Abstract
Membrane trafficking is regulated in part by small GTP-binding proteins of the ADP-ribosylation factor (Arf) family. Arf function depends on the controlled exchange and hydrolysis of GTP. We have purified and cloned two variants of a 130-kDa phosphatidylinositol 4,5-biphosphate (PIP2)-dependent Arf1 GTPase-activating protein (GAP), which we call ASAP1a and ASAP1b. Both contain a pleckstrin homology (PH) domain, a zinc finger similar to that found in another Arf GAP, three ankyrin (ANK) repeats, a proline-rich region with alternative splicing and SH3 binding motifs, eight repeats of the sequence E/DLPPKP, and an SH3 domain. Together, the PH, zinc finger, and ANK repeat regions possess PIP2-dependent GAP activity on Arf1 and Arf5, less activity on Arf6, and no detectable activity on Arl2 in vitro. The cDNA for ASAP1 was independently identified in a screen for proteins that interact with the SH3 domain of the tyrosine kinase Src. ASAP1 associates in vitro with the SH3 domains of Src family members and with the Crk adapter protein. ASAP1 coprecipitates with Src from cell lysates and is phosphorylated on tyrosine residues in cells expressing activated Src. Both coimmunoprecipitation and tyrosine phosphorylation depend on the same proline-rich class II Src SH3 binding site required for in vitro association. By directly interacting with both Arfs and tyrosine kinases involved in regulating cell growth and cytoskeletal organization, ASAP1 could coordinate membrane remodeling events with these processes.
Membrane traffic, the transfer of material between membrane-bound compartments, is needed for such diverse cellular processes as secretion, endocytosis, and changes in cell shape that accompany cell growth, division, and migration (reviewed in references 84, 85, and 87). It is mediated by transport vesicles that are formed by budding from a donor membrane. The process of budding is driven by the assembly of a proteinaceous coat. Once the vesicle is formed, the coat must dissociate to permit fusion with an acceptor membrane and the consequent delivery of the vesicle’s contents. These steps are regulated in part by the Arf family of small GTP-binding proteins (reviewed in references 8, 23, 61, and 63). Arfs are highly conserved and are found in eukaryotes ranging from yeast to humans. The mammalian Arf family is divided into several classes based largely on sequence similarity: class I (Arfs 1 through 3), class II (Arfs 4 and 5), class III (Arf6), and the more distantly related Arf-like (Arl) class. By linking GTP binding and hydrolysis to coat assembly and disassembly, Arfs regulate membrane trafficking at a number of sites. Arf1 has been implicated in endoplasmic reticulum-to-Golgi and intra-Golgi transport, endosome-to-endosome fusion, and synaptic vesicle formation (8, 23, 28, 61, 63, 66). Arf6 has been implicated in regulation of membrane traffic between the plasma membrane and a specialized endocytic compartment, and its function has been linked to cytoskeletal reorganization (25, 26, 71, 73, 74). The specific sites of action of the other Arf family members are not known.
The hydrolysis of GTP on Arf requires a GTPase-activating protein (GAP) (19, 61). With multiple Arfs and multiple sites of action, the existence of several unique Arf GAPs had been anticipated. A number of activities have been purified or partially purified from mammalian sources, including rat liver (19, 57, 77), rat spleen (21), and bovine brain (79), and two Arf GAP activities from rat liver have been resolved (77). They have similar Arf specificities but differ in their lipid dependencies. One of the Arf GAPs (ArfGAP/ArfGAP1, hereafter referred to as ArfGAP1) which functions in the Golgi is activated by dioleoglycerols (3, 4, 19, 40). ArfGAP1, in common with a yeast Arf GAP, GCS1 (72), contains a zinc finger domain which is required for activity (19). The second Arf GAP (ArfGAP2) is specifically activated by phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidic acid (PA). Based on lipid requirements, ArfGAP2 was speculated to function at the plasma membrane and be regulated independently of ArfGAP1 (77). ArfGAP1 and ArfGAP2 were antigenically distinct and, therefore, likely to be distinct gene products; however, prior to this study, only ArfGAP1 had been cloned (19).
Src, a cytoplasmic tyrosine kinase with N-terminal Src homology 3 (SH3) and SH2 domains, transduces signals important for cell growth and cytoskeletal organization (12, 68, 91). A number of studies suggest that Src is also involved in regulating membrane traffic. Src associates primarily with endosomal membranes and in several cell types has been localized to specialized secretory vesicles, including synaptic vesicles (5, 20, 34, 46, 54, 69, 81). Overexpression of Src accelerates endocytosis (95). In addition, Src associates with or phosphorylates several proteins involved in membrane trafficking (5, 31, 43, 65).
Here, we report the purification and cloning of a PIP2-dependent Arf GAP, ASAP1. ASAP1 contains a zinc finger domain similar to that required for GAP activity in ArfGAP1 and GCS1. ASAP1 also contains a number of domains that are likely to be involved in regulation and/or localization: a pleckstrin homology (PH) domain, three ankyrin (ANK) repeats, a proline-rich region with SH3 binding motifs, and an SH3 domain. In addition, ASAP1 was identified independently as a binding protein for Src and was found to be phosphorylated on tyrosine in cells that express activated Src. ASAP1 also associated with the adapter protein c-Crk in vitro. ASAP1 was localized to the cytoplasm and the cell edge likely associated with the plasma membrane. We propose that ASAP1, by binding both Src and PIP2, could coordinate membrane trafficking with cell growth or actin cytoskeleton remodeling.
MATERIALS AND METHODS
Purification of the PIP2-dependent Arf GAP.
Arf1-GTP was used as a substrate to detect activity. Purification was performed at 4°C except where noted otherwise. Three bovine brains were homogenized in 4 volumes of 10 mM Tris (pH 8.0)–10% sucrose at 4°C, filtered through cheesecloth, and centrifuged at 20,000 × g for 60 min. The supernatant was brought to 35% ammonium sulfate. The precipitate was collected by centrifugation (10,000 × g for 10 min) and resuspended in 500 ml of 20 mM Tris (pH 8.0), 1 mM β-mercaptoethanol, and 10% (vol/vol) glycerol (buffer A) containing 25 mM NaCl, dialyzed two times against 10 liters of the same buffer, and clarified by centrifugation at 100,000 × g for 60 min. The supernatant was loaded onto a 250-ml column of DEAE-Sephacel. The column was developed with a 50 to 325 mM NaCl gradient in 1 liter of buffer A. Fractions containing PIP2-dependent GAP activity were pooled, and protein was adsorbed to a 40-ml column of hydroxylapatite (Bio-Rad) equilibrated in 10 mM KPi, 100 mM KCl, 1 mM MgCl2, 1 mM β-mercaptoethanol, and 20% glycerol. The column was developed in a 10 to 400 mM KPi (pH 7.0) gradient in a buffer containing 100 mM KCl, 1 mM MgCl2, 1 mM β-mercaptoethanol, and 20% glycerol over 200 ml. Fractions containing activity were diluted 1:2 with 10 mM KPi, 100 mM KCl, 1 mM MgCl2, 1 mM β-mercaptoethanol, and 20% glycerol and adsorbed to a hydroxylapatite column with a 4-ml bed volume. The column was developed by isocratic elution with 100 mM KPi, 2 mM KCl, 1 mM MgCl2, 1 mM β-mercaptoethanol, and 20% (vol/vol) glycerol. Fractions containing activity were pooled, adjusted to 3 M KCl, and adsorbed at room temperature to a 1-ml Phenyl-Sepharose HP column (Pharmacia, Uppsala, Sweden) equilibrated with 100 mM KPi, 3 M KCl, 1 mM MgCl2, 1 mM β-mercaptoethanol, and 20% (vol/vol) glycerol. The column was developed with a descending salt gradient. The remaining steps were performed at 4°C. The activity was then gel filtered on a 120-ml column of Sephacryl S-300 (Pharmacia) in 20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM MgCl2, and 1 mM β-mercaptoethanol followed by chromatography on a 1-ml Resource Q (Pharmacia) column with a 100 to 600 mM NaCl gradient in 20 mM Tris (pH 8.0), 1 mM MgCl2, 1 mM β-mercaptoethanol, and 20% (vol/vol) glycerol over 12 ml. The purification is summarized in Table 1.
TABLE 1.
Purification of a PIP2-dependent Arf GAP from bovine braina
| Step | Protein (mg) | Specific activity (pmol/min/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|
| S20 | 12,000 | 0.42 | 1 | 100 |
| (NH4)2SO4 | 4,300 | 0.78 | 1.9 | 68 |
| DEAE | 400 | 8.0 | 19 | 64 |
| HAP1 | 20 | 56 | 134 | 23 |
| HAP2 | 7.2 | 118 | 280 | 17 |
| Phenyl-Sepharose | 1.1 | 590 | 1,404 | 13 |
| Sephacryl 300 | 0.35 | 944 | 2,246 | 6.7 |
| Resource Q | 0.09 | 1,875 | 4,464 | 3.3 |
GAP was purified as described in Materials and Methods. S20 is the soluble fraction from the 20,000 × g centrifugation of the crude brain homogenate. HAP1, hydroxylapatite column developed with a KPi gradient; HAP2, hydroxylapatite column developed with a KCl gradient. Activity was determined using 15 nM Arf1-GTP as substrate and 1 mg of crude phosphoinositide/ml as a source of phospholipid.
Trypsin digestion, HPLC separation, and microsequencing.
Purified Arf GAP protein was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by copper staining (Bio-Rad). The band of interest was subjected to in gel tryptic digestion as described by Hellman et al. (37) but without the addition of 0.02% Tween. The resulting peptide mixture was separated by microbore high-performance liquid chromatography (HPLC) with a Zorbax C18 1.0- by 150-mm reverse-phase column on a Hewlett-Packard 1090 HPLC/1040 diode array detector. Optimum fractions were chosen based on differential UV aborbance at 205, 277, and 292 nm, peak symmetry, and resolution and then further screened for length and homogeneity by matrix-assisted laser desorption time-of-flight mass spectrometry on a Lasermat 2000 (Finnigan, Hemel, England). Strategies for peak selection, reverse-phase separation, and Edman microsequencing have been previously described (52). Tryptic peptides were subjected to automated Edman degradation on a 494A HT or 494A cLC protein sequencer (Applied Biosystems, Foster City, Calif.).
Identification of cDNAs for ASAP1.
A partial murine ASAP1 cDNA clone (accession no. W89336) was identified by using the tryptic peptide sequences from purified PIP2-dependent Arf GAP to scan an expressed-sequence tag (EST) database. Anchored PCR was used to extend the clone to the 5′ end of the open reading frame, using as templates mouse embryo and brain cDNA libraries (Marathon cDNA; Clontech) and using methods described in the manufacturer’s manual. The full-length open reading frames and part of the 3′ untranslated region were amplified from a mouse embryo and a brain cDNA library by PCR using one primer incorporating the initiating methionine (GAT GTG ACG GCT GAG ACA TGA GAT CTT CAG) and one of two antisense primers from the 3′ untranslated region (CTA CCA TGA GTT CTT GGT CTG TAA CAG CAG C and GCA ATC TTG TAA CTT CTG CTT TAA TGG CAA TC). The DNA fragments obtained were ligated into pCR2.1 (Topo cloning kit; Invitrogen) by following the manufacturer’s protocol. All but the 5′ end of the open reading frame (codons 1 to 136) was cloned independently by screening a mouse mixed-spermatocyte lambda cDNA library (a gift from E. M. Eddy) with a probe derived from a two-hybrid screen (see below). Two alternative splice forms were identified by both approaches. Both strands of the PCR-amplified cDNA and of the spermatocyte library clones were sequenced by dye terminator sequencing.
