ABSTRACT
Superinfection exclusion (SIE) is a phenomenon in which a preexisting infection prevents a secondary infection. SIE has been described for several flaviviruses, such as West Nile virus vs Nhumirim virus and Dengue virus vs yellow fever virus. Zika virus (ZIKV) is an emerging flavivirus posing threats to human health. The SIE between ZIKV and Japanese encephalitis virus (JEV) is investigated in this study. Our results demonstrate for the first time that JEV inhibits ZIKV infection in both mammalian and mosquito cells, whether co-infects or subsequently infects after ZIKV. The exclusion effect happens at the stage of ZIKV RNA replication. Further studies show that the expression of JEV NS2B protein is sufficient to inhibit the replication of ZIKV, and the outer membrane region of NS2B (46–103 aa) is responsible for this SIE. JEV infection and NS2B expression also inhibit the infection of the vesicular stomatitis virus. In summary, our study characterized a SIE caused by JEV NS2B. This may have potential applications in the prevention and treatment of ZIKV or other RNA viruses.
IMPORTANCE
The reemerged Zika virus (ZIKV) has caused severe symptoms in humans and poses a continuous threat to public health. New vaccines or antiviral agents need to be developed to cope with possible future pandemics. In this study, we found that infection of Japanese encephalitis virus (JEV) or expression of NS2B protein well inhibited the replication of ZIKV. It is worth noting that both the P3 strain and vaccine strain SA14-14-2 of JEV exhibited significant inhibitory effects on ZIKV. Additionally, the JEV NS2B protein also had an inhibitory effect on vesicular stomatitis virus infection, suggesting that it may be a broad-spectrum antiviral factor. These findings provide a new way of thinking about the prevention and treatment of ZIKV.
KEYWORDS: Japanese encephalitis virus, Zika virus, superinfection exclusion, NS2B protein
INTRODUCTION
Zika virus (ZIKV), a member of the genus Flavivirus in the family Flaviviridae, is transmitted principally by mosquitoes but can also be transmitted through blood transfusions and mother-to-fetus. The ZIKV genome comprises an 11 kb single-strand of positive-sense RNA. ZIKV was discovered in Uganda in 1947, and the first large-scale outbreak was on Yap Island in 2007 (1). Until the outbreaks in French Polynesia in 2013–2014 (2) and northeastern Brazil in 2015 (3), ZIKV had been confined to a narrow equatorial belt running across Africa into Asia (4). The reemergence of ZIKV is associated with severe neurological complications, including Guillain-Barré syndrome in adults and microcephaly in neonates (5). To date, although there are a number of vaccines in clinical trials, there is neither vaccine available for ZIKV nor are there any antiviral treatments (6, 7).
Superinfection exclusion (SIE), also termed homologous interference, is a phenomenon in which a cell infected with one virus is resistant to subsequent infection with homologous or heterologous viruses (8). Superinfection exclusion has been observed for many viruses, such as vaccinia virus (9), hepatitis C virus (HCV) (10, 11), influenza A virus (12), Sindbis virus (SINV) (13), vesicular stomatitis virus (VSV) (14), and human papillomavirus (15). Superinfection exclusion has also been found among different members of flaviviruses (16, 17). Infection of an insect-specific flavivirus, Nhumirim virus (NHUV), suppresses the multiplication of West Nile virus (WNV) (18), ZIKV (19), Dengue virus 2 (DENV-2) (19), Japanese encephalitis virus (JEV), and St. Louis encephalitis virus (SLEV) (20) in C6/36 cells. Palm Creek virus suppresses subsequent replication of WNV and Murray Valley encephalitis viruses in co-infected mosquito cells (21).
Superinfection exclusion can be induced by restricting any steps in the life cycle of the secondary virus (22–24). The vaccinia proteins A56 and K2 form heterodimers on the cell membrane (25), these then interact with the vaccinia proteins A16 and G9 on the cell surface, resulting in the suppression of spontaneous syncytium formation (26). Semliki Forest virus (SFV) can inhibit further infection by more of itself by blocking the entry and uncoating of the superinfecting virus (27). In flavivirus, replication of WNV RNA suppresses subsequent WNV infection (28). A valine-to-methionine substitution at position nine of the WNV 2K peptide (2 K-V9M) confers the ability to overcome superinfection exclusion (29). Superinfection exclusion has promising implications for the development of antiviral strategies (8).
RESULTS
JEV infection excludes superinfection of ZIKV
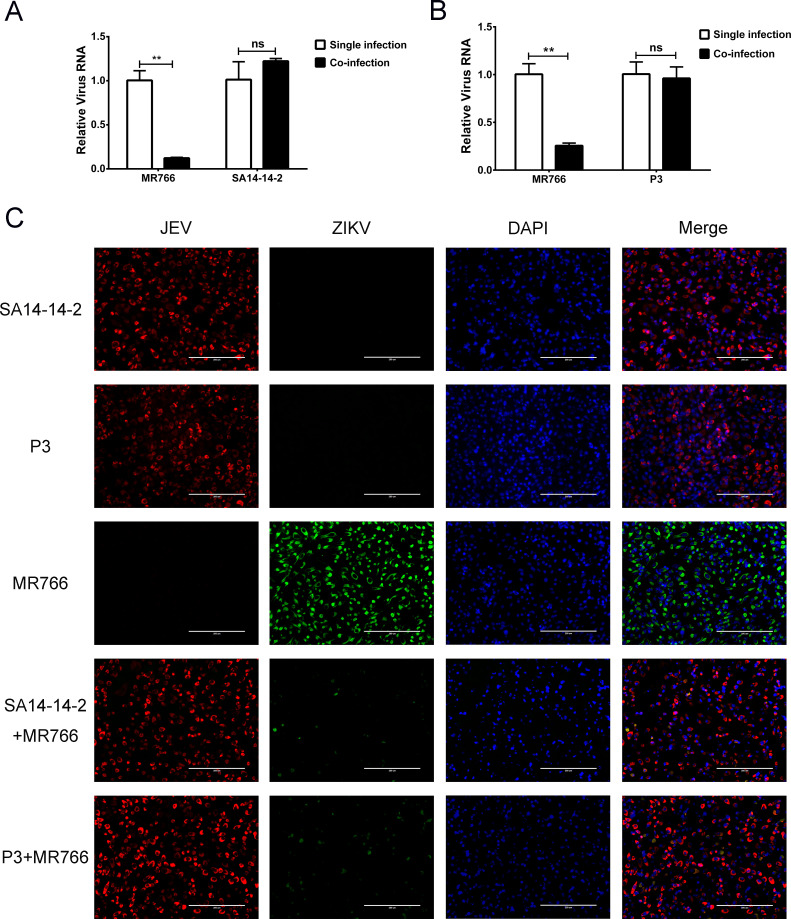
To examine the superinfection exclusion between JEV and ZIKV, we co-infected BHK-21 and Vero cells with JEV and ZIKV. Cells were infected with the JEV P3 strain at an multiplicity of infection (MOI) of 1, the JEV SA14-14-2 strain at an MOI of 5, and the ZIKV MR766 strain at an MOI of 5. The MOI we used for each virus resulted in nearly 100% infection at 30 h post infection (Fig. S1 and S2). The RNA levels of JEV and ZIKV were quantitated by quantitative reverse transcriptase PCR (qRT-PCR) after 30 h incubation. In both cell lines, there was significantly less ZIKV RNA in co-infected cells (P < 0.01) than in cells infected with ZIKV alone (Fig. 1A; Fig. S3A). There was, however, no significant change in the level of JEV RNA in co-infected cells (P > 0.05) compared with those infected with JEV alone (Fig. 1A; Fig. S3A). We found the same level of ZIKV exclusion caused by JEV SA14-14-2 strain (attenuated vaccine strain) and P3 strain (virulent strain; Fig. 1A and B; Fig. S3A and B).
Fig 1.
