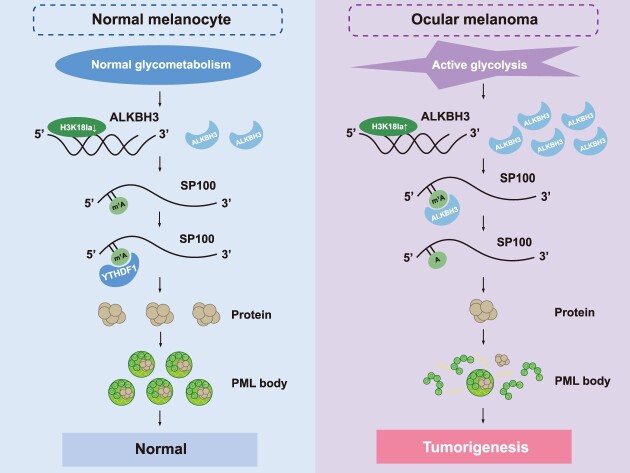
Abstract
Albeit N1-Methyladenosine (m1A) RNA modification represents an important regulator of RNA metabolism, the role of m1A modification in carcinogenesis remains enigmatic. Herein, we found that histone lactylation enhances ALKBH3 expression and simultaneously attenuates the formation of tumor-suppressive promyelocytic leukemia protein (PML) condensates by removing the m1A methylation of SP100A, promoting the malignant transformation of cancers. First, ALKBH3 is specifically upregulated in high-risk ocular melanoma due to excessive histone lactylation levels, referring to m1A hypomethylation status. Moreover, the multiomics analysis subsequently identified that SP100A, a core component for PML bodies, serves as a downstream candidate target for ALKBH3. Therapeutically, the silencing of ALKBH3 exhibits efficient therapeutic efficacy in melanoma both in vitro and in vivo, which could be reversed by the depletion of SP100A. Mechanistically, we found that YTHDF1 is responsible for recognition of the m1A methylated SP100A transcript, which increases its RNA stability and translational efficacy. Conclusively, we initially demonstrated that m1A modification is necessary for tumor suppressor gene expression, expanding the current understandings of dynamic m1A function during tumor progression. In addition, our results indicate that lactylation-driven ALKBH3 is essential for the formation of PML nuclear condensates, which bridges our knowledge of m1A modification, metabolic reprogramming, and phase-separation events.
Graphical Abstract
Graphical Abstract.
Introduction
RNA modifications are modified or removed by a variety of enzymes, and these modifications play roles in essential molecular mechanisms (1). The precise temporal and spatial regulation of N1-methyladenosine (m1A) RNA modification is essential for epigenome homeostasis (2). Importantly, m1A RNA methylation blocks the Watson-Crick interface and thereby influences RNA secondary structures and protein-RNA interactions, which are observed in tRNAs, rRNAs, mRNAs and mitochondrial RNAs (3,4). Dynamic m1A methylation is mediated by methyltransferases (writers: TRMT6, TRMT61A, TRMT61B, and TRMT10C), demethylases (erasers: ALKBH1 and ALKBH3) and RNA-binding proteins (readers: YTHDFs) (5,6). To date, emerging evidence has demonstrated that dynamic m1A methylation plays vital roles in the biological processes, including gene expression, RNA stability and translational efficacy (7–9). For example, ALKBH3-mediated m1A demethylation is essential for Aurora A expression, which regulates ciliogenesis in mammalian cells (10).
Notably, m1A methylation participates in carcinogenesis of diversified cancers, including gastrointestinal cancer (11), oral squamous cell carcinoma (12), cervical cancer (13) and Hodgkin lymphoma (14). Moreover, aberrant m1A methylation triggers several oncogenic events, including metabolic reprogramming (8), collagen production (14) and the generation of tRNA-derived small RNAs (15). Notably, the increased expression of the m1A demethylase ALKBH3 is frequently observed in many cancers and accelerates malignant proliferation and invasion (9). Notably, several genome-wide screening studies have demonstrated over ∼500 salient m1A binding sites (16,17); however, the underlying functions of these m1A methylation modifications in carcinogenesis remain poorly understood.
Ocular melanoma, including conjunctival melanoma (CoM) and uveal melanoma (UM), is the most frequent and life-threatening form of ocular malignancy and is resistant to common chemotherapy. For CoM, gain-of-function mutations of BRAF and NRAS have been identified as driver oncogenic events (18). Most UM patients harbor activating mutations in G protein subunit alpha Q (GNAQ) and G protein subunit alpha 11 (GNA11) (19). Moreover, several epigenetic abnormalities are involved in the pathogenesis of ocular melanoma, including aberrant histone modifications, m6A modifications and 3D chromosomal conformations (20–22). For example, ALKBH5-mediated m6A demethylation inhibits the translation of histidine triad nucleotide-binding protein 2 (HINT2), which promotes the malignant proliferation and metastasis of ocular melanoma cells (20). However, the mechanism of m1A modification in ocular melanoma tumorigenesis remains enigmatic.
We thus aimed to identify the molecular mechanisms and potential clinical application of m1A modification targets in ocular melanoma. In this study, we revealed for the first time that global m1A modification is specifically decreased in ocular melanoma, which is attributed to the increased ALKBH3 expression by histone lactylation. Moreover, genome-wide proteomic analysis, transcriptome screening and m1A-methylated RNA immunoprecipitation sequencing (MeRIP-seq) were performed. We revealed that ALKBH3 removes the m1A modification on SP100A and inhibits its expression. Importantly, the silencing of ALKBH3 exhibits great therapeutic efficacy in melanoma both in vitro and in vivo, and these effects are attenuated by the depletion of SP100A. Furthermore, YTHDF1 is responsible for recognition of the m1A modification of SP100A, which enhances its RNA stability and translational efficacy. Overall, our results demonstrate that ALKBH3-dependent m1A demethylation is essential for the formation of PML nuclear bodies, thereby providing a novel therapeutic strategy in which the ‘targeted m1A reprogramming strategy’ is efficient for tumor therapy.
Materials and methods
Patients and tissue specimens
We collected 82 human ocular melanoma tissues and 28 human normal nevus tissues from the Department of Ophthalmology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine. The clinicopathological and prognostic information of these tissue specimens was described previously (20,21).
Cell culture
The human UM cell lines 92.1, MUM2B and OCM1 were provided by Professor John F. Marshall (Tumor Biology Laboratory, Cancer Research UK Clinical Center, John Vane Science Centre, London, UK). The human UM cell lines MEL290, OMM2.3, and OMM1 and the human conjunctival melanoma cell lines CRMM1, CRMM2, and CM2005.1 were kind gifts from Professor Martine J. Jager (Department of Ophthalmology, Leiden University Medical Center, Leiden, The Netherlands). The human normal melanocyte cell line PIG1 was kindly provided by the Department of Ophthalmology, Peking University Third Hospital. HEK293T cells were purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were authenticated by short tandem repeat (STR) profiling. MUM2B, OCM1 and HEK293T cells were cultured in DMEM (Gibco, USA). 92.1, MEL290, OMM2.3, OMM1 and PIG1 cells were cultured in RPMI 1640 medium (Gibco, USA), and CRMM1, CRMM2, CM2005.1 cells were cultured in Ham's F-12 K (Gibco, USA). All medium types were supplemented with 10% certified heat-inactivated fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 mg/ml), and the cells were cultured at 37°C in a humidified 5% CO2 atmosphere.
Dot blot assay
Total RNA was extracted using TRIzol Reagent (Invitrogen, USA), and then was quantified and diluted in 10 mM Tris-EDTA buffer. Equivalent amounts of RNA samples were loaded onto Amersham Hybond-N + membranes (GE Healthcare, USA). The membrane was UV-crosslinked, blocked with 5% milk for 1 h and incubated with an anti-m1A antibody (ab208196, Abcam, USA) at 4°C overnight, followed by HRP-conjugated anti-rabbit IgG (CST, USA) for 1 h at room temperature (RT), and then detected with ECL SuperSignal Western Blotting Detection Reagent (Thermo Fisher Scientific, USA).
Immunofluorescence (IF)
Paraffin-embedded tissues were deparaffinized, rehydrated, fixed with 4% formaldehyde for 30 min, blocked with 5% normal goat serum for 1 h, and permeabilized with 0.5% Triton X-100 for 15 min. Then, they were incubated with the primary antibodies at 4°C overnight and the secondary antibodies for 1 h. Next, DAPI (Sigma-Aldrich, USA) was employed to stain nuclei for 10 min. A ZEISS Axio Scope A1 upright microscope (Germany) and a Nikon Eclipse 80i microscope (Japan) were used to acquire digital images. The following antibodies were used in this study: anti-ALKBH3 (ab251697, Abcam, USA), anti-Pan Kla (1401RM, PTM, China), anti-H3K18la (1406RM, PTM, China), anti-SP100A (NBP1-89457, Novus Biologicals, USA), and anti-PML (ab96051, Abcam, USA).
Western blot
Cell and tissue were lysed with RIPA lysis buffer supplemented with protease and phosphatase inhibitor cocktails (NCM Biotech, China) for 30 min at 4°C and then centrifuged at 13000 × g for 30 min. Protein supernatants were separated by 7.5% (wt/vol) SDS-PAGE and transferred to PVDF membranes (Millipore Corporation, USA). After blocking with 5% milk for 1 h at RT, the membranes were incubated with the primary antibodies at 4°C overnight and then with secondary antibodies conjugated to a fluorescent tag (Invitrogen, USA). The band signals were visualized by an Odyssey Infrared Imaging System (LI-COR, USA). The following primary antibodies were used in the study: anti-ALKBH3 (ab251697, Abcam, USA), anti-LDHA (19987–1-AP, Proteintech, China), anti-LDHB (14824–1-AP, Proteintech, China), anti-SP100 (PA5-53476, ThermoFisher, USA), anti-PML (ab179466, Abcam, USA), anti-YTHDF1 (57530, CST, USA), anti-Pan Kla (1401RM, PTM, China), anti-H3K18la (1406RM, PTM, China), anti-Flag (2368S, CST, USA), anti-ACTB (3700, CST, USA) and anti-Histone H3 (4499, CST, USA). Unprocessed western blot figures were performed in Supplementary Figures S14 and S15.
RNA isolation, reverse transcription, and real-time quantitative PCR (qPCR)
Total RNA was extracted from cultured cells using TRIzol Reagent (Invitrogen, USA), and cDNA was synthesized using PrimeScript RT Reagent Mix (Takara Bio, USA) according to the manufacturer's instructions. qPCR was performed using SYBR Green PCR Master Mix (Life Technologies, USA) and an ABI 7500 real-time PCR system (Applied Biosystems, USA). The levels of targets were normalized to those of ACTB and to those of control samples. The primers for qPCR are listed in Supplementary Table S1.
