Abstract
The expression of human T-cell leukemia virus type 1 (HTLV-1) is activated by interaction of a viral transactivator protein, Tax, and cellular transcription factor, CREB (cyclic AMP response element binding protein), which bind to a 21-bp enhancer in the long terminal repeats (LTR). THP (Tax-helping protein) was previously determined to enhance the transactivation by Tax protein. Here we report novel forms of the human homolog of a member of the Gli oncogene family, Gli2 (also termed Gli2/THP), an extended form of a zinc finger protein, THP, which was described previously. Four possible isoforms (hGli2 α, β, γ, and δ) are formed by combinations of two independent alternative splicings, and all the isoforms could bind to a DNA motif, TRE2S, in the LTR. The longer isoforms, α and β, were abundantly expressed in various cell lines including HTLV-1-infected T-cell lines. Fusion proteins of the hGli2 isoforms with the DNA-binding domain of Gal4 activated transcription when the reporter contained a Gal4-binding site and one copy of the 21-bp sequence, to which CREB binds. This activation was observed only in the presence of Tax. The 21-bp sequence in the reporter was also essential for the activation. These results suggest that simultaneous binding of hGli2 and CREB to the respective sites in the reporter seems to be critical for Tax protein to activate transcription. Consequently, it is probable that the LTR can be regulated by two independent signals through hGli2 and CREB, since the LTR contains the 21-bp and TRE2S sequences in the vicinity.
Human T-cell leukemia virus type 1 (HTLV-1) (11, 21, 31) is the etiologic agent of adult T-cell leukemia (11, 32) and HTLV-1-associated myelopathy/tropical spastic paraparesis (10, 20). The expression of the HTLV-1 genome is strongly enhanced by its own product Tax through activation of transcription (2, 4, 6, 28). Tax binds directly to unphosphorylated cyclic AMP response element binding protein (CREB) (29, 34, 35) or CREM (29) and also binds to another transcription factor, CREB binding protein (CBP), forming CREB-Tax-CBP (19). In uninfected cells, the CREB-CBP complex is formed only when CBP is phosphorylated (3, 18). The CREB-Tax-CBP complex binds to the 21-bp sequence, which is repeated three times in the long terminal repeat (LTR) (24, 25) and activates transcription without any external signals (19). At least two copies of the 21-bp sequence are required for efficient activation by Tax; thus, transactivation of the LTR has been thought to be fully explained by the 21-bp sequence (7, 27).
One copy of the 21-bp enhancer, on the other hand, has been found to be insufficient for efficient activation (8, 27). However, we and others (1, 7) previously noted that some deletion mutants with mutations of the LTR that had only one copy of the 21-bp sequence were fully activated by Tax. These mutants had an additional DNA motif termed TRE2S adjacent to the 3′-proximal 21-bp sequence, and deletion of the sequence drastically diminished the activity in response to Tax activation (1, 30). Therefore, it was suggested that TRE2S binding protein is involved in Tax-mediated activation of transcription. We previously identified the zinc finger proteins THP-1 and THP-2, which bind to the TRE2S sequence (30).
The THP protein (30) has five zinc finger motifs at the N-terminal region, and the motifs are highly homologous to the Gli oncogene family, Gli1 and Gli3 (8, 22, 23). However, other structural features including the size were rather different from those of the other Gli family proteins. Therefore, we tried to isolate other possible clones that encode TRE2S binding protein. We report here four novel isoforms of the cDNAs, which are formed by a combination of two independent splicings. These clones contain sequences identical to two partial exon sequences of human Gli2 (hGli2) (22), but they show only 48 and 55% homology to hGli1 (16) and hGli3 (23), respectively. Furthermore, the longest cDNA can encode a protein with 80% homology to the mouse Gli2 (mGli2) oncogene product (14). Thus, we concluded that these isoforms are hGli2 isoforms, and we have called them hGli2 α, β, γ, and δ. These isoforms encode 133-, 131-, 88-, and 86-kDa proteins, respectively. They can bind to the TRE2S DNA sequence, and their Gal4 fusion proteins strongly activate Tax-dependent transcription when the reporter contains the Gal4 binding site and one copy of the 21-bp sequence. These observations suggest that simultaneous binding of hGli2 and CREB to the respective binding sites in the reporter enhances Tax-mediated transactivation.
MATERIALS AND METHODS
Cells, plasmids, and antibodies.
An HTLV-1-infected T-cell line, Hut102, was maintained in RPMI 1640 with 10% fetal calf serum. Cells of the human embryonic kidney cell line, 293T, and the human amnion cell line, FL, and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum.
