Gates imprinted on a paper program capillary flow of reagents to create frugal dipstick tests with advanced functionalities.
Abstract
Lateral flow assays (LFAs) use capillary flow of liquids for simple detection of analytes. While useful for spontaneously wicking samples, the capillary flow inherently limits performing complex reactions that require timely application of multiple solutions. Here, we introduce a technique to control capillary flow on paper by imprinting roadblocks on the flow path with water-insoluble ink and using the gradual formation of a void between a wetted paper and a sheath polymer tape to create timers. Timers are drawn at strategic nodes to hold the capillary flow for a desired period and thereby enable multiple liquids to be introduced into multistep chemical reactions following a programmed sequence. Using our technique, we developed (i) an LFA with built-in signal amplification to detect human chorionic gonadotropin with an order of magnitude higher sensitivity than the conventional assay and (ii) a device to extract DNA from bodily fluids without relying on laboratory instruments.
INTRODUCTION
The standard platform for point-of-care (POC) testing in resource-limited settings has long been the lateral flow assays (LFAs) (1–4). LFAs offer unique advantages, such as rapid analysis, low cost, portability, ease of use, instrument-free operation, and compatibility with various biological samples (e.g., blood, plasma, serum, urine, sweat, and saliva), all of which make LFAs the predominant diagnostic format for POC applications (5–11). LFAs readily satisfy most of the ASSURED (Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free, and Deliverable to end-users) criteria reported by the World Health Organization in 2004 to establish capabilities of the POC devices (8–10). On the other hand, conventional LFAs are less prudent in performing multiplexed assays (i.e., simultaneously screening for multiple analytes within a sample) and are less sensitive in detecting various analytes of clinical importance (3–5).
Microfluidic paper-based analytical devices (μPADs) (12–14) combine the capillary-driven flow of LFAs with deterministic liquid routing enabled by microfluidic channels. In μPADs, a liquid-confining channel geometry is created on paper by patterning a hydrophobic material [e.g., photoresist (14), wax (15, 16), polydimethylsiloxane (17), and alkyl ketene dimer (18)], inkjet printing/etching (19, 20), plasma or laser treatment (21–23), or stamping (24). While channels formed on paper can readily direct multiple liquids to multiple reagents in a μPAD to perform multiplexed assays in parallel, spontaneous capillary flow in paper hinders execution of multistep bioassays that require timely application of different reagents or buffers in multiple steps, such as immunoassay, enzyme-linked immunosorbent assay (ELISA), and sample purification (9–12).
The lack of an intrinsic mechanism to control capillary-driven liquid flow remains a major bottleneck for LFAs. Recent approaches to control liquid flow in LFAs such as modified paper geometry (25, 26), volume-limited source pad (9), tunable-delay shunts (10), sugar deposition (11), pressurized paper (27), valving function via electromagnet (28, 29), fluidic diodes (30), and volume-metered actuation (7, 31) are highly effective but require either exotic and unscalable fabrication processes hindering their use in practice or large dead volumes limiting their use on manipulating small-volume samples.
In this work, we introduce a simple and scalable method to control capillary flow in LFAs by exploiting the gradual delamination of a wetted paper from a sheath polymer tape in producing controlled delays. Adhesive polymer tapes are integral parts of LFAs not only for laminating the capillary flow layer to limit external interference but also for isolating stacked layers for three-dimensional (3D) flow routing. Here, we purposely block the capillary flow path on a laminated paper with a water-insoluble ink, which leaves the void to be formed at the interface of the ink-infused paper and the polymer tape as the only path for the flow to proceed. By modifying the imprint geometry, we set the time it takes for a void to form and effectively create timers that hold capillary flow for a desired duration. Because the ink is specially formulated to infuse into and remain on paper, our timers can be constructed in a practical and reliable manner throughout an LFA. By imprinting these timers at strategic nodes, we program LFAs to coordinate different capillary flows, sequentially introduce different reagents into a reaction leaving optimal incubation times in between, and autonomously perform complex assays that could otherwise not be possible with conventional LFAs.
RESULTS
Capillary flow control via sheath tape delamination
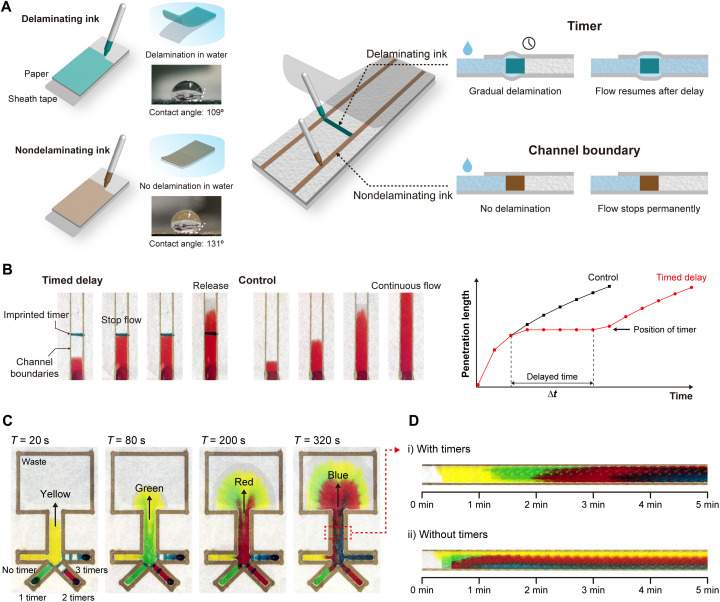
To manipulate capillary flow on a laminated paper, we created patterns by imprinting with two water-insoluble inks with different levels of hydrophobicity: The less hydrophobic one was expected to eventually separate from the adhesive sheath tape when wetted (i.e., delaminating ink) and hence was used to mark spots where we aimed to temporarily hold the flow. The more hydrophobic ink was required to remain permanently tethered to the sheath tape during the assay, leaving no voids for flow (i.e., nondelaminating ink), and was used to create channel boundaries to guide the flow (Fig. 1A). Those inks with desired properties were identified among commercially available permanent markers by measuring water contact angle on marked paper (Fig. 1A) and by verifying desired delamination response in water (fig. S1). The main compositional difference between the two inks was that the nondelaminating ink contained a hydrophobic resin, which formed a water-impenetrable bond with adhesive polymer on the sheath tape.
