Abstract
Transcriptional factors play a crucial role in regulating cellular functions. Understanding and altering the dynamic behavior of the transcriptional factor-based biosensors will expand our knowledge in investigating biomolecular interactions and facilitating biosynthetic applications. In this study, we characterized and engineered a TrpR-based tryptophan repressor system in Escherichia coli. We found that the reconstructed TrpR1-PtrpO1 biosensor system exhibited low basal expression and narrow dynamic range in the presence of tryptophan or its analog 5-hydroxytryptophan (5-HTP). Given the application potential of the biosensor, we introduced engineering approaches in multiple levels to optimize its dynamic behavior. Firstly, the I57 and V58 residues in the ligand-binding pocket were rationally mutated in search of variants with altered ligand specificity. Two TrpR1 variants, V58E and V58K, successfully acquired ligand preference towards tryptophan and 5-HTP, respectively. The biosensor-induced expression levels were increased up to 10-fold with those variants. Furthermore, to pursue broader operational range, we tuned the regulator-operator binding affinity by mutating the binding box of TrpR. Collectively, we demonstrated that the biosynthesis-significant biosensor TrpR1-PtrpO1 can be engineered to acquire extended dynamic ranges and improved ligand preference. The engineered biosensor variants with remarkable dynamic behavior can serve as key genetic elements in high-throughput screening and dynamic regulation in biosynthetic scenarios.
Keywords: TrpR, tryptophan, 5-hydroxytryptophan, biosensor, dynamic behavior
Graphical Abstract
The main strategies used in this study included the rational mutagenesis of TrpR1 regulator protein and trpO1 operator. The variants screenings were conducted separately. The additive effect of the selected TrpR1 and trpO1 variants exhibited improved dynamic behavior and altered ligand preference.
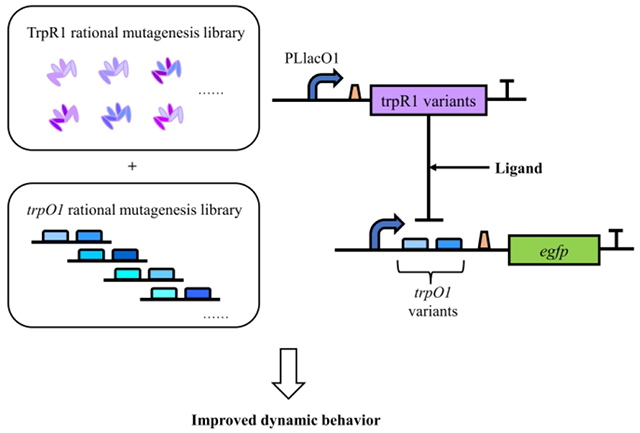
Introduction
Transcriptional factor-based (TF-based) biosensors have been extensively studied to reveal cellular regulation mechanisms and to stimulate biotechnological applications, such as cellular function modulating and metabolites sensing 1–4. The most well-known system is the lactose operon 5, which regulates β-galactosidase expression and detects lactose abundance and won the Nobel Prize in Physiology or Medicine in 1965. More biosensors that sense different signals, such as metabolites, light, temperature, or oxygen, have been identified since then 6–8. However, available biosensors are still greatly outnumbered by the population of cellular metabolites (over 2600 metabolites in E. coli) 9. Therefore, the exploration and modification of biosensors for varied sensing capabilities is a promising but challenging task. A typical TF-based biosensor consists of three components: a transcriptional factor, a ligand molecule, and a responsive nucleic acid sequence (namely operator) 1. On the one hand, some TF-based biosensors have been extensively characterized and engineered, as the regulator-ligand interaction resemble enzyme-substrate binding, thus their studies could be guided by protein engineering knowledge 10. On the other hand, the fragility of regulator binding sites and the diversity of regulator-operator recognition mechanisms make the engineering of TF-based biosensors difficult but feasible 11. Nevertheless, the engineering of TF-based biosensors will have a broad impact on programmable regulation of cellular activities, making it an attractive topic in synthetic biology and metabolic engineering.
One of the major applications of biosensors is the dynamic regulation of cell factories for enhanced bioproduction. In conventional metabolic engineering strategies, cells are statically modified for metabolic pathway reconstruction, foreign gene expression, and metabolites accumulation. However, cell growth is a dynamic process, pursuing bioproduction efficiency might easily overlook the burden cast upon cells during vulnerable stages of cell growth. As opposed to conventional static regulation, a concept of dynamic regulation has been coined and implemented 12. Essentially, dynamic regulation relies on the self-adjustments of cells based on introduced biosensors, with the goal of resolving the conflict between native cellular activities and target biochemical generations. Biosensor-based dynamic regulation has been proved beneficial in biosynthesis of fatty acids 13, 14, biofuel 15, terpene 16, and aromatic compounds 17. Characterization and development of TF-based biosensors represent a major effort in decoding protein-ligand and protein-DNA interactions, as well as a coordinative view of achieving optimal biosynthesis. For the precise control of cellular functions and metabolism, the dynamic behavior optimization is required for adopting biosensors under distinct conditions.
Tryptophan is a commercial amino acid which is widely used as ingredient in food industry, feedstocks and healthcare products; and its derivatives, such as 5-hydroxytryptophan (5-HTP) and monoterpene indole alkaloids (MIA), are valuable compounds in drug discovery 18, 19. Thus, obtaining tunable tryptophan biosensors is highly desired for the balanced and high-yield bioproduction of tryptophan and its derivatives. TrpR has been found to regulate a number of genes in E. coli, including trpEDCBA operon 20, aroH 21, trpR 22, aroL 23 and the tryptophan:H+ symporter mtr 24. As a typical TF-based biosensor, this system is composed of three components: a transcriptional factor TrpR, a ligand molecule, and a responsive nucleic acid sequence trpO 1. For the regulation principle, TrpR works as a dimer repressor that binds to the corresponding operators in the presence of tryptophan and subsequently represses the downstream gene expression. The trpO of trpEDCBA operon is the most studied operator sequence, containing a symmetric pair of binding sequences that interact with the helix-turn-helix (HTH) domain of TrpR 25, 26. To date, several studies has focused on investigating its regulation functions or applications in biosynthesis. Ellefson and colleagues used a replication-based screening strategy to identify key residues of TrpR for ligand recognition 27. They also investigated the orthogonality and modularity of the regulatory system, obtaining numerous TrpR-trpO variants with minimal crosstalk to the native components. Zhang and colleagues converted E. coli TrpR into an activating biosensor in yeast for monitoring cellular tryptophan concentrations by fusing with the Gal4 activation domain 28. However, the dynamic behavior tweaking of the TrpR-based biosensor in E. coli has not been achieved yet.
