Abstract
Genomic rearrangements are a hallmark of most childhood tumors, including medulloblastoma, one of the most common brain tumors in children, but their causes remain largely unknown. Here, we show that PiggyBac transposable element derived 5 (Pgbd5) promotes tumor development in multiple developmentally accurate mouse models of Sonic Hedgehog (SHH) medulloblastoma. Most Pgbd5-deficient mice do not develop tumors, while maintaining normal cerebellar development. Ectopic activation of SHH signaling is sufficient to enforce cerebellar granule cell progenitor–like cell states, which exhibit Pgbd5-dependent expression of distinct DNA repair and neurodevelopmental factors. Mouse medulloblastomas expressing Pgbd5 have increased numbers of somatic structural DNA rearrangements, some of which carry PGBD5-specific sequences at their breakpoints. Similar sequence breakpoints recurrently affect somatic DNA rearrangements of known tumor suppressors and oncogenes in medulloblastomas in 329 children. This identifies PGBD5 as a medulloblastoma mutator and provides a genetic mechanism for the generation of oncogenic DNA rearrangements in childhood cancer.
Induction of somatic oncogenic mutations by the domesticated transposase-derived gene PGBD5 promotes medulloblastoma development.
INTRODUCTION
Cancer development is caused by the acquisition of somatic mutations affecting tumor suppressors and oncogenes that encode factors regulating cell proliferation, differentiation, and other hallmarks of cancer (1). Compared to aging-associated cancers, childhood tumors are characterized by significantly lower total numbers of genetic mutations, although they exhibit chromosomal deletions, amplifications, translocations, and other complex oncogenic genomic rearrangements (2). The origins of these oncogenic mutations in childhood and young adult tumors remain obscure. A quintessential example is medulloblastoma (MB), one of the most frequent childhood brain tumors. For example, MBs with activation of Sonic Hedgehog (SHH) signaling are caused by the aberrant proliferation of cerebellar granule cell precursors (GCPs) due to activating mutations in SMO or somatic deletions of the tumor suppressors PTCH1, SUFU, and TP53. MBs also exhibit frequent amplifications of oncogenes such as GLI2, MYC, and MYCN, and other complex genomic rearrangements. MBs can be caused by Li-Fraumeni syndrome with germline deficiency of TP53, which dysregulates chromosome replication and repair, thereby causing complex oncogenic DNA rearrangements known as chromothripsis. However, the causes of somatic DNA rearrangements in sporadic MBs are not known, despite being a defining feature of this and most other childhood and young adult tumors.
Recently, we found that PGBD5 (PiggyBac transposable element derived 5), the most evolutionarily conserved domesticated DNA transposase–derived gene in vertebrates, can mediate sequence-specific DNA rearrangements dependent on its putative nuclease activity and end-joining DNA repair in human cells (3–5). Although PGBD5 can support genomic DNA integration in cells, its cellular activity predominantly involves double-strand DNA breaks, deletions, and other DNA rearrangements. While the specific enzymatic mechanisms of PGBD5-induced genome rearrangements need to be defined, PGBD5 can mediate rearrangements of both heterologous transposon substrates as well as sequence elements in the human and mouse genomes (3, 4), as validated by Helou et al. (6, 7) and recently confirmed by Bigot et al. (8).
RESULTS
In surveying the expression of PGBD5 across human cancers, we found that most human MBs express high levels of PGBD5 (4). Therefore, we hypothesized that dysregulation of PGBD5 might contribute to the somatic induction of oncogenic DNA rearrangements. Among the four major types of MBs, PGBD5 gene expression is highest in tumors with constitutive SHH signaling (4). Here, we used mouse models of sporadic SHH MB, induced by constitutive activation of SHH signaling in developing mouse cerebellar GCPs, to investigate the possibility of developmental mutator activity of PGBD5 (Fig. 1A).
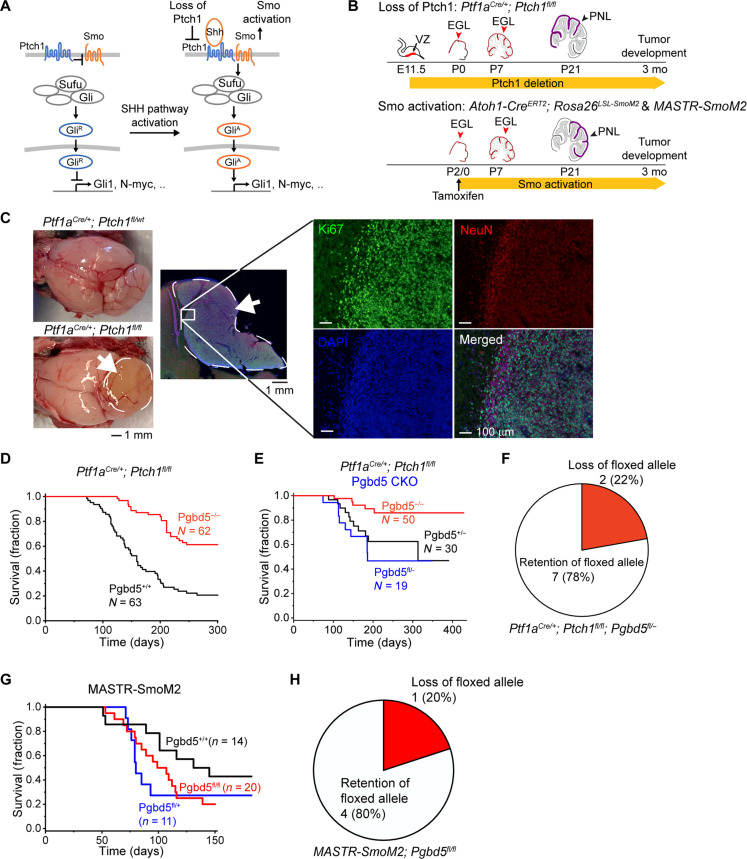
Fig. 1. Pgbd5 promotes tumorigenesis in diverse developmentally accurate mouse models of SHH MB.
(A) Schematic of aberrant mechanisms of SHH signaling in cerebellar GCPs in MB development (left). In Ptch1-mutant Ptf1aCre/+;Ptch1fl/fl MB, by deletion of Ptch1 encoding a receptor for SHH, SMO signaling is disinhibited and highly activated, leading to the generation of activated GLI (GLIA). In Smo-mutant MASTR-SmoM2 or Atoh1-CreERT2;Rosa26LSL-SmoM2 MB, oncogenic constitutively activated form of SMO results in GLI activation and aberrant SHH signaling. (B) Schematic of cerebellar tumor development in Ptch1- (top) and SmoM2-mutant (bottom) MB. Red arrowheads mark conditionally gene-targeted cell populations. PNLs (purple) lead to MB development. E, embryonic day; P, postnatal day. (C) Representative photographs of dissected brains of Ptf1aCre/+;Ptch1fl/fl (bottom left) mice that develop MBs marked by white arrows and dashed circles, as compared to Ptf1aCre/+;Ptch1fl/wt mice (top left) that do not develop tumors. Immunofluorescence microscopy (right) shows high Ki67 (green) and low NeuN (red) expression in MB tumors, with nuclei marked with DAPI (blue). The edge of the tumor (white inset) is magnified with NeuN-positive cells on tumor edge corresponding to normal cerebellum. (D) Survival of Ptf1aCre/+;Ptch1fl/fl;Pgbd5+/+ (black) and Ptf1aCre/+;Ptch1fl/fl;Pgbd5−/− (red) mice. (E) Survival of Ptf1aCre/+;Ptch1fl/fl mice with conditional knockout (CKO) of Pgbd5fl/− (blue) or control Pgbd5+/− (black) or Pgbd5−/− (red) littermates. (F) Genomic PCR analysis of conditional Pgbd5 excision in Pgbd5fl/− CKO Ptch1-mutant tumors demonstrates that seven of nine (79%) analyzed tumors retain intact Pgbd5, as detailed in fig. S1D. (G) Survival of MASTR-SmoM2 mice with CKO of Pgbd5fl/fl (red), as compared to control Pgbd5+/+ (black) or Pgbd5fl/+ (blue) littermates. (H) Genomic PCR analysis of conditional Pgbd5 excision in MASTR-SmoM2; Pgbd5fl/fl tumors demonstrates that four of five (80%) analyzed tumors retain Pgbd5 floxed alleles, as detailed in fig. S2B.
First, we engineered mice with loxP sites flanking exon 4 of mouse Pgbd5, generating Pgbd5fl/fl and knockout Pgbd5−/− mice in which translation of the protein is out of frame after exon 4 (fig. S1A). In situ hybridization (ISH) with a Pgbd5 exon 4–specific probe set confirmed complete loss of exon 4 transcripts in Pgbd5−/− mice (figs. S1B and S3B). We then crossed the Pgbd5−/− alleles into Ptf1aCre/+;Ptch1fl/fl mice, which develop tumors with the highest known penetrance among mouse models of sporadic SHH MB (Fig. 1, B and C) (9). Mosaic loss of Ptch1 leads to preneoplastic hyperplasia of developing cerebellar GCPs due to the constitutive activation of SHH signaling, similar to sporadic human SHH MBs with somatic mutations of PTCH1, which affect more than 40% of patients with SHH MBs (10). Analysis of three independent cohorts of Pgbd5+/+;Ptf1aCre/+;Ptch1fl/fl and Pgbd5−/−;Ptf1aCre/+;Ptch1fl/fl mice showed that Pgbd5 significantly promoted MB development (Fig. 1D and fig. S1C). As observed with other mouse cancer models (11), tumorigenesis was more penetrant in mice with mixed SW genetic background as compared to the congenic C57BL/6J strain. Among Pgbd5-knockout tumor model mice, as many as 70% of animals did not develop tumors after 1 year of life (mean 61%, log-rank P = 1.4 × 10−8), whereas most Pgbd5+/+ mice (79%) rapidly succumbed to MBs with a median latency of 5 months.
To exclude the possibility of tumor cell–extrinsic effects of germline Pgbd5−/− deletion, we used Pgbd5-floxed mice in which Pgbd5 loss was primarily confined to cerebellar progenitor cells due to conditional Cre expression and loxP recombination. Both Pgbd5fl/−;Ptf1aCre/+;Ptch1fl/fl mice and their Pgbd5+/−;Ptf1aCre/+;Ptch1fl/fl littermates developed MBs with similar penetrance and latency (Fig. 1E). However, genomic polymerase chain reaction (PCR) analysis using primers specific for the Pgbd5-floxed allele demonstrated that seven of nine analyzed tumors (78%) retained a substantial amount of intact Pgbd5, indicating a selective advantage for Pgbd5-expressing tumor progenitor cells (Fisher’s exact test P = 2.3 × 10−3; Fig. 1F and fig. S1D).
We performed a similar analysis using a developmentally accurate mouse model of sporadic SMO-mutant MB, corresponding to a mutation of SMO that aberrantly activates SHH signaling in human patients (10, 12). This model leverages a system for mosaic mutagenesis with spatial and temporal control of recombination (MASTR) (12, 13). In this system, green fluorescent protein (GFP)–Cre is induced by tamoxifen at postnatal day 0 (P0) in cerebellar GCPs and further induces oncogenic SmoM2 expression and concurrent deletion of Pgbd5 (fig. S2A). Both Pgbd5fl/fl and their Pgbd5fl/+ littermates developed tumors with similar latencies (Fig. 1G). However, genomic PCR analysis again showed that most analyzed Pgbd5fl/fl tumors (four of five) retained intact Pgbd5 alleles, indicating that Pgbd5 enhances SmoM2-mutant SHH MB development (Fisher’s exact test P = 2.1 × 10−2; Fig. 1H and fig. S2B). To exclude the possibility that apparent Pgbd5 expression was due to the infiltration of stromal or immune cells, we confirmed the requirement for Pgbd5 in MB tumor cell development using three of the same SmoM2-mutant tumors for ISH with a Pgbd5 exon 4–specific probe and Cre-specific probe as a positive control for tumor cells (fig. S3, C to F). This revealed specific Pgbd5 transcript expression in tumor cells for two of three analyzed MASTR-SmoM2;Pgbd5fl/fl tumors. In all, these results indicate that Pgbd5 promotes tumor development in diverse developmentally accurate mouse models of SHH MBs.
SHH MBs are thought to originate in developing cerebellar GCPs, which in turn are dependent on SHH signaling (13–20). To exclude the possibility that PGBD5-induced tumorigenesis is due to its control of normal cerebellar development, we analyzed the cerebellar cytoarchitecture of Pgbd5−/− mice. We observed grossly intact medial cerebellar vermis and lateral hemispheres, including normal cytoarchitecture and morphology, which are essential hallmarks of cerebellar development (Fig. 2A). To examine the effects of Pgbd5 deficiency on SHH signaling directly, we isolated GCPs from the cerebella of 5-day-old mice, when SHH signaling is required for cerebellar development, and measured SHH pathway activity using quantitative reverse transcription PCR of the canonical SHH signaling biomarker Gli1 (17). We observed no significant differences in Gli1 expression between Pgbd5−/− and wild-type developing cerebellar GCPs (P = 0.87; Fig. 2B). Thus, the requirement of Pgbd5 for SHH MB development cannot be explained by the effects of SHH signaling on growth and survival of mutant GCPs, which initiate tumor progression.