Fig. 1. Capillary flow control via sheath tape delamination.
(A) Manipulation of capillary flow on paper by imprinting patterns to modulate the paper-tape adhesion. The timer is drawn with a water-insoluble ink that is free of hydrophobic resin (delaminating ink). When the paper is wetted, a void gradually forms at the paper-tape interface and eventually resumes the flow. The flow boundaries are defined with a water-insoluble hydrophobic ink (nondelaminating). The resin remains attached to the tape and permanently blocks the flow. (B) Time-lapse images of a dye solution progressing on paper with a timer (left) and without a timer (right). The timer is imprinted as a discrete line normal to the capillary flow. The plot shows the measured penetration distance as a function of delayed and control flow. The timer acted like a relay, temporarily stopping the flow and releasing it after a certain duration. The width of each channel is 5 mm. (C) Time-lapse images of a test paper platform with four dye solutions simultaneously introduced from different inlets. Different number of timers drawn in different inlet branches differentially delay the capillary flows resulting in a sequential delivery. Shared channel width is 5 mm. (D) Change of observed color at the middle of the shared channel as a function of time with (top) and without timers (bottom). Discrete color transitions observed with timers demonstrate sequential routing of different capillary flows. Photo Credit: Dohwan Lee, Georgia Institute of Technology.
To test the functionality of our approach for capillary flow control, we used dye solutions and tracked the leading edge of the flow (Fig. 1B). In these experiments, the timer was created as a discrete line normal to the flow direction within channel boundaries that were imprinted by the nondelaminating ink. The advance of the dye on paper was observed to completely stop when it reached the timer and only resumed its progress after a delay. Moreover, once the timer was cleared, the flow showed no memory effect and advanced as if there were no obstacles in the first place. Last, the colorants within the timer were not washed away with the flow, a result that suggested the wetted timer retained its integrity as intended.
Using the same experimental setup, we investigated the mechanism of action for our timer. We first compared the timer response on laminated versus naked paper and observed that the capillary flow was permanently blocked by the same timer when it was imprinted on a naked paper (movie S1). These results invalidated dissolving of the timer or flow leaking through it as potential causes behind the delay observed in the laminated paper. Considering that the formation of a void between the polymer tape and the paper would necessitate mechanical displacement, we next attempted to constrain the paper-polymer tape stack by a compression force. When the timer in the laminated paper was placed in between two attracting magnets, the capillary flow indeed stopped permanently (fig. S2). In contrast, the control setup that only included the compressing magnets but no timer resulted in a continuous capillary flow. All in all, these analyses established the delamination of polymer tape from the paper as the mechanism of action for delaying capillary flow.
Next, we attempted to coordinate multiple capillary flow streams through a group of imprinted timers on paper. As a test platform, we created a simple flow layout, where four inlet branches merged into a single channel (Fig. 1C). In each branch, we placed a varying number of timers to differentially delay the flow between different branches. To distinguish between different capillary flows on the device, each inlet received a distinctly colored dye solution (movie S2). Observed color patterns clearly showed four fluids, simultaneously introduced to the device, reached the shared channel sequentially, with the flow from the branch without any timers arriving the first (yellow) and the one from the branch equipped with most timers arriving the last (blue) (Fig. 1D). In contrast, a control experiment on an otherwise identical device with no timers resulted in the simultaneous arrival of all dyed fluids to the shared channel, thereby proving the role of timers in achieving a sequential flow pattern. Together, these results demonstrated the feasibility of performing multistep assays in an automated fashion with imprinted timers that delaminate from the sheath tape.
