Abstract
During development, cells make switch-like decisions to activate new gene programs specifying cell lineage. The mechanisms underlying these decisive choices remain unclear. Here, we show that the cardiovascular transcriptional coactivator myocardin (MYOCD) activates cell identity genes by concentration-dependent and switch-like formation of transcriptional condensates. MYOCD forms such condensates and activates cell identity genes at critical concentration thresholds achieved during smooth muscle cell and cardiomyocyte differentiation. The carboxyl-terminal disordered region of MYOCD is necessary and sufficient for condensate formation. Disrupting this region’s ability to form condensates disrupts gene activation and smooth muscle cell reprogramming. Rescuing condensate formation by replacing this region with disordered regions from functionally unrelated proteins rescues gene activation and smooth muscle cell reprogramming. Our findings demonstrate that MYOCD condensate formation is required for gene activation during cardiovascular differentiation. We propose that the formation of transcriptional condensates at critical concentrations of cell type–specific regulators provides a molecular switch underlying the activation of key cell identity genes during development.
Myocardin activates cardiovascular gene expression by formation of transcriptional condensates.
INTRODUCTION
Through the precise and decisive activation of different gene programs, a single genome can give rise to the large diversity of cellular phenotypes and functions found in multicellular organisms. Cell type–specific transcriptional regulators determine cell fate during development, reprogram cell states, and perform these functions at critical concentrations leading to switches in cell state (1–3). During differentiation, the concentrations of competing, lineage-specifying transcriptional regulators gradually increase until reaching a critical threshold when a switch-like, lineage-specifying decision is made (4–6). Such decisions have been proposed to be controlled by a dosage-dependent and hyper-cooperative molecular switch (7). The mechanisms underlying the switch-like activation in cell type–specific gene programs are unclear (3). It has been suggested that the cooperativity of transcription factor–DNA binding underlies such switches (3, 8, 9). While DNA binding transcription factors are required to specify genomic loci, transcription factor binding alone is not sufficient to activate genes (10–12). Gene activation also requires the recruitment of coactivators, RNA polymerase II (RNA Pol II), and its various allosteric regulators (10–12). Furthermore, several coactivators lacking any DNA binding domains are capable of determining cell fate (1).
Recent studies have shown that general transcriptional coactivators activate genes in part through the formation of biomolecular condensates (13–15), dynamic assemblies formed at critical concentrations through cooperative, weak, multivalent interactions, often among disordered regions (16–19). However, quantitative analysis of condensate formation and its correlation with lineage-specific gene activation has, thus far, been lacking. While the formation of coactivator condensates requires DNA binding transcription factors (20, 21), these condensates activate genes by creating high local concentrations of RNA Pol II and its positive allosteric regulators (13). The threshold concentration required for condensate formation provides a potential mechanism for the switch-like activation of cell lineage–specific gene programs and efficient transition of cell fate during development. Several lineage-specifying transcriptional coactivators exhibit large changes in concentration during development (1), prompting us to investigate whether the physiologically regulated changes in coactivator concentration lead to condensate-driven activation of cell identity genes.
To investigate whether cell identity gene activation is coupled to condensate formation of cell type–specific transcriptional regulators, we chose the smooth muscle cell (SMC)–specific and cardiomyocyte (CM)–specific transcriptional coactivator myocardin (MYOCD). MYOCD is expressed at the earliest stages of cardiac lineage specification (22) before many critical lineage-specific genes are expressed (23), and it is responsible for activating SMC- and CM-specific gene expression programs during development (22, 24–26). MYOCD lacks DNA binding activity and activates specific genes by its association with the ubiquitously expressed serum response factor (SRF) (22, 27). SRF is critical for MYOCD-mediated gene activation (28, 29). MYOCD provides an especially informative model to test the relationship between condensate formation and cell lineage specification because (i) MYOCD forms nuclear puncta when ectopically expressed in noncardiovascular cells (22), (ii) loss of MYOCD causes severe defects in vascular smooth muscle development, as well as defects in CM survival and postnatal function (30–32), (iii) expression of MYOCD is sufficient to reprogram fibroblasts into SMCs (33), and (iv) reduction in MYOCD expression has been linked to defects in smooth muscle development in mice and humans (34).
Here, we find that MYOCD condensate formation is required for its gene activation and lineage specification functions. By modeling the physiological changes in MYOCD concentration during cell differentiation, coupled with quantitative fluorescence microscopy, single-cell resolution reporter assays, and cellular reprogramming assays, we demonstrate that condensate formation is directly linked to the activation of cell identity genes and cell lineage specification. During SMC and CM differentiation, MYOCD condensates are present at sites of cell identity gene transcription. MYOCD condensates form, activate gene expression, and specify cell state at critical concentration thresholds, dependent on the C-terminal disordered region of MYOCD. Disrupting condensate formation by manipulating the sequence of this region impairs gene activation and lineage specification, which can be partially rescued by replacing this region with condensate-forming disordered regions from functionally unrelated proteins. MYOCD condensates lead to gene activation at least in part by the selective partitioning of p300 and RNA Pol II to target genes through its C-terminal disordered region. These results demonstrate that coactivator condensation enables switch-like changes in gene expression programs crucial for lineage specification.
RESULTS
MYOCD nuclear condensates are associated with components of active transcription
We previously observed that MYOCD forms nuclear puncta when ectopically expressed in COS-7 cells (22). We confirmed that MYOCD forms nuclear puncta upon ectopic expression by multiple imaging methods including indirect immunofluorescence of a Myc-epitope–tagged construct (Fig. 1, A and B) and direct fluorescence of a monomeric enhanced green fluorescent protein (mEGFP)–tagged construct in both fixed (fig. S1, A and B) and live cells (fig. S1C). In addition, fluorescence recovery after photobleaching (FRAP) experiments revealed that MYOCD puncta exhibit similar dynamic rearrangement, as observed for other nuclear condensates (fig. S1, C and D) (14). Given these results, we will refer to the MYOCD puncta as MYOCD condensates.
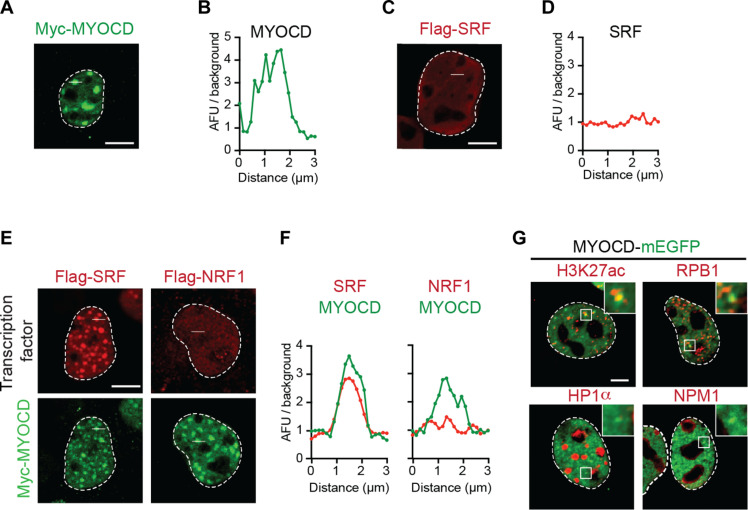
Fig. 1. MYOCD nuclear condensates are associated with components of active transcription.
(A) Immunofluorescence (IF) imaging of Myc-tagged MYOCD ectopically expressed in COS-7 cells with Myc antibody targeting the epitope. Scale bar, 5 μm. (B) Line profile plot presented as arbitrary fluorescence units (AFU)/background across the 3-μm white line shown in (A). (C) IF imaging of Flag-tagged SRF ectopically expressed in COS-7 cells with Flag antibody targeting the epitope. Scale bar, 5 μm. (D) Line profile plot presented as AFU/background across the 3-μm white line shown in (C). (E) IF imaging of Myc-tagged MYOCD and other transcription factors (Flag-tagged SRF and Flag-tagged NRF1) coexpressed in COS-7 cells. Scale bar, 5 μm. (F) Line profile plot presented as AFU/background for either MYOCD (green) or the indicated coexpressed transcription factor (red) across the 3-μm white line shown in (E). (G) Representative micrographs of COS-7 cells expressing MYOCD-mEGFP (green) and co-IF for factors, including H3K27ac, RPB1, HP1α, and NPM1 (red). Scale bar, 5 μm.
To investigate whether MYOCD condensates are relevant to the function of MYOCD as a transcriptional coactivator, we first investigated how they interact with known MYOCD-associated transcription factors. MYOCD lacks intrinsic DNA binding activity and is recruited to cardiovascular gene regulatory regions by association with DNA binding transcription factors like SRF (22). SRF, when expressed alone, displayed diffuse intranuclear distributions with no indication of condensate formation (Fig. 1, C and D), but when coexpressed with MYOCD, SRF became concentrated within MYOCD condensates (Fig. 1, E and F). Similarly, myocyte enhancer factor 2C, another transcription factor associated with MYOCD (35), was diffuse when expressed alone and concentrated in MYOCD condensates when coexpressed with MYOCD (fig. S1, E to H). In contrast, functionally unrelated transcription factors [nuclear respiratory factor 1 (NRF1) and nuclear transcription factor Y subunit α (NFYA)] were not concentrated in nuclear condensates when coexpressed with MYOCD (Fig. 1, E and F, and fig. S1, E to H). These results show that MYOCD nuclear condensates selectively concentrate functionally relevant DNA binding transcription factors and exclude unrelated factors.
To test whether MYOCD condensates correspond to sites of gene activation, we next performed immunofluorescence to investigate whether the endogenous gene regulatory machinery is concentrated within MYOCD condensates. Expression of MYOCD followed by immunofluorescence revealed that MYOCD condensates in COS-7 cells concentrate endogenous RNA Pol II (by probing its largest subunit, RPB1) and colocalize with histone H3 lysine 27 acetylation (H3K27ac), a histone modification associated with active chromatin (Fig. 1G and fig. S1, I to L). As controls, we also performed immunofluorescence for heterochromatin protein 1α (HP1α), a marker of transcriptionally repressed regions of chromatin, and nucleophosmin 1 (NPM1), a marker of the nucleolus (Fig. 1G). Line profile and average image analysis centered on MYOCD condensates revealed that RPB1 (the largest subunit of RNA Pol II) and H3K27ac were significantly enriched within MYOCD condensates, while HP1α and NPM1 were not (fig. S1, I to L). Together, these results demonstrate that ectopic expression of MYOCD leads to the formation of condensates that selectively concentrate specific crucial DNA binding transcription factors and RNA Pol II at active regions of chromatin, thereby linking MYOCD condensates with gene activation.
MYOCD forms condensates during lineage specification of hiPSCs
To investigate whether MYOCD forms condensates at endogenous concentrations during cardiovascular development, we modeled SMC and CM differentiation in vitro using human induced pluripotent stem cells (hiPSCs). We generated an isogenic hiPSC line with homozygous in-frame knock-in of an mEGFP expression cassette within the MYOCD open reading frame (Fig. 2A; fig. S2, A to C; and Materials and Methods). Using this cell line, we monitored MYOCD concentration and condensate formation by fluorescence microscopy before and after differentiation into SMCs or CMs (Fig. 2A, fig. S2D, and Materials and Methods). MYOCD is not expressed in the stem cell state, but in mature SMCs and CMs, we observed nuclear condensates by both fixed-cell and live-cell imaging (Fig. 2, B and C). These results demonstrate that MYOCD forms condensates at endogenous concentrations.
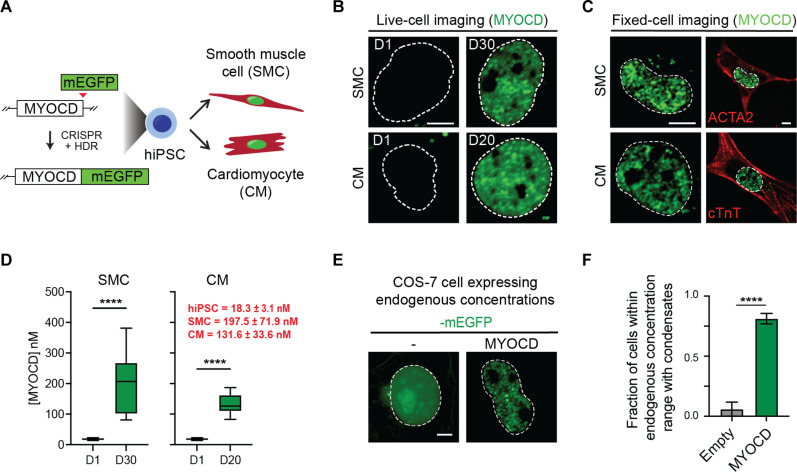
Fig. 2. MYOCD forms condensates during hiPSC differentiation into SMCs and CMs.
(A) Schematic of the creation of MYOCD-mEGFP knock-in hiPSC line and differentiation of hiPSCs into SMCs and CMs. (B) Live-cell imaging of endogenously tagged MYOCD-mEGFP in differentiation day 1 (D1) and mature hiPSC-derived SMC (D30) and CM (D20) nuclei. Scale bar, 5 μm. Dotted line denotes the nucleus, drawn using Hoechst 33342 staining (not shown). (C) Co-IF of MYOCD-mEGFP with smooth muscle α-actin (ACTA2) or cardiac troponin T (cTnT) in either mature hiPSC-derived SMCs and CMs, as labeled. Left column is zoomed in to highlight MYOCD condensates, and only the GFP channel is presented. Scale bar, 5 μm. Right column is zoomed out of the same cell to highlight cell body with cell marker stain. Scale bar, 5 μm. (D) Box plot (10 to 90%) of the concentration of MYOCD in hiPSCs compared to mature SMCs (left) or mature CMs (right). Red text denotes concentration of MYOCD as mean ± SD. P values from t test, ****P ≤ 0.0001. n = 10. (E) Representative micrographs (max projections) of COS-7 cells expressing mEGFP or MYOCD-mEGFP. The nuclear concentration of MYOCD-mEGFP in this micrograph is 124.6 nM, equivalent to physiological concentrations in SMCs and CMs, as shown in (D). The nuclear concentration of the mEGFP control is 718.9 nM. Dotted line defines nucleus from Hoechst 33342 staining (not shown). Scale bar, 5 μm. (F) Bar chart of the fraction of cells with condensates between the concentration range of MYOCD in SMCs and CMs, 98.0 to 268.4 nM [± SD of mean concentration measured in SMC and CM in (D)]. Condensates are identified by automated analysis pipeline. Data are mean ± SEM. P values from t test, ****P ≤ 0.0001. n ~ 25.
