Abstract
Three new genera, six new species, three combinations, six epitypes, and 25 interesting new host and / or geographical records are introduced in this study. New genera: Neoleptodontidium (based on Neoleptodontidium aquaticum), and Nothoramularia (based on Nothoramularia ragnhildianicola). New species: Acremonium aquaticum (from cooling pad water, USA, Cladophialophora laricicola (on dead wood of Larix sp., Netherlands), Cyphellophora neerlandica (on lichen on brick wall, Netherlands), Geonectria muralis (on moss growing on a wall, Netherlands), Harposporium illinoisense (from rockwool, USA), and Neoleptodontidium aquaticum (from hydroponic water, USA). New combinations: Cyphellophora deltoidea (based on Anthopsis deltoidea), Neoleptodontidium aciculare (based on Leptodontidium aciculare), and Nothoramularia ragnhildianicola (based on Ramularia ragnhildianicola). Epitypes: Cephaliophora tropica (from water, USA), Miricatena prunicola (on leaves of Prunus serotina, Netherlands), Nothoramularia ragnhildianicola (on Ragnhildiana ferruginea, parasitic on Artemisia vulgaris, Germany), Phyllosticta multicorniculata (on needles of Abietis balsamea, Canada), Thyronectria caraganae (on twigs of Caragana arborescens, Ukraine), and Trichosphaeria pilosa (on decayed Salix branch, Netherlands). Furthermore, the higher order phylogeny of three genera regarded as incertae sedis is resolved, namely Cephaliophora (Ascodesmidaceae, Pezizales), Miricatena (Helotiales, Leotiomycetes), and Trichosphaeria (Trichosphaeriaceae, Trichosphaeriales), with Trichosphaeriaceae being an older name for Plectosphaerellaceae.
Citation: Crous PW, Akulov A, Balashov S, Boers J, Braun U, Castillo J, Delgado MA, Denman S, Erhard A, Gusella G, Jurjević Ž, Kruse J, Malloch DW, Osieck ER, Polizzi G, Schumacher RK, Slootweg E, Starink-Willemse M, van Iperen AL, Verkley GJM, Groenewald JZ (2023). New and Interesting Fungi. 6. Fungal Systematics and Evolution 11: 109–156. doi: 10.3114/fuse.2023.11.09
Keywords: biodiversity, ITS barcodes, multi-gene phylogeny, new taxa, systematics, typification
INTRODUCTION
Although the Fungi are highly diverse and estimated to represent between 2.2 and 3.8 million species, only around 150 000 species have been described to date. Fungi are essential for ecosystem processes, and are of economic importance as plant, human or animal pathogens, or as agents for industrial or pharmaceutical industries (Lücking et al. 2021). In spite of their importance, only 2 000–2 500 species of fungi are described annually, illustrating a great challenge to understand and preserve this important biodiversity resource. Resolving this problem relies on increased efforts to collect, preserve and describe novel species (Cheek et al. 2020). In order to facilitate the description of novel species, the New and Interesting Fungi (NIF) series is published annually in the journal Fungal Systematics and Evolution. Papers include the description of new species, report new host or geographical records, and new sexual-asexual connections. These and the present study also include validations (typifications) of fungal taxa and list interesting observations relating to fungi and their biology.
MATERIALS AND METHODS
Isolates
Samples (see Table 1) were treated as previously detailed (Crous et al. 2019c). Single conidial colonies were established on Petri dishes containing 2 % malt extract agar (MEA) as described by Crous et al. (1991), and single ascospore cultures were established following the method described by Crous (1998). Colonies were sub-cultured on 2 % potato dextrose agar (PDA), oatmeal agar (OA), MEA (Crous et al. 2019c), or autoclaved pine needles on 2 % tap water agar (PNA) (Smith et al. 1996), and incubated at 25 °C under continuous near-ultraviolet light to promote sporulation. Reference strains and specimens of the studied fungi are maintained in the culture collection and fungarium (CBS) of the Westerdijk Fungal Biodiversity Institute (WI), Utrecht, the Netherlands.
Table 1.
Collection details and GenBank accession numbers of isolates treated in this study, and associated ex-type strains where available. Species for which additional sequences were generated during the course of this study are also listed here. Novel GenBank accession numbers are indicated in bold font.
| Species | Culture or voucher accession number(s)1 | Locality and Substrate | Collector(s) and collection date |
GenBank accession number2
|
||||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | rpb2 | tub2 | Other loci | ||||
| Acremonium aquaticum, sp. nov. | CBS 149454 = CPC 42867, ex-type | USA: Cooling pad water from greenhouse | Z. Jurjević, 4 Oct. 2021 | OQ990087 | OQ990041 | OQ989208 | OQ989251 | actA: OQ989189, tef1 (second part): OQ989237 |
| Acrostalagmus luteoalbus | CBS 149685 = CPC 43187 | South Africa: Portulacaria afra, leaf | P.W. Crous, 27 Feb. 2022 | OQ990088 | OQ990042 | OQ989209 | – | tef1 (second part): OQ989238 |
| Appendopyricularia juncicola | CBS 149232 = CPC 41278, ex-type | Netherlands: Juncus effusus, dead culms | E.R. Osieck, 25 Feb. 2021 | NR_182605 | NG_149075 | – | ON605635 | actA: ON605619, tef1 (first part): ON605627 |
| CBS 149686 = CPC 44053 | Netherlands: Carex elongata, culms | E.R. Osieck, 20 Mar. 2022 | OQ990091 | OQ990045 | – | – | tef1 (first part): OQ989225 | |
| CPC 44055 | Netherlands: Carex elongata, culms | E.R. Osieck, 20 Mar. 2022 | OQ990092 | OQ990046 | – | OQ989254 | tef1 (first part): OQ989226 | |
| CPC 44106 | Netherlands: Juncus effusus, culms | E.R. Osieck, 28 Apr. 2022 | OQ990093 | OQ990047 | – | OQ989255 | tef1 (first part): OQ989227 | |
| CPC 44107 | Netherlands: Juncus effusus, culms | E.R. Osieck, 28 Apr. 2022 | OQ990094 | OQ990048 | – | OQ989256 | – | |
| CPC 42686 | Netherlands: Juncus effusus, culms | E.R. Osieck, 9 Dec. 2021 | OQ990095 | OQ990049 | – | OQ989257 | tef1 (first part): OQ989228 | |
| Biscogniauxia anceps | CBS 149687 = CPC 43197 | Spain: Eucalyptus sp., bark | M.A. Delgado, 25 Mar. 2022 | OQ990096 | – | – | – | – |
| Cephaliophora tropica | CBS 149457 = CPC 42877, ex-epitype | USA: Pan water of crocodile farm | Z. Jurjević, 24 Nov. 2021 | OQ990097 | OQ990050 | OQ989210 | – | rpb1: OQ989203, SSU: OQ990136, tef1 (second part): OQ989239 |
| Ceratocystis ficicola | CBS 149669 = CPC 44213 | Italy (Sicily): Trunk necrosis in Ficus carica | G. Polizzi, 2022 | OQ990098 | OQ990051 | OQ989211 | OQ989258 | tef1 (second part): OQ989240 |
| CBS 149670 = CPC 44214 | Italy (Sicily): Trunk necrosis in Ficus carica | G. Polizzi, 2022 | OQ990099 | – | – | – | – | |
| MAFF 625119, ex-type | Japan: Ficus carica, twigs | Y. Kajitani, Nov. 1990 | NR_119410 | – | – | – | – | |
| Chloridium caudigerum | CBS 149688 = CPC 42899 | Netherlands: Ulmus sp., branch | E.R. Osieck, 19 Feb. 2022 | OQ990100 | OQ990052 | – | OQ989259 | tef1 (second part): OQ989241 |
| Chloridium gamsii | CBS 667.75, ex-type | Belgium: Decaying wood | W. Gams, Sep. 1975 | OP455415 | OP455522 | – | OP465095 | tef1 (second part): OP464990 |
| CBS 149043 = CPC 41933 | Netherlands: Cladonia portentosa | J. Boers, 7 Jun. 2021 | OQ990101 | OQ990053 | – | – | tef1 (second part): OQ989242 | |
| Cladophialophora laricicola, sp. nov. | CBS 148944 = CPC 41384, ex-type | Netherlands: Larix sp., dead wood | J. Boers, 16 Mar. 2021 | OQ990102 | OQ990054 | – | OQ989260 | tef1 (first part): OQ989229 |
| Cylindromonium eugeniicola | CBS 146075 = CPC 37170, ex-type | South Africa: Eugenia capensis, leaf litter | M.J. Wingfield, 2010 | NR_166338 | NG_068337 | – | – | – |
| CBS 149689 = CPC 43326 | Spain (Gran Canaria): Eucalyptus sp., dead twig | A.L. van Iperen, 1 Apr. 2022 | OQ990103 | OQ990055 | – | – | – | |
| Cyphellophora deltoidea, comb. nov. | CBS 263.77 = CMT 1111.74, ex-type | Italy: Soil | Unknown | NR_153555 | NG_057113 | – | – | – |
| Cyphellophora neerlandica, sp. nov. | CBS 149512 = CPC 42634, ex-type | Netherlands: Lichen on brick wall | J. Boers, 12 Nov. 2021 | OQ990089 | OQ990043 | – | OQ989252 | – |
| CPC 42641 | Netherlands: Lichen on brick wall | J. Boers, 12 Nov. 2021 | OQ990090 | OQ990044 | – | OQ989253 | – | |
| Didymella brevipilosa | CBS 148654 = FMR 17415, ex-type | Spain: Plant debris submerged in freshwater | V. Magaña-Dueñas, May. 2018 | OU612373 | OU612372 | OU612359 | OU612358 | – |
| CBS 149049 = CPC 41600 | Canada: Abies balsamea, buds | D. Malloch, 4 May 2021 | OQ990104 | OQ990056 | OQ989212 | OQ989261 | actA: OQ989190 | |
| Drepanopeziza populi-albae | CBS 149510 = CPC 42336 | Russia: Populus alba | T.S. Bulgakov, 26 Jun. 2021 | OQ990105 | OQ990057 | – | – | – |
| Endoconidioma populi | CBS 149070 = CPC 41602 | Canada: Abies balsamea, buds | D. Malloch, 4 May 2021 | OQ990106 | OQ990058 | – | – | – |
| UAMH 10297, ex-type | USA: Populus tremuloides, twig | A. Tsuneda, 7 Aug. 2001 | NR_121303 | NG_059198 | – | – | – | |
| Fusariella atrovirens | CBS 149690 = CPC 43304 | Namibia: Lichenicolous on unknown lichen growing on rock | P.W. Crous, 4 Apr. 2022 | OQ990107 | OQ990059 | OQ989213 | – | tef1 (second part): OQ989243 |
| Fusariella hughesii | CBS 149074 = CPC 41594 | Ukraine: Adonis vernalis, overwintered stems | A. Akulov, 11 Apr. 2021 | OQ990108 | OQ990060 | – | – | – |
| Geonectria muralis, sp. nov. | CBS 149515 = CPC 42404, ex-type | Netherlands: Moss growing on the bottom part of wall | J. Boers, 7 Sep. 2021 | OQ990109 | OQ990061 | – | – | – |
| CPC 42405 | Netherlands: Moss growing on the bottom part of wall | J. Boers, 7 Sep. 2021 | OQ990110 | OQ990062 | – | – | – | |
| CPC 42406 | Netherlands: Moss growing on the bottom part of wall | J. Boers, 7 Sep. 2021 | OQ990111 | – | – | – | – | |
| Harposporium illinoisensis, sp. nov. | CBS 149456 = CPC 42872, ex-type | USA: Rockwool | Z. Jurjević, Oct. 2021 | OQ990112 | OQ990063 | OQ989214 | OQ989262 | actA: OQ989191, tef1 (second part): OQ989244 |
| Hysterobrevium rosae | CBS 149699 = CPC 42948 | Netherlands: Bamboo stick | E.R. Osieck, 11 Feb. 2022 | OQ990113 | OQ990064 | OQ989215 | – | tef1 (second part): OQ989245 |
| MFLUCC 14-0551, ex-type | Italy: Rosa canina, dead aerial branch | E. Camporesi, 15 Jun. 2014 | – | MH535897 | – | – | tef1 (second part): MH535879 | |
| Microcera physciae | CBS 148283 = CPC 41284, ex-type | Netherlands: Physcia tenella | J. Boers, 10 Mar. 2021 | NR_175225 | NG_081335 | OK651168 | OK651208 | rpb1: OK651153, tef1 (first part): OK651190 |
| CBS 149570 = CPC 42638 | Spain: Phragmites australis | M. Delgado, 8 Oct. 2021 | OQ990114 | OQ990065 | OQ989216 | OQ989263 | rpb1: OQ989204, tef1 (first part): OQ989230 | |
| Miricatena prunicola | CBS 149448 = CPC 42627, ex-epitype | Netherlands: Prunus serotina, leaves | E. Slootweg, 7 Nov. 2021 | OQ990115 | OQ990066 | – | – | tef1 (first part): OQ989231 |
| Neoleptodontidium aquaticum, gen. et sp. nov. | CBS 149455 = CPC 42868, ex-type | USA: Hydroponic water in greenhouse | Z. Jurjević, 4 Oct. 2021 | OQ990116 | OQ990067 | – | – | – |
| CPC 42875 | USA: Greenhouse peet | Z. Jurjević, 4 Oct. 2021 | OQ990117 | OQ990068 | – | – | – | |
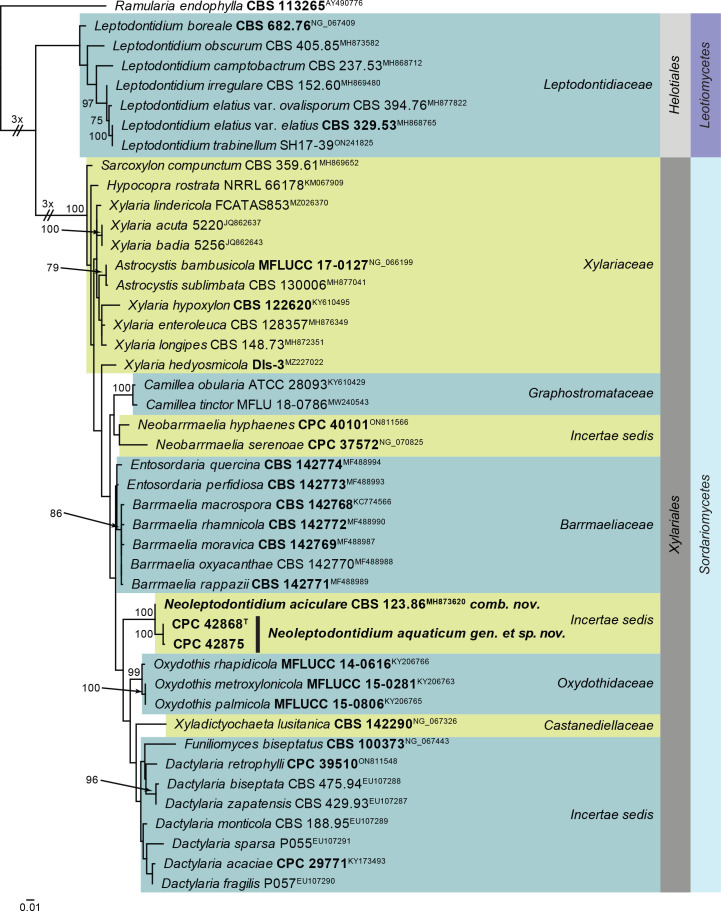
| Neoleptodontidium aciculare, comb. nov. | CBS 123.86, ex-type | India: Rotten wood | V. Rao, Jan. 1984 | MH861931 | MH873620 | – | – | – |
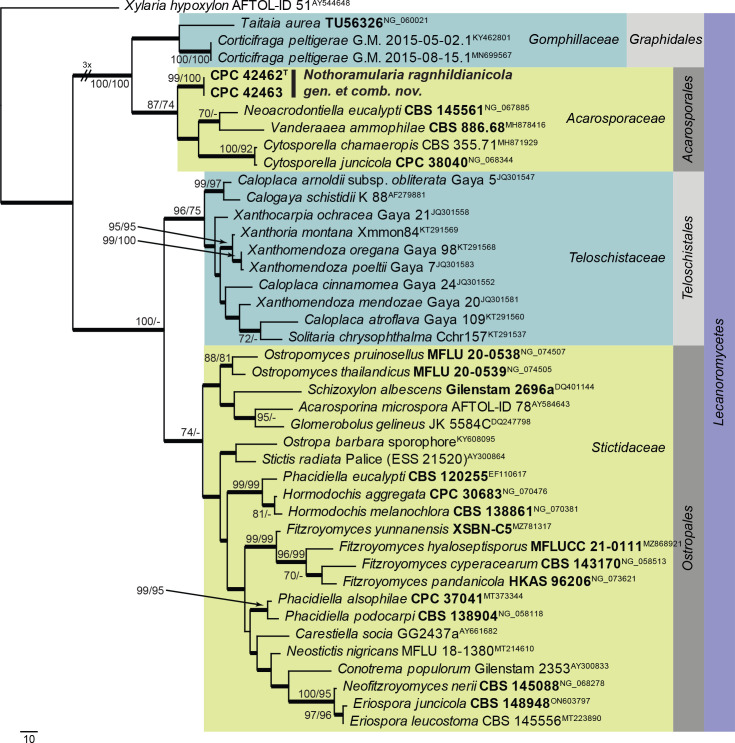
| Nothoramularia ragnhildianicola, gen. et comb. nov. | CBS 149075 = CPC 42463 | Germany: On Ragnhildiana ferruginea, parasitic on Artemisia vulgaris | J. Kruse, 7 Sep. 2021 | OQ990119 | OQ990070 | – | – | – |
