Abstract
Wound healing is an intricate and fine regulatory process. In diabetic patients, advanced glycation end products (AGEs), excessive reactive oxygen species (ROS), biofilm formation, persistent inflammation, and angiogenesis regression contribute to delayed wound healing. Epigenetics, the fast-moving science in the 21st century, has been up to date and associated with diabetic wound repair. In this review, we go over the functions of epigenetics in diabetic wound repair in retrospect, covering transcriptional and posttranscriptional regulation. Among these, we found that histone modification is widely involved in inflammation and angiogenesis by affecting macrophages and endothelial cells. DNA methylation is involved in factors regulation in wound repair but also affects the differentiation phenotype of cells in hyperglycemia. In addition, noncodingRNA regulation and RNA modification in diabetic wound repair were also generalized. The future prospects for epigenetic applications are discussed in the end. In conclusion, the study suggests that epigenetics is an integral regulatory mechanism in diabetic wound healing.
Keywords: Diabetic wound, Epigenetics, Histone modification, DNA methylation, N6- methyladenosine, Non-coding RNA
1. Introduction
Diabetes is a chronic disease characterized by disturbances in blood glucose metabolism. Global estimates of diabetes patients in 2015 put the number at 415 million, with type 2 diabetes accounting for the majority. By 2040 [1], it is anticipated that the number would rise to 642 million [1]. The International Diabetes Federation projects that by 2021, there will be 536.6 million individuals worldwide who have diabetes, or 10.5% of the world's population. By 2045, the prevalence of diabetes is predicted to increase to 12.2% [2]. This demonstrates the growing social burden of diabetes. Diabetes mellitus can be associated with various complications, including retinopathy, neuropathy, renal failure, cardiovascular disease, and diabetic foot ulcers (DFUs) [3]. The incidence of DFUs is approximately 15–25% or even more [4]. Among patients with diabetes, these ulcers are linked to a higher likelihood of mortality, which occur before 80% of maputations in the lower extremities [5]. Currently, the main treatments for DFUs include debridement, wound dressing, skin grafting, hyperbarics, gene therapy [6]. Despite their prevalence and detrimental effects, therapy for diabetic wounds is often unsatisfactory [7]. Consequently, investigating the diabetes wound healing mechanism is essential.
One of the main causes for delayed diabetic wound healing is hyperglycemia. In diabetic patients, "metabolic memory" exists: diabetic complications are proceeding even after blood sugar control [8]. This may be related to epigenetic alterations of the cells due to chronic exposure to high glucose. T2DM pathogenesis is caused by a confluence of environmental and genetic factors [9], and epigenetics bridges the gap between genetics and the environment, playing an important role in many physiologic and pathologic conditions [10]. According to certain recent studies, epigenetic control of structural and immunological cells within wounds may affect the healing process and cell phenotypes in individuals with diabetes [11]. Epigenetics will help with precision medicine approaches to diabetic wound healing [12,13]. Epigenetics is the study of non-heritable and heritable genetic coding [14]. Epigenetics is a concept relative to genetics. Here, the definition from Giacomo is adopted, “the study of molecules and mechanisms that can perpetuate alternative gene activity states in the context of the same DNA sequence” [15]. In this concept, the study focuses on the involvement of histone modification, DNA methylation, noncoding RNA (ncRNA) regulation, and RNA methylation in diabetic wound repair. Furthermore, we discuss future prospects for epigenetic applications, which are expected to bring new insights into precision medicine approaches to diabetic wound healing.
2. Normal and diabetic wound healing
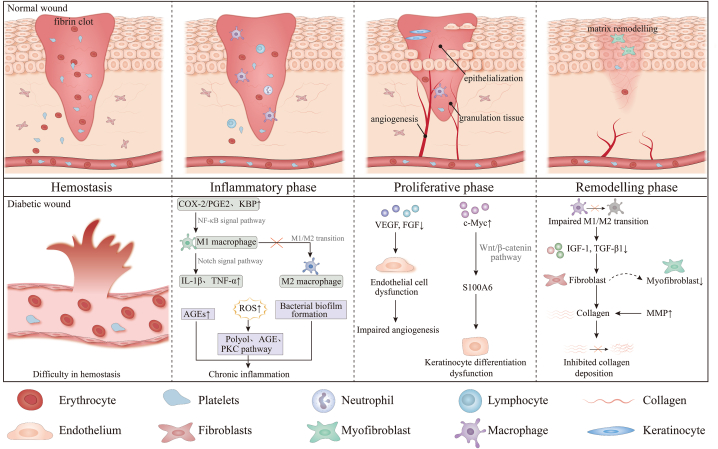
Hemostasis, inflammation, proliferation, and remodeling are the four stages of wound healing [16]. The following is a description of each stage's physiological processes (Fig. 1).
Fig. 1.
The molecular and cellular mechanisms in normal and diabetic wound healing. Abbreviations: AGEs:Advanced glycation end products; VEGF: Vascular endothelial growth factor; TNF-α: Tumor necrosis factor alpha; MMP: Matrix metalloproteinase; kallikrein-binding protein (KBP); ROS: reactive oxygen species; IL: Interleukin; COX-2/PGE2: Cyclooxygenase 2/prostaglandin E2; IGF-1: Insulin growth faction-1; TGF-β: Transforming growth factor; FGFs: Fibroblast growth factors
Wound healing is composed of four stages: hemostasis, inflammation, proliferation, and remodeling. In the hemostasis process, platelets are activated and aggregate, forming fibrin clot with the fibrin network. During the inflammatory phase, neutrophils are drawn to the wound region to remove germs, followed by macrophages and lymphocytes. In the proliferative stage, the characteristic changes are angiogenesis, development of granulation tissue and re-epithelialization. Endothelial cells, fibroblasts, and keratinocytes are active cells in this stage. In the remodeling phase, apoptosis and collagen remodeling are predominant. The diabetic wound healing process is more complex compared to normal wounds. An environment with high glucose levels might hinder wound healing by causing problems with hemostasis, persistent inflammation, poor angiogenesis, delayed re-epithelialization, and inhibited collagen deposition. Chronic hyperglycemia leads to persistent inflammation in diabetic wounds by specific mechanisms including increased AGEs, increased intracellular reactive oxygen species (ROS), bacterial biofilm formation, and abnormalities in M1/M2 macrophage polarization. Hyperglycemia also increases intracellular reactive oxygen species (ROS), leading to chronic inflammation via the polyol pathway, AGE pathway, and PKC pathway. Cyclooxygenase 2/prostaglandin E2 (COX-2/PGE2) and kallikrein-binding protein (KBP) are increased in diabetic wounds and promotes M1 polarization through NF-κB signaling. Notch signaling mediates increased secretion of IL-1β and TNF-α by M1 macrophages in diabetic wounds, leading to chronic inflammation. VEGF, FGF and other cytokines secretion is reduced, leading to endothelial cell dysregulation and impaired angiogenesis. The high glucose microenvironment increases c-Myc expression and activates S100A6 transcription through activation of the Wnt/β-catenin signaling pathway, leading to keratinocyte differentiation dysregulation. Decreased polarization of M1//M2 macrophages leads to decreased secretion of insulin growth faction-1 (IGF-1) and transforming growth faction-β1 (TGF-β1), which in turn leads to fewer myofibroblasts and reduced collagen deposition. At the same time, elevated matrix metalloproteinase (MMP) exacerbates collagen degradation and inhibits collagen deposition. In summary, a combination of above factors ultimately inhibits diabetic wound repair.
2.1. Hemostasis
The hemostatic phase begins as soon as the skin is injured [17]. At the site of vascular injury, platelets are activated and aggregate due to the exposure of subendothelial collagen, forming "platelet plugs" [18]. At the same time, this activates the coagulation cascade. With thrombin, fibrinogen cleaves into fibrin and forms a fibrin mesh that strengthens the platelet plug into a stable clot [19]. Moreover, the adhering platelets degranulate and produce cytokines, including vascular endothelial growth factor (VEGF), basic fibroblast growth factor (bFGF), transforming growth factor-β (TGF-β), and platelet-derived growth factor (PDGF). They participate in the subsequent stages of wound repair [[20], [21], [22], [23], [24], [25]].
2.2. Inflammatory phase
The aggregation of inflammatory cells is the main feature of this stage. Neutrophils and macrophages are two important cells. Interleukin-8 (IL-8) and other chemotactic cytokines attract neutrophils to the wound site [26,27]. When arriving at a wound, neutrophils initiate wound debridement by releasing diverse factors such as proteolytic enzymes, eicosanoids, ROS, and antimicrobial peptides [[28], [29], [30], [31]], initiating phagocytic function and generating neutrophil extracellular traps (NETs). Macrophages are another essential cell type in the inflammatory phase. The majority of studies are on macrophages that stem from monocytes, although tissue-resident macrophages exist. PDGF, TGF-β, catabolic elastin, collagen products, and so on are most times the main focus. Macrophages are able to be polarized to two distinct types in different microenvironments: proinflammatory (M1) and anti-inflammatory (M2). In early inflammation, IFN-γ and TNF-α polarize macrophages to M1 with microbicidal functions; in late inflammation, IL-4, IL-10, and IL-13polarize macrophages to M2, reducing inflammation and promoting skin cell proliferation, collagen deposition, and angiogenesis. Macrophages are known to interact with other structural cells and can serve as antigen presenting cells [32]. One way macrophages can spread to areas of infection or tissue injury is by adhering to endothelial cells that line blood vessels. This process is called extravasation [33]. In addition, macrophages can interact with fibroblasts to regulate tissue repair and remodeling [34]. More specifically, macrophages secrete growth factors such as TGF-α, epidermal growth factor, and keratinocyte growth factor, which stimulate the growth of fibroblasts and keratinocytes as well as the synthesis of collagen and extracellular matrix (ECM), which in turn causes wounds to granulate and reepithelialize [35]. Furthermore, macrophages can communicate with epithelial cells to coordinate immune responses and tissue repair [36]. Macrophages generate chemokines (such as TNF-α, VEGF, and TGF-β) to promote endothelial cell migration and aid in the angiogenesis of microvascular endothelial cells, as well as proteases like MMP to break down dense fibrin networks [37]. Besides, macrophages are professional antigen-presenting cells (APCs). They can engulf pathogens or particles through phagocytosis and break down these substances within lysosomes, thus presenting them to helper T cells (CD4+ T cells) [38]. This interaction is essential for activating and coordinating adaptive immune responses. As a result, macrophages are essential for the healing process as wounds move from the inflammatory to the proliferative phases [39].
2.3. Proliferative phase
Angiogenesis, granulation tissue formation, and re-epithelialization make up the proliferative phase. At this stage, the active cells are keratinocytes, fibroblasts, and endothelial cells. VEGF stimulates angiogenesis and boosts endothelial cell motility [40]. Fibroblasts proliferate, migrate to the damaged location, and create copious amounts of collagen I and III [41,42]. Granulation tissue is produced by extracellular matrix, inflammatory cells, and newly formed capillaries to fill up tissue defects and establish the environment for the covering of epidermal cells [43,44]. Additionally, the fibroblasts differentiate into myofibroblasts pulling the wound edges together, reducing the area to be re-epithelialization [45]. Keratinocytes are connected by the cell-cell junction to re-establish epidermal barrier function [22].
2.4. Remodeling phase
The final phase consists of cell apoptosis and collagen remodeling. New vessels are regressed. Myofibroblasts undergo apoptosis to make room for fibroblasts. A more stable ECM is synthesized. Type III collagen undergoes conversion into mature type I collagen [[46], [47], [48]]. These modifications increase the skin's tensile strength.
2.5. Characteristics of diabetic wound healing
Diabetes-related wounds may heal slowly or not at all as a result of hypoxia, tissue ischemia, and an environment with high glucose levels impeding the progression of these physiological healing processes.
Compared to normal wounds, diabetic wound healing processes are more intricate. In diabetes, chronic hyperglycemia can raise levels of AGEs [49]. Reduced sugars combined nonenzymatically with proteins, phospholipids, or DNAs result in the diverse class of substances known as AGEs. Prior research has demonstrated that AGE buildup in tissues results in diabetic wounds with reduced proliferation and chronic inflammation [50]. Oxidative stress is another factor involved in diabetic wound repair. Biological oxidative stress promotes wound repair and disinfection, but excessive oxidative stress results in sustained inflammation and nonhealing wounds. In diabetes, hyperglycemia also augments intracellular ROS, resulting in excess oxidative stress [51]. By interfering with several metabolic pathways, including the polyol, AGE, and PKC pathways, ROS impede the healing of wounds [52]. Bacterial biofilm formation further hinders diabetic wound healing [53,54]. Biofilms cause constant inflammatory response, and most anti-inflammatory antibiotics are ineffective [54]. Another characteristic of diabetic wounds is dyspolarization of macrophages, which prevents M1 macrophages from polarizing into M2 macrophages. The recruitment of macrophages in the wound is determined by the production of monocyte chemoattractant protein-1 (MCP-1) and its receptor, CC-chemokine receptor 2 (CCR2) [55]. Anti-MCP-1 injections into diabetic mice's wounds can inhibit the growth of M1 macrophages and encourage the healing of wounds [56]. Furthermore, M1 polarization is facilitated by the classical pathway known as the NF-κB signaling pathway. Using single-cell RNA sequencing, Wolf's group showed that diabetic wounds had higher NF-κB-mediated inflammation than control wounds, partly because of higher cyclooxygenase 2/prostaglandin E2 (COX-2/PGE2). The M1 phenotype was then reversed and wound healing was enhanced by blocking the COX-2/PGE2 pathway in mouse models by genetic engineering or by employing COX-2 nanotherapy specifically targeted at macrophages in mouse tissue macrophages [57]. An additional investigation revealed that obstructing elevated amounts of kallikrein-binding protein (KBP) in DFUs reduced the NF-κB signaling pathway, which in turn reduced M1 polarization and improved the management of diabetic wounds [58]. One important regulating element in the biological operations of macrophages is Notch signaling. According to experimental evidence, M1 macrophages secrete less IL-1β and TNF-α in pre-diabetic mice with genetic abnormalities in Notch signaling, which promotes wound healing and lowers inflammation levels [59]. Furthermore, in TLR-stimulated macrophages, the PI3K/AKT pathway's activation has an anti-inflammatory effect. Insulin-regulated macrophages changed from an M1 to an M2 phenotype and took part in the anti-inflammatory process via the PI3K/AKT signaling pathway, according to a research performed on diabetic burn wounds, which improved chronic wound healing [60]. In conclusion, numerous studies have demonstrated that reversing inflammation mediated by macrophages can improve diabetes healing. Besides, hyperglycemia impairs micro and macrovascular, which affects angiogenesis during wound healing [61,62]. Meanwhile, macrophages, neutrophils, and other cells produce excessive pro-inflammatory cytokines and fewer pro-angiogenesis factors in the hyperglycemic environment [63,64]. In addition, a high glucose microenvironment elevated the levels of c-Myc, inhibiting keratinocyte differentiation by S100A6 transcriptional activation via the Wnt/β-catenin signaling pathway in diabetic wound [65]. Reduced M1 macrophage transformation into M2 macrophages resulted in decreased insulin growth factor-1 (IGF-1) and transforming growth factor-β1 (TGF-β1) secretion, which in turn caused a decrease in myofibroblasts and decreased collagen synthesis. Meanwhile, increased matrix metalloproteinase (MMP) exacerbates collagen degradation and ultimately inhibits collagen deposition [66]. These changes present a significant challenge to diabetic wound healing.
3. Epigenetic modification in diabetic wound healing
In this review, we outlined the aberrant changes and processes underlying diabetic wounds from an epigenetics viewpoint in recent years. The study of epigenetics is predicated on the expression of genes without alterations to DNA sequences. It has garnered significant interest in the 21st century and is involved in numerous diseases [67]. As follows, we introduced abnormal regulatory mechanisms of histone modification, DNA methylation, ncRNA and N6-methyladenosine (m6A) in diabetic wounds in recent years from gene transcription to posttranscriptional regulation (Table 1).
Table 1.
