Abstract
The hepatitis B virus X protein (HBx) is a broadly acting transactivator implicated in the development of liver cancer. Recently, HBx has been reported to interact with several different cellular proteins, including our report of its binding to XAP-1, the human homolog of the simian repair protein UVDDB. In the present study, several HBx mutants were used to localize the minimal domain of HBx required for binding to XAP-1/UVDDB to amino acids 55 to 101. The normal function of XAP-1/UVDDB is thought to involve binding to damaged DNA, the first step in nucleotide excision repair (NER); therefore, we hypothesized that this interaction may affect the cell’s capacity to correct lesions in the genome. When tested in two independent assays that measure NER (unscheduled DNA synthesis and host cell reactivation), the expression of HBx significantly inhibited the ability of cells to repair damaged DNA. Under the assay conditions, HBx was expressed at a level similar to that previously observed during natural viral infection and was able to transactivate several target reporter genes. These results are consistent with a model in which HBx acts as a cofactor in hepatocarcinogenesis by preventing the cell from efficiently repairing damaged DNA, thus leading to an accumulation of DNA mutations and, eventually, cancer. An adverse effect on cellular DNA repair processes suggests a new mechanism by which a tumor-associated virus might contribute to carcinogenesis.
Chronic infection with hepatitis B virus (HBV) is a major risk factor for the development of hepatocellular carcinoma (HCC). The virus encodes a 17-kDa protein, HBx, that is thought to be involved in the development of HBV-associated HCC. Studies with transgenic mice provide conflicting results. Some HBx transgenic mice develop liver cancer (32), while others do not (7, 17, 38). However, HBx can serve as a cofactor for HCC in those transgenic mice that do not develop spontaneous tumors (17, 51, 55). A cofactor role for the X protein is also observed in woodchuck hepatitis virus transgenic mice that do not spontaneously develop liver tumors (14). In vitro data further suggest that HBx may play a role in tumor formation. HBx functions as an oncoprotein in hepatocyte cells immortalized with the large-T antigen from simian virus 40 (SV40) and in the immortal cell line NIH 3T3 (23, 48). HBx is a promiscuous transactivator, and it has been shown to transactivate several viral and cellular targets, including oncogenes such as c-fos and c-myc (reviewed in reference 10).
Although its exact role in the viral life cycle is not clear, HBx is essential for viral replication in vivo (11, 60). Several functions have been attributed to the HBx protein. It has been reported to possess ribo-deoxyATPase activity (15) and to activate cellular protein kinase C signaling (30) and Ras-Raf-MAP kinase (6, 13, 41) pathways. HBx has also been reported to bind to several different cellular proteins, including CREB and ATF2 (40), the protease tryptase TL2 (53), p53 (19, 56, 57), TATA-binding protein (44), RNA polymerase subunit RBP5 (12), a regulatory α subunit of a proteasome complex (20, 24), and a novel cytoplasmic protein that inhibits HBx transactivation (36). Although the functional significance of these protein-protein interactions is not fully understood, HBx has been found in both the nucleus and cytoplasm of transfected cells, indicating that the protein may interact with cellular proteins localized in both of these compartments (16).
Recently, HBx was shown to interact with X-associated protein 1 (XAP-1) (37, 49), a probable DNA repair protein. XAP-1 is more than 99% homologous with the simian UV-damaged DNA binding protein (UVDDB) (54), the component believed to be defective in some patients with xeroderma pigmentosum complementation group E (XPE) (25, 26, 29). All XP patients show deficiencies in nucleotide excision repair (NER) and are at a greatly increased risk of developing cancer. The normal function of UVDDB appears to be recognition and binding of certain types of damaged DNA (27, 42, 45), and the protein has been shown to possess an auxiliary role in DNA repair in vitro (1). Importantly, microinjection of UVDDB into some DNA repair-deficient human XPE fibroblasts can rescue repair to normal levels (28), suggesting that XAP-1/UVDDB can complement the defective XPE binding factor.
In the study reported here, a panel of HBx mutants was employed to map the domain of HBx required for binding to XAP-1/UVDDB. Although HBx most likely binds XAP-1/UVDDB to benefit some aspect of virus replication, we considered the possibility that this interaction may incidentally interfere with the normal role of UVDDB in the repair of damaged DNA. Functional assays were used to measure the effect of HBx expression on cellular DNA repair, and several HBx mutant proteins were used to evaluate whether the inhibitory effect of HBx requires the binding of XAP-1/UVDDB. The significance of these results to HBV-mediated HCC is also discussed.
MATERIALS AND METHODS
Generation of mutant HBx genes.
The X open reading frame (subtype adw2) was cloned in frame with the Saccharomyces cerevisiae GAL4 DNA binding domain to create the vector pASX as previously described (37). Point mutations were introduced into pASX with mutated oligonucleotides and a site-directed mutagenesis kit (Clontech, Palo Alto, Calif.) to alter the following amino acids (aa): HBx7 (aa 7; Cys changed to Ser), HBx61 (aa 61; Cys changed to Leu), HBx69 (aa 69; Cys changed to Leu), and HBx90–91 (aa 90; Pro changed to Val and aa 91; Lys changed to Leu). Deletion mutants were created within pASX by PCR amplification of the desired nucleotides followed by the introduction of a TAA stop sequence and included HBx1–67, HBx1–80, HBx1–101, HBx1–140, HBx43–154, HBx15–101, and HBx55–101 (the numbers indicate the portion of the HBx protein retained in the mutant product). Restriction enzyme sites were added to the 5′ ends of the oligonucleotides used for PCR to allow digestion and ligation of the amplified product directly into the appropriate vector. In the yeast system, the deletion mutants were cloned in frame with the GAL4 DNA binding domain into the pAS1 vector. Also, the full-length X gene and selected X genes containing point mutations or deletions were subcloned into the HindIII site of the mammalian expression plasmid pSV2neo (52). All constructs were DNA sequenced to confirm the presence of the expected mutations (Sequenase kit; United States Biochemical Corp., Cleveland, Ohio). Full-length XAP-1/UVDDB was cloned into the yeast prey vector, in frame with the GAL4 activating domain, generating pACT-XAP-1 as previously reported (37).