Herein, we successfully reconstructed the TrpR1-PtrpO1 biosensor system and discovered the relaxed ligand specificity of TrpR1 which allowed tryptophan and its analog 5-HTP to respond. Upon initial validation, we found that the original biosensor version had narrow operational and dynamic ranges to both tryptophan and 5-HTP, which would limit the applicability. To improve the operational and dynamic ranges, as well as explore the ligand specificity, we employed regulator and operator engineering approaches to obtain biosensor variants. Some variants possessed greater dynamic ranges with up to 90% repression efficiency via mutating the crucial amino acid residue V58 in TrpR1. Among them, TrpR1(V58E) and TrpR1(V58K) variants successfully altered ligand preference, responding to tryptophan and 5-HTP, respectively. The additive effect of mutating the TrpR1 binding box had led to regulator variants with optimized dynamic behavior. TrpR1(V58E)-PtrpO1(A4C) variant was more favorable to tryptophan sensing, and TrpR1(V58K)-PtrpO1(A4C) variant preferred 5-HTP sensing with a broader operational range. We believe that this study enriched our knowledge in biosensor engineering. The biosensor variants with enhanced dynamic behavior can be applied to various biosynthetic settings, including high-throughput screening and dynamic regulation.
Materials and Methods
Media and chemicals
Luria-Bertani (LB) medium containing 5 g/L yeast extract, 10 g/L tryptone, and 10 g/L NaCl was used for strain reviving and initial inoculation. The pure modified M9 minimal medium and modified M9 minimal medium with 5 g/L yeast (M9Y-5) were used for biosensor sensor validation, containing 0 g/L or 5 g/L yeast extract, 20 g/L glycerol, 6 g/L Na2HPO4, 0.5 g/L NaCl, 3 g/L KH2PO4, 1 g/L NH4Cl, 1 mM MgSO4 and 0.1 mM CaCl2, respectively. The modified M9 minimal medium with 0.5 g/L yeast extract (M9Y-0.5) was mainly used for fluorescence assays, containing 0.5 g/L yeast extract, 20 g/L glycerol, 6 g/L Na2HPO4, 0.5 g/L NaCl, 3 g/L KH2PO4, 1 g/L NH4Cl, 1 mM MgSO4 and 0.1 mM CaCl2, respectively. Standard chemicals including glycerol, tryptophan (Trp), 5-hydroxytryptophan (5-HTP), isopropyl β-D-1-thiogalactopyranoside (IPTG), ampicillin (Amp), kanamycin (Kan) and chloramphenicol (Cl) were purchased from Sigma-Aldrich unless otherwise noted. While added, Amp, Kan and Cl were used at the concentration of 100, 50 and 34 μg/ml, respectively. Phusion High-Fidelity DNA polymerase, restriction endonucleases and Quick Ligation Kit were purchased from New England Biolabs (Beverly, MA, USA). Plasmid Miniprep Kit, Gel Recovery Kit, and DNA Cleanup kit were purchased from Zymo Research (Irvine, CA, USA).
Strain and plasmid construction
E. coli XL1-blue (Stratagene) was used for plasmid construction and storage. The E. coli BW25113 ΔtnaA ΔtrpR was developed via P1 transduction process 29 and was used for fluorescence assays in this study. The removal of antibiotic resistance markers was conducted by electroporation of pCP20 plasmid into the cells according to standard protocols 30. The plasmids pCS27 and pSA74 were used as the medium- and low-copy number vectors, respectively 31. To construct pSA-pLlacO1-TrpRwt, trpR was amplified from E. coli genome and inserted between Acc65I/XbaI sites of pSA74 upon digestion. Similarly, to construct pSA-pLlacO1-TrpR1, trpR1 was inserted into the same sites of pSA74, after amplification through the primer including the mutations. The primers that include PtrpOwt or PtrpO1 sequences were used to amplify the promoter and egfp gene. The amplified DNA fragments were then inserted into pCS27 vector between XhoI/BamHI sites, yielding pCS-PtrpOwt-eGFP and pCS-PtrpO1-eGFP, respectively. For base mutations in TrpR1 binding box, degenerate primers were used to replace operator sequence and egfp gene between ClaI/BamHI sites of pCS27, yielding operator mutagenesis library. The list of strain and plasmid details are included in Table S1.
TrpR1 rational mutagenesis library construction and screening
To introduce amino acid mutations at I57 and V58 of TrpR1, firstly we introduced a HindIII site at A50 of trpR1 sequence. A 3-fragment cloning was conducted by amplifying and digesting upper and lower part of trpR1 by Acc65I/HindIII and HindIII/XbaI, respectively, and then inserted into pSA74 plasmid after digestion, yielding pSA-pLlacO1-TrpR1_HindIII. The rational mutations at I57 and V58 were then introduced on primers for trpR1 variants. The amplified fragments were inserted between HindIII/BamHI sites of pSA74 vector, yielding a series of plasmid variants (Table S1). To screen the variants, pCS-PtrpO1-eGFP and pSA-pLlacO1-TrpR1_mutation plasmids were co-transformed into E. coli BW25113 ΔtnaA ΔtrpR for testing. For each variant, three single colonies were selected for optical density and fluorescence assays with or without 500 mg/L tryptophan or 5-HTP. Finally, the variants were selected by comparing their repression efficiency.
Optical density and fluorescence assays
The quantification of cell density and fluorescence intensities was measured with Synergy HT microplate reader (BioTek). E. coli BW25113 ΔtnaA ΔtrpR was transformed with corresponding plasmids and cultured in 3 ml LB medium with appropriate antibiotics at 37°C and 270 rpm overnight. The subculture was conducted by adding 1% of the cultures into 3 ml pure M9, M9Y-0.5 or M9Y-5 medium with appropriate antibiotics. Then, the cells were cultured at 37°C and 270 rpm, feeding with specific ligand concentrations, and sampled at 24 h. For the measurement, cells were diluted 10 folds in 200 μL volume and were loaded into 96-well plates to read optical density at 600 nm (OD600) and eGFP fluorescence under excitation wavelength of 485 nm and emission wavelength of 528 nm. All fluorescence intensity results were normalized by respective cell growth (OD600) and eliminated the background. All experimental results were obtained with three biological replicates (n=3). Error bars indicate standard deviations of the presented results.