ZIKV proliferation is decreased by co-infection with JEV on BHK-21 cells. BHK-21 cells were co-infected with ZIKV (MOI of 5) and JEV (A) strain SA14-14-2 (MOI of 5) or (B) strain P3 (MOI of 1). ZIKV and JEV RNA levels were determined by qRT-PCR at 30 h post infection. Compared to single-infected cells, ZIKV RNA levels were significantly reduced (P < 0.01) in co-infected cells. Levels of JEV RNA in co-infected cells were not statistically different than in cells infected with JEV alone. (C) Viral protein expression in single- and co-infected BHK-21 cells. JEV NS1 protein (red) and ZIKV NS2B-NS3 protease protein (green) were detected by indirect immunofluorescence assay (IFA) at 30 h post infection. Nuclei were stained with 4'-6-diamidino-2-phenylindole (DAPI) (blue). JEV and ZIKV proteins were detected in single-infected cells. In co-infected cells, the number of JEV-positive cells did not observably change, while the number of ZIKV-infected cells was greatly reduced. ns, non-significant. P > 0.05; **, very significant, P < 0.01.
Levels of JEV NS1 and ZIKV NS2B-NS3 protease expression were determined by IFA in co-infected cells. Viral proteins were detected in most cells infected with JEV or ZIKV alone (Fig. 1C; Fig. S3C), but in co-infected cells, there was a clear decrease in the number of ZIKV-positive cells (Fig. 1C; Fig. S3C). These results indicated that JEV infection inhibits the proliferation of ZIKV in both BHK-21 and Vero cells. It should be noted that Vero cells are IFN-α and IFN-β deficient, indicating that this exclusion is not caused by an innate immune response triggered by JEV.
As JEV and ZIKV are both mosquito-borne viruses, superinfection exclusion between JEV and ZIKV was further explored in mosquito cells. C6/36 cells were co-infected with JEV (SA14-14-2 strain at an MOI of 5 and P3 strain at an MOI of 1) and ZIKV (MOI of 5). The expression of JEV NS1 and ZIKV NS2B-NS3 was determined by IFA at 3 days post infection. The lower level of ZIKV protein was found in co-infected cells than in cells infected with ZIKV alone, whereas there was no observable difference in JEV protein levels in co-infected cells vs cells infected with JEV alone (Fig. S4). Taken together, these data demonstrated that JEV infection can exclude ZIKV in both mammalian and mosquito cells.
ZIKV infectivity is not recovered by the addition of excess ZIKV virions
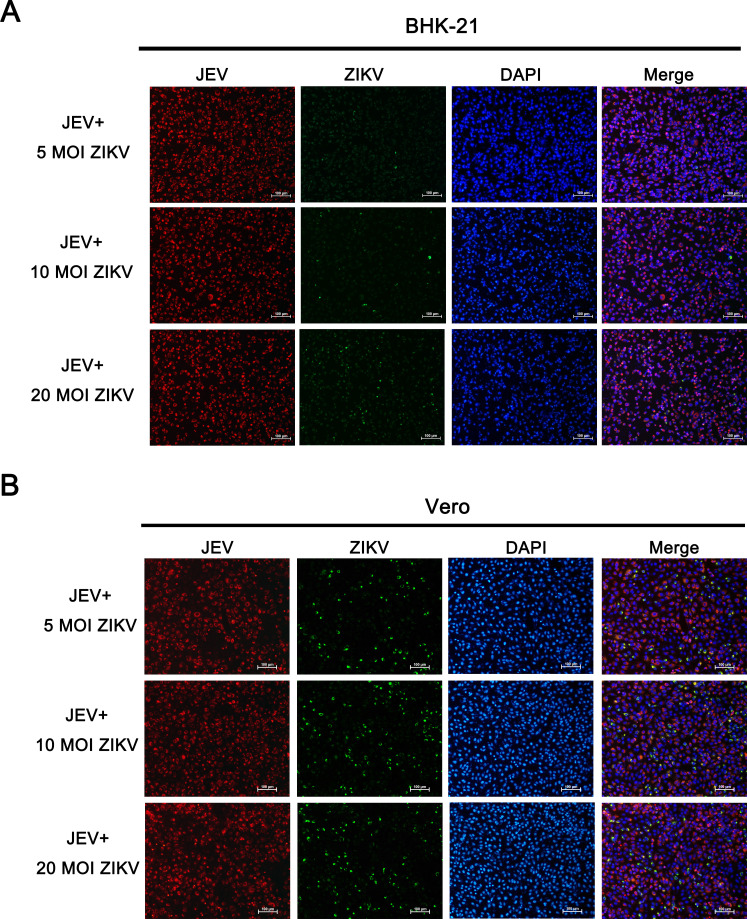
To determine if a higher dose of ZIKV could restore the infection blocked by JEV co-infection, BHK-21 or Vero cells were co-inoculated with a constant MOI of JEV SA14-14-2 (MOI of 5) and different doses of ZIKV (MOI of 5, 10, and 20). JEV NS1 and ZIKV NS2B-NS3 protease protein were stained by IFA at 30 h post infection. Increasing the dose of ZIKV in co-infected cells did not affect the proportion of JEV-positive cells, and ZIKV infection was still inhibited (Fig. 2).
Fig 2.
ZIKV infection inhibited by JEV in co-infected cells could not be recovered by increasing the dose of ZIKV. (A) BHK-21 or (B) Vero cells were co-infected with a constant dose of JEV SA14-14-2 (MOI of 5) and different doses of ZIKV (MOI of 5,10, and 20). The infection levels of JEV (red) and ZIKV (green) were accessed by IFA at 30 h post infection. Nuclei were stained with DAPI (blue). Increasing the dose of ZIKV in co-infection did not improve ZIKV infection.
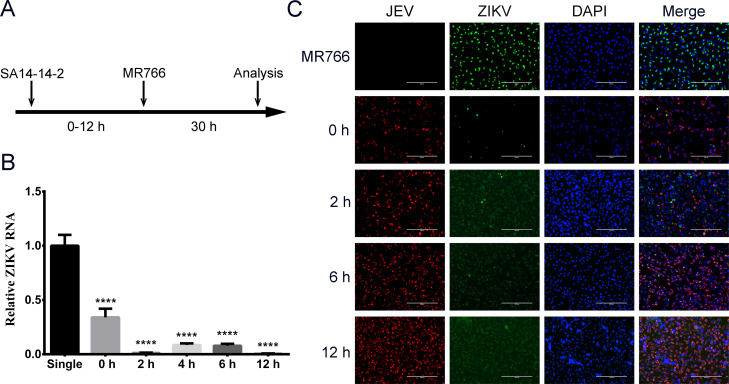
Superinfection exclusion of ZIKV during JEV replication
ZIKV (MOI of 5) was inoculated onto Vero cells at 0, 2, 6, and 12 h post JEV infection (MOI of 5; Fig. 3A). Levels of ZIKV RNA and protein were analyzed by qRT-PCR and IFA at 30 h after ZIKV infection, respectively. Cells that were infected first with JEV had significantly lower (P < 0.0001) levels of ZIKV RNA than cells infected with ZIKV alone, as well as cells simultaneously co-infected with JEV and ZIKV (0 h; Fig. 3B). In the IFA, high levels of ZIKV protein were seen only in cells infected by ZIKV alone (Fig. 3C). These results showed that superinfection exclusion of ZIKV by JEV happens at any stage of JEV replication.
Fig 3.
Superinfection exclusion at different stages of JEV replication. (A) Diagram of the infection timeline. Vero cells were infected with ZIKV (MOI of 5) at 0, 2, 4, 6, and 12 h after JEV infection (MOI of 5). The qRT-PCR and IFA were performed to determine RNA replication and protein expression at 30 h after ZIKV infection. (B) ZIKV RNA levels were determined by qRT-PCR 30 h post-ZIKV infection. (C) The viral proteins were detected by IFA. ZIKV was stained with NS2B-NS3 protease antibody (green), and JEV was stained with NS1 antibody (red). Nuclei were stained with DAPI (blue). ****, extremely significant, P < 0.0001.
