Abstract
Mutations in PINK1 and parkin highlight the mitochondrial axis of Parkinson’s disease (PD) pathogenesis. PINK1/parkin regulation of the transcriptional repressor PARIS bears direct relevance to dopamine neuron survival through augmentation of PGC-1α–dependent mitochondrial biogenesis. Notably, knockout of PARIS attenuates dopaminergic neurodegeneration in mouse models, indicating that interventions that prevent dopaminergic accumulation of PARIS could have therapeutic potential in PD. To this end, we have identified the deubiquitinase cylindromatosis (CYLD) to be a regulator of PARIS protein stability and proteasomal degradation via the PINK1/parkin pathway. Knockdown of CYLD in multiple models of PINK1 or parkin inactivation attenuates PARIS accumulation by modulating its ubiquitination levels and relieving its repressive effect on PGC-1α to promote mitochondrial biogenesis. Together, our studies identify CYLD as a negative regulator of dopamine neuron survival, and inhibition of CYLD may potentially be beneficial in PD by lowering PARIS levels and promoting mitochondrial biogenesis.
CYLD operates in the PINK1/parkin pathway to regulate proteasomal degradation of PARIS controlling dopamine neuron survival.
INTRODUCTION
Genetic mutations in the E3 ubiquitin ligase parkin or the Ser/Thr kinase PINK1 play a causal role in autosomal recessive Parkinson’s disease (PD) (1, 2). PINK1 facilitates parkin activation by phosphorylating both ubiquitin and parkin phosphorylation at S65 (3, 4). Several substrates of PINK1 and parkin are now known to link these proteins in common pathways regulating multiple aspects of mitochondrial quality control (5, 6). Of these, the pathogenic substrate, PARIS (ZNF746), which is subject to proteasomal regulation through sequential phosphorylation and ubiquitination by PINK1 and parkin, respectively, plays a pivotal role in dopamine neuron survival (7–9). Accumulation of PARIS is evident in sporadic and familial PD brains and also leads to loss of dopamine neurons in Drosophila models of conditional parkin or PINK1 knockdown (KD) (10), a mouse model of parkin inactivation (11, 12), and adult conditional mouse models of parkin knockout (7, 8) or PINK1 KD (9). The underlying molecular mechanism involves PARIS-mediated transcriptional repression of peroxisome proliferator-activated receptor-gamma coactivator-1a (PGC-1α) that leads to impaired mitochondrial biogenesis, declines in mitochondrial mass, and deficits in respiratory capacity of mitochondrial pools within the dopamine neurons. PARIS also plays a key role in α-synuclein (α-syn)–induced neurodegeneration under conditions of parkin inactivation in two A53T α-syn transgenic models of familial PD and in the α-syn preformed fibril model of sporadic PD (13). Collectively, these studies indicate that PARIS is the key mediator of neurodegeneration in familial and sporadic PD through regulation of PGC-1α and provide a strong rationale to identify strategies that reduce or inhibit PARIS to prevent neurodegeneration in PD.
Ubiquitination is a reversible modification, and the actions of E3 ligases such as parkin are countered by deubiquitinating enzymes (DUBs) that catalyze the removal of ubiquitin from substrates. The human genome encodes ~100 DUBs belonging to six families (14). However, the identity of DUBs that function in the PINK1/parkin pathway to regulate cellular levels of PARIS is currently unknown. Identification of these DUBs could nevertheless have immense therapeutic potential as inhibition of these DUBs could counter PARIS accumulation by promoting protein destabilization. We recently reported that transgenic expression of human PARIS (hPARIS) in Drosophila causes selective progressive degeneration of dopamine neurons and motor deficits that result from PARIS-dependent repression of PGC-1α and its downstream transcription factors nuclear respiratory factor 1 (NRF1) and transcription factor A, mitochondrial (TFAM), ultimately impinging on mitochondrial biogenesis. Notably, short hairpin RNA (shRNA)–mediated reductions in Drosophila PINK1 (dPINK1) or parkin activity in the fly dopamine neurons lead to up-regulation of the Drosophila homolog of PARIS and phenocopy the mitochondrial biogenesis defects (10). Given this highly conserved nature of PARIS regulation by the PINK1/parkin pathway, we reasoned that these Drosophila models would serve as powerful systems to screen for DUBs that function as part of the PINK1/parkin pathway to regulate PARIS protein stability and thereby promote dopaminergic (DA) mitochondrial health via the PGC-1α axis. To this end, we conducted a genome-wide DUB-specific RNA interference (RNAi) screen in the Drosophila model of hPARIS to identify DUBs whose loss of function in vivo suppressed the PARIS-induced motor deficits. Such DUB candidates were further examined to identify DUBs that not only suppressed motor deficits under conditions of PINK1 or parkin insufficiency but also promoted survival of fly dopamine neurons. This led to the identification of the DUB cylindromatosis (CYLD) that effectively suppressed DA neurotoxicity incurred under conditions of PARIS accumulation.
Here, we show that the neuroprotective effects of CYLD KD are mediated through its DUB activity. DA neuron KD of CYLD in Drosophila increases PARIS ubiquitination, thereby accelerating its proteasomal degradation. CYLD KD also relieves the repressive effect PARIS has on PGC-1α and alleviates the mitochondrial biogenesis defects that result from PARIS accumulation in the fly dopamine neurons. Our studies further identify mammalian PARIS to be a DUB substrate of CYLD. Depletion of CYLD activity ameliorates DA neurodegeneration and behavioral deficits observable in adult conditional parkin KD mouse models. In human embryonic stem (hES) cell–derived midbrain dopamine neurons, reducing CYLD activity in the parkin-deficient background facilitates PGC-1α–dependent gene expression changes that promote mitochondrial protein synthesis to sustain mitochondrial biogenesis. Notably, the neuroprotective effects mediated by CYLD KD are abrogated in the absence of PGC-1α. Together, these observations uncover a previously unidentified role for CYLD in promoting mitochondrial health within dopamine neurons via the PARIS/ PGC-1α axis and highlight its utility as a potential therapeutic target for small molecule inhibitors.
RESULTS
Genome-wide DUB-specific RNAi screen in Drosophila identifies candidates that function in the PINK1/parkin pathway
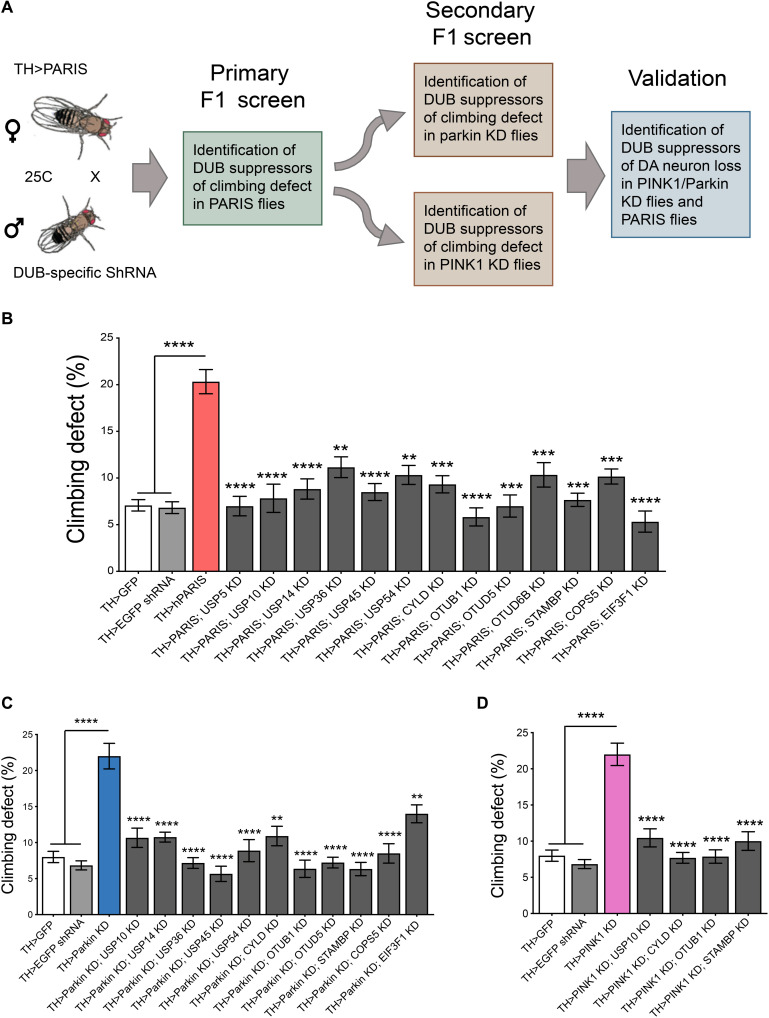
To identify DUBs that regulate PARIS neurotoxicity via the PINK1/parkin pathway, we devised a genome-wide in vivo RNAi screen in Drosophila targeting all the DUBs encoded in the fly genome. An inventory of DUBs encoded in the fly genome has been described (15) and includes 45 putative DUBs belonging to five typical DUB subfamilies, of which 43 have direct orthologs in humans (table S1). We have previously shown that DA overexpression of hPARIS in Drosophila leads to adult onset progressive loss of dopamine neurons and climbing deficits that phenocopy the effect DA KD of dPINK1 or parkin has (10). We therefore used DUB-specific RNAi fly lines in a series of sequential primary and secondary F1 screens to identify DUBs that, when knocked down, rescued the pronounced climbing defects under these conditions (Fig. 1A). A primary F1 screen was conducted in flies that overexpressed hPARIS in the dopamine neurons under the control of the DA neuron–specific tyrosine hydroxylase (TH)–Gal4 driver (TH>hPARIS). RNAi fly lines targeting each of the 43 DUBs with human orthologs were crossed to TH>hPARIS flies and DUB KDs that suppressed PARIS-induced climbing defects in the F1 progeny were scored as positive hits in this primary screen. As control, each of the 43 DUB-specific RNAi lines was crossed to the TH-Gal4 driver to ensure that KD of the respective DUBs did not in itself lead to climbing defects that could potentially obscure readout in the TH>hPARIS flies (table S2). Of the 43 DUBs screened in the TH>PARIS flies, KD of 13 individual DUBs effectively suppressed the PARIS-induced climbing defects (Fig. 1B and fig. S1). To identify DUBs that function in the PINK1/parkin pathway to regulate PARIS, these 13 DUB candidates were then subjected to additional secondary screens to identify DUBs that, when knocked-down, were also able to rescue climbing defects that stem from DA KD of Drosophila parkin (dparkin) or dPINK1. RNAi fly lines targeting each of the above 13 DUBs were crossed to flies that exhibit DA KD of dparkin (TH>dparkin KD) or dPINK1 (TH>dPINK1 KD), and climbing performance was scored in the F1 progeny. Of the 13 DUBs screened simultaneously in the TH>dparkin KD and TH>dPINK1 KD flies, 11 DUBs suppressed climbing defects under conditions of dparkin KD (Fig. 1C and fig. S2A), while 4 DUBs suppressed climbing defects under conditions of dPINK1 KD (Fig. 1D and fig. S2B). Of note, the four DUBs, namely, USP10, CYLD, OTUB1, and STAMBP, identified in the dPINK1 KD screen were represented in all three rounds of the F1 screens and are thus likely to function in a common pathway with PINK1 and parkin to regulate PARIS-mediated neurotoxicity in vivo.
Fig. 1. Genome-wide RNAi screen to identify candidate DUBs that function in PINK1/parkin pathway.
(A) Schematic of the screen. TH-Gal4–driven DUB-specific RNAi fly lines were used in an F1 primary screen to identify DUBs that rescued hPARIS-induced climbing defects on day 20. Such candidate DUBs were subjected to additional secondary F1 screens probing for DUBs that rescued climbing defects under conditions of DA parkin or PINK1 KD on day 20, respectively. Hits from the secondary screen were then examined for their ability to promote dopamine neuron survival in 20-day-old parkin or PINK1 KD flies. (B) Primary F1 screen based on rescue of PARIS-induced climbing defect identified 13 DUBs. (C) Summary of candidate DUBs from primary screen that rescued climbing defects in parkin KD flies. (D) Summary of candidate DUBs from primary screen that suppressed climbing defects in PINK1 KD flies and progressed for further validation. TH-Gal4/+ flies served as control. N = 60 flies per genotype for both primary and secondary screens. Quantitative data = means ± SEM. One-way analysis of variance (ANOVA); **P < 0.01, ***P < 0.001, ****P < 0.0001. See also figs. S1 and S2 and table S1.
