ABSTRACT
In bacteria, the Sec translocase mediates the translocation of proteins into and across the cytoplasmic membrane. It consists of a protein conducting channel SecYEG, the ATP-dependent motor SecA, and the accessory SecDF complex. Here we discuss the function and structure of the Sec translocase.
INTRODUCTION
Protein transport occurs in all domains of life (1). Proteins that function outside the cytosol are translocated across membranes. The general system for protein translocation is formed by the Sec translocase at its core the translocon: SecYEG in bacteria (2), SecYEβ in archaea (3), and Sec61αβγ in the endoplasmic reticulum of eukaryotes (4, 5). The translocon forms a protein conducting channel in the membrane for unfolded preproteins (6) but also mediates cotranslational insertion of nascent membrane proteins into the membrane (Fig. 1).
FIGURE 1.
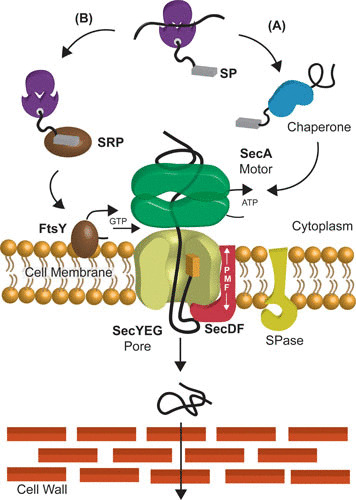
The Sec pathway. (A) Posttranslational pathway: after complete synthesis at the ribosome, the unfolded preprotein is recognized by the molecular chaperone SecB (blue) and targeted to SecA (green). SecA guides the preprotein through the SecYEG pore (lime), employing the energy from ATP binding and hydrolysis. The signal peptide is cleaved by the signal peptidase (SPase [yellow]). SecDF (pink) pulls the preprotein across the membrane at the expense of the PMF. (B) Cotranslational pathway: once a hydrophobic transmembrane domain of a nascent membrane protein emerges from the ribosomes, signal recognition particle (SRP) (brown) binds to the ribosome nascent chain (RNC) and guides the complex to the SR receptor FtsY (dark brown) at the membrane. Upon the binding of GTP to the SRP:FtsY heterodimer, the RNC is released from SRP and transferred to the SecYEG channel, where chain elongation at the ribosome is directly coupled to membrane insertion of the nascent membrane protein.
During posttranslational translocation, preproteins are synthesized at the ribosome with a cleavable N-terminal signal sequence and bound by the molecular chaperone SecB, which stabilizes the preprotein in an unfolded state (7). SecB targets preproteins to the SecYEG-bound SecA (8–10). SecA is an ATPase (11, 12) that directs preproteins in a stepwise manner into the pore (2). The SecDF complex (13) aids this process by facilitating proton motive force (PMF)-dependent translocation (14). In the cotranslational pathway, nascent membrane proteins are guided to the translocon by signal recognition particle (SRP) to the SRP receptor, FtsY, at the membrane. Subsequently, GTP binding to the SRP-FtsY heterodimer results in release of the nascent chain from SRP to the translocon. Eukaryotes may use this pathway for both translocation and membrane insertion, whereas in bacteria, it is mostly used for insertion (15). This review focuses on bacterial protein translocation.
SecYEG, THE PROTEIN CONDUCTING CHANNEL
Structural analysis of SecYEG provides strong support for its role as a protein conducting channel. SecY, the major subunit, consists of two halves formed by transmembrane segments (TMS) 1 to 5 and 6 to 10 (16) (Fig. 2A). The two halves are connected by a loop of TMS 5/6, resulting in a clamshell-like structure of the translocon (17). SecY is shaped like an hourglass with a funnel-like entrance and a subcentral constriction (Fig. 2D). At the front of SecY, a lateral gate between TMS 2 and 7 can open to the lipid bilayer (16). The exit site of the pore is closed by an α-helical plug (TMS2a) that folds back into the channel (18, 19). The Methanococcus jannaschii SecYEβ structure concerns a resting state with a sealed pore where six hydrophobic residues close the constriction ring and the plug closes the exit funnel (16).
FIGURE 2.
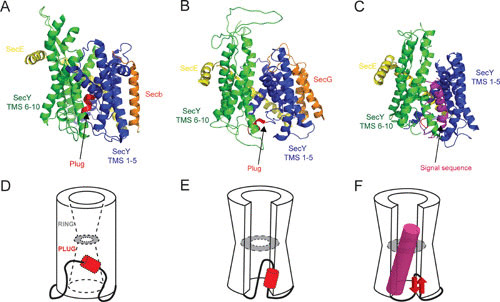
Structural stages of the translocation channel. (A to C) The SecYEG/β crystal structures viewed from the membrane: SecY TMS 1 to 5 (blue), TMS 6 to 10 (green), plug domain (red), SecE (yellow), and SecG/β (orange). (D to F) Cartoon illustration of SecYEG/β. The illustrations depict the opening of the constriction and movement of the plug domain depending on the state of the translocon. (A and D) Methanococcus jannaschii SecYEβ (PDB entry 1RH5), known as the closed or resting conformation. (B and E) Thermotoga maritima SecYEG cocrystallized with SecA (not shown) in an Mg-ADP-BeFx-bound transition state (PDB entry 3DIN) as a preopen conformation. (C and F) Geobacillus thermodenitrificans SecYEG cocrystallized with SecA (not shown) and a signal sequence (magenta) latched into the lateral gate (PDB entry 5EUL), resembling an actively engaged translocation channel.
SecE surrounds SecY at its back and embraces the SecY clamshell structure with a long transmembrane helix that via a hinge is connected to a surface-exposed amphipathic helix that contacts loops of SecY (Fig. 2A). The third subunit SecG is peripherally bound to SecY. This subunit is not essential for cell viability (20, 21) but stabilizes the closed channel (22) whereby the cytosolic loop of SecG folds back into the channel at the cis-side of the membrane (23).
SecYEG Channel Opening
The lateral gate creates a pathway for the insertion of membrane proteins (24) and provides a binding site for the signal sequence of preproteins (25–27). The signal sequence intercalates into the lateral gate, causing a conformational change in the pore region (26). Three of the six pore ring residues are located on TMS 2 and 7 of the lateral gate, and thus, intercalation of the signal sequence between these TMS is directly coupled to channel opening (26). SecE presumably stabilizes the two halves of SecY when the channel opens and when the plug is displaced from its subcentral position (16, 19). Channel opening is also influenced by SecA, as shown in the Thermotoga maritima SecA-SecYEG complex structure (Fig. 2B and E) (28), where a partial opening of lateral gate by ∼5 Å provides a gap that may allow an inserting signal sequence to sample the phospholipid bilayer. For translocation, the lateral gate needs to open up to 10 to 12 Å (25, 29).