Recombinant Arf GAP.
Amino acid residues 325 to 724 of an ASAP1 cDNA cloned from a mouse spermatocyte lambda library was amplified by using primers that incorporated a 5′ NdeI site and a 3′ XhoI site and ligated into the His-tag vector pET19 (Novagen). The resulting recombinant PZA protein was expressed in Escherichia coli BL21(DE3). A 250-ml culture was lysed in 10 ml of buffer A containing 25 mM NaCl and one tablet of Complete protease inhibitor cocktail (Boehringer Mannheim) by using a Piranha press (Tesla Inc., Paxton, Ill.). The lysate was clarified by centrifugation at 100,000 × g for 60 min and fractionated on a HiLoad Q (5-ml bed volume) (Pharmacia) with a 50 to 600 mM NaCl gradient in 30 ml of buffer A. The protein was greater than 95% pure as estimated by fractionating a sample by SDS-PAGE and staining with Coomassie blue. The protein was adsorbed to Ni2+ bound to a HiTrap chelating column (Pharmacia). The column was developed with a gradient of 5 to 500 mM imidazole (pH 7.0), in 20 mM Tris (pH 8.0), 500 mM NaCl, and 10% glycerol over 10 ml.
Arf GAP assays.
Arf GAP activity was assayed as described previously (79) by using nonmyristoylated Arf1 as a substrate except when comparing Arf specificities, in which case all Arfs were myristoylated. Crude phosphoinositides (P-6023; Sigma) at a concentration of 1 mg/ml were the source of phosphoinositides except where otherwise indicated. The concentration of Arf-GTP was less than 20 nM. The rate of GTP hydrolysis by Arf1 induced by Arf GAP was linear to 200 nM Arf1-GTP, and none of the Arf proteins used in this paper competed with Arf1-GTP, consistent with the substrate concentration being nonsaturating and much less than the Km. This is in agreement with previous work (77) in which the Km for Arf1 using the native GAP as enzyme was determined to be 5 μM. With Arf at concentrations much less than the Km, GTP hydrolysis occurred with a first-order rate. The rate constant is proportional to Vmax/Km. For single-point assays, time points and enzyme concentrations were chosen such that less than 80% of the GTP on Arf was hydrolyzed, and the first-order rate was calculated as k = ln(Arf-GTP at time 0/Arf-GTP remaining)/time, as has been described previously (79). Nonmyristoylated Arf1 and Arl2 and myristoylated Arf1 and Arf5 were prepared as described previously (77, 80). Myristoylated Arf6 expressed in bacteria was purified from a particulate fraction of cells lysed with a Piranha press and centrifuged at 100,000 × g for 60 min at 4°C. Arf6 was extracted into 20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM dithiothreitol (DTT), 10% glycerol, and 1% Triton X-100. The extract was clarified by centrifugation. The proteins were precipitated with 35% ammonium sulfate, dissolved in 20 mM Tris (pH 8.0), 25 mM NaCl, 10 μM GDP, 1 mM DTT, 10% glycerol, and 1% Triton X-100. The solubilized proteins were fractionated on a 1-ml DEAE-Sephacel column equilibrated with the same buffer. Arf6 that was greater than 90% pure eluted from the column in the equilibration buffer. To determine GAP activity in total cell homogenates, cells were lysed in 10 mM Tris (pH 7.2), 158 mM NaCl, 1% Triton X-100, 1% aprotonin, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.1% β-mercaptoethanol.
Mammalian expression.
The ASAP1a and ASAP1b open reading frames were amplified using primers that placed a FLAG tag (MDYKDDDDK) in frame with the initiating methionine and ligated into pcDNA3 (Invitrogen). Alternatively, the ASAP1b coding sequence was cloned into pCS3+MT, which carries six copies of the Myc epitope in frame with the initiator methionine (92). The P1, P2, and P3 mutations were introduced into CS3+MT−ASAP1b by Pfu site-directed mutagenesis. Wild-type and activated mutant (Y527F) c-Src cloned into the retroviral vector pLXSH were gifts from C. Sachsenmaier. Transient transfections were performed by using the calcium phosphate method (293, HeLa, and NIH 3T3 cells) or Lipofectamine (COS cells) as described by the manufacturer (GIBCO BRL).
Production of anti-ASAP1 antibodies.
The ASAP1a Src-interacting domain (SID) (amino acids 697 to 850) was cloned into pGEX-3X to create a glutathione S-transferase (GST)-ASAP1a-SID fusion protein, which was used to immunize female New Zealand White rabbits. The resulting antiserum, called 3820J, was precleared with GST-glutathione-Sepharose (Pharmacia) and then affinity purified on cyanogen bromide-activated Sepharose (Sigma) that had been cross-linked to GST-ASAP1a-SID. Antisera 551 was from rabbits immunized with a recombinant protein containing the PH domain, zinc finger, and ANK repeats of ASAP1 conjugated to KLH.
Two-hybrid screen.
A mouse 9.5-day-old-embryo cDNA library was screened by using a LexA-TRP1/VP16-LEU2 two-hybrid system in the Saccharomyces cerevisiae L40 (MATa trp1 leu2 his3 LYS::lexA-HIS3 URA3::lexA-lacZ) (38, 94). The two-hybrid bait plasmid LexA-c-Src-SH3 was created by inserting the chicken c-Src SH3 domain (residues 81 to 144) into the LexA vector BTM116. The bait plasmid was slightly transactivating. By including 3-aminotriazole or limiting amounts of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside in either the histidine prototrophy or β-galactosidase assays, background could be abolished without eliminating production of a clear, positive signal by LexA-c-Src-SH3 in combination with a VP16 fusion to residues 484 to 551 of 3BP1, a protein that binds directly to Src SH3 in vitro (15). A total of 5 × 107 yeast transformants were screened for growth on synthetic medium containing 1 to 6 mM 3-aminotriazole and lacking histidine, tryptophan, and leucine. Approximately 5 × 103 transformants expressed the HIS3 reporter gene at levels at least as great as yeast carrying LexA-Src-SH3 and VP16-3BP1. Of these, 618 were screened for expression of the second reporter gene by a β-galactosidase filter assay, and 522 were positive. Selected clones expressing both reporter genes and not interacting with the unrelated, negative control bait plasmid LexA-lamin (7) were also tested against full-length wild-type or various mutant forms of c-Src (39) or with the n-Src variant SH3 domain. Seventeen percent of clones interacting with LexA-c-Src-SH3 also interacted with LexA-c-Src (full length). LexA-n-Src and n-Src-SH3 plasmids were constructed by inserting either the entire coding sequence or amino acids 81 to 150 of chicken n-Src into BTM116. Approximately 70 clones were sequenced. Four nonidentical but overlapping clones containing ASAP1 sequence were identified.
In vitro binding.
CS3+MT−ASAP1a−SID or CS3+MT−ASAP1b DNA was added to a coupled in vitro transcription-translation system (TNT Kit; Promega) in the presence of [35S]methionine to generate labeled SID or ASAP1b. GST-SH3 fusion proteins were expressed in E. coli and purified on glutathione-Sepharose (Pharmacia). Approximately 2 μg of GST fusion protein was mixed with the in vitro translation mix in 500 μl of radioimmunoprecipitation assay (RIPA) buffer (158 mM NaCl, 5 mM EDTA, 10 mM Tris [pH 7.2], 0.1% SDS, 1% Na-deoxycholate, 1% Triton X-100, 1% aprotinin, 1 mM PMSF, 0.1% β-mercaptoethanol). Samples were incubated for 3 h at 4°C and washed three times in RIPA buffer. Proteins associating with the fusion protein-Sepharose pellet were examined by SDS-PAGE and detected by autoradiography. Binding was quantified with a PhosphorImager (Molecular Dynamics). GST-SH3 fusion plasmids for chicken c-Src, bovine p85 PI 3-kinase, human p120 RasGAP, and mouse Efs/Sin were constructed by inserting, respectively, sequences encoding amino acids 81 to 144, 6 to 83, 281 to 346, and 5 to 70 into pGEX1 (Pharmacia). Additional GST fusion plasmids were obtained for n-Src SH3, Fyn SH3, Lyn SH3, Csk SH3, Grb2-N and Grb2-C SH3, Abl SH3, and full-length c-Crk.
Immunoprecipitations, Western blotting, and kinase assays.
Twenty-four hours following transfection, 293 cells were lysed in RIPA buffer containing 2 mM Na3VO4 and centrifuged for 45 min at 15,000 × g. The supernatants were incubated with primary antibodies, secondary antibodies (goat anti-mouse for monoclonal antibodies), and a 1:1 mix of Staphylococcus-Streptococcus cell fragments bearing proteins A and G (Pansorb and Omnisorb; CalBiochem-Novabiochem Corp., La Jolla, Calif.) for 2 h at 4°C. Immunoprecipitating antibodies used were 3060 rabbit antiserum raised to residues 519 to 533 of the c-Src C terminus (17) and 9E10, an anti-Myc-tag monoclonal antibody (27). Negative control immunoprecipitations were done with either preimmune rabbit antiserum or mock antibody mixes containing all components except for primary antibody. Proteins were electrophoresed through SDS-acrylamide gels containing 10 to 15% polyacrylamide and transferred to nitrocellulose (Schleicher & Schuell, Keene, N.H.). For Western blotting, monoclonal primary antibodies 9E10, antiphosphotyrosine 4G10 (24), and 327 anti-Src (55) were detected with a secondary horseradish-peroxidase linked anti-mouse immunoglobulin F(ab′) fragment (Amersham) and enhanced chemiluminescence (Renaissance reagents; NEN, Boston, Mass.). For in vitro tyrosine kinase assays, 1 U of recombinant c-Src (Upstate Biotechnology, Inc., Lake Placid, N.Y.) was incubated with 40 ng of ASAP1, purified from bovine brain, in 10 mM Tris (pH 7.4), 12.5 mM MgCl2, 8 mM MnCl2, 0.2 mM EGTA, 25 μM Na2VO4, 0.2 mM DTT, 5 μM [γ-32P]ATP, and 1 mg of crude phosphoinositides/ml for 20 min at 30°C. Reaction products were fractionated by SDS-PAGE on a 7.5% polyacrylamide gel. Radioisotope was detected with a PhosphorImager.
Immunofluorescence.