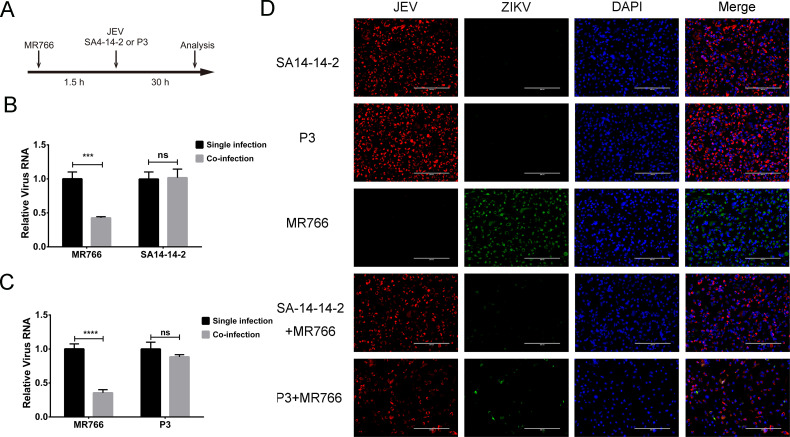
Superinfection exclusion does not occur during ZIKV attachment and entry
ZIKV was incubated with cells at an MOI of 5 for 1.5 h at 37°C, which allowed the virus to enter the cells. Cells were then washed three times with Dulbecco’s Modified Eagle’s Medium (DMEM) to remove any unbound particles and infected with JEV (SA14-14-2 strain at an MOI of 5 or P3 strain at an MOI of 1) for 30 h (Fig. 4A). The proliferation of ZIKV and JEV was assessed by qRT-PCR and IFA. Compared with cells infected with ZIKV alone, those superinfected with JEV had significantly less ZIKV RNA (P < 0.01; Fig. 4B and C), while levels of JEV RNA in the co-infected cells were not changed significantly (P > 0.05; Fig. 4B and C). Similar results were seen by IFA. The number of cells expressing ZIKV protein was sharply decreased in the presence of JEV (Fig. 4D). These results suggested that the exclusion of ZIKV by JEV does not occur during attachment and entry of ZIKV.
Fig 4.
The JEV exclusion of ZIKV at the stage of attachment and entry. (A) Diagram of infection timeline. Vero cells were infected with ZIKV (MOI of 5) 1.5 h prior to JEV (P3 strain at an MOI of 1 and SA14-14-2 strain at an MOI of 1). ZIKV and JEV infection were analyzed by qRT-PCR and IFA at 30 h post JEV infection. (B and C) Viral RNA replication. The replication of ZIKV was significantly inhibited by infection of JEV SA14-14-2 or P3 strain (P < 0.01). (D) IFA. JEV protein was detected by NS1 monoclonal antibody (red), and ZIKV protein was detected by NS2B-NS3 protease polyclonal antibody (green). Nuclei were stained with DAPI (blue). The ZIKV protein was barely detectable in the co-infected cells. ns, non-significant, P > 0.05; ***, very significant, P < 0.01; ****, extremely significant, P < 0.0001.
JEV inhibits ZIKV at the stage of RNA replication
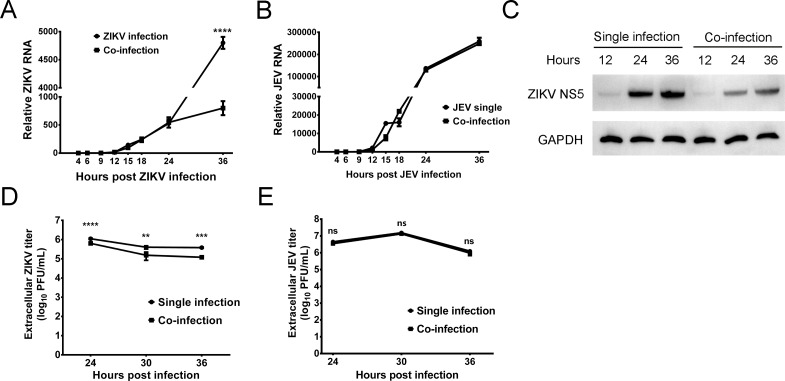
Cells were co-infected with ZIKV (MOI of 5) and JEV (SA14-14-2 strain at an MOI of 5), and the relative viral RNA levels were determined at 4, 6, 9, 12, 15, 18, 24, and 36 h post infection. RNA replication for both viruses began after 12 h (Fig. 5A and B). Up to 24 hpi, there was no significant difference in ZIKV RNA levels between single- and co-infected cells (P > 0.05; Fig. 5A). However, at 36 hpi, ZIKV RNA replication was significantly decreased in co-infected cells (P < 0.0001; Fig. 5A). In contrast, JEV RNA levels had no significant difference between single and co-infection groups at all time points (P > 0.05; Fig. 5B). Viral protein levels in single- and co-infected cells were determined by Western blot at 12, 24, and 36 hpi (Fig. 5C). In ZIKV single-infected cells, the NS5 protein was just detectable at 12 hpi, and as expected, the level increased through 36 hpi. The same profile was seen in co-infected cells, except that the level of NS5 protein in co-infected cells was lower than that in ZIKV single-infected cells (Fig. 5C). The difference in viral titers between co-infected cells and single-infected cells was examined by plaque assay. The titer of ZIKV in co-infected cells was significantly lower than that in ZIKV single-infected cells at 24 (P < 0.0001), 30 (P < 0.01), and 36 (P < 0.001) hpi. There was no significant difference at any time point in JEV titer between single- and co-infected cells (P > 0.05; Fig. 5E). Collectively, these results indicated that JEV inhibits ZIKV at the stage of RNA replication.
Fig 5.
RNA replication, viral protein expression, and packaging in co-infected cells. Vero cells were co-infected with ZIKV (MOI of 5) and JEV (SA14-14-2 strain at an MOI of 5), and JEV or ZIKV single-infected cells were used as a control. The single- and co-infected cells were harvested at 4, 6, 9, 12, 15, 18, 24, and 36 h post infection. (A) Levels of ZIKV RNA from 4 to 36 hpi. The levels of RNA were not significantly different from 4 to 24 hpi, and by 36 hpi, ZIKV RNA replication was significantly inhibited in the co-infected cells (P < 0.0001). (B) Levels of JEV RNA from 4 to 36 hpi. JEV RNA levels were not significantly different between single- and co-infected cells (P > 0.05). (C) Protein expression in single- and co-infected cells determined by Western blot. The protein loading was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) The titer of extracellular ZIKV in single- and co-infected cells. The titer of extracellular ZIKV in co-infected cells was significantly lower than that in ZIKV-only infected cells. (P < 0.01). (E) The titer of extracellular JEV in single- and co-infected cells. There was no significant difference in the titer of extracellular JEV between the single- and co-infected cells.
