Abstract
Alternative splicing of pre-mRNAs plays a critical role in maximizing the coding capacity of the small parvovirus genome. The small-intron region of minute virus of mice (MVM) pre-mRNAs undergoes an unusual pattern of overlapping alternative splicing—using two donors (D1 and D2) and two acceptors (A1 and A2) within a region of 120 nucleotides—that determines the steady-state ratios of the various viral mRNAs. In this report, we show that the determinants that govern excision of the small intron are complex and are also required for efficient definition of the upstream exon. For the MVM small intron in its natural context, the two donors appear to compete for the splicing machinery: the position of D1 favors its usage, while the primary sequence of D2 must be more like the consensus sequence than is D1 to be used efficiently. We have genetically defined the branch points that are used for generation of the major and minor spliced forms and show that recognition of components of the small-intron acceptors is likely to be the dominant determinant in alternative small-intron excision. We have also identified a G-rich intronic enhancer sequence within the small intron that is essential for splicing of the minor form (D2 to A2) but not the major form (D1 to A1) of MVM mRNAs and is required for efficient definition of the upstream NS2-specific exon. In its natural context, the small intron appears to be excised by a mechanism consistent with intron definition. When the MVM small intron is expanded, various parameters of its excision are altered, indicating that critical cis-acting signals are context dependent. Relative use of the donors and acceptors is altered, and the upstream NS2-specific exon is no longer efficiently defined. The fact that definition of the upstream NS2-specific exon can be achieved by the MVM small intron in its natural context, but not when it is expanded, suggests that the multiple determinants that govern definition and excision of the small intron are required, in concert, for upstream exon definition. Our data are consistent with a model in which alternative splicing of the MVM P4-generated pre-mRNAs is governed by a hybrid of intron- and exon-defining mechanisms.
Splicing of mRNA precursors (pre-mRNAs) plays a fundamental role in gene regulation and is a critical mechanism for the generation of genetic diversity in many eukaryotic and viral systems (6). Splicing of complex pre-mRNAs is complicated by the fact that cis-acting splicing signals which define intron and exon borders are short and often poorly conserved. The splicing machinery must accurately discriminate between exons and introns while maintaining enough flexibility to allow for alternative splicing. In vertebrates, the primary unit of recognition by the splicing machinery is thought to be the exon (1). Vertebrate exons are usually small with respect to vertebrate introns. Current evidence suggests that multiple weak interactions between splicing factors and sequence elements in and around vertebrate exons cause pairs of splice sites to be recognized across exons rather than introns (1, 21). Very small vertebrate introns, however, appear to be recognized by interactions which span the intron, but this is currently poorly understood (12).
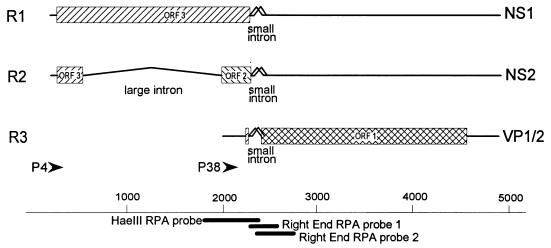
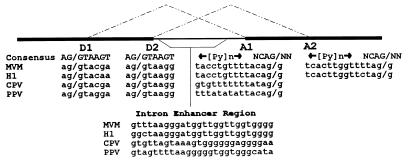
The parvovirus minute virus of mice (MVM) generates three classes of mature mRNA during infection (Fig. 1). mRNAs generated from the promoter at map unit 4 (R1 and R2) code for two viral nonstructural proteins, NS1 and NS2, while mRNAs generated by a promoter at map unit 38 encode the viral capsid proteins VP1 and VP2 (3). Maintenance of the relative steady-state levels of these messages is critical to the parvovirus life cycle (20). A small, downstream, overlapping intron is located between map units 44 and 46 and is common to pre-mRNAs generated from both promoters (Fig. 1) (19). This small intron is very unusual in that it is extremely small (82 to 119 nucleotides [nt]) and utilizes two donors, D1 and D2, at nt 2280 and 2317, respectively, and two acceptors, A1 and A2, at nt 2377 and 2399, respectively, to produce three species of spliced RNAs from each transcript class, resulting in nine steady-state mRNAs produced during MVM infection (10, 14, 19, 24). The major spliced product is detected in approximately 75 to 80% of the spliced molecules of each message class and joins D1 to A2 (Fig. 1 and 2a) (14, 19, 24). The minor spliced product is detected in approximately 20% of the spliced molecules of each message class and joins D2 to A2 (Fig. 1 and 2a) (14, 19, 24). The rare spliced product is detected in fewer than 5% of the spliced molecules of each message class and joins D1 to A2 (Fig. 1 and 2a) (24). The fourth potential splicing pattern, D2 to A2, is not detected in vivo (2, 10, 24), presumably because the distance between these sites (60 nt) is below the minimum distance at which mammalian introns are efficiently spliced (20). Both donors of the small intron are very similar to the consensus donor sequence (Fig. 2a) and differ from each other at only two positions. Both acceptors have well-conserved AG cleavage sites but poor polypyrimidine tracts (PPTs) with several purine interruptions much like those found in lower eukaryotes. Determinants which govern the very unusual alternative splicing pattern of this intron have not been addressed until now.
FIG. 1.
Genetic map of MVM. The three major transcript classes and protein-encoding open reading frames (ORFs) are shown. The two promoters are indicated by arrows, and the large and small introns are indicated. The thick lines at the bottom indicate approximate locations for the three probes used for RNase protection assays as described fully in Materials and Methods.
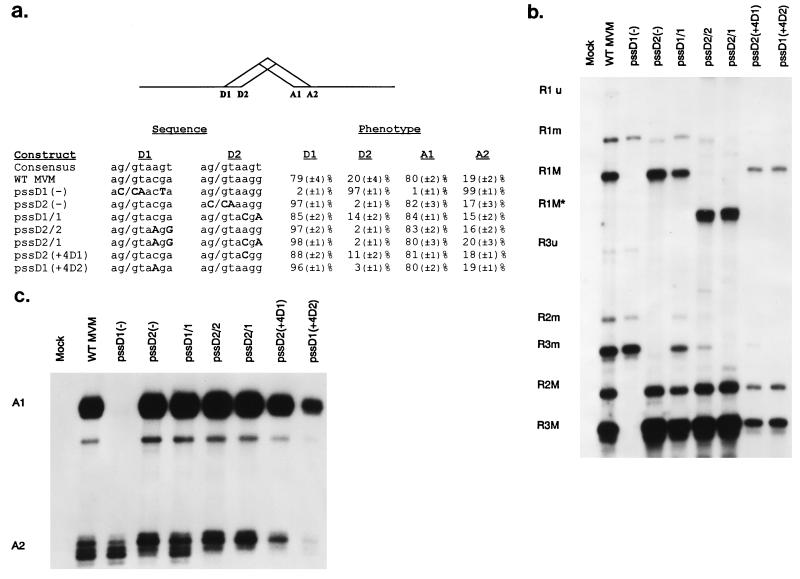
FIG. 2.