A cDNA library of HTLV-1-infected Hut102 cells (33), a cDNA clone of THP (30), and the Tax expression vector pCG-Tax (9) was previously described. The entire coding sequence of each hGli2 isoform was inserted into pCG vector (30) under the control of the promoter of the cytomegalovirus early genes to construct pCG-hGli2 isoforms. Similarly, the same coding sequences were also inserted into the pCG-Gal4 cassette by using the sites for XbaI and BamHI or HindIII to express fusion proteins of hGli2 isoforms with the Gal4 DNA binding domain (positions 1 to 147).
Antiserum against Tax was raised against the C-terminal peptide as described previously (29). Antibodies against hGli2 were raised by inoculating a bacterially produced fragment of hGli2β (amino acids 1 to 506), which was fused to a six-histidine track and purified with a nickel column as described elsewhere (12, 29).
Isolation and sequencing of hGli2 cDNA clones.
A λgt11 cDNA library containing 2 × 106 phages, which was prepared from Hut102 (33), was screened with the coding sequence of THP-1 (30) as a probe. The cDNA inserts of positive clones were subcloned into Bluescript KS+ and sequenced by the dideoxy method (33). By using the 3′ fragment of the longest cDNA, the original library was further screened to isolate the cDNAs downstream of the mRNA. By overlapping these sequences, four possible full-sized cDNAs were constructed.
Reverse transcriptase PCR (RT-PCR) of cellular RNA.
cDNA was synthesized from the total cytoplasmic RNA isolated from Hut102 cells. The specific sequence was amplified by 30 cycles of PCR at 72°C for 2 min, 98°C for 0.5 min, and 55°C for 1 min, and the primers were a (positions 130 to 154 [with the ATG as +1]) and b (positions 284 to 308) to detect the first alternative splicing site and c (positions 2267 to 2291) and d (positions 3729 to 3753) to detect the second alternative splicing site. The products were analyzed by agarose gel electrophoresis.
Gel retardation assay.
The reaction mixture was incubated for 20 min at 25°C and then subjected to electrophoresis in 4% nondenaturing polyacrylamide gel as previously described (30). The 10-μl mixture contained nuclear extract (5 μg of protein), radiolabeled oligonucleotide probe (5 × 104 cpm; 1 ng), poly(dI-dC) (2.5 μg), 12 mM HEPES (pH 7.5), 0.6 mM EDTA, 0.6 mM dithiothreitol, 60 mM KCl, and 12% glycerol.
CAT assay.
Expression vectors for hGli2 or the fusion protein of Gal4-hGli2 (0.025, 0.1, or 1 μg) were cotransfected with either reporter plasmid, pTRE2S-21bp-CAT or pGal4-21bp-CAT, into FL cells by the calcium phosphate procedure (6). The Tax expression vector pCG-Tax (0.05 μg) was also included when specified. After culture for 40 h, the cells were harvested and subjected to a chloramphenicol acetyltransferase (CAT) assay as described previously (6). Under the conditions used, the activity was linearly proportional to the incubation time and the protein concentration. The CAT activity was expressed as the percent acetylation of chloramphenicol per 100 μg of protein in 30 min at 37°C or as the ratio to that with the reporter alone.
DNA affinity precipitation assay.
Cells were harvested, lysed in lysis buffer containing Nonidet P-40, and centrifuged at 1,000 rpm for 10 min. The pellet was washed with lysis buffer twice and used as the nuclear fraction. The nuclear proteins were extracted with 0.4 M sodium chloride as described previously (17). DNA probes with the TRE2S sequence were prepared by PCR with biotinylated primers. The nuclear extract and DNA probe were incubated at 25°C for 20 min, and DNA-protein complexes were isolated with strepavidin beads. The complexes were then directly analyzed by Western blotting (13, 29) with anti-Tax or anti-hGli2 antibodies.
Nucleotide sequence accession numbers.
The accession numbers for the sequences of the four isoforms are AB007295, AB007296, AB007297, and AB007298.
RESULTS
Structure of cDNA isoforms of hGli2.