Characterization and modeling of the timer delay
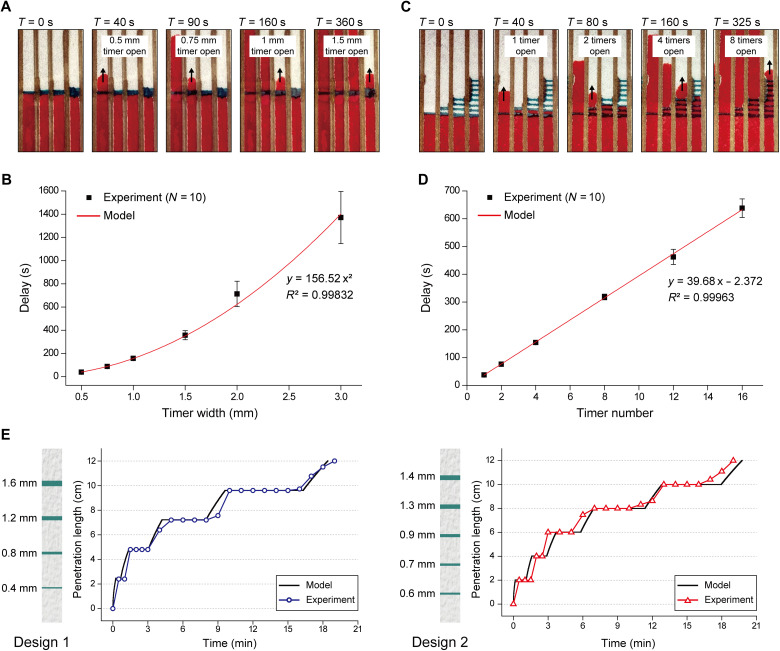
We studied the effect of timer geometry on the amount of time delay introduced into the capillary flow on a paper. The amount of delays generated by timers of different widths (0.5 to 3 mm) was measured in independent experiments by monitoring the progress of dye solutions through these timers (Fig. 2A). We found that the delay introduced by the timer increased with its width, which was expected as wider timers comprise a larger bonding interface with the polymer tape making it harder to form a void. Furthermore, the increase in delay was nonlinear as a 0.5-mm-wide timer resulted in an average flow delay of 37.6 s, while a 3-mm-wide timer produced an average flow delay of ~23 min (Fig. 2B). We experimentally realized capillary flow delays ranging from 30 s to >18 hours using imprinted timers. The minimum achievable delay was limited by the smallest feature size (~0.5 mm) that we could reliably imprint on the paper using our experimental setup. On the other hand, we could arbitrarily increase the flow delay in our experiments as long as the space permitted for wider or more timers. The maximum achievable delay will likely be limited by eventual disintegration of wetted paper, dissolution of permanent ink, or sample evaporation. Together, we concluded that the width of the timer, a parameter that can easily be tweaked, is a viable lever to set specific time delays for capillary flow on paper.
Fig. 2. Characterization and modeling of the timer delay.
(A) Time-lapse images taken of dye solution advancing on a paper test platform consisting of four lanes, each equipped with a timer of different width. The images show the capillary flow resuming after varying delays with wider timers resulting in longer delays. (B) Measured flow delays generated by timers (N = 10) as a function of their width. The measurements are overlaid with a quadratic regression model that was used to estimate delay when designing timers in this work. (C) Time-lapse images taken of dye solution advancing on a paper test platform consisting of four lanes, each equipped with a different number of cascaded timers. The images show the capillary flow resuming after varying delays with more timers resulting in longer delays. (D) Measured flow delays produced by different numbers of cascaded timers (1 to 16 timers) (N = 10). The total delay in flow linearly increases with the number of timers on the flow path. (E) Plots showing measured versus model-predicted flow delays in two test designs, each equipped with multiple timers of varying widths. For the first design, four timers of 0.4-, 0.8-, 1.2-, 1.6 mm widths were imprinted on a 12-cm-long flow path at 2.4-cm intervals. For the second design, five timers of 0.6-, 0.7-, 0.9-, 1.3-, 1.4 mm widths were imprinted on a 12-cm-long channel with 2-cm intervals. Photo Credit: Dohwan Lee, Georgia Institute of Technology.
While all tested timers produced relatively reproducible delays over repeated measurements (N = 10), we found that wider timers led to more noise in the produced time delay (with a coefficient of variation of ~16.1% for a 3-mm-wide timer), likely due to higher entropy in the delamination process (fig. S3). Inevitable variations of the timer geometry from imprinting on paper also produced variability in time delays, which were more pronounced for narrower timers. Variations in delamination duration for individual timers can be treated as noise and ultimately determined the temporal resolution of flow control using our technology.
While the nonlinear increase in delay with wider timers is advantageous when long delays are to be implemented, we also studied cascaded timer arrangements with the goal of achieving linear control over capillary flow delay (Fig. 2C). Our measurements confirmed the linear dependence of the total flow delay on the number of timers imprinted in the channel and showed high repeatability (N = 10) (Fig. 2D). The ability to modulate capillary flow delay only through creating new instances of the same delay element is important as it simplifies the LFA manufacturing process by eliminating the need to tune the timer geometry at different spots on a device. In addition, less variability from narrower timers can potentially be used to achieve the same time delay with a higher temporal resolution with cascaded timers compared to a single wider timer.
We also investigated the effect of liquid viscosity and temperature on the timer response. To manipulate the liquid viscosity, we added sucrose into deionized (DI) water and repeated the measurements with DI water mixed with glycerol to ensure against ingredient-specific artifacts. In both studies, we consistently measured longer delays with increasing viscosity (fig. S4). As for investigating the temperature effect, we compared the flow through the timers at room temperature versus when it was on a thermoelectric plate set at 4° and 60°C. At higher temperatures, the timer delay was reduced indicating a more efficient delamination process potentially contributed by increased evaporation of wicked liquid trapped under the sheath tape (fig. S5).
On the basis of the characterization data, we attempted to model the capillary flow in channels equipped with our timers. First, we empirically derived a quadratic relation that relates the flow delay to the width of the timer through regression analysis (Fig. 2B), which allowed us to predict the flow delay per timer as
| (1) |
where td is the flow delay (in s) and d is the width of the timer (in mm). To model the capillary flow on paper, we used the Lucas-Washburn equation (2, 32), which depicts the capillary flow penetration distance (l) into a porous material at a given time point as
| (2) |
where γ is the liquid-air surface tension, r is the effective pore radius, θ is the contact angle, μ is the dynamic viscosity of the fluid, and t is the wicking time. We then combined the two models to estimate the time it takes for the capillary flow to progress in a paper channel equipped with our timers (see Materials and Methods). Through tests on channels with different timer arrangements, we found our model to be highly accurate (Fig. 2E) and used it for the rest of this work in designing complex LFAs that sequentially routed different flow streams with preset delays to perform multistep chemical processes.
