Abstract
The chemokine receptor CCR5 and to a lesser extent CCR3 and CCR2b have been shown to serve as coreceptors for human immunodeficiency virus type 1 (HIV-1) entry into blood- or tissue-derived macrophages. Therefore, we examined the expression of the chemokine receptors CCR1, CCR2b, CCR3, CCR5, and CXCR4 as RNAs or as membrane-expressed antigens in monocytes maturing into macrophages and correlated these results with the susceptibility of macrophages to HIV-1 infection, as measured by their concentrations of extracellular p24 antigen and levels of intracellular HIV DNA by quantitative PCR. There was little change in levels of CCR1, CCR2b, and CCR5 RNAs. CCR3 RNA and surface antigen were undetectable throughout maturation of adherent monocytes over 10 days. CXCR4 RNA and membrane antigen were strongly expressed in newly adherent monocytes, but their levels declined at day 7. The amounts of CCR5 RNA remained stable, but the amounts of CCR5 antigen increased from undetectable to peak levels at day 7 and then declined slightly at day 10. Levels of susceptibility to laboratory (HIV-1BaL) and clinical strains of HIV-1 showed parallel kinetics, peaking at day 7 and then decreasing at days 10 to 14. The concordance of levels of HIV DNA and p24 antigen suggested that the changes in susceptibility with monocyte maturation were at or immediately after entry and correlated well with CCR5 expression and inversely with CXCR4 expression.
The chemokine receptor CCR5 was recently identified as the major coreceptor for the entry of non-syncytium-inducing (NSI) human immunodeficiency virus (HIV) strains into primary CD4 lymphocytes and macrophages but not into T-cell lines (1, 11, 15). These data were supported by the relative refractoriness to HIV infection of CD4 lymphocytes from patients who are homozygous for the CCR5 Δ32 mutation (10, 21, 27, 39). In contrast, T-cell-line-tropic strains of HIV-1 use the chemokine receptor CXCR4 as an entry coreceptor (17) while dual-tropic strains can use both CCR5 and CXCR4 (14, 43). These findings correlate with inhibition of infection of T-cell lines with SI isolates by the natural ligand for CXCR4, SDF1 (3, 33), and also by the inhibition of infection of primary lymphocytes or macrophages with NSI HIV strains by the β-chemokines RANTES, MIP1β, and MIP1α, all of which bind to CCR5 (8).
HIV binds to CCR5 and CXCR4 via gp120, especially its V3 loop, which has been previously described to determine lymphocyte/macrophage tropism (6, 22, 42). Studies of chimeric chemokine receptors suggest that macrophage and dual-tropic HIV strains may differ in their relative affinities for the amino-terminal and extracellular domains of CCR5 (2, 12, 37). SI strains appear to bind to the extracellular domains of CXCR4. In addition, different strains of HIV appear to bind to different amino acids within the extracellular domains of CCR5 (13). Binding of gp120 to CCR5 is enhanced by prior binding to CD4 (46, 47). Furthermore, laboratory-adapted strains appear to be able to bind to CD4 and thence probably to chemokine receptors with higher affinities than clinical strains (25).
However, some clinical or laboratory-adapted HIV strains, including the dual-tropic strain 89.6, also use other chemokine coreceptors, either CCR3, CCR2b (5, 7, 14), or other orphan coreceptors, in addition to CCR5 for entry into primary T lymphocytes and blood-derived macrophages.
In addition, the chemokine receptors used during HIV entry of blood-derived and some tissue macrophages differ. For example CCR3 appears to be as important as CCR5 for entry into microglial cells (20).
The role of CCR5 and to a lesser extent CCR3 and CCR2b as HIV coreceptors for infection of blood-derived macrophages by most clinical and laboratory strains raises the question of whether differences in the susceptibilities of maturing monocytes to HIV-1 infection are determined by differential levels of expression of this coreceptor. Therefore, we tested the hypothesis that increasing permissiveness of differentiating monocytes to productive infection with M-tropic HIV-1 is related to changes in levels of expression of CCR5 or other chemokine receptors such as CCR1, CCR2b, CCR3, and CXCR4.
