Abstract
The hepatitis B virus (HBV) X protein is essential for viral infectivity, and evidence indicates that it is a strong contributor to HBV-mediated oncogenesis. X has been shown to transactivate a wide variety of RNA polymerase (Pol) II-dependent, as well as RNA Pol III-dependent, promoters. In this study, we have investigated the possibility that X modulates RNA Pol I-dependent rRNA transcription. In both human hepatoma Huh7 and Drosophila Schneider S2 cell lines, X expression stimulated rRNA promoter activity. Extracts prepared from X-expressing cells stably transfected with an X gene also exhibited an increased ability to transcribe the rRNA promoter. The mechanism for X transactivation was examined by determining whether this regulatory event was dependent on Ras activation and increased TATA-binding protein (TBP) levels. Our previous studies have demonstrated that X, and the activation of Ras, produces an increase in the cellular levels of TBP (H.-D. Wang, A. Trivedi, and D. L. Johnson, Mol. Cell. Biol. 17:6838–6846, 1997). Expression of a dominant negative form of Ras blocked the X-mediated induction of the rRNA promoters, whereas expression of a constitutively activated form of Ras mimicked the enhancing effect of X on rRNA promoter activity. When TBP was overexpressed in either Huh7 or S2 cells, a dose-dependent increase in rRNA promoter activity was observed. To analyze whether the increase in TBP was modulating rRNA promoter activity indirectly, by increasing activity of RNA Pol II-dependent promoters, a Drosophila TBP cDNA was constructed with a mutation that eliminated its ability to stimulate RNA Pol II-dependent promoters. Transient expression of wild-type TBP in S2 cells increased the activities of specific RNA Pol I- and Pol II-dependent promoters. Expression of the mutant TBP protein failed to enhance the activity of the RNA Pol II-dependent promoters, yet the protein completely retained its ability to stimulate the rRNA promoter. Furthermore, the addition of recombinant TBP to S2 extracts stimulated rRNA promoter activity in vitro. Together, these results demonstrate that the HBV X protein up-regulates RNA Pol I-dependent promoters via a Ras-activated pathway in two distinct cell lines. The enhanced promoter activity can, at least in part, be attributed to the X- and Ras-mediated increase in cellular TBP, a limiting transcription component.
The hepatitis B virus (HBV) X protein is a transcriptional activator present in the mammalian hepadnaviruses. X has been shown to be required for viral replication in animal hosts (8, 69), and its expression is correlated with viral replication (14). Evidence indicates that X has an important role in the development of hepatocellular carcinoma in individuals chronically infected with HBV. In certain transgenic mouse strains, X was found to induce tumors (27, 31, 53), whereas in other strains, no tumors were found, yet these mice were significantly more sensitive to the tumorigenic effects of hepatocarcinogens (50). Although the specific biochemical events by which X contributes to viral replication and tumorigenesis have not been defined, X has been widely shown to transactivate a large and diverse number of viral and cellular promoters. X has been shown to activate specific RNA polymerase (Pol) III-dependent promoters (2, 62) and RNA Pol II-dependent promoters which contain recognition sequences for ATF/CREB, AP-1, AP-2, c/EBP, NF-κB, SRF, and a variety of acidic activator proteins (for a review, see reference 66).
In some cases, X may function as a transactivator by directly participating in the transcription process. X has no affinity for DNA, yet it has been shown to bind to various transcription components, such as the RPB5 subunit of RNA Pol (9, 36), the TATA-binding protein (TBP) (44), members of the ATF/CREB family (38, 65), TFIIH (25, 45), and TFIIB (23, 36). Studies by one group suggest that X can act as a coactivator and substitute for the TBP-associated factors in RNA Pol II transcription in vitro (24) and in vivo (22). In contrast to these studies, other studies have suggested that X may function indirectly to regulate gene activity. X has been shown to interact directly with a subunit of the proteasome complex (19, 28) and a DNA repair protein (35). In addition, X has been shown to associate with the tumor suppressor p53 to inactivate its function (18, 63). Since many of these interactions have only been demonstrated in vitro, the biological relevance of these associations remains to be elucidated.
A major function of X that contributes to its transactivation capacity is its ability to activate cellular signaling pathways (3, 5, 13, 30, 37, 42, 61, 62). Activation of the Ras–Raf–mitogen-activated protein kinase signal transduction pathway has been shown to be required for X transactivation of both AP-1 and NF-κB-dependent promoters (3, 5, 13, 42). Consistent with these studies, X has been found to be largely localized to the cytoplasm, and targeting it to the nucleus abolishes X activation of these promoters (17). In addition to its effect on RNA Pol II-dependent promoters, the X-mediated activation of the Ras-Raf signaling pathway is also responsible for inducing RNA Pol III-dependent promoter activity (61). Thus, the ability of X to activate Ras plays an important role in the X-mediated induction of a variety of cellular promoters. In addition to its effect on transcription, the X-mediated activation of Ras stimulates cells to proliferate by deregulating cell cycle checkpoint controls (4). Although a direct cytoplasmic target of X that mediates the activation of the cellular signaling events observed has not been identified, X-mediated activation of Ras has been shown to require activation of the Src family of nonreceptor tyrosine kinases (32).
Our previous studies have shown that X-expressing cells produce an increase in the cellular levels of TBP (62) via Ras activation (61). TBP is a factor essential for the transcription of all cellular promoters, and it is associated with other proteins to form at least three distinct TBP–TBP-associated factor complexes, SL1, TFIID, and TFIIIB, which are involved in the transcription of RNA Pol I-, Pol II-, and Pol III-dependent promoters, respectively (26). Evidence indicates that the X- and Ras-mediated increases in cellular TBP could have pronounced effects on cellular gene activity. The activity of TFIIIB is increased in X-expressing cells (62), and overexpression of cellular TBP is sufficient to induce RNA Pol III-dependent promoter activity (52, 62). In the case of RNA Pol II-dependent promoters, TBP is limiting for certain TATA-containing promoters in Drosophila L-2 cells, since they can be stimulated to various extents by overexpression of TBP (11). In contrast, TATA-lacking promoters are either unaffected or repressed by TBP overexpression. In mammalian cells, overexpression of TBP can potentiate the effect of certain activators, such as VP16, while inhibiting others, such as Sp1 or NF-1 (48). Thus, increasing cellular TBP levels appears to differentially regulate the activities of various RNA Pol II-dependent promoters. To date, the effect of increasing cellular TBP on RNA Pol I-dependent promoter activity has not been addressed.
In the study presented here, we have investigated whether RNA Pol I-dependent promoters are responsive to X. RNA Pol I is responsible for the transcription of the three large rRNAs which are synthesized as a single precursor RNA from tandemly repeated units in the nucleolus (for a review, see references 29 and 41). In vertebrates, rRNA transcription requires at least two transcription factors, the upstream binding factor, UBF, and the selectivity factor, SL1. rRNA transcription plays an essential role in ribosome biosynthesis, and transcription by RNA Pol I has been shown to be tightly coordinated with the growth rate of cells. Therefore, increased RNA Pol I-dependent transcription could play an important role in the ability of the HBV X protein to stimulate cell proliferation.
