Abstract
A GroEL homolog with a molecular mass of 60 kDa, produced by the primary endosymbiotic bacterium (a Buchnera sp.) of Myzus persicae and released into the hemolymph, has previously been shown to be a key protein in the transmission of potato leafroll virus (PLRV). Like other luteoviruses and pea enation mosaic virus, PLRV readily binds to extracellular Buchnera GroEL, and in vivo interference in this interaction coincides with reduced capsid integrity and loss of infectivity. To gain more knowledge of the nature of the association between PLRV and Buchnera GroEL, the groE operon of the primary endosymbiont of M. persicae (MpB groE) and its flanking sequences were characterized and the PLRV-binding domain of Buchnera GroEL was identified by deletion mutant analysis. MpB GroEL has extensive sequence similarity (92%) with Escherichia coli GroEL and other members of the chaperonin-60 family. The genomic organization of the Buchnera groE operon is similar to that of the groE operon of E. coli except that a constitutive promoter sequence could not be identified; only the heat shock promoter was present. By a virus overlay assay of protein blots, it was shown that purified PLRV bound as efficiently to recombinant MpB GroEL (expressed in E. coli) as it did to wild-type MpB GroEL. Mutational analysis of the gene encoding MpB GroEL revealed that the PLRV-binding site was located in the so-called equatorial domain and not in the apical domain which is generally involved in polypeptide binding and folding. Buchnera GroEL mutants lacking the entire equatorial domain or parts of it lost the ability to bind PLRV. The equatorial domain is made up of two regions at the N and C termini that are not contiguous in the amino acid sequence but are in spatial proximity after folding of the GroEL polypeptide. Both the N- and C-terminal regions of the equatorial domain were implicated in virus binding.
Potato leafroll virus (PLRV; genus Luteovirus), a positive-stranded RNA virus, mainly replicates in the phloem tissue of a plant and is transmitted by aphids in a persistent and circulative manner (28, 41, 46). When they feed on the phloem sap, aphids ingest virus particles, which are subsequently transported from the digestive tube into the hemolymph (24) and from there across the basal lamina that surrounds the accessory salivary cells into the salivary gland (25). Virus particles that reach the salivary gland are eventually released in the phloem sap of the plant as the aphid feeds (25). The hemolymph acts as a reservoir in which PLRV is retained in an infective form during the aphid’s lifespan without replication (19).
It has previously been demonstrated that the primary endosymbiotic bacterium (a Buchnera sp.) of Myzus persicae, the principal vector of PLRV, plays a crucial role in determining the persistent nature of PLRV in the aphid hemolymph (50). Buchnera spp. abundantly produce a protein which is highly homologous to the Escherichia coli chaperonin GroEL (5, 23, 35, 50). GroEL of the Buchnera sp. of M. persicae (MpB GroEL) was found to be released in the hemolymph, most likely as a result of the lysis of endosymbiotic bacteria (50). After antibiotic treatment of the aphid, MpB GroEL could no longer be detected in the hemolymph and PLRV transmission was greatly reduced due to degradation of virus capsid proteins (50). Since in vitro studies have previously shown that PLRV exhibits specific affinity for MpB GroEL, it was suggested that virus particles associate with MpB GroEL in the hemolymph of the aphid to retard proteolytic breakdown of virus particles (50).
Buchnera spp. are common to all major aphid groups but the Phylloxeridae (9). These intracellular bacteria are gram-negative and closely related to members of the Enterobacteriaceae family (36, 48). Buchnera spp. are harbored in specialized cells, mycetocytes, localized in the abdomen of the aphid (50) and are maternally inherited (9). Comparisons of rRNA sequences of Buchnera spp. and morphological features of aphid hosts provide strong evidence that a single aphid ancestor was infected by the bacterium about 250 million years ago (37).
GroEL of E. coli is a heat shock protein (Hsp60) with 60-kDa subunits; it is involved in intracellular folding and assembly of nonnative proteins in an ATP-dependent manner (18). Hsp60s are common to prokaryotes, mitochondria, and chloroplasts (18, 26). Crystallography of E. coli GroEL demonstrated that the protein forms a homo-oligomer of 14 subunits, which are arranged in two heptameric rings stacked back to back, and that each subunit consists of the following three domains: the equatorial domain, the apical domain, and the small intermediate domain (7). In general, the apical domain of GroEL has previously been implicated in polypeptide binding (22), a process which may require ATP hydrolysis. The ATPase activity of GroEL is regulated by GroES (34, 53), a single heptameric ring of 10-kDa subunits also encoded by the groE operon (14, 47). The structural and functional characteristics of Buchnera GroELs are highly similar to those of E. coli GroEL (23, 27, 38). However, unlike E. coli GroEL, Buchnera GroEL is not restricted to the cytosol of the bacterium; it also occurs extracellularly in the hemolymph of an aphid (23, 50, 51).
In this study, the nucleotide sequence of the gene encoding MpB GroEL was determined and structural and functional domains were identified by sequence comparison to the other GroELs. In addition, the regions upstream and downstream of this gene were sequenced and compared with the corresponding regions of E. coli. To gain a better understanding of the molecular basis of the association between PLRV and MpB GroEL, the protein was expressed in E. coli and mutational analysis was carried out to identify the domain of MpB GroEL implicated in PLRV binding.
MATERIALS AND METHODS
Isolation of genomic DNA from the Buchnera sp. of M. persicae.
Approximately 1 g of M. persicae aphids was collected and surface sterilized with 70% ethanol containing 0.5% Tween 20 and 0.5% hypochlorite. Sterilized aphids were rinsed with water and homogenized in 3 ml of isolation medium (8). Subsequently, the homogenate was filtered through cheesecloth and centrifuged at 5,000 × g for 15 min. Either bacterial genomic DNA was isolated directly from the resulting pellet (lysis buffer method) or further purification steps were undertaken to enrich for bacterial cells (Ficoll procedure). In the lysis buffer method, the pellet was incubated for 1 h at 56°C in 0.7 ml of lysis buffer (150 mM Tris-HCl [pH 8.0] containing 150 mM EDTA, 3% sodium dodecyl sulfate [SDS], and 1.5 to 2% sodium lauroyl sarcosine). After 5 min of incubation on ice, 0.5 ml of Tris-EDTA buffer was added, the suspension was gently mixed, and the debris was allowed to precipitate. Genomic DNA was extracted with phenol-chloroform from the supernatant. In the Ficoll method, the pellet was resuspended in 2 ml of 100-fold-diluted isolation medium and layered on a 2 to 10% Ficoll gradient in 0.01 M phosphate buffer (pH 7.2). After centrifugation at 400 × g for 10 min, the fraction containing bacterial cells was collected. To this fraction, 5 volumes of saline-EDTA (0.15 M sodium chloride, 0.1 M EDTA [pH 8.0]) was added and the mixture was centrifuged at 1,000 × g for 12 min. The pellet was resuspended in 1 ml of saline-EDTA containing 8% SDS and incubated at 60°C for 10 min, and DNA was extracted as mentioned above.
