Abstract
Hepatitis delta virus (HDV) RNA replicates in the nuclei of virus-infected cells. The mechanism of nuclear import of HDV RNA is so far unknown. Using a fluorescein-labeled HDV RNA introduced into partially permeabilized HeLa cells, we found that HDV RNA accumulated only in the cytoplasm. However, in the presence of hepatitis delta antigen (HDAg), which is the only protein encoded by HDV RNA, the HDV RNA was translocated into the nucleus, suggesting that nuclear import of HDV RNA is mediated by HDAg. Deletion of the nuclear localization signal (NLS) or RNA-binding motifs of HDAg resulted in the failure of nuclear import of HDV RNA, indicating that both the NLS and an RNA-binding motif of HDAg are required for the RNA-transporting activity of HDAg. Surprisingly, any one of the three previously identified RNA-binding motifs was sufficient to confer the RNA-transporting activity. We have further shown that HDAg, via its NLS, interacts with karyopherin α2 in vitro, suggesting that nuclear import of the HDAg-HDV RNA complex is mediated by the karyopherin α2β heterodimer. The nuclear import of HDV RNA may be the first biological function of HDAg in the HDV life cycle.
Hepatitis delta virus (HDV) genome is a circular, single-stranded, viroid-like RNA of 1.7 kb which has a large number of intramolecular complementary sequence (23, 25, 29, 44), resulting in a rod-like RNA structure (23, 25). Hepatitis delta antigen (HDAg), the only protein encoded by HDV RNA, is localized almost exclusively in the nuclei of virus-infected cells (6, 41). It usually consists of two protein species, a large HDAg of 214 amino acids and a small HDAg of 195 amino acids, the latter being identical to the former except for a truncation of 19 amino acids at the C terminus. The small HDAg is required for HDV RNA replication (24), while the large HDAg inhibits RNA replication (7, 14) but is required for virion assembly (5, 42). Both antigens have several structural domains, including a nuclear localization signal (NLS) and RNA-binding domains (25). The NLS is located between amino acids 68 and 88 from the N terminus (46), consisting of a bipartite, basic amino acid-rich region, which is required for nuclear localization of the HDAg (46). The main RNA-binding domain consists of two stretches of arginine-rich motifs (ARMs) (amino acids 97 to 107 and 136 to 146), both of which are required for in vitro binding of HDAg to HDV RNA (26). Another stretch of sequence located between amino acids 2 and 27 from the N terminus also contains a cryptic RNA-binding activity, as demonstrated by peptide binding (39). The RNA-binding properties of HDAg, at least its major RNA-binding domain, have been demonstrated to be specific for HDV RNA (27) and are required for its trans-acting activity for HDV RNA replication.
The genome of HDV is presumed to replicate in the nuclei of infected cells, since both HDAg and HDV RNA are localized mainly in the nucleus (18). However, it is not clear whether HDV RNA is transported into the nucleus independently or together with the HDAg. Considering that HDV RNA is a viroid-like RNA, which generally can enter the nucleus without any viral proteins (11, 19), it is plausible that HDV RNA has an intrinsic nuclear importing capability. This would be advantageous for the virus because HDV virion contains both the small and large HDAg, the latter of which inhibits HDV RNA replication (7, 14); thus, the nuclear import of HDV RNA independent of HDAg will ensure the successful replication of RNA. However, it has been shown that nuclear import of influenza virus RNA is mediated by the viral nucleoprotein and nuclear transport factors of host cells (36). A number of transport factors required for nuclear import of cellular proteins have been identified (9, 10, 31, 32, 38). Targeting of proteins to the cell nucleus is usually mediated by interactions between the NLS contained within proteins and the karyopherin (or importin) αβ heterodimeric complex (34, 35). Either karyopherin α1 or karyopherin α2 serves as the NLS-binding subunit (34, 35, 45), whereas karyopherin β serves as an adaptor subunit that mediates docking of the NLS-karyopherin complex with the nuclear pore complex (8, 15, 17, 22).
To understand the mechanism of HDV RNA transport into the nucleus, we have used a nuclear import assay for HDV RNA in digitonin-permeabilized cells. We have demonstrated that nuclear import of HDV RNA is mediated by the HDAg, and both the NLS and RNA-binding motif of HDAg are required for the RNA-transporting activity of HDAg. Thus, the HDV RNA-transporting activity is another new function associated with HDAg. Furthermore, we show that HDAg interacts with karyopherin α2 in vitro, suggesting that the nuclear import of HDAg-HDV RNA complex is mediated by karyopherin α2β. Since HDV RNA has to be transported to the nucleus for RNA replication upon HDV infection, the nuclear import of HDV RNA may be the first biological function of HDAg in the HDV life cycle.