Regulation of histone modification and DNA methylation in normal and diabetic wound healing.
| Types of epigenetic modifications | The type of enzymes | enzymes | Sites of modification | Target genes | Regulation of target gene (up/down) | Effect in wound healing | Refs |
|---|---|---|---|---|---|---|---|
| Histone methylation | Histone methyltransferases (HMTs) | Mixed-lineage leukemia 1 (MLL1) | H3K4me3 | TLR4 | up | Promoted wound macrophage-mediated inflammation | [68] |
| H3K4me3 | NF-κB inflammatory genes | up | Promoted wound macrophage-mediated inflammation | [69] | |||
| Setdb2 | H3K9me3 | NF-κB inflammatory genes | down | Promoted wound macrophage-mediated inflammation | [70] | ||
| SET domain containing 2 (SETD2) | H3K36me3 | AKT and mTOR | down | Inhibited the proliferation and migration of HaCaT | [71] | ||
| SET7/9 | – | HIF-1α | down | Contributed to hyperglycemia-induced inflammation and reduces angiogenesis | [72] | ||
| Enhancer of Zeste Homolog 2 (EZH2) | H3K4me3 | HIF-1α | up | accelerated the activation of dermal fibroblasts | [73] | ||
| H3K27me3 | ZNF24 and Runx1 | down | Promoted angiogenesis | [74] | |||
| Histone demethylases (HDMs) | Lysine Specific Demethylase 1 (LSD1) | – | – | – | Resulted in dysregulation of endothelial cell proliferation | [75] | |
| JMJD 3 | H3K27me3 | NF-κB inflammatory genes | up | Elevated levels of inflammatory cytokines | [76,77] | ||
| Histone acetylation | Histone acetyltransferases (HATs) | MOF | H4K16 | TNF-α/NF-κB inflammatory genes | up | Promoted wound macrophage-mediated inflammation | [78] |
| Histone deacetylases (HDACs) | HDAC5 | – | KLFs | down | Regulated macrophage activation | [79] | |
| sirtuins | H3K27 | IL-6 and TNF-α | down | Reduced the number of M1 macrophages that produce cytokines | [80] | ||
| HDAC2 | – | HO-1/Sirt1 pathway | down | caused decreased tube formation and proliferation of endothelial progenitor cells. | [81] | ||
| Histone lactylation | Acyltransferase | – | – | – | – | enhanced the expression of genes in M1 macrophages that support wound healing. | [82] |
| Histone phosphorylation | – | – | H3 | – | – | Prevented type 2 diabetic cardiomyopathy in mice. | [83] |
| Histone ubiquitination | – | – | H2AK119, H2BK120 | – | – | stopped mice from developing type 2 diabetic cardiomyopathy. | [83] |
| DNA methylation | DNA methyltransferases (DNMTs) | DNMT1 | – | Notch1, PU.1, and Klf4 | down | Diminished wound macrophages and biases their polarization toward M1 | [84] |
| – | Ang-1 | down | caused NF-κB to become permanently activated, which in turn caused endothelial dysfunction. | [85] | |||
| – | TGF-β, WAKMAR1, E2F1 | down | impeded the migration, adhesion, and proliferation of keratinocytes | [86] | |||
| DNA demethylase | Ten-eleven translocation-2 (TET2) | – | MMP-9 | up | disrupted the equilibrium between synthesis and degradation of the extracellular matrix. | [87,88] | |
| thymine-DNA glycosylase (TDG) | – | MMP-9 | up | Inhibited migration of keratinocytesed | [89] |
Abbreviations: HMTs: Histone methyltransferasesand; HDMs: Histone demethylases; MLL1: Mixed-lineage leukemia 1; EZH2: Enhancer of Zeste Homolog 2; TLR4: Toll-like receptor 4; SETD2: SET domain containing 2; LSD1: Lysine specific demethylase 1; HIF-1α: Hypoxia-inducible factor-1α; HATs: Histone acetyltransferases; NF-κB: nuclear factor kappa-B; JMJD: JmjC domain-containing family; TNF-α: Tumor necrosis factor alpha; KLFs: Kruppel-like factors; TET2: Ten-eleven translocation-2; HDACs: Histone deacetylases; TDG: thymine-DNA glycosylase; TGF-β: Transforming growth factor; MMP-9: Matrix metalloproteinase-9; WAKMAR1: wound and keratinocyte migration-associated lncRNA 1; E2F1: E2F Transcription Factor 1; IL: Interleukin.
3.1. Histone modification
There are five different forms of histones, which are tiny, incredibly basic proteins: H1, H2A, H2B, H3, and H4. The nucleosome, the basic chromatin unit, is formed by the octamers of H2A, H2B, H3, and H4 combined with DNA sequences [90].The N-terminal tail of each histone subunit can be altered by phosphorylation, ubiquitination, acetylation, lactylation, or methylation, which can activate or inhibit transcription [91].
3.1.1. Histone methylation/de-methylation
Histone methylation is the methyl transfer from S-adenosylmethionine (SAM) to specific amino acid residues on histones. It often manifests at the lysine (Lys or K) and arginine (Arg or R) residues of H3 and H4 [92]. Histone methyltransferases (HMTs) and histone demethylases (HDMs) can catalyze dynamically reversible histone methylation, which offers a route for modulating gene expression [93].
It has been demonstrated that HMTs control macrophage-mediated inflammation during wound healing in diabetic individuals. A methyltransferase specific to H3K4me3 that causes gene activation is known as mixed-lineage leukemia 1 (MLL1). Studies have demonstrated that MLL1 can drive TLR4 (Toll-like receptor 4) expression by increasing H3K4me3 at the TLR4 promoter, leading to persistent inflammation in diabetic mice [94,68]. Furthermore, by raising H3K4me3 at the NF-κB binding spots on promoters of inflammatory genes in macrophages, MLL1 may potentially stimulate the expression of inflammatory cytokines, postponing the healing of wounds [69]. Like MLL1, Setdb2 regulates inflammatory cytokine genes by binding to the NF-κB region on their promoter. However, Setdb2 is a specific enzyme to H3K9me3, which helps maintain chromatin in a state where the promoter cannot bind to transcription factors, silencing gene transcription. Hence, the reduction in Setdb2 expression results in persistent inflammation in macrophages, which is not conducive to wound healing [70]. These findings indicate that Setdb2 and MLL1 may be therapeutic targets to reduce chronic inflammation in diabetic wounds.
In the proliferative stage of diabetic wound healing, HMTs also take part. The only HMT identified to trimethylate histone 3 lysine 36 (H3K36me3) is SET domain containing 2 (SETD2). Researchers discovered that via activating the AKT/mTOR signaling pathway, SETD2 knockdown enhances the migration and proliferation of human immortalized keratinocyte cell lines (HaCaT), hence boosting wound healing [71]. Proangiogenic factor expression is another thing that HMTs can influence. For instance, in hypoxic and hyperglycemic conditions, SET7/9 can methylate HIF-1α at lysine residue 32 and suppress the levels of hypoxia-inducible factor-1α (HIF-1α), which delays the healing of wounds [72]. Enhancer of zeste homolog 2 (EZH2), on the other hand, can stimulate HIF-1α H3K4me3 to raise HIF-1α expression, improve wound repair in diabetic rats wounds, and increase dermal fibroblast viability [73]. In a different investigation, EZH2 repressed ZNF24 and Runx1 transcription through H3K27me3, thereby promoting VEGFA expression and angiogenesis and accelerating wound healing [74].
HDMs include lysine specific demethylase 1 (LSD1) and the JmjC domain-containing family (JMJD) [95]. LSD1 knockdown results in the dysregulation of endothelial cell proliferation and delays wound healing [75]. By deleting inhibitory H3K27me3 from the promoters of NF-κB-regulated genes, JMJD3 can upregulate inflammatory cytokine levels, which is harmful to wound healing. According to one study, the most common circulating SFA in T2D, palmitate (C16:0), increased JMJD3 expression through a signaling pathway that is dependent on TLR4 and MyD88 [76]. In a related study, the JAK1/3/STAT3 pathway governed the constant elevation of JMJD3 expression in diabetic wound macrophages during the late inflammatory phase. Additionally, researchers have discovered that the primary regulator of this signaling pathway in diabetic wounds is IL-6. In summary, JMJD3 targeting offers a workable therapeutic strategy for enhancing diabetic tissue regeneration and lowering NF-κB inflammatory cytokines [77].
3.1.2. Histone acetylation/deacetylation
Histone deacetylases (HDACs) and histone acetyltransferases (HATs) regulate the acetylation of histones [96,97]. The acetyl group (CH3COO-) of acetyl coenzyme A is transferred by HATs to the ε-amino group of a particular lysine residue at the N-terminal end of histones [98]. Histone acetylation can neutralize the positive charge on the lysine residue, impair histone binding to DNA, and facilitate the binding of transcriptional regulatory proteins or transcription factors to DNA, hence boosting gene transcription [99,100]. HDACs work in the opposite way [101]. There are 18 HDACs in mammals, including HDAC1-11 and SIRT1-7 [102].
Histone acetylation/deacetylation influences in wound healing inflammation. MOF is a HAT that promotes the expression of TNF-α/NF-κB inflammatory genes by increasing H4K16 acetylation of inflammatory gene promoters in macrophages, leading to delayed wound healing [78]. HDACs can regulate inflammation by influencing monocyte-macrophage phenotype. HDAC5 regulates macrophage activation by suppressing zinc finger protein Kruppel-like factor (KLF) expression [79]. A greater M1 macrophage phenotype that is detrimental to wound healing is produced when sirtuins are inhibited because this leads to an increase in the levels of IL-6 and TNF-α via H3K27 acetylation [80]. In addition, a study found that anti-inflammatory Ly6Clow monocyte subtypes were enriched in HDAC inhibitor (HDACi) cultured bone marrow progenitor cells (BMMP) [103].
The role of HDACs in angiogenesis depends on the type. SIRT1/3/6 deficiency inhibits angiogenesis in wound healing [104]. However, HDAC2/8/11 plays a negative role in angiogenesis. Patients with chronic DFUs had higher levels of HDAC2 expression in their endothelial progenitor cells (EPCs). By protecting EPCs from high glucose-induced impairment, HDAC2 inhibitors can enhance wound healing by encouraging proliferation and tube formation. The mechanism is that HDAC2 can down-regulate the HO-1/Sirt1 pathway [81]. Human umbilical endothelial cells' (HUVECs') ability to form tubes and migrate has been shown to be decreased by HDAC8, which inhibits angiogenesis [105]. In addition, the latest study found that HDAC11 promotes HUVEC pyroptosis [106].
3.1.3. Other histone modifications
Moreover, histones can be ubiquitinated, lactylated, and phosphorylated, but there are few studies on how these modifications affect diabetic wound healing. In 2019, Zhang et al. first discovered the existence of lactate-derived histone lactylation in M1 macrophage polarization [82]. The "Warburg effect," or aerobic glycolysis, is a metabolic characteristic of M1 macrophages that results in the generation of lactate when large amounts of glucose are ingested even in a condition of oxygen [107]. Lactic acid can produce lactoyl-CoA (lactyl-CoA), which acyltransferase uses to add a lactoyl group to the lysine tail of histones [108]. During the investigation, they found that histone lysine lactylation (Kla) increased in M1 polarization triggered by lipopolysaccharide (LPS) and interferon-gamma (IFN-γ) as lactate accumulated. Subsequent analysis revealed that Kla was positively correlated with these genes rather than inflammation and was located in the promoter areas of genes relevant to wound healing. Thus, histone lactylation in the late stages of M1 macrophages is regarded as a “switch” to activate gene expression associated with wound healing to promote homeostasis [82,109]. The phrase "histone phosphorylation" describes the phosphorylation of amino acid residues at the histone's N terminus, primarily affecting histones H1, H2A/H2B, H3, and H4. Histone phosphorylation contributes in DNA repair, gene transcription, apoptosis, and chromosomal condensation, and it regulates a number of biological processes such as cell growth and division, along with other changes such as histone acetylation [110]. Current studies on histone ubiquitination mainly focus on the ubiquitination of H2A and H2B. In general, ubiquitinated H2A occurs mostly in silenced chromatin, while ubiquitinated H2B occurs mostly in activated chromatin [111]. Mice with insulin resistance (IR) and type 2 diabetes had higher levels of ubiquitination of H2AK119 and H2BK120 in their hearts than in any other study. Esculetin prevented type 2 diabetic cardiomyopathy by reversing ubiquitination of histone H2A/H2B, dimethylation, acetylation, and phosphorylation of H3 [83]. The mechanism is not entirely known, though. There is further work to be done to determine how these histone alterations impact the healing of diabetic wounds.
3.2. DNA methylation
For gene expression, not only must histone modification sites be activated, but the promoter region must also be hypomethylated. The process by which the methyl group of S-adenyl methionine (SAM) moves to the fifth carbon of a cytosine residue is known as DNA methylation, or m5C [112]. This process can suppress transcription, causing gene silencing. DNA methylation can alter DNA geometry, and mechanical properties and affect chromatin accessibility, thereby inducing DNA to pack more tightly around histones and ultimately inhibiting gene transcription [113]. DNA methylation is mediated by a class of DNA methyltransferases (DNMTs) that includes DNMT1, DNMT3A, and DNMT3B, while demethylases, such as ten eleven translocation (TET) proteins, can eliminate the modification [114]. During the 5 mC to cytosine conversion, intermediate products are generated, such as 5caC, 5 fC, and 5hmC. However, recent studies have found that certain products exist stably and modulate diseases [115]. However, due to the paucity of literature on intermediates, the focus of this study will be on 5 mC in diabetic wound repair.
Diabetes is a complicated systemic disease. Hyperglycemia has a far-reaching impact on systemic cells, for which the DNA methylation mechanism may account. Diabetic wound healing is hampered in large part by the persistent inflammatory response in a hyperglycemic environment. Bahu et al. discovered that the promoters of pro-inflammatory M1 genes were hypomethylated in hyperlipidemia and T2DM ischemic muscles, but the promoters of anti-inflammatory M2 genes were hypermethylated [116]. Additionally, the oxidative stress caused by NOX-2 in diabetic rats' bone marrow-derived hematopoietic stem cells (HSCs) resulted in the overexpression of DNMT1, which in turn upregulated the methylation of PU.1, kruppel-like factor (Klf4), and Notch1, decreased the quantity of wound macrophages, and polarized them toward M1 [84]. Pasquier et al. examined the differences in DNA methylation within circulating monocytes between the acute diabetic Charcot foot (CF) and non-CF groups. According to data, genes with variable DNA methylation were involved in monocyte migration and osteoclast differentiation [117]. These dysregulations of DNA methylation in systemic cells may be a contribution to refractory wounds diabetic. These new inflammatory mechanisms may account for hard-to-healing wound healing in T2D.
Impaired angiogenesis is largely caused by vascular endothelial cell dysfunction brought on by hyperglycemia. Under high-glucose, the miR-200b promoter is hypomethylated in human microvascular endothelial cells, which results inan increase in gene expression and impairs wound healing [118]. Furthermore, Zhao et al. discovered that momentary hyperglycemia increased the production of DNMT1 in vascular endothelial cells, which resulted in hypermethylation and decreased Ang-1 expression. This, in turn, caused long-term activation of NF-κB and endothelial dysfunction [85].
Matrix metalloproteinase-9 (MMP-9) is a kind of IV collagenase that regulates the re-epithelialization process and is released by keratinocytes. It is involved in the remodeling phase. The equilibrium between the creation of extracellular matrix and catabolism is upset in diabetic patients due to increased MMP-9 expression in keratinocytes, which impairs diabetic wound repair [119]. It has been discovered that in AGE-BSA-induced keratinocytes, AGEs up-regulate the expression of demethylase ten-eleven translocation-2 (TET2), which results in DNA demethylation of the MMP-9 promoter and higher MMP-9 levels [87]. TET2-interacting long ncRNA (TETILA) was discovered in another study to impede wound repair in diabetic skin by indirectly activating MMP-9 promoter demethylation, which results in MMP-9 hyperactivation [88]. Thymine-DNA glycosylase (TDG), which is also attracted by TETILA, aids in the demethylation of the MMP-9 promoter.Additionally, it has been discovered that the DNA damage-inducible 45a (GADD45a), a DNA demethylation regulatory protein, stimulates MMP-9 transcription, promotes MMP-9 DNA demethylation via TDG, and prevents keratinocyte migration in diabetic patients [89]. Furthermore, "wound and keratinocyte migration-associated lncRNA 1" (WAKMAR1) was found in much lower amounts in human wounds that do not heal. Mechanistically, WAKMAR1 enhances the functions of keratinocyte proliferation and migration by inhibiting E2F1 (E2F Transcription Factor 1) DNA methylation, thus promoting diabetic wound healing [86].
Aside from coding genes, studies have revealed that DNA methylation in regulatory elements is involved in wound healing. B1 (in rodents) and Alu elements (in humans) are the most common retrotransposons in the genome. They are generally methylated and maintain DNA stability by forming heterochromatin. In both diabetic and second-degree burn rats, researchers have found that B1siRNA therapy promotes wound healing [120,121]. siRNA therapy can directly add a methyl group at B1 elements, a process known as RNA-directed DNA methylation (RdDM) [122].