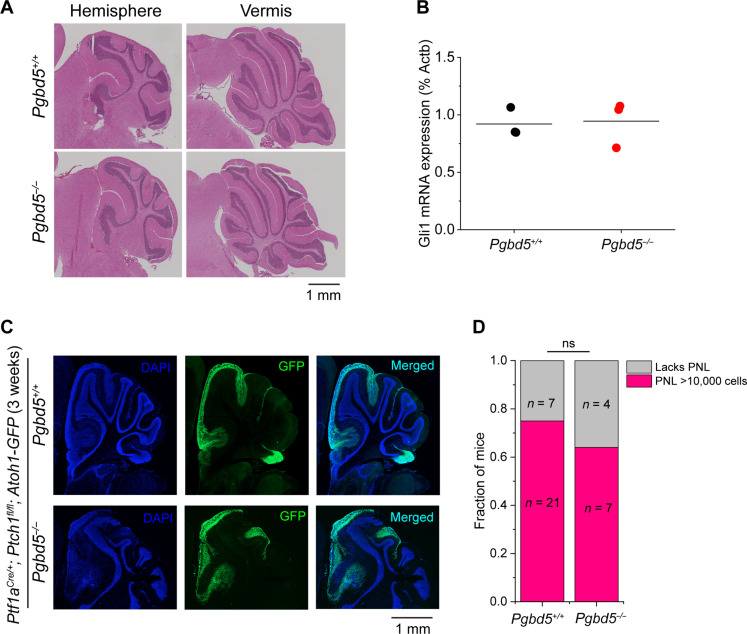
Fig. 2. Pgbd5 is dispensable for normal SHH signaling and cerebellar development.
(A) Representative immunohistochemistry micrographs of sagittal sections of cerebellum of Pgbd5+/+ (top) and Pgbd5−/− (bottom) mice at 6 weeks of age show normal cytoarchitecture and morphology of cerebellar hemispheres (left) and vermis (right). (B) Expression of Gli1 mRNA in purified cerebellar GCPs from 5-day-old Pgbd5+/+ (black) and Pgbd5−/− (red) mice. Bars represent means of three biologic replicates (P = 0.87). (C) Representative fluorescence images of cerebellar hemispheres of 3-week-old Pgbd5+/+ (top) versus Pgbd5−/− (bottom) Ptf1aCre/+;Ptch1fl/fl;Atoh1-GFP mice showing PNLs (green) marked by Atoh1-GFP expression, with nuclei marked with DAPI (blue). (D) Fraction of mice harboring PNLs (red) in Pgbd5wt/wt;Ptf1aCre/+;Ptch1fl/fl;Atoh1-GFP and Pgbd5−/−;Ptf1aCre/+;Ptch1fl/fl;Atoh1-GFP mice between 3 and 8 weeks of age. Both groups harbor similar fractions of PNLs that are defined as at least 10,000 Atoh1-GFP–positive cells (Fisher’s exact test P = 0.69); ns, not significant.
Oncogenic SHH signaling leads to hyperplasia of cerebellar GCPs. These preneoplastic cells express Atoh1 and persist in the external granule layer between 3 and 8 weeks of age (20, 21). To examine the effects of Pgbd5 on preneoplastic GCP hyperplasia, we used an Atoh1-GFP reporter transgene and fluorescence-activated cell sorting (FACS) to isolate preneoplastic GCPs developing in the cerebellar external granule layer in 3- to 8-week-old Pgbd5−/−;Ptf1aCre/+;Ptch1fl/fl;Atoh1-GFP mice (22). We found that 21 of 28 (75%) of Pgbd5+/+;Ptf1aCre/+;Ptch1fl/fl;Atoh1-GFP mice harbored preneoplastic cells in their cerebella, which was similar to Pgbd5-knockout mice (Fig. 2, C and D). The preneoplastic populations also showed no significant differences in the numbers of Atoh1-expressing cells (P = 0.16; fig. S4). Thus, Pgbd5 is dispensable for normal cerebellar development, physiologic SHH signaling, and the growth of preneoplastic SHH MB progenitor cells.
Vertebrate PGBD5 is derived from piggyBac DNA transposases, which induce double-strand DNA breaks and rearrangements at specific sequences via their ribonuclease H (RNase H)–like domain, which is highly conserved among vertebrate PGBD5 genes (3, 4, 23, 24). Does PGBD5 promote MB development by inducing sequence-specific somatic mutations? To test this hypothesis, we performed whole-genome PCR-free DNA sequencing of MBs from both Ptch1- and SmoM2-mutant tumors, as compared to their matched normal tissues. We analyzed resultant sequencing data using recently developed methods optimized for the accurate detection of somatic cancer genome rearrangements (25).
Pgbd5-expressing MBs from Ptf1aCre/+;Ptch1fl/fl mice exhibited nearly twice as many somatic DNA rearrangements, including deletions, insertions, translocations, and amplifications, as compared to those from Pgbd5-deficient mice (mean 69 and 44, respectively; t test P = 0.036; Fig. 3A). This increase could not be attributed to the age of tumor development, as Pgbd5-expressing MBs were significantly younger (mean 140 and 201 days, respectively; t test P = 0.024; Fig. 3B). There was no correlation between the number of somatic genome rearrangements and tumor age (r = −0.18, P = 0.55; fig. S5A), consistent with a distinct somatic mutational process responsible for tumor development. Pgbd5+/+ and Pgbd5−/− tumors exhibited similar types of somatic structural rearrangements (fig. S5B). Similar to human MBs, both Ptch1- and SmoM2-mutant mouse MBs exhibited relatively low numbers of single-nucleotide variants (SNVs), consistent with their early embryonal age of onset, regardless of Pgbd5 expression (mean 1.6 and 0.28 mutations/megabase, respectively; fig. S6, A and B). There were also no significant differences in single- or double-nucleotide mutational signatures between Pgbd5−/− and Pgbd5+/+ tumors, with the predominance of SBS5 and SBS18 signatures, currently attributed to chronological age and radical oxygen stress damage, respectively (fig. S6C) (26).
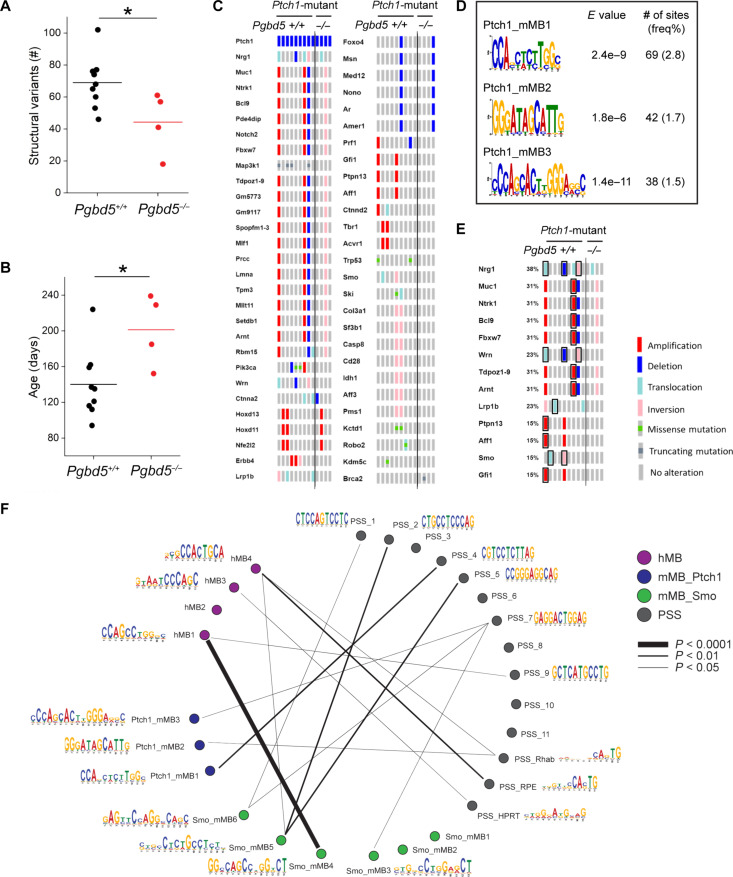
Fig. 3. Pgbd5 promotes somatic mutagenesis of recurrently mutated tumor suppressor and oncogenes in mouse SHH MBs.
(A) Numbers of SVs in Ptf1aCre/+;Ptch1fl/fl tumors. Pgbd5+/+ tumors (black, n = 9) harbor more SVs than Pgbd5−/− (red, n = 4). Lines indicate mean (69 and 44, respectively), and significance is measured using t test (*P = 0.036). (B) Age of tumors (days) in Ptch1-mutant tumors. Pgbd5+/+ tumors (black, n = 9) are younger than Pgbd5−/− tumors (red, n = 4; mean 140 and 201 days, *P = 0.024). (C) Oncoprint showing genes recurrently affected by SVs and SNVs in independent Ptch1-mutant tumors. Genes are curated based on likelihood that SVs or SNVs affect gene function (see Materials and Methods for details). The left nine and right four columns indicate tumors from Pgbd5+/+ and Pgbd5−/− mice, respectively. Red, blue, light blue, pink, and gray symbols indicate amplifications, deletions, translocations, inversions, and no alteration, respectively. Green and dark gray squares in gray symbols indicate missense and truncating mutations, respectively. (D) Three Pgbd5+/+-specific motifs are identified at SV breakpoints in Ptch1-mutant tumors, using discriminative MEME with Pgbd5−/− tumors as controls. E values indicate MEME discriminative algorithm significance (see Materials and Methods for details). The frequency shown was calculated by dividing the number of sites by total numbers of 50-bp breakpoint sequences extracted from SVs. (E) These motifs in (D) and the previously identified PSS_RPE and PSS_Rhab motifs were identified at SV breakpoints affecting known tumor suppressor and oncogenes in six of nine (66%) of Pgbd5+/+ tumors. The affected tumor suppressors and oncogenes are boxed black. (F) Circos plot showing similarities among all motifs (see Materials and Methods for details).
We identified putative MB tumor suppressor and oncogenes arising from Pgbd5-induced genomic rearrangements by analyzing their recurrence in independent mouse tumors, as compared to genes recurrently mutated in human MBs (Fig. 3C, fig. S7, and data S1, S2, and S5). This identified several genes, including Fbxw7, Tbr1, Gfi1, Pik3ca, and others known to be recurrently mutated in human SHH and non-SHH MBs (data S8) (10). The many of the same genes were also recurrently mutated in SmoM2-mutant mouse MB tumors, but not in the rare Pgbd5-deficient tumors (fig. S8A). Mutated genes were distributed across multiple distinct chromosomal regions, consistent with the specific genome-wide activity of their Pgbd5-dependent somatic mutagenesis (fig. S9 and data S20). This suggests that mouse SHH tumors model salient mutational features of human MBs, including specific developmental PGBD5-induced mutations.
To elucidate the specific mutational processes responsible for PGBD5-induced somatic genomic rearrangements in SHH MBs, we extracted 50–base pair (bp) sequences flanking all somatic DNA rearrangement breakpoints and analyzed their composition using supervised and de novo sequence motif analysis algorithms (fig. S10, A and B, and data S6). This analysis showed that most of the somatic DNA rearrangements contained repetitive sequences at their breakpoints, without apparent differences in rearrangement lengths (fig. S11), consistent with involvement of nonallelic homologous recombination or microhomology-mediated end-joining, which showed modest but not significant differences between Pgbd5-expressing and Pgbd5-deficient SHH MBs.
In contrast, specific nonrepetitive sequence breakpoints were significantly enriched at breakpoints of somatic DNA rearrangements in Pgbd5-expressing SHH mouse MBs as compared to Pgbd5-deficient tumors (mean 18 versus 6 per tumor, respectively; t test P = 2.7 × 10−3; fig. S11, A and B). Of the 2480 breakpoint sequences derived from somatic DNA rearrangements among nine Ptch1-mutant Pgbd5+/+ MB tumors, 149 exhibited distinct sequence motifs (Fig. 3D). Some of these breakpoint sequences exhibited significant similarity to the PGBD5-specific signal sequence (PSS) motifs previously observed to be rearranged by PGBD5 in genomic transposition and forward genetic assays (P = 7.6 × 10−3, 4.5 × 10−2, and 2.4 × 10−2, respectively; Fig. 3F and table S1) (4, 24). We confirmed the specificity of this PSS breakpoint detection using shuffled sequences that showed no significant associations despite having identical sequence composition (fig. S13 and table S1). We observed similar results in SmoM2-mutant MB tumors (252 of 5036 PSS-like breakpoints; P = 5.2 × 10−3 to 3.4 × 10−2; Fig. 3E, figs. S12 and S13, and table S1). The relatively modest association of MB genomic rearrangement breakpoints with previously observed PSS motifs suggests that other developmental mutational processes remain to be found.
