Abstract
Immune checkpoint inhibitor-mediated colitis (IMC) is a common adverse event of treatment with immune checkpoint inhibitors (ICI). We hypothesize that genetic susceptibility to Crohn’s disease (CD) and ulcerative colitis (UC) predisposes to IMC. In this study, we first develop a polygenic risk scores for CD (PRSCD) and UC (PRSUC) in cancer-free individuals and then test these PRSs on IMC in a cohort of 1316 patients with ICI-treated non-small cell lung cancer and perform a replication in 873 ICI-treated pan-cancer patients. In a meta-analysis, the PRSUC predicts all-grade IMC (ORmeta=1.35 per standard deviation [SD], 95% CI = 1.12–1.64, P = 2×10−03) and severe IMC (ORmeta=1.49 per SD, 95% CI = 1.18–1.88, P = 9×10−04). PRSCD is not associated with IMC. Furthermore, PRSUC predicts severe IMC among patients treated with combination ICIs (ORmeta=2.20 per SD, 95% CI = 1.07–4.53, P = 0.03). Overall, PRSUC can identify patients receiving ICI at risk of developing IMC and may be useful to monitor patients and improve patient outcomes.
Subject terms: Outcomes research, Non-small-cell lung cancer, Cancer genetics, Cancer epidemiology
Colitis is one of the most common immune-related adverse events in patients with cancer treated with immune checkpoint inhibitors. Here the authors show that a polygenic risk score for ulcerative colitis can predict immune checkpoint inhibitor-mediated colitis in patients with cancer.
Introduction
Immunotherapy with immune checkpoint inhibitors (ICI) has substantially improved clinical outcomes in patients with advanced cancers such as melanoma, non-small cell lung cancer (NSCLC), bladder, renal, breast, and other cancers1–7. ICIs block the ability of malignant cells to escape detection through immune checkpoints such as programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) or cytotoxic T-lymphocyte associated protein 4 (CTLA-4). Blockade of these checkpoints restores host immunosurveillance in some tumors by stimulating cytotoxic T-cells to induce cancer cell apoptosis2,8–11.
Despite ICIs being a paradigm-shifting breakthrough in cancer treatment, enhanced activation of the immune system can lead to immune-related adverse events (irAEs) that can result in permanent discontinuation of ICIs, severe morbidity, and even patient death12–14. The most severe irAEs include hypophysitis, diabetes, colitis, hepatitis, and pneumonitis, with other common irAEs including rash and thyroiditis12,15–17. The incidence of immune checkpoint inhibitor-mediated colitis (IMC) ranges from 1%-25% and varies by ICI therapy18,19. The incidence of IMC is higher in patients treated with combined anti-PD-1/PD-L1 and anti-CTLA4 therapy20,21. Nearly 15-20% of patients receiving combination therapy develop severe IMC, which is the leading cause of hospitalization and treatment cessation13,14,18,20,21. Endoscopic and histological findings suggest that the presentation of IMC mimics autoimmune colitis such as ulcerative colitis (UC), a form of inflammatory bowel disease (IBD)22,23. Despite the phenotypic similarities between IBD and IMC, it is unclear if the underlying mechanism is shared or distinct.
In this study, we sought to characterize the relationship between genetic predisposition to common types of autoimmune colitis (ulcerative colitis (UC) and Crohn’s disease (CD)), and IMC in a cohort of NSCLC patients receiving ICI treatment. We first develop polygenic risk scores (PRS) for UC and CD using individuals not diagnosed with cancer at baseline in UK Biobank (UKB) and validate these PRSs in an independent dataset of cancer-free participants in Vanderbilt University Medical Center biobank (BioVU)24. Next, we evaluate the association between each of these PRS and the development of IMC in a cohort of patients with NSCLC receiving ICI therapy and conduct an independent replication in a cohort of patients with diverse cancer types treated with ICI therapy in BioVU24. We also investigate the association between human leukocyte antigen (HLA) alleles known to affect UC risk with IMC. Additionally, we examine the role of IMC and PRS for UC, and CD, respectively, on progression-free survival (PFS) and overall survival (OS).
Results
Patient characteristics
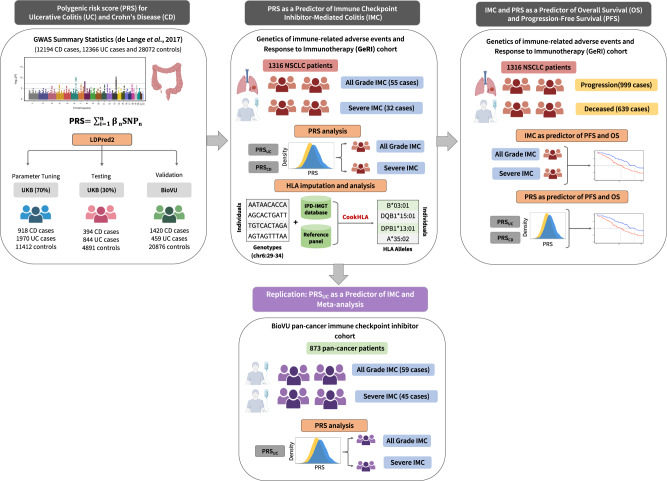
We analyzed data from 1316 study participants included in the GeRI cohort, which included four sites (Supplementary Table 1 and see “Methods”). The GeRI cohort comprised approximately 50% men and the mean age at lung cancer diagnosis was 65 years (+/−10.3). The study was composed of 69.5% of individuals who self-reported as White followed by 6.7% identifying as Asian, and 5.3% as Black. A small proportion (9%) received the combined anti-PD-1/PD-L1 and anti-CTLA-4 inhibitor therapy and the remainder received either anti-PD-1 or PD-L1 inhibitor monotherapy (91%). The cumulative incidence of IMC was ~4% (55 events); it was ~2% (32 events) for severe IMC in the GeRI cohort. The rates were similar across all study sites. The analytic strategy of our study is illustrated in Fig. 1.
Fig. 1. Overview of the analytical pipeline.
Development and validation of the polygenic risk scores (PRSs) for ulcerative colitis and Crohn’s disease was conducted in cancer-free individuals using UK Biobank and BioVU. LDPred2 method was used to tune the parameters for the PRS for ulcerative colitis and Crohn’s disease (PRSUC, PRSCD) in 70% of the UK Biobank, using the summary statistics from the largest genome-wide association study of UC and CD. The PRSs were then tested in the remaining 30% of the UK Biobank and validated in BioVU. In the next step, the role of PRSUC and PRSCD on all-grade and severe immune checkpoint inhibitor-mediated colitis (IMC) was evaluated in a cohort of 1316 non-small cell lung cancer patients who received at least one dose of immune checkpoint inhibitor therapy. Furthermore, replication was conducted using 873 pan-cancer patients treated with immune checkpoint inhibitors obtained from BioVU. Finally, associations of all-grade and severe IMC along with PRSUC and PRSCD on progression-free survival (PFS) and overall survival (OS) were assessed. Figure created with BioRender.com.
