Abstract
Isogenic, E3-deleted adenovirus vectors defective in E1, E1 and E2A, or E1 and E4 were generated in complementation cell lines expressing E1, E1 and E2A, or E1 and E4 and characterized in vitro and in vivo. In the absence of complementation, deletion of both E1 and E2A completely abolished expression of early and late viral genes, while deletion of E1 and E4 impaired expression of viral genes, although at a lower level than the E1/E2A deletion. The in vivo persistence of these three types of vectors was monitored in selected strains of mice with viral genomes devoid of transgenes to exclude any interference by immunogenic transgene-encoded products. Our studies showed no significant differences among the vectors in the short-term maintenance and long-term (4-month) persistence of viral DNA in liver and lung cells of immunocompetent and immunodeficient mice. Furthermore, all vectors induced similar antibody responses and comparable levels of adenovirus-specific cytotoxic T lymphocytes. These results suggest that in the absence of transgenes, the progressive deletion of the adenovirus genome does not extend the in vivo persistence of the transduced cells and does not reduce the antivirus immune response. In addition, our data confirm that, in the absence of transgene expression, mouse cellular immunity to viral antigens plays a minor role in the progressive elimination of the virus genome.
Replication-deficient human adenoviruses (Ad) have been widely investigated as ex vivo and in vivo gene delivery systems for human gene therapy. The ability of these vectors to mediate the efficient expression of candidate therapeutic or vaccine genes in a variety of cell types, including postmitotic cells, is considered an advantage over other gene transfer vectors (3, 28, 49). However, the successful application of currently available E1-defective Ad vectors in human gene therapy has been hampered by the fact that transgene expression is only transient in vivo (2, 15, 16, 33, 36, 46). This short-lived in vivo expression of the transgene has been explained, at least in part, by the induction in vivo of cytotoxic immune responses to cells infected with the Ad vector. Studies with rodent systems have suggested that cytotoxic T lymphocytes (CTLs) directed against virus antigens synthesized de novo in the transduced tissues play a major role in eliminating cells containing the E1-deleted viral genome (56–58, 61). Consistent with the concept of cellular antiviral immunity, expression of transgenes is significantly extended in experimental rodent systems that are deficient in various components of the cellular immune system or that have been rendered immunocompromised by administration of pharmacological agents (2, 33, 37, 48, 60, 64).
Based on the assumption that further reduction of viral antigen expression may lower the immune response and thus extend persistence of transgene expression, previous studies have investigated the consequences of deleting both E1 and an additional viral regulatory region, such as E2A or E4. The E2A region encodes a DNA binding protein (DBP) with specific affinity for single-stranded Ad DNA. The DNA binding function is essential for the initiation and elongation of viral DNA synthesis during the early phase of Ad infection. During the late phase of infection, DBP plays a central role in the activation of the major late promoter (MLP) (for a recent review, see reference 44). The E4 region, located at the right end of the viral genome, encodes several regulatory proteins with pleiotropic functions which are involved in the accumulation, splicing, and transport of early and late viral mRNAs, in DNA replication, and in virus particle assembly (reviewed in reference 44). The simultaneous deletion of E1 and E2A or of E1 and E4 should therefore further reduce the replication of the virus genome and the expression of early and late viral genes. Such multidefective vectors have been generated and tested in vitro and in vivo (9, 12, 17, 19–21, 23, 24, 26, 34, 40, 52, 53, 59, 62, 63). Recombinant vectors with E1 deleted and carrying an E2A temperature-sensitive mutation (E2Ats) have been shown in vitro to express much smaller amounts of virus proteins, leading to extended transgene expression in cotton rats and mice (19, 20, 24, 59). To eliminate the risks of reversion of the E2Ats point mutation to a wild-type phenotype, improved vectors with both E1 and E2A deleted were subsequently generated in complementation cell lines coexpressing E1 and E2A genes (26, 40, 63). In vitro analysis of human cells infected by these viruses demonstrated that the double deletion completely abolished viral DNA replication and late protein synthesis (26). Similarly, E1/E4-deleted vectors have been generated in various in vitro complementation systems and tested in vitro and in vivo (9, 17, 23, 45, 52, 53, 62). These studies showed that deletion of both E1 and E4 did indeed reduce significantly the expression of early and late virus proteins (17, 23), leading to a decreased anti-Ad host immune response (23), reduced hepatotoxicity (17, 23, 52), and improved in vivo persistence of the transduced liver cells (17, 23, 52).
Interpretation of these results is difficult, however, since all tested E1- and E1/E4-deleted vectors encoded the bacterial β-galactosidase (βgal) marker, whose strong immunogenicity is known to influence the in vivo persistence of Ad-transduced cells (32, 37). Moreover, the results described above are not consistent with the conclusions from other studies showing, in various immunocompetent mouse models, that cellular immunity to Ad antigens has no detectable impact on the persistence of the transduced cells (37, 40, 50, 51). Furthermore, in contrast to results of earlier studies (19, 20, 59), Fang et al. (21) demonstrated that injection of E1-deleted/E2Ats vectors into immunocompetent mice and hemophilia B dogs did not lead to an improvement of the persistence of transgene expression compared to that with isogenic E1-deleted vectors. Similarly, Morral et al. (40) did not observe any difference in persistence of transgene expression in mice injected with either vectors deleted in E1 only or vectors deleted in both E1 and E2A. Finally, the demonstration that some E4-encoded products can modulate transgene expression (1, 17, 36a) makes the evaluation of E1- and E1/E4-deleted vectors even more complex when persistence of transgene expression is used for direct comparison of the in vivo persistence of cells transduced by the two types of vectors.
The precise influence of the host immune response to viral antigens on the in vivo persistence of the transduced cells, and hence the impact of further deletions in the virus genome, therefore still remains unclear. To investigate these questions, we generated a set of isogenic vectors with single deletions (AdE1°) and double deletions (AdE1°E2A° and AdE1°E4°) and their corresponding complementation cell lines and compared the biologies and immunogenicities of these vectors in vitro and in vivo. To eliminate any possible influence of transgene-encoded products on the interpretation of the in vivo results, we used E1-, E1/E2A-, and E1/E4-deleted vectors with no transgenes.
MATERIALS AND METHODS
E2A and E4 expression plasmids.
All cloning steps were performed by using standard molecular biology techniques (43). The E2A and E4 expression plasmids, derived from the plasmid vector ppoly II (35), are schematically depicted in and described in the legends to Fig. 1 and 3. The DBP expression plasmid pTG9595 (see Fig. 1) contains the entire DBP-coding region (nucleotides [nt] 24334 to 22440) (throughout this paper, Ad type 5 [Ad5] nucleotide numbering is according to reference 13) inserted into a mouse mammary tumor virus (MMTV) promoter-driven expression cassette (22). The E4 expression plasmid pTG1653 (see Fig. 3) contains the entire Ad5 E4 region (nt 32800 to 35826), including the E1A-inducible E4 promoter. The E4ORF6+7 expression plasmid pTG5606 (see Fig. 3) contains the E4 open reading frame 6 (ORF6) and -7 genes (nt 32800 to 34219).
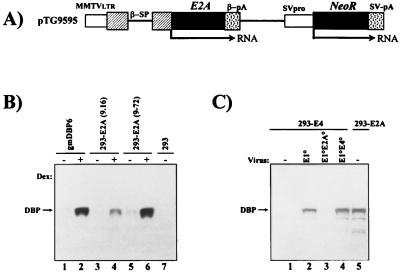
FIG. 1.
Structure of the E2A expression plasmid and steady-state levels of DBP protein in stable cell lines and after viral infection. (A) Schematic representation of the DBP expression plasmid pTG9595. Ad5 E2A sequences (nt 22440 to 24334) were inserted into an MMTV promoter-driven expression cassette (22) containing the rabbit β-globin splicing (β-SP) and polyadenylation (β-pA) signals. Expression of the neomycin resistance gene (NeoR) is regulated by the Simian virus 40 early promoter (SVpro) and late polyadenylation signal (SV-pA). LTR, long terminal repeat. (B) Western blot analysis of DBP protein in stable E1/E2A complementation cell lines. 293-E2A clones (9-16 and 9-72), established with pTG9595, and the control E2A complementation cell line gmDBP-6 (9) were compared for DBP expression in the presence (+) or absence (−) of dexamethasone (Dex). Total protein was extracted at 24 h postinduction, polypeptides (10 μg of protein) were separated on a 12% polyacrylamide–SDS gel, and the DBP protein was detected with the B6α72K anti-DBP monoclonal antibody (41) combined with enhanced chemiluminescence. (C) Comparison of DBP expression in 293-E2A cells (clone 9-72) and 293-E4 cells (clone 5-19; see Fig. 3) infected at an MOI of 6 IU/cell with AdE1°, AdE1°E2A°, and AdE1°E4° vectors. Total protein was extracted at 16 h p.i. (lanes 1 to 4) and 24 h after induction with dexamethasone (lane 5). DBP analysis was as described above.
