Abstract
Cleavage and DNA joining reactions, carried out by human immunodeficiency virus type 1 (HIV-1) integrase, are necessary to effect the covalent insertion of HIV-1 DNA into the host genome. For the integration of HIV-1 DNA into the cellular genome to be completed, short gaps flanking the integrated proviral DNA must be repaired. It has been widely assumed that host cell DNA repair enzymes are involved. Here we report that HIV-1 integrase multimers possess an intrinsic DNA-dependent DNA polymerase activity. The activity was characterized by its dependence on Mg2+, resistance to N-ethylmaleimide, and inhibition by 3′-azido-2′,3′-dideoxythymidine-5′-triphosphate, coumermycin A1, and pyridoxal 5′-phosphate. The enzyme efficiently utilized poly(dA)-oligo(dT) or self-annealing oligonucleotides as a template primer but displayed relatively low activity with gapped calf thymus DNA and no activity with poly(dA) or poly(rA)-oligo(dT). A monoclonal antibody binding specifically to an epitope comprised of amino acids 264 to 273 near the C terminus of HIV-1 integrase severely inhibited the DNA polymerase activity. A deletion of 50 amino acids at the C terminus of integrase drastically altered the gel filtration properties of the DNA polymerase, although the level of activity was unaffected by this mutation. The DNA polymerase efficiently extended a hairpin DNA primer up to 19 nucleotides on a T20 DNA template, although addition of the last nucleotide occurred infrequently or not at all. The ability of integrase to repair gaps in DNA was also investigated. We designed a series of gapped molecules containing a single-stranded region flanked by a duplex U5 viral arm on one side and by a duplex nonviral arm on the other side. Molecules varied structurally depending on the size of the gap (one, two, five, or seven nucleotides), their content of T’s or C’s in the single-stranded region, whether the CA dinucleotide in the viral arm had been replaced with a nonviral sequence, or whether they contained 5′ AC dinucleotides as unpaired tails. The results indicated that the integrase DNA polymerase is specifically designed to repair gaps efficiently and completely, regardless of gap size, base composition, or structural features such as the internal CA dinucleotide or unpaired 5′-terminal AC dinucleotides. When the U5 arm of the gapped DNA substrate was removed, leaving a nongapped DNA template-primer, the integrase DNA polymerase failed to repair the last nucleotide in the DNA template effectively. A post-gap repair reaction did depend on the CA dinucleotide. This secondary reaction was highly regulated. Only two nucleotides beyond the gap were synthesized, and these were complementary to and dependent for their synthesis on the CA dinucleotide. We were also able to identify a specific requirement for the C terminus of integrase in the post-gap repair reaction. The results are consistent with a direct role for a heretofore unsuspected DNA polymerase function of HIV-1 integrase in the repair of short gaps flanking proviral DNA integration intermediates that arise during virus infection.
Integration of human immunodeficiency virus type 1 (HIV-1) DNA is an essential step in the replicative cycle of the virus (6, 13, 16, 29, 41). The initial steps whereby HIV-1 DNA becomes covalently associated with the host DNA are mediated by the viral integrase protein. Two distinct chemical reactions are involved. In a processing step, integrase cleaves viral DNA endonucleolytically, resulting in the removal of a GT dinucleotide from the 3′ ends of the DNA (15, 48, 51). Once in the nucleus, concerted cleavage and DNA strand transfer reactions, involving viral and host DNA, enable the processed 3′ termini to become covalently joined to a host DNA target site. The intermediate produced in this manner contains unpaired 5′ ends adjacent to five-base gaps. Completion of integration requires the repair of these gaps and the joining of the 5′ ends of viral DNA to the host DNA (2). The relatively rapid kinetics of 5′-end joining in vivo has been used as a basis on which to argue in favor of a role for integrase in this step of integration (40). Although integrase can catalyze the latter reaction in vitro, albeit inefficiently (28), it has been generally assumed that host cell enzymes perform gap repair and 5′-end joining.
Structural, functional, and mutational studies have defined integrase as a 32-kDa protein that can be divided into three distinct functional domains (50). The catalytic core, including amino acids 50 to 212, contains a triad of acidic amino acids (Asp 64, Asp 116, and Glu 152) that form a highly conserved D,D-35-E motif. In the three-dimensional crystal structure, these amino acids are in close proximity (10). Mutation of any one of these acidic residues severely hampers the ability of integrase to catalyze endonucleolytic cleavage and DNA strand transfer (5, 9, 12, 13, 27, 31, 32). The C terminus binds DNA nonspecifically and is required for cleavage and integration activity (47, 49, 52, 53). The amino terminus contains a zinc finger or HHCC motif, which coordinates a molar equivalent of zinc (4). This domain influences DNA binding (21, 25, 47), although it does not bind DNA on its own (26, 38).
In the functional integration complex, integrase is believed to act as a multimer (11, 24, 46). Transcomplementation, in which DNA strand transfer and cleavage activities are restored by mixing nonfunctional mutants, implies that the active form of integrase is minimally a dimer (46). Integrase can exist in equilibrium between dimeric and tetrameric forms, and multimerization determinants can be identified within the integrase protein (1). Association of one molar equivalent of zinc with a soluble mutant of integrase favored the formation of the tetrameric form of the protein (54).
The present study was undertaken to further characterize HIV-1 integrase by searching for novel enzymatic activities that may be associated with this viral protein. We chose specifically to look for an associated DNA polymerase activity in an attempt to elucidate the final steps in integration, namely, gap repair and 5′-end joining.
MATERIALS AND METHODS
Materials.
Calf thymus DNA was from Worthington. Poly(dA)-oligo(dT), poly(rA)-oligo(dT), poly(dA), dATP, TTP, HindIII linker DNA 5′-pCCCAAGCTTGGG-3′ in duplex form, and Sephacryl S-300 were from Pharmacia-PL Biochemicals. Coumermycin A1, pyridoxal 5′-phosphate, N-ethylmaleimide (NEM), and protein gel filtration standards were from Sigma. CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate} and Escherichia coli DNA polymerase I (Klenow fragment) and the holoenzyme were from Boehringer Mannheim. 3′-Azido-2′,3′-dideoxythymidine-5′-triphosphate (AZT-TP) was from Moravek Chemicals Inc., Loma Linda, Calif. Nickel-nitrilotriacetate (NTA) Superflow resins and plasmid kits were obtained from Qiagen. Radioactive nucleotides [α-32P]dATP and [α-32P]TTP (specific activity, 3,000 Ci/mmol) were from New England Nuclear-Dupont, and [α-32P]ddATP and [α-32P]dGTP were from Amersham. Guanidine hydrochloride and urea were obtained from BioShop. Synthetic oligonucleotides were from Sheldon Biotechnologies Inc. or from Gibco/BRL Life Technologies. The oligonucleotides used were DISPOL 17 (5′T20ACTGCTAGAGATTTTAAAATCTCTAGCAGT3′), DISPOL 10 (5′T20TTTTTACTGCTAGAGATATCTCTAGCAGTAAAAA3′), DISPOL 16NV (5′T20AAGCCGGGGTACCCCGGCTT3′), G1 (5′ACTGCTAGAGATTTATCTCTAGCATTTTTGTAGCTGATCCGGTATACCGGATCAGCTAC3′), G2 (5′TGCTAGAGATTTATCTCTAGCATTTTTGTAGCTGATCCGGTATAC CGGATCAGCTAC3′), G1T1 (5′ACTGCTAGAGATTTAAATCTCTAGCATGTAGCTGATCCGGTATACCGGATCAGCTAC3′), G1T7 (5′ACTGCTAGAGATTTAAATCTCTAGCATTTTTTTGTAGCTGATCCGGTATACCGGAT CAGCTAC3′), G1T2 (5′ACTGCTAGAGATTTAAATCTCTAGCATTGTAGCTGATCCGGTATACCGGATCAGCTAC3′), G3C5 (5′ACTGCTAGAGATTTATCTCTAGCACCCCCGTAGCTGATCCGGTATACCGGATCAGCTAC 3′), G1H (5′ACCTCCGGAGATTTAAATCTCCGGAGTTTTTGTAGCTGATCCGGTATACCGGATCAGCTAC3′), T5H (5′TTTTTGTAGCTGATCCGGTATACCGGATCAGCTAC3′), 17-mer (5′CGGATCAGCTACAAAAA3′), 14-mer (5′ATCAGCTACAAAAA3′), 12-mer, (5′TAGCAGTAAAAA3′), 8-mer (5′TAGCAGTA3′), 36-mer (5′ATCAGCTACAAAAATGCTAGAGATTTATCTCTAGCA3′), and 48-mer (5′ATCAGCTACAAAAATGCTAGAGATTTATCTCTCTAGCATTTTTGTAGCTG3′). HEPES, IPTG (isopropylthio-β-d-galactoside), and DNA ligase were from Gibco/BRL Life Technologies. DNA-grade glass fiber filters (GF/B) were from Whatman. Microcon filtration units were from Amicon. Ecolume scintillation fluid was from ICN. Stop solution containing formamide, bromophenol blue, and xylene cyanol FF was from U.S. Biochemicals. Goat anti-rabbit and anti-mouse antibodies conjugated to alkaline phosphatase were obtained from Bio-Rad.