PCR amplification procedure.
PCR amplification was performed in a final volume of 100 μl of 10 mM Tris-HCl (pH 8.3) containing 0.4 mM (total) deoxynucleoside triphosphates, 3 mM MgCl2, 50 mM KCl, 1 μg of DNA, 0.25 μM (each) primers, and 2.5 U of Taq polymerase (Boehringer Mannheim). Mixtures were incubated for 2 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, with a final incubation of 10 min at 72°C. Samples were stored at 4°C until used. PCR products were analyzed on agarose gels.
Sequencing strategy.
Clones containing the MpB groEL sequence were generated by PCR with primers F1 and R1 (Table 1). The primer sequences were based on the N-terminal amino acid sequence of MpB GroEL (50) and the 3′-terminal nucleotide sequence of the Buchnera groEL gene of Acyrthosiphon pisum (38). The resulting 1,732-bp PCR product was cloned by using a TA cloning kit (Invitrogen), yielding plasmid pCR[Buchnera GroEL]. Overlapping restriction fragments were subcloned into pBluescript KS (Stratagene), and their nucleotide sequences were determined at the sequence facilities of the Department of Molecular Biology, Wageningen Agricultural University, with a sequencing kit and AmpliTaq DNA polymerase (Applied Biosystems), universal and sequence-specific primers, and an automated sequencer (model 373; Applied Biosystems).
TABLE 1.
Oligonucleotides used for the construction of MpB GroEL deletion mutants
| Oligonucleotide | Sequence (5′–3′)a | Corresponding positionsb |
|---|---|---|
| F1 | ccggatccATGGCCGCTTAAAGATGTA | 1–6 |
| F2 | ggatccatgAAAGCTGTTATTAGTGCG | 122–127 |
| F3 | ccatggatcCGTTAAAGGTATGCAG | 189–194 |
| F4 | ggatccatgGTTGCAGTACTTAAAGTAG | 376–385 |
| F5 | ggatccatgGAAGGTGTAGTTGCTGG | 409–413 |
| F6 | ggtgaagcttAACTATGGTTATAATGCAGC | 475–480 |
| R1 | acggatccTTACATCATTCCaCCC | 545–548 |
| R2 | gaattcttaACCTTTTCCATCTTTTTACG | 470–474 |
| R3 | caataagcttTTCAACAGCTGCACCAGT | 404–408 |
| R4 | gaattcttaTTCAACAGCTGCACGAG | 404–408 |
| R5 | gaattcagatcaCCTCCTGATAATTTAGC | 370–374 |
| R6 | ggatccttaAACGACTTCTAGTTCATTTTG | 184–190 |
Uppercase letters indicate Buchnera groEL sequences, and lowercase letters indicate sequences which are not part of the gene. Restriction sites (EcoRI, BamHI, and HindIII) are in boldface. Start codons are double underlined, and termination codons are single underlined.
Numbering refers to the corresponding positions of the amino acid residues of MpB GroEL.
To determine the sequence of the entire MpB groE operon, a genomic DNA library was constructed by using a λ ZAP II cloning kit and Gigapack III Gold packaging extract (Stratagene) according to the manufacturer’s instructions. Genomic DNA from the Buchnera sp. of M. persicae was isolated by the lysis buffer method and digested with XbaI. Fragments were ligated into XbaI-digested λ ZAP II vector arms. Two radiolabeled probes of 569 and 521 bp, corresponding to the 5′ and 3′ ends of the open reading frame (ORF) coding for MpB GroEL, respectively, were used to screen for recombinant clones. After the excision of positive plaques, the nucleotide sequences of phagemids pSK2500 and pSK3500 (see Fig. 2) were determined.
FIG. 2.
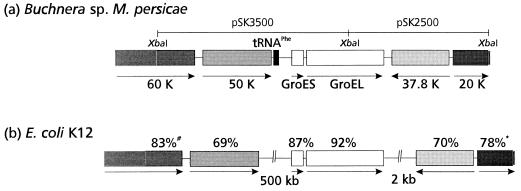
Schematic representation of the MpB groE operon and comparison of the chromosomal arrangements of the regions flanking the groE operons of the Buchnera sp. of M. persicae (a) and E. coli (b) (11, 12). Identical shading indicates high amino acid sequence similarity of gene products. The percentage noted above a box indicates the similarity of the MpB ORF product to the E. coli homolog. #, percentage of similarity of the C-terminal 295 amino acids of the 60-kDa (60 K) gene; ∗, percentage of similarity of the N-terminal 186 amino acids of the 20-kDa (20 K) protein. The arrow below each gene indicates the direction of translation.
Southern blot analysis.
Genomic DNA from the Buchnera sp. of M. persicae was isolated by the Ficoll method (see above), and 5 μg of DNA was digested with either PstI, XbaI, or XhoI. Samples were run on a 1% agarose gel and transferred to HybondN (Amersham). The 1,732-bp PCR product containing the MpB groEL gene mentioned above was radiolabeled and used as the probe for hybridization.
GroEL isolation from the Buchnera sp. of M. persicae and from E. coli.
GroEL was isolated from the endosymbiotic bacteria of 6-day-old M. persicae nymphs and from heat-shocked E. coli cells as described before (51).
Cloning and expression of Buchnera GroEL deletion mutants.
