ABSTRACT
The increased detection of H3 C-IVA (1990.4.a) clade influenza A viruses (IAVs) in US swine in 2019 was associated with a reassortment event to acquire an H1N1pdm09 lineage nucleoprotein (pdmNP) gene, replacing a TRIG lineage NP (trigNP). We hypothesized that acquiring the pdmNP conferred a selective advantage over prior circulating H3 viruses with a trigNP. To investigate the role of NP reassortment in transmission, we identified two contemporary 1990.4.a representative strains (NC/19 and MN/18) with different evolutionary origins of the NP gene. A reverse genetics system was used to generate wild-type (wt) strains and swap the pdm and TRIG lineage NP genes, generating four viruses: wtNC/19-pdmNP, NC/19-trigNP, wtMN/18-trigNP, and MN/18-pdmNP. The pathogenicity and transmission of the four viruses were compared in pigs. All four viruses infected 10 primary pigs and transmitted to five indirect contact pigs per group. Pigs infected via contact with MN/18-pdmNP shed virus 2 days earlier than pigs infected with wtMN/18-trigNP. The inverse did not occur for wtNC/19-pdmNP and NC/19-trigNP. This suggests that pdmNP reassortment resulted in a combination of genes that improved transmission efficiency when paired with the 1990.4.a hemagglutinin (HA). This is likely a multigenic trait, as replacing the trigNP gene did not diminish the transmission of a wild-type IAV in swine. This study demonstrates how reassortment and evolutionary change of internal genes can result in more transmissible viruses that influence HA clade detection frequency. Thus, rapidly identifying novel reassortants paired with dominant hemagglutinin/neuraminidase may improve the prediction of strains to include in vaccines.
IMPORTANCE
Influenza A viruses (IAVs) are composed of eight non-continuous gene segments that can reassort during coinfection of a host, creating new combinations. Some gene combinations may convey a selective advantage and be paired together preferentially. A reassortment event was detected in swine in the United States that involved the exchange of two lineages of nucleoprotein (NP) genes (trigNP to pdmNP) that became a predominant genotype detected in surveillance. Using a transmission study, we demonstrated that exchanging the trigNP for a pdmNP caused the virus to shed from the nose at higher levels and transmit to other pigs more rapidly. Replacing a pdmNP with a trigNP did not hinder transmission, suggesting that transmission efficiency depends on interactions between multiple genes. This demonstrates how reassortment alters IAV transmission and that reassortment events can provide an explanation for why genetically related viruses with different internal gene combinations experience rapid fluxes in detection frequency.
KEYWORDS: influenza A virus, nucleoprotein, reassortment, viral transmission, reverse genetics
INTRODUCTION
The interactions between the eight genes of influenza A virus (IAV) contribute to pathogenesis and transmission in swine. The two surface genes, hemagglutinin (HA) and neuraminidase (NA), have a well-defined role in IAV biology, but the contributions of the remaining six genes are less clearly defined. Colloquially, these six genes are called “internal genes” to differentiate them from the surface glycoproteins, and they encode the viral polymerase (PB2, PB1, and PA), nucleoprotein (NP), matrix protein (M), and non-structural protein (NS). IAV has a segmented genome and the ability to reassort, leading to different combinations of the eight gene segments and a vast potential for genetic diversity. Similar to the previously described requirement of balance between the HA and NA (1–3), there appear to be specific gene segments and/or combinations of internal genes, known as internal gene constellations, that may be preferentially selected among viral populations (4). These combinations of internal genes are permissive to different HA and NA combinations and demonstrate sustained transmission in swine populations (5).
In swine, the internal genes are classified based on their evolutionary lineage: the triple reassortant (TRIG) lineage, the human 2009 pandemic (pdm) lineage, or the live-attenuated influenza virus (LAIV) vaccine lineage. The internal gene constellation is abbreviated using the first letter of each lineage (T, P, or L) to form a six-character sequence in the following order: PB2, PB1, PA, NP, M, and NS. The TRIG lineage emerged in the late 1990s and consists of PB1 gene segments derived from human seasonal H3N2; PB2 and PA gene segments from avian IAV; and NP, M, and NS gene segments from classical swine H1N1 (6–8). Prior to the 2009 pandemic, all endemic viruses in the US swine population contained the TRIG internal gene constellation, TTTTTT (5). Following the H1N1 pandemic in 2009, the virus was repeatedly introduced into the pig population by humans (9–11). The eight gene segments in their entirety generally do not persist in US pig populations for extended periods of time, but onward transmission of the pdm internal genes through reassortment was common (11). From 2009 to 2012, the pdmM gene increased in abundance from 38% to 70%, making the TTTTPT constellation the second most commonly detected during this time period (12). The pdmPA and pdmNP genes were also detected during this period but did not increase in abundance like the M gene (4, 13, 14).
In a previous work, we demonstrated a rapid change in the detection frequency of the 1990.4.a clade of H3 IAV that began in 2019 (15). Following the same general genome constellation patterns described above, the 1990.4.a internal gene constellation pre-pandemic was TTTTTT. The 1990.4.a viruses then acquired a pdmM gene, and this TTTTPT constellation became the most common in the clade between 2011 and 2018 (4). However, the internal gene constellation found in the 1990.4.a major clade that expanded in detection frequency beginning in 2019 acquired a pdmNP gene (15). Prior to 2019, this TTTPPT constellation was sporadically observed but was uncommon (22 of 368 isolates detected between 2009 and 2016) (4). Despite sharing a common ancestor with the classical swine H1N1 lineage, the trigNP and pdmNP have evolved into distinct genetic clades over the past decade. The pdmNP acquired by the 1990.4.a clade in 2019 was derived from a human-to-swine spillover event during the 2009–2010 human influenza season that subsequently reassorted multiple times with endemic swine IAV (15). The NP gene codes for a structural RNA-binding protein that, along with the trimeric polymerase complex and viral RNA, forms the ribonucleoprotein (RNP) particle. The NP is known to regulate multiple processes, including nuclear import/export of vRNP and transcription/replication (16). In swine, results from a transmission study in pigs demonstrated that H3 viruses with the TTTPPT constellation were more effective in transmission when compared to H3 strains with a TTTTPT constellation (4). Based on the function of the NP and the increasing detection of the TTTPPT constellation, we hypothesized that the success of the expanding clade of 1990.4.a H3 viruses could be explained by differences in the genetic lineage of the NP acquired following reassortment. We tested this hypothesis using a reverse genetics system to swap the respective pdm or TRIG lineage NP between two sustained 1990.4.a viruses. We demonstrated that the exchange of the pdmNP in the context of a genome previously containing a trigNP has the potential to improve transmission efficiency, but replacing a pdmNP with a trigNP does not necessarily have the opposite effect. Our results suggest that viral transmission is dependent on multiple genes and the context of the other seven genes of the genome.