Drosophila CYLD functions in the PINK1/parkin pathway
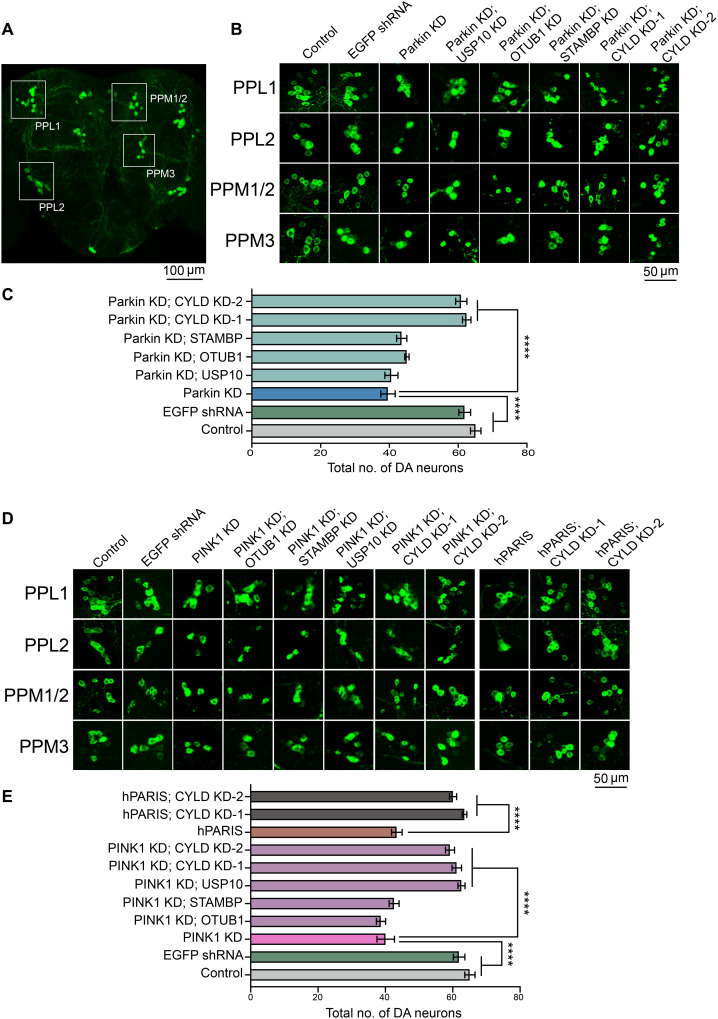
DA KD of parkin or PINK1 in Drosophila leads to progressive loss of dopamine neurons within all major DA neuronal clusters (10). The four candidate DUBs (USP10, CYLD, OTUB1, and STAMBP) identified from the primary and secondary F1 screens were therefore examined for their ability to promote dopamine neuron survival under conditions of dparkin or dPINK1 KD. Among the four DUBs examined in the TH>dparkin KD flies, only KD of the DUB CYLD effectively suppressed dopamine neuron loss within all the major dopamine neuronal clusters PPL1, PPL2, PPM1/2, and PPM3 (Fig. 2, A to C, and fig. S3A). In the TH>PINK1 KD flies, in addition to CYLD, KD of USP10 also promoted neuronal survival within the major DA clusters (Fig. 2, D and E, and fig. S3B). The opposite effects USP10 KD has on dopamine neuronal survival in the dparkin KD and dPINK1 KD flies suggest that USP10-mediated effects likely occur in a parkin-independent fashion. Since we set out to conduct the genome-wide RNAi screen to identify DUBs that function in a common pathway with PINK1 and parkin, the DUB CYLD (hereafter referred to as dCYLD) that suppressed both the climbing defects and dopamine neuron loss in the parkin KD and PINK1 KD flies emerged as a strong candidate from the series of our sequential F1 screens. Following the outcome of this initial screen, the effect of dCYLD KD was further verified using a second independent dCYLD-specific RNAi line, which similarly rescued climbing defects (fig. S2, A and B, and movie S1) and dopamine neuron loss (Fig. 2, B to E, and fig. S3, A and B) in the dparkin and dPINK1 KD flies. DA KD of dCYLD using both the dCYLD RNAi lines (dCYLD KD-1 and dCYLD KD-2) also rescued the DA phenotypes in the TH>hPARIS flies (Fig. 2, B to E, and figs. S1, S2, A and B, and S3, A and B).
Fig. 2. Validation of candidate DUBs identified in primary and secondary RNAi screens places Drosophila cylindromatosis (dCYLD) in the PINK1/parkin pathway.
(A) Representative confocal image of the Drosophila whole brain showing the PPL1, PPL2, PPM1/2, and PPM3 DA neuron clusters. Scale bar, 100 μM. (B) Representative confocal images of individual dopamine neuron clusters visualized using TH immunofluorescence in the indicated genotypes under conditions of parkin KD. Scale bar, 50 μM. N = 10 flies per genotype. (C) Quantification of neuronal numbers within individual dopamine neuron clusters in the indicated genotypes. (D) Representative confocal images of individual dopamine neuron clusters visualized using TH immunofluorescence in the indicated genotypes under conditions of PINK1 KD or hPARIS overexpression. Scale bar, 50 μM. (E) Summary of dopamine neuron quantifications in the indicated genotypes. TH-Gal4/+ flies served as control. N = 10 flies per genotype. Quantitative data = means ± SEM. One-way ANOVA; ****P < 0.0001. See also fig. S3.
We have previously shown that DA accumulation of the Drosophila homolog of PARIS (dPARIS) under conditions of parkin or PINK1 KD leads to adult onset progressive loss of DA neurons and climbing defects similar to hPARIS (10). To examine whether KD of dCYLD exerts neuroprotective effects under conditions of dPARIS accumulation, dCYLD was knocked down using the two independent dCYLD RNAi lines (dCYLD KD-1 and dCYLD KD-2) in flies that exhibit DA overexpression of dPARIS (TH>dPARIS flies). dCYLD KD using both the dCYLD RNAi lines promoted dopamine neuronal survival in the TH>dPARIS flies and suppressed climbing defects as assessed in 30-day-old flies (fig. S4, A to D, and movie S2). Together, these findings uncover a novel role for CYLD in the PINK1/parkin pathway and further indicate that the neuroprotective effects CYLD KD has toward alleviating PARIS-dependent neurotoxicity are highly conserved.
DA KD of dCYLD promotes mitochondrial biogenesis
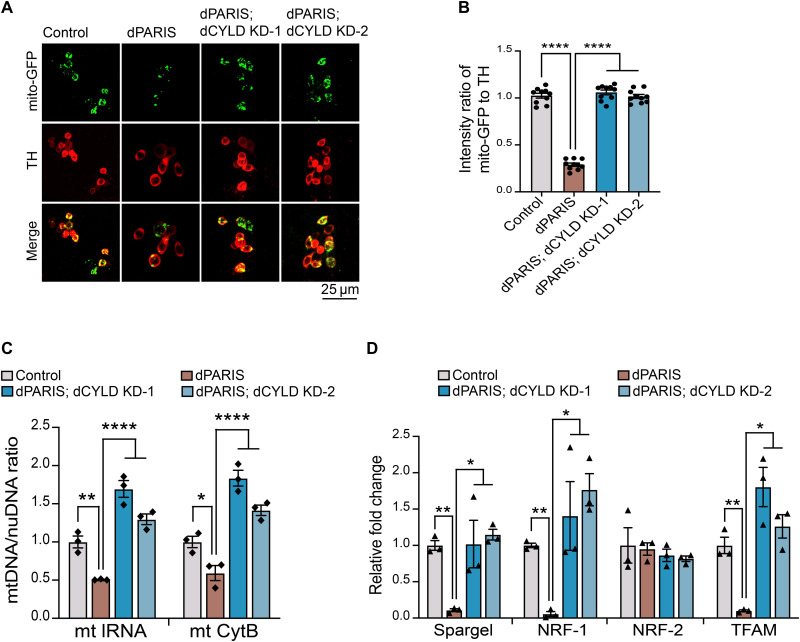
DA accumulation of dPARIS leads to defects in mitochondrial biogenesis that stem from dPARIS-mediated transcriptional repression of the Drosophila homolog of PGC-1α (Spargel) and its downstream transcription factors NRF-1 (ewg) and mitochondrial transcription factor (TFAM) (10). We therefore examined whether dCYLD KD alleviates the mitochondrial biogenesis defects that result from dPARIS accumulation. Quantification of mitochondria labeled within the DA neurons using mitochondrial targeted green fluorescent protein fluorescence (mito-GFP) reveals that dCYLD KD under conditions of DA overexpression of dPARIS abrogates the detrimental effect dPARIS accumulation has and leads to a significant increase in mitochondrial abundance within the dopamine neurons. Of note, both the dCYLD RNAi lines (dCYLD KD-1 and dCYLD KD-2) similarly rescue dPARIS-mediated mitotoxicity (Fig. 3, A and B). Assessment of mitochondrial DNA (mtDNA) copy number in dopamine neurons isolated from fly brains using fluorescence-activated cell sorting (FACS) also reveals a significant increase in mtDNA copy numbers in the dPARIS overexpressing flies under conditions of dCYLD KD (Fig. 3C). Quantitative reverse transcription polymerase chain reaction (RT-PCR) measurements of transcript levels of Drosophila homologs of PGC-1α (Spargel), NRF-1 (ewg), and TFAM show up-regulation of these genes under conditions of dCYLD KD (Fig. 3D). Transcript levels of the Drosophila homolog of NRF-2 (Delg), also a known target of PGC-1α, remain unaffected under dPARIS overexpression (Fig. 3D).
Fig. 3. dCYLD KD promotes mitochondrial biogenesis in dopamine neurons.
(A) Representative confocal images showing mito-GFP (green)–labeled mitochondria within dopamine neurons immunostained for TH (red) in the indicated genotypes. Scale bar, 25 μM. (B) Quantification of intensity ratio of mito-GFP to TH immunofluorescence in the indicated genotypes. TH>mito-GFP flies served as control, N = 10 flies per genotype. (C) Assessment of mitochondrial DNA copy number in FACS-sorted dopamine neurons in 30-day-old flies of the indicated genotypes. Mean ratio of mtDNA to nuDNA from three independent FACS experiments, each using 50 whole fly brains shown. TH>GFP flies served as control. (D) Quantitative RT-PCR analysis of Drosophila homologs of PGC-1α (Spargel), NRF1 (ewg), NRF-2 (Delg), and TFAM in FACS-sorted DA neurons from 30-day-old flies. Mean values from three independent FACS experiments shown. TH>GFP flies served as control. Quantitative data = means ± SEM. One-way ANOVA; *P < 0.05, **P < 0.01, and ****P < 0.0001.
Neuroprotective effects of dCYLD KD are mediated by its DUB activity
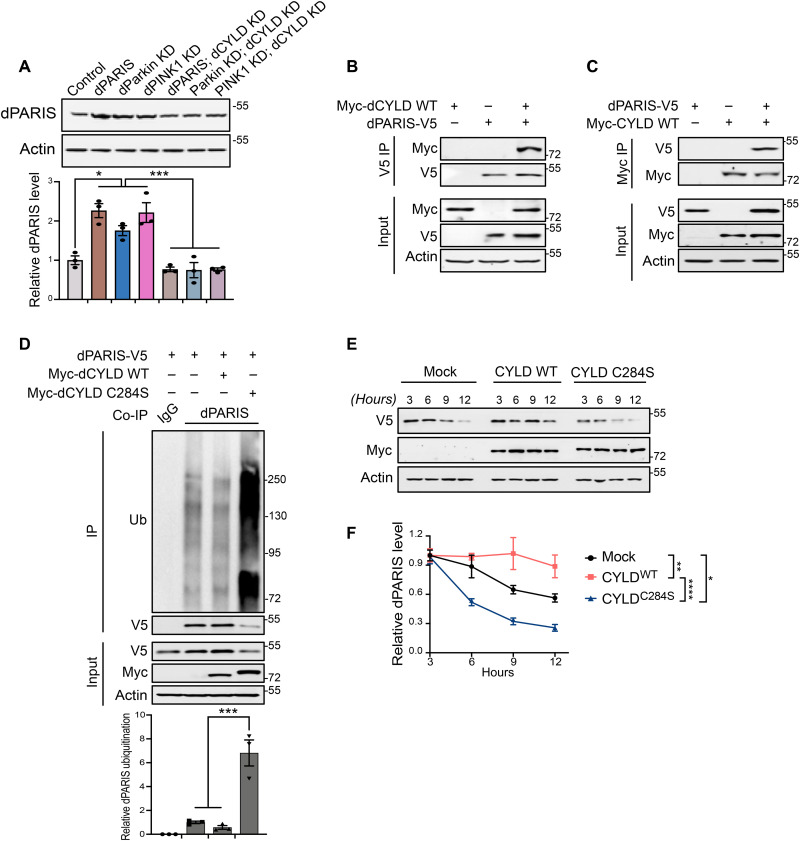
To examine the potential mechanism underlying the neuroprotective effects dCYLD KD has under conditions of dparkin or dPINK1 KD, we examined the effect of dCYLD KD on cellular levels of PARIS. Immunoblot analyses show that DA KD of dCYLD prevents dPARIS accumulation in flies exhibiting DA KD of dparkin or dPINK1, indicating that CYLD regulates PARIS protein stability (Fig. 4A). We next sought to determine whether dPARIS is a deubiquitination substrate of dCYLD. Toward this end, reciprocal coimmunoprecipitation experiments in Drosophila S2 cells indicate that CYLD interacts with PARIS (Fig. 4, B and C). Furthermore, coimmunoprecipitation of dPARIS in S2 cells using a dPARIS-specific antibody shows that the dCYLD catalytic mutant (dCYLD C284S) but not wild-type (WT) dCYLD leads to increased ubiquitination of dPARIS. Increased ubiquitination of dPARIS is observable in the form of a higher molecular weight smear in immunoblots of immunoprecipitated dPARIS, characteristic of polyubiquitinated proteins and leads to substantially reduced cellular levels of dPARIS (Fig. 4D). This indicates that dCYLD targets dPARIS for deubiquitination. To investigate the effect of dCYLD on proteasomal degradation of dPARIS, a C-terminal V5-tagged dPARIS was ectopically expressed in S2 cells under the control of Drosophila metallothionein gene promoter whose expression could be induced by addition of copper sulfate (CuSo4). This inducible dPARIS expression system was then used to monitor the steady-state levels of dPARIS-V5 in the presence of dCYLD WT or the dCYLD C284S catalytic mutant. Forty-eight hours after dPARIS induction, CuSo4 was removed from the S2 culture medium to block further dPARIS expression. Assessment of dPARIS over a period of 12 hours indicates that the dCYLD C284S catalytic mutant accelerates degradation of dPARIS, while dCYLD WT sustains consistently high levels of dPARIS (Fig. 4, E and F). This indicates that under conditions of dCYLD KD, increased ubiquitination of dPARIS targets it for proteasomal degradation, thereby preventing its accumulation and promoting DA neuron survival in vivo.