| CBS 149076 = CPC 42462, ex-epitype | Germany: On Ragnhildiana ferruginea, parasitic on Artemisia vulgaris | J. Kruse, 7 Sep. 2021 | OQ990118 | OQ990069 | – | – | – | |
| Ophiognomonia setacea | CBS 859.79, ex-epitype | Switzerland: Quercus sp. | M. Monod, 8 May 1979 | AY818958 | AY818962 | – | – | tef1 (second part): JQ414154 |
| CBS 149691 = CPC 43206 | Spain: Quercus robur, leaves | J. Castillo, 21 Jan. 2022 | OQ990120 | OQ990071 | – | – | tef1 (first part): OQ989232, tef1 (second part): OQ989246 | |
| Paradissoconium narthecii | CBS 148449 = CPC 41970, ex-type | Netherlands: Narthecium ossifragum, dead leaves | J. Boers, 4 Jul. 2021 | NR_175214 | NG_081323 | – | – | actA: OK651125, rpb1: OK651151 |
| CBS 149692 = CPC 42494 | Netherlands: Narthecium ossifragum, dead leaves | J. Boers, 26 Sep. 2021 | OQ990121 | OQ990072 | – | – | actA: OQ989192, rpb1: OQ989205 | |
| Paraeutypella citricola | CBS 149693 = CPC 43208 | Spain: Bark of unknown tree | 26 Jan. 2022, J. Castillo | OQ990122 | OQ990073 | OQ989217 | OQ989264 | – |
| Phomatospora endopteris | CBS 149073 = CPC 41832 | Netherlands: Pteridium aquilinum, stems | P.W. Crous, 24 May 2021 | OQ990123 | OQ990074 | OQ989218 | – | SSU: OQ990137 |
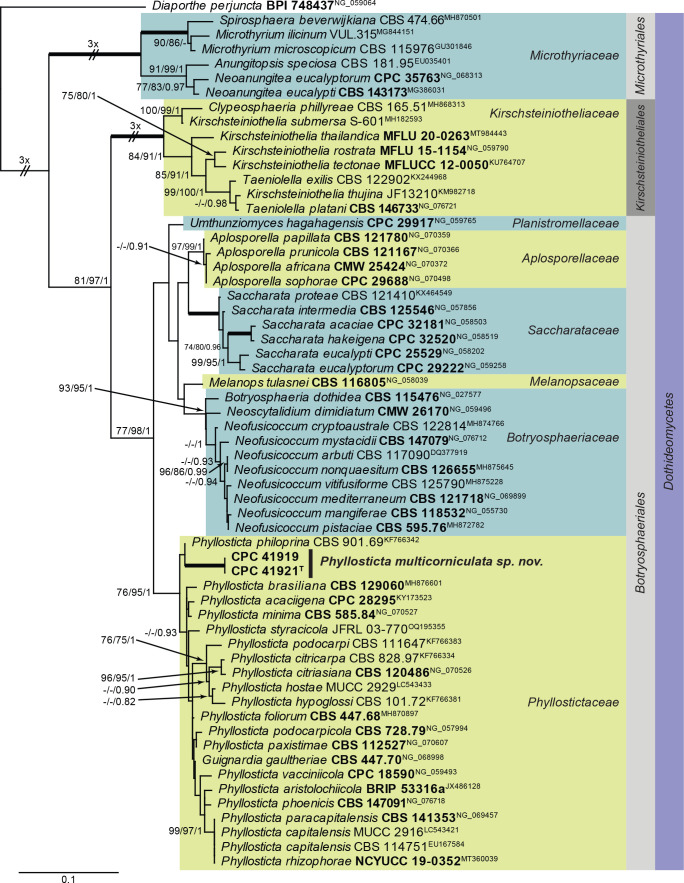
| Phyllosticta multicorniculata | CBS 149077 = CPC 41921, ex-epitype | Canada: Abies balsamea, buds | D. Malloch, 4 May 2021 | OQ990124 | OQ990075 | – | – | actA: OQ989193, gapdh: OQ989198, tef1 (first part): OQ989233 |
| CBS 149078 = CPC 41919 | Canada: Abies balsamea, buds | D. Malloch, 4 May 2021 | OQ990125 | OQ990076 | – | – | actA: OQ989194, gapdh: OQ989199, tef1 (first part): OQ989234 | |
| Pleurotheciella aquatica | CBS 149694 = CPC 44105 | UK: Allium schoenoprasum | P.W. Crous, May 2022 | OQ990126 | OQ990077 | OQ989219 | – | – |
| MFLUCC 17-0464, ex-type | China: Saprobic on decaying wood submerged in Jinsha River | H.Y. Su, Apr. 2015 | NR_160591 | NG_066193 | MF401405 | – | SSU: MF399220 | |
| Ramularia pistaciae | CBS 145564 = CPC 35443, ex-type | Italy: Pistacia lentiscus, leaves | P.W. Crous, 13 Apr. 2018 | NR_165576 | – | – | – | actA: MK876462, gapdh: MK876473 |
| CBS 149696 = CPC 44067 | UK: Arbutus unedo, leaf spot | P.W. Crous & S. Denman, 14 May 2022 | OQ990127 | OQ990078 | OQ989220 | – | actA: OQ989195, gapdh: OQ989200, his3: OQ989201, tef1 (first part): OQ989235 | |
| Ruptoseptoria unedonis | CBS 149697 = CPC 44069 | UK: Arbutus unedo, leaf spot | P.W. Crous & S. Denman, 14 May 2022 | OQ990128 | OQ990079 | OQ989221 | – | – |
| Schizothecium conicum | CBS 149695 = CPC 44110 | Netherlands: Juncus effusus, stems | E.R. Osieck, 28 Apr. 2022 | OQ990129 | OQ990080 | – | – | – |
| Sporidesmiella pini | CBS 148302 = CPC 40067, ex-type | Netherlands: Pinus sylvestris | A.L. van Iperen, 1 Nov. 2020 | OK664747 | NG_081347 | OK651177 | – | – |
| CBS 149045 = CPC 41495 | Netherlands: Juncus effusus, dead culms | E.R. Osieck, 9 Mar. 2021 | OQ990131 | OQ990082 | – | – | – | |
| CPC 41494 | Netherlands: Juncus effusus, dead culms | E.R. Osieck, 9 Mar. 2021 | OQ990130 | OQ990081 | OQ989222 | – | tef1 (second part): OQ989247 | |
| Thyronectria caraganae | CBS 148949 = CPC 41504, ex-epitype | Ukraine: Caragana arborescens, twigs | A. Akulov, 11 Apr. 2021 | OQ990132 | OQ990083 | – | OQ989265 | actA: OQ989196, rpb1: OQ989206 |
| CBS 149509 = CPC 42342 | Ukraine: Caragana arborescens, dead branch | A. Akulov, 1 Aug. 2021 | OQ990133 | OQ990084 | OQ989223 | OQ989266 | rpb1: OQ989207, tef1 (second part): OQ989248 | |
| TCA1, ex-type | Ukraine: Caragana arborescens, dead branch | L.V. Smyk, 6 May 1990 | NR_155911 | KX514385 | – | KX514399 | actA: KX514382, rpb1: KX514390, tef1 (second part): KX514396 | |
| Trichosphaeria pilosa | CBS 149698 = CPC 42927, ex-epitype | Netherlands: Salix sp., decayed branch | E.R. Osieck, 28 Jan. 2022 | OQ990134 | OQ990085 | OQ989224 | OQ989267 | actA: OQ989197, his3: OQ989202, tef1 (first part): OQ989236, tef1 (second part): OQ989249 |
| Zaanenomyces versatilis | CBS 148312 = CPC 41224, ex-type | Netherlands: Juncus inflexus, dead culms | E.R. Osieck, 4 Feb. 2021 | NR_175227 | NG_081336 | – | – | – |
| CBS 149453 = CPC 42831 | Netherlands: Juncus effusus, dead stems | E.R. Osieck, 9 Mar. 2021 | OQ990135 | OQ990086 | – | – | tef1 (second part): OQ989250 | |
1 C.P.K.: Collection maintained at the University of Technology Vienna, Austria; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CPC: Culture collection of Pedro Crous, housed at CBS; FMR: Facultat de Medicina, Universitat Rovira i Virgili, Reus, Spain; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; UAMH: University of Alberta Microfungus Collection and Herbarium, Edmonton, Alberta, Canada.
2 ITS: internal transcribed spacers and intervening 5.8S nrDNA; LSU: large subunit (28S) of the nrRNA gene operon; act: partial actin gene; gapdh: partial glyceraldehyde-3-phosphate dehydrogenase gene; his3: partial histone H3 gene; rpb1: partial DNA-directed RNA polymerase II largest subunit gene; rpb2: partial DNA-directed RNA polymerase II second largest subunit gene; SSU: small subunit (18S) of the nrRNA gene operon; tef1: partial translation elongation factor 1-alpha gene; tub2: partial beta-tubulin gene.
DNA extraction, amplification (PCR) and phylogeny
Fungal mycelium (Table 1) was scraped from the surface of agar cultures with a sterile scalpel and the genomic DNA was isolated using the Wizard® Genomic DNA Purification Kit (Promega Corporation, WI, USA) following the manufacturers’ protocols. All loci were amplified following previously published protocols. The first part of the 28S nrRNA gene (LSU) and complete internal transcribed spacer regions with intervening 5.8S nrRNA gene (ITS) of the nrDNA operon were sequenced for all the isolates included in this study (for amplification conditions, see Fan et al. 2018). Other loci were sequenced for various species or genera using primers and conditions specific for those groups of fungi. Amplification of the partial DNA-directed RNA polymerase II second largest subunit gene (rpb2), the partial translation elongation factor 1-alpha gene (tef1, first part) and the partial beta-tubulin gene (tub2) followed Braun et al. (2018), while amplification of the partial actin gene (actA), the partial glyceraldehyde-3-phosphate dehydrogenase gene (gapdh) and the partial histone H3 gene (his3) followed Videira et al. (2016). Amplification of the partial DNA-directed RNA polymerase II largest subunit gene (rpb1) followed Klaubauf et al. (2014), and the partial translation elongation factor 1-alpha gene (tef1, second part) followed Réblová et al. (2020). The first part of the 18S nrRNA gene (SSU) was amplified as described by Hernández-Restrepo et al. (2020). The resulting fragments were sequenced in both directions using the respective PCR primers and the BigDye Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems Life Technologies, Carlsbad, CA, USA); DNA sequencing amplicons were purified through Sephadex G-50 Superfine columns (Sigma-Aldrich, St. Louis, MO) in MultiScreen HV plates (Millipore, Billerica, MA). Purified sequence reactions were analysed on an Applied Biosystems 3730xl DNA Analyzer (Life Technologies, Carlsbad, CA, USA). The DNA sequences were analysed and consensus sequences were computed using Geneious Prime v. 2022.0.2 (http://www.geneious.com, Kearse et al. 2012).
The sequences for each gene region were subjected to megablast searches (Zhang et al. 2000) to identify closely related sequences in the NCBI’s GenBank nucleotide database. The results are provided as part of the species notes or as selected phylogenetic trees. Maximum-likelihood (ML) phylogenetic trees were constructed generated using IQ-TREE v. 2.1.3 (Nguyen et al. 2015) and branch support values were calculated with 1 000 non-parametric bootstrap replicates and optimal modelfinding using the TESTNEW option using ModelFinder (Kalyaanamoorthy et al. 2017) as implemented in IQ-TREE. Bayesian analyses were performed with MrBayes v. 3.2.7a (Ronquist et al. 2012) as explained in Braun et al. (2018), while RAxML v. 8.0.0.0 (Stamatakis 2014) was used with default parameters to provide additional ML support values for selected trees, while parsimony analyses using PAUP* v. 4.0a build 168 (Swofford et al.) were performed as explained in Videira et al. (2016). All resulting trees were printed with Geneious Prime v. 2022.0.2 and the layout of the trees was done using Adobe Illustrator 2022 v. 26.3.1. Sequences derived in this study were submitted to GenBank (Table 1) and the alignments and phylogenetic trees in figshare.com (doi: 10.6084/m9.figshare.23447330). The optimal identity thresholds to discriminate filamentous fungal species followed Vu et al. (2019).
Morphology
Slide preparations were mounted in lactic acid, Shear’s mounting fluid, Melzer’s solution, or water, from colonies sporulating on MEA, PDA, PNA or OA. Observations were made with a Nikon SMZ25 dissection microscope, and with a Zeiss Axio Imager 2 light microscope using differential interference contrast (DIC) illumination and images recorded on a Nikon DS-Ri2 camera with associated software. Colony characters and pigment production were noted after 2–4 wk of growth on MEA, PDA and OA (Crous et al. 2019c) incubated at 25 deg;C. Colony colours (surface and reverse) were scored using the colour charts of Rayner (1970). Taxonomic novelties were submitted to MycoBank (www.MycoBank.org; Crous et al. 2004).
RESULTS
Phylogeny
Phylogenetic trees were generated for the taxonomic novelties, or to better clarify the position of a taxon in a broader context where needed. These trees are discussed in the species notes and the statistics associated with the phylogenetic analyses presented in this study are provided in supplementary Table S1.
Taxonomy
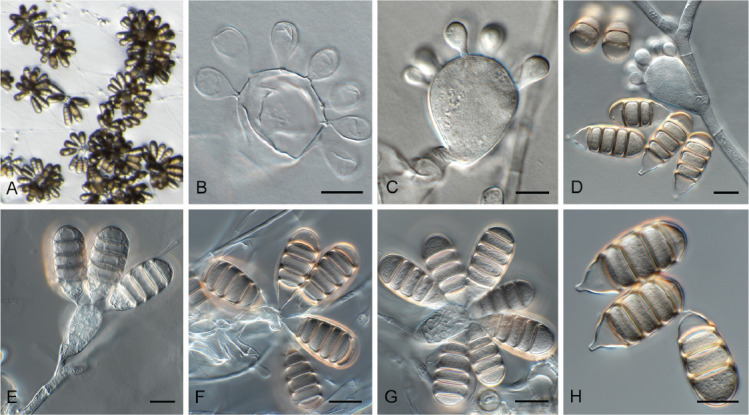
Acremonium aquaticum Crous & Jurjević, sp. nov. MycoBank MB 848820. Fig. 1.
Fig. 1.
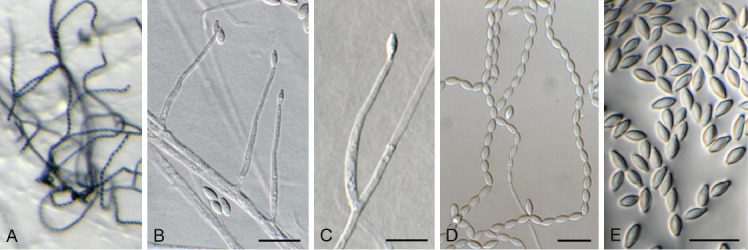
Acremonium aquaticum (CPC 42867). A. Colony on SNA. B–D. Conidiogenous cells giving rise to chains of conidia. E. Conidia. Scale bars = 10 μm.
Etymology: Name refers to the fact that it was isolated from water.
Mycelium consisting of hyaline, smooth, branched, septate, 1.5–2 μm diam hyphae. Conidiophores reduced to conidiogenous cells, solitary, erect, subcylindrical with apical taper, hyaline, smooth, phialidic, 20–30 × 1.5–2 μm. Conidia in long, unbranched chains, aseptate, smooth, fusoid-ellipsoid, initially hyaline, but becoming olivaceous with age, (4–)5–6 × (2–)2.5 μm; forming olivaceous mucoid droplets with age.
Culture characteristics: Colonies flat, spreading, with sparse to moderate aerial mycelium and smooth, lobate margin, reaching 40 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface and reverse dirty white, but sectoring with age to form olivaceous zones, most prominent on MEA.
Typus: USA, North Carolina, Durham, cooling pad water, greenhouse, Oct. 2021, Z. Jurjević 5662 (holotype CBS H-25167, culture ex-type CPC 42867 = CBS 149454).
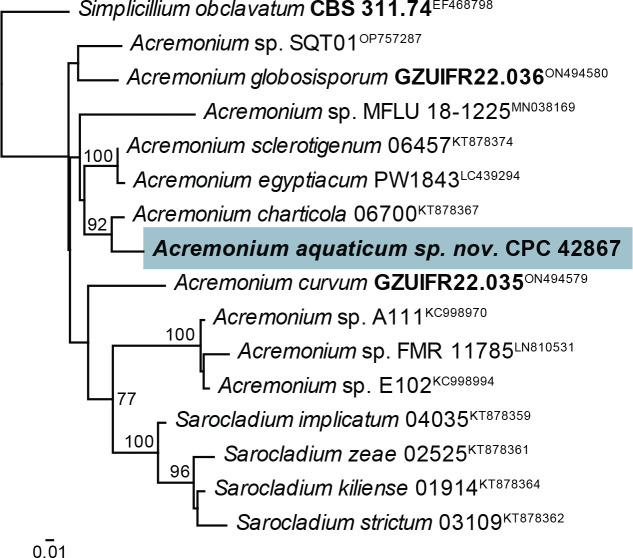
Notes: Acremonium and allied genera were recently revised by Hou et al. (2023). Acremonium aquaticum is phylogenetically (92 % bootstrap support; Fig. 2) closely related to A. charticola (conidia in mucoid heads, 3.2–4.5 × 1.4–2.0 μm; Gams 1971) but is morphologically distinct in having larger conidia that are formed in chains.