The occurrence of somatic genomic rearrangements with specific PSS motif breakpoints frequently involved known genetic drivers of MB, including Smo, Nrg1, Wrn, Ntrk1, Fbxw7, and Gfi1 (Fig. 3E and table S2). In some cases, we observed recurrently mutated regions, such as, for example, mutations in chromosome 8 affecting both Nrg1 and Wrn genes with distinct breakpoints in three independent mouse MB tumors (table S2). In total, six of nine (67%) mouse MB tumors exhibit recurrent Pgbd5-associated genetic somatic mutations affecting known MB tumor suppressor and oncogenes (table S2). These findings indicate that Pgbd5-induced sequence-specific somatic mutagenesis contributes to mouse SHH MB development.
To examine the contribution of PGBD5-induced sequence-specific somatic mutagenesis in human MBs, we used de novo local sequence assembly–based methods to analyze 329 tumor genomes isolated from patients with all four major MB tumor subtypes, including SHH MBs (Fig. 4, A and B) (27). We found that nearly one in five somatic MB DNA rearrangement breakpoints contained PSS motifs, previously observed to be rearranged by PGBD5 in genomic transposition and forward genetic assays (Fig. 4C and data S9 to S12). This enrichment was significant when compared to somatic breakpoints in non–PGBD5-expressing but highly somatically rearranged human breast carcinomas (χ2 test P = 7.7 × 10−117; Fig. 4C) (4, 28). Using unsupervised de novo sequence motif analysis, we also identified four sequence motifs, termed hMB1 to hMB4, which were also specifically and significantly enriched in breakpoint sequences of somatic human DNA MB rearrangements, but not in those from human breast carcinomas that are also highly somatically rearranged (χ2 test P = 1.5 × 10−155; Fig. 4D and figs. S14 and S15, A and B).
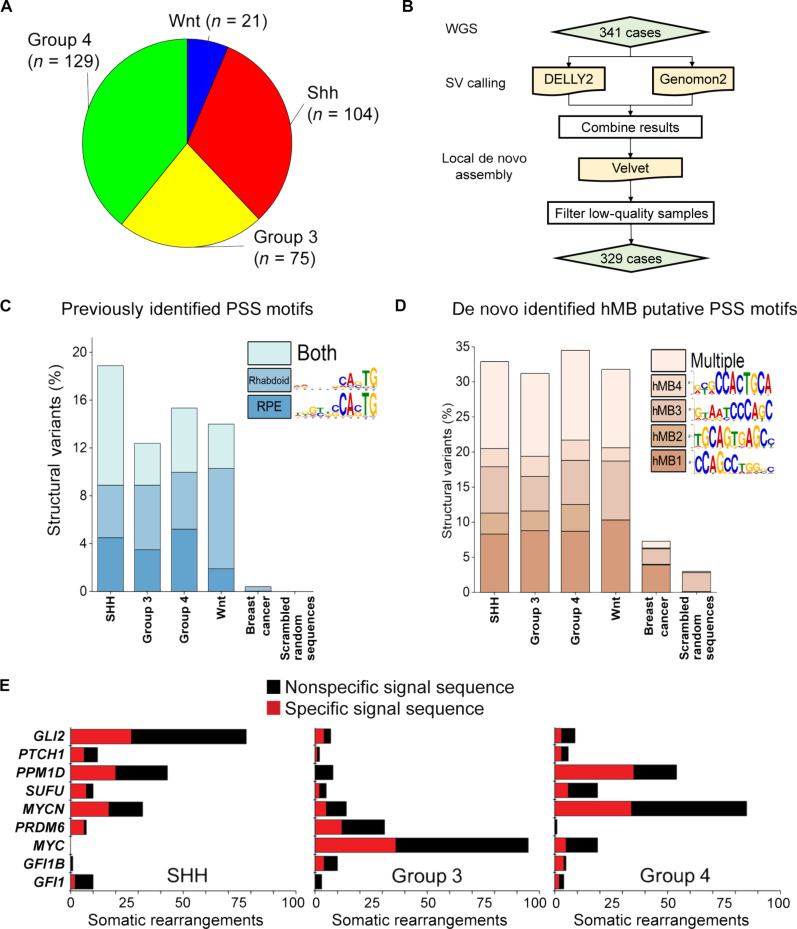
Fig. 4. PGBD5-associated sequence breakpoints recurrently affect somatic DNA rearrangements of known tumor suppressor and oncogenes in human MBs.
(A) Representation of human patient cohort showing the four major subgroups of MB that were included in the analysis (n = 329). (B) Pipeline to identify somatic SVs in human MB. (C) Previously identified PSS sequences (4, 24) are enriched at SV breakpoints in human MB as compared to somatic SVs in human breast carcinomas (χ2 test P = 7.7 × 10−117). Percentages represent the frequency of SVs (each SV has two breakpoints and 4 × 50-mers) where the motif was identified using a FIMO q value threshold of 0.3 based on a ROC curve analysis (fig. S14, B and C). Multiple indicates that more than one motif was identified at one SV, in either distinct or the same 50-mers. Scrambled sequences showed no enrichment and represent the background of the FIMO algorithm. (D) A set of four de novo motifs identified at SV breakpoints in human MB is enriched relative to breast carcinoma and scrambled sequences. hMB1 to hMB4 were identified as being specific using MEME and eliminating repetitive motifs. In addition, discriminative MEME, where control sequences were a set of 50,000 randomly selected 50-mers from the hg19 reference genome, was used to determine whether the motif was enriched at breakpoints relative to the genome (fig. S14). Percentages represent motif frequency among SVs as in (C) and are compared to SV breakpoints in human breast carcinoma (χ2 test P = 1.5 × 10−155) and scrambled sequences, which represent the background of the FIMO algorithm. (E) Recurrently mutated MB tumor suppressors and oncogenes in diverse tumor subtypes involve somatic DNA rearrangements with specific (red) sequence breakpoints, including PSS motifs. Numbers refer to the SVs detected in human patient cohort described in (A).
In total, nearly one in three somatic human MB DNA rearrangements exhibited specific sequence breakpoints (Fig. 4D). Multiple human tumor DNA rearrangement breakpoints were similar to those detected in Pgbd5-induced mouse SHH MBs (P = 5.3 × 10−6 and 1.0 × 10−2 for hMB1 versus Smo_mMB4 and hMB4 versus Smo_mMB5, respectively; Fig. 3F, fig. S13, and table S2). In particular, hMB4 motif showed significant similarity to PSS_Rhab and PSS_RPE sequences identified as PGBD5 substrates in cellular assays (P = 3.3 × 10−2 and 6.0 × 10−4, respectively; Fig. 3F, figs. S13 and S15C, and table S1). Consistent with the oncogenic activity of PGBD5-induced somatic mutagenesis, many recurrently mutated key SHH and non-SHH MB tumor suppressors and oncogenes, including GLI2, PPM1D, and MYC (29, 30), involved breakpoints with specific PSS-like sequences (Fig. 4E and data S5). Therefore, human MBs are defined by somatic DNA deletions, amplifications, and other chromosomal rearrangements, marked in part by specific breakpoints with similarity to PSSs, which recurrently affect MB tumor suppressors and oncogenes.
We reasoned that PGBD5 may induce somatic mutations in preneoplastic lesions (PNLs) in transformed neuronal progenitor cells and/or their progeny, as they develop into fulminant MB tumors. To investigate this process, we analyzed γH2AX as a surrogate marker of DNA damage signaling in Pgbd5+/+ and Pgbd5−/− cerebellar GCP cells transformed by oncogenic SHH signaling in Atoh1-CreERT2; R26SmoM2 mice. We observed prominent preneoplastic expansions in both Pgbd5+/+and Pgbd5−/− mice at P22 and P23 of development (fig. S16A). We used the external cell layer structure to identify the PNLs and 5-ethynyl-2′-deoxyuridine (EdU) pulse-chase labeling to identify proliferating cells (fig. S16, A and B). Pgbd5+/+ preneoplastic cells did not exhibit higher level of γH2AX nuclear foci in both dividing EdU-positive and nondividing EdU-negative cells compared with Pgbd5−/− preneoplastic cells (fig. S16, C and D). This suggests that Pgbd5-dependent mutagenesis occurs upon later tumorigenesis of PNLs and/or its early activity in transformed GCPs does not lead to global DNA damage signaling.
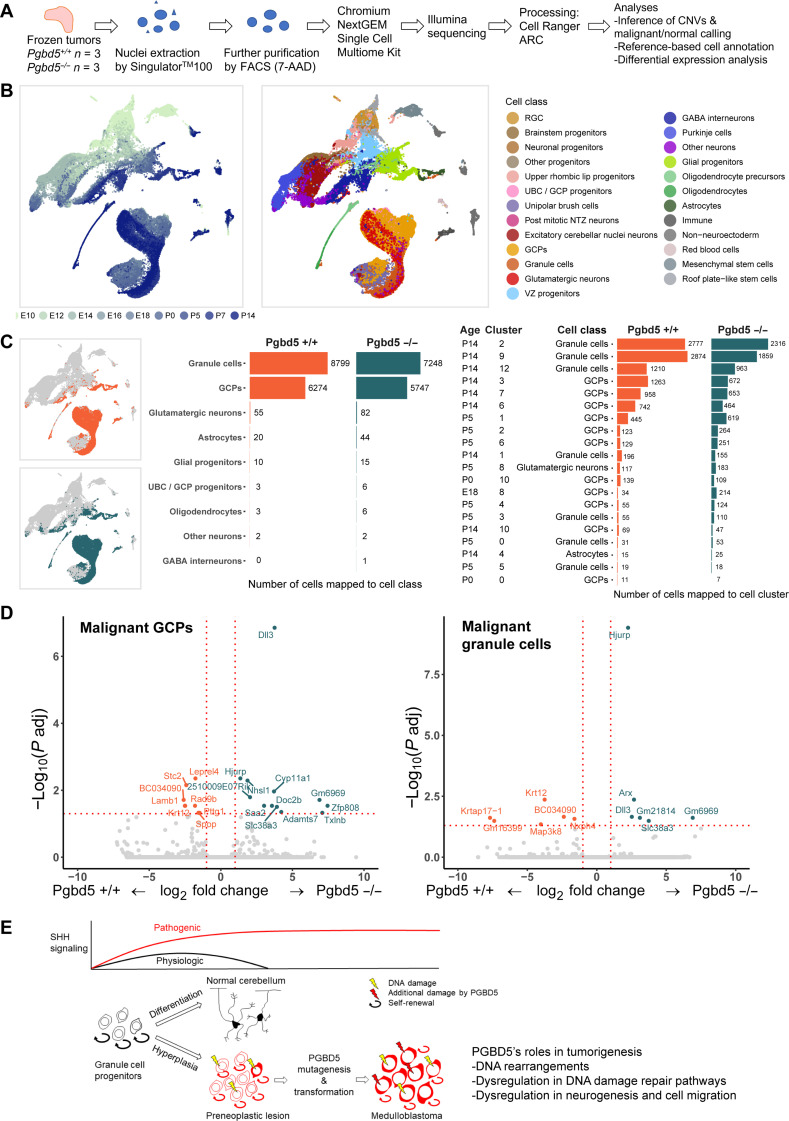
To define potential Pgbd5-dependent mechanisms in cerebellar GCP transformation induced by oncogenic SHH signaling, we performed single-nucleus RNA-sequencing (snRNA-seq) gene expression analysis of Pgbd5+/+ and Pgbd5−/− MBs isolated from Ptf1aCre/+;Ptch1fl/fl mice (Fig. 5A). Upon identifying MB tumor cells by detecting their DNA copy number alterations induced by somatic mutations, we mapped the observed MB tumor cell gene expression onto the developmental ontogeny of normal mouse cerebellum (31). We observed that gene expression states of both Pgbd5+/+ and Pgbd5−/− MB tumor cells resembled developing cerebellar GCPs and granule cells, with similar abundance of inferred cell states between Pgbd5+/+ and Pgbd5−/− MB tumors (Fig. 5, B and C; figs. S17 and S18; and data S21). Thus, Pgbd5-dependent neuronal progenitor cell transformation involves GCP-like cells and their subsequent tumorigenic evolution (Fig. 5E).
Fig. 5. Pgbd5-dependent mechanisms of cerebellar GCP transformation.