Development and validation of PRS for UC and CD
We used 70% of the cancer-free UKB dataset to tune parameters for PRS using LDpred225. We then obtained effect estimates for the PRS for CD and UC in the remaining 30% (testing data). In the UKB testing data, the area under the receiver operating curve (AUROC) for the PRSUC was 0.66 (95% CI = 0.64–0.68), and the AUROC for PRSCD was 0.72 (95% CI = 0.69–0.74) (Supplementary Fig. 1). In the adjusted model, PRSUC was strongly associated with UC with an odds ratio (OR) of 1.84 per standard deviation (SD) (95% CI = 1.76–1.93, p < 1.0 × 10−12). Similarly, PRSCD was positively associated with CD with OR of 1.83 per SD (95% CI = 1.72–1.95, p < 1.0 × 10−12). We observed an intermediate correlation between the two PRSs (Pearson correlation = 0.38). Additionally, the AUROC for PRSUC on CD was 0.58 (95% CI: 0.57−0.60), while PRSCD on UC yielded an AUROC of 0.58 (95% CI: 0.56−0.59). These results suggest the presence of some shared genetic susceptibility between UC and CD. However, the distinct genetic factors influencing each phenotype remain the primary drivers of the individual PRS effects. The two PRSs were validated in another sample of cancer-free individuals from BioVU21. Similar to the UKB results, the individual PRSCD and PRSUC were also strongly associated with CD and UC in BioVU. We observed an OR of 2.18 per SD (95% CI = 2.05–2.32, p < 1.0 × 10−12) for PRSCD, and an OR of 1.75 per SD (95% CI = 1.59–1.92, p < 1.0 × 10−12) for PRSUC. The AUROC for PRSCD and PRSUC were 0.72 (95% CI = 0.70–0.73) and 0.65 (95% CI = 0.62–0.68), respectively (Supplementary Fig. 2).
PRS of autoimmune colitis as a predictor of IMC
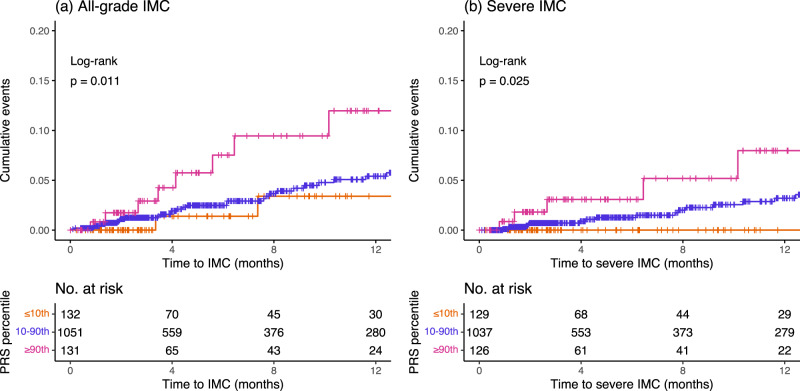
The mean PRSUC was significantly higher in patients who developed IMC (Supplementary Fig. 3). We examined the cumulative incidence of IMC (all-grade and severe) in the top 10th percentile (high genetic risk), 10–90th percentile (average genetic risk), and lowest 10th percentile (low genetic risk) of the PRSUC. Individuals in the top 10th percentile of the PRSUC had higher rates of IMC (all-grade: p = 0.01 and severe: p = 0.03) compared to the other two categories (Fig. 2). Using the Cox proportional hazards model and adjusting for genetic ancestry, recruiting site, age, sex, cancer histology, and type of therapy, we observed that the PRSUC was significantly associated with any diagnosis of IMC in the GeRI cohort with a hazard ratio (HR) of 1.34 per SD (95% CI = 1.02–1.76, p = 0.04). For a diagnosis of severe IMC, the HR per SD was 1.62 (95% CI = 1.12–2.35, p = 0.01) (Table 1). We found no significant association between PRSCD and IMC or severe IMC (Table 1).
Fig. 2. Cumulative incidence curves of all-grade and severe immune checkpoint inhibitor-mediated colitis by polygenic risk score of ulcerative colitis in the GeRI cohort.
Cumulative incidence curves of a All-grade immune checkpoint inhibitor-mediated colitis (IMC) and b Severe IMC by categories of polygenic risk score of ulcerative colitis (PRSUC) in the entire GeRI cohort. Cumulative incidence curves are unadjusted, and PRSUC is categorized as ≤10th percentile (low genetic risk), 10–90th percentile (average genetic risk), and >90th percentile (high genetic risk). The p-values included on each plot are the results of a log-rank test for the difference between the curves (two-sided). Underneath each set of curves is the number of study participants at risk beyond that time point for each of the PRS groups. Source data are provided as a Source Data file.
Table 1.
Polygenic risk score (PRS) of ulcerative colitis (UC) and Crohn’s disease (CD) as a predictor of time to development of all-grade and severe immune checkpoint inhibitor-mediated colitis (IMC) in the entire GeRI cohort, using Cox proportional hazards models and stratified analysis assessing the association between PRSUC and all-grade/severe IMC by type of therapy and lung cancer histology
| PRSa | All-grade IMC | Severe IMC | ||||
|---|---|---|---|---|---|---|
| HR per SD | 95% CI | p | HR per SD | 95% CI | p | |
| PRSUC | 1.34 | 1.02–1.76 | 0.04 | 1.62 | 1.12–2.35 | 0.01 |
| PRSCD | 0.97 | 0.72–1.32 | 0.87 | 0.99 | 0.66–1.46 | 0.94 |
| Stratified analysis (Restricted to PRSUC) | ||||||
| Therapyb | All-grade IMC | Severe IMC | ||||
| Anti-PD1/Anti-PD-L1 monotherapy | 1.33 | 0.99–1.78 | 0.06 | 1.51 | 1.01–2.27 | 0.04 |
| Anti-PD1/Anti-PD-L1 + Anti-CTLA4 | 1.64 | 0.67–4.03 | 0.28 | 4.31 | 1.08–17.24 | 0.03 |
| Histologyc | All-grade IMC | Severe IMC | ||||
| Adenocarcinoma | 1.43 | 1.06–1.93 | 0.02 | 2.12 | 1.37–3.26 | 6×10−04 |
| Squamous cell carcinoma | 0.79 | 0.16–3.78 | 0.76 | 0.79 | 0.16–3.78 | 0.76 |
All p-values are two-sided.
Statistically significant results are highlighted in bold.
PRS polygenic risk score, IMC immune checkpoint inhibitor-mediated colitis, HR hazard ratio, SD standard deviation, CI confidence interval, UC ulcerative colitis, CD Crohn’s disease.
aModels are adjusted for age at diagnosis, sex, histology, type of therapy, recruiting site, and 5 principal components.
bModels are adjusted for age at diagnosis, sex, histology, recruiting site, and 5 principal components.
cModels are adjusted for age at diagnosis, sex, type of therapy, recruiting site, and 5 principal components.
Additionally, we conducted stratified analysis by type of therapy and histology of lung cancer to further characterize the association between PRSUC and IMC (all-grade and severe). For all-grade IMC, the results showed little attenuation and nominal significance when stratified by type of therapy (Table 1). However, for severe IMC we observed an HR per SD of 1.51 (95% CI = 1.01–2.27, p = 0.04) in patients receiving anti-PD1/anti-PD-L1 monotherapy versus a HR per SD of 4.31 (95% CI = 1.08–17.24, p = 0.03) in those patients receiving a combined therapy. Patients with adenocarcinoma had an HR per SD of 1.43 (95% CI = 1.06–1.93, p = 0.02) for all-grade IMC and an HR per SD of 2.12 (95% CI = 1.37–3.26, p = 6 × 10−04) for severe IMC. We also performed association analyses between ulcerative colitis PRS and IMC using different previously published PRSUC and noted consistent and robust trends toward the association (Supplementary Table 2).
Replication of the association between PRSUC and IMC
Replication was conducted within an independent study of 873 patients from a pan-cancer cohort in BioVU24 who underwent treatment with either anti-PD1/PD-L1 monotherapy or combination ICI therapy. The characteristics of the replication cohort are shown in Supplementary Table 2. Briefly, the replication study consisted of 63% males and 37% females. Among 873 ICI-treated patients, approximately 95% of the patients received anti-PD1/PD-L1 monotherapy and 5% of patients received combined anti-PD-1/PD-L1 and anti-CTLA4 therapy. An additional 274 cancer patients were identified and were treated with anti-CTLA4 monotherapy.
