ABSTRACT
Acinetobacter baumannii is a Gram-negative bacterial pathogen that poses a major health concern due to increasing multidrug resistance. The Gram-negative cell envelope is a key barrier to antimicrobial entry and includes an inner and outer membrane. The maintenance of lipid asymmetry (Mla) system is the main homeostatic mechanism by which Gram-negative bacteria maintain outer membrane asymmetry. Loss of the Mla system in A. baumannii results in attenuated virulence and increased susceptibility to membrane stressors and some antibiotics. We recently reported two strain variants of the A. baumannii type strain ATCC 17978: 17978VU and 17978UN. Here, ∆mlaF mutants in the two ATCC 17978 strains display different phenotypes for membrane stress resistance, antibiotic resistance, and pathogenicity in a murine pneumonia model. Although allele differences in obgE were previously reported to synergize with ∆mlaF to affect growth and stringent response, obgE alleles do not affect membrane stress resistance. Instead, a single-nucleotide polymorphism (SNP) in the essential gene encoding undecaprenyl pyrophosphate (Und-PP) synthase, uppS, results in decreased enzymatic rate and decrease in total Und-P levels in 17978UN compared to 17978VU. The UppSUN variant synergizes with ∆mlaF to reduce capsule and lipooligosaccharide (LOS) levels, increase susceptibility to membrane stress and antibiotics, and reduce persistence in a mouse lung infection. Und-P is a lipid glycan carrier required for the biosynthesis of A. baumannii capsule, cell wall, and glycoproteins. These findings uncover synergy between Und-P and the Mla system in maintaining the A. baumannii cell envelope and antibiotic resistance.
IMPORTANCE
Acinetobacter baumannii is a critical threat to global public health due to its multidrug resistance and persistence in hospital settings. Therefore, novel therapeutic approaches are urgently needed. We report that a defective undecaprenyl pyrophosphate synthase (UppS) paired with a perturbed Mla system leads to synthetically sick cells that are more susceptible to clinically relevant antibiotics and show reduced virulence in a lung infection model. These results suggest that targeting UppS or undecaprenyl species and the Mla system may resensitize A. baumannii to antibiotics in combination therapies. This work uncovers a previously unknown synergistic relationship in cellular envelope homeostasis that could be leveraged for use in combination therapy against A. baumannii.
KEYWORDS: Acinetobacter, antibiotic resistance, membrane stress, isoprenoid, Und-P, maintenance of lipid asymmetry, lipooligosaccharide, LOS, capsule
INTRODUCTION
Acinetobacter baumannii is a Gram-negative bacterial pathogen that is a major cause of healthcare-associated infections. Clinical isolates of A. baumannii demonstrate resistance to first line antibiotics such as meropenem and last resort antibiotics such as colistin (1). The World Health Organization and the Centers for Disease Control name A. baumannii as an urgent threat, calling for the development of novel antimicrobials (2–4). A. baumannii has multiple intrinsic mechanisms for resisting antibiotics and host stressors. The first line of defense is the cellular envelope including capsule, a peptidoglycan cell wall, and a dual-membrane system conserved among Gram-negative bacteria (5, 6). The inner membrane (IM) and outer membrane (OM) protect Gram-negative bacteria from environmental stress (7, 8). The Gram-negative OM is asymmetric with phospholipids composing the inner leaflet and lipopolysaccharides (LPS) or lipooligosaccharides (LOS) composing the outer leaflet. A. baumannii does not encode the gene required for O-antigen elaboration, resulting in LOS rather than LPS (9, 10). Additionally, A. baumannii is able to survive without LOS, whereas in most Gram-negative bacteria, LPS is essential (11–13). In all Gram-negative bacteria, the asymmetric bilayer of the OM is critical for resistance to membrane stressors and antimicrobials (14, 15).
The maintenance of lipid asymmetry (Mla) system is thought to be the primary homeostatic mechanism to maintain OM lipid asymmetry by removing mislocalized phospholipids from the outer leaflet of the OM (16). MlaBDEF forms an IM ATP-binding cassette (ABC) transporter with ATPase activity, MlaC is a periplasmic protein, and MlaA is an OM lipoprotein that forms a complex with OmpC/F (16–19,20). While the Mla system is not required for growth in lysogeny broth, bacteria lacking a functional Mla system are more sensitive to the membrane stress SDS/EDTA (16, 17, 21–23). Inactivating mlaF or mlaC in multiple species results in the loss of function of the Mla system and increased sensitivity to membrane stressors, antibiotics, and the host (21, 24–26). Additionally, dominant negative mutations in mlaA (mlaA*) have been shown to increase outer membrane permeability and susceptibility to erythromycin and rifampicin in Escherichia coli (27). The Mla system is necessary for the virulence of multiple pathogenic species including Shigella flexneri, Burkholderia pseudomallei, and Pseudomonas aeruginosa (28–32). By contrast, mutations in the Mla system have been shown to increase virulence in E. coli and Neisseria gonorrhoeae (33, 34). In summary, the Mla system is critical for Gram-negative outer membrane maintenance, stress resistance, and virulence.
While the directionality of lipid transport by the Mla system has been debated, the preponderance of evidence supports a retrograde transport model of phospholipid movement from the OM to the IM (35–37). Genetic evidence from E. coli, A. baumannii, and chloroplasts supports a retrograde transport model in which phospholipids are removed from the outer leaflet of the OM and transported to the IM (16, 21, 27, 38–41). Additionally, crystallographic and cryo-EM structural data from Klebsiella pneumoniae, Serratia marcescens, and E. coli support a model in which phospholipids are removed from the OM by the MlaA-OmpF complex and transported toward MlaBDEF based on the orientation of MlaA-OmpF in the OM (17, 42–44). Recent studies identifying AsmA-like proteins facilitating anterograde lipid transport machinery in E. coli further support the retrograde transport model for the Mla system (45, 46). By contrast, in vitro work in E. coli as well as cryo-EM, molecular dynamics, and pulse-chase studies in A. baumannii described an anterograde transport model where newly synthesized phospholipids are transported to the inner leaflet of the OM (19, 24, 47, 48). However, Mann et al., whose structural work in A. baumannii supports an anterograde lipid transport model, speculate that nucleotide state or solubilization approach of the MlaBDEF studies may explain the differences in conclusions (48). Recent structures of MlaBDEF from E. coli and A. baumannii and E. coli MlaC in complex with MlaA or MlaD were solved and provide mechanistic insight on lipid binding but do not provide clear evidence for either direction of lipid transport (22, 49, 50).
In A. baumannii, the Mla system synergizes with essential pathways to promote growth. A previous study reported synergy between the Mla system and the essential GTPase ObgE in promoting ΔmlaF growth and stringent response (51, 52). We previously reported a suppressor mutation in the isoprenoid biosynthetic pathway that restored resistance of an A. baumannii ΔmlaF mutant to membrane stress, some antibiotics, and host stressors (21). The suppressor is an ISAba11 transposition in the 5′ untranslated region of ispB that results in ispB downregulation (21). We hypothesized that the downregulation of ispB increased flux of the branchpoint substrate farnesyl-pyrophosphate (FPP) to undecaprenyl pyrophosphate (Und-PP) synthase, UppS. Und-PP is a precursor to the essential glycan carrier undecaprenyl phosphate (Und-P), also known as bactoprenol phosphate (BP), that transfers glycans across the plasma membrane for biosynthesis of peptidoglycan, capsular polysaccharides, and the O-antigen in LPS-producing bacteria (53). This finding suggested a role for Und-P biosynthesis in membrane stress resistance in the absence of the Mla system in A. baumannii. Thus, multiple reports have identified synergy between the Mla system and other pathways in A. baumannii.
Recently, we reported two variants of the commonly used laboratory strain A. baumannii 17978 distributed by ATCC (54). These variants, A. baumannii ATCC 17978VU and 17978UN, have distinct genotypes with six protein-encoding single-nucleotide polymorphisms (SNPs) and a 44 kb accessory locus (AbaAL44) that is present in 17978UN but absent in 17978VU (54). The protein-coding SNPs encode variants of the lipooligosaccharide transporter LptD and the essential proteins ObgE and UppS (55). Our previous work used isogenic ∆mlaF strains in the 17978UN (AbaAL44+) background (21). Upon reconstructing ∆mlaF strains in the A. baumannii ATCC 17978VU strain background, we discovered that the 17978VU ∆mlaF mutant was more resistant to SDS/EDTA membrane stress than the 17978UN ∆mlaF mutant. Here, we describe genetic synergy between the maintenance of lipid asymmetry and Und-P biosynthesis uncovered through genetic dissection and comparison of ATCC 17978VU and 17978UN ∆mlaF strains. These findings suggest an underlying relationship between the Mla system and undecaprenyl biosynthesis in A. baumannii that function together to maintain LOS abundance and promote membrane stress resistance, virulence, and antimicrobial resistance.
