Abstract
The protein corona, a dynamic biomolecular layer that forms on nanoparticle (NP) surfaces upon exposure to biological fluids is emerging as a valuable diagnostic tool for improving plasma proteome coverage analyzed by liquid chromatography-mass spectrometry (LC-MS/MS). Here, we show that spiking small molecules, including metabolites, lipids, vitamins, and nutrients, into plasma can induce diverse protein corona patterns on otherwise identical NPs, significantly enhancing the depth of plasma proteome profiling. The protein coronas on polystyrene NPs when exposed to plasma treated with an array of small molecules (n=10) allowed for detection of 1793 proteins marking an 8.25-fold increase in the number of quantified proteins compared to plasma alone (218 proteins) and a 2.63-fold increase relative to the untreated protein corona (681 proteins). Furthermore, we discovered that adding 1000 μg/ml phosphatidylcholine could singularly increase the number of unique proteins within the protein corona (897 proteins). This specific concentration of phosphatidylcholine selectively depleted the four most abundant plasma proteins, including albumin, thus reducing concentration dynamic range of plasma proteome and boosting LC-MS/MS sensitivity for detection of proteins with lower abundance. By employing an optimized data-independent acquisition (DIA) approach, the inclusion of phosphatidylcholine led to the detection of 1436 proteins in plasma. This significant achievement is made utilizing only a single NP type and one small molecule to analyze a single plasma sample, setting a new standard in proteomic depth of the plasma sample. Given the critical role of plasma proteomics in biomarker discovery and disease monitoring, we anticipate widespread adoption of this methodology for identification and clinical translation of proteomic biomarkers into FDA approved diagnostics.
Introduction
The quest to comprehensively analyze the plasma proteome has become crucial for advancing disease diagnosis and monitoring, as well as biomarker discovery.1, 2 Yet, obstacles like identifying low-abundance proteins remain owing to the prevalence of high-abundance proteins in plasma where the seven most abundant proteins collectively represent 85% of the total protein mass.3, 4 Peptides from these high-abundance proteins, especially those of albumin, tend to dominate mass spectra impeding the detection of proteins with lower-abundance.
To address this challenge, techniques such as affinity depletion, protein equalizer, and electrolyte fractionation have been developed to reduce the concentration of these abundant proteins, thereby enhancing the detection of proteins with lower-abundance.5, 6, 7 Additionally, a range of techniques has been developed to enhance the throughput and depth of protein detection and identification, from advanced acquisition modes to methods that concentrate low-abundance proteins or peptides for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.5, 8, 9, 10, 11, 12, 13 For instance, in the affinity depletion strategy14, affinity chromatography columns are used with specific ligands that bind to high-abundance proteins such as albumin, immunoglobulins, and haptoglobin. However, cost and labor associated with such depletion strategies hampers their application for large cohorts. As another example, salting-out technique15 is used to add reagents (e.g., ammonium sulfate) to selectively precipitate high-abundance proteins, leaving the lower-abundance proteins in the supernatant. However, these methods can introduce biases in precipitating lower-abundance proteins as well, and therefore, additional robust strategies are needed to ensure low-abundance proteins with high diagnostic potential are not missed in biomarker discovery studies.
Recently, nanoparticles (NPs) have gained attention for their ability to enhance biomarker discovery through analysis of the spontaneously-forming protein/biomolecular corona (i.e., a layer of biomolecules, primarily proteins, that forms on NPs when exposed to plasma or other biological fluids)16, 17, 18, 19. The NP protein corona can contain a unique ability to concentrate proteins with lower abundance, easily reducing the proteome complexity for LC-MS/MS analysis.5, 16, 20
Application of single NPs for biomarker discovery has limitations in achieving deep proteome coverage, typically enabling the detection of only hundreds of proteins.21 To enhance proteome coverage and quantify a higher number of plasma proteins, the use of a protein corona sensor array or multiple NPs with distinct physicochemical properties can be implemented. This approach leverages the unique protein corona that forms on each NP to increase proteome coverage, but carries the drawback of having to analyze multiple NP samples and needing to test many NP types to increase proteome coverage.5, 20, 22
Small molecules native to human biofluids play a significant role in regulating human physiology, often through interactions with proteins. Therefore, we hypothesize that small molecules might influence the formation of the NP protein corona and serve to enrich specific proteins including biomarkers or lower abundance proteins. Recent findings have reported that high levels of cholesterol results in a protein corona with enriched apolipoproteins and reduced complement proteins, which is due to the changes in the binding affinity of the proteins to the NPs in the presence of cholesterol.23 Accordingly, we hypothesized that small molecules endogenous to human plasma may affect the composition of the NP protein corona differently depending on whether these molecules act individually or collectively.24
Our work presents an efficient methodology that harnesses the influence of various small molecules in creating diverse protein coronas on otherwise identical NPs. Our primary hypothesis, corroborated by our findings, posits that introducing small molecules into plasma alters the way these proteins engage with NPs. This alteration, in turn, modulates the protein corona profile of the NPs. As a result, when NPs are incubated with plasma pre-treated with an array of small molecules at diverse concentrations, these small molecules significantly enhance the detection of a broad spectrum of low-abundance proteins through LC-MS/MS analyses. The selected small molecules include essential biological metabolites, lipids, vitamins, and nutrients consisting of glucose, triglyceride, diglycerol, phosphatidylcholine (PtdChos), phosphatidylethanolamine (PE), L-α-phosphatidylinositol (PtdIns), inosine 5′-monophosphate (IMP), and B complex and their combinations. Our findings confirm that the addition of these small molecules in plasma generates distinct protein corona profiles on otherwise identical NPs, significantly expanding the dynamic range of the plasma proteome that can be captured and detected by simple LC-MS/MS analysis. Notably, we discover that the addition of specific small molecules, such as PtdChos, leads to a substantial increase in proteome coverage, which is attributed to the unique ability of PdtChos to bind albumin and reduce its participation in protein corona formation. Therefore, PtdChos coupled with NP protein corona analysis can replace the expensive albumin depletion kits and accelerate the plasma analysis workflow by reducing processing steps. Furthermore, our single small molecule-single NP platform reduces the necessity for employing multiple NP workflows in plasma proteome profiling. This approach can seamlessly integrate with existing LC-MS/MS workflows to further enhance the depth of plasma proteome analysis for biomarker discovery.
