Abstract
Recent GWASs have demonstrated that comorbid disorders share genetic liabilities. But whether and how these shared liabilities can be used for the classification and differentiation of comorbid disorders remains unclear. In this study, we use polygenic risk scores (PRSs) estimated from 42 comorbid traits and the deep neural networks (DNN) architecture to classify and differentiate schizophrenia (SCZ), bipolar disorder (BIP) and major depressive disorder (MDD). Multiple PRSs were obtained for individuals from the schizophrenia (SCZ) (cases = 6,317, controls = 7,240), bipolar disorder (BIP) (cases = 2,634, controls 4,425) and major depressive disorder (MDD) (cases = 1,704, controls = 3,357) datasets, and classification models were constructed with and without the inclusion of PRSs of the target (SCZ, BIP or MDD). Models with the inclusion of target PRSs performed well as expected. Surprisingly, we found that SCZ could be classified with only the PRSs from 35 comorbid traits (not including the target SCZ and directly related traits) (accuracy 0.760 ± 0.007, AUC 0.843 ± 0.005). Similar results were obtained for BIP (33 traits, accuracy 0.768 ± 0.007, AUC 0.848 ± 0.009), and MDD (36 traits, accuracy 0.794 ± 0.010, AUC 0.869 ± 0.004). Furthermore, these PRSs from comorbid traits alone could effectively differentiate unaffected controls, SCZ, BIP, and MDD patients (average categorical accuracy 0.861 ± 0.003, average AUC 0.961 ± 0.041). These results suggest that the shared liabilities from comorbid traits alone may be sufficient to classify SCZ, BIP and MDD. More importantly, these results imply that a data-driven and objective diagnosis and differentiation of SCZ, BIP and MDD may be feasible.
Introduction
It is well known in psychiatry that comorbidities and overlapping phenomenology are common between different disorders, some symptoms are observed in multiple disorders, including physical diseases and behavioral traits.1,2 Over the years of genome wide association studies (GWASs), it has been clear that many of these comorbid disorders and traits share genetic liabilities.3–6 These shared liabilities suggest that not all genetic variants identified by a GWAS are specific to the disease, but it remains unclear to what extent that the liabilities shared with comorbid conditions account for the liability specific to the disease. From a clinical perspective, comorbidity increases the difficulties and challenges in disease diagnosis and treatment. Major psychiatric disorders such as schizophrenia (SCZ), bipolar disorder (BIP), and major depressive disorder (MDD) are often “misdiagnosed,” especially in the early stage of the disorders when typical symptoms have not been fully manifested. Understanding the genetic architecture of comorbid conditions could provide new insights into the underlying mechanisms and open new windows for a data driven, biology-based diagnosis and separation of these comorbid conditions.
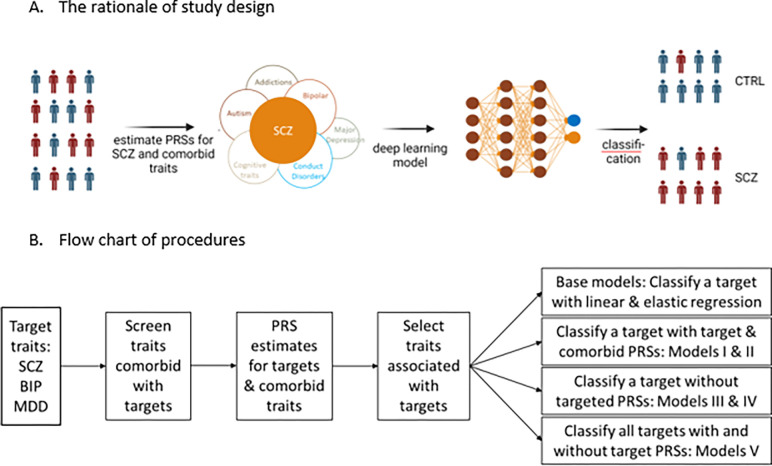
Conceptually, for an individual, we can consider that his/her genetic liability to a psychiatric disorder consists of a core of liability specific to the disorder and a peripheral shell made of shared liabilities from comorbid disorders and traits. For a given individual, the shared patterns and extents in the peripheral shell can be different from another individual. Therefore, we may be able to take advantage of these differences between people to separate affected individuals from unaffected individuals. Figure 1A illustrates this concept using SCZ as an example. Since polygenic risk score (PRS) has been established as a reliable approximation of genetic liability,7,8 we can consider that the total genetic liability of a disorder for a given individual is the sum of PRS of the targeted disorder and the PRSs of all other comorbid disorders and traits. Similarly, we can extend the concept of overlapping genetic liabilities to several disorders with common clinical symptoms. Due to the differences in the degree of overlaps among these disorders, we can utilize these differences to distinguish major psychiatric disorders that have substantial overlaps in both genetic liabilities and clinical symptoms. Based on these rationales, for a specific disorder, we can construct a classification model that integrates the PRS of targeted disorder with the PRSs of all other comorbid disorders and traits, and this model may have a better performance than the model that uses the PRS of the targeted disorder alone. Similarly, we can also build models with these PRSs to differentiate several different but symptomatically overlapped disorders.
Figure 1.
The rationale and study design. A. The rationale. Classification of a target trait, SCZ is used as an example. Conceptually, we can consider that the genetic liability of SCZ consists of a core of genetic factors that are specific to SCZ and a varying number of genetic factors from comorbid disorders and traits. The extent and variation of the sharing of genetic risks between individuals are explored with a deep neural network model to classify SCZ. B. A flow chart of procedures used in the study.
We have performed this study as a demonstration of these principles. We searched diseases and traits that are comorbid with SCZ, BIP, and MDD from the literature, and matched these traits with those reported in the GWAS catalog 9,10 (https://www.ebi.ac.uk/gwas/). We then calculated the PRSs for SCZ, BIP, MDD, and the comorbid traits, and evaluated their genetic correlations. We selected those traits with PRSs statistically associated with the targeted disorders (SCZ, BIP, and MDD), and constructed deep neural networks (DNN) models with these PRSs to evaluate their utilities in the classification and differentiation of the targeted disorders (Figure 1B). The results obtained from the models could help us understand the genetic architecture of comorbid conditions and provide strategies for biology-based diagnosis and distinction of these comorbid conditions.