Fortunately, some studies report that certain physical therapy can alter DNA methylation modification and promote wound healing. For example, negative pressure wound therapy was proven the clinical effect may be owing to the inhibition of complement activation via methylation change [123]. Likewise, methylation of DNA and histones can be induced in non-thermal atmospheric pressure plasma (NTAPP) by NO, which in turn can stimulate the production of growth factors and cytokines. Low NO levels increase the activity of pluripotent genes Sox2, Oct4, and Nanog in human embryonic stem cells (ESCs), according to a previous study [124], but high NO levels encourage ESC differentiation. This could be the process by which NO functions [125]. The gaseous signaling molecule hydrogen sulfide (H2S), which enters cells and tissues quickly, is becoming more and more studied for its potential to aid in wound healing. It has been discovered that H2S increases the expression of miR-126-3p and subsequently increases angiogenesis in diabetic rats via lowering the methylation level of miR-126-3p upstream gene sequences and DNMT1 protein levels that are caused by high glucose [126].
3.3. Noncoding RNA(ncRNA)
ncRNA is a type of nonprotein-coding RNA that is engaged in numerous biological processes and is crucial in the regulation of transcription and posttranscription. ncRNAs can be classified as small nuclear RNAs (snRNAs) or long noncoding RNAs (lncRNAs) based on the quantity of nucleotides they contain. snRNAs can be separated into circular RNAs (circRNAs) and microRNAs (miRNAs) based on distinct production pathways [127]. Since the discovery of the RNA interference process, researchers have paid increasing attention to the processes of non-coding RNA (ncRNA) regulation [128]. Diabetic wound healing is regulated by an intricate modulatory network that is created by the interactions of lncRNAs, miRNAs, and circRNAs (Fig. 2).
Fig. 2.
Regulation of the non-coding RNA in in normal and diabetic wound healing
The diabetic wound microenvironment causes abnormal expression of non-coding RNAs (ncRNAs), which in turn controls the expression of genes downstream that are involved in inflammation, angiogenesis, proliferation, and remodeling during different phases of diabetic wound repair. The figure shows the expression of ncRNAs and the downstream genes they regulate in normal and diabetic wounds.
3.3.1. microRNA (miRNA)
miRNAs, which have an average length of 22 nucleotides, are involved in the regulation of posttranscription.By attaching to the 3′ or 5′ untranslated region of the target mRNA, miRNAs suppress the expression of genes at the post-transcriptional stage by either translational silencing or mRNA degradation [129,130]. According to a review [131], miRNAs affect hemostasis, inflammation, angiogenesis, re-epithelialization, and remodeling phases of diabetic wound repair. The basic prerequisite for wound repair is an inflammatory response. The imbalance of pro- and anti-inflammatory signals on the site caused by abnormal miRNA expression has an impact on wound repair. Previous studies have reported that significantly lower miRNA-129-2-3p expression in type 2 diabetic rats may be associated with significantly higher neutrophil counts and delayed wound healing compared to nondiabetic mice [132]. Furthermore, miRNAs play a role in controlling the expression of chemokines and inflammatory proteins. Studies conducted in vitro and in vivo have demonstrated that miRNA-497 can promote wound epithelialization and granulation tissue development in diabetic mice by reducing pro-inflammatory factors such as TNF-α, IL-1, and IL-6 [133]. By blocking the TLR4/NF-κB axis, miR-146a facilitated M2 macrophage polarization, decreasing inflammatory response and hastening diabetic wound healing, as shown by Peng et al. [134].
Several investigations have demonstrated that miRNAs regulate angiogenesis and have a role in the healing of diabetic wounds by stimulating the release of growth factors and angiogenic cytokines, as well as the migration and proliferation of topical microvascular endothelial cells. Extracellular vesicles (EVs) recovered from diabetic foot ulcer wound fluid contain miR-195-5p and miR-205-5p, which limit VEGFA production to reduce angiogenesis [135]. By enhancing fibroblast growth factor 4 (FGF4) and VEGF expression for angiogenesis via the FGF4/p38MAPK pathway, the miR-106a-5p generated from mmu_circ_0001052-modified ADSC -Exos accelerated diabetic wound repair [136]. Additionally, by inhibiting Homeodom-Interacting protein kinase 2 (HIPK2), miR-221-3p improved wound repair in diabetic mice and enhanced the motility and migration of HUVECs [137]. Similar findings were observed in another work, which found that via regulating the PTEN/AKT/HIF-17α/VEGF pathway, miR-17-5p in EVs generated from human umbilical cord mesenchymal stem cells enhanced diabetic wound healing and exhibited regenerative and protective functions on HG-induced endothelial cells [138].
In the term of miRNA, much more was found in diabetic wound healing, particularly with more growth factors such as FGF7, also the interaction with the microbiota. According to one study, miR-155 prevented type 2 diabetes mice's wounds from reepithelializing by suppressing the production of FGF-7 in diabetic wounds, which in turn prevented KC (Keratinocyte) from proliferating and migrating [139]. In recent years, gut microbiota–miRNA interaction has received increasing attention [140]. miRNAs can influence the makeup of the gut microbiota, along with the gut microbiota can modulate host miRNA expression through metabolites such as lipopolysaccharide (LPS), amyloids, and butyrate, ultimately altering host physiology and pathology [141]. Through its interactions with the gut microbiota, miR-155 has been shown to have anti-inflammatory effects. Bitar et al. discovered miR-155 and miR-144a in IECs (major cells that engage with gut bacteria and maintain intestinal homeostasis) in patients with acute Vibrio cholerae infection, suppressing the immune system at the gut level and lowering cytokine production [142]. In summary, research on the relationship between gut microbiota and miRNA may offer a fresh approach to the management of diabetic wounds.
The epithelial-to-mesenchymal transition (EMT), in which cells change from epithelial to mesenchymal, is a critical element in wound healing. E-cadherin (E-cad, Cdh1) is downregulated and N-cadherin (N-cad, Cdh2) is upregulated, which are key features of EMT [143]. There are three primary subtypes of EMT: Type 1 EMT, which is linked to embryogenesis; Type 2 EMT, which is linked to tissue regeneration and fibrosis; and Type 3 EMT, which is linked to metastasis [144]. To repair tissues damaged by inflammation and trauma, epithelial cells perform Type II EMT, where they develop into motile, fibroblast-like cells [145]. Through negative post-transcriptional regulation of target gene expression, miRNA can control EMT.
In comparison to normal skin tissue, diabetic skin tissue exhibits a significantly greater expression level of miR-203 [146], which has a positive correlation with the severity of diabetes mellitus. Mice with decreased wound repair as a result of miRNA-203's downregulation of IL-8 production, inhibition of the EMT process, and inhibition of keratinocyte proliferation and migration [147].
3.3.2. Long noncoding RNA (lncRNA)
LncRNAs are a class of RNAs with over 200 nucleotides that do not encode proteins. Studies have shown that differential lncRNAs have been detected in diabetic foot wounds and the expression network analysis has been performed [148,149]. Mechanistically, lncRNAs can regulate nearby chromatin structure and/or gene expression in cis and perform cellular functions in trans [150]. Compared to the lncRNA locus (in cis), there are more studies of lncRNA transcripts (in trans) function in diabetic wound. In AGE-treated rat primary dermal fibroblasts, lncRNA lncURIDS has been identified to stabilize procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1(Plod1) protein, which dysregulates collagen production and delays wound healing [151]. Furthermore, through their interactions with transcription factors, lncRNAs influence the transcriptional regulation of target genes like HIF-1α and connective tissue growth factor (CTGF), which in turn affects the healing of diabetic wounds [152,153]. Furthermore, as competing endogenous RNAs (ceRNAs), lncRNAs can bind to base-matched miRNAs in a competitive manner and slow down the degradation of mRNAs [[154], [155], [156], [157]].
Many recent studies have demonstrated the critical role lncRNAs play in the inflammatory process that occurs during the diabetic wound repair. The aberrant increase in ROS in the wound of diabetic rats promotes lymphocyte apoptosis and causes the body to up- and down-regulate pro- and anti-apoptotic proteins, which increases the apoptosis of local wound cells and delays healing [158]. The concentration of serum ROS can be controlled by lncRNA-Lethe and lncRNA-neutrophin 5 (NT5), respectively. Through the nuclear factor-κB (NF-κB) signaling pathway, lncRNA-Lethe controls the production of decreased nicotinamide adenine dinucleotide phosphate (NADPH) oxidases 2, which in turn affects the quantity of ROS in the serum. However, via raising blood ROS and IL-1 concentrations, lncRNA-NT5 suppresses IL-10 and insulin release, delaying wound repair in diabetic rats [159]. Furthermore, loss of WAKMAR2, a long non-coding RNA involved with wound and keratinocyte migration, stimulates the NF-KB signaling pathway and encourages an excessive amount of inflammation during diabetic wound recovery [160]. Furthermore, it has been shown that lncRNAs can modulate diabetic wound inflammation by regulating the M1/M2 polarization [161]. By encouraging the production of STAT1, the lncRNA GAS5 converts M1 macrophages into M2 macrophages, which in turn enhances diabetic wound healing [162]. Human keratinocyte-derived exosomal MALAT1 upregulated MFGE8 through miRNA-1914-3p, thereby enhancing macrophage phagocytosis, converting to M2 phenotype and reducing apoptosis to promote diabetic wound healing [163].
By controlling several signaling elements involved in wound healing, lncRNAs can control angiogenesis. Research has demonstrated that long noncoding RNAs (lncRNAs) stimulate the growth and migration of topical microvascular endothelial cells, which in turn stimulates angiogenesis in diabetic wounds. The upregulation of exosome MALAT1 from macrophages treated with high glucose inhibited the expression of miR-150-5p, significantly increased the expression of resistin in macrophages, and promotes vascular disease [164]. In HG-treated HUVECs, GAS5 overexpression aided in tubule formation, cell proliferation, and the repair of diabetic wounds. Furthermore, GAS5 promoted angiogenesis in DFUs mice by activating the HIF1A/VEGF pathway, which sped up wound healing [153]. lncRNA H19 is associated with angiogenesis and its expression level is remarkably lower in ulcerated tissues of diabetic patients compared to normal tissues. Research has demonstrated that via attracting serum response factor (SRF) to the CTGF promoter region, lncRNA H19 up-regulates the production of CTGF, encouraging angiogenesis and accelerating DFU repair in diabetic rats [152].
LncRNA can suppress fibroblast and keratinocyte apoptosis, increase their migration and proliferation, and hence aid in the healing of diabetic wounds. LncRNA H19 in ADSC-derived exosomes increased SOX9 expression via miR-19b, which accelerated human skin fibroblast proliferation, migration, and invasion [165]. Moreover, it was discovered that MALAT1 binds to miR-106a-5p in diabetic mouse wound models to raise ZNF148 levels, which in turn promotes HaCaT cell migration, proliferation, and apoptosis [166].
LncRNA could play an impact in the extracellular matrix's (ECM) remodeling during wound healing. In comparison to non-diabetic individuals, the expression level of lncRNA (lncRNA-TETILA), which interacts with tet methylcytosine dioxygenase 2 (TET2), was considerably higher in the skin tissue of diabetes patients. A high quantity of lncRNA-TETILA may encourage matrix metalloproteinase 9 (MMP-9) promoter demethylation and lower MMP-9 expression levels, which would degrade ECM and slow wound healing. Diabetes wound healing can be sped up by blocking the production of lncRNA-TETILA in wound tissue, which can also promote HaCaT cell migration [88]. lncRNA and miRNA interactions are involved in ECM reconstruction. The lncRNA-H19/miRNA-152-3p/phosphatase and tensin homolog (PTEN) axis has been shown to regulate Fb viability and secretory capacity and affect DFU healing [155]. Additionally, in humans with heat-damaged tissues, lncRNA XIST targets miR-29b-3p/COL1A1 to enhance extracellular matrix production [167]. What's more, Liang et al. found that downregulation of lnc RNA NEAT1 inhibited scar fibroblast viability, migration, and EMC formation by regulating miR-196b-5p/FGF2 (fibroblast growth factor 2) [168].
3.3.3. Circular RNA (circRNA)
CircRNA can regulate the inflammatory response of diabetic wounds by targeting miRNAs. Research has demonstrated that through controlling the miR-384/LIN28B axis, CircBPTF can lessen oxidative stress brought on by hyperglycemia and inflammation of wound tissue [169]. Within a different investigation, circ-Snhg11 in anoxic ADSC-derived exosomes attenuated inflammation in wounds of diabetic rats by targeting miR-144-3p and HIF-1α and inducing M2 macrophage polarization [170]. Additionally, mmu_circ_0000250 in ADSC-derived exosomes activated autophagy to prevent apoptosis and enhanced wound repair in diabetic rats by sponging miR-128-3p and upregulating SIRT1 [171].
Through the regulation of autophagy and apoptosis, circRNA can impact the viability of endothelial cells and angiogenesis. By focusing on the miR-212-3p/SIRT5 axis and triggering autophagy to prevent hyperglycemia-induced damage to endothelial progenitor cells (EPCs), Circ-Klhl8 overexpression facilitates diabetic wound repair [172]. Research has demonstrated that by adsorbing miRNA-128-3p, the up-regulation of circRNA-0000250 in diabetic rats can up-regulate the expression of SIRT1, hence enhancing wound repair, angiogenesis, autophagy, and inhibition of cell death [173]. In a diabetic mouse model of posterior limb ischemia, Circ-ADAM7 promotes autophagy and apoptosis of diabetic endothelial progeners and inhibits angiogenesis by targeting PTEN and ATG9 via sponge miR-20a-5p [174]. Furthermore, by sponging the miR-106a-5p, mmu_circ_0001052 in exosomes produced from ADSCs promotes DFU angiogenesis via the FGF4/p38MAPK pathways [136]. circ-Gcap14 promotes angiogenesis and accelerates diabetic wound healing by regulating miR-18a-5p/HIF-1α expression [175].
NcRNAs are also associated with histone modification and DNA methylation. Immortalized human epidermal cells treated with high glucose (HG-Exos) produce exosomes that can impede the healing of wounds. Further research has shown that HG-Exos-Linc01435 upregulates histone deacetylase 8 (HDAC8) expression by combining and recruiting transcription factors (Yin Yang 1) YY1 into the nucleus, resulting in reduced tube formation and migration of HUVECs [105]. Another study found that by enlisting zeste homolog 2 (EZH2), lncRNA H19 enhanced HIF-1α histone H3K4me3 methylation and HIF-1α expression in diabetic mice [73]. And as mentioned above, siRNA-directed DNA methylation of Alu elements promotes wound repair. In summary, epigenetics mechanisms are interactional rather than completely independent in diabetic wound regulation.
3.4. RNA methylation
In eukaryotic cells, N6-methyladenosine (m6A) is the most widespread and prevalent kind of posttranscriptional RNA modification at the nitrogen-6 position of adenine. Two classes of proteins mediate it dynamically and reversibly: demethylases (also known as "erasers"), such as FTO and ALKBH5, and methyltransferases (sometimes known as "writers"), such as METTL3, METTL14, WTAP, and KIAA1429. The former is responsible for the modification and the latter for removal m6A mark from the RNAs. The modification is detected by m6A binding proteins ("readers") including YT, HDF1-3, YTHDC1, YTHDC2, and HNRNPA2B1 etc., which influence the fate of RNAs. The modification is recognized by the m6A binding proteins (“readers”, such as YT, HDF1-3, YTHDC1, YTHDC2, and HNRNPA2B1), which impact the destiny of RNAs [176]. Diabetic wound healing is affected by inflammatory cytokines, macrophages, autophagy, angiogenesis and other factors. An increasing number of studies have proven that m6A is closely related to these factors. Researchers found that m6A deficiency impairs skin morphogenesis. METTL3 knockdown affects the later stages of epidermal differentiation in mice. METTL3 (“writer”) conditional knockout mice were challenged in that spinous cells transitioned to the granular layer in the later stages of differentiation. The downregulation of m6A methylation damages the normal physiological proliferation and differentiation ability of epidermal cells, thus affecting wound repair. In addition, a study unexpectedly observed that METTL3 knockout skin epithelial progenitors fail to form colonies in vitro [177]. Another study of METTL14 (“writer”) cKO mice had a similar finding, which elucidated that METTL14-mediated Pvt1 methylation contributed to the stemness of epidermal progenitor cells. Mechanically, m6A of Pvt1 facilitates the interaction with the MYC protein, thus inhibiting MYC phosphorylation-mediated degradation. MYC is an oncogene, with a central role in the stemness of basal progenitor cells [178]. Furthermore, research has demonstrated that angiogenesis is impacted by mRNA m6A methylation, which is found in cytokines linked to angiogenesis. The METTL3/IGF2BP2- m6A pathway was discovered to influence the production of VEGF-C, increasing wound healing, in a study on the treatment of DFU using adipose-derived mesenchymal stem cells (ADSC) (Fig. 3) [179].