HeLa cells were grown on glass coverslips in six-well dishes and transfected with an expression vector for FLAG-tagged ASAP1b or FLAG-tagged ASAP1a (in pcDNA3) (10 μg/well) by using the calcium phosphate procedure. Cells were fixed with 2% formaldehyde in phosphate-buffered saline for 10 min at room temperature followed by a 30-s exposure to methanol. After rinsing with 10% fetal bovine serum and 0.02% azide in phosphate-buffered saline, cells were incubated with primary and secondary antibodies as described previously (74). Transferrin uptake was performed as described previously (74) except that uptake was for 10 min and cells were briefly rinsed with 0.5 M NaCl–0.5% acetic acid and washed with media immediately before fixing. Fluorescein- and rhodamine-conjugated donkey anti-mouse and anti-rabbit antibodies were from Jackson ImmunoResearch Laboratories (West Grove, Pa.). The hybridoma M3A5, used for preparing the antibody to β-COP, was a gift from Thomas Kreis. A rabbit polyclonal antiserum raised against human transferrin was from Boehringer Mannheim, and human transferrin was from Sigma Chemicals.
RESULTS
Cloning of murine homologs of a 130-kDa Arf GAP-associated protein.
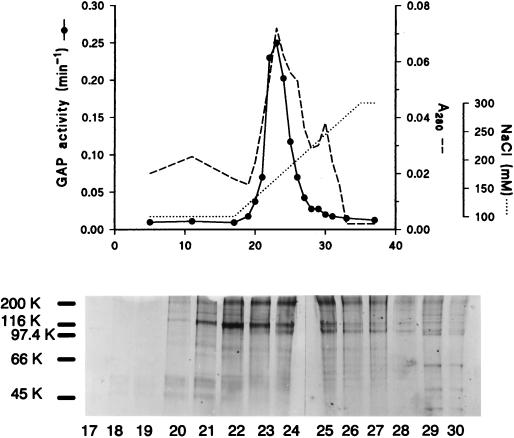
A PIP2-dependent Arf GAP was purified 4,000-fold from a soluble fraction of bovine brain (Table 1, see Materials and Methods). The PIP2-dependent GAP was the major GAP activity in the brain homogenate. Over 90% of the recovered GAP activity was PIP2 dependent and eluted as a single peak from each column. Fractions from the last two chromatographic steps, gel filtration (results not shown) and ion exchange (Fig. 1), were electrophoresed in SDS through a 7.5% polyacrylamide gel, and the proteins were visualized. A 130-kDa polypeptide coeluted with the activity.
FIG. 1.
Polypeptides coeluting with GAP activity on anion exchange. Activity eluting from the Sephacryl S-300 column was adsorbed to a Resource Q column and eluted as described in Materials and Methods. Samples from the indicated fractions were electrophoresed in SDS on a 7.5% polyacrylamide gel, and proteins were visualized with a colloidal blue stain.
The 130-kDa protein was excised from the polyacrylamide gel and trypsinized, and the peptide sequences of several of the tryptic fragments were determined to be TLLK, EEALTMAFR, LGTSELLLAK, and VGNSF??IMEAN. The latter two peptides were found encoded within a mouse EST clone (W89336). Anchored PCR was used to determine a 5′ sequence that was not in the EST clone. The remainder of the open reading frame was cloned as described earlier (see Materials and Methods). All but the extreme N terminus was also cloned from a mouse spermatocyte lambda library by using a two-hybrid probe (see below) and found to match the cDNA derived by rapid amplification of cDNA ends. Two variants that differed by a 57-amino-acid insertion were identified in several libraries. We call these two proteins ASAP1a (127,396 Da) and ASAP1b (121,649 Da) for Arf GAP containing SH3, ANK repeat, and PH domains.
Sequence and expression of ASAP1.
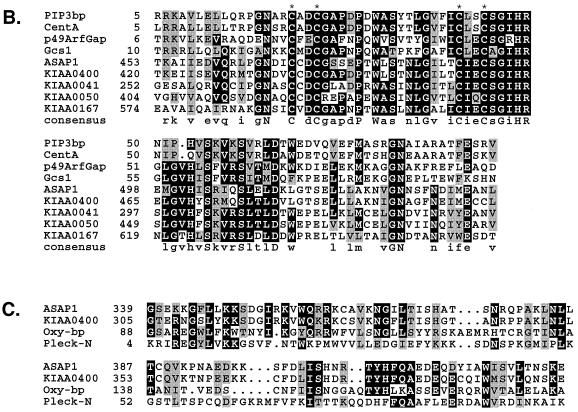
The ASAP1a cDNA encodes a 1,147-amino-acid polypeptide. ASAP1a and ASAP1b both contain a PH domain (residues 339 to 431), a zinc finger motif of the type CXXCX16CXXC (residues 469 to 492), three ANK repeats (residues 615 to 715), a proline-rich region containing several SH3 ligand motifs (residues 798 to 913), eight consecutive repeats of E/DLPPKP (residues 946 to 1011), and an SH3 domain (residues 1085 to 1147) (Fig. 2A). The variants differ only in the proline-rich region. ASAP1a contains 57 residues (817 to 873) that are absent in ASAP1b.
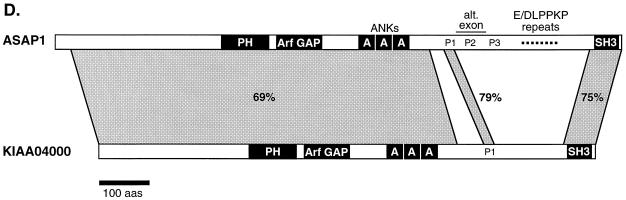
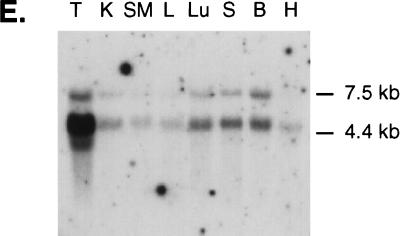
FIG. 2.
ASAP1 sequence, alignment with related proteins, and expression. (A) Predicted amino acid sequence of ASAP1a and ASAP1b. ASAP1 was conceptually translated from the cDNA sequence (GenBank accession no. AF075461 and AF075462). Shaded boxes indicate the PH, GCS-type zinc finger, ANK repeat, and SH3 domains, and the alternate exon found only in ASAP1a. Potential SH3 ligand sites P1, P2, and P3 are underlined with solid lines, E/DLPPKP repeats are underlined with a broken line, and peptide sequences of tryptic fragments from the purified bovine protein are underlined with a dotted line. The SID is indicated in boldface type. (B) Alignment of zinc-finger-containing domains. Conserved cysteine residues of the CXXCX16CXXC zinc finger motif are indicated by asterisks. Sequences were aligned by using PILEUP (University of Wisconsin Genetics Computer Group package). (C) Alignment of PH domains. PH domains were aligned by using PILEUP and secondary structure predictions. Databases and accession numbers: ArfGAP1 (GenBank U35776), GCS1 (Swiss Protein P35197), PIP3bp (DDBJ D89940), CentA (Centaurin-α; GenBank U51013), KIAA0400 (DDBJ AB007860), KIAA0041 (DDBJ D26069), KIAA0050 (DDBJ D30758), KIAA0167 (DDBJ D79989), Oxy-bp (oxysterol binding protein; Swiss Protein P22059), Pleck-N (N-terminal PH domain of pleckstrin; Swiss Protein P08567). (D) Schematic structure of ASAP1 and similarity to human KIAA0400. Numbers in boldface indicate the percentages of identity between the two proteins for the specified regions. Drawn to scale; bar = 100 amino acids. (E) Hybridization of ASAP1 cDNA to mouse poly(A) RNA. A radiolabeled probe corresponding to the ASAP1a SID was hybridized to poly (A) RNA on nylon membranes (obtained from Clontech) as recommended by Clontech. Each lane contains approximately 2 μg of RNA. H, heart; B, brain; S, spleen; Lu, lung; L, liver; SM, skeletal muscle; K, kidney; T, testis.
The zinc finger motif is contained within a larger conserved region of about 90 amino acids (residues 457 to 527) that is similar to sequences found in ArfGAP1 and GCS1, the phosphatidylinositol 3,4,5-triphosphate (PIP3) binding proteins PIP3bp and centaurin-α, and a number of other proteins from mammals, plants, and lower eukaryotes (Fig. 2B) (19, 35, 72, 89). In ArfGAP1 and GCS1, the zinc finger is essential for GAP activity (19, 72). Zinc finger domains showing the strongest similarity to ASAP1 include those of several proteins that, like ASAP1, have PH domains located just N-terminal to the zinc finger, including the human proteins KIAA0400 (86% similar, 75% identical), KIAA0041 (67% similar, 48% identical), KIAA050 (63% similar, 43% identical), and KIAA0167 (60% similar, 44% identical) (Fig. 2B). The region between the PH and zinc finger domains is also conserved in these proteins, and they possess adjacent C-terminal ANK repeats. The PH-Zn-finger-ANK (PZA) combination is also found in plants, worms, and flies and therefore appears to define an evolutionarily conserved module.
The PH domain of ASAP1 is most similar to the PH domains of several PZA proteins, to Dictyostelium rac-α (Akt/PKB) serine-threonine kinase homolog, and to oxysterol binding protein (Fig. 2C). The latter PH domain has been shown to bind PIP2 and PIP3 with equal affinity in vitro (75). The protein whose zinc finger and PH domains are most closely related to ASAP1 is the PZA protein KIAA0400. This human protein is a true ASAP family member; it shows strong similarity to ASAP1 over three regions encompassing approximately 74% of the proteins (Fig. 2D). The N-terminal 728 amino acids are 69% identical to residues 21 to 759 of ASAP1; a 14-amino-acid proline-rich SH3 ligand motif (P1) is also strongly conserved (79% identity), and the C terminus containing the SH3 domain shows 75% identity between the two proteins. The next most closely related SH3 domain to ASAP1 is Acanthamoeba myosin I (48% identity). Conservation of the N-terminal 330 residues of ASAP1 and KIAA0400 suggests that this region, which lacks identifiable sequence motifs, is also functionally important.
Northern analysis revealed three mRNAs in mouse tissues (Fig. 2E). mRNAs of 7.5 and 5 kb were expressed in all tissues examined but were most abundant in testis, brain, lung, and spleen. Testis also contained an mRNA of 4 kb that hybridized with the probe. An antibody raised in rabbits to residues 697 to 850 of mouse ASAP1a reacted with 130-kDa proteins in homogenates of these tissues (not shown). The relative abundance of the 130-kDa species correlated with the mRNA levels. Several cross-reacting proteins were also detected. It remains to be determined whether these proteins are related to ASAP1.
ASAP1 contains GAP activity.