Although the primary sequence of donor 2 is more consensus, the position of donor 1 is preferred by the splicing machinery. (a) The sequences of a consensus donor and the wild-type and mutant MVM donors (changes are in bold, capital letters) are shown, with quantitations of donor and acceptor usage at each donor and acceptor position for each. Relative donor and acceptor position usage values were calculated as a percentage of total spliced products and are the average of at least four separate experiments, except the values of pssD2(+4D1) and pssD2(+4D1), which are the average of three. Standard deviations are indicated in parentheses. (b) A representative RNase protection assay detecting donor usage of RNAs generated by wild-type MVM and the donor mutants is shown. The identities of the protected bands are at the left. As described in Materials and Methods, the pssD2/2 and pssD2/1 mutants were analyzed with an RNase protection probe spanning nt 1886 to 2350 which generated R1-protected fragments (designated R1M*) shorter than those generated by the HaeIII probe (nt 1852 to 2378). (c) A representative RNase protection assay detecting acceptor usage of RNAs generated by wild-type MVM and the donor mutants, with right-end RPA probe 1, is shown (as described in Materials and Methods). The identities of the protected bands are at the left. Multiple minor bands within the A1 and A2 bands are likely due to incomplete RNase digestion; the prominent band running slightly below the A1 band is a breakdown product of A1 that is seen inconsistently. WT, wild type.
A large upstream intron, located between map units 10 and 39, is present in the pre-mRNAs generated from the promoter at map unit 4. This intron utilizes a nonconsensus donor at nt 514 (AA/GCAAGT) and has a poor PPT at its 3′ splice site which overlaps the TATA element of the capsid gene promoter at map unit 38. This large intron is either retained (in the R1 message class) or excised (in the R2 message class) in mature mRNA at a ratio of 1:2, respectively (20). Previous work in our laboratory has demonstrated that efficient excision of this large intron requires elements contained in the downstream intron (31). The small intron appears to play a primary role in efficient excision of the large intron, perhaps as the initial entry site for elements of the spliceosome which help to define the intervening exon (20, 31). Improvement of the PPT of the upstream large intron relieves this requirement, and prior excision of the downstream small intron is unnecessary (31). These results suggest that the large intron is spliced via an exon-defined pathway (1).
In this study, we have performed extensive mutational analysis of the small intron in order to investigate cis-acting factors which control alternative splicing of the small intron and, to a lesser extent, alternative splicing of the large intron. Our results show that complex interactions of both donor and acceptor sequences, as well as size constraints, define and govern alternative small-intron splicing. We have also identified an intronic splicing enhancer sequence which appears to function as both an intron- and an exon-defining element. Further, our results show that in P4-generated MVM pre-mRNAs, definition of the small intron is required for efficient definition of the upstream NS2-specific exon. Our data are consistent with a model in which alternative splicing of MVM P4-generated pre-mRNAs is governed by a composite of intron- and exon-defining mechanisms.
MATERIALS AND METHODS
Plasmid constructs.
Wild-type MVM is an infectious plasmid clone of MVM and has been described previously as pMVM (29).
(i) Donor mutations.
Construction of pMVM, pssD1(−), and pssD2(−) has been described previously (29). pssD1/1, pssD2/2, pssD2/1, pssD2(+4 D1), and pssD1(+4 D2) were constructed by m13-based oligonucleotide mutagenesis as previously described (31). Mutant oligonucleotides varied between 23 and 33 nt in length and were homologous to the viral DNA except at the nucleotides which were to be altered. The changes made in these mutants are shown in Fig. 2a.
(ii) Acceptor mutations.
pssPBP-1 knockout (ko), pssPBP-2 ko, pssPBP-3 ko, pssPBP1/2 ko, pssPBP1/3 ko, pssA1-PPT ko, and pssA2-PPT ko were constructed by PCR as described by Innis et al. (9). Briefly, two PCR products (right and left) containing the desired mutation(s) at the 3′ and 5′ ends, respectively, were generated such that they overlapped at their 3′ and 5′ ends. Four oligonucleotides were used for these reactions: two complementary mutagenic oligonucleotides which were homologous to the viral DNA except for the desired nucleotide alteration(s) (“inside primers”) and two external oligonucleotides A and B which contained no mutations (“outside primers”). The external A oligonucleotide was used with one of the inside primers which was homologous to the bottom strand of MVM DNA to generate the 5′-most PCR product, and the B oligonucleotide was used with one of the inside primers which was homologous to the top strand of MVM DNA to generate the 3′-most PCR product. Next, both PCR products were annealed to create a heteroduplex containing the desired mutation and reamplified with only the outside primers to create a PCR fragment containing the desired mutation. From this fragment, a smaller fragment containing the mutation was subcloned back into the pMVM plasmid by standard techniques. The inside primer sets varied in length from 26 to 38 nt and were homologous to the viral DNA except at the nucleotides which were to be altered. The changes made in these mutants are shown in Fig. 3a. Outside primer A spanned MVM 2028 to 2053. Outside primer B spanned MVM 4081 to 4057.
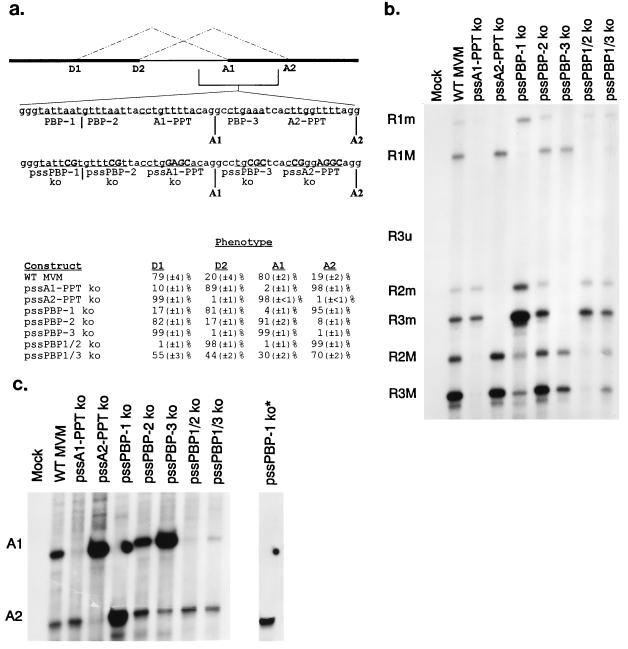
FIG. 3.
