Abstract
Background.
Among all biopsies in the Trifecta-Kidney Study (ClinicalTrials.gov NCT04239703), elevated plasma donor-derived cell-free DNA (dd-cfDNA) correlated most strongly with molecular antibody–mediated rejection (AMR) but was also elevated in other states: T cell–mediated rejection (TCMR), acute kidney injury (AKI), and some apparently normal biopsies. The present study aimed to define the molecular correlates of plasma dd-cfDNA within specific states.
Methods.
Dd-cfDNA was measured by the Prospera test. Molecular rejection and injury states were defined using the Molecular Microscope system. We studied the correlation between dd-cfDNA and the expression of genes, transcript sets, and classifier scores within specific disease states, and compared AMR, TCMR, and AKI to biopsies classified as normal and no injury (NRNI).
Results.
In all 604 biopsies, dd-cfDNA was elevated in AMR, TCMR, and AKI. Within AMR biopsies, dd-cfDNA correlated with AMR activity and stage. Within AKI, the correlations reflected acute parenchymal injury, including cell cycling. Within biopsies classified as MMDx Normal and archetypal No injury (NRNI), dd-cfDNA still correlated significantly with rejection- and injury-related genes. TCMR activity (eg, the TCMRProb classifier) correlated with dd-cfDNA, but within TCMR biopsies, top gene correlations were complex and not the top TCMR-selective genes.
Conclusions.
In kidney transplants, elevated plasma dd-cfDNA is associated with 3 distinct molecular states in the donor tissue: AMR, recent parenchymal injury (including cell cycling), and TCMR, potentially complicated by parenchymal disruption. Moreover, subtle rejection- and injury-related changes in the donor tissue can contribute to dd-cfDNA elevations in transplants considered to have no rejection or injury.
INTRODUCTION
Plasma donor-derived cell-free DNA (dd-cfDNA) levels are widely used to screen for rejection in organ transplants.1-15 We previously explored the relationship between dd-cfDNA and the molecular state in 300 kidney transplant indication biopsies in the Trifecta-Kidney Study (ClinicalTrials.gov NCT04239703) to define the relationship between the dd-cfDNA level (taken at the time of indication biopsies) and the rejection and injury states in the biopsy as defined by the Molecular Microscope Diagnostic System (MMDx).16,17 The strongest population-wide correlations were with molecular antibody–mediated rejection (AMR)-related processes, but dd-cfDNA levels were also elevated in T cell–mediated rejection (TCMR) and acute kidney injury (AKI). This could be because TCMR and AKI release dd-cfDNA, or because of subtle subthreshold AMR-like activity (defined by AMRProb classifier score and expression of transcripts selective for AMR), which is found in some biopsies previously considered to have no rejection.18,19 Moreover, some biopsies with no rejection or injury also had unexplained elevations of dd-cfDNA, which could be “false positives” but could also reflect other disease states.
The present study sought to define the underlying molecular associations of dd-cfDNA within specific disease and injury states, namely AMR, TCMR, and AKI, and to clarify the interpretation of elevated dd-cfDNA in kidneys classified as normal and no injury (NRNI). Discovering the molecular associations of dd-cfDNA within specific disease states is critical to optimizing the diagnostic applications of dd-cfDNA, as well as its use to track the effect of treatment. Moreover, we hoped that the correlations of dd-cfDNA with the molecular changes in NRNI biopsies would clarify whether these apparent false positives could be safely ignored. We examined 604 indication biopsies from the Trifecta-Kidney Study assessed by both MMDx genome-wide analyses and plasma dd-cfDNA measured by the Prospera test. We examined molecules expressed within the biopsies grouped by their disease states for their correlations with plasma dd-cfDNA elevation. We were particularly interested in whether subtle subthreshold AMR-related activity could contribute to some apparent false-positive dd-cfDNA tests in kidneys with NRNI.
MATERIALS AND METHODS
Population
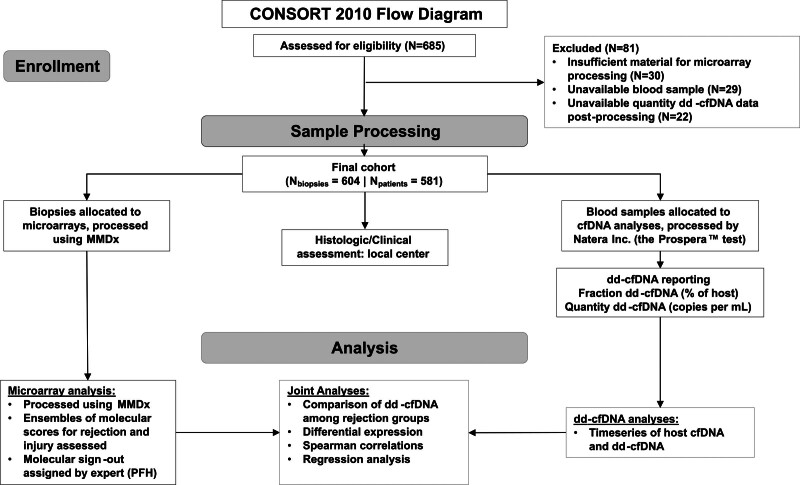
The Trifecta-Kidney Study (ClinicalTrials.gov NCT04239703) is a prospective multicenter study of consenting patients involving 68 investigators from 30 transplant institutions (Table S1, SDC, http://links.lww.com/TP/C935) under local institutional review board-approved protocols.16,20,21 Blood for dd-cfDNA was always taken before the biopsy to avoid detecting dd-cfDNA released by the biopsy collection procedure. MMDx assessments and dd-cfDNA measurements were blinded to each other and both were blinded to the biopsy’s histology and clinical data. The study design is shown in Figure 1.
FIGURE 1.
CONSORT 2010 flow diagram and the study design. CONSORT, Consolidated Standards of Reporting Trials; dd-cfDNA, donor-derived cell-free DNA; MMDx, Molecular Microscope Diagnostic System; PFH, Philip F. Halloran.
Biopsy Collection
Six hundred and four kidney transplant biopsies collected from 598 patients in the Trifecta-Kidney Study were processed for MMDx and had estimated fraction and quantity dd-cfDNA. This study adheres to the Declaration of Helsinki. Biopsies were collected with informed consent per the institutional review board at each local center and approved in Edmonton by the University of Alberta (No. Pro00022226). The clinical and research activities being reported are consistent with the Principles of the Declaration of Istanbul as outlined in the “Declaration of Istanbul on Organ Trafficking and Transplant Tourism.”
Clinical Diagnostics
Of the 604 biopsies, 545 had available local histological diagnoses assigned by the center. Histological and clinical data and donor-specific antibody status were collected at each center per standard-of-care as approved by institutional review boards as per the published Trifecta-Kidney protocol. The histology findings were recorded by local standard-of-care assessment following Banff 2019 guidelines. Histology diagnoses were interpreted as “no rejection (NR),” “AMR,” “possible AMR,” “TCMR,” “possible TCMR,” and “mixed rejection” with no knowledge of MMDx or dd-cfDNA results. There was general agreement between MMDx and histology, but the dd-cfDNA results have higher agreement with MMDx rejection.16,22 The present work focused on MMDx assessments.
dd-cfDNA Analysis
Blood samples were collected before biopsy and sent to Natera, Inc., (Austin, TX) for central dd-cfDNA assessment using the Prospera test.8 Estimated dd-cfDNA quantity (genomic copies per mL [cp/mL]) and fraction (percentage of total cfDNA) were reported (SDC, http://links.lww.com/TP/C935).
Molecular Diagnoses of Rejection and Injury
Biopsies were diagnosed by their MMDx signouts and automated archetypal assignments of rejection and injury.17,23-29 Biopsies with no molecular rejection taken within 6-wk posttransplant were classified as no-rejection–clinical AKI (cAKI), and the remaining NR biopsies were classified as Normal. A group of “pristine” biopsies was classified as NRNI based on intersection of MMDx Normal and archetypal no injury in the whole population (SDC, http://links.lww.com/TP/C935).
Multigene Scores and Molecular Classifiers
Molecular interpretations were made for the top correlated transcripts, transcript sets, and classifier scores. Multigene scores included previously annotated pathogenesis-based transcript sets related to allograft rejection and injury, and cell cycle and apoptosis sets from the Kyoto Encyclopedia of Genes and Genomes (Table S2, SDC, http://links.lww.com/TP/C935). Normalized multigene scores were assigned to each biopsy as the mean log expression of all transcripts within the set, normalized to the mean expression of a set of 4 control nephrectomies. Ten previously derived molecular classifier scores trained on histology or clinical features were also considered17,23 (Table S3, SDC, http://links.lww.com/TP/C935).