Plasmid construction
The PGMLV-CMV-MCS-EF1-ZsGreen1-T2A-Puro, PGMLV-hU6-MCS-CMV-ZsGreen1-PGK-Puro-WPRE, ZV502 and ZV102 vectors were used in our study. ALKBH3 shRNAs, SP100A shRNA, YTHDF1 shRNA and their corresponding verified negative control sequences were amplified by PCR and inserted into the PGMLV-hU6-MCS-CMV-ZsGreen1-PGK-Puro-WPRE vectors. The SP100A ORF was generated by PCR and cloned into the PGMLV-CMV-MCS-EF1-ZsGreen1-T2A-Puro vector. The sequences of the constructed plasmids constructed are listed in Supplementary Table S1.
Lentiviral packaging and generation of stable cell lines
A mixture of 3 μg of the indicated plasmid, 3 μg of pMD2.D plasmid, and 6 μg of PsPax plasmid was transfected into HEK239T cells by Lipofectamine 2000 (Invitrogen, USA) in Opti-MEM I Reduced Serum Medium (Gibco, USA). The medium was substituted with fresh complete medium 6 h after transfection. Forty-eight hours and 72 h after transfection, the virus-containing supernatant was collected, filtered by 0.45-mm cellulose acetate filters, and concentrated with a Lenti-X Concentrator (Takara Bio, USA). Medium comprising 25 μL/ml lentivirus and 8 ng/ml polybrene (Sigma-Aldrich, USA) was used to replace the original medium for cells seeded 24 h before transduction. After 48 h, the selection of a stable cell line was performed by incubation with 4 μg/ml puromycin (InvivoGen, USA) for 2 weeks, and a maintenance dose of 1 μg/ml puromycin was used thereafter.
Cell proliferation assays
CCK-8 colorimetric assays were employed to assess cell proliferation capability. A total of 2000–3000 cells were seeded into 96-well plates (Corning, USA) with 100 μL medium. For 2.5 h prior to detection, 10 μL of CCK-8 solution (Dojindo, Japan) was added and the samples were incubated at 37°C. To measure the absorbance of the samples at 450 nm, a microplate reader (ELX800, BioTek, USA) was employed.
Colony formation assay
A total of 2 ml of medium comprising 1000 cells was seeded in a 6-well plate, and the medium was replaced every 3–4 days. After 14–18 days, the colonies were stained with 0.25% crystal violet.
Transwell assay
A 24-well Transwell system with polycarbonate filters (8 μm pores) was employed to evaluate the cell migration capability. A total of 5.0 × 104 –1.0 × 105 cells in medium containing 2% FBS was added to the upper compartment, and 900 μl of complete medium was placed in the lower chamber. After a 24 h incubation, the Transwell system was stained with 0.25% crystal violet. The cells migrating to the lower chamber were imaged.
Xenograft models
The xenograft models were established in a specific pathogen-free (SPF) animal room. Four-week-old male BALB/c nude mice were anesthetized, and their scleras were preperforated with a sharp 30-gauge injection needle. A total of 2 × 106 UM cells were injected into the choroid. The infected eyes were protected with ophthalmic bacitracin ointment. In vivo small animal imaging systems were employed to detect bioluminescence after 28 days. The tumors were fixed with 4% formaldehyde and embedded in paraffin. Then histologic analysis of hematoxylin and eosin (H&E)-stained sections was used to examine tumor formation.
Chromatin immunoprecipitation sequencing (ChIP-seq)
One hundred million cells were fixed with 1% formaldehyde and sonicated for 8 min (10 s on and 15 s off) on ice with a 2-mm microtip with a 40% output control and 90% duty cycle setting. To perform ChIP, sonicated chromatin fragments (150 μl) were diluted 10-fold, and Protein G agarose (60 μl) (Millipore, USA) was added and shaken at 4°C for 2 h. Then, the mixture was briefly centrifuged at 1000 rpm for 5 min at 4°C, and then, the supernatant was collected into a new tube. Anti-H3K18la (PTM-1406) was added to the supernatant and incubated overnight at 4°C. Protein A and protein G magnetic beads (60 μl) (Millipore, USA) were retained in the supernatant for 6 h to pull down the protein at 4°C. The DNA was released from the bound chromatin after cross-linking reversal and proteinase K treatment, precipitated, and diluted in 100 μl of 0.2 M glycine. Purified DNA fragments were constructed and added to ChIP-seq libraries, amplified, and sequenced on an HiSeq 2500 platform (Illumina). The primers used for the quantitative real-time PCR analysis in this study are listed in Supplementary Table S1.
Cleavage under targets and tagmentation (CUT&Tag) and data analysis
The CUT&Tag assay was performed as described previously with modifications (23). In brief, 1 × 105 cells were washed gently with wash buffer (20 mM HEPES pH 7.5; 150 mM NaCl; 0.5 mM spermidine and 1× protease inhibitor cocktail) two times. Next, 10 μl concanavalin A-coated magnetic beads (Bangs Laboratories) was added to each sample and incubated at RT for 10 min. The unbound supernatant and resuspended cells were removed with Dig Wash Buffer (20 mM HEPES, pH 7.5; 150 mM NaCl; 0.5 mM spermidine; 1× protease inhibitor cocktail; 0.05% digitonin, and 2mM EDTA), and the cells were incubated with a 1:50 dilution of an anti-H3K18la (1406RM, PTM, China) primary antibody or IgG control antibody (12–370, Millipore, USA) overnight at 4 °C. The cells were then incubated first with a 1:100 dilution of a secondary antibody (AP132, Millipore, USA) at RT for 60 min, subsequently with a 1:100 dilution of pA-Tn5 adapter complex at RT for 60 min and with tagmentation buffer (10 mM MgCl2 in Dig-med Buffer) at 37°C for 60 min. DNA was purified using phenol–chloroform–isoamyl alcohol extraction and ethanol precipitation. For library amplification, 21 μl of DNA was mixed with 2 μl of a universal i5 primer and a uniquely barcoded i7 primer. A 25-μl volume of NEBNext HiFi 2 × PCR Master Mix was added, and the sample was placed in a thermocycler with a heated lid and processed with the following thermal cycling conditions: 72°C for 5 min (gap filling); 98°C for 30 s; 14 cycles of 98°C for 10 s and 63°C for 30 s; 72°C for 1 min (final extension); and holding at 8°C. Sequencing was performed on an Illumina NovaSeq 6000 system using 150-bp paired-end sequencing following the manufacturer's instructions. The criteria of fold change >2.0 (>1 in the log2 ratio value) and P value <0.05 were employed to identify differentially accessible peaks.
MeRIP-seq and data analysis
MeRIP-Seq, referred to here as m1A-MeRIP-seq, was performed by Cloudseq Biotech Inc. (Shanghai, China) as per published protocols with minor modifications (16). In brief, RNA was randomly fragmented to approximately 200 nt through RNA Fragmentation Reagents and protein A/G beads were connected to an m1A antibody (202003, Synaptic Systems, Germany) by rotating at RT for 1 h. The RNA fragments were incubated with the beads at 4°C for 4 h. Then, the captured RNA was eluted from the beads and extracted with TRIzol Reagent (Invitrogen, USA). Both the input sample and the m1A IP sample were subjected to library generation with the NEBNext Ultra RNA Library Prep Kit (New England Biolab, UK). Libraries were qualified by an Agilent 2100 bioanalyzer (Agilent, USA) and sequenced on a NovaSeq 6000 platform (Illumina, USA). The harvested paired-end reads were subjected to quality control using Q30 and 3′ adaptor trimming by cutadapt software (v1.9.3) to remove low-quality reads (24). Clean reads of all libraries were aligned to the reference genome (HG19) using HISAT2 software (v2.0.4) (25). Methylated sites on RNAs (peaks) were identified through MACS software and differentially methylated sites were identified by diffReps (26,27). The peaks identified overlapping with exons of mRNA were chosen. In addition, GO and pathway enrichment analyses were performed using the differentially methylated protein coding genes.
RNA sequencing (RNA-seq) and data analysis
Total RNA was extracted from the cultured cells by TRIzol Reagent (Invitrogen, USA). A 2100 Bioanalyzer (Agilent Technologies, USA) was used to confirm the integrity of the RNA and a Qubit 2.0 fluorometer with a Qubit RNA Assay Kit (Life Technologies, USA) was employed to measure the concentration. Then, the Illumina TruSeq RNA Sample Prep Kit (San Diego, USA) was used to prepare sequencing libraries and an Illumina HiSeq 2500 platform (San Diego, USA) was used for sequencing. The mRNA levels of the unigenes were identified by TopHat v2.0.9 and Cufflinks and then normalized to FPKM. The criteria of FDR < 0.01 and fold change <0.5 or >2.0 (<−1 or >1 in the log2 ratio value, P value < 0.05) were used to identify differentially expressed genes.
Isobaric tags for relative and absolute quantitation (iTRAQ) proteomic analysis
iTRAQ quantitative proteomic analysis was performed by Applied Protein Technology (Shanghai, China) as previously described (28). In brief, wild-type and ALKBH3-deficient cells were lysed with SDT buffer (4% SDS, 100 mM Tris–HCl, 1 mM DTT, pH 7.6). Protein supernatants were subjected to protein digestion, tandem mass tag (TMT) labeling, fractionation, LC–MS/MS analysis, protein identification, and protein quantitation.
RNA-binding protein immunoprecipitation (RIP)-qPCR
The Magana RIP Quad kit (17-704, Millipore, USA) was employed to examine RNA-binding proteins on specific genes according to the manufacturer's protocols. Briefly, a total of 1.0 × 107 cells were lysed to obtain 200 μl RIP lysate supernatant, of which 15 μl was kept as input control and 150 μl was enriched with antibody- or rabbit IgG-conjugated Protein A/G Magnetic Beads in IP buffer supplemented with RNase inhibitor at 4°C overnight. The immunoprecipitated RNA was digested and purified with the beads. The retrieved RNA and the input control were further analyzed through qPCR. The following primary antibodies were used in the study: anti-YTHDF1 (57530, CST, USA), anti-YTHDF2 (71283, CST, USA) and anti-YTHDF3 (24206, CST, USA).
Luciferase reporter assay
The DNA fragments of the SP100A-5′UTR containing the wild-type m1A motifs and mutant motifs (potential m1A was replaced by T) were synthesized and inserted upstream of firefly luciferase of the pmirGLO vector. Cultured 92.1 cells were transfected with pmirGLO-SP100A-WT-luc or pmirGLO-SP100A-MUT-luc. Relative luciferase activity was evaluated 48 h after transfection by the Dual-Luciferase Reporter Assay System (Promega, USA). The sequences are listed in Supplementary Table S2.
Polysome profiling
Polysome profiling was performed with optimization following the reported protocol (https://www.jove.com/video/51455). In brief, 1.5 × 108 cells were cultured with medium containing 0.1 mg/ml cyclohexamide (CHX) (Sigma-Aldrich, USA) for 5 min at 4°C to freeze the translating ribosomes. Then, the cells were harvested, rinsed with 0.1 mg/ml CHX in precooled PBS and frozen in liquid nitrogen. Next, the cells were homogenized by a liquid nitrogen grinder, lysed with lysis buffer (10 mM Tris–HCl pH 7.5, 100 mM NaCl, 30 mM MgCl2, 0.1 mg/ml CHX, 40 U/ml RNase Inhibitor and 1:100 protease inhibitor) and centrifuged. The supernatant was gently added to a linear 10% to 40% w/v sucrose gradient, ultracentrifuged through a Rotor SW41Ti (Beckman, USA), fractioned by a Gradient Station (BioCamp, USA) equipped with an ECONOUV monitor (Bio-Rad, USA) and collected with a Gilson FC203B fraction collector (Mandel Scientific, Canada). RNA was purified from fractions 2 to 17 and subjected to qPCR analysis.
