Abstract
The host immune system status remains an unresolved mystery among several malignancies. An immune-compromised state or smart immune-surveillance tactics orchestrated by cancer cells are the primary cause of cancer invasion and metastasis. Taking a closer look at the tumour-immune microenvironment, a complex network and crosstalk between infiltrating immune cells and cancer cells mediated by cytokines, chemokines, exosomal mediators and shed ligands are present. Cytokines such as interleukins can influence all components of the tumour microenvironment (TME), consequently promoting or suppressing tumour invasion based on their secreting source. Interleukin-10 (IL-10) is an interlocked cytokine that has been associated with several types of malignancies and proved to have paradoxical effects. IL-10 has multiple functions on cellular and non-cellular components within the TME. In this review, the authors shed the light on the regulatory role of IL-10 in the TME of several malignant contexts. Moreover, detailed epigenomic and pharmacogenomic approaches for the regulation of IL-10 were presented and discussed.
Keywords: Cancer, Cytokines, IL-10, Interleukins, Non-coding RNAs, Pharmacogenomics, Tumour-microenvironment
Introduction
The use of immunotherapy as a novel therapeutic approach in preventing cancer has become widespread (Ref. 1). Immune checkpoint blockade modalities targeting PD-1 and CTLA-4 provide long-lasting immune responses with established therapeutic benefits for some cancer patients (Refs 2–6). Although, targeting cytokines is considered a crucial approach in immunotherapy as evidenced in the treatment of solid tumours, such as renal cell carcinoma (RCC) and melanoma, only interferons (IFNs) and IL-2 have been approved by Food and Drug Administration (FDA) for use as cancer therapies (Ref. 7).
IL-10 is considered one of the very promising targets for immunotherapy; however, its controversial role in carcinogenesis hinders the applicability of benefiting from its blockade in cancer treatment (Ref. 8). IL-10 has been shown to possess both anti- and pro-inflammatory roles in cancer (Ref. 9). The intensity of the immunological response to both self and foreign antigens is reduced by IL-10. In light of this, IL-10 signalling blockage improves vaccine-induced T-cell responses and tumour growth inhibition (Ref. 10). On the other hand, tumour regression is also induced by exogenous IL-10, particularly PEGylated (PEG)-IL-10 (Ref. 11). This paradoxical data urges the need to investigate the role of pharmacogenomics, epigenetics and genetic variants in IL-10 and its receptor to identify those patients that might benefit from IL-10 targeted therapies. In this review, the authors will address the role of IL-10 in cancer, the currently available IL-10-based immunotherapy, the epigenetic regulation of IL-10 and the single nucleotide polymorphisms (SNPs) present in IL-10 that might influence patient responses to therapy.
The tumour microenvironment
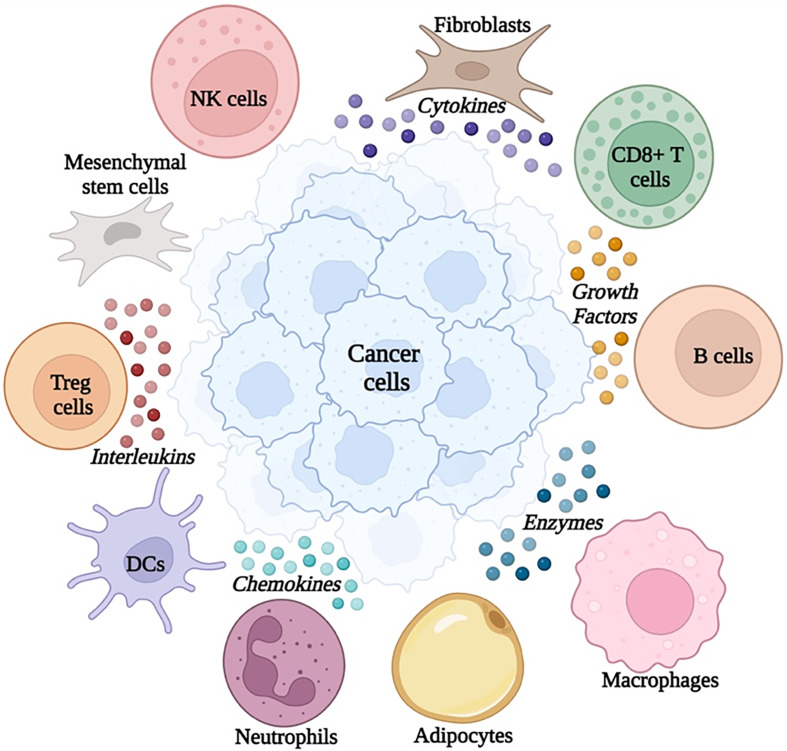
Cancer definition has been revolutionized over the past few decades from the concept of being abnormal cells to a plethora of complex network that is made up of both neoplastic cells with their surrounding stroma (Refs 1, 4, 6, 12). The multifaceted dynamic milieu of cellular components along with non-cellular compartments portrays what is now known as the tumour microenvironment (TME) (Refs 6, 13, 14). Such a microenvironment could control the aggressiveness, rate of growth and metastatic potential of the tumour (Refs 15–18). These cellular components include immune cells such as T lymphocytes (Refs 19–24), regulatory T cells (Tregs) (Ref. 25), B lymphocytes, natural killer (NK) cells (Refs 16, 26–29), mesenchymal stem cells (Refs 30, 31), tumour-associated-macrophages (Refs 32, 33), tumour-associated neutrophils (Refs 34, 35), dendritic cells (DCs) (Ref. 36) and non-immune cells such as pericytes (Ref. 37), adipocytes (Refs 38, 39), myeloid-derived suppressor cells (MDSCs) (Refs 40–42) and cancer-associated fibroblastic cells (Refs 43, 44). Interestingly, these immune cells drive the production of soluble components that include cytokines, chemokines, growth factors and extra-cellular remodelling enzymes (Refs 27, 28). Such mediators, particularly cytokines, assist in the communication between the cellular TME components and cancer cells as shown in Figure 1 (Refs 45, 46).
Figure 1.
Snapshot of cellular and non-cellular components of the tumour microenvironment
Interleukin-10 (IL-10)
One of these cytokines is the paradoxical interleukin ‘IL-10’, which remains an integral part of several malignancies, and regulates the secretion of other cytokines. This pleiotropic cytokine was characterized early in the late 1980s and was named cytokine synthesis inhibitory factor (Refs 47, 48). Later on, six immune mediators (IL-10, IL-19, IL-20, IL-22, IL-24 and IL-26) were grouped into the IL-10 family of cytokines based on their similarities with respect to the structure and location of their encoding genes, their primary and secondary protein structures and the receptor complexes (Refs 49–51). Out of these six members, IL-10 has been recognized as a major member mediating different functions within the immune system and cancer cells (Ref. 52).
Paradoxical role of IL-10 in oncology
IL-10 produced by immune cells
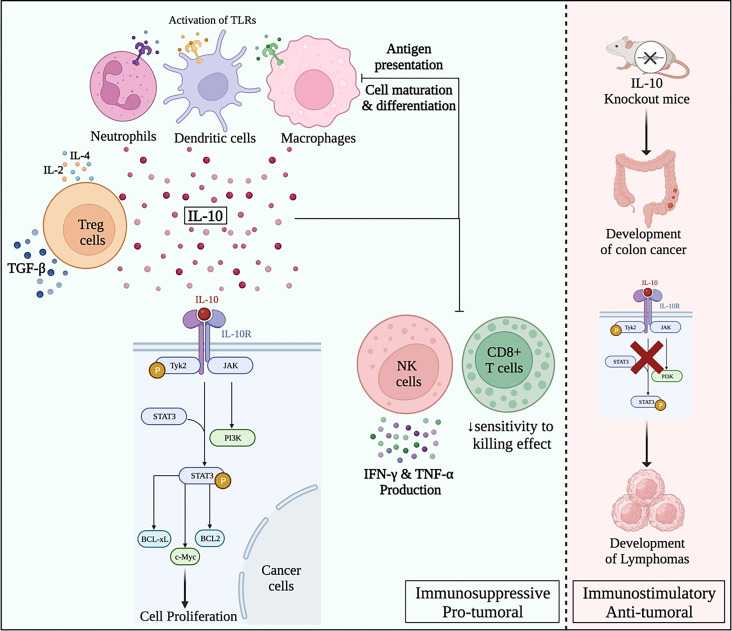
IL-10 has also been causally linked to immunity in both the innate and adaptive immune arms. Different triggers have been shown to induce IL-10 production in various immune cells (Ref. 53). The main source of IL-10 appears to be monocytes, and different T-cell subsets (Ref. 54). Moreover, DCs, B cells, NK cells, mast cells, as well as neutrophils, and eosinophils can also synthesize IL-10 (Ref. 54). During infection, macrophages are considered a major source of IL-10. Several toll-like receptors (TLRs), including TLR2, TLR4, TLR5, TLR7 and TLR9 have been shown to induce IL-10 production in macrophages and DCs (Refs 55–63). Also, IL-10 production in DCs is enhanced by the co-activation of TLR2 and Dectin-1 (Ref. 64). Following exposure to IL-10, DCs can initiate the development of regulatory T cells (Tregs) that limit these effector responses (Refs 65, 66). B cells also express several TLRs which have been shown to promote IL-10 production including TLR2, TLR4 or TLR9 (Refs 67–69). Nonetheless, it is also worth mentioning that IFN-α augments IL-10 production if combined with TLR agonists from B cells (Refs 70, 71). Additionally, neutrophils produce IL-10 in response to TLR and C-type lectin co-activation through myeloid differentiation primary response 88 (MyD88) and spleen tyrosine kinase (SYK), respectively (Ref. 72).
The key producer of IL-10 is Treg cells that produce other immunoregulatory cytokines, such as TGF-β (Ref. 73). The production and action of both cytokines IL-10 and TGF-β are involved in a positive feedback loop (Ref. 74). Concerning the mechanism of IL-10 production from Tregs, it has been shown that IL-2 and IL-4 induce IL-10 production from Tregs (Refs 75–77). Additionally, a study concluded that TGF-β is required for the differentiation and production of IL-10 from Tregs (Ref. 78). IL-2 and IL-27 are responsible for inducing IL-10 expression in cytotoxic CD8+ T cells (Ref. 79). However, IL-12 and IL-23 prime CD8+ and CD4+ T cells for IL-10 production (Refs 80–82).
Some studies reported IL-10 immunosuppressive effects such as inhibiting IFN-γ and TNF-α production by NK cells in-vitro (Ref. 83). However, other studies reported IL-10 immunostimulatory effects via the promotion of NK cell cytotoxicity in preclinical models (Refs 9, 84). Adding to the complexity of this master cytokine, one of the studies has shown that the exposure of malignant cells to IL-10 resulted in a reduction in their sensitivity to cytotoxic T cells but an increase in NK cell cytotoxicity (Ref. 85). This might suggest that IL-10 contributes to fighting malignant cells by stimulating the immune innate arm (Ref. 86).