(A) Schematic of experimental procedure for snRNA-seq of three Pgbd5+/+ and three Pgbd5−/− fresh-frozen Ptf1aCre/+;Ptch1fl/fl SHH MBs; same tumors as were analyzed by whole-genome DNA sequencing were used. (B) Uniform Manifold Approximation and Projection (UMAP) plots of the cerebellar reference (N = 62,040) used to annotate the cells from Pgbd5+/+ and Pgbd5−/− Ptf1aCre/+;Ptch1fl/fl SHH MBs. Cells are colored by age (left) and by refined cell class ontology (right). (C) Cell class and cluster consensus predictions for Pgbd5+/+ (orange) and Pgbd5−/− (blue) MB tumor cells. Left: UMAP plots of the cerebellar reference highlighting the mapped clusters of malignant cells of each genotype. Middle: Bar plot depicting the cell class consensus predictions for the tumor cells for each genotype. Right: Bar plot depicting the cluster-specific consensus predictions for tumor cells for each genotype. Only clusters with more than 10 cells mapped to by either genotype are shown. (D) Volcano plot showing differential gene expression between Pgbd5+/+ and Pgbd5−/− malignant GCPs and granule cells. Genes significantly up-regulated in Pgbd5+/+ MBs are highlighted in orange, while significantly up-regulated genes in Pgbd5−/− tumor cells are highlighted in blue (log2FC > 0.25, adjusted P < 0.05). (E) Model of PGBD5-dependent tumorigenesis, illustrating how pathogenic SHH signaling is associated with hyperplasia of cerebellar granule cell progenitor cells, leading to PNLs that undergo PGBD5-dependent somatic mutagenesis and malignant transformation.
Can Pgbd5 contribute to the transformation of cerebellar GCP transformation induced by oncogenic SHH signaling? To explore this question, we compared single-nucleus gene expression profiles of Pgbd5+/+ and Pgbd5−/− for each cell population within the tumors. We observed significantly differentially expressed Pgbd5-associated genes known to encode regulators of DNA damage repair, neurogenesis, and cell migration (Fig. 5D and data S22). This included Hjurp, which can regulate double-strand DNA repair (32–34), neurogenesis factors Dll3 and Arx (35, 36), as well as neuronal cell migration regulator Nhsl1a (Fig. 5D and data S22) (37). Thus, in addition to its oncogenic mutator functions via sequence-specific somatic mutagenesis, Pgbd5 may also regulate neuronal progenitor cell development and DNA damage repair.
DISCUSSION
Although somatic mutational mechanisms have been extensively documented in aging-associated cancers, how oncogenic DNA rearrangements occur during dysregulated development in childhood and young-onset tumors is poorly understood. Here, we demonstrate that a domesticated DNA transposase–derived gene causes oncogenic sequence-specific DNA rearrangements in medulloblastomas, a common childhood brain tumor (38). We provide evidence that oncogenic developmental signaling not only can induce preneoplastic cell expansion but also involves endogenous mutators to generate genomic rearrangements that are predicted to affect tumor suppressors and oncogenes. We also derived the first principles of how a domesticated DNA transposase can promote somatic genome mutagenesis, providing a foundation for the identification of other sequence-specific mutational processes in other cancers. This raises the possibility that incorporation of sequence-specific DNA rearrangements into cancer mutational profiling could improve diagnosis and treatment (39, 40).
PGBD5-mediated somatic mutagenesis offers a genetic mechanism for sequence-specific DNA rearrangements in developmental cancers, including neuroblastomas, Ewing sarcomas, desmoplastic small round cell tumors, rhabdomyosarcomas, small cell carcinomas, and many other young-onset cancers that express PGBD5. Aberrant PGBD5 activity also offers a plausible mechanism for the generation of complex DNA rearrangements typically observed in these tumors, including chromoplexy, and other complex DNA rearrangements (41).
Many studies have implicated replication stress as a cause of somatic mutations in cancer, including that induced by high-level SHH signaling in cerebellar GCPs (42). However, this concept does not explain how sequence-specific mutations, including those affecting tumor suppressors and oncogenes, initially occur. The results presented here offer a mechanism by which dysregulated SHH developmental signaling can activate PGBD5 mutagenic activity and/or impair its efficient repair. It is also possible that dysregulation of PGBD5 and other sequence-specific nucleases can induce not only somatic mutagenesis by virtue of their cellular nuclease activities but also epigenetic dysregulation via its interactions with chromatin and cellular factors, such as those identified in single-nucleus gene expression profiling of specific tumor cells.
While we favor the idea that PGBD5 acts directly on DNA (3, 4, 6–8), direct biochemical and structural studies will be needed to define the exact enzymatic mechanisms of PGBD5 cellular activities and their developmental regulatory factors, including the possibility that PGBD5 promotes somatic DNA rearrangements through recruitment of other nucleases and chromatin remodeling factors. This is an important direction of future work because two recent publications reported an inability to detect DNA transposition of PGBD5 in cellular assays (43, 44). While PGBD5 can support genomic DNA integration in cells, its cellular activity predominantly involves double-strand DNA breaks, deletions, and other DNA rearrangements, with relatively few precise transposon-specific excisions and insertions (8). Thus, human PGBD5 is divergent from “cut-and-paste” transposases such as Trichoplusia ni piggyBac, but rather should be considered a transposase-derived protein with domesticated genomic integration and DNA rearrangement activities in human cells.
Although we focused on the domesticated DNA transposase–derived PGBD5 in this study, we propose that the principles and implications of somatic developmental mutators revealed herein extend beyond cancer development (38). First, PGBD5 has been recently implicated in normal brain development, where PGBD5 is required for the induction of double-strand DNA breaks in developing neurons and proper neuronal migration and development (45, 46). Second, transposable element–derived genes are among the most ubiquitously present genes in living organisms (47), with many transposases domesticated in diverse genetic species and somatic tissues (48). One can imagine how domesticated transposases can provide molecular mechanisms for somatic genetic diversification during normal tissue development and, when dysregulated, cause somatic mutations that are increasingly being found as causes of diverse sporadic human diseases.
MATERIALS AND METHODS
Mice
Ptf1a-Cre/+, Ptch1fl/fl, and Atoh1-GFP mice were obtained from M. Hoshino (49), B. Wainwright (50), and J. Johnson (22), respectively. All three lines were initially maintained on mixed backgrounds (SW and C57BL/6J) and subsequently backcrossed with C57BL/6J mice to generate C57BL/6J-background mice. Atoh1-CreERT2 (Math1-CreERT2) (007684) and R26SmoM2 (005130) mice were obtained from the Jackson Laboratory (51, 52). Atoh1-CreERT2 mice were maintained on SW background. Pgbd5-floxed mice were generated by targeting exon 4 of mouse Pgbd5 (InGenious Targeting Laboratory; fig. S1A). Targeting vector consisting of LacZ and NeoR cassettes was electroporated in C57BL/6 embryonic stem cells, and targeted clones were microinjected into Balb/c blastocysts. Resulting chimeras were crossed with C57BL/6 FLP mice to remove the Neo cassette to generate Pgbd5-floxed mice, as confirmed using genotyping with SC2 (GAGAGCACCGTTGGTGCATATCAG) and SC4 (AGAGTATGAGCGGGAGAGGAGCAG) (fig. S1A). Pgbd5-floxed mice were then crossed with B6.FVB-Tg(EIIa-cre)C5379Lmgd/J (EIIa-Cre) mice to generate global Pgbd5-deficient mice, as confirmed by genotyping with SC2 and SC5 (TTTCTCAGCTGTCCCCAGCATAGC) primers. Pgbd5-deficient mice were backcrossed with C57BL/6J mice for six generations. The MASTR model was adapted from Wojcinski et al. (53) and Lao et al. (13) (fig. S2A). Atoh1-FlpoER/+, R26MASTR (MA), and R26SmoM2 (SmoM2) mice were maintained on a mixed background and subsequently outbred into an SW background. Genotyping of Atoh1-FlpoER/+ and R26MASTR were done by PCR using following pairs of forward and reverse primers: GCTCTACTTCATCGCATTCCTTGC (forward) and ATTATTTTTGACACCAGACCAAC (reverse) and GATATCTCACGTACTGACGG (forward) and TGACCAGAGTCATCCTTAGC (reverse), respectively. Offspring were obtained by crossing either Atoh1-FlpoER/+; SmoM2/SmoM2; Pgbd5fl/+ with MA/+; Pgbd5fl/fl, Atoh1-FlpoER/+; SmoM2/SmoM2; Pgbd5fl/+ with MA/MA or Atoh1-FlpoER/+; SmoM2/SmoM2; Pgbd5fl/+ with MA/MA; Pgbd5fl/+. All experiments were conducted in compliance with protocols approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee.
Tumor induction
On P2, pups from Atoh1-CreERT2 × R26SmoM2 and crossings from the MASTR model were injected with tamoxifen (2 and 200 mg/kg; Sigma-Aldrich, T5648), respectively, in corn oil subcutaneously. Some pups from the MASTR model were injected on P0. For the γH2AX analysis using p22/23 Atoh1-CreERT; R26SmoM2, pups were injected with tamoxifen (200 mg/kg) on P0. Study end points included reduced activity, ataxia, and/or domed skulls.
Histology, immunohistochemistry, and immunofluorescence of mouse cerebellum and tumors
Under deep anesthesia, animals were perfused with intracardiac 0.9% saline followed by 4% paraformaldehyde (PFA)/0.1 M phosphate buffer (PB). Brains were dissected, further fixed in 4% PFA/0.1 M PB overnight, and embedded in paraffin blocks. Sagittal sections of cerebella were stained with hematoxylin and eosin. Immunohistochemistry was performed using anti-Ki67 (ab15580, Abcam) and anti-NeuN (A60, EMD Millipore), respectively. For γH2AX staining of the PNLs, cryosections were used. On P22/23, cerebella were dissected and fixed in 4% PFA/0.1 M PB overnight. After cryopreservation in sucrose, cerebella were embedded in OCT (Thermo Fisher Scientific), and blocks were sectioned sagittally in 10-μm sections using a rotary microtome cryostat (Leica). Sections were stored at −20°C. Antigen retrieval was done in sodium citrate buffer [10 mM sodium citrate and 0.05% Tween20 (pH6.0)] for an hour at 99°C. Anti–phospho-histone H2A.X (Ser139) antibody (JBW301, MilliporeSigma) was used in combination with an Alexa Fluor 488–conjugated anti-mouse immunoglobulin G antibody (Invitrogen).
EdU staining
On P22/23, at 2 hours before sacrifice, EdU (50 mg/kg, Invitrogen) diluted in 0.9% saline was injected intraperitoneally. Cryosections were prepared as detailed above. For double staining with γH2AX, antigen retrieval was performed before EdU staining. Click-iT Plus EdU Alexa647 Imaging kit (Invitrogen) was used to stain EdU+ nuclei.
Counting of γH2AX foci
The PNLs between lobule VI and VII of the vermis from Atoh1-CreERT2; R26SmoM2; Pgbd5+/+ and Pgbd5−/− mice were imaged using LSM800 (Zeiss) with a 63× oil lens. A total of 90 nuclei per animal (3 images × 30 nuclei) were counted for γH2AX foci and for the presence of EdU.
Copy number analysis
Upon euthanasia and tumor dissection, genomic DNA was extracted from tumors and matched normal tissues (Transnetyx). Genomic DNA real-time PCR was performed using probes for Pgbd5 wild-type (Pgbd5-1 WT), Pgbd5-floxed (Pgbd5-1 FL), and Pbgd5-null (Pgbd5-1 EX) alleles, with probes for jun as reference (Transnetyx).
Isolation of mouse cerebellar GCPs
The protocol was adapted from Nakashima et al. (54) and Lee et al. (55). Briefly, P5 cerebella were trypsinized at 37°C, followed by deoxyribonuclease (DNase) I treatment. GCPs were then isolated by a Percoll gradient consisting of 60 and 35% Percoll solutions. After centrifugation at 2000g, cells at the interface between the 35 and 60% Percoll were collected. The cells were washed twice in phosphate-buffered saline (PBS) before analysis.
Isolation and sorting of PNLs
After euthanasia, cerebella at 3 to 8 weeks were isolated and dissociated by trypsin followed by DNase I treatment (55). The cells were resuspended in 0.1% bovine serum albumin/PBS with DNase I.The GFP-positive population representing PNLs were collected by FACS using the FACSMelody system (BD Biosciences). After doublet/triplet and dead cells were excluded, GFP-positive fractions were collected for analysis. Data were analyzed using FlowJo version 10 (BD Biosciences).
Gene expression analysis
Total RNA was extracted from P5 GCPs using RNeasy Plus Micro Kit using on-column DNA digestion protocol (Qiagen). Reverse transcription was performed using qScript (Quanta Biosciences) followed by real-time PCR using KAPA SYBR FAST ROX Low (Roche) on the ViiA 7 Real-Time PCR instrument (Applied Biosystems). The following primers were used for Gli1 (forward, GAGGTTGGGATGAAGAAGCA; reverse, CTTGTGGTGGAGTCATTGGA), Pgbd5 (forward, GCGGCCGGAAAGAACTATATC; reverse, CACAGCAGTAGATCCCTTGC), and Actb (forward, GAGAAGATCTGGCACCACACC; reverse, GGTCTCAAACATGATCTGGGTC).