Fig. 2.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE v. 2.1.3 of the Acremonium tef1 (second part) nucleotide alignment. Bootstrap support values (> 75 %) from 1 000 non-parametric bootstrap replicates are shown at the nodes. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Simplicillium obclavatum (culture CBS 311.74; GenBank EF468798) and the species treated here is highlighted with bold face. The scale bar indicates the expected number of changes per site.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Acremonium sp. from indoor plaster in Russia (strain tk2, GenBank LT549084.1; Identities = 537/537 (100 %), no gaps), Acremonium charticola (strain CBS 881.73, GenBank AJ621774.1; Identities = 472/506 (93 %), 11 gaps (2 %)), and Acremonium alternatum (strain NIOSN M-120, GenBank MG589592.1; Identities = 412/446 (92 %), six gaps (1 %)). It is also identical to isolates from a pine tree in South Korea (GenBank MK848676.1), a tempera painting on canvas in Slovenia (GenBank MZ687371.1), soil collected in a 40-yr-old Pinus merkusii forest in Viet Nam (GenBank MW504687.1), from Ageratina adenophora in China (GenBank MK304178.1), and marine sediment in China (GenBank KX098125.1). Closest hits using the LSU sequence were Acremonium charticola (strain CBS 881.73, GenBank MH872552.1; Identities = 817/823 (99 %), two gaps (0 %)), Acremonium sordidulum (strain SP17, GenBank MZ269296.1; Identities = 814/822 (99 %), one gap (0 %)), and Acremonium alternatum (strain MUT<ITA> 6246, GenBank MN947574.1; Identities = 792/800 (99 %), one gap (0 %)). Closest hits using the actA sequence had highest similarity to Tilachlidium brachiatum (strain CBS 505.67, GenBank KM231249.1; Identities =543/637 (85 %), 20 gaps (3 %)), Acremonium chrysogenum (no strain number specified, GenBank AF056976.1; Identities = 558/658 (85 %), 35 gaps (5 %)), and Acremonium sp. from Leymus chinensis in China (strain 324, GenBank JN836732.1; Identities = 587/633 (93 %), 15 gaps (2 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Acremonium alternatum (strain AFTOL-ID 1396, GenBank FJ238366.1; Identities = 573/676 (85 %), 11 gaps (1 %)), and Caespitomonium euphorbiae (culture CPC 39083, GenBank OK651157.1; Identities = 627/866 (72 %), 42 gaps (4 %)). Closest hits using the tef1 (second part) sequence had highest similarity to Acremonium charticola (strain 06700, GenBank KT878367.1; Identities = 722/753 (96 %), no gaps), Amphichorda guana (strain LC5819, GenBank KX855212.1; Identities = 696/748 (93 %), no gaps), and Acremonium sclerotigenum (strain 06239, GenBank KT878358.1; Identities = 712/769 (93 %), five gaps (0 %)). No significant hits were obtained using the tub2 sequence.
Authors: P.W. Crous, J.Z. Groenewald, Z. Jurjević & S. Balashov
Acrostalagmus luteoalbus (Link) Zare et al. [as ‘luteo-albus’], Mycol. Res. 108: 581. 2004. Fig. 3.
Fig. 3.

Acrostalagmus luteoalbus (CPC 43187). A–G. Conidiophores with conidiogenous cells giving rise to conidia. H. Conidia. Scale bars = 10 μm.
Description and illustration: Zare et al. (2004).
Material examined: South Africa, Western Cape Province, Kirstenbosch, on leaf of Portulacaria afra (Portulacaceae), 27 Feb. 2022, P.W. Crous, HPC 3862, culture CPC 43187 = CBS 149685.
Notes: Acrostalagmus luteoalbus was recently reported as a major constituent of the mixed mycobiota in the wet cork liner of a water-damaged outdoor wall, and also from indoor dust in Finland (Andersson et al. 2021). The present record represents a new report from South Africa, where it was isolated from a leaf of Portulacaria afra.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to numerous sequences of Acrostalagmus luteoalbus (e.g. EF_332, GenBank MT528981.1; Identities = 502/502 (100 %), no gaps), and Nectria inventa (e.g. strain CBS 388.65, GenBank MH858627.1; Identities = 502/502 (100 %), no gaps). Closest hits using the LSU sequence were Nectria inventa (strain CBS 236.55, GenBank MH869007.1; Identities = 855/855 (100 %), no gaps), Acrostalagmus luteoalbus (strain MUT<ITA> 4778, GenBank KP671745.1; Identities = 855/855 (100 %), no gaps), and Acrostalagmus annulatus (strain CBS 121213, GenBank LR025806.1; Identities = 812/812 (100 %), no gaps). Closest hits using the rpb2 (first part) sequence had highest similarity to Acrostalagmus luteoalbus (strain CBS 112.16, GenBank LR026101.1; Identities = 742/743 (99 %), no gaps), and Acrostalagmus annulatus (strain CBS 121213, GenBank LR026108.1; Identities = 738/743 (99 %), no gaps). Closest hits using the tef1 (second part) sequence had highest similarity to Acrostalagmus luteoalbus (strain CBS 388.65, GenBank LR026372.1; Identities = 787/787 (100 %), no gaps), Acrostalagmus annulatus (strain CBS 121213, GenBank LR026378.1; Identities = 782/786 (99 %), no gaps), and Verticillium zaregamsianum (strain V202, GenBank KJ443225.1; Identities = 856/904 (95 %), four gaps (0 %)).
Authors: P.W. Crous & J.Z. Groenewald
Appendopyricularia juncicola Crous & Osieck, Persoonia 48: 265. 2022. Fig. 4.
Fig. 4.
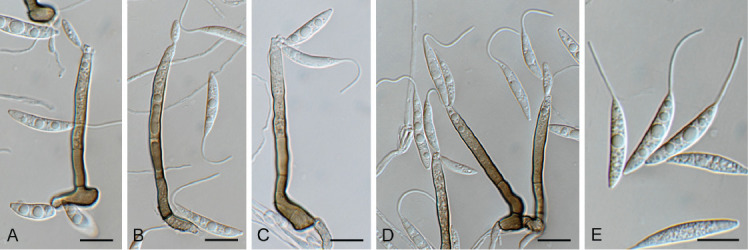
Appendopyricularia juncicola (CPC 44053). A–D. Conidiophores with conidiogenous cells giving rise to conidia. E. Conidia. Scale bars = 10 μm.
Description and illustration: Crous et al. (2022b).
Materials examined: Netherlands, Overijsel Province, Reutum, Reutumerveen, 22.5 m a.s.l., 52°23’43’’N, 06°49’24’’E, on dead culms of Carex elongata (Cyperaceae), 20 Mar. 2022, E.R. Osieck, HPC 3952 = WI-50#4449, cultures CPC 44053 = CBS 149686, CPC 44055; Overijssel Province, Witte Veen, Haaksbergen, 39 m a.s.l., 52°08’25’’N, 06°52’20’’E, on dead culms of Juncus effusus (Juncaceae), 28 Apr. 2022, E.R. Osieck, HPC 3962 = WI-55#4461, cultures CPC 44107, 44106; Utrecht Province, Nieuw-Wulven, north of Houten, 1.5 m a.s.l., 52°02’46’’N, 05°10’34’’E, on dead culms of J. effusus, 9 Dec. 2021, E.R. Osieck, HPC 3812 = WI-42#4355, culture CPC 42686.
Notes: Appendopyricularia, based on A. juncicola, was introduced as a new hyphomycete genus occurring on culms of Juncus effusus in the Netherlands (Crous et al. 2022b). This is the first record of this taxon also occurring on culms of Carex elongata.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 42686 had highest similarity to Appendopyricularia juncicola (strain CPC 41278, GenBank NR_182605.1; Identities = 500/501 (99 %), one gap (0 %)), Thyridium pluriloculosum (strain GZUIFR21.876, GenBank OK493561.1; Identities = 402/481 (84 %), 25 gaps (5 %)), and Phialemonium dimorphosporum (strain SLE, GenBank DQ403199.1; Identities = 412/495 (83 %), 30 gaps (6 %)). The ITS sequence of CPC 42686 is identical to those of CPC 44053, 44055, 44106 and 44107 (508/508, 508/508, 502/502 and 508/508 nucleotides, respectively). Closest hits using the LSU sequence of CPC 42686 were Appendopyricularia juncicola (strain CPC 41278, GenBank NG_149075.1; Identities = 808/808 (100 %), no gaps), Paradiplococcium singulare (strain CBS 126091, GenBank NG_066271.1; Identities = 797/839 (95 %), three gaps (0 %)), and Barbatosphaeria varioseptata (strain CBS 137797, GenBank NG_058674.1; Identities = 798/840 (95 %), five gaps (0 %)). The LSU sequence of CPC 42686 is identical to those of CPC 44053, 44055, 44106 and 44107 (792/792, 825/825, 804/804 and 838/838 nucleotides, respectively). Closest hits using the tef1 (first part) sequence had highest similarity to Appendopyricularia juncicola (strain CPC 41278, GenBank ON605627.1; Identities = 338/345 (98 %), one gap (0 %)), Madurella fahalii (strain CBS 129176, GenBank MN078441.1; Identities = 141/149 (95 %), no gaps), and Podospora comata (strain Wa139-, GenBank CP071493.1; Identities = 151/165 (92 %), four gaps (2 %)). The tef1 sequence of CPC 42686 is 98 % similar to those of CPC 44053, 44055 and 44106 (338/345, 337/344, and 338/345 nucleotides, respectively; all including one gap). The closest hits using the tub2 sequence of CPC 42686 had highest similarity to Appendopyricularia juncicola (strain CPC 41278, GenBank ON605635.1; Identities = 671/690 (97 %), three gaps (0 %)); while the tub2 sequence of CPC 44055 is identical to that of Appendopyricularia juncicola (strain CPC 41278, GenBank ON605635.1; Identities = 690/690 (100 %), no gaps). The tub2 sequence of CPC 42686 is 97 % similar to those of CPC 44055, 44106 and 44107 (677/696, 672/691 and 677/696, nucleotides, respectively; all three includes an indel of three nucleotides and are identical to Appendopyricularia juncicola strain CPC 41278, GenBank ON605635.1).
Authors: P.W. Crous, J.Z. Groenewald & E.R. Osieck
Biscogniauxia anceps (Sacc.) J.D. Rogers et al., Mycol. Res. 100: 669. 1996. Fig. 5.
Fig. 5.
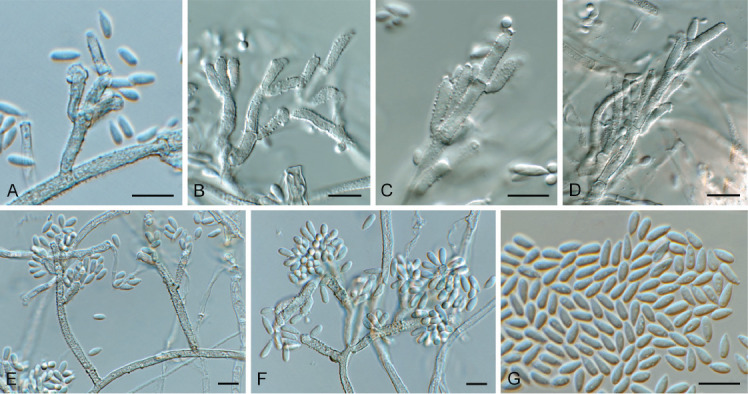
Biscogniauxia anceps (CPC 43197). A–F. Conidiophores with conidiogenous cells giving rise to conidia. G. Conidia. Scale bars = 10 μm.
Description and illustration: Rogers et al. (1996).
Material examined: Spain, Pontevedra, O Grove, on bark of Eucalyptus sp. (Myrtaceae), 25 Mar. 2022, M.A. Delgado, HPC 3867 = RKS 1164, culture CPC 43197 = CBS 149687.
Notes: Species of Biscogniauxia are found as endophytes and opportunistic pathogens on old and stressed trees (Bahmani et al. 2021).Biscogniauxia anceps is known to occur on bark of various tree hosts in Europe and is reported here from Eucalyptus bark in Spain.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Biscogniauxia anceps (strain 123, GenBank EF026132.1; Identities = 481/481 (100 %), no gaps), Biscogniauxia nummularia (strain MUCL 51395, GenBank NR_153649.1; Identities = 534/585 (91 %), 16 gaps (2 %)), and Digitodochium amoenum (strain LA, GenBank KC774569.1; Identities = 534/585 (91 %), 16 gaps (2 %)).
Authors: P.W. Crous, J.Z. Groenewald, M.A. Delgado & R.K. Schumacher
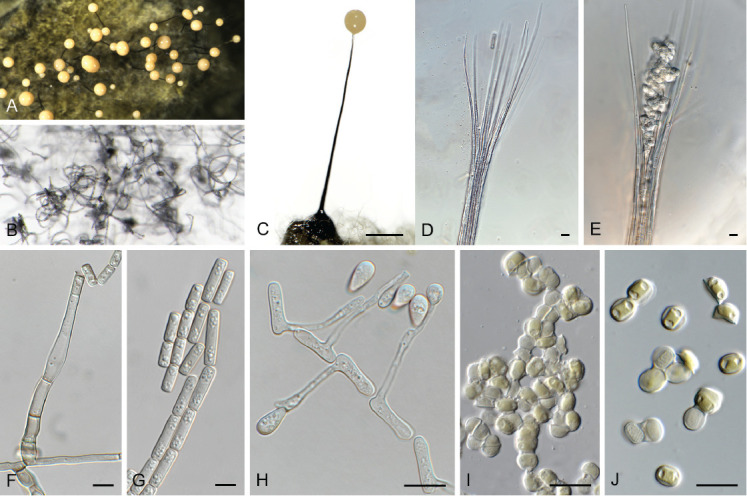
Cephaliophora tropica Thaxt., Bot. Gaz. 35: 158. 1903. Fig. 6.
Fig. 6.
Cephaliophora tropica (CPC 42877). A–G. Conidiophores with conidiogenous cells giving rise to conidia. H. Conidia. Scale bars = 10 μm.
Classification: Pezizomycetes, Pezizales, Ascodesmidaceae.
Description and illustration: Ruszkiewicz-Michalska et al. (2017).
Typus: Lectotype here designated, Bot. Gaz. 35: 158. 1903, plate V, figs 11–16, MBT 10013412. USA, Louisiana, crocodile farm, pan water, 24 Nov. 2021, Z. Jurjević 5686 (epitype here designated CBS H-25171, MBT 10013411, culture ex-epitype CPC 42877 = CBS 149457).
Notes: Cephaliophora tropica is a pantropical and occasionally temperate species (Seifert et al. 2011). It is commonly isolated from dung, soil, and water (Ruszkiewicz-Michalska et al. 2017). Because no type was indicated in the original description, a lectotype and epitype are designated here to fix the application of the name. The genus Cephaliophora is considered incertae sedis in MycoBank and Index Fungorum, but was shown by Hansen et al. (2013) and confirmed here to belong to Ascodesmidaceae.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Cephaliophora tropica (strain CBS 133.33, GenBank MH855385.1; Identities = 529/529 (100 %), no gaps), and Cephaliophora irregularis (strain YG-C22, GenBank KX683420.1; Identities = 504/514 (98 %), no gaps). Closest hits using the LSU sequence were Cephaliophora tropica (strain CBS 315.66, GenBank MH870444.1; Identities = 830/830 (100 %), no gaps), Cephaliophora irregularis (strain CBS 218.62, GenBank KC012668.1; Identities = 826/830 (99 %), no gaps), and Ascodesmis rosicola (voucher GUCC 190035.1, GenBank MZ221605.1; Identities = 815/830 (98 %), no gaps). Closest hits using the SSU sequence were Cephaliophora tropica (strain JCM 6019, GenBank AB001112.1; Identities = 1 000/1 000 (100 %), no gaps), Cephaliophora irregularis (strain IFO 6778, GenBank AB001109.2; Identities = 998/1 000 (99 %), no gaps), and Eleutherascus lectardii (strain CBS 626.71, GenBank NG_062685.1; Identities = 994/1 000 (99 %), no gaps). Closest hits using the rpb1 sequence had highest similarity to Cephaliophora tropica (strain CBS 133.33, GenBank JX943656.1; Identities = 727/729 (99 %), no gaps), Cephaliophora irregularis (strain CBS 218.62, GenBank JX943655.1; Identities = 674/719 (94 %), five gaps (0 %)), and Ascodesmis nigricans (strain CBS 428.91, GenBank JX943653.1; Identities = 582/690 (85 %), nine gaps (1 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Cephaliophora tropica (strain CBS 133.33, GenBank JX943763.1; Identities = 598/600 (99 %), no gaps), Cephaliophora irregularis (strain CBS 218.62, GenBank JX943762.1; Identities = 686/732 (94 %), two gaps (0 %)), and Ascodesmis rosicola (voucher GUCC 190204.1, GenBank MZ333140.1; Identities = 728/882 (83%), three gaps (0 %)). Closest hits using the tef1 (second part) sequence had highest similarity to Cephaliophora tropica (strain CBS 133.33, GenBank KC109224.1; Identities = 901/903 (99 %), no gaps), Cephaliophora irregularis (strain CBS 218.62, GenBank KC109223.1; Identities = 899/958 (94 %), 11 gaps (1 %)), and Ascodesmis nigricans (strain CBS 389.68, GenBank KC109221.1; Identities = 781/887 (88 %), 24 gaps (2 %)).