Fig. 4. KD of dCYLD promotes dopamine neuron survival by destabilizing dPARIS through its DUB activity.
(A) Representative immunoblot and dPARIS quantification in flies expressing the indicated transgenes under the control of TH-Gal4 driver. TH-Gal4/+ flies served as control, N = 3. (B) Coimmunoprecipitation using anti-V5 antibodies shows interaction between C-terminal V5-tagged dPARIS and N-terminal Myc-tagged dCYLD in Drosophila S2 cells. Similar results were observed in three independent experiments. (C) Reciprocal coimmunoprecipitation experiments using anti-Myc antibodies verify dPARIS interaction with dCYLD in S2 cells transfected with indicated constructs in three independent experiments, N = 3. (D) Deubiquitination of dPARIS by dCYLD as assessed in S2 cells transfected with indicated constructs. Immunoblot analysis and relative quantification of immunoprecipitated (IP) dPARIS shows increased ubiquitination (Ub) of dPARIS in the presence of the dCYLD C284S catalytic mutant, thereby enhancing its proteasomal degradation, N = 3. (E) DUB activity of dCYLD affects the turnover rate of dPARIS. (F) Immunoblot analysis and dPARIS quantification in S2 cells at the indicated time points shows that while the dCYLD catalytic mutant accelerates dPARIS turnover, overexpression of WT dCYLD increases its half-life, N = 3. Quantitative data = means ± SEM. One-way ANOVA; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See also fig. S4.
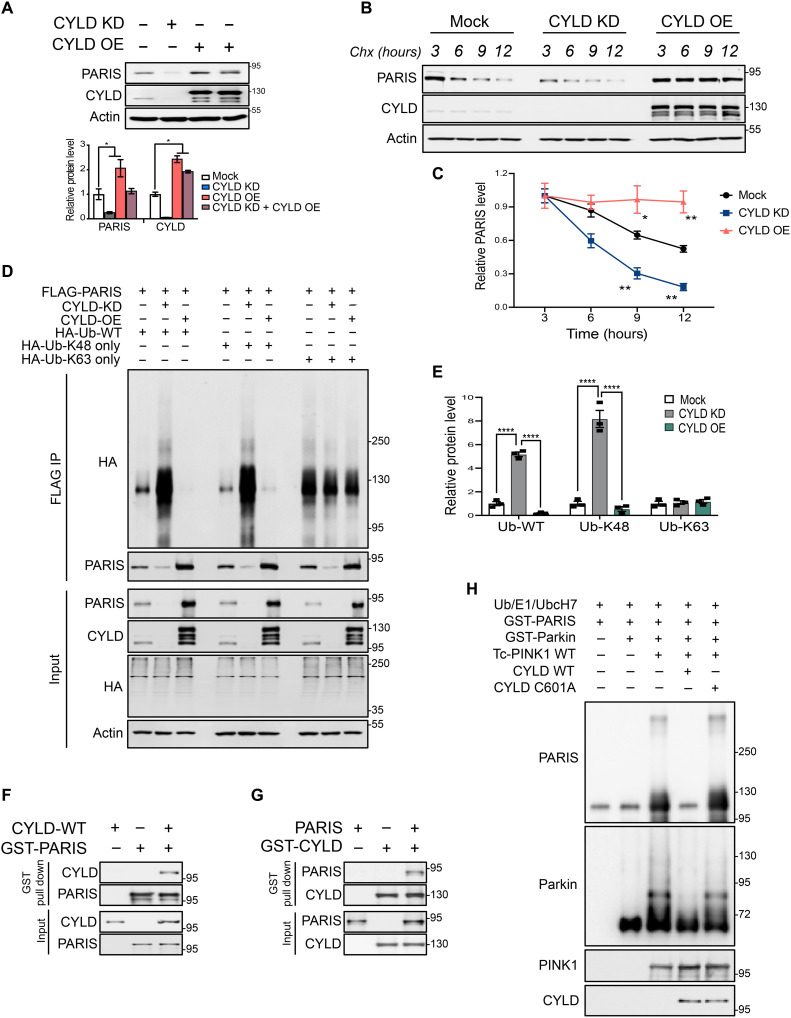
Mammalian PARIS is a deubiquitination substrate of CYLD
We next sought to determine whether the DUB activity of CYLD regulates proteasomal degradation of PARIS in mammalian cells similar to that observed in Drosophila. In human SH-SY5Y neuroblastoma cells, CRISPR-cas9 gene editing–mediated KD of CYLD leads to a reduction in endogenous PARIS levels that can be restored upon reintroduction of WT CYLD. CYLD overexpression, on the other hand, leads to PARIS accumulation, indicating that CYLD specifically regulates PARIS protein stability (Fig. 5A). This effect is further demonstrable in cycloheximide chase assays that were conducted to follow the rate of PARIS turnover in SH-SY5Y cells. While CYLD KD accelerates proteasomal degradation of PARIS, overexpression of CYLD increases PARIS protein stability, thereby sustaining consistently high levels of PARIS at all the time points assessed (Fig. 5, B and C). Reciprocal coimmunoprecipitation experiments in SH-SY5Y cells using V5-tagged CYLD and FLAG-tagged PARIS indicate that CYLD and PARIS interact similarly to their Drosophila counterparts (fig. S5, A and B). To further characterize this interaction, SH-SY5Y cells were transfected with full-length V5-CYLD and FLAG PARIS or FLAG-tagged deletion mutants of PARIS (fig. S5C). Coimmunoprecipitation experiments indicate that the N-terminal region PARIS harboring the Kruppel-associated box domain interacts with CYLD (fig. S5D). The CYLD binding region in PARIS was further verified in reciprocal coimmunoprecipitation experiments (fig. S5E). To map the PARIS binding region in CYLD, V5-tagged full-length CYLD or its deletion mutants (fig. S5F) were expressed in SH-SY5Y cells together with FLAG-PARIS. Reciprocal coimmunoprecipitation experiments show that PARIS binds CYLD at its third cytoskeleton-associated protein (CAP) domain located in close proximity to the CYLD USP (catalytic) domain (fig. S5, G and H).
Fig. 5. Mammalian PARIS is a deubiquitination substrate of CYLD.
(A) Immunoblot analysis and quantification of endogenous PARIS in SH-SY5Y neuroblastoma cell line following CRISPR-Cas9–mediated KD or overexpression (OE) of CYLD, N = 3. (B) Cycloheximide chase experiment at the indicated time points showing PARIS turnover rate under conditions of CYLD KD or OE in SH-SY5Y cells. (C) Relative quantification of endogenous PARIS shows accelerated protein turnover under conditions of CYLD KD, whereas CYLD OE increases PARIS half-life. N = 3 independent experiments. (D) CYLD promotes deubiquitination of PARIS through hydrolysis of K48 ubiquitin chains. Ubiquitination of immunoprecipitated FLAG-tagged PARIS in SH-SY5Y cells monitored by immunoblot analysis under conditions of CYLD KD or OE in the presence of HA-tagged WT ubiquitin or ubiquitin mutants that can only append either K48 or K63 ubiquitin chains. (E) Quantification of PARIS ubiquitination showing enrichment of WT or K48 ubiquitin chains under conditions of CYLD KD but not CYLD OE. No changes in levels of K63 ubiquitin chains on PARIS observable under the different conditions assayed. N = 3. (F) GST pull-down assays using purified recombinant GST-tagged PARIS and WT CYLD indicate a robust interaction between PARIS and CYLD. Similar results were observed in three independent pull-down experiments. (G) Reciprocal GST pull-down assays using GST-tagged CYLD and PARIS confirm the direct interaction between PARIS and CYLD. N = 3. (H) In vitro deubiquitination of PARIS by CYLD. Immunoblot analysis of purified recombinant GST-PARIS ubiquitinated in the presence of PINK1 and activated parkin showing decreased ubiquitination in the presence of WT CYLD. The CYLD C601A catalytic mutant, however, has no impact on PARIS ubiquitination. Similar results were observed in three independent experiments. Quantitative data = means ± SEM. One-way ANOVA; *P < 0.05, **P < 0.01, and ****P < 0.0001. See also fig. S5.
To determine whether CYLD regulates PARIS protein stability through its DUB activity, ubiquitination of ectopically expressed FLAG-PARIS was monitored in SH-SY5Y cells cotransfected with hemagglutinin (HA)–tagged WT ubiquitin under conditions of CYLD KD and overexpression. CYLD KD that reduces PARIS protein levels leads to significant enrichment of WT ubiquitin chains on PARIS evident in the form of high molecular weight smears. This effect is abrogated by CYLD overexpression that leads to PARIS accumulation (Fig. 5, D and E). To examine the specific type of ubiquitin linkage on PARIS that CYLD targets for deubiquitination, ubiquitination of FLAG-PARIS was examined as before in SH-SY5Y cells expressing either HA-K48 or HA-K63 ubiquitin mutants that only append either K48 or K63 ubiquitin chain linkages, respectively. Under conditions of CYLD KD, immunoprecipitated PARIS exhibits substantial enrichment of K48 ubiquitin chain linkages that are hydrolyzed by CYLD overexpression resulting in PARIS ubiquitination pattern similar to that observed with WT ubiquitin. However, no discernible changes in PARIS ubiquitination were observed in cells coexpressing the HA-K63 ubiquitin mutants (Fig. 5, D and E). This indicates that CYLD predominantly targets K48 ubiquitin chain modifications on PARIS for deubiquitination, thereby facilitating its proteasomal degradation. To further confirm whether PARIS is a DUB substrate of CYLD, purified recombinant proteins were used to verify the interaction between CYLD and PARIS followed by in vitro deubiquitination assays. In in vitro glutathione S-transferase (GST) pull-down assays, GST-tagged PARIS pulls down WT CYLD (Fig. 5F), indicating that the interaction between these proteins is direct, which is further evident in reciprocal pull-down assays that used GST-CYLD (Fig. 5G). In vitro ubiquitination reactions wherein PARIS was ubiquitinated using ubiquitin, E1 and UbcH7 (E2) in the presence of WT Tc PINK1 and activated parkin, the presence of WT CYLD leads to substantial reduction in PARIS ubiquitination, whereas the CYLD catalytic mutant (CYLD C601A) has no effect. These studies provide additional evidence that PARIS is a DUB substrate of CYLD.
