Abstract
Objective
Synovial fibrosis contributes to osteoarthritis (OA) pathology, but the underlying mechanisms remain unknown. We have observed increased microRNA‐27b‐3p (miR‐27b‐3p) levels in synovial fluid of patients with late‐stage radiographic knee OA. Here, we investigated the contribution of miR‐27b‐3p to synovial fibrosis in patients with severe knee OA and in a mouse model of knee OA.
Methods
We stained synovium sections obtained from patients with radiographic knee OA scored according to the Kellgren/Lawrence scale and mice that underwent destabilization of the medial meniscus (DMM) for miR‐27b‐3p using in situ hybridization. We examined the effects of intraarticular injection of miR‐27b‐3p mimic into naive mouse knee joints and intraarticular injection of a miR‐27b‐3p inhibitor into mouse knee joints after DMM. We performed transfection with miR‐27b‐3p mimic and miR‐27b‐3p inhibitor in human OA fibroblast‐like synoviocytes (FLS) using reverse transcriptase–quantitative polymerase chain reaction (RT‐qPCR) array, RNA sequencing, RT‐qPCR, Western blotting, immunofluorescence, and migration assays.
Results
We observed increased miR‐27b‐3p expression in the synovium from patients with knee OA and in mice with DMM‐induced arthritis. Injection of the miR‐27b‐3p mimic in mouse knee joints induced a synovial fibrosis‐like phenotype, increased synovitis scores, and increased COL1A1 and α‐smooth muscle actin (α‐SMA) expression. In the mouse model of DMM‐induced arthritis, injection of the miR‐27b‐3p inhibitor decreased α‐SMA but did not change COL1A1 expression levels or synovitis scores. Transfection with the miR‐27b‐3p mimic in human OA FLS induced profibrotic responses, including increased migration and expression of key extracellular matrix (ECM) genes, but transfection with the miR‐27b‐3p inhibitor had the opposite effects. RNA sequencing identified a PPARG/ADAMTS8 signaling axis regulated by miR‐27b‐3p in OA FLS. Human OA FLS transfected with miR‐27b‐3p mimic and then treated with the PPARG agonist rosiglitazone or with ADAMTS8 small interfering RNA exhibited altered expression of select ECM genes.
Conclusion
Our findings demonstrate that miR‐27b‐3p has a key role in ECM regulation associated with synovial fibrosis during OA.
INTRODUCTION
Osteoarthritis (OA), the most common form of arthritis, is a leading cause of disability worldwide (1). Research on OA has frequently focused on articular cartilage degradation and subchondral bone sclerosis, but the synovium also undergoes pathologic changes (2). The synovium, a thin connective tissue layer consisting largely of 2 types of cells (fibroblast‐like synoviocytes [FLS] and tissue‐resident macrophages), produces synovial fluid, which nourishes articular chondrocytes and removes products of tissue metabolism (3). As OA progresses, the synovium becomes inflamed (synovitis) and hypertrophies as a result of increased vascularization, infiltration of inflammatory cells, aberrant OA FLS proliferation, and accumulation of excessive extracellular matrix (ECM) (3, 4). These changes contribute to synovial fibrosis and promote joint stiffness and pain (5). Although synovial pathology was originally thought to be a secondary reaction to evolving damage in neighboring cartilage and bone tissues, it is now recognized as an active participant of OA pathogenesis (6); however, little is known about the underlying regulatory signaling networks, particularly those responsible for the altered ECM regulation contributing to synovial fibrosis.
As potent regulators of gene expression, microRNAs (miRNAs) in knee OA have been studied but mainly for their roles in cartilage homeostasis (7, 8). Less is known about the contribution of these important transcriptional regulators to synovitis, particularly fibrotic responses (5, 9, 10). We previously reported that miRNA‐27b‐3p (miR‐27b‐3p) levels are higher in the synovial fluid of patients with advanced radiographic knee OA and identified the synovium as the major source of miR‐27b‐3p, as stimulation of synovial explants with interleukin‐1β (IL‐1β) elicited its secretion (11). However, the specific role of miR‐27b‐3p to OA synovial pathology is not known. We hypothesized that miR‐27b‐3p helps modulate synovial ECM regulatory networks involved in synovial fibrosis during OA. In this study, we used in vivo and in vitro models, RNA sequencing analysis, and computational analysis to determine, for the first time to our knowledge, the role and mechanisms of miR‐27b‐3p in ECM regulation and synovial fibrotic responses during OA.
PATIENTS AND METHODS
Study design and OA patients
All patients provided written informed consent for inclusion in the University Health Network Research Ethics Board–approved biomarker exploration studies (16‐5969‐AE and 14‐7592‐AE). All animal studies were approved by the University Health Network's Animal Care Committee (animal use protocol 3729) and were conducted in accordance with relevant guidelines and regulations. A minimum of 6 animals were needed to detect a 25% difference by histology with 80% power (sigma = 0.15, alpha level of 0.05). Full sequencing data sets are available through Gene Expression Omnibus (GEO accession no. GSE152638).
We obtained synovial tissue samples from patients with radiographic knee OA (Kellgren/Lawrence [K/L] grades 3 and 4) who were undergoing total knee replacement and from patients who were undergoing knee arthroscopy (K/L grades 1 and 2) at the Toronto Western Hospital (Toronto, Canada). Synovial tissue samples were either used fresh for retrieval of FLS for culture studies or were fixed and processed for histology or for in situ hybridization (ISH). Patient anthropometric and demographic data and the patient's experimental group are listed in Supplementary Table 1, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285.
Mouse model of surgical destabilization of the medial meniscus (DMM) and intraarticular injections
We housed 10‐ to 12‐week‐old C57BL/6J male mice (The Jackson Laboratory) in the Krembil Research Institute (Toronto, Ontario, Canada) vivarium on a 12‐hour light/dark cycle in a temperature‐controlled room (21°C ± 1°C) with food provided ad libitum for 1 week before we performed DMM or sham surgery (12). Knee joints (n = 6–10 for each group [DMM and sham]) were collected at 2, 5, or 10 weeks after surgery and processed for ISH, immunohistochemistry (IHC), or histology.
For the miR‐27b‐3p mimic injection experiments, naive 12‐week‐old C57BL/6J male mice were intraarticularly injected twice, 2 weeks apart, under isoflurane anesthesia with 5 μg of mirVana miR‐27b‐3p mimic (ThermoFisher catalog no. 4464066) in the right knee or mirVana miRNA mimic negative control 1 (ThermoFisher catalog no. 4464061) in the left knee. Knee joints were retrieved 5 weeks after injection (n = 6 knee joints per each treatment group) and processed for IHC and histology.
For the miR‐27b‐3p inhibitor injection experiments, 12‐week‐old C57BL/6J male mice were subjected to DMM or sham surgery (right knee) and intraarticularly injected with 5 μg of custom in vivo–grade miRCURY locked nucleic acid (LNA) mmu‐miR‐27b‐3p inhibitor (Qiagen catalog no. 339204‐YCI201647‐FZA) at 1 week and 3 weeks after surgery, with knee joints retrieved at 5 weeks after surgery (n = 10). Control animals (9 animals that had sham surgery and received control inhibitor or 10 animals that had DMM surgery and received control inhibitor) underwent the same procedure but received injections of a scrambled negative control inhibitor (Qiagen catalog no. 339204‐YCI0201821‐FZA).
Histology and IHC analyses of synovium
Human synovial samples and mouse knee joints were processed, sectioned, and stained with Safranin O (Sigma‐Aldrich catalog no. S2255) and fast green (Bio Basic catalog no. FB0452), Masson's trichrome for connective tissue (Electron Microscopy Sciences catalog series no. 26367), or hematoxylin and eosin (Vector catalog no. H‐3404), all in accordance with manufacturers' recommendations. Articular cartilage and synovial samples were scored in a blinded manner by 2 observers who used the Osteoarthritis Research Society International (OARSI) grading for cartilage damage and degree of synovitis (13). For IHC, sections were stained for ADAMTS8 (5 μg/ml; Novus Biologicals catalog no. NBP2‐46494), type V collagen (COL5A1; 0.5 μg/ml; Abcam catalog no. ab7046), type XIV collagen alpha 1 chain (COL14A1; 1:200; Novus Biologicals catalog no. NBP2‐15940), COL1A1 (1:200; Abcam catalog no. ab34710), α‐smooth muscle actin (α‐SMA; 1:400; Sigma‐Aldrich catalog no. A2547), or PPARG (1:750; Novus Biologicals catalog no. NBP2‐22106SS). After sections were stained, we used 3,3′‐diaminobenzidine substrate (Vector catalog no. SK‐4105) for visualization. We determined percentage of stained area or percentage of positive cells using ImageJ version 1.53c (14).