The results from the analysis in the replication study are presented in Table 2. In our analysis of 873 patients, we found a trend toward association between PRSUC and all-grade IMC (OR per SD = 1.29, 95% CI = 0.98–1.69, p = 0.07). However, for PRSUC and severe IMC, we observed statistically significant replication with an OR per SD of 1.39 (95% CI = 1.02–1.90, p = 0.04, Table 2). Within our stratified analysis by type of therapy, for anti-PD1/PD-L1 monotherapy, we observed an OR per SD of 1.25 (95% CI = 0.88−1.78, p = 0.21) for all-grade IMC, while a slightly stronger and nominally significant association was seen for severe IMC (OR per SD = 1.47, 95% CI = 0.96−2.25, p = 0.08). For those receiving dual therapy, we observed an OR per SD of 2.04 (95% CI = 0.79−5.28, p = 0.14) for all-grade IMC and an OR per SD of 1.89 (95% CI = 0.74−4.86, p = 0.19) for severe IMC. Furthermore, we conducted an adjusted logistic regression model within the anti-CTLA4 monotherapy (N = 274) and found an OR per SD of 0.92 (95% CI = 0.67–1.26, p = 0.59) for all-grade IMC. For severe IMC in the anti-CTLA4 monotherapy group, we observed an OR per SD of 1.00 (95% CI = 0.71–1.40, p = 0.99).
Table 2.
Polygenic risk score (PRS) of ulcerative colitis (UC) as a predictor of all-grade and severe immune checkpoint inhibitor-mediated colitis (IMC) in the replication cohort (BioVU) and meta-analysis (GeRI and BioVU), using logistic regression model and stratified analysis assessing the association between PRSUC and all-grade/severe IMC by type of therapy
| IMCa | Replication cohort BioVU | Meta-analysis GeRI+ BioVU | ||||
|---|---|---|---|---|---|---|
| OR per SD | 95% CI | p | OR per SD | 95% CI | p | |
| All-grade | 1.29 | 0.98–1.69 | 0.07 | 1.35 | 1.12–1.64 | 2 x 10−3 |
| Severe | 1.39 | 1.02–1.90 | 0.04 | 1.49 | 1.18–1.88 | 9 x 10−4 |
| Stratified analysis by type of therapy: All-grade IMC | ||||||
| Therapyb | Replication cohort BioVU | Meta-analysis GeRI + BioVU | ||||
| Anti-PD1/Anti-PD-L1 monotherapy | 1.25 | 0.88–1.78 | 0.21 | 1.35 | 1.07–1.69 | 0.01 |
| Anti-PD1/Anti-PD-L1 + Anti-CTLA4 | 2.04 | 0.79–5.28 | 0.14 | 1.80 | 0.95-3.41 | 0.07 |
| Anti-CTLA4 monotherapy | 0.92 | 0.67–1.26 | 0.59 | - | - | - |
| Stratified analysis by type of therapy: Severe IMC | ||||||
| Therapyb | Replication cohort BioVU | Meta-analysis GeRI + BioVU | ||||
| Anti-PD1/Anti-PD-L1 monotherapy | 1.47 | 0.96–2.25 | 0.08 | 1.48 | 1.10–1.98 | 9 x 10−3 |
| Anti-PD1/Anti-PD-L1 + Anti-CTLA4 | 1.89 | 0.74–4.86 | 0.19 | 2.20 | 1.07–4.53 | 0.03 |
| Anti-CTLA4 monotherapy | 1.00 | 0.71–1.40 | 0.99 | - | - | - |
All p-values are two-sided.
Statistically significant results are highlighted in bold.
IMC immune checkpoint inhibitor-mediated colitis, OR odds ratio, SD standard deviation, CI confidence interval.
aModels are adjusted for age at diagnosis, sex, type of therapy, and 5 principal components.
bModels are adjusted for age at diagnosis, sex, and 5 principal components.
Meta-analysis of PRSUC and IMC associations in discovery and replication studies
Next, we performed a meta-analysis using fixed-effect inverse-variance weighting, combining the logistic regression models from the initial GeRI cohort and BioVU replication cohort (Table 2). Our findings show a significantly positive association between PRSUC and all-grade IMC with an ORmeta per SD of 1.35 (95% CI = 1.12–1.64, p = 2 × 10−03). Similarly, a robust association of PRSUC and severe IMC was observed with an ORmeta per SD of 1.49 (95% CI = 1.18–1.88, p = 9 × 10−04).
For patients who received anti-PD1/PD-L1 monotherapy, PRSUC demonstrated a significant association with all-grade IMC, showing an ORmeta per SD of 1.35 (95% CI = 1.07−1.69, p = 0.01). Similarly, a stronger association was observed with severe IMC, with an ORmeta per SD of 1.48 (95% CI = 1.10−1.98, p = 9 × 10-3). Among patients treated with combination or dual therapy, a trend towards association with all-grade IMC was seen (ORmeta per SD = 1.80, 95% CI = 0.95−3.41, p = 0.07); however, a robust and pronounced association was found in relation to severe IMC (ORmeta per SD = 2.20, 95% CI = 1.07−4.53, p = 0.03).
Role of known UC-HLA associations on IMC in GeRI cohort
We assessed the association between all-grade IMC and HLA markers known to be associated with ulcerative colitis26,27 (Supplementary Fig. 4). Out of 12 known UC-associated HLA markers, we observed an OR of 2.63 (95% CI = 1.08–6.40, p = 0.03) for HLA-DRB1*12:01 and all-grade IMC. However, at a false-discovery rate (FDR) < 0.05 none of the known HLA markers were associated with all-grade IMC in the GeRI cohort.
IMC and PRS of autoimmune colitis as a predictor of PFS and OS
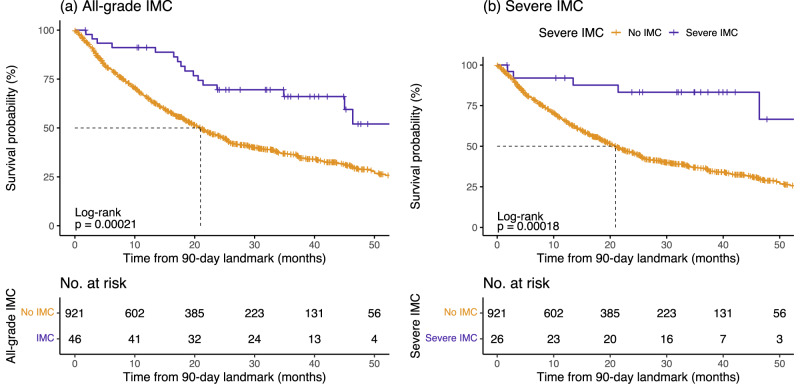
To assess the role of IMC on clinical outcomes, we conducted a Cox proportional hazards model with a 90-day treatment landmark in the GeRI cohort (Table 3 and Fig. 3). We observed the effect of all-grade IMC on OS with an HR of 0.40 (95% CI = 0.24–0.66, p = 3.0 × 10−04) and of severe IMC on OS with an HR of 0.23 (95% CI = 0.09–0.55, p = 9.0 × 10−04). However, we observed no significant association between PFS and IMC (Table 3 and Supplementary Fig. 5).
Table 3.
All-grade and severe immune checkpoint inhibitor-mediated colitis (IMC) as predictors of progression-free survival (PFS) and overall survival (OS) in the entire GeRI cohort, using Cox proportional hazards models with 90-day landmark
| IMC | PFS | OS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| All-grade | 0.80 | 0.55–1.17 | 0.26 | 0.40 | 0.24–0.66 | 3 x 10-04 |
| Severe | 0.61 | 0.34–1.09 | 0.09 | 0.23 | 0.09–0.55 | 9 x 10-04 |
All p-values are two-sided.
Statistically significant results are highlighted in bold.
IMC immune checkpoint inhibitor-mediated colitis, PFS progression-free survival, OS overall survival, HR hazards ratio, CI confidence interval.
All models are adjusted for age at diagnosis, sex, histology, type of therapy, recruiting site, and 5 principal components.