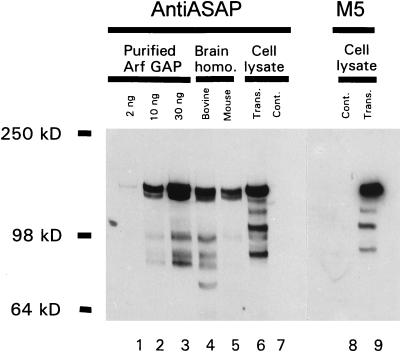
Transient transfection of mammalian cells with either the cDNA for ASAP1a or ASAP1b resulted in expression of proteins that comigrated with the 130-kDa bovine protein that copurified with Arf GAP activity. Anti-ASAP1a antiserum was used to detect recombinant ASAP1b and purified bovine Arf GAP (Fig. 3). The antibody bound to 125- and 130-kDa proteins present in the purified bovine Arf GAP and in homogenates of bovine and mouse brain (lanes 1 to 5). As little as 2 ng of the purified bovine protein was detected by using the antibody (lane 1). The larger immunoreactive protein comigrated with FLAG-tagged ASAP1a (not shown) and FLAG-tagged ASAP1b expressed in COS7 cells and detected with either the rabbit polyclonal antibody (lane 6) or with a monoclonal antibody to the epitope tag (lane 8). On longer exposures, the 130-kDa protein was also detected in untransfected COS7 extracts (not shown). Our results cannot distinguish whether both ASAP1a and ASAP1b proteins are expressed in brain and present in the purified preparation of Arf GAP.
FIG. 3.
An antibody raised to mouse ASAP1 recognizes Arf GAP purified from bovine brain. The indicated quantity of purified bovine brain Arf GAP (lanes 1 to 3), a bovine brain homogenate (30 μg of protein), a mouse brain homogenate (25 μg of protein), and lysates (0.75 μg of protein) from COS7 cells transfected with a FLAG-tagged ASAP1b expression vector (trans) and COS7 cells that had not been transfected (cont) were fractionated by SDS-PAGE and transferred to nitrocellulose. The blots were probed with 3820J anti-ASAP1 antiserum (lanes 1 to 7) or a monoclonal antibody (M5) to the FLAG epitope (lanes 8 and 9).
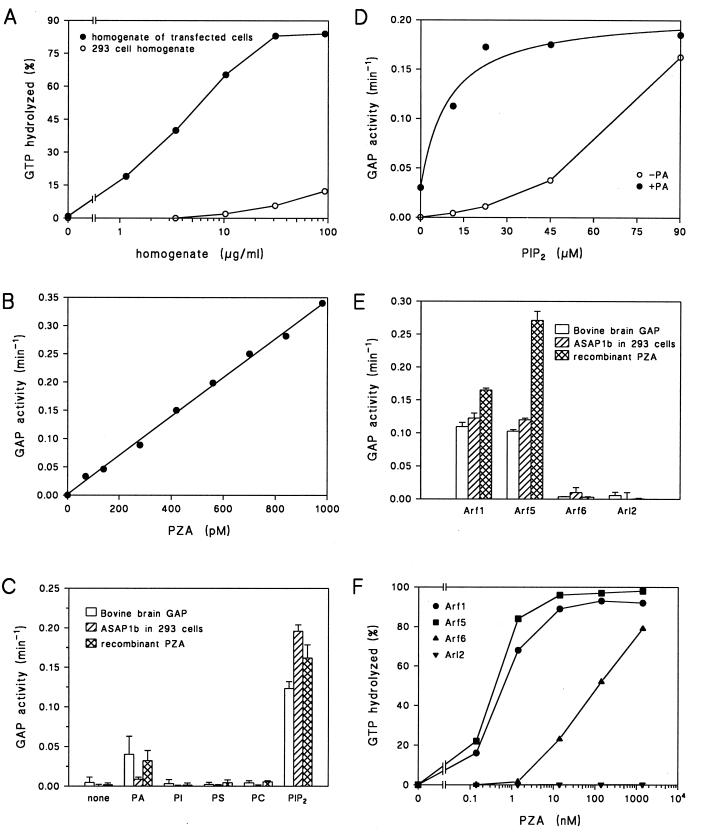
ASAP1 expressed in two contexts was tested as an Arf GAP. First, the Arf1 GAP activity in homogenates prepared from cells transiently transfected with cDNA for Myc-tagged ASAP1b was compared to that in homogenates from nontransfected cells (Fig. 4A). The activity was 100-fold greater in the transfected cells than in the controls. Second, a recombinant protein consisting of the ASAP1 PZA region was expressed in E. coli (see Materials and Methods). The fragment PZA induced GTP hydrolysis on Arf1 with a first-order rate that was proportional to the concentration of PZA (Fig. 4B).
FIG. 4.
Recombinant ASAP1 has Arf GAP activity. (A) Increased Arf GAP activity in lysates of cells overexpressing ASAP1b. The Arf GAP activity in lysates of 293 cells that were transfected with an expression vector for ASAP1b (closed circles) or cells that were not transfected (open circles) was determined. (B) Arf GAP activity of PZA. The recombinant protein PZA was incubated at the indicated concentrations with approximately 10 nM Arf1-GTP in the presence of crude phosphoinositides for 3 min at 30°C. (C) Phospholipid dependence of purified Arf GAP, recombinant protein ASAP1, and PZA. Arf-GTP was incubated with either 0.9 nM Arf GAP purified from bovine brain, 0.03 μg of protein from the lysate of 293 cells transiently transfected with a ASAP1b expression vector per ml, or 0.7 nM bacterially expressed recombinant protein PZA. PIP2, 180 μM PIP2; PA, 750 μM phosphatidic acid; PI, 720 μM phosphatidylinositol; PS, 720 μM phosphatidylserine; PC, 720 μM phosphatidylcholine. (D) PIP2-dependence of recombinant PZA. Arf-GTP was incubated with 0.7 nM PZA and the indicated concentrations of PIP2 in the presence or absence of 750 μM PA. (E) Substrate specificity of purified Arf GAP, recombinant protein ASAP1, and PZA. MyrArf1-GTP, myrArf5-GTP, myrArf6-GTP and Arl2-GTP were used as substrates in reactions containing 1 mg of crude phosphoinositide/ml as a source of PIP2 and 2 nM purified bovine brain Arf GAP, lysates of 293 cells transiently transfected with an expression vector for ASAP1b (0.03 μg of protein/ml), or 1.4 nM bacterially expressed PZA. The means ± standard deviations of quadruplicate determinations are shown for bovine Arf GAP and PZA. The means ± the ranges for duplicates are shown for the cell lysates. (F) Substrate specificity of PZA. Conditions were the same as for panel E, but the indicated concentrations of PZA were used.
The phospholipid dependence of the recombinant proteins was similar to that of the native protein. As with the partially purified protein (77, 79), 180 μM PIP2 increased the activity of purified bovine ASAP1, ASAP1b expressed in 293 cells, and the bacterially expressed fragment PZA (Fig. 4C). PA at a fourfold-greater concentration than PIP2 enhanced activity but to a lesser extent than did PIP2. No activity was detected with 720 μM phosphatidylinositol (PI), phosphatidylserine, or phosphatidylcholine. As has been previously reported for Arf GAP from bovine brain (79) and rat liver (77), the effect of PIP2 was saturable (Fig. 4D), and the concentration of PIP2 required for activation was reduced in the presence of PA, from 74 to 10 μM. A protein containing the zinc finger and ANK domains but not the PH domain had negligible Arf GAP activity (data not shown), implying that the PH domain is critical for activity.
The Arf specificities of the native and recombinant proteins were also similar. Four Arf family members were examined as substrates (Fig. 4E). As had been found with a partially purified GAP2 from rat liver (77), the purified bovine protein, epitope-tagged ASAP1b, and bacterially expressed PZA induced GTP hydrolysis by a class I Arf (Arf1) and a class II Arf (Arf5), weakly induced a class III Arf (Arf6), and was inactive on Arl2 (Fig. 4E). The Arf specificity was further examined by titrating recombinant PZA into the reaction (Fig. 4F). Arf1 and Arf5 GAP activity had similar concentration dependencies with 50% of the GTP bound on Arf hydrolyzed in 4 min in the presence of 0.55 ± 0.05 and 0.36 ± 0.08 nM PZA. Hydrolysis of GTP by Arf6 was detected with ∼200-fold more PZA than for Arf1. No activity using Arl2 as a substrate was seen at the highest concentration of recombinant PZA tested, 1.4 μM.
ASAP1 associates with the Src SH3 domain.
To identify possible targets or regulators of Src, a mouse embryo cDNA library (38, 94) was screened by using the yeast two-hybrid system and the c-Src SH3 domain as bait. Partial cDNAs for four known Src substrates, Sam68, p130CAS, AFAP110, and Efs/Sin (1, 30, 33, 42, 86, 90), were identified, together with a number of novel SH3-interacting proteins, including ASAP1. Four overlapping cDNA clones that contained sequences corresponding to amino acids 697 to 850 of the ASAP1a cDNA variant were isolated. As noted above, we have termed this region the Src-interacting domain (SID; Fig. 2A). Full-length c-Src also bound to the ASAP1 SID in the two-hybrid assay (Table 2). The ASAP1a SID also bound to a kinase-inactive mutant of Src but not to either a Src SH3 deletion mutant or to n-Src, which contains a six-residue insertion into the SH3 domain (59). Thus, in the yeast system, the SH3 domain of Src is necessary and sufficient for association, and tyrosine phosphorylation is not required for interaction. The insertion in the n-Src SH3 domain blocks interaction.
TABLE 2.
Yeast two-hybrid characterization of the ASAP1-Src interactiona
| LexA plasmid | VP16 plasmid
|
|
|---|---|---|
| VP16 | VP16-ASAP1-SID | |
| LexA | − | − |
| LexA-lamin | − | − |
| LexA-c-Src-SH3 | − | + |
| LexA-c-Src | − | + |
| LexA-c-SrcΔSH3 | − | − |
| LexA-c-SrcKD | − | + |
| LexA-n-Src | − | − |
| LexA-n-Src-SH3 | − | − |
Two-hybrid assays for protein-protein interactions were carried out by transformation of LexA and VP16 plasmids into the L40 yeast strain, which carries a lacZ reporter gene. β-Galactosidase filter assays on dual transformants were scored + (strong interaction) or − (no interaction). The LexA fusions tested include the following: lamin (negative control); c-Src-SH3 (wild-type c-Src SH3 domain); c-Src (wild-type full-length c-Src); c-SrcΔSH3 (c-Src with SH3 domain deletion); c-SrcKD (kinase dead c-Src K295R mutant); n-Src (full-length neuronal Src); n-Src-SH3 (SH3 domain of neuronal Src). VP16-ASAP1-SID contains amino acids 697 to 850 from ASAP1a.
In vitro binding of ASAP1 to SH3 domains.
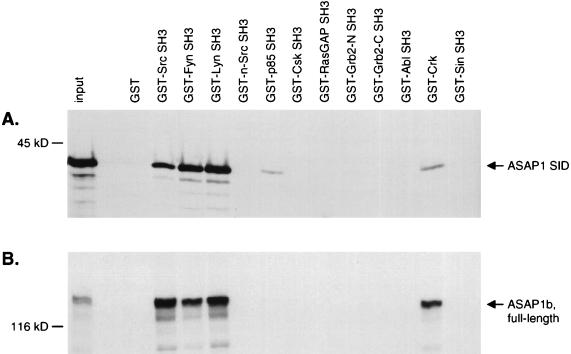
To determine the specificity of ASAP1 for the Src SH3 domain, 13 SH3 domains were expressed as GST fusion proteins in E. coli and tested for in vitro binding to the ASAP1a SID (Fig. 5A) or full-length ASAP1b (Fig. 5B). The ASAP1 SID interacted most strongly with SH3 domains of the Src family members Src, Fyn, and Lyn. The SID also bound weakly to the SH3 domain of the p85 regulatory subunit of PI 3-kinase and to a fusion of GST to full-length c-Crk (containing two SH3 domains). No binding was detected to the SH3 domains of n-Src, Csk, p120 RasGAP, Grb2, Abl, or Efs/Sin.
