Abstract
Diagnostic coding, or ICD coding, is the task of assigning diagnosis codes defined by the ICD (International Classification of Diseases) standard to patient visits based on clinical notes. The current process of manual ICD coding is time-consuming and often error-prone, which suggests the need for automatic ICD coding. However, despite the long history of automatic ICD coding, there have been no standardized frameworks for benchmarking ICD coding models.
We open-source an easy-to-use tool named AnEMIC, which provides a streamlined pipeline for preprocessing, training, and evaluating for automatic ICD coding. We correct errors in preprocessing by existing works, and provide key models and weights trained on the correctly preprocessed datasets. We also provide an interactive demo performing real-time inference from custom inputs, and visualizations drawn from explainable AI to analyze the models. We hope the framework helps move the research of ICD coding forward and helps professionals explore the potential of ICD coding. The framework and the associated code are available here.
1. Introduction
Diagnostic coding is the task of assigning alphanumeric codes to diagnoses and procedures after a patient visits a healthcare provider. These codes are typically specified by a medical classification standard called the International Classification of Diseases (ICD). Diagnostic coding, or ICD coding, is an integral component of medical billing, and integral to claims paid by health insurance carriers. The diagnostic coding process alone accounts for approximately 21% of medical administrative costs in the US (Tseng et al., 2018). During this process, a professional coder reviews the patient’s medical records, including clinical narratives, and manually selects ICD codes. Since the task requires in-depth clinical knowledge and understanding of medical records, and importantly, due to the fact that there are a large number of ICD codes, the task is labor-intensive and error-prone (Manchikanti, 2002).
These difficulties motivate the need for automatic ICD coding systems which perform diagnosis classification given a patient’s health record (Kaur et al., 2021; Yan et al., 2022). This has been the subject of considerable research, with some of the early work dating back to the 1990s (Larkey and Croft, 1996), to more recent deep neural NLP approaches. There are a few outstanding and major challenges in the diagnostic coding task. Firstly, the label space, the set of all ICD codes, is large, and the label distribution is highly imbalanced. Secondly, the input text, i.e., the discharge summaries, is noisy and can contain abstruse medical terms, lesser-known abbreviations, misspelt words, etc. Also, they are much longer than what most state-of-the-art models take as input.
Along with those challenges, the absence of a benchmark has impeded the progress of research. Due to privacy restrictions that limit access to even publicly available clinical databases, researchers have to create datasets manually from these, and this results in discrepancies in the actual datasets used in individual papers. For instance, the label set of MIMIC-III top-50 dataset varies among the literature, and some of them are even used incorrectly. Inconsistency in processing the dataset and the inevitable errors introduced as a result of this makes it hard to compare different methods.
In this paper, we introduce a framework for benchmarking automatic ICD coding with the MIMIC clinical database. We name our framework AnEMIC, for An Error-reduced MIMIC ICD Coding benchmark. To the best of our knowledge, AnEMIC is the first attempt to collate and benchmark different deep learning approaches for automatic ICD coding with a configurable pipeline.
Our contributions can be summarized as follows:
We provide a pipeline covering the entire process of automatic ICD coding, including preprocessing, training, and evaluation. The whole process is easily configurable with the use of YAML files. We additionally provide key deep learning-based ICD coding models.
We correct errors in the most widely used datasets and provide benchmark results of the key models on the new datasets.
We open-source an easy-to-use interactive demo that enables researchers to test their models on custom inputs and visualize input attribution scores for explainability.
The remainder of the paper is organized as follows. In Section 2, we discuss popular automatic ICD coding approaches and datasets. Section 3 details our approaches for preprocessing, training, evaluation, and our demo application. In Section 4, we perform a quantitative and qualitative analysis of AnEMIC. Finally, we conclude with discussion and future work in Section 5.
2. Related Work
2.1. ICD Coding
Over the history of automatic diagnosis coding, approaches have ranged from classical methods such as rule-based approaches (Farkas and Szarvas, 2008), traditional ML models such as SVMs (Perotte et al., 2014), to more recent Deep Learning-based methods. A neural network-based approach was first attempted by Prakash et al. (2017). A prominent deep learning approach is CAML (Mullenbach et al., 2018), which uses a CNN encoder with a unique per-label attention mechanism. Since CAML, there have been many other CNN and RNN-based approaches (Yu et al., 2019; Vu et al., 2020). A few notable CNN based approaches include using dilated convolutional layers (Ji et al., 2020) and multi-filter convolutional layers (Li and Yu, 2020; Luo et al., 2021).