In this study, we demonstrated that X induces RNA Pol I-dependent promoter activity in both human and Drosophila cells. The X-mediated induction is dependent on the activation of the Ras signaling pathway. Since our previous studies have shown that one consequence of X expression and Ras activation is an increase in the cellular levels of TBP (61, 62), we examined the possibility that this increase could also modulate RNA Pol I-dependent promoter activity. We found that rRNA promoter activity is up-regulated with the overexpression of TBP. Furthermore, we showed that the increase in RNA Pol I-dependent promoter activity by TBP overexpression is not due to alterations in RNA Pol II-dependent promoter activity. Together, these results are the first to demonstrate that rRNA promoter activity can be induced by the HBV X protein and oncogenic Ras. Although other changes in the RNA Pol I-dependent transcriptional machinery may occur in this regulatory event, the X- and Ras-mediated increase in cellular TBP, by itself, is sufficient to up-regulate promoter activity.
MATERIALS AND METHODS
Cell culture.
The Drosophila stable cell lines S2, X-S2 (62), and TBP-S2, previously designated F-TBP (52), were constructed from a Schneider S-2 cell line as previously described. These cell lines were propagated in Schneider’s Drosophila medium (Gibco) supplemented with 10% fetal bovine serum (Gemini Bio-Products, Inc.), 500 U of penicillin/ml; 500 μg of streptomycin/ml (Gibco), and 250 μM hygromycin B (Boehringer Mannheim). The human hepatoma Huh7 cell line was maintained in F-12 medium (Gibco) with 10% fetal bovine serum 500 U of penicillin/ml, and 500 μg of streptomycin/ml (Gibco).
Plasmid DNAs.
Plasmids pR119B, pT7B, and pDmr19 were kindly provided by Maria Pellegrini. pDmr19 contains a Drosophila ribosomal DNA (rDNA) sequence of −150 to +680 derived from pDmr275c2 and cloned into the EcoRI/BamHI sites of pBR322 (7). pR119B contains the 234-nucleotide (nt) DNA fragment from the T7 bacteriophage B arm which was ligated downstream from the −150 to +680 Drosophila rDNA sequence in pUC. pT7B contains the 234-nt T7B fragment inserted between the BamHI and HindIII sites of the pGEM vector (Promega). To construct human ribosomal reporter plasmid phRR, a 1,650-bp human rDNA sequence of −150 to +1500 from prHu3 (34) was ligated at the 3′ end to the 234-nt T7B DNA fragment and then subcloned into pBluescript SK+ (pSK) vector DNA (Stratagene). To generate pCMV-X, the HBV X gene was subcloned into the pcDNA3 expression vector (Invitrogen) under the control of the cytomegalovirus immediate-early gene promoter.
Drosophila expression plasmids pAct-Ras-val12 and pAct-Ras-ala15 and mammalian expression plasmids pCMV-Ras-val12 and pCMV-Ras-ala15, containing human Ras cDNAs, were constructed as described previously (61). pLTReTBP contains a human TBP cDNA under the control of the Rous sarcoma virus long terminal repeat promoter (68). pAct-TBP contains a Drosophila wild-type TBP cDNA driven by the Drosophila actin 5C distal promoter, and pADH-Luc and pSV40-Luc contain the firefly luciferase gene driven by the alcohol dehydrogenase (ADH) promoter and the simian virus 40 (SV40) minimal promoter, respectively (52). To construct the Drosophila RNA Pol II-defective TBP, pAct-TBPA338V, the pAct-flu-TBP DNA was used with the MORPH Site-Specific Plasmid DNA Mutagenesis Kit (5 Prime → 3 Prime, Inc.) together with primers 5′-CAGGAGATCTACGATGTGTTCGACAAGATATTC-3′ and 5′-TTTTATCCGCATAGTGCTC-3′. The resultant change produced a cDNA which changed the coding sequence from an alanine residue to a valine residue at amino acid position 338 of the Drosophila TBP. The pcopia-lacZ construct contains a β-galactosidase gene under the control of the Drosophila copia promoter (21). pSK vector DNA (Stratagene) was used to maintain a constant amount of DNA in the transient-transfection experiments.
Transient transfections.
Transient-transfection assays were performed by using a calcium phosphate precipitation technique (62). For each transfection assay of Drosophila Schneider S2, X-S2, and TBP-S2 cells, 0.5 × 106 to 1.0 × 106 cells/ml in a total of 5 ml were cotransfected with 4 μg of pR119B and 2 μg of pcopia-lacZ. Other DNAs were used as indicated, and the final DNA concentration was maintained at 20 μg by using pSK. Twenty-four hours after transfection, the cells were placed in fresh medium and induced 4 h later with CuSO4 (where indicated). After an additional 24 h, the cells were harvested. Half of the cells were used for isolation of RNA to analyze the transcription activity of the rRNA reporter plasmid (pR119B), and the other half of the cells were used to prepare protein lysates for determination of β-galactosidase activities as previously described (52).
Transient transfections of Huh7 cells were carried out by using the calcium phosphate precipitation method at a cell density of 0.5 × 105 to 1.0 × 105/ml and by using 20 μg of total DNA per 10 ml of cell culture. For each assay, 4 μg of phRR was used together with other DNAs as indicated. After 24 h, the cells were washed with Dulbecco’s phosphate-buffered saline (Gibco), placed in fresh medium, and collected after an additional 24 h, and RNA was isolated for RNase protection assays.
Runoff in vitro transcription assays and RNase protection assays.
The runoff in vitro transcription assays were carried out by using nuclear extracts derived from S2 and X-S2 cells prepared by the method described by Chao and Pellegrini (7). For each reaction, the indicated amount of BamHI-linearized pDmr19 was used as the template, together with 17 μg of nuclear extract. The runoff transcription assays were preformed as described by Chao and Pellegrini (7). The resultant labeled RNA transcripts were resolved by using 4% acrylamide–8 M urea gel electrophoresis. The gel was exposed to X-ray film at −80°C, and the resultant autoradiographs were quantitated by using a Bioimage Scanner.
For determination of the amount of ribosomal reporter transcript generated in the transient-transfection assays, RNA was extracted by using TRIzol (Gibco) and following the protocol provided by the vendor. RNase protection assays were carried out by using the RPA II kit (Ambion). The isolated RNAs (0.3 μg of Drosophila RNA and 2 μg of Huh7 RNA) were hybridized with an excess of 32P-labeled antisense transcript at 45°C overnight. The antisense transcript was generated from pT7B by using a Maxiscript kit (Ambion). pT7B was linearized with NdeI and used as a template to make the antisense T7B riboprobe. The DNA was transcribed with SP6 RNA polymerase in the presence of [32P]CTP (specific activity, >600 Ci/mmol; ICN). The resultant riboprobe was treated with DNase I and ethanol precipitated. For each reaction, 0.5 × 106 to 1 × 106 cpm was used. The hybridized RNA was digested with 200 μl of a 1:1,000 dilution of highly concentrated RNase T1 (1,000 U/μl) at 37°C for 30 min. The reaction was terminated by adding 300 μl of stop buffer and 200 μl of ethanol, and the RNA products were precipitated and resuspended in 8 μl of RNA loading dye and electrophoresed on 5% polyacrylamide–8 M urea gels. The gels were exposed to X-ray film at −80°C, and the resultant autoradiographs were quantitated by using a Bioimage Scanner.