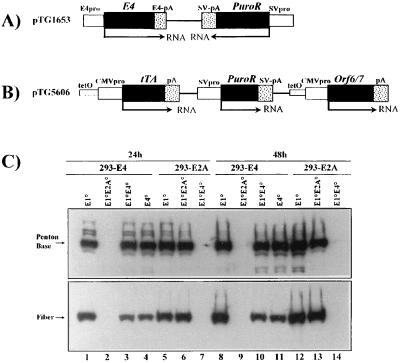
FIG. 3.
Structure of E4 expression plasmids and analysis of late viral proteins in 293-E2A and 293-E4 complementation cells. (A) Schematic representation of the E4 expression plasmid, pTG1653, containing the entire E4 region (Ad5 nt 32800 to 35826), including the E4 promoter (pro) and polyadenylation signal (pA). Expression of the puromycin resistance gene (PuroR) is regulated by the Simian virus 40 (SV) early promoter. (B) Schematic representation of the E4ORF6+7 expression plasmid, pTG5606. The tTA gene (26) as well as the E4ORF6+7 (Ad5 nt 32800 to 34219) genes are under the control of the minimal CMV immediate-early promoter fused to a heptameric tet operator (26), while the puromycin resistance gene is regulated by the simian virus 40 early promoter. (C) Analysis of late viral proteins in infected 293-E2A (clone 293/9-72; lanes 5 to 7 and 12 to 14) and 293-E4 (clone 293/5-19; lanes 1 to 4 and 8 to 11) complementation cells. Cells were infected with the indicated viruses at an MOI of 5 IU/cell (E1°, AdTG4656; E1°E4°, AdTG8595; E4°, AdTG9572) or 5 BU/cell (E1°E2°, AdTG9542). 293-E2A cells were induced with dexamethasone. All viruses contained the lacZ gene in place of the E1 region (Table 1), except for the vector AdTG9572, which contained an intact E1 region. Total protein extraction, electrophoresis, and Western blot analysis were performed as described in the legend to Fig. 1. Late viral proteins were detected with polyclonal antisera directed against the knob domain of fiber (serum E642; obtained from R. Gerard, Leuven, Belgium [31]) or against the penton base (serum SE262; obtained from P. Boulanger, Montpellier, France).
Generation of E1/E2A, E1/E4, and E1/E4ORF6+7 cell lines.
The respective expression plasmids were transfected into 293 cells (29) by standard calcium phosphate precipitation methods (30). All cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Selection, isolation, and expansion of stable cell lines were done by standard procedures (43). 293-E2A cells and 293-E4ORF6+7 cells were selected in the absence of dexamethasone and in the presence of tetracycline, respectively.
Screening of complementation cell lines.
293-E2A cell lines were screened by Western blot analysis for the expression of DBP (see below) in the presence of dexamethasone. Clone 9-72 was chosen among the clones with the highest steady-state level of DBP protein to generate the doubly deleted AdE1°E2° vectors (see Table 1). The ability of 293-E4 clones to provide E4 functions in trans was initially tested with an E4 deletion mutant of Ad2, H2dl808 (10). Growth of H2dl808 on pTG1653 cells was monitored by a microinfection-microtitration procedure. In brief, individual pTG1653 clones were infected with H2dl808 at a multiplicity of infection (MOI) of 2. Viral progeny was recovered at 48 h postinfection (p.i.) and titrated by serial dilution in 96-well microtiter plates on W162 cells, an indicator Vero cell line containing and complementing the E4 functions (54). 293 cells infected with H2dl808 were used as a negative control, whereas 293 cells infected with an E1-deleted Ad vector served as a positive control. pTG1653 clones scoring positive in this assay were selected, and the best clone (293/1653il) was used to establish the doubly deleted AdE1°E4° vectors (see below). Similarly, individual pTG5606 clones were screened for E4 complementation in the presence and absence of tetracycline, using AdE1°E4° vectors previously generated on pTG1653 clones.
TABLE 1.
Genomic organization of the recombinant Ad vectorsa
| Vector | Virus | Expression cassette | E1 | E2A | E4 |
|---|---|---|---|---|---|
| AdE1° | AdRSVβgal | RSV-lacZ | Deleted | wtb | wt |
| AdE1° | AdTG4656 | MLP-lacZ | Deleted | wt | wt |
| AdE1°E2A° | AdTG9542 | MLP-lacZ | Deleted | Deleted | wt |
| AdE1°E4° | AdTG8595 | MLP-lacZ | Deleted | wt | Deleted |
| AdE1°E4° | AdTG5643 | CMV-CFTRc | Deleted | wt | Deleted |
| AdE1° | AdTG6401 | Deleted | wt | wt | |
| AdE1°E2A° | AdTG9592 | Deleted | Deleted | wt | |
| AdE1°E4° | AdTG9546 | Deleted | wt | Deleted | |
| AdE1+E4° | AdTG9572 | wt | wt | Deleted |
All vectors contain a deletion (nt 28592 to 30480) in the viral E3 region.
wt, wild type.
CFTR, cystic fibrosis transmembrane conductance regulator.
Viral vectors.
The viral vectors are shown in Table 1. All viral genomes described in this study were constructed as infectious plasmids by homologous recombination in Escherichia coli as described by Chartier et al. (11). In brief, all vectors except AdTG9572 contain a deletion (nt 459 to 3327) in E1 (Table 1), and all vectors contain a deletion (nt 28592 to 30470) in E3. Where indicated, vectors contain a transgene in place of E1. The E2A deletion in the AdE1°E2° vectors comprises nt 22440 to 24036. The E4 deletion in the AdE1°E4° vectors is identical to the H2dl808 deletion in Ad2 (10), removing most of the E4 coding sequences (nt 32994 to 34998) but not E4 ORF1. For the generation of viruses, the viral genomes were released from the respective plasmids by PacI digestion and transfected into the appropriate complementation cell lines, as described previously (11). Viral stocks were prepared from the transfected cells, and viruses were purified by standard procedures (27) and stored in viral storage buffer (1 M sucrose, 10 mM Tris-HCl [pH 8.5], 1 mM MgCl2, 150 mM NaCl, 0.005% [vol/vol] Tween 80).
Viral growth and titration.
The efficiency of E1/E2 and E1/E4 complementation in the respective cell lines was assessed with single-step growth curves. 293, 293-E2A, 293-E4, and 293-E4ORF6+7 cells were infected with AdE1°, AdE1°E2°, and AdE1°E4° vectors as indicated in the legend to Fig. 2. Viral progeny was recovered at various time points by three rounds of freezing and thawing of the infected cells, and titers were determined. PFU were determined by standard plaque assays (27) with the appropriate indicator cells. Plaques were scored for AdE1° and AdE1°E2° vectors at 14 days p.i. and for AdE1°E4° vectors at 21 to 24 days p.i. Titers of infectious viral progeny were determined as infectious units (IU) by quantitative DBP immunofluorescence or as βgal transducing units (BU) by quantitative βgal staining. To determine the IU titer, 293 cells were infected with serial dilutions of virus; this was followed by immunofluorescence staining at 16 h p.i. with B6α72K, an anti-DBP monoclonal antibody (41), and quantitation. Similarly, the BU titer was determined by infection of 293 cells with serial virus dilutions followed by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining at 24 h p.i. and quantitation.
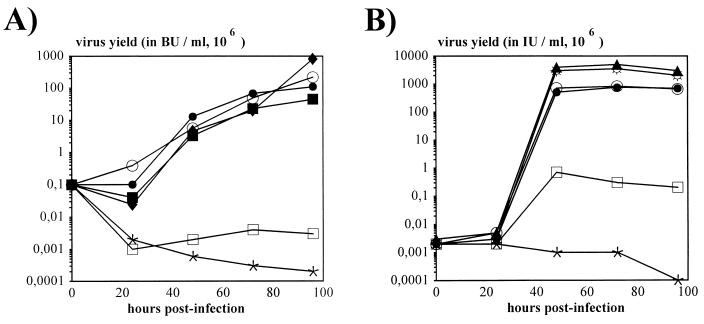
FIG. 2.
Kinetics of viral growth. (A) Kinetics of AdE1° and AdE1°E2A° virus propagation in 293 and 293-E2A cells. Cells were infected at an MOI of 0.2 BU/cell. Titration of infectious viral progeny (in BU) at the indicated times p.i. was performed on 293-E2A cells. AdE1° (AdTG4656) (⧫, ○, and •) was used to infect 293 cells (⧫) and 293-E2A-9-72 cells in the presence (•) and absence (○) of dexamethasone induction. Similarly, AdE1°E2A° (AdTG9542) (▪, □, and ∗) was used to infect 293 cells (∗) and 293-E2A-9-72 cells in the presence (▪) and absence (□) of dexamethasone induction. (B) Kinetics of AdE1° and AdE1°E4° virus propagation in 293, 293-E4 (clone 293/1653il), and 293-E4ORF6+7 (clone 293/5-19) cells. Cells were infected at an MOI of 0.05 IU/cell. Titration of infectious viral progeny (in IU) at the indicated times p.i. was performed on 293 cells. AdE1° (AdTG4656) (⧫ and ☼) was used to infect 293 cells (⧫) and 239-E4ORF6+7 cells (☼); similarly, AdE1°E4° (AdTG8595 [•, □, and ∗] or AdTG5643 [○]) was used to infect 239-E4ORF6+7 (• and ○), 293-E4 (□), and 293 (∗) cells.