Additionally, researchers have leveraged the hierarchy of ICD codes (Cao et al., 2020; Xie et al., 2019), used external knowledge sources like Wikipedia (Bai and Vucetic, 2019), and knowledge graphs such as UMLS (Yuan et al., 2022) and Freebase (Teng et al., 2020), etc. More recently, there has been an effort to use Transformer-based language models pretrained on clinical datasets, albeit without much success (Pascual et al., 2021; Zhang et al., 2020; Ji et al., 2021). Instead, using a few Transformer encoder layers trained from scratch has proven to be more effective (Biswas et al., 2021).
Kaur et al. (2021) and Yan et al. (2022) perform extensive literature reviews of automatic ICD coding approaches. The reader is referred to these surveys for a more detailed description of various architectures and approaches.
2.2. ICD Coding Datasets and Benchmark
Typical ICD coding dataset consists of discharge summaries and the corresponding sets of ICD codes. There are many ICD coding datasets in various languages, but not all are publicly available. The most widely used datasets are from MIMIC-III1 and MIMIC-II2 databases. The MIMIC-III clinical database (Johnson et al., 2016) is a collection of medical records from an intensive care unit (ICU) at a hospital between 2001 and 2012. MIMIC-III consists of multiple tables containing diagnosis, procedures, clinical notes, etc., and each patient admission is indicated with an HADM_ID identifier. MIMIC-II is a subset of the MIMIC-III dataset and contains medical records between 2001 and 20083.
CAML (Mullenbach et al., 2018) published the preprocessing code of their MIMIC-III full and top-50 datasets, and since then, these have been the most widely used datasets. We correct some errors in preprocessing of CAML and make the process easily configurable. Also, compared to a leader-board that only manages reported performance, our work provides a framework for benchmarking, i.e., users can run the code to reproduce the results and further perform research on top of it.
3. ICD Coding Benchmark
AnEMIC has been designed so that researchers can easily configure the overall process with config files and therefore, easily start research on ICD coding with minimal code. Also, the architecture has modularity at the center of its design so that researchers can replace one module with another or with their own implementation. Such design enables easy comparison between models and reduces burden while developing new models.
Our system also provides an interactive demo for visualizing model predictions with input attribution scores. This demo will help users analyze the performance and interpretability of their models.
In the following subsections, we explain each stage in the pipeline. From now on, we will focus on ICD coding dataset from MIMIC-III since it is the most widely used dataset for this task. Figure 1 illustrates the overall pipeline.
Figure 1:
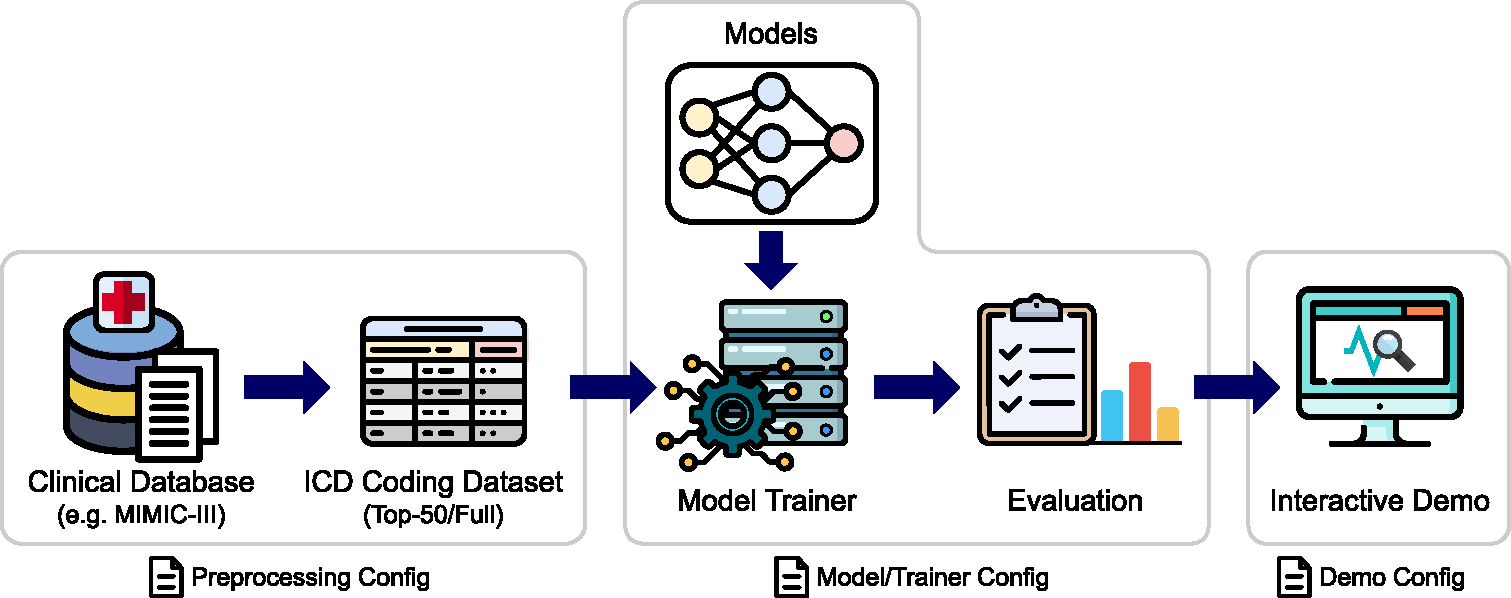