In situ hybridization
After samples were subjected to proteinase K digestion (with 7.5 μg/ml for 10 minutes for mouse knee joints and 20 μg/ml for 30 minutes for human synovium), sections were incubated for 1 hour at 55°C with denatured LNA‐modified and 5′‐end and 3′‐end digoxigenin–labeled antisense oligonucleotides, which included the miR‐27b‐3p probe (80 nM for mouse and 160 nM for human; Qiagen catalog no. YD00619142), U6 small nuclear RNA (1 nM; positive control), or scrambled control probe (40 nM; negative control). Nuclei were counterstained with nuclear fast red (Sigma‐Aldrich catalog no. N3020‐100ML). We allowed coverslipped sections to dry overnight before visualization under brightfield illumination (Leica catalog no. ICC50W5021). We counted a minimum of 3 random fields to measure the percentage of positive cells in each section.
FLS cultures
We isolated FLS by enzymatic digestion of human OA synovium obtained at time of surgery (total knee replacement or knee arthroscopy). Cells were cultured at 37°C and at 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Gibco catalog no. 11995073) supplemented with 10% fetal bovine serum (Wisent catalog no. 080‐150) and 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were passaged when they reached 85% confluency. Passages 3–5 were used for all culture experiments. OA FLS cultures were serum‐starved (0.5% fetal bovine serum) for 3 hours before addition of 3 μg/ml Lipofectamine RNAiMAX transfection reagent (Life Technologies, ThermoFisher catalog no. 13778‐075) with either 5 nM Homo sapiens (hsa)‐miR‐27b‐3p miRCURY LNA miRNA mimic (Qiagen catalog no. 339173 YM00470553) or the corresponding control mimic (Cel‐miR‐39‐3p; Qiagen catalog no. YM00479902) or 50 nM hsa‐miR‐27b‐3p miRCURY LNA Power inhibitor (Qiagen catalog no. 339131) or the corresponding control inhibitor (negative control A; Qiagen catalog no. 339136), after which cultures remained for 48 hours in serum‐starved media. For some experiments, OA FLS culture medium was replaced 24 hours later with fresh serum‐starved DMEM containing either 1) 20 μM rosiglitazone (Tocris catalog no. 5325) or vehicle (dimethyl sulfoxide; Sigma‐Aldrich catalog no. D2650) or 2) 10 nM ADAMTS8 small interfering RNA (siRNA; Sigma‐Aldrich catalog no. EHU129241‐20UG) or MISSION siRNA universal negative control (Sigma‐Aldrich catalog no. SIC001) in 3 μg/ml Lipofectamine and then incubated for an additional 24 hours before cells were harvested. Cells were either fixed and stained with 0.3% crystal violet (Transwell migration assay) or for immunofluorescence, or extracts were collected for protein (Western blot) or RNA analysis. For RNA analysis, we used the 834‐well RT2 Profiler and polymerase chain reaction (PCR) arrays for human ECM and adhesion molecules (Qiagen catalog no. PAHS‐013ZE‐1), conventional reverse transcriptase–quantitative PCR (RT‐qPCR) (primers listed in Supplementary Table 2, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285) and RT‐qPCR for detection of mmu‐miR‐27b‐3p and U6 (Qiagen catalog no. 339306), and RNA sequencing (Truseq stranded total RNA; Illumina catalog no. 20020596).
RNA sequencing and pathway analysis
RNA was extracted from 48‐hour mimic– or control–transfected OA FLS, and libraries were prepared (Truseq stranded total RNA). We used the Illumina NextSeq 550 system (2 × 75 bp) at the Centre for Arthritis Diagnostic and Therapeutic Innovation (Schroeder Arthritis Institute, Toronto, Ontario, Canada) for sequencing analysis, followed by bioinformatics analysis to determine gene expression levels per sample (15). Genes with ≥10 counts per million in ≥2 samples were retained for the analysis. We identified differentially expressed genes (DEGs) by using the negative binomial generalized linear model with trended dispersion and trimmed means normalization. Dispersion and treatment effect estimates were adjusted for sample pairs and genes. Significance was set at unadjusted P < 0.05, as no significant differences in gene expression remained after multiple testing correction (false discovery rate). We used R (version 3.6.0) and the edgeR (version 3.25.8) package for analysis.
We obtained putative gene targets of hsa‐miR‐27b‐3p from mirDIP version 4.1 (http://ophid.utoronto.ca/mirDIP) (16) and retrieved physical protein–protein interactions from the Integrated Interactions Database version 2020‐05 (http://ophid.utoronto.ca/iid) (17). We obtained pathway annotations from pathDIP version 4 (http://ophid.utoronto.ca/pathDIP) (18). The transcription regulatory network was downloaded from Catalogue of Transcriptional Regulatory Interactions, Catrin version 1 (http://ophid.utoronto.ca/Catrin). All networks were integrated, annotated, visualized, and analyzed using NAViGaTOR version 3.0.16 (19). The final networks were exported in an SVG format, and the final images (300 dpi in PNG format) with legends were prepared with Adobe Illustrator version 26.0.2. Gene nodes were color coded by top Gene Ontology molecular function terms identified using Uniprot and Gene Ontology slim.
Statistical analysis
Data analyses were completed in Excel 2010 and R 3.1.0. With the exception of RNA sequencing, we conducted all statistical analyses using GraphPad PRISM 9 software. P or q values (FDR‐adjusted P) less than 0.05 were considered significant for analysis between groups.
Additional details of methodology can be found in Supplementary Methods, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285.
RESULTS
Association between synovial expression of miR‐27b‐3p and human and mouse knee OA severity
We previously showed that miR‐27b‐3p expression levels were higher in synovial fluid of patients with radiographic knee OA and K/L grades 3 and 4 compared with patients with K/L grades 1 and 2 and that ex vivo human OA synovium treated with the proinflammatory mediator IL‐1β had increased secretion of miR‐27b‐3p (11). Because the synovium is a major contributor to synovial fluid, we first examined expression of miR‐27b‐3p in synovium of patients with varying K/L grades of knee OA, with the small nuclear RNA U6 used as a positive control (Supplementary Figure 1A, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285). With increased radiographic knee OA severity, we observed increased collagen fiber density under a more stratified synovial lining in patients with knee OA and K/L grades 3 and 4 relative to patients with K/L grades 1 and 2 (Figure 1A). Concomitantly, the proportion of miR‐27b‐3p–positive cells relative to the total number of cells quantified in the synovial lining increased with K/L grade (Figure 1A). Thus, consistent with expression levels of miR‐27b‐3p in synovial fluid (11), the proportion of synovial lining cells expressing miR‐27b‐3p increased with radiographic knee OA severity.
Figure 1.
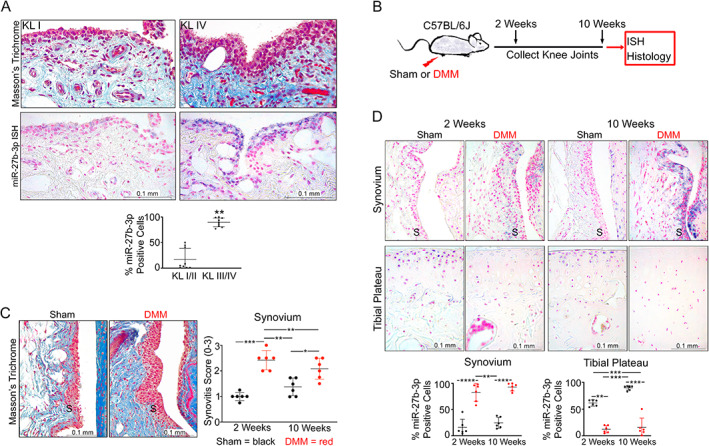
Expression of microRNA‐27b‐3p (miR‐27b‐3p) in samples obtained from human and mouse knee osteoarthritis (OA) synovium. A, Top, Representative images of synovium (original magnification ×40) stained with Masson's trichrome or miR‐27b‐3p in situ hybridization (ISH; purple regions) in samples from radiographic knee OA patients with Kellgren/Lawrence (K/L) radiographic severity grades 1 and 2 or K/L grades 3 and 4. Bottom, Scatter plot showing mean ± SD proportion of miR‐27b‐3p–labeled synovial lining cells in synovium from patients with K/L grade 1 or 2 radiographic knee OA or K/L grade 3 or 4 radiographic knee OA (n = 9/group). B, Destabilization of the medial meniscus (DMM) was conducted on right knees of 10‐ to 12‐week‐old C57BL/6 mice and knee joints were collected at 2 weeks or 10 weeks after surgery. C, Left, Representative images of synovium at 10 weeks after sham or DMM surgery; collagen accumulation in DMM model is shown in blue (Masson's trichrome). Right, Scatter plot showing mean ± SD synovitis severity scores of mouse synovium 2 weeks or 10 weeks after sham or DMM surgery (n = 6/group). D, Top, ISH images of the synovium (S) or tibial plateau (original magnification ×40) (miR‐27b‐3p indicated in purple, nuclei indicated in pink [nuclear fast red]). Bottom, Scatter plots of mean ± SD percentage of miR‐27b‐3p–labeled cells in the synovium or tibial chondrocytes at 2 weeks and 10 weeks after surgery. Relative data were log‐transformed before analyses using Student's unpaired t‐test (A), Kruskal‐Wallis test followed by 2‐stage linear step‐up procedure of Benjamini, Krieger, and Yekutieli (C), or 2‐way analysis of variance with Tukey's post hoc test (D). * = P/q < 0.05; ** = P/q < 0.01; *** = P/q < 0.001; **** = P < 0.0001.