MATERIALS AND METHODS
Genetic analysis
In 2019, a NP reassortment event associated with increased detections of an H3 1990.4.a clade of viruses was detected (15). There was a shift from a whole genome constellation of TTTTPT to the constellation including a pdmNP (TTTPPT) in the majority of viruses from this H3 HA clade. We collected all available complete whole genome sequences (WGSs) of 1990.4.a viruses from 2016 to 2021 (n = 177) from the USDA passive surveillance database, octoFLUshow (17), to quantify the frequency of detection of the internal gene constellations in this expanding clade over time. To determine the detection frequency of the pdmNP in viruses of other HA clades, we also collated all complete swine H1 and H3 WGS from the USDA surveillance system (n = 1,720). Differences in frequency of each whole genome constellation between two time periods (2016–2018 and 2019–2021) were plotted using R Statistical Software (v4.0.5; R Core Team 2021). The Nextstrain (18) platform was used to analyze the genetic diversity of the pdmNP genes (n = 68) that paired with the H3 1990.4.a clade from 2010 to 2021. Augur commands translate and ancestral (19) were used to annotate amino acid substitutions on the backbone of the time-scaled maximum likelihood tree.
Strain selection
Two IAVs in swine viruses, A/swine/North Carolina/A02245294/2019 (NC/19) (GenBank accession numbers MN646208.1, MN646209.1, and MT233822.1–MT233827.1) and A/swine/Minnesota/A02266068/2018 (MN/18) (GenBank accession numbers MK122759.1, MK122760.1, and MW308842.1–MW308847.1), were identified that had representative HA genes of the minor (TTTTPT) and major (TTTPPT) clades and were associated with the 1990.4.a expansion in 2019 (Fig. 1) (15). The remaining seven gene segments of these two strains were confirmed to represent the TTTTPT and TTTPPT constellations circulating in swine. First, we subsetted the larger 1990.4.a data set described above to only include viruses with the internal gene constellations TTTTPT (n = 21) and TTTPPT (n = 58) since 2018. Geneious Prime v2021.2 was used to construct two consensus sequences for each of the seven gene segments representing the TTTTPT and TTTPPT constellations. We used mafft v7.490 to confirm that the genes in NC/19 and MN/18 had at least 99.0% amino acid (AA) identity to the consensus sequence of the constellation they represented (20).
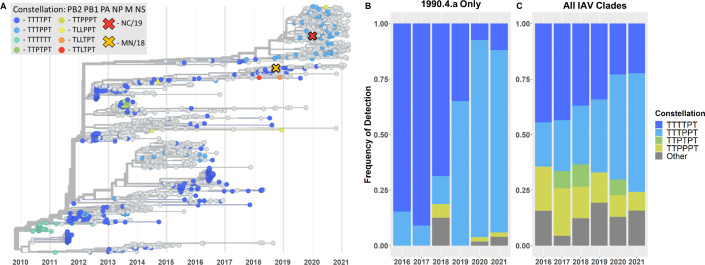
Fig 1.
Summary of change in predominant internal gene constellation showed a shift from TRIG to pdm lineage NP over time. (A) Time-scaled phylogenetic tree of H3 1990.4.a IAV from 2010 to 2021. Chosen clade representative viruses, NC/19 and MN/18, are represented by an “X” on the tree. When possible, strains are colored by their internal gene constellation. (B) Frequency of detection of the most common internal gene constellations in H3 1990.4.a IAV from 2016 to 2021. (C) Frequency of detection of the most common internal gene constellation in all IAV clades from 2016 to 2021. The “other” category in (B) and (C) includes any gene constellations not explicitly listed.
Reverse genetics
Reverse genetics was used to generate four viruses using an eight-plasmid system as previously described (21, 22). All the genes of the two representative viruses (NC/19 and MN/18) were synthesized (Twist Bioscience, San Francisco, CA) and subsequently cloned into the bidirectional plasmid vector pDP2002. For the transfection, cocultures of 9 × 105 HEK293T and 1.5 × 105 Madin-Darby canine kidney (MDCK) cells were seeded per well in a six-well plate. The following day, 1 µg of each plasmid was mixed with 18 µL of TransIT-LT1 transfection reagent (Mirus Bio LLC, Madison, WI). The mixture was incubated for 45 min and then used to overlay the 293T/MDCK cells overnight. The next day, the transfection mixture was replaced with fresh Opti-MEM media containing 1% AB (Life Technologies, Carlsbad, CA), and 24 h post-transfection, the media was supplemented with 1 µg/mL of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Worthington Biochemicals, Lakewood, NJ). Wild-type viruses were generated with their respective NP genes (wtNC/19-pdmNP and wtMN/18-trigNP). Reassortants of NC/19 and MN/18 were created by swapping only the NP genes between the two viruses (NC/19-trigNP and MN/18-pdmNP) and maintaining the remaining seven genes from the respective parental strains. Viral sequences were confirmed by Sanger sequencing and propagated in MDCK cells.
