Abstract
In the Saccharomyces cerevisiae Msh2p-Msh6p complex, mutations that were predicted to disrupt ATP binding, ATP hydrolysis, or both activities in each subunit were created. Mutations in either subunit resulted in a mismatch repair defect, and overexpression of either mutant subunit in a wild-type strain resulted in a dominant negative phenotype. Msh2p-Msh6p complexes bearing one or both mutant subunits were analyzed for binding to DNA containing base pair mismatches. None of the mutant complexes displayed a significant defect in mismatch binding; however, unlike wild-type protein, all mutant combinations continued to display mismatch binding specificity in the presence of ATP and did not display ATP-dependent conformational changes as measured by limited trypsin protease digestion. Both wild-type complex and complexes defective in the Msh2p ATPase displayed ATPase activities that were modulated by mismatch and homoduplex DNA substrates. Complexes defective in the Msh6p ATPase, however, displayed weak ATPase activities that were unaffected by the presence of DNA substrate. The results from these studies suggest that the Msh2p and Msh6p subunits of the Msh2p-Msh6p complex play important and coordinated roles in postmismatch recognition steps that involve ATP hydrolysis. Furthermore, our data support a model whereby Msh6p uses its ATP binding or hydrolysis activity to coordinate mismatch binding with additional mismatch repair components.
In organisms ranging from Escherichia coli to humans, mismatch repair pathways that recognize and repair both base pair mismatches and small insertion/deletion mismatches have been identified. Such mismatches can result from DNA replication errors, genetic recombination, and DNA damage; if uncorrected, these errors become fixed in the genome upon DNA replication (reviewed in references 15, 34, 41, and 42). In E. coli, the MutHLS system directs repair of base pair mismatches that result from DNA replication errors so that the newly replicated DNA strand is excised (reviewed in references 15, 34, 41, and 42). Our understanding of MutHLS repair was aided by the development of an E. coli in vitro mismatch repair reaction that was reconstituted from purified components (36). This assay showed that three components, MutSp, MutLp, and MutHp, played critical roles in initiating the repair process. A MutSp dimer appears to be the key mismatch recognition protein, as it binds to heteroduplex DNA containing base pair mismatches and up to 3-nucleotide (nt) insertions/deletions (41, 45, 54, 55). MutLp is thought to play the role of a molecular matchmaker by binding to the MutSp-mismatch DNA complex and mediating the activation of MutHp, an endonuclease that nicks only the unmethylated strand of hemimethylated GATC sites that are present immediately after passage of the replication fork (7, 20, 41, 50). Strand incision by MutHp, which can occur 3′ or 5′ to a mismatch, is then followed by excision, resynthesis, and ligation steps, resulting in the removal of the mismatch on the newly replicated strand, with the parental DNA strand serving as a template for repair (7).
Components of the mismatch repair reaction appear to be highly conserved in prokaryotes and eukaryotes, as homologs of E. coli MutSp and MutLp have been identified in Saccharomyces cerevisiae, Xenopus, Drosophila, mouse, and human cells (reviewed in references 34 and 42). In S. cerevisiae, six mutS homologs (MSH1 to MSH6) and four mutL homologs (PMS1, MLH1, MLH2, and MLH3) have been identified (reviewed in references 15, 34, and 42). It is unclear how parental and replicated DNA strands are distinguished in eukaryotes, as methylation does not appear to play a role in strand discrimination and no eukaryotic homologs of mutH have been identified (15, 34, 42). It is also unclear why there are so many MutSp and MutLp homologs in eukaryotes whereas only a single homolog of each protein is found in E. coli. One possibility is that multiple homologs evolved to allow for specialized mismatch recognition functions. Studies of both yeast and human cells support this idea: in both organisms, a heterodimer of Msh2p and Msh6p forms to repair mismatches resulting from nucleotide substitution and single-nucleotide insertion/deletion mutations, and a heterodimer of Msh2p and Msh3p forms to repair DNA slippage events that result in 2- to 4-nt insertion/deletion mismatches (1, 2, 17, 23–25, 29, 31, 33, 38, 43, 44).
While all of the components in the E. coli mismatch repair system have been identified, little is known at the mechanistic level about how mismatch recognition by MutSp and its homologs results in the activation of downstream components. Studies to address this issue have focused on biochemical interactions between purified components and structure-function analyses of the MutSp and MutLp homologs (5, 7, 16, 20, 25, 30, 36, 47). These studies have indicated that ATP binding, hydrolysis, or both function in key control points in the mismatch repair reaction. Of the three components required for mismatch recognition and incision of the newly replicated strand in E. coli, only MutSp and its homologs have been shown to bind and hydrolyze ATP via a highly conserved Walker type A nucleotide binding motif (5, 22, 30). Mutant MutS and Msh2 proteins (referred to as mutSp and msh2p, respectively) that contain amino acid substitutions in the nucleotide binding domain are defective in mismatch repair and confer a dominant negative phenotype when overexpressed in wild-type strains (5, 22, 30, 59).
A series of biochemical and genetic studies of bacteria, S. cerevisiae, and humans have suggested that MutSp homolog-ATP interactions play an important role in multiple steps in the mismatch repair reaction. These studies have shown that ATP is important for the modulation of mismatch recognition, the recruitment of additional mismatch repair factors, in some cases the translocation of the mismatch repair complex along DNA, and the activation of proteins such as MutHp that play a role in strand discrimination steps (5–7, 17, 19–21, 25, 30, 36, 44, 57). Studies that demonstrated the ATP requirement in mismatch repair included the following. First, in the bacterial, yeast, and human systems, ATP or the nonhydrolyzable analog ATPγS was shown to be required for the formation of MutSp homolog-MutLp homolog complexes at a mismatch site (20, 21, 25). Second, in the presence of ATP or ATPγS, the mismatch binding specificity of the bacterial, yeast, and human MutSp homologs was dramatically decreased (2, 17, 19, 20, 31). Finally, in the presence of ATP, MutSp and MutLp formed α-shaped looped structures on linear DNA substrates containing a mismatch (6). The positioning of the MutSp dimer at the base of the loop was consistent with the idea that ATP hydrolysis by MutSp enabled a MutSp-MutLp complex to translocate bidirectionally away from a mismatch site (6). Such a proposed activity is attractive because it can also explain how a complex of MutSp and MutLp could, in a step requiring ATP hydrolysis, encounter and activate MutHp at hemimethylated GATC sites located several kilobase pairs away from a mismatch (6, 7, 36).
While the amino acid sequences of the MutSp homologs are highly conserved between prokaryotes and eukaryotes, the organization of the eukaryotic mismatch binding factors into Msh2p-Msh6p and Msh2p-Msh3p complexes that display different mismatch binding specificities raises questions about the role of each subunit in mismatch recognition. While ATP appears to modulate the mismatch binding specificity of the eukaryotic and prokaryotic MutS homolog proteins in similar ways, it is unclear whether the two ATPases that are present in MutSp homolog complexes are coordinated and whether eukaryotic homologs display an in vitro translocation activity similar to that observed for MutSp. The role that each subunit plays in mediating interactions between MutSp and MutLp homologs and between factors involved in strand discrimination is also unclear.
The differences between the prokaryotic and eukaryotic systems encouraged us to examine the eukaryotic MutSp homolog subunit’s interactions with ATP. We focused on the S. cerevisiae Msh2p-Msh6p complex as a model because genetic analysis of msh2 and msh6 null mutations indicated that the Msh2p-Msh6p complex plays a major role in recognizing base pair and single-nucleotide insertion/deletion mismatches (33, 38) and biochemical analyses indicated that the Msh2p-Msh6p complex displayed both mismatch binding and ATP hydrolysis activities (2, 31). In this study, we used genetic and biochemical analyses to show that the ATPase activity of each subunit of Msh2p-Msh6p is coordinated during at least two discrete steps in mismatch repair. First, this analysis indicated that both the Msh2p and Msh6p subunits appear to be equally required in mediating ATP-dependent conformational changes in the complex that result in the modulation of mismatch binding specificity. Second, in mutant complex analyses, we observed that the Msh6p ATPase activity was responsive to the presence of mismatched DNA substrates whereas the Msh2p ATPase appeared unresponsive. Third, genetic analyses of strains overexpressing msh2 or msh6 gene products that contain ATP binding domain mutations resulted in a dominant negative phenotype. Taken together, our data suggest a role for the Msh6p subunit in relaying mismatch binding signals and a role for both subunits in downstream mismatch repair functions once the signaling event has been completed. A model consistent with the data obtained is presented.
MATERIALS AND METHODS
Strains and genetic procedures.
