Abstract
To modulate transcription, regulatory factors communicate with basal transcription factors and/or RNA polymerases in a variety of ways. Previously, it has been reported that RNA polymerase II subunit 5 (RPB5) is one of the targets of hepatitis B virus X protein (HBx) and that both HBx and RPB5 specifically interact with general transcription factor IIB (TFIIB), implying that RPB5 is one of the communicating subunits of RNA polymerase II involved in transcriptional regulation. In this context, we screened for a host protein(s) that interacts with RPB5. By far-Western blot screening, we cloned a novel gene encoding a 508-amino-acid-residue RPB5-binding protein from a HepG2 cDNA library and designated it RPB5-mediating protein (RMP). Expression of RMP mRNA was detected ubiquitously in various tissues. Bacterially expressed recombinant RMP strongly bound RPB5 but neither HBx nor TATA-binding protein in vitro. Endogenous RMP was immunologically detected interacting with assembled RPB5 in RNA polymerase in mammalian cells. The central part of RMP is responsible for RPB5 binding, and the RMP-binding region covers both the TFIIB- and HBx-binding sites of RPB5. Overexpression of RMP, but not mutant RMP lacking the RPB5-binding region, inhibited HBx transactivation of reporters with different HBx-responsive cis elements in transiently transfected cells. The repression by RMP was counteracted by HBx in a dose-dependent manner. Furthermore, RMP has an inhibitory effect on transcriptional activation by VP16 in the absence of HBx. These results suggest that RMP negatively modulates RNA polymerase II function by binding to RPB5 and that HBx counteracts the negative role of RMP on transcription indirectly by interacting with RPB5.
In eukaryotes, nuclear RNA polymerases I, II, and III are highly conserved multisubunit enzymes involved in the synthesis of rRNAs, mRNAs, and tRNAs, respectively (48). All these nuclear RNA polymerases share the function of RNA synthesis but utilize different promoters and require different transcription factors. For example, promoter-specific transcription initiation from protein-coding genes requires the concerted action of a complex array of factors involving RNA polymerase II and general transcriptional factors (TFIID, TFIIA, TFIIB, TFIIE, TFIIF, and TFIIH) (14, 46). Transcriptional activators and repressors bind distal elements of a promoter and modulate transcription through communication with components of a preinitiation complex, such as TATA-binding protein (TBP) and its binding proteins, TFIIB, TFIIA, and TFIIH (18, 19, 24, 42). Recently, another group of proteins, cofactors (also called mediators), have been demonstrated to affect transcription positively or negatively by communicating with promoter-specific regulatory factors and the transcriptional machinery. These proteins include global coactivator p300/CBP (20, 25, 31, 34), nuclear silence mediators (2, 12, 52), and a large number of mediator proteins, or SRBs (10, 28, 39, 47, 54). RNA polymerase subunits may be additional targets for transcriptional regulators, because RNA polymerases are the ultimate target of transcriptional modulation. In this context, several subunits have been recently reported to interact with the regulators (8, 9), in addition to the well-documented regulatory role of the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (28, 30).
Hepatitis B virus (HBV) X protein (HBx) is essential for HBV infection and plays an important role in HBV-associated hepatocellular carcinoma (11, 17, 26, 51). Many reports have shown that HBx transactivates viral and cellular genes through a wide variety of cis-acting elements. However, the mechanism of this effect has not been well elucidated (3, 5, 7, 15, 16, 22, 23, 50). It has been previously demonstrated that HBx directly interacts with RNA polymerase II subunit 5 (RPB5), a common RNA polymerase subunit (13), that both RPB5 and HBx communicate with TFIIB but through different sites (21), and that the trimeric interaction of these three factors is involved in HBx transactivation (35). These observations suggest that RPB5 is the communicating subunit of RNA polymerase that interacts with transcriptional regulators (9). To gain insight into the role of RPB5 in transcriptional regulation, we screened for proteins that interact with RPB5. Here, we report a novel protein, RPB5-mediating protein (RMP), which specifically binds RPB5 and whose overexpression inhibits the transactivation function of HBx.
MATERIALS AND METHODS
cDNA cloning of RPB5-binding protein.
Approximately 2 × 106 plaques of a λgt11 cDNA library of HepG2 cells (HL1105b; Clontech Laboratories) were screened by far-Western blotting, essentially as previously reported (13). Briefly, nitrocellulose filters were incubated in modified GBT buffer (10% glycerol, 50 mM HEPES-NaOH [pH 7.5], 175 mM KCl, 7.5 mM MgCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol, 1% Triton X-100) and subjected to far-Western blotting. Purified glutathione S-transferase (GST)–RPB5 was labeled in vitro and used as a probe with [γ-32P]ATP by using the catalytic subunit of cyclic-AMP-dependent protein kinase (Sigma). Protein-protein binding reactions (far-Western) were performed with an RPB5 probe (40 to 100 ng of protein per μl, 2 × 106 to 4 × 106 cpm of protein per μg) in modified GBT buffer supplemented with 1% bovine serum albumin, 2 mM unlabeled ATP, and sonicated supernatants of Escherichia coli JM109 transformed with pGENK1 containing 1 mg of GST per ml (final concentration). Filters were rotated in the reaction mixture for 1 h at room temperature, washed four times with modified GBT buffer, and then exposed on X-ray films (XAR Omat; Kodak) for 1 to 2 days. At the second screening, several pairs of positive and negative plaques, the latter closely juxtaposed on the plate, were examined for inserts of identical size by PCR with λgt11 forward and reverse primers. After the third screening, seven positive clones were selected. PCR products of these clones were sequenced by the dideoxy sequence method and were subcloned into the EcoRI site of pBluescript II SK(+) (Stratagene). The putative full-length cDNA with an initiation codon at the 5′ end and a stop codon at the 3′ end was obtained with a Marathon cDNA amplification kit (Clontech Laboratories Inc.) according to the manufacturer’s protocol.
Plasmid constructions.
The plasmid pSG5UTPL was a mammalian expression vector derived from pSG5 (Stratagene) (38, 45). With pSG5UTPL, another FLAG-tagged mammalian expression vector, pNKFLAG, was constructed. The FLAG tag insert fragment was generated from pFLAGHis/p53 by PCR with a primer pair, one primer containing an artificial NotI site and a consensus translation initiation site and the other containing a BamHI site. The plasmids pGENK1 and pGENKS were used as vectors for GST fusion proteins, and they contained a thrombin cleavage site and a phosphorylation site for cyclic-AMP-dependent protein kinase (13, 44, 45). pYFLAG, used as the E. coli expression vector for FLAG-tagged proteins, was derived from pLHis (36) by replacing the NdeI-BamHI fragment with an insert containing a multicloning site to generate the EcoRI, SacI, and BamHI digestion sites.