METHODS
3.1. Datasets and genotype imputation
In this study, we used datasets obtained from multiple sources. For SCZ datasets, we used the Molecular Genetics of Schizophrenia (MGS) 11 (accession phs000167.v1.p1, cases = 2,681, controls = 2,653) and the Swedish Case-Control Study of Schizophrenia (SCCSS) 12 ( accession phs000473.v2.p2, cases = 2,895, controls = 3,836) from dbGaP (https://www.ncbi.nlm.nih.gov/gap/), and the Clinical Antipsychotic Trials of Intervention Effectiveness 13,14 (CATIE, cases = 741, controls =751) from NIMH’s genetic repository (https://www.nimhgenetics.org/). These SCZ datasets were combined with PLINK software.15,16 For BIP datasets, we used samples from the Psychiatric Genomics Consortium (PGC) (TOP3, cases = 203, controls = 349; DUB, cases = 150, controls = 797; EDI, cases = 282 controls = 275) and the Wellcome Trust Case Control Consortium (https://www.wtccc.org.uk/) (cases = 1998, controls = 3004). The BIP datasets were also combined as a single sample using PLINK. The MDD data was obtained from dbGaP, accession phs000486.v1.p1, with 1,704 cases and 3,357 controls.17,18 For all datasets, we obtained the genotype and phenotype information from the sources, conducted genotype quality assessments, and removed SNPs with minor frequency less than 0.01 and Hardy-Weinberg equilibrium p-value less than 0.0001. We then conducted genotype imputations for each dataset separately using the Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html#!) using the 1000 Genomes Phase 3 as reference and default parameters. After the imputation, SNPs with INFO value less than 0.4 were removed. For analyses that required combining the datasets (i.e., main models V), we used PLINK to merge the datasets. For phenotypes, we used the same definitions as defined in the original studies.
3.2. Selection of comorbid traits and PRS calculation
We reviewed literature on comorbid traits of psychiatric disorders and searched the GWAS catalog9,10to find whether the traits had GWAS performed. If a GWAS was found, the summary statistics would be downloaded. Of note, a trait here was defined as a phenotype that GWAS was performed for. For a psychiatric disorder, if multiple related phenotypes were used for GWAS, all of these phenotypes were considered individual traits and were included in our study. With the summary statistics of GWASs, we used the default settings of PRSice2 package 19,20to calculate the PRSs for the best-fit p-value threshold (bestPRS, hereafter) for the SCZ, BIP, and MDD samples for each candidate trait. For a candidate trait, if the association p-value of the bestPRS with any one of our targeted disorders was ≤ 0.050, we would include it in our study. With this procedure, a total of 42 traits were obtained (supplementary Table S1). For all included traits, we then calculated PRSs at six p-value thresholds (5e-8, 1e-5, 1e-3, 0.1, 1 and the best-fit p-value) for all subjects of the datasets used in this study. For all PRSs, we rescaled them to the range between 0 and 1 and stored them as individual by feature matrices for model inputs.
3.3. Model definition, training, and optimization
We used 6 main models in this study. For convenience, we used this convention to name our models: main_model.submodel.target_trait. If the main models did not have submodels, then they would be named as main_model.target_trait. The details of models were listed in Table 1. We used logistic and elastic regressions to establish a baseline (main model B) to compare to DNN models. For the elastic models, we used a grid search to find the optimal alpha value and L1 ratio (alpha of 0.010 and L1 ratio of 0.920) for submodels B.II and B.III.
Table 1.
Model information*
| Main Model | Submodel | Target | Structure | Traits (# Predictors) |
|---|---|---|---|---|
| Baseline | I | SCZ; BIP; MDD | logistic regression | target bestPRS (1) |
| Baseline | II | SCZ; BIP; MDD | elastic regression | all bestPRSs (42) |
| Baseline | III | SCZ; BIP; MDD | elastic regression | exclusion of target bestPRSs, SCZ (35); BIP (33); MDD (36) |
| I | SCZ; BIP; MDD | deep neural network | all PRSs, SCZ (237); BIP (237); MDD (237) | |
| II | SCZ; BIP; MDD | deep neural network | all bestPRSs, SCZ (42), BIP (42); MDD (42) | |
| III | SCZ; BIP; MDD | deep neural network | exclusion of target PRSs, SCZ (196); BIP (184); MDD (202) | |
| IV | SCZ; BIP; MDD | deep neural network | exclusion of target bestPRSs, SCZ (35); BIP (33); MDD (36) | |
| V | I | CTRL, SCZ, BIP and MDD | deep neural network | all PRSs (237) |
| V | II | CTRL, SCZ, BIP and MDD | deep neural network | all bestPRSs (42) |
| V | III | CTRL, SCZ, BIP and MDD | deep neural network | exclusion of target PRSs (166) |
| V | IV | CTRL, SCZ, BIP and MDD | deep neural network | exclusion of target bestPRSs (30) |
Model nomenclature: Main_model.submodel.target_disorder. If a main model does not have a submodel, then main_model.target_disorder. For example, B.I.SCZ, I.SCZ and V.I.4C.
Main models I to IV were binary models designed to classify the target disorders using various PRS combinations. Main model V was a multiclass model, and we used it to classify the 4 classes of CTRL, SCZ, BIP and MDD by combining the datasets from the 3 targeted diseases together. To account for batch effects among the 3 datasets, we conducted batch correction with the pyComBat package.21,22 Since the number of subjects in each class was substantially different, the combined dataset from the 3 diseases was imbalanced for the 4 classes, therefore, we used oversampling techniques (ADASYN 23 and borderline SMOTE 24,25) to balance the classes and train the models.
For models that needed to remove targeted disease and related phenotypes, i.e., models B.III, III, IV, V.III, and V.IV (see model definitions below), PRSs obtained for these phenotypes would be removed from the models (see supplementary Table S1, columns 5–8). For models that used multiple levels of PRSs for the same traits, i.e., models III and V.III, when a trait was removed from the model, all levels of PRS of that trait would be removed.
W used the TensorFlow (version 2.5.0; www.tensorflow.org/),26,27 keras (version 2.9.0; https://keras.io/api/) and DNN architecture to construct the models. An example of the models (model II.SCZ) was shown in supplementary Figure S1, and the Python scripts for main models could be found in our Github site (https://github.com/mdsamchen/scz_bip_mdd). For each model, we used the leave one out cross validation (LOOCV) procedure to conduct validation. All results reported were obtained from the 20% left-out samples that were not seen by any of the models during the training processes. For binary classification (main models B, I to IV), we reported the binary accuracy, precision, recall, F1 score, and AUC as defined in the scikit-learn package (version 0.23.2).28 For multiclass model (i.e., model V), we reported the average of categorical accuracies from all classes, and the AUCs were also averaged across the classes. The precision, recall, and F1 score were reported for each class.