E. coli and yeast strains were grown under conditions described previously (5, 40, 49). E. coli RKY1400 (thr leuB6 thi thyA trpC1117 hsrk12 hsmk12 Strr recA13) was kindly provided by R. Kolodner and was used to amplify and manipulate all plasmids described in this report. S. cerevisiae FY23 (MATa ura3-52 leu2Δ1 trp1Δ63 [58]) and the FY23 derivatives EAY252 (MATa ura3-52 leu2Δ1 trp1Δ63 msh2Δ::TRP1), EAY420 (MATa ura3-52 leu2Δ1 trp1Δ63 msh3Δ::hisG), and EAY337 (MATa ura3-52 leu2Δ1 trp1Δ63 msh6Δ::hisG) were used in dominance and complementation studies. These strains were transformed with the following episomal vectors individually or in pairs: pEAE51 (GAL10-MSH6 TRP1 2μm [5]), pEAE84 (GAL10-msh6-GD987 TRP1 2μm [this report]), pEAE20 (GAL10-MSH2 URA3 2 μm [3]), pEAE27 (GAL10-msh2GD693 URA3 2μm [5]), pEAE86 (GAL10-MSH2 TRP1 2μm [this report]), and pEAE87 (GAL10-msh2-GD693 TRP1 2μm [this report]). S. cerevisiae BJ5464 (MATα ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) was obtained from the Yeast Genetic Stock Center and was the parental strain used for the overexpression and purification of Msh2p-Msh6p and the mutant derivative complexes. Msh2p-Msh6p complex was purified from BJ5464 transformed with pEAE20 and pEAE51 (EAY359), msh2-GD693p-Msh6p complex was purified from BJ5464 transformed with pEAE27 and pEAE51 (EAY360), Msh2p-msh6-GD987p complex was purified from BJ5464 transformed with pEAE20 and pEAE84 (EAY532), and msh2-GD693p-msh6-GD987p complex was purified from BJ5464 transformed with pEAE27 and pEAE84 (EAY533).
Yeast strains were transformed with episomal vectors by the lithium acetate method described by Geitz and Schiestl (18). Mutation rates in FY23, EAY420, and EAY337 strains containing wild-type and mutant MSH2 and MSH6 overexpression plasmids were determined by measuring forward mutations to canavanine resistance (4, 48). DNA slippage events were measured by detecting frameshift events resulting in resistance to 5-fluoro-orotic acid (5-FOA) in FY23-derived strains containing pEAA69 [(GT)16-URA3 ARSH4 CEN6 LEU2], a LEU2 plasmid derived from pSH44 (27, 53). In both the mutator and DNA slippage studies, tested strains were streaked to form single colonies on selective minimal plates containing 2% galactose and 2% sucrose. Eleven independent colonies were suspended in water, and appropriate dilutions were then plated onto minimal medium containing 2% each galactose and sucrose with or without canavanine (Sigma, St. Louis, Mo.) for the mutator assays or with or without 5-FOA (U.S. Biological, San Antonio, Tex.) for the DNA slippage studies (27, 48, 49). The median frequency of canavanine and 5-FOA resistance was determined for each strain. Each experiment was repeated 2 to 10 times. We chose to analyze our genetic data by the Wilcoxon rank-sum test to avoid making assumptions about the shape of the population distribution. Most data were evaluated by this rank-sum test, where P values of <0.05 were considered significant (8). The DNA slippage data were analyzed by the χ2 test, where P values of <0.05 were considered significant (8).
Media, reagents, and chemicals.
Trypsin and endo-Glu proteases were a gift from the Cornell Biotechnology analytical-synthesis facility. ATP, GTP, and deoxynucleoside triphosphates (dNTPs) were purchased from Pharmacia (Uppsala, Sweden); [γ-32P]ATP was obtained from Amersham (Arlington Heights, Ill.), and ADP and AMP-PNP (adenylyl-imidodiphosphate) were purchased from Sigma and Boehringer Mannheim (Indianapolis, Ind.), respectively. BA85 0.45-μm-pore-size nitrocellulose filters were purchased from Schleicher & Schuell (Keene, N.H.). Protein concentrations were determined by the Bradford dye method, using bovine serum albumin (BSA) as a standard (12), and reagents were obtained from Bio-Rad (Richmond, Calif.). Purified antihemagglutinin (anti-HA) mouse monoclonal antibody (clone 12CA5) was purchased from Boehringer Mannheim. For column chromatography, PBE94 and Resource Q were purchased from Pharmacia, and single-stranded DNA (ssDNA)-cellulose (catalog no. D8273) was purchased from Sigma; all resins were precycled according to the manufacturers’ instructions.
To obtain anti-Msh2p and anti-Msh6p polyclonal antibodies, rabbits were immunized with sodium dodecyl sulfate (SDS)-polyacrylamide gel-isolated Msh2p and Msh6p by previously described methods (26). A single rabbit was injected with one initial and two booster injections for each antigen (∼100 μg of each polypeptide per injection). Rabbits were housed and handled by members of the Center for Research Animal Resources, Cornell University. Western blotting was performed according to the manufacturer’s instructions, using the Immun-blot alkaline phosphatase assay system (Bio-Rad). Polypeptides were transferred to nitrocellulose by using a Bio-Rad semidry electrophoretic transfer system and incubated with primary antibody at a 1:5,000 dilution overnight and anti-rabbit immunoglobulin G secondary antibody at a 1:3,000 dilution for 2 h.
Nucleic acid techniques.
All restriction endonucleases, T4 polynucleotide kinase, T4 DNA ligase, T4 DNA polymerase, and Vent polymerase were from New England Biolabs and used according to manufacturer’s specifications. Oligonucleotide synthesis and double-stranded DNA sequencing of the entire subcloned fragment used to make the msh6-GD987 allele was performed at the Cornell Biotechnology analytical-synthesis facility. High-pressure liquid chromatography-purified oligonucleotides used in the filter binding studies were purchased from Operon Technologies, Inc. (Alameda, Calif.). Oligonucleotide concentration determination, annealing conditions, and 5′ labeling using [γ-32P]ATP and T4 polynucleotide kinase were performed as described previously (3, 13). The DNA sequences of the +1 and homoduplex oligonucleotides used in the filter binding studies are the same as described by Alani et al. (5).
Site-directed mutagenesis of MSH6 to create the msh6-GD987 allele in pEAE84 was performed by the overlap extension PCR method (28). pEAA69 [LEU2 promoter-(GT)16-URA3 ARSH4-CEN6 LEU2] was derived from pSH44 (27) and was constructed by inserting the 2.0-kb HindIII LEU2 promoter-(GT)16-URA3 fragment from pSH44 into the HindIII site of pRS305 (11a, 14). Plasmid DNA was isolated by alkaline lysis, and all DNA manipulations were performed as described previously (37).
Biochemical techniques.
Overexpression, purification, and gel filtration analysis of Msh2p-Msh6p was performed as described previously (2). Msh2p-msh6-GD987p, msh2-GD693p-Msh6p, and msh2-GD693p-msh6-GD987p complexes were purified and analyzed by the same procedures. For the trypsin proteolysis studies, the procedure for purification of the Msh2p-Msh6p complex was modified as follows. After elution from ssDNA-cellulose, Msh2p-Msh6p fractions were applied in 200 mM NaCl–1× buffer A (25 mM Tris [pH 7.5], 1 mM EDTA, 10 mM β-mercaptoethanol) to a 0.67-cm2 by 1.0-cm Source 30Q column (Pharmacia) at 10 ml/h and washed with 5 volumes of 200 mM NaCl–buffer A. The column was then eluted with 30 ml of a linear gradient from 0.20 to 1.0 M NaCl run in buffer A at 10 ml/h. Peak fractions containing Msh2p-Msh6p protein eluted at ∼400 mM NaCl. These fractions were pooled and concentrated up to 5 mg/ml with a Microcon 50 concentrator as instructed by the manufacturer (Amicon). The purity of protein preparations was monitored by SDS-polyacrylamide gel electrophoresis (PAGE) with 8% polyacrylamide gels (35) and by measuring binding to +1 and homoduplex oligonucleotide substrates (see Fig. 1 and 2) (2).
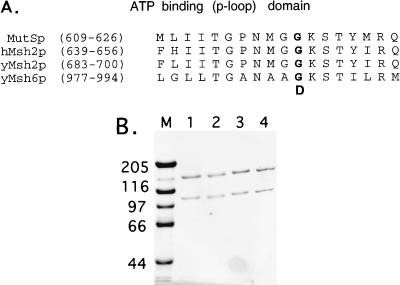
FIG. 1.
(A) Alignment of the P-loop motif of purine nucleotide binding proteins found in E. coli MutSp, human Msh2p, and S. cerevisiae Msh2p and Msh6p. The amino acid substitutions resulting in the msh2-GD693 and msh6-GD987 alleles are indicated in bold. (B) SDS-PAGE (8% gel) analysis of purified Msh2p-Msh6p (lane 1), msh2p-GD693p-Msh6p (lane 2), Msh2p-msh6-GD987p (lane 3), and msh2-GD693p-msh6-GD987p (lane 4) complexes. M, molecular weight standards; relative molecular masses are indicated in kilodaltons.