Viral gene expression in complementing and noncomplementing cell lines.
The ability of 293-E2A and 293-E4 cells to efficiently complement the doubly deleted viral vectors was monitored by the analysis of early and late viral proteins. 293-E2A and 293-E4 cells were infected with the indicated vectors. At various times, whole-cell extracts were prepared in lysis buffer (5 mM KCl, 50 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 1 mM EDTA, 150 mM NaCl, 1% [vol/vol] Triton X-100). Protein concentration was measured by the Bradford assay (Bio-Rad, Ivry sur Seine, France) with bovine serum albumin as the standard. Polypeptides (10 μg of total protein) were denatured and resolved on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels according to the instructions of the manufacturer (Novex-prolabo, Fontenay-sous-Bois, France). The B6α72K monoclonal anti-DBP antibody (41), a polyclonal antiserum directed against the knob domain of the Ad5 fiber (serum E642; obtained from R. Gerard, Leuven, Belgium [31]), and a polyclonal antiserum directed against the Ad5 penton base (serum SE262; obtained from P. Boulanger, Montpellier, France), combined with the enhanced chemiluminescence detection system (ECL; Amersham, Les-Ulis, France), were used to detect the respective proteins on Western blots. To monitor viral gene expression in noncomplementing cells, human lung epithelial A549 cells were infected with the vectors and protein analysis was performed as described above.
Animal studies.
The mice used in this study were 6- to 8-week-old female immunocompetent CBA/J (H-2k) and immunodeficient C.B17-scid/scid (5) mice (IFFA Credo, Lyon, France). The mice were administered the vectors without transgenes via tail vein injection in 100 μl of viral storage buffer at various doses. The ratios of total virus particles to IU were 38:1 for the AdE1° and AdE1°E4° vectors and 175:1 for the AdE1°E2A vector. Animals were sacrificed at the times indicated in the figure legends. Organs were removed, cut into equal pieces, and immediately frozen in liquid nitrogen until analysis.
DNA analysis.
Total DNA was extracted from the organs as described previously (38). Briefly, the tissues were digested overnight with proteinase K solution (1 mg of proteinase K in 1% SDS) in DNA lysis buffer (10 mM Tris-HCl [pH 7.4], 400 mM NaCl, 2 mM EDTA). The DNA was isolated by phenol-chloroform extraction followed by ethanol precipitation. DNA (10 μg) was digested with BamHI and analyzed by Southern blot analysis (43) with a 32P-labeled EcoRI-HindIII restriction fragment purified from Ad5 genomic DNA (nt 27331 to 31993). DNA signals were quantitated by densitometry scanning of the autoradiographs with a GS-700 Imaging Densitometer (Bio-Rad), followed by analysis of the data with Molecular Analyst/PC Software (Bio-Rad). All data are presented as means ± standard errors (SE).
CTL and lymphoproliferation assays.
Six-week-old female CBA/J mice were immunized intraperitoneally at days 0 and 14 with AdE1°, AdE1°E2A°, and AdE1°E4° vectors in 100 μl of saline buffer. Negative control mice were mock injected with saline alone. All mice were sacrificed 4 days after the second immunization, and their spleens were removed. Stimulation of splenocytes and determination of cytolytic activity by the standard 4-h chromium release assay were performed as previously described (37). To assay lymphoproliferation, CBA/J mice were immunized as described above. Proliferation of splenocytes was as previously described (37).
Anti-Ad antibody assay.
Anti-Ad antibodies were measured by enzyme-linked immunosorbent assay (ELISA). Preparations of ELISA plates, incubations with mouse sera followed by a biotinylated second antibody, and streptavidin amplification were as described previously (37). Substrate conversion was initiated with 3,3′,5,5′-tetramethyl benzidine (Sigma) (stock solution; 100 mg in 10 ml of dimethyl sulfoxide) diluted 100-fold, immediately before use, in 0.1 M citric acid–0.1 M sodium acetate (pH 4.0) containing H2O2 (1.5 μl of 30% H2O2 per 10 ml of substrate solution). The reaction was stopped by the addition of 100 μl of 0.3 M H2SO4, and absorbance was read at 450 nm in an ELISA reader. Each plate contained the same positive and negative control sera.
RESULTS
Generation of AdE1°E2A° vectors and packaging cells coexpressing E1 and E2A.
An E2A expression plasmid (pTG9595) (Fig. 1A) containing the Ad5 E2A sequences (nt 22440 to 24334) under the control of the glucocorticoid-inducible MMTV long terminal repeat was transfected into 293 cells. G418-resistant clones were selected in the absence of dexamethasone and screened for E2A gene expression in the presence of dexamethasone. As shown in Fig. 1B for two individual clones (293/9-16 and 293/9-72), steady-state levels of DBP protein were low in the absence (Fig. 1B, lanes 3 and 5) and efficiently induced in the presence (Fig. 1B, lanes 4 and 6) of dexamethasone. The induced level of DBP expression was comparable to that observed in control HeLa cells transfected with an MMTV-E2A expression cassette (gmDBP6 cells [8]) after dexamethasone induction. Clone 293/9-72 was chosen for the remainder of the study to explore its ability to complement and propagate doubly deleted AdE1°E2A° vectors. In these cells, the induced steady-state level of DBP protein was similar (Fig. 1C, lane 5) to that observed in 293 cells 24 h after infection with an AdE1° vector (Fig. 1C, lane 2).
Deletion of the entire E2A coding region (nt 22440 to 24036) from an E1-defective Ad was engineered by homologous recombination in E. coli (11) with an infectious plasmid containing an E1/E3-deleted Ad5 genome carrying the bacterial βgal (lacZ) gene, in place of E1, under the control of the Ad2 MLP. Infectious viral DNA was released from the resulting plasmid backbone (pTG9542) and transfected into 293/9-72 cells in the presence of dexamethasone. Viral progeny (AdE1°E2A°/MLP-lacZ [AdTG9542]) was isolated and amplified in 293/9-72 cells. Analysis of the purified AdTG9542 viral DNA confirmed the E1 and E2A deletions. The kinetics of growth of AdE1°/Rous sarcoma virus (RSV) βgal (48) (Table 1) and AdE1°E2A°/MLP-lacZ (AdTG9542) viruses, measured as BU (see Materials and Methods), were compared in 293 and 293/9-72 cells (Fig. 2A). This in vitro analysis showed that (i) the growth of AdE1°/RSV βgal was not affected by expression of E2A, indicating that introducing E2A sequences into 293 cells did not interfere with E1 complementation; (ii) the growth kinetics of AdE1°E2A°/MLP-lacZ in dexamethasone-induced 293/9-72 cells was similar to that seen with AdE1°/RSV βgal; and (iii) as expected, no growth of AdE1°E2A°/MLP-lacZ was observed in 293 and noninduced 293/9-72 cells. However, the final yield of infectious AdE1°E2A° vector in induced 293/9-72 cells was 5- to 30-fold reduced compared to that of the AdE1° vector. Efficient complementation and viral growth were confirmed by plaque assays. AdE1°E2A°/MLP-lacZ could readily form plaques on monolayers of 293/9-72 cells in the presence but not in the absence of dexamethasone (data not shown). From these results we conclude that 293/9-72 cells can efficiently complement the growth of AdE1°E2A° vectors.
Generation of AdE1°E4° vectors and packaging cells coexpressing E1 and E4.