MATERIALS AND METHODS
Construction of plasmids.
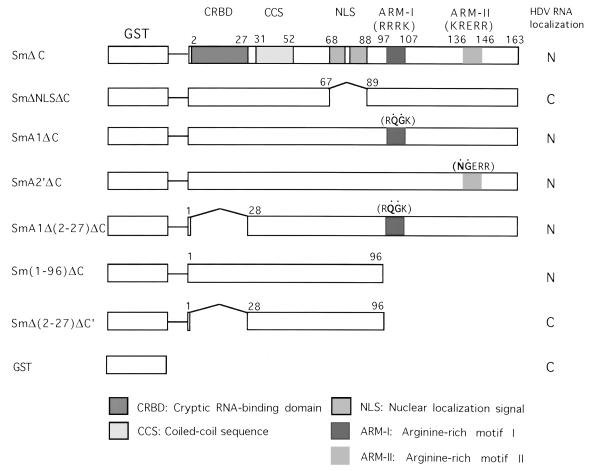
Glutathione S-transferase (GST) expression vector pGEX (Pharmacia) was used for the construction of various plasmids expressing GST-HDAg fusion proteins (Fig. 1). The cDNA fragment corresponding to the small HDAg-coding region was amplified by PCR using plasmid S29 (27) as the template and cloned into the BamHI site of pGEX to generate pGEX-Sm. To improve the expression of GST fusion protein, the C-terminal hydrophobic region (amino acids 164 to 195) of the small HDAg was removed by treating pGEX-Sm with restriction enzyme SmaI and self-religation to generate SmΔC. The SmA1ΔC and SmA2′ΔC constructs, which have two different mutations within the RNA-binding motifs of HDAg (26), were constructed by replacing the SphI-StuI fragment of pGEX-Sm with the corresponding fragments from pECE-A1 and pECE-A2′, respectively (26); the C termini of the resulting clones were removed by SmaI digestion as described above to yield SmA1ΔC and SmA2′ΔC. To construct plasmids SmA1Δ(2-27)ΔC and SmΔ(2-27)ΔC′, primers containing sequences corresponding to the initial amino acid sequence ATG and the desired amino acid sequence plus the BamHI site at both ends were used to amplify HDAg-coding region from amino acids 28 to 163 and 28 to 96, respectively. The PCR fragments were then inserted into the BamHI site of pGEX. To obtain SmΔNLSΔC, inverse PCR (36) was used to amplify the entire SmΔC sequences except the nuclear localization sequences (amino acids 68 to 88), using pGEX-SmΔC as the template. After 5′-end phosphorylation of the PCR fragment, the fragment was self-ligated. Fusions of GST to karyopherins α1 and α2 (also named influenza virus nucleoprotein-interacting proteins 1 and 3, respectively) (37, 38) were kindly provided by R. O’Neill and P. Palese, Mount Sinai School of Medicine, New York, N.Y.
FIG. 1.
Schematic diagram of GST fusion protein constructs. The mutated sequences in the ARMs are represented by dots. The deleted sequences are indicated by bent lines, and the single horizontal lines represent vector sequences. The various functional domains of HDAg are shown. The subcellular localizations of HDV RNA when coexpressed with the indicated proteins are shown. N, nucleus; C, cytoplasm.
Expression and purification of GST fusion proteins.
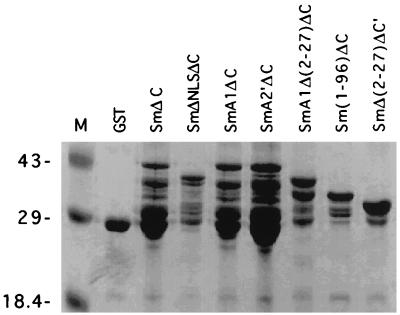
All GST fusion proteins were expressed in Escherichia coli BL21(DE3) and purified by standard procedures (43). Briefly, bacteria were grown in LB medium to an optical density at 600 nm of 0.8 at 37°C. Expression of GST fusion proteins was induced by the addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubation for 3 h. The bacterial pellet was collected and sonicated. GST fusion proteins were purified by incubating bacterial lysates with glutathione-agarose beads and eluted with reduced glutathione. The identity of the GST-fused proteins was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5% polyacrylamide gels (Fig. 2).