As mentioned earlier, one of the main drivers of IL-10 expression in many immune cells is TLR signalling (Ref. 56). TLR ligation leads to the activation of several downstream pathways, including the mitogen-activated protein kinase (MAPK) pathway and the phosphoinositide 3-kinases (PI3K)/AKT pathways (Ref. 87). Activation of the MAPK and downstream extracellular-signal-regulated kinase (ERK1 and ERK2) are critical for IL-10 production in macrophages and DCs in response to several TLR activators (Refs 58, 62, 88, 89). The MAPK pathway eventually results in the activation of several transcription family members such as the activator protein-1 (AP-1) which activates IL-10 transcription (Refs 55, 58, 62, 90). Moreover, ERK and p38 also contribute to IL-10 production in TLR-stimulated macrophages, monocytes, and DCs (Refs 57, 89–92). Both ERK and p38 may function cooperatively in their regulation of IL-10 production, through their joined activation of mitogen and stress-activated protein kinases (MSK1 and MSK2) which promote IL-10 production in TLR-stimulated macrophages. Downstream of MSK1 and MSK2 are the transcription factors, cAMP-response element binding protein (CREB), and AP-1, which also bind and transactivate the IL-10 promoter (Refs 93–95). Moreover, it is worth mentioning that both ERK and p38 were shown also to directly phosphorylate Sp1, one of the IL-10 transcription factors (Refs 96, 97).
The phosphatidylinositol-3-kinase (PI3K/AKT) pathway also contributes to IL-10 expression in myeloid cells either by antagonizing glycogen synthase kinase 3 beta (GSK3-β), a constitutively active kinase that inhibits the production of IL-10 or through ERK and mammalian target of rapamycin (mTOR) and STAT-3 activation (Refs 98–100).
IL-10 produced by cancer cells
IL-10 has been linked to many types of cancers such as gastric cancer (Ref. 101), cervical cancer (Ref. 102), lung cancer (Ref. 103), breast cancer (Ref. 104), colon adenocarcinoma (Ref. 105), head and neck cancer (Ref. 106), oesophageal cancer, nasopharyngeal cancer, oral cancer (Ref. 107) and colorectal cancer (Ref. 108). Its role in tumourigenesis is reported to be controversial where it could be a tumour suppressor or promoter. However, due to the complex nature of IL-10, its role in shaping the TME remains a gap that needs further research. Most of the literature is directed towards presenting the pro-tumoural activity of IL-10 in different oncological settings. This could be through the positive feedback loop with STAT-3, as IL-10 has been shown to activate STAT-3 resulting in the upregulation of B-cell lymphoma 2 (BCL-2) or B-cell lymphoma-extra-large (BCL-xL), and stimulation of cell proliferation by cyclins D1, D2, B, and proto-oncogene c-Myc, thus contributing to cancer progression (Ref. 93). On the other hand, IL-10 immunosuppressive activity has been reported on macrophages and DCs, where it was found to dampen antigen presentation, cell maturation, and differentiation resulting in tumour immune evasion as shown in Figure 2 (Ref. 109). Several studies have examined the role of IL-10 in different types of malignancies as listed in Table 1 below.
Figure 2.
Paradoxical pro- and anti-tumour roles of IL-10 in oncology
Table 1.
Role of IL-10 in different solid malignancies
| Cancer type | Effect of IL-10 | Reference |
|---|---|---|
| Colon cancer | Serum level of IL-10 was correlated with reduced cytotoxic activity of CD8 + T cells in MC38- mouse colon cancer model | (Ref. 110) |
| Gastric cancer | Induction of the autocrine secretion of IL-10 from DCs resulted in the maturation of DCs yet the antigen delivery activity was inhibited, leading to evasion of the host immune surveillance and the development of gastric cancer. | (Ref. 111) |
| Breast cancer | Neutralizing IL-10 using anti-IL-10 antibody resulted in the attenuation of STAT3 activation and decreased Bcl-2 mRNA expression and reduced BC cells chemoresistance | (Refs 112, 113) |
| Ovarian cancer | Recombinant IL-10 enhanced cellular migration verifying the pro-tumoural activities of IL-10 in ovarian cancer. | (Ref. 114) |
| Bladder cancer | Paracrine secretion of IL-10 by bladder cancer cells was found to paralyse most of the host immune-regulatory actions to concur the tumour. | (Ref. 115) |
| Cervical cancer | Evaluation of IL-10 expression in cervical lesions, IL-10 mRNA was detected positive only in precancerous and invasive cervical cancers. None of the patients with normal cervical cytology expressed the IL-10 mRNA. IL-10 was also shown to contribute to human Papillomavirus persistency to establish a low-grade squamous intraepithelial neoplasia (LGSIL), then high-grade squamous intraepithelial neoplasia (HGSIL) and finally, progression to cervical cancer. |
(Ref. 116) |
Previous studies highlighted a significant correlation between IL-10 and the percentage of plasma cells in multiple myeloma patients as it induces the proliferation of plasma cells (Refs 117–119). Other studies indicated an elevation of IL-10 in different haematological malignancies such as Hodgkin lymphoma and non-Hodgkin lymphoma (Refs 120, 121). High IL-10 levels were reported to be associated with a shorter survival rate among patients with diffuse large-cell lymphoma (Ref. 120). Similarly, high IL-10 levels was found to be a prognostic factor in peripheral T cell lymphoma, which can lead to worsening of overall survival, low complete response rate, and higher early relapse rate (Ref. 122). Moreover, elevated IL-10 at diagnosis was found to be an independent prognostic marker in adult hemophagocytic lymphohistiocytosis patients in order to find the right treatment strategy (Ref. 123).
The riddle of IL-10 at the tumour-immune cell synapse
The balance between pro-inflammatory and anti-inflammatory signals is generally crucial for the maintenance of normal physiology and the prevention of cancer and a wide variety of diseases (Refs 14, 124–126). In the context of IL-10, it plays a dual function acting either as a pro-inflammatory or an anti-inflammatory mediator (Ref. 127). Regarding its role in cancer, studies have reported that IL-10, secreted by tumours or tumour-infiltrating immune cells, has allowed malignant cells to escape from the immune surveillance (Refs 128–130). In a study by Neven et al., IL-10 knockout in mice promoted the development of colon cancer. Moreover, the same study showed that humans deficient in IL-10 signalling molecules were more prone to develop lymphomas at a younger age (Ref. 131). As an anti-inflammatory cytokine, IL-10 is considered crucial for the homoeostasis of the anti-inflammatory Tregs and the suppression of proinflammatory IL-17-expressing T cells. However, IL-10 action depends on multiple factors such as targeted cells, other stimuli, and the time and duration of its effect (Ref. 132). Though, with many rationales presented, a question mark continues to rise to explore the nature of this complex cytokine.
Is IL-10 blockade a possible option as a novel immunotherapeutic approach for cancer patients?
Controversial data exists regarding the effectiveness of IL-10 immunotherapy in cancer (Ref. 133). Cancer vaccines that utilized monoclonal antibody (mAb) against IL-10 receptors succeeded to increase CD8+ T cell responses and to inhibit tumour growth whether injected intraperitoneally or subcutaneously (Refs 134, 135). The beneficial effect of IL-10 blockade is best explained through the inhibition of IL-10-induced suppression of DCs and prevention of their antigen presentation capacity by decreasing the expression of MHC class II and co-stimulator molecules (Ref. 136). Thus, DC-based vaccinations that disrupt IL-10 signalling provide more potent anti-tumour responses (Ref. 136). On the contrary, others claimed that antibodies targeting IL-10R had no protective effect against tumour growth when used with vaccines containing adjuvants that do not induce IL-10, such as the TLR3 ligand poly (I: C) or anti-CD40 agonistic antibodies (Ref. 137). Such a controversy regarding the effectiveness of therapeutic immunization could be explained and summarized by vaccine-induced IL-10 rather than IL-10 produced by tumours (Ref. 137).
It was previously reported that the prognosis of cancer patients is inversely correlated with elevated serum and tumour IL-10 levels (Ref. 138). Despite that, exogenous administration of IL-10 was tested in clinical studies, and resulted in immunological activation, as evidenced by higher granzymes and IFN in the serum of those patients receiving treatment. Pegylated recombinant (PEG) murine IL-10 promoted rejection of tumours and metastases by enhancing CD8 + T cell-mediated immune responses (Ref. 139). In addition, PEG-IL-10 exhibited immunologic and clinical advantages in solid tumours in clinical trials, particularly in RCC and uveal melanoma (Ref. 140). CD8 + tumour-infiltrating lymphocytes (TILs) in metastatic melanoma co-upregulate IL-10R and PD-1. While PD-1 blockade or IL-10 neutralization as monotherapies were insufficient to produce anti-tumour activity, combination therapies of PD-L1 blockers with IL-10R blockers were shown to exert anti-tumour effects by enhancing T cell responses, thereby suppressing the tumour growth (Ref. 141). Similarly, mice with ovarian tumours treated with PD-1 blocking antibodies have higher levels of IL-10 in their serum and ascites. Moreover, infiltration of immunosuppressive MDSCs was reduced, and the immunological activity was increased when IL-10 and PD-1 blockers were used together (Ref. 142). On the other hand, a multi-centred trial involving 111 patients with advanced malignant solid tumours unresponsive to previous therapies revealed that anti-PD-1 treatment (pembrolizumab or nivolumab) in combination with PEG-IL-10 offered a new therapeutic option (Ref. 143).
Most of the immune cells express IL-10 receptors and can activate subsequent downstream signalling pathways. Therefore, the paradox underlying the IL-10 blockade and whether it carries a beneficial or detrimental role in cancer treatment might be deciphered if we understood how exactly these cells react to IL-10 signalling through comprehensive genomic, epigenomic, and proteomic analysis.
Epigenomic approach
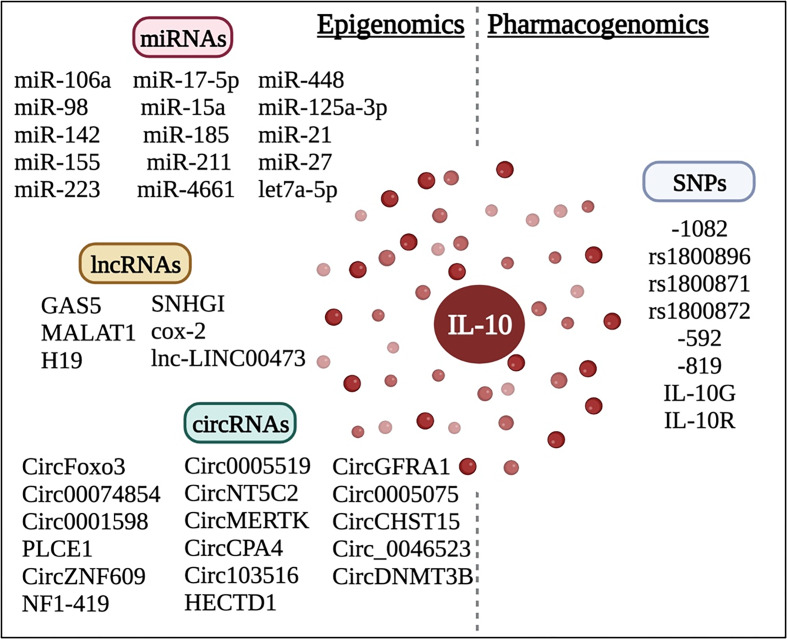
Epigenetic regulations include DNA methylation, histone modifications, histone acetylation, and the action of non-coding RNAs (ncRNAs) (Refs 144–146). Epigenetics arising from an alteration in the chromatin usually leads to alterations in gene expression. Moreover, epigenetic changes could either activate or suppress an oncogene or a tumour suppressor gene (Refs 147–150). It has been recently revealed that IL-10 is highly epigenetically regulated (Refs 93, 151). It is worth noting that such a level of post-transcriptional regulation of IL-10 expression might be a relevant explanation for the differential expression and effects of IL-10 in different cells at the TME despite the existence of common pathways for IL-10 induction as previously mentioned in this review, via the action of non-coding RNAs including microRNAs (miRNAs) (Refs 152, 153), long non-coding RNAs (lncRNAs) (Refs 154–156), and circular RNAs (circRNAs) (Refs 144, 156, 157).