3.4. Evaluation of feature importance
For DL models, permutations had been used as a method to evaluate the importance of the features.29 We use permutations to estimate the importance of the features by the following procedures: a). define the feature importance as the change of model performance. For binary classification models, we used r2 score, as defined in the scikit-learn package, as the measurement of model performance. For multiclass classification models, we used class weighted average of AUC as the measurement. To estimate the feature importance, we permuted each feature 100 times for the trained model and took the average of these 100 permutations as the performance of the permuted feature. The importance of the feature was the difference between the performance of the trained model and the permuted feature:
Where is the importance of feature is the performance score (r2 score for binary model and AUC for multiclass model) of the trained model, is the number of permutations. We used one sample t-test to evaluate whether the change was statistically significant assuming that the performance changes from permutations followed normal distribution. The changes of the permuted performances were plotted using Seaborn (version 0.12.2) 30 and Matplot (version 3.5.2; https://www.mathworks.com/help/stats/index.html) libraries.
3.5. UMAP plotting of model embeddings
Model embeddings were extracted from the layer immediately before the classification layer. For direct comparison between the models with and without the inclusion of target specific PRSs, all models had 32 dimensions at this embedding layer. Then the embeddings were projected into a 2-dimensional space by umap-learn (version 0.5.4) 31 and plotted by classes with the Matplot library.
RESULTS
4.1. Selection of comorbid traits
Based on the survey of the literature 3,32–35 and our test of association, we selected a total of 42 diseases/traits in this study, including the targeted disorders (see Supplementary Table S1). From Table S1, it was clear that the associations between the targeted disorders and comorbid PRSs vary substantially. All selected traits are associated with at least one of our targeted disorders, i.e., SCZ, BIP, or MDD.
4.2. Comparison between baseline models with and without the inclusion of PRSs from target disorder and comorbid traits
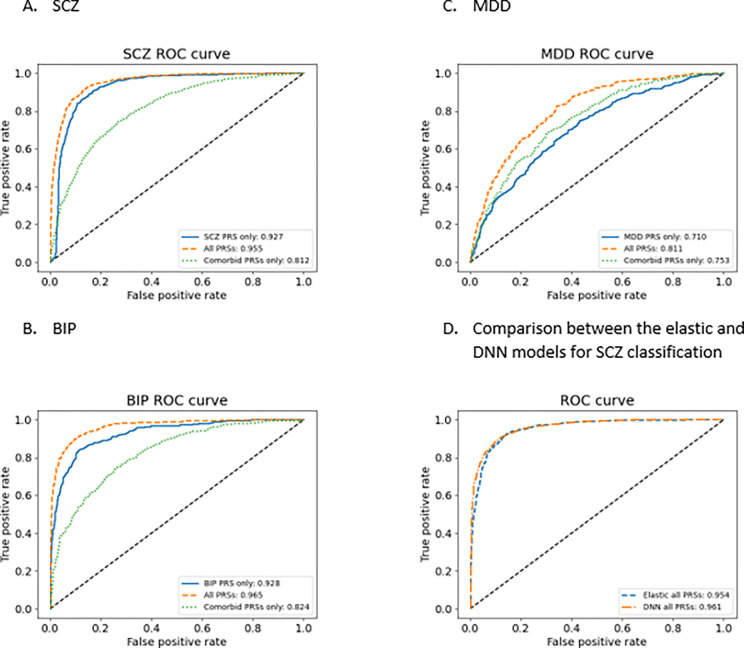
We started our study by building models using the bestPRSs and sex to classify targeted disorders and treated them as the baseline models for comparison with all other models. For each targeted disorder, there were 3 baseline models, B.I, B.II, and B.III, which used targeted bestPRS, all best PRSs, and all bestPRSs but targeted bestPRS as predictors respectively (Table 1). The performances of these baseline models for SCZ were summarized in Tables S2. Inclusion of PRSs from comorbid traits did improve model performance (AUC with about 2.8% improvement) (Figure 2A, Table S2), but the improvement was modest. Similar improvements were also observed in class specific precision, recall, and F1-score (Table S2, models B.I.SCZ and B.II.SCZ). Model B.III used only the PRSs from comorbid traits, and surprisingly, model B.III.SCZ achieved an accuracy of 0.734 ± 0.001 and AUC of 0.812 ± 0.001. While the performance of model B.III.SCZ was worse (Figure 2A, Table S2) than that of models B.I.SCZ and B.II.SCZ, the results were intriguing given the fact that no bestPRSs of SCZ and directly related traits were included in model B.III.SCZ.
Figure 2.
Base model performances for SCZ (A), BIP (B) and MDD (C). The plots show the ROC curves for the baseline models that use only the bestPRSs from the targeted disorders (blue, Model B.I), bestPRSs from all traits (targeted disorders and comorbid traits, gold, Model B.II), and bestPRSs from only the comorbid traits (green, Model B.III). The results indicate that models with the inclusion of PRSs from comorbid traits have better performances than that of using only the PRSs from targeted traits. It is also clear that models using only the PRSs from comorbid traits can also have decent performance. Panel D is a comparison between the baseline elastic model (Model B.II.SCZ) and DNN for SCZ classification (Model II.SCZ).
For BIP, we observed a similar trend as observed in SCZ, i.e., model B.II.BIP performance > model B.I.BIP performance > model B.III.BIP performance (Figure 2B, Table S3). But for MDD, model B.III.MDD had a better performance than model B.I.MDD (Figure 2C, Table S4). Overall, these results indicated that inclusion of PRSs from comorbid traits improved model performances, and the use of only the PRSs of comorbid traits could predict disease status for SCZ, BIP, and MDD.
We evaluated the performances between the elastic regression models and DNN models using the bestPRS dataset of SCZ (comparison between models B.II.SCZ and II.SCZ), and the two models performed virtually the same (Figure 2D).