FIG. 2.
SDS-PAGE analysis of GST fusion proteins. The partially purified GST fusion proteins were separated by SDS-PAGE on 12.5% polyacrylamide gels and stained with Coomassie blue. The molecular markers (M) are indicated in kilodaltons.
Synthesis of fluorescein-labeled RNA.
Plasmid S29 (27), which contains a 1.7-kb HDV cDNA in genomic orientation under the control of T7 promoter, was linearized with restriction enzyme HindIII. The HDV RNA genome was transcribed in vitro from the linearized plasmid in a 20-μl transcription reaction mixture containing 0.5 mM each ATP, GTP, and CTP, 0.3 mM UTP, 0.2 mM fluorescein-12-UTP (Boehringer Mannheim), 40 mM Tris-HCl (pH 8.0), 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine, 10 mM NaCl, 1 U of RNasin, 10 U of T7 RNA polymerase (Ambion), and 1 μg of linearized plasmid DNA at 37°C for 2 h. After reaction, the DNA template was digested with 2 U of DNase I at 37°C for 15 min. To remove unincorporated nucleotides, RNA was precipitated with either LiCl or ammonium acetate-ethanol.
Nuclear import assay.
The assay was performed as previously described (1, 33) with slight modifications. Briefly, HeLa cells grown on 22-mm-square coverslips were treated with 40 μg of digitonin (Sigma) per ml on ice for 5 min. After two washes with buffer A (20 mM HEPES-KOH [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 5 mM sodium acetate, 1 mM EGTA, 2 mM dithiothreitol, 1 μg each of aprotinin, leupeptin, and pepstatin per ml), the coverslip was transferred to transport buffer (buffer A supplemented with 1 mg of bovine serum albumin per ml, 1 mM ATP, 5 mM creatine phosphate, 20 U of creatine phosphokinase per ml) containing fluorescein-labeled HDV RNA and various GST fusion proteins on a sheet of Parafilm for 15 min at room temperature. The coverslip was transferred back to the original plate and washed twice with buffer A. Then 2% formaldehyde was added to the plate to fix cells. After washing, the coverslip was removed from the plate and excess moisture was wiped off. The mounting solution was added to the coverslip, and then the coverslip was examined under a confocal microscope. Initially, 25 μl of rabbit reticulocyte lysate (Promega) or HeLa cell cytosol was added to the nuclear transport buffer to make a final volume of 50 μl in each assay as described previously (1). However, we subsequently noted that under the permeabilization conditions used, such lysates were not necessary for the nuclear transport assay. Therefore, in all experiments reported here, neither rabbit reticulocyte lysates nor HeLa cytoplasmic extracts were used.
Immunofluorescent staining.
Digitonin-permeabilized HeLa cells were incubated with the various GST fusion proteins in the nuclear transport buffer, fixed with 2% formaldehyde for 30 min at room temperature, blocked with phosphate-buffered saline containing 1% heat-inactivated fetal bovine serum, and incubated with the monoclonal antibody (1:200) specific for HDAg (21) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (Boehringer Mannheim) as the primary and secondary antibodies, respectively. After staining, HeLa cells were mounted with the mounting solution. Coverslips were sealed with nail polish and examined by confocal microscopy.
RESULTS
Nuclear import of HDV RNA is mediated by HDAg.