JEV NS2B is involved in superinfection exclusion
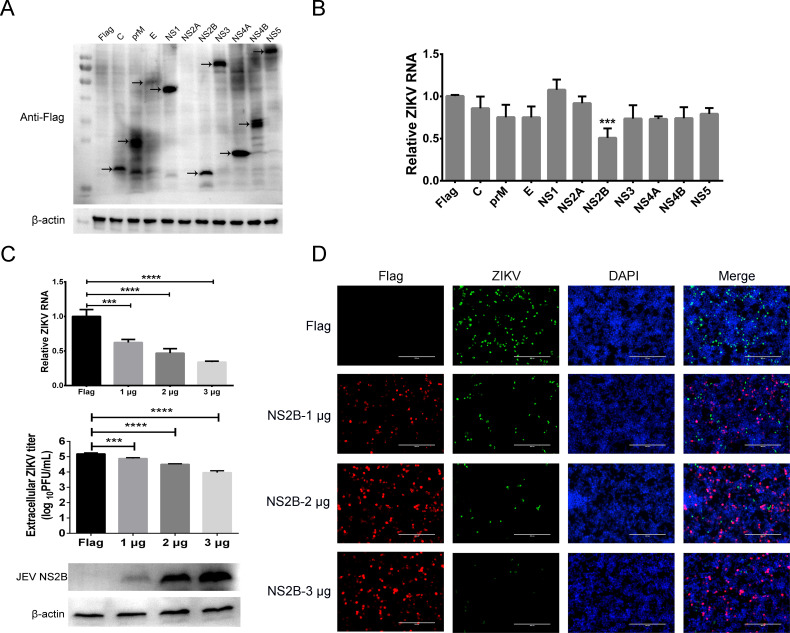
To clarify the roles of JEV proteins in the superinfection exclusion, structural and non-structural proteins C, prM, E, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 were over expressed in cells. The transfected cells were then infected with ZIKV at an MOI of 5, and the ZIKV RNA levels were quantitated by qRT-PCR. Western blot showed that all JEV proteins, except NS2A, were expressed in cells (Fig. 6A). The levels of ZIKV RNA significantly decreased in JEV NS2B expressed cells compared with the control cells (P < 0.001; Fig. 6B). Although the possibility of JEV NS2A cannot be ruled out, these results demonstrated that JEV NS2B was a major factor causing ZIKV superinfection exclusion. To further study the roles of NS2B in superinfection exclusion, 1, 2, and 3 µg of the NS2B expression plasmid was transfected into cells. Western blot showed that the expression of JEV NS2B increased with the amount of transfected plasmid, and the levels of ZIKV RNA and extracellular ZIKV titer decreased significantly (P < 0.001; Fig. 6C). ZIKV protein expression in JEV NS2B expressing cells was then assessed by IFA. Cells were transfected with 1, 2, and 3 µg of NS2B expressing plasmid for 24 h, then infected with ZIKV. JEV NS2B and ZIKV proteins were stained with anti-FLAG and ZIKV NS2B-NS3 protease antibodies, respectively. The results showed that the number of cells expressing ZIKV protein decreased with the increase of JEV NS2B (Fig. 6D). It should be noted that no JEV NS2B and ZIKV NS2B-NS3 protease double-positive cells are there in merged images (Fig. 6D). As control, different amounts of JEV NS1 and NS4B expression plasmid were also transfected into cells for 24 h, and then the cells were infected with ZIKV. Contrary to the superinfection exclusion exhibited by JEV NS2B, the expression of JEV NS1 or NS4B had no significant influence on the level of ZIKV RNA and extracellular viral titer (Fig. S5). Collectively, these results demonstrated that the expression of JEV NS2B could inhibit the proliferation of ZIKV and is involved in the superinfection exclusion of ZIKV.
Fig 6.
JEV NS2B is involved in the superinfection exclusion of ZIKV. (A) Overexpression of JEV proteins in cells. Eukaryotic expression plasmids carrying JEV genes were transfected into HEK-293T cells, and expression was identified by Western blot using anti-FLAG antibody. Arrows indicate expressed proteins. (B) Replication of ZIKV RNA in JEV proteins-transfected cells. HEK-293T cells expressing JEV proteins were infected with ZIKV (MOI of 5) at 24 h post-transfection, and ZIKV RNA was quantitated by qRT-PCR. Only cells transfected JEV NS2B had significantly reduced levels of ZIKV RNA compared to the empty vector control (P < 0.001). (C) ZIKV RNA replication level and extracellular ZIKV titer in different amounts of JEV NS2B expressing cells. HEK-293T cells transfected with 1, 2, and 3 µg JEV NS2B expression plasmid were then infected with ZIKV (MOI of 5) at 24 h post-transfection for 30 h. Western blot shows that expression of NS2B increased with the increase of plasmid transfected. The level of ZIKV RNA and extracellular ZIKV titer decreased with the increase of NS2B expression. (D) ZIKV protein expression in JEV NS2B expressing cells. The JEV NS2B (red) and ZIKV NS2B-NS3 protease (green) were detected by IFA. Nuclei were stained with DAPI (blue). Exclusion of ZIKV increased with increased input of JEV NS2B. ZIKV protein could not be detected in JEV NS2B-positive cells.
The superinfection exclusion dominant region in JEV NS2B
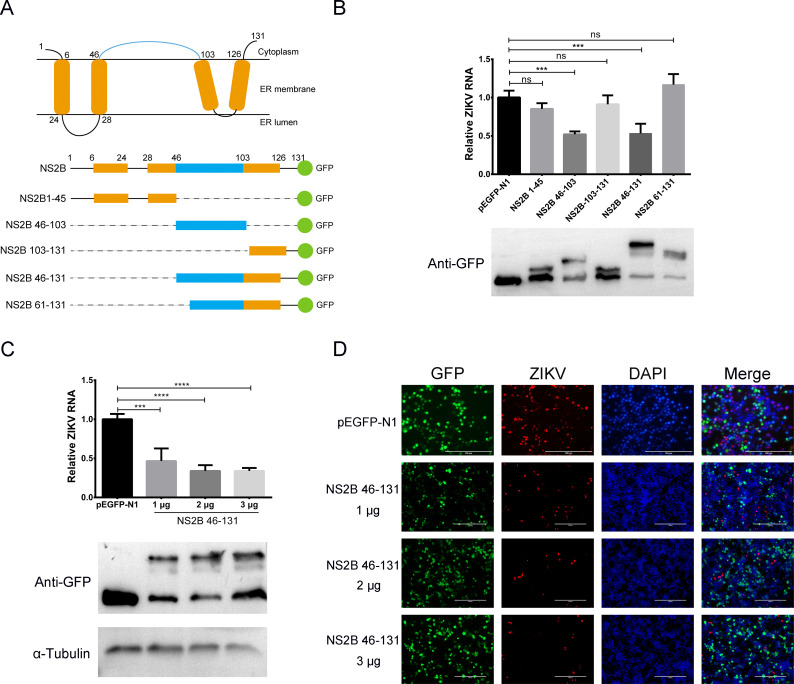
To determine the superinfection exclusion dominant region in JEV NS2B protein, truncation mutants of NS2B were constructed, and the exclusion of ZIKV was evaluated by qRT-PCR and IFA. JEV NS2B is a transmembrane protein with four intramembrane regions and one large outer membrane region (46–103 aa; Fig. 7A). Based on the structure of NS2B, five truncated mutants were constructed and transfected into cells (Fig. 7A). Cells were inoculated with ZIKV (MOI of 5) 24 h post-transfection. RNA replication and protein expression were determined by qRT-PCR and Western blot, respectively. Western blot showed that all truncated mutants were expressed in the transfected cells (Fig. 7B, lower). ZIKV RNA replication in NS2B 46–103 aa and NS2B 46–131 aa expressing cells was significantly reduced compared with the control cells (P < 0.01), while the viral RNA replication did not change significantly in other groups (P > 0.05; Fig. 7B, upper). 1, 2, and 3 µg of NS2B 46–131 aa were transfected into cells and then infected with ZIKV. The inhibition of viral RNA replication and protein expression exhibited a dose-dependent effect (Fig. 7C and D). These results indicated that the outer membrane area (46–103 aa) of JEV NS2B was the dominant region in the superinfection exclusion of ZIKV.
Fig 7.
Superinfection exclusion dominant region of JEV NS2B. (A) Schematic of JEV NS2B structure and diagram of the truncated NS2B constructs. Orange represents the transmembrane areas, and blue shows the outer-membrane region of NS2B. Green dots are green fluorescent protein (GFP) proteins. Dash lines indicate the deleted areas in NS2B. The names of constructs are listed on the left. (B) Inhibition of ZIKV RNA replication by NS2B truncated mutants. Different NS2B truncated mutants were transfected into HEK-293T cells, and ZIKV (MOI of 5) was infected at 24 h post-transfection for 30 h. ZIKV RNA was quantitated by qRT-PCR. Western blot shows that all the NS2B truncated mutants are expressed. The ZIKV RNA levels in NS2B 46–103 and NS2B 46–131 expressing cells were significantly lower than in control cells (P < 0.001). (C) The effect of NS2B 46–131 expression on ZIKV RNA replication. Different amounts of NS2B 46–131 expression plasmids were transfected into HEK-293T cells. ZIKV (MOI of 5) was infected at 24 h post-transfection for 30 h. ZIKV RNA was quantitated by qRT-PCR. Western blot shows that the NS2B 46–131 expression increased with the increase of transfected plasmid. The replication of ZIKV RNA decreased with increased NS2B 46–131 expression. (D) Effect of NS2B 46–131 on ZIKV protein expression. HEK-293T cells were transfected with increasing amounts (1, 2, and 3 µg) of plasmid encoding JEV NS2B 46–131 for 24 h and then infected with ZIKV for another 30 h. The expression of JEV NS2B 46–131 (green, GFP autofluorescence) and ZIKV NS2B-NS3 protease protein (red) were analyzed by IFA. Nuclei were stained with DAPI (blue).
