Abstract
We have studied the breadth and potency of the inhibitory actions of the CC chemokines macrophage inhibitory protein 1α (MIP-1α), MIP-1β, and RANTES against macrophage-tropic (M-tropic) primary isolates of human immunodeficiency virus type 1 (HIV-1) and of the CXC chemokine stromal cell-derived factor 1α against T-cell-tropic (T-tropic) isolates, using mitogen-stimulated primary CD4+ T cells as targets. There was considerable interisolate variation in the sensitivity of HIV-1 to chemokine inhibition, which was especially pronounced for the CC chemokines and M-tropic strains. However, this variation was not obviously dependent on the genetic subtype (A through F) of the virus isolates. Peripheral blood mononuclear cell donor-dependent variation in chemokine inhibition potency was also observed. Among the CC chemokines, the rank order for potency (from most to least potent) was RANTES, MIP-1β, MIP-1α. Some M-tropic isolates, unexpectedly, were much more sensitive to RANTES than to MIP-1β, whereas other isolates showed sensitivities comparable to those of these two chemokines. Down-regulation of the CCR5 and CXCR4 receptors occurred in cells treated with the cognate chemokines and probably contributes to anti-HIV-1 activity. Thus, for CCR5, the rank order for down-regulation was also RANTES, MIP-1β, MIP-1α.
The CC chemokines macrophage inhibitory protein-1α (MIP-1α), MIP-1β, and RANTES inhibit the replication of certain human immunodeficiency virus type 1 (HIV-1) strains in CD4+ T cells (10, 11, 29, 43, 46). The HIV-1 isolates that are most sensitive to these CC chemokines have the macrophage-tropic (M-tropic) phenotype and do not form syncytia in MT-2 cells (29). These viruses are therefore alternatively described as non-syncytium inducing (NSI). M-tropic viruses enter CD4+ T cells by fusion at the plasma membrane in a pathway that involves the CD4 molecule and the CC chemokine receptor CCR5 (1, 9, 15, 17, 18), for which the known ligands are MIP-1α, MIP-1β, and RANTES (45, 48). These CC chemokines inhibit HIV-1 replication because they are antagonists of HIV-1 entry (or env-mediated membrane fusion) (1, 11, 15, 42). Competition for CCR5 binding between the above-mentioned CC chemokines and the HIV-1 surface glycoprotein gp120 contributes to the inhibitory mechanism (27, 55, 59). Taken together, these studies suggest that the development of inhibitors of viral entry based on the CC chemokines might be a viable approach to antiviral therapy against HIV-1. Indeed, derivatives of RANTES have been shown to be more effective than RANTES itself, in vitro (4, 52).
The CXC chemokine stromal cell-derived factor (SDF)-1α has been shown to inhibit the replication of T-cell-tropic (T-tropic) primary isolates, or T-cell line-adapted strains, at the level of virus entry (7, 41). Usually, these viruses form syncytia in MT-2 cells, and they are often called syncytium-inducing (SI) strains. T-tropic viruses can enter CD4+ T cells by using the CXC chemokine receptor CXCR4 (22), of which the only known ligand is SDF-1α (7, 41). However, many T-tropic primary isolates can use both CCR5 and CXCR4 and so are considered dual tropic (12, 53). T-tropic viruses are usually relatively insensitive to the above-mentioned CC chemokines (10, 29).
A decade ago, gp120 antagonists that inhibit the binding of HIV-1 to its primary receptor, CD4, such as the soluble CD4 molecule, were found to be highly effective at neutralizing the infectivity of the T-cell line-adapted HIV-1 strains on which they were tested initially (23, 39). However, it was later apparent that primary viruses were relatively resistant to the effects of soluble CD4 (8, 13, 38). With this lesson in mind, we set out to investigate the broadness of the spectrum of HIV-1 strains on which the CC and CXC chemokines were active. A second consideration was to determine whether there were genetic subtype-dependent patterns in the sensitivity of HIV-1 strains to these chemokines, since therapeutic agents should ideally not act on only a single genetic subtype. Finally, we wished to know which of the CC chemokines was the most active inhibitor of HIV-1 replication (hence, the best template for therapeutic development) and whether we could discern subtleties in the mechanisms of action of the individual CC chemokines.
We therefore assembled panels of M- and T-tropic HIV-1 strains from genetic subtypes A through F and tested the effects of MIP-1α, MIP-1β, RANTES, and SDF-1α on their replication in primary CD4+ T cells. Our conclusions are that there is no obvious restriction on the actions of the CC chemokines or SDF-1α that relates to the genetic subtypes, that RANTES was the most potent inhibitor among the three CC chemokines we tested, and that RANTES is better than MIP-1β and MIP-1α at down-regulating CCR5, which probably contributes to its actions in vitro.
MATERIALS AND METHODS
Viruses and chemokines.
Many virus isolates were obtained as part of the National Institute of Allergy and Infectious Disease HIV-1 Antigenic Variation study or from similar programs organized by the U.S. Department of Defense or the World Health Organization. Their precise origins and their use in previous studies have been described previously (36, 37, 56). Other isolates were obtained as follows: isolate C 7/86 was from R. Connor (12), the molecular clone of SF-2 was from C. Cheng-Mayer (34), the biologically cloned isolate HC4 was from S. Forte and J. Sullivan (24), the molecular clone DH123 was from R. Shibata and M. Martin (50), and the biologically cloned isolate 2076 clone 3 was from P. Clapham (53).
The recombinant human CC chemokines MIP-1α, MIP-1β, and RANTES were purchased from R&D Systems Inc. (Minneapolis, Minn.). Synthetic SDF-1α stocks were provided by Gryphon Sciences (M.A.S. and D.A.T.) (51) and Berlex Biosciences (R.H.) (30). These proved to be of comparable purity and potency in blocking HIV-1 replication. Initial experiments were performed with both stocks, but only data derived from using the Gryphon preparation are shown. The CXCR4 down-regulation experiments (see Fig. 5b and 6b) were only performed with Gryphon SDF-1α.
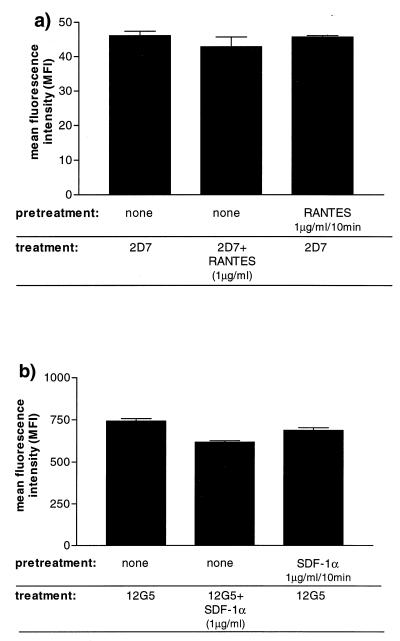
FIG. 5.
Determination of surface expression of CCR5 and CXCR4 on chemokine-treated cells. Chemokine receptor expression on CD4+ T cells was determined by MAb staining on untreated cells, on cells pretreated for 10 min with 1 μg of chemokine per ml and then washed, and in the continuous presence of 1 μg of chemokine per ml, as indicated. Shown are mean fluorescence intensities obtained for CCR5 staining with MAb 2D7 in the presence and absence of RANTES (a) and for CXCR4 with MAb 12G5 in the presence and absence of SDF-1α (b). Error bars, standard error of the mean.