Epigenetic modulation of IL-10 on the post-transcriptional has been highly evident in several reports via DNA methylation, histone modifications and histone acetylation, which have been extensively studied before in several studies (Refs 158, 159) and recently reviewed in (Ref. 158). However, the epigenetic regulation of IL-10 via ncRNAs, miRNAs, lncRNAs and circRNAs is recently being explored. Therefore, a closer approach to exploring the epigenetic regulation of IL-10 via ncRNAs could aid in understanding the complex nature of this cytokine.
microRNAs (miRNAs) regulating IL-10
miRNAs are short ncRNAs around 18–25 nucleotides long that widely exist in plants, viruses and animals (Refs 29, 150, 160, 161). These miRNAs can regulate gene expression by either degrading the mRNA target or by suppressing mRNA translation and reducing mRNA stability by binding to the 3′UTR (untranslated region) of a target gene (Refs 125, 153). Thus, a miRNA could therefore inhibit or activate the expression of tumour suppressors or oncogenes. Generally, oncogenic miRNAs (oncomiRs) are found to be over-expressed in cancers, whereas miRNAs with tumour-suppressive function are found to be under-expressed (Refs 124, 146, 150). When these oncomiRs or tumour suppressor miRNAs are inhibited or stimulated, respectively, cancer cell metastasis, proliferation and survival may be reduced, depending on the specific miRNA being affected and the type of cancer (Refs 28, 29, 133). Moreover, some cancers are dependent on specific oncomiRs, and suppressing such oncomiRs could completely regress cancer growth (Refs 149, 151, 162).
Few studies have presented miRNAs that could modulate IL-10 expression. In a study, testing for the possible post-transcriptional modulation of IL-10Rα and IL-10Rβ expression by miRNAs, three miRNAs were shown to have seed regions that target the 3′UTR of IL-10Rα; miR-15a, miR-185 and miR-211. These miRNAs were shown to inhibit the proliferation of IL-10-treated melanoma cells, while their inhibitors caused an increase in cell proliferation in melanoma (Ref. 163). IL-10 was also shown to be targeted by several other miRNAs (Ref. 164). Another study showed that miR-106a could bind to the 3′UTR of IL-10 and significantly downregulate its expression in-vitro (Ref. 165). Two transcription factors; early growth response 1 (Egr1) and Sp1 were implicated in the induction of miR-106a, which consequently reduced IL-10 levels (Ref. 164). Furthermore, an inverse relation was reported between Egr1-stimulated miR-106a and IL-10 levels. It is also worth mentioning that miR-106a is part of a cluster that is known to be dysregulated in 46% of human T-cell leukaemias. Thus, it was deduced that the promotion of leukaemic cell survival by IL-10 might be through its modulation via miR-106a (Ref. 164).
Another miRNA reported to positively regulate IL-10 was miRNA-4661. The miR-4661 binding to the 3′UTR of IL-10 resulted in a net increase in the half-life of IL-10. This action was favoured by preventing tristetraprolin (TTP) from binding to the IL-10 mRNA (Ref. 166). TTP is an RNA binding protein that plays a critical role in regulating proinflammatory immune responses by destabilizing target mRNAs via binding to their AU-rich elements (AREs) in the 3′-UTRs of mRNAs (Ref. 167). Moreover, miRNA/IL-10 interactions were reported in a study by Liu et al. revealing that miR-98-mediated post-transcriptional control could potentially be involved in fine-tuning IL-10 production in endotoxin tolerance (Refs 168, 169). On the other hand, IL-10 was reported to upregulate miRNAs that contribute towards an anti-inflammatory response such as miR-187 or downregulate those that are highly pro-inflammatory, such as miR-155 (Ref. 164). IL-10 was able to downregulate the induction of miR-155 induced by LPS (Ref. 170). Moreover, in-vivo studies on mice deficient in miR-155, could not generate a protective immune response (Ref. 171). Whereas in IL-10 mice-deficient cells, miR-155 levels were shown to highly increase. It was previously known that miR-155 could target a number of genes involved in the immune response, such as suppressor of cytokine signalling (SOCS), inhibitor of NK-κB kinase subunit epsilon (IKBKE) and Fas-associated death domain (FADD). Thus, targeting this miRNA by IL-10 is likely to elucidate key mechanisms through which IL-10 exerts control in the cell. Another study uncovered details of the IL-10 pathway by examining the effect of IL-10 on miRNAs, using IL-10 deficient mice for expression. Ten miRNAs were found to be upregulated in IL-10 deficient mice (miR-19a, miR-21, miR-31, miR-101, miR-223, miR-326, miR-142-3p, miR-142-5p, miR-146a and miR-155) (Ref. 172). miR-223 could hinder Roquin ubiquitin ligase by binding to its 3′UTR, eventually regulating IL-17 production and its inhibitor IL-10. Thus, this suggested a mechanism by which IL-10 could modulate the expression of IL-17 through miR-223. As previously mentioned, IL-10 can also induce the expression of anti-inflammatory miRNAs, such as miR-198 which is known to suppress TNF-α and IL-6. Consequently, this resulted in the promotion of an anti-inflammatory environment (Ref. 173). Collectively, such interesting findings of the mutual interaction between IL-10 and miRNAs discussed in the previous section highlighted an important role in the miRNA-mediated regulation of IL-10 expression and provided new insights into the intertwined mechanistic details of such immunomodulatory cytokine.
LncRNAs regulating IL-10
Long transcripts of RNA having more than 200 nucleotides, and not involved in protein translation are regarded as lncRNAs (Refs 16, 18, 154). LncRNAs play a significant role in the occurrence and development of cancer and thus, regulate the expression of cytokines such as IL-10 and IFN-γ as reported in a study by Tang et al. on non-small cell lung cancer (NSCLC) (Ref. 174). A large number of lncRNAs has been associated with cancer as recognized by genome-wide association studies on numerous tumours (Ref. 126). They are believed to exhibit functions such as tumour suppression and promotion, hence depicting to have a promising novel approach as biomarkers and therapeutic targets for cancers (Ref. 175). An increased expression of lncRNA SNHGI in cancerous breast cells of CD4 + TILs was also reported, whereas the expression of FOX and IL-10 was seen to be greatly reduced by siRNA SNHGI (Ref. 176). Moreover, silencing the lncRNA cox-2 was believed to increase the expression of IL-10, Arg-1 and Fizz-1 in M2 macrophages (Ref. 177). A study conducted by Zhou et al. reported reduced expression of IL-10 via suppression of lnc-LINC00473 (Ref. 178). Additionally, increased expression of IL-10 has been associated with the knockdown of lncRNA growth arrest-specific transcript 5 (GAS5) and reduced CRC cell proliferation while knockout of GAS5 promoted CRC colony formation and proliferation (Ref. 179). LncRNAs are known to regulate various signalling pathways such as TGF-β, STAT3, Hippo, EGF, Wnt, PI3 K/AKT and p53, whilst IL-10 is mostly involved in T-cell immune surveillance and suppression of cancer-associated inflammation. The expression of interleukins is regulated by lncRNAs that are known to be involved in various types of cancer. For instance, previous work by our group highlighted the potential of miRNA and lncRNA in the regulation of IL-10 in breast cancer, where miR-17-5p was identified as a dual regulator of TNF-α and IL-10. Additionally, knocking down the lncRNAs MALAT1 and/or H19 induced miR-17-5p and decreased TNF-α and IL-10 expression levels (Ref. 8). Such reports ed the immune-activator potential of miRNAs and the oncogenic potential of lncRNAs in cancers by regulating immunological targets in the TME. Hence, the extensive research on the relationship between the lncRNAs regulating IL-10 in various cancer needs to be validated further to establish a valid therapeutic link (Ref. 180).
CircRNAs regulating IL-10
CircRNAs are recognized as special ncRNA molecules with a distinctive ring structure and play significant roles as gene regulators and are considered one of the recently discovered epigenetic factors (Refs 153, 157). Abnormal production of circRNAs was found to influence the onset, progression and metastasis of cancer by acting as either tumour-suppressive or oncogenic factors (Refs 152, 181–183). This happens via interactions with proteins, miRNA sponge function and posttranscriptional regulation (Refs 155, 157, 184). Moreover, a line of evidence showed that circRNAs play pivotal roles in the chemoresistance (Refs 157, 185). Recently, specific circRNAs were found to possess an immunomodulatory function and alter the response of the TME by regulating the functions of tumour-infiltrating immune cells. For instance, CD4 + T cells activity is enhanced by circ0005519 through promoting the expression of IL-13 and IL-6 via affecting the expression of hsa-let-7a-5p (Ref. 186). On the other hand, circNT5C2 could attenuate the immune response by targeting miR-448 and serve as an oncogene via promoting tumour proliferation and metastasis (Ref. 187).
