Abstract
Resistive pulse nanopore sensing enables label-free single-molecule analysis of a wide range of analytes. An increasing number of studies have demonstrated the feasibility and usefulness of nanopore sensing for protein and peptide characterization. Nanopores offer the potential to study a variety of protein-related phenomena that includes unfolding kinetics, differences in unfolding pathways, protein structure stability, and free-energy profiles of DNA–protein and RNA–protein binding. In addition to providing a tool for fundamental protein characterization, nanopores have also been used as highly selective protein detectors in various solution mixtures and conditions. This review highlights these and other developments in the area of nanopore-based protein and peptide detection.
Keywords: biosensors, nanopores, nanotechnology, peptide sensing, protein identification
1. Introduction
Coulter counters are based on the idea that micron-sized particles can block the flow of ionic current through micron-sized channels and these current blockades can be used to characterize the particles.[1] The Coulter method approached nanoscale dimensions with nuclear track-etched membranes in the early 1970s,[2] but it was not until researchers demonstrated resistive pulse sensing with pore forming proteins alpha hemolysin (αHL) from Staphylococcus aureus[3] and alamethicin from Trichoderma viride[4] that nanopore sensing began in earnest. The discovery that DNA could be translocated through αHL pores and give rise to current blockades that correlate with the length[5] and possibly the sequence of polynucleotides[6] led to a concentrated effort to develop next-generation sequencing applications based on nanopore sensors. Many reviews on the principle of operation for nanopore sensing are available.[7–14] In addition to DNA sequencing, nanopore sensing has also been used to explore other analytes of interest. The discovery by Krasilnikov that manipulating the ionic strength of the electrolyte solution could extend the residence times of synthetic polymers from submicrosecond to millisecond ranges[15] enabled the nanopore to be refashioned as a spectrometer that can effectively size molecules that partition completely into the pore.[16] Homopolymer systems offer unique spectral fingerprints that were used to develop comprehensive theories that describe the characteristic blockade depth and residence time using simplified thermodynamic models.[17]
Proteins and peptides offer complexity from their size and structure that is not typically associated with synthetic polymers and nucleic acids. Nevertheless, an increasing number of researchers have developed methods for nanopore sensing to enable better characterization of proteins and peptides at the single-molecule limit. This growth suggests that nanopore sensing is a viable and important tool for characterizing a large number of proteins and peptides. Several reviews on nanopore sensing applied to protein detection have been reported.[18,19] The purpose of this review article is to provide a thorough and up-to-date summary of the various avenues that have been explored for protein and peptide sensing with a variety of different nanopore configurations.
The review begins with a brief introduction to the operating principals of resistive pulse nanopore sensing. This is followed by two major sections that focus on large amino acid polymers (proteins) and small peptides. The protein portion of this review begins with early efforts to detect full-length proteins with a variety of different nanopore-based sensors. This focuses on the detection of proteins outside and inside the pore, the study of protein–protein interactions, and the demonstration of practical applications of protein sensing that include detection of post-translational modifications and mechanisms for neurodegenerative disease development. We also focus on protein unfolding studies both outside and inside the pore and discuss the role of DNA aptamers for improving nanopore-based protein detection. This is followed by a review of both fundamental and biosensing-based studies of DNA and RNA binding to proteins. The second major part of the review focuses on peptide detection and begins with a review of nanopore-based proteolysis. This is followed by a section on peptide detection inside the pore with an emphasis on peptide–nanopore kinetics, peptide translocation through outer membrane channels, and direct observation of peptide–metal binding. Finally, we conclude with a discussion of more recent developments that demonstrate nanopore-based peptide identification in a manner analogous to mass spectrometry.
2. Resistive Pulse Nanopore Sensing
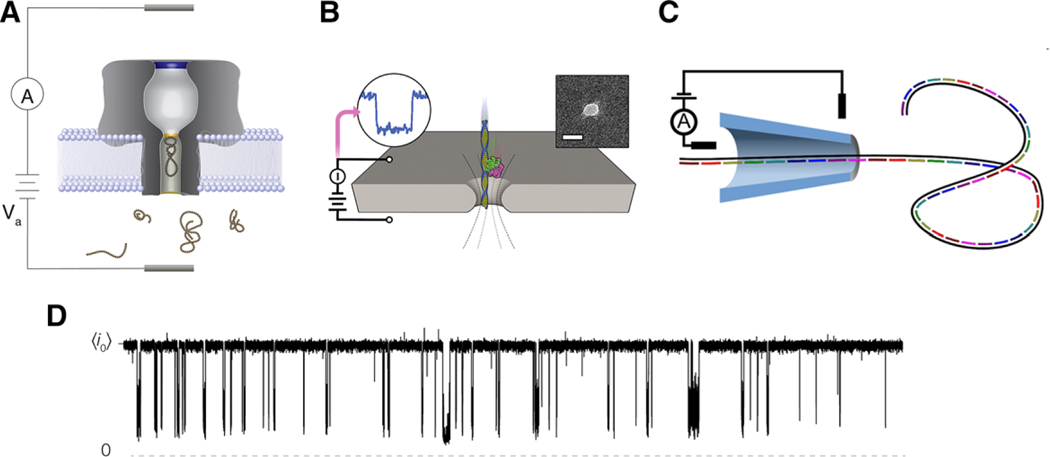
Figure 1 shows several illustrations of typical nanopore sensing experiments with a variety of different configurations utilized for protein and peptide detection. Because of the wide range of sizes and structures in peptides and proteins, both chemistry and geometry of the pores must be carefully chosen for optimal sensing. Figure 1A–C shows three characteristic pores used for nanopore sensing. Oftentimes smaller analytes utilize the precise chemistry available in protein-based nanopores (Figure 1A), while larger analytes require the geometric flexibility of solid-state pores in nanofabricated semiconductor membranes (Figure 1B) or pulled glass capillaries (Figure 1C). Regardless of the type of pore used, all nanopore experiments operate by isolating a nanopore in a membrane that separates two electrolyte solutions (Figure 1D). Applying a potential across the membrane drives ionic current through the pore, which is interrupted when analyte molecules partition into the pore. Generally speaking, the analyte size, concentration, and interaction with the pore can be inferred from the magnitude of the resistive pulses, the time between pulses (i.e., on time), and the duration of the pulses (i.e., off time), respectively. Additionally, some more intricate characterization methods may also be applied and are described throughout the review.
Figure 1.
Schematic illustrations of the different nanopore sensing configurations discussed herein. A) Biological pores are embedded in lipid bilayer membranes. B) Etched pores are nanoscale holes formed in solid-state supports such as silicon nitride or graphene. The length scales for these systems are typically on the order of 5–10 nm. C) Nanopore sensing has also been demonstrated with glass nanopipettes where length scales tend to be on the order of 50 nm. In all the three examples, target analyte enters into the sensing region of the pore and reduce the flow of current across the boundary. D) This gives rise to sizable current blockades whose depth (with respect to the open pore current ) and duration can inform about the physical and chemical attributes of the analyte under investigation. Data shown here is a 20 s current trace of 20 μm neurotensin interacting with αHL under a 70 mV applied transmembrane potential in 3 m KCl at pH 7.2. Images B and C are reproduced with permission.[98,101] Copyright 2014, ACS Publications and Copyright 2015, American Chemical Society.
3. Protein Sensing
3.1. Full-Length Proteins
Full-length proteins present a challenging sample for direct detection by protein-based nanopores because their size often prevents entry into the small pore volumes.[20,21] Therefore, many protein sensing studies rely on fabricated solid-state pores, which offer a wider range of pore sizes despite the lack of precise control over the surface chemistry.[10] As discussed in the following sections,thesignalisgeneratedanytimetheioniccurrentisinterrupted either through protein partitioning into the pore, or when the protein is held fixed in the vicinity of the pore. The critical element is that the analyte molecule needs to be held in place for a sufficiently long time to record the corresponding changes to current.
3.1.1. Outside the Pore
It is often easier to detect proteins through an interaction outside the pore. One can take advantage of the natural binding affinity of AB toxins, such as the anthrax toxins, to devise a binding assay sensor.[22,23] Rostovsteva and colleagues used a co-localization assay to identify a likely sensor for the tubulin, which has disordered c-terminal tails (CTT) that differ between isotypes. Each CTT produces a unique ion current signature based on the distribution of negative charges along the sequence.[24] Several systems have been devised that anchor recognition sites within the pore, such that a portion of the anchor is inside the pore either through a specific biological recognition element (i.e., biotin–streptavidin),[25] or by integrating binding sites to the pore that capture the natural ligand of the protein analyte.[26] By anchoring proteins to the pore, binding events can be observed that occur completely outside the channel.[27,28] Often, segments of a protein can be captured by a pore[29] and manipulated with proteins outside the pore.[30] While the above papers use rigid pore forming toxins, it is sometimes advantageous to use channels that produce significant conformational fluctuations like OmpG.[31–33] By anchoring a tether with recognition elements to OmpG, characteristic fluctuations can be observed for several proteins with different binding strength[31] including antibodies.[32]
3.1.2. Direct Detection in Cylindrical Pores
Much of the work related to direct detection of proteins with nanopores has evolved from using the pioneering work of DeBlois and Bean,[2] to estimate the size of proteins based on the magnitude of the current blockade. Indeed, Han showed that BSA could be estimated to have a hydrodynamic diameter from 7 to 9 nm using pores with diameters as large as 55 nm.[34] Detecting proteins based on size alone is often insufficient for identification. Fortunately, it is possible to estimate charge as well from the residence time of the molecule.[35,36] Fologea and Li used a combination of blockade depth and translocation time to determine pH dependent properties of BSA and fibrinogen. Here, BSA is modeled as a charged ellipsoid and shifts in both blockade depth and translocation time of the protein suggest that dimerization reactions can be observed with pH changes consistent with known BSA behavior.[36] Furthermore, the translocation of BSA was confirmed by a sensitive immunosensor assay in the cis-side chamber. Translocation, however, cannot be directly inferred from the signals as Krasniqi and Lee show that RNase, which produces similar resistive pulses to those of Fologea and Lee, does not translocate through the αHL channel.[37] Beyond the analysis of a single protein, the pore was used to classify different signals from BSA and Fib, pointing to the possibility of using a nanopore as a qualitative analytical tool in proteomics. Utilizing avidin translocating through Si3N4 nanopores,[38] Rant and colleagues expand on the theme of electrophoresis, electroosmosis, and zeta potential measurements that were introduced by Fologea’s[36] and Sexton’s[35] work. Here, pH dependent analysis of avidin was performed in Si3N4 membranes over a wide range of pH (2–10) and ionic strength values. By utilizing such a wide range of conditions, the relative contributions of electroosmosis and electrophoresis can be used to estimate protein charge with better fidelity than previous work. The resolution (or signal processing) limited some of the more detailed biological work of Fologea and Talaga,[73] but nevertheless Rant and colleagues present a compelling update for understanding the mechanism of transport in solid-state pores.