Statistical Analyses
Estimated host cfDNA quantity, and dd-cfDNA quantity and fraction, over time posttransplant were analyzed with binscatter regression.30,31 Differences in mean plasma dd-cfDNA among MMDx subgroups, rejection archetypes, and injury archetypes27-29 with and without biopsies diagnoses as MMDx rejection were assessed by ANOVA followed by post-hoc tests with all pairwise contrasts. The probability of MMDx rejection given the corresponding quantity and fraction dd-cfDNA was assessed by logistic regression. Relationships of the top genes, multigene and classifier scores, and estimated glomerular filtration rate (eGFR) with dd-cfDNA were assessed by Spearman correlation coefficients (SCC). The effect of sample size on correlation patterns of individual genes was evaluated in 10 random subsamplings of 37 AMR (ie, the same number of TCMR biopsies). Paired differential expression/correlation analyses in AMR + NRNI (N = 305), TCMR + NRNI (N = 171), and cAKI + NRNI (N = 184) were used to compare top genes by dd-cfDNA correlations and fold change within the same populations. Regression analyses were carried out using the top 3 multigene scores (by SCC) to determine their relationships with dd-cfDNA quantity in all biopsies and NRNI biopsies using Akaike information criterion–weighted model averaging. All statistical analyses were carried out in R v4.3.1. P values for SCC and fold change were corrected using the false discovery rate method. Further details on statistical analyses can be found in the Supplemental Material (SDC, http://links.lww.com/TP/C935).
RESULTS
Demographics and biopsy diagnoses (Table 1) were comparable to our previous study.16
TABLE 1.
Description of study population demographics, N = 604
| Patients | N = 581 |
|---|---|
| Recipient sex (% of known) | |
| Female | 218 (38) |
| Male | 360 (62) |
| Missing (% of total) | 3 (1) |
| Ethnicity (% of known) | |
| African American | 84 (15) |
| Not African American | 489 (85) |
| Missing (% of total) | 8 (1) |
| Graft status (% of known) | |
| Tx failed after death | 3 (1) |
| Tx failed before death | 35 (7) |
| Tx functioning | 489 (93) |
| Missing (% of total) | 54 (9) |
| Biopsies | N = 604 |
| Indication for biopsy (% of known) | |
| For cause | 550 (92) |
| Surveillance | 47 (8) |
| Missing (% of total) | 7 (1) |
| Days from transplant to biopsy (median, range) | 580 (3–13 441) |
| Days from biopsy to follow-up (median, range) | 67 (1–621) |
| Days from biopsy to failure (median, range) | 29.5 (0–364) |
| DSA status (% of known) | |
| DSA negative | 243 (64) |
| DSA positive | 137 (36) |
| Missing (% of total) | 224 (37) |
| PRA status (% of known) | |
| PRA negative | 99 (37) |
| PRA positive | 171 (63) |
| Missing (% of total) | 334 (55) |
| C4d (% of known) | |
| Negative | 410 (77) |
| Positive | 121 (23) |
| Missing (% of total) | 73 (12) |
| MMDx diagnosis (% of total) | |
| AMR | 140 (23) |
| pAMR | 31 (5) |
| Mixed | 34 (6) |
| TCMR | 28 (5) |
| pTCMR | 9 (1) |
| cAKI | 50 (8) |
| Normal | 312 (52) |
| Histological rejection (% of known) | |
| AMR | 90 (17) |
| pAMR | 53 (10) |
| Mixed | 21 (4) |
| TCMR | 59 (11) |
| pTCMR | 45 (8) |
| cAKI | 29 (5) |
| BK | 20 (4) |
| No rejection | 228 (42) |
| Missing (% of total) | 59 (10) |
AMR, antibody-mediated rejection; BK, BK polyomavirus; cAKI, clinical acute kidney injury; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; pAMR, possible antibody-mediated rejection; PRA, panel reactive antibodies; pTCMR, possible TCMR; TCMR, T cell–mediated rejection.
Comparison of Donor- and Host-derived cfDNA Levels Overtime
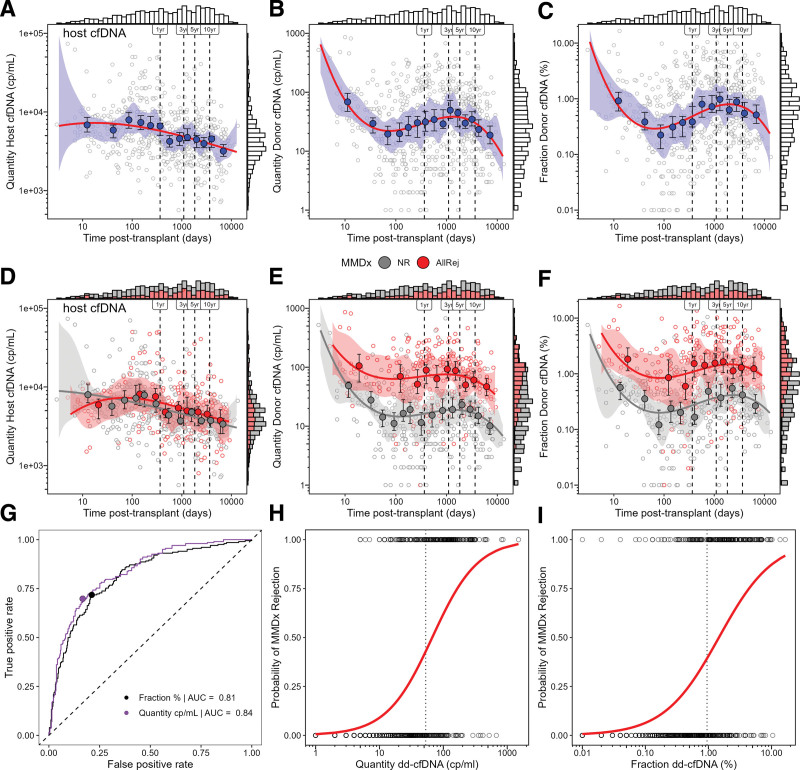
Host cfDNA, dd-cfDNA quantity, and dd-cfDNA fraction varied with time posttransplant (Figure 2A–C). Host cfDNA decreased steadily after ~100 d posttransplant, perhaps reflecting recovery from the injury of donation implantation (Figure 2A). Dd-cfDNA quantity or fraction was initially high (presumably reflecting AKI in the donor tissue), decreased in the first 100 d, then increased between 100 and 1000 d when rejection is common (Figure 2B and C). For host cfDNA, the relationship between time posttransplant and cfDNA was linear (Phost = 0.430; Figure 2A), but for dd-cfDNA, it was nonlinear (PdonorQuantity < 0.0001; PdonorFraction < 0.0001; Figure 2B and C).
FIGURE 2.
Time series of estimated quantity of host cell-free DNA (cp/mL), dd-cfDNA (cp/mL), and fraction dd-cfDNA (%) following transplant (A–F). Temporal patterns were estimated using binscatter regression. Shaded blue, gray, and red bands represent the confidence bands estimated by the binscatter method (A–C). Open circles represent individual biopsies and closed blue, gray, and red circles represent the conditional means dd-cfDNA for each bin and their confidence intervals (A–F). Solid lines represent the predicted dd-cfDNA (A–F). Bars at the top and right margins of the plots represent stacked histograms of the distribution biopsies according to estimated plasma cfDNA and time posttransplant (A–F). Performance of dd-cfDNA in predicting MMDx all-rejection was assessed by ROC curves with diagonal dashed lines representing the 1:1 ratio of sensitivity and specificity (G). The probability of rejection was modeled by logistic regression predicting MMDx all-rejection with open circles represent individual biopsies and solid lines representing the probability of MMDx rejection predicted by logistic regression (H and I). Vertical dashed lines represent defined time points (ie, 1, 3, 5, and 10 y; A–F), and the optimal cutoffs (H and I). dd-cfDNA, donor-derived cell-free DNA; MMDx, Molecular Microscope Diagnostic System.
Host cfDNA did not differ between MMDx rejection and NR (Phost = 0.120; Figure 2D). However, dd-cfDNA in MMDx rejection was higher compared with NR for quantity (P < 0.0001; Figure 2E) and fraction (P < 0.0001; Figure 2F). Quantity (t = 7.05) had slightly greater separation between rejection and NR than fraction (t = 6.26).