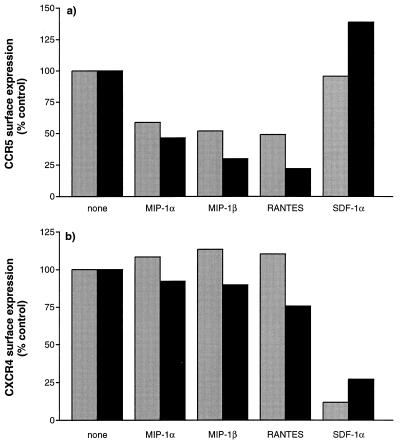
FIG. 6.
Surface expression of CCR5 and CXCR4 after exposure to CC and CXC chemokines. CD4+ T cells were treated for 2 h (gray bars) and 3 days (black bars) with CC or CXC chemokines (1 μg/ml) and the stained for surface expression of CCR5 (a) and CXCR4 (b) with MAbs 2D7 and 12G5, respectively. The median fluorescence intensities of the CCR5- and CXCR4-positive populations are shown as percentages of the levels for untreated control cells. The data shown were derived from one of two independent experiments on cells from two different donors.
Determination of viral phenotype and coreceptor use.
The phenotypes of many of the test viruses have been described previously (36). Others were tested by the same method: their ability to form syncytia in MT-2 cells. These cells were cultured in RPMI 1640 medium containing 10% fetal calf serum (FCS), glutamine, and antibiotics and split twice a week. For infection assays, 105 cells were incubated with virus for 16 h and then unbound virus was removed by two washes in culture medium. From days 3 through 7 postinfection, the cultures were examined microscopically for syncytium formation and the supernatant was analyzed for p24 antigen production by an in-house enzyme-linked immunosorbent assay, as described previously (56).
U87MG-CD4 cell lines stably transfected with the CCR5 or CXCR4 genes were a gift from Dan Littman (Skirball Institute for Molecular Medicine, New York, N.Y.) (27). These cells were maintained in Dulbecco’s minimal essential medium containing 10% FCS, glutamine, antibiotics, puromycin (1 μg/ml; Sigma Chemicals), and neomycin (300 μg/ml; G418; Sigma) and split twice a week. For HIV-1 infection experiments, 5 × 104 cells were incubated with virus for 16 h, and then unbound virus was removed by two washes in culture medium. On days 3 and 6 postinfection, the cultures were examined microscopically for syncytium formation and the supernatant was analyzed for the presence of p24 antigen.
Chemokine inhibition of HIV-1 replication in primary CD4+ T cells.
Peripheral blood mononuclear cells (PBMC) were isolated from healthy blood donors by Ficoll-Hypaque centrifugation and then stimulated for 2 to 3 days with phytohemagglutinin (5 μg/ml) and interleukin-2 (IL-2) (100 U/ml) (a gift from Hoffmann-LaRoche, Nutley, N.J.). CD4+ T cells were purified from the activated PBMC by positive selection with anti-CD4 immunomagnetic beads (DYNAL Inc.). The purified lymphocytes were cultured for at least 3 days at 2 × 106/ml in medium containing IL-2 (200 U/ml) before being used in the 125I-chemokine binding assay and for at least 1 day before being used in infection assays. The cells were screened for CCR5-defective alleles (32), and only cells from wild-type donors were used (except when specified).
Inhibition of infection by chemokines was assessed as follows: 2 × 105 CD4+ T cells in 100 μl of assay medium (RPMI 1640, 10% FCS, 100 U/ml IL-2, glutamine, and antibiotics) were incubated with serial dilutions of the chemokines (50 μl) for 1 h at 37°C. The virus inoculum was adjusted to 400 to 1,000 50% tissue culture infectious doses/ml, and a 50-μl aliquot was added to each culture. The calculated inhibitory doses refer to the final concentration of chemokine in the culture on day 0. On days 4 and 6 postinfection, 50 μl of supernatant was assayed for p24 antigen. As the virus inoculum was not washed out at any stage of the experiment, we also measured the residual input p24 concentration, which was subtracted from all test results. If virus production in the cultures had not reached its peak on day 6, the cultures were fed with 100 μl of medium without adding fresh chemokines and then reanalyzed for p24 production on days 8, 10, and 12. Virus production in the absence of chemokine was designated as 100%, and the ratios of p24 antigen production in chemokine-containing cultures were calculated relative to this. The chemokine concentrations (in picograms per milliliter) causing 50% and 90% reduction in p24 antigen production were determined by linear regression analysis. If the appropriate degree of inhibition was not achieved at the highest or lowest chemokine concentration, a value with “>” or “<” was recorded.
Competition between gp120 and labeled chemokines.
These experiments, using activated CD4+ T cells (2 × 106 in 200 μl), were performed as described previously (55). 125I-RANTES and 125I-MIP-1β (specific activity, 2,200 Ci/mmol; Dupont-NEN) were used at 220 μCi/ml (0.1 nM). Monomeric gp120 from the M-tropic JR-FL strain was a gift from Paul Maddon (Progenics Pharmaceuticals Inc.) (55).
Fluorescence-activated cell sorter analysis of CCR5 and CXCR4 expression levels.
Phytohemagglutinin- and IL-2-stimulated CD4+ T cells were adjusted to 2 × 106/ml in RPMI 1640 medium containing 10% FCS, 100 mM glutamine, antibiotics, and IL-2 (200 U/ml). The cells were incubated for 3 days with or without RANTES (1 μg/ml), MIP-1α, MIP-1β, or SDF-1α. On day 3, the cells were washed twice with Dulbecco’s phosphate-buffered saline (PBS) containing 1% bovine serum albumin and 0.05% sodium azide (staining buffer) and then incubated for 20 min at room temperature with the murine anti-CCR5 monoclonal antibody (MAb) 2D7 (10 μl of hybridoma supernatant) (60, 61), the murine anti-CXCR4 MAb 12G5 (10 μg/ml in staining buffer; 10 μl) (19), or murine immunoglobulin G1 (IgG1) and IgG2a isotype control MAbs (Becton Dickinson). The cells were then washed three times with staining buffer and resuspended in 25 μl of R-phycoerythrin-labeled goat-anti mouse IgG (1:50 in staining buffer; DAKO). After 20 min at room temperature, the cells were again washed three times with staining buffer, resuspended in 50 μl of PBS, and fixed with 200 μl of PBS containing 1% formaldehyde. Surface staining was analyzed with a FACScalibur machine (Becton Dickinson). Mean and median fluorescence intensity values were derived by using CellQuest software.
RESULTS
Characterization of test panels of HIV-1 isolates.