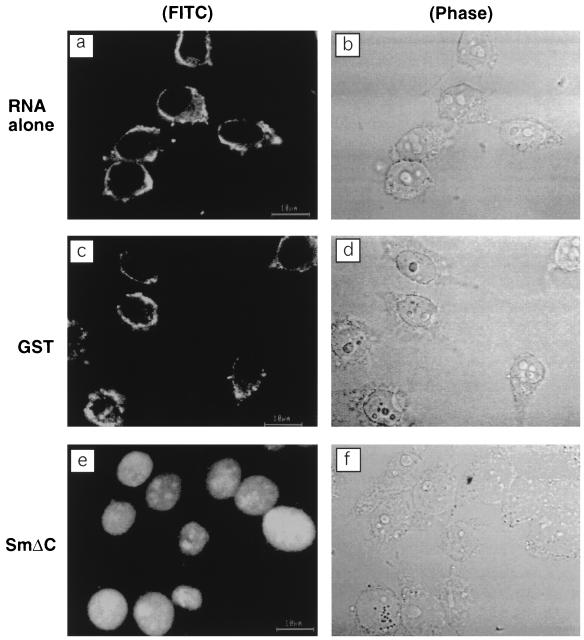
Since HDV RNA is predominantly localized in the nuclei of HDV-infected cells (18), we first examined whether HDV RNA alone could be transported to the nuclei when it was introduced into the cytoplasm of HeLa cells. For this purpose, digitonin-permeabilized cells were incubated with FITC-labeled HDV genomic RNA alone. Figure 3a shows that all of the labeled HDV RNA was localized exclusively in the cytoplasm. Since HDV RNA is expected to complex with HDAg, which is a nuclear protein (6, 46), we next examined whether the addition of HDAg would allow the HDV RNA to be transported to the nucleus. The HDAg was expressed as a GST fusion protein. Because the C-terminal hydrophobic domain (amino acids 164 to 195) of the HDAg caused the protein to be insoluble and affected the expression level of the protein, we deleted this domain, which does not have demonstrable functions (25), from all of the GST-HDAg fusion proteins (Fig. 1). These proteins were partially purified and added together with the FITC-labeled HDV RNA to the permeabilized cells. The results (Fig. 3e) showed that HDV RNA was completely transported to the nucleus. In contrast, the addition of GST protein did not cause HDV RNA to be transported to the nuclei (Fig. 3c). These results indicate that the nuclear import of HDV RNA is mediated by HDAg.
FIG. 3.
Nuclear import of HDV RNA is mediated by HDAg. Digitonin-permeabilized cells were incubated at room temperature for 15 min with FITC-labeled RNA only (a) or in the presence of either GST (c) or SmΔC (e). Panels b, d, and f are the phase-contrast images of panels a, c, and e, respectively.
NLS of HDAg is required for the nuclear import of HDV RNA.
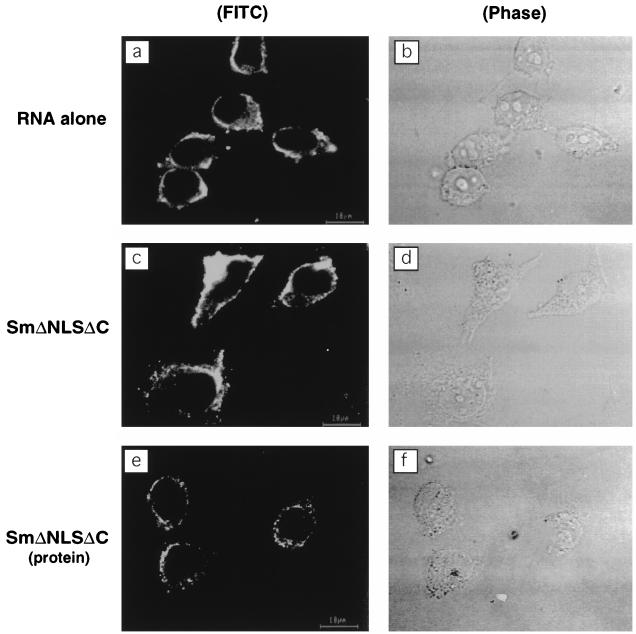
It has been shown that the NLS of HDAg is responsible for its nuclear transport and localization (46). To determine whether the NLS is required for the nuclear import of HDV RNA, we constructed plasmid SmΔNLSΔC, in which the NLS located between amino acids 68 and 88 of HDAg was deleted. Digitonin-permeabilized cells were incubated with FITC-labeled RNA alone (Fig. 4a) or together with SmΔNLSΔC (Fig. 4c). The results showed that HDV RNA was not transported into the nucleus even in the presence of SmΔNLSΔC. To establish that the failure of HDV RNA to be transported into the nucleus correlated with the failure of SmΔNLSΔC protein to gain entry into the nucleus, immunofluorescent staining of SmΔNLSΔC was performed with the monoclonal antibody specific for HDAg (Fig. 4e). The results showed that the SmΔNLSΔC protein accumulated exclusively in the cytoplasm. Although we cannot rule out completely the possibility that the deletion in the NLS region caused a major conformational change of the HDAg, these data strongly suggest that the NLS of HDAg is required for the nuclear import of HDAg and that the nuclear import of HDV RNA is dependent on the nuclear import of HDAg.
FIG. 4.