Conditional KD of CYLD in adult mice prevents DA neurodegeneration under conditions of parkin deficiency
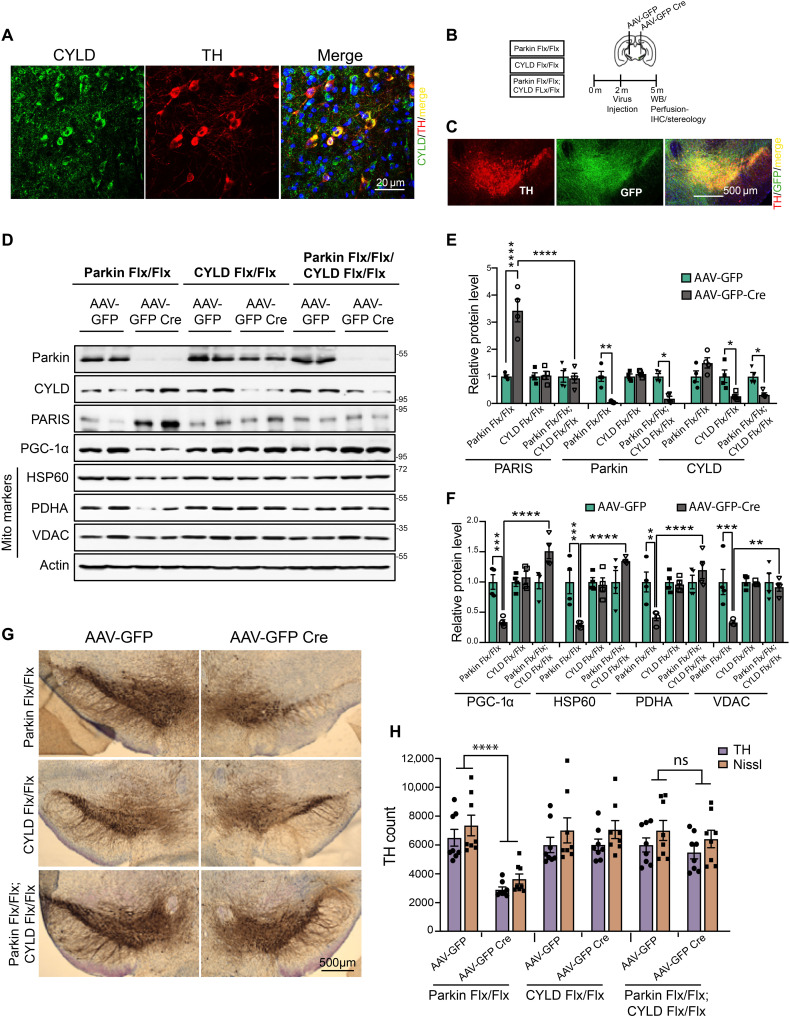
Conditional knockout of parkin in adult mice leads to degeneration of dopamine neurons that is dependent on PARIS (8, 9). CYLD is expressed in multiple regions of the mouse brain (16–18), including the substantia nigra (SN) dopamine neurons, where it predominantly localizes to the cytosol (Fig. 6A). We therefore investigated whether CYLD inactivation in adult mice could ameliorate PARIS-dependent DA degeneration. CYLD was knocked down along with parkin by stereotactically injecting adeno-associated virus serotype 2 (AAV2) expressing GFPCre into the ventral midbrain of 2-month-old parkin Flx/Flx; CYLD Flx/Flx mice (Fig. 6B). Stereotactic injection with AAV2-GFPCre, which efficiently transduces TH-positive dopamine neurons (Fig. 6C), leads to over 80% reduction in CYLD and parkin protein levels in these mice that were assessed 3 months after the stereotactic injections (Fig. 6, D and E). While PARIS levels are markedly increased in the absence of optimal levels of parkin activity, depletion of CYLD in the conditional parkin KD animals significantly suppressed such toxic accumulations of PARIS (Fig. 6, D and E). Furthermore, CYLD KD relieves the repressive effect PARIS accumulation has on PGC-1α–dependent mitochondrial protein synthesis in the parkin KD mice (Fig. 6, D and F). To examine whether CYLD KD abrogates the detrimental effects PARIS accumulation has on dopamine neuron survival in the parkin KD mice, the number of dopamine neurons was assessed via stereological analysis of TH- and Nissl-positive neurons in the SN pars compacta (SNpc) of parkin Flx/Flx, CYLD Flx/Flx, and parkin Flx/Flx; CYLD Flx/Flx mice. Significant dopamine neuron loss in the parkin KD mice becomes discernible 3 months after injection with AAV2-GFP Cre. However, conditional KD of CYLD in the parkin Flx/Flx mice protects against dopamine neuron loss (Fig. 6, G and H). There was no loss of dopamine neurons in the AAV-GFP Cre–injected CYLD Flx/Flx mice that was comparable to that in the AAV-GFP–injected CYLD Flx/Flx control group (Fig. 6, G and H). Since the AAV injections were done unilaterally, amphetamine-induced rotation was used as a functional behavioral readout of DA degeneration. Consistent with the loss of dopamine neurons, conditional parkin KD led to a significant increase in amphetamine-induced rotational behavior that was rescued by CYLD KD (fig. S6A). In assessments of motor performance by the pole test, CYLD KD also rescued motor deficits in the conditional parkin KD mice evident from the decreased time to turn and time to reach the base (fig. S6B and movie S3). In grip strength analyses, CYLD KD improved forelimb and all limbs grip strength in the conditional parkin KD mice (fig. S6C).
Fig. 6. CYLD KD prevents DA neurodegeneration in adult conditional parkin KD mice.
(A) Representative confocal images showing CYLD coexpression in TH-immunostained SN dopamine neurons in midbrain sections from WT C57BL/6 mice. Scale bar, 20 μM. (B) Schematic of experimental schedule following stereotaxic injection with AAV-GFP or AAV-GFP Cre in SN of 2-month-old C57BL6 mice of indicated genotype. (C) Representative immunofluorescence images verifying stereotaxic injection quality. Scale bar, 500 μM. (D) Representative immunoblots of indicated proteins in ventral midbrain lysates from adult mice harboring conditional KD of parkin, CYLD, or a combination of parkin and CYLD. (E) Relative quantification of PARIS, parkin, and CYLD in ventral midbrain lysates from floxed mice of indicated genotypes stereotaxically injected with AAV-GFP or AAV-GFP-Cre. Mean values from four independent stereotaxic injections in each case shown. (F) Relative quantification of PGC-1α and the indicated mitochondrial proteins in ventral midbrain lysates as shown in (C). N = 4 independent stereotaxic injections with AAV-GFP or AAV-GFP-Cre. (G) Representative TH immunostaining of midbrain sections from SN of mice homozygous for the floxed parkin, CYLD, or parkin; CYLD alleles injected with AAV-GFP or AAV-GFP-Cre. (H) Stereotaxic assessment of TH- and Nissl-positive neurons in the SN pars compacta (SNpc) of indicated injection groups (N = 8 unilaterally injected mice per group). Quantitative data = means ± SEM. One-way ANOVA with Tukey post hoc test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. See also fig. S6.
CYLD down-regulation in hES cell–derived midbrain dopamine neurons attenuates PARIS neurotoxicity under conditions of parkin deficiency
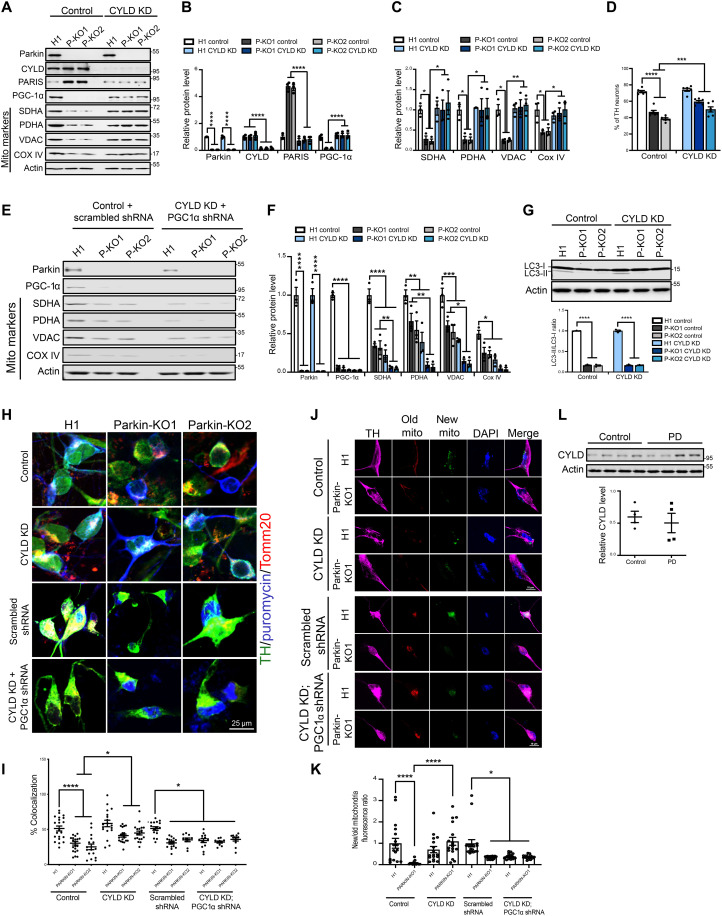
In differentiated midbrain dopamine neurons derived from hES cells, loss of parkin leads to defects in mitochondrial biogenesis stemming from toxic accumulations of PARIS (19). We therefore examined whether inactivation of CYLD in the parkin-deficient hES cell–derived midbrain dopamine neurons could abrogate the mitotoxic effects of PARIS accumulation. To this end, CYLD was down-regulated in the H1 control and parkin-deficient human midbrain dopamine neurons using lentiviral CRISPR-Cas9–mediated gene editing technique. Intriguingly, an ~50% reduction in endogenous CYLD levels was sufficient to prevent PARIS accumulation in the parkin-deficient human midbrain dopamine neurons (Fig. 7, A and B). CYLD KD also restored the neuronal protein levels of PGC-1α and other mitochondrial markers that are substantially reduced in the parkin-deficient midbrain dopamine neurons under conditions of PARIS accumulation (Fig. 7, A to C). The absence of parkin leads to substantial loss of TH-positive dopamine neurons, and CYLD KD partially rescues this neuronal loss (Fig. 7D). Thus, CYLD KD facilitates PGC-1α–dependent gene expression changes that promote mitochondrial protein synthesis and dopamine neuron survival. To examine whether CYLD specifically affects PGC-1α–dependent mitochondrial biogenesis, we knocked down PGC-1α in the parkin-deficient midbrain dopamine neurons using shRNA under conditions of CYLD KD. Relative to the scrambled RNA–treated control, shRNA KD of PGC-1α led to over 90% reduction in PGC-1α protein levels and abrogated the rescue effect CYLD KD has on the mitochondrial markers in the parkin-deficient neurons. (Fig. 7, E and F). In addition to defects in mitochondrial biogenesis and respiratory capacity, parkin-deficient human midbrain neurons also exhibit deficiencies in mitophagy (19). To examine whether CYLD KD affects mitophagy, we evaluated levels of the autophagic markers LC3-I and LC3-II as a measure of mitophagy in the parkin-deficient human midbrain neurons. Conversion of LC3-I to LC3-II occurs during autophagy, and an increase in LC3-II protein levels with a concomitant decrease in LC3-I levels serves as a good indicator of autophagosome formation (20). Consistent with previous observations, a significant reduction in LC3-II to LC3-I ratio is observable in the parkin-deficient human DA (hDA) neurons compared to the control condition (Fig. 7G). However, the LC3-II to LC3-I ratio in the parkin KO hDA neurons remains unaffected under conditions of CYLD KD (Fig. 7G). This indicates that the protective effects mediated by CYLD KD are predominantly through prevention of PARIS accumulation and the resultant augmentation of mitochondrial biogenesis. To further verify the effect of CYLD on mitochondrial biogenesis, we used surface sensing of translation (SUnSET), a protein-labeling method based on incorporation of puromycin into newly synthesized proteins and its detection with anti-puromycin antibodies (21). To measure the rate of mitochondrial protein synthesis, the H1 control and parkin-deficient human midbrain dopamine neurons in culture were pulsed with puromycin. Anti-puromycin and anti-Tomm20 antibodies were then used to fluorescently tag puromycin-labeled mitochondrial peptides within TH-positive dopamine neurons. Quantitative measurements of puromycin-labeled peptides that colocalized with the mitochondrial Tomm20 marker reveal a significant reduction in mitochondrial protein synthesis in the parkin-deficient human midbrain dopamine neurons that is effectively restored by CYLD KD (Fig. 7, H and I). However, shRNA KD of PGC-1α in addition to reducing mitochondrial protein synthesis in the H1 control also abolished the rescue effects mediated by CYLD KD in the parkin-deficient human midbrain neurons (Fig. 7, H and I). We next monitored mitochondrial turnover using soluble SNAP-tag, a self-labeling protein tag that allows covalent fusion of specific ligands to any protein of interest (22). Specifically, we used SNAP-Cox8A that consists of the inner mitochondrial protein, cytochrome C oxidase, subunits 8-2 (COX8-2) fused to the N terminus of the SNAP-tag. This COX8-2–SNAP fusion protein when labeled with distinct SNAP substrates allows mitochondria to be labeled with different fluorescence tags and offers the feasibility of measuring turnover rates of labeled mitochondria over time. Human midbrain dopamine neurons were transduced with the COX8-2–SNAP fusion tag, and old versus newly formed mitochondria in TH-positive dopamine neurons were distinguished using a red (TMR-Star) and a green (Oregon Green) SNAP-tag substrate, respectively. As previously reported (19), parkin-deficient dopamine neurons (P-KO1) exhibit a significant reduction in the proportion of newly formed mitochondria. However, this is restored to control levels under conditions of CYLD KD (Fig. 7, J and K), indicating that reduction in CYLD levels promotes mitochondrial biogenesis. Furthermore, shRNA KD of PGC-1α reduces formation of new mitochondria by ~50% in the H1 control and also abrogates the rescue effect CYLD KD has in the parkin-deficient dopamine neurons (Fig. 7, J and K), providing further support that CYLD KD facilitates PGC-1α–dependent transcriptional events that promote mitochondrial biogenesis. We next examined whether CYLD levels are altered in postmortem PD samples. Immunoblot analysis of ventral midbrain samples from postmortem PD brains shows no significant difference in CYLD protein levels compared to age-matched controls (Fig. 7L).