The ICD coding benchmark pipeline of AnEMIC. We provide a pipeline covering the entire process of ICD coding. All steps in the pipeline can be easily configured with YAML files.
3.1. Data Preprocessing
The first step of the pipeline is to preprocess the available clinical dataset, i.e., the MIMIC-III database. As with other parts of the pipeline, we specify preprocessing-related options in a YAML config file.
Many of the preprocessing steps are inspired by CAML’s preprocessing pipeline. However, an important observation to be noted here is that there are errors in CAML’s preprocessing pipeline. Unfortunately, many subsequent works use CAML’s code, and hence, the results obtained by most papers are on the incorrectly preprocessed dataset. This will be discussed later in this subsection and Appendix A.
3.1.1. ICD Code Preprocessing
In the MIMIC-III database, the DIAGNOSES_ICD and PROCEDURES_ICD tables contain the ICD-9 diagnosis and procedure codes, respectively, of every admission. Since MIMIC-III has ICD-9 codes without the period punctuation (e.g. 4019 instead of 401.9), we reformat those ICD codes to their original format adopting the method of CAML, and use them as labels. ICD-9 codes can have leading and trailing zeros, so care must be taken to retain them when processing. However, in CAML’s preprocessing code, some of ICD codes are implicitly treated as integer or floating point numbers4, resulting in an incorrect set of ICD-9 labels. While correcting this error, we provide an option incorrect_code_loading to reproduce the behavior of CAML for researchers who want to make a comparison with previous works.
In addition to the above option, we also provide an option code_type to use either diagnosis, procedure, or both types of ICD codes. We set “both” as the default.
3.1.2. Clinical Note Preprocessing
From the NOTEEVENTS table of MIMIC-III containing clinical notes in various categories, we select notes belonging to the Discharge_Summary category. We provide several options of standard NLP preprocessing for the discharge summary. These can be turned on/off from the config file.
Convert text to lowercase.
Remove punctuation marks using \w+ as the RegEx expression, i.e., retain only alphanumeric characters.
Either remove numeric characters, or replace all numeric characters with “n”.
Remove stopwords; we use the list of stop-words provided by NLTK, and add common medical terms like “hospital”, “admission”, “history”, etc. to the list.
Stem or lemmatize the text; we provide popular choices for these such as “WordNet Lemmatizer” and “Porter Stemmer”.
Truncate the text to a maximum length.
After note preprocessing, we build the vocabulary and train a Word2Vec model on preprocessed discharge summaries using the Gensim library (Řehůřek and Sojka, 2010). Word2Vec embeddings are used to initialize the embedding layers of models.
3.1.3. Top-k Codes and Data Splitting
Many works report results on two datasets – “MIMIC-III full” and “MIMIC-III top-50”. The latter contains the top-50 frequent ICD codes as labels and examples with at least one of these labels.