Results
Protein corona and small molecules enable deep profiling of the plasma proteome
We assessed the effect of eight distinct sets of small molecules, namely, glucose, triglyceride, diglycerol, PtdChos, PE, PtdIns, IMP, and vitamin B complex, on the protein corona formed around polystyrene NPs. The selection of these molecules was based on their ability to interact with a broad spectrum of proteins, which significantly influences the composition of the protein corona surrounding NPs. For example B complex components can interact with a wide range of proteins including albumin25, 26, hemoglobin25, myoglobin27, pantothenate permease28, acyl carrier protein29, lactoferrin30, prion31, β-amyloid precursor32, and niacin-responsive repressor33. Additionally, to assess the collective effects of these molecules, we analyzed two representative “molecular sauces.” Molecular sauce 1 contained a blend of glucose, triglyceride, diglycerol, PtdChos, and molecular sauce 2 consisted of PE, PtdIns, IMP, and vitamin B complex. The workflow of the study is outlined in Supplementary Scheme 1.
Commercially available plain polystyrene NPs, averaging 80 nm in size, were purchased. Each small molecule, at varying concentrations (10, 100, and 1000 μg/ml), was first incubated with commercial pooled healthy human plasma at 37°C for 1 h allowing the small molecules to interact with the biological matrix. Subsequently, NPs at a concentration of 0.2 mg/ml were introduced into the plasma containing small molecules or sauces and incubated for an additional hour at 37°C with agitation. These methodological parameters were refined from previous studies to guarantee the formation of a distinct protein corona around the NPs. Supplementary Fig. 1 offers further details on our methodologies, showcasing dynamic light scattering (DLS), zeta potential, and transmission electron microscopy (TEM) analyses for both the untreated NPs and those covered by a protein corona34. The untreated polystyrene NPs exhibited excellent monodispersity, with an average size of 78.8 nm with the polydispersity index of 0.026 and a surface charge of −30.1 ± 0.6 mV. Upon formation of the protein corona, the average size of NPs expanded to 113 nm, and the surface charge shifted to −10 mV ± 0.4 mV. TEM analysis further corroborated the size and morphology alterations of the NPs before and after protein corona formation (Supplementary Fig. 1).
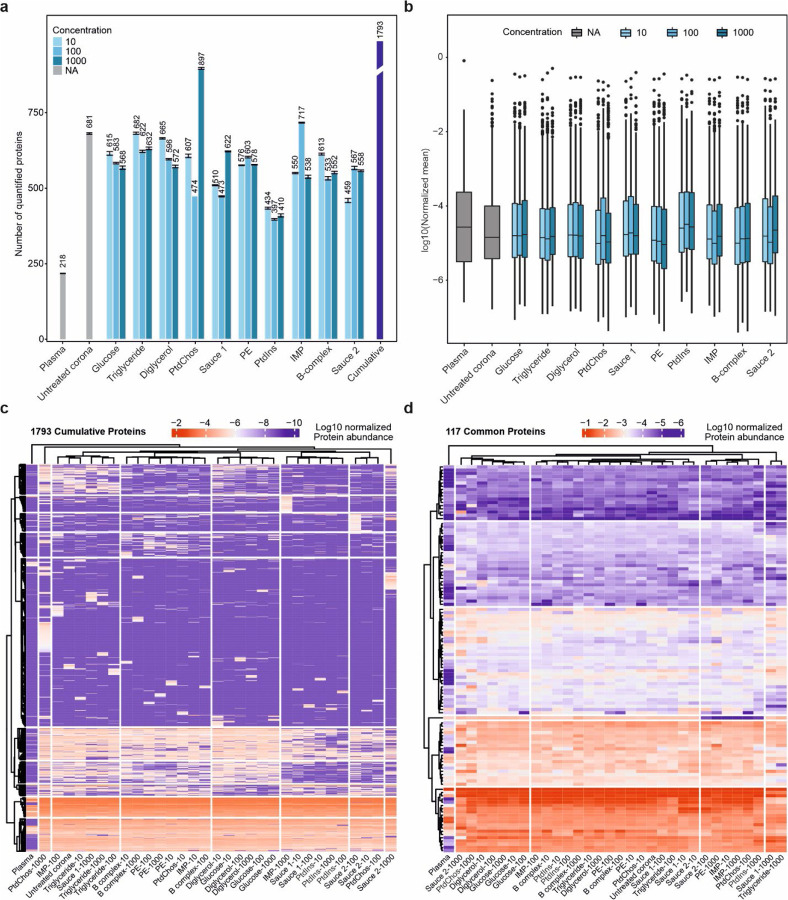
To investigate how spiking different concentrations of small molecules can influence the molecular composition of the protein corona, samples were subjected to high-resolution LC-MS/MS analysis. While the analysis of plasma alone led to quantification of 218 unique proteins, analysis of the protein corona formed on the polystyrene NPs significantly enhanced the depth of plasma proteome sampling to enable quantification of 681 unique proteins. Furthermore, the inclusion of small molecules further deepened plasma proteome sampling to enable quantification of between 397 and up to 897 unique proteins, depending on the small molecules added to plasma prior to corona formation. When comparing the use of protein coronas, both with and without the inclusion of small molecules, to the analysis of plasma alone (Fig. 1a and Supplementary Data 1), there is a notable increase—approximately a threefold rise—in the number of proteins that can be quantified.
