Abstract
Background:
Heart failure (HF) is a complex clinical syndrome with high mortality. Current risk stratification approaches lack precision. High-throughput proteomics could improve risk prediction. Their use in clinical practice to guide the management of patients with HF depends on validation and evidence of clinical benefit.
Objective:
To develop and validate a protein risk score for mortality in patients with HF.
Design:
Community-based cohort.
Setting:
Southeast Minnesota.
Participants:
Patients with HF enrolled between 2003–2012 and followed through 2021.
Measurements:
We measured 7,289 plasma proteins in 1,351 HF patients, using the SomaScan assay. Using least absolute shrinkage and selection operator (LASSO) regression and temporal validation, we derived a protein risk score in patients enrolled between 2003–2007 (development cohort) and 2008–2012 (validation cohort). Multivariable Cox regression was used to examine the association between the protein risk score and mortality. The performance of the protein risk score to predict 5-year mortality risk was assessed using calibration plots, decision curves, and relative utility analyses and compared with a clinical model including the Meta-Analysis Global Group in Chronic Heart Failure (MAGGIC) mortality risk score and N-terminal pro-brain natriuretic peptide (NT-proBNP).
Results:
The development (N, 855; median age, 78 years; 50% female; 29% with ejection fraction <40%) and validation cohorts (N, 496; median age, 76 years; 45% female; 33% with ejection fraction < 40%) were mostly similar. In the development cohort, 38 unique proteins were selected for the protein risk score. Independently of ejection fraction, the protein risk score demonstrated good calibration, reclassified mortality risk particularly at the extremes of the risk distribution and greater clinical utility compared with the clinical model.
Limitation:
Participants were predominantly of European ancestry, potentially limiting the generalizability of the findings to different patient populations.
Conclusion:
Validation of the protein risk score demonstrated good calibration and evidence of predicted benefits to stratify the risk of death in HF superior to that of clinical methods. Further studies are needed to prospectively evaluate the score performance in diverse populations and determine risk thresholds for interventions.
Keywords: Heart failure, proteomics, mortality, risk prediction, risk stratification
Introduction
Heart failure (HF) is a highly prevalent and heterogeneous clinical syndrome (1). Despite therapeutic advances, survival after the diagnosis remains poor (2–6). The 2022 American Heart Association/American College of Cardiology/ Heart Failure Society of America (AHA/ACC/HFSA) Guideline for the Management of Heart Failure recommends the development and evaluation of omics-based strategies to improve HF risk stratification beyond current guideline-recommended risk-prediction tools such as the Meta-Analysis Global Group in Chronic Heart Failure (MAGGIC) score and natriuretic peptides (7). Risk stratification approaches that capture the biological complexity of the HF syndrome and show clinical utility are needed (6, 8–10). High-throughput proteomics is an attractive and novel approach to address this need and can be evaluated in large epidemiologic studies (11, 12). Prior studies examining the prognostic value of proteomics in HF (13–21) have provided an important proof of concept for its use in HF risk stratification. However, these studies have notable limitations, including methodological heterogeneity, selection biases, left ventricular ejection fraction (EF) restrictions, and in some cases, small sample sizes (13–20, 22). These limitations underscore the critical need to evaluate the prognostic value of proteomics in unselected populations such as community cohorts which have the advantage of high clinical relevance. In assembling such cohorts, it is critical to include the entire spectrum of HF encountered in clinical practice regardless of EF (12, 23).
Our primary goal was to assess if proteomics improved mortality risk prediction, beyond clinical factors and across the spectrum of HF syndrome. A secondary objective was to explore the potential clinical utility, including among patients with preserved and reduced EF. Accordingly, we assembled a large community cohort of patients with confirmed HF in which we developed and validated a protein risk score to predict mortality.
Methods
Patient Population
This HF community cohort was derived from the record linkage system of the Rochester Epidemiology Project, which is an ideal setting to conduct population research as it captures nearly all clinical diagnoses, procedures, results, and outcomes in its catchment area (24, 25). The approach for case identification, cohort assembling, and data collection has been previously reported (26, 27). In brief, patients with HF were identified with natural language processing of the text of the electronic medical record. Patients 20 years and older who were residents of Olmsted, Dodge, and Fillmore Counties in Minnesota were identified. This approach yielded 100% sensitivity compared with billing data, a reference method for case finding (28). Research nurses reviewed and validated HF diagnosis using the Framingham criteria (29). Patients were approached in the hospital or after an outpatient encounter to provide written consent, including a blood draw, between 2003 and 2012. The Mayo Clinic and Olmsted Medical Center Institutional Review Boards approved this study.
Data Collection
Clinical information including inpatient and outpatient provider records was collected by nurse abstractors (25). Body mass index was calculated using weight (kilograms) from the last outpatient prior to enrollment divided by the square of the earliest recorded adult height (meters). Patients were classified as either HF with preserved EF (HFpEF; EF≥40) or HF with reduced EF (HFrEF; EF<40). MAGGIC scores were calculated using sex, age, EF, systolic blood pressure, body mass index, creatinine, New York Heart Association class, current smoking status, diabetes, chronic obstructive pulmonary disease, HF diagnosis ≥18 months ago, and use of beta blockers, angiotensin-converting enzyme inhibitors or angiotensin-receptor blockers (30). The estimated glomerular filtration rate was calculated using the Chronic Kidney Disease Epidemiology Collaboration 2021 creatinine equation (31). N-terminal pro-brain natriuretic peptide (NT-proBNP) levels were measured at the National Institute of Diabetes and Digestive and Kidney Diseases Clinical Laboratory Core using a multiplex immunoassay (Meso Scale Diagnostics).
Mortality Ascertainment
Patients were followed through March 31, 2021. Death information was obtained from the healthcare providers that participated in the Rochester Epidemiology Project, from Minnesota death certificates, and patient record linkage to the National Death Index Plus. All-cause mortality and cardiovascular-related mortality (International Classification of Diseases, Tenth Revision, codes I00–178) were considered. Patients alive at the end of follow-up were censored as of March 31, 2021, or the date of last known healthcare contact, whichever was earlier.
Plasma Protein Measurements and Quality Control
Ethylenediaminetetraacetic acid-plasma samples, collected using a standardized protocol through the Rochester Epidemiology Project at enrollment (24, 25), were shipped to the National Institutes of Health (Bethesda, Maryland, USA) and stored at −80°C until they were aliquoted and shipped to SomaLogic, Inc. (Boulder, CO, USA) for proteomic analysis, using the SomaScan 7K version 4.1 assay.
The SomaScan platform, technology, assay, sample stability assessment, and annotations for proteomic measurements have been previously described (11, 32, 33). Briefly, the SomaScan 7K assay consists of 7,596 slow off-rate chemically modified single-stranded DNA aptamer reagents (SOMAmer), which are oligonucleotides of ~50 base pairs in length that are capable of binding target proteins or peptides with high specificity and affinity. This includes 7,289 SOMAmers targeting human proteins, 261 non-human proteins, 12 hybridization control elution, 4 binding mouse Fc-fusion, 4 non-cleavable, 6 ephrin-type receptors, and 20 spuriomers.
SomaScan measurements were normalized by SomaLogic using results from 51 vendor quality control samples derived from a pool of healthy participants. Specifically, normalization included a series of standardization techniques, including hybridization of controls to mitigate variations within the runs, median signal normalization across pooled calibrator replicates to account for signal variation using adaptive normalization by maximum likelihood, plate scaling to adjust for overall signal intensity differences, and SOMAmer calibration to correct for reagent-specific assay differences between runs. Samples in which the calibration factors fell outside the accepted range (hybridization scale factors, 0.4–2.5; plate scale/median scale and median normalization scale factors, 0.4–2.5; plate calibration scale factors, 0.6–14, SOMAmer calibration, 0.8–12) were flagged and excluded from the analysis.
