Summary
The existence of neural stem cells (NSCs) in adult human brain neurogenic regions remains unresolved. To address this, we created a cell atlas of the adult human subventricular zone (SVZ) derived from fresh neurosurgical samples using single-cell transcriptomics. We discovered 2 adult radial glia (RG)-like populations, aRG1 and aRG2. aRG1 shared features with fetal early RG (eRG) and aRG2 were transcriptomically similar to fetal outer RG (oRG). We also captured early neuronal and oligodendrocytic NSC states. We found that the biological programs driven by their transcriptomes support their roles as early lineage NSCs. Finally, we show that these NSCs have the potential to transition between states and along lineage trajectories. These data reveal that multipotent NSCs reside in the adult human SVZ.
Subject areas: Molecular biology, Cell biology, Omics, Transcriptomics
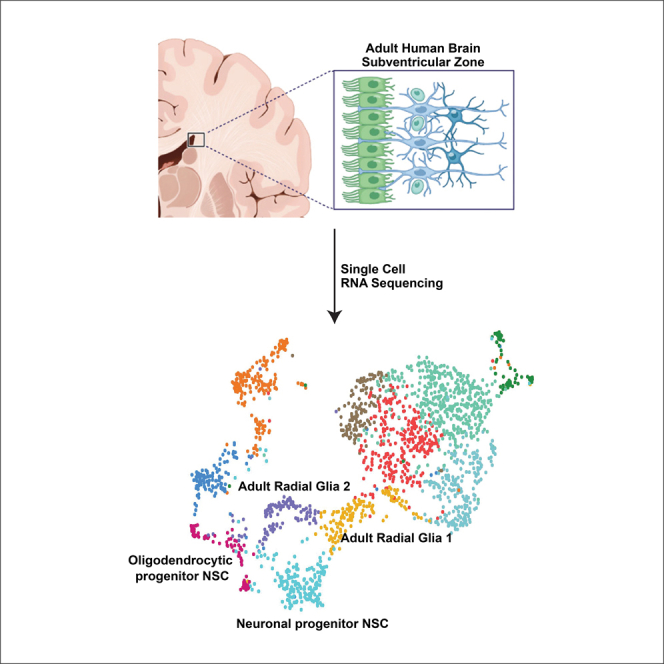
Graphical abstract
Highlights
-
•
We created a single-cell atlas of the adult human SVZ derived from fresh samples
-
•
We discovered 2 adult radial glia cell types similar to fetal outer radial glia
-
•
We also captured early neuronal and oligodendrocytic neural stem cell states
-
•
We show NSCs can transition between states and along lineage trajectories
Molecular biology; Cell biology; Omics; Transcriptomics
Introduction
The existence of active neurogenic sites in the human adult brain has not been resolved. Evidence for adult neurogenesis in the hippocampus1 and neuroepithelium2 suggests some degree of regenerative capacity.
During early human development, the subventricular zone (SVZ) emerges as a neurogenic niche containing molecularly diverse populations of radial glia (RG) that give rise to cells of the neocortex.3,4,5 With aging, the SVZ undergoes cytoarchitectural changes maturing into four cellular layers,6 and the fetal RG are thought to acquire astroglial characteristics becoming potentially adult NSCs of the SVZ.7
Extensively characterized in the adult mouse brain, these SVZ NSCs, known as B cells, retain regenerative potential and can generate a variety of new-born neurons and glia.8,9,10 While the adult human brain is believed to retain NSCs of embryonic origin,11,12 evidence for their existence in situ, particularly in the SVZ, the largest germinal region in the adult brain, is less certain. This paucity of data is primarily due to access to high-quality specimens.
To address this, we created a single-cell transcriptomic atlas of the adult human SVZ from freshly derived neurosurgical specimens. We found 4 NSC populations. Two NSC types, aRG1 and aRG2, are transcriptomically and biologically similar to fetal early RG (eRG) and fetal outer RG (oRG), respectively. We also discovered early neuronal and oligodendrocytic NSC states showing lineage emergence and maturation while retaining developmental features. Biological programs underlying these states revealed neurodevelopmental, neuroinflammatory, and injury response programs. Lastly, we show that these NSCs have the potential to transition between populations and along lineage trajectories.
Results
Adult human subventricular zone cell atlas
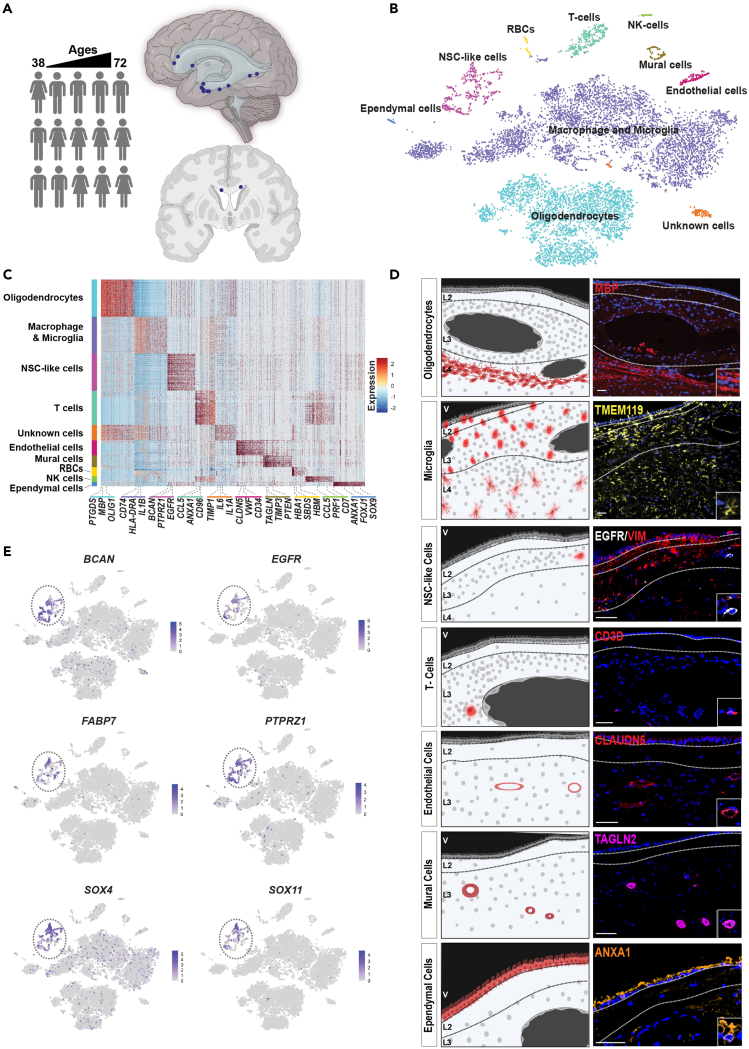
SVZ samples were acquired from 15 patients undergoing brain tumor surgery. Their ages ranged from 38 to 72 years (Figure 1A and S1). SVZ regions that were normal on MRI and were to be removed during the course of surgery were sampled. The transcriptomes of 10,834 cells were acquired by single-cell RNA sequencing (scRNA-seq). Cells containing cancer-type-specific copy number aberrations were removed from further analysis (Figure S2).
Figure 1.
Adult human subventricular zone cell atlas
(A) Schematic diagram illustrating the sources and brain regions of adult human SVZ neurosurgical samples. Samples were derived from patients with ages ranging from 38 to 72 years old. Eight samples were derived from females and 7 from males. The marks on the right panels show the 15 subventricular zone sample sites.
(B) t-distributed stochastic neighbor embedding visualization (tSNE) of adult SVZ cells colored by cell types. NSC—neural stem cell, NK—natural killer, RBC—red blood cell.
(C) Heatmap of top 100 genes expressed in each SVZ cell type.
(D) Localization of cell types within normotypic autopsy SVZs using cell-type-specific protein markers. Scale bar, 100mm.
(E) Expression of indicated genes in all SVZ cell type clusters visualized on tSNE.
The Louvain algorithm resolved 10 clusters (Figure 1B). Cell types were labeled based on expression of canonical gene markers: oligodendrocytes; macrophage/microglia; neural stem cell (NSC)-like; T cells; endothelial cells; mural cells; red blood cells; natural killer cells; ependymal cells; and an unidentified cell cluster (Figure 1C and Table S1). Cell-type-specific protein markers were used to spatially locate these cell types in human adult SVZ samples derived from normotypic autopsy brain tissue (Figure 1D). Consistent with the known cellular composition of the SVZ,6 oligodendrocytes were most abundant in layers 4 and 3, respectively. NSC-like cells were detected in the astrocytic band, layer 3. The astrocytic band in the human adult SVZ has been reported to contain NSCs.7
Thirteen of the 15 patients contributed cells to the NSC-like cluster. Cells in this cluster expressed canonical neural progenitor and RG genes such as BCAN (64% of cells), SOX4 (68% of cells), SOX11 (47% of cells), PTPRZ1 (69% of cells), FABP7 (43% of cells), and EGFR (43% of cells) (Figure 1E).3,4,5,13 However, cells in this cluster showed different patterns of gene expression, suggesting cellular diversity within this cell cluster.