Exclusion of other viruses by JEV
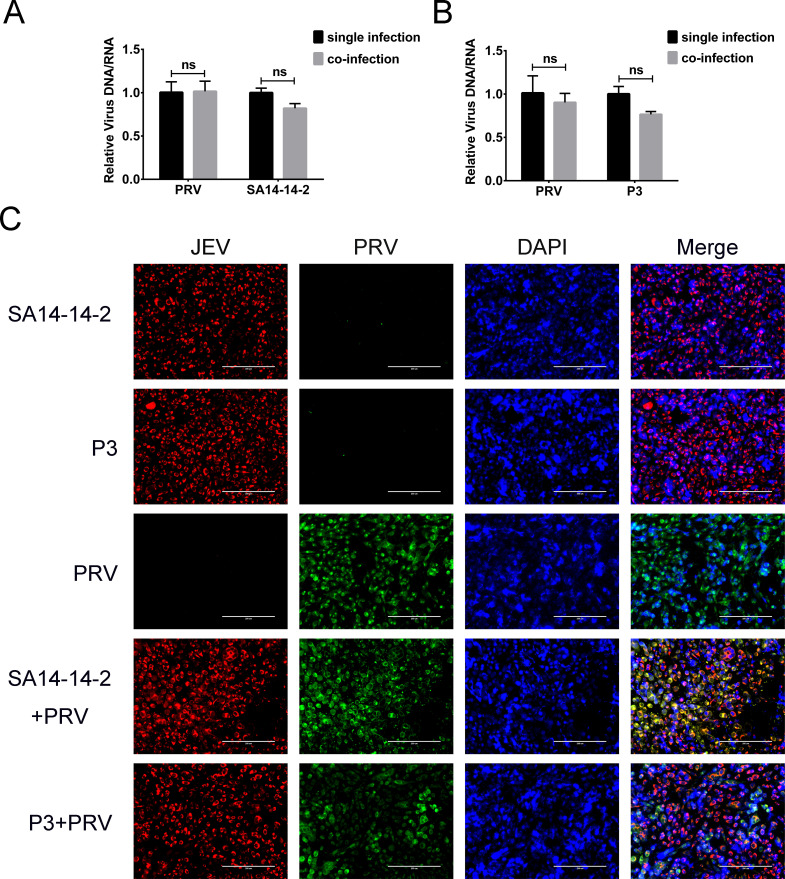
Vero cells were co-infected with JEV and pseudorabies virus (PRV, a DNA virus) or VSV (a negative-strand RNA virus). The proliferation of these viruses was determined by qPCR (or qRT-PCR) and IFA. The levels of both PRV and JEV genomes had no significant difference between single- and co-infected cells (Fig. 8A and B). IFA also showed that there was no reduction in the number of PRV-positive cells in the co-infected cells (Fig. 8C). In addition, the PRV and JEV proteins were co-expressed in many cells. These results indicated that there was no superinfection exclusion between JEV and PRV.
Fig 8.
Co-infection of JEV and PRV in cells. (A and B) Relative levels of viral genomes in single- or co-infected Vero cells at 30 h post infection. Both the JEV and PRV genomes showed no significant change in co-infected cells compared with single-infected cells. (C) IFA of viral proteins of PRV and JEV at 30 h post infection. The nuclei were stained with DAPI (blue). PRV (green) and JEV (red) specific proteins were detected in most cells, and the proteins of the two viruses could be expressed in the same cells in merged panels. ns, non-significant (P > 0.05).
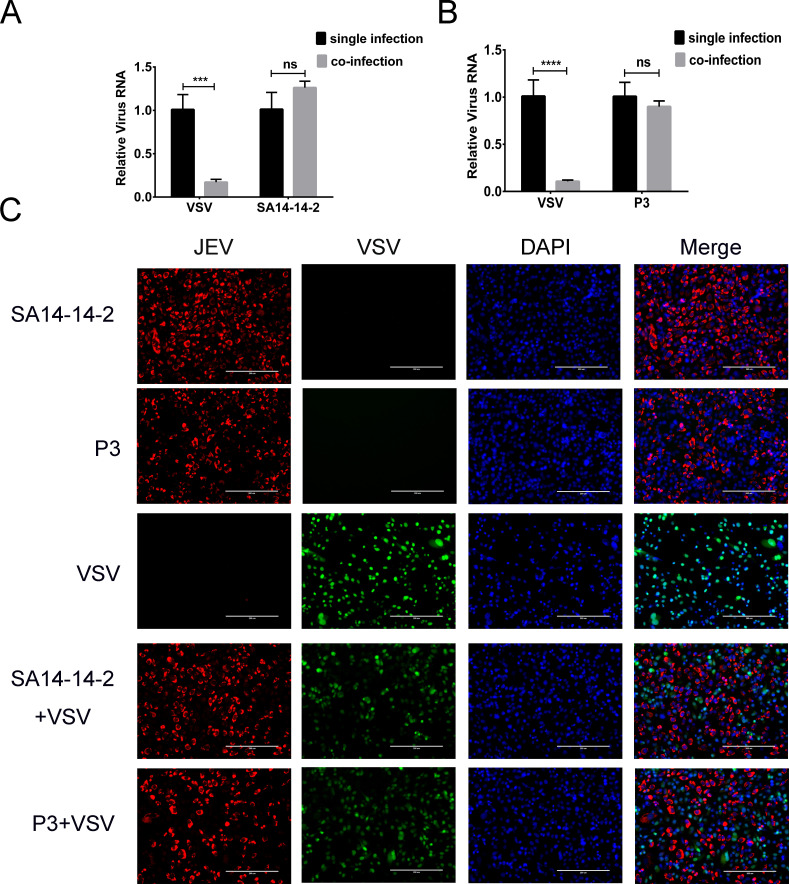
In JEV and VSV co-infected cells, VSV RNA replication was significantly inhibited by both JEV SA14-14-2 and P3 strains (P < 0.001; Fig. 9A and B). Protein expression of both viruses was detected by IFA. VSV-positive cells were decreased after JEV co-infection, and there were no JEV and VSV co-infected cells (Fig. 9C). These results suggested that the JEV and VSV could not proliferate simultaneously in the same cell.
Fig 9.
Co-infection of JEV and VSV in cells. (A and B) Relative levels of viral genomes in single- or co-infected cells. VSV RNA levels were extremely significantly reduced in co-infected cells (P < 0.001). (C) Detection of viral proteins by IFA. The nuclei were stained with DAPI (blue). VSV protein (green) expression was inhibited by the JEV (red). Proteins from two viruses were not detected in the same cells in the merged panel. ns, non-significant, P > 0.05; ***, extremely significant, P < 0.001; ****, extremely significant, P < 0.0001.
