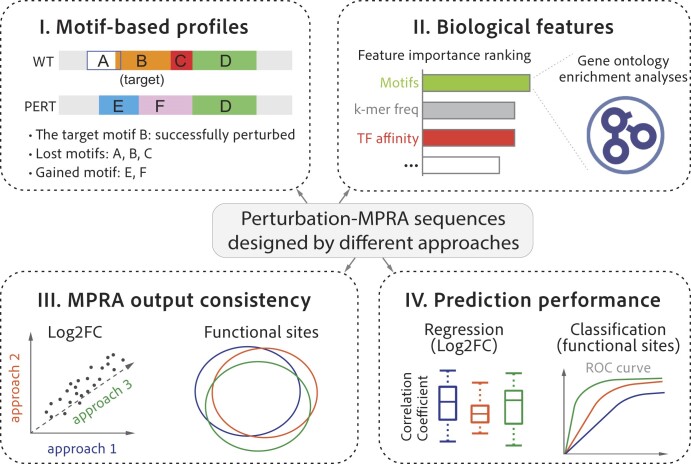
Abstract
The advent of perturbation-based massively parallel reporter assays (MPRAs) technique has facilitated the delineation of the roles of non-coding regulatory elements in orchestrating gene expression. However, computational efforts remain scant to evaluate and establish guidelines for sequence design strategies for perturbation MPRAs. In this study, we propose a framework for evaluating and comparing various perturbation strategies for MPRA experiments. Within this framework, we benchmark three different perturbation approaches from the perspectives of alteration in motif-based profiles, consistency of MPRA outputs, and robustness of models that predict the activities of putative regulatory motifs. While our analyses show very similar results across multiple benchmarking metrics, the predictive modeling for the approach involving random nucleotide shuffling shows significant robustness compared with the other two approaches. Thus, we recommend designing sequences by randomly shuffling the nucleotides of the perturbed site in perturbation-MPRA, followed by a coherence check to prevent the introduction of other variations of the target motifs. In summary, our evaluation framework and the benchmarking findings create a resource of computational pipelines and highlight the potential of perturbation-MPRA in predicting non-coding regulatory activities.
Graphical Abstract
Graphical Abstract.
Introduction
Advances in high-throughput technologies have allowed a detailed characterization of the human genome, encompassing regulatory elements such as enhancers. These enhancers, housing binding motifs for transcription factors (TFs), play a central role in the transcriptional regulation of gene expression. Aberrations in the non-coding regions of the genome have been linked to numerous polygenic disorders such as cancer, heart, and neurological disorders (1–3), making the study of non-coding regions a pivotal area of research.
However, linking the non-coding genome to the etiology of diseases is largely limited by the low throughput of conventional ‘luciferase reporter assays’, especially when investigating numerous non-coding regions of interest. To address this challenge, massively parallel reporter assays (MPRAs) were developed to simultaneously measure the activity of thousands of regulatory elements and their variants in a single experiment (4–14). Furthermore, a perturbation-based MPRA approach was introduced to elucidate the regulatory effects of transcription factor (TF) binding motifs instead of single nucleotide variants (15–17). The essence of this technique is to analyze the change in the transcription activity of reporter genes after altering the DNA sequence of putative functional regulatory regions.
In our recent studies, we have utilized the perturbation MPRA technique to successfully identify over 500 non-coding genomic regions that temporally regulate gene transcription during neural differentiation (18,19). However, insufficient attention has been given to the comprehensive evaluation of various perturbation approaches. As a result, a gold standard of perturbation sequence design strategies remains scant.
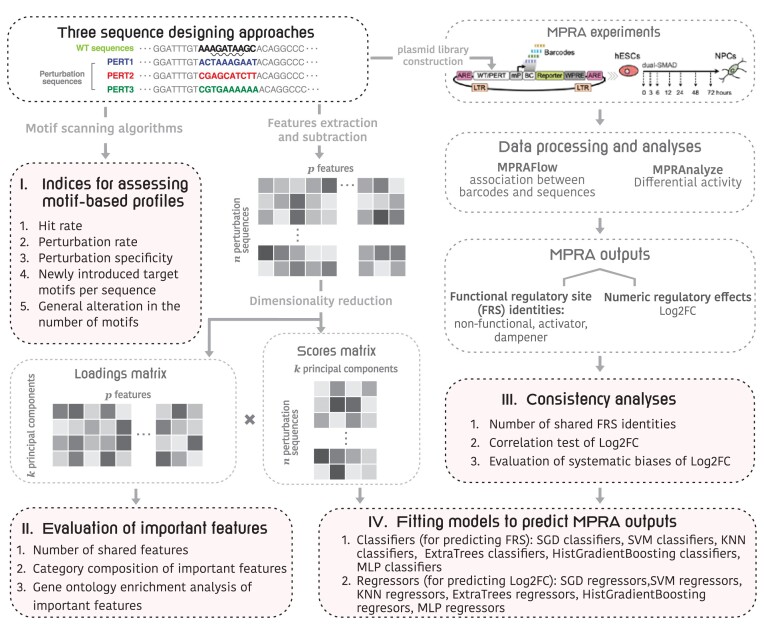
Motivated by the scarcity of the gold standard for DNA sequence designing strategies for the MPRAs technique, we propose a framework for assessing and comparing perturbation strategies (Figure 1). Within this framework, we benchmark three different perturbation approaches using a publicly available dataset recently generated by our team (18,19). This dataset includes 591 wild-type (WT) sequences, 2144 motif perturbation sequences, with each sequence perturbed using three different perturbation approaches, and 591 negative control sequences. Perturbation approaches 1 and 2 (PERT1 and PERT2) involve replacing the target motifs with a constant ‘non-motif’ sequence that is respectively identified; the perturbation approach 3 (PERT3) shuffles the nucleotides of target motifs (Supplementary Methods).
Figure 1.
An outline of the framework for evaluation of perturbation-based massively parallel assays technique. In the ‘Three sequence designing approaches’ box, we used the ‘GATA_known9’ motif as an example. In detail, the GATA motifs are a group of sequences conforming to the consensus WGATAR (W = A or T and R = A or G) (marked by the wavy underline), that can be recognized and bound by GATAbinding transcription factors (45).
For benchmarking, we first define five metrics to evaluate the achievement of the perturbation goals comprehensively. These metrics include, for example, the perturbation rate that indicates the impact on the target motifs both in-situ and ex-situ, and the perturbation specificity metric indicating the proportion of WT motifs that ‘survive’ the perturbation processes. Our analysis reveals that PERT3 exhibits the highest specificity with the lowest perturbation rate. Additionally, we compare the consistency of MPRA outputs, both in functional regulatory site (FRS) identities and numeric regulatory effects. Although our analyses revealed a high correlation among the three perturbation approaches, we also found a constant bias in the results of PERT1 and PERT2. This is likely attributed to their insertion of fixed sequences, which may introduce systematic biases to the assayed regions.
Finally, we extract multiple genomic features for each tested sequence, and we use the difference in the features between the perturbation sequences and their WT equivalents as independent variables to fit predictive machine-learning models. Our results for these predictive models demonstrate the robustness of both classifiers and regressors based on PERT3 data.
To the best of our knowledge, this is the first study that assesses and compares different perturbation approaches of MPRA experiments. Our study fills this gap by constructing a blueprint evaluation framework for perturbation sequence designing strategies. Additionally, our results provide guidance for establishing a gold standard of perturbation MPRA techniques, and our prediction pipeline holds great promise for further computational identification of functional genomic regulatory regions.
Materials and methods
Dataset overview
We utilized a publicly available dataset of perturbation MPRA that was recently generated by our team (18). The MPRA experiment was conducted in the human embryonic stem cell line across seven time points after neural differentiation induction (0, 3, 6, 12, 24, 48 and 72 h).
Description of the assayed sequences
The experiment assayed four groups of genomic sequences:
The wild-type group consists of 591 wild-type sequences (denoted as ‘WT’). Each WT sequence represents a 171-nucleotide genomic region whose regulatory activity differs over time (see Supplementary Methods for the selection procedure of region and motif combinations) (19);
-
The motif perturbation group consists of 2144 sequences. Each sequence houses a single-perturbed motif within the genomic region of its WT equivalent . Furthermore, each sequence is perturbed using three different perturbation approaches, denoted as ‘motif_PERT1’, ‘motif_PERT2’ and ‘motif_PERT3’ (Figure 1, see details in Supplementary Methods):
motif_PERT1: A target motif is substituted with the artificially scrambled motif so that the number of motifs is the least within the region extending from 3 bp upstream of the motif’s start position to 3 bp downstream of the motif’s end position. (Details regarding the selection of the scrambled motif are described in Supplementary Methods)
motif_PERT2: similar to PERT1, A target motif is substituted with the artificially scrambled motif so that the number of motifs is minimized across the entire genomic region of the p sequence. (Details regarding the selection of the scrambled motif are described in Supplementary Methods.)
motif_PERT3: the target motif is scrambled by randomly shuffling its nucleotides.