Results
Reconstruction of the synthetic TrpR system
TrpR-mediated regulation is an important mechanism in aromatic amino acid biosynthesis in microbes. In E. coli, TrpR could repress the expression of the trpEDCBA operon 20, aroH 21, trpR 22 and aroL 23 by up to 100-folds when bound with tryptophan. Although the regulation mechanism of genes by TrpR has been extensively elucidated, its synthetic reconstruction and application in biosynthesis are rarely studied. The TrpR repressor system contains TrpR regulatory protein, regulated trpO operator and tryptophan as ligand. In the absence of tryptophan, TrpR is an inactive repressor and unable to occupy trpO operator. In the presence of tryptophan, TrpR forms an active complex with tryptophan and binds to trpO operator, inhibiting the expression of downstream operons (Figure 1a).
Figure 1. The mechanism of TrpR-PtrpO biosensor system and its orthogonality verification.
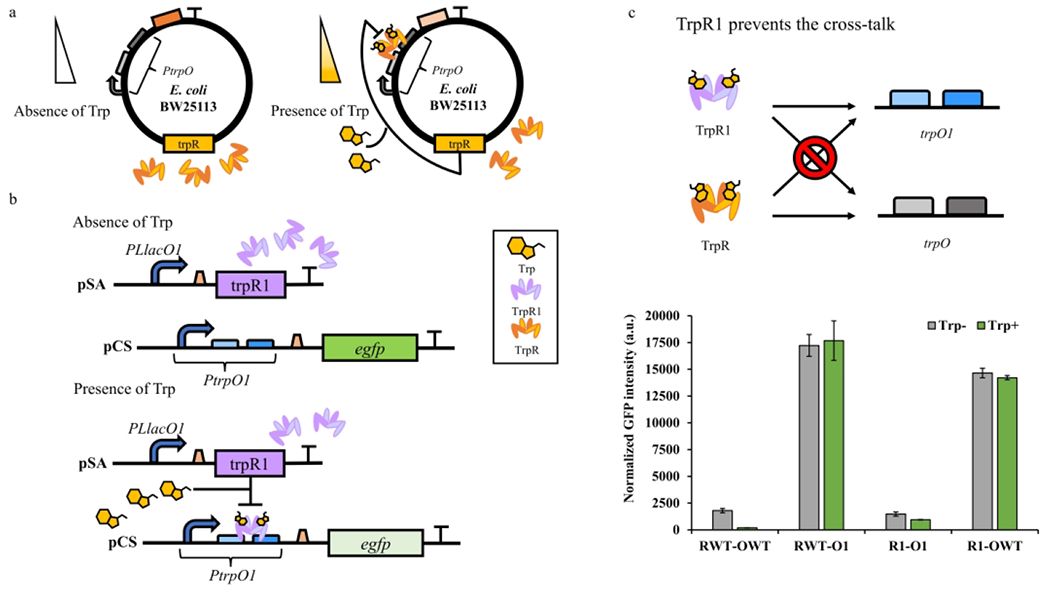
(a) The mechanism of wild type TrpR-PtrpO in E. coli genome. In the absence of tryptophan, TrpR will not bind to trpO. In the presence of tryptophan, TrpR will form a complex with two molecular tryptophan and bind to trpO to inhibit the expression of downstream genes. (b) The genetic circuit of TrpR1-PtrpO1. In the absence of tryptophan, TrpR1 will not bind to trpO1 and egfp will express normally. In the presence of tryptophan, TrpR1 will form a complex with two molecular tryptophan and bind to trpO1 to inhibit the expression of egfp. (c) TrpR1-trpO1 prevents the crosstalk between the native genetic elements. There is no interaction between trpO1 and trpO or trpO and trpO1. The regulatory proteins only bind to their paired operator, i.e., wild type TrpR-PtrpO and TrpR1-PtrpO1. RWT: wild type TrpR; R1: TrpR1 variant; OWT: wild type PtrpO promoter; O1: PtrpO1 variant. Trp−: no tryptophan supplementation; Trp+: 1000 mg/L tryptophan supplementation. All experimental data was collected from three independent biological replicates (n=3). The standard deviations were present as error bars.
In this study, we sought to rebuild the TrpR-based repressor system in the laboratory settings and explore its dynamic behavior. Recently, by evolving the regulator-operator binding region, a series of orthogonal TrpR-trpO variants have been developed with dramatically reduced cross-talk to the native TrpR system 27. To minimize the interference of unexpected regulation, we adopted one set of the genetic elements TrpR1 (K72S, G78N, I79W, A80R, and T83K) and trpO1 (C6G and A7T) for further engineering. To reassemble the TrpR1-PtrpO1 biosensor system, we put trpR1 in a low-copy plasmid (pSA-pLlacO1-TrpR1, designated as R1) controlled by an IPTG inducible PLlacO1 promoter, and the egfp expression was under the control of PtrpO1 in a medium-copy plasmid (pCS-PtrpO1-egfp, designated as O1) as a reporter (Figure 1b).
To demonstrate the feasibility of the TrpR1-PtrpO1 biosensor system, the tryptophanase (tnaA) responsible for tryptophan degradation was knocked out from E. coli BW25113 to prevent intracellular tryptophan degradation (E. coli BW25113 ΔtnaA). The native trpR in the genome was further knocked out (resultant strain RZ1, E. coli BW25113 ΔtnaA ΔtrpR) to establish a clear background strain for the repression system validation. M9Y-0.5 medium with 0.5 g/L yeast extract was used for the system verification by minimizing the influence of tryptophan from the culture media and supporting cell growth (Figure S1 and Table S2). After transferring R1 and O1 plasmids into strain RZ1, only about 36% repression efficiency was observed in 1000 mg/L tryptophan presence, showing a quite narrow dynamic range compared to the wild-type TrpR-PtrpO biosensor (RWT-OWT, 89% repression efficiency) (Figure 1c). However, both systems exhibited low expression levels under 1500 a.u. To validate the orthogonality of the adopted biosensor, trpO operator were not paired with the corresponding regulator protein, i.e., RWT-O1 and R1-OWT. As expected, the tryptophan supply did not lead to egfp repression (Figure 1c). Interestingly, the high eGFP fluorescence (exceeded 14000 a.u.) was observed in those unpaired groups, indicating that PtrpO and PtrpO1 were strong promoters, and their expression capabilities were inhibited in the biosensor system. Overall, these results confirmed the feasibility of our reconstructed synthetic TrpR1-PtrpO1 biosensor system. However, the limitations associated with this biosensor system, such as low expression level and low repression efficiency, needed to be overcome by further engineering.