Fig. 3.
The effects of m6A methylation on normal and diabetic wound healing
The figure demonstrates that methyltransferases (e.g., METTL3, METTL14) and demethylases (e.g., FTO) and m6A binding proteins (e.g., YTHDC1, YTHDC2) mediate N6-methyladenosine (m6A) related to the inflammation, angiogenesis, EPC (endothelial progenitor cells) and autophagy in normal and diabetic wound healing.
3.4.1. m6 A and inflammation
Compared to normoglycemic wounds, diabetic wounds exhibited greater expression of inflammatory cytokines, including tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-8 (IL-8). The activation of several genes linked to the inflammatory response as well as the rise in several inflammatory cytokines can both be inhibited by METTL3 knockdown. Researchers found that METTL3 knockdown enhanced an inflammatory response and phosphorylated p38, ERK, JNK, and p65 in MAPK and NF-κB signaling pathways in a mouse osteoblast inflammatory model produced by LPS [180]. METTL3 overexpression inhibited NF-κB activation and decreased inflammatory responses in models of macrophage inflammation [181]. When LPS was used to stimulate human dental pulp cells (HDPCs), METTL3 knockdown made it easier for MyD88S, a splice variation of MyD88 that prevents the generation of inflammatory cytokines, to be expressed [182]. The above studies indicate that METTL3 can dynamically regulate inflammatory response by affecting several different molecules, thus producing different results in the regulation of inflammation. Furthermore, in an endothelial cell inflammation model generated by TNFα, METTL14 enhances the m6A alteration of FOXO1 and induces an inflammatory response, hence promoting FOXO1 expression [183].
The transition of macrophages from pro-inflammatory to anti-inflammatory phenotype is disrupted in diabetic wounds, leading to an accumulation of M1macrophages in the tissues and an upregulation of inflammatory cytokines such as MCP-1, IL-12, TNF-α, and IL-1β. Researchers have found that Up‐regulated METTL3 increased the expression level of the signal transducers and activators of transcription 1 (STAT1), which in turn encouraged M1 macrophage polarization [184]. It's interesting to note that different enzymes have distinct effects on the up-regulation of STAT1 methylation. Interaction between RBM4 and YTHDF2 reduces the amounts of m6A-modified STAT1 mRNA and prevents M1 macrophage polarization [185]. In a different investigation, methylase FTO knockdown made STAT1 and peroxysomal activator γ (PPAR-γ) mRNA less stable, which made M1 and M2 macrophage polarization easier, respectively [186]. Because M1 macrophages play a crucial role in diabetic wound healing, METTL3-mediated macrophage polarization may influence the course of diabetic wound repair and, as a result, present a promising target for anti-inflammatory therapy.
3.4.2. m6A and autophagy
One possible explanation for the delayed wound repair of diabetic ulcers is the inhibition of autophagy of wound tissue by the diabetes condition [[187], [188], [189]]. According to a recent study, YTHDC1 ("reader") knockdown can decrease autophagy by targeting the autophagy receptor sequestosome 1 (SQSTM1), which can impact keratinocyte biological function and impede the healing of diabetic wounds [190]. In a different research, METTL14 knockdown results in decreased AMPK activity and consequent autophagy suppression by stabilizing the mRNA of calcium/calmodulin-dependent protein kinase 2 (CAMKK2) [191]. In conclusion, diabetic wound repair may be impacted by m6A's function in autophagy.
4. Future prospects for epigenetic applications
Taking advantage of epigenetic theory, epigenetic drugs have become a new area of research. To date, many drugs targeting epigenetic sites have been developed. Histone deacetylase inhibitors (HDACis) and DNA methyltransferase inhibitors (DNMTis) are the representative medications. Research on the effects of epidrugs on wound healing has been conducted in animal models. Zebularine, a DNMTi, has been demonstrated to successfully enhance auricle wound closure in rats [192]. In diabetic mice, HDACi tubastatin A and trichostatin A (TSA) can both quicken wound healing [79,193]. Another study devised a novel technique to improve diabetic wound healing by employing MNs to distribute TSA patches via the skin [194].
4.1. Combination with artificial intelligence (AI)
Artificial intelligence (AI) has greatly improved the possibilities of epigenetic therapies. AI is driving drug discovery, dramatically reducing the time it takes to reach the preclinical stage of drug development. Based on existing data, AI provides new methods and strategies for estimating unknown epigenomes. First, AI collects and collates published important epigenetic data and modification sites, and then uses deep learning algorithms to extract feature sequences, learn and train feature sequences, and build intelligent prediction models. Then, it scans genome sequences to discover potential epigenetic modification sites. Epigenetics in the field of AI medicine is still in the stage of exploration and data accumulation. The core difficulty is how to establish the causal relationship between different modified signals and diseases, and how to clearly understand the working principle of complex machine learning models [195,196].
4.2. Epigenetic CRISPR
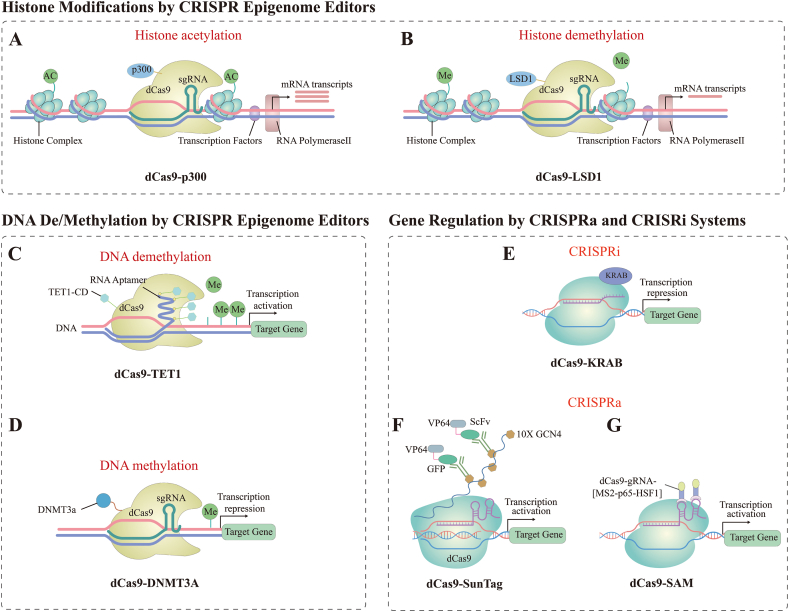
Classical CRISPR/Cas9 gene editing technology uses small guide RNAs (sgRNAs) to guide Cas9 endonuclease to the target site and cut the DNA double strand, on which the gene can be cut and edited [110]. However, CRISPR/Cas9 has limitations, such as off-target effects, unplanned loss of DNA fragments, and activated DNA repair mechanisms that may cause complex cellular stress responses [197,198]. A novel technology that is reversible, does not modify DNA sequences, and is not off-target is the epigenetic CRISPR. Although epigenetic editing does not directly alter gene sequences, it uses histone methylation, acetylation, and DNA methylation to control target gene expression at the transcriptional level [199]. Through the mechanism of gene activation controlled by the CRISPR/Cas9 system, novel therapeutic approaches utilizing particular inhibitors of histone post-translational modifications or proteins controlling DNA methylation are made possible. In recent years, scientists have fused dCas9 with epigenetic modifier enzymes to form the dCas9 epigenetic editing system, which realizes epigenetic modifications at specific genomic loci. CRISPR-based epigenome editors (CRISPR epi-editors) are composed of dCas9 and epigenetic effectors that can activate or repress gene transcription, according to the type of epigenetic effector [200]. The development of epigenetic CRISPR is summarized in Fig. 4 (Fig. 4). Histone modification systems include dCas9-p300 (histone acetylation system), dCas9-LSD1 (histone demethylation system). dCas9-p300 enables histone acetylation, which in turn enhances gene expression. dCas9-LSD1 is a complementary gene repressor system to the dCas9-p300 system, which induces histone demethylation of enhancers, which can down-regulate target gene expression. DNA modification systems include dCas9-TET1 (DNA demethylation system), dCas9-DNMT3A (DNA methylation system). The use of dCas9-TET1 enables rapid demethylation of the promoter and induces up-regulation of the expression of target genes. dCas9-DNMT3A targets DNA methylation at the promoter, thereby repressing gene expression. In addition, dCas9 can bind to transcriptional activation or repression domains, expanding the ability of histone code epi-editors to activate or repress target genes, including dCas9-SunTag (CRISPR activation, CRISPRa system), dCas9-SAM (CRISPRa system), and dCas9-KRAB (CRISPR interferencem system, CRISPRi system) [200,201].
Fig. 4.
CRISPR Epigenome Editors
(A) dCas9 protein with effector domain of p300 to activate gene expression through histone acetylation; (B) dCas9 protein with effector domain of LSD1 to repress gene expression through histone demethylation; (C) dCas9 protein with effector domain of TET1 to activate gene expression through DNA demethylation; (D) dCas9 protein with effector domain of DNMT3A to repress gene expression through DNA methylation; (E) CRISPRi, dCas9 with KRAB to repress gene expression; (F) CRISPRa, dCas9- SunTag system to activate gene expression; (G) CRISPRa, dCas9-SAM to activate gene expression.
An article published in Cell reports a novel CRISPR-based epigenetic editing technique [198]: CRISPRoff. The researchers took advantage of the ability of sgRNA to direct Cas9 to the target site, but disregarded Cas9's DNA splicing ability. Rather, they employed a combination of the catalytically inactive Cas9 (dCas9) and CRISPRoff-V1, which is made up of the protein domains ZNF10 KRAB, Dnmt3A (D3A), and Dnmt3L (D3L), to produce DNA methylation at the target region and so decrease target gene expression. When a gene is silenced with CRISPRoff, the effect lasts for up to 450 generations. Surprisingly, this was still true in maturing stem cells. In addition, researchers have developed matching CRISPRon that can reverse CRISPRoff editing. In one work, a dCas9 coupled with KRAB was directed to a specific genomic region using sgRNAs. Genes are repressed as a result of targeted H3K9 tri-methylation caused by KRAB binding to these particular genomic locations [202].
With epigenetic editing, DNA is not directly altered, meaning that the effects of the treatment may be reversible, allowing more subtle regulation of gene expression. It offers a number of potential advantages: (1)Epigenome editing can increase or decrease gene expression [203]. (2) Epigenome editing does not induce double or single strand breaks in DNA, nor does it rewrite or make permanent changes to the DNA sequence. (3)Epigenome editing can simultaneously control the expression levels of multiple genes. These advantages are expected to enable epigenetic editing techniques to treat more common and complex diseases.
4.3. Develop engineered extracellular vesicle delivery platform
EV-ncRNA has been shown to support wound repair in diabetes at different phases [204]. Extracellular vesicles (EVs) have a high degree of safety and stability and can enhance the effectiveness of diabetic wound healing by artificially modulating the ncRNAs they carry [205]. For instance, MFGE8 is upregulated by the human keratinocyte-derived exosome MALAT1 via miRNA-1914-3p, which facilitates macrophage polarization to M2 phenotype and diabetic wound healing [163]. Similarly, circ-Snhg11 in exosomes derived from hypoxic ADSCs affects HIF-1α expression via miR-144-3p, promotes wound M2 macrophage polarization and attenuates inflammatory responses in diabetic mice [170]. Moreover, mmu_circ_0001052 in exosomes produced from ADSCs promotes DFU angiogenesis by sponging miR-106a-5p via the FGF4/p38MAPK pathway [136]. Many new technologies have been invented to facilitate the production of large-scale EVs [206]. For instance, mesenchymal stem cells (MSCs) may be produced in large batches and continuously homogenized thanks to Emstein's unique two-step cell differentiation technology. MSC spheroids can be delivered and maintained at room temperature for up to 10 days [207]. Above all, epigenetic medications can be delivered by EVs to aid in the healing of diabetic wounds.In diabetic patients, aberrant ncRNA expression is directly linked to the advancement of the disease and can be used as a novel indicator and diagnostic biomarker for DFU. When compared to healthy persons, serum miRNA-217 levels were considerably elevated in DFU patients, and they progressively rose as Wagner grading increased [208]. According to another study, the degree of severity of DFU was positively correlated with the expression level of miRNA-203, which was found to be much greater in the skin of people with DFU compared to normal skin tissues [139]. When blood levels of miRNA-200b and miRNA-191 were lower in circulating people with type 2 diabetes mellitus than in healthy individuals, pro-inflammatory cytokine levels and C-reactive protein were elevated [148].
Furthermore, research has revealed that most medications contain underlying ncRNA binding sites, which lays the groundwork for the creation of medications that use ncRNAs as therapeutic targets. For instance, ginsenoside decreases the inhibitory impact of miRNA-23a on interferon regulatory factor 2 (IRF2), upregulates the amount of inducible nitric oxide synthase (NOS), which stimulates angiogenesis, and speeds up wound healing in diabetic rats [209]. Mevastatin used topically speeds up diabetic wound healing. One possible explanation for this is that mevastatin stimulates the production of lncRNA-neurotrophic factor 5, which suppresses c-myc expression and encourages KC migration and proliferation (Table 2) [210].
Table 2.
Prospective application of ncRNAs in the treatment of diabetic wounds.
|
Application in diabetic wound healing |
Related ncRNA | Effect | Refs |
|---|---|---|---|
|
NcRNA delivered by extracellular vesicle delivery platform |
MALAT1, miRNA-1914-3p | MALAT1 in human keratinocyte-derived exosomes upregulates MFGE8 via miRNA-1914-3p, thereby promoting macrophage polarization to M2 phenotype and diabetic wound healing. | [163] |
| circ-Snhg11 | circ-Snhg11 in hypoxic ADSCS-derived exosomes induces polarization of M2 macrophages by targeting miR-144-3p and HIF-1α, alleviating diabetic wound inflammation. | [170] | |
| mmu_circ_0001052 | mmu_circ_0001052 in exosomes produced from ADSCs promotes DFU angiogenesis by sponging miR-106a-5p via the FGF4/p38MAPK pathway. | [136] | |
| As a diagnostic marker | miRNA-217 | In comparison with healthy individuals, patients with DFU had considerably higher serum miRNA-217 levels, which also increased with Wagner grading. | [208] |
| miRNA-203 | When comparing the skin of DFU patients to normal skin tissue, the expression level of miRNA-203 was considerably greater, and it was strongly correlated with the severity of DFU. | [139] | |
| miRNA-191, miRNA-200b | Compared to individuals in general, those with type 2 diabetes had greater amounts of pro-inflammatory cytokines and fewer amounts of miRNA-191 and miRNA-200b in their blood. | [148] | |
| Clinical studies of ncRNA-based drugs | miRNA-23a | Ginsenoside promotes diabetic wound angiogenesis by down-regulating miRNA-23a expression and up-regulating inducible NOS levels | [209] |
| lncRNA-neurotrophic factor 5 | Mevastatin induces lncRNA-neurotrophic factor 5 expression, inhibits c-myc expression, and regulates KC proliferation and migration, thereby promoting wound healing | [210] |
Abbreviations: miRNA: microRNA; ADSC: adipose-derived mesenchymal stem cells; HIF-1α: Hypoxia-inducible factor-1α; DFU: Diabetic foot ulcer; KC: Keratinocyte; ncRNA: noncoding RNA; NOS: nitric oxide synthase.
5. Conclusion
From the past to the future, epigenetics is a topic to be studied. After more than decades of development, basic research on epigenetics has yielded fruitful results, and epigenetic drugs have shown a rapidly increasing trend in recent years. If there are so many epigenetic targets why has it taken decades for them to develop a method? Until recently, only a few have been available. The development of this industry faces relatively large challenges. One is the complexity of epigenetics. Each mutation results in a change in the structure of DNA, histone, or chromatin. These epigenetic changes, in turn, affect one or more different cellular activities, such as transcription, replication, or DNA repair. Epigenetic factors do not only modify a gene, a section of DNA, histones and chromatin, the result can be one-to-one or one-to-many. In addition, there is a security window. Epigenetic inhibitors typically have a narrow safety window. The reasons for this result are closely related to the complexity and wide influence mentioned in the Introduction. To solve this problem, in addition to the study of biological mechanisms, more drugs need to be designed to increase the selectivity and specificity of compounds. Determining the ideal dosage and method of administration is also necessary. Although there are problems that need to be addressed, epigenetic drug therapy still has clear advantages. Currently, medication combinations, immune escape suppression, and drug resistance resolution are the three main ways that epigenetic medicines are advantageous in the treatment of cancer. In the future, the development direction of epigenetic inheritance may lead to new targets, new mechanisms and drug combinations.