RNA ISH/FISH
BaseScope hybridization probes specific for Pgbd5 exon 4 were generated as per the manufacturer’s instructions (ACD, catalog no. 1181898-C1). Upon cardiac perfusion and fixation in 4% PFA/0.1 M PB overnight, dissected brains were washed twice with 30% sucrose/PBS and incubated overnight at 4°C. After cryopreservation in sucrose, brains were embedded in OCT (Thermo Fisher Scientific), and blocks were sectioned sagittally in 10-μm sections using a rotary microtome cryostat (Leica). Sections were stored at −80°C.Cryosections were baked for 1 hour at 60°C and fixed in 4% PFA for 15 min followed by washing in PBS. After dehydration, epitope retrieval treatment with ER2 for 5 min at 95°C and subsequent Protease III treatment for 15 min at 40°C were performed. The probe set was hybridized for 2 hours at 42°C. Signal amplification steps were performed according to the manufacturer’s protocol. Fast Red (Leica Bond Polymer Refine Red Detection kit DS9390) was used as chromogen. Hematoxylin was used as a counterstain. Mouse Ppib (ACD, catalog no. 701078) and Bacillus subtilis dapB (ACD, catalog no. 701018) probe sets were used as positive and negative controls, respectively. An adjacent section of the Pgbd5 ISH section was used to identify Cre-expressing cells, i.e., tumor cells. A Cre-specific probe set (ACD, catalog no. 312288-C2) was used for fluorescence ISH (FISH). Cryosections from frozen samples were baked for 1 hour at 60°C. Sections were fixed in 4% PFA for 15 min and washed in PBS followed by dehydration. Epitope retrieval treatment with ER2 for 10 min at 95°C, followed by 10 min of incubation with ACD 2.5 LS Hydrogen Peroxide, was performed. The probe set was hybridized for 2 hours at 42°C. Mouse Ppib (ACD, catalog no. 313918) and bacterial dapB (ACD, catalog no. 312038) probes were used as positive and negative controls, respectively. The hybridized probes were detected using the RNAscope 2.5 LS Reagent Kit–Brown (ACD, catalog no. 322100) according to the manufacturer’s instructions with the following modifications. 3,3′-diaminobenzidine application was omitted and replaced with Alexa Fluor 488 Tyramide signal amplification reagent for 20 min at room temperature (Life Technologies, B40953). After staining, slides were washed in PBS and incubated in 4′,6-diamidino-2-phenylindole (DAPI; 5 μg/ml; Sigma-Aldrich) in PBS for 5 min, rinsed in PBS, and mounted in Mowiol 4-88 (Calbiochem). Slides were stored at −20°C before imaging.
Whole-genome sequencing of mouse MBs
Tissues, including tumor and matched skin or spleen, were harvested from symptomatic mice and flash-frozen using a dry ice and ethanol bath. DNA and RNA were extracted using the Qiagen AllPrep kit, according to the manufacturer’s instructions. Paired end libraries (2 × 150 bp) were prepared using Illumina TruSeq DNA PCR-Free kit and sequenced using Illumina HiSeq X at a depth of 80× for tumors and 40× for matched normal tissues. Reads were aligned to the mm10 reference genome with BWA-MEM (version 0.7.15) and processed by eliminating duplicate reads with NovoSort MarkDuplicates (version 3.08.02). SNVs were detected using Mutect2 (version 4.0.5.1), Strelka2 (version 2.9.3), and Lancet (exonic) (version 1.0.7). High-confidence SNVs included those that were detected by more than one caller. SVs were detected using Manta, SvABA (version 0.2.1), and Lumpy (version 0.2.13). For SVs, all those that passed quality filters were included. Sequencing data are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA).
Single-nucleotide mutational signature analysis of mouse MBs
Mutational signature analysis was performed using three main steps: de novo extraction, assignment, and fitting (56). For the first step, we ran hierarchical Dirichlet process (https://github.com/nicolaroberts/hdp, c78989b). Then, all extracted signatures were assigned to the COSMIC reference (https://cancer.sanger.ac.uk/signatures/sbs/) (39) to define which known mutational processes are active. Last, we applied our recently developed fitting algorithm (https://github.com/UM-Myeloma-Genomics/mmsig, df771ba) to estimate both the presence and the contribution of each mutational signature in each sample (57).
Identification of genes affected by SVs in mouse MBs
Genes analyzed included those from the COSMIC cancer gene census (58). We considered genes to be putatively affected by SVs if (i) the gene was affected by an SV breakpoint; (ii) the gene overlapped with a duplication, inversion, or deletion; or (iii) the gene resided within 25 kb of a translocation breakpoint. For the Oncoprint depicted in Fig. 3C, we curated putative driver alterations arising from Pgbd5-induced SVs by analyzing for recurrence, as well as comparing to genes known to be recurrently mutated in human SHH MBs. Specifically, the putative tumor suppressor and oncogenes were identified as (i) genes affected by SVs in ≥4 Ptch1-mutant tumors, (ii) genes affected by point mutations in ≥2 tumors, (iii) genes affected by SVs in ≥2 tumors that were also affected by SVs in ≥10% of human SHH MBs, or (iv) genes affected by SVs that were demonstrated to be recurrently mutated by Northcott et al. (10). These SVs were further analyzed for their chromosomal loci. By using the UCSC cytoband annotation file, the cytobands corresponding to the region in which each breakpoint of SVs resides were annotated (fig. S9 and data S20).
Whole-genome sequencing of human MBs
All patient material was collected after receiving written informed consent, which includes consent to publish the data, as approved by the Medulloblastoma Advanced Genomics International Consortium and the International Cancer Genome Consortium (27). Somatic SVs were called using Genomon-SV (v0.4.1) and DELLY2 (v0.7.5) as described by Skowron et al. (27). For each SV detected by either algorithm, we used Velvet to assemble reads around the detected breakpoints, and the resultant contigs were then remapped locally using human genome reference sequences with and without detected SVs using blat. This approach ensures detection of heterozygous SVs, which would be mapped both to reference sequence without incorporated variant contigs and to reference sequence that incorporates them. Subsequently, we selected variants for which assembled contigs could be mapped to the reference sequence containing somatic SVs, and variants from matched normal tissue that could not be mapped to the reference sequence that incorporates variant contig sequences. Sequencing data are available from European Genome-phenome Archive (EGA) using the following accession numbers: EGAD00001003125, EGAD00001004347, and EGAD00001003127.
Identification of breakpoint sequence motifs in mouse MBs
Sequences ±50 bp flanking both breakpoints from identified SVs were extracted with bedtools (version 2.29.2) using the mm10 reference genome. To identify motifs that are putatively associated with Pgbd5 activity in mouse tumors, we used de novo motif analysis of the breakpoint sequences from Pgbd5-expressing tumors and the breakpoint sequences from Pgbd5-deficient tumors as background. We used discriminative MEME (v5.4.1) (59), which determines relative sequence enrichment in an experimental set of sequences relative to a control set. Pgbd5-deficient breakpoint sequences were used as controls. Discriminative MEME was used with default parameters but limited to between 11 and 16 bp. Repetitive motifs were eliminated, and the remainder were chosen as putative Pgbd5-associated motifs. For SmoM2-mutant tumors, motifs were identified using classic MEME with default parameters and limited to between 11 and 16 bp.
Identification of breakpoint sequence motifs in human MBs
Sequences ±50 bp flanking both breakpoints from identified SVs were extracted with bedtools (version 2.29.2) using the hg19 reference genome. To identify de novo motifs, classic MEME (version 5.1.1) (60) with default parameters but limited to 11 bp was used (http://meme-suite.org/). Putative PSS motifs were selected by eliminating repetitive motifs and by determining whether candidate motifs were found when discriminative MEME was run relative to 50,000 randomly selected 50-mers from the hg19 reference genome without repeat masking. To quantify the abundance of motifs at SV breakpoints, we used FIMO (version 5.1.1) (60) with default parameters (fig. S14). Q-value cutoffs for quantitation were determined by identifying a threshold where previously identified PSS motifs (Rhabdoid and RPE) could be specifically detected, but negative control motifs (RAG1/2 and scrambled) were not. This cutoff was characterized by a receiver operating characteristic (ROC) curve and chosen for sensitivity of 100% and specificity of 75% for the previously identified PSS motifs. For the quantifications, each SV is counted only once (i.e., each SV has 4 × 50-mers and 2× breakpoints). As a comparator, we included human breast carcinoma SVs (4, 28). To mimic the background distribution of the FIMO algorithm, we used randomly scrambled sequences.
Sequence motif comparisons
The similarity of motifs was evaluated using TomTom (https://meme-suite.org/meme/tools/tomtom) (61). The first set of query motifs that included those from three Ptch1-mutant tumors and six SmoM2-mutant tumors were compared with previously identified target motifs [PSS motifs: including 11 motifs (PSS_1 to 11), PSS_Rhab, PSS_RPE, and PSS_HPRT]. To test for similarities, 10,000 shuffled target motifs were generated and their P values were examined. If the P value of the target motif ranked within the lowest 5% of all 10,001 P values, motif was deemed significantly similar. In the same way, the second set of query motifs (hMB1 to hMB4) were compared with nine mouse motifs and their shuffled sequences. Last, the query motif sets (hMB1 to hMB4) were compared with the previously identified 14 PSS motifs and their shuffled sequences. A Circos plot (Fig. 3F) was generated to show similarities among all motifs. Line thickness is proportional to degree of similarity, as indicated. The mouse MB motifs include three Ptch1-mutant motifs in blue (Ptch1_mMB1-3) and six SmoM2-mutant motifs in green (Smo_mMB1-6). The human MB motifs are colored purple (hMB1 to hMB4). The previously identified PSS motifs include 14 motifs in gray (PSS_1-11, PSS_Rhab, PSS_RPE, and PSS_HPRT) (4, 24).
Single-nuclei multiome ATAC + gene expression of mouse MBs
Nuclei extraction
Frozen pieces (~50 mg) of tumors from Ptf1a-Cre/+; Ptch1fl/fl; Pgbd5+/+ (n = 3) and Pgbd5−/− (n = 3) were used for nuclei isolation. Nuclei extraction was performed according to the protocol from Masilionis et al. (62). Singlulator100 (S2 Genomics) was used for nuclei extraction. Extracted nuclei were stained with 7-aminoactinomycin D (Invitrogen) and FACS-sorted for further nuclei purification. Ten thousand nuclei were targeted for library construction using ChromiumNextGEM Multiome ATAC + Gene Expression kit (10x Genomics).
Single-cell multiome data processing and QC
Cell Ranger ARC v2.0.0 (10x Genomics) was used (“count” option with default parameters) to filter and align raw reads, identify transposase cut sites, detect accessible chromatin peaks, call cells, and generate raw count matrices for scMultiome samples. Alignment was performed using the mm10 reference genome build coupled with the Ensembl 98 gene annotation. Reads that mapped to the intronic regions were excluded for the RNA modality.
Quality control (QC) and data processing steps were performed using Signac v1.3.0 (63) and Seurat v4.3.0 (64). QC metrics for RNA and Assay for Transposase-Accessible Chromatin (ATAC) modalities were calculated independently but were jointly used to filter cells. A combination of thresholds was established for each sample based on hard cutoffs or on the distribution of each metric within the sample (data S21). In the RNA modality, cells were filtered on the number of genes, unique molecular identifiers (UMIs), and mitochondrial content. In the ATAC modality, cells were filtered on the number of peaks detected, transcription start site enrichment, and nucleosome signal.
Normalization and dimensionality reduction in single-cell multiome data
For the RNA modality, libraries were scaled to 10,000 UMIs per cell and log-normalized. UMI counts and mitochondrial content were regressed out from normalized gene counts, and the residuals were z-scored gene-wise. Dimensionality reduction was performed using principal components analysis (PCA) on the top 2000 most variable features. For the ATAC modality, peaks were called using MACS2 (v2.2.7.1) (65) using the CallPeaks function from Signac library with default parameters. ATAC reads were quantified in each peak per cell, and a resulting count matrix was generated. Dimensionality reduction was performed using latent semantic indexing (LSI) (66).
A weighted nearest neighbor graph was constructed between all cells using the first 30 principal components from the RNA data and the top six dimensions of the LSI reduction from the ATAC data with the following default parameters: 20 multimodal neighbors, 200 approximate neighbors, and L2 normalization enabled. This weighted nearest neighbor graph was used as input for projection into two dimensions using Uniform Manifold Approximation and Projection (UMAP) (67) and for clustering using a shared nearest-neighbor (SNN) algorithm (64) based on the Louvain algorithm on a k-nearest neighbor graph with k = 20 and resolution 0.2.