To understand the actual concentration changes that occur during differentiation, we sought to measure the nuclear concentrations of MYOCD during SMC and CM differentiation. To convert fluorescence intensity measurements to concentrations, we imaged a standard curve of known concentrations of recombinant purified mEGFP. The fluorescence intensity was linear within the tested mEGFP concentration range of 0 to 500 nM (fig. S2, E and F). Using this standard curve, we calculated the nuclear concentration of MYOCD in hiPSCs (18.3 ± 3.1 nM), mature SMCs (197.5 ± 71.9 nM), and mature CMs (131.6 ± 33.6 nM) (Fig. 2D). To relate these endogenous nuclear concentrations found in mature SMCs and CMs to those found upon ectopic expression of MYOCD in COS-7 cells, we performed a similar analysis in COS-7 cells expressing mEGFP-tagged MYOCD (fig. S2, G and H). We confirmed that MYOCD formed condensates in COS-7 cells within the range of concentrations observed in mature SMCs and CMs (Fig. 2, E and F). These results demonstrate that MYOCD is sufficient to form condensates at endogenous concentrations found in SMCs and CMs.
MYOCD condensates are sites of cell type–specific transcription
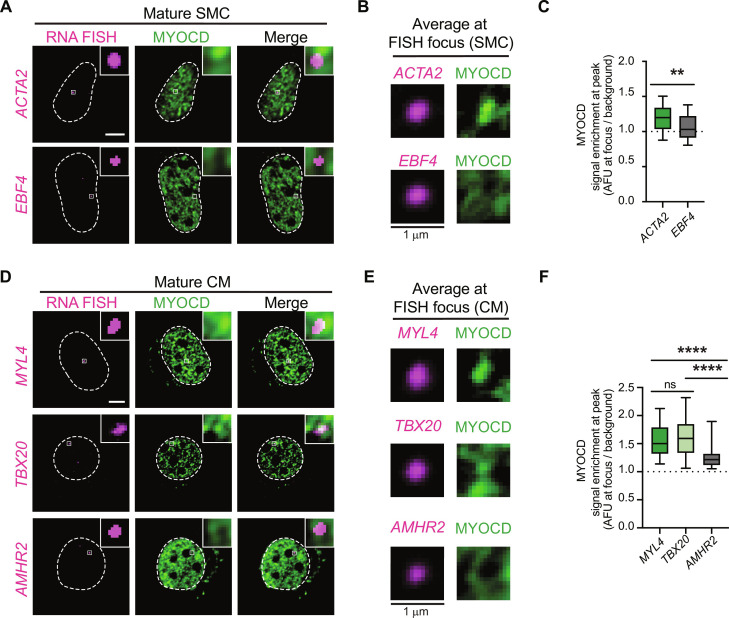
To examine whether MYOCD condensates formed in SMCs or CMs are sites of cell-identity gene transcription, we performed nascent RNA fluorescence in situ hybridization (FISH) using intronic probes, combined with imaging of MYOCD (Materials and Methods). To define the expected targets of MYOCD condensates, we performed chromatin immunoprecipitation assays with sequencing (ChIP-seq) and RNA sequencing (RNA-seq) in cells during differentiation (Materials and Methods). Analysis of MYOCD ChIP-seq showed the expected results that MYOCD was bound to putative enhancers and promoters of key SMC and CM genes (fig. S3, A to C) and that peaks had DNA motif enrichment for the CArG box, which is bound by SRF (fig. S3C) (22, 36). Using our ChIP-seq and RNA-seq data, we identified genes that are activated during differentiation with either high MYOCD occupancy as expected targets of MYOCD condensates [smooth muscle α-actin (ACTA2) for SMCs] [myosin light chain 4 (MYL4) and T-box transcription factor 20 (TBX20) for CMs] or with no observed MYOCD occupancy as negative controls [EBF family member 4 (EBF4) for SMCs and anti-Mullerian hormone receptor type 2 (AMHR2) for CMs] (fig. S3, D and E). In SMCs, we found that MYOCD condensates consistently overlapped with RNA-FISH foci of ACTA2, but not EBF4 (Fig. 3, A to C). In CMs, MYOCD condensates overlapped with RNA-FISH foci of MYL4, but not AMHR2 (Fig. 3, D to F). Average image analysis (Materials and Methods) for MYOCD signal, centered on FISH foci or randomly selected nuclear locations, showed that MYOCD fluorescence intensity was higher at the center of expected target FISH foci than at FISH foci of the negative controls (Fig. 3, A to F). These findings demonstrate that MYOCD condensates colocalize with sites of cell lineage transcription during lineage specification.
Fig. 3. MYOCD condensates are sites of cell identity gene transcription.
(A) RNA FISH and IF in mature SMC nuclei for indicated nascent transcript (magenta) and MYOCD (GFP). Square box denotes location of crop shown in top right. Scale bar, 5 μm. (B) Average signal of indicated nascent transcript (magenta) or MYOCD (GFP) centered on the nascent transcript focus (1.5 μm2) in SMC. n ~ 25. (C) Boxplot (mean ± 10 to 90%) showing enrichment of MYOCD at either the center of the nascent transcript foci or at random sites in SMCs. P values represent results of t test followed (P values: **P ≤ 0.01). (D) RNA FISH and IF in mature CM nuclei for RNA of indicated transcript (magenta) and MYOCD (GFP). Square box denotes location of crop shown in top right. Scale bar, 5 μm. (E) Average signal of indicated nascent transcript (magenta) or MYOCD (GFP) centered on the nascent transcript focus (1.5 μm2) in CM. n ~ 25. (F) Boxplot (mean ± 10 to 90%) showing enrichment of MYOCD either at the center of the nascent transcript foci or at random sites in CMs. P values represent results of Kruskal-Wallis test followed by Dunn’s test for multiple comparisons (P values: ns P > 0.05, ****P ≤ 0.0001).
Formation of MYOCD condensates activates reporter gene expression
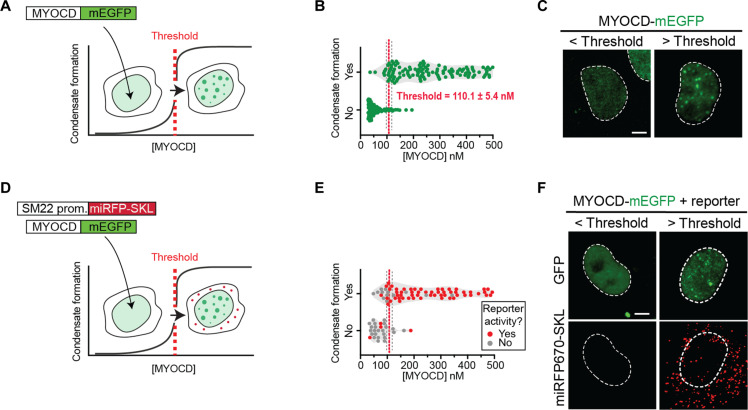
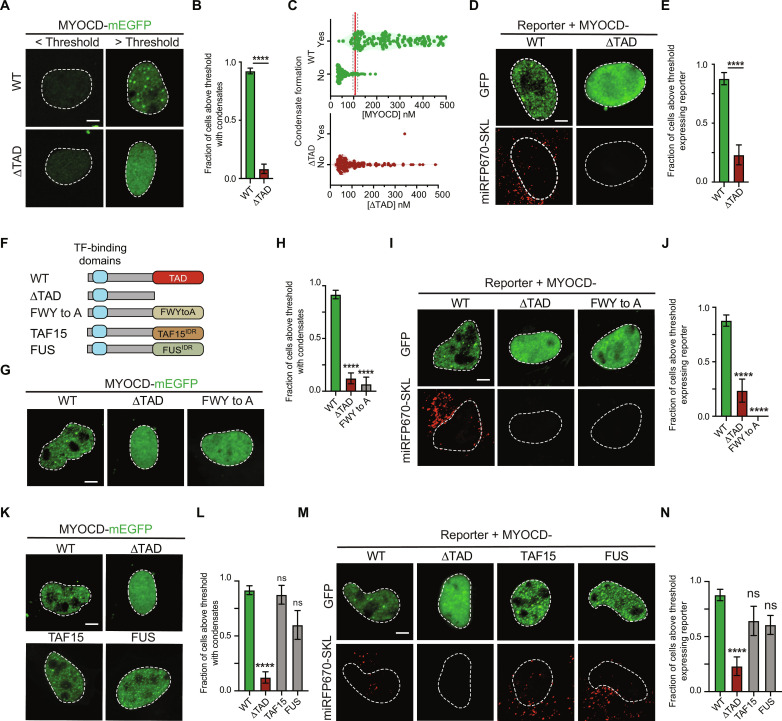
Given that MYOCD condensates are coincident with sites of gene activity, we next sought to test the extent to which condensate formation and gene activation were linked processes. Because of the highly cooperative nature of the weak multivalent interactions underlying condensates, their formation is expected to occur at sharp concentration thresholds (17). Identifying the concentration threshold for condensate formation and measuring reporter gene activity above and below this concentration would enable us to assess the extent to which the formation of MYOCD condensates is coupled to MYOCD’s gene activation function. We first set out to define the critical concentration for condensate formation by taking advantage of the wide range of MYOCD expression achieved upon transient transfection of MYOCD-mEGFP in COS-7 cells (Fig. 4A). Using a standard curve (fig. S2G), we measured the nuclear concentration of MYOCD in 292 individual cells across five replicates and scored whether or not MYOCD condensates were detectable in each of these cells (fig. S4, A and B, and Materials and Methods). By fitting a simple logistic regression to these data points, we defined a concentration threshold (Materials and Methods) for MYOCD condensate formation of 110.1 ± 5.4 nM (Fig. 4B). Nearly all cells above the threshold contained condensates (95.0%) and only a few cells below the threshold contained condensates (11.5%) (Fig. 4C and fig. S4C). Notably, this threshold is above the endogenous MYOCD concentration measured in hiPSCs and below the endogenous MYOCD concentration measured in mature SMCs and CMs (Fig. 2D). These results demonstrate that MYOCD condensates form at critical threshold concentrations of MYOCD in cells that are comparable to the endogenous concentrations of MYOCD we observed in mature SMCs and CMs.
Fig. 4. Formation of MYOCD condensates activates reporter gene expression.
(A) Schematic showing that MYOCD-mEGFP forms nuclear condensates (green) in COS-7 cells in a concentration-dependent manner. (B) Violin plot of MYOCD concentration and condensate formation in COS-7 cells where each dot represents one nucleus. Threshold is determined by a simple logarithmic regression analysis and represented by X at 50% ± SD. P values from t test, ****P ≤ 0.0001. Five biological replicates total. n = 292. (C) Representative micrographs (max projections) of COS-7 cells expressing MYOCD-mEGFP below or above the critical concentration threshold demonstrated in (B). Scale bar, 5 μm. (D) Schematic showing that the formation of condensates (green) is coupled to the expression of the SM22 promoter-driven fluorescent reporter trafficked to peroxisomes (miRFP670-SKL, red). (E) Violin plot of MYOCD concentration and condensate formation where each dot represents one nucleus. Cells expressing reporter shown in red and cells not expressing reporter shown in gray. n = 48. (F) Representative micrographs (max projections) of COS-7 cells expressing MYOCD-mEGFP below or above the critical concentration threshold showing either MYOCD (GFP) or the fluorescent reporter (miRFP670-SKL, shown in red). Brightness and contrast of displayed micrographs are equivalent unless otherwise stated. Scale bar, 5 μm.
To test whether MYOCD condensate formation is coupled to gene activation, we measured the expression of a reporter gene as a function of MYOCD concentration at single-cell resolution. To perform this analysis, we modified a previously described MYOCD-dependent SM22 luciferase reporter (22) in two ways. First, we reduced the size of the SM22 regulatory sequence by selecting only the region corresponding to MYOCD ChIP-seq peaks (fig. S4D), which increased the reporter’s responsiveness to MYOCD expression (fig. S4E). Second, to achieve single-cell resolution, we replaced luciferase with a far-red fluorescent protein (miRFP670) fused to a peroxisomal targeting sequence (SKL) (37). With this construct, we could detect far-red fluorescence in peroxisomes and GFP fluorescence in the nucleus of the same cell, allowing us to measure reporter activity, MYOCD concentration, and condensate formation at single-cell resolution (Fig. 4D). As expected, activation of the reporter gene was dependent on expression of MYOCD (fig. S4, F and G). Reporter activation almost exclusively occurred in cells above the critical concentration threshold for condensate formation (Fig. 4, E and F). Nearly all cells above the threshold activated reporter expression (90.5%) and only a few cells below the threshold showed evidence of reporter expression (18.8%) (fig. S4H). These results demonstrate that reporter gene activation is directly linked to MYOCD condensate formation.
Disrupting MYOCD condensate formation disrupts gene activation
We next sought to understand the molecular features of MYOCD that underlie its ability to form condensates with the goal of manipulating the formation of MYOCD condensates to test the effect on gene activation. MYOCD has several annotated domains in its N-terminal half that are required for its interaction with SRF and other DNA binding transcription factors (fig. S5A) (22). In the C terminus of MYOCD, there is a disordered region (fig. S5A) empirically defined as a transactivation domain (TAD) due to its necessity for reporter activity and its sufficiency as a Gal4 fusion to activate UAS reporters (22). To test whether MYOCDTAD has an intrinsic ability to form condensates, we purified this domain as a GFP fusion and found that it formed condensates in vitro (fig. S5, B and C). To test whether MYOCDTAD was required for condensate formation in cells, we expressed wild-type (WT) MYOCD and a deletion mutant lacking this domain (ΔTAD) in cells and found that the ΔTAD mutant did not readily form condensates within the concentration range tested (Fig. 5, A to C, and fig. S5D). As expected, deletion of MYOCDTAD also disrupted activation of our fluorescent reporter construct (Fig. 5, D and E). We noticed that MYOCDTAD contained 14 well-dispersed aromatic residues throughout its sequence (fig. S5E). Given the documented role of aromatic residues in condensate formation (38, 39), we tested whether substituting these residues to alanine would prevent condensate formation. To do that, we created a mutant MYOCD construct in which all 14 aromatic amino acids within the TAD were changed to alanine (FWY to A). We purified the FWY to A TAD mutant as a GFP fusion and found that it was unable to form condensates in vitro (fig. S5, F and G). When expressed in cells, MYOCD with the FWY to A mutant TAD (Fig. 5F) was also unable to form condensates within the concentration range tested (Fig. 5, G and H) and failed to activate the fluorescent reporter gene (Fig. 5, I and J). These results demonstrate that the condensate-forming capacity of MYOCDTAD and its ability to activate genes are linked.