The development and improvement of epigenetics has opened up a new door for precision medicine. The application of epigenetic medications in the treatment of diabetic wounds has a bright future. Nevertheless, further studies must be done before clinal application. However, a number of experiments have already given us a glimpse into the future of disease treatment, and we believe that epigenetic therapy could one day be the next big thing.
Data availability statement
No data was used for the research described in the article.
CRediT authorship contribution statement
Cong-Cong Ju: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Data curation, Conceptualization. Xiao-Xiao Liu: Writing – original draft, Validation, Resources, Methodology, Investigation, Data curation, Conceptualization. Li-hua Liu: Writing – review & editing. Nan Guo: Writing – review & editing. Le-wei Guan: Writing – review & editing. Jun-xian Wu: Writing – review & editing. De-Wu Liu: Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China under grant number 81460293 and Chongqing Traditional Chinese Medicine Inheritance and Innovation Team Project (2023090006KJZX2022WJW008).
References
- 1.Chatterjee S., Khunti K., Davies M.J. Type 2 diabetes. Lancet. 2017;389(10085):2239–2251. doi: 10.1016/s0140-6736(17)30058-2. [DOI] [PubMed] [Google Scholar]
- 2.Sun H., Saeedi P., Karuranga S., Pinkepank M., Ogurtsova K., Duncan B.B., Stein C., Basit A., Chan J.C.N., Mbanya J.C., Pavkov M.E., Ramachandaran A., Wild S.H., James S., Herman W.H., Zhang P., Bommer C., Kuo S., Boyko E.J., Magliano D.J. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022;183 doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Jiang J., Zhao C., Shan H., Shao Z., Wang C., Guan J., Xie Z., Li S. The protective effect of theaflavins on the kidney of mice with type II diabetes mellitus. Nutrients. 2022;15(1) doi: 10.3390/nu15010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong D.G., Boulton A.J.M., Bus S.A. Diabetic foot ulcers and their recurrence. N. Engl. J. Med. 2017;376(24):2367–2375. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong D.G., Tan T.W., Boulton A.J.M., Bus S.A. Diabetic foot ulcers: a review. JAMA. 2023;330(1):62–75. doi: 10.1001/jama.2023.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teena R., Dhamodharan U., Ali D., Rajesh K., Ramkumar K.M. Gene expression profiling of multiple histone deacetylases (HDAC) and its correlation with NRF2-mediated redox regulation in the pathogenesis of diabetic foot ulcers. Biomolecules. 2020;10(10) doi: 10.3390/biom10101466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dogruel H., Aydemir M., Balci M.K. Management of diabetic foot ulcers and the challenging points: an endocrine view. World J. Diabetes. 2022;13(1):27–36. doi: 10.4239/wjd.v13.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyko E.J., Zelnick L.R., Braffett B.H., Pop-Busui R., Cowie C.C., Lorenzi G.M., Gubitosi-Klug R., Zinman B., de Boer I.H. Risk of foot ulcer and lower-extremity amputation among participants in the diabetes control and complications trial/epidemiology of diabetes interventions and complications study. Diabetes Care. 2022;45(2):357–364. doi: 10.2337/dc21-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin L., Feng G., Huang C., Cai W. Proteomic analysis of serum lysine acetylation in Uyghur patients with T2DM. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.787885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S., Liu H., Ruan Z., Guo R., Sun C., Tang Y., Huang X., Gao T., Hao S., Li H., Song N., Su Y., Ning F., Li Z., Chang T. Landscape analysis of m6A modification regulators related biological functions and immune characteristics in myasthenia gravis. J. Transl. Med. 2023;21(1):166. doi: 10.1186/s12967-023-03947-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Dekker A., Davis F.M., Kunkel S.L., Gallagher K.A. Targeting epigenetic mechanisms in diabetic wound healing. Transl. Res. : J. Lab. Clin. Med. 2019;204:39–50. doi: 10.1016/j.trsl.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Effendi W.I., Nagano T. Epigenetics approaches toward precision medicine for idiopathic pulmonary fibrosis: focus on DNA methylation. Biomedicines. 2023;11(4) doi: 10.3390/biomedicines11041047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng J.Y., Wu X.Q., He W.J., Liao X., Tang M., Nie X.Q. Targeting DNA methylation and demethylation in diabetic foot ulcers. J. Adv. Res. 2023 doi: 10.1016/j.jare.2023.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorincz A.T. Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytol. 2016;60(6):501–512. doi: 10.1159/000450595. [DOI] [PubMed] [Google Scholar]
- 15.Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571(7766):489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 16.Kim H., Wang S.Y., Kwak G., Yang Y., Kwon I.C., Kim S.H. Exosome-guided phenotypic switch of M1 to M2 macrophages for cutaneous wound healing. Adv. Sci. 2019;6(20) doi: 10.1002/advs.201900513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piperigkou Z., Götte M., Theocharis A.D., Karamanos N.K. Insights into the key roles of epigenetics in matrix macromolecules-associated wound healing. Adv. Drug Deliv. Rev. 2018;129:16–36. doi: 10.1016/j.addr.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Mailer R.K., Allende M., Heestermans M., Schweizer M., Deppermann C., Frye M., Pula G., Odeberg J., Gelderblom M., Rose-John S., Sickmann A., Blankenberg S., Huber T.B., Kubisch C., Maas C., Gambaryan S., Firsov D., Stavrou E.X., Butler L.M., Renné T. Xenotropic and polytropic retrovirus receptor 1 regulates procoagulant platelet polyphosphate. Blood. 2021;137(10):1392–1405. doi: 10.1182/blood.2019004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao S., He X., Qin L., He M., Yang Y., Liu Z., Mao W. Anticoagulant and antithrombotic properties in vitro and in vivo of a novel sulfated polysaccharide from marine green alga monostroma nitidum. Mar. Drugs. 2019;17(4) doi: 10.3390/md17040247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopsinis G., Tsoukanas D., Kopsini D., Filippopoulos T. Intracameral bevacizumab versus sub-tenon's mitomycin C as adjuncts to trabeculectomy: 3-year results of a prospective randomized study. J. Clin. Med. 2021;10(10) doi: 10.3390/jcm10102054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long C.Y., Lin K.L., Shen C.R., Ker C.R., Liu Y.Y., Loo Z.X., Hsiao H.H., Lee Y.C. A pilot study: effectiveness of local injection of autologous platelet-rich plasma in treating women with stress urinary incontinence. Sci. Rep. 2021;11(1):1584. doi: 10.1038/s41598-020-80598-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigues M., Kosaric N., Bonham C.A., Gurtner G.C. Wound healing: a cellular perspective. Physiol. Rev. 2019;99(1):665–706. doi: 10.1152/physrev.00067.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahramian K., Abdulmajeed A., Kangasniemi I., Söderling E., Närhi T. TiO(2) coating and UV photofunctionalization enhance blood coagulation on zirconia surfaces. BioMed Res. Int. 2019;2019 doi: 10.1155/2019/8078230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi X., Jiang L., Zhao X., Chen B., Shi W., Cao Y., Chen Y., Li X., He Y., Li C., Liu X., Li X., Lu H., Chen C., Liu J. Adipose-derived stromal cell-sheets sandwiched, book-shaped acellular dermal matrix capable of sustained release of basic fibroblast growth factor promote diabetic wound healing. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.646967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Q., Lu C., Jiang X., Yao Q., Jiang X., Huang Z., Jiang Y., Peng L., Fu H., Zhao Y. Using recombinant human collagen with basic fibroblast growth factor to provide a simulated extracellular matrix microenvironment for the revascularization and attachment of islets to the transplantation region. Front. Pharmacol. 2019;10:1536. doi: 10.3389/fphar.2019.01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei L., Fukutani K.F., Tibúrcio R., Rupert A., Dahlstrom E.W., Galindo F., Laidlaw E., Lisco A., Manion M., Andrade B.B., Sereti I. Plasma metabolomics reveals dysregulated metabolic signatures in HIV-associated immune reconstitution inflammatory syndrome. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.693074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H., Lo T.H., Ma D., Condor B., Lesmana B., Parungao R.J., Tsai K.H., Kim S., Chen H.T., Silveira P.A., Li Z., Cooper M.S., Simanainen U., Handelsman D.J., Maitz P.K., Wang Y. Dihydrotestosterone (DHT) enhances wound healing of major burn injury by accelerating resolution of inflammation in mice. Int. J. Mol. Sci. 2020;21(17) doi: 10.3390/ijms21176231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skoglund C., Appelgren D., Johansson I., Casas R., Ludvigsson J. Increase of neutrophil extracellular traps, mitochondrial DNA and nuclear DNA in newly diagnosed type 1 diabetes children but not in high-risk children. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.628564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garg M., Johri S., Sagar S., Mundhada A., Agrawal A., Ray P., Chakraborty K. Cardiolipin-mediated PPARγ S112 phosphorylation impairs IL-10 production and inflammation resolution during bacterial pneumonia. Cell Rep. 2021;34(6) doi: 10.1016/j.celrep.2021.108736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W.C., Chen N.J., Chen H.P., Yu W.K., Su V.Y., Chen H., Wu H.H., Yang K.Y. Nintedanib reduces neutrophil chemotaxis via activating GRK2 in bleomycin-induced pulmonary fibrosis. Int. J. Mol. Sci. 2020;21(13) doi: 10.3390/ijms21134735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkinson H.N., Hardman M.J. Wound healing: cellular mechanisms and pathological outcomes. Open Biol. 2020;10(9) doi: 10.1098/rsob.200223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosser D.M., Hamidzadeh K., Goncalves R. Macrophages and the maintenance of homeostasis. Cell. Mol. Immunol. 2021;18(3):579–587. doi: 10.1038/s41423-020-00541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flood-Nichols S.K., Kazanjian A.A., Tinnemore D., Gafken P.R., Ogata Y., Napolitano P.G., Stallings J.D., Ippolito D.L. Aberrant glycosylation of plasma proteins in severe preeclampsia promotes monocyte adhesion. Reprod. Sci. 2014;21(2):204–214. doi: 10.1177/1933719113492210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang P., Xu J., Xie L., Gao G., Chen S., Gong Z., Lao X., Shan Z., Shi J., Zhou Z., Chen Z., Cao Y., Wang Y., Chen Z. Improving hard metal implant and soft tissue integration by modulating the "inflammatory-fibrous complex" response. Bioact. Mater. 2023;20:42–52. doi: 10.1016/j.bioactmat.2022.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzyszczyk P., Schloss R., Palmer A., Berthiaume F. The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front. Physiol. 2018;9:419. doi: 10.3389/fphys.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Z., Petree J.R., Lee F.E., Fan X., Salaita K., Guidot D.M., Sadikot R.T. Macrophages exposed to HIV viral protein disrupt lung epithelial cell integrity and mitochondrial bioenergetics via exosomal microRNA shuttling. Cell Death Dis. 2019;10(8):580. doi: 10.1038/s41419-019-1803-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Cheyne C., Tay H., De Spiegelaere W. The complex TIE between macrophages and angiogenesis. Anat. Histol. Embryol. 2020;49(5):585–596. doi: 10.1111/ahe.12518. [DOI] [PubMed] [Google Scholar]
- 38.Basri F., Jung S., Park S.H., Park S.H. CD1d deficiency limits tolerogenic properties of peritoneal macrophages. BMB Rep. 2021;54(4):209–214. doi: 10.5483/BMBRep.2021.54.4.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Guan M., Ren R., Gao C., Cheng H., Li Y., Gao B., Wei Y., Fu J., Sun J., Xiong W. Improved immunoregulation of ultra-low-dose silver nanoparticle-loaded TiO(2) nanotubes via M2 macrophage polarization by regulating GLUT1 and autophagy. Int. J. Nanomed. 2020;15:2011–2026. doi: 10.2147/ijn.S242919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim S., Lyu H.Z., Lee J.R., Han S.H., Lee J.H., Kim B.S. Umbilical cord mesenchymal stem cell-derived nanovesicles potentiate the bone-formation efficacy of bone morphogenetic protein 2. Int. J. Mol. Sci. 2020;21(17) doi: 10.3390/ijms21176425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan B.T., Qu F., Wang S.X., Qi W., Shen X.Z., Li C.B., Liu Y.J. Histology and molecular pathology of iliotibial tract contracture in patients with gluteal muscle contracture. Biosci. Rep. 2019;39(9) doi: 10.1042/bsr20181351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee B.C., Song J., Lee A., Cho D., Kim T.S. Erythroid differentiation regulator 1 promotes wound healing by inducing the production of C-C motif chemokine ligand 2 via the activation of MAP kinases in vitro and in vivo. Int. J. Mol. Med. 2020;46(6):2185–2193. doi: 10.3892/ijmm.2020.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monsuur H.N., van den Broek L.J., Koolwijk P., Niessen F.B., Gibbs S. Endothelial cells enhance adipose mesenchymal stromal cell-mediated matrix contraction via ALK receptors and reduced follistatin: potential role of endothelial cells in skin fibrosis. J. Cell. Physiol. 2018;233(10):6714–6722. doi: 10.1002/jcp.26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S.T., Chang C.C., Pang J.S., Huang H.S., Chou S.C., Kao M.C., You H.L. Drynaria fortunei promoted angiogenesis associated with modified MMP-2/TIMP-2 balance and activation of VEGF ligand/receptors expression. Front. Pharmacol. 2018;9:979. doi: 10.3389/fphar.2018.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bian X., Li B., Yang J., Ma K., Sun M., Zhang C., Fu X. Regenerative and protective effects of dMSC-sEVs on high-glucose-induced senescent fibroblasts by suppressing RAGE pathway and activating Smad pathway. Stem Cell Res. Ther. 2020;11(1):166. doi: 10.1186/s13287-020-01681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arif S., Larochelle S., Moulin V.J. PLGF-1 contained in normal wound myofibroblast-derived microvesicles stimulated collagen production by dermal fibroblasts. Journal of cell communication and signaling. 2020;14(4):427–438. doi: 10.1007/s12079-020-00572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henriksen J.L., Sørensen N.B., Fink T., Zachar V., Porsborg S.R. Systematic review of stem-cell-based therapy of burn wounds: lessons learned from animal and clinical studies. Cells. 2020;9(12) doi: 10.3390/cells9122545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Słoniecka M., Danielson P. Acetylcholine decreases formation of myofibroblasts and excessive extracellular matrix production in an in vitro human corneal fibrosis model. J. Cell Mol. Med. 2020;24(8):4850–4862. doi: 10.1111/jcmm.15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simard E., Sollradl T., Maltais J.S., Boucher J., D'Orleans-Juste P., Grandbois M. Receptor for advanced glycation end-products signaling interferes with the vascular smooth muscle cell contractile phenotype and function. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0128881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peppa M., Stavroulakis P., Raptis S.A. Advanced glycoxidation products and impaired diabetic wound healing. Wound Repair Regen. 2009;17(4):461–472. doi: 10.1111/j.1524-475X.2009.00518.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W., Chen L., Xiong Y., Panayi A.C., Abududilibaier A., Hu Y., Yu C., Zhou W., Sun Y., Liu M., Xue H., Hu L., Yan C., Xie X., Lin Z., Cao F., Mi B., Liu G. Antioxidant therapy and antioxidant-related bionanomaterials in diabetic wound healing. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.707479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng L., Du C., Song P., Chen T., Rui S., Armstrong D.G., Deng W. The role of oxidative stress and antioxidants in diabetic wound healing. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/8852759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nouvong A., Ambrus A.M., Zhang E.R., Hultman L., Coller H.A. Reactive oxygen species and bacterial biofilms in diabetic wound healing. Physiol. Genom. 2016;48(12):889–896. doi: 10.1152/physiolgenomics.00066.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q., Qiu W., Li M., Li N., Li X., Qin X., Wang X., Yu J., Li F., Huang L., Wu D. Multifunctional hydrogel platform for biofilm scavenging and O(2) generating with photothermal effect on diabetic chronic wound healing. J. Colloid Interface Sci. 2022;617:542–556. doi: 10.1016/j.jcis.2022.03.040. [DOI] [PubMed] [Google Scholar]