GST-HBx, GST-TFIIB, GST-CTD, and the full-length and truncated GST-RPB5 expression plasmids have been described (13, 35). cDNA covering the long open reading frame was prepared by PCR with pBluescript II SK(+) plasmid containing the full-length RMP cDNA as a template and a primer set, GCGAGCTCCATGAGGCTAGGAAATGTA and GCGGATCCGTCTTTCTGTTGCAA, generating an artificial SacI site at the 5′ end and a BamHI site at the 3′ end, respectively. The PCR products were inserted into the SacI and BamHI sites of pSG5UTPL. The resultant plasmid, pSG5UTPL-RMP, was used as the template to construct truncated versions of RMP by PCR cloning. The sequences encoding RMP-D1, -D2, -D6, -D11, -D12, -D13, -D14, -D15, and -D16 have an initiation codon followed by codons corresponding to amino acids (aa) 137 to 231, 232 to 431, 137 to 315, 232 to 508, 88 to 136, 88 to 150, 175 to 231, 151 to 231, and 151 to 198 of RMP, respectively, and the sequences encoding RMP-D8, RMP-D7, and RMP-14D2 have codons corresponding to aa 1 to 87, 1 to 150, and 1 to 231, respectively. The internal-deletion mutants RMP-Id150 and RMP-Id175, lacking amino acid residues 151 to 231 and 175 to 231, respectively, were constructed by splicing PCR. The sequences encoding full-length or truncated fragments of RMP were inserted into the SacI or EcoRI and the BamHI sites of pSG5UTPL, pNKFLAG, pGENKS, and pYFLAG. All the constructs were sequenced by the dideoxy method with Taq sequencing kits and a DNA sequencer (370A; Applied Biosystems).
Preparation of recombinant proteins.
GST-fused proteins were expressed in E. coli by induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for 3 h. Cells were harvested and sonicated in PBST buffer (phosphate-buffered saline containing 1% Triton X-100). After centrifugation, the extracts (supernatants) were collected and stored at −80°C. For purification, the extracts were incubated with glutathione-Sepharose 4B (Pharmacia) at room temperature for 1 h. The beads were precipitated, washed four times with a 50-fold volume of PBST buffer, and then eluted with 10 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0). The eluted proteins were divided into aliquots and stored at −80°C.
Plasmid pLHis-RMP was expressed in BL21 (DE3)/pLys cells by induction with 0.4 mM IPTG at 30°C. Cells were harvested 3 h postinduction and sonicated in native binding buffer (20 mM sodium phosphate, 500 mM NaCl [pH 7.8]). Histidine-tagged RMPs were purified by incubating the sonication supernatant with nickel-resin (Invitrogen), followed by extensive washing and elution with imidazole elution buffer (300 mM imidazole, 20 mM sodium phosphate, 500 mM NaCl [pH 6.3]).
FLAG-tagged wild-type and mutant RMPs were expressed in BL21 by induction with 0.4 mM IPTG at 30°C for 3 to 6 h. Cells were harvested and sonicated in 50 mM Tris-HCl (pH 8.0)–150 mM NaCl–0.1% Triton X-100. After centrifugation, the supernatant was stored at −80°C. FLAG-tagged proteins were purified by incubating the sonication supernatant with anti-FLAG M2 resin (Kodak Scientific Imaging Systems), followed by several washes and elution with buffer containing FLAG peptide (0.2 mg of FLAG peptide per ml, 50 mM Tris-HCl [pH 8.0], 150 mM NaCl).
Protein-protein binding assays.
GST resin pull-down assays were carried out as reported previously (35). Approximately 1 μg of GST or of GST-fused RPB5 and its truncated mutants immobilized on 15 μl of glutathione-Sepharose 4B preblocked in 0.5% nonfat milk and 0.05% bovine serum albumin was incubated with 0.2 μg of full-length or mutated versions of FLAG-RMP, purified from E. coli lysates, in 0.5 ml of modified GBT buffer for 1 to 2 h on a rotator at 4°C. After being washed four times with modified GBT buffer, the bound proteins were eluted and subjected to Western blot analysis with anti-FLAG monoclonal antibody (M2).
Far-Western blotting was performed with ∼0.2 μg of purified GST fusion protein and GST control protein. The proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes. Proteins on membranes were denatured, renatured, and far-Western blotted as described for library screening.
Antibodies.
His-tagged RMP (aa 1 to 150) was used to raise rabbit anti-RMP antibodies. Briefly, approximately 200 μg of His-tagged RMP mixed with incomplete Freund’s adjuvant was injected into two animals subcutaneously. Booster injections consisting of 100 μg of proteins with a complete Freund’s adjuvant were given three times at 2-week intervals. Sera were taken by bleeding from the ear artery. Immunoglobulin G fractions were purified by affinity chromatography with protein A-Sepharose (Pharmacia LKB). Anti-RPB6 antibody was a generous gift from R. G. Roeder, and anti-CTD monoclonal antibody (7G5) was kindly provided by M. Vigneron. Anti-RPB5 antibody was reported previously (35), and anti-FLAG M2 antibody was commercially purchased (Kodak Scientific Imaging Systems).
Immunoprecipitation and Western blot analysis.
Transient transfection of COS1, HeLa, and HepG2 cells was carried out as reported previously (45). The cells were harvested; washed; sonicated in LAC buffer, which contained 10% glycerol, 20 mM HEPES (pH 7.9), 50 mM KCl, 0.4 M NaCl, 10 mM MgCl2, 0.1 mM dithiothreitol, 0.1 mM EDTA, 9 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 0.5 mM phenylmethylsulfonyl fluoride, and 10 μg each of aprotinin and leupeptin per ml; and centrifuged. The total cell lysates were stored at −80°C. Approximately 200 to 800 μg of total cell lysates in an appropriate volume of TBST buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20) was clarified by incubation with 50 μl of 50% swollen agarose beads for 30 min followed by centrifugation. The supernatants with FLAG-tagged proteins were immunoprecipitated with 20 μl of 50% anti-FLAG M2 resin and rotated for 2 h, followed by four washes with washing buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl). For precipitation of endogenous proteins, HepG2 cell lysates were incubated with the primary antibodies for several hours at 4°C, and then 20 μl of 50% swollen protein A resin was added. After being rotated for another 2 h and washed four times with washing buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% SDS, 1 mM EDTA), the bound proteins were eluted, fractionated by SDS-PAGE, transferred onto nitrocellulose membranes, and subjected to Western blot analysis with the secondary antibodies. The proteins were visualized by enhanced chemiluminescence according to the manufacturer’s instructions (Amersham).
Northern blot analysis.
An RMP cDNA probe was prepared with [α-32P]dCTP by using a random primer kit (Stratagene). Poly(A) RNA fractions of various human tissues (Clontech Laboratories) were subjected to Northern blotting with the RMP cDNA probe. The blots were hybridized with 32P-labeled probes for 1 to 2 h with ExpressHyb hybridization solution (Clontech Laboratories), washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 58°C, and exposed on X-ray film at −70°C for 4 h. Each lane contained approximately 2 μg of poly(A)+ RNA from each tissue. To control the relative amount of RNA in each lane, after hybridization with RMP, blots were stripped by incubation in 0.5% SDS at 95°C and reprobed with a human β-actin cDNA probe.
Transfection and CAT assays.
