Summary
Neurodevelopmental disorders (NDDs) result from impaired development and functioning of the brain. Here, we identify loss-of-function (LoF) variation in ZFHX3 as a cause for syndromic intellectual disability (ID). ZFHX3 is a zinc-finger homeodomain transcription factor involved in various biological processes, including cell differentiation and tumorigenesis.
We describe 42 individuals with protein-truncating variants (PTVs) or (partial) deletions of ZFHX3, exhibiting variable intellectual disability and autism spectrum disorder, recurrent facial features, relative short stature, brachydactyly, and, rarely, cleft palate. ZFHX3 LoF associates with a specific methylation profile in whole blood extracted DNA. Nuclear abundance of ZFHX3 increases during human brain development and neuronal differentiation. ZFHX3 was found to interact with the chromatin remodeling BRG1/Brm-associated factor complex and the cleavage and polyadenylation complex, suggesting a function in chromatin remodeling and mRNA processing. Furthermore, ChIP-seq for ZFHX3 revealed that it predominantly binds promoters of genes involved in nervous system development. We conclude that loss-of-function variants in ZFHX3 are a cause of syndromic ID associating with a specific DNA methylation profile.
Keywords: neurodevelopmental disorder, ZFHX3, chromatin remodeling complex, mRNA polyadenylation and cleavage complex
Graphical abstract

We identify loss-of-function (LoF) variation in ZFHX3 as a cause for syndromic intellectual disability. We show that LoF is associated with a specific methylation signature and that ZFHX3 targets promoters of genes implicated in neural development. Proteomics analysis indicates a contribution of ZFHX3 to chromatin remodeling and pre-messenger RNA processing.
Introduction
Neurodevelopmental disorders (NDDs) are a heterogeneous group of early-onset conditions that impact development and functioning of the central nervous system (CNS) affecting approximately 2%–5% of children worldwide. The clinical and genetic heterogeneity of NDDs make it challenging to find a molecular diagnosis for individual cases.1 Although recent technological improvements led to a significant increase in diagnostic yield and the identification of several new NDD associated genes, many associated genes remain to be discovered.2 Here, we identify loss-of-function (LoF) defects in zinc finger homeobox 3 (ZFHX3) in an NDD.
ZFHX3 (MIM: 104155), previously known as AT motif-binding transcription factor 1 (ATBF1), encodes one of the largest transcription factors (404 kDa) containing 23 zinc finger motifs and four homeodomains. It was originally identified as a transcriptional suppressor that binds an AT-rich motif in the alfa-fetoprotein enhancer in human hepatoma cells.3 Subsequently, ZFHX3 was found to function in multiple biological processes including mammary gland development,4,5 myogenic differentiation,6 and pathological processes such as tumorigenesis7,8,9 and atrial fibrillation (MIM: 613055).10,11,12,13 In mice, expression of Zfhx3 is high during embryonic stages of CNS development but ceases upon brain maturation.14 Nevertheless, expression remains enriched in the adult suprachiasmatic nucleus where it is involved in circadian function.15
Zfhx3 expression also continues in cortical excitatory neurons and appears in late differentiating, nonoverlapping neuron populations.16 These results correlate with previous work from Moreau et al., showing that cells expressing Zfhx3 belong to the Pbx3 neuron cluster, an early cortical excitatory neuron subtype.17 Zfhx3 is further selectively expressed in immature dopamine receptor expressing medium spiny neurons within mice striatum.18 In further support of a role in neuronal differentiation, Zfhx3 expression increases upon neuronal differentiation of pluripotent mouse embryonal carcinoma P19 cells.19,20 Similarly, in the zebrafish, zfhx3 expression coincides with regions of neurogenesis at critical developmental time points.21
ZFHX3 is predicted to be intolerant to protein-truncating variants (PTVs), reflected by the probability of being loss-of-function intolerant (pLI)-score of 1 and a constraint score of observed/expected (oe) of 0.19 in the gnomAD database (v4.0.0).22 Moreover, it belongs to the top 5% of genes implicated in monoallelic autism spectrum disorder (ASD) and/or developmental delay (DD) according to the NDD risk gene prediction tool mantis-ml (https://nddgenes.com)23 and was suggested as an NDD candidate gene susceptible to de novo likely gene disruptive variation by the shallow neural network developed by Chow & Hormozdiari.24
In this study, we define the phenotypic spectrum in an extensive cohort of 42 individuals with LoF variants (18 [partial] deletions and 24 truncating variants) affecting ZFHX3. We report the spatiotemporal and subcellular ZFHX3 levels during human brain and neuronal stem cell development and show that its target genes are implicated in neuron and axon development. Furthermore, proteomic analysis indicates that ZFHX3 contributes to pre-messenger RNA (pre-mRNA) processing, chromatin remodeling and cytoskeleton reorganization. Overall, our data identify ZFHX3 as an important regulator in neuronal differentiation.
Material and methods
Recruitment of affected individuals
Data sharing through GeneMatcher,25 DECIPHER,26 or personal communication identified 42 probands with truncating variants in ZFHX3 (n = 24) and (micro)deletions affecting ZFHX3 (n = 18). Inheritance of ZFHX3 variants was de novo (n = 33), inherited from a (mildly) affected parent (n = 2) or unknown (n = 7). All affected probands were investigated by their referring physicians, and all genetic analyses were performed in a diagnostic setting. An overview of the clinical presentation of all probands and family members is given in Tables S1 and S2. A summary of the clinical characteristics is presented in Table 1. Legal guardians of affected probands gave informed consent for genomic investigations and publication of pseudonymized data following the rules of the Helsinki declaration and according to EC2019/1430 of Ghent University Hospital and/or local ethical committees of the referring centers and the Western University Research Ethics Board (REB 106302). Fisher’s exact test (RStudio) was used for comparative studies between the clinical characteristics of individuals with microdeletions and truncating variants, p values <0.05 were considered statistically significant.
Table 1.
Clinical characteristics of 33 individuals and 6 affected family members with a microdeletion or a protein-truncating variant affecting ZFHX3
| Microdeletions only affecting ZFHX3 (n = 13a) | PTVs (n = 26b) | Total (n = 39) | Fisher’s exact p value | Multigenic deletions (n = 4) | Overall total (n = 43) | |
|---|---|---|---|---|---|---|
| Neurological | ||||||
| Global developmental delay | 6/13 | 18/26 | 24/39 | 0.19 | 4/4 | 27/43 |
| Delayed language development | 7/13 | 10/26 | 16/39 | 0.50 | 3/4 | 19/43 |
| ID | 5/13 | 14/26 | 18/39 | 0.50 | 3/4 | 21/43 |
| Hypotonia | 7/13 | 15/26 | 22/39 | 0.33 | 1/4 | 23/43 |
| Behavioral problems | 5/13 | 11/26 | 16/39 | 1 | 1/4 | 17/43 |
| Skeletal | ||||||
| Postnatal growth retardation | 8/13 | 12/26 | 20/39 | 0.50 | 2/4 | 22/43 |
| Brachydactyly | 3/13 | 12/26 | 15/39 | 0.30 | 2/4 | 17/43 |
| Broad first rays | 3/13 | 8/26 | 11/39 | 0.72 | 0/4 | 11/43 |
| Gastro-intestinal | ||||||
| Feeding difficulties | 9/13 | 15/26 | 24/39 | 0.73 | 3/4 | 27/43 |
| Ocular | ||||||
| Strabism/other | 3/13 | 3/26 | 6/39 | 0.38 | 1/4 | 7/43 |
| Craniofacial | ||||||
| Thin hair | 5/13 | 15/26 | 20/39 | 0.32 | 4/4 | 24/43 |
| Receding anterotemporal hairline | 6/13 | 17/26 | 23/39 | 0.31 | 4/4 | 27/43 |
| High and broad forehead | 11/13 | 18/26 | 29/39 | 0.45 | 4/4 | 33/43 |
| Laterally sparse eyebrows | 4/13 | 9/26 | 13/39 | 1 | 3/4 | 16/43 |
| Upslanted palpebral fissures | 7/13 | 6/26 | 13/39 | 0.08 | 3/4 | 16/43 |
| Epicanthal folds | 6/13 | 10/26 | 16/39 | 0.76 | 0/4 | 16/43 |
| Bulbous nasal tip | 9/13 | 15/26 | 24/39 | 0.73 | 2/4 | 26/43 |
| Low hanging columella | 7/13 | 11/26 | 18/39 | 0.52 | 3/4 | 21/43 |
| Short philtrum | 4/13 | 5/26 | 9/39 | 0.45 | 3/4 | 12/43 |
| Flat philtrum | 9/13 | 11/26 | 20/39 | 0.18 | 2/4 | 22/43 |
| Thin upper lip | 8/13 | 10/26 | 18/39 | 0.20 | 3/4 | 21/43 |
| Downturned corners of the mouth | 6/13 | 10/26 | 16/39 | 0.74 | 3/4 | 19/43 |
| Low-set ears | 5/13 | 5/26 | 10/39 | 0.25 | 3/4 | 13/43 |
| Protruding upper helix of ears | 6/13 | 7/26 | 13/39 | 0.29 | 2/4 | 15/43 |
| Cleft lip/palate | 0/13 | 2/26 | 2/39 | 0.54 | 1/4 | 3/43 |
The Fisher’s exact p value was calculated for individuals with a microdeletion vs. those with a protein-truncating variant.
11 probands and two affected family members.
22 probands and four affected family members.
Facial features analysis
Frontal facial images of 17 individuals with either truncating variants in ZFHX3 or microdeletions only affecting ZFHX3 were uploaded to the Face2Gene Research app (FDNA Inc., Boston, MA, USA), de-identified, and analyzed through the use of the DeepGestalt image analysis technology, a deep-learning computer algorithm.27 The resulting artificial composite image was compared to that of a control cohort matched by age, sex, and ethnicity in the same way as described by Mak et al.28 This resulted in a score distribution and a receiver operating characteristic (ROC) curve.
Cell culture
SH-SY5Y and HEK293T
SH-SY5Y and HEK293T cells were cultured in RPMI (Roswell Park Memorial Institute) 1640 medium (52400041, Gibco) supplemented with 10% fetal bovine serum (FBS), 2 mM L-Glutamine, and 100 IU/mL penicillin/streptavidin at 37°C in a 5% CO2 humid atmosphere.
Human embryonic stem cells (hESCs)
The naive hESCs were cultured on CF-1 irradiated mouse embryonic fibroblasts (MEFs) 2M (A34180, Gibco) in conditioned GDN (glia-derived nexin) medium consisting of KO-DMEM (KnockOut D-MEM, 10829018, Invitrogen) containing 20% knockout serum replacement (10828028, Invitrogen), 1% non-essential amino acids (11140050, Invitrogen), 1% penicillin/streptomycin (15140122, Invitrogen), 0.1 mM L-glutamine (25030024, Invitrogen), and 0.1 mM β-mercaptoethanol (31350010, Invitrogen). The medium is supplemented with 12 ng/mL bFGF (basic fibroblast growth factor; 100–18B, Peprotech), 1,000 U recombinant human LIF (L5283, Sigma), 1 μM PD0325903 (13034, Sanbio), 3μM CHIR99021 (1386, Axon Medchem), 10 μM Forskolin (F6886, Sigma), and 50 ng/mL ascorbic acid (A8960, Sigma). The naive colonies were passaged as single cells using 0.05% trypsin/EDTA (25300054, Invitrogen) every three days and were re-plated on inactivated MEFs. To convert the hESCs on feeder cells to feeder-free conditions, the naive hESCs were passaged with 0.05% trypsin/EDTA (25300054, Invitrogen) onto Matrigel coated plates with conditioned GDN medium (protocol described above) and cultured at hypoxic conditions (37°C, 5% CO2, and 5% O2). On day one, half of the GDN medium was replaced by the RSeT Feeder-Free medium (STEMCELL Technologies). Every second day, a full medium change was performed with RSeT Feeder-Free medium. On day seven, colonies were passaged with Accutase (Thermo Fisher, A1110501) onto freshly made Matrigel-coated plates. Every other day, fresh RSeT Feeder-Free medium was added, and cells were passaged every third or fourth day (depending on the cell density).