- 55.Kolattukudy P.E., Niu J. Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circ. Res. 2012;110(1):174–189. doi: 10.1161/circresaha.111.243212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimball A., Schaller M., Joshi A., Davis F.M., denDekker A., Boniakowski A., Bermick J., Obi A., Moore B., Henke P.K., Kunkel S.L., Gallagher K.A. Ly6C(Hi) blood monocyte/macrophage drive chronic inflammation and impair wound healing in diabetes mellitus. Arterioscler. Thromb. Vasc. Biol. 2018;38(5):1102–1114. doi: 10.1161/atvbaha.118.310703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis F.M., Tsoi L.C., Wasikowski R., denDekker A., Joshi A., Wilke C., Deng H., Wolf S., Obi A., Huang S., Billi A.C., Robinson S., Lipinski J., Melvin W.J., Audu C.O., Weidinger S., Kunkel S.L., Smith A., Gudjonsson J.E., Moore B.B., Gallagher K.A. Epigenetic regulation of the PGE2 pathway modulates macrophage phenotype in normal and pathologic wound repair. JCI insight. 2020;5(17) doi: 10.1172/jci.insight.138443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng J., Dong C., Long Y., Mai L., Ren M., Li L., Zhou T., Yang Z., Ma J., Yan L., Yang X., Gao G., Qi W. Elevated Kallikrein-binding protein in diabetes impairs wound healing through inducing macrophage M1 polarization. Cell Commun. Signal. : CCS. 2019;17(1):60. doi: 10.1186/s12964-019-0376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimball A.S., Joshi A.D., Boniakowski A.E., Schaller M., Chung J., Allen R., Bermick J., Carson W.F.t., Henke P.K., Maillard I., Kunkel S.L., Gallagher K.A. Notch regulates macrophage-mediated inflammation in diabetic wound healing. Front. Immunol. 2017;8:635. doi: 10.3389/fimmu.2017.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jere S.W., Houreld N.N., Abrahamse H. Role of the PI3K/AKT (mTOR and GSK3β) signalling pathway and photobiomodulation in diabetic wound healing. Cytokine Growth Factor Rev. 2019;50:52–59. doi: 10.1016/j.cytogfr.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Okonkwo U.A., DiPietro L.A. Diabetes and wound angiogenesis. Int. J. Mol. Sci. 2017;18(7) doi: 10.3390/ijms18071419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu M., Liu W., Li J., Lu J., Lu H., Jia W., Liu F. Exosomes derived from atorvastatin-pretreated MSC accelerate diabetic wound repair by enhancing angiogenesis via AKT/eNOS pathway. Stem Cell Res. Ther. 2020;11(1):350. doi: 10.1186/s13287-020-01824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dasari N., Jiang A., Skochdopole A., Chung J., Reece E.M., Vorstenbosch J., Winocour S. Updates in diabetic wound healing, inflammation, and scarring. Semin. Plast. Surg. 2021;35(3):153–158. doi: 10.1055/s-0041-1731460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu T., Gao M., Yang P., Liu D., Wang D., Song F., Zhang X., Liu Y. Insulin promotes macrophage phenotype transition through PI3K/Akt and PPAR‐γ signaling during diabetic wound healing. J. Cell. Physiol. 2018;234(4):4217–4231. doi: 10.1002/jcp.27185. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J., Yang P., Liu D., Gao M., Wang J., Wang X., Liu Y., Zhang X. c-Myc upregulated by high glucose inhibits HaCaT differentiation by S100A6 transcriptional activation. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.676403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang M., Jiang X., Li H., Zhang C., Zhang Z., Wu C., Zhang J., Hu J., Zhang J. The role of mesenchymal stem cell-derived EVs in diabetic wound healing. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1136098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang J., Daines J.R., Warren A.N., Cowan M.L. Epigenetics for the 21st-century biology student. J. Microbiol. Biol. Educ. 2019;20(3) doi: 10.1128/jmbe.v20i3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis F.M., Kimball A., denDekker A., Joshi A.D., Boniakowski A.E., Nysz D., Allen R.M., Obi A., Singer K., Henke P.K., Moore B.B., Kunkel S.L., Gallagher K.A. Histone methylation directs myeloid TLR4 expression and regulates wound healing following cutaneous tissue injury. Journal of immunology (Baltimore, Md. : 1950) 2019;202(6):1777–1785. doi: 10.4049/jimmunol.1801258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimball A.S., Joshi A., Carson W.F.t., Boniakowski A.E., Schaller M., Allen R., Bermick J., Davis F.M., Henke P.K., Burant C.F., Kunkel S.L., Gallagher K.A. The histone methyltransferase MLL1 directs macrophage-mediated inflammation in wound healing and is altered in a murine model of obesity and type 2 diabetes. Diabetes. 2017;66(9):2459–2471. doi: 10.2337/db17-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kimball A.S., Davis F.M., denDekker A., Joshi A.D., Schaller M.A., Bermick J., Xing X., Burant C.F., Obi A.T., Nysz D., Robinson S., Allen R., Lukacs N.W., Henke P.K., Gudjonsson J.E., Moore B.B., Kunkel S.L., Gallagher K.A. The histone methyltransferase Setdb2 modulates macrophage phenotype and uric acid production in diabetic wound repair. Immunity. 2019;51(2):258–271.e5. doi: 10.1016/j.immuni.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X., Liu C., Zhu Y., Rao H., Liu M., Gui L., Feng W., Tang H., Xu J., Gao W.Q., Li L. SETD2 epidermal deficiency promotes cutaneous wound healing via activation of AKT/mTOR Signalling. Cell Prolif. 2021;54(6) doi: 10.1111/cpr.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li G., Li D., Wu C., Li S., Chen F., Li P., Ko C.N., Wang W., Lee S.M., Lin L., Ma D.L., Leung C.H. Homocysteine-targeting compounds as a new treatment strategy for diabetic wounds via inhibition of the histone methyltransferase SET7/9. Exp. Mol. Med. 2022;54(7):988–998. doi: 10.1038/s12276-022-00804-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo J.R., Yin L., Chen Y.Q., Jin X.J., Zhou X., Zhu N.N., Liu X.Q., Wei H.W., Duan L.S. Autologous blood transfusion augments impaired wound healing in diabetic mice by enhancing lncRNA H19 expression via the HIF-1α signaling pathway. Cell Commun. Signal. : CCS. 2018;16(1):84. doi: 10.1186/s12964-018-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang M., Huang X., Jiang B., Zhang P., Guo L., Cui X., Zhou S., Ren L., Zhang M., Zeng J., Huang X., Liang P. linc00174-EZH2-ZNF24/Runx1-VEGFA regulatory mechanism modulates post-burn wound healing, molecular therapy. Nucleic acids. 2020;21:824–836. doi: 10.1016/j.omtn.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wojtala M., Dąbek A., Rybaczek D., Śliwińska A., Świderska E., Słapek K., El-Osta A., Balcerczyk A. Silencing lysine-specific histone demethylase 1 (LSD1) causes increased HP1-positive chromatin, stimulation of DNA repair processes, and dysregulation of proliferation by Chk1 phosphorylation in human endothelial cells. Cells. 2019;8(10) doi: 10.3390/cells8101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis F.M., denDekker A., Joshi A.D., Wolf S.J., Audu C., Melvin W.J., Mangum K., Riordan M.O., Kunkel S.L., Gallagher K.A. Palmitate-TLR4 signaling regulates the histone demethylase, JMJD3, in macrophages and impairs diabetic wound healing. Eur. J. Immunol. 2020;50(12):1929–1940. doi: 10.1002/eji.202048651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Audu C.O., Melvin W.J., Joshi A.D., Wolf S.J., Moon J.Y., Davis F.M., Barrett E.C., Mangum K.D., Deng H., Xing X., Wasikowski R., Tsoi L.C., Sharma S.B., Bauer T.M., Shadiow J., Corriere M.A., Obi A.T., Kunkel S.L., Levi B., Moore B.B., Gudjonsson J.E., Smith A.M., Gallagher K.A. Macrophage-specific inhibition of the histone demethylase JMJD3 decreases STING and pathologic inflammation in diabetic wound repair. Cell. Mol. Immunol. 2022;19(11):1251–1262. doi: 10.1038/s41423-022-00919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.denDekker A.D., Davis F.M., Joshi A.D., Wolf S.J., Allen R., Lipinski J., Nguyen B., Kirma J., Nycz D., Bermick J., Moore B.B., Gudjonsson J.E., Kunkel S.L., Gallagher K.A. TNF-α regulates diabetic macrophage function through the histone acetyltransferase MOF. JCI insight. 2020;5(5) doi: 10.1172/jci.insight.132306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karnam K., Sedmaki K., Sharma P., Routholla G., Goli S., Ghosh B., Venuganti V.V.K., Kulkarni O.P. HDAC6 inhibitor accelerates wound healing by inhibiting tubulin mediated IL-1β secretion in diabetic mice, Biochimica et biophysica acta. Molecular basis of disease. 2020;1866(11) doi: 10.1016/j.bbadis.2020.165903. [DOI] [PubMed] [Google Scholar]
- 80.Collins S.L., Oh M.H., Sun I.H., Chan-Li Y., Zhao L., Powell J.D., Horton M.R. mTORC1 signaling regulates proinflammatory macrophage function and metabolism. Journal of immunology (Baltimore, Md. : 1950) 2021;207(3):913–922. doi: 10.4049/jimmunol.2100230. [DOI] [PubMed] [Google Scholar]
- 81.Gao J., Wang Y., Li W., Zhang J., Che Y., Cui X., Sun B., Zhao G. Loss of histone deacetylase 2 inhibits oxidative stress induced by high glucose via the HO-1/SIRT1 pathway in endothelial progenitor cells. Gene. 2018;678:1–7. doi: 10.1016/j.gene.2018.07.072. [DOI] [PubMed] [Google Scholar]
- 82.Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M., Ding J., Czyz D., Hu R., Ye Z., He M., Zheng Y.G., Shuman H.A., Dai L., Ren B., Roeder R.G., Becker L., Zhao Y. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574(7779):575–580. doi: 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kadakol A., Malek V., Goru S.K., Pandey A., Gaikwad A.B. Esculetin reverses histone H2A/H2B ubiquitination, H3 dimethylation, acetylation and phosphorylation in preventing type 2 diabetic cardiomyopathy. J. Funct.Foods. 2015;17:127–136. doi: 10.1016/j.jff.2015.05.017. [DOI] [Google Scholar]
- 84.Yan J., Tie G., Wang S., Tutto A., DeMarco N., Khair L., Fazzio T.G., Messina L.M. Diabetes impairs wound healing by Dnmt1-dependent dysregulation of hematopoietic stem cells differentiation towards macrophages. Nat. Commun. 2018;9(1):33. doi: 10.1038/s41467-017-02425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao J., Yang S., Shu B., Chen L., Yang R., Xu Y., Xie J., Liu X., Qi S. Transient high glucose causes persistent vascular dysfunction and delayed wound healing by the DNMT1-mediated ang-1/NF-κB pathway. J. Invest. Dermatol. 2021;141(6):1573–1584. doi: 10.1016/j.jid.2020.10.023. [DOI] [PubMed] [Google Scholar]
- 86.Li D., Kular L., Vij M., Herter E.K., Li X., Wang A., Chu T., Toma M.A., Zhang L., Liapi E., Mota A., Blomqvist L., Gallais Sérézal I., Rollman O., Wikstrom J.D., Bienko M., Berglund D., Ståhle M., Sommar P., Jagodic M., Landén N.X. Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration. Proc. Natl. Acad. Sci. U.S.A. 2019;116(19):9443–9452. doi: 10.1073/pnas.1814097116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J., Yang C., Wang C., Liu D., Lao G., Liang Y., Sun K., Luo H., Tan Q., Ren M., Yan L. AGE-induced keratinocyte MMP-9 expression is linked to TET2-mediated CpG demethylation. Wound Repair Regen. 2016;24(3):489–500. doi: 10.1111/wrr.12426. [DOI] [PubMed] [Google Scholar]
- 88.Zhou L., Ren M., Zeng T., Wang W., Wang X., Hu M., Su S., Sun K., Wang C., Liu J., Yang C., Yan L. TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death Dis. 2019;10(11):813. doi: 10.1038/s41419-019-2047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou L., Wang W., Yang C., Zeng T., Hu M., Wang X., Li N., Sun K., Wang C., Zhou J., Ren M., Yan L. GADD45a promotes active DNA demethylation of the MMP-9 promoter via base excision repair pathway in AGEs-treated keratinocytes and in diabetic male rat skin. Endocrinology. 2018;159(2):1172–1186. doi: 10.1210/en.2017-00686. [DOI] [PubMed] [Google Scholar]
- 90.Ivic N., Potocnjak M., Solis-Mezarino V., Herzog F., Bilokapic S., Halic M. Fuzzy interactions form and shape the histone transport complex. Mol. Cell. 2019;73(6):1191–1203.e6. doi: 10.1016/j.molcel.2019.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen K., Bennett S.A., Rana N., Yousuf H., Said M., Taaseen S., Mendo N., Meltser S.M., Torrente M.P. Neurodegenerative disease proteinopathies are connected to distinct histone post-translational modification landscapes. ACS Chem. Neurosci. 2018;9(4):838–848. doi: 10.1021/acschemneuro.7b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y.J., Sun Z.X., Jia J.Q., Du T.J., Zhang N.C., Tang Y., Fang Y., Fang D. In: Histone Mutations and Cancer. Fang D., Han J., editors. 2021. Overview of histone modification; pp. 1–16. [DOI] [PubMed] [Google Scholar]