Fig. 5. Aromatic residues are required for MYOCD condensate formation and reporter gene expression.
(A) Representative micrographs (max projections) of COS-7 cells expressing MYOCD-mEGFP WT or ΔTAD below or above the critical concentration threshold. Scale bar, 5 μm. (B) Bar chart of the fraction of cells with condensates. Data are mean ± SEM. P values from Mann-Whitney test, ****P ≤ 0.0001. Three biological replicates total, n = 292 (WT) and 106 (ΔTAD). (C) Violin plot of MYOCD concentration and condensate formation in COS-7 cells expressing MYOCD WT or ΔTAD, where each dot represents one nucleus. (D) Representative micrographs (max projections) of COS-7 cells expressing MYOCD-mEGFP WT or ΔTAD above the critical concentration threshold. Scale bar, 5 μm. (E) Bar chart of the fraction of cells with condensates. Data are mean ± SEM. P values from Mann-Whitney test, ****P ≤ 0.0001. n ~ 40. (F) Schematic of MYOCD WT, ΔTAD, aromatic substitution to alanine (FWYtoA) mutants, and TAD domain chimeras with indicated disordered region (FUS and TAF15) with transcription factor (TF)–binding domains (blue box). See fig. S5A for detailed domain architecture of MYOCD. (G and K) Representative micrographs of COS-7 cells expressing indicated MYOCD variants above the critical concentration threshold. (H and L) Bar chart of the fraction of cells with condensates. Data are mean ± SEM. P values represent results of one-way analysis of variance (ANOVA) with Dunn’s multiple comparison test (P values: ns P ≥ 0.05, ****P ≤ 0.0001). (H) n ~ 20. (l) n ~ 15. (I and M) Representative micrographs (max projections) of COS-7 cells expressing the fluorescent reporter and the indicated MYOCD variant above the critical concentration threshold. Scale bar, 5 μm. (J and N) Bar chart of the fraction of cells expressing reporter. Data are mean ± SEM. P values from one-way ANOVA with Dunn’s multiple comparison test (P values: ns P ≥ 0.05, ****P ≤ 0.0001). (I) n ~ 40. (N) n ~ 20.
Restoring MYOCD condensate formation promotes gene activation
Having shown that removing the TAD prevented both condensate formation and gene activation functions of MYOCD, we next sought to test whether MYOCD function could be rescued by replacing the TAD with condensate-promoting sequences from other proteins. To do this, we swapped MYOCDTAD with the N-terminal disordered regions of the FUS RNA binding protein (FUS) and TATA box–binding protein–associated factor 15 (TAF15). FUS and TAF15 are members of the FET family of RNA binding proteins implicated in diverse nuclear processes from mRNA processing to the DNA damage response. The N-terminal disordered regions of FUS and TAF15 are well-established to form condensates and have been associated with gene regulation (40, 41). While the disordered regions of TAF15 and FUS do not share positional sequence homology with MYOCDTAD (fig. S6A), they do share an intrinsic capacity to form condensates dependent on aromatic residues (39). Chimeric fusions of MYOCD in which its TAD was swapped for disordered regions of FUS and TAF15 (Fig. 5F) were ectopically expressed in COS-7 cells together with WT MYOCD and ΔTAD. While ΔTAD did not readily form condensates, both chimeric fusions of FUS and TAF15 rescued condensate formation to levels comparable to WT (Fig. 5, K and L). Both chimeric fusions also maintained SRF partitioning (fig. S6, B and C). Unexpectedly, while ΔTAD had significantly reduced activation of the fluorescent reporter gene, both chimeric fusions of FUS and TAF15 rescued reporter gene activation to levels comparable to WT (Fig. 5, M and N). These results demonstrate that condensate formation is necessary for MYOCD’s ability to activate genes.
Formation of MYOCD condensates drives activation of SMC genes
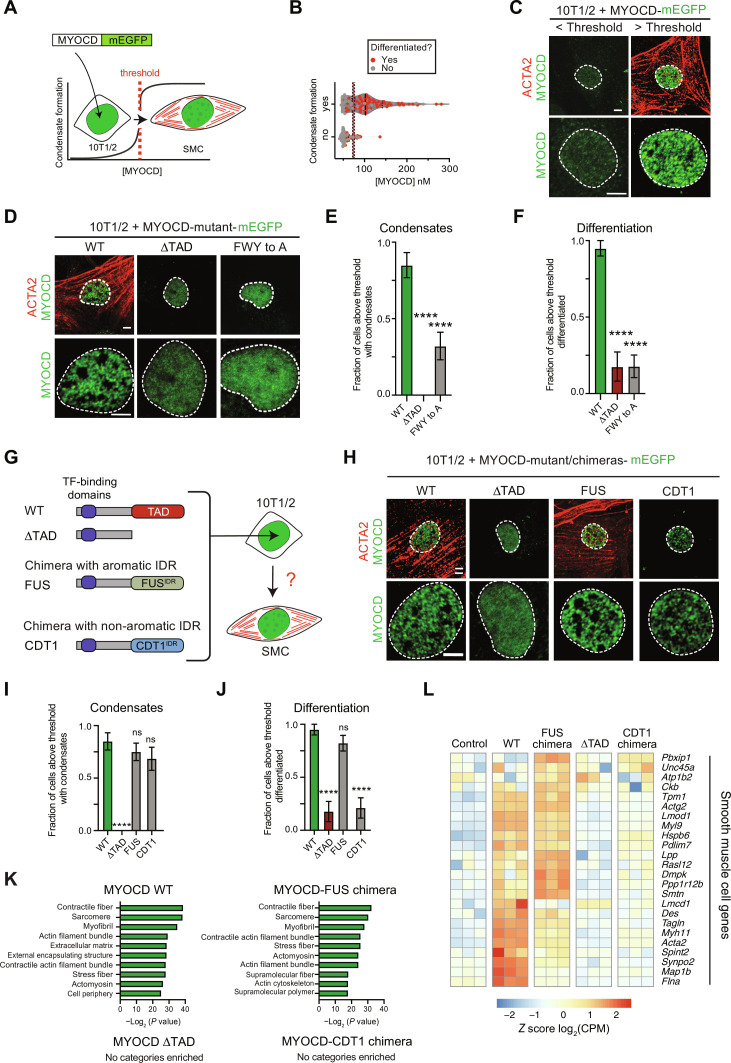
Ectopic expression of MYOCD is sufficient to reprogram 10T1/2 fibroblasts into SMCs (33), providing a powerful assay for us to test the relationship between MYOCD condensate formation and activation of cell identity genes. To determine the extent to which MYOCD condensate formation drives lineage-specific gene activation, we first examined the relationship between MYOCD condensate formation and SMC reprogramming. mEGFP-tagged MYOCD and its variants were expressed in 10T1/2 fibroblasts by retroviral transduction, allowing us to quantify nuclear concentrations and monitor condensate formation as a function of SMC reprogramming. To determine SMC reprogramming efficiency, we performed immunofluorescence for ACTA2, a marker of SMCs not expressed in 10T1/2 fibroblasts (33). Using this strategy, we could monitor MYOCD nuclear concentrations, MYOCD condensate formation, and reprogramming efficiency in individual cells (Fig. 6A). Using our standard curve method (fig. S7A) to calculate nuclear concentrations and our method to determine critical concentration (Fig. 6B), as previously described, we found that most cells above the critical concentration were reprogrammed, while the most cells below the critical concentration were not (Fig. 6, B and C, and fig. S7, B and C). The critical concentration for MYOCD in 10T1/2 cells is slightly lower than what we measured for COS-7 cells, suggesting that cellular context can influence threshold concentrations, likely due to the presence of more relevant binding partners.
Fig. 6. Multivalent interactions of aromatic residues are required for MYOCD condensate formation and 10T1/2 reprogramming to SMC.
(A) Schematic showing the correlation of MYOCD-mEGFP nuclear condensate formation (green) to the reprogramming of 10T1/2 cells (fibroblasts) into SMCs (ACTA2 in red). (B) Violin plot of MYOCD concentration and condensate formation in 10T1/2 cells, where each dot represents one nucleus. Dots shown in red denote cells that differentiated; dots shown in gray denote cells that did not differentiate. Threshold is determined by simple logarithmic regression analysis and represented by X at 50% ± SD. Two biological replicates, total n = 188. (C) Representative micrographs (max projections) of 10T1/2 cells expressing MYOCD-mEGFP (green) below or above the critical concentration threshold demonstrated in (B). IF for ACTA2 (red) denotes SMC differentiation. Scale bar, 5 μm. (D and H) Representative micrographs (max projections) of 10T1/2 cells expressing MYOCD-mEGFP (green) and indicated variants. Scale bar, 5 μm. (E, F, I, and J) Bar chart (mean ± SEM) displaying the fraction of cells above the critical MYOCD concentration threshold that form condensates (E and I) or differentiated (F and J). P value from one-way ANOVA with Dunnett’s multiple comparison test (P values: ns P ≥ 0.05, ****P ≤ 0.0001). n = 40. (G) Schematic of MYOCD WT, ΔTAD, and TAD domain chimeras with indicated disordered regions (FUS and CDT1) with transcription factor–binding domains. (K) Panther GO analysis for cellular component of differentially expressed genes from 10T1/2 cells expressing indicated construct with greater than or equal to a threefold increase of expression as compared to the empty vector control. (L) Heatmap of the differential expression [shown as z score of log2(CPM)] of crucial cardiovascular SMC genes. Data are from RNA-seq for indicated constructs upon overexpression in 10T1/2 cells.
We next sought to test whether disrupting condensate formation also disrupted activation of cell identity genes. We transduced 10T1/2 cells with retrovirus encoding mEGFP-tagged WT MYOCD, MYOCD with TAD deleted (ΔTAD), or MYOCD with the aromatic residues within the TAD substituted for alanine (FWY to A). In agreement with experiments in COS-7 cells, both ΔTAD and FWY to A constructs had reduced levels of condensate formation compared to WT [Fig. 6, D (bottom row) and E]. Analysis of ACTA2 staining showed that both ΔTAD and FWY to A constructs also had reduced reprogramming efficiency compared to WT [Fig. 6, D (top row) and F, and fig. S7, D and E]. While the FWY to A mutant showed some increase in condensate formation compared to ΔTAD (Fig. 6E), this modest increase did not lead to any measurable change in differentiation (Fig. 6F), suggesting that these condensates were not functional. These results demonstrate that disrupting condensate formation disrupts activation of SMC genes in 10T1/2 cells. Together, these results show that MYOCD condensate formation drives lineage specification and gene activation during 10T1/2 reprogramming.
MYOCD condensate formation is required for activation of SMC genes
Having shown that disrupting MYOCD condensate formation disrupts its ability to reprogram 10T1/2 cells, we next investigated whether rescuing condensate formation would rescue SMC reprogramming. Since the disordered regions of FUS and TAF15 are implicated in transcription as oncogenic fusions with various transcription factors (42), we added two additional condensate-promoting disordered regions from the proteins DEAD-box helicase 4 (DDX4) and Nephrin (NPHS1), which have no reported functional association with transcription. DDX4 is a germline-specific RNA helicase (43), and NPHS1 is the renal cell adhesion receptor Nephrin (44). Disordered regions from these two proteins do not share positional sequence homology with MYOCDTAD (fig. S8, A and B) but, like MYOCDTAD, promote condensate formation dependent on aromatic residues (43, 44). mEGFP-tagged MYOCD chimeras with the TAD swapped for the disordered regions of FUS, TAF15, DDX4, or NPHS1 were expressed in 10T1/2 cells by retroviral transduction (Fig. 6G and fig. S8A). WT MYOCD and the ΔTAD mutant were included as positive and negative controls, respectively. All constructs were expressed at equivalent levels relative to WT (fig. S8C). As expected, the ΔTAD mutant failed to form condensates, but chimeric fusions with disordered regions of FUS, TAF15, DDX4, and NPHS1 all rescued condensate formation to levels comparable with WT [Fig. 6, H (bottom row) and I, and fig. S8, D and E]. Analysis of ACTA2 staining showed that while the ΔTAD mutant had reduced reprogramming efficiency, chimeric fusions with disordered regions of FUS, TAF15, DDX4, and NPHS1 all rescued reprogramming to levels comparable to WT [Fig. 6, H (top row) and J, and fig. S8, D and F]. Analysis of bulk mRNA abundance for Acta2 by quantitative reverse transcription polymerase chain reaction (qRT-PCR) also showed that while the ΔTAD mutant was unable to activate Acta2 expression, chimeric fusions of FUS, TAF15, DDX4, and NPHS1 all activated Acta2 expression to comparable or higher levels relative to WT (fig. S8I). These results demonstrate that MYOCD condensate formation is required for the activation of SMC-specific genes.
Given the ability of four different condensate-promoting disordered regions to functionally replace MYOCDTAD, we next asked whether any condensate-promoting disordered region would rescue this function or if there was any specificity. Given that all four disordered regions tested require multivalent interactions of aromatic residues for condensate formation, we tested whether the disordered region of chromatin licensing and DNA replication factor 1 (CDT1), which does not require aromatics to form condensates, could rescue SMC reprogramming (45, 46). CDT1 is a DNA replication initiation factor, and its disordered region forms condensates independently of aromatic residues (46). As with the other chimeric fusions, we expressed an mEGFP-tagged MYOCD chimera with the TAD swapped for the disordered region of CDT1. The construct was expressed at equivalent levels to WT and the other chimeric fusions (fig. S8C). As expected, the chimeric fusion with the disordered region of CDT1 rescued condensate formation to levels comparable to WT and the other chimeric fusions [Fig. 6, H (bottom row) and I, and fig. S8, F and G]. Unexpectedly, the CDT1 chimeric fusion was unable to rescue SMC reprogramming as measured by ACTA2 staining [Fig. 6, H (bottom row) and J, and fig. S8, D to H] and Acta2 expression (fig. S8I). The functional difference between condensates formed by these two classes of disordered regions (aromatic-dependent or independent) highlights the specificity of condensates formed by different disordered regions.