Since IL-10 function represents an unresolved enigma in cancer therapy, and since circRNAs also have dual roles in cancer therapy, the comprehensive understanding of circRNAs regulating IL-10 expression and function might be the key to answering numerous questions. Therefore, several studies that shed the light on novel circRNAs regulating IL-10 in different oncological and non-oncological contexts are highlighted. Some circRNAs can either enhance or inhibit IL-10 production and consequently could either promote or inhibit carcinogenesis. For example, circMERTK was reported to inhibit IL-10 production in colorectal cancer. The same study came to the conclusion that circMERTK knockdown reduced the activity of CD8 + T cells, suggesting that circMERTK may affect immunosuppressive activity through the circMERTK/miR-125a-3p/IL-10 axis (Ref. 188). According to another in vitro study, the downregulation of secreted PD-L1 by non-small cell lung cancer cells upon knockdown of circCPA4 resulted in the activation of CD8 + T cells in the TME (Ref. 188). In addition, the study found that PD-L1 abrogation reduced the expression of IL-10 in CD8 + T cells (Ref. 189). Circ103516 expression was found to be inversely correlated with IL-10 in inflammatory bowel diseases and thus it was postulated to play a proinflammatory role by sponging miR-19b. Additionally, it was discovered that circRNA HECTD1 contributed to the development of acute ischaemic stroke and that it was inversely linked with IL-10 production, suggesting that IL-10 played a protective function in acute ischaemic stroke (Ref. 190). In another cardiac context, the synthesis of IL-10 was decreased as a result of the overexpression of circFoxo3, a circRNA that is crucial in avoiding cardiac dysfunction brought on by myocardial infarction (Ref. 191). Downregulation of circ00074854 was reported to prevent polarization of M2 macrophages, which consequently alleviated the invasion and migration of hepatocellular carcinoma cells. According to the same study, macrophages exposed to exosomes produced by HepG2 cells that contained lower amounts of circ00074854 had significantly lower levels of IL-10 than those exposed to exosomes produced by HepG2 cells, demonstrating the direct relationship between Circ00074854 and IL-10 in different cancer settings (Ref. 192). Furthermore, a recent study emphasized the potential of CircSnx5 as a therapeutic target for immunological disorders since it has the ability to regulate the immunity and tolerance induced by DCs. It is interesting to note that knockdown of CircSnx5 led to a significant drop in IL-10, whilst overexpression of CircSnx5 was found to block DC maturation and boost IL-10 expression (Ref. 193). Another study focused on Circ0001598 as a potential target for treating breast cancer. It was discovered that circ0001598 regulates miR-1184 and PD-L1 via significantly increasing breast cancer proliferation, chemo-resistance and escape from immune surveillance. According to the same study mentioned above, depletion of circ0001598 increased breast cancer cells' susceptibility to Tratuzumab-induced CD8 + T cell cytotoxicity while decreasing the production of IL-10 (Ref. 194). Another study showed that the knockdown of circRNA PLCE1 ablated IL-10 production from macrophages while PLCE1 encouraged the transformation of epithelial cells into mesenchymal tissue, thus aiding glycolysis in colorectal cancer (Ref. 195). Another recently identified circRNA; circZNF609 has been linked to the pathogenesis of coronary artery disease, and forced overexpression of circZNF609 resulted in augmenting IL-10 expression (Ref. 196). It is also worth mentioning that a recent study discovered that circRNA NF1-419 attenuated inflammatory factors such as IL-10 and aging markers to postpone the onset of senile dementia (Ref. 197). Also, circGFRA1 has been indicated as a potential therapeutic target in prostate cancer; where Meng et al. reported that through a reduction in IL-10, knocking down circGFRA1 lessens the tumourigenic and immune-evading characteristics of prostate cancer cells (Ref. 198). Zhang et al. also discovered the role of circ0005075 in mediating neuroinflammation where silencing of circ0005075 in rat models resulted in a decrease in IL-10 production and protected against neuro-inflammation (Ref. 199). Another in vitro study revealed that circCdr1 overexpression enhanced the transcription of IL-10 both in naïve and pro-inflammatory macrophages (Ref. 200). CircCHST15 was recently reported to possess an oncogenic role by promoting immune escape through upregulating the expression of IL-10 and a sponging effect on miR-155 and miR-194 in lung cancer (Ref. 201). Additionally, circ_0046523 was found to promote carcinogenesis, mediate immunosuppression and abrogate CD8 + T cells function in pancreatic cancer via enhancing the secretion of IL-10 and TGF-β (Ref. 202). Furthermore, silencing circDNMT3B was discovered to decrease cell survival, promote apoptosis and increase IL-10 production in rat intestinal tissue (Ref. 203).
Collectively, it is quite clear that the circRNAs that inhibit IL-10 production from tumour cells act as tumour suppressors, while those that increase the production of IL-10 from tumour cells promote oncogenesis, cell survival, drug resistance and mediate immunosuppression. This highlights the promising role of such circRNAs as novel immunotherapeutic molecules that could ablate IL-10 production and act as a powerful immunomodulatory anti-cancer treatment for several cancer patients.
Pharmacogenomic approach: single nucleotide polymorphisms in IL-10 and its receptor
IL-10 gene
A very important basis for studies and research in IL-10 regulation is the examination of its genomic location and promoter structure. IL-10 gene encodes a protein, 178 amino acids long, which is secreted after cleavage to be comprised of 18 amino acids (Ref. 54). At the proximal promoter sequence of IL-10 in the human genome, there is a TATA box located upstream of the translation start site, for several transcription family members, including nuclear factor-κB (NF-κB), STAT, specificity protein (Sp), CREB, CCATT enhancer/binding protein (C/EBP), c-musculoaponeurotic fibrosarcoma factor (c-MAF), which have been characterized as ‘critical’ factors in regulating IL-10 expression (Ref. 204).
IL-10 signalling
Next, it is necessary to understand how IL-10 can signal through its receptor. IL-10R is a heterodimeric receptor complex composed of two chains (IL-10Rα ‘R1’ and IL-10Rβ ‘R2’). The α-chain binds directly to IL-10, while the β-chain is subsequently recruited into the IL-10/IL-10Rα complex (Ref. 205). The binding of IL-10 to IL-10Rα induces a conformational change in the receptor, allowing it to dimerize with IL-10Rβ. This dimerization leads to signal transduction in target cells (Ref. 206). When the IL-10 complex is formed, tyrosine kinases Tyk2 and Jak1 become activated and phosphorylate specific tyrosine residues. This phosphorylation further activates the cytoplasmic inactive transcription factor; STAT-3 resulting in the translocation and transcriptional activation (Ref. 207). IL-10 rapidly activates STAT-3 and remains phosphorylated over a sustained period, unlike the transient phosphorylation of IL-6 (Ref. 208). The STAT-3 docking sites in IL-10R1 appear to be sufficient to induce IL-10-mediated proliferative responses (Ref. 209). While IL-10R2 intracellular domain seems to provide the docking site for Tyk2. Thus, most IL-10-specific cellular functions appear to reside in the IL-10R1 chain, whereas IL-10R2 recruits the downstream signalling kinases (Ref. 210).
SNPs affecting IL-10
The IL-10 gene promoter and IL-10R have been found to include a significant number of SNPs (Refs 145, 211). There is strong evidence that several of these polymorphisms are linked to the differential expression of IL-10 in vitro and in some situations, in vivo (Refs 161, 212, 213). Some of these IL-10 variants have been associated with either low or high expression in several cancer types. For example, some genotypes have been evidenced to be correlated with a decreased expression of IL-10 and a higher risk to develop prostate cancer or non-Hodgkin's lymphoma (Refs 214, 215). On the other hand, other evidence concluded that some IL-10 variants are associated with higher expression of IL-10 and consequently, an elevated risk for cancer development of multiple myeloma, cervical cancer and gastric cancer in patients harbouring a particular IL-10 variant (Refs 216–218). Also, it has been demonstrated that the IL-10 gene transcription and translation were impacted by the SNPs in the IL-10 promoter region, leading to aberrant cell division and emergence of breast cancer (Ref. 219). Table 2 summarizes most of the IL-10 polymorphisms documented in the literature and their association with cancer development and risk.
Table 2.
IL-10 polymorphisms and their association with cancer development and risk
| Type of cancer | IL-10 polymorphism | Variant | Contribution | References |
|---|---|---|---|---|
| Chronic lymphocytic leukaemia (CLL) | −1082 | 1082 G/A and A/A | Increased risk to CLL | (Ref. 220) |
| Prostate cancer | −1082 | −1082 AA | High risk factor and susceptibility | (Ref. 221) |
| Cervical cancer | rs1800896 | AG/AA genotypes | High risk factor and susceptibility | (Ref. 222) |
| Breast cancer | rs1800896 rs1800871 rs1800872 |
AA genotypes | High risk factor and susceptibility | (Ref. 223) |
| Gastric carcinoma | −1082, −592 −819 |
GCC, ATA, AG haplotype | Advanced stage, high risk factor and susceptibility, | (Ref. 224) |
| Oral cancer | −1082 | −1082 G allele | High susceptibility to oral carcinoma | (Ref. 225) |
| Multiple myeloma | IL-10G IL-10R |
IL-10 G 136/136, IL-10R 112/114 | Increased susceptibility | (Ref. 226) |
| Lung cancer | −592 | A > C | Increased risk | (Ref. 227) |
| Non-Hodgkin's lymphoma | −1082 −592 −819 |
−1082 AA, ATA, ACC haplotypes | More aggressive form of the disease. | (Ref. 228) |
| Acute lymphoblastic leukaemia | −1082 | −1082 GG | Low possibility of poor response to prednisolone | (Ref. 229) |
| Acute myeloid leukaemia | −819 | −819 T/C | Increased risk of AML | (Ref. 230) |
| Colorectal cancer | −819 | T > C genotypes | Increased risk of colorectal cancer | (Ref. 231) |
| Cutaneous malignant melanoma | −1082 −592 −819 |
High risk, larger tumour thickness, disease progression and shorter survival | 1082 AA, 1082 GG, ACC/ACC, ACC/ATA, ATA/ATA | (Refs 232, 233) |
Since IL-10 has a role in malignancy, it is regarded to be the subject of numerous disputes in the literature, whether it has a positive or negative effect. As a result, whether IL-10 blockage is effective as an immunotherapeutic strategy is another unsolved puzzle. This opens the door to a crucial query that might provide the answer. However, it has not yet been addressed in the literature. It remains unclear whether SNPs in the IL-10 or its receptor account for the varying effects of IL-10 inhibition on cancer treatment. A clinical investigation addressing the existence of SNPs in IL-10 or its receptors and their impact on the response to IL-10 therapy is necessary. These pharmacogenomic investigations will aid in the development of immunotherapeutic modalities by identifying the most qualified individuals to provide these cutting-edge drugs.
Conclusions
This review highlighted the controversial functions of IL-10 in oncology. Such contradictory information prevented researchers from determining whether exogenous IL-10 administration or blockage will boost the immune system and combat changes at the TME. This could be explained by the fact that IL-10 has two distinct functions depending on which immune cell and which receptor would be activated. Also, epigenetic regulation of IL-10 in cancer via ncRNAs is quite complex (Fig. 3). Also, the relationship between IL-10 SNPs will help us better understand the precise function of IL-10 in the TME and will help us develop more individualized immunotherapeutic approaches by classifying patients into responders and non-responders.
Figure 3.
Epigenomic and pharmacogenomic regulation of IL-10 in oncology
Authors's contributions
Conceptualization, R. Y. M., N. M. E. and R. A. Y.; writing – original draft preparation, R. Y. M., R. A. S., G. R. and R. A. Y.; writing – review and editing, R. Y. M., N. M. E., M. B. and R. A. Y. All authors have read and agreed to the published version of the manuscript.
Funding statement
This work is not supported by any funds or grants.
Competing interests
None.