In many protein detection papers (and nanopore detection papers more broadly), it is critical to understand that the ability to detect an analyte is dependent not only on the capture efficiency of the pore, but also on the analyte’s residence time that enables the necessary signal-to-noise (S/N) ratio needed to identify a resistive pulse. Plesa and colleagues offer a quantitative treatment of the capture rates of proteins using mass transport governed by diffusion (Smoluchowski rate equation) and explores the limitations on protein detection as a function of the size of the protein (in the absence of specific interactions).[39] This work suggests that a Smoluchowski approach provides a reasonable estimation for capture rates, assuming that the diffusion constant of the protein in question is small enough to detect most of the events. Thus, for a 540 kDa protein, the predicted behavior is observed but not for a 37 kDa protein (similar behavior was predicted and explained for polymer molecules,[17] and alluded to by Mayer [lipid-walled pores,[40,41] discussed below]). Wanunu and colleagues extend the Smoluchowski rate arguments of Plesa by measuring resistive pulses of <30 kDa proteins (RNase A, see Krasiniqi below, and ProtK) using hafnium oxide and silicon oxide solid-state pores.[42] Using thin membranes increases the magnitude of the resistive pulse allowing event detection with higher bandwidth (250 kHz in this work). This work clearly identifies the regime where capture rates are voltage dependent (<|125 mV|) and where translocation causes signal to be lost (>|125 mV|). Using a 1D drift–diffusion model for translocation, the confined diffusion coefficients and electrophoretic mobility were estimated from the drift velocity dependence on the applied voltage. Under most conditions, these appear to be fixed values for each protein/pore system studied offering a pathway to uniquely identify proteins from mixtures in the future. Similar efforts suggest that these same approaches can work with conical glass nanopores as well as with nanofabricated cylindrical pores.[43,44]
In the above work, all molecular species that can partition into the pore will generate a resistive pulse–provided that they are retained in the pore for a sufficiently long time. To detect specific analytes from a mixture, it is often necessary to add selectivity to the nanopores by embedding selective molecular recognition elements.[45,46] Using antibodies immobilized in a conical glass nanopore, Siwy showed that nanopore sensing can be used to detect ricin using anti-ricin.[45] With strong molecular interactions, the pores do not offer resistive pulses, but a global reduction in current. By using weaker interactions, protein–antibody interactions can produce resistive pulses[46] which allows for single-molecule detection.
Using a molecular recognition approach, a protein rotaxane[47] was fabricated that mimics the functionality shown for DNA-based molecular rulers.[48] The protein rotaxane offers a unique force spectroscopy, which can be used to estimate the directionality of protein transport as a function of applied potential.
A critical limitation of the solid-state systems, on which many of these studies rely, is the proclivity of solid-state pores to foul. To overcome this issue, pores can be coated with an antifouling layer such as polyethylene glycol.[49] Yusko and colleagues offer a completely novel nanopore system where a solid-state nanopore is coated with a solid-supported lipid nanopore.[40,41] The lipid molecules serve two purposes: they are antifouling and resist the irreversible binding and clogging that plague other solid-state nanopore systems and they offer dynamic molecular recognition elements. By anchoring analyte molecules (i.e., biotin) the translocation is controlled by the lipid-diffusion rates rather than bulk solute diffusion rates. Thus, the resistive pulses are slowed and are more easily resolvable. Building on the electrophoretic arguments of Sexton[35] and Rant[38] and modified for the novel geometry, the size and charge of streptavidin, anti-biotin Fab, and anti-biotin antibodies were estimated. Because translocation can be controlled through the lipids and the protein attachment to the wall can be specifically orientated, and the electric fields in the nanopore are high, Yusko and colleagues were able to estimate size, geometry, and dipole moments from a number of natively folded proteins.[40]
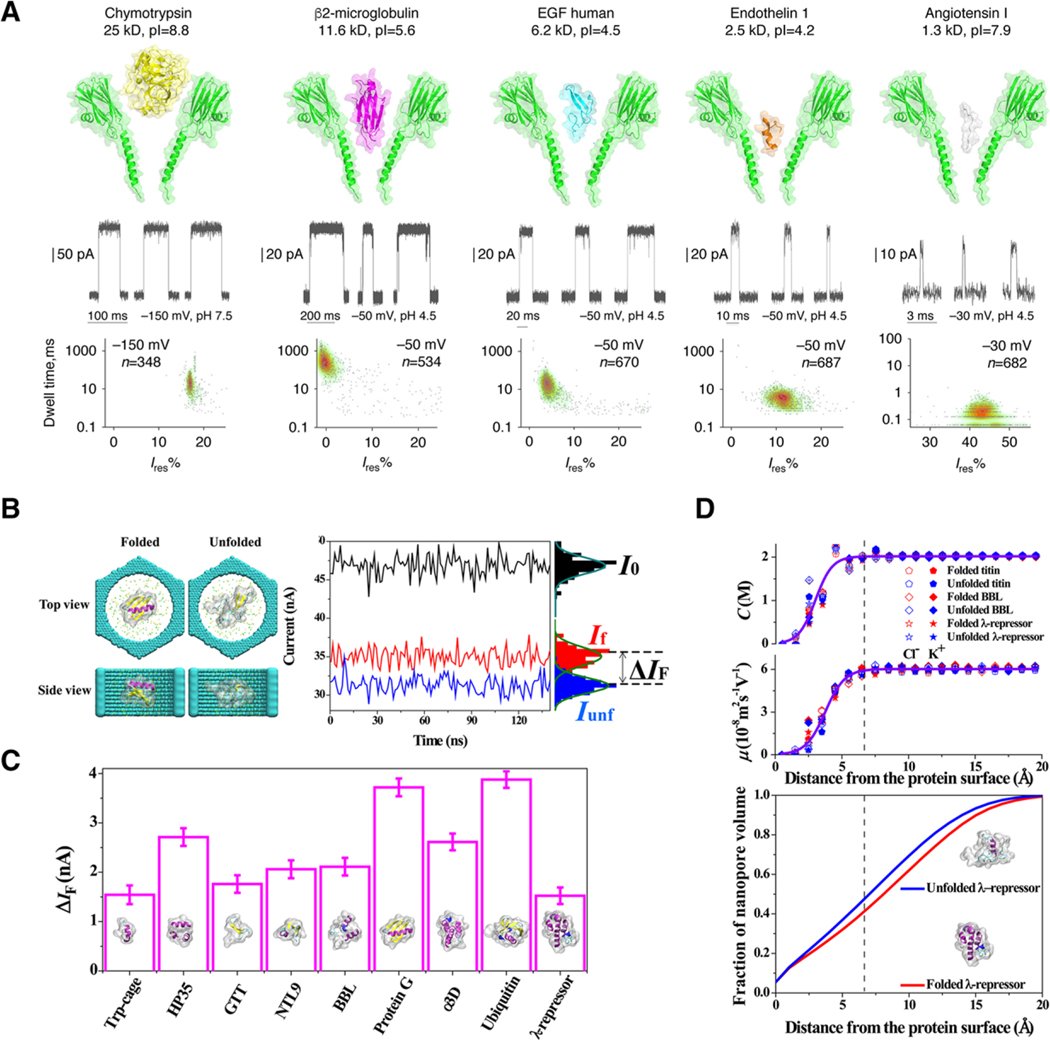
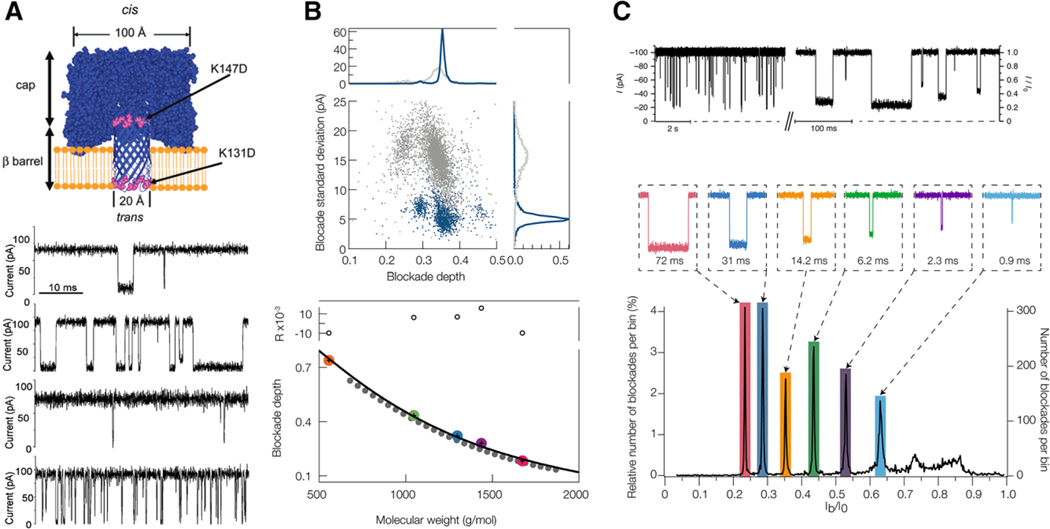
Although most of the above work relied on solid-state pores for their geometrical attributes, Maglia and colleagues have developed several different protein–pore systems that allow the detection and characterization of a wide variety of protein analytes.[50,51] One compelling pore is an engineered version of fragaceatoxin C (Fra C), which allows the detection of a wide range of intact, folded proteins (Figure 2A).[51] FraC can not only discriminate between intact proteins through simple scatter plots of dwell time and residual current (or blockade depth), but also shows promise for detecting subtle difference in primary structure—demonstrated by the discrimination of endothelin 1 and 2 which differ by a single tryptophan residue. The choice of a protein nanopore is critical to the success of the analysis. Wantanabe and colleagues propose a classification scheme based on the pore forming attributes and noise characteristics of several different pore forming proteins.[52] They used the results of their study to detect granzyme B with perforin nanopores.