Neural stem cell diversity within the adult subventricular zone
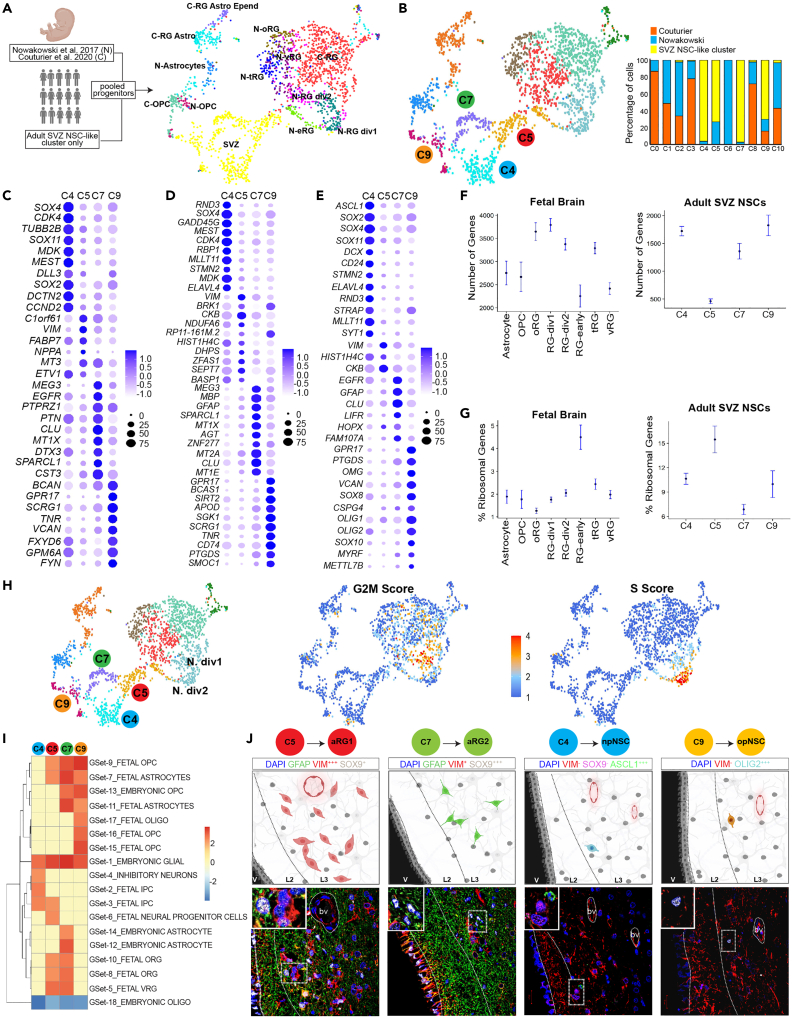
Since differentiated cells in the SVZ cell dataset forced diverse NSC-like cells into one cluster, we created a progenitor-only dataset using the adult NSC-like cluster and enriched it with fetal progenitor/stem cells from Nowakowski et al. (2017) and Couturier et al. (2020)4,9 (Figure 2A). In this merged dataset the adult NSC-like cluster resolved into 4 clusters (C4, C5, C7, and C9) (Figure 2B). C5 contained cells from the adult SVZ and fetal eRG, and C9 contained cells from the adult SVZ and fetal oligodendrocyte progenitor cells (OPCs). To preserve the pure adult gene signatures and account for the transcriptomic differences between adult and fetal cells, all fetal cells were removed from further downstream analysis of the 4 SVZ NSC-like cell clusters.
Figure 2.
Neural stem cell diversity within the adult subventricular zone
(A) Schematic diagram illustrating sources of samples from adult human SVZ regions and fetal brains, and uniform manifold approximation and projection (UMAP) visualization of the merged progenitor cells selected from the adult SVZ NSC-like cluster and the fetal datasets colored by original cell annotations.
(B) UMAP visualization of the merged progenitor cells selected from the adult SVZ NSC-like cluster and fetal datasets by clusters. Bar graph shows the dataset source of the cells contributing to each cluster.
(C) Dot plot of the top 10 genes expressed in each of the 4 NSC clusters compared to all SVZ clusters.
(D) Dot plot of the top 10 genes expressed in each of the 4 NSC clusters compared to NSC clusters. Ribosomal genes are not shown (see STAR Methods). Redundant genes are shown only once.
(E) Expression of curated known gene markers of RG, NSCs, neuronal progenitors and oligodendrocyte progenitors.
(F) Transcriptome size of adult NSC-like and fetal brain cells. Confidence interval = 95%.
(G) Ribosomal gene content of adult NSC-like and fetal brain cells. Confidence interval = 95%.
(H) Cell cycle phase scores calculated based on expression of G2M and S markers.
(I) Summary of gene set enrichment analysis using curated human brain cell types published single-cell RNA-seq datasets. Dendrogram shows unsupervised hierarchical clustering of the data. Color bar indicates normalized enrichment score. Red and blue colors represent significant enrichment or down-regulation (FDR < 0.0001), respectively, yellow indicates no significant result.
(J) Localization of aRG1, aRG2, npNSCs, and opNSCs within normotypic autopsy SVZs using cell type-specific protein markers. Scale bar, 50μm.
Differential gene expression analysis was then used to determine NSC cluster-specific expression patterns (Figures 2C and S3; Table S2). The most highly differentially expressed genes in each of the 4 adult NSC clusters, compared to all other SVZ cells, were C5—C1orf61, VIM, FABP7, NPPA, MT3; C7—MEG3, EGFR, PTPRZ1, PTN, CLU; C4—SOX4, CDK4, TUBB2B, SOX11, MDK; and C9—BCAN, GPR17, SCRG1, TNR, VCAN.
Transcriptomic differences between NSC clusters show radial glial identity and lineage-specific programs
To explore the transcriptomic differences between the NSC clusters, we identified genes that were differentially expressed between these clusters. C5 showed high expression of classical RG markers VIM and CKB (Figures 2C–2E; Table S3).3,4,5 Ribosomal genes RPS3A, RPLP1, RPS14, RPL23A, RPS27A, and RPL39 were also highly expressed, as were the mouse radial precursors ribosomal genes RPL31, RPS15, and RPL3.14 No lineage-specific genes were expressed. Since C5 contained adult SVZ NSCs and fetal eRG from gestational week 8,4 we compared transcriptomic features of these cells to other SVZ and fetal cell types. We found that both adult and fetal C5 cells shared low transcriptome sizes (Figure 2F) and high expression of ribosomal genes (Figure 2G). The uncoupling of protein synthesis and ribosome biogenesis has been reported in activated and quiescent stem cells, albeit in opposite directions.15 Likewise, RG precursors from mouse embryos at embryonic week E11.5 have been shown to express higher levels of ribosomal biogenesis genes.14 Some cells in C5 expressed a cell cycle gene signature (Figure 2H). Cycling in C5 suggests that these cells have the capacity for division. That they may exist across the lifespan suggests a capacity for self-renewal.
The C7 transcriptome contained classical astroglial marker genes expressed in human embryonic and murine adult NSCs such as ALDOC, ID3, GFAP, and GLAST.14,16,17,18 Fetal oRG genes HES1, HOPX, LIFR, SLC1A3, HEPACAM, FAM107A, and TNC were also highly and uniquely expressed in C7 (Figures 2D and 2E; Table S3).4,5 Adult SVZ NSCs have been reported to retain astroglial characteristics.16,19 C7 also expressed extracellular matrix-related genes PTN, BCAN, NCAN, and SPARCL1,20 and gap/tight junction genes CLDN1/12, COX43/GJA1, and TJP1 (Table S3).3,21,22
C4 expressed genes associated with embryonic development and neuronal programs such as ASCL1, SOX2, CD24, SOX4, and SOX11 (Figures 2C–2E; Table S3),23,24,25 and neurogenic oRG transcription factors HES6, NHLH1, and CBFA2T2 (Table S3).26 C4 also expressed BEST3, STMN2, ELAVL4, and DCX, gene markers of fetal intermediate neuronal progenitors/new-born neurons, and ZIC2, MOXD1, and C1orf61, markers of ventral telencephalic RG.4 These cells did not express classical fetal newborn interneuron markers PAX6, TBR2, or SATB2, suggesting differences between fetal and adult-born neurons. RND3, MLLT11, STRAP, and SYT1, markers of neuronal trajectory maturation critical in cortical evolution for neurite outgrowth and axonogenesis, were differentially highly expressed in C4,23,27,28,29,30,31,32 whereas expression of RG/neural progenitor markers VIM, GFAP, and HES1 were downregulated. Markers of adult human neurons CALB2, CCK, PVALB, SST, and VIP were also absent.33 C4 also expressed markers of cell cycle activity including CCND1, CCND2, TOP3A, and CDK4, and some cells expressed a cell cycle gene signature (Figure 2H).