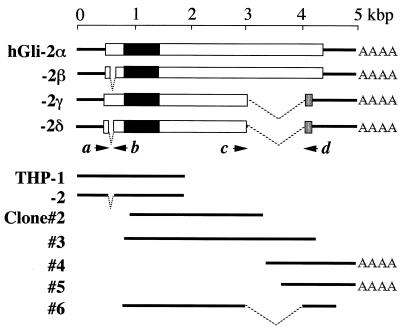
Previously, we reported a THP protein that binds to a Tax-responsive element (TRE2S) in the LTR (30). To isolate more cDNA clones related to THP, we further surveyed a cDNA library of Hut102 cells with THP-1 cDNA as a probe. The sequences of some positive clones, for example clones 2 and 3, revealed the presence of further extended sequence at the 3′ end of the overlapping sequence with THP (Fig. 1). However, none of them had a poly(A) stretch reflecting the poly(A) sequence at the 3′ end of the mRNA. To isolate clones with the sequence downstream, we used the 3′ fragment of clone 3 as a probe and isolated clones 4, 5, and 6, two of which had an A stretch at the 3′ end. By overlapping the sequences of these clones with the others, four possible cDNA isoforms were constructed. The clones thus constructed could be categorized into four subtypes (α, β, γ, and δ), which were generated by combinations of two independent splicings (summarized in Fig. 1). The sequence of the longest isoform is shown in Fig. 2A.
FIG. 1.
Construction of full-sized cDNAs of the hGli2 isoforms. Thick lines represent cDNA sequences, and boxes represent open reading frames. Solid and shaded regions in the boxes are the zinc finger motifs and amino acid sequences alternatively translated from different frames, respectively. Full-sized cDNA isoforms of hGli2 were derived from overlapping sequences of THP-1 and -2 previously reported (30) and clones 2 to 6 isolated in the present study.
FIG. 2.
(A) Nucleotide and amino acid sequence of hGli2α. The sequence between the solid triangles is deleted in β and δ isoforms by alternative splicing, and that between the open triangles is deleted in the γ and δ isoforms. (B) The second splicing induced a frameshift in translation, and the altered amino acid sequence is shown. The underline indicates five zinc finger motifs, and arrows with a, b, c, and d indicate the primers for RT-PCR.
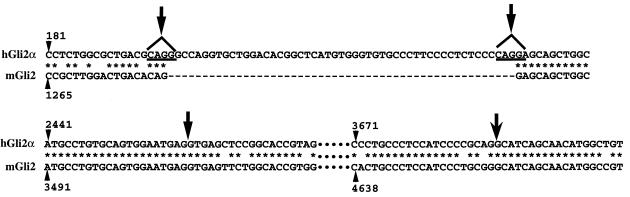
The longest clone consists of 4,961 bp and encodes 1,258 amino acids. Alternative splicings were detected at two sites: a 51-base deletion at the 5′ region of the coding frame in the β and δ isoforms, and a 1,231-base deletion at the 3′ region in the γ and δ isoforms (Fig. 1). The sites for the first splicing correspond to the identical sequences of CAGG; therefore, the site of the splicing could not be assigned accurately. The termini of the sequence between the splicing sites do not contain any sequence to follow the GT/AG rule for the intron, suggesting that the sequence may be an alternative exon in the α and γ forms (Fig. 3, top). The sites for the second splicing also correspond to repeated sequences; however, the splicing sites can be predicted so that GT/AG can be allocated at the termini (Fig. 3, bottom). Therefore, the sequence between the second splicing sites is probably an intron in the genome. By the second splicing, the reading frame is shifted into an alternative one, resulting in a different amino acid sequence in the C termini of the γ and δ isoforms. The altered amino acid sequence is shown in Fig. 2B.
FIG. 3.
Sequence comparison of hGli2 α with mGli2. Sequences around possible splicing sites are compared. The numbers with the arrowheads indicate the positions in hGli2α and mGli2 (14), respectively, and * indicates identical nucleotides. The sequence between the arrows in the top panel was deleted in the hGli2 β and γ isoforms, and that in the lower panel was deleted in the hGli2 γ and δ isoforms.
The nucleotide sequences of these isoforms contained sequences identical to two genomic fragments of hGli2 (22), indicating that these isoforms are the hGli2. On the other hand, the amino acid sequence encoded by the α isoform is 80% homologous to mGli2 (14). Furthermore, the nucleotide sequence of the zinc finger motifs was identical to that of mGli2 except for 1 of 159 bases (14). The amino acid sequence of the α form, on the other hand, showed 48 and 55% homology to hGli1 (16) and hGli3 (23), respectively, although the nucleotide sequence of the zinc fingers showed 86 and 93% homology to hGli1 and hGli3, respectively (16, 23) (Fig. 3). Therefore, it was concluded that these novel isoforms correspond to hGli2.