The negative control group 1 includes 591 scrambled sequences (denoted as ‘SCRAM’). Scrambled sequences are based on WT sequences with shuffled nucleotides, creating a set of negative controls;
The Negative control group 2 is a set of all the 591 WT sequences where we perturbed a sub-sequence in the length of the average motif (12 bp) in a random location within the WT sequence using the same three perturbation approaches (denoted as ‘RAND_PERT1’, ‘RAND_PERT2’ and ‘RAND_PERT3’). The RAND sequences are perturbed using the same three perturbation approaches as described above.
Description of the MPRA output
The experimental read-out of the perturbed sequences is then subjected to the MPRAnalyze (20) and the MPRAflow (21) tools to assess the motifs’ regulatory effect over time, represented by the Log2-fold changes (Log2FC) of PERT read-outs compared to WT and SCRAM at each time point. Sequences are further classified as activating (Log2FC > 0) or repressing (Log2FC < 0).
To identify the functional regulatory sites (FRS), we applied a set of four filters to the PERT sequences using MPRAnalyze (18,20):
At one or more time points, the activity of a PERT sequence significantly deviates from its WT equivalent.
The temporal activity of a PERT sequence significantly deviates from its WT equivalent.
The activity of either a PERT sequence (at one or more time points) or a WT sequence (across all the time points) is significantly higher than its corresponding SCRAM negative control sequence.
The temporal activity of either a PERT or a WT sequence is significantly higher than its corresponding SCRAM negative control sequence.
The target motif of a sequence will be labeled as an FRS if the sequence passes all four filters and shares consistent effects (either activating or repressing) in PERT3 and either PERT1 or PERT2.
In summary, the MPRA output consists of the numeric regulatory effect (Log2FC) and the multi-class FRS identities at seven time points. These two modalities of MPRA outputs are used as input variables for training the prediction models.
Metrics for assessing motif-based profiles
The motif-based profiles of each sequence were constructed using the Find Individual Motif Occurrences (FIMO) program. In brief, FIMO systematically scans a given sequence to identify individual matches to each motif from the #ENCODE/CIS-BP motif databases, treating each motif independently. Subsequently, we extracted motif occurrences of a given sequence and their reverse complements, selecting those with a P value less than 10−4. These sequence-specific motif-based profiles were then used to calculate the following five metrics:
Hit rate (HR)
A ‘hit’ sequence indicates the in-situ elimination of the target motif (in-situ elimination = ‘genomic-position-specific elimination’). In detail, we regard a sequence as ‘hit’ if all potential variations of its target motif are absent at the target genomic location in the scanning results of the Find Individual Motif Occurrences (FIMO) program (22), matched by the motif name, DNA strands, and genomic coordinates; otherwise, it’s a ‘fail.’ The hit rate of PERTi is denoted as HRi:
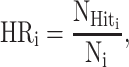 |
(1) |
where 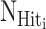 is the number of ‘Hit’ sequences and Ni is the total number of designed sequences in PERTi.
is the number of ‘Hit’ sequences and Ni is the total number of designed sequences in PERTi.
Perturbation rate (PR)
A ‘perturbed’ sequence indicates that all possible variations of the target motif are removed within the designated genomic region. In detail, we define a sequence as ‘perturbed’ if none of the variations of target motif is found in its FIMO scanning results, regardless of its genomic position. It encompasses both ex-situ and in-situ motif perturbation, regardless of the specific genomic position. To this end, the perturbation rate of PERTi is formulated as:
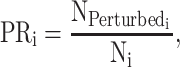 |
(2) |
where 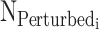 is the number of ‘perturbed’ sequences and Ni is the total number of designed sequences in PERTi.
is the number of ‘perturbed’ sequences and Ni is the total number of designed sequences in PERTi.
Perturbation specificity (PS)
To assess how many WT motifs are impacted by the perturbation, we introduce the ‘perturbation specificity’ metric. For the designed sequence j of PERTi, its perturbation specificity is formulated as:
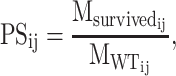 |
(3) |
where 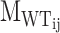 is the number of motifs that overlap with the target motif in the corresponding WT sequence of designed sequence j of PERTi, and
is the number of motifs that overlap with the target motif in the corresponding WT sequence of designed sequence j of PERTi, and 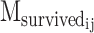 is the occurrence of wild-type motifs that are still present within the designed sequence j of PERTi. Both
is the occurrence of wild-type motifs that are still present within the designed sequence j of PERTi. Both 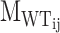 and
and 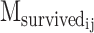 are obtained from FIMO scanning results.
are obtained from FIMO scanning results.
Newly introduced target motifs per sequence (NTM)
Of note, motifs in the context of genomic sequences do not necessarily correspond to a single, fixed nucleotide sequence. This means that motifs of the same name found in different genomic positions can exhibit some degree of variability or degeneracy. For example, motif ‘BHLHE40_disc2’ has 432 variations, including CCCGCGCCCGGGCGCGC, GGGACAGCCCGGAGGCC, CCCCCGCGCCCGGGCGC, etc. Since the perturbation process alters the orders of nucleotides, it becomes possible that the newly introduced motifs are variations of the target motif, thus retaining the functionality of the ‘supposedly perturbed’ motifs. To assess the impact of the perturbation approaches, we calculated and compared the ‘number of newly introduced target motifs per sequence’ among the three perturbation approaches. For PERTi, its ‘newly introduced target motifs per sequence’ metric is formulated as:
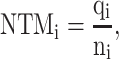 |
(4) |
where qi is the number of newly introduced motifs that are identical to the target motif names in PERTi, and ni is the total number of designed sequences in PERTi.
General alteration in the number of motifs
To assess the non-specific impacts of the perturbation, we obtained and compared these metrics among the three perturbation approaches:
The number of gained motifs
The number of lost motifs
The net change in the number of motifs
Consistency analysis of MPRA outputs
The MPRA outputs consist of two parts: the multi-labeled FRS identities and the numerical regulatory effects. To analyze the consistency of FRS identities, we counted the number of overlapped and unique activators/repressors that are specific to their genomic coordinates and DNA strands across three perturbation approaches. And the results are visualized by an UpSet plot (23). As for the agreement in numerical regulatory effects, we tested the correlation of Log2FCs between any two of the three perturbation approaches using three correlation tests: Pearson r correlation, Spearman’s rank correlation, and Kendall’s rank correlation test.
Features extraction for designed sequences
The features are a major determinant of the performance of predictive models (24,25). The features used in this work can be grouped into two main categories: sequence-based features and time-specific features.
Group A: sequence-based features
Since this group of features is based on the nucleotide sequences, each assayed sequence, either WT or perturbed, has its own set of features:
DNA 5-mer frequencies: 1024 features indicating the counts of all possible nucleotide 5-mers.
#5-mers: a single feature summarizing the number of distinct 5-mers.
DeepBind scores: 515 predicted scores of all pre-trained DeepBind models for transcription factor (TF) binding (26).
#DeepBind-top: a single feature summarizing the number of models above the 90th percentile across all the DeepBind models for TF binding (26).
DeepSEA scores: 21 907 chromatin profiles (transcription factor, histone marks, and chromatin accessibility profiles across a wide range of cell types) from the underlying DeepSEA learning model (25).
#DeepSea-top: a single feature summarizing the number of chromatin profiles above the 90th percentile across all the DeepSEA profiles (25).
DNA shape metrics: 13 predicted DNA shape features, that are: helix twist (HelT), Rise, Roll, Shift, Slide, Tilt, Buckle, Opening, propeller twist (ProT), Shear, Stagger, Stretch, and minor groove width (MGW) (27,28).
Max polyA/polyT lengths: two features indicating the length of the longest polyA and polyT subsequences, respectively.
#ENCODE/CIS-BP motifs: 4,706 features, showing the number of significant DNA-binding ENCODE/CIS-BP (29–31) motifs from simple DNA-binding motif scoring using the Find Individual Motif Occurrences (FIMO) tool (22).
ENCODE/CIS-BP motif summaries: four features indicating the number of motifs, and the maximum number of ENCODE/CIS-BP motifs within a 20 bp window in the sequence, as determined by FIMO scanning algorithm (22,30,31).