Yeast two-hybrid system and β-galactosidase assay.
To study the interaction of HBx mutant proteins with XAP-1/UVDDB, plasmid pASX (or derivative mutant forms) was used to transform yeast strain Y153. Resulting transformants were then cotransformed with pACT-XAP-1 and grown on plates containing 3-aminotriazole. Protein-protein interactions were confirmed by the β-galactosidase assay, and positive interactions were indicated by the appearance of blue colonies in ≤24 h, as described previously (39).
Cell culture and transfection of cells with plasmid DNA.
Human hepatoblastoma HepG2 cells (2) were grown in RPMI 1640 medium (Irvine Scientific, Santa Ana, Calif.) supplemented with 10% fetal calf serum (Life Technologies, Gaithersburg, Md.) and antibiotics. Normal fibroblast cells (CCD27Sk) and cells from XPE and XPC patients (XP2RO and GOR DO cells, respectively) were obtained from the American Type Culture Collection (Rockville, Md.) and maintained in Dulbecco’s modified Eagle’s medium (Life Technologies) supplemented with 10% fetal calf serum and antibiotics. Plasmid DNAs encoding wild-type or mutant forms of HBx proteins were introduced into HepG2 cells by Lipofectin (Life Technologies)-mediated DNA transfer according to protocols supplied by the manufacturer. Briefly, a total of 6 μg of DNA mixed with 20 μl of Lipofectin was added to each 60-mm-diameter plate and incubated at 37°C in serum-free medium for 6.5 h, after which the medium was replaced with RPMI containing 10% serum.
Immunoprecipitation and Western blot detection of HBx proteins.
Protein expression for each of the plasmid constructs was confirmed as described previously (51). Briefly, cells transfected with plasmids encoding wild-type or mutant HBx were extracted 48 h posttransfection in buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1% Nonidet P-40, 1% aprotinin) and analyzed for HBx protein by a combination immunoprecipitation and Western blot procedure with rabbit polyclonal anti-HBx serum. Other antibodies used for protein detection included rabbit antiserum to an amino- or carboxy-terminal peptide of XAP-1 (37). Negative control antisera included rabbit polyclonal anti-mouse mammary tumor virus p28 (50) and rabbit preimmune sera.
UDS assay.
The ability of cells to repair UV-induced damage was measured by a modification of previously reported techniques (18, 34, 43). Five micrograms of plasmid DNA encoding HBx or a control protein under the control of the SV40 early promoter (described above) were cotransfected with 1 μg of pGL2-control luciferase reporter plasmid (Promega, Madison, Wis.) into HepG2 cells. At 48 h posttransfection, cell growth was suppressed in medium containing 0.5% serum for 4 h, with hydroxyurea added during the last hour (final concentration, 20 mM), and the cells were treated with 0 (control) or 15 J of 254-nm UV light per m2 with a Stratalinker cross-linker (Stratagene, La Jolla, Calif.). The cells were then labeled for 2 h in the growth suppression medium supplemented with 5 μCi of [3H]thymidine/ml and lysed by freeze-thaw. A 50-μl aliquot of each sample was analyzed for luciferase activity to control the efficiency of transfection, and the remaining cellular DNA was precipitated with trichloroacetic acid. The amount of unscheduled DNA synthesis (UDS) was determined for each group of cells transfected with a given plasmid by comparing 3H counts incorporated into the repair patches of the UV-damaged cells versus the background incorporation seen in the mock-treated controls containing the same plasmid. The amount of UDS measured for cells transfected with pSV2neo (encoding neomycin resistance, Neo) was set to 100%, and the level of repair specific for each test plasmid was compared to that value.
Host cell reactivation (HCR) assay.
The cell’s ability to repair a UV-damaged reporter plasmid was measured as reported previously (5, 58) with the same pSVX and derivative plasmid DNA constructs. Luciferase reporter plasmid DNA (pGL2-control) was either damaged with 1,000 J of UV light per m2 or mock treated, and then 1 μg was cotransfected into HepG2 cells with 4 μg of the construct to be tested and 1 μg of an undamaged β-galactosidase reporter (pSVβ-gal; Promega) to control for transfection efficiency. Fifty hours posttransfection, cells were harvested in reporter lysis buffer (Promega), additionally lysed by one cycle of freeze-thaw, and assayed for luciferase and β-galactosidase activities following the manufacturer’s protocols.
Statistical analysis.
For the UDS and HCR assays, data from at least three independent experiments were averaged to determine the mean effect of the test plasmid on DNA repair and the standard deviation. The Student t test was used to compare the effect of the test plasmid to that of the pSV2neo control. Calculations were performed with the Statistical Package for the Social Sciences software (3).
RESULTS
Identification of HBx domains important for binding XAP-1/UVDDB.
The binding of full-length HBx protein to XAP-1/UVDDB (referred to hereafter as UVDDB) has been reported (37, 49). To identify the region(s) of HBx important for binding to UVDDB, a panel of X mutants was created in the yeast two-hybrid HBx bait plasmid, pASX (37). Binding with UVDDB prey protein was measured by activation of the reporter gene by the β-galactosidase assay. Comparison of β-galactosidase activities among the deletion mutants revealed that the minimal region of HBx required for binding to UVDDB in yeast was localized to aa 55 to 101 (Fig. 1B).