Comparison of these novel forms of hGli2 and mGli2 indicates that mGli2, previously published by Hughs et al. (14), might correspond to a β form of mGli2 (Fig. 3). On the other hand, the 5′ half of hGli2 was identical to the previously described THP (30), except for two deletions at position 1277 and 1294. Careful analysis of the sequence of THP clones showed that the clone originally sequenced had deletions at two sites described above; thus, the sequence previously described (30) was an artifact at these two sites. The corrected sequence allowed extension of the translation further downstream. The Gli oncogene family was originally identified as an oncogene amplified in human glioblastoma cells (16) and characterized as a transcription factor that binds to DNA through zinc finger motifs (14, 15).
DNA binding of hGli2 isoforms.
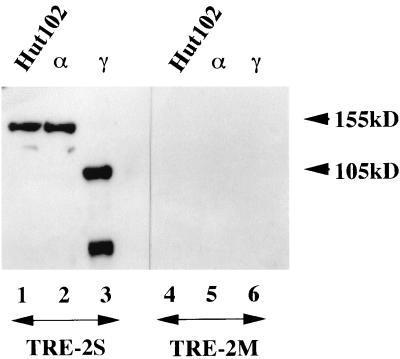
To confirm the DNA binding activity of hGli2, expression vectors for α and γ isoforms were transfected into 293T cells and the nuclear extracts were examined by a DNA affinity precipitation assay. In a parallel experiment, a nuclear extract of Hut102 cells was similarly assayed to determine which isoform is the major species in DNA binding in HTLV-1-infected T cells. The double-stranded DNA probe for these assays was TRE2S containing biotinylated nucleotides at both 5′ termini. Each nuclear extract was incubated with the DNA probe, and the DNA-protein complexes were isolated with streptavidin beads. The isolated complexes were analyzed by Western blotting with antibodies against hGli2 (Fig. 4). Both hGli2α and hGli2γ were shown to bind to TRE2S DNA (lanes 2 and 3) but not to a mutant probe (lanes 5 and 6), clearly demonstrating that the hGli2 isoforms are proteins that bind to a specific DNA sequence. A faster-migrating band detected in lane 3 might be a degraded fragment of hGli2γ containing the zinc finger motifs, which is thought to be the DNA binding domain of hGli2.
FIG. 4.
Binding of the hGli2 isoforms to the TRE2S DNA sequence and of the endogenous proteins in HTLV-1-infected cells. Expression vectors, pCG-hGli2 α (lanes 2 and 5) and γ (lanes 3 and 6), were separately transfected into 293T cells, and the nuclear extracts were incubated with TRE2S DNA (lanes 1 to 3) or TRE2M, an inactive mutant (lanes 4 to 6), which were labeled with biotin. The DNA-protein complexes were isolated with avidin beads and analyzed by Western blotting with antiserum against hGli2. A nuclear extract from Hut102 cells, an HTLV-1-infected T-cell line, was also analyzed similarly (lanes 1 and 4). TRE2S, CCGGGAAGCCACCGGGAACCACCCA; TRE2M, CCGGGAAGCCACCGGGAACAAATTA.
In these assays, we included a Tax expression plasmid, pCG-Tax, to examine whether Tax protein can bind to the hGli2-TRE2S complex. However, we could not see a significant binding of Tax to the hGli2-TRE2S complex, although we detected a very weak binding only when a large excess of pCG-Tax was used (data not shown). Therefore, Tax does not bind effectively to hGli2 protein on the TRE2S DNA sequence.
A similar assay with a nuclear extract of HTLV-1-infected Hut102 cells gave a single band migrating with the same mobility as hGli2 isoform α (Fig. 4, lane 1). Therefore, it is concluded that the endogenous Gli2 α and β isoforms are the major components of hGli2 to bind to TRE2S in vivo.
Expression of hGli2 isoforms in cell lines.
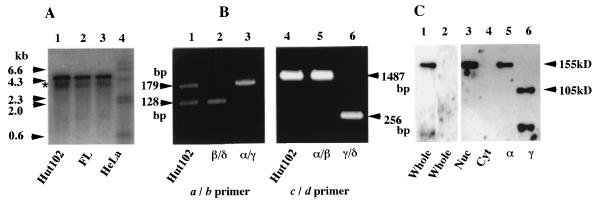
The sequences of the hGli2 isoforms predicted the sizes of mRNA as 5.1 kb for the α and β isoforms and 3.9 kb for the γ and δ isoforms, including poly(A). To examine which species are expressed in cell lines, Northern blot analysis was carried out. As shown in Fig. 5A, a major band of 5.1 kb was detected in Hut102 cells, a T-cell line infected with HTLV-1 (lane 1), FL cells, a human amnion cell line (lane 2), and HeLa cells, a human epithelial cell line (lane 3). A faint band just below the major band was also detected in all the cell lines (Fig. 5A). These observations may indicate that hGli2 α and/or β was the major species expressed in various types of cell lines and that the γ and/or δ isoforms were minor.