To establish an E1/E4 complementation system, 293 cells were initially transduced with an E4 expression plasmid (pTG1653) (Fig. 3A) containing the entire E4 region (nt 32800 to 35826) under the control of the E1A-inducible homologous E4 promoter. One clone, 293/1653il, supporting the growth of an AdE1+E4° vector (Ad2H2dl808 [10]) with the highest efficiency, was selected to generate a doubly deleted vector, AdE1°E4°/MLP-lacZ. An E4 deletion (nt 32994 to 34998), equivalent to that of H2dl808, was introduced into pTG4656 by homologous recombination in E. coli (11) (see above). This E4 deletion removed all E4 ORFs except ORF1. Infectious viral DNA was released from the resulting plasmid (pTG8595) and transfected into 293/1653il cells to generate and propagate the doubly deleted viral genome AdE1°E4°/MLP-lacZ (AdTG8595). In the course of these studies, we noted that the final yields of infectious AdE1°E4° vectors produced in 293/1653il cells were always much lower (1,000- to 10,000-fold) than the yields of AdE1° viruses (Fig. 2B). E4-mediated cytotoxicity (6) could have resulted in the survival of a population of 293-E4 cells (293/1653il) with levels of E4 proteins insufficient for optimal complementation of AdE1°E4° vectors. Alternatively, a disturbance of the appropriate balance between the various E4 ORFs might have occurred through integration of the E4 region into the host cell chromosome. For example, E4 ORF4 was shown to efficiently inhibit the E1A-mediated transactivation of the E4 promoter, leading to a down-regulation of the E4 ORF3 and ORF6 genes, which are essential for optimal virus propagation (4, 7). A comparative determination of the ratio of infectious viruses (IU) (see Materials and Methods) to productive viruses (PFU) revealed an IU/PFU ratio of 1:1 to 2:1 for AdE1° and 200:1 to 1,000:1 for AdE1°E4° (Table 2). This indicates that the majority of the AdE1°E4° viruses produced on 293/1653il cells were infectious but were unable to efficiently replicate and generate new virus progeny. Therefore, the total viral yields of AdE1°E4° vectors in 293-E4 cells were significantly reduced compared to those obtained with AdE1° viruses.
TABLE 2.
Physical characteristics of the Ad vectors
| Vector | Cells | Total virus particles/IU (BU) | IU (BU)/PFU |
|---|---|---|---|
| Ad5 (wild type) | 293 | ≥10:1 | 1 |
| AdE1° | 293 | ≥20:1 | 1:1–2:1 |
| AdE1°E2° | 293-E2 | ≥100:1 | 1:1–2:1 |
| AdE1°E4° | 293-E4ORF6+7 | ≥30:1 | 5:1–10:1 |
| AdE1°E4° | 293-E4 | NDa | 200:1–1,000:1 |
ND, not determined.
To improve the E1/E4 complementation system, we took advantage of the observation that the E4 ORF6 gene product is sufficient for efficient virus growth in vitro (6). In addition, in order to bypass possible E4-mediated cytotoxicity, we designed an inducible E4 ORF6 expression plasmid, pTG5606 (Fig. 3B), containing the E4 ORF6 and ORF7 genes (nt 32800 to 34219) under the control of a minimal cytomegalovirus (CMV) promoter linked to seven copies of the tet operator. The VP16-tetR chimeric gene (tTA), whose expression is itself autoregulated by the tetO-CMV promoter, was inserted in cis downstream of the E4 genes. Thus, transcription of the E4 genes should be turned off in the presence of tetracycline (25). Stable 293-E4ORF6+7 clones were established with pTG5606 in the presence, and screened for the complementation of AdE1°E4° vectors in the absence, of tetracycline. Of 137 clones, 13 scored positive in this assay. One clone, designated 293/5-19, was selected to further compare the growth kinetics of AdE1°E4° vectors (Fig. 2B). In contrast to 293-E4 cells, 293-E4ORF6+7 (293/5-19) cells could efficiently support the growth of AdE1°E4°/MLP-lacZ and AdE1°E4°/cytomegalovirus immediate-early promoter-cystic fibrosis transmembrane conductance regulator (CMV-CFTR) (AdTG5643 [Table 1]), with a final virus yield only 5- to 10-fold lower than that of AdE1° vectors. This improved complementation was reflected by an IU-to-PFU ratio reaching values similar to the ratio determined for AdE1° viruses (Table 2). Growth of AdE1°/MLP-lacZ was identical in 293/5-19 and 293 cells, indicating that introducing E4 genes into 293 cells did not impair E1 complementation. However, 293/5-19 cells did not allow clear plaque formation when infected with AdE1°E4° viruses. We therefore used another 293-E4ORF6+7 clone (293/5-38) for PFU titration of AdE1°E4° vectors.
Analysis of the steady-state levels of late viral proteins in 293-E2A (clone 293/9-72) and 293/E4 (clones 293/1653il and 293/5-19) cells infected with AdE1°, AdE1°E2A°, AdE1°E4°, or AdE1+E4° (AdTG5672 [Table 1]) vectors showed that the reproducible lower AdE1°E4° virus yields in 293-E4 cells (clone 293/1653il) correlated with a strong reduction in the accumulation of the viral fiber protein (data not shown). In contrast, accumulation of fiber in AdE1°E4°-infected 293-E4ORF6+7 cells (clone 293/5-19) was markedly augmented, although it was still lower than that observed for AdE1° viruses (Fig. 3C, lanes 1, 3, and 4 and lanes 8, 10, and 11). As expected, production of fiber was similar in 293-E4 and 293-E2A cells infected with AdE1° vectors (Fig. 3C, lanes 1, 5, 8, and 12). Accumulation of fiber protein was also similar in 293-E2A cells infected with AdE1°E2A° or AdE1° viruses (Fig. 3C, lanes 5, 6, 12, and 13). These observations are consistent with those reported by Brough et al. (9), who in addition showed that insertion of a transcriptional cassette in place of the E4 region could rescue the fiber defect. Expression of penton base was efficient for all vectors in their respective complementation cells (Fig. 3B).
An unexpected observation was the finding that the propagation of AdE1°E4° vectors was not affected by tetracycline. While the reason(s) for the loss of tetracycline regulation in 293-E4ORF6+7 cells is not clear, we noted that expression of E4 genes incorporated into the host cell genome could be regulated by tetracycline over a 1,000-fold range when the tTA gene was provided in trans by an Ad vector (36b).
In vitro expression of early and late viral antigens.
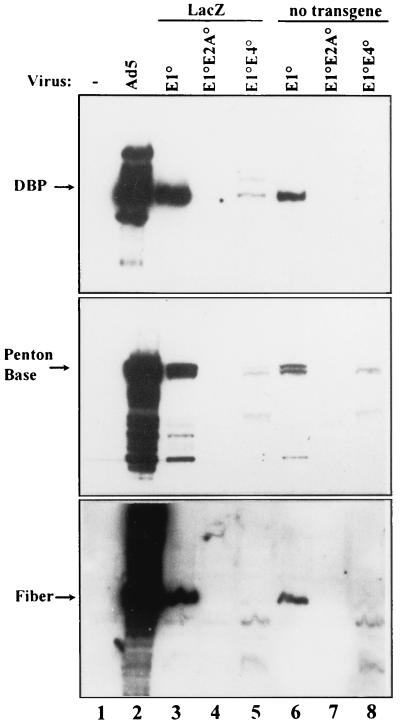
The consequences of the E2A and E4 deletions for early and late viral antigen expression were assessed in vitro. Two series of isogenic E1-defective vectors, with (MLP-lacZ) and without transgenes (Table 1), differing only in the E2A and E4 deletions, were compared in noncomplementing cells. Human A549 cells were infected with the vectors at an MOI of 500 IU/cell (for vectors without transgenes) or 500 BU/cell (for vectors with MLP-lacZ). For comparison, wild-type Ad5 was used to infect the cells at an MOI of 0.1 IU/cell. Infected cells were collected at 3 days p.i., and steady state levels of early and late viral proteins (Fig. 4) and mRNAs (data not shown) were monitored. Early and late viral proteins could readily be detected in A549 cells infected with AdE1° vectors (Fig. 4, lanes 3 and 6), albeit at reduced levels compared to wild-type Ad5 (Fig. 4, lane 2). Consistent with our previous data (42), additional deletion of E2A or E4 genes markedly reduced the expression of early and late proteins: no DBP, penton base, or fiber proteins could be detected in A549 cells infected with AdE1°E2A° vectors (Fig. 4, lanes 4 and 7), while barely detectable fiber and markedly reduced levels of DBP and penton base were observed in A549 cells infected with AdE1°E4° vectors (Fig. 4, lanes 5 and 8). Northern blot analysis of early and late mRNAs from these infected cells confirmed these results. In addition, DNA replication in A549 cells was abolished by the simultaneous deletion of E1 and E2A or of E1 and E4 (data not shown). At lower MOIs early and late viral gene expression could not be detected with either AdE1°E2A° or AdE1°E4° vectors, and therefore, differences could not be scored.
FIG. 4.
Expression of early and late viral proteins in noncomplementing human A549 cells. A549 cells were infected with the indicated vectors at an MOI of 500 BU/cell (lanes 3, 4, and 5) or 500 IU/cell (lanes 6, 7, and 8). Wild-type Ad5 was infected with an MOI of 0.5 IU/cell. At 72 h p.i. total protein was extracted and processed as described in the legend to Fig. 1.
Nonspecific elimination of AdE1°, AdE1°E2A°, and AdE1°E4° genomes.