- 93.Pu J., Wang J., Wei H., Lu T., Wu X., Wu Y., Shao Z., Luo C., Lu Y. lncRNA MAGI2-AS3 prevents the development of HCC via recruiting KDM1A and promoting H3K4me2 demethylation of the RACGAP1 promoter, molecular therapy. Nucleic acids. 2019;18:351–362. doi: 10.1016/j.omtn.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davis F.M., denDekker A., Kimball A., Joshi A.D., El Azzouny M., Wolf S.J., Obi A.T., Lipinski J., Gudjonsson J.E., Xing X., Plazyo O., Audu C., Melvin W.J., Singer K., Henke P.K., Moore B.B., Burant C., Kunkel S.L., Gallagher K.A. Epigenetic regulation of TLR4 in diabetic macrophages modulates immunometabolism and wound repair. Journal of immunology (Baltimore, Md. : 1950) 2020;204(9):2503–2513. doi: 10.4049/jimmunol.1901263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mocavini I., Pippa S., Licursi V., Paci P., Trisciuoglio D., Mannironi C., Presutti C., Negri R. JARID1B expression and its function in DNA damage repair are tightly regulated by miRNAs in breast cancer. Cancer Sci. 2019;110(4):1232–1243. doi: 10.1111/cas.13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meng Y., Qian X., Zhao L., Li N., Wu S., Chen B., Sun T., Wang X. Trichostatin A downregulates bromodomain and extra-terminal proteins to suppress osimertinib resistant non-small cell lung carcinoma. Cancer Cell Int. 2021;21(1):216. doi: 10.1186/s12935-021-01914-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hou X., Shi X., Zhang W., Li D., Hu L., Yang J., Zhao J., Wei S., Wei X., Ruan X., Zheng X., Gao M. LDHA induces EMT gene transcription and regulates autophagy to promote the metastasis and tumorigenesis of papillary thyroid carcinoma. Cell Death Dis. 2021;12(4):347. doi: 10.1038/s41419-021-03641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chelladurai P., Dabral S., Basineni S.R., Chen C.N., Schmoranzer M., Bender N., Feld C., Nötzold R.R., Dobreva G., Wilhelm J., Jungblut B., Zhao L., Bauer U.M., Seeger W., Pullamsetti S.S. Isoform-specific characterization of class I histone deacetylases and their therapeutic modulation in pulmonary hypertension. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-69737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Azevedo H., Pessoa G.C., de Luna Vitorino F.N., Nsengimana J., Newton-Bishop J., Reis E.M., da Cunha J.P.C., Jasiulionis M.G. Gene co-expression and histone modification signatures are associated with melanoma progression, epithelial-to-mesenchymal transition, and metastasis. Clin. Epigenet. 2020;12(1):127. doi: 10.1186/s13148-020-00910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Slaughter M.J., Shanle E.K., Khan A., Chua K.F., Hong T., Boxer L.D., Allis C.D., Josefowicz S.Z., Garcia B.A., Rothbart S.B., Strahl B.D., Davis I.J. HDAC inhibition results in widespread alteration of the histone acetylation landscape and BRD4 targeting to gene bodies. Cell Rep. 2021;34(3) doi: 10.1016/j.celrep.2020.108638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu J., Livingston M.J., Dong G., Tang C., Su Y., Wu G., Yin X.M., Dong Z. Histone deacetylase inhibitors protect against cisplatin-induced acute kidney injury by activating autophagy in proximal tubular cells. Cell Death Dis. 2018;9(3):322. doi: 10.1038/s41419-018-0374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rahhal R., Seto E. Emerging roles of histone modifications and HDACs in RNA splicing. Nucleic Acids Res. 2019;47(10):4911–4926. doi: 10.1093/nar/gkz292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cabanel M., da Costa T.P., El-Cheikh M.C., Carneiro K. The epigenome as a putative target for skin repair: the HDAC inhibitor Trichostatin A modulates myeloid progenitor plasticity and behavior and improves wound healing. J. Transl. Med. 2019;17(1):247. doi: 10.1186/s12967-019-1998-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beegum F., A. P V., George K.T., D. K P., Begum F., Krishnadas N., Shenoy R.R. Sirtuins as therapeutic targets for improving delayed wound healing in diabetes. J. Drug Target. 2022:1–16. doi: 10.1080/1061186x.2022.2085729. [DOI] [PubMed] [Google Scholar]
- 105.Fu W., Liang D., Wu X., Chen H., Hong X., Wang J., Zhu T., Zeng T., Lin W., Chen S., Yan L., Ren M. Long noncoding RNA LINC01435 impedes diabetic wound healing by facilitating YY1-mediated HDAC8 expression. iScience. 2022;25(4) doi: 10.1016/j.isci.2022.104006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yao F., Jin Z., Zheng Z., Lv X., Ren L., Yang J., Chen D., Wang B., Yang W., Chen L., Wang W., Gu J., Lin R. HDAC11 promotes both NLRP3/caspase-1/GSDMD and caspase-3/GSDME pathways causing pyroptosis via ERG in vascular endothelial cells. Cell Death Dis. 2022;8(1):112. doi: 10.1038/s41420-022-00906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Galvan-Pena S., O'Neill L.A. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liberti M.V., Locasale J.W. Histone lactylation: a new role for glucose metabolism. Trends Biochem. Sci. 2020;45(3):179–182. doi: 10.1016/j.tibs.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 109.Notarangelo G., Haigis M.C. Sweet temptation: from sugar metabolism to gene regulation. Immunity. 2019;51(6):980–981. doi: 10.1016/j.immuni.2019.11.008. [DOI] [PubMed] [Google Scholar]
- 110.Sar P., Dalai S. CRISPR/Cas9 in epigenetics studies of health and disease. Prog Mol Biol Transl Sci. 2021;181:309–343. doi: 10.1016/bs.pmbts.2021.01.022. [DOI] [PubMed] [Google Scholar]
- 111.Wang J., Qiu Z., Wu Y. Ubiquitin regulation: the histone modifying enzyme's story. Cells. 2018;7(9) doi: 10.3390/cells7090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nishiyama A., Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021;37(11):1012–1027. doi: 10.1016/j.tig.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 113.Li S., Peng Y., Panchenko A.R. DNA methylation: precise modulation of chromatin structure and dynamics. Curr. Opin. Struct. Biol. 2022;75 doi: 10.1016/j.sbi.2022.102430. [DOI] [PubMed] [Google Scholar]
- 114.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zsindely N., Siagi F., Bodai L. DNA methylation in huntington's disease. Int. J. Mol. Sci. 2021;22(23) doi: 10.3390/ijms222312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Babu M., Durga Devi T., Mäkinen P., Kaikkonen M., Lesch H.P., Junttila S., Laiho A., Ghimire B., Gyenesei A., Ylä-Herttuala S. Differential promoter methylation of macrophage genes is associated with impaired vascular growth in ischemic muscles of hyperlipidemic and type 2 diabetic mice: genome-wide promoter methylation study. Circ. Res. 2015;117(3):289–299. doi: 10.1161/circresaha.115.306424. [DOI] [PubMed] [Google Scholar]
- 117.Pasquier J., Spurgeon M., Bradic M., Thomas B., Robay A., Chidiac O., Dib M.J., Turjoman R., Liberska A., Staudt M., Fakhro K.A., Menzies R., Jayyousi A., Zirie M., Suwaidi J.A., Malik R.A., Talal T., Rafii A., Mezey J., Rodriguez-Flores J., Crystal R.G., Abi Khalil C. Whole-methylome analysis of circulating monocytes in acute diabetic Charcot foot reveals differentially methylated genes involved in the formation of osteoclasts. Epigenomics. 2019;11(3):281–296. doi: 10.2217/epi-2018-0144. [DOI] [PubMed] [Google Scholar]
- 118.Singh K., Pal D., Sinha M., Ghatak S., Gnyawali S.C., Khanna S., Roy S., Sen C.K. Epigenetic modification of MicroRNA-200b contributes to diabetic vasculopathy. Mol. Ther. 2017;25(12):2689–2704. doi: 10.1016/j.ymthe.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liang Y., Yang C., Lin Y., Parviz Y., Sun K., Wang W., Ren M., Yan L. Matrix metalloproteinase 9 induces keratinocyte apoptosis through FasL/Fas pathway in diabetic wound. Apoptosis. 2019;24(7–8):542–551. doi: 10.1007/s10495-019-01536-w. [DOI] [PubMed] [Google Scholar]
- 120.Meevassana J., Nacharoenkul P., Wititsuwannakul J., Kitkumthorn N., Hamill K.J., Angspatt A., Mutirangura A. B1 repetitive sequence methylation enhances wound healing of second-degree burns in rats. Biomedical reports. 2022;16(3):20. doi: 10.3892/br.2022.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yasom S., Khumsri W., Boonsongserm P., Kitkumthorn N., Ruangvejvorachai P., Sooksamran A., Wanotayan R., Mutirangura A. B1 siRNA Increases de novo DNA Methylation of B1 Elements and Promotes Wound Healing in Diabetic Rats. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.802024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chalertpet K., Pin-On P., Aporntewan C., Patchsung M., Ingrungruanglert P., Israsena N., Mutirangura A. Argonaute 4 as an effector protein in RNA-directed DNA methylation in human cells. Front. Genet. 2019;10:645. doi: 10.3389/fgene.2019.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ludwig-Slomczynska A.H., Borys S., Seweryn M.T., Hohendorff J., Kapusta P., Kiec-Wilk B., Pitera E., Wolkow P.P., Malecki M.T. DNA methylation analysis of negative pressure therapy effect in diabetic foot ulcers. Endocr Connect. 2019;8(11):1474–1482. doi: 10.1530/EC-19-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mora-Castilla S., Tejedo J.R., Hmadcha A., Cahuana G.M., Martín F., Soria B., Bedoya F.J. Nitric oxide repression of Nanog promotes mouse embryonic stem cell differentiation. Cell Death Differ. 2010;17(6):1025–1033. doi: 10.1038/cdd.2009.204. [DOI] [PubMed] [Google Scholar]
- 125.Park J., Suh D., Tang T., Lee H.J., Roe J.S., Kim G.C., Han S., Song K. Non-thermal atmospheric pressure plasma induces epigenetic modifications that activate the expression of various cytokines and growth factors in human mesoderm-derived stem cells. Free Radic. Biol. Med. 2020;148:108–122. doi: 10.1016/j.freeradbiomed.2019.12.035. [DOI] [PubMed] [Google Scholar]
- 126.Xue W., Zhang Q., Chen Y., Zhu Y. Hydrogen sulfide improves angiogenesis by regulating the transcription of pri-miR-126 in diabetic endothelial cells. Cells. 2022;11(17) doi: 10.3390/cells11172651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cao W., Dong Y., Geng Y., Bi S., Liu Z., Zhou L., Sun X., Xia S., Chi C., Wu B. Comprehensive analysis of whole-transcriptome profiles in response to acute hypersaline challenge in Chinese razor clam sinonovacula constricta. Biology. 2023;12(1) doi: 10.3390/biology12010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peixoto P., Cartron P.F., Serandour A.A., Hervouet E. From 1957 to nowadays: a brief history of epigenetics. Int. J. Mol. Sci. 2020;21(20) doi: 10.3390/ijms21207571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mohr A.M., Mott J.L. Overview of microRNA biology. Semin. Liver Dis. 2015;35(1):3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ozdemir D., Feinberg M.W. MicroRNAs in diabetic wound healing: pathophysiology and therapeutic opportunities. Trends Cardiovasc. Med. 2019;29(3):131–137. doi: 10.1016/j.tcm.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Umehara T., Mori R., Mace K.A., Murase T., Abe Y., Yamamoto T., Ikematsu K. Identification of specific miRNAs in neutrophils of type 2 diabetic mice: overexpression of miRNA-129-2-3p accelerates diabetic wound healing. Diabetes. 2019;68(3):617–630. doi: 10.2337/db18-0313. [DOI] [PubMed] [Google Scholar]
- 133.Ban E., Jeong S., Park M., Kwon H., Park J., Song E.J., Kim A. Accelerated wound healing in diabetic mice by miRNA-497 and its anti-inflammatory activity. Biomed. Pharmacother. 2020;121 doi: 10.1016/j.biopha.2019.109613. [DOI] [PubMed] [Google Scholar]
- 134.Peng X., He F., Mao Y., Lin Y., Fang J., Chen Y., Sun Z., Zhuo Y., Jiang J. miR-146a promotes M2 macrophage polarization and accelerates diabetic wound healing by inhibiting the TLR4/NF-κB axis. J. Mol. Endocrinol. 2022;69(2):315–327. doi: 10.1530/jme-21-0019. [DOI] [PubMed] [Google Scholar]
- 135.Liu J., Wang J., Fu W., Wang X., Chen H., Wu X., Lao G., Wu Y., Hu M., Yang C., Yan L., Ren M. MiR-195-5p and miR-205-5p in extracellular vesicles isolated from diabetic foot ulcer wound fluid decrease angiogenesis by inhibiting VEGFA expression. Aging (Albany NY) 2021;13(15):19805–19821. doi: 10.18632/aging.203393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liang Z.H., Pan N.F., Lin S.S., Qiu Z.Y., Liang P., Wang J., Zhang Z., Pan Y.C. Exosomes from mmu_circ_0001052-modified adipose-derived stem cells promote angiogenesis of DFU via miR-106a-5p and FGF4/p38MAPK pathway. Stem Cell Res. Ther. 2022;13(1):336. doi: 10.1186/s13287-022-03015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu Q., Liu L., Zhang X., Chang H., Ma S., Xie Z., Tang S., Ju X., Zhu H., Shen B., Zhang Q. MiR-221-3p targets HIPK2 to promote diabetic wound healing. Microvasc. Res. 2022;140 doi: 10.1016/j.mvr.2021.104306. [DOI] [PubMed] [Google Scholar]
- 138.Wei Q., Wang Y., Ma K., Li Q., Li B., Hu W., Fu X., Zhang C. Extracellular vesicles from human umbilical cord mesenchymal stem cells facilitate diabetic wound healing through MiR-17-5p-mediated enhancement of angiogenesis. Stem Cell Rev Rep. 2022;18(3):1025–1040. doi: 10.1007/s12015-021-10176-0. [DOI] [PubMed] [Google Scholar]
- 139.Moura J., Sørensen A., Leal E.C., Svendsen R., Carvalho L., Willemoes R.J., Jørgensen P.T., Jenssen H., Wengel J., Dalgaard L.T., Carvalho E. microRNA-155 inhibition restores Fibroblast Growth Factor 7 expression in diabetic skin and decreases wound inflammation. Sci. Rep. 2019;9(1):5836. doi: 10.1038/s41598-019-42309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ionescu R.F., Enache R.M., Cretoiu S.M., Cretoiu D. The interplay between gut microbiota and miRNAs in cardiovascular diseases. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li M., Chen W.D., Wang Y.D. The roles of the gut microbiota-miRNA interaction in the host pathophysiology. Mol. Med. 2020;26(1):101. doi: 10.1186/s10020-020-00234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bitar A., Aung K.M., Wai S.N., Hammarström M.L. Vibrio cholerae derived outer membrane vesicles modulate the inflammatory response of human intestinal epithelial cells by inducing microRNA-146a. Sci. Rep. 2019;9(1):7212. doi: 10.1038/s41598-019-43691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 144.Kar R., Jha N.K., Jha S.K., Sharma A., Dholpuria S., Asthana N., Chaurasiya K., Singh V.K., Burgee S., Nand P. A "NOTCH" deeper into the epithelial-to-mesenchymal transition (EMT) program in breast cancer. Genes. 2019;10(12) doi: 10.3390/genes10120961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Desai P., Yang J., Tian B., Sun H., Kalita M., Ju H., Paulucci-Holthauzen A., Zhao Y., Brasier A.R., Sadygov R.G. Mixed-effects model of epithelial-mesenchymal transition reveals rewiring of signaling networks. Cell. Signal. 2015;27(7):1413–1425. doi: 10.1016/j.cellsig.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu L., Chen R., Jia Z., Li X., Tang Y., Zhao X., Zhang S., Luo L., Fang Z., Zhang Y., Chen M. Downregulation of hsa-miR-203 in peripheral blood and wound margin tissue by negative pressure wound therapy contributes to wound healing of diabetic foot ulcers. Microvasc. Res. 2022;139 doi: 10.1016/j.mvr.2021.104275. [DOI] [PubMed] [Google Scholar]
- 147.Yuan L., Sun Y., Xu M., Zeng F., Xiong X. miR-203 acts as an inhibitor for epithelial-mesenchymal transition process in diabetic foot ulcers via targeting interleukin-8. Neuroimmunomodulation. 2019;26(5):239–249. doi: 10.1159/000503087. [DOI] [PubMed] [Google Scholar]
- 148.Xu Y.X., Pu S.D., Li X., Yu Z.W., Zhang Y.T., Tong X.W., Shan Y.Y., Gao X.Y. Exosomal ncRNAs: novel therapeutic target and biomarker for diabetic complications. Pharmacol. Res. 2022;178 doi: 10.1016/j.phrs.2022.106135. [DOI] [PubMed] [Google Scholar]
- 149.Yu P., Guo J., Li J., Chen W., Zhao T. Co-expression network analysis revealing the key lncRNAs in diabetic foot ulcers. Arch. Med. Sci. : AMS. 2019;15(5):1123–1132. doi: 10.5114/aoms.2019.84699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hu M., Wu Y., Yang C., Wang X., Wang W., Zhou L., Zeng T., Zhou J., Wang C., Lao G., Yan L., Ren M. Novel long noncoding RNA lnc-URIDS delays diabetic wound healing by targeting Plod1. Diabetes. 2020;69(10):2144–2156. doi: 10.2337/db20-0147. [DOI] [PubMed] [Google Scholar]
- 152.Li B., Zhou Y., Chen J., Wang T., Li Z., Fu Y., Bi C., Zhai A. Long non-coding RNA H19 contributes to wound healing of diabetic foot ulcer. J. Mol. Endocrinol. 2020 doi: 10.1530/jme-19-0242. [DOI] [PubMed] [Google Scholar]
- 153.Peng W.X., He P.X., Liu L.J., Zhu T., Zhong Y.Q., Xiang L., Peng K., Yang J.J., Xiang G.D. LncRNA GAS5 activates the HIF1A/VEGF pathway by binding to TAF15 to promote wound healing in diabetic foot ulcers. Lab. Invest. 2021;101(8):1071–1083. doi: 10.1038/s41374-021-00598-2. [DOI] [PubMed] [Google Scholar]