Neural stem cells (NSCs)
The BJ fibroblast cell line from human foreskin (#CRL-2522, ATCC) was reprogrammed to human induced pluripotent stem cells (iPSCs) using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Invitrogen). Pluripotency of iPSC clones was validated with immunostaining for pluripotent markers and an embryoid body assay. These iPSCs were differentiated to NSCs using PSC Neural Induction Medium (Gibco, Thermo Fisher). These NSCs were kindly provided by the group of Peter Ponsaerts, University of Antwerp. The NSC line was further expanded in Neural Expansion Medium (Gibco, Thermo Fisher) on coated cultured vessels with Geltrex LDEV-Free hESC-Qualified Reduced Growth Factor Basement Membrane Matrix (Gibco, Thermo Fisher), according to the manufacturer’s instructions. The NSC line was cultured at 37°C in a 5% CO2 humid atmosphere.
Neural differentiation using BrainPhys media (STEMCELL Technologies)
NSCs were cultured in BrainPhys Neuronal Medium (STEMCELL Technologies), supplemented with NeuroCultTM SM1 Neuronal Supplement (STEMCELL Technologies), N2 Supplement-A (STEMCELL Technologies), recombinant human brain-derived neurotrophic factor (BDNF, PeproTech, 20 ng/mL), recombinant human glial-derived neurotrophic factor (GDNF, PeproTech, 20 ng/mL), dibutyryl cAMP ( cyclic adenosine monophosphate; 1 mM, Sigma), and ascorbic acid (200 nM, Sigma). Half of the total volume was replaced every 2–3 days.
Immunofluorescence
SH-SY5Y and NSCs were cultured onto glass coverslips in 12-well plates and fixed with 3.7% formaldehyde for 20 min at room temperature. Firstly, for the NSCs, a specific coating (Geltrex LDEV-Free, hESC-qualified, Reduced Growth Factor Basement Membrane Matrix, Gibco, Thermo Fisher) was embedded into the coverslips. Phosphate buffered saline (PBS) was used for all washing steps on all cell types. Autofluorescence quenching of the cells was performed with 50 mM NH4Cl for 10 min, followed by permeabilization with 0.2% Triton X-100 for 10 min. Non-specific antibody binding was avoided by means of a blocking buffer containing 3% goat serum (GS) (Agilent cat. no. X0907), as well as 1% bovine serum albumin (BSA) in a PBS and 0.1% Tween 20 (PBS-T) solution for 30 min at room temperature. The primary antibody, rabbit anti-ZFHX3 (HPA059353, Sigma-Aldrich, 1/100), was added to the cells and incubated overnight at 4°C. The fluorescently labeled secondary antibody, 2 μg of goat anti-rabbit IgG Alexa Fluor 488 (A-11034, Thermo Fisher) diluted in a Hoechst (Life Technologies) solution with blocking buffer, was then added for 1 h at room temperature while avoiding light exposure. Cover slips were mounted onto glass slides, and propyl gallate mounting media (Sigma-Aldrich) were added onto the coverslip. For each cell line, three technical replicates were included. Imaging was performed by a Zeiss Axio Observer Z1 fluorescent microscope (Carl Zeiss, Australia). The magnification of the images was 100×.
Expression profiling
RNA of cultured cells was isolated using the Direct-zol RNA MiniPrep Kit (Zymo Research), according to manufacturer’s instructions. NanoDrop (Thermo Fisher) was used to determine the RNA concentration. cDNA synthesis was performed using the iScript Advanced cDNA synthesis kit (BioRad), according to manufacturer’s instructions. Subsequently, RT-qPCR (real-time quantitative PCR)was performed with a PCR mix containing 5 ng cDNA, 2.5 μL of SsoAdvanced SYBR qPCR mix (BioRad), and 0.25 μL of forward and reverse primers (250 μM concentration, Integrated DNA Technologies). The RT-qPCR was performed on an LC-480 device (Roche), and gene expression levels were analyzed using the qBase+ software 3.2 (Biogazelle). The software qBase+ uses a generalized model of the delta-delta-Ct approach, thereby supporting the use of gene-specific amplification efficiencies and normalization with multiple reference genes. All formulas of this model are detailed in Hellemans et al.29 The RT-qPCR primers used in this study can be found in Table S12. Three biological and two technical replicates were collected per timepoint.
Immunoprecipitation followed by mass spectrometry (IP-MS)
Protein from 2 × 107 cells were extracted with freshly made RIPA (radioimmunoprecipitation assay) buffer. Next, 2 μg antibody (ZFHX3 antibody [D1-120, MBL Life Science], anti-human IgG antibody [ab2410, Abcam]) was added, and after 4 h, 20 μL extensively washed protein A Ultralink resin beads (Thermo Fisher) were added and further rotated overnight. The samples were centrifuged and washed five times with a 50 mM digestion buffer (50 mM TrisHCl pH 8, 2 mM CaCl2) to remove unbound proteins. Beads containing antibody and proteins of interest were resuspended in 175 μL digestion buffer. For IP, 25 μL of samples were incubated at 95°C for 10 min with 40 μL of 2× laemmli buffer at 1,000 rpm to elute the proteins. Supernatants containing the proteins of interest were used for western blot analysis. For IP-MS, the remaining 150 μL of the samples was incubated for 4 h with 1 μg trypsin (Promega) at 37°C, while shaking. Beads were removed by centrifugation at maximum speed and proteins were digested again with 1 μg of trypsin, overnight at 37°C. The peptide mixture was acidified with 1% TFA (trifluoroacetic acid) and purified on Omix C18 tips (Agilent). Next, the samples were dried and re-dissolved in 20 μL 0.1% formic acid in water/acetonitrile (98:2, v/v) of which 2 μL was injected for LC-MS/MS analysis on an Ultimate 3000 RSLC nano LC (Thermo Fisher, Bremen, Germany) in-line connected to a Q Exactive mass spectrometer (Thermo Fisher). For each cell line, three biological replicates were included. The data analysis is further explained in the supplemental information.
Expression constructs, transfection, cell harvesting, and protein extraction
The plasmid encoding ZFHX3-FLAG was ordered from AddGene (pcDNA3.1-FLAG-ATFB1, #40927). SMARCB1 and CPSF2 constructs were ordered from Genecopoeia as shuttle vectors. Subsequently, Gateway cloning (Gateway Technology, Gateway LR reaction, Invitrogen) was performed according to the manufacturer’s instructions to insert the shuttle vectors’ open reading frame (ORF) (SMARCB1 or CPSF2) into a destination vector pMH-HA (human influenza hemagglutinin) N-terminal tag vector. An N-terminal 3xHA-tag followed the ORF in the designed SMARCB1-3xHA or CPSF2-3xHA expression vector. All constructs were Sanger sequenced (GATC sequencing, Eurofins Genomics) bidirectionally for their entire ORF. The vector maps can be found in Figure S7.
To confirm protein-protein interaction via co-immunoprecipitation (co-IP) (see below), the ZFHX3-FLAG construct was co-transfected with the SMARCB1-3xHA or the CPSF2-3xHA expression vector in HEK293T cells. As control conditions, we performed single transfections of ZFHX3-FLAG, SMARCB1-3xHA, or CPSF2-3xHA alone. Transfection complexes were prepared according to the Lipofectamine 3000 manufacturer’s instructions. Appropriate amounts of the above-described plasmid DNA and P3000 buffer (Lipofectamine 3000 protocol) were diluted and added to the Lipofectamine 3000 solution (Thermo Fisher) in a 1:1 ratio, depending on the amount of desired transfected cells. At room temperature and within a 15-min incubation period, the complex was added to the corresponding amount of HEK293T cells, which were seeded for 24 h before the transfection. Cells were incubated upon normoxic conditions at 37°C.
Upon 48 h incubation time, 2.5 × 106 cells were harvested per condition and lysed in freshly made RIPA buffer for protein extraction. Following centrifugation (10 min, 4°C, 8,000 g), proteins were extracted, and the concentration was determined using the BCA protein assay kit (Thermo Fisher). Production of the protein of interest was confirmed by western blot.
co-IP
Immunoprecipitation was performed according to the Dynabeads Protein G (Invitrogen) manufacturer’s instructions. 300 μL lysate was added to the antibody beads complex and incubated overnight at 4°C. Protein complexes were eluted with 20 μL glycine buffer (pH 2.8) and determined using western blot. As a negative control for the co-immunoprecipitation (co-IP), 300 μL per cell lysate was added to Dynabeads protein G, coupled to non-specific IgG antibodies.
The anti-HA.11 Epitope Tag antibody (1/1000, Clone 16B12, Biolegend) was used to enrich the capture of the SMARCB1-HA-tagged protein and the CPSF2-HA-tagged protein within the co-IP. Each experiment was performed twice under the same conditions.
Western blot
Western blot was performed according to an in-house protocol. Either protein lysates or eluted proteins after co-IP were subjected to NuPage gel electrophoresis, subsequently transferred to a nitrocellulose membrane, and detected using the appropriate antibodies: anti-ZFHX3 (custom antibody [Parsons et al.15] kindly provided by Prof. Patrick Nolan), anti-HA.11 Epitope Tag antibody (1/1000, Clone 16B12, Biolegend), anti-ATBF1 (1/1000, AT-6 PD011, MBL) and anti-Flag M2 (1/2000, F3165-.2MG, Sigma-Aldrich). Blots were stripped twice. Imaging was performed with an Amersham Imager 600 CCD camera (GE Healthcare, Chicago, Illinois, USA).
DNA methylation analysis
The workflow used to establish whether a specific DNA methylation profile is found within our LoF cohort in comparison with other neurodevelopmental syndromes is described in detail in Aref-Eshghi et al.30 Further description can be found in the supplemental information.
ChIP-sequencing and data analysis
ChIP-seq (Chromatin Immunoprecipitation Sequencing) for ZFHX3 has been performed on 10 million NSCs (with endogenous ZFHX3 levels) and 10 million NSCs with ZFHX3 overexpression. These experiments were conducted by the Diagenode ChIP-seq/ChIP-qPCR Profiling service (Diagenode Cat# G02010000). Material and methods are described in the supplemental information.
Using ChIPSeeker,31 the ChIP peaks called in both the sample with endogenous ZFHX3 expression and the sample with ectopic ZFHX3 expression were annotated to the nearest gene. The annotation is based on the distance of the peak to the nearest transcription start site (TSS) with a max distance cut-off of 1 kb using the annotations from the TxDb.Hsapiens.UCSC.hg38.knownGene package (https://doi.org/10.18129/B9.bioc.TxDb.Hsapiens.UCSC.hg38.knownGene). To identify predominant biological processes (BPs), we performed functional enrichment analysis on the intersect peaks using the BP gene ontology (GO) terms of the annotated genes. The top 20 of enriched GO terms (q value < 0.05) is visualized as a dot plot in Figure 6B. In Figure 6C, the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis provides the annotation of biological pathways. The top 10 of enriched pathways (q value < 0.05) is visualized as a dot plot.
Figure 6.
ZFHX3 binds primarily to promoter regions of genes associated with pathways involved in neuron and axon development
(A) Left: annotation of the 22,094 overlapping ChIP-seq peaks. 57.18% of these peaks correspond to promoter regions. The pie chart visualizes the genomic annotation and the percentage of the peaks that reside in/contain the transcription start site (TSS), exonic sequences, 5′UTR, 3′UTR, intronic, or intergenic regions, as displayed with the corresponding colors. Color code is used to assign each region. Right: distribution of the ZFHX3-binding loci relative to TSSs. The majority of the peaks falls within 0–1 kb of the TSS, also suggesting that ZFHX3 mainly binds to promoter sequences. Color code is used to represent the distance from TSS in kb.
(B) The target genes of ZFHX3 are involved in axonogenesis, axon development, and the regulation of the nervous system, including neurogenesis. The gene ontology (GO) dot plot displays the top 20 enriched biological processes (BPs) ranked by gene ratio (# of genes related to GO term/total number of significant genes) and the p-adjusted values for these terms (color). The size of the dot represents the gene counts per BP.
(C) ZFHX3 plays a key role in neuron and axon developmental pathways. The Kyoto Encyclopedia of Genes and Genomes (KEGG) dot plot provides all enriched biological pathways ranked by gene ratio (# of genes related to GO term/total number of significant genes) and the p-adjusted values for these terms (color). The size of the dot represents the gene counts per pathway.