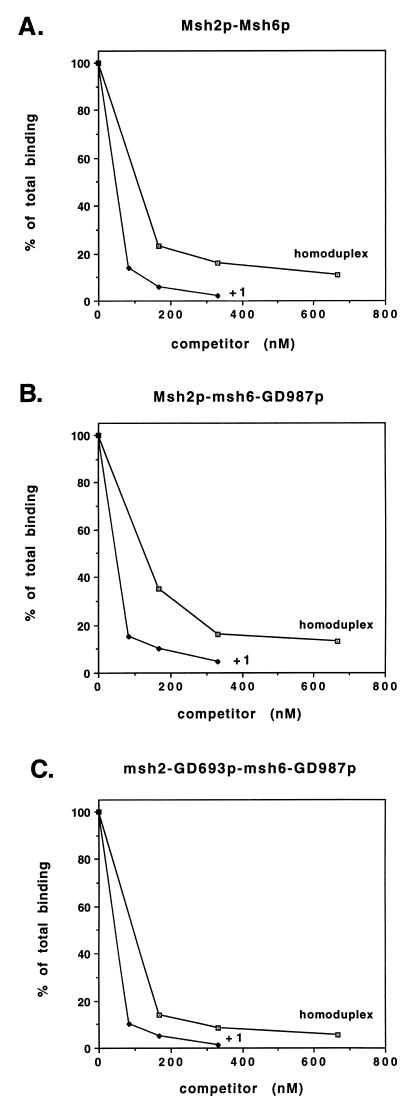
FIG. 2.
Mismatch binding assays performed with Msh2p-Msh6p (A), Msh2p-msh6-GD987p (B), and msh2-GD693p-msh6-GD987p (C) complexes. Binding was performed at 30°C for 15 min in 60-μl reaction mixtures containing 25 mM Tris (pH 7.5), 0.1 mM DTT, 0.01 mM EDTA, 40 μg of BSA per ml, 0.30 μg of wild-type or mutant complex, 16.7 nM 32P-labeled +1 substrate, and the indicated amount of unlabeled +1 and homoduplex competitor substrate. After a 15-min incubation, the amount of 32P-labeled +1 substrate that remained bound to protein complexes was measured by filter binding. Binding data are presented relative to binding observed in the absence of competitor (normalized to 100%). The proportions of input +1 substrate bound for the complexes in the absence of competitor were 26% for Msh2p-Msh6p, 18% for Msh2p-msh6-GD987p, and 20% for msh2-GD693p-msh6-GD987p.
Immunoprecipitation reactions were performed at 4°C as follows. Msh2p-Msh6p and mutant derivative complexes (26 μg of each) were incubated with 55 μg of 12CA5 affinity-purified antibody in 200 μl of 0.5 M NaCl–1× buffer A. After a 1-h rocking incubation, 84 μl of a 1:1 mixture of protein A-Sepharose beads and 0.5 M NaCl–1× buffer A was added. After incubation for an additional hour, samples were centrifuged in an Eppendorf centrifuge at 3,000 rpm for 30 s. The supernatant was removed, and the protein-Sepharose beads were successively washed four times with 200 μl of 0.5 M NaCl–1× buffer A and twice with a 200-μl solution containing 0.1 M NaCl, 25 mM Tris (pH 7.5), 2.0 mM MgCl2, 0.1 mM dithiothreitol (DTT), 0.01 mM EDTA, and 40 μg of BSA per ml (0.1 M NaCl–1× ATPase buffer). The protein A-Sepharose beads were then resuspended with 42 μl of 0.1 M NaCl–1× ATPase buffer and stored on ice prior to use in the ATPase assays described below.
Trypsin protease digestions were performed at room temperature in 20-μl reaction mixtures containing 3.0 μg of Msh2p-Msh6p complex, 0.03 μg of trypsin, 25 mM Tris (pH 7.5), 0.01 mM EDTA, 0.1 mM DTT, and 2 mM MgCl2. Endo-Glu digestion was performed under the same conditions except that 0.3 μg of endo-Glu was substituted for trypsin. When specified, NTPs, dNTPs, ADP, and AMP-PNP were included at 400 μM and homoduplex, +1, and single-stranded oligonucleotides were included at 250 nM. These reagents were added prior to protease addition, and the reaction mixture was preincubated for 15 min at 30°C. Protease was then added, and the reaction mixtures were incubated at room temperature for 60 min. After the incubation, samples were immediately boiled for 3 min in SDS-PAGE sample buffer and loaded onto an 8% polyacrylamide gel for analysis by SDS-PAGE.
DNA binding assays.
DNA binding assays were performed as described previously (3, 13). The 37-mer homoduplex and +1 oligonucleotide substrates used in this study were identical to those described previously (5). Following incubation, samples were analyzed by filter binding to KOH-treated nitrocellulose filters (39), using a Hoefer Scientific Instruments (San Francisco, Calif.) model FH225V filtering unit.
ATPase assays.
ATPase assays were performed in 60-μl reaction mixtures containing 0.3 μg of Msh2p-Msh6p or mutant derivative, 1.2 to 100 μM [γ-32P]ATP, 25 mM Tris (pH 7.5), 2.0 mM MgCl2, 0.1 mM DTT, 0.01 mM EDTA, and 40 μg of BSA per ml. When specified, homoduplex and +1 oligonucleotide substrates were included at 167 nM. The reactions were incubated for 15 min at 30°C, and the amount of ATP hydrolyzed was determined in Norit A absorption assays (13). Km and Vmax measurements were determined from Eadie-Scatchard and Lineweaver-Burk plots (51), using data obtained in the standard ATPase assay in which the concentration of [γ-32P]ATP was varied from 1.2 to 33.3 μM. The two plotting methods yielded identical Km and Vmax values in the ATPase studies performed in the absence of DNA substrate. ATPase assays were performed on immunoprecipitated Msh2p-Msh6p and mutant derivative complexes as follows. A 0.3-μg aliquot of immunoprecipitated Msh2p–Msh6p–protein A-Sepharose bead complex (determined by comparing the concentration of immunoprecipitated protein after SDS-PAGE with that of purified Msh2p-Msh6p) was added to 60-μl reaction mixtures containing 100 μM [γ-32P]ATP, 25 mM Tris (pH 7.5), 2.0 mM MgCl2, 0.1 mM DTT, 0.01 mM EDTA, and 40 μg of BSA per ml. Reaction mixtures were incubated at room temperature for 60 min on a Varimix rocker. The amount of ATP hydrolyzed was then determined in Norit A absorption assays (13).
RESULTS
Genetic analysis indicates that both msh6 and msh2 P-loop mutants display a dominant negative phenotype.
A key feature of MutS homolog proteins is that they contain a highly conserved phosphate binding loop (P-loop) that is found in purine nucleotide binding proteins (Fig. 1A) (22). Substitutions in highly conserved residues of Salmonella typhimurium MutSp, E. coli MutSp, and S. cerevisiae Msh2p indicated that the P-loop motif is required for mismatch repair function and overexpression of these mutant proteins in the corresponding wild-type organism resulted in a dominant negative phenotype (5, 22, 59). Biochemical and genetic analyses of these mutant proteins suggest that they are defective in post-mismatch recognition steps that require ATP hydrolysis (5, 22).
Genetic analysis of the S. cerevisiae msh2Δ and msh6Δ mutations indicated that each confers a strong mutator phenotype (Table 1) (reviewed in references 15 and 34). We assessed these phenotypes in a canavanine resistance assay that measured the frequency of base pair and single-nucleotide frameshift mutations in the CAN1 gene and in a DNA slippage assay that measures poly(GT) tract alterations (principally 2- and 4-nt loop insertions/deletions) that disrupt the URA3 open reading frame and render cells resistant to the uracil analog 5-FOA (27, 53). As described previously and shown in Table 1, msh2Δ strains display a strong mismatch repair defect in both the canavanine and DNA slippage assays, msh6Δ strains display a repair defect primarily in the canavanine assay, and msh3Δ strains display a repair defect primarily in the DNA slippage assay. These data are consistent with the Msh2p-Msh6p pathway repairing base pair and single-nucleotide insertion/deletion mismatches (reviewed in reference 34).
TABLE 1.