Exploration of the dynamic range and ligand specificity of TrpR1-PtrpO1 system
In the TrpR1-PtrpO1 biosensor system, TrpR1 abundance was controlled by an IPTG-inducible PLlacO1 promoter, and TrpR1 regulated the expression of the reporter plasmid via regulating PtrpO1 (Figure 2a). Therefore, we hypothesized that tuning the expression level of TrpR1 with gradient IPTG concentrations would change the basal expression of the biosensor. To investigate the influence of the reduced TrpR1 expression on egfp expression, strain RZ1 carrying plasmids R1 and O1 was subjected to a gradient IPTG concentration from 0 to 50 μM and cultured in M9Y-0.5 medium. As a result, the eGFP fluorescence was increased by decreasing the IPTG concentrations (25 to 2.5 μM) in a gradient manner (Figure 2b). This validated that PtrpO1 promoter was sensitive to the TrpR1 abundance. We then chose 5, 10 and 25 μM IPTG to investigate the dynamic behavior of TrpR1-PtrpO1 biosensor system in the range of 0 to 1000 mg/L tryptophan (Figure 2c). Each group had a repression efficiency of 47.6%, 36.2%, and 26.2%, respectively. The dynamic ranges decreased as the concentrations of IPTG increased. The operational ranges for all groups were around 500 mg/L tryptophan. These results indicated that the 5 μM IPTG concentration was sufficient to induce adequate TrpR1 abundance, enable a proper basal expression, and trigger a modest repression efficiency. Hence, 5 μM IPTG concentration was applied to subsequent experiments.
Figure 2. TrpR1 expression level tuning and ligand specificity exploration.
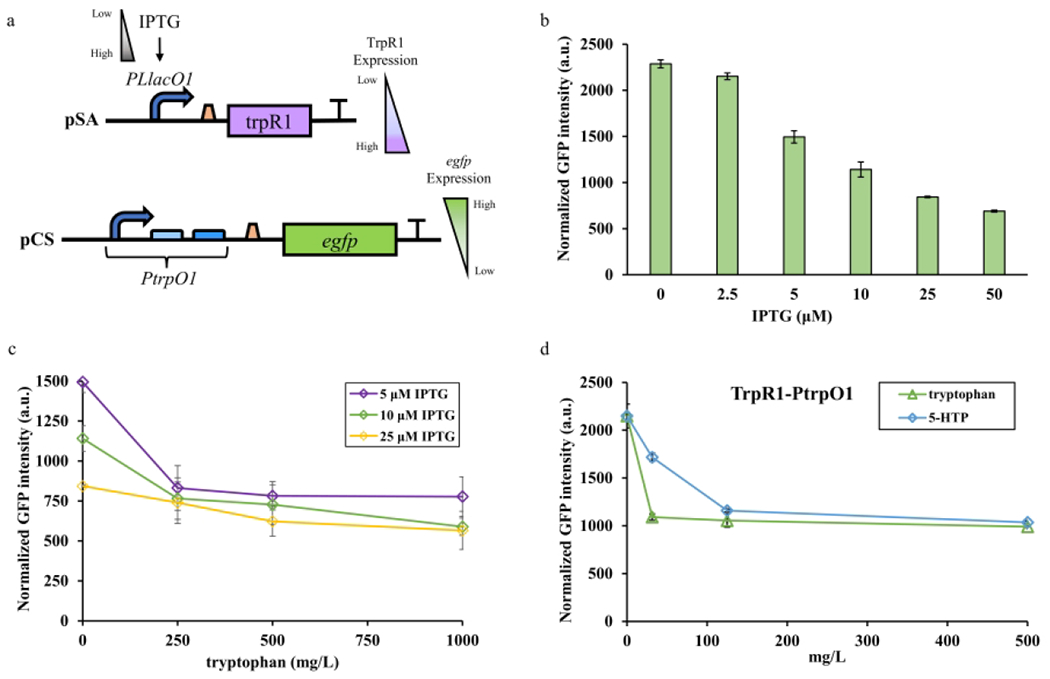
(a) The expression of TrpR1 was under the control of the IPTG-inducible PLlacO1 promoter. The expression of egfp was under the control of PtrpO1 promoter, while the abundance of TrpR1 would influence the egfp expression by regulating PtrpO1. (b) The influence of gradient IPTG concentrations on the basal expression level of TrpR1-PtrpO1 biosensor system. (c) The dynamic behavior of TrpR1-PtrpO1 with 5, 10, or 25 μM IPTG concentrations under 0 to 1000 mg/L tryptophan. (d) The ligand specificity test for TrpR1-PtrpO1 with both tryptophan (0 to 500 mg/L) and 5-HTP (0 to 500 mg/L). All experimental data was collected from three independent biological replicates (n=3). The standard deviations were present as error bars.
In order to understand the ligand specificity of the biosensor, we then explored its sensing ability towards the tryptophan analog 5-HTP, a high-value natural product with pharmaceutical applications in the treatments of depression, sleep disorder and myoclonus 32, 33. No natural 5-HTP responsive biosensor has so far been reported. To evaluate the applicability of TrpR1-PtrpO1 biosensor system, strain RZ1 with plasmids R1 and O1 was employed to characterize its dynamic behavior in the presence of 0 to 500 mg/L tryptophan or 5-HTP (Figure 2d). The fluorescence assay indicated that TrpR1 biosensor could serve as a promiscuous biosensor for both tryptophan and 5-HTP. When tryptophan was added as the ligand, 2.17-fold dynamic range was observed with about only 30 mg/L operational range. While 5-HTP was added as the ligand, a 2.07-fold dynamic range was observed with around 125 mg/L operational range. These results suggested that TrpR1 can accept both ligands but prefer tryptophan over 5-HTP. To better suit in metabolic engineering applications for biosynthesis, it’s important to alter the ligand specificity and broaden the dynamic range of the biosensor.