#TF family: fourteen features indicating the frequency of major TF families based on the FIMO scanning results against ENCODE/CIS-BP databases, which are: Basic Domain Group, Beta-Scaffold Factors, Helix-turn-helix, Other Alpha-Helix Group, Unclassified Structure and Zinc-Coordinating Group (32).
For each perturbed sequence, we subtract its sequence-specific features from that of its WT equivalent. Additionally, we calculate the Levenshtein similarity scores between the perturbed sequences and their respective correspondent WT sequences (33,34). In total, 28 189 features are yielded from group A.
These differences in features (denoted as Δ‘[feature name]’, e.g., Δ#5-mers), along with the Levenshtein similarity scores, are then subject to the feature normalization process (see Section ‘Feature normalization’).
Group B: time-specific features
The time-specific features used in this study are the experimental read-outs of WT sequences (19). These features include the signals of three genomic assays at seven time points (0, 3, 6, 12, 24, 48 and 72 h):
ATAC-seq: the normalized number of reads using DESeq2 (35) from overlapping ATAC-seq peaks within the designed genomic region
H3K27ac ChIP-seq, the normalized number of reads using DESeq2 (35) from an overlapping H3K27ac peak within the designed genomic region
RNA-seq: mRNA expression of the nearest gene to the designed region
In total, three features are yielded from group B. For each perturbed sequence, we use the time-specific feature of its corresponding WT sequence as its feature to fit prediction models.
Feature normalization
Performing principal component analysis (PCA) is a common technique to reduce the number of features in high-dimensional data to avoid over-fitting and improve the generalization performance of machine learning models. In this study, PCA was applied to the large number of group A features (28,189) to reduce them into a smaller set of principal components (PCs) that capture the maximum amount of variability in the data. By selecting the number of PCs such that they explain at least 99% of the variance in the data, the most important information in the original features is retained while reducing their dimensionality.
In this study, we employed PCA to transform the 28 189 group A features into 1500 PCs for each perturbation approach. Together with the time-specific features of group 2, a total of 1503 features were used as input for subsequent prediction tasks. This approach helps to prevent over-fitting and improves the accuracy of the machine learning models.
Calculation of the feature importance scores
We first defined the importance score I of feature i as the largest loading score of feature i across 1,500 PCs. In particular, from the PCA step, we obtain a matrix L to denote the loadings matrix that explains the correlations between the original features and the PCs. L is a 28, 189 × 1, 500 matrix with rows representing features and columns representing 1500 PCs. For feature i, its loading score on the jth dimension is denoted as Lij. We then define the importance score I of feature i as its largest loading score across the 1500 PCs:
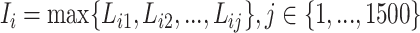 |
(5) |
Gene ontology analysis
We conducted the Gene ontology (GO) over-representation analysis using the genes corresponding to the top 2500 important TF binding features. The results were determined using the R package ClusterProfiler (36). The significance of GO terms was defined as an FDR-adjusted P < 0.05.
Model training
Classification models
We utilized six classification models to predict the FRS identity of perturbation sequences:
SGD: linear SVM classifiers with stochastic gradient descent (SGD) training (37)
SVC: C-Support vector classifiers (38)
KNN: classifiers based on k-nearest neighbors voting (39)
ET: ExtraTrees classifiers (40)
HGB: histogram-based gradient boosting classifiers (41)
MLP: multilayer perceptron classifiers (42)
All classifiers were run with the default settings of the scikit-learn package (43). The 1503 normalized feature values were used as input. To generate target values, the FRS identity labels at seven time points were concatenated and stacked into a single variable.
Regression models
We utilized six regressors to predict the Log2FC of perturbation sequences:
SGD: SGD: linear regressors fitted by minimizing a regularized empirical loss with SGD training (37)
SVC: SVR: Epsilon-Support vector regressors (38)
KNN: regressors based on k-nearest neighbors voting (39)
ET: ExtraTrees regressors (40)
HGB: histogram-based gradient boosting regressors (41)
MLP: multilayer perceptron regressors (42)
All regressors were run with the default settings of the scikit-learn package (43). The 1503 normalized feature values were used as input. The Log2FCs at seven time points were concatenated and stacked into a single variable, and then regarded as target values.
The randomized 10-fold cross-validation
We performed 10-fold cross-validation tests to evaluate the performances of different models. A 10-fold cross-validation test was chosen as it provides a good balance between minimizing bias and reducing variance. In detail, the dataset is randomly partitioned into 10 subsets, with one subset utilized as the testing dataset and the other nine together as the training data set. This procedure was conducted 10 times, with each subset being used once as a testing dataset to generate ten models. The average performance of these ten models was used to evaluate the performance of the different models.
To ensure a fair and objective comparison among the models, we strictly implemented their algorithms and optimized parameters to build models on the same training dataset and subsequently benchmark their performance on the independent test datasets.
Model performance measures
The performance of classification models is evaluated using the area under the receiver-operating characteristic curve (AUROC). For the regression models, we evaluated their performance using three correlation tests: Pearson, Spearman, and Kendall. Specifically, we tested the correlation between the predicted Log2FC values and the observed Log2FC values for each fold.
Statistical tests
For the motif-based profile metrics, the Kruskal–Wallis one-way analysis of variance and post-hoc pairwise Dunn’s multiple comparisons test was used to identify statistically significant differences in continuous variables, including the perturbation specificity and the number of gained/lost motifs. Moreover, the pairwise Fisher’s exact test was conducted to compare the count data, including hit and perturbation rates. The pairwise exact binomial test was performed to compare newly introduced target motifs per sequence (NTM).
For the consistency analyses, the correlation of Log2FCs was indicated by three correlation coefficients: Pearson’s r, Spearman’s ρ, and Kendall’s τ coefficient. The P values of correlation tests were subsequently adjusted for multiple comparisons at seven different time points by the Benjamini–Hochberg method.
For the performance evaluation of prediction models, we performed pairwise Wilcoxon rank sum tests on the AUROC and correlation coefficients. For all pairwise tests, a threshold of 0.05 was applied to the P values adjusted by the Benjamini-Hochberg method. An α level was considered 0.05 for all statistical tests in this study.
Results
To evaluate the three perturbation approaches, we first defined five motif-based metrics: (i) hit rate, representing the rate of in-situ motif perturbation, defined as the proportion of designed sequences that successfully eliminate the target motif at the target genomic locale, (ii) perturbation rate, which represents the rate of both ex-situ and in-situ motif perturbation and is defined as the proportion of designed sequences that eliminate all the motifs that match the target motif name within the 171-nucleotide genomic region, (iii) perturbation specificity, indicating the global impact of perturbation on all the motifs that lie within the perturbed sequence, and is defined as the proportion of WT motifs that are still found in the perturbation sequence, (iv) newly introduced target motifs per sequence, which reflects the occurrence of gained motifs that are identical to the target motif name, and is calculated by dividing the total number of such gained motifs by the total number of perturbation sequences, (v) non-specific changes in the number of motifs, which include the number of gained, lost motifs, as well as the net change in the number of motifs within the perturbation sequence. We then assess the differences in these aforementioned metrics across the three perturbation approaches (Figure 1, part I). Next, we assess the important features representing variability among all perturbation approaches (Figure 1, part II). Third, we compare the consistency of MPRA outputs (Figure 1, part III). Finally, to evaluate the generalizability in referencing non-coding regulatory activity across different perturbation approaches, we compare the performance of different prediction models across the three perturbation approaches (Figure 1, part IV).
Three perturbation approaches show similar hit rates and perturbation rates
Fundamentally, the primary goal of motif perturbation is the precise elimination of the target motif from its designated genomic position. To assess how well each perturbation approach is in reaching this goal, we computationally identified the occurrences of the motifs in the sequences, using the FIMO (22) scanning results and matching the motif names, DNA strands, and genomic coordinates (section ‘Materials and methods’).
