-sigma factors (Dma) that play a role in controlling vesiculation. We show that the Dma family are proteins shown to span both the inner and outer membrane of a gram-negative bacterium. This work provides insights regarding the regulation of vesiculation among Bacteroidota.
Keywords: Bacteroides, microbiota, regulation, signaling, vesicles
Abstract
Bacteroidota are abundant members of the human gut microbiota that shape the enteric landscape by modulating host immunity and degrading dietary- and host-derived glycans. These processes are mediated in part by Outer Membrane Vesicles (OMVs). Here, we developed a high-throughput screen to identify genes required for OMV biogenesis and its regulation in Bacteroides thetaiotaomicron (Bt). We identified a family of Dual membrane-spanning anti-sigma factors (Dma) that control OMV biogenesis. We conducted molecular and multiomic analyses to demonstrate that deletion of Dma1, the founding member of the Dma family, modulates OMV production by controlling the activity of the ECF21 family sigma factor, Das1, and its downstream regulon. Dma1 has a previously uncharacterized domain organization that enables Dma1 to span both the inner and outer membrane of Bt. Phylogenetic analyses reveal that this common feature of the Dma family is restricted to the phylum Bacteroidota. This study provides mechanistic insights into the regulation of OMV biogenesis in human gut bacteria.
Vesiculation is a process by which cells utilize membranous compartments to traffic cellular contents (1, 2). In eukaryotes, vesiculation, in the form of the trans-Golgi network, exosomes, and other extracellular vesicles, has been extensively studied (2–5). However, much less is known about vesiculation in bacteria (6–8). Outer Membrane Vesicles (OMVs) are small, spherical structures generated by the active blebbing of the outer membrane (OM) in gram-negative bacteria. Due to their OM origin, OMVs are composed of lipopolysaccharides (LPS), phospholipids, OM proteins, and periplasmic contents (6–8). OMVs have been studied in many gram-negative bacteria and are reported to mediate key bacterial processes, including pathogenesis, quorum sensing, immunomodulation, nutrient uptake, and envelope stress response (9–15). In spite of this, no general mechanisms for OMV biogenesis in gram-negative bacteria have been established (1, 6–8). The current consensus is that vesicles can be derived from active processes or passive phenomena involving membrane destabilization. Recent reports have shown that explosive cell lysis is the primary source of vesicles in Pseudomonas aeruginosa (16). It is hinted that this process occurs in other closely related species (17), which suggests that OMVs may not be actively produced in these organisms. In contrast, a growing body of literature has demonstrated that OMV biogenesis in Bacteroides spp. is a highly regulated process (18–20). Nonetheless, the mechanism of OMV biogenesis in Bacteroidota remains poorly understood.
Bacteroides spp. are gut commensals that comprise ~40% of the bacterial species in the human gastrointestinal tract (21–23). Bacteroides shape the intestinal environment by producing immunogenic compounds and degrading dietary- and host-derived glycans (24, 25). Studies have shown that OMVs produced by Bacteroides spp. help facilitate many of their functions in the gut (11, 12, 14, 15, 18, 26). Transmission electron microscopy (TEM) revealed that Bacteroides thetaiotaomicron (Bt) and Bacteroides fragilis (Bf) produce large quantities of OMVs of uniform size (19, 20). Mass spectrometry (MS) analyses revealed that OMV protein cargo is primarily composed of lipoproteins that function as glycosidases or proteases, and this cargo is tailored according to the available glycan landscape (18, 20). In addition, OMV-enriched lipoproteins contain a negatively charged motif (S-D/E3), called the Lipoprotein Export Sequence (LES), that is absent from OM-retained lipoproteins (18, 20, 27). These features are unprecedented and distinguish Bacteroides OMVs from the vesicles isolated by other gram-negative bacteria. By exploiting these characteristics, we previously labeled OM- and OMV-specific proteins with fluorescent markers to visualize OMV biogenesis in Bt. Fluorescence microscopy was employed to observe OMVs actively blebbing from the OM of Bt in the absence of cell lysis (18). Altogether, these findings demonstrate that OMV biogenesis in Bacteroides spp. is the result of an active, regulated process and not the result of cell lysis, as has been suggested for other bacteria. In this work, we developed a high-throughput screen to identify components of the machinery involved in OMV biogenesis and its regulation in Bt. We identified and characterized a family of structurally unique Dual membrane-spanning anti-sigma factors (Dma) and investigated their role in modulating OMV biogenesis in Bt.
Results
Screening for Genes Involved in OMV Biogenesis.
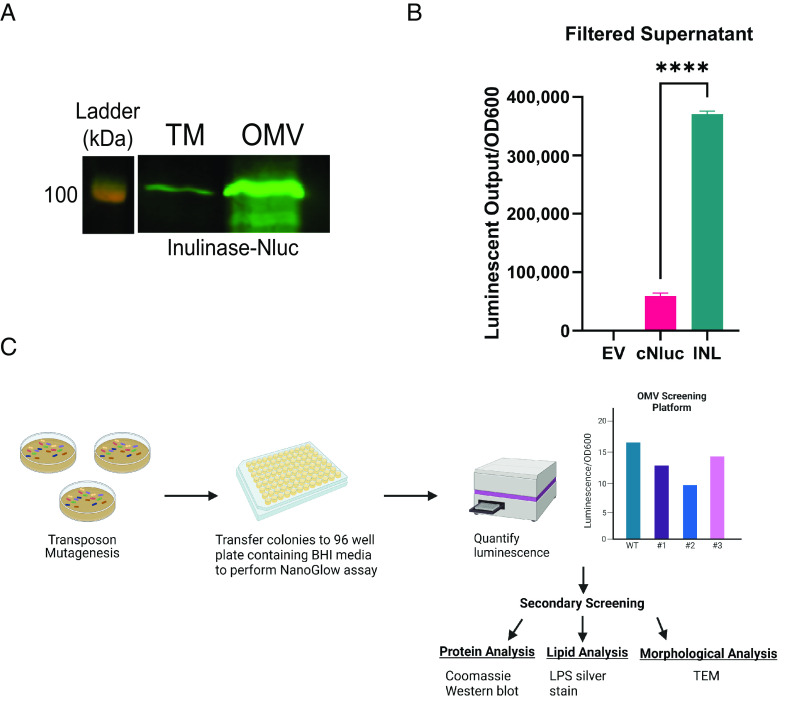
Vesiculation has been studied in gram-negative bacteria for ~60 y; however, our understanding of OMV biogenesis is still in its infancy. To identify bacterial proteins involved in OMV biogenesis in Bt, we developed a high-throughput assay to quantify OMV production in vitro. We constructed an OMV reporter consisting of Bacteroides ovatus inulinase (BACOVA_04502; INL), a protein previously shown to be enriched in OMVs (18, 19), fused to Nanoluciferase (NLuc) (28). Western blotting confirmed that the fusion protein was stable and almost exclusively present in OMVs (Fig. 1A). Filtered supernatants from cells constitutively expressing INL-NLuc exhibited significantly higher luminescent output than a lysis control strain expressing cytoplasmic NLuc (cNLuc) (Fig. 1B). This demonstrates that luminescence in culture supernatants can be used as an easily quantifiable proxy for OMV production. Next, we created a transposon library in the strain constitutively expressing the OMV reporter, INL-NLuc. Since the reporter is almost exclusively trafficked to OMVs, we anticipated that mutants displaying abnormal levels of NLuc activity in their supernatants corresponded to abnormal levels of OMV production. The strategy employed for the screening is described in Fig. 1C. We screened ~5,300 colonies and detected several mutants displaying abnormally high or low levels of NLuc activity (SI Appendix, Fig. S1). Secondary screening, which consisted of TEM, western blotting, and LPS and protein analysis, was performed to validate potential candidates (SI Appendix, Fig. S2). The genomes of mutants displaying atypical levels of vesiculation were sequenced to identify the transposon insertion sites. Altogether, these analyses identified several hyper- and hypovesiculating strains, summarized in SI Appendix, Table S2.
Fig. 1.
Identification of genes involved in OMV biogenesis. (A) Western blot against anti-polyHis showing total membrane (TM) and OMV fractions from BT WT constitutively expressing Inulinase fused to Nluc with a C-terminal 6xHis tag (INL-Nluc). This blot demonstrates that INL-Nluc is stably expressed and properly localizes to the OMV fraction. Samples were normalized by OD600 and ran on 10% SDS-PAGE. (B) Filtered supernatants from cells expressing INL-Nluc display higher luminescent output than those expressing Nluc in the cytoplasm. Filtered supernatants from overnight cultures (~20 h) were used for NanoGlow assays to quantify OMV production in vitro. Total luminescent output was normalized by OD600. (C) Schematic of OMV screening methodology (Created with https://BioRender.com).