SOMAmer measurements were log2 transformed to account for skewness. SOMAmer measurements outside ±5 standard deviations of the sample mean were winsorized (34). SOMAmer measurements were standardized to have a mean of 0 and a standard deviation of 1. Sensitivity analyses were performed considering rank-based inverse normal transformation (35). Intra-assay Bland-Altman coefficient of variation was calculated using SOMAmer measurements from thirty randomly selected duplicate patient samples. SOMAmer measurements with a coefficient of variation greater than or equal to 50% were excluded from further analyses.
Protein Risk Score Development
The development and validation of the protein risk score is summarized in Supplementary Figure 1. Briefly, using temporal validation (36), patients enrolled between 2003 and 2007 (n=855) were used as the protein risk score development cohort, and patients enrolled between 2008 and 2012 (n=496) were used as the protein risk score validation cohort. For comparability, patient follow-up was administratively censored at 10 years in both cohorts. Associations between individual SOMAmers and mortality were assessed using Cox proportional hazard model adjusted for age, sex, and estimated glomerular filtration rate, as renal function has been reported to affect plasma protein levels (37, 38). SOMAmers with a Benjamini-Hochberg false discovery rate of less than 1% were selected for least absolute shrinkage and selection operator (LASSO) penalized Cox regression model. The LASSO penalty parameter was optimized as the value within 1 standard error to minimize the 10-fold cross-validation error. Then using this lambda, the LASSO model was fitted. The beta coefficients from this model were used to generate the weighted linear sum of the protein risk score, further details are provided in the Supplemental Methods. Given the high dimensionality of the predictors, LASSO was used to avoid overfitting (39). Under the assumption the expected R-square of a continuous predictor (protein risk score) ranged from 0.2 to 0.5, we were powered to detect a minimum hazard ratio of 1.18 to 1.24 with 80% power with a two-sided type I error rate of 5% in the validation cohort.
Statistical Analysis
Baseline characteristics are reported as frequencies (percentages) for categorical variables or median (interquartile range [IQR]) for continuous variables. NT-proBNP values were log2 transformed for analyses. Multiple imputations by chained equations, using Rubin’s rule with 10 imputed data sets (40), were performed to account for missing clinical data used to calculate MAGGIC scores, including body mass index (2.9%), New York Heart Association class (0.4%), HF duration (0.1%), and EF (1.8%).
Multivariable Cox proportional hazard regression was used to examine the association between a 1 standard deviation change in protein risk score and all-cause mortality, with and without adjustment for MAGGIC score and NT-proBNP. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated for the protein risk score in both the development and validation cohorts. Secondary analyses included stratification by EF groups (< or ≥40%) and HF duration at enrollment (< or ≥18 months). The association between the protein risk score and cardiovascular disease-related mortality was also examined using Fine and Gray sub-distribution hazard modeling (41).
The performance of the protein risk score to predict a patient’s 5-year mortality risk was assessed in the validation cohort using calibration plots (42, 43), decision curves (44, 45), and relative utility (46) analyses. Performance was compared with models containing MAGGIC score and NT-proBNP (“Clinical Model”) across a range of predicted risk thresholds including four predefined risk groups (≤25%, 26–50%, 51–75%, and >75%). HRs and 95% CIs were estimated for each model across risk groups and compared to the reference risk group of ≤25%. Decision curves were plotted to examine the predicted net benefit of the protein risk score model compared with the clinical model. The predicted net benefit is the difference between true positives and false positives adjusted for the relative harms of false positive classifications across risk thresholds (44, 45). Relative utility curves plot the predicted net benefit of a model compared to that of a perfect prediction across a range of risk thresholds (46). We examined relative utility curves of the protein risk score model and the clinical models.
All the analyses were performed using R statistical software v4.0.2 (R Core Team, Vienna, Austria) with a two-sided p-value < .05 considered statistically significant. Our approach follows the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) guidelines (47).
Role of the Funding Source
The investigators were supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute of the National Institutes of Health (ZIAHL006279). This study also used in part the resources from the Rochester Epidemiology Project medical records-linkage system, which is supported by the National Institute on Aging, the Mayo Clinic Research Committee, and fees paid annually by Rochester Epidemiology Project users. The funding institution did not play a role in the design, conduct, analysis, or reporting nor in the decision to submit this manuscript for publication.
Results
Cohort Characteristics
After excluding one patient with insufficient plasma volume and 37 patients whose samples failed SomaLogic quality control, we analyzed data from 1,351 patients. Baseline clinical characteristics are summarized in Table 1. In brief, the patient population was 48% female and had a median age of 78 years. Thirty percent of the patient population had an EF <40%, the median MAGGIC score was 25 (IQR 20 – 29), and the median NT-proBNP level was 8,903 (IQR 4,211 – 16,384) pg/ml. A total of 1,013 deaths occurred during follow-up with an overall 5-year mortality rate of 52.1% (95% CI 49.3 – 54.7%). Baseline characteristics of the development and validation cohorts were mostly similar in terms of demographics, medical history, and clinical presentation of HF.
Table 1.
Cohort Characteristics
| Variable | Total (N=1,351) | Development Cohort (N=855) | Validation Cohort (N=496) |
|---|---|---|---|
|
| |||
| Demographics | |||
| Age (years) | 78 (68, 84) | 78 (69, 85) | 76 (65, 84) |
| Male | 701 (52) | 427 (50) | 274 (55) |
| Body Mass Index (kg/m2) | 28 (24, 33) | 28 (24, 33) | 29 (25, 34) |
| Medical History | |||
| Current Smoker | 138 (10) | 85 (10) | 53 (11) |
| Diabetes | 481 (36) | 288 (34) | 193 (39) |
| Hypertension | 1,234 (91) | 774 (91) | 460 (93) |
| COPD | 380 (28) | 257 (30) | 123 (25) |
| Cerebrovascular Disease | 385 (28) | 265 (31) | 120 (24) |
| Atrial Fibrillation | 486 (36) | 282 (33) | 204 (41) |
| Ischemic Etiology | 678 (50) | 436 (51) | 242 (49) |
| Clinical Presentation | |||
| NYHA Class (III/IV) | 930 (69) | 584 (68) | 346 (70) |
| Ejection Fraction <40% | 415 (30) | 250 (29) | 165 (33) |
| MAGGIC Score | 25 (20, 29) | 25 (20, 29) | 23 (19, 27) |
| HF Duration (≥ 18 months) | 486 (36) | 310 (36) | 176 (36) |
| Laboratory Measurements | |||
| eGFR (mL/min/1.73m2) | 57 (42, 73) | 54 (41, 68) | 62 (45, 82) |
| NT-proBNP (pg/mL) | 8903 (4211, 16384) | 9345 (4182, 17080) | 8481 (4240, 15181) |
| Medications | |||
| ACEI/ARB | 868 (64) | 541 (63) | 327 (66) |
| Beta-Blocker | 1,024 (76) | 614 (72) | 410(83) |
| Diuretics | 1,047 (77) | 639 (75) | 408 (82) |
| Mortality per 100 Person-Years | |||
| All-Cause | 14.7 (13.8, 15.6) | 16.0 (14.8, 17.2) | 12.6 (11.3, 14.0) |
| Cardiovascular-Related | 7.4 (6.8, 8.1) | 8.2 (7.3, 9.1) | 6.3 (5.4, 7.3) |
Values are reported as the frequency (percent) or median (interquartile range).