For the present analysis, we elected to use quantity rather than fraction because quantity was slightly numerically superior to fraction by AUC (Figure 2G) and regression (Figure 2H and I), but the results for both were very similar (data not shown).
Estimated dd-cfDNA in MMDx and Molecular Rejection and Injury Subgroups
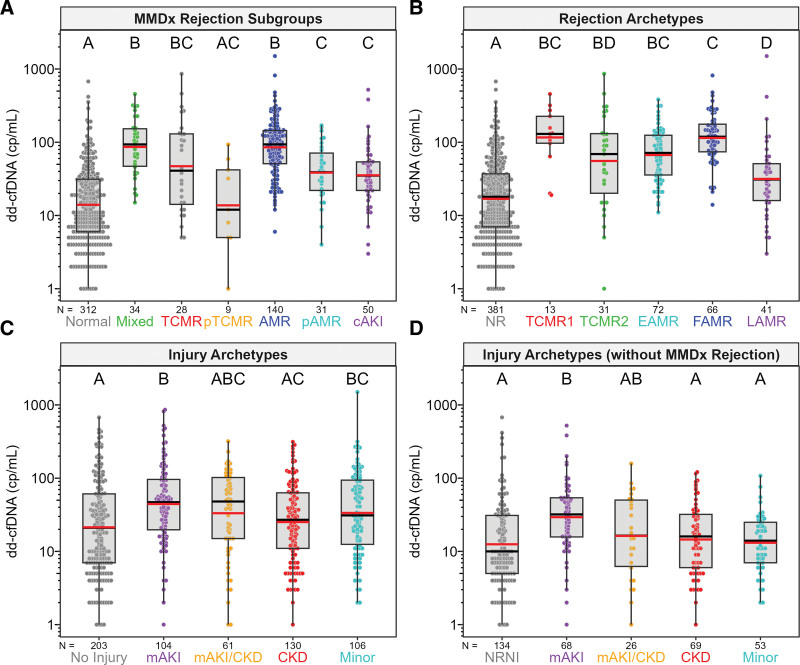
Plasma dd-cfDNA was elevated in MMDx AMR (P < 0.0001), MMDx TCMR (P < 0.0001), and cAKI (P < 0.0001) compared with MMDx Normal biopsies (Figure 3A), similar to our previous findings,16 in agreement with histology diagnoses (Figure S1, SDC, http://links.lww.com/TP/C935). Plasma dd-cfDNA in rejection archetypes also agreed with MMDx results and was elevated in TCMR1 (P < 0.0001; many of which also have AMR activity27), TCMR2 (P < 0.0001), early stage AMR (P < 0.0001), fully developed AMR (FAMR; P < 0.0001), and late-stage AMR (P = 0.012) compared with NR (Figure 3B). Some TCMR2 biopsies had low dd-cfDNA, possibly because TCMR2 is less intense and has more fibrosis than TCMR1.27
FIGURE 3.
Measured dd-cfDNA (cp/mL) in MMDx subgroups (A), rejection archetypes (B), injury archetypes (C), and injury archetypes without MMDx rejection (D). Plasma dd-cfDNA varied significantly across MMDx subgroups, rejection archetypes, injury archetypes, and injury archetypes without MMDx rejection. Colored circles represent dd-cfDNA results for individual biopsies. Gray boxes represent the interquartile range, with whiskers representing 1.5× the interquartile range. Median and geometric mean values for each subgroup/archetype are represented by horizontal black and red bars, respectively. Lettering display above each subgroup/archetype represents post-hoc test results among groups. Groups/archetypes sharing the same letter, within each panel, are not significantly different from one another. AMR, antibody-mediated rejection; cAKI, clinical acute kidney injury; CKD, chronic kidney disease; dd-cfDNA, donor-derived cell-free DNA; EAMR, early-stage AMR; FAMR, fully-developed AMR; LAMR, late-stage AMR; mAKI, molecular acute kidney injury; MMDx, Molecular Microscope Diagnostic System; NR, no rejection; NRNI, MMDx Normal and archetypal No injury; pAMR, possible AMR; pTCMR, possible TCMR; TCMR, T cell–mediated rejection.
Among injury archetype groupings,28,29 dd-cfDNA was highest in molecular AKI (mAKI; P < 0.0001; Figure 3C). Removing biopsies diagnosed as rejection from the injury archetype set resulted in lower dd-cfDNA in all groups. Only mAKI had elevated dd-cfDNA compared with no injury (Figure 3D). However, the dd-cfDNA was sometimes high even in biopsies with NRNI.
There were only 20 biopsies with histological BK nephropathy in the Trifecta-Kidney cohort, limiting the power available for assessing BK-dd-cfDNA relationships. Dd-cfDNA was elevated when AMR was present, but not in biopsies with BK without rejection or injury (Figure S2, SDC, http://links.lww.com/TP/C935).
Top Genes Correlated With dd-cfDNA Among All Biopsies and Within MMDx Subgroups
We wanted to understand what determines variability in plasma dd-cfDNA within specific states. Therefore, in addition to correlations across all biopsies (N = 604), we calculated correlations within groups representing specific states: AMR (N = 171), TCMR (N = 37), cAKI (N = 50), Normal (N = 312), and a molecularly pristine set of biopsies classified as normal and no injury (NRNI; N = 134; Table 2; see Tables S4–S10, SDC [http://links.lww.com/TP/C935] for detailed summaries).
TABLE 2.
Summary of top genes by P value correlating with dd-cfDNA (cp/mL) in all 604 biopsies and within subgroups
| Subgroup | Gene | Gene name | SCC | P | Inference |
|---|---|---|---|---|---|
| All N = 604 |
WARS | Tryptophanyl-tRNA synthetase | 0.58 | 2E-55 | AMR activity: NK cells and IFNG effects |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 0.57 | 4E-54 | ||
| CCL4 | Chemokine (C-C motif) ligand 4/CCL4-like1/CCL4-lik2 | 0.57 | 2E-53 | ||
| GNLY | Granulysin | 0.56 | 2E-51 | ||
| GBP1 | Guanylate binding protein 1, interferon-inducible | 0.55 | 1E-49 | ||
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | 0.55 | 1E-49 | ||
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | 0.55 | 2E-48 | ||
| PLA1A | Phospholipase A1 member A | 0.54 | 2E-47 | ||
| IDO1 | Indoleamine 2,3-dioxygenase 1 | 0.53 | 2E-45 | ||
| GBP4 | Guanylate binding protein 4 | 0.53 | 9E-45 | ||
| AMR N = 171 |
CCL3 | Chemokine (C-C motif) ligand 3 | 0.50 | 6E-12 | AMR activity and AMR stage (microcirculation endothelium, eg, ROBO4) |
| WARS | Tryptophanyl-tRNA synthetase | 0.47 | 9E-11 | ||
| CCL3L1 | Chemokine (C-C motif) ligand 3-like 1/CCL3-like 3 | 0.46 | 2E-10 | ||
| ROBO4 | Roundabout guidance receptor 4 | 0.46 | 2E-10 | ||
| CCL4 | Chemokine (C-C motif) ligand 4/CCL4-like1/CCL4-lik2 | 0.45 | 4E-10 | ||
| ICAM2 | Intercellular adhesion molecule 2 | 0.45 | 6E-10 | ||
| STX11 | Syntaxin 11 | 0.44 | 1E-09 | ||
| IDO1 | Indoleamine 2,3-dioxygenase 1 | 0.44 | 1E-09 | ||
| LYPD5 | LY6/PLAUR domain containing 5 | 0.44 | 1E-09 | ||
| CCL4 | Chemokine (C-C motif) ligand 4/CCL4-like1/CCL4-lik2 | 0.44 | 2E-09 | ||
| TCMR N = 37 |
AKT3 | v-akt murine thymoma viral oncogene homolog 3 | −0.71 | 1E-06 | Complex: not TCMR activity |
| ZNF367 | Zinc finger protein 367 | 0.68 | 4E-06 | ||
| VCPKMT | Valosin-containing protein lysine (K) methyltransferase | −0.66 | 1E-05 | ||
| CD9 | CD9 molecule | −0.65 | 1E-05 | ||
| TAF9B | RNA polymerase II, TATA box associated factor, 31kDa | −0.65 | 2E-05 | ||
| CXCR2 | Chemokine (C-X-C motif) receptor 2/CXCR2 pseudogene 1 | 0.64 | 2E-05 | ||
| PDCD1 | Programmed cell death 1 | 0.64 | 2E-05 | ||
| RTN3 | Reticulon 3 | −0.63 | 3E-05 | ||
| RARRES3 | Retinoic acid receptor responder (tazarotene induced) 3 | 0.63 | 3E-05 | ||
| KIF23 | Kinesin family member 23 | 0.63 | 3E-05 | ||
| cAKI N = 50 |
DENR | Density-regulated protein | 0.61 | 3E-06 | AKI: (Response to wounding) |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | 0.59 | 7E-06 | ||
| FKBP1A | FK506 binding protein 1A///FK506 binding protein 1Cs | 0.59 | 7E-06 | ||
| PARPBP | PARP1 binding protein | 0.55 | 3E-05 | ||
| LPGAT1 | Lysophosphatidyl glycerol acyltransferase 1 | 0.55 | 4E-05 | ||