Median frequencies of spontaneous mutations and DNA slippage events in msh2Δ, msh3Δ, and msh6Δ strains bearing the msh2-GD693 and msh6-GD987 alleles on GAL10 2μm plasmidsa
| Relevant genotype | Frequency
|
|||
|---|---|---|---|---|
| Canavanine resistance assay
|
Poly(GT) tract alteration assay
|
|||
| Median (10−6) | Relative to wt | Median (10−4) | Relative to wt | |
| wt | 0.98 | 1 | 3.6 | 1 |
| msh2Δ | 43 | 44 | 118 | 33 |
| msh6Δ | 15 | 15 | 6.5 | 1.8 |
| msh6Δ, vector | 28 | 19 | NT | NT |
| msh6Δ, pGAL10-MSH2 | 18 | 12 | NT | NT |
| msh6Δ, pGAL10-MSH6 | 6.7 | 4.4 | NT | NT |
| msh6Δ, pGAL10-msh2-GD693 | 26 | 17 | NT | NT |
| msh6Δ, pGAL10-msh6-GD987 | 57 | 38 | NT | NT |
| msh3Δ | 1.6 | 1.6 | 59 | 16 |
| msh3Δ, vector | 1.6 | 1.6 | NT | NT |
| msh3Δ, pGAL10-MSH2 | 3.8 | 2.5 | NT | NT |
| msh3Δ, pGAL10-MSH6 | 4.2 | 2.8 | NT | NT |
| msh3Δ, pGAL10-msh2-GD693 | 52 | 35 | NT | NT |
| msh3Δ, pGAL10-msh6-GD987 | 37 | 25 | NT | NT |
Wild-type (wt; FY23), msh2Δ (EAY252), msh3Δ (EAY420), and msh6Δ (EAY337) strains were transformed individually with pEAE51 (pGAL10-MSH6), pEAE20 (pGAL10-MSH2), pEAE27 (pGAL10-msh2-GD693), and pEAE84 (pGAL10-msh6-GD987) and tested for forward mutations to Canr. Vector refers to control plasmids that lacked MSH2 or MSH6 sequences. Strains were also transformed with pEAA69 [(GT)16-URA3 ARS CEN] and, where indicated, tested in the poly(GT) tract alteration assay described in Materials and Methods. The median frequency was determined from 11 independent colonies for each experiment. The averages of 2 to 10 independent experiments are presented. NT, not tested.
Previous analysis indicated that overexpression of the msh2-GD693 P-loop allele in wild-type strains resulted in a mutator phenotype that was similar to that observed in msh2Δ strains (Tables 1 and 2) (5). Genetic analysis of the msh2-GD693 allele, coupled with biochemical analysis of the msh2-GD693p-Msh6p complex, suggested that mutant complexes defective in the Msh2p ATPase activity were unresponsive to ATP (5). To determine whether an analogous mutation in the Msh6p ATP binding domain would confer a similar phenotype, we constructed an msh6 allele containing an analogous P-loop mutation. As shown in Fig. 1, the msh6-GD987 allele contains a glycine-to-aspartic acid change in the same position of the P-loop as is present in the msh2-GD693 allele.
TABLE 2.
Median frequencies of spontaneous mutations and DNA slippage events in wild-type strains bearing the msh2-GD693 and msh6-GD987 alleles on GAL10 2μm plasmidsa
| Relevant plasmid | Frequency
|
|||
|---|---|---|---|---|
| Canavanine resistance assay
|
Poly(GT) tract alteration assay
|
|||
| Median (10−6) | Relative to wt | Median (10−4) | Relative to wt | |
| Vector | 1.5 | 1 | 3.4 | 1 |
| pGAL10-MSH2 | 3.2 | 2 | 3.4 | 1 |
| pGAL10-MSH6 | 4.0 | 2.7 | 5.7 | 1.7 |
| pGAL10-msh2-GD693 | 93 | 62 | 69 | 20 |
| pGAL10-msh6-GD987 | 17 | 11 | 17 | 5.0 |
| Vector, vector | 1.2 | 1 | NT | NT |
| pGAL10-MSH2, vector | 1.8 | 1.5 | NT | NT |
| pGAL10-MSH6, vector | 2.8 | 2.3 | NT | NT |
| pGAL10-MSH2, MSH6 | 2.3 | 1.9 | NT | NT |
| pGAL10-msh6-GD987, vector | 17 | 14 | NT | NT |
| pGAL10-MSH2, msh6-GD987 | 16 | 13 | NT | NT |
| pGAL10-msh2-GD693, vector | 44 | 37 | NT | NT |
| pGAL10-msh2-GD693, MSH6 | 169 | 141 | NT | NT |
| pGAL10-msh2GD693, msh6GD987 | 193 | 161 | NT | NT |
Strain FY23 (wild type [wt]) was transformed individually or in pairs with pEAE51 (pGAL10-MSH6), pEAE86 (pGAL10-MSH2), pEAE87 (pGAL10-msh2-GD693), and pEAE84 (pGAL10-msh6-GD987) and tested for forward mutations to Canr. Vector refers to control plasmids that lacked MSH2 or MSH6 sequences. These strains were also transformed with pEAA69 [(GT)16-URA3 ARS CEN] and, where indicated, tested in the poly(GT) tract alteration assay described in the Materials and Methods. The median frequency was determined from 11 independent colonies for each experiment. The averages of 2 to 10 independent experiments are presented. NT, not tested.
A GAL10 2μm overexpression plasmid containing the msh6-GD987 allele was transformed into a msh6Δ strain to assess complementation and into wild-type and msh3Δ strains to determine whether the allele confers a dominant negative phenotype. Galactose induction studies indicated that msh2-GD693p and msh6-GD987p were expressed to similar levels by the GAL10 promoter, as each displayed ∼1% of total protein in wild-type strains (reference 3 and data not shown). As shown in Table 1, the msh6-GD987 mutation failed to complement the msh6Δ phenotype and instead displayed an enhanced mutator phenotype. Overexpression of the msh6-GD987 allele in a wild-type strain resulted in an 11-fold increase in the mutation frequency as measured in the canavanine resistance assay (P = 0.008) (Table 2). In the same assay, the msh2-GD693 allele displayed a 62-fold increase (P = 0.008) in mutation frequency over the wild type (Table 2); this increase in frequency was significantly greater than that observed for the msh6-GD987 allele (P = 0.019). It is important to note that overexpression of Msh6p did not result in the complete complementation of the msh6Δ strain (Table 1); we believe that overexpression of Msh6p decreases the efficiency of mismatch repair because lower expression of MSH6 allows for complete complementation of a msh6Δ strain and overexpression of MSH6 in a wild-type strain results in a weak dominant negative phenotype (Table 2 and reference 53a).
We hypothesize that the dominant negative phenotype conferred by the msh6-GD987 allele is primarily restricted to defects in the repair of MSH6- but not MSH3-dependent repair events. Consistent with this idea were these observations. First, the weaker dominant negative phenotype observed for the msh6-GD987 allele than observed for the msh2-GD693 allele paralleled the respective mutator phenotypes conferred by the msh6Δ and msh2Δ mutations but not the msh3Δ mutation (Tables 1 and 2). Second, overexpression of the msh6-GD987 allele in a wild-type strain resulted in a smaller increase in the frequency of poly(GT) tract alterations compared to overexpression of the msh2-GD693 allele (5- versus 20-fold [Table 2]; P << 0.05, χ2 test). The increase in tract alteration due to overexpression of the msh6-GD987 allele was also lower than the frequency of tract alteration observed in msh3Δ strains (5- versus 16-fold [Tables 1 and 2]; P << 0.05, χ2 test). Third, we observed a mutator phenotype in msh3Δ strains overexpressing msh6-GD987p that was higher than that observed for wild-type strains overexpressing msh6-GD987p (Tables 1 and 2). This observation is reminiscent of the higher mutation rate observed in msh3 msh6 strains than in msh6 strains (33, 39).
The two dominant negative alleles displayed different phenotypes when the corresponding wild-type partner subunit was also overexpressed. As shown in Table 2, the dominant negative phenotype exhibited by the msh2-GD693 allele was enhanced by cooverexpression of Msh6p (P = 0.007) whereas the msh6-GD987 dominant negative phenotype was unchanged by co-overexpression of Msh2p. Co-overexpression of msh6-GD987p and msh2-GD693p in a wild-type strain resulted in a dominant negative phenotype that was similar to that observed when msh2-GD693p and Msh6p were co-overexpressed.
Biochemical purification of wild-type and mutant Msh2p-Msh6p complexes.
Previously, we showed that the msh2-GD693p-Msh6p complex displayed a mismatch binding activity that was indistinguishable from that of the wild-type complex with respect to its discrimination between homoduplex and mismatch substrates (5). However, the wild-type and mutant proteins displayed different properties when binding assays were performed in the presence of ATP. In the presence of ATP, bacterial, yeast, and human mismatch binding complexes were unable to discriminate between homoduplex and mismatch substrates; however, specific mismatch binding activity was still observed when reactions were performed in the presence of ADP or the nonhydrolyzable analog AMP-PNP (see Fig. 3) (2, 17, 19, 20, 30). Previously we observed that the msh2-GD693p-Msh6p complex, which would be expected to be defective in ATP binding, hydrolysis, or both, retained mismatch binding activity in the presence of ATP (5). While the ATPase activity of this complex was less than that of the wild-type complex, there was still a significant residual ATPase activity (∼67% of the wild-type level), suggesting that the residual activity was due to the Msh6p ATPase (Table 3) (5).