NLS of HDAg is required for nuclear import of HDV RNA. Digitonin-permeabilized cells were incubated at room temperature for 15 min with FITC-labeled RNA alone (a) or in the presence of SmΔNLSΔC (c). (e) After digitonin-permeabilized cells were incubated with SmΔNLSΔC, immunostaining was performed with the monoclonal antibody specific for HDAg. Panels b, d, and f are the phase-contrast images of panels a, c, and e, respectively.
RNA-binding motifs of HDAg are required for the nuclear import of HDV RNA.
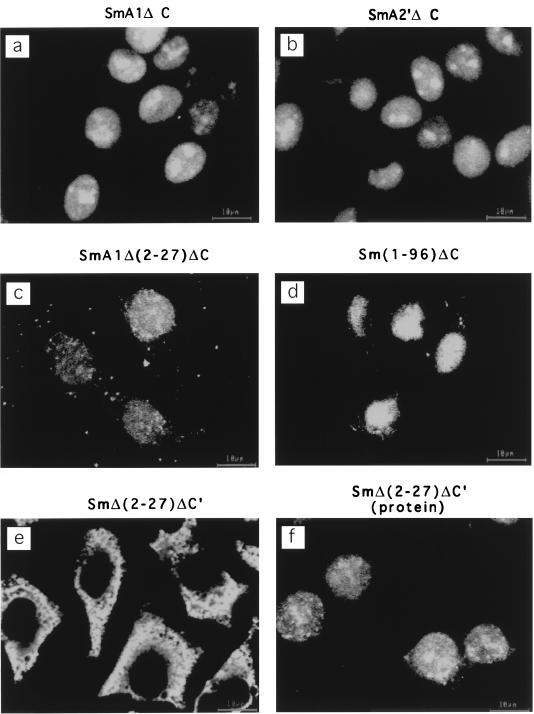
HDV RNA has been shown to bind to HDAg specifically (27). Two ARMs in the HDAg are required for this binding (26). Another stretch of sequence (amino acids 2 to 27) has also been shown to contain a cryptic RNA-binding activity (39). If the nuclear import of HDV RNA is mediated by HDAg, then it is likely that this activity requires the RNA-binding activity of HDAg. To test this possibility, we constructed several truncation and site-specific mutants of HDAg, which have mutated sequences in the various reported RNA-binding domains (Fig. 1). These proteins had at least one of the three RNA-binding sequences mutated. For examples, SmA1ΔC contained Arg-104→Gln and Arg-105→Gly mutations in the ARM I. SmA2′ΔC contained Lys-139→Asn and Arg-140→Gly mutations in ARM II, SmA1Δ(2-27)ΔC contained the same point mutations as in SmA1ΔC plus deletion of amino acids 2 to 27, and Sm(1-96)ΔC had the two ARMs deleted. Surprisingly, all of these proteins mediated nuclear import of HDV RNA (Fig. 5a to d). In contrast, SmΔ(2-27)ΔC′, which has all of the three reported RNA-binding motifs deleted, failed to mediate HDV RNA import (Fig. 5e). To rule out the possibility that SmΔ(2-27)ΔC′ was not transported to the nucleus and thus failed to mediate HDV RNA nuclear import, we performed immunofluorescent staining of SmΔ(2-27)ΔC′ protein. Figure 5f shows that this protein was localized in the nucleus, whereas HDV RNA remained in the cytoplasm (Fig. 5e). These data indicated that HDAg binds to HDV RNA and mediates nuclear import of HDV RNA and that either one of the ARMs (26) or the N-terminal amino acids 2 to 27, which contain a cryptic RNA-binding activity (39), is sufficient for nuclear import of HDV RNA. Thus, the requirement for RNA-binding in vivo appears to be less stringent than in vitro, because both ARM sequences are required for RNA binding in vitro (26).
FIG. 5.
RNA-binding motifs of HDAg are required for nuclear import of HDV RNA. Digitonin-permeabilized cells were incubated at room temperature for 15 min with FITC-labeled RNA in the presence of various HDAg constructs containing mutations in the RNA-binding motifs. (a) SmA1ΔC; (b) SmA2′ΔC; (c) SmA1Δ(2-27)ΔC; (d) Sm(1-96)ΔC (e) SmΔ(2-27)ΔC′. (f) After digitonin-permeabilized cells were incubated at room temperature for 15 min with SmΔ(2-27)ΔC′ alone, immunostaining was performed with the antibody specific for HDAg.
The NLS of HDAg interacts with karyopherin α2.