FIG. 5.
Expression of hGli2 isoforms at the mRNA and protein levels. (A) Northern blot of poly(A)+ RNA from Hut102 (lane 1), FL (lane 2), and HeLa (lane 3) cells with the coding sequence of hGli2 α as a probe. A HindIII digest of λ phage DNA is shown as a molecular size marker (lane 4). (B) Detection of the endogenous Gli2 isoforms expressed in Hut102 cells. Total RNA of Hut102 cells was used as the template for reverse transcription with random primers, and the cDNA sequences obtained were amplified with two sets of primers, A and B (lane 1) or C and D (lane 4), which encompass each splicing site (Fig. 1 and 2). As the standards, hGli2 α (lanes 2 and 5), β (lane 3) and γ (lane 6) were also amplified with the same primers. Fragments of 179 and 128 bp are from RNA without and with splicing at the first site, respectively, and fragments of 1,487 and 256 bp are those from RNA without and with splicing at the second site, respectively. (C) Identification of protein isoforms of hGli2 in Hut102 cells by Western blot analysis. Lanes: 1 and 2, whole-cell extract; 3, nuclear fraction; 4, cytoplasmic fraction; 5, hGli2 α; 6, hGli2 γ. Antiserum against hGli2 was used for lanes 1 and 3 to 6, and preimmune serum was used for lane 2.
To distinguish the α and β or the γ and δ isoforms, we carried out reverse RT-PCR. RNA isolated from Hut102, an HTLV-1-infected T-cell line, was converted into cDNA with RT and the cDNA was amplified with primers a and b or c and d as indicated in Fig. 1 and 2. The sequence amplified with primers a and b encompasses the first splicing site, and the α and γ and the β and δ isoforms would produce 179- and 128-bp DNA fragments, respectively. These fragments, amplified with primers c and d, encompass the second splicing site, and the α and β and the γ and δ isoforms would produce 1,487- and 256-bp DNA fragments, respectively (Fig. 1). The results shown in Fig. 5B demonstrate that primers a and b gave two bands expected from the α and β isoforms with almost equal intensity (lane 1), clearly indicating an equal expression of the α and γ and the β and δ isoforms. On the other hand, primers c and d gave a very dominant band of 1,487-bp DNA (lane 4) but extremely low levels of the 256-bp DNA band, demonstrating that the vast majority of hGli2 mRNA expressed in Hut102 cells is made up of α and β isoforms and that there is only a very minor fraction of γ and δ isoforms. A similar assay was also performed with RNA from another HTLV-1-infected cell line, MT2, and almost identical results were obtained (data not shown), suggesting that a dominant expression of the α and β isoforms of hGli2 might be a general feature in HTLV-1-infected T-cell lines.
The expression of hGli2 isoform in Hut102 cells was also confirmed at the protein level (Fig. 5C). Western blot analysis detected a major band with a molecular mass of 155 kDa with antibodies against the N-terminal region (amino acids 1 to 506), but not with a preimmune serum (Fig. 5C, lanes 1 and 2). The size of the band was similar to that of the α isoform (lane 5). When the nuclear and cytoplasmic fractions were separated by brief centrifugation, the band was detected only in the pellet defined as a nuclear fraction (lanes 3 and 4). In the pellet, severe contamination of undisrupted cells was excluded by microscopic examination. Therefore, the α and β isoforms of hGli2 are expressed almost exclusively in the nucleus.
Enhancement of Tax-dependent transactivation by hGli2.
The TRE2S sequence was identified in the LTR as a cis element to enhance Tax-dependent transactivation (30). To examine whether the TRE2S binding protein hGli2 is able to enhance Tax-dependent transactivation, the expression vector for hGli2 α and a reporter plasmid were transfected into FL cells in the presence or absence of the Tax-expressing vector. We used FL cells for these assays simply because these cells took up DNA efficiently in our transfection assay. The reporter gene CAT was under the control of one copy each of TRE2S and the 21-bp sequence. However, the reporter plasmid alone was almost fully activated by Tax as previously described (30). Overexpression of hGli2 α by exogenous plasmid induced significant suppression of CAT expression (Table 1).