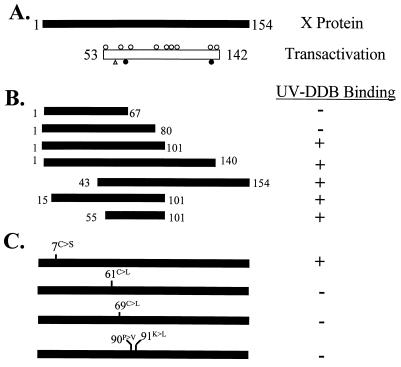
FIG. 1.
Mutant X proteins used for identifying regions of HBx required for interaction with XAP-1/UVDDB. (A) The 154-amino-acid HBx protein (black bar). The region known to be important for HBx transactivation of the SV40 early promoter in HepG2 cells (10) is shown as an open box. Open circles above the transactivation domain bar indicate point mutants (codons 58, 64, 74, 82, 107, 111, 114, 126, and 134) whose alteration has no effect on transactivation by HBx in HepG2 cells; an open triangle indicates the point mutation (residue 61) whose alteration reduces or completely abolishes transactivation, depending on the amino acid change; and filled circles below the bar indicate point mutations (codons 69 and 132) whose alteration completely abolishes HBx activity (4, 24, 33). (B) Deletion mutants of HBx. Black bars represent regions of HBx retained in the deletion mutants. Numbers represent amino acids retained in the mutant. XAP-1 binding was measured in the yeast two-hybrid system, with a positive interaction indicated by β-galactosidase activity as described in Materials and Methods. (C) HBx proteins containing point mutations. Mutations were introduced into pAS1-X by site-directed mutagenesis and mutated oligonucleotides. Mutations included a conserved Cys (position 7) changed to Ser; Cys at positions 61 and 69 each converted to Leu; and amino acids at positions 90 and 91 changed to eliminate a Pro and a charged (Lys) residue.
Point mutant HBx proteins were then created to more precisely define the domain(s) of HBx required for binding to UVDDB. Results of the β-galactosidase assay to detect binding of UVDDB to the HBx point mutants revealed that mutations at two different conserved Cys residues (HBx61 and HBx69) eliminated UVDDB binding (Fig. 1C). In addition, a double point mutation in HBx, predicted to result in an altered protein conformation by eliminating both a Pro residue and a basic charge (HBx90–91), no longer binds to UVDDB (Fig. 1C). Interestingly, the domain of HBx responsible for binding to UVDDB in yeast (aa 55 to 101) overlaps with the region of HBx previously shown to be important for HBx-mediated transactivation of the SV40 promoter-enhancer element in HepG2 cells (Fig. 1A) (10). These results also demonstrate that amino acids previously shown to be important for maximal HBx transactivation ability (aa 61 and 69) (4, 24) also appear to be important for HBx binding to UVDDB.
The UDS assay for DNA repair.
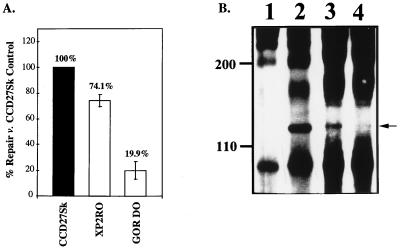
The ability of a nondividing cell to repair damaged DNA can be measured in the UDS assay (18, 34, 43). This assay has been used extensively to examine DNA repair capabilities in primary hepatocytes (18) and in repair-deficient fibroblasts from XP patients (28, 34). Normal (CCD27Sk), XPE (XP2RO), and XPC (GOR DO) fibroblasts were first analyzed in the UDS assay to establish the experimental parameters. As expected, repair of UV-damaged DNA was deficient in cells from XP patients compared to the levels of repair in normal fibroblasts (Fig. 2A). Repair in the XP2RO cells was decreased to ∼70% of repair in normal fibroblasts, and repair in the GOR DO cells was decreased to ∼20% of normal repair. These results are consistent with the levels of repair deficiency observed for these cells in previous studies (reviewed in reference 22). As UVDDB is thought to be the DNA-binding factor defective in the XP2RO cells (5, 26, 29) and because XAP-1 appears to be the human homolog of UVDDB, we hypothesized that XP2RO cells would be deficient in the UVDDB protein. Extracts of XP2RO and GOR DO cells were analyzed by combined immunoprecipitation and Western blotting with anti-XAP-1 peptide antibodies (37). The 127-kDa XAP-1/UVDDB protein was easily detected in HeLa (positive control) (37) and XPC GOR DO cells (Fig. 2B, lanes 2 and 3) but was not observed in the XP2RO cells under these assay conditions (Fig. 2B, lane 4). This result provides further support for the hypothesis that XAP-1 is the equivalent to UVDDB, the binding factor deficient in XP2RO cells (28).
FIG. 2.
The UDS assay measures repair ability. (A) DNA repair in fibroblasts. Normal (CCD27Sk), XPE (XP2RO), and XPC (GOR DO) fibroblasts were plated in 60-mm-diameter dishes and subjected to the UDS protocol as described in Materials and Methods. Repair seen in the normal cells was set to 100%, and repair for the XPC and XPE cells was compared to this value (numbers above the bars). (B) XAP-1/UVDDB is not detected in XP2RO cells. HeLa cells and XP fibroblasts were extracted and immunoprecipitated with either preimmune rabbit antiserum or antiserum produced against a carboxy-terminal peptide of XAP-1/UVDDB (37). Following fractionation by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis and Western transfer, the proteins were detected with antiserum produced against an amino-terminal peptide of XAP-1/UVDDB (37) and enhanced chemiluminescence (Amersham, Arlington Heights, Ill.). The 127-kDa protein (arrowhead at right) was detected in HeLa cells (lane 2), XPC cells (lane 3), and normal fibroblasts (data not shown) but not in XPE fibroblasts (lane 4) or HeLa cells immunoprecipitated with preimmune rabbit serum (lane 1). The original autoradiogram was photographed with the U.V.P. documentation system (SW2000; U.V.P. Inc., San Gabriel, Calif.).