The Cancer Genome Atlas (TCGA) dataset
To validate the prognostic significance of ALKBH3 and SP100A and determine the correlations between LDHA and ALKBH3, LDHB and ALKBH3, and ALKBH3 and SP100A levels in melanoma, we queried the GEPIA (gepia.cancerpku.cn) and R2 (http://hgserver1.amc.nl/cgibin/r2/main.cgi) to acquire the transcriptional landscape and follow-up information.
Statistics
Prism 8.0 software (GraphPad, USA) was used to perform statistical analysis. Quantification data are presented as the mean ± SD, and the differences between two groups were compared by unpaired Student's t test. Kaplan–Meier curves and log-rank tests were performed for survival analysis. A P value <0.05 was considered statistically significant and asterisks denote statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001).
Results
ALKBH3 is specifically increased in ocular melanoma and is associated with an unfavorable outcome
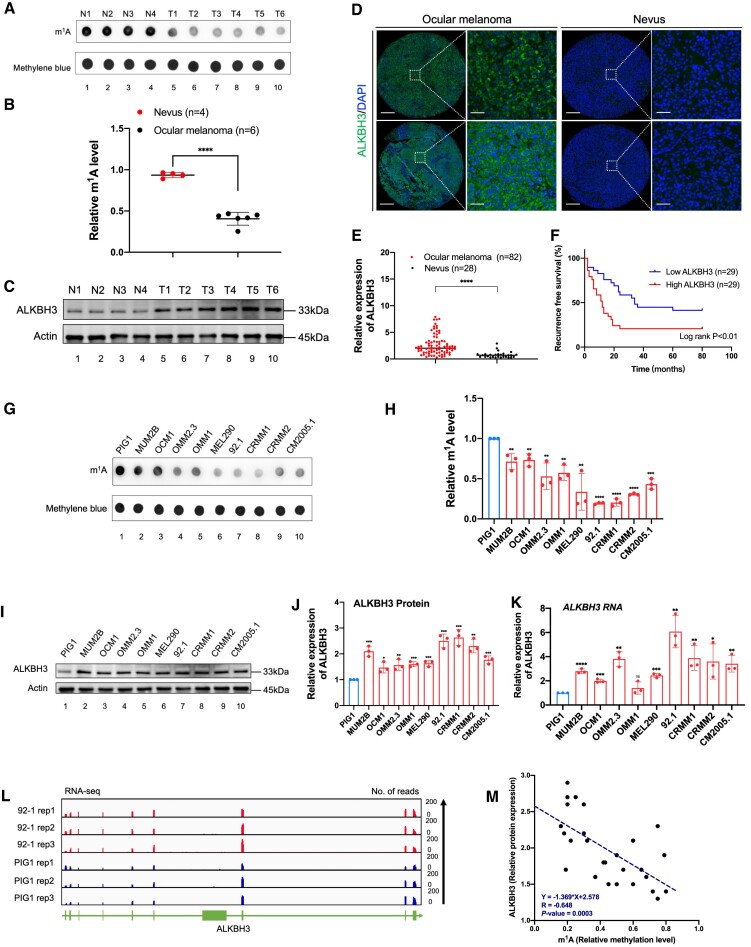
To determine the role of m1A modification in the pathogenesis of ocular melanoma, we first compared the global m1A modification levels between ocular melanoma samples (3 CoM samples and 3 UM samples) and 4 control nevus samples. Notably, ocular melanoma samples presented significantly attenuated m1A levels, as demonstrated by the anti-m1A dot-blot assay (Figure 1A and B). In addition, the demethylase ALKBH3 exhibited increased protein expression in tumors (Figure 1C-E, Supplementary Tables S4 and S5), which is in agreement with the finding of decreased m1A levels in ocular melanomas. Moreover, elevated ALKBH3 is associated with unfavorable outcomes (Figure 1F and Supplementary Tables S4 and S6), further underscoring the importance of ALKBH3 in the carcinogenesis of ocular melanoma.
Figure 1.
Ocular melanoma exhibits increased ALKBH3 expression and decreased m1A levels, which are associated with poor survival. (A) Dot blot showing the m1A signal relative to the methylene blue signal in tumor and normal samples. The data are representative of experimental triplicates. (B) Densitometric analysis showing the expression of m1A relative to methylene blue in tumor and normal samples. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ****P < 0.0001. (C) Western blot showing ALKBH3 expression relative to ACTB expression in tumor and normal samples. The data are representative of experimental triplicates. (D) Immunofluorescence of ALKBH3 (green) and DAPI (blue) in tumor and normal samples. Scale bars: left panel, 100 μm; right panel, 20 μm. (E) Statistical results of ALKBH3 levels in normal and tumor tissues. Significance was determined by unpaired two-tailed Student's t test. ****P < 0.0001. (F) Kaplan–Meier curves of tumor recurrence showing the difference between ocular melanoma patients with low ALKBH3 levels (n = 29) and high ALKBH3 levels (n = 29). log rank test, P < 0.01. (G) Dot blot showing the m1A signal relative to the methylene blue signal in ocular melanoma cells and normal melanocytes. The data are representative of experimental triplicates. (H) Densitometric analysis showing the expression of m1A relative to methylene blue in ocular melanoma cells and normal melanocytes. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (I) Western blot showing ALKBH3 expression relative to ACTB expression in ocular melanoma cells and normal melanocytes. The data are representative of experimental triplicates. (J) Densitometric analysis showing the protein expression of ALKBH3 relative to that of ACTB in ocular melanoma cells and normal melanocytes. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. *P < 0.05, **P < 0.01, ***P < 0.001. (K) qPCR data showing ALKBH3 expression in ocular melanoma cells relative to PIG1 cells. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (L) Integrative Genomics Viewer (IGV) tracks for ALKBH3 from RNA-seq data in an ocular melanoma cell line (92.1) and a normal melanocyte cell line (PIG1). Data were obtained from biological triplicates. (M) Correlation analysis of the relative protein expression of ALKBH3 and the m1A methylation level in ocular melanoma cells and normal melanocytes. Significance was determined by Pearson correlation analysis (R = –0.648, P= 0.0003).
Consistently, in most melanoma cell lines (MUM2B, OCM1, OMM2.3, OMM1, MEL290, 92.1, CRMM1, CRMM2, CM2005.1), a significant decrease in m1A levels (Figure 1G and H) and the increase of ALKBH3 levels (Figure 1I–K) were observed when compared with normal pigmented cells (PIG1). Furthermore, high-throughput transcriptome sequencing further confirmed that ALKBH3 is upregulated in ocular melanoma cell lines (Figure 1L). Importantly, among these cells, ALKBH3 presented a significant negative correlation with m1A levels, suggesting that upregulated ALKBH3 is responsible for the decreased m1A level (Figure 1M).
ALKBH3 accelerates ocular melanoma oncogenesis in vitro and in vivo
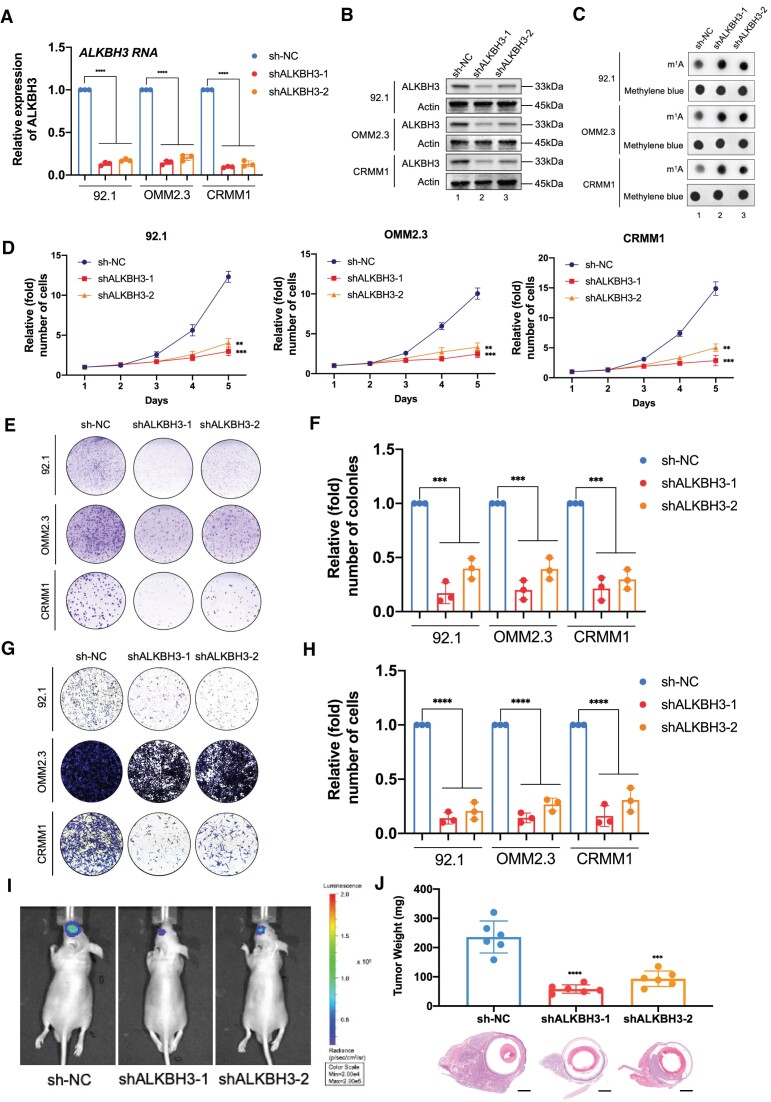
To explore the oncogenic function of ALKBH3, we silenced ALKBH3 expression using two individual shRNAs (Figure 2A and B). Concordantly, a significant increase of m1A levels was observed in ALKBH3-deficient cells (Figure 2C). Moreover, ALKBH3 silencing resulted in significant attenuation of cell growth (Figure 2D) and colony formation capacity (Figure 2E and F) in all tested ocular melanoma cells. In addition, the knockdown of ALKBH3 led to decreased migration capacity, as demonstrated by the Transwell assay (Figure 2G and H). These data support the fact that ALKBH3 serves as a necessary oncogenic factor for malignant proliferation and metastasis of ocular melanoma in vitro. To assess their tumor formation ability in vivo, we injected control and ALKBH3-silenced 92.1 melanoma cells (luciferase-labeled) into nude mice and monitored tumor growth in the orthotopic xenograft model (Supplementary Figure S1A and B). Bioluminescence imaging showed a weaker signal intensity in ALKBH3-deficient ocular melanoma cells than in control cells (Figure 2I). In addition, an ∼80% decrease was noted in the average weight of the xenografts in the ALKBH3-silenced group (Figure 2J). Taken together, these experiments demonstrated that ALKBH3 plays an oncogenic role in the tumorigenesis of ocular melanoma in vitro and in vivo.