To determine whether similar expression differences could be detected in an animal model of knee OA, we examined miR‐27b‐3p expression by ISH at 2 weeks and 10 weeks after DMM‐induced OA in C57BL/6 male mice (Figure 1B). Similar to our findings in human knee OA, we observed increased collagen deposition using Masson's trichrome stain in synovium from mice with DMM‐induced OA, which coincided with increased synovitis scores compared with that shown in control mice that received sham surgery (Figure 1C). DMM surgery also induced cartilage degeneration that increased in severity from 2 weeks to 10 weeks, as reflected by the increases in OARSI scores of the medial femoral condyle and medial tibial plateau (Supplementary Figure 1B). These changes in matrix organization were accompanied by increased miR‐27b‐3p staining in the synovium as early as 2 weeks and persisted through 10 weeks after surgery (Figure 1D). In contrast, the mouse cartilage tissue showed a decrease in miR‐27b‐3p expression in tibial articular chondrocytes at both time points, similar to the decreases in miR‐27b‐3p observed in human OA cartilage cultured with IL‐1β (11) or as reported in fresh cartilage from patients with either OA or rheumatoid arthritis compared with that obtained from trauma patients without a history of disease (20, 21). In sham‐operated animals, miR‐27b‐3p was primarily detected in tibial articular chondrocytes, with minimal cells labeled in the synovium. In both human and mouse knee OA, synovial miR‐27b‐3p expression coincided with increased ECM deposition.
Induction of a synovial fibrosis‐like phenotype after intraarticular injection of miR‐27b‐3p mimic into the mouse knee joint
The increases in miR‐27b‐3p expression observed in human and mouse OA synovium suggested a causal relationship with disease severity and prompted us to examine the effect of modifying miR‐27b‐3p expression in the knee joints of naive mice (Figure 2A) and of mice with DMM‐induced OA using miR‐27b‐3p mimic and inhibitor, respectively. In synovium from miR‐27b‐3p mimic–injected naive knee joints, we observed consistently higher synovitis scores, as reflected by synovial thickening, increased collagen deposition, and cell infiltration (Figure 2B). These changes were paralleled by a miR‐27b‐3p mimic–mediated increase in the percentage of synovial cells labeled with COL1A1, a major structural collagen component of synovial ECM (22), and α‐SMA, a marker of activated fibroblasts (23, 24). Of note, cartilage integrity remained mostly unchanged in both the miR‐27b‐3p mimic– and control mimic–injected mouse knee joints (Supplementary Figure 2, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285). Thus, intraarticular injection of miR‐27b‐3p mimic elicited a fibrosis‐like phenotype in the synovium of healthy mouse knee joints.
Figure 2.
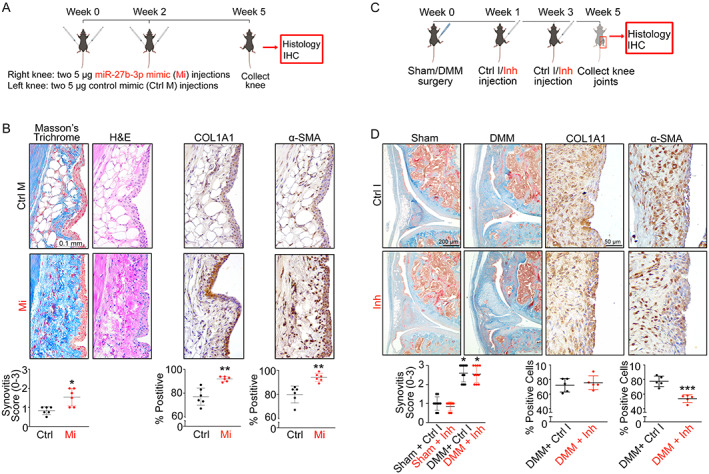
Overexpression of microRNA‐27b‐3p (miR‐27b‐3p) promotes synovial fibrosis‐like responses in vivo. A, Schematic of miR‐27b‐3p mimic or control mimic injections in mouse knees. B, Images of synovia (original magnification ×40) from mouse knee joints (n = 6) injected with miR‐27b‐3p mimic or with control mimic and stained with Masson's trichrome or hematoxylin and eosin (H&E) for immunohistochemical (IHC) analysis or immunolabeled with COL1A1 or α‐smooth muscle actin (α‐SMA) (shown in brown, with nuclei counterstained in blue). C, Schematic of injection of miR‐27b‐3p inhibitor (Inh) or control inhibitor (Ctrl Inh) in mouse knees (destabilization of the medial meniscus [DMM] vs. sham surgery). D, Top, Images of synovia (original magnification ×10) from sham‐operated or DMM‐operated mouse knee joints injected with miR‐27b‐3p inhibitor or with control inhibitor and stained with Masson's trichrome or DMM‐operated knee joint immunolabeled with COL1A1 or α‐SMA (shown in brown, with nuclei counterstained in blue). Bottom, Results quantified as synovitis severity scores, and percentage of cells staining positive for COL1A1 or α‐SMA. Symbols represent individual samples; bars show the mean ± SD. * = P < 0.05; ** = P < 0.01; *** = P < 0.001, by Mann‐Whitney unpaired U test. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42285/abstract.
In mice that received intraarticular injection of miRCURY LNA miR‐27b‐3p inhibitor at 1 week and 3 weeks after DMM surgery (Figure 2C), we observed no obvious differences in the histologic changes in cartilage or in synovial pathology (OARSI and synovitis scores) compared with results shown with the control inhibitor at 5 weeks after surgery (Figure 2D and Supplementary Figure 3, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285) or in the percentage of synovial cells expressing COL1A1. However, the percentage of synovial cells expressing α‐SMA decreased in the miR‐27b‐3p inhibitor–treated group compared with the group that received the control inhibitor.
Increased expression of ECM markers and migration of human OA FLS with miR‐27b‐3p overexpression
Given that intraarticular injection of miR‐27b‐3p mimic induced a synovial fibrosis–like phenotype with increased expression of COL1A1 in the synovium in vivo, we next examined the effect of miR‐27b‐3p overexpression on the production of COL1A1 in cultures of FLS, the major cell type isolated from human OA synovium obtained during total knee replacement surgeries. Transfection of OA FLS with miR‐27b‐3p mimic resulted in a mean ± SD 199 ± 53‐fold increase in miR‐27b‐3p expression (Supplementary Figure 4A, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285). In addition, transfection with miR‐27b‐3p mimic in OA FLS resulted in an increased change of COL1A1 transcript levels of mean ± SD 4.43 ± 1.73‐fold (Figure 4B) and protein levels of 1.27 ± 0.22‐fold (Figure 3A). We observed opposite effects when cells were transfected with miR‐27b‐3p inhibitor, with a mean ± SD 0.73 ± 0.14‐fold change in transcript levels (Supplementary Figure 4B) and a mean ± SD 0.47 ± 0.19‐fold change in protein levels (Figure 3B). Immunofluorescence experiments demonstrated that the number of cultured OA FLS transfected with miR‐27b‐3p mimic expressing COL1A1 also increased (Figure 3C and Supplementary Figure 5, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285), with cell numbers comparable in OA FLS that were transfected with either miR‐27b‐3p mimic (mean ± SD 53.0 ± 11.1) or control mimic (mean ± SD 52.5 ± 7.9). Overall, miR‐27b‐3p mimic transfection increased COL1A1 expression in human OA FLS, while inhibition of miR‐27b‐3p had the opposite effect.
Figure 4.