The structure of Geobacillus thermodenitrificans SecYE-SecA complex with a covalently linked signal sequence in the channel further shows large conformational changes in the lateral gate region (30) (Fig. 2C). Now also the plug has shifted to the back of the channel in close proximity to the TMS of SecE, in line with cross-linking studies (19, 31) (Fig. 2C and F). Compared to the T. maritima SecA-SecYEG structure (Fig. 2B), TMS 7 of the G. thermodenitrificans SecY is tilted 10° relative to the membrane and the periplasmic ends of TMS 3 and 7 are now in close proximity (Fig. 2C). This results in a large opening in the lateral gate that allows for signal sequence intercalation (30). The plug of G. thermodenitrificans SecY adopted a β-strand structure, which differs from the resting α-helical structure (16, 32, 33). The plug domain is poorly conserved (16), and its deletion only reduces the efficiency of translocation (34, 35). It is important for signal sequence recognition (34, 36, 37) and appears to sample the hydrophobicity of the incoming polypeptide (38) to coordinate channel opening. A very large movement of the plug, by ∼20 to 27 Å, creates an unobstructed path for protein translocation (19, 31), but this large displacement is not critical for translocation per se (39). The plug stabilizes the closed state of SecY (34) and acts as a periplasmic seal to prevent ion leakage.
Pore Constriction and Width
Protein localization (Prl) mutations in sec genes allow the translocation of preproteins with a defective or even missing signal sequence. The most dominant prl variants are found in SecY (40–43), and these destabilize the closed state of the channel (22, 36, 40), possibly mimicking the function of the signal sequence, SecA, or the ribosome (16). The PrlA4 mutant (44) exhibits a tighter binding of SecA (45), allowing more efficient translocation (45–47) and a lower PMF dependence (48). Overall, this can be understood as a reduced proofreading activity (49–52), as signal sequence recognition is less stringent in PrlA mutants, likely because the channel is already in a partially opened state.
Many of the PrlA mutants cluster around the pore constriction and cause increased ion leakage (53). The hydrophobic constriction ring functions as a gasket around the translocating preprotein to seal the pore (16, 18). The pore exhibits a high plasticity and even supports translocation of preproteins with an internal disulfide bridge, a stable fold induced by chemical cross-linking (54, 55) or bulky fluorophores (56). Structures with a cross section of up to ∼22 Å can be translocated (57).
Oligomeric State of SecYEG
The oligomeric state of the SecYEG translocon remains a topic of debate. SecYEG can be purified as a monomer (16, 28) but may also form dimers and higher oligomers (58–60). Crystallography and cross-linking experiments have suggested that SecYEG is dimeric (58, 59, 61–63), but the functional role of the dimer has remained obscure, as only one channel is used to translocate proteins (58, 64). Single SecYEG complexes reconstituted into nanodiscs show that the monomer is sufficient for translocation, as well as for ribosome nascent chain (RNC) binding (47, 64, 65). Further, the cryo-electron microscopic structure of the RNC-SecYEG complex (28, 30) defines the monomer as the minimal functional unit.
SecA, AN ATP-DEPENDENT MOTOR PROTEIN
SecA is a molecular motor that drives protein translocation at the expense of ATP hydrolysis (66). SecA associates with SecYEG but also binds to the phospholipid bilayer and to ribosomes (67, 68).
SecA Structure
SecA is a relatively large protein with a subunit mass of about 102 kDa. It consists of functional and structural subdomains (Fig. 3). The nucleotide binding domain (NBD), comprising NBD1 and NBD2 (also termed intramolecular regulator of ATPase 2 [IRA2]), is essential for ATP binding and hydrolysis (69). The ATPase activity occurs at the interface of NBD1 and NBD2 (70). These NBDs form the so-called DEAD motor, which is also found in DNA/RNA helicases, and contain the highly conserved Walker A and B motifs (71).
FIGURE 3.
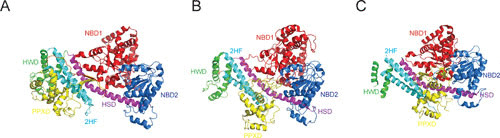
Conformational states of SecA. Structures of SecA from Bacillus subtilis (PDB entry 1M6N) (A), Mg-ADP-BeFx-bound SecA cocrystallized with SecYEG (not shown) from T. maritima (PDB 3DIN) (B), and Mg-ADP-BeFx-bound SecA from B. subtilis engaged with the G. thermodenitrificans SecYEG and a signal sequence (not shown) (PDB entry 5EUL) (C). The locations of the PPXD domain (yellow), NBD1 (red), NBD2 (blue), HWD (green), HSD (purple), and 2HF (cyan) are indicated. A large movement of the PPXD domain (yellow) suggests a closed (A) or open (B and C) conformation of SecA.
Preprotein can cross-link to the PPXD domain (72), which plays an important role in the activation of the ATPase activity (73, 74). The C-terminal domain of SecA can be divided into four subdomains: the α-helical scaffold domain (HSD), which interconnects all other SecA domains (75); the α-helical wing domain (HWD); the two-helix finger (2HF), which is part of the intramolecular regulator of ATP hydrolysis 1 (IRA1) (76); and the C-terminal linker domain (CTL). In Escherichia coli, the CTL harbors a zinc finger, which plays a role in the interaction with SecB (7, 9) and phospholipids (77). SecA exhibits a low basal ATPase activity (78), which is allosterically stimulated by binding of SecB, SecYEG, and preprotein and by anionic phospholipids (79–84). The ATPase activity of cytosolic SecA is inhibited by IRA1, or 2HF, which forms a helix-loop-helix structure of the HSD that contacts both NBD2 and PPXD (85).
Oligomeric State of SecA
The functional oligomeric state of SecA is a major topic of controversy. SecA purified from cells is mainly dimeric (86). Although SecA appears to function as a dimer (87–90), the monomer-dimer equilibrium is affected by ligands of SecA (91, 92). SecA is highly thermolabile in the presence of phospholipids, but inactivation is prevented by preproteins (93).