COPD, Chronic Obstructive Pulmonary Disease; NYHA, New York Heart Association; MAGGIC, Meta-Analysis Global Group in Chronic Heart Failure; HF, Heart Failure; eGFR, Estimated Glomerular Filtration Rate; NT-proBNP, N-Terminal Pro–B-Type Natriuretic Peptide; ACEI, Angiotensin-converting enzyme inhibitor; ARB, Angiotensin receptor blocker;.
Protein Risk Score Development
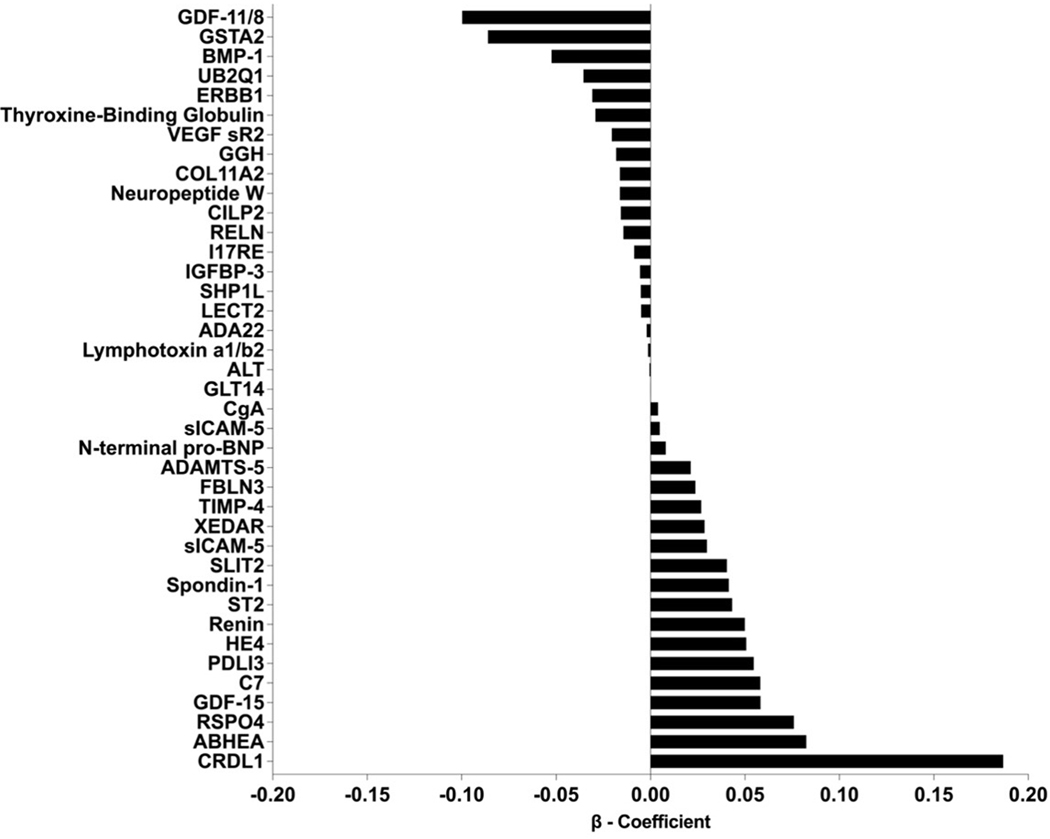
Out of 7,289 SOMAmers measured, 7,151 were available after quality control (138 SOMAmers were excluded with a Bland-Altman coefficient of variation greater than or equal to 50%). The median intra-assay coefficient of variation was 8.8% (IQR 6.7 – 12.9%). In univariate analyses, 1,342 SOMAmers were associated with mortality with FDR less than 1%. Thirty-nine SOMAmers targeting 38 unique proteins were selected by the LASSO model to generate the protein risk score (Figure 1). Twenty SOMAmers were negatively associated with mortality, and 19 were positively associated with mortality. Further details of the 39 SOMAmers and their possible biological links (48) are provided in Supplementary Table 1 and Supplementary Figure 2 respectively.
Figure 1. Beta Coefficients of the 39 SOMAmers (38 unique proteins) in the Protein Risk Score.
GDF-11/8, Growth/differentiation factor 11/8; GSTA2, Glutathione S-transferase A2; BMP-1, Bone morphogenetic protein 1; UB2Q1, Ubiquitin-conjugating enzyme E2 Q1; ERBB1, Epidermal growth factor receptor; THBG, Thyroxine-binding globulin; VEGF sR2, Vascular endothelial growth factor receptor 2; GGH, Gamma-glutamyl hydrolase; COL11A2, Collagen alpha-2(XI) chain; NPW, Neuropeptide W; CILP2, Cartilage intermediate layer protein 2; RELN, Reelin; I17RE, Interleukin-17 receptor E; IGFBP-3, Insulin-like growth factor-binding protein 3; SHP1L, Testicular spindle-associated protein SHCBP1L; LECT2, Leukocyte cell-derived chemotaxin-2; ADA22, Disintegrin and metalloproteinase domain-containing protein 22; TNFC, Lymphotoxin alpha1:beta2; ALT, Alanine aminotransferase 1; GLT14, Polypeptide N-acetylgalactosaminyltransferase 14; CgA, Chromogranin-A; sICAM-5, Intercellular adhesion molecule 5; NPPB, N-terminal pro-BNP; ADAMTS-5, A disintegrin and metalloproteinase with thrombospondin motifs 5; FBLN3, EGF-containing fibulin-like extracellular matrix protein 1; TIMP-4, Metalloproteinase inhibitor 4; XEDAR, Tumor necrosis factor receptor superfamily member 27; sICAM-5, Intercellular adhesion molecule 5; SLIT2, Slit homolog 2 protein; SPON1, Spondin-1; ST2, Suppression of Tumorigenicity 2 protein; REN, Renin; HE4, Human Epididymis protein; PDLI3, PDZ and LIM domain protein 3; C7, Complement component C7; GDF-15, Growth/differentiation factor 15; RSPO4, R-spondin-4; ABHEA, Protein ABHD14A; CRDL1, Chordin-like protein 1.
The protein risk score had a standard deviation of 0.663 and 0.659 in the development and validation cohorts, respectively. After adjustment for the MAGGIC score, a 1 standard deviation increase in protein risk score was associated with an increased risk of mortality in both the development (HR: 2.62, 95% CI, 2.34 – 2.93) and validation cohorts (HR: 2.01, 95% CI, 1.75 – 2.32). Improved risk stratification persisted after further adjustment for NT-proBNP (Supplementary Figure 3).
The association between the protein risk score and increased mortality persisted in analyses stratified by HF duration, in sensitivity analyses considering rank-based inverse normal transformation, and after excluding either of the SOMAmers that targeted the same protein were retained in the protein risk score. In the validation cohort, the association between the protein risk score and mortality persisted when restricted to cardiovascular disease causes in the crude model (HR: 1.55, 95% CI, 1.34 – 1.79) but was attenuated after adjustment for the MAGGIC score (HR: 1.12, 95% CI, 0.93 – 1.35).