Fig. 3. Immune checkpoint inhibitor-mediated colitis (IMC) as a predictor of overall survival (OS) in the entire GeRI cohort.
a All-grade IMC and b Severe IMC. Kaplan–Meier survival curves are unadjusted with 90-day landmark and compare those who had an IMC (all-grade or severe) with those who did not have an IMC (No IMC). The p-values in the graph represent the log-rank p-values (two-sided), and the dotted line represents the median survival time. Underneath each set of curves is the number of study participants at risk beyond that time point for the IMC and No IMC groups. Source data are provided as a Source Data file.
Despite the association between PRSUC and IMC, PRSUC was not associated with PFS (HR per SD = 1.00, 95% CI = 0.94–1.07, p = 0.99) and OS (HR per SD = 1.01, 95% CI = 0.93–1.09, p = 0.91) in the GeRI cohort (Table 4). Similarly, we observed no association between PRSCD and PFS (HR per SD = 0.98, 95% CI = 0.91–1.05, p = 0.50) and OS (HR per SD = 1.02, 95% CI = 0.93–1.11, p = 0.68), respectively (Table 4).
Table 4.
Polygenic risk scores of ulcerative colitis (PRSUC) and Crohn’s disease (PRSCD) as predictors of progression-free survival (PFS) and overall survival (OS) in the GeRI cohort, using Cox proportional hazards models
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| PRS | HR per SD | 95% CI | p-value | HR per SD | 95% CI | p-value |
| PRSUC | 1.00 | 0.94–1.07 | 0.99 | 1.01 | 0.93–1.09 | 0.91 |
| PRSCD | 0.98 | 0.91–1.05 | 0.50 | 1.02 | 0.93–1.11 | 0.68 |
All p-values are two-sided.
All models are adjusted for age at diagnosis, sex, histology, type of therapy, recruiting site, and 5 principal components.
PRS polygenic risk score, PFS progression-free survival, OS overall survival, HR hazards ratio, SD standard deviation, CI confidence interval, UC ulcerative colitis, CD Crohn’s disease.
Discussion
Immune checkpoint inhibitors are part of standard regimens to treat many advanced cancers and are used in adjuvant and neoadjuvant settings for early-stage diseases in multiple cancers3,4,10,28–32. Immune-related adverse events are common complications from ICI, and there are few predictors of irAEs33,34. We sought to identify genetic predictors of immune checkpoint inhibitor-mediated colitis which frequently results in hospitalization and ICI discontinuation and can occasionally lead to death18,19,35. Specifically, we evaluated the relationship between genetic predisposition for autoimmune colitis (UC, CD) and IMC, and found that the PRSUC can predict IMC. The association was stronger when analyses were restricted to individuals with severe IMC—an important finding as the most important clinical cases to identify were best predicted by PRSUC. Furthermore, we investigated the role of HLA markers associated with UC on the development of IMC. However, we did not have HLA typing for these individuals, and therefore, the imputation of HLA was not validated. Furthermore, our study was not well-powered to detect the effect of many different HLA alleles after multiple hypothesis testing. Future studies will need to analyze HLA effects on IMC.
Our findings significantly contribute to our understanding of the biological underpinnings of IMC and may also impact the management of patients treated with ICIs. First, we demonstrate that IMC has some genetic overlap with UC, but we found no evidence for overlap with CD. This is notable despite the correlation observed in our PRS for UC and CD, signifying that the genetic factors associated with IMC align more closely with the distinct genetic markers associated with UC. Our finding is also consistent with clinical reports in which the most frequent phenotype of IMC resembles UC most closely22,23,36. Our results also suggest that as the genetic risk prediction of UC improves, the genetic risk of IMC may also be improved. In particular, rare variants in certain genes substantially increase the risk of UC and we hypothesize may also affect IMC risk37–39. Prior reports on ICI-induced hypothyroidism40,41 and rash42 demonstrated that PRS for autoimmune disorders predict irAEs, suggesting that ICI may unmask autoimmune syndromes in some genetically predisposed individuals.
We also found that individuals who developed IMC had improved survival outcomes when compared to those who did not develop IMC, including in a landmark sensitivity analysis, which is concordant with previously published literature43–48. However, PRSUC and PRSCD were not associated with PFS or OS, suggesting that the genetic basis of autoimmune disease susceptibility is distinct from genetic factors influencing survival outcomes. It has been postulated that both anti-tumor responses to ICIs, and the development of irAEs are representative of a robust immune response; however, one possible explanation for our finding is that the genetic contributions captured in the autoimmune PRSs are probably capturing the cross-presentation of shared antigens which may not be associated with clinical outcomes. This suggests there could be other genetic and environmental factors driving the association between IMC and overall survival.
Our study has several implications that may impact the care of cancer patients treated with ICIs. For example, our results suggest that germline genotyping could help assist selection of patients at high risk of IMC in a clinical trial setting to assess the role of preventative measures such as the commencement of concurrent anti-TNFα therapies or anti-integrin α4β7 antibodies49,50 along with ICI treatment in patients at high risk for IMC and toxicity-related early treatment cessation. Additionally, these findings may also help facilitate clinical decision-making. Combination immunotherapies are more effective but are also associated with a substantially increased risk of irAEs45,51–55. Our stratified analysis by type of therapy demonstrated the association between PRSUC and severe IMC in individuals receiving anti-PD-1/PD-L1 and anti-CTLA4 combination therapy. Among patients who may be candidates for combination immunotherapies but have a high genetic risk based on PRSUC, oncologists may consider monotherapy, particularly in clinical situations in which the benefits of dual therapy on disease control may be modest. Conversely, patients who are at relatively low risk based on PRSUC may be better candidates for combination therapy. In addition, the use of PRSUC might also be considered to assist with treatment decisions in clinical settings where ICI therapy is approved but there is substantial clinical equipoise; for example, in the adjuvant setting for patients with resectable NSCLC56,57 and low PD-L1 expression or adjuvant setting for resected stage II melanoma58. Our analysis within the anti-CTLA4 monotherapy subgroup did not reveal any significant association between PRSUC and IMC. These results should be interpreted cautiously since the sample size was limited in this subgroup. However, anti-CTLA4 as monotherapy has become less common in contemporary clinical practice, with its predominant use being in combination with anti-PD1/PD-L1 therapy, and our PRSUC did predict IMC in these patients. Our initial findings were observed in a cohort of NSCLC patients. However, our replication study included a broader array of pan-cancer studies and demonstrated the generalizability of PRSUC to predict IMC.
Although our study has important clinical implications and strengths, it also has some limitations. While PRS effectively captures established variants associated with UC, it may not account for unidentified genetic contributors (missing heritability). Nevertheless, as we unveil the missing heritability of UC, we expect to further improve the polygenic prediction of IMC. Furthermore, we developed these PRSs in a predominantly European ancestry cohort (UK Biobank) and the GeRI cohort and BioVU replication study were also predominantly of European ancestry; more work is needed to generalize these results to other ancestries. In addition, there may be other limitations to implementing PRS in the clinic including cost, rapidity of return of results, and reliability and consistency across different algorithms59–62. Although we included one replication cohort, additional studies of more ICI-treated patients will help strengthen our findings and, in particular, may give better power to evaluate HLA associations and other individual loci that may improve our understanding of the genetic similarities and potential differences between IMC and UC. For most complex traits, including autoimmune disorders and, likely for irAEs, environmental factors also play an important role. Our study does not address how environmental factors affect the risk of IMC. For example, the gut microbiome may modify susceptibility to and severity of IBD63 and, therefore, may also contribute to the susceptibility of developing IMC in cancer patients who have undergone ICI treatment. To determine the joint associations between PRSUC and environmental risk factors, further studies are necessary.
We also found an association between IMC and OS. This result could be due to survivor bias64,65, where patients who respond to therapy and are on therapy longer are at an increased risk of developing irAEs. We used a 90-day landmark analysis66 to account for this bias for both PFS and OS, although this may not completely eliminate the survivor bias.