Mutation of Dma1 (BT_4721) Leads to Hypervesiculation.
Genomic sequencing of potential candidates revealed four independent mutants containing transposon insertions in the gene BT_4721, which we renamed Dma1 (Fig. 2A). In the initial screening, these four strains appeared to display a hypovesiculating phenotype (SI Appendix, Fig. S1 and Table S2). However, western blots showed that these mutants had increased abundance of INL-NLuc in the OMV fraction (Fig. 2B). To further analyze the role of Dma1, we generated a Dma1 deletion mutant (Δdma1) and its corresponding complemented strain (Δdma1Comp). Growth curves confirmed that the fitness of Δdma1 is not attenuated in this context (SI Appendix, Fig. S3). Next, we isolated and quantified OMVs by TEM. This analysis confirmed the western blot data showing that Δdma1 produces significantly more OMVs than the WT (Fig. 2C). Admittedly, our ability to accurately quantify OMVs using TEM and manual counting is limited. However, since Bacteroides OMVs contain lipopolysaccharide (LPS), membrane lipids (phospholipids, sphingolipids, and amino lipids), and protein cargo (29), we aimed to confirm the phenotype of Δdma1 biochemically by quantifying the relative amount of these components in total membrane (inner and outer; TM) and OMV preparations from WT, Δdma1, and Δdma1Comp. We began by analyzing their respective protein content by SDS-PAGE. Surprisingly, the electrophoretic profile of the OMV fraction from Δdma1 exhibited a significant distortion in the low molecular weight region of the gel, which was reverted by complementation (Fig. 2D). We hypothesized that the distortion was due to the presence of high amounts of LPS in the samples due to Δdma1 hypervesiculation. Analysis of LPS from TM and OMV fractions via SDS-PAGE followed by silver stain confirmed that Δdma1 secretes significantly more LPS than the wild-type and complemented strains (Fig. 2E). The OMV fraction of Δdma1 also contained more proteins than the WT and complemented strains (Fig. 2F). Finally, we performed comparative lipidomics on TM and OMV fractions from WT, Δdma1, and Δdma1Comp. While the lipid composition for both fractions remained relatively unperturbed, we detected a general increase in secreted phospho, sphingo-, and amino lipid content in the Δdma1 OMVs (Fig. 2G and SI Appendix, Fig. S4). Altogether, our findings demonstrate that Δdma1 produces significantly more OMVs than its parental strain.
Fig. 2.
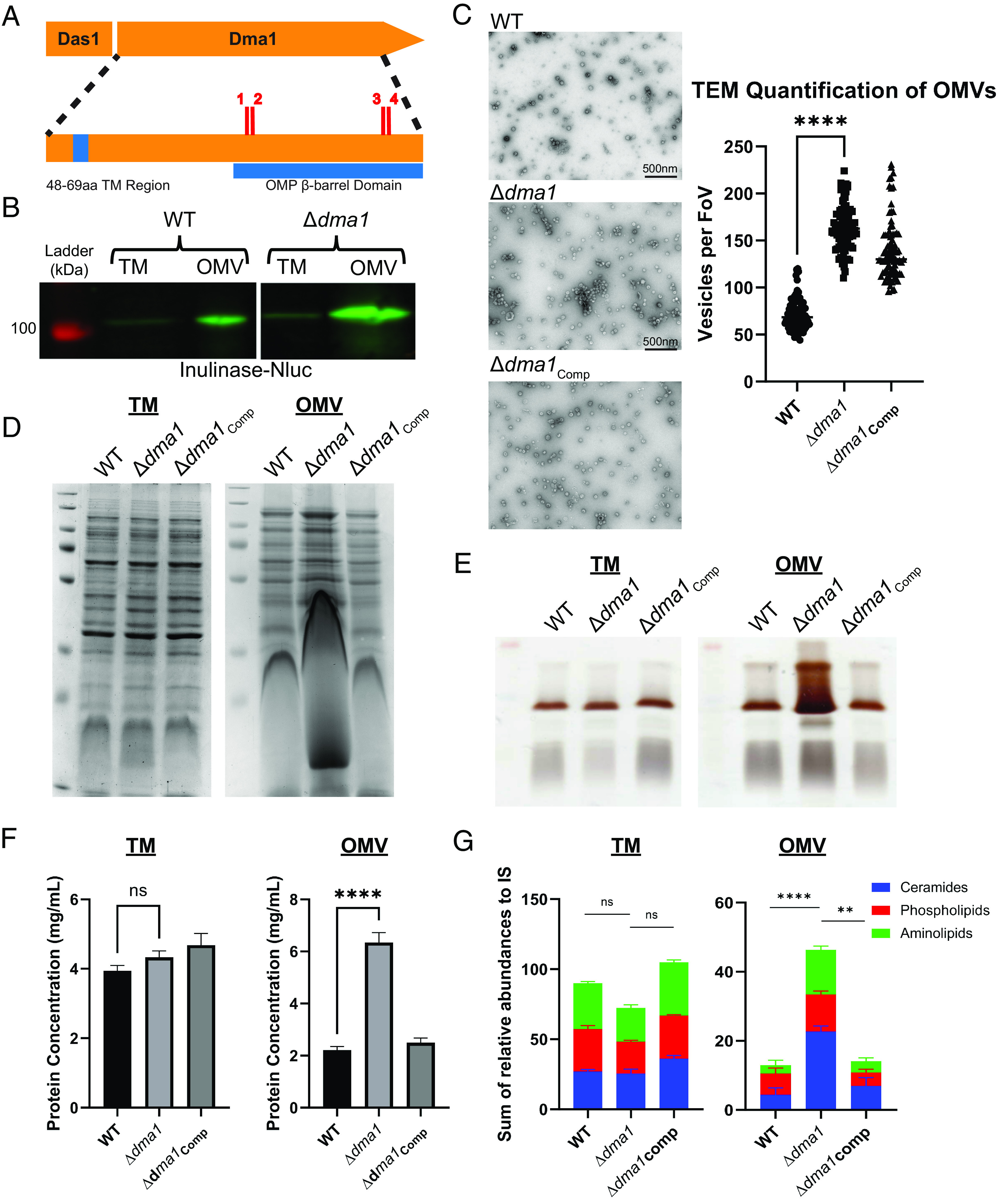
Deletion of Dma1 leads to hypervesiculation. (A) Genomic sequencing of screening candidates revealed four independent transposon mutants in Dma1. Red dashes denote transposon insertions (1: 674nt, 2: 677nt, 3: 1,139nt, 4: 1,143nt) (Created with https://BioRender.com). (B) Western blot using anti-polyHis shows that Dma1 transposon mutants contain more INL-Nluc in the OMV fraction than the wild type. Samples were normalized by OD600 and run on 10% SDS-PAGE. (C) Transmission electron microscopy (TEM) reveals that Δdma1 produces significantly more OMVs than the wild type. Left: Representative TEM images of OMVs from each strain. Right: Quantification of TEM images from each strain (FoV: field of view). Three biological replicates of samples from each strain were fixed onto grids in triplicate (in Materials and Methods). Ten random images were taken from each grid (n = 90 per strain), and OMVs were counted manually. (D) SDS-PAGE followed by staining with Coomassie Blue suggests that ∆dma1 produces more OMVs than the WT (E) SDS-PAGE followed by LPS Silver Staining demonstrates that ∆dma1 secretes more LPS into the OMV fraction. (F) Lowry Protein Assay shows that Δdma1 secretes significantly more protein into the OMV fraction than the WT. (G) Total lipids were extracted from TMs and OMVs from WT, Δdma1, and Δdma1Comp demonstrating that Δdma1 secretes significantly more membrane lipids as a result of hypervesiculation. Samples were relativized to an internal standard (IS). In all cases, samples were normalized to OD600.
Deletion of Dma1 Modulates OMV Production but Not Composition.
To demonstrate that the hypervesiculation phenotype of Δdma1 is due to an increase in the production of bona fide OMVs and not the result of a generalized destabilization of the outer membrane (OM), we performed comparative proteomic analyses of TM and OMV fractions from the WT and Δdma1 strains. A principal component analysis shows that the TM and OMV fractions of the WT and mutant contain similar protein content (SI Appendix, Fig. S5). Bacteroides OMVs are enriched with LES motif containing lipoproteins, while other OM proteins are typically excluded (18, 20). Volcano plots show that similar to the WT, the OMVs of Δdma1 are primarily enriched with lipoproteins containing the LES motif (18, 20), while porins and other proteins were retained predominantly in the bacterial membranes (SI Appendix, Fig. S6). Thus, OMV cargo selection is maintained in the Δdma1, which rules out that the increased amount of OMVs is the result of increased cell lysis or membrane instability, which would result in the presence of cytosolic and inner membrane (IM) proteins in the OMV proteome. Further supporting this conclusion, western blots show that the partitioning of the OMV protein SusG and the OM protein BT_0587 (19, 20) is not affected in Δdma1 (SI Appendix, Fig. S7).