Fig. 1. Small molecules affect the plasma proteome sampling.
a, The number of quantified proteins in plasma, untreated protein corona and protein coronas in the presence of small molecules and molecular sauces (mean ± SD of three technical replicates). The cumulative number of unique proteins identified across all conditions is also shown using the purple bar. For fair comparison, the database was performed individually for each small molecule. b, The distribution of normalized intensities for proteins quantified in the plasma, untreated protein corona and protein coronas in the presence of small molecules and molecular sauces (center line, median; box limits contain 50%; upper and lower quartiles, 75 and 25%; maximum, greatest value excluding outliers; minimum, least value excluding outliers; outliers, more than 1.5 times of upper and lower quartiles). c, The hierarchical clustering of all proteins quantified across all samples. d, The clustering of 117 shared proteins across all samples. Experiments were performed in three technical replicates and protein abundances were averaged for each condition.
Interestingly, the concentration of small molecules did not significantly affect the number of quantified proteins; only a small stepwise reduction in the number of quantified proteins was noted with increasing concentrations of glucose and diglycerol. Cumulatively, the incorporation of small molecules and molecular sauces into the protein corona of NPs led to a significant increase in protein quantification, with a total of 1793 proteins identified, marking an 8.25-fold increase compared to plasma alone. Specifically, the addition of small molecules resulted in the quantification of 1573 additional proteins compared to plasma alone, and 1037 more proteins than the untreated protein corona. Strikingly, spiking 1000 μg/ml of PtdChos singlehandedly increased the number of quantified proteins to 897 (1.3-fold of quantified proteins in untreated plasma). This observation prompted a detailed investigation into the influence of PtdChos on plasma proteome coverage, which is elaborated in the following sections. We note that our analysis focuses on the fold changes of quantified proteins compared to plasma and the untreated corona, rather than the absolute numbers of quantified proteins. This is mainly because the numbers of quantified proteins are strongly dependent to the workflow of mass spectrometry; for example we recently revealed that analysis of identical corona-coated polystyrene NPs by various mass spectrometry centers resulted in varying numbers of quantified proteins, ranging from 235 to more than a thousand.35 Therefore, our fold-change analysis is more conservative and less subject to mass spectrometry workflow biases.36
The distribution of normalized protein intensities for the samples is shown in Fig. 1b. The median value in the plasma group was notably higher than in the other samples, although the overall distribution did not differ significantly. In general, the proteomes obtained from protein corona profiles in the presence of small molecules showed a good correlation (generally a Pearson correlation above 0.6 for most small molecule comparisons) demonstrating the faithful relative representation of proteins after treatment with different small molecules (Supplementary Fig. 2).
Small molecules diversify the protein corona composition
We next investigated if the addition of small molecules would change the type and number of proteins detected by LC-MS/MS. Indeed, each small molecule and the molecular sauces generated a proteomic fingerprint that was distinct from untreated protein corona or those of other small molecules (Fig. 1c). Spiking small molecules led to detection of a diverse set of proteins in the plasma. Interestingly, even different concentrations of the same small molecules or molecular sauces produced unique fingerprints. A similar analysis was performed for the 117 shared proteins across the samples (Fig. 1d). The Venn diagrams in Supplementary Fig. 3a and 3b show the number of unique proteins that were quantified in the respective group across all concentrations which were not quantified in the plasma or in the untreated protein corona. These results suggest that spiking small molecules into human biofluids can diversify the range of proteins that are quantifiable in protein corona profiles, effectively increasing proteomic coverage to lower abundance proteins. Such an enrichment or depletion of a specific subset of proteins can be instrumental in biomarker discovery focused on a disease area. This feature can also be used for designing assays where the enrichement of a known biomarker is facilitated by using a given small molecule. As representative examples, a comparison of enriched and depleted proteins for molecular sauce 1 and 2 against the untreated protein corona is shown in Supplementary Fig. 3c and 3d, respectively (Supplementary Data 2). In certain cases, the enrichment or depletion was drastic, spanning several orders of magnitude. The enriched and depleted proteins for the molecular sauce 1 and 2 were mapped to KEGG pathways and biological processes in StringDB (Supplementary Fig. 3c–d). While most of the enriched pathways were shared, some pathways were specifically enriched for a given molecular sauce. For example, systemic lupus erythematosus (SLE) was only enriched among the top pathways for molecular sauce 2. Therefore, the small molecules can be potentially used for facilitating the discovery of biomarkers for specific diseases, or for assaying the abundance of a known biomarker in disease detection.
Similar analyses were performed for all the small molecules and the volcano plots for the highest concentration of each molecule (i.e., 1000 μg/ml) are demonstrated in Supplementary Fig. 4 (Supplementary Data 2). A pathway analysis was also performed for all the significantly changing proteins for each small molecule at all concentrations (Supplementary Fig. 5). To facilitate comparison, we have combined the enrichement analysis for all the samples vs. the untreated protein corona in Supplementary Fig. 6.
To demonstrate how small molecules affect the composition and functional categories of proteins in the protein corona, potentially aiding in early diagnosis of diseases (since proteins enriched in the corona are pivotal in conditions like cardiovascular and neurodegenerative diseases), we utilized bioanalytical methods37 to categorize the identified proteins based on their blood-related functions namely complement activation, immune response, coagulation, acute phase response, and lipid metabolism (Supplementary Fig. 7). In our analysis, apolipoproteins were major protein types that were found in the small molecule treated protein corona, and their types and abundance were heavily dependent to the type and concentrations of the employed small molecules (Supplementary Fig. 8). Similarly, the enrichment of other specific protein categories on NPs surfaces was influenced by the type and concentration of small molecules used (Supplementary Fig. 8). For example, antithrombin-III in coagulation factors plays a significant role in the protein corona composition of all tested small molecules, but this effect is observed only at their highest concentration. At lower concentrations, or in the untreated protein corona, this considerable participation is not evident (Supplementary Fig. 8). This ability of small molecules to modify the protein composition on NPs highlights their potential for early disease diagnosis (e.g., apolipoprtoeins in cardiovascular and neurodegenerative disorders)38, 39, where these protein categories are crucial in disease onset and progression 38.