Figure 2.
Nanopore characterization of full-length proteins provides unique “spectral” fingerprints for size, chemistry and folded state. A) The flexible FraC pore can be used as a sensor for a wide range of proteins in part due to the conical structure of its transmembrane domain. Proteins are driven into the pore by electroosmotic forces and produce blockades characteristic of the size and depth of the protein partition. Reproduced under the terms and conditions of the Creative Commons Attribution license 4.0.[51] Copyright 2017, the authors, published by Springer Nature. B) Molecular dynamic simulations suggest that discrete folding states can be readily resolved in solid-state nanopores. C) The folded proteins create current blockades based on the hydrodynamic size of the protein, as it is oriented inside the pore. Error bars were calculated from the standard error of the current over 150 ns in 1.2 ns segments. D) Volume exclusion is calculated as a gradient that extends from the “surface” of the protein, which alters the mobility of ions inside the pore leading to a clear signal based on the secondary structure of the protein in folded and unfolded states. Reproduced with permission.[60] Copyright 2017, American Chemical Society.
3.1.3. Protein–Protein Interactions
Specific protein–protein interactions offer another sensing scheme that is reminiscent of molecular recognition.[53,54] Of particular interest, Japrung and colleagues show that intrinsically disordered proteins can be characterized by their ability to affect changes in the ionic current based on their orientation as they adsorb to the nanopore wall.[54] Two such proteins with similar zeta potentials, disordered activator of thyroid hormone and retinoid receptors (ACTR) with nuclear coactivator binding domain (NCBD) of CREB-binding protein and their complex, can either enhance the ionic current or reduce the ionic current. Some evidence is presented for the existence of at least two structural states of the ACTR and NCBD protein based on apparent population densities in blockade depth versus residence time scatter plots. However, the complex appears to only resolve a single state. This report highlights the need to consider ionic strength as an important variable for optimizing nanopore characterization of proteins.
The pore systems have also produced studies that highlight structural variations of proteins in nanopores.[55,56] Waduge and colleagues used a series of proteins with different alpha helix:beta sheet ratios, which correlate to the stiffness of a protein to study secondary structure fluctuations and conformational changes of proteins. They determined that large flexible proteins produce a broader distribution of resistive pulses than their stiffer counterparts. Some correlation was shown with simulation and nanopore measurements which were used to detect and confirm a conformational change within calmodulin, a calcium binding protein.
Beyond secondary and tertiary structures, there is now some indication that the primary sequence may be detectible with a solid-state nanopore.[57,58] In this work, subnanometer pores were used which force single-file translocation. The subtly different volumes of each amino acid residue can offer at least a fingerprint of the primary structure of the protein. With further refinements, in chemistry and computational algorithms, complete sequencing may be achievable. Molecular modeling suggests that the charge density along a protein may further complicate the directional translocation required by such a method particularly since amino acids can adopt positive, negative, or neutral charges with highly variable charge densities.[59]
To facilitate a deeper understanding of all of these systems, Si and Aksimentiev (Figure 2 B–D) offer a comprehensive molecular dynamics (MD) simulation that unambiguously tracks calculated ionic current signals to conformational changes of proteins inside a 12-layer graphene nanopore.[60] This work uses ultra-long time (i.e. milliseconds) all atom simulations of protein folding trajectories, and estimates the change in current of several proteins in their native and unfolded state (Figure 2 B,C) Critically, the simulations show that the protein ionic current blockade is due in part to reduced ion mobility in the vicinity of the protein (Figure 2D). This approach allows a more refined volume dependence for proteins in the pore that specifically incorporates the 3D structure of the protein.
3.1.4. Practical Applications: Detection of Posttranslational Modification
Posttranslational modification is a high-impact target for nanopore detection in protein systems. Bayley and colleagues developed a method for using αHL channels to detect phosphorylation of denatured thioredoxin.[61,62] In this work, thioredoxin was modified with an oligonucleotide to capture the protein with hemolysin and begin an electric-field-induced force on the protein to unfold and pull the molecule through the pore. By characterizing the blockade depth (residual current) and noise during the translocation step, mutant proteins would be identified by their phosphorylation state (0, 1, or 2) and location of the modification. In general, phosphorylation decreases the observed noise and results in shallower blockades.
Two systems have been published for the direct detection of ubiquitin and ubiquitination of proteins using either solid-state pores[63] or engineered cytolysin A (ClyA) pores.[64] In the former, the S/N ratio of a solid-state nanopore was improved by a fine adjustment of pH to maximize the residence time of ubiquitin (Ub MW 8.5 kDA) resulting in discrimination of UB, diUB, and pentaUB with 3 nm diameter pores. Impressively, the work was also able to detect signal difference between dimers formed by linkage between two different sites leading to subtly different 3D structures. In the latter paper, ClyA pore was modified with a tryptophan substitute to create a protein sensor for real-time ubiquitination. This mutation increases the retention time of a ubiquitin-conjugating enzyme (E2) by greater than tenfold. The modified pore was then used to detect Ub addition to E2. When ATP is added to the solution E2 takes on Ub in two different orientations, which are uniquely detected and characterized. Because of the single-molecule nature of the experiment, the cascading ubiquitination steps were observed in real time.
3.1.5. Practical Application: Mechanism for Neurodegenerative Disease
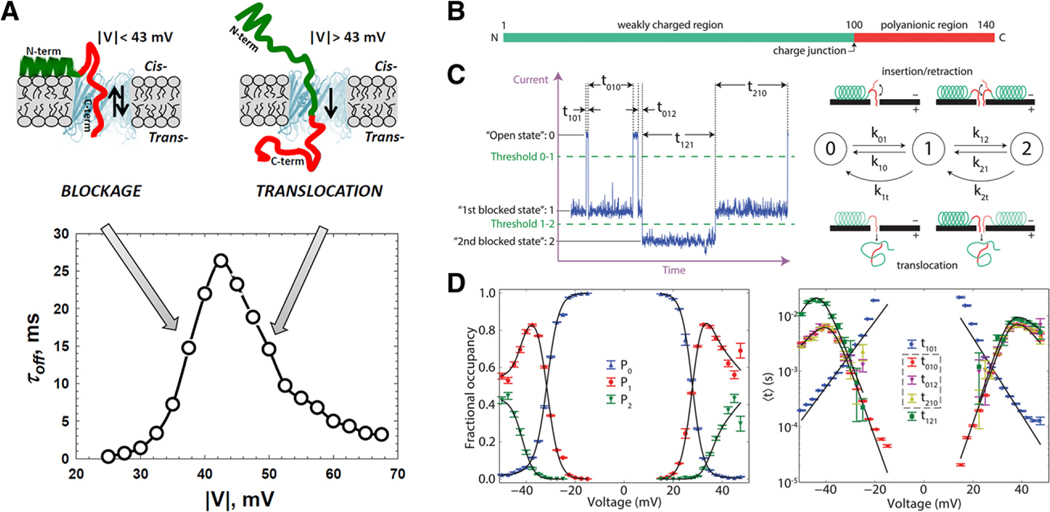
The detailed progression of a translocation event is often difficult to identify due to the lack of information in a single nanopore event (Figure 3). This is not the case for alpha-synuclein (α-syn) at the outer mitochondrial membrane as it interacts with VDAC. α-syn (a small intrinsically disordered protein) has been implicated in a number of neurodegenerative diseases, including Parkinson’s disease, through its interaction with mitochondria. One hypothesis is that α-syn interacts with regulatory channels such as VDAC. Rostovtseva and colleagues presented a single-molecule study of α-syn/VDAC, which demonstrated that α-syn can translocate the VDAC channel when the transmembrane potential exceeds approx. 40 mV gaining access to the respiratory chain of a cell under potentials well within the observed potentials of excitable membranes (Figure 3A).[65] By observing the detailed trajectory of the resistive pulses and applying a rigorous Markov modeling analysis to the events (Figure 3C), a full energy landscape was produced, which details the translocation process with probabilistic trajectories as a function of the membrane potential (Figure 3D).[66] Such analysis has direct implication in the molecular mechanism for diseases such as Parkinson’s, but it also offers a generalizable mechanism for detection of proteins modified with selectivity tags.[67]
Figure 3.
α-Synuclein regulates the permeability of VDAC pores in the outer mitochondrial membrane. A) The voltage dependence of the residence time of α-Syn in VDAC can be used as an estimate for the membrane potential necessary to drive translocation. α-Syn can be modeled as a diblock polymer with a B) polyanionic region and a C) weakly charged region which presents characteristic states during a resistive pulse event. D) Modeling the state transitions with a three-state Markov chain model allows for the potential dependent occupancy and mean state times to be modeled resulting in a detailed thermodynamic description of the translocation process. Error bars represent one standard deviation from the mean using a bootstrap analysis. Reproduced with permission.[65,66] Copyright 2015, American Society for Biochemistry and Molecular Biology and Copyright 2017, The Royal Society of Chemistry.