Fig. 7. CYLD down-regulation attenuates PARIS-mediated neurotoxicity in hES cell–derived midbrain dopamine neurons deficient for parkin activity.
(A) Immunoblot analysis of the indicated proteins in control (H1) and two independent parkin knockout (P-KO1 and P-KO2) human midbrain dopamine neurons. (B) Quantification of indicated proteins in the different conditions. Mean values from three independent experiments shown. (C) Quantification of indicated mitochondrial markers in the different conditions. N = 3 independent experiments. (D) Quantification of TH-positive dopamine neurons in differentiated human midbrain cultures immunostained with TH and neuronal TUJ1 marker. N = 6 independent experiments (E) Immunoblot analysis of mitochondrial biomarkers in indicated conditions. (F) Quantification of indicated proteins under respective conditions. N = 3 independent experiments. (G) Immunoblot analysis and quantification of LC3II/I ratio in differentiated human midbrain neurons. N = 3 independent experiments. (H) Representative confocal images of human midbrain dopamine neurons immunostained for TH (green) and puromycin. Puromycin-labeled mitochondrial proteins detected by colocalization of anti-puromycin immunofluorescence (magenta) with the mitochondrial marker Tomm20 (red). (I) Quantification of anti-puromycin colocalization with Tomm20 in TH-positive midbrain dopamine neurons. Mean values from at least 15 TH-positive midbrain dopamine neurons from three independent differentiation shown. (J) Representative confocal images of human midbrain dopamine neurons expressing SNAP-Tag-Cox8a fusion protein immunostained for TH (magenta). Existing mitochondrial pool (old mito) shown in red and mitochondria newly assembled (new mito) shown in green. Nucleus stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (K) Ratio of newly assembled to old mitochondria in TH-positive midbrain dopamine neurons. Mean values from at least 15 TH-positive midbrain dopamine neurons from three independent differentiations shown. (L) Immunoblot analysis and quantification of CYLD expression in SN from postmortem PD and age-matched controls. N = 4 per group. Quantitative data = means ± SEM. One-way ANOVA; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
DISCUSSION
Dysfunction in mitochondrial quality control is emerging as a common feature in both familial and sporadic forms of PD. Involvement of PINK1 and parkin in multiple aspects of mitochondrial quality control suggests that PINK1/parkin signaling could be a point of convergence of common pathogenic mechanisms likely at play in PD pathogenesis. Furthermore, the pathogenic substrate, PARIS, which is subject to proteasomal regulation by PINK1 and parkin, directly ties these proteins to dopamine neuron survival and thus represents a nodal point in the PINK1/parkin pathway around which novel therapeutics could be developed to augment mitochondrial health. To this end, using a sequential series of DUB-focused RNAi screens, we have identified CYLD to be a deubiquitinase that functions as part of the PINK1/parkin pathway to regulate cellular levels of PARIS. We show here that DA KD of CYLD in Drosophila promotes neuron survival under conditions of PINK1 or parkin insufficiency. We also show that CYLD interacts with PARIS and reverses PARIS ubiquitylation to increase protein stability, an effect that is highly conserved across species. Reducing CYLD activity has neuroprotective effects in adult conditional parkin KD mice or differentiated human dopamine neurons deficient for parkin activity. In the parkin-deficient human dopamine neurons, such CYLD KD–mediated neuroprotective effects are abrogated in the absence of PGC-1α, a crucial regulator of mitochondrial biogenesis. These findings indicate that suppression of PARIS accumulation under conditions of CYLD KD effectively relieves the repressive effect PARIS has on PGC-1α–dependent transcriptional programs with a net effect of promoting mitochondrial biogenesis to sustain optimal mitochondrial functions crucial for dopamine neuron survival.
CYLD, originally identified as a tumor suppressor, is implicated in numerous polyubiquitin-dependent signaling pathways critical to the regulation of inflammatory responses (23, 24), cell proliferation (25–27), and cell death (28–32). Although CYLD has not been extensively studied in a neuronal context, it is highly expressed in the brain (33, 34), promotes dendritic and spine morphogenesis (35), and is enriched at the striatal synapses where it interacts with numerous proteins of the striatal synaptosome (16). CYLD is also localized to the hippocampal postsynaptic densities where it regulates activity-dependent clustering of PSD-95 with potential implications for synaptic organization, function, and plasticity (36–38). Maintenance of functional synaptic connections also crucially depends on the synaptic availability of mitochondria to meet their adenosine triphosphate (ATP) requirements (39). In this regard, the increased mitochondrial biogenesis observable under conditions of CYLD KD would in essence increase the pool of functional mitochondria for synaptic trafficking by augmenting the synthesis of mitochondrial proteins. This becomes even more crucial in SN dopamine neurons that have proportionally greater energy requirements owing to their remarkably extensive axonal arbors and numerous synaptic connections (40). Thus, our observation of the role CYLD plays within the dopamine neurons to promote mitochondrial network homeostasis via the PARIS/PGC-1a axis adds to its repertoire of neuronal functions and indicates that modulation of CYLD activity could ameliorate nigral synaptic transmission deficits.
Several other DUBs are now implicated in PD etiology (41–43) and some are also known to affect PINK1/parkin-mediated mitophagy in cultured mammalian cells (43, 44), raising the possibility that modulating the activity of such DUBs could hold therapeutic potential in PD. In support of this notion, studies in dPINK1 or parkin null mutants show that genetic or pharmacological inhibition of mitophagy-related DUBs like USP8, USP14, USP15, or USP30 ameliorates mitochondrial dysfunctions in adult flight muscles and/or rescues locomotor deficiencies observable in the PINK1/parkin-deficient flies (44–46). However, there is no direct evidence to date to indicate that defects in mitophagy persist specifically within the dopamine neurons in Drosophila or any other in vivo PD model (47). Moreover, reports on dopamine neuron degeneration in the dPINK1 and parkin Drosophila mutants are highly variable (48–53), making any inferences drawn from these models on the link between mitochondrial dysfunctions and DA neurodegeneration rather inconclusive. In our fly DUB screen, USP10 was one other DUB that, when knocked down, rescued climbing defects in the PINK1/parkin-deficient flies and PARIS-overexpressing flies. USP10 activation has recently been shown to promote dopamine neuron survival by stimulating the antioxidant Nrf2 activity (54). However, whether USP10 has a role in mitophagy or has mitochondrial functions remains unclear. Moreover, in our follow-up studies, USP10 KD rescued dopamine neuron loss only in the PINK1-deficient flies, suggesting that USP10 could function in a parkin-independent manner. Hence, the pharmacological utility of the mitophagy-related DUBs in PD therapeutics remains a conjecture. Conditional KD of PINK1 or parkin in adult animals, on the other hand, leads to progressive loss of dopamine neurons and behavioral abnormalities that directly stem from PARIS-mediated repression of mitochondrial biogenesis (7–10). Notably, this effect is highly conserved from Drosophila to mouse models, and the inverse relationship between PARIS and PGC-1α observed in these models is similar to what occurs in human postmortem PD brain tissues. Moreover, PARIS plays an important role in neurodegeneration in sporadic models of PD (11, 13). Notably, CYLD KD prevents PARIS accumulation under conditions of PINK1 or parkin inactivation and promotes DA neuronal survival and motor performances in Drosophila and mouse models of PARIS accumulation. The fact that CYLD KD mediates similar neuroprotective effects in human midbrain dopamine neurons indicates that modulating CYLD activity could be a viable therapeutic avenue to alter the progression of PD resulting from reduced PINK1 or parkin function. Mitochondrial biogenesis not only augments mitochondrial protein synthesis to meet cellular bioenergetic demands but also serves to replace depleted parts. Thus, the PINK1/parkin pathway could promote proteasomal degradation of PARIS to counter mitochondria that are eliminated via mitophagy as a homeostatic mechanism. In the absence of optimal PINK/parkin activity, CYLD KD could serve to replenish a healthy pool of functional mitochondria by preventing PARIS accumulation and promoting biogenesis through PGC-1α. The downstream targets of PGC-1α include genes encoding for all five respiratory complex (RC) subunits. Hence, activation of PGC-1α–dependent transcriptional events under conditions of CYLD KD could facilitate the synthesis and assembly of RC components with feed-forward effects on mitochondrial energy metabolism.
While we do not find any evidence for changes in CYLD protein levels in postmortem PD samples, our studies in multiple model systems indicate that reducing CYLD activity is beneficial under conditions of suboptimal PINK1/parkin functions. CYLD inactivation is shown to have beneficial effects in other models of neuronal toxicity as well. For instance, CYLD depletion prevents glutamate toxicity in vitro and exhibits significant protection against traumatic neuronal injury in vivo (32). Recently, a novel missense variant in CYLD (M719V) was found to be associated with autosomal dominant inheritance of frontotemporal dementia and amyotrophic lateral sclerosis (34). Intriguingly, compared to the WT protein, mutant CYLDM719V exhibits increased deubiquitinase activity and leads to impaired autophagy. Together, these studies provide a strong rationale to design and test specific CYLD inhibitors that could have translational potential as therapy for PD and other neurodegenerative diseases.
MATERIALS AND METHODS
Experimental design
Cell lines
Drosophila S2 cells (DGRC 181) were cultured in M3 + BYPE medium (pH 6.6) supplemented with fetal bovine serum (HyClone, SH30070.03HI) and penicillin-streptomycin solution (Sigma-Aldrich, P4333). The M3 + BYPE medium contained Shields and Sang powdered medium (39.4 g/liter; Sigma-Aldrich, S8398-1), potassium bicarbonate (0.5 g/liter; Sigma-Aldrich, 367877), yeast extract (1 g/liter; Sigma-Aldrich, Y-1000), and bactopeptone (2.5 g/liter; Difco, 211677). Cell cultures and transfected cells were maintained at 25°C without CO2. Human neuroblastoma SH-SY5Y cells were cultured in 1× Dulbecco’s modified Eagle’s medium (DMEM; Gibco-Invitrogen), supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA), penicillin (1000 U/ml), and streptomycin solution (1 mg/ml; Biochrom AG, Berlin, Germany). Cultured and transfected cells were grown in a humidified incubator at 37°C, in a 5% CO2 atmosphere. Transfections of S2 and SHSY-5Y cells were carried out using X-tremeGENE HP DNA Transfection Reagent (Sigma-Aldrich, 6366244001) following the manufacturer’s protocol.
Fly stocks and maintenance
Drosophila melanogaster fly stocks were handled using standard protocols, maintained in a 12-hour light/dark cycle, and fed Drosophila-standard diet consisting of cornmeal, agar, yeast, sucrose, and dextrose. All experimental crosses were kept at 25°C. In all experiments, both male and female flies were used. Transgenic stocks, DUB-specific RNAi lines, and Gal4 lines were obtained from the Bloomington Drosophila Stock Center (BDSC), the Vienna Drosophila RNAi stock Center, the FlyORF, or were gifts (see table S1 for details). In each experiment, the relevant Gal4 heterozygous flies were used as control. Flies expressing shRNA-targeting GFP (BDSC, 41556) were used as control for all shRNA induction studies.
Plasmids and transfection
Plasmids used are as follows: Cloning of pMT-dPARIS is described in (10), pAMW-Myc-dCYLD WT and pAMW-Myc-dCYLD C284S plasmids are described in (55) and were obtained from F. Ikeda’s laboratory. Human CYLD (hCYLD) CRISPR guide RNAs (gRNAs) were computationally designed using gRNA design software (Benchling, San Francisco, CA, USA) and cloned into the lentiCRISPR v2 plasmid (Addgene, catalog no. 52961) at Bsm BI restriction enzyme sites upstream of the scaffold sequence of the U6-driven gRNA cassette. The lentiCRISPR v2 vector also includes SpCas9 and a puromycin resistance selection marker that was used to select for stably transfected SHSY-5Y cells. The lentiCRISPR V2 vector encoding the hCYLD gRNA was packaged as described previously (56). pCMV6-AC-CYLD-GFP plasmid to express C-terminal GFP-tagged hCYLD was obtained from Origene (catalog no. RG219629). N-terminal V5-tagged full-length hCYLD or truncated forms of hCYLD were PCR-amplified and cloned into the pSF-CMV-NH2-V5-EKT-Ncol vector (Oxford Genetics, catalog no. OG91) at the Xho I-Nhe I restriction sites using standard cloning techniques. Coding sequence of full-length hPARIS or its truncation domains was PCR-amplified and cloned into pCMV-3Tag-3 mammalian expression vector (Stratagene, catalog no. 240197) using standard cloning techniques. For recombinant protein purifications, full-length hCYLD WT, hCYLD C601A mutant, or hPARIS WT was cloned into pGEX-6P-1 bacterial expression vector using standard cloning techniques. pRK5-HA-Ubiquitin-WT, pRK5-HA-Ubiquitin-K48 only, and pRK5-HA-Ubiquitin-K63 only plasmids have been previously described (57). PGC-1α (PPARGC1A) human shRNA lentiviral particles were obtained from Origene (catalog no. TL310260V). Cloning and characterization of pMT-V5/His-dPARIS plasmid are described in (10).