C9 cells highly expressed OPC markers BCAN, CSPG4, OLIG1/2, and SOX10 (Figures 2C–2E; Table S3).34,35,36 OPC, preOPC, and differentiation-committed OPC markers VCAN, SOX6, PCDH15, MEGF1, and GPR17 were uniquely expressed in C9, as were migration-related genes TNS3 and FYN.33,37,38 Premyelinating oligodendrocyte genes BCAS1, SGK1, TCF7L2, and SCRG1 were also expressed in C9,39,40 as were genes involved in neuro-immune processes such as CD74, HLA-DRB1, and CXCR4.41 Demyelination has been shown to cause adult murine OPCs to express immune cues contributing to the post-injury inflammatory milieu and support migration.42
Adult neural stem cell gene signature comparison using gene set enrichment analysis
We next compared gene signatures of the 4 NSC clusters to curated genesets of human embryonic and fetal brain cell types sourced from published scRNA-seq studies using gene set enrichment analysis.4,43,44,45 All 4 NSC clusters showed enrichment for an NSC-like signature and low expression for differentiated oligodendrocytes (Figure 2I). C4 showed enrichment for 3 neuronal progenitor programs. C9 showed enrichment for oligodendrocyte progenitor programs. C7 is most strongly enriched for embryonic astrocytes and fetal RGs. C5 was enriched for RG programs and a neural progenitor program.
Based on these gene expression patterns and comparisons to available datasets, we labeled C5 cells as adult radial glia 1 (aRG1) and C7 cells as aRG2. C4 and C9 cells have acquired lineage specificity, and because of this potential transient nature, we labeled these as progenitor states, C4 as neuronal progenitor NSCs (npNSCs) and C9 as oligodendrocytic progenitor NSCs (opNSCs).
Using the differentially expressed genes as a guide, we selected protein markers to locate these 4 NSC populations within the SVZ from normotypic autopsy adult human brain samples (Figure 2J). aRG1 (VIM+++, SOX9+) and aRG2(VIM+, SOX9+++) were distributed in layer 37. opNSCs (OLIG2+++, VIM−) and npNSCs (ASCL1+++, VIM−, SOX9-) were found in layers 3 and 2, respectively.
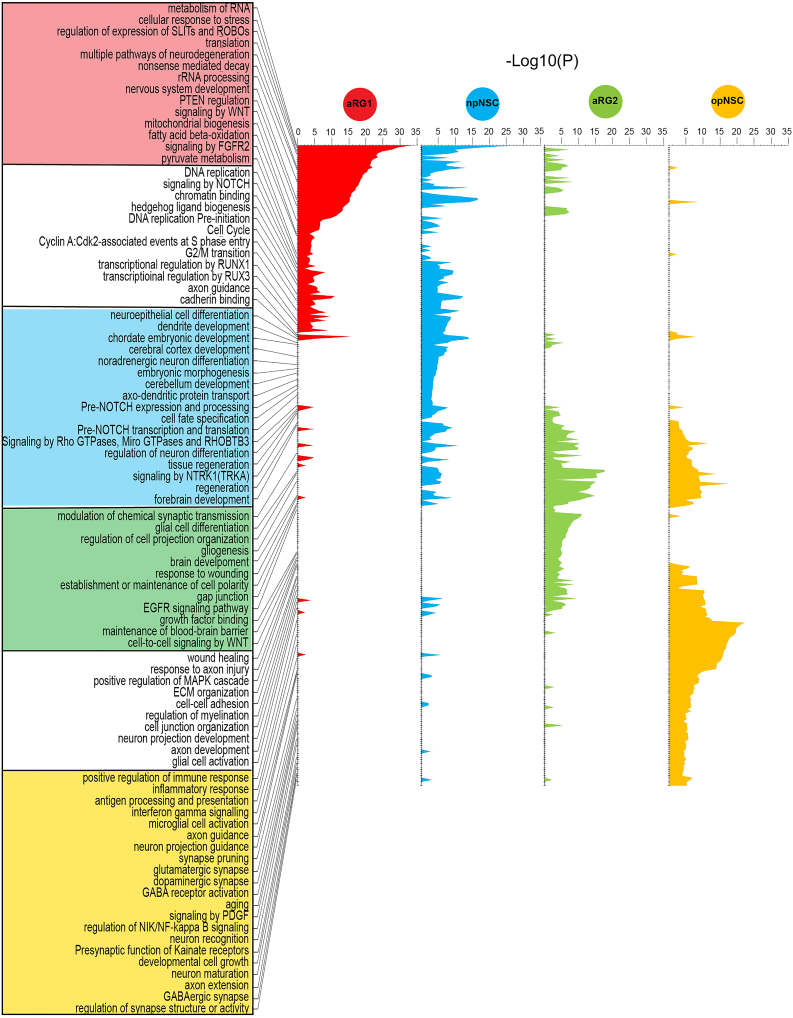
Uncovering neural stem cell programs using biological program analysis
We also characterized the biological programs inherent in aRG1, aRG2, opNSCs, and npNSCs using their most differentially expressed genes (Figure 3 and Table S4). npNSCs showed enrichment for programs such as forebrain development, cerebellar cortex development, transcriptional regulation by RUNX1 and 3, pre-NOTCH expression and processing, hedgehog ligand biogenesis, and NOTCH signaling. Cell cycle processes were also active, consistent with their cell cycle score (Figure 2H), as were neuronal processes including dendrite development, neuroepithelial cell differentiation, cell fate determination, regulation of neuron differentiation, embryonic morphogenesis, cadherin binding, and cell fate specification. These programs have been reported to be required for neuronal differentiation and maturation.27,29,30
Figure 3.
Enriched neural stem cell biological programs
Biological program analysis performed for aRG1, npNSCs, aRG2, and opNSCs, using a maximum of the top 500 differentially expressed genes. Unique pathways/process are boxed and colored according to their state. Shared pathways/processes are boxed white.
opNSCs were enriched for processes such as gliogenesis, regulation of myelination, extracellular matrix organization, PDGF signaling, GABA B receptor activation, presynaptic function of kainate receptor, neuron recognition and maturation, axon development, synapse organization, and response to axon injury (Figure 3). While embryonic OPCs lack ion channels or glutamatergic receptors, these channels have been shown to emerge in OPCs postnatally.46 opNSCs also expressed neuroimmune processes including glial cell activation, cytokine signaling, antigen processing and presentation, inflammatory response, and microglial activation. OPCs have been shown to modulate neuroinflammation, immune response, and central nervous system repair.47,48 Cell motility processes, regulation of locomotion and cell motility, RAC1 GTPase cycle, and cell junction disassembly were enriched in npNSCs and opNSCs.
Although aRG1 and aRG2 shared astroglial markers, their biological programs differed (Figure 3 and Table S4). aRG1 were enriched for rRNA processing, mRNA splicing, translation, ribosome biogenesis, and nonsense mediated decay. Ribosome biogenesis has been shown to be crucial for controlling stem cell homeostasis. In adult stem cells the level of ribosome biogenesis and protein synthesis determines the switch between quiescent and activated states with self-renewal decisions,15,49,50 while senescence is associated with shutdown of ribosome biogenesis.15,50 In addition, active metabolic processes such as oxidative phosphorylation, mitochondrial biogenesis, pyruvate metabolism, fatty acid beta-oxidation, and stress-related processes were also enriched in aRG1. Quiescent neural stem cells maintain high levels of mitochondrial fatty acid β-oxidation and express proteins involved in mitochondrial metabolism.49,51,52 Mitochondrial dynamics have been reported to affect self-renewal and fate choices.53,54 aRG1 also showed enrichment for cell cycle, consistent with their cell cycle score (Figure 2H), and DNA replication, chromatin binding, and axon guidance, albeit to a lesser degree than npNSCs.