In vivo animal study
A total of 65 healthy 3-week-old, crossbred pigs were obtained from a herd free of IAV, porcine reproductive and respiratory syndrome virus, and Mycoplasma hyopneumoniae. Upon arrival, pigs were confirmed to be seronegative to IAV NP antibodies by a commercial enzyme-linked immunosorbent assay kit (Swine Influenza Virus Antibody Test, IDEXX, Westbrook, ME). They were also treated with ceftiofur crystalline-free acid and tulathromycin (Zoetis Animal Health, Florham Park, NJ) to reduce the risk of bacterial contaminants. Pigs were randomly selected for the non-challenge control (n = 5) or four primary challenge groups (n = 10/group), implanted with a LifeChip with Bio-Thermo Technology (Destron Fearing, DFW Airport, Texas), and moved to separate isolation rooms by group. The remaining pigs were assigned to be indirect contacts to the primary challenge pigs (n = 5/group). All pigs were housed in biosafety level 2 containment and cared for in compliance with the Institutional Animal Care and Use Committee of the USDA National Animal Disease Center.
At approximately 5 weeks of age, the primary challenge pigs were inoculated with one of the four viruses: wtNC/19-pdmNP, NC/19-trigNP, wtMN/18-trigNP, and MN/18-pdmNP. Pigs in the assigned group were inoculated intranasally (0.5 mL/nostril) and intratracheally (2 mL) with 1 × 105 50% tissue culture infectious dose (TCID50) per milliliter. For inoculation, pigs were anesthetized with an intramuscular injection of ketamine (8 mg/kg body weight; Phoenix, St. Joseph, MO), xylazine (4 mg/kg; Lloyd Inc., Shenandoah, IA), and tiletamine HCL/zolazepam HCL (Telazol; 6 mg/kg; Zoetis Animal Health, Florham Park, NJ). At 2 days post-inoculation (DPI), indirect contact pigs were placed in separate raised decks in each of the four isolated challenge rooms. Nasal swabs (NS) and temperature readings were collected from the primary pigs on 0–5 DPI. NSs were collected from contact pigs on 0–5, 7, and 9 days post-contact (DPC). Nasal swabs were placed in tubes containing 2 mL minimum essential media (MEM) with 1 µL/mL TPCK trypsin and stored at −80°C.
On 5 DPI, primary pigs were humanely euthanized with a lethal dose of pentobarbital sodium (Fatal Plus; Vortech Pharmaceuticals, Dearborn, MI) and necropsied, at which time the lungs were removed, scored for macroscopic lesions, and lavaged with 50 mL of MEM containing 1% bovine serum albumin to collect bronchoalveolar lavage fluid (BALF). BALF samples were plated on casein and blood agar plates to evaluate for bacterial contamination prior to storing at −80°C. The distal trachea and right middle or affected lung lobe were collected and fixed in 10% buffered formalin, then processed using routine histologic methods to score microscopic lesions. On 11 DPC, contact pigs were bled and humanely euthanized using the same method described above.
Virologic assays
Nasal swabs were filtered through a 0.2-µ syringe filter prior to virus isolation (VI). MDCK cells were grown to confluency in 48-well plates, which were then washed twice with phosphate-buffered saline. The filtered NS samples were then plated onto confluent MDCK cells and cultured at 37°C for 48 h. Plates were fixed at 48 h and stained via immunocytochemistry as previously described (23). To calculate a virus titer for NS that was positive by VI and all BALF samples, 10-fold serial dilutions in MEM with 1:1,000 TPCK trypsin were performed in triplicate onto confluent MDCK cells in 96-well plates. After 48 h of incubation at 37°C, the plates were fixed and stained with the same protocol utilized for VI. Titers were calculated using the Reed and Muench method, and TCID50/mL titers were log10-transformed for statistical analysis (24).
Pathology
When the lungs were removed at necropsy, the percentage of lung surface affected by the purple-red consolidation typical of IAV infection was immediately recorded for each lobe. The weighted proportion of each lobe to the total lung volume was estimated to calculate the affected percentage of the entire lung (25). The fixed lung and trachea sections were routinely processed and stained with hematoxylin and eosin. A veterinary pathologist blinded to the treatment groups scored the microscopic lesions using previously described parameters (26). The parameters of each tissue were summed to calculate a total microscopic lesion score for the lung (0–22) and trachea (0–8).
Serologic assays
Serum samples collected from the indirect contact pigs at 11 DPC were prepared for the hemagglutination inhibition (HI) assay by receptor-destroying enzyme (RDE II; Hardy Diagnostics, Santa Maria, CA) treatment, heat inactivation, and the addition of 50% turkey red blood cells to remove non-specific hemagglutination inhibitors. HI assays were performed using test antigens from the respective wild-type challenge viruses with the same HA gene using previously described techniques (27).
Statistical analysis
All analyses were performed using R Statistical Software (v4.0.5; R Core Team 2021). One-way analysis of variance followed by a Tukey–Kramer test was performed on the following variables using the core statistics package in R (v4.0.5; R Core Team): macroscopic pneumonia, microscopic lung and trachea lesions, and log10-transformed BALF and NS titers. Compact letter displays were generated when significant differences in group means (P-value <0.05) were present using the multcomp R package (v1.4.16; Bretz 2010).
