Abstract
Although the tools based on split proteins have found broad applications, ranging from controlled biological signaling to advanced molecular architectures, many of them suffer from drawbacks such as background reassembly, low thermodynamic stability, and static structural features. Here, we present a chemically inducible protein assembly method enabled by the dissection of the carboxyl-terminal domain of a B12-dependent photoreceptor, CarHC. The resulting segments reassemble efficiently upon addition of cobalamin (AdoB12, MeB12, or CNB12). Photolysis of the cofactors such as AdoB12 and MeB12 further leads to stable protein adducts harboring a bis-His–ligated B12. Split CarHC enables the creation of a series of protein hydrogels, of which the mechanics can be either photostrengthened or photoweakened, depending on the type of B12. These materials are also well suited for three dimensional cell culturing. Together, this new protein chemistry, featuring negligible background autoassembly, stable conjugation, and phototunability, has opened up opportunities for designing smart materials.
Development of a B12-induced protein assembly system has enabled the design of smart, cytocompatible hydrogels.
INTRODUCTION
Functional dissection of proteins into complementing fragments has led to a number of molecular tools for biological studies. Some early tools such as split ribonuclease S (1), ubiquitin (2), and green fluorescent protein (GFP) (3, 4) were initially developed to probe protein-protein interactions (5–7). Recently developed tools such as split esterase (5), kinase (8), protease (7, 9), and Cas9 (10–13) have strengthened our abilities to control molecular function and signaling in complex biological systems (14). Splitting a protein into two halves often results in loss of function (e.g., catalysis, fluorescence, luminescence, and stimuli responsiveness), which can be restored by spontaneous or induced reassembly of the two fragments. This gain of function upon reassembly has proven to be very useful, insofar as precise control over or within a complex biological system is concerned. For example, delicately controlled genome editing and transcriptional modulation in living cells have been achieved by genetically fusing split Cas9 with the chemically inducible dimerization domains, FKBP/FRB, where rapamycin-induced protein reassembly restores the activity of Cas9 (10). However, because many split protein systems exhibit a high level of self-assembly that is independent of inducers, this uncontrollability constrains their wider use (15, 16). It is therefore highly desirable to have a tightly controlled protein assembly tool with a minimal background reactivity, which would enrich the arsenal for biological regulation.
Split proteins have also gained traction with materials scientists in recent years because of their great diversity and dynamic features. Split GFP and its variants have enabled the creation of several uncommon protein architectures (4, 17). To overcome the low thermodynamic and mechanical stability of these noncovalent split protein systems, Howarth and coworkers developed genetically encoded SpyTag/SpyCatcher chemistry—a pair of peptide/protein that is derived from dissected bacterial adhesin protein and can spontaneously form an Asp-Lys isopeptide bond under mild physiological conditions (18). The product of the Spy chemistry is thermodynamically and mechanically stable because of its covalent nature and unique folded β-barrel structure. In addition to being fully genetically encoded, this protein chemistry, alongside its later optimized versions (e.g., Spy002 and Spy003) (19, 20), is also noted for its high efficiency and specificity, reminiscent of the powerful Cu (I)–catalyzed azide/alkyne cycloaddition click reaction (21–23). These prototypical genetically encoded click chemistries (GECCs) have found numerous applications in creating advanced protein architectures, subunit vaccines, and functional biomaterials (17, 21, 24–31). On the other hand, since these GECC pairs react spontaneously and invariably form static and rigid covalent adducts, we envisioned that an alternative reaction mode, which is inducible and forms a stable yet dynamically tunable product, would be complementary to the existing GECCs and thus open the door to diverse smart materials.
The photoreceptor CarH protein has originally been identified as a bacterial transcriptional regulator that controls the biosynthesis of carotenoid (32–34). Its C-terminal domain, CarHC, has been shown to tetramerize upon binding to adenosylcobalamin (AdoB12) and disassemble into monomers on exposure to green (522 nm) or white light. This photolysis is accompanied with the cleavage of the C─Co bond in B12, the removal of the adenosyl moiety, and the formation of a stable bis-His–ligated B12 complex (Fig. 1A) (32, 33, 35). This AdoB12/light-induced assembly/disassembly mechanism has enabled bacterial cells to turn on and off the DNA binding activity of CarH and modulate gene expression (32–34).
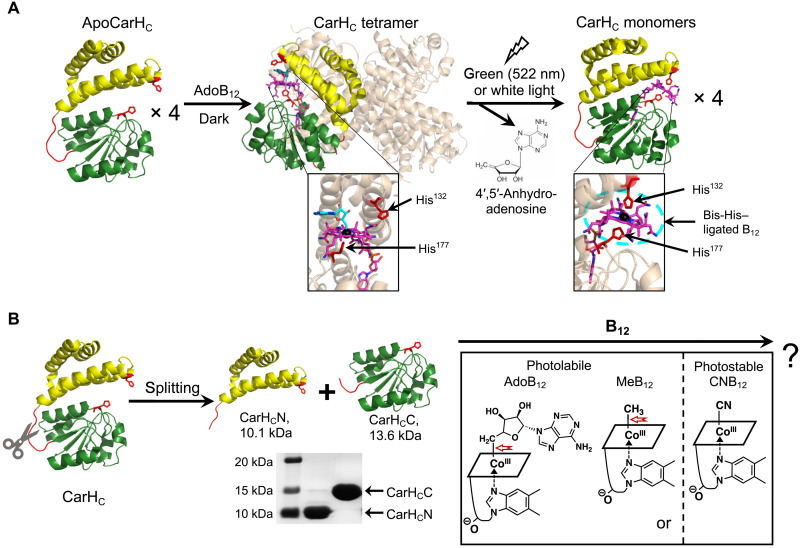
Fig. 1. Dissection of the B12-dependent photoresponsive CarHC protein.
(A) Photochemistry of CarHC. CarHC monomers, upon binding to AdoB12, assemble into a tetramer, which further disassembles into monomers on light exposure, accompanied with the photolysis of AdoB12, the release of 4′,5′-anhydroadenosine, and the coordination of His132 to the cobalt center in B12. The bottom panel shows close-up views of the cofactor binding pocket. For clarity, three monomers in the CarHC tetramer complex are in high transparency, with AdoB12 hidden. Monomeric CarHC is shown with the N-terminal four-helix bundle in yellow, the unstructured loop region in red, and the C-terminal B12-binding domain in green. (B) Schematic showing the dissection of CarHC at the loop region into CarHCN and CarHCC, and their possible reassembly with various cobalamins, such as AdoB12, MeB12, or CNB12. The SDS-PAGE analysis shows successful production of CarHCN (calculated MW, 10.1 kDa) and CarHCC (calculated MW, 13.6 kDa) by E. coli expression. The structure of B12 is presented as a simplified parallelogram for clarity. The light-sensitive cobalt-carbon bond is highlighted by a red arrow. ApoCarHC and monomeric CarHC: Protein Data Bank (PDB) ID code, 5C8F; tetrameric CarHC: PDB ID code, 5C8A.
CarHC has proven to be a powerful molecular tool; it not only has served as an optogenetic tool to control biological signaling in vivo (36) but it also has enabled the creation of photoresponsive materials for therapeutic delivery and tissue regeneration (17, 24, 37). In this study, we demonstrate that CarHC can be split into two smaller polypeptides, CarHCN and CarHCC, which, upon addition of B12, reassemble and regain the B12-dependent photoresponsiveness. The integrated use of split CarHC and Spy chemistries led to the synthesis of several entirely protein-based networks, of which mechanics can be either photoweakened or photostrengthened, depending on the nature of the cobalamin cofactor. This tightly controlled split CarHC system is characterized by several important features, including negligible background autoassembly, highly efficient reconstitution upon induction, optical tunability, and a markedly stable complex after photolysis. It is for these reasons that we regard this split CarHC chemistry as an important alternative to the existing GECCs, which will open up great opportunities for optogenetics and materials synthetic biology.