4.3. Classification of SCZ with DNN models
We constructed 4 DNN models to classify SCZ diagnosis with the PRSs obtained from the selected traits. For all models, sex was included as a predictor. For model I.SCZ, using a 5-fold LOOCV scheme, we obtained accuracy and AUC for the left-out samples of 0.913 ± 0.004 and 0.974 ± 0.002, respectively. The class specific precision, recall, and F1-score for SCZ were 0.915 ± 0.004, 0.912 ± 0.005, and 0.913 ± 0.005, respectively (Table 2, model I.SCZ). Please note that while there were some differences between SCZ and CTRL for class specific precision, recall, and F1-score matrix, the results for the two classes were comparable.
Table 2.
Classification of schizophrenia
| Model | Class | Accuracy | AUC | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|
| I.SCZ | CTRL | 0.913 ± 0.004 | 0.974 ± 0.002 | 0.912 ± 0.005 | 0.915 ± 0.004 | 0.914 ± 0.004 |
| SCZ | 0.915 ± 0.004 | 0.912 ± 0.005 | 0.913 ± 0.005 | |||
| II.SCZ | CTRL | 0.880 ± 0.005 | 0.956 ± 0.003 | 0.884 ± 0.004 | 0.875 ± 0.009 | 0.879 ± 0.006 |
| SCZ | 0.876 ± 0.008 | 0.885 ± 0.003 | 0.881 ± 0.006 | |||
| III.SCZ | CTRL | 0.760 ± 0.007 | 0.843 ± 0.005 | 0.740 ± 0.014 | 0.802 ± 0.014 | 0.770 ± 0.006 |
| SCZ | 0.784 ± 0.009 | 0.719 ± 0.023 | 0.749 ± 0.012 | |||
| IV.SCZ | CTRL | 0.710 ± 0.008 | 0.789 ± 0.011 | 0.713 ± 0.014 | 0.702 ± 0.005 | 0.707 ± 0.007 |
| SCZ | 0.706 ± 0.005 | 0.718 ± 0.020 | 0.712 ± 0.012 |
For model II.SCZ, we built a DNN model (Figure S1) and used the same LOOCV procedures for model training and validation. The performance was very close to that of model I.SCZ, the accuracy and AUC were 0.880 ± 0.005 and 0.956 ± 0.003, respectively (Table 2, model II.SCZ). The SCZ specific precision, recall, and F1-score were close to those of model I.SCZ as well. Of note, model II.SCZ (Table 2) and model B.II.SCZ (Table S2) used exactly the same predictors, the two models performed virtually the same for accuracy, AUC, and class specific precision, recall, and F1-score.
For model III.SCZ, after removing all PRSs of the traits that included SCZ phenotypes directly in their prospective GWASs, we were surprised to find that the model performed reasonably well, with a validation accuracy of 0.760 ± 0.007 and a validation AUC of 0.843 ± 0.005 (Table 2, model III.SCZ). The SCZ specific precision, recall, and F1-score were 0.784 ± 0.009, 0.719 ± 0.023, and 0.749 ± 0.012. Although the accuracy and AUC, and the class specific matrices were substantially lower than that of model I.SCZ (Table 2, comparing models I.SCZ and III.SCZ), the results remained significant.
For model IV.SCZ, the accuracy and AUC were significantly lower than that of model II.SCZ, but the results were decent, with validation accuracy of 0.710 ± 0.008 and validation AUC of 0.789 ± 0.011 (Table 2, model IV.SCZ vs. model II.SCZ). Similar trends were also observed for the class specific matrices.
4.4. Classification of BIP and MDD with DNN models
We pursued similar strategies for the classification of BIP and MDD with DNN models. The results for BIP were summarized in Table 3, which closely mirrored those of SCZ reported in Table 2 except that models III.BIP and IV.BIP had virtually the same performance.
Table 3.
Classification of bipolar disorder
| Model | Class | Accuracy | AUC | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|
| I.BIP | CTRL | 0.895 ± 0.020 | 0.965 ± 0.003 | 0.904 ± 0.066 | 0.892 ± 0.056 | 0.894 ± 0.018 |
| BIP | 0.896 ± 0.040 | 0.900 ± 0.084 | 0.894 ± 0.031 | |||
| II.BIP | CTRL | 0.904 ± 0.014 | 0.965 ± 0.001 | 0.924 ± 0.027 | 0.883 ± 0.056 | 0.901 ± 0.021 |
| BIP | 0.890 ± 0.042 | 0.925 ± 0.030 | 0.906 ± 0.012 | |||
| III.BIP | CTRL | 0.768 ± 0.007 | 0.848 ± 0.009 | 0.760 ± 0.013 | 0.782 ± 0.016 | 0.771 ± 0.007 |
| BIP | 0.775 ± 0.010 | 0.752 ± 0.021 | 0.764 ± 0.010 | |||
| IV.BIP | CTRL | 0.782 ± 0.006 | 0.852 ± 0.004 | 0.787 ± 0.005 | 0.770 ± 0.011 | 0.778 ± 0.008 |
| BIP | 0.775 ± 0.009 | 0.792 ± 0.004 | 0.783 ± 0.006 |
For MDD, while there were some differences among the 4 models, the performances were generally on a par with one another, including class specific matrices (Table 4). This was different from the trend observed from both SCZ and BIP. Additionally, the overall performance of MDD models was worse than that of SCZ and BIP. This could be due to the difference in heritability between these disorders.
Table 4.