Motif enrichment analysis (i.e., enrichment analysis of known motifs and de novo motif finding) on the intersect peaks was performed with HOMER (v4.11)32 with 200 bp around the peak summit as input. The top 20 de novo and known motifs detected from the analyses are found in the Table S11.
Human expression datasets
Expression levels in terms of transcripts per million (TPM) were retrieved from Cardoso-Moreira et al. (https://apps.kaessmannlab.org/evodevoapp/).33 We examined the expression of ZFHX3 in seven human tissues (i.e., brain, cerebellum, heart, kidney, liver, ovary, and testis) and in up to 23 developmental stages (see Figure S1B). Furthermore, for the developmental data of these seven human organs, Spearman’s correlations were computed between expression values of ZFHX3 and expression values of all other genes. Subsequently, these data were ranked based on the Rho value (>0.8) and a p value < 0.01. This ranked list was used to perform a preranked GSEA (Gene Set Enrichment Analysis, https://www.gsea-msigdb.org/gsea/index.jsp) with the following settings: Gene Set “C5 GO:MF”; number of permutations, 1,000; min/max number of genes per gene set, 15/200. The GO terms of the top 20 enriched gene sets (all with normalized enrichment score > 1.8 and FDR < 0.10) of this GSEA preranked analysis were subsequently visualized.
In addition, the ZFHX3 expression levels in terms of TPM were retrieved from GTEx (https://www.gtexportal.org/home/). We examined the expression of ZFHX3 and its different isoforms in 54 non-diseased adult tissues (see Figures S1A and S3). Furthermore, the ZFHX3 expression levels in terms of reads per kilobase million (RPKM) from six brain regions during neurodevelopment and adulthood were retrieved from Brainspan (http://www.brainspan.org, see Figure 3B). Moreover, single-cell ZFHX3 expression data during neuronal differentiation from stem cells to neural progenitor cells and mature neurons was retrieved from https://bioinf.eva.mpg.de/shiny/sample-apps/scApeX/ 34 (Figure S1C), and single-cell ZFHX3 expression data from one- or five-months undirected cerebral organoids was retrieved from https://shcheglovitov.shinyapps.io/u_brain_browser/ 35 (Figure S2).
Figure 3.
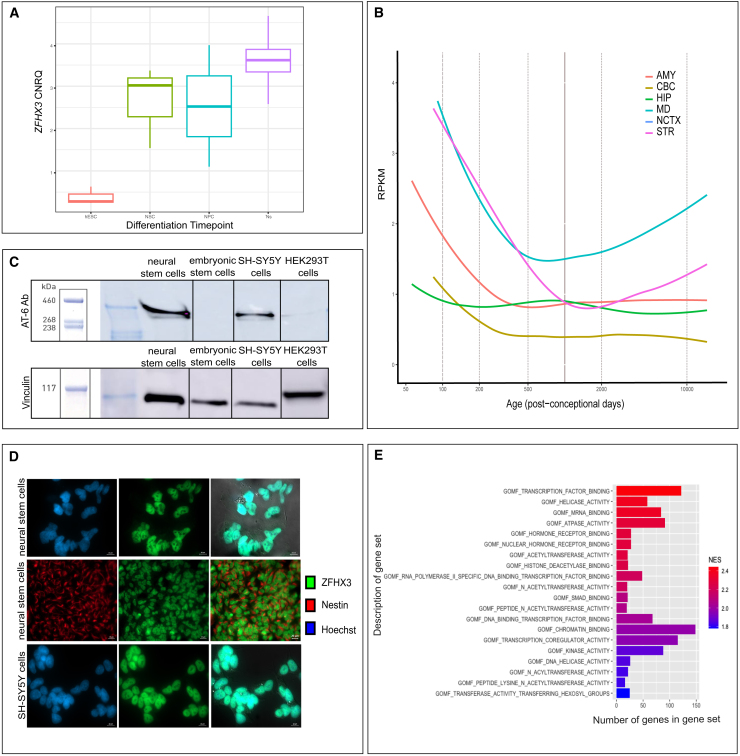
ZFHX3 is increasingly abundant during neural differentiation and localizes to the nucleus
(A) ZFHX3 levels during in vitro neural differentiation. RNA was sampled at the following stages: hESCs, NSCs, neural progenitor cells (NPC), and direct differentiated neurons (Ns). An increasing expression is observed upon differentiation. The median expression value is indicated by the middle horizontal line. The whiskers indicate the minimum and maximum values. CNRQ, calibrated normalized relative quantity.
(B) ZFHX3 levels in developing human brain. ZFHX3 expression in the developing human brain (normalized RPKM data) showing higher expression during early prenatal development followed by decreased expression upon brain maturation. Data obtained from BrainSpan: http://www.brainspan.org. Abbreviations: AMY, amygdala; CBC, cerebellum; HIP, hippocampus; MD: medial dorsal nucleus of the thalamus; NCTX: neocortex; STR: striatum. The dashed vertical lines indicate the age post-conception to the corresponding expression level.
(C) Endogenous ZFHX3 (404 kDa) accumulation in NSCs, hESCs, SH-SY5Y, and HEK293T as detected with western blot using the AT-6 antibody (top) and a vinculin antibody as a loading control (bottom).
(D) ZFHX3 is localized in the nucleus of SH-SY5Y cells and NSCs. Indirect immunofluorescence staining for ZFHX3 (SH-SY5Y and NSCs) and nestin (cytoplasmic neuronal marker, NSCs) was performed. DNA in the nucleus was counterstained with Hoechst dye. Images were merged to observe the contrast between the compartments. Scale bar: 10μm (upper row and bottom row), 20 μm (middle row); NSC, neural stem cells.
(E) Molecular functions of the top 20 positive correlated gene sets in the data by Cardoso et al..33 All have an FDR < 0.10 and a normalized enrichment score (NES) > 1.8. Gene sets are ranked based on NES-value.
Results
Individuals harboring a ZFHX3 deletion or protein-truncating variant present with an NDD phenotype
Molecular characterization
Eighteen probands harbor a microdeletion in chromosome band 16q22.2–16q22.3 ranging in size from 53.04 kb to 8.51 Mb (probands 1–18; Table S1) resulting in either partial or complete deletion of ZFHX3 (Figure 1A). The deletion in probands 4–14 and 16–18 only affects one protein-coding gene, i.e., ZFHX3. In probands 4–8, 17, and 18, the neighboring long-noncoding RNA (lncRNA) genes LINC01572, LINC01568, and HCCAT5 are also affected. The function of these lncRNAs is hitherto unknown. The deletion in probands 1, 2, 3, and 15 involves additional genes (see Table S9). Inheritance could be determined for fifteen deletions: fourteen occurred de novo (probands 1–7; 12–18), and one was inherited from the similarly affected father (proband 10; also present in similarly affected sister). Parental segregation data were incomplete for proband 8, 9, and 11; the deletion in proband 11 is absent in the mother but could not be investigated in the father.
Figure 1.
Overview of ZFHX3 aberrations in 42 probands
(A) (Micro)deletions identified in probands 1–18 (#) are represented by the red bars, all affecting ZFHX3 (gray box). RefSeq coding and non-coding genes are indicated respectively in blue and green. Genomic positions are according to hg38; ZFHX3 is located on the reverse strand.
(B) Nonsense and frameshift variants in ZFHX3 are identified in probands 19–42. Gene and protein structure of the canonical transcript (GenBank: NM_006885.4; GenBank: NP_008816.3) are shown in the top and middle part, respectively. The majority of PTVs are located in exon 9. Top: coding exons are indicated as dark gray boxes, while UTR regions are indicated with a light gray bar. Middle: LoF variants are present in the ZFHX3 cohort and located in different domains of ZFHX3. Bottom: Frameshift and nonsense variants present in gnomAD (v3.1.2) are enriched at the C-terminus, i.e., 25 of the 44 reported variants are located in exon 10.
(C) Clinical presentation of individuals with ZFHX3 aberration. Left: de-identified face mask of images from 17 individuals with either a microdeletion or truncating variant affecting ZFHX3 (left) and matched healthy individuals (right). Note a high and broad forehead, laterally sparse eyebrows, upslanted palpebral fissures, a low-hanging columella, flat philtrum, a relatively short midface, and a thin upper lip. Right: score distribution plots of the ZFHX3 cohort (green) and the control cohort (orange) and the receiver operating characteristic (ROC) curve to determine the capacity of Face2Gene to identify individuals in the ZFHX3 cohort against controls.
Twenty-four probands harbor a protein-truncating variant (PTV) in ZFHX3 of which 19 PTVs were confirmed de novo (probands 20–29; 31–36; 38–40), and one nonsense ZFHX3 variant segregates with the phenotype (proband 37 and four affected family members) (Figure 1B; Table S2). Inheritance could not be determined for the variants in probands 19, 30 (not maternal), 41, and 42. However, the same frameshift variant identified in proband 30 was also observed de novo in proband 31.
All variants are absent from the public variant database gnomAD (release v3.1.2) and are predicted to result in protein truncation or to initiate nonsense mediated decay (NMD). Furthermore, all eleven nonsense variants have a CADD (combined annotation dependent depletion) score greater than 36 (Table S2), indicating they are all predicted to be within the 0.1% most deleterious possible substitutions in the human genome. Eleven missense variants present in gnomAD affect the same codon as the truncating variant in probands 21, 28, 36, 37, 41, and 42 (Table S3), suggesting these missense variations do not result in LoF outcome. Missense variation seems tolerated based on the Z score of −0.2, while the pLI score of 1.0 and loss-of-function observed/expected upper bound fraction (LOEUF) score of 0.19 clearly indicate intolerance of LoF variation (gnomAD v4.0.0). Almost half of the truncating variants in the cohort are located in the large and penultimate exon 9 (11/24). Furthermore, all PTVs present in our cohort are at least 50 base pairs upstream of the last exon-exon boundary (Figure 1B). No truncating variants are observed at the C-terminus in the cohort, while the predicted LoF variants present in gnomAD are enriched at the C-terminus (i.e., exon 10), which could imply NMD escape. Nevertheless, it remains possible that individuals with LoF variants present in gnomAD exhibit a milder phenotype, considering the wide neurodevelopmental clinical spectrum observed in our cohort (see phenotypic characterization).
Phenotypic characterization
Probands 1, 2, 3, and 15 harboring multigenic deletions were excluded for the characterization of the ZFHX3-associated phenotype to avoid a possible bias due to haploinsufficiency of other genes (Tables 1, S1, S2, and S9). In addition, besides neurodevelopmental delay, no detailed clinical information could be obtained for probands 8, 9, 14, 19, and 25. Therefore, the phenotypic characterization is based on 33 probands and six (similarly) affected family members. Early neurological development is characterized by global neurodevelopmental delay (24/39) and delayed language (17/39) and/or motor (26/39) development, the latter often preceded by hypotonia (22/39). Intellectual capacities vary from normal with or without some learning problems (n = 14) to mild and moderate intellectual impairment (n = 19). Behavioral problems (16/39) are variable. ASD was noted in nine individuals but is the sole neurological manifestation in proband 27, 38, and 39. Seizures were reported in two individuals (probands 28 and 35). Frequently associated features are postnatal growth retardation (20/39) and feeding difficulties (24/39) due to oropharyngeal dysphagia, eosinophilic esophagitis (n = 2), or food allergies. Three individuals needed a gastrostomy tube. Hands and feet may be small and show brachydactyly (mostly of the distal phalanges, n = 15), clinodactyly (mostly of the 4th or 5th rays, n = 6), tapering fingers (n = 3), and/or abnormal dermal creases (n = 6). Other skeletal features include scoliosis and kyphosis (n = 3), pectus deformity (n = 3), hip dysplasia (n = 3), and osteoporosis/fractures. Five individuals reported musculoskeletal pains, joint laxity, and/or fatigue. Cardiovascular features include septal defects in four individuals, and two individuals showed arterial stenoses. Sporadic urogenital defects include pyeloureteral junction stenosis (proband 4), vesicoureteral reflux (proband 27), renal cysts (proband 26), and cryptorchidism (probands 15 and 38). Hearing is reportedly normal, but several probands received transtympanic drains (n = 7/39).