| ARNTL2 | Aryl hydrocarbon receptor nuclear translocator-like 2 | 0.55 | 4E-05 | ||
| NUP50 | Nucleoporin 50kDa | 0.54 | 5E-05 | ||
| DERL1 | Derlin 1 | 0.54 | 5E-05 | ||
| PAK2 | p21 protein (Cdc42/Rac)-activated kinase 2 | 0.54 | 6E-05 | ||
| CHEK2 | Checkpoint kinase 2///uncharacterized LOC101929031, etc | 0.54 | 6E-05 | ||
| Normal N = 312 |
ATAD2 | ATPase family, AAA domain containing 2 | 0.31 | 1E-08 | AKI: (Response to wounding), including cell cycle (eg, MKI67) |
| BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B | 0.31 | 2E-08 | ||
| ASPM | Abnormal spindle microtubule assembly | 0.31 | 4E-08 | ||
| MYBL1 | v-myb avian myeloblastosis viral oncogene homolog-like 1 | 0.30 | 5E-08 | ||
| SLITRK2 | SLIT and NTRK-like family, member 2 | −0.30 | 5E-08 | ||
| TMPRSS4 | Transmembrane protease, serine 4 | −0.30 | 6E-08 | ||
| MKI67 | Marker of proliferation Ki-67 | 0.30 | 7E-08 | ||
| PRC1 | Protein regulator of cytokinesis 1 | 0.30 | 8E-08 | ||
| TOP2A | Topoisomerase (DNA) II alpha | 0.29 | 2E-07 | ||
| KIAA0101 | KIAA0101 | 0.29 | 3E-07 | ||
| NRNI N = 134 |
CLEC7A | C-type lectin domain family 7, member A | 0.40 | 2E-06 | All rejection (eg, CLEC7A, MYBL1), inflammation (CARD8), apoptosis (NLRP1, EIF5A) |
| MYBL1 | v-myb avian myeloblastosis viral oncogene homolog-like 1 | 0.40 | 2E-06 | ||
| CARD8 | Caspase recruitment domain family, member 8 | 0.39 | 4E-06 | ||
| CLEC7A | C-type lectin domain family 7, member A | 0.38 | 5E-06 | ||
| PIK3CD | Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta | 0.38 | 5E-06 | ||
| CLEC7A | C-type lectin domain family 7, member A | 0.37 | 9E-06 | ||
| GMFG | Glia maturation factor, gamma | 0.37 | 1E-05 | ||
| NLRP1 | NLR family, pyrin domain containing 1 | 0.37 | 1E-05 | ||
| MYO1F | Myosin IF | 0.37 | 1E-05 | ||
| EIF5A | Eukaryotic translation initiation factor 5A | −0.36 | 2E-05 |
Gray shading highlights genes with negative SCCs. For details, see Tables S4–S10 (SDC, http://links.lww.com/TP/C935).
AKI, acute kidney injury; AMR, molecular antibody-mediated rejection; cAKI, clinical acute kidney injury; dd-cfDNA, donor-derived cell-free DNA; IFNG, interferon gamma; NK, natural killer; NRNI, MMDx Normal and archetypal No injury; SCC, standard-of-care; TCMR, T cell–mediated rejection.
Across all biopsies, dd-cfDNA release correlated with AMR activity, particularly interferon gamma effects and expression of natural killer (NK) cell genes, confirming the previous analysis.16 Within AMR biopsies, dd-cfDNA release also correlated with expression of AMR-selective genes (eg, WARS, CCL3), as well as genes related to stage (eg, ROBO4, STX11, ICAM2, LYPD5). These genes were highest in FAMR likely reflecting time-dependent microcirculation changes.
Within TCMR biopsies, no clear molecular pattern was found, and the top genes correlating with dd-cfDNA were not clearly associated with TCMR activity (ie, transcripts most selective for TCMR)32 and had weak associations by P value. For example, the top gene was AKT3, a pan-rejection gene, which was negatively correlated with dd-cfDNA (SCC = −0.71, P = 1E-06) but was unchanged in TCMR versus Normal (Table S6, SDC, http://links.lww.com/TP/C935). The pattern was similarly complex in 30 mixed rejection biopsies (Table S7, SDC, http://links.lww.com/TP/C935), suggesting a dominant effect of the TCMR-induced parenchymal damage in the donor tissue.
To address the possibility that failure to observe correlations with TCMR activity was because of small sample size, we evaluated if the correlations with AMR activity we observed within AMR would be detectable within random subsamplings of 37 AMR biopsies (Table 3). Despite the smaller sample size, dd-cfDNA correlations within AMR subsamplings were consistent with those in 171 AMR biopsies, suggesting that limited sample size is likely not responsible for the noise observed within TCMR.
TABLE 3.
Top 10 genes correlated with dd-cfDNA (cp/mL) from random subsamplings of 37 biopsies within AMR (N = 171)
| Genes by all AMR biopsies | Subsampling | Genes within subsampling | Interpretation |
|---|---|---|---|
|
CCL3
WARS CCL3L1 ROBO4 CCL4 ICAM2 STX11 IDO1 LYPD5 CCL4 |
1 | ROBO4, SH2D1B, LTF, IDO1, CCL4, PLA1A, LYPD5, EGR1, ZBED2, WARS | Genes related to AMR (eg, WARS, ROBO4) and IFNG effects (eg, CXCL11) are represented across all subsamplings |
| 2 | CCL4, IDO1, PLA1A, WARS, S1PR5, PRF1, FGFBP2, GNLY, GBP4, LYPD5 | ||
| 3 | LYPD5, WARS, ROBO4, GBP4, LTF, CXCL11, CCL4, IDO1, PLA1A, BATF | ||
| 4 | WARS, CXCL11, CXCL10, PRF1, ROBO4, LYPD5, CXCL9, IDO1, CCL4, GBP4 | ||
| 5 | LYPD5, PLA1A, ROBO4, WARS, KLRF1, IDO1, CXCL11, SH2D1B, CCL4, CXCL10 | ||
| 6 | GNLY, SERPINA3, WARS, CXCL10, ROBO4, CXCL9, PRF1, CCL4, GBP4, LYPD5 | ||
| 7 | EGR1, GNLY, S1PR5, FGFBP2, ROBO4, LTF, PRF1, PTX3, KLRF1, TIGIT | ||
| 8 | WARS, SH2D1B, LYPD5, GBP4, CCL4, CXCL11, GNLY, LTF, CXCL9, ROBO4 | ||
| 9 | CCL4, IDO1, GNLY, LYPD5, GBP4, CXCL10, IFNG, CXCL11, CXCL9, AKAP12 | ||
| 10 | GNLY, CXCL11, WARS, IDO1, KLRF1, CXCL10, PRF1, GBP4, CCL4, ROBO4 |
AMR, antibody-mediated rejection; dd-cfDNA, donor-derived cell-free DNA; IFNG, interferon gamma.
Within cAKI (ie, MMDx NR ≤6 wk posttransplant), all the top genes had positive SCCs, albeit with low P values, and depicted a molecular response to wounding. Although not in the top 10, mitosis gene MKI67 was significantly correlated with dd-cfDNA (Table S8, SDC, http://links.lww.com/TP/C935). The molecular profile in Normal biopsies (ie, MMDx NR >6 wk posttransplant) was similar to that of cAKI, probably because there is considerable injury in biopsies with no rejection beyond 6 wk.28,29 Five of the top 10 genes in Normal biopsies were related to cell division: MKI67 (a mitosis marker), BUB1B, ASPM, TOP2A, and KIAA0101.
After excluding injury from the Normal group, leaving 134 NRNI biopsies, the top gene associated with dd-cfDNA was CLEC7A, a macrophage-expressed gene that is associated with rejection and injury. The SCC was also high for MYBL1, an NK-expressed gene annotated as increased in AMR (Table S10, SDC, http://links.lww.com/TP/C935). The molecular profile of dd-cfDNA in NRNI was compatible with subthreshold rejection and injury-induced inflammation, but does not exclude other potential sources of dd-cfDNA.