Classification of major depressive disorder
| Model | Class | Accuracy | AUC | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|
| I.MDD | CTRL | 0.782 ± 0.015 | 0.854 ± 0.010 | 0.784 ± 0.046 | 0.785 ± 0.053 | 0.782 ± 0.007 |
| MDD | 0.787 ± 0.029 | 0.778 ± 0.083 | 0.779 ± 0.034 | |||
| II.MDD | CTRL | 0.782 ± 0.004 | 0.848 ± 0.007 | 0.794 ± 0.020 | 0.763 ± 0.030 | 0.778 ± 0.007 |
| MDD | 0.773 ± 0.017 | 0.801 ± 0.033 | 0.786 ± 0.010 | |||
| III.MDD | CTRL | 0.794 ± 0.010 | 0.869 ± 0.004 | 0.800 ± 0.017 | 0.767 ± 0.025 | 0.783 ± 0.016 |
| MDD | 0.783 ± 0.018 | 0.813 ± 0.021 | 0.797 ± 0.013 | |||
| IV.MDD | CTRL | 0.753 ± 0.019 | 0.822 ± 0.010 | 0.739 ± 0.047 | 0.793 ± 0.057 | 0.762 ± 0.011 |
| MDD | 0.779 ± 0.031 | 0.712 ± 0.093 | 0.740 ± 0.043 |
4.5. Classification of control, SCZ, BIP, and MDD
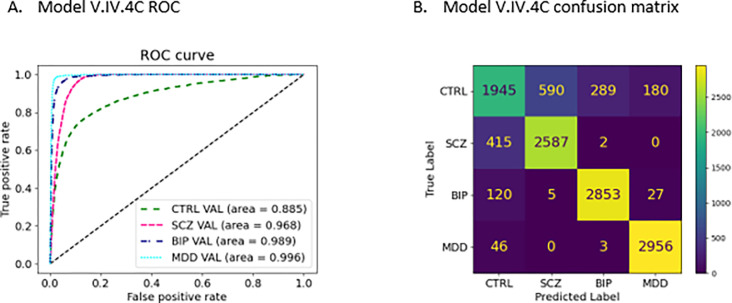
We built a multiclass model, model V, to classify and differentiate the 4 classes (CTRL, SCZ, BIP and MDD). The results were summarized in Table 5. Among the models, model V.I.4C had a similar performance as model V.II.4C and model V.III.4C had a similar performance as model V.IV.4C. For example, the average AUC of model V.I.4C (0.986 ± 0.015) was very close to that of model V.II.4C (0.990 ± 0.010), and so forth (Table 4). As expected, models with the inclusion of PRSs from targeted diseases, i.e., models V.I.4C and V.II.4C, had better performances than that of models without PRSs from targeted diseases (models V.III.4C and V.IV.4C). For all models, the class specific measures for the CTRL were the worst (Table 5, Figure 3A). When we examined the confusion matrices more carefully, it was apparent that most of the misclassifications occurred with the controls (Figure 3B).
Table 5.
Classification of controls and patients diagnosed with schizophrenia, bipolar and major depression
| Model | Class | Accuracy* | AUC** | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|
| V.I.4C | CTRL | 0.911 ± 0.009 | 0.986 ± 0.015 | 0.856 ± 0.076 | 0.821 ± 0.065 | 0.834 ± 0.008 |
| SCZ | 0.938 ± 0.025 | 0.942 ± 0.044 | 0.939 ± 0.012 | |||
| BIP | 0.954 ± 0.022 | 0.932 ± 0.054 | 0.941 ± 0.023 | |||
| MDD | 0.907 ± 0.026 | 0.948 ± 0.041 | 0.926 ± 0.009 | |||
| V.M.4C | CTRL | 0.938 ± 0.003 | 0.990 ± 0.010 | 0.914 ± 0.024 | 0.841 ± 0.029 | 0.875 ± 0.008 |
| SCZ | 0.942 ± 0.007 | 0.962 ± 0.013 | 0.952 ± 0.003 | |||
| BIP | 0.959 ± 0.015 | 0.972 ± 0.010 | 0.965 ± 0.004 | |||
| MDD | 0.936 ± 0.015 | 0.977 ± 0.006 | 0.956 ± 0.005 | |||
| V.NI.4C | CTRL | 0.850 ± 0.017 | 0.960 ± 0.041 | 0.733 ± 0.064 | 0.659 ± 0.046 | 0.690 ± 0.016 |
| SCZ | 0.823 ± 0.013 | 0.857 ± 0.057 | 0.838 ± 0.023 | |||
| BIP | 0.902 ± 0.011 | 0.927 ± 0.039 | 0.914 ± 0.016 | |||
| MDD | 0.938 ± 0.006 | 0.956 ± 0.018 | 0.947 ± 0.010 | |||
| V.IV.4C | CTRL | 0.861 ± 0.003 | 0.961 ± 0.041 | 0.768 ± 0.013 | 0.655 ± 0.029 | 0.706 ± 0.013 |
| SCZ | 0.825 ± 0.009 | 0.868 ± 0.020 | 0.846 ± 0.007 | |||
| BIP | 0.893 ± 0.011 | 0.944 ± 0.004 | 0.918 ± 0.005 | |||
| MDD | 0.941 ± 0.008 | 0.976 ± 0.005 | 0.959 ± 0.003 |
average categorical accuracy from all classes
average AUC from all classes
Figure 3.
The ROCs and Confusion matrices for Models V. The worst performance (A), and the confusion matrix showed that misclassifications largely came from the CTRL group (B), the classification of the other 3 groups were good.
4.6. Evaluation of contribution of comorbid traits to SCZ, BIP and MDD
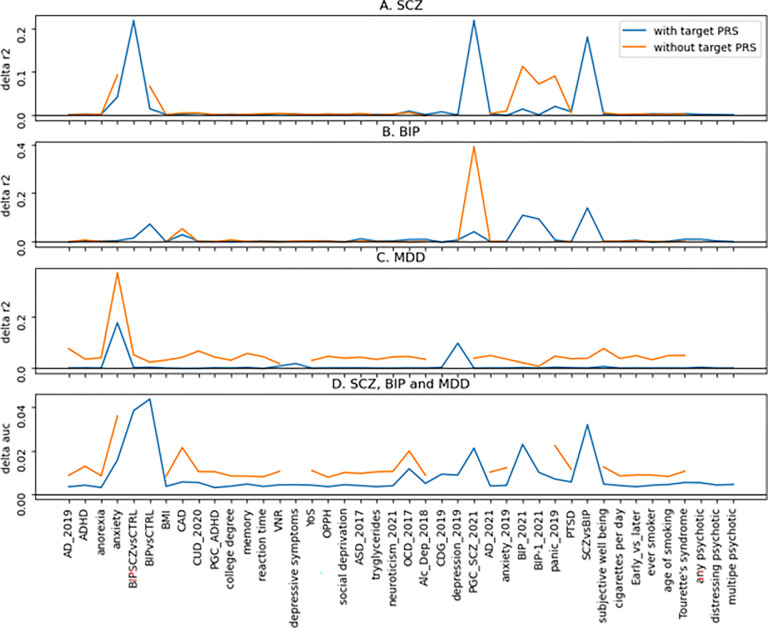
Based on the results from the models that did not include the PRSs from the target and directly related traits, PRSs from comorbid traits could substitute for the target PRSs, but we did not know what comorbid traits were involved. To answer this question, we implemented a permutation procedure to evaluate the importance of the features in the models. Figure 4 shows the results as measured by the changes of r2 score (delta r2) and AUC (delta auc) between models II and IV for SCZ, BIP and MDD, and between models V.II.4C and V.IV.4C. From the first panel, the SCZ panel, for model II.SCZ, 3 predictors/traits, i.e., BIP and SCZ vs CTRL, PGC_SCZ_2021, and SCZ vs BIP, made the most contribution to the model (Figure 5A). When these traits were removed from the model, i.e., model IV.SCZ, anxiety, BIP versus CTRL, BIP_2021, BIP-1_2021 and panic_2019 became the most important contributors of the model. In other words, these 5 traits could largely account for the effects of SCZ specific traits in model II.SCZ. Similarly, for BIP and MDD (Figure 4B and 4C), PGC_SCZ_2021 and anxiety could largely account for the effects of BIP- and MDD-specific traits respectively. For the multiclass classification model V.II.4C, in addition to those traits directly involved in SCZ, BIP and MDD (BIP and SCZ vs CTRL, BIP vs CTRL, depression_2019, PGC_SCZ_2021, BIP_2021 and SCZ vs BIP), anxiety and OCD_2017 also made substantial contributions to the model. After removing target specific PRSs, anxiety, CAD (coronary artery disease), OCD_2017, anxiety_2019 and panic_2019 became the major players in model V.IV.4C. Intriguingly, a physical disease, coronary artery disease, became a prominent contributor in the classification and differentiation of CTRL, SCZ, BIP and MDD.