RESULTS AND DISCUSSION
Dissection of CarHC
The structural study of CarHC revealed a “sandwich” configuration, in which the cofactor AdoB12 is wrapped by a four-helix bundle domain (Pro80-Ala159) and a Rossmann-fold B12-binding domain (Val167-Leu276). The Rossmann-fold domain binds to the lower face of B12 via His177 coordination to the cobalt ion (33). Between these two domains lies an unstructured Gly- and Pro-rich loop, GFPPGPP (Gly160-Pro166) (Fig. 1A and fig. S1). We envisioned that CarHC could be dissected at this loop region, yielding two smaller polypeptides, CarHCN (Pro80-Pro166) and CarHCC (Phe161-Leu276), while maintaining their respective folding (Fig. 1B).
To test this hypothesis, we designed and cloned the two genes encoding the corresponding recombinant proteins, thioredoxin (Trx)–His6–CarHCN and Trx-His6-CarHCC (fig. S2). Throughout the study, the native loop region PPGPP was kept in both CarHCN and CarHCC as a flexible spacer to minimize the steric hindrance when fusing them with other folded domains. The proteins were produced using Escherichia coli expression and purified using Ni–nitrilotriacetic acid (Ni-NTA) chromatography. After removal of the Trx-His6 tag by 3C protease digestion, the resulting neat constructs, CarHCN [molecular weight (MW), 10.1 kDa] and CarHCC (MW, 13.6 kDa) (Fig. 1B and fig. S3), remained highly soluble in tris-buffered saline [TBS; 50 mM tris and 300 mM NaCl (pH 8)], showing no apparent aggregation or precipitation. Circular dichroism (CD) spectra of both proteins exhibited double dips at 208 and 222 nm, highly indicative of structures rich in α helix (Fig. 2A). Moreover, the mix of CarHCN and CarHCC at an equimolar ratio exhibited a CD spectrum that closely resembles that of apoCarHC (Fig. 2, A and B). Together, these results corroborated that the photoreceptor CarHC protein can be split into two small folded segments at its loop region (Gly160-Pro166).
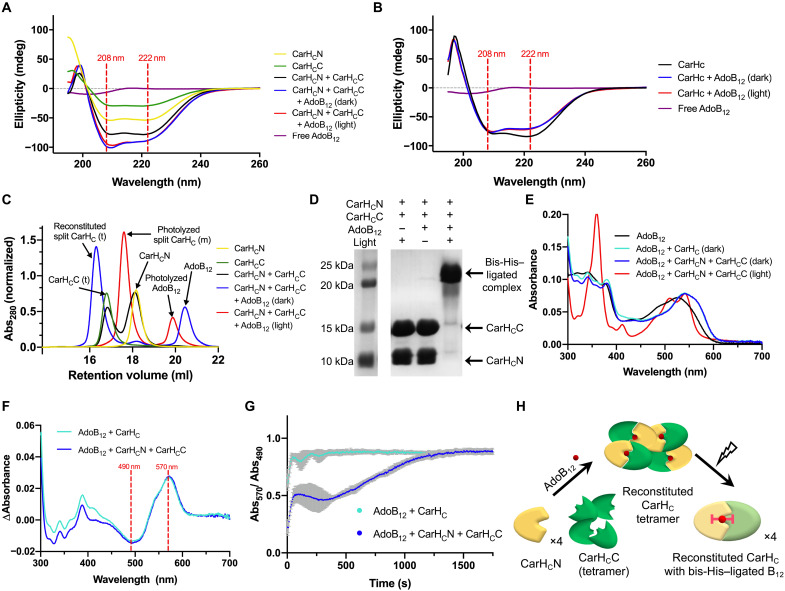
Fig. 2. AdoB12-induced reconstitution of split CarHC.
(A) CD spectra of free AdoB12, CarHCN, CarHCC, CarHCN + CarHCC, CarHCN + CarHCC + AdoB12 (dark), and CarHCN + CarHCC + AdoB12 (light). All protein samples exhibit distinctive dips at 208 and 222 nm, the characteristic of an α helix–rich structure. (B) CD spectra of intact CarHC with AdoB12. (C) SEC analyses. “m” and “t” denote “monomer” and “tetramer,” respectively. (D) SDS-PAGE analysis. Protein samples were denatured using the 4% SDS loading dye (2×) at room temperature. (E) UV-vis spectra. The spectrum of CarHCN + CarHCC + AdoB12 (dark) resembles that of intact CarHC + AdoB12 (dark), both showing a peak shift to 550 from 525 nm of free AdoB12. The spectrum of CarHCN + CarHCC + AdoB12 (light) shows a dual-peak structure between 500 and 550 nm, a characteristic of bis-imidazole–ligated B12. (F) Difference spectra showing the change in AdoB12 absorbance as a function of wavelength upon addition of intact CarHC or split CarHC. The minimum and the maximum of both difference spectra appeared at 490 and 570 nm, respectively. (G) Binding kinetics of AdoB12 (50 μM) to split CarHC or intact CarHC (50 μM). Evolution of the ratio of the absorbance at 570 to 490 nm was recorded as a function of time. The corresponding Abs570/Abs490 of free AdoB12 was subtracted as background. Data are presented as means ±SD (n = 5), with error bars in gray. (H) Model of AdoB12-induced split CarHC reconstitution. In the dark, CarHCN and CarHCC reassemble into a tetrameric complex upon addition of AdoB12. Light exposure leads to a monomeric one, accompanied with the photolysis of AdoB12 and the bis-His ligation of B12.
AdoB12-induced reconstitution of Split CarHC
To examine whether split CarHC retains the ability of the cofactor binding, we mixed CarHCN and CarHCC at an equimolar ratio with AdoB12 in the dark. The products of CarHCN + CarHCC + AdoB12, before and after light exposure, exhibited CD spectra that largely retained the double-dip pattern (Fig. 2A), which is typical for α helix–rich structures and resembling that of the AdoB12-bound intact CarHC (Fig. 2B); the dip at 208 nm appeared to be deeper than at 222 nm because of the presence of AdoB12, of which the CD spectrum contained a single dip at 195 to 210 nm (Fig. 2, A and B).
To verify that the CarHCN and CarHCC proteins reassembled upon AdoB12 binding, we performed a series of size exclusion chromatography (SEC) and dynamic light scattering (DLS) experiments (Fig. 2C and figs. S4 and S5). CarHCN (theoretical MW, 10.1 kDa) alone exhibited a retention volume of 18.2 ml in SEC and an MW of ~9.6 kDa as determined by DLS, highly indicative of a monomeric protein, while the retention volume and the measured MW of CarHCC are 16.8 ml and 45.9 kDa, respectively, the latter of which was substantially larger than the theoretical MW of monomeric CarHCC (13.6 kDa), suggesting the occurrence of a tetrameric CarHCC in the absence of the cofactor. This outcome raised the possibility that tetramerization is a property inherent to this Rossmann-fold domain, which is just inhibited by the adjacent N-terminal four-helix bundle when in proximity. The previous structural study of AdoB12-bound CarH showed that the four residues (Arg176, Asp201, Gly160, and Gly192), which all reside in the Rossmann-fold domain, are critical for its tetramerization; the Arg176/Asp201 interaction stabilizes the head-to-tail dimer interface, while the Gly160/Gly192 pair at the dimer-dimer interface helps minimize the steric clash within the tetramer (33). The fact that these four residues all reside in CarHCC is unlikely to be coincidental with the observation of a tetrameric CarHCC, independent of the cofactor.