A complex arrangement of cis-acting acceptor components governs alternative small-intron acceptor usage. (a) The location and sequences of small-intron acceptor components, mutant sequences (changes are shown below in bold, capital letters), and quantitations of donor and acceptor usage for RNAs generated by each are shown. Relative donor and acceptor usage values were calculated as a percentage of total spliced products and are the average of at least three separate experiments (except the value for pssPBP1/3 ko, which is the average of two). Standard deviations are indicated in parentheses. Mutants are described fully in Materials and Methods. (b) A representative RNase protection assay detecting donor usage of RNAs generated by wild-type MVM and acceptor mutants with the HaeIII RPA probe is shown. The identities of the protected bands are designated on the left. (c) A representative RNase protection assay detecting acceptor usage of RNAs generated by wild-type MVM and acceptor mutants with the right-end RPA 2 probe is shown. The lane to the extreme right, pssPBP-1 ko*, is a shorter exposure of the pssPBP-1 ko lane, showing no protected fragment at A1. The identities of the protected bands are at the left. WT, wild type.
(iii) Intronic enhancer mutations.
pMVM(2322–2348) has been previously described. pIES-ko (1) was constructed as follows: pAfl-2(2324), constructed by m13-based oligonucleotide mutagenesis as described above, was linearized with AflII and then partially digested with AseI. Single-stranded, complementary oligonucleotides, containing the mutations shown in Fig. 4a and including AflII and AseI overhangs, were annealed and cloned between the AflII (2324) and AseI (2350) sites of pAfl-2. pIES-ko (3) contained three copies of the annealed oligonucleotides concatemerized in a head-to-tail-to-head arrangement.
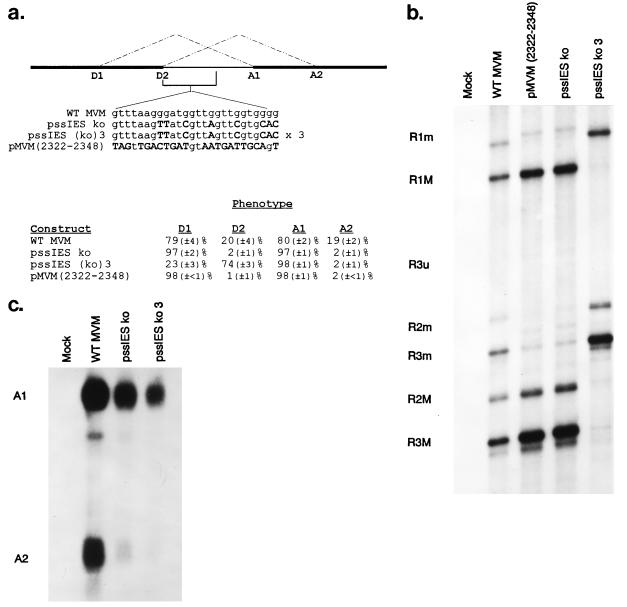
FIG. 4.
A G-rich intronic enhancer is required for generation of the major but not the minor spliced form. (a) The location and sequence of the wild-type G-rich intronic enhancer and mutant sequences (changes are shown in bold, capital letters) and quantitations of donor and acceptor usage for RNA generated by each are shown. Relative donor and acceptor usage values were calculated as a percentage of total spliced products and are the average of at least four separate experiments. Standard deviations are indicated in parentheses. (b) A representative RNase protection assay detecting donor usage of RNAs generated by wild-type MVM and IES mutants with the HaeIII RPA probe is shown. The identities of the protected bands are at the left. (c) A representative RNase protection assay detecting acceptor usage of RNAs generated by wild-type MVM and IES mutants with right-end RPA probe 1 is shown. The identities of the protected bands are designated on the left. Multiple minor bands within the A1 and A2 bands are likely due to incomplete RNase digestion; the prominent band running slightly below the A1 band in the wild-type lane is a breakdown product of A1 that is seen inconsistently. WT, wild type.
(iv) Intron expansion mutations.
pAfl-2(+217) was made by inserting a fragment spanning MVM 543 to 760 into the engineered AflII site (2324) by standard techniques. pAfl-2(+1344) was made by inserting a fragment spanning MVM 543 to 1886 into the engineered AflII site by standard techniques. The 543 to 760 and 543–1886 fragments, from the MVM large-intron region, were selected for their apparent lack of cryptic splice sites based upon data gathered in our lab.
(v) Other mutations.
pNS2-1989 (15) and pD1/2(−) (30) have been previously described. All mutations used in this study were sequenced to verify the changes that were made.
Cells, transfections, and RNA isolation.
Murine A92L cells, the normal tissue culture host of MVM(p), were grown and transfected with wild-type and mutant plasmids by using DEAE-dextran as described previously (31). RNA was typically isolated 48 h posttransfection, after lysis in guanidinium hydrochloride and centrifugation through CsCl exactly as previously described (31).
RNA analysis. (i) RNase protection.
RNase protection assays were performed on 10 μg of total RNA generated by wild-type MVM or mutant constructs 48 h posttransfection with homologous probes, as previously described (31). The probes used for RNase protection which detected small-intron donor usage were [α-32P]UTP-labeled Sp6-generated antisense MVM RNAs from nt 1852 to 2378 (corresponding to an MVM HaeIII restriction fragment [Fig. 1]) or [α-32P]UTP-labeled Sp6-generated antisense MVM RNA from nt 1886 to 2350 of pssD2(+4D1) and pssD1(+4D2) (Fig. 2, lanes 6 and 7). These probes extend from before the acceptor site of the large intron to within the small intron common to all MVM pre-mRNAs and identify RNA species with either of the small-intron donors (24). Unspliced P4 and unspliced P38 products are designated R1u and R3u, respectively. R1, R2, and R3 RNAs utilizing D1 at 2280 are designated R1M, R2M, and R3M, respectively. R1, R2, and R3 RNAs utilizing D2 at 2317 are designated R1M, R2M, and R3M, respectively. For analysis of RNA produced by other mutant constructs, HaeIII probes homologous to the mutants being analyzed were used. The probe used for RNase protection which detected acceptor usage, shown in Fig. 2c, 4c, and 5c, was [α-32P]UTP-labeled, T7-generated antisense MVM RNA from nt 2288 to 2505 (Fig. 1, right-end RPA probe 1). This probe extends from just after the small-intron D1 to well beyond A2 of the MVM small intron and distinguishes processed RNAs which use A1 from those which use A2. The probe used for RNase protection which detected acceptor usage in Fig. 3c was [α-32P]UTP-labeled SP6-generated antisense MVM RNA from nt 2350 to 2652 (Fig. 1, right-end RPA probe 2). This probe extends from the center of the MVM small intron to well beyond A2 of the MVM small intron and distinguishes processed RNAs which use A1 from those which use A2. For analysis of RNA produced by constructs which contained mutations between A1 and A2, homologous probes were used. RNase protection assays were analyzed on a Betagen B-scanning phosphorimage analyzer, and molar ratios of MVM RNA were determined by standardization to the number of uridines in each protected fragment.