Preparation of nuclear extracts.
For the preparation of nuclear extracts for runoff in vitro transcription assays, S2 and X-S2 cells were collected and washed once with cold phosphate-buffered saline. The cell pellets were used to prepare nuclear extracts by the method of Chao and Pellegrini (7). The cell pellet was rinsed with 10 volumes of buffer A (15 mM KCl, 10 mM HEPES [pH 7.9], 2 mM MgCl2, 0.1 mM EDTA). Before Dounce homogenization, the cell pellet was resuspended in 10 ml of buffer A containing 1 mM dithiothreitol (DTT) per liter of cells and incubated on ice for 20 min. After centrifugation of the cell suspension, the nuclear pellet was resuspended in 4.5 ml of buffer A and 0.5 ml of buffer B (1 M KCl, 50 mM HEPES [pH 7.9], 30 mM MgCl2, 0.1 mM EDTA, 1 mM DTT). To the crude nuclear suspension, 4 M ammonium sulfate (pH 7.9) was added to a final concentration of 0.36 M. The viscous suspension was Dounce homogenized and sedimented by centrifugation. The nuclear protein was precipitated by adding 0.35 g of ammonium sulfate/ml of supernatant. The protein pellet was dissolved in 1 ml of buffer C (20% glycerol, 100 mM KCl, 20 mM HEPES [pH 7.9], 0.2 mM EDTA, 0.5 mM DTT)/109 cells and dialyzed against buffer C for 4 h. Protein concentrations of the resultant nuclear extracts were measured by the Bradford method by using a Bio-Rad protein assay reagent.
RESULTS
The HBV X protein induces RNA Pol I-dependent promoter activity.
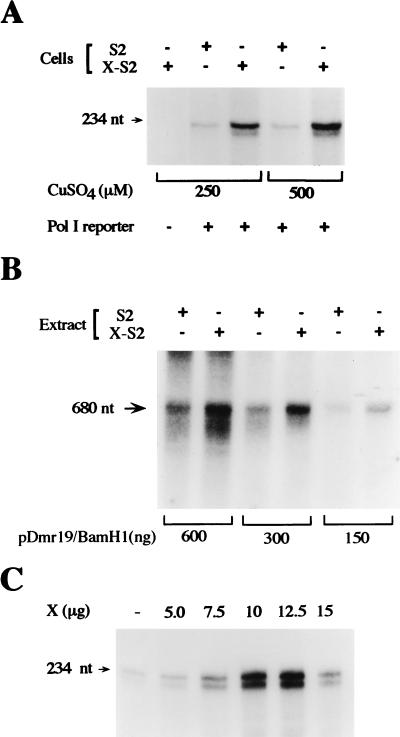
To examine whether the X protein regulates rRNA promoter activity, we first took advantage of two previously constructed stable cell lines, S2 and X-S2, which were derived from a Drosophila S-2 cell line (62). These cell lines were previously used to examine the mechanism of X-mediated induction of RNA Pol III-dependent promoters (61, 62). Both cell lines were stably transfected with a hygromycin B resistance gene, and the X-S2 line additionally contains the X gene under the control of the metallothionein promoter. To examine whether X could also induce RNA Pol I-dependent promoter activity, these cell lines were transiently transfected with a reporter plasmid containing the Drosophila RNA Pol I-dependent promoter previously characterized by Chao and Pellegrini (7). After transfection, the cells were induced to express X by incubation with CuSO4. An RNase protection assay was carried out by using RNAs isolated from the transfected cells to determine the amount of transcript generated from the reporter plasmid. To distinguish the reporter transcripts produced from the endogenous rRNAs produced, a 234-bp sequence from the T7B gene was inserted into the reporter plasmid at position +680 within the 18S rRNA coding sequence. In these assays, we generally observed two labeled transcripts that hybridized to the T7B riboprobe, one of the expected 234 nt and another that was approximately 3 nt shorter. Since the ratio of the two fragments generated varied between different RNase protection assays, and it was not dependent on the RNA preparation, it is likely that the two fragments were generated by differences in the RNase cleavage reaction. As shown in Fig. 1A, these fragments were only produced when the RNA Pol I reporter plasmid was transfected into the cells. A significant increase in rRNA promoter activity was observed in the cells expressing X compared to the control cells. Maximum stimulation was observed when the cells were induced with 500 μM CuSO4. Under these conditions, the X protein caused an approximately fivefold increase in reporter gene transcription (Table 1).
FIG. 1.
HBV X protein induces RNA Pol I-dependent promoter activity. (A) X expression stimulates the transcription of the Drosophila rRNA promoter in S2 cells. Transient transfections of S2 and X-S2 cells were performed as described in Materials and Methods, with or without 4 μg of reporter plasmid pR119B and CuSO4, as shown. RNase protection assays were carried out with equal amounts (0.3 μg) of extracted RNA from the transfected cells and an excess of a 32P-labeled antisense T7B riboprobe. The resultant RNA was separated by gel electrophoresis and visualized by autoradiography. (B) Extracts derived from X-expressing S2 cells exhibit an increased ability to transcribe the Drosophila RNA Pol I-dependent promoter. Runoff in vitro transcription assays were carried out as described in Materials and Methods, by using 17 μg of protein from nuclear extracts derived from either S2 or X-S2 cells and the designated amounts of BamHI-linearized pDmr19 as the template. (C) Human RNA Pol I-dependent promoter activity is induced by X expression. Huh7 cells were cotransfected with increasing amounts of an expression plasmid containing the HBV X gene (pCMV-X), as indicated, together with 4 μg of reporter plasmid phRR, as described in Materials and Methods. RNA was extracted from the transfected cells, RNase protection assays were performed, and the protection labeled RNA products were visualized by gel electrophoresis and autoradiography.
TABLE 1.
Enhancement of RNA Pol I-dependent promoter activity by HBV X protein is dependent on Ras activation
| Transfected gene | Mean fold enhancement ± SDa
|
||
|---|---|---|---|
| S2 | X-S2 | Huh7 | |
| None | 1 | 4.8 ± 0.8b | 1 |
| Ras-ala15 | 1.1 ± 0.1 | 0.9 ± 0.2b | ND |
| Ras-val12 | 2.9 ± 0.7 | 4.7 ± 0.8b | 4.8 ± 1.8c |
| HBV X | ND | ND | 7.9 ± 0.2c |
| HBV X + Ras-ala15 | ND | ND | 1.4 ± 0.1c |
Transient-transfection assays were performed with the indicated DNAs, which were cotransfected with either the Drosophila or human rRNA promoter-reporter plasmid into S2, X-S2, or Huh7 cells as described in Materials and Methods. Where indicated, 2 μg of Ras gene expresion plasmid and 10 μg of the X gene expression plasmid were used in each transfection. For S2 and X-S2 cells, 2 μg of the pcopia-lacZ plasmid was also transfected to normalize for transfection efficiency. For each determination, at least four independent experiments were performed. ND, not determined.
The data shown were derived by comparison with Drosophila rRNA promoter-reporter gene activity in S2 cells that were not transfected with either a Ras or X gene epxressoin plasmid.