To further assess the extent to which MYOCD condensate formation was required to activate SMC gene programs, we performed RNA-seq experiments with 10T1/2 cells expressing WT MYOCD, ΔTAD MYOCD, the FUS chimera, the CDT1 chimera, and GFP alone as a control. Gene ontology (GO) enrichment analysis of genes up-regulated by these different constructs relative to control revealed that genes up-regulated by the expression of WT MYOCD are mainly related to SMC cytoskeletal structure, confirming the reprogramming of fibroblast into SMCs by WT MYOCD (Fig. 6K). Intriguingly, the FUS chimera was the only variant tested that recapitulated the up-regulation of SMC-related genes (Fig. 6K). In contrast, we did not identify any relevant gene categories enriched with the ΔTAD or CDT1 chimera.
To specifically investigate the extent to which physiologically relevant cardiovascular SMC gene programs were activated in our cellular reprogramming system, we investigated the expression of a cluster of heart and aortic SMC genes identified by a single-cell RNA-seq analysis on mouse heart and aorta (47). Heatmap representations of the expression changes across our different conditions show that WT MYOCD robustly activates this SMC gene program in 10T1/2 cells (Fig. 6L and fig. S8J). Deletion of MYOCD TAD, which disrupts condensate formation, disrupts the activation of SMC gene program, which can be partially rescued by the FUS chimera but not the CDT1 chimera (Fig. 6L and fig. S8F). These results demonstrate that condensate formation driven by aromatic residues from functionally diverse proteins can partially replace MYOCDTAD and support the activation of cell identity gene programs. Together, our results demonstrate that MYOCD-driven gene activation requires condensate formation, enabling switch-like activation of cell identity genes underlying lineage specification.
Functional MYOCD condensates partition components of gene expression machinery
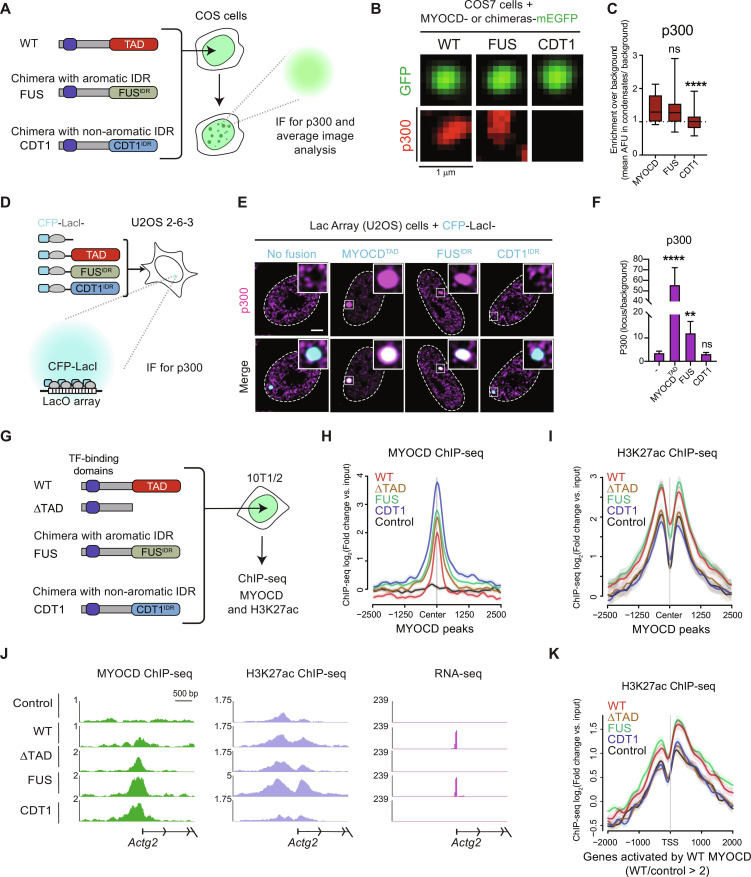
We next sought to investigate the mechanism underlying gene activation by MYOCD condensates. A major mechanism of condensate function is selective compartmentalization of functionally related proteins (13, 16, 18, 48). Having shown that condensates formed by the FUS chimera rescue function, but the condensates formed by the CDT1 chimera do not rescue function, we reasoned that by identifying what is commonly partitioned by WT MYOCD and the FUS chimera condensates but not by the CDT1 chimera condensates would highlight the mechanisms underlying MYOCD condensate function. We took a candidate approach to investigate whether condensates formed by WT MYOCD, the FUS chimera, or the CDT1 chimera had differential partitioning of p300 or RNA Pol II. We chose p300 because it has previously been implicated as a key regulator of MYOCD function (49), and we chose RNA Pol II because it is the enzyme ultimately responsible for transcription. We performed immunofluorescence for p300 or RPB1 in COS-7 cells ectopically expressing WT MYOCD, the FUS chimera, or the CDT1 chimera (Fig. 7A). Average image analysis (Materials and Methods) for immunofluorescence signal, centered on WT, FUS chimera, or CDT1 chimera condensates, showed that both p300 and RPB1 fluorescence intensity was higher at the center of WT MYOCD and FUS chimera condensates than at CDT1 chimera condensates (Fig. 7, B and C, and fig. S9, A to C). These findings demonstrate that WT MYOCD and FUS chimera condensates colocalize with components of active gene transcription.
Fig. 7. Condensates formed by MYOCD TAD domain and FUS-IDR selectively partition the histone acetyltransferase p300, but CDT1-IDR does not.
(A) Schematic of experimental design for colocalization of p300 in MYOCD condensates. (B) Average projection of WT MYOCD or chimeric condensates in COS-7 cells with IF for endogenous p300. Scale bar, 1 μm. (C) Box plot (10 to 90%) displaying the enrichment of p300 at the center of the MYOCD condensates normalized to background intensity. P value is from one-way ANOVA with Dunnett’s multiple comparison test (P values: ns P > 0.05, ***P ≤ 0.001, ****P ≤ 0.0001). n ~ 25. (D) Schematic of Lac array cells. (E) IF for p300 (magenta) in Lac array cells expressing indicated CFP-LacI fusion. Inset is LacO locus (white shows magenta and cyan overlap). Scale bar, 5 μm. (F) IF bar chart (mean ± SD) quantifying enrichment of p300 at CFP-LacI (no fusion) or CFP-LacI with fusion (x-axis label). P values from one-way ANOVA with Dunnett’s test. (P values: ns P > 0.05, **P ≤ 0.01, ****P ≤ 0.0001). n = 22. (G) Schematic of experimental design for ChIP-seq in reprogrammed 10T1/2 cells. (H) Metagene plot of the log2fold change compared to input control of MYOCD occupancy centered on WT MYOCD peaks compared to the empty vector control. (I) Metagene plot of the log2fold change compared to input control of H3K27ac occupancy centered on WT MYOCD peaks compared to the empty vector control. (J) Gene tracks of MYOCD and H3K27ac ChIP-seq and RNA-seq from differentiated 10T1/2 cells expressing the indicated constructs at Actg2 locus. (K) Metagene plot of the log2fold change compared to input control of H3K27ac occupancy centered on the TSS of genes activated by WT MYOCD.
To investigate whether the MYOCDTAD, the FUSIDR or the CDT1IDR are sufficient to partition positive regulators of gene expression when recruited to a specific genomic locus, we used a secondary assay that uses an orthogonal DNA binding domain (LacI) to recruit these protein regions to a defined genomic locus. To do this, we used the U2OS 2-6-3 cell line (Lac array cells) (13), which contains an integrated LacO array and compared cells transfected with either cyan fluorescent protein (CFP)–LacI or CFP-LacI-IDR (Fig. 7D). In the Lac array cells, the immunofluorescence signal for the positive regulators of transcription, p300 and RNA Pol II, was enriched at CFP-LacI-MYOCDTAD and CFP-LacI-FUSIDR foci relative to the control. CFP-LacI-CDT1IDR showed the opposite result (Fig. 7, E and F, and fig. S9, D to F). While MYOCDTAD showed greater enrichment of endogenous p300 than the FUSIDR, both enriched comparable levels of RNA Pol II, and both had significantly more enrichment of both factors relative to the control or CDT1IDR. These results demonstrate that high local concentrations of MYOCDTAD and FUSIDR are both sufficient for the local concentration of crucial gene regulatory enzymes.
To test the relationship between MYOCD chromatin occupancy and histone acetylation, we next performed ChIP-seq for H3K27ac and MYOCD or MYOCD variants. Using 10T1/2 cells expressing mEGFP-tagged WT MYOCD, ΔTAD MYOCD, the FUS chimera, the CDT1 chimera, and GFP alone, we performed ChIP-seq using GFP and H3K27ac antibodies (Fig. 7G). As expected, WT MYOCD showed binding at putative regulatory elements of its target gene Lmod1 marked by H3K27ac (fig. S10, A and B). In agreement with our results connecting MYOCD with p300, MYOCD bound regions showed an increase in H3K27ac relative to the GFP only control (fig. S10, C and D) and an increase in gene expression (fig. S10F). Focusing on all MYOCD peaks, we compared the occupancy of ΔTAD and the chimeras. As expected, given that they all share the same N-terminal portion of MYOCD responsible for interacting with transcription factors, all constructs bound at these MYOCD peaks (Fig. 7H). Unexpectedly, all constructs showed greater occupancy relative to WT MYOCD with the CDT1 chimera having the highest average signal. Despite this common occupancy, H3K27ac signal surrounding these MYOCD peaks only showed an increase relative to the control for WT MYOCD and the FUS chimera (Fig. 7I). Although, on average, ΔTAD MYOCD and the CDT1 chimera had comparable or even higher occupancy relative to WT MYOCD and the FUS chimera, H3K27ac levels were unchanged relative to the control. These results agree with our experiments measuring partitioning of p300 in COS-7 cells (Fig. 7, A to C) and Lac array cells (Fig. 7, D to F). Together, these data suggest that MYOCD condensate formation leads to local activation of chromatin by p300-mediated acetylation.
To better understand the relationship between MYOCD occupancy to gene activation, we identified MYOCD direct target genes (DTGs) by first annotating MYOCD peaks to the gene with the nearest transcription start site (TSS) and then designated the genes as DTGs if the associated gene exhibited a twofold or greater increase of gene expression. GO analysis of this subset of genes revealed that MYOCD DTGs are enriched for cellular components related to cytoskeletal components of smooth muscle genes (fig. S10E). To test the relationship between MYOCD chromatin occupancy, histone acetylation, and gene activation, we compared the ChIP-seq data with RNA-seq from 10T1/2 cells expressing the same constructs. For many of the genes activated by MYOCD expression, we could identify MYOCD peaks at associated H3K27ac-marked regulatory elements (metagene of other constructs and H3K27ac at all MYOCD peaks). While these regions were also occupied by the other constructs, only WT MYOCD or the FUS chimera were capable of activating gene expression (Fig. 7J). Analysis of H3K27ac centered on the TSS of all genes up-regulated by WT MYOCD (defined by RNA-seq, WT/control > 2) revealed the expected increase in H3K27ac relative to control (fig. S10G). Unexpectedly, among the other constructs tested, only the FUS chimera led to a comparable increase in H3K27ac at MYOCD-activated genes with ΔTAD and the CDT1 chimera being indistinguishable from the control (Fig. 7K). Together, these results connect the condensate formation of MYOCD and promotion of genomic accessibility to the activation of cell identity gene programs.
DISCUSSION
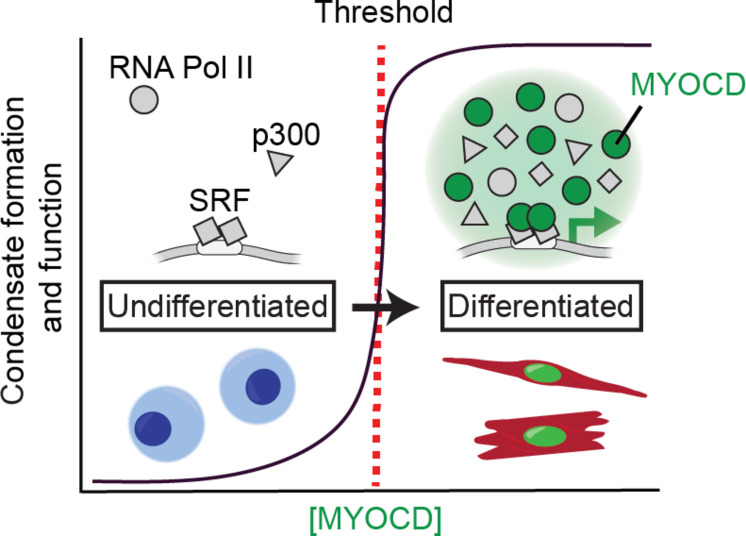
The results of this study demonstrate that MYOCD condensate formation is required for gene activation and cardiovascular cell lineage specification. Coupling gene activation to condensate formation enables robust and switch-like changes to gene expression programs defined by critical threshold concentrations (schematized in Fig. 8). Specifically, we show that MYOCD forms condensates at endogenous concentrations and that cells containing condensates express MYOCD target genes. Cells with MYOCD concentration above the threshold concentrations for condensate formation had concomitant activation of gene expression. Disrupting the ability of MYOCD to form condensates disrupted its ability to activate genes and drive reprogramming of cell states. Rescuing MYOCD condensate formation with four different disordered regions rescued its ability to activate genes and drive reprogramming. Our findings reveal a mechanism governing the activation of lineage-specific gene programs, highlighting the role of concentration-triggered condensate formation in this process. Last, we demonstrate that MYOCD condensates mediated by biochemically distinct IDRs confer different functional outcomes, which we have attributed to the ability of these distinct condensates to selectively partition positive regulators of gene expression.
Fig. 8. MYOCD coactivator condensation drives cardiovascular cell lineage specification.
Schematic illustrating the relationship between natural changes in MYOCD (green) concentration during differentiation, condensate formation, and switch-like gene activation. In undifferentiated cells, although DNA binding transcription factors like SRF (gray squares) and other components of the transcriptional machinery including RNA Pol II and p300 (gray shapes) are present, MYOCD is below the threshold concentration for condensate formation (red dotted line) and cell identity genes are not expressed. When the concentration of MYOCD exceeds the threshold for condensate formation, MYOCD forms nuclear condensates that concentrate the transcriptional machinery (gray shapes) to drive cell identity gene expression, facilitating lineage specification of SMC and CM.