Rational engineering of TrpR1 for improved dynamic range and altered ligand preference
The dynamic behavior of the biosensor systems can be affected by multiple factors, such as regulator and operator abundance, regulator-ligand affinity, and regulator-operator affinity. As the TrpR1-PtrpO1 biosensor system has been successfully reconstructed, we confirmed that the regulator protein TrpR1 with mutations in the groove of the DNA binding region would have no obvious influence on its repression response and ligand binding. In pursuit of altering the ligand specificity, we hypothesized that mutations of TrpR1 residues within the ligand binding pocket would shift its ligand specificity. First of all, the regulator-ligand interaction region of the crystallized TrpR protein structure was referred in order to find the mutation targets 34, 35. The tryptophan binding pocket of TrpR was formed by the hydrophobic interaction between amino acid residues R54, I57, V58, R84, G85, and S88 (Figure 3a, purple blue region). Previous research has shown that mutations of the ligand proximal residues I57 and V58 can help flip the ligand specificity 27. Therefore, we conducted scanning mutagenesis of TrpR1 by targeting I57 and V58. Since 5-HTP contains a hydroxyl group, we reasoned that the amino acid residues with small side chains (e. g., glycine) would probably enlarge the binding pocket to accommodate the extra hydroxy group of 5-HTP, while amino acid residues with longer side chains (e.g., lysine) could potentially form extra hydrogen bonds to the ligand. All variants were evaluated against 500 mg/L tryptophan and 5-HTP in strain RZ1 harboring plasmids O1 and R1 or R1 mutations in M9Y-0.5 medium (Figure 3b). The variants I57T, I57A and I57V exhibited ligand preference to tryptophan with repression efficiency of 73.5%, 47.6%, 76.9%, respectively. Unfortunately, most of the I57 variants had an impaired sensing capability to both ligands. On the other hand, V58T, V58G, V58A, V58N, V58S and V58Q variants obtained significantly enhanced dynamic range to both ligands (Figure 3b). Among them, V58G variant had the highest 94.6% repression efficiency with tryptophan, and V58Q variant had the highest 94.8% repression efficiency to 5-HTP. Moreover, there were two variants that displayed obvious ligand preference to one of the ligands. The V58E variant showed a substantial repression (96.4%) to tryptophan, but only showed 46.1% repression to 5-HTP. On the contrary, the V58K variant showed a strong repression (90.1%) to 5-HTP, but only 13.5% repression to tryptophan. To further test the combinatorial effects of I57 and V58 mutants, we created double mutants by introducing the V58E or V58K into I57T, I57A and I57V variants. However, none of the double variants exhibited better repression efficiency towards any ligands than the counterpart I57 or V58 variants in the fluorescence assay (Figure S2). These results suggested the efficiency of our protein engineering strategy on V58 residue, allowing a variety of dynamic behavior. However, comprehensive dynamic behavior of the variants was needed to evaluate their applicability.
Figure 3. Rational mutagenesis library screening for TrpR1 variants.
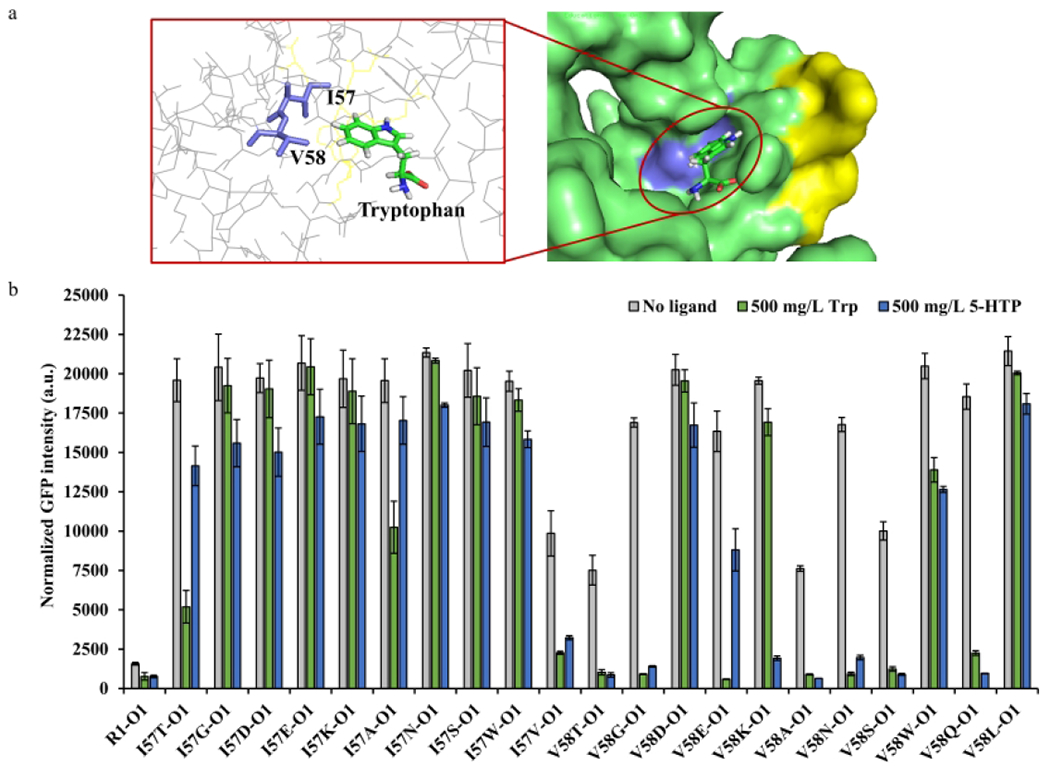
(a) The crystal structure analysis of wild type TrpR (PDB entry: 2OZ9) 35. In the left panel, the purple blue amino acid residues I57 and V58 that interact with the indole side chain of tryptophan are identified as essential residuals for ligand binding. In the right panel, the purple blue region showed the ligand binding region of TrpR. And the yellow region shows the evolved DNA binding region of TrpR for TrpR1 generation. (b) The TrpR1 I57 and V58 variants screening. The TrpR1 variants were selected and compared by calculating the repression efficiency of each ligand. All experimental data was collected from three independent biological replicates (n=3). The standard deviations were present as error bars.
For further characterization, we proceeded to test the selected variants, V58G, V58Q, V58E, and V58K, with 0 to 500 mg/L ligand concentrations for both tryptophan and 5-HTP. The plasmids O1 and R1, or R1 variants were co-transferred into strain RZ1 and grew in M9Y-0.5 medium. In comparison to R1, the variants V58G and V58Q exhibited significantly improved dynamic ranges to both ligands (Figure 2d, Figure 4a and 4b). The V58G variant possessed a 17.7-fold dynamic range towards tryptophan with about 30 mg/L operational range, and a 11.1-fold dynamic range towards 5-HTP with about 125 mg/L operational range. The V58Q variant had around 125 mg/L operational ranges to two ligands. And the dynamic range of V58Q variant was 10.5-fold and 23.7-fold towards tryptophan and 5-HTP, respectively. Instead, the variants V58E and V58K exhibited their own ligand preference. The V58E variant showed a 32.6-fold dynamic range to tryptophan with about 30 mg/L operational range and just a 1.9-fold dynamic range to 5-HTP, which largely reduced the 5-HTP sensing capability (Figure 4c). The V58K variant was the most interesting one that almost flipped the ligand preference from tryptophan to 5-HTP with a 10.0-fold dynamic range and about 125 mg/L operational range to 5-HTP (Figure 4d). To summarize, some TrpR1 variants had improved dynamic ranges and altered ligand preference, but they were too sensitive to respond to low-concentrated ligands, which might limit their applications. Therefore, the operational ranges of the variants needed to be further optimized to accommodate various application scenarios.