LFA with built-in signal amplification
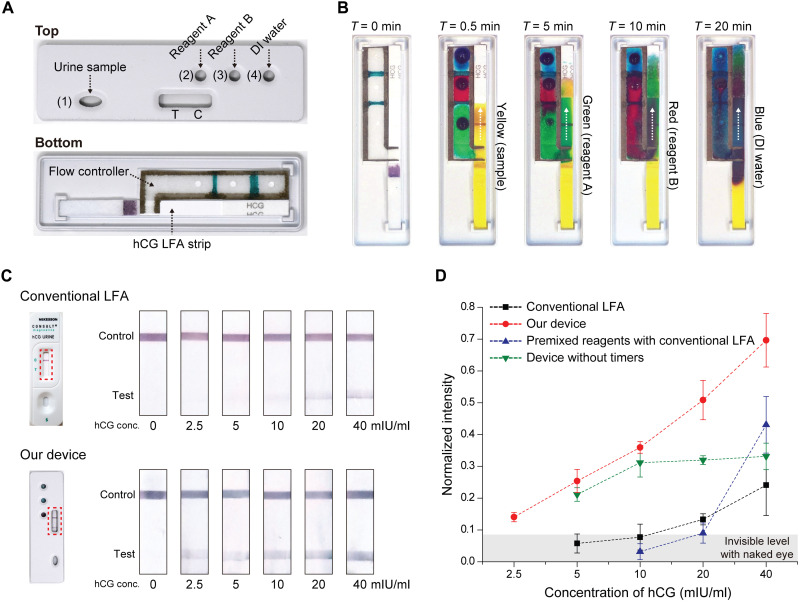
Because conventional LFAs are limited to assays that can be performed in a single step, they cannot benefit from already available chemistries to amplify their colorimetric results. Given our ability to coordinate multiple capillary flows on paper, we designed an LFA programmed to automatically apply amplifying reagents to enhance the sensitivity. Without loss of generality, we built an assay to detect human chorionic gonadotropin (hCG) (Fig. 3A). The device was built by incorporating a commercial hCG LFA strip with our paper-based flow controller (see fig. S6 for the detailed device layout and integration and alignment of different layers) followed by sealing it in a 3D-printed enclosure (see Materials and Methods). The commercial LFA strip (McKesson, Irving, TX) harbored recognition antibodies conjugated with gold nanoparticles (AuNPs), which produced a visible color change on the test line (fig. S7). The flow controller consisted of two timers placed in between three auxiliary inlets, which supplied chemical reagents designed to increase the size of AuNPs for enhanced visibility. Coordinating the flow of solutions loaded into these auxiliary inlets, the timers ensured their orderly and timely arrival to the reaction spot for maximal signal amplification.
Fig. 3. LFA with built-in signal amplification.
(A) A photo of our device taken from the top and bottom. The device consists of a commercial hCG LFA strip integrated with a flow controller. The 3D-printed case has a sample inlet 1, three auxiliary inlets 2, 3, and 4, and a detection window for observing test (T) and control (C) lines. In the flow controller, two timers (1 and 1.5 mm wide) are placed in between three auxiliary inlets. (B) Time-lapse images taken of the device when dye solutions of different colors, instead of actual solutions, were introduced from four inlets. The images confirm as-programmed sequential delivery of sample (yellow), signal amplification reagent A (green), B (red), and DI water (blue) to the test spot. (C) Assay results corresponding to samples with different hCG concentrations obtained using the conventional LFA and our device. (D) Plots showing the measured test line intensity as functions of the hCG concentration. In addition to our device and the conventional LFA from (C), results are shown for two other measurement settings: Premixed reagents with conventional LFA results correspond to a urine sample premixed with two signal amplification reagents (A and B) and DI water and processed using the conventional LFA. The device without timers corresponds to results obtained from a device that is identical to our device except that it lacks the timers to coordinate the flow between reagents. Photo Credit: Dohwan Lee, Georgia Institute of Technology.
First, to confirm automatic, as-programmed routing of liquids within the fabricated device, we input dye solutions in lieu of actual chemicals and recorded the time each dye solution arrived at the test spot from changes in color (Fig. 3B). The sample (yellow) reached the test spot in ~30 s, while the signal amplification reagents A (green) and B (red) took 5 and 10 min, respectively. Last, the test spot was washed with DI water (blue) at 20 min to quench the amplification for consistent readings (see fig. S7 and Materials and Methods).
We tested the assay by loading a 60 μl of urine sample spiked with a known amount of hCG hormone into the inlet 1 and 20 μl of each signal amplification reagents (A: enhancer + activator, B: initiator + buffer), along with 40 μl of DI water into auxiliary inlets 2, 3, and 4, respectively (see Materials and Methods). At the completion of the reaction after 20 min, the color change on the test line indicated the presence of hCG and a positive control line ensured against potential artifacts (Fig. 3C). The intensity of the test line, which was quantified by digital image processing (see Materials and Methods), linearly increased with the hCG concentration changing 0 to 40 mIU/ml (Fig. 3D, red dotted line). The limit of detection (LOD) was determined to be 2.5 mIU/ml, a level that corresponds to approximately an order of magnitude higher sensitivity than the conventional LFA with its manufacturer-specified LOD of 20 mIU/ml (Fig. 3D, black dotted line). In direct comparisons using matched samples, we found the conventional LFA signal level at a concentration of hCG (20 mIU/ml) was indeed comparable to the signal from our assay obtained at 2.5 mIU/ml (Fig. 3D).