The data shown were derived by comparison with the human rRNA promoter-reporter gene activity in Huh7 cells that were not transfected with either a Ras or X gene expression plasmid.
To determine if this result could be observed in vitro, nuclear extracts were derived from S2 and X-S2 cells and transcription runoff assays were performed by using a linearized RNA Pol I-dependent promoter template (Fig. 1B). Specific full-length RNA transcripts of 680 nt were produced that corresponded to the expected size based on the previously mapped transcription start site and the site where the plasmid was linearized (7). To further determine whether the RNA transcripts generated were accurately initiated, transcription templates were linearized with different restriction enzymes and used to generate runoff transcription products. In each case, the sizes of the RNA fragments produced were as predicted (data not shown). Irrespective of the amount of template added to the reaction mixtures, the extracts derived from the X-S2 cells exhibited a 3.4-fold ± 0.2-fold increase in transcriptional capacity compared to the control cell-derived extracts (Fig. 1B). The RNA transcripts generated by the reactions were shown to be RNA Pol I dependent, as high concentrations of α-amanitin did not inhibit RNA synthesis (data not shown). Together, these results demonstrate that X expression in S-2 cells is able to induce transcription from the Drosophila rRNA promoter.
We next analyzed whether the X protein could stimulate human RNA Pol I-dependent promoter activity in human hepatoma cell line Huh7. A plasmid containing the X cDNA under the control of the cytomegalovirus promoter was transiently cotransfected with a human rRNA promoter construct containing the T7B sequence. As shown in Fig. 1C, expression of X resulted in dose-dependent induction of the rRNA promoter. Maximum stimulation was observed when 10 μg of the X expression plasmid was transfected, which produced approximately eightfold induction of RNA Pol I-dependent promoter activity compared to cells that did not express X (Table 1). Together, these results demonstrate that X is capable of increasing RNA Pol I-dependent promoter activity. The observation that this regulatory event occurs in two distinct cell lines further indicates that X activation of rRNA promoters is not cell type specific.
X-mediated induction of RNA Pol I-dependent promoter activity is dependent on activation of the Ras signal transduction pathway.
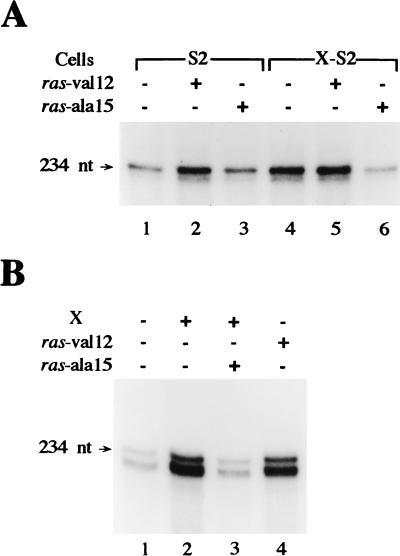
To examine the mechanism for the X-mediated induction of the RNA Pol I-dependent promoters observed, we considered the possibility that this regulatory event was dependent on the activation of cellular signaling pathways. Previous studies have shown that rRNA transcription is up-regulated in both mammalian (1, 51) and Drosophila (54, 64) cells by the phorbol ester tetradecanoyl phorbol acetate (TPA), a potent activator of protein kinase C (PKC). X has been shown to activate both PKC (30) and Ras (3), and activation of these signaling proteins is required for X transactivation of certain RNA Pol II (3, 5, 13, 30, 42)- and RNA Pol III (61, 62)-dependent promoters. Therefore, we examined whether the X-mediated activation of Ras is necessary for induction of the rRNA promoters. The S2 and X-S2 cell lines were transiently transfected with the RNA Pol I-dependent promoter construct together with an expression plasmid containing a cDNA encoding a mutant form of Ras, and the level of transcription resulting from the rRNA promoter was determined. As shown in Fig. 2A, when a dominant negative form of Ras, Ras-ala15, was expressed in X-S2 cells, the X-mediated increase in RNA Pol I-dependent transcription was completely inhibited (compare lanes 4 and 6). Expression of the dominant negative Ras mutant in S2 cells had no effect on rRNA promoter activity (compare lanes 1 and 3). When a constitutively activated form of Ras, Ras-val12, was expressed in S2 cells, rRNA promoter activity was significantly enhanced (compare lanes 1 and 2). Expression of Ras-val12 in X-S2 cells did not further enhance rRNA promoter activity (compare lanes 4 and 5). Similar results were obtained when we examined whether X induction of the human rRNA promoter was dependent on Ras activation in Huh7 cells (Fig. 2B). X-dependent induction of the RNA Pol I-dependent promoter was abolished by inhibition of Ras activation (compare lanes 2 and 3), and activated Ras was able to mimic the ability of X to induce RNA Pol I-dependent promoter activity (compare lanes 1 and 4). Thus, these results demonstrate that rRNA promoters are induced by the activation of the Ras signaling pathway, and the X-mediated enhancement of RNA Pol I-dependent promoter activity is dependent on Ras activation. These results are summarized in Table 1.
FIG. 2.
X-mediated induction of RNA Pol I-dependent promoter activity is dependent on Ras activation. (A) The X-mediated increase in Drosophila rRNA promoter activity requires Ras activation. S2 and X-S2 cells were transiently transfected with 4 μg of pR119B and, where indicated, 2 μg of expression plasmids containing either the Ras-val12 gene or the Ras-ala15 gene. RNase protection assays were carried out with equal amounts (0.3 μg) of extracted RNA and an excess of a 32P-labeled antisense T7B riboprobe as described in Materials and Methods. The resultant RNA was separated by gel electrophoresis and visualized by autoradiography. (B) The X-mediated induction of human rRNA promoter activity is modulated by Ras. Huh7 cells were cotransfected with 4 μg of phRR and, where indicated, 10 μg of pCMV-X and 2 μg of either pCMV-Ras-ala15 or pCMV-Ras-val12. RNase protection assays were performed with RNA extracted from transfected cells, and the labeled RNA was visualized by gel electrophoresis and autoradiography as described in Materials and Methods.
Overexpression of TBP increases RNA Pol I-dependent promoter activity without requiring activation of Ras.