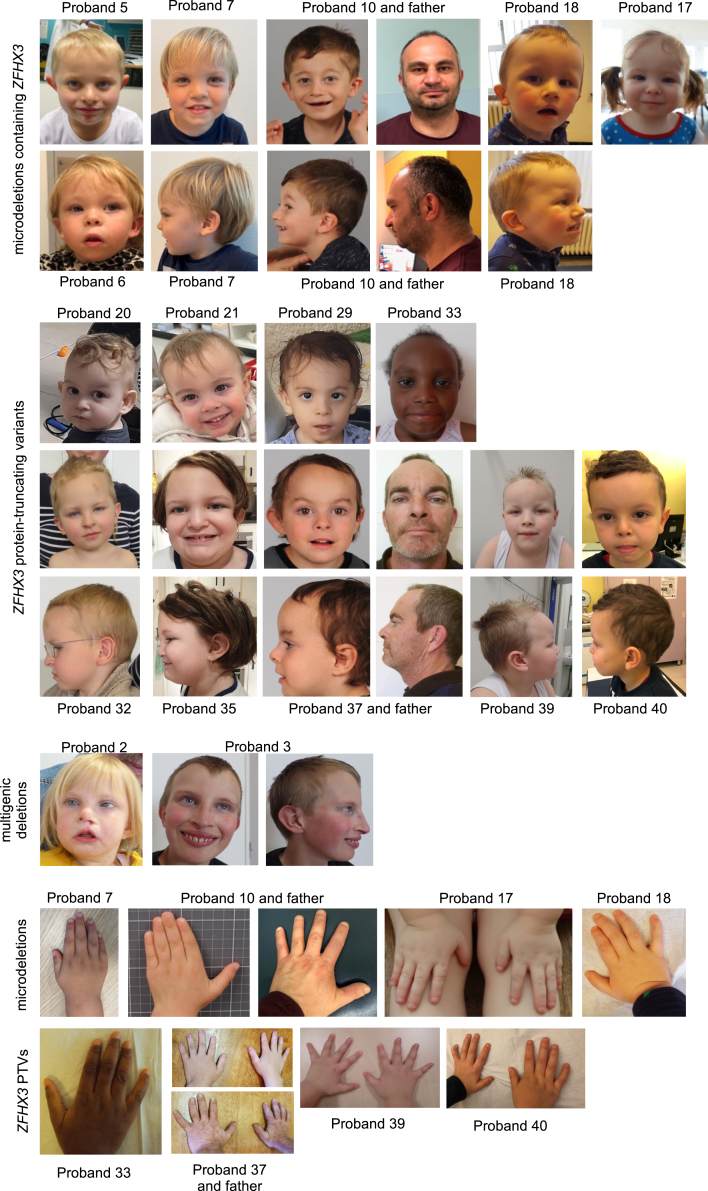
Facial gestalt comprises a receding anterotemperal hairline (23/39), high and broad forehead (29/39), bulbous nasal tip (24/39), low hanging columella (18/39), flat philtrum (17/39), thin upper lip (18/39), and thin vermillion border (18/39). Less-common facial features include protruding ears (13/39), laterally sparse eyebrows (13/39), upslanted palpebral fissures (13/39), epicanthal folds (16/39), a short philtrum (9/39), and downturned corners of the mouth (16/39). Cleft lip and/or palate is present in three individuals (i.e., two individuals with a PTV and one individual with a multigenic deletion). Ocular abnormalities (ptosis, strabismus, nystagmus, tear duct aplasia, coloboma) are nonspecific and uncommon. Figure 1C shows a facial composite image of 17 individuals with a ZFHX3 aberration compared to a healthy cohort matched for age and ethnicity. The ROC curves in Figure 1C show that Face2Gene was able to accurately categorize the ZFHX3 cohort vs. controls 87.7% of the time (area under the curve [AUC] = 0.877) with a p value of 0.018. The score distribution shows that there is a subtle but clear difference between the facial features of our ZFHX3 cohort in comparison with those of the control cohort.
The individuals with multiple deleted genes share most of the clinical characteristics but show a more severe developmental phenotype (Table 1). Sex (22 males and 20 females) is equally distributed and does not influence the clinical presentation. Age ranges from 10 months to 63 years. Overall, no clear differences could be observed between individuals with a microdeletion only affecting ZFHX3 and a truncating variant in ZFHX3 (Table 1, Fisher’s exact test). Detailed clinical characteristics per individual can be found in Table S1 (Deletions) and Table S2 (PTVs). Pictures of front and side profiles as well as hands and digits of several probands (and affected family members) can be found in Figure 2.
Figure 2.
Front and side profile pictures, as well as hand and digits pictures of individuals with a deletion or PTV affecting ZFHX3
Rows 1–2: frontal and profile photographs of probands with a microdeletion containing ZFHX3. Rows 3–5: frontal and profile photographs of probands with a ZFHX3 protein-truncating variant (PTV). Row 6: frontal and profile photographs of probands with a multigenic deletion containing ZFHX3. Rows 7–8: photographs showing the hands of probands with a microdeletion (row 7) or PTV (row 8) affecting ZFHX3.
ZFHX3 expression increases during human neurodevelopment
Publicly available RNA-sequencing data from 54 non-disease tissue sites, derived from mostly older donors (GTEx, release v8), indicates abundant ZFHX3 expression throughout the human adult body with highest expression in arteries (Figure S1A). Expression in various adult brain regions is relatively low. Evaluation of ZFHX3 expression profiles across seven organs and up to 23 developmental stages33 shows highest expression of ZFHX3 during the prenatal period, especially in the human brain (Figure S1B). This is also confirmed by expression data from Brainspan, showing highest ZFHX3 expression in the brain prenatally, especially in the striatum, dorsal thalamus, and mediodorsal nucleus of the thalamus (Figure 3B). This prenatally enriched expression pattern indicates a role during early (brain) development.
We observed increased expression of ZFHX3 during in vitro neural differentiation starting from hESCs, with the highest expression in early developed neurons (Figure 3A). Kanton et al.34 confirmed our observation with single-cell RNA-sequencing data during cerebral organoid differentiation starting from pluripotency (Figure S1C). Furthermore, single-cell sequencing data generated from single neural rosette (SNR)-derived organoids by the Shcheglovitov lab revealed that ZFHX3 is particularly expressed in 1-month-old organoids in inhibitory neurons (Figure S2).35
To screen for potential functional and regulatory interactions of ZFHX3, we performed a correlation analysis between expression of ZFHX3 and all other genes across the seven organs and 23 developmental stages assessed by Cardoso-Moreira.33 Table S4 contains the rho and p values with significant correlation (p value < 0.05). Top correlated genes appear to play a role in transcription factor binding, acetyltransferase activity, kinase activity, chromatin binding, and mRNA binding (Figure 3E).
ZFHX3 has two isoforms and is localized in the nucleus of different neuronal cell lines
There are two main ZFHX3 isoforms, previously called ATBF1-A (encoded by GenBank: NM_006885.4; ENST00000268489.10) and ATBF1-B (encoded by GenBank: NM_001164766.2; ENST00000397992.5), generated by alternative splicing and promotor usage. ATBF1-A is a 404-kDa protein and contains 23 zinc finger and four homeodomains. ATBF1-B (306-kDa) contains the same homeodomains, but lacks five zinc finger domains due to the absence of 920 amino acids at the N-terminus.19 In 18/24 of the individuals with a ZFHX3 PTV, the aberration affects both the short and long isoform. The variants identified in probands 19–24 only affect the longest isoform.
According to GTEx data, the 306-kDa isoform shows higher expression at mRNA level in all adult tissues (Figure S3). Nevertheless, the public data we leveraged indicates that ZFHX3 is particularly abundant in the prenatal brain and that its levels in the adult brain are much lower33 (Figures 3B and S1B). We therefore evaluated the abundance of ZFHX3 in hESCs, two immature neuronal-like cell lines (SH-SY5Y a neuroblastoma cell line, often used as neuronal in vitro model and human NSCs), and a non-neuronal cell line (human embryonic kidney cells [HEK293T]) at protein level, showing that the long 404-kDa isoform is mainly present in immature neuronal-like cell lines (i.e., SH-SY5Y and NSCs) (Figure 3C). We then investigated the location of ZFHX3 within these two cell lines to correlate subcellular localization with its potential functions. Immunocytochemistry for ZFHX3 in these two cell lines shows its presence in the nucleus correlating with its function as a transcription factor (Figures 3D and S4).
ZFHX3 interacts with members of the BAF and CP complexes
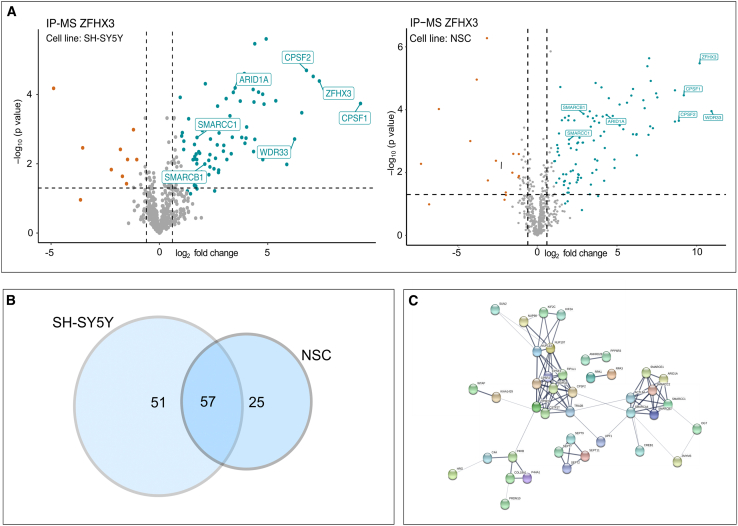
We performed a discovery screen for potential interaction partners of ZFHX3 using immunoprecipitation (IP) of endogenous ZFHX3 in SH-SY5Y and NSCs followed by mass spectrometry (MS). This showed, respectively, enrichment of 83 and 109 proteins compared to an IgG isotype control sample (t test, FDR = 0.05) (Figure 4A; Tables S5 and S6). An overlap of 57 ZFHX3 interaction partners was observed in both cell lines (Figure 4B). Among these 57 interactors, SPECC1L is the only known interaction partner of ZFHX3 according to a reference map of human binary protein-protein interactions.36 This protein is involved in actin-cytoskeleton reorganization and is crucial for proper facial morphogenesis.37 Mapping the 56 remaining identified ZFHX3 binding partners against the STRING database38 reveals several ZFHX3-containing protein complexes, such as the mammalian SWI/SNF (mSWI/SNF) complex, also known as the BRG1/BRM associated factor (BAF) complex,39 the cleavage and polyadenylation (CP) complex, the nuclear pore complex (NPC), and the septin complex (Figure 4C). In agreement with the function of these complexes, we show enrichment of the following processes by the GO enrichment analysis of the ZFHX3 interactome using Metascape40: nucleosome assembly, mRNA transport from nucleus to cytoplasm, and cytoskeleton-dependent cytokinesis (Figure S5). This supports the finding of chromatin remodeling and mRNA binding as enriched biological processes in our transcriptome data mining (see Figure 3E).
Figure 4.
ZFHX3 has a robust protein-protein interaction network of 57 interaction partners
(A) Volcano plot showing the enriched ZFHX3 interactor proteins in SH-SY5Y (left: n = 83) and NSCs (right: n = 109). For each detected protein, the log2 fold change (FC) between the ZFHX3 IP-enriched samples and the IgG controls is shown on the x axis, while the -log10 of the p value is shown on the y axis. Vertical dashed lines indicated a fold change of abs(0.6). The horizontal dashed line indicates the -log10 value of a p value equal to 0.05. Proteins with an FDR < 0.05, log2 FC > 0.6/<-0.6 and S0 = 1 are, respectively, highlighted in blue and red. Important members of the BAF and CP complexes are also highlighted, as well as ZFHX3 itself.
(B) Overlap of ZFHX3 interaction factors between SH-SY5Y and NSCs IP results. A total of 57 proteins interacts in both cell lines with ZFHX3. A total of respectively 51 or 25 interaction partners for ZFHX3 were found in SH-SY5Y or NSCs.