Protein Risk Score Performance
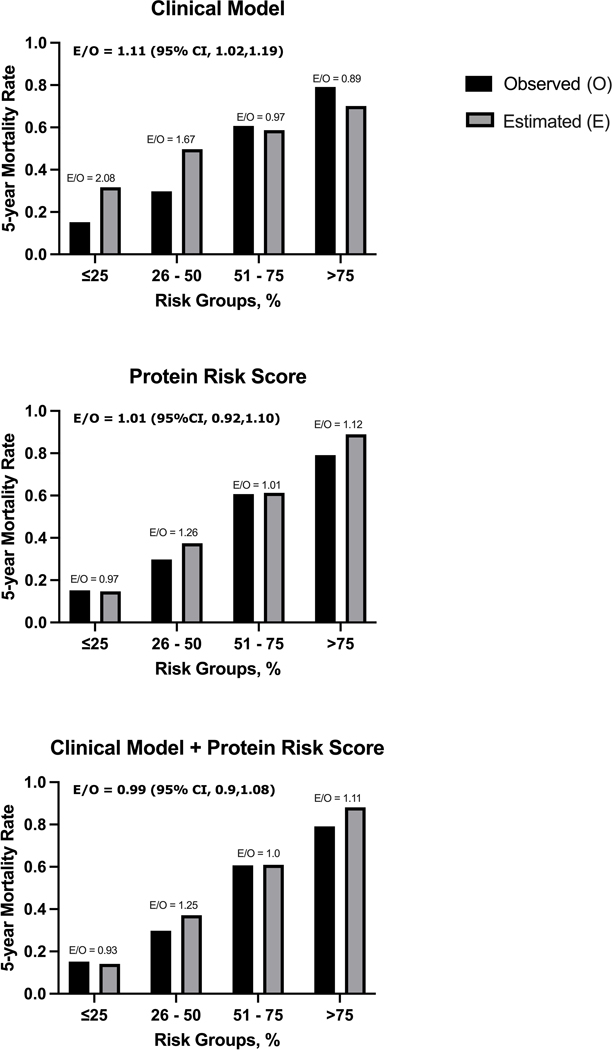
The calibration in the validation cohort was excellent for the protein risk score with an estimated to observed mortality ratio of 1.01 (95% CI, 0.92 – 1.10) and appeared superior to that of the clinical models, particularly at the extremes of the risk distribution (Figure 2, Supplemental Figure 4). The protein risk score showed strong discrimination across risk groups, which persisted after adjustment for MAGGIC score and NT-proBNP (Table 2). Addition of the protein risk score to the clinical model classified more patients at the extremes of the risk distribution: 27.8% compared to 12.5% for the lowest risk group and 22.4% compared to 13.3% in the highest risk group (Table 3). The protein risk score performed similarly among HFrEF and HFpEF patients (Supplementary Table 2).
Figure 2. Observed and Estimated Risk of 5-Year Mortality by Predicted Risk Groups in Validation Cohort.
The calibration of the Protein Risk Score model is excellent across all risk groups. Improved calibration above that of the clinical models is evident at both extremes of the risk distribution.
Table 2.
5-year Mortality Hazard Ratios and 95% Confidence Intervals by Risk Groups and Multivariable Model; (A) Protein Risk Score; (B) Clinical Model (MAGGIC + NTproBNP) and (C) Clinical Model plus Protein Risk Score
| A. Clinical Model | Risk Groups | ||||
|---|---|---|---|---|---|
|
| |||||
| ≤25% | 26 – 50% | 51 – 75% | >75% | Total | |
|
| |||||
| Patients (N) | 62 | 164 | 204 | 66 | 496 |
| Mortality Rate | 0.13 (0.04,0.21) | 0.3 (0.23,0.37) | 0.53 (0.46,0.59) | 0.83 (0.71,0.90) | 0.45 |
| (0.4, 0.49) | |||||
| Mean Prediction | 0.17 | 0.39 | 0.62 | 0.84 | 0.45 |
| HR | 1 |
2.55
(1.21, 5.38) |
5.34
(2.60, 10.94) |
10.20
(6.27,16.60) |
|
|
| |||||
| B. Protein Risk Score | |||||
|
| |||||
| ≤25% | 26 – 50% | 51 – 75% | >75% | Total | |
|
| |||||
| Patients (N) | 133 | 131 | 117 | 115 | 496 |
| Mortality Rate | 0.15 (0.09,0.21) | 0.3 | 0.61 | 0.79 (0.7,0.85) | 0.45 |
| (0.21, 0.37) | (0.51, 0.69) | (0.4, 0.49) | |||
| Mean Prediction | 0.14 | 0.37 | 0.61 | 0.89 | 0.45 |
| HR | 1 |
2.13
(1.24,3.64) |
5.75
(3.50,9.45) |
10.20
(6.27,16.60) |
|
|
| |||||
| C. Clinical Model + Protein Risk Score | |||||
|
| |||||
| ≤25% | 26 – 50% | 51 – 75% | >75% | Total | |
|
| |||||
| Patients (N) | 138 | 131 | 116 | 111 | 496 |
| Mortality Rate | 0.15 (0.09,0.20) | 0.31 (0.23,0.39) | 0.6 (0.50,0.68) | 0.81 (0.72,0.87) | 0.45 (0.4, 0.49) |
| Mean Prediction | 0.14 | 0.38 | 0.62 | 0.89 | 0.45 |
| HR | 1 |
2.37
(1.39,4.04) |
5.84
(3.55, 9.61) |
11.44
(7.02, 18.64) |
|
Table 3.
Reclassification of 5-year Mortality Risk by Adding the Protein Risk Score to the Clinical Model
| Clinical Model | Clinical Model + Protein Risk Score | Reclassified by Protein Risk Score | ||||||
|---|---|---|---|---|---|---|---|---|
| ≤25% | 26 – 50% | 51 – 75% | >75% | Total N (%) | Lower | Higher | ||
|
| ||||||||
| ≤25% | Patients, N (%) | 54 (87.1) | 7 (11.3) | 1 (1.6) | 0 (0.0) | 62 (12.5) | − | 8 (12.9%) |
| Mortality (%) | 7.6 | 42.9 | 100 | − | 13.1 | − | 50 | |
| 26 – 50% | Patients, N (%) | 69 (42.1) | 62 (37.8) | 25 (15.2) | 8 (4.9) | 164 (33.1) | 69 (42.1%) | 33 (20.1%) |
| Mortality (%) | 20.3 | 29 | 56 | 50 | 30.5 | 20.3 | 54.5 | |
| 51 – 75% | Patients, N (%) | 15 (7.4) | 58 (28.4) | 76 (37.3) | 55 (27.0) | 204 (41.1) | 73 (35.8%) | 55 (27.0%) |
| Mortality (%) | 14.3 | 31 | 59.2 | 78.2 | 53.1 | 27.7 | 78.2 | |
| >75% | Patients, N (%) | 0 (0.0) | 4 (6.1) | 14 (21.2) | 48 (72.7) | 66 (13.3) | 18 (27.3%) | − |
| Mortality (%) | 50 | 71.4 | 89.6 | 83.3 | 66.7 | |||
| Total | Patients, N (%) | 138 (27.8) | 131 (26.4) | 116 (23.4) | 111 (22.4) | 496 (100.0) | ||
| Mortality (%) | 14.7 | 31.3 | 60.3 | 81.1 | 44.7 | |||
Adding the Protein Risk Score to the clinical model places more patients at the extremes of the risk distribution.