Comparison of Gene Correlations With dd-cfDNA to Genes Associated With Disease States
In addition to understanding molecular relationships of dd-cfDNA within isolated subgroups, we also wanted to understand what determines dd-cfDNA along a gradient from NRNI to each disease state. This was accomplished by pooling AMR + NRNI (N = 305), TCMR + NRNI (N = 171), and cAKI + NRNI (N = 184) groupings and carrying out correlation and differential expression analyses for each gene within these groupings (Tables S11–S13, SDC, http://links.lww.com/TP/C935).
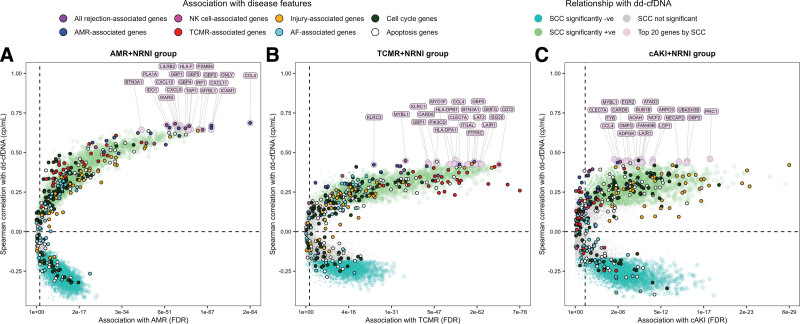
Within the AMR + NRNI group, there was strong convergence in the top genes correlated with dd-cfDNA and those most associated with AMR versus NRNI, supporting the conclusion that AMR activity was the major molecular driver of dd-cfDNA release in AMR (Figure 4A). The SCCs with dd-cfDNA were also strongest compared with other groupings.
FIGURE 4.
Differential expression and correlation genescape demonstrating gradients in gene associations with dd-cfDNA by MMDx subgroups. Biopsies called AMR (A), TCMR (B), and cAKI (C) were pooled with biopsies called NRNI in attempt to isolate the gradient moving from low intensity to high intensity for each disease state. Differential expression is represented FDR in FC in AMR, TCMR, and cAKI vs NRNI. Correlations are expressed as SCC of gene expression with dd-cfDNA within each grouping. Individual genes are represented as colored circles. Circles are first colored to show genes with significantly positive SCCs (green), significantly negative SCCs (turquoise), and nonsignificant SCCs (gray). Circles/genes are then colored by their annotation with major disease phenotypes as described in the Supplemental Material (SDC, http://links.lww.com/TP/C935). Finally, the top 20 genes by SCC are labeled and colored (mauve). AF, atrophy-fibrosis; AMR, antibody–mediated rejection; cAKI, clinical acute kidney injury; dd-cfDNA, donor-derived cell-free DNA; FC, fold change; FDR, false discovery rate; MMDx, Molecular Microscope Diagnostic System; NK, natural killer; NRNI, MMDx Normal and archetypal No injury; SCC, Spearman correlation coefficients; TCMR, T cell–mediated rejection.
Within the TCMR + NRNI group, the top dd-cfDNA SCCs were lower compared with AMR + NRNI (<0.30) and there was poor agreement between the top dd-cfDNA correlated genes and the top TCMR-selective genes (Figure 4B). Some of the top correlated genes could potentially be related to AMR or all-rejection (eg, CCL4; interferon gamma-inducible gene GBP1; and NK cell and macrophage-expressed gene LAT2). Some genes most induced by TCMR (eg, ADAMDEC1 and CXCL13) had lower correlations with dd-cfDNA. Thus, TCMR activity was only modestly correlated with dd-cfDNA and was not the main driver of dd-cfDNA release in TCMR.
Within cAKI + NRNI, the top dd-cfDNA SCCs were lower than those in AMR + NRNI (eg, injury-inducible gene PRC1; SCC = 0.45). There was limited convergence between the top genes correlating with dd-cfDNA and the top genes increased in AKI, but many were related to injury and cell cycle (eg, BUB1B) (Figure 4C). Some of the top genes correlated with dd-cfDNA by SCC could also potentially reflect subthreshold AMR activity (eg, CCL4 and MYBL1).
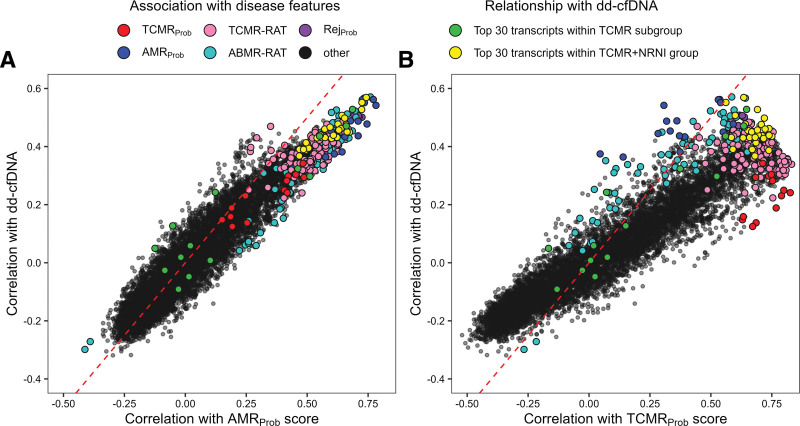
We visualized the dd-cfDNA SCCs for each probeset with AMR activity (the AMRProb classifier score) or TCMR activity (the TCMRProb classifier score) in all 604 biopsies (Figure 5). The AMR relationships were highly consistent between the AMRProb classifier and dd-cfDNA (Figure 5A). In contrast, the TCMR relationships were weaker. TCMR-associated genes were not among the most strongly associated with dd-cfDNA (Figure 5B).
FIGURE 5.
Spearman correlation gene expression with dd-cfDNA in IQR-filtered probesets compared with Spearman correlations with AMRProb (A) and TCMRProb (B) classifier scores. Individual probesets are represented as colored circles. Circles are first colored to show probesets with significantly positive SCCs (green), significantly negative SCCs (turquoise), and nonsignificant SCCs (gray). Circles/probesets are then colored by their gene annotation with major disease phenotypes as described in the Supplemental Material (SDC, http://links.lww.com/TP/C935). AMR, antibody–mediated rejection; dd-cfDNA, donor-derived cell-free DNA; IQR, interquartile range; SCC, Spearman correlation coefficients; TCMR, T cell–mediated rejection.
Correlations of dd-cfDNA With Multigene Scores
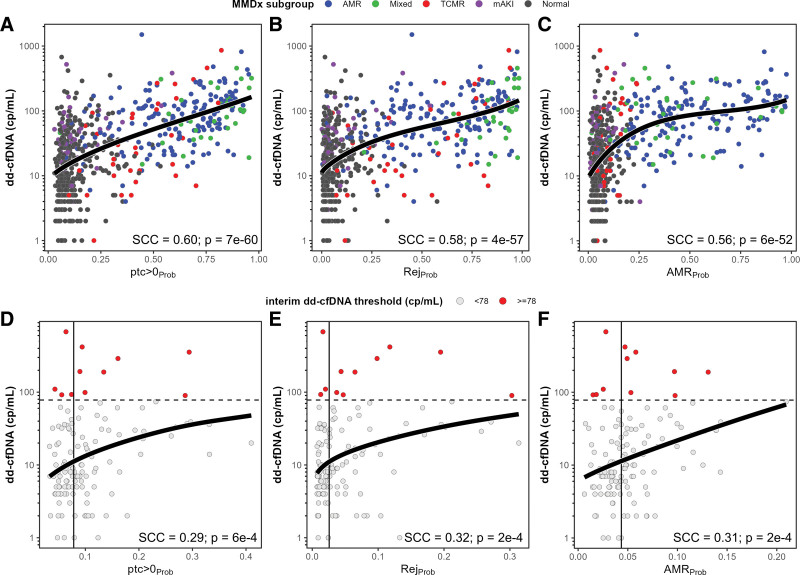
To capture bulk changes in expression of genes describing the same biological processes, we assessed the correlation of multigene scores with dd-cfDNA (Table 4; see Tables S14–S19, SDC [http://links.lww.com/TP/C935] for detailed summaries). Across all biopsies, the top 3 multigene scores correlated with dd-cfDNA were ptc > 0Prob (SCC = 0.60), RejProb (SCC = 0.58), and AMRProb (SCC = 0.56). Within AMR biopsies, the top multigene scores depicted AMR activity (AMRProb, g > 0Prob, PC1Rej). Within TCMR biopsies, the best multigene scores depicted overall rejection (ptc > 0Prob SCC = 0.57; RejProb SCC = 0.56), with lower correlations with TCMR activity (TCMRProb SCC = 0.26) and AMR activity (AMRProb SCC = 0.30). Within cAKI biopsies, the best multigene scores depicted injury (IRRAT SCC = 0.41) and cell cycle (SCC = 0.40). Within Normal biopsies, the top 3 multigene scores correlated with dd-cfDNA depicted all-rejection (ptc > 0Prob and RejProb, both SCC = 0.25) and AMR activity (AMRProb SCC = 0.23). Within NRNI biopsies, the top multigene scores were similar to those in Normal, including all-rejection (ptc > 0Prob SCC = 0.29, RejProb SCC = 0.32) and AMR activity (AMRProb SCC = 0.31).