Next, we ran the assay without the timers to test whether the LFA signal was enhanced solely due to the extra reagents introduced to the reaction or from the sequential delivery of those reagents coordinated by embedded timers. First, we premixed the urine sample with two signal amplification reagents (A and B) and DI water and then processed the mixture using the conventional LFA (fig. S8). Premixing reagents resulted in less sensitivity than our timer-controlled assay and gave a nonlinear response (Fig. 3D, blue dotted line). The signal levels were particularly low (even less than the conventional LFA) for hCG concentrations of ≤20 mIU/ml, likely due to the reducing agent interfering with the antigen/antibody interaction (33), which produced a pronounced effect at lower concentrations of hCG. Next, we ran the assay as we did on our device, but this time, without the timers imprinted on paper (fig. S9). Removing timers resulted in premature mixing of reagents in locations external to the test spot, which resulted in limited signal amplification for hCG concentrations of ≥10 mIU/ml (Fig. 3D, green dotted line). Reduced sensitivity at higher concentrations was due to the decrease in the amount of gold ions still available for signal amplification as some were already reduced by the concurrently running reagents before reaching the test spot. Together, the results demonstrated the crucial role played by the imprinted timers on paper in running a multistep chemical reaction in an optimal manner by coordinating timely delivery of reagents into the reaction.
Automated human DNA purification on paper
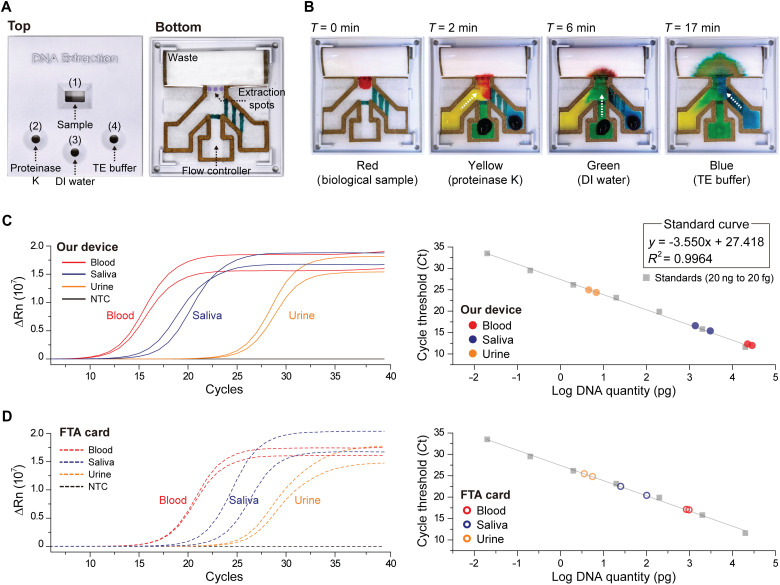
To demonstrate the utility of our timers for automating a sample preparation process, we developed a device for human DNA purification from bodily fluids (Fig. 4A). Our paper device consisted of flow channels defined by the nondelaminating ink and 1-mm-wide timers imprinted with the delaminating ink. The sample collection zone consisted of three DNA extraction spots (pink color) punched out from a Whatman FTA Elute Card, a specialized filter paper pretreated with chemicals for lysing the cells and preserving the extracted DNA. The device was designed to purify the DNA extracted by the FTA card through sequentially delivering solutions to the extraction spots from three different inlets. Specifically, the extraction spots were first subjected to a proteinase K solution for denaturing proteins; then to DI water for washing the denatured proteins, cell debris, and potential inhibitors away; and lastly to the Tris-EDTA (TE) buffer for stabilizing the purified DNA for downstream analysis. This sequence was programmed onto the device by imprinting different numbers of timers on different flow paths.
Fig. 4. Automated human DNA purification on paper.
(A) A photo of our device taken from the top and bottom. The device is composed of three DNA extraction spots (punched out of an FTA Elute Card) integrated with a flow controller where a single timer and three timers, all 1 mm-wide, are placed at middle and right channels, respectively. The case for the assay has four openings: a sample inlet 1 and three auxiliary inlets 2, 3, and 4. (B) Time-lapse images taken of the device when dye solutions of different colors, representative of actual solutions, were introduced from four inlets. The images confirm that once the sample (red) is loaded, dye solutions representing proteinase K (yellow), DI water (green), and TE buffer (blue) arrive at the extraction spots sequentially following the programmed order. Total process time for DNA purification with our assay is 20 min. Plots showing the quantitative polymerase chain reaction results (left) and the corresponding cycle threshold (Ct) values (right) of DNA extracted from blood, saliva, and urine sample using (C) our device and (D) the conventional FTA card. Photo Credit: Dohwan Lee, Georgia Institute of Technology.
We verified automated and timely routing of different fluids within the device by introducing dye solutions from all inlets and measuring their times of arrival to the extraction spots (Fig. 4B). We observed that after loading the sample (red), it took the proteinase K solution (yellow) ~2 min to reach the extraction spots. The DI water (green) surpassed a single timer and reached the extraction spots at ~6 min. Last, the TE buffer (blue) had to flow through three cascaded timers, and it took ~17 min to reach the extraction spots. With this design, we aimed for a total process time of 20 min.