RESULTS
TTTPPT replaced TTTTPT as the majority IAV internal gene constellations in 2021
Since 2010, eight different internal gene constellations were paired with the H3 1990.4.a clade (Fig. 1A), and the TTTTPT constellation comprised the majority of detections until 2019. There were sporadic TTTPPT detections during this timeframe, but only the detections after 2018 were sustained in the swine population. The TTTPPT constellation increased in frequency of detection in the H3 1990.4.a clade from 12.5% in 2018 to 65% in 2019 (Fig. 1B). In 2020, the TTTPPT constellation further increased in detection making up 88.7% of the detections within the H3 1990.4.a clade. When evaluating all IAV H1 and H3 clades, the TTTPPT constellation detection frequency increased steadily, with 20.0% of detections in 2016 increasing to 53.3% of detections by 2021 (Fig. 1C). The increased detection of the TTTPPT constellation (27% increase between 2016–2018 and 2019–2021) was associated with a decreased detection of the TTTTPT constellation that demonstrated a 16.5% decrease between 2016–2018 and 2019–2021 (Fig. S1). In addition to H3 1990.4.a, the TTTPPT constellation also increased in the H1 1A.3.3.3 (gamma) and H1 1B.2.1 (delta2) clades (Fig. S1).
Two distinct clades of pdmNP were detected in the 1990.4.a clade between 2010 and 2021. The detections associated with the 2019 HA clade expansion were of the same lineage, which was the result of a human-to-swine spillover in the 2009–2010 human influenza season and subsequent reassortment in the swine population (15). Three amino acid substitutions differentiate the lineage of pdmNP associated with the clade expansion from the lineage of previously detected pdmNP (I100V, K452R, and N377S). All three substitutions were estimated to have occurred between 2012 and 2014. There were no recent amino acid substitutions in the pdmNP associated with the increased detection of the TTTPPT constellation in the 1990.4.a clade beginning in 2019.
Test strains represented contemporary detections and were significantly different
Two strains, NC/19 and MN/18, were chosen to represent the two contemporary H3 1990.4.a clades (Fig. 1A). Both consisted of internal gene segments that were 99.0%–100% identical at the amino acid level to the constellation consensus of the clade they were chosen to represent (Table 1). The two representative strains differed across their genomes, with gene segments ranging from 91.7% to 98.5% identical at the amino acid level (Table 1). The NP genes were 97.2% AA identical between the pdmNP of NC/19 and the trigNP of MN/18, with their amino acid sequences differing at 14 positions (38, 70, 109, 217, 231, 301, 313, 353, 373, 377, 426, 456, and 473).
TABLE 1.
Percent pairwise AA identity for each pair of IAV gene segments
| % AA identity | PB2 | PB1 | PA | HA | NP | NA | M | NS |
|---|---|---|---|---|---|---|---|---|
| NC/19 vs MN/18a | 97.2% | 98.3% | 98.5% | 95.6% | 97.2% | 91.7% | 97.6% | 92.5% |
| NC/19 vs H3 1990.4.a TTTPPT 2018–2020 consensus sequence (n = 58)b | 99.7% | 99.7% | 99.0% | 100.0% | 100.0% | 100.0% | ||
| MN/18 vs H3 1990.4.a TTTTPT 2018–2020 consensus sequence (n = 21)b | 99.9% | 99.9% | 100.0% | 99.6% | 99.7% | 99.6% |
Comparison between the two representative field strains, A/swine/North Carolina/A02245294/2019 and A/swine/Minnesota/A02266068/2018.
Comparisons between one of the representative strains (NC/19 or MN/18) and the consensus sequence of the strains it was chosen to represent.
NP lineage did not affect pathology
All pigs infected with the four experimental strains selected to represent the NP reassortment exhibited limited to mild clinical signs consistent with experimental IAV infection in swine. The non-challenged control pigs showed no clinical signs nor evidence of lung pathology indicative of IAV infection. All four of the challenge strains induced mild pneumonia in the primary pigs, with percentages of total lung affected that were not dependent on NP lineage (Table 2). There were no significant differences in the mean microscopic lung or trachea scores between challenge groups (one-way ANOVA, P = 0.96, 0.80).
TABLE 2.
Characteristics of infection with four challenge strainsa
| Group | Macroscopic pneumonia (%) | Microscopic lung scores (0–22) | Microscopic trachea scores (0–8) | BALF virus titer (log10 TCID50) |
|---|---|---|---|---|
| Control | 0.2 ± 0.1b | 0.0 ± 0.0b | 0.0 ± 0.0b | 0.0 ± 0.0b |
| wtNC/19-pdmNP | 6.4 ± 1.4d | 6.4 ± 0.7c | 3.3 ± 0.5c | 4.1 ± 0.3c |
| NC/19-trigNP | 5.0 ± 0.6cd | 6.3 ± 1.0c | 3.7 ± 0.4c | 5.4 ± 0.2d |
| wtMN/18-trigNP | 3.6 ± 0.6bcd | 6.0 ± 0.6c | 3.2 ± 0.3c | 3.8 ± 0.4c |
| MN/18-pdmNP | 3.2 ± 0.5bc | 5.8 ± 1.1c | 3.2 ± 0.4c | 4.4 ± 0.2cd |
Shown are the mean (±SEM) percentages of macroscopic lung consolidation, microscopic lung and trachea composite scores, and BALF virus titers of pigs experimentally inoculated with the challenge viruses or negative control pigs. Superscript letters b, c, and d depict statistically significant differences between values in the same column (P < 0.05).
Inclusion of a pdmNP into the MN/18 gene combination improved transmission efficiency
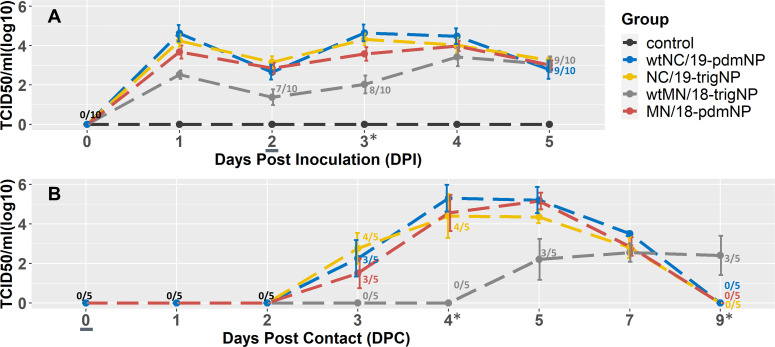
IAV was isolated from the NS of all primary challenge pigs and the BALF of all but one pig, which was in the wtMN/18-trigNP challenge group. No virus was detected in the NS or BALF of non-challenged negative control pigs. Nasal shedding of all primary challenged groups began on 1 DPI, decreased slightly on 2 DPI, and then was sustained at levels similar to 1 DPI until euthanasia on 5 DPI. The wtMN/18-trigNP challenge group had a trend for lower NS viral titers between 1 and 4 DPI, with a statistically significant difference compared to the other groups on 3 DPI (Fig. 2; one-way ANOVA, P < 0.001). The amount of virus in the BALF did not differ significantly between the two groups challenged with the two MN/18 viruses, but the NC/19-trigNP group had significantly higher BALF viral titers than the wtNC/19-pdmNP group (Table 3; one-way ANOVA, P = 0.50, P < 0.01).