TABLE 4.
Summary of molecular score SCCs with dd-cfDNA (cp/mL) within subgroup
| Category | Molecular score | AMR N = 171 |
TCMR N = 37 |
cAKI N = 50 |
Normal N = 312 |
NRNI N = 134 |
All N = 604 |
|---|---|---|---|---|---|---|---|
| All rejection–related | Peritubular capillaritis classifier (ptc > 0Prob) | 0.44* | 0.57* | 0.31 | 0.25* | 0.29* | 0.60* |
| Rejection classifier (RejProb) | 0.40* | 0.56* | 0.30 | 0.25* | 0.32* | 0.58* | |
| Interferon gamma-inducible (GRIT3) | 0.28* | 0.32 | 0.30 | 0.23* | 0.29* | 0.49* | |
| AMR-related | AMR classifier (AMRProb) | 0.48* | 0.30 | 0.22 | 0.23* | 0.31* | 0.56* |
| Glomerulitis classifier (g > 0Prob) | 0.47* | 0.39 | 0.26 | 0.18* | 0.21 | 0.56* | |
| TCMR-related | TCMR classifier (TCMRProb) | 0.03 | 0.26 | −0.04 | 0.13 | 0.10 | 0.25* |
| t-score classifier (t > 1Prob) | 0.04 | 0.33 | 0.05 | 0.14 | 0.12 | 0.27* | |
| Recent injury–related | Injury repair–associated transcripts (IRRAT30) | −0.04 | −0.11 | 0.41* | 0.16* | 0.10 | 0.26* |
| Cell cycle–related | Cell cycle KEGG pathway (cell cycle KEGG) | 0.10 | 0.02 | 0.40* | 0.22* | 0.20 | 0.32* |
| Apoptosis-related | Apoptosis KEGG pathway (apoptosis KEGG) | 0.17 | −0.10 | 0.29 | 0.19* | 0.25* | 0.37* |
| No injury-related | Injury archetype no injury score | 0.02 | 0.26 | −0.26 | −0.21* | −0.25* | −0.25* |
| No rejection-related | Rejection archetype no rejection score | −0.26* | −0.41 | −0.30 | −0.27* | −0.27* | −0.57* |
*P value < 0.01. Gray shading highlights absolute SCCs ≥ 0.4.
AMR, antibody–mediated rejection; cAKI, clinical acute kidney injury; dd-cfDNA, donor-derived cell-free DNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; SCC, Spearman correlation coefficients; TCMR, T cell–mediated rejection.
A summary of the molecular association with dd-cfDNA within all biopsies and major subgroups is provided in Table 5.
TABLE 5.
Summary of molecular associations with dd-cfDNA release in subgroups
| Subgroup | Associations with dd-cfDNA | Conclusions | |
|---|---|---|---|
| Spearman correlations | |||
| Individual genes | Multigene scores | ||
| All N = 604 |
AMR-related | AMR-related | Across the whole population, AMR activity was the dominant correlate of dd-cfDNA release |
| AMR N = 171 |
AMR-related | AMR-related | Within AMR, AMR activity processes were the dominant driver of dd-cfDNA release. The top genes correlated with dd-cfDNA within AMR have reflected AMR activity (NK and IFNG-inducible) and AMR stage (eg, ROBO4) probably reflecting endothelial changes in FAMR. These findings were consistent for the continuum from NRNI to AMR as well as within AMR |
| TCMR N = 37 |
Complex. Injury and all rejection–related | Complex. Peritubular capillaritis classifier was strongest correlation | Within TCMR, TCMR-selective transcripts were not the dominant correlates of dd-cfDNA release. The top genes correlated with dd-cfDNA within TCMR were broadly expressed, decreased as well as increased in expression, probably reflecting parenchymal effects of TCMR (visible as tubulitis) |
| cAKI N = 50 |
Injury-related | Injury-related and cell cycle–related | Within AKI, injury-related processes were the dominant driver of dd-cfDNA release. The top PBTs correlated with dd-cfDNA within AKI were cell cycle related. Also, potential contribution of mild rejection effects. These findings were consistent for the continuum from NRNI to AKI as well as within AKI |
| NRNI N = 134 |
Rejection and injury-related | Rejection and injury-related | Within NRNI, subtle AMR activity and all rejection were significantly correlated with dd-cfDNA. The no-injury archetype score correlated negatively with dd-cfDNA, indicating that subtle injury could contribute |
AKI, acute kidney injury; AMR, antibody–mediated rejection; cAKI, clinical acute kidney injury; dd-cfDNA, donor-derived cell-free DNA; FAMR, fully-developed AMR; IFNG, interferon gamma; NK, natural killer; NRNI, MMDx Normal and archetypal No injury; PBTs, pathogenesis-based transcript sets; TCMR, T cell–mediated rejection.
Regression Analysis
The relationships between the top 3 multigene scores and dd-cfDNA in the whole population were further explored with regression analyses using Akaike information criterion–weighted model averaging (Tables S20–S28, SDC, http://links.lww.com/TP/C935). Across the whole population, plasma dd-cfDNA increased as a function of ptc > 0Prob, RejProb, and AMRProb (Figure 6A–C). We then modeled the relationships of these multigene scores in NRNI biopsies, and found that increases in ptc > 0Prob, RejProb, and AMRProb were also related to increases in dd-cfDNA (Figure 6D–F). NRNI biopsies were then separated by their median molecular scores and the interim quantity cutoff (78 cp/mL). Only 11 of 134 NRNI biopsies (8.9%) had elevated dd-cfDNA above the cutoff. Among these 11, the majority had molecular scores above the median: 7 ptc > 0Prob, 8 RejProb, and 8 AMRProb.
FIGURE 6.
Relationships of estimated quantity dd-cfDNA (cp/mL) with the top selected molecular scores by Spearman correlation coefficient in all biopsies (A–C) compared with NRNI biopsies (D–F). Colored circles represent individual biopsies and their MMDx (A–C) and dd-cfDNA (D–F) diagnoses. Black solid curves represent the AIC-weighted model-averaged predictions from all model fits (excluding those that did not converge). Dashed horizontal lines in D–F indicate the interim dd-cfDNA threshold of 78 cp/mL. Vertical solid lines in D–F represent median molecular scores within NRNI. See Supplemental Material (SDC, http://links.lww.com/TP/C935) for detailed summary of the model select and averaging method, including statistics for each model used in the model average. AIC, Akaike information criterion; AMR, antibody-mediated rejection; dd-cfDNA, donor-derived cell-free DNA; mAKI, molecular acute kidney injury; MMDx, Molecular Microscope Diagnostic System; NRNI, MMDx Normal and archetypal No injury; SCC, Spearman correlation coefficients; TCMR, T cell–mediated rejection.
Relating dd-cfDNA to eGFR
There was no relationship between dd-cfDNA and eGFR across all biopsies (SCC = −0.06, P = 0.145), TCMR (SCC = −0.26, P = 0.118), or within Normal (SCC = 0.04, P = 0.0462). Within AMR, there was a weak positive relationship between dd-cfDNA and eGFR (SCC = 0.16, P = 0.036), compatible with the higher dd-cfDNA signal in the early stage and FAMR compared with late-stage late-stage AMR. Of interest, there is a weak positive correlation between AMR activity and eGFR in the larger INTERCOMEX (International Collaborative Microarray Study Extension) study (Halloran et al, 2023, unpublished results). Within cAKI, dd-cfDNA decreased numerically as eGFR increased, but low sample size limited power for detecting a statistical relationship (SCC = −0.27, P = 0.062).