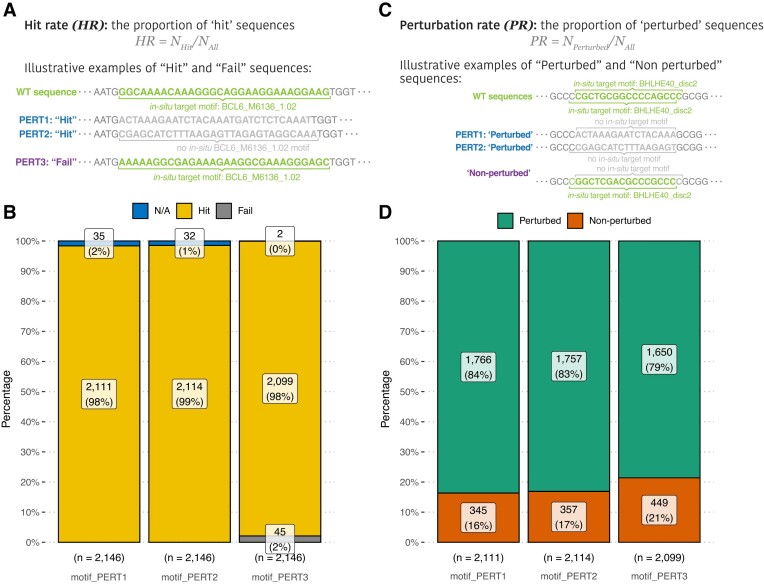
Prior to our analyses, we excluded the sequences that didn’t pass the quality check of the library processing (marked as ‘N/A’ in Figure 2, Supplementary Notes). After this exclusion of low-quality sequences, a sequence yielding a ‘non-occurrent’ result is defined as a ‘hit’ indicating a successful perturbation; otherwise it is labeled as a ‘fail’ (refer to the ‘Materials and methods’ section; Figure 2A). Then, we calculated and compared the proportion of hit and fail sequences for each perturbation approach (equation 1). Although all three perturbation approaches exhibit high hit rates (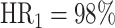 ,
, 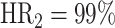 ,
, 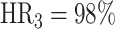 ), the PERT3 is significantly lower than the other two approaches (pairwise Fisher’s exact test, PERT1 versus PERT2, P = 1.00; PERT1 versus PERT3, P = 1.42 × 10−3; PERT2 versus PERT3, P = 1.42 × 10−3).
), the PERT3 is significantly lower than the other two approaches (pairwise Fisher’s exact test, PERT1 versus PERT2, P = 1.00; PERT1 versus PERT3, P = 1.42 × 10−3; PERT2 versus PERT3, P = 1.42 × 10−3).
Figure 2.
Evaluations of perturbation-wise metrics. (A) Examples of ‘hit’ and ‘fail’ sequences. Please refer to the Supplementary Notes for the full perturbation sequences. (B) A comparison of hit rates among three perturbation approaches. The ‘N/A’ category represents the sequences that are excluded from this study because their barcodes failed the sequencing quality check (Supplementary Notes). (C) Examples of ‘perturbed’ and ‘non-perturbed’ sequences. (D) A comparison of perturbation rates among three perturbation approaches.
The expected high hit rates in both the PERT1 and PERT2 approaches stem from their fundamental replacement of target motifs with ‘non-motifs’ (refer to Supplementary Methods). Conversely, the observed disparity in the hit rate of PERT3 can be attributed to the inherent nucleotide sequence variability of specific motifs. As an example, consider the motif ‘BCL6_M6136_1.02.’ In its native or wild-type sequence, this motif is represented as ‘CAAAGAGAGAAGGGGAAGGGGGTTGGGGAA’. Upon subjecting this motif to the randomly stuffing approach (PERT3), one of the resultant sequences becomes ‘AGGGGAGAGGGGGAAGAGAGAATCGAGATG.’ Notably, this perturbed sequence exhibits a discernible variation from the original ‘BCL6_M6136_1.02’ motif while retaining the physiological activity of the same motif ‘BCL6_M6136_1.02.’ Thus, this sequence designed by PERT3 is rendered a ‘Fail’ sequence.
This inherent motif diversity underscores the challenges encountered in achieving precise genomic-position motif perturbations, particularly when utilizing the PERT3 approach. Despite the high hit rate of PERT3, such motif variability emphasizes the importance of coherence checks when simply shuffling nucleotides for MPRA experiments.
Apart from the primary goal, one of the advanced goals of motif perturbation is to reduce the regulatory activity of the target motif to the baseline, that is, to eliminate all the motifs that are identical to the target motif name within the 171-nucleotide genomic region of perturbation sequence. Hence, we further quantified the occurrence of the target motif in each ‘hit’ sequence using the FIMO scanning results by matching only the motif name and not its position. Sequences were defined as ‘perturbed’ if no designed target motif was found within their genomic region, and the perturbation rate was then calculated as the proportion of ‘perturbed’ sequences (Figure 2C). In simple words, this metric indicates the rate of both ex-situ and in-situ motif perturbation that is not specific to the target genomic position (section ‘Materials and methods’, equation 2).
Comparing the perturbation rate of the three PERTs, we found that PERT1 and PERT2 possess similar perturbation rates of over 80%. Although the perturbation rate of PERT3 is significantly lower than those of the other two, it is still as high as 79% (Figure 2D,  ; pairwise Fisher’s exact test, PERT1 versus PERT2, P = 0.649; PERT1 versus PERT3, P = 8.79 × 10−5; PERT2 versus PERT3, P = 4.58 × 10−4). These results indicate that the strategic design of perturbation sequences (PERT1 and PERT2), instead of simply shuffling the nucleotide sequences (PERT3), leads to a higher chance of perturbing non-position-specific target motifs within genomic regions.
; pairwise Fisher’s exact test, PERT1 versus PERT2, P = 0.649; PERT1 versus PERT3, P = 8.79 × 10−5; PERT2 versus PERT3, P = 4.58 × 10−4). These results indicate that the strategic design of perturbation sequences (PERT1 and PERT2), instead of simply shuffling the nucleotide sequences (PERT3), leads to a higher chance of perturbing non-position-specific target motifs within genomic regions.
Perturbation specificity is similar among the three approaches
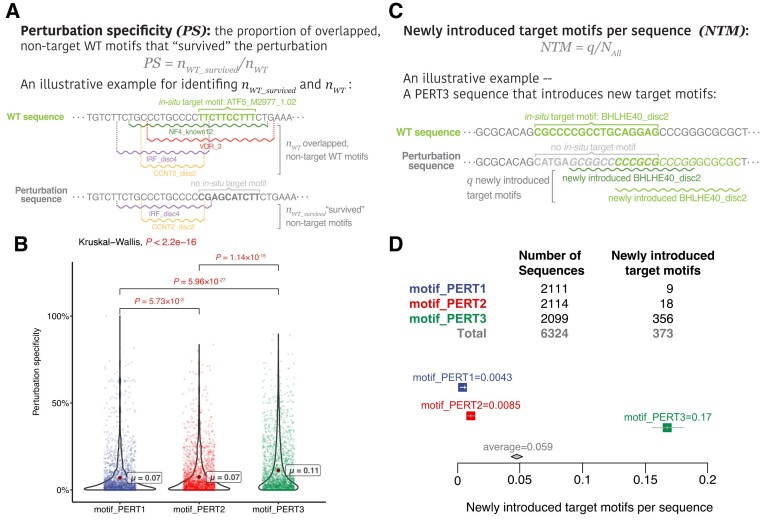
Another advanced goal of motif perturbation is to keep the impact on the overall motifs as low as possible—since the perturbation process essentially alters the DNA sequence within a certain range of the genome, the motifs that overlap with the target motifs are likely to be affected. To assess such a global impact of the perturbation on all the motifs that lie within the perturbation sequence, we introduced the perturbation specificity metric. It is defined as ‘the proportion of WT motifs that are still present within the genomic region after perturbation’ (section ‘Materials and methods’, equation 3, Figure 3).
Figure 3.
Evaluations of motif-based metrics. (A) An example of calculating perturbation specificity. Refer to the Supplementary Notes for the full perturbation sequences. (B) A comparison of perturbation specificity among three perturbation approaches. Significant P values (P < 0.05) are shown in red. (C) An example of calculating ‘newly introduced target motifs per sequence’. Please refer to the Supplementary Notes for the full perturbation sequences. (D) A comparison of ‘newly introduced target motifs per sequence’ among three perturbation approaches.
Comparing the perturbation specificity among three PERTs, we found that all three perturbation approaches vastly affect the WT motifs. Namely, only 10% of the overlapping WT motifs ‘survived’ the perturbation processes. Specifically, PERT3 has the highest perturbation specificity, which implies that randomly shuffling nucleotides exerts the least overall impact within the genomic regions of perturbed sequences (Figure 3B, 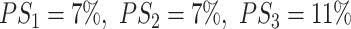 ; pairwise Dunn’s test, PERT1 versus PERT2, P = 5.73 × 10−3; PERT1 versus PERT3, P = 8.95 × 10−27; PERT2 versus PERT3, P = 1.14 × 10−15).
; pairwise Dunn’s test, PERT1 versus PERT2, P = 5.73 × 10−3; PERT1 versus PERT3, P = 8.95 × 10−27; PERT2 versus PERT3, P = 1.14 × 10−15).