An important point to note is that MIMIC-III has some duplicate ICD codes, i.e., an ICD code can be repeated multiple times in one admission. These duplicate codes need to be removed when counting the ICD code occurrence. This is another source of error in CAML’s code: they do not remove the duplicate codes while counting the ICD codes occurrence, resulting in a change in the top-50 ICD codes. While we correctly select the top-50 ICD codes, we also provide an option count_duplicate_codes to reproduce the behavior of CAML.
For data splitting, we use the splits of HADM_IDs provided by CAML. They provide separate sets of splits for the full and top-50 datasets, and the split for top-50 dataset has substantially smaller number of examples. To make full use of MIMIC-III, we use the splits of the CAML’s full dataset for both versions of our dataset.
As a result of data preprocessing, we have four main variants of the dataset – “MIMIC-III full”, “MIMIC-III top-50”, “MIMIC-III full (old)”, and “MIMIC-III top-50 (old)”. Here “(old)” refers to the CAML variants.
3.2. Supported Models
This subsection describes the models we provide in the framework and the criteria for choosing models. To provide researchers with good baselines for ICD coding research, we selected models based on novelty or superior performance. For now, we have chosen a subset of models for which the code is publicly available, but we do plan on implementing other approaches in the near future which have not been open-sourced. The models and the trainer are based on PyTorch.
The models currently supported by the framework are as follows:
CAML (Mullenbach et al., 2018) is a landmark model in automatic ICD coding which uses a label attention layer. We also implement the vanilla CNN model in the paper and refer to it as CNN.
MultiResCNN (Li and Yu, 2020) uses multiple CNNs with different filter sizes in parallel.
DCAN (Ji et al., 2020) uses dilated convolutional layers for ICD coding.
TransICD (Biswas et al., 2021) is the first Transformer-based approach that achieved results comparable to the CNN-based model.
Fusion (Luo et al., 2021) uses multi-CNN, Transformer encoder, and label attention.
To replicate the author’s work in our own system, we re-wired the model from the author’s code to make it compatible with our framework. This allows users to also easily tweak the model and its hyperparameters with the config files.
3.3. Training and Evaluation
To train and evaluate the models, we implement a trainer module that manages training and evaluation, with sub-modules for the additional functionalities related to training, such as objective functions, logging, and managing checkpoints. Following the design principle of the framework, the trainer module is also highly configurable so the users can easily customize training and visualize metrics by modifying config files. This also applies to evaluation metrics, and we provide all major evaluation metrics adopted by the automatic ICD coding literature.
3.4. Interactive Demo
In order to enable users to use trained models off-the-shelf, we open source an interactive web application based on Streamlit. Using the app, users can feed in a new discharge summary and get the ICD code predictions in real time without writing code to preprocess the input text and to run the models. The app also allows users to change the models and toggle the preprocessing options on the fly so that they can compare models and change preprocessing options.
A major highlight of the app is explainability visualization, i.e., the attribution or importance scores for each word present in the input clinical note. We provide two methods – Integrated Gradients (Sundararajan et al., 2017) and attention scores. Upon choosing the attribution method with an ICD code, the app displays the input tokens with important words highlighted. Note that this interpretability feature is model-agnostic because the explainable AI techniques we use such as integrated gradients are in turn model-agnostic.
A screenshot of the app running on a discharge summary is shown in Figure 2. The bottom of Figure 2 shows the integrated gradient (IG) visualization of ICD code 250.00 “Type II diabetes”. We can see that important terms like “diabetes mellitus” exhibit high IG scores5. Overall, we expect the interactive demo will be helpful for both researchers who want to validate models, and professionals who want explanations of the model’s predictions.