DISCUSSION
We studied the molecular rejection and injury states in 604 kidney transplant biopsies with paired estimates of plasma dd-cfDNA for insights into the underlying processes that release dd-cfDNA within specific molecular states in the donor tissue: AMR, TCMR, AKI, and Normal, and a molecularly pristine set of biopsies defined as NRNI (ie, MMDx Normal without any archetypal injury). The molecular patterns of dd-cfDNA were characterized along a continuum from pristine (ie, NRNI) to each disease state as well as within each isolated disease state. We then studied why dd-cfDNA was sometimes elevated in patients whose biopsies lacked apparent rejection and injury. As in our earlier analysis,16 mean dd-cfDNA in biopsies with AMR, TCMR, and AKI was elevated compared with Normal biopsies. Subgroup analyses identified 3 distinct molecular processes involved in dd-cfDNA release: AMR activity, TCMR-related parenchymal damage in donor tissue, and AKI, particularly cell cycling. Moreover, analysis of isolated NRNI biopsies indicated that subtle rejection- and injury-related changes in patients without apparent rejection or injury can still contribute to dd-cfDNA levels.
There were significant molecular associations with rejection and injury linked to dd-cfDNA in NRNI biopsies. The top 2 genes correlated with dd-cfDNA in NRNI were CLEC7a (expressed in macrophages) and MYBL1 (expressed in NK cells and macrophages). Although dd-cfDNA is generally low in NRNI, low-level molecular rejection and injury process are nonetheless active and associated with dd-cfDNA. Given the prevalence of subtle AMR in kidneys that do not meet molecular or histological definitions of AMR is now recognized,18,19 it is plausible that elevated dd-cfDNA in nonrejecting biopsies could also indicate the presence of subthreshold molecular AMR processes. Whether these subtle states are of significance to the long-term kidney health is not known, particularly because subtle AMR may not progress over time. Further studies are needed to establish the impact of the subtle subthreshold rejection and injury and their contributions to the cases of high dd-cfDNA in apparently normal kidney transplants. Moreover, while we believe that rejection and injury are diffuse processes, it remains possible that focal changes may release dd-cfDNA but not be represented in the core biopsy. The significant dd-cfDNA associations with rejection- and injury-related genes in NRNI indicate that consistent dd-cfDNA elevation in kidney transplants that fail to meet the diagnostic thresholds for rejection or injury should be taken as evidence for the need of heightened surveillance.
Within AMR biopsies, the gradient in dd-cfDNA levels correlated with AMR activity and stage, peaking in FAMR, and with AMR-related multigene scores such as the AMR and ptc-lesion classifiers and the all-rejection classifier. Regression analyses of the top multigene scores correlated with AMR-related dd-cfDNA release support that suppression of dd-cfDNA can be followed to indicate suppression of AMR activity in the kidney, and lack of response (ie, failure to reduce molecular AMR activity) indicates lack of success, although this needs to be proven in clinical trials.
As in previous studies, we defined AKI in 2 ways: biopsies in the first 6 wk posttransplant having MMDx NR (cAKI), and biopsies with mAKI in injury archetype analysis (mAKI).28 Plasma dd-cfDNA was increased by AKI compared with Normal biopsies, and was most correlated with cell cycle and injury-induced transcripts, a conclusion supported by assessment of single genes (eg, BUB1B) and multigene scores (eg, Kyoto Encyclopedia of Genes and Genomes cell cycle geneset), along a gradient from NRNI to AKI as well as within AKI biopsies. Cell cycle has long been recognized as a key process in the early stages of AKI,33,34 in which an early wave of DNA synthesis is followed by persistence of other injury-related changes. Cell cycle transcripts were also associated with dd-cfDNA release in Normal biopsies (eg, MKI67) but less so when injury was removed (ie, NRNI biopsies), supporting the overall relationship between dd-cfDNA with cell cycling in the injury states. In contrast, dd-cfDNA levels within AMR and TCMR biopsies were not related to cell cycle. However, in AMR, cell cycling could be localized to the microcirculation compartment (ie, with direct access to the plasma), which would not be adequately represented by a biopsy core in which only a small fraction of the nuclei are from the microcirculation compartment.
Plasma dd-cfDNA correlations in the NRNI + TCMR groups were strongest with all-rejection genes (eg, CLEC7A) and classifiers (eg, RejProb and ptc > 0Prob) as well as some injury-inducible genes (eg, PTPRC), and not the genes and classifiers strongly selective for TCMR activity (eg, ADAMDEC1 and TCMRProb). Within TCMR, the range of dd-cfDNA varied considerably, but the molecular associations with dd-cfDNA within TCMR did not reveal a consistent molecular pattern. The top correlations with dd-cfDNA were both negative (eg, AKT3) and positive (eg, NF367) and had weak false discovery rates, suggesting that they represented noise. The complexity within TCMR was similar for mixed biopsies, perhaps in-part reflecting the disruption of the donor tissue by tubulitis. We entertained the possibility that the limited sample size of the TCMR biopsies (N = 37) could account for the lack of clear signal within TCMR, but random subsamplings of 37 AMR biopsies retained strong and consistent dd-cfDNA correlations, suggesting that the complexity within TCMR was not simply a statistical aberration associated with sample size but potentially a real biological phenomenon. The TCMR process causes AKI-like changes (eg, elevated injury scores) but the epithelial changes are more severe than in AKI and rapidly progress to atrophy-fibrosis.27 The dd-cfDNA signal from donor cells may reflect the balance between recent/ongoing injury and injury progressing to atrophy-fibrosis. We hypothesize that once TCMR has manifested in the kidney, the complex mixture of recent injury and atrophy-fibrosis in the donor epithelium (tubulitis) obscures the molecular gradient associated with dd-cfDNA in TCMR.
A clearer picture about the complexity of plasma dd-cfDNA among different disease states will be critical to optimizing the interpretation of dd-cfDNA not only as a screening tool but also as a tool for monitoring disease activity during treatment and follow-up. The distinct molecular associations of dd-cfDNA within specific disease states versus across the whole population is an example of Simpson paradox, a phenomenon in which the trends across a population change when they are analyzed within subgroups of that population. For example, population-wide, dd-cfDNA correlated with AMR activity, but within cAKI, dd-cfDNA correlated most strongly with genes related to injury-induced genes (eg, IRRATs) and cell cycle genes. Moreover, while the goals of treatment of TCMR are to extinguish both dd-cfDNA and TCMR activity, more information will be needed about the relationship of dd-cfDNA to TCMR and the donor parenchyma, including how to interpret persistence of dd-cfDNA after treatment of TCMR (ie, measuring persistent TCMR activity versus persisting parenchymal injury in the donor tissue). Finally, in the NRNI group, subtle rejection and subthreshold AMR in-part contributed to elevated dd-cfDNA, indicating that close monitoring should be considered for these patients.
ACKNOWLEDGMENTS
The authors thank their valued clinicians in the Trifecta study group who partnered with them for this study by contributing biopsies and feedback. They also thank Martina Mackova and Anna Hutton for biopsy processing for the Microarray biopsy assessment component of this study and Natera Inc for blood sample processing and the Prospera test results.
The Trifecta-Kidney Investigators: Justyna Fryc, Beata Naumnik, Jonathan Bromberg, Matt Weir, Nadiesa Costa, Daniel Brennan, Sam Kant, Vignesh Viswanathan, Milagros Samaniego-Picota, Iman Francis, Anita Patel, Alicja Dębska-Ślizień, Joanna Konopa, Andrzej Chamienia, Andrzej Więcek, Grzegorz Piecha, Željka Veceric-Haler, Miha Arnol, Nika Kojc, Maciej Glyda, Katarzyna Smykal-Jankowiak, Ondrej Viklicky, Petra Hruba, Silvie Rajnochová Bloudíčkova, Janka Slatinská, Marius Miglinas, Marek Myślak, Joanna Mazurkiewicz, Marta Gryczman, Leszek Domański, Mahmoud Kamel, Agnieszka Perkowska-Ptasińska, Dominika Dęborska-Materkowska, Michal Ciszek, Magdalena Durlik, Ryszard Grenda, Miroslaw Banasik, Mladen Knotek, Ksenija Vucur, Zeljka Jurekovic, Thomas Müller, Thomas Schachtner, Andrew Malone, Tarek Alhamad, Arksarapuk Jittirat, Emilio Poggio, Richard Fatica, Ziad Zaky, Kevin Chow, Peter Hughes, Sanjiv Anand, Gaurav Gupta, Layla Kamal, Dhiren Kumar, Irfan Moinuddin, Sindhura Bobba.