Figure 2.
ALKBH3 knockdown increased the m1A levels and suppressed ocular melanoma tumorigenesis. (A) qPCR data showing ALKBH3 expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. The data are presented as the mean± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ****P < 0.0001. (B) Western blot showing ALKBH3 expression relative to ACTB expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. (C) Dot blot showing the m1A signal relative to the methylene blue signal in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. (D) A CCK-8 assay was employed to evaluate the proliferation of ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001. (E) A colony formation assay was employed to evaluate the growth of ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. Representative images from three experimental replicates are shown. (F) Statistical analysis of the colony formation assay data in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001. (G) A transwell assay was employed to evaluate the migration of ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. Representative images from three experimental replicates are shown. (H) Statistical analysis of the transwell assay data in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ****P < 0.0001. (I) Images acquired with an in vivo small animal imaging system showing the suppression of bioluminescent signals in orthotopic xenografts derived from ALKBH3-deficient 92.1 cells. Representative images from six biological replicates are shown. The remaining images are provided in Supplementary Figure 1B. (J) Histograms of the weights of orthotopic xenografts derived from ALKBH3-deficient 92.1 cells. H&E staining was used to visualize tumor tissues. Representative images from six biological replicates are shown. The data are presented as the mean ± SD. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001, ****P< 0.0001. Scale bar: 200 μm.
Histone lactylation potentiates the excessive expression of ALKBH3
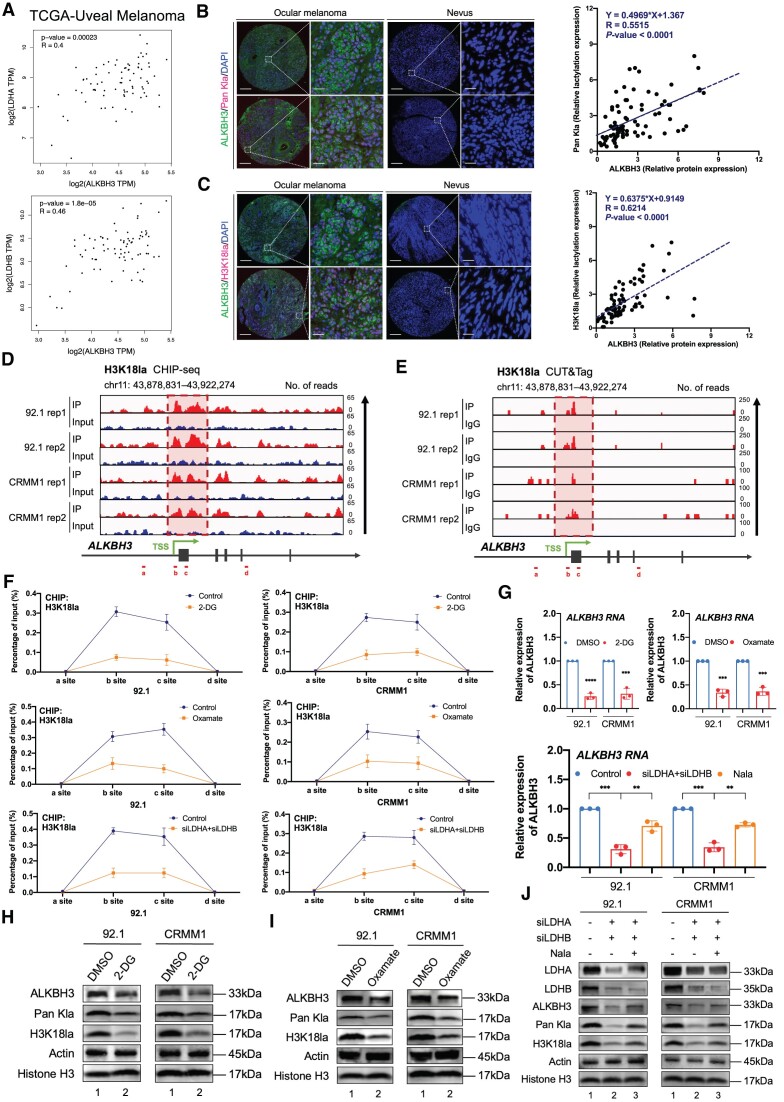
To determine the molecular basis underlying the increased expression of ALKBH3, we have queried TCGA database to screen the genes sharing paralleled expression pattern with ALKBH3. According to GO and KEGG analysis, we found ALKBH3-related genes are enriched in several metabolic terms, including oxidative phosphorylation (P = 1.36e-05), cellular metabolic process (P = 1.50e-05) and carbohydrate derivative metabolic process (P = 4.18e-05), suggesting that elevated ALKBH3 RNA expression could be contributed from the metabolic reprogramming (Supplementary Figure S2A and B). Moreover, the lactate producing enzyme LDHA and LDHB showed significant positive correlation with ALKBH3, which indicates the association between the ALKBH3 and lactate (Figure 3A and B, Supplementary Figure S2C and D). Since previous studies have indicated histone lactylation contributes to the activation of oncogenes (29) and ocular melanoma harbor an increased lactylation level (21), we assume that the increase of ALKBH3 levels could be related to the histone lactylation.
Figure 3.
Histone lactylation potentiates the expression of ALKBH3. (A) Correlation analysis of ALKBH3 expression and LDHA or LDHB expression in the TCGA-UM cohort (n = 80). Significance was determined by Pearson correlation analysis (LDHA: R = 0.4, P = 0.00023, LDHB: R = 0.46, P = 0.000018). (B) Immunofluorescence of ALKBH3 (green), Pan Kla (red) and DAPI (blue) in tumor and normal samples. Scale bars: left panel, 100 μm; right panel, 20 μm. Correlation analysis of the relative protein expression of ALKBH3 and Pan Kla level in ocular melanoma cells and normal melanocytes. Significance was determined by Pearson correlation analysis (R= 0.5515, P < 0.0001). (C) Immunofluorescence of ALKBH3 (green), H3K18la (red) and DAPI (blue) in tumor and normal samples. Scale bars: left panel, 100 μm; right panel, 20 μm. Correlation analysis of the relative protein expression of ALKBH3 and H3K18la level in ocular melanoma cells and normal melanocytes. Significance was determined by Pearson correlation analysis (R= 0.6214, P < 0.0001). (D) IGV tracks from ChIP-seq analysis showing H3K18la enrichment at the promoter of ALKBH3. Sites a–d are distributed in the ALKBH3 genomic region, and sites b and c are the H3K18la peaks. Biological duplicates were analyzed. (E) IGV tracks from CUT&Tag analysis showing H3K18la enrichment at the promoter of ALKBH3. Sites a–d are distributed in the ALKBH3 genomic region, and sites b and c are the H3K18la peaks. Biological duplicates were analyzed. (F) ChIP-qPCR assay of H3K18la status in the ALKBH3genomic region in ocular melanoma cells (92.1 and CRMM1) upon treatment with histone lactylation inhibitors (2-DG or oxamate) or LDHA/B inhibition. The data are presented as the mean ± SD of experimental triplicates. (G) qPCR data showing ALKBH3 expression in ocular melanoma cells (92.1 and CRMM1) upon treatment with histone lactylation inhibitors (2-DG or oxamate) or LDHA/B inhibition. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (H and I) Western blot showing ALKBH3 expression relative to ACTB expression and Pan Kla, H3K18la expression relative to Histone H3 expression in ocular melanoma cells (92.1 and CRMM1) upon histone lactylation inhibitors (2-DG or oxamate). (J) Western blot showing LDHA, LDHB, ALKBH3 expression relative to ACTB expression and Pan Kla, H3K18la expression relative to Histone H3 expression in ocular melanoma cells (92.1 and CRMM1) upon LDHA/B inhibition. Nala, sodium lactate.
We then verified the expression pattern between pan-lactylation and histone lactylation marker (H3K18la) in the ocular melanoma cohorts, which showed a remarkable positive correlation with ALKBH3 protein expression (Figure 3B and C). More importantly, both CUT&Tag and ChIP-seq of H3K18la analysis demonstrated a robust signal of histone lactylation signal, captured in the promoter region of ALKBH3 (deposited in GEO database: GSE242019, Figure 3D and E). Moreover, both histone lactylation inhibitors (oxamate and 2-DG) and LDHA/B inhibition have resulted in a dramatic decreased histone lactylation levels in the promoter region of ALKBH3 (Figure 3F), which subsequently abrogates the RNA (Figure 3G) and protein levels (Figure 3H-J) of ALKBH3 in all tested melanoma cells.
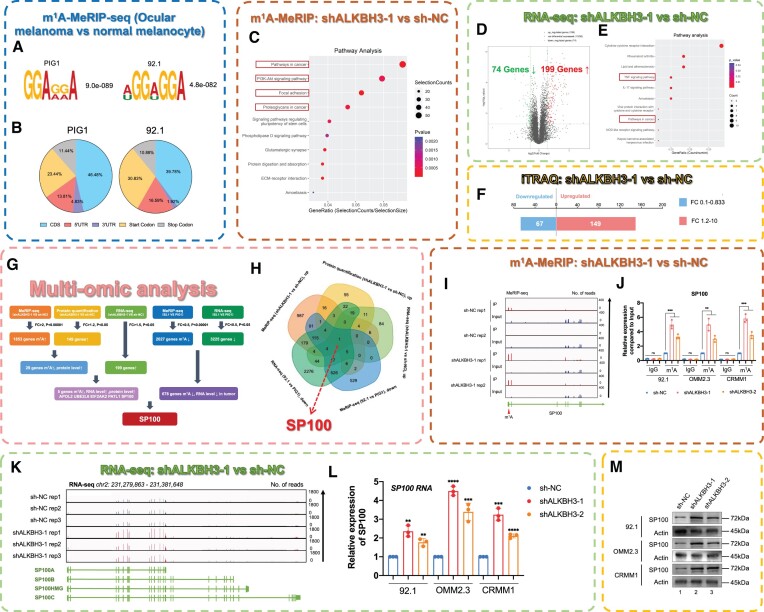
Multi-omic screening identified SP100A as the downstream candidate of ALKBH3
We then explored the mechanism underlying the inhibitory effect induced by ALKBH3 silencing in ocular melanoma cells. Since ALKBH3 is responsible for removing m1A modifications from RNA, we first performed m1A-MeRIP-seq of both ocular melanoma cells and normal melanocytes (deposited in GEO database: GSE213748, Figure 4A). As a result, an average of 16 864 and 10 212 m1A peaks were identified from m1A-MeRIP-seq libraries generated from normal and tumor cells (Figure 4A and B, blue box). Consistent with the previous m1A-meRIP-seq results (30), the m1A peaks were enriched in the 5′ UTR, especially near the start codons (Supplementary Figure S3A). Notably, the differentially expressed m1A-modified genes were associated with multiple melanoma-related pathways, including DNA replication, mTOR/AMPK signaling and melanin synthesis, suggesting a regulatory role of m1A modification in the pathogenesis of ocular melanoma (Supplementary Figure S3B and C).
Figure 4.