PtdChos increases proteome coverage by depleting the abundant plasma proteins
To understand whether the quantification of a higher number of proteins in protein corona profiles was due to a lower dynamic range of proteins available in human plasma for NP binding, we plotted the maximum protein abundance vs. minimum protein abundance for plasma alone, and plasma treated with small molecules in Supplementary Fig. 9. The plasma alone showed the highest dynamic range, suggesting that quantification of low-abundance proteins would be most difficult from plasma alone. Conversely, addition of small molecules was shown to reduce plasma protein dynamic range, thereby allowing for detection of more peptides and quantification of proteins with lower abundance through the NP protein corona.
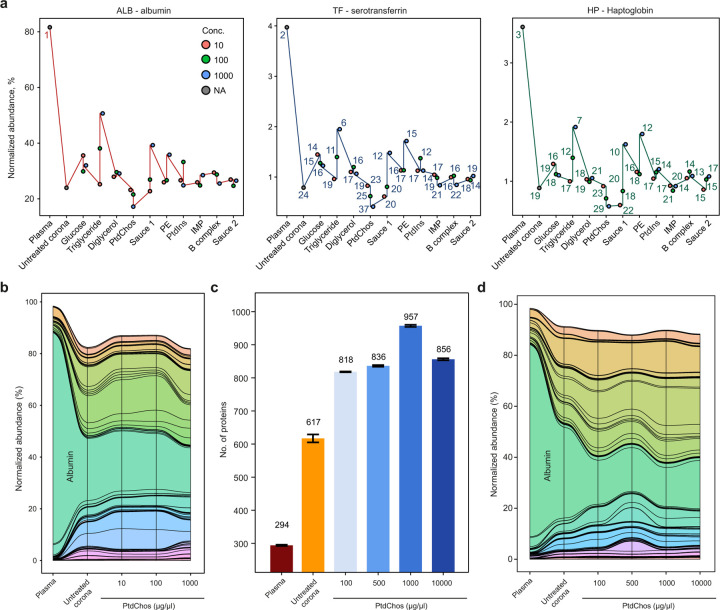
Notably, while albumin accounted for over 81% of our plasma sample, its representation was significantly lowered to an average of 29% in the protein coronas, both with and without small molecule modifications. This reduction was most pronounced with PtdChos treatment at 1000 μg/ml, where albumin levels dropped to around 17% of plasma proteins (Fig. 2a). Despite these changes, albumin remained the most abundant protein in all samples. A similar diminishing trend was observed for the second and third most abundant proteins, serotransferrin (TF) and haptoglobin (HB), which made up about 3.9% and 3.6% of plasma protein abundance, respectively. The rankings of these proteins’ abundance in each sample are depicted above their corresponding circles in Fig. 2a. From this analysis, it is evident that the protein corona, both in its native form and when altered by small molecules, can drastically reduce the combined representation of the top three proteins from about 90% to roughly 29%. The most substantial reduction was observed with PtdChos at 1000 μg/ml, reducing the top three proteins’ cumulative representation from 90% to under 17%. PtdChos treatment also effectively reduced the levels of the fourth most abundant plasma protein IGHA1.This significant decrease in the abundance of highly prevalent plasma proteins explains the marked increase in the number of unique proteins detected from NP corona samples treated with PtdChos. These results indicate that high concentrations of PtdChos can be strategically employed to enable more comprehensive plasma protein sampling by specifically targeting and depleting the most abundant plasma proteins, especially albumin.
Fig. 2. PtdChos can deplete the most abundant plasma proteins in protein corona.
a, Comparative analysis of protein abundance and rankings in untreated plasma versus protein corona profiles, both with and without the addition of small molecules (note that albumin ranked 1st in all samples). b, A stream (or alluvial) diagram illustrating the significant depletion of abundant plasma proteins, particularly albumin, following the incubation of plasma with NPs and PtdChos (only shared proteins with plasma are included). c, The total count of proteins identified in plasma samples incubated with NPs, comparing treatments with and without PtdChos at various concentrations (mean ± SD of three technical replicates). d, A stream diagram demonstrating the depletion pattern of abundant plasma proteins, especially albumin, in response to NP addition and enhanced with escalating concentrations of PtdChos (only shared proteins with plasma are included).
The stream (or alluvial) diagram in Fig 2b shows the overall changes in the representation of proteins found in plasma upon incubation of protein corona with different concentrations of PtdChos. To validate this discovery, we prepared fresh samples treated with a series of PtdChos concentrations ranging from 100 to 10,000 μg/ml (Supplementary Data 3). As shown in Fig 2c, 957 proteins could be quantified in the protein corona treated with PtdChos at 1000 μg/ml, while neither lower concentration nor further addition of PtdChos enhanced the number of quantified proteins. The stream diagram in Fig 2d shows the specific depletion of albumin and a number of other abundant proteins in plasma upon addition of PtdChos, allowing for more robust detection of other proteins with lower abundance.
To confirm that the improved proteome coverage achieved with PtdChos treatment is independent of the LC-MS platform used, we prepared new samples of plasma, untreated protein corona, and protein corona treated with 1000 μg/ml PtdChos, and analyzed them using data-independent acquisition (DIA) LC-MS in another core facility. We identified 322 proteins in the plasma alone, 1011 proteins in the untreated protein corona samples, and 1436 proteins in the protein corona treated with PtdChos (1.4-fold increase over the untreated corona) (Supplementary Data 4). These findings not only validate the enhancement of plasma proteome coverage by PtdChos but also illustrate the capability of PtdChos to facilitate the in-depth profiling of the plasma proteome associated with protein corona formed on the surface of a single type of NP. Since the ratio of the number of quantified proteins through PtdChos spiking is generally around 1.4-fold higher than in the NP corona alone, PtdChos can be incorporated into any LC-MS workflow aiming to boost plasma proteome profiling. More optimized plasma proteomics pipelines or high-end mass spectrometers such as Orbitrap Astral are envisioned to quantify an even higher number of proteins that those reported in the current study.