Full-length MpB GroEL and deletion mutants of MpB GroEL in fusion with glutathione S-transferase (GST) were expressed in E. coli with plasmid pGEX-2T (Pharmacia). GST fusion proteins were affinity purified with glutathione-Sepharose (Pharmacia) according to the manufacturer’s recommendations. To remove the GST moiety, fusion proteins were incubated with thrombin for 3 h at 10°C. Cleaved products were analyzed on SDS-polyacrylamide gel electrophoresis (PAGE) gels and by Western blot analysis with anti-MpB GroEL immunoglobulin G (IgG). To ensure that similar quantities of deletion mutants were tested for their virus-binding capacities (described below), they were diluted to yield bands of similar intensities, as assessed by amido black staining after electroblotting. Each mutant was named after the positions of the first and last amino acids bordering the included fragment.
Full-length MpB GroEL was obtained by digesting pCR[Buchnera GroEL] with BamHI and cloning the BamHI fragment containing the MpB groEL gene into the BamHI sites of pGEX-2T, resulting in pGEX[Buchnera GroEL]. Constructs for the expression of MpB GroEL(1-121) and MpB GroEL(1-314) were derived by digesting plasmid pGEX[Buchnera GroEL] with SmaI (located downstream of the BamHI site in the multiple cloning site of pGEX-2T) and ClaI or XbaI. Protruding 5′ ends were filled in with the Klenow fragment of DNA polymerase I and by religation of constructs. pGEX-2T constructs for the expression of all other truncated mutants of GroEL were generated by PCR. The primers used were complementary or identical to the border sequences of the three domains recognized in MpB GroEL and included additional restriction sites (BamHI, EcoRI, or HindIII sites) for cloning purposes (Table 1). Plasmid pCR[Buchnera GroEL] served as the template. All PCR products were first cloned into the pCRII vector (TA cloning kit; Invitrogen), digested with BamHI or BamHI/EcoRI, and subsequently religated into the BamHI or BamHI/EcoRI sites of pGEX-2T. For the expression of MpB GroEL(122-408/475-548), a pGEX-2T construct was synthesized with primer pair F2 and R3 and primer pair F6 and R1 (Table 1). The amplified fragments of 850 (F2 and R3) and 225 (F6 and R1) bp were cloned into pCRII and digested with BamHI/HindIII and HindIII/EcoRI, respectively. The HindIII-cleaved ends of both fragments were ligated, and the ligated product was cloned into the BamHI/EcoRI sites of pGEX-2T. All constructs were verified by nucleotide sequence analysis.
The pGEX constructs mentioned above were introduced into E. coli JM101, DH5α, or protease-deficient BL21 (Stratagene). For expression, overnight cultures were diluted 1:10 in Luria broth containing ampicillin (100 μg/ml) and incubated at 37°C for 3 h. Subsequently, 1 mM isopropyl-β-d-thiogalactosidase was added to induce protein synthesis of the pGEX plasmid and cultures were allowed to grow at room temperature. After 7 h, cells were pelleted at 4,000 × g for 10 min and resuspended in 50 mM Tris-HCl (pH 7.5) containing 10 mM MgCl2. Cells were lysed by one cycle of freeze-thaw and sonication. Insoluble debris was removed by centrifugation, and the supernatant containing the soluble protein was collected.
Virus overlay assay.
PLRV (52) was maintained on Physalis floridana as previously described and purified from leaf material by a modified enzyme-assisted procedure (49). The virus overlay was performed essentially as described before (50). Similar amounts of various MpB GroEL polypeptides were run on denaturing polyacrylamide gels for SDS-PAGE. After electrophoresis, gels were conditioned in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid (pH 11.0) containing 10% methanol for 1 h and proteins were electrotransferred onto nitrocellulose. Protein blots were incubated overnight with purified PLRV (10 μg per ml), after which immunodetection with anti-PLRV IgG and alkaline phosphatase-conjugated anti-rabbit IgG was carried out (50).
Nucleotide sequence accession number.
The sequence data of the groEL operon and its flanking regions have been submitted to the GenBank database under accession no. AF003957.
RESULTS
Characterization of the groE operon of the Buchnera sp. of M. persicae.
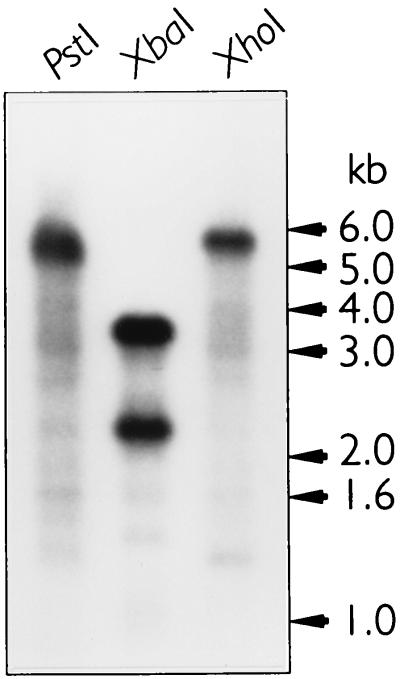
The nucleotide sequence of the groE operon of the primary endosymbiont (a Buchnera sp.) of M. persicae (MpB groE) and its flanking sequences were determined both by PCR and with clones derived from a genomic library. Southern blot analysis revealed that one copy of the MpB groEL gene was present on the genome (Fig. 1). The genomic organization of the MpB groE operon (Fig. 2) is similar to that of the groE and sym operons of E. coli and the intracellular symbiont of A. pisum, respectively (29, 38). The operon accomodates two ORFs encoding 10- and 60-kDa proteins, which have 72 and 73% homologies at the nucleotide level with E. coli groES and groEL, respectively. The MpB groE genes are also highly homologous to symS (89%) and symL (91%) from A. pisum. However, sequence comparisons of the promoter regions of the groE operons of various Buchnera spp. with that of E. coli revealed the only conserved element to be the heat shock promoter. A constitutive promoter similar to the one in the E. coli groE operon (and reported to be present in the A. pisum groE operon) could not be identified. A terminator sequence comparable to the one in the E. coli groE operon was not present either. Most likely, the GC-rich inverted repeat at the end of a Buchnera groE operon performs this function.
FIG. 1.
Southern blot analysis of the Buchnera sp. of M. persicae to determine the copy number of the groEL gene. DNA from the Buchnera sp. of M. persicae was digested with XbaI, PstI, or XhoI. XbaI recognizes a single restriction site within the MpB groEL gene, whereas PstI and XhoI restriction sites are present only outside this gene. A radiolabeled PCR fragment comprising the gene encoding MpB GroEL was used as the probe.