Figure 2:
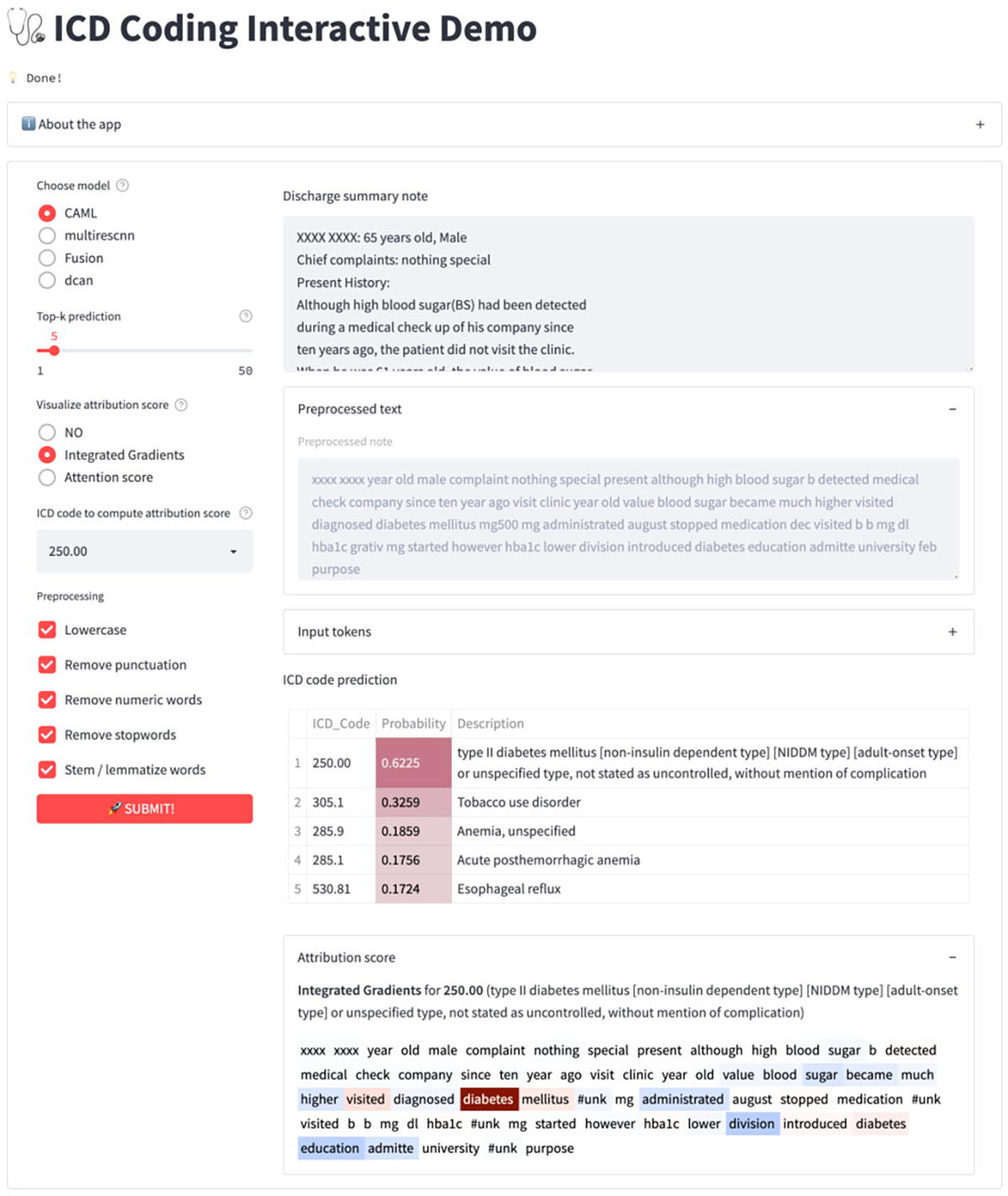
A snapshot of ICD coding interactive demo showing ICD code predictions and the integrated gradient. Input text is extracted from Tsumoto et al. (2019).
4. Results
In this section, we discuss the quantitative and qualitative results of AnEMIC. On quantitative aspects, we discuss the brief statistics of the datasets and the benchmark results on the our ICD coding datasets. For the qualitative results, we present and analyze some example of interpretability visualization from our demo application.
4.1. Quantitative Results
Dataset Statistics
Table 1 shows brief statistics of our ICD coding datasets and the CAML’s datasets (old). Our full dataset contains the same number of examples as CAML’s full dataset since we used the same data split. However, it has a different set of labels since we corrected the preprocessing of CAML. Our top-50 dataset has the same number of labels as CAML’s top-50 dataset, but the label set differs6. Also, our top-50 dataset has substantially more examples since the data split of the full dataset is used to make full use of MIMIC-III. It has a slightly less number of examples than the full dataset since examples without any of the top-50 codes are removed.