References
- 1.Elemam NM et al. (2023) Editorial: understanding the crosstalk between immune cells and the tumor microenvironment in cancer and its implications for immunotherapy. Frontiers in Medicine (Lausanne) 10, 1202581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sponghini A et al. (2017) Complete response to anti-PD-1 nivolumab in massive skin metastasis from melanoma: efficacy and tolerability in an elderly patient. Anti-Cancer Drugs 28, 808–810. [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Latif M and Youness RA (2020) Why natural killer cells in triple negative breast cancer? World Journal of Clinical Oncology 11, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramzy A et al. (2022) Drugless nanoparticles tune-up an array of intertwined pathways contributing to immune checkpoint signaling and metabolic reprogramming in triple-negative breast cancer. Biomedical Materials 18. doi: 10.1088/1748-605X/aca85d. [DOI] [PubMed] [Google Scholar]
- 5.Soliman AH et al. (2023) Phytochemical-derived tumor-associated macrophage remodeling strategy using Phoenix dactylifera L. Boosted photodynamic therapy in melanoma via H19/iNOS/PD-L1 axis. Photodiagnosis and Photodynamic Therapy 44, 103792. [DOI] [PubMed] [Google Scholar]
- 6.Selem NA et al. (2023) Let-7a/cMyc/CCAT1/miR-17-5p circuit re-sensitizes atezolizumab resistance in triple negative breast cancer through modulating PD-L1. Pathology Research and Practice 248, 154579. [DOI] [PubMed] [Google Scholar]
- 7.Conlon KC, Miljkovic MD and Waldmann TA (2019) Cytokines in the treatment of cancer. Journal of Interferon and Cytokine Research 39, 6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soliman R-A et al. (2022) Uncoupling tumor necrosis factor-α and interleukin-10 at tumor immune microenvironment of breast cancer through miR-17-5p/MALAT-1/H19 circuit. BIOCELL 46, 769–783. [Google Scholar]
- 9.Mocellin S et al. (2003) The dual role of IL-10. Trends in Immunology 24, 36–43. [DOI] [PubMed] [Google Scholar]
- 10.Shen L et al. (2018) Local blockade of interleukin 10 and C-X-C motif chemokine ligand 12 with nano-delivery promotes antitumor response in murine cancers. ACS Nano 12, 9830–9841. [DOI] [PubMed] [Google Scholar]
- 11.Mumm JB and Oft M (2013) Pegylated IL-10 induces cancer immunity. BioEssays 35, 623–631. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D and Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 13.Ocana MC et al. (2019) Metabolism within the tumor microenvironment and its implication on cancer progression: an ongoing therapeutic target. Medicinal Research Reviews 39, 70–113. [DOI] [PubMed] [Google Scholar]
- 14.Youness RA et al. (2023) Heat shock proteins: central players in oncological and immuno-oncological tracks. Advances in Experimental Medicine and Biology 1409, 193–203. [DOI] [PubMed] [Google Scholar]
- 15.Nyberg P, Salo T and Kalluri R (2008) Tumor microenvironment and angiogenesis. Frontiers in Bioscience 13, 6537–6553. [DOI] [PubMed] [Google Scholar]
- 16.Mekky RY et al. (2023) MALAT-1: immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Translational Oncology 31, 101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdel-Latif M et al. (2022) MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Molecular and Cellular Biochemistry 477, 1281–1293. [DOI] [PubMed] [Google Scholar]
- 18.Youness RA et al. (2018) A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide 80, 12–23. [DOI] [PubMed] [Google Scholar]
- 19.Fridman WH et al. (2012) The immune contexture in human tumours: impact on clinical outcome. Nature Reviews Cancer 12, 298–306. [DOI] [PubMed] [Google Scholar]
- 20.Strauss L et al. (2007) The frequency and suppressor function of CD4 + CD25highFoxp3 + T cells in the circulation of patients with squamous cell carcinoma of the head and neck. Clinical Cancer Research 13, 6301–6311. [DOI] [PubMed] [Google Scholar]
- 21.Chen L et al. (2018) IL-6 influences the polarization of macrophages and the formation and growth of colorectal tumor. Oncotarget 9, 17443–17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaimala S et al. (2018) Attenuated bacteria as immunotherapeutic tools for cancer treatment. Frontiers in Oncology 8, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JC et al. (2018) Metformin's antitumour and anti-angiogenic activities are mediated by skewing macrophage polarization. Journal of Cellular and Molecular Medicine 22, 3825–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan B et al. (2018) Inhibition of rspo-Lgr4 facilitates checkpoint blockade therapy by switching macrophage polarization. Cancer Research 78, 4929–4942. [DOI] [PubMed] [Google Scholar]
- 25.Tan YS et al. (2018) Mitigating SOX2-potentiated immune escape of head and neck squamous cell carcinoma with a STING-inducing nanosatellite vaccine. Clinical Cancer Research 24, 4242–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baek JH et al. (2018) Knockdown of end-binding protein 1 induces apoptosis in radioresistant A549 lung cancer cells via p38 kinase-dependent COX-2 upregulation. Oncology Reports 39, 1565–1572. [DOI] [PubMed] [Google Scholar]
- 27.Youness RA et al. (2016) Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors 34, 128–140. [DOI] [PubMed] [Google Scholar]
- 28.Rahmoon MA et al. (2017) MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors 35, 76–87. [DOI] [PubMed] [Google Scholar]
- 29.Awad AR et al. (2021) An acetylated derivative of vitexin halts MDA-MB-231 cellular progression and improves its immunogenic profile through tuning miR- 20a-MICA/B axis. Natural Product Research 35, 3126–3130. [DOI] [PubMed] [Google Scholar]
- 30.Hall B, Andreeff M and Marini F (2007) The participation of mesenchymal stem cells in tumor stroma formation and their application as targeted-gene delivery vehicles. Handbook of Experimental Pharmacology 180, 263–283. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez G et al. (2018) Mesenchymal stem cell's secretome promotes selective enrichment of cancer stem-like cells with specific cytogenetic profile. Cancer Letters 429, 78–88. [DOI] [PubMed] [Google Scholar]
- 32.Guerriero JL (2018) Macrophages: the road less traveled, changing anticancer therapy. Trends in Molecular Medicine 24, 472–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Overmeire E et al. (2014) Mechanisms driving macrophage diversity and specialization in distinct tumor microenvironments and parallelisms with other tissues. Frontiers in Immunology 5, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurt B et al. (2017) Cancer-promoting mechanisms of tumor-associated neutrophils. American Journal of Surgery 214, 938–944. [DOI] [PubMed] [Google Scholar]
- 35.Mensurado S et al. (2018) Tumor-associated neutrophils suppress pro-tumoral IL-17 + gammadelta T cells through induction of oxidative stress. PLoS Biology 16, e2004990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen M and Andersen MH (2017) The role of dendritic cells in cancer. Seminars in Immunopathology 39, 307–316. [DOI] [PubMed] [Google Scholar]
- 37.Guerra DAP et al. (2018) Targeting glioblastoma-derived pericytes improves chemotherapeutic outcome. Angiogenesis 21, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reina-Campos M et al. (2018) Metabolic reprogramming of the tumor microenvironment by p62 and its partners. Biochimica et Biophysica Acta, Reviews on Cancer 1870, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akutagawa T et al. (2018) Cancer-adipose tissue interaction and fluid flow synergistically modulate cell kinetics, HER2 expression, and trastuzumab efficacy in gastric cancer. Gastric Cancer: Official Journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association 21, 946–955. [DOI] [PubMed] [Google Scholar]
- 40.Kumar V et al. (2016) The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends in Immunology 37, 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin Z et al. (2019) Myeloid-derived suppressor cells: roles in the tumor microenvironment and tumor radiotherapy. International Journal of Cancer 144, 933–946. [DOI] [PubMed] [Google Scholar]
- 42.Shen M et al. (2018) A novel MDSC-induced PD-1(-)PD-L1(+) B-cell subset in breast tumor microenvironment possesses immuno-suppressive properties. Oncoimmunology 7, e1413520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahirwar DK et al. (2018) Fibroblast-derived CXCL12 promotes breast cancer metastasis by facilitating tumor cell intravasation. Oncogene 37, 4428–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liotta LA and Kohn EC (2001) The microenvironment of the tumour-host interface. Nature 411, 375–379. [DOI] [PubMed] [Google Scholar]
- 45.Burkholder B et al. (2014) Tumor-induced perturbations of cytokines and immune cell networks. Biochimica et Biophysica Acta 1845, 182–201. [DOI] [PubMed] [Google Scholar]
- 46.Zigrino P, Loffek S and Mauch C (2005) Tumor-stroma interactions: their role in the control of tumor cell invasion. Biochimie 87, 321–328. [DOI] [PubMed] [Google Scholar]
- 47.Fiorentino DF, Bond MW and Mosmann TR (1989) Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. Journal of Experimental Medicine 170, 2081–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore KW et al. (1990) Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science (New York, N.Y.) 248, 1230–1234. [DOI] [PubMed] [Google Scholar]
- 49.Kotenko SV (2002) The family of IL-10-related cytokines and their receptors: related, but to what extent? Cytokine & Growth Factor Reviews 13, 223–240. [DOI] [PubMed] [Google Scholar]
- 50.Volk H et al. (2001) IL-10 and its homologs: important immune mediators and emerging immunotherapeutic targets. Trends in Immunology 22, 414–417. [DOI] [PubMed] [Google Scholar]
- 51.Fickenscher H et al. (2002) The interleukin-10 family of cytokines. Trends in Immunology 23, 89–96. [DOI] [PubMed] [Google Scholar]
- 52.Mannino MH et al. (2015) The paradoxical role of IL-10 in immunity and cancer. Cancer Letters 367, 103–107. [DOI] [PubMed] [Google Scholar]
- 53.Nagpal G et al. (2017) Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Scientific Reports 7, 42851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabat R et al. (2010) Biology of interleukin-10. Cytokine & Growth Factor Reviews 21, 331–344. [DOI] [PubMed] [Google Scholar]
- 55.Agrawal S et al. (2003) Cutting edge: different Toll-like receptor agonists instruct dendritic cells to induce distinct Th responses via differential modulation of extracellular signal-regulated kinase-mitogen-activated protein kinase and c-Fos. Journal of Immunology 171, 4984–4989. [DOI] [PubMed] [Google Scholar]
- 56.Boonstra A et al. (2006) Macrophages and myeloid dendritic cells, but not plasmacytoid dendritic cells, produce IL-10 in response to MyD88- and TRIF-dependent TLR signals, and TLR-independent signals. Journal of Immunology 177, 7551–7558. [DOI] [PubMed] [Google Scholar]