By measuring concentration, condensate formation, and gene activity at single-cell resolution, we found that the formation of MYOCD condensates is directly linked to gene activation. By swapping MYOCDTAD with four different condensate-promoting regions, we found that the ability of MYOCD to form condensates is required for its ability to activate genes. Showing that the activity of a protein is directly linked to condensate formation at physiological concentrations in cells has been a major challenge for the field, leading some to debate whether condensates are functional (50). A few key examples using in vitro reconstituted condensates have shown that biochemical activity (e.g., nucleation of actin and microtubule polymerization) is increased upon condensate formation (51–54). Cell-based approaches to investigate the link between condensates and gene transcription have mainly used engineered, synthetic, or otherwise artificial approaches at supraphysiological concentrations, leading to contradictory conclusions with some reporting no link (55, 56) and others finding a link (57, 58). Here, we show, at endogenous protein concentrations, in both COS-7 cells with a reporter gene and 10T1/2 cells with a cell identity gene, that the gene activation function of MYOCD is coupled to and requires its formation of condensates.
The requirement of condensate formation for MYOCD activity is demonstrated by our disordered region swapping experiments, in which four condensate-promoting disordered regions were able to partially replace the functions of MYOCDTAD. Swapping the condensate-forming disordered regions from CDT1 did not fully rescue function, suggesting functional specificity among disordered regions and the condensates they help form. We recently demonstrated that condensates formed by the disordered region of the transcription coactivator MED1 concentrated specific positive regulators of transcription while excluding negative regulators (13). This functional specificity was found to depend on charge patterning within the disordered regions of MED1 and its partitioned partners. The functional specificity between different IDRs for rescuing MYOCD activity, seemingly due to aromatic content, suggests that there are different types of sequence and structural features leading to condensate-mediated specificity.
The ability of condensate-promoting disordered regions from proteins with diverse functions to replace the TAD of MYOCD suggests that a primary function of the TAD is to promote condensate formation. In so doing, the TAD enables the coalescence of multiple components of the transcriptional apparatus at regulatory elements of target genes (schematized in Fig. 8). Such a mechanism also provides stability to cell-specific gene programs during lineage specification in development. The relationship between the sequence and structural features of TADs required for condensate formation (38, 39, 59, 60) and those required for transactivation (61–63) and whether they are separable is an exciting topic for future investigation.
In addition to concentration changes that occur on a developmental timescale, the formation of condensates can be more dynamically regulated by posttranslational modifications (PTMs). MYOCD is known to be modified by phosphorylation, acetylation, and SUMOylation, which regulate its function. Given that many of these reversible modifications occur in the TAD (64, 65), it is possible that these modifications have an impact on MYOCD condensate formation and its stage-specific activity in cardiovascular development. The role of these PTMs in regulating MYOCD-mediated condensate formation and gene activation remains to be explored.
Our results demonstrate that the formation of MYOCD condensates is necessary for the activation of cell identity genes during lineage specification. Many lineage-specifying factors display similar changes to nuclear concentration during development. Many of these factors are highly dosage dependent, as exemplified by developmental disorders caused by haploinsufficiency or by small changes in expression due to regulatory element mutations (3). This strong concentration dependence suggests that the gene activation functions of these factors might also be linked to condensate formation. Furthermore, aberrant increases in the concentration of transcriptional regulators are a hallmark of many cancers, exemplified by genetic amplification, overexpression, or dysregulation of cytonuclear trafficking. Our work leads us to propose that transcriptional condensate formation may represent a general mechanism for precise and decisive activation of gene programs crucial for cell lineage specification, suggesting that changes in the nuclear concentration of transcriptional regulators contribute to disease as a consequence of dysregulated condensate formation.
MATERIALS AND METHODS
Generation of an isogenic MYOCD-mEGFP knock-in hiPSC line
The isogenic mEGFP knock-in hiPSC line was generated by CRISPR-Cas9–mediated homology-directed repair. Single-guide RNAs (sgRNAs) near the stop codon in exon 14 of the MYOCD gene were selected from CRISPR 10K Genome Browser Track, cloned into pX458 (Addgene, no. 48138), transfected into human embryonic kidney (HEK) 293T cells, and fluorescence-activated cell sorting (FACS)–sorted, and cutting efficiency was assessed by sequencing. The sgRNA with the highest indel-causing efficiency (TGGACCTTCACTTGCAGCAG) was used for subsequent knock-in assays. To construct the donor template, mEGFP sequence was cloned in between 5′ and 3′ homology arms [~1000 base pairs (bp) each] into a donor plasmid vector (pENTR/D-TOPO) by infusion cloning. HC01-5, male human iPSCs were cultured in mTeSR1 medium to reach 95% confluency. One hour before nucleofection, hiPSCs were treated with 10 μM ROCK inhibitor (Tocris, Y-27632) and dissociated into single cells using Accutase (Innovative Cell Technologies Inc., NC9839010). hiPSCs (1 × 106) were mixed with 2.5 μg of pX458-MYOCD-sgRNA-2A-GFP plasmid and 2.5 μg of donor plasmid and nucleofected using the P3 Primary Cell 4D Nucleofector X Kit (Lonza, V4XP-3024) according to the manufacturer’s protocol. After nucleofection, hiPSCs were cultured in mTeSR1 medium supplemented with 10 μM ROCK inhibitor for 24 hours and changed to mTeSR1 medium the next day. Two days after nucleofection, the medium was changed into mTeSR1 medium supplemented with 10 μM ROCK inhibitor and Primocin (100 mg/ml) (InvivoGen, ant-pm-05) 1 hour before FACS. GFP (+) and (−) cells were sorted by FACS and seeded back into 12-well plates. Single clones derived from GFP (+) iPSCs were picked and sequenced for in-frame mEGFP knock-in. After screening more than 200 clones, one isogeneic hiPSC clone was identified with homozygous MYOCD-mEGFP knock-in and we established a cell line for subsequent experiments.
Differentiation of hiPSC to CMs and SMCs
Differentiation of hiPSC to CMs was performed as previously stated (66). Briefly, cells were cultured in mTeSR1 medium until they reached 80 to 90% confluency and then cultured in CDM3-C medium consisting of RPMI 1640 (Life Technologies, 11875), Oryza sativa–derived recombinant human albumin (500 μg/ml; Sigma-Aldrich, A0237), and l ascorbic acid 2-phosphate (213 μg/ml) supplemented with 6 μM CHIR-99021 (Selleckchem, S2924) for 2 days. The cells were then cultured in CDM3-C medium, supplemented with 2 μM WNT-C59 (Selleckchem, S7037) for 2 days. The cells were cultured in BASAL medium (RPMI 1640 with B27 supplement, Thermo Fisher Scientific, 17504044) for 6 days, and the medium was changed every 2 days. CMs were selected by culturing in SELECTIVE medium [RPMI 1640, no glucose (Gibco, 11879-020) with B27 supplement (Thermo Fisher Scientific, 17504044)] for 6 days. Then, purified CMs were dissociated using TrypLE Express Enzyme (Gibco, 12604021) and replated.
The differentiation protocol for SMCs was adapted from an established protocol (67). hiPSCs were cultured in mTeSR1 medium on Matrigel-coated plates, with daily medium changes to reach 90% confluency. The differentiation into mesodermal-lineage cells was initiated on day 0 by culturing the cells with CHIR99021 (5 μM) and BMP-4 (10 ng/ml, PeproTech, 120-05ET) in RPMI 1640 medium and 2% B27. The culture medium was replenished with CHIR99021 and BMP-4 on day 1.5. The expression of MYOCD is high in contractile SMCs. Contractile SMCs were produced by culturing the cells with vascular endothelial growth factor A (VEGF-A) and fibroblast growth factor β (FGFβ) (25 ng/ml) in RPMI 1640 and 2% B27 minus insulin from days 3 to 7, replenishing the culture medium with VEGF-A (PeproTech, 100-20) and FGFβ (PeproTech, AF-100-18B) on day 4. Then, the cells were treated with platelet-derived growth factor β (PDGF; 5 ng/ml; PeproTech, AF-100-14B) and transforming growth factor β (TGFβ; 2.5 ng/ml; PeproTech, 100-21) in RPMI 1640 and 2% B27 from days 7 to 14, and the culture medium was replenished with PDGFβ and TGFβ on days 9 and 11. The differentiated cells were enriched for SMCs by maintaining them in 4 mM lactate RPMI 1640 metabolic medium for 4 to 6 days. Then, purified SMCs were dissociated using TrypLE Express Enzyme and replated.
Microscopy
All micrographs in figures are representative for the respective experimental sample. Unless otherwise noted, images were acquired with a CSU-W1 Yokogawa Spinning Disk Field Scanning Confocal System coupled to a Hamamatsu ORCA-Fusion C14440 sCMOS camera using a Nikon 60× Plan Apo Lamba Oil Immersion objective [numerical aperture (NA), 1.40]. Images in Fig. 1 (A, C, and E) and figs. S1 (A, E, and G) and S6B were acquired using a Zeiss LSM 800 point scanning confocal using a Zeiss 63× Plan Apo oil immersion objective (NA, 1.40) with Airy Scan SR mode activated. Raw Airyscan images were processed using Zeiss ZEN 2.3 software.
Live-cell imaging
Cells were grown on glass plates (Mattek Corporation, P35G-1.5-20-C) coated with Matrigel (hESC-qualified, Corning, 354277) in phenol red–free Dulbecco’s modified Eagle’s medium (DMEM)/F-12 (1:1) (Gibco, 21041-025) to reduce background autofluorescence. hiPSC-CMs and SMCs were differentiated and cultured in phenol red–free RPMI 1640 medium (Gibco, 11835-030). Before imaging, the medium was replaced by 37°C phosphate-buffered saline (PBS) with 1:5000 Hoechst 33342 for nuclear staining and cell debris removal. After 5-min incubation, Hoechst-containing PBS was replaced with fresh 37°C PBS. The cells were imaged on a heated stage top incubator (Tokai Hit) and supplemented with warmed (37°C), humidified air with 5% CO2.
Immunofluorescence
Cells grown on coated glass were fixed in 4% paraformaldehyde (PFA) (VWR, BT140770) in PBS for 10 min at room temperature. After three washes in PBS for 5 min, the cells were stored at 4°C or processed for immunofluorescence. Cells were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich, X100) in PBS for 5 min at room temperature. Following three washes in PBS for 5 min, the cells were blocked with 4% immunoglobulin G (IgG)–free bovine serum albumin (BSA) (VWR, 102643-516) for 30 min at room temperature and then incubated with primary antibodies in 4% IgG-free BSA overnight at 4°C. Primary antibodies used in the experiments were anti-GFP (Thermo Fisher Scientific, A-11122, 1:500 dilution), anti–cardiac troponin T (Abcam, ab8295, 1:1000 dilution), anti–α smooth muscle actin (Abcam, ab7817, 1:1000 dilution), anti-Flag M2 (Sigma-Aldrich, F3165, 1:1000 dilution), anti-V5 (Cell Signaling Technology, 13202S, 1:500), anti-H3K27ac (Active Motif, 39133, 1:500), anti–RNA Pol II C-terminal domain (Millipore, 04-1572, 1:500), anti-HP1a (Abcam, ab109028, 1:500), and anti-NPM1 (Thermo Fisher Scientific, 32-5200). Samples were subsequently washed with PBS three times and incubated with corresponding secondary antibodies conjugated to Alexa Fluor 488, 555, or 647 (Invitrogen) prepared in blocking solution at room temperature in the dark for 3 hours. Cells were washed three times with PBS. Hoechst (Thermo Fisher Scientific, 62249, 1:5000) was used to stain nuclei for 5 min at room temperature in the dark. Glass slides were mounted onto slides with Vectashield (VWR, 101098-042). Coverslips were sealed with transparent nail polish (Electron Microscopy Science Nm, 72180) and stored at 4°C.
RNA FISH with immunofluorescence
RNA FISH probes were designed using Stellaris probe designer with intronic sequences from genes of interest (Biosearch Technologies Inc.). Libraries of 48 intron-targeting probes for each gene conjugated to Quasar 670 fluorophore were used for nascent RNA FISH experiments. Immunofluorescence was performed as previously described (14) with the following modifications. Immunofluorescence was performed in a ribonuclease (RNase)–free environment, and pipettes and bench were treated with RNaseZap (Life Technologies, AM9780). RNase-free PBS was used, and antibodies were always diluted in RNase-free PBS. After immunofluorescence completion, the cells were post-fixed with 4% PFA in PBS for 10 min at room temperature. The cells were washed twice with RNase-free PBS. The cells were washed once with 20% Stellaris RNA FISH Wash Buffer A (Biosearch Technologies Inc., SMF-WA1-60) and 10% deionized formamide (EMD Millipore, S4117) in RNase-free water (Life Technologies, AM9932) for 5 min at room temperature. The cells were hybridized with 90% Stellaris RNA FISH Hybridization Buffer (Biosearch Technologies, SMF-HB1-10), 10% deionized formamide, and 12.5 μM Stellaris RNA FISH probes designed to hybridize introns of the transcripts of SE-associated genes. Hybridization was performed overnight at 37°C. The cells were then washed with wash buffer A for 30 min at 37°C, and nuclei were stained with 1:5000 Hoechst (Thermo Fisher Scientific, 62249, 1:1,000) in wash buffer A for 5 min at room temperature. After one 5-min wash with Stellaris RNA FISH Wash Buffer B (Biosearch Technologies, SMF-WB1-20) at room temperature, coverslips were mounted as described for immunofluorescence.
Colocalization image analysis
Analyzing colocalization of co-immunofluorescence and RNA FISH was done by creating 1.5 μm × 1.5 μm crops of either the MYOCD puncta for coimmunofluorescence or the RNA FISH focus. The average images of colocalization were created using average projections of z-stacks in FIJI at foci compared to random sites in the nucleus. Intensity line profiles of GFP puncta or RNA FISH foci were drawn using FIJI and saved in region of interest (ROI) manager for use in all samples (≈1.5 μm). Values outside the focus of interest were used to normalize intensity by averaging the first and last two values across the line plot. The data point at the center of the line for each sample was used for statistical analysis against the control group.