Using our device, we extracted DNA from blood, saliva, and urine samples collected from consenting participants according to Institutional Review Board (IRB)–approved protocols and quantified the extracted amount of DNA using quantitative polymerase chain reaction (qPCR) (Fig. 4C). In these experiments, the DNA purification was performed by loading 3 to 5 μl of the specimen to the sample inlet 1 and 30 μl of proteinase K solution, 90 μl of DI water, and 30 μl of TE buffer to the reagent inlets 2, 3, and 4, respectively. Once the assay was complete, the purified DNA was first recovered from the device by removing the extraction spots and then amplified in a thermal cycler through qPCR (see Materials and Methods). The amount of DNA extracted per each sample was calculated on the basis of a calibration curve constructed from processing standard DNA templates (see Materials and Methods). Also, each sample was processed twice to demonstrate the consistency of the purification process on the device. The qPCR results confirmed the successful extraction of DNA from blood, saliva, and urine samples (Fig. 4C). As anticipated, the extracted DNA amount was found to be the highest from the blood sample and the lowest from the urine sample (Table 1). The fact that qPCR could successfully amplify the DNA from our device confirmed that capillary flow through our delaminating timers did not introduce inhibitor contaminants to the reaction.
Table 1. The amount of purified DNA by our device and the FTA card.
|
Biological
sample |
Our device | FTA Elute Card | ||
| Ct value |
Mean
amount of DNA |
Ct value |
Mean
amount of DNA |
|
| Blood | 11.587 | 25.8 ng | 16.790 | 953.6 pg |
| 11.949 | 16.894 | |||
| Saliva | 15.022 | 2.3 ng | 20.280 | 63.9 pg |
| 16.257 | 22.430 | |||
| Urine | 24.430 | 5.8 pg | 24.867 | 4.3 pg |
| 25.044 | 25.545 | |||
We compared our assay results with those obtained from the conventional FTA card DNA extraction protocol applied on matched samples (Fig. 4D). For all samples, but particularly for blood and saliva samples, our device was able to extract more purified DNA than the FTA card using less sample (5 μl versus 50 μl) and in less time (20 min versus 3 hours) (Table 1). The higher purified DNA extraction efficiency of our device was likely due to gentler sample handling compared to vortex washing steps required for the conventional processing of FTA cards. Moreover, our device offers a simpler solution (fig. S10) that can be carried out by nonspecialists to purify DNA samples in POC settings compared to conventional methods, such as filter-based ultracentrifugation, magnetic purification, or processing of FTA cards. Together, these results demonstrated the feasibility of performing a molecular biology protocol, traditionally performed using laboratory instrumentation, on a paper-based assay with built-in flow control to run multistep chemical reactions.
DISCUSSION
To control capillary flow in LFAs, we introduced a technique that used local voids forming between a wetted paper and a sheath polymer tape. By selectively modulating the paper-tape adhesion through imprinting patterns on the paper, we transformed the long-overlooked polymer sheath tape into an extrinsic flow path that can be programmed to manipulate the flow within the paper. While we demonstrated the concept using a specific set of materials, the technique can potentially be implemented with different combinations of paper, ink, and sheath tapes. With the expansion of available materials, we envision that the delamination process within timers could be manipulated through a variety of physical and chemical processes, which would extend the capabilities of the introduced technology beyond nondiscriminatively delaying capillary flows and introduce complex gating mechanisms to LFAs integrated with delaminating timers.
The ability to sequentially deliver different reagents into a reaction via programmable timers imprinted on paper makes it possible to automate multistep assays, which otherwise could only be performed in laboratories or with manual intervention. By simply drawing patterns on paper, we created immunoassays that were an order of magnitude more sensitive than commercial counterparts or POC devices that can extract, purify, and store DNA from a variety of bodily fluids with higher efficiency than the established laboratory protocols. Hence, the capabilities provided by the presented technology can make it possible to develop LFAs that can rival ELISA- or PCR-based assays for sensitive and specific detection of pathogenic targets such as Zika virus, HIV, hepatitis B virus, or diseases like malaria. Considering how prominent LFAs are for biomedical testing, our work has the potential to make complex and traditionally labor-intensive assays available in assay formats as simple and frugal as conventional LFAs.
MATERIALS AND METHODS
Fabrication of imprinted paper-based device
The assays were fabricated by drawing lines on tissue paper (Kimtech Science Kimwipes delicate task wipers, Kimberly-Clark, Irving, TX) with two types of permanent inks. The nondelaminating ink (Sharpie metallic permanent marker, manufacturer part number SHP 1823889) was used for drawing channel boundaries, and the delaminating ink (Sharpie ultrafine point marker, manufacturer part number SHP 37243) was used to draw timers. Before drawing the lines, one side of the tissue paper was covered with clear tape (manufacturer part number 3850-60), and the drawing was conducted manually or using an automatic drawing machine (Silhouette CAMEO, Silhouette America, Lindon, Utah, TX). The machine was only used to fabricate the devices when there was a need to precisely control the line geometry. After drawing all lines with these two markers, the remaining side of the paper was also covered by tape with holes punched to be used as reagent inlets.
Fabrication of device case
The device cases (Figs. 3 and 4) were designed using SolidWorks software (SolidWorks Corp., Waltham, MA) and printed using a 3D printer (ProJet 3510 HD) with VisiJet M3-X as a structural material. After printing, the device was immersed in mineral oil (Durvet, Blue Springs, MO) at 65°C for the dewaxing of the uncured supporting material and washed with soapy water, DI water, and ethanol sequentially. Then, filler primer (Rust-Oleum, Automotive Filler Primer Spray) was sprayed on the device to render the surface smooth. After drying for 10 min, white paint (Rust-Oleum, Painters Touch 2X Spray Paint Matte White) was sprayed and fully dried.