Authors: P.W. Crous, J.Z. Groenewald & Z. Jurjević
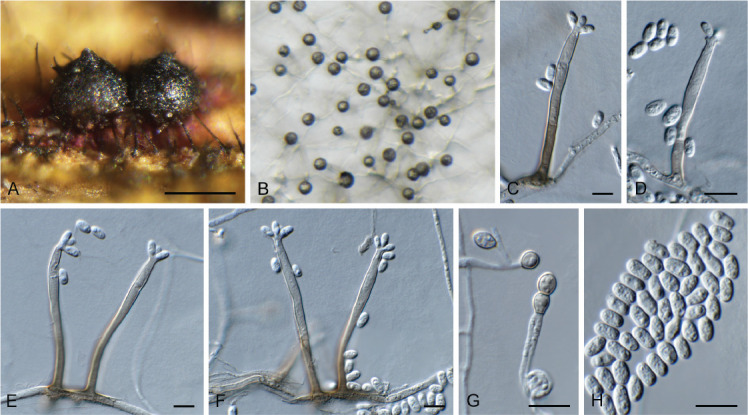
Ceratocystis ficicola Kajitani & Masuya, Mycoscience 52: 351. 2011. Fig. 7.
Fig. 7.
Ceratocystis ficicola (CPC 44213). A. Colony on PDA. B. Colony on SNA. C. Ascoma exuding ascospores. D, E. Ostiolar hyphae. F, G. Subcylindrical endoconidia. H. Aleuroconidia. I, J. Ascospores. Scale bars: C = 300 μm, all others = 10 μm.
Ascomata perithecial, solitary with brown to black, globose base, 180–220 μm diam, with erect, brown neck, 1 000–1 300 μm long, becoming paler brown toward apex, 40–50 μm diam at base, 18–20 μm diam at apex. Ostiolar hyphae divergent, subhyaline, 130–200 μm long. Asci not observed. Ascospores hyaline, galeate, aseptate, 7–8 × 6–7 μm in top view, 4–5 μm high in side view, accumulating in creamy mucoid masses at apices of perithecia. Thielaviopsis asexual morph: endoconidiophores solitary on mycelium, pale brown to brown, smooth, tapering to truncate apex, 80–160 μm long, 5–7 μm diam at base, 1–4-septate. Conidiogenous cells phialidic, cylindrical, 500–75 μm long, 4–5 μm diam at base, 4 μm diam at apex. Endoconidia hyaline, becoming pale brown, smooth, guttulate, aseptate, subcylindrical with obtuse to truncate ends, (11–)15–17(–20) × 4–5(–6) μm, occurring in unbranched chains. Aleuroconidia solitary, aseptate, clavate to obovoid, hyaline, smooth, (9–) 12–14(–15) × 4–5(–6) μm, frequently forming via microcyclic conidiation from endoconidia. Culture characteristics: Colonies erumpent, spreading, with moderate aerial mycelium and feathery, lobate margin, covering dish in 2 wk at 25 °C. On MEA, PDA and OA surface olivaceous grey, and reverse iron grey.
Materials examined: Sicily, Noto, from trunk necrosis in Ficus carica (Moraceae), 2022, G. Polizzi, CERA30 = CBS H-25213, culture CPC 44213 = CBS 149669; CERA20 = CBS H-25214, culture CPC 44214 = CBS 149670.
Notes — Ceratocystis ficicola causes vascular wilt of fig trees in Japan, and has also recently been reported from Greece (Tsopelas et al. 2021). This is the first record of the pathogen from Italy. Based on ITS alone, the present collections might represent a novel species. However, this was not supported by the secondary barcodes (see below). A closer inspection of the ITS blast alignment revealed a similarity of 577/600 nucleotides, with the 19 of the 23 mismatches being accounted for by gaps caused mainly by differences in repeat length repeats in T- or A-rich parts of the sequences.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 44213 had highest similarity to Ceratocystis ficicola (strain MAFF 625119, GenBank NR_119410.1; Identities = 577/600 (96 %), 19 gaps (3 %)), Ceratocystis cercfabiensis (strain CMW 42512, GenBank KP727589.1; Identities = 541/598 (90 %), 20 gaps (3 %)), and Ceratocystis uchidae (strain CBS 115164, GenBank NR_164012.1; Identities = 560/620 (90 %), 23 gaps (3 %)). The ITS sequences of CPC 44213 and 44214 are identical (603/603 nucleotides). Closest hits using the LSU sequence of CPC 44213 were Ceratocystis ficicola (strain CMW38543, GenBank KM495342.1; Identities = 808/809 (99 %), one gap (0 %)), Ceratocystis polychroma (strain CMW11424, GenBank KM495368.1; Identities = 805/808 (99 %), no gaps), and Ceratocystis obpyriformis (strain CBS 122511, GenBank MH874746.1; Identities = 817/821 (99 %), one gap (0 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Ceratocystis ficicola (strain CMW38543, GenBank KY685082.1; Identities = 961/961 (100 %), no gaps), Ceratocystis cercfabiensis (strain CMW42795, GenBank KY644022.1; Identities = 902/913 (99 %), no gaps), and Ceratocystis corymbiicola (strain CMW29120, GenBank KY644037.1; Identities = 908/920 (99 %), no gaps). Closest hits using the tef1 (second part) sequence of CPC 44213 had highest similarity to Ceratocystis ficicola (strain C1355, GenBank KY316544.1; Identities = 897/897 (100 %), no gaps), Ceratocystis fimbriata (strain C3372, GenBank KY982682.1; Identities = 892/897 (99 %), no gaps), and Ceratocystis uchidae (strain C1714, GenBank KY982680.1; Identities = 892/897 (99 %), no gaps). Closest hits using the tub2 sequence of CPC 44213 had highest similarity to Ceratocystis huliohia (strain B, GenBank KU043268.1; Identities = 1 241/1 327 (94 %), 27 gaps (2 %)), Ceratocystis uchidae (strain CBS 114720, GenBank KU043266.1; Identities = 1 241/1 327 (94 %), 27 gaps (2 %)), and Ceratocystis populicola (strain CBS 114725, GenBank KC589392.1; Identities = 522/621 (84 %), 35 gaps (5 %)). There was no overlap between our tub2 sequence and the two sequences available on GenBank for Ceratocystis ficicola (GenBank KY685077 and KY685078).
Authors: P.W. Crous, J.Z. Groenewald, G. Gusella & G. Polizzi
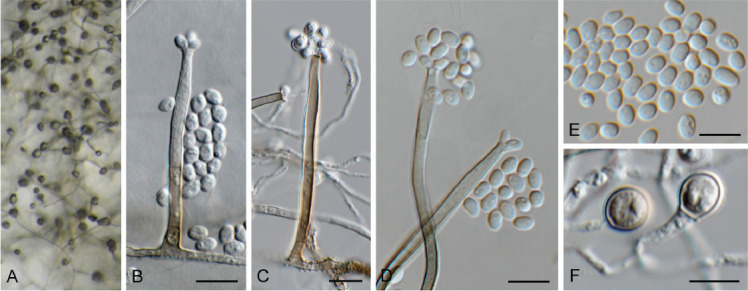
Chloridium caudigerum (Höhn.) S. Hughes, Canad. J. Bot. 36: 748. 1958. Fig. 8.
Fig. 8.
Chloridium caudigerum (CPC 42899). A. Ascomata. B–F. Conidiophores with conidiogenous cells giving rise to conidia. G. Chlamydospores. H. Conidia. Scale bars: A = 300 μm, all others = 10 μm.
Description and illustration: Réblová et al. (2022).
Material examined: Netherlands, Utrecht Province, Houten, Nieuw Wulven, 1.5 m a.s.l., 52°02’53’’N, 05°09’42’’E, on branch of Ulmus laevis (Ulmaceae), 28 Jan. 2022, E.R. Osieck, HPC 3827 = WI-46#4391, culture CPC 42899 = CBS 149688.
Notes: Chloridium caudigerum represents a common European species, found especially on decaying wood of deciduous trees. This species closely resembles Chl. chlamydosporum and Chl. virescens (Réblová et al. 2022). The sexual morph of Chl. virescens is also known as Melanopsammella vermicularioides. Melanopsammella is characterised by ascospores already fragmenting in the ascus. The sexual morph of Chl. caudigerum (also present in the collection) is similar but differs in having setose ascomata, which are glabrous in Chl. virescens (Réblová loc. cit.).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Chloridium caudigerum (strain ICMP 22547, GenBank OP455384.1; Identities = 491/491 (100 %), no gaps), Chloridium virescens (strain CBS 127310, GenBank MH864519.1; Identities = 498/520 (96 %), one gap (0 %)), and Chloridium jilinense (strain NN046507, GenBank OL627659.1; Identities = 457/469 (97 %), two gaps (0 %)). Closest hits using the LSU sequence were Chloridium caudigerum (strain ICMP 22547, GenBank OP455491.1; Identities = 844/845 (99 %), one gap (0 %)), Chloridium jilinense (strain NN046507, GenBank OL655058.1; Identities = 834/840 (99 %), no gaps), and Chloridium virescens (strain CBS 127627, GenBank MH876080.1; Identities = 836/845 (99 %), one gap (0 %)). Closest hits using the tef1 (second part) sequence had highest similarity to Chloridium caudigerum (strain CBS 145490, GenBank OP464953.1; Identities = 859/860 (99 %), no gaps), Chloridium moratum (strain FMR 11343, GenBank OP464997.1; Identities = 838/861 (97 %), two gaps (0 %)), and Chloridium detriticola var. detriticola (strain ICMP 15144, GenBank OP464977.1; Identities = 836/860 (97 %), no gaps). Closest hits using the tub2 sequence had highest similarity to Chloridium caudigerum (strain FMR 12411, GenBank OP465062.1; Identities = 702/702 (100 %), no gaps), Chloridium moratum (strain FMR 11343, GenBank OP465102.1; Identities = 612/703 (87 %), 12 gaps (1 %)), and Chloridium bellum var. luteum (strain CBS 141.54, GenBank OP465041.1; Identities = 613/718 (85 %), 28 gaps (3 %)).
Authors: P.W. Crous, J.Z. Groenewald & E.R. Osieck
Chloridium gamsii Réblová & Hern.-Restr., Stud. Mycol. 103: 143. 2022. Fig. 9.
Fig. 9.
Chloridium gamsii (CPC 41933). A–D. Conidiophores with conidiogenous cells giving rise to conidia. E. Conidia. F. Chlamydospores. Scale bars = 10 μm.
Description and illustration: Réblová et al. (2022).
Mycelium consisting of hyaline, smooth, branched, septate, 1.5–2 μm diam hyphae, which become brown and verruculose adjacent to conidiophores, up to 3 μm diam. Conidiophores solitary, erect, straight, flexuous, 1–3-septate, subcylindrical, medium brown, smooth, 35–80 × 3–3.5 μm. Conidiogenous cells integrated, terminal, subcylindrical, medium brown, smooth, with flared collarette; apex incl. collarette 3–5 μm diam, 20–30 × 2–3 μm. Conidia solitary, aggregated in mucoid mass, emerging in sympodial arrangement at apex, hyaline, smooth, guttulate, ellipsoid to ovoid, aseptate, 3.5–6 × 3–3.5 μm. Chlamydospores solitary, terminal on hyphae, medium brown, smooth, thick-walled, aseptate, guttulate, ellipsoid to ovoid, 5–7 × 4–5 μm.
Culture characteristics: Colonies erumpent, spreading, with moderate aerial mycelium and smooth, even margin, reaching 15 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface olivaceous grey and reverse iron-grey.
Material examined: Netherlands, Friesland Province, Terschelling, on Cladonia portentosa (Cladoniaceae), 7 Jun. 2021, J. Boers, HPC 3646 = CBS H-24962, culture CPC 41933 = CBS 149043.
Notes: Chloridium gamsii was recently described from decaying wood collected in Belgium and is reported here from a lichen in the Netherlands. Based on published data, this species appears to be common in Europe, Australasia, with a few records from Canada and the USA (Réblová et al. 2022).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Chloridium gamsii (strain CBS 667.75, GenBank OP455415.1; Identities = 481/484 (99 %), no gaps), Chloridium virescens (strain CBS 127310, GenBank MH864519.1; Identities = 489/508 (96 %), two gaps (0 %)), and Chloridium biforme (strain ICMP 23429, GenBank OP455363.1; Identities = 472/485 (97 %), two gaps (0 %)). Closest hits using the LSU sequence were Chloridium gamsii (strain CBS 667.75, GenBank OP455522.1; Identities = 808/809 (99 %), no gaps), Chloridium virescens var. chlamydosporum (strain CBS 126074, GenBank MH875525.1; Identities = 802/809 (99 %), no gaps), and Chloridium peruense (strain CBS 126074, GenBank OP455531.1; Identities = 802/809 (99 %), no gaps). Closest hits using the tef1 (second part) sequence had highest similarity to Chloridium gamsii (strain CBS 667.75, GenBank OP464990.1; Identities = 814/833 (98 %), no gaps), Chloridium biforme (strain ICMP 23429, GenBank OP464937.1; Identities = 813/833 (98 %), no gaps), and Chloridium caudigerum (strain FMR 12411, GenBank OP464956.1; Identities = 807/834 (97 %), two gaps (0 %)).
Authors: P.W. Crous, J.Z. Groenewald & J. Boers
Cladophialophora laricicola Crous & Boers, sp. nov. MycoBank MB 848823. Fig. 10.
Fig. 10.

Cladophialophora laricicola (CPC 41384). A–E, G. Conidiophores with conidiogenous cells giving rise to conidia. F, H, I. Conidia. Scale bars = 10 μm.
Etymology: Name refers to the host genus Larix from which it was isolated.
Mycelium consisting of medium brown, smooth, septate, branched, 2.5–5 μm diam hyphae. Conidiophores erect, flexuous, subcylindrical, medium brown, smooth, with terminal and at times intercalary conidiogenous cells. Conidiogenous cells medium brown, subcylindrical, smooth, holoblastic, 10–15 × 3–4 μm. Conidia occurring in cylindrical chains (5–12), brown, smooth to roughened, aseptate, broadly ellipsoid, guttulate, becoming thick-walled, encased in mucoid sheath, individual conidia (6–)8–9(–10) × (4–)5(–6) μm; at time cylindrical conidial chains form lateral chains, and in extreme cases arranged in hand-like, penicillate configuration.
Culture characteristics: Colonies erumpent, spreading, with folded surface, lobate, feathery margin, and medium aerial mycelium, reaching 10 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface and reverse iron-grey.
Typus: Netherlands, Drenthe Province, Dwingelderveld National Park, 52.829188, 6.432495, on dead wood of Larix sp. (Pinaceae), 16 Mar. 2021, J. Boers, HPC 3608 (holotype CBS H-24954 culture ex-type CPC 41384 = CBS 148944).
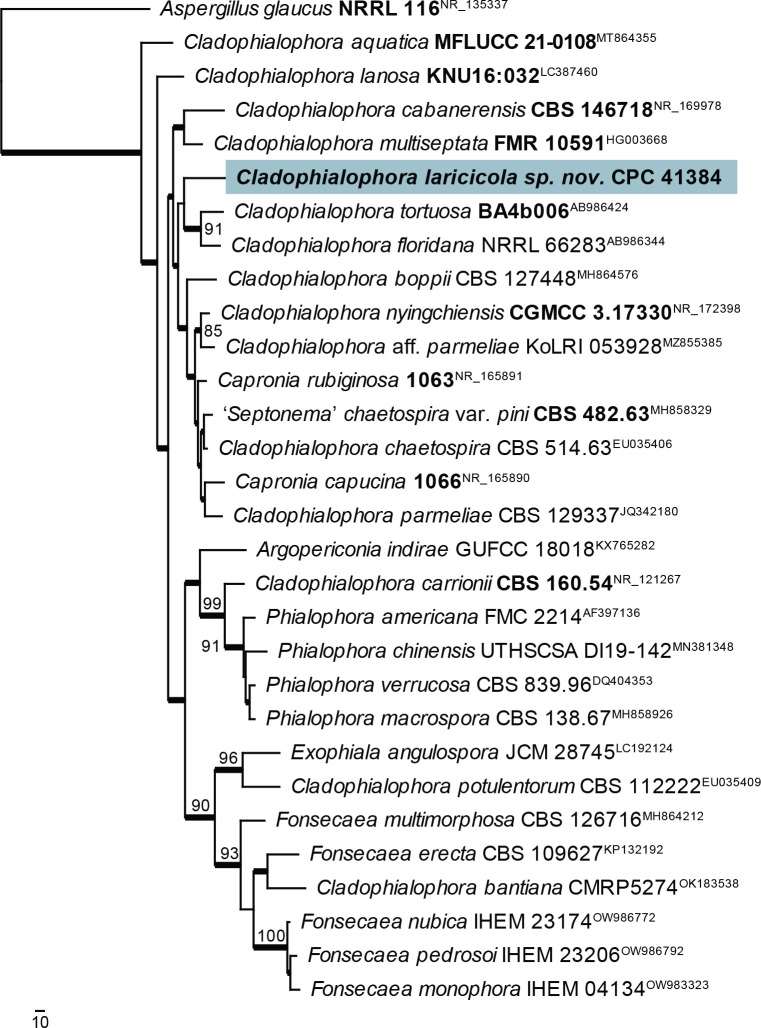
Notes: Cladophialophora laricicola is phylogenetically distinct from other species presently known from their DNA sequence data. In the maximum parsimony phylogenetic tree (Fig. 11), it clustered sister to Cl. tortuosa and Cl. floridana but with no support. A maximum likelihood analysis conducted with IQ-TREE placed it basal to all ingroup species (data not shown). All sequenced loci indicated some affinity with Cladophialophora but not a tight association with any other sequenced species (see below).