Only a few studies have addressed the oligomeric state of SecA while bound to SecYEG. SecA remains dimeric during translocation (90, 94) and is active as a dimer (88, 89, 94, 95), likely as a discrete anti-parallel dimer (96). Dimeric SecA binds the SecYEG with high affinity, where one of the protomers binds tightly to SecYEG and the other protomer is bound to the SecYEG-bound SecA (88). Mutation-induced monomerization abolishes SecA activity (97), but this defect can be overcome by high concentrations of SecA that restore the dimer (88, 98). SecB interacts with the SecYEG-bound dimeric SecA (99).
Binding Partners of SecA
The N-terminal signal sequence of preproteins functions as a targeting signal (100, 101) and induces channel opening; also, the targeting function of signal sequences has been challenged (102–104). Prior to translocation, the preprotein is stabilized in an unfolded state by SecB. SecYEG-bound SecA binds SecB, and this interaction results in a transfer of the preprotein from SecB to SecA (7). SecB is a homotetramer arranged as a dimer of dimers (105). SecB contains two peptide binding grooves that run along either side of the tetramer (106, 107), where preproteins are bound in their folding core (108). During the ATP-dependent initiation of translocation, SecB is released into the cytosol to bind another preprotein.
SecA binds to SecYEG via a phospholipid-bound intermediate (93, 109–111) that involves its amphipathic positively charged N terminus (93). This region serves to tether SecA to the membrane (109), but the membrane interaction also enforces a conformational change which primes SecA for high-affinity SecYEG binding (93). Phospholipid-bound SecA likely functions as a membrane queue of SecA-preprotein complexes before they are delivered to SecYEG for translocation.
Structural Mechanism of SecA Function
SecA undergoes a multitude of ATP-dependent conformation changes during translocation (28, 70, 112) (Fig. 3). In the T. maritima SecA-SecYEG structure, where SecA is in an open transition state stabilized by ADP beryllium fluoride, the PPXD domain has moved towards the NBD2 and away from the HWD (28, 113), while the NBDs are in close proximity with the PPXD domain (Fig. 3B). In contrast, the Bacillus subtilis SecA structure (Fig. 3A) represents a closed state in which the PPXD domain is located near the HWD (70). The formation of the nucleotide-binding pocket between NBD1 and NBD2 allows the preprotein to bind in a groove between NBD2 and PPXD. This opening stabilizes the preprotein-SecA interaction, which allows for an increased rate of nucleotide exchange, resulting in activation of the ATPase activity (114, 115). The structure of the G. thermodenitrificans SecYE engaged with B. subtilis SecA (30) and a signal peptide suggests that SecA does not undergo further dramatic conformational changes compared to the T. maritima SecA-SecYEG complex (Fig. 3C). It has been proposed that SecA in its ATP-bound state prevents the two halves of SecY from moving further apart.
The 2HF of SecA is inserted into the cytoplasmic opening of the SecY channel (Fig. 4), where it is in close proximity to the translocating preprotein (28). The 2HF makes contact with C4 loop of SecY, and the insertion may result in the opening of lateral gate by a rigid body movement of the two halves of SecY (29). The tip of the loop of the 2HF contains a highly conserved tyrosine residue which is crucial for translocation (76). It has been suggested that the 2HF associates with the unfolded preprotein through hydrophobic side chain interactions, but this model does not explain how SecA can mediate the translocation of stretches of glycine residues (116), which would only allow for main chain interactions. Alternatively, the 2HF acts by opening the channel through its interaction with the SecY C4 loop. Strikingly, chemical cross-linking of the 2HF with the C4 loop did not interfere with translocation (117), suggesting that the 2HF does not function as an ATP-dependent lever to push preproteins through the channel but rather serves to push the two halves of SecY apart. Additionally, the 2HF of SecA may act as a template by inserting the hairpin formed by the signal peptide and the early mature region of the preprotein (118).
FIGURE 4.
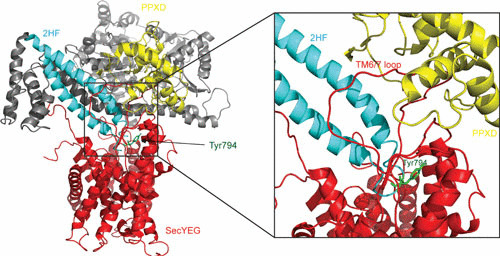
Structure of T. maritima SecA-SecYEG complex. SecA penetrates into the SecYEG channel (red) via the so-called two-helix finger (2HF [light blue]). The SecA PPXD domain (yellow) also binds to TMS6/7 loop of SecYEG. The conserved tyrosine 794 is depicted in green.
TRANSLOCATION MODELS
Various models for SecA-mediated translocation have been proposed as outlined below (Fig. 5).
FIGURE 5.
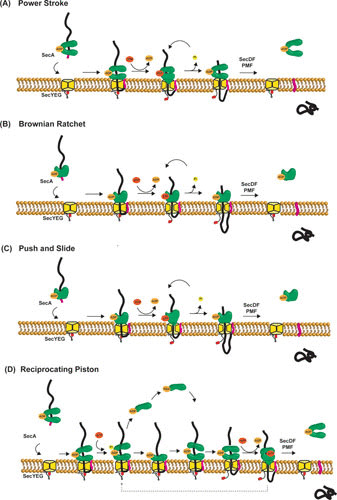
Proposed models of SecA-mediated protein translocation. (A) Power stroke: ATP binding and hydrolysis induce conformational changes of SecA that result in a mechanical force on the preprotein, pushing it through the SecYEG channel. In this model, oligomerization of SecA is required to prevent backsliding of the preprotein. (B) Brownian ratchet: SecA regulates the SecYEG channel opening via the 2HF of SecA, allowing the protein translocation via diffusion. Trapping and release of the translocating preprotein at the cis-side result in translocation, while SecA may fulfill an additional function by opening the translocation channel. The oligomeric state of SecA is not explicitly shown in this model. (C) Push and slide: this model uses both SecA-dependent pushing and Brownian motion. The oligomeric state of SecA is not explicitly shown in this model. (D) Reciprocating piston: this model is a combination of a power stroke mechanism with the conversion of dimeric-monomeric SecA. Repeated cycles of SecA monomerization-rebinding and ATP binding-hydrolysis yield a stepwise translocation process. In none of these models is the exact role of the PMF and SecDF included, but they contribute to efficient translocation.