FIG. 5.
In vitro association of ASAP1 with SH3 domains. Myc-tagged ASAP1a SID (A) or full-length ASAP1b (B) was labeled with [35S]methionine and mixed with ∼2 μg of immobilized GST-SH3 fusion proteins (see Materials and Methods). Bound proteins were eluted and separated by SDS-PAGE. The first lane of each gel contains 1/30 the amount of the radioactive protein that was added to each binding reaction. Grb2-N, N-terminal SH3 domain of mouse Grb2; Grb2-C, C-terminal Grb2 SH3 domain; p85, p85 subunit of PI3-kinase; Ras-GAP, p120 RasGAP.
To confirm these interactions and check for binding to regions outside the ASAP1a SID, the SH3 domains were tested for binding to full-length ASAP1b (Fig. 5B). Due to alternative splicing, ASAP1b lacks the 34 most-C-terminal amino acids of the ASAP1a SID. However, full-length ASAP1b also bound to the Src, Fyn, and Lyn SH3 domains. Indeed, full-length ASAP1b was bound approximately 10-fold more efficiently, with 35 to 45% of [35S]methionine-labeled ASAP1b bound. This suggests that regions outside the residues 697 to 816 of ASAP1 contribute to binding to Src family SH3 domains. Similar to Src, Crk bound the full-length ASAP1b more efficiently than the isolated SID.
ASAP1 binds to Src and becomes tyrosine phosphorylated in mammalian cells.
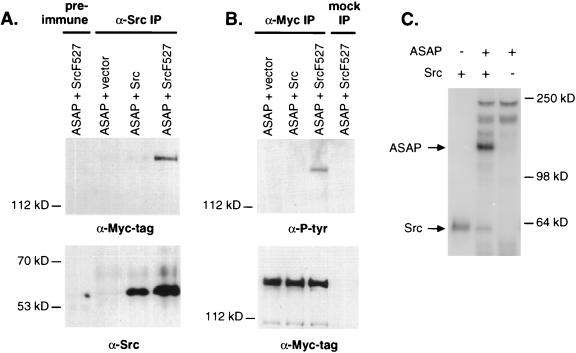
Wild-type or mutant Src and Myc-tagged ASAP1b were expressed in 293 cells. Lysates were immunoprecipitated with either anti-Src rabbit antiserum or pre-immune rabbit serum. ASAP1 coprecipitation was detected by Western blotting with 9E10 anti-Myc tag monoclonal antibody (Fig. 6A). ASAP1 coprecipitated with an activated Src variant containing a phenylalanine substitution at tyrosine 527 (SrcF527; lane 4). Weaker association was also detected with wild-type Src (lane 3) but not in the absence of Src (lane 2). Similar results could be obtained by using another Src antibody or when ASAP1b was detected with anti-ASAP1 antiserum in the Western blots (data not shown). Increased binding to activated mutant Src is consistent with binding to the Src SH3 domain, since the Src SH3 domain is more available for intermolecular interactions in the activated mutant (12).
FIG. 6.
ASAP1 associates with Src in 293 cells and becomes tyrosine phosphorylated both in Src-expressing cells and by Src in vitro. Lysates were prepared from 293 cells cotransfected with CS3+MT−ASAP1b and either LXSH, LXSH-c-Src, or LXSH-c-SrcF527. (A) Coimmunoprecipitation of ASAP1 with Src. Lysates were immunoprecipitated with 3060 anti-Src rabbit antiserum or preimmune rabbit serum. Twin blots were probed with either 9E10 anti-Myc monoclonal antibody (top panel) or 327 anti-Src monoclonal antibody (bottom panel). (B) Tyrosine phosphorylation of ASAP1 in vivo. 293 lysates were immunoprecipitated with either 9E10 or a mock antibody mix lacking 9E10. Identical blots were probed with anti-phosphotyrosine 4G10 (top panel) or 9E10 (bottom panel) monoclonal antibodies. (C) In vitro phosphorylation of native ASAP1 by Src. Recombinant c-Src (1 U) was incubated with 40 ng of purified ASAP1 from bovine brain and 5 μM [γ-32P]ATP.
Proteins that associate with the regulatory domains of Src family kinases frequently serve as their substrates (12). To test whether ASAP1 is a Src substrate, tyrosine phosphorylation of transfected Myc-ASAP1b was assayed in 293 cells that were also transfected with Src. ASAP1 was immunoprecipitated from cell lysates with anti-Myc tag 9E10, and phosphotyrosine was detected by Western blotting. ASAP1b contained phosphotyrosine when coexpressed with active SrcF527 but not when coexpressed with Src (Fig. 6B, lanes 2 and 3). Tyrosine-phosphorylated ASAP1b was also detected among proteins from SrcF527-expressing 293 cells even without immunoprecipitation and was one of the major phosphotyrosine-containing proteins (data not shown). The appearance of tyrosine-phosphorylated ASAP1b in response to active Src suggests that ASAP1 is a substrate for either Src or for another tyrosine kinase that is activated by Src. To further test ASAP1 as a direct substrate for Src, native ASAP1 (Fig. 6C) or PZA (not shown) was incubated with radioactive ATP and purified Src in vitro. Both forms of ASAP1 were phosphorylated by Src, suggesting that the tyrosine phosphorylation of ASAP1 in cells expressing activated Src is direct.
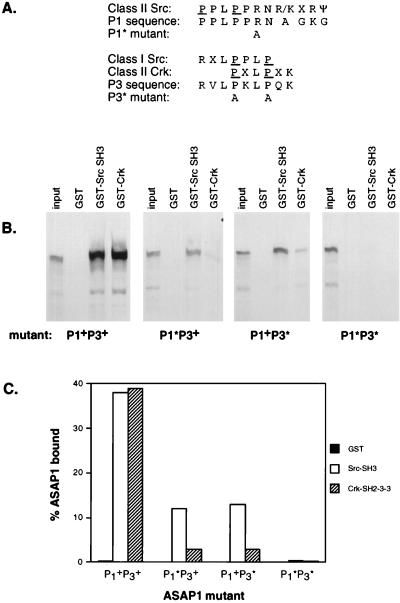
Identification of proline-containing ASAP1 sequences required for binding Src and Crk.
ASAP1a contains several proline-rich sequences that match the minimal SH3 domain binding motif consensus PXXP, including the three sites labeled in Fig. 2A as P1, P2, and P3. P1 (805PPLPPRNAGKG) closely resembles the consensus site for binding Src SH3 in the class II orientation, PPLPPRNR/KXRΨ (where Ψ is a hydrophobic residue, X is any residue, and underlining indicates essential proline and basic residues) (83). P3 (907RVLPKLPQK) is similar to both the optimal class I Src SH3 binding site RXLPPLP (97) and the consensus for binding the N-terminal SH3 domain of Crk in the class II orientation, PXLPXK (49, 50). The P2 (839KKRPPPPPPGHKR) sequence contains six sequential prolines in the context of both N- and C-terminal basic residues but is not a close match to the optimal binding site for any SH3 domains that have been tested. P1 and P2 lie within the SID of ASAP1a, but P2 is spliced out of the mRNA for ASAP1b. P3 lies C-terminal to the SID.
To determine which proline-rich sequences in ASAP1 were involved in Src binding, we made mutations in the P1, P2, and P3 regions of ASAP1. The resulting mutants were tested for binding to full-length Crk and the Src SH3 domain in vitro (Fig. 7). In the context of full-length ASAP1b, which lacks P2, an R811A mutation in P1 (P1*) significantly decreased binding to the Src SH3 and to Crk in vitro, as did a P910A, P913A double mutation in P3 (P3*) (Fig. 7A and B). A P1*P3* double mutant completely abolished binding of ASAP1b to the Src SH3 and to Crk (Fig. 7B). Src binding to either P1* or P3* was decreased approximately threefold, whereas Crk binding was decreased 13-fold (Fig. 7C). The greater decrease in Crk binding after mutation of either P1 or P3 suggests that these proline-rich regions may cooperate to bind Crk, possibly one engaging the first Crk SH3 domain and the other engaging the second Crk SH3 domain. In the ASAP1a SID context, the P1* mutation also eliminated binding to Src SH3 and Crk, whereas a P2 P842A, P843A, P844A triple mutant bound Src family SH3 domains as well as wild-type SID (data not shown). This suggests that the P2 region, which is found only in ASAP1a, is not involved in Src binding.
FIG. 7.
ASAP1 proline-rich sequences are required for binding to Src and Crk in vitro. (A) P1 and P3 amino acid sequences, alignment with SH3 consensus binding motifs, and residues changed by site-directed mutagenesis. Underlined residues indicate conserved prolines in the canonical SH3 binding motif PXXP. (B) In vitro binding of ASAP1b to GST-Src SH3 and GST-Crk. Binding reactions were as described in Fig. 5 and Materials and Methods. The first lane of each of the four gels contains 1/30 the amount of [35S]methionine-labeled ASAP1b added to each binding reaction. (C) Quantitation of ASAP1 mutant binding efficiencies. The fraction of ASAP1 bound by immobilized GST fusion protein was quantitated by PhosphorImager analysis.
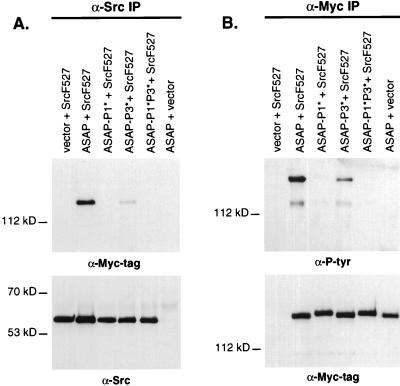
The binding and phosphorylation of the P1* and P3* ASAP1b mutants were also assayed in ASAP1 and SrcF527-cotransfected 293 cells (Fig. 8). Binding of ASAP1 to SrcF527 was completely eliminated by the P1* mutation (Fig. 8A, lane 3), while binding of the P3* mutant was reduced but still detectable (Fig. 8A, lane 4). The P1*P3* double mutant did not bind to SrcF527 in vivo (Fig. 8A, lane 5). ASAP1b tyrosine phosphorylation was completely inhibited by the P1* mutation (Fig. 8B, lane 3) and reduced by the P3* mutation (lane 4). In contrast to the approximately equal binding of the P1* and P3* mutants to Src in vitro, the P1 sequence may play a greater role in binding to Src in vivo. The low level of tyrosine phosphorylation of the P3* mutant may allow binding to Src via the Src SH2 domain, contributing to the greater binding of this mutant than the P1* mutant to full-length SrcF527 in vivo. The P1*P3* double mutant was not tyrosine phosphorylated. The extent of phosphotyrosine incorporation by ASAP1 mutants parallels their ability to associate with Src in cells. The tyrosine phosphorylation of ASAP1, therefore, is dependent on its ability to associate with Src via the Src SH3 domain.