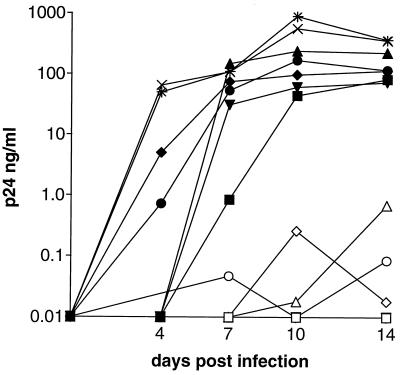
To assess the anti-HIV-1 activity of the CC chemokine ligands of CCR5, it was necessary to assemble a suitable panel of isolates, using viruses from multiple genetic subtypes. We first chose those known from our previous studies to have the NSI phenotype (36, 37, 56), in that they did not form syncytia in MT-2 cells and so were unlikely to use the CXCR4 coreceptor efficiently (12, 53). These isolates were then tested for their ability to replicate in PBMC from individuals with two wild-type or two defective (Δ-32) CCR5 alleles (depicted for representative isolates in (Fig. 1). An inability of HIV-1 to replicate efficiently in cells from Δ-32 CCR5 homozygotes indicates a dependency on CCR5 for their entry and replication (18, 32, 43), although some caveats as to the interpretation of these experiments are noted below (see Discussion section).
FIG. 1.
Replication of representative M-tropic isolates in CD4+ T cells from donors homozygous for either wild-type or Δ32 CCR5 alleles. T cells from individuals homozygous for wild-type or Δ-32 CCR5 alleles are indicated by closed and open symbols respectively. Results for replication of the M-tropic isolates 92RW026 (▪, □), JRCSF (▴, ▵), DJ259 (▾, ▿), BZ162 (•, ○), and CM235 (⧫, ◊) and the T-tropic isolate NL4/3 (wild type [×]; Δ-32 [*]) are shown.
The final test panel comprised two isolates from each of the genetic subtypes A, D, E, and F, three isolates from subtype C, and four isolates from subtype B (Table 1). We tested the coreceptor usage of these isolates with U87MG-CD4 cells stably expressing either CCR5 or CXCR4 (27). Each isolate could use only CCR5 under these circumstances (Table 1). This panel is referred to, for convenience, as the M-tropic panel, although we have not tested the ability of all the isolates to replicate in macrophages.
TABLE 1.
Panels of M- and T-tropic HIV-1 isolatesa
| Virus | Genetic subtype | Phenotypeb | Replication in CD4+ T cells
|
Replication in U87MG-CD4 transfectantse
|
|||
|---|---|---|---|---|---|---|---|
| CCR5 wild typec | CCR5 wild-type p24 (ng/ml)d | CCR5 d32/d32c | CCR5 | CXCR4 | |||
| M-tropic isolates | |||||||
| 92RW026 | A | NSI | + | 18 | − | + | − |
| DJ258 | A | NSI | + | 32 | − | + | − |
| JRFL | B | NSI | + | 16.5 | − | + | − |
| JRCSF | B | NSI | + | 64 | − | + | − |
| SF162 | B | NSI | + | 22.5 | − | + | − |
| 92US657 | B | NSI | + | 65 | − | + | − |
| DJ259 | C | NSI | + | 18 | − | + | − |
| 94ZW103 | C | NSI | + | 59.5 | − | + | − |
| 94ZW109 | C | NSI | + | 89.5 | − | + | − |
| 94KE102 | D | NSI | + | 23.5 | − | + | − |
| 94KE103 | D | NSI | + | 24.5 | − | + | − |
| CM235 | E | NSI | + | 22 | − | + | − |
| 92TH001 | E | NSI | + | 14 | − | + | − |
| BZ162 | F | NSI | + | 10 | − | + | − |
| R1 | F | NSI | + | 17.5 | − | + | − |
| T-tropic isolates | |||||||
| 92UG029 | A | SI | + | 7 | + | − | + |
| 2076 cI.3 | B | SI | + | 27.5 | + | + | + |
| C 7/86 | B | SI | + | 23.5 | + | + | + |
| DH123 | B | SI | + | 21 | + | + | + |
| SF2 | B | SI | + | 12 | + | + | + |
| HC4 | B | SI | + | 16 | + | − | + |
| NL4-3 | B | SI | + | 11 | + | − | + |
| ZAM20 | C | SI | + | 14.5 | + | − | + |
| 94ZW106 | C | SI | + | 4 | + | − | + |
| UG270 | D | SI | + | 15 | + | − | + |
| 92UG046 | D | SI | + | 36 | + | − | + |
| 92UG024 | D | SI | + | 34.5 | + | − | + |
| 94TH304 | E | SI | + | 6.5 | + | − | + |
The designations and properties of M-tropic and T-tropic primary HIV-1 isolates are recorded.
Phenotype determined by ability of isolate to form (SI) or not form (NSI) syncytia in MT-2 cells.
Ability (+) or inability (−) of isolate to replicate efficiently in activated CD4+ T cells from individuals homozygous for either wild-type or Δ-32 CCR5 alleles.
Amount of p24 antigen produced in wild-type CD4+ T cells. Values are derived from a single experiment typical of several performed.
Ability of isolate to replicate in U87MG-CD4 cells stably expressing either CCR5 or CXCR4.
We also assembled a panel of CXCR4-using viruses for studies of the antiviral effect of CXC chemokine SDF-1α. The viruses chosen were all of the SI phenotype, in that they formed syncytia in MT-2 cells, and all the SI strains that were tested replicated in PBMC from individuals homozygous for Δ-32 CCR5 alleles (Table 1). Each SI virus could replicate in U87MG-CD4 cells stably expressing CXCR4, but some also replicated in CCR5-expressing U87MG-CD4 cells (27), indicating that they were dual tropic (Table 1). This panel is referred to as the T-tropic panel. It comprised one isolate from each of the genetic subtypes A and E, two isolates from subtype C, three isolates from subtype D, and six isolates from subtype B; we were unable to identify a T-tropic primary virus from subtype F (Table 1).
CC chemokine sensitivity of M-tropic isolates.
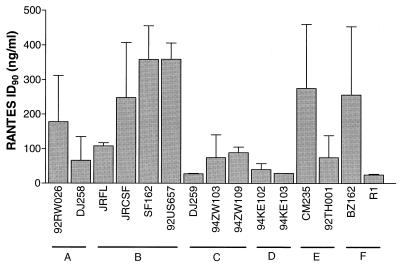
The M-tropic isolates were tested for sensitivity to inhibition by each of the CC chemokines MIP-1α, MIP-1β, and RANTES and by a 1:1:1 mixture of the three, in mitogen-stimulated PBMC (Table 2; Fig. 2 and 3). Recorded in Table 2 are the median 50% inhibitory doses (ID50s) and ID90s for each CC chemokine and the equimolar mixture against each virus. There was, clearly, considerable variation in the sensitivity of different isolates to inhibition by CC chemokines; for example, the virus most sensitive to RANTES was the Romanian subtype F isolate R1 (ID90, 25 ng/ml); the one least sensitive was the North American subtype B strain SF162 (ID90, 413 ng/ml [a 16.5-fold higher concentration]).
TABLE 2.