ALKBH3 suppresses SP100 expression by removing its m1A modification. (A) m1A-MeRIP-seq data showing the top enriched motifs within m1A peaks identified in ocular melanoma cells and normal melanocytes. (B) Pie charts showing the m1A peak distribution in different RNA regions (CDS, 5′ UTR, 3′ UTR, start codon and stop codon) in ocular melanoma cells and normal melanocytes. (C) KEGG pathway analysis of m1A-modified genes in wild-type and ALKBH3-deficient ocular melanoma cells. (D) Volcano plots showing ALKBH3-regulated genes in wild-type and ALKBH3-deficient ocular melanoma cells. (E) KEGG pathway analysis of ALKBH3-regulated genes in wild-type and ALKBH3-deficient ocular melanoma cells. (F) Histogram showing the protein expression of ALKBH3-regulated genes in wide-type and ALKBH3-deficient ocular melanoma cells. (G and H) Multiomics analysis identified SP100 as a downstream target of ALKBH3. (I) IGV tracks from m1A-meRIP-seq analysis showing m1A enrichment at the 5′UTR of SP100A. Biological duplicates were analyzed. (J) m1A-MeRIP-qPCR assay of m1A status in SP100 in wild-type and ALKBH3-deficient ocular melanoma cells (92.1, OMM2.3 and CRMM1). The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001. (K) IGV tracks for SP100 from RNA-seq data in wild-type and ALKBH3-deficient ocular melanoma cells. Biological triplicates were analyzed. (L) qPCR data showing SP100 RNA expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 knockdown. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (M) Western blot showing SP100 expression relative to ACTB expression in wild-type and ALKBH3-deficient ocular melanoma cells (92.1, OMM2.3 and CRMM1).
Additionally, after silencing ALKBH3, we performed a series of comprehensive high-throughput screenings, including m1A-MeRIP-seq (deposited in GEO database: GSE213748, Figure 4C, brown box), RNA-seq (deposited in GEO database: GSE213681, Figure 4D and E, Supplementary Figure S4, green box) and proteomic analysis (iTRAQ, Figure 4F) for the 92.1 melanoma cell line. Concordantly, we noticed that the change in m1A modification sites was significantly enriched in tumor-related pathways, including PI3K-Akt signaling, focal adhesion and proteoglycan synthesis (Figure 4C). Moreover, the silencing of ALKBH3 results in a remarkable change of the gene expression levels, with 199 upregulated genes and 74 downregulated genes (Figure 4D and E, deposited in GEO database: GSE213681). Consistently, a dramatic genome-wide change in the proteomic levels was observed (149 upregulated and 67 downregulated proteins, Supplementary Table S3, Figure 4F), further underscoring the importance of ALKBH3 in the pathogenesis of ocular melanoma.
Interestingly, combining these multiomics data, we noticed that nuclear autoantigen speckled protein 100 (SP100) was upregulated at both the mRNA and protein levels after silencing ALKBH3 in ocular melanoma cells, following a dramatic change in the m1A modification level (Figure 4G-M). Notably, SP100 is responsible for the formation of PML nuclear bodies, predominantly serving as a suppressor of tumorigenesis across various cancer types, encompassing melanoma, glioblastoma, leiomyosarcoma, breast cancer, and laryngeal cancer (31–35). This observation aligns with the findings regarding the inhibitory efficacy observed in ALKBH3-deficient cells. Importantly, SP100 showed decreased m1A methylation in ocular melanoma cells compared with normal melanocytes (Supplementary Figure S3D). Interestingly, the decrease in m1A modification levels was restored after ALKBH3 inhibition in ocular melanoma cells, as demonstrated by both m1A-MeRIP-seq (deposited in GEO database: GSE213748, Figure 4I) and m1A-MeRIP-qPCR (Figure 4J). Since recent studies have observed that m1A modification of mRNA enhances gene expression and translational efficacy, it is possible that ALKBH3-mediated m1A modification is essential for the expression of the tumor suppressor SP100. We then validated that SP100 was significantly upregulated at the mRNA level after inhibiting ALKBH3, as demonstrated by both RNA-seq (deposited in GEO database: GSE213681, Figure 4K and Supplementary Figure S4) and qPCR (Figure 4L). Consistently, SP100 also presented increased protein levels in ALKBH3-inhibited cells, as shown by iTRAQ (Supplementary Figure S5A and B) and Western blot assays (Figure 4M). Consistently, ALKBH3 exhibited a significant negative correlation with SP100 in a group of 87 metastatic melanoma samples (deposited in GEO database: GSE7553, R = –0.227, P= 0.035, Supplementary Figure S6). Taken together, these data suggest that ALKBH3 may inhibit SP100 expression levels by removing its m1A modification.
SP100A serves as a tumor suppressor in ocular melanoma
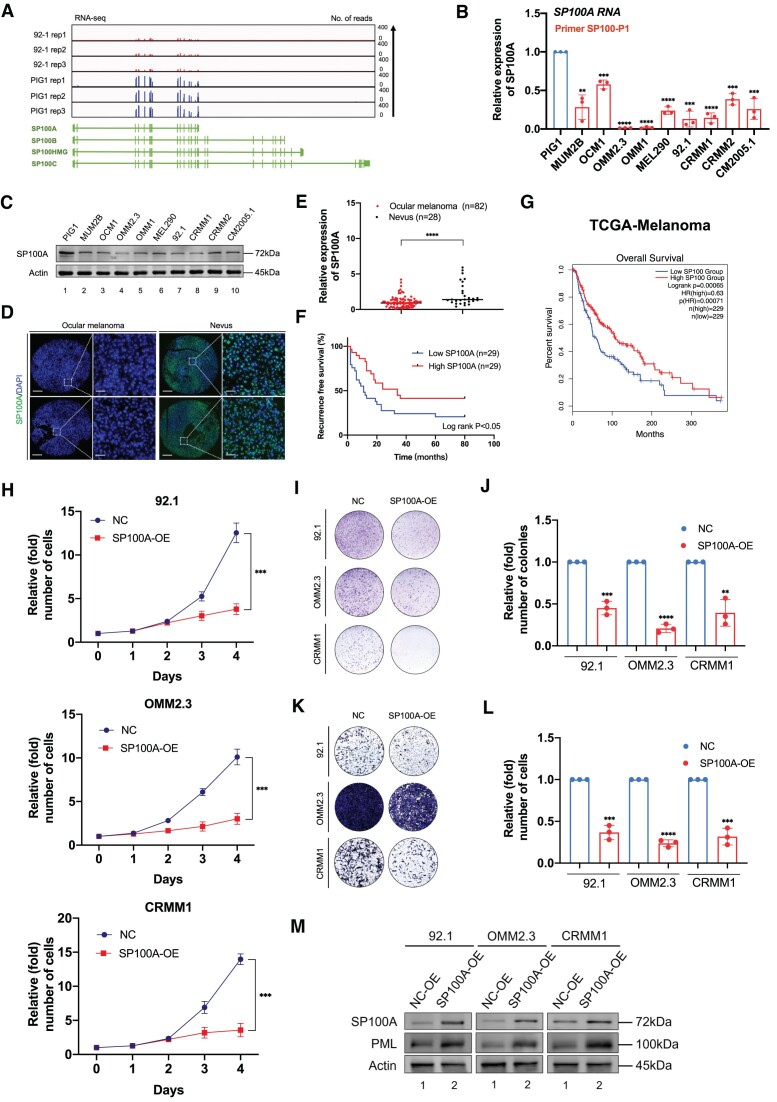
Significantly, in the context of viral infection response, four prominent isoforms of SP100 (namely SP100A, SP100B, SP100C, and SP100HMG) have been discerned (36), prompting us to undertake a comparative analysis of the expression levels exhibited by these transcripts. It is worth mentioning that SP100A exhibits a commonality with SP100B, SP100C and SP100HMG in terms of the initial 15 exons (1–1562 bp), while a disparity of 395-bp in length is observed in the final exon (Supplementary Figure S7A). The RNA-seq data obtained from ocular melanoma cells (92.1) and normal melanocytes (PIG1) revealed a prominent signal in SP100A, while the signal in the exons not included in SP100A was negligible (Figure 5A). Furthermore, we proceeded to conduct qPCR using various primers. These primers encompassed a set designed specifically for SP100A (referred to as SP100-P1), and another set that detects SP100B, SP100C, and SP100HMG (referred to as SP100-P2). The RNA expression of SP100A was observed in ocular melanoma cells and normal pigmented cells (Figure 5B). Since human breast cancer cell line ZR-75-1 was reported to express SP100HMG, we employed ZR-75–1 cells as the positive control (37). Importantly, SP100B/SP100C/SP100HMG could be only detected in ZR-75-1, while undetected in other cell lines (Supplementary Figure S7B). Moreover, the four isoforms have discrepancies in the molecular weight (Supplementary Figure S8A). The full western blots revealed the specific protein expression of SP100A (∼72 kDa) in all tested cell lines (Supplementary Figure S8B and C), which is consistent with the molecular weight of SP100A in the previous studies (18). The findings suggest that the SP100A is abundantly expressed in our experimental context, whereas the expression of other isoforms (SP100B, SP100C and SP100HMG) is negligible.
Figure 5.
SP100A functions as a tumor suppressor in ocular melanoma. (A) IGV tracks for SP100 from RNA-seq data in an ocular melanoma cell line (92.1) and a normal melanocyte cell line (PIG1). Data were obtained from biological triplicates. (B) qPCR data showing the SP100A expression in ocular melanoma cells relative to PIG1 cells. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (C) Western blot showing SP100A expression relative to ACTB expression in ocular melanoma cells and normal melanocytes. The data are representative of experimental triplicates. (D) Immunofluorescence of SP100A (green) and DAPI (blue) in tumor and normal samples. Scale bars: left panel, 100 μm; right panel, 20 μm. (E) Statistical results of SP100A levels in normal and tumor tissues. Significance was determined by unpaired two-tailed Student's t test. ****P < 0.0001. (F) Kaplan–Meier curves of tumor recurrence showing the difference between ocular melanoma patients with low SP100A levels (n = 29) and high SP100A levels (n = 29). log rank test, P < 0.05. (G) Kaplan–Meier analysis of the correlations between SP100 expression and overall survival in TCGA melanoma patients stratified by the SP100 expression level: high (top 50th percentile, n = 229) and low (bottom 50th percentile, n = 229). Significance was determined by a two-sided log-rank test. (H) A CCK8 assay was employed to evaluate the proliferation of ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A overexpression. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001. (I) A colony formation assay was employed to evaluate the growth of ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A overexpression. Representative images from three experimental replicates are shown. (J) Statistical analysis of the colony formation assay data in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A overexpression. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (K) A transwell assay was employed to evaluate the migration of ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A overexpression. Representative images from three experimental replicates are shown. (L) Statistical analysis of the transwell assay data in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A overexpression. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001, ****P < 0.0001. (M) Western blot showing PML expression relative to ACTB expression in wild-type and SP100A-overexpressing ocular melanoma cells (92.1, OMM2.3 and CRMM1).