Power Stroke Model
A large class of ATPases contains a RecA-like structural domain and uses the energy of ATP binding and hydrolysis to move proteins or nucleic acids (71). SecA has a DEAD box typically found in helicase, and therefore a DNA helicase molecular mechanism has been proposed (119). In this power stoke mechanism, SecA acts as a mechanical device that pushes preproteins into the pore (82). The 2HF may function as an ATP-dependent lever to support such a power stroke mechanism causing stepwise translocation (120). To apply the DNA helicase principle, SecA is required to multimerize in order to have multiple substrate binding sites since monomeric SecA appears to have only one substrate binding site (81, 121). One SecA protomer may act as a clamp and move the preprotein into the channel, while the other SecA protomer traps the chains to prevent retrograde transport, as SecYEG is not able to make a stable anchor for preproteins (122). This implies that a high degree of cooperativity is needed between the two protomers of SecA to ensure that the preprotein is bound to one of the protomers at any given time. Currently, it is unclear how a small movement of the 2HF can drive translocation of polypeptide segments ∼20 amino acids long.
Brownian Ratchet Model
In the Brownian ratchet model, SecA acts as the regulator for channel opening of SecYEG (123), while translocation occurs by Brownian movement of the unfolded preprotein through the channel. Because of the contact of the 2HF of SecA with SecYEG (28), movement of the 2HF could potentially result in an opening of the channel. Backsliding of the preprotein would be prevented by the SecA association and provide directionality to the process, which might be further facilitated by folding of the polypeptide at the cis-side of the membrane and/or binding by SecDF (124). This model explains the promiscuity of the system for diverse preprotein substrates (125) but does not explain stepwise translocation (11, 82, 122).
Push-and-Slide Model
The “push-and-slide” mechanism (109) combines the power stroke and Brownian ratchet models and explains earlier observations that SecA-mediated translocation occurs stepwise, whereas in the absence of SecA association, the preprotein may slide within the pore (122). Again, this model depends on the 2HF for the power stroke (76, 109). Once ATP is hydrolyzed, the 2HF would return to its pretranslocation position and dissociate from the preprotein to allow passive sliding of the protein into the channel. This model, however, does not explain that a complex of SecA-SecYEG wherein the SecA 2HF is cross-linked to the C4 loop of SecY is functional in translocation (117). Alternatively, stepwise translocation may arise from binding and release of SecA to and from SecYEG (122, 126).
Reciprocating Piston Model
SecA exists as a dimer during translocation (89, 90, 94, 127), but monomeric states have also been reported (28, 97, 127, 128). The reciprocating piston model combines the power stroke model with the SecA monomer-dimer transition (29). Translocation is initiated by binding of dimeric SecA to SecYEG. Next, ATP hydrolysis induces SecA monomerization where one of the SecA monomers remains anchored to SecYEG to prevent backsliding of the partially translocated preprotein, while the other monomer is released from the membrane. Rebinding of another SecA monomer to SecYEG-SecA-preprotein complex then promotes ATP-independent translocation of a preprotein segment, while subsequent binding of ATP drives the translocation by a power stroke. These steps are repeated until the preprotein is fully translocated. This model explains the two consecutive translocation stages observed biochemically, i.e., translocation driven by SecA binding to the preprotein and by ATP binding (82, 122). Complete dissociation of SecA from SecYEG may allow translocation by Brownian diffusion and enable PMF-driven translocation.
ROLE OF THE SecDF COMPLEX
The aforementioned models do not take the role of the PMF into account. Although ATP suffices for translocation in vitro, in vivo it strongly depends on the PMF. SecA may mainly serve to initiate and direct translocation by releasing a looped structure of the signal sequence and early mature protein domain into the pore, whereupon translocation is further driven by the PMF (14). Indeed, in vitro translocation at low SecA concentrations is highly PMF dependent (129, 130). Late stages of translocation allow large unfolded regions of the preprotein to be translocated without ATP and are SecDF and PMF dependent (131).
SecDF is a subcomplex that associates with the translocon to form the holo-translocon complex (124). The crystal structure of SecDF shows a single polypeptide with 12 TMS, 6 TMS each in both SecD and SecF (132, 133). SecDF also contains 6 periplasmic domains (P1 to P6); P1 and P4 form a periplasmic protruding structure. P1 has been proposed to interact with the polypeptide substrate, and movement of P1 may result in a PMF-dependent pulling action by SecDF at the periplasmic side of the membrane (133, 134).
CONCLUDING REMARKS
Integrating biochemical, biophysical, and structural studies has led to a basic understanding of the molecular mechanism of protein translocation. However, still many mechanistic questions remain unresolved. Although translocation exhibits power stroke- and Brownian diffusion-like mechanistic features, it remains unclear how translocation is linked to the SecA dimer. To unify potentially conflicting results, the process needs to be examined at the single-molecule level to reveal the dynamic interplay between the components and identify their roles at the different stages of the process.
ACKNOWLEDGMENTS
This work is supported by the incentive scheme of the Zernike Institute of Advanced Materials and by the Foundation of Life Sciences of the Netherlands Organisation for Scientific Research (ALW-NWO).
Contributor Information
Amalina Ghaisani Komarudin, Molecular Microbiology, Groningen Biomolecular Sciences and Biotechnology Institute, and the Zernike Institute of Advanced Materials, University of Groningen, Nijenborgh 7, 9747AG Groningen, The Netherlands.