3.2. Protein Unfolding
3.2.1. Unfolding: Outside the Pore
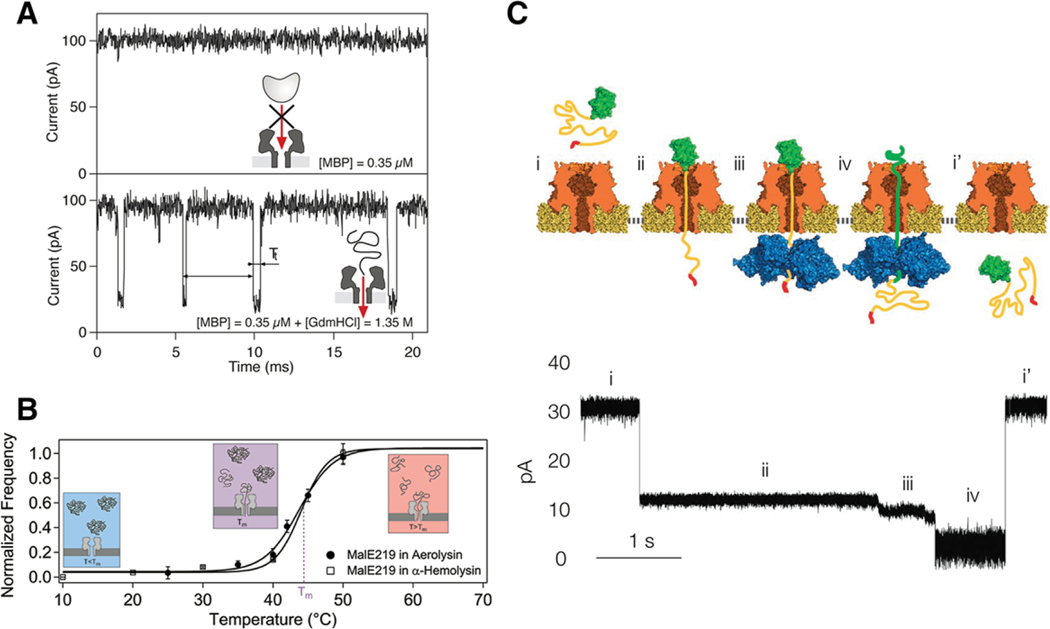
To detect and characterize proteins and peptides in biological nanopores it is often necessary to denature the analytes to accommodate the dimensions of the cavity. This method was pioneered by Auvray and colleagues over a number of years using chemical denaturants (guanidinium hydrochloride; Figure 4A)[68–71] or temperature (Figure 4B).[72] Similar treatments of BSA with urea,[73,74] temperature,[74] and electric fields[74] also display characteristic denaturation in solid-state systems. The unfolding shows striking effects on the resistive pulse signal. First, the event frequency is nearly 100-fold faster for the folded protein when normalized for concentration and the dwell time is nearly tenfold longer for the folded protein and both effects are exponentially dependent on the applied voltage suggesting a free-energy barrier model for transport, consistent with other molecular systems such as polyethylene glycol.[17] By reducing the pore diameter from ≈20 to ≈3 nm the retention mechanism for denatured proteins becomes less dependent on electroosmotic flow and presumably more dependent on entropy changes.[75,76] However, Cressiot and colleagues do not present temperature dependent analysis to solidify the arguments for entropy. The question of whether or not denatured protein translocates through a nanopore is an important and difficult one to address given the small molecular flux through nanopores and the lack of methods for amplifying signal on the trans-side of the pore. Although Pastoriza-Gallego et al. describe an interesting approach to addressing this problem by tagging MBP with a 60 bp DNA molecule with which they can perform protease chain reaction (PCR) amplification on molecules that move through the aerolysin pore.[77]
Figure 4.
Examples of protein unfolding experiments outside and inside the nanopore. A) Denaturing MBP with Gdm-HCl unfolds the protein outside the pore while maintaining the structural integrity of the αHL pore. The unfolded protein threads into the pore yielding numerous current blockades. B) Mutated MBP can also be unfolded with heat. In a similar fashion as the chemical denaturing, the protein unfolds outside the pore and leads to sizable current blockades. The relative frequency of blockades correlates with the unfolding population for both αHL and aerolysin pores. C) Unfolding has also been reported within the pore volume as was the case for the ubiquitin-like protein Smt3. An ssrA tag was attached to the Smt3 as a means for capture by a ClpX unfoldase on the trans-side of an αHL pore. The molecular motor unwinds the protein across the pore volume, which yields several substates corresponding to different stages of the unfolding process. Reproduced with permission.[71,72,78] Copyright 2007, American Physical Society; Copyright 2012, American Chemical Society; Copyright 2013, Springer Nature.
3.2.2. Unfolding: Inside the Pore
In addition to unfolding proteins outside the pore to facilitate pore entry, some studies demonstrated direct observation of protein unfolding inside the pore. As described above, this was shown for a DNA-tagged thioredoxin protein where an applied voltage could pull on the DNA tag and unfold the protein through the nanopore sensor.[61] A similar report from Nivala et al. showed that ClpX unfolding protein, located on the trans-side of an αHL pore, could capture a polyanionic tail attached to a target protein entering from the cis-side of the pore.[78] Figure 4C illustrates the principle of operation where the unfolding molecular motor unwinds the protein in a manner similar to the applied voltage study by Rodriguez-Larrea and Bayley. Freedman et al. showed that high electric field strengths, accessible with a silicon nitride pore, could be used to unfold ubiquitin protein[79] More recently, Meervelt et al.[80] reported the use of ClyA nanopores to study protein conformation fluctuations of the substrate binding domains (SBD1 and SBD2) of the ABD importer Glop. These proteins undergo conformational changes upon binding with ligands (asparagine for SBD 1 and glutamine for SBD2) and they report similar conformational change kinetics inside the ClyA pore to those measured in the bulk. It is worth noting that these unfolding studies have opened up an important class of single-molecule protein experiments that utilize the pore’s ability to monitor unfolding kinetics and pathways while applying forces to the protein as it unfolds.
3.3. Aptamer-Based Detection
Given the role nanopore sensing plays in DNA detection, it is natural to take advantage of the binding of peptides and proteins to a variety of aptamer sensors for protein detection. DNA aptamers are short oligonucleotide sequences that exhibit specific binding to a wide variety of protein molecules.[81] Early nanopore studies with DNA aptamers started in 2011 with Kawano et al.’s development of aptamer-based cocaine detection.[82] The aptamer-based selectivity approach was used for protein detection by Rotem et al. who demonstrated the use of aptamer attachment to the cis-side entry of an αHL pore.[83] They chose a thrombin binding aptamer (aptamerT4) and showed they could detect thrombin through variations in the transmembrane current. This approach enables thrombin protein detection at nanomolar levels. The large vestibule volume of ClyA nanopores provides an optimal tool for incorporating DNA aptamers for enhanced protein detection as demonstrated for various protein ensembles (Lysozyme, Dendra2 M159A, Human Thrombin and Bovine Thrombin).[84,85] Aptamer-based nanopore detection has also been demonstrated with aptamers outside the pore. Li et al. detected a variety of proteins and molecules (VEGF, thrombin, and cocaine) with a duplex DNA structure where one strand of the duplex is an aptamer that will selectively bind to one of the aforementioned molecules.[86] Without the molecule there, the duplex molecule does not interact with the pore, but in the presence of the target, the aptamer is removed and the DNA probe interacts with the pore to give rise to unique current blockades (Figure 5A). Magnetic bead–based analyte extraction enabled picomolar level detection. Aptamer-based detection has also been used in conjunction with glass nanopipette sensors. Lin et al. functionalized 5 nm gold particles with DNA aptamers for targeted detection of lysozyme.[87] Figure 5B illustrates the principle of operation where aptamer labelled gold nanoparticles give rise to current blockades, but when lysozyme is bound to the aptamer this gives rise to sizable and unique current blockades. They used this approach to detect lysozyme, cytochrome C, and trypsin and to show that the selectivity of the DNA aptamer enables detection from various background mixtures. Sze et al. developed a rapid and simple aptamer assay that grafts a selective binding region to a long λ-DNA carrier.[88] This work is also noteworthy because it demonstrates the possibility of nanopore sensing in complex physiological fluids. In all cases, combining the aptamer selectivity both in and outside the pore enables accurate sensing of target analyte in various mixture conditions.
Figure 5.
Aptamer-based protein sensing performed indirectly with biological nanopores and directly with a glass nanopipette. A) Schematic illustration of a protein probe that utilizes a weakly hybridized aptamer and DNA probe. When the protein analyte interacts with the aptamer construct, the DNA aptamer binds to the protein and the DNA probe (modified with CB7) yields unique current block signatures. B) Attaching specific aptamers to gold nanoparticles enables clear discrimination between several different proteins with a nanopipette sensor (lysozyme, cytochrome C, trypsin). Both figures shown here are reproduced with permission.[86,87] Copyright 2015, Wiley-VCH and Copyright 2017, The Royal Society of Chemistry.
3.4. Proteins Bound to DNA and RNA
3.4.1. Fundamental Characterization
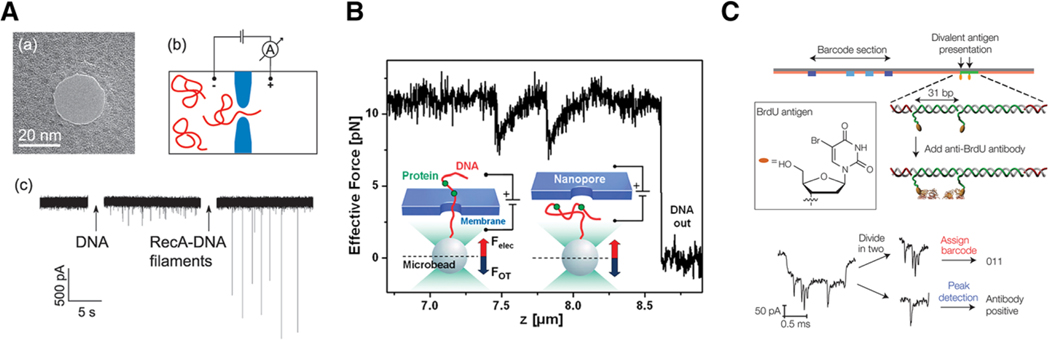
One of the most fundamental interactions in biology is the binding of proteins to DNA and RNA.[89,90] As reported in the unfolding subsection, DNA is oftentimes used as a handle to feed and/or pull larger molecules through the pore. This has motivated a large number of studies to characterize protein–DNA/RNA interactions with nanopore sensors. Most of these efforts require the protein to remain in its functional form meaning that many of the reports in this section utilize solid-state pores. Early efforts begin with the work of Smeets et al. who reported RecA-DNA binding through solid-state pores.[91] Figure 6A shows how RecA-DNA conjugates give rise to sizable and long-lived current blockades (1–100 ms). Further refinements from more detailed analysis of substate transitions enabled identification of low and high coverage regions of RecA,[92] and optical tweezers (see Figure 6B) were used to distinguish between different proteins (RecA and EcoRI) bound to DNA.[93] A more recent optical tweezer–based experiment was reported by Bulushev et al., which studied RNA polymerase, and dCas9 proteins bound to dsDNA. They were able to move the DNA–protein construct through the pore and localize where the protein binds from the steps in the current and force versus time curves.[94] Moving beyond RecA experiments, Raillon et al. demonstrated RNA polymerase–DNA interactions,[95] and Squires et al. showed transcription factor (zif268) binding with DNA and showed that nanopores could distinguish between nonspecific and specific binding.[96] Graphene has also been used to characterize RecA-DNA conjugates, which highlights the important role graphene may play in the development of future nanopore sensors.[97]
Figure 6.