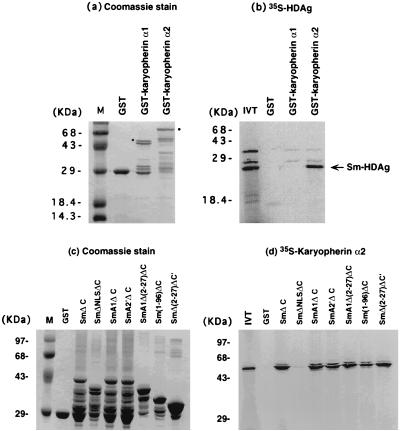
It has been shown that nuclear import of some NLS-containing proteins requires the interaction between the NLS and karyopherin α1β or α2β (37). To investigate whether nuclear import of HDAg, and therefore HDV RNA, involves these pathways, an in vitro GST fusion protein binding assay was performed. Either GST-karyopherin α1 or GST-karyopherin α2 was incubated with 35S-labeled HDAg, and the bound complex was separated by SDS-PAGE (Fig. 6a and b). The results showed that HDAg bound karyopherin α2 but not karyopherin α1. The reciprocal experiment was also performed. The various GST-HDAg fusion proteins (Fig. 6c) were incubated with 35S-labeled karyopherin α2. The results showed that karyopherin α2 bound to the wild-type HDAg (SmΔC) but not the HDAg mutant without NLS (SmΔNLSΔC) (Fig. 6d). Furthermore, all of the other HDAg mutants, which are defective in one or all of the RNA-binding motifs but retain NLS, were found to bind karyopherin α2. Most significantly, SmΔ(2-27)ΔC′, which failed to mediate nuclear import of HDV RNA, still bound karyopherin α2, consistent with the finding that this protein was localized in the nucleus (Fig. 5f). These data suggest that karyopherin α2 is involved in nuclear import of HDAg and HDV RNA. In addition, the NLS is required for the interaction between HDAg and karyopherin α2.
FIG. 6.
Binding between HDAg and karyopherin. (a and c) Coomassie blue stain of the GST-karyopherin α1 and α2 (a) and GST-HDAg (c) mutants after SDS-PAGE. (b) GST-karyopherin α1 and α2 were incubated with [35S]Met-labeled small HDAg. The bound proteins were separated by SDS-PAGE and visualized by autoradiography. The bands corresponding to full-length GST fusion protein products are indicated by dots. (d) The reciprocal experiment using various GST-HDAg clones and [35S]Met-labeled karyopherin α2. M, molecular markers; IVT, input in vitro-translated HDAg (b) or karyopherin α2 (d).
DISCUSSION
This report shows that HDV RNA cannot be transported into the nucleus by itself and that its nuclear import is mediated by HDAg. This finding expands the long list of the potential biological activities of HDAg (25). This RNA-importing function requires both the NLS and RNA-binding properties of HDAg. Since HDV RNA replication occurs in the nucleus, nuclear import function for HDV RNA likely represents the first biological activity of HDAg in the HDV life cycle. Previously, it has been shown that transfection of HDV RNA into mammalian cells does not lead to HDV replication unless a preexisting HDAg is present in the cells (13, 20), but transfection of HDV ribonucleoprotein can lead to HDV replication (3). These observations are consistent with the interpretation that HDAg is crucial for the early steps, including import of HDV RNA into the nucleus, of HDV replication cycle. This nuclear importing function at least partially accounts for the requirement of a functional HDAg for HDV RNA replication (24, 26). Whether HDAg is also involved in other processes of HDV replication is not certain. It is commonly implied that HDAg is required for HDV RNA replication per se (24, 25); however, several in vitro RNA replication studies demonstrated that HDV RNA replication can take place even in the absence of HDAg (2, 12, 28). Thus, MacNaughton et al. suggested that HDAg may be needed only to transport HDV RNA into the nucleus, where RNA replication occurs, but not for RNA replication per se (28). However, our present study showed that several RNA-binding mutants, which have been shown to be defective in transactivating HDV RNA replication (26), could mediate the nuclear import of HDV RNA. Therefore, HDAg likely has direct functions in HDV RNA replication.