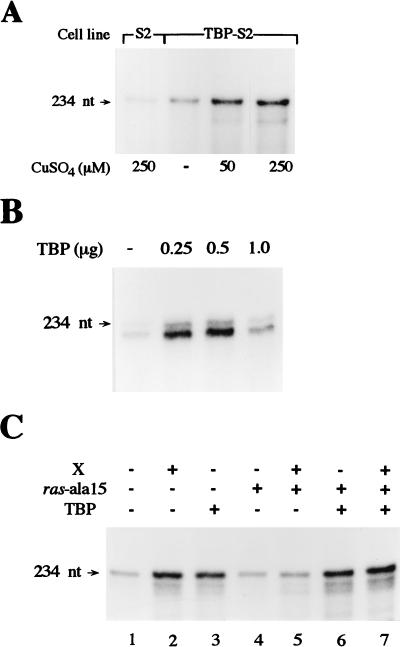
Our previous studies have shown that X expression, and the activation of Ras, produces an increase in TBP levels in both Drosophila and mammalian cell lines (61, 62). Our results described above revealed that Ras activation is necessary for X-mediated induction of RNA Pol I-dependent promoter activity. Since TBP is a subunit of RNA Pol I transcription factor SL1, it is conceivable that the X- and activated-Ras-dependent increase in the cellular concentrations of TBP could affect RNA Pol I-dependent promoter activity. To determine if this increase in TBP was contributing to the rRNA promoter induction observed, we next examined whether directly increasing cellular levels of TBP could augment RNA Pol I-dependent promoter activity. We have previously constructed and analyzed a cell line derived from S-2 cells, TBP-S2, that contains a stably transfected Drosophila TBP cDNA under the control of the metallothionein promoter (52). Overexpression of TBP can be induced from the introduced gene in a copper-dependent manner without any alterations in the steady-state levels of TBP from the expressed endogenous TBP gene (52). When 500 μM CuSO4 was used to induce expression of the stably introduced TBP cDNA, immunoblot analysis revealed an approximately 10-fold increase in the total cellular levels of TBP in the resultant cell lysates compared to the control S2 cell lysates (52, 55). By using the S2 and TBP-S2 cell lines, we examined the effect of TBP overexpression on a transiently transfected rRNA promoter (Fig. 3A). Since there is a low level of TBP expression from the metallothionein promoter even in the absence of CuSO4 (52), the TBP-S2 cells also exhibited an approximately twofold increase in promoter activity without copper induction. When cells were incubated with CuSO4, a significant increase in RNA Pol I-dependent promoter activity was observed compared to the control cells. At 250 μM CuSO4, a 5.8-fold ± 1.4-fold increase was obtained. In addition, extracts derived from the induced TBP-S2 cells exhibited an approximately fivefold increase in RNA Pol I-dependent transcription activity compared to extracts derived from control S2 cells (data not shown). To next examine whether the human rRNA promoter was similarly responsive to the overexpression of TBP, Huh7 cells were transiently cotransfected with the RNA Pol I-dependent reporter together with increasing amounts of an expression plasmid containing a human TBP cDNA driven by a retroviral promoter (Fig. 3B). A dose-dependent increase in rRNA activity was observed in which the maximum induction was obtained at 0.5 μg of TBP plasmid, resulting in a 4.6-fold ± 1.3-fold increase in transcription. Together, these results indicate that increasing cellular TBP, by itself, is sufficient to increase RNA Pol I-dependent promoter activity.
FIG. 3.
Overexpression of TBP increases RNA Pol I-dependent promoter activity. (A) Overexpression of Drosophila TBP in S2 cells augments RNA Pol I-dependent promoter activity. S2 and TBP-S2 cells were transfected with 4 μg of pR119B as described in Materials and Methods and treated with CuSO4 as indicated. RNA was extracted, RNase protection assays were performed with equal amounts of RNA, and the resultant labeled RNA digestion products were separated by gel electrophoresis and visualized by autoradiography. (B) Overexpression of human TBP in Huh7 cells augments RNA Pol I-dependent promoter activity. Huh7 cells were transiently cotransfected with increasing amounts of an expression plasmid containing a human TBP gene (pLTReTBP), as indicated, together with 4 μg of reporter plasmid phRR as described in Materials and Methods. RNase protection assays were performed with equal amounts of RNA from the transfected cells, and the resultant labeled RNA digestion products were separated by gel electrophoresis and visualized by autoradiography. (C) Inhibition of Ras activation does not block induction of RNA Pol I-dependent promoters in cells overexpressing TBP. S2 cells were transiently transfected with expression plasmids containing a ADH promoter-driven X cDNA (10 μg), an actin 5C promoter-driven TBP cDNA (0.5 μg), and an actin 5C promoter-driven dominant negative Ras (Ras-ala15) cDNA (2 μg), as shown, together with 4 μg of pR119B, as described in Materials and Methods. RNase protection assays were performed with equal amounts of RNA from the transfected cells, and the resultant labeled RNA digestion products were separated by gel electrophoresis and visualized by autoradiography.
To further ascertain whether the increase in TBP mediated by the activation of Ras was responsible for the induction of the rRNA promoters observed or whether other Ras-mediated events are required to enhance promoter activity, we examined whether expression of dominant negative Ras would affect the induction of the rRNA promoter in S2 cells overexpressing TBP (Fig. 3C). Consistent with the results obtained by using the X-S2 and TBP-S2 stable cell lines, transient expression of either an X or TBP expression plasmid increased RNA Pol I-dependent promoter activity (compare lanes 1 and 2 and lanes 1 and 3). Expression of Ras-ala15 specifically inhibited the X-mediated stimulation of rRNA transcription (compare lanes 1 and 5), but it was unable to inhibit the TBP-mediated stimulation of rRNA transcription (compare lanes 3 and 6). Furthermore, the ability of Ras-ala15 to block induction of the rRNA promoter by X was abrogated by coexpression of the TBP expression plasmid (compare lanes 5 and 7). These results demonstrate that the increase in TBP generated by the X-mediated activation of Ras is sufficient to induce rRNA promoter activity and that additional events mediated by the activation of the Ras signaling pathway are not required.
Induction of rRNA promoter activity by increased TBP levels is not mediated through alterations in RNA Pol II-dependent promoter activity.
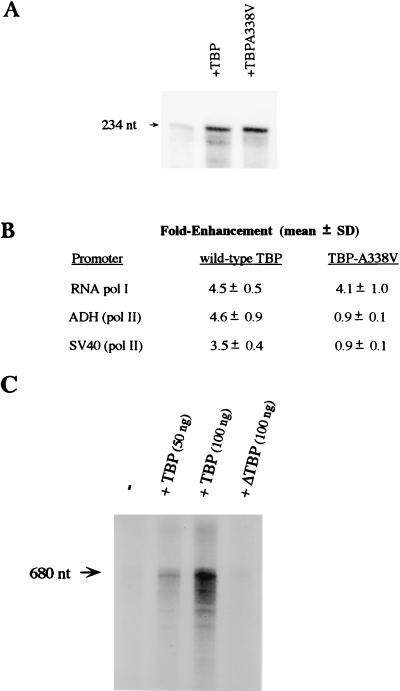
The results described above are consistent with the notion that rRNA promoter activity is limiting for TBP in both the Drosophila and human cell lines. However, increased cellular TBP levels have been previously shown to enhance the activity of certain RNA Pol II promoters in both Drosophila (11) and mammalian (48) cell lines. Therefore, increasing cellular TBP could potentially mediate an increase in RNA Pol I-dependent promoter activity by indirectly augmenting RNA Pol II-dependent transcription, leading to increased production of limiting rRNA transcription factor subunits. Therefore, we addressed the question of whether the observed TBP-mediated increase in rRNA promoter activity was dependent on the ability of TBP to stimulate RNA Pol II-dependent transcription. Previous studies by Cormack and Struhl (12) identified several temperature-sensitive mutations in Saccharomyces cerevisiae TBP that were specifically defective for RNA Pol II-dependent transcription. One of these mutant proteins contained an alanine-to-valine amino acid change at position 226. Based on these results and the strong sequence conservation within the carboxy-terminal domain of TBP, we constructed an analogous mutation in the Drosophila TBP cDNA which resulted in an alanine-to-valine change at amino acid position 338. This mutant TBP cDNA was transiently expressed in S2 cells under the control of the actin 5C promoter and analyzed for the ability to stimulate both RNA Pol I- and Pol II-dependent promoters compared to overexpression of a wild-type TBP cDNA (Fig. 4). As expected, transient overexpression of wild-type TBP augmented rRNA promoter activity. Both the Drosophila ADH promoter and the SV40 promoter were also stimulated by the overexpression of TBP (Fig. 4B). When the mutant TBP, TBP-A338V, was expressed in these cells, however, it failed to stimulate either RNA Pol II-dependent promoter, consistent with the notion that it is defective for RNA Pol II-dependent transcription. Regardless of the amount of TBP-A338V transfected into the S2 cells, no change in RNA Pol II-dependent promoter activity was observed (data not shown). However, expression of TBP-A338V fully retained its ability to stimulate the rRNA promoter. Thus, these results reveal that the TBP-mediated induction of rRNA promoter activity does not require enhanced RNA Pol II-dependent promoter activity.