Heparinized peripheral blood was obtained from healthy HIV-seronegative donors. Peripheral blood mononuclear cells (PBMCs) were separated from blood by density gradient sedimentation on Ficoll-Paque (Pharmacia, Uppsala, Sweden). Monocytes were separated by countercurrent elutriation as previously described (30). The fractions containing monocytes were collected, and contaminating lymphocytes were depleted by complement-dependent lysis with monoclonal anti-CD3 antibody (OKT3; Ortho Diagnostics, Raritan, N.J.). Residual contamination with T lymphocytes in this population was <1% as determined by flow cytometry with anti-CD3 monoclonal antibody. More than 96% of the cells were nonspecific esterase positive. Monocytes were either used immediately after isolation (day 0) or cultured on 24-well tissue culture plates for 1, 3, 5, 7, and 10 days at 37°C and 5% CO2. Cells were then used for infection with NSI HIV-1 isolates, RNA extraction, or flow cytometry.
Cells were infected with low-number-passage clinical isolate WM1101 and the laboratory macrophage-tropic strain HIV-1BaL, which was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Infectivities of HIV isolates were quantified by calculation of their 50% tissue culture infective doses in phytohemagglutinin-stimulated PBMCs. Monocytes were inoculated with cell-free HIV isolates at 105 cpm of reverse transcriptase (RT) activity per ml (105 tissue culture infective doses per ml on PBMCs and at a multiplicity of infection of 0.025/cell) and allowed to adsorb for 4 h before complete aspiration of medium, washing, and addition of fresh medium. Cultures were replenished with fresh medium every three days, and culture supernatants were stored for quantification of HIV p24 antigen with an enzyme-linked immunosorbent assay kit (Organon Teknika, Sydney, Australia). The limit of detection of HIV p24 antigen was 12 pg/ml. Virus inocula which were used in DNA experiments were first filtered through a 0.22-μm-pore-size filter (Millex-GS; Millipore, Bedford, Mass.) and then treated with DNase (20 μg/ml) for 1 h at room temperature in the presence of 10 mM MgCl2 to decontaminate the inoculum of HIV-1 DNA. Cell cultures were treated with zidovudine (20 mM) for 30 min at 37°C to control for de novo DNA synthesis. Cell lysate preparation and PCR conditions were as previously described (30). Primers M667 (49) and gag1 (30) were used to amplify a 320-bp region extending from the R region within the 5′ long terminal repeat to the gag region, representing full-length or nearly full-length HIV-1 cDNA. Primers M667 and AA55 (49) were also used to amplify a 140-bp region flanking the R and U3 regions of the 5′ long terminal repeat, representing the sequence of the initiation product of HIV reverse transcription. Hot PCR was used to increase sensitivity where M667 was 5′ end labeled with [γ32P]ATP, and then the modified M667 was added to the PCR mixture at a ratio to cold primer of 1:2. Samples were subjected to 30 cycles of amplification as follows: 1 min at 95°C, 2 min at 60°C, and 3 min at 72°C with a final extension at 72°C for 7 min. Concurrent reactions were also performed with primers PCO3 and PCO4 (38) to amplify a 110-bp DNA fragment of the human β-globin gene to ensure that equivalent amounts of DNA were used in each sample reaction. HIV DNA standards were prepared from 8E5 cells (18), containing one integrated copy of HIV-1 DNA per cell, and PBMCs were used to optimize cell number. Amplified products were run on an agarose gel, dried, exposed to X-ray film for 6 h, and autoradiographed.