Inhibition of M-tropic viruses by CC-chemokinesa
| Virus | Subtype | RANTES
|
MIP-1α
|
MIP-1β
|
Mix
|
||||
|---|---|---|---|---|---|---|---|---|---|
| ID90 | ID50 | ID90 | ID50 | ID90 | ID50 | ID90 | ID50 | ||
| 92RW026 | A | 175 | 53.8 | >456* | 177 | 272 | 61.4 | 201 | 94.5 |
| DJ258 | A | 66 | 28.7 | 281.9 | 142 | 339 | 55.9 | 54.7 | <12.2† |
| JRFL | B | 107 | 32.0 | >499* | 236 | 439 | 156 | 152 | 75.5 |
| JRCSF | B | 223 | 99.2 | >485* | 369 | 463 | 295 | 122 | 59.3 |
| SF162 | B | 413 | 50.8 | >500 | >500 | >500 | >500* | >500* | 296 |
| 92US657 | B | 359 | 97.9 | >348* | 281* | 467 | 86.8 | >294* | 15.5 |
| DJ259 | C | 27.3 | 11.4 | 255 | 27.5† | 403 | 69.3 | 27 | 13.8 |
| 94ZW103 | C | 74.2 | 42.0 | 303 | 79.4 | 297 | 153 | 198 | 54.0 |
| 94ZW109 | C | 88.9 | <13.9† | 460 | 205 | 381* | <192† | 226 | 15.1 |
| 94KE102 | D | 39.9 | 16.3 | 376 | 60.1 | 190 | 101 | 83.4 | 18.4 |
| 94KE103 | D | 28.9 | 19.7 | 91.3 | 19.6 | 27.7 | 5.9 | 24 | 9.6 |
| CM235 | E | 227 | 70.6 | >500 | >500 | >500 | >500* | >500* | 313 |
| 92TH001 | E | 74.6 | 51.2 | >500 | 168 | 275.2 | 176 | 118 | 87.5 |
| BZ162 | F | 256 | 49.2 | >500 | 455 | 246.2 | 184 | 408 | 108 |
| R1 | F | 24.6 | <1.95† | 376 | <1.95 | 310.5 | <1.95 | 255 | <1.95 |
The median ID50s and ID90s (in nanograms per milliliter) for the CC chemokines RANTES, MIP-1α, and MIP-1β, and a 1:1:1 mixture of all three (Mix) against each M-tropic isolate are recorded. A value of >500 indicates that 50 or 90% inhibition was not achieved at a CC chemokine concentration of 500 ng/ml (the highest tested) in any experiment. A value of <1.95 indicates that 50 or 90% inhibition was always achieved at a CC chemokine concentration of 1.95 ng/ml (the lowest tested). Some median values are marked with an asterisk or dagger because in some of several experiments with this virus and CC chemokine combination, the ID50 or ID90 recorded was <1.95 ng/ml or >500 ng/ml. For the purposes of calculating the medians of multiple determinations, the off-scale values were set at 1.95 or 500, and the resulting median is considered a lower or upper limit, respectively. The “<” or “>” symbols by these values also indicate this.
FIG. 2.
Inhibition of M-tropic primary isolates by RANTES. The genetic subtypes of the test isolates are recorded below the isolate designations. The values shown are the medians + standard deviations (error bars) of two to five determinations, each on CD4+ T cells from different donors who were each wild type for CCR5.
FIG. 3.
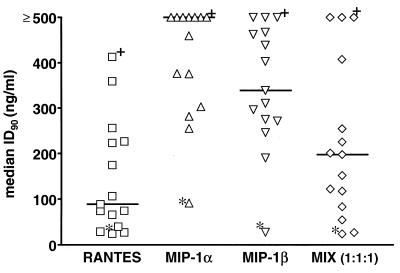
Inhibition of M-tropic viruses by CC chemokines. The individual median ID90s for each isolate and each CC chemokine (Table 2) are presented, with the overall median values indicated by bars. The data points marked with an asterisk and a dagger represent, respectively, the isolates most (94KE103, subtype D) and least (SF162, subtype B) sensitive to inhibition. MIX (1:1:1), 1:1:1 mixture of all three CC chemokines.
In principle, differences in virus replication rates could affect the degree of inhibition by CC chemokines, since several rounds of replication take place in the PBMC cultures. To limit this effect, we took care to record HIV-1 antigen production when virus replication initially peaked, by sampling the cultures repeatedly from 4 days after infection onwards. Rapidly replicating viruses were therefore harvested earlier than slowly replicating ones, which means that we measured the inhibitory effects of the CC chemokines after a similar number of replication rounds irrespective of the replication kinetics. Typical p24 values at the time of harvest for each isolate are recorded in Table 1. We found no correlation between the replication efficiency of an isolate and its sensitivity to CC chemokine inhibition (Tables 1 and 2).
There was also no obvious relationship between the genetic subtype of the test isolate and CC chemokine sensitivity, a point illustrated for RANTES in Fig. 2. Although the number of test isolates from each individual subtype was small, some isolates from each subtype were RANTES sensitive (ID90s, <100 ng/ml). Relatively insensitive isolates (ID90, >100 ng/ml) were identified from subtypes A, B, E, and F (Fig. 2; Table 2). The absence of insensitive subtype C and D strains from our panel is probably attributable to chance.
Figure 2 also illustrates another point: interdonor variability in the CC chemokine sensitivity of HIV-1 replication in mitogen-stimulated PBMC. Thus, the standard deviations of the median values can be quite large (also for MIP-1α and MIP-1β [not shown]). Our experience is that this is due to the use of cells from different donors in repeat experiments (within an individual experiment, variation among replicates is much less profound). Overall, HIV-1 replication in some donors’ cells is quite sensitive to these three CC chemokines and that in others is quite insensitive, and the range of variation in ID90s can exceed 1 log. This does not obscure interisolate variation in sensitivity but obviously complicates analysis of it. Because of the anonymous nature of the blood donors whose cells we used, we were unable to explore any gender, racial, or other personal factors that could impact the variation observed.
When we compared the three CC chemokines for their individual potencies as HIV-1 inhibitors, RANTES was clearly the most effective, MIP-1α was the least active, and MIP-1β had intermediate potency. This is best illustrated in Fig. 3, in which the individual median ID90s for each isolate and each CC chemokine (Table 2) are presented as a scatter plot, with the overall median values indicated by bars. The equimolar mixture of the three CC chemokines was actually less effective than RANTES alone, presumably reflecting the dilution of the most active agent by less active ones. Thus, the three CC chemokines are not synergistic (or even additive) in their actions. The data points in Fig. 3 that are marked with an asterisk and a dagger represent, respectively, the isolates most (94KE103, subtype D) and least (SF162, subtype B) sensitive to CC chemokines (Table 2). Note that both these isolates replicated with similar efficiencies (Table 1). Why there should be interisolate variation to this extent remains obscure, but 94KE103 was particularly unusual in its sensitivity to MIP-1α. Note also that RANTES and MIP-1β had very similar effects on some isolates (e.g., 94KE103, sensitive; BZ162 and 92US657, insensitive) but widely differing effects on many others (e.g., R1 and DJ259, RANTES sensitive, MIP-1β insensitive) (Table 2).
Interactions of chemokines with their receptors: gp120 competition and receptor down-regulation.