Nanopores enable clear detection of protein–DNA binding for improved characterization and sensor development. A) RecA protein, bound to dsDNA, yields deep current blockades as compared to blank DNA alone. B) Optical tweezers can be used to controllably thread the DNA-bound protein molecules through the pore. This example shows two EcoRI proteins bound to a DNA strand and the tweezers allow one to measure the force required to move the protein through the pore. C) Targeted attachment of streptavidin, along with hairpin-based DNA barcoding, enables high efficiency and specific antibody detection from heterogeneous mixtures. All figures shown here are reproduced with permission.[91,93,101] Copyright 2008, 2011, 2015, American Chemical Society.
3.4.2. DNA–Protein Binding for Biosensing
In addition to better understanding the fundamental phenomenon behind DNA–protein binding, other groups have utilized DNA–protein constructs to improve nanopore sensing to enable identification and characterization of proteins in a variety of mixtures. Streptavidin tagged to biotin modified DNA gives rise to clear current blockades and suggests a means for developing a single-molecule immunoprecipitation technique.[98] Single-stranded DNA protein binding was also used to improve ssDNA sensing with solid-states pores,[99] and to study the binding of Escherichia coli derived SSB with ssDNA.[100] To further improve nanopore sensing, protein binding has also been used to improve the sensing and identification of proteins from a background mixture. Bell et al. used a cut version of the m13mp18 virus genome to attach linkers capable of attaching various binding proteins. These individual proteins can be detected as short downward spikes while the DNA backbone yields longer-lived blockade states. They showed clear detection of proteins separated by approx. 600 nm along the DNA backbone and their system noise suggests they should be able to resolve proteins separated by approximately 100 nm. They also demonstrated specific detection of streptavidin in a background with four different proteins.[101] Figure 6C shows how this approach was expanded by attaching DNA hairpin constructs to the DNA to enable “barcode” identification that allows them to attach specific antibodies to a particular barcode. They use this to detect and identify four different proteins in various mixtures.[102] In a similar experiment, Plesa et al. describe experiments where anti-DNA antibodies were attached to DNA.[103] The antibodies are positively charged so they will not enter the pore under the applied potential needed to force DNA through the pore. However, these antibodies exhibit strong nonspecific binding to DNA and give rise to short-lived current blockade spikes as the DNA moves through the pore. Interestingly, they can resolve multiple antibodies bound to a single DNA molecule moving through the pore. The authors suggest that this could be used as a sensor to target specific proteins that would bind to specific antibodies. In a slight modification to the DNA–protein binding approach, Lee et al. modified the inner surface of glass nanocapillary tips with U6snRNA that is known to bind SART 3 protein. RNA-coated glass tips enhance the SART 3 residence times approx. sixfold when compared to DNA modified glass nanopipettes.[104] The application of DNA–protein binding for biosensing points toward the near-term development of rapid and accurate detection of proteins from complex mixtures. The coded nature of the analyte enables simplified fabrication processes, such as those found in glass nanopipettes, which offer a cost-effective and facile approach to nanopore sensing that may enable rapid and accurate protein identification in a clinical setting.
4. Peptide Sensing
4.1. Protease Activity
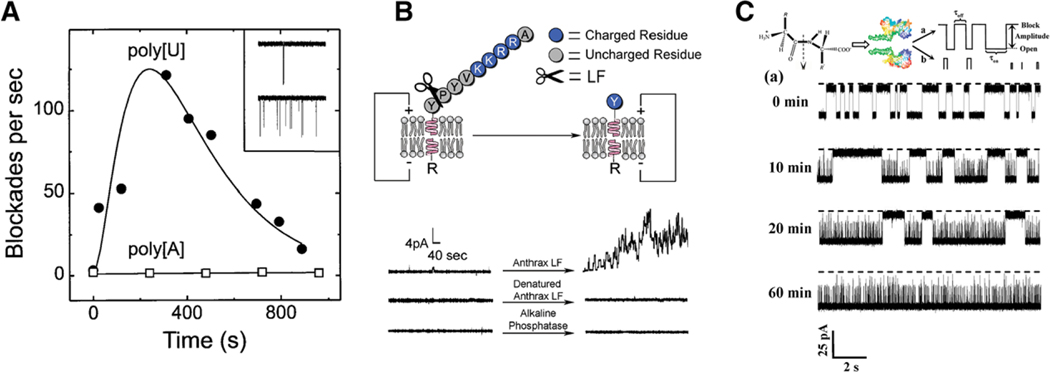
Proteolysis is an important and critical phenomenon responsible for forming numerous peptides from larger proteins. In addition, proteins are routinely broken down into smaller peptides for mass spectroscopy characterization. Given the smaller sensing volume of the bionanopore sensors, cleavage proteins have found wide use in nanopore sensing. These efforts can be traced back to the seminal paper on DNA nanopore sensing, where Kasianowicz et al. demonstrated enzymatic hydrolysis of poly U with ribonuclease A proteins.[5] Figure 7A shows selective cleavage of polyU molecules as evidenced by increasing on-rate with time. Hydrolysis was also used as a means for detecting low levels of the anthrax secreted lethal factor (LF). Macrae et al. first used proof-of-principle to demonstrate alkaline phosphatase would cleave the c-terminus of gramicidin A ion channels.[105] Figure 7B shows how this changes the current signature of the gA channel and shows direct evidence of cleavage. They then attached a peptide to gA that cleaves with LF so that changes in the current correspond to interactions with the LF in solution. Interestingly, before cleavage they saw no current, then under LF cleavage they saw current spikes. They demonstrated LF detection down to 10 nM. Trypsin-based cleavage studies have also been reported by several groups.[106–108] Figure 7C shows nanopore detection of trypsin activity on amyloid beta peptides as detected by a mutated form of the αHL pore (M113F)7.[106] Aerolysin pores were used to study the depolymerization of hyaluronic acid with the addition of hyaluronidase.[109] Wang et al. used cleavage-based nanopore sensing to develop a botulinum neurotoxin type B sensor (BoNT-B) by taking advantage of the fact that BoNT-B acts as a cleavage enzyme on synaptobrevin peptides.[110] The smaller fragment cleaved from this peptide gives rise to measurable current blockades in an aerolysin nanopore and indicates the presence of the toxin. Rauf et al. used a cleavage-based experiment to measure DNA methyltransferase activity. An adenine methyltransferase protein (Dam) adds methyl groups to target DNA and an MboI restriction endonuclease cleaves the CH2-free version, but not the methylated DNA.[111] The cleaved molecules give rise to detectable blockades in a wild type αHL pore while the methylated DNA gives rise to long-lived blockades. Recently, Shang et al. described the development of a cleavage-based sensing protocol for human 8-oxoguanine DNA glycosylase (hOGG1) activity, which is indicative of disease onset. The protocol requires synthesis of a DNA hybrid complex that contains Goxo and cleaves in the presence of hOGG1, the nanopore sensor detects the release of the DNA via deep and long-lived blockades.[112]
Figure 7.
Nanopore-based detection of protease activity. A) Early efforts demonstrated specific hydrolysis of polyU and not polyA with the addition of ribonuclease A through the detected increase of blockade events. Reproduced with permission.[5] Copyright 1996, The National Academy of Sciences of the USA. B) Binding a specific peptide sequence to the cis-side of the gramicidin A pore yielded clear current fluctuations upon cleavage with the lethal factor (LF) protein. This demonstrates the possibility of developing sensitive and rapid sensors for numerous transmembrane forming toxins. C) Trypsin-based cleavage of the A-β(10–20) peptide is easily detected with the onset of two distinct, shorter-lived, blockade states. Detection in this case was performed with a mutated form of the αHL pore (M113F)7. B,C) are reproduced with permission.[105,106] Copyright 2009, American Chemical Society.
4.2. Peptides Inside the Pore
Peptides are biopolymers with a small number of amino acid (aa) residues (<50 aa). These smaller molecules exhibit a wide degree of functionality and present ideal candidates with which to study protein kinetics and proteolysis activity. Their reduced size makes them ideal targets for nanopore-based characterization of a wide degree of behaviors seen in larger proteins. While tertiary and quaternary structures are nonexistent in peptides, primary and secondary structures are prevalent and are ideally suited for nanopore study.