Fig. 11.
The first of four equally most parsimonious trees obtained from a ‘Cladophialophora’ ITS sequence alignment. The scale bar indicates the number of changes and the numbers at the nodes represent bootstrap support values (>74 %) based on 1 000 resamplings. Branches that appear in the strict consensus tree are highlighted by thickened lines. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Aspergillus glaucus (culture NRRL 116; GenBank NR_135337) and the species treated here is highlighted with bold face.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Cladophialophora aff. parmeliae (strain KoLRI_053928, GenBank MZ855385.1; Identities = 497/546 (91 %), 12 gaps (2 %)), Cladophialophora chaetospira (strain CBS 114747, GenBank EU035403.1; Identities = 551/607 (91 %), 14 gaps (2 %)), and Capronia rubiginosa (strain BBB 536, GenBank NR_165891.1; Identities = 534/589 (91 %), 17 gaps (2 %)). Closest hits using the LSU sequence were Capronia semiimmersa (strain AFTOL-ID 658, GenBank FJ358226.1; Identities = 773/782 (99 %), no gaps), Phialophora americana (strain MUCL 40613, GenBank AF050280.1; Identities = 773/782 (99 %), no gaps), and Argopericonia indirae (strain GUFCC 18018, GenBank KY977982.1; Identities = 772/782 (99 %), no gaps). Closest hits using the tef1 (first part) sequence had highest similarity to Cladophialophora sp. (strain SYPF 8340, GenBank MF588932.1; Identities = 282/339 (83 %), 17 gaps (5 %)), Exophiala bergeri (strain RBG7236, GenBank OP066900.1; Identities = 252/303 (83 %), 15 gaps (4 %)), and Cladophialophora carrionii (strain CBS 114399, GenBank KJ609515.1; Identities = 228/269 (85 %), 12 gaps (4 %)). Closest hits using the tub2 sequence had highest similarity to Cladophialophora chaetospira (strain CBS 114747, GenBank KF928578.1; Identities = 308/377 (82 %), ten gaps (2 %)), Cladophialophora carrionii (strain CBS 114393, GenBank KF928580.1; Identities = 306/382 (80 %), 21 gaps (5 %)), and Phialophora americana (strain CBS 221.97, GenBank KU306350.1; Identities = 283/355 (80 %), ten gaps (2 %)).
Authors: P.W. Crous, J.Z. Groenewald & J. Boers
Cylindromonium eugeniicola Crous, Persoonia 43: 313. 2019. Fig. 12.
Fig. 12.
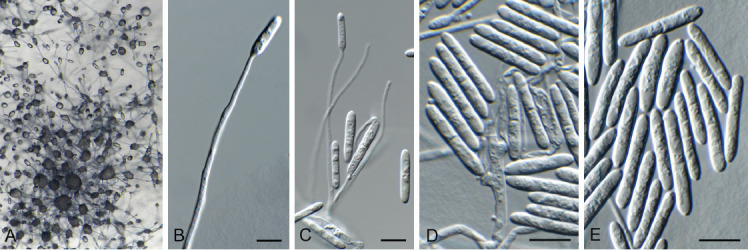
Cylindromonium eugeniicola (CPC 43326). A. Colony on SNA. B–D. Conidiophores with conidiogenous cells giving rise to conidia. E. Conidia. Scale bars = 10 μm.
Description and illustration: Crous et al. (2019c).
Material examined: Spain, Gran Canaria, on dead twig of Eucalyptus sp. (Myrtaceae), 1 Apr. 2022, A.L. van Iperen, HPC 3904, culture CPC 43326 = CBS 149689.
Notes: The hyphomycete genus Cylindromonium (based on Cy. eugeniicola) was described from leaf litter of Eugenia capensis collected in South Africa (Crous et al. 2019c). This is the first record of the fungus occurring on twigs of a Eucalyptus sp. in Spain, which is interesting as both host genera are members of Myrtaceae.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Cylindromonium eugeniicola (strain CPC 37170, GenBank NR_166338.1; Identities = 563/567 (99 %), one gap (0 %)), Cylindromonium lichenicola (strain CBS 188.70, GenBank MH859549.1; Identities = 514/570 (90 %), 14 gaps (2 %)), and Cylindromonium dirinariae (strain FAO006, GenBank LC731277.1; Identities = 513/569 (90 %), 11 gaps (1 %)). Closest hits using the LSU sequence were Cylindromonium eugeniicola (strain CPC 37170, GenBank NG_068337; Identities = 828/830 (99 %), no gaps), Trichonectria setadpressa (voucher A.F.28886, GenBank MT154012; Identities = 819/842 (97 %), no gaps), and Cylindrocladiella lanceolata (strain CBS 129565, GenBank MH876849; Identities = 833/861 (97 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & A.L. van Iperen
Cyphellophora neerlandica Crous & Boers, sp. nov. MycoBank MB 848821. Fig. 13.
Fig. 13.
Cyphellophora neerlandica (CPC 42634). A, B. Conidiogenous cells giving rise to conidia. C. Conidia. Scale bars = 10 μm.
Etymology: Name refers to the Netherlands where it was collected.
Sporulating poorly on SNA. Mycelium consisting of pale brown, smooth, branched, septate, 1.5–2 μm diam hyphae. Conidiophores reduced to conidiogenous cells aggregated in clusters, pale brown, smooth, ampulliform to ellipsoid, phialidic, 4–6 × 3–4 μm, with cylindrical collarette, 1–2 μm long. Conidia solitary, pale brown, smooth, subcylindrical, straight to slightly curved, apex subobtuse, base truncate, 3-septate, (27–)30–33(–36) × 2 μm.
Culture characteristics: Colonies flat, spreading, with moderate aerial mycelium and smooth, lobate margin, reaching 30 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface and reverse iron grey.
Typus: Netherlands, Limburg Province, Eys, brick wall, on lichen, 12 Nov. 2021, J. Boers, HPC 3805 (holotype CBS H-25164, culture ex-type CPC 42634 = CBS 149512); culture CPC 42641.
Cyphellophora deltoidea (Fil. March. et al.) Crous, comb. nov. MycoBank MB 848831.
Basionym: Anthopsis deltoidea Fil. March. et al., Canad. J. Bot. 55: 117. 1977.
Material examined: Italy, Torino, Botanical Garden, from soil, Jun. 1974, A.M. Fontana, holotype CMT 1111.74, ex-type culture CBS 263.77.
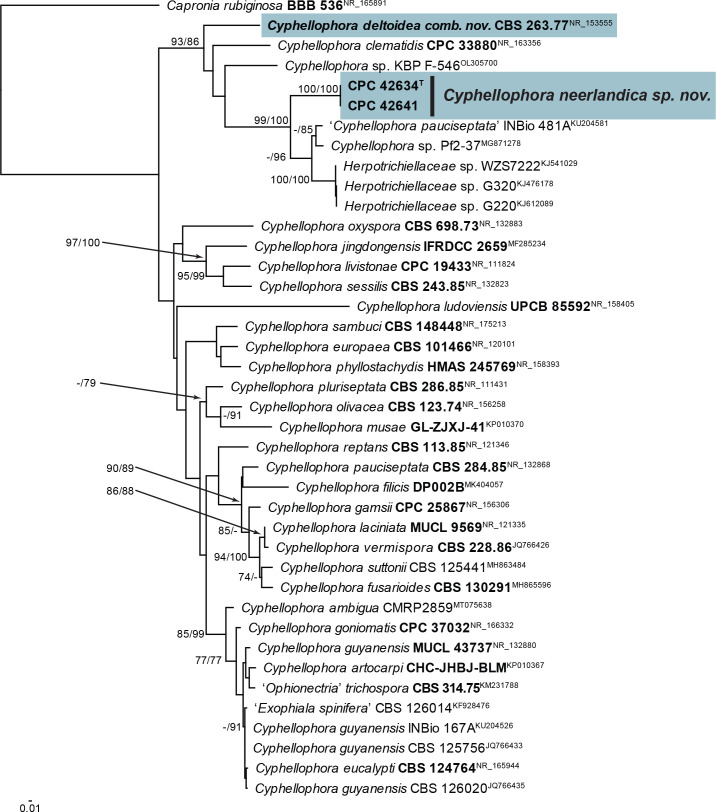
Notes: Cyphellophora neerlandica is related to Cyp. clematidis (conidia aseptate, ellipsoid, (3–)4–5(–6.5) × (1.5–)2(–2.5) μm; Crous et al. 2019b), and Cyp. jingdongensis (only known from its sexual morph; Yang et al. 2018), but is phylogenetically and morphologically distinct. In the phylogenetic tree (Fig. 14), Cyp. neerlandica clusters in a lineage containing sequences from the ex-type cultures of Cyp. clematidis and Anthopsis deltoidea (conidia aseptate, deltoid) as well as several unnamed species. Anthopsis deltoidea is the type species of the genus Anthopsis (Marchisio et al. 1977) and should be reduced to synonymy under Cyphellophora (De Vries 1962), as this clade now contains species with aseptate, as well as septate conidia. Cyphellophora clematidis and Cyp. deltoidea also form a well-supported basal lineage in Cyphellophoraceae in the phylogeny of Quan et al. (2020) (clade 2 in fig. 3). Two additional species of Anthopsis are known, namely A. catenata and A. microspora. Only the former is known from molecular data, and the ITS and LSU sequences of its ex-type culture (GenBank NR_159623 and MH873124) blast distant to Dactylospora, indicating an association with Dactylosporaceae (Sclerococcales, Eurotiomycetes).
Fig. 14.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE v. 2.1.3 of the Anthopsis / Cyphellophora ITS nucleotide alignment. Maximum likelihood (> 74 %) and maximum parsimony (> 74 %) bootstrap support values from 1 000 non-parametric bootstrap replicates are shown at the nodes. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Capronia rubiginosa (culture BBB 536; GenBank NR_165891) and the species treated here is highlighted with bold face. The scale bar indicates the expected number of changes per site.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 42634 had highest similarity to Cyphellophora ‘pauciseptata’ (voucher INBio 481A, GenBank KU204581.1; Identities = 540/601 (90 %), 16 gaps (2 %)), Cyphellophora clematidis (strain CBS 144983, GenBank NR_163356.1; Identities = 469/545 (86 %), 39 gaps (7 %)), and Anthopsis deltoidea (strain CBS 263.77, GenBank NR_153555.1; Identities = 398/452 (88 %), 22 gaps (4 %)). The ITS sequence of CPC 42634 is identical to that of CPC 42641 (591/591 nucleotides). Closest hits using the LSU sequence of CPC 42634 were Cyphellophora clematidis (strain CBS 144983, GenBank NG_068614.1; Identities = 818/850 (96 %), 11 gaps (1 %)), Xenobotrytis acaducospora (strain CBS 219.95, GenBank NG_067437.1; Identities = 816/848 (96 %), eight gaps (0 %)), and Cyphellophora jingdongensis (strain IFRDCC 2659, GenBank MF285236.1; Identities = 796/830 (96 %), ten gaps (1 %)). The LSU sequence of CPC 42634 differs with a single substitution from that of CPC 42641 (814/815 nucleotides). Closest hits using the tub2 sequence of CPC 42634 had highest similarity to Cyphellophora oxyspora (strain CBS 698.73, GenBank KC455232.1; Identities = 309/403 (77 %), 22 gaps (5 %)), and Cyphellophoraceae sp. (strain not specified, GenBank MN913418.1; Identities = 269/338 (80 %), 22 gaps (6 %)). The tub2 sequence of CPC 42634 is identical to that of CPC 42641 (495/495 nucleotides).
Authors: P.W. Crous, J.Z. Groenewald & J. Boers
Didymella brevipilosa Magaña-Dueñas et al., J. Fungi 7: 4. 2021. Fig. 15.
Fig. 15.
Didymella brevipilosa (CPC 41600). A. Conidiomata on SNA. B. Conidiomata on OA. C. Conidioma with ostiole. D, E. Conidiogenous cells. F. Conidia. Scale bars = 10 μm.
Description and illustration: Magaña-Dueñas et al. (2021).
Conidiomata pycnidial, solitary, eustromatica, brown, 200–250 μm diam, with one to several apical ostioles; wall of 3–4 layers of brown textura angularis. Conidiophores reduced to conidiogenous cells, hyaline, smooth, ampulliform, phialidic, 4–6 × 4–5 μm. Conidia solitary, aseptate, fusoid-ellipsoid, apex subobtuse, base truncate, hyaline, smooth, guttulate, (4.5–)5–6(–8) × (2–)2.5–3(–3.5) μm.
Culture characteristics: Colonies flat, spreading, with moderate aerial mycelium and smooth, lobate margin, reaching 45 mm diam after 2 wk at 25 °C. On MEA surface amber, reverse ochreous; on PDA surface and reverse isabelline; on OA surface isabelline.
Material examined: Canada, New Brunswick, Charlotte Co., 1.5 km SW of Little Lepreau, 45.135614° -66.492269°, on buds of Abies balsamea (Pinaceae), 4 May 2021, D. Malloch, HPC 3633 = CBS H-24968, culture CPC 41600 = CBS 149049.
Notes: Didymella brevipilosa was recently described from submerged plant debris collected in freshwater in Spain (conidia aseptate, hyaline, smooth, bacilliform to kidney-shaped, 4–5 × 2–3 μm; Magaña-Dueñas et al. 2021). This is the first record of this fungus from Canada.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Didymella brevipilosa (as Didymella sp.; strain FMR 17415, GenBank OU612373.1; Identities = 498/499 (99 %), no gaps), Didymella americana (strain 8907, GenBank MK646045.1; Identities = 524/531 (99 %), one gap (0 %)), Didymella keratinophila (strain UTHSC DI16-200, GenBank NR_158275.1; Identities = 524/531 (99 %), one gap (0 %)), and Peyronellaea pomorum (strain F115, GenBank KM979827.1; Identities = 522/529 (99 %), one gap (0 %)). Closest hits using the LSU sequence were Didymella brevipilosa (as Didymella sp.; strain FMR 17415, GenBank OU612372.1; Identities = 812/812 (100 %), no gaps), Ascochyta medicaginicola (strain CBS 111.53, GenBank MH868649.1; Identities = 811/812 (99 %), no gaps), Didysimulans mezzanensis (strain MFLUCC 15-0067, GenBank KY496733.1; Identities = 810/812 (99 %), no gaps), and Didysimulans italica (strain MFLUCC 15-0059, GenBank KY496730.1; Identities = 810/812 (99 %), no gaps). Closest hits using the actA sequence had highest similarity to are Didymella finnmarkica (strain CBS 145572, GenBank MK876458.1; Identities = 573/607 (94 %), no gaps), Didymella combreti (strain CBS 137982, GenBank KJ869228.1; Identities = 559/600 (93 %), no gaps), and Didymella rabiei (strain AR628, GenBank KM244530.1; Identities = 559/610 (92 %), five gaps (0 %)). Closest hits using the rpb2 sequence had highest similarity to are Didymella brevipilosa (as Didymella sp.; strain FMR 17415, GenBank OU612359.1; Identities = 482/489 (99 %), no gaps), Didymella aliena (strain JZB380013, GenBank MH645899.1; Identities = 524/575 (91 %), no gaps), Didymella microchlamydospora (strain CBS 140543, GenBank MN983514.1; Identities = 458/504 (91 %), no gaps), and Didymella subrosea (strain CBS 733.79, GenBank MT018174.1; Identities = 457/504 (91 %), no gaps). Closest hits using the tub2 sequence had highest similarity to are Didymella brevipilosa (as Didymella sp.; strain FMR 17415, GenBank no gaps; Identities = 450/454 (99 %), no gaps), Didymella glomerata (strain ATCC MYA-2373, GenBank MZ073910.1; Identities = 447/483 (93 %), four gaps (0 %)) and Didymella combreti (strain CBS 137982, GenBank KJ869246.1; Identities = 448/488 (92 %), six gaps (1 %)).
Authors: P.W. Crous, J.Z. Groenewald & D. Malloch
Drepanopeziza populi-albae (Kleb.) Nannf., Nova Acta R. Soc. Scient. upsal., Ser. 4 8(no. 2): 170. 1932. Fig. 16.
Fig. 16.
Drepanopeziza populi-albae (CPC 42336). A. Colony on MEA. B–G. Conidiophores with conidiogenous cells giving rise to conidia (note germinating conidia in D). Scale bars = 10 μm.
Description and illustration: Spiers (1998), Spiers & Hopcroft (1998).
Typus: Russia, Rostov region, Shakhty, Donetsk, on Populus alba (Salicaceae), 26 Jun. 2021, T.S. Bulgakov, HPC 3708 = CBS H-25157, culture CPC 42336 = CBS 149510.
Notes: Drepanopeziza populi-albae is a common foliar pathogen of Populus alba (Spiers 1998).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Drepanopeziza populi-albae (strain CBS 152.66, GenBank MH858754.1; Identities = 547/548 (99 %), one gap (0 %)), Erysiphe adunca (strain 3_23, GenBank KY660741.1; Identities = 546/548 (99 %), one gap (0 %)), and Drepanopeziza brunnea f. sp. ‘monogermtubi’ (as Marssonina brunnea f. sp. ‘monogermtubi’, strain RBHB1, GenBank KM246347.1; Identities = 532/548 (97 %), one gap (0 %)). Closest hits using the LSU sequence were Drepanopeziza populi-albae (strain CBS 153.66, GenBank MH870387.1; Identities = 836/836 (100 %), no gaps), Drepanopeziza tremulae (strain CBS 408.64, GenBank MH870103.1; Identities = 812/824 (99 %), three gaps (0 %)), and Mastigosporium rubricosum (strain CBS 405.66, GenBank MH870478.1; Identities = 829/845 (98 %), no gaps).