The observation that MIP-1α is a relatively weak inhibitor of HIV-1 replication is consistent with reports that this ligand has the lowest ability among MIP-1α, MIP-1β, and RANTES to activate signal transduction through CCR5 (3, 45). However, these three CC chemokines have similar affinities for CCR5 in radioligand binding assays (3, 45, 48); indeed, RANTES actually had the lowest CCR5 affinity among them, in one study (3). To explore why RANTES was, despite this, often significantly the most potent at suppressing HIV-1 replication (Table 2, Fig. 3), we first tested whether there was a difference in the ability of an M-tropic gp120 to compete with radiolabeled RANTES and MIP-1β for binding to CCR5 on mitogen-stimulated primary CD4+ T cells (55). We were not able to test MIP-1α under the same conditions, as the level of specific binding of 125I–MIP-1α to activated CD4+ T cells was too low at the radioligand concentrations used for the experiments with RANTES and MIP-1β (data not shown).
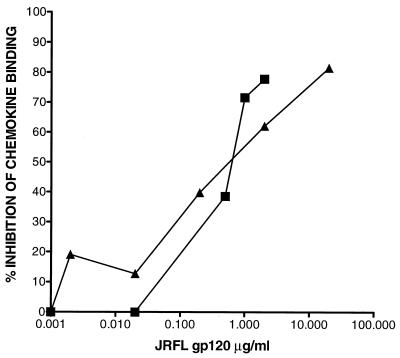
The rationale for the gp120 competition experiment was that ligand affinities for CCR5 had been determined previously with CCR5-transfected nonlymphoid cells and that the cellular context might conceivably impact on RANTES or MIP-1β binding. In fact, we found no significant difference in the interactions of RANTES or MIP-1β with CCR5 on activated CD4+ T cells, in that gp120 was able to block the binding of each of them equivalently (Fig. 4). This suggested that the greater efficacy of RANTES as an inhibitor of HIV-1 replication could be attributable to another mechanism(s).
FIG. 4.
Inhibition of RANTES and MIP-1β binding to CCR5 by gp120 from the M-tropic strain, JRFL. JRFL gp120 at the concentrations indicated was used to compete for the binding of 125I-labeled RANTES (▴) or MIP-1β (▪) to CCR5 on activated CD4+ T cells. Each value recorded represents the percentage inhibition of radioligand binding at each gp120 concentration. The experiments shown are representative for two to three experiments performed with cells from different donors.
We took into consideration that, although RANTES can interact with receptors other than CCR5 (44, 57), the viruses in our M-tropic test panel replicated poorly in CD4+ T cells from individuals lacking a functional CCR5 protein (Table 1). This focused our attention on CCR5. Because ligand binding can cause receptor down-regulation as part of a desensitization mechanism (3, 21, 25, 54), we investigated whether this occurred when chemokines interacted with CCR5 and CXCR4 on activated CD4+ T cells. We therefore used the CCR5-specific MAb 2D7 (60, 61) and the CXCR4-specific MAb 12G5 (19) to measure the levels of coreceptor expression on the surface of these cells, before and after exposure to chemokines.
Both MAbs have been shown to bind to epitopes which overlap with the chemokine binding sites on these receptors (19, 60, 61). It was therefore necessary to demonstrate that residual chemokines did not interfere with the binding of 2D7 and 12G5 to CCR5 and CXCR4 under the assay conditions we used. Thus, we compared 2D7 binding to activated CD4+ T cells in the presence of CC chemokines (1 μg/ml) with that in the absence of CC chemokines. When RANTES (1 μg/ml) was added simultaneously with 2D7, only a 7.2% reduction in 2D7 binding was observed (Fig. 5a). When cells were pretreated for 10 min with RANTES, followed by two washes (to mimic more closely our staining protocol), and then stained with 2D7, there was only a 0.9% decrease in 2D7 binding. Analogous experiments were performed with 12G5 and SDF-1α (Fig. 5b). Simultaneous addition of the chemokine and the MAb resulted in a 16.8% decrease in 12G5 binding, but when the cells were preincubated with SDF-1α and then washed, there was only a 7.4% reduction in 12G5 binding. Residual chemokines do not, therefore, significantly interfere with the use of the 2D7 and 12G5 MAbs to monitor CCR5 and CXCR4 expression.
We then treated activated CD4+ T cells for 2 h and 3 days with MIP-1α, MIP-1β, RANTES, or the CXC chemokine SDF-1α. We used a chemokine concentration of 1 μg/ml, which is saturating for each CC chemokine in terms of CCR5 binding (3, 45, 48) and which is comparable with the highest concentration used in the infection-inhibition studies. All three CC chemokines reduced the surface expression of CCR5 significantly within 2 h and to a greater extent after 3 days (Fig. 6a). In a separate experiment, significant down-regulation of CCR5 was also observed after a 1-h incubation (data not shown). Among the three CC chemokines, RANTES was the most potent at down-regulating CCR5, whereas SDF-1α had no effect (Fig. 6a and data not shown). Similar results were obtained with a mouse pre-B lymphoma cell line (L1.2) expressing human CCR5 (60).
On the CD4+ T cells, the rank order for the extent of CCR5 down-regulation (from greatest to least) was reproducibly RANTES, MIP-1β, MIP-1α, which is the same as the rank order for their inhibition of HIV-1 replication (cf. Fig. 3 and 5a) and also for their activation of signal transduction (3, 45). However, in absolute terms, RANTES was only a little more potent than MIP-1α at down-regulating CCR5 (Fig. 5a). Whether the additional down-regulation of CCR5 by RANTES is sufficient to account for the greater HIV-1-inhibitory effect of this ligand in the CD4+ T-cell cultures remains to be resolved. Although the cells were preincubated with CC chemokines for only 1 h before HIV-1 addition and there is less CCR5 down-regulation at this time (Fig. 6a and data not shown), the chemokines were not subsequently removed from the cultures. A more sustained down-regulation of CCR5 might significantly impact the efficiency of subsequent rounds of HIV-1 replication in these cultures.
Effect of CXC chemokine SDF-1α on replication of T-tropic strains.
The CXC chemokine SDF-1α is a ligand for CXCR4 and inhibits the infection of CD4+ T cells by T-tropic HIV-1 strains (7, 41). We tested the breadth of SDF-1α activity by using the T-tropic HIV-1 panel described in Table 1. As found with the CC chemokines, there could be significant variation in the potency with which SDF-1α inhibited the replication of different HIV-1 isolates in activated primary CD4+ T cells with wild-type CCR5 alleles (Table 3). Thus, the ID90 of SDF-1α for the most-sensitive strain (ZAM20, subtype C) was 0.2 μg/ml, whereas the least-sensitive strain (DH123, subtype B) was not inhibited by 90% at 5 μg/ml, a 20-fold higher SDF-1α concentration. However, most of the test isolates had ID90s between 2.7 and 4.9 μg/ml, a relatively narrow range (Table 3). Analogous to what was observed with the M-tropic isolates, replication efficiency was not correlated with sensitivity to SDF-1α inhibition (Tables 1 and 2). Although some of the test isolates were dual tropic, in that they were able to use both CCR5 and CXCR4 to enter transfected U87MG-CD4 cells, they were not, on average, differentially sensitive to SDF-1α compared to the isolates that could use only CXCR4 (Table 3).
TABLE 3.