To study the RNA expression of CCR1, CCR2b, CCR3, CCR5, and CXCR4 in differentiating monocytes (0, 1, 3, and 7 days postadherence), RT-PCR was performed on these cultures with oligonucleotide primer pairs specific for each chemokine receptor. Briefly, total RNA was isolated with Trizol reagents and 2 μg was used for cDNA synthesis with Superscript II RNase H-reverse transcriptase (Life Technologies/Gibco-BRL, Melbourne, Australia) and oligo(dT) primer (Boehringer, Mannheim, Germany) at 42°C for 1 h. One-tenth of this product was used as a template for PCR amplification with Taq polymerase (Perkin-Elmer, Sydney, Australia). PCR was carried out for 30 cycles at 94°C for 1 min, 55°C for 2 min, and 72°C for 2 min. The primer pairs used in RT-PCR were CCR1 (296 bp) (sense, TGGAAACTCCAAACACCACAG; antisense, CCCAGTCATCCTTCAACTTG) (32), CCR2b (351 bp) (sense, CCAACGAGAGCGGTGAAGAAGTC; antisense, GCCAAAATAACCGATGTGATA) (48), CCR3 (539 bp) (sense, TGACAACCTCACTAGATACAGTTG; antisense, CTCTTCAAACAACTCTTCAGTCTC) (36), CCR5 (280 bp) (sense, AATAATTGCAGTAGCTCTAACAGG; antisense, TTGAGTCCGTGTCACAAGCCC) (40), CXCR4 (381 bp) (sense, TGACTCCATGAAGGAACCCTG; antisense, CTTGGCCTCTGACTGTTGGTG) (26), and GAPDH (196 bp) (sense, ATGGAGAAGGCTGGGGCTC; antisense, AAGTTGTCATGGATGACCTTG) (19).
Flow cytometry was carried out to examine the cell surface expression of CCR3, CCR5, CXCR4, and CD4 in differentiating monocytes (days 0, 1, 5, and 10 after adherence). After cells were removed from the plastic surface with 5 mM EDTA in phosphate-buffered saline, they were washed twice with cold fluorescence-activated cell sorter buffer containing 1% fetal bovine serum and 0.01% sodium azide in phosphate-buffered saline and then resuspended in 50 μl of human serum and labeled with specific antibody as previously described (29). Cells were examined with a Becton Dickinson (Franklin Lakes, N.J.) FACScan flow cytometer. Monoclonal antibodies to CCR3 (7B11) and CCR5 (2F9) were obtained from LeukoSite Inc. (Cambridge, Mass.), and monoclonal antibody to CXCR4 (12G5) (16) was purchased from R & D Systems (Minneapolis, Minn.). Monoclonal antibody to CD4 (Q4120) was a gift from Q. Sattentau (Inserm, Paris, France), and anti-Leu-M3 (CD14) phycoerythrin conjugate and anti-Leu3a fluorescein isothiocyanate conjugate were purchased from Becton Dickinson.