TABLE 1.
Enhancement of Tax-dependent transactivation by hGli2 isoformsa
| Effector | CAT activityb with reporter:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| TRE2S-21bp-CAT
|
Gal4-21bp-CAT
|
Gal4-CAT
|
21bp-CAT
|
|||||
| −Tax | +Tax | −Tax | +Tax | −Tax | +Tax | −Tax | +Tax | |
| None | 1.8 | 128.0 | 1.2 | 11.7 | ||||
| hGli2 α | 1.5 | 41.2 | ||||||
| hGli2 β | 1.6 | 58.7 | 1.1 | 2.8 | ||||
| hGli2 γ | 1.8 | 38.9 | ||||||
| hGli2 δ | 1.6 | 31.6 | 1.2 | 4.8 | ||||
| None | 1.2 | 10.7 | 0.8 | 1.2 | 1.4 | 14.2 | ||
| Gal4 | 1.2 | 8.1 | 1.0 | 0.8 | 1.8 | 13.2 | ||
| Gal4-hGli2 α | 2.5 | 74.6 | 0.6 | 1.4 | 2.4 | 12.1 | ||
| Gal4-hGli2 β | 1.8 | 82.6 | ||||||
| Gal4-hGli2 γ | 2.8 | 87.2 | ||||||
| Gal4-hGli2 δ | 1.8 | 88.8 | ||||||
Each expression vector, pCG-hGli2 isoform (0.1 μg) or pCG-Gal4-hGli2 isoform (0.05 μg), was transfected into FL cells together with the respective reporter plasmid (0.05 μg) with or without pCG-Tax (0.1 μg), and CAT activity was assayed on day 2.
CAT activity is expressed as percent acetylation of chloramphenicol per 100 μg of protein in 30 min at 37°C. Bold type indicates significantly activated values by Tax.
To avoid the effects of endogenous Gli2 α and β, we used a fusion protein of hGli2 α with the DNA binding domain of Gal4 protein (Table 1). The reporter CAT construct contained five copies of the Gal4 binding site instead of TRE2S and one copy of the 21-bp sequence. In the presence of Tax, the Gal4-hGli2 α fusion protein activated CAT expression very efficiently; however, in the absence of Tax, it did not affect CAT expression significantly (Table 1). Tax alone induced a six- to eightfold enhancement of CAT expression but was not sufficient for full activation. Furthermore, the activation by Gal4-hGli2 α was dose dependent, as shown in Fig. 6. When a reporter did not contain the Gal4-binding site, no such activation was observed (Table 1). From these observations, it was concluded that DNA binding of the hGli2 α fusion protein through the Gal4 binding site enhanced Tax-dependent transactivation. All the other isoforms of hGli2 also showed similar enhancement of Tax-dependent activation (Table 1).
FIG. 6.
Dose-dependent activation of CAT expression by pCG-Gal4-hGli2 α. The reporter plasmid, pGal4-21bp-CAT, contained the Gal4 binding site and one copy of the 21-bp sequence. The assay was carried out similarly to that described for Table 1.
DISCUSSION
In this study, we identified four novel isoforms of hGli2 α, β, γ, and δ, that bind to a Tax response element, TRE2S, in the LTR of HTLV-1. The novel isoforms had the same sequence at the 5′ region as THP previously described (30), except for two deletions, which allowed further extension of the translation. Careful reexamination indicated that the sequence of the previous THP was a cloning artifact. hGli2 α showed 80% homology of amino acid sequence to mGli2 (14). The isoforms were formed by combinations of two independent alternative splicings in the coding sequence, and the larger isoforms, α and β, were abundantly expressed in HTLV-1-infected T-cell lines and also in other cell lines so far tested. All these isoforms were found to bind to TRE2S with conservation of the zinc finger motifs. Comparing the sequence of mGli2 reported by Hughes et al. (14) with the isoforms of hGli2 described in this paper, mGli2 appeared to correspond to the β form. It underwent splicing at the first site but not at the second site, although the site showed very high homology, as shown in Fig. 3.
Fusion proteins of hGli2 with the DNA binding domain of Gal4 enhanced the Tax-dependent activation of gene expression when the reporter contained the Gal4 binding site and the 21-bp sequence. From these observations, we concluded that binding of hGli2 to the TRE2S sequence enhanced Tax-dependent transactivation of transcription. However, the free form of hGli2 did not enhance but, instead, rather efficiently inhibited Tax-dependent transcription, probably because the endogenous Gli2 gene is abundantly expressed in the cell line used and the expression of the reporter gene had been activated by Tax alone at the maximum level. The mechanism of this inhibition is not well understood, but two possibilities can be assumed: (i) overexpressed hGli2 is squelching Tax-mediated transactivation, or (ii) it should be modified to be active, but, it might not be sufficiently modified in transfected cells.