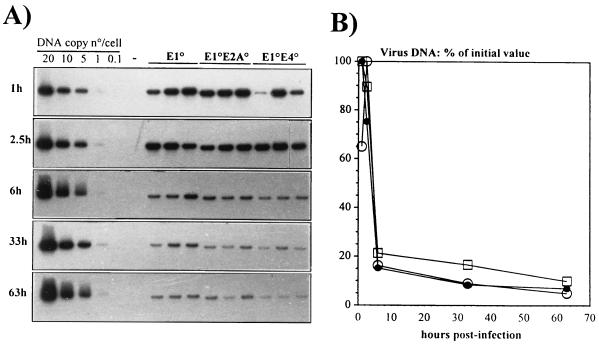
According to previous hypothesis (56–58, 61), reduced viral antigen expression should lead to a blunted immune response and, hence, to extended viral DNA persistence and transgene expression in vivo. However, reduced antigen expression should not influence the early elimination of the Ad genome mediated by the innate immune system (55). In order to investigate the contribution of the innate immune response to the in vivo loss of recombinant Ad genomes, 4 × 1010 virus particles of the vectors were injected into the tail veins of immunocompetent CBA/J mice, and the fate of the Ad DNA in the liver, spleen, and lung was monitored at 1, 2.5, 6, 33, and 63 h postinjection. Southern blot analysis showed no significant differences between the three types of vectors: 80% of all viral genomes were eliminated from the liver during the first 6 h after administration of the vectors (Fig. 5). A similar rapid, nonspecific elimination was observed for all vectors in the lung and spleen (reference 12 and data not shown). These findings are consistent with those of Worgall et al. (55) and further indicate that early vector clearance by innate immune mechanisms is not influenced by the vector backbone.
FIG. 5.
Short-term stability of the vector DNAs in the livers of CBA/J mice. (A) Southern blot analysis of liver DNAs of CBA/J mice following intravenous administration of 4 × 1010 total viral particles of the indicated vectors. The experiment involved three mice per vector per time point. Total genomic liver DNA was extracted at the indicated times, digested with the restriction endonuclease BamHI, and analyzed by using a 32P-labeled restriction fragment from the E3-E4 region as probe. Control lanes contain 20, 10, 5, 1, and 0.1 viral genome copies, each mixed with 10 μg of mouse liver DNA (1 viral genome copy is equivalent to 30 pg of viral DNA). (B) Quantitative analysis of Ad vector DNA from the autoradiogram shown in panel A. The Southern blot was evaluated by densitometry scanning. The data are expressed as the percentage of viral DNA with respect to the initial value at 1 h p.i. □, E1°; •, E1°E2°; ○, E1°E4°.
Long-term in vivo persistence of AdE1°, AdE1°E2A°, and AdE1°E4° genomes.
In order to determine the impact of multiple viral gene deletions on long-term in vivo viral DNA stability and persistence, similar doses of infectious AdE1°, AdE1°E2A°, and AdE1°E4° vectors were administered to immunocompetent CBA/J and immunodeficient SCID mice by tail vein injection. The fate of the viral genomes in the liver and lungs was monitored over time. To eliminate all bias introduced by the expression of immunogenic transgene-encoded product, the vectors without transgenes (Table 1) were compared.
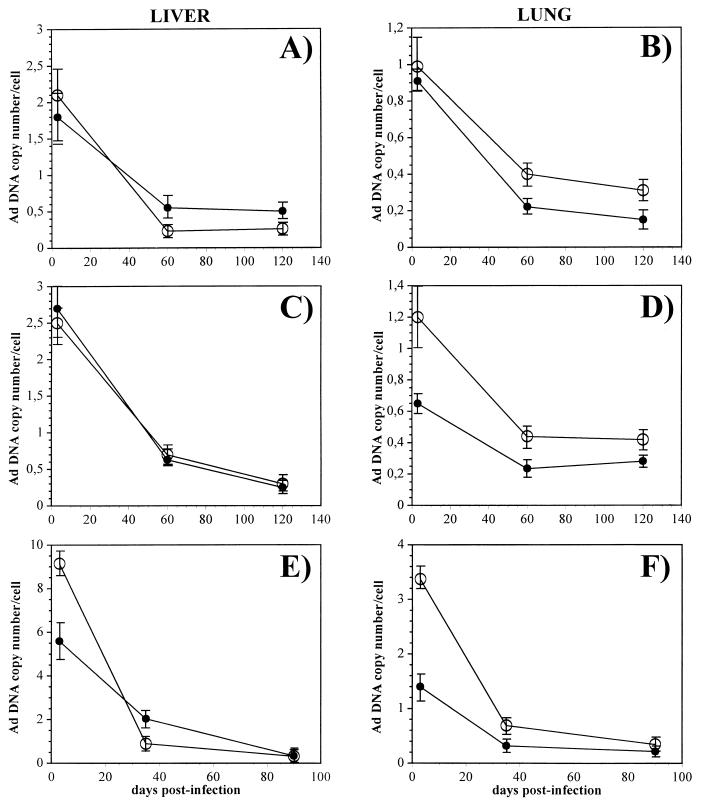
In a first study, 2 × 109 IU (7.6 × 1010 virus particles) of AdE1° (AdTG6401) and AdE1°E4° (AdTG9546) vectors was injected in the tail vein of each animal, and the persistences of the viral genomes were compared at 3, 60, and 120 days postinjection. Twenty-four CBA/J mice were used for both vectors (eight animals per time point), while 18 and 15 SCID mice were used for the AdE1° and AdE1°E4 vectors, respectively (six and five animals per time point, respectively). Southern blot analysis was performed with total liver and lung DNAs at the indicated times, and the intensities of the specific Ad DNA were quantitated by densitometry scanning (Fig. 6A to D). Two major observations were made in this experiment. (i) The viral DNA copy numbers of the AdE1° and AdE1°E4° vectors declined at approximately the same rate in transduced liver and lung cells of CBA/J and SCID animals. At 120 days postinjection, the viral copy numbers of the AdE1° and AdE1°E4° vectors were still at about 10 to 30% of the initial values in both liver and lung, irrespective of the immune status of the tested animals (Fig. 6A to D). These results suggest that loss of both viral genomes over time was not significantly influenced by the immune status of the animals. (ii) The persistences of E1- and E1/E4-deleted vectors were similar in the immunocompetent mice, indicating that additional deletion of the viral E4 region combined with reduction of virus antigen expression did not improve the persistence of the transduced cells in immunocompetent backgrounds.
FIG. 6.
Long-term persistence of vector DNA in the livers and lungs of CBA/J and SCID mice. Totals of 2 × 109 IU of AdE1° (A and B) and AdE1°E4 (C and D) and 109 PFU of AdE1°E2A° (E and F) were injected intravenously into SCID (•) and CBA/J (○) mice. DNA from livers (A, C, and E) and lungs (B, D, and F) was prepared at the indicated times and analyzed by Southern blotting and densitometry scanning. Symbols represent the means ± SE for 8 (A to D) and 10 (E and F) CBA/J animals and for 6 (A and B), 5 (C and D), and 10 (E and F) SCID mice.
In a second study, the viral DNA persistences of AdE1° (AdTG6401), AdE1°E2A° (AdTG9592), and AdE1°E4° (AdTG9546) vectors were compared by using the same strains of immunocompetent and immunodeficient mice. In this experiment, 30 CBA/J and 30 SCID mice were used for each virus (10 animals per time point); 109 PFU (AdE1°E2A°) and 109 IU (AdE1° and AdE1°E4°) of the vectors were injected in the tail veins of all mice, and the fate of the viral genomes in liver and lung was monitored by Southern blot analysis over a 3-month period (Fig. 6E and F and data not shown). Similar to the case for the first experiment, no significant differences in the declines of all viral genomes were observed, irrespective of the immune status of the mice. The AdE1°E2A° viral genome declined at 3 months postinjection to approximately 5% of the initial copy number in the livers of both strains of mice and to 10 to 20% in the lungs. The more rapid decline of this virus genome in the livers of both SCID and CBA/J mice compared to the AdE1° and AdE1°E4° vectors might be related to the higher initial AdE1°E2A° DNA copy number: the ratio of total virus particles to PFU of this vector preparation was 175:1, compared to 38:1 for the AdE1° and AdE1°E4° viruses. Thus, at the indicated doses, the amount of total AdE1°E2A° particles injected into mice was 4.6-fold higher than that for the other two vectors, resulting in the higher initial copy number observed. This observation is consistent with previous reports (14, 40) showing, in mice, a positive correlation between the rate of elimination of the Ad genome in liver and the hepatotoxicity induced by injection of high virus doses.
Host cellular and humoral immune responses to AdE1°-, AdE1°E2A°-, and AdE1°E4°-infected cells.