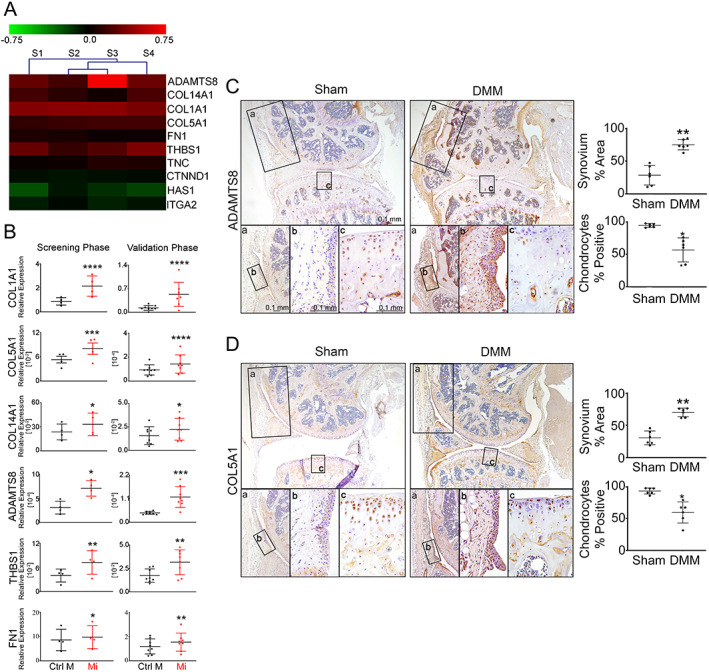
MicroRNA‐27b‐3p (miR‐27b‐3p) regulates the expression of multiple extracellular matrix (ECM)–related genes. A, Heatmap of genes differentially expressed in reverse transcriptase–quantitative polymerase chain reaction (RT‐qPCR) array among 84 ECM‐related genes found in fibroblast‐like synoviocytes (FLS) isolated from osteoarthritis (OA) synovium that were transfected with miR‐27b‐3p mimic or with control mimic and cultured for 48 hours (n = 4). B, Scatter plots obtained by RT‐qPCR array (screening phase (n = 4) and RT‐qPCR (validation phase, n = 8) comparing relative expression (mean ± SD) of ECM‐related genes (normalized to GAPDH) in OA FLS transfected with miR‐27b‐3p mimic (Mi) or control mimic (Ctrl M) and cultured for 48 hours. Data were log‐transformed before analysis by Student's paired 2‐tailed t‐test. C and D, Left, Immunohistochemical analysis (DAB, brown) of ADAMTS8 (C) and type V collagen (COL5A1) (D) expression in mouse knee joints 10 weeks after destabilization of the medial meniscus (DMM) or sham surgery. Nuclei are counterstained in blue (original magnification × 10). Bottom panels a–c are higher‐magnification views of the boxed areas, with panels a and b showing synovium (original magnification ×20 and ×40) and panel c showing medial tibial plateau cartilage (original magnification ×40). Right, Results plotted as the mean ± SD percentage of synovium or chondrocytes staining positive for ADAMTS8 or COL5A1 in 3 digital images of ×40 fields of view (n = 6 samples/group). * = P < 0.05; ** = P < 0.01; *** = P < 0.001; **** = P < 0.0001, by Student's unpaired 2‐tailed t‐test with Welch's correction. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42285/abstract.
Figure 3.
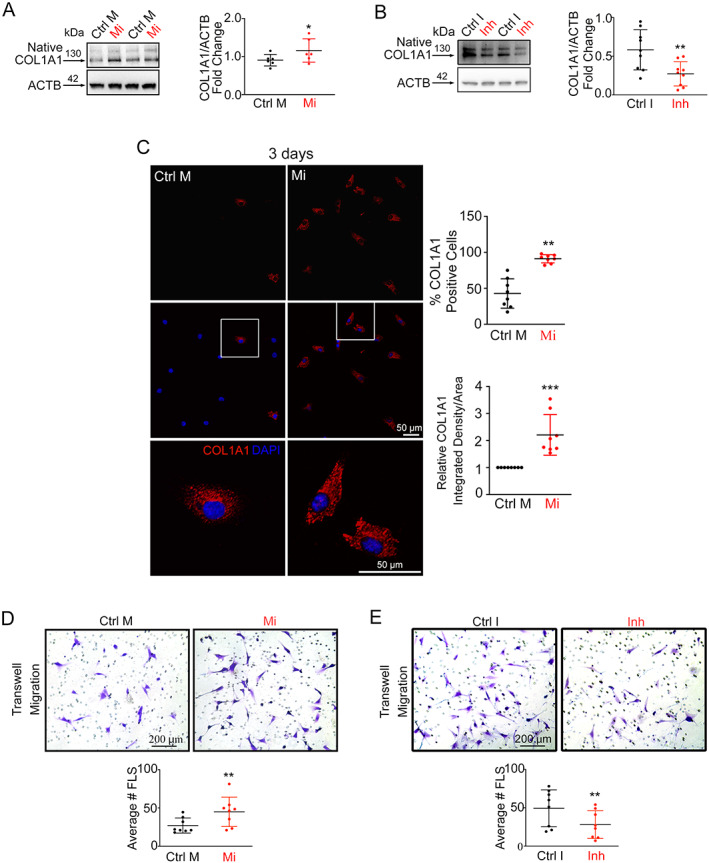
MicroRNA‐27b‐3p (miR‐27b‐3p) promotes synovial fibrosis–like responses in vitro. A and B, Left, Western blots of COL1A1 expression (compared to β‐actin) in osteoarthritis (OA) fibroblast‐like synoviocytes (FLS) transfected with miR‐27b‐3p mimic (Mi) or control mimic (Ctrl M) (A) or with miRCURY locked nucleic acid (LNA) miR‐27b‐3p inhibitor (Inh) or a control inhibitor (Ctrl I) (B) and cultured for 48 hours (n = 6–9). Right, Log‐transformed data plotted as the mean ± SD fold change in relative expression. C, Left, Immunofluorescent images of COL1A1 expression in OA FLS transfected with miR‐27b‐3p mimic or control mimic and cultured for 3 days. Boxed areas in middlle panels are shown at higher magnification in bottom panels (original magnification ×20). Right, Results plotted as the mean ± SD percentage of COL1A1‐positive cells and as relative COL1A1 integrated density per area (n = 8 paired cultures). D and E, Top, Crystal violet–stained images of Transwell‐migrated OA FLS 24 hours after transfection with miR‐27b‐3p mimic or control mimic (D) or with miR‐27b‐3p inhibitor (Inh) or control inhibitor (Ctrl I) (E). Bottom, Results plotted as the mean ± SD number of OA FLS migrating in the Transwells (n = 8 paired cultures). Data were analyzed with Student's paired 2‐tailed t‐tests (A–C) or Wilcoxon's tests (D and E). * = P < 0.05; ** = P < 0.01; *** = P < 0.001. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42285/abstract.
Because increased migration is a key process associated with a profibrotic response of OA FLS (25, 26), we next investigated whether miR‐27b‐3p influenced OA FLS migration using a Transwell migration assay. We found that transfection of cells with miR‐27b‐3p mimic increased the number of OA FLS that migrated to the underside of Transwell membranes relative to cells transfected with control mimic (Figure 3D). As with COL1A1 expression, transfection with the miR‐27b‐3p inhibitor had the inverse effect, reducing OA FLS migration (Figure 3E). These data suggested that miR‐27b‐3p influences migration in OA FLS. However, when we examined the effect of miR‐27b‐3p overexpression on talin and vinculin, cytoskeletal mediators of cell adhesion (27), and the cytoskeletal component vimentin, we detected no differences in the protein levels of these molecules (Supplementary Figure 6A, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285) or in the organization of F‐actin filaments or vinculin (Supplementary Figure 6B).
The effect of miR‐27b‐3p mimic on OA FLS COL1A1 expression prompted us to investigate whether miR‐27b‐3p regulated the expression of other ECM genes. Among the 84 matrix‐related genes (listed in Supplementary Table 3, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285) that we investigated in OA FLS using ECM‐specific qPCR array, 17 genes had responses to the miR‐27b‐3p mimic that were below the assay's detection limits and were excluded from further analysis. Of the remaining genes, when compared to transfection with control mimic, transfection with miR‐27b‐3p mimic significantly increased the expression of 7 genes (COL1A1, COL5A1, COL14A1, thrombospondin 1 [THBS1], ADAMTS8, tenascin C [TNC], and fibronectin [FN1]) and decreased the expression of 3 genes (catenin delta 1 [CTNND1], hyaluronan synthase 1 [HAS1], and integrin alpha 2 ITGA2]) (Figures 4A and B, screening phase). Validation of the array screening by RT‐qPCR using mimic‐transfected FLS from 4 additional patients with advanced radiographic knee OA confirmed increases in COL1A1, COL14A1, COL5A1, FN1, ADAMTS8, and THBS1 (Figure 4B, validation phase). No marked differences were observed for TNC, ITGA2, HAS1, and CTNND1 (Supplementary Figure 4C, available at https://onlinelibrary.wiley.com/doi/10.1002/art.42285). Two genes confirmed to be modified by miR‐27b‐3p mimic transfection in OA FLS, ADAMTS8 and COL5A1, showed increased staining in cells in the synovium and reduced staining in articular chondrocytes of DMM‐operated mouse knee joints (Figures 4C and D). Similar results were observed with COL14A1 staining (Supplementary Figure 4D). Together, miR‐27b‐3p overexpression promoted OA FLS expression of key ECM genes and increased migration capacity, both important events associated with synovial fibrosis pathology during OA.
Identification of ECM‐related putative target genes of miR‐27b‐3p using RNA sequencing and computational analyses
To better understand the complex regulatory effects of miR‐27b‐3p in OA FLS, we performed RNA sequencing and computational analysis to create a comprehensive signaling network of miR‐27b‐3p gene targets in OA FLS (Figure 5A). RNA sequencing data showed that transfection with miR‐27b‐3p mimic elicited significant (P < 0.05) expression changes in 2,295 DEGs compared with that shown in OA FLS transfected with control mimic, with increased expression in 1,428 DEGs and decreased expression in 867 DEGs (Figure 5B; see Supplementary Table 4, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285, for the full list and segregated up‐/down‐regulated DEGs). Using computational approaches, we compared predicted gene targets of miR‐27b‐3p using mirDIP (16) with the list of DEGs identified by RNA sequencing (Supplementary Figure 7, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285). Of the 2,295 DEGs identified by RNA sequencing in miR‐27b‐3p mimic–treated OA FLS, 1,862 genes (81%) overlapped with miR‐27b‐3p putative gene targets: 1,203 genes were up‐regulated (red edges), and 659 were down‐regulated (green edges). Of note, many of the miR‐27b‐3p–modulated DEGs were related to ECM pathways (reactome ECM‐related pathway nodes outlined in purple in Supplementary Figure 7, as obtained from pathDIP) (18).