Purification of His-tagged HIV-1 integrase.
Integrase expression from plasmid pQE30 Δ IN or pProEX IN was induced by IPTG in E. coli M15pREP or JM109, respectively, and purified under denaturing conditions as described previously (18), with some modifications. pProEX IN was produced by cloning the small HindIII fragment of pCMV IN (17) at the HindIII site of pProEX. The amount of nickel-NTA resin used was 1 ml of a 50% slurry for extract obtained from 200 ml of E. coli. Renaturation from 8 M urea in 0.1 M phosphate buffer (pH 7.5) was performed by stepwise dialysis over several days in 10 volumes of 4 and 2 M deionized urea in 50 mM HEPES-HCl (pH 7.5)–1 M NaCl–1 mM dithiothreitol (DTT) followed by two overnight changes of final dialysis buffer (FDB; 50 mM HEPES-HCl [pH 7.5], 1 mM DTT, 1.0 M NaCl, 10% glycerol, 1 mM CHAPS, 0.1 mM EDTA). The renatured sample (5 ml) was concentrated to a final volume of 1 ml in a Microcon 30 filtration unit.
Sephacryl S-300 chromatography.
The integrase sample purified by adsorption on a nickel-NTA resin was applied to a 92-ml bed of Sephacryl S-300 in a volume of 1 ml. The column of S-300 (Bio-Rad Econocolumn; 1 by 120 cm) was equilibrated in FDB and developed at room temperature at a flow rate of 5 to 6 ml/h. Fractions (1 ml) were collected with an ISCO Retriever II fraction collector and assayed for DNA polymerase activity.
Sedimentation velocity analysis using a glycerol gradient.
Glycerol gradients were formed from 10 to 30% glycerol in FDB. Samples of the integrase DNA polymerase were diluted 1:1 with FDB lacking glycerol to a final volume of 180 μl prior to loading on the glycerol gradient. Sedimentation was performed in a Beckman L8-70 ultracentrifuge at 41,000 rpm for 66 h at 4°C, using a Beckman SW41 rotor.
DNA polymerase reactions.
DNA polymerase reaction mixtures (25 μl) contained 10 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 5 mM DTT, bovine serum albumin (200 μg/ml), poly(dA)-oligo(dT) (10 μg/ml) or DISPOL 17 DNA (0.25 μg/ml), and either 1 μM TTP with 4 μCi of [α-32P]TTP or 1 μM dATP with 4 μCi of [α-32P]dATP, respectively. Alternatively, reactions with T5H or gapped DNA substrate G1, G1H, G1T1, G1T2, G1T7, or G2 were performed with 1 μM dATP and 1 μCi of [α-32P]dATP. Reactions with G1 were also conducted with 1 μM ddATP and 4 μCi of [α-32P]ddATP. Reactions with G3C5 were conducted with 1 μM dATP and 1 μCi of [α-32P]dATP or with 1 μM dGTP and 1 μCi of [α-32P]dGTP. Purified integrase multimers or Ni2+-NTA-purified integrase was added in 1 ml of FDB to a final concentration of 0.5 nM (0.5 to 1.0 U of DNA polymerase activity) or 30 nM, respectively. Reaction mixtures were incubated at 37°C for 1 h. Reactions were terminated in 10% trichloroacetic acid containing 25 mM sodium pyrophosphate. Radiolabeled DNA precipitates were collected on glass fiber filters, and radioactivity was quantified in a Canberra Packard scintillation counter, using Ecolume. For gel analysis reactions were terminated by using formamide-dye mix and treated as described below prior to polyacrylamide gel electrophoresis (PAGE).
Construction and expression of a C-terminal deletion mutant of HIV-1 integrase.
The integrase plasmid pCMV-IN RRE (17) was subjected to partial digestion with AvaII followed by digestion with SmaI. The recessed ends were repaired by using E. coli DNA polymerase I (Klenow fragment). HindIII linker DNA was added, and the linearized plasmid was recircularized with DNA ligase. After transformation of E. coli JM109, plasmid pCMV IN Δ713, deleted for a C-terminal portion of the integrase gene, was identified by DNA sequencing. The C-terminal amino acid sequence of the truncated protein is predicted to be GPKLN. The first two amino acids correspond to residues 237 and 238 of wild-type integrase, whereas the last three amino acids in this C-terminal sequence are derived from the plasmid DNA sequence and replace the integrase amino acids AKL. To prepare an expression clone, the small HindIII fragment derived from pCMV IN Δ713 was cloned into the HindIII site of the modified Qiagen expression vector pQE30 Δ, using E. coli M15/pREP. Colonies were inoculated in 2-ml cultures, induced with IPTG, and screened for expression by immunoblot analysis using a rabbit polyclonal antibody directed to the N terminus of integrase as described previously (18). Mouse monoclonal antibody (MAb) 35 (3) was used in immunoblots in conjunction with a goat anti-mouse secondary antibody conjugated with alkaline phosphatase.
PAGE.
DNA polymerase reaction mixtures were adjusted with 40% formamide and 0.1% bromophenol blue and xylene cyanol FF. Samples were heated to 80°C for 3 min and applied to a 20% polyacrylamide denaturing sequencing gel containing 7 M urea (0.4-mm thickness). Gels were preelectrophoresed for 0.5 h prior to loading of the DNA samples. Electrophoresis was done at 12 W for 2 h at constant power. Following electrophoresis, gels were soaked in 15% methanol–5% acetic acid for 15 min and then in water for 5 min. Gels were dried under vacuum and placed against Kodak Biomax film at −80°C.
RESULTS
Copurification of DNA polymerase activity with integrase multimers.
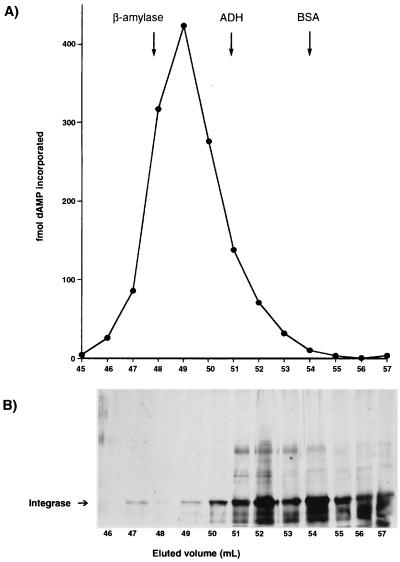
A His6-tagged recombinant HIV-1 integrase fusion protein of 35 kDa was purified from bacterial extracts under denaturing conditions using a Ni2+-NTA affinity resin as described in Materials and Methods. Following renaturation, a DNA polymerase activity was detected in the purified integrase preparation (Table 1). The DNA polymerase was purified further by gel filtration using an S-300 column. The DNA polymerase eluted as a single peak of activity ahead of the β-amylase marker (Mr = 200,000) coinciding with multimeric forms of integrase (Fig. 1). The symmetrical elution profile was suggestive of a monodisperse, homogeneous enzyme. Dimeric forms of integrase eluted ahead of the bovine serum albumin standard, as expected. Although integrase dimers constituted the major form of integrase in the sample, little or no DNA polymerase activity coeluted with this form of integrase (Fig. 1).