- 57.Chi H et al. (2006) Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proceedings of the National Academy of Sciences of the USA 103, 2274–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dillon S et al. (2004) A Toll-like receptor 2 ligand stimulates Th2 responses in vivo, via induction of extracellular signal-regulated kinase mitogen-activated protein kinase and c-Fos in dendritic cells. Journal of Immunology 172, 4733–4743. [DOI] [PubMed] [Google Scholar]
- 59.Geijtenbeek TB et al. (2003) Mycobacteria target DC-SIGN to suppress dendritic cell function. Journal of Experimental Medicine 197, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hacker H et al. (2006) Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207. [DOI] [PubMed] [Google Scholar]
- 61.Higgins SC et al. (2003) Toll-like receptor 4-mediated innate IL-10 activates antigen-specific regulatory T cells and confers resistance to Bordetella pertussis by inhibiting inflammatory pathology. Journal of Immunology 171, 3119–3127. [DOI] [PubMed] [Google Scholar]
- 62.Kaiser F et al. (2009) TPL-2 negatively regulates interferon-beta production in macrophages and myeloid dendritic cells. Journal of Experimental Medicine 206, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogers NC et al. (2005) Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity 22, 507–517. [DOI] [PubMed] [Google Scholar]
- 64.Dennehy KM et al. (2009) Reciprocal regulation of IL-23 and IL-12 following co-activation of Dectin-1 and TLR signaling pathways. European Journal of Immunology 39, 1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kapsenberg ML (2003) Dendritic-cell control of pathogen-driven T-cell polarization. Nature Reviews Immunology 3, 984–993. [DOI] [PubMed] [Google Scholar]
- 66.Mills KH (2004) Regulatory T cells: friend or foe in immunity to infection? Nature Reviews Immunology 4, 841–855. [DOI] [PubMed] [Google Scholar]
- 67.Agrawal S and Gupta S (2011) TLR1/2, TLR7, and TLR9 signals directly activate human peripheral blood naive and memory B cell subsets to produce cytokines, chemokines, and hematopoietic growth factors. Journal of Clinical Immunology 31, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lampropoulou V et al. (2008) TLR-activated B cells suppress T cell-mediated autoimmunity. Journal of Immunology 180, 4763–4773. [DOI] [PubMed] [Google Scholar]
- 69.Sayi A et al. (2011) TLR-2-activated B cells suppress Helicobacter-induced preneoplastic gastric immunopathology by inducing T regulatory-1 cells. Journal of Immunology 186, 878–890. [DOI] [PubMed] [Google Scholar]
- 70.Shaw MH et al. (2006) Tyk2 negatively regulates adaptive Th1 immunity by mediating IL-10 signaling and promoting IFN-gamma-dependent IL-10 reactivation. Journal of Immunology 176, 7263–7271. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X et al. (2007) Type I interferons protect neonates from acute inflammation through interleukin 10-producing B cells. Journal of Experimental Medicine 204, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X et al. (2009) Coactivation of Syk kinase and MyD88 adaptor protein pathways by bacteria promotes regulatory properties of neutrophils. Immunity 31, 761–771. [DOI] [PubMed] [Google Scholar]
- 73.Peppa D et al. (2010) Blockade of immunosuppressive cytokines restores NK cell antiviral function in chronic hepatitis B virus infection. PLoS Pathogens 6, e1001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cottrez F and Groux H (2001) Regulation of TGF-beta response during T cell activation is modulated by IL-10. Journal of Immunology 167, 773–778. [DOI] [PubMed] [Google Scholar]
- 75.Barthlott T et al. (2005) CD25 + CD4 + T Cells compete with naive CD4 + T cells for IL-2 and exploit it for the induction of IL-10 production. International Immunology 17, 279–288. [DOI] [PubMed] [Google Scholar]
- 76.Cretney E et al. (2011) The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nature Immunology 12, 304–311. [DOI] [PubMed] [Google Scholar]
- 77.de la Rosa M et al. (2004) Interleukin-2 is essential for CD4 + CD25 + regulatory T cell function. European Journal of Immunology 34, 2480–2488. [DOI] [PubMed] [Google Scholar]
- 78.Maynard CL et al. (2007) Regulatory T cells expressing interleukin 10 develop from Foxp3 + and Foxp3- precursor cells in the absence of interleukin 10. Nature Immunology 8, 931–941. [DOI] [PubMed] [Google Scholar]
- 79.Sun J et al. (2011) CD4 + T Cell help and innate-derived IL-27 induce blimp-1-dependent IL-10 production by antiviral CTLs. Nature Immunology 12, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerosa F et al. (1996) Interleukin-12 primes human CD4 and CD8 T cell clones for high production of both interferon-gamma and interleukin-10. Journal of Experimental Medicine 183, 2559–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanden Eijnden S et al. (2005) IL-23 up-regulates IL-10 and induces IL-17 synthesis by polyclonally activated naive T cells in human. European Journal of Immunology 35, 469–475. [DOI] [PubMed] [Google Scholar]
- 82.Meyaard L et al. (1996) IL-12-induced IL-10 production by human T cells as a negative feedback for IL-12-induced immune responses. Journal of Immunology 156, 2776–2782. [PubMed] [Google Scholar]
- 83.Moore KW et al. (2001) Interleukin-10 and the interleukin-10 receptor. Annual Review of Immunology 19, 683–765. [DOI] [PubMed] [Google Scholar]
- 84.Mocellin S et al. (2004) IL-10 stimulatory effects on human NK cells explored by gene profile analysis. Genes and Immunity 5, 621–630. [DOI] [PubMed] [Google Scholar]
- 85.Abd Razak NA et al. (2016) Patient-specific interface pressure case study at transradial prosthetic socket: comparison trials between ICRC polypropylene socket and air splint socket. European Journal of Physical and Rehabilitation Medicine. PMID: 26771916. [PubMed] [Google Scholar]
- 86.Kelly JM et al. (2002) Induction of tumor-specific T cell memory by NK cell-mediated tumor rejection. Nature Immunology 3, 83–90. [DOI] [PubMed] [Google Scholar]
- 87.Kawai T and Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunology 11, 373–384. [DOI] [PubMed] [Google Scholar]
- 88.Banerjee A et al. (2006) Diverse Toll-like receptors utilize Tpl2 to activate extracellular signal-regulated kinase (ERK) in hemopoietic cells. Proceedings of the National Academy of Sciences of the USA 103, 3274–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yi AK et al. (2002) Role of mitogen-activated protein kinases in CpG DNA-mediated IL-10 and IL-12 production: central role of extracellular signal-regulated kinase in the negative feedback loop of the CpG DNA-mediated Th1 response. Journal of Immunology 168, 4711–4720. [DOI] [PubMed] [Google Scholar]
- 90.Hu X et al. (2006) IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity 24, 563–574. [DOI] [PubMed] [Google Scholar]
- 91.Foey AD et al. (1998) Regulation of monocyte IL-10 synthesis by endogenous IL-1 and TNF-alpha: role of the p38 and p42/44 mitogen-activated protein kinases. Journal of Immunology 160, 920–928. [PubMed] [Google Scholar]
- 92.Kim L et al. (2005) p38 MAPK autophosphorylation drives macrophage IL-12 production during intracellular infection. Journal of Immunology 174, 4178–4184. [DOI] [PubMed] [Google Scholar]
- 93.Saraiva M and O'Garra A (2010) The regulation of IL-10 production by immune cells. Nature Reviews Immunology 10, 170–181. [DOI] [PubMed] [Google Scholar]
- 94.Ananieva O et al. (2008) The kinases MSK1 and MSK2 act as negative regulators of toll-like receptor signaling. Nature Immunology 9, 1028–1036. [DOI] [PubMed] [Google Scholar]
- 95.Kallies A et al. (2009) Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity 31, 283–295. [DOI] [PubMed] [Google Scholar]
- 96.D'Addario M, Arora PD and McCulloch CA (2006) Role of p38 in stress activation of Sp1. Gene 379, 51–61. [DOI] [PubMed] [Google Scholar]
- 97.Tan NY and Khachigian LM (2009) Sp1 phosphorylation and its regulation of gene transcription. Molecular and Cellular Biology 29, 2483–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Asnagli H, Afkarian M and Murphy KM (2002) Cutting edge: identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. Journal of Immunology 168, 4268–4271. [DOI] [PubMed] [Google Scholar]
- 99.Martins GA et al. (2006) Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nature Immunology 7, 457–465. [DOI] [PubMed] [Google Scholar]
- 100.Oliver PM et al. (2006) Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25, 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanchez-Zauco N et al. (2017) Circulating blood levels of IL-6, IFN-gamma, and IL-10 as potential diagnostic biomarkers in gastric cancer: a controlled study. BMC Cancer 17, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Berti FCB et al. (2017) The role of interleukin 10 in human papilloma virus infection and progression to cervical carcinoma. Cytokine & Growth Factor Reviews 34, 1–13. [DOI] [PubMed] [Google Scholar]
- 103.Vahl JM et al. (2017) Interleukin-10-regulated tumour tolerance in non-small cell lung cancer. British Journal of Cancer 117, 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alotaibi MR et al. (2018) Characterization of apoptosis in a breast cancer cell line after IL-10 silencing. Asian Pacific Journal of Cancer Prevention: APJCP 19, 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Townsend MH et al. (2018) Metastatic colon adenocarcinoma has a significantly elevated expression of IL-10 compared with primary colon adenocarcinoma tumors. Cancer biology & therapy 19, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bornstein S et al. (2016) IL-10 and integrin signaling pathways are associated with head and neck cancer progression. BMC Genomics 17, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li YF, Yang PZ and Li HF (2016) Functional polymorphisms in the IL-10 gene with susceptibility to esophageal, nasopharyngeal, and oral cancers. Cancer Biomarkers: Section A of Disease Markers 16, 641–651. [DOI] [PubMed] [Google Scholar]
- 108.Abtahi S et al. (2017) Dual association of serum interleukin-10 levels with colorectal cancer. Journal of Cancer Research and Therapeutics 13, 252–256. [DOI] [PubMed] [Google Scholar]
- 109.Shrihari TG (2017) Dual role of inflammatory mediators in cancer. Ecancermedicalscience 11, 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanikawa T et al. (2012) Interleukin-10 ablation promotes tumor development, growth, and metastasis. Cancer Research 72, 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rizzuti D et al. (2015) Helicobacter pylori inhibits dendritic cell maturation via interleukin-10-mediated activation of the signal transducer and activator of transcription 3 pathway. Journal of Innate Immunity 7, 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Y et al. (2014) [prognostic value of IL-10 expression in tumor tissues of breast cancer patients]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 30, 517–520. [PubMed] [Google Scholar]