Assessment of Potential for Clinical Application
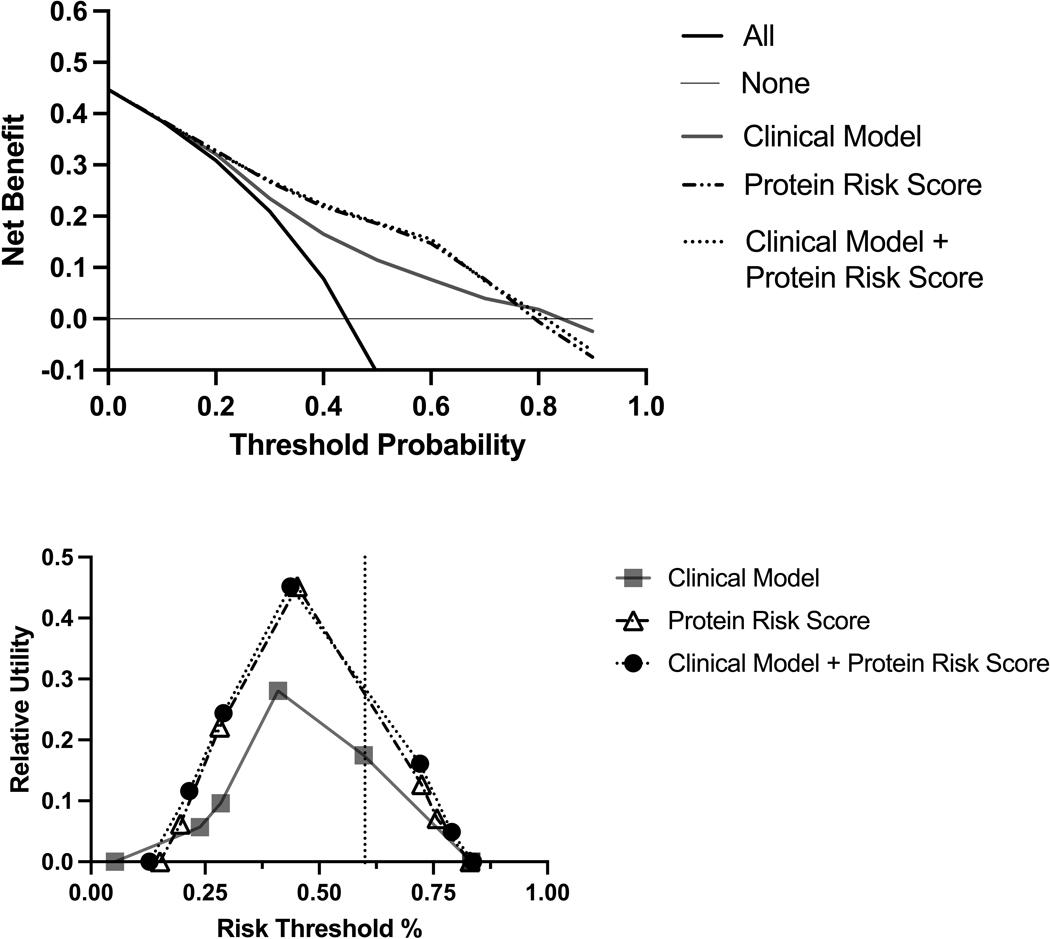
Decision curve analysis showed that the protein risk score provided a greater net benefit compared to the clinical model across a wide range (17–76%) of risk thresholds (Figure 3A). The relative utility for the protein risk score was higher compared with the clinical model, across risk thresholds for probability of mortality (Figure 3B). At a risk threshold of 50%, the relative utility of the protein risk score only model (0.39) and protein risk score plus clinical model (0.39) were both notably superior to that of the clinical model alone (0.23).
Figure 3. Decision Curves (A) and Relative Utility Curve (B) of the Protein Risk Score Compared with the Clinical Model for Heart Failure Management Decisions.
Panel A: In the decision curve analysis, the protein risk score shows greater net benefit compared with the clinical model alone across a wide range of risk thresholds (17–76%).
Panel B: The relative utility for the protein risk score is higher compared to the clinical model, across risk thresholds for probability of mortality.
Discussion
In this community cohort, using high-throughput proteomics, we derived a novel protein risk score, which greatly contributed to mortality risk prediction across a wide range of risk thresholds. By integrating proteomics measurements with extensive clinical data from medical records, we demonstrated that the risk prediction information, measured using proteomics, was independent of and superior to clinical variables (MAGGIC score and NTproBNP), which have been the cornerstone of HF risk stratification (7). Importantly, the excess risk of mortality conferred by the protein risk score did not differ by EF. These findings provide clinically significant new evidence in support of the important role of proteomics in stratifying risk across the HF syndrome.
Risk Stratification in HF: Need for a Precision Approach
Despite advancements in medical therapy, mortality risk remains large in HF (6, 49). Clinical risk stratification is currently centered on the use of scores, including the MAGGIC score as well as the natriuretic peptides (7, 30, 50). The performance of these risk markers typically declines once disseminated to settings and populations different from the initial validation populations (51, 52). This degradation in predictive performance negatively impacts the utility of scores in clinical practice.
Proteins and Risk Scores in Heart Failure
There are few studies of protein risk scores for HF risk stratification. Two studies that used the same aptamer-based method as used herein, were both restricted to HF with reduced EF (16, 20). The first study utilized data from a voluntary registry to derive an 8-protein risk score that improved mortality risk stratification, independent of the MAGGIC score and NT-proBNP (16). The second study conducted a secondary analysis of clinical trial data to derive a 64-protein risk score which improved the prediction of the composite outcome of cardiovascular mortality or HF hospitalization, over the MAGGIC score (20). Our protein risk score shared two proteins (renin and epidermal growth factor receptor) with the 8-protein risk score (16), and five proteins (growth differentiation factor 15, collagen alpha-2[XI] chain, renin, NT-proBNP, and cartilage intermediate layer protein 2) with the 64-protein risk score (20). Renin was the only protein included in the two published risk scores (16, 20), and in ours. Renin is associated with left ventricular dysfunction, cardiac dilatation, fluid retention, and cachexia (53). While the prognostic value of renin has been questioned (54), the present study underscores the need for further studies examining its role in clinical practice.
Our protein risk score included several additional proteins known to be associated with HF, including NT-proBNP, growth differentiation factor-15, and interleukin-1 receptor-like 1 (commonly known as ST2) (55). NT-proBNP increases in response to left ventricular wall stress and is involved in the regulation of blood pressure, blood volume, and sodium balance (56, 57). Growth differentiation factor-15 is a prognostic biomarker in HF associated with inflammation and apoptosis (58). Similarly, interleukin-1 receptor-like 1, a marker of myocardial and vascular strain and remodeling (59, 60) is associated with mortality in HF (61, 62). Identification of these markers in our protein risk score supports its biological plausibility.
An advantage of a non-targeted proteomic approach is the ability to discover novel proteins and examine their possible biological links. Three of the top predictors in our protein risk score (chordin-like protein 1, growth differentiation factor 11/8, and R-spondin-4) have been previously reported as associated with mortality in aptamer assay studies of HF with reduced EF (16, 20). Our findings extend these observations across the entire spectrum of HF, independently of EF. Chordin-like protein 1 is a transforming growth factor beta 1 antagonist, which inhibits fibrosis and cardiac remodeling (63). Growth differentiation factor 11/8 is associated with left ventricular hypertrophy and cardiovascular events in large cohorts (64). R-spondin-4 is linked with cardiac remodeling and fibrosis (65). The novel findings of the prognostic value of these proteins identified in our protein risk score provide a blueprint for future mechanistic studies relevant to the entire spectrum of the HF syndrome, regardless of EF.
The predictive capabilities of sparse combinations of proteins open the door to assays, that cover several mechanistic domains with prognostic value in HF, potentially accessible at a lower cost than discovery assays such as the one used herein. However, clinical translation will likely require the development of assays with absolute quantification as opposed to the relative quantification provided by current tests.