(C) ZFHX3 protein-protein interaction network extracted from the STRING 11.0 database. Shown are interactors that are connected within a network. Line thickness indicates the strength of data support. The BAF complex, CPSF (cleavage and polyadenylation specificity factor) and CSTF (cleavage stimulation factor), nuclear pore complex (NPC), and septin complex are ZFHX3-containing protein complexes. CPSF and CSTF are two multi-subunit protein complexes responsible for cleavage and polyadenylation.41
To validate the ZFHX3 protein-protein interactions with the BAF and CP complex, we selected SMARCB1 (subunit of the BAF complex) and CPSF2 (subunit of CP complex), respectively, to perform co-immunoprecipitation, since both were identified in the IP-MS screen as ZFHX3 interactors in SH-SY5Y and NSCs (Tables S5 and S6). In HEK293T, overexpression of a construct encoding FLAG-tagged ZFHX3 (±404 kDa) with overexpression of another construct, either containing 3xHA-tagged SMARCB1 (±50 kDa), or 3xHA-tagged CPSF2 (±91 kDa), followed by immunoprecipitation with an HA-tagged antibody and western blot for ZFHX3 appears to corroborate the presence of ZFHX3 among the SMARCB1-3xHA and CPSF2-3xHA binding protein extracts (Figure S6).
LoF variants affecting ZFHX3 are associated with a methylation profile
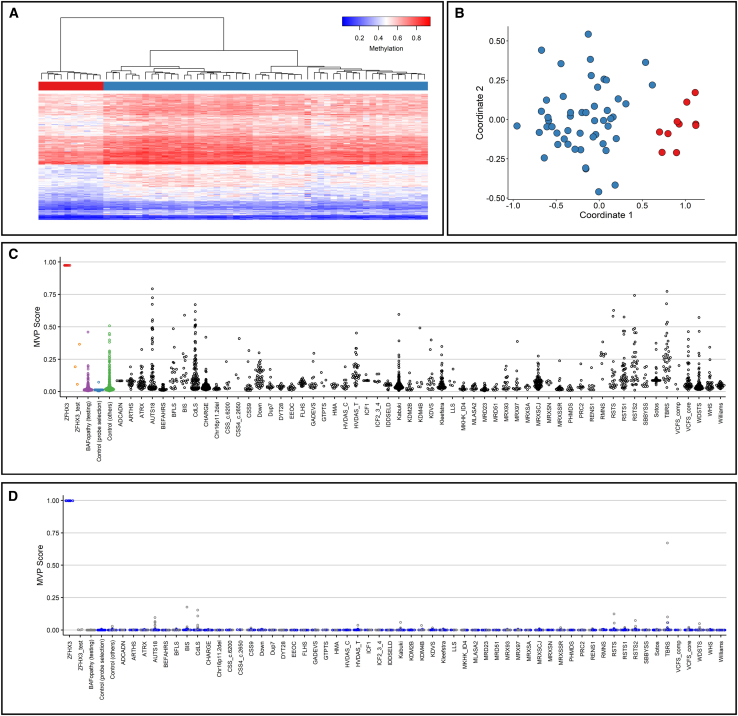
Mutations in genes encoding BAF subunits are associated with Coffin-Siris syndrome (CSS; MIM: 135900, 614607, 614608, 614609, 615866, 616938, 617808, 618027, 618362, 618506, 618779, and 619325), Nicolaides-Baraitser syndrome (NCBRS; MIM: 601358), Kleefstra syndrome (MIM: 610253 and 617768) and ASD, commonly referred to as BAFopathies.42 Aref-Eshgi et al. described specific DNA methylation epi-signatures in blood from individuals with BAFopathies.42 Since ZFHX3 appears to interact with the BAF complex, we investigated whether ZFHX3 haploinsufficiency associates with a specific methylation profile. To this end, 10 individuals from our cohort (six individuals with ZFHX3 PTVs and four with a [partial] deletion of ZFHX3, listed in Table S7) were used for probe selection and model construction. We selected 50 control samples matched to the ZFHX3 aberration samples by age, sex, and array type, and a set of 208 probes were identified to be differentially methylated between the cohort and control groups.
The robustness of the selected probes was verified using hierarchical clustering and multidimensional scaling (MDS) models (Figures 5A and 5B). The set of 208 probes were then used in order to construct support vector machine (SVM) models in order to classify the DNA samples of ZFHX3 haploinsufficient individuals with more accuracy. Using these models, two methylation variant pathogenicity (MVP) plots were generated where the model was constructed by training 10 samples of affected individuals against matched controls and the second model by training the 10 samples of affected individuals against matched control samples and 75% of other control samples and samples of individuals with other disorders from the EpiSign Knowledge Database (EKD; https://episign.lhsc.on.ca/index.html) to increase the specificity of the model to ZFHX3 haploinsufficiency. In both models, the remaining samples from the EKD, BAFopathy samples, and three samples with ZFHX3 missense variants (classified as variant of unknown significance, Table S8) were used for testing. The samples with missense variants (ZFHX3_test in Figures 5C and 5D) and the BAFopathy samples (BAFopathy [testing] in Figures 5C and 5D) received low MVP scores, demonstrating that they did not match the methylation profile identified for ZFHX3 haploinsufficiency.
Figure 5.
Deletion of ZFHX3 and ZFHX3 PTVs are associated with a specific methylation profile
(A) Hierarchical clustering using the selected CpG sites, where rows represent selected probes and columns indicate samples. ZFHX3 samples are illustrated with red, and control individuals with blue on the heatmap pane. The heatmap color scale demonstrates methylation levels ranging from blue (no methylation or 0) to red (full methylation or 1). Clear separation between ZFHX3 and control samples is observed.
(B) MDS demonstrating different methylation pattern between the cohort and control groups using the selected CpG sites. Red and blue circles represent cohort and control samples, respectively.
(C) MVP scores generated by the SVM classifier trained using only the 10 ZFHX3 samples and the 56 matched control samples, where the majority of the other control samples and individuals with other disorders received low scores, demonstrating the high specificity of the methylation pattern to ZFHX3 haploinsufficiency.
(D) MVP scores generated by the SVM constructed by training the 10 ZFHX3 samples against matched control samples and 75% of other control samples and samples from other disorders (blue). The remaining 25% of the database samples used for testing (gray) received very low MVP scores, illustrative of the significant improvement in the specificity of the model. The 3 samples with missense variants in ZFHX3 (ZFHX3_test) and the BAFopathy samples (BAFopathy [testing]) were also supplied into the model as testing samples and received low MVP scores, indicating that their methylation pattern is different from that of ZFHX3 haploinsufficiency. Abbreviations: ADCADN, cerebellar ataxia, deafness, and narcolepsy, autosomal dominant; ARTHS, Arboleda-Tham syndrome; ATRX, alpha-thalassemia/intellectual development syndrome, X-linked; AUTS18, autism, susceptibility to 18; BEFAHRS, Beck-Fahrner syndrome; BFLS, Börjeson-Forssman-Lehmann syndrome; BIS, blepharophimosis intellectual disability SMARCA2 syndrome; CdLS, Cornelia de Lange syndrome 1–4; CHARGE, CHARGE syndrome; 16p11.2del, 16p11.2 deletion syndrome; CSS_c.6200, Coffin-Siris syndrome (c.6232G>A [GenBank: NM_006015.4 (ARID1A)]; p.Glu2078Lys) (c.6254T>G [GenBank: NM_006015.4 (ARID1A)]; p.Leu2085Arg) (c.6133T>C [GenBank: NM_017519.2 (ARID1B)]; p.Cys2045Arg); CSS4_c.2650, Coffin-Siris syndrome (c.2656A>G [GenBank: NM_001128849.1 (SMARCA4)]; p.Met886Val); CSS9, Coffin-Siris syndrome-9; Down, Down syndrome; Dup7, Williams-Beuren duplication syndrome (7q11.23 duplication syndrome); DYT28, Dystonia-28, childhood onset; EEOC, epileptic encephalopathy, childhood onset; FLHS, Floating-Harbour syndrome; GADEVS, Gabriele de Vries syndrome; GTPTS, Genitopatellar syndrome; HMA, HVDAS_C, Helsmoortel-Van der Aa syndrome (ADNP syndrome [central]); HVDAS_T, Helsmoortel-Van der Aa syndrome (ADNP syndrome [Terminal]); ICF1 Immunodeficiency, centromeric instability, facial anomalies syndrome 1, ICF2_3_4, immunodeficiency, centromeric instability, facial anomalies syndrome 2, 3, and 4; IDDSELD, intellectual developmental disorder with seizures and language delay; Kabuki, Kabuki syndromes 1 and 2; KDM2B, KDM2B-related syndrome; KDM4B, KDM4B-related syndrome; KDVS, Koolen de Vries syndrome; Kleefstra, Kleefstra syndrome 1; LLS, Luscan-Lumish syndrome; MKHK_ID4, Menke-Hennekam syndrome-1, 2; MLASA2, myopathy, lactic acidosis, and sideroblastic anemia-2; MRD23, intellectual developmental disorder, autosomal dominant 23; MRD51, intellectual developmental disorder, autosomal dominant 51; MRX93, intellectual developmental disorder, X-linked, XLID93; MRX97, intellectual developmental disorder, X-linked 97, XLID97; MRXSA, Armfield syndrome; MRXSCJ, syndromic X-linked intellectual disability, Claes-Jensen type; MRXSN, syndromic X-linked intellectual disability, Nascimento type; MRXSSR, syndromic X-linked intellectual disability, Snyder-Robinson type; PHMDS, Phelan-McDermid syndrome; PRC2, RENS1, Renpenning syndrome; RMNS, Rahman syndrome; RSTS, Rubinstein-Taybi syndrome-1, 2; RSTS1, Rubinstein-Taybi syndrome-1; RSTS2, Rubinstein-Taybi syndrome-2; SBBYSS, Say-Barber-Biesecker-Young-Simpson syndrome; Sotos, Sotos syndrome; TBRS, Tatton-Brown-Rahman syndrome; VCFS_comp, velocardiofacial syndrome; VCFS_core, velocardiofacial syndrome; WDSTS, Wiedemann-Steiner syndrome; WHS, Wolf-Hirschhorn syndrome; and Williams, Williams-Beuren deletion syndrome (7q11.23 deletion syndrome).
ZFHX3 predominantly binds promoters of genes involved in nervous system development
ChIP-seq for ZFHX3 was performed in NSCs with endogenous ZFHX3 levels and NSCs with ZFHX3 overexpression (see material and methods) for which 24,674 and 57,886 peaks were called, respectively. Of the 24,674 peaks identified with ChIP-seq for ZFHX3 in NSCs, 22,094 (or 89.5%) were also called in NSCs with overexpression of ZFHX3 (Table S11). Functional enrichment analysis of these 22,094 overlapping peaks revealed that ZFHX3 predominantly binds to promoter regions (57.18%) (Figures 6A and 6B). Furthermore, GO analysis revealed that ZFHX3 binds to genes involved in axonogenesis, axon development, cell growth, and development regulation of the nervous system and neurogenesis (Figure 6B). KEGG pathway enrichment analysis showed that the proteins encoded by these genes are involved in axon guidance, the Wnt, Hippo, and mTOR signaling pathways, all pathways strongly involved the nervous system development43,44,45 (Figure 6C). These results again provide clear indications of a key role for ZFHX3 in neuron and axon development. De novo as well as known motif analysis was performed for the overlapping ChIP-seq peaks. Both analyses show that homeodomain and zinc finger motifs (i.e., ZFHX3 contains multiple DNA-binding zinc fingers and homeodomains) were indeed enriched. The top 20 motifs identified via de novo and known enrichment motif analysis are listed in the Table S11. In addition to zinc finger and homeobox motifs, the known motif analysis revealed an enrichment of motifs associated with the SOX (SRY-related HMG box) family. The BORIS (CTCFL) and CTCF zinc finger motifs were identified with the highest percentage in the intersect peaks in both analyses.
Discussion
We identified LoF variation in ZFHX3 as the cause for an NDD, characterized by developmental delay and/or intellectual impairment and/or behavioral problems, feeding difficulties, postnatal growth retardation, and recognizable facial features. Individuals with larger microdeletions containing ZFHX3 and other coding genes present with a similar phenotype, albeit with more pronounced developmental delay, pointing toward a primary role for ZFHX3 in these microdeletions.
Proband 1 harbors a multigenic deletion also including AP1G1 that encodes the AP1γ1 protein, important for the formation of clathrin-coated vesicles. De novo PTVs and missense variants, as well as bi-allelic missense variants affecting AP1G1, have recently been associated with the Usmani-Riazuddin syndrome (MIM: 619467).46 As the consequences on AP1γ1 protein levels, protein aggregation, and clathrin formation are divergent for the different variants, it remains unclear if the disease mechanisms result from pure AP1G1 LoF. Moreover, since the phenotype associated with de novo AP1G1 mutations seems relatively similar to the phenotype observed for ZFHX3 LoF, a potential effect of the AP1G1 deletion cannot be excluded in proband 1.