FIG. 8.
ASAP1 mutants show impaired Src association and tyrosine phosphorylation in mammalian cells. 293 cells were cotransfected with LXSH or LXSH-c-Src F527 and either CS3+MT, CS3+MT−ASAP1b, CS3+MT−ASAP1b-P1*, CS3+MT−ASAP1b−P3*, or CS3+MT−ASAP1b−P1*P3*. (A) Cell lysates were immunoprecipitated (IP) with 3060 anti-Src antiserum. Identical Western blots were probed with either 9E10 anti-Myc tag (top panel) or 327 anti-Src (bottom panel). (B) Lysates were immunoprecipitated with 9E10. 4G10 anti-phosphotyrosine (top panel) or 9E10 (bottom panel) was used to probe twin Western blots.
Subcellular localization of ASAP1.
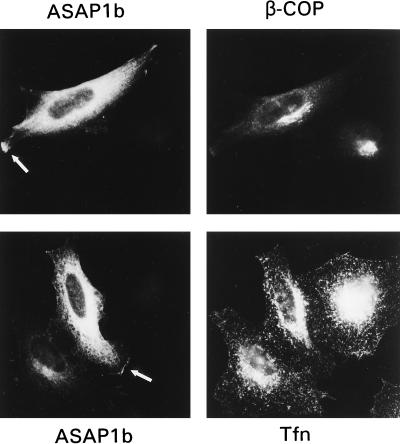
Epitope-tagged ASAP1b expressed in COS7, HeLa, and NIH 3T3 cells and epitope-tagged ASAP1a expressed in HeLa cells were localized by immunofluorescence by using either anti-ASAP1 or anti-epitope tag antibodies. The distribution was similar for both proteins and in all three cell types. HeLa cells expressing tagged ASAP1b are shown in Fig. 9. ASAP1 was detected in the cytoplasm in a perinuclear, reticulate network and at the cell edge, likely associated with the plasma membrane. ASAP1 neither colocalized with nor affected the distribution of a marker of the Golgi or β-COP or that of an endosome marker, transferrin (Fig. 9). Preliminary cell fractionation studies suggest that >95% of ASAP1 is not associated with membranes, since it is mostly in a high-speed supernatant of cells lysed in the absence of detergent (not shown). Purification of ASAP1 from a high-speed supernatant of brain also indicates a lack of membrane association. Thus, most overexpressed ASAP1 is cytosolic with a subpopulation associated with membranes.
FIG. 9.
Localization of epitope-tagged ASAP1b in tissue culture cells. ASAP1b was detected in HeLa cells transfected with a FLAG-tagged ASAP1b expression vector by using either a rabbit polyclonal antibody 551 raised to ASAP1 (upper panels) or a monoclonal antibody against the FLAG tag (lower left panel). Arrows indicate ASAP1 that was detected at the cell edge, likely associated with the plasma membrane.
DISCUSSION
ASAP1 was purified and cloned on the basis of being an Arf GAP that was PIP2 dependent and, therefore, a potential link between the phosphoinositide signalling pathway and membrane traffic. ASAP1 was independently identified by screening for proteins that interact with another signalling molecule, the cytoplasmic tryosine kinase Src that is involved in the regulation of cell growth and cytoskeletal organization. ASAP1 contains a GCS1-type zinc finger, the only region of homology with ArfGAP1. In addition, ASAP1 contains a type II SH3 binding site that mediates interaction with Src and a number of other conserved domains that frequently occur in components of established signal transduction pathways. These include a PH domain, an SH3 domain, ANK repeats, and several proline-rich SH3-binding motifs (9, 53, 70). Based on our data, it is possible that ASAP1 activity or localization is regulated by Src. Furthermore, through interaction with Src as well as phosphoinositides, ASAP1 could coordinate membrane traffic with other cellular responses mediated by these signalling molecules.
Many studies have demonstrated Src’s involvement in regulating cytoskeletal architecture and cell adhesion. Evidence for a role for Src in regulating membrane traffic is accumulating as well. Src has been localized to several membrane compartments in the secretory and endocytic pathways, including endocytic, exocytic, and synaptic vesicles (5, 20, 34, 46, 54, 69, 81). The activities of Src and its close relative Fyn are modulated during secretagogue-stimulated exocytosis of chromaffin cells (2, 64), and tyrosine kinase activity is required for exocytosis (18). Src stimulates epidermal growth factor receptor internalization (95). Src associates with or phosphorylates various proteins implicated in vesicle transport, including synapsin I, dynamin, synaptophysin, synaptogyrin, and cellugyrin (5, 31, 43, 65). Like ASAP1, synapsin I and dynamin bind Src through the Src SH3 domain, but neither protein is phosphorylated on tyrosine (31, 65). The clathrin adapter protein α-adaptin is also found in Src-dynamin complexes (31). Synaptophysin, synaptogyrin, and cellugyrin all appear to be Src substrates (5, 43). Src may also influence membrane traffic by interacting with or activating enzymes involved in phospholipid metabolism such as phosphatidylinositol 3-kinase and phospholipase D (12, 44). We are currently examining stimuli that activate Src to identify upstream signals that result in ASAP1 phosphorylation.
ASAP1 is broadly expressed and may regulate Arf activity in a variety of cell types. The two cDNA clones isolated, ASAP1a and ASAP1b, are indicative of alternative splicing of the mRNA. The exon deleted in ASAP1b encodes part of the proline-rich Src interaction domain and includes several PXXP motifs that might serve as SH3 binding sites, e.g., 817PTGPPSTLP, 841RPPPPPPGHK (P2), and 856PPSPLPHGPP. However, this exon is not required for binding Src. Both an ASAP1a fragment lacking the 856PPSPLPHGPP sequence and an ASAP1a construct with proline mutations in 841RPPPPPPGHK were able to bind Src family SH3 domains in vitro. Although not important for Src SH3 binding, the ASAP1a-specific exon may have another role.
In addition to interacting with Src in vivo and in vitro, ASAP1 also interacted in vitro with Crk, an adapter protein consisting of one SH2 and two SH3 domains (82). Mutated or activated forms of Src and Crk cause changes in cytoskeletal architecture that can result in cellular transformation. In addition, Src and Crk associate with some of the same partners, including proteins of cell adhesion signalling pathways such as the focal adhesion proteins p130CAS and paxillin (reviewed in reference 36). Since at least two proline-rich sites in ASAP1 contributed to binding Crk and Src SH3 domains, it is possible that both proteins could bind ASAP1 simultaneously or compete for the same sites. The role that Src and Crk interactions with ASAP1 play in ASAP1 localization and activity is currently being investigated.
Arfs are known to function at a number of intracellular sites, including the plasma membrane (25, 26, 28, 66, 71, 73, 74). Because the first mammalian Arf GAP cloned, ArfGAP1, is confined to the Golgi apparatus (4, 19), the regulation of Arfs at other sites would require unique GAPs. Consistent with this, several distinct Arf GAP activities have been purified (19, 21, 57, 77). Of these, ASAP1 is the second mammalian GAP to be cloned and shown to be a gene product distinct from ArfGAP1. ArfGAP1 and ASAP1 are divergent proteins with homology limited to 38% identity over the 86 amino acids that include the Arf GAP domain–GCS-like zinc finger domain (Fig. 2B). Despite these structural differences, the proteins have similar Arf specificities (77); therefore, rather than being GAPs for different Arfs, the structural divergence likely provides differential localization and regulation. The different lipid requirements of the native proteins, as previously reported (77), are consistent with independent regulation, and the immunofluorescence reported here supports the idea of differential localization. In contrast to ArfGAP1, ASAP1 did not colocalize with markers of the Golgi apparatus, and overexpression of ASAP1 had no detectable effect on Golgi morphology (Fig. 9). Instead, ASAP1 was found mostly in the cytosol, with a smaller population at the plasma membrane (Fig. 9). This localization pattern is consistent with ASAP1 being both a target for Src and regulated by phosphoinositides. ASAP1 could also be an effector for Arf. Because ASAP1 must bind to Arf-GTP, it could transmit a signal from Arf-GTP.
Based on the specificity of ASAP1 in vitro, ASAP1 is expected to use class I and class II Arfs as substrates. The localizations of Arf1, a class I Arf, and Arf6, a class III Arf, have been determined (13, 22, 71, 73). Arf1 is considered to be Golgi associated, whereas Arf6 is at the plasma membrane; however, a number of studies support a role outside of the Golgi for class I and class II Arfs, including Arf1. In vitro studies have shown that class I Arfs affect diverse processes including intra-Golgi transport, endoplasmic reticulum-to-Golgi transport, endosome-to-endosome fusion, and synaptic-vesicle maturation (23, 28, 61). In cell fractionation studies, all of the class I and II Arfs have been found associated with endocytic vesicles (96). In vivo, class I and class II Arfs have been implicated in a number of specialized endocytic events, including synaptic vesicle maturation (6, 28, 41, 66). We are now testing the endocytic compartment as a possible target site of ASAP1 action. We are also attempting to identify the Arf family member(s) that is the in vivo substrate for ASAP1, which could be restricted by subcellular localization, specific cofactors, or conditions not reproduced in our in vitro assay.
The PH domain of ASAP1 likely contributes to the phosphoinositide-dependent regulation of Arf. Regulation of a number of proteins by phosphoinositide binding to their PH domains has been demonstrated (53). PIP2 stimulates the activity of ASAP1 and of a recombinant fragment of ASAP1 containing the PH, zinc finger, and ANK repeat domains (PZA). Our preliminary studies suggest that PIP2 binds to the PZA fragment but not to a protein containing only the zinc finger and ankyrin repeats (45), and this latter protein has no detectable activity. Therefore, the PH domain of ASAP1 may allow phosphoinositide-dependent activation of the Arf GAP domain. However, PIP2 binding to the substrate, Arf, is also important for the GAP reaction (76). Thus, phosphoinositide likely binds to both the enzyme (PZA) and the substrate (Arf), similar to the dual role of phosphoinositides in regulating the phosphorylation of protein kinase B (88).
In the cell, phosphoinositides have complex effects on Arf. Arf has been found to activate both PIP-kinase (29, 58) and phospholipase D (10, 16); therefore, the comodulation of GAP activity by PA and PIP2 could be involved in a system of feedforward and feedback loops that control the time that Arf spends in the GTP state (56, 76, 77). In addition, phosphoinositides contribute to Arf activation. Three Arf exchange factors, ARNO, GRP-1, and cytohesin, contain PH domains and function at the plasma membrane (14, 32, 47, 48, 60). ARNO has a PIP2-dependent exchange activity on myristoylated Arf but is able to act on nonmyristoylated Arf independently of PIP2, suggesting a role for the PH domain and PIP2 in recruiting the enzyme (ARNO) and substrate (Arf) into the same complex (67). The PH domains of these molecules actually appear to favor binding to PIP3 over PIP2 (47, 48, 93). In contrast, PIP3 had no effect on ASAP1 activity in preliminary studies (45), raising the possibility that regulating PIP2 and PIP3 levels could order the inactivation and activation of Arf. Consistent with this idea, insulin-induced PI 3-kinase stimulation causes a PH domain-dependent translocation of ARNO to the plasma membrane (93). The PH domain of cytohesin, an exchange factor that binds β2 integrin, is also required for membrane recruitment and is essential for PI 3-kinase activation of β2 integrin adhesion (51, 62).