Cell culture, transient transfection, and chloramphenicol acetyltransferase (CAT) assays were carried out as reported previously (38, 44). The CAT reporters pHECx2CAT, pNFκBx3CAT, and pGalCAT have two tandem repeats of the HBV enhancer 1 (Enh1) core, three tandem repeats of the NFκB-binding site, and five tandem repeats of the Gal4-binding site, respectively, as binding sites for transcriptional activators. Total cell lysates were prepared from cells harvested 48 h after transfection. CAT assay reactions were performed for 60 min at 37°C with 20 μg of protein from the transfected cell lysates (44). The fractionated thin-layer chromatography plates were exposed on imaging plates, and the CAT activities were measured as the percentage of conversion to acetylated forms of [14C]chloramphenicol (Amersham) with a Bioimage analyzer (BAS1000; Fuji). Transfection and CAT assays were performed at least three times with each combination of transactivator and CAT reporter construct.
Nucleotide sequence accession number.
The nucleotide sequence of the cDNA clone has been deposited in the DNA Data Bank of Japan (DDBJ) and assigned no. AB 006572.
RESULTS
Cloning of RPB5-binding protein by far-Western blotting.
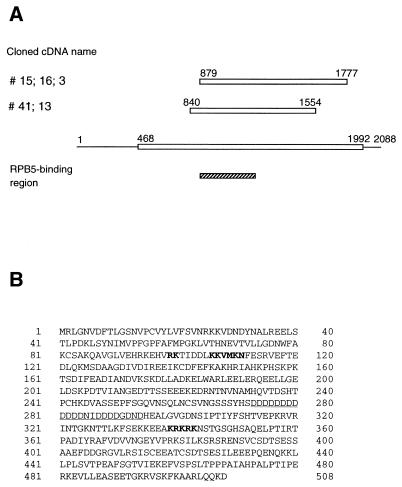
To identify cDNA encoding proteins that bind to RPB5, we applied far-Western blotting, a cloning method to detect protein-protein interaction in situ (13). A HepG2 cDNA library in a λ phage expression vector was screened with a labeled recombinant human RPB5 probe. After the third screening, seven positive clones were selected from a total of 107 plaques and subjected to sequence analysis of the insert termini. Of the five clones two groups had identical sequences in the inserts (Fig. 1A). The remaining two clones with short stretches of sequence in frame have not been subjected to further analysis.
FIG. 1.
DNA and amino acid sequence of RMP. (A) Schematic representation of five positive RMP cDNA clones screened by far-Western blotting. Open boxes indicate the predicted open reading frames. The RPB5-binding region was included in all the inserts of the positive clones. (B) Deduced amino acid sequence of RMP. The two different putative NLS are in bold letters, and the Asp-rich region is underlined.
All inserts of the five positive clones were subcloned and completely sequenced. Two different fragments covered 1.2 kb of cDNA encoding 306 continuous amino acids in frame with the LacZ gene. 5′ and 3′ rapid amplification of cDNA ends with HepG2 mRNA was applied to clone the full-length cDNA (see Materials and Methods), and a putative full-length clone of 2,088 bp was obtained. The cDNA harbors an open reading frame of 1.5 kb, beginning at a consensus sequence for initiation at nucleotide 468 and ending at nucleotide 1992. A nucleotide homology search of the GenBank and EMBL databases revealed that several human and rodent sequences of cDNA from different tissues matched different parts of the cloned cDNA with high scores. The 508-aa polypeptide RMP is a novel protein which has no homology to known sequences in protein databases. There is an Asp-rich region, including 13 contiguous Asp residues, in the central part of RMP which shows a high degree of homology to several unrelated proteins with different functions. RMP has no known motif except for two putative nuclear localization signals (NLS) from aa 98 to 111 and from aa 339 to 344 (Fig. 1B).
Expression of RMP mRNA and protein in mammalian cells.
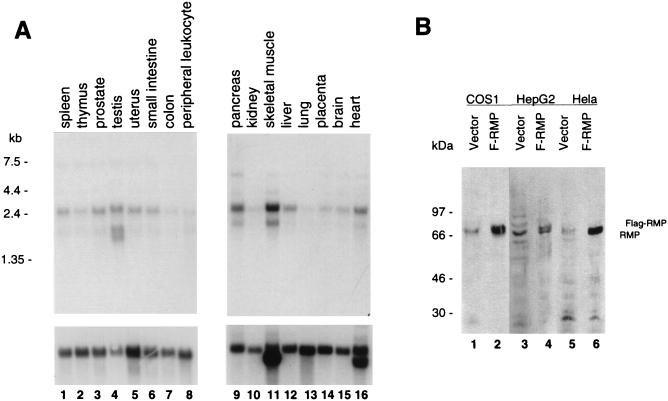
The expression of RMP mRNA was examined in RNA samples from various sources by Northern blot analysis (Fig. 2A). RMP mRNA (approximately 2.4 kb in length) was detected at different levels in all human tissues examined. An additional weak band of 1.6 kb was detected in some tissues, such as testis (Fig. 2, lane 4). Low levels of RMP expression were observed in peripheral tissue, leukocytes, and lung. Under high-stringency conditions, RMP mRNA was also detected in RNA samples of mouse and rat tissues (data not shown). These results suggest that the RMP gene is conserved among mammals and ubiquitously expressed in various tissues.
FIG. 2.
RMP is ubiquitously expressed in mammalian cells. (A) Poly(A) RNA fractions of various human tissues were subjected to Northern blotting with RMP (upper panels) and human β-actin cDNA probes (lower panels) as described in Materials and Methods. Each lane contained 2 μg of poly(A)+ RNA from each tissue. (B) Detection of endogenous and overexpressed RMPs. Total cell lysates of COS1, HepG2, and HeLa cells transiently transfected with the FLAG-RMP expression vector (F-RMP) or the pSG5UTPL vector alone were fractionated by SDS–10% PAGE and subjected to Western blotting with anti-RMP antibody.
The presence of RMP was examined in different cell lines by immunoblotting with polyclonal antibody raised against bacterially expressed recombinant His-tagged RMP. The anti-RMP antibody recognized a 69-kDa protein in human cell lines, such as HepG2 and HeLa, and also in the monkey cell line COS1. To confirm the specificity of the endogenous RMP detected with anti-RMP antibody, these cells were transiently transfected with FLAG-tagged RMP. A protein migrating slightly slower than the endogenous RMP was detected by anti-RMP antibody in the FLAG-RMP-transfected cell lysates but not in cells transfected with the vector alone (Fig. 2B). This protein was also detected with anti-FLAG M2 antibody (data not shown). The apparent molecular sizes of the endogenous and FLAG-tagged RMPs were much larger than the calculated molecular mass (57 kDa). The migration behavior of RMP detected by SDS-PAGE was due not to modification of the protein but probably to the high content of charged amino acid residues, since similar discrepancies were observed with bacterial recombinant RMPs.
RMP specifically interacts with RPB5 in vitro.
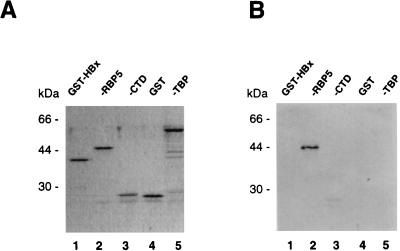
Since the positive clones of RMP cDNA were selected from a λgt11 library with a GST-RPB5 probe, it remained possible that the LacZ-fused RMP portion but not the RMP moiety was recognized by RPB5. To exclude this possibility, GST-RMP was used as a probe to examine the interaction between RPB5 and RMP by far-Western blot analysis under the same binding conditions as those used to isolate the RMP clones (Fig. 3). Comparable amounts of purified bacterial recombinant proteins were fractionated by SDS-PAGE (Fig. 3A) and subjected to far-Western blot analysis (Fig. 3B). Only GST-RPB5 (Fig. 3B, lane 2) bound to the RMP probe. The other GST-fused proteins, such as CTD and TBP, showed no binding to RMP. Interestingly, RMP did not bind HBx (Fig. 3, lanes 1). These results indicated the specificity of the binding of RMP and RPB5 in vitro.