Notably, a significantly decreased RNA expression level of SP100A was also observed in ocular melanoma cells when compared to the normal pigmented cells, as demonstrated by RNA-seq (Figure 5A), qPCR (Figure 5B) and Western blot assays (Figure 5C). To fully uncover the function of SP100A in ocular melanoma, we then determined SP100A expression in clinical samples of ocular melanoma. Notably, we found that SP100A was significantly decreased in the ocular melanoma samples (Figure 5D and E, Supplementary Tables S4 and S5). More importantly, the loss of SP100A was associated with an unfavorable outcome in both our cohort (Figure 5F and Supplementary Tables S4 and S7) and the TCGA cohort (Figure 5G).
Moreover, according to the single-cell analysis of ocular melanoma samples (Supplementary Figure S9A, GSE139829, using CancerSEA platform (38)), SP100 expression is related to a decreased score of cancer-activation hallmarks, including invasion (R = –0.33, P< 0.001), metastasis (R = –0.28, P< 0.001), cell cycle activation (R = –0.19, P< 0.001), proliferation (R = –0.16, P< 0.001) and epithelial-to-mesenchymal transition (R = –0.16, P< 0.001). Collectively, these data indicate that SP100A is downregulated and potentially inhibits several oncogenic events in ocular melanoma.
Since SP100A was downregulated in ocular melanoma, we exogenously overexpressed SP100A in three ocular melanoma cell lines (Supplementary Figure S9B). Interestingly, all tested ocular melanoma cells presented with attenuated proliferation ability after overexpressing SP100A (Figure 5H). Moreover, SP100A-overexpressing melanoma cells formed smaller and fewer colonies than the control group (Figure 5I and J). In addition, a significant inhibition of cancer metastasis capacity was observed after introducing SP100A in ocular melanoma cells (Figure 5K and L).
Most importantly, since SP100A serves as the molecular scaffold within PML bodies (39), we then assessed the PML expression in SP100A-overexpressing cells. As a result, the exogenous overexpression of SP100A resulted in a noteworthy increase in the protein level of PML (Figure 5M). Concordantly, the IF analysis demonstrated a notable augmentation in the presence of PML bodies in SP100A-overexpressing cells (Supplementary Figure S10A). Collectively, this data further confirmed that SP100A is essential for PML expression, consistent with previous research (40). Moreover, ALKBH3-deficient cells exhibited a substantial elevation in PML bodies (Supplementary Figure S10B), which is in alignment with the pivotal role of ALKBH3 in the regulation of SP100A expression. Taken together, these gain-of-function data revealed that SP100A is responsible for the formation of PML nuclear condensates, serving as a tumor suppressor in ocular melanoma.
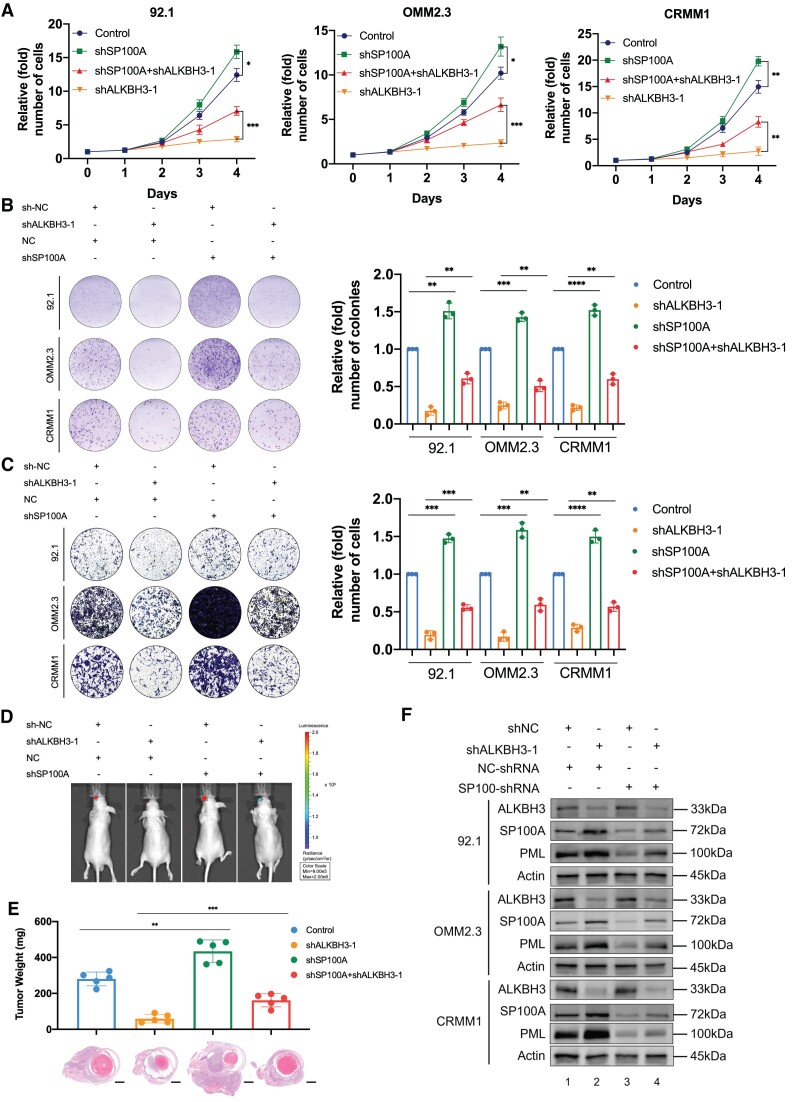
Silencing of SP100A partially compromised tumor inhibition efficacy in ALKBH3-deficient cells
To further verify the relationship between ALKBH3 and SP100A expression, we further altered SP100A expression after ALKBH3 inhibition in ocular melanoma cells by transfecting a reported shRNA against SP100A (41). As expected, SP100A expression was significantly decreased after transfecting SP100A-shRNA into three ocular melanoma cells at both the RNA (Supplementary Figure S11A) and protein (Supplementary Figure S11B) levels. In cell growth, the depletion of SP100A partially rescued (∼50–60%) the inhibition guided by ALKBH3 inhibition (Figure 6A, red line), and SP100A-deficient cells were much more resistant to the deprivation of ALKBH3 (Figure 6A). Similarly, SP100A-silenced cells presented with more colonies than the control group (Figure 6B, red and orange column). Moreover, SP100A inhibition significantly enhanced cell migration ability and compromised the inhibitory efficacy in ALKBH3-deficient melanoma cells (Figure 6C). Most importantly, the knockdown of SP100A further rescued orthotopic tumor formation in ALKBH3-depleted melanoma cells (Figure 6D and E, Supplementary Figure S12A and B). Consistently, the stable knockdown of SP100A resulted in a compromised expression of PML expression in both wild-type (Figure 6F, lanes 1 and 3) and ALKBH3-deficient ocular melanoma cells (Figure 6F, lanes 2 and 4). Together, these results suggested that ALKBH3 promotes ocular melanoma through the decrease of SP100A-mediated PML bodies both in vivo and in vitro.
Figure 6.
The anticancer effects of ALKBH3 knockdown were partially blocked by SP100A silencing. (A) A CCK8 assay was performed to assess the proliferation of ALKBH3-deficient ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A silencing. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. *P < 0.05, **P < 0.01, ***P < 0.001. (B) A colony formation assay was performed to assess the growth of ALKBH3-deficient ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A silencing. Representative images from three experimental replicates are shown. The data are presented as the mean ± SD. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (C) A transwell assay was performed to assess the migration of ALKBH3-deficient ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon SP100A silencing. Representative images from three experimental replicates are shown. The data are presented as the mean ± SD. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001, ****P < 0.0001. (D) Images acquired with an in vivo small animal imaging system showing the suppression of bioluminescent signals in orthotopic xenografts derived from ALKBH3-deficient 92.1 cells upon SP100A silencing. Representative images from five biological replicates are shown. The remaining images are provided in Supplementary Figure S12B. (E) Histograms of the weights of orthotopic xenografts derived from ALKBH3-deficient 92.1 cells upon SP100A silencing. H&E staining was used to visualize tumor tissues. Representative images from five biological replicates are shown. The data are presented as the mean ± SD values. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001. Scale bar: 200 μm. (F) Western blot showing ALKBH3, SP100A, PML expression relative to ACTB expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 and SP100A knockdown.
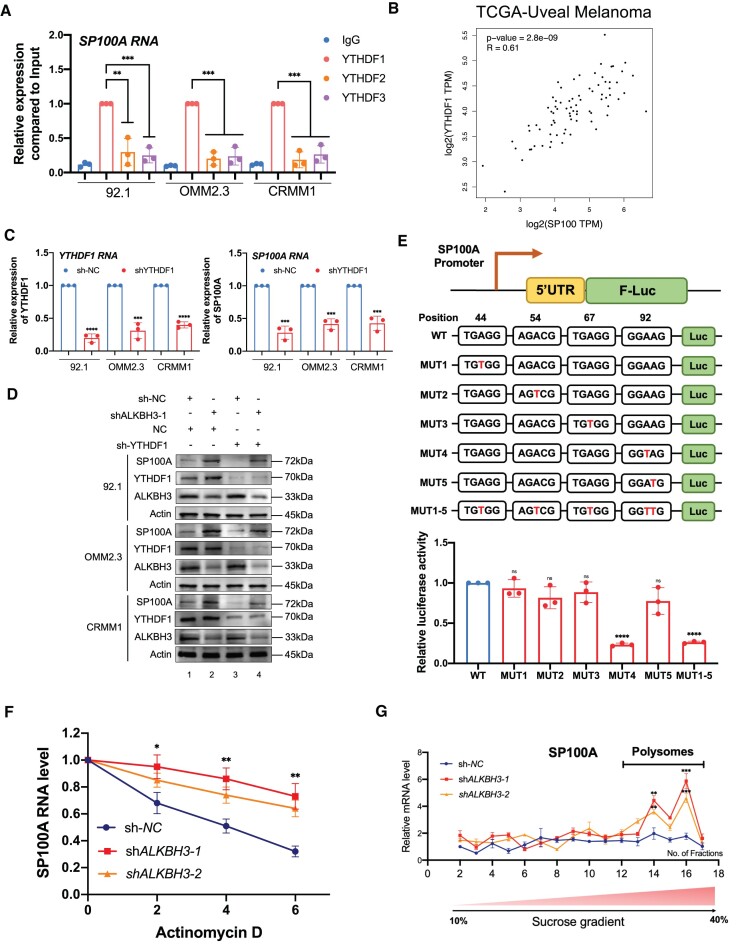
m1A modification of SP100A enhances its RNA stability and translational efficacy
We then explored the epigenetic mechanisms underlying the m1A modification of SP100A. Since previous studies have demonstrated that YTHDF proteins are responsible for the recognition of m1A methylation, we first tested the RNA-binding status of SP100A mRNA with YTHDF1, YTHDF2 and YTHDF3. RNA-immunoprecipitation analysis demonstrated that YTHDF1 specifically recognizes SP100A mRNA; however, YTHDF2 and YTHDF3 only presented with limited interaction intensities (Figure 7A). Moreover, YTHDF1 exhibited a salient positive correlation (R = 0.61, P< 0.0001) with SP100A expression in the TCGA cohort (Figure 7B), which is in perfect alignment with the hypothesis that YTHDF1 is necessary for the recognition of SP100A. Moreover, YTHDF1 silencing dramatically inhibited SP100A expression and fully rescued ALKBH3 silencing-mediated SP100A levels elevation (Figure 7C and D). Taken together, these data showed that YTHDF1 functions as the reader protein of SP100A.