To determine the degree of conservation of the genomic region flanking the MpB groE operon, the regions upstream and downstream were compared with those of E. coli (Fig. 2), the only well-characterized free-living relative of Buchnera spp. (36). Upstream of the MpB groE operon, three ORFs which show similarities to genes on the E. coli genome were identified. The ORF immediately adjacent to the groE operon shows homology to the tRNAPhe gene (11). The other ORFs display 69% similarity to the E. coli 50-kDa thiophene and furan oxidation protein (ThdF) and 83% similarity to the C-terminal part of the gene that encodes the 60-kDa inner membrane protein of E. coli (2, 11). These genes are present at similar sites on the genome of Buchnera aphidicola, the primary endosymbiont of Schizaphis graminum (4), although the tRNAPhe gene has not been previously reported. Interestingly, on the E. coli genome, the groE operon is separated by approximately 500 kbp from the genes encoding ThdF and the inner membrane protein (11, 12). Downstream of the MpB groE operon, two genes which display 70% similarity with a 37.8-kDa protein of E. coli of unknown function and 78% similarity with the N-terminal sequence of elongation factor P of E. coli (3, 12) were identified. In E. coli, an additional segment of approximately 2 kbp harboring two ORFs with unknown functions is located between the terminator sequence of the groE operon and the gene encoding the 37.8-kDa protein (12).
Analysis of the groEL gene of the Buchnera sp. of M. persicae.
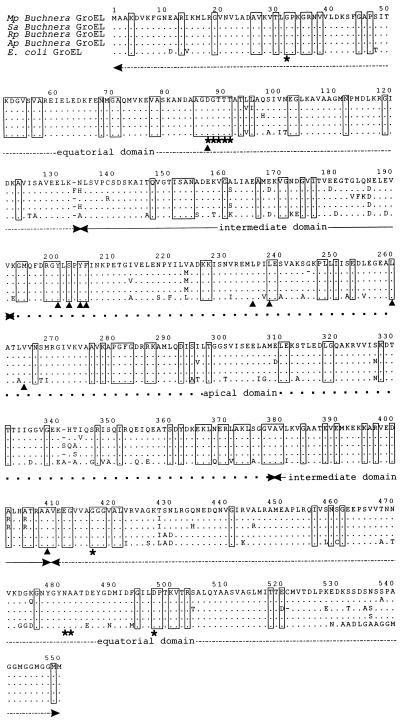
To ascertain whether MpB GroEL has structural and functional similarities to E. coli GroEL, the deduced amino acid sequence of the MpB groEL product was compared with those of E. coli and other Buchnera spp. GroELs (Fig. 3). This disclosed that MpB GroEL is 98 to 99% similar to the GroELs of Buchnera spp. of the aphids Sitobion avenae, Rhopalosiphum padi, and A. pisum and 92% similar to E. coli GroEL. A comparison of the MpB GroEL sequence with conserved residues in 50 prokaryotic Hsp60/GroEL homologs (22) showed that all of these residues except for the alanine at position 294 (Ala294) are identical. In all of the Buchnera spp. analyzed, Ala294 is replaced by serine (Fig. 3). Since the Buchnera GroEL of A. pisum has previously been demonstrated to fully complement E. coli GroEL in groE mutants of E. coli (38), this substitution seems to be of minor importance to GroEL’s functioning as a molecular chaperon in vivo. Moreover, in vitro experiments showed that the replacement of Ala294 by glutamic acid in E. coli GroEL did not affect polypeptide binding, folding, or its ATPase activity (22). The highly conserved amino acid residues of E. coli and Buchnera spp. GroELs are shown in Fig. 3. They are evenly distributed over the three domains of GroEL and are involved in polypeptide binding and folding (mainly located in the apical domain), ATP binding and hydrolysis (equatorial domain), maintaining inter- and intrasubunit interactions, and movement of the GroEL domains relative to each other (13, 22, 30). Amino acid residues in less-conserved regions which are known to mediate polypeptide binding (Leu238 and Val264) (22) or which have previously been reported to be essential for ATP binding in E. coli GroEL (Ala482 and Asp497) (6) are also identical in MpB GroEL.
FIG. 3.
Amino acid sequence alignment of the GroELs of E. coli (29) and Buchnera spp. from M. persicae (Mp), S. avenae (Sa) (23), R. padi (Rp) (23), and A. pisum (Ap) (38). Conserved regions in other GroEL/Hsp60 chaperonins (7) are boxed. Amino acids that are involved in polypeptide binding are indicated by arrow heads, and those that are involved in ATP binding are indicated by asterisks. Identical residues and gaps are indicated by periods and dashes, respectively. The equatorial domain is indicated by a dashed line, the intermediate domain is indicated by a continuous line, and the apical domain is indicated by dots. The sequence alignment was carried out by using the program PILEUP (Genetics Computer Group, Madison, Wis.) (17).
Binding of PLRV to Buchnera GroEL deletion mutants.
To determine which of the three domains of the MpB GroEL molecule are implicated in the interaction with PLRV in vitro, MpB GroEL was expressed in fusion with GST and affinity purified. After the GST moiety was removed by thrombin, the recombinant protein was tested for its PLRV-binding capacity by a virus overlay assay of protein blots which had previously been used to show that PLRV displayed a high and specific affinity for the 60-kDa subunit of MpB GroEL (50). The in vitro binding assay clearly established that full-length recombinant MpB GroEL bound PLRV as readily as wild-type MpB GroEL did (data not shown).