TABLE 1.
Reaction requirements for DNA polymerase activity
| Reaction conditiona | % Activityb (mean ± SEM) |
|---|---|
| Complete | 100.0 ± 7.7 |
| −Mg2+/+Mn2+ | 5.7 ± 2.1 |
| +10 mM NEM | 92.4 ± 8.9 |
| Gapped calf thymus DNA | 6.6 ± 1.7 |
| Poly(rA)-oligo(dT) | 0.5 ± 0.2 |
| Poly(dA)-oligo(dT) | 64.0 ± 11.9 |
| Poly(dA) | 0.3 ± 0.05 |
Complete reaction mixtures contained DISPOL 17 DNA, 10 mM Tris-Cl (pH 7.5), 5 mM DTT, and 10 mM MgCl2. Other template-primers listed were added in place of DISPOL 17 DNA.
Normalized to 100% for the complete reaction mixture. After incubation at 37°C for 45 min, complete reaction mixtures yielded 2.4 pmol of dAMP incorporated into DNA (8 × 105 dpm).
FIG. 1.
Analysis of DNA polymerase activity by molecular exclusion chromatography. Integrase was purified by elution from a nickel chelate resin and applied to a Sephacryl S-300 column as described in Materials and Methods. The top panel shows the DNA polymerase activity profile obtained with DISPOL 17 DNA and 1 μl of each column fraction in reaction mixtures. The elution positions of standard proteins (ADH, alcohol dehydrogenase; BSA, bovine serum albumin) used to calibrate the column are indicated by the arrows. The bottom panel shows an SDS-PAGE analysis of proteins in individual fractions eluted from the S-300 column. Prior to analysis, samples were concentrated 10-fold by centrifugation in a Microcon 30 unit. Proteins were detected by silver staining. The position of integrase is indicated at the left.
When analyzed further by chromatography on n-butyl–Sepharose and hydroxylapatite or by sedimentation in a glycerol gradient, the DNA polymerase was recovered as a single peak of activity, confirming the presence of a single homogeneous DNA polymerase in our integrase preparations. We also purified the DNA polymerase sequentially through Ni2+-NTA, n-butyl–Sepharose, Sephacryl S-300, hydroxylapatite, and sedimentation in glycerol gradient. The overall recovery of DNA polymerase activity with this complicated purification protocol was 50%. However, the purity did not increase significantly beyond the Sephacryl S-300 step. Purification of integrase monomers by sodium dodecyl sulfate (SDS)-PAGE followed by renaturation resulted in a relatively low but detectable level of activity. Removal of the His tag had no effect on the level of DNA polymerase activity. We therefore performed the experiments presented below with His-tagged integrase DNA polymerase obtained by using the simplified S-300 purification protocol.
Determination of the molecular weight of the DNA polymerase.
The DNA polymerase sedimented faster than bovine serum albumin (Mr = 67,000) and alcohol dehydrogenase (Mr = 150,000; 7.3S) but slower than β-amylase (Mr = 200,000) in a glycerol gradient and had a sedimentation coefficient (S20,w × 1013 s) of 9.1. A Stokes radius of 4.8 nm was determined based on the gel filtration data in comparison with bovine serum albumin, alcohol dehydrogenase, and catalase standards. The molecular weight calculated according to Siegl and Monty (43) was 172,100.
Time course of the DNA polymerase reaction and dependence on DNA concentration.
When the purified DNA polymerase was incubated at 37°C with poly(dA)-oligo(dT), DTT, MgCl2, and radioactive TTP as the nucleotide precursor, radiolabeled DNA was readily recovered as trichloroacetic acid-precipitable material. The reaction kinetics were linear for up to 40 min. A self-annealing oligonucleotide template-primer designed to fold into a 15-bp hairpin primer stem attached to a homopolymeric T tail was also used to promote DNA synthesis (Table 1). The DNA polymerase reaction with this template-primer increased in a DNA-dependent manner. The optimal DNA concentration was 5 nM. Oligonucleotide template-primers with a 15-bp hairpin primer stem containing either a nonviral sequence or the HIV-1 U5 long terminal repeat sequence gave identical results, indicating that the HIV-1 U5 sequence was not essential for activity. A template-primer without U5 sequences but with a 10-bp primer stem (DISPOL 16NV) exhibited relatively low activity.
Analysis of template-primer and divalent cation preference and effect of NEM.
The DNA polymerase activity obtained with a self-annealing oligonucleotide containing a homopolymeric [poly(dT)] template was completely dependent on the use of dATP as the complementary nucleotide precursor; absolutely no DNA synthesis occurred when the noncomplementary nucleotide dCTP, dGTP, or TTP was used in place of dATP. In terms of template-primer preference, the DNA polymerase was not active with poly(dA) but was highly active with poly(dA)-oligo(dT) (Table 1). Therefore, like all true DNA polymerases, the integrase DNA polymerase requires a DNA primer for activity. Poly(rA)-oligo(dT) failed to yield any detectable activity (Table 1), ruling out the presence of a bacterial reverse transcriptase (30). The DNA-dependent DNA polymerase activity was reduced 20-fold when Mn2+ was used in place of Mg2+ (Table 1) and therefore was Mg2+ dependent. The optimum Mg2+ concentration was 10 mM. E. coli DNA polymerase I, the major bacterial DNA polymerase, was able to utilize both divalent cations almost interchangeably at this concentration. Also, gapped calf thymus DNA was used relatively poorly as a template-primer by integrase (Table 1), whereas E. coli DNA polymerase I was equally active with gapped calf thymus DNA or oligonucleotide hook template-primers. In addition, the integrase-associated DNA polymerase activity was completely resistant to inhibition by the sulfhydryl reagent NEM, whereas bacterial DNA polymerases II and III are both extremely (100%) sensitive to NEM (19, 23, 35). DNA polymerase II is also sensitive to aphidicolin (Ki = 50 μM) (7, 22), whereas the integrase DNA polymerase activity was aphidicolin resistant.
Effect of a C-terminal deletion in integrase on DNA polymerase activity.
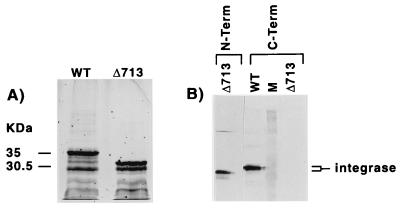
A truncated form of integrase lacking 50 amino acids at the C terminus was purified from a bacterial extract as described for wild-type recombinant integrase. The mutant integrase was produced in the same quantity as wild-type integrase and was approximately 5 kDa smaller than the wild-type protein when analyzed by SDS-PAGE, as expected (Fig. 2A). As shown by immunoblotting, the truncated form of integrase was missing an antibody epitope located at the C terminus of the protein (Fig. 2B). Lower-molecular-weight proteins evident in the SDS-PAGE analysis represent N-terminal fragments of integrase as confirmed by Western analysis.
FIG. 2.
SDS-PAGE and Western blot analysis of wild-type and deletion mutant Δ713 integrase. The panel on the left shows a silver-stained SDS-PAGE profile of wild-type (WT) and mutant (Δ713) integrase purified on a Ni2+-NTA resin; the panel on the right shows an immunoblot analysis of Δ713 and WT integrase. Nitrocellulose filters were incubated with antibody directed at the amino terminus of integrase (N-Term) or the carboxy terminus of integrase (C-Term) as described in Materials and Methods. The lane marked M contained Rainbow colored protein standards.