Figure 4. Dynamic behavior of selected TrpR1 variants.
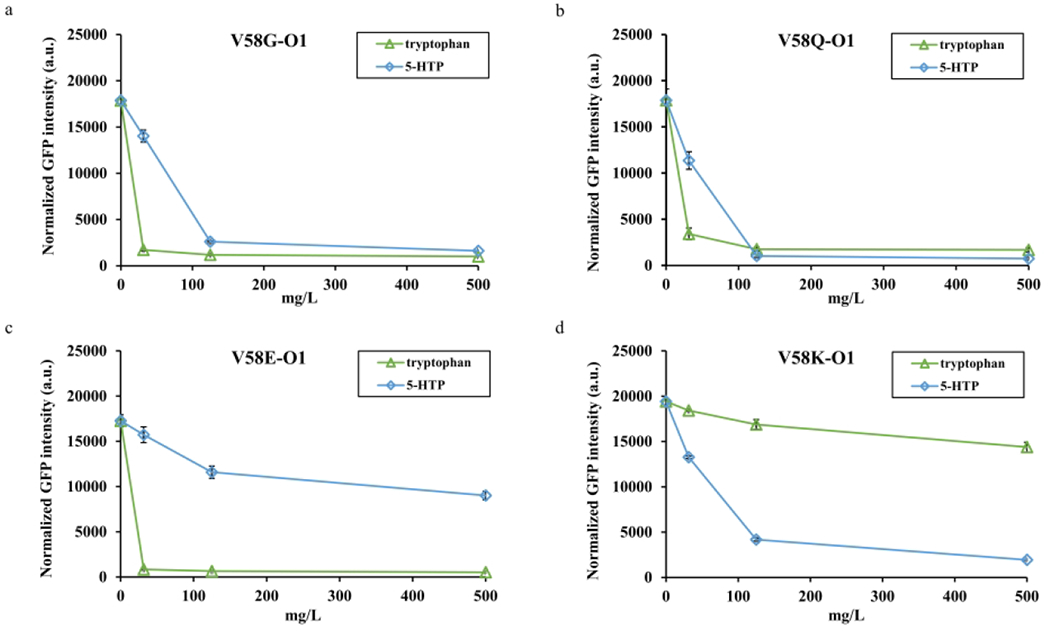
(a) The dynamic behavior of V58G variant under 0 to 500 mg/L tryptophan and 5-HTP, respectively. (b) The dynamic behavior of V58Q variant under 0 to 500 mg/L tryptophan and 5-HTP, respectively. (c) The dynamic behavior of V58E variant under 0 to 500 mg/L tryptophan and 5-HTP, respectively. (d) The dynamic behavior of V58K variant under 0 to 500 mg/L tryptophan and 5-HTP, respectively. All experimental data was collected from three independent biological replicates (n=3). The standard deviations were present as error bars.
TrpO1 operator mutations enable tuning of the dynamic behavior of the biosensor system
Although some interesting TrpR1 variants were obtained, their current operational ranges were too narrow to use. Consequently, we hypothesized that mutating the TrpR1 binding boxes within the PtrpO1 promoter would affect the regulator-operator affinity, resulting in a change of the operational ranges. The trpO1 operator is a part of the PtrpO1 promoter and consists of left and right symmetric binding boxes (Figure 5a). The mutations of nucleic acids in loci 5 and 6 of left binding box of wild type trpO operator were proved to cause severe impact on wild type TrpR binding, while the nucleic acid mutations in loci 2, 3, and 4 had mild impact 36 (Figure 5a). We then introduced saturated mutations in loci 2, 3, and 4 of trpO1 separately to investigate their effects on the dynamic behavior. The pre-screening was conducted by co-transferring TrpR1 and trpO1 variants into strain RZ1 in M9Y-0.5 medium, with both tryptophan and 5-HTP as ligands (Figure 5b). According to the pre-screening results, the mutations in loci 2 (designated as R1-T2A, R1-T2C, and R1-T2G) resulted in the fully abolished dynamic behavior, indicating the potential significance of loci 2 in maintaining the normal function of the PtrpO1 promoter. The variants in loci 3 (designated as R1-G3A, R1-G3C, and R1-G3T) acquired compromised dynamic behavior with low expression level. The variants in loci 4 (designated as R1-A4C, R1-A4G, and R1-A4T) possessed varied dynamic behavior and higher expression levels.
Figure 5. The operator engineering of trpO1.
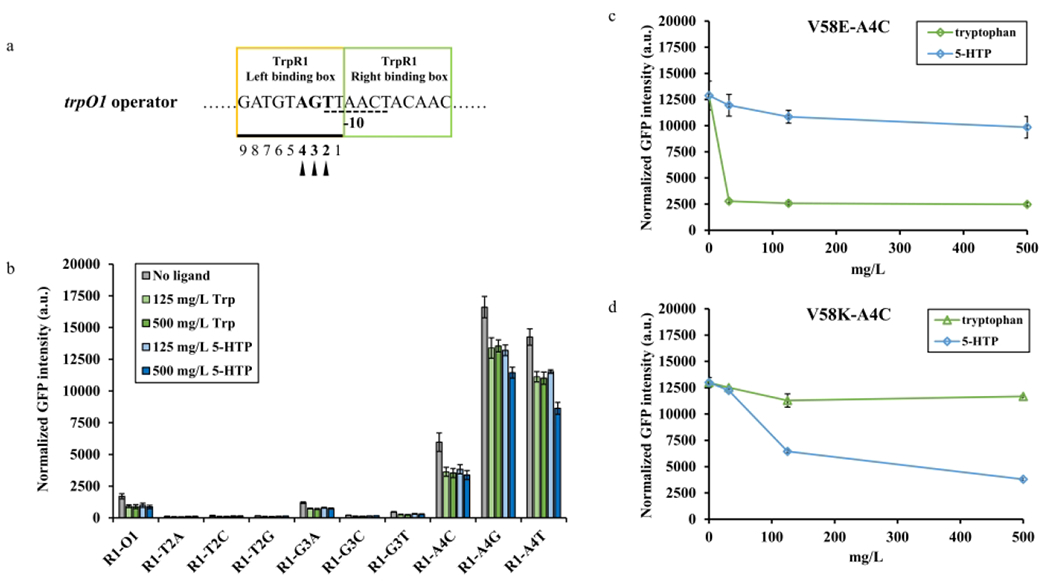
(a) The DNA sequence of trpO1 operator. The yellow and green squares indicated the TrpR1 left and right binding boxes, respectively. The nucleic acids with dash underline indicated the −10 region of the PtrpO1 promoter. The nucleic acids in the TrpR1 left binding box are underlined and numbered. Among them, the bold nucleic acids with triangle markers are mutated separately for operator engineering. (b) The trpO1 mutations screening. Each trpO1 variant was paired with TrpR1 for first round screening against both ligands. (c) The dynamic behavior of TrpR1(V58E)-PtrpO1(A4C) biosensor variant under 0 to 500 mg/L tryptophan and 5-HTP, respectively. (d) The dynamic behavior of TrpR1(V58K)-PtrpO1(A4C) biosensor variant under 0 to 500 mg/L tryptophan and 5-HTP, respectively. All experimental data was collected from three independent biological replicates (n=3). The standard deviations were present as error bars.