Supplementary Material
Footnotes
A full list of Trifecta-Kidney Investigators can be found in the Acknowledgments.
P.F.H. has shares in Transcriptome Sciences Inc (TSI), a University of Alberta research company with an interest in molecular diagnostics, and is a consultant to Natera Inc. All Natera Inc authors are employees and own equity at Natera Inc. The other authors declare no conflicts of interest.
The Trifecta study is an investigator-initiated study supported by a grant from Natera Inc to TSI/ATAGC. The Microarray biopsy assessment project is supported in part by a licensing agreement with One Lambda/Thermo Fisher.
P.T.G. was responsible for data analysis and interpretation, writing, editing, and reviewing the article. P.F.H. was the principal investigator, edited and reviewed the article, and was responsible for data interpretation and study design. K.S.M.-T. edited and reviewed the article and assisted with data interpretation. Z.D., A.P., and P.G. were responsible for the measurement of dd-cfDNA (the Prospera test) and helped in data interpretation and discussion, as well as article preparation and editing. The Trifecta Investigators were responsible for biopsy and blood sample collection.
CEL files will be available on Gene Expression Omnibus on publication.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
Contributor Information
Collaborators: Justyna Fryc, Beata Naumnik, Jonathan Bromberg, Matt Weir, Nadiesa Costa, Daniel Brennan, Sam Kant, Vignesh Viswanathan, Milagros Samaniego-Picota, Iman Francis, Anita Patel, Alicja Dębska-Ślizień, Joanna Konopa, Andrzej Chamienia, Andrzej Więcek, Grzegorz Piecha, Željka Veceric-Haler, Miha Arnol, Nika Kojc, Maciej Glyda, Katarzyna Smykal-Jankowiak, Ondrej Viklicky, Petra Hruba, Silvie Rajnochová Bloudíčkova, Janka Slatinská, Marius Miglinas, Marek Myślak, Joanna Mazurkiewicz, Marta Gryczman, Leszek Domański, Mahmoud Kamel, Agnieszka Perkowska-Ptasińska, Dominika Dęborska-Materkowska, Michal Ciszek, Magdalena Durlik, Ryszard Grenda, Miroslaw Banasik, Mladen Knotek, Ksenija Vucur, Zeljka Jurekovic, Thomas Müller, Thomas Schachtner, Andrew Malone, Tarek Alhamad, Arksarapuk Jittirat, Emilio Poggio, Richard Fatica, Ziad Zaky, Kevin Chow, Peter Hughes, Sanjiv Anand, Gaurav Gupta, Layla Kamal, Dhiren Kumar, Irfan Moinuddin, and Sindhura Bobba.
REFERENCES
- 1.Moinuddin I, Kumar D, Kamal L, et al. Calibration of donor-derived cell-free DNA criteria for rejection with molecular diagnoses of kidney transplant biopsies. Am J Transplant. 2020;20:680–680. [Google Scholar]
- 2.Huang E, Sethi S, Peng A, et al. Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am J Transplant. 2019;19:1663–1670. [DOI] [PubMed] [Google Scholar]
- 3.Dengu F. Next-generation sequencing methods to detect donor-derived cell-free DNA after transplantation. Transplant Rev (Orlando). 2020;34:100542. [DOI] [PubMed] [Google Scholar]
- 4.Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant. 2019;19:3087–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agbor-Enoh S, Wang Y, Tunc I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. EBioMedicine. 2019;40:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharon E, Shi H, Kharbanda S, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput Biol. 2017;13:e1005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altug Y, Liang N, Ram R, et al. Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation. 2019;103:2657–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moinuddin I, Kumar D, Halloran P, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Am J Transplant. 2019;19:521–521. [DOI] [PubMed] [Google Scholar]
- 10.Sigdel TK, Archila FA, Constantin T, et al. Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med. 2018;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thongprayoon C, Vaitla P, Craici IM, et al. The use of donor-derived cell-free DNA for assessment of allograft rejection and injury status. J Clin Med. 2020;9:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kataria A, Kumar D, Gupta G. Donor-derived cell-free DNA in solid-organ transplant diagnostics: indications, limitations, and future directions. Transplantation. 2021;105:1203–1211. [DOI] [PubMed] [Google Scholar]
- 13.Bloom RD, Augustine JJ. Beyond the biopsy: monitoring immune status in kidney recipients. Clin J Am Soc Nephrol. 2021;16:1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agbor-Enoh S, Shah P, Tunc I, et al. ; GRAfT Investigators. Cell-free DNA to detect heart allograft acute rejection. Circulation. 2021;143:1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: a prospective multicenter study. Am J Transplant. 2019;19:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators. The Trifecta Study: comparing plasma levels of donor-derived cell-free DNA with the molecular phenotype of kidney transplant biopsies. J Am Soc Nephrol. 2022;33:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halloran PF, Madill-Thomsen KS, Reeve J. The molecular phenotype of kidney transplants: insights from the MMDx project. Transplantation. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madill-Thomsen KS, Bohmig GA, Bromberg J, et al. ; INTERCOMEX Investigators. Donor-specific antibody is associated with increased expression of rejection transcripts in renal transplant biopsies classified as no rejection. J Am Soc Nephrol. 2021;32:2743–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosales IA, Mahowald GK, Tomaszewski K, et al. Banff human organ transplant transcripts correlate with renal allograft pathology and outcome: importance of capillaritis and subpathologic rejection. J Am Soc Nephrol. 2022;33:2306–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators*. Combining donor-derived cell-free DNA fraction and quantity to detect kidney transplant rejection using molecular diagnoses and histology as confirmation. Transplantation. 2022;106:2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; the Trifecta Investigators. Antibody-mediated rejection without detectable donor-specific antibody releases donor-derived cell-free DNA: results from the Trifecta study. Transplantation. 2023;107:709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation. 2022;106:1061–1070. [DOI] [PubMed] [Google Scholar]
- 23.Reeve J, Bohmig GA, Eskandary F, et al. ; INTERCOMEX MMDx-Kidney Study Group. Generating automated kidney transplant biopsy reports combining molecular measurements with ensembles of machine learning classifiers. Am J Transplant. 2019;19:2719–2731. [DOI] [PubMed] [Google Scholar]
- 24.Madill-Thomsen K, Perkowska-Ptasinska A, Bohmig GA, et al. ; MMDx-Kidney Study Group. Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Am J Transplant. 2020;20:1341–1350. [DOI] [PubMed] [Google Scholar]
- 25.Reeve J, Bohmig GA, Eskandary F, et al. ; MMDx-Kidney Study Group. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight. 2017;2:e94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halloran PF, Madill-Thomsen KS, Pon S, et al. ; INTERCOMEX Investigators. Molecular diagnosis of AMR with or without donor-specific antibody in kidney transplant biopsies: differences in timing and intensity but similar mechanisms and outcomes. Am J Transplant. 2022;22:1976–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madill-Thomsen KS, Bohmig GA, Bromberg J, et al. ; the INTERCOMEX Investigators. Relating molecular T cell-mediated rejection activity in kidney transplant biopsies to time and to histologic tubulitis and atrophy-fibrosis. Transplantation. 2023;107:1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halloran PF, Bohmig GA, Bromberg JS, et al. ; INTERCOMEX Investigators. Discovering novel injury features in kidney transplant biopsies associated with TCMR and donor aging. Am J Transplant. 2021;21:1725–1739. [DOI] [PubMed] [Google Scholar]
- 29.Halloran PF, Bohmig GA, Bromberg J, et al. ; INTERCOMEX Investigators. Archetypal analysis of injury in kidney transplant biopsies identifies two classes of early AKI. Front Med (Lausanne). 2022;9:817324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cattaneo D, Crump RK, Farrell MH, et al. On Binscatter. Available at https://arxiv.org/abs/1902.09608. Published 2019. 2023.
- 31.Cattaneo MD, Crump RK, Farrell MH, et al. Binscatter Regressions. Available at https://arxiv.org/abs/1902.09615. Published 2019. 2023.
- 32.Venner JM, Famulski KS, Badr D, et al. Molecular landscape of T cell-mediated rejection in human kidney transplants: prominence of CTLA4 and PD ligands. Am J Transplant. 2014;14:2565–2576. [DOI] [PubMed] [Google Scholar]
- 33.Price PM, Safirstein RL, Megyesi J. The cell cycle and acute kidney injury. Kidney Int. 2009;76:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan MGM, Wang Y. Cell cycle-related clinical applications. Methods Mol Biol. 2022;2579:35–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.