Figure 7.
m1A modification of SP100A increases its RNA stability and translational efficacy. (A) RIP-qPCR assay of SP100A expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) by YTHDF1, YTHDF2 and YTHDF3. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. **P < 0.01, ***P < 0.001. (B) Correlation analysis of SP100A expression and YTHDF1 expression in the TCGA-UM cohort (n = 80). Significance was determined by Pearson correlation analysis (R = 0.61, P < 0.0001). (C) qPCR data showing YTHDF1 RNA expression and SP100A RNA in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon YTHDF1 knockdown. The data are presented as the mean ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001, ****P< 0.0001. (D) Western blot showing YTHDF1, SP100A, ALKBH3 expression relative to ACTB expression in ocular melanoma cells (92.1, OMM2.3 and CRMM1) upon ALKBH3 and YTHDF1 knockdown. (E) The luciferase reporter gene assay demonstrated the relative luciferase activity of the wild-type and six mutant SP100A 5′UTR reporter vectors. The data are presented as the means ± SD of experimental triplicates. Significance was determined by unpaired two-tailed Student's t test. ****P < 0.0001. (F) Half-life of SP100A in wild-type and ALKBH3-deficient 92.1 cells treated with actinomycin (5 g/ml) for 0–6 h. (G) Polysome profiling assays of 92.1 cells with or without ALKBH3 knockdown. RNAs in different ribosome fractions were extracted and subjected to qPCR analysis. Data are shown as the mean ± SD. Significance was determined by unpaired two-tailed Student's t test. ***P < 0.001, ****P < 0.0001.
We then determined the specific m1A modification site of SP100A mRNA. In the 5′ UTR of SP100A, we found five potential m1A sites according to the identified peak (Figure 4I, Supplementary Figure S3D) [c.44A (TGTGG), c.54A (AGACG) and c.67A (TGAGG), and c. 92A/c. 93A (GGAAG)] and mutated each A into a T. We then cloned the corresponding wild-type and mutated 5′ UTRs into the pmirGLO vector (Figure 7E, upper panel). The luciferase reporter gene assay demonstrated that c.A92T presented a decreased signal, while the signals in the other mutated groups remained unchanged (Figure 7E). Furthermore, we performed RIP-qPCR to identify the interaction frequency between YTHDF1 and reporter transcripts (F-luc). We observed that pmirGLO-MUT4 (c.A92T) transcript presented a decreased binding affinity between YTHDF1 and F-Luc, while other transcripts remained unchanged (Supplementary Figure S13A). This result agrees with the previous observation that YTHDF1 directly interacts with m1A methylated sequences, which was regarded as ‘Reader’ for m1A in RNA (42,43). Moreover, we found that ALKBH3-deficient cells presented with enhanced RNA stability of SP100A, which agrees with the increase of SP100A RNA expression after ALKBH3 depletion (Figure 7F). Importantly, polysome profiling in 92.1 cells showed that stable knockdown of ALKBH3 resulted in pronounced SP100A mRNA abundance in polysome portions (Figure 7G, Supplementary Figure S13B), which generally harbor effective translation capacity (44). This observation is in alignment with a previous conclusion that m1A modification in early exons leads to enhanced translational capacity (30). Notably, the nascent SP100A RNA expression remained unaltered upon ALKBH3 knockdown (Supplementary Figure S13C). Collectively, these results demonstrated that the m1A RNA methylation of SP100A mRNA contributes to increased RNA stability and translational capacity post-transcriptionally.
Discussion
In this study, we described that lactylation-driven ALKBH3 elevation contributes to the SP100A m1A demethylation and inhibits its RNA and protein expression. The decreased expression of SP100A further results in the loss of PML bodies, which initiate the progression of ocular melanoma. Notably, PML bodies are pan-cancer pleiotropic tumor suppressors that form phase-separated liquid-like condensates and are organized by multivalent interactions among proteins and RNA molecules. Albeit m1A RNA modification plays key roles in the regulation of RNA metabolism and gene expression, the relationship between m1A modification and PML formation remains enigmatic. In this study, we revealed for the first time that ALKBH3-mediated m1A modification plays an inhibitory role in the formation of the PML body, which bridges our understanding of dynamic RNA modifications and phase-separation events in cancers.
Loss of homeostasis of the m1A modification has been associated with several oncogenic hallmarks. For example, ALKBH3-mediated m1A demethylation of ATP5D facilitates its expression in metabolic reprogramming during carcinogenesis (9). Moreover, ALKBH3 loss induces m1A hypermethylation of the transcriptome in Hodgkin lymphoma, which is associated with poor clinical outcome (14). In addition, m1A modification regulates colony-stimulating factor 1 expression in breast and ovarian cancer cells, which indicates that m1A modification is responsible for regulating immune surveillance in cancer (45). m1A modification also modulates tRNA stability, which plays an important role in regulating apoptosis (15). In this study, we show that the m1A modification of SP100A is responsible for the formation of tumor-suppressive PML bodies. To our knowledge, this is the first study to demonstrate that m1A is necessary for tumor suppressor gene expression, which expands the current understandings of dynamic m1A function.
Histone lactylation, serving as a metabolic stress-related histone modification, promotes cardiac repair, macrophage polarization and facilitates carcinogenesis and progression (29,46). For example, histone lactylation fuels early activation of the reparative transcriptional response in monocytes, regulating immune homeostasis in cardiac repair process (47). Moreover, enhanced histone lactylation also contributes M1 macrophage polarization during bacterial infection (29). In this study, we revealed that histone lactylation is also responsible for the oncogenic activation of the ALKBH3, contributing to the hypo-m1A status in cancers. While lactylation inhibitors such as oxamate and 2-DG, along with LDHA/B inhibition, demonstrate effective regulation of lactylation levels, they also elicit additional undesirable consequences (48). Furthermore, due to the shared regulators (eg. EP300 and HDACs) between histone lactylation and other forms of histone modifications (49), the ability to specifically modulate histone lactylation level is constrained.
SP100 was first identified as a nuclear autoimmune antigen and is a constituent of the nuclear body, serving as an important tumor suppressor in diverse cancers, including pancreatic cancer, leukemia and glioma (32,39,50). SP100 plays a vital role in the response to interferon, regulating immune surveillance in carcinogenesis (51,52). Moreover, SP100 interacts with the ETS1 transcription factor, reducing ETS1-DNA binding and impairing ETS1 transcriptional activity on the promoter of several oncogenes (34,53). SP100A, as an important functional isoform of SP100, is a tumor suppressor that activates p53-dependent transcription and counteracts E1A/E1B-55K-mediated transformation (54).
In addition, SP100 was discovered to exert its tumor-suppressive effects when it functions as a constituent of PML nuclear bodies. Notably, PML nuclear bodies have been recognized as tumor suppressors due to their ability to impede the progression of the cell cycle, initiate apoptosis, and inhibit angiogenesis in cancerous cells (55). For example, PML nuclear bodies mediate the p53 response to oncogenic signals and induce senescence. The integrity of the PML nuclear bodies is essential for p53-dependent senescence (56). Moreover, PML nuclear bodies regulate TNFα-induced cell death in cancer cells and suppress capillary tube formation and migration in endothelial cells (57). In this study, we revealed for the first time that SP100A also function as a tumor suppressor in ocular melanoma, providing a novel therapeutic target for vision- and life- threatening ocular melanoma.
Notably, loss of SP100A only partially rescues the ALKBH3-knockdown inhibitory effect, which may be attributed to the diversified functions of ALKBH3 in different RNA categories. ALKBH3, as the demethylase of m1A methylation, mediates m1A methylation in tRNAs, rRNAs, mRNAs, and mitochondrial RNAs (3). For example, ALKBH3-mediated tRNA demethylation efficiently promotes cancer progression via induction of tRNA-derived small RNAs, which prevents apoptosis induced by cytochrome C (15). Moreover, the m1A modification of tRNA also leads to liver tumorigenesis by regulating cholesterol metabolism (8). However, the function of ALKBH3-mediated m1A demethylation of tRNAs/rRNAs in the oncogenesis of ocular melanoma remains enigmatic, which warrants further explorations.
In summary, our results reveal a completely novel model of tumorigenesis in which m1A modification of SP100A mRNA promotes the formation of PML nuclear condensates. Additionally, this study for the first time reveals that m1A is responsible for the activation of tumor suppressor gene and unveils crosstalk between histone lactylation, mRNA m1A modification, and phase-separation-mediated condensate formation, thereby providing a novel therapeutic method of targeted m1A reprogramming for efficient for tumor treatment.
Supplementary Material
Acknowledgements
We thank all the ocular melanoma patients enrolled in our study and wish them good health.
Author contributions: R.J., X.F. and P.C. conceived and supervised the whole project. X.G., J.Y. and L.Y. performed the experiments. X.G. and A.Z. performed data analysis. S.G. and J.R. performed sample collection. X.G. and P.C. wrote the manuscript. All authors have read and approved the manuscript.
Contributor Information
Xiang Gu, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Ai Zhuang, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Jie Yu, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Ludi Yang, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Shengfang Ge, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Jing Ruan, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Renbing Jia, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Xianqun Fan, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Peiwei Chai, Department of Ophthalmology, Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, People's Republic of China.
Data availability
The raw sequence data reported in this paper, including CUT&Tag, ChIPseq, m1A-meRIP-seq and RNA-seq data, have been deposited in the Gene Expression Omnibus database (GSE176345, 213681, 213748 and 242019).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Funding for open access charge: National Natural Science Foundation of China [82373298, 82103240, 82388101, U23A20466]; Science and Technology Commission of Shanghai [20DZ2270800, 23ZR1438400, 23ZR1480100, 23YF1422400]; China Postdoctoral Science Foundation [2023M732290]; Shanghai Key Clinical Specialty, Shanghai Eye Disease Research Center [2022ZZ01003]; Innovative Research Team of High-level Local Universities in Shanghai [SHSMU-ZDCX20210900, SHSMU-ZDCX20210902]; Shanghai Municipal Health Commission (GWVI-11.2-YQ19).
Conflict of interest statement. None declared.