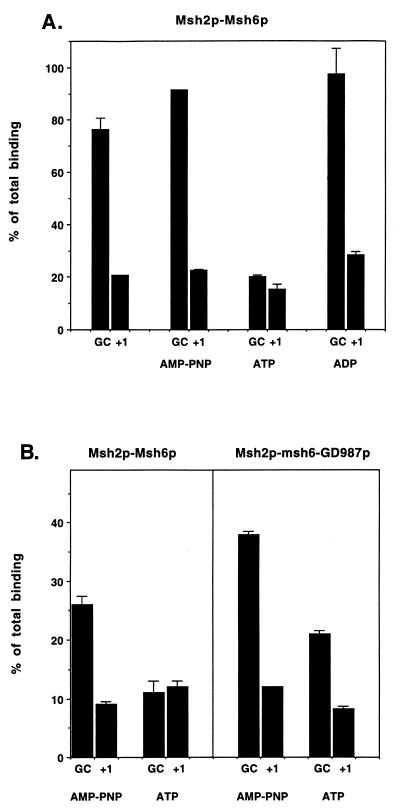
FIG. 3.
Addition of ATP to mismatch binding reactions eliminated the mismatch binding specificity of the Msh2p-Msh6p but not the Msh2p-msh6-GD987p complex. (A) Binding reactions were performed at 30°C for 15 min in 60-μl volumes containing 25 mM Tris (pH 7.5), 0.1 mM DTT, 0.01 mM EDTA, 40 μg of BSA per ml, 0.30 μg of wild-type complex, 16.7 nM 32P-labeled +1 substrate, and 83 nM unlabeled +1 or homoduplex competitor substrate; 1.6 mM ATP, ADP, or AMP-PNP was included as indicated. The amount of 32P-labeled +1 substrate that remained bound to Msh2p-Msh6p was measured by filter binding. Binding data are presented relative to binding observed in the absence of competitor (normalized to 100%). The results from duplicate experiments were averaged, and the range between the two values is shown. The proportion of input +1 substrate bound for the Msh2p-Msh6p complex in the absence of competitor was 16%. (B) Filter binding reactions were performed for both wild-type and Msh2p-msh6-GD987p complexes under the same binding conditions with the exception that MgCl2 was included at 2 mM; 1.6 mM ATP or AMP-PNP was included as indicated. Binding data are presented relative to binding observed in the absence of competitor, ATP, and AMP-PNP (normalized to 100%). The results from duplicate experiments were averaged, and the range between the two values is shown. The proportions of input +1 substrate bound for the complexes in the absence of competitor and ATP and AMP-PNP were 18% for Msh2p-Msh6p and 18% for Msh2p-msh6-GD987p.
TABLE 3.
ATPase activities of Msh2p-Msh6p and mutant derivativesa
| Complex | Km (μM) | Vmax (nM ADP/min)b | kcat (min−1) | kcat/Kmc |
|---|---|---|---|---|
| Msh2p-Msh6p | 7.6, 6.8, 8.0 | 253, 142, 156 | 12.2, 6.9, 7.5 | 1.1 |
| msh2-GD693p-Msh6p | 17, 13, 15 | 120, 123, 126 | 5.8, 6.0, 6.1 | 0.40 |
| Msh2p-msh6-GD987p | 17, 27 | 63, 56 | 3.0, 2.7 | 0.13 |
| msh2-GD693p-msh6-GD987p | 24 | 12 | 0.58 | 0.024 |
Km and Vmax measurements for the ATPase activities of wild-type and mutant complexes were determined in the absence of DNA substrates as described in Materials and Methods. Measurements were made on several independently prepared preparations, and the values determined for each preparation are shown.
Per 0.3 μg of complex.
Determined by using the average values for Km and kcat for each preparation.
The residual ATPase activity that was observed in the msh2-GD693p-Msh6p complex encouraged us to examine the DNA binding and ATPase activities of Msh2p-msh6-GD987p complexes. We previously purified the Msh2p-Msh6p and msh2-GD693p-Msh6p complexes from yeast using strains that co-overexpressed MSH2 and MSH6 from GAL10 2μm vectors (references 2 and 5; Materials and Methods). The yield and purity of the msh2-GD693p-Msh6p, Msh2p-msh6-GD987p, and msh2-GD693p-msh6-GD987p complexes were similar to those obtained for the Msh2p-Msh6p complex (Fig. 1; Materials and Methods). Like the Msh2p-Msh6p and msh2-GD693p-Msh6p complexes, both subunits of the Msh2p-msh6-GD987p and msh2-GD693p-msh6-GD987p complexes copurified during purification and eluted in a Superose 6HR gel filtration column as a single complex (reference 5 and data not shown). Finally, the ratio of Msh2p to Msh6p upon SDS-PAGE was indistinguishable for wild-type and mutant complexes immunoprecipitated with an antibody specific to the HA epitope present in Msh2p (reference 2; data not shown; Materials and Methods).
Mismatch binding specificity of mutant complexes is similar to wild-type specificity.
In filter binding assays, the Msh2p-Msh6p complex displayed an approximately five- to sevenfold-higher binding specificity for an oligonucleotide DNA substrate containing a +1 base pair mismatch compared to a homoduplex oligonucleotide (Fig. 2). Briefly, this assay measured the binding of Msh2p-Msh6p to a 32P-labeled +1 duplex oligonucleotide mismatch substrate in the presence or absence of unlabeled +1 and homoduplex competitors. The +1 substrate contains a single adenine nucleotide insertion in position 16 of a 37-mer duplex oligonucleotide (5). A similar binding specificity for bacterial, yeast, and human mismatch repair complexes was demonstrated in both filter binding and gel shift assays (1, 2, 9, 16, 17, 32, 44, 54, 55).
To compare the binding specificities of wild-type and mutant complexes, we performed competitive filter binding assays in which 0.3 μg (1.2 pmol) of Msh2p-Msh6p, Msh2p-msh6-GD987p, or msh2-GD693p-msh6-GD987p was incubated in a 1:1 molar ratio with 32P-labeled +1 substrate plus various amounts of unlabeled competitor (Fig. 2). In a previous study (2), we measured the stoichiometry of binding of the Msh2p-Msh6p complex to the +1 mismatch substrate by incubating a constant concentration of Msh2p-Msh6p in the presence of increasing amounts of 32P-labeled +1 substrate. Surprisingly, a biphasic curve was observed. At low concentrations of DNA substrate, a linear relationship was observed. When the stoichiometry of DNA substrate to Msh2p-Msh6p reached 1:1, the slope of the curve decreased but remained constant even at DNA substrate concentrations that were sixfold greater than the concentration of Msh2p-Msh6p. This complex mode of binding prevents us from determining the KD.
Because we cannot measure the KD for binding, we tested whether Msh2p-Msh6p specifically recognized mispaired bases by performing competition assays under conditions where Msh2p-Msh6p was incubated with 32P-labeled +1 mismatched substrate at a ratio of 1:1. At this concentration of DNA substrate, the binding of Msh2p-Msh6p complex to substrate was typically 15 to 25% of total input counts. The addition of specific amounts of unlabeled +1 competitor substrate resulted in the expected corresponding decrease in binding to the 32P-labeled +1 substrate. Theoretical and experimental considerations have demonstrated that the discrimination between two competitors can be determined by measuring the maximal horizontal separation between the binding curves resulting from such titrations (13). This measurement is best made at the maximum concentration of competitor that still allows accurate determination of substrate binding because the horizontal separation between the binding curves is constant at high degrees of competition. As shown in Fig. 2, for the Msh2p-Msh6p, Msh2p-msh6-GD987p, and msh2-GD693p-msh6-GD987p complexes, approximately sevenfold-higher levels of homoduplex competitor were required to achieve the same degree of competition for the 32P-labeled +1 substrate as was observed with the +1 competitor when high levels of competition were achieved. This finding indicates that P-loop mutations in either Msh2p or Msh6p do not dramatically affect the mismatch binding specificity of the Msh2p-Msh6p complex in vitro. It is important to note that while the overall levels of binding of all of the complexes to the +1 substrate in the absence of competitor were similar (see the legend to Fig. 2), the binding of the msh2-GD693p-msh6-GD987p complex to the 32P-labeled +1 substrate appeared more sensitive to competition when incubated with either unlabeled +1 or homoduplex substrate. A similar finding was observed with a mutS protein containing a P-loop mutation (22), suggesting that the P-loop class of mutations may also destabilize the mutS homolog complexes.
We then tested the effect of ATP, ADP, and the nonhydrolyzable ATP analog AMP-PNP on the mismatch binding properties of Msh2p-msh6-GD987p by incubating the complex in the standard DNA binding assay in the presence and absence of 1.6 mM ATP, ADP, or AMP-PNP. Like the human Msh2p-Msh6p complex, the binding specificity of the yeast Msh2p-Msh6p complex for mismatch substrates was observed in reactions containing ADP or AMP-PNP but was not observed in reactions containing ATP in the presence or absence of magnesium (Fig. 3) (2, 19, 25). However, like the msh2-GD693p-Msh6p complex, mismatch binding specificity of the Msh2p-msh6-GD987p complex was still observed in the presence of ATP (Fig. 3) (5). A similar result was observed with the msh2-GD693-msh6-GD987p complex (data not shown). Taken together, these results suggest that both subunits of the Msh2p-Msh6p complex are required for the ATP-dependent modulation of mismatch binding.