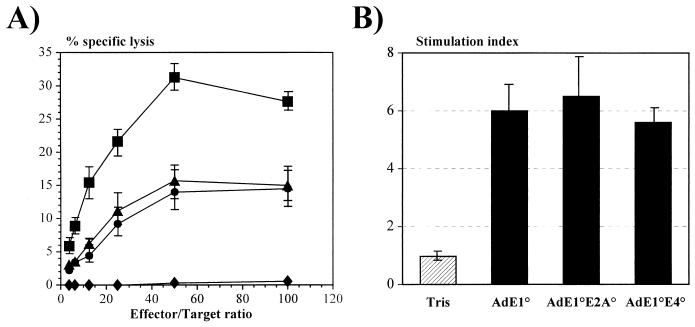
To evaluate the impact of the E2A and E4 deletions on the host anti-Ad immune response, CBA/J mice were immunized intraperitoneally with 5 × 108 IU of AdE1° (1.9 × 1010 particles), AdE1°E4° (1.9 × 1010 particles), and AdE1°E2A° (8.7 × 1010 particles) vectors carrying no transgenes and were tested for the Ad-specific cellular immune responses. As we previously reported (37), injection of E1-deleted vectors induced a consistent anti-Ad CTL response. Administration of AdE1°E2A° and AdE1°E4° vectors did not result in a detectable decrease of the antiviral CTL activity (Fig. 7A). The CTL activity was even slightly higher in animals injected with the AdE1°E2A° vector, but this might be due to the higher total virus particles/IU ratio of the AdE1°E2A viruses: injection of similar numbers of infectious AdE1°, AdE1°E2A°, and AdE1°E4° virions led to the administration of five times more virus particles for AdE1°E2A°. While these results do not support the hypothesis that reduction of virus protein synthesis should decrease the anti-Ad CTL response, they are consistent with a recent report showing that in human cells endogenous virus gene expression is not required to sensitize cells to lysis by Ad-specific CTLs (45). Taken together, these results suggest that infected cells can process virus particles to present target epitopes for lysis by CTLs without de novo synthesis of virus antigens. Alternatively, the reduced levels of viral antigen synthesis in vivo could still be sufficient to induce the immune response observed.
FIG. 7.
Induction of cellular immune response in CBA/J mice. (A) Mice were injected intraperitoneally with Tris (⧫) or with 109 IU of AdE1° (▴) and AdE1°E4° (•) and 109 PFU of AdE1°E2A° (▪) vectors. Splenocytes of the treated mice were tested for CTL activity against Ad-infected syngeneic cells. CTL activity was measured in a 4-h 51Cr assay. (B) Splenocytes from mock-infected (Tris) or Ad-infected mice were analyzed for their T-cell proliferative response to Ad particles applied to the culture plates. The stimulation index is the ratio between the values of [3H]thymidine incorporation by the stimulated cells and the unstimulated cells. Results are expressed as the means ± SE for four animals per group.
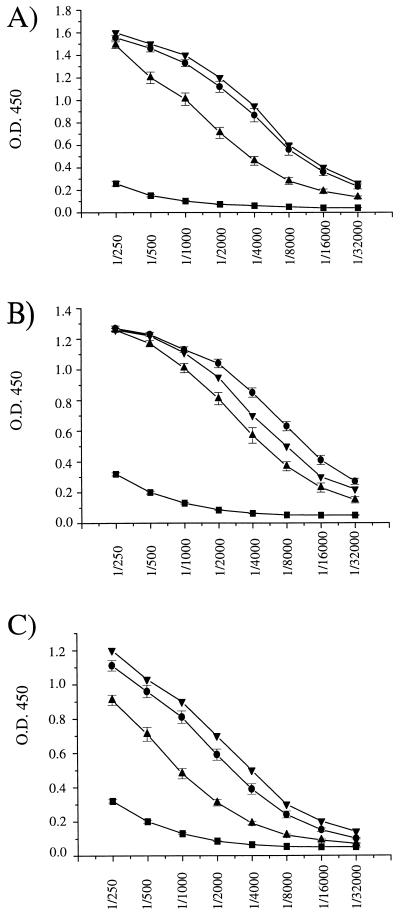
As expected, inoculation of AdE1°, AdE1°E4°, and AdE1°E2A° vectors stimulated similar virus-specific proliferation of the splenocytes of the treated CBA/J mice (Fig. 7B), reflecting the activation of CD4 lymphocytes recognizing virus epitopes presented in the context of major histocompatibility complex class II molecules. Consistent with this observation, high circulating levels of total anti-Ad antibodies were detected in the sera of CBA/J mice injected with the three types of vectors (Fig. 8).
FIG. 8.
Anti-Ad antibodies in CBA/J mice. Mice were injected intravenously with 109 IU of AdE1° (A) and AdE1°E4° (C) and 109 PFU of AdE1°E2A° (B) vectors. Sera were collected at days 4 (▪), 35 (▴), and 90 (•) postinjection and analyzed by ELISA as previously described (37). Data are expressed as the means of the optical density at 450 nm (O.D. 450) ± SE determined for successive serial dilutions of the sera recovered from 5 to 10 mice per experimental group. Each plate contained the same positive control serum (▾).
DISCUSSION
Previous studies have indicated that current generations of E1-deleted Ad vectors still express low levels of early and late viral proteins in infected target cells, leading to the immunological destruction of the transduced tissues (56–58, 61). To specifically evaluate the contribution of viral antigens to the host antiviral immune response and the impact of this response on the in vivo persistence of the transduced cells, Ad vectors with deletions only in E1 or with deletions in E1 and E2A or in E1 and E4 were generated, and their biological and immunological properties were analyzed in vitro and in vivo. To eliminate all bias introduced by the expression of immunogenic transgene-encoded products, we focused on a comparison of vectors devoid of any transgenes. For these studies, a series of isogenic singly and doubly deleted vectors was produced by homologous recombination in E. coli (11). This method allows the construction of identical viral genomes differing only in the defined deletions, thus limiting the risk of genetic variations between the various vectors to be compared.
We show that AdE1°E2A° and AdE1°E4° vectors can be produced efficiently in 293 complementation cells expressing the E2A or the E4ORF6+7 genes, respectively. In contrast, generation of high-titer, productive AdE1°E4° vectors in 293 cells expressing the whole E4 region was not possible, despite an efficient production of virus particles. The molecular mechanisms responsible for the altered life cycle of AdE1°E4° viruses in 293-E4 cells remain unclear, but this appears to be associated with a strong reduction of fiber protein synthesis. Consistent with this concept, AdE1°E4° viruses generated in 293-E4ORF6+7 cells synthesized much higher, albeit not optimal, concentrations of fiber proteins and could replicate to levels close to those of AdE1° vectors.
Expression of fiber, penton base, and DBP was abolished in noncomplementing A549 cells infected with AdE1°E2A° vectors at a high multiplicity (e.g., 500 BU/cell), in agreement with previous observations (26, 42). Similarly, the AdE1°E4° vectors were also markedly impaired for DNA replication and in their ability to express early and late viral genes. However, low levels of DBP and penton base could still be detected at a high multiplicity of infection (e.g., 500 IU/cell) but not at lower multiplicities (data not shown). Taken together, these results confirm that deletion of E1 alone is not sufficient to prevent expression of early and late virus genes.
The consequences of E1/E2A and E1/E4 deletions for the in vivo persistence and immunogenicity of the vectors with no transgenes were evaluated with immunocompetent CBA/J mice and with immunodeficient SCID mice. To limit the experimental variations observed from animal to animal, we performed two independent series of experiments and injected the vectors in a large number of mice (100 mice for AdE1° and AdE1°E4° vectors and 60 mice for AdE1°E2A° vectors). This study showed that the short-term maintenance and long-term (4-month) persistence of the virus genomes were comparable in the livers and lungs of immunocompetent or immunodeficient mice. After a rapid elimination of 80% of all vector genomes during the first 24 h, a progressive decline of viral genomes was observed in all animals, irrespective of the virus type and of the mouse immune status. Interestingly, 10 to 30% of the day 3 DNA value was still found 4 months after the inoculation of the vectors. This is consistent with our previous studies showing that E1-deleted vectors can persist and express a transgene for several months in immunocompetent mice tolerant for that particular transgene product (37). Our data also imply that similar long-term persistence of the singly and doubly deleted viral genomes should not lead to significant differences in the long-term persistence of transgene expression from these vectors, if other parameters influencing transgene expression are optimized.
Administration of AdE1°E2A° and AdE1°E4° vectors to immunocompetent animals resulted in antiviral CTL activity comparable to that observed with AdE1° vectors. This observation supports our previous results indicating that cellular immunity to viral antigens plays a minor role in controlling the persistence of the virus genomes (37) and is in agreement with recent data from Wadsworth et al. showing that Ad-infected cells can escape Ad-specific CTLs (51). Furthermore, the similar in vivo persistences of E1-deleted and multiply deleted viruses are consistent with a report from Smith et al. (45) showing that synthesis of virus protein is not required to sensitize target cells to CTL recognition. The evaluation of the antiviral CTL response was performed with vectors administered intraperitoneally, whereas the persistence of viral genomes was monitored after intravenous administration. While differences due to the route of administration cannot be excluded, this is unlikely since a recent study demonstrated no significant differences in the antiviral CTL responses for different routes of administration of E1-deleted vectors (47). Moreover, in all our studies, the E1-deleted vector was used as the reference vector for the evaluation of the multiply deleted viruses. As expected, all mice developed a strong antibody response to the E1-, E1/E4-, and E1/E2A-deleted vectors. While the in vivo parameters investigated in this study did not reveal any significant differences between the singly and doubly deleted vectors studied, we cannot exclude the possibility that these vectors might behave differently in their ability to induce an inflammatory response in the animal hosts (17, 23, 52).