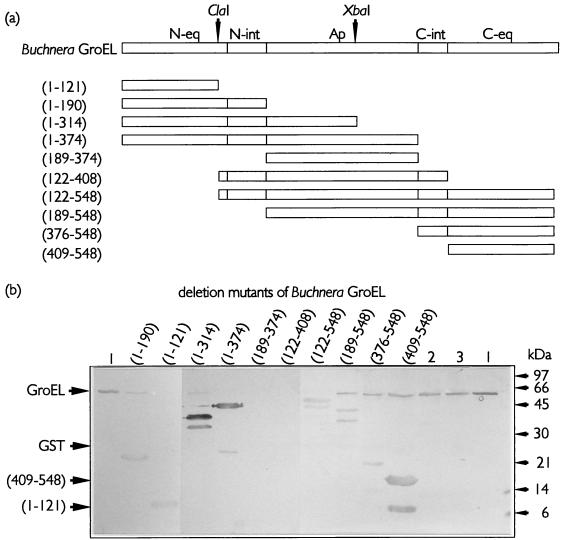
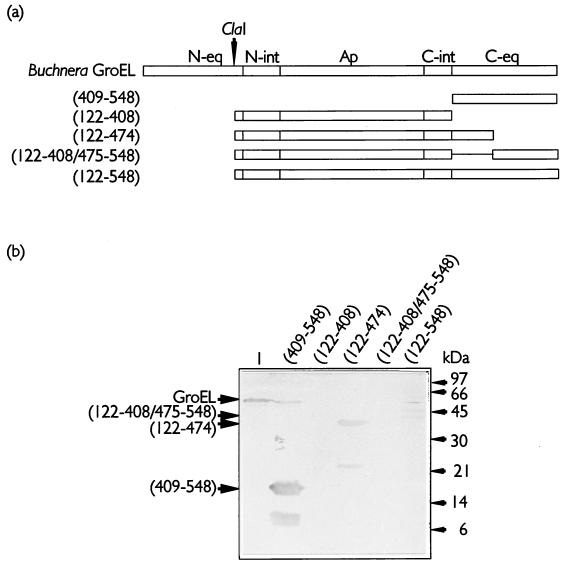
By utilizing the sequence similarity between Buchnera and E. coli GroELs in areas that are relevant for intrasubunit interactions, the first set of deletion mutants, based on the primary structure of the different domains on the GroEL molecule, was generated (Fig. 4a). The crystal structure of GroEL shows that the individual subunits are folded into three distinct domains (7), of which only the apical domain is continuous on the primary structure. The equatorial and intermediate domains are discontinuous, with regions located in both N- and C-terminal halves of the molecule (Fig. 4a). Testing similar amounts of MpB GroEL deletion mutants in virus overlay assays revealed that purified PLRV displayed affinities for all mutants containing the N- or C-terminal region of the equatorial domain (Fig. 4). Extending the N-terminal equatorial domain [MpB GroEL(1-121)], but not the C-terminal equatorial domain, with sequences of the intermediate and apical domains [MpB GroEL(1-314) and MpB GroEL(1-374)] improved the efficiency of virus binding (Fig. 4). Strikingly, PLRV binding to polypeptides containing the apical domain alone [MpB GroEL(189-374)] or the entire region between the ClaI site (amino acid residue 122) and the C terminus of the intermediate domain (amino acid residue 408) did not occur (Fig. 4). The smallest deletion mutants that showed binding to PLRV harbored the N-terminal 121 amino acid residues [MpB GroEL(1-121)] or the C-terminal 139 residues [MpB GroEL(409-548)] (Fig. 4). The presence of at least one of these regions is required for the virus-binding capacities of MpB GroEL deletion mutants. The virus overlay assay also showed that PLRV interacted with E. coli GroEL. E. coli GroEL was copurified with mutants expressed in the protease-deficient strain E. coli BL21. As E. coli GroEL was insensitive to the thrombin treatment (Fig. 4b; compare lanes 2 and 3) and its presence did not interfere with the migrations of MpB GroEL mutant polypeptides during SDS-PAGE, no steps were undertaken to remove endogenous GroEL from suspensions.
FIG. 4.
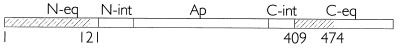
PLRV binding to deletion derivatives of MpB GroEL. (a) Schematic representations of MpB GroEL deletion mutants. The numbers in parentheses correspond to the positions of amino acid residues of MpB GroEL (Fig. 3) and mark the borders of the deletion mutants. The ClaI and XbaI restriction sites are indicated by arrowheads. N-eq, N-terminal region of the equatorial domain; N-int, N-terminal region of the intermediate domain; Ap, apical domain; C-int, C-terminal region of the intermediate domain; C-eq, C-terminal region of the equatorial domain. (b) Virus overlay assays. Lanes: 1, wild-type MpB GroEL; 2, GroEL of E. coli treated with thrombin; 3, GroEL of E. coli. All other lanes contain the indicated deletion mutants of MpB GroEL, as depicted in panel a. The positions of GroEL (60 kDa), GST (28 kDa), and the smallest truncated MpB GroEL fragments that bind PLRV [(409-548) and (1-121)] are indicated by arrowheads.
It is noteworthy that all MpB GroEL constructs harboring the C-terminal region of the equatorial domain produced smaller fragments for which PLRV showed affinities [MpB GroEL(122-548), MpB GroEL(189-548), MpB GroEL(376-548), and MpB GroEL(409-548)] (Fig. 4). These truncated products were approximately 8.5 kDa smaller than the corresponding mutants. Protein microsequencing by automated Edman degradation of the truncated products MpB GroEL(376-548) and MpB GroEL(409-548) revealed that the N-terminal residues are identical to those expected in the corresponding full-length polypeptides. This implies that the 8.5-kDa fragment was cleaved from the C terminus and that this fragment is dispensable for PLRV binding. Based on the relative molecular masses of the truncated products, the approximate position of the truncation mapped between amino acid residues 471 and 476 of MpB GroEL. The region N terminal of the truncation site, which is involved in PLRV binding, is characterized by the presence of three α-helices (7). To investigate more firmly the role of these structural elements in PLRV binding, the following two additional mutants were constructed: MpB GroEL(122-474), which contained the α-helices (between residues 408 and 475), and MpB GroEL(122-408/475-548), from which these elements were deleted (Fig. 5a). Purified PLRV clearly demonstrated an in vitro binding affinity for MpB GroEL(122-474) but not for MpB GroEL(122-408/475-548). Thus, the determinant for PLRV binding is located between amino acids 408 and 475 (Fig. 5b) of the C-terminal region of the equatorial domain.
FIG. 5.
Localization of the PLRV-binding site in the C-terminal part of the equatorial domain of MpB GroEL. (a) Schematic representations of the C-terminal deletion mutants of MpB GroEL. The numbering and abbreviations used are explained in the legend to Fig. 4. (b) Virus overlay assay of MpB GroEL deletion mutants. Lane 1, wild-type MpB GroEL; all other lanes contain the indicated MpB GroEL mutants, as depicted in panel a. The positions of GroEL (60 kDa), Buchnera GroEL(122-408/475-548), MpB GroEL(122-474), and MpB GroEL(409-548) are indicated by arrowheads.