Chromatin immunoprecipitation with sequencing
ChIP-seq was carried out as previously described (68), with modifications. Cells (WT and MYOCD-mEGFP knock-in hiPSCs and reprogrammed 10T1/2 cells) were cross-linked with 4% PFA in PBS for 15 min and quenched with 0.125 M glycine for 10 min at room temperature. For mesodermal CMs, we used hiPSCs at cardiac lineage differentiation day 5. For mesodermal SMCs, we used hiPSCs at smooth muscle lineage differentiation day 8. For reprogrammed 10T1/2 cells, we collected cells with different retrovirus infections after 5-day culturing in differentiation medium. Cross-linked samples were then washed with cold PBS. Samples were collected by a brief spin and treated with 10 mM tris-HCl (pH 8.0), 10 mM NaCl, and 0.2% NP-40 for 30 min to collect nuclei. After nuclear extraction, chromatin was sheared on a Bioruptor Pico (Diagenode) for 16 cycles (30-s on/30-s off for each cycle) at 4°C in sonication buffer [0.1% SDS, 1% Triton X-100, 10 mM tris-HCl, 1 mM EDTA, 0.1% sodium deoxycholate, 0.25% sarkosyl, 1 mM dithiothreitol (DTT), 1× cOmplete protease inhibitor cocktail (Sigma-Aldrich, 11836170001), and 200 μM phenylmethylsulfonyl fluoride (PMSF), pH 8.0]. After sonication, 1% of the sonicated chromatin from each sample was taken out as “input” samples, and 20 μl of sonicated sample from each group was used for electrophoresis on the 1% agarose gel for sonication efficiency and quality control. The remaining sonicated chromatin was evenly split for MYOCD-GFP or H3k27ac ChIP. NaCl was then added to a final concentration of 300 mM for histone ChIP and 150 mM for MYOCD-GFP ChIP. H3k27ac antibody (1 μg/ml; Diagenode, C15410196) or GFP antibody (10 μg/ml; Thermo Fisher Scientific, A-11122) was added to each sample and incubated at 4°C overnight with gentle rotation. The next day, prewashed Dynabeads Protein G (30 μl/ml; Invitrogen, 10004D) was added to each sample for a 2-hour incubation. After that, the beads were washed twice with 1 ml of radioimmunoprecipitation assay (RIPA) 0 buffer [0.1% SDS, 1% Triton X-100, 10 mM tris-HCl, 1 mM EDTA, and 0.1% sodium deoxycholate (pH 8.0)], twice with 1 ml of RIPA 0.3 buffer [0.1% SDS, 1% Triton X-100, 10 mM tris-HCl, 1 mM EDTA, 0.1% sodium deoxycholate, and 300 mM NaCl (pH 8.0)], twice with 1 ml of LiCl wash buffer [250 mM LiCl, 0.5% IGEPAL CA-630, 0.5% sodium deoxycholate, 1 mM EDTA, and 10 mM tris-HCl (pH 8.0)], and finally twice with 1 ml of TE buffer [10 mM tris-HCl and 1 mM EDTA (pH 8.0)]. For each ChIP sample or input sample, 100 μl of SDS elution buffer [1% SDS, 10 mM EDTA, and 50 mM tris-HCl (pH 8.0)] was added and incubation was done at 65°C overnight on a ThermoMixer (Eppendorf) at 1000 rpm. The next day, supernatant was collected and further treated with 0.5 μg of RNaseA (Sigma-Aldrich, 11119915001) for 30 min at 37°C, followed by 20 μg of proteinase K (NEB, P8107S) treatment at 37°C for 2 hours. DNA was recovered using MinElute PCR Purification Kit (QIAGEN, 28004) according to the manufacturer’s protocol. ChIP-seq libraries were prepared using the NEBNext Ultra II DNA Library Preparation Kit complemented with NEBNext Multiplex Oligos for Illumina from NEB (NEB, E6177S). Ten nanograms of DNA was used as starting material for input samples and 1 ng for IP samples. Libraries were amplified using 12 cycles for input samples and 15 cycles for IP samples on the thermocycler. Post-amplification libraries were size-selected at 250 to 450 bp in length using SPRISelect beads from Beckman Coulter. Libraries were validated using the Agilent D1000 ScreenTape. ChIP-seq was performed by the Sequencing Facility of University of Texas Southwestern Medical Center Children’s Research Institute using the P2-100cycle kit.
Analysis of ChIP-seq data
For analysis of ChIP-seq data from hiPSC-derived samples, reads were aligned to the hg19 genome assembly using bowtie (v1.0.0) (69) with parameters –k 1 –m 1 –best and –strata. Subsequent analyses were performed using a combination of samtools (v1.6) and BEDtools (v2.29.2). Counts per million normalized bigwig files were displayed on the UCSC genome browser for the visualization of ChIP-seq tracks. Peak calling was done with MACS (v1.4.2) (70). We performed a de novo motif search using HOMER (v4.0) (71) on the ±20-bp peak summit regions of both CM and SMC samples at days 5 and 8, respectively. The top five enriched known motifs were presented in the figures. To annotate the peak regions and associate them with nearby expressed genes, we used HOMER’s annotatePeaks.pl function. This process considered the peak summit and used custom gene annotation definitions using GTF files. When multiple peak regions were annotated with the same gene, we summed the ChIP RPM (reads per million mapped reads) values to identify and prioritize genes highly bound by MYOCD. GO enrichment analysis was performed for the top 10% genes with the highest summed ChIP RPM values for both CM and SMC samples at days 5 and 8, respectively, using PANTHER overrepresentation test (v16.0) (72) with slim biological process categories and Fisher’s exact test.
Analysis of ChIP-seq from 10T1/2-derived samples was carried out as previously described (73). Briefly, raw sequencing reads with >30% nucleotide with phred quality scores <20 were filtered. Single-end sequencing reads were then aligned to the mouse reference genome GRCm38 (mm10) using bowtie2 aligner (v 2.3.4.3) with default parameters. For transcription factor ChIP-seq data, peaks were called using HOMER software package (version 4.9) findpeaks command with parameter “-style factor,” peaks were called with >2-fold enrichment over input controls and >4-fold enrichment over local tag counts, and false discovery rate (FDR) threshold was set to 1 × 10−3. For histone marker ChIP-seq data, peaks were called by findpeaks command with parameter “–style histone,” peaks were called with >2-fold enrichment over input controls and >4-fold enrichment over local tag counts, and FDR threshold was set to 1 × 10−3. ChIP-seq peaks within a 1000-bp range were stitched together to form broad regions. ChIP-seq signal heatmap was generated using ngs.plot.r software.
RNA-seq library preparation
RNA from iPSCs, SMC-meso (day 8) cells, CM-meso (day 5) cells, and reprogrammed 10T1/2 cells were extracted using TRIzol reagents (300 μl per well of a six-well plate) and Direct-zol RNA Miniprep Kits (Zymo Research, R2052) according to the manufacturer’s protocol. Stranded mRNA-seq libraries were generated using KAPA mRNA HyperPrep Kit (Roche, KK8581) following the manufacturer’s protocol. Sequencing was performed on an Illumina Nextseq 500 system for 75-bp single-end sequencing.
Analysis of RNA-seq data
For analysis of RNA-seq data from hiPSC-derived samples, analysis was performed as follows. For differentiation conditions, eight mRNA-seq data samples were analyzed, comprising two biological replicates of day 0 hiPSC cells and three biological replicates each of CM (day 5) and SMC (day 8) samples. For reprogramming assay, 15 mRNA-seq data samples were analyzed, comprising three biological replicates of each group of reprogrammed 10T1/2 cells (GFP, MYOCD, FUS, CDT1, and ΔTAD). To ensure quality, FastQC (version 0.11.8.1) (74) was used to confirm the quality of the Fastq files from the RNA-seq samples, and only reads with a minimum length of more than one base were selected for further analysis. The demultiplexed mRNA-seq reads were mapped to the human reference genome version hg19 using STAR (v2.7.3a) (75), and subsequent analyses were performed with samtools (v1.6) (76) and BEDtools (v2.29.2) (77). Bigwig files were generated using deeptools (v.3.5.0) (78) and normalized with the following parameters: -bs 50 -normalize using counts per million (CPM). These files were displayed in the UCSC genome browser. For each transcript, expression was quantified as fragments per kilobase million (FPKMs) using featureCounts from the subread (v.1.6.3) (79) package and annotated to the genes.gtf file of the human reference genome version hg19 or the mouse reference genome version mm10. Expressed transcripts were defined as those with average FPKM > 10 in three replicates.
For analysis of RNA-seq data from 10T1/2 cell–derived samples, analysis was performed as follows. Quality control of RNA-seq data was performed using FastQC Tool (version 0.11.4). Single-end reads were aligned to the mouse reference genome GRCm38 (mm10) using the HiSAT2 (version 2.1.0) aligner using default settings. Aligned reads were counted using featureCounts (version 1.6.0) per gene ID. Differential gene expression analysis was done using the R package DESeq (version 1.12.1). For principal components analysis, genes with 0 count across samples were removed and counts were normalized using rlog function in R. Principal components were calculated using R function prcomp. For heatmap of SMC genes, we transformed the raw count into log2CPM values using the cpm function of edgeR package and pheatmap function was used to generate the final heatmap, with row values scaled as z score.
For identification of MYOCD DTGs, we first annotated MYOCD-GFP ChIP peaks and assign each peak to the gene with nearest TSS. Differential expressed genes (Padj < 0.05 by DESeq using RNA-seq data) with an associated peak that has >2-fold change between MYOCD-GFP and GFP-control were designated as DTGs. Enrichment analysis of gene sets was performed using the Metascape (https://metascape.org/) with supply of DTGs.
Calculating nuclear concentration from fluorescence microscopy
Purified recombinant mEGFP was diluted to desired concentration in protein storage buffer (50 mM tris, 75 mM NaCl, 5% glycerol, 1 mM PMSF, and 1 mM DTT) and immediately loaded into chamber slide. The chamber slide consists of a coverslip attached to a glass slide by parallel strips of double-sided tape. mEGFP solution was imaged using a Hamamatsu ORCA-Fusion C14440 digital camera and a CSU-W1 Yokogawa Spinning Disk Field Scanning Confocal System. Images were acquired using 60× Plan Apo Lambda Oil Immersion objective (NA, 1.40), focusing the objective on the tape-GFP solution boundary, and then imaging +4 and −3 μm from the center of the chamber slides at steps of 1 μm at three separate xy coordinates. The brightest image from each stack was chosen for analysis. These images were acquired with the same acquisition parameters used to acquire images from cells. Different standard curves were therefore derived for different acquisition parameters used for different cells. The total fluorescence intensity for images of purified mEGFP at known concentrations was measured using the CellProfiler Measure Image Intensity module. Known concentration of mEGFP (x axis) and total fluorescence intensity of mEGFP per μm2 (y axis) for three replicates each were correlated, and linear regression analysis revealed the following relationship where x equals concentration of GFP in nM and y equals total fluorescence intensity per μm2: mature CM (1.419 × 10−4x + 0.001060), mature SMC (1.609 × 10−4x + 0.0004057), COS-7 (4.704 × 10−5x + 0.0008428), and 10T1/2 (5.510 × 10−5x + 0.0003226). Using these standard curve-derived equations, we could calculate the nuclear concentration where y equals the nuclear fluorescence [arbitrary fluorescence units (AFU)] divided by the nuclear area in μm2 of the nucleus, and x represents the MYOCD concentration in nM.
COS-7 and 10T1/2 cell culture
COS-7 and 10T1/2 cells were grown in full medium (DMEM; Fisher Scientific, 119950) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, F0926), 1× GlutaMAX (Fisher Scientific, 35050), and 1× penicillin-streptomycin (Fisher Scientific, 15140). Cells were grown at 37°C with 5% CO2 in a humidified, sterile incubator. 10T1/2 cells were maintained at 50 to 60% confluency.
U2OS (Lac array cells) cell culture
Cells were cultured as previously described (80). Briefly, U2OS 2-6-3 cells (Lac array cells) were grown in DMEM supplemented with 10% Tet system–approved FBS (Takara Bio USA 631105), 1% penicillin-streptomycin (Fisher Scientific, 15-140-122), and 1% GlutaMAX supplement (Fisher Scientific, 35050061). Cells were grown at 37°C with 5% CO2 in a humidified sterile incubator.
Cloning MYOCD, mutants, and chimeras
Mouse cDNA encoding MYOCD was used in these studies. The TAD was previously defined (33) as amino acids 712 to 934. MYOCD cDNA was modified by using a PCR + HiFi (NEB, E2621L) strategy followed by sequence verification by Sanger sequencing (GeneWiz) or whole-plasmid nanopore sequencing (Plasmidsaurus). For ΔTAD, the DNA encoding the N-terminal region of MYOCD was PCR-amplified and inserted using HiFi. For the FWY to A substitution mutant, a custom DNA fragment was ordered from Twist Bioscience as a gene fragment and inserted using HiFi. For the chimeras, the DNA encoding the IDR regions from TAF15 (Addgene, no. 26379), FUS (Addgene, no. 26374), DDX4 (Addgene, no. 101225), NPHS1 (a gift from the Rosen laboratory), and CDT1 (a gift from the Parker laboratory) were PCR-amplified and inserted in-frame with the N-terminal (ΔTAD) region of MYOCD.
Cell transfections
COS-7 cells were seeded in six-well plates at 70% confluency and transfected on the same day using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, L3000015). All transfections of samples with MYOCD-mEGFP, mutant, chimeric fusions, or controls plasmids were performed at approximately 250 ng/ml. Cotransfections of MYOCD plasmids, tdTomato internal control, and SM22-miRFP670-SKL reporter construct were performed at 250, 125, and 500 ng/ml, respectively. The medium was changed approximately 16 hours after transfections, and the cells were allowed to recover for approximately 8 hours before fixation. Imaging was performed 1 to 2 days after fixing cells.
Lac array cells were seeded on coverslips in six-well plates at 50% confluency and transfected on the same day using ViaFect Transfection Reagent (Promega, E4982). All transfections for CFP-LacI–containing plasmids were performed at approximately equimolar concentrations following the manufacturer’s guidelines. The cells were fixed the morning after transfection.
Retrovirus production and 10T1/2 reprogramming
Platinum E cells (Cell Biolabs, NC0066908) were cultured in DMEM with 10% FBS, 1× penicillin-streptomycin, puromycin (1 μg/ml; Takara Bio, 631305), and blasticidin (10 μg/ml; Thermo Fisher Scientific, A1113903). Cells in 10-cm dish (around 50% confluency) were transfected with 10 μg of plasmid using 30 μl of Fugene 6 transfection reagent (Promega, E2691). Forty-eight hours after transfection, the supernatants were collected and filtered through a 0.45-μm syringe filter. Virus was concentrated with Retro-X concentrator (Takara, 631456). After 16 hours of viral concentration, viral soup was centrifuged at 1500g for 45 min, and the pellet was resuspended in fresh growth medium supplemented with polybrene (Sigma-Aldrich, H9268) at a final concentration of 8 μg/ml.