Expression of JEV NS2B excludes VSV infection
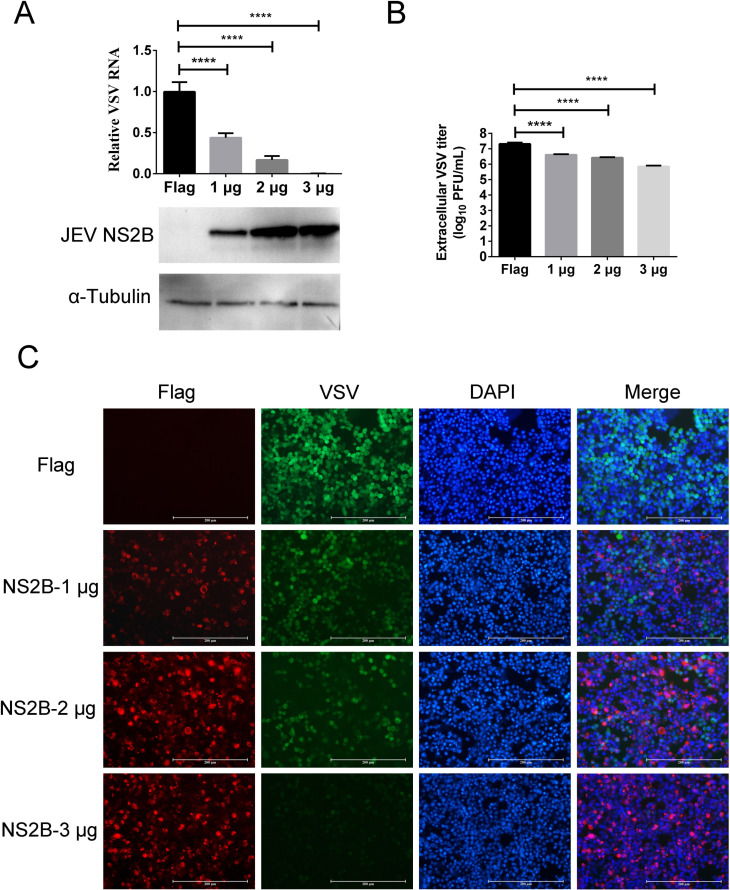
To explore the effect of JEV NS2B on VSV superinfection exclusion, 1, 2, and 3 µg of the NS2B expression plasmid were transfected into Vero cells. The expression of NS2B increased with the increase of plasmid transfected, while levels of VSV RNA decreased significantly with the increase of NS2B expression (P < 0.0001; Fig. 10A). VSV titers in culture supernatants showed a dose-dependent decrease as the expression of NS2B expression increased (P < 0.0001; Fig. 10B). IFA results showed that the VSV protein expression was also inhibited in NS2B expressing cells. There were no VSV and NS2B double-positive cells (Fig. 10C). These results suggested that the JEV NS2B protein also had interference effects on VSV proliferation.
Fig 10.
Expression of JEV NS2B on VSV replication, protein expression, and virus packaging. (A) VSV RNA replication in JEV NS2B expressing HEK-293T cells. Levels of JEV NS2B were assessed by Western blot; protein loading was normalized to α-tubulin. The replication of VSV RNA was inhibited by the expression of JEV NS2B. (B) VSV titer in culture supernatants. The mature virus particles were decreased in NS2B-transfected cells. (C) The effect of JEV NS2B on VSV protein expression. The VSV protein expression was interfered with by the expression of JEV NS2B. No VSV protein and NS2B double-positive cells were found in the merged panel. ****, extremely significant, P < 0.0001.
DISCUSSION
Superinfection exclusion has been reported in a broad range of viruses and is considered a selective advantage for virus evolution. This phenomenon is especially common in the members of Flaviviridae. NHUV inhibits the production of WNV, SLEV, JEV, DENV-2, and ZIKV in cells with a concurrent or prior infection (19, 20). The DENV-2 and yellow fever virus interfere with the replication of each other, and this interference depends on the infection sequence, i.e., the primary-infecting virus will inhibit the replication of the secondary-infecting virus (30). In this study, we report for the first time that JEV excludes superinfection of ZIKV, but ZIKV does not exclude superinfection of JEV, even when cells are infected by ZIKV before JEV.
Several mechanisms for the modulation of superinfection exclusion have been suggested, for example, competition for limited cellular resources, activation of specific cellular defenses that produce interferon or interferon-like substances, activation of the cellular RNAi defense system, receptor interference by viral proteins, or the production of a specific viral protein (31, 32). Superinfection exclusion can occur at any step in the viral life cycle, such as entry, replication, translation, or virus budding. WNV replicon can inhibit subsequent WNV infection. This interference is related to two amino acid sites of NS4A (K124 and 2KV9), indicating that the exclusion of WNV superinfection occurs during the process of virus replication (28). HCV also establishes superinfection exclusion to other HCV genotypes at the stage of replication (11). In alpha herpesviruses, the exclusion between PRV and herpes simplex virus 1 (HSV-1) depends on the manner of infection, and the mechanisms are different in these two viruses. PRV infection inhibits viral entry or fusion, while HSV-1 inhibits infection through a post-entry mechanism (33, 34).
Previous studies have shown that some viral proteins were responsible for the superinfection exclusion. For example, the citrus tristeza virus excludes the same or a closely related virus through its p33 protein (35). The coat protein and NIa-Pro encoded by wheat streak mosaic virus and Triticum mosaic virus exert superinfection exclusion to the cognate viruses (36). Interestingly, alphaviruses exhibit different molecular mechanisms in superinfection exclusion. SFV non-structural protein 1 (nsp1) expression interferes with SFV genomic RNA synthesis (37). While another two alphaviruses, Chikungunya virus and SINV, exclude homologous or heterologous alphaviruses through its nsp2 protein (38). These results suggest that the mechanism of superinfection exclusion may be different even in the same genus viruses. The mechanism of JEV excluding ZIKV is also investigated in the current study. The results show that JEV NS2B protein can inhibit the replication of ZIKV, and the out-membrane region of NS2B is responsible for this exclusion.
Flavivirus NS2B is a transmembrane protein, interacting with NS3 and recruiting it to the endoplasmic reticulum (39). As a cofactor, the main function of JEV NS2B protein is to bind NS3 protease to activate the cleaving activity (40). Our results show that the expression of JEV NS3 protein does not affect ZIKV replication (Fig. 6B). Meanwhile, the expression of NS2B protein also inhibits the replication of VSV, a negative-sense RNA virus that does not encode a viral protease (Fig. 10). These results imply that the superinfection exclusion caused by JEV NS2B may be independent of its role in protease activation. Apart from serving as a cofactor for NS3 protease, JEV NS2B protein has been found to affect viral RNA replication and virion formation (41). Moreover, NS2B is one of the components of the flavivirus replication complex, and the outer membrane hydrophilic region of NS2B (46–103 aa) is oriented to the cytoplasmic viral replication complex (42). We hypothesize that the JEV NS2B protein, especially its extracellular region, inhibits the replication of ZIKV by competing for cellular resources. We also compare the amino acid sequence of the different NS2B proteins. With three amino acid differences, JEV SA14-14-2 and P3 strains have high similarity in their sequences of NS2B. However, the NS2B of ZIKV shows low homology to JEV (46.46% with SA14-14-2 and 47.62% with P3; Fig. S6). The difference between JEV and ZIKV NS2B protein may cause different affinity to the essential cellular components. However, more work is needed to clarify the exclusion mechanism of NS2B protein.
In this study, we report a superinfection exclusion caused by JEV NS2B protein. The antiviral effect of JEV NS2B protein has potential value as a new antiviral strategy.
MATERIALS AND METHODS
Cells and viruses
Baby hamster kidney (BHK-21) cells, African green monkey kidney (Vero) cells, and HEK-293T cells were cultured in DMEM (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; GIBCO, Grand Island, NY, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin (Beyotime Biotechnology, Shanghai, China) in 5% CO2 at 37°C. Aedes albopictus mosquito cell line C6/36 was propagated in Roswell Park Memorial Institute 1640 medium (GIBCO), supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin and maintained at 28°C with no CO2.
JEV (strain SA14-14-2, accession number MK585066.1, and strain P3, accession number U47032.1) and ZIKV (strain MR766, accession number MK105975.1) were propagated on Ae. albopictus (C6/36) cells and titrated by plaque assay on BHK-21 cells and Vero cells, respectively.
Virus titration
BHK-21 or Vero cells were infected with SA14-14-2 (MOI of 5), P3 (MOI of 1), and MR766 (MOI of 5) virus. At 30 h post-infection, culture supernatants were harvested and serially 10-fold diluted with DMEM. The viral dilutions were inoculated onto BHK-21 or Vero cells in 24-well plates. After incubating for 1.5 h at 37°C, the cells were washed with phosphate-buffered saline (PBS) and overlaid with 0.8% carboxymethyl cellulose in DMEM (Sigma, St. Louis, MO, USA) containing 2% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin. Plates were incubated at 37°C for 4 days, and infectious centers were then detected by immunostaining as previously described (28, 43). Briefly, cell monolayers were fixed in 100% methanol for 10 min and blocked with 1% bovine serum albumin (BSA) in PBS (pH 7.2) for 2 h at room temperature. Monoclonal antibodies to JEV NS1 (1:500) (44) or ZIKV NS5 (1:500) (45) were added to each well followed by a 1 h incubation at room temperature. After removing the primary antibody and washing three times with PBS, cells were incubated with peroxidase-labeled goat-anti-mouse IgG (1:300; Boster, Wuhan, China) for 1 h at room temperature. After washing with PBS, SuperSignal West Pico PLUS reagent (Thermo Fisher, MA, USA) was added to each well. Infectious centers were visualized with a Tanon-5100 chemiluminescence system (Tanon, Shanghai, China).