FIG. 4.
Increased cellular TBP stimulates RNA Pol I-dependent promoter activity in an RNA Pol II-independent manner. (A) The mutant TBP, TBPA338V, retains the capacity to stimulate RNA Pol I promoter activity. Drosophila S-2 cells were transiently transfected with 4 μg of pR119B and, where designated, 0.5 μg of Drosophila wild-type TBP or 0.5 μg of an expression plasmid containing the mutant TBP cDNA, TBPA338V. RNase protection assays were carried out with equal amounts of RNA from the transfected cells, and the resultant RNA was separated by gel electrophoresis and visualized by autoradiography. (B) Analysis of RNA pol I- and II-dependent promoter activity in S-2 cells transiently transfected with wild-type and mutant TBP genes. The data shown were derived by comparison of the promoter activities measured in the absence and in the presence of a cotransfected TBP expression plasmid, as indicated. At least three independent experiments were performed with each promoter. Two micrograms of either pADH-Luc (ADH) or pSV40-Luc (SV40) or 4 μg of pR119B (RNA Pol I) was transfected into S2 cells without or with 0.5 μg of either the pAct-TBP or pAct-TBPA338V expression vector. Resultant luciferase activity was measured as described by Trivedi et al. (52) after preparing lysates from pADH-Luc- or pSV40-Luc-transfected cells. For S2 cells transfected with rRNA promoter-reporter plasmid pR119B, RNase protection assays were carried out as described in Materials and Methods. (C) Addition of recombinant TBP to S2 cell extracts stimulates rRNA transcription in vitro. Transcription runoff assays were performed as described in Materials and Methods by using 25 μg of nuclear extract derived from S2 cells and 300 ng of BamHI-linearized pDmr19 as the template. Where specified, 50 or 100 ng of bacterially produced and purified Drosophila TBP was added to the reaction mixtures. Δ denotes that the recombinant TBP was heat treated at 95°C for 3 min and chilled on ice prior to its addition to the transcription assay.
To further determine whether the increase in TBP was directly participating at the rRNA promoter to augment its activity, we examined whether the addition of bacterium-derived Drosophila TBP to S2 cell extracts could stimulate rRNA promoter activity in vitro (Fig. 4C). The addition of recombinant TBP to the transcription assay enhanced the transcriptional capacity of the S2 cell extracts in a dose-dependent manner, by which approximately 10- to 20-fold enhancement of rRNA transcription was obtained with the addition of 100 ng of recombinant TBP. However, the addition of heat-inactivated TBP was not able to stimulate transcription of the rRNA promoter. Similar results were obtained with four different preparations of nuclear extracts (data not shown). These results reveal that TBP is able to directly enhance transcription of rRNA promoter activity in vitro. Together, these results establish that the TBP-mediated induction of rRNA transcription does not depend on alterations in RNA Pol II-dependent transcription.
DISCUSSION
In this study, we demonstrated that the HBV X protein regulates RNA Pol I-dependent transcription. The regulation of transcription of rRNA by RNA Pol I is a key mechanism that controls the abundance of ribosomes in response to environmental stimuli and the physiological state of the cell (for a review, see references 29 and 41). It has been well established that the cytoplasmic content of ribosomes correlates with the growth rate of eucaryotic cells (59). In human cancer cells, rRNA transcriptional activity and nucleolar size have been shown to be inversely related to cell doubling time (16). The abilities of X to induce tumors in certain transgenic mouse strains (27, 31, 53) and to stimulate DNA synthesis (3, 33) and cell cycle progression (4, 33) are evidence that the X protein contributes to the development of hepatocellular carcinoma in individuals chronically infected with HBV. Thus, it is likely that the X-mediated increase in RNA Pol I-dependent transcription is needed to increase ribosome production to sustain the enhanced rates of cell proliferation during tumorigenesis.
Viral infection has been shown to have a substantial effect on rRNA transcription. Infection of host cells by adenovirus (46), herpes simplex virus (58), or poliovirus (15) causes a shutdown of rRNA synthesis, whereas SV40 and polyomavirus produce an increase in rRNA transcription in infected cells (43). Little is known, however, regarding the RNA Pol I transcription factors that are targeted in these responses and the mechanisms by which transcription is regulated. Poliovirus protease 3C is thought to be responsible for the repression of RNA Pol I-dependent transcription observed (47), although the rRNA transcription factor(s) that is targeted by the protease has not been identified. Interestingly, human TBP has been shown to be a substrate for protease 3C, and cleavage of TBP results in significant inhibition of RNA Pol II-dependent transcription (10). Whether the poliovirus protease 3C-directed cleavage of TBP also mediates its ability to decrease RNA Pol I-dependent transcription remains to be determined. Recently, the mechanism of activation of rRNA transcription by the SV40 large T antigen was examined (67). These studies demonstrated that the large T antigen binds to the SL1 complex through direct interaction with all three TBP-associated factors and that the recruitment of large T antigen to the RNA Pol I-dependent promoter by SL1 is necessary for transcription induction. Thus, the large T antigen directly participates in the RNA Pol I transcription complex to activate transcription. Our studies indicate that the HBV X protein functions indirectly to stimulate RNA Pol I-dependent transcription via activation of the Ras signaling pathway. One consequence of X expression and Ras activation is an increase in the cellular levels of TBP (61, 62). Our studies show that by directly overexpressing TBP, RNA Pol I promoter activity is induced. In addition, by adding recombinant TBP to cell extracts, the RNA Pol I-dependent transcriptional capacity of the extracts is increased. Although we cannot rule out the possibility that there are other changes in the RNA Pol I transcription machinery caused by X or Ras that contribute to the increase in promoter activity observed, these results indicate that the X- and Ras-mediated increase in TBP, by itself, is capable of inducing RNA Pol I-dependent promoter activity. These results provide new evidence that TBP is a limiting transcription component for RNA Pol I-dependent transcription.