4.2.1. Peptide Kinetics
Early work with peptide characterization focused on studying on and off rate kinetics of alpha helical peptide structures with the αHL pore (GPPn,[113] and (AAKAA)n[114]) and the aerolysin pore (DxAyKz).[115] Beta hairpin folded peptides (G41 peptide from the B1 domain of G protein, along with several mutations) represent another important class of secondary structure and these were studied by Goodrich et al. with αHL.[116] Singh et al. used a hetero-oligomeric protein channel similar to MspA to study peptide binding and translocation through the pore in order to expand the biosensing of peptides beyond aerolysin and αHL.[117] This pore was cation selective with a large negative charge density on the trans-side. Mereuta et al. performed a series of experiments with CAMA 1 and CAMA 3 peptides under various pH and ionic strength conditions.[118] They found that adjusting the interaction time of the CAMA peptides through pH, enables the identification of different substates inside the pore.[119] Solid-state pores have also been used to characterize peptide folding states. Niedzwiecki et al. used a 10 nm thick silicon nitride membrane and a low noise amplifier (25 pA rms at 100 kHz) to sense different states of a helical peptide (GCN4-p1).[120]
4.2.2. Peptide Translocation Through Outer Membrane Pores
Peptide transit through outer membrane channels plays an integral part of the trafficking of molecules across cellular and organelle membranes. Most methods for characterizing peptide transit lack the ability to observe single-molecule trajectories, but nanopore sensing enables direct observation of these processes as demonstrated by Wolfe et al. who describe the transport of positively charged polypeptides through a mutated αHL pore. Figure 8A illustrates how placing aspartic acid rings in place of lysine rings at the trans-side entry and constriction ring of the αHL pore modifies the on-rate and off-rate of the cationic peptides and suggests a mechanism for transport through outer membrane mitochondrial channels.[121,122] Peptide transport through mitochondrial membranes is mostly carried through the Tom40 channel, which Mahendran et al. studied with the pF1beta presequence peptide.[123,124] Further translocation studies through the MOMP porin with short polyarginine peptides were carried out and voltage dependent kinetics suggested translocation through the pore.[125]
Figure 8.
Peptide sensing inside the pore volume for peptide translocation and identification studies. A) Mutated forms of the pore can be used to facilitate peptide transport. Here Syn B2 polypeptides enter and move through the trans-side of the pore more easily in the presence of charged rings placed at the pore entrance and pinch point. B) Denaturing peptides greatly reduces current blockade fluctuations and narrows the corresponding peak in the blockade distribution for angiotensin 1 (top). This enables accurate peptide identification for the near neutral peptides ranging in size from 500–1500 Da (bottom). Several different peptides (angiotensin 1, angiotensin 2, leu-enkephalen, neurotensin, and QBP1) are all well characterized with a linear ohmic model that connects peptide size to blockade depth. C) Aerolysin pores enable high-resolution detection of polydisperse mixtures of homopolymer peptides ranging in size from 4 to 10 repeat units. All figures are reproduced with permission.[121,132] Copyright 2007, American Chemical Society; Copyright 2018, American Chemical Society and reproduced under the terms and conditions of the Creative Commons Attribution license 4.0.[134] Copyright 2018, the authors, published by Springer Nature.
4.2.3. Peptide–Metal Binding
Nanopore sensing can also characterize metal folding peptides as shown for the case of the Zn-finger peptide (Zif268),[126] copper-based CAMA peptides,[127] and the HH14 uranyl binding peptide.[128] In all cases, the addition of specific metals cause the peptides to fold which changes the current blockade characteristics. Enantiomer-based peptide sensing was demonstrated by Boersma et al. who attached a Cu+2 ion receptor into the lumen of an αHL channel.[129] The copper binds d and l forms of amino acids differently and it was shown that this could be used to detect differences in blockade current from tyrosine, phenylalanine, cysteine, and aspartic acid. They also detected differences in the d and l forms of tryptophan through the blockade noise and demonstrated simultaneous detection of d and l types of Phe and Tyr.
4.2.4. Nanopore-Based Peptide Identification
Peptide identification with nanopore sensing has been the focus of more recent efforts as demonstrated by Asandei et al. in a paper describing how the constriction ring in αHL can sense differences between three different 6 amino acid length portions of a polypeptide. The peptides are engineered to contain 12 aa positive and negative charge handles (arginine and glutamic acid) and they find different current signatures between the three different sequences studied.[130] Ji et al. used phi29 DNA packaging motor nanopore to sense larger peptides TAT, MAR, and EpCAM.[131] They found distinct blockade distributions for each and they confirmed translocation of the TAT peptide by labelling it with Cy3 and using fluorescence to indicate an increasing number of translocated molecules with time. Chavis et al. utilized gold clusters and various solution modifications (Gdm-HCl and pH) to optimize detection of a series of neutral and cationic peptides.[132] As can be seen in Figure 8B, the presence of Gdm-HCl removes secondary structure in the angiotensin 1 peptide, which leads to a significant decrease in the blockade current noise and a corresponding reduction in the width of the current blockade distribution. It was also shown that a variety of water soluble peptides ranging from 550 to 1600 Da could be identified from the corresponding depth of their current blockades. FraC nanopores have been shown to enable detection and characterization of peptides and proteins between 2.5 and 25 kDa (see Fig. 2A).[51] Taken together, these papers demonstrate the wide range of protein and peptide analytes that can be effectively sized with a simple nanopore measurement. Lysenin channels have been used to detect angiotensin 2 peptides as described by Shreshta et al.[133] They verified translocation through the pore by performing a 36 h run with 22700 channels and then performed LC-MS on the trans-side solution to prove analyte was present. This represents a rare demonstration of direct peptide translocation evidence. Figure 8C shows that aerolysin can be used to clearly distinguish between different homopolymer repeats of arginine (5–10 amino acids in length).[134] They confirm with trypsin activity over a several hour period detecting the evolution of 9-mers to 4-mers. They find similar behavior for polylysine peptides as well, and they showed the ability to discriminate between different, 10 aa length, synthetic peptides. This suggests that aerolysin may be an optimal channel with which to perform small peptide characterization. The mass of a large number of important peptides falls within an ideal range for nanopore detection (≈300–5000 Da) and the ability to accurately identify and characterize these peptides in solution represents a critical sensing problem in the field of peptidomics. Future efforts in this area will continue and will most likely focus on improving the nanopore’s ability to distinguish between similarly sized peptides from mixtures of ever-increasing complexity and polydispersity.
5. Conclusion
Nanopore-based resistive pulse sensing began in earnest with the observation that ssDNA gives rise to unique and descriptive blockades, and this motivated the initial pursuit of DNA sequencing. Protein analysis with nanopores began with small peptide characterization and unfolding-based studies, but with the advent of solid-state nanopore sensors, protein sensing and characterization flourished. More recently, reports of bionanopore sensors being used to characterize a variety of protein and peptide-related phenomenon has demonstrated that nanopore-based sensing and characterization of proteins is entirely possible and provides useful information at the single-molecule limit. Future efforts in this area will most likely continue to pursue aptamer-based nanopore protein sensors and the development of optimized sensing geometries (i.e., the discovery of new protein nanopores,[50] improvements to graphene-based sensors, and controlled synthesis of solid-state pores via dielectric breakdown,[135] and DNA origami[136,137]) for development of portable sensing devices and sensors capable of sequencing proteins. The continued growth of nanopore sensors for protein and peptide detection should continue for years to come.
Acknowledgements
This article has been contributed to by US Government employees and their work is in the public domain in the USA.
Biographies

Joseph W. F. Robertson is a staff scientist in the Physical Measurement Laboratory at the National Institute of Standards and Technology. He received his Ph.D. in Chemistry from the University of Arizona in 2004 working under the guidance of Jeanne Pemberton. He then moved to the Max Planck Institute for Polymer Research where he studied biological charge transport through model lipid bilayer systems. His current research interests include advancing measurement science through biomimetic surface development, single-molecule biosensing, and new methodologies for membrane protein structure and function.

Joseph E. Reiner is an Associate Professor in the Physics department at Virginia Commonwealth University. He has been at VCU since 2011, and he worked at the National Institute of Standards and Technology from 2003–2011.Hereceived his physics Ph.D. from the University of Stony Brook in 2003 working under the guidance of Luis Orozco on cavity quantum electrodynamics. His primary research interests include single-molecule and single-cell detection and manipulation with an emphasis on nanopore sensing and optical tweezers.
Footnotes
Conflict of Interest
The authors declare no conflict of interest. Certain commercial materials, equipment, and instruments may be identified in this work to describe the experiments as completely as posible. In no case does such an identification imply a recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials, equipment, or instrument identified are necessarily the best available for the purpose.
Contributor Information
Dr. Joseph W. F. Robertson, Physical Measurement Laboratory, National Institute of Standards and Technology, Gaithersburg, MD, 20899, USA
Dr. Joseph E. Reiner, Department of Physics, Virginia Commonwealth University, Richmond, VA, 23284, USA
References
- [1].Coulter WH, US Patent 2656508, 1953.