Study Limitations, Strengths, and Clinical Implications
Our cohort was predominantly of European ancestry, potentially limiting the generalizability to ethnically and racially diverse populations. This emphasizes the importance of replication in diverse populations. While patients were recruited several years ago to allow event accrual, survival after HF diagnosis has remained disappointingly poor (6), such that the outcomes observed herein are relevant to current practice. Although our cohort contained both incident and prevalent cases of HF, reflective of a community cohort, the protein risk score performed independently of HF duration. We acknowledge the conceptual importance of comorbid conditions and/or drug therapies. Future studies of proteomic expression according to HF endophenotypes and comorbidities should be conducted in cohorts large enough to allow robust inference. Finally, we acknowledge that, as with any observational study, we cannot rule out potential residual confounding related to unmeasured characteristics and measurement error.
This study has several important strengths. Firstly, we studied a population-based cohort, which represents the complete and consecutive experience of a geographically defined community of patients with validated HF including all EF categories. This, coupled with the availability of rich medical record data from in-patient and out-patient encounters and extensive follow-up with complete mortality ascertainment provides strong clinical relevance to our findings. Secondly, we used a high-throughput proteomics assay with the largest number of proteins measured to date. Our agnostic discovery approach enabled the discovery of signatures that exhibit strong associations with mortality. Thirdly, we used statistical methods designed to optimize reproducibility and validity such as cross-validation, and temporal validation per the TRIPOD guideline (47).
By identifying protein-based signatures that can stratify the risk of death in HF in a manner beyond current clinical tools, our findings foreshadow the clinical utility of large-scale proteomic assays for precision risk prediction in HF. Examples within the heterogenous HF syndrome, include selection of candidates for rapid drug titration (66) or patients with advanced HF (67), at particularly high risk for adverse outcomes, regardless of EF that should be considered for mechanical circulatory support or transplantation. The need to better identify candidates for referral to advanced HF specialists is recognized (68). To assess the clinical application of proteomics, model performance should be formally evaluated for clinical translation and practice implementation. Clinical decision support must be examined separately, ideally by randomized studies (69). These considerations notwithstanding, our results provide the proof of concept that proteomics can address this need with greater precision than current clinical methods. By illustrating the potential of high-throughput omics to improve the clinical management of the HF syndrome, our findings are directly relevant to clinical practice and strongly support the pursuit of the evaluation of proteomics for this purpose.
In conclusion, our study responds to the recognized need to evaluate how “omics” data can be used for HF risk stratification. We developed and validated a protein risk score that stratified the risk of death in HF and showed clinical utility beyond current clinical tools in a community cohort that includes all forms of the HF syndrome and is therefore of high clinical relevance. Collectively, our findings bring important new evidence in support of the value of proteomics for risk stratification in HF.
Supplementary Material
Primary Funding Source:
Division of Intramural Research at the National Heart, Lung, and Blood Institute of the National Institutes of Health (ZIAHL006279).
Abbreviations and Acronyms
- HF
Heart Failure
- HFrEF
Heart Failure with Reduced Ejection Fraction
- HFpEF
Heart Failure with Preserved Ejection Fraction
- HR
Hazard Ratio
- CI
Confidence Interval
- EF
Ejection Fraction
- AHA/ACC/HFSA
American Heart Association/American College of Cardiology/ Heart Failure Society of America
- MAGGIC
Meta-Analysis Global Group in Chronic Heart Failure
- SOMAmers
Slow Off-Rate Modified Aptamers
- LASSO
Least Absolute Shrinkage and Selection Operator
- IQR
Interquartile Range
References
- 1.Tsao CW, Aday AW, Almarzooq ZI, Alonso A, Beaton AZ, Bittencourt MS, et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation. 2022;145(8):e153–e639. Epub 2022/01/27. doi: 10.1161/CIR.0000000000001052. [DOI] [PubMed] [Google Scholar]
- 2.Levy D, Kenchaiah S, Larson MG, Benjamin EJ, Kupka MJ, Ho KK, et al. Long-term trends in the incidence of and survival with heart failure. N Engl J Med. 2002;347(18):1397–402. doi: 10.1056/NEJMoa020265. [DOI] [PubMed] [Google Scholar]
- 3.Taylor CJ, Ordóñez-Mena JM, Roalfe AK, Lay-Flurrie S, Jones NR, Marshall T, et al. Trends in survival after a diagnosis of heart failure in the United Kingdom 2000–2017: population based cohort study. BMJ (Clinical research ed). 2019;364:l223–l. doi: 10.1136/bmj.l223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah KS, Xu H, Matsouaka RA, Bhatt DL, Heidenreich PA, Hernandez AF, et al. Heart Failure With Preserved, Borderline, and Reduced Ejection Fraction: 5-Year Outcomes. J Am Coll Cardiol. 2017;70(20):2476–86. Epub 20171112. doi: 10.1016/j.jacc.2017.08.074. [DOI] [PubMed] [Google Scholar]
- 5.Cowie MR, Wood DA, Coats AJ, Thompson SG, Suresh V, Poole-Wilson PA, et al. Survival of patients with a new diagnosis of heart failure: a population based study. Heart. 2000;83(5):505–10. doi: 10.1136/heart.83.5.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roger VL. Epidemiology of Heart Failure. Circulation Research. 2021;128(10):1421–34. doi: doi: 10.1161/CIRCRESAHA.121.318172. [DOI] [PubMed] [Google Scholar]
- 7.Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, et al. 2022. AHA/ACC/HFSA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation.0(0): 10.1161/CIR.0000000000001063. doi: doi: 10.1161/CIR.0000000000001063. [DOI] [PubMed] [Google Scholar]
- 8.Chirinos JA, Orlenko A, Zhao L, Basso MD, Cvijic ME, Li Z, et al. Multiple Plasma Biomarkers for Risk Stratification in Patients With Heart Failure and Preserved Ejection Fraction. Journal of the American College of Cardiology. 2020;75(11):1281–95. doi: doi: 10.1016/j.jacc.2019.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francis GS, Cogswell R, Thenappan T. The heterogeneity of heart failure: will enhanced phenotyping be necessary for future clinical trial success? J Am Coll Cardiol. 2014;64(17):1775–6. Epub 20141021. doi: 10.1016/j.jacc.2014.07.978. [DOI] [PubMed] [Google Scholar]
- 10.Mann DL. Is It Time for a New Taxonomy for Heart Failure? Journal of Cardiac Failure. 2016;22(9):710–2. doi: 10.1016/j.cardfail.2016.07.432. [DOI] [PubMed] [Google Scholar]