Overall, our findings suggest a shared genetic basis between ulcerative colitis and immune checkpoint inhibitor-mediated colitis among patients undergoing ICI treatment. Prediction of IMC using genetic information should create new opportunities for better risk stratification and ultimately for better management and possibly prevention of this common and important side effect from immunotherapy.
Methods
This research complies with all relevant ethical regulations. Institutional Review Board approvals were obtained at each site individually, and written informed consent was acquired from all study participants prior to inclusion in the study.
Study population
Genetics of immune-related adverse events and Response to Immunotherapy (GeRI) cohort is comprised of 1316 advanced Stage IIIB/IV NSCLC patients who received ICI therapy (PD-1 or PD-L1 inhibitors as monotherapy or in combination with either CTLA-4 inhibitors and/or chemotherapy) and were recruited from four different institutions: Memorial Sloan Kettering Cancer Center (MSKCC), Vanderbilt University Medical Center (VUMC), Princess Margaret Cancer Center (PM), and University of California, San Francisco (UCSF).
A total of 752 individuals were treated with ICIs at MSKCC between 2011 and 2018 and had an available blood sample. Clinical data were extracted from a manual review of medical and pharmacy records for demographics, lung cancer histology, and ICI treatment history, including detailed information on immune-related adverse events (irAEs). The VUMC cohort is composed of 267 patients who received ICI therapy at the medical center between 2009 and 2019. Patients participated in BioVU21, Vanderbilt’s biomedical repository of DNA that is linked to de-identified health records. Treatment dates and irAEs were extracted using manual chart review by a trained thoracic oncology nurse. The PM cohort included 266 advanced NSCLC patients who received ICI therapy between 2011 and 2022; all provided a blood sample and completed a questionnaire. Clinical data were manually extracted by trained abstractors and supplemented by the PM Cancer Registry. From UCSF, 31 patients who had received ICIs were identified by thoracic oncologists between 2019 and 2021 and provided either a blood or saliva sample after informed consent. Clinical data including, demographics, history of lung cancer and ICI treatment, and irAEs were extracted after a manual review of electronic health records.
Immune checkpoint inhibitor-mediated colitis (IMC)
After the initiation of ICI therapy, immune checkpoint inhibitor-mediated colitis (IMC) was defined based on clinical chart review and documentation of IMC by the primary oncologist, gastroenterologist, and/or other clinicians treating the patient based on clinical features and/or radiologic/histologic evidence suggesting colitis due to ICI. Participants who were diagnosed with infectious causes of colitis including Clostridium difficile, or a pathogen on a gastrointestinal pathogen panel or ova and parasite test were excluded. To assess the severity of IMC, we used 2 metrics based on NCI Common Terminology Criteria for Adverse Events Version 5 (NCI-CTCAE) that capture grade 3 IMC or above (i) hospitalization for management of IMC and/or (ii) permanent cessation of ICI therapy due to the adverse event.
IMC was coded as a dichotomous variable (1: all IMC, 0: no IMC) and time-to-IMC was assessed from the start of the ICI therapy to the date of onset of IMC or the date of ICI discontinuation due to IMC. Patients who did not experience IMC were censored either at the end of treatment due to any reason or last follow-up date if the treatment was ongoing. Based on the severity criteria, severe IMCs were also coded as binary variables (1: severe IMC, 0: no IMC).
Ascertainment of clinical outcomes
Progression-free survival (PFS) and overall survival (OS) were evaluated from the date of initiation of ICI therapy to the date of progression and death, respectively, at MSK, PM, and UCSF sites. At VUMC, time-to-discontinuation of therapy due to progression from therapy initiation was used as a surrogate. If the treatment was ongoing, patients were censored at the date of the last follow-up. The VUMC cohort is de-identified and not linked to the National Death Index; therefore, all-cause mortality (overall survival) information is unavailable for VUMC participants (n = 267).
Quality control, genotyping, and imputation of the GeRI cohort
DNA from blood or saliva was extracted and genotyped using Affymetrix Axiom Precision Medicine Diversity Array. Samples with a call rate <95% were excluded from the analysis and SNPs with missing rates >5% were also excluded from the analysis. Genetic ancestry was calculated using principal component analysis in PLINK after linkage disequilibrium pruning (R2 < 0.1). Imputation was performed using the Michigan Imputation Server with the 1000 Genomes phase3 v5 reference panel. Standard genotyping and quality control procedures were implemented. Variants with minor allele frequency <0.01 were excluded from the analysis.
Development and validation of polygenic risk score (PRS) for autoimmune colitis
We developed PRS for CD (1312 CD cases and 16,303 controls) and UC (2814 UC cases and 16,303 controls), separately using UK Biobank (UKB) data, where we divided the data into two parts: 70% for hyperparameter tuning and 30% of the remaining data for testing the PRS. Genetic data from both the UKB Affymetrix Axiom array (89%) or the UK BiLEVE array (11%)67 which have been imputed using the Haplotype Reference Consortium and the UK10K and 1000 Genomes phase3 reference panels67 were utilized in the analysis. Analyses were restricted to European ancestry individuals based on self-reported White ethnicity and genetic ancestry PCs within five standard deviations of the population mean. Samples with discordant self-reported and genetic sex were excluded. Additionally, we also excluded one sample from each pair of first-degree relatives. Samples with greater than five standard deviations from the mean heterozygosity were further excluded from the analysis. Information from both self-report and ICD9/10 codes were used to capture CD (1312 cases) and UC (2814 cases) phenotypes in UKB.
We used the LDPred225 method to develop PRS of CD and UC. LDpred2 estimates the posterior effect sizes based on summary statistics from genome-wide association studies while taking into account the linkage disequilibrium between variants and assuming a prior on the markers. To derive PRS, summary statistics were obtained from the previously published largest genome-wide association study of CD, and UC68. We restricted the analysis to HapMap3 variants and implemented LDPred2-auto function to evaluate the posterior effect sizes for each variant. LDPred2-auto first estimates the proportion of causal variants and heritability for the trait under evaluation. Next, it determines the posterior effects estimates for the included variants. The final PRS weights are available at PGS catalog (See Data Availability). Briefly, PRSUC included 744,575 variants, whereas PRSCD comprised 744,682 variants.
PRS was constructed using the formula: PRS = β1 x SNP1 + β2 x SNP2 +………+ βn x SNPn, where β was estimated using LDPred2-auto function. Each PRS was standardized to have a mean of zero and a standard deviation of 1. The association of PRSCD and PRSUC with each respective target phenotype was assessed using logistic regression models, adjusted for age at diagnosis for cases and age at enrollment for controls, sex, genotyping array, and the top 10 genetic ancestry principal components (PCs). Area under the receiver operating characteristic (AUROC) curves were calculated in the testing dataset and used to assess the overall prediction accuracy of each PRS in UKB.
We validated the two PRSs in a sample of cancer-free individuals (1420 CD cases, 459 UC cases, and 20,876 controls in the VUMC BioVU24. All analyses were restricted to individuals of European ancestry and adjusted for age, sex, and ten principal components. AUROC curves were used to estimate the prediction of the PRSs.
Assessment of autoimmune colitis PRS to predict IMC in GeRI cohort
Using the weights generated from LDPred2 for CD, and UC, we separately calculated two weighted PRSs (PRSCD, PRSUC) for the GeRI participants. The cumulative incidence of IMC (all-grade and severe) was assessed by categories of PRS percentiles. Individuals in the top 10% of the PRS distribution (PRS > 90th percentile) were classified as having high genetic risk, those in the bottom 10% (PRS ≤ 10th percentile) were classified as low risk, and the middle category (>10th to ≤90th percentile) classified as average genetic risk. Additionally, to evaluate the performance of each potential PRS on either time-to-IMC or time-to-severe IMC, we used Cox proportional hazards models, adjusted for age at diagnosis, sex, lung cancer histology, type of therapy, recruiting site, and the first 5 genetic ancestry PCs. To further understand the differential effects of type of therapy and histology on the association between PRSUC and IMC, we conducted stratified analysis by type of ICI therapy and histology of lung cancer.