Figure 5.
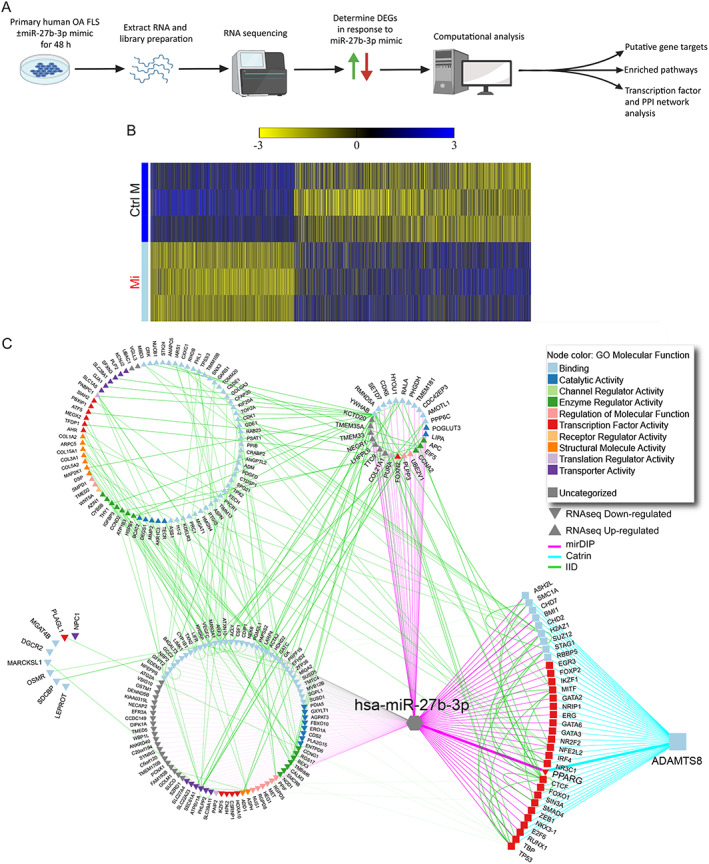
Expression profile and extracellular matrix–related network analysis of fibroblast‐like synoviocytes (FLS) isolated from osteoarthritis synovium overexpressing microRNA‐27b‐3p (miR‐27b‐3p). A, Schematic of workflow for determination of differentially expressed genes (DEGs) and putative networks regulated by miR‐27b‐3p mimic treatment of human OA FLS in vitro using RNA sequencing and computational analyses. PPI = protein–protein interactions. B, Heatmap of the DEGs (unadjusted P < 0.05) determined by RNA sequencing of OA FLS transfected with miR‐27b‐3p mimic (Mi) or control mimic (Ctrl M) and cultured for 48 hours. Columns indicate treatment‐paired replicates (n = 3). Genes are ordered based on log fold change (largest to smallest), and values are scaled by row (mean centered and divided by SD). C, Integrated network analysis of miR‐27b‐3p putative gene targets predicted to modulate ADAMTS8. The assembled network contains previously predicted Homo sapiens (hsa)‐miR‐27b‐3p gene targets (microRNA Data Integration Portal [mirDIP], purple edges), ADAMTS8‐associated transcription factors (Catalogue of Transcriptional Regulatory Interactions [Catrin], turquoise edges), and physical PPIs (Integrated Interactions Database [IID], green edges) of DEGs identified by RNA sequencing (RNAseq) of OA FLS transfected with miR‐27b‐3p (direction of expression change relative to control mimic–transfected cells is indicated by up and down triangles). Node color indicates associated Gene Ontology (GO) molecular functions. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42285/abstract.
To better understand and visualize the ECM‐specific signaling network of miR‐27b‐3p in OA FLS, we used the Gene Ontology cellular component annotations to extract the ECM‐related miR‐27b‐3p putative gene targets from all potential targets (Supplementary Figures 7 and 8, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285). The association of DEGs identified by RNA sequencing to the ECM are highlighted by blue, green, and purple gene names. Of note, several of the ECM‐related target genes down‐regulated by miR‐27b‐3p transfection were involved in vascular health, including the genes for aspartate beta‐hydroxylase (ASPH) (28), cadherin EGF LAG seven‐pass G‐type receptor 1 (CELSR1) (29), heart development protein with EGF‐like domain 1 (HEG1) (30), laminin subunit alpha 1 (LAMA1) and laminin subunit beta 1 (LAMB1) (31), nidogen 2 (NID2) (32), neuropilin 2 (NRP2) (33), plexin domain containing 1 (PLXDC1) (34), syndecan binding protein (SDCBP) (35), secreted protein acidic and cysteine rich (SPARC) modular calcium‐binding protein 1 (SMOC1) (36), and vascular endothelial growth factor c (VEGFC) (37) or bone remodeling gene, including oncostatin M receptor (OSMR) (38), SMOC1 (39), and SMOC2 (40). In contrast, expression of many structurally related ECM miR‐27b‐3p target genes, including collagen genes (COL1A2, COL3A1, COL5A2, COL5A3, COL8A2, COL11A1, COL15A1, COL21A1) and peptidylprolyl isomerase B gene (PPIB), an important regulator of collagen folding (41), members of the ADAMTS family (ADAMTS1, ADAMTS8, ADAMTS9), and matricellular proteins (SPARC, secreted phosphoprotein 1 [SPP1]) were up‐regulated in miR‐27b‐3p–transfected OA FLS. Overall, RNA sequencing coupled with computational analysis identified multiple ECM‐related miR‐27b‐3p gene targets in OA FLS.
Identification of a miR‐27b‐3p/PPARG/ADAMTS8 signaling axis regulating select ECM genes in OA FLS
To identify the gene(s) consistently differentially expressed across all 3 investigations (qPCR array, RT‐qPCR, and RNA sequencing), we took a focused approach to specifically compare the DEGs identified by qPCR array and RT‐qPCR with the ECM‐related putative targets identified through RNA sequencing. Although most of the genes identified by qPCR array and RT‐qPCR followed similar trends in RNA sequencing (e.g., COL1A1 showed a 2‐fold increase), only ADAMTS8 (3.4‐fold increase) was significantly up‐regulated (P < 0.05) in miR‐27b‐3p–transfected OA FLS compared with control across all 3 investigations.
Because ADAMTS8 is reported to regulate signal transduction, ECM remodeling, and fibrosis (42, 43), we focused on the contribution of ADAMTS8 in miR‐27b‐3p signaling in OA FLS. To examine the relationship of miR‐27b‐3p and ADAMTS8 in further detail, we assembled an integrated predictive network combining miR‐27b‐3p putative gene targets, ADAMTS8‐associated transcription factors, and physical protein–protein interactions (Figure 5C). From this analysis, multiple putative miR‐27b‐3p–regulated transcription factors were predicted to modulate ADAMTS8. When we applied a more stringent approach by restricting miR‐27b‐3p putative gene targets and ADAMTS8‐associated transcription factors to the top 1% of the prediction scores in the network analysis, we identified 14 predicted miR‐27b‐3p–regulated transcription factors of ADAMTS8, with only PPARG down‐regulated in our RNA sequencing data.
To investigate whether PPARG was indeed involved in miR‐27b‐3p regulation of ADAMTS8, we first used OA FLS to determine whether modification of miR‐27b‐3p levels by mimic and inhibitor transfection alters PPARG expression. Our results showed that miR‐27b‐3p overexpression reduced the expression of PPARG (Figure 6A), while inhibition of miR‐27b‐3p had the reverse effect, increasing PPARG expression (Figure 6D). We found that miR‐27b‐3p mimic reduced and miR‐27b‐3p inhibitor increased protein levels of PPARG in OA FLS in vitro (Figures 6B and 6E).
Figure 6.