There are several possibilities for how an increase in TBP might regulate RNA Pol I-dependent transcription. The increase in TBP could produce an increase in the number of functional TFIID complexes which could stimulate RNA Pol II-dependent gene activity and indirectly enhance rRNA promoter activity. However, our results argue against this possibility, since we found that (i) directly increasing the amount of TBP in extracts enhances rRNA transcription in vitro and (ii) the overexpression of a TBP mutant that cannot support RNA Pol II-dependent promoter activity is still capable of augmenting RNA Pol I-dependent promoter activity. Our analysis of Drosophila mutant TBP-A338V supports earlier results obtained with yeast that demonstrated that the highly conserved alanine residue in the helix H2 region of TBP is important for RNA Pol II-dependent, but not RNA Pol I-dependent, promoter activity (12). This mutation appears to impair basal transcription, yet the contacts that the residue potentially makes with TFIIA, TFIIB, or other RNA Pol II-specific components have not been defined.
Another possible mechanism contributing to TBP-mediated enhanced transcription could involve antirepression by TBP through its interaction with molecules such as the retinoblastoma protein (pRb), which is known to inhibit rRNA transcription (6, 56). The mechanism by which pRb represses RNA Pol I-dependent transcription has been shown to involve its direct binding to UBF, which subsequently inhibits UBF-DNA interactions and transcription complex formation (56). If the increase in TBP were to enhance rRNA transcription by complexing with pRb, and thereby increasing the amount of available UBF, we might expect to observe an increase in UBF DNA binding activity in extracts that support an increase in RNA Pol I-dependent transcription. Our preliminary analysis, however, indicates that there is no apparent qualitative or quantitative change in the DNA binding activity of UBF in X-expressing cell extracts (60). In addition, evidence indicates that pRb is inactivated by Ras signaling and that this function of Ras promotes cell proliferation (40). These results suggest that pRb would be inactivated in X-expressing cells that contain activated Ras, rendering it incapable of binding to UBF. Alternatively, or in addition to an antirepression mechanism, the increase in TBP could generate an increase in the number of functional SL1 complexes. Our results are most consistent with this mechanism, as addition of exogenous recombinant TBP to cell extracts is capable of mediating an increase in transcription in vitro. In the case of the TBP-mediated increase in RNA Pol III-dependent transcription, evidence indicates that this also occurs via an increase in the amount of TBP-containing TFIIIB complexes (52, 55, 61, 62). Thus, these regulatory events mediated by TBP represent the first example of coregulation of RNA Pol I- and Pol III-dependent promoter activity by the same transcription component.
Our results show that X transactivation of rRNA promoters is dependent on the activation of the Ras cellular signaling pathway. This represents the first demonstration that oncogenic Ras can up-regulate RNA Pol I-dependent promoter activity. The fact that we observed induction of rRNA promoters by both X and oncogenic Ras in two distinct cell lines suggests that this regulatory mechanism is conserved and it is not organism or cell type dependent. Previous studies have shown that the phorbol ester TPA, a potent activator of PKC, can stimulate endogenous rRNA transcription in both mammalian (1, 51) and Drosophila (54, 64) cells. X has been shown to stimulate both PKC (30) and Ras activation (3), and evidence indicates that, depending on the stimuli, PKC activation and Ras activation converge to activate the same downstream signaling events (39, 49). Therefore, it is likely that these two signaling pathways are connected. Consistent with this notion, our previous work has shown that the activation of cellular signaling by treatment of cells with the phorbol ester TPA (20) or expression of HBV X (62) or oncogenic Ras (61) produces an increase in the cellular levels of TBP. Since TBP is a central transcription factor, it is important to identify the specific changes in cellular gene activity that are mediated by this response. We have previously shown that increased cellular concentrations of TBP results in the activation of RNA Pol III-dependent promoters (52, 62). Our present studies reveal that RNA Pol I-dependent promoters are also up-regulated by increasing cellular TBP. Although studies have only begun to examine how RNA Pol II-dependent promoters are affected by the overexpression of TBP, initial studies have demonstrated that certain promoters, depending on their architecture, are either activated or repressed (11, 48). Thus, the overexpression of TBP has profound effects on cellular gene expression, affecting all three major classes of eucaryotic promoters. Interestingly, TBP mRNA has been found to be highly overexpressed in human lung and breast carcinomas compared to normal tissue (57). How the TBP-mediated changes in gene activity contribute to the ability of the HBV X protein and oncogenic Ras to transform cells is an important issue to be addressed.
ACKNOWLEDGMENTS
We thank Adrian Vilalta for helpful discussions and Michael Stallcup and Lucio Comai for critical review of the manuscript. We also thank Chiou-Hwa Yuh for help in the constructions of the expression plasmids; Maria Pellegrini for providing pR119B, pT7B, and pDmr19 DNAs; Lucio Comai for providing prHu3; James Ou for providing Huh7 cells; and Michael Holmes and Robert Tjian for providing recombinant TBP.
This work was supported by National Institutes of Health grant CA74138 to D.L.J. and by a predoctoral fellowship from the Pharmaceutical Research and Manufacturers of America Foundation to H.D.W.
REFERENCES
- 1.Allo S N, McDermott P J, Carl L L, Morgan H E. Phorbol ester stimulation of protein kinase C activity and ribosomal DNA transcription. J Biol Chem. 1991;266:22003–22009. [PubMed] [Google Scholar]
- 2.Aufiero B, Schneider R J. The hepatitis B virus X gene product transactivates both RNA polymerase II and III promoters. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benn J, Schneider R J. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benn J, Su F, Doria M, Schneider R J. Hepatitis B virus Hbx protein induces transcription factor AP-1 by activation of extracellular signal-related and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 7.Chao Y, Pellegrini M. In vitro transcription of Drosophila rRNA genes shows stimulation by a phorbol ester and serum. Mol Cell Biol. 1993;13:934–941. doi: 10.1128/mcb.13.2.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H-S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis B virus X gene is important for the establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheong J H, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eucaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:142–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark M E, Lieberman P M, Berk A J, Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol Cell Biol. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colgan J, Manley J L. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA pol II promoters. Genes Dev. 1992;6:304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- 12.Cormack B P, Struhl K. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science. 1993;262:244–248. doi: 10.1126/science.8211143. [DOI] [PubMed] [Google Scholar]
- 13.Cross J C, Wen P, Rutter W J. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen-activated cellular serine/threonine kinases. Proc Natl Acad Sci USA. 1993;90:8078–8082. doi: 10.1073/pnas.90.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandri M, Schirmacher P, Rogler C E. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darnell J E, Girard M, Baltimore D, Summers D F, Maizel J V. The synthesis and translation of poliovirus RNA. In: Colter J, Paranchych W, editors. The molecular biology of viruses. New York, N.Y: Academic Press, Inc.; 1967. pp. 375–401. [Google Scholar]
- 16.Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs R L. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 17.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus Hbx protein is a dual specificity cytoplasmic activator of ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feitelson M A, Zhu M, Duan L X, London W T. Hepatitis B x antigen and p53 are associated in vitro and in liver tissue from patients with primary hepatocellular carcinoma. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 19.Fisher M, Runkel L, Schaller H. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteosome alpha subunit. Virus Genes. 1995;10:99–102. doi: 10.1007/BF01724303. [DOI] [PubMed] [Google Scholar]