FIG. 5.
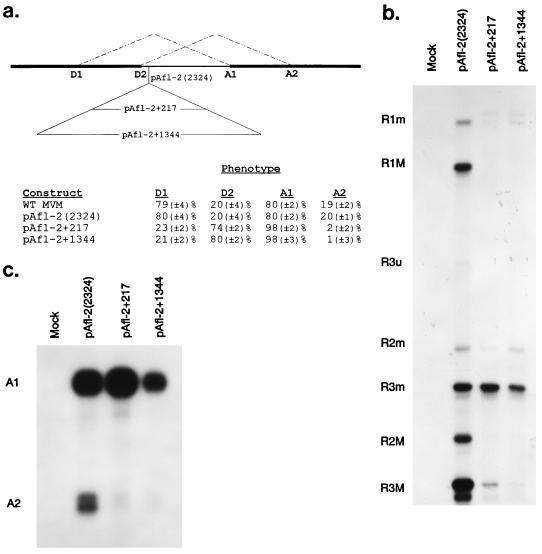
Expanding the small intron alters the relative usage of the donors and acceptors. (a) The location and sizes of the insertions used to expand the small intron as well as the parent pAfl-2(2324) mutant (as described fully in Materials and Methods) are shown. Quantitations of donor and acceptor usage for RNAs generated by wild-type (WT) MVM and each expansion construct are also shown. Relative donor and acceptor usage values were calculated as a percentage of total spliced products and are the average of at least four separate experiments. Standard deviations are indicated in parentheses. (b) A representative RNase protection assay detecting donor usage for intron expansion constructs from RNAs generated by the pAfl-2(2324) parent construct and expansion constructs with the HaeIII RPA probe is shown. Splicing of the pAfl-2(2324) parent construct is indistinguishable from wild-type MVM. The identities of the protected bands are at the left. (c) A representative RNase protection assay detecting acceptor usage for RNAs generated by the pAfl-2(2324) parent construct and expansion constructs with right-end RPA probe 1 is shown. The identities of the protected bands are at the left. Splicing of the pAfl-2(2324) parent construct is indistinguishable from wild-type MVM. Multiple minor bands within the A1 and A2 bands are likely due to incomplete RNase digestion.
(ii) Reverse transcription (RT)-PCRs.
First-strand cDNA synthesis used 5 μg of total RNA isolated after transfection and oligonucleotide T primer by standard techniques (9). PCR detection of R2 and exon-skipped products was performed with primers described previously (30).
RESULTS
Although the primary sequence of donor 2 is preferred by the splicing machinery, the position of donor 1 is preferred in the context of the wild-type small intron.
Since the two small-intron donors have different sequences, we first examined whether these sequence differences play a role in alternative 5′ splice site selection. When D1 was debilitated by point mutation [Fig. 2a, pssD1(−)], only the minor spliced form (D2/A2) was detected (Fig. 2). When D2 was debilitated by point mutation [Fig. 2a, pssD2(−)], only the major spliced form (D1/A1) was detected (Fig. 2). In neither case did we observe a compensating increase in the relative levels of unspliced RNA or detect usage of cryptic donors or acceptors (data not shown). Thus, the preferential use of D1 relative to D2 during small-intron excision does not appear to be because the sequence of D2 is intrinsically deficient. In addition, although our results address the issue only indirectly, it appears that production of the rare spliced form (D1/A2) may occur significantly less than 5% of the time.
When constructs were made in which both donor positions had the primary sequence of D1 (Fig. 2a, pssD1/1), the relative usage of both donors and acceptors remained similar to that for the wild type; although there was a slight increase in the use of the donor in the upstream position, there was still significant use of the donor in the downstream position (Fig. 2). However, in constructs in which both donor positions were given the primary sequence of donor 2 (Fig. 2a, pssD2/2), the donor in the upstream position was used almost exclusively, while acceptor usage remained relatively unchanged (Fig. 2). In addition, when the primary sequences of the two donors were switched, such that the sequence of the upstream donor site was replaced with the sequence normally used for D2, and vice versa (Fig. 2a, pssD2/1), the upstream donor was again used almost exclusively, while the acceptor usage remained unchanged (Fig. 2). In no case did we detect a compensating increase in the relative abundance of unspliced RNA or the usage of cryptic donors or acceptors (data not shown). These results suggest that while the primary sequence of donor 2 is preferred by the splicing machinery, the relative position of donor 1 is preferred in the context of the wild-type small intron. Further, they imply that to achieve wild-type levels of the minor spliced form (D2/A2), the downstream donor position must have a donor sequence that is more like the consensus sequence. In addition, there is likely another determinant that facilitates generation of the minor spliced form because, when both donors were given the primary sequence of D1, significant production of the minor form was observed. Finally, our results suggest that acceptor usage is not governed by relative donor strengths because the A1/A2 ratio remains relatively unchanged unless D1 is destroyed.
We noted that D1 is nonconsensus at positions 4 and 6 in the intron, while D2 is unlike the consensus sequence only at position 6 (Fig. 2a). In addition, while position 4 of D2 is conserved among other, similar parvoviruses (canine parvovirus [CPV], porcine parvovirus [PPV], and H1), position 4 of D1 is not (see Fig. 7), suggesting that this position might play a pivotal role in the relative strength of the two donors. Making only position 4 in the primary sequence of D1 consensus [Fig. 2a (C to A), pssD1(+4D2)] had the same effect as changing the entire primary sequence of D1 to that of D2; i.e., D1 was used almost exclusively, while acceptor usage was relatively unchanged (Fig. 2). Furthermore, changing position 4 of D2 to nonconsensus [Fig. 2a (A to C), pssD2(+4D1)] had the same effect as changing the entire primary sequence of D2 to that of D1, causing a slight increase in usage of D1 with no alteration in acceptor usage (Fig. 2). Finally, altering position 6 of either donor had no appreciable effect on relative donor usage (data not shown). In no case did we detect an increase in the relative abundance of unspliced RNA or usage of cryptic donors or acceptors (data not shown). We conclude that homology to the consensus donor sequence at position 4 in the intron, which is known to interact with the U1 snRNA, is critical for the apparent preference of the splicing machinery for the primary sequence of D2.
FIG. 7.
The small-intron region of related parvoviruses contains sequence elements similar to those of the small-intron region of MVM. Sequence elements of the small-intron region of MVM are compared to those of related parvoviruses. CPV and PPV utilize a single acceptor.
Definition of small-intron acceptor components.