Authors: P.W. Crous & J.Z. Groenewald
Endoconidioma populi Tsuneda et al., Mycologia 96: 1129. 2004. Fig. 17.
Fig. 17.
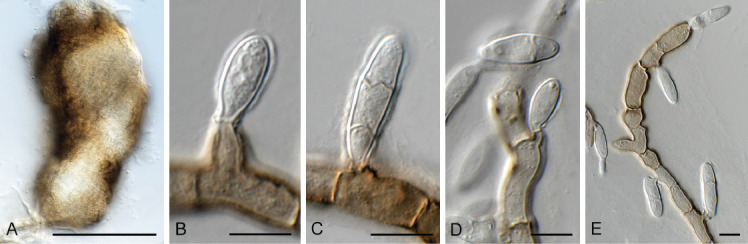
Endoconidioma populi (CPC 41602). A. Sclerotium-like body on SNA. B–E. Conidiogenous cells giving rise to conidia. Scale bars = 10 μm.
Description and illustration: Tsuneda et al. (2004).
On OA forming immersed sclerotium-like structures, 70–250 μm diam, brown, ellipsoid to globose, remaining sterile. On SNA mycelium brown, covered in mucoid layer, roughened, 3–5 μm diam. Conidiophores reduced to solitary or aggregated conidiogenous loci on hyphae, denticulate, 2–3 μm diam, blastic, giving rise to solitary conidia. Conidia fusoid-ellipsoid, apex subobtuse, base truncate, 0(–1)-septate, hyaline, smooth, becoming pale brown, covered in mucoid layer, (13–)16–18(–22) × (5–)6–6.5(–7) μm.
Culture characteristics: Colonies flat, spreading, with sparse aerial mycelium and feathery, lobate margin, reaching 40 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface and reverse iron-grey.
Material examined: Canada, New Brunswick, Charlotte Co., 1.5 km SW of Little Lepreau, 45.135614° -66.492269°, on buds of Abies balsamea (Pinaceae), 4 May 2021, D. Malloch, HPC 3633 = CBS H-24972, culture CPC 41602 = CBS 149070.
Notes: Endoconidioma populi was originally described from twigs of Populus tremuloides collected in Canada (Tsuneda et al. 2004). It produces a yeast-like morph in culture, as well as endoconidia, and a coelomycetous, coniothyrium-like morph (Crous et al. 2020).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to “Hormonema carpetanum” (strain 235J14, GenBank KU516485.1; Identities = 570/570 (100 %), no gaps), Endoconidioma populi (strain IRAN2350C, GenBank KX180155.1; Identities = 563/563 (100 %), no gaps), Endoconidioma leucospermi (as Coniozyma leucospermi; strain CBS 111289, GenBank EU552113.1; Identities = 578/581 (99 %), no gaps), and Endoconidioma populi (strain UAMH 10297, GenBank NR_121303.1; Identities = 553/556 (99 %), no gaps). Closest hits using the LSU sequence were “Hormonema carpetanum” (strain ATCC 74360, GenBank MF611880.1; Identities = 826/830 (99 %), no gaps), Endoconidioma leucospermi (as Coniozyma leucospermi; strain CBS 111289, GenBank EU552113.1; Identities = 825/830 (99 %), no gaps), Endoconidioma euphorbiae (strain CPC 38583, GenBank MW175391.1; Identities = 823/830 (99 %), no gaps), and Endoconidioma populi (strain UAMH 10297, GenBank NG_059198.1; Identities = 823/830 (99 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & D. Malloch
Fusariella atrovirens (Berk.) Sacc., Atti dell′Istituto Veneto Scienze 2: 463. 1884. Fig. 18.
Fig. 18.
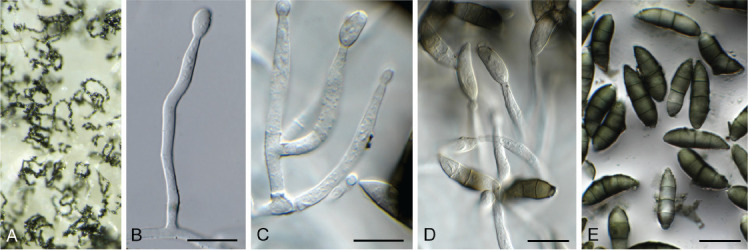
Fusariella atrovirens (CPC 43304). A. Colony on OA. B–D. Conidiogenous cells giving rise to conidia. E. Conidia. Scale bars = 10 μm.
Description and illustration: Lin et al. (2016).
Material examined: Namibia, Gobabeb Namib Research Institute, salt pan, lichenicolous on unknown lichen growing on rock, 4 Apr. 2022, P.W. Crous, HPC 3888, culture CPC 43304 = CBS 149690.
Notes: Fusariella atrovirens is cosmopolitan, occurring on various plant hosts, leaf litter, dung and in soil (Seifert et al. 2011). It is here recorded as lichenicolous, growing on an unknown lichen in Namibia.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Fusariella atrovirens (strain CBS 311.73, GenBank MH860688.1; Identities = 536/536 (100 %), no gaps), Hydropisphaera erubescens (strain I-10, GenBank KF813068.1; Identities = 455/456 (99 %), no gaps), and Fusariella sinensis (strain OUCMBI110148, GenBank KP269058.1; Identities = 533/536 (99 %), no gaps). Closest hits using the LSU sequence were Fusariella bizzozeriana (strain CBS 306.73, GenBank MH878365.1; Identities = 810/810 (100 %), no gaps), Fusariella concinna (strain CBS 312.73, GenBank MH878376.1; Identities = 853/857 (99 %), no gaps), and Fusariella atrovirens (strain CBS 311.73, GenBank MH872395.1; Identities = 841/845 (99 %), two gaps (0 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Fusariella atrovirens (strain AK18-21, GenBank OX335003.1; Identities = 857/868 (99 %), no gaps), Hydropisphaera peziza (strain CBS 102038, GenBank DQ522444.1; Identities = 716/838 (85 %), two gaps (0 %)), and Heleococcum aurantiacum (strain CBS 201.35, GenBank JX158463.1; Identities = 734/863 (85 %), two gaps (0 %)). Closest hits using the tef1 (second part) sequence had highest similarity to Fusariella sp. (strain MFLUCC 15-0844, GenBank KX025155.1; Identities = 899/915 (98 %), no gaps), Hydropisphaera erubescens (strain AFTOL-ID 186, GenBank DQ518174.1; Identities = 867/916 (95 %), no gaps), and Heleococcum japonense (strain CBS 397.67, GenBank JX158398.1; Identities = 859/916 (94 %), no gaps). No tef1 sequences of Fusariella atrovirens are available for comparison.
Authors: P.W. Crous & J.Z. Groenewald
Fusariella hughesii Chab.-Frydm., Canad. J. Bot. 42: 1485. 1964. Fig. 19.
Fig. 19.
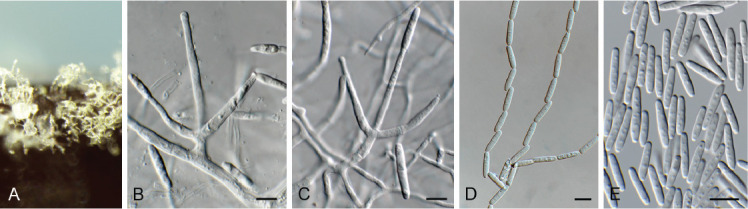
Fusariella hughesii (CPC 41594). A. Sporulation on PNA. B, C. Conidiophores and conidiogenous cells giving rise to conidia. D, E. Conidia. Scale bars = 10 μm.
Description and illustration: Lin et al. (2016).
Mycelium consisting of hyaline, smooth, branched, septate, 2.5–3 μm diam hyphae, forming hyphal swellings (up to 10 μm diam) in older hyphae on MEA and PDA. Conidiophores solitary or aggregated, erect, arising from superficial hyphae, subcylindrical, branched, up to 5-septate, 100 μm tall. Conidiogenous cells terminal and intercalary, subcylindrical with apical taper, hyaline, smooth, 20–35 × 2.5–3 μm, phialidic, giving rise to basipetal conidial chains. Conidia subcylindrical, 3-septate, guttulate, hyaline to pale greenish, smooth, apex obtuse, base obconically truncate, hilum 2–2.5 μm diam, straight, occurring in long, unbranched chains, (8–)16–19(–22) × (2.5–)3(–3.5) μm.
Culture characteristics: Colonies erumpent, spreading, surface folded, with sparse aerial mycelium and smooth, lobate margin, reaching 20 mm diam after 2 wk at 25 °C. On MEA surface ochreous, reverse umber; on PDA surface and reverse pale luteous; on OA surface pale luteous.
Material examined: Ukraine, Dvorichna district, Kharkiv region, Krasne Pershe village, National Park Dvorichanskyi, on overwintered stems of Adonis vernalis (Ranunculaceae), 11 Apr. 2021, A. Akulov, CWU (MYC) AS 8121 = HPC 3630 = CBS H-24976, culture CPC 41594 = CBS 149074.
Notes: Fusariella hughesii, which was originally isolated from seeds of Trigonella arabica and Phalaris minor in Israel (Lin et al. 2016), is reported here from dead stems of Adonis vernalis in Ukraine.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Fusariella hughesii (strain CBS 435.70, GenBank MH859784.1; Identities = 569/576 (99 %), no gaps), Fusariella sinensis (strain OUCMBI110131, GenBank KP269041.1; Identities = 545/569 (96 %), four gaps (0 %)), and Fusariella atrovirens (strain CBS 311.73, GenBank MH860688.1; Identities = 554/579 (96 %), four gaps (0 %)).Closest hits using the LSU sequence were Fusariella hughesii (strain CBS 435.70, GenBank MH871547.1; Identities = 722/724 (99 %), no gaps), Hydropisphaera erubescens (strain CBS 128364, GenBank MH876356.1; Identities = 721/724 (99 %), no gaps), and Fusariella concinna (strain CBS 312.73, GenBank MH878376.1; Identities = 720/724 (99 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & A. Akulov
Geonectria muralis Crous & Boers, sp. nov. MycoBank MB 848824. Fig. 20.
Fig. 20.
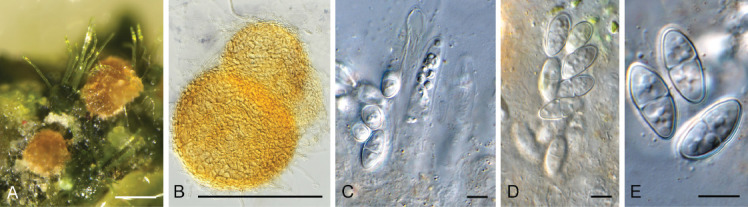
Geonectria muralis (CPC 42404). A. Ascomata in vivo. B. Ascomata in vitro. C, D. Asci with ascospores. E. Ascospores. Scale bars: A, B = 180 μm, all others = 10 μm.
Etymology: Name refers to the phycoparasitic habit of the fungus on an old church wall.
Perithecia globose, 150–180 μm diam, orange, arising from substratal hyphae, not changing colour in KOH, with central ostiole, 15 μm diam; wall 15–18 μm thick, of 3–4 layers of textura angularis; outer wall smooth, with hyphal outgrowths; hyphae smooth, branched, septate, 3–4 μm diam. Asci 8-spored, stipitate, subcylindrical, unitunicate with apical mechanism, 40–65 × 9–11 μm. Ascospores bi- to triseriate, hyaline, becoming pale brown with age, smooth, guttulate, broadly ellipsoid, medianly 1-septate, constricted at septum, (12–)14–16(–17) × (5–)8–9 μm. Asexual morph not seen.
Culture characteristics: Colonies flat, spreading, with sparse to moderate aerial mycelium and smooth, lobate margin, reaching 35 mm diam after 2 wk at 25 °C. On MEA and PDA surface and reverse sienna; on OA surface apricot. Homothallic, with perithecia also forming in culture.
Typus: Netherlands, Gelderland Province, Dodewaard, church, on algae growing on the bottom part of wall, 7 Sep. 2021, J. Boers, HPC 3749 (holotype CBS H-25161, culture ex-type CPC 42404 = CBS 149515); cultures CPC 42405, 42406.
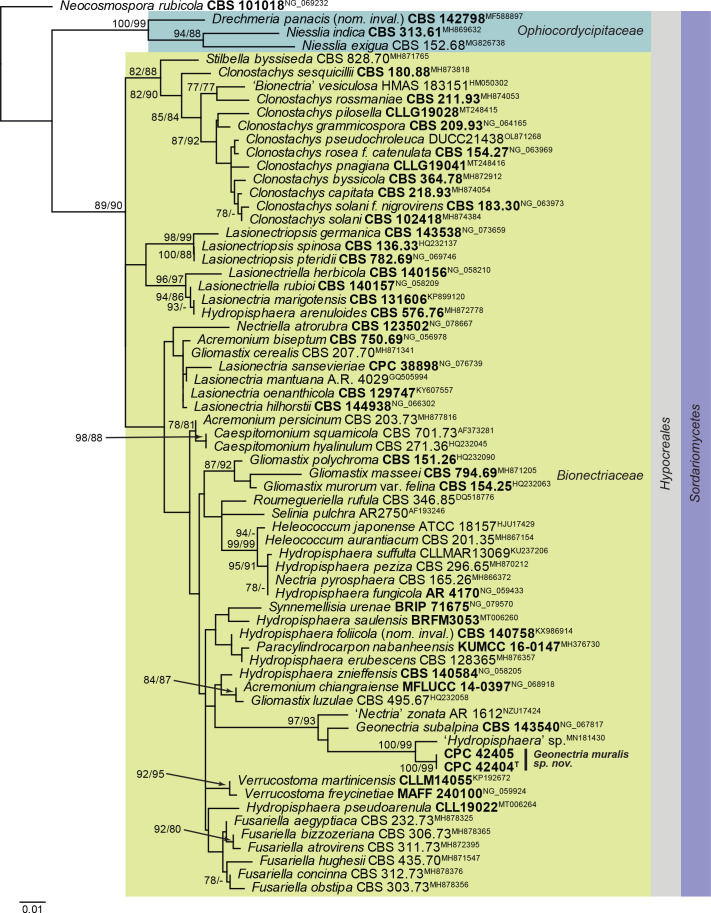
Notes: Geonectria muralis is related to Nectria pyrosphaera (CBS 165.26), Nectria zonata (AR 1612) and Geonectria subalpina (CBS 143540). In the phylogenetic tree (Fig. 21), it clustered with 97 % ML bootstrap support and 93 % parsimony bootstrap support with ‘Nectria zonata’, a ‘Hydropisphaera sp.’ and the LSU sequence of the ex-type strain of Geonectria subalpina, the type species of the monotypic genus Geonectria. Geonectria, based on G. subalpina, was described from bare soil collected in the subalpine region, and is characterised by orange perithecia that do not change colour in 3 % KOH, striate, 1-septate, hyaline, finely striate ascospores, and an acremonium-like asexual morph (Lechat et al. 2018).
Fig. 21.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE v. 2.1.3 of the Hypocreales LSU nucleotide alignment. Maximum likelihood (> 74 %) and maximum parsimony (> 74 %) bootstrap support values from 1 000 non-parametric bootstrap replicates are shown at the nodes. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Neocosmospora rubicola (culture CBS 101018; GenBank NG_069232) and the species treated here is highlighted with bold face. The families, order and class are shown in coloured blocks to the right of the tree. The scale bar indicates the expected number of changes per site.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 42404 had highest similarity to Hydropisphaera sp. BGL-2019a (voucher BDNA-L-0000095, GenBank MN187059.1; Identities = 501/554 (90 %), 28 gaps (5 %)), Nectria pyrosphaera (strain CBS 165.26, GenBank MH854877.1; Identities = 476/569 (84 %), 41 gaps (7 %)), and Heleococcum aurantiacum (strain CBS 201.35, GenBank MH855645.1; Identities = 472/568 (83 %), 36 gaps (6 %)). The ITS sequence of CPC 42404 is identical to those of CPC 42405 and CPC 42406 (both 552/552 nucleotides). Closest hits using the LSU sequence of CPC 42404 were Hydropisphaera sp. BGL-2019a (voucher BDNA-L-0000095, GenBank MN181431.1; Identities = 828/842 (98 %), three gaps (0 %)), Geonectria subalpina (strain CBS 143540, GenBank NG_067817.1; Identities = 808/842 (96 %), four gaps (0 %)), and Nectria zonata (strain AR 1612, GenBank U17424.1; Identities = 809/845 (96 %), 11 gaps (1 %)). The LSU sequence of CPC 42404 differs from that of CPC 42405 at three nucleotide positions (796/799 nucleotides, all in repeat motifs).
Authors: P.W. Crous, J.Z. Groenewald & J. Boers
Harposporium illinoisense Crous & Jurjević, sp. nov. MycoBank MB 848825. Fig. 22.
Fig. 22.
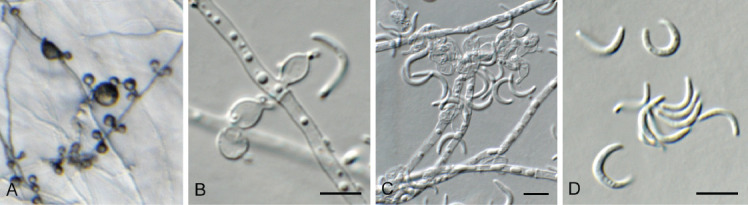
Harposporium illinoisense (CPC 42872). A. Colony on SNA. B, C. Conidiogenous cells giving rise to conidia. D. Conidia. Scale bars = 10 μm.