Expression of HBx protein inhibits cellular repair of UV-damaged DNA.
UVDDB has been shown to be involved in the repair of UV-damaged DNA (1, 28). To determine if HBx expression alters the ability of cells to repair damaged DNA, plasmid DNAs encoding HBx or control proteins were introduced into HepG2 cells with liposomes. Because of the high transfection efficiency obtained with the liver-derived HepG2 cells, all subsequent experiments were conducted in these cells. HepG2 cells express the UVDDB protein (37). At 48 h posttransfection, cells were UV damaged and analyzed by the UDS assay as described in Materials and Methods for their ability to repair the DNA lesions.
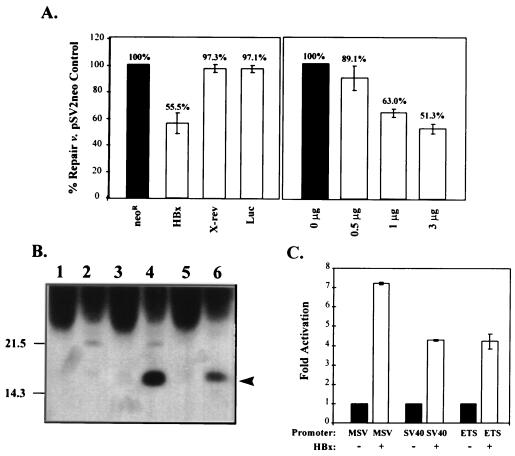
Cells transfected with a plasmid expressing HBx from the SV40 early promoter (pSV-X) revealed a significantly decreased capacity to repair UV-damaged DNA compared to cells transfected with a control plasmid (Fig. 3A, left graph). HBx-expressing cells could repair damage to only 55.5% of the repair level in pSV2neo-transfected control cells (P < 0.001). The repair inhibition observed in the presence of HBx was slightly stronger than the repair deficiency observed in fibroblasts from an XPE patient (Fig. 2A). The inhibitory effect of HBx was dose dependent (Fig. 3A, right graph) and was also observed with another subtype of HBx (adr4) expressed from the native X promoter (data not shown). At the same 48-h time point, HBx protein could be detected in the transfected HepG2 cells by a combination of immunoprecipitation and Western blotting (Fig. 3B). At least 5 × 106 pSV-X-transfected cells, pooled from duplicate 60-mm-diameter plates, were required for detection of HBx protein by this technique. This level of HBx expression is similar to that observed during natural woodchuck hepatitis virus infection, where X protein was detectable in 2 × 106 hepatocytes (14). Although an identical combination immunoprecipitation and Western immunoblotting procedure was used for both studies, the use of different anti-X antibodies prevents an absolute comparison of X protein expression levels. Importantly, the expressed protein was functional, as evidenced by its ability to transactivate several reporter constructs (Fig. 3C).
FIG. 3.
Expression of HBx protein interferes with cellular UDS. (A) Repair of damaged DNA in HBx-expressing cells. HepG2 cells were cotransfected with the pSV plasmid including the gene indicated below each bar and a luciferase reporter to control for transfection efficiency. Neor, neomycin resistance gene; HBx, X gene from HBV subtype adw2; X-rev, X in the reverse orientation; Luc, luciferase reporter gene alone. The number of micrograms of pSV-X transfected is shown below each bar in the righthand graph. Control pSV2neo plasmid was used to standardize the total amount of DNA introduced into the cells to 6 μg per plate. Repair for cells transfected with pSV2neo was set to 100%, and the repair measured for other transfected cells was compared to that value (numbers above each bar). Values shown are the means of at least three independent experiments; the error bars represent the corresponding standard deviations. (B) HBx expression. Duplicate 60-mm-diameter plates of cells transfected with pSV2neo (lanes 1 and 2) or pSV-X (lanes 3 and 4) were harvested 48 h posttransfection, pooled, and immunoprecipitated with anti-p28 negative control (odd-numbered lanes) or anti-X (even-numbered lanes). Liver extracts (25%, wt/vol) from ATX transgenic mice (38) were diluted 150-fold and then similarly immunoprecipitated and used as controls (lanes 5 and 6). Recovered proteins were transferred to nitrocellulose membrane for detection with anti-X by Western blotting. Migration of the 17-kDa HBx protein is shown at the right (arrowhead). (C) Transactivation by HBx. Cells were cotransfected with 5 μg of pSV-X and 1 μg of luciferase reporter under the control of one of several regulatory elements, noted below each bar: MSV, murine sarcoma virus; SV40, SV40 early promoter; ETS, regulatory element for the ets oncogene. The activation of each luciferase construct in the absence of HBx was normalized to 1.0 (black bars), and the fold activation of that basal level in the presence of HBx was calculated. Error bars represent the standard errors of the means from triplicate samples.
In contrast, no repair inhibition was observed for cells transfected with the X gene cloned in the reverse orientation (Fig. 3A, X-rev) or with the luciferase reporter alone (Fig. 3A, Luc). Together, these data demonstrate that expression of HBx protein inhibits the ability of HepG2 cells to repair UV-damaged DNA.