Discussion
The protein corona is a layer of proteins that spontaneously adsorbs on the surface of nanomaterials when exposed to biological fluids.40 The composition and dynamic evolution of the protein corona is critically important as it can impact the interactions of NPs with biological systems (e.g., activate the immune system), can cause either positive or adverse biocompatibility outcomes, and can greatly affect NP biodistribution in vivo.40 The specific proteins that adsorb on the surface of the NPs depend on various factors, including the physicochemical properties of the NPs and the composition of the surrounding biological fluid.35 Metabolites, lipids, vitamins, nutrition, and other types of small biomolecules present in the biological fluid can interact with proteins in these fluids and influence their behavior, including their adsorption onto NPs. For example, it was shown that addition of glucose and cholesterol to plasma can alter the composition of protein corona on the surface of otherwise identical NPs.23, 34 Small molecules can alter the protein corona of NPs, after interaction with plasma proteins, due to various mechanisms such as i) their competition with proteins for binding to the surface of NPs; ii) altering proteins’ binding affinities to NPs; and iii) changing protein conformation.23, 34 For example, previous studies revealed that triglyceride, PtdChos, PE , and PtdIns can interact with lipovitellin41, C-reactive protein42, protein Z43, and myelin basic protein (MBP)44, respectively. Each individual small molecule and their combinations interrogates tens to hundreds of additional proteins across a broad dynamic range in an unbiased and untargeted manner. Therefore, any changes in the level of the small molecules in the body can alter the overall composition of the protein corona, leading to variations in the types and number of proteins that bind to NPs and consequently their corresponding interactions with biosystems.23, 24
Among the various employed small molecules, we discovered that PtdChos alone demonstrates a remarkably high ability to reduce the participation of the most highly abundant proteins in protein corona composition. PtdChos is the most common class of phospholipids in the majority of eukaryotic cell membranes.45 For a long time, it has been established that PtdChos can engage in specific interactions with serum albumin through hydrophobic processes.46, 47, forming distinct protein-lipid complexes.46, 48 As a result, we found that the simple addition of PtdChos to plasma can significantly reduce albumin’s affinity for the surface of polystyrene NPs, thereby creating unique opportunities for the involvement of a broader range of proteins with lower-abundance in the corona. Not only PtdChos is an economical and simple alternative for conventional albumin depletion strategies, but it can also deplete several other highly abundant proteins as an added advantage. This approach reduces the necessity for employing NP arrays in plasma proteome profiling, and the cost and biases that can occur with albumin depletion or multistep protocols. Additionally, PtdChos can help accelerate plasma analysis workflows by reducing the sample preparation steps.
In summary, our study highlights the tremendous potential of leveraging small molecules in enhancing the capabilities of protein corona profiles for broader plasma proteome analysis. By introducing individual small molecules and their combinations into plasma, we have successfully created distinct protein corona patterns on single identical NPs, thereby expanding the repertoire of attached proteins. Using LC-MS/MS, we quantified an additional 1573 unique proteins that would otherwise remain undetected in plasma. This enhanced depth in protein coverage can be attributed, in part, to the unique interactions of each small molecule, allowing for representation of a diverse set of proteins in the corona. Moreover, our findings underscore the influence of small molecules on the types and categories of proteins in the protein corona shell. This feature opens exciting possibilities for early disease diagnosis, particularly in conditions such as cardiovascular and neurodegenerative disorders, where enriched proteins, such as apolipoproteins, play pivotal roles. Overall, our research not only enhances the depth and scope of plasma proteome analysis but also suggests promising avenues for the development of novel biomarkers and diagnostic approaches.
In summary, our platform is capable of quantifying up to 1793 proteins when using a single NP with an array of small molecules, while only 218 and 681 proteins could be quantified using the plasma or the NP protein corona alone. Finally, we showed the possibility of quantifying up to 1436 proteins using a single NP and PtdChos alone using a single plasma sample. The number of detected proteins will therefore dramatically increase if this platform is applied to a cohort of patient samples with individual variabilities. Expectedly, with progressive development of LC-MS platforms, the depth of analysis can further increase towards the ultimate goal of comprehensive human proteome coverage. We envision the widespread use of this platform in plasma proteome profiling, holding huge potential in disease diagnostics and monitoring.
Materials and Methods
Materials.
Healthy human plasma protein was obtained from Innovative Research (www.innov-research.com) and diluted to a final concentration of 55% using phosphate buffer solution (PBS, 1X). Plain polystyrene NPs (~ 80 nm) were provided by Polysciences. (www.polysciences.com). Small molecules were ordered from Sigma, Abcam, Fisher Scientific, VWR, Beantown, and diluted with PBS (1X) to the desired concentration.
Protein corona formation on the surface of NPs in the presence of small molecules.
For protein corona formation in the presence of small molecules, human plasma diluted to 55% by PBS was first incubated with individual small molecules or in combination by preparing two molecular sauces of individual small molecules at different concentration (i.e., 10, 100, and 1000 μg/ml) for 1 h at 37 °C. Then, polystyrene NPs were added to the mixture of plasma and small molecules solution so that the final concentration of the NPs was 0.2 mg/ml and incubated for another 1 h at 37 °C. It is noteworthy that all experiments are designed in a way that the concentration of NPs, human plasma, and small molecules were 0.2 mg/ml, 55%, and 10, 100, 1000 μg/ml, respectively. To remove unbound and plasma proteins only loosely attached to the surface of NPs, protein-NPs complexes were then centrifuged at 14,000 ´ g for 20 minutes, the collected NPs’ pellets were washed three times with cold PBS under the same conditions, and the final pellet was collected for further analysis. For the PtdChos concentration study, we used various concentrations of PtdChos (i.e., 100, 500, 1000, and 10000 μg/ml) and used the above protocol for preparation of the samples for mass spectrometry analysis.