RESULTS
UppS synergizes with the Mla system under membrane stress
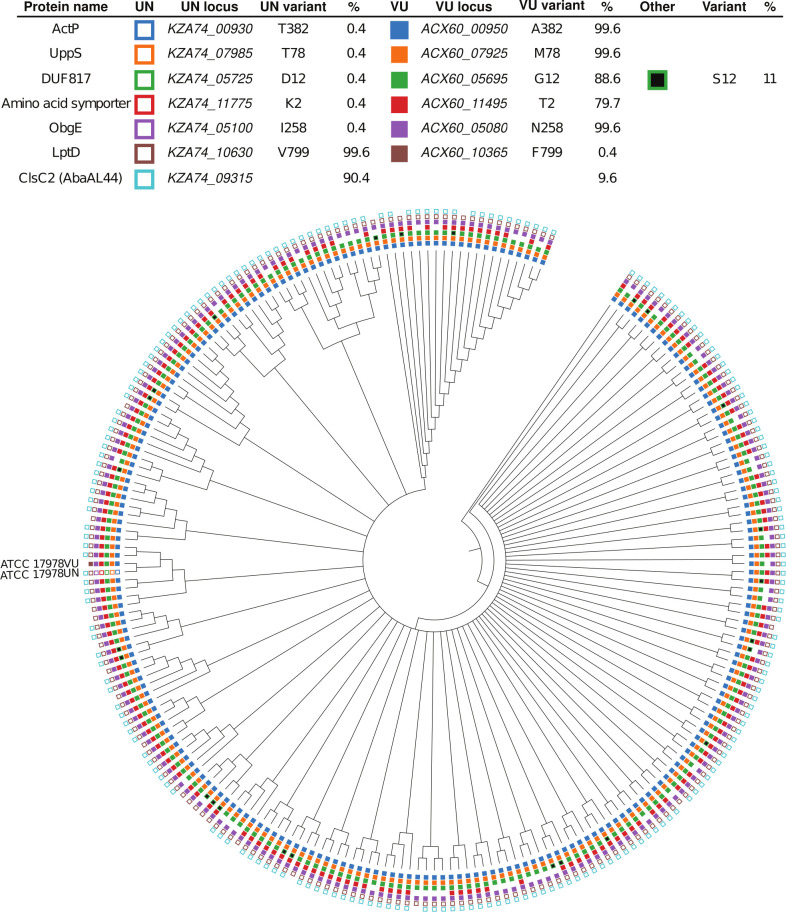
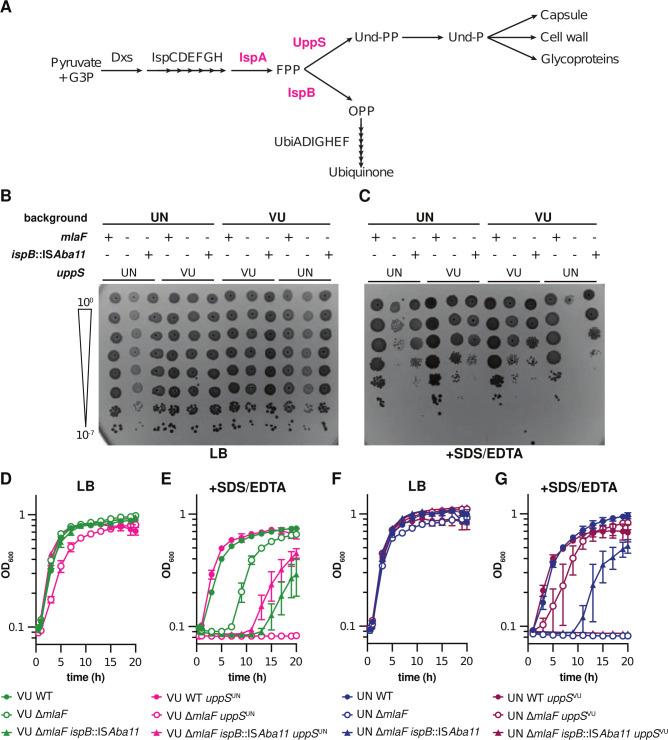
The 17978VU and 17978UN strain variants of ATCC 17978 contain SNPs in multiple protein coding genes (54). We first assessed the prevalence of the 17978VU and 17978UN alleles across A. baumannii genomes. A set of 5945 genomes from NCBI were de-duplicated to remove near-clonal lineages and the predicted proteomes of the resulting 230 genomes were analyzed by OrthoFinder to deduce orthologues within A. baumannii (56, 57). The 17978UN predicted protein variants were more common for LptD (99%) and ClsC2 (indicative of the presence of the AbaAL44 cluster; 90%) (Fig. 1). The 17978VU predicted protein variants were more common for ObgE (99%), UppS (99%), ActP (99%), the amino acid symporter ACX60_11495 (79%), and DUF817 (89%) (Fig. 1). Thus, both 17978VU and 17978UN contain alleles representing the majority of published A. baumannii genomes. Next, the ∆mlaF::Kn (∆mlaF) mutation was reconstructed in A. baumannii ATCC 17978VU and SDS/EDTA membrane stress resistance was compared in the 17978VU (AbaAL44−) and 17978UN (AbaAL44+) strain backgrounds. Neither the wild-type strains nor the ΔmlaF mutants demonstrated a growth defect in lysogeny broth (LB) regardless of strain background (Fig. 2A and B). However, in the presence of SDS/EDTA membrane stress, the 17978UN ΔmlaF mutant exhibited a greater growth defect than the 17978VU ΔmlaF mutant (Fig. 2F and G), suggesting synergy between mlaF and genetic differences between the strains. Therefore, we reasoned that the closely related strain variants ATCC 17978VU and 17978UN could be used as a tool to uncover this synergy.
Fig 1.
Phylogenetic tree of Acinetobacter baumannii strains depicting prevalence of variants encoded by ATCC 17978VU and 17978UN. Filled, non-black boxes indicate the presence of a protein variant identical to A. baumannii ATCC 17978VU, while unfilled boxes denote the presence of the A. baumannii ATCC 17978UN variant. The absence of a box indicates that a predicted ortholog was not found in the genome. Black boxes indicate the presence of a variant different from both the 17978VU and 17978UN strains. The presence of ClsC2, indicated by an unfilled box of the 17978UN strain, is representative of the presence of the 44 kb accessory locus AbaAL44.
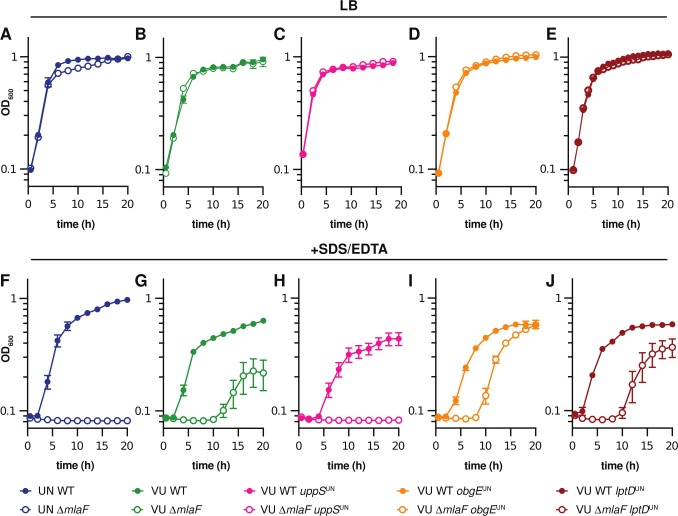
Fig 2.
UppSUN protein variant results in increased membrane stress sensitivity in A. baumannii ATCC 17978 ∆mlaF mutants. (A–E) 17978UN and 17978VU wild-type, ΔmlaF, and isogenic mutant strains were grown in LB. (F–J) 17978UN and 17978VU wild-type, ΔmlaF, and isogenic mutant strains were grown in LB with 0.01% SDS and 0.175 mM EDTA. Data are means ± SEM, n = 3.
To determine the genetic cause behind the differences in membrane stress resistance between ATCC 17978VU and 17978UN ΔmlaF strains, point mutants were constructed for three candidate genes: obgE, lptD, and uppS. Candidate genes were chosen based on literature findings and known function. First, Powers et al. previously reported that obgE alleles in 17978 strains maintained at University of Washington (UW) and University of Georgia (UGA) synergized with mlaF to affect growth and stringent response. Specifically, the ∆mlaF strain with ObgEI258 (ObgEUN) had defects in LB growth and stringent response compared to the ∆mlaF strain with ObgEN258 (ObgEVU) (51). The strains were otherwise isogenic, suggesting that one strain was a derivative of 17978VU or 17978UN. Second, LptD is a β-barrel OM protein responsible for the translocation of LPS/LOS in Gram-negative bacteria (58). Third, we previously reported a potential role for undecaprenyl pyrophosphate, synthesized by UppS, in promoting A. baumannii envelope integrity in 17978UN ∆mlaF (21). The 17978UN allele for each candidate gene (encoding ObgE I258; LptD V799; UppS T78) was substituted in the endogenous locus (encoding ObgE N258; LptD F799; UppS M78) in 17978VU wild-type and 17978VU mlaF strains. We reasoned that if the candidate gene contributes to the contrasting phenotypes, then the 17978VU mlaF mutant containing the 17978UN candidate allele would exhibit the 17978UN mlaF phenotype of increased membrane stress sensitivity. As expected, none of the strains displayed a growth defect when grown in LB alone (Fig. 2C through E). 17978VU mlaF uppSUN was unable to grow in SDS/EDTA similar to 17978UN mlaF, suggesting that the uppS allele determines membrane stress sensitivities of 17978VU and 17978UN mlaF strains (Fig. 2H). By contrast, neither the obgE nor lptD 17978UN alleles altered membrane stress sensitivity in the 17978VU ∆mlaF strain (Fig. 2I and J). We observed similar results in an independent dilution spotting experiment (Fig. S1A and B). To test allele prevalence in the ATCC culture stock, we screened 46 isolates from the ATCC 17978 culture received in 2021 for the uppS allele and found that one was indeterminant, 43/45 contained uppSUN and 2/45 contained uppSVU(Fig. S1C). Together, these data show that the Mla system is synthetic with uppS alleles in A. baumannii for resistance to SDS/EDTA membrane stress.