Arnold J. M. Driessen, Molecular Microbiology, Groningen Biomolecular Sciences and Biotechnology Institute, and the Zernike Institute of Advanced Materials, University of Groningen, Nijenborgh 7, 9747AG Groningen, The Netherlands
REFERENCES
- 1.Tsirigotaki A, De Geyter J, Šoštaric N, Economou A, Karamanou S. 2017. Protein export through the bacterial Sec pathway. Nat Rev Microbiol 15:21–36. 10.1038/nrmicro.2016.161. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Driessen AJM, Nouwen N. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77:643–667. 10.1146/annurev.biochem.77.061606.160747. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis A. 2004. The archaeal Sec-dependent protein translocation pathway. Philos Trans R Soc Lond B Biol Sci 359:919–927. 10.1098/rstb.2003.1461. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bondar A-N, del Val C, Freites JA, Tobias DJ, White SH. 2010. Dynamics of SecY translocons with translocation-defective mutations. Structure 18:847–857. 10.1016/j.str.2010.04.010. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pohlschröder M, Prinz WA, Hartmann E, Beckwith J. 1997. Protein translocation in the three domains of life: variations on a theme. Cell 91:563–566. 10.1016/S0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 6.Natale P, Brüser T, Driessen AJM. 2008. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—distinct translocases and mechanisms. Biochim Biophys Acta 1778:1735–1756. 10.1016/j.bbamem.2007.07.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 7.Fekkes P, van der Does C, Driessen AJ. 1997. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J 16:6105–6113. 10.1093/emboj/16.20.6105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bechtluft P, Nouwen N, Tans SJ, Driessen AJM. 2010. SecB—a chaperone dedicated to protein translocation. Mol Biosyst 6:620–627. 10.1039/B915435C. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Fekkes P, Driessen AJ. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev 63:161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabelli RJ, Chen L, Tai PC, Oliver DB. 1988. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell 55:683–692. 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 11.Economou A, Wickner W. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78:835–843. 10.1016/S0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 12.du Plessis DJF, Nouwen N, Driessen AJM. 2011. The Sec translocase. Biochim Biophys Acta 1808:851–865. 10.1016/j.bbamem.2010.08.016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Duong F, Wickner W. 1997. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J 16:4871–4879. 10.1093/emboj/16.16.4871. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukazaki T, Nureki O. 2011. The mechanism of protein export enhancement by the SecDF membrane component. Biophysics (Nagoya-Shi) 7:129–133. 10.2142/biophysics.7.129. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller M, Koch HG, Beck K, Schäfer U. 2001. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog Nucleic Acid Res Mol Biol 66:107–157. 10.1016/S0079-6603(00)66028-2. [DOI] [PubMed] [Google Scholar]
- 16.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44. 10.1038/nature02218. [PubMed] [DOI] [PubMed] [Google Scholar]
- 17.Gumbart J, Schulten K. 2007. Structural determinants of lateral gate opening in the protein translocon. Biochemistry 46:11147–11157. 10.1021/bi700835d. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Park E, Rapoport TA. 2011. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 473:239–242. 10.1038/nature10014. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam PCK, Maillard AP, Chan KKY, Duong F. 2005. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J 24:3380–3388. 10.1038/sj.emboj.7600804. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. 1990. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62:649–657. 10.1016/0092-8674(90)90111-Q. [DOI] [PubMed] [Google Scholar]
- 21.Hanada M, Nishiyama KI, Mizushima S, Tokuda H. 1994. Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE, and SecG (p12). J Biol Chem 269:23625–23631. [PubMed] [Google Scholar]
- 22.Belin D, Plaia G, Boulfekhar Y, Silva F. 2015. Escherichia coli SecG is required for residual export mediated by mutant signal sequences and for SecY-SecE complex stability. J Bacteriol 197:542–552. 10.1128/JB.02136-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka Y, Sugano Y, Takemoto M, Mori T, Furukawa A, Kusakizako T, Kumazaki K, Kashima A, Ishitani R, Sugita Y, Nureki O, Tsukazaki T. 2015. Crystal structures of SecYEG in lipidic cubic phase elucidate a precise resting and a peptide-bound state. Cell Rep 13:1561–1568. 10.1016/j.celrep.2015.10.025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 24.Heinrich SU, Mothes W, Brunner J, Rapoport TA. 2000. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell 102:233–244. 10.1016/S0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 25.du Plessis DJF, Berrelkamp G, Nouwen N, Driessen AJM. 2009. The lateral gate of SecYEG opens during protein translocation. J Biol Chem 284:15805–15814. 10.1074/jbc.M901855200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corey RA, Allen WJ, Komar J, Masiulis S, Menzies S, Robson A, Collinson I. 2016. Unlocking the bacterial SecY translocon. Structure 24:518–527. 10.1016/j.str.2016.02.001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. 1998. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94:795–807. 10.1016/S0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 28.Zimmer J, Nam Y, Rapoport TA. 2008. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455:936–943. 10.1038/nature07335. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusters I, Driessen AJM. 2011. SecA, a remarkable nanomachine. Cell Mol Life Sci 68:2053–2066. 10.1007/s00018-011-0681-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Park E, Ling J, Ingram J, Ploegh H, Rapoport TA. 2016. Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531:395–399. 10.1038/nature17163. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris CR, Silhavy TJ. 1999. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J Bacteriol 181:3438–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. 2008. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455:988–991. 10.1038/nature07421. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egea PF, Stroud RM. 2010. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci U S A 107:17182–17187. 10.1073/pnas.1012556107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maillard AP, Lalani S, Silva F, Belin D, Duong F. 2007. Deregulation of the SecYEG translocation channel upon removal of the plug domain. J Biol Chem 282:1281–1287. 10.1074/jbc.M610060200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 35.Junne T, Schwede T, Goder V, Spiess M. 2006. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol Biol Cell 17:4063–4068. 10.1091/mbc.e06-03-0200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA. 2007. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell 26:511–521. 10.1016/j.molcel.2007.05.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Junne T, Schwede T, Goder V, Spiess M. 2007. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem 282:33201–33209. 10.1074/jbc.M707219200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, Miller TF III. 2010. Hydrophobically stabilized open state for the lateral gate of the Sec translocon. Proc Natl Acad Sci U S A 107:5399–5404. 