- 113.Yang C et al. (2015) Increased drug resistance in breast cancer by tumor-associated macrophages through IL-10/STAT3/bcl-2 signaling pathway. Medical Oncology 32, 352. [DOI] [PubMed] [Google Scholar]
- 114.Lane D et al. (2018) Ascites IL-10 promotes ovarian cancer cell migration. Cancer Microenvironment 11, 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X et al. (2017) Bladder cancer cells induce immunosuppression of T cells by supporting PD-L1 expression in tumour macrophages partially through interleukin 10. Cell Biology International 41, 177–186. [DOI] [PubMed] [Google Scholar]
- 116.Bermudez-Morales VH et al. (2008) Correlation between IL-10 gene expression and HPV infection in cervical cancer: a mechanism for immune response escape. Cancer Investigation 26, 1037–1043. [DOI] [PubMed] [Google Scholar]
- 117.Shekarriz R, Janbabaei G and Abedian Kenari S (2018) Prognostic value of IL-10 and Its relationship with disease stage in Iranian patients with multiple myeloma. Asian Pacific Journal of Cancer Prevention: APJCP 19, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alexandrakis MG et al. (2013) Relationship between circulating BAFF serum levels with proliferating markers in patients with multiple myeloma. Biomed Research International 2013, 389579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kovacs E (2010) Interleukin-6 leads to interleukin-10 production in several human multiple myeloma cell lines. Does interleukin-10 enhance the proliferation of these cells? Leukemia Research 34, 912–916. [DOI] [PubMed] [Google Scholar]
- 120.Gupta M et al. (2012) Elevated serum IL-10 levels in diffuse large B-cell lymphoma: a mechanism of aberrant JAK2 activation. Blood 119, 2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Visco C et al. (2004) Elevated serum levels of IL-10 are associated with inferior progression-free survival in patients with Hodgkin's disease treated with radiotherapy. Leukemia & Lymphoma 45, 2085–2092. [DOI] [PubMed] [Google Scholar]
- 122.Zhang Y et al. (2020) Increased serum level of interleukin-10 predicts poor survival and early recurrence in patients with peripheral T-cell lymphomas. Frontiers in Oncology 10, 584261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou Y et al. (2021) Increased levels of serum interleukin-10 are associated with poor outcome in adult hemophagocytic lymphohistiocytosis patients. Orphanet Journal of Rare Diseases 16, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abdallah RM et al. (2022) Hindering the synchronization between miR-486-5p and H19 lncRNA by hesperetin halts breast cancer aggressiveness through tuning ICAM-1. Anti-Cancer Agents in Medicinal Chemistry 22, 586–595. [DOI] [PubMed] [Google Scholar]
- 125.Youness RA et al. (2021) Targeting hydrogen sulphide signaling in breast cancer. Journal of Advanced Research 27, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Youness RA et al. (2019) The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. Journal of Cellular Physiology 234, 20286–20297. [DOI] [PubMed] [Google Scholar]
- 127.Hatanaka H et al. (2001) Significant correlation between interleukin 10 expression and vascularization through angiopoietin/TIE2 networks in non-small cell lung cancer. Clinical Cancer Research 7, 1287–1292. [PubMed] [Google Scholar]
- 128.Marincola FM et al. (2000) Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Advances in Immunology 74, 181–273. [DOI] [PubMed] [Google Scholar]
- 129.Yang L and Carbone DP (2004) Tumor-host immune interactions and dendritic cell dysfunction. Advances in Cancer Research 92, 13–27. [DOI] [PubMed] [Google Scholar]
- 130.Mapara MY and Sykes M (2004) Tolerance and cancer: mechanisms of tumor evasion and strategies for breaking tolerance. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology 22, 1136–1151. [DOI] [PubMed] [Google Scholar]
- 131.Neven B et al. (2013) A Mendelian predisposition to B-cell lymphoma caused by IL-10R deficiency. Blood 122, 3713–3722. [DOI] [PubMed] [Google Scholar]
- 132.Couper KN, Blount DG and Riley EM (2008) IL-10: the master regulator of immunity to infection. Journal of Immunology 180, 5771–5777. [DOI] [PubMed] [Google Scholar]
- 133.Nafea H et al. (2021) LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. Journal of Cellular Physiology 236, 5362–5372. [DOI] [PubMed] [Google Scholar]
- 134.Ni G et al. (2020) Targeting interleukin-10 signalling for cancer immunotherapy, a promising and complicated task. Human Vaccines & Immunotherapeutics 16, 2328–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen S et al. (2014) IL-10 signalling blockade at the time of immunization inhibits human papillomavirus 16 E7 transformed TC-1 tumour cells growth in mice. Cellular Immunology 290, 145–151. [DOI] [PubMed] [Google Scholar]
- 136.Llopiz D et al. (2018) Enhancement of antitumor vaccination by targeting dendritic cell-related IL-10. Frontiers in Immunology 9, doi: 10.3389/fimmu.2018.01923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Llopiz D et al. (2016) Vaccine-induced but not tumor-derived interleukin-10 dictates the efficacy of interleukin-10 blockade in therapeutic vaccination. Oncoimmunology 5, e1075113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Soliman R et al. (2021) Uncoupling tumor necrosis factor-α and interleukin-10 at tumor immune microenvironment of breast cancer through miR-17-5p/MALAT-1/H19 circuit.
- 139.Mumm JB and Oft M (2013) Pegylated IL-10 induces cancer immunity: the surprising role of IL-10 as a potent inducer of IFN-γ-mediated CD8(+) T cell cytotoxicity. Bioessays 35, 623–631. [DOI] [PubMed] [Google Scholar]
- 140.Naing A et al. (2016) Safety, antitumor activity, and immune activation of pegylated recombinant human interleukin-10 (AM0010) in patients with advanced solid tumors. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology 34, 3562–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun Z et al. (2015) IL10 And PD-1 cooperate to limit the activity of tumor-specific CD8 + T cells. Cancer Research 75, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lamichhane P et al. (2017) IL10 Release upon PD-1 blockade sustains immunosuppression in ovarian cancer. Cancer Research 77, 6667–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Naing A et al. (2019) Pegilodecakin combined with pembrolizumab or nivolumab for patients with advanced solid tumours (IVY): a multicentre, multicohort, open-label, phase 1b trial. The Lancet. Oncology 20, 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li L et al. (2016) Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Scientific Reports 6, 26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.El Din GS et al. (2020) miRNA-506-3p directly regulates rs10754339 (A/G) in the immune checkpoint protein B7-H4 in breast cancer. MicroRNA 9, 346–353. [DOI] [PubMed] [Google Scholar]
- 146.El-Daly SM et al. (2023) Editorial: recent breakthroughs in the decoding of circulating nucleic acids and their applications to human diseases. Frontiers in Molecular Biosciences 10, 1203495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Reis AH, Vargas FR and Lemos B (2016) Biomarkers of genome instability and cancer epigenetics. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine 37, 13029–13038. [DOI] [PubMed] [Google Scholar]
- 148.Ahmed Youness R et al. (2020) A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Natural Product Research 34, 1475–1480. [DOI] [PubMed] [Google Scholar]
- 149.Shaalan YM et al. (2018) Destabilizing the interplay between miR-1275 and IGF2BPs by Tamarix articulata and quercetin in hepatocellular carcinoma. Natural Product Research 32, 2217–2220. [DOI] [PubMed] [Google Scholar]
- 150.Kilany E, et al. FH (2021) miR-744/eNOS/NO axis: a novel target to halt triple negative breast cancer progression. Breast Disease 40, 161–169. [DOI] [PubMed] [Google Scholar]
- 151.Fahmy SA et al. (2022) Molecular engines, therapeutic targets, and challenges in pediatric brain tumors: a special emphasis on hydrogen sulfide and RNA-based nano-delivery. Cancers (Basel) 14, 5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.ElKhouly AM, Youness RA and Gad MZ (2020) MicroRNA-486-5p and microRNA-486-3p: multifaceted pleiotropic mediators in oncological and non-oncological conditions. Non-Coding RNA Research 5, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.ZeinElAbdeen YA, AbdAlSeed A and Youness RA (2022) Decoding insulin-like growth factor signaling pathway from a non-coding RNAs perspective: a step towards precision oncology in breast cancer. Journal of Mammary Gland Biology and Neoplasia 27, 79–99. [DOI] [PubMed] [Google Scholar]
- 154.Youness RA and Gad MZ (2019) Long non-coding RNAs: functional regulatory players in breast cancer. Non-coding RNA Research 4, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Selem NA, Youness RA and Gad MZ (2021) What is beyond LncRNAs in breast cancer: a special focus on colon cancer-associated transcript-1 (CCAT-1). Non-Coding RNA Research 6, 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.El-Aziz MKA et al. (2023) Decoding hepatocarcinogenesis from a noncoding RNAs perspective. Journal of Cellular Physiology 238, 1982–2009. [DOI] [PubMed] [Google Scholar]
- 157.Dawoud A et al. (2023) Circular RNAs: new layer of complexity evading breast cancer heterogeneity. Non-Coding RNA Research 8, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang H and Kuchroo V (2019) Epigenetic and transcriptional mechanisms for the regulation of IL-10. Seminars in Immunology 44, 101324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Larsson L et al. (2012) Influence of epigenetic modifications of the interleukin-10 promoter on IL10 gene expression. European Journal of Oral Sciences 120, 14–20. [DOI] [PubMed] [Google Scholar]
- 160.Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 161.Youssef SS et al. (2022) miR-516a-3P, a potential circulating biomarker in hepatocellular carcinoma, correlated with rs738409 polymorphism in PNPLA3. Personalized Medicine 19, 483–493. [DOI] [PubMed] [Google Scholar]
- 162.Medina PP, Nolde M and Slack FJ (2010) OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467, 86–90. [DOI] [PubMed] [Google Scholar]
- 163.Venza I et al. (2015) IL-10Ralpha expression is post-transcriptionally regulated by miR-15a, miR-185, and miR-211 in melanoma. BMC Medical Genomics 8, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fillatreau S and O'Garra A and Springer-Verlag Gmb H (2016) Interleukin-10 in Health and Disease.