- 20.Garber M, Vilalta A, Johnson D L. Induction of Drosophila RNA polymerase III gene expression by the phorbol ester, TPA, is mediated by TFIIIB. Mol Cell Biol. 1994;14:339–347. doi: 10.1128/mcb.14.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han K, Levine M S, Manley J L. Synergistic activation and repression of transcription by Drosophila homeobox protein. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- 22.Haviv I, Matza Y, Shaul Y. pX, the HBV-encoded activator, suppresses the phenotypes of TBP and TAFII250 mutants. Genes Dev. 1998;12:1217–1226. doi: 10.1101/gad.12.8.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haviv I, Vaizel D, Shaul Y. The X protein of hepatitis B virus coactivates potent activation domains. Mol Cell Biol. 1995;15:1079–1085. doi: 10.1128/mcb.15.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haviv I, Vaizel D, Shaul Y. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 1996;15:3413–3420. [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 27.Hohne M, Schaefer S, Seifer M, Feitelson M A, Paul D, Gerlich W H. Malignant transformation of immortalized transgenic hepatocytes after transfection with hepatitis B virus DNA. EMBO J. 1990;9:1137–1145. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Kwong J, Sun E C Y, Liang T J. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob S T. Regulation of ribosomal gene transcription. Biochem J. 1995;306:617–626. doi: 10.1042/bj3060617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kekule A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Hepatitis B virus transactivator uses a tumor promoter signalling pathway. Science. 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 31.Kim C M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 32.Klein N P, Schneider R J. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signal to Ras. Mol Cell Biol. 1997;17:6427–6436. doi: 10.1128/mcb.17.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koike K, Moriya K, Yotsuyanagi H, Iino S, Kurokawa K. Induction of cell cycle progression by hepatitis B virus HBx gene expression in quiescent mouse fibroblasts. J Clin Invest. 1994;94:44–49. doi: 10.1172/JCI117343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Learned R M, Tjian R. In vitro transcription of human ribosomal RNA genes by RNA polymerase I. J Mol Appl Genet. 1982;1:575–584. [PubMed] [Google Scholar]
- 35.Lee T H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y, Nomura T, Cheong J, Dorjsuren D, Iida K, Murakami S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 37.Lucito R, Schneider R J. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J Virol. 1992;66:983–991. doi: 10.1128/jvi.66.2.983-991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 39.Marais R, Light Y, Mason C, Paterson H, Olson M F, Marshall C J. Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by protein kinase C. Science. 1998;280:109–112. doi: 10.1126/science.280.5360.109. [DOI] [PubMed] [Google Scholar]
- 40.Mittnacht S, Paterson H, Olson M F, Marshall C J. Ras signalling is required for inactivation of the tumour suppressor pRB cell-cycle control protein. Curr Biol. 1997;7:219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- 41.Moss T, Stefanovsky V Y. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- 42.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Ras- and Raf-dependent activation of c-Jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 43.Pöckl E, Wintersberger E. Increased rate of RNA synthesis: early reaction of primary mouse kidney cells to infection with polyoma virus and simian virus 40. J Virol. 1980;35:8–19. doi: 10.1128/jvi.35.1.8-19.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qadri I, Maguire H F, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qadri I, Conaway J W, Conaway R C, Schaack J, Siddiqui A. Hepatitis B virus transactivator protein, Hbx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci USA. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raskas H, Thomas D, Green M. Biochemical studies on adenovirus multiplication. XVII. Ribosome synthesis in uninfected and infected KB cells. Virology. 1970;40:893–902. doi: 10.1016/0042-6822(70)90135-2. [DOI] [PubMed] [Google Scholar]
- 47.Rubinstein S J, Hammerle T, Wimmer E, Dasgupta A. Infection of HeLa cells with poliovirus results in modification of a complex that binds to the rRNA promoter. J Virol. 1992;66:3062–3068. doi: 10.1128/jvi.66.5.3062-3068.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadovsky Y, Webb P, Lopez G, Baxter J D, Fitzpatrick P M, Gizang-Ginsberg E, Cavailles V, Parker M G, Kushner P J. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schonwasser D C, Marais R M, Marshall C J, Parker P J. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol. 1998;18:790–798. doi: 10.1128/mcb.18.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slage B L, Lee T H, Medina D, Finegold M J, Butel J S. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 51.Soprano K J, Dev V G, Croce C M, Baserga R. Reactivation of silent rRNA genes by simian virus 40 in human-mouse hybrid cells. Proc Natl Acad Sci USA. 1979;76:3885–3889. doi: 10.1073/pnas.76.8.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trivedi A, Vilalta A, Gopalan S, Johnson D L. TATA-binding protein is limiting for both TATA-containing and TATA-lacking RNA polymerase III promoters in Drosophila cells. Mol Cell Biol. 1996;16:6909–6916. doi: 10.1128/mcb.16.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueda H, Ullrich S J, Gangemi J D, Kappel C A, Ngo L, Feitelson M A, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41–47. doi: 10.1038/ng0195-41. [DOI] [PubMed] [Google Scholar]
- 54.Vallett A M, Brudnak M, Pellegrini M, Weber H W. In vivo regulation of rRNA transcription occurs rapidly in nondividing and dividing Drosophila cells in response to a phorbol ester and serum. Mol Cell Biol. 1993;13:928–933. doi: 10.1128/mcb.13.2.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vilalta A, Trivedi A, Wang Z, Roeder R G, Johnson D L. An RNA polymerase III-defective mutation in TATA-binding protein disrupts its interaction with a transcription factor IIIB subunit in Drosophila cells. J Biol Chem. 1997;272:18087–18092. doi: 10.1074/jbc.272.29.18087. [DOI] [PubMed] [Google Scholar]
- 56.Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wada C, Kasai K, Kameya T, Ohtani H. A general transcription factor, human transcription factor IID, overexpressed in human lung and breast carcinoma and rapidly induced with serum stimulation. Cancer Res. 1992;52:307–313. [PubMed] [Google Scholar]
- 58.Wagner E, Roizman B. Ribonucleic acid synthesis in cells infected with herpes simplex virus. I. Patterns of ribonucleic acid synthesis in productively infected cells. J Virol. 1969;4:36–57. doi: 10.1128/jvi.4.1.36-46.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122:855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, H.-D., and D. L. Johnson. Unpublished data.
- 61.Wang H-D, Trivedi A, Johnson D L. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases the cellular TATA-binding protein by activating the Ras signaling pathway. Mol Cell Biol. 1997;17:6838–6846. doi: 10.1128/mcb.17.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H-D, Yuh C-H, Dang C V, Johnson D L. The hepatitis B virus increases the cellular level of TATA-binding protein which mediates transactivation of RNA polymerase III genes. Mol Cell Biol. 1995;15:6720–6728. doi: 10.1128/mcb.15.12.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber H W, Vallett S, Neilson L, Grotke M, Chao Y, Brudnak M, Juan A S, Pellegrini M. Serum, insulin and phorbol esters stimulate rRNA and tRNA gene expression in both dividing and non-dividing Drosophila cells. Mol Cell Biochem. 1993;104:201–207. doi: 10.1007/BF00229821. [DOI] [PubMed] [Google Scholar]
- 65.Williams J S, Andrisani O M. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc Natl Acad Sci USA. 1995;92:3819–3823. doi: 10.1073/pnas.92.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yen T S B. Hepadnaviral X protein: review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 67.Zhai W, Tuan J, Comai L. SV40 large T antigen binds to the TBP-TAF1 complex SL1 and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]
- 69.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis B virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]