10.1073/pnas.0914752107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lycklama a Nijeholt JA, Bulacu M, Marrink SJ, Driessen AJM. 2010. Immobilization of the plug domain inside the SecY channel allows unrestricted protein translocation. J Biol Chem 285:23747–23754. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duong F, Wickner W. 1999. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J 18:3263–3270. 10.1093/emboj/18.12.3263. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. 2005. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol 187:6454–6465. 10.1128/JB.187.18.6454-6465.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silhavy TJ, Mitchell AM. 25 January 2019. Genetic analysis of protein translocation. Protein J 38:217–228. 10.1007/s10930-019-09813-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osborne RS, Silhavy TJ. 1993. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J 12:3391–3398. 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emr SD, Hanley-Way S, Silhavy TJ. 1981. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell 23:79–88. 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- 45.de Keyzer J, van der Does C, Swaving J, Driessen AJM. 2002. The F286Y mutation of PrlA4 tempers the signal sequence suppressor phenotype by reducing the SecA binding affinity. FEBS Lett 510:17–21. 10.1016/S0014-5793(01)03213-6. [DOI] [PubMed] [Google Scholar]
- 46.van der Wolk JP, Fekkes P, Boorsma A, Huie JL, Silhavy TJ, Driessen AJ. 1998. PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA-SecY interaction during the initiation of translocation. EMBO J 17:3631–3639. 10.1093/emboj/17.13.3631. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taufik I, Kedrov A, Exterkate M, Driessen AJM. 2013. Monitoring the activity of single translocons. J Mol Biol 425:4145–4153. 10.1016/j.jmb.2013.08.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Nouwen N, de Kruijff B, Tommassen J. 1996. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc Natl Acad Sci U S A 93:5953–5957. 10.1073/pnas.93.12.5953. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fikes JD, Bassford PJ, Jr. 1989. Novel secA alleles improve export of maltose-binding protein synthesized with a defective signal peptide. J Bacteriol 171:402–409. 10.1128/jb.171.1.402-409.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stader J, Gansheroff LJ, Silhavy TJ. 1989. New suppressors of signal-sequence mutations, prlG, are linked tightly to the secE gene of Escherichia coli. Genes Dev 3:1045–1052. 10.1101/gad.3.7.1045. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Flower AM, Doebele RC, Silhavy TJ. 1994. PrlA and PrlG suppressors reduce the requirement for signal sequence recognition. J Bacteriol 176:5607–5614. 10.1128/jb.176.18.5607-5614.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prinz WA, Spiess C, Ehrmann M, Schierle C, Beckwith J. 1996. Targeting of signal sequenceless proteins for export in Escherichia coli with altered protein translocase. EMBO J 15:5209–5217. 10.1002/j.1460-2075.1996.tb00906.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saparov SM, Erlandson K, Cannon K, Schaletzky J, Schulman S, Rapoport TA, Pohl P. 2007. Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell 26:501–509. 10.1016/j.molcel.2007.03.022. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Tani K, Tokuda H, Mizushima S. 1990. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J Biol Chem 265:17341–17347. [PubMed] [Google Scholar]
- 55.Tani K, Mizushima S. 1991. A chemically cross-linked nonlinear proOmpA molecule can be translocated into everted membrane vesicles of Escherichia coli in the presence of the proton motive force. FEBS Lett 285:127–131. 10.1016/0014-5793(91)80741-K. [DOI] [PubMed] [Google Scholar]
- 56.De Keyzer J, Van Der Does C, Driessen AJM. 2002. Kinetic analysis of the translocation of fluorescent precursor proteins into Escherichia coli membrane vesicles. J Biol Chem 277:46059–46065. 10.1074/jbc.M208449200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Bonardi F, Halza E, Walko M, Du Plessis F, Nouwen N, Feringa BL, Driessen AJM. 2011. Probing the SecYEG translocation pore size with preproteins conjugated with sizable rigid spherical molecules. Proc Natl Acad Sci U S A 108:7775–7780. 10.1073/pnas.1101705108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hizlan D, Robson A, Whitehouse S, Gold VA, Vonck J, Mills D, Kühlbrandt W, Collinson I. 2012. Structure of the SecY complex unlocked by a preprotein mimic. Cell Rep 1:21–28. 10.1016/j.celrep.2011.11.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I. 2002. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418:662–665. 10.1038/nature00827. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, III, Ban N, Frank J, Frank J. 2005. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438:318–324. 10.1038/nature04133. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deville K, Gold VAM, Robson A, Whitehouse S, Sessions RB, Baldwin SA, Radford SE, Collinson I. 2011. The oligomeric state and arrangement of the active bacterial translocon. J Biol Chem 286:4659–4669. 10.1074/jbc.M110.175638. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Das S, Oliver DB. 2011. Mapping of the SecA·SecY and SecA·SecG interfaces by site-directed in vivo photocross-linking. J Biol Chem 286:12371–12380. 10.1074/jbc.M110.182931. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Z, Blum A, Banerjee T, Wang Q, Dantis V, Oliver D. 2016. Determination of the oligomeric state of SecYEG protein secretion channel complex using in vivo photo- and disulfide cross-linking. J Biol Chem 291:5997–6010. 10.1074/jbc.M115.694844. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osborne AR, Rapoport TA. 2007. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell 129:97–110. 10.1016/j.cell.2007.02.036. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Kedrov A, Kusters I, Krasnikov VV, Driessen AJM. 2011. A single copy of SecYEG is sufficient for preprotein translocation. EMBO J 30:4387–4397. 10.1038/emboj.2011.314. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomkiewicz D, Nouwen N, Driessen AJM. 2007. Pushing, pulling and trapping—modes of motor protein supported protein translocation. FEBS Lett 581:2820–2828. 10.1016/j.febslet.2007.04.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Findik BT, Smith VF, Randall LL. 2018. Penetration into membrane of amino-terminal region of SecA when associated with SecYEG in active complexes. Protein Sci 27:681–691. 10.1002/pro.3362. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huber D, Rajagopalan N, Preissler S, Rocco MA, Merz F, Kramer G, Bukau B. 2011. SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell 41:343–353. 10.1016/j.molcel.2010.12.028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Sato K, Mori H, Yoshida M, Mizushima S. 1996. Characterization of a potential catalytic residue, Asp-133, in the high affinity ATP-binding site of Escherichia coli SecA, translocation ATPase. J Biol Chem 271:17439–17444. 10.1074/jbc.271.29.17439. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297:2018–2026. 10.1126/science.1074424. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Ye J, Osborne AR, Groll M, Rapoport TA. 2004. RecA-like motor ATPases—lessons from structures. Biochim Biophys Acta 1659:1–18. 10.1016/j.bbabio.2004.06.