A second experimental approach, the HCR assay (5, 58), was used to confirm and extend the UDS results. This assay avoids the possible confounding effect(s) of UV irradiation on the cell by measuring the repair of a reporter plasmid that is damaged prior to transfection into the cell. The ability of the cells to repair the damaged reporter plasmid was determined as described in Materials and Methods. Compared with cells transfected with control plasmids, cells expressing HBx protein demonstrated decreased repair of the damaged reporter plasmid (Fig. 4). HBx-expressing cells could only repair the damaged reporter plasmid to 57.1% of the level measured in cells transfected with the pSV2neo control (P < 0.001). The decrease in DNA repair measured in the HCR assay was specific for cells expressing HBx; no inhibition was observed in cells transfected with control plasmids pSVX-rev or pSVβ-gal (Fig. 4). The results from the HCR assay confirm those of the UDS assay: expression of HBx protein interferes with the ability of the cell to repair UV-damaged DNA.
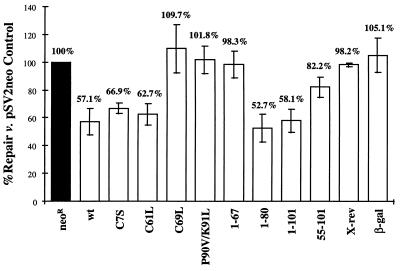
FIG. 4.
Interference with repair by HBx and mutant derivatives. HepG2 cells were assayed by the HCR assay described in Materials and Methods with the test plasmid construct listed under each bar: Neor, neomycin-resistance; wt, wild-type HBx subtype adw2; amino acid numbers, residues mutagenized (point mutants with the changed codon indicated by the standard single-letter abbreviation) or retained (deletion constructs); X-rev, X gene in the reverse orientation; β-gal, β-galactosidase reporter. Repair for the pSV2neo control (black bar) was set to 100%, and repair for cells transfected with the other constructs was compared to that value (numbers above each bar). Values shown are the means of at least three independent experiments, and the error bars represent the standard deviations.
Identification of HBx domains required for inhibition of DNA repair.
Given the proposed role of UVDDB protein in the repair of UV-damaged DNA, we hypothesized that the inhibition of DNA repair by HBx may be mediated by its ability to bind UVDDB. To test this hypothesis, several mutant X genes tested for binding to UVDDB in yeast (Fig. 1) were subcloned into a mammalian expression vector and tested for the ability to inhibit DNA repair in HepG2 cells. Four HBx deletion mutants were examined. HBx1–67 did not interfere with HCR repair of the damaged luciferase reporter, while HBx1–80 and HBx1–101 both reduced repair of the luciferase reporter to levels similar to the inhibition observed for full-length HBx (Fig. 4). The minimal UVDDB binding domain identified in the yeast studies, aa 55 to 101 (Fig. 1), also interfered with repair of damaged reporter to a level intermediate between those of full-length HBx and negative control proteins (Fig. 4).
Selected HBx proteins containing point mutations were next used to further map the domain(s) required for inhibition of DNA repair. Two different point mutant HBx proteins (C7S and C61L) were able to reduce DNA repair to levels similar to that observed for full-length HBx (Fig. 4). Mutant HBx with an amino acid substitution at codon 69 (C69L) or a double point mutant at residues 90 and 91 (P90V-K91L), however, did not interfere with the ability of cells to repair damaged DNA (Fig. 4). Expression of the deletion and point mutant HBx proteins used in these studies was confirmed by a combination of immunoprecipitation and Western blot analysis (data not shown).
Correlation of HBx-UVDDB binding with inhibition of DNA repair.
Results from the UDS and HCR assays demonstrate that full-length HBx, as well as certain point and deletion mutant derivatives, was able to inhibit the ability of HepG2 cells to repair UV-damaged DNA. Comparison of mutant HBx binding to UVDDB in yeast with the ability of those mutants to inhibit DNA repair in HepG2 cells revealed that all three HBx mutants that retain the ability to bind UVDDB in yeast are also able to inhibit DNA repair in the mammalian cells (Table 1; HBx7, HBx1–101, and HBx55–101). Of the five HBx mutants no longer able to bind UVDDB in yeast, three have also lost the ability to inhibit NER (Table 1; HBx69, HBx90–91, and HBx1–67) while two are still able to interfere with DNA repair in HepG2 cells (Table 1; HBx61 and HBx1–80). Thus, these results suggest a close, but not complete, correlation between HBx binding UVDDB in yeast and HBx interference with mammalian NER.
TABLE 1.
Correlation of HBx binding to UVDDB and HBx-induced inhibition of DNA repair
| HBV X proteina | Binding to UVDDBb | Inhibition of DNA repairc |
|---|---|---|
| HBx1–154 | + | + |
| Point mutations | ||
| HBx7 | + | + |
| HBx61 | − | + |
| HBx69 | − | − |
| HBx90–91 | − | − |
| Deletion mutations | ||
| HBx1–67 | − | − |
| HBx1–80 | − | + |
| HBx1–101 | + | + |
| HBx55–101 | + | + |
HBx proteins tested in the assays. Full-length HBx (aa 1 to 154) and HBx proteins containing point and deletion mutations were created as described in Materials and Methods.
Binding of HBx to UVDDB measured by yeast two-hybrid assay as described previously (37). +, binding; −, no binding.
Inhibition of DNA repair was measured in the host cell reactivation assay (see Materials and Methods). +, inhibition; −, no inhibition.