Table 1:
Statistics of the MIMIC-III full and top-50 datasets. Mean # labels refers to the average number of labels per example.
| Dataset | AnEMIC | CAML (old) | ||
|---|---|---|---|---|
|
| ||||
| Full | Top-50 | Full | Top-50 | |
|
| ||||
| # labels | 8930 | 50 | 8922 | 50 |
| Mean # labels | 15.88 | 5.73 | 16.10 | 5.78 |
| # examples | ||||
| - Train set | 47723 | 44728 | 47723 | 8066 |
| - Val set | 1631 | 1569 | 1631 | 1573 |
| - Test set | 3372 | 3234 | 3372 | 1729 |
Benchmark Results
To provide the benchmark of our ICD coding datasets, we trained the models introduced in Section 3.2. Hyper-parameters for each model are chosen as reported in the respective paper or code. Note that these hyper-parameters are tuned to CAML datasets, so may not be optimal for our datasets, especially for the top-50 dataset. For DCAN and TransICD model, only the MIMIC-III top-50 experiments was performed, so we use the hyper-parameters for the top-50 dataset in the full dataset experiment. For each model, we ran the experiment three times and computed the mean and variance of the results. Table 2 and 3 shows the benchmark results. Among the models that we implemented, MultiResCNN and Fusion achieved the best test performance on the MIMIC-III full dataset, and DCAN performed best on the MIMIC-III top-50 dataset.
Table 2:
Test set results on the MIMIC-III full dataset. The results are shown using the mean±standard deviation format.
| Model | Macro AUC | Micro AUC | Macro F1 | Micro F1 | P@8 | P@15 |
|---|---|---|---|---|---|---|
|
| ||||||
| CNN | 0.835±0.001 | 0.974±0.000 | 0.034±0.001 | 0.420±0.006 | 0.619±0.002 | 0.474±0.004 |
| CAML | 0.893±0.002 | 0.985±0.000 | 0.056±0.006 | 0.506±0.006 | 0.704±0.001 | 0.555±0.001 |
| MultiResCNN | 0.912±0.004 | 0.987±0.000 | 0.078±0.005 | 0.555±0.004 | 0.741±0.002 | 0.589±0.002 |
| DCAN | 0.848±0.009 | 0.979±0.001 | 0.066±0.005 | 0.533±0.006 | 0.721±0.001 | 0.573±0.000 |
| TransICD | 0.886±0.010 | 0.983±0.002 | 0.058±0.001 | 0.497±0.001 | 0.666±0.000 | 0.524±0.001 |
| Fusion | 0.910±0.003 | 0.986±0.000 | 0.081±0.002 | 0.560±0.003 | 0.744±0.002 | 0.589±0.001 |
Table 3:
Test set results on the MIMIC-III top-50 dataset. The results are shown using the mean±standard deviation format.
| Model | Macro AUC | Micro AUC | Macro F1 | Micro F1 | P@5 |
|---|---|---|---|---|---|
|
| |||||
| CNN | 0.913±0.002 | 0.936±0.002 | 0.627±0.001 | 0.693±0.003 | 0.649±0.001 |
| CAML | 0.918±0.000 | 0.942±0.000 | 0.614±0.005 | 0.690±0.001 | 0.661±0.002 |
| MultiResCNN | 0.928±0.001 | 0.950±0.000 | 0.652±0.006 | 0.720±0.002 | 0.674±0.001 |
| DCAN | 0.934±0.001 | 0.953±0.001 | 0.651±0.010 | 0.724±0.005 | 0.682±0.003 |
| TransICD | 0.917±0.002 | 0.939±0.001 | 0.602±0.002 | 0.679±0.001 | 0.643±0.001 |
| Fusion | 0.932±0.001 | 0.952±0.000 | 0.664±0.003 | 0.727±0.003 | 0.679±0.001 |
To validate the implementation of key models and the CAML version of dataset, we also ran the same experiments on the CAML version of the datasets. Overall, the results display similar level of performance as reported in the papers. Please see Appendix C for the full results and details of the reproduction experiments.
4.2. Qualitative Analysis
Explainability Visualization
Figure 3 shows some examples of explainability visualization from the demo app. For each example, we extract the window around the word with the highest attribution score. In the left figure, for a fixed discharge summary and an ICD code (599.0, Urinary tract infection, site not specified), we examine the integrated gradients of various models. From the figure, we can observe that all models correctly attribute their prediction to the words relevant to the diagnosis. In the right figure, for a fixed discharge summary and a model (CAML), we visualize the integrated gradients of some ICD codes that are predicted as positive. As the figure shows, different parts of the input are attributed and they are all semantically relevant to the corresponding ICD code. As both figures illustrate, our interactive demo provides an effective visualization tool for explaining the model’s predictions.