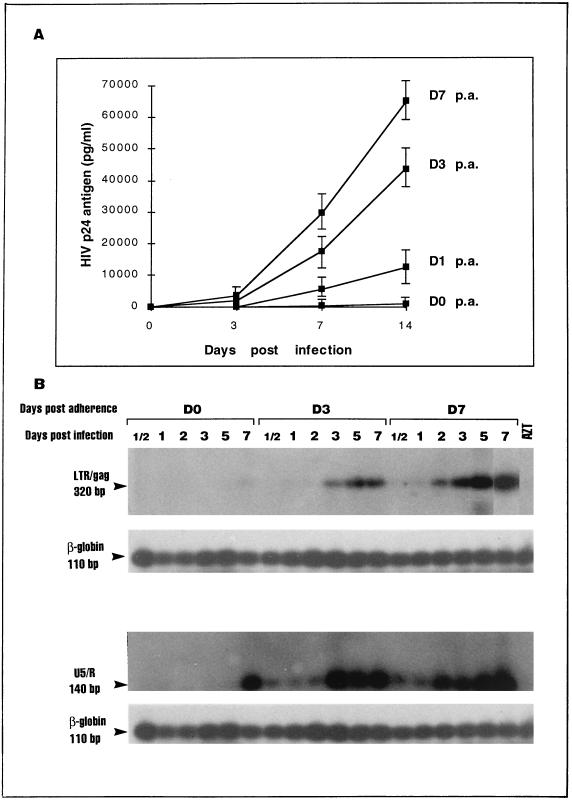
Monocytes differentiating into macrophages (i.e., monocyte-derived macrophages [MDM]) over 7 days of adherence to plastic became increasingly permissive to productive infection with a laboratory-adapted (BaL) and a clinical (NSI) strain of HIV, as shown by levels of extracellular HIV p24 antigen (Fig. 1A) and particle-associated reverse transcription (data not shown). A corresponding increase in HIV DNA was also observed by using quantitative sensitive hot PCR and primers to detect the full-length cDNAs (Fig. 1B, first gel) or the initiation products (Fig. 1B, third gel) of HIV reverse transcription. These results demonstrate clearly that restriction of both tropism and the level of productive infection in fresh uncultured monocytes occurs mainly at the level of entry (or uncoating). This confirms previous reports from our own group and other groups demonstrating that tropism of laboratory-adapted (34, 44) and clinical (4) isolates increases with monocyte maturation. The use of highly sensitive hot PCR supports the notion that this increased tropism restricts HIV entry into monocytes rather than restricting the spread of HIV from infected foci as a result of the occasional permissiveness of monocytes. Furthermore, susceptibility to productive infection of MDM reached a peak at approximately 7 days of adherence (Fig. 1). Infection of 10- and 14-day (data not shown)-old MDM resulted in lower levels of HIV p24 antigen (Fig. 1A) and DNA (data not shown). At day 7 the cells were also susceptible to infection with a greater number of clinical isolates and the mean level of extracellular p24 antigen was greatest (31).
FIG. 1.
Permissiveness to HIV-1 infection increased as monocytes differentiated into MDM. Cells were infected with HIV-1 WM1101 primary isolate (or HIV-1BaL [data not shown]) either immediately after isolation (day 0 [D0]) or after culture on plastic for 1, 3, 7, and 10 days (D1, D3, D7, and D10, respectively). Replication kinetics of HIV-1 WM1101 by cultured monocytes was measured by determining amounts of the extracellular p24 antigen in culture supernatants (A) with an enzyme-linked immunosorbent assay commercial kit (Organon Teknika) and by quantitative hot PCR with 32P-labeled primer for the detection of full-length cDNA (320 bp) (panel B, first gel) or initiation products (140 bp) (panel B, third gel) of HIV reverse transcription. Culture supernatants were collected 0, 3, 7, and 14 days after infection for the detection of HIV p24 antigen. DNA was extracted from cells at 12 h and 1, 2, 3, 5, and 7 days after infection and used in hot PCR to detect HIV DNA. β-Globin DNA (110 bp) was amplified concurrently with HIV DNA, and results are shown underneath each DNA sample (panel B, second and fourth gels). p.a., postadherence; AZT, zidovudine.