Simply mixing CarHCN and CarHCC at an equimolar ratio failed to alter the oligomerization of either protein, showing the absence of background autoassembly. The spontaneous tetramerization of CarHCC is very likely to pose an additional hurdle that prevents the autoassembly of CarHCN and CarHCC in the absence of a cofactor. On the other hand, addition of AdoB12 in the dark led to a nearly quantitative conversion of the mix into a complex, even larger than tetrameric CarHCC, as evidenced by a retention volume (16.3 ml) that was smaller than that of CarHCC (16.8 ml). Moreover, this resulting complex exhibited almost the same retention volume as the AdoB12-bound intact CarHC (tetramer) in the dark did (fig. S4). These results reflect the combined contributions from the split CarHC reconstitution and tetramerization in the presence of AdoB12. The reconstituted protein complex regained AdoB12-dependent photoresponsiveness, as light exposure thoroughly disassembled the tetrameric complex into monomeric ones, exhibiting an increased retention volume (17.6 ml) in the SEC analysis and a reduced MW (~22.9 kDa) according to the DLS assay (Fig. 2C and fig. S5). Moreover, the photolyzed complex turned out to be quite stable and remained conjugated throughout the SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis at room temperature, which can be explained by the stable bis-His ligation to B12 (Fig. 2D and fig. S6) (33). However, the reconstituted product disintegrated in 4% SDS after boiling for 10 min (fig. S7), suggesting that these adducts were not as stable under extreme conditions as those covalently stitched by SpyTag/SpyCatcher chemistry (18). This bis-His/Co ligation in the photolyzed complex was corroborated by its ultraviolet-visible (UV-vis) spectrum (Fig. 2E), in which the pronounced dual-peak structure between 500 and 550 nm is a signature feature of the bis-imidazole ligated cobalamin (38). Fast reaction kinetics has become increasingly important for the use of a protein chemistry in complex biological systems. A previous study has already shown that the photoreaction of AdoB12-bound CarH, despite the involvement of multiple intermediates, is fast, in a time scale of milliseconds (32). In the case of split CarHC chemistry, the binding of AdoB12 to proteins, an intermolecular event that has to overcome the diffusion barrier, is more likely to be the rate-limiting step than the other such as photolysis or the subsequent His displacement. In this sense, the rate of AdoB12 protein binding could serve as an approximation of the overall efficiency of this split CarHC chemistry. As the AdoB12 protein binding altered the absorbance pattern of AdoB12 between 490 and 570 nm substantially (Fig. 2F), alongside a peak shift from 525 (free AdoB12) to 550 nm (protein bound) (Fig. 2E) (32), this pronounced spectral change enabled us to estimate the binding kinetics between AdoB12 and the protein; upon mixing AdoB12 with split or intact CarHC at a 1:1 molar ratio at the concentration of 50 μM, we recorded the evolution of the absorbance values at 570 and 490 nm, as well as their ratios, as a function of time (Fig. 2G).
It turned out that the binding of AdoB12 to split CarHC is efficient; Abs570/Abs490 reached maximum within ~20 min (Fig. 2G and figs. S8 and S9). The binding curves of AdoB12 and the protein consistently exhibited a two-step feature, in line with the conceived two-step mechanism by which AdoB12 interacts with CarHC (fig. S10) (33). The interaction between AdoB12 and the protein can be viewed as a two-step process consisting of the initial docking that is reversible and forms a loose and dynamic complex and the ensuing His177 displacement of the B12 dimethylbenzimidazole base (fig. S10) (33), leading to a more stable, His177-ligated Co adduct. According to the binding curves (Fig. 2G and figs. S8 and S9), the ensuing His177 displacement appears to be rate limiting for the binding of AdoB12 to the split CarHC, which, alongside the larger diffusion barrier expected for this three-molecule assembly system, made the overall binding substantially slower than that of AdoB12 to the intact CarHC, the latter of which completed within 200 s (Fig. 2G).
This split CarHC reconstitution was robust not only in TBS but also in other solution systems such as phosphate-buffered saline (PBS) and Dulbecco’s modified Eagle’s medium (DMEM; the cell culture medium used in this study). It was also insensitive to altered protein concentration (10 to 100 μM) (fig. S11). These results together demonstrated that the AdoB12-induced reassembly of split CarHC is an efficient and robust process and that AdoB12 is essential for this protein assembly, as well as the consequent photoresponsiveness, whereas no spontaneous reassembly occurred in the absence of the cofactor, pointing to the potential of this new protein chemistry in exerting precise control within complex systems (Fig. 2H).
Reconstitution of split CarHC with MeB12 and CNB12
In addition to AdoB12, other cobalamins such as photolabile methylcobalamin (MeB12) and photoinert cyanocobalamin (CNB12) have also been shown to bind to CarH, though unable to elicit its tetramerization (34). This discrepancy in protein oligomerization caused by different cofactors prompted us to investigate the influence of MeB12 and CNB12 on the reconstitution of split CarHC. SEC analyses showed that addition of MeB12 in the dark led to a thorough conversion of monomeric CarHCN and tetrameric CarHCC into a monomeric protein complex, the latter of which remained unchanged in its retention volume after light exposure, accompanied with the complete photolysis of the excessive portion of free MeB12 (Fig. 3A). In contrast to the reconstituted complex before light exposure, which disintegrated into two bands corresponding to CarHCN and CarHCC, respectively, on SDS-PAGE, the photolyzed product turned out to be stable and remained intact throughout the SDS-PAGE analysis, reminiscent of the bis-His ligation of B12 observed in the photolyzed AdoB12 complex (Fig. 3B and fig. S7). This bis-His ligation was corroborated by the UV-vis spectra, where the photolyzed product with MeB12 (Fig. 3C) was almost indistinguishable from that with AdoB12 (Fig. 2E), both also in good agreement with the spectrum of the bis-imidazole ligated cobalamin as previously reported (32, 33).
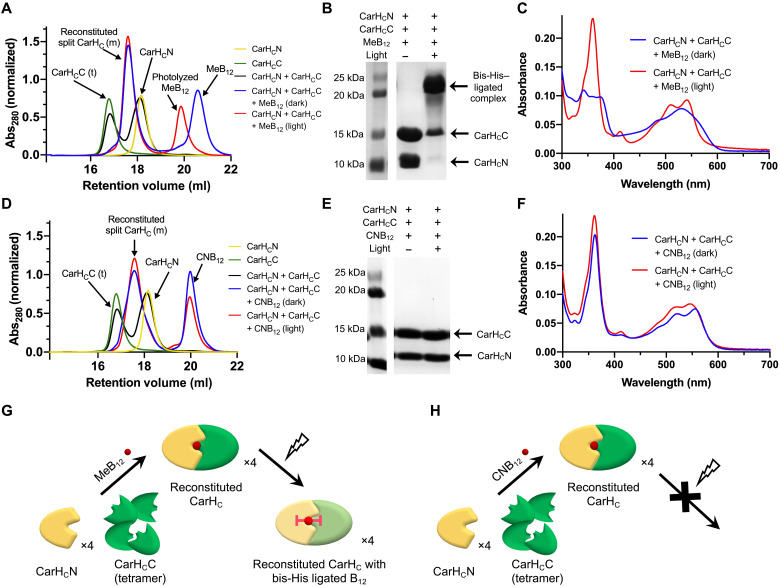
Fig. 3. Reconstitution of split CarHC induced by MeB12 and CNB12.
(A and D) SEC analyses of the split-CarHC reconstitution induced by MeB12 (A) or CNB12 (D). “m” and “t” denote “monomer” and “tetramer,” respectively. (B and E) SDS-PAGE analyses of the split CarHC reconstitution induced by MeB12 (B) or CNB12 (E). Protein samples were denatured using the 4% SDS loading dye (2×) at room temperature before the electrophoresis. (C and F) UV-vis spectra of split CarHC in the presence of MeB12 (C) or CNB12. (G and H) Models of the reconstitution of split CarHC induced by photolabile MeB12 (G) and photostable CNB12 (H). In the dark, CarHCN and CarHCC reassemble into a monomeric complex upon addition of MeB12 or CNB12. Photolysis of MeB12 leads to the formation of a stable, monomeric complex harboring a bis-His–ligated B12, while the CNB12-bound complex remains unchanged on light exposure.