Figure 3:

Interpretability visualization examples. Left: the integrated gradients of various models on a fixed input and a fixed ICD code (HADM_ID=100020, ICD-9 599.0). Right: the integrated gradients of CAML for various ICD codes on a fixed input (HADM_ID=139574).
5. Conclusions and Future Work
In this work, we present AnEMIC, a comprehensive framework for automatic diagnostic coding. It serves as a standardized benchmark for ICD coding on MIMIC-III by correcting errors in existing datasets and providing popular deep learning-based models. Our framework has a modularized and easy-to-use config-based design, and researchers can easily experiment by writing config files or adding custom submodules. We also provide an interactive app for performing real-time inference and visualization for model explainability.
AnEMIC is under active development and welcomes contributions from the community. Upcoming updates to our pipelines include adding more recent approaches and models, especially those that incorporate additional sources of external knowledge, as well as supporting other datasets like the MIMIC-II dataset.
Supplementary Material
Acknowledgements
We acknowledge the support of Center for Machine Learning and Health at Carnegie Mellon University. This work was supported by the intramural research program at the U.S. National Library of Medicine, National Institutes of Health.
Footnotes
There is also the recently released MIMIC-IV database, but clinical notes for this are currently not yet available.
Due to not specifying data types when loading tables
Red and blue color in the visualization represent positive and negative scores, respectively.
References
- Bai Tian and Vucetic Slobodan. 2019. Improving medical code prediction from clinical text via incorporating online knowledge sources. In The World Wide Web Conference, WWW ‘19, page 72–82, New York, NY, USA. Association for Computing Machinery. [Google Scholar]
- Biswas Biplob, Pham Thai-Hoang, and Zhang Ping. 2021. Transicd: Transformer based code-wise attention model for explainable icd coding.
- Cao Pengfei, Chen Yubo, Liu Kang, Zhao Jun, Liu Shengping, and Chong Weifeng. 2020. HyperCore: Hyperbolic and co-graph representation for automatic ICD coding. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, pages 3105–3114, Online. Association for Computational Linguistics. [Google Scholar]
- Farkas Richárd and Szarvas György. 2008. Automatic construction of rule-based icd-9-cm coding systems. In BMC bioinformatics, volume 9, pages 1–9. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Shaoxiong, Cambria Erik, and Marttinen Pekka. 2020. Dilated convolutional attention network for medical code assignment from clinical text. In Proceedings of the 3rd Clinical Natural Language Processing Workshop, pages 73–78, Online. Association for Computational Linguistics. [Google Scholar]
- Ji Shaoxiong, Hölttä Matti, and Marttinen Pekka. 2021. Does the magic of bert apply to medical code assignment? a quantitative study. Comput. Biol. Med, 139(C). [DOI] [PubMed] [Google Scholar]
- Johnson Alistair EW, Pollard Tom J, Shen Lu, Lehman Li-wei H, Feng Mengling, Ghassemi Mohammad, Moody Benjamin, Szolovits Peter, Celi Leo Anthony, and Mark Roger G. 2016. Mimic-iii, a freely accessible critical care database. Scientific data, 3(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur Rajvir, Ginige Jeewani Anupama, and Obst Oliver. 2021. A systematic literature review of automated ICD coding and classification systems using discharge summaries. CoRR, abs/2107.10652. [Google Scholar]
- Larkey Leah S and Croft W Bruce. 1996. Combining classifiers in text categorization. In Proceedings of the 19th annual international ACM SIGIR conference on Research and development in information retrieval, pages 289–297. [Google Scholar]
- Li Fei and Yu Hong. 2020. Icd coding from clinical text using multi-filter residual convolutional neural network. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 34, pages 8180–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Junyu, Xiao Cao, Glass Lucas, Sun Jimeng, and Ma Fenglong. 2021. Fusion: Towards automated ICD coding via feature compression. In Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021, pages 2096–2101, Online. Association for Computational Linguistics. [Google Scholar]