UppSUN has decreased enzymatic activity, leading to lower cellular Und-P levels
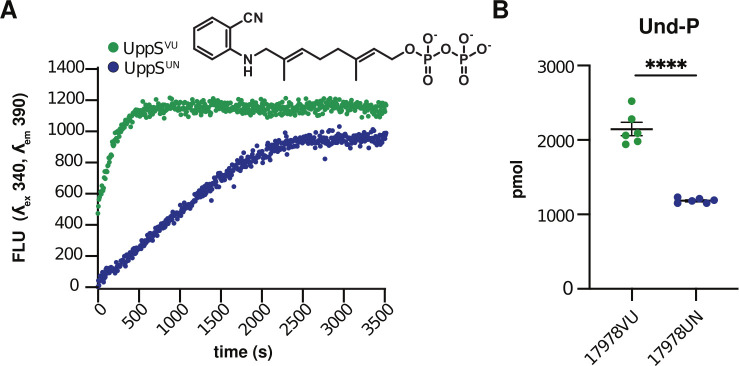
Based on results thus far, we hypothesized that UppSUN (T78) was defective compared to UppSVU (M78). First, protein secondary structure was compared by circular dichroism analysis which showed that there were no structural differences between the UppS variants (Fig. S2A and B). Next, enzymatic activity was compared using purified protein and a fluorescent analog (2-nitrileanilinogeranyl diphosphate; 2CNA-GPP) of the UppS substrate farnesyl-pyrophosphate (FPP). UppS from E. coli, Vibrio vulnificus, Staphylococcus aureus, and Bacteroides fragilis have previously been shown to catalyze the extension of the fluorescent substrate analog 2CNA-GPP (59, 60). Upon elongation of the analog, a concomitant increase in fluorescence can be readily monitored via a microplate assay. By this assay, UppSUN was found to have a fivefold decrease in the enzymatic rate compared to UppSVU (Fig. 3A). To determine if the decreased enzymatic rate of UppSUN results in decreased levels of the essential glycan carrier Und-P in A. baumannii, cellular pools of Und-P in the 17978VU and 17978UN wild-type strains were quantified. The 17978UN strain contained twofold lower levels of Und-P compared to the 17978VU wild-type strain (Fig. 3B). Transcript abundance of uppS was assessed via reverse transcription quantitative PCR (RT-qPCR) and revealed no differences in abundance between 17978VU and 17978UN (Fig. S2C). Therefore, these data suggest that the UppSUN enzyme has decreased enzyme function that results in decreased cellular levels of Und-P.
Fig 3.
UppSUN demonstrates decreased enzymatic rate and results in decreased cellular Und-P compared to UppSVU. (A) UppS activity with 2CNA-GPP, a fluorescent analog of the endogenous substrate farnesyl pyrophosphate (FPP). Activity of purified UppS from strains A. baumannii ATCC 17978VU (green) and 17978UN (blue) was measured with the 2CNA-GPP substrate analogue and monitoring fluorescence increase upon elongation at 340 nm excitation and 390 nm emission. Data are n = 1. Experiments were conducted three independent times with similar results. (B) Mass spectrometry quantitation of bacterial C55 undecaprenyl phosphate (Und-P) in ATCC 17978VU and 17978UN wild-type strains. Significance is by unpaired t-test. ****P < 0.0001. Data are mean ± SEM, n = 6.
The uppSUN allele increases envelope permeability in ∆mlaF strains
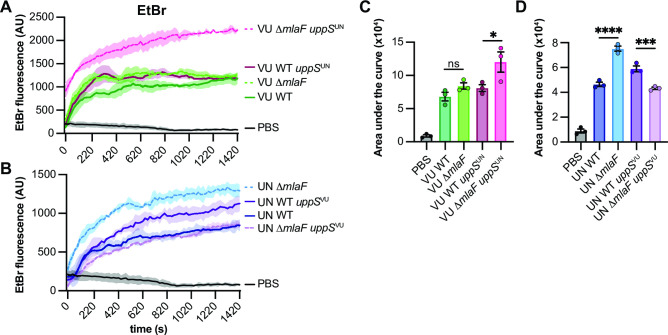
Given the role of UppS and the Mla system in cell envelope biosynthesis and integrity, we hypothesized that the uppSUN allele would increase envelope permeability in ∆mlaF mutants. Likewise, we predicted that the uppSVU allele would reduce envelope permeability in a 17978UN ΔmlaF background. Therefore, the uppSVU allele was introduced into 17978UN wild-type and 17978UN mlaF strains. An ethidium bromide (EtBr) uptake assay was used to test the effect of the uppS alleles on envelope permeability in wild-type and ∆mlaF strains (24, 61). In the 17978VU background, mlaF uppSUN had significantly increased EtBr permeability compared to wild-type, mlaF, and wild-type uppSUN (Fig. 4A and C). Similarly, in the 17978UN background, introducing the uppSVU reduced EtBr permeability in a mlaF mutant (Fig. 4B and D). Together, these data suggest that uppS and the Mla system synergize to promote cell envelope integrity.
Fig 4.
UppSUN increases membrane permeability in ΔmlaF mutants. (A and B) A. baumannii ATCC 17978VU (A) and 17978UN (B) wild-type and ΔmlaF strains with the endogenous or alternate uppS allele were incubated with efflux pump inhibitor CCCP and ethidium bromide and fluorescence was measured. (C and D) Quantified area under the curve for A and B, respectively. Significance is by one-way ANOVA with Tukey’s multiple comparisons test. All strains were significantly higher than the PBS control (P < 0.001). Comparisons between ΔmlaF strains with their respective wild-type strain are shown. Significant differences are indicated by *P < 0.05, ***P < 0.001, ****P < 0.0001. Data are means ± SEM, n = 3. Experiments were conducted two independent times with similar results.
Isoprenoid pathway mutations suppress ∆mlaF membrane stress sensitivity only in the presence of uppSUN
We previously identified a suppressor mutation that restored virulence, membrane stress, and antibiotic resistance to wild-type levels in a 17978UN mlaF background by decreasing transcript abundance of ispB (21). This 17978UN mlaF ispB::ISAba11 suppressor was included to test the contribution of the isoprenoid biosynthesis pathway to the contrasting phenotypes (outlined in Fig. 5A). Thus, to determine if the isoprenoid biosynthetic pathway impacting ΔmlaF phenotypes depends on the uppS allele present, 17978UN mlaF ispB::ISAba11 and 17978VU mlaF ispB::ISAba11 strains were constructed with the endogenous or alternate uppS allele. Dilution spotting to LB agar plates with and without SDS/EDTA determined colony formation efficiency in the presence of membrane stress. As expected, none of the strains exhibited defects in colony formation on LB alone (Fig. 5B). Notably, the 17978UN ∆mlaF mutant displayed decreased opacity that was reversed by the presence of ispB::ISAba11, as previously reported (21), or uppSVU. In the presence of SDS/EDTA membrane stress, mlaF strains encoding UppSUN exhibited a clear defect in colony formation efficiency compared to mlaF strains encoding UppSVU (Fig. 5C). This further supports the conclusion that uppS is the genetic cause behind the differences in membrane stress resistance between ΔmlaF strains in ATCC 17978VU and 17978UN. Similarly, the ispB suppressor restored colony formation on SDS/EDTA in ΔmlaF strains encoding the uppSUN allele, regardless of strain background (Fig. 5C). This suggests that the ispB suppressor function is associated specifically with uppSUN. We noted that the 17978UN ∆mlaF strain often had increased colony formation on SDS/EDTA plates than the 17978VU ∆mlaF uppSUN strain which was unexpected; we isolated several colonies of 17978UN ∆mlaF from the SDS/EDTA plate and determined that 1/8 appeared to be a stable suppressor but it did not encode the ispB::ISAba11 allele and the strain was not further investigated. This suggests that uppSUN defects may be partially compensated by additional alleles in the 17978UN background.
Fig 5.
Isoprenoid biosynthesis suppressor mutations only confer increased resistance to SDS/EDTA in A. baumannii ATCC 17978 ΔmlaF strains with UppSUN. (A) Schematic of the isoprenoid biosynthetic pathway with key genes highlighted in red. (B and C) Wild-type and mutant strains were serially diluted before spotting on LB plates without (B) and with (C) 0.01% SDS and 0.15 mM EDTA. Experiments were conducted four independent times (n = 8) with similar results. (D–G) 17978VU (D and E) and 17978UN (F and G) wild-type and mutant strains were grown in LB without (D and F) or with (E and G) 0.01% SDS and 0.175 mM EDTA. Data are means ± SEM, n = 3.
To examine the effect of the uppS alleles on ΔmlaF growth over time, we performed growth curves in LB with or without SDS/EDTA. Again, none of the strains showed overt growth defects when grown in LB alone (Fig. 5D and F). However, in the ΔmlaF background, the strain with the uppSUN allele was unable to grow in media containing SDS/EDTA. Conversely, the uppSVU allele conferred partial resistance to SDS/EDTA in the ΔmlaF background (Fig. 5E and G). Interestingly, 17978UN ΔmlaF ispB::ISAba11 uppSVU was unable to grow in the presence of membrane stressors SDS/EDTA (Fig. 5G). This result suggests that the uppSVU allele in a 17978UN mlaF background may be hindered by the ispB suppressor, perhaps due to a decreased flux toward ubiquinone production.
While constructing the 17978VU mlaF uppSUN strain, one isolate displayed a distinct phenotype resembling that of 17978VU mlaF. Upon whole-genome sequencing, a mutation was discovered in ispA that resulted in a G223E substitution. This mutation was then reconstructed in the 17978UN mlaF background to assess its function as a suppressor of the mlaF uppSUN phenotype. A dilution spotting assay was used to characterize both ispA and ispB suppressors in the 17978UN background. All four strains grew similarly on LB medium (Fig. S3). 17978UN mlaF displayed a strong growth defect that was partially rescued by suppressor mutations in ispA and ispB (Fig. S3). These data support a model in which the isoprenoid biosynthetic pathway is synthetic with the Mla system in A. baumannii. We predict that the ispA and ispB ∆mlaF suppressors function by increasing the metabolic flux toward UppS and the production of Und-P, which promotes ∆mlaF mutant growth in the presence of the uppSUN allele.