A unique property of the enhancement was that binding of hGli2 alone to TRE2S had almost no effect on transcription. Two additional factors were required for the enhancement; these were the 21-bp sequence as a cis element and Tax protein as a trans-acting factor. These requirements for transcriptional activation suggest that hGli2 might function through a unique mechanism, although the mechanism is not well understood. A possible mechanism, however, could be as follows: hGli2 and CREB bind to each cis element in the vicinity and interact with each other on the DNA. Such an interaction could be enhanced by the Tax protein, since Tax binds to CREB to activate the enhancer activity of the 21-bp repeats (29, 35). A putative interaction of hGli2 and CREB might be supported by the previous observations on YY1, a human Gli-Kruppel-related protein (26). YY1 interacts with CREB/ATF and exerts repression of specific transcription (36). Furthermore, the repression exerted by YY1 is relieved by adenovirus E1A (26). Another factor, UCRBP, a zinc finger protein of the Gli family, bound to the upstream conserved region of the murine leukemia virus LTR and down-regulated the MuLV promoter activity (5). These observations on other factors related to Gli proteins allow speculation that hGli2 is a transcriptional repressor and that HTLV-1 Tax can prevent the repression. However, this mechanism seems unlikely, because hGli2 did not show any inhibitory activity on transcription in the absence of Tax.
Whatever the mechanism, the presence of the TRE2S sequence adjacent to the 3′ proximal 21-bp sequence in the LTR (1, 30) and our observations reported in this paper suggest that the binding of hGli2 and CREB to TRE2S and 21-bp sequences, respectively, in the LTR would be an alternative pathway to augment the capacity of Tax to transactivate viral gene expression.
ACKNOWLEDGMENT
This work was supported in part by a special grant for Advanced Research on Cancer from the Ministry of Education, Culture and Science of Japan.
REFERENCES
- 1.Brady J N, Jeang K-T, Duvall J, Khoury G. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J Virol. 1987;61:2175–2181. doi: 10.1128/jvi.61.7.2175-2181.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen I S Y, Cann A J, Shah N P, Gaynor R B. Functional relation between HTLV-2 x and adenovirus I1A proteins in transcriptional activation. Science. 1985;230:570–573. doi: 10.1126/science.2996140. [DOI] [PubMed] [Google Scholar]
- 3.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 4.Felber B K, Paskalis H, Kleinman-Ewing D, Wong-Staal F, Pavlakis G N. The pX protein of HTLV-1 is a transcriptional activator of its long terminal repeats. Science. 1985;229:675–679. doi: 10.1126/science.2992082. [DOI] [PubMed] [Google Scholar]
- 5.Flanagan J R, Becker K G, Ennist D L, Gleason S L, Driggers P H, Levi B-Z, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol Cell Biol. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujisawa J, Seiki M, Kiyokawa T, Yoshida M. Functional activation of long terminal repeat of human T-cell leukemia virus type I by trans-activator. Proc Natl Acad Sci USA. 1985;82:2277–2281. doi: 10.1073/pnas.82.8.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujisawa J, Seiki M, Sato M, Yoshida M. A transcriptional sequence of HTLV-1 is responsible for trans-activation mediated by p40x of HTLV-1. EMBO J. 1986;5:713–718. doi: 10.1002/j.1460-2075.1986.tb04272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujisawa J, Toita M, Yoshida M. A unique sequence element for the trans-activator (p40tax) of human T-cell leukemia virus type I from cyclic AMP- and TPA-responsive elements. J Virol. 1989;63:3234–3239. doi: 10.1128/jvi.63.8.3234-3239.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujisawa J, Toita M, Yoshimura T, Yoshida M. The indirect association of human T-cell leukemia virus tax fusion protein with DNA results in transcriptional activation. J Virol. 1991;65:4525–4528. doi: 10.1128/jvi.65.8.4525-4528.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T lymphotropic virus type 1 in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 11.Hinuma Y, Nagata K, Misaka M, Nakai M, Matsumoto T, Kinoshita K, Shirakawa S, Miyoshi I. Adult T cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981;78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai H, Suzuki T, Fujisawa J, Inoue J, Yoshida M. Tax protein of human T-cell leukemia virus type I binds to the ankyrin motifs of inhibitory factor kappa B and induces nuclear translocation of transcription factor NF-κB proteins for transcriptional activation. Proc Natl Acad Sci USA. 1994;91:3584–3588. doi: 10.1073/pnas.91.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirai H, Fujisawa J, Suzuki T, Ueda K, Muramatsu M, Tsuboi A, Arai N, Yoshida M. Trans-activator Tax of HTLV-1 binds to the NF-κB precursor p105. Oncogene. 1992;7:1737–1742. [PubMed] [Google Scholar]