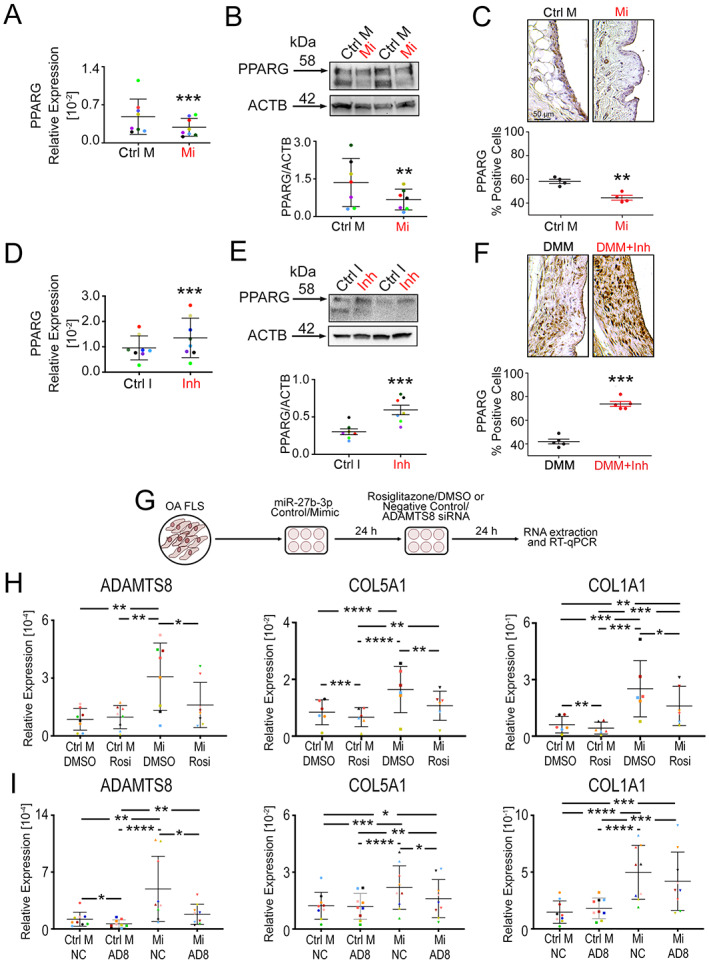
Regulation of select extracellular matrix (ECM) genes through a microRNA‐27b‐3p (miR‐27b‐3p)/PPARG/ADAMTS8 signaling axis. PPARG transcript levels (A, D) and protein levels (B, E) were determined in fibroblast‐like synoviocytes (FLS) isolated from osteoarthritis (OA) synovium transfected with either miR‐27b‐3p mimic (Mi) or control mimic (Ctrl M) or transfected with miR‐27b‐3p inhibitor (Inh) or control inhibitor (Ctrl I) and cultured for 48 hours. Results were determined by reverse transcriptase–quantitative polymerase chain reaction (RT‐qPCR) normalized to GAPDH (n = 8) or by Western blot densitometry measured from upper bands appearing at ~58 kd (n = 7). PPARG expression was determined in the synovium of naive mice injected intraarticularly with Mi or Ctrl M (C) or in mice that underwent destabilization of the medial meniscus (DMM) surgery and injected intraarticularly with Inh or Ctrl I (F), as determined by immunohistochemistry. See negative controls in Supplementary Figure 11 (available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285). G, Schematic showing transfection of OA FLS with control mimic (Ctrl M) or miR‐27b‐3p mimic (Mi), followed by treatment with rosiglitazone (Rosi) or DMSO as vehicle control or with small interfering RNA (siRNA) targeting ADAMTS8 (AD8) or a negative control siRNA (NC). H and I, OA FLS were transfected with control mimic or miR‐27b‐3p and treated with Rosi/DMSO (H) or ADAMTS8 siRNA/NC siRNA (I). RNA were extracted and assessed by RT‐qPCR for expression of ADAMTS8, COL5A1, and COL1A1. Colored data points in A, D, H, and I highlight biologic replicates (n = 6–9). Scatter plots show mean ± SD, with relative data log‐transformed before analysis by Student's paired 2‐tailed t‐test (A–F) or repeated measures 2‐way analysis of variance followed by Tukey's multiple comparisons post hoc test (H, I). * = P < 0.05; ** = P < 0.01; *** = P < 0.001; **** = P < 0.0001. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/doi/10.1002/art.42285/abstract.
We next examined whether injection of miR‐27b‐3p mimic or inhibitor had any effect on the expression of PPARG in the synovium in vivo. The number of PPARG‐labeled cells was reduced in the synovium of naive mouse joints treated with intraarticular injection of miR‐27b‐3p mimic, whereas PPARG‐labeled cells in the synovium of DMM‐operated mice were increased with injection of miR‐27b‐3p inhibitor, as shown by IHC (Figures 6C and 6F). However, no significant changes in PPARG staining were observed in the articular cartilage in response to the miR‐27b‐3p inhibitor (Supplementary Figure 9, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285).
We next used the PPARG agonist, rosiglitazone, in miR‐27b‐3p–transfected OA FLS to investigate the role of PPARG in miR‐27b‐3p–regulated expression of ADAMTS8 (Figures 6G and 6H). In support of our predictive network analysis, treatment with rosiglitazone counteracted the miR‐27b‐3p mimic–mediated up‐regulation of ADAMTS8 (Figure 6H). We also investigated whether some of the ECM genes that we identified by qPCR array and RT‐qPCR were also regulated by PPARG. Increases in COL5A1, COL1A1, and THBS1 mediated by miR‐27b‐3p mimic were inhibited by rosiglitazone, whereas COL14A1 and FN1 were not (Figure 6H, Supplementary Figure 10A, available on the Arthritis & Rheumatology website at https://onlinelibrary.wiley.com/doi/10.1002/art.42285), suggesting a link between miR‐27b‐3p and PPARG activity in regulation of select ECM genes.
To further investigate the involvement of ADAMTS8 in miR‐27b‐3p–mediated ECM regulation, we also examined the effect of ADAMTS8 knockdown on the expression of key ECM genes in miR‐27b‐3p mimic–transfected cells (Figures 6G and 6I). ADAMTS8 siRNA treatment effectively reduced ADAMTS8 transcript levels by mean ± SD 58 ± 38% in OA FLS transfected with miR‐27b‐3p mimic and by mean ± SD 65 ± 18% in OA FLS transfected with control. Of note, the miR‐27b‐3p mimic–induced expression of COL5A1 but not COL1A1 was reduced (Figure 6I). Similarly, miR‐27b‐3p mimic–mediated COL14A1 expression was also decreased by ADAMTS8 siRNA, but THBS1 or FN1 were not (Supplementary Figure 10B). Together, these findings suggest that, in OA FLS, miR‐27b‐3p regulates a subset of ECM genes, in part, through a PPARG/ADAMTS8 signaling axis.
DISCUSSION
This study showed that miR‐27b‐3p expression is elevated in human knee OA and mouse knee OA synovia and plays a crucial role in the regulation of key synovial ECM components. Injection of miR‐27b‐3p mimic in naive mouse knee joints induced a fibrosis‐like phenotype, and the transfection of human OA FLS with miR‐27b‐3p mimic increased the migratory capacity of FLS and up‐regulated the expression of multiple ECM genes, including COL1A1, COL5A1, and FN1, which are among the top 10 genes expressed in human synovium most closely related to OA (44). RNA sequencing coupled with computational analysis identified a complex ECM‐specific signaling network with multiple targets of miR‐27b‐3p in OA FLS. Furthermore, this study identified one of the signaling arms of miR‐27b‐3p involving a PPARG/ADAMTS8 signaling axis that, in part, regulates the expression of select ECM components in OA FLS.
The fibrosis‐modifying effects of miR‐27b‐3p have been demonstrated in other pathologic conditions. For instance, miR‐27b‐3p was found to promote cardiac fibrosis, in part through up‐regulation of profibrotic ECM genes in atrial fibroblasts (45). In contrast, miR‐27b‐3p has been found to negatively regulate lung fibrosis (46). Thus, it is possible that the effects of miR‐27b‐3p on fibrotic responses may be tissue‐ and/or disease‐specific. We observed that miR‐27b‐3p increased in the synovium and decreased in the cartilage in mice after DMM surgery, similar to our previous observations of miR‐27b‐3p expression changes in cultured human synovium and in cartilage explants in response to IL‐1β (11). Although our study did not focus on changes in cartilage, further studies that examine the role of miR‐27b‐3p in cartilage homeostasis are warranted. It is intriguing that miR‐27b‐3p levels have been shown to be lower in cartilage tissue of patients with OA and patients with rheumatoid arthritis than in control tissues without these diseases (20, 21). We also showed that intraarticular injection of miR‐27b‐3p mimic into healthy mouse knee joints largely spared cartilage from degeneration while promoting a fibrosis‐like phenotype in the synovium with increased percentages of COL1A1‐ and α‐SMA–expressing cells. We did not observe the inverse, that is, a reduced severity of synovial pathology in the mouse model of DMM that was treated with a miR‐27b‐3p inhibitor. However, of note, the number of cells in the synovium expressing α‐SMA was reduced. Although the number of activated fibroblasts (α‐SMA–positive cells) was reduced in response to the miR‐27b‐3p inhibitor, the persistence of DMM‐induced synovitis and the observation of no significant differences in the COL1A1 expression suggested that other compensatory mechanisms may be sufficient to sustain synovial pathology.
Although we did not focus on cartilage degradation, one surprising observation was the overt expression of COL5A1 in chondrocytes of sham‐operated animals. COL5A1 is a minor component of healthy cartilage (47), with expression of COL5A1 increased after cartilage damage; recently, COL5A1 has been identified as a hub gene elevated in damaged knee cartilage (48). Knee surgery itself, even without meniscus destabilization (sham surgery), poses risks to the joint (e.g., during incision) and could explain the COL5A1 expression detected in chondrocytes of sham‐operated animals. Given that COL5A1 expression declined in chondrocytes of mice subjected to DMM surgery, it is likely that meniscus destabilization elicits additional changes in the joint.