- Manchikanti Laxmaiah. 2002. Implications of fraud and abuse in interventional pain management. Pain Physician, 5(3):320. [PubMed] [Google Scholar]
- Mullenbach James, Wiegreffe Sarah, Duke Jon, Sun Jimeng, and Eisenstein Jacob. 2018. Explainable prediction of medical codes from clinical text. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), pages 1101–1111, New Orleans, Louisiana. Association for Computational Linguistics. [Google Scholar]
- Pascual Damian, Luck Sandro, and Wattenhofer Roger. 2021. Towards BERT-based automatic ICD coding: Limitations and opportunities. In Proceedings of the 20th Workshop on Biomedical Language Processing, pages 54–63, Online. Association for Computational Linguistics. [Google Scholar]
- Perotte Adler, Pivovarov Rimma, Natarajan Karthik, Weiskopf Nicole, Wood Frank, and Elhadad Noémie. 2014. Diagnosis code assignment: models and evaluation metrics. Journal of the American Medical Informatics Association, 21(2):231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash Aaditya, Zhao Siyuan, Hasan Sadid A, Datla Vivek, Lee Kathy, Qadir Ashequl, Liu Joey, and Farri Oladimeji. 2017. Condensed memory networks for clinical diagnostic inferencing. In Thirty-first AAAI conference on artificial intelligence. [Google Scholar]
- Řehůřek Radim and Sojka Petr. 2010. Software Framework for Topic Modelling with Large Corpora. In Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks, pages 45–50, Valletta, Malta. ELRA. [Google Scholar]
- Sundararajan Mukund, Taly Ankur, and Yan Qiqi. 2017. Axiomatic attribution for deep networks. In P roceedings of the 34th International Conference on Machine Learning - Volume 70, ICML’17, page 3319–3328. JMLR.org. [Google Scholar]
- Teng Fei, Yang Wei, Chen Li, Huang LuFei, and Xu Qiang. 2020. Explainable prediction of medical codes with knowledge graphs. Frontiers in Bioengineering and Biotechnology, 8:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Phillip, Kaplan Robert S, Richman Barak D, Shah Mahek A, and Schulman Kevin A. 2018. Administrative costs associated with physician billing and insurance-related activities at an academic health care system. Jama, 319(7):691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumoto Shusaku, Kimura Tomohiro, Iwata Haruko, and Hirano Shoji. 2019. Estimation of disease code from electronic patient records. In 2019 IEEE International Conference on Big Data (Big Data), pages 2698–2707. IEEE. [Google Scholar]
- Vu Thanh, Nguyen Dat Quoc, and Nguyen Anthony. 2020. A label attention model for icd coding from clinical text. In Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, IJCAI-20, pages 3335–3341. International Joint Conferences on Artificial Intelligence Organization. Main track. [Google Scholar]
- Xie Xiancheng, Xiong Yun, Yu Philip S., and Zhu Yangyong. 2019. Ehr coding with multi-scale feature attention and structured knowledge graph propagation. In Proceedings of the 28th ACM International Conference on Information and Knowledge Management, CIKM ‘19, page 649–658, New York, NY, USA. Association for Computing Machinery. [Google Scholar]
- Yan Chenwei, Fu Xiangling, Liu Xien, Zhang Yuanqiu, Gao Yue, Wu Ji, and Li Qiang. 2022. A survey of automated international classification of diseases coding: development, challenges, and applications. Intelligent Medicine. [Google Scholar]
- Yu Ying, Li Min, Liu Liangliang, Fei Zhihui, Wu Fang-Xiang, and Wang Jianxin. 2019. Automatic icd code assignment of chinese clinical notes based on multilayer attention birnn. Journal of Biomedical Informatics, 91:103114. [DOI] [PubMed] [Google Scholar]
- Yuan Zheng, Tan Chuanqi, and Huang Songfang. 2022. Code synonyms do matter: Multiple synonyms matching network for automatic ICD coding. In Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), pages 808–814, Dublin, Ireland. Association for Computational Linguistics. [Google Scholar]
- Zhang Zachariah, Liu Jingshu, and Razavian Narges. 2020. BERT-XML: Large scale automated ICD coding using BERT pretraining. In Proceedings of the 3rd Clinical Natural Language Processing Workshop, pages 24–34, Online. Association for Computational Linguistics. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