FIG. 3.
RMP specifically binds RPB5 in vitro. Partially purified bacterial recombinant proteins were fractionated by SDS–12.5% PAGE and stained with Coomassie brilliant blue (A) or transferred onto nitrocellulose membranes for far-Western blot analysis (B). Coomassie brilliant blue staining showed that comparable amounts of GST-fused HBx, human RPB5, TBP, CTD, and GST proteins were applied. (B) The GST-RMP probe bound specifically to RPB5 but not to HBx, CTD, TBP, or GST.
The RPB5-binding region resides in the region shared by the positive clones.
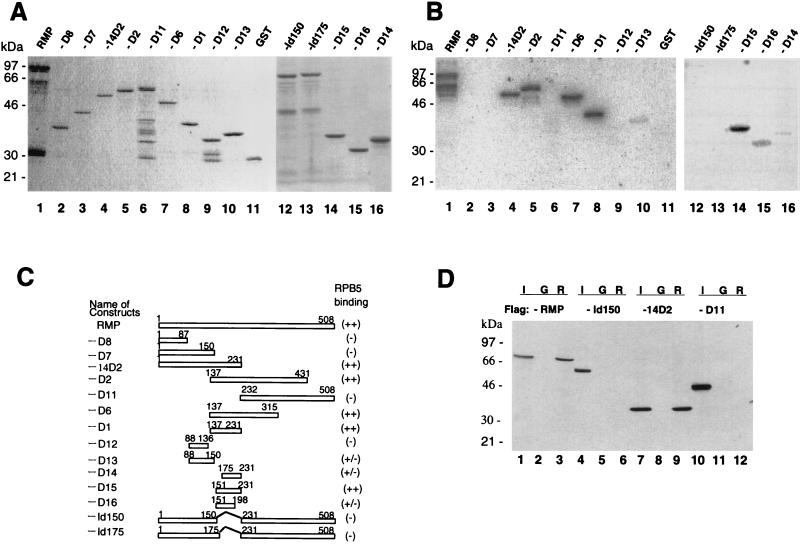
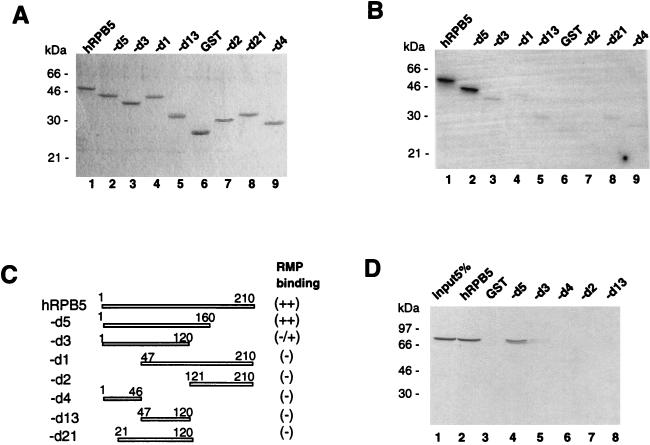
To further confirm the specific binding of RPB5, mapping of the RPB5-binding site in RMP was carried out with a GST-RPB5 probe (Fig. 4B). The GST-fused full-length and truncated RMPs were bacterially expressed and purified (Fig. 4A and C). The RPB5 probe bound strongly to the full-length RMP (Fig. 4A and B, lanes 1) as well as RMP-D1, -D2, -D6, -14D2, and -D15 (lanes 8, 5, 7, 4, and 14, respectively) and weakly to RMP-D13, -D14, and -D16 (lanes 10, 16, and 15, respectively) but not to the other truncated proteins or GST (lanes 2, 3, 6, and 11). Since RMP-D1 strongly bound RPB5, the RPB5-binding site was within the region from aa 137 to 231, the sequence of which was included in the two different inserts of the positive λgt11 phage clones (Fig. 1A). Further examination of the region around RMP-D1 with constructs RMP-D13 (aa 88 to 150), RMP-D14 (aa 176 to 231), RMP-D15 (aa 150 to 231), and RMP-D16 (aa 151 to 198) showed that only RMP-D15 had strong GST-RPB5-binding ability. RMP-D13, -D14, and -D16 had much weaker RPB5-binding ability than RMP-D15. Internal-deletion mutants of RMP, RMP-Id150 and RMP-Id175, did not exhibit RPB5 binding (Fig. 4A and B, lanes 12 and 13). These results indicated that the region covered by RMP-D16 (aa 151 to 198) is essential for RPB5 binding and that the region from aa 198 to 231 may have an accessory role in this binding (Fig. 4B, lane 14).
FIG. 4.
Delineation of the RPB5-binding region of RMP. (A) Coomassie brilliant blue staining of the fractionated full-length and truncated RMPs in GST-fused forms. (B) GST-RMP and truncated mutant proteins were subjected to far-Western blot analysis with a GST-RPB5 probe as described in the legend to Fig. 3. (C) Map of various RMP deletion constructs. The expression plasmids were constructed as described in Materials and Methods. The sequences encoding RMP-D1, -D2, -D6, -D11, -D12, and -D14 have an initiation codon followed by codons for various fragments of RMP. RMP-Id150 and RMP-Id175 have an internal deletion from aa 150 to 231 and from 175 to 231, respectively. The relative RPB5-binding abilities detected by far-Western blot analysis in panel B are shown at the right. (D) GST resin pull-down assay. Bacterially expressed FLAG-tagged full-length RMP and truncated mutants RMP-Id150, -14D2, and -D11 were incubated with GST (G) or GST-RPB5 (R). Approximately 1 μg of GST or GST-fused protein immobilized on glutathione resin was incubated with 100 ng of partially purified, bacterially expressed FLAG-RMP in GBT buffer for 2 h at 4°C. After being washed extensively, the bound proteins were eluted, fractionated by SDS–12.5% PAGE, and Western blotted with anti-FLAG M2 antibody. Lanes 1, 4, 7, and 10 show 5% of the input (I).
The mapping results by far-Western blot analysis were further confirmed by GST resin pull-down assay (Fig. 4D). Bacterially expressed FLAG-RMP (Fig. 4D, lane 3) and FLAG-RMP-14D2 (aa 1 to 231 [lane 9]) harboring the RPB5-binding region were efficiently recovered by the immobilized GST-RPB5 resin, whereas neither a C-terminal part, RMP-D11 (aa 232 to 508 [lane 12]), nor RMP-Id150 (lane 6) (lacking aa 151 to 231) was recovered. The negative-control GST bound none of the RMPs. From the consistent results obtained by the two different methods in vitro, we concluded that RMP specifically interacts with RPB5 through its central region.
The RMP-binding region covers the TFIIB- and HBx-binding sites in RPB5.