DISCUSSION
HBx has been shown to bind to several different cellular proteins (12, 19, 20, 24, 36, 40, 44, 56, 57); however, the biological significance of these interactions to the virus life cycle is not clear. We previously described the interaction of HBx with XAP-1/UVDDB (37), a cellular protein believed to be involved in the NER pathway for repairing bulky lesions in DNA (26–28). Recently, UVDDB was also shown to interact with the X proteins from other mammalian hepadnaviruses (49). Although the importance of HBx binding to UVDDB for the virus life cycle remains unknown, we considered the possibility that the HBx-UVDDB interaction may incidentally alter the normal function of the cellular protein. The results of this study demonstrate that HBx expression is associated with an approximately 45% decrease in DNA repair capacity, as measured by two independent DNA repair assays. In comparison, fibroblasts from a cancer-prone XPE patient demonstrate a similar decrease in the ability to repair UV-damaged DNA, suggesting that the repair inhibition mediated by HBx may have biological relevance.
The mechanism by which HBx inhibits DNA repair is not known. Recently, HBx has been shown to preferentially bind UV-damaged DNA in the presence of a nuclear extract (9). Because HBx interacts with UVDDB, a protein involved in NER, we predicted that HBx might interfere with DNA repair through a UVDDB pathway. Toward this end, a panel of HBx mutants was used to map the inhibitory function of HBx. Three HBx mutants that retain the ability to bind UVDDB are able to inhibit DNA repair, while three HBx mutants that no longer bind UVDDB have lost the ability to interfere with DNA repair. However, two additional HBx mutant proteins gave discordant results in that they no longer bind UVDDB but continue to inhibit DNA repair. These latter results may indicate that UVDDB binding is not required for HBx-induced inhibition of DNA repair. Alternatively, we must consider that the binding studies were performed in yeast and utilized a GAL4 fusion protein, while the repair assays were performed in mammalian HepG2 cells with HBx protein (or mutant derivatives). As there may be different requirements for binding in the two systems, the ability of the discordant HBx mutants to interact with UVDDB in mammalian cells is currently under investigation. Taken together, the mapping data are suggestive, but do not prove, that inhibition of DNA repair by HBx occurs via a UVDDB pathway.
There are additional molecular pathways through which HBx may be influencing DNA repair. HBx has been reported to interact with the p53 tumor suppressor protein (19, 56, 57), and several studies suggest a role for p53 in the repair of UV-damaged DNA (reviewed in reference 46). Therefore, it is possible that HBx may affect NER through its effect on p53. Secondly, the ability of HBx to inhibit DNA repair may be related to the broadly acting transactivation function of HBx. Many cellular promoters have been reported to be up-regulated by HBx (10), and perhaps one (or more) of these proteins may subsequently alter NER. It is unclear whether any of the other cellular proteins reported to interact with HBx are involved in the inhibition of NER, as a role for those cell factors in DNA repair has not been established.
The region of HBx required for interaction with UVDDB, aa 55 to 101, is compatible with a recent report in which the minimal UVDDB-binding domain of HBx was further narrowed to aa 66 to 101 (49). The point mutation at HBx codon 61 abolished binding to UVDDB even though aa 61 lies outside the minimal domain defined by Sitterlin et al. (49). We hypothesize that the elimination of this highly conserved Cys at codon 61 may have altered the conformation of HBx, thereby preventing an interaction with UVDDB. The UVDDB-binding site of HBx is highly conserved among HBV subtypes (31) and is within the region identified as the minimal domain required for transactivation of the SV40 promoter in HepG2 cells (10).
There is evidence that, in addition to binding damaged DNA, UVDDB may have a role in transcription, as has been described for other NER components (reviewed in reference 21). A recent analysis of BRF-2, a binding factor from rat livers that transactivates the apolipoprotein B promoter (59), has revealed high protein homology and antigen cross-reactivity with XAP-1/UVDDB (35). These results would suggest that HBx-mediated transactivation may proceed through a UVDDB pathway and that mutants of HBx which no longer bind UVDDB should no longer possess transactivation function. Using a panel of HBx mutants containing a series of two amino acid insertions, Sitterlin et al. found a correlation between the ability of HBx to bind UVDDB and the ability of HBx to transactivate an AP1-containing promoter-enhancer-driven reporter construct (49).
Our mapping results obtained with HBx point mutants are also consistent with the hypothesis that UVDDB binding may be important for HBx transactivation. In a previous study, alteration of codon 69 abolished HBx transactivation (4). In the present study, a mutation at this position also abrogated binding to UVDDB. A possible association between UVDDB binding and transactivation is less apparent with the HBx construct containing a point mutation at codon 61. Alteration of residue 61 from Cys to Leu abolished binding to UVDDB; however, the role of codon 61 in transactivation is unclear. Mutation of this residue to two different amino acids (both different than the change to Leu used in the present study) has been shown to reduce transactivation to about half the activity seen with wild-type HBx (4) or to completely disrupt transactivation (24). Future transactivational analysis of this panel of HBx mutants will clarify these issues.
Several observations suggest that the HBx-induced interference with DNA repair is biologically significant. Our preliminary studies demonstrate that expression of HBx also inhibits DNA repair in the HepG2-derivative 2.2.15 cell line, which expresses the other HBV proteins and produces virions (data not shown; see reference 47 for a description of the 2.2.15 cells). This result suggests that HBx would retain the capacity to inhibit DNA repair in infected hepatocytes during natural infection. Importantly, the level of repair inhibition associated with HBx expression in HepG2 and 2.2.15 cells was similar to that observed in fibroblasts from cancer-prone XP patients, suggesting that such a level of repair interference by HBx may be associated with increased risk of HCC. Although UV damage was the experimental tool in this study, the NER pathway repairs many bulky lesions, including those introduced by carcinogens (42, 46). Given the known role of carcinogens in the etiology of subsets of HBV-positive HCCs, we predict that HBx-induced inhibition of DNA repair may contribute to the accumulation of DNA errors, eventually leading to HCC (8).