Extracytoplasmic Function (ECF) Sigma Factor, Das1 (BT_4720), Is Required for Dma1-Mediated Hypervesiculation.
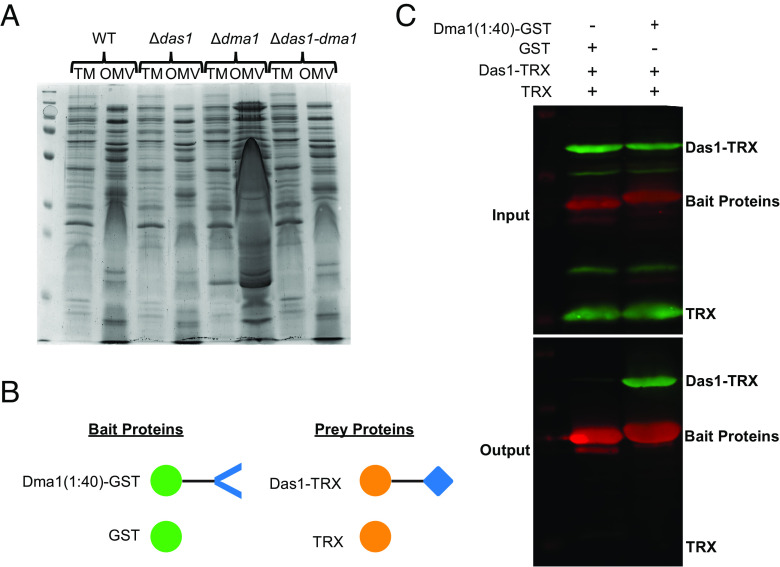
Dma1 is encoded in an operon with the ECF21 family sigma factor, BT_4720 (Das1, for Dma-associated sigma factor 1. ECF-type sigma factors are a family of transcriptional regulators that modulate gene expression in response to extracytoplasmic signals. These are typically encoded adjacent to their cognate anti-sigma factor, which negatively regulates the activity of the sigma factor (30, 31). ECF21 family sigma factors are found solely among Bacteroidota, but very little is known regarding their function (31). An ortholog of Dma1 in Bf, Reo, was shown to control the activity of its cognate sigma factor, ecfO, by sequestering it at the IM via its N-terminal region in the cytoplasm (32). There is strong consensus between the amino acid sequences from Reo and Dma1 (SI Appendix, Fig. S8), which suggests that Dma1 possesses a cytoplasmic domain capable of directly regulating the activity of Das1. To provide support for the interaction between Dma1 and Das1, we mutated Das1 in the Δdma1 background (Δdas1-dma1). A single deletion mutant lacking Das1 (Δdas1) was included as control. Growth curves show that the fitness of these mutants is not impacted in vitro (SI Appendix, Fig. S9). By employing SDS-PAGE, we determined that removal of Das1 reverts the distortion in the electrophoretic pattern seen for Δdma1 OMVs (Fig. 3A), indicating that the hypervesiculation observed in Δdma1 requires Das1. Next, we performed targeted pulldowns to determine whether Dma1 and Das1 physically interact. For this, we engineered a chimeric protein containing the N-terminal 40 amino acids of Dma1 fused to Glutathione-S transferase (GST), Dma1(1:40)-GST, as the bait protein. The prey protein consisted of Das1 fused to thioredoxin (TRX), Das1-TRX. Controls expressing GST and TRX alone were included (Fig. 3B). Each protein was individually expressed in Escherichia coli BL21. Lysates from strains expressing each bait protein were mixed with lysates containing the prey proteins and subjected to affinity chromatography by employing a column containing anti-GST resin. Our results demonstrate that Dma1(1:40)-GST specifically interacts with Das1-TRX, indicating that the N-terminal region of Dma1 is sufficient to sequester Das1 (Fig. 3C). Together, these findings demonstrate that Das1 and Dma1 form a sigma/anti-sigma pair and that the unregulated activity of Das1 is responsible for hypervesiculation observed in Δdma1. A previous study has shown that deletion of Reo, in Bf, increases fitness in response to oxidative stressors, while deletion of ecfO renders them more susceptible (32). We assessed the fitness of WT, Δdas1, Δdma1, and their corresponding complemented strains in response to prolonged aerobic stress but observed no changes between the mutants and the wild type (SI Appendix, Fig. S10). This suggests that Dma1 and Reo control different responses in Bt and Bf, respectively.
Fig. 3.
ECF21 family sigma factor, Das1, is required for hypervesiculation. (A) Coomassie blue stain comparing protein profiles between WT, Δdas1, Δdma1, and Δdas1-dma1. This shows that deletion of Das1 in the Δdma1 background restores WT levels of vesiculation. Samples were normalized by OD600 values and run on 10% SDS-PAGE gel. (B) Schematic of constructs used for targeted pulldown assay (created with https://BioRender.com). (C) Western blots using anti-polyHis (Green) and anti-GST (Red). Top: Gel shows that equivalent amounts of bait and prey proteins were mixed prior to passage through a column containing anti-GST resin. Bottom: Gel showing the proteins present after the elution step, which demonstrates that the N terminus of Dma1 is sufficient to sequester Das1.
NigD1, Part of the Dma1-Das1 Regulon, Is Necessary for Hypervesiculation.
To understand how Dma1 controls vesiculation, we performed RNA sequencing to compare transcriptomes of the wild-type and Δdma1 strains. We found that three genes, BT_1287, NigD1 (BT_4005), and NigD2 (BT_4719), were dramatically up-regulated in the Δdma1 mutant (Fig. 4A and SI Appendix, Table S3). There were other genes differentially expressed (SI Appendix, Table S3), but their changes were much less pronounced. Comparative proteomic analyses of WT and Δdma1 identified the same three proteins as the most differentially expressed proteins between these strains (Fig. 4B and SI Appendix, Table S4). No function has been assigned to any of these three proteins; however, both NigD1 and NigD2 are annotated as NigD-like proteins. NigD-like proteins are a family of uncharacterized lipoproteins proteins found solely in Bacteroidota. We determined that deletion of NigD1 (but not BT_1287 or NigD2) in the Δdma1 background restores wild-type levels of vesiculation, indicating that NigD1 is involved in controlling OMV production in Δdma1 (Fig. 4C). NigD1 is not predicted to be encoded within an operon; however, it is encoded nearby genes required for LPS biosynthesis and regulation (LpxB and FtsH) and phospholipid synthesis (CdsA). Our transcriptomic and proteomic analyses failed to identify these, or any other known genes involved in the biosynthesis of LPS or phospholipids, as differentially regulated between WT and Δdma1. However, our results suggest that NigD1 acts as a molecular switch for OMV biogenesis.
Fig. 4.