Figure 4.
Comparison of feature importance between models with and without the inclusion of the targeted PRSs. Permutation based feature importance estimates, as measured by delta R2 scores and delta AUC, were plotted for the two models. The features not included in the models were replaced with NA, and plotted as broken lines. A. SCZ, models II.SCZ and IV.SCZ. B. BIP, models II.BIP and IV.BIP. C. MDD, models II.MDD and IV.MDD. D. SCZ, BIP, and MDD, models V.II.4C and V.IV.4C.
Figure 5.

Comparisons of the embedding structures between models with and without the inclusion of targeted PRSs. The embedding layers immediately before the classification layer of the models were extracted and projected into a 2-dimension space using the UMAP method. Panels on the left side were from models with the inclusion of targeted PRSs (II.SCZ, II.BIP, II.MDD and V.II.4C), and panels on the right side were from models without the inclusion of targeted PRSs (IV.SCZ, IV.BIP, IV.MDD and V.IV.4C). For binary classification models, there were no apparent differences in cluster structures. For the multiclass models, model V.IV.4C did not have a CTRL cluster (green), and BIP became the connecting cluster between the SCZ and MDD clusters.
We also examined the embedding projections of these models to see whether there were significant differences in cluster structures. The results are presented in Figure 5. For the binary classification models, with or without the use of target specific PRSs, the models had similar embedding structures. For example, for models II.SCZ and IV.SCZ, shown as the top two panels in Figure 5, while there were significant differences in model performance, i.e., classification accuracy, there was no apparent difference in cluster structure and were no apparent gaps in embedding projections. The difference between models V.II.4C and V.IV.4C, the multiclass classification models, was the disappearance of the CTRL group, and BIP became the group connecting the SCZ and MDD groups.
DISCUSSION
In this study, we used PRSs from multiple comorbid traits to classify SCZ, BIP and MDD. Our study shows that PRSs from both target traits and comorbid traits can consistently predict the disease status of the targets. The results from models I and II might be inflated because the samples we used are part of the GWASs for SCZ, BIP, and MDD. The original reason we designed models III and IV was to address the sample independence issue. We reasoned that if we excluded all PRSs obtained from the targeted disorder and directly related phenotypes, we could establish the lower bound of the model performance. The results we observed exceeded our expectations and were exciting. Based on our reading of the literature, there is no report that PRSs from comorbid traits alone can classify major psychiatric disorders, such as SCZ, BIP, and MDD. Although we 36 and others 37,38 have reported that inclusion of comorbid traits could improve the classification of targeted disorders, but there are no reports of classification that exclude the PRSs of targeted disorders.
Our study has two major findings. One is that the PRSs from the comorbid traits alone, i.e., without the inclusion of PRSs of targeted diseases, can classify the targeted diseases, and these models perform reasonably well. This observation is true for the 3 psychiatric disorders studied in this article. This finding is consistent with recent reports that major psychiatric disorders share significant genetic liabilities,3,4,39 suggesting that many of the risk alleles found in disorder specific GWASs may be the same alleles found in a different GWAS. Our study explicitly shows that a combination of PRSs from comorbid traits can replace the target specific PRSs to predict the disease status of the targets, and we can quantify the effects of target specific risks by comparing the performance between models II and IV. Furthermore, by examining the feature importance of these models, we can find which traits can be used to replace the targets in perspective models. In the case of BIP, PRSs from coronary artery disease (CAD) and SCZ can substitute the BIP specific PRSs (Figure 4B).
This finding raises an interesting question, that is how many genetic risk variants found in a GWAS are specific to the target disease, say, SCZ? In this study, we used a total of 42 traits, and the 35 traits not directly related to SCZ could predict SCZ status reasonably well. From the feature importance analyses, anxiety, BIP and panic can largely compensate the effects of SCZ specific PRSs (Figure 4A). The differences in model performance between model II.SCZ and IV.SCZ were 0.170 (accuracy) and 0.147 (AUC) (Table 2). The differences between II.BIP and IV.BIP and between II.MDD and IV.MDD were even smaller. Since the comorbid traits used in our models were far from exhaustive, other potential traits could be included. For examples, breast cancer,40 migraine41 and amyotrophic lateral sclerosis42 have been reported to have genetic correlation with SCZ, they could be good candidates to expand our list of comorbid traits. Should more genetically correlated traits be included, we would reasonably expect that the gap between models II.SCZ and IV.SCZ would decrease, i.e., the number of variants specific to SCZ would decrease, and this observation can be extended to BIP and MDD as well. In other words, the risk variants specific to SCZ, BIP or MDD are limited, most variants found in disease specific GWAS are shared between comorbid traits. The consequence is that, if we believe that the main roles of genetic risks on SCZ are disruption of normal biological process/functions and our finding that many, perhaps, a majority, of the genetic risks are not specific to SCZ, it would lead to a conclusion that unless we know that a drug is designed to target variants specific to SCZ, the drug is unlikely to have a specific effect to SCZ, and it is more likely to be interchangeable among multiple disorders. This is consistent with current clinical practice that several drugs are exchangeable in treating SCZ, BIP and MDD because they all target the same or similar mechanisms.43
Another potentially important finding is that with PRSs from multiple comorbid traits, we can effectively differentiate CTRL, SCZ, BIP, and MDD. The results remained significant with or without the inclusion of the PRSs of targeted diseases (Tables 5). When we examine the results more carefully, we find that the CTRL group has the lowest AUC and significant misclassifications (Fig. 3A and 3B), leading to lower precision, recall and F1-score for this group. A possible reason could be that the controls used in the datasets were not likely super controls who did not have any symptoms or diagnoses for all comorbid traits included in this study. The PRSs from comorbid traits help to differentiate each patient group (SCZ, BIP, or MDD), but they may blur the line between controls and case groups (SCZ, BIP, or MDD). This is because controls in the SCZ, BIP, and MDD datasets might not have been screened against all the comorbid traits, such as years of school attended ,44 body mass index,45 and memory and neural function measures,46 traits for which GWAS based PRSs were used in the models.