In this report, we examined only the truncated forms of the small HDAg. For unknown technical reasons, the full-length small HDAg and large HDAg were difficult to be expressed efficiently in the bacteria. Furthermore, the expressed proteins were difficult to purify and did not retain the biological activities because they aggregated under physiological conditions. The most likely reason for this is that the C-terminal domain of HDAg is hydrophobic, which may have interfered with the protein expression and/or its biochemical properties. Regardless, the truncated forms of both the large and small HDAg contain both RNA-binding motifs (26) and NLS (46) of their full-length counterparts. Since the truncated form of the large HDAg is identical to that of the small HDAg, there is little doubt that the large HDAg will be found to be able to import the HDV RNA into the nucleus as well. Both the large and small HDAg are present in the HDV virion particles, although the large HDAg is the protein that initiates the virion assembly process (5, 42). The findings presented in this report suggested that the HDV RNA is transported into the nucleus in the form of an RNA-protein complex. This raises a conceptually difficult issue: since the large HDAg inhibits HDV RNA replication (7, 14), import of HDV RNA together with the large HDAg should inhibit HDV replication. Understanding of how HDV RNA replicates in the presence of the large HDAg will require future studies.
Using RNA import as a marker for the RNA-binding property of HDAg, surprisingly, we found that the requirement for binding of HDV RNA to HDAg is less stringent than that in vitro. In RNA mobility shift assay or Northwestern RNA-protein binding assay, both of the ARMs of HDAg are required for its binding to HDV RNA (26). However, either one of these two ARMs is sufficient for RNA transport in vivo. Furthermore, another potential RNA-binding domain (amino acids 2 to 27), identified only by peptide binding in vitro (39), was demonstrated to be sufficient for HDV RNA import as well. Thus, all three potential RNA-binding domains may be functional in the HDV life cycle. The presence of the redundant RNA-binding functions ensures that HDV RNA can be transported into the nucleus, in case that some domains of the HDAg are concealed, as has been demonstrated for the native form of HDV ribonucleoproteins (4).
Several different nuclear transport mechanisms of proteins have been identified (16, 30, 40). The HDAg appears to contain the classical type I NLS (46), which consists of basic amino acid residues. This type of proteins are usually transported into the nucleus by interacting with karyopherin α1 or α2. The HDAg appears to be mediated by karyopherin α2. It is interesting that HDV RNA, which is similar to viroids, requires a viral protein to interact with the karyopherin. In contrast, viroids, which do not encode proteins, can themselves be transported into the nucleus when microinjected into the cytoplasm of plant cells (11). Whether there are fundamental differences between the two types of RNA or between plant and animal cells in the mechanism of nuclear transport is an interesting question. This question is currently being studied.
ACKNOWLEDGMENTS
We thank Robert O’Neill and Peter Palese (Mount Sinai School of Medicine, New York) for providing plasmids pGST-34 and pGST-39, which express GST-karyopherin α1 and GST-karyopherin α2, respectively, and for comments on the manuscript. We also thank Robert Schneider for editorial assistance. H.-C. Chou is most grateful to J.-C. Sheu, Director of Liver Disease Prevention and Treatment Research Foundation, Taiwan, for his constant support.
This work was partially supported by NIH grant AI40038. M.M.C.L. is an Investigator of Howard Hughes Medical Institute.
REFERENCES
- 1.Adam S A, Marr R S, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beard M R, MacNaughton T B, Gowans E J. Identification and characterization of a hepatitis delta virus RNA transcriptional promoter. J Virol. 1996;70:4986–4995. doi: 10.1128/jvi.70.8.4986-4995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bichko V, Netter H J, Taylor J. Introduction of hepatitis delta virus into animal cell lines via cationic liposomes. J Virol. 1994;68:5247–5252. doi: 10.1128/jvi.68.8.5247-5252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bichko V V, Lemon S M, Wang J G, Hwang S, Lai M M C, Taylor J M. Epitopes exposed on hepatitis delta virus ribonucleoproteins. J Virol. 1996;70:5807–5811. doi: 10.1128/jvi.70.9.5807-5811.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang F L, Chen P J, Tu S J, Wang C J, Chen D S. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc Natl Acad Sci USA. 1991;88:8490–8494. doi: 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang M-F, Baker S C, Soe L H, Kamahora T, Keck J G, Makino S, Govindarajan S, Lai M M C. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J Virol. 1988;62:2403–2410. doi: 10.1128/jvi.62.7.2403-2410.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao M, Hsieh S Y, Taylor J. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64:5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi N C, Adam E J, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes P, Ye Z S, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuomo C A, Kirch S A, Gyuris J, Brent R, Oettinger M A. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding B, Kwon M-O, Hammond R, Owens R A. Cell-to-cell movement of potato spindle tuber viroid. Plant J. 1997;12:931–936. doi: 10.1046/j.1365-313x.1997.12040931.x. [DOI] [PubMed] [Google Scholar]