Because miR‐27b‐3p appeared to be an important mediator of synovial fibrosis in our study, we used RNA sequencing to examine the effects of its overexpression on OA FLS transcription profiles. A large proportion (81%) of DEGs identified overlapped with mirDIP‐predicted putative gene targets. In addition, many of the up‐regulated DEGs were associated with the ECM; however, many of the down‐regulated DEGs were associated with vascular health and bone remodeling. Intriguingly, ADAMTS8 was the only gene consistently and significantly increased in OA FLS with qPCR array, RT‐qPCR, and RNA sequencing in response to the miR‐27b‐3p mimic. Its knockdown in OA FLS inhibited the miR‐27b‐3p–mediated induction of COL5A1, but not COL1A1, suggesting that ADAMTS8 contributes to the miR‐27b‐3p regulation of select ECM genes. When the top predicted transcription factors of ADAMTS8 and miR‐27b‐3p targets were examined and cross‐referenced, PPARG was the only one identified by RNA sequencing to be down‐regulated by miR‐27b‐3p transfection of OA FLS.
PPARG, a previously identified target of miR‐27b‐3p (49, 50) with a miR‐27b–targeted sequence in its 3′ UTR (51), helps regulate fibroblast ECM production (52). Consistent with these previous observations, we showed that the number of PPARG‐positive cells was reduced in the synovium of naive mouse joints treated with miR‐27b‐3p mimic, whereas the number of PPARG‐positive cells in the synovium of mice subjected to DMM surgery was increased with injection of miR‐27b‐3p inhibitor in vivo. Similarly, we showed that miR‐27b‐3p mimic decreased and miR‐27b‐3p inhibitor increased PPARG expression in human OA FLS in vitro. Intriguingly, we showed that rosiglitazone, a PPARG agonist, inhibited miR‐27b‐3p–induced ADAMTS8 as well as COL5A1 and COL1A1 expression in OA FLS. These data further suggest that regulation of select ECM genes occurs in part through a miR‐27b‐3p/PPARG/ADAMTS8 signaling axis in OA FLS.
One limitation of the present study was that we did not have access to synovial samples from healthy individuals with no musculoskeletal disease; thus, we do not know whether miR‐27b‐3p is expressed or whether it has an influence on ECM production in human synovium under homeostatic conditions. Obtaining synovial specimens continues to be challenging, especially from those with early stages of OA. Consequently, we are limited in our ability to investigate the influence of demographic and anthropometric variables, like sex, on the miR‐27b‐3p effects reported in our present study. Furthermore, our cultures of OA FLS are enriched for FLS but are likely not pure. Even after a minimum of 3 passages used in our study, it is possible that some macrophages remained and contributed to the observed effects, although their contribution is likely minimal (53, 54, 55).
In summary, we showed for the first time a key role of miR‐27b‐3p and its downstream signaling mediators in ECM regulation associated with synovial fibrosis during OA.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Kapoor had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Tavallaee, Lively, Rockel, Ali, Hinz, Jurisica, Kapoor.
Acquisition of data
Tavallaee, Lively, Im, Sarda, Mitchell, Rossomacha, Nakamura, Gabrial, Ratneswaran, Perry.
Analysis and interpretation of data
Tavallaee, Lively, Rockel, Potla, Matelski, Hinz, Gandhi, Jurisica, Kapoor.
Supporting information
Disclosure Form
Supplementary Table 1 Description of demographic and anthropometric variables, and experimental use of synovial samples from patients exhibiting KL1/KL2 or KL3/KL4 radiographic OA.
Supplementary Table 2 Primers used for RT‐qPCR.
Supplementary Table 3 Genes significantly upregulated or downregulated (p < 0.05), or not significantly changed (p > 0.05) by miR‐27b‐3p mimic‐treated compared to control mimic‐treated OA FLS, as determined by RT‐qPCR array. Genes undedected by array analysis are also indicated.
Supplementary Table 4 Lists of significantly upregulated genes or significantly downregulated genes from miR‐27b‐3p mimic‐treated compared to control mimic‐treated OA FLS, as determined by RNA sequencing.
Figure S1 Supplementary Figure
Figure S2 Supplementary Figure
Figure S3 Supplementary Figure
Figure S4 Supplementary Figure
Figure S5 Supplementary Figure
Figure S6 Supplementary Figure
Figure S7 Supplementary Figure
Figure S8 Supplementary Figure
Figure S9 Supplementary Figure
Figure S10 Supplementary Figure
Figure S11 Supplementary Figure
Appendix S1 Supplementary Methods
ACKNOWLEDGMENTS
We acknowledge the Schroeder Arthritis Institute's Division of Orthopedics clinical research team for help with patient consent and collecting clinical data. We thank Brian Wu for help with tissue histology scoring. We thank Dr. Khalid Syed for help with the clinical data. We thank the staff of the Collaborative Advanced Microscopy Labs of Dentistry (Faculty of Dentistry, University of Toronto) for training and help with image acquisition and data analysis. Figures 2A and 2C and Figure 5A were created, in part, with BioRender.com.
Dr. Tavallaee was recipient of a PhD scholarship from the Arthritis Society and a Queen Elizabeth II/Canadian Arthritis Network Graduate Scholarship in Science and Technology. Dr. Ali's work was supported by an Arthritis Society Postdoctoral Fellowship. Dr. Ratneswaran's work was supported by Canadian Institute of Health Research and Krembil Research Institute postdoctoral fellowships. Dr. Hinz's work was supported by the Canadian Institutes of Health Research (foundation grant 375597) and the John R. Evans Leadership Fund (awards 36050, 38861), as well as innovation funds (Fibrosis Network, award 36349) from the Canada Foundation for Innovation and the Ontario Research Fund. Dr. Jurisica's computational analysis was supported in part by the Natural Sciences Research Council (award 203475), the Canada Foundation for Innovation (awards 225404, 30865), the Ontario Research Fund (award 34876), IBM, and the Ian Lawson van Toch Fund. Dr. Kapoor's work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Schroeder Arthritis Institute via the Toronto General and Western Hospital Foundation; Dr. Kapoor was also recipient of a Tier 1 Canada Research Chair and the Tony and Shari Fell Chair in Arthritis Research.
Drs. Tavallaee and Lively contributed equally to this work.
Author disclosures are available at https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fart.42285&file=art42285‐sup‐0001‐Disclosureform.pdf.
REFERENCES
- 1. Neogi T. The epidemiology and impact of pain in osteoarthritis. Osteoarthritis Cartilage 2013;21:1145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Felson DT, Niu J, Neogi T, et al. Synovitis and the risk of knee osteoarthritis: the MOST study. Osteoarthritis Cartilage 2016;24:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scanzello CR, Goldring SR. The role of synovitis in osteoarthritis pathogenesis. Bone 2012;51:249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maglaviceanu A, Wu B, Kapoor M. Fibroblast‐like synoviocytes: role in synovial fibrosis associated with osteoarthritis. Wound Repair Regen 2021;29:642–9. [DOI] [PubMed] [Google Scholar]
- 5. Rim YA, Ju JH. The role of fibrosis in osteoarthritis progression. Life (Basel) 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mathiessen A, Conaghan PG. Synovitis in osteoarthritis: current understanding with therapeutic implications. Arthritis Res Ther 2017;19:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Endisha H, Rockel J, Jurisica I, et al. The complex landscape of microRNAs in articular cartilage: biology, pathology, and therapeutic targets. JCI Insight 2018;3:e121630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malemud CJ. MicroRNAs and osteoarthritis. Cells 2018;7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tavallaee G, Rockel JS, Lively S, et al. MicroRNAs in synovial pathology associated with osteoarthritis. Front Med (Lausanne) 2020;7:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L, Xing R, Huang Z, et al. Synovial fibrosis involvement in osteoarthritis. Front Med (Lausanne) 2021;8:684389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li YH, Tavallaee G, Tokar T, et al. Identification of synovial fluid microRNA signature in knee osteoarthritis: differentiating early‐ and late‐stage knee osteoarthritis. Osteoarthritis Cartilage 2016;24:1577–86. [DOI] [PubMed] [Google Scholar]
- 12. Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage 2007;15:1061–9. [DOI] [PubMed] [Google Scholar]
- 13. Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative–recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthritis Cartilage 2010;18 Suppl 3:S17–23. [DOI] [PubMed] [Google Scholar]