NP characterization.
DLS and zeta potential analyses were performed to measure the size distribution and surface charge of the NPs before and after protein corona formation using a Zetasizer nano series DLS instrument (Malvern company). A Helium Neon laser with a wavelength of 632 nm was used for size distribution measurement at room temperature. TEM was carried out using a JEM-2200FS (JEOL Ltd.) operated at 200kV. The instrument was equipped with an in-column energy filter and an Oxford X-ray energy dispersive spectroscopy (EDS) system. 20 μl of the bare NPs were deposited onto a copper grid and used for imaging. For protein corona–coated NPs, 20 μl of sample was negatively stained using 20 μl uranyl acetate 1%, washed with DI water, deposited onto a copper grid, and used for imaging. PC composition was also determined using LC-MS/MS.
LC-MS/MS sample preparation for the screening and concentration series experiments
Initial processing:
NPs coated with protein corona were first washed with PBS and then resuspended in 30 μl of PBS, enriched with 15 mM phosphate (pH 7.4). The bound total protein content was estimated to be around 1 μg per sample, determined via a micro BCA assay. The samples were reduced with 2 mM dithiothreitol (DTT) incubated at 50°C for 45 minutes with shaking at 700 rpm. Thereafter, proteins were alkylated with iodoacetamide (IAA) at 8 mM in the dark at room temperature. LysC/Trypsin enzyme mix was added at a concentration of 0.02 μg/μl and samples were incubated overnight at 37°C.
Samples were centrifuged at 16,000xg for 20 minutes at room temperature to pellet the NPs. The supernatant, containing the peptide digest, was collected, vacuum dried, and desalted using Pierce C18 spin tips (Thermo, 84850) following the manufacturer’s instructions. Samples were vacuum dried again and stored in the refrigerator until LC-MS analysis.
LC-MS/MS Analysis:
Dried samples were reconstituted with 1 μg of peptides in 25 μl of LC loading buffer (3% ACN, 0.1% TFA) and analyzed using LC-MS/MS. A 60-minute gradient was applied in LFQ mode, with 5 μl aliquots injected in triplicate. Control samples (55% human plasma) were prepared with 8 μg of peptides in 200 μl of loading buffer and analyzed similarly. An Ultimate 3000RSLCnano (Thermo Fisher) HPLC system was used with predefined columns, solvents, and gradient settings. Data Dependent Analysis (DDA) was performed with specific MS and MS2 scan settings, followed by data analysis using Proteome Discoverer 2.4 (Thermo Fisher), applying the protocols detailed in our earlier publication (center #9)35. The PtdChos concentration series experiment was performed using the same protocol, and the samples were analyzed over a 120 min gradient.
LC-MS analysis by DIA
The samples were centrifuged at 14,000 g for 20 min to remove the unbound proteins. The collected NP pellets were washed three times with cold PBS under the same conditions. The samples were resuspended in 20 μl of PBS, and the proteins were reduced with 2 mM DTT (final concentration) for 45 min and then alkylated using 8 mM IAA (final concentration) for 45 min in the dark. Subsequently, 5 μl of LysC at 0.02 μg/μl was added for 4h, followed by the same concentration and volume of trypsin overnight. The samples were then centrifuged at 16,000g for 20 min at room temperature to remove the NPs then cleaned using C18 cartridges and vacuum dried.
Dried peptides were resuspended in 0.1% aqueous formic acid and subjected to LC–MS/MS analysis using an Exploris 480 Mass Spectrometer fitted with an Vanquish Neo (both Thermo Fisher Scientific) and a custom-made column heater set to 60°C. Peptides were resolved using a RP-HPLC column (75 μm × 30 cm) packed in-house with C18 resin (ReproSil-Pur C18–AQ, 1.9 μm resin; Dr. Maisch GmbH) at a flow rate of 0.2 μL/min. The following gradient was used for peptide separation: from 4% B to 10% B over 7.5 min to 35% B over 67.5 min to 50% B over 15 min to 95% B over 1 min followed by 10 min at 95% B to 5% B over 1 min followed by 4 min at 5% B. Buffer A was 0.1% formic acid in water and buffer B was 80% acetonitrile, 0.1% formic acid in water.
The mass spectrometer was operated in DIA mode with a cycle time of 3 seconds. MS1 scans were acquired in the Orbitrap in centroid mode at a resolution of 120,000 FWHM (at 200 m/z), a scan range from 390 to 910 m/z, normalized AGC target set to 300 % and maximum ion injection time mode set to Auto. MS2 scans were acquired in the Orbitrap in centroid mode at a resolution of 15,000 FWHM (at 200 m/z), precursor mass range of 400 to 900, quadrupole isolation window of 7 m/z with 1 m/z window overlap, a defined first mass of 120 m/z, normalized AGC target set to 3000% and a maximum injection time of 22 ms. Peptides were fragmented by higher-energy collisional dissociation (HCD) with collision energy set to 28% and one microscan was acquired for each spectrum.
The acquired raw-files were searched individually using the Spectronaut (Biognosys v18.6) directDIA workflow against a Homo sapiens database (consisting of 20,360 protein sequences downloaded from Uniprot on 2022/02/22) and 392 commonly observed contaminants. Default settings were used.
Data analysis
First, data were normalized by total protein intensity in each technical replicate. Data analysis was performed using R project version 4.1.0. Missing values were replaced by the constant value of 10^−10 for all conditions.
Statistics and reproducibility
All measurements were performed as a triplicate analysis of a given aliquot. The DIA analysis was performed in one replicate.
Supplementary Material
Acknowledgement
MM gratefully acknowledge financial support from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (grant DK131417). A.A.S. was supported by an Ambizione Fellowship from the Swiss National Science Foundation (SNSF grant number: PZ00P3_216203) and a grant from Karolinska Institutet (2–188/2022). L.S. thanks the support from the National Cancer Institute (NCI) through the grant R01CA247863.