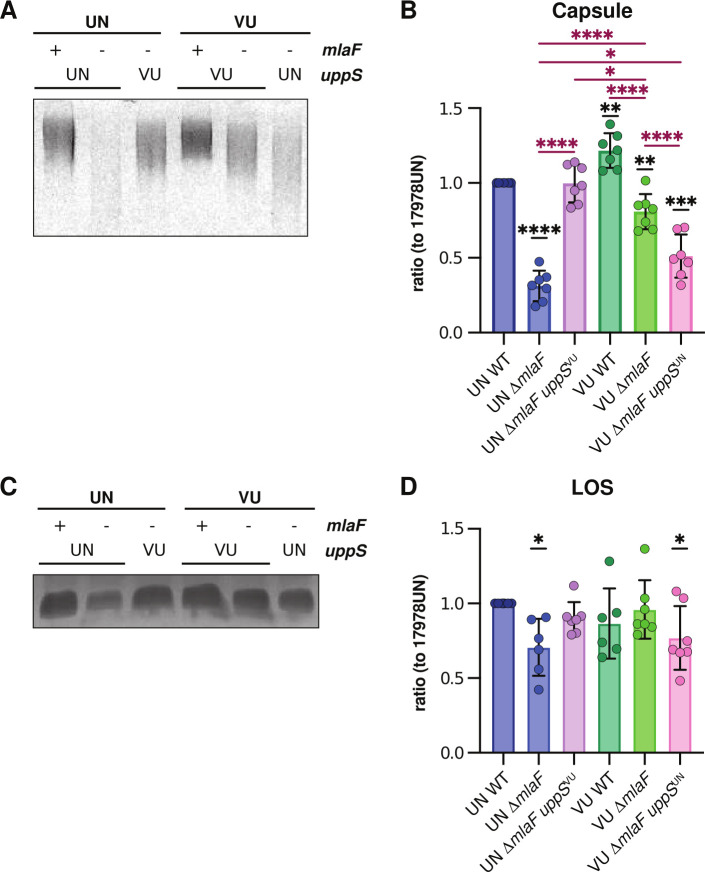
UppSUN results in reduced capsule and LOS abundance in the ∆mlaF background
Und-P has an established role in capsule, peptidoglycan, and glycoprotein biosynthesis; therefore, we tested whether cell wall or capsule is altered by the reduced activity of UppSUN. Peptidoglycan staining by 3-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-d-alanine hydrochloride (NADA) staining showed no dramatic differences among strains (Fig. S4A). However, capsule staining by Maneval suggested there may be subtle differences in the capsule among 17978UN and 17978VU ΔmlaF strains (Fig. S4B). Therefore, capsule polysaccharide was visualized and quantified by Alcian blue staining. Alcian blue revealed that ΔmlaF strains with uppSUN alleles had dramatically different polysaccharide distributions which were more diffuse and consistent with decreased molecular weight (Fig. 6A; Fig. S4C). Gel densitometry quantification showed that ΔmlaF strains with uppSUN alleles also had decreased capsule abundance compared to 17978UN wild type (Fig. 6B). This analysis also showed that 17978VU wild type had increased capsule compared to 17978UN (Fig. 6A and B). Overall, ΔmlaF strains expressing uppSUN had further reduced capsule when compared to the 17978UN wild type than the same strain background expressing uppSVU (Fig. 6A and B); however, the uppS allele did not fully restore these differences, suggesting other genetic differences between 17978UN and 17978VU also contribute to capsule abundance.
Fig 6.
UppSUN results in reduced capsule and LOS abundance. (A) Alcian blue stain of capsular polysaccharides from cell lysates. Cell lysates were normalized to total protein. Image is representative of seven biological replicates from three independent experiments. (B) Densitometric quantification of capsule bands as a ratio to 17978UN wild type. Data are means ± SEM, n = 7 from three independent experiments, significance is by one-sample t-test compared to 1 (black asterisk) and one-way ANOVA with Sidak’s multiple comparisons (maroon asterisk). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (C) Silver stain of proteinase K-treated cell lysates to stain for LOS. Cell lysates were normalized to total protein. Image is representative of seven biological replicates from three independent experiments. (D) Densitometric quantification of LOS bands as a ratio (to 17978UN wild type). Data are means ± SEM, n = 6–7 from three independent experiments, significance is by one-sample t-test compared to 1. *P < 0.05.
Next, we tested whether the UppSUN variant was required for the decreased LOS abundance in the 17978UN ΔmlaF mutant we previously reported (21). Although Und-P has an established role in the production of LPS as the lipid carrier of O-antigen precursors in other Gram-negative organisms (15), there is no known role for Und-P in the biosynthesis of LOS in A. baumannii. We hypothesized that reduced LOS abundance in the 17978UN ∆mlaF mutant would depend on the presence of the uppSUN allele. Indeed, when LOS was quantified by silver staining of proteinase K-treated cell lysates, ΔmlaF strains with uppSUN had reduced LOS abundance (Fig. 6C and D; Fig. S4D). The wild-type strains and the ΔmlaF strains with uppSVU showed no reduction in LOS (Fig. 6C and D; Fig. S4D). Together, these results suggest that UppS activity and the Mla system are important for maintaining capsule and LOS abundance in A. baumannii.
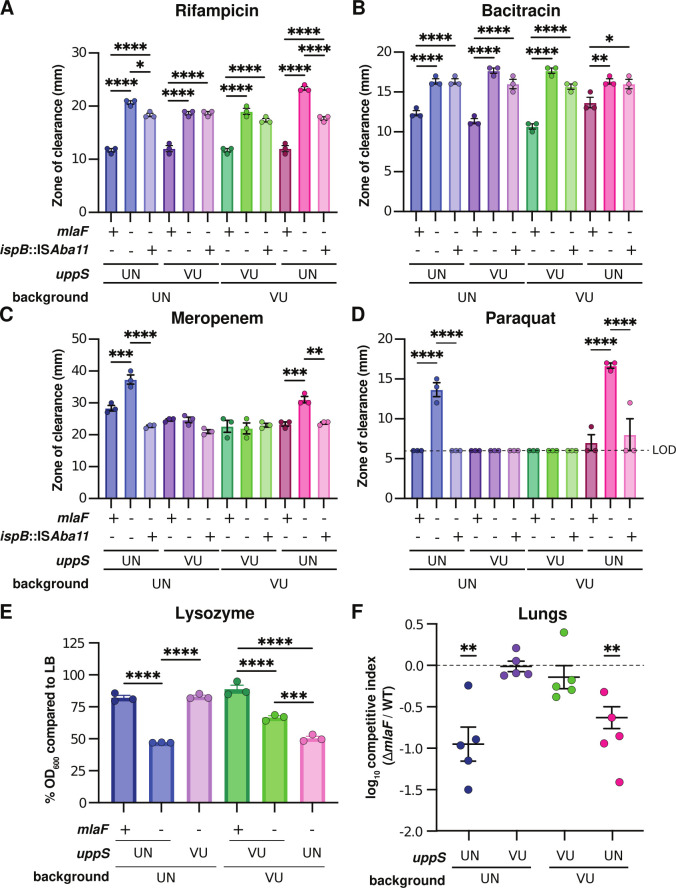
Mla and UppS synergy influences clinically relevant phenotypes
Previous reports showed that A. baumannii mla mutants have increased susceptibility to antimicrobials such as gentamicin, novobiocin, rifampicin, meropenem, and the superoxide donor paraquat (21, 24, 51). We hypothesized that in a mlaF mutant background, the presence of the uppSUN allele would result in increased susceptibility to antimicrobials compared to strains with the uppSVU allele. In a disk diffusion assay, the presence of uppSUN in a mlaF mutant resulted in increased susceptibility to antimicrobials from multiple classes regardless of the ATCC 17978 background, represented by increased zone of clearance diameters (Fig. 7A through D; Fig. S5A and B). These antimicrobials included first line antibiotics such as meropenem and imipenem. These results show that UppS synergizes with the Mla system to promote A. baumannii antimicrobial resistance. Consistent with above findings, the ispB suppressor primarily restored resistance to strains containing the uppSUN allele. We further hypothesized that the uppSUN allele would confer decreased resistance of ∆mlaF strains to lysozyme, a host antimicrobial enzyme that cleaves the peptidoglycan cell wall (62). During growth in LB with 1 mg/mL lysozyme, the 17978UN mlaF exhibited severely reduced resistance to lysozyme compared to the 17978UN wild-type strain (Fig. 7E; Fig. S5A). Resistance was restored to 17978UN mlaF by the uppSVU allele. Similarly, the uppSUN allele conferred decreased lysozyme resistance to 17978VU mlaF (Fig. 7E; Fig. S5A). Together, these data show that the uppSUN allele decreases resistance to multiple antimicrobial stresses in a mlaF mutant.
Fig 7.
In an ΔmlaF background, UppSUN decreases antimicrobial resistance and virulence in a murine model of pneumonia. (A–D) Antimicrobial susceptibility of A. baumannii ATCC 17978VU and 17978UN wild-type and mutant strains was determined by a disk diffusion assay and measuring the zone of clearance. Experiments were conducted three independent times with similar results. Data are mean ± SEM, n = 3. Significance is by one-way ANOVA with Tukey’s multiple comparisons. Limit of detection (LOD) = 6 mm. (E) Lysozyme susceptibility of 17978VU and 17978UN wild-type and mutant strains at 12 h during growth with a final concentration of 1 mg/mL lysozyme. Data are mean ± SEM, n = 3. Significance is by one-way ANOVA with Tukey’s multiple comparisons. (F) 17978VU and 17978UN wild-type and ΔmlaF strains with the endogenous or alternate uppS allele were used to intranasally infect C57BL/6 mice in a 1:1 wild-type:mutant ratio. After 48 h post infection, the lungs were harvested and bacterial burdens were enumerated. Data are mean ± SEM, n = 5. Normality of log10-transformed competitive index data was determined by the Kolmogorov-Smirnov test, and significance was determined by a one-sample t-test compared to 0. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We previously reported that the mlaF mutant in the ATCC 17978UN background had a virulence defect in a murine model of pneumonia (21). Based on decreased resistance to host stresses, we hypothesized that the mlaF uppSUN strain would have a greater virulence defect than the mlaF uppSVU strain. Therefore, we compared bacterial burdens in a competitive infection with wild-type 17978VU and 17978UN strains and mlaF mutants where the mutant expressed either the endogenous or alternate uppS allele. At 40 h post infection, the mlaF mutants containing the uppSUN allele had a significant defect in virulence compared to wild-type regardless of strain background (Fig. 7F; Fig. S5B through E). The mlaF mutant strains with uppSVU showed no virulence defect compared to wild type (Fig. 7F; Fig. S5B through E). This demonstrates that the virulence defect observed in mlaF is due to synergy with the uppSUN allele. Together, these data suggest that the Mla system and Und-P levels synergize to promote antimicrobial resistance and virulence in A. baumannii.