Mathematical model of the timer delay
The model to predict the capillary flow in channels equipped with our timer was established in MATLAB by using the following two equations as
| (1) |
| (2) |
where γ = 72 mN/m, θ = 81.82, and μ = 0.89 mPa·s (34). For the effective pore radius r, we chose 6.5 μm by averaging the pore sizes obtained from microscopy images of the paper. Initially, l follows the Eq. 2 for a duration of t1 until it reaches the first timer. Upon reaching it, l remains constant for the assigned delay td1 according to the Eq. 1. After the delay, l resumes to progress following Eq. 2, with t = t − td1 until it reaches the next timer and repeats this process whenever encountering the subsequent timers with t = t − td1 − td2⋯.
hCG signal amplification
Urine samples were collected according to a protocol approved by IRB of Georgia Institute of Technology. The hCG-positive control (McKesson, Irving, TX) (urine matrix, 200 mIU/ml) was serially diluted with urine to set the concentration from 2.5 to 40 mIU/ml. The samples were directly loaded into the AuNP-based hCG pregnancy test kit (McKesson, Irving, TX) embedded in our device from the designated sample inlet concurrently with gold enhancement solution (Nanoprobes, Yaphank, NY) and DI water from auxiliary inlets. The hCG-AuNPs complex and unbound AuNPs were captured at the test and control lines, respectively, and produced a red color initially (fig. S7, i to iii). Following the delivery of gold enhancement reagents, AuNPs were enlarged because of the autometallographic reaction, which made the test and control line colors darker, yielding better contrast and higher signal intensity (fig. S7, iv to vi).
Quantification of color intensity
The test and control line images captured before and after the signal amplification were individually analyzed using the ImageJ program (http://imagej.nih.gov/ij/). The raw images were first converted into monochrome. Measured gray tones at the test and control lines as well as in the background were then used to calculate the normalized intensity based on the following equation
where It, Ic, and Ib are the measured gray values at test, control line, and background, respectively.
DNA purification by the conventional FTA card
Whole blood, saliva, and urine samples were each collected from healthy donors according to protocols approved by IRB of Georgia Institute of Technology. To purify DNA by the FTA card, Whatman FTA Elute Card (GE Healthcare, UK) was used according to manufacturer-provided protocol. Samples (50 μl) were deposited onto the FTA cards within the circular spot, and the samples were dried thoroughly for at least 3 hours at room temperature. The dried samples on the FTA card were punched out using a 3-mm biopsy punch, and each of them was placed into sterile microcentrifuge tubes for the following washing steps. The punched discs were then triple-washed in 500 μl of DI water through vortexing and centrifugation. Following the final wash, the extracted water was aspirated from the tube, and the washed discs were directly used for qPCR analysis.
Measurement of purified DNA with qPCR
To measure the amount of DNA purified by our device and by the FTA card, qPCR was conducted using the Femto Human DNA Quantification Kit (Zymo Research, Orange, CA) on QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA). Whenever qPCR was conducted, standards were operated together in duplicates with 10-fold serial dilutions of standard DNA templates (20 ng to 20 fg), which enabled us to construct a calibration curve (Fig. 4, C and D) between the amount of human DNA and Ct (cycle threshold) values. The calibration curve was y = −3.55 x + 27.418, and the amount of DNA purified by our device and the FTA card were estimated on the basis of the calibration curve.
Acknowledgments
Funding: This work was supported by the start-up funds provided to A.F.S. by Georgia Institute of Technology. Author contributions: D.L. and A.F.S. designed the research and overall direction. D.L., T.O.-A., C.H.-C., and R.L. fabricated the devices. D.L., T.O.-A., and M.B. performed experiments. All authors conducted data analysis. D.L. and A.F.S. mainly wrote the manuscript. All authors discussed and commented on refining the final manuscript. Competing interests: D.L., T.O.-A., and A.F.S. are the inventors for “Capillary flow control in lateral flow assays via delaminating timers,” filed by Georgia Institute of Technology (U.S. Provisional Patent Application No. 63/197,711, filed on 07 June 2021). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Legends for movies S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Sackmann E. K., Fulton A. L., Beebe D. J., The present and future role of microfluidics in biomedical research. Nature 507, 181–189 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Gong M. M., Sinton D., Turning the page: Advancing paper-based microfluidics for broad diagnostic application. Chem. Rev. 117, 8447–8480 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Yetisen A. K., Akram M. S., Lowe C. R., Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 13, 2210–2251 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Posthuma-Trumpie G. A., Korf J., van Amerongen A., Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 393, 569–582 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Lee D., Shin Y., Chung S., Hwang K. S., Yoon D. S., Lee J. H., Simple and highly sensitive molecular diagnosis of Zika virus by lateral flow assays. Anal. Chem. 88, 12272–12278 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Banerjee R., Jaiswal A., Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst 143, 1970–1996 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Kong T., Flanigan S., Weinstein M., Kalwa U., Legner C., Pandey S., A fast, reconfigurable flow switch for paper microfluidics based on selective wetting of folded paper actuator strips. Lab Chip 17, 3621–3633 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Verma M. S., Tsaloglou M. N., Sisley T., Christodouleas D., Chen A., Milette J., Whitesides G. M., Sliding-strip microfluidic device enables ELISA on paper. Biosens. Bioelectron. 