Inhibition of T-tropic viruses by SDF-1α
| Genetic subtype | Virus | Use ofa:
|
Wild typeb
|
Δ-32/Δ-32b
|
|||
|---|---|---|---|---|---|---|---|
| CCR5 | CXCR4 | ID90 | ID50 | ID90 | ID50 | ||
| A | 92UG029 | − | + | >4.8 | 3.5 | ND | ND |
| B | 2076 cI.3 | + | + | 3.2 | 0.3 | 1.7 | 0.2 |
| B | C 7/86 | + | + | >4.5 | 0.2 | 2.6 | 1.2 |
| B | DH123 | + | + | >5.0 | >5.0 | 4.7 | 2.4 |
| B | SF2 | + | + | >4.6 | 0.3 | 3.0 | <0.3 |
| B | HC4 | − | + | 4.3 | 1.3 | 3.9 | 1.3 |
| B | NL4-3 | − | + | 3.5 | 0.4 | 0.7 | 0.4 |
| C | ZAM20 | − | + | 0.2 | <0.2 | ND | ND |
| C | 94ZW106 | − | + | 2.7 | 1.5 | ND | ND |
| D | UG270 | − | + | 3.9 | <1.8 | ND | ND |
| D | 92UG046 | − | + | 4.9 | 3.2 | ND | ND |
| D | 92UG024 | − | + | 4.5 | 2.3 | ND | ND |
| E | 94TH304 | − | + | >5.0 | >3.8 | ND | ND |
The test isolates’ abilities (+) or inabilities (−) to use CXCR4 and CCR5 to enter transfected U87MG-CD4 cells are recorded.
The median ID50s and ID90s (in micrograms per milliliter) for the CXC chemokine SDF-1α against T-tropic isolates in cells from individuals homozygous for wild-type or Δ-32 CCR5 alleles are recorded. The data represent the means of two independent experiments. A value marked by “>” indicates that 50 or 90% inhibition was not achieved at the highest SDF-1α concentration tested. A value marked by “<” indicates that 50 or 90% inhibition was always achieved at the lowest SDF-1α concentration tested. ND, not done.
The limited number of T-tropic isolates from outside subype B limits any conclusion that can be drawn about the relationship between SDF-1α sensitivity and the genetic subtypes, but no pattern was obvious from inspection of the available data (Table 3). The subtype B strains were also tested for SDF-1α sensitivity with CD4+ T cells from individuals homozygous for Δ-32 CCR5 alleles, to assess whether the absence of CCR5 affected entry via CXCR4. Any differences observed with these cells compared to CCR5 wild-type cells were minor (Table 3). SDF-1α was also tested for its effects on the replication of M-tropic isolates in CD4+ T cells. Under certain circumstances, it was observed to cause enhancement of HIV-1 replication. The results of these studies will be described elsewhere.
We also assessed whether CXCR4 was down-regulated after SDF-1α binding, by staining with the CXCR4-specific MAb 12G5 (19). SDF-1α, but none of the three CC chemokines, significantly reduced surface expression of CXCR4 after 3 days of culture (Fig. 6b). In contrast to what was observed with the CC chemokines and CCR5, down-regulation of CXCR4 expression was greater after 2 h of incubation with SDF-1α than it was after 3 days (Fig. 6b). This is consistent with observations that after 2 h of SDF-1α treatment of CXCR4-expressing cell lines, up to 90% of surface CXCR4 is down-regulated (28). Thus, CXCR4 down-regulation could contribute to the mechanism by which SDF-1α inhibits HIV-1 entry and replication.
DISCUSSION
The CC chemokines quite broadly inhibit primary, M-tropic HIV-1 viruses; most isolates were sensitive to, at least, RANTES. However, there was a wide spectrum of sensitivity, even to RANTES, and some isolates were only inhibited at relatively high chemokine concentrations. Certain isolates were far more sensitive to RANTES than to MIP-1α and MIP-1β. It would be prudent to include both sensitive and relatively insensitive isolates, such as those identified here, when testing the activity of CC chemokine-based inhibitors targeted at CCR5. Similarly, we also observed variation in the sensitivity of T-tropic isolates to SDF-1α; one isolate (ZAM20) was unusually sensitive.
It is not clear why HIV-1 isolates are differentially sensitive to CC or CXC chemokines. Replication efficiency was clearly not correlated with the sensitivity of chemokine inhibition. Most of the viruses in the M-tropic panel were uncloned isolates (as opposed to molecular clones), because few clones are available. It is appropriate to use uncloned isolates in vitro, because antiviral agents have to be effective in the face of quasispecies variation in vivo to be of any value. However, the use of uncloned isolates does impact analyses of the mechanisms of chemokine insensitivity. We were careful to check that the M-tropic isolates did not replicate efficiently in CD4+ T cells from individuals homozygous for Δ-32-CCR5 alleles (18, 32, 43). Thus, the presence of CCR5 is necessary for entry of these viruses into human CD4+ T cells. The simplest explanation is that CCR5 is the only coreceptor used efficiently for entry into these cells, but an alternative possibility is discussed below. None of the M-tropic isolates induced syncytia in MT-2 cells, indicating they have the NSI phenotype and do not use CXCR4 efficiently. Nonetheless, some CC chemokine-insensitive entry, via CXCR4 perhaps, of some quasispecies of certain M-tropic isolates could occur, reducing the inhibitory effect of CC chemokines.
Similar arguments could be made to explain the variable sensitivity of T-tropic isolates to SDF-1α. However, whether a T-tropic isolate could use CCR5 as well as CXCR4 in transfected cells (i.e., whether it was capable of dual tropism) did not have a major effect on SDF-1α inhibition among the limited number of isolates we tested. Furthermore, although there were outliers, most T-tropic isolates were inhibited by SDF-1α in a concentration range that was relatively narrow compared to that found for (e.g.) RANTES.
Notwithstanding the mechanisms that could contribute to the efficiency of inhibition, most HIV-1 isolates were sensitive to some degree to either CC or CXC chemokines. We could find no evidence for a genetic subtype-dependent component to the inhibitory mechanism(s). Thus, although the number of isolates we could test from each individual subtype was limited (especially for T-tropic viruses), no subtype was either chemokine sensitive or insensitive. For M-tropic isolates, this is consistent with observations that all subtypes require CCR5 for entry into CD4+ T cells (62) and can use the cloned CCR5 coreceptor (9, 62). Thus, genetic variation at the subtype level is unlikely to be a major limitation to the development of antiviral therapies aimed at CCR5 and CXCR4. However, genetic variation on a less-profound scale cannot be ignored; the possible presence of CC chemokine-insensitive viruses in uncloned M-tropic isolates suggests that escape mutants may develop.
Among the three CC chemokines we tested, RANTES clearly had the greatest breadth and potency of action, whereas MIP-1α was only weakly active (Fig. 3). Assuming that this is not an artifact of the use of recombinant proteins in vitro (35, 58), drug development based on the CC-chemokines should focus on the RANTES structure and not on MIP-1α. This conclusion was, perhaps, anticipated by the creators of the Met-RANTES and AOP-RANTES derivatives (4, 52). But why is RANTES the most potent CC chemokine and MIP-1α the least potent? Since gp120 and CC chemokines mutually compete for binding to CCR5 (27, 55, 59), variations in ligand affinities for CCR5 could be relevant. However, MIP-1α, MIP-1β, and RANTES have comparable affinities for CCR5 (3, 45, 48). In one study, MIP-1α was actually found to have a 10-fold-higher affinity than RANTES for CCR5 (3), yet RANTES is much more potent at inhibiting HIV-1 replication. Furthermore, an M-tropic gp120 competed equally well for the binding of MIP-1β and RANTES to CCR5 (Fig. 4), implying that a factor other than CCR5 affinity may contribute to the more potent antiviral activity of RANTES. Together, these observations suggest that competitive inhibition of the gp120-CCR5 interaction, although it occurs (27, 55, 59), is not the sole mechanism by which CC chemokines inhibit HIV-1 replication.