- 11.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. Epub 2010/12/18. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JG, Gerszten RE. Emerging Affinity-Based Proteomic Technologies for Large-Scale Plasma Profiling in Cardiovascular Disease. Circulation. 2017;135(17):1651–64. doi: 10.1161/CIRCULATIONAHA.116.025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuvelliez M, Vandewalle V, Brunin M, Beseme O, Hulot A, de Groote P, et al. Circulating proteomic signature of early death in heart failure patients with reduced ejection fraction. Sci Rep. 2019;9(1):19202. Epub 20191216. doi: 10.1038/s41598-019-55727-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egerstedt A, Berntsson J, Smith ML, Gidlof O, Nilsson R, Benson M, et al. Profiling of the plasma proteome across different stages of human heart failure. Nat Commun. 2019;10(1):5830. Epub 20191220. doi: 10.1038/s41467-019-13306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira JP, Verdonschot J, Collier T, Wang P, Pizard A, Bär C, et al. Proteomic Bioprofiles and Mechanistic Pathways of Progression to Heart Failure. Circ Heart Fail. 2019;12(5):e005897-e. doi: 10.1161/CIRCHEARTFAILURE.118.005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui H, She R, Luzum J, Li J, Bryson TD, Pinto Y, et al. Plasma Proteomic Profile Predicts Survival in Heart Failure With Reduced Ejection Fraction. Circ Genom Precis Med. 2021;14(3):e003140. Epub 2021/05/18. doi: 10.1161/CIRCGEN.120.003140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nayor M, Short MI, Rasheed H, Lin H, Jonasson C, Yang Q, et al. Aptamer-Based Proteomic Platform Identifies Novel Protein Predictors of Incident Heart Failure and Echocardiographic Traits. Circ Heart Fail. 2020;13(5):e006749. Epub 20200515. doi: 10.1161/circheartfailure.119.006749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regan JA, Truby LK, Tahir UA, Katz DH, Nguyen M, Kwee LC, et al. Protein biomarkers of cardiac remodeling and inflammation associated with HFpEF and incident events. Scientific Reports. 2022;12(1):20072. doi: 10.1038/s41598-022-24226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders-van Wijk S, Tromp J, Beussink-Nelson L, Hage C, Svedlund S, Saraste A, et al. Proteomic Evaluation of the Comorbidity-Inflammation Paradigm in Heart Failure With Preserved Ejection Fraction: Results From the PROMIS-HFpEF Study. Circulation. 2020;142(21):2029–44. Epub 20201009. doi: 10.1161/CIRCULATIONAHA.120.045810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Cunningham JW, Claggett BL, Jacob J, Mendelson MM, Serrano-Fernandez P, et al. Aptamer Proteomics for Biomarker Discovery in Heart Failure With Reduced Ejection Fraction. Circulation. 2022;146(18):1411–4. doi: doi: 10.1161/CIRCULATIONAHA.122.061481. [DOI] [PubMed] [Google Scholar]
- 21.Girerd N, Levy D, Duarte K, Ferreira JP, Ballantyne C, Collier T, et al. Protein Biomarkers of New-Onset Heart Failure: Insights From the Heart Omics and Ageing Cohort, the Atherosclerosis Risk in Communities Study, and the Framingham Heart Study. Circ Heart Fail. 2023;16(5):e009694. Epub 20230516. doi: 10.1161/circheartfailure.122.009694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamo L, Yu J, Rocha-Resende C, Javaheri A, Head RD, Mann DL. Proteomic Signatures of Heart Failure in Relation to Left Ventricular Ejection Fraction. Journal of the American College of Cardiology. 2020;76(17):1982–94. doi: 10.1016/j.jacc.2020.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad T, Lund LH, Rao P, Ghosh R, Warier P, Vaccaro B, et al. Machine Learning Methods Improve Prognostication, Identify Clinically Distinct Phenotypes, and Detect Heterogeneity in Response to Therapy in a Large Cohort of Heart Failure Patients. J Am Heart Assoc. 2018;7(8). Epub 20180412. doi: 10.1161/jaha.117.008081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rocca WA, Yawn BP, St Sauver JL, Grossardt BR, Melton LJ, 3rd. History of the Rochester Epidemiology Project: half a century of medical records linkage in a US population. Mayo Clinic proceedings. 2012;87(12):1202–13. Epub 2012/11/28. doi: 10.1016/j.mayocp.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocca WA, Grossardt BR, Brue SM, Bock-Goodner CM, Chamberlain AM, Wilson PM, et al. Data Resource Profile: Expansion of the Rochester Epidemiology Project medical records-linkage system (E-REP). Int J Epidemiol. 2018;47(2):368-j. doi: 10.1093/ije/dyx268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunlay SM, Gerber Y, Weston SA, Killian JM, Redfield MM, Roger VL. Prognostic value of biomarkers in heart failure: application of novel methods in the community. Circ Heart Fail. 2009;2(5):393–400. Epub 20090729. doi: 10.1161/circheartfailure.109.849299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chamberlain AM, St Sauver JL, Gerber Y, Manemann SM, Boyd CM, Dunlay SM, et al. Multimorbidity in heart failure: a community perspective. Am J Med. 2015;128(1):38–45. Epub 20140916. doi: 10.1016/j.amjmed.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pakhomov SV, Buntrock J, Chute CG. Prospective recruitment of patients with congestive heart failure using an ad-hoc binary classifier. J Biomed Inform. 2005;38(2):145–53. doi: 10.1016/j.jbi.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 29.McKee PA, Castelli WP, McNamara PM, Kannel WB. The Natural History of Congestive Heart Failure: The Framingham Study. New England Journal of Medicine. 1971;285(26):1441–6. doi: 10.1056/nejm197112232852601. [DOI] [PubMed] [Google Scholar]
- 30.Pocock SJ, Ariti CA, McMurray JJ, Maggioni A, Køber L, Squire IB, et al. Predicting survival in heart failure: a risk score based on 39 372 patients from 30 studies. Eur Heart J. 2013;34(19):1404–13. Epub 20121024. doi: 10.1093/eurheartj/ehs337. [DOI] [PubMed] [Google Scholar]
- 31.Inker LA, Eneanya ND, Coresh J, Tighiouart H, Wang D, Sang Y, et al. New Creatinine- and Cystatin C-Based Equations to Estimate GFR without Race. N Engl J Med. 2021;385(19):1737–49. Epub 20210923. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS One. 2011;6(10):e26332. Epub 2011/10/25. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hensley P. SOMAmers and SOMAscan – A Protein Biomarker Discovery Platform for Rapid Analysis of Sample Collections From Bench Top to the Clinic. J Biomol Tech. 2013;24(Suppl):S5-S. [Google Scholar]
- 34.Reifman AKKWIS, Neil J. Encyclopedia of research design: SAGE Publications; 2010. 1636–8 p. [Google Scholar]
- 35.McCaw ZR, Lane JM, Saxena R, Redline S, Lin X. Operating characteristics of the rank-based inverse normal transformation for quantitative trait analysis in genome-wide association studies. Biometrics. 2020;76(4):1262–72. Epub 20200113. doi: 10.1111/biom.13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altman DG, Royston P. What do we mean by validating a prognostic model? Statistics in Medicine. 2000;19(4):453–73. doi: . [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Brody EN, Murthy AC, Mehler RE, Weiss SJ, DeLisle RK, et al. Impact of Kidney Function on the Blood Proteome and on Protein Cardiovascular Risk Biomarkers in Patients With Stable Coronary Heart Disease. J Am Heart Assoc. 2020;9(15):e016463. Epub 2020/07/23. doi: 10.1161/JAHA.120.016463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keshawarz A, Hwang SJ, Lee GY, Yu Z, Yao C, Kottgen A, et al. Cardiovascular disease protein biomarkers are associated with kidney function: The Framingham Heart Study. PLoS One. 2022;17(5):e0268293. Epub 2022/05/12. doi: 10.1371/journal.pone.0268293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J Stat Softw. 2011;39(5):1–13. Epub 2011/03/01. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubin DB. Multiple imputation. Flexible Imputation of Missing Data, Second Edition: Chapman and Hall/CRC; 2018. p. 29–62. [Google Scholar]