DISCUSSION
In the present study, the groE operon of the Buchnera sp. from the major aphid vector of PLRV, M. persicae, was characterized and the PLRV-binding domain of MpB GroEL was identified by deletion mutant analysis. PLRV-binding studies revealed that virus particles exhibited in vitro affinities for all deletion mutants of MpB GroEL containing parts of the N-terminal (amino acid residues 1 to 121) (Fig. 3) or C-terminal (amino acids 409 to 474) (Fig. 3) regions of the equatorial domain (Fig. 4 through 6). Computer-generated structural predictions of the monomer of MpB GroEL (39) showed that these two regions assemble in the tertiary structure. It is therefore suggested that the residues involved in PLRV binding from either region join to compose a single PLRV-binding site. These results are remarkable, as previous single amino acid replacement studies of E. coli GroEL have demonstrated that residues in the apical domain are generally involved in polypeptide binding and folding (7, 22). Thus far, the equatorial domain has been implicated only in the in vitro binding of two multimeric proteins, ribulose-1,5-biphosphate carboxylase-oxygenase and malate dehydrogenase (54). Apparently, protein-binding sites are not necessarily located in the apical region of the central cavity of the GroEL cylinder but may be located in the equatorial domain as well. Large multimeric proteins and luteoviruses may employ these sites to overcome the size limitations (50 to 80 Å wide [15]) imposed by the central cavity of the GroEL molecule. The equatorial domain also accommodates the ATP-binding site on its external envelope (7, 22, 42). Like the putative PLRV-binding site, this site is composed of amino acid residues from both C- and N-terminal regions of the discontinuous equatorial domain (Fig. 3) (6). All of the MpB GroEL mutants in this study contain deletions known to impair intersubunit interactions (13, 22, 30) and are unable to assemble into the multimeric form of GroEL which prevails in the aphid hemolymph (51). Single amino acid replacements in the PLRV-binding regions which do not affect GroEL assembly are required to verify the role of the equatorial domain of the native molecule in the interaction with PLRV.
FIG. 6.
Summary of the PLRV-binding regions in MpB GroEL. The PLRV-binding regions in the N- (amino acids 1 to 121) and C- (amino acids 409 to 475) terminal parts of the equatorial domain (N-eq and C-eq, respectively) are shaded. N-int, N-terminal region of the intermediate domain; Ap, apical domain; C-int, C-terminal region of the intermediate domain.
Based on the differential binding of subgroup I and II luteoviruses and pea enation mosaic virus to Buchnera GroELs from vector and nonvector species and to GroEL of E. coli (23, 51), it was concluded that the basic virus-binding capacity resides in a conserved part of the GroEL molecule (51). Indeed, the regions in the equatorial domain of MpB GroEL which mediate PLRV binding are highly conserved among Buchnera GroEL homologs. However, regions of variability in these or other parts of the GroEL molecule may potentially influence the efficiency of binding, thus explaining the observed differences in the affinity of luteoviruses for GroEL homologs (23, 51).
While GroEL is abundantly produced by Buchnera spp. in aphids (5, 27), Buchnera GroES is difficult to detect (5, 32). Undoubtedly, Buchnera GroES is an important cofactor for cellular protein folding (38), but its potential role in extracellular protein interactions in the hemolymph of an aphid is yet to be investigated. Bacterial symbionts and pathogens, like Rhizobium meliloti, Pseudomonas aeruginosa, the X bacteria of Amoeba proteus, and Agrobacterium tumefaciens, have adopted different strategies at the transcriptional and translational levels to overproduce GroEL homologs relative to GroES production (1, 16, 21, 43, 45). P. aeruginosa and Rhizobium meliloti employ three groEL genes, of which there are one and two copies, respectively, on operons that also encode GroES (43). In the symbiotic bacteria of Amoeba proteus, GroEL is overproduced by an additional promoter in the GroES-encoding part in front of the groEL gene (1); in Agrobacterium tumefaciens, two mRNA fragments are produced and the mRNA fragment containing the gene encoding GroES is rapidly degraded (45). We detected only one copy of the gene encoding MpB GroEL (Fig. 1), located on the same operon that harbors the groES gene (Fig. 2). This suggests that overproduction of MpB GroEL occurs through the mechanisms found in the X bacteria of Amoeba proteus or Agrobacterium tumefaciens. Alternatively, it may well be that Buchnera GroEL is more stable than is GroES and readily accumulates while GroES is rapidly degraded.
To further identify potential genetic elements that may explain the high level of GroEL accumulation, sequences upstream of the coding regions were compared with the consensus sequences involved in transcription and translation of the groE operons of E. coli and the Buchnera sp. of A. pisum (38, 44, 55). This comparison disclosed the presence of sequences highly homologous to the E. coli heat shock promoter and Shine-Dalgarno sequences (Fig. 7). Although the MpB groE operon sequence is nearly identical to that of the Buchnera sp. of A. pisum in this region, we were not able to identify the constitutive promoter sequence of the groE operon of E. coli, which was previously reported to be present on the groE operon of the Buchnera sp. from A. pisum (38). Our observation corroborates recent findings that the only conserved promoter sequences of the groE operons of the Buchnera spp. of A. pisum and Schizaphis graminum are those recognized by ς32, a factor involved in the heat shock response (5), and that the heat shock promoter alone is responsible for transcription of the groE operon of the Buchnera sp. from A. pisum (44). An AT-rich nucleotide sequence upstream of this promoter, which may enhance promoter activity (10), is present in the groE operons of both E. coli and Buchnera spp. (Fig. 7). These observations are of interest, since GroEL expression in Buchnera spp. is similar to that in E. coli cells growing under stress (5).
FIG. 7.
Sequence comparison of regions involved in transcription and translation of the groE operons of E. coli (29, 55) and the Buchnera spp. of A. pisum (Ap) (38) and M. persicae (Mp). Conserved regions of the putative heat shock promoter and Shine-Dalgarno (SD) sequences are boxed, and the inverted repeats of putative transcription terminator sites are indicated by arrows. The localization of the constitutive promoter within the groE operon of E. coli is underlined. Gaps are indicated by dashes, stop codons are indicated by asterisks, and start codons are indicated by M.