DISCUSSION
The A. baumannii cell envelope is the first line of defense against antibiotic stress and the host. Therefore, it is critical to understand the biological processes that uphold this barrier. The Mla system is an important factor in maintaining outer membrane lipid asymmetry to promote envelope integrity in Gram-negative bacteria. Multiple reports have demonstrated envelope defects in the absence of the Mla system in A. baumannii (16, 24, 51). Here, we found that the deletion of mlaF in two closely related A. baumannii ATCC 17978 strains results in different phenotypes. Genetic dissection uncovered synergy between Und-P abundance and the Mla system in A. baumannii.
A. baumannii ATCC 17978 is a commonly used type strain. We recently discovered that ATCC 17978 was a mixed culture of two closely related strains that differed by the accessory locus AbaAL44 and multiple SNPs (54). Depending on the time of order, the ratio of 17978UN to 17978VU isolates received from ATCC varied. In 2009, 4/6 isolates screened were 17978UN and 2/6 were 17978VU; by contrast, in an ATCC order from 2021, 35/36 were 17978UN and 1/36 were 17978VU (54). This suggests that while the 17978UN allele of uppS is uncommon in circulating A. baumannii strains (Fig. 1), the majority of recent 17978 isolates from ATCC are likely 17978UN. Differences in ∆mlaF phenotypes of ATCC 17978UN and 17978VU presented the opportunity to use these variants as a genetic tool to investigate synthetic gene pairs. Ultimately, uppS was shown to synergize with ∆mlaF in resistance to membrane stresses, envelope permeability, antibiotic and host stress resistance, and virulence in a mouse lung infection model. This result was surprising as a previous report identified SNPs in obgE, which encodes an essential GTPase involved in the stringent response, as the critical determinant of differences in growth and the stringent response in two isogenic ATCC 17978 variants (51). While we did not observe overt differences in growth based on the obgE allele, we speculate that is likely due to differences in growth conditions such as aeration. Interestingly, among the protein-encoding genetic differences between 17978VU and 17978UN, the 44 kb AbaAL44 locus and lptDUN are the only 17978UN alleles that are more common than 17978VU alleles (Fig. 1). This may suggest that there may have been selective pressure to maintain lptDUN in the 17978UN background. Together, these findings exemplify that closely related strains can be leveraged to uncover underlying integration of essential biological process.
As an essential glycan carrier, Und-P plays a role in the biosynthesis of multiple bacterial cell envelope components including peptidoglycan, glycoproteins, LPS O-antigen, lipid A, and capsular polysaccharides (53, 63, 64). However, A. baumannii does not synthesize the LPS O-antigen and does not encode LpxT that uses Und-PP as a phosphate donor for lipid A. Imaging of isogenic strains with UppS variants showed no overt difference in peptidoglycan (Fig. S3A). Staining for capsular polysaccharide from whole cell lysates revealed reduced capsule in ΔmlaF mutants and further reduced capsule in ΔmlaF mutants expressing uppSUN. Differences between ΔmlaF strains expressing the same uppS allele but in opposing 17978 backgrounds suggest that there may be additional synergistic relationships impacting capsule content. UppSUN has a decreased enzymatic rate and confers increased membrane sensitivity in an ΔmlaF background. However, there were no defects in the wild-type strains for membrane stress susceptibility, suggesting that the ΔmlaF defects synergize with reduced Und-P to result in a weakened cell envelope. We previously observed that A. baumannii ATCC 17978UN ∆mlaF had decreased LOS abundance (21), suggesting the Mla system is important for LOS abundance in A. baumannii. Here, we show that the reduction in LOS within 17978UN ∆mlaF is due to UppSUN, as an isogenic mutant with UppSVU displays wild-type-like LOS levels. This suggests that Und-P may have an uncharacterized role in LOS synthesis in A. baumannii. For example, Und-P could serve as a glycan carrier for LOS such as for the sugar molecules in the core oligosaccharides or a non-homologous enzyme may use Und-PP as a phosphate donor for lipid A similar to LpxT. In many gammaproteobacteria, the uppS gene is in an operon with a phosphatidate cytidylyltransferase, an enzyme important in phospholipid biosynthesis, suggesting there may be a broader synergistic relationship between the transport and biosynthesis of phospholipids and other envelope components such as capsule, peptidoglycan, and glycoproteins. In subpopulations of A. baumannii that have been evolved to be LOS deficient, the Mla system hinders fitness (39). However, multiple A. baumannii strains with LOS deficiency showed upregulation of Mla genes (12, 65). This demonstrates a distinction between populations that have adapted to LOS deficiency and non-adapted populations with LOS deficiency. In our work, UppSUN reduces but does not fully deplete LOS in the ∆mlaF strain background. We, therefore, show that the Mla system is important for maintaining cell envelope integrity in the presence of reduced Und-P.
Suppressor mutants in genes encoding isoprenoid biosynthetic pathway enzymes restored membrane stress resistance to ΔmlaF uppSUN strains. We found that the ΔmlaF uppSUN phenotype was suppressed by mutations in isoprenoid biosynthetic genes ispA and ispB. The ISAba11 transposition to the 5′ untranslated region of ispB was also found in an extensively drug resistant clinical isolate of A. baumannii, demonstrating potential clinical implications for this insertion (66). Here, we identified another suppressor in an isoprenoid biosynthetic gene encoding IspAG223E that arose when constructing the 17978VU ΔmlaF uppSUN strain. In E. coli, ispA null mutants had reduced isoprenoid quinone levels that was rescued by the overexpression of either ispB or ispU (uppS) (67, 68). This suggests that there may be low-level functional redundancy between IspA, IspB, and UppS. We, therefore, hypothesize that IspAG223E enhances low-level Und-PP synthesis performed by IspA. Similarly, in E. coli, a defective UppS variant resulted in growth defects that were suppressed by mutations in isoprenoid pathway genes (69). This suggests that there may be a conserved mechanism to promote the production of undecaprenyl species in the presence of a defective UppS.
The structure of UppS has been solved from multiple organisms, including A. baumannii (70–72). The substitution in UppS that differs between 17978VU (M78) and 17978UN (T78) is at the 78th amino acid position, located on the α3 helix. The α3 helix borders the active site and surrounds the open cavity the product chain would occupy. Residues along the α3 helix are largely conserved (73). The methionine at this position on the α3 helix is conserved in prenyl-transferases from E. coli, Saccharomyces cerevisiae, Mycobacterium tuberculosis, Arabidopsis thaliana, and humans (74). This suggests that the methionine is important for the structure or function of prenyl-transferases. In the 17978UN UppS variant, the methionine at position 78 is replaced with a threonine. While not highlighted as a critical residue, the surrounding hydrophobic residues near E. coli UppS M86 (M78 in A. baumannii) are implicated in interactions with the substrate (74). Importantly, there were no major structural differences in UppS between the M78 and T78 variants by circular dichroism analysis (Fig. S1A and B). The substitution to a threonine in 17978UN may, therefore, perturb internal hydrophobic interactions between the α3 helix and substrate. We found that 17978UN UppS has a reduced enzymatic rate compared to 17978VU UppS. This suggests that the T78 encoded by ATCC 17978UN may perturb crucial interactions with the substrate hydrophobic tail required for efficient Und-PP production.
Isoprenoid biosynthesis is essential for diverse processes in bacteria ranging from metabolism to virulence at the host-pathogen interface. As such, proteins within isoprenoid biosynthesis have been considered a drug target for novel therapeutics (75, 76). Furthermore, UppS specifically is a proposed drug target for A. baumannii (77). The synthetically sensitive ΔmlaF mutants in ATCC 17978UN suggest that targeting UppS or depleting undecaprenyl species in a combination therapy may represent an effective therapeutic strategy. The cell envelope is the largest barrier to effective antibiotic treatment in Gram-negative bacteria. The epistatic interaction between the A. baumannii Mla system and UppS identifies a potential strategy to circumvent the envelope barrier for difficult-to-treat infections. Our findings provide a deeper understanding into how A. baumannii maintains cell envelope integrity to survive in hostile environments.
MATERIALS AND METHODS
Bacterial strains and growth
All bacterial strains and plasmids used in this study are listed in Tables S1 and S2, respectively. Strains were grown in LB or on LB plates with 1.5% (wt/vol) agar. Antibiotics were used at the following concentrations: carbenicillin, 75 mg/L; kanamycin, 40 mg/L; chloramphenicol, 15 mg/L. Overnight cultures were started in 3–5 mL LB, inoculated with a single colony, and incubated at 37°C while shaking at 180 rpm for 8–16 h. Growth curves were conducted in 100 µL media in a flat bottom 96-well plate, inoculated with 1 µL overnight culture, and incubated at 37°C with shaking. Bacterial growth was monitored by optical density at 600 nm (OD600) in an EPOCH2 BioTek (Winooski, VT) plate reader. For assays on membrane stress, SDS/EDTA was included in the media at varying concentrations and the concentrations used are noted in each figure due to variability. For lysozyme susceptibility assays, lysozyme was included in the media at a final concentration of 1 mg/mL and the OD600 at 12 h was used to calculate the %OD600 compared to the average of the OD600 at 12 h in LB for each strain.
Plasmid construction and allelic exchange mutants
All oligonucleotides used are listed in Table S3. DNA was amplified using 2× Phusion Master Mix (ThermoFisher, Waltham, MA), Q5 High Fidelity 2× Master Mix [New England Biolabs (NEB), Ipswich, MA], or GoTaq Green Master Mix (NEB, Ipswich, MA). The pFLP2 vector was used for all allelic exchange mutants. Using ATCC 17978VU or 17978UN as the template, 1,000 bp upstream and downstream of the mutation of interest was amplified. For pFLP2-obgE, HN1 and HN2 were used. For pFLP2-lptD, HN23 and HN24 were used. For pFLP2-uppS, HN25 and HN26 were used. For ispA* reconstruction, HN85 and HN86 were used with strain LP546 as the template. The PCR product was incorporated into a digested pFLP2 backbone using KpnI and BamHI restriction sites and HiFi ligation mix (NEB, Ipswich, MA). All restriction enzymes are from NEB (Ipswich, MA). Strains containing the ispB suppressor mutation and mlaF knockout were generated using pLDP70 and pLDP8, respectively, (21). A. baumannii was transformed through conjugation by triparental mating with E. coli strain HB101 containing pRK2013 as the helper strain. Merodiploids containing the integrated pFLP2 vector were screened on plates containing 10% sucrose and 75 mg/L carbenicillin and SucS and CarbR colonies were selected. Merodiploids with the appropriate phenotype were screened by PCR to confirm plasmid incorporation at the correct site. Merodiploids were grown on LB agar, resuspended in LB, and plated to LB agar containing 10% sucrose to select for second crossover events, and the SucR strains were screened for CarbS. Genotypes were confirmed via Sanger Sequencing (UIC Genomics Research Core) and/or whole-genome sequencing (SeqCoast Genomics, Portsmouth New Hampshire; SeqCenter, Pittsburg PA) to confirm no additional mutations in relevant loci.