Western blot
Cells were lysed in RIPA buffer (Beyotime Biotechnology, Shanghai, China) for 30 min at 4°C. Protein concentrations of the whole cell lysates were determined by using a BCA protein quantification kit (Beyotime Biotechnology). For each sample, the same amount of protein was separated by 12.5% SDS-PAGE and electroblotted onto polyvinylidene fluoride (PVDF) membranes. Blots were incubated with relevant primary antibodies at room temperature for 2 h, washed three times, and then incubated with horseradish peroxidase-labeled goat anti-mouse or goat anti-rabbit IgG at room temperature for 45 min. Bands were visualized with a chemiluminescence system.
Immunofluorescence assay
BHK-21 or Vero cells were seeded into wells of 12-well plates, allowed to attach for 12 h, and then infected with the virus. At 30 h post-infection, the supernatants were removed, and cells were fixed with methanol for 10 min at room temperature, then blocked with 1% BSA in PBS for 2 h. Cells were then incubated with primary antibodies [anti-JEV NS1 (1:500) monoclonal antibody (44) and anti-ZIKV NS2B-NS3 protease (1:100) polyclonal antibody (prepared by our laboratory)] for 1 h at room temperature. Cells were washed three times with PBS and then incubated with an Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 549 goat anti-mouse IgG for 1 h. Cell nuclei were stained with DAPI, and cells were observed and imaged using a fluorescence microscope (Zeiss, Germany).
Quantitative RT-PCR
Total cellular RNA was extracted using TRIzol reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. The RNA was transcribed to cDNA using a ReverTra Ace qPCR RT kit (Toyobo, Japan). The level of cDNA was determined by quantitative PCR using SYBR green real-time PCR master mix (Toyobo). The results were normalized to the expression of β-actin in each sample. Primer pairs used were as follows: JEV NS4A gene (forward primer: 5′-AAGCACACCGAATGGCTCTCGAAGAG-3′; reverse primer: 5′-AGAAGGTAGCTAGTGTGAGCACTAGAGC-3′), ZIKV prM gene (forward primer: 5′-CGACATCAACTTGGGTTGTGGCCGG-3′; reverse primer: 5′-GCGACCGCGTTTGCAACTTCCTTGTAG-3′), VSV gene (forward primer: 5’- ATGATACAGTACAATTATTTTGGGACA-3′; reverse primer: 5’- CAAAGACATGCCCGACACT-3′), and PRV gC gene (forward primer: 5’- GTGCTGTTGGTCACGAAGG-3′; reverse primer: 5’- AACGTCTCGCTCCTCCTGTA-3′).
Effect of JEV protein expression on ZIKV replication
Each of the 10 JEV protein-encoding genes was amplified by RT-PCR and then cloned into pCMV-3×FLAG expression plasmids. HEK-293T cells were seeded in 12-well plates and incubated for 12 h. Cells were transfected with plasmids using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s protocol. At 24 h post-transfection, cells were infected with ZIKV MR766 and incubated for another 30 h. The level of ZIKV RNA was determined by qRT-PCR, and the expression of viral proteins was detected by Western blot.
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China (2022YFD1800100 and 2021YFD1800303) and the National Natural Science Foundation of China (31972658).
Contributor Information
Yunfeng Song, Email: syf@mail.hzau.edu.cn.
Mark T. Heise, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the manuscript and its supplementary materials.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01859-23.
JEV and ZIKV dose trial on BHK-21 cells.
JEV and ZIKV dose trial on Vero cells.
ZIKV proliferation is decreased by co-infection of JEV on Vero cells.
Superinfection exclusion of JEV and ZIKV in C6/36 cells.
The expression of JEV NS1 or NS4B protein did not affect the superinfected ZIKV infection.
Homology analysis of NS2B protein among ZIKV MR766 strain, JEV P3 strain, and JEV SA14-14-2 strain.
Legends for Fig. S1 to S6.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. 2009. Zika virus outbreak on yap island, federated states of micronesia.. N Engl J Med 360:2536–2543. doi: 10.1056/NEJMoa0805715 [DOI] [PubMed] [Google Scholar]
- 2. Cao-Lormeau VM, Roche C, Teissier A, Robin E, Berry AL, Mallet HP, Sall AA, Musso D. 2014. Zika virus, French polynesia, South Pacific, 2013. Emerg Infect Dis 20:1085–1086. doi: 10.3201/eid2006.140138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zanluca C, Melo VCA de, Mosimann ALP, Santos GIVD, Santos CNDD, Luz K. 2015. First report of autochthonous transmission of Zika virus in Brazil. Mem Inst Oswaldo Cruz 110:569–572. doi: 10.1590/0074-02760150192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fauci AS, Morens DM. 2016. Zika virus in the Americas--yet another arbovirus threat. N Engl J Med 374:601–604. doi: 10.1056/NEJMp1600297 [DOI] [PubMed] [Google Scholar]
- 5. Musso D, Gubler DJ. 2016. Zika virus. Clin Microbiol Rev 29:487–524. doi: 10.1128/CMR.00072-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Plourde AR, Bloch EM. 2016. A literature review of Zika virus. Emerg Infect Dis 22:1185–1192. doi: 10.3201/eid2207.151990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poland GA, Ovsyannikova IG, Kennedy RB. 2019. Zika vaccine development: current status. Mayo Clin Proc 94:2572–2586. doi: 10.1016/j.mayocp.2019.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laureti M, Paradkar PN, Fazakerley JK, Rodriguez-Andres J. 2020. Superinfection exclusion in mosquitoes and its potential as an arbovirus control strategy. Viruses 12:1259. doi: 10.3390/v12111259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laliberte JP, Moss B. 2014. A novel mode of poxvirus superinfection exclusion that prevents fusion of the lipid bilayers of viral and cellular membranes. J Virol 88:9751–9768. doi: 10.1128/JVI.00816-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Webster B, Ott M, Greene WC. 2013. Evasion of superinfection exclusion and elimination of primary viral RNA by an adapted strain of hepatitis c virus. J Virol 87:13354–13369. doi: 10.1128/JVI.02465-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tscherne DM, Evans MJ, von Hahn T, Jones CT, Stamataki Z, McKeating JA, Lindenbach BD, Rice CM. 2007. Superinfection exclusion in cells infected with hepatitis c virus. J Virol 81:3693–3703. doi: 10.1128/JVI.01748-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang IC, Li W, Sui J, Marasco W, Choe H, Farzan M. 2008. Influenza a virus neuraminidase limits viral superinfection. J Virol 82:4834–4843. doi: 10.1128/JVI.00079-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karpf AR, Lenches E, Strauss EG, Strauss JH, Brown DT. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with sindbis virus. J Virol 71:7119–7123. doi: 10.1128/JVI.71.9.7119-7123.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whitaker-Dowling P, Youngner JS, Widnell CC, Wilcox DK. 1983. Superinfection exclusion by vesicular stomatitis virus. Virology 131:137–143. doi: 10.1016/0042-6822(83)90540-8 [DOI] [PubMed] [Google Scholar]