On the other hand, another advanced goal is to avoid ‘creating’ target motifs in the perturbation sequences. In detail, motifs are typically short, conserved sequences that represent a binding site for transcription factors or other regulatory proteins. Found in multiple positions within a genome, motifs can exhibit some degree of variability or degeneracy. This variability allows motifs to occur in various sequence contexts while still maintaining their functional significance.
In the context of experiment design, while people can attempt to synthesize different shuffled sequences for cases where new motif instances are created, it’s important to recognize that these newly synthesized sequences may still contain those motifs or motif-like elements due to the inherent variability of motifs. In this case, the newly introduced target motifs could retain the functionality of the ‘supposedly perturbed’ motifs.
To this end, we sought to investigate which perturbation approach introduces the highest number of new motifs that are identical to the target motif name. We defined the newly introduced target motifs per sequence metric, which is calculated by dividing the total number of ‘newly introduced target motifs’ by the total number of sequences for each perturbation approach (section ‘Materials and methods’, equation 4, Figure 3C). The highest metric is produced by PERT3, indicating that shuffling the nucleotides increases the probability of generating the same motifs as the target ones (Figure 3D, NTM1 = 0.0043, NTM2 = 0.0085, NTM3 = 0.17; pairwise exact binomial test, PERT1 versus PERT2, P = 0.122; PERT1 versus PERT3, P = 2.35 × 10−92; PERT2 versus PERT3, P = 1.73 × 10−82).
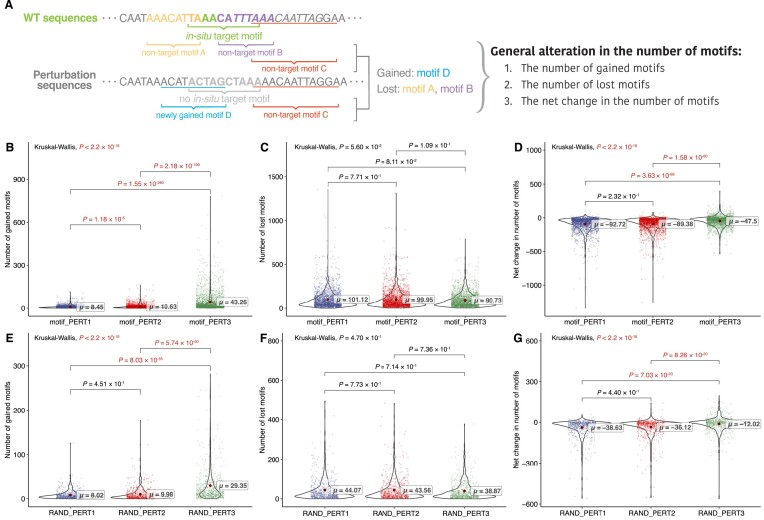
All three perturbation approaches vary in motif gain/loss
To gain a better perturbation effect, the impacts that are non-specific to the target motifs should also be minimized as much as possible. To address such impacts, we evaluated the overall motifs gained or lost across motif perturbation approaches (Figure 4A) and found that PERT3 gains significantly over 30 more motifs on average than PERT1 and PERT2 (Figure 4B, PERT1 ∼ =8.45, PERT2 ∼ =10.63, PERT3 ∼ =43.26; pairwise Dunn’s test, PERT1 versus PERT2, P = 1.18 × 10−5; PERT1 versus PERT3, P = 1.55 × 10−206; PERT2 versus PERT3, P = 2.18 × 10−199). However, the number of motifs lost was similar among the three approaches (Figure 4C, PERT1 ∼ =101.12, PERT2 ∼ =99.95, PERT3 ∼ =90.73; pairwise Dunn’s test, PERT1 versus PERT2, P = 0.771; PERT1 versus PERT3, P = 0.0811; PERT2 versus PERT3, P = 0.109).
Figure 4.
Evaluation of general alteration in the number of motifs. (A) Toy examples of calculating general alteration in the number of motifs. (B–D) The results for motif perturbations: (B) the number of gained motifs, (C) the number of lost motifs and (D) the net change in the number of motifs. Significant P values (P < 0.05) are shown in red. (E–G) The results for random perturbation sequences: (E) number of gained motifs, (F) number of lost motifs and (G) net change in the number of motifs. Significant P values (P < 0.05) are shown in red.
We also compared the net change in the number of motifs for each perturbation approach. We observed that PERT3 resulted in a significantly greater net change compared to the other two approaches. In contrast, there was no significant difference between PERT1 and PERT2 (Figure 4D, PERT1 ∼ = − 92.72, PERT2 ∼ = − 89.38, PERT3 ∼ = − 47.50; pairwise Dunn’s test, PERT1 versus PERT2, P = 0.232; PERT1 versus PERT3, P = 3.63 × 10−69; PERT2 versus PERT3, P = 1.58 × 10−60).
We then compared these non-specific metrics for the RAND perturbation sequences. We found similar results to the motif perturbation group: PERT3 resulted in the most motif gains (Figure 4E, PERT1 ∼ =8.02, PERT2 ∼ =9.98, PERT3 ∼ =29.35; pairwise Dunn’s test, PERT1 versus PERT2, P = 0.451; PERT1 versus PERT3, P = 8.03 × 10−55; PERT2 versus PERT3, P = 5.74 × 10−50), with no significant difference in the number of lost motifs (Figure 4F, PERT1 ∼ =44.07, PERT2 ∼ =43.56, PERT3 ∼ =38.67). In addition, the net change in the number of motifs of PERT3 is negative but the highest (Figure 4G, PERT1 ∼ = − 38.63, PERT2 ∼ = − 36.12, PERT3 ∼ = − 12.02; pairwise Dunn’s test, PERT1 versus PERT2, P = 0.44; PERT1 versus PERT3, P = 7.03 × 10−23; PERT2 versus PERT3, P = 8.26 × 10−20). These findings further support that the differences in the non-specific impacts are due to the perturbation approach used.
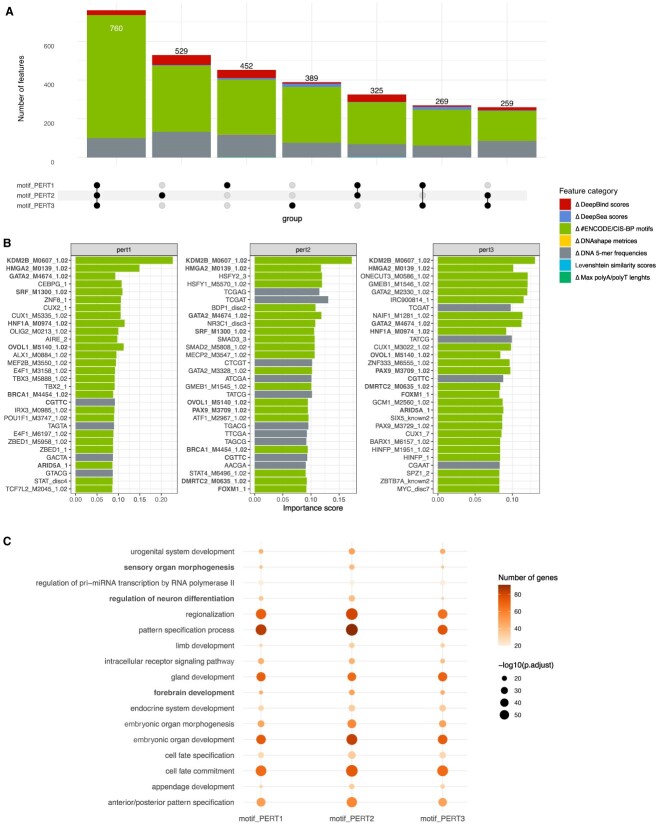
The three perturbation approaches share similar important features, specifically neural developmental features
We then set out to investigate which innate features represent the variances among perturbation sequences, and whether these features differ using different perturbation approaches. First, we queried the top 10% of the features (∼2500) that explain the variability among perturbed sequences (section ‘Materials and methods’). We found that a majority of these features (1601) are shared by at least two perturbation approaches (Figure 5A). Notably, these features mainly fall into ‘the change in the number of ENCODE/CIS-BP motifs’ and ‘5-mers frequencies’ categories.
Figure 5.
Assessment of the important features representing perturbation sequences. (A) The number of important features shared by three perturbation approaches. (B) Top 30 important features of each perturbation approach. The names of features that are shared by at least two perturbation approaches are marked in bold. (C) Gene ontology enrichment analysis of the top 2500 genes represented by the TF binding factors.
Further scrutiny of the top 30 features revealed a substantially large overlap among the three perturbation approaches (Figure 5B). Since a majority of the shared features are transcription factor (TF) binding motifs, we conducted gene ontology analysis on the TFs corresponding to the top 2500 binding motifs. The analysis revealed consistent enrichment of early embryonic development ontologies, including neural development pathways among three perturbation approaches (Figure 5C). These findings suggest that the three perturbation approaches share important features related to neural development, as expected.