In vivo interference with the interactions among extracellular MpB GroEL, PLRV, and beet western yellows luteovirus led to the suggestion that a transient association is required to protect luteoviruses in the hemolymph of an aphid from proteolysis (50, 51). Clearly, these interactions differ from the usual intracellular polypeptide-GroEL interactions; the mechanisms and potential roles of cofactors, including that of GroES, have not yet been revealed. It should be noted, however, that a functional extracellular GroEL was also observed in Helicobacter pylori, a gram-negative bacterium which causes chronic gastritis. It produces a GroEL homolog (HspB) which protects against inactivation of urease outside the bacterial cell in the hostile environment of the stomach of a vertebrate host (20, 40). Urease and HspB are released, probably by cell autolysis, and adhere to the surfaces of intact bacteria (40). Moreover, surface-associated Hsp60 fractions were also found in P. aeruginosa and Legionella pneumophila (31, 33). In this respect, it is interesting that GroEL proteins of Buchnera spp. were found to be more related to Hsp60s of pathogenic bacteria, such as L. pneumophila, than to E. coli GroEL (26).
ACKNOWLEDGMENTS
This research was financed in part by Priority Program Crop Protection grant 45.014 from the Ministry of Agriculture, Nature Management and Fisheries (LNV) and the Netherlands Organisation for Scientific Research (NWO).
Our discussions with K. E. Richards were greatly appreciated.
REFERENCES
- 1.Ahn T I, Lim S T, Leeu H K, Lee J E, Jeon K W. A novel strong promoter of the groEx operon of symbiotic bacteria in Amoeba proteus. Gene. 1994;128:43–49. doi: 10.1016/0378-1119(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 2.Alam K W, Clark D P. Molecular cloning and sequence of the thdF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J Bacteriol. 1991;173:6018–6024. doi: 10.1128/jb.173.19.6018-6024.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki H, Adams S L, Chung D G, Yaguchi M, Chuang S E, Ganoza M C. Cloning, sequencing, and overexpression of the gene for prokaryotic factor EF-P involved in peptide bond synthesis. Nucleic Acids Res. 1991;19:6215–6220. doi: 10.1093/nar/19.22.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P, Baumann L, Lai C-Y, Rouhbakhsh D, Moran N, Clark M. Genetics, physiology and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 5.Baumann P, Baumann L, Clark M A. Levels of Buchnera aphidicola chaperonin GroEL during growth of the aphid Schizaphis graminum. Curr Microbiol. 1996;32:279–285. [Google Scholar]
- 6.Boisvert D C, Wang J, Otwinowski Z, Horwich A L, Sigler P. The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATPγS. Nat Struct Biol. 1996;3:170–177. doi: 10.1038/nsb0296-170. [DOI] [PubMed] [Google Scholar]
- 7.Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 8.Bruening G, Criddle R, Preiss J, Rudert F. Biochemical experiments. New York, N.Y: John Wiley & Sons; 1971. p. 309. [Google Scholar]
- 9.Buchner P. Endosymbionts of animals with plant micro-organisms. New York, N.Y: John Wiley & Sons; 1965. pp. 210–332. [Google Scholar]
- 10.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 11.Burland V, Plunkett III G, Daniels D L, Blattner F R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 12.Burland V, Plunkett III G, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. VI. DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnett B P, Horwich A L, Low K B. A carboxy-terminal deletion impairs the assembly of GroEL and confers a pleiotropic phenotype in Escherichia coli K-12. J Bacteriol. 1994;176:6980–6985. doi: 10.1128/jb.176.22.6980-6985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrasekhar G N, Tilly K, Woolford C, Hendrix R, Georgopoulos C. Purification and properties of the groES morphogenetic protein of Escherichia coli. J Biol Chem. 1986;261:12414–12419. [PubMed] [Google Scholar]
- 15.Chen S, Roseman A M, Hunter A S, Wood S P, Burston S G, Ranson N A, Clarke A R, Saibil H R. Localisation of a folding protein and shape charges in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature. 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 16.Choi E Y, Ahn G S, Jeon K W. Elevated levels of stress proteins associated with bacterial symbiosis in Amoeba proteus and soybean root nodule cells. BioSystems. 1991;25:205–212. doi: 10.1016/0303-2647(91)90006-7. [DOI] [PubMed] [Google Scholar]
- 17.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis R J, van der Vies S M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- 19.Eskandari F, Sylvester E S, Richardson J. Evidence for lack of propagation of potato leaf roll virus in its aphid vector, Myzus persicae. Phytopathology. 1979;69:45–47. [Google Scholar]
- 20.Evans D J, Evans D G, Engstrand L, Graham D Y. Urease-associated heat shock proteins of Helicobacter pylori. Infect Immun. 1992;60:2125–2127. doi: 10.1128/iai.60.5.2125-2127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farinha M A, Mockett R, Went C J, Jardine S, Naczynski L M, Kropinski A M. Physical mapping of several heat-shock genes in Pseudomonas aeruginosa and the cloning of the MopA (GroEL) gene. Can J Microbiol. 1996;42:326–334. doi: 10.1139/m96-048. [DOI] [PubMed] [Google Scholar]
- 22.Fenton W A, Kashi Y, Furtak K, Horwich A L. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 23.Filichkin S A, Brumfield S, Filichkin T P, Young M J. In vitro interactions of the aphid endosymbiotic SymL chaperonin with barley yellow dwarf virus. J Virol. 1997;71:569–577. doi: 10.1128/jvi.71.1.569-577.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garret A, Kerlan C, Thomas D. The intestine is a site of passage for potato leafroll virus from the gut lumen into the haemocoel in the aphid vector, Myzus persicae Sulz. Arch Virol. 1993;131:377–392. doi: 10.1007/BF01378639. [DOI] [PubMed] [Google Scholar]
- 25.Gildow F S, Gray S M. The aphid salivary gland basal lamina as a selective barrier associated with vector-specific transmission of barley yellow dwarf luteovirus. Phytopathology. 1993;83:1293–1302. [Google Scholar]
- 26.Gupta R S. Evolution of the chaperonin families (Hsp60, Hsp60 and TCP-1) of protein and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 27.Hara E, Ishikawa H. Purification and partial characterization of symbionin, an aphid endosymbiont-specific protein. Insect Biochem. 1990;20:421–427. [Google Scholar]