Fig 2.
Mean (±SEM) viral titers over the (A) 5 days post-inoculation for 10 primary pigs and (B) 9 days post-contact for five indirect contact pigs. The contacts were added on 2 DPI, which corresponds with 0 DPC, denoted by an underline. Days when the viral titer of the wtMN/18-trigNP challenge group significantly differed from the other challenge groups are denoted by an asterisk (*). Days when <100% of pigs were shedding are labeled as n/10 (A) or n/5 (B) and colored by the group to which they belong. Black coloring indicates this number applies to all groups.
TABLE 3.
Hemagglutination inhibition titers in serum from contact pigs at 11 DPC
| HI assay | wtNC/19-pdmNP | NC/19-trigNP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pig | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 |
| wtNC/19-pdmNP | 160 | 160 | 80 | 160 | 160 | 640 | 160 | 160 | 80 | 160 |
| wtMN/18-trigNP | MN/18-pdmNP | |||||||||
| Pig | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 |
| wtMN/18-trigNP | 10 | 80 | 40 | 10 | 80 | 80 | 80 | 80 | 80 | 160 |
The indirect contact groups were added on 2 DPI/0 DPC; all four challenge viruses successfully transmitted to indirect contact pigs, i.e., the virus was detected by titration from NS and BALF. Virus was detected in the NS of at least three out of five pigs in each group by 3 DPC (Fig. 2); however, IAV was not detected in the wtMN/18-trigNP contact group NS until 5 DPC (three out of five). By 9 DPC, the virus was no longer detectable in the NS of the wtNC/19-pdmNP, NC/19-trigNP, and MN/18-pdmNP contact pigs. At the same time point, 3/5 of the wtMN/18-trigNP contact pigs were still positive by NS but did not reach the same maximum mean group titer as the contacts in the other virus groups. These data suggest that the transmission curve for the wtMN/19-trigNP was delayed by approximately 2 days along with a reduction in group mean titers in nasal secretions, while the MN/19-pdmNP transmission dynamics were comparable with the two NC/19 viruses.
Reduced transmission efficiency delayed seroconversion
HI assays against the homologous virus were performed on the terminal serum of each indirect contact pig. Using a cutoff of HI titer ≥20, all indirect contact pigs seroconverted by the time of euthanasia at 11 DPC for the wtNC/19-pdmNP, NC/19-trigNP, and MN/18-pdm NP contact groups (Table 3). Two pigs (#622 and #625) in the wtMN/18-trigNP contact group did not seroconvert by this time point, and the HI titers of the wtMN/18-trigNP contact group were the lowest of the four groups.
DISCUSSION
The observed genetic diversity of IAV in swine has been substantially shaped by the emergence of the H1N1 pandemic in 2009 and subsequent human-to-swine transmission events. Following interspecies transmission, the internal genes of the human H1N1pdm09 virus reassort with endemic swine IAV, and these endemic swine IAVs with H1N1pdm09-origin gene segments continue to diffuse through the swine IAV population. In 2011, the pdm lineage M gene became more commonly detected in IAV isolates from swine than the TRIG lineage M gene (28). However, the majority of the other internal genes sequenced from swine isolates have remained in the TRIG lineage. We previously reported that H3 1990.4.a viruses first expanded the TTTPPT constellation in 2019 (15). Here, we report that in 2021, 12 years after the 2009 pandemic, the majority of all H1 and H3 IAV in swine in the US contained pdmNP genes. To investigate the expansion of the H3 1990.4.a clade, we used a reverse genetics approach to generate two wild-type viruses and then exchanged the pdm and TRIG lineage NP genes between them. We evaluated the pathogenesis and transmission of these four viruses in commercial weaned pigs. Our results showed that the acquisition of the pdmNP improved transmission efficiency in the context of the MN/18 virus genome, but replacing a pdmNP gene alone may not diminish the transmission of an otherwise successful virus. Though this is a subtle difference, a virus that transmits 2 days sooner may “beat” other viruses to infect the remainder of the susceptible population within a swine production system and effectively outcompete them, causing certain genes to achieve dominance in the population.
It was expected that all four viruses would infect the primary challenged pigs and successfully transmit to the indirect contacts as we demonstrated. We used two wild-type viruses that were representative of H3N2 viruses routinely isolated from field cases submitted for diagnostic investigation, indicating that both viruses were fit for the swine host. Additionally, these two strains contained two of the most common internal gene constellations found in all H1 and H3 IAVs circulating in swine in the USA, TTTTPT and TTTPPT. When compared to the wtNC/19-pdmNP virus, the wtMN/18-trigNP virus had decreased nasal shedding in the primary pigs at 1 and 3 DPI, and infection of the indirect contact pigs was delayed by approximately 2 days. When the trigNP was replaced with a pdmNP in the MN/18 virus (MN/18-pdmNP), the magnitude of nasal shedding and transmission kinetics were comparable to the wtNC/19-pdmNP. Because the NP gene was the only difference between these two viruses, we conclude that the acquisition of the pdmNP gene in the context of the other genes comprising the MN/18 virus could improve transmission efficiency. The NP protein is expressed in the early stage of infection and regulates multiple processes in the virus lifecycle, including nuclear import and export of viral RNP and transcription and replication of viral RNA (16, 29). These roles in viral replication affect transmission efficiency by the impact on nascent virus production.