According to the above results, we hypothesize that mutations in loci 3 would function as low expression level biosensors whereas mutations in loci 4 would serve as medium and high expression level biosensors. In search of superior operational range, we did a second-round screening with combinations of representative TrpR1 variants and trpO1 variants under 0 to 500 mg/L tryptophan and 5-HTP, respectively (Figure S3 and S4). Following this round of screening, these operator variants firstly showed a different range of promoter strength as expected, triggering different expression levels. Then, the binding affinity between TrpR1-5-HTP complex and trpO1 operator variants was reduced by observing the wider operational ranges (Figure S3 and S4). When 5-HTP was added as the ligand, egfp expression was gradually repressed between 0 and 500 mg/L 5-HTP among operator mutations, showing an extended operational range. The loci 3 operator mutations combined with TrpR1 variants resulted in decreased basal expression and partially eliminated the leaky expression of the biosensor system. Two loci 4 mutations (A4G, A4T) coupled with TrpR1 variants showed increased basal expression and reduced dynamic ranges. The other one (A4C) had a medium expression strength and an acceptable dynamic behavior. Although most of the variant combinations exposed unsatisfied dynamic behavior, such as damaged dynamic ranges or unchanged operational ranges, there were still two notable combinations selected from the screening. The V58E-A4C variant was more favorable to sensing tryptophan than 5-HTP, showing an 80.7% repression efficiency to tryptophan and only 23.5% repression efficiency to 5-HTP (Figure 5c). However, the dynamic range towards tryptophan was decreased to a 5.2-fold change, which was lower than V58E-O1. Surprisingly, the V58K-A4C variant greatly reduced the tryptophan sensing capacity with only 10.1% repression efficiency (Figure 5d). Meanwhile, this variant well preserved the 5-HTP sensing capacity but with a modest dynamic range (a 10-fold change), and displayed a broadened operational range than V58K-O1 (Figure 4d and 5d). Overall, these new biosensor variants showed more distinct ligand preference but narrower dynamic ranges. Mutagenesis of the operator was shown to be a feasible technique for adjusting the dynamic behavior of the biosensor in this study.
Conclusion
The TrpR-based regulation in metabolic pathways of E. coli has been identified in previous studies. The native TrpR mainly regulates the enzymes expression of aromatic amino acid pathway by high-affinity binding to the operators in the presence of ligands. However, its engineering and application efforts have not emerged until recently. Within the scope of metabolic engineering, we applied synthetic biology methodologies to customize the dynamic behavior of TrpR1-PtrpO1 biosensor. Herein, we employed protein and operator engineering strategies to improve the dynamic and operational ranges. The key protein residue (I57 and V58) variants were screened for higher dynamic ranges and ligand specificity. The binding affinity of TrpR1 binding box was tuned by introducing base mutations within the trpO1 operator. Notably, operator mutations affected the expression levels and dynamic behavior of biosensor variants, implying an additive influence of regulator and operator engineering.
In conclusion, the genetic elements (regulator TrpR1 and operator trpO1) were engineered to create the optimized TrpR-based biosensor variants. The protein engineering of TrpR1 successfully improved the dynamic ranges of most variants. The V58 residue was identified as an important engineering target for accommodating larger side chain of 5-HTP due to the improvement of 5-HTP sensing ability among most TrpR1 V58 variants. While altered ligand preference was observed in TrpR1 variants V58E and V58K. These two variants preferred to sense tryptophan and 5-HTP, respectively. The operator engineering of trpO1 showed an addictive effect based on TrpR1 variant which led to a 4-fold broader operational range (from about 125mg/L to about 500 mg/L) in TrpR1(V58K)-PtrpO1(A4C) biosensor variant. This study comprehensively investigated the dynamic behavior and ligand preference of a series of TrpR-based biosensor variants, which could imply further applications in synthetic biology.
Supplementary Material
Acknowledgement
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM128620. We also acknowledge the support from the College of Engineering, The University of Georgia, Georgia, Athens, United States.
Footnotes
The authors declare no competing financial interest.
List of plasmids and strains used in this study, data of tryptophan content in different media, data of cell growth and repression efficiency of TrpR1-PtrpO1 biosensor in different media, data of TrpR1 double variants, data of the second round trpO1 variants screening.