Etymology: Name refers to Illinois, the state in the USA where it was isolated.
Mycelium consisting of hyaline, smooth, branched, septate, 2–3 μm diam hyphae, constricted at septa with age. Conidiophores reduced to conidiogenous cells; phialides solitary on hyphae, or in clusters, ampulliform, 5–7 × 2.5–3 μm, with cylindrical necks, 1.5–2 × 1 μm. Conidia solitary, aggregating in mucoid mass, hyaline, smooth, aseptate, sickle-shaped, widest in middle, apex subobtuse, base truncate, 6–10 × 1–2 μm.
Culture characteristics: Colonies erumpent, spreading, with moderate aerial mycelium and feathery, lobate margin, reaching 20 mm diam after 2 wk at 25 °C. On MEA surface amber, reverse sienna; on PDA surface amber, reverse sienna; Czapek Yeast Autolysate Agar (CYA) surface amber, reverse sienna; on OA surface hazel. It also shows antibacterial properties on CYA and MEA. No growth at 37 °C, on CYA.
Typus: USA, Illinois, marijuana greenhouse, Rockwool, Oct. 2021, Z. Jurjević 5670 (holotype CBS H-25170, culture ex-type CPC 42872 = CBS 149456).
Notes: Species of Harposporium parasitise free-living nematodes and rotifers. Most species of Harposporium infect nematodes via ingested conidia (Glockling 1998). Harposporium illinoisense was isolated from rockwool in a greenhouse, and was probably was associated with nematodes. Phylogenetically, H. illinoisense is quite distinct from other presently known from DNA data (Fig. 23) and also the blast results (see below) are insufficiently conclusive to provide a proper placement for this species.
Fig. 23.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE v. 2.1.3 of the Ophiocordycipitaceae LSU nucleotide alignment. Bootstrap support values (> 75 %) from 1 000 non-parametric bootstrap replicates are shown at the nodes. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Falcocladium eucalypti (culture CPC 38019; GenBank NG_068318) and the species treated here is highlighted with bold face. The family, order and class are shown in coloured blocks to the right of the tree. The scale bar indicates the expected number of changes per site.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Purpureocillium takamizusanense (strain PT3, GenBank CP086360.1; Identities = 476/554 (86 %), 46 gaps (8 %)), Harposporium helicoides (strain CBS 944.70, GenBank MH860014.1; Identities = 490/558 (88 %), 29 gaps (5 %)), and Drechmeria balanoides (as Haptocillium balanoides, strain Harp5, GenBank EF546660.1; Identities = 475/540 (88 %), 27 gaps (5 %)). Closest hits using the LSU sequence were Harposporium harposporiferum (strain CBS 213.86, GenBank MH873635.1; Identities = 814/836 (97 %), three gaps (0 %)), Drechmeria zeospora (strain CBS 335.80, GenBank NG_063928.1; Identities = 787/809 (97 %), two gaps (0 %)), and Drechmeria panacis (nom. inval.) (strain SYPF 8335, GenBank MF588897.1; Identities = 810/833 (97 %), two gaps (0 %)). Closest hits using the actA sequence had highest similarity to Purpureocillium lilacinum (strain PLFJ-1, GenBank XM_018325012.1; Identities = 314/329 (95 %), no gaps), and Purpureocillium takamizusanense (strain PT3, GenBank XM_047986424.1; Identities = 317/338 (94 %), one gap (0 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Isaria takamizusanensis (strain NHJ 3497, GenBank EU369074.1; Identities = 697/788 (88 %), no gaps), Purpureocillium lilacinum (strain G406, GenBank KJ443162.1; Identities = 717/813 (88 %), no gaps), and Cordyceps gunnii (strain OSC 76404, GenBank DQ522426.1; Identities = 674/767 (88 %), no gaps). Closest hits using the tef1 (second part) sequence had highest similarity to Ophiocordyceps myrmicarum (strain CG1361, GenBank MG922555.1; Identities = 827/877 (94 %), no gaps), Hirsutella citriformis (voucher ARSEF 1446, GenBank KM651990.1; Identities = 840/892 (94 %), no gaps), and Ophiocordyceps delicatula (strain ARSEF 14442, GenBank MZ246828.1; Identities = 840/892 (94 %), no gaps). Closest hits using the tub2 sequence had highest similarity to Sarocladium kiliense (strain CBS 400.52, GenBank KM232119.1; Identities = 321/418 (77 %), 25 gaps (5 %)), Sarocladium junci (strain CPC 41107, GenBank OK651210.1; Identities = 317/407 (78 %), 26 gaps (6 %)), and Paecilomyces wawuensis (strain GZU-BCECYN4-3, GenBank JQ965117.1; Identities = 317/408 (78 %), 25 gaps (6 %)).
Authors: P.W. Crous, J.Z. Groenewald & Z. Jurjević
Hysterobrevium rosae Jayasiri et al., Mycosphere 9: 818. 2018.
Description and illustration: Jayasiri et al. (2018).
Material examined: Netherlands, Utrecht Province, Nieuw Wulven, near Houten, 1.5 m a.s.l., 52°02’53’N, 05°10’09’’E, on bamboo stick (used as support post, not locally grown), 11 Feb. 2022, E.R. Osieck, HPC 3837 = WI-48/ #4404, culture CPC 42948 = CBS 149699.
Notes: The genus Hysterobrevium is characterised by hysteriaceous ascomata and muriform spores, which are less than 25 μm in length (Boehm et al. 2009). Hysterobrevium rosae is morphologically similar to H. mori (Boehm loc. cit.), but distinguished in having smaller hysterothecia, shorter asci and hyaline ascospores (Jayasiri et al. 2018).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Hysterobrevium mori (strain A1045A, GenBank MT230464.1; Identities = 710/710 (100 %), no gaps), Hysterobrevium constrictum (strain JCM 2753, GenBank NR_175063.1; Identities = 676/716 (94 %), four gaps (0 %)), and Hysterobrevium sp. MR-2017a (strain MFLUCC 16-2163, GenBank MZ467050.1; Identities = 393/422 (93 %), six gaps (1 %)). No ITS sequence of Hysterobrevium rosae was available for comparison. Closest hits using the LSU sequence were Hysterobrevium rosae (strain MFUCC 14-0551, GenBank MH535897.1; Identities = 844/844 (100 %), no gaps), Hysterobrevium mori (strain CBS 123335, GenBank FJ161202.2; Identities = 866/869 (99 %), no gaps), and Gloniopsis praelonga (strain SMH5280, GenBank GQ221912.2; Identities = 865/869 (99 %), no gaps). Closest hits using the rpb2 (first part) sequence had highest similarity to Hysterobrevium smilacis (as Gloniopsis smilacis, strain CBS 114601, GenBank FJ161114.1; Identities = 833/882 (94 %), no gaps), and Hysterobrevium constrictum (strain HKAS 121127, GenBank OK506220.1; Identities = 808/876 (92 %), no gaps). Closest hits using the tef1 (second part) sequence had highest similarity to Hysterobrevium rosae (strain MFUCC 14-0551, GenBank MH535879.1; Identities = 733/733 (100 %), no gaps), Hysterobrevium mori (strain EB 0315, GenBank FJ161106.1; Identities = 745/771 (97 %), no gaps), and Hysterobrevium smilacis (strain CBS 114601, GenBank FJ161091.1; Identities = 728/763 (95 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & E.R. Osieck
Microcera physciae Crous & Boers, Persoonia 47: 233. 2021. Fig. 24.
Description and illustration: Crous et al. (2021b).
Material examined: Netherlands, Limburg Province, Eys, brick wall, on lichen, 12 Nov. 2021, J. Boers, HPC 3805, culture CPC 42638.
Notes: Microcera physciae is a lichenicolous fungus described from Physcia tenella in the Netherlands (Crous et al. 2021b), while M. lichenicola was described from Parmelia sulcata, also in the Netherlands (Crous et al. 2022a).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Microcera lichenicola (strain CBS 149169, GenBank ON811502.1; Identities = 530/530 (100 %), no gaps), Microcera physciae (strain CBS 148283, GenBank NR_175225.1; Identities = 496/497 (99 %), no gaps), and Microcera larvarum (strain CBS 738.79, GenBank KM231825.1; Identities = 519/531 (98 %), three gaps (0 %)). Closest hits using the LSU sequence were Microcera physciae (strain CBS 148283, GenBank NG_081335.1; Identities = 814/814 (100 %), no gaps), Microcera lichenicola (strain CBS 149169, GenBank ON811561.1; Identities = 814/815 (99 %), no gaps), and Microcera rubra (strain CBS 638.76, GenBank NG_058100.1; Identities = 805/816 (99 %), no gaps). Closest hits using the rpb1 sequence had highest similarity to Microcera physciae (strain CPC 41284, GenBank OK651153.1; Identities = 717/720 (99 %), two gaps (0 %)), Microcera lichenicola (strain CBS 148313, GenBank ON803533.1; Identities = 690/720 (96 %), two gaps (0 %)), and Microcera coccophila (strain MAFF 241482, GenBank KC291895.1; Identities = 629/689 (91 %), two gaps (0 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Microcera physciae (strain CPC 41284, GenBank OK651168.1; Identities = 871/874 (99 %), no gaps), Microcera lichenicola (strain CBS 148313, GenBank ON803543.1; Identities = 840/876 (96 %), one gap (0 %)), and Microcera larvarum (strain NRRL 20473, GenBank JX171587.1; Identities = 766/850 (90 %), one gap (0 %)). Closest hits using the tef1 (first part) sequence had highest similarity to Microcera physciae (strain CPC 41284, GenBank OK651190.1; Identities = 460/470 (98 %), two gaps (0 %)), Microcera lichenicola (strain CBS 148313, GenBank ON803570.1; Identities = 449/483 (93 %), six gaps (1 %)), and Microcera larvarum (strain CBS 738.79, GenBank KM231957.1; Identities = 375/470 (80 %), 24 gaps (5 %)). Closest hits using the tub2 sequence had highest similarity to Microcera physciae (strain CPC 41284, GenBank OK651208.1; Identities = 526/526 (100 %), no gaps), Microcera lichenicola (strain CBS 149169, GenBank ON803591.1; Identities = 524/540 (97 %), two gaps (0 %)), and Microcera larvarum (strain CBS 169.30, GenBank AB587032.1; Identities = 274/304 (90 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & J. Boers
Miricatena prunicola Punith. & Spooner, Kew Bull. 66: 638. 2011. Fig. 25.
Fig. 25.

Miricatena prunicola (CPC 42627). A, B. Leaf spots. C. Conidiogenous cells giving rise to secondary conidia. D. Secondary conidia. E, F. Primary conidia. G, H. Primary conidia giving rise to secondary conidia. Scale bars -= 10 μm.
Classification: Leotiomycetes, Helotiales, incertae sedis.
Description and illustration: Punithalingam & Spooner (2011).
The compound conidia and conidiogenesis of M. prunicola was discussed and fully described by Punithalingham & Spooner (2011). In this paper they comment on the terminal cells being ampulliform to lageniform to obpyriform. What they did not observe, however, is that these terminal cells become fertile, acting as conidiogenous cells, or give rise to 2–4 smaller ampulliform phialides, 5–10 × 3–4.5 μm, that again give rise to solitary conidia, ellipsoid, aseptate, hyaline, smooth, guttulate, 3–4 × 2.5–3 μm, aggregating in a mucoid conidial mass.
Culture characteristics: Colonies erumpent, spreading, with sparse to moderate aerial mycelium and even, lobate margin, reaching 35 mm diam after 2 wk at 25 °C. On malt extract agar (MEA) surface and reverse isabelline; on potato dextrose agar (PDA) surface cinnamon, reverse isabelline; on oatmeal agar (OA) surface sienna to orange.
Typus: Netherlands, Gelderland Province, Ede, Kreelsche Zand, on leaves of Prunus serotina (Rosaceae), 7 Nov. 2021, E. Slootweg, HPC 3800 (epitype designated here CBS H-25163, MBT 10013416, culture ex-epitype CPC 42627 = CBS 149448). UK, Surrey, Frensham, Little Pond (near), on living leaves of Prunus serotina, 8 Jul. 2007, B.M. Spooner (holotype K(M) 155328).
Notes: The monotypic genus Miricatena was introduced for M. prunicola, a foliar pathogen of Prunus serotina in the UK (Punithalingam & Spooner 2011). Due to the lack of cultures and DNA data, the phylogenetic position of Miricatena has remained unknown. The present record of M. prunicola from the Netherlands made it possible to designate an epitype for this pathogen, and also resolve its placement in the Helotiales, Leotiomycetes based on the high similarity of the ITS and LSU sequences to species of Cadophora and Pyrenopeziza.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Cadophora prunicola (voucher JKI-Cad64, GenBank MW960107.1; Identities = 490/547 (90 %), 16 gaps (2 %)), Cadophora malorum (strain CBS 101359, GenBank DQ404350.1; Identities = 488/548 (89 %), 17 gaps (3 %)), and Cadophora orientoamericana (voucher JKI-Cad62, GenBank MW804716.1; Identities = 417/448 (93 %), six gaps (1 %)). Closest hits using the LSU sequence were Cadophora luteo-olivacea (strain CBS 128576, GenBank MH876422.1; Identities = 836/850 (98 %), no gaps), Cadophora fastigiata (strain CBS 872.69, GenBank MH871250.1; Identities = 836/850 (98 %), no gaps), and Pyrenopeziza lonicerae (strain CBS 332.58, GenBank MH869339.1; Identities = 836/850 (98 %), no gaps). No significant hits were obtained using the tef1 (first part) sequence.
Authors: P.W. Crous, J.Z. Groenewald & E. Slootweg
Neoleptodontidium Crous & Jurjević, gen. nov. MycoBank MB 848826.
Etymology: Name refers to its morphological similarity to Leptodontidium.
Classification: Sordariomycetes, Xylariomycetidae, Xylariales, incertae sedis.
Mycelium consisting of hyaline, smooth, branched, septate hyphae. Conidiophores solitary, erect, subcylindrical, medium brown, thick-walled, lower part finely roughened, septate, frequently rejuvenating through terminal phialide, forming a new phialide above the older phialide, where a rosette of conidia remains attached in a mucoid mass. Conidiogenous cells terminal, subcylindrical, medium brown, somewhat paler to the rest of the conidiophore, smooth, terminal phialidic opening with flared collarette, at times also with lateral phialidic openings on conidiogenous cell. Conidia solitary, hyaline, smooth, guttulate, aseptate, subcylindrical, apex obtuse, straight to slightly curved, tapering to subobtuse hilum, aggregating in mucoid mass.
Type species: Neoleptodontidium aquaticum Crous & Jurjević
Neoleptodontidium aquaticum Crous & Jurjević, sp. nov. MycoBank MB 848827. Fig. 26.
Fig. 26.
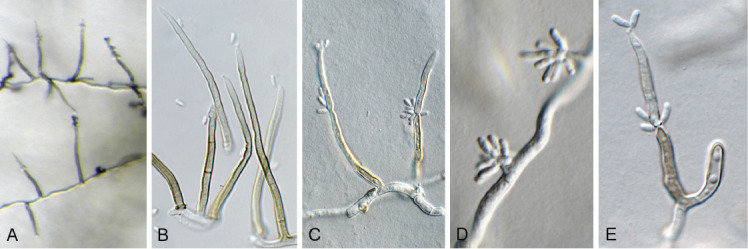
Neoleptodontidium aquaticum (CPC 42868). A. Colony on SNA. B–E. Conidiophores and conidiogenous cells giving rise to conidia. Scale bars = 10 μm.
Etymology: Name refers to the fact that it was isolated from water.
Mycelium consisting of hyaline, smooth, branched, septate, 2.5–3 μm diam hyphae. Conidiophores solitary, erect, subcylindrical, medium brown, thick-walled, lower part finely roughened, 1–3-septate, frequently rejuvenating through terminal phialide, forming a new phialide above the older phialide, where a rosette of conidia remains attached in a mucoid mass, 30–60 × 2.5–3.5 μm. Conidiogenous cells terminal, subcylindrical, medium brown, somewhat paler to the rest of the conidiophore, smooth, terminal phialidic opening with flared collarette, 1.5–2 μm diam, at times also with lateral phialidic openings on conidiogenous cell, 10–30 × 2–2.5 μm. Conidia solitary, hyaline, smooth, guttulate, aseptate, subcylindrical, apex obtuse, straight to slightly curved, tapering to subobtuse hilum, 3–4 × 1.5 μm, aggregating in mucoid mass.
Culture characteristics: Colonies flat, spreading, with sparse to moderate aerial mycelium and feathery, lobate margin, reaching 25 mm diam after 2 wk at 25 °C. On MEA surface isabelline, reverse honey; on PDA surface and reverse honey; on OA surface olivaceous grey.
Typus: USA, North Carolina, Durham, greenhouse, hydroponic water, Oct. 2021, Z. Jurjević 5663 (holotype CBS H-25168, culture ex-type CPC 42868 = CBS 149455).
Additional isolate examined: USA, greenhouse, peet, Z. Jurjević 5683, culture CPC 42875.
Neoleptodontidium aciculare (V. Rao & de Hoog) Crous, comb. nov. MycoBank MB 848828.
Basionym. Leptodontidium aciculare V. Rao & de Hoog, Stud. Mycol. 28: 37. 1986
Typus: India, Karnataka, Dt. Bidar, Maniknagar, on rotten wood, Jan. 1984, V. Rao, culture ex-type CBS 123.86.