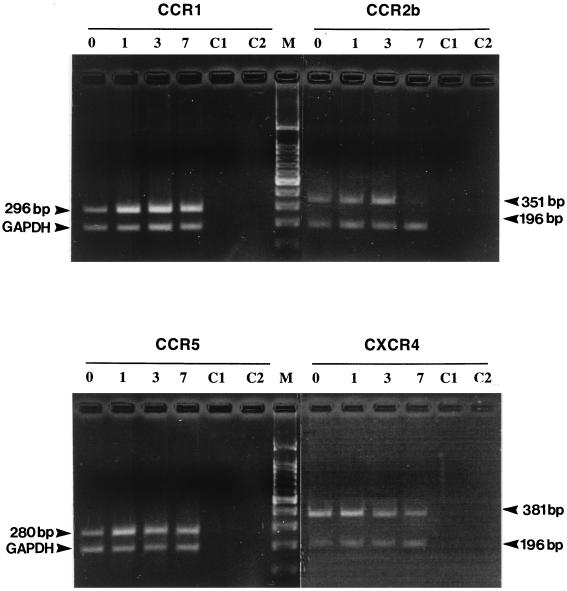
The expression of chemokine receptors (CCR1, CCR2b, CCR3, CCR5, and CXCR4) in differentiating monocytes was determined by measuring the specific levels of expression of mRNA or surface membrane antigen by semiquantitative RT-PCR or flow cytometry, respectively. In three different donors of blood monocytes, mRNAs for CCR1, CCR2b, CCR5, and CXCR4 were detected in both freshly isolated monocytes and differentiated macrophages (Fig. 2). CCR3 RNA was below the detectable level in all donors. CCR1 mRNA expression was relatively higher than that of other coreceptors and constant, irrespective of the stage of cell differentiation. CCR2b RNA expression in monocytes was variable between donors but showed a significant and consistent decline as cell differentiation proceeded at 7 days of adherence (Fig. 2). Whether this change is also reflected in the production of cell surface antigen is clarified when specific monoclonal antibodies become available. The CCR5 transcript was detected in fresh monocytes, with a slight increase at day 1 of monocyte adherence, and its level then remained constant to day 7 (Fig. 2). In addition, the levels of expression of CXCR4 (fusin) RNA were relatively high in fresh and newly adherent (day 1) monocytes but declined significantly when cells differentiated into MDM at day 7 of adherence (Fig. 2). Residual T-cell contamination in freshly isolated monocytes was <1%, as determined by flow cytometry with anti-CD3 antibody, and 86% of monocytes were stained with monoclonal antibody against CD14. The identities of all CCR transcripts were confirmed by DNA sequencing of the PCR products (data not shown). RNA preparations without reverse transcriptase were used in PCR analysis and shown consistently to be negative for chemokine receptors and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA (Fig. 2).
FIG. 2.
Expression of the chemokine receptors CCR1, CCR2b, CCR3, CCR5, and CXCR4 in maturing monocytes. Total RNAs from nonadherent cells (day 0; lanes 0) and adherent cells at days 1, 3, and 7 (lanes 1, 3, and 7, respectively) were extracted, treated with RNase-free DNase (Boehringer), and used in RT-PCR. Products were amplified with primer pairs specific for each chemokine receptor or for GAPDH (see the text). Amplified DNA fragments were visualized on ethidium bromide-stained agarose gels under UV transillumination and photographed. To exclude DNA contamination, mock PCRs were run in parallel without reverse transcriptase (lanes C1) or without the cDNA template (lanes C2).
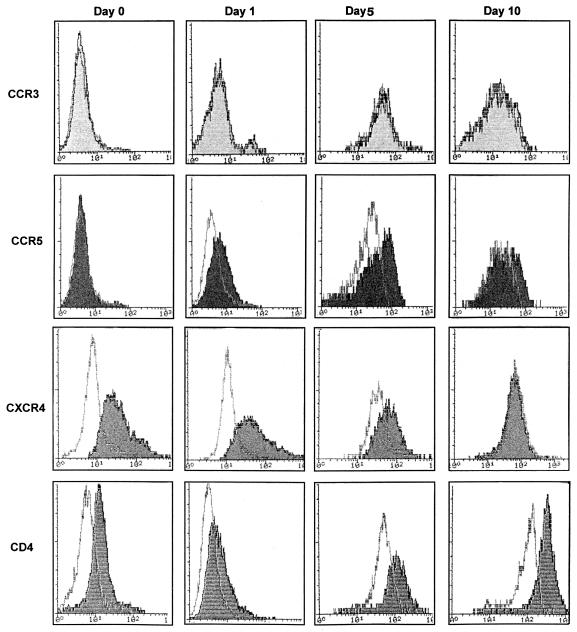
The levels of expression of CCR3, CCR5, and CXCR4 on cell surfaces at different stages of differentiation were then examined by flow cytometry with monoclonal antibodies against these coreceptors. The expression of CCR5 on cell surfaces significantly increased as monocytes matured to MDM over 1 to 7 days (Fig. 3, second row). In repeated experiments, surface CCR5 was not detected immediately after monocyte isolation but increased progressively from day 1 and was maximal at day 5, plateauing until day 7, and then decreased to lower levels at days 10 to 14 of adherence (Fig. 3, second row). This decrease in CCR5 surface expression of MDM was also correlated with a decrease in CCR5 RNA (data not shown) and a decrease in the susceptibility of MDM to productive infection at the same time points (Fig. 1A). CXCR4 membrane expression