- 14. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to imageJ: 25 years of image analysis. Nat Methods 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Potla P, Ali SA, Kapoor M. A bioinformatics approach to microRNA‐sequencing analysis. Osteoarthr Cartil Open 2021;3:100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tokar T, Pastrello C, Rossos AE, et al. mirDIP 4.1‐integrative database of human microRNA target predictions. Nucleic Acids Res 2018;46:D360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kotlyar M, Pastrello C, Malik Z, et al. IID 2018 update: context‐specific physical protein‐protein interactions in human, model organisms and domesticated species. Nucleic Acids Res 2019;47:D581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rahmati S, Abovsky M, Pastrello C, et al. pathDIP 4: an extended pathway annotations and enrichment analysis resource for human, model organisms and domesticated species. Nucleic Acids Res 2020;48:D479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown KR, Otasek D, Ali M, et al. NAViGaTOR: Network analysis, visualization and graphing Toronto. Bioinformatics 2009;25:3327–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Y, Li S, Chen P, et al. MicroRNA‐27b‐3p inhibits apoptosis of chondrocyte in rheumatoid arthritis by targeting HIPK2. Artif Cells Nanomed Biotechnol 2019;47:1766–71. [DOI] [PubMed] [Google Scholar]
- 21. Akhtar N, Rasheed Z, Ramamurthy S, et al. MicroRNA‐27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum 2010;62:1361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kapoor M, McCann M, Liu S, et al. Loss of peroxisome proliferator–activated receptor γ in mouse fibroblasts results in increased susceptibility to bleomycin‐induced skin fibrosis. Arthritis Rheum 2009;60:2822–9. [DOI] [PubMed] [Google Scholar]
- 23. Hinz B, Celetta G, Tomasek JJ, et al. Alpha‐smooth muscle actin expression upregulates fibroblast contractile activity. Mol Biol Cell 2001;12:2730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schuster R, Rockel JS, Kapoor M, et al. The inflammatory speech of fibroblasts. Immunol Rev 2021;302:126–46. [DOI] [PubMed] [Google Scholar]
- 25. Vaamonde‐Garcia C, Malaise O, Charlier E, et al. 15‐Deoxy‐δ‐12, 14‐prostaglandin J2 acts cooperatively with prednisolone to reduce TGF‐β‐induced pro‐fibrotic pathways in human osteoarthritis fibroblasts. Biochem Pharmacol 2019;165:66–78. [DOI] [PubMed] [Google Scholar]
- 26. Qadri M, Jay GD, Zhang LX, et al. Proteoglycan‐4 regulates fibroblast to myofibroblast transition and expression of fibrotic genes in the synovium. Arthritis Res Ther 2020;22:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Pascalis C, Etienne‐Manneville S. Single and collective cell migration: the mechanics of adhesions. Mol Biol Cell 2017;28:1833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanwal M, Smahel M, Olsen M, et al. Aspartate β‐hydroxylase as a target for cancer therapy. J Exp Clin Cancer Res 2020;39:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhan YH, Luo QC, Zhang XR, et al. CELSR1 Is a positive regulator of endothelial cell migration and angiogenesis. Biochemistry (Mosc) 2016;81:591–9. [DOI] [PubMed] [Google Scholar]
- 30. De Kreuk BJ, Gingras AR, Knight JD, et al. Heart of glass anchors Rasip1 at endothelial cell‐cell junctions to support vascular integrity. Elife 2016;5:e11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aumailley M, Krieg T. Laminins: a family of diverse multifunctional molecules of basement membranes. J Invest Dermatol 1996;106:209–14. [DOI] [PubMed] [Google Scholar]
- 32. Mokkapati S, Bechtel M, Reibetanz M, et al. Absence of the basement membrane component nidogen 2, but not of nidogen 1, results in increased lung metastasis in mice. J Histochem Cytochem 2012;60:280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harman JL, Sayers J, Chapman C, et al. Emerging roles for neuropilin‐2 in cardiovascular disease. Int J Mol Sci 2020;21:5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamaji Y, Yoshida S, Ishikawa K, et al. TEM7 (PLXDC1) in neovascular endothelial cells of fibrovascular membranes from patients with proliferative diabetic retinopathy. Invest Ophthalmol Vis Sci 2008;49:3151–7. [DOI] [PubMed] [Google Scholar]
- 35. Pradhan AK, Maji S, Das SK, et al. MDA‐9/Syntenin/SDCBP: new insights into a unique multifunctional scaffold protein. Cancer Metastasis Rev 2020;39:769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Awwad K, Hu J, Shi L, et al. Role of secreted modular calcium‐binding protein 1 (SMOC1) in transforming growth factor β signalling and angiogenesis. Cardiovasc Res 2015;106:284–94. [DOI] [PubMed] [Google Scholar]
- 37. Gonzalez‐Loyola A, Petrova TV. Development and aging of the lymphatic vascular system. Adv Drug Deliv Rev 2021;169:63–78. [DOI] [PubMed] [Google Scholar]
- 38. Walker EC, McGregor NE, Poulton IJ, et al. Oncostatin M promotes bone formation independently of resorption when signaling through leukemia inhibitory factor receptor in mice. J Clin Invest 2010;120:582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takahata Y, Hagino H, Kimura A, et al. Smoc1 and Smoc2 regulate bone formation as downstream molecules of Runx2. Commun Biol 2021;4:1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morkmued S, Clauss F, Schuhbaur B, et al. Deficiency of the SMOC2 matricellular protein impairs bone healing and produces age‐dependent bone loss. Sci Rep 2020;10:14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pyott SM, Schwarze U, Christiansen HE, et al. Mutations in PPIB (cyclophilin B) delay type I procollagen chain association and result in perinatal lethal to moderate osteogenesis imperfecta phenotypes. Hum Mol Genet 2011;20:1595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Badshah, II , Brown S, Weibel L, et al. Differential expression of secreted factors SOSTDC1 and ADAMTS8 cause profibrotic changes in linear morphoea fibroblasts. Br J Dermatol 2019;180:1135–49. [DOI] [PubMed] [Google Scholar]
- 43. Omura J, Satoh K, Kikuchi N, et al. ADAMTS8 promotes the development of pulmonary arterial hypertension and right ventricular failure: a possible novel therapeutic target. Circ Res 2019;125:884–906. [DOI] [PubMed] [Google Scholar]
- 44. Zhu Z, Zhong L, Li R, et al. Study of osteoarthritis‐related hub genes based on bioinformatics analysis. Biomed Res Int 2020;2020:2379280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang Z, Xiao Z, Guo H, et al. Novel role of the clustered miR‐23b‐3p and miR‐27b‐3p in enhanced expression of fibrosis‐associated genes by targeting TGFBR3 in atrial fibroblasts. J Cell Mol Med 2019;23:3246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cui H, Banerjee S, Xie N, et al. MicroRNA‐27a‐3p is a negative regulator of lung fibrosis by targeting myofibroblast differentiation. Am J Respir Cell Mol Biol 2016;54:843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bielajew BJ, Hu JC, Athanasiou KA. Collagen: quantification, biomechanics, and role of minor subtypes in cartilage. Nat Rev Mater 2020;5:730–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aoqierbatu, Luo A , Shi Y, et al. Microarray analysis of hub genes and pathways in damaged cartilage tissues of knee. Medicine (Baltimore) 2021;100:e27183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu Y, Han YF, Ye B, et al. miR‐27b‐3p is involved in doxorubicin resistance of human anaplastic thyroid cancer cells via targeting peroxisome proliferator‐activated receptor γ. Basic Clin Pharmacol Toxicol 2018;123:670–7. [DOI] [PubMed] [Google Scholar]
- 50. Shen SJ, Song Y, Ren XY, et al. MicroRNA‐27b‐3p promotes tumor progression and metastasis by inhibiting peroxisome proliferator‐activated receptor γ in triple‐negative breast cancer. Front Oncol 2020;10:1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jennewein C, von Knethen A, Schmid T, et al. MicroRNA‐27b contributes to lipopolysaccharide‐mediated peroxisome proliferator‐activated receptor γ (PPARγ) mRNA destabilization. J Biol Chem 2010;285:11846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu HY, Bai WD, Wang HT, et al. Peroxisome proliferator‐activated receptor‐γ agonist inhibits collagen synthesis in human keloid fibroblasts by suppression of early growth response‐1 expression through upregulation of miR‐543 expression. Am J Cancer Res 2016;6:1358–70. [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao J, Ouyang Q, Hu Z, et al. A protocol for the culture and isolation of murine synovial fibroblasts. Biomed Rep 2016;5:171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rosengren S, Boyle DL, Firestein GS. Acquisition, culture, and phenotyping of synovial fibroblasts. Methods Mol Med 2007;135:365–75. [DOI] [PubMed] [Google Scholar]
- 55. Seidel MF, Koch FW, Vetter H. Macrophage‐like synoviocytes display phenotypic polymorphisms in a serum‐free tissue‐culture medium. Rheumatol Int 2006;26:244–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disclosure Form
Supplementary Table 1 Description of demographic and anthropometric variables, and experimental use of synovial samples from patients exhibiting KL1/KL2 or KL3/KL4 radiographic OA.
Supplementary Table 2 Primers used for RT‐qPCR.
Supplementary Table 3 Genes significantly upregulated or downregulated (p < 0.05), or not significantly changed (p > 0.05) by miR‐27b‐3p mimic‐treated compared to control mimic‐treated OA FLS, as determined by RT‐qPCR array. Genes undedected by array analysis are also indicated.
Supplementary Table 4 Lists of significantly upregulated genes or significantly downregulated genes from miR‐27b‐3p mimic‐treated compared to control mimic‐treated OA FLS, as determined by RNA sequencing.
Figure S1 Supplementary Figure
Figure S2 Supplementary Figure
Figure S3 Supplementary Figure
Figure S4 Supplementary Figure
Figure S5 Supplementary Figure
Figure S6 Supplementary Figure
Figure S7 Supplementary Figure
Figure S8 Supplementary Figure
Figure S9 Supplementary Figure
Figure S10 Supplementary Figure
Figure S11 Supplementary Figure
Appendix S1 Supplementary Methods