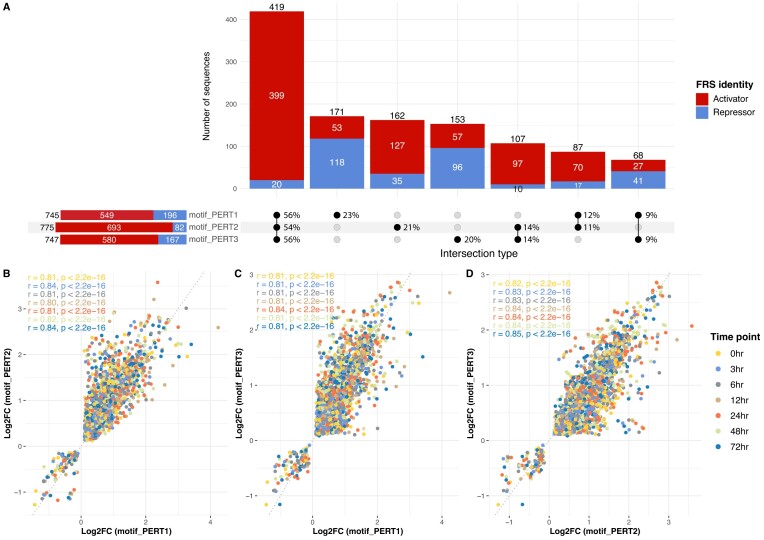
The MPRA outputs are largely consistent across different perturbations
After assessing the basic and advanced goals of perturbation approaches, we next evaluated the consistency of MPRA outputs among three perturbation approaches. The MPRA output consists of two parts: the multi-class FRS identities, and the numeric regulatory effect (Log2FC) at seven time points of neural differentiation (section ‘Materials and methods’).
For the FRS identities, the activities of 419 functional regulatory sites are consistent across three perturbation approaches, and 95% (399) of them are activators (Figure 6A). Additionally, 262 sites are consistent in any of two of the approaches but not in the remaining one (Figure 6A).
Figure 6.
Consistency of MPRA outputs among three perturbations. (A) Number of sequences that share the same FRS identities. The bars are colored by activators (red) and repressors (blue). In the ‘intersection type’ matrix. The percentages are row-normalized, indicating the proportion of sequences belonging to different intersection types within each perturbation approach. (B) The correlation of Log2FC between motif_PERT1 and motif_PERT2. Each dot is a perturbation sequence and is colored by the time point. (C) The correlation of Log2FC between motif_PERT1 and motif_PERT3. (D) The correlation of Log2FC between motif_PERT2 and motif_PERT3.
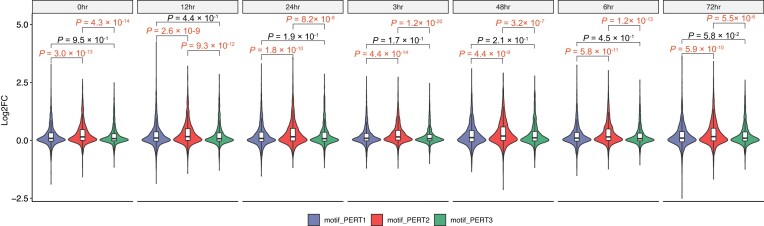
In terms of the Log2FC, we found a high correlation among all three perturbations across all time points (Figure 6B–D). However, we found that PERT2 yielded the highest Log2FC than the other two approaches across all the seven time points (Figure 7, Supplementary Figure S1). Regarding the RAND sequences, PERT2 exhibits the highest, Log2FC while PERT1 exhibits the lowest Log2FC across all time points. This indicates that using a perturbation approach where a constant sequence replaces the target motifs can introduce a constant bias in the results (e.g. higher Log2FC for PERT2). This higher Log2FC also explains the higher proportion of activators identified using PERT2 (Figure 6A).
Figure 7.
Comparison of the Log2FC among three perturbation approaches. The Log2FC values are separated by time point before being compared among three perturbation approaches.
Predictive models of MPRA activity perform the best in PERT3
The perturbation MPRA technique, if designed appropriately, has the potential to predict the activity of non-coding regulatory genomic regions (24). Namely, it is feasible to predict the regulatory activity of a motif by fitting predictive models using the difference in the features between its WT sequence and perturbation sequence. Consequently, this leads to a critical question: which sequence design approach for motif perturbation could yield the best performance of such prediction models? This suggests that by designing the perturbation sequences, we may expand the applicability of perturbation MPRA from experimentally identifying regulatory motifs only within designed genomic regions to computationally predicting regulatory elements throughout the non-coding genome and under different cellular contexts. In light of this, we further compared the performances of three perturbation approaches using the supervised models as described in the Materials and methods section.
Briefly, we use the difference of features between perturbation sequences and their equivalent WT sequence as the independent variables to fit both classification and regression models. Next, we perform a 10-fold cross-validation for each perturbation data. To benchmark the performance of the models, we statistically compared the AUROC for classifiers and the Pearson correlation coefficient for regressors on the independent test data sets in each fold.
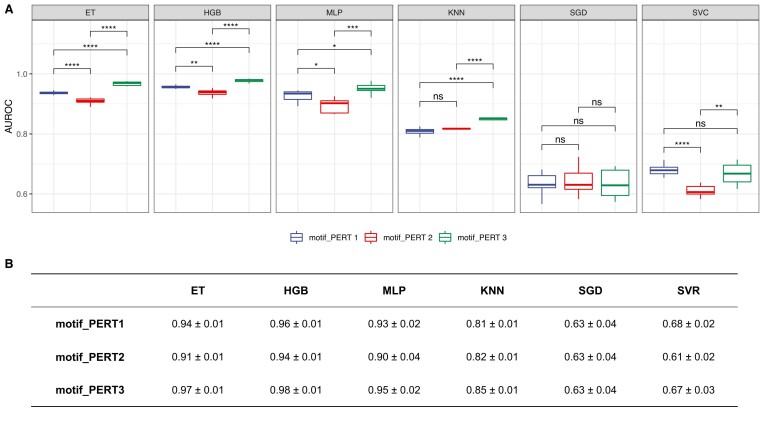
For the classification models that predict the measure of motif FRS identities, we report the receiver-operating characteristic curve (AUROC) of three perturbation approaches (Figure 8). We found that three non-linear models (ET, HGB, and MLP) exhibit high robustness in predicting the FRS identities in the three perturbations. Furthermore, using the results from ET models, we found that PERT3 significantly outperforms PERT2 and PERT1, and PERT1 significantly outperforms PERT2 (pairwise Wilcoxon rank sum test, PERT1 versus PERT2, P = 5.58 × 10−5; PERT1 versus PERT3, P = 3.24 × 10−5; PERT2 versus PERT3, P = 3.24 × 10−5).
Figure 8.
Performance of classification models. (A) The area under the receiver-operating characteristic curve (AUROC) of different classification models. Asterisks/ns indicate levels of statistical significance, calculated by pairwise Wilcoxon rank sum tests (P-value < 0.05*, < 0.01**, < 0.001***, < 0.0001****; ns, non-significant). (B) A summary of the mean ± standard deviation values for AUROCs of classification models.
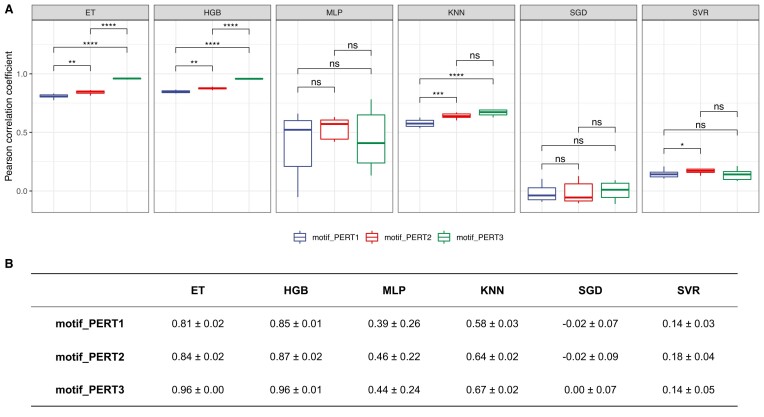
For the regression models that predict the quantitative measure of motif regulatory effect, we report the Pearson correlation coefficients for the three perturbation approaches (Figure 9, Supplementary Figures S2 and S3). Similarly, the model-wise comparison shows the robustness of the ET and HGB model, and PERT3 significantly outperforms the other two approaches, while PERT2 outperforms PERT1 (pairwise Wilcoxon rank sum tests, PERT1 versus PERT2, P = 2.57 × 10−3; PERT1 versus PERT3, P = 3.89 × 10−5; PERT2 versus PERT3, P = 3.89 × 10−5).