All identified single-nucleotide and indel variants are truncating and likely activate the NMD pathway, as they are situated more than 50–55 nucleotides upstream of the final exon-exon junction. Probands with a microdeletion affecting ZFHX3 only are clinically indistinguishable from probands with ZFHX3 truncating variants. Moreover, preliminary data provided promising evidence for the existence of a shared methylation profile for individuals harboring either a microdeletion including ZFHX3 or a PTV in ZFHX3, suggesting haploinsufficiency of ZFHX3 as the underlying mechanism in both. All PTVs in our cohort are absent in gnomAD v2.1.1 and v3.1.2. We observed that the variants in probands 24, 29, and 41 are present in gnomAD v4.0.0, albeit at an extremely low frequency of one or two alleles. Since gnomAD has removed cohorts for individuals evaluated for pediatric disease, these individuals in gnomAD v4.0.0 might exhibit a mild phenotype that might merge with normal variation. Nevertheless, gnomAD v4.0.0 gene constraint scores pLI = 1 and LOEUF = 0.19 confirm that ZFHX3 is under selection against protein-truncating variants, and similar to gnomAD v3.1.2, the LoF variants present in gnomAD v4.0.0 appear to be enriched at the region encoding the C-terminus. Samples from three individuals harboring missense variants did not show a similar methylation profile, suggesting that the molecular consequences of the different protein variants are not similar. Moreover, in one of the individuals with a missense variant, a structural variant disrupting SON was later identified via optical mapping, matching the phenotype (no further information on variant available). In previous studies, (compound) heterozygous missense variants of ZFHX3 were reported in probands with epilepsy21,47 and behavioral issues, but with normal IQ.48,49 Consequently, we cannot rule out that missense variation in ZFHX3 could associate with epilepsy, rather than with a primary cognitive phenotype, despite the observation that missense variation in ZFHX3 is not constrained (Z score: −0.2 [gnomAD release v4.0.0]). Moreover, recently, He et al. identified compound heterozygous variants in ZFHX3 in eight unrelated individuals with childhood epilepsy. The eight pairs of compound heterozygous variants consisted of eleven missense variants, one in-frame deletion and a frameshift variant in the last exon (suggestive for NMD escape).50 In 2020, Zhang et al. reported two de novo ZFHX3 missense variants, respectively, in an individual with non-syndromic esophageal atresia (c.1601C>G [GenBank: NM_006885.3]; p.Pro534Arg) with or without tracheoesophageal fistula (EA/TEF) and in an individual with EA/TEF as part of a VATER/VACTERL association (c.6377C>T [GenBank: NM_006885.3]; p.Ala2126Val).51 They suggest ZFHX3 as a putative candidate gene for EA/TEF since it has a high expression in mouse embryonic foregut tissue and in the pharyngeal arches in zebrafish larvae. EA/TEF was however not observed in our cohort. Recently, Jameson et al. showed that the intronic single-nucleotide polymorphism (SNP) rs12931021 modulates the expression of ZFHX3, and they provided evidence implicating ZFHX3 variants as the causative mutations at the 16q22 locus for atrial fibrillation.13 In our cohort, none of the individuals was reported to have atrial fibrillation. In addition, very recently, three groups reported that a GGC expansion in the last exon of ZFHX3 appears to be associated with spinocerebellar ataxia type 4 (SCA4; MIM: 600223).52,53,54 Further work on the different types of variants in ZFHX3, including phenotypic delineation, is ongoing to enable development of a sensitive and specific diagnostic biomarker, that would be beneficial for the interpretation of ZFHX3 aberrations.
Functional enrichment analysis of ZFHX3 ChIP-seq data from human NSCs show that ZFHX3 predominantly binds to promoter regions. The motif enrichment analysis conducted on the ZFHX3 ChIP-seq peaks unveiled a significant enrichment of zinc finger and homeodomain motifs, which can be attributed to the presence of the 23 zinc fingers and four homeodomains within ZFHX3. Furthermore, we also observed enrichment of motifs associated with the SOX-family of transcription factors, several of which are known to play pivotal roles in neuronal development.55These results warrant further exploration of an interplay between ZFHX3 and SOX family members in neurodevelopmental processes.
The functional enrichment analysis in the ChIP-seq dataset indicates ZFHX3 binds genes involved in neural development and migration, including genes encoding key members of the Hippo pathway.45 Other important pathways and processes influenced by ZFHX3 functioning are Wnt/β-catenin and mTOR signaling and cell cycling. Wnt/β-catenin signaling is crucial during neurodevelopment and function of the CNS, including neurogenesis, cell pluripotency, and fate decisions,43,56,57,58,59 while dysregulated mTOR signaling has been associated with impaired neurogenesis and neurodevelopmental and neuropsychiatric disorders.44,60,61 In addition, ZFHX3 has already been shown to modulate cell cycling, triggering cell-cycle arrest in neuronal progenitor cells to induce neuronal differentiation.62
In agreement with the ChIP-seq data, the spatiotemporal expression of ZFHX3 in the developing mouse,63 rat,62 fruit fly,64 and human brain, as well as in differentiating neuronal progenitor cells, supports a role for neuronal differentiation.14,65 In the single-cell data from the recently published Fly Cell Atlas,66 zfh2—the putative Drosophila ortholog of ZFHX3—was mainly localized in neurons, sensory neurons, and glial cells in fly adult brains. The temporal patterning is similar to that of Zfhx3 within mice striatum from previously published single-cell sequencing data.17
In mice, a heterozygous truncating deletion of exon 7 and 8 in Zfhx3 is associated with growth retardation and preweaning mortality.67 Moreover, defects in other members of the ZFHX family have been associated with neuropsychiatric or neurodevelopmental phenotypes in humans and mice. In humans, the paralog ZFHX4 is likely the major contributing gene in the 8q21.11 microdeletion syndrome (MIM: 614230) associated with intellectual disability.2,68 Zfhx2-deficient mice show behavioral abnormalities (hyperactivity, depression, and anxiety-related behavior) but normal cognition.42
The complex structure of ZFHX3, containing 4 homeodomains, suggests dynamic functions in diverse biological processes.5 In line with this, we observed several major interaction partners. Its interaction with several subunits of the BAF complex suggests a role in chromatin remodeling. The BAF complex is a chromatin remodeling complex that consists of up to 15 subunits. It mediates DNA accessibility by modulating nucleosomes in an ATP-dependent manner, hence regulating gene expression. Not surprisingly, pathogenic variants in several subunits of the BAF complex were found in NDDs within the spectrum of CSS and NCBRS69,70 commonly coined BAFopathies. Messenger RNA transcripts undergo several processing events including addition of a 5′-cap, splicing, and 3′-end cleavage and polyadenylation before transportation to the cytoplasm.71 In addition to their role in transcription regulation, chromatin remodelers may influence pre-mRNA splicing and 3′-end processing.72 Previous studies showed a physical and functional interaction between the BAF complex and cleavage and polyadenylation factors.73 Furthermore, many 3′ end processing factors (CPSF, CSTF, and PABP [poly(A)-binding protein]), nucleoporins, and a subset of the SWI/SNF chromatin remodeling complex are identified in supraspliceosomes (or polyspliceosomes) in HeLa and chicken cells.74 Since we identify ZFHX3 as an interactor with these 3′ end processing factors, this raises the intriguing hypothesis that ZFHX3 facilitates interconnection of these mRNA processing activities.
Furthermore, IP-MS revealed a possible interaction between ZFHX3 and the septin complex. The cytosolic septin complex serves an important role in various cellular processes, pertinent for several neuronal functions, such as axon dynamics and growth and dendrite formation.75 This is in line with the cytoskeletal function of the previously identified interaction partner SPECC1L.36 How and if ZFHX3 plays a role in these processes, needs further investigation.
In conclusion, we report LoF variants in ZFHX3 as a cause for syndromic ID, associated with a specific DNA methylation profile. ZFHX3 seems to play a significant role in neuronal differentiation and brain development. Preliminary data suggests an association with chromatin remodeling and mRNA processing. Nevertheless, additional research is necessary to fully understand and elucidate its precise functions in these processes.
Data and code availability
The analyzed IP-MS data and the overlapping peaks between endogenous and ectopic ZFHX3 ChIP-seq, as well as the results of the de novo and known motif analyses are available in the supplemental information.
Some of the DNA methylation datasets used in this study are publicly available and may be obtained from gene expression omnibus (GEO) using the following accession numbers. GEO: GSE116992, GSE66552, GSE74432, GSE97362, GSE116300, GSE95040, GSE104451, GSE125367, GSE55491, GSE108423, GSE116300, GSE89353, GSE52588, GSE42861, GSE85210, GSE87571, GSE87648, GSE99863, and GSE35069. These include DNA methylation data from individuals with Kabuki syndrome (MIM: 147920 and 300867) (GSE97362 and GSE116300), Sotos syndrome (MIM: 117550) (GSE74432), CHARGE syndrome (MIM: 214800) (GSE97362), immunodeficiency-centromeric instability-facial anomalies (ICF) syndrome (MIM: 242860) (GSE95040), Williams-Beuren syndrome (MIM: 194050), 7q11.23 duplication syndrome (MIM: 609757) (GSE66552), BAFopathies(GSE116992, GSE125367), Down syndrome (MIM: 190685) (GSE52588), Silver-Russell syndrome (MIM: 180860, 618905, 616489, 618907, and 618908) (GSE104451 and GSE55491), X-linked syndromic intellectual developmental disorder, Claes-Jensen type (MIM: 300534) (GSE108423), a large cohort of (unresolved) subjects with developmental delays and congenital abnormalities (GSE89353), and several large cohorts of DNA methylation data from the general population (GSE42861, GSE85210, GSE87571, GSE87648, GSE99863, and GSE35069). Remaining methylation data is not publicly available due to institutional and ethics restrictions.
Consortia
Members of the ZFHX3 Consortium: Pankaj Agrawal, Daryl Armstrong Scott, Elizabeth Barkoudah, Melissa Bellini, Claire Beneteau, Kathrine Bjørgo, Alice Brooks, Natasha Brown, Alison Castle, Diana Castro, Odelia Chorin, Mark Cleghorn, Emma Clement, David Coman, Carrie Costin, Koen Devriendt, Dexin Dong, Annika Dries, Tina Duelund Hjortshøj, David Dyment, Christine Eng, Casie Genetti, Siera Grano, Peter Henneman, Delphine Heron, Katrin Hoffmann, Jason Hom, Haowei Du, Maria Iascone, Bertrand Isidor, Irma E. Järvelä, Julie Jones, Boris Keren, Mary Kay Koenig, Jürgen Kohlhase, Seema Lalani, Cedric Le Caignec, Andi Lewis, Pengfei Liu, Alysia Lovgren, James R. Lupski, Mike Lyons, Philippe Lysy, Melanie Manning, Carlo Marcelis, Scott Douglas McLean, Sandra Mercie, Mareike Mertens, Arnaud Molin, Mathilde Nizon, Kimberly Margaret Nugent, Susanna Öhman, Melanie O'Leary, Rebecca Okashah Littlejohn, Florence Petit, Rolph Pfundt, Lorraine Pottocki, Annick Raas-Rotschild, Kara Ranguin, Nicole Revencu, Jill Rosenfeld, Lindsay Rhodes, Fernando Santos Simmaro, Karen Sals, Jolanda Schieving, Isabelle Schrauwen, Janneke H.M. Schuurs-Hoeijmakers, Eleanor G. Seaby, Ruth Sheffer, Lot Snijders Blok, Kristina P. Sørensen, Siddharth Srivastava, Zornitza Stark, Radka Stoeva, Chloe Stutterd, Natalie B. Tan, Pernille Mathiesen Torring, Olivier Vanakker, Liselot van der Laan, Athina Ververi, Pablo Villavicencio-Lorini, Marie Vincent, Dorothea Wand, Marja Wessels, Sue White, Monica H Wojcik, Nan Wu, and Sen Zhao.
A list of consortia member affiliations can be found in Document S1.