Footnotes
Competing interests
Morteza Mahmoudi discloses that (i) he is a co-founder and director of the Academic Parity Movement (www.paritymovement.org), a non-profit organization dedicated to addressing academic discrimination, violence and incivility; (ii) he is a co-founder of Targets’ Tip; and (iii) he receives royalties/honoraria for his published books, plenary lectures, and licensed patent.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information and data files. All relevant data are available from the corresponding authors (A.A.S. and M.M.).
References
- 1.Schwenk JM, Omenn GS, Sun Z, Campbell DS, Baker MS, Overall CM, et al. The human plasma proteome draft of 2017: building on the human plasma PeptideAtlas from mass spectrometry and complementary assays. Journal of proteome research 2017, 16(12): 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zubarev RA. The challenge of the proteome dynamic range and its implications for in‐depth proteomics. Proteomics 2013, 13(5): 723–726. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Q, Faca V, Hanash S. Mining the plasma proteome for disease applications across seven logs of protein abundance. Journal of proteome research 2011, 10(1): 46–50. [DOI] [PubMed] [Google Scholar]
- 4.Pernemalm M, Sandberg A, Zhu Y, Boekel J, Tamburro D, Schwenk JM, et al. In-depth human plasma proteome analysis captures tissue proteins and transfer of protein variants across the placenta. Elife 2019, 8: e41608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blume JE, Manning WC, Troiano G, Hornburg D, Figa M, Hesterberg L, et al. Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nature Communications 2020, 11(1): 3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu G, Zhao P, Deng N, Tao D, Sun L, Liang Z, et al. Single Chain Variable Fragment Displaying M13 Phage Library Functionalized Magnetic Microsphere-Based Protein Equalizer for Human Serum Protein Analysis. Analytical Chemistry 2012, 84(18): 7633–7637. [DOI] [PubMed] [Google Scholar]
- 7.Fonslow BR, Stein BD, Webb KJ, Xu T, Choi J, Park SK, et al. Digestion and depletion of abundant proteins improves proteomic coverage. Nature Methods 2013, 10(1): 54–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer PE, Voytik E, Treit PV, Doll S, Kleinhempel A, Niu L, et al. Plasma Proteome Profiling to detect and avoid sample‐related biases in biomarker studies. EMBO molecular medicine 2019, 11(11): e10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ignjatovic V, Geyer PE, Palaniappan KK, Chaaban JE, Omenn GS, Baker MS, et al. Mass spectrometry-based plasma proteomics: considerations from sample collection to achieving translational data. Journal of proteome research 2019, 18(12): 4085–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pattipeiluhu R, Crielaard S, Klein-Schiphorst I, Florea BI, Kros A, Campbell F. Unbiased Identification of the Liposome Protein Corona using Photoaffinity-based Chemoproteomics. ACS Central Science 2020, 6(4): 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saei AA, Beusch CM, Chernobrovkin A, Sabatier P, Zhang B, Tokat ÜG, et al. ProTargetMiner as a proteome signature library of anticancer molecules for functional discovery. Nature communications 2019, 10(1): 5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo J, Zhang Q. A Streamlined High-Throughput Plasma Proteomics Platform for Clinical Proteomics with Improved Proteome Coverage, Reproducibility, and Robustness. Journal of the American Society for Mass Spectrometry 2023, 34(4): 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viode A, van Zalm P, Smolen KK, Fatou B, Stevenson D, Jha M, et al. A simple, time- and cost-effective, high-throughput depletion strategy for deep plasma proteomics. Science Advances 2023, 9(13): eadf9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palstrøm NB, Rasmussen LM, Beck HC. Affinity capture enrichment versus affinity depletion: a comparison of strategies for increasing coverage of low-abundant human plasma proteins. International journal of molecular sciences 2020, 21(16): 5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pringels L, Broeckx V, Boonen K, Landuyt B, Schoofs L. Abundant plasma protein depletion using ammonium sulfate precipitation and Protein A affinity chromatography. Journal of Chromatography B 2018, 1089: 43–59. [DOI] [PubMed] [Google Scholar]
- 16.Hadjidemetriou M, Al-Ahmady Z, Buggio M, Swift J, Kostarelos K. A novel scavenging tool for cancer biomarker discovery based on the blood-circulating nanoparticle protein corona. Biomaterials 2019, 188: 118–129. [DOI] [PubMed] [Google Scholar]
- 17.Papafilippou L, Claxton A, Dark P, Kostarelos K, Hadjidemetriou M. Protein corona fingerprinting to differentiate sepsis from non-infectious systemic inflammation. Nanoscale 2020, 12(18): 10240–10253. [DOI] [PubMed] [Google Scholar]
- 18.Papafilippou L, Claxton A, Dark P, Kostarelos K, Hadjidemetriou M. Nanotools for sepsis diagnosis and treatment. Advanced Healthcare Materials 2021, 10(1): 2001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, et al. Physical-Chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. Journal of the American Chemical Society 2011, 133(8): 2525–2534. [DOI] [PubMed] [Google Scholar]
- 20.Caracciolo G, Safavi-Sohi R, Malekzadeh R, Poustchi H, Vasighi M, Chiozzi RZ, et al. Disease-specific protein corona sensor arrays may have disease detection capacity. Nanoscale Horizons 2019, 4(5): 1063–1076. [Google Scholar]
- 21.Hajipour MJ, Safavi-Sohi R, Sharifi S, Mahmoud N, Ashkarran AA, Voke E, et al. An Overview of Nanoparticle Protein Corona Literature. Small, n/a(n/a): 2301838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Meyer JG. 1.4 min Plasma Proteome Profiling via Nanoparticle Protein Corona and Direct Infusion Mass Spectrometry. bioRxiv 2024: 2024.2002. 2006.579213. [DOI] [PubMed]