The nature of the gp120-CCR5 interaction could, however, contribute to interisolate variation in CC chemokine sensitivity. If CC chemokines inhibit HIV-1 binding to CCR5 by a truly competitive mechanism, then the affinity of the gp120 ligand will be an important variable: a low-affinity gp120-CCR5 interaction would be more efficiently blocked by a CC-chemokine than a high-affinity one. Because we used uncloned isolates, we cannot readily test this. A related, but subtly distinct, scenario is that different gp120s interact with nonidentical sites on CCR5, a concept for which there is some support (5, 6, 16, 33, 47). The degree of overlap between the different gp120 binding sites and the CC chemokine site(s) might, therefore, differ, which could impact the efficiency of what would be a noncompetitive mechanism of inhibition (parallel arguments can be made for CXCR4 and SDF-1α). Highly detailed competitive binding studies with pure reagents (i.e., clonal gp120s and coreceptor-transfected cells) could resolve these issues, but they are beyond the scope of the present work.
The rank order we observed among the CC chemokines for inhibition of HIV-1 replication might be explained by sustained CCR5 desensitization after ligand binding. Indeed, the rank order for CCR5 down-regulation on CD4+ T cells by the three CC chemokines after 3 days in culture (Fig. 6a) was reproducibly the same as the rank order for their median antiviral activity (Fig. 3). Other studies have found the same rank order for the abilities of RANTES, MIP-1β, and MIP-1α to activate signal transduction through CCR5 (3, 45). In particular, the ineffectiveness of MIP-1α against HIV-1 may be related to its limited ability to cause signal transduction (3, 45); indeed, MIP-1α has been described as being only a partial agonist of CCR5 (3). Signaling via CCR5 is clearly not necessary for HIV-1 entry (3, 21, 25) but might contribute to the mechanism by which CC chemokines inhibit HIV-1 replication. There is now good evidence that CXCR4 down-regulation contributes to the antiviral action of SDF-1α (2). Down-regulation of CCR5 could help reduce HIV-1 spread through the PBMC cultures, which were not maintained under single-cycle replication conditions. However, MIP-1α is also the weakest among the three CC chemokines at blocking HIV-1 entry in a single-cycle assay (18), suggesting that effects on viral spread are not the entire story.
The extra extent of CCR5 down-regulation induced by RANTES compared to MIP-1α and MIP-1β may not be sufficient to account for the far greater antiviral effect shown by RANTES against some viruses (e.g., isolates R1 and DJ259 were RANTES sensitive, but both were MIP-1α and MIP-1β insensitive [Table 2]). Furthermore, MIP-1β and RANTES are almost indistinguishable in their activation of signal transduction through CCR5 (3, 45, 48). Hence, there may be additional complexities to the antiviral action of RANTES, at least for some isolates.
The interactions of RANTES with receptors other than CCR5 might impact the efficiency with which HIV-1 replication (as opposed to entry) is inhibited. CD4+ T cells express another coreceptor(s) that can be used by some M-tropic viruses (14, 20, 31). In principle, this could help account for both the relative insensitivity of some isolates to MIP-1α, MIP-1β, and RANTES (these isolates would use an additional coreceptor unable to bind some or all of these ligands) and the superiority of RANTES against a subset of isolates (the additional coreceptor might bind only RANTES). But how could this scenario be squared with the observations that M-tropic isolates replicate poorly in CD4+ T cells from individuals homozygous for Δ-32 CCR5 alleles? A possible explanation is that CD4+ T cells from these individuals oversecrete CC chemokines, especially RANTES (18, 43). In principle, this could cause autocrine inhibition of HIV-1 entry via another coreceptor normally used as well as CCR5 (or perhaps even in concert with it, as has been proposed for CCR3 and CCR5 on brain monocytes [26]). If the hypothetical coreceptor were a RANTES, but not MIP-1β, receptor, this could account for the more potent antiviral activity of RANTES against some isolates (both putative coreceptors, not just CCR5, would be blocked). However, none of the recently identified new coreceptors is a RANTES receptor (14, 20, 31).
It is a curious complexity that the apparent insensitivity to CC chemokines of HIV-1 replication in monocytes/macrophages observed by most (but not all [1]) groups (18, 40, 49, 52) is overcome by the use of a RANTES derivative that is unable to activate signal transduction (52). In these cells, signalling via CCR5 may activate HIV-1 replication at a postentry stage, whereas in T cells, signaling that leads to CCR5 down-regulation may contribute to the inhibition of HIV-1 replication. In PBMC cultures, where both T cells and monocytes/macrophages are present, multiple, possibly opposing effects may occur. The point at which activating and inhibitory effects of the chemokines on HIV-1 replication balance out may be both donor and isolate dependent, as we have observed. Much remains to be discovered about precisely how the CC chemokines affect HIV-1 replication in CD4+ T cells and macrophages. Such knowledge could contribute to the rational development of antiviral drugs aimed at blocking virus entry, via CCR5 and/or other coreceptors, and should be sought.
ACKNOWLEDGMENTS
We thank Audrey Pomales, Shab Alipanah, Tom Ketas, Fred Endorf, and Stan Kang for technical assistance during the course of this study. We are very grateful to Paul Maddon for provision of recombinant gp120 (JRFL) and to Dan Littman for his gift of U87MG-CD4 cells stably expressing the CCR5 or CXCR4 coreceptors. We thank R. Connor, C. Cheng-Mayer, S. Forte, J. Sullivan, R. Shibata, M. Martin, and P. Clapham for HIV-1 isolates and clones.