Replication of PRSUC and IMC in an independent study
We performed an independent replication to further characterize the association between PRSUC and IMC. Our replication study comprises of 873 patients enrolled in BioVU24, across all cancer types and treated with either anti-PD-1/PD-L1 monotherapy or a combination of anti-PD1/PD-L1 and anti-CTLA4 therapy. There was no overlap of samples between individuals from BioVU included in the GeRI cohort (discovery) and the replication dataset from BioVU. Immune checkpoint inhibitor-mediated colitis was ascertained by a manual review of the electronic health records. An IMC case was defined as either biopsy-confirmed colitis or the occurrence of diarrhea in ICI patients, not attributable to any other cause, that required treatment with steroids and subsequently showed improvement with steroid therapy. All samples were genotyped using Illumina Expanded Multi-Ethnic Genotyping Array (MEGA-EX) and imputed to 1000 Genomes reference panel (version 3)24. Post-imputation standard quality control procedures were employed to exclude low-quality variants and samples. In short, samples with a call rate <95% and SNPs with missing rates >2% were excluded from the analysis. Additionally, all SNPs with minor allele frequency <1% and Hardy–Weinberg p-value < 1e-06, and INFO < 0.95 were excluded.
We performed unconditional logistic regression to assess the association between PRSUC and all-grade IMC and severe IMC, respectively. All models were adjusted for age at diagnosis, sex, type of therapy, and 5 principal components. In addition, we conducted stratified logistic regression by type of therapy, and the models were adjusted for age at diagnosis, sex, and 5 principal components. This study had an additional 274 patients who received anti-CTLA4 monotherapy, and we further evaluated the association between PRSUC and IMC separately in this group.
Meta-analysis of the association between PRSUC and IMC
For meta-analysis, we conducted standard logistic regression adjusted for age at diagnosis, sex, type of therapy, site, and 5 PCs in the GeRI study. Next, we carried out an inverse-variance weighted fixed-effect meta-analysis between our discovery and replication studies. Additionally, we conducted a meta-analysis of the stratified results by type of therapy in the GeRI cohort and the replication study from BioVU.
Role of HLA markers associated with UC and CD on IMC in GeRI cohort
To elucidate the role of known UC-associated HLA markers on IMC, we performed HLA imputation using CookHLA69 and HATK70. HLA alleles were imputed at 2-field resolution against the Type 1 Diabetes Genetics Consortium reference panel71 and using the nomenclature from IPD-IMGT/HLA database v3.51. Association analysis with all-grade IMC was conducted using logistic regression models adjusted for age at diagnosis, sex, lung cancer histology, type of therapy, recruiting site, and 5 PCs. Analyses were restricted to common HLA alleles (frequency ≥0.01) known to be associated with ulcerative colitis26.
Impact of IMC and PRS of autoimmune colitis on PFS and OS in GeRI cohort
The association of IMC (all-grade and severe) on PFS and OS was examined using the Cox proportional hazards model by examining only the patients who had PFS and OS longer than 90 days (90-day landmark)66. All models were adjusted for age at diagnosis, sex, lung cancer histology, type of therapy, and 5 PCs. Survival curves and rates were estimated using the Kaplan–Meier method. To investigate the association between PRSCD, PRSUC on PFS, and OS, we conducted Cox proportional hazards models, adjusted for age at diagnosis, sex, histology, type of therapy, and 5 PCs. All p-values are two-sided, and analyses were conducted using Plink2, R v4.2.2 (R Foundation for Statistical Computing) with RStudio v2022.12.0.353.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by the National Institutes of Health R01-CA227466 and K24-CA169004 to E.Ziv; C.M.Lovly was supported in part by NIH NCI UG1CA233259, P01CA129243, and P30CA068485; R.Thummalapalli was supported by T32-CA009207; The Lusi Wong Fund, Posluns Fund, Alan Brown Chair in Molecular Genomics, Princess Margaret Cancer Foundation were awarded to G. Liu for this work; M.C. Aldrich was supported in part by R01-CA227466, U01CA253560, R01CA251758 and the Vanderbilt Institute for Clinical and Translational Research (UL1TR002243); Z. Quandt was supported NIDDK DiabDocs K12DK133995 and a Larry L Hillblom Foundation Start-Up Grant; A.J.Schoenfeld, D.Faleck were supported by the Memorial Sloan Kettering Cancer Center Support Grant/Core (P30-CA008748), the Druckenmiller Center for Lung Cancer Research at Memorial Sloan Kettering Cancer Center. Justin M. Balko was supported by NIH/NCI R01CA227481; Douglas B Johnson was supported by NIH/NCI R01CA227481 and NIH/NHLBI R01HL156021. The samples and/or dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH-funded Shared Instrumentation Grant S10OD017985 and S10RR025141; and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vumc.org/biovu-funding/. Megan H. Murray’s work on this project was completed in August 2022 while she was working at Vanderbilt University Medical Center.
Author contributions
Concept and design: P.M., M.C.A., A.J.S., E.Z.; Data acquisition: P.M., R.T., Z.Q., K.B., C.A.B., E.C., C.J.F., P.M.L.G., M.A.G., S.H., D.J.B., L.K., K.K., M.L., C.M.L., M.H.M., D.P., K.W., Y.X., L.J.Z., J.M.B., G.L., M.C.A., A.J.S., E.Z.; Analysis or interpretation of data: P.M., R.T., M.J.B., L.Y., Z.Q., D.M.F., L.K., C.M.L., G.L., M.C.A, A.J.S., E.Z.; Drafting of the manuscript: P.M., R.T., M.J.B., L.Y., Z.Q., K.B., C.A.B., E.C., C.J.F., D.M.F., P.M.L.G, M.A.G., S.H., D.B.J., L.K., K.K., M.L., C.M.L., M.H.M., D.P., K.W., Y.X., L.J.Z., J.M.B., G.L., M.C.A., A.J.S., E.Z. Critical revision of the manuscript for important intellectual content: P.M., R.T., G.L., M.C.A., A.J.S., E.Z.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
UK Biobank data are publicly available by request from https://www.ukbiobank.ac.uk. Scoring files for Crohn’s disease and ulcerative colitis are available from the PGS catalog http://www.pgscatalog.org/score/PGS004253/ and http://www.pgscatalog.org/score/PGS004254/. De-identified data along with outcomes used for this work are available at https://github.com/PoojaMiddha/GeRI_colitis/tree/GeRI_colitis_manuscript72. BioVU data is available to Vanderbilt-affiliated members subject to approval by the BioVU Review Committee from https://victr.vumc.org/how-to-use-biovu/24. External Users will need a Vanderbilt PI for all BioVU projects, and contracts will need to be in place before data can be shared. Investigators can reach out to BioVU (biovu@vumc.org) if they would like more information or help establishing a collaboration. The remaining data are available within the Article, Supplementary Information, or Source Data file. Source data are provided in this paper.
Code availability
We used R programming language (v4.2.2), survival R package (v3.4.0), ggplot2 package (v3.4.2), pROC package (v1.18.0), bigsnpr package (v1.11.6), and PLINK2 for PRS calculations. Analysis scripts can be found at https://github.com/PoojaMiddha/GeRI_colitis/tree/GeRI_colitis_manuscript72.
Competing interests
J.M.B. receives research support from Genentech/Roche and Incyte Corporation, has received advisory board payments from AstraZeneca and Mallinckrodt, and is an inventor on patents regarding immunotherapy targets and biomarkers in cancer. The remaining authors declare no other competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Elad Ziv, Email: Elad.Ziv@ucsf.edu.
Princess Margaret Lung Group:
Natasha B. Leighl, Penelope A. Bradbury, Frances A. Shepherd, Adrian G. Sacher, and Lawson Eng
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-44512-4.