In summary, we have shown that expression of the HBx protein interferes with the ability of HepG2 cells to repair damaged DNA. The 50 to 60% reduction is similar to that observed in XPE cells from patients who have a greatly increased risk of cancer, suggesting that this level of interference is biologically significant. Although the repair inhibition mediated by HBx may be due to its interaction with the cellular repair protein UVDDB, further studies of HBx-UVDDB binding in mammalian cells must be performed to confirm this observation. Recent evidence that UVDDB may be involved in transcription suggests a novel mechanism by which a tumor-associated virus could usurp a cellular pathway for the benefit of the virus and incidentally contribute to cancer formation.
ACKNOWLEDGMENTS
This work was supported in part by grant CA 54557 (B.L.S.) and Research Training Grant CA 09197 (S.A.B.) from the National Cancer Institute.
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Aden D P, Fogel A, Plotkin S, Damjanov I, Knowles B B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature (London) 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. Statistical Package for the Social Sciences (SPSS) for Windows, version 7. Chicago, Ill: SPSS, Inc.; 1995. [Google Scholar]
- 4.Arii M, Takada S, Koike K. Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene. 1992;7:397–403. [PubMed] [Google Scholar]
- 5.Athas W F, Hedayati M A, Matanoski G M, Farmer E R, Grossman L. Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res. 1991;51:5786–5793. [PubMed] [Google Scholar]
- 6.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billet O, Grimber G, Levrero M, Seye K A, Briand P, Joulin V. In vivo activity of the hepatitis B virus core promoter: tissue specificity and temporal regulation. J Virol. 1995;69:5912–5916. doi: 10.1128/jvi.69.9.5912-5916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butel J S, Lee T-H, Slagle B L. Is the DNA repair system involved in hepatitis-B-virus-mediated hepatocellular carcinogenesis? Trends Microbiol. 1996;4:119–124. doi: 10.1016/0966-842X(96)81529-0. [DOI] [PubMed] [Google Scholar]
- 9.Capovilla A, Carmona S, Arbuthnot P. Hepatitis B virus X-protein binds damaged DNA and sensitizes liver cells to ultraviolet irradiation. Biochem Biophys Res Commun. 1997;232:255–260. doi: 10.1006/bbrc.1997.6269. [DOI] [PubMed] [Google Scholar]
- 10.Caselmann W H. Trans-activation of cellular genes by hepatitis B virus proteins: a possible mechanism of hepatocarcinogenesis. Adv Virus Res. 1996;47:253–302. doi: 10.1016/s0065-3527(08)60737-x. [DOI] [PubMed] [Google Scholar]
- 11.Chen H-S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheong J-H, Yi M-K, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross J C, Wen P, Rutter W J. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen-activated cellular serine/threonine kinases. Proc Natl Acad Sci USA. 1993;90:8078–8082. doi: 10.1073/pnas.90.17.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandri M, Schirmacher P, Rogler C E. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De-Medina T, Haviv I, Noiman S, Shaul Y. The X protein of hepatitis B virus has a ribo-deoxy ATPase activity. Virology. 1994;202:401–407. doi: 10.1006/viro.1994.1356. [DOI] [PubMed] [Google Scholar]
- 16.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragani T A, Manenti G, Farza H, Della Porta G, Tiollais P, Pourcel C. Transgenic mice containing hepatitis B virus sequences are more susceptible to carcinogen-induced hepatocarcinogenesis. Carcinogenesis. 1989;11:953–956. doi: 10.1093/carcin/11.6.953. [DOI] [PubMed] [Google Scholar]
- 18.Fautz R, Husein B, Efstathiou E, Hechenberger-Freudl C. Assessment of the relation between the initial viability and the attachment of freshly isolated rat hepatocytes used for the in vivo/in vitro DNA repair assay (UDS) Mutat Res. 1993;291:21–27. doi: 10.1016/0165-1161(93)90013-p. [DOI] [PubMed] [Google Scholar]
- 19.Feitelson M A, Zhu M, Duan L X, London W T. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 20.Fischer M, Runkel L, Schaller H. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes. 1995;10:99–102. doi: 10.1007/BF01724303. [DOI] [PubMed] [Google Scholar]
- 21.Friedberg E C. Relationships between DNA repair and transcription. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- 22.Hoeijmakers J H J. Nucleotide excision repair. II. From yeast to mammals. Trends Genet. 1993;9:211–217. doi: 10.1016/0168-9525(93)90121-w. [DOI] [PubMed] [Google Scholar]
- 23.Hohne M, Schaefer S, Seifer M, Feitelson M A, Paul D, Gerlich W H. Malignant transformation of immortalized transgenic hepatocytes after transfection with hepatitis B virus DNA. EMBO J. 1990;9:1137–1145. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Kwong J, Sun E C-Y, Liang T J. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang B J, Chu G. Purification and characterization of a human protein that binds to damaged DNA. Biochemistry. 1993;32:1657–1666. doi: 10.1021/bi00057a033. [DOI] [PubMed] [Google Scholar]
- 26.Kataoka H, Fujiwara Y. UV damage-specific DNA-binding protein in xeroderma pigmentosum complementation group E. Biochem Biophys Res Commun. 1991;175:1139–1143. doi: 10.1016/0006-291x(91)91684-5. [DOI] [PubMed] [Google Scholar]
- 27.Keeney S, Chang G J, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 28.Keeney S, Eker A P M, Brody T, Vermeulen W, Bootsma D, Hoeijmakers J H J, Linn S. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc Natl Acad Sci USA. 1994;91:4053–4056. doi: 10.1073/pnas.91.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keeney S, Wein H, Linn S. Biochemical heterogeneity in xeroderma pigmentosum complementation group E. Mutat Res DNA Repair. 1992;273:49–56. doi: 10.1016/0921-8777(92)90049-9. [DOI] [PubMed] [Google Scholar]