As to CNB12, while this photoinert molecule was able to reassemble CarHCN and CarHCC into a monomeric complex, though less efficiently (~77% conversion) (fig. S12), the resulting product exhibited no responsiveness to light (Fig. 3D). The CNB12-bound complex, whether light-exposed or not, was unstable and disintegrated on SDS-PAGE into two segments, correspondingly CarHCN and CarHCC (Fig. 3E and fig. S7). Its UV-vis spectra remained unchanged throughout light exposure, showing its photoinertness (Fig. 3F). These results together showed that although both cobalamins induced the reassembly of split CarHC, only the photolabile MeB12, but not the photoinert CNB12, enabled the formation of a highly stable adduct harboring bis-His–ligated B12 on light exposure (Fig. 3, G and H).
Note that MeB12 and CNB12 induced the assembly of CarHCN and CarHCC into a monomeric complex, in contrast to AdoB12-induced formation of a tetrameric complex. As aforementioned, the SEC and DLS analyses of CarHCC showed that its ability of tetramerization is inherent to this Rossmann-fold domain (Fig. 2B and fig. S5). This tetramerization is inhibited by the four-helix bundle CarHCN when the two domains are in close contact. This proximity-induced inhibition can be found not only in the MeB12- and CNB12-bound proteins/complexes but also in the photolyzed products harboring the bis-His–ligated cobalamin. In any of these cases, a small ligand like the methyl or cyanide group or the direct cobalamin ligation by His132 of CarHCN ensures sufficient proximity between CarHCN and CarHCC, thus inhibiting the latter’s tetramerization. By contrast, the AdoB12-bound protein, either intact CarHC or reconstituted split CarHC complex, adopts an upright conformation (35), in which the bulky 5′-dAdo moiety separates the two domains and thus preserves the CarHCC tetramerization. The removal of 5′-dAdo on light exposure can bring the two domains into proximity, thus disintegrating the tetramers. The disparate effects of different cobalamins on CarHC reconstitution, oligomerization, and photoresponsiveness have also led us to a better understanding of the molecular mechanism by which this bacterial transcriptional regulator senses and responds to light.
Photostrengthening and photoweakening protein hydrogels
Hydrogels with dynamically tunable mechanics are increasingly important for biomedical applications ranging from stem cell fate decision control to scarless wound regeneration (39–41). Split CarHC, with its dual responsiveness to cobalamins and light, could serve to construct and control such dynamic materials. Prompted by our previous success with Spy network hydrogels—a collection of engineered protein networks covalently assembled by SpyTag/SpyCatcher chemistry (24, 25), we envisioned that direct assembly of split CarHC in a similar fashion could also lead to macroscopic protein materials while enabling faithful transfer of the multi-stimuli responsiveness from the molecular level to the macroscopic material level.
We created four gene constructs that encode the corresponding telechelic proteins including SpyTag-ELP-CarHCC-ELP-SpyTag (ACCA; 33.4 kDa), SpyCatcher-ELP-CarHCN-ELP-SpyCatcher (BCNB; 56.0 kDa), SpyTag-ELP-CarHCN-ELP-SpyTag (ACNA; 29.8 kDa), and SpyCatcher-ELP-CarHCC-ELP-SpyCatcher (BCCB; 59.5 kDa) (fig. S13). The elastin-like polypeptide (ELP) domain consists of repeating pentapeptides (VPGXG)15 (X represents valine or glutamate at a ratio of 4:1), which has been noted for its excellent expression yield and solubility under physiological conditions (42). This ELP domain is also intended as a spacer to minimize the steric hindrance between the flanking folded globular domains.
These four proteins, all of which are His6-tagged, were produced by E. coli and purified by Ni-NTA chromatography (figs. S14 and S15). They were further dialyzed against distilled water and lyophilized into white powders at −80°C. Throughout the study, we arbitrarily chose the combination of ACCA and BCNB, which is essentially no different from the alternative pair, ACNA + BCCB (fig. S26), to illustrate the use of split CarHC chemistry for material design. The protein solutions, ACCA and BCNB [10 weight (wt) % in TBS], were mixed at an equimolar ratio, followed by addition of a stoichiometric amount of the cofactor, AdoB12, MeB12, or CNB12, in the dark to initiate gelation (Fig. 4, A and B, and fig. S16). A red gel-like material emerged in the mixture supplemented with AdoB12 within 5 min, while the reaction mixture with MeB12 or CNB12 remained liquid like—only slight solidification occurred to the MeB12 reaction after prolonged incubation (>10 min).
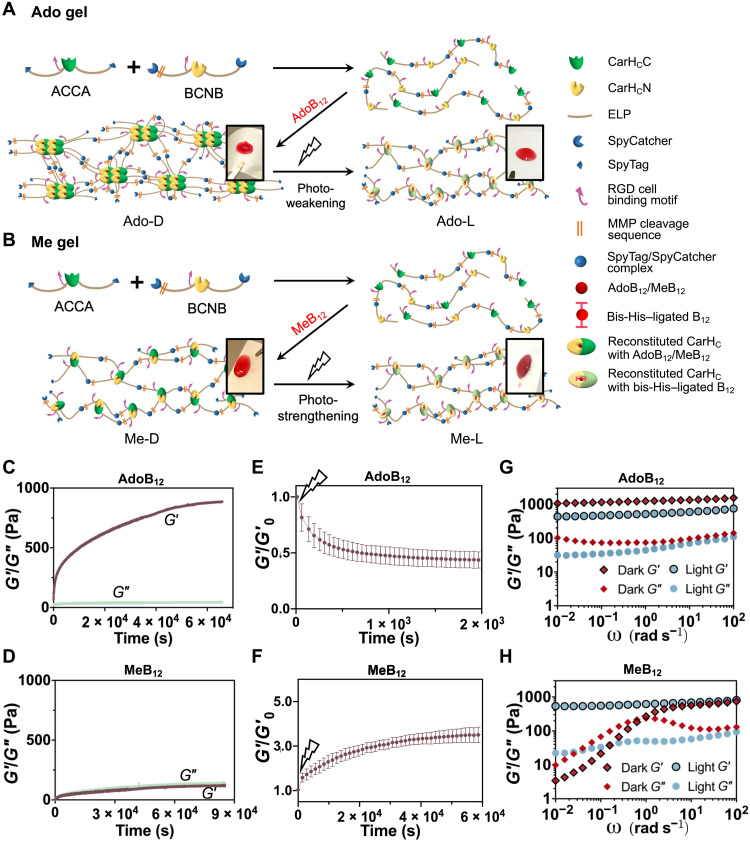
Fig. 4. Synthesis of photoresponsive hydrogels enabled by the integrated use of split CarHC and Spy chemistries.
(A and B) Schematic showing the synthesis of AdoB12- and MeB12-dependent photoresponsive protein hydrogels, Ado gel (A) and Me gel (B), of which the mechanics can be photoweakened and photostrengthened, respectively. Insets are photographs of corresponding materials. (C and D) Evolution of storage (G′) and loss (G″) modulus of the reaction of SpyTag-ELP-CarHCC-ELP-SpyTag (ACCA) + SpyCatcher-ELP-CarHCN-ELP-SpyCatcher (BCNB) in the dark after addition of AdoB12 (C) or MeB12 (D) as a function of time. (E and F) Normalized storage modulus (G′/G′0) of photoweakening Ado gel (E) and photostrengthening Me gel (F) monitored by time sweep tests at a fixed frequency of 1 rad/s and strain of 5% at 23°C. Data are presented as means ± SD (n = 3). (G and H) Frequency sweep tests of the Ado (G) and Me (H) gels at a fixed strain of 10% at 23°C before and after photolysis with 30-klux light.