UppS expression plasmids for protein purification were generated using pET-15b digested with BamHI and NdeI. UppS was amplified from either 17978VU or 17978UN with HN94 and HN95 and ligated into the pET-15b backbone by HiFi ligation. All plasmid sequences were confirmed with Sanger sequencing by the UIC Genomics Research Core or whole plasmid sequencing by Primordium (Monrovia, CA).
To distinguish the uppSUN allele from the uppSVU allele, primers HN31 (UN) and HN32 (VU) were used with HN30, 2× Green Gotaq (Promega, Madison, WI), and an annealing temperature of 61.9°C and extension time of 2 min.
Serial dilution spotting assays
Overnight cultures were 10-fold serially diluted in 96-well plates in 1× PBS to 10−7. Dilutions were spotted in 3 or 5 µL drops on LB agar and LB agar containing SDS/EDTA and incubated at 37°C overnight. Plates were imaged with a BioTek (Winooski, VT) ChemiDoc MP imager.
Antibiotic susceptibility assay
Melted soft agar (0.8% agar, 0.8% NaCl) was brought to 50°C in 9.5 mL aliquots before inoculation with 270 µL overnight culture and plating to a prewarmed 15 cm LB agar plate. Once solidified, pre-loaded antibiotic discs [BD (Becton Dickinson), Franklin Lakes, NJ] were placed using the BD automatic disc dispenser onto the agar overlay, and the plates were incubated at 37°C overnight. The following day, diameters of the zones of clearance were measured in mm.
Ethidium bromide uptake assay
The EtBr uptake assay was adapted from previous studies (24, 61). Bacteria were grown in 3 mL LB to mid-log phase before centrifuging and normalizing to ~1 × 1010 CFU in PBS. Normalized cells were plated to confirm equal CFU across strains. In 200 µL final volume, normalized cells were combined with 200 µM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) and 1.2 µM EtBr. EtBr uptake was monitored in a black 96-well plate with reads every 15 s on a BioTek Synergy H1 (Winooski, VT) using excitation and emission wavelengths of 530 nm and 590 nm, respectively.
UppS purification
UppSVU and UppSUN were purified from BL21 ArcticExpress DE3 RIL cells (VWR, Radnor, PA). Cells expressing pET-15b-UppSVU or pET-15b-UppSUN were grown in 10 mL LB containing carbenicillin overnight at 37°C. The following day, cells were subcultured into 1 L LB containing carbenicillin in a 2.8-L Fernbach flask and grown to mid-log phase at 37°C. At OD600 of 0.6, IPTG was added to a final concentration of 0.5 mM to induce protein expression. Cells were incubated overnight while shaking (180 rpm) at 25°C. The following day cells were centrifuged at 2,000 × g for 10 min and the cell pellet was lysed using 8 mL of B-PER Bacterial Protein Extraction Reagent (Thermo Scientific, Waltham, MA) per gram of pellet with gentle shaking for 1 h. The lysate was pelleted by centrifugation at 4,300 × g for 5 min. The soluble fraction in the supernatant was mixed with an equal part lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole) and was added to 8 mL Ni-NTA resin (Qiagen, Hilden, Germany) preequilibrated with lysis buffer and rocked for 1 h at 4°C. Protein-bound resin was then applied to a 10 mL chromatography column (BioRad, Hercules, CA) and washed twice with 10 mL wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 20 mM imidazole). Protein was eluted using elution buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 250 mM imidazole) in serial 2 mL elution volumes. Samples were separated by SDS-PAGE and stained with SimplyBlue Safe Stain (Invitrogen, Waltham, MA) to verify protein purification. Protein was desalted using PD-10 desalting columns (Cytiva, Marlborough, MA) into circular dichroism and UppS assay buffers (see SI and below).
Undecaprenyl-phosphate quantification
Cultures of 17978UN and 17978VU in 5 mL LB were grown to mid-log phase with a final OD600 of 0.5. Cultures were pelleted and stored at −80°C. Pellets were subjected to Bligh and Dyer extraction and dried overnight. The crude cell lysate was then resuspended in 200 µL of n-propanol and 0.1% ammonium hydroxide (1:3) (78). After sonicating the cell suspension using water bath, 5 µL of sample was injected into C18 column and analyzed for C55 BP/Und-P m/z ratio of 845.7 by liquid chromatography/mass spectrometry (LC-MS). Area under the curve of each Und-P peak of all samples was recorded and used to calculate Und-P (pmol) using a standard curve generated from known Und-P concentrations.
UppS microplate enzyme assays
2CNA-GPP was prepared as described previously (59, 69). In each well of a black-walled 96 well plate, reactions were prepared with 2.5 mM 2CNA-GPP, 0.5 mM MgCl2, 5 mM KCl, 0.1% Triton-X-100, and 100 nM recombinant UppS from 17978UN or 17978VU. The plate was incubated in the plate reader at 30°C for 5 min and then, the reaction was initiated with the addition of 1 mM IPP (final concentration). Fluorescence was monitored at 30°C over 1 h at an excitation wavelength of 340 nm and emission wavelength of 390 nm. The reaction rate was determined based on the linear fluorescence increase over the first 8 min for proteins from both strains (n = 3).
Capsule
Samples were prepared similarly as previously described (79). Briefly, 3-mL overnight cultures were diluted 1:100 in 3 mL LB and grown to early stationary phase with an OD600 of 1.4. Two 1-mL aliquots were harvested. One aliquot was boiled at 85°C for 15 min and subjected to a Pierce 660 (Thermo Fisher, Waltham, MA) total protein quantification following manufacturer instructions. The second aliquot was pelleted and resuspended in lysis buffer (60 mM Tris, pH 8, 10 mM MgCl2, 50 µM CaCl2, 3 mg/mL lysozyme, 60 U/mL DNase, 10 µg/mL RNase) normalized to 5 mg/mL total protein, which was determined to be in the linear range of the capsule densitometry analysis by a dilution series experiment and incubated at 37°C for 1 h. Samples were subjected to three freeze/thaw cycles followed by further DNase and RNase treatment and incubation for 30 min at 37°C. Samples were then treated with SDS to a final concentration of 0.5% and incubated at 37°C for 30 min, followed by 40 g proteinase K treatment and incubation at 60°C for 1 h. Laemmli sample buffer (Bio-Rad, Hercules, CA) supplemented with DTT to 54 mg/mL was then added before gel electrophoresis on a 4%–12% Bis-Tris gel (MilliporeSigma, Burlington, MA). Fifteen micrograms of total protein was loaded for each sample. Gels were then stained with 0.1% Alcian blue in 40% ethanol/60% 20 mM sodium acetate pH 4.75. Images were captured on a BioTek (Winooski, VT) ChemiDoc MP imager.
LOS silver stain
Samples were prepared similar to previous descriptions (80, 81). Three milliliters of overnight cultures was diluted 1:100 in 5 mL fresh LB and grown to mid-log with an OD600 of 0.5. Two 1mL aliquots were harvested. One aliquot was boiled at 85°C for 15 min and subjected to a Pierce 660 (Thermo Fisher, Waltham, MA) total protein quantification following manufacturer instructions. The second aliquot was pelleted and resuspended in 1× Novex SDS sample buffer (Invitrogen, Waltham, MA) so that the final total protein concentration was 6 mg/mL, which was determined to be in the linear range of the LOS densitometry analysis by a dilution series experiment. Samples were lysed via boiling for 15 min at 85°C and treated with proteinase K at a final concentration of 0.16 µg/µL. Gels were loaded with samples containing 10 µg total protein before electrophoresis on a 16.5% Tris Tricine gel (Bio-Rad, Hercules, CA). Gels were stained with the Pierce Silver Stain kit (Thermo Fisher, Waltham, MA) following manufacturer instructions. Images were captured on a BioTek (Winooski, VT) ChemiDoc MP imager.
Densitometry analysis of capsule and LOS gels
Densitometry analysis was performed using ImageJ. Images were converted to an 8-bit image, and the background was subtracted using a 50-pixel rolling ball radius. Using the gel analysis tool, bands were selected and peaks were quantified. Data are presented as a ratio to the density of 17978UN wild type for each independent gel.
Murine model of A. baumannii lung infection
Six-week-old female C57BL/6 mice were purchased from Jackson Laboratory. Mice were housed in a temperature-controlled environment with 12 h light/dark cycles and food and water were provided as needed and were acclimated to the facility to 1 week prior to infection. Mice were anesthetized with ketamine/xylazine and inoculated intranasally with 35 µL bacterial suspension 1:1 mixture A. baumannii ATCC 17978 wild-type and ΔmlaF mutant derivatives, containing approximately 3 × 108 CFU as described previously (21). The inoculum dose was confirmed by serial dilution and plating on selective agar media. Mice were euthanized at 48 h post infection by CO2 asphyxiation, and the lungs were excised aseptically. Tissues were homogenized, and all samples were serially diluted and plated on LB and kanamycin selective agar plates for bacterial enumeration. The competitive index of mutant/wild-type strains was calculated by dividing the mutant CFU ratio (CFUoutput/CFUinput) by the wild-type CFU ratio. All animal care protocols were approved by the University of Illinois Chicago Institutional Animal Care and Use Committee (IACUC; protocol number 20–165) in accordance with the Animal Care Policies of UIC, the Animal Welfare Act, the National Institutes of Health, and the American Veterinary Medical Association (AVMA). Animals were humanely euthanized consistent with the AVMA guidelines.