Figure 9.
Performance of regression models. (A) The Pearson correlation coefficients of different regression models. Asterisks/ns indicate levels of statistical significance, calculated by pairwise Wilcoxon rank sum tests (P-value < 0.05*, < 0.01**, < 0.001***, < 0.0001****; ns, non-significant). (B) A summary of the mean ± standard deviation values for Pearson correlation coefficients of regression models.
Discussion
Comprehensively deciphering the regulatory activity of non-coding loci is crucial to the understanding of gene expression dynamics. Shedding light on this, the perturbation-based MPRA technique has enabled the identification of regulatory elements such as enhancers, promoters, and silencers (15,18,19). However, insufficient attention has been given to the comprehensive evaluation of various perturbation approaches. As a result, a gold standard of perturbation sequence design strategies remains scant.
Motivated by this scarcity, we proposed a framework for assessing different perturbation approaches, with the aim of better identifying regulatory elements using the perturbation-based MPRA technique. Further, we took advantage of a publicly available data set, which contains the MPRA results acquired from three perturbation approaches (PERT1, PERT2 and PERT3), to conduct an all-inclusive characterization and comparison of these approaches. In short, PERT1 and PERT2 replaced the target motif with two different ‘non-motif’ sequences identified respectively (Supplementary Methods and Supplementary Notes), and PERT3 shuffled the nucleotides of target motifs.
Starting from the essential ideas of perturbation, which is to eliminate the regulatory effects from target motif(s) within a certain genomic region, we first defined five metrics for assessing the impact of different perturbation approaches (hit rate, perturbation rate, perturbation specificity, newly introduced target motifs per sequence, and general alteration in the number of motifs, see Section ‘Methods’). These metrics allowed us to scrutinize the overall modification of motif-based profiles within perturbation sequences from different perspectives. Based on our findings, the three approaches exhibit consistently high rates of removing the target motifs at their targeted positions, which indicates success in in-situ motif perturbation. Additionally, the perturbation rate is kept high across the three perturbation approaches (80%), with PERT3 being the lowest (79%), while not significantly different. This implies a further achievement in both in-situ and ex-situ removal of target motifs of the three approaches. We note that PERT3 shows a higher probability of introducing target-identical motifs. Despite these, PERT3 brings minimal alterations to the WT motifs within the sequence region, implying that the perturbation specificity of PERT3 is the highest. Moreover, PERT3 leads to the least non-specific motif changes. So far, our observation suggests that the selection of perturbation approaches is a trade-off: for the researchers, it becomes a question of whether to sacrifice the perturbation specificity to achieve a high perturbation rate, or whether to pursue a higher specificity at the cost of a lower perturbation rate.
The next part of our framework is the comparison of MPRA outputs since they are crucial for inferring the activity of target motifs. Particularly, MPRA outputs consist of two parts: (i) the functional regulatory site (FRS) identities that indicate whether the target motif is a non-functional, repressing, or activating element; (ii) the numeric regulatory effects (Log2FC) that quantify the FRS motifs. According to our results, the FRS identities are largely consistent, and the Log2FC are highly correlated among all three perturbations. Yet, we also observed a constant skew in the results of PERT2, which indicates that inserting repeated/fixed sequences across the assayed regions is likely to introduce systematic biases in downstream results. The results of this part demonstrated that PERT3 is less likely to introduce systematic biases in MPRA outputs, albeit the high-consistency and high-accuracy profiling for the regulatory activity across all three perturbation approaches.
The final part of the framework is to evaluate the potential of perturbation-MPRA in predicting the regulatory activity of non-coding motifs since our previous works have shown robustness in predicting the activity of putative regulatory elements (24,44). Specifically, by adequately designing perturbation sequences, the MPRA outputs could be computationally predicted by machine-learning models using the biological features of designed sequences as predictor variables. This approach, in some cases, can efficiently identify functional regulatory regions so as to reduce the time and cost of wet lab experiments. Therefore, we developed data-driven models to predict the regulatory activity of target motifs by using the difference in over 28 000 predictive features between perturbation and wild-type sequences. Comparing the performance of models that are built upon the three perturbation approaches, we found that PERT3 significantly outperforms the other two in both classification and regression tasks. These findings further support the notion that using a perturbation approach where the nucleotides are being shuffled randomly works generally better than approaches that replace the target motif with a constant ‘non-motif’ sequence (see Materials and methods, Supplementary Methods, and Supplementary Notes).
In summary, we proposed a framework for the evaluation of perturbation sequence design strategies for MPRA experiments, and we utilized this framework to compare three perturbation-based MPRA approaches. From a computational perspective, this study is the first to evaluate the library design of the MPRA technique comprehensively. From an experimental perspective, our results provide deep insights into understanding the impacts of motif perturbation in MPRA experiments. Given the inherent challenge of providing precise guidance in the absence of verifiable in-vivo ground truth, we advocate for a prudent approach to perturbation MPRA sequence design. In the context of unbiased prediction of non-coding functional genomics, we recommend a design strategy that involves random nucleotide shuffling within the perturbed site. Additionally, we advise prioritizing sequences that introduce the fewest new target motifs to maintain the fidelity of the experimental setup.
In conclusion, our study has the potential to catalyze a new era in non-coding genomic research utilizing MPRA techniques and foster the development of innovative, comprehensive computational methodologies. By providing a robust framework and shedding light on the intricacies of sequence perturbations in MPRA experiments, our findings hold significant promise for advancing not only the field of non-coding genomics but also related domains such as enhancer characterization, regulatory genomics, and computational biology. This interdisciplinary value, situated at the convergence of molecular biology, genomics, and computational biology, broadens the scope of our work, making it relevant to a diverse audience keen to unravel the intricate interplay between sequence perturbations and transcription factor binding. As researchers across these disciplines continue to harness the insights from our study, they will contribute to an increasingly refined understanding of the functional impacts of non-coding regulatory elements, driving the progress of genomics research.
Supplementary Material
Acknowledgements
We acknowledge the Office of Advanced Research Computing (OARC) at Rutgers, The State University of New Jersey, for providing access to the Amarel cluster and associated research computing resources that have contributed to the results reported here. URL: https://oarc.rutgers.edu. We also thank the anonymous reviewers for their insightful comments.
Contributor Information
Jiayi Liu, Graduate Program in Cell & Developmental Biology, Rutgers, The State University of New Jersey, 604 Allison Rd, Piscataway, NJ 08854, USA; Department of Biochemistry and Molecular Biology, Rutgers, The State University of New Jersey, 604 Allison Road, Piscataway, NJ 08854, USA; Center for Advanced Biotechnology and Medicine, Rutgers, The State University of New Jersey, 679 Hoes Lane West, Piscataway, Piscataway, NJ 08854, USA.
Tal Ashuach, Department of Electrical Engineering and Computer Sciences and Center for Computational Biology, University of California, Berkeley, 387 Soda Hall, Berkeley, CA 94720, USA.
Fumitaka Inoue, Institute for the Advanced Study of Human Biology (WPI-ASHBi), Kyoto University, Faculty of Medicine Building B, Yoshidatachibanacho, Sakyo Ward, Kyoto 606-8303, Japan.
Nadav Ahituv, Department of Bioengineering and Therapeutic Sciences, University of California, 1700 4th Street, San Francisco, CA 94158, USA; Institute for Human Genetics, University of California, 513 Parnassus Ave, San Francisco, CA 94143, USA.
Nir Yosef, Department of Systems Immunology, Weizmann Institute of Science, 234 Herzl Street, Rehovot 7610001, Israel; Chan-Zuckerberg Biohub, 499 Illinois St, San Francisco, CA 94158, USA; Department of Systems Immunology, Ragon Institute of MGH, MIT, and Harvard Institute of Science, 400 Technology Square, Cambridge, MA 02139, USA.
Anat Kreimer, Department of Biochemistry and Molecular Biology, Rutgers, The State University of New Jersey, 604 Allison Road, Piscataway, NJ 08854, USA; Center for Advanced Biotechnology and Medicine, Rutgers, The State University of New Jersey, 679 Hoes Lane West, Piscataway, Piscataway, NJ 08854, USA.