- 165.Sharma A et al. (2009) Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proceedings of the National Academy of Sciences of the USA 106, 5761–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ma F et al. (2010) MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. Journal of Immunology 184, 6053–6059. [DOI] [PubMed] [Google Scholar]
- 167.Patial S and Blackshear PJ (2016) Tristetraprolin as a therapeutic target in inflammatory disease. Trends in Pharmacological Sciences 37, 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu Y et al. (2011) MicroRNA-98 negatively regulates IL-10 production and endotoxin tolerance in macrophages after LPS stimulation. FEBS Letters 585, 1963–1968. [DOI] [PubMed] [Google Scholar]
- 169.Swaminathan S et al. (2012) Differential regulation of the Let-7 family of microRNAs in CD4 + T cells alters IL-10 expression. Journal of Immunology 188, 6238–6246. [DOI] [PubMed] [Google Scholar]
- 170.McCoy CE et al. (2010) IL-10 inhibits miR-155 induction by toll-like receptors. Journal of Biological Chemistry 285, 20492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rodriguez A et al. (2007) Requirement of bic/microRNA-155 for normal immune function. Science 316, 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schaefer JS et al. (2011) Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. Journal of Immunology 187, 5834–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rossato M et al. (2012) IL-10-induced microRNA-187 negatively regulates TNF-alpha, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proceedings of the National Academy of Sciences of the USA 109, E3101–E3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tang XD et al. (2018) lncRNA AFAP1-AS1 promotes migration and invasion of non-small cell lung cancer via up-regulating IRF7 and the RIG-I-like receptor signaling pathway. Cellular Physiology and Biochemistry 50, 179–195. [DOI] [PubMed] [Google Scholar]
- 175.Bhan A, Soleimani M and Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Research 77, 3965–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pei X, Wang X and Li H (2018) LncRNA SNHG1 regulates the differentiation of Treg cells and affects the immune escape of breast cancer via regulating miR-448/IDO. International Journal of Biological Macromolecules 118, 24–30. [DOI] [PubMed] [Google Scholar]
- 177.Ye Y et al. (2018) Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. Journal of Cellular Biochemistry 119, 2951–2963. [DOI] [PubMed] [Google Scholar]
- 178.Zhou WY et al. (2019) Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. Journal of Cellular Physiology 234, 23176–23189. [DOI] [PubMed] [Google Scholar]
- 179.Li Y et al. (2017) Long non-coding RNA growth arrest specific transcript 5 acts as a tumour suppressor in colorectal cancer by inhibiting interleukin-10 and vascular endothelial growth factor expression. Oncotarget 8, 13690–13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Elton TS et al. (2013) Regulation of the MIR155 host gene in physiological and pathological processes. Gene 532, 1–12. [DOI] [PubMed] [Google Scholar]
- 181.Sun YM et al. (2019) circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 134, 1533–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yu J et al. (2018) Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. Journal of Hepatology 68, 1214–1227. [DOI] [PubMed] [Google Scholar]
- 183.Wang S et al. (2019) Circular RNA FOXP1 promotes tumor progression and Warburg effect in gallbladder cancer by regulating PKLR expression. Molecular Cancer 18, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Memczak S et al. (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- 185.Kun-Peng Z et al. (2018) Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics 10, 1327–1346. [DOI] [PubMed] [Google Scholar]
- 186.Huang Z et al. (2019) Hsa_circ_0005519 increases IL-13/IL-6 by regulating hsa-let-7a-5p in CD4 + T cells to affect asthma. Clinical & Experimental Allergy 49, 1116–1127. [DOI] [PubMed] [Google Scholar]
- 187.Liu X et al. (2017) Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget 8, 114829–114838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhu M et al. (2023) CircMERTK modulates the suppressive capacity of tumor-associated macrophage via targeting IL-10 in colorectal cancer. Human Cell 36, 276–285. [DOI] [PubMed] [Google Scholar]
- 189.Hong W et al. (2020) Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). Journal of Experimental & Clinical Cancer Research 39, 149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 190.Peng X et al. (2019) The role of circular RNA HECTD1 expression in disease risk, disease severity, inflammation, and recurrence of acute ischemic stroke. Journal of Clinical Laboratory Analysis 33, e22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun G et al. (2021) Circular RNA Foxo3 relieves myocardial ischemia/reperfusion injury by suppressing autophagy via inhibiting HMGB1 by repressing KAT7 in myocardial infarction. Journal Of inflammation Research 14, 6397–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang Y et al. (2021) Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. International Journal of Nanomedicine 16, 2803–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen Q et al. (2020) Circular RNA circSnx5 controls immunogenicity of dendritic cells through the miR-544/SOCS1 axis and PU.1 activity regulation. Molecular Therapy 28, 2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huang L, Ma J and Cui M (2021) Circular RNA hsa_circ_0001598 promotes programmed death-ligand-1-mediated immune escape and trastuzumab resistance via sponging miR-1184 in breast cancer cells. Immunologic Research 69, 558–567. [DOI] [PubMed] [Google Scholar]
- 195.Yi B et al. (2022) Circular RNA PLCE1 promotes epithelial mesenchymal transformation, glycolysis in colorectal cancer and M2 polarization of tumor-associated macrophages. Bioengineered 13, 6243–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liang B et al. (2020) CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Annals of Translational Medicine 8, 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ghafouri-Fard S, Gholipour M and Taheri M (2021) The emerging role of long Non-coding RNAs and circular RNAs in coronary artery disease. Frontiers in Cardiovascular Medicine 8, 632393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Meng M and Wu YC (2022) LMX1B Activated circular RNA GFRA1 modulates the tumorigenic properties and immune Escape of prostate cancer. Journal of Immunology Research 2022, 7375879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhang Y et al. (2021) Circ_0005075 targeting miR-151a-3p promotes neuropathic pain in CCI rats via inducing NOTCH2 expression. Gene 767, 145079. [DOI] [PubMed] [Google Scholar]
- 200.Gonzalez C et al. (2022) Role of circular RNA cdr1as in modulation of macrophage phenotype. Life Sciences 309, 121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yang J et al. (2021) Circular RNA CHST15 sponges miR-155-5p and miR-194-5p to promote the immune Escape of lung cancer cells mediated by PD-L1. Frontiers in Oncology 11, doi: 10.3389/fonc.2021.595609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fu X et al. (2022) Hsa_circ_0046523 mediates an immunosuppressive tumor microenvironment by regulating MiR-148a-3p/PD-L1 axis in pancreatic cancer. Frontiers in Oncology 12, 877376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu J et al. (2020) Down-regulation of circDMNT3B is conducive to intestinal mucosal permeability dysfunction of rats with sepsis via sponging miR-20b-5p. Journal of Cellular and Molecular Medicine 24, 6731–6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Iyer SS and Cheng G (2012) Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Critical Reviews in Immunology 32, 23–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Asadullah K, Sterry W and Volk HD (2003) Interleukin-10 therapy--review of a new approach. Pharmacological Reviews 55, 241–269. [DOI] [PubMed] [Google Scholar]
- 206.Josephson K, Logsdon NJ and Walter MR (2001) Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity 15, 35–46. [DOI] [PubMed] [Google Scholar]
- 207.Finbloom DS and Winestock KD (1995) IL-10 induces the tyrosine phosphorylation of tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. Journal of Immunology 155, 1079–1090. [PubMed] [Google Scholar]
- 208.Williams LM et al. (2004) Interleukin-10 suppression of myeloid cell activation--a continuing puzzle. Immunology 113, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Riley JK et al. (1999) Interleukin-10 receptor signaling through the JAK-STAT pathway. Requirement for two distinct receptor-derived signals for anti-inflammatory action. Journal of Biological Chemistry 274, 16513–16521. [DOI] [PubMed] [Google Scholar]
- 210.Walter MR (2014) The molecular basis of IL-10 function: from receptor structure to the onset of signaling. Current Topics in Microbiology and Immunology 380, 191–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Youssef SS et al. (2022) PNPLA3 and IL 28B signature for predicting susceptibility to chronic hepatitis C infection and fibrosis progression. Archives of Physiology and Biochemistry 128, 483–489. [DOI] [PubMed] [Google Scholar]
- 212.Li G and Li D (2016) Relationship between IL-10 gene polymorphisms and the risk of non-Hodgkin lymphoma: a meta-analysis. Human Immunology 77, 418–425. [DOI] [PubMed] [Google Scholar]
- 213.Chagas BS et al. (2013) An interleukin-10 gene polymorphism associated with the development of cervical lesions in women infected with human papillomavirus and using oral contraceptives. Infection Genetics and Evolution 19, 32–37. [DOI] [PubMed] [Google Scholar]
- 214.Faupel-Badger JM et al. (2008) Association of IL-10 polymorphisms with prostate cancer risk and grade of disease. Cancer Causes & Control 19, 119–124. [DOI] [PubMed] [Google Scholar]
- 215.Cunningham LM et al. (2003) Polymorphisms in the interleukin 10 gene promoter are associated with susceptibility to aggressive non-Hodgkin's Lymphoma. Leukemia & Lymphoma 44, 251–255. [DOI] [PubMed] [Google Scholar]
- 216.Shahzad MN et al. (2020) Association between interleukin gene polymorphisms and multiple myeloma susceptibility. Molecular and Clinical Oncology 12, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stanczuk GA et al. (2001) Cancer of the uterine cervix may be significantly associated with a gene polymorphism coding for increased IL-10 production. International Journal of Cancer 94, 792–794. [DOI] [PubMed] [Google Scholar]
- 218.El-Omar EM et al. (2003) Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124, 1193–1201. [DOI] [PubMed] [Google Scholar]
- 219.Moghimi M et al. (2018) Association of IL-10 rs1800871 and rs1800872 polymorphisms with breast cancer risk: a systematic review and meta-analysis. Asian Pacific Journal of Cancer Prevention: APJCP 19, 3353–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Gamaleldin M, Moussa M and Eldin Imbaby S (2022) Role of interleukin-10 (1082G/A) and splicing factor 3B subunit 1 (2098A/G) gene polymorphisms in chronic lymphocytic leukemia. Journal of Applied Hematology 13, 76–83. [Google Scholar]
- 221.McCarron SL et al. (2002) Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Research 62, 3369–3372. [PubMed] [Google Scholar]
- 222.Khorrami S et al. (2022) Association of a genetic variant in interleukin-10 gene with increased risk and inflammation associated with cervical cancer. Gene 807, 145933. [DOI] [PubMed] [Google Scholar]
- 223.Li L et al. (2022) Association of interleukin-10 polymorphism (rs1800896, rs1800871, and rs1800872) with breast cancer risk: an updated meta-analysis based on different ethnic groups. Frontiers in Genetics 13, 10.3389/fgene.2022.829283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Namazi A et al. (2018) Association of interleukin-10 −1082 A/G (rs1800896) polymorphism with susceptibility to gastric cancer: meta-analysis of 6,101 cases and 8,557 controls. Arquivos de Gastroenterologia 55, 33–40. [DOI] [PubMed] [Google Scholar]
- 225.Li F et al. (2020) Association between interleukin-10 gene polymorphisms and risk of oral carcinoma: a meta-analysis. Histology and Histopathology 35, 1329–1336. [DOI] [PubMed] [Google Scholar]
- 226.Zheng C et al. (2001) Interleukin-10 gene promoter polymorphisms in multiple myeloma. International Journal of Cancer 95, 184–188. [DOI] [PubMed] [Google Scholar]
- 227.Jafari-Nedooshan J et al. (2019) Association of promoter region polymorphisms of IL-10 gene with susceptibility to lung cancer: systematic review and meta-analysis. Asian Pacific Journal of Cancer Prevention: APJCP 20, 1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Cunningham LM et al. (2003) Polymorphisms in the interleukin 10 gene promoter are associated with susceptibility to aggressive non-Hodgkin's lymphoma. Leukemia & Lymphoma 44, 251–255. [DOI] [PubMed] [Google Scholar]
- 229.Lauten M et al. (2002) Association of initial response to prednisone treatment in childhood acute lymphoblastic leukaemia and polymorphisms within the tumour necrosis factor and the interleukin-10 genes. Leukemia 16, 1437–1442. [DOI] [PubMed] [Google Scholar]
- 230.Rashed R et al. (2018) Associations of interleukin-10 gene polymorphisms with acute myeloid leukemia in human (Egypt). Journal of Cancer Research and Therapeutics 14, 1083–1086. [DOI] [PubMed] [Google Scholar]
- 231.Mirjalili SA et al. (2018) Association of promoter region polymorphisms of interleukin-10 gene with susceptibility to colorectal cancer: a systematic review and meta-analysis. Arquivos de Gastroenterologia 55, 306–313. [DOI] [PubMed] [Google Scholar]
- 232.Martínez-Escribano JA et al. (2002) Interleukin-10, interleukin-6 and interferon-gamma gene polymorphisms in melanoma patients. Melanoma Research 12, 465–469. [DOI] [PubMed] [Google Scholar]
- 233.Howell WM et al. (2001) IL-10 promoter polymorphisms influence tumour development in cutaneous malignant melanoma. Genes and Immunity 2, 25–31. [DOI] [PubMed] [Google Scholar]