This study was supported by NIH grant AI41420, by the Pediatric AIDS Foundation, and by NIAID contract NO1 AI35168 (Antigenic variation of HIV-1 and related lentiviruses). A.T. was a Fellow of the Fonds zur Förderung der wissenschaftlichen Forschung (award J01165-MED) and the Austrian Program for Advanced Research and Technology; J.P.M. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
ADDENDUM IN PROOF
All the studies we report in this paper were performed using a single stock of RANTES (R&D Systems; Lot D012) that was available between 1996 and 1997. Preliminary studies on new preparations of RANTES from the same manufacturer, and also on stocks from Gryphon, indicate that there can be stock-dependent variations in the efficiency with which RANTES inhibits HIV-1 replication. Although these variations are relatively minor in magnitude, they do need to be taken into account when comparing data sets in this paper with those obtained using other preparations of RANTES.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Amara A, Le Gall S, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier J-L, Arenzana-Seisdedos F. HIV coreceptor downregulation as antiviral principle: SDF-1α-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aramori I, Ferguson S S G, Bieniasz P D, Cullen B R, Caron M G. Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signalling and internalization are dissociable from its role as an HIV-1 co-receptor. EMBO J. 1997;16:4606–4616. doi: 10.1093/emboj/16.15.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arenzana-Seisedos F, Virelizier J-L, Rousset D, Clark-Lewis I, Loetscher P, Moser B, Baggiolini M. HIV blocked by chemokine antagonist. Nature. 1996;383:400. doi: 10.1038/383400a0. [DOI] [PubMed] [Google Scholar]
- 5.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 6.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1 induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., and D. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):587–598. [PubMed]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;86:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi F, DeVico A L, Garzino-Derno A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha and MIP-1 beta as the major HIV suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 12.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daar E S, Li X L, Moudgil T, Ho D D. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci USA. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng H, Unutmaz D, Kewal-Ramani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 15.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 16.Doranz B J, Lu Z-H, Rucker J, Zhang T-J, Sharron M, Cen Y-H, Wang Z-X, Guo H-H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S, Parmentier M, Collman R G, Doms R W. A dual-tropic, primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;86:1149–1159. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 18.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J M, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 19.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 20.Farzan M, Choe M, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1β-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 23.Fisher R. CD4 molecules: rationale and potential as antiviral agents. AIDS Res Rev. 1991;1:289–300. [Google Scholar]
- 24.Forte S E, Sullivan J L, Somasundaran M. In vitro characterization of HIV type 1 biological clones from asymptomatic and symptomatic pediatric patients. AIDS Res Hum Retroviruses. 1996;12:1585–1592. doi: 10.1089/aid.1996.12.1585. [DOI] [PubMed] [Google Scholar]
- 25.Gosling J, Monteclaro F S, Atchison R E, Arai H, Tsou C-L, Goldsmith M A, Charo I F. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 27.Hill C M, Deng H-K, Unutmaz D, Kewalramani V, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horuk, R. Unpublished data.
- 29.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, Albert J, Wigzell H. Sensitivity to inhibition by β-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacey F C, McDanal C B, Horuk R, Greenberg M L. The CXC chemokine stromal cell-derived factor 1 is not responsible for CD8+ T cell suppression of syncytia-inducing strains of HIV-1. Proc Natl Acad Sci USA. 1997;94:9842–9847. doi: 10.1073/pnas.94.18.9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao F, Alkhatib G, Peden K C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–378. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 33.Lu Z-H, Berson J F, Chen Y-H, Turner J D, Zhang T-Y, Sharron M, Jenks M H, Wang Z-X, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 co-receptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luciw P A, Potter S J, Steimer K, Dian D, Levy J A. Molecular cloning of AIDS-associated retrovirus. Nature. 1984;312:760–763. doi: 10.1038/312760a0. [DOI] [PubMed] [Google Scholar]
- 35.Mantel C, Kim Y J, Cooper S, Kwon B, Broxmeyer H E. Polymerization of murine macrophage inflammatory protein 1a inactivates its myelosuppressive effects in vitro: the active form is a monomer. Proc Natl Acad Sci USA. 1993;90:2232–2236. doi: 10.1073/pnas.90.6.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore J P, Cao Y, Leu J, Qin L, Korber B, Ho D D. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J Virol. 1996;70:427–444. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore J P, McCutchan F E, Poon S-W, Mascola J, Liu J, Cao Y, Ho D D. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J Virol. 1994;68:8350–8364. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore J P, McKeating J A, Huang Y, Ashkenazi A, Ho D D. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore J P, Sweet R W. The HIV gp120-CD4 interaction: a target for pharmacological and immunological intervention? Perspect Drug Discov Des. 1993;1:235–250. [Google Scholar]
- 40.Moriuchi M, Moriuchi H, Combadiere C, Murphy P M, Fauci A S. CD8+ T cell-derived factor(s), but not β-chemokines RANTES, MIP-1α, and MIP-1β, suppress HIV-1 replication in monocyte/macrophages. Proc Natl Acad Sci USA. 1996;93:15341–15345. doi: 10.1073/pnas.93.26.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Mosser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 42.Oravecz T, Pall M, Norcross M A. β-Chemokine inhibition of monocytotropic HIV-1 infection. Interference with a postbinding fusion step. J Immunol. 1996;157:1329–1332. [PubMed] [Google Scholar]
- 43.Paxton W A, Martin S R, Tse D, O’Brien T R, Skurnick J, VanDevanter N L, Padian N, Braun J F, Kotler D P, Wolinsky S M, Koup R A. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposures. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 44.Premack B A, Schall T J. Chemokine receptors: gateways to inflammation and infection. Nat Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 45.Raport C J, Gosling J, Schweickart V L, Gray P W, Charo I F. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for Rantes, MIP-1beta, and MIP-1alpha. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 46.Rubbert A, Weissman D, Combadiere C, Pettrone K A, Daucher J A, Murphy P M, Fauci A S. Multifactorial nature of noncytolytic CD8+ T-cell-mediated suppression of HIV replication: β-chemokine-dependent and -independent effects. AIDS Res Hum Retroviruses. 1997;13:63–69. doi: 10.1089/aid.1997.13.63. [DOI] [PubMed] [Google Scholar]
- 47.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 48.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new CC-chemokine receptor gene, CC-CKR5. Biochemistry. 1996;11:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 49.Schmidtmayerova H, Sherry B, Bukrinsky M. Chemokines and HIV replication. Nature. 1996;382:767. doi: 10.1038/382767a0. [DOI] [PubMed] [Google Scholar]
- 50.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siani, M. A., D. A. Thompson, L. E. Canne, G. M. Figliozzi, S. B. H. Kent, R. Simon, J. Cyster, P. E. Kennedy, E. A. Smith, and E. A. Berger. 1997. Rapid modular synthesis of SDF-1α with full biological activities, abstr. 5. National Managed Health Care Congress (NMHCC): chemokine receptors and host cell interaction.
- 52.Simmons G, Clapham P R, Picard C, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E I. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonists. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 53.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solari R, Offord R E, Remy S, Aubry J-P, Wells T N C, Whitehorn E, Oung T, Proudfoot A E I. Receptor-mediated endocytosis of CC-chemokines. J Biol Chem. 1997;272:9617–9620. doi: 10.1074/jbc.272.15.9617. [DOI] [PubMed] [Google Scholar]
- 55.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody sensitive interactions between HIV-1 and its co-receptor CCR5. Nature. 1996;384:184–186. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 56.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P J, Allaway G P, Katinger H, Barbas III C F, Burton D R, Ho D D, Moore J P. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wells T N C, Power C A, Lusti-Narasimhan M, Hoogewerf A J, Cooke R M, Chung C-W, Peitsch M C, Proudfoot A E I. Selectivity and antagonism of chemokine receptors. J Leukocyte Biol. 1996;59:53–60. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 58.Wolpe S D, Davatelis G, Sherry B, Beutler B, Hesse D G, Nguyen H T, Moldawer L L, Nathan C F, Lowry S F, Cerami A. Macrophages secrete a novel heparin-binding protein with inflammatory and neutrophil chemokinetic properties. J Exp Med. 1988;167:570–581. doi: 10.1084/jem.167.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 60.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore J P, Mackay C R. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C. CCR5 levels and expression pattern correlate with infectability by macrophage tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second-receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]