In contrast to the significant impact of the pdmNP on the MN/18 combination of genes, nasal shedding and transmission to indirect contacts did not differ between the NC/19 viruses when the pdmNP was replaced with a trigNP. While the contemporary pdmNP and trigNP differ by approximately 3% (14 amino acids), there were other larger differences across the genomes of the NC/19 and MN/18 viruses, such as those in the NA and NS genes, that could explain why the impacts of the NP were not bi-directional. Thus, it is likely that transmission efficiency is a multigenic trait. A parallel idea exists when we consider the transmission of IAV within a new host species. Supporting this proposition was an in vivo pathogen and transmission study performed with reassortant viruses created to assess the contributions of gene combinations to the replication and transmission of IAV in a novel host species (30). This study used a human-seasonal H3 virus and a swine H3N1 virus and demonstrated that a swine-adapted HA gene alone could confer the ability to replicate in the lungs in the context of human NA and internal genes but that the ability to replicate in the upper respiratory tract was dependent on the interaction between the HA gene and the NA or internal genes. Thus, transmission and viral phenotype rely upon individual gene segments and the interactions between all gene segments in the virus. Characterization of a diversity of viruses that capture whole genome constellations that have increased or decreased in detection frequency, alongside those that have gone extinct, should be investigated to understand how gene–gene interactions are impacting IAV transmission in swine. In our study, reassortment to acquire a pdmNP may not be the only mechanism explaining the 2019 H3N2 1990.4.a expansion. In our previous investigation, the emergence of this clade was associated with the pdmNP reassortment and substitution in a key HA antigenic site (N156H) of the expanding H3 genes (15). Additionally, population immunity to the diversity of H3 viruses in swine was also a likely contributor to the detection frequency of the H3 1990.4.a genetic clade (15). Though we documented earlier transmission of the virus with the pdmNP in the MN/18 genome, there was no statistically significant difference between the wtNC/19-pdmNP and NC/19-trigNP viruses in the parameters we measured. Furthermore, the NC/19 representative of the expanding clade was capable of replicating and transmitting effectively in pigs and was robust to changes in the NP gene, i.e., inserting the trigNP did not diminish the transmission efficiency of this combination of genes. These data further support the proposition that replication and transmission efficiency of IAV in the swine host, and likely virulence, are multigenic traits. Our study of the expansion of the H3 1990.4.a clade highlights the need for better tools for determining the net effect of multiple gene pairings and mutations within genes (e.g., mutations within the HA and NP genes as seen here) on virus phenotype.
Genomic surveillance of influenza A virus in swine has been conducted systematically for over a decade in the US (17), but most of the effort has been guided toward analyzing the HA and, to a lesser extent, the NA gene(s). These surveillance data have been used to determine spatial and temporal trends in IAV in swine genetic diversity (14, 31, 32), support public health pandemic preparedness (8, 33), and facilitate the selection of strains for use as antigens in vaccines (34–36). However, our data reveal that understanding reassortment and the interactions between internal genes and the surface proteins is necessary to understand the dynamics of HA/NA clade detection frequency and turnover. Several veterinary diagnostic laboratories now offer whole genome sequencing for IAV, and the USDA IAV in swine surveillance system routinely generates whole genome sequences of swine IAV isolates. Based on current knowledge, the high genetic diversity and interplay between genes have made sequencing the entire viral genome a necessity. We show the impact that the NP gene can have on the transmission efficiency of an isogenic virus and hypothesize that the NP gene provided a competitive advantage to certain viruses. We also show that the TTTPPT constellation has increased in detection frequency for other H1 and H3 genetic clades: this suggests that other surface proteins may also be compatible with this gene combination and may also have experienced increases in transmission efficiency. Routine whole genome sequencing can facilitate in predicting when an HA/NA combination will increase or decrease in detection frequency through the identification of internal genes or constellations that have evidence for impact on transmission, or represent unique pairings, thus improving the efficacy and utility of monitoring and surveillance programs.
An unresolved question within our study is why it took 12 years for this pdmNP gene that has demonstrated a benefit to transmission to become dominant in the genomes of current IAV viruses in swine. When H1N1pdm2009 viruses were originally transmitted from humans into swine as a sequela of the pandemic, they rarely maintained purely H1N1pdm09 genomes: the surface proteins tended to persist when they underwent reassortment events that replaced human pdm internal genes with those currently endemic in US swine (10, 37). However, within 2 years, the pdm M gene provided a selective advantage in the swine host and became the primary M gene in endemic IAV. Perhaps, the difference is in how similar the individual genes were to those that were currently circulating in the swine population. The H1N1 2009 pandemic marks a historical moment in time from which influenza researchers have classified the descendant internal genes, but this classification scheme is not equally reflective of differences in identity. The trigNP, pdmNP, and trig M gene are all descendants of classical swine H1N1, while the pdm M gene is of Eurasian avian-like swine origin. At the time of the H1N1 2009 pandemic, the pdmNP gene was 99%–100% identical to what was currently circulating in US swine (38). The pdmNP described in this study can be epidemiologically linked to a human-to-swine spillover event in the 2009–2010 influenza season (15). Despite this early spillover, the pdmNP did not demonstrate significant changes in detection frequency like the pdm M gene and was maintained at relatively low levels. Over 12 years, the gene has evolved independently of the trigNP and has reassorted with different swine IAV viruses; thus, the contemporary pdmNP now differs from the contemporary trigNP by approximately 3% (14 amino acids). It is plausible that the evolution of this specific pdmNP in swine over time resulted in a NP with a selective advantage, which it did not originally have at the time of spillover. This analysis highlights a need for a fine-scale characterization of the NP and other IAV genes to better reflect differences in evolutionary origin, genetic similarity, and phenotype.