aRG2 were significantly enriched for brain development, EGFR signaling, response to metal ions, gap junction, cell polarity, transepithelial transport, cell-cell adhesion, and maintenance and transport across the blood-brain barrier (Figure 3 and Table S4).55,56,57,58,59 SVZ astrocytic NSCs have been reported to regulate the niche.60 Developmental processes such as cell-cell signaling by WNT, nervous system development, signaling by NTRK1, regulation of neurogenesis, regulation of gliogenesis and glial cell differentiation, neuron projection development, kinase activity and regulation of growth, and wound healing and tissue regeneration processes were also enriched.56,58,61,62,63,64
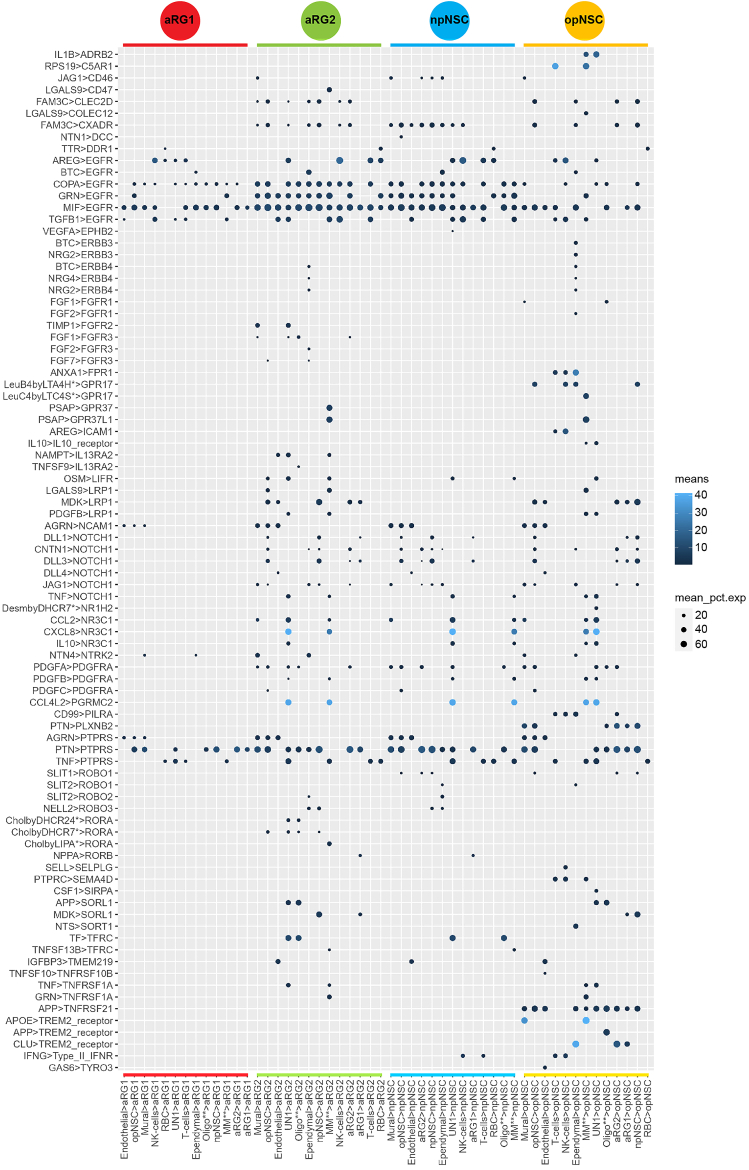
Subventricular zone cell to cell interactions
Cell-to-cell interaction analysis using CellPhoneDB revealed interactions involving receptors expressed on NSCs known to be important in neurodevelopment, including EGFR, NOTCH, PDGFRA and PTPRS (Figure 4 and Table S5). Corresponding ligands were expressed by NSCs and other niche cells. Consistent with the transcriptomic analyses, aRG1 showed relatively fewer interactions compared to aRG2, npNSCs, and opNSCs.
Figure 4.
Subventricular zone cellular interactions
Dot plot showing the ligand-receptor interactions from all niche cell types to NSCs. Scale bar is the mean expression of the ligand-receptor pair. Dot size is the mean of percentage of expressed cells for each cell type pair. MM∗∗—Macrophage and Microglia, Oligo∗∗—Oligodendrocyte, UN1—unidentified.
EGFR expression was common across NSCs but most abundant in aRG2 and npNSCs. Six ligands interacted with EGFR. Betacellulin (BTC), expressed by ependymal cells, has been reported to be involved in regulating various processes ranging from reproduction to the control of NSCs.65 Amphiregulin (AREG), expressed mainly by NK cells and T cells, is a mitogen for astrocytes and has been shown to be involved in embryonic morphogenesis and wound healing by interacting with EGFR.66 Progranulin (GRN) is associated with processes including embryogenesis, tumorigenesis, inflammation, wound repair, and neurodegeneration.67,68 The macrophage migration inhibitory factor (MIF) interacted with all NSCs. Although its activity has not been studied extensively, it has been reported to block EGFR activation.69 The actions of TGFβ1 and COPA on EGFR are also not well understood.
Pleiotrophin (PTN), known to be associated with new-born neuron development,70 was expressed by many niches cell types and interacted with the protein tyrosine-phosphatase receptor S (PTPRS) expressed on all NSC types. PTPRS has been shown to be involved in axonogenesis, axon guidance, and adult nerve repair.71,72
Interactions exclusive to opNSCs were apolipoprotein E (APOE) from mural cells and macrophage/microglia, and clusterin (CLU) from ependymal cells, interacting with the triggering receptor expressed on myeloid cells 2 (TREM2), implicating opNSCs in response to injury, neuroprotection, and degenerative pathologies.73,74
Neural stem cell transition capacity
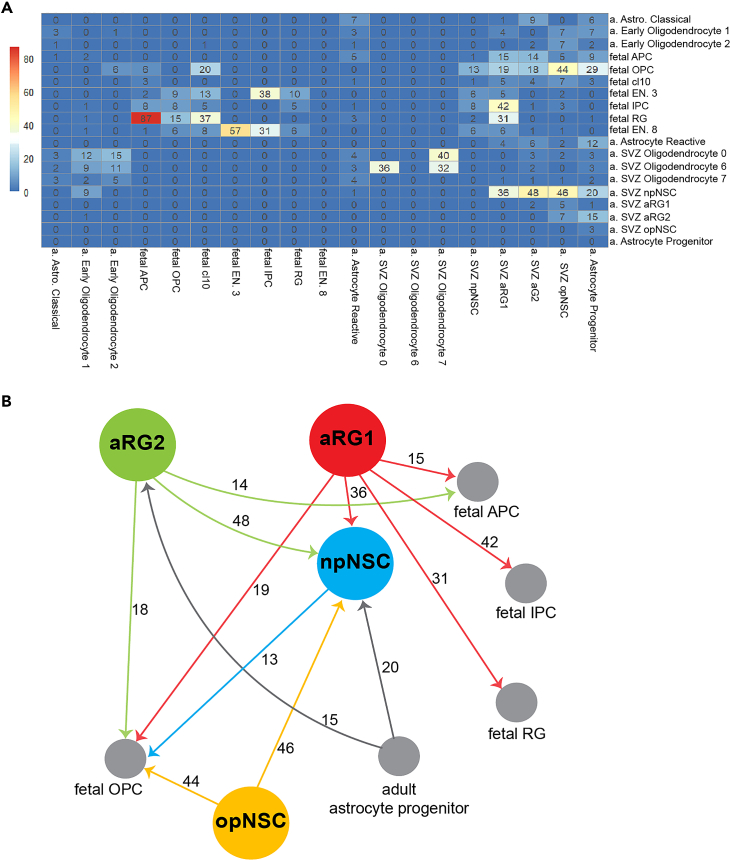
We used RNA velocity to investigate trajectories between SVZ NSCs and their progeny. To reconstruct the spectrum of neuronal cell differentiation, we created a merged scRNA-seq dataset containing adult SVZ cells, fetal brain cells,9 and adult brain astrocytes75 and oligodendrocytes75 along the spectrum of differentiation. Three groups of state transition patterns were found (Figures 5A and 5B). Group 1 cells show multistate transition capacity. All SVZ-derived NSCs were in group 1. We found that aRG1 can transition to fetal intermediate neuronal progenitor cells (IPCs), npNSCs, fetal RG, fetal OPCs, and fetal astrocyte progenitor cells (APCs). aRG2 can transition to npNSCs, fetal APCs, fetal OPCs, and adult astrocytes. opNSCs showed transition capacity to npNSCs and fetal OPCs, and adult early oligodendrocytes to a lesser degree. npNSCs showed weak transition capacity to fetal IPCs and fetal OPCs. Group 2 cells show only intra-lineage transition capacity. For example, fetal IPCs can give rise to newborn excitatory neurons. Group 3 cells show no transition capacity (Figure 5A). For example, oligodendrocyte cluster 6 derived from the adult SVZ, a fully differentiated cell type, did not give rise to any other cell type in this dataset.
Figure 5.
Neural stem cell transition capacity
(A) Cluster-based velocity analysis (PAGA, see STAR Methods) for adult and fetal brain cell types. Numbers are transition confidence ranging from 0 to 100.
(B) Summary schematic showing cell transitions with confidences greater than 10. a—adult, astro—astrocyte. APC—astrocyte progenitor cell, OPC—oligodendrocyte progenitor cell, EN—excitatory neuron, IPC—intermediate progenitor cell, RG—radial glia, SVZ—subventricular zone.