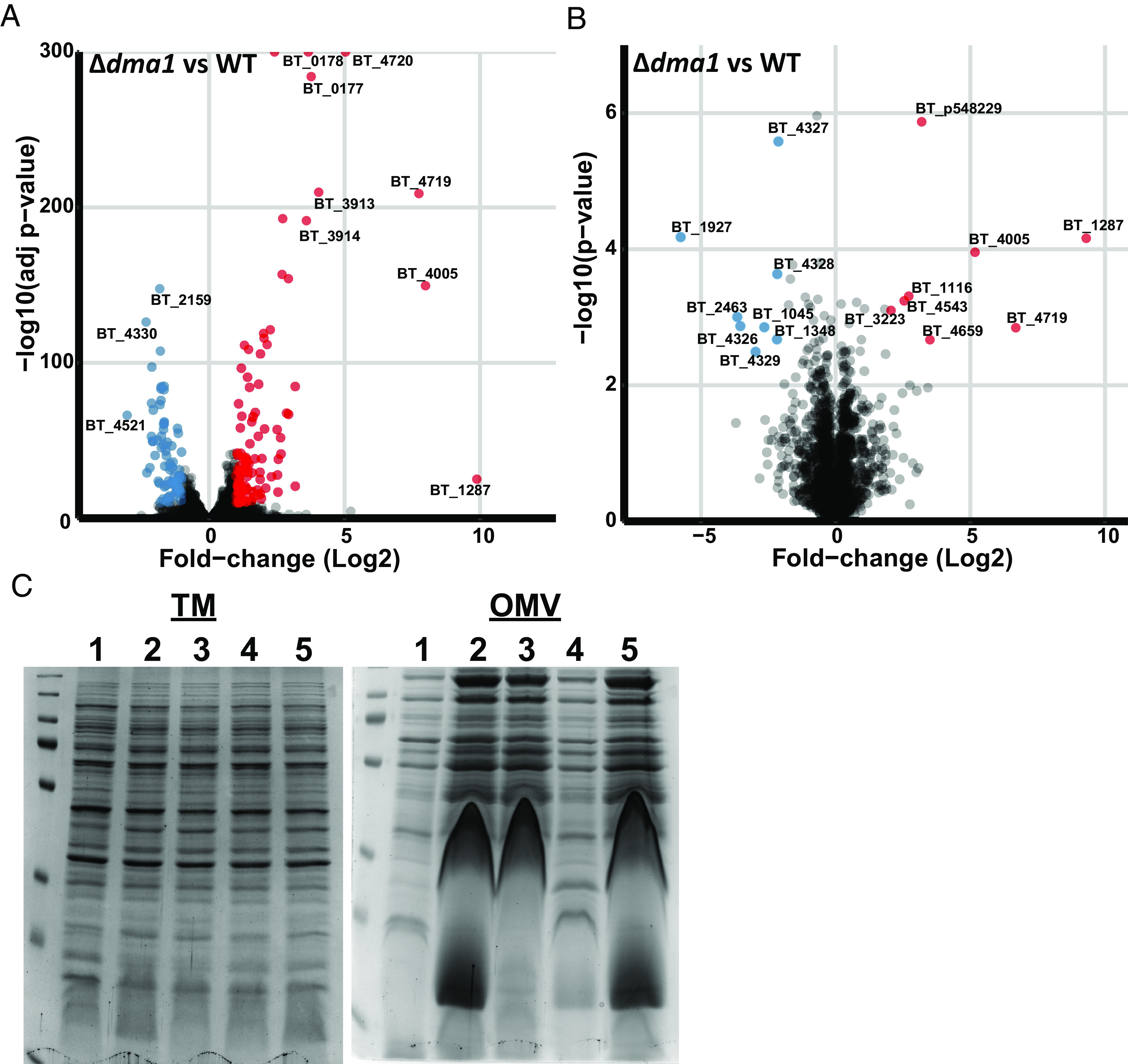
Member of the Dma1 regulon, NigD1 (BT_4005), is necessary to induce vesiculation. Volcano plot representations of (A) transcriptome and (B) proteome data comparing WT and Δdma1. (C) Coomassie blue stain of TM and OMV fractions isolated from 1. WT, 2. Δdma1, 3. Δ1287/dma1, 4. ΔnigD1/dma1, and 5. ΔnigD2/dma1. Left: The protein profile of mutant TMs is the same as WT, indicating that all changes are specific to the OMV fraction. Right: Deletion of NigD1 in the Δdma1 background restores the protein profile to WT. Samples were normalized by OD600 and run on 10% SDS-PAGE.
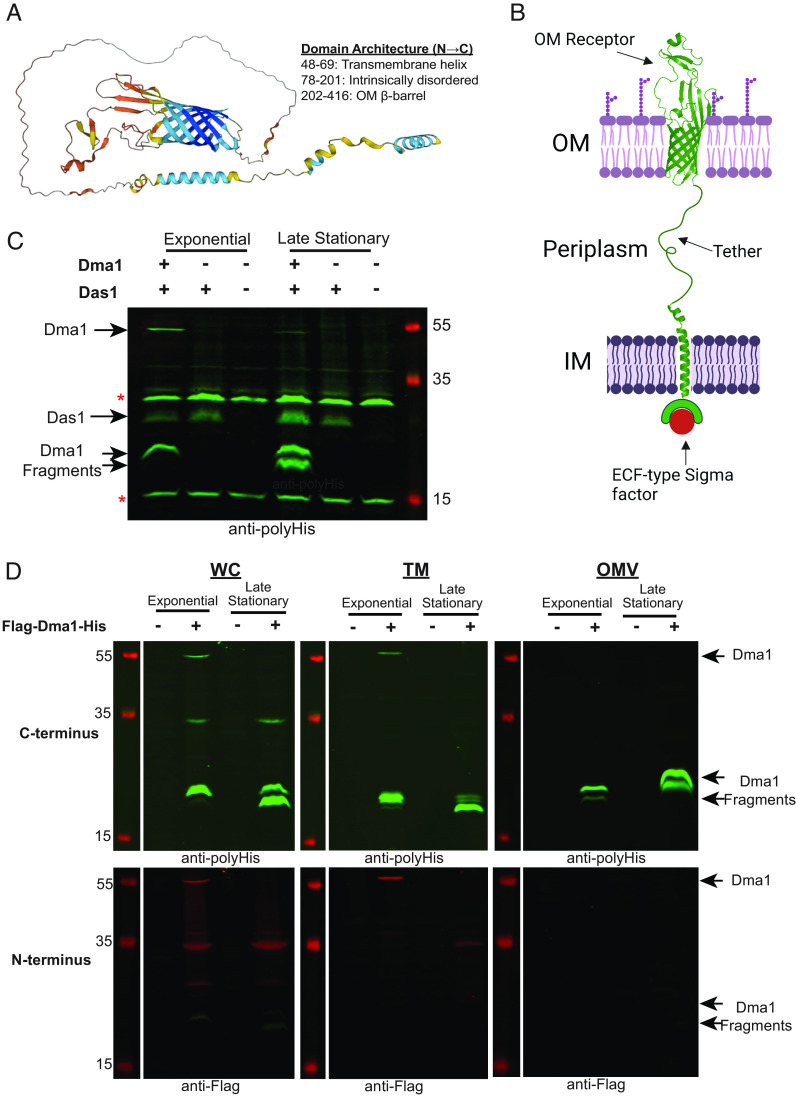
Dma1 Represents an Unprecedented Class of Anti-Sigma Factor That Spans Both the IM and OM.
Canonical anti-sigma factors contain a cytoplasmic domain, responsible for the sequestration of their cognate sigma-factors at the IM, connected via a transmembrane region to a periplasmic sensor domain (30). Dma1 is annotated as an OMP_b-brl_2 domain-containing protein. Structural modeling with AlphaFold (33) predicts that Dma1 contains an N-terminal alpha-helical transmembrane region and a C-terminal eight stranded β-barrel domain connected via a long intrinsically disordered region (Fig. 5A). Eight stranded β-barrel domain proteins are found in the OM of gram-negative bacteria. In Bf, Reo, was shown to directly sequester its cognate sigma factor via its N-terminal region in the cytoplasm (32), and we show that this is also the case for Dma1. Based on these observations, we propose an unprecedented domain architecture for Dma1, in which the C-terminal β-barrel is embedded in the OM; the N-terminal domain that interacts with the cognate sigma factor is in the cytoplasm anchored to the IM via an alpha-helical transmembrane helix; and both domains are tethered via the intrinsically disordered region that traverses the periplasm (Fig. 5B). Proteins with similar domain organizations have been suggested for Reo in B. fragilis, and M0N98_03760 in P. vulgatus; however, the orientation of these proteins has not been experimentally addressed (32, 34). To provide support for this model, we performed western blots of Bt cells coexpressing C-terminally His-tagged Dma1 and Das1. Cells were harvested during the exponential and late stationary phase. Expression of Dma1 was enhanced when expressed alongside Das1. Wild-type Bt and cells expressing only Das1 were employed as controls. During exponential phase growth, a band corresponding to full-length (~55 kDa) and a C-terminal-containing fragment (~20 kDa) of Dma1 were detected (Fig. 5C). At stationary phase, full-length Dma1 was no longer detected, with the concomitant appearance of an additional smaller fragment (~17 kDa). These results suggest that Dma1 is temporally regulated (Fig. 5C). Next, we generated cells expressing Dma1 tagged with both a C-terminal 10×His tag and an N-terminal 3×FLAG tag. Whole cells (WCs), TMs, and OMVs were analyzed by western blot. Full-length Dma1 (~55 kDa) was detected in the WC and TM fractions with both antibodies, confirming that Dma1 is translated and translocated as a complete and single polypeptide (Fig. 5D). Only C-terminal fragments were detected in the OMVs using the anti-His antibodies, indicating that, as predicted, the β-barrel domain is localized in the OM (Fig. 5D). In addition, our conclusion is supported by the MS analysis of wild-type OMVs, which only detected peptides containing the predicted C-terminal β-barrel of Dma1 (SI Appendix, Table S5). Taken together, our results provide strong evidence that Dma1 is an anti-sigma factor that spans both membranes, directly connecting the exterior of the cell with its cytoplasm. Based on this, we propose that Dma1 is the founding member of the Dual Membrane-spanning Anti-sigma factor (Dma) family.