Acknowledgments
First of all, we would like to thank the reported families for their cooperation in this study. We are grateful for the group of Peter Ponsaerts, University of Antwerp, by providing us iPSC-derived NSCs in order to start the culture on our own at our lab. We would also like to thank the research group of Prof. Patrick Nolan (MRC Harwell, Harwell Science and Innovation Campus, Oxfordshire, UK) and Dr. Rebecca Dumbell (Nottingham Trent University, Nottingham, UK) for providing us their ZFHX3 custom antibody.
Financial support has been provided by grants 1520518N, G044615N, and G055422N from the Research Foundation – Flanders (FWO) and BOF/STA/201909/009 from the Special Research Fund (BOF) from Ghent University. B.S. received funding from the government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-188). M.d.R.P.B is supported by a doctoral grant of the Marguerite-Marie Delacroix foundation. E.Z.J. was supported by a doctoral grant of the Research Foundation Flanders. B.C. is a senior clinical investigator of the Research Foundation Flanders.
M.H.W. is supported by NIH/NICHD K23 HD102589, NIH/NHGRI R21 HG012397 and an Early Career Award from the Thrasher Research Fund. Sequencing and analysis for individual 15 were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung, and Blood Institute grant UM1 HG008900, and in part by National Human Genome Research Institute grants U01 HG0011755 and R01 HG009141.
Declaration of interests
L.R. is an employee of GeneDx, LLC. X.W. is a co-founder and employee of AiLife Diagnostics.
Published: February 26, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.01.013.
Contributor Information
Sarah Vergult, Email: sarah.vergult@ugent.be.
Bert Callewaert, Email: bert.callewaert@ugent.be.
ZFHX3 consortium:
Pankaj Agrawal, Daryl Armstrong Scott, Elizabeth Barkoudah, Melissa Bellini, Claire Beneteau, Kathrine Bjørgo, Alice Brooks, Natasha Brown, Alison Castle, Diana Castro, Odelia Chorin, Mark Cleghorn, Emma Clement, David Coman, Carrie Costin, Koen Devriendt, Dexin Dong, Annika Dries, Tina Duelund Hjortshøj, David Dyment, Christine Eng, Casie Genetti, Siera Grano, Peter Henneman, Delphine Heron, Katrin Hoffmann, Jason Hom, Haowei Du, Maria Iascone, Bertrand Isidor, Irma E. Järvelä, Julie Jones, Boris Keren, Mary Kay Koenig, Jürgen Kohlhase, Seema Lalani, Cedric Le Caignec, Andi Lewis, Pengfei Liu, Alysia Lovgren, James R. Lupski, Mike Lyons, Philippe Lysy, Melanie Manning, Carlo Marcelis, Scott Douglas McLean, Sandra Mercie, Mareike Mertens, Arnaud Molin, Mathilde Nizon, Kimberly Margaret Nugent, Susanna Öhman, Melanie O'Leary, Rebecca Okashah Littlejohn, Florence Petit, Rolph Pfundt, Lorraine Pottocki, Annick Raas-Rotschild, Kara Ranguin, Nicole Revencu, Jill Rosenfeld, Lindsay Rhodes, Fernando Santos Simmaro, Karen Sals, Jolanda Schieving, Isabelle Schrauwen, Janneke H.M. Schuurs-Hoeijmakers, Eleanor G. Seaby, Ruth Sheffer, Lot Snijders Blok, Kristina P. Sørensen, Siddharth Srivastava, Zornitza Stark, Radka Stoeva, Chloe Stutterd, Natalie B. Tan, Pernille Mathiesen Torring, Olivier Vanakker, Liselot van der Laan, Athina Ververi, Pablo Villavicencio-Lorini, Marie Vincent, Dorothea Wand, Marja Wessels, Sue White, Monica H. Wojcik, Nan Wu, and Sen Zhao
Web resources
Evo-devo mammalian organs app, https://apps.kaessmannlab.org/evodevoapp/
GTEx portal browser, https://www.gtexportal.org/home/
BrainSpan Atlas of the Developing Human Brain, http://www.brainspan.org
ScApeX browser, https://bioinf.eva.mpg.de/shiny/sample-apps/scApeX/
Human Organoid Single-Cell Browser (Shcheglovitov Lab), https://shcheglovitov.shinyapps.io/u_brain_browser/
gnomAD browser, https://gnomad.broadinstitute.org/about
Supplemental information
References
- 1.Wilfert A.B., Sulovari A., Turner T.N., Coe B.P., Eichler E.E. Recurrent de novo mutations in neurodevelopmental disorders: properties and clinical implications. Genome Med. 2017;9 doi: 10.1186/s13073-017-0498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F., et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nat. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morinaga T., Yasuda H., Hashimoto T., Higashio K., Tamaoki T. A human alpha-fetoprotein enhancer-binding protein, ATBF1, contains four homeodomains and seventeen zinc fingers. Mol. Cell Biol. 1991;11:6041–6049. doi: 10.1128/MCB.11.12.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma G., Gao A., Yang Y., He Y., Zhang X., Zhang B., Zhang Z., Li M., Fu X., Zhao D., et al. Zfhx3 is essential for progesterone/progesterone receptor signaling to drive ductal side-branching and alveologenesis in mouse mammary glands. J. Genet. Genomics. 2019;46:119–131. doi: 10.1016/j.jgg.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Zhao D., Ma G., Zhang X., He Y., Li M., Han X., Fu L., Dong X.Y., Nagy T., Zhao Q., et al. Zinc finger homeodomain factor Zfhx3 is essential for mammary lactogenic differentiation by maintaining prolactin signaling activity. J. Biol. Chem. 2016;291:12809–12820. doi: 10.1074/jbc.M116.719377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry F.B., Miura Y., Mihara K., Kaspar P., Sakata N., Hashimoto-Tamaoki T., Tamaoki T. Positive and Negative Regulation of Myogenic Differentiation of C2C12 Cells by Isoforms of the Multiple Homeodomain Zinc Finger Transcription Factor ATBF1. J. Biol. Chem. 2001;276:25057–25065. doi: 10.1074/jbc.M010378200. [DOI] [PubMed] [Google Scholar]
- 7.Sun X., Frierson H.F., Chen C., Li C., Ran Q., Otto K.B., Cantarel B.L., Vessella R.L., Gao A.C., Petros J., et al. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat. Genet. 2005;37:407–412. doi: 10.1038/ng1528. [DOI] [PubMed] [Google Scholar]
- 8.Mori Y., Kataoka H., Miura Y., Kawaguchi M., Kubota E., Ogasawara N., Oshima T., Tanida S., Sasaki M., Ohara H., et al. Subcellular localization of ATBF1 regulates MUC5AC transcription in gastric cancer. Int. J. Cancer. 2007;121:241–247. doi: 10.1002/ijc.22654. [DOI] [PubMed] [Google Scholar]
- 9.Kataoka H., Miura Y., Kawaguchi M., Suzuki S., Okamoto Y., Ozeki K., Shimura T., Mizoshita T., Kubota E., Tanida S., et al. Expression and subcellular localization of at motif binding factor 1 in colon tumours. Mol. Med. Rep. 2017;16:3095–3102. doi: 10.3892/mmr.2017.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin E.J., Rice K.M., Arking D.E., Pfeufer A., Van Noord C., Smith A.V., Schnabel R.B., Bis J.C., Boerwinkle E., Sinner M.F., et al. Variants in ZFHX3 are associated with a trial fibrillation in individuals of European ancestry. Nat. Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudbjartsson D.F., Holm H., Gretarsdottir S., Thorleifsson G., Walters G.B., Thorgeirsson G., Gulcher J., Mathiesen E.B., Njølstad I., Nyrnes A., et al. A sequence variant in ZFHX3 on 16q22 associates with a trial fibrillation and ischemic stroke. Nat. Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Ni B., Lin Y., Chen X.G., Fang Z., Zhao L., Hu Z., Zhang F. Genetic polymorphisms in ZFHX3 are associated with atrial fibrillation in a Chinese Han population. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jameson H.S., Hanley A., Hill M.C., Xiao L., Ye J., Bapat A., Ronzier E., Hall A.W., Hucker W.J., Clauss S., et al. Loss of the Atrial Fibrillation-Related Gene, Zfhx3 , Results in Atrial Dilation and Arrhythmias. Circ. Res. 2023;133:313–329. doi: 10.1161/CIRCRESAHA.123.323029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe M., Miura Y., Ido A., Sakai M., Nishi S., Inoue Y., Hashimoto T., Tamaoki T. Developmental changes in expression of the ATBF1 transcription factor gene. Mol. Brain Res. 1996;42:344–349. doi: 10.1016/S0169-328X(96)00204-5. [DOI] [PubMed] [Google Scholar]
- 15.Parsons M.J., Brancaccio M., Sethi S., Maywood E.S., Satija R., Edwards J.K., Jagannath A., Couch Y., Finelli M.J., Smyllie N.J., et al. The Regulatory Factor ZFHX3 Modifies Circadian Function in SCN via an at Motif-Driven Axis. Cell. 2015;162:607–621. doi: 10.1016/j.cell.2015.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sagner A., Zhang I., Watson T., Lazaro J., Melchionda M., Briscoe J. A shared transcriptional code orchestrates temporal patterning of the central nervous system. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreau M.X., Saillour Y., Cwetsch A.W., Pierani A., Causeret F. Single-cell transcriptomics of the early developing mouse cerebral cortex disentangle the spatial and temporal components of neuronal fate acquisition. Dev. 2021;148:dev197962. doi: 10.1242/DEV.197962. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z., Wei S., Du H., Su Z., Wen Y., Shang Z., Song X., Xu Z., You Y., Yang Z. Zfhx3 is required for the differentiation of late born D1-type medium spiny neurons. Exp. Neurol. 2019;322 doi: 10.1016/j.expneurol.2019.113055. [DOI] [PubMed] [Google Scholar]
- 19.Miura Y., Tam T., Ido A., Morinaga T., Miki T., Hashimoto T., Tamaoki T. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J. Biol. Chem. 1995;270:26840–26848. doi: 10.1074/jbc.270.45.26840. [DOI] [PubMed] [Google Scholar]
- 20.Ido A., Miura Y., Tamaoki T. Activation of ATBF1, a Multiple-Homeodomain Zinc-Finger Gene, during Neuronal Differentiation of Murine Embryonal Carcinoma Cells. Dev. Biol. 1994;163:184–187. doi: 10.1006/dbio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 21.Fuller T.D., Westfall T.A., Das T., Dawson D.V., Slusarski D.C. High-throughput behavioral assay to investigate seizure sensitivity in zebrafish implicates ZFHX3 in epilepsy. J. Neurogenet. 2018;32:92–105. doi: 10.1080/01677063.2018.1445247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhindsa R.S., Weido B., Dhindsa J.S., Shetty A.J., Sands C., Petrovski S., Vitsios D., Zoghbi A.W. Genome-wide prediction of dominant and recessive neurodevelopmental disorder risk genes. bioRxiv. 2022 doi: 10.1101/2022.11.21.517436. Preprint at. [DOI] [Google Scholar]
- 24.Chow J.C., Hormozdiari F. Prediction of Neurodevelopmental Disorders Based on De Novo Coding Variation. J. Autism Dev. Disord. 2023;53:963–976. doi: 10.1007/s10803-022-05586-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurovich Y., Hanani Y., Bar O., Nadav G., Fleischer N., Gelbman D., Basel-Salmon L., Krawitz P.M., Kamphausen S.B., Zenker M., et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019;25:60–64. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 28.Mak B.C., Sanchez Russo R., Gambello M.J., Fleischer N., Black E.D., Leslie E., Murphy M.M., Emory 3q29 Project. Mulle J.G., Aberizk K., et al. Craniofacial features of 3q29 deletion syndrome: Application of next-generation phenotyping technology. Am. J. Med. Genet. 2021;185:2094–2101. doi: 10.1002/ajmg.a.62227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aref-Eshghi E., Kerkhof J., Pedro V.P., Groupe DI France. Barat-Houari M., Ruiz-Pallares N., Andrau J.C., Lacombe D., Van-Gils J., Fergelot P., et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G., Wang L.-G., He Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 32.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cardoso-Moreira M., Halbert J., Valloton D., Velten B., Chen C., Shao Y., Liechti A., Ascenção K., Rummel C., Ovchinnikova S., et al. Gene expression across mammalian organ development. Nature. 2019;571:505–509. doi: 10.1038/S41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Sanchís-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D., et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nat. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Chiola S., Yang G., Russell C., Armstrong C.J., Wu Y., Spampanato J., Tarboton P., Ullah H.M.A., Edgar N.U., et al. Modeling human telencephalic development and autism-associated SHANK3 de fi ciency using organoids generated from single neural rosettes. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-33364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luck K., Kim D.K., Lambourne L., Spirohn K., Begg B.E., Bian W., Brignall R., Cafarelli T., Campos-Laborie F.J., Charloteaux B., et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saadi I., Alkuraya F.S., Gisselbrecht S.S., Goessling W., Cavallesco R., Turbe-Doan A., Petrin A.L., Harris J., Siddiqui U., Grix A.W., et al. Deficiency of the cytoskeletal protein SPECC1L leads to oblique facial clefting. Am. J. Hum. Genet. 2011;89:44–55. doi: 10.1016/j.ajhg.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfert A., Moreno N., Kerl K. The BAF complex in development and disease. Epigenet. Chromatin. 2019;12 doi: 10.1186/s13072-019-0264-y. 19–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Giammartino D.C., Nishida K., Manley J.L. Mechanisms and Consequences of Alternative Polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aref-Eshghi E., Bend E.G., Hood R.L., Schenkel L.C., Carere D.A., Chakrabarti R., Nagamani S.C.S., Cheung S.W., Campeau P.M., Prasad C., et al. BAFopathies’ DNA methylation epi-signatures demonstrate diagnostic utility and functional continuum of Coffin-Siris and Nicolaides-Baraitser syndromes. Nat. Commun. 2018;9:4885. doi: 10.1038/s41467-018-07193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caracci M.O., Avila M.E., Espinoza-Cavieres F.A., López H.R., Ugarte G.D., De Ferrari G.V. Wnt/β-Catenin-Dependent Transcription in Autism Spectrum Disorders. Front. Mol. Neurosci. 2021;14:1–16. doi: 10.3389/fnmol.2021.764756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa-Mattioli M., Monteggia L.M. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat. Neurosci. 2013;16:1537–1543. doi: 10.1038/nn.3546. [DOI] [PubMed] [Google Scholar]
- 45.Hindley C.J., Condurat A.L., Menon V., Thomas R., Azmitia L.M., Davis J.A., Pruszak J. The Hippo pathway member YAP enhances human neural crest cell fate and migration. Sci. Rep. 2016;6:23208. doi: 10.1038/srep23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usmani M.A., Ahmed Z.M., Magini P., Pienkowski V.M., Rasmussen K.J., Hernan R., Rasheed F., Hussain M., Shahzad M., Lanpher B.C., et al. De novo and bi-allelic variants in AP1G1 cause neurodevelopmental disorder with developmental delay, intellectual disability, and epilepsy. Am. J. Hum. Genet. 2021;108:1330–1341. doi: 10.1016/j.ajhg.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epi4K Consortium. Epilepsy Phenome/Genome Project. Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toma C., Torrico B., Hervás A., Valdés-Mas R., Tristán-Noguero A., Padillo V., Maristany M., Salgado M., Arenas C., Puente X.S., et al. Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol. Psychiatry. 2014;19:784–790. doi: 10.1038/mp.2013.106. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto R., Nakazawa T., Tsurusaki Y., Yasuda Y., Nagayasu K., Matsumura K., Kawashima H., Yamamori H., Fujimoto M., Ohi K., et al. Whole-exome sequencing and neurite outgrowth analysis in autism spectrum disorder. J. Hum. Genet. 2016;61:199–206. doi: 10.1038/jhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He M., Liu L., Luo S., Wang J., Guo J., Zhai Q., He S., Zou D., Liu X., Zhou P., et al. ZFHX3 Associated with Partial Epilepsy/Spasms and Correlation between Outcome & Gene Expression Stage. medRxiv. 2023 doi: 10.1101/2023.07.16.23292551. Preprint at. [DOI] [Google Scholar]
- 51.Zhang R., Gehlen J., Kawalia A., Melissari M.-T., Dakal T.C., Menon A.M., Höfele J., Riedhammer K., Waffenschmidt L., Fabian J., et al. Human exome and mouse embryonic expression data implicate ZFHX3, TRPS1, and CHD7 in human esophageal atresia. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paucar M., Nilsson D., Engvall M., Laffita-Mesa J., Söderhäll C., Skorpil M., Halldin C., Fazio P., Lagerstedt-Robinson K., Solders G., et al. Spinocerebellar ataxia type 4 is caused by a GGC expansion in the ZFHX3 gene and is associated with prominent dysautonomia and motor neuron signs. medRxiv. 2023 doi: 10.1101/2023.10.03.23296230. Preprint at. [DOI] [PubMed] [Google Scholar]
- 53.Figueroa K.P., Gross C., Atienza E.B., Paul S., Gandelman M., Haack T.B., Kakar N., Sturm M., Casadei N., Admard J., et al. GGC expansion in ZFHX3 causes SCA4 and impairs autophagy. medRxiv. 2023 doi: 10.1101/2023.10.26.23297560. Preprint at. [DOI] [Google Scholar]
- 54.Wallenius J., Kafantari E., Jhaveri E., Gorcenco S., Ameur A., Karremo C., Dobloug S., Karrman K., de Koning T., Ilinca A., et al. Exonic trinucleotide repeat expansions in ZFHX3 cause spinocerebellar ataxia type 4: A poly-glycine disease. Am. J. Hum. Genet. 2024;111:82–95. doi: 10.1016/j.ajhg.2023.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wegner M., Stolt C.C. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 56.Hollis E.R., Zou Y. Reinduced Wnt signaling limits regenerative potential of sensory axons in the spinal cord following conditioning lesion. Proc. Natl. Acad. Sci. USA. 2012;109:14663–14668. doi: 10.1073/pnas.1206218109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lie D.C., Colamarino S.A., Song H.J., Désiré L., Mira H., Consiglio A., Lein E.S., Jessberger S., Lansford H., Dearie A.R., Gage F.H. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi S., Watanabe T., Okada M., Inoue K., Ueda T., Takada I., Watabe T., Yamamoto Y., Fukuda T., Nakamura T., et al. Noncanonical Wnt signaling mediates androgen-dependent tumor growth in a mouse model of prostate cancer. Proc. Natl. Acad. Sci. USA. 2011;108:4938–4943. doi: 10.1073/pnas.1014850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mardones M.D., Andaur G.A., Varas-Godoy M., Henriquez J.F., Salech F., Behrens M.I., Couve A., Inestrosa N.C., Varela-Nallar L. Frizzled-1 receptor regulates adult hippocampal neurogenesis. Mol. Brain. 2016;9 doi: 10.1186/s13041-016-0209-3. 29–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J.Y., Duan X., Liu C.Y., Jang M.H., Guo J.U., Pow-anpongkul N., Kang E., Song H., Ming G.l. DISC1 Regulates New Neuron Development in the Adult Brain via Modulation of AKT-mTOR Signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J.Y., Liu C.Y., Zhang F., Duan X., Wen Z., Song J., Feighery E., Lu B., Rujescu D., St Clair D., et al. Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell. 2012;148:1051–1064. doi: 10.1016/j.cell.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung C.G., Kim H.J., Kawaguchi M., Khanna K.K., Hida H., Asai K., Nishino H., Miura Y. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–5145. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- 63.Ido A., Miura Y., Watanabe M., Sakai M., Inoue Y., Miki T., Hashimoto T., Morinaga T., Nishi S., Tamaoki T. Cloning of the cDNA encoding the mouse ATBF1 transcription factor. Gene. 1996;168:227–231. doi: 10.1016/0378-1119(95)00740-7. [DOI] [PubMed] [Google Scholar]
- 64.Isshiki T., Pearson B., Holbrook S., Doe C.Q. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/S0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 65.Ouhaz Z., Fleming H., Mitchell A.S. Cognitive functions and neurodevelopmental disorders involving the prefrontal cortex and mediodorsal thalamus. Front. Neurosci. 2018;12:33. doi: 10.3389/fnins.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H., Janssens J., de Waegeneer M., Kolluru S.S., Davie K., Gardeux V., Saelens W., David F.P.A., Brbić M., Spanier K., et al. Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science. 2022;375:eabk2432. doi: 10.1126/science.abk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun X., Fu X., Li J., Xing C., Martin D.W., Zhang H.H., Chen Z., Dong J.-T. Heterozygous deletion of Atbf1 by the Cre-loxP system in mice causes preweaning mortality. genesis. 2012;50:819–827. doi: 10.1002/dvg.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palomares M., Delicado A., Mansilla E., De Torres M.L., Vallespín E., Fernandez L., Martinez-Glez V., García-Miñaur S., Nevado J., Simarro F.S., et al. Characterization of a 8q21.11 microdeletion syndrome associated with intellectual disability and a recognizable phenotype. Am. J. Hum. Genet. 2011;89:295–301. doi: 10.1016/j.ajhg.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bögershausen N., Wollnik B. Mutational Landscapes and Phenotypic Spectrum of SWI/SNF-Related Intellectual Disability Disorders. Front. Mol. Neurosci. 2018;11:252. doi: 10.3389/fnmol.2018.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C.A., Lattier J., Zhu W., Rosenfeld J., Wang L., Scott T.M., Du H., Patel V., Dang A., Magoulas P., et al. Retrospective analysis of a clinical exome sequencing cohort reveals the mutational spectrum and identi fi es candidate disease – associated loci for BAFopathies. Genet. Med. 2022;24:364–373. doi: 10.1016/j.gim.2021.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelly S.M., Corbett A.H. Messenger RNA Export from the Nucleus: A Series of Molecular Wardrobe Changes. Traffic. 2009;10:1199–1208. doi: 10.1111/j.1600-0854.2009.00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dargemont C., Babour A. Novel functions for chromatin dynamics in mRNA biogenesis beyond transcription. Nucleus. 2017;8:482–488. doi: 10.1080/19491034.2017.1342916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jordán-Pla A., Yu S., Waldholm J., Källman T., Östlund Farrants A.K., Visa N. SWI/SNF regulates half of its targets without the need of ATP-driven nucleosome remodeling by Brahma. BMC Genom. 2018;19 doi: 10.1186/s12864-018-4746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y.I.G., Moore R.E., Ge H.Y., Young M.K., Lee T.D., Stevens S.W. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ageta-Ishihara N., Miyata T., Ohshima C., Watanabe M., Sato Y., Hamamura Y., Higashiyama T., Mazitschek R., Bito H., Kinoshita M. Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun. 2013;4:2532. doi: 10.1038/ncomms3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed IP-MS data and the overlapping peaks between endogenous and ectopic ZFHX3 ChIP-seq, as well as the results of the de novo and known motif analyses are available in the supplemental information.
Some of the DNA methylation datasets used in this study are publicly available and may be obtained from gene expression omnibus (GEO) using the following accession numbers. GEO: GSE116992, GSE66552, GSE74432, GSE97362, GSE116300, GSE95040, GSE104451, GSE125367, GSE55491, GSE108423, GSE116300, GSE89353, GSE52588, GSE42861, GSE85210, GSE87571, GSE87648, GSE99863, and GSE35069. These include DNA methylation data from individuals with Kabuki syndrome (MIM: 147920 and 300867) (GSE97362 and GSE116300), Sotos syndrome (MIM: 117550) (GSE74432), CHARGE syndrome (MIM: 214800) (GSE97362), immunodeficiency-centromeric instability-facial anomalies (ICF) syndrome (MIM: 242860) (GSE95040), Williams-Beuren syndrome (MIM: 194050), 7q11.23 duplication syndrome (MIM: 609757) (GSE66552), BAFopathies(GSE116992, GSE125367), Down syndrome (MIM: 190685) (GSE52588), Silver-Russell syndrome (MIM: 180860, 618905, 616489, 618907, and 618908) (GSE104451 and GSE55491), X-linked syndromic intellectual developmental disorder, Claes-Jensen type (MIM: 300534) (GSE108423), a large cohort of (unresolved) subjects with developmental delays and congenital abnormalities (GSE89353), and several large cohorts of DNA methylation data from the general population (GSE42861, GSE85210, GSE87571, GSE87648, GSE99863, and GSE35069). Remaining methylation data is not publicly available due to institutional and ethics restrictions.