To quantitatively assess and compare these materials harboring different cobalamins, we performed dynamic rheological measurements in time, frequency, and strain sweep modes. The reaction product of ACCA + BCNB in the absence of cobalamins remained as a viscous liquid throughout the time sweep experiments, accompanied with a very low storage modulus (G′; <10 Pa) and loss modulus (G″; <10 Pa), as expected for linear protein polymers (fig. S17), so did the product of ACCA + BCNB + CNB12. Despite the proven ability of CNB12 to induce the split CarHC reassembly, the resulting complex was too weak to maintain a robust network (fig. S18). On the other hand, the reaction product of ACCA + BCNB + AdoB12 in the dark, namely, the Ado gel or Ado-D, exhibited a rapidly increasing G′, which reached a plateau of ~0.85 kPa after 16 hours and was substantially higher than the corresponding G″ (~40 Pa), suggesting the formation of a solid (Fig. 4C and fig. S19A). By contrast, the reaction of ACCA + BCNB + MeB12 in the dark, namely, the Me gel or Me-D, exhibited a gradually increasing G′, which plateaued at ~0.12 kPa and was much lower than that of the Ado gel, pointing to the importance of CarHC tetramerization in supporting gel mechanics (Fig. 4D and fig. S19B) (17, 24).
Both Ado and Me gels were photoresponsive, as light exposure altered the structural patterns of their UV-vis spectra in a fashion that is no different from the corresponding split CarHC reconstitution in solutions (Figs. 2D and 3C and fig. S20). Light exerted opposite influence over the mechanics of the Ado and Me gels; exposing the Ado gel to continuous white light (~30 klux) decreased G′, (43.7 ± 7.2)% of its initial value, showing a photoweakening effect, while the Me gel under the same irradiation condition was strengthened mechanically, with a (3.5 ± 0.4)–fold increase of G′, according to the time sweep tests at a fixed frequency (1 rad/s) and strain (5%) (Fig. 4, E and F, and fig. S21). Frequency sweep tests at the strain of 10% further confirmed this contrast in their response to light (Fig. 4, G and H, and fig. S22). The Ado gel became a softened elastic solid after photolysis; over an angular frequency of 0.01 to 100 rad/s, the G′ values of the gel decreased from ~1.0 to 1.5 kPa before light exposure to ~0.44 to 0.75 kPa after light exposure yet remained substantially higher than the corresponding G″ (0.04 to 0.11 kPa). The decreased G′ after photolysis reflected the reduced crosslinking density, which can be attributed to the light-induced disassembly of CarHC tetramers within the molecular network. In contrast to the Ado gel, the Me gel exhibited a photostrengthening behavior. Before photolysis, its G′ (<0.01 to 0.74 kPa) and G″ (0.01 to 0.23 kPa) were frequency dependent and exhibited a crossover point at the angular frequency of ~2 rad/s, which coincided with the local maximum of G″ (~0.23 kPa). This material in the dark exhibited liquid-like (G′ < G″) and solid-like (G′ > G″) properties at low (<2 rad/s) and high (>2 rad/s) frequency, respectively, as expected for a physical network, in which the MeB12-mediated complexation of CarHCN and CarHCC constitutes the major interchain interaction. On light exposure, the Me gel transitioned rapidly from a viscoelastic liquid to an elastic solid. The resulting product exhibited a steady G′ of 0.53 to 0.81 kPa and G″ of 0.02 to 0.09 kPa over the frequency range of 0.01 to 100 rad/s, comparable to those of the photolyzed Ado gel. The similarity in their mechanical properties after photolysis came as no surprise, because both products, in principle, share a nearly identical network composition and structure, with the bis-His–ligated cobalamins as the chief interchain crosslinks. In addition, the high-frequency G′ (0.74 kPa) of the Me gel before light exposure (Me-D), which reflects the contributions from all interchain interactions combined, was close to that of the photolyzed gel (Me-L; ~0.81 kPa) (Fig. 4H and fig. S21B), suggesting that photolysis merely stabilized the interchain interactions, leaving the network structure largely unchanged.
Last but not least, according to the strain sweep tests at the frequency of 10 rad/s—a value determined by the frequency sweep tests to be within the shared viscoelastic range, these materials, including Ado-D, Ado-L, Me-D, and Me-L, were all able to maintain a steady G′, showing the stability of these molecular networks under mechanical deformation (fig. S23). However, the strain sweep tests still revealed noticeable effects of light illumination on the mechanics of the gels. The MeB12 gel, a physical molecular network, before photostrengthening can self-heal. The step strain test revealed that the Me-D gel under low strain (5%) quickly recovered from the rupture caused by high strain (500%) and that G′ and G″ remain close to the original values even after five cycles of rupture and recovery, which is highly indicative of its self-healing ability (fig. S24). Large amplitude strain sweep tests showed that photostrengthening of the MeB12 gels increased their strain linearity limit; Me-D exhibited a linearity limit (yield point, γy) at ~200% and flow point (γf), where G′ and G″ cross over, at ~400%, while the photostrengthened gel, Me-L, exhibited a much extended linear viscoelastic region, with the yield point approaching 1000% (fig. S25).
These elastic gels, including Ado-D, Ado-L, and Me-L, were remarkably stable in aqueous solution; about 10% of the materials eroded in excess PBS in 1 day and less than 30% in 9 days (fig. S27). Moreover, the time sweep test of Ado-D over a prolonged period (up to 3 days) showed that G′ and G″ leveled off within half a day and remained steady afterward, suggesting its stable mechanical properties in the dark with the passage of time (fig. S28). By contrast, Me-D, which was essentially a viscoelastic liquid, was unstable and eroded rapidly in the dark; half of the material eroded in 1 day and was fully dissolved in a week (fig. S27).
Cell encapsulation
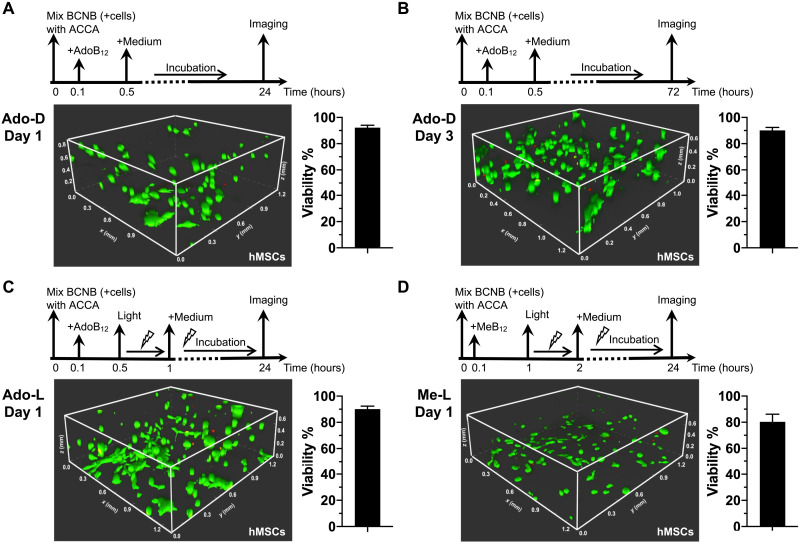
We examined the feasibility of using these stable hydrogels, i.e., Ado-D, Ado-L, and Me-L, to encapsulate and culture mouse 3T3 fibroblasts and human mesenchymal stem cells (hMSCs)—two representative cell lines widely used in tissue engineering. The gelation precursors, ACCA and BCNB, have been designed to contain the putative arginine-glycine-aspartate (RGD) cell adhesion ligand and the matrix metalloproteinase (MMP) cleavage sequence, GPQG↓IWGQ, the latter of which facilitates the matrix remodeling by encapsulated cells (Fig. 4, A and B, and fig. S13). To examine the cytocompatibility of our hydrogel composition, we added to the fibroblasts and hMSCs cultured on petri dishes the gel precursors including ACCA, BCNB, AdoB12, and MeB12. The cells turned out to be highly viable (≥90%) in the presence of these proteins and cofactors under various dark and bright conditions, which was comparable to the cell cultures free of those gel precursors, thus confirming the cytocompatibility of the gel components (fig. S29). Encapsulations were achieved by suspending the cells in a solution of BCNB [10 weight % (wt %)] in the corresponding culture medium, followed by addition of ACCA and a suitable cofactor (AdoB12 or MeB12) to initiate gelation (Fig. 5). The viability of encapsulated cells was assessed using the standard live-dead staining assay. For Ado-D, a relatively stiff matrix, encapsulated fibroblasts and hMSCs exhibited consistently not only high viability after 24 hours [(92.9 ± 1.4)% and (92.2 ± 1.9)%, respectively] but also pronounced proliferation after prolonged 72-hour incubation, with a viability of (91.3 ± 2.7)% and (90.1 ± 2.3)%, respectively (Fig. 5 and figs. S30 and S31). As to the photolyzed gels, i.e., the photoweakened Ado-L and photostrengthened Me-L, (91.2 ± 3.8)% and (88.8 ± 6.0)%, respectively, of encapsulated fibroblasts and (91.2 ± 4.7)% and (80.3 ± 5.8)% of hMSCs remained viable after 24 hours, suggesting that the photodegradation of the cofactors caused negligible cytotoxicity (Fig. 5 and figs. S30 and S31). Note that these gels used for cell encapsulation before (Ado-D) and after irradiation (Ado-L and Me-L) exhibited stiffness (~1.0 and ~0.3 to 0.4 kPa, respectively), which was comparable to that of bone marrow (0.5 to 1.5 kPa), fat (0.5 to 1.0 kPa), and brain tissues (0.1 to 1.0 kPa) (43–45). Given their resemblance to native biological tissues, these photoresponsive materials hold great promise for materials biology and tissue regeneration.