Fig. 5.
Dual membrane-spanning 1 (Dma1) has the properties to span both the inner and outer membranes of a gram-negative bacterium. (A) AlphaFold predicted stucture of Dma1. (B) Proposed orientation of Dma1 in the membrane of Bt (Created with https://BioRender.com). (C) Western blots were performed using anti-polyHis against cells constitutively expressing either 1. Dma1 and Das1 together, 2. Das1 alone, and 3. empty vector control. Bands adjacent to stars (*) are nonspecific bands. This demonstrates that full-length Dma1 is present in WCs, and the state of the protein is growth-phase dependent. (D) Western blots of WCs, TMs, and OMVs collected from Bt strain constitutively expressing Dma1 containing a C-terminal His tag and an N-terminal Flag tag (Flag-Dma1-His). The green channel is anti-polyHis, while the red channel is anti-Flag. Full-length Dma1 is present solely in the WC and TM fraction, while only C-terminal fragments are detected in the OMVs.
Dual Membrane-Spanning Anti-Sigma Factors Are Present throughout Bacteroidota.
To determine whether proteins with similar domain architecture exist in Bt, we searched for sigma/anti-sigma pairs containing anti-sigma factors encoding a predicted β-barrel domain. We identified two additional proteins in Bt, Dma2 (BT_1558) and Dma3 (BT_2778), that are structurally similar to Dma1 (Fig. 6A). Dma2 is encoded adjacent to an ECF21 family sigma factor, BT_1559 (Das2), while Dma3 is more complex, as it is predicted to contain the sigma factor fused to the rest of the polypeptide at the N terminus (Fig. 6A). A bioinformatic analyses showed that Dma1 is present in almost all Bacteroidota, while Dma2 and Dma3 are less prevalent. Fig. 6B shows a phylogenetic analysis containing select members of the Bacteroidota representing various classes ranging from mammalian gut commensals to soil-dwelling microbes. Dma1 is present in almost all Bacteroidota chosen (27/29), while Dma2 (5/29) and Dma3 (10/29) were less prevalent (Fig. 6B). Interestingly, Dma2 is only present in members of the genus Bacteroides, suggesting that this protein was recently acquired in the genus. Dma3 on the other hand is predominantly found in Bacteroides and Prevotella (Fig. 6B and SI Appendix, Fig. S11). We were unable to identify any structurally similar proteins in gram-negative bacteria outside of Bacteroidota. To determine whether Dma2 and Dma3 may also modulate OMV biogenesis in Bt, we generated clean deletion mutants in each gene and analyzed their protein profiles by SDS-PAGE. Mutation of dma2 produced a phenotype similar to that of Δdma1, which suggests that Dma2 also potentially modulates OMV biogenesis. Deletion of its cognate sigma factor das2 also restores the WT electrophoretic profile (SI Appendix, Fig. S12). Mutation of dma3 revealed no detectable phenotype by SDS-PAGE (SI Appendix, Fig. S12). Additional studies are needed to validate the potential impacts of Dma2 and Dma3 on OMV biogenesis; however, these are outside the scope of this manuscript.
Fig. 6.
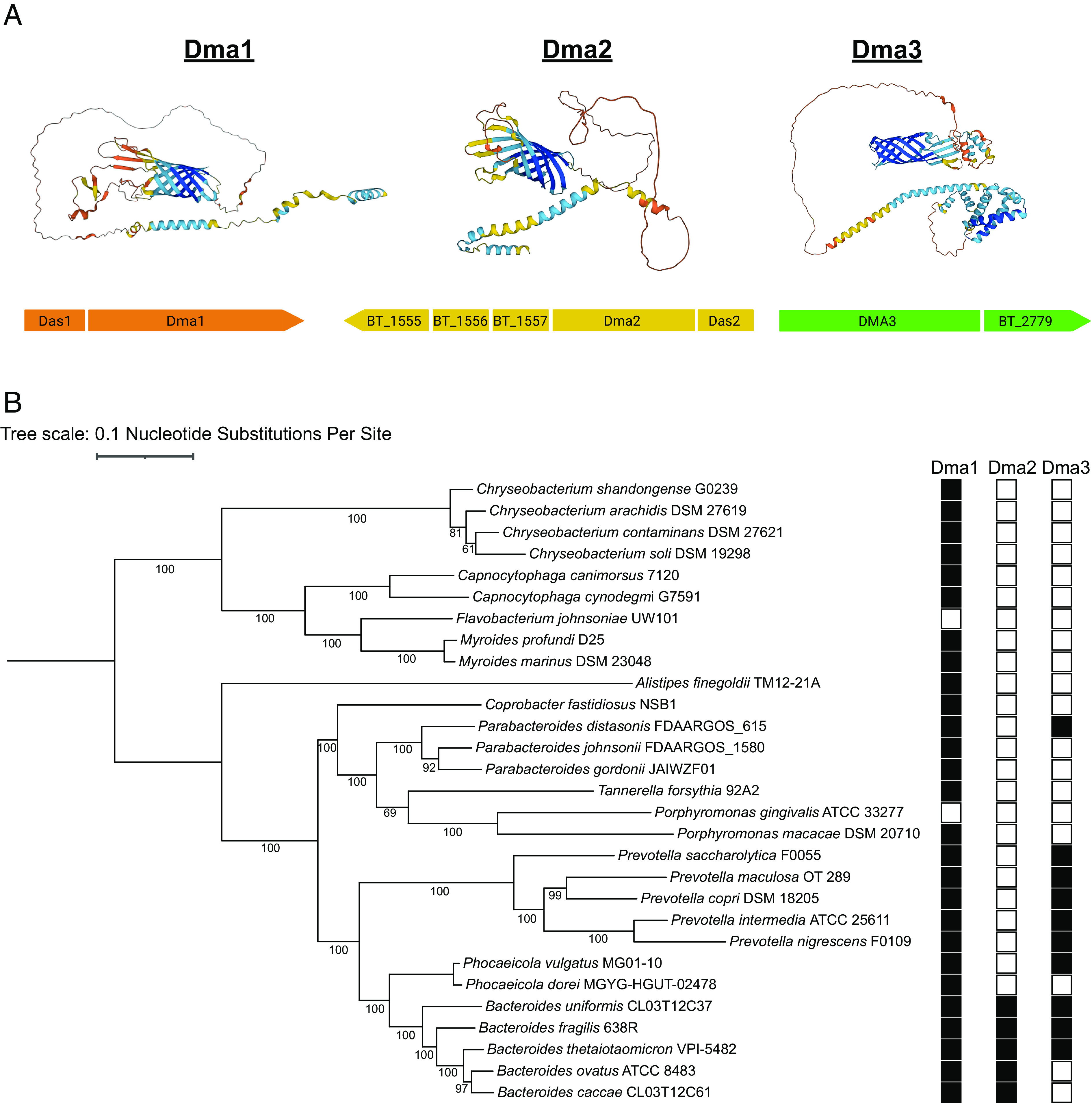
Members of the Dma family are present throughout Bacteroidota. (A) Genome analysis identified two additional proteins, Dma2 and Dma3, with structural similarity to Dma1. The AlphaFold predicted structure of each Dma, along with their respective operons, are presented (Created with https://BioRender.com). (B) A maximum likelihood phylogenetic tree was constructed from 29 core genomes of various Bacteroidota. Amino acid sequences of Dma1 (WP_008766767.1), Dma2 (WP_008762208.1), and Dma3 (WP_011108473.1) from Bt served as references for identification in the other genomes.
Discussion
Since their discovery in the 1960s, many important roles have been proposed for OMVs (35). However, to date, very little is known about the mechanism(s) of bacterial vesiculation and its regulation. In this study, we aimed to advance our understanding of the mechanism of OMV biogenesis in Bacteroidota. To this aim, we developed a screening methodology to identify genes involved in OMV biogenesis. We identified the Dma family, which we show to be a class of anti-sigma factors with an unprecedented domain organization. These proteins span both membranes, possessing an extracellular and a cytoplasmic domain connected via a large, intrinsically disordered region that crosses the periplasm. We show that inactivation of Dma1 (BT_4721) or Dma2 (BT_1558) results in hypervesiculation in Bt.