Our study has some limitations. One is potential overlap of controls between the targeted GWASs (i.e., SCZ, BIP, and MDD) and the GWASs of the traits included in our study. Some GWASs use consortium data that include control subjects from multiple sources. While these overlapping control subjects may not impact these separate GWASs, it might lead to some dependences between our targeted disorders and those comorbid traits if their GWASs share some control subjects. Since our study used a substantial number of GWASs, it was difficult for us to know whether and to what extent these GWASs have overlapping controls. Therefore, it would be difficult to estimate their impact on our model’s performance. Independent studies may be needed to validate our findings. Another potential issue is the differentiation of the targeted disorders. The samples for SCZ, BIP, and MDD studies come from different sources, even though we conducted batch effect correction before using the data in our model, it may not be sufficient to account for the stratification, leading to inflated performance. Further studies with the same genotype platform and samples coming from the same populations are necessary to validate our findings.
In summary, we find that without the use of PRSs of targeted disorders, we can predict the status of these disorders. These results suggest that most genetic risk variants found by GWASs may not be specific to the disorders. Furthermore, PRSs from comorbid conditions, with or without the inclusion of PRSs from targeted disorders, can be used to differentiate the targeted disorders, indicating that it may be feasible to obtain a data-driven and biology-based diagnosis for these disorders.
Acknowledgements
This work was supported in part by NIH grant P20GM121325 and R01LM012806.
The genetic and clinical data for the MGS, SCCSS, and CATIE studies were obtained from the Genomics Repository of National Institute of Mental Health (https://www.nimhgenetics.org/). We thank the patients, control subjects, and the investigators involved in these studies. The investigators and co-investigators for the MGS were: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V. Gejman, M.D. (Collaboration Coordinator; PI), Alan R. Sanders, M.D.; Emory University School of Medicine, Atlanta, GA, MH59587, Farooq Amin, M.D. (PI); Louisiana State University Health Sciences Center; New Orleans, LA, MH067257, Nancy Buccola APRN, B.C., M.S.N. (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, M.D. (PI); Washington University, St. Louis, MO, U01, MH060879, C. Robert Cloninger, M.D. (PI); University of Iowa, Iowa, IA, MH59566, Raymond Crowe, M.D. (PI), Donald Black, M.D.; University of Colorado, Denver, CO, MH059565, Robert Freedman, M.D. (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson, M.D. (PI); University of Queensland, QLD, Australia, MH059588, Bryan Mowry, M.D. (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, Ph.D. (PI). The SCCSS was supported by funding provided by the NIMH (R01 MH077139 to Patrick F. Sullivan and R01 MH095034 to Pamela Sklar), the Stanley Center for Psychiatric Research, the Sylvan Herman Foundation, the Friedman Brain Institute, Icahn School of Medicine at Mount Sinai at the Mount Sinai School of Medicine, the Karolinska Institutet, Karolinska University Hospital, the Swedish Research Council, the Swedish County Council, the Sö derström Kö nigska Foundation, and the Netherlands Scientific Organization (NWO 645–000-003). Co-principal investigators involved in this study were Pamela Sklar (Mount Sinai School of Medicine), Christina M. Hultman (Karolinska Institutet, Stockholm, Sweden), and Patrick F. Sullivan (University of North Carolina and Karolinska Institutet, Stockholm, Sweden). We are deeply grateful for the participation of all subjects contributing to this research and to the collection team that worked to recruit them. The principal investigators of the CATIE (Clinical Antipsychotic Trials of Intervention Effectiveness) trial were Jeffrey A. Lieberman, M.D., T. Scott Stroup, M.D., M.P.H., and Joseph P. McEvoy, M.D. The CATIE trial was funded by a grant from the National Institute of Mental Health (N01 MH900001) along with MH074027 (PI PF Sullivan). Genotyping was funded by Eli Lilly and Company.
Footnotes
Declaration of conflict of interest
The authors declare no conflict of interest.
Data and code availability
This study used data from NIH dbGaP (https://www.ncbi.nlm.nih.gov/gap/) and NIMH genetic data repository (https://www.nimhgenetics.org/) via controlled access. Qualified investigators can obtain access to these datasets by applying to these organizations.
The Python script codes used in the study are available at the Xiangning Chen’s Github site (https://github.com/mdsamchen/scz_bip_mdd).
Contributor Information
Xiangning Chen, The university of Texas Health Science Center at Houston.
Yimei Liu, Director and CEO, Lieber Institute for Brain Development, Johns Hopkins School of Medicine: Departments of Psychiatry, Neurology, Neuroscience and Genetic Medicine.
Joan Cue, Director and CEO, Lieber Institute for Brain Development, Johns Hopkins School of Medicine: Departments of Psychiatry, Neurology, Neuroscience and Genetic Medicine.
Mira Han Vishwajit Nimgaonkar, Director and CEO, Lieber Institute for Brain Development, Johns Hopkins School of Medicine: Departments of Psychiatry, Neurology, Neuroscience and Genetic Medicine.
Daniel Weinberger, Director and CEO, Lieber Institute for Brain Development, Johns Hopkins School of Medicine: Departments of Psychiatry, Neurology, Neuroscience and Genetic Medicine.
Shizhong Han, Lieber Institute for Brain Development; Johns Hopkins School of Medicine Department of Psychiatry and Behavioral Sciences.