was relatively high at the early stage of monocyte maturation (days 0 to 3) and then decreased during the later stages of maturation (7 days after adherence) (Fig. 3, third row). These results correlated well with mRNA levels quantified by RT-PCR and Northern analysis (data not shown). CCR3 surface antigen expression was not detectable in monocytes or MDM (Fig. 3, first row), as was expected from the results with CCR3 mRNA. The level of CD4 expression (Fig. 3, fourth row) declined after initial monocyte adherence and then was slowly upregulated, as previously reported by us and others (9, 23), but it correlated neither with permissiveness of monocytes to infection nor with CCR5 expression. In addition, we have now examined CCR5 expression on maturing monocytes in more than the three cases mentioned above, verifying the results shown in Fig. 3 but also demonstrating that there was variability in the levels of expression of CCR5 on MDM on days 3 to 5 which was quite considerable (three- to fivefold) and which was independent of the CCR5Δ32 heterozygous states of individuals (data not shown). Despite this variability in levels of CCR5 expression, there was always a significant increase during maturation from an undetectable level at day 0. Furthermore, monocyte maturation or activation was induced with macrophage colony-stimulating factor over 3 days of culture. Surface expression of CCR5 was upregulated by 35% ± 6% (mean ± standard deviation), while CXCR4 was downregulated by 20% ± 3%, a pattern similar to that observed with differentiation by adherence to plastic.
FIG. 3.
Cell surface expression of CCR3, CCR5, CXCR4, and CD4 on maturing monocytes. CCR3, CCR5, CXCR4, and CD4 cell surface proteins in differentiating monocytes were detected by immunofluorescence and flow cytometric analysis. Monoclonal antibodies to CCR3 and CCR5 (7B11 and 2F9, respectively; LeukoSite), CXCR4 (12G5; R & D Systems), and CD4 (Q4120) were used. The immunofluorescence profile obtained for each antibody was compared to that of its corresponding isotype (immunoglobulin G1 or G2a) control, and the result is shown as an open curve. Note that the mean fluorescence intensity of control cells increases with maturation to day 5 and then plateaus. In each instance, the intensity of a specific receptor is shown with a shaded curve. The level of expression is determined by differences between the level of staining with a specific receptor monoclonal antibody and the variable baseline of the isotype control. These data are representative of similar profiles obtained from three different donors.
Therefore, the level of expression of CCR5, as a major coreceptor, corresponded well to increasing permissiveness for infection by NSI viruses at early stages of monocyte maturation (1 to 7 days after adherence) and then with a decrease in productive infection at later stages of maturation (10 to 14 days). The significance of the CXCR4 decline (which appears to be inversely proportional to the rise in CCR5) is not yet clear. Freshly isolated monocytes express significant levels of CXCR4 on their membranes and bind SDF1α but are still refractory to infection with SI isolates. Therefore, it appears that CCR5 but not CXCR4 is functional as an HIV coreceptor in blood monocytes, perhaps because of differences in the association of CXCR4 (28) with CD4 or conformational differences in the coreceptors of lymphocyte and monocyte membranes.