We previously showed that labeling OM and OMV-specific proteins with distinct fluorescent markers is an effective way to visualize OMVs and distinguish them from by-products of cell lysis (18). In this work, we labeled an OMV marker with NLuc instead of fluorescent proteins, which allowed us to quantify OMV production in vitro in a high throughput format (Fig. 1C). Screens attempting to identify genes involved in OMV biogenesis have been performed (36) however, these were unable to establish causal associations between specific genes and OMV formation. We speculate that this is due to their use of nonspecific markers, like LPS, OmpA, or phospholipids, as a readout for OMV production. Many of the genes identified in these studies led to membrane destabilization, which confounded the interpretation of the results, due to their inability to differentiate genuine OMVs from cell lysis. Although OMV cargo selection is not common in all bacteria, the methods developed here can be adapted to conduct OMV screens in other bacterial species.
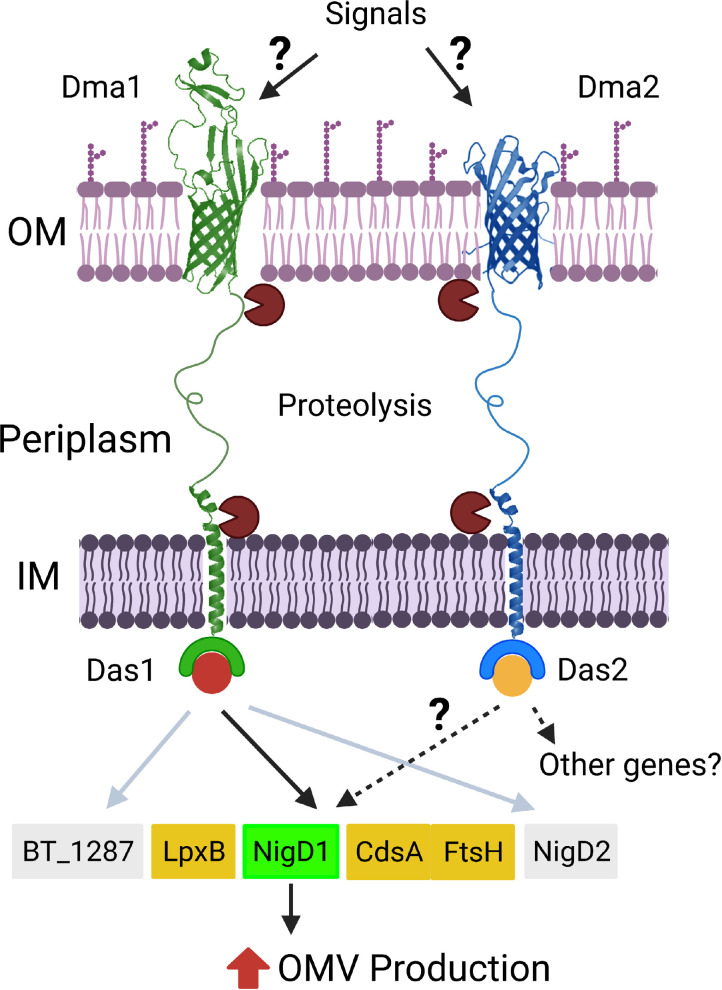
Our model for regulation of OMV biogenesis in Bt is summarized in Fig. 7. We propose that members of the Dma family sense extracellular stimuli or perturbations in the OM via their C-terminal β-barrel domain, which triggers a series of proteolytic events that liberates their cognate sigma factors to modulate gene expression and induce OMV production (Fig. 7). By employing transcriptomic and proteomic analyses, we demonstrated that NigD1 (BT_4005) is required for the induction of vesiculation in the absence of Dma1 NigD-like proteins are ubiquitous among Bacteroidota, but their functions have not been defined. NigD1 is encoded adjacent to genes required for biosynthesis and regulation of LPS and phospholipids. Our transcriptomic and proteomic analyses indicate that these genes/proteins are not differentially regulated between the wild type and Δdma1. This suggests the presence of currently unknown mechanisms where NigD1 controls the amount of LPS, proteins, and lipids allocated to OMVs. However, Bacteroidota membranes are rich in sphingolipids and other lipids not commonly found in bacteria. More knowledge about the regulation of LPS and lipid biosynthesis will be required to fully understand how Dma1, Dma2 and NigD1 control OMV biogenesis in Bacteroides. NigD2 (BT_4719) and BT_1287 are part of the Dma1 regulon, but they are dispensable for the hypervesiculation phenotype. This strongly suggests that Dma1 regulates other processes in Bt.
Fig. 7.
Schematic of Dma1- and Dma2-mediated induction of OMV biogenesis. In our model, Dma1 and Dma2 sense extracellular stimuli and/or perturbations in the OM. This leads to the proteolytic degradation of the proteins to release their cognate sigma factors and subsequently modulate gene expression to induce OMV biogenesis. Dma1/Das1-mediated hypervesiculation is dependent upon NigD1, while the regulon of Dma2 is unclear (created with https://BioRender.com).
Previous studies have investigated orthologs of Dma1. In B. fragilis, deletion of Reo, Dma1 ortholog, was shown to increase fitness in response to oxidative stressors (32). However, we found that Dma1 does not seem to play the same role in Bt (SI Appendix, Fig. S10). In Phocaeicola vulgatus, mutation of the Dma1 ortholog, M098_03760, conferred increased protection against the antimicrobial toxin BcpT. Authors hypothesize that this occurs by increasing LPS O-antigen length and preventing BcpT from binding lipid A core and destabilizing the OM (34). We did not analyze the structure of LPS in this work, but reports suggest that Bt lacks LPS, and instead makes lipooligosaccharide (LOS) (37, 38). Therefore, Dma1 is not likely to induce a similar change. In Bt, the Dma family impacts OMV biogenesis; however, members of the Dma family likely play diverse roles in different species within Bacteroidota. Since OMVs have been implicated in stress response, it would be interesting to determine whether the fitness advantages observed in other studies are somehow related to increased vesiculation.
Dma1 is the first protein shown to span both membranes of a gram-negative bacteria (Fig. 5). As anti-sigma factors, the Dma family represents a unique class of regulatory protein. The canonical cell surface signaling (CSS) system, like the Fec system in E. coli, consists of a TonB-dependent OM receptor (FecA), which senses external stimuli, an anti-sigma factor (FecR), to relay the signal, and a sigma factor (FecI), to modulate gene expression (30, 39). Members of the Dma family are distinct from this model because they encode the OM receptor and anti-sigma factor in a single polypeptide (Fig. 5). To date, such structures have yet to be reported. Bacteroides spp. distinctively encode an expanded repertoire of transcriptional regulators, specifically hybrid two-component systems and ECF-type sigma factors, many of which are uncharacterized (40). The identification of the Dma family in Bacteroidota suggests that these organisms have evolved additional modes of transcriptional regulation, many of which are yet to be described.
The unprecedented domain organization of the Dma family raises fundamental questions regarding their translocation and assembly. In gram-negative bacteria, β-barrel Outer Membrane Proteins (OMPs) are trafficked to the OM by translocation through the Sec system, where periplasmic chaperones, like surA, shuttle the unfolded OMPs to the β-barrel Assembly Machine (BAM) for insertion into the OM (41, 42). Since Dma1 has domains inserted into both membranes, it is unclear whether Dma1 is localized via the BAM complex. It is tempting to speculate that Bacteroidota have evolved additional systems specifically to ensure the proper localization of members of the Dma family, but future studies are required to validate this hypothesis.
The presence of Dma1 proteolytic fragment provides clues about Dma1 regulation. In E. coli and similar organisms, there are typically two proteolytic events that occur at the inner and outer leaflet of the IM to inactivate anti-sigma factors (30, 43). Two IM proteases, DegS and RseP, involved in RIP have been characterized (44, 45). Potential orthologs of these IM proteases are predicted to be present in Bt. Future work will be focused on understanding what are the signals that trigger Dma1 proteolysis, and the enzymes involved in the process.
Our screening led us to the identification of the Dma family. However, we also identified multiple mutants displaying hyper- and hypovesiculation phenotypes. The investigation of these mutants will further our understanding of the bacterial vesiculation mechanisms and will lead to identification of the elusive machinery responsible for OMV biogenesis.
Materials and Methods
Bacterial Strains and Growth Conditions.
Strains, oligonucleotides, and plasmids are described in SI Appendix, Table S1. E. coli was grown aerobically at 37 °C in Luria-Bertani (LB) medium. Bacteroides strains were grown in an anaerobic chamber (Coy Laboratories) at 37 °C containing an atmosphere of 10% H2, 5% CO2, 85% N2. Bacteroides were cultured in brain heart infusion (BHI) medium (Fisher Scientific) supplemented with 5 µg/mL hemin and 1 µg/mL vitamin K3. When applicable, antibiotics were used as follows: 100 µg/mL ampicillin, 200 µg/mL gentamicin, 25 µg/mL erythromycin, and 10 μg/mL tetracycline.
Genetic Manipulation of Bt.