- 14.Hughes D C, Allen J, Morley G, Sutherland K, Ahmed W, Prosser J, Lettice L, Allen G, Mattei M-G, Farrall M, Hill R E. Cloning and sequencing of the mouse Gli2 gene: localization to the dominant hemimelia critical region. Genomics. 1997;39:205–215. doi: 10.1006/geno.1996.4468. [DOI] [PubMed] [Google Scholar]
- 15.Kinzler K W, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinzler K W, Ruppert J M, Bigner S H, Vogelstein B. The GLI gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 17.Kiyokawa T, Seiki M, Iwashita S, Imagawa K, Shimizu F, Yoshida M. P27x-III and p21x-III, proteins encoded by the pX sequence of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1985;82:8359–8363. doi: 10.1073/pnas.82.24.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok R P S, Lundblad J R, Chrivia J C, Richard J P, Bächinger H P, Brennan R G, Roberts S G E, Green M, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 19.Kwok R P S, Laurence M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriott S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 20.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-1 associated myelopathy, a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 21.Poiesz B J, Ruscetti R W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruppert J M, Kinzler K W, Wong A J, Bingner S H, Kao F-T, Law M L, Seuanes H N, O’Brien S J, Vogelstein B. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruppert J M, Vogelstein B, Arheden K, Kinzler K W. GLI3 encodes a 190-kilodalton protein with multiple regions of GLI similarity. Mol Cell Biol. 1990;10:5408–5415. doi: 10.1128/mcb.10.10.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seiki M, Hattori S, Yoshida M. Human adult T-cell leukemia virus: Molecular cloning of the provirus DNA and the unique terminal structure. Proc Natl Acad Sci USA. 1982;79:6899–6902. doi: 10.1073/pnas.79.22.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 27.Shimotohno K, Takano M, Teruuchi T, Miwa M. Requirement of multiple copies of a 21-nucleotide sequence in the U3 regions of human T-cell leukemia virus type I and type II long terminal repeats for trans-acting activation of transcription. Proc Natl Acad Sci USA. 1986;83:8112–8116. doi: 10.1073/pnas.83.21.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sodroski J G, Rosen C A, Goh W C, Haseltine W A. Trans-acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science. 1984;226:177–179. doi: 10.1126/science.6330891. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, Fujisawa J, Toita M, Yoshida M. A trans-activator Tax of human T-cell leukemia virus type 1 (HTLV-1) interacts with AP-responsive element (CRE) binding and CRE modulator proteins that bind to the 21-base-pair sequence of HTLV-1. Proc Natl Acad Sci USA. 1993;90:610–614. doi: 10.1073/pnas.90.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanimura A, Teshima H, Fujisawa J, Yoshida M. A new regulatory element that augments the Tax-dependent sequence of human T-cell leukemia virus type 1 and cloning of cDNAs encoding its binding protein. J Virol. 1993;67:5375–5382. doi: 10.1128/jvi.67.9.5375-5382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida M, Seiki M, Yamaguchi K, Takatsuki K. Monoclonal integration of HTLV in all primary tumors of adult T-cell leukemia suggests causative role of HTLV in the disease. Proc Natl Acad Sci USA. 1984;81:2534–2537. doi: 10.1073/pnas.81.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimura T, Fujisawa J, Yoshida M. Multiple cDNA clones encoding nuclear proteins that bind to the Tax-dependent sequence of HTLV-1: all contain a leucine zipper structure and basic amino acid domain. EMBO J. 1990;9:2537–2542. doi: 10.1002/j.1460-2075.1990.tb07434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, Giam C. Interaction of the human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-1 enhancer. Proc Natl Acad Sci USA. 1991;88:11445–11449. doi: 10.1073/pnas.88.24.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L, Giam C. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc Natl Acad Sci USA. 1992;89:7070–7074. doi: 10.1073/pnas.89.15.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Q, Gedrich R W, Engel D A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]