Zhongming Zhao, University of Texas HSC Houston.
References
- 1.McGrath JJ, Lim CCW, Plana-Ripoll O, Holtz Y, Agerbo E, Momen NC et al. Comorbidity within mental disorders: a comprehensive analysis based on 145 990 survey respondents from 27 countries. Epidemiol Psychiatr Sci 2020; 29: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Šprah L, Dernovšek MZ, Wahlbeck K, Haaramo P. Psychiatric readmissions and their association with physical comorbidity: a systematic literature review. BMC Psychiatry 2017; 17: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romero C, Werme J, Jansen PR, Gelernter J, Stein MB, Levey D et al. Exploring the genetic overlap between twelve psychiatric disorders. Nat Genet 2022; 54: 1795–1802. [DOI] [PubMed] [Google Scholar]
- 4.Cross-Disorder Group of the Psychiatric Genomics Consortium. Electronic address: plee0@mgh.harvard.edu, Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019; 179: 1469–1482.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium Brainstorm, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J et al. Analysis of shared heritability in common disorders of the brain. Science 2018; 360. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baselmans BML, Yengo L, van Rheenen W, Wray NR. Risk in Relatives, Heritability, SNP-Based Heritability, and Genetic Correlations in Psychiatric Disorders: A Review. Biol Psychiatry 2021; 89: 11–19. [DOI] [PubMed] [Google Scholar]
- 7.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med 2020; 12: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legge SE, Cardno AG, Allardyce J, Dennison C, Hubbard L, Pardiñas AF et al. Associations Between Schizophrenia Polygenic Liability, Symptom Dimensions, and Cognitive Ability in Schizophrenia. JAMA Psychiatry 2021;78: 1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017; 45: D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019; 47: D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009; 460: 753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergen SE, O’Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry 2012; 17: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stroup TS, McEvoy JP, Swartz MS, Byerly MJ, Glick ID, Canive JM et al. The National Institute of Mental Health Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) project: schizophrenia trial design and protocol development. Schizophr Bull 2003; 29:15–31. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan PF, Lin D, Tzeng J-Y, van den Oord E, Perkins D, Stroup TS et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry 2008; 13: 570–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan PF, de Geus EJC, Willemsen G, James MR, Smit JH, Zandbelt T et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry 2009; 14: 359–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet 2014; 46: 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinforma Oxf Engl 2015; 31: 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi SW, O’Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. GigaScience 2019; 8. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat Oxf Engl 2007; 8: 118–127. [DOI] [PubMed] [Google Scholar]
- 22.Behdenna A, Haziza J, Azencott C-A, Nordor A. pyComBat, a Python tool for batch effects correction in high-throughput molecular data using empirical Bayes methods. 2020; : 2020.03.17.995431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He H, Bai Y, Garcia EA, Li S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. 2008. IEEE Int Jt Conf Neural Netw IEEE World Congr Comput Intell 2008. doi: 10.1109/IJCNN.2008.4633969. [DOI] [Google Scholar]
- 24.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: Synthetic Minority Over-sampling Technique. J Artif Intell Res 2002; 16: 321–357. [Google Scholar]
- 25.Han H, Wang W-Y, Mao B-H. Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning. In: Huang D-S, Zhang X-P, Huang G-B (eds). Advances in Intelligent Computing. Springer: Berlin, Heidelberg, 2005, pp 878–887. [Google Scholar]
- 26.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. ArXiv160304467 Cs 2016.http://arxiv.org/abs/1603.04467 (accessed 28 Jan2019). [Google Scholar]
- 27.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J et al. TensorFlow: A system for large-scale machine learning. ArXiv160508695 Cs 2016.http://arxiv.org/abs/1605.08695 (accessed 28 Jan2019). [Google Scholar]
- 28.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011; 12: 2825–2830. [Google Scholar]
- 29.Mi X, Zou B, Zou F, Hu J. Permutation-based identification of important biomarkers for complex diseases via machine learning models. Nat Commun 2021; 12: 3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waskom ML. seaborn: statistical data visualization. J Open Source Softw 2021; 6: 3021. [Google Scholar]
- 31.McInnes L, Healy J, Saul N, Großberger L. UMAP: Uniform Manifold Approximation and Projection. J Open Source Softw 2018; 3: 861. [Google Scholar]
- 32.Cross-Disorder Group of the Psychiatric Genomics Consortium, Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin J, Taylor MJ, Lichtenstein P. Assessing the evidence for shared genetic risks across psychiatric disorders and traits. Psychol Med 2018; 48: 1759–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grotzinger AD. Shared genetic architecture across psychiatric disorders. Psychol Med 2021; 51: 2210–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng S, Guan F, Ma M, Zhang L, Cheng B, Qi X et al. An atlas of genetic correlations between psychiatric disorders and human blood plasma proteome. Eur Psychiatry 2020; 63: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Wu J-S, Mize T, Shui D, Chen X. Prediction of Schizophrenia Diagnosis by Integration of Genetically Correlated Conditions and Traits. J Neuroimmune Pharmacol Off J Soc NeuroImmune Pharmacol 2018; 13: 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, Lu Q, Liu W, Zhang Y, Li M, Zhao H. Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction. PLoS Genet 2017; 13: e1006836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maier R, Moser G, Chen G-B, Ripke S, Cross-Disorder Working Group of the Psychiatric Genomics Consortium, Coryell W et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am J Hum Genet 2015; 96: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee PH, Feng Y- CA, Smoller JW. Pleiotropy and Cross-Disorder Genetics Among Psychiatric Disorders. Biol Psychiatry 2021; 89: 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang M, Wu X, Zhang W, Cui H, Zhang L, Yan P et al. Epidemiological and Genetic Analyses of Schizophrenia and Breast Cancer. Schizophr Bull 2023; : sbad106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bahrami S, Hindley G, Winsvold BS, O’Connell KS, Frei O, Shadrin A et al. Dissecting the shared genetic basis of migraine and mental disorders using novel statistical tools. Brain 2021; 145: 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaughlin RL, Schijven D, van Rheenen W, van Eijk KR, O’Brien M, Kahn RS et al. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat Commun 2017; 8: 14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul SM, Potter WZ. Finding new and better treatments for psychiatric disorders. Neuropsychopharmacology 2024; 49: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016; 533: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L et al. Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults. PLoS Genet 2017; 13: e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry 2011; 16: 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]