Deletion mutants were constructed using the pSIE1 vector described in Bencivenga-Barry et al. (46). Briefly, ~750 base pair regions flanking genes of interest were cloned into pSIE1. Vectors were conjugated into Bt, and positive conjugants were identified by selection on BHI plates containing gentamicin and erythromycin. Counterselection was performed on BHI plates in the presence or absence of 125 ng/mL anhydrotetracycline (aTc). Mutants were identified by PCR prior to whole genome sequencing. Complemented strains were made using vectors from ref. 47.
OMV Isolation.
OMVs were purified by ultracentrifugation from cell-free culture supernatants. Briefly, 50 mL of Bt culture grown to late stationary phase was centrifuged twice at 6,500 rpm at 4 °C for 10 min. Supernatants were filtered using a 0.22-µm-pore membrane (Millipore) to remove residual cells. The filtrate was subjected to ultracentrifugation at 200,000 × g for 2 h (Optima L-100 XP ultracentrifuge; Beckman Coulter). Supernatants were discarded, and pellets were resuspended in PBS standardized to OD600. When performing MS analysis, purified OMV preparations were lyophilized.
Subcellular Fractionation.
Total Membrane (TM) preparations were isolated by cell lysis and ultracentrifugation. Briefly, late stationary phase cultures were harvested by centrifugation at 6,500 rpm at 4 °C for 10 min. The pellets were gently resuspended in a mixture of phosphate-buffered saline (PBS) containing complete EDTA-free protease inhibitor mixture (Roche Applied Science). Cells were then lysed using two passes through a cell disruptor at 35 kPa. Next, centrifugation at 8,500 rpm at 4 °C for 8 min was performed to remove unbroken cells. Total membranes were collected by ultracentrifugation at 200,000 × g for 1 h at 4 °C. Supernatants were discarded, and pellets were resuspended in PBS standardized to OD600. TM fractions were lyophilized for MS analysis.
SDS-PAGE and Western Blot Analyses.
Total membrane and vesicle fractions were analyzed by standard 10% Tris-glycine SDS-PAGE. Samples were normalized by OD600, and equivalent volumes were loaded onto an SDS-PAGE gel. Coomassie Blue staining was employed to analyze protein profiles. When applicable, samples were transferred onto a nitrocellulose membrane for western blot analysis. Membranes were blocked using Tris-buffered saline (TBS)-based Odyssey blocking solution (LI-COR). Primary antibodies used in this study were rabbit polyclonal anti-His (ThermoFisher) and mouse monoclonal anti-FLAG (Sigma). Secondary antibodies used were IRDye anti-rabbit 780 (LI-COR). Imaging was performed using an Odyssey CLx scanner (LI-COR).
Lipopolysaccharide (LPS) Silver Stain.
Abundance of LPS was measured according to Tsai and Frasch (48). Briefly, samples were normalized by OD600, then diluted and treated with Proteinase K, prior to loading equal amounts onto a 15% SDS-PAGE gel. After running, gels were fixed overnight in 200 mL of 40% ethanol in 5% acetic acid. Next, gels were oxidized for 5 min in 100 mL of 0.7% fresh periodic acid in 40% ethanol and 5% acetic acid. Upon completion, gels underwent three washes (15 min each) in milliQ H2O. The gels were then stained for 10 min in the dark with 28 mL 0.1 M NaOH, 2 mL NH4OH, 5 mL 20% AgNO3, and 115 milliQ H2O. Gels underwent three additional washes prior to developing in 200 mL H2O with 10 mg citric acid and 100 μL formaldehyde.
OMV Reporter Screen.
Transposon mutagenesis was performed by adapting the protocol of Veeranagouda et al. 2013 (49, 50) on Bt constitutively expressing BACOVA_04502 (Inulinase; INL) fused to Nanoluciferase (NLuc) at the C terminus. Briefly, INL-NLuc was cloned in the pWW3867 vector (47) and expressed in Bt. Next, cells were conjugated with pSAM-Bt_Tet (SI Appendix, Table S1) to create the transposon mutant library. Selection was performed on BHI agar plates containing 25 µg/mL erythromycin and 10 μg/mL tetracycline. Upon plating, individual colonies were isolated and transferred to 200 μL of BHI media in clear, round-bottom 96-well plates (Corning) and incubated for 24 h. After incubation, samples were subcultured and diluted (1:20) in the same volume of BHI media and transferred to a second 96-well plate for a 20-h incubation. Next, we measured the OD600 of cultures in each plate with a BioTek microplate reader. Plates then underwent centrifugation at 4,000 rpm to pellet cells. Supernatants were collected and transferred to 0.22 μm hydrophilic low protein binding 96-well filter plates (Millipore) and centrifuged at 4,000 rpm to remove residual cells. Quantification of OMV production in 96-well plate format was done by performing Nano-Glo assays using the Nano-Glo Live Cell Assay System (Promega). Briefly, 100 μL of filtered supernatants was transferred to white bottom 96-well plates (Corning) and 20 μL of Nano-Glo Live Cell Reagent was added to each well. Plates were shaken for 20 s in a BioTek microplate reader prior to quantifying luminescent output. Luminescence was normalized to OD600. Transposon mutants displaying a >1.5-fold increase relative to the wild type were considered hypervesiculating strains, while those displaying <0.5-fold decrease were deemed hypovesiculating strains. These candidates of interest then underwent secondary screening to further characterize the phenotype. Transposon insertions from candidates of interest were identified by genomic DNA extraction and sequencing.
Protein Expression for In Vitro Pulldown Assay.
Here, in vitro pulldown assays were adapted from Ndamukong et al. (32). Briefly, we cloned the N-terminal 40 amino acids of Dma1 into the pGSTag vector (51) to serve as a bait protein, while Das1 was cloned into the pET32a-TRXtag vector (52) to function as the prey protein. Constructs were expressed in E. coli BL21 by induction of mid-log phase cells for 3 h with 1 mM isopropyl-β-D-thio-galactopyranoside (IPTG). Cells were extracted in 20 mM Tris-HCl pH 8.0, 300 mM NaCl, and supplemented with protease inhibitor (53). Cells were lysed by a single passage through a cell disruptor at 35 kPa. After disruption, lysates were incubated in 1% n-dodecyl-β-D-maltoside (DDM) for at least 3 h. Protein stability was checked by affinity purifying GST fusion proteins with glutathione–Sepharose 4B resin and TRX-His fusion proteins with Ni-nitrilotriacetic acid (NTA) resin, followed by SDS-PAGE. Upon confirmation of stability, lysates from the bait and prey proteins were used during the pulldown assays.
In Vitro Pulldown Assay.
Pulldown assays were performed by collecting lysates from cells expressing either bait or prey proteins. Next, lysates containing the GST-fused bait protein were mixed in a 1:1 ratio with those of the TRX-His tagged prey proteins and incubated in the presence of 10% glycerol, protein extraction buffer, and glutathione Sepharose resin at 4 °C overnight while shaking. Controls were included to rule out nonspecific interactions with the affinity tags. Next, we collected an aliquot of the mixture prior to passage through a column (Input). The resin underwent three successive washes with extraction buffer supplemented with 0.2% DDM. Bound proteins were eluted in 50 mM Tris-HCI pH 8.0 and 10 mM reduced glutathione (Output). Equivalent amounts of sample from the input and output were analyzed by western blotting with mouse monoclonal GST (Millipore Sigma) and rabbit polyclonal His (Invitrogen) antibodies.
Phylogenetic Analysis of the Dma Family.
Whole-genome fasta files were obtained from NCBI Assembly on 06/28/23. Genomes were annotated for open reading frames with prokka v1.14.6 using the command “prokka ${ID}_*.fna --outdir ${ID} --locustag ${ID} --mincontiglen 500 --prefix ${ID} --force --notrna --norrna” {24642063}. The core genome alignment of the 29 genomes was created using panaroo v1.2.10 on the .gff files from prokka with the command “panaroo -i ${indir}/*.gff -o ${outdir} --clean-mode moderate -a core -c .8 -f .5 --core_threshold .99 -t 12 --search_radius 10 --refind_prop_match 100 -t ${SLURM_CPUS_PER_TASK}” {32698896}. Alignment of the 29 core genes identified by panaroo was performed using mafft v7 {23329690}. The core genome alignment from panaroo was constructed into a maximum likelihood phylogenetic tree using RAxML v8.2.12 with the command “raxmlHPC -s core_gene_alignment.aln -n EP_raxml -m GTRGAMMA -f a -T 4 -N 100 -p 12345 -x 54321” {24451623}. Identification of Dma1, Dma2, and Dma3 homologs outside of B. thetaiotamicron was performed with the NCBI tblastn webserver using the protein sequences for each gene as query and the whole-genome fasta files as the subject with 95% query cover threshold as a positive result for gene presence. AlphaFold structural predictions were used to confirm genes identified during the blast search. The newick file was viewed and analyzed with metadata on Dma1, Dma2, Dma3, related gene information in iTOL webserver{33885785}.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank all members of the Feldman lab for critically reading the manuscript. This work was supported by NIH grants R21AI168719 and R21AI151873 to M.F.F. N.E.S. is supported by an Australian Research Council Future Fellowship (FT200100270) and an ARC Discovery Project Grant (DP210100362). We thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute for access to MS instrumentation.