Climbing assay
Climbing assays were performed as described previously (58). Flies were collected immediately following eclosion and maintained in batches of 20° at 25°C in vials containing standard food media. Each batch of flies was transferred to an empty 9-cm vial (Genesee Scientific, 32-113) with a line drawn 2 cm from the bottom. Flies were tapped to the bottom to induce an innate climbing response. The number of flies that crossed the 2-cm mark in 15 s was counted. Five to ten technical replicates were performed for each batch to ensure an accurate reading at each time point, and average of the independent trials was used to calculate the percentage of flies with climbing defects.
Immunostaining of Drosophila brains
Drosophila brain dissection and immunostaining were performed as described previously with modifications (59). Briefly, adult Drosophila brains were fixed in 4% paraformaldehyde (PFA) for 45 min at room temperature (RT), washed three times in 1× phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (wash buffer), and blocked in PBS containing 5% normal goat serum and 0.2% Triton X-100 overnight at 4°C. Brains were then incubated in primary antibody in blocking solution for at least 48 hours at 4°C, followed by three 10-min washes at RT. Secondary antibody was applied in blocking solution, and samples were incubated for 48 hours at 4°C. Following five 10-min washes at RT, brains were mounted on glass slides using ProLong Gold antifade mountant (Life Technologies, P36930).
Drosophila brain imaging and quantification
Confocal microscopy and image acquisition were performed with a Zeiss LSM710 laser scanning confocal microscope at a magnification of ×40 using procedures described previously (10). Images were scanned at 1024 × 1024 pixels, with a slice thickness of 1 μM and line and frame average of 4. Sequential scanning of the different channels was used to avoid bleed-through. Imaging settings were first determined for the control conditions in each experiment, which were then maintained for the other experimental conditions. Stacked Z-projections of images were created for DA neuron counts, and cell bodies stained with anti-TH were manually counted. The investigator was blinded to the individual genotype information while performing the neuronal counts. ImageJ software [National Institutes of Health (NIH)] was used to measure the integrated intensity of mito-GFP relative to TH. TH intensity in the 568-nm channel (marking DA neurons) was used to select regions of interest (ROIs). These ROIs were then transferred to the 488-nm channel to measure fluorescence intensity of mito-GFP–labeled mitochondria within the same ROIs.
FACS of TH neurons
FACS of TH neurons was performed following procedures described previously in (10). Transgenic fly lines carrying the TH-Gal4; UAS-GFP constructs were crossed with flies carrying the respective UAS construct. F1 progeny that expresses GFP and the respective transgene in the DA neurons in each case were collected and aged for 20 days to be used for FACS analysis as previously described with modifications (60). Fifty adult brains were dissected and mechanically dissociated in TrypLE Express (Gibco) by incubating at 37°C for 40 min followed by passage through a syringe fitted with a 29-gauge needle 5 to 10 times. Flies were maintained at RT during brain dissections to avoid temperature-induced induction of gene expression in the DA neurons. Following dissociation, cells were pelleted at low speed, resuspended in DMEM (Gibco) supplemented with 10% fetal bovine serum (Hyclone), and passed through a 70-μM mesh strainer. To label live cells, the membrane permeant Vybrant DyeCycle Ruby stain (Life Technologies) was applied to the cell suspension before sorting. Cells were maintained at 4°C until flow analysis and sorting that was always performed within 30 min of completion of the protocol. Cells were subjected to FACS using a SH800 (Sony, IL) flow cytometer fitted with a 100-μM nozzle at 20 psi. GFP and DyeCycle Ruby detection was done using a 488- and 635-nm lasers, respectively. Samples were sorted at 4°C and the sorted cells were collected in 0.5 ml of TRIzol-LS (Invitrogen) or DMEM media supplemented with 10% fetal bovine serum (Hyclone) for RNA and DNA isolation, respectively. The yield of GFP-positive cells ranged from 2000 to 2500 cells per sort per 50 adult fly brains.
RNA and DNA extraction from FACS neurons
RNA was extracted from DA neurons after cell sorting using the TRIzol LS Reagent (Invitrogen) according to the manufacturer’s instructions. Following deoxyribonuclease I (Invitrogen) treatment, RNA was used for cDNA preparation using the Superscript III first-strand synthesis system (Thermo Fisher Scientific) following the manufacturer’s protocol. Genomic DNA extraction from DA neurons after cell sorting was performed as previously described (10) by pelleting FACS-sorted neurons by centrifuging at 4000 rpm for 10 min. The pellet was resuspended in 100 μl of 1× PBS containing 20 mM EDTA followed by addition of 100 μl of 2× proteinase K buffer [100 mM tris-HCl (pH 7.5), 50 mM EDTA, and 2% SDS]. After vortexing, proteinase K was added to the sample to final concentration of 500 μg/ml and incubated overnight at 56°C. An equal volume of phenol/chloroform/isoamyl alcohol was added to the DNA solution, following which DNA was precipitated by adding 1:10 volume of 3 M sodium acetate (pH 5.2) and 1 volume of 100% EtOH and centrifuged at 13000g for 10 min. The DNA pellet was then washed with 70% ethanol, air-dried, and resuspended in 20 μl of distilled water.
Real-time quantitative PCR
Real-time quantitative PCR was performed on an Applied Biosystems ViiA 7 Real-Time PCR System using the Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific) as described in (10). Real-time PCR reactions were carried out in duplicates in 20 μl reaction volumes using 2 μl of cDNA template and 1.5 μM each of forward and reverse primer. Reactions were performed in 384-well plates for 40 amplification cycles at 95°C for 15 s, 60°C for 45 s, and 72°C for 1 min with plate readings recorded after each cycle. Threshold cycle (Ct) values were obtained, and the ΔΔCT method was used to calculate the fold change in the transcript level of the sample relative to the control. RP49 was used as an internal standard and reference gene. Primers used are described in (10).
Measurement of mtDNA density
The relative number of mtDNA genomes per diploid nuclear genome in the different genotypes was determined using quantitative real-time PCR as described previously (10). FACS-sorted DA neurons were used for genomic DNA isolation. A 105–base pair (bp) region of the mitochondrial large ribosomal RNA gene (CR34094) and a 249-bp region of the mitochondrial cytochrome B gene (CG34090) were used for mtDNA quantification. A 215-bp region of the single-copy nuclear gene that encodes the 215-kDa subunit of RNA polymerase II (CG1554) was used for nuclear genomic DNA quantification. The amount of mtDNA was assessed by calculating the ratio of mtDNA to nuclear DNA (nuDNA) copy number. Primers used are described in (10).
Coimmunoprecipitation
Drosophila S2 cells were lysed in S2 lysis buffer [50 mM tris (pH 7.5), 150 mM NaCl, 5 mM EDTA, and 1% NP-40] supplemented with EDTA-free protease inhibitor cocktail (Roche). Following a 15-min incubation on ice, samples were centrifuged at 12000g for 10 min. Human neuroblastoma SHSY-5Y cells were lysed in Triton lysis buffer [50 mM tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton and supplemented with EDTA-free protease inhibitor cocktail (Roche)] and incubated on ice for 30 min to ensure complete lysis. Samples were then centrifuged at 12000g for 10 min. Protein concentrations in the lysates (supernatant) were estimated using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific), and equivalent amounts of lysates were used in coimmunoprecipitation experiments. Anti-FLAG M2 magnetic beads (Sigma-Aldrich), anti-Myc beads (Chromotek), and anti-V5 tag monoclonal antibody magnetic beads (MBL International) were used as per the manufacturer’s protocol for FLAG or V5 pull-down experiments, respectively. Lysates were incubated with 40 μl of FLAG/V5 beads or 20 μl of Myc beads overnight at 4°C in a rotating wheel. The respective beads were washed thrice with S2 or Triton lysis buffer (for S2 or SHSY-5Y cells, respectively), and bound proteins were eluted in 1× Laemmli sample buffer (Bio-Rad, 1610737) containing 2-mercaptoethanol. A fraction of the lysates was also mixed in a 1:1 ratio with 2× Laemmli sample buffer containing 2-mercaptoethanol to be used as input.
GST pull-down assays
For in vitro GST pull down, purified recombinant GST-PARIS or GST-CYLD fusion protein was immobilized on glutathione-agarose beads (Thermo Fisher Scientific) and incubated with recombinant CYLD WT or PARIS in reaction buffer [250 mM tris-HCl (pH 7.6), 500 mM NaCl, 25 mM MgCl2, 5 mM dithiothreitol (DTT), 0.5 mM EGTA, and 20 mM freshly prepared ATP] for 2 hours at 4°C on a rotating wheel. After washing the glutathione-agarose beads thrice in reaction buffer, samples were eluted in 1× Laemmli buffer containing 2-mercaptoethanol.
Immunoblot analysis
Drosophila heads collected from cold anaesthetized flies were homogenized in lysis buffer (20 mM Hepes, 100 mM KCl, 5% glycerol, 10 mM EDTA, 0.1% Triton, and 1 mM DTT) supplemented with EDTA-free protease inhibitor cocktail (Roche) using a micro-tissue grinder. Following a 30-min incubation on ice, samples were centrifuged at 12000g for 10 min. S2 or SHSY-5Y cell lysates were prepared as described in the previous section. For mouse studies and postmortem brain sample analysis, lysates from ventral midbrain tissues from indicated genotypes were prepared as previously described (9). All experiments that included human postmortem brain samples were carried out in accordance with the human Institutional Review Board at the Johns Hopkins University. Protein concentrations in each case were estimated using a BCA protein assay kit. Lysates in each case were mixed in a 1:1 ratio with 2× Laemmli sample buffer (Bio-Rad) containing 2-mercaptoethanol. Equivalent amounts of protein were electrophoresed in 10% tris-glycine SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to Hybond enhanced chemiluminescence nitrocellulose membrane (GE Healthcare). Membranes were blocked for 1 hour using 1× tris-buffered saline (TBS) supplemented with 0.05% Tween-20 (1× TBST) and 5% nonfat milk followed by overnight incubation at 4°C in block containing corresponding primary antibody. The next day, following three 10-min washes in 1× TBST, membranes were incubated in a corresponding secondary antibody (1:5000) in 1× TBST with 5% nonfat dry milk for at least 1 hour at RT. Membranes were thereafter washed thrice with 1× TBST and imaged by electrochemiluminescence. Images were collected digitally, and band intensities were quantified by densitometry using ImageJ software (NIH) and normalized to the loading control.
Antibodies
Primary antibodies used for immunoblot analyses include rabbit antibody to hPARIS (1:2000; Proteintech, catalog no. 24543-1-AP), rabbit antibody to Drosophila PARIS [1:1000; (10)], rabbit antibody to CYLD (1:1000, Cell Signaling Technology, catalog no. 8462), rabbit antibody to CYLD (1:1000; Sigma-Aldrich, SAB4200060), mouse antibody to parkin (Prk8) (1:1000; Cell Signaling Technology, catalog no. 4211), and rabbit antibody to PGC-1a (1:1000; Novus Biologicals, catalog no. NBP1-04676). Anti-FLAG M2 (Sigma-Aldrich, catalog no. F3165), horseradish peroxidase (HRP)–conjugated anti-Myc (Cell Signaling Technology, catalog no. 2040), anti-V5 (Cell Signaling Technology, catalog no. 13202S), or anti-HA antibody (Sigma-Aldrich, catalog no. H9658) antibodies were used for detection of FLAG, Myc, V5, or HA-tagged proteins, respectively. Anti-ubiquitin antibody (1B4-UB, Abcam, catalog no. ab122) was used to detect polyubiquitin modifications of PARIS. Anti–maltose binding protein (MBP) (1:1000; New England Biolabs, E8032S) was used for detecting MBP-tagged recombinant Tc PINK1. The Mitochondrial Marker Antibody Sampler Kit (Cell Signaling Technology, catalog no. 8674) was used to detect indicated mitochondrial proteins. Actin used as loading control was detected using anti–β-actin antibody (1:10000; Abcam, catalog no. ab49900). Anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG HRP–conjugated secondary antibody (1:2000; Cell Signaling Technology, catalog nos. 7076 and 7074) was used to detect protein bands by electrochemiluminescence.
Primary antibodies used for immunohistochemistry/immunocytochemistry are as follows: mouse anti-TH antibody (1:1000; Immunostar, catalog no. 22941) and chicken anti-GFP antibody (1:500; Millipore, catalog no. AB10145) for Drosophila brains, rabbit anti-TH antibody (1:1000; Novus Biologicals, catalog no. NB300-109) for mouse tissues, and mouse anti-Tomm20 antibody (1:1000; Abcam, catalog no. ab56783) and anti-puromycin (1:1000l; Sigma-Aldrich, catalog no. MABE343) for human dopamine neurons. Secondary antibodies were goat anti-mouse, anti-rabbit, or anti-chicken Alexa fluorochromes at 1:1000 (Thermo Fisher Scientific, catalog nos. A-11032, A32731, and A-11039).