- 23.Tang H, Zhang Y, Yang T, Wang C, Zhu Y, Qiu L, et al. Cholesterol modulates the physiological response to nanoparticles by changing the composition of protein corona. Nature Nanotechnology 2023. [DOI] [PubMed]
- 24.Mahmoudi N, Mahmoudi M. Effects of cholesterol on biomolecular corona. Nature Nanotechnology 2023. [DOI] [PubMed]
- 25.Fonda ML. Vitamin B6 metabolism and binding to proteins in the blood of alcoholic and nonalcoholic men. Alcoholism: Clinical and Experimental Research 1993, 17(6): 1171–1178. [DOI] [PubMed] [Google Scholar]
- 26.Panja S, Khatua DK, Halder M. Simultaneous Binding of Folic Acid and Methotrexate to Human Serum Albumin: Insights into the Structural Changes of Protein and the Location and Competitive Displacement of Drugs. ACS Omega 2018, 3(1): 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh R, Thomas DS, Arcot J. Molecular Recognition Patterns between Vitamin B12 and Proteins Explored through STD-NMR and In Silico Studies. Foods 2023, 12(3): 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackowski S, Alix J-H. Cloning, sequence, and expression of the pantothenate permease (panF) gene of Escherichia coli. Journal of bacteriology 1990, 172(7): 3842–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musa TL, Ioerger TR, Sacchettini JC. The Tuberculosis Structural Genomics Consortium: A Structural Genomics Approachto Drug Discovery. In: Joachimiak A (ed). Advances in Protein Chemistry and Structural Biology, vol. 77. Academic Press, 2009, pp 41–76. [DOI] [PubMed] [Google Scholar]
- 30.Adhel E, Ha Duong N-T, Vu TH, Taverna D, Ammar S, Serradji N. Interaction between carbon dots from folic acid and their cellular receptor: a qualitative physicochemical approach. Physical Chemistry Chemical Physics 2023, 25(20): 14324–14333. [DOI] [PubMed] [Google Scholar]
- 31.Lonsdale D. Thiamin and protein folding. Medical Hypotheses 2019, 129: 109252. [DOI] [PubMed] [Google Scholar]
- 32.Mkrtchyan G, Aleshin V, Parkhomenko Y, Kaehne T, Luigi Di Salvo M, Parroni A, et al. Molecular mechanisms of the non-coenzyme action of thiamin in brain: biochemical, structural and pathway analysis. Scientific Reports 2015, 5(1): 12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee DW, Park YW, Lee MY, Jeong KH, Lee JY. Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans. Scientific Reports 2020, 10(1): 21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavakol M, Montazeri A, Naghdabadi R, Hajipour MJ, Zanganeh S, Caracciolo G, et al. Disease-related metabolites affect protein–nanoparticle interactions. Nanoscale 2018, 10(15): 7108–7115. [DOI] [PubMed] [Google Scholar]
- 35.Ashkarran AA, Gharibi H, Voke E, Landry MP, Saei AA, Mahmoudi M. Measurements of heterogeneity in proteomics analysis of the nanoparticle protein corona across core facilities. Nature Communications 2022, 13(1): 6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gharibi H, Ashkarran AA, Jafari M, Voke E, Landry MP, Saei AA, et al. A uniform data processing pipeline enables harmonized nanoparticle protein corona analysis across proteomics core facilities. Nature Communications 2024, 15(1): 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nature nanotechnology 2013, 8(10): 772. [DOI] [PubMed] [Google Scholar]
- 38.Mahley RW. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. Journal of molecular medicine 2016, 94: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudi N, Mahmoudi M. Effects of cholesterol on biomolecular corona. Nature Nanotechnology 2023, 18(9): 974–976. [DOI] [PubMed] [Google Scholar]
- 40.Sheibani S, Basu K, Farnudi A, Ashkarran A, Ichikawa M, Presley J, et al. Nanoscale Characterization of the Biomolecular Corona by Cryo-Electron Microscopy, Cryo-Electron Tomography, and Image Simulation. Nature Communications 2021, 12: 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biterova EI, Isupov MN, Keegan RM, Lebedev AA, Sohail AA, Liaqat I, et al. The crystal structure of human microsomal triglyceride transfer protein. Proceedings of the National Academy of Sciences 2019, 116(35): 17251–17260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volanakis JE, Wirtz KW. Interaction of C-reactive protein with artificial phosphatidylcholine bilayers. Nature 1979, 281(5727): 155–157. [DOI] [PubMed] [Google Scholar]
- 43.Sengupta T, Manoj N. Phosphatidylserine and phosphatidylethanolamine bind to protein Z cooperatively and with equal affinity. PLoS One 2016, 11(9): e0161896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boggs JM, Rangaraj G, Dicko A. Effect of phosphorylation of phosphatidylinositol on myelin basic protein-mediated binding of actin filaments to lipid bilayers in vitro. Biochimica et Biophysica Acta (BBA) - Biomembranes 2012, 1818(9): 2217–2227. [DOI] [PubMed] [Google Scholar]
- 45.Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nature Structural Biology 2002, 9(7): 507–511. [DOI] [PubMed] [Google Scholar]
- 46.Jonas A. Interaction of phosphatidylcholine with bovine serum albumin. Specificity and properties of the complexes. Biochimica et Biophysica Acta (BBA)-Protein Structure 1976, 427(1): 325–336. [DOI] [PubMed] [Google Scholar]
- 47.Zborowski J, Roerdink F, Scherphof G. Leakage of sucrose from phosphatidylcholine liposomes induced by interaction with serum albumin. Biochimica et Biophysica Acta (BBA)-General Subjects 1977, 497(1): 183–191. [DOI] [PubMed] [Google Scholar]
- 48.Morrisett J, Jackson R, Gotto A Jr. Lipid-protein interactions in the plasma lipoproteins. Biochimica et Biophysica Acta (BBA)-Reviews on Biomembranes 1977, 472(2): 93–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information and data files. All relevant data are available from the corresponding authors (A.A.S. and M.M.).