- [2].DeBlois R, Bean C, Rev. Sci. Instrum 1970, 41, 909. [Google Scholar]
- [3].Bezrukov S, Kasianowicz JJ, Phys. Rev. Lett 1993, 70, 2352. [DOI] [PubMed] [Google Scholar]
- [4].Bezrukov SM, Vodyanoy I, Parsegian VA, Nature 1994, 370, 279. [DOI] [PubMed] [Google Scholar]
- [5].Kasianowicz JJ, Brandin E, Branton D, Deamer DW, Proc. Natl. Acad. Sci. U. S. A 1996, 93, 13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW, Biophys. J 1999, 77, 3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shi W, Friedman AK, Baker LA, Anal. Chem 2017, 89, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Feng Y, Zhang Y, Ying C, Wang D, Du C, Genomics Proteomics Bioinf. 2015, 13, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haque F, Li J, Wu H-C, Liang X-J, Guo P, Nano Today 2013, 8, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miles BN, Ivanov AP, Wilson KA, Doğan F, Japrung D, Edel JB, Chem. Soc. Rev 2013, 42, 15. [DOI] [PubMed] [Google Scholar]
- [11].Howorka S, Siwy Z, Chem. Soc. Rev 2009, 38, 2360. [DOI] [PubMed] [Google Scholar]
- [12].Dekker C, Nat. Nanotech 2007, 2, 209. [DOI] [PubMed] [Google Scholar]
- [13].Kasianowicz JJ, Robertson JWF, Chan ER, Reiner JE, Stanford VM, Annu. Rev. Anal. Chem 2008, 1, 737. [DOI] [PubMed] [Google Scholar]
- [14].Reiner JE, Balijepalli A, Robertson JWF, Campbell J, Suehle J, Kasianowicz JJ, Chem. Rev 2012, 112, 6431. [DOI] [PubMed] [Google Scholar]
- [15].Krasilnikov OV, Rodrigues CG, Bezrukov SM, Phys. Rev. Lett 2006, 97, 018301. [DOI] [PubMed] [Google Scholar]
- [16].Robertson JWF, Rodrigues CG, Stanford VM, Rubinson KA, Krasilnikov OV, Kasianowicz JJ, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reiner JE, Kasianowicz JJ, Nablo BJ, Robertson JWF, Proc. Natl. Acad. Sci. U. S. A 2010, 107, 12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Movileanu L, Trends Biotechnol. 2009, 27, 333. [DOI] [PubMed] [Google Scholar]
- [19].Oukhaled A, Bacri L, Pastoriza-Gallego M, Betton J-M, Pelta J, ACS Chem. Biol 2012, 7, 1935. [DOI] [PubMed] [Google Scholar]
- [20].Ying Y-L, Cao C, Long Y-T, Analyst 2014, 139, 3826. [DOI] [PubMed] [Google Scholar]
- [21].Kasianowicz JJ, Balijepalli AK, Ettedgui J, Forstater JH, Wang H, Zhang H, Robertson JWF, Biochim. Biophys. Acta 2016, 1858, 593. [DOI] [PubMed] [Google Scholar]
- [22].Halverson KM, Panchal RG, Nguyen TL, Gussio R, Little SF, Misakian M, Bavari S, Kasianowicz JJ, J. Biol. Chem 2005, 280, 34056. [DOI] [PubMed] [Google Scholar]
- [23].Silin V, Kasianowicz JJ, Michelman-Ribeiro A, Panchal RG, Bavari S, Robertson JWF, Membranes 2016, 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rostovtseva TK, Gurnev PA, Hoogerheide DP, Rovini A, Sirajuddin M, Bezrukov SM, J. Biol. Chem 2018, jbc.RA117.001569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Movileanu L, Howorka S, Braha O, Bayley H, Nat. Biotechnol 2000, 18, 1091. [DOI] [PubMed] [Google Scholar]
- [26].Howorka S, Nam J, Bayley H, Kahne D, Angew. Chem. Int. Ed 2004, 43, 842. [DOI] [PubMed] [Google Scholar]
- [27].Xie H, Braha O, Gu L-Q, Cheley S, Bayley H, Chem. Biol 2005, 12, 109. [DOI] [PubMed] [Google Scholar]
- [28].Cheley S, Xie H, Bayley H, ChemBioChem 2006, 7, 1923. [DOI] [PubMed] [Google Scholar]
- [29].Mohammad MM, Prakash S, Matouschek A, Movileanu L, J. Am. Chem. Soc 2008, 130, 4081. [DOI] [PubMed] [Google Scholar]
- [30].Nivala J, Mulroney L, Li G, Schreiber J, Akeson M, ACS Nano 2014, 8, 12365. [DOI] [PubMed] [Google Scholar]
- [31].Fahie MA, Chen M, J. Phys. Chem. B 2015, 119, 10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fahie M, Chisholm C, Chen M, ACS Nano 2015, 9, 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Perez-Rathke A, Fahie MA, Chisholm C, Liang J, Chen M, J. Am. Chem. Soc 2018, 140, 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Han A, Schürmann G, Mondin G, Bitterli RA, Hegelbach NG, De Rooij NF, Staufer U, Appl. Phys. Lett 2006, 88, 093901. [Google Scholar]
- [35].Sexton LT, Horne LP, Sherrill SA, Bishop GW, Baker LA, Martin CR, J. Am. Chem. Soc 2007, 129, 13144. [DOI] [PubMed] [Google Scholar]
- [36].Fologea D, Ledden B, McNabb DS, Li J, Appl. Phys. Lett 2007, 91, 053901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Krasniqi B, Lee JS, PLoS One 2014, 9, e88004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Firnkes M, Pedone D, Knezevic J, Döblinger M, Rant U, Nano Lett. 2010, 10, 2162. [DOI] [PubMed] [Google Scholar]
- [39].Plesa C, Kowalczyk SW, Zinsmeester R, Grosberg AY, Rabin Y, Dekker C, Nano Lett. 2013, 13, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yusko EC, Bruhn BR, Eggenberger OM, Houghtaling J, Rollings RC, Walsh NC, Nandivada S, Pindrus M, Hall AR, Sept D, Li J, Kalonia DS, Mayer M, Nat. Nanotech 2017, 12, 360. [DOI] [PubMed] [Google Scholar]
- [41].Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M, Nat. Nanotech 2011, 6, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Larkin J, Henley RY, Muthukumar M, Rosenstein JK, Wanunu M, Biophys. J 2014, 106, 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li W, Bell NAW, Hernández-Ainsa S, Thacker VV, Thackray AM, Bujdoso R, Keyser UF, ACS Nano 2013, 7, 4129. [DOI] [PubMed] [Google Scholar]
- [44].Steinbock LJ, Krishnan S, Bulushev RD, Borgeaud S, Blokesch M, Feletti L, Radenovic A, Nanoscale 2014, 6, 14380. [DOI] [PubMed] [Google Scholar]
- [45].Siwy ZS, Trofin L, Kohli P, Baker L, Trautmann C, Martin C, J. Am. Chem. Soc 2005, 127, 5000. [DOI] [PubMed] [Google Scholar]
- [46].Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U, Nat. Nanotech 2012, 7, 1. [DOI] [PubMed] [Google Scholar]
- [47].Biesemans A, Soskine M, Maglia G, Nano Lett. 2015, 15, 6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Henrickson SE, DiMarzio EA, Wang Q, Stanford VM, Kasianowicz JJ, J. Chem. Phys 2010, 132, 135101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Giamblanco N, Coglitore D, Janot J-M, Coulon PE, Charlot B, Balme S, Sens. Actuators B 2018, 260, 736. [DOI] [PubMed] [Google Scholar]
- [50].Willems K, Van Meervelt V, Wloka C, Maglia G, Philos. Trans. Royal Soc. Lond. B Biol. Sci 2017, 372, 20160230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Huang G, Willems K, Soskine M, Wloka C, Maglia G, Nat. Commun 2017, 8, 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Watanabe H, Gubbiotti A, Chinappi M, Takai N, Tanaka K, Tsumoto K, Kawano R, Anal. Chem 2017, 89, 11269. [DOI] [PubMed] [Google Scholar]
- [53].Han A, Creus M, Schürmann G, Linder V, Ward TR, De Rooij NF, Staufer U, Anal. Chem 2008, 80, 4651. [DOI] [PubMed] [Google Scholar]
- [54].Japrung D, Dogan J, Freedman KJ, Nadzeyka A, Bauerdick S, Albrecht T, Kim MJ, Jemth P, Edel JB, Anal. Chem 2013, 85, 2449. [DOI] [PubMed] [Google Scholar]
- [55].Waduge P, Hu R, Bandarkar P, Yamazaki H, Cressiot B, Zhao Q, Whitford PC, Wanunu M, ACS Nano 2017, 11, 5706. [DOI] [PubMed] [Google Scholar]
- [56].Varongchayakul N, Huttner D, Grinstaff MW, Meller A, Sci. Rep 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kennedy E, Dong Z, Tennant C, Timp G, Nat. Nanotech 2016, 11, 968. [DOI] [PubMed] [Google Scholar]
- [58].Kolmogorov M, Kennedy E, Dong Z, Timp G, Pevzner PA, PLoS Comp. Biol 2017, 13, e1005356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen H, Li L, Zhang T, Qiao Z, Tang J, Zhou J, J. Phys. Chem. C 2018, 122, 2070. [Google Scholar]
- [60].Si W, Aksimentiev A, ACS Nano 2017, 11, 7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rodriguez-Larrea D, Bayley H, Nat. Nanotech 2013, 8, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rosen CB, Rodriguez-Larrea D, Bayley H, Nat. Biotechnol 2014, 32, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nir I, Huttner D, Meller A, Biophys. J 2015, 108, 2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wloka C, Van Meervelt V, van Gelder D, Danda N, Jager N, Williams CP, Maglia G, ACS Nano 2017, 11, 4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rostovtseva TK, Gurnev PA, Protchenko O, Hoogerheide DP, Yap TL, Philpott CC, Lee JC, Bezrukov SM, J. Biol. Chem 2015, 290, 18467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hoogerheide DP, Gurnev PA, Rostovtseva TK, Bezrukov SM, Nanoscale 2017, 9, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hoogerheide DP, Gurnev PA, Rostovtseva TK, Bezrukov SM, Biophys. J 2018, 114, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Merstorf C, Cressiot B, Pastoriza-Gallego M, Oukhaled A, Betton J-M, Auvray L, Pelta J, ACS Chem. Biol 2012, 7, 652. [DOI] [PubMed] [Google Scholar]
- [69].Oukhaled A, Cressiot B, Bacri L, Pastoriza-Gallego M, Betton J-M, Bourhis E, Jede R, Gierak J, Auvray L, Pelta J, ACS Nano 2011, 5, 3628. [DOI] [PubMed] [Google Scholar]