It is important to note that the lack of mismatch binding specificity for the Msh2p-Msh6p complex in reactions containing ATP was not the result of a reduced affinity of the Msh2p-Msh6p complex for DNA containing or lacking a mismatch. In DNA binding titrations that involved incubating a constant amount of DNA with increasing amounts of Msh2p-Msh6p, the presence of ATP increased the binding of the Msh2p-Msh6p complex to homoduplex substrate but decreased the binding of the complex to the +1 mismatch (11a). This observation is consistent with the failure to observe mismatch discrimination for the Msh2p-Msh6p complex in the presence of ATP (Fig. 3) and is inconsistent with the idea that ATP caused a reduction of Msh2p-Msh6p binding to DNA irrespective of DNA substrate.
Limited trypsin proteolysis of Msh2p-Msh6p indicates that both Msh2p and Msh6p subunits undergo a conformational change in the presence of ATP.
The DNA binding studies described above suggested that the ATP binding domains present in both subunits of the Msh2p-Msh6p complex are required to alter the mismatch binding specificity of the complex in response to ATP. Previous analysis of the yeast Msh2p-Msh6p complex suggested that ATP binding, ATP hydrolysis, or both resulted in a conformational change in the complex that was mediated through a domain that was important for Msh2p-Msh6p interactions (5). We examined whether the Msh2p-Msh6p complex undergoes an ATP-dependent conformational change by performing trypsin and endo-Glu protease digestion experiments. Limited treatment of Msh2p-Msh6p with trypsin and endo-Glu was performed in the presence and absence of homoduplex and +1 DNA substrates and the nucleotides dATP, GTP, ATP, ADP, and AMP-PNP (Materials and Methods). When the Msh2p-Msh6p complex was preincubated with ATP, dATP, or ADP prior to trypsin digestion, four species, A to D, ranging from ∼92 to ∼48 kDa (Fig. 4A) were specifically protected from proteolysis. Incubation with GTP did not affect the proteolysis pattern of the Msh2p-Msh6p complex (Fig. 4A). A similar set of experiments was performed with the protease endo-Glu. Only a single species was observed to be protected by protease digestion in the presence of ATP, and the protection of this species was less dramatic than observed with trypsin (data not shown). Western blot analysis with Msh2p- and Msh6p-specific antibodies indicated that species A (∼92 kDa) and B (∼87 kDa) were derived from Msh6p and that species C (∼55 kDa) and D (∼48 kDa) were derived from Msh2p (Fig. 4B). When trypsin digestion was performed in the presence of the nonhydrolyzable analog AMP-PNP, a weaker protection pattern was observed: species A and C were observed at a similar intensity, but species B was detected at a lower intensity and species D could not be detected.
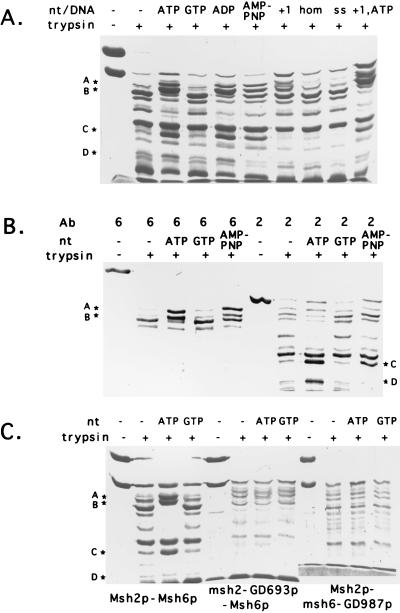
FIG. 4.
Limited trypsin proteolysis analysis reveals that the Msh2p-Msh6p complex undergoes an ATP-dependent conformational change. (A) SDS-PAGE analysis of Msh2p-Msh6p proteolytic products. Trypsin protease digestions were performed for 60 min at 23°C in 20-μl reaction mixtures containing 0.03 μg of trypsin and 3.0 μg of Msh2p-Msh6p complex as described in Materials and Methods. After incubation, samples were analyzed by SDS-PAGE. When indicated, 400 μM ATP, GTP, ADP, or AMP-PNP was preincubated with Msh2p-Msh6p prior to protease digestion; 250 nM +1, homoduplex (hom), and ssDNA(ss) substrates were included in the reaction mixtures prior to protease digestion as indicated. Bands A (∼92 kDa), B (∼87 kDa), C (∼55 kDa), and D (∼48 kDa), which were resistant to proteolytic cleavage in reactions preincubated with ATP, AMP-PNP, or ADP, are indicated by asterisks. (B) Complexes incubated without nucleotide and in the presence of ATP and AMP-PNP prior to protease treatment were separated by SDS-PAGE and analyzed by Western blotting using Msh2p (lanes 2)- and Msh6p (lanes 6)-specific antibodies (Ab). The assignment of bands A to D is indicated. (C) Trypsin protease digestion of Msh2p-Msh6p, msh2-GD693p-Msh6p, and Msh2p-msh6-GD987p complexes preincubated in the absence and presence of ATP. Bands A to D are indicated.
When the Msh2p-Msh6p complex was incubated with +1 and homoduplex DNA substrates, a band corresponding to full-length Msh2p and a set of fragments distinct from those observed in the presence of ATP were protected from trypsin digestion (Fig. 4A and data not shown). While a higher yield of these fragments was observed in incubations involving the +1 compared to the homoduplex substrate, there was no apparent difference in the size of fragments protected. When both ATP and oligonucleotide substrate were incubated with Msh2p-Msh6p and then subjected to trypsin proteolysis, we observed a pattern of cleavage that appeared to be a combination of the ATP and +1 substrate patterns. Unfortunately, these experiments do not allow us to determine whether this pattern represented the simultaneous binding of ATP and DNA to the Msh2p-Msh6p complex or represented a mixed population of ATP-bound and DNA-bound complexes.
We tested whether the mutant complexes were capable of displaying complete or partial protection from trypsin digestion when incubated with ATP. As shown in Fig. 4C, none of the four species was specifically protected from trypsin digestion in either the msh2-GD693p-Msh6p or Msh2p-msh6-GD987p complexes. The mutant complexes appeared more sensitive to trypsin in the absence of ATP than the wild-type complexes, suggesting that the nucleotide binding site mutations may alter the stability or structure of the Msh2p-Msh6p complex. Taken together, these results suggested that functional ATP binding domains for each subunit are required to induce an ATP-dependent conformational change in the Msh2p-Msh6p complex. In addition, the finding that both ADP and ATP were capable of inducing a conformational change in the complex (based on the similar sizes of tryptic fragments) suggests that both nucleotides were capable of binding to the Msh2p-Msh6p complex.
ATPase activity in wild-type and mutant Msh2p-Msh6p complexes.
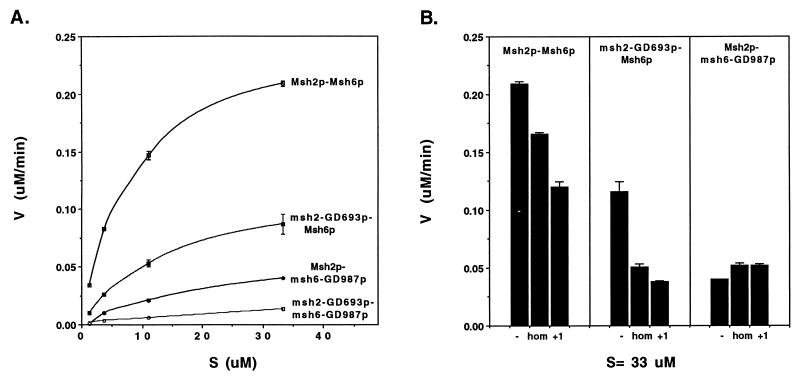
The biochemical and genetic assays outlined above suggested that the ATP binding domains of both subunits of the Msh2p-Msh6p complex were required for the interaction of the Msh2p-Msh6p complex with mismatch DNA substrates. These observations encouraged us to measure the ATPase activity of each of the mutant complexes to determine the relative contributions of the Msh2p and Msh6p ATPase activities in the presence and absence of DNA substrates. As shown in Fig. 5 and Table 3, in the absence of DNA substrate, the msh2-GD693p-Msh6p complex displayed about two-thirds of the wild-type ATPase activity and about twice the activity of the Msh2p-msh6-GD987p complex. Km and Vmax values for the wild-type and mutant complexes were determined from Eadie-Scatchard and Lineweaver-Burk plots (51, 56a) (Table 3). The data in Table 3 were obtained from Lineweaver-Burk plots; these values were indistinguishable from those determined from Eadie-Scatchard plots (data not shown). The sum of the individual Vmax values of the Msh2p-msh6-GD987p (60 nM/min) and msh2-GD693p-Msh6p (123 nM/min) complexes was approximately equal to the Vmax value observed for the Msh2p-Msh6p (184 nM/min) complex. This observation suggests that the inactivation of one ATPase activity did not affect the activity of the other (Fig. 5). A similar effect of P-loop mutations on the ATPase activity of Msh2p-Msh6p complexes was observed in a recently published analysis of human Msh2p-Msh6p, where it was also shown that these mutations inhibited ATP binding to the subunit containing the P-loop mutation (30). Analogous to the yeast complex, complexes bearing a P-loop mutation in hMsh2p displayed a stronger ATPase activity than those bearing a P-loop mutation in hMsh6 (30). In addition, the kcat/Km values for the wild-type and P-loop mutant complexes were similar in the yeast and human systems (Table 3 and reference 30). A residual ATPase activity (∼2%) was observed in the msh2-GD693p-msh6-GD987p fraction IV preparations that was also observed in immunoprecipitated msh2p-GD693p-msh6-GD987p complexes (data not shown). This activity was unexpected, as analogous mutations in other purine nucleotide binding proteins almost completely eliminated ATPase activity (22, 56). Results from control experiments involving analysis of immunoprecipitated complexes suggested that the low level of ATP hydrolysis observed is intrinsic to the mutant complex (Materials and Methods; data not shown).