We next wished to locate the small intron’s acceptor components and determine the role which they played in its alternative splicing. Three potential branch point sequences (Fig. 3a) can be identified based on close homology to the weak mammalian branch point consensus sequence (16). Two of these putative branch point sequences (PBP1 and PBP2) are located upstream of the A1 cleavage site, and the other (PBP3) lies between A1 and A2 (Fig. 3a). While branch points in mammalian introns can diverge widely from the consensus sequence, and cryptic branch points can be activated when authentic branch points are mutated, we believed the small size of the intron and the lack of additional PBPs within the intron would permit the use of a mutational approach to identify and characterize these signals.
When PBP1 was destroyed by point mutation (Fig. 3a, pssPBP-1 ko), splicing to A1 was dramatically reduced. Pre-mRNAs generated by this construct predominantly used the D2/A2 pair (Fig. 3), and there was no compensating increase in the relative abundance of unspliced RNA (data not shown). There was, however, a significant level of detectable splicing from D1 which appeared to be joined to A2 (Fig. 3). Joining of D1 to A2 is normally observed at low frequency during a viral infection (previously described as the rare spliced form [14, 19, 24]). When PBP2 was destroyed by point mutation (Fig. 3a, pssPBP-2 ko), small-intron excision was essentially unaltered; however, there was a slight, reproducible increase in the major (D1/A1) spliced form and a concomitant small decrease in the minor (D2/A2) spliced form (Fig. 3). When both PBP1 and PBP2 were destroyed by point mutation (Fig. 3a, pssPBP1/2 ko), there was no longer detectable usage of A1 or D1; D2 and A2 (minor spliced form) were used exclusively (Fig. 3), without a compensating increase in the relative abundance of unspliced RNA or in the usage of cryptic donors or acceptors (data not shown). When PBP3 was destroyed by point mutation (Fig. 3a, pssPBP-3 ko) neither D2 nor A2 was used and the minor and rare spliced forms were no longer detectable (Fig. 3). We conclude from these results that BP-1 is the primary branch point for A1 and PBP3 is the primary branch point for A2. When both PBP1 and PBP3 were destroyed by point mutation (Fig. 3a, pssPBP1/3 ko), both donors and acceptors were used, with a slight preference for D1 and A2 (Fig. 3). Thus, under these experimental conditions, A2 appeared to be able to use PBP2 to make the rare (D1/A2) spliced form. PBP3 apparently cannot be utilized to make the rare (D1/A2) spliced form, since no D1 usage was detected when both PBP1 and PBP2 were destroyed.
Both small-intron acceptors have what appear to be poor PPTs. Both are very short and have several purine interruptions (Fig. 3a). A PPT of at least five consecutive uridines or nine consecutive pyrimidines is normally a critical part of the 3′ splice site in mammalian introns (23) but does not appear to be a required feature in lower eukaryotic (4) or minor class 3′ splice sites (5). The requirement for PPTs in the excision of small vertebrate introns has not been well characterized. Kennedy and Berget (11) recently determined that pyrimidine tracts between the 5′ splice site and the branch point sequence, rather than between the branch point sequence and the 3′ splice site, may be required for small-intron recognition, suggesting that the PPT requirement in small introns may be different from that in large introns. Therefore, to gain a clearer picture of the cis-acting acceptor components of the MVM small intron, the PPTs, associated with each acceptor, were modified (Fig. 3a). When the PPT of A1 was made highly purine rich, the usage of A1 was no longer detectable and there was a dramatic decrease in D1 usage with a concomitant increase in D2 usage (Fig. 3, pssA1-PPT ko) but no compensating increase in the relative abundance of unspliced RNA (data not shown). There also appeared to be an increase in the abundance of the rare (D1/A2) spliced form, suggesting that the rare (D1/A2) spliced form utilizes the A2 PPT. When the PPT of A2 was made highly purine rich, the usage of A2 was no longer detectable, and there was an increase in D1 usage, no detectable D2 usage (Fig. 3, pssA2-PPT ko), and no compensating increase in the relative abundance of unspliced RNA (data not shown). Thus, the A1 PPT appears to be required for efficient splicing to A1, and the A2 PPT appears to be required for efficient splicing to A2. Further, the presence of the A1 PPT in its wild-type position is not able to compensate for the loss of the A2 PPT. We conclude that although both A1 and A2 PPTs appear to be unlike the consensus sequence, both are required for proper function of the MVM small-intron acceptors. Our results also show that although small-intron donor mutations have relatively small effects on acceptor usage, acceptor mutations have significant effects on relative donor usage.
A G-rich sequence located between D2 and PBP1 of the small intron is required for production of the minor (D2/A2) but not the major (D1/A1) spliced form.
The sequence between D2 and PBP1 in MVM pre-mRNAs is over 40% G (Fig. 4a), and the G-rich nature of this area is conserved among all similar parvoviruses (although in the DNA this region is a strong binding site for the viral NS1 protein). Recently, several intronic G-rich sequences which act as splicing enhancer elements for alternatively spliced exons have been identified (12, 13, 25). When this region of the MVM small intron was altered to reduce its G-rich nature [Fig. 4a, pssIES ko, pMVM(2322–2348)], there was no detectable decrease in the total amount of steady-state accumulated MVM RNAs; yet, the minor spliced form (D2/A2) was no longer detectable (Fig. 4). Surprisingly, generation of the major spliced form (D1/A1) was not adversely affected by alteration of the G-rich sequence but, rather, seemed to compensate for the loss of the minor (D2/A2) spliced product. We conclude that the area between nucleotides 2322 and 2348, although dispensable for generation of the major spliced form, is required for efficient generation of the minor spliced form and therefore may be a member of the group of intron-splicing enhancers (12, 13, 25). Thus, both greater homology of D2 to the consensus donor sequence and the presence of this G-rich intronic enhancer are required for efficient generation of the minor spliced form of MVM mRNAs (D2/A2). Interestingly, we noted that when the intronic enhancer sequence (IES) is replaced by an insert that also enlarged the small intron by 60 nt [Fig. 4a, pssIES (ko) 3], D2 was joined primarily to A1 (Fig. 4), suggesting that expansion of the small intron leads to a change in the relative use of the small-intron donors and acceptors, as addressed more fully below.
When the small intron is expanded, relative use of the donors and acceptors is altered.