Together, our results clearly challenge the hypothesis that progressive deletions of Ad vectors combined with reduced viral antigen expression should increase their in vivo persistence and should reduce the antiviral immune response. Our data are in contrast to those from other reports which demonstrated an extended persistence of transduced liver cells in animals injected with vectors defective in both E1 and E2A or in both E1 and E4 (17, 19, 20, 23, 24, 52, 59), which was correlated with a strongly reduced antigen expression in vivo (23). However, our results are consistent with previous reports indicating that mouse cellular immunity to viral antigens has a negligible influence on the in vivo persistence of transduced cells (37, 40, 51). All previous animal studies were performed with viral vectors expressing bacterial or human proteins that are recognized as foreign by the host immune system (17, 23). Depending on the type of immune response elicited in the treated hosts, the immunogenicity of such transgene products might therefore significantly modify the in vivo persistence of the transduced cells (32, 37, 40, 50). While the influence of the reporter gene might be minimized by using mice transgenic for the transgene-encoded product (17, 23), tolerance to the transgene may be either partial (40) or even broken by transgene overexpression (50). Thus, in those studies, interpretation of the respective contributions of the host antiviral and antitransgene responses to the in vivo behavior of the vector genomes is complex. Moreover, the existence of an interplay between the anti-Ad and antitransgene immune responses and the biological consequences of such a phenomenon on the transduced cells remain unclear. Although we believe that the results reported here have important implications for the design of recombinant vectors for human applications, we cannot extrapolate these conclusions to other animals or to humans.
Despite similar immunogenicities, multiply deleted vectors nevertheless have several clear advantages over E1-deleted viruses. (i) Emergence of replication-competent Ad is highly unlikely, since this would require simultaneous reversions in the E1 and E2 or E4 regions; moreover, reversions at the E2 or E4 loci would require illegitimate recombination events, since overlapping sequences between the E2 and E4 genes in the packaging cells and the E2- and E4-deleted vectors are present on only one side of the deletion breakpoint in the viral genomes. (ii) Deletion of E4 should theoretically improve the safety profile of the Ad vectors, given the oncogenic potential of the E4 ORF6 protein (18, 39). (iii) Together with deletion of E3, deletion of E1 and E2A or E4 allows the insertion of larger foreign DNA sequences. However, the impact of such multiple deletions on transgene expression is unpredictable (35a) and should be carefully investigated, as illustrated by several recent reports (1, 17).
ACKNOWLEDGMENTS
We thank L. Zenner (CDTA, Orléans, France) and C. Pêcheur for cooperation in the animal studies and M. Courtney for critical reading of the manuscript.
This work was supported in part by the Association Française contre la Mucoviscidose (AFLM) and the Association Française contre les Myopathies (AFM).
REFERENCES
- 1.Armentano D, Zabner J, Sacks C, Sookdeo C C, Smith M P, St. George J A, Wadsworth S C, Smith A E, Gregory R J. Effect of the E4 region on the persistence of transgene expression from adenovirus vectors. J Virol. 1997;71:2408–2416. doi: 10.1128/jvi.71.3.2408-2416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr D, Tubb J, Fergusson D, Scaria A, Lieber A, Wilson C, Perkins J, Kay M A. Strain related variations in adenovirally mediated transgene expression from mouse hepatocytes in vivo: comparisons between immunocompetent and immunodeficient inbred strains. Gene Ther. 1995;2:151–155. [PubMed] [Google Scholar]
- 3.Berkner K L. Development of adenovirus vectors for the expression of heterologous genes. BioTechniques. 1988;6:616–629. [PubMed] [Google Scholar]
- 4.Bondesson M, Ohman K, Mannervik M, Fan S, Aküsjarvi G. Adenovirus E4 open reading frame 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J Virol. 1996;70:3844–3851. doi: 10.1128/jvi.70.6.3844-3851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosma G C, Custer M C, Bosma M J. A severe combined immunodeficiency mutation in the mouse. Nature (London) 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 6.Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge E, Medghalchi S, Ubol S, Leesong M, Ketner G. Adenovirus early region 4 and viral DNA synthesis. Virology. 1993;193:794–801. doi: 10.1006/viro.1993.1188. [DOI] [PubMed] [Google Scholar]
- 8.Brough D E, Cleghon V, Klessig D F. Construction, characterization, and utilization of cell lines which inducibly express the adenovirus DNA-binding protein. Virology. 1992;190:624–634. doi: 10.1016/0042-6822(92)90900-a. [DOI] [PubMed] [Google Scholar]
- 9.Brough D E, Lizonova A, Hsu C, Kulesa V A, Kovesdi I. A gene transfer vector-cell line system for complete functional complementation of adenovirus early regions E1 and E4. J Virol. 1996;70:6197–6501. doi: 10.1128/jvi.70.9.6497-6501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challberg S S, Ketner G. Deletion mutants of adenovirus 2: isolation and initial characterization of virus carrying mutations near the right end of the viral genome. Virology. 1981;114:196–209. doi: 10.1016/0042-6822(81)90265-8. [DOI] [PubMed] [Google Scholar]
- 11.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christ M, Lusky M, Stoeckel F, Dreyer D, Dieterle A, Michou A I, Pavirani A, Mehtali M. Gene therapy with recombinant adenovirus vectors: evaluation of the host immune response. Immunol Lett. 1997;57:19–25. doi: 10.1016/s0165-2478(97)00049-7. [DOI] [PubMed] [Google Scholar]
- 13.Chroboczek J, Bieber F, Jacrot B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology. 1992;186:280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- 14.Connelly S, Gardner J M, Lyons R M, McLelland A, Kaleko M. Sustained expression of therapeutic levels of human factor VIII in mice. Blood. 1996;87:4671–4677. [PubMed] [Google Scholar]
- 15.Connelly S, Smith T A G, Dhir G, Gardner J M, Mehaffey M G, Zaret K S, McLelland A, Kaleko M. In vivo gene delivery and expression of physiological levels of functional human factor VIII in mice. Hum Gene Ther. 1995;6:185–193. doi: 10.1089/hum.1995.6.2-185. [DOI] [PubMed] [Google Scholar]
- 16.Dai Y, Schwartz E M, Gu D, Zhang W W, Sarvetnick N, Verma I M. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc Natl Acad Sci USA. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dedieu J F, Vigne E, Torrent C, Jullien C, Mahfouz I, Caillaud J M, Aubailly N, Orisini C, Guillaume J M, Opolon P, Delaere P, Perricaudet M, Yeh P. Long-term gene delivery into the livers of immunocompetent mice with E1/E4-defective adenoviruses. J Virol. 1997;71:4626–4637. doi: 10.1128/jvi.71.6.4626-4637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4-ORF6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 19.Engelhardt J F, Litzky L, Wilson J M. Prolonged transgene expression in cotton rat lung with recombinant adenovirus defective in E2a. Hum Gene Ther. 1994;5:1217–1229. doi: 10.1089/hum.1994.5.10-1217. [DOI] [PubMed] [Google Scholar]
- 20.Engelhardt J F, Ye X, Doranz B, Wilson J M. Ablation of E2a in recombinant adenoviruses improves transgene persistence and decreases inflammatory response in mouse liver. Proc Natl Acad Sci USA. 1994;91:6196–6200. doi: 10.1073/pnas.91.13.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang B, Wang H, Gordon G, Bellinger D A, Read M S, Brinkhous K M, Woo S L C, Eisensmith R C. Lack of persistence of E1− recombinant adenoviral vectors containing a temperature-sensitive E1A mutation in immunocompetent mice and hemophilia B dogs. Gene Ther. 1996;3:217–222. [PubMed] [Google Scholar]
- 22.Fasel N, Pearson K, Buetti E, Diggelmann H. The region of mouse mammary tumor virus DNA containing the long terminal repeat includes a long coding sequence and signals for hormonally regulated transcription. EMBO J. 1982;1:3–7. doi: 10.1002/j.1460-2075.1982.tb01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao G P, Yang Y, Wilson J M. Biology of adenovirus vectors with E1 and E4 deletions for liver-directed gene therapy. J Virol. 1996;70:8934–8943. doi: 10.1128/jvi.70.12.8934-8943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldman M J, Litzky L A, Engelhardt J F, Wilson J M. Transfer of the CFTR gene to the lung of non-human primates with E1-deleted, E2A-defective recombinant adenovirus: a preclinical toxicology study. Hum Gene Ther. 1995;6:839–851. doi: 10.1089/hum.1995.6.7-839. [DOI] [PubMed] [Google Scholar]
- 25.Gossen M, Freundlieb S, Bender G, Muller G, Hille W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 26.Gorziglia M I, Kadan M J, Yei S, Lim J, Lee G M, Luthra R, Trapnell B C. Elimination of both E1 and E2A from adenovirus vectors further improves prospects for in vivo human gene therapy. J Virol. 1996;70:4173–4178. doi: 10.1128/jvi.70.6.4173-4178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham F L, Prevec L. Manipulation of adenovirus vectors. In: Murray E J, editor. Methods in molecular biology. Vol. 7. Clifton, N.J: The Humana Press Inc.; 1991. pp. 109–128. [DOI] [PubMed] [Google Scholar]
- 28.Graham F L, Prevec L. Adenovirus based expression vectors and recombinant vaccines. In: Ellis R W, editor. Vaccines: new approaches to immunological problems. London, United Kingdom: Butterworth-Heinemann; 1992. pp. 363–390. [DOI] [PubMed] [Google Scholar]
- 29.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 30.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 31.Henry L, Xia D, Wilke M, Deisenhofer J, Gerard R. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–5246. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juillard V, Villefroy P, Godfrin D, Pavirani A, Venet A, Guillet J G. Long term humoral and cellular immunity induced by a single immunization with replication-defective adenovirus recombinant vector. Eur J Immunol. 1995;25:3467–3473. doi: 10.1002/eji.1830251239. [DOI] [PubMed] [Google Scholar]
- 33.Kass-Eisler A, Falck-Pedersen E, Elfenbein D H, Alvira M, Buttrick P M, Leinwand L A. The impact of development stage, route of administration, and the immune system on adenovirus-mediated gene transfer. Gene Ther. 1994;1:395–402. [PubMed] [Google Scholar]
- 34.Krougliak V, Graham F L. Development of cell lines capable of complementing E1, E4 and protein IX defective adenovirus type 5 mutants. Hum Gene Ther. 1995;6:1575–1586. doi: 10.1089/hum.1995.6.12-1575. [DOI] [PubMed] [Google Scholar]
- 35.Lathe R, Vilotte J L, Clark A J. Plasmid and bacteriophage vectors for excision of intact inserts. Gene. 1987;57:193–201. doi: 10.1016/0378-1119(87)90122-3. [DOI] [PubMed] [Google Scholar]
- 35a.Leroy, P., A. I. Michou, and M. Lusky. Unpublished observations.