Analysis of protein turnover
To monitor dPARIS turnover, dPARIS was cotransfected in S2 cells with WT or mutant (C284S) dCYLD. dPARIS was expressed under the control of a Drosophila metallothionein (MT) gene promoter (pMT-V5/His-dPARIS plasmid) that allows for protein expression upon addition of CuSo4 to the culture medium. Twenty-four hours after transfection, CuSo4 was added to the culture medium to a final concentration of 500 μM to induce dPARIS expression. After another 48 hours, culture medium was replaced with fresh media without CuSo4 to inhibit dPARIS expression. dPARIS turnover was then monitored in the presence of WT or mutant dCYLD at the indicated time points. hPARIS turnover rates were monitored in SHSY-5Y cells by addition of cycloheximide to a final concentration of 50 μM, and lysates were assayed at the indicated time points.
Deubiquitination assays
In cell deubiquitination, assays were performed using SH-SY5Y cells under CYLD KD or overexpressing conditions. CYLD KD was achieved using CRISPR-/Cas9–mediated gene editing. Transient transfection using pCMV6-CYLD-GFP plasmid was used to overexpress CYLD. Cells were cotransfected with N-terminal FLAG-tagged hPARIS with either WT ubiquitin or ubiquitin mutants that can append only K48 (HA-Ub-K48 only) or K63 (HA-Ub-K63 only) Ub chains under CYLD KD or overexpressing conditions. Following coimmunoprecipitation of PARIS using anti-FLAG M2 magnetic beads (Sigma-Aldrich) as described above, polyubiquitin modifications of PARIS were detected in immunoblots using anti-HA antibody.
In vitro deubiquitination assays were performed as previously described (9). Recombinant GST, GST-PARIS, GST-parkin, GST-CYLD WT, and GST-CYLD C601A mutant were expressed in Escherichia coli and purified using glutathione-agarose beads. GST tag on parkin, CYLD, and CYLD C601A mutant was cleaved using PreScission protease. Two micrograms of GST-PARIS was incubated with 40 ng of UBE1 (E1, Boston Biochem, catalog no. E-305), 200 ng of UbcH7 (E2, Boston Biochem, catalog no. A-640), 1 μg of His-Ub (Boston Biochem, catalog no. U-530), 500 ng of parkin, and 500 ng of His/MBP-PINK1 (Boston Biochem, catalog no. AP-180) together with 500 ng of CYLD WT or CYLD C601A mutant in reaction buffer [250 mM tris-HCl (pH 7.6), 500 mM NaCl, 25 mM MgCl2, 5 mM DTT, 0.5 mM EGTA, and 20 mM freshly prepared ATP] at 37°C for 2 hours with agitation. The reaction mixture was then subjected to SDS-PAGE and Western blot analysis. Polyubiquitination of GST-PARIS was detected using anti-PARIS antibody. Autoubiquitination of parkin was detected using anti-parkin antibody.
Maintenance and differentiation of hES cells into dopamine neurons
H1 hES cells (Wi Cell, Madison, WI) were cultured using standard protocol on inactivated mouse embryonic fibroblast. All experiments that included hES cells were carried out in accordance with the human Institutional Review Board and the Institutional Stem Cell Research Oversight Committee at the Johns Hopkins University. Differentiation of hES cells to dopamine neurons was done as described in (61). Single hES cells were cultured on Matrigel-coated plate at a density of 40,000 cells/cm2 in serum replacement medium (SRM) containing growth factor and small molecules, including FGF8a (100 ng/ml), SHH C25II (100 ng/ml), 100 nM LDN193189, 10 μM SB431542, 3 μM CHIR99021, and 2 μM purmorphine for the first 5 days. Cells were maintained for the next 6 days in neurobasal medium containing B27 minus vitamin A, N2 supplement, along with LDN193189 and CHIR99021. Last, cells were made into a single-cell suspension and seeded at a density of 400,000/cm2 on polyornithine- and laminin-coated plates in neurobasal media containing B27 minus vitamin A, brain-derived neurotrophic factor (20 ng/ml), glial cell line–derived neurotrophic factor (20 ng/ml), 0.2 mM transforming growth factor–β ascorbic acid (1 ng/ml), 0.5 mM cyclic adenosine 3′,5′-monophosphate (cAMP), and 10 μM DAPT until maturation (approximately 60 days). Generation and characterization of parkin knockout hES cells have been previously described in detail (19).
SUnSET assay
Differentiated human dopamine neurons cultured on glass coverslips precoated with polyornithine and laminin were incubated in media containing 1 μM puromycin for 1 hour and subsequently washed once with PBS. Cells were replaced with media containing no puromycin and cultured for another hour to remove the traces of unlabeled puromycin. Labeled cells were then fixed using 4% PFA for further immunocytochemistry.
SNAP-tag labeling of mitochondria
The SNAP-Tag-Cox8a fusion sequence was subcloned from the pSNAPf-Cox8A control plasmid (New England Biolabs) into cFUGw lentiviral vector using the Gibson assembly cloning kit (New England Biolabs). Differentiated human dopamine neurons cultured on coated glass coverslips were infected with the SNAP-Tag-Cox8a lentivirus for 5 days under culture conditions as described in (19). Transduced dopamine neurons in culture were then treated with a SNAP-tag substrate conjugated to 488 fluorophore (substrate 1) for 1 hour to label the existing mitochondrial pool. Neurons were then washed with media and cultured for an additional 24 hours. Thereafter, neurons were treated for 1 hour with a second SNAP-tag substrate conjugated to 568 fluorophore (substrate 2) to label mitochondria assembled since treatment with substrate 1. Neurons were washed again with media and cultured for an additional 1 hour before fixing with 4% PFA for immunocytochemistry.
Immunocytochemistry and microscopy
Following puromycin labeling or SNAP-tag treatment, cells were fixed for 15 min with 4% PFA and washed with 1× PBS. Cells were permeabilized with 0.3% Triton X-100 for 15 min, washed three times in 1× PBS, blocked for 1 hour with 4% bovine serum albumin/10% serum in PBS, and incubated with primary antibody overnight at 4°C. Following washing, cells were treated with secondary antibody for 1 hour in dark at RT. Cells were washed, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in PBS (1 μg/ml; Roth) for 3 min, and mounted with ProLong Gold antifade mountant (Life Technologies, P36930). Samples were imaged soon after the experiment and stored at 4°C. All images were taken for analysis with a Zeiss LSM 880 AiryScan laser scanning confocal microscope at a magnification of ×40. Images were scanned at 1024 × 1024 pixels, with a slice thickness of 1 μM and line and frame average of 4.
Animals
All experiments that included animals (8-week-old C57BL/6J mice; Jackson Laboratories, Bar Harbor, Maine, USA) were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee at the Johns Hopkins University. All efforts were made to minimize suffering.
Generation of virally induced conditional KD mouse models
Exon 7 floxed parkin mice (Parkin Flx/Flx) have been previously described (8). Exon 9 floxed CYLD mice (CYLD Flx/Flx) were obtained from G. Mosialos, Aristotle University of Thessaloniki, Greece. The CYLD Flx/Flx mice were cross-bred to Parkin Flx/Flx mice to generate CYLD Flx/+; Parkin Flx/+ heterozygous mice, which were then cross-bred to each other to generate Parkin Flx/Flx; CYLD Flx/Flx homozygous mice. Conditional KD of parkin, CYLD, or both parkin and CYLD was achieved by stereotactically introducing an AAV2 vector encoding the GF-fused Cre recombinase (AAV2-GFPCre) into SN of 6- to 8-week-old mice of the relevant genotype. AAV2-GFP injections served as control and SN coordinates (1.3 mm lateral, 3.2 mm caudal, and 4.3 mm ventral relative to Bregma) were used for the stereotactic injections as described previously in (8, 9).
Immunohistochemistry and stereological assessment
Mice were perfused with ice-cold PBS solution followed by ice-cold 4% PFA/PBS solution (pH 7.4) and prepared for immunohistochemical staining as previously described (62). Unbiased stereological counting of TH-positive and Nissl-positive neurons was performed as previously described (9, 63).
Behavioral tests
Pole test
The pole test was performed as described previously (64). A 9-mm-diameter 2.5-foot metal rod wrapped with bandage gauze was used as the pole. Mice were placed on the top of the pole (7.62 cms from the top of the pole) facing head up. The time taken to turn and total time taken to reach the base of the pole were recorded. Before the actual test, the mice were trained for two consecutive days, and each training session consisted of three test trials. The maximum cutoff time to stop the test was 120 s. Results were expressed in turn and total time (in seconds) (64).
Grip strength
The grip strength test was performed as described before (64). Neuromuscular strength was measured by maximum holding force generated by the mice (Bioseb). Mice were allowed to grasp a metal grid with either their fore and/or hindlimbs or both. The tail was gently pulled, and the maximum holding force was recorded by the force transducer when the mice released their grasp on the grid. The peak holding strength was digitally recorded and displayed as force (in grams) (64).
Amphetamine-induced stereotypic rotation
Three months after mice received the AAV-GFP or AVV-GFP Cre intranigral injection, amphetamine (5 mg/kg body weight; Sigma-Aldrich) was administered intraperitonially. Mice were placed into a white paper cylinder of 20 cm in diameter and monitored for 30 min. The behavior of mice was recorded for at least 5 min between 20 and 30 min following amphetamine administration. Full-body ipsilateral rotations (clockwise) were counted for each mouse from the video recordings. The animal genotypes were blinded and randomly used during the experiment, and the blind was removed for final quantification and statistical assessment.
Statistical analysis
Statistical analyses were performed using GraphPad statistical software (GraphPad Prism, version 8, San Diego, CA) as described in (10). The criteria for significance was: ns (not significant) P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Significant differences between two groups were analyzed using a two-tailed Student’s t test. For more than two groups, a one-way analysis of variance (ANOVA) (with Tukey’s post hoc correction for multiple comparisons) was used. For mixed groups, a two-way ANOVA (with Tukey’s post hoc correction for multiple comparisons) was used. The graph representation, definition of n, and which statistical test was performed are indicated in the figure legends. Error bars show SEM or SD, as indicated in the figure legends. Sample size was chosen according to that used for similar experiments in the literature.
Acknowledgments
We thank G. Mosialos, Aristotle University of Thessaloniki, Greece for providing Exon 9 floxed CYLD mice and F. Ikeda, Institute of Molecular Biotechnology, Vienna, Austria for providing Drosophila plasmid CYLD constructs.
Funding: This work was supported by grants from the NIH/NINDS NS38377 and K99AG066862 and the JPB Foundation. The authors acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation and the Diana Helis Henry Medical Research Foundation through direct engagement in the continuous active conduct of medical research in conjunction with the Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation’s Parkinson’s Disease Programs M-1, M-2, M-2014, and H-2104.
Author contributions: Conceptualization: S.K.P., V.L.D., and T.M.D. Methodology: S.K.P., H.W., M.K., V.L.D., and T.M.D. Formal analysis: S.K.P. Investigation: S.K.P., H.W., N.P., M.K., S.N., M.A.D., E.L., and B.G.K. Writing—original draft: S.K.P., V.L.D., and T.M.D. Writing—review and editing: S.K.P., H.W., N.P., M.K., S.N., M.D., E.L., B.G.K., J.C.T., V.L.D., and T.M.D. Reagents and materials: J.R.-O., J.C.T., V.L.D., and T.M.D. Funding acquisition: V.L.D. and T.M.D. Supervision: V.L.D. and T.M.D. All authors have read and approved the final manuscript.