We next examined the role that the extremely small size of the small intron plays in its alternative splicing. We created a unique restriction site just downstream of D2 which by itself did not alter splicing of the small intron [Fig. 5, pAfl-2(2324)]. Then we made insertions of several different sizes at this site (Fig. 5a) with portions of the large intron that did not appear to contain regulatory elements or cryptic splice sites. These expansions separate the IES, identified above, from its wild-type position adjacent to D2. These mutant constructs generated dramatically decreased levels of both R1 and R2 and, for this reason, donor usage analysis is predominantly for R3. We found that when the small intron was expanded by an arbitrary 247 nt, splicing was dramatically shifted to the internal (D2/A1) donor/acceptor pair (Fig. 5, pAfl-2+217). When the small intron was expanded by 1,344 nt, the shift was even more pronounced (Fig. 3, pAfl-2+1344). In addition, as described above, when the intronic enhancer region was debilitated by mutation that also expanded the intron by 60 nt [Fig. 4a, pssIES ko (3)], splicing was also shifted toward the internal donor/acceptor pair (Fig. 4). We draw several conclusions from these experiments. First, these results are consistent with our previous conclusion that the more consensus primary sequence of D2 is preferred by the splicing machinery. Second, they support the hypothesis that A1 is the intrinsically stronger acceptor. Third, they imply that D2 is used at lower efficiency in the wild type because of a constraint related to the small size of the intron. Perhaps because it is too close to the best acceptor, A1, D2 used the poorer acceptor (A2). Our results also suggest that, since D2 functions well when the enhancer region is far removed, in the context of the wild-type small intron, the G-rich enhancer may somehow act to improve A2, facilitating production of the minor spliced form. Expansion of the small intron also resulted in low steady-state levels of R1 and R2 (Fig. 5b; note the lack of R2). This result suggested that the P4 products that were generated may have skipped the internal NS2-specific exon, implying that the size of the small intron may be critical for facilitation of large-intron splicing by the small intron.
Complex interactions with components of the small intron affect definition of the upstream exon.
We have previously shown that the small intron facilitates definition of the intervening NS2-specific exon and excision of the upstream large intron by overcoming the poor PPT at the large-intron 3′ splice site (31). For exon-defined systems, it is postulated that initial splice site recognition is accomplished through multiple weak interactions within and around the exon (1, 21). Thus, strong downstream donors can often compensate for a poor upstream 3′ splice site in the definition of an intervening exon (1, 22). Efficient excision of the large, upstream intron in MVM P4-generated pre-mRNAs, however, requires more than just a downstream donor; at least the small-intron internal donor and acceptor pair and the intervening sequence are required (29).
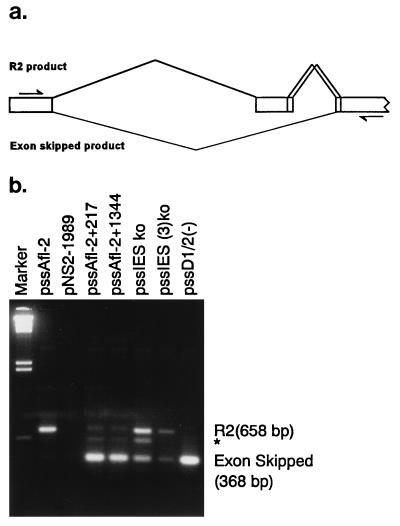
As can be seen in Fig. 5, when the small intron was expanded, the levels of R1 and R2 were dramatically reduced. RT-PCR analysis, shown in Fig. 6, demonstrated that the upstream NS2-specific exon was no longer well defined in RNAs generated by small-intron expansion mutants. A portion of the P4-generated pre-mRNAs joined the large-intron donor to the small-intron acceptor, A1, skipping the intervening exon [the parent construct, pAfl-2(2324), does not generate exon-skipped product (data not shown)]. Thus, wild-type levels of excision of the small intron are, in the context of P4-generated pre-mRNAs, dependent upon its small size (when it is expanded, a portion of the P4 product exon skips) and imply that excision of the wild-type small intron is governed by interactions contained wholly within the intron, i.e., via an intron-defined model as has been suggested for small introns (27). These results are also consistent with a model in which the wild-type small intron is required, as a unit, to overcome the poor upstream 3′ splice site and efficiently facilitate intervening exon definition and large-intron excision.
FIG. 6.
When the small intron is expanded or the IES is disrupted, the upstream NS2-specific exon is poorly defined. (a) Diagram of R2 and the exon-skipped splicing patterns of P4-generated pre-mRNAs. The normal R2 product (top) includes the NS2-specific exon. The exon-skipped product (bottom) skips the NS2-specific exon and joins the large-intron donor to the small-intron acceptor A1. The arrows designate the approximate positions of the PCR primers used. (b) A representative RT-PCR analysis detecting R2 (658 bp) and exon-skipped (368 bp) products (described fully in Materials and Methods) for RNAs generated by wild-type MVM and mutant constructs (as shown in Fig. 4a and 5a) is shown. The minor band designated by an asterisk has been characterized previously (29) and utilizes a cryptic donor site within the NS2-specific exon. The marker is a HindIII digest of λ DNA.
As mentioned above, previous results in our lab suggested that the sequence between D2 and A1 is required for efficient excision of the upstream intron (29); in the absence of these sequences, the NS2-specific exon is poorly defined (30). Therefore, we suspected that the IES, in its natural context, may play a role in definition of the upstream exon and concomitant excision of the large intron. Further examination of IES mutants showed this to be the case. When the IES was altered by point mutation (Fig. 4A, pssIES ko), the NS2-specific exon was less well defined, and both skipping and retention of the R2-specific exon were detected (Fig. 6). In addition, when the IES was altered by mutations which also enlarged it, the small intron was enlarged by 60 nt (Fig. 4a, pssIES ko [3]). Exon-skipped products were also detectable (Fig. 6). This result suggests that, in the context of the wild-type small intron, at least three elements within the small intron are required for both small-intron excision and efficient upstream-exon definition: the donors, the acceptors, and the IES. Since expansion of the intron causes both exon skipping and preferential usage of D2, the role of each of these elements appears to be highly context sensitive.
DISCUSSION
Pre-mRNAs of the autonomous parvovirus MVM undergo alternative splicing which determines the relative ratio of the accumulated levels of viral mRNAs and, in turn, critically influences the relative steady-state levels of the essential viral nonstructural and capsid proteins. No viral proteins participate in this process, which must, therefore, be accomplished solely by the interactions between cellular factors and viral cis-acting elements (20).
The small intron of MVM undergoes a very unusual alternative splicing pattern utilizing two donors and two acceptors to generate three spliced isoforms. Alternative splicing of P38-generated R3 products results in mRNAs that encode the two capsid proteins VP1 and VP2. Alternative splicing of the small intron from P4-generated pre-mRNAs results in R2 mRNAs that encode the three different isoforms of NS2 (20). Further, it is now clear that the small intron plays a pivotal role in excision of the upstream large intron, which determines the relative accumulated ratios of R1 and R2, which encode the viral nonstructural proteins NS1 and NS2, respectively.
Determinants that govern excision of the small intron.