Notes: Neoleptodontidium is reminiscent of the genus Leptodontidium (based on L. trabinellum), in having erect conidiophores and conidiogenous cells with a long rachis with denticles (Hernández-Restrepo et al. 2017). However, Neoleptodontidium is distinct in that it forms minute, terminal and lateral exophiala-like phialides. Phylogenetically it is allied to Leptodontidium aciculare, which also forms similar phialides (Rao & De Hoog 1986), and is therefore also placed in Neoleptodontidium (Fig. 27). Neoleptodontidium clusters in the Xylariales, but with unclear familial association; the closest families are Oxydothidaceae, Castanediellaceae and Barrmaeliaceae.
Fig. 27.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE v. 2.1.3 of the Xylariales LSU nucleotide alignment. Maximum likelihood (> 74 %) bootstrap support values from 1 000 non-parametric bootstrap replicates are shown at the nodes. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Ramularia endophylla (culture CBS 113265; GenBank AY490776) and the species treated here are highlighted with bold face. The families, orders and classes are shown in coloured blocks to the right of the tree. The scale bar indicates the expected number of changes per site.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 42868 had highest similarity to Leptodontidium aciculare (strain CBS 123.86, GenBank MH861931.1; Identities = 498/513 (97 %), one gap (0 %)), Castanediella cagnizarii (strain CBS 542.96, GenBank MH862597.1; Identities = 376/418 (90 %), nine gaps (2 %)), and Linteromyces quintiniae (strain CBS 146792, GenBank NR_171989.1; Identities = 389/433 (90 %), 11 gaps (2 %)). The ITS sequences of CPC 42868 and 42875 are identical (510/510 nucleotides). Closest hits using the LSU sequence of CPC 42868 were Leptodontidium aciculare (strain CBS 123.86, GenBank MH873620.1; Identities = 817/824 (99 %), no gaps), Entosordaria quercina (strain RQ, GenBank MF488994.1; Identities = 794/826 (96 %), two gaps (0 %)), and Entosordaria perfidiosa (strain EPE, GenBank MF488993.1; Identities = 791/826 (96 %), two gaps (0 %)). The LSU sequences of CPC 42868 and 42875 are identical (822/822 nucleotides).
Authors: P.W. Crous, J.Z. Groenewald, Z. Jurjević & A. Erhard
Nothoramularia Crous, J. Kruse & U. Braun, gen. nov. MycoBank MB 848829.
Etymology: Name refers to the fact that it resembles Ramularia in morphology, but is phylogenetically distinct from that genus.
Classification: Lecanoromycetes, Acarosporomycetidae, Acarosporales, Acarosporaceae.
Colonies hyperparasitic on colonies caused by the leaf-spotting Ragnhildiana ferruginea. Mycelium hyaline, composed of branched, hyaline, septate, smooth hyphae. Conidiophores solitary, arising from hyphae, lateral, occasionally terminal, erect, conidiophores usually aseptate, i.e., reduced to conidiogenous cells, rarely with a single septum, subcylindrical to usually conical, straight to somewhat geniculate-sinuous caused by sympodial proliferation, unbranched, hyaline, thin-walled, with a single or 2–3 minute conidiogenous loci, sometimes almost denticle-like, slightly thickened and darkened, or conidia arising from minute peg-like protuberances of the hyphae, somewhat attenuated towards a truncated tip. Conidia solitary or usually in simple or branched acropetal chains, ellipsoid-ovoid, fusiform, subcylindrical, usually straight, 0–1-septate, thin-walled, hyaline, smooth or almost so to finely rough-walled, ends rounded to attenuated, with a single basal hilum and 1–2 hila at the apex, minute, barely thickened and darkened (adapted from Braun & Kruse 2021).
Type species: Nothoramularia ragnhildianicola (J. Kruse & U. Braun) Crous, J. Kruse & U. Braun
Nothoramularia ragnhildianicola (J. Kruse & U. Braun) Crous, J. Kruse & U. Braun, comb. nov. MB 848830. Fig. 28.
Fig. 28.
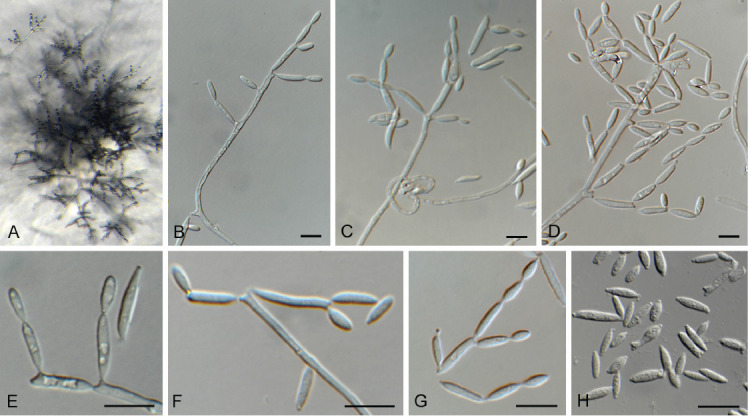
Nothoramularia ragnhildianicola (CPC 42462). A. Colony on SNA. B–D, F. Conidiophores and conidiogenous cells giving rise to conidia. E, G, H. Conidia. Scale bars = 10 μm.
Basionym: Ramularia ragnhildianicola J. Kruse & U. Braun, Schlechtendalia 38: 165. 2021.
Description and illustration: Braun & Kruse (2021).
Culture characteristics: Colonies erumpent, spreading, with moderate aerial mycelium and feathery, lobate margin, reaching 6 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface dirty white, reverse luteous with patches of ochreous.
Typus: Germany, Rheinland-Pfalz, Landkreis Bad Dürkheim, Birkenheide, 1.6 km northwest of Eyersheimer Hof, meadow, 49°29’41’N, 8°15’29’’E, ca. 105 m alt, on Ragnhildiana ferruginea (Mycosphaerellaceae) parasitic on Artemisia vulgaris (Asteraceae), 8 Sep. 2020, J. Kruse (holotype POLL 9793), isotype HAL 3375 F; Rheinland-Pfalz, Landkreis Bad Dürkheim, Birkenheide, 49°29’41’’N, 8°15’29’’E, on Ragnhildiana ferruginea, parasitic on Artemisia vulgaris, 7 Sep. 2021, J. Kruse, POLL 9802 = HPC 3779 (epitype designated here CBS H-24978, MBT 10013418, culture ex-epitype CPC 42462 = CBS 149076).
Additional material examined: Germany, Rheinland-Pfalz, Landkreis Bad Dürkheim, Birkenheide, 49°29’41’’N, 8°15’29’’E, on Ragnhildiana ferruginea, parasitic on Artemisia vulgaris, 7 Sep. 2021, J. Kruse, POLL 9802 = HPC 3779, culture CPC 42463 = CBS 149075.
Notes: In culture, conidial loci are somewhat thickened, darkened, but not refractive. Ramoconidia commonly have numerous loci, giving rise to branched conidial chains. Conidiophores are erect, subcylindrical, straight, with terminal and intercalary conidiogenous cells, up to 150 μm tall, or reduced to conidiogenous loci on hyphae. Phylogenetically (Fig. 29), No. ragnhildianicola forms a lineage basal to Neoacrodontiella, Neospermospora and Cytosporella, with which it clusters with moderate support (87 % maximum parsimony and 74 % maximum likelihood).
Fig. 29.
The first of two equally most parsimonious trees obtained from the Lecanoromycetes LSU sequence alignment. The scale bar indicates the number of changes and the numbers at the nodes represent maximum parsimony (>70 %) and maximum likelihood (> 70 %) bootstrap support values from 1 000 non-parametric bootstrap replicates. Branches that appear in the strict consensus tree are highlighted by thickened lines. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Xylaria hypoxylon (AFTOL-ID 51; GenBank AY544648) and the species treated here is highlighted with bold face. The families, orders and class are shown in coloured blocks to the right of the tree.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 42462 had highest similarity to Neoacrodontiella eucalypti (strain CBS 145561, GenBank NR_165567.1; Identities = 473/549 (86 %), 43 gaps (7 %)), Cytosporella juncicola (strain CPC 38040, GenBank MN562153.1; Identities = 379/436 (87 %), 27 gaps (6 %)), Vanderaaea ammophilae (strain CBS 886.68, GenBank NR_178155.1; Identities = 465/539 (86 %), 46 gaps (8 %)), and Corticifraga ramalinae (voucher Pinault, GenBank ON569808.1; Identities = 432/508 (85 %), 34 gaps (6 %)). The ITS sequence of CPC 42462 is 99 % similar to that of CPC 42463 (883/887 nucleotides, including four gaps from a single indel in an A-repeat region of the sequence). Closest hits using the LSU sequence of CPC 42462 were Neoacrodontiella eucalypti (strain CBS 145561, GenBank NG_067885.1; Identities = 622/651 (96 %), six gaps (0 %)), Neospermospora avenae (strain CBS 886.68, GenBank MH878416.1; Identities = 607/652 (93 %), eight gaps (1 %)), and Cytosporella juncicola (strain CPC 38040, GenBank NG_068344.1; Identities = 597/651 (92 %), six gaps (0 %)). The LSU sequence of CPC 42462 is identical to that of CPC 42463 (739/739 nucleotides).
Authors: P.W. Crous, J.Z. Groenewald, U. Braun & J. Kruse
Ophiognomonia setacea (Pers.) Sogonov, Stud. Mycol. 62: 64. 2008.
Description and illustration: Sogonov et al. (2008).
Material examined: Spain, Pontevedra, O Grove, on leaves of Quercus robur (Fagaceae), 21 Jan. 2022, J. Castillo, HPC 3874 = RKS 1161b, culture CPC 43206 = CBS 149691.
Notes: Known from overwintered leaves of Fagaceae in Canada, Europe, and the USA (Sogonov et al. 2008).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Ophiognomonia setacea (strain CBS 859.79, GenBank AY818958.1; Identities = 534/535 (99 %), no gaps), Ophiognomonia asiatica (strain CBS 131351, GenBank NR_120057.1; Identities = 501/512 (98 %), five gaps (0 %)), and Ophiognomonia sogonovii (strain CBS 131341, GenBank NR_120053.1; Identities = 500/513 (97 %), five gaps (0 %)). Closest hits using the LSU sequence were Ophiognomonia setacea (strain CBS 859.79, GenBank AY818962.1; Identities = 830/830 (100 %), no gaps), Gnomonia gnomon (strain CBS 829.79, GenBank AY818964.1; Identities = 819/830 (99 %), no gaps), and Amphiporthe hranicensis (strain AR3651, GenBank DQ323521.1; Identities = 817/831 (98 %), two gaps (0 %)). Closest hits using the tef1 (first part) sequence had highest similarity to Ophiognomonia setacea (strain CBS 859.79, GenBank JQ414154.1; Identities = 564/564 (100 %), no gaps), Ophiognomonia sogonovii (strain CBS 131341, GenBank JQ414161.1; Identities = 267/278 (96 %), one gap (0 %)), and Ophiognomonia alni-cordatae (strain CBS 131353, GenBank JQ414175.1; Identities = 226/237 (95 %), no gaps). Closest hits using the tef1 (second part) sequence had highest similarity to Ophiognomonia setacea (strain CBS 859.79, GenBank JQ414154.1; Identities = 308/308 (100 %), no gaps), Melanconis stilbostoma (strain AFTOL-ID 936, GenBank DQ836910.1; Identities = 864/925 (93 %), two gaps (0 %)), Cryptodiaporthe aesculi (strain AFTOL-ID 1238, GenBank DQ836914.1; Identities = 861/924 (93 %), no gaps), and Gnomonia gnomon (strain AFTOL-ID 952, GenBank DQ471094.1; Identities = 859/924 (93 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald, J. Castillo & R.K. Schumacher
Paradissoconium narthecii Crous & Boers, Persoonia 47: 211. 2021. Fig. 30.
Fig. 30.
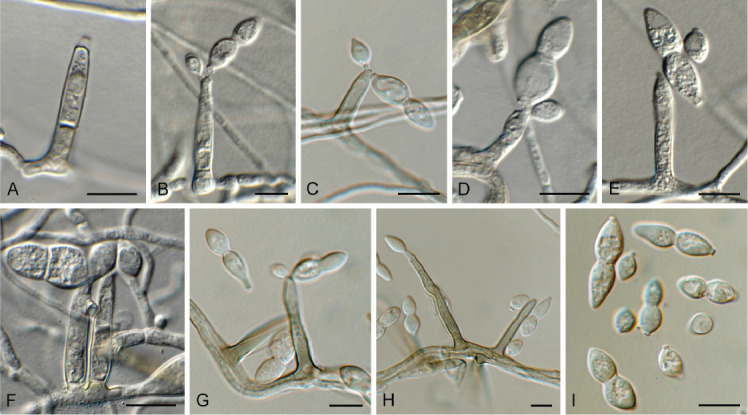
Paradissoconium narthecii (CPC 42494). A. Conidiophore. B–H. Conidiophores and conidiogenous cells giving rise to primary and secondary conidia. I. One-septate primary and aseptate secondary conidia. Scale bars = 10 μm.
Description and illustration: Crous et al. (2021b).
Material examined: Netherlands, Drenthe Province, Dwingeloo, on dead leaves of Narthecium ossifragum (Melanthiaceae), 26 Sep. 2021, J. Boers, HPC 3781, culture CPC 42494 = CBS 149692.
Notes: Paradissoconium narthecii was described from dead leaves of Narthecium ossifragum in the Netherlands (Crous et al. 2021b), for which this represents a second collection of this fungus.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Paradissoconium narthecii (strain CBS 148449, GenBank NR_175214.1; Identities = 497/497 (100 %), no gaps), Ramichloridium punctatum (strain NTOU 4892, GenBank MK448255.1; Identities = 458/502 (91 %), 14 gaps (2 %)), and Dissoconium proteae (strain CBS 122900, GenBank NR_156213.1; Identities = 462/507 (91 %), 17 gaps (3 %)). Closest hits using the LSU sequence were Paradissoconium narthecii (strain CBS 148449, GenBank NG_081323.1; Identities = 804/804 (100 %), no gaps), Ramichloridium eucleae (strain CBS 138000, GenBank NG_058086.1; Identities = 813/827 (98 %), two gaps (0 %)), and Ramichloridium indicum (strain CBS 171.96, GenBank EU041856.1; Identities = 795/811 (98 %), no gaps). Closest hits using the actA sequence had highest similarity to Paradissoconium narthecii (strain CPC 41970, GenBank OK651125.1; Identities = 539/539 (100 %), no gaps), Cladosporium ramotenellum (strain CBS 145592, GenBank MT223748.1; Identities = 382/407 (94 %), no gaps), and Davidiellomyces australiensis (strain CBS 142165, GenBank KY979853.1; Identities = 382/407 (94 %), no gaps). The rpb1 sequence is identical to that of Paradissoconium narthecii (strain CPC 41970, GenBank OK651151.1; Identities = 518/518 (100 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & J. Boers
Paraeutypella citricola (Speg.) L.S. Dissan. et al., Biodivers. Data J. 9: e63864 [14]. 2021.
Description and illustration: Dissanayake et al. (2021).
Material examined: Spain, Cambrils, on bark of unknown tree, 26 Jan. 2022, J. Castillo, HPC 3869 = RKS 1158, culture CPC 43208 = CBS 149693.
Notes: Paraeutypella citricola was originally described (as Eutypella citricola) from twigs of Citrus in Argentina, and recently reported from dead twigs of Acer palmatum in China (Dissanayake et al. 2021). It is recorded here from bark of a dead, unknown tree in Spain. This species has inoperculate asci that are distinctly amyloid (mounted in Lugol).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Paraeutypella citricola (strain ICMP 17420, GenBank MH497577.1; Identities = 529/529 (100 %), no gaps), and Paraeutypella vitis (strain 59, GenBank KU320620.1; Identities = 513/530 (97 %), three gaps (0 %)). Closest hits using the LSU sequence were Paraeutypella citricola (as Eutypella citricola, strain CBS 128334, GenBank MH876333.1; Identities = 867/867 (100 %), no gaps), Libertella faginea f. minor (strain CBS 196.30, GenBank MH866558.1; Identities = 862/868 (99 %), one gap (0 %)), and Eutypella microtheca (strain CBS 128336, GenBank NG_064278.1; Identities = 830/837 (99 %), no gaps). Closest hits using the rpb2 (first part) sequence had highest similarity to Paraeutypella citricola (strain GMBC0053, GenBank MW814898.1; Identities = 898/899 (99 %), no gaps), Allocryptovalsa sichuanensis (voucher HKAS 107017, GenBank MW658624.1; Identities = 770/886 (87 %), eight gaps (0 %)), and Eutypella quaternata (voucher MFLU 15-2605, GenBank MT432247.1; Identities = 715/860 (83 %), 14 gaps (1 %)). Closest hits using the tub2 sequence had highest similarity to Paraeutypella citricola (voucher HUEFS 131041, GenBank KT175565.1; Identities = 666/671 (99 %), no gaps), Diatrypella longiasca (strain KUMCC 20-0021, GenBank MW239658.1; Identities = 564/663 (85 %), 26 gaps (3 %)), and Eutypella microtheca (strain AD53, GenBank KR822701.1; Identities = 548/667 (82 %), 39 gaps (5 %)).
Authors: P.W. Crous, J.Z. Groenewald, J. Castillo & R.K. Schumacher
Phomatospora endopteris W. Phillips & Plowr., Grevillea 13(no. 67): 76. 1885. Fig. 31.
Fig. 31.