Joint sample integration and visualization in single-nuclei RNA data
RNA libraries from all samples were merged and processed as described above for normalization and dimensionality reduction with a minor change (no variables were regressed out). To visualize the samples in a shared UMAP space, Harmony (v0.1.1) (68) was run to integrate the samples using the first 30 principal components as input and regressing out the differences between samples. The resulting batch-corrected embedding and the top 30 dimensions were used as input for projection into two dimensions (UMAP) and for clustering (SNN algorithm) with k = 20 and resolution 0.5.
Cell type annotation in single-nuclei RNA data
Annotation of cell types was performed using four different reference-based annotation tools: three machine learning–based prediction methods [SciBet (69), SingleCellNet (70), and SingleR (71)] and a statistical model [Spearman correlation as previously described (72)]. A consensus label annotation was assigned when at least two methods agreed.
Cell type annotation was performed using a murine cerebellar developmental atlas (31) [nine developmental samples ranging from time points E10 (embryonic day 10) to P14; N = 62,040 cells] as the reference. To obtain more granular clusters in the reference, we reprocessed and clustered each individual reference sample as described above, resulting in 209 new clusters. To label these new clusters, we used a combination of age, new cluster number, and cell class information from the original publication (31). Cell class was assigned on the basis of the proportion of the original cell type in the new cluster. If the proportion of one original cell type was greater than 75% in the new cluster, that label was used. In cases where this was not achieved, a label consisting of two prominent cell type labels was used. The new cluster labels were used to perform annotation of cell types in the mouse MB samples.
Malignant cell identification in single-nuclei RNA data
Copy number variants (CNVs) were inferred on a sample basis using inferCNV (v1.10.1) (73) with the following parameters: cutoff = 0.1, window_length = 101, analysis_mode = “samples,” cluster_by_groups = FALSE, sd_amplifier = 1.5, HMM = FALSE, and denoise = TRUE. The mitochondrial chromosome (defined as having gene symbols starting with “mt”-), ribosomal genes (defined as having gene symbols matching “Rps,” “Rpl,” “Mrps,” “Mrpl”), and MHC genes (defined as having gene symbols starting with “H2”-) were excluded from the CNV inference. A normal reference was assembled selecting clusters composed of a single cell type and originating from multiple samples (immune and oligodendrocyte precursor cell clusters) in the integrated snRNA-seq space. Last, hierarchical clustering of cells based on their CNV profiles was used to identify subtrees of malignant cells with prominent copy number signal, distinguishing them from normal cells lacking CNV signal.
Differential expression in single-nuclei RNA data
To identify differentially expressed genes (DEGs) between different genotypes within malignant cell populations, we used two different strategies. The first approach used a Wilcoxon rank sum test applied through the FindMarkers function from the Seurat package (logFC.threshold = 0, min.pct = 0.05). For the second approach, pseudobulk RNA files were generated and processed for the targeted cell populations of each sample. Adaptor sequences and the first four nucleotides of each read were removed from the pseudobulk read sets using Trimmomatic (v0.39) (74). Reads were scanned from the 5′ end and truncated when the average quality of a 4-nucleotide sliding window fell below a threshold (phred33 < 30). Short reads after trimming (< 30 bp) were discarded. High-quality reads were aligned to the mouse reference genome build mm10 using STAR (v2.7.9a) (75) with default parameters. Multimapping reads (MAPQ < 1) were discarded from downstream analysis. Gene expression levels were estimated by quantifying reads mapping to at most two locations (MAPQ ≥ 3) to exonic regions (the maximal genomic locus of each gene and its known isoforms) using featureCounts (v2.0.3) (76) and the mm10 ensGene annotation set from Ensembl. Gene expression levels were estimated by quantifying primary alignments mapping to at most two locations (MAPQ ≥ 3) to exonic regions (the maximal genomic locus of each gene and its known isoforms) using featureCounts (v2.0.3) (76) and the mm10 ensGene annotation set from Ensembl. Normalization (mean of ratios) of the data and differential gene expression analysis were performed using DESeq2 (v1.30.1) (77) with the Wald test. Genes that pass significance in both strategies (log2FC > 0.25 and adjusted P value > 0.05) are DEGs with strongest confidence.
Acknowledgments
We thank Y. Bigot, A. Henssen, G. Casalena, H. Mueller, S. Takao, S. Cheng, M. Kharas, H. Zhu, A. Gutierrez, and M. Mansour for helpful suggestions; M. Gönen for statistical advice; W. Kang, E. Rosiek, K. Manova-Todorova, V. Morell, the MSK Molecular Cytology, Integrated Genomics and Bioinformatics Core facilities, and the Center for Comparative Medicine and Pathology for technical assistance; and N. Kentsis for editing help. Data analyses were enabled by compute and storage resources provided by the Digital Research Alliance of Canada. A.K. is a Scholar of the Leukemia & Lymphoma Society.
Funding: This work was supported by NIH grants R01 CA214812 (A.K.), R01 CA192176 (A.L.J.), and P30 CA008748 (A.K., A.L.J., and N.D.S.); St. Baldrick’s Foundation (A.K.); Burroughs Wellcome Fund (A.K.); Rita Allen Foundation (A.K.); Pershing Square Sohn Cancer Research Alliance and the G. Harold and Leila Y. Mathers Foundation (A.K.); Starr Cancer Consortium (A.K.); Cookies for Kids’ Cancer (A.K. and G.P.R.); MSK Brain Tumor Center (A.K. and G.P.R.); MSK Functional Genomics Initiative (A.L.J.); MSK Cycle for Survival (A.L.J.); and Canadian Institutes of Health Research grant PJT-190271 (C.L.K.).
Author contributions: Conceptualization: A.K., G.P.R., A.L.J., M.D.T., R.R.K., and M.Y. Methodology: R.R.K., M.Y., D.C., H.S., R.S., J.V., J.G., F.M., W.H., M.S., N.R., P.D., N.S.B., L.J.Z., C.R., I.M., R.C., N.D.S., S.H., R.L.G., and C.L.K. Investigation: R.R.K., M.Y., D.C., H.S., R.S., J.V., J.G., F.M., W.H., M.S., N.R., P.D., N.S.B., L.J.Z., C.R., N.D.S., R.L.G., and C.L.K. Visualization: M.Y., R.R.K., F.M., H.S., J.V., N.S.B., and R.L.G. Funding acquisition: A.K., G.P.R., A.L.J., and M.D.T. Project administration: A.K. Supervision: A.K., G.P.R., A.L.J., and M.D.T. Writing—original draft: A.K., M.Y., and R.R.K. Writing—review and editing: all authors.
Competing interests: A.K. is a consultant for Novartis, Rgenta, Blueprint, and Syndax. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data are available from the NCBI SRA (mouse) and the EGA (human) using respective accession numbers listed in Materials and Methods. All processed data, including specific structural variants and their breakpoint sequences, are openly available via Zenodo (https://zenodo.org/records/10666885). Atoh1-FlpoER/+ and R26MASTR can be requested from A.L.J.; all other strains are available from the Jackson Laboratory.
Supplementary Materials
This PDF file includes:
Materials and Methods
Figs. S1 to S18
Tables S1 and S2
Legends for data S1 to S22
References
Other Supplementary Material for this manuscript includes the following:
Data S1 to S22
REFERENCES AND NOTES
- 1.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Anderson N. D., de Borja R., Young M. D., Fuligni F., Rosic A., Roberts N. D., Hajjar S., Layeghifard M., Novokmet A., Kowalski P. E., Anaka M., Davidson S., Zarrei M., Id Said B., Schreiner L. C., Marchand R., Sitter J., Gokgoz N., Brunga L., Graham G. T., Fullam A., Pillay N., Toretsky J. A., Yoshida A., Shibata T., Metzler M., Somers G. R., Scherer S. W., Flanagan A. M., Campbell P. J., Schiffman J. D., Shago M., Alexandrov L. B., Wunder J. S., Andrulis I. L., Malkin D., Behjati S., Shlien A., Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 361, eaam8419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henssen A. G., Henaff E., Jiang E., Eisenberg A. R., Carson J. R., Villasante C. M., Ray M., Still E., Burns M., Gandara J., Feschotte C., Mason C. E., Kentsis A., Genomic DNA transposition induced by human PGBD5. eLife 4, e10565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henssen A. G., Koche R., Zhuang J., Jiang E., Reed C., Eisenberg A., Still E., MacArthur I. C., Rodríguez-Fos E., Gonzalez S., Puiggròs M., Blackford A. N., Mason C. E., de Stanchina E., Gönen M., Emde A. K., Shah M., Arora K., Reeves C., Socci N. D., Perlman E., Antonescu C. R., Roberts C. W. M., Steen H., Mullen E., Jackson S. P., Torrents D., Weng Z., Armstrong S. A., Kentsis A., PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat. Genet. 49, 1005–1014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henssen A. G., Reed C., Jiang E., Garcia H. D., von Stebut J., MacArthur I. C., Hundsdoerfer P., Kim J. H., de Stanchina E., Kuwahara Y., Hosoi H., Ganem N. J., dela Cruz F., Kung A. L., Schulte J. H., Petrini J. H., Kentsis A., Therapeutic targeting of PGBD5-induced DNA repair dependency in pediatric solid tumors. Sci. Transl. Med. 9, eaam9078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helou L., Beauclair L., Dardente H., Arensburger P., Buisine N., Jaszczyszyn Y., Guillou F., Lecomte T., Kentsis A., Bigot Y., The C-terminal domain of piggyBac transposase is not required for DNA transposition. J. Mol. Biol. 433, 166805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helou L., Beauclair L., Dardente H., Piégu B., Tsakou-Ngouafo L., Lecomte T., Kentsis A., Pontarotti P., Bigot Y., The piggyBac-derived protein 5 (PGBD5) transposes both the closely and the distantly related piggyBac-like elements Tcr-pble and Ifp2. J. Mol. Biol. 433, 166839 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y. Bigot, M. Yamada, H. Mueller, V. Morell, S. Alves, T. Lecomte, A. Kentsis, Analysis of DNA transposition by DNA transposases in human cells. bioRxiv 2023.04.26.538406 [Preprint] (2023). 10.1101/2023.04.26.538406. [DOI]
- 9.Suero-Abreu G. A., Praveen Raju G., Aristizábal O., Volkova E., Wojcinski A., Houston E. J., Pham D., Szulc K. U., Colon D., Joyner A. L., Turnbull D. H., In vivo Mn-enhanced MRI for early tumor detection and growth rate analysis in a mouse medulloblastoma model. Neoplasia 16, 993–1006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Northcott P. A., Buchhalter I., Morrissy A. S., Hovestadt V., Weischenfeldt J., Ehrenberger T., Gröbner S., Segura-Wang M., Zichner T., Rudneva V. A., Warnatz H. J., Sidiropoulos N., Phillips A. H., Schumacher S., Kleinheinz K., Waszak S. M., Erkek S., Jones D. T. W., Worst B. C., Kool M., Zapatka M., Jäger N., Chavez L., Hutter B., Bieg M., Paramasivam N., Heinold M., Gu Z., Ishaque N., Jäger-Schmidt C., Imbusch C. D., Jugold A., Hübschmann D., Risch T., Amstislavskiy V., Gonzalez F. G. R., Weber U. D., Wolf S., Robinson G. W., Zhou X., Wu G., Finkelstein D., Liu Y., Cavalli F. M. G., Luu B., Ramaswamy V., Wu X., Koster J., Ryzhova M., Cho Y. J., Pomeroy S. L., Herold-Mende C., Schuhmann M., Ebinger M., Liau L. M., Mora J., McLendon R. E., Jabado N., Kumabe T., Chuah E., Ma Y., Moore R. A., Mungall A. J., Mungall K. L., Thiessen N., Tse K., Wong T., Jones S. J. M., Witt O., Milde T., von Deimling A., Capper D., Korshunov A., Yaspo M. L., Kriwacki R., Gajjar A., Zhang J., Beroukhim R., Fraenkel E., Korbel J. O., Brors B., Schlesner M., Eils R., Marra M. A., Pfister S. M., Taylor M. D., Lichter P., The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reilly K. M., The effects of genetic background of mouse models of cancer: Friend or foe? Cold Spring Harb. Protoc. 