References
- 1.Bai RL, Chen NF, Li LY, Cui JW. A brand new era of cancer immunotherapy: breakthroughs and challenges. Chin. Med. J. (Engl.). 2021;134:1267–1275. doi: 10.1097/CM9.0000000000001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 3.Twomey JD, Zhang B. Cancer immunotherapy update: FDA-approved checkpoint inhibitors and companion diagnostics. AAPS J. 2021;23:39. doi: 10.1208/s12248-021-00574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpe AH. Introduction to checkpoint inhibitors and cancer immunotherapy. Immunol. Rev. 2017;276:5–8. doi: 10.1111/imr.12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020;11:3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 2018;50:1–11. doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchbinder EI, Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am. J. Clin. Oncol. 2016;39:98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Hogg GD, DeNardo DG. Rethinking immune checkpoint blockade: ‘Beyond the T cell. J. Immunother. Cancer. 2021;9:e001460. doi: 10.1136/jitc-2020-001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribas A. Tumor immunotherapy directed at PD-1. N. Engl. J. Med. 2012;366:2517–2519. doi: 10.1056/NEJMe1205943. [DOI] [PubMed] [Google Scholar]
- 12.Byun DJ, Wolchok JD, Rosenberg LM, Girotra M. Cancer immunotherapy—immune checkpoint blockade and associated endocrinopathies. Nat. Rev. Endocrinol. 2017;13:195–207. doi: 10.1038/nrendo.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N. Engl. J. Med. 2018;378:158–168. doi: 10.1056/NEJMra1703481. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Treatment-related adverse events of PD-1 and PD-L1 inhibitors in clinical trials. JAMA Oncol. 2019;5:1008–1019. doi: 10.1001/jamaoncol.2019.0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okiyama N, Tanaka R. Immune-related adverse events in various organs caused by immune checkpoint inhibitors. Allergol. Int. 2022;71:169–178. doi: 10.1016/j.alit.2022.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Shankar B, et al. Multisystem immune-related adverse events associated with immune checkpoint inhibitors for treatment of non-small cell lung cancer. JAMA Oncol. 2020;6:1952–1956. doi: 10.1001/jamaoncol.2020.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conroy M, Naidoo J. Immune-related adverse events and the balancing act of immunotherapy. Nat. Commun. 2022;13:392. doi: 10.1038/s41467-022-27960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Som A, et al. Immune checkpoint inhibitor-induced colitis: a comprehensive review. World J. Clin. Cases. 2019;7:405–418. doi: 10.12998/wjcc.v7.i4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pocha C, Roat J, Viskocil K. Immune-mediated colitis: important to recognize and treat. J. Crohn’s Colitis. 2014;8:181–182. doi: 10.1016/j.crohns.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Pauken KE, Dougan M, Rose NR, Lichtman AH, Sharpe AH. Adverse events following cancer immunotherapy: obstacles and opportunities. Trends Immunol. 2019;40:511–523. doi: 10.1016/j.it.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly-Goss MR, Badran YR, Dougan M. Update on immune checkpoint inhibitor Enterocolitis. Curr. Gastroenterol. Rep. 2022;24:171–181. doi: 10.1007/s11894-022-00852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JH, Pezhouh MK, Lauwers GY, Masia R. Histopathologic features of colitis due to immunotherapy with anti-PD-1 antibodies. Am. J. Surg. Pathol. 2017;41:643–654. doi: 10.1097/PAS.0000000000000829. [DOI] [PubMed] [Google Scholar]
- 23.Nahar KJ, et al. Clinicopathological characteristics and management of colitis with anti-PD1 immunotherapy alone or in combination with ipilimumab. J. Immunother. Cancer. 2020;8:e001488. doi: 10.1136/jitc-2020-001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharm. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Privé F, Arbel J, Vilhjálmsson BJ. LDpred2: better, faster, stronger. Bioinformatics. 2020;36:5424–5431. doi: 10.1093/bioinformatics/btaa1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashton JJ, Latham K, Beattie RM, Ennis S. Review article: the genetics of the human leucocyte antigen region in inflammatory bowel disease. Aliment. Pharmacol. Ther. 2019;50:885–900. doi: 10.1111/apt.15485. [DOI] [PubMed] [Google Scholar]
- 27.Goyette P, et al. High density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet. 2015;47:172–179. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marin-Acevedo JA, Kimbrough EO, Lou Y. Next generation of immune checkpoint inhibitors and beyond. J. Hematol. Oncol. 2021;14:45. doi: 10.1186/s13045-021-01056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang WF, et al. Adjuvant immunotherapy in early-stage resectable non–small cell lung cancer: a new milestone. Front. Oncol. 2023;13:1063183. doi: 10.3389/fonc.2023.1063183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittendorf, E. A., Burgers, F., Haanen, J. & Cascone, T. Neoadjuvant immunotherapy: leveraging the immune system to treat early-stage disease. Am. Soc. Clin. Oncol. Educ. Book. 189–203 10.1200/EDBK_349411 (2022) [DOI] [PubMed]
- 31.Krishnamoorthy M, Lenehan JG, Maleki Vareki S. Neoadjuvant immunotherapy for high-risk, resectable malignancies: scientific rationale and clinical challenges. J. Natl Cancer Inst. 2021;113:823–832. doi: 10.1093/jnci/djaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu HG, Li YM. Emerging adjuvants for cancer immunotherapy. Front. Chem. 2020;8:601. doi: 10.3389/fchem.2020.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Biomarkers and risk factors for the early prediction of immune-related adverse events: a review. Hum. Vaccin. Immunother. 2022;18:2018894. doi: 10.1080/21645515.2021.2018894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hommes JW, Verheijden RJ, Suijkerbuijk KPM, Hamann D. Biomarkers of checkpoint inhibitor induced immune-related adverse events—a comprehensive review. Front Oncol. 2021;10:585311. doi: 10.3389/fonc.2020.585311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy LC, Grivas P. Immunotherapy-related colitis: an emerging challenge and a quest for prospective data. JCO Oncol. Pract. 2020;16:464–465. doi: 10.1200/OP.20.00620. [DOI] [PubMed] [Google Scholar]
- 36.Westdorp, H. et al. Mechanisms of immune checkpoint inhibitor-mediated colitis. Front. Immunol. 2021;12. Accessed September 23, 2022. https://www.frontiersin.org/articles/10.3389/fimmu.2021.768957. [DOI] [PMC free article] [PubMed]
- 37.Luo Y, et al. Exploring the genetic architecture of inflammatory bowel disease by whole-genome sequencing identifies association at ADCY7. Nat. Genet. 2017;49:186–192. doi: 10.1038/ng.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onoufriadis A, et al. Exome sequencing and genotyping identify a rare variant in NLRP7 gene associated with ulcerative colitis. J. Crohn’s Colitis. 2018;12:321–326. doi: 10.1093/ecco-jcc/jjx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellinghaus D, Bethune J, Petersen BS, Franke A. The genetics of Crohn’s disease and ulcerative colitis–status quo and beyond. Scand. J. Gastroenterol. 2015;50:13–23. doi: 10.3109/00365521.2014.990507. [DOI] [PubMed] [Google Scholar]
- 40.Luo J, et al. Immunotherapy-mediated thyroid dysfunction: genetic risk and impact on outcomes with PD-1 blockade in non–small cell lung cancer. Clin. Cancer Res. 2021;27:5131–5140. doi: 10.1158/1078-0432.CCR-21-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan Z, et al. Genetic variation associated with thyroid autoimmunity shapes the systemic immune response to PD-1 checkpoint blockade. Nat. Commun. 2021;12:3355. doi: 10.1038/s41467-021-23661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan Z, et al. Polygenic risk for skin autoimmunity impacts immune checkpoint blockade in bladder cancer. Proc. Natl Acad. Sci. USA. 2020;117:12288–12294. doi: 10.1073/pnas.1922867117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, et al. Are immune-related adverse events associated with the efficacy of immune checkpoint inhibitors in patients with cancer? A systematic review and meta-analysis. BMC Med. 2020;18:87. doi: 10.1186/s12916-020-01549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, Z. et al. Immune-related adverse events associated with outcomes in patients with NSCLC treated with anti-PD-1 inhibitors: a systematic review and meta-analysis. Front. Oncol. 2021;11. Accessed June 1, 2022. https://www.frontiersin.org/article/10.3389/fonc.2021.708195. [DOI] [PMC free article] [PubMed]