Competing interests: The value of patents owned by Valted LLC could be affected by the study described in this article. V.L.D. and T.M.D. are founders of Valted, LLC and hold ownership equity interest in the company. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The Exon 9 floxed CYLD mice can be provided by Aristotle University of Thessaloniki, Greece pending scientific review and a completed material transfer agreement. Requests for the Exon 9 floxed CYLD mice should be submitted to G. Mosialos. Requests for other materials should be submitted to T.M.D.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N., Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., Gonzalez-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W., Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Kane L. A., Lazarou M., Fogel A. I., Li Y., Yamano K., Sarraf S. A., Banerjee S., Youle R. J., PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., Endo T., Fon E. A., Trempe J. F., Saeki Y., Tanaka K., Matsuda N., Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Scarffe L. A., Stevens D. A., Dawson V. L., Dawson T. M., Parkin and PINK1: Much more than mitophagy. Trends Neurosci. 37, 315–324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truban D., Hou X., Caulfield T. R., Fiesel F. C., Springer W., PINK1, Parkin, and mitochondrial quality control: What can we learn about Parkinson’s disease pathobiology? J. Parkinsons Dis. 7, 13–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin J. H., Ko H. S., Kang H., Lee Y., Lee Y. I., Pletinkova O., Troconso J. C., Dawson V. L., Dawson T. M., PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell 144, 689–702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens D. A., Lee Y., Kang H. C., Lee B. D., Lee Y. I., Bower A., Jiang H., Kang S. U., Andrabi S. A., Dawson V. L., Shin J. H., Dawson T. M., Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc. Natl. Acad. Sci. U.S.A. 112, 11696–11701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y., Stevens D. A., Kang S. U., Jiang H., Lee Y. I., Ko H. S., Scarffe L. A., Umanah G. E., Kang H., Ham S., Kam T. I., Allen K., Brahmachari S., Kim J. W., Neifert S., Yun S. P., Fiesel F. C., Springer W., Dawson V. L., Shin J. H., Dawson T. M., PINK1 primes parkin-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Rep. 18, 918–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirooznia S. K., Yuan C., Khan M. R., Karuppagounder S. S., Wang L., Xiong Y., Kang S. U., Lee Y., Dawson V. L., Dawson T. M., PARIS induced defects in mitochondrial biogenesis drive dopamine neuron loss under conditions of parkin or PINK1 deficiency. Mol. Neurodegener. 15, 17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddiqui A., Bhaumik D., Chinta S. J., Rane A., Rajagopalan S., Lieu C. A., Lithgow G. J., Andersen J. K., Mitochondrial quality control via the PGC1α-TFEB signaling pathway is compromised by Parkin Q311X mutation but independently restored by rapamycin. J. Neurosci. 35, 12833–12844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddiqui A., Rane A., Rajagopalan S., Chinta S. J., Andersen J. K., Detrimental effects of oxidative losses in parkin activity in a model of sporadic Parkinson’s disease are attenuated by restoration of PGC1alpha. Neurobiol. Dis. 93, 115–120 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Brahmachari S., Lee S., Kim S., Yuan C., Karuppagounder S. S., Ge P., Shi R., Kim E. J., Liu A., Kim D., Quintin S., Jiang H., Kumar M., Yun S. P., Kam T. I., Mao X., Lee Y., Swing D. A., Tessarollo L., Ko H. S., Dawson V. L., Dawson T. M., Parkin interacting substrate zinc finger protein 746 is a pathological mediator in Parkinson’s disease. Brain 142, 2380–2401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lork M., Verhelst K., Beyaert R., CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: So similar, yet so different. Cell Death Differ. 24, 1172–1183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Liu M., Su Y., Du J., Zhu A. J., A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling. G3 2, 1563–1575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin C., Kim S., Kang H., Yun K. N., Lee Y., Zhang Y., Kim Y., Kim J. Y., Han K., Shank3 regulates striatal synaptic abundance of Cyld, a deubiquitinase specific for Lys63-linked polyubiquitin chains. J. Neurochem. 150, 776–786 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Han Y. Y., Jin K., Pan Q. S., Li B., Wu Z. Q., Gan L., Yang L., Long C., Microglial activation in the dorsal striatum participates in anxiety-like behavior in Cyld knockout mice. Brain Behav. Immun. 89, 326–338 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Li H. D., Li D. N., Yang L., Long C., Deficiency of the CYLD impairs fear memory of mice and disrupts neuronal activity and synaptic transmission in the basolateral amygdala. Front. Cell. Neurosci. 15, 740165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar M., Acevedo-Cintron J., Jhaldiyal A., Wang H., Andrabi S. A., Eacker S., Karuppagounder S. S., Brahmachari S., Chen R., Kim H., Ko H. S., Dawson V. L., Dawson T. M., Defects in mitochondrial biogenesis drive mitochondrial alterations in PARKIN-deficient human dopamine neurons. Stem Cell Rep. 15, 629–645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanida I., Waguri S., Measurement of autophagy in cells and tissues. Methods Mol. Biol. 648, 193–214 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Schmidt E. K., Clavarino G., Ceppi M., Pierre P., SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Hussain A. F., Amoury M., Barth S., SNAP-tag technology: A powerful tool for site specific conjugation of therapeutic and imaging agents. Curr. Pharm. Des. 19, 5437–5442 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Sun S. C., CYLD: A tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 17, 25–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uematsu A., Kido K., Takahashi H., Takahashi C., Yanagihara Y., Saeki N., Yoshida S., Maekawa M., Honda M., Kai T., Shimizu K., Higashiyama S., Imai Y., Tokunaga F., Sawasaki T., The E3 ubiquitin ligase MIB2 enhances inflammation by degrading the deubiquitinating enzyme CYLD. J. Biol. Chem. 294, 14135–14148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stegmeier F., Sowa M. E., Nalepa G., Gygi S. P., Harper J. W., Elledge S. J., The tumor suppressor CYLD regulates entry into mitosis. Proc. Natl. Acad. Sci. U.S.A. 104, 8869–8874 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Liu M., Li D., Ran J., Gao J., Suo S., Sun S. C., Zhou J., CYLD regulates spindle orientation by stabilizing astral microtubules and promoting dishevelled-NuMA-dynein/dynactin complex formation. Proc. Natl. Acad. Sci. U.S.A. 111, 2158–2163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johari T., Maiti T. K., Catalytic domain mutation in CYLD inactivates its enzyme function by structural perturbation and induces cell migration and proliferation. Biochim. Biophys. Acta Gen. Subj. 1862, 2081–2089 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R., Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797–801 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Xue L., Igaki T., Kuranaga E., Kanda H., Miura M., Xu T., Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev. Cell 13, 446–454 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Hitomi J., Christofferson D. E., Ng A., Yao J., Degterev A., Xavier R. J., Yuan J., Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135, 1311–1323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Donnell M. A., Perez-Jimenez E., Oberst A., Ng A., Massoumi R., Xavier R., Green D. R., Ting A. T., Caspase 8 inhibits programmed necrosis by processing CYLD. Nat. Cell Biol. 13, 1437–1442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganjam G. K., Terpolilli N. A., Diemert S., Eisenbach I., Hoffmann L., Reuther C., Herden C., Roth J., Plesnila N., Culmsee C., Cylindromatosis mediates neuronal cell death in vitro and in vivo. Cell Death Differ. 25, 1394–1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bignell G. R., Warren W., Seal S., Takahashi M., Rapley E., Barfoot R., Green H., Brown C., Biggs P. J., Lakhani S. R., Jones C., Hansen J., Blair E., Hofmann B., Siebert R., Turner G., Evans D. G., Schrander-Stumpel C., Beemer F. A., van Den Ouweland A., Halley D., Delpech B., Cleveland M. G., Leigh I., Leisti J., Rasmussen S., Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25, 160–165 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Dobson-Stone C., Hallupp M., Shahheydari H., Ragagnin A. M. G., Chatterton Z., Carew-Jones F., Shepherd C. E., Stefen H., Paric E., Fath T., Thompson E. M., Blumbergs P., Short C. L., Field C. D., Panegyres P. K., Hecker J., Nicholson G., Shaw A. D., Fullerton J. M., Luty A. A., Schofield P. R., Brooks W. S., Rajan N., Bennett M. F., Bahlo M., Landers J. E., Piguet O., Hodges J. R., Halliday G. M., Topp S. D., Smith B. N., Shaw C. E., McCann E., Fifita J. A., Williams K. L., Atkin J. D., Blair I. P., Kwok J. B., CYLD is a causative gene for frontotemporal dementia - amyotrophic lateral sclerosis. Brain 143, 783–799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Sekine-Aizawa Y., Ebrahimi S., Tanaka S., Okabe S., Tumor suppressor protein CYLD regulates morphogenesis of dendrites and spines. Eur. J. Neurosci. 50, 2722–2739 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Dosemeci A., Thein S., Yang Y., Reese T. S., Tao-Cheng J. H., CYLD, a deubiquitinase specific for lysine63-linked polyubiquitins, accumulates at the postsynaptic density in an activity-dependent manner. Biochem. Biophys. Res. Commun. 430, 245–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thein S., Pham A., Bayer K. U., Tao-Cheng J. H., Dosemeci A., IKK regulates the deubiquitinase CYLD at the postsynaptic density. Biochem. Biophys. Res. Commun. 450, 550–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Q., Ruan H., Peng L., Zhang M., Gack M. U., Yao W. D., Proteasome-independent polyubiquitin linkage regulates synapse scaffolding, efficacy, and plasticity. Proc. Natl. Acad. Sci. U.S.A. 114, E8760–E8769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rangaraju V., Lauterbach M., Schuman E. M., Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell 176, 73–84.e15 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Haddad D., Nakamura K., Understanding the susceptibility of dopamine neurons to mitochondrial stressors in Parkinson’s disease. FEBS Lett. 589, 3702–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durcan T. M., Kontogiannea M., Thorarinsdottir T., Fallon L., Williams A. J., Djarmati A., Fantaneanu T., Paulson H. L., Fon E. A., The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 20, 141–154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexopoulou Z., Lang J., Perrett R. M., Elschami M., Hurry M. E., Kim H. T., Mazaraki D., Szabo A., Kessler B. M., Goldberg A. L., Ansorge O., Fulga T. A., Tofaris G. K., Deubiquitinase Usp8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proc. Natl. Acad. Sci. U.S.A. 113, E4688–E4697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rott R., Szargel R., Haskin J., Bandopadhyay R., Lees A. J., Shani V., Engelender S., α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc. Natl. Acad. Sci. U.S.A. 108, 18666–18671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornelissen T., Haddad D., Wauters F., Van Humbeeck C., Mandemakers W., Koentjoro B., Sue C., Gevaert K., De Strooper B., Verstreken P., Vandenberghe W., The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum. Mol. Genet. 23, 5227–5242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chakraborty J., von Stockum S., Marchesan E., Caicci F., Ferrari V., Rakovic A., Klein C., Antonini A., Bubacco L., Ziviani E., USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Mol. Med. 10, e9014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Stockum S., Sanchez-Martinez A., Corra S., Chakraborty J., Marchesan E., Locatello L., Da Re C., Cusumano P., Caicci F., Ferrari V., Costa R., Bubacco L., Rasotto M. B., Szabo I., Whitworth A. J., Scorrano L., Ziviani E., Inhibition of the deubiquitinase USP8 corrects a Drosophila PINK1 model of mitochondria dysfunction. Life Sci. Alliance 2, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ge P., Dawson V. L., Dawson T. M., PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 15, 20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greene J. C., Whitworth A. J., Kuo I., Andrews L. A., Feany M. B., Pallanck L. J., Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U.S.A. 100, 4078–4083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pesah Y., Pham T., Burgess H., Middlebrooks B., Verstreken P., Zhou Y., Harding M., Bellen H., Mardon G., Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 131, 2183–2194 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Whitworth A. J., Theodore D. A., Greene J. C., Benes H., Wes P. D., Pallanck L. J., Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 102, 8024–8029 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M., Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J., Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., Lu B., Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U.S.A. 103, 10793–10798 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sango J., Kakihana T., Takahashi M., Katsuragi Y., Anisimov S., Komatsu M., Fujii M., USP10 inhibits the dopamine-induced reactive oxygen species–dependent apoptosis of neuronal cells by stimulating the antioxidant Nrf2 activity. J. Biol. Chem. 298, 101448 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asaoka T., Almagro J., Ehrhardt C., Tsai I., Schleiffer A., Deszcz L., Junttila S., Ringrose L., Mechtler K., Kavirayani A., Gyenesei A., Hofmann K., Duchek P., Rittinger K., Ikeda F., Linear ubiquitination by LUBEL has a role in Drosophila heat stress response. EMBO Rep. 17, 1624–1640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M., Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: Implications for Lewy body formation. J. Neurosci. 25, 2002–2009 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nichols C. D., Becnel J., Pandey U. B., Methods to assay Drosophila behavior. J. Vis. Exp. 3795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J. S., Luo L., A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat. Protoc. 1, 2110–2115 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Gonzalo-Gomez A., Turiegano E., Leon Y., Molina I., Torroja L., Canal I., Ih current is necessary to maintain normal dopamine fluctuations and sleep consolidation in Drosophila. PLOS ONE 7, e36477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kriks S., Shim J. W., Piao J., Ganat Y. M., Wakeman D. R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A., Yang L., Beal M. F., Surmeier D. J., Kordower J. H., Tabar V., Studer L., Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480, 547–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong Y., Neifert S., Karuppagounder S. S., Stankowski J. N., Lee B. D., Grima J. C., Chen G., Ko H. S., Lee Y., Swing D., Tessarollo L., Dawson T. M., Dawson V. L., Overexpression of Parkinson’s disease-associated mutation LRRK2 G2019S in mouse forebrain induces behavioral deficits and α-synuclein pathology. eNeuro 4, ENEURO.0004–ENEU17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karuppagounder S. S., Xiong Y., Lee Y., Lawless M. C., Kim D., Nordquist E., Martin I., Ge P., Brahmachari S., Jhaldiyal A., Kumar M., Andrabi S. A., Dawson T. M., Dawson V. L., LRRK2 G2019S transgenic mice display increased susceptibility to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-mediated neurotoxicity. J. Chem. Neuroanat. 76, 90–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao X., Ou M. T., Karuppagounder S. S., Kam T. I., Yin X., Xiong Y., Ge P., Umanah G. E., Brahmachari S., Shin J. H., Kang H. C., Zhang J., Xu J., Chen R., Park H., Andrabi S. A., Kang S. U., Goncalves R. A., Liang Y., Zhang S., Qi C., Lam S., Keiler J. A., Tyson J., Kim D., Panicker N., Yun S. P., Workman C. J., Vignali D. A., Dawson V. L., Ko H. S., Dawson T. M., Pathological α-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science 353, aah3374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 to S3
Movies S1 to S3