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Bauer BW, Rapoport TA. 2009. Mapping polypeptide interactions of the SecA ATPase during translocation. Proc Natl Acad Sci U S A 106:20800–20805. 10.1073/pnas.0910550106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ding H, Mukerji I, Oliver D. 2003. Nucleotide and phospholipid-dependent control of PPXD and C-domain association for SecA ATPase. Biochemistry 42:13468–13475. 10.1021/bi035099b. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Chada N, Chattrakun K, Marsh BP, Mao C, Bariya P, King GM. 2018. Single-molecule observation of nucleotide induced conformational changes in basal SecA-ATP hydrolysis. Sci Adv 4:eaat8797. 10.1126/sciadv.aat8797. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Papanikolau Y, Papadovasilaki M, Ravelli RBG, McCarthy AA, Cusack S, Economou A, Petratos K. 2007. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol 366:1545–1557. 10.1016/j.jmb.2006.12.049. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Erlandson KJ, Miller SBM, Nam Y, Osborne AR, Zimmer J, Rapoport TA. 2008. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature 455:984–987. 10.1038/nature07439. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. 1995. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem 270:7902–7907. 10.1074/jbc.270.14.7902. [PubMed] [DOI] [PubMed] [Google Scholar]
- 78.Gold VAM, Robson A, Clarke AR, Collinson I. 2007. Allosteric regulation of SecA: magnesium-mediated control of conformation and activity. J Biol Chem 282:17424–17432. 10.1074/jbc.M702066200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Lill R, Dowhan W, Wickner W. 1990. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60:271–280. 10.1016/0092-8674(90)90742-W. [DOI] [PubMed] [Google Scholar]
- 80.Miller A, Wang L, Kendall DA. 2002. SecB modulates the nucleotide-bound state of SecA and stimulates ATPase activity. Biochemistry 41:5325–5332. 10.1021/bi025639p. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. 2007. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131:756–769. 10.1016/j.cell.2007.09.039. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Wolk JPW, de Wit JG, Driessen AJ. 1997. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J 16:7297–7304. 10.1093/emboj/16.24.7297. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corey RA, Pyle E, Allen WJ, Watkins DW, Casiraghi M, Miroux B, Arechaga I, Politis A, Collinson I. 2018. Specific cardiolipin-SecY interactions are required for proton-motive force stimulation of protein secretion. Proc Natl Acad Sci U S A 115:7967–7972. 10.1073/pnas.1721536115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gold VAM, Robson A, Bao H, Romantsov T, Duong F, Collinson I. 2010. The action of cardiolipin on the bacterial translocon. Proc Natl Acad Sci U S A 107:10044–10049. 10.1073/pnas.0914680107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. 1999. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol 34:1133–1145. 10.1046/j.1365-2958.1999.01686.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Woodbury RL, Hardy SJ, Randall LL. 2002. Complex behavior in solution of homodimeric SecA. Protein Sci 11:875–882. 10.1110/ps.4090102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang H, Na B, Yang H, Tai PC. 2008. Additional in vitro and in vivo evidence for SecA functioning as dimers in the membrane: dissociation into monomers is not essential for protein translocation in Escherichia coli. J Bacteriol 190:1413–1418. 10.1128/JB.01633-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kusters I, van den Bogaart G, Kedrov A, Krasnikov V, Fulyani F, Poolman B, Driessen AJM. 2011. Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure 19:430–439. 10.1016/j.str.2010.12.016. [PubMed] [DOI] [PubMed] [Google Scholar]
- 89.Driessen AJ. 1993. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry 32:13190–13197. 10.1021/bi00211a030. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.de Keyzer J, van der Sluis EO, Spelbrink REJ, Nijstad N, de Kruijff B, Nouwen N, van der Does C, Driessen AJM. 2005. Covalently dimerized SecA is functional in protein translocation. J Biol Chem 280:35255–35260. 10.1074/jbc.M506157200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Benach J, Chou YT, Fak JJ, Itkin A, Nicolae DD, Smith PC, Wittrock G, Floyd DL, Golsaz CM, Gierasch LM, Hunt JF. 2003. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J Biol Chem 278:3628–3638. 10.1074/jbc.M205992200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Bu Z, Wang L, Kendall DA. 2003. Nucleotide binding induces changes in the oligomeric state and conformation of Sec A [sic] in a lipid environment: a small-angle neutron-scattering study. J Mol Biol 332:23–30. 10.1016/S0022-2836(03)00840-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koch S, de Wit JG, Vos I, Birkner JP, Gordiichuk P, Herrmann A, van Oijen AM, Driessen AJM. 2016. Lipids activate SecA for high affinity binding to the SecYEG complex. J Biol Chem 291:22534–22543. 10.1074/jbc.M116.743831. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jilaveanu LB, Zito CR, Oliver D. 2005. Dimeric SecA is essential for protein translocation. Proc Natl Acad Sci U S A 102:7511–7516. 10.1073/pnas.0502774102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karamanou S, Sianidis G, Gouridis G, Pozidis C, Papanikolau Y, Papanikou E, Economou A. 2005. Escherichia coli SecA truncated at its termini is functional and dimeric. FEBS Lett 579:1267–1271. 10.1016/j.febslet.2005.01.025. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Banerjee T, Lindenthal C, Oliver D. 2017. SecA functions in vivo as a discrete anti-parallel dimer to promote protein transport. Mol Microbiol 103:439–451. 10.1111/mmi.13567. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Or E, Boyd D, Gon S, Beckwith J, Rapoport T. 2005. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem 280:9097–9105. 10.1074/jbc.M413947200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Gouridis G, Karamanou S, Sardis MF, Schärer MA, Capitani G, Economou A. 2013. Quaternary dynamics of the SecA motor drive translocase catalysis. Mol Cell 52:655–666. 10.1016/j.molcel.2013.10.036. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Fekkes P, de Wit JG, Boorsma A, Friesen RHE, Driessen AJM. 1999. Zinc stabilizes the SecB binding site of SecA. Biochemistry 38:5111–5116. 10.1021/bi982818r. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Hegde RS, Bernstein HD. 2006. The surprising complexity of signal sequences. Trends Biochem Sci 31:563–571. 10.1016/j.tibs.2006.08.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. 2018. A comprehensive review of signal peptides: structure, roles, and applications. Eur J Cell Biol 97:422–441. 10.1016/j.ejcb.2018.06.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Chatzi KE, Sardis MF, Tsirigotaki A, Koukaki M, Šoštarić N, Konijnenberg A, Sobott F, Kalodimos CG, Karamanou S, Economou A. 2017. Preprotein mature domains contain translocase targeting signals that are essential for secretion. J Cell Biol 216:1357–1369. 10.1083/jcb.201609022. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fessl T, Watkins D, Oatley P, Allen WJ, Corey RA, Horne J, Baldwin SA, Radford SE, Collinson I, Tuma R. 2018. Dynamic action of the Sec machinery during initiation, protein translocation and termination. eLife 7:e35112. 