We used a similar approach to delineate the RMP-binding region of RPB5 by far-Western blot analysis (Fig. 5). The GST-RMP probe strongly bound to full-length RPB5 and RPB5-d5 (aa 1 to 160) but showed no or only weak binding to all other truncated forms of RPB5 (Fig. 5B, lanes 3 to 9). The region of RPB5 necessary for RMP binding was confirmed by GST resin pull-down assay (Fig. 5D). In this experiment, bacterially expressed and purified FLAG-RMP was incubated with immobilized GST fusion proteins harboring different portions of RPB5, and the recovered FLAG-tagged RMP was detected with an anti-FLAG antibody. Consistent with the results of far-Western blotting, RPB5-d5 as well as RPB5, but not the other truncated forms, associated with FLAG-RMP. These results suggest that RMP binding requires a region of more than two-thirds of RPB5, which completely covers both the TFIIB- and HBx-binding sites (aa 1 to 46 and 47 to 120, respectively) (13, 35).
FIG. 5.
Mapping of the RMP-binding region of RPB5. (A) Coomassie brilliant blue staining of the applied GST-RPB5 and its mutant proteins. (B) GST-fused RPB5 and its truncated mutants were subjected to far-Western blot analysis with the GST-RMP probe. (C) Schematic map of RPB5 deletion constructs. GST-fused forms have been reported previously (13, 28). The relative RPB5-binding abilities detected by far-Western blot analysis in panel B are shown at the right. (D) GST resin pull-down assay. Bacterial recombinant FLAG-RMP was incubated with immobilized GST, GST-RPB5, or GST-RPB-d5, -d3, -d4, -d2, or -d13 on resin as described in the legend to Fig. 4. The bound FLAG-RMP was detected by anti-FLAG M2 antibody. Lane 1 shows 5% of the input of the FLAG-RMP. h, human.
RMP binds RPB5 in vivo.
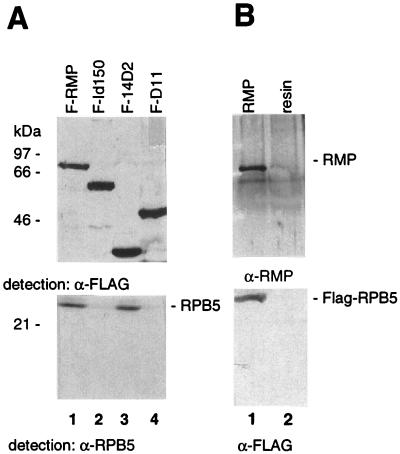
Next, we determined whether the interaction of RMP and RPB5 occurs in vivo by immunoprecipitation and Western blot analysis. COS1 cells were transiently cotransfected with mammalian expression vectors of RPB5 and FLAG-tagged RMP, and the cell lysates were precipitated with anti-FLAG M2 resin (Fig. 6A). The nontagged RPB5 protein was efficiently recovered in the immunoprecipitates with FLAG-RMP and the FLAG-RMP-14D2 mutant (Fig. 6A, lanes 1 and 3) but was not coprecipitated at all with FLAG-RMP-Id150 or RMP-D11 lacking the RPB5-binding region (lanes 2 and 4). Conversely, when the lysate of the COS1 cells cotransfected with FLAG-tagged RPB5 and RMP was precipitated with anti-FLAG M2 resin, FLAG-RPB5 also coimmunoprecipitated RMP (Fig. 6B, lane 1). RMP was barely recovered in the precipitates with anti-FLAG M2 resin in lysates of nontransfected cells (Fig. 6B, lane 2). These results clearly indicate that RMP interacts with RPB5 in mammalian cells and that the RPB5-binding region of RMP delineated in vivo is consistent with the mapping results obtained in vitro (Fig. 4).
FIG. 6.
RMP binds RPB5 in vivo. (A) COS1 cells were cotransfected with mammalian expression plasmids of RPB5 and FLAG-RMP (F-RMP), FLAG-RMP-Id150 (F-Id150), FLAG-RMP-14D2 (F-14D2), or FLAG-RMP-D11 (F-D11). Total cell lysates were immunoprecipitated with anti-FLAG M2 antibody-bound resin. After being washed, the bound proteins were eluted, fractionated by SDS–12.5% PAGE, and subjected to Western blotting with anti-FLAG antibody (α-FLAG) or anti-RPB5 antibody (α-RPB5). (B) COS1 cells were cotransfected with FLAG-RPB5 and RMP mammalian expression plasmids. Total cell lysates were precipitated as described for panel A and detected with anti-FLAG and anti-RMP (α-RMP) antibodies.
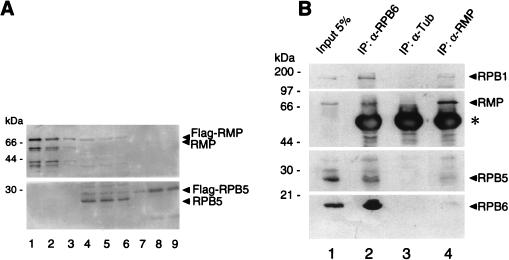
The interaction of RMP and RPB5 described above was detected in the lysates of cells overexpressing both proteins, but it may not reflect the interaction of endogenous RMP and RPB5. Therefore, we examined whether endogenous RMP interacts with RPB5 (Fig. 7). At first, relative amounts of endogenous RMP and RPB5 in HepG2 cells were determined immunologically (Fig. 7A). As determined by comparison of the band intensities of the various amounts of the purified recombinant RMP and RPB5, the amount of endogenous RMP in 60 μg of the total proteins is close to 2.5 ng of the recombinant RMP (Fig. 7A, lanes 3 and 4), and the amount of endogenous RPB5 in 20 μg of the total protein is close to 5 ng of the recombinant RPB5 (lanes 6 and 9). Therefore, the relative molecular ratio of endogenous RMP to RPB5 is approximately 1:10, indicating that RMP is substoichiometric to RPB5 or to RNA polymerases. Next, the possible interaction of endogenous RMP and RPB5 in HepG2 cells was examined by coimmunoprecipitation with anti-RMP antibody. RPB5 was weakly detected in the anti-RMP immunoprecipitates, and two other polymerase II subunits, RPB1 and RPB6, were also faintly detected in the immunoprecipitates, whereas all of the examined subunits were not recovered in the control immunoprecipitates (Fig. 7B, lanes 3 and 4). This result suggests that RMP, at least in part, is complexed with RNA polymerase II. To confirm this notion, coimmunoprecipitation with anti-RPB6 antibody was used to detect the RMP-polymerase II interaction, since the anti-RPB5 antibody we raised was not able to coimmunoprecipitate RMP and RNA polymerase subunits. Anti-RPB6 coimmunoprecipitated RMP and the other RNA polymerase subunits (Fig. 7B, lane 2). This result indicates that endogenous RMP is complexed with RNA polymerase II through RPB5 in HepG2 cells. The RPB5-binding region of RMP mapped in vitro is necessary for the association to RNA polymerase II, since anti-RPB6 antibody coimmunoprecipitated RMP but not RMP-Id150 in the overexpressed cell lysates (data not shown).
FIG. 7.