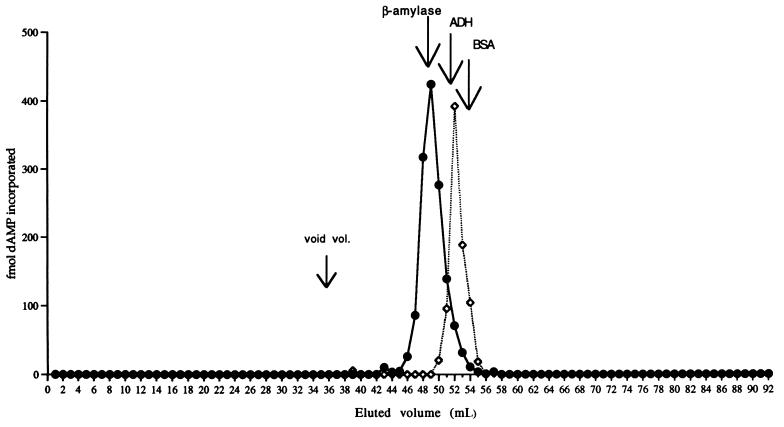
The truncated form of integrase displayed approximately the same level of DNA polymerase activity as wild-type integrase, indicating that the C-terminal 50 amino acids of integrase are not essential for DNA polymerase activity. According to gel filtration analysis, the DNA polymerase activity in this mutant integrase preparation eluted just after the alcohol dehydrogenase marker protein (Mr = 150,000), clearly later than the DNA polymerase associated with wild-type integrase (Fig. 3). This alteration in the gel filtration properties of the DNA polymerase caused specifically by a deletion mutation in the integrase protein argues strongly against contamination with a free bacterial DNA polymerase and points instead to an intrinsic association of the DNA polymerase activity with integrase.
FIG. 3.
Sephacryl S-300 DNA polymerase activity elution profile of wild-type (•) and deletion mutant Δ713 (◊) integrase. The mutant elution profile was superimposed on the wild-type profile given in Fig. 1. Fractions of 1 ml were collected, and aliquots of 1 μl were assayed for DNA polymerase activity as described in Materials and Methods.
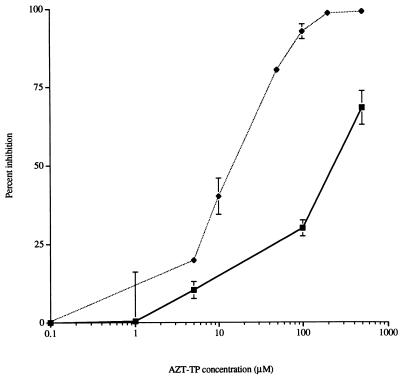
AZT-TP inhibits the integrase DNA polymerase activity.
Next, we investigated the effects of the nucleotide analog AZT-TP on the integrase DNA polymerase. AZT-TP inhibited the integrase DNA polymerase with a 50% inhibitory concentration (IC50) of 220 μM (Fig. 4). This value was reduced to 15 μM in the case of polymerization of TMP, possibly because incorporation of AZT-5′-monophosphate into growing DNA chains caused chain termination. Azidothymidine did not affect DNA polymerase activity up to 500 μM. We also found that E. coli DNA polymerase I was insensitive to AZT-TP up to a concentration of 500 μM. Thus, the effect of AZT-TP on DNA polymerase activity (when polymerizing dAMP residues into DNA) was identical to the effect on the other known activities of integrase (36). The sensitivity of the putative integrase DNA polymerase activity to AZT-TP (when polymerizing TMP residues into DNA) serves to further distinguish this activity from E. coli DNA polymerase I.
FIG. 4.
Effect of AZT-TP on DNA polymerase activity. DNA polymerase reactions were conducted in the presence of various concentrations of AZT-TP, using either DISPOL 17 DNA (▪) or poly(dA)-oligo(dT) (⧫) as a template primer. The DNA polymerase assays were done in triplicate.
The nucleotide polymerizing capability of HIV-1 integrase was also inhibited by coumermycin A1 (IC50 = 50 μM) and by pyridoxal 5′-phosphate (IC50 = 480 μM), compounds that inhibit other functions of HIV-1 integrase at somewhat lower concentrations (36).
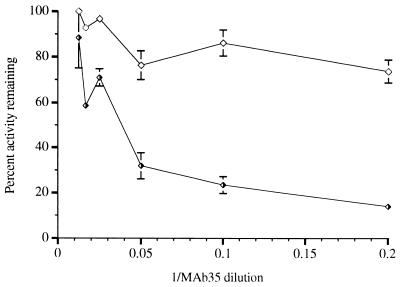
Neutralization of DNA polymerase activity by a MAb.
To obtain further evidence that integrase has an intrinsic DNA polymerase activity, we used MAb 35, directed against the sequence KAKIIRDYGK (amino acids 264 to 273) (3), an epitope near the C terminus of integrase. This antibody reacted with the full-length integrase protein in immunoblots but failed to recognize a C-terminal deletion mutant lacking 50 amino acids, as mentioned earlier (Fig. 2B). This antibody inhibited integrase DNA polymerase by up to 85% while having a significantly weaker effect on DNA polymerase I (Klenow fragment) (Fig. 5) and no effect on HIV-1 reverse transcriptase. In addition, a pool of purified MAbs directed against HIV-1 reverse transcriptase had no effect on integrase DNA polymerase activity under conditions that completely inactivated HIV-1 reverse transcriptase.
FIG. 5.
Neutralization of DNA polymerase activity by an anti-integrase MAb. Dilutions of a mouse ascites fluid containing MAb 35 were prepared in phosphate-buffered saline, and 1 μl of the diluted antibody was added to each 25-μl reaction mixture. Half-filled triangles represent reactions with integrase; open triangles represent reactions with E. coli DNA polymerase I (Klenow fragment).
Analysis of the products of the DNA polymerase reaction.
Products of DNA polymerase reactions performed on a self-annealing template-primer with a 20-mer homopolymeric T tail (DISPOL 10) ranged in size from +1 to +19, but +18 and +19 products predominated. However, no +20 products were detected, implying that a template strand consisting of one nucleotide could not be utilized by integrase (see below). Reaction products generated under identical conditions with E. coli DNA polymerase I (Klenow fragment) were analyzed in parallel. The E. coli enzyme added 20 nucleotides to the template-primer, resulting in the production of a single major DNA product.
Gap repair by integrase DNA polymerase.