Author contributions
E.J.P. and M.F.F. designed research; E.J.P., M.G.S., B.J., N.E.S., W.L.B., J.C.O.-M., and F.-F.H. performed research; N.E.S., J.C.O.-M., T.V.O., and F.-F.H. contributed new reagents/analytic tools; E.J.P., M.G.S., N.E.S., J.C.O.-M., F.-F.H., and R.F.P. analyzed data; and E.J.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Mass Spectrometry Proteomics data has been deposited in the Proteome Xchange Consortium via the PRIDE partner repository (54) with the dataset identifier PXD043360 (55). RNA sequencing data has been deposited with links to BioProject accession number PRJNA994135 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) (56).
Supporting Information
References
- 1.Deatherage Brooke L., Cookson Brad T., Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 80, 1948–1957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill S., Catchpole R., Forterre P., Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 43, 273–303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Blume J., Hausser A., Lipid-dependent coupling of secretory cargo sorting and trafficking at the trans-Golgi network. FEBS Lett. 593, 2412–2427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalluri R., LeBleu V. S., The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tricarico C., Clancy J., D’Souza-Schorey C., Biology and biogenesis of shed microvesicles. Small GTPases 8, 220–232 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avila-Calderón E. D., et al. , Outer membrane vesicles of gram-negative bacteria: An outlook on biogenesis. Front. Microbiol. 12, 557902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulp A., Kuehn M. J., Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sartorio M. G., Pardue E. J., Feldman M. F., Haurat M. F., Bacterial outer membrane vesicles: From discovery to applications. Annu. Rev. Microbiol. 75, 609–630 (2021), 10.1146/annurev-micro-052821-031444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bomberger J. M., et al. , Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5, e1000382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke A. C., et al. , Pseudomonas quinolone signal-induced outer membrane vesicles enhance biofilm dispersion in Pseudomonas aeruginosa. mSphere 5, e01109-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durant L., et al. , Bacteroides thetaiotaomicron-derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome 8, 88–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickey C. A., et al. , Colitogenic bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe 17, 672–680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBroom A. J., Kuehn M. J., Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63, 545–558 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rakoff-Nahoum S., Coyne M. J., Comstock L. E., An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. CB 24, 40–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Y., et al. , Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbull L., et al. , Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7, 11220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyofuku M., Schild S., Kaparakis-Liaskos M., Eberl L., Composition and functions of bacterial membrane vesicles. Nat. Rev. Microbiol. 21, 415–430 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Sartorio M. G., Pardue E. J., Scott N. E., Feldman M. F., Human gut bacteria tailor extracellular vesicle cargo for the breakdown of diet- and host-derived glycans. Proc. Natl. Acad. Sci. U.S.A. 120, e2306314120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elhenawy W., Debelyy M. O., Feldman M. F., Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5, e00909-00914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valguarnera E., Scott N. E., Azimzadeh P., Feldman M. F., Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in bacteroides. mSphere 3, e00559-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckburg P. B., et al. , Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faith J. J., et al. , The long-term stability of the human gut microbiota. Science 341, 1237439–1237439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wexler A. G., Goodman A. L., An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2, 17026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wexler H. M., Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zafar H., Saier M. H., Gut Bacteroides species in health and disease. Gut Microbes 13, 1848158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakoff-Nahoum S., Foster K. R., Comstock L. E., The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauber F., Cornelis G. R., Renzi F., Identification of a new lipoprotein export signal in gram-negative bacteria. mBio 7, e01232-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.England C. G., Ehlerding E. B., Cai W., NanoLuc: A small luciferase is brightening up the field of bioluminescence. Bioconjug. Chem. 27, 1175–1187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sartorio Mariana G., Valguarnera Ezequiel F.-F., Hsu M. F., Feldman, Lipidomics analysis of outer membrane vesicles and elucidation of the inositol phosphoceramide biosynthetic pathway in bacteroides thetaiotaomicron. Microbiol. Spectr. 10, e00634-21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otero-Asman J. R., Wettstadt S., Bernal P., Llamas M. A., Diversity of extracytoplasmic function sigma (σECF) factor-dependent signaling in Pseudomonas. Mol. Microbiol. 112, 356–373 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Staroń A., et al. , The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 74, 557–581 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Ndamukong I. C., Gee J., Smith C. J., The extracytoplasmic function sigma factor EcfO protects Bacteroides fragilis against oxidative stress. J. Bacteriol. 195, 145–155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varadi M., et al. , AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans J. C., et al. , A proteolytically activated antimicrobial toxin encoded on a mobile plasmid of Bacteroidales induces a protective response. Nat. Commun. 13, 4258 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop D. G., Work E., An extracellular glycolipid produced by Escherichia coli grown under lysine-limiting conditions. Biochem. J. 96, 567–576 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulp A. J., et al. , Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One 10, e0139200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobson Amy N., Choudhury Biswa P., Fischbach Michael A., The biosynthesis of lipooligosaccharide from bacteroides thetaiotaomicron. mBio 9 (2018), 10.1128/mbio.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pither M. D., et al. , Bacteroides thetaiotaomicron rough-type lipopolysaccharide: The chemical structure and the immunological activity. Carbohydr. Polym. 297, 120040 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Braun V., Mahren S., Sauter A., Gene regulation by transmembrane signaling. Biometals 19, 103–113 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Xu J., et al. , A genomic view of the human-bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Knowles T. J., Scott-Tucker A., Overduin M., Henderson I. R., Membrane protein architects: The role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7, 206–214 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Noinaj N., Gumbart J. C., Buchanan S. K., The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15, 197–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D. Y., Two stress sensor proteins for the expression of sigmaE regulon: DegS and RseB. J. Microbiol. 53, 306–310 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Dartigalongue C., Loferer H., Raina S., EcfE, a new essential inner membrane protease: Its role in the regulation of heat shock response in Escherichia coli. EMBO J. 20, 5908–5918 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., et al. , Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proc. Natl. Acad. Sci. U.S.A. 106, 14837–14842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bencivenga-Barry N. A., Lim B., Herrera C. M., Trent M. S., Goodman A. L., Genetic manipulation of wild human gut bacteroides. J. Bacteriol. 202, e00544-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitaker W. R., Shepherd E. S., Sonnenburg J. L., Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell 169, 538–546.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai C.-M., Frasch C. E., A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119, 115–119 (1982). [DOI] [PubMed] [Google Scholar]
- 49.Veeranagouda Y., Husain F., Wexler H. M., Transposon mutagenesis of Bacteroides fragilis using a mariner transposon vector. Anaerobe 22, 126–129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veeranagouda Y., Husain F., Wexler H. M., Transposon mutagenesis of Bacteroides fragilis. Methods Mol. Biol. 2016, 105–116 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Ron D., Dressler H., pGSTag–a versatile bacterial expression plasmid for enzymatic labeling of recombinant proteins. Biotechniques 13, 866–869 (1992). [PubMed] [Google Scholar]
- 52.Bennett M. J., et al. , A linear lattice model for polyglutamine in CAG-expansion diseases. Proc. Natl. Acad. Sci. U.S.A. 99, 11634–11639 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White J. B. R., et al. , Outer membrane utilisomes mediate glycan uptake in gut Bacteroidetes. Nature 618, 583–589 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pardue E. J., Scott N. E., Feldman M. F., Bacteroides thetaiotaomicron OMV, TM and WC comparsion of delta BT4720, delta BT_4721, delta BT_4720_4721 and WT. PRIDE. https://www.ebi.ac.uk/pride/archive/projects/PXD043360.Deposited 28 June 2023.
- 56.Pardue E. J., Ortiz-Marquez J. C., Feldman M. F., Dual membrane-spanning anti-sigma factors regulate vesiculation in gut bacteroidota. NCBI BioProject. https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/bioproject/?term=PRJNA994135. Deposited 12 July 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Mass Spectrometry Proteomics data has been deposited in the Proteome Xchange Consortium via the PRIDE partner repository (54) with the dataset identifier PXD043360 (55). RNA sequencing data has been deposited with links to BioProject accession number PRJNA994135 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) (56).