Endogenous RMP associates with RNA polymerase II complex. (A) Western blot analysis of molar ratio of endogenous RMP to RPB5 in HepG2 cells. Proteins were loaded in lanes as follows: lanes 4 to 6, 60, 40, and 20 μg, respectively, of HepG2 total cell lysate; lanes 1 to 3, 15, 5, and 2.5 ng, respectively, of bacterially expressed recombinant FLAG-RMP; and lanes 7 to 9, 2, 10, and 5 ng, respectively, of FLAG-RPB5. Proteins were fractionated by SDS-PAGE and subjected to Western blot analysis with anti-RMP (upper panel) and anti-RPB5 (lower panel) antibodies. (B) Coimmunoprecipitation of RMP with RNA polymerase II subunits. HepG2 cell lysates were precipitated with the indicated antibodies. The immunoprecipitates (IP) together with an aliquot of the lysate (Input 5%), were separated by SDS-PAGE and detected with antibodies directed against RPB1 (anti-CTD), RMP (α-RMP), RPB5, and RPB6 (α-RPB6). The asterisk shows the heavy chain of the first antibody used in the immunoprecipitation. α-Tub, antitubulin.
RMP counteracts transactivation by HBx.
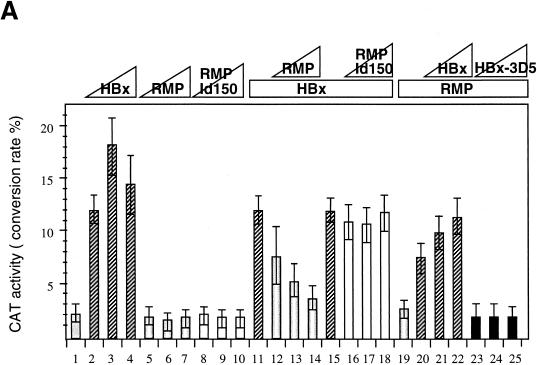
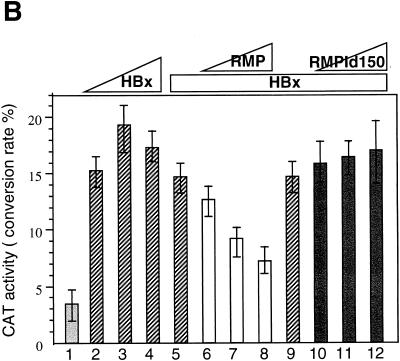
Because the RMP-binding region of RPB5 (aa 1 to 160) covers the regions necessary for TFIIB binding (aa 1 to 46) and HBx binding (aa 47 to 120), RMP may interfere with the trimeric interaction of RPB5, TFIIB, and HBx. Overexpression of RMP, therefore, may inhibit HBx transactivation. To examine this possibility, HepG2 cells were transiently cotransfected with RMP and HBx expression plasmids together with pHECx2CAT, an HBx-responsive CAT reporter, under the control of the X-responsive element derived from the core sequence in HBV Enh1 (Fig. 8A). Cotransfection of RMP inhibited the transactivation by HBx (Fig. 8A, bars 12 to 14), although the inhibition was not complete in the presence of excess RMP. Expression levels of HBx immunologically detected by anti-HBx antibody were similar among cotransfected cell lysates in the absence or presence of RMP (data not shown). Coexpression of RMP-Id150 and RMP-Id175, which lack 57 and 82 aa in the RPB5-binding region, respectively, exhibited no inhibitory effect on the transactivation by HBx (Fig. 8A, bars 16 to 18, and data not shown), implying that RPB5 binding is essential for the inhibitory effect. Because HBx has been reported to transactivate a wide variety of cis elements (16, 37, 38), we examined the possibility that RMP may inhibit transactivation of HBx through the other X-responsive cis element. A similar inhibitory effect of RMP was observed when pNFκBx3CAT, harboring three tandem repeats of the NFκB-binding site, another X-responsive cis element, was used as the reporter (Fig. 8B). This result suggests that RMP inhibits transactivation by HBx of different X-responsive cis elements.
FIG. 8.
RMP inhibits HBx transactivation. (A) Transfection of HepG2 cells and CAT assays were described in Materials and Methods. Various amounts of HBx and RMP plasmids were transfected together with the reporter plasmid, pHECx2CAT, in which the CAT gene is under the control of the dimeric sequence of the HBV Enh1 core. The transfected plasmid DNA was 5 μg of pHECx2CAT together with the following: bar 1, 5 μg of pSG5UTPL; bars 2 to 4, 1, 2, and 4 μg of pSG5UTPL-HBx, respectively; bars 5 to 7, 1, 2, and 4 μg of pSG5UTPL-RMP, respectively; bars 8 to 10, 1, 2, and 4 μg of pSG5UTPL-RMP-Id150, respectively; bars 11 to 14, 0, 1, 2, and 4 μg of pSG5UTPL-RMP, respectively, plus 1 μg of pSG5UTPL-HBx; and bars 15 to 18, 0, 1, 2, and 4 μg of pSG5UTPL-RMP-Id150, respectively, plus 1 μg of pSG5UTPL-HBx. HBx counteracted the corepressor activity of RMP in the system in a dose-dependent manner: bars 19 to 22, 0, 1, 3, and 4 μg of pSG5UTPL-HBx, respectively, plus 1 μg of pSG5UTPL-RMP; and bars 23 to 25, 1, 3, and 4 μg of pSG5UTPL-HBx-3D5, respectively, plus 1 μg of pSG5UTPL-RMP. The total amount of DNA added per transfection was adjusted to 10 μg with the control vector, pSG5UTPL. (B) RMP inhibits the transactivation by HBx of the reporter. The NFκBx3CAT harbors a trimeric repeat of the NFκB-binding site, which is responsive to HBx. The transfected plasmid DNA was 5 μg of pNFκBx3CAT together with the following: bar 1, 5 μg of pSG5UTPL; bars 2 to 4, 1, 2, and 3 μg of pSG5UTPL-HBx, respectively; bars 5 to 8, 0, 1, 2, and 3 μg of pSG5UTPL-RMP, respectively, plus 1 μg of pSG5UTPL-HBx; and bars 9 to 12, 0, 1, 2, and 3 μg of pSG5UTPL-RMP-Id150, respectively, plus 1 μg of pSG5UTPL-HBx. The total amount of DNA added per transfection was adjusted to 10 μg with the control vector, pSG5UTPL. Error bars show standard deviations.
HBx counteracts the inhibitory effect of RMP.
The inhibitory function of RMP on activated transcription implies that HBx may counteract the negative regulatory role of RMP, which may be involved in HBx transactivation. To address this possibility, we examined whether HBx can release the corepressor effect of RMP in the pHECx2CAT reporter system, in which the CAT gene is under the control of the dimeric sequence of the HBV Enh1 core. HBx counteracted the corepressor activity of RMP in this system in a dose-dependent manner (Fig. 8A, bars 19 to 22). The anticorepressor function of HBx resides in its transactivation domain, since HBx-5D1 bearing the transactivation domain similarly counteracted the corepressor function of RMP (data not shown). Overexpression of HBx-3D5 lacking the RPB5 binding site had no counteracting effect on RMP (Fig. 8A, bars 23 to 25).