Discussion
We have provided a cell atlas of the adult human brain SVZ. We have identified 4 NSC populations using transcriptomic and biological program analyses. aRG1, like fetal eRG, showed hallmarks of dormancy and localized to the SVZ astrocytic band (layer 3). aRG2 were similar to fetal oRG, showing features consistent with mouse NSC populations.8,9,10 npNSCs and opNSCs showed early lineage specification toward neuronal and oligodendroglial differentiation, respectively. We have also provided a trajectory analysis that demonstrates the intercellular and differentiation transition capacity of these NSC populations.
These data shed light on the potential differences between developmental and regenerative programs in adult SVZ NSCs and uncover the post-developmental potential of the adult human brain. These data have important implications for our understanding of neurodegenerative, neuroinflammatory, neuroimmune, and neurooncological processes.75,76
Importantly, although the cells captured in this study were isolated from SVZ regions that are normal on MRI, they may have been impacted by their proximity to brain tumors. It is possible that signals derived from the tumors may be relevant. If true, these signals may reveal the potential of NSCs to become activated, divide, and differentiate in the adult brain.
Finally, aRG1 cells exhibited a low transcriptome size, which can be indicative of a low-quality sample or biologically real. We believe it is biologically real here for the following reasons. First, the aRG1 (C5) cells in the adult SVZ dataset that have a low transcriptome size co-cluster in the merged dataset with radial glial cells are derived from a separate fetal dataset (Nowakowski et al., 2017).4 These cells, from both datasets, share a low transcriptome size and gene expression pattern, and they are both radial glia cells. Small transcriptome size is a feature of quiescent radial glial cells. Second, the aRG1 (C5) cells are derived from multiple samples, and they have less than 8 percent mitochondrial RNA, similar to other cells in this study. Last, we have confirmed the expression of aRG1 cells in the adult SVZ using proteomic marks in an immunolabelling experiment (Figure 2J).
Limitations of the study
A limitation of this study is the limited cell counts in the NSC population. This may be an accurate representation, or it may be artificially low because of technological limitations inherent in the single-cell RNA sequencing approach. Other single-cell RNA sequencing techniques or spatial transcriptomics may resolve this question.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Vimentin | EMD Millipore | BL202 |
| Sox9 | Abcam | ab185966 |
| GFAP | Abcam | ab4674 |
| Olig2 | EMD Millipore | ab9610 |
| ASCL1 | Abcam | ab74065 |
| TMEM1110 | Cell Signaling | E3E4T |
| EGFR | Abcam | ab32198 |
| CLDN5 | LS Bio | C352946 |
| ANXA1 | Abcam | ab214486 |
| MBP | Abcam | ab40390 |
| TAGLN2 | Abcam | Ab121146 |
| CD3D | ThermoFisher | MA5-32462 |
| Deposited data | ||
| Normalized data and raw counts, and loom files for the velocity analyses | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE248995 |
| Software and algorithms | ||
| Analysis Codes | This paper | https://github.com/kpetrecca/iscience2023/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kevin Petrecca (kevin.petrecca@mcgill.ca).
Data and code availability
-
•
Single cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication.
-
•
All original codes have been deposited of Github and are publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
All samples were obtained from surgeries performed at the Montreal Neurological Institute-Hospital under a research ethics board approved protocol NEU-10-066. Consent was given by all patients. Preoperative magnetic resonance imaging (MRI) was performed for surgical planning. SVZ regions that were normal on MRI and were to be removed during the brain tumor resection surgery were sampled.
Human normotypic rapid postmortem brain tissue samples were obtained from the Neuroimmunology Research Laboratory, Center de Recherche du Center Hospitalier de l'Université de Montréal under ethics approval protocol BH07.001. Brains were cut in the coronal plane and immersed in 3% paraformaldehyde or formalin for 1–2 weeks and then portions of their lateral ventricular walls were excised.
Method details
For each sample, an aliquot of cells was assessed for viability with calcein-AM and ethidium-homodimer1 (P/N L3224 ThermoFisher Scientific). Single-cell capture was performed following the Single Cell 3′ Reagent Kits v2 User Guide (CG0052 10X Genomics). The viability was >70% each sample. Cell barcodes and unique molecular identifiers (UMI) barcodes were demultiplexed and single-end reads aligned to the reference genome, GRCh38, using the CellRanger pipeline (10X Genomics).
Immunohistochemistry
Slides with 5μm thick sections were baked overnight at 60°C then de-paraffinized and rehydrated using a graded series of xylene and ethanol, respectively. For heat-mediated antigen retrieval, slides were incubated in citrate buffer at 125°C for 20 minutes in a decloaking chamber (BioCare Medical), followed by a cool-down period. Slides were rinsed in distilled water and phosphate buffered saline (PBS) and blocked using Protein Block (Spring Bioscience) for 30 minutes. Sections were incubated with primary antibodies diluted in 2% BSA in PBS overnight in a humid chamber at 4°C, washed using the immunofluorescence (IF) buffer (0.05% Tween-20 and 0.2% Triton X-100 in PBS) and incubated with secondary antibodies diluted in 2% BSA in PBS for 1 hour at room temperature. Following additional wash steps in IF buffer, the slides were mounted with ProLong™ Diamond Antifade Mountant with DAPI (Invitrogen) to counterstain cell nuclei. Fluorescent images were acquired using a ZEISS LSM 700 laser scanning confocal microscope.
Quantification and statistical analysis
Single-cell RNA sequencing data processing
We identified potential doublets using Scrublet, per sample with default parameters. Scrublet identified 3 cells in the NSC-like cluster and 0 cells in the UN1 cluster. The rate of potential doublets was low across the dataset (<%0.6), and they were not removed. The number of doublets per cluster are shown in Table S6. We applied similar approaches for adult SVZ (S, 10 834 cells), fetal (F, 12544 cells) and merged (M, 1965 cells) datasets. In the merged dataset C4 contained 185 cells, C5 contained 141 cells, C7 contained 128 cells, and C9 contained 80 cells. For each cell, counts (number of UMIs) per gene were normalized to the total counts of the cell and scaled by 10000. The data were natural-log transformed (log(counts+1); using Seurat::NormalizeData). All cells expressing more than 200 and less than 3500 genes, with lower than 8% expression for mitochondrial genes, were included for further analyses. The top most significant PCAs were selected (S:35;F:30;M:20) using jackstraw method and elbow visualization. We used ElbowPlot to find elbow points. In addition, JackStraw was used to confirm the elbow points. Cells were clustered based on a shared nearest neighbor (SNN), applying the Louvain algorithm as described in77 and implemented in,78 with the resolution of 0.6–0.8 (S:0.6;F:0.6;M:0.8). Markers, for each cluster/program compared to all other programs, were identified using the FindAllMarkers function with default Wilcoxon test.78 We also applied the FindAllMarkers function within the 4 NSC clusters to highlight the difference between the 4 clusters. Cell types annotations were assigned to each cluster using known markers shown in the Figure 1C, which were also consistent with markers previously reported.4 For visualization purposes, t-distributed stochastic neighbor embedding (t-SNE), and Uniform Manifold Approximation and Projection (UMAP) were used. Regarding the M dataset, since the single-cell platforms were different (Nowakowski et al., 2017 - Fluidigm C1, Couturier et al., 2020 and adult SVZ datasets - 10x), we regress out the effect of platforms (Seurat: vars.to.regress = platform). Regarding the velocity dataset, where we added progenitor cells from a non-cancer adult brain dataset (n = 322), we did not perform clustering and we only used the previous cell type annotations for each cell (n total = 4593; SVZ = 940, fetal = 3331).
Copy number alteration analysis
InferCNV (V.1.4.0)79 was used to detect large-scale chromosomal copy number alterations (CNAs) at single-cell level using gene expression data. Gene expression values were smoothed on a moving window of 101 genes and CNAs were analyzed relative to the reference cells. The three largest clusters, Oligodendrocyte, Macrophage/Microglia, were used as reference cells. The most variable chromosomes, those with highest number of alterations, were chromosomes 7 and 10, consistent with glioblastoma. Cells with any chromosomal alterations on chromosome 7 (gain) and chromosome 10 (loss) were defined as pathological cells and were excluded from further analyses.
Cell cycle analysis
We used gene signatures for G2M and S phase, as reported by Tirosh et al.,80 to quantify cell cycle phase score. Cells scoring low for both phases were considered to be in G1/G0 phase. We used CellCycleScoring function of Seurat with default parameters.
Ribosomal gene quantification
The proportion of ribosomal protein genes (Ribosomal Protein Large/Small subunit) were calculated using Seurat (PercentageFeatureSet (pattern = “RPS|RPL”)). Since the datasets were generated using different single-cell platforms, the analyses were performed separately for each dataset.