- 30.Kekulé A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Hepatitis B virus transactivator HBx uses a tumour promoter signaling pathway. Nature (London) 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 31.Kidd-Ljunggren K, Oberg M, Kidd A H. The hepatitis B virus X gene: analysis of functional domain variation and gene phylogeny using multiple sequences. J Gen Virol. 1995;76:2119–2130. doi: 10.1099/0022-1317-76-9-2119. [DOI] [PubMed] [Google Scholar]
- 32.Kim C-M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature (London) 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 33.Kim Y H, Kang S K, Lee Y I. Functional analysis of hepatitis B virus transactivator X: implication of the leucine zipper-like region and C-terminal 7 conserved amino acids in functional regions. Biochem Biophys Res Commun. 1993;197:894–903. doi: 10.1006/bbrc.1993.2563. [DOI] [PubMed] [Google Scholar]
- 34.Kleijer W J, De Weerd-Kastelein A, Sluyter M L, Keijzer W, De Wit J, Bootsma D. UV-induced DNA repair synthesis in cells of patients with different forms of xeroderma pigmentosum and of heterozygotes. Mutat Res. 1973;20:417–428. doi: 10.1016/0027-5107(73)90062-6. [DOI] [PubMed] [Google Scholar]
- 35.Krishnamoorthy R R, Lee T-H, Butel J S, Das H K. Apolipoprotein B gene regulatory factor-2 (BRF-2) is structurally and immunologically highly related to hepatitis B virus X associated protein-1 (XAP-1) Biochemistry. 1997;36:960–969. doi: 10.1021/bi961407c. [DOI] [PubMed] [Google Scholar]
- 36.Kuzhandaivelu N, Cong Y-S, Inouye C, Yang W-M, Seto E. XAP2, a novel hepatitis B virus X-associated protein that inhibits X transactivation. Nucleic Acids Res. 1996;24:4741–4750. doi: 10.1093/nar/24.23.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee T-H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee T-H, Finegold M F, Shen R-F, DeMayo J L, Woo S L C, Butel J S. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J Virol. 1990;64:5939–5947. doi: 10.1128/jvi.64.12.5939-5947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacGregor G R, Nolan G P, Fiering S, Roederer M, Herzenberg L A. Use of E. coli lacZ (B-galactosidase) as a reporter gene. In: Murray J, editor. Methods in molecular biology. Clifton, N.J: Humana Press, Inc.; 1991. p. 217E. [DOI] [PubMed] [Google Scholar]
- 40.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 41.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Ras- and Raf-dependent activation of c-Jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 42.Payne A, Chu G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat Res. 1994;310:89–102. doi: 10.1016/0027-5107(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 43.Pietras R J, Fendly B M, Chazin V R, Pegram M D, Howell S B, Slamon D J. Antibody to HER-2/neu receptor blocks DNA repair after cisplatin in human breast and ovarian cancer cells. Oncogene. 1994;9:1829–1838. [PubMed] [Google Scholar]
- 44.Qadri I, Maguire H F, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J-S, Linn S, Sancar A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 46.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 47.Sells M A, Chen M-L, Acs G. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirakata Y, Kawada M, Fujiki Y, Sano H, Oda M, Kobayashi M, Koike K. The X gene of hepatitis B virus induced growth stimulation and tumorigenic transformation of mouse NIH3T3 cells. Jpn J Cancer Res. 1989;80:617–621. doi: 10.1111/j.1349-7006.1989.tb01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sitterlin D, Lee T-H, Prigent S, Tiollais P, Butel J S, Transy C. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J Virol. 1997;71:6194–6199. doi: 10.1128/jvi.71.8.6194-6199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slagle B L, Lanford R E, Medina D, Butel J S. Expression of mammary tumor virus proteins in preneoplastic outgrowth lines and mammary tumors of BALB/cV mice. Cancer Res. 1984;44:2155–2162. [PubMed] [Google Scholar]
- 51.Slagle B L, Lee T-H, Medina D, Finegold M J, Butel J S. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 52.Southern P J, Berg J. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 53.Takada S, Kido H, Fukutomi A, Mori T, Koike K. Interaction of hepatitis B virus X protein with a serine protease, tryptase TL2, as an inhibitor. Oncogene. 1994;9:341–348. [PubMed] [Google Scholar]
- 54.Takao M, Abramic M, Moss M, Jr, Otrin V R, Wootton J C, McLenigan M, Levine A S, Protic M. A 127 kDa component of a UV-damaged DNA-binding complex, which is defective in some xeroderma pigmentosum group E patients, is homologous to a slime mold protein. Nucleic Acids Res. 1993;21:4111–4118. doi: 10.1093/nar/21.17.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terradillos O, Billet O, Renard C-A, Levy R, Molina T, Briand P, Buendia M A. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 56.Truant R, Antunovic J, Greenblatt J, Prives C, Cromlish J A. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J Virol. 1995;69:1851–1859. doi: 10.1128/jvi.69.3.1851-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei Q, Matanoski G M, Farmer E R, Hedayati M A, Grossman L. DNA repair related to multiple skin cancers and drug use. Cancer Res. 1994;54:437–440. [PubMed] [Google Scholar]
- 59.Zhuang H, Chuang S S, Das H K. Transcriptional regulation of the apolipoprotein B100 gene: purification and characterization of trans-acting factor BRF-2. Mol Cell Biol. 1992;12:3183–3191. doi: 10.1128/mcb.12.7.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]