Our data suggest that the rapid increase of the TTTPPT constellation in the H3 1990.4.a clade in swine also reflects the multigenic nature of IAV transmission: specifically, the pdmNP was only detected within the H3 1990.4.a clade at substantial levels beginning in 2019, and prior to the reassortment event, the pdmNP had acquired three amino acid substitutions. It is plausible that the impact of the pdmNP on transmission was only evident following the acquisition of these mutations, when paired with specific surface proteins, and under a specific immunological landscape reflecting vaccination and exposure to different cocirculating IAVs in swine. IAVs in swine are constantly evolving, and reassortment to acquire divergent gene segments generates a vast amount of genetic diversity upon which selection can act to drive adaptation to the swine host and influence transmission and virulence. Future studies on the diversity and evolution of the pdmNP and implications of acquisition into swine IAV genomes are necessary to understand whether this transmission benefit is a general property of the gene or amino acids within the gene or reflects specific gene pairings for IAV in swine.
ACKNOWLEDGMENTS
We gratefully acknowledge pork producers, swine veterinarians, and laboratories for participating in the USDA Influenza A Virus in Swine Surveillance System and publicly sharing sequences. We thank USDA-ARS NADC animal resources unit caretakers and staff for assistance with animal studies, Blake Inderski for assistance managing the USDA IAV in swine surveillance database, and Celeste Snyder, Katharine Young, and Nicholas Otis for technical assistance.
This work was supported in part by the Iowa State University Presidential Interdisciplinary Research Initiative; the Iowa State University Veterinary Diagnostic Laboratory; the US Department of Agriculture (USDA) Agricultural Research Service (ARS project number 5030-32000-231-000-D); the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract numbers 75N93021C00015 and 75N93021C00014); and the SCINet project and the AI Center of Excellence of the USDA Agricultural Research Service (ARS project numbers 0201-88888-003-000D and 0201-88888-002-000D). The funders had no role in study design, data collection and interpretation, or decision to submit the work for publication. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA, NIH, or ISU. USDA is an equal opportunity provider and employer.
Contributor Information
Tavis K. Anderson, Email: tavis.anderson@usda.gov.
Anice C. Lowen, Emory University School of Medicine, Atlanta, Georgia, USA
DATA AVAILABILITY
All sequence data are publicly available in NCBI GenBank, and the code, data, and supplementary material associated with this study are provided at https://github.com/flu-crew/datasets.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01703-23.
Difference in gene constellations detected in U.S. swine between two time periods (2016-2018 and 2019-2021).
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. de Vries E, Du W, Guo H, de Haan CAM. 2020. Influenza A virus hemagglutinin–neuraminidase–receptor balance: preserving virus motility. Trends Microbiol 28:57–67. doi: 10.1016/j.tim.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wagner R, Wolff T, Herwig A, Pleschka S, Klenk HD. 2000. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J Virol 74:6316–6323. doi: 10.1128/jvi.74.14.6316-6323.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352 [DOI] [PubMed] [Google Scholar]
- 4. Rajão DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, Vincent AL. 2017. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 91:e01763-16. doi: 10.1128/JVI.01763-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vincent A. 2008. Chapter 3 swine influenza viruses. In A North American perspective, advances in virus research. Elsevier. [DOI] [PubMed] [Google Scholar]
- 6. Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. 2000. Evolution of swine H3N2 influenza viruses in the United States. J Virol 74:8243–8251. doi: 10.1128/jvi.74.18.8243-8251.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K j, Krauss S, Webster RG. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73:8851–8856. doi: 10.1128/JVI.73.10.8851-8856.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, Lewis NS, Davis CT, Vincent AL. 2021. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med 11:a038737. doi: 10.1101/cshperspect.a038737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pasma T, Joseph T. 2010. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg Infect Dis 16:706–708. doi: 10.3201/eid1604.091636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Markin A, Ciacci Zanella G, Arendsee ZW, Zhang J, Krueger KM, Gauger PC, Vincent Baker AL, Anderson TK. 2023. Reverse-zoonoses of 2009 H1N1 pandemic influenza A viruses and evolution in United States swine results in viruses with zoonotic potential. PLoS Pathog 19:e1011476. doi: 10.1371/journal.ppat.1011476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. 2015. Continual reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol 89:6218–6226. doi: 10.1128/JVI.00459-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Vir 7:42–51. doi: 10.1111/irv.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kitikoon P, Nelson MI, Killian ML, Anderson TK, Koster L, Culhane MR, Vincent AL. 2013. Genotype patterns of contemporary reassorted H3N2 virus in US swine. J Gen Virol 94:1236–1241. doi: 10.1099/vir.0.51839-0 [DOI] [PubMed] [Google Scholar]
- 14. Walia RR, Anderson TK, Vincent AL. 2019. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respir Viruses 13:262–273. doi: 10.1111/irv.12559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neveau MN, Zeller MA, Kaplan BS, Souza CK, Gauger PC, Vincent AL, Anderson TK. 2022. Genetic and antigenic characterization of an expanding H3 influenza A virus clade in U.S. swine visualized by nextstrain. mSphere 7:e0099421. doi: 10.1128/msphere.00994-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hagiwara K, Kondoh Y, Ueda A, Yamada K, Goto H, Watanabe T, Nakata T, Osada H, Aida Y. 2010. Discovery of novel antiviral agents directed against the influenza A virus nucleoprotein using photo-cross-linked chemical arrays. Biochem Biophys Res Commun 394:721–727. doi: 10.1016/j.bbrc.2010.03.058 [DOI] [PubMed] [Google Scholar]