Data availability
The datasets are available at the NCBI Gene Expression Omnibus (GEO) as accession number GEO: GSE115046. The raw sequences have been deposited at DDBJEMBLGenBank under the accession KIBZ00000000. The version described in this paper is the first version, KIBZ01000000. The source code for data processing and analysis for this study has been deposited in Zenodo (URL: https://zenodo.org/records/10028417).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institute of Mental Health [R00MH117393 to A.K.]. Funding for open access charge: National Institute of Mental Health [R00MH117393 to A.K.].
Conflict of interest statement. T.A. is currently an employee of Vevo Therapeutics but pursued this work independently of the organization. N.A. is a co-founder and on the scientific advisory board of Regel Therapeutics. N.A. receives funding from BioMarin Pharmaceutical Incorporate. All other authors declare no competing interests.
References
- 1. Rheinbay E., Nielsen M.M., Abascal F., Wala J.A., Shapira O., Tiao G., Hornshøj H., Hess J.M., Juul R.I., Lin Z. et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020; 578:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agarwal V., Inoue F., Schubach M., Martin B.K., Dash P.M., Zhang Z., Sohota A., Noble W.S., Yardimci G.G., Kircher M. et al. Massively parallel characterization of transcriptional regulatory elements in three diverse human cell types. 2023; bioRxiv doi:06 March 2023, preprint: not peer reviewed 10.1101/2023.03.05.531189. [DOI]
- 3. Koesterich J., An J.-Y., Inoue F., Sohota A., Ahituv N., Sanders S.J., Kreimer A. Characterization of de novo promoter variants in autism spectrum disorder with massively parallel reporter assays. Int. J. Mol. Sci. 2023; 24:3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deng C., Whalen S., Steyert M., Ziffra R., Przytycki P.F., Inoue F., Pereira D.A., Capauto D., Norton S., Vaccarino F.M. et al. Massively parallel characterization of psychiatric disorder-associated and cell-type-specific regulatory elements in the developing human cortex. 2023; bioRxiv doi:16 February 2023, preprint: not peer reviewed 10.1101/2023.02.15.528663. [DOI] [PMC free article] [PubMed]
- 5. Koh K.D., Bonser L.R., Eckalbar W.L., Yizhar-Barnea O., Shen J., Zeng X., Hargett K.L., Sun D.I., Zlock L.T., Finkbeiner W.E. et al. Genomic characterization and therapeutic utilization of IL-13-responsive sequences in asthma. Cell Genom. 2022; 3:100229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Melnikov A., Murugan A., Zhang X., Tesileanu T., Wang L., Rogov P., Feizi S., Gnirke A., Callan C.G., Kinney J.B. et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 2012; 30:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mogno I., Kwasnieski J.C., Cohen B.A. Massively parallel synthetic promoter assays reveal the in vivo effects of binding site variants. Genome Res. 2013; 23:1908–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patwardhan R.P., Hiatt J.B., Witten D.M., Kim M.J., Smith R.P., May D., Lee C., Andrie J.M., Lee S.-I., Cooper G.M. et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012; 30:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patwardhan R.P., Lee C., Litvin O., Young D.L., Pe’er D., Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 2009; 27:1173–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peters D.T., Musunuru K. Functional evaluation of genetic variation in complex human traits. Hum. Mol. Genet. 2012; 21:R18–R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharon E., Kalma Y., Sharp A., Raveh-Sadka T., Levo M., Zeevi D., Keren L., Yakhini Z., Weinberger A., Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012; 30:521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F. et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016; 165:1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu Q., Wu J., Karim K., Chen X., Wang T., Iwama S., Carobbio S., Keen P., Vidal-Puig A., Kotter M.R. et al. Massively parallel characterization of CRISPR activator efficacy in human induced pluripotent stem cells and neurons. Mol. Cell. 2023; 83:1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Akhtar W., de Jong J., Pindyurin A.V., Pagie L., Meuleman W., de Ridder J., Berns A., Wessels L.F.A., van Lohuizen M., van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013; 154:914–927. [DOI] [PubMed] [Google Scholar]
- 15. Kheradpour P., Ernst J., Melnikov A., Rogov P., Wang L., Zhang X., Alston J., Mikkelsen T.S., Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013; 23:800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. White M.A., Myers C.A., Corbo J.C., Cohen B.A. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:11952–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X., He L., Goggin S.M., Saadat A., Wang L., Sinnott-Armstrong N., Claussnitzer M., Kellis M. High-resolution genome-wide functional dissection of transcriptional regulatory regions and nucleotides in human. Nat. Commun. 2018; 9:5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kreimer A., Ashuach T., Inoue F., Khodaverdian A., Deng C., Yosef N., Ahituv N. Massively parallel reporter perturbation assays uncover temporal regulatory architecture during neural differentiation. Nat. Commun. 2022; 13:1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inoue F., Kreimer A., Ashuach T., Ahituv N., Yosef N. Identification and massively parallel characterization of regulatory elements driving neural induction. Cell Stem Cell. 2019; 25:713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashuach T., Fischer D.S., Kreimer A., Ahituv N., Theis F.J., Yosef N. MPRAnalyze: statistical framework for massively parallel reporter assays. Genome Biol. 2019; 20:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordon M.G., Inoue F., Martin B., Schubach M., Agarwal V., Whalen S., Feng S., Zhao J., Ashuach T., Ziffra R. et al. lentiMPRA and MPRAflow for high-throughput functional characterization of gene regulatory elements. Nat. Protoc. 2020; 15:2387–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017; 33:2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kreimer A., Yan Z., Ahituv N., Yosef N. Meta-analysis of massively parallel reporter assays enables prediction of regulatory function across cell types. Hum. Mutat. 2019; 40:1299–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alipanahi B., Delong A., Weirauch M.T., Frey B.J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015; 33:831–838. [DOI] [PubMed] [Google Scholar]
- 26. Chen K.M., Wong A.K., Troyanskaya O.G., Zhou J. A sequence-based global map of regulatory activity for deciphering human genetics. Nat. Genet. 2022; 54:940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chiu T.-P., Comoglio F., Zhou T., Yang L., Paro R., Rohs R. DNAshapeR: an R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics. 2016; 32:1211–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou T., Yang L., Lu Y., Dror I., Dantas Machado A.C., Ghane T., Di Felice R., Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013; 41:W56–W62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwasnieski J.C., Fiore C., Chaudhari H.G., Cohen B.A. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014; 24:1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kheradpour P., Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014; 42:2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu H., Miao Y.-R., Jia L.-H., Yu Q.-Y., Zhang Q., Guo A.-Y. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019; 47:D33–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winkler W.E. String comparator metrics and enhanced decision rules in the fellegi-sunter model of record linkage. Proceedings of the Section on Survey Research. 1990; [Google Scholar]
- 34. Sariyar M., Borg A. The recordlinkage package: detecting errors in data. The R. Journal. 2010; 2:61. [Google Scholar]
- 35. Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021; 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bottou L. Lechevallier Y, Saporta G Large-scale machine learning with stochastic gradient descent. Proceedings of COMPSTAT’2010. 2010; Heidelberg: Physica-Verlag HD; 177–186. [Google Scholar]
- 38. Cristianini N., Ricci E.. Kao M.-Y. Support vector machines. Encyclopedia of Algorithms. 2008; US Boston, MA: Springer; 928–932. [Google Scholar]
- 39. Zhang Z. Introduction to machine learning: k-nearest neighbors. Ann. Trans. Med. 2016; 4:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Geurts P., Ernst D., Wehenkel L. Extremely randomized trees. Mach. Learn. 2006; 63:3–42. [Google Scholar]
- 41. Sipper M., Moore J.H. AddGBoost: a gradient boosting-style algorithm based on strong learners. Mach. Learn. Appl. 2022; 7:100243. [Google Scholar]
- 42. He K., Zhang X., Ren S., Sun J. Delving deep into rectifiers: surpassing human-level performance on imagenet classification. Proc. IEEE Int. Conf. Comput. Vis. 2015; 1026–1034. [Google Scholar]
- 43. Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011; 12:2825–2830. [Google Scholar]
- 44. Kreimer A., Zeng H., Edwards M.D., Guo Y., Tian K., Shin S., Welch R., Wainberg M., Mohan R., Sinnott-Armstrong N.A. et al. Predicting gene expression in massively parallel reporter assays: a comparative study. Hum. Mutat. 2017; 38:1240–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Merika M., Orkin S.H. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 1993; 13:3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets are available at the NCBI Gene Expression Omnibus (GEO) as accession number GEO: GSE115046. The raw sequences have been deposited at DDBJEMBLGenBank under the accession KIBZ00000000. The version described in this paper is the first version, KIBZ01000000. The source code for data processing and analysis for this study has been deposited in Zenodo (URL: https://zenodo.org/records/10028417).