Alternative splicing of the small intron is controlled by a complex interaction of splicing factors acting within a very small size constraint. Our results show that, within the context of the wild-type small intron, the two donors appear to compete for the splicing machinery. To generate appreciable amounts of the minor (D2/A2) spliced forms of MVM mRNAs, the primary sequence of D2 must be more like the consensus sequence than the primary sequence of D1. Initial 5′ splice site identification requires interactions with the U1 snRNP mediated by members of the SR protein family of splicing factors (21). Because the relative strength of MVM small-intron donor sequences is controlled by the +4 position, which is known to interact with the U1 snRNA, discrimination between D1 and D2 is presumably mediated by interactions between the donors and the U1 snRNA; however, interactions with specific SR proteins may also be involved. As discussed in more detail below, the preference for the D1 that is less like the consensus sequence, which is most distal to the acceptors, is linked to the small size of the intron. This suggests that the critical determinant in the choice of the donor that is less like the consensus sequence (D1) in MVM pre-mRNAs may be the preferential recognition of A1, which is probably too close to D2 for efficient splicing (28).
A1 appears to utilize a branch point at nt 2354 (PBP1) preferentially over the adjacent potential branch point (PBP2) at nt 2361. In vertebrate introns, the branch point is usually located 18 to 40 nt upstream of the 3′ splice site (18). This may explain why PBP2, which is only 15 nt from A1, is not efficiently used by A1 when only PBP1 is destroyed. However, in its natural context, the branch point for A2 is at nt 2383 (PBP3), which is only 14 nt from the 3′ splice site. Although under certain experimental conditions, PBP2 can be used for A2 (Fig. 3, pssPBP1/3 ko), it clearly is not able to compensate for the loss of PBP3 when PBP1 is present (Fig. 3, pssPBP-3 ko). These data are consistent with a model in which the primary branch point for the major spliced form (D1/A1) is PBP1 and the primary branch point for the minor spliced form (D2/A2) is PBP3.
The role of PBP2 in splicing of the small intron in its natural context is unclear. In the absence of both PBP1 and PBP2, there is no detectable D1 or A1 usage; however, in the absence of both PBP1 and PBP3, there is significant usage of both donors and acceptors. Thus, in the absence of competing branch points, it appears that PBP2 can be utilized by both acceptors (D1 is used slightly more than D2, and A2 is used approximately twice as efficiently as A1). Perhaps when present, PBP1 precludes the use of PBP2. Loss of both PBP1 and PBP3 may allow the use of PBP2, which, in an intermediate position, results in the use of both A1 and A2. These data raise the possibility that PBP2 could serve as the branch point for the rare spliced form (D1/A2). The utilization by different donors of separate branch points which splice to a single acceptor has been observed in simian virus 40 large- and small-T- antigen splicing (17). If, for the MVM small intron, the branch point for the rare mRNA form (D1/A2) is upstream of A1, this arrangement would be quite unusual and may affect the efficiency of its usage. In any case, our results demonstrate that the presence of three potential branch points is required to ensure correct stoichiometry of all alternatively spliced mRNAs.
The reasons for the preferential use of A1 over A2 remain unclear; however, it may be that the cis-acting factors present at A1 (this area contains two potential branch points and perhaps a better PPT) serve to favor assembly of splicing factors at this site. This would also explain the preferential use of D1, since D2 appears to be too close to A1 for efficient splicing (28). This explanation is consistent with the observation that D2 is joined to A1 when the small intron is expanded and the size constraints are removed, as described more fully below. While the requirement for a PPT in small vertebrate introns remains unclear (12), both MVM small-intron 3′ splice sites require their respective PPTs, even though the PPTs are both poor. This may reflect a need to compensate for the relatively nonconsensus branch point sequence (BPS) of each small-intron acceptor (23) or a need to enforce the 3′ boundary of the small intron. Since donor mutations have a relatively small effect on acceptor usage, and acceptor mutations have a relatively great effect on donor usage, recognition of components in the small-intron acceptors may be the primary determinants in alternative small-intron excision.
We have also identified a G-rich sequence (IES) within the small intron that is required for efficient generation of the minor spliced form when the small intron is in its natural context. Several G-rich intronic enhancers have been identified recently by others (12, 13, 25). Our system appears to most closely resemble that described by McCullough and Berget (12). In that system, several G-rich enhancers enforce the small-intron borders, both 5′ and 3′, which flank the enhancer sequence. At the acceptor, the effect appeared to be related to the ability of the enhancer to compensate for a poor PPT. In the MVM small intron, the G-rich IES is required for generation of the minor (D2/A2) spliced form, which uses the 5′ splice site which is adjacent to the enhancer and the 3′ splice site which is most distal to the enhancer. The mechanism of IES action is unclear; however, since D2 is used almost exclusively when the size constraint is removed, it is unlikely that the IES exerts its influence directly at the 5′ splice sites. The IES could potentially compensate for the relatively poor PPT of A2, much like what was observed in another system (12); however, because of its proximity to A1, it is equally likely that the function of the IES is to hinder assembly of splicing factors at A1.
We also show here that skipping of the NS2-specific exon results when the IES is mutated. Thus, the IES also appears to play a role in efficient definition of the upstream exon. We have previously demonstrated that at least one donor, one acceptor, and an undefined element within the intervening sequence of the small intron are required for efficient splicing of the upstream exon (29). We have now identified the IES as an additional requirement for this process. Thus, the IES is required both for generation of the minor (D2/A2) spliced form (intron defining) and for efficient definition of the upstream exon (exon defining). Intronic enhancers have not been previously shown to act simultaneously as intron- and exon-defining factors. We are currently investigating the mechanism by which the IES aids in the definition of the upstream exon.
Intron and exon definition are linked for MVM P4-generated pre-mRNAs.
Initial splice site recognition in very small mammalian introns, like that in the typically small introns of lower eukaryotes, appears to occur via a mechanism most accurately described as intron rather than exon definition (26, 27). Data discussed above as well as the observation that the MVM small intron, but not the large intron, is efficiently spliced in lower eukaryotes (7) suggest that the MVM small intron may be defined and excised in a manner more aptly described by an intron-defined model. Consistent with this model, when the MVM small intron is expanded, various parameters of its excision are altered. The internal donor (D2), which is more like the consensus sequence, is used preferentially over the external donor (D1) and shows no requirement for the IES. This indicates that, in its natural context, the small size of the intron somehow constrains the splicing machinery to choose the poorer donor (D1), which would be more consistent with an intron-defined system. As mentioned above, this may be due to preferential recognition of A1, which is used most often in both normal and expanded intron constructs. Also, and perhaps most dramatically, when the small intron is expanded, the upstream intervening NS2-specific exon is no longer well defined, and a significant amount of P4-generated product joins the large-intron donor and the small-intron A1. These results support recent proposals that many cis-acting splicing signals are highly context dependent (8). That is, when the MVM small intron is made large, the relative use of the donors is altered, since the cis-acting features of the small intron that define the upstream exon are no longer capable of functioning in that capacity.