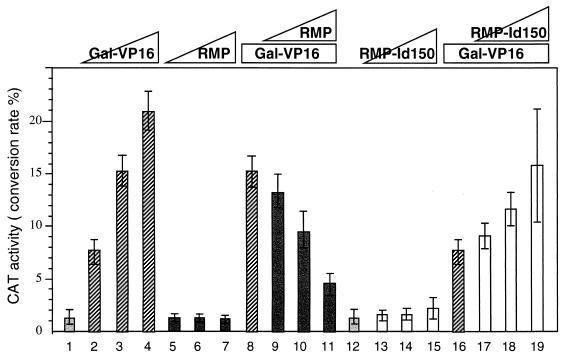
RMP inhibits activated transcription in mammalian cells.
Since RMP has no HBx-binding ability, the negative effect of RMP on transcription may occur in the absence of HBx. Therefore, we examined the effects of RMP on activated transcription by Gal-VP16, a chimeric activator with the Gal4 DNA-binding domain fused to the VP16 activation domain. pGalCAT, a CAT reporter with the simian virus 40 promoter driven by five Gal4-binding sites, was used as the reporter (Fig. 9). The transactivation of Gal-VP16 was inhibited twofold by the coexpression of RMP, indicating that RMP negatively affects a wide variety of activated transcriptions (Fig. 9, bars 9 to 11). No such effect was observed with pSG9RMP in which the RMP cDNA was inserted in the reverse orientation (data not shown). If RMP inhibits activated transcription through the RPB5 binding of RMP, deletion mutants lacking RPB5-binding abilities may have no such inhibitory effect. The internal-deletion mutant RMP-Id150, lacking the 82 amino acid residues in the RPB5-binding region, had no effect on pGalCAT in the absence of Gal-VP16 (Fig. 9, bars 12 to 15) but augmented CAT activity in the presence of Gal-VP16 (lanes 16 to 19). These results suggest that RMP has a corepressor activity and that the mutant RMP lacking the RPB5-binding ability acts positively, probably by perturbing the negative regulation in which endogenous RMP is involved.
FIG. 9.
RMP inhibits activated transcription by Gal-VP16. HepG2 cells were cotransfected with pGalCAT and Gal-VP16 together with HBx or RMP constructs. The transfected plasmid DNA was 5 μg of pGalx5CAT together with the following: bars 1 to 4, 0, 5, 10, and 20 ng of pGal-VP16, respectively; bars 5 to 7, 1, 2, and 3 μg of pSG5UTPL-RMP, respectively; bars 8 to 11, 0, 1, 2, and 3 μg of pSG5UTPL-RMP, respectively, plus 10 ng of pGal-VP16; bars 12 to 15, 0, 1, 2, and 3 μg of pSG5UTPL-RMP-Id150, respectively; and bars 16 to 19, 0, 1, 2, and 3 μg of pSG5UTPL-RMP-Id150, respectively, plus 5 ng of pGal-VP16. The total amount of DNA added per transfection was adjusted to 10 μg with the control vector, pSG5UTPL. Error bars show standard deviations.
DISCUSSION
Most RNA polymerase subunits are unique to their respective RNA polymerases, while some subunits are common among RNA polymerases I, II, and III. With the exception of RPB5, all of the common subunits in human RNA polymerases I, II, and III can substitute for their yeast homologues, although RPB5 is highly conserved among humans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe, and is essential in yeast (40, 41, 55). Yeast RPB5 has been reported to interact with RPB3 in vitro, and both RPB5 and RPB3 are present in two molar amounts in RNA polymerase II (4, 55) and seem to play an important role in subunit assembly (29, 43), as does human RPB5 (1). In a recent study using photo-cross-linking of RNA polymerase II preinitiation complex on DNA, RPB5 was suggested as one of the subunits of RNA polymerase II positioned close to DNA (27). Although anti-RPB5 antibody has been reported to block transcription in vitro, the actual role of RPB5 in transcription remains unclear. It has previously been demonstrated that human RPB5 directly interacts with HBx and TFIIB and that the trimeric interaction seems to be necessary for HBx transactivation (13, 35). Here we report that a novel protein, RMP, specifically binds to RPB5 both in vitro and in vivo and negatively modulates transcription through binding to RPB5. Endogenous RMP associates, at least partially, with the assembled RPB5 in RNA polymerase II in HepG2 cells as detected immunologically. The results with RMP further support the hypothesis that RPB5 is a communicating subunit of RNA polymerase II for transcriptional modulators (13, 35). Evidence for transcriptional regulation through the interaction of RNA polymerase subunits and modulators has been accumulating (6, 8, 9). Interestingly, recent studies showed that hTAFII68 and its homologue pro-oncoproteins, TLS/FUS and EWS, complexed with TFIID and RNA polymerase II. These oncoproteins seem to modulate RNA polymerase II activity by interacting with RNA polymerase II subunits, including RPB3 and RPB5 (8, 9). Therefore, the multiple interactions of RPB5 with the transcriptional regulators may be reminiscent of the activator-binding property of the prokaryotic α subunit, provided that the core assembly of the two molars of RPB5 and RPB3 has a short stretch of homology, or “α-motif” (1, 29, 32, 43).
RMP has inhibitory effects on various types of activated transcription, but they seem not to be global, since activated transcription by p53 seems to be unaffected by RMP (data not shown). The inhibitory effect of RMP requires the RPB5-binding region, and RMP may interfere with the association of TFIIB and RPB5, since the TFIIB-binding region is overlapped by the RMP-binding region of RPB5 (35). Because overexpression of HBx released the inhibitory effect of RMP on activated transcription, it remains to be determined whether HBx replaces RMP complexed with RPB5 and facilitates the interaction of TFIIB and RPB5. The negative effect of RMP on the activated transcription by Gal-VP16 suggests that RMP behaves as a corepressor. Such a corepressor function of RMP seems to be substantial, since HBx acts as a coactivator in the same artificial reporter system (21, 23) but could not act as a transactivator in the Gal4-fused form (36a). Taken together, these results imply that HBx may be a functional antagonist of RMP and that both RMP and HBx act as transcriptional cofactors which require direct communication with RPB5. However, this notion should be evaluated carefully, since RMP is substoichiometric to RPB5 and even overexpressed RMP has a moderate inhibitory effect on activated transcription. Interestingly, the internal-deletion mutant RMP-Id150 lacking the RPB5-binding ability positively augmented Gal-VP16-activated transcription but had no effect on transactivation by HBx. RMP-Id150 may augment transcription by competitively binding to the partner(s) of endogenous RMP, whereas HBx by itself competes out endogenous RMP for RPB5 binding. It is possible that RMP requires signaling processes for its function (2, 12, 53) or that RMP negatively modulates genes in the chromatin structure (49). Additionally, RMP may require a functional partner(s) for its function in addition to the RPB5 binding.
RMP has no known motifs except for two different putative NLS, the roles of which are not yet clear (unpublished data). Interestingly, RMP has several long helix-rich regions in which many charged amino acid residues are clustered. These characteristics are also found in RPB5. The amino acid sequence of RMP harbors 13 contiguous Asp residues in its middle region, in which 18 Asp residues are clustered with 24 amino acid residues. Interestingly, the 13-Asp stretch is encoded by two different stretches of the triplets GAC and GAT. Thus, the Asp-rich region seems to avoid destabilizing the DNA structure by a long triplet repeat (33), although this region is dispensable for the RPB5 binding and the inhibitory effect on activated transcription.