Gene set enrichment analysis
Differentially expression genes were identified for each of the 4 NSC clusters compared to all other SVZ cells using the FindAllMarkers function with default Wilcoxon test. The genes were ranked based on average fold change followed by gene set enrichment analysis (GSEA).81 We used curated gene sets of known human brain cell types from single-cell RNA-Seq studies as reported in Molecular Signatures Database (MsigDB,C8),82 as well as all signatures in fetal brains reported by Nowakowski et al.4 We used GSEA-Preranked (V4.1.0; Build 27) with default parameters and 10000 permutations.
Cell-cell interaction analysis
We used CellPhoneDB,83 a repository of curated receptors, ligands, and their interactions, to infer cell-cell communication networks between cell types. We provided CellPhoneDB v3.1.0 the RC-normalized count matrix by Seurat using genes with ENSEMBL ID along with cell type identities and ran the statistical analysis with the following parameters: –iteractions 10000 –result-precision 10 –threads 10 –threshold 0.1. We selected interactions with p value <0.05, and selected ligand/receptor interactions. Only interactions to NSCs were considered.
RNA velocity analysis
We performed velocity analysis using partition-based graph abstraction.84 In the first step, reads were aligned to the reference genome, GRCh38, using the CellRanger pipeline form 10X Genomics as previously described.75 Spliced and un-spliced counts were calculated and saved in loom file format using Velocyto package.85 The loom files from single samples were combined using scVelo package86 (V 0.1.16) and were normalized with default parameters using “scVelo.pp.filter_and_normalize”. First and second-order moments were calculated for each cell across its 30 nearest neighbors in PCA space (scVelo.pp.moments(n_pcs = 15, n_neighbors = 30). Velocity was estimated and the velocity graph was constructed using default parameters in “stochastic” mode (scv.tl.velocity(mode = ‘stochastic’); scv.tl.velocity_graph). PAGA was used to quantify the transition between clusters (partitions) using PAGA algorithm as implemented in scVelo with the default parameters (scv.tl.paga(groups = ‘clusters’,use_rna_velocity = True)). Transitions usually ranged between 0 and 1, and a higher value means a better transition confidence. For visualization we multiplied these values by 100. All transitions are shown in Figure 5A, and those passing a transition confidence threshold (>10; default = 5) for nodes from or to NSCs are in Figure 5B.
Pathway and biological process enrichment analysis
Pathway and biological process enrichment analysis was performed using Metascape (multi-list enrichment analysis).87 For each gene list a maximum of the top 500 differentially expressed genes were used for each cell type. Pathway and biological process enrichment analysis was carried out with the following ontology sources: KEGG Pathway, GO Biological Processes, Reactome Gene Sets, Canonical Pathways, and WikiPathways. All genes in the genome were used as the enrichment background. Terms with a p value <0.01, a minimum count of 3, and an enrichment factor >1.5 were collected. p values were calculated based on the cumulative hypergeometric distribution, and q-values were calculated using the Benjamini-Hochberg procedure to account for multiple tests.
Additional resources
Antibodies
The following antibodies were used for immunolabelling: VIM (BL202 – EMD Millipore); SOX9 (ab185966 - Abcam); GFAP (ab4674 – Abcam); OLIG2 (ab9610 – EMD Millipore); ASCL1 (ab74065 – Abcam); TMEM119 (E3E4T – Cell Signaling); EGFR (ab32198 – Abcam), CLDN5 (LS-C352946 – LS Bio); ANXA1 (ab214486 – Abcam); MBP (ab40390 – Abcam); TAGLN2 (ab121146 – Abcam); CD3D (MA5-32462 – ThermoFisher).
Acknowledgments
We would like to thank Carmen Sabau for her contribution to the administrative work of the project, and Rozica Bolovan for technical support.
Funding
Cancer Research Society70813 (KP).
Canadian Cancer Research Institute702411 (KP).
Brain Tumour Foundation of Canada62766 (KP).
TARGiT Foundation (KP).
A Brilliant Night Foundation (KP).
Argento Family Group Ercole (KP).
KP is supported by a clinician-scientist salary award from the Fonds de recherche du Québec and the William Feindel Chair in Neuro-Oncology.
Author contributions
Conceptualization: S.B., K.P.; methodology: S.B., J.N., R.A., K.P.; investigation: S.B., J.N., R.A., P.L., M.L., A.D., A.N.M., M.S., A.P., J.A., M.C.G., K.P.; visualization: S.B., J.N., P.L., A.D., K.P.; funding acquisition: K.P.; project administration: K.P.; supervision: K.P.; writing – original draft: S.B., J.N., P.L., K.P.; writing – review & editing: S.B., J.N., P.L., M.L., K.P.
Declaration of interests
The authors declare no competing interests.
Published: February 27, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109342.
Supplemental information
References
- 1.Boldrini M., Fulmore C.A., Tartt A.N., Simeon L.R., Pavlova I., Poposka V., Rosoklija G.B., Stankov A., Arango V., Dwork A.J., et al. Human Hippocampal Neurogenesis Persists throughout Aging. Cell Stem Cell. 2018;22:589–599.e5. doi: 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durante M.A., Kurtenbach S., Sargi Z.B., Harbour J.W., Choi R., Kurtenbach S., Goss G.M., Matsunami H., Goldstein B.J. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat. Neurosci. 2020;23:323–326. doi: 10.1038/s41593-020-0587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eze U.C., Bhaduri A., Haeussler M., Nowakowski T.J., Kriegstein A.R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 2021;24:584–594. doi: 10.1038/s41593-020-00794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D., et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science (New York, N.Y.) 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollen A.A., Nowakowski T.J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C.R., Shuga J., Liu S.J., Oldham M.C., Diaz A., et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quiñones-Hinojosa A., Sanai N., Soriano-Navarro M., Gonzalez-Perez O., Mirzadeh Z., Gil-Perotin S., Romero-Rodriguez R., Berger M.S., Garcia-Verdugo J.M., Alvarez-Buylla A. Cellular composition and cytoarchitecture of the adult human subventricular zone: a niche of neural stem cells. J. Comp. Neurol. 2006;494:415–434. doi: 10.1002/cne.20798. [DOI] [PubMed] [Google Scholar]
- 7.Sanai N., Tramontin A.D., Quiñones-Hinojosa A., Barbaro N.M., Gupta N., Kunwar S., Lawton M.T., McDermott M.W., Parsa A.T., Manuel-García Verdugo J., et al. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427:740–744. doi: 10.1038/nature02301. [DOI] [PubMed] [Google Scholar]
- 8.Cebrian-Silla A., Nascimento M.A., Redmond S.A., Mansky B., Wu D., Obernier K., Romero Rodriguez R., Gonzalez-Granero S., García-Verdugo J.M., Lim D.A., Álvarez-Buylla A. Single-cell analysis of the ventricular-subventricular zone reveals signatures of dorsal and ventral adult neurogenesis. Elife. 2021;10 doi: 10.7554/eLife.67436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couturier C.P., Ayyadhury S., Le P.U., Nadaf J., Monlong J., Riva G., Allache R., Baig S., Yan X., Bourgey M., et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 2020;11:3406. doi: 10.1038/s41467-020-17186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delgado A.C., Maldonado-Soto A.R., Silva-Vargas V., Mizrak D., von Känel T., Tan K.R., Paul A., Madar A., Cuervo H., Kitajewski J., et al. Release of stem cells from quiescence reveals gliogenic domains in the adult mouse brain. Science (New York, N.Y.) 2021;372:1205–1209. doi: 10.1126/science.abg8467. [DOI] [PubMed] [Google Scholar]
- 11.Berg D.A., Su Y., Jimenez-Cyrus D., Patel A., Huang N., Morizet D., Lee S., Shah R., Ringeling F.R., Jain R., et al. A Common Embryonic Origin of Stem Cells Drives Developmental and Adult Neurogenesis. Cell. 2019;177:654–668.e15. doi: 10.1016/j.cell.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonfanti L., Peretto P. Radial glial origin of the adult neural stem cells in the subventricular zone. Prog. Neurobiol. 2007;83:24–36. doi: 10.1016/j.pneurobio.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y., Yang M., Yu H., Wang Y., Wu X., Yong J., Mao Y., Cui Y., Fan X., Wen L., et al. Heterogeneity of glial progenitor cells during the neurogenesis-to-gliogenesis switch in the developing human cerebral cortex. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108788. [DOI] [PubMed] [Google Scholar]