- [70].Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, van der Goot FG, Auvray L, Betton J-M, Pelta J, J. Am. Chem. Soc 2011, 133, 2923. [DOI] [PubMed] [Google Scholar]
- [71].Oukhaled G, Mathe J, Biance A-L, Bacri L, Betton J-M, Lairez D, Pelta J, Auvray L, Phys. Rev. Lett 2007, 98, 158101. [DOI] [PubMed] [Google Scholar]
- [72].Payet L, Martinho M, Pastoriza-Gallego M, Betton J-M, Auvray L, Pelta J, Mathe J, Anal. Chem 2012, 84, 4071. [DOI] [PubMed] [Google Scholar]
- [73].Talaga DS, Li J, J. Am. Chem. Soc 2009, 131, 9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Freedman KJ, Jurgens M, Prabhu A, Ahn CW, Jemth P, Edel JB, Kim MJ, Anal. Chem 2011, 83, 5137. [DOI] [PubMed] [Google Scholar]
- [75].Cressiot B, Braselmann E, Oukhaled A, Elcock AH, Pelta J, Clark PL, ACS Nano 2015, 9, 9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cressiot B, Oukhaled A, Patriarche G, Pastoriza-Gallego M, Betton J-M, Auvray L, Muthukumar M, Bacri L, Pelta J, ACS Nano 2012, 6, 6236. [DOI] [PubMed] [Google Scholar]
- [77].Pastoriza-Gallego M, Breton M-F, Discala F, Auvray L, Betton J-M, Pelta J, ACS Nano 2014, 8, 11350. [DOI] [PubMed] [Google Scholar]
- [78].Nivala J, Marks DB, Akeson M, Nat. Biotechnol 2013, 31, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Freedman KJ, Haq SR, Edel JB, Jemth P, Kim MJ, Sci. Rep 2013, 3, 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Van Meervelt V, Soskine M, Singh S, Schuurman-Wolters GK, Wijma HJ, Poolman B, Maglia G, J. Am. Chem. Soc 2017, 139, 18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wilson DS, Keefe AD, Szostak JW, Proc. Natl. Acad. Sci. U. S. A 2001, 98, 3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kawano R, Osaki T, Sasaki H, Takinoue M, Yoshizawa S, Takeuchi S, J. Am. Chem. Soc 2011, 133, 8474. [DOI] [PubMed] [Google Scholar]
- [83].Rotem D, Jayasinghe L, Salichou M, Bayley H, J. Am. Chem. Soc 2012, 134, 2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Soskine M, Biesemans A, Moeyaert B, Cheley S, Bayley H, Maglia G, Nano Lett. 2012, 12, 4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Van Meervelt V, Soskine M, Maglia G, ACS Nano 2014, 8, 12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Li T, Liu L, Li Y, Xie J, Wu H-C, Angew. Chem. Int. Ed 2015, 54, 7568. [DOI] [PubMed] [Google Scholar]
- [87].Lin X, Ivanov AP, Edel JB, Chem. Sci 2017, 8, 3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sze JYY, Ivanov AP, Cass AEG, Edel JB, Nat. Commun 2017, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Travers AA, Annu. Rev. Biochem 1989, 58, 427. [DOI] [PubMed] [Google Scholar]
- [90].Lunde BM, Moore C, Varani G, Nat. Rev. Mol. Cell Bio 2007, 8, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Smeets RMM, Kowalczyk SW, Hall AR, Dekker NH, Dekker C, Nano Lett. 2009, 9, 3089. [DOI] [PubMed] [Google Scholar]
- [92].Kowalczyk SW, Hall AR, Dekker C, Nano Lett. 2010, 10, 324. [DOI] [PubMed] [Google Scholar]
- [93].Spiering A, Getfert S, Sischka A, Reimann P, Anselmetti D, Nano Lett. 2011, 11, 2978. [DOI] [PubMed] [Google Scholar]
- [94].Bulushev RD, Marion S, Petrova E, Davis SJ, Maerkl SJ, Radenovic A, Nano Lett. 2016, 16, 7882. [DOI] [PubMed] [Google Scholar]
- [95].Raillon C, Cousin P, Traversi F, Garcia-Cordero E, Hernandez N, Radenovic A, Nano Lett. 2012, 12, 1157. [DOI] [PubMed] [Google Scholar]
- [96].Squires A, Atas E, Meller A, Sci. Rep 2015, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Venkatesan BM, Estrada D, Banerjee S, Jin X, Dorgan VE, Bae M-H, Aluru NR, Pop E, Bashir R, ACS Nano 2011, 6, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Carlsen AT, Zahid OK, Ruzicka JA, Taylor EW, Hall AR, Nano Lett. 2014, 14, 5488. [DOI] [PubMed] [Google Scholar]
- [99].Japrung D, Bahrami A, Nadzeyka A, Peto L, Bauerdick S, Edel JB, Albrecht T, J. Phys. Chem. B 2014, 118, 11605. [DOI] [PubMed] [Google Scholar]
- [100].Marshall MM, Ruzicka J, Zahid OK, Henrich VC, Taylor EW, Hall AR, Langmuir 2015, 31, 4582. [DOI] [PubMed] [Google Scholar]
- [101].Bell NAW, Keyser UF, J. Am. Chem. Soc 2015, 137, 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bell NAW, Keyser UF, Nat. Nanotech 2016, 11, 645. [DOI] [PubMed] [Google Scholar]
- [103].Plesa C, Ruitenberg JW, Witteveen MJ, Dekker C, Nano Lett. 2015, 15, 3153. [DOI] [PubMed] [Google Scholar]
- [104].Lee C, Park JK, Youn Y, Kim JH, Lee K-S, Kim N-K, Kim E, Kim EE, Yoo K-H, Anal. Chem 2017, 89, 2390. [DOI] [PubMed] [Google Scholar]
- [105].Macrae MX, Blake S, Jiang X, Capone R, Estes DJ, Mayer M, Yang J, ACS Nano 2009, 3, 3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhao Q, de Zoysa RSS, Wang D, Jayawardhana DA, Guan X, J. Am. Chem. Soc 2009, 131, 6324. [DOI] [PubMed] [Google Scholar]
- [107].Kukwikila M, Howorka S, J. Phys. Condens. Mat 2010, 22, 454103. [DOI] [PubMed] [Google Scholar]
- [108].Zhou S, Wang L, Chen X, Guan X, ACS Sens. 2016, 1, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Fennouri A, Daniel R, Pastoriza-Gallego M, Auvray L, Pelta J, Bacri L, Anal. Chem 2013, 85, 8488. [DOI] [PubMed] [Google Scholar]
- [110].Wang Y, Montana V, Grubišić V, Stout RF Jr., Parpura V, Gu L-Q, ACS Appl. Mater. Interfaces 2014, 7, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Rauf S, Zhang L, Ali A, Ahmad J, Liu Y, Li J, Anal. Chem 2017, 89, 13252. [DOI] [PubMed] [Google Scholar]
- [112].Shang J, Li Z, Liu L, Xi D, Wang H, ACS Sens. 2018, 3, 512. [DOI] [PubMed] [Google Scholar]
- [113].Sutherland TC, Long Y-T, Stefureac R-I, Bediako-Amoa I, Kraatz H-B, Lee JS, Nano Lett. 2004, 4, 1273. [Google Scholar]
- [114].Movileanu L, Schmittschmitt JP, Martin Scholtz J, Bayley H, Biophys. J 2005, 89, 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Stefureac R, Long Y-T, Kraatz H-B, Howard P, Lee JS, Biochemistry 2006, 45, 9172. [DOI] [PubMed] [Google Scholar]
- [116].Goodrich CP, Kirmizialtin S, Huyghues-Despointes BM, Zhu A, Scholtz JM, Makarov DE, Movileanu L, J. Phys. Chem. B 2007, 111, 3332. [DOI] [PubMed] [Google Scholar]
- [117].Singh PR, Bárcena-Uribarri I, Modi N, Kleinekathöfer U, Benz R, Winterhalter M, Mahendran KR, ACS Nano 2012, 6, 10699. [DOI] [PubMed] [Google Scholar]
- [118].Mereuta L, Asandei A, Seo CH, Park Y, Luchian T, ACS Appl. Mater. Interfaces 2014, 6, 13242. [DOI] [PubMed] [Google Scholar]
- [119].Mereuta L, Roy M, Asandei A, Lee J-K, Park Y, Andricioaei I, Luchian T, Sci. Rep 2014, 4, 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Niedzwiecki DJ, Lanci CJ, Shemer G, Cheng PS, Saven JG, Drndic M, ACS Nano 2015, 9, 8907. [DOI] [PubMed] [Google Scholar]
- [121].Wolfe AJ, Mohammad MM, Cheley S, Bayley H, Movileanu L, J. Am. Chem. Soc 2007, 129, 14034. [DOI] [PubMed] [Google Scholar]
- [122].Bikwemu R, Wolfe AJ, Xing X, Movileanu L, J. Phys. Condens. Mat 2010, 22, 454117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Mahendran KR, Lamichhane U, Romero-Ruiz M, Nussberger S, Winterhalter M, J. Phys. Chem. Lett 2012, 4, 78. [DOI] [PubMed] [Google Scholar]
- [124].Mahendran KR, Romero-Ruiz M, Schlösinger A, Winterhalter M, Nussberger S, Biophys. J 2012, 102, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Dhanasekar NN, Aliouane S, Winterhalter M, Pagès J-M, Bolla J-M, Biochem. Biophys. Rep 2017, 11, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Stefureac RI, Lee JS, Small 2008, 4, 1646. [DOI] [PubMed] [Google Scholar]
- [127].Mereuta L, Schiopu I, Asandei A, Park Y, Hahm K-S, Luchian T, Langmuir 2012, 28, 17079. [DOI] [PubMed] [Google Scholar]
- [128].Roozbahani GM, Chen X, Zhang Y, Xie R, Ma R, Li D, Li H, Guan X, ACS Sens. 2017, 2, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Boersma AJ, Bayley H, Angew. Chem. Int. Ed 2012, 51, 9606. [DOI] [PubMed] [Google Scholar]
- [130].Asandei A, Rossini AE, Chinappi M, Park Y, Luchian T, Langmuir 2017, 33, 14451. [DOI] [PubMed] [Google Scholar]
- [131].Ji Z, Wang S, Zhao Z, Zhou Z, Haque F, Guo P, Small 2016, 12, 4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Chavis AE, Brady KT, Hatmaker GA, Angevine CE, Kothalawala N, Dass A, Robertson JWF, Reiner JE, ACS Sens. 2017, 2, 1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Shrestha N, Bryant SL, Thomas C, Richtsmeier D, Pu X, Tinker J, Fologea D, Sci. Rep 2017, 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Piguet F, Ouldali H, Pastoriza-Gallego M, Manivet P, Pelta J, Oukhaled A, Nat. Commun 2018, 9, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Kwok H, Briggs K, Tabard-Cossa V, PLoS One 2014, 9, 10.1371/journal.pone.0092880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Bell NAW, Engst CR, Ablay M, Divitini G, Ducati C, Liedl T, Keyser UF, Nano Lett. 2012, 12, 512. [DOI] [PubMed] [Google Scholar]
- [137].Burns JR, Seifert A, Fertig N, Howorka S, Nat. Nanotech 2016, 11, 152. [DOI] [PubMed] [Google Scholar]