Fig. 5. 3D culturing of hMSCs.
(A) One-day culturing with Ado gels in the dark. (B) Three-day culturing with Ado gels in the dark. (C) One-day culturing with photoweakened Ado gels. (D) One-day culturing with photostrengthened Me gels. Cell viability was assessed by the standard live (green)/dead (red) staining assay. Three independent cell encapsulation experiments were performed, each time done in triplicate (with three identical gels prepared for each corresponding encapsulation condition). Cell viability under each condition was calculated as a ratio of the live-cell population to the entire cell population based on the three parallel experiments and presented as means ± SD (n = 3).
In summary, we have reported the development of a B12-induced, photoresponsive protein assembly system by dissecting the C-terminal domain of the bacterial photoreceptor CarH protein, which offers several major advantages over its traditional counterparts, including negligible background autoassembly, high reaction efficiency, and optically controlled stable conjugation. We have used this tool to create a variety of photoresponsive protein hydrogels, with tunable mechanics and well suited for three dimensional (3D) cell culturing. This new protein chemistry, with its marked efficiency, specificity, and photoresponsiveness, might also provide an alternative to the existing optogenetic tools for controlling biological signaling in vivo.
MATERIALS AND METHODS
Gene construction and protein expression
The carHCN and carHCC genes were polymerase chain reaction (PCR)–amplified from the previously reported construct, SpyTag-ELP-carHC-ELP-SpyTag (24), and cloned into the pET32m3C plasmid (46) that enabled the fusion with a Trx tag, a His6 tag, and a human rhinovirus (HRV 3C) protease cleavage sequence, using Eco RI and Hind III restriction enzymes. The CarHCN and CarHCC proteins were expressed in E. coli BL21 (DE3) and purified using Ni-NTA affinity chromatography. The Trx-His6 tag was removed by incubation with HRV 3C protease, followed by further purification with SEC. TBS [50 mM tris and 300 mM NaCl (pH 8)] was used to elute the proteins. The purified proteins were flash-frozen in liquid nitrogen and stored at −80°C.
The genes encoding hydrogel precursors, including SpyTag-ELP-carHCN-ELP-SpyTag (ACNA), SpyTag-ELP-carHCC-ELP-SpyTag (ACCA), SpyCatcher-ELP-carHCN-ELP-SpyCatcher (BCNB), and SpyCatcher-ELP-carHCC-ELP-SpyCatcher (BCCB), were created by inserting the carHCN or carHCC gene correspondingly into the previously reported construct, pQE80l::SpyTag-ELP-SpyTag-ELP-SpyTag or pQE80l::SpyCatcher-ELP-RGD-ELP-SpyCatcher, using Sac I and Spe I restriction enzymes (25). These recombinant proteins were expressed in E. coli BL21 (DE3) and purified using Ni-NTA affinity chromatography. Purified proteins were dialyzed against Milli-Q water (4 liters × 5), lyophilized, and stored at −80°C.
Cobalamin-induced protein reconstitution and photolysis
Cobalamins, including AdoB12, MeB12, and CNB12, were dissolved in PBS to make 10 mM stock solutions. All reconstitution experiments were accomplished by mixing proteins with the designated cobalamin (AdoB12, MeB12, or CNB12) in the dark for 1 hour at room temperature. The reconstitution was performed in TBS. Gentle mixing (pipetting three to five times) was needed to ensure the thorough binding of cobalamins to split CarHC. Photolysis was performed by exposing the reconstituted samples to continuous white light-emitting diode (LED) light (30 klux) for 1 hour.
White LED (Warsun, 60 W) was the major light source used in this study. The distance between the samples and the light source was ~20 cm. The intensity (30 klux), which was determined using Extech Light Meter LT300, was comparable to that of the low end of sunlight (~10 to 100 klux), according to the Engineering ToolBox (2004), Illuminance - Recommended Light Level, which is available at www.engineeringtoolbox.com/light-level-rooms-d_708.html.
CD spectroscopy
CD spectra were obtained on a Chirascan-plus CD spectrometer at room temperature. The ellipticity values of CarHC, CarHCN, CarHCC, CarHCN + CarHCC, and CarHCN + CarHCC + AdoB12 were recorded. The concentrations of each protein and cobalamin, if there was, in a low-salt tris buffer [50 mM NaCl and 10 mM tris (pH 8)] were 25 and 50 μM, respectively. Excessive cobalamin (2 equiv) was used to ensure maximum protein reassembly.
UV-vis spectroscopy
UV-vis spectra were obtained at room temperature using a Thermo Scientific Varioskan LUX multimode microplate reader. The concentration of each protein and cobalamin in samples, except for the AdoB12 binding kinetics study, was 100 and 50 μM, respectively. Note that excessive proteins (2 equiv) were used here to ensure the maximum binding of cobalamins to proteins so that the spectral changes, predominantly attributed to these cofactors, could be fully revealed. In the data analysis, the spectrum of TBS served as a background baseline, which was subtracted from the other spectra. The samples containing photolabile cobalamins such as AdoB12 and MeB12 were prepared in the dark. Repeated measurements led to a consistent spectral pattern, indicating negligible photolysis throughout the experiments. In the AdoB12 binding kinetics study, the absorbance values at 490 and 570 nm were recorded immediately upon addition of AdoB12 into protein solutions (intact CarHC or split CarHC) at a 1:1 molar ratio and the concentration of 50 μM. Abs570/Abs490 of free AdoB12 solution was used for background subtraction.
Analytical SEC
The experiments were performed on an ÄKTA Pure system (GE Healthcare) equipped with a Superose 6 Increase 10/300 GL SEC column. The designated protein sample (50 μM) with or without cobalamin (100 μM) was loaded into the column pre-equilibrated by TBS. Two equivalent cofactors were used to ensure maximum protein reassembly and reduce the sensitivity of the assays to possible off-stoichiometry caused by measurement inaccuracies.