In this in vitro system, β-chemokine production in monocyte/macrophage cultures was influenced by three different stimuli. Initial plating of monocytes and culture manipulation resulted in a temporary surge of β-chemokine production to levels of 1 to 4 ng/ml, such that production of RANTES, MIP1α, and MIP1β by cultured monocytes peaked at 1 day after adherence and then declined to undetectable levels at day 3 and thereafter. HIV infection itself can specifically upregulate β-chemokine expression in monocytes and MDM, reaching levels four- to sixfold higher than those found in uninfected cultures (24). Similarly, it has been demonstrated that β-chemokine expression is enhanced in lymph nodes of HIV-infected patients (45) and in in vitro-infected MDM (41). Hence, there was no simple relationship between endogenous chemokine levels and the relative refractoriness of monocytes to HIV infection.
The above-described data supports the notion that increasing expression of CCR5 antigen on monocytes maturing into macrophages may account for at least a significant component of the inhanced susceptibility of MDM to infection with M-tropic HIV-1 isolates. Furthermore, it has been shown that monocyte susceptibility to HIV-1 infection increased despite declining expression of the CD4 receptor during the first day of adherence in blood monocytes (9, 23). However, other chemokine receptors may also be involved in this mechanism, as a recent study has suggested that at least one other unidentified coreceptor may be active in MDM, especially where infection with M-tropic isolates cannot be determined solely by utilization of CCR5 (5). As reagents for the other coreceptors become available, similar studies of expression on maturing monocytes will be necessary, as will determination of the chemokine receptor utilization of the infecting HIV strains used in such studies.
Acknowledgments
We thank the Australian National Centre for HIV Virology Research and the National Health and Medical Research Council for great support.
We thank LeukoSite Inc. (Cambridge, Mass.) for kindly providing us with antibodies to CCR3 and CCR5 and Q. Sattentau (Inserm) for antibody to CD4. We also thank Claire Wolczak for the preparation of the manuscript.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-α, MIP-β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Atchinson R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 3.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 4.Chang J, Naif H M, Li S, Sullivan J S, Randle C M, Cunningham A L. Twin studies demonstrated a host cell genetic effect on productive HIV infection of human monocytes and macrophages in vitro. J Virol. 1996;70:7792–7803. doi: 10.1128/jvi.70.11.7792-7803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, We L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 8.Cocchi F, De Vico A L, Grazino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1816. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 9.Collman R G, Godfrey B, Cutilli J, Rhodes A, Hassan N F, Sweet R, Douglas S D, Friedman H, Nathanson N, Gonzalez-Scarano F. Macrophage-tropic strains of human immunodeficiency virus type 1 utilize the CD4 receptor. J Virol. 1990;64:4468–4476. doi: 10.1128/jvi.64.9.4468-4476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 11.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.Doms R W. AIDS pathogenesis conference. Keystone, Colo: Keystone Symposia on Molecular and Cellular Biology; 1997. Structure-function studies of chemokine receptors and their role in HIV and SIV infection; p. 1. [Google Scholar]
- 13.Doranz B J, Lu Z-H, Rucker J, Zhang T, Berson J F, Wang Z, Sharron M, Doms R W, Peiper S C. AIDS pathogenesis conference. Keystone, Colo: Keystone Symposia on Molecular and Cellular Biology; 1997. Structural determinants of CCR5 coreceptor function for HIV-1, abstr. 116-12. [Google Scholar]
- 14.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Pieper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 15.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 16.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-dependent infection by HIV-2 is determined by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y, Broder C, Kennedy P, Berger E. HIV-1 entry cofactor: functional cDNA cloning of seven-transmembrane, G-protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 18.Folks T M, Powel D, Lightfoote M, Koenig S, Fauci A S, Hogan S, Venatesan S, Martin M A. Biological and biochemical characterization of a cloned Leu-3-cell surviving infection with the acquired immunodeficiency syndrome retrovirus. J Exp Med. 1986;164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanauer A, Mandel J L. The glyceraldehyde-3-phosphate dehydrogenase gene family: structure of a human cDNA and an X chromosome linked pseudogene; amazing complexity of the gene family in mouse. EMBO J. 1984;3:2627–2633. doi: 10.1002/j.1460-2075.1984.tb02185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection in microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 22.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–73. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 23.Kazazi F, Mathijs J-M, Foley P, Cunningham A L. Variations in CD4 expression by human monocytes and macrophages and their relationship to infection with the human immunodeficiency virus. J Gen Virol. 1989;70:2661–2672. doi: 10.1099/0022-1317-70-10-2661. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, M., and H. Naif. Unpublished observations.