- 28.Harrison B D. Studies on the behavior of potato leaf roll and other viruses in the body of their aphid vector Myzus persicae (Sulz.) Virology. 1985;6:265–277. doi: 10.1016/0042-6822(58)90074-6. [DOI] [PubMed] [Google Scholar]
- 29.Hemmingsen S M, Woolford C, van der Vies S M, Tilly K, Dennis D T, Georgopoulos C P, Hendrix R W, Ellis J R. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- 30.Horovitz A, Bochkareva E S, Kovalenko O, Girshovich A S. Mutation Ala2→Ser destabilizes intersubunit interactions in the molecular chaperone GroEL. J Mol Biol. 1993;231:58–64. doi: 10.1006/jmbi.1993.1256. [DOI] [PubMed] [Google Scholar]
- 31.Jensen P, Fomsgaard A, Shand G, Hindersson P, Hoiby N. Antigenic analysis of Pseudomonas aeruginosa and Pseudomonas cepacia GroEL proteins and demonstration of a lipopolysaccharide-associated GroEL fraction in P. aeruginosa. APMIS. 1993;101:621–630. doi: 10.1111/j.1699-0463.1993.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 32.Kakeda K, Ishikawa H. Molecular chaperon produced by an intracellular symbiont. J Biochem. 1991;110:583–587. doi: 10.1093/oxfordjournals.jbchem.a123623. [DOI] [PubMed] [Google Scholar]
- 33.Lema M W, Brown A. Legionella pneumophila has two 60-kilodalton heat-shock proteins. Curr Microbiol. 1995;31:332–335. doi: 10.1007/BF00294694. [DOI] [PubMed] [Google Scholar]
- 34.Martin J, Larger T, Boteva R, Schramel A, Horwich A L, Hartl F-U. Chaperonin-mediated protein folding at the surface of GroEL through a ‘molten-globule‘-like intermediate. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 35.Morioka M, Ishikawa H. Mutualism based on stress: selective synthesis and phosphorylation of a stress protein by an intracellular symbiont. J Biochem. 1992;111:431–435. doi: 10.1093/oxfordjournals.jbchem.a123774. [DOI] [PubMed] [Google Scholar]
- 36.Munson M A, Baumann P, Clark M A, Baumann L, Moran N A, Voegtlin D J, Campbell B C. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 1991;173:6321–6324. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munson M A, Baumann P, Kinsey M G. Buchnera gen. nov. and Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol. 1991;41:566–568. [Google Scholar]
- 38.Ohtaka C, Nakamura H, Ishikawa H. Structures of chaperonins from an intracellular symbiont and their functional expression in Escherichia coli groE mutants. J Bacteriol. 1992;174:1869–1874. doi: 10.1128/jb.174.6.1869-1874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peitsch M C. ProMod and Swiss-model: internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 40.Phadnis S H, Parlow M H, Levy M, Ilver D, Caulkins C M, Connors J B, Dunn B E. Surface localization of Helicobacter pylori urease and a heat shock homolog requires bacterial autolysis. Infect Immun. 1996;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Randles J W, Rathjen J P. Genus Luteovirus. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 379–383. [Google Scholar]
- 42.Roseman A M, Chen S, White H, Braig K, Saibil H. The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell. 1996;87:241–251. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- 43.Rusanganwa E, Gupta R S. Cloning and characterization of multiple groEL chaperonin-encoding genes in Rhizobium meliloti. Gene. 1993;126:67–75. doi: 10.1016/0378-1119(93)90591-p. [DOI] [PubMed] [Google Scholar]
- 44.Sato S, Ishikawa H. Expression and control of an operon from an intracellular symbiont which is homologous to the groE operon. J Bacteriol. 1997;179:2300–2304. doi: 10.1128/jb.179.7.2300-2304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segal G, Ron E Z. The groESL operon of Agrobacterium tumefaciens: evidence for heat shock-dependent mRNA cleavage. J Bacteriol. 1995;177:750–757. doi: 10.1128/jb.177.3.750-757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sylvester E S. Circulative and propagative virus transmission by aphids. Annu Rev Entomol. 1980;25:257–286. [Google Scholar]
- 47.Tilly K, Murialdo H, Georgopoulos C. Identification of a second Escherichia coli groE gene whose product is necessary for bacteriophage morphogenesis. Proc Natl Acad Sci USA. 1981;78:1629–1633. doi: 10.1073/pnas.78.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unterman B M, Baumann P, McLean L D. Pea aphid symbiont relationships established by analysis of 16S rRNA. J Bacteriol. 1989;171:2970–2674. doi: 10.1128/jb.171.6.2970-2974.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van den Heuvel J F J M, Boerma T M, Peters D. Transmission of potato leafroll virus from plants and artificial diets by Myzus persicae. Phytopathology. 1991;81:150–154. [Google Scholar]
- 50.Van den Heuvel J F J M, Verbeek M, van der Wilk F. Endosymbiotic bacteria associated with circulative transmission of potato leafroll virus by Myzus persicae. J Gen Virol. 1994;75:2559–2565. doi: 10.1099/0022-1317-75-10-2559. [DOI] [PubMed] [Google Scholar]
- 51.Van den Heuvel J F J M, Bruyère A, Hogenhout S A, Ziegler-Graff V, Brault V, Verbeek M, van der Wilk F, Richards K. The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J Virol. 1997;71:7258–7265. doi: 10.1128/jvi.71.10.7258-7265.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Wilk F, Huisman M J, Cornelissen B J C, Huttinga H, Goldbach R. Nucleotide sequence and organization of potato leafroll virus genomic RNA. FEBS Lett. 1989;245:51–56. doi: 10.1016/0014-5793(89)80190-5. [DOI] [PubMed] [Google Scholar]
- 53.Viitanen P V, Lubben T H, Reed J, Goloubinoff P, O’Keefe D P, Lorimer G H. Chaperonin-facilitated refolding of ribulosebisphosphate carboxylase and ATP hydrolysis by chaperonin 60 (groEL) are K+ dependent. Biochemistry. 1990;29:5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- 54.Weiss C, Goloubinoff P. A mutant at position 87 of the GroEL chaperonin is affected in protein binding and ATP hydrolysis. J Biol Chem. 1995;270:13956–13960. doi: 10.1074/jbc.270.23.13956. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y-N, Kusukawa N, Erickson J W, Gross C A, Yura T. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor ς32. J Bacteriol. 1988;170:3640–3649. doi: 10.1128/jb.170.8.3640-3649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]