Hydrogel preparation
The protein powders, ACNA, ACCA, BCNB, and BCCB, were weighed and dissolved in PBS to yield 10 wt % solutions. The two reactants, ACNA + BCCB or ACCA + BCNB, were mixed at an equimolar ratio, followed by addition of 1.1 equiv cobalamin (i.e., AdoB12, MeB12, or CNB12) at room temperature in the dark.
Dynamic shear rheology
Dynamic rheological measurements in time, strain, and frequency sweep modes were conducted on a TA Instruments ARES-RFS strain-controlled rheometer with a parallel steel plate geometry (top, 8 mm in diameter; bottom, 25 mm in diameter). Because AdoB12 and MeB12 were stable under long-wavelength light (>600 nm), a red-light lamp was used if photolysis needed to be avoided. Hydrogel samples were prepared by mixing 16.6 μl of ACCA (10 wt %), 27.8 μl of BCNB (10 wt %), and 5.6 μl of cobalamin (AdoB12, MeB12, or CNB12) (10 mM) or, alternatively, 14.8 μl of ACNA (10 wt %), 29.6 μl of BCCB (10 wt %), and 5.6 μl of cobalamin (10 mM) on the bottom plate. Note that the ratio of the two proteins, ACCA and BCNB, to cobalamin is 1:1:1.1, with the cofactor in slight excess, to ensure the complete reassembly of split CarHC and minimize the sensitivity of the system to possible off-stoichiometry caused by experimental inaccuracies. The loaded samples were sealed with silicone oil to prevent water evaporation. Those photolabile samples were shielded in a closed box to avoid light throughout the experiments. Time sweep tests were performed at 23°C, with the strain and frequency fixed at 5% and 1 rad/s, respectively. Strain sweep tests were performed over the strain ranging from 0.1 to 100% and at a fixed frequency of 10 rad/s at 23°C. Frequency sweep tests were performed over the frequency ranging from 100 to 0.01 rad/s and at a constant strain of 10% at 23°C. Photolysis was monitored using dynamic time sweep tests while exposing the loaded gels to continuous white LED light (30 klux). At least three independent batches of samples were used for each set of rheological tests to ensure consistency.
Culturing and encapsulation of fibroblasts and hMSCs
NIH 3T3 fibroblasts were cultured in high-glucose DMEM (Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco) and 1% (v/v) penicillin-streptomycin (Sangon Biotech) in a 5% CO2 atmosphere at 37°C and passaged every third day. hMSCs were cultured in minimum essential medium (MEM α) (Gibco) supplemented with 10% (v/v) FBS (Gibco), 1% (v/v) penicillin-streptomycin (Sangon Biotech), and 2 mM l-glutamine in a 5% CO2 atmosphere at 37°C and passaged every week. At 70 to 80% confluence, cells were detached with 1 ml of 0.25% trypsin solution (Sangon Biotech) followed by addition of 2 ml of full medium to neutralize trypsin. About 50,000 cells were pelleted and resuspended with 27.8 μl of the 10 wt % BCNB solution in the corresponding culture medium. The resulting cell suspension was then placed on a cell culture confocal dish with a coverslip bottom and mixed with 16.6 μl of ACCA (10 wt % in culture medium). Gelation was initiated by adding 5.6 μl of AdoB12 or MeB12 (10 mM in PBS). For the encapsulation with Ado-D gels, the gels were cured in the dark for 20 min, immersed with the corresponding culture medium (1 ml), covered by aluminum foil, and then placed in a cell culture incubator (5% CO2, 37°C) for 24 or 72 hours. As to the encapsulation with photolyzed Ado-L gels, a similar procedure was followed, except that the cell-laden gels were exposed to white light (30 klux) for 30 min before the supplementation of the corresponding culture medium (1 ml). Cell encapsulation with Me-L gels was similar to that with the Ado-L gels, except of prolonged gelation—1-hour curation in the dark and 1-hour light exposure. After incubation, standard live/dead staining assays (Thermo Fisher Scientific) were performed, followed by confocal fluorescence z-stack imaging, to determine cell viability. For each cell encapsulation experiment, three identical gels from the same batch of proteins were synthesized and tested under the same condition. At least three independent experiments were performed for each set of cell culture studies to ensure consistency.
Acknowledgments
We thank Jiguang Wang from the Hong Kong University of Science and Technology (HKUST) for constructive comments.
Funding: This work was supported by Ministry of Science and Technology 2020YFA0908100 (F.S.); Natural Science Foundation of China Excellent Young Scientists Fund NSFC21EG08 (F.S.); Guangdong Natural Science Foundation GDST19EG22 (F.S.); Research Institute of Tsinghua, Pearl River Delta #RITPRD21EG01 (F.S.); Science, Technology and Innovation Commission of Shenzhen Municipality Basic Research Program Grant #JCYJ20190813094601656 (F.S.) and Key Research Program Grant #JCYJ20200109141241950 (F.S.); and Research Grants Council of Hong Kong SAR GRF Grant #16103519 and #16103421 (F.S.). MALDI-TOF service: Biosciences Central Research Facility, HKUST.
Author contributions: Conceptualization: F.S. Methodology: Z.Y. and F.S. Investigation: Z.Y., H.K.F.F., J.L., Y.Y., R.W., and X.H. Funding acquisition: F.S. Writing—original draft: Z.Y. and F.S. Writing—review and editing: All authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The plasmids, including pET32m3C-carHCN, pET32m3C-carHCC, pQE-ACNA, pQE-ACCA, pQE-BCNB, and/or pQE-BCCB, can be provided by the HKUST pending scientific review and a completed material transfer agreement. Requests for these materials should be submitted to F.S.