- 25.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loetscher M, Geiser T, O’Reilly T, Zwahlen R, Baggiolini M, Moser B. Cloning of a human seven-transmembrane domain receptor, LESTR, that is highly expressed in leukocytes. J Biol Chem. 1994;269:232–237. [PubMed] [Google Scholar]
- 27.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 28.McKnight A, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naif H M, Li S, Ho-Shon M, Mathijs J-M, Williamson P, Cunningham A L. The state of maturation of monocytes into macrophages determines the effects of interleukin 4 and interleukin 13 on HIV replication. J Immunol. 1997;158:501–511. [PubMed] [Google Scholar]
- 30.Naif H M, Chang J, Ho-Shon M, Li S, Cunningham A L. Inhibition of human immunodeficiency virus replication in differentiating monocytes by interleukin 10 occurs in parallel with inhibition of cellular RNA expression. AIDS Res Hum Retroviruses. 1996;12:1237–1245. doi: 10.1089/aid.1996.12.1237. [DOI] [PubMed] [Google Scholar]
- 31.Naif, H. M. Unpublished observations.
- 32.Neote K, DiGregorio D, Mak J Y, Horuk R, Schall T J. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 33.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien W A, Koyanagi Y, Namazi A, Zhao J-Q, Diagne A, Idler K, Zack J, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 35.Paxton W A, Martin S R, Tse D, O’Brien T R, Skurnick J, Van Devanter N L, Padian N, Braun J F, Kotler D P, Wolinsky S M, Koup R A. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 36.Ponath P D, Qin S, Post T W, Wang J, Wu L, Gerard N P, Newman W, Gerarad C, Mackay C R. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med. 1996;183:2437–2448. doi: 10.1084/jem.183.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 38.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullins K B, Erlich H A. Primer-directed enzyme amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 39.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 40.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 41.Schmidtmayerova H, Nottet H S, Nuovo G, Raabe T, Flanagan C R, Dubrovsky L, Gendelman H E, Cerami A, Bukrinsky M, Sherry B. Human immunodeficiency virus type 1 infection alters chemokine β peptide expression in human monocytes: implications for recruitment of leukocytes into brain and lymph nodes. Proc Natl Acad Sci USA. 1996;93:700–704. doi: 10.1073/pnas.93.2.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T-cell line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 43.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonza S, Maerz A, Deacon N, Meanger J, Mills J, Crowe S. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J Virol. 1996;70:3863–3869. doi: 10.1128/jvi.70.6.3863-3869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tedla N, Palladinetti P, Kelly M, Kumar R K, DiGirolamo N, Chattophadhay U, Cooke B, Truskett P, Dwyer J, Wakefield D, Lloyd A. Chemokines and T lymphocyte recruitment to lymph nodes in HIV infection. Am J Pathol. 1996;148:1367–1373. [PMC free article] [PubMed] [Google Scholar]
- 46.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 47.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 48.Yamagami S, Tokuda Y, Ishii K, Tanaka H, Endo N. cDNA cloning and functional expression of a human monocytes chemoattractant protein 1 receptor. Biochem Biophys Res Commun. 1994;202:1156–1162. doi: 10.1006/bbrc.1994.2049. [DOI] [PubMed] [Google Scholar]
- 49.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]