Data reporting, statistical analysis, and figure preparation
Each measurement was taken from a distinct biological sample (e.g., bacterial culture from a single colony or an individual mouse). Data processing and statistical analyses were performed using Microsoft Excel 16.77.1 and GraphPad Prism 10. Statistical tests used are indicated in each figure legend. Figures were prepared in Adobe Illustrator 28.1.
ACKNOWLEDGMENTS
We thank the UIC Biophysics Core and the Center for Structural Biology for assisting with the circular dichroism experiment and analysis. We thank members of the Palmer laboratory for critical reading of the manuscript, Zachery Lonergan for review of the manuscript, and Matthew Jorgenson for helpful discussion.
This work was supported by startup funds from the University of Illinois Chicago and National Institutes of Health (NIH) Awards R00HL143440 to L.D.P. and R01GM123251 to J.M.T.
Contributor Information
Lauren D. Palmer, Email: ldpalmer@uic.edu.
Indranil Biswas, The University of Kansas Medical Center, Kansas City, Kansas, USA.
DATA AVAILABILITY
Whole genome sequencing data is available in the National Center for Biotechnology Information (NCBI) sequence read archive (SRA) under BioProject: PRJNA1020123 and PRJNA656143 (22).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02804-23.
Figures S1-S5, Tables S1-3, and supplemental text.
Proteomes and alleles depicted in Figure 1.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Clark NM, Zhanel GG, Lynch JPI. 2016. Emergence of antimicrobial resistance among Acinetobacter species: a global threat. Curr Opin Crit Care 22:491–499. doi: 10.1097/MCC.0000000000000337 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization . 2019. No time to wait: securing the future from drug-resistant infections. Report to the Secretary-General of the United Nations. World Health Organization and Interagency Coordination Group on Antimicrobial Resistance [Google Scholar]
- 3. Weiner LM, Webb AK, Limbago B, Dudeck MA, Patel J, Kallen AJ, Edwards JR, Sievert DM. 2016. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011-2014. Infect Control Hosp Epidemiol 37:1288–1301. doi: 10.1017/ice.2016.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 5. Geisinger E, Huo W, Hernandez-Bird J, Isberg RR. 2019. Acinetobacter baumannii: envelope determinants that control drug resistance, virulence, and surface variability. Annu Rev Microbiol 73:481–506. doi: 10.1146/annurev-micro-020518-115714 [DOI] [PubMed] [Google Scholar]
- 6. Islam N, Kazi MI, Kang KN, Biboy J, Gray J, Ahmed F, Schargel RD, Boutte CC, Dörr T, Vollmer W, Boll JM. 2022. Peptidoglycan recycling promotes outer membrane integrity and carbapenem tolerance in Acinetobacter baumannii. mBio 13:e0100122. doi: 10.1128/mbio.01001-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu L, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. 2018. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559:617–621. doi: 10.1038/s41586-018-0344-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kenyon JJ, Nigro SJ, Hall RM. 2014. Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS One 9:e107833. doi: 10.1371/journal.pone.0107833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simpson BW, Nieckarz M, Pinedo V, McLean AB, Cava F, Trent MS. 2021. Acinetobacter baumannii can survive with an outer membrane lacking lipooligosaccharide due to structural support from elongasome peptidoglycan synthesis. mBio 12:e0309921. doi: 10.1128/mBio.03099-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boll JM, Crofts AA, Peters K, Cattoir V, Vollmer W, Davies BW, Trent MS. 2016. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc Natl Acad Sci U S A 113:E6228–E6237. doi: 10.1073/pnas.1611594113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chong Z-S, Woo W-F, Chng S-S. 2015. Osmoporin OmpC forms a complex with MlaA to maintain outer membrane lipid asymmetry in Escherichia coli. Mol Microbiol 98:1133–1146. doi: 10.1111/mmi.13202 [DOI] [PubMed] [Google Scholar]
- 18. Thong S, Ercan B, Torta F, Fong ZY, Wong HYA, Wenk MR, Chng S-S. 2016. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. eLife 5:e19042. doi: 10.7554/eLife.19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ercan B, Low W-Y, Liu X, Chng S-S. 2019. Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC-Mla system. Biochemistry 58:114–119. doi: 10.1021/acs.biochem.8b00897 [DOI] [PubMed] [Google Scholar]
- 20. Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. 2017. Architectures of lipid transport systems for the bacterial outer membrane. Cell 169:273–285. doi: 10.1016/j.cell.2017.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palmer LD, Minor KE, Mettlach JA, Rivera ES, Boyd KL, Caprioli RM, Spraggins JM, Dalebroux ZD, Skaar EP. 2020. Modulating isoprenoid biosynthesis increases lipooligosaccharides and restores Acinetobacter baumannii resistance to host and antibiotic stress. Cell Rep 32:108129. doi: 10.1016/j.celrep.2020.108129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. MacRae MR, Puvanendran D, Haase MAB, Coudray N, Kolich L, Lam C, Baek M, Bhabha G, Ekiert DC. 2023. Protein-protein interactions in the Mla lipid transport system probed by computational structure prediction and deep mutational scanning. J Biol Chem 299:104744. doi: 10.1016/j.jbc.2023.104744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Isom GL, Davies NJ, Chong Z-S, Bryant JA, Jamshad M, Sharif M, Cunningham AF, Knowles TJ, Chng S-S, Cole JA, Henderson IR. 2017. MCE domain proteins: conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci Rep 7:8608. doi: 10.1038/s41598-017-09111-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamischke C, Fan J, Bergeron J, Kulasekara HD, Dalebroux ZD, Burrell A, Kollman JM, Miller SI. 2019. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife 8:e40171. doi: 10.7554/eLife.40171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Jonge EF, Vogrinec L, van Boxtel R, Tommassen J. 2022. Inactivation of the Mla system and outer-membrane phospholipase A results in disrupted outer-membrane lipid asymmetry and hypervesiculation in Bordetella pertussis. Curr Res Microb Sci 3:100172. doi: 10.1016/j.crmicr.2022.100172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bernier SP, Son S, Surette MG. 2018. The Mla pathway plays an essential role in the intrinsic resistance of Burkholderia cepacia complex species to antimicrobials and host innate components. J Bacteriol 200:e00156-18. doi: 10.1128/JB.00156-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ. 2016. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113:E1565–E1574. doi: 10.1073/pnas.1601375113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki T, Murai T, Fukuda I, Tobe T, Yoshikawa M, Sasakawa C. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol 11:31–41. doi: 10.1111/j.1365-2958.1994.tb00287.x [DOI] [PubMed] [Google Scholar]
- 29. Hong M, Gleason Y, Wyckoff EE, Payne SM. 1998. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect Immun 66:4700–4710. doi: 10.1128/IAI.66.10.4700-4710.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carpenter CD, Cooley BJ, Needham BD, Fisher CR, Trent MS, Gordon V, Payne SM. 2014. The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect Immun 82:660–669. doi: 10.1128/IAI.01057-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PCF, Titball RW, Wren BW. 2007. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect Immun 75:1186–1195. doi: 10.1128/IAI.01240-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munguia J, LaRock DL, Tsunemoto H, Olson J, Cornax I, Pogliano J, Nizet V. 2017. The Mla pathway is critical for Pseudomonas aeruginosa resistance to outer membrane permeabilization and host innate immune clearance. J Mol Med (Berl) 95:1127–1136. doi: 10.1007/s00109-017-1579-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nasu H, Shirakawa R, Furuta K, Kaito C. 2022. Knockout of mlaA increases Escherichia coli virulence in a silkworm infection model. PLoS One 17:e0270166. doi: 10.1371/journal.pone.0270166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baarda BI, Zielke RA, Le Van A, Jerse AE, Sikora AE. 2019. Neisseria gonorrhoeae MlaA influences gonococcal virulence and membrane vesicle production. PLoS Pathog 15:e1007385. doi: 10.1371/journal.ppat.1007385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abellon-Ruiz J. 2022. Forward or backward, that is the question: phospholipid trafficking by the Mla system. Emerg Top Life Sci 7:125–135. doi: 10.1042/ETLS20220087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Powers MJ, Trent MS. 2019. Intermembrane transport: glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc Natl Acad Sci U S A 116:17147–17155. doi: 10.1073/pnas.1902026116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, Herrera CM, Trent MS. 2016. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol 70:255–278. doi: 10.1146/annurev-micro-102215-095308 [DOI] [PubMed] [Google Scholar]
- 38. Nagy E, Losick R, Kahne D. 2019. Robust suppression of lipopolysaccharide deficiency in Acinetobacter baumannii by growth in minimal medium. J Bacteriol 201:e00420-19. doi: 10.1128/JB.00420-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Powers MJ, Trent MS. 2018. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc Natl Acad Sci U S A 115:E8518–E8527. doi: 10.1073/pnas.1806714115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Low W-Y, Thong S, Chng S-S. 2021. ATP disrupts lipid-binding equilibrium to drive retrograde transport critical for bacterial outer membrane asymmetry. Proc Natl Acad Sci U S A 118:e2110055118. doi: 10.1073/pnas.2110055118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Awai K, Xu C, Tamot B, Benning C. 2006. A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc Natl Acad Sci U S A 103:10817–10822. doi: 10.1073/pnas.0602754103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abellón-Ruiz J, Kaptan SS, Baslé A, Claudi B, Bumann D, Kleinekathöfer U, van den Berg B. 2017. Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2:1616–1623. doi: 10.1038/s41564-017-0046-x [DOI] [PubMed] [Google Scholar]