In addition to its conserved domains, ASAP1 has an unusual sequence feature, a series of repeats of E/DLPPKP, many of which are separated by the tripeptide QLG. The sequence is repeated eight times, with an additional five degenerate repeats. A database search did not identify other occurrences of tandem repeats of this sequence. Although the repeat region has high proline content and contains seven PXXP sequences, it did not bind any of 10 different SH3 domains tested (11). One possible role of this domain is to mediate homodimerization of ASAP1 that we have detected in vitro and in vivo (45).
ASAP1 has a number of relatives, the closest being the human KIAA0400 protein. The strong sequence similarity and conservation of domain order between mouse ASAP1 and human KIAA0400 suggest they could be species homologs; however, we have three reasons in particular why we think they are different family members. First, using primers complementary to ASAP1 mouse sequence, partial cDNAs were amplified from human and bovine cDNA library by PCR. The predicted amino acid sequences of the human and bovine clones were 99 and 94% identical to ASAP1, respectively (78). The same region from KIAA0400 is 67% identical to human ASAP1. Second, KIAA0400 lacks the E/DLPPKP repeat region contained in ASAP1. Third, ASAP1 and KIAA0400 sequences diverge in the proline-rich region except for the class II Src SH3 binding sequence. Thus, ASAP1 and KIAA0400 define a new protein family. Both may bind Src, but they could be differentially localized or regulated by interaction with other SH3-containing proteins. There are three additional proteins in GenBank with 40% or greater overall identity to ASAP1 (Fig. 2B). These proteins, ASAP1, and KIAA0400 all contain a conserved region consisting of, in order from the amino terminus, consecutive PH, zinc finger, and ANK repeat domains. The ordered PZA region, therefore, defines a PZA superfamily of proteins, of which ASAP1 and KIAA0400 constitute a subgroup. Given the diverse sites of Arf action and the number of Arf family members, ASAP1 and other PZA family members could provide site-specific or Arf-specific regulation of membrane traffic.
ACKNOWLEDGMENTS
We thank Douglas Lowy, Dan Cassel, James Battey, and Patrick Donohue for discussions and advice; W. S. Lane, R. Robinson, J. Neveu, and D. Arnelle of the Harvard Microchemistry Facility for expertise in HPLC, mass spectrometry, and peptide sequencing; Jenny Clark for expertise in nucleotide sequencing; S. Stauffer and J. Kam for technical assistance; S. Hollenberg, B. Mayer, F. Gertler, B. Howell, C. Sachsenmaier, S. M. Thomas, P. Cicchetti, E. M. Eddy, R. Braun, J. Wang, S. Feller, G. Myles, and R. Bourette for libraries, strains, and plasmids; Douglas Lowy for expertise in etymology; and Dan Cassel for initiating contact between our groups.
This work was supported by grant CA41072 from the U.S. Public Health Service (J.A.C.) and the Division of Basic Sciences, National Cancer Institute (P.A.R.). M.T.B. was supported by a postdoctoral fellowship from the National Institutes of Health (CA62598).
REFERENCES
- 1.Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel p130Cas-related protein, Sin. Genes Dev. 1996;10:1341–1355. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- 2.Allen C M, Ely C M, Juaneza M A, Parsons S J. Activation of Fyn tyrosine kinase upon secretogogue stimulation of bovine chromaffin cells. J Neurosci Res. 1996;44:421–429. doi: 10.1002/(SICI)1097-4547(19960601)44:5<421::AID-JNR2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 3.Antonny B, Huber I, Paris S, Chabre M, Cassel D. Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J Biol Chem. 1997;272:30848–30851. doi: 10.1074/jbc.272.49.30848. [DOI] [PubMed] [Google Scholar]
- 4.Aoe T, Cukierman E, Lee A, Cassel D, Peters P J, Hsu V W. The KDEL receptor, ERD2, regulates intracellular traffic by recruiting a GTPase-activating protein for ARF1. EMBO J. 1997;16:7305–7316. doi: 10.1093/emboj/16.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnekow A, Jahn R, Schartl M. Synaptophysin: a substrate for the protein tyrosine kinase pp60c-src in intact synaptic vesicles. Oncogene. 1990;5:1019–1024. [PubMed] [Google Scholar]
- 6.Barroso M, Sztul E S. Basolateral to apical transcytosis in polarized cells is indirect and involves BFA and trimeric G protein sensitive passage through the apical endosome. J Cell Biol. 1994;124:83–100. doi: 10.1083/jcb.124.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel P, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 8.Boman A L, Kahn R A. Arf proteins: the membrane traffic police? Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- 9.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 10.Brown H A, Gutowski S, Moomaw C R, Slaughter C, Sternweis P C. ADP-ribosylation factor, a small GTP-dependent regulatory protein, activates phospholipase D. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 11.Brown, M. T. Unpublished results.
- 12.Brown M T, Cooper J A. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 13.Cavenagh M M, Whitney J A, Carroll K, Zhang C, Bowman A L, Rosenwald A G, Mellman I, Kahn R A. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- 14.Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson C L, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 15.Cicchetti P, Mayer B J, Thiel G, Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- 16.Cockcroft S, Thomas G M H, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty N F, Truong O, Hsuan J J. Phospholipase D: a downstream effector of Arf in granulocytes. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- 17.Cooper J A, Gould K L, Cartwright C A, Hunter T. Tyr527 is phosphorylated in pp60c-Src: implications for regulation. Science. 1986;231:1431–1434. doi: 10.1126/science.2420005. [DOI] [PubMed] [Google Scholar]
- 18.Cox M E, Ely C M, Catling A D, Weber M J, Parsons S J. Tyrosine kinases are required for catecholamine secretion and mitogen-activated protein kinase activation in bovine adrenal chromaffin cells. J Neurochem. 1996;66:1103–1112. doi: 10.1046/j.1471-4159.1996.66031103.x. [DOI] [PubMed] [Google Scholar]
- 19.Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 20.David-Pfeuty T, Nouvian-Dooghe Y. Immunolocalization of the cellular src protein in interphase and mitotic NIH c-src overexpresser cells. J Cell Biol. 1990;111:3097–3116. doi: 10.1083/jcb.111.6.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding M, Vitale N, Tsai S C, Adamik R, Moss J, Vaughan M. Characterization of a GTPase-activating protein that stimulates GTP hydrolysis by both ADP-ribosylation factor (ARF) and ARF-like proteins. Comparison to the ARD1 gap domain. J Biol Chem. 1996;271:24005–24009. doi: 10.1074/jbc.271.39.24005. [DOI] [PubMed] [Google Scholar]
- 22.Donaldson J G, Kahn R A, Lippincott-Schwartz J, Klausner R D. Binding of ARF and beta-COP to Golgi membranes: possible regulation by a trimeric G protein. Science. 1991;254:1197–1199. doi: 10.1126/science.1957170. [DOI] [PubMed] [Google Scholar]
- 23.Donaldson J G, Klausner R D. ARF: a key regulatory switch in membrane traffic and organelle structure. Curr Opin Cell Biol. 1994;6:527–532. doi: 10.1016/0955-0674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 24.Druker B J, Mamon H J, Roberts T M. Oncogenes, growth factors, and signal transduction. N Engl J Med. 1989;321:1383–1391. doi: 10.1056/NEJM198911163212007. [DOI] [PubMed] [Google Scholar]
- 25.D’Souza-Schorey C, Li G, Colombo M I, Stahl P D. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 26.D’Souza-Schorey C, van Donselaar E, Hsu V W, Yang C, Stahl P D, Peters P J. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faundez V, Horng J T, Kelly R B. ADP ribosylation factor 1 is required for synaptic vesicle budding in PC12 cells. J Cell Biol. 1997;138:505–515. doi: 10.1083/jcb.138.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fensome A, Cunningham I, Prosser S, Tan S K, Swigart P, Thomas G, Hsuan J, Cockcroft S. ARF and PITP restore GTP-gamma-S-stimulated protein secretion from cytosol-depleted HL60 cells by promoting PIP2 synthesis. Curr Biol. 1996;6:730–738. doi: 10.1016/s0960-9822(09)00454-0. [DOI] [PubMed] [Google Scholar]
- 30.Flynn D C, Li T-H, Reynolds A, Parsons J T. Identification and sequence analysis of cDNAs encoding a 110-kilodalton actin filament-associated pp60src substrate. Mol Cell Biol. 1993;13:7892–7900. doi: 10.1128/mcb.13.12.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foster-Barber A, Bishop J M. Src interacts with dynamin and sysnapsin in neuronal cells. Proc Natl Acad Sci USA. 1998;95:4673–4677. doi: 10.1073/pnas.95.8.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank S, Upender S, Hansen S H, Casanova J E. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 33.Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 34.Grandori C, Hanafusa H. p60c-src is complexed with a cellular protein in subcellular compartments involved in exocytosis. J Cell Biol. 1988;107:2125–2135. doi: 10.1083/jcb.107.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammonds-Odie L P, Jackson T R, Profit A A, Blader I J, Turck C W, Prestwich G D, Theibert A B. Identification and cloning of centaurin-α. J Biol Chem. 1996;271:18859–18868. doi: 10.1074/jbc.271.31.18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanks S K, Polte T R. Signaling through focal adhesion kinase. Bioessays. 1997;19:137–145. doi: 10.1002/bies.950190208. [DOI] [PubMed] [Google Scholar]
- 37.Hellman U, Wernstedt C, Gonez J, Heldin C H. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 38.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howell B W, Gertler F B, Cooper J A. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 1997;16:121–131. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber I, Cukierman E, Rotman M, Aoe T, Hsu V W, Cassel D. Requirement for both the amino terminal catalytic domain and a “non-catalytic” domain for in vivo activity of ADP-ribosylation factor GTPase-activating protein. J Biol Chem. 1998;273:24786–24791. doi: 10.1074/jbc.273.38.24786. [DOI] [PubMed] [Google Scholar]
- 41.Hunziker W, Whitney J A, Mellman I. Selective inhibition of transcytosis by brefeldin A in MDCK cells. Cell. 1991;67:617–627. doi: 10.1016/0092-8674(91)90535-7. [DOI] [PubMed] [Google Scholar]
- 42.Ishino M, Ohba T, Sasaki H, Sasaki T. Molecular cloning of a cDNA encoding a phosphoprotein, Efs, which contains a Src homology 3 domain and associates with Fyn. Oncogene. 1995;11:2331–2338. [PubMed] [Google Scholar]
- 43.Janz R, Sudhof T C. Cellugyrin, a novel ubiquitous form of synaptogyrin that is phosphorylated by pp60c-src. J Biol Chem. 1998;273:2851–2857. doi: 10.1074/jbc.273.5.2851. [DOI] [PubMed] [Google Scholar]
- 44.Jiang H, Lu Z, Luo J-Q, Wolfman A, Foster D A. Ras mediates the activation of phospholipase D by v-Src. J Biol Chem. 1995;270:6006–6009. doi: 10.1074/jbc.270.11.6006. [DOI] [PubMed] [Google Scholar]
- 45.Kam, J., R. Aneja, J. Andrade, and P. A. Randazzo. 1998. Unpublished results.