References
- 1. Barbieri I., Kouzarides T. Role of RNA modifications in cancer. Nat. Rev. Cancer. 2020; 20:303–322. [DOI] [PubMed] [Google Scholar]
- 2. Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boriack-Sjodin P.A., Ribich S., Copeland R.A. RNA-modifying proteins as anticancer drug targets. Nat. Rev. Drug Discov. 2018; 17:435–453. [DOI] [PubMed] [Google Scholar]
- 4. Choi J., Indrisiunaite G., DeMirci H., Ieong K.W., Wang J., Petrov A., Prabhakar A., Rechavi G., Dominissini D., He C. et al. 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 2018; 25:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han X., Wang M., Zhao Y.L., Yang Y., Yang Y.G. RNA methylations in human cancers. Semin. Cancer Biol. 2021; 75:97–115. [DOI] [PubMed] [Google Scholar]
- 6. Safra M., Sas-Chen A., Nir R., Winkler R., Nachshon A., Bar-Yaacov D., Erlacher M., Rossmanith W., Stern-Ginossar N., Schwartz S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017; 551:251–255. [DOI] [PubMed] [Google Scholar]
- 7. Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G. et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016; 167:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y., Wang J., Li X., Xiong X., Wang J., Zhou Z., Zhu X., Gu Y., Dominissini D., He L. et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021; 12:6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu Y., Chen Z., Xie G., Zhang H., Wang Z., Zhou J., Chen F., Li J., Chen L., Niu H. et al. RNA m(1)A methylation regulates glycolysis of cancer cells through modulating ATP5D. Proc. Natl. Acad. Sci. U.S.A. 2022; 119:e2119038119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuang W., Jin H., Yang F., Chen X., Liu J., Li T., Chang Y., Liu M., Xu Z., Huo C. et al. ALKBH3-dependent m(1)A demethylation of Aurora A mRNA inhibits ciliogenesis. Cell Discov. 2022; 8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao Y., Zhao Q., Kaboli P.J., Shen J., Li M., Wu X., Yin J., Zhang H., Wu Y., Lin L. et al. m1A regulated genes modulate PI3K/AKT/mTOR and ErbB pathways in gastrointestinal cancer. Transl Oncol. 2019; 12:1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao L., Chen R., Sugimoto M., Mizuta M., Kishimoto Y., Omori K. The impact of m1A methylation modification patterns on tumor immune microenvironment and prognosis in oral squamous cell carcinoma. Int. J. Mol. Sci. 2021; 22:10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pier G.B., Thomas D.M. Characterization of the human immune response to a polysaccharide vaccine from Pseudomonas aeruginosa. J. Infect. Dis. 1983; 148:206–213. [DOI] [PubMed] [Google Scholar]
- 14. Esteve-Puig R., Climent F., Piñeyro D., Domingo-Domènech E., Davalos V., Encuentra M., Rea A., Espejo-Herrera N., Soler M., Lopez M. et al. Epigenetic loss of m1A RNA demethylase ALKBH3 in Hodgkin lymphoma targets collagen, conferring poor clinical outcome. Blood. 2021; 137:994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Z., Qi M., Shen B., Luo G., Wu Y., Li J., Lu Z., Zheng Z., Dai Q., Wang H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019; 47:2533–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016; 12:311–316. [DOI] [PubMed] [Google Scholar]
- 17. Zhou H., Rauch S., Dai Q., Cui X., Zhang Z., Nachtergaele S., Sepich C., He C., Dickinson B.C. Evolution of a reverse transcriptase to map N(1)-methyladenosine in human messenger RNA. Nat. Methods. 2019; 16:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Griewank K.G., Westekemper H., Murali R., Mach M., Schilling B., Wiesner T., Schimming T., Livingstone E., Sucker A., Grabellus F. et al. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin. Cancer Res. 2013; 19:3143–3152. [DOI] [PubMed] [Google Scholar]
- 19. Jager M.J., Shields C.L., Cebulla C.M., Abdel-Rahman M.H., Grossniklaus H.E., Stern M.H., Carvajal R.D., Belfort R.N., Jia R., Shields J.A. et al. Uveal melanoma. Nat. Rev. Dis. Primers. 2020; 6:24. [DOI] [PubMed] [Google Scholar]
- 20. Jia R., Chai P., Wang S., Sun B., Xu Y., Yang Y., Ge S., Jia R., Yang Y.G., Fan X. m(6)A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol. Cancer. 2019; 18:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu J., Chai P., Xie M., Ge S., Ruan J., Fan X., Jia R. Histone lactylation drives oncogenesis by facilitating m(6)A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021; 22:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chai P., Yu J., Jia R., Wen X., Ding T., Zhang X., Ni H., Jia R., Ge S., Zhang H. et al. Generation of onco-enhancer enhances chromosomal remodeling and accelerates tumorigenesis. Nucleic Acids Res. 2020; 48:12135–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019; 10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kechin A., Boyarskikh U., Kel A., Filipenko M. cutPrimers: a New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017; 24:1138–1143. [DOI] [PubMed] [Google Scholar]
- 25. Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu T. Use model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol. Biol. 2014; 1150:81–95. [DOI] [PubMed] [Google Scholar]
- 27. Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013; 8:e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xi Y., Liu M., Xu S., Hong H., Chen M., Tian L., Xie J., Deng P., Zhou C., Zhang L. et al. Inhibition of SERPINA3N-dependent neuroinflammation is essential for melatonin to ameliorate trimethyltin chloride-induced neurotoxicity. J. Pineal Res. 2019; 67:e12596. [DOI] [PubMed] [Google Scholar]
- 29. Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M. et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019; 574:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X., Xiong X., Zhang M., Wang K., Chen Y., Zhou J., Mao Y., Lv J., Yi D., Chen X.W. et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell. 2017; 68:993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jinesh G.G., Smallin M.T., Mtchedlidze N., Godwin I., Napoli M., Hackel N., Phadke M.S., Deshpande A.A., Li X., Lockhart J.H. et al. C19MC miRNA-520G induces SP100 antiviral gene transcription and inhibits melanin production in skin cutaneous melanoma. Genes Dis. 2024; 11:60–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Held-Feindt J., Hattermann K., Knerlich-Lukoschus F., Mehdorn H.M., Mentlein R. SP100 reduces malignancy of human glioma cells. Int. J. Oncol. 2011; 38:1023–1030. [DOI] [PubMed] [Google Scholar]
- 33. Chudasama P., Mughal S.S., Sanders M.A., Hübschmann D., Chung I., Deeg K.I., Wong S.H., Rabe S., Hlevnjak M., Zapatka M. et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat. Commun. 2018; 9:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yordy J.S., Li R., Sementchenko V.I., Pei H., Muise-Helmericks R.C., Watson D.K. SP100 expression modulates ETS1 transcriptional activity and inhibits cell invasion. Oncogene. 2004; 23:6654–6665. [DOI] [PubMed] [Google Scholar]
- 35. Li W., Shang C., Guan C., Zhang Y., Sun K., Fu W. Low expression of Sp100 in laryngeal cancer: correlation with cell differentiation. Med. Sci. Monit. 2010; 16:Br174–BR178. [PubMed] [Google Scholar]
- 36. Negorev D.G., Vladimirova O.V., Ivanov A., Rauscher F. 3rd, Maul G.G. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J. Virol. 2006; 80:8019–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seeler J.S., Marchio A., Sitterlin D., Transy C., Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:7316–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Durante M.A., Rodriguez D.A., Kurtenbach S., Kuznetsov J.N., Sanchez M.I., Decatur C.L., Snyder H., Feun L.G., Livingstone A.S., Harbour J.W. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020; 11:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corpet A., Kleijwegt C., Roubille S., Juillard F., Jacquet K., Texier P., Lomonte P. PML nuclear bodies and chromatin dynamics: catch me if you can!. Nucleic Acids Res. 2020; 48:11890–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu P., Roizman B. The SP100 component of ND10 enhances accumulation of PML and suppresses replication and the assembly of HSV replication compartments. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3823–E3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Milovic-Holm K., Krieghoff E., Jensen K., Will H., Hofmann T.G. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 2007; 26:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ren W., Yuan Y., Li Y., Mutti L., Peng J., Jiang X. The function and clinical implication of YTHDF1 in the human system development and cancer. Biomark. Res. 2023; 11:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dai X., Wang T., Gonzalez G., Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal. Chem. 2018; 90:6380–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu Y., He X., Wang S., Sun B., Jia R., Chai P., Li F., Yang Y., Ge S., Jia R. et al. The m(6)A reading protein YTHDF3 potentiates tumorigenicity of cancer stem-like cells in ocular melanoma through facilitating CTNNB1 translation. Oncogene. 2022; 41:1281–1297. [DOI] [PubMed] [Google Scholar]
- 45. Woo H.H., Chambers S.K. Human ALKBH3-induced m(1)A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim Biophys Acta Gene Regul Mech. 2019; 1862:35–46. [DOI] [PubMed] [Google Scholar]
- 46. Izzo L.T., Wellen K.E. Histone lactylation links metabolism and gene regulation. Nature. 2019; 574:492–493. [DOI] [PubMed] [Google Scholar]
- 47. Wang N., Wang W., Wang X., Mang G., Chen J., Yan X., Tong Z., Yang Q., Wang M., Chen L. et al. Histone lactylation boosts reparative gene activation post-myocardial infarction. Circ. Res. 2022; 131:893–908. [DOI] [PubMed] [Google Scholar]
- 48. Martin-Requero A., Ayuso M.S., Parrilla R. Rate-limiting steps for hepatic gluconeogenesis. Mechanism of oxamate inhibition of mitochondrial pyruvate metabolism. J. Biol. Chem. 1986; 261:13973–13978. [PubMed] [Google Scholar]
- 49. Zeng Q., Wang K., Zhao Y., Ma Q., Chen Z., Huang W. Effects of the acetyltransferase p300 on tumour regulation from the novel perspective of posttranslational protein modification. Biomolecules. 2023; 13:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nakayama H., Ohuchida K., Yonenaga A., Sagara A., Ando Y., Kibe S., Takesue S., Abe T., Endo S., Koikawa K. et al. S100P regulates the collective invasion of pancreatic cancer cells into the lymphatic endothelial monolayer. Int. J. Oncol. 2019; 55:211–222. [DOI] [PubMed] [Google Scholar]
- 51. Stepp W.H., Meyers J.M., McBride A.A. Sp100 provides intrinsic immunity against human papillomavirus infection. mBio. 2013; 4:e00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Collados Rodríguez M. The fate of speckled protein 100 (Sp100) during herpesviruses infection. Front. Cell. Infect. Microbiol. 2020; 10:607526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yordy J.S., Moussa O., Pei H., Chaussabel D., Li R., Watson D.K. SP100 inhibits ETS1 activity in primary endothelial cells. Oncogene. 2005; 24:916–931. [DOI] [PubMed] [Google Scholar]
- 54. Berscheminski J., Brun J., Speiseder T., Wimmer P., Ip W.H., Terzic M., Dobner T., Schreiner S. Sp100A is a tumor suppressor that activates p53-dependent transcription and counteracts E1A/E1B-55K-mediated transformation. Oncogene. 2016; 35:3178–3189. [DOI] [PubMed] [Google Scholar]
- 55. Bernardi R., Pandolfi P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007; 8:1006–1016. [DOI] [PubMed] [Google Scholar]
- 56. Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P.P. et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000; 406:207–210. [DOI] [PubMed] [Google Scholar]
- 57. Hsu K.S., Guan B.J., Cheng X., Guan D., Lam M., Hatzoglou M., Kao H.Y. Translational control of PML contributes to TNFα-induced apoptosis of MCF7 breast cancer cells and decreased angiogenesis in HUVECs. Cell Death Differ. 2016; 23:469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper, including CUT&Tag, ChIPseq, m1A-meRIP-seq and RNA-seq data, have been deposited in the Gene Expression Omnibus database (GSE176345, 213681, 213748 and 242019).