- 43. Tang X, Chang S, Qiao W, Luo Q, Chen Y, Jia Z, Coleman J, Zhang K, Wang T, Zhang Z, Zhang C, Zhu X, Wei X, Dong C, Zhang X, Dong H. 2021. Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB. Nat Struct Mol Biol 28:81–91. doi: 10.1038/s41594-020-00532-y [DOI] [PubMed] [Google Scholar]
- 44. Yeow J, Tan KW, Holdbrook DA, Chong Z-S, Marzinek JK, Bond PJ, Chng S-S. 2018. The architecture of the OmpC-MlaA complex sheds light on the maintenance of outer membrane lipid asymmetry in Escherichia coli. J Biol Chem 293:11325–11340. doi: 10.1074/jbc.RA118.002441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Douglass MV, McLean AB, Trent MS. 2022. Absence of YhdP, TamB, and YdbH leads to defects in glycerophospholipid transport and cell morphology in Gram-negative bacteria. PLoS Genet 18:e1010096. doi: 10.1371/journal.pgen.1010096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruiz N, Davis RM, Kumar S. 2021. YhdP, TamB, and YdbH are redundant but essential for growth and lipid homeostasis of the Gram-negative outer membrane. mBio 12:e0271421. doi: 10.1128/mBio.02714-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hughes GW, Hall SCL, Laxton CS, Sridhar P, Mahadi AH, Hatton C, Piggot TJ, Wotherspoon PJ, Leney AC, Ward DG, Jamshad M, Spana V, Cadby IT, Harding C, Isom GL, Bryant JA, Parr RJ, Yakub Y, Jeeves M, Huber D, Henderson IR, Clifton LA, Lovering AL, Knowles TJ. 2019. Evidence for phospholipid export from the bacterial inner membrane by the Mla ABC transport system. Nat Microbiol 4:1692–1705. doi: 10.1038/s41564-019-0481-y [DOI] [PubMed] [Google Scholar]
- 48. Mann D, Fan J, Somboon K, Farrell DP, Muenks A, Tzokov SB, DiMaio F, Khalid S, Miller SI, Bergeron JRC. 2021. Structure and lipid dynamics in the maintenance of lipid asymmetry inner membrane complex of A. baumannii. Commun Biol 4:817. doi: 10.1038/s42003-021-02318-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Coudray N, Isom GL, MacRae MR, Saiduddin MN, Bhabha G, Ekiert DC. 2020. Structure of bacterial phospholipid transporter MlaFEDB with substrate bound. Elife 9:e62518. doi: 10.7554/eLife.62518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chi X, Fan Q, Zhang Y, Liang K, Wan L, Zhou Q, Li Y. 2020. Structural mechanism of phospholipids translocation by MlaFEDB complex. Cell Res 30:1127–1135. doi: 10.1038/s41422-020-00404-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Powers MJ, Simpson BW, Trent MS. 2020. The Mla pathway in Acinetobacter baumannii has no demonstrable role in anterograde lipid transport. Elife 9:e56571. doi: 10.7554/eLife.56571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Persky NS, Ferullo DJ, Cooper DL, Moore HR, Lovett ST. 2009. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 73:253–266. doi: 10.1111/j.1365-2958.2009.06767.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Manat G, Roure S, Auger R, Bouhss A, Barreteau H, Mengin-Lecreulx D, Touzé T. 2014. Deciphering the metabolism of undecaprenyl-phosphate: the bacterial cell-wall unit carrier at the membrane frontier. Microb Drug Resist 20:199–214. doi: 10.1089/mdr.2014.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wijers CDM, Pham L, Menon S, Boyd KL, Noel HR, Skaar EP, Gaddy JA, Palmer LD, Noto MJ. 2021. Identification of two variants of Acinetobacter baumannii strain ATCC 17978 with distinct genotypes and phenotypes. Infect Immun 89:e0045421. doi: 10.1128/IAI.00454-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lundstedt E, Kahne D, Ruiz N. 2021. Assembly and maintenance of lipids at the bacterial outer membrane. Chem Rev 121:5098–5123. doi: 10.1021/acs.chemrev.0c00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157. doi: 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. doi: 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Dehò G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469. doi: 10.1128/JB.00270-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dodbele S, Martinez CD, Troutman JM. 2014. Species differences in alternative substrate utilization by the antibacterial target undecaprenyl pyrophosphate synthase. Biochemistry 53:5042–5050. doi: 10.1021/bi500545g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reid AJ, Scarbrough BA, Williams TC, Gates CE, Eade CR, Troutman JM. 2020. General utilization of fluorescent polyisoprenoids with sugar selective phosphoglycosyltransferases. Biochemistry 59:615–626. doi: 10.1021/acs.biochem.9b01026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lonergan ZR, Nairn BL, Wang J, Hsu Y-P, Hesse LE, Beavers WN, Chazin WJ, Trinidad JC, VanNieuwenhze MS, Giedroc DP, Skaar EP. 2019. An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep 26:2009–2018. doi: 10.1016/j.celrep.2019.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ragland SA, Criss AK. 2017. From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog 13:e1006512. doi: 10.1371/journal.ppat.1006512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jorgenson MA, Kannan S, Laubacher ME, Young KD. 2016. Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol 100:1–14. doi: 10.1111/mmi.13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Touzé T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. 2008. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol 67:264–277. doi: 10.1111/j.1365-2958.2007.06044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. 2012. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob Agents Chemother 56:59–69. doi: 10.1128/AAC.05191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Loewen PC, Alsaadi Y, Fernando D, Kumar A. 2014. Genome sequence of an extremely drug-resistant clinical isolate of Acinetobacter baumannii strain AB030. Genome Announc 2:e01035-14. doi: 10.1128/genomeA.01035-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fujisaki S, Takahashi I, Hara H, Horiuchi K, Nishino T, Nishimura Y. 2005. Disruption of the structural gene for farnesyl diphosphate synthase in Escherichia coli. J Biochem 137:395–400. doi: 10.1093/jb/mvi049 [DOI] [PubMed] [Google Scholar]
- 68. Takahashi H, Aihara Y, Ogawa Y, Murata Y, Nakajima K-I, Iida M, Shirai M, Fujisaki S. 2018. Suppression of phenotype of Escherichia coli mutant defective in farnesyl diphosphate synthase by overexpression of gene for octaprenyl diphosphate synthase. Biosci Biotechnol Biochem 82:1003–1010. doi: 10.1080/09168451.2017.1398066 [DOI] [PubMed] [Google Scholar]
- 69. MacCain WJ, Kannan S, Jameel DZ, Troutman JM, Young KD. 2018. A defective undecaprenyl pyrophosphate synthase induces growth and morphological defects that are suppressed by mutations in the isoprenoid pathway of Escherichia coli. J Bacteriol 200:e00255-18. doi: 10.1128/JB.00255-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Allen CM. 1985. Purification and characterization of undecaprenylpyrophosphate synthetase. Methods Enzymol 110:281–299. doi: 10.1016/s0076-6879(85)10085-6 [DOI] [PubMed] [Google Scholar]
- 71. Fujihashi M, Zhang YW, Higuchi Y, Li XY, Koyama T, Miki K. 2001. Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase. Proc Natl Acad Sci U S A 98:4337–4342. doi: 10.1073/pnas.071514398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ko TP, Huang CH, Lai SJ, Chen Y. 2018. Structure of undecaprenyl pyrophosphate synthase from Acinetobacter baumannii. Acta Crystallogr F Struct Biol Commun 74:765–769. doi: 10.1107/S2053230X18012931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. TouzÉ T, Mengin-Lecreulx D. 2008. Undecaprenyl phosphate synthesis. EcoSal Plus 3. doi: 10.1128/ecosalplus.4.7.1.7 [DOI] [PubMed] [Google Scholar]
- 74. Guo R-T, Ko T-P, Chen AP-C, Kuo C-J, Wang AH-J, Liang P-H. 2005. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. J Biol Chem 280:20762–20774. doi: 10.1074/jbc.M502121200 [DOI] [PubMed] [Google Scholar]
- 75. Heuston S, Begley M, Gahan CGM, Hill C. 2012. Isoprenoid biosynthesis in bacterial pathogens. Microbiology (Reading) 158:1389–1401. doi: 10.1099/mic.0.051599-0 [DOI] [PubMed] [Google Scholar]
- 76. Concha N, Huang J, Bai X, Benowitz A, Brady P, Grady LC, Kryn LH, Holmes D, Ingraham K, Jin Q, Pothier Kaushansky L, McCloskey L, Messer JA, O’Keefe H, Patel A, Satz AL, Sinnamon RH, Schneck J, Skinner SR, Summerfield J, Taylor A, Taylor JD, Evindar G, Stavenger RA. 2016. Discovery and characterization of a class of pyrazole inhibitors of bacterial undecaprenyl pyrophosphate synthase. J Med Chem 59:7299–7304. doi: 10.1021/acs.jmedchem.6b00746 [DOI] [PubMed] [Google Scholar]
- 77. Thorpe JH, Wall ID, Sinnamon RH, Taylor AN, Stavenger RA. 2020. Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii. Acta Crystallogr F Struct Biol Commun 76:40–46. doi: 10.1107/S2053230X19017199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Reid AJ, Eade CR, Jones KJ, Jorgenson MA, Troutman JM. 2021. Tracking colanic acid repeat unit formation from stepwise biosynthesis inactivation in Escherichia coli. Biochemistry 60:2221–2230. doi: 10.1021/acs.biochem.1c00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. doi: 10.1371/journal.ppat.1004691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bai J, Raustad N, Denoncourt J, van Opijnen T, Geisinger E. 2023. Genome-wide phage susceptibility analysis in Acinetobacter baumannii reveals capsule modulation strategies that determine phage infectivity. PLoS Pathog 19:e1010928. doi: 10.1371/journal.ppat.1010928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Geisinger E, Mortman NJ, Dai Y, Cokol M, Syal S, Farinha A, Fisher DG, Tang AY, Lazinski DW, Wood S, Anthony J, van Opijnen T, Isberg RR. 2020. Antibiotic susceptibility signatures identify potential antimicrobial targets in the Acinetobacter baumannii cell envelope. Nat Commun 11:4522. doi: 10.1038/s41467-020-18301-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S5, Tables S1-3, and supplemental text.
Proteomes and alleles depicted in Figure 1.
Data Availability Statement
Whole genome sequencing data is available in the National Center for Biotechnology Information (NCBI) sequence read archive (SRA) under BioProject: PRJNA1020123 and PRJNA656143 (22).