- 36.Li Q, Kay M A, Finegold M, Stratford-Perricaudet L D, Woo S L. Assessment of recombinant adenoviral vectors for hepatic gene therapy. Hum Gene Ther. 1993;4:403–409. doi: 10.1089/hum.1993.4.4-403. [DOI] [PubMed] [Google Scholar]
- 36a.Lusky, M., and M. Christ. Unpublished observations.
- 36b.Lusky, M., and B. Mourot. Unpublished observations.
- 37.Michou A I, Santoro L, Christ M, Juillard V, Pavirani A, Mehtali M. Adenovirus-mediated gene therapy: influence of transgene, mouse strain and type of immune response on persistence of transgene expression. Gene Ther. 1997;4:473–482. doi: 10.1038/sj.gt.3300412. [DOI] [PubMed] [Google Scholar]
- 38.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore M N, Horikoshi S, Shenk T. Oncogenic potential of the adenovirus E4-ORF6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morral N, O’Neal W, Zhou H, Langston C, Beaudet A. Immune response to reporter proteins and high viral dose limit duration of expression with adenoviral vectors: comparison of E2A wild-type and E2A-deleted vectors. Hum Gene Ther. 1997;8:1275–1286. doi: 10.1089/hum.1997.8.10-1275. [DOI] [PubMed] [Google Scholar]
- 41.Reich N C P, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 42.Rittner K, Schultz H, Pavirani A, Mehtali M. Conditional repression of the E2 transcription unit in E1-E3-deleted adenovirus vectors is correlated with a strong reduction in viral DNA replication and late gene expression in vitro. J Virol. 1997;71:3307–3311. doi: 10.1128/jvi.71.4.3307-3311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. Philadelphia, Pa: Raven Press; 1996. pp. 2111–2148. [Google Scholar]
- 45.Smith C A, Woodruff L S, Kitchingman G R, Rooney C M. Adenovirus-pulsed dendritic cells stimulate human virus-specific T-cell responses in vitro. J Virol. 1996;70:6733–6740. doi: 10.1128/jvi.70.10.6733-6740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith T A G, Mehaffey M G, Kayda D B, Saunders J M, Yei S, Trapnell B C, McLelland A, Kaleko M. Adenovirus mediated expression of therapeutic plasma levels of human factor IX in mice. Nat Genet. 1993;5:392–402. doi: 10.1038/ng1293-397. [DOI] [PubMed] [Google Scholar]
- 47.Song W, Kong H-L, Traktman P, Crystal R G. Cytotoxic T lymphocyte responses to proteins encoded by heterologous transgenes transferred in vivo by adenoviral vectors. Hum Gene Ther. 1997;8:1207–1217. doi: 10.1089/hum.1997.8.10-1207. [DOI] [PubMed] [Google Scholar]
- 48.Stratford-Perricaudet L D, Makeh I, Perricaudet M, Briand P. Widespread long-term gene transfer to mouse skeletal muscles and heart. J Clin Invest. 1992;90:626–630. doi: 10.1172/JCI115902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell B C. Adenoviral vectors for gene transfer. Adv Drug Deliv Rev. 1993;12:185–199. [Google Scholar]
- 50.Tripathy S K, Black H B, Goldwasser E, Leiden J M. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 51.Wadsworth S C, Zhou H, Smith A E, Kaplan J M. Adenovirus vector-infected cells can escape adenovirus antigen-specific cytotoxic T lymphocyte killing in vivo. J Virol. 1997;71:5189–5196. doi: 10.1128/jvi.71.7.5189-5196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q, Greenburg C, Bunch D, Farson D, Finer M H. Persistent transgene expression in mouse liver following in vivo gene transfer with ΔE1/ΔE4 adenovirus vectors. Gene Ther. 1997;4:393–400. doi: 10.1038/sj.gt.3300404. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, Jia X C, Finer M H. A packaging cell line for the propagation of recombinant adenovirus vectors containing two lethal gene-region deletions. Gene Ther. 1995;2:775–783. [PubMed] [Google Scholar]
- 54.Weinberg D H, Ketner G. A cell line that supports the growth of a defective early region 4 mutant of human adenovirus type 2. Proc Natl Acad Sci USA. 1983;80:5383–5386. doi: 10.1073/pnas.80.17.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worgall S, Wolff G, Falck-Pederson E, Crystal R G. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum Gene Ther. 1997;8:37–44. doi: 10.1089/hum.1997.8.1-37. [DOI] [PubMed] [Google Scholar]
- 56.Yang Y, Ertl H C J, Wilson J M. MHC class I restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Jooss K U, Su Q, Ertl H C, Wilson J M. Immune responses to viral antigens versus transgene product in the elimination of recombinant adenovirus-infected hepatocytes in vivo. Gene Ther. 1996;3:137–144. [PubMed] [Google Scholar]
- 58.Yang Y, Nunes F A, Berencsi K, Furth E, Gönczöl E, Wilson J M. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y, Nunes F A, Berencsi K, Gönczöl E, Engelhardt J F, Wilson J M. Inactivation of E2A in recombinant adenoviruses improves the prospect for gene therapy in cystic fibrosis. Nat Genet. 1994;7:362–369. doi: 10.1038/ng0794-362. [DOI] [PubMed] [Google Scholar]
- 60.Yang Y, Su Q, Grewal I S, Schiltz R, Flavell R A, Wilson J M. Transient subversion of CD40 ligand function diminishes immune response to adenovirus vectors in mouse liver and lung tissues. J Virol. 1996;70:6370–6377. doi: 10.1128/jvi.70.9.6370-6377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y, Qin I, Ertl H J C, Wilson J M. Cellular and humoral immune response to viral antigens create barriers to lung-directed gene therapy with recombinant adenoviruses. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeh P, Dedieu J F, Orsini C, Vigne E, Denefle P, Perricaudet M. Efficient dual transcomplementation of adenovirus E1 and E4 regions from a 293-derived cell line expressing a minimal E4 functional unit. J Virol. 1996;70:559–565. doi: 10.1128/jvi.70.1.559-565.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou H, O’Neal W, Morral N, Beaudet A L. Development of a complementation cell line and a system for construction of adenovirus with E1 and E2A deleted. J Virol. 1996;70:7030–7038. doi: 10.1128/jvi.70.10.7030-7038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zsengeller Z K, Wert S E, Hull W M, Hu X, Yei S, Trapnell B C, Whitsett J A. Persistence of replication-deficient adenovirus-mediated gene transfer in lungs of immune-deficient (nu/nu) mice. Hum Gene Ther. 1995;6:457–467. doi: 10.1089/hum.1995.6.4-457. [DOI] [PubMed] [Google Scholar]