References
- [1].Mitchler MM, Garcia JM, Montero NE, and Williams GJ (2021) Transcription factor-based biosensors: a molecular-guided approach for natural product engineering, Curr. Opin. Biotechnol 69, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hossain GS, Saini M, Miyake R, Ling H, and Chang MW (2020) Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms, Trends Biotechnol. 38, 797–810. [DOI] [PubMed] [Google Scholar]
- [3].Koch M, Pandi A, Borkowski O, Batista AC, and Faulon JL (2019) Custom-made transcriptional biosensors for metabolic engineering, Curr. Opin. Biotechnol 59, 78–84. [DOI] [PubMed] [Google Scholar]
- [4].Yang Y, Lin Y, Wang J, Wu Y, Zhang R, Cheng M, Shen X, Wang J, Chen Z, Li C, Yuan Q, and Yan Y (2018) Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis, Nat. Commun 9, 3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jacob F, and Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins, J. Mol Biol 3, 318–356. [DOI] [PubMed] [Google Scholar]
- [6].Zhang F, and Keasling J (2011) Biosensors and their applications in microbial metabolic engineering, Trends Microbiol. 19, 323–329. [DOI] [PubMed] [Google Scholar]
- [7].Liu D, Evans T, and Zhang F (2015) Applications and advances of metabolite biosensors for metabolic engineering, Metab. Eng 31, 35–43. [DOI] [PubMed] [Google Scholar]
- [8].Conrad KS, Manahan CC, and Crane BR (2014) Photochemistry of flavoprotein light sensors, Nat. Chem. Biol 10, 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guo AC, Jewison T, Wilson M, Liu Y, Knox C, Djoumbou Y, Lo P, Mandal R, Krishnamurthy R, and Wishart DS (2013) ECMDB: the E. coli Metabolome Database, Nucleic Acids Res. 41, 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jiang T, Li C, and Yan Y (2021) Optimization of a p-Coumaric Acid Biosensor System for Versatile Dynamic Performance, ACS Synth. Biol 10, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zou Y, Li C, Zhang R, Jiang T, Liu N, Wang J, Wang X, and Yan Y (2021) Exploring the Tunability and Dynamic Properties of MarR-PmarO Sensor System in Escherichia coli, ACS Synth. Biol 10, 2076–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Farmer WR, and Liao JC (2000) Improving lycopene production in Escherichia coli by engineering metabolic control, Nat. Biotechnol 18, 533–537. [DOI] [PubMed] [Google Scholar]
- [13].Xu P, Li L, Zhang F, Stephanopoulos G, and Koffas M (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control, Proc. Natl. Acad. Sci. U. S. A 111, 11299–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang F, Carothers JM, and Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids, Nat. Biotechnol 30, 354–359. [DOI] [PubMed] [Google Scholar]
- [15].Wang J, Zhang R, Zhang J, Gong X, Jiang T, Sun X, Shen X, Wang J, Yuan Q, and Yan Y (2021) Tunable hybrid carbon metabolism coordination for the carbon-efficient biosynthesis of 1,3-butanediol in Escherichia coli, Green Chem. 23, 8694–8706. [Google Scholar]
- [16].Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, and Keasling JD (2013) Engineering dynamic pathway regulation using stress-response promoters, Nat. Biotechnol 31, 1039–1046. [DOI] [PubMed] [Google Scholar]
- [17].Hartline CJ, Schmitz AC, Han Y, and Zhang F (2021) Dynamic control in metabolic engineering: Theories, tools, and applications, Metab. Eng 63, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin Y, Sun X, Yuan Q, and Yan Y (2014) Engineering bacterial phenylalanine 4-hydroxylase for microbial synthesis of human neurotransmitter precursor 5-hydroxytryptophan, ACS Synth. Biol 3, 497–505. [DOI] [PubMed] [Google Scholar]
- [19].Wang J, Guleria S, Koffas MAG, and Yan Y (2016) Microbial production of value-added nutraceuticals, Curr. Opin. Biotechnol 37, 97–104. [DOI] [PubMed] [Google Scholar]
- [20].Kumamoto AA, Miller WG, and Gunsalus RP (1987) Escherichia coli tryptophan repressor binds multiple sites within the aroH and trp operators, Genes Dev. 1, 556–564. [DOI] [PubMed] [Google Scholar]
- [21].Grove CL, and Gunsalus RP (1987) Regulation of the aroH operon of Escherichia coli by the tryptophan repressor, J. Bacteriol 169, 2158–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bogosian G, Somerville RL, Nishi K, Kano Y, and Imamoto F (1984) Transcription of the trpR gene of Escherichia coli: an autogeneously regulated system studied by direct measurements of mRNA levels in vivo, Mol. Gen. Genet 193, 244–250. [DOI] [PubMed] [Google Scholar]
- [23].Lawley B, and Pittard AJ (1994) Regulation of aroL expression by TyrR protein and Trp repressor in Escherichia coli K-12, J. Bacteriol 176, 6921–6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jeeves M, Evans PD, Parslow RA, Jaseja M, and Hyde EI (1999) Studies of the Escherichia coli Trp repressor binding to its five operators and to variant operator sequences, Eur. J. Biochem 265, 919–928. [DOI] [PubMed] [Google Scholar]
- [25].Zhao D, Arrowsmith CH, Jia X, and Jardetzky O (1993) Refined solution structures of the Escherichia coli trp holo- and aporepressor, J. Mol. Biol 229, 735–746. [DOI] [PubMed] [Google Scholar]
- [26].Bass S, Sugiono P, Arvidson DN, Gunsalus RP, and Youderian P (1987) DNA specificity determinants of Escherichia coli tryptophan repressor binding, Genes Dev. 1, 565–572. [DOI] [PubMed] [Google Scholar]
- [27].Ellefson JW, Ledbetter MP, and Ellington AD (2018) Directed evolution of a synthetic phylogeny of programmable Trp repressors, Nat. Chem. Biol 14, 361–367. [DOI] [PubMed] [Google Scholar]
- [28].Zhang J, Petersen SD, Radivojevic T, Ramirez A, Pérez-Manríquez A, Abeliuk E, Sánchez BJ, Costello Z, Chen Y, Fero MJ, Martin HG, Nielsen J, Keasling JD, and Jensen MK (2020) Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism, Nat. Commun 11, 4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Thomason LC, Costantino N, and Court DL (2007) E. coli genome manipulation by P1 transduction, Curr. Protoc. Mol. Biol Chapter 1, 1.17.11–11.17.18. [DOI] [PubMed] [Google Scholar]
- [30].Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products, Proc. Natl. Acad. Sci. U. S. A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lutz R, and Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements, Nucleic Acids Res. 25, 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jacobsen JPR, Krystal AD, Krishnan KRR, and Caron MG (2016) Adjunctive 5-Hydroxytryptophan Slow-Release for Treatment-Resistant Depression: Clinical and Preclinical Rationale, Trends Pharmacol. Sci 37, 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Maffei ME (2020) 5-Hydroxytryptophan (5-HTP): Natural Occurrence, Analysis, Biosynthesis, Biotechnology, Physiology and Toxicology, Int. J. Mol. Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schevitz RW, Otwinowski Z, Joachimiak A, Lawson CL, and Sigler PB (1985) The three-dimensional structure of trp repressor, Nature. 317, 782–786. [DOI] [PubMed] [Google Scholar]
- [35].Lawson CL, Zhang RG, Schevitz RW, Otwinowski Z, Joachimiak A, and Sigler PB (1988) Flexibility of the DNA-binding domains of trp repressor, Proteins 3, 18–31. [DOI] [PubMed] [Google Scholar]
- [36].Bass S, Sorrells V, and Youderian P (1988) Mutant Trp repressors with new DNA-binding specificities, Science 242, 240–245 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