- 46.Kaplan K B, Swedlow J R, Varmus H E, Morgan D O. Association of pp60c-src with endosomal membranes in mammalian fibroblasts. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klarlund J K, Guilherme A, Holik J J, Virbasius J V, Chawla A, Czech M P. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 48.Klarlund J K, Rameh L E, Cantley L C, Buxton J M, Holik J J, Sakelis C, Patki V, Corvera S, Czech M P. Regulation of GRP1-catalyzed ADP ribosylation factor guanine nucleotide exchange by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- 49.Knudsen B S, Feller S M, Hanafusa H. Four proline-rich sequences of the guanine-nucleotide exchange factor C3G bind with unique specificity to the first Src homology 3 domain of Crk. J Biol Chem. 1994;269:32781–32787. [PubMed] [Google Scholar]
- 50.Knudsen B S, Zheng J, Feller S M, Mayer J P, Burrell S K, Coxburn D, Hanafusa H. Affinity and specificity requirements for the first Src homology 3 domain of the Crk proteins. EMBO J. 1995;14:2191–2198. doi: 10.1002/j.1460-2075.1995.tb07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockinger H, Seed B. αLβ2 Integrin/LFA-1 binding to ICAM-1 induced by cytohesin-1, a cytoplasmic regulatory molecule. Cell. 1996;86:233–242. doi: 10.1016/s0092-8674(00)80095-1. [DOI] [PubMed] [Google Scholar]
- 52.Lane W S, Galat A, Harding M W, Schreiber S L. Complete amino acid sequence of the FK506 and rapamycin binding protein, FKBP, isolated from calf thymus. J Protein Chem. 1991;10:151–160. doi: 10.1007/BF01024778. [DOI] [PubMed] [Google Scholar]
- 53.Lemmon M A, Ferguson K M. Pleckstrin homology domains. Curr Top Microbiol Immunol. 1998;228:39–74. doi: 10.1007/978-3-642-80481-6_3. [DOI] [PubMed] [Google Scholar]
- 54.Linstedt A D, Vetter M L, Bishop J M, Kelly R B. Specific association of the proto-oncogene product pp60c-src with an intracellular organelle, the PC12 synaptic vesicle. J Cell Biol. 1992;117:1077–1084. doi: 10.1083/jcb.117.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lipsich L A, Lewis A J, Brugge J S. Isolation of monoclonal antibodies that recognize the transforming proteins of avian sarcoma viruses. J Virol. 1983;48:352–360. doi: 10.1128/jvi.48.2.352-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liscovitch M, Cantley L C. Signal transduction and membrane traffic: the PITP/phosphoinositide connection. Cell. 1995;81:659–662. doi: 10.1016/0092-8674(95)90525-1. [DOI] [PubMed] [Google Scholar]
- 57.Makler V, Cukierman E, Rotman M, Admon A, Cassel D. ADP-ribosylation factor-directed GTPase-activating protein. Purification and partial characterization. J Biol Chem. 1995;270:5232–5237. doi: 10.1074/jbc.270.10.5232. [DOI] [PubMed] [Google Scholar]
- 58.Martin A, Brown F D, Hodgkin M N, Bradwell A J, Cook S J, Hart M, Wakelam M J O. Activation of phospholipase D and phosphatidylinositol 4-phosphate 5-kinase in HL60 membranes is mediated by endogenous Arf but not Rho. J Biol Chem. 1996;271:17397–17903. doi: 10.1074/jbc.271.29.17397. [DOI] [PubMed] [Google Scholar]
- 59.Martinez R, Mathey-Prevot B, Bernards A, Baltimore D. Neuronal pp60c-src contains a six-amino-acid insertion relative to its non-neuronal counterpart. Science. 1987;237:411–415. doi: 10.1126/science.2440106. [DOI] [PubMed] [Google Scholar]
- 60.Meacci E, Tsai S C, Adamik R, Moss J, Vaughan M. Cytohesin-1, a cytosolic guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc Natl Acad Sci USA. 1997;94:1745–1748. doi: 10.1073/pnas.94.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moss J, Vaughan M. Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 62.Nagel W, Zeitlmann L, Schilcher P, Geiger C, Kolanus J, Kolanus W. Phosphoinositide 3-OH kinase activates the β2 integrin adhesion pathway and induces membrane recruitment of cytohesin-1. J Biol Chem. 1998;273:14853–14861. doi: 10.1074/jbc.273.24.14853. [DOI] [PubMed] [Google Scholar]
- 63.Nuoffer C, Balch W E. GTPases: multifunctional molecular switches regulating vesicular traffic. Annu Rev Biochem. 1994;63:949–990. doi: 10.1146/annurev.bi.63.070194.004505. [DOI] [PubMed] [Google Scholar]
- 64.Oddie K M, Litz J S, Balserak J C, Payne D M, Creutz C E, Parsons S J. Modulation of pp60c-src tyrosine kinase activity during secretion in stimulated bovine adrenal chromaffin cells. J Neurosci Res. 1989;66:1103–1112. doi: 10.1002/jnr.490240107. [DOI] [PubMed] [Google Scholar]
- 65.Onofri F, Giovedi S, Vaccaro P, Czernik A J, Valtorta F, De C P, Greengard P, Benfenati F. Synapsin I interacts with c-Src and stimulates its tyrosine kinase activity. Proc Natl Acad Sci USA. 1997;94:12168–12173. doi: 10.1073/pnas.94.22.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ooi C E, Dell’Angelica E C, Bonifacino J S. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paris S, Beraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P. Role of protein-phospholipid interaction in the activation of Arf1 by the guanine nucleotide exchange factor Arno. J Biol Chem. 1997;272:22221–22226. doi: 10.1074/jbc.272.35.22221. [DOI] [PubMed] [Google Scholar]
- 68.Parsons J T, Parsons S J. Src family protein tyrosine kinases: cooperating with growth factor and adhesion signaling pathways. Curr Opin Cell Biol. 1997;9:187–192. doi: 10.1016/s0955-0674(97)80062-2. [DOI] [PubMed] [Google Scholar]
- 69.Parsons S J, Creutz C E. p60c-src activity detected in the chromaffin granule membrane. Biochem Biophys Res Commun. 1986;134:736–742. doi: 10.1016/s0006-291x(86)80482-x. [DOI] [PubMed] [Google Scholar]
- 70.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 71.Peters P J, Hsu V W, Ooi C E, Finazzi D, Teal S B, Oorschot V, Donaldson J G, Klausner R D. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poon P P, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singer R A, Johnston G C. Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA. 1996;93:10074–10077. doi: 10.1073/pnas.93.19.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Radhakrishna H, Donaldson J G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Radhakrishna H, Klausner R D, Donaldson J G. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rameh L, Arvidsson A-K, Carraway III K L, Couvillon A D, Rathbun G, Crompton A, Van Renterghem B, Czech M P, Ravichandran K S, Burakoff S J, Wang D-S, Chen C-S, Cantley L C. A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- 76.Randazzo P A. Functional interaction of ADP-ribosylation factor 1 with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1997;272:7688–7692. [PubMed] [Google Scholar]
- 77.Randazzo P A. Resolution of two ADP-ribosylation factor 1 GTPase-activating proteins from rat liver. Biochem J. 1997;324:413–419. doi: 10.1042/bj3240413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Randazzo, P. A. Unpublished results.
- 79.Randazzo P A, Kahn R A. GTP hydrolysis by ADP-ribosylation factor is dependent on both an ADP-ribosylation factor GTPase-activating protein and acid phospholipids. J Biol Chem. 1994;269:10758–10763. [PubMed] [Google Scholar]
- 80.Randazzo P A, Weiss O, Kahn R A. Preparation of recombinant ADP-ribosylation factor. Methods Enzymol. 1995;257:128–135. doi: 10.1016/s0076-6879(95)57018-7. [DOI] [PubMed] [Google Scholar]
- 81.Redmond T, Brott B K, Jove R, Welsh M J. Localization of the viral and cellular src kinases to perinuclear vesicles in fibroblasts. Cell Growth Differ. 1992;3:567–576. [PubMed] [Google Scholar]
- 82.Reichmann C T, Mayer B J, Keshav S, Hanafusa H. The product of the cellular Crk gene consists primarily of SH2 and SH3 regions. Cell Growth Differ. 1992;3:451–460. [PubMed] [Google Scholar]
- 83.Rickles R J, Botfield M C, Zhou X-M, Henry P A, Brugge J S, Zoller M J. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc Natl Acad Sci USA. 1995;92:10909–10913. doi: 10.1073/pnas.92.24.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rothman J E. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 85.Rothman J E. The protein machinery of vesicle budding and fusion. Protein Sci. 1996;5:185–194. doi: 10.1002/pro.5560050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994;16:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 88.Stokoe D, Stephens L, Copeland T, Gafney P R J, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Dual role of phosphatidylinositol 3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 89.Tanaka K, Imajoh-Ohmi S, Sawada T, Shirai R, Hashimoto Y, Iwasaki S, Kaibuchi K, Kanaho Y, Shirai T, Terada Y, Kimura K, Nagata S, Fukui Y. A target of phosphatidylinositol 3,4,5-trisphosphate with a zinc finger motif similar to that of the ADP-ribosylation-factor GTPase-activating protein and two pleckstrin homology domains. Eur J Biochem. 1997;245:512–519. doi: 10.1111/j.1432-1033.1997.00512.x. [DOI] [PubMed] [Google Scholar]
- 90.Taylor T C, Kahn R A, Melancon P. Two distinct members of the ADP-ribosylation factor family of GTP-binding proteins regulate cell-free intra-Golgi transport. Cell. 1992;70:69–79. doi: 10.1016/0092-8674(92)90534-j. [DOI] [PubMed] [Google Scholar]
- 91.Thomas S M, Brugge J S. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 92.Turner D L, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1995;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 93.Venkateswarlu K, Oatey P B, Tavare J M, Cullen P J. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol. 1998;8:463–466. doi: 10.1016/s0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- 94.Vojtek A B, Hollenberg S, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 95.Ware M F, Tice D A, Parsons S J, Lauffenburger D A. Overexpression of cellular Src in fibroblasts enhances endocytic internalization of epidermal growth factor receptor. J Biol Chem. 1997;272:30185–30190. doi: 10.1074/jbc.272.48.30185. [DOI] [PubMed] [Google Scholar]
- 96.Whitney J A, Gomez M, Sheff D, Kreis T E, Mellman I. Cytoplasmic coat proteins involved in endosome function. Cell. 1995;83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 97.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]