- 45.Petrelli F, et al. Immune-related adverse events and survival in solid tumors treated with immune checkpoint inhibitors: a systematic review and meta-analysis. J. Immunother. 2020;43:1–7. doi: 10.1097/CJI.0000000000000300. [DOI] [PubMed] [Google Scholar]
- 46.Toi Y, et al. Association of immune‐related adverse events with clinical benefit in patients with advanced non‐small‐cell lung cancer treated with nivolumab. Oncologist. 2018;23:1358–1365. doi: 10.1634/theoncologist.2017-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morimoto K, et al. Immune-related adverse events are associated with clinical benefit in patients with non-small-cell lung cancer treated with immunotherapy plus chemotherapy: a retrospective study. Front. Oncol. 2021;11:630136. doi: 10.3389/fonc.2021.630136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abu-Sbeih H, et al. Immune checkpoint inhibitor-induced colitis as a predictor of survival in metastatic melanoma. Cancer Immunol. Immunother. 2019;68:553–561. doi: 10.1007/s00262-019-02303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abu-Sbeih H, et al. Outcomes of vedolizumab therapy in patients with immune checkpoint inhibitor–induced colitis: a multi-center study. J. Immunother. Cancer. 2018;6:142. doi: 10.1186/s40425-018-0461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergqvist V, et al. Vedolizumab treatment for immune checkpoint inhibitor-induced enterocolitis. Cancer Immunol. Immunother. 2017;66:581–592. doi: 10.1007/s00262-017-1962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park, R., Lopes, L., Cristancho, C. R., Riano, I. M. & Saeed, A. Treatment-related adverse events of combination immune checkpoint inhibitors: systematic review and meta-analysis. Front. Oncol. 2020;10. Accessed March 15, 2023. https://www.frontiersin.org/articles/10.3389/fonc.2020.00258. [DOI] [PMC free article] [PubMed]
- 52.Paz-Ares L, et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): an international, randomised, open-label, phase 3 trial. Lancet Oncol. 2021;22:198–211. doi: 10.1016/S1470-2045(20)30641-0. [DOI] [PubMed] [Google Scholar]
- 53.Da L, et al. Organ-specific immune-related adverse events associated with immune checkpoint inhibitor monotherapy versus combination therapy in cancer: a meta-analysis of randomized controlled trials. Front. Pharmacol. 2020;10:1671. doi: 10.3389/fphar.2019.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carretero-González A, et al. Comparative safety analysis of immunotherapy combined with chemotherapy versus monotherapy in solid tumors: a meta-analysis of randomized clinical trials. Oncotarget. 2019;10:3294–3301. doi: 10.18632/oncotarget.26908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhuo M, Chi Y, Wang Z. The adverse events associated with combination immunotherapy in cancers: Challenges and chances. Asia-Pac. J. Clin. Oncol. 2020;16:e154–e159. doi: 10.1111/ajco.13365. [DOI] [PubMed] [Google Scholar]
- 56.Felip E, et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB–IIIA non-small-cell lung cancer (IMpower010): a randomised, multicentre, open-label, phase 3 trial. Lancet. 2021;398:1344–1357. doi: 10.1016/S0140-6736(21)02098-5. [DOI] [PubMed] [Google Scholar]
- 57.O’Brien M, et al. Pembrolizumab versus placebo as adjuvant therapy for completely resected stage IB–IIIA non-small-cell lung cancer (PEARLS/KEYNOTE-091): an interim analysis of a randomised, triple-blind, phase 3 trial. Lancet Oncol. 2022;23:1274–1286. doi: 10.1016/S1470-2045(22)00518-6. [DOI] [PubMed] [Google Scholar]
- 58.Luke JJ, et al. Pembrolizumab versus placebo as adjuvant therapy in completely resected stage IIB or IIC melanoma (KEYNOTE-716): a randomised, double-blind, phase 3 trial. Lancet. 2022;399:1718–1729. doi: 10.1016/S0140-6736(22)00562-1. [DOI] [PubMed] [Google Scholar]
- 59.Klein RJ, Gümüş ZH. Are polygenic risk scores ready for the cancer clinic?-a perspective. Transl. Lung Cancer Res. 2022;11:910–919. doi: 10.21037/tlcr-21-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis ACF, Green RC. Polygenic risk scores in the clinic: new perspectives needed on familiar ethical issues. Genome Med. 2021;13:14. doi: 10.1186/s13073-021-00829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis ACF, Green RC, Vassy JL. Polygenic risk scores in the clinic: translating risk into action. HGG Adv. 2021;2:100047. doi: 10.1016/j.xhgg.2021.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin AR, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome—searching the crime scene for clues. Gastroenterology. 2021;160:524–537. doi: 10.1053/j.gastro.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson JR, Cain KC, Gelber RD. Analysis of survival by tumor response. J. Clin. Oncol. 1983;1:710–719. doi: 10.1200/JCO.1983.1.11.710. [DOI] [PubMed] [Google Scholar]
- 65.Anderson JR, Cain KC, Gelber RD. Analysis of survival by tumor response and other comparisons of time-to-event by outcome variables. J. Clin. Oncol. 2008;26:3913–3915. doi: 10.1200/JCO.2008.16.1000. [DOI] [PubMed] [Google Scholar]
- 66.Morgan CJ. Landmark analysis: a primer. J. Nucl. Cardiol. 2019;26:391–393. doi: 10.1007/s12350-019-01624-z. [DOI] [PubMed] [Google Scholar]
- 67.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cook S, et al. Accurate imputation of human leukocyte antigens with CookHLA. Nat. Commun. 2021;12:1264. doi: 10.1038/s41467-021-21541-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi W, Luo Y, Raychaudhuri S, Han B. HATK: HLA analysis toolkit. Bioinformatics. 2021;37:416–418. doi: 10.1093/bioinformatics/btaa684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown WM, et al. Overview of the MHC fine mapping data. Diabetes Obes. Metab. 2009;11:2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Middha, P. PoojaMiddha/GeRI_colitis: Polygenic risk score for ulcerative colitis predicts immune checkpoint inhibitor-mediated colitis. Published online November 20, 2023. 10.5281/ZENODO.10162343. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are publicly available by request from https://www.ukbiobank.ac.uk. Scoring files for Crohn’s disease and ulcerative colitis are available from the PGS catalog http://www.pgscatalog.org/score/PGS004253/ and http://www.pgscatalog.org/score/PGS004254/. De-identified data along with outcomes used for this work are available at https://github.com/PoojaMiddha/GeRI_colitis/tree/GeRI_colitis_manuscript72. BioVU data is available to Vanderbilt-affiliated members subject to approval by the BioVU Review Committee from https://victr.vumc.org/how-to-use-biovu/24. External Users will need a Vanderbilt PI for all BioVU projects, and contracts will need to be in place before data can be shared. Investigators can reach out to BioVU (biovu@vumc.org) if they would like more information or help establishing a collaboration. The remaining data are available within the Article, Supplementary Information, or Source Data file. Source data are provided in this paper.
We used R programming language (v4.2.2), survival R package (v3.4.0), ggplot2 package (v3.4.2), pROC package (v1.18.0), bigsnpr package (v1.11.6), and PLINK2 for PRS calculations. Analysis scripts can be found at https://github.com/PoojaMiddha/GeRI_colitis/tree/GeRI_colitis_manuscript72.