- 41.Fine JP, Gray RJ. A Proportional Hazards Model for the Subdistribution of a Competing Risk. Journal of the American Statistical Association. 1999;94(446):496–509. doi: 10.1080/01621459.1999.10474144. [DOI] [Google Scholar]
- 42.Cook NR, Ridker PM. Advances in measuring the effect of individual predictors of cardiovascular risk: the role of reclassification measures. Ann Intern Med. 2009;150(11):795–802. Epub 2009/06/03. doi: 10.7326/0003-4819-150-11-200906020-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demler OV, Paynter NP, Cook NR. Tests of calibration and goodness-of-fit in the survival setting. Stat Med. 2015;34(10):1659–80. Epub 2015/02/17. doi: 10.1002/sim.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–74. Epub 2006/11/14. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. Epub 2008/11/28. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker SG. Putting risk prediction in perspective: relative utility curves. J Natl Cancer Inst. 2009;101(22):1538–42. Epub 2009/10/22. doi: 10.1093/jnci/djp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–73. doi: 10.7326/m14-0698. [DOI] [PubMed] [Google Scholar]
- 48.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D12. Epub 2020/11/26. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savarese G, Becher PM, Lund LH, Seferovic P, Rosano GMC, Coats AJS. Global burden of heart failure: a comprehensive and updated review of epidemiology. Cardiovascular Research. 2022;118(17):3272–87. doi: 10.1093/cvr/cvac013. [DOI] [PubMed] [Google Scholar]
- 50.Rich JD, Burns J, Freed BH, Maurer MS, Burkhoff D, Shah SJ. Meta-Analysis Global Group in Chronic (MAGGIC) Heart Failure Risk Score: Validation of a Simple Tool for the Prediction of Morbidity and Mortality in Heart Failure With Preserved Ejection Fraction. Journal of the American Heart Association. 2018;7(20):e009594. doi: doi: 10.1161/JAHA.118.009594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawano M, Shiraishi Y, Kohsaka S, Nagai T, Goda A, Mizuno A, et al. Performance of the MAGGIC heart failure risk score and its modification with the addition of discharge natriuretic peptides. ESC Heart Failure. 2018;5(4):610–9. doi: 10.1002/ehf2.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michaels A, Aurora L, Peterson E, Liu B, Pinto YM, Sabbah HN, et al. Risk Prediction in Transition: MAGGIC Score Performance at Discharge and Incremental Utility of Natriuretic Peptides. Journal of Cardiac Failure. 2020;26(1):52–60. doi: 10.1016/j.cardfail.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.George J, Struthers AD, Lang CC. Modulation of the renin-angiotensin-aldosterone system in heart failure. Curr Atheroscler Rep. 2014;16(4):403. doi: 10.1007/s11883-014-0403-7. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi M, Stienen S, Ter Maaten JM, Dickstein K, Samani NJ, Lang CC, et al. Clinical determinants and prognostic implications of renin and aldosterone in patients with symptomatic heart failure. ESC Heart Fail. 2020;7(3):953–63. Epub 20200313. doi: 10.1002/ehf2.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magnussen C, Blankenberg S. Biomarkers for heart failure: small molecules with high clinical relevance. Journal of Internal Medicine. 2018;283(6):530–43. doi: 10.1111/joim.12756. [DOI] [PubMed] [Google Scholar]
- 56.Kinnunen P, Vuolteenaho O, Ruskoaho H. Mechanisms of atrial and brain natriuretic peptide release from rat ventricular myocardium: effect of stretching. Endocrinology. 1993;132(5):1961–70. [DOI] [PubMed] [Google Scholar]
- 57.Mueller C, McDonald K, de Boer RA, Maisel A, Cleland JGF, Kozhuharov N, et al. Heart Failure Association of the European Society of Cardiology practical guidance on the use of natriuretic peptide concentrations. European Journal of Heart Failure. 2019;21(6):715–31. doi: 10.1002/ejhf.1494. [DOI] [PubMed] [Google Scholar]
- 58.Sarhene M, Wang Y, Wei J, Huang Y, Li M, Li L, et al. Biomarkers in heart failure: the past, current and future. Heart Fail Rev. 2019;24(6):867–903. doi: 10.1007/s10741-019-09807-z. [DOI] [PubMed] [Google Scholar]
- 59.Pascual-Figal DA, Pérez-Martínez MT, Asensio-Lopez MC, Sanchez-Más J, García-García ME, Martinez CM, et al. Pulmonary Production of Soluble ST2 in Heart Failure. Circulation: Heart Failure. 2018;11(12):e005488. doi: doi: 10.1161/CIRCHEARTFAILURE.118.005488. [DOI] [PubMed] [Google Scholar]
- 60.Vallejo-Vaz AJ. Novel Biomarkers in Heart Failure Beyond Natriuretic Peptides - The Case for Soluble ST2. Eur Cardiol. 2015;10(1):37–41. doi: 10.15420/ecr.2015.10.01.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaggin HK, Szymonifka J, Bhardwaj A, Belcher A, De Berardinis B, Motiwala S, et al. Head-to-head comparison of serial soluble ST2, growth differentiation factor-15, and highly-sensitive troponin T measurements in patients with chronic heart failure. JACC Heart Fail. 2014;2(1):65–72. Epub 20140125. doi: 10.1016/j.jchf.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 62.Emdin M, Aimo A, Vergaro G, Bayes-Genis A, Lupón J, Latini R, et al. sST2 Predicts Outcome in Chronic Heart Failure Beyond NT−proBNP and High-Sensitivity Troponin T. Journal of the American College of Cardiology. 2018;72(19):2309–20. doi: 10.1016/j.jacc.2018.08.2165. [DOI] [PubMed] [Google Scholar]
- 63.Ruozi G, Bortolotti F, Mura A, Tomczyk M, Falcione A, Martinelli V, et al. Cardioprotective factors against myocardial infarction selected in vivo from an AAV secretome library. Sci Transl Med. 2022;14(660):eabo0699. Epub 20220831. doi: 10.1126/scitranslmed.abo0699. [DOI] [PubMed] [Google Scholar]
- 64.Olson KA, Beatty AL, Heidecker B, Regan MC, Brody EN, Foreman T, et al. Association of growth differentiation factor 11/8, putative anti-ageing factor, with cardiovascular outcomes and overall mortality in humans: analysis of the Heart and Soul and HUNT3 cohorts. Eur Heart J. 2015;36(48):3426–34. Epub 20150820. doi: 10.1093/eurheartj/ehv385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduction and Targeted Therapy. 2022;7(1):3. doi: 10.1038/s41392-021-00762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma A, Verma S, Bhatt DL, Connelly KA, Swiggum E, Vaduganathan M, et al. Optimizing Foundational Therapies in Patients With HFrEF: How Do We Translate These Findings Into Clinical Care? JACC Basic Transl Sci. 2022;7(5):504–17. Epub 2022/06/07. doi: 10.1016/j.jacbts.2021.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morris AA, Khazanie P, Drazner MH, Albert NM, Breathett K, Cooper LB, et al. Guidance for Timely and Appropriate Referral of Patients With Advanced Heart Failure: A Scientific Statement From the American Heart Association. Circulation. 2021;144(15):e238–e50. Epub 2021/09/11. doi: 10.1161/CIR.0000000000001016. [DOI] [PubMed] [Google Scholar]
- 68.Dunlay SM, Roger VL, Killian JM, Weston SA, Schulte PJ, Subramaniam AV, et al. Advanced Heart Failure Epidemiology and Outcomes: A Population-Based Study. JACC Heart Fail. 2021;9(10):722–32. Epub 2021/08/16. doi: 10.1016/j.jchf.2021.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pencina MJ, Goldstein BA, D’Agostino RB. Prediction Models - Development, Evaluation, and Clinical Application. N Engl J Med. 2020;382(17):1583–6. Epub 2020/04/23. doi: 10.1056/NEJMp2000589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.