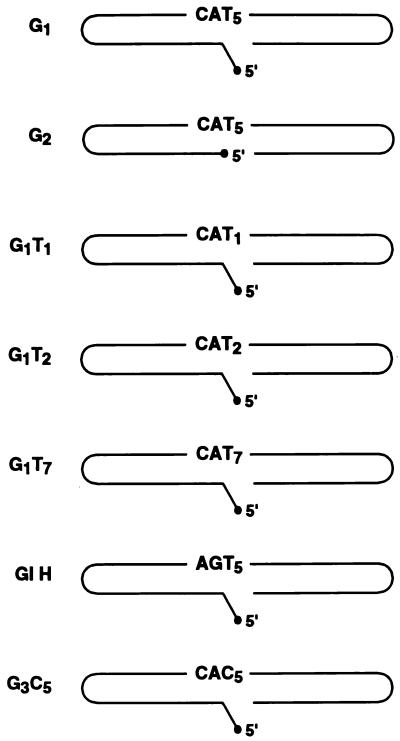
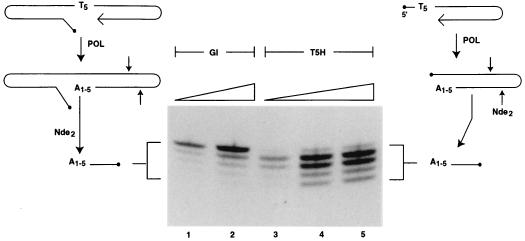
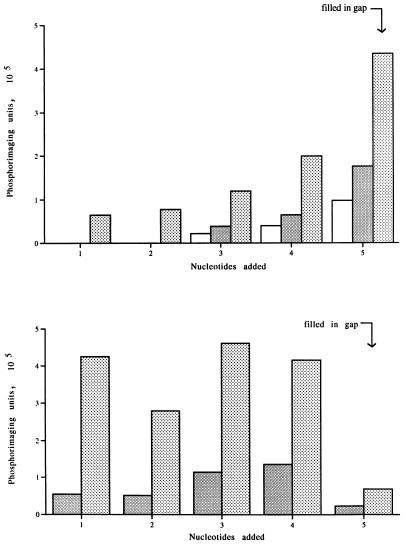
Synthetic oligonucleotides predicted to fold into gapped DNA molecules resembling proviral integration intermediates were used to investigate gap repair by the putative polymerase function of HIV-1 integrase (Fig. 6). The DNA molecule G1 is predicted to fold so as to provide a gap of five nucleotides comprised solely of TMP template residues situated immediately adjacent to a DNA primer. The gap is flanked on one side by a 15-bp region representing host DNA and on the other side by 10 bp of DNA matching the end of the U5 region of the HIV-1 long terminal repeat. There is a mismatched 5′ AC dinucleotide tail adjacent to the gap. Both ends of the 59-mer molecule are hairpinned, and the duplex regions contain cleavage sites for several restriction enzymes. The extent of DNA polymerization in the gapped region was assessed by measuring an increase in the length of the DNA primer strand, following cleavage of radiolabeled DNA products with NdeII upstream of the polymerization start site at the 3′ end of the DNA molecule. When a DNA polymerase reaction was conducted with G1 DNA as a template-primer in the presence of [α-32P] dATP as the sole deoxynucleoside 5′-triphosphate (dNTP), polymerization of between one and five nucleotides at the 3′ end of the gapped oligonucleotide occurred (Fig. 7). Phosphorimaging analysis revealed that gaps were filled to completion (polymerization of five nucleotides) approximately 50% of the time (Fig. 8). We also investigated the ability of integrase to copy a five-nucleotide tail that was not part of a gapped structure. We constructed a molecule, T5H, which was identical to the gapped molecule referred to above but without the viral DNA U5 arm. When the experiment outlined above was repeated with T5H as the template-primer, five radiolabeled DNA products were once again obtained (Fig. 7). The overall level of DNA synthesis with T5H DNA was about the same as with the gapped DNA substrate. However, polymerization of five nucleotides at the 3′ end of the T5H molecule occurred only about 3% of the time (Fig. 8). Therefore, both arms of a gapped DNA substrate are required for efficient polymerization of the last nucleotide of a growing DNA chain by the integrase DNA polymerase.
FIG. 6.
Schematic representation of the gapped DNA molecules used in this study. Torsional strain in the region of the hairpin termini probably prevents base pairing at the ends of the molecule. The molecules feature a U5 hairpin contiguous with the CA dinucleotide to the left of the gap (except for G1H, where five nucleotides adjacent to the gapped region were changed to a nonviral sequence), a 5′ unpaired AC tail (except for G2), and a gapped region which varies in length and base composition adjacent to the CA dinucleotide. The right hairpin in each molecule is composed of a nonviral sequence which contains an NdeII cleavage site 10 nucleotides from the 3′ end of the DNA.
FIG. 7.
Repair products obtained in an integrase DNA polymerase reaction using gapped and nongapped DNAs as template primers. The diagrams on the left and right illustrate polymerase reactions with gapped (G1) and nongapped (T5H) DNA substrates, respectively. Following digestion with NdeII, samples were analyzed on a denaturing DNA sequencing gel as described in Materials and Methods. The radiolabeled DNA fragments seen in the autoradiogram range from 11 to 15 nucleotides in length as determined by using 5′-end labeled oligonucleotides of known length as markers (oligonucleotides of 8, 12, 14, and 17 nucleotides were used; see Fig. 11 for an in-gel comparison). A1-5 refers to the number of dAMP residues polymerized at the 3′ end of the respective DNA template-primers. The inclined planes lying over the photo of the autoradiogram represent increases in the amount of DNA added to reaction mixtures ranging from 10 to 20 ng (lanes 1 and 2) or 10, 20, and 50 ng (lanes 3 to 5).
FIG. 8.
Quantitative analysis of repair reactions conducted with gapped (G1; top) and nongapped (T5H; bottom) DNA. The radiolabeled DNA fragments seen by autoradiography were imaged by using a detection screen. The data were scanned into a Bio-Rad phosphorimaging unit and quantified by computer analysis of the scanned images. The numbers 1 to 5 on the abscissa of each graph refer to the number of dAMP residues added to the 3′ end of the DNA template-primer. The data were corrected for cumulative increases in band intensity caused by differences in fragment size. □, 5 ng; ░⃞, 10 ng; , 20 ng.
Utilization of gapped DNA substrates is template dependent.
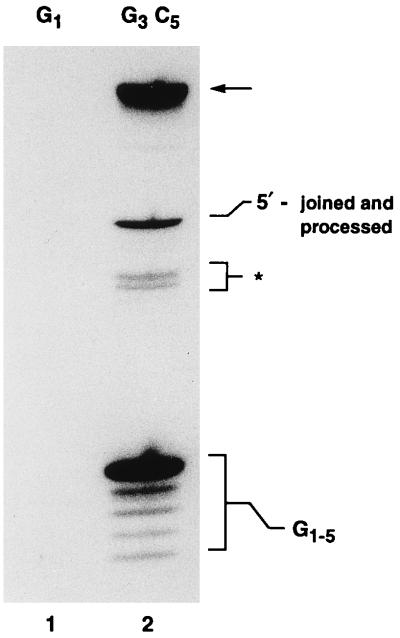
To determine whether gap repair by the integrase DNA polymerase was influenced by base composition, a gapped DNA molecule containing a C5 gap (G3C5), but otherwise identical to the G1 molecule described earlier, was incubated with the integrase DNA polymerase in the presence of [α-32P]dGTP as the only dNTP. In the case of dGTP reactions, five radiolabeled DNA products, corresponding to the addition of between one and five nucleotides at the 3′ end of the DNA substrate, were obtained after NdeII digestion (Fig. 9). Gaps were completely filled (addition of five nucleotides) 65% of the time. In contrast, no radiolabeled products were observed when reactions were conducted with G1 DNA (contains a T5 gap) in the presence of [α-32P]dGTP. Therefore, virtually all DNA synthesis by the integrase DNA polymerase is template dependent and the incorporation of G-C base pairs into the gapped region serves to increase the relative frequency with which gaps are filled to completion.
FIG. 9.
Influence of base composition on gap repair. Integrase DNA polymerase reactions were conducted in the presence of dGTP with either G1 (lane 1) or G3C5 (lane 2) DNA. The repair products released from the G3C5 DNA substrate by NdeII digestion are indicated by G1-5 at the lower right. The zone marked by the asterisk marks the position of the products of incomplete digestion with NdeII. A 36-mer marker oligonucleotide, of the structure expected if G3 molecules participating in repair underwent 5′ joining followed by cleavage at the CA dinucleotide adjacent to the C5 gap, comigrated with the band marked 5′-joined and processed. The structure of this repair product was not investigated further. The arrow marks the position of repaired DNA that is not treated with NdeII.