- 17. Arendsee ZW, Chang J, Hufnagel DE, Markin A, Janas-Martindale A, Vincent AL, Anderson TK. 2021. octoFLUshow: an interactive tool describing spatial and temporal trends in the genetic diversity of influenza A virus in U.S. swine. Microbiol Resour Announc 10:e0108121. doi: 10.1128/MRA.01081-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huddleston J, Hadfield J, Sibley TR, Lee J, Fay K, Ilcisin M, Harkins E, Bedford T, Neher RA, Hodcroft EB. 2021. Augur: a bioinformatics toolkit for phylogenetic analyses of human pathogens. J Open Source Softw 6:2906. doi: 10.21105/joss.02906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cáceres CJ, Cardenas-Garcia S, Jain A, Gay LC, Carnaccini S, Seibert B, Ferreri LM, Geiger G, Jasinskas A, Nakajima R, Rajao DS, Isakova-Sivak I, Rudenko L, Vincent AL, Davies DH, Perez DR. 2021. Development of a novel live attenuated influenza A virus vaccine encoding the IgA-inducing protein. Vaccines (Basel) 9:703. doi: 10.3390/vaccines9070703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rajao DS, Zanella GC, Wymore Brand M, Khan S, Miller ME, Ferreri LM, Caceres CJ, Cadernas-Garcia S, Souza CK, Anderson TK, Gauger PC, Vincent Baker AL, Perez DR. 2023. Live attenuated influenza A virus vaccine expressing an IgA-inducing protein protects pigs against replication and transmission. Front Virol 3:1042724. doi: 10.3389/fviro.2023.1042724 [DOI] [Google Scholar]
- 23. Gauger PC, Loving CL, Vincent AL. 2014. Enzyme-linked immunosorbent assay for detection of serum or mucosal Isotype-specific IgG and IgA whole-virus antibody to influenza A virus in swine. Methods Mol Biol 1161:303–312. doi: 10.1007/978-1-4939-0758-8_25 [DOI] [PubMed] [Google Scholar]
- 24. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]
- 25. Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. 1995. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the lelystad virus. Vet Pathol 32:648–660. doi: 10.1177/030098589503200606 [DOI] [PubMed] [Google Scholar]
- 26. Gauger PC, Vincent AL, Loving CL, Henningson JN, Lager KM, Janke BH, Kehrli ME, Roth JA. 2012. Kinetics of lung lesion development and pro-inflammatory cytokine response in pigs with vaccine-associated enhanced respiratory disease induced by challenge with pandemic (2009) A/H1N1 influenza virus. Vet Pathol 49:900–912. doi: 10.1177/0300985812439724 [DOI] [PubMed] [Google Scholar]
- 27. Kitikoon P, Gauger PC, Vincent AL. 2014. Hemagglutinin inhibition assay with swine sera, in animal influenza virus, p 295–301. Springer. [DOI] [PubMed] [Google Scholar]
- 28. Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7:42–51. doi: 10.1111/irv.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boulo S, Akarsu H, Ruigrok RWH, Baudin F. 2007. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res 124:12–21. doi: 10.1016/j.virusres.2006.09.013 [DOI] [PubMed] [Google Scholar]
- 30. Rajão DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL. 2015. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol 89:11213–11222. doi: 10.1128/JVI.01675-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hufnagel DE, Young KM, Arendsee ZW, Gay LC, Caceres CJ, Rajão DS, Perez DR, Vincent Baker AL, Anderson TK. 2023. Characterizing a century of genetic diversity and contemporary antigenic diversity of N1 neuraminidase in influenza A virus from North American swine. Virus Evol 9:vead015. doi: 10.1093/ve/vead015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeller MA, Chang J, Vincent AL, Gauger PC, Anderson TK. 2021. Spatial and temporal coevolution of N2 neuraminidase and H1 and H3 hemagglutinin genes of influenza A virus in US swine. Virus Evol 7:veab090. doi: 10.1093/ve/veab090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun X, Belser JA, Pulit-Penaloza JA, Brock N, Pappas C, Zanders N, Jang Y, Jones J, Tumpey TM, Davis CT, Maines TR. 2023. Pathogenesis and transmission assessment of three swine-origin influenza A(H3N2) viruses with zoonotic risk to humans isolated in the U.S from 2017-2020. J Infect Dis:jiad359. doi: 10.1093/infdis/jiad359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vincent AL, Perez DR, Rajao D, Anderson TK, Abente EJ, Walia RR, Lewis NS. 2017. Influenza A virus vaccines for swine. Vet Microbiol 206:35–44. doi: 10.1016/j.vetmic.2016.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu J, Sreenivasan C, Sheng Z, Zhai S-L, Wollman JW, Luo S, Huang C, Gao R, Wang Z, Kaushik RS, Christopher-Hennings J, Nelson E, Hause BM, Li F, Wang D. 2023. A recombinant chimeric influenza virus vaccine expressing the consensus H3 hemagglutinin elicits broad hemagglutination inhibition antibodies against divergent swine H3N2 influenza viruses. Vaccine 41:6318–6326. doi: 10.1016/j.vaccine.2023.09.007 [DOI] [PubMed] [Google Scholar]
- 36. Petro-Turnquist E, Pekarek M, Jeanjaquet N, Wooledge C, Steffen D, Vu H, Weaver EA. 2023. Adenoviral-vectored epigraph vaccine elicits robust, durable, and protective immunity against H3 influenza A virus in swine. Front Immunol 14:1143451. doi: 10.3389/fimmu.2023.1143451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Lin X, Holmes EC, Detmer SE. 2014. Introductions and evolution of human-origin seasonal influenza a viruses in multinational swine populations. J Virol 88:10110–10119. doi: 10.1128/JVI.01080-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM, Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team . 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 360:2605–2615. doi: 10.1056/NEJMoa0903810 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Difference in gene constellations detected in U.S. swine between two time periods (2016-2018 and 2019-2021).
Data Availability Statement
All sequence data are publicly available in NCBI GenBank, and the code, data, and supplementary material associated with this study are provided at https://github.com/flu-crew/datasets.