2016, pdb top076273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao J., Ligon K. L., Rakhlin E. Y., Thayer S. P., Bronson R. T., Rowitch D., McMahon A. P., A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 66, 10171–10178 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lao Z., Raju G. P., Bai C. B., Joyner A. L., MASTR: A technique for mosaic mutant analysis with spatial and temporal control of recombination using conditional floxed alleles in mice. Cell Rep. 2, 386–396 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan I. L., Wojcinski A., Rallapalli H., Lao Z., Sanghrajka R. M., Stephen D., Volkova E., Korshunov A., Remke M., Taylor M. D., Turnbull D. H., Joyner A. L., Lateral cerebellum is preferentially sensitive to high sonic hedgehog signaling and medulloblastoma formation. Proc. Natl. Acad. Sci. U.S.A. 115, 3392–3397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson P., Tong Y., Robinson G., Thompson M. C., Currle D. S., Eden C., Kranenburg T. A., Hogg T., Poppleton H., Martin J., Finkelstein D., Pounds S., Weiss A., Patay Z., Scoggins M., Ogg R., Pei Y., Yang Z. J., Brun S., Lee Y., Zindy F., Lindsey J. C., Taketo M. M., Boop F. A., Sanford R. A., Gajjar A., Clifford S. C., Roussel M. F., McKinnon P. J., Gutmann D. H., Ellison D. W., Wechsler-Reya R., Gilbertson R. J., Subtypes of medulloblastoma have distinct developmental origins. Nature 468, 1095–1099 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jessa S., Blanchet-Cohen A., Krug B., Vladoiu M., Coutelier M., Faury D., Poreau B., de Jay N., Hébert S., Monlong J., Farmer W. T., Donovan L. K., Hu Y., McConechy M. K., Cavalli F. M. G., Mikael L. G., Ellezam B., Richer M., Allaire A., Weil A. G., Atkinson J., Farmer J. P., Dudley R. W. R., Larouche V., Crevier L., Albrecht S., Filbin M. G., Sartelet H., Lutz P. E., Nagy C., Turecki G., Costantino S., Dirks P. B., Murai K. K., Bourque G., Ragoussis J., Garzia L., Taylor M. D., Jabado N., Kleinman C. L., Stalled developmental programs at the root of pediatric brain tumors. Nat. Genet. 51, 1702–1713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wechsler-Reya R. J., Scott M. P., Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22, 103–114 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Goodrich L. V., Milenkovic L., Higgins K. M., Scott M. P., Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Corrales J. D., Blaess S., Mahoney E. M., Joyner A. L., The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development 133, 1811–1821 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Flora A., Klisch T. J., Schuster G., Zoghbi H. Y., Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science 326, 1424–1427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliver T. G., Read T. A., Kessler J. D., Mehmeti A., Wells J. F., Huynh T. T. T., Lin S. M., Wechsler-Reya R. J., Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 132, 2425–2439 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Lumpkin E. A., Collisson T., Parab P., Omer-Abdalla A., Haeberle H., Chen P., Doetzlhofer A., White P., Groves A., Segil N., Johnson J. E., Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389–395 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Pavelitz T., Gray L. T., Padilla S. L., Bailey A. D., Weiner A. M., PGBD5: A neural-specific intron-containing piggyBac transposase domesticated over 500 million years ago and conserved from cephalochordates to humans. Mob. DNA 4, 23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henssen A. G., Jiang E., Zhuang J., Pinello L., Socci N. D., Koche R., Gonen M., Villasante C. M., Armstrong S. A., Bauer D. E., Weng Z., Kentsis A., Forward genetic screen of human transposase genomic rearrangements. BMC Genomics 17, 548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arora K., Shah M., Johnson M., Sanghvi R., Shelton J., Nagulapalli K., Oschwald D. M., Zody M. C., Germer S., Jobanputra V., Carter J., Robine N., Deep whole-genome sequencing of 3 cancer cell lines on 2 sequencing platforms. Sci. Rep. 9, 19123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexandrov L. B., Nik-Zainal S., Wedge D. C., Aparicio S. A. J. R., Behjati S., Biankin A. V., Bignell G. R., Bolli N., Borg A., Børresen-Dale A.-L., Boyault S., Burkhardt B., Butler A. P., Caldas C., Davies H. R., Desmedt C., Eils R., Eyfjörd J. E., Foekens J. A., Greaves M., Hosoda F., Hutter B., Ilicic T., Imbeaud S., Imielinski M., Jäger N., Jones D. T. W., Jones D., Knappskog S., Kool M., Lakhani S. R., López-Otín C., Martin S., Munshi N. C., Nakamura H., Northcott P. A., Pajic M., Papaemmanuil E., Paradiso A., Pearson J. V., Puente X. S., Raine K., Ramakrishna M., Richardson A. L., Richter J., Rosenstiel P., Schlesner M., Schumacher T. N., Span P. N., Teague J. W., Totoki Y., Tutt A. N. J., Valdés-Mas R., van Buuren M. M., van ‘t Veer L., Vincent-Salomon A., Waddell N., Yates L. R.; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain, Zucman-Rossi J., Futreal P. A., Dermott U. M., Lichter P., Meyerson M., Grimmond S. M., Siebert R., Campo E., Shibata T., Pfister S. M., Campbell P. J., Stratton M. R., Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skowron P., Farooq H., Cavalli F. M. G., Morrissy A. S., Ly M., Hendrikse L. D., Wang E. Y., Djambazian H., Zhu H., Mungall K. L., Trinh Q. M., Zheng T., Dai S., Stucklin A. S. G., Vladoiu M. C., Fong V., Holgado B. L., Nor C., Wu X., Abd-Rabbo D., Bérubé P., Wang Y. C., Luu B., Suarez R. A., Rastan A., Gillmor A. H., Lee J. J. Y., Zhang X. Y., Daniels C., Dirks P., Malkin D., Bouffet E., Tabori U., Loukides J., Doz F. P., Bourdeaut F., Delattre O. O., Masliah-Planchon J., Ayrault O., Kim S. K., Meyronet D., Grajkowska W. A., Carlotti C. G., de Torres C., Mora J., Eberhart C. G., van Meir E. G., Kumabe T., French P. J., Kros J. M., Jabado N., Lach B., Pollack I. F., Hamilton R. L., Rao A. A. N., Giannini C., Olson J. M., Bognár L., Klekner A., Zitterbart K., Phillips J. J., Thompson R. C., Cooper M. K., Rubin J. B., Liau L. M., Garami M., Hauser P., Li K. K. W., Ng H. K., Poon W. S., Yancey Gillespie G., Chan J. A., Jung S., McLendon R. E., Thompson E. M., Zagzag D., Vibhakar R., Ra Y. S., Garre M. L., Schüller U., Shofuda T., Faria C. C., López-Aguilar E., Zadeh G., Hui C. C., Ramaswamy V., Bailey S. D., Jones S. J., Mungall A. J., Moore R. A., Calarco J. A., Stein L. D., Bader G. D., Reimand J., Ragoussis J., Weiss W. A., Marra M. A., Suzuki H., Taylor M. D., The transcriptional landscape of Shh medulloblastoma. Nat. Commun. 12, 1749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhuang J., Weng Z., Local sequence assembly reveals a high-resolution profile of somatic structural variations in 97 cancer genomes. Nucleic Acids Res. 43, 8146–8156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pugh T. J., Weeraratne S. D., Archer T. C., Pomeranz Krummel D. A., Auclair D., Bochicchio J., Carneiro M. O., Carter S. L., Cibulskis K., Erlich R. L., Greulich H., Lawrence M. S., Lennon N. J., McKenna A., Meldrim J., Ramos A. H., Ross M. G., Russ C., Shefler E., Sivachenko A., Sogoloff B., Stojanov P., Tamayo P., Mesirov J. P., Amani V., Teider N., Sengupta S., Francois J. P., Northcott P. A., Taylor M. D., Yu F., Crabtree G. R., Kautzman A. G., Gabriel S. B., Getz G., Jäger N., Jones D. T. W., Lichter P., Pfister S. M., Roberts T. M., Meyerson M., Pomeroy S. L., Cho Y. J., Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kool M., Jones D. T., Jäger N., Northcott P. A., Pugh T. J., Hovestadt V., Piro R. M., Esparza L. A., Markant S. L., Remke M., Milde T., Bourdeaut F., Ryzhova M., Sturm D., Pfaff E., Stark S., Hutter S., Seker-Cin H., Johann P., Bender S., Schmidt C., Rausch T., Shih D., Reimand J., Sieber L., Wittmann A., Linke L., Witt H., Weber U. D., Zapatka M., König R., Beroukhim R., Bergthold G., van Sluis P., Volckmann R., Koster J., Versteeg R., Schmidt S., Wolf S., Lawerenz C., Bartholomae C. C., von Kalle C., Unterberg A., Herold-Mende C., Hofer S., Kulozik A. E., von Deimling A., Scheurlen W., Felsberg J., Reifenberger G., Hasselblatt M., Crawford J. R., Grant G. A., Jabado N., Perry A., Cowdrey C., Croul S., Zadeh G., Korbel J. O., Doz F., Delattre O., Bader G. D., McCabe M., Collins V. P., Kieran M. W., Cho Y. J., Pomeroy S. L., Witt O., Brors B., Taylor M. D., Schüller U., Korshunov A., Eils R., Wechsler-Reya R. J., Lichter P., Pfister S. M.; ICGC PedBrain Tumor Project , Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25, 393–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vladoiu M. C., el-Hamamy I., Donovan L. K., Farooq H., Holgado B. L., Sundaravadanam Y., Ramaswamy V., Hendrikse L. D., Kumar S., Mack S. C., Lee J. J. Y., Fong V., Juraschka K., Przelicki D., Michealraj A., Skowron P., Luu B., Suzuki H., Morrissy A. S., Cavalli F. M. G., Garzia L., Daniels C., Wu X., Qazi M. A., Singh S. K., Chan J. A., Marra M. A., Malkin D., Dirks P., Heisler L., Pugh T., Ng K., Notta F., Thompson E. M., Kleinman C. L., Joyner A. L., Jabado N., Stein L., Taylor M. D., Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato T., Sato N., Hayama S., Yamabuki T., Ito T., Miyamoto M., Kondo S., Nakamura Y., Daigo Y., Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 67, 8544–8553 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Foltz D. R., Jansen L. E. T., Bailey A. O., Yates J. R. III, Bassett E. A., Wood S., Black B. E., Cleveland D. W., Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunleavy E. M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G., HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Lutolf S., Radtke F., Aguet M., Suter U., Taylor V., Notch1 is required for neuronal and glial differentiation in the cerebellum. Development 129, 373–385 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Kitamura K., Yanazawa M., Sugiyama N., Miura H., Iizuka-Kogo A., Kusaka M., Omichi K., Suzuki R., Kato-Fukui Y., Kamiirisa K., Matsuo M., Kamijo S. I., Kasahara M., Yoshioka H., Ogata T., Fukuda T., Kondo I., Kato M., Dobyns W. B., Yokoyama M., Morohashi K. I., Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 32, 359–369 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Law A. L., Jalal S., Pallett T., Mosis F., Guni A., Brayford S., Yolland L., Marcotti S., Levitt J. A., Poland S. P., Rowe-Sampson M., Jandke A., Köchl R., Pula G., Ameer-Beg S. M., Stramer B. M., Krause M., Nance-Horan Syndrome-like 1 protein negatively regulates Scar/WAVE-Arp2/3 activity and inhibits lamellipodia stability and cell migration. Nat. Commun. 12, 5687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kentsis A., Frank S. A., Developmental mutators and early onset cancer. Front. Pediatr. 8, 189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov L. B., Kim J., Haradhvala N. J., Huang M. N., Ng A. W. T., Wu Y., Boot A., Covington K. R., Gordenin D. A., Bergstrom E. N., Islam S. M. A., Lopez-Bigas N., Klimczak L. J., McPherson J. R., Morganella S., Sabarinathan R., Wheeler D. A., Mustonen V.; PCAWG Mutational Signatures Working Group, Getz G., Rozen S. G., Stratton M. R.; PCAWG Consortium , The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degasperi A., Zou X., Amarante T. D., Martinez-Martinez A., Koh G. C. C., Dias J. M. L., Heskin L., Chmelova L., Rinaldi G., Wang V. Y. W., Nanda A. S., Bernstein A., Momen S. E., Young J., Perez-Gil D., Memari Y., Badja C., Shooter S., Czarnecki J., Brown M. A., Davies H. R.; Genomics England Research Consortium, Nik-Zainal S., Substitution mutational signatures in whole-genome–Sequenced cancers in the UK population. Science 376, science.abl.9283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadi K., Yao X., Behr J. M., Deshpande A., Xanthopoulakis C., Tian H., Kudman S., Rosiene J., Darmofal M., DeRose J., Mortensen R., Adney E. M., Shaiber A., Gajic Z., Sigouros M., Eng K., Wala J. A., Wrzeszczyński K. O., Arora K., Shah M., Emde A. K., Felice V., Frank M. O., Darnell R. B., Ghandi M., Huang F., Dewhurst S., Maciejowski J., de Lange T., Setton J., Riaz N., Reis-Filho J. S., Powell S., Knowles D. A., Reznik E., Mishra B., Beroukhim R., Zody M. C., Robine N., Oman K. M., Sanchez C. A., Kuhner M. K., Smith L. P., Galipeau P. C., Paulson T. G., Reid B. J., Li X., Wilkes D., Sboner A., Mosquera J. M., Elemento O., Imielinski M., Distinct classes of complex structural variation uncovered across thousands of cancer genome graphs. Cell 183, 197–210.e32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamayo-Orrego L., Gallo D., Racicot F., Bemmo A., Mohan S., Ho B., Salameh S., Hoang T., Jackson A. P., Brown G. W., Charron F., Sonic hedgehog accelerates DNA replication to cause replication stress promoting cancer initiation in medulloblastoma. Nat. Cancer 1, 840–854 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Beckermann T. M., Luo W., Wilson C. M., Veach R. A., Wilson M. H., Cognate restriction of transposition by piggyBac-like proteins. Nucleic Acids Res. 49, 8135–8144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolacsek O., Wachtl G., Fóthi Á., Schamberger A., Sándor S., Pergel E., Varga N., Raskó T., Izsvák Z., Apáti Á., Orbán T. I., Functional indications for transposase domestications—Characterization of the human piggyBac transposase derived (PGBD) activities. Gene 834, 146609 (2022). [DOI] [PubMed] [Google Scholar]