FIG. 5.
(A) ATP hydrolysis activity exhibited by wild-type and mutant Msh2p-Msh6p complexes. Fraction IV of Msh2p-Msh6p, msh2-GD693p-Msh6p, Msh2p-msh6-GD987p, and msh2-GD693p-msh6-GD987p preparations (0.3 μg in each case) was incubated in the presence of 1.2 to 33.3 μM [γ-32P]ATP, and the rate of ATP hydrolysis (V) was determined for duplicate reactions after a 15-min incubation at 30°C (Materials and Methods). The results from duplicates were averaged, and the range between the two values is shown. (B) Comparison of ATP hydrolysis activities for wild-type and mutant Msh2p-Msh6p complexes in the presence of homoduplex and mismatch substrates. Msh2p-Msh6p, msh2-GD693p-Msh6p, and Msh2p-msh6-GD987p complexes (0.3 μg of each) were incubated with 33.3 μM [γ-32P]ATP and 167 nM each indicated +1 or homoduplex (hom) substrate. The rate of ATP hydrolysis (V) was determined after a 15-min incubation at 30°C. The results from duplicate reactions were averaged, and the range between the two values is shown.
The effect of homoduplex and +1 DNA oligonucleotide substrates on the ATPase activities of the wild-type and mutant complexes was tested under the same conditions as described for Fig. 5 except that a 10-fold stoichiometric excess of DNA substrate was included in indicated reactions. As shown previously and in Fig. 5, the Msh2p-Msh6p ATPase activity was reduced by homoduplex substrate and reduced even further by the +1 substrate. A similar modulation of ATPase activity by these substrates was observed for the msh2-GD693p-Msh6p complex. The ATPase activity of the Msh2p-msh6-GD987p complex, however, did not appear to be significantly affected by the presence of these substrates. In Fig. 5B, the ATPase activities of wild-type and mutant complexes are presented at only a single ATP concentration (33.3 μM); it is important to note that the effects of DNA substrate on the ATPase activity of wild-type and mutant complexes were qualitatively similar at all ATP concentrations tested (1.2 to 33.3 μM).
DISCUSSION
We used several approaches to define the role of the ATP binding domains in the Msh2p-Msh6p complex. Genetic analysis revealed that mutations in the ATP binding domains of both subunits conferred a dominant negative phenotype when their respective gene products were overexpressed. In mismatch binding assays, both Msh2p-msh6-GD987p and msh2-GD693p-Msh6p complexes displayed mismatch recognition properties similar to those of the wild type; however, unlike the case for the wild type, the mismatch binding specificity of the two mutant complexes was not eliminated by ATP. Protease digestion analysis revealed that the Msh2p-Msh6p complex undergoes an ATP-dependent conformational change that requires the ATP binding domains of both subunits. Finally, we showed that Msh2p and Msh6p contain distinct ATPase activities that respond differently to the presence of mismatched substrate.
While the above observations support an equivalent role for the Msh2p and Msh6p nucleotide binding domains in ATP-dependent release from a mismatch site, additional studies presented here on the ATPase activity of the Msh2p-Msh6p complex suggest that the Msh6p subunit plays a unique role in earlier steps in mismatch repair. In genetic studies, overexpression of msh6-GD987p resulted in a dominant negative phenotype that was unaffected by co-overexpression with Msh2p; however, in the reciprocal experiment, co-overexpression of msh2-GD693p and Msh6p resulted in a stronger dominant negative phenotype than was observed when msh2-GD693p was overexpressed by itself (Table 2). In biochemical studies that measured the ATPase activity of Msh2p-msh6-GD987p and msh2-GD693p-Msh6p complexes in the presence of homoduplex and mismatch substrates, the Msh2p ATPase was insensitive to DNA substrate, while the Msh6p ATPase displayed a modulation of ATPase activity by homoduplex and mismatch DNA substrates that was similar to that observed for the Msh2p-Msh6p complex (Fig. 5).
The above data are consistent with the proposal that the Msh6p subunit ATPase activity plays an important role in steps that occur prior to the proposed release step of MutS homolog proteins from a mismatch site (41). Previously we argued that the Msh6p subunit acts as a specificity factor for mismatch recognition (2). This proposal was based on the observation that Msh6p and Msh3p subunits provide different mismatch binding specificities to the Msh2p subunit and neither the Msh2p nor Msh6p subunit independently displays the mismatch binding properties displayed by the Msh2p-Msh6p complex (reviewed in references 34 and 41). This information, taken together with the ATPase analysis described here, suggests that the Msh6p subunit also acts as a specificity factor in postrecognition steps by sensing mismatch through its ATPase activity and then relaying the information to downstream mismatch repair components. An attractive candidate to receive these signals is the yeast Mlh1p-Pms1p complex, as studies of both bacteria and yeast suggest that the MutLp homologs specifically interact with MutSp homologs bound to a mismatch in steps that require ATP (20, 21, 25, 46, 47). A prediction of this proposal is that the msh2p-GD693p-Msh6p but not the Msh2p-msh6-GD987p complex is still capable of interacting with the Mlh1p-Pms1p complex. Experiments to address this question are in progress.
A model for mismatch recognition by the yeast Msh2p-Msh6p complex.
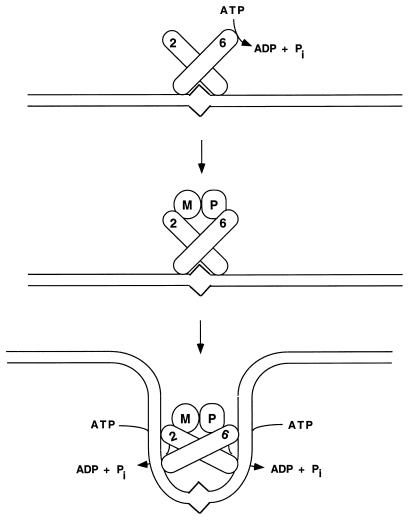
The data presented in this paper are consistent with the model shown in Fig. 6. In this model, binding of the Msh2p-Msh6p complex to a mismatch substrate triggers a change in the ATPase activity of the Msh6p subunit that allows the Msh2p-Msh6p-mismatch complex to interact with Mlh1p-Pms1p. The formation of this complex then stimulates the ATPase activity of both subunits of the Msh2p-Msh6p heterodimer, inducing a conformational change in the complex that facilitates release and bidirectional translocation of the complex away from the mismatch.
FIG. 6.
Model describing Msh2p-Msh6p interactions with ATP and DNA containing base pair mismatches. Binding of Msh2p-Msh6p to a mismatch substrate modulates the ATPase activity of the Msh6p subunit so that Msh2p-Msh6p can interact with Mlh1p-Pms1p. This complex then undergoes an ATP-dependent conformational change that requires the ATP binding domain functions of both Msh2p and Msh6p. The conformational change allows the Msh2p-Msh6p-Mlh1p-Pms1p complex to leave the mismatch and translocate along DNA to participate in subsequent mismatch repair steps. The numbers 2 and 6 refer to Msh2p and Msh6p, respectively, and the letters M and P refer to Mlh1p and Pms1p, respectively.