- 14.Yuzwa S.A., Borrett M.J., Innes B.T., Voronova A., Ketela T., Kaplan D.R., Bader G.D., Miller F.D. Developmental Emergence of Adult Neural Stem Cells as Revealed by Single-Cell Transcriptional Profiling. Cell Rep. 2017;21:3970–3986. doi: 10.1016/j.celrep.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez C.G., Teixeira F.K., Czech B., Preall J.B., Zamparini A.L., Seifert J.R.K., Malone C.D., Hannon G.J., Lehmann R. Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell. 2016;18:276–290. doi: 10.1016/j.stem.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulken B.W., Leeman D.S., Boutet S.C., Hebestreit K., Brunet A. Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cell Rep. 2017;18:777–790. doi: 10.1016/j.celrep.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borrett M.J., Innes B.T., Jeong D., Tahmasian N., Storer M.A., Bader G.D., Kaplan D.R., Miller F.D. Single-Cell Profiling Shows Murine Forebrain Neural Stem Cells Reacquire a Developmental State when Activated for Adult Neurogenesis. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108022. [DOI] [PubMed] [Google Scholar]
- 18.Li Z., Tyler W.A., Zeldich E., Santpere Baró G., Okamoto M., Gao T., Li M., Sestan N., Haydar T.F. Transcriptional priming as a conserved mechanism of lineage diversification in the developing mouse and human neocortex. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doetsch F., Caillé I., Lim D.A., García-Verdugo J.M., Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 20.Long K.R., Huttner W.B. How the extracellular matrix shapes neural development. Open Biol. 2019;9 doi: 10.1098/rsob.180216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watters A.K., Rom S., Hill J.D., Dematatis M.K., Zhou Y., Merkel S.F., Andrews A.M., Cena J., Potula R., Skuba A., et al. Identification and dynamic regulation of tight junction protein expression in human neural stem cells. Stem Cells Dev. 2015;24:1377–1389. doi: 10.1089/scd.2014.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga B.V., Faiz M., Pivonkova H., Khelifi G., Yang H., Gao S., Linderoth E., Zhen M., Karadottir R.T., Hussein S.M., Nagy A. Signal requirement for cortical potential of transplantable human neuroepithelial stem cells. Nat. Commun. 2022;13:2844. doi: 10.1038/s41467-022-29839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basak O., Krieger T.G., Muraro M.J., Wiebrands K., Stange D.E., Frias-Aldeguer J., Rivron N.C., van de Wetering M., van Es J.H., van Oudenaarden A., et al. Troy+ brain stem cells cycle through quiescence and regulate their number by sensing niche occupancy. SA. 2018;115:E610–E619. doi: 10.1073/pnas.1715911114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao Q., Hill M.C., Chen F., Mo Q., Ku A.T., Ramos C., Sock E., Lefebvre V., Nguyen H. SOX11 and SOX4 drive the reactivation of an embryonic gene program during murine wound repair. Nat. Commun. 2019;10:4042. doi: 10.1038/s41467-019-11880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Lin L., Lai H., Parada L.F., Lei L. Transcription factor Sox11 is essential for both embryonic and adult neurogenesis. Dev. Dyn. 2013;242:638–653. doi: 10.1002/dvdy.23962. [DOI] [PubMed] [Google Scholar]
- 26.Johnson M.B., Wang P.P., Atabay K.D., Murphy E.A., Doan R.N., Hecht J.L., Walsh C.A. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anantha J., Goulding S.R., Wyatt S.L., Concannon R.M., Collins L.M., Sullivan A.M., O'Keeffe G.W. STRAP and NME1 Mediate the Neurite Growth-Promoting Effects of the Neurotrophic Factor GDF5. iScience. 2020;23 doi: 10.1016/j.isci.2020.101457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farioli-Vecchioli S., Micheli L., Saraulli D., Ceccarelli M., Cannas S., Scardigli R., Leonardi L., Cinà I., Costanzi M., Ciotti M.T., et al. Btg1 is Required to Maintain the Pool of Stem and Progenitor Cells of the Dentate Gyrus and Subventricular Zone. Front. Neurosci. 2012;6:124. doi: 10.3389/fnins.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur N., Han W., Li Z., Madrigal M.P., Shim S., Pochareddy S., Gulden F.O., Li M., Xu X., Xing X., et al. Neural Stem Cells Direct Axon Guidance via Their Radial Fiber Scaffold. Neuron. 2020;107:1197–1211.e9. doi: 10.1016/j.neuron.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanton-Turcotte D., Hsu K., Moore S.A., Yamada M., Fawcett J.P., Iulianella A. Mllt11 Regulates Migration and Neurite Outgrowth of Cortical Projection Neurons during Development. J. Neurosci. 2022;42:3931–3948. doi: 10.1523/jneurosci.0124-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kase Y., Sato T., Okano Y., Okano H. The GADD45G/p38 MAPK/CDC25B signaling pathway enhances neurite outgrowth by promoting microtubule polymerization. iScience. 2022;25 doi: 10.1016/j.isci.2022.104089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsunaga E., Nambu S., Oka M., Iriki A. Comparative analysis of developmentally regulated expressions of Gadd45a, Gadd45b, and Gadd45g in the mouse and marmoset cerebral cortex. Neuroscience. 2015;284:566–580. doi: 10.1016/j.neuroscience.2014.10.032. [DOI] [PubMed] [Google Scholar]
- 33.Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. SA. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chamling X., Kallman A., Fang W., Berlinicke C.A., Mertz J.L., Devkota P., Pantoja I.E.M., Smith M.D., Ji Z., Chang C., et al. Single-cell transcriptomic reveals molecular diversity and developmental heterogeneity of human stem cell-derived oligodendrocyte lineage cells. Nat. Commun. 2021;12:652. doi: 10.1038/s41467-021-20892-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marques S., van Bruggen D., Vanichkina D.P., Floriddia E.M., Munguba H., Väremo L., Giacomello S., Falcão A.M., Meijer M., Björklund Å.K., et al. Transcriptional Convergence of Oligodendrocyte Lineage Progenitors during Development. Dev. Cell. 2018;46:504–517.e7. doi: 10.1016/j.devcel.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang S.H., Fukaya M., Yang J.K., Rothstein J.D., Bergles D.E. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marques S., Zeisel A., Codeluppi S., van Bruggen D., Mendanha Falcão A., Xiao L., Li H., Häring M., Hochgerner H., Romanov R.A., et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science (New York, N.Y.) 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Bruggen D., Pohl F., Langseth C.M., Kukanja P., Lee H., Albiach A.M., Kabbe M., Meijer M., Linnarsson S., Hilscher M.M., et al. Developmental landscape of human forebrain at a single-cell level identifies early waves of oligodendrogenesis. Dev. Cell. 2022;57:1421–1436.e5. doi: 10.1016/j.devcel.2022.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Chapman T.W., Olveda G.E., Bame X., Pereira E., Hill R.A. Oligodendrocyte death initiates synchronous remyelination to restore cortical myelin patterns in mice. Nat. Neurosci. 2023;26:555–569. doi: 10.1038/s41593-023-01271-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fard M.K., van der Meer F., Sánchez P., Cantuti-Castelvetri L., Mandad S., Jäkel S., Fornasiero E.F., Schmitt S., Ehrlich M., Starost L., et al. BCAS1 expression defines a population of early myelinating oligodendrocytes in multiple sclerosis lesions. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aam7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrington E.P., Bergles D.E., Calabresi P.A. Immune cell modulation of oligodendrocyte lineage cells. Neurosci. Lett. 2020;715 doi: 10.1016/j.neulet.2019.134601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moyon S., Dubessy A.L., Aigrot M.S., Trotter M., Huang J.K., Dauphinot L., Potier M.C., Kerninon C., Melik Parsadaniantz S., Franklin R.J.M., Lubetzki C. Demyelination causes adult CNS progenitors to revert to an immature state and express immune cues that support their migration. J. Neurosci. 2015;35:4–20. doi: 10.1523/jneurosci.0849-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao J., O'Day D.R., Pliner H.A., Kingsley P.D., Deng M., Daza R.M., Zager M.A., Aldinger K.A., Blecher-Gonen R., Zhang F., et al. Vol. 370. Science; New York, N.Y: 2020. A Human Cell Atlas of Fetal Gene Expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan X., Dong J., Zhong S., Wei Y., Wu Q., Yan L., Yong J., Sun L., Wang X., Zhao Y., et al. Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res. 2018;28:730–745. doi: 10.1038/s41422-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong S., Zhang S., Fan X., Wu Q., Yan L., Dong J., Zhang H., Li L., Sun L., Pan N., et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–528. doi: 10.1038/nature25980. [DOI] [PubMed] [Google Scholar]