10.7554/eLife.35112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sardis MF, Tsirigotaki A, Chatzi KE, Portaliou AG, Gouridis G, Karamanou S, Economou A. 2017. Preprotein conformational dynamics drive bivalent translocase docking and secretion. Structure 25:1056–1067.e6. 10.1016/j.str.2017.05.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Xu Z, Knafels JD, Yoshino K. 2000. Crystal structure of the bacterial protein export chaperone SecB. Nat Struct Biol 7:1172–1177. 10.1038/82040. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Crane JM, Suo Y, Lilly AA, Mao C, Hubbell WL, Randall LL. 2006. Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J Mol Biol 363:63–74. 10.1016/j.jmb.2006.07.021. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van der Sluis EO, Driessen AJM. 2006. Stepwise evolution of the Sec machinery in Proteobacteria. Trends Microbiol 14:105–108. 10.1016/j.tim.2006.01.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Bechtluft P, van Leeuwen RGH, Tyreman M, Tomkiewicz D, Nouwen N, Tepper HL, Driessen AJM, Tans SJ. 2007. Direct observation of chaperone-induced changes in a protein folding pathway. Science 318:1458–1461. 10.1126/science.1144972. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Bauer BW, Shemesh T, Chen Y, Rapoport TA. 2014. A “push and slide” mechanism allows sequence-insensitive translocation of secretory proteins by the SecA ATPase. Cell 157:1416–1429. 10.1016/j.cell.2014.03.063. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hendrick JP, Wickner W. 1991. SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem 266:24596–24600. [PubMed] [Google Scholar]
- 111.Floyd JH, You Z, Hsieh Y-H, Ma Y, Yang H, Tai PC. 2014. The dispensability and requirement of SecA N-terminal aminoacyl residues for complementation, membrane binding, lipid-specific domains and channel activities. Biochem Biophys Res Commun 453:138–142. 10.1016/j.bbrc.2014.09.080. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sianidis G, Karamanou S, Vrontou E, Boulias K, Repanas K, Kyrpides N, Politou AS, Economou A. 2001. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J 20:961–970. 10.1093/emboj/20.5.961. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen Y, Bauer BW, Rapoport TA, Gumbart JC. 2015. Conformational changes of the clamp of the protein translocation ATPase SecA. J Mol Biol 427:2348–2359. 10.1016/j.jmb.2015.05.003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gold VAM, Whitehouse S, Robson A, Collinson I. 2013. The dynamic action of SecA during the initiation of protein translocation. Biochem J 449:695–705. 10.1042/BJ20121314. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fak JJ, Itkin A, Ciobanu DD, Lin EC, Song X-J, Chou Y-T, Gierasch LM, Hunt JF. 2004. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry 43:7307–7327. 10.1021/bi0357208. [PubMed] [DOI] [PubMed] [Google Scholar]
- 116.Nouwen N, Berrelkamp G, Driessen AJM. 2009. Charged amino acids in a preprotein inhibit SecA-dependent protein translocation. J Mol Biol 386:1000–1010. 10.1016/j.jmb.2009.01.031. [PubMed] [DOI] [PubMed] [Google Scholar]
- 117.Whitehouse S, Gold VA, Robson A, Allen WJ, Sessions RB, Collinson I. 2012. Mobility of the SecA 2-helix-finger is not essential for polypeptide translocation via the SecYEG complex. J Cell Biol 199:919–929. 10.1083/jcb.201205191. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang Q, Lahiri S, Banerjee T, Sun Z, Oliver D, Mukerji I. 2017. Alignment of the protein substrate hairpin along the SecA two-helix finger primes protein transport in Escherichia coli. Proc Natl Acad Sci U S A 114:9343–9348. 10.1073/pnas.1702201114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Osborne AR, Clemons WM Jr, Rapoport TA. 2004. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci U S A 101:10937–10942. 10.1073/pnas.0401742101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Catipovic MA, Bauer BW, Loparo JJ, Rapoport TA. 2019. Protein translocation by the SecA ATPase occurs by a power-stroke mechanism. EMBO J 38:e101140. 10.15252/embj.2018101140. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Papanikou E, Karamanou S, Baud C, Frank M, Sianidis G, Keramisanou D, Kalodimos CG, Kuhn A, Economou A. 2005. Identification of the preprotein binding domain of SecA. J Biol Chem 280:43209–43217. 10.1074/jbc.M509990200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Schiebel E, Driessen AJM, Hartl FU, Wickner W. 1991. Δ mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64:927–939. 10.1016/0092-8674(91)90317-R. [DOI] [PubMed] [Google Scholar]
- 123.Allen WJ, Corey RA, Oatley P, Sessions RB, Baldwin SA, Radford SE, Tuma R, Collinson I. 2016. Two-way communication between SecY and SecA suggests a Brownian ratchet mechanism for protein translocation. eLife 5:e15598. 10.7554/eLife.15598. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Botte M, Zaccai NR, Nijeholt JL, Martin R, Knoops K, Papai G, Zou J, Deniaud A, Karuppasamy M, Jiang Q, Roy AS, Schulten K, Schultz P, Rappsilber J, Zaccai G, Berger I, Collinson I, Schaffitzel C. 2016. A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion. Sci Rep 6:38399. 10.1038/srep38399. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Simon SM, Peskin CS, Oster GF. 1992. What drives the translocation of proteins? Proc Natl Acad Sci U S A 89:3770–3774. 10.1073/pnas.89.9.3770. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Young J, Duong F. 2019. Investigating the stability of the SecA-SecYEG complex during protein translocation across the bacterial membrane. J Biol Chem 294:3577–3587. 10.1074/jbc.RA118.006447. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Duong F. 2003. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J 22:4375–4384. 10.1093/emboj/cdg418. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Or E, Navon A, Rapoport T. 2002. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J 21:4470–4479. 10.1093/emboj/cdf471. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mori H, Ito K. 2003. Biochemical characterization of a mutationally altered protein translocase: proton motive force stimulation of the initiation phase of translocation. J Bacteriol 185:405–412. 10.1128/JB.185.2.405-412.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nishiyama K, Fukuda A, Morita K, Tokuda H. 1999. Membrane deinsertion of SecA underlying proton motive force-dependent stimulation of protein translocation. EMBO J 18:1049–1058. 10.1093/emboj/18.4.1049. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tsukazaki T. 2018. Structure-based working model of SecDF, a proton-driven bacterial protein translocation factor. FEMS Microbiol Lett 365:fny112. 10.1093/femsle/fny112. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Furukawa A, Yoshikaie K, Mori T, Mori H, Morimoto YV, Sugano Y, Iwaki S, Minamino T, Sugita Y, Tanaka Y, Tsukazaki T. 2017. Tunnel formation inferred from the I-form structures of the proton-driven protein secretion motor SecDF. Cell Rep 19:895–901. 10.1016/j.celrep.2017.04.030. [PubMed] [DOI] [PubMed] [Google Scholar]
- 133.Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, Ito K, Nureki O. 2011. Structure and function of a membrane component SecDF that enhances protein export. Nature 474:235–238. 10.1038/nature09980. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Park E, Rapoport TA. 2012. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys 41:21–40. 10.1146/annurev-biophys-050511-102312. [PubMed] [DOI] [PubMed] [Google Scholar]