ACKNOWLEDGMENTS
We are grateful to R. G. Roeder for anti-RPB6 antibody, to M. Vigneron for anti-CTD monoclonal antibody, and to R. G. Roeder and A. Ishihama for encouraging discussions. We thank C. Matsushima, F. Momoshima, M. Yasukawa, and K. Kuwabara for their technical assistance.
This study was partly supported by a Science Grant from the Ministry of Education and Culture of Japan.
REFERENCES
- 1.Acker J, Graaf D M, Cheynel I, Khazak V, Kedinger C, Vigneron M. Interactions between the human RNA polymerase II subunits. J Biol Chem. 1997;272:16815–16821. doi: 10.1074/jbc.272.27.16815. [DOI] [PubMed] [Google Scholar]
- 2.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Agus N S, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 3.Antunovic J, Lemieux N, Cromlish J A. The 17 kDa HBx protein encoded by hepatitis B virus interacts with the activation domains of Oct-1, and functions as a coactivator in the activation and repression of a human U6 promoter. Cell Mol Biol Res. 1993;39:463–482. [PubMed] [Google Scholar]
- 4.Archambault J, Friesen J D. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aufiero B, Schneider R J. The hepatitis B virus X-gene product transactivates both RNA polymerase II and III promoters. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awrey D E, Weilbaecher R G, Hemming S A, Orlicky S M, Kane C M, Edwards A M. Transcriptional elongation through DNA arrest sites: a multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J Biol Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 7.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertolotti A, Lutz Y, Heard D J, Chambon P, Tora L. hTAF68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS, is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 9.Bertolotti A, Melot T, Acker J, Vigneron M, Delattre O, Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol Cell Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen H-S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 13.Cheong J H, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conaway R C, Conaway J W. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 15.Cross J C, Wen P, Rutter J W. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen-activated cellular serine/threonine kinase. Proc Natl Acad Sci USA. 1993;90:8078–8082. doi: 10.1073/pnas.90.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and a nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feitelson M A, Zhu M, Duan L X, London W T. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 18.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 19.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 21.Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haviv I, Vaizel D, Shaul Y. pX, the HBV-encoded coactivator, interacts with components of the transcription machinery and stimulates transcription in a TAF-independent manner. EMBO J. 1996;15:3413–3420. [PMC free article] [PubMed] [Google Scholar]
- 23.Haviv I, Vaizel D, Shaul Y. The X protein of hepatitis B virus coactivates potent activation domains. Mol Cell Biol. 1995;15:1079–1085. doi: 10.1128/mcb.15.2.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inostroza J M, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 25.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 26.Kim C M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 27.Kim T K, Lagrange T, Wang Y H, Griffith J D, Reinberg D, Ebright R H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y-J, Bjorklund S, Li S, Sayer M H, Kornberg R D. A multiprotein complex mediator of transcriptional activation and its interaction with C-terminal domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 29.Kimura M, Ishiguro A, Ishihama A. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 30.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 31.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 32.Lalo D, Carles C, Sentenac A, Thuriaux P. Interactions between three common subunits of yeast RNA polymerases I and III. Proc Natl Acad Sci USA. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La-Spada A R, Paulson H L, Fischbeck K H. Trinucleotide repeat expansion in neurological disease. Ann Neurol. 1994;36:814–822. doi: 10.1002/ana.410360604. [DOI] [PubMed] [Google Scholar]
- 34.Lill N L, Grossman S R, Ginberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y, Nomura T, Cheong J H, Dorjsuren D, Iida K, Murakami S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcriptional factor IIB and RNA polymerase II subunit 5. J Biol Chem. 1997;272:7132–7139. doi: 10.1074/jbc.272.11.7132. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y, Nomura T, Yamashita T, Dorjsuren D, Tang H, Murakami S. The transactivation and p53-interacting functions of hepatitis B virus X protein are mutually interfering but distinct. Cancer Res. 1997;57:5137–5142. [PubMed] [Google Scholar]
- 36a.Lin Y, Tang H, Nomura T, Dorjsuren D, Hayashi N, Wei W, Ohta T, Roeder R, Murakami S. The hepatitis B virus X protein is a co-activator of activated transcription that modulates the transcription machinery and distal binding activators. J Biol Chem. 1998;273:27097–27103. doi: 10.1074/jbc.273.42.27097. [DOI] [PubMed] [Google Scholar]
- 37.Lucito R, Schneider R J. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J Virol. 1992;66:983–991. doi: 10.1128/jvi.66.2.983-991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahe Y, Mukaida N, Kuno K, Akiyama M, Ikeda N, Matsushima K, Murakami S. Hepatitis B virus X protein transactivates human interleukin-8 gene through acting on nuclear factor kB and CCAAT/enhancer-binding protein-like cis-elements. J Biol Chem. 1991;266:13759–13763. [PubMed] [Google Scholar]
- 39.Maldonaldo E, Siekhatter R, Shelton M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 40.McKune K, Moore P A, Hull M W, Woychik N A. Six human RNA polymerase subunits functionally substitute for their yeast counterparts. Mol Cell Biol. 1995;15:6895–6900. doi: 10.1128/mcb.15.12.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKune K, Woychik N A. Functional substitution of an essential yeast RNA polymerase subunit by a highly conserved mammalian counterpart. Mol Cell Biol. 1994;14:4155–4159. doi: 10.1128/mcb.14.6.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meisterernst M, Roeder R G. Family of proteins that interacts with TFIID and regulates promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 43.Miyao T, Yasui K, Sakurai H, Yamaguchi M, Ishihama A. Molecular assembly of RNA polymerase II from fission yeast Schizosaccharomyces pombe: subunit-subunit contact network involving RPB5. Genes Cells. 1996;1:843–854. doi: 10.1046/j.1365-2443.1996.730274.x. [DOI] [PubMed] [Google Scholar]
- 44.Murakami S, Cheong J, Kaneko S. Human hepatitis B virus X gene encodes a regulatory domain that represses transactivation of X protein. J Biol Chem. 1994;269:15118–15123. [PubMed] [Google Scholar]
- 45.Murakami S, Cheong J, Ohno S, Matsushima K, Kaneko S. Transactivation of human hepatitis B virus X protein, HBx, operates through a mechanism distinct from protein kinase C and okadaic acid activation pathways. Virology. 1994;199:243–246. doi: 10.1006/viro.1994.1119. [DOI] [PubMed] [Google Scholar]
- 46.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 47.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 48.Sentenac A. Eukaryotic RNA polymerases. Crit Rev Biochem. 1985;18:31–91. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- 49.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 50.Su F, Schneider R J. Hepatitis B virus HBx protein activates transcription factor NF-κB by acting on multiple cytoplasmic inhibitors of rel-related proteins. J Virol. 1996;70:4558–4566. doi: 10.1128/jvi.70.7.4558-4566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terradillos O, Billet O, Renard C A, Levy R, Molina T, Briand P, Buendia M A. The hepatitis B virus X gene potentiates c-myc induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 52.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 53.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R. RNA polymerase holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 55.Woychik N A, Liao S M, Kolodziej P A, Young R A. Subunits shared by eukaryotic nuclear RNA polymerases. Genes Dev. 1990;4:313–323. doi: 10.1101/gad.4.3.313. [DOI] [PubMed] [Google Scholar]