Supplementary Materials
This PDF file includes:
Figs. S1 to S31
Table S1
Appendix
REFERENCES AND NOTES
- 1.Richards F. M., On the enzymatic activity of subtilisin-modified ribonuclease. Proc. Natl. Acad. Sci. U.S.A. 44, 162–166 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnsson N., Varshavsky A., Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U.S.A. 91, 10340–10344 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh I., Hamilton A. D., Regan L., Antiparallel leucine zipper-directed protein reassembly: Application to the green fluorescent protein. J. Am. Chem. Soc. 122, 5658–5659 (2000). [Google Scholar]
- 4.Romei M. G., Boxer S. G., Split green fluorescent proteins: Scope, limitations, and outlook. Annu. Rev. Biophys. 48, 19–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones K. A., Kentala K., Beck M. W., An W. W., Lippert A. R., Lewis J. C., Dickinson B. C., Development of a split esterase for protein-protein interaction-dependent small-molecule activation. ACS Cent. Sci. 5, 1768–1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulmurugan R., Umezawa Y., Gambhir S. S., Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. U.S.A. 99, 15608–15613 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehr M. C., Reinecke L., Botvinnik A., Rossner M. J., Analysis of transient phosphorylation-dependent protein-protein interactions in living mammalian cells using split-TEV. BMC Biotechnol. 8, 55 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camacho-Soto K., Castillo-Montoya J., Tye B., Ghosh I., Ligand-gated split-kinases. J. Am. Chem. Soc. 136, 3995–4002 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Gao X. J., Chong L. S., Kim M. S., Elowitz M. B., Programmable protein circuits in living cells. Science 361, 1252–1258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zetsche B., Volz S. E., Zhang F., A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nihongaki Y., Kawano F., Nakajima T., Sato M., Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Nihongaki Y., Otabe T., Ueda Y., Sato M., A split CRISPR-Cpf1 platform for inducible genome editing and gene activation. Nat. Chem. Biol. 15, 882–888 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Yu Y., Wu X., Guan N., Shao J., Li H., Chen Y., Ping Y., Li D., Ye H., Engineering a far-red light-activated split-Cas9 system for remote-controlled genome editing of internal organs and tumors. Sci. Adv. 6, eabb1777 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fink T., Lonzaric J., Praznik A., Plaper T., Merljak E., Leben K., Jerala N., Lebar T., Strmsek Z., Lapenta F., Bencina M., Jerala R., Design of fast proteolysis-based signaling and logic circuits in mammalian cells. Nat. Chem. Biol. 15, 115–122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagliyan O., Krokhotin A., Ozkan-Dagliyan I., Deiters A., Der C. J., Hahn K. M., Dokholyan N. V., Computational design of chemogenetic and optogenetic split proteins. Nat. Commun. 9, 4042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolberg T. B., Meger A. T., Boucher J. D., Corcoran W. K., Schauer E. E., Prybutok A. N., Raman S., Leonard J. N., Computation-guided optimization of split protein systems. Nat. Chem. Biol. 17, 531–539 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z., Yang Y., Wang M., Wang T., Fok H. K. F., Jiang B., Xiao W., Kou S., Guo Y., Yan Y., Deng X., Zhang W.-B., Sun F., Dynamically tunable, macroscopic molecular networks enabled by cellular synthesis of 4-arm star-like proteins. Matter 2, 233–249 (2020). [Google Scholar]
- 18.Zakeri B., Fierer J. O., Celik E., Chittock E. C., Schwarz-Linek U., Moy V. T., Howarth M., Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keeble A. H., Banerjee A., Ferla M. P., Reddington S. C., Anuar I., Howarth M., Evolving accelerated amidation by SpyTag/SpyCatcher to analyze membrane dynamics. Angew. Chem. Int. Ed. 56, 16521–16525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeble A. H., Turkki P., Stokes S., Khairil Anuar I. N. A., Rahikainen R., Hytonen V. P., Howarth M., Approaching infinite affinity through engineering of peptide-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 26523–26533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W. B., Sun F., Tirrell D. A., Arnold F. H., Controlling macromolecular topology with genetically encoded SpyTag-SpyCatcher chemistry. J. Am. Chem. Soc. 135, 13988–13997 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Sun F., Zhang W. B., Genetically encoded click chemistry. Chin. J. Chem. 38, 894–896 (2020). [Google Scholar]
- 23.Sun F., Zhang W. B., Unleashing chemical power from protein sequence space toward genetically encoded “click” chemistry. Chin. Chem. Lett. 28, 2078–2084 (2017). [Google Scholar]
- 24.Wang R., Yang Z., Luo J., Hsing I. M., Sun F., B12-dependent photoresponsive protein hydrogels for controlled stem cell/protein release. Proc. Natl. Acad. Sci. U.S.A. 114, 5912–5917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun F., Zhang W. B., Mahdavi A., Arnold F. H., Tirrell D. A., Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc. Natl. Acad. Sci. U.S.A. 111, 11269–11274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X., Fang J., Xue B., Fu L., Li H., Engineering protein hydrogels using SpyCatcher-SpyTag chemistry. Biomacromolecules 17, 2812–2819 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Wei J., Wu W.-H., Wang R., Yang Z., Sun F., Zhang W.-B., B12-dependent protein oligomerization facilitates layer-by-layer growth of photo/thermal responsive nanofilms. ACS Macro Lett. 7, 514–518 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Obana M., Silverman B. R., Tirrell D. A., Protein-mediated colloidal assembly. J. Am. Chem. Soc. 139, 14251–14256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z., Kou S., Wei X., Zhang F., Li F., Wang X.-W., Lin Y., Wan C., Zhang W.-B., Sun F., Genetically programming stress-relaxation behavior in entirely protein-based molecular networks. ACS Macro Lett. 7, 1468–1474 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Tan T. K., Rijal P., Rahikainen R., Keeble A. H., Schimanski L., Hussain S., Harvey R., Hayes J. W. P., Edwards J. C., McLean R. K., Martini V., Pedrera M., Thakur N., Conceicao C., Dietrich I., Shelton H., Ludi A., Wilsden G., Browning C., Zagrajek A. K., Bialy D., Bhat S., Stevenson-Leggett P., Hollinghurst P., Tully M., Moffat K., Chiu C., Waters R., Gray A., Azhar M., Mioulet V., Newman J., Asfor A. S., Burman A., Crossley S., Hammond J. A., Tchilian E., Charleston B., Bailey D., Tuthill T. J., Graham S. P., Duyvesteyn H. M. E., Malinauskas T., Huo J., Tree J. A., Buttigieg K. R., Owens R. J., Carroll M. W., Daniels R. S., McCauley J. W., Stuart D. I., Huang K.-Y. A., Howarth M., Townsend A. R., A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 12, 542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J., Sun F., Calcium-responsive hydrogels enabled by inducible protein–protein interactions. Polym. Chem. 11, 4973–4977 (2020). [Google Scholar]
- 32.Kutta R. J., Hardman S. J. O., Johannissen L. O., Bellina B., Messiha H. L., Ortiz-Guerrero J. M., Elias-Arnanz M., Padmanabhan S., Barran P., Scrutton N. S., Jones A. R., The photochemical mechanism of a B12-dependent photoreceptor protein. Nat. Commun. 6, 7907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jost M., Fernandez-Zapata J., Polanco M. C., Ortiz-Guerrero J. M., Chen P. Y., Kang G., Padmanabhan S., Elias-Arnanz M., Drennan C. L., Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 526, 536–541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortiz-Guerrero J. M., Polanco M. C., Murillo F. J., Padmanabhan S., Elias-Arnanz M., Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 108, 7565–7570 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camacho I. S., Black R., Heyes D. J., Johannissen L. O., Ramakers L. A. I., Bellina B., Barran P. E., Hay S., Jones A. R., Interplay between chromophore binding and domain assembly by the B12-dependent photoreceptor protein, CarH. Chem. Sci. 12, 8333–8341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kainrath S., Stadler M., Reichhart E., Distel M., Janovjak H., Green-light-induced inactivation of receptor signaling using cobalamin-binding domains. Angew. Chem. Int. Ed. 56, 4608–4611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang B., Liu X., Yang C., Yang Z., Luo J., Kou S., Liu K., Sun F., Injectable, photoresponsive hydrogels for delivering neuroprotective proteins enabled by metal-directed protein assembly. Sci. Adv. 6, eabc4824 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marques H. M., Marsh J. H., Mellor J. R., Munro O. Q., The coordination of imidazole and its derivatives by aquocobalamin. Inorg. Chim. Acta 170, 259–269 (1990). [Google Scholar]
- 39.Burdick J. A., Murphy W. L., Moving from static to dynamic complexity in hydrogel design. Nat. Commun. 3, 1269 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Langer R., Tirrell D. A., Designing materials for biology and medicine. Nature 428, 487–492 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Mascharak S., desJardins-Park H. E., Davitt M. F., Griffin M., Borrelli M. R., Moore A. L., Chen K., Duoto B., Chinta M., Foster D. S., Shen A. H., Januszyk M., Kwon S. H., Wernig G., Wan D. C., Lorenz H. P., Gurtner G. C., Longaker M. T., Preventing Engrailed-1 activation in fibroblasts yields wound regeneration without scarring. Science 372, eaba2374 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen B. D., Kornfield J. A., Tirrell D. A., Yielding behavior in injectable hydrogels from telechelic proteins. Macromolecules 43, 9094–9099 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler A. J., Sen S., Sweeney H. L., Discher D. E., Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Levental K. R., Yu H., Kass L., Lakins J. N., Egeblad M., Erler J. T., Fong S. F., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D. L., Weaver V. M., Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handorf A. M., Zhou Y., Halanski M. A., Li W. J., Tissue stiffness dictates development, homeostasis, and disease progression. Organogenesis 11, 1–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu C., Feng W., Wei Z., Miyanoiri Y., Wen W., Zhao Y., Zhang M., Myosin VI undergoes cargo-mediated dimerization. Cell 138, 537–548 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S31
Table S1
Appendix