The model in Fig. 6 is based on the studies presented here and interpreted in light of recent reports from the Prakash and the Griffith-Modrich laboratories (6, 25). Habraken et al. (25) observed that ATP was required for the assembly of a ternary complex consisting of a DNA mismatch substrate and the yeast Msh2p-Msh6p and Mlh1p-Pms1p complexes. This ternary complex could not form if ADP or AMP-PNP was substituted for ATP. Based on electron microscopy studies involving MutSp, MutLp, and mismatch substrates, Allen et al. (6) proposed that an ATP-dependent conformational change in MutS was required for the observed bidirectional translocation of the MutSp-MutLp complex away from a mismatch site. The bidirectional translocation activity observed for MutSp could function in a manner similar to a bind-release switch mechanism proposed for the E. coli dimeric Rep helicase (10, 11). In this model, a functional asymmetry exists between the two subunits of Rep helicase such that ATP hydrolysis stimulates the rate of DNA exchange. Changes in protein conformation and DNA affinity that accompany ATP hydrolysis are thought to allow the dimer to translocate along DNA (11). Although this mechanism may apply only to certain classes of DNA helicases, an analogous mechanism may allow MutSp to translocate bidirectionally along DNA in a manner whereby one ADP-bound subunit of the dimer is bound to DNA while the other ATP-bound subunit transiently dissociates from DNA (10, 11, 19). This paradigm provides an attractive way to analyze the ATPase functions of the Msh2p-Msh6p complex because it suggests that each subunit of the Msh2p-Msh6p complex displays a mismatch release activity that requires ATP hydrolysis and exchange. The observations that the msh2p-GD693p-Msh6p and Msh2p-msh6-GD987p complexes remained bound to a mismatch substrate in the presence of ATP and that the ATPase activities of the msh2-GD693p-Msh6p and Msh2p-msh6-GD987p were similar in the presence of homoduplex DNA provide support for this idea (Fig. 5).
The model shown in Fig. 6 can also explain why overexpression of msh6-GD987p did not dramatically interfere with the Msh2p-Msh3p-dependent repair of small loop insertions/deletions that resulted from DNA slippage events (Table 2). A similar lack of interference was also observed when Msh6p, which cannot complement the slippage defect when overexpressed in msh3Δ strains, was overexpressed in wild-type strains (52). These results can be explained if the Mlh1p-Pms1p complex is required in a commitment step to stabilize Msh2p-Msh6p or Msh2-Msh3p binding at a mismatch site. Prior to such a step, Msh2p can rapidly switch between Msh3p and Msh6p subunits so that the presence of a high level of one Msh2p partner will not prevent Msh2p from interacting with a partner present at lower levels.
Comparison of the ATPase activities of the yeast and human Msh2p-Msh6p complexes.
Recently Gradia et al. (19) showed that the ATPase activity of the human Msh2p-Msh6p complex was stimulated by the addition of mismatch DNA substrate: compared to reactions performed in the presence of homoduplex DNA, the kcat for ATP hydrolysis by the human Msh2p-Msh6p complex increased from 7.4 to 26 min−1; the Km, however, increased from 23 to 46 μM. These results contrasted with those observed with yeast Msh1p and Msh2p-Msh6p where the ATPase activity of these proteins was lower in reactions containing mismatch substrate than in reactions containing homoduplex substrate (reference 13 and this study). For example, in experiments involving Msh1p, Chi and Kolodner (13) found that the Km was unaffected by DNA substrate whereas the kcat for ATP hydrolysis was lower in reactions containing mismatch DNA substrate than in reactions containing homoduplex substrate. It is important to note that in their steady-state analysis of human Msh2p-Msh6p, Gradia et al. (19) showed that ATP-ADP exchange but not γ-phosphate hydrolysis was rate limiting. However in single-step γ-phosphate hydrolysis studies, they observed that mismatch substrates did not stimulate and in fact stoichiometrically inhibited γ-phosphate hydrolysis. Based on these observations, Gradia et al. (19) speculated that the inhibition of γ-phosphate hydrolysis was due to the inability of ATP to bind to a Msh2p-Msh6p complex that was already bound to a mismatch. This observation can reconcile the differences between the yeast and human ATPase activities if ATP binding to the yeast MutSp homolog complexes is severely inhibited in the case where complexes are bound to a mismatch. Experiments to test this idea are in progress.
ACKNOWLEDGMENTS
We thank Elizabeth Evans for extensive comments on the manuscript; Barbara Baird, Jeff Brodsky, Phillip Cole, Elizabeth Evans, Cara Olsen, Jeff Roberts, and Stanley Zahler for helpful discussions; and Mark Berryman, Liz Evans, Jinlin Peng, and Tanya Sokolsky for providing reagents or technical advice. We are also appreciative of the insights provided by Tanya Sokolsky in some of the initial dominant negative studies.
E.A. was supported by National Institutes of Health grant GM53085 and USDA Hatch grant NYC-186424, B.S. was supported by a State University of New York fellowship and a Cornell University anonymous donor fellowship, and T.Q. was supported by an undergraduate summer research fellowship from the Howard Hughes Medical Institute awarded to Cornell University.
REFERENCES
- 1.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E. The Saccharomyces cerevisiae Msh2p and Msh6p form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alani E, Chi N W, Kolodner R D. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and loop insertions. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 4.Alani E, Reenan R A G, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alani E, Sokolsky T, Studamire B, Miret J J, Lahue R S. Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol Cell Biol. 1997;17:2436–2447. doi: 10.1128/mcb.17.5.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen D J, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith J D. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Au K G, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 8.Bhattacharyya G K, Johnson R A. Statistical concepts and methods. New York, N.Y: John Wiley & Sons; 1977. [Google Scholar]
- 9.Biswas I, Hsieh P. Identification and characterization of a thermostable MutS homolog from Thermus aquaticus. J Biol Chem. 1996;271:5040–5048. doi: 10.1074/jbc.271.9.5040. [DOI] [PubMed] [Google Scholar]
- 10.Bjornson K P, Wong I, Lohman T M. ATP hydrolysis stimulates binding and release of single stranded DNA from alternating subunits of the dimeric E. coli Rep helicase: implications for ATP-driven helicase translocation. J Mol Biol. 1996;263:411–422. doi: 10.1006/jmbi.1996.0585. [DOI] [PubMed] [Google Scholar]
- 11.Bjornson K P, Hsieh J, Amaratunga M, Lohman T M. Kinetic mechanism for the sequential binding of two single-stranded oligodeoxynucleotides to the Escherichia coli Rep helicase dimer. Biochemistry. 1998;37:891–899. doi: 10.1021/bi9719307. [DOI] [PubMed] [Google Scholar]
- 11a.Bowers, J., and E. Alani. Unpublished data.
- 12.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 13.Chi N, Kolodner R D. The effect of DNA mismatches on the ATPase activity of Msh1, a protein in yeast mitochondria that recognizes DNA mismatches. J Biol Chem. 1994;269:29993–29997. [PubMed] [Google Scholar]
- 14.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 15.Crouse G F. Mismatch repair systems in Saccharomyces cerevisiae. In: Nickoloff J, Hoekstra M, editors. DNA damage and repair—biochemistry, genetics and cell biology. Clifton, N.J: Humana Press; 1996. pp. 411–448. [Google Scholar]
- 16.Drotschmann K, Aronshtam A, Fritz H J, Marinus M G. The E. coli MutL protein stimulates binding of VSR and MutS to heteroduplex DNA. Nucleic Acids Res. 1998;26:948–953. doi: 10.1093/nar/26.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummond J T, Li G-M, Longley M J, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 18.Geitz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 19.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 20.Grilley M, Welsh K M, Su S-S, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 21.Gu L, Hong Y, McCulloch S, Watanabe H, Li G M. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haber L T, Walker G C. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 24.Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 25.Habraken Y, Sung P, Prakash L, Prakash S. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. J Biol Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 27.Henderson S T, Petes T D. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Hughes M J, Jiricny J. The purification of a human mismatch-binding protein and identification of its associated ATPase and helicase activities. J Biol Chem. 1992;267:23876–23882. [PubMed] [Google Scholar]
- 30.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSα. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iaccarino I, Palombo F, Drummond J, Totty N F, Hsuan J J, Modrich P, Jiricny J. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 32.Jiricny J, Su S, Wood S G, Modrich P. Mismatch-containing oligonucleotide duplexes bound by the E. coli mutS-encoded protein. Nucleic Acids Res. 1988;16:7843–7853. doi: 10.1093/nar/16.16.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2 dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 34.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Lahue R S, Au K G, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 38.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 39.McEntee K, Weinstock G, Lehman I R. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci USA. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 41.Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 42.Modrich P, Lahue R S. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 43.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. GTBP, a 160 kD protein essential for mismatch binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 44.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 45.Parker B O, Marinus M G. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prolla T A, Christie D M, Liskay R M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prolla T A, Pang Q, Alani E, Kolodner R D, Liskay R M. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 48.Reenan R A G, Kolodner R D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 50.Sancar A, Hearst J E. Molecular matchmakers. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 51.Segel I H. Biochemical calculations. New York, N.Y: John Wiley & Sons; 1976. [Google Scholar]
- 52.Sokolsky, T., and E. Alani. Unpublished data.
- 53.Strand M, Earley M C, Crouse G F, Petes T D. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Studamire, B., and E. Alani. Unpublished data.
- 54.Su S-S, Lahue R S, Au K G, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- 55.Su S-S, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung P, Higgins D, Prakash L, Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988;7:3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Tipton K F. Principles of enzyme assay and kinetic studies. In: Eisenthal R, Danson M J, editors. Enzyme assays: a practical approach. Oxford, England: Oxford University Press; 1992. pp. 1–58. [Google Scholar]
- 57.Welsh K M, Lu A-L, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 58.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 59.Wu T-H, Marinus M G. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]