99, 77–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu E., Liang T., Spicar-Mihalic P., Houghtaling J., Ramachandran S., Yager P., Two-dimensional paper network format that enables simple multistep assays for use in low-resource settings in the context of malaria antigen detection. Anal. Chem. 84, 4574–4579 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toley B. J., McKenzie B., Liang T., Buser J. R., Yager P., Fu E., Tunable-delay shunts for paper microfluidic devices. Anal. Chem. 85, 11545–11552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutz B., Liang T., Fu E., Ramachandran S., Kauffman P., Yager P., Dissolvable fluidic time delays for programming multi-step assays in instrument-free paper diagnostics. Lab Chip 13, 2840–2847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng C.-M., Martinez A. W., Gong J., Mace C. R., Phillips S. T., Carrilho E., Mirica K. A., Whitesides G. M., Paper-based ELISA. Angew. Chem. Int. Ed. 49, 4771–4774 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Martinez A. W., Phillips S. T., Butte M. J., Whitesides G. M., Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem. Int. Ed. 46, 1318–1320 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez A. W., Phillips S. T., Whitesides G. M., Three-dimensional microfluidic devices fabricated in layered paper and tape. Proc. Natl. Acad. Sci. U.S.A. 105, 19606–19611 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y., Shi W., Jiang L., Qin J., Lin B., Rapid prototyping of paper-based microfluidics with wax for low-cost, portable bioassay. Electrophoresis 30, 1497–1500 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Lu Y., Shi W., Qin J., Lin B., Fabrication and characterization of paper-based microfluidics prepared in nitrocellulose membrane by wax printing. Anal. Chem. 82, 329–335 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Bruzewicz D. A., Reches M., Whitesides G. M., Low-cost printing of poly(dimethylsiloxane) barriers to define microchannels in paper. Anal. Chem. 80, 3387–3392 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X., Tian J., Garnier G., Shen W., Fabrication of paper-based microfluidic sensors by printing. Colloids Surf. B Biointerfaces 76, 564–570 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Abe K., Suzuki K., Citterio D., Inkjet-printed microfluidic multianalyte chemical sensing paper. Anal. Chem. 80, 6928–6934 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Abe K., Kotera K., Suzuki K., Citterio D., Inkjet-printed paperfluidic immuno-chemical sensing device. Anal. Bioanal. Chem. 398, 885–893 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Li X., Tian J., Nguyen T., Shen W., Paper-based microfluidic devices by plasma treatment. Anal. Chem. 80, 9131–9134 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Li X., Tian J., Shen W., Progress in patterned paper sizing for fabrication of paper-based microfluidic sensors. Cellul. 17, 649–659 (2010). [Google Scholar]
- 23.Chitnis G., Ding Z., Chang C. L., Savran C. A., Ziaie B., Laser-treated hydrophobic paper: An inexpensive microfluidic platform. Lab Chip 11, 1161–1165 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Dornelas K. L., Dossi N., Piccin E., A simple method for patterning poly(dimethylsiloxane) barriers in paper using contact-printing with low-cost rubber stamps. Anal. Chim. Acta 858, 82–90 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Fu E., Lutz B., Kauffman P., Yager P., Controlled reagent transport in disposable 2D paper networks. Lab Chip 10, 918–920 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apilux A., Ukita Y., Chikae M., Chailapakul O., Takamura Y., Development of automated paper-based devices for sequential multistep sandwich enzyme-linked immunosorbent assays using inkjet printing. Lab Chip 13, 126–135 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Shin J. H., Park J., Kim S. H., Park J. K., Programmed sample delivery on a pressurized paper. Biomicrofluidics 8, 054121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Zwanenburg P., Liu X., Magnetic timing valves for fluid control in paper-based microfluidics. Lab Chip 13, 2609–2614 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Fratzl M., Chang B. S., Oyola-Reynoso S., Blaire G., Delshadi S., Devillers T., Ward T., Dempsey N. M., Bloch J.-F., Thuo M. M., Magnetic two-way valves for paper-based capillary-driven microfluidic devices. ACS Omega 3, 2049–2057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H., Cogswell J., Anagnostopoulos C., Faghri M., A fluidic diode, valves, and a sequential-loading circuit fabricated on layered paper. Lab Chip 12, 2909–2913 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Toley B. J., Wang J. A., Gupta M., Buser J. R., Lafleur L. K., Lutz B. R., Fu E., Yager P., A versatile valving toolkit for automating fluidic operations in paper microfluidic devices. Lab Chip 15, 1432–1444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cummins B. M., Chinthapatla R., Ligler F. S., Walker G. M., Time-dependent model for fluid flow in porous materials with multiple pore sizes. Anal. Chem. 89, 4377–4381 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Doña V. V., Fossati C. A., Chirdo F. G., Interference of denaturing and reducing agents on the antigen/antibody interaction. Impact on the performance of quantitative immunoassays in gliadin analysis. Eur. Food Res. Technol. 226, 591–602 (2008). [Google Scholar]
- 34.Jahanshahi-Anbuhi S., Chavan P., Sicard C., Leung V., Hossain S. M. Z., Pelton R., Brennan J. D., Filipe C. D. M., Creating fast flow channels in paper fluidic devices to control timing of sequential reactions. Lab Chip 12, 5079–5085 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Telis V. R. N., Telis-Romero J., Mazzotti H. B., Gabas A. L., Viscosity of aqueous carbohydrate solutions at different temperatures and concentrations. Int. J. Food Prop. 10, 185–195 (2007). [Google Scholar]
- 36.Segur J. B., Oberstar H. E., Viscosity of glycerol and its aqueous solutions. Ind. Eng. Chem. Res. 43, 2117–2120 (1951). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Legends for movies S1 and S2
References
Movies S1 and S2