- 46.Spitzer S.O., Sitnikov S., Kamen Y., Evans K.A., Kronenberg-Versteeg D., Dietmann S., de Faria O., Jr., Agathou S., Káradóttir R.T. Oligodendrocyte Progenitor Cells Become Regionally Diverse and Heterogeneous with Age. Neuron. 2019;101:459–471.e5. doi: 10.1016/j.neuron.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boccazzi M., Raffaele S., Fumagalli M. Not only myelination: the immune-inflammatory functions of oligodendrocytes. Neural Regen. Res. 2022;17:2661–2663. doi: 10.4103/1673-5374.342678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Psenicka M.W., Smith B.C., Tinkey R.A., Williams J.L. Connecting Neuroinflammation and Neurodegeneration in Multiple Sclerosis: Are Oligodendrocyte Precursor Cells a Nexus of Disease? Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.654284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrelli F., Scandella V., Montessuit S., Zamboni N., Martinou J.C., Knobloch M. Mitochondrial pyruvate metabolism regulates the activation of quiescent adult neural stem cells. Sci. Adv. 2023;9 doi: 10.1126/sciadv.add5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharifi S., da Costa H.F.R., Bierhoff H. The circuitry between ribosome biogenesis and translation in stem cell function and ageing. Mech. Ageing Dev. 2020;189 doi: 10.1016/j.mad.2020.111282. [DOI] [PubMed] [Google Scholar]
- 51.Knobloch M., Pilz G.A., Ghesquière B., Kovacs W.J., Wegleiter T., Moore D.L., Hruzova M., Zamboni N., Carmeliet P., Jessberger S. A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates Adult Neural Stem Cell Activity. Cell Rep. 2017;20:2144–2155. doi: 10.1016/j.celrep.2017.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wani G.A., Sprenger H.G., Ndoci K., Chandragiri S., Acton R.J., Schatton D., Kochan S.M.V., Sakthivelu V., Jevtic M., Seeger J.M., et al. Metabolic control of adult neural stem cell self-renewal by the mitochondrial protease YME1L. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110370. [DOI] [PubMed] [Google Scholar]
- 53.Iwata R., Casimir P., Vanderhaeghen P. Mitochondrial dynamics in postmitotic cells regulate neurogenesis. Science (New York, N.Y.) 2020;369:858–862. doi: 10.1126/science.aba9760. [DOI] [PubMed] [Google Scholar]
- 54.Khacho M., Clark A., Svoboda D.S., Azzi J., MacLaurin J.G., Meghaizel C., Sesaki H., Lagace D.C., Germain M., Harper M.E., et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 55.Andrews M.G., Subramanian L., Salma J., Kriegstein A.R. How mechanisms of stem cell polarity shape the human cerebral cortex. Nat. Rev. Neurosci. 2022;23:711–724. doi: 10.1038/s41583-022-00631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta M.K., Papay R.S., Jurgens C.W.D., Gaivin R.J., Shi T., Doze V.A., Perez D.M. alpha1-Adrenergic receptors regulate neurogenesis and gliogenesis. Mol. Pharmacol. 2009;76:314–326. doi: 10.1124/mol.109.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samarasinghe R.A., Di Maio R., Volonte D., Galbiati F., Lewis M., Romero G., DeFranco D.B. Nongenomic glucocorticoid receptor action regulates gap junction intercellular communication and neural progenitor cell proliferation. SA. 2011;108:16657–16662. doi: 10.1073/pnas.1102821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tavazoie M., Van der Veken L., Silva-Vargas V., Louissaint M., Colonna L., Zaidi B., Garcia-Verdugo J.M., Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lacar B., Young S.Z., Platel J.C., Bordey A. Gap junction-mediated calcium waves define communication networks among murine postnatal neural progenitor cells. Eur. J. Neurosci. 2011;34:1895–1905. doi: 10.1111/j.1460-9568.2011.07901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urbán N., Blomfield I.M., Guillemot F. Quiescence of Adult Mammalian Neural Stem Cells: A Highly Regulated Rest. Neuron. 2019;104:834–848. doi: 10.1016/j.neuron.2019.09.026. [DOI] [PubMed] [Google Scholar]
- 61.Aguirre A., Gallo V. Postnatal neurogenesis and gliogenesis in the olfactory bulb from NG2-expressing progenitors of the subventricular zone. J. Neurosci. 2004;24:10530–10541. doi: 10.1523/jneurosci.3572-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paez-Gonzalez P., Asrican B., Rodriguez E., Kuo C.T. Identification of distinct ChAT+ neurons and activity-dependent control of postnatal SVZ neurogenesis. Nat. Neurosci. 2014;17:934–942. doi: 10.1038/nn.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lie D.C., Colamarino S.A., Song H.J., Désiré L., Mira H., Consiglio A., Lein E.S., Jessberger S., Lansford H., Dearie A.R., Gage F.H. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 64.Sun F.L., Wang W., Zuo W., Xue J.L., Xu J.D., Ai H.X., Zhang L., Wang X.M., Ji X.M. Promoting neurogenesis via Wnt/β-catenin signaling pathway accounts for the neurorestorative effects of morroniside against cerebral ischemia injury. Eur. J. Pharmacol. 2014;738:214–221. doi: 10.1016/j.ejphar.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 65.Dahlhoff M., Wolf E., Schneider M.R. The ABC of BTC: structural properties and biological roles of betacellulin. Semin. Cell Dev. Biol. 2014;28:42–48. doi: 10.1016/j.semcdb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 66.Singh S.S., Chauhan S.B., Kumar A., Kumar S., Engwerda C.R., Sundar S., Kumar R. Amphiregulin in cellular physiology, health, and disease: Potential use as a biomarker and therapeutic target. J. Cell. Physiol. 2022;237:1143–1156. doi: 10.1002/jcp.30615. [DOI] [PubMed] [Google Scholar]
- 67.He Z., Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J. Mol. Med. 2003;81:600–612. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- 68.He Z., Ong C.H.P., Halper J., Bateman A. Progranulin is a mediator of the wound response. Nat. Med. 2003;9:225–229. doi: 10.1038/nm816. [DOI] [PubMed] [Google Scholar]
- 69.Zheng Y., Li X., Qian X., Wang Y., Lee J.H., Xia Y., Hawke D.H., Zhang G., Lyu J., Lu Z. Secreted and O-GlcNAcylated MIF binds to the human EGF receptor and inhibits its activation. Nat. Cell Biol. 2015;17:1348–1355. doi: 10.1038/ncb3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang C., Wang M., Wang P., Wang L., Wu Q., Guo W. Neural Stem Cells Behave as a Functional Niche for the Maturation of Newborn Neurons through the Secretion of PTN. Neuron. 2019;101:32–44.e6. doi: 10.1016/j.neuron.2018.10.051. [DOI] [PubMed] [Google Scholar]
- 71.Coles C.H., Mitakidis N., Zhang P., Elegheert J., Lu W., Stoker A.W., Nakagawa T., Craig A.M., Jones E.Y., Aricescu A.R. Structural basis for extracellular cis and trans RPTPσ signal competition in synaptogenesis. Nat. Commun. 2014;5:5209. doi: 10.1038/ncomms6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lang B.T., Cregg J.M., DePaul M.A., Tran A.P., Xu K., Dyck S.M., Madalena K.M., Brown B.P., Weng Y.L., Li S., et al. Modulation of the proteoglycan receptor PTPσ promotes recovery after spinal cord injury. Nature. 2015;518:404–408. doi: 10.1038/nature13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ayyubova G. TREM2 signalling as a multifaceted player in brain homoeostasis and a potential target for Alzheimer's disease treatment. Eur. J. Neurosci. 2023;57:718–733. doi: 10.1111/ejn.15914. [DOI] [PubMed] [Google Scholar]
- 74.Mazaheri F., Snaidero N., Kleinberger G., Madore C., Daria A., Werner G., Krasemann S., Capell A., Trümbach D., Wurst W., et al. TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep. 2017;18:1186–1198. doi: 10.15252/embr.201743922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Couturier C.P., Nadaf J., Li Z., Baig S., Riva G., Le P., Kloosterman D.J., Monlong J., Nkili Meyong A., Allache R., et al. Glioblastoma scRNAseq Shows Treatment-induced, Immune-dependent Rise In Mesenchymal Cancer Cells, and Structural Variants in Distal Neural Stem Cells. Neuro Oncol. 2022;24:1494–1508. doi: 10.1093/neuonc/noac085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee J.H., Lee J.E., Kahng J.Y., Kim S.H., Park J.S., Yoon S.J., Um J.-Y., Kim W.K., Lee J.-K., Park J., et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560:243–247. doi: 10.1038/s41586-018-0389-3. [DOI] [PubMed] [Google Scholar]
- 77.Waltman L., van Eck N.J. A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B. 2013;86:471. doi: 10.1140/epjb/e2013-40829-0. [DOI] [Google Scholar]
- 78.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tickle T., Tirosh I., Georgescu C., Brown M., Haas B. Klarman Cell Observatory, Broad Institute of MIT and Harvard; 2019. inferCNV of the Trinity CTAT Project. [Google Scholar]
- 80.Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science (New York, N.Y.) 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. SA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 84.Wolf F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Göttgens B., Rajewsky N., Simon L., Theis F.J. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M.E., Lönnerberg P., Furlan A., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 87.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication.
-
•
All original codes have been deposited of Github and are publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.