- 154.Chen X., Peng Y., Xue H., Liu G., Wang N., Shao Z. MiR-21 regulating PVT1/PTEN/IL-17 axis towards the treatment of infectious diabetic wound healing by modified GO-derived biomaterial in mouse models. J. Nanobiotechnol. 2022;20(1):309. doi: 10.1186/s12951-022-01516-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li B., Luan S., Chen J., Zhou Y., Wang T., Li Z., Fu Y., Zhai A., Bi C. The MSC-derived exosomal lncRNA H19 promotes wound healing in diabetic foot ulcers by upregulating PTEN via MicroRNA-152-3p, molecular therapy. Nucleic acids. 2020;19:814–826. doi: 10.1016/j.omtn.2019.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li B., Zhou Y., Chen J., Wang T., Li Z., Fu Y., Zhai A., Bi C. Long noncoding RNA H19 acts as a miR-29b sponge to promote wound healing in diabetic foot ulcer. Faseb. J. : official publication of the Federation of American Societies for Experimental Biology. 2021;35(1) doi: 10.1096/fj.201900076RRRRR. [DOI] [PubMed] [Google Scholar]
- 157.Li Y., Zhi K., Han S., Li X., Li M., Lian W., Zhang H., Zhang X. TUG1 enhances high glucose-impaired endothelial progenitor cell function via miR-29c-3p/PDGF-BB/Wnt signaling. Stem Cell Res. Ther. 2020;11(1):441. doi: 10.1186/s13287-020-01958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ding J., Gao B., Chen Z., Mei X. An NIR-triggered Au nanocage used for photo-thermo therapy of chronic wound in diabetic rats through bacterial membrane destruction and skin cell mitochondrial protection. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.779944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zgheib C., Hodges M.M., Hu J., Liechty K.W., Xu J. Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Herter E.K., Li D., Toma M.A., Vij M., Li X., Visscher D., Wang A., Chu T., Sommar P., Blomqvist L., Berglund D., Ståhle M., Wikstrom J.D., Xu Landén N. WAKMAR2, a long noncoding RNA downregulated in human chronic wounds, modulates keratinocyte motility and production of inflammatory chemokines. J. Invest. Dermatol. 2019;139(6):1373–1384. doi: 10.1016/j.jid.2018.11.033. [DOI] [PubMed] [Google Scholar]
- 161.Wu X., He W., Mu X., Liu Y., Deng J., Liu Y., Nie X. Macrophage polarization in diabetic wound healing. Burns Trauma. 2022;10 doi: 10.1093/burnst/tkac051. tkac051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu J., Zhang L., Liechty C., Zgheib C., Hodges M.M., Liechty K.W., Xu J. Long noncoding RNA GAS5 regulates macrophage polarization and diabetic wound healing. J. Invest. Dermatol. 2020;140(8):1629–1638. doi: 10.1016/j.jid.2019.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kuang L., Zhang C., Li B., Deng H., Chen R., Li G. Human keratinocyte-derived exosomal MALAT1 promotes diabetic wound healing by upregulating MFGE8 via microRNA-1914-3p. Int. J. Nanomed. 2023;18:949–970. doi: 10.2147/ijn.S399785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shyu K.G., Wang B.W., Fang W.J., Pan C.M., Lin C.M. Exosomal MALAT1 derived from high glucose-treated macrophages up-regulates resistin expression via miR-150-5p downregulation. Int. J. Mol. Sci. 2022;23(3) doi: 10.3390/ijms23031095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Qian L., Pi L., Fang B.R., Meng X.X. Adipose mesenchymal stem cell-derived exosomes accelerate skin wound healing via the lncRNA H19/miR-19b/SOX9 axis. Lab. Invest. 2021;101(9):1254–1266. doi: 10.1038/s41374-021-00611-8. [DOI] [PubMed] [Google Scholar]
- 166.Kuang L.W., Zhang C.C., Li B.H., Liu H.Z., Wang H., Li G.C. Identification of the MALAT1/miR-106a-5p/ZNF148 feedback loop in regulating HaCaT cell proliferation, migration and apoptosis. Regen. Med. 2023;18(3):239–258. doi: 10.2217/rme-2022-0189. [DOI] [PubMed] [Google Scholar]
- 167.Cao W., Feng Y. LncRNA XIST promotes extracellular matrix synthesis, proliferation and migration by targeting miR-29b-3p/COL1A1 in human skin fibroblasts after thermal injury. Biol. Res. 2019;52(1):52. doi: 10.1186/s40659-019-0260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang J., Deng P., Qi Y., Feng X., Wen H., Chen F. NEAT1 knockdown inhibits keloid fibroblast progression by miR-196b-5p/FGF2 Axis. J. Surg. Res. 2021;259:261–270. doi: 10.1016/j.jss.2020.09.038. [DOI] [PubMed] [Google Scholar]
- 169.Zhang W., Sui Y. CircBPTF knockdown ameliorates high glucose-induced inflammatory injuries and oxidative stress by targeting the miR-384/LIN28B axis in human umbilical vein endothelial cells. Mol. Cell. Biochem. 2020;471(1–2):101–111. doi: 10.1007/s11010-020-03770-2. [DOI] [PubMed] [Google Scholar]
- 170.Shi R., Jin Y., Zhao S., Yuan H., Shi J., Zhao H. Hypoxic ADSC-derived exosomes enhance wound healing in diabetic mice via delivery of circ-Snhg11 and induction of M2-like macrophage polarization. Biomed. Pharmacother. 2022;153 doi: 10.1016/j.biopha.2022.113463. [DOI] [PubMed] [Google Scholar]
- 171.Shi R., Jin Y., Hu W., Lian W., Cao C., Han S., Zhao S., Yuan H., Yang X., Shi J., Zhao H. Exosomes derived from mmu_circ_0000250-modified adipose-derived mesenchymal stem cells promote wound healing in diabetic mice by inducing miR-128-3p/SIRT1-mediated autophagy. Am. J. Physiol. Cell Physiol. 2020;318(5):C848–c856. doi: 10.1152/ajpcell.00041.2020. [DOI] [PubMed] [Google Scholar]
- 172.Shang B., Xu T., Hu N., Mao Y., Du X. Circ-Klhl8 overexpression increased the therapeutic effect of EPCs in diabetic wound healing via the miR-212-3p/SIRT5 axis. J. Diabet. Complicat. 2021;35(11) doi: 10.1016/j.jdiacomp.2021.108020. [DOI] [PubMed] [Google Scholar]
- 173.Zheng J., Mao Y., Dong P., Huang Z., Yu F. Long noncoding RNA HOTTIP mediates SRF expression through sponging miR-150 in hepatic stellate cells. J. Cell Mol. Med. 2019;23(2):1572–1580. doi: 10.1111/jcmm.14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tian D., Xiang Y., Tang Y., Ge Z., Li Q., Zhang Y. Circ-ADAM9 targeting PTEN and ATG7 promotes autophagy and apoptosis of diabetic endothelial progenitor cells by sponging mir-20a-5p. Cell Death Dis. 2020;11(7):526. doi: 10.1038/s41419-020-02745-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang Z., Feng C., Liu H., Meng T., Huang W., Long X., Wang X. Hypoxic pretreatment of adipose-derived stem cells accelerates diabetic wound healing via circ-gcap14 and HIF-1α/VEGF mediated angiopoiesis. Int J Stem Cells. 2021;14(4):447–454. doi: 10.15283/ijsc21050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xi L., Carroll T., Matos I., Luo J.D., Polak L., Pasolli H.A., Jaffrey S.R., Fuchs E. m6A RNA methylation impacts fate choices during skin morphogenesis. Elife. 2020;9 doi: 10.7554/eLife.56980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee J., Wu Y., Harada B.T., Li Y., Zhao J., He C., Ma Y., Wu X. N(6) -methyladenosine modification of lncRNA Pvt1 governs epidermal stemness. EMBO J. 2021;40(8) doi: 10.15252/embj.2020106276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhou J., Wei T., He Z. ADSCs enhance VEGFR3-mediated lymphangiogenesis via METTL3-mediated VEGF-C m(6)A modification to improve wound healing of diabetic foot ulcers. Mol. Med. 2021;27(1):146. doi: 10.1186/s10020-021-00406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang Y., Gu X., Li D., Cai L., Xu Q. METTL3 regulates osteoblast differentiation and inflammatory response via smad signaling and MAPK signaling. Int. J. Mol. Sci. 2019;21(1) doi: 10.3390/ijms21010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang J., Yan S., Lu H., Wang S., Xu D. METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-κB signaling pathway. Mediat. Inflamm. 2019;2019 doi: 10.1155/2019/3120391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Feng Z., Li Q., Meng R., Yi B., Xu Q. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell Mol. Med. 2018;22(5):2558–2568. doi: 10.1111/jcmm.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jian D., Wang Y., Jian L., Tang H., Rao L., Chen K., Jia Z., Zhang W., Liu Y., Chen X., Shen X., Gao C., Wang S., Li M. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics. 2020;10(20):8939–8956. doi: 10.7150/thno.45178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liu Y., Liu Z., Tang H., Shen Y., Gong Z., Xie N., Zhang X., Wang W., Kong W., Zhou Y., Fu Y. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am. J. Physiol. Cell Physiol. 2019;317(4):C762–c775. doi: 10.1152/ajpcell.00212.2019. [DOI] [PubMed] [Google Scholar]
- 185.Huangfu N., Zheng W., Xu Z., Wang S., Wang Y., Cheng J., Li Z., Cheng K., Zhang S., Chen X., Zhu J. RBM4 regulates M1 macrophages polarization through targeting STAT1-mediated glycolysis. Int Immunopharmacol. 2020;83 doi: 10.1016/j.intimp.2020.106432. [DOI] [PubMed] [Google Scholar]
- 186.Gu X., Zhang Y., Li D., Cai H., Cai L., Xu Q. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell. Signal. 2020;69 doi: 10.1016/j.cellsig.2020.109553. [DOI] [PubMed] [Google Scholar]
- 187.Li L., Zhang J., Zhang Q., Zhang D., Xiang F., Jia J., Wei P., Zhang J., Hu J., Huang Y. High glucose suppresses keratinocyte migration through the inhibition of p38 MAPK/autophagy pathway. Front. Physiol. 2019;10:24. doi: 10.3389/fphys.2019.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Qiang L., Yang S., Cui Y.H., He Y.Y. Keratinocyte autophagy enables the activation of keratinocytes and fibroblastsand facilitates wound healing. Autophagy. 2021;17(9):2128–2143. doi: 10.1080/15548627.2020.1816342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xiao M., Li L., Hu Q., Ma L., Liu L., Chu W., Zhang H. Rapamycin reduces burn wound progression by enhancing autophagy in deep second-degree burn in rats. Wound Repair Regen. 2013;21(6):852–859. doi: 10.1111/wrr.12090. [DOI] [PubMed] [Google Scholar]
- 190.Liang D., Lin W.J., Ren M., Qiu J., Yang C., Wang X., Li N., Zeng T., Sun K., You L., Yan L., Wang W. m(6)A reader YTHDC1 modulates autophagy by targeting SQSTM1 in diabetic skin. Autophagy. 2022;18(6):1318–1337. doi: 10.1080/15548627.2021.1974175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen Y., Wang J., Xu D., Xiang Z., Ding J., Yang X., Li D., Han X. m(6)A mRNA methylation regulates testosterone synthesis through modulating autophagy in Leydig cells. Autophagy. 2021;17(2):457–475. doi: 10.1080/15548627.2020.1720431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sass P., Sosnowski P., Podolak-Popinigis J., Gornikiewicz B., Kaminska J., Deptula M., Nowicka E., Wardowska A., Ruczynski J., Rekowski P., Rogujski P., Filipowicz N., Mieczkowska A., Peszynska-Sularz G., Janus L., Skowron P., Czupryn A., Mucha P., Piotrowski A., Rodziewicz-Motowidlo S., Pikula M., Sachadyn P. Epigenetic inhibitor zebularine activates ear pinna wound closure in the mouse. EBioMedicine. 2019;46:317–329. doi: 10.1016/j.ebiom.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xue Y., Chen C., Tan R., Zhang J., Fang Q., Jin R., Mi X., Sun D., Xue Y., Wang Y., Xiong R., Lu H., Tan W. Artificial intelligence-assisted bioinformatics, microneedle, and diabetic wound healing: a "new deal" of an old drug. ACS Appl. Mater. Interfaces. 2022 doi: 10.1021/acsami.2c08994. [DOI] [PubMed] [Google Scholar]
- 194.Xue Y., Chen C., Tan R., Zhang J., Fang Q., Jin R., Mi X., Sun D., Xue Y., Wang Y., Xiong R., Lu H., Tan W. Artificial intelligence-assisted bioinformatics, microneedle, and diabetic wound healing: a "new deal" of an old drug. ACS applied materials & interfaces. 2022;14(33):37396–37409. doi: 10.1021/acsami.2c08994. [DOI] [PubMed] [Google Scholar]
- 195.De Riso G., Cocozza S. Artificial intelligence for epigenetics: towards personalized medicine. Curr. Med. Chem. 2021;28(32):6654–6674. doi: 10.2174/0929867327666201117142006. [DOI] [PubMed] [Google Scholar]
- 196.Hamamoto R., Komatsu M., Takasawa K., Asada K., Kaneko S. Epigenetics analysis and integrated analysis of multiomics data, including epigenetic data, using artificial intelligence in the era of precision medicine. Biomolecules. 2019;10(1) doi: 10.3390/biom10010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nishiga M., Liu C., Qi L.S., Wu J.C. The use of new CRISPR tools in cardiovascular research and medicine. Nat. Rev. Cardiol. 2022;19(8):505–521. doi: 10.1038/s41569-021-00669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nuñez J.K., Chen J., Pommier G.C., Cogan J.Z., Replogle J.M., Adriaens C., Ramadoss G.N., Shi Q., Hung K.L., Samelson A.J., Pogson A.N., Kim J.Y.S., Chung A., Leonetti M.D., Chang H.Y., Kampmann M., Bernstein B.E., Hovestadt V., Gilbert L.A., Weissman J.S. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184(9):2503–2519.e17. doi: 10.1016/j.cell.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jogam P., Sandhya D., Alok A., Peddaboina V., Allini V.R., Zhang B. A review on CRISPR/Cas-based epigenetic regulation in plants. Int. J. Biol. Macromol. 2022;219:1261–1271. doi: 10.1016/j.ijbiomac.2022.08.182. [DOI] [PubMed] [Google Scholar]
- 200.Syding L.A., Nickl P., Kasparek P., Sedlacek R. CRISPR/Cas9 epigenome editing potential for rare imprinting diseases: a review. Cells. 2020;9(4) doi: 10.3390/cells9040993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xie N., Zhou Y., Sun Q., Tang B. Novel epigenetic techniques provided by the CRISPR/Cas9 system. Stem Cell. Int. 2018;2018 doi: 10.1155/2018/7834175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Stover J.D., Farhang N., Berrett K.C., Gertz J., Lawrence B., Bowles R.D. CRISPR epigenome editing of AKAP150 in DRG neurons abolishes degenerative IVD-induced neuronal activation. Mol. Ther. 2017;25(9):2014–2027. doi: 10.1016/j.ymthe.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hofacker D., Broche J., Laistner L., Adam S., Bashtrykov P., Jeltsch A. Engineering of effector domains for targeted DNA methylation with reduced off-target effects. Int. J. Mol. Sci. 2020;21(2) doi: 10.3390/ijms21020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ju C., Liu D. Exosomal microRNAs from mesenchymal stem cells: novel therapeutic effect in wound healing. Tissue Eng Regen Med. 2023;20(5):647–660. doi: 10.1007/s13770-023-00542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lu S., Lu L., Liu Y., Li Z., Fang Y., Chen Z., Zhou J. Native and engineered extracellular vesicles for wound healing. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.1053217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hahm J., Kim J., Park J. Strategies to enhance extracellular vesicle production. Tissue Eng Regen Med. 2021;18(4):513–524. doi: 10.1007/s13770-021-00364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jiang B., Yan L., Wang X., Li E., Murphy K., Vaccaro K., Li Y., Xu R.H. Concise review: mesenchymal stem cells derived from human pluripotent cells, an unlimited and quality-controllable source for therapeutic applications. Stem Cell. 2019;37(5):572–581. doi: 10.1002/stem.2964. [DOI] [PubMed] [Google Scholar]
- 208.Lin C.J., Lan Y.M., Ou M.Q., Ji L.Q., Lin S.D. Expression of miR-217 and HIF-1α/VEGF pathway in patients with diabetic foot ulcer and its effect on angiogenesis of diabetic foot ulcer rats. J. Endocrinol. Invest. 2019;42(11):1307–1317. doi: 10.1007/s40618-019-01053-2. [DOI] [PubMed] [Google Scholar]
- 209.Cai H.A., Huang L., Zheng L.J., Fu K., Wang J., Hu F.D., Liao R.Y. Ginsenoside (Rg-1) promoted the wound closure of diabetic foot ulcer through iNOS elevation via miR-23a/IRF-1 axis. Life Sci. 2019;233 doi: 10.1016/j.lfs.2019.05.081. [DOI] [PubMed] [Google Scholar]
- 210.Sawaya A.P., Jozic I., Stone R.C., Pastar I., Egger A.N., Stojadinovic O., Glinos G.D., Kirsner R.S., Tomic-Canic M. Mevastatin promotes healing by targeting caveolin-1 to restore EGFR signaling. JCI insight. 2019;4(23) doi: 10.1172/jci.insight.129320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.