- 45.A. Simi, F. Ansaloni, D. Damiani, A. Codino, D. Mangoni, P. Lau, D. Vozzi, L. Pandolfini, R. Sanges, S. Gustincich, The Pgbd5 DNA transposase is required for mouse cerebral cortex development through DNA double-strand breaks formation. bioRxiv 2023.05.09.539730 [Preprint] (2023). 10.1101/2023.05.09.539730. [DOI]
- 46.L. J. Zapater, E. Rodriguez-Fos, M. Planas-Felix, S. Lewis, Daniel, Cameron, P. Demarest, A. Nabila, J. Zhao, P. Bergin, C. Reed, Makiko, Yamada, N. D. Socci, M. Hayes, R. Rabadan, D. Torrents, Michael C., Kruer, M. Toth, A. Kentsis, A transposase-derived gene required for human brain development. bioRxiv 2023.04.28.538770 [Preprint] (2023). 10.1101/2023.04.28.538770. [DOI]
- 47.Aziz R. K., Breitbart M., Edwards R. A., Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 38, 4207–4217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cosby R. L., Judd J., Zhang R., Zhong A., Garry N., Pritham E. J., Feschotte C., Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371, eabc6405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoshino M., Nakamura S., Mori K., Kawauchi T., Terao M., Nishimura Y. V., Fukuda A., Fuse T., Matsuo N., Sone M., Watanabe M., Bito H., Terashima T., Wright C. V. E., Kawaguchi Y., Nakao K., Nabeshima Y. I., Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron 47, 201–213 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Ellis T., Smyth I., Riley E., Graham S., Elliot K., Narang M., Kay G. F., Wicking C., Wainwright B., Patched 1 conditional null allele in mice. Genesis 36, 158–161 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Machold R., Fishell G., Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48, 17–24 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Jeong J., Mao J., Tenzen T., Kottmann A. H., McMahon A. P., Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937–951 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wojcinski A., Lawton A. K., Bayin N. S., Lao Z., Stephen D. N., Joyner A. L., Cerebellar granule cell replenishment postinjury by adaptive reprogramming of Nestin+ progenitors. Nat. Neurosci. 20, 1361–1370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakashima K., Umeshima H., Kengaku M., Cerebellar granule cells are predominantly generated by terminal symmetric divisions of granule cell precursors. Dev. Dyn. 244, 748–758 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Lee H. Y., Greene L. A., Mason C. A., Manzini M. C., Isolation and culture of post-natal mouse cerebellar granule neuron progenitor cells and neurons. J. Vis. Exp., 990 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maura F., Degasperi A., Nadeu F., Leongamornlert D., Davies H., Moore L., Royo R., Ziccheddu B., Puente X. S., Avet-Loiseau H., Campbell P. J., Nik-Zainal S., Campo E., Munshi N., Bolli N., A practical guide for mutational signature analysis in hematological malignancies. Nat. Commun. 10, 2969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rustad E. H., Yellapantula V., Leongamornlert D., Bolli N., Ledergor G., Nadeu F., Angelopoulos N., Dawson K. J., Mitchell T. J., Osborne R. J., Ziccheddu B., Carniti C., Montefusco V., Corradini P., Anderson K. C., Moreau P., Papaemmanuil E., Alexandrov L. B., Puente X. S., Campo E., Siebert R., Avet-Loiseau H., Landgren O., Munshi N., Campbell P. J., Maura F., Timing the initiation of multiple myeloma. Nat. Commun. 11, 1917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sondka Z., Bamford S., Cole C. G., Ward S. A., Dunham I., Forbes S. A., The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 18, 696–705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey T. L., Elkan C., Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994). [PubMed] [Google Scholar]
- 60.Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S., MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S., Quantifying similarity between motifs. Genome Biol. 8, R24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.I. Masilionis, O. Chaudhary, B. M. Urben, R. Chaligne, Nuclei extraction for 10x Genomics Single Cell Multiome ATAC + Gene Expression from frozen tissue using Singulator™ 100 or 200 (S2 Genomics) V2.0. protocols.io (2023).
- 63.Stuart T., Srivastava A., Madad S., Lareau C. A., Satija R., Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.L. McInnes, J. Healy, J. Melville, UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426 [stat.ML] (2018).
- 68.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P. R., Raychaudhuri S., Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li C., Liu B., Kang B., Liu Z., Liu Y., Chen C., Ren X., Zhang Z., SciBet as a portable and fast single cell type identifier. Nat. Commun. 11, 1818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan Y., Cahan P., SingleCellNet: A computational tool to classify single cell RNA-seq data across platforms and across species. Cell Syst. 9, 207–213.e2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aran D., Looney A. P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R. P., Wolters P. J., Abate A. R., Butte A. J., Bhattacharya M., Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jessa S., Mohammadnia A., Harutyunyan A. S., Hulswit M., Varadharajan S., Lakkis H., Kabir N., Bashardanesh Z., Hébert S., Faury D., Vladoiu M. C., Worme S., Coutelier M., Krug B., Faria Andrade A., Pathania M., Bajic A., Weil A. G., Ellezam B., Atkinson J., Dudley R. W. R., Farmer J. P., Perreault S., Garcia B. A., Larouche V., Blanchette M., Garzia L., Bhaduri A., Ligon K. L., Bandopadhayay P., Taylor M. D., Mack S. C., Jabado N., Kleinman C. L., K27M in canonical and noncanonical H3 variants occurs in distinct oligodendroglial cell lineages in brain midline gliomas. Nat. Genet. 54, 1865–1880 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Tickle, I. Tirosh, C. Georgescu, M. Brown, B. Haas, inferCNV of the Trinity CTAT Project. Klarman Cell Observatory, Broad Institute of MIT and Harvard, Cambridge, MA, USA (2019).
- 74.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Northcott P. A., Shih D. J. H., Peacock J., Garzia L., Sorana Morrissy A., Zichner T., Stütz A. M., Korshunov A., Reimand J., Schumacher S. E., Beroukhim R., Ellison D. W., Marshall C. R., Lionel A. C., Mack S., Dubuc A., Yao Y., Ramaswamy V., Luu B., Rolider A., Cavalli F. M. G., Wang X., Remke M., Wu X., Chiu R. Y. B., Chu A., Chuah E., Corbett R. D., Hoad G. R., Jackman S. D., Li Y., Lo A., Mungall K. L., Ming Nip K., Qian J. Q., Raymond A. G. J., Thiessen N., Varhol R. J., Birol I., Moore R. A., Mungall A. J., Holt R., Kawauchi D., Roussel M. F., Kool M., Jones D. T. W., Witt H., Fernandez-L A., Kenney A. M., Wechsler-Reya R. J., Dirks P., Aviv T., Grajkowska W. A., Perek-Polnik M., Haberler C. C., Delattre O., Reynaud S. S., Doz F. F., Pernet-Fattet S. S., Cho B. K., Kim S. K., Wang K. C., Scheurlen W., Eberhart C. G., Fèvre-Montange M., Jouvet A., Pollack I. F., Fan X., Muraszko K. M., Yancey Gillespie G., di Rocco C., Massimi L., Michiels E. M. C., Kloosterhof N. K., French P. J., Kros J. M., Olson J. M., Ellenbogen R. G., Zitterbart K., Kren L., Thompson R. C., Cooper M. K., Lach B., McLendon R. E., Bigner D. D., Fontebasso A., Albrecht S., Jabado N., Lindsey J. C., Bailey S., Gupta N., Weiss W. A., Bognár L., Klekner A., van Meter T. E., Kumabe T., Tominaga T., Elbabaa S. K., Leonard J. R., Rubin J. B., Liau L. M., van Meir E. G., Fouladi M., Nakamura H., Cinalli G., Garami M., Hauser P., Saad A. G., Iolascon A., Jung S., Carlotti C. G., Vibhakar R., Shin Ra Y., Robinson S., Zollo M., Faria C. C., Chan J. A., Levy M. L., Sorensen P. H. B., Meyerson M., Pomeroy S. L., Cho Y. J., Bader G. D., Tabori U., Hawkins C. E., Bouffet E., Scherer S. W., Rutka J. T., Malkin D., Clifford S. C., Jones S. J. M., Korbel J. O., Pfister S. M., Marra M. A., Taylor M. D., Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gilbertson R. J., Clifford S. C., MacMeekin W., Meekin W., Wright C., Perry R. H., Kelly P., Pearson A. D., Lunec J., Expression of the ErbB-neuregulin signaling network during human cerebellar development: Implications for the biology of medulloblastoma. Cancer Res. 58, 3932–3941 (1998). [PubMed] [Google Scholar]
- 80.Aldaregia J., Errarte P., Olazagoitia-Garmendia A., Gimeno M., Uriz J. J., Gershon T. R., Garcia I., Matheu A., Erbb4 is required for cerebellar developmentand malignant phenotype of medulloblastoma. Cancers 12, 997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu C., Yokota N., Gao Y., Yamamoto J., Tokuyama T., Namba H., Gene expression of growth signaling pathways is up-regulated in CD133-positive medulloblastoma cells. Oncol. Lett. 2, 357–361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fukushima H., Suzuki R., Yamaki Y., Hosaka S., Inaba M., Muroi A., Tsurubuchi T., Morii W., Noguchi E., Takada H., Cancer-predisposition genetic analysis in children with brain tumors treated at a single institution in Japan. Oncology 100, 163–172 (2022). [DOI] [PubMed] [Google Scholar]
- 83.Kloosterman W. P., Koster J., Molenaar J. J., Prevalence and clinical implications of chromothripsis in cancer genomes. Curr. Opin. Oncol. 26, 64–72 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Thomaz A., Jaeger M., Brunetto A. L., Brunetto A. T., Gregianin L., de Farias C. B., Ramaswamy V., Nör C., Taylor M. D., Roesler R., Neurotrophin signaling in medulloblastoma. Cancers 12, 2542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Northcott P. A., Jones D. T. W., Kool M., Robinson G. W., Gilbertson R. J., Cho Y. J., Pomeroy S. L., Korshunov A., Lichter P., Taylor M. D., Pfister S. M., Medulloblastomics: The end of the beginning. Nat. Rev. Cancer 12, 818–834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Northcott P. A., Lee C., Zichner T., Stütz A. M., Erkek S., Kawauchi D., Shih D. J. H., Hovestadt V., Zapatka M., Sturm D., Jones D. T. W., Kool M., Remke M., Cavalli F. M. G., Zuyderduyn S., Bader G. D., VandenBerg S., Esparza L. A., Ryzhova M., Wang W., Wittmann A., Stark S., Sieber L., Seker-Cin H., Linke L., Kratochwil F., Jäger N., Buchhalter I., Imbusch C. D., Zipprich G., Raeder B., Schmidt S., Diessl N., Wolf S., Wiemann S., Brors B., Lawerenz C., Eils J., Warnatz H. J., Risch T., Yaspo M. L., Weber U. D., Bartholomae C. C., von Kalle C., Turányi E., Hauser P., Sanden E., Darabi A., Siesjö P., Sterba J., Zitterbart K., Sumerauer D., van Sluis P., Versteeg R., Volckmann R., Koster J., Schuhmann M. U., Ebinger M., Grimes H. L., Robinson G. W., Gajjar A., Mynarek M., von Hoff K., Rutkowski S., Pietsch T., Scheurlen W., Felsberg J., Reifenberger G., Kulozik A. E., von Deimling A., Witt O., Eils R., Gilbertson R. J., Korshunov A., Taylor M. D., Lichter P., Korbel J. O., Wechsler-Reya R. J., Pfister S. M., Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511, 428–434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs. S1 to S18
Tables S1 and S2
Legends for data S1 to S22
References
Data S1 to S22