- 12.Fu T B, Taylor J. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J Virol. 1993;67:6965–6972. doi: 10.1128/jvi.67.12.6965-6972.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glenn J S, Taylor J M, White J M. In vitro-synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J Virol. 1990;64:3104–3107. doi: 10.1128/jvi.64.6.3104-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glenn J S, White J M. trans-dominant inhibition of human hepatitis delta virus genome replication. J Virol. 1991;65:2357–2361. doi: 10.1128/jvi.65.5.2357-2361.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 17.Gorlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 18.Gowans E J, Baroudy B M, Negro F, Ponzetto A, Purcell R H, Gerin J L. Evidence for replication of hepatitis delta virus RNA in hepatocyte nuclei after in vivo infection. Virology. 1988;167:274–278. doi: 10.1016/0042-6822(88)90078-5. [DOI] [PubMed] [Google Scholar]
- 19.Harders J, Lukacs N, Robert-Nicoud M, Jovin T M, Riesner D. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 1989;8:3941–3949. doi: 10.1002/j.1460-2075.1989.tb08577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang S B, Jeng K-S, Lai M M C. The unique hepatitis delta virus. In: Dinter-Gottlieb G, editor. Studies of functional roles of hepatitis delta antigen in delta virus RNA replication. R. G. Austin, Tex: Landes, Co.; 1995. pp. 95–106. [Google Scholar]
- 21.Hwang S B, Lai M M C. A unique conformation at the carboxyl terminus of the small hepatitis delta antigen revealed by a specific monoclonal antibody. Virology. 1993;193:924–931. doi: 10.1006/viro.1993.1201. [DOI] [PubMed] [Google Scholar]
- 22.Imamoto N, Shimamoto T, Kose S, Takao T, Tachibana T, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- 23.Kos A, Dijkema R, Arnberg A C, van der Merde P H, Schellekens H. The HDV possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 24.Kuo M Y-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai M M C. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 26.Lee C-Z, Lin J-H, Chao M, McKnight K, Lai M M C. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J Virol. 1993;67:2221–2227. doi: 10.1128/jvi.67.4.2221-2227.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J H, Chang M F, Baker S C, Govindarajan S, Lai M M C. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J Virol. 1990;64:4051–4058. doi: 10.1128/jvi.64.9.4051-4058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacNaughton T B, Gowans E J, McNamara S P, Burrell C J. Hepatitis delta antigen is necessary for access of hepatitis delta virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology. 1991;184:387–390. doi: 10.1016/0042-6822(91)90855-6. [DOI] [PubMed] [Google Scholar]
- 29.Makino S, Chang M F, Shieh C K, Kamahora T, Vannier D M, Govindarajan S, Lai M M C. Molecular cloning and sequencing of a human hepatitis delta virus RNA. Nature. 1987;329:343–346. doi: 10.1038/329343a0. [DOI] [PubMed] [Google Scholar]
- 30.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 32.Moore M S, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore M S, Blobel G. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- 34.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Neill R E, Jaskunas R, Blobel G, Palese P, Moroianu J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem. 1995;270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 38.O’Neill R E, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 39.Poisson F, Roingeard P, Baillou A, Dubois F, Bonelli F, Calogero R A, Goudeau A. Characterization of RNA-binding domains of hepatitis delta antigen. J Gen Virol. 1993;74:2473–2478. doi: 10.1099/0022-1317-74-11-2473. [DOI] [PubMed] [Google Scholar]
- 40.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 41.Rizzetto M, Canese M G, Arico S, Crivelli O, Bonino F, Trepo C G, Verme G. Immunofluorescence detection of a new antigen-antibody system (δ/anti-δ) associated to the hepatitis virus in the liver and in the serum of HBsAg carriers. Gut. 1977;18:997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryu W S, Bayer M, Taylor J. Assembly of hepatitis delta virus particles. J Virol. 1992;66:2310–2315. doi: 10.1128/jvi.66.4.2310-2315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 44.Wang K-S, Choo Q-L, Weiner A J, Ou J-H, Najarian R C, Thayer R M, Mullenbach G T, Denniston K J, Gerin J L, Houghton M. Structure, sequence and expression of the hepatitis delta viral genome. Nature. 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 45.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 46.Xia Y-P, Yeh C-T, Ou J-H, Lai M M C. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J Virol. 1992;66:914–921. doi: 10.1128/jvi.66.2.914-921.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]