- 15. Biryukov J, Meyers C. 2018. Superinfection exclusion between two high-risk human papillomavirus types during a coinfection. J Virol 92:e01993-17. doi: 10.1128/JVI.01993-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blitvich BJ, Firth AE. 2015. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 7:1927–1959. doi: 10.3390/v7041927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Torres FJ, Parry R, Hugo LE, Slonchak A, Newton ND, Vet LJ, Modhiran N, Pullinger B, Wang X, Potter J, Winterford C, Hobson-Peters J, Hall RA, Khromykh AA. 2022. Reporter flaviviruses as tools to demonstrate homologous and heterologous superinfection exclusion. Viruses 14:1501. doi: 10.3390/v14071501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goenaga S, Kenney JL, Duggal NK, Delorey M, Ebel GD, Zhang B, Levis SC, Enria DA, Brault AC. 2015. Potential for co-infection of a mosquito-specific flavivirus, nhumirim virus, to block west nile virus transmission in mosquitoes. Viruses 7:5801–5812. doi: 10.3390/v7112911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romo H, Kenney JL, Blitvich BJ, Brault AC. 2018. Restriction of Zika virus infection and transmission in aedes aegypti mediated by an insect-specific flavivirus. Emerg Microbes Infect 7:181. doi: 10.1038/s41426-018-0180-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kenney JL, Solberg OD, Langevin SA, Brault AC. 2014. Characterization of a novel insect-specific flavivirus from Brazil: potential for inhibition of infection of arthropod cells with medically important flaviviruses. J Gen Virol 95:2796–2808. doi: 10.1099/vir.0.068031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hobson-Peters J, Yam AWY, Lu JWF, Setoh YX, May FJ, Kurucz N, Walsh S, Prow NA, Davis SS, Weir R, Melville L, Hunt N, Webb RI, Blitvich BJ, Whelan P, Hall RA. 2013. A new insect-specific flavivirus from northern Australia suppresses replication of west nile virus and murray valley encephalitis virus in co-infected mosquito cells. PLoS One 8:e56534. doi: 10.1371/journal.pone.0056534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hall-Mendelin S, McLean BJ, Bielefeldt-Ohmann H, Hobson-Peters J, Hall RA, van den Hurk AF. 2016. The insect-specific palm creek virus modulates west nile virus infection in and transmission by Australian mosquitoes. Parasit Vectors 9:414. doi: 10.1186/s13071-016-1683-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang XF, Zhang S, Guo Q, Sun R, Wei T, Qu F. 2018. A new mechanistic model for viral cross protection and superinfection exclusion. Front Plant Sci 9:40. doi: 10.3389/fpls.2018.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perdoncini Carvalho C, Ren R, Han J, Qu F. 2022. Natural selection, intracellular bottlenecks of virus populations, and viral superinfection exclusion. Annu Rev Virol 9:121–137. doi: 10.1146/annurev-virology-100520-114758 [DOI] [PubMed] [Google Scholar]
- 25. Wagenaar TR, Moss B. 2009. Expression of the a56 and k2 proteins is sufficient to inhibit vaccinia virus entry and cell fusion. J Virol 83:1546–1554. doi: 10.1128/JVI.01684-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wagenaar TR, Ojeda S, Moss B. 2008. Vaccinia virus a56/k2 fusion regulatory protein interacts with the a16 and g9 subunits of the entry fusion complex. J Virol 82:5153–5160. doi: 10.1128/JVI.00162-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh IR, Suomalainen M, Varadarajan S, Garoff H, Helenius A. 1997. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting semliki forest virus. Virology 231:59–71. doi: 10.1006/viro.1997.8492 [DOI] [PubMed] [Google Scholar]
- 28. Zou G, Zhang B, Lim PY, Yuan Z, Bernard KA, Shi PY. 2009. Exclusion of west nile virus superinfection through RNA replication. J Virol 83:11765–11776. doi: 10.1128/JVI.01205-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Campbell CL, Smith DR, Sanchez-Vargas I, Zhang B, Shi PY, Ebel GD. 2014. A positively selected mutation in the wnv 2k peptide confers resistance to superinfection exclusion in vivo. Virology 464–465:228–232. doi: 10.1016/j.virol.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abrao EP, da Fonseca BAL. 2016. Infection of mosquito cells (c6/36) by dengue-2 virus interferes with subsequent infection by yellow fever virus. Vector Borne Zoonotic Dis 16:124–130. doi: 10.1089/vbz.2015.1804 [DOI] [PubMed] [Google Scholar]
- 31. Pepin KM, Lambeth K, Hanley KA. 2008. Asymmetric competitive suppression between strains of dengue virus. BMC Microbiol 8:28. doi: 10.1186/1471-2180-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang XF, Sun R, Guo Q, Zhang S, Meulia T, Halfmann R, Li D, Qu F. 2017. A self-perpetuating repressive state of a viral replication protein blocks superinfection by the same virus. PLoS Pathog 13:e1006253. doi: 10.1371/journal.ppat.1006253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cwick JP, Owen JE, Kochetkova I, Hain KS, Horssen NV, Taylor MP. 2022. Superinfection exclusion of alphaherpesviruses interferes with virion trafficking. Microbiol Spectr 10:e0068422. doi: 10.1128/spectrum.00684-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Criddle A, Thornburg T, Kochetkova I, DePartee M, Taylor MP. 2016. Gd-independent superinfection exclusion of alphaherpesviruses. J Virol 90:4049–4058. doi: 10.1128/JVI.00089-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Folimonova SY. 2012. Superinfection exclusion is an active virus-controlled function that requires a specific viral protein. J Virol 86:5554–5561. doi: 10.1128/JVI.00310-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tatineni S, French R. 2016. The coat protein and nia protease of two potyviridae family members independently confer superinfection exclusion. J Virol 90:10886–10905. doi: 10.1128/JVI.01697-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kiiver K, Tagen I, Žusinaite E, Tamberg N, Fazakerley JK, Merits A. 2008. Properties of non-structural protein 1 of semliki forest virus and its interference with virus replication. J Gen Virol 89:1457–1466. doi: 10.1099/vir.0.2008/000299-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cherkashchenko L, Rausalu K, Basu S, Alphey L, Merits A. 2022. Expression of alphavirus nonstructural protein 2 (nsp2) in mosquito cells inhibits viral RNA replication in both a protease activity-dependent and -independent manner. Viruses 14:1327. doi: 10.3390/v14061327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xing H, Xu S, Jia F, Yang Y, Xu C, Qin C, Shi L. 2020. Zika ns2b is a crucial factor recruiting ns3 to the er and activating its protease activity. Virus Res 275:197793. doi: 10.1016/j.virusres.2019.197793 [DOI] [PubMed] [Google Scholar]
- 40. Falgout B, Pethel M, Zhang YM, Lai CJ. 1991. Both nonstructural proteins ns2b and ns3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol 65:2467–2475. doi: 10.1128/JVI.65.5.2467-2475.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li XD, Deng CL, Ye HQ, Zhang HL, Zhang QY, Chen DD, Zhang PT, Shi PY, Yuan ZM, Zhang B. 2016. Transmembrane domains of ns2b contribute to both viral RNA replication and particle formation in Japanese encephalitis virus. J Virol 90:5735–5749. doi: 10.1128/JVI.00340-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Apte-Sengupta S, Sirohi D, Kuhn RJ. 2014. Coupling of replication and assembly in flaviviruses. Curr Opin Virol 9:134–142. doi: 10.1016/j.coviro.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Durbin AP, Karron RA, Sun W, Vaughn DW, Reynolds MJ, Perreault JR, Thumar B, Men R, Lai CJ, Elkins WR, Chanock RM, Murphy BR, Whitehead SS. 2001. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3'-untranslated region. Am J Trop Med Hyg 65:405–413. doi: 10.4269/ajtmh.2001.65.405 [DOI] [PubMed] [Google Scholar]
- 44. Zhou D, Jia F, Li Q, Zhang L, Chen Z, Zhao Z, Cui M, Song Y, Chen H, Cao S, Ye J. 2018. Japanese encephalitis virus ns1' protein antagonizes interferon beta production. Virol Sin 33:515–523. doi: 10.1007/s12250-018-0067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao Z, Tao M, Han W, Fan Z, Imran M, Cao S, Ye J. 2021. Nuclear localization of Zika virus ns5 contributes to suppression of type I interferon production and response. J Gen Virol 102:001376. doi: 10.1099/jgv.0.001376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
JEV and ZIKV dose trial on BHK-21 cells.
JEV and ZIKV dose trial on Vero cells.
ZIKV proliferation is decreased by co-infection of JEV on Vero cells.
Superinfection exclusion of JEV and ZIKV in C6/36 cells.
The expression of JEV NS1 or NS4B protein did not affect the superinfected ZIKV infection.
Homology analysis of NS2B protein among ZIKV MR766 strain, JEV P3 strain, and JEV SA14-14-2 strain.
Legends for Fig. S1 to S6.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the manuscript and its supplementary materials.