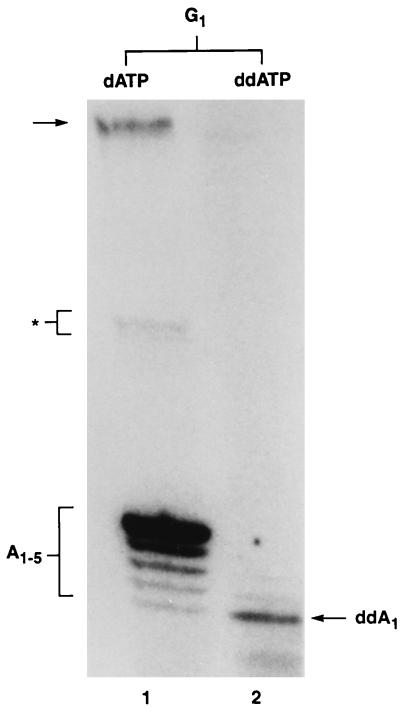
The results of this experiment also argue against the possibility that radiolabeled nucleotides are used by integrase to perform a nucleophilic attack on the DNA substrate. To rule out this possibility, we performed an additional experiment in which [α-32P]ddATP was used as the radiolabeled nucleotide. According to the DNA polymerase model, one nucleotide should be added to the G1 DNA substrate in this case, whereas in the nucleophilic attack model, radiolabeling of G1 DNA should not occur. The results clearly demonstrate that radiolabeled DNA products were obtained when ddATP was the only nucleotide present in the reaction mixtures. Only a single nucleotide was added to the 3′ end of G1 DNA under these conditions (Fig. 10) compared with reactions containing dATP, which resulted in efficient gap repair (addition of five nucleotides). This result also demonstrates clearly that the integrase DNA polymerase utilizes dideoxynucleotides as chain terminators.
FIG. 10.
Utilization of ddATP by the integrase DNA polymerase. The DNA repair products obtained with a G1 gapped DNA substrate were compared for reactions conducted either with dATP (lane 1) or ddATP (lane 2) as the nucleotide precursor. The repair products released from the G1 DNA substrate by NdeII digestion are indicated by A1-5 and ddA1 for the dATP and ddATP reactions, respectively. The radiolabeled DNA repair products ranged in size from 11 to 15 nucleotides.
Extension of DNA chains beyond a gapped region is highly regulated.
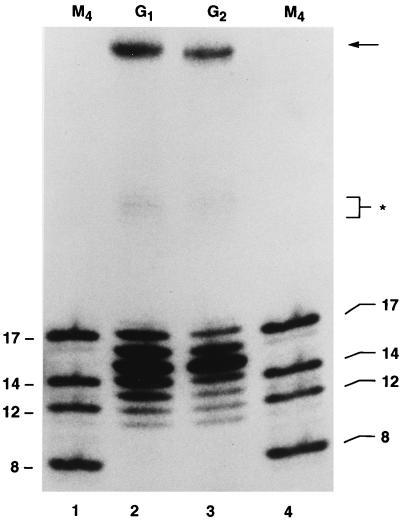
When all four dNTPs were added to reaction mixtures in the presence of [α-32P]dATP, gaps in G1 molecules were filled to completion about 65% of the time (Table 2). In about half of these molecules, 1 or 2 nucleotides were added beyond the 5-nucleotide gap, as demonstrated by the production of DNA molecules 16 and 17 nucleotides in length following digestion of the repair products with NdeII (Fig. 11; Table 2). The addition of these nucleotides was highly regulated. We obtained no evidence for the synthesis of DNA chains longer than that expected for the addition of seven nucleotides to the 3′ end of the gapped DNA substrate. The addition of two nucleotides most probably occurs by a mechanism in which the CA dinucleotide in the U5 region is used as a template. The two nucleotides must therefore be T and G. This conclusion was further verified by radiolabeling the nascent DNA with [α-32P]dGTP. In this case, only the DNA fragment containing the G residue at the seventh position beyond the start of the gap became radiolabeled, as expected. Similar results were obtained in assays using gapped DNA molecules that were identical in every respect to the G1 molecule but did not possess a 5′ AC tail (Fig. 6 and 11; Table 2). Therefore, neither gap repair nor the addition of two nucleotides beyond the gap require a 5′ tail. Interestingly, by changing the last five nucleotides of the U5 segment, including the CA dinucleotide, to a nonviral sequence (G1H), the frequency of synthesis beyond the gap was reduced from 34% to about 6% (Table 2). Furthermore, the C-terminal truncation mutant of integrase, Δ713, referred to earlier, was similarly impaired in its ability to extend DNA chains two nucleotides beyond the gap (Table 2). Therefore, polymerization of nucleotides beyond the gap may be regulated by the viral U5 sequence, most probably by the CA dinucleotide that is situated immediately adjacent to the gap, and may involve an interaction of the U5 sequence with a C-terminal domain of integrase.
TABLE 2.
Effects of U5 sequences and a C-terminal truncation on the distribution frequency of integrase DNA repair products
| Nucleotide position | Relative frequency of
DNAa
|
||||
|---|---|---|---|---|---|
| Wild-type
IN
|
Δ713 IN
|
||||
| G1 | G1H | G2 | G1 | G2 | |
| 7 | 12.6 | 1.8 | 8.3 | 2.7 | 1.7 |
| 6 | 21.2 | 4.4 | 16.7 | 14.4 | 7.5 |
| 5 | 28.8 | 31.2 | 41.0 | 36.5 | 45.9 |
| 4 | 14.1 | 28.0 | 14.6 | 21.4 | 16.8 |
| 3 | 8.6 | 13.4 | 7.6 | 11.5 | 10.8 |
| 2 | 6.6 | 9.2 | 5.4 | 6.0 | 7.5 |
| 1 | 8.6 | 10.9 | 7.8 | 7.4 | 10.1 |
The relative frequency of radiolabeled nascent DNA corresponding to the addition of one to seven nucleotides to the DNA substrate was quantified by phosphorimaging and expressed as a percentage of the total radiolabel in repair products.
FIG. 11.
Effect of a 5′ AC unpaired dinucleotide on the DNA repair reaction. Repair reactions were conducted as described in the text, using either tailed (G1; lane 2) or untailed (G2; lane 3) gapped DNA as a template-primer. The lanes marked M4 (lanes 1 and 4) contain 5′-32P-labeled oligonucleotides whose lengths (between 8 and 17 nucleotides) are indicated at the sides. The significance of the asterisk and the arrow is explained in the legend to Fig. 9.
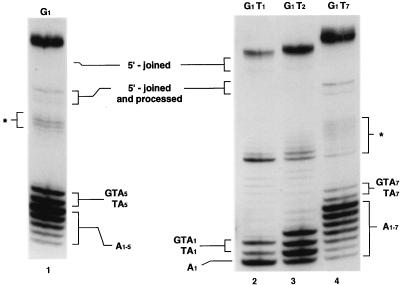
Similar results were obtained using G1-type molecules with T1, T2, and T7 gaps (Fig. 6 and 12), except that in the case of molecules with one- and two-nucleotide gaps, polymerization continued beyond the gapped region for up to seven or eight nucleotides, albeit at a relatively low level. Therefore, T1 and T2 molecules appeared to have partially lost the regulation inherent in the structure of the T5 and T7 gapped molecules.
FIG. 12.
Effect of gap size on DNA repair by the integrase DNA polymerase. The major repair products range in size from 11 to 13, 11 to 14, 11 to 17, and 11 to 19 nucleotides in the cases of G1T1 (lane 2), G1T2 (lane 3), G1 (lane 1), and G1T7 (lane 4), respectively. Fragments of DNA corresponding to the complete fill-in of each gap are 11, 12, 15, and 17 in the cases of G1T1 (lane 2), G1T2 (lane 3), G1 (lane 1), and G1T7 (lane 4), respectively. The size of each DNA fragment was determined in comparison with oligonucleotide size markers (M4 in Fig. 11). Except for lane 3, the presumed nucleotide composition of the added nucleotides is shown opposite the repair products. In lane 3, the four bands at the bottom represent the addition of A1, A2, TA2, or GTA2. The sequences (read from right to left) represent the addition of nucleotides to the gapped DNA substrates in a 5′-3′ direction. The position of a 48-mer oligonucleotide corresponding to the structure expected for a 5′-joined product after NdeII cleavage is shown near the top. The significance of the asterisk and the identity of the bands near the top of the photograph are as described above.