Efficient definition of the NS2-specific exon and excision of the upstream large intron require sequences within the small intron that overcome a poor 3′ splice site (31). However, contrary to the predictions that one would make for a purely exon-defined system (1), definition of the NS2-specific exon requires a small-intron donor and acceptor, as well as the sequences between them (including the IES), all within the context of a very small intron. The fact that definition of the upstream NS2-specific exon can be achieved by the MVM small intron in its natural context, but not when it is expanded, is consistent with the interpretation that the MVM small intron is excised via an intron-defined mechanism (as described above) but also suggests that those multiple determinants that govern excision of this small intron are required, in concert, for upstream-exon definition and cannot function in that regard when the relative positions of these elements are altered. Thus, in P4-generated MVM pre-mRNAs, definition of the NS2-specific exon is linked to definition of the small intron. However, because destruction of both donors of the MVM small intron results in upstream-exon skipping (30) rather than in small-intron inclusion, the MVM small intron also has characteristics of an exon-defined system (1). Therefore, the MVM small intron exhibits characteristics of both intron- and exon-defined systems, and determinants which govern splicing of both the small and the large intron cannot be accurately described by either a purely exon-defined or intron-defined model. For excision of the large intron, we must use a hybrid of both exon and intron definition models to accurately represent generation of the doubly spliced P4 message.
It is interesting that the small intron has acquired such a complex mechanism to determine the frequencies at which major, minor, and rare spliced forms are produced. Such a mechanism may be required to generate protein diversity in a genome as compact as that of MVM. The requirement for two large genes, each of which occupies essentially one-half of the viral genome, may preclude additional internal exons. Clearly, the ratio of VP1 to VP2 and the relative ratios of the NS2 isoforms must be critical for the life cycle of this virus. It is possible that the virus has evolved this complex machinery in order to tightly control the relative ratios of these proteins.
Other related parvoviruses have somewhat different small-intron structures, as shown in Fig. 7. For both CPV and PPV, the NS protein terminates prior to the small splice region. Alternative splicing in this region affects only capsid protein ratios (20). CPV and PPV have two donors which are used at relative efficiencies similar to that of MVM. In each case, the internal donor is more like the consensus sequence than the external, preferred donor, suggesting that some of the cis-acting regulatory determinants in MVM may be conserved among these other, related viruses. While CPV and PPV each have only one acceptor, they both have two adjacent potential BPSs like PBP1 and PBP2 of MVM. In addition, each has a G-rich region which is adjacent to the most-5′ potential BPS. These similarities suggest that these viruses have evolved similar mechanisms to gain tight control over the relative steady-state levels of their mRNAs, and analysis of various related parvoviruses will likely yield important information concerning the determinants that govern alternative splicing.
ACKNOWLEDGMENTS
This work was supported by PHS grant RO1 AI21302 from NIAID and a grant from the Council for Tobacco Research, USA. D.D.H. was supported by T32 AI07276 during a portion of this work.
We thank members of the laboratory for helpful discussions and especially Anand Gersappe for critical review of the manuscript. Thanks to Lisa Burger for expert technical assistance.
REFERENCES
- 1.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2444. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 2.Clemens K E, Pintel D J. Minute virus of mice (MVM) mRNAs predominantly polyadenylate at a single site. Virology. 1987;160:511–514. doi: 10.1016/0042-6822(87)90028-6. [DOI] [PubMed] [Google Scholar]
- 3.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 4.Guo M, Lo P C, Mount S M. Species-specific signals for the splicing of a short Drosophila intron in vitro. Mol Cell Biol. 1993;13:1104–1118. doi: 10.1128/mcb.13.2.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall S L, Padgett R A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 6.Hampson R K, La Follette L, Rottman F M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989;9:1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haut, D., and D. J. Pintel. 1995. Unpublished data.
- 8.Hertel K, Lynch K, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 9.Innis M A, GelFand D H, Sninski J J, White T J. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 10.Jongeneel C V, Sahli R, McMaster G K, Hirt B. A precise map of splice junctions in the mRNAs of minute virus of mice, an autonomous parvovirus. J Virol. 1986;59:564–573. doi: 10.1128/jvi.59.3.564-573.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy C F, Berget S M. Pyrimidine tracts between the 5′ splice site and branch point facilitate splicing and recognition of a small Drosophila intron. Mol Cell Biol. 1997;17:2774–2780. doi: 10.1128/mcb.17.5.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCullough A J, Berget S M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 14.Morgan W R, Ward D C. Three splicing patterns are used to excise the small intron common to all minute virus of mice RNAs. J Virol. 1986;60:1170–1174. doi: 10.1128/jvi.60.3.1170-1174.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naeger L K, Schoborg R V, Zhao Q, Tullis G E, Pintel D J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1111. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- 16.Nilsen T W. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- 17.Noble J C, Prives C, Manley J L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988;2:1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- 18.Padgett R A, Grabowski P J, Konarska M M, Seiler S R, Sharp P A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 19.Pintel D, Dadachanji D, Astell C R, Ward D C. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 1983;11:1019–1038. doi: 10.1093/nar/11.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pintel D J, Gersappe A, Haut D, Pearson J. Determinants that govern alternative splicing of parvovirus pre-mRNAs. Semin Virol. 1995;6:283–290. [Google Scholar]
- 21.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 22.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roscigno R F, Weiner M, Garcia-Blanco M A. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J Biol Chem. 1993;268:11222–11229. [PubMed] [Google Scholar]
- 24.Schoborg R V, Pintel D J. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology. 1991;181:22–34. doi: 10.1016/0042-6822(91)90466-o. [DOI] [PubMed] [Google Scholar]
- 25.Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sterner D A, Carlo T, Berget S M. Architectural limits on split genes. Proc Natl Acad Sci USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talerico M, Berget S M. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wieringa B, Hofer E, Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984;37:915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q, Gersappe A, Pintel D J. Efficient excision of the upstream large intron from P4-generated pre-mRNA of the parvovirus minute virus of mice requires at least one donor and the 3′ splice site of the small downstream intron. J Virol. 1995;69:6170–6179. doi: 10.1128/jvi.69.10.6170-6179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Q, Mathur S, Burger L R, Pintel D J. Sequences within the parvovirus minute virus of mice NS2-specific exon are required for inclusion of this exon into spliced steady-state RNA. J Virol. 1995;69:5864–5868. doi: 10.1128/jvi.69.9.5864-5868.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, Schoborg R V, Pintel D J. Alternative splicing of pre-mRNAs encoding the nonstructural proteins of minute virus of mice is facilitated by sequences within the downstream intron. J Virol. 1994;68:2849–2859. doi: 10.1128/jvi.68.5.2849-2859.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]