The 10T1/2 differentiation procedure was conducted as described previously (33). Briefly, 10T1/2 cells were infected by retrovirus at 50% confluency. Fresh medium containing retrovirus was supplemented with polybrene (Sigma-Aldrich, H9268) at a final concentration of 8 μg/ml and then added to 10T1/2 cells. Two days after infection, differentiation was induced by treatment of full medium supplemented with 2% horse serum (Sigma-Aldrich, H1270-100ML). Cells were treated with this differentiation medium for 5 days before fixation for microscopy or harvesting for Western blot or qRT-PCR. 10T1/2 cells used for microscopy were seeded on coverslips (VWR, 48366-067). Lower magnification imaging of 10T1/2 cells shown in fig. S7D was acquired using 40× Plan Apo water immersion objection (NA, 1.15).
Imaging of GFP-MYOCD in COS-7 and 10T1/2 cells
Cells were fixed in 4% PFA (VWR, BT140770) in PBS for 10 min at room temperature in the dark. Samples were washed three times for 5 min each in PBS at room temperature. Samples were permeabilized for 10 min with 0.5% Triton X-100 (Sigma-Aldrich, T9284) in PBS at room temperature and then washed three times for 5 min each in PBS. Later, cells were incubated with 1:5000 Hoechst 33342 (Thermo Fisher Scientific, 62249) in double-distilled water for 5 min at room temperature. Cells were washed once more with water, and coverslips were mounted on slides (VWR, 10144-820) with Vectashield (VWR, 101098-042). Coverslips were sealed with nail polish (VWR, 100491-940) and stored at 4°C.
Lac array immunofluorescence
Immunofluorescence was performed as formerly described with certain modifications (13). In brief, cells were grown on coverslips (VWR, 48366-067) in a six-well plate and fixed in 4% PFA (VWR, BT140770) in PBS for 10 min at room temperature. After three washes in PBS for 10 min, samples were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich, T9284) in PBS for 10 min at room temperature. After washing three times in PBS for 5 min, cells were blocked with 4% IgG-free BSA (VWR 102643-516) for 2 hours at room temperature and incubated overnight at room temperature with primary antibodies in 4% IgG-free BSA (samples were kept in the dark). Following three washes in PBS for 10 min each, samples were incubated with secondary antibodies for 2 hours in the dark. After washing with PBS three times, cells were incubated with 1:5000 Hoechst 33342 (Thermo Fisher Scientific, 62249) in double-distilled water at room temperature in the dark (5 min). After washing once more with water, coverslips were mounted on slides (VWR, 10144-820) with Vectashield (VWR, 101098-042). Coverslips were sealed with nail polish (VWR, 100491-940) and stored at 4°C in the dark. All primary antibodies for immunofluorescence were diluted 1:500. All secondary antibodies were diluted 1:5000.
Analysis for condensate formation and threshold detection
Nuclei of GFP-positive cells were cropped into 30 μm–by–30 μm boxes and rendered as a max projection of a z-stack in FIJI. Images were then processed using a modified CellProfiler pipeline from the published “Speckle Counter” pipeline (81). Briefly, nuclei are identified on the basis of Hoechst 33342 staining and the adaptive, minimum cross-entropy image thresholding model. Only nuclei that do not touch the border of the image and are within a diameter of 100 to 400 pixel units are used in analysis. Nuclei boundaries are applied as a mask in the GFP channel, and puncta are identified using the global Otsu image thresholding model, which identifies puncta between the size of 2 and 50 pixels. Total fluorescence intensity and area are measured for both image, nuclei, and puncta.
To classify the nuclei into groups of having puncta, the following was done. The fraction of GFP signal within puncta compared to GFP within the nucleus and total apparent concentration of GFP in the nucleus were plotted on an XY plot in Prism, and a relative threshold of puncta formation was estimated. The threshold for puncta formation in COS-7 cells was set to a fraction of GFP in puncta of 0.0075, and in 10T1/2 cells, this threshold was set to 0.0045, where any fractional GFP fluorescence greater than or equal to this value is denoted as 1 (signifying the presence of puncta) and any value less is denoted as 0 (signifying no puncta) to reduce noise from puncta identified as an artifact of the CellProfiler pipeline. To calculate the apparent threshold concentration of MYOCD for both puncta formation and reporter expression, a simple logarithmic regression of the dataset was used where the best-fit value of X at 50% is the x-axis threshold (i.e., the concentration threshold where there is a 1:2 probability of either state).
Fluorescence recovery after photobleaching
After transfection, FRAP was performed on live COS-7 cells with 405-nm laser. Bleaching was performed at condensate ROI using 50% laser power, and images were collected every 1 s. Fluorescence intensity was measured using FIJI. FRAP was analyzed using FRAP Profiler (v2) plugin for FIJI where fluorescence recovery is normalized to background to account for photobleaching over time and fits a one-component exponential curve to estimate fraction of fluorescent signal that is mobile (82).
Luciferase assay
For the luciferase assay presented in fig. S4E, COS-7 cells were seeded at 70% confluency in 12-well plate and transfected the same day using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, L3000015). For the luciferase assay, transfections of the MYOCD-mEGFP plasmid were performed at approximately 50 ng/ml, cotransfected with luciferase reporter construct under control of the minimal SM22 promoter (250 ng/ml) and NanoLuc (50 ng/μl) internal control. Fresh medium was added after 16 hours, and cells were harvested at 20 hours. Luciferase assay was performed as per the manufacturer’s recommendations using the Nano-Glo Dual-Luciferase Reporter Assay system with control NanoLuc vector (Promega, N1521). Luminescence was measured using CLARIOstar microplate reader (BMG Labtech).
Protein expression and purification
Protein expression and purification were conducted following our previous protocols (13). The DNA fragments encoding the protein ROI were cloned into a modified T7 pET expression vector, resulting in the protein having N-terminal 6×His mEGFP followed by the 14–amino acid linker sequence “GAPGSAGSAAGGSG” before the protein ROI. NEBuilder HiFi DNA Assembly Master Mix (NEB, E2621L) was used to insert the DNA sequence in-frame with the linker sequence. The generated construct was transformed into NEB 5-alpha Competent Escherichia coli (NEB, C2987H), and plasmids were isolated from a selected bacterial colony. Sequence identity was confirmed using Sanger sequencing.
NiCo21(DE3) Competent E. coli cells (NEB, C2529H) were transformed with the bacterial expression plasmids described above. Transformed colonies were grown in LB medium at 37°C on a rotating shaker (250 rpm) until OD600 (optical density at 600 nm) of 0.6 was reached. Following 1 mM of isopropyl ß-d-1-thiogalactopyranoside (IPTG; Sigma-Aldrich, 70527) induction for protein expression, the bacterial culture was incubated at room temperature for 16 hours. All bacterial pellets were collected by spinning the culture at 14,000 rpm at 4°C for 30 min in a Sorvall Rc6+ centrifuge. The pellet was lysed and dissolved using 30 ml of lysis buffer (50 mM tris, 500 mM NaCl, and 5 mM β-mercaptoethanol) supplemented with cOmplete EDTA free 1× protease inhibitor cocktail (Sigma-Aldrich, 11873580001). Dissolved pellets were subjected to sonication on ice using Branson Digital Sonifier 250 at 50% amplitude with 5-s bursts followed by 10-s cooling period until homogeneous lysate was obtained. The lysate was centrifuged at 18,000 rpm at 4°C for 30 min. During the lysate centrifugation step, HisPur Ni-NTA resin (Fisher Scientific, PI88222) was equilibrated with lysis buffer. The supernatant from the bacterial lysate was collected and incubated with the equilibrated resin for 1 hour at 4°C, after which the mixture was poured into empty polypropylene columns (Bio-Rad, 7311550). Wash buffer [50 mM tris, 2.5 M NaCl, 5 mM β-mercaptoethanol (BME). 1× cOmplete protease inhibitor cocktail (Sigma-Aldrich, 11873580001)] was applied to the column before elution with elution buffer (50 mM tris, 500 mM NaCl, 5 mM BME, 500 mM imidazole, and 1× cOmplete protease inhibitor cocktail). mEGFP was purified by cation exchange chromatography on UNO S6 column (Bio-Rad, 7200023) using buffer A (50 mM tris and 5 mM BME) and buffer B (50 mM tris, 5 mM BME, and 1 M NaCl). Fractions (0.5 ml each) containing purified protein were collected. For purifying MYOCD-TAD WT and FWY-A mutant proteins, size exclusion chromatography was performed by fast protein liquid chromatography (Bio-Rad NGC quest) with an ENrich SEC 650 10 × 300 column (Bio-Rad, 7801650) in buffer containing 50 mM tris, 75 mM NaCl, and 5 mM BME. Next, all proteins were dialyzed against protein storage buffer (50 mM tris, 75 mM NaCl, 5% glycerol, 1 mM PMSF, and 1 mM DTT) overnight at 4°C. The concentration of dialyzed protein was calculated by reading A280 on a spectrophotometer (DeNovix, DS-11 FX+). The purified proteins were either used immediately or stored at −80°C.
In vitro condensate formation assay
To examine the ability of MYOCD-TAD and its FWYtoA mutant protein to form droplets, protein (50 mM tris, 75 mM NaCl, 5% glycerol, 1 mM PMSF, and 1 mM DTT) was mixed with a 20% polyethylene glycol–8000 (PEG-8000) solution (prepared with 50 mM tris, 75 mM NaCl, 5% glycerol, 1 mM PMSF, and 1 mM DTT) to obtain the concentrations indicated in fig. S5 (B, C, F, and G). The resulting mixture was immediately loaded onto a custom-made chamber consisting of a coverslip attached to a glass slide using parallel strips of double-sided tape. The droplets that settled on the coverslip were captured using a Hamamatsu ORCA-Fusion C14440 digital camera and a CSU-W1 Yokogawa Spinning Disk Field Scanning Confocal System equipped with a Super Resolution by Optical Pixel Reassignment (SoRa) module. Images were acquired using a 60× Plan Apo Lambda Oil Immersion objective (NA 1.40) with SoRa engaged.
Analysis of in vitro condensates
To analyze in vitro droplet experiments, we used a Python script (version of 10 June 2021) obtained from https://github.com/jehenninger/in_vitro_droplet_assay, as described previously (13). The script was written to identify droplets and characterize their size, shape, and intensity. Besides using default parameters to identify droplets, parameters of circ, max_a, and min_a were adjusted to identify the maximum number of droplets. The output file obtained for individual droplets (individual_output.xlsx) with unique IDs for each droplet was further analyzed and plotted using GraphPad Prism version 9.3.1(350). Briefly, condensed fraction (CF) of channel (GFP) in each field of view was plotted, and the differences between groups of a family were statistically tested using unpaired t test. As described previously (21), condensed fraction is defined as
where Ichannel,droplet is the total intensity in the condensed droplet phase and Ichannel,bulk is the total background intensity outside droplets.
Quantitative real-time PCR analysis
RNA was extracted from 10T1/2 cells after 5-day reprogramming using TRIzol (Thermo Fisher Scientific, 15596026) and reverse-transcribed using iScript Reverse Transcription Supermix (Bio-Rad, 1708840) with random primers. The qPCRs were assembled using KAPA SYBR Fast qPCR Master Mix (KAPA, KK4605). Assays were performed using a 7900HT Fast Real-Time PCR machine (Applied Biosystems). Expression values were normalized to 18S ribosomal RNA and mEGFP and were represented as fold change. The following oligonucleotides were ordered from Integrated DNA Technologies to measure transcript abundance by qRT-PCR: mouse Acta2 (forward: GTCCCAGACATCAGGGAGTAA, reverse: TCGGATACTTCAGCGTCAGGA), mouse 18S (forward: ACCGCAGCTAGGAATGGA, reverse: GCCTCAGTTCCGAAAACCA), and mEGFP (forward: ACCACTACCAGCAGAACACC, reverse: GGGTCTTTGCTCAGCTTGGA).
Two-dimensional analysis for IF at CFP foci in Lac array cells
Following an established protocol (13), the relative fraction of immunofluorescence (IF) intensity described in Fig. 8 was performed considering the integrated background fluorescence in the cytoplasm and the integrated fluorescence at the LacO locus core (highest fluorescence intensity range on the Z-slice) using FIJI 2D measurement tools and ROI tools. The same ROI area was used to measure the fluorescence at both background and CFP foci. The quotient used in this analysis (arbitrary units) was calculated as raw intensity at CFP foci divided by raw intensity at background (cytoplasm) using the color channel for the IF of interest. Despite this internal control, it must be noted that images were processed together using the same IF treatment, exposure time, and laser intensity.
Quantification and statistical analysis
GraphPad Prism software was used for statistical analysis. Error bars, n values, number of replicates, P values, and statistical tests used for analysis are indicated in figure legends. Parametric or nonparametric tests were determined after performing normality test on samples. GraphPad denotes P values in our data as follows: ns P > 0.05, **P ≤ 0.05, **P ≤ 0.01 ***P ≤ 0.001, ****P ≤ 0.0001. Multiple comparison tests were Dunnett’s test or Dunn’s test for nonparametric samples against a control group. No statistical method was used for predetermining sample size.
Acknowledgments
We thank R. Bassel-Duby for advice and guidance in many aspects of this work, M. Parker for sharing Cdt1 cDNA, M. K. Rosen for sharing NPHS1 cDNA, A. Dall’Agnese for help in design of RNA FISH experiments, N. Munshi and S. Banani for thoughtful discussions, the Children’s Research Institute Sequencing Facility for performing Illumina sequencing, and the Moody Foundation Flow Cytometry core for performing FACS on hiPSCs.
Funding: This work was supported by the following: Cancer Prevention and Research Institute of Texas grant RR190090 (B.R.S.); NIH grants AR071980 (N.L.), HL130253 (E.N.O.), HL157281 (E.N.O.), and GM147583 (B.R.S.); Robert A. Welch Foundation grants 1-0025 (E.N.O.) and 1062707 (B.R.S.); and American Heart Association postdoctoral fellowship 825635 (P.G.).
Author contributions: Conceptualization: P.G., M.E., E.N.O., and B.R.S. Methodology: P.G., M.E., N.D.L.C., and B.R.S. Software: M.E. and R.T.V. Formal analysis: M.E., H.L., R.T.V., K.C., and L.X. Investigation: P.G., M.E., N.D.L.C., H.L., R.T.V., A.M.S., P.P., and S.B. Writing—original draft preparation: P.G., M.E., and B.R.S. Writing—review and editing: P.G., M.E., N.D.L.C., R.T.V., N.L., E.N.O., and B.R.S. Visualization: M.E., N.D.L.C., H.L., R.T.V., and B.R.S. Supervision: E.N.O. and B.R.S. Funding acquisition: E.N.O. and B.R.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq and ChIP-seq data from hiPSC presented here are available for download at GEO under the accession number GSE235928. RNA-seq and ChIP-seq data from 10T1/2 cells presented here are available for download at GEO under accession number GSE250206. All microscopy images used for analysis in this paper are available for download through the Dryad repository under doi:10.5061/dryad.r2280gbkf. These files can be accessed using the following link: https://datadryad.org/stash/share/RWRnD5YyEFUxAUTHHWIPA1GYwBveiWxSO3vYpJTRlu4. All data needed to evaluate the conclusions in the paper are present in the paper, the Supplementary Materials, or publicly available data repositories as detailed above. All code or software packages used in this study are publicly available. Correspondence and requests for materials should be addressed to E.N.O. or B.R.S.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
REFERENCES AND NOTES
- 1.Spiegelman B. M., Heinrich R., Biological control through regulated transcriptional coactivators. Cell 119, 157–167 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Graf T., Enver T., Forcing cells to change lineages. Nature 462, 587–594 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Zug R., Developmental disorders caused by haploinsufficiency of transcriptional regulators: A perspective based on cell fate determination. Biol. Open 11, bio058896 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palii C. G., Cheng Q., Gillespie M. A., Shannon P., Mazurczyk M., Napolitani G., Price N. D., Ranish J. A., Morrissey E., Higgs D. R., Brand M., Single-cell proteomics reveal that quantitative changes in co-expressed lineage-specific transcription factors determine cell fate. Cell Stem Cell 24, 812–820.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faure L., Wang Y., Kastriti M. E., Fontanet P., Cheung K. K. Y., Petitpre C., Wu H., Sun L. L., Runge K., Croci L., Landy M. A., Lai H. C., Consalez G. G., de Chevigny A., Lallemend F., Adameyko I., Hadjab S., Single cell RNA sequencing identifies early diversity of sensory neurons forming via bi-potential intermediates. Nat. Commun. 11, 4175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soldatov R., Kaucka M., Kastriti M. E., Petersen J., Chontorotzea T., Englmaier L., Akkuratova N., Yang Y., Haring M., Dyachuk V., Bock C., Farlik M., Piacentino M. L., Boismoreau F., Hilscher M. M., Yokota C., Qian X., Nilsson M., Bronner M. E., Croci L., Hsiao W. Y., Guertin D. A., Brunet J. F., Consalez G. G., Ernfors P., Fried K., Kharchenko P. V., Adameyko I., Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Ferrell J. E. Jr., Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14, 140–148 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Spitz F., Furlong E. E. M., Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Morgunova E., Taipale J., Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol. 47, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Cramer P., Organization and regulation of gene transcription. Nature 573, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Biggin M. D., Animal transcription networks as highly connected, quantitative continua. Dev. Cell 21, 611–626 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Slattery M., Zhou T., Yang L., Machado A. C. D., Gordân R., Rohs R., Absence of a simple code: How transcription factors read the genome. Trends Biochem. Sci. 39, 381–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyons H., Veettil R. T., Pradhan P., Fornero C., De La Cruz N., Ito K., Eppert M., Roeder R. G., Sabari B. R., Functional partitioning of transcriptional regulators by patterned charge blocks. Cell 186, 327–345.e28 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabari B. R., Dall'Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cissé I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., Young R. A., Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L., Gao Z., Wu J., Zhong B., Xie Y., Huang W., Lin Y., Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Mol. Cell 81, 1682–1697.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittag T., Pappu R. V., A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell 82, 2201–2214 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabari B. R., Dall'Agnese A., Young R. A., Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabari B. R., Biomolecular condensates and gene activation in development and disease. Dev. Cell 55, 84–96 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Boija A., Klein I. A., Sabari B. R., Dall'Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., Young R. A., Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrinivas K., Sabari B. R., Coffey E. L., Klein I. A., Boija A., Zamudio A. V., Schuijers J., Hannett N. M., Sharp P. A., Young R. A., Chakraborty A. K., Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75, 549–561.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D.-Z., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N., Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105, 851–862 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Ruzicka D. L., Schwartz R. J., Sequential activation of alpha-actin genes during avian cardiogenesis: Vascular smooth muscle alpha-actin gene transcripts mark the onset of cardiomyocyte differentiation. J. Cell Biol. 107, 2575–2586 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J., Kitchen C. M., Streb J. W., Miano J. M., Myocardin: A component of a molecular switch for smooth muscle differentiation. J. Mol. Cell. Cardiol. 34, 1345–1356 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Pipes G. C. T., Creemers E. E., Olson E. N., The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 20, 1545–1556 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Miano J. M., Myocardin in biology and disease. J. Biomed. Res. 29, 3–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Wang D.-Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N., Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428, 185–189 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Niu Z., Yu W., Zhang S. X., Barron M., Belaguli N. S., Schneider M. D., Parmacek M., Nordheim A., Schwartz R. J., Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J. Biol. Chem. 280, 32531–32538 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Niu Z., Iyer D., Conway S. J., Martin J. F., Ivey K., Srivastava D., Nordheim A., Schwartz R. J., Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc. Natl. Acad. Sci. U.S.A. 105, 17824–17829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S., Wang D.-Z., Wang Z., Richardson J. A., Olson E. N., The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl. Acad. Sci. U.S.A. 100, 9366–9370 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Lu M. M., Cheng L., Yuan L.-J., Zhu X., Stout A. L., Chen M., Li J., Parmacek M. S., Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc. Natl. Acad. Sci. U.S.A. 106, 18734–18739 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J., Wang T., Wright A. C., Yang J., Zhou S., Li L., Yang J., Small A., Parmacek M. S., Myocardin is required for maintenance of vascular and visceral smooth muscle homeostasis during postnatal development. Proc. Natl. Acad. Sci. U.S.A. 112, 4447–4452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Wang D.-Z., Pipes G. C. T., Olson E. N., Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. U.S.A. 100, 7129–7134 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houweling A. C., Beaman G. M., Postma A. V., Gainous T. B., Lichtenbelt K. D., Brancati F., Lopes F. M., van der Made I., Polstra A. M., Robinson M. L., Wright K. D., Ellingford J. M., Jackson A. R., Overwater E., Genesio R., Romano S., Camerota L., D'Angelo E., Meijers-Heijboer E. J., Christoffels V. M., McHugh K. M., Black B. L., Newman W. G., Woolf A. S., Creemers E. E., Loss-of-function variants in myocardin cause congenital megabladder in humans and mice. J. Clin. Invest. 129, 5374–5380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creemers E. E., Sutherland L. B., Oh J., Barbosa A. C., Olson E. N., Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol. Cell 23, 83–96 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Shore P., Sharrocks A. D., The MADS-Box family of transcription factors. Eur. J. Biochem. 229, 1–13 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Gould S. J., Keller G. A., Hosken N., Wilkinson J., Subramani S., A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108, 1657–1664 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vernon R. M., Chong P. A., Tsang B., Kim T. H., Bah A., Farber P., Lin H., Forman-Kay J. D., Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Choi J.-M., Holehouse A. S., Lee H. O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., Poser I., Pappu R. V., Alberti S., Hyman A. A., A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H., Han T., Xie S., Corden J. L., McKnight S. L., Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei M.-T., Chang Y.-C., Shimobayashi S. F., Shin Y., Brangwynne C. P., Nucleated transcriptional condensates amplify gene expression. Nat. Cell Biol. 22, 1187–1196 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Quiroga I. Y., Ahn J. H., Wang G. G., Phanstiel D., Oncogenic fusion proteins and their role in three-dimensional chromatin structure, phase separation, and cancer. Curr. Opin. Genet. Dev. 74, 101901 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., Baldwin A. J., Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pak C. W., Kosno M., Holehouse A. S., Padrick S. B., Mittal A., Ali R., Yunus A. A., Liu D. R., Pappu R. V., Rosen M. K., Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker M. W., Bell M., Mir M., Kao J. A., Darzacq X., Botchan M. R., Berger J. M., A new class of disordered elements controls DNA replication through initiator self-assembly. eLife 8, e48562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker M. W., Kao J. A., Huang A., Berger J. M., Botchan M. R., Molecular determinants of phase separation for Drosophila DNA replication licensing factors. eLife 10, e70535 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muhl L., Mocci G., Pietila R., Liu J., He L., Genove G., Leptidis S., Gustafsson S., Buyandelger B., Raschperger E., Hansson E. M., Bjorkegren J. L. M., Vanlandewijck M., Lendahl U., Betsholtz C., A single-cell transcriptomic inventory of murine smooth muscle cells. Dev. Cell 57, 2426–2443.e6 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Holehouse A. S., Pappu R. V., Functional implications of intracellular phase transitions. Biochemistry 57, 2415–2423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao D., Wang C., Tang R., Chen H., Zhang Z., Tatsuguchi M., Wang D. Z., Acetylation of myocardin is required for the activation of cardiac and smooth muscle genes. J. Biol. Chem. 287, 38495–38504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narlikar G. J., Myong S., Larson D., Maeshima K., Francis N., Rippe K., Sabari B., Strader L., Tjian R., Is transcriptional regulation just going through a phase? Mol. Cell 81, 1579–1585 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J. V., King D. S., Banani S. F., Russo P. S., Jiang Q.-X., Nixon B. T., Rosen M. K., Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peeples W., Rosen M. K., Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 17, 693–702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodruff J. B., Gomes B. F., Widlund P. O., Mahamid J., Honigmann A., Hyman A. A., The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Case L. B., Zhang X., Ditlev J. A., Rosen M. K., Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trojanowski J., Frank L., Rademacher A., Mücke N., Grigaitis P., Rippe K., Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e10 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Chong S., Graham T. G. W., Dugast-Darzacq C., Dailey G. M., Darzacq X., Tjian R., Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol. Cell 82, 2084–2097.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Kim Y. J., LeeJr M., Lee Y.-T., Jing J., Sanders J. T., Botten G. A., He L., Lyu J., Zhang Y., Mettlen M., Ly P., Zhou Y., Xu J., Light-activated macromolecular phase separation modulates transcription by reconfiguring chromatin interactions. Sci. Adv. 9, eadg1123 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuo L., Zhang G., Massett M., Cheng J., Guo Z., Wang L., Gao Y., Li R., Huang X., Li P., Qi Z., Loci-specific phase separation of FET fusion oncoproteins promotes gene transcription. Nat. Commun. 12, 1491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin E. W., Holehouse A. S., Peran I., Farag M., Incicco J. J., Bremer A., Grace C. R., Soranno A., Pappu R. V., Mittag T., Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin E. W., Holehouse A. S., Intrinsically disordered protein regions and phase separation: Sequence determinants of assembly or lack thereof. Emerg. Top. Life Sci. 4, 307–329 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Sanborn A. L., Yeh B. T., Feigerle J. T., Hao C. V., Townshend R. J. L., Lieberman-Aiden E., Dror R. O., Kornberg R. D., Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to mediator. eLife 10, e68068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erijman A., Kozlowski L., Sohrabi-Jahromi S., Fishburn J., Warfield L., Schreiber J., Noble W. S., Söding J., Hahn S., A high-throughput screen for transcription activation domains reveals their sequence features and permits prediction by deep learning. Mol. Cell 78, 890–902.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Staller M. V., Ramirez E., Kotha S. R., Holehouse A. S., Pappu R. V., Cohen B. A., Directed mutational scanning reveals a balance between acidic and hydrophobic residues in strong human activation domains. Cell Syst. 13, 334–345.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J., Li A., Wang Z., Feng X., Olson E. N., Schwartz R. J., Myocardin sumoylation transactivates cardiogenic genes in pluripotent 10T1/2 fibroblasts. Mol. Cell. Biol. 27, 622–632 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taurin S., Sandbo N., Yau D. M., Sethakorn N., Kach J., Dulin N. O., Phosphorylation of myocardin by extracellular signal-regulated kinase. J. Biol. Chem. 284, 33789–33794 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gan P., Wang Z., Morales M. G., Zhang Y., Bassel-Duby R., Liu N., Olson E. N., RBPMS is an RNA-binding protein that mediates cardiomyocyte binucleation and cardiovascular development. Dev. Cell 57, 959–973.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang L., Geng Z., Nickel T., Johnson C., Gao L., Dutton J., Hou C., Zhang J., Differentiation of human induced-pluripotent stem cells into smooth-muscle cells: Two novel protocols. PLOS ONE 11, e0147155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z., Cui M., Shah A. M., Ye W., Tan W., Min Y.-L., Botten G. A., Shelton J. M., Liu N., Bassel-Duby R., Olson E. N., Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. Proc. Natl. Acad. Sci. U.S.A. 116, 18455–18465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mi H., Ebert D., Muruganujan A., Mills C., Albou L.-P., Mushayamaha T., Thomas P. D., PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49, D394–D403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramirez-Martinez A., Zhang Y., Chen K., Kim J., Cenik B. K., McAnally J. R., Cai C., Shelton J. M., Huang J., Brennan A., Evers B. M., Mammen P. P. A., Xu L., Bassel-Duby R., Liu N., Olson E. N., The nuclear envelope protein Net39 is essential for muscle nuclear integrity and chromatin organization. Nat. Commun. 12, 690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.S. Andrews, FASTQC. A quality control tool for high throughput sequence data (2010); https://bioinformatics.babraham.ac.uk/projects/fastqc/.
- 75.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., DeepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Janicki S. M., Tsukamoto T., Salghetti S. E., Tansey W. P., Sachidanandam R., Prasanth K. V., Ried T., Shav-Tal Y., Bertrand E., Singer R. H., Spector D. L., From silencing to gene expression: Real-time analysis in single cells. Cell 116, 683–698 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jones T. R., Kang I. H., Wheeler D. B., Lindquist R. A., Papallo A., Sabatini D. M., Golland P., Carpenter A. E., CellProfiler analyst: Data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9, 482 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.J. Hardin, FRAP Profiler FIJI plugin (2018); https://worms.zoology.wisc.edu/research/4d/4d.html#frap.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10