DISCUSSION
To the best of our knowledge, there has been no previous report of a retroviral integrase exhibiting DNA-dependent DNA polymerase activity. In the present study, we obtained results indicating that the integrase protein of HIV-1, a lentivirus, possesses such an intrinsic DNA polymerase activity. Due to the novelty of this conclusion, several criteria were applied to the catalytic activity of the putative HIV-1 integrase DNA polymerase, in an attempt to distinguish it from the known bacterial DNA polymerases. We found that the HIV-1 integrase DNA polymerase behaved differently from E. coli DNA polymerase I according to no less than six independent criteria. Neither divalent cation or template-primer preference, sensitivity to AZT-TP or an anti-integrase MAb, or the inability to copy the last nucleotide in a DNA template of a nongapped substrate or to extend nascent DNA chains through a duplex region matched the behavior of E. coli DNA polymerase I (or its counterpart, the Klenow fragment), a potential source of contamination in our integrase preparations. E. coli DNA polymerases II and III could be ruled out as well on the basis of their extreme sensitivity to NEM, while failure to detect RNA-dependent DNA polymerase activity ruled out the presence of a bacterial reverse transcriptase.
Perhaps the inhibitory effect of the well-characterized anti-integrase antibody MAb 35 provided the strongest evidence in favor of the intrinsic association of a DNA polymerase activity with the HIV-1 integrase protein. Also, the fact that a C-terminal truncation mutation in integrase altered the gel filtration properties of the DNA polymerase attests to the intrinsic association of a DNA polymerase with the HIV-1 integrase protein.
Separation of dimeric and multimeric forms of integrase on a Sephacryl S-300 column revealed that the DNA polymerase activity coeluted exclusively with integrase multimers. A molecular weight determination for the DNA polymerase of 172,100 is consistent with the suggestion that the integrase polymerase holoenzyme is a multimer, possibly a hexamer. The more abundant dimeric forms of integrase failed to display any DNA polymerase activity, at least under the conditions tested so far. It is unclear why dimers do not catalyze DNA synthesis. Since dimeric forms of integrase possess a nucleotide binding site (33) and bind DNA (14, 20, 34, 39, 45, 49, 52, 53), other requirements for DNA polymerization must be lacking. It would be of interest to further purify multimeric forms of integrase to be able to demonstrate copurification with the DNA polymerase activity through several chromatography steps. This was difficult to do in the present study due to the extremely low abundance of multimers relative to other forms of integrase, mainly dimers.
Inhibition of the integrase DNA polymerase activity by AZT-TP (when measuring polymerization of the cognate nucleotide TTP) occurred in the 10 μM range, at least 2 orders of magnitude greater than for HIV-1 reverse transcriptase. The DNA polymerase function of integrase is therefore probably not an in vivo target for AZT-TP. Interestingly, when the noncognate nucleotide dATP was polymerized, sensitivity of the DNA polymerase to this inhibitor was considerably reduced and fell in the same range observed previously for inhibition of 3′ processing and DNA strand transfer (36). The mechanism of inhibition of all three functions of integrase by AZT-TP could therefore be similar under these conditions. In view of these results, studies done previously with other nucleotide analog-inhibitors of integrase (37) should be reevaluated to determine their effects on the DNA polymerase function of integrase compared with their effects on the 3′-processing or DNA strand transfer reactions. As shown here for AZT-TP, other nucleotide analogs may display a greater inhibitory effect on the DNA polymerase function of integrase than on the other integrase-mediated reactions.
The HIV-1 integrase DNA polymerase is capable of carrying out efficient gap repair in vitro and has the ability to extend a DNA chain up to two nucleotides beyond a gap. A viral U5 sequence downstream of the gap is not required for gap repair but significantly enhances the frequency of post-gap synthesis, indicating that recognition of a U5 viral sequence in the DNA substrate, possibly the CA dinucleotide, may be required. Notably, post-gap repair involves only the polymerization of viral sequences. The 5-bp repeats that flank the integrated provirus are derived entirely from the adjacent host cell DNA and are not lengthened by post-gap repair. There is no net gain of viral DNA either since the nascent DNA (one to two nucleotides) produced in the post-gap repair reaction displaces a preexisting viral sequence; the displaced 5′ tail would theoretically be removed by integrase in vivo in a subsequent DNA strand transfer reaction. Post-gap repair may play a physiologically relevant role in HIV-1 replication by providing a mechanism for creating a new 5′ tail at the virus-host DNA junction in the event that the existing 5′ tail was removed by a host cell exonuclease.
Failure to copy the last nucleotide of a DNA template that is not part of a gapped structure is a most remarkable and defining feature of the integrase DNA polymerase. This unusual property may be physiologically relevant in regard to favoring the maintenance of the 3′-processed ends of HIV-1 DNA. Yet, a one-nucleotide template is effectively copied when it is part of a gapped DNA substrate. As far as we know, no other DNA polymerase has been reported to behave in this way. The mechanism for gap repair may therefore involve a requirement for the binding of integrase to both arms of duplex DNA lying across a single-stranded gap. In any case, integrase shows a remarkable ability to repair even very small gaps (one nucleotide) as well as larger gaps of at least seven nucleotides. The integrase DNA polymerase is well suited for a presumed in vivo role as a gap repair enzyme since it can copy only homopolymers or short stretches of natural DNA where it is unlikely to encounter a region of secondary structure.
Although they do influence post-gap repair, the C-terminal 50 amino acids of integrase are not required for DNA polymerase activity per se and therefore likely do not harbor active site amino acids. In view of this result, the inhibitory effect of the C-terminal antibody MAb 35 on DNA polymerase activity may be explained by a structural alteration at a distal site required for polymerase activity caused by binding of the antibody to its C-terminal epitope. Clearly, the antibody did not affect the DNA polymerase active site directly since deletion of the antibody binding site did not affect DNA polymerase activity. Significantly, MAb 35 has no inhibitory effects on the 3′-processing or DNA strand transfer activities of integrase, whose active site is located in the core domain (3). Its effects on the DNA polymerase are therefore not due to a general disruption of integrase structure. It is important to note that the mechanism of inhibition of polymerase activity was not established by the studies presented here and may not be related to the mechanism established previously for the effect of the Fab fragment of MAb 35 on integration activity (3). In that instance, C-terminus-specific integrase functional multimerization was blocked by the Fab fragment, and MAb 35 was shown to be incapable of doing this. We note that the integrase we are dealing with here is already in a multimeric, possibly hexameric, state when the antibody is added to reaction mixtures. This is not the case in previous work (3), where integrase is in a dimeric form initially and is believed to multimerize upon interacting with DNA in the presence of divalent cations.
The results of the present study do not allow us to draw a conclusion as to whether the polymerase active site is related to the core domain active site established previously for integration and 3′ processing. Preliminary studies have indicated that mutations in the core domain active site amino acids do not affect polymerase activity whereas a P109S mutation inactivates polymerase (42). The latter mutation is known to block the 3′-processing and integration activities of purified integrase and to abolish viral infectivity. The mutation also exerts a profound effect on the structure of integrase since these mutants form large aggregates in solution (9, 44). The P109S mutation also abrogates the ability of integrase to cross-link DNA (8). Either of these phenotypic effects of the P109 mutation could account for its inhibitory effect on polymerase as well. Further experiments are therefore necessary to identify the active site amino acid(s) for the DNA polymerase activity of HIV-1 integrase, and this remains a major goal of our research.
ACKNOWLEDGMENTS
We thank M. Parniak for providing MAbs to HIV-1 reverse transcriptase and for many helpful suggestions throughout the course of this work. We are grateful to Avi Shtevi for suggesting the use of hairpinned gapped DNA substrates and for analysis of the P109S integrase mutant. MAb 35 was generously provided by S. H. Hughes.
This work was supported by grants from the National Health and Research Development Program of Health and Welfare Canada and the Canadian Foundation for AIDS Research awarded to E. A. Faust.
REFERENCES
- 1.Andrake M D, Skalka A M. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 2.Andrake M D, Skalka A M. Retroviral integrase, putting the pieces together. J Biol Chem. 1996;271:19633–19636. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 3.Barsov E V, Huber W E, Marcotrigiano J, Clark P K, Clark A D, Arnold E A, Hughes S H. Inhibition of human immunodeficiency virus type 1 integrase by the Fab fragment of a specific monoclonal antibody suggests that different multimerization states are required for different enzymatic functions. J Virol. 1996;70:4484–4494. doi: 10.1128/jvi.70.7.4484-4494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke C J, Sanyal G, Bruner M W, Ryan J A, LaFemina R L, Robbins H L, Zeft A S, Middaugh C R, Cordingley M G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- 5.Bushman F, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon P M, Wilson W, Byles E, Kingsman S M, Kingsman A J. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Lawrence C B, Bryan S K, Moses R E. Aphidicolin inhibits DNA polymerase II of Escherichia coli, an alpha-like DNA polymerase. Nucleic Acids Res. 1990;18:7185–7186. doi: 10.1093/nar/18.23.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drelich M, Haenggi M, Mous J. Conserved residues Pro-109 and Asp-116 are required for interaction of the human immunodeficiency virus type 1 integrase protein with its viral DNA substrate. J Virol. 1993;67:5041–5044. doi: 10.1128/jvi.67.8.5041-5044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 10.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 11.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman A, Hickman A B, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;61:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 16.Englund G, Theodore T S, Freed E O, Engelman A, Martin M A. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faust E A, Acel A, Udashkin B, Wainberg M A. Human immunodeficiency virus type 1 integrase stabilizes a model HIV-1 LTR plasmid in vivo. Biochem Mol Biol Int. 1993;36:745–758. [PubMed] [Google Scholar]
- 18.Faust E A, Garg A, Small L, Acel A, Wald R, Udashkin B. Enzymatic capability of HIS-tagged HIV-1 integrase using oligonucleotide disintegration substrates. J Biomed Sci. 1996;3:254–265. doi: 10.1007/BF02253705. [DOI] [PubMed] [Google Scholar]
- 19.Firshein W, Strumph P, Benjamin P, Burnstein K, Kornacki J. Replication of a low-copy-number plasmid by a plasmid DNA-membrane complex extracted from minicells of Escherichia coli. J Bacteriol. 1982;150:1234–1243. doi: 10.1128/jb.150.3.1234-1243.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugan I R, Nilsen B M, Worland S, Olsen L, Helland D E. Characterization of the DNA-binding activity of HIV-1 integrase using a filter binding assay. Biochem Biophys Res Commun. 1995;217:802–810. doi: 10.1006/bbrc.1995.2843. [DOI] [PubMed] [Google Scholar]
- 21.Hazuda D J, Wolfe A L, Hastings J C, Robbins H L, Graham P L, LaFemina R L, Emini E A. Viral long terminal repeat substrate binding characteristics of the human immunodeficiency virus type 1 integrase. J Biol Chem. 1994;269:3999–4004. [PubMed] [Google Scholar]
- 22.Ishino Y, Iwasaki H, Fukui H, Mineno J, Kato I, Shinagawa H. Aphidicolin inhibits DNA polymerizing activity but not nucleolytic activity of Escherichia coli DNA polymerase II. Biochimie. 1992;74:131–136. doi: 10.1016/0300-9084(92)90036-e. [DOI] [PubMed] [Google Scholar]
- 23.Iwasaki H, Nakata A, Walker G C. The Escherichia coli polBgene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol. 1990;172:6268–6273. doi: 10.1128/jb.172.11.6268-6273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones K S, Coleman J, Merkel G W, Laue T M, Skalka A M. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992;267:16037–16040. [PubMed] [Google Scholar]
- 25.Jonsson C B, Donzella G A, Roth M J. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993;268:1462–1469. [PubMed] [Google Scholar]
- 26.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkosky J, Jones K S, Katz R, Mack J, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkosky J, Katz R A, Merkel G, Skalka A M. Activities and substrate specificity of the evolutionarily conserved central domain of retroviral integrase. Virology. 1995;206:448–456. doi: 10.1016/s0042-6822(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 29.Lafemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lampson B C, Viswanathan M, Inouye M, Inouye S. Reverse transcriptase from Escherichia coli exists as a complex with ms DNA and is able to synthesize double-stranded DNA. J Biol Chem. 1990;265:8490–8496. [PubMed] [Google Scholar]
- 31.Leavitt A D, Rose R B, Varmus H E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leavitt A D, Shuie L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 33.Lipford J R, Worland S T, Farnet C M. Nucleotide binding by the HIV-1 integrase protein in vitro. J Acquired Immune Defic Syndr. 1994;7:1215–1223. [PubMed] [Google Scholar]
- 34.Lutzke R A P, Vink C, Plasterk R H A. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maki H, Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985;260:12987–12992. [PubMed] [Google Scholar]
- 36.Mazumder A, Cooney D, Agbaria R, Gupta M, Pommier Y. Inhibition of human immunodeficiency virus type 1 integrase by 3′-azido-3′-deoxythymidylate. Proc Natl Acad Sci USA. 1994;91:5771–5775. doi: 10.1073/pnas.91.13.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazumder A, Neamati N, Sommadossi J P, Gosselin G, Schinazi R F, Imbach J L, Pommier Y. Effects of nucleotide analogues on human immunodeficiency virus type 1 integrase. Mol Pharmacol. 1996;49:621–628. [PubMed] [Google Scholar]
- 38.Mumm S R, Grandgenett D P. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pemberton I K, Buckle M, Buc H. The metal ion-induced cooperative binding of HIV-1 integrase to DNA exhibits a marked preference for Mn(II) rather than Mg(II) J Biol Chem. 1996;271:1498–1506. doi: 10.1074/jbc.271.3.1498. [DOI] [PubMed] [Google Scholar]
- 40.Roe T, Chow S A, Brown P O. 3′-end processing and kinetics of 5′-end joining during retroviral integration in vivo. J Virol. 1997;71:1334–1340. doi: 10.1128/jvi.71.2.1334-1340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai H, Kawamura M, Sakuragi J-I, Sakuragi R, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shtevi, A., R. Wald, and E. A. Faust. 1997. Unpublished observations.
- 43.Siegl L M, Monty K J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 44.Taddeo B, Carlini F, Verani P, Engelman A. Reversion of a human immunodeficiency virus type 1 integrase mutant at a second site restores enzyme function and virus infectivity. J Virol. 1996;70:8277–8284. doi: 10.1128/jvi.70.12.8277-8284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Gent D C, Elgersma Y, Bolk M W, Vink C, Plasterk R H. DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2. Nucleic Acids Res. 1991;19:3821–3827. doi: 10.1093/nar/19.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Gent D C, Vink C, Groeneger A M O, Plasterk R H A. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coliand analysis of variants with amino terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vink C. Site-specific hydrolysis and alcoholysis of human immunodeficiency viral DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991;19:6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vink C, Groeneger A A M O, Plasterk R H A. Identification of the catalytic and DNA binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vink C, Plasterk R H A. The human immunodeficiency virus integrase protein. Trends Genet. 1993;9:433–437. doi: 10.1016/0168-9525(93)90107-s. [DOI] [PubMed] [Google Scholar]
- 51.Vink C, van Gent D C, Elgersma Y, Plasterk R H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woerner A M, Klutch M, Levin J G, Marcus-Sekura C J. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992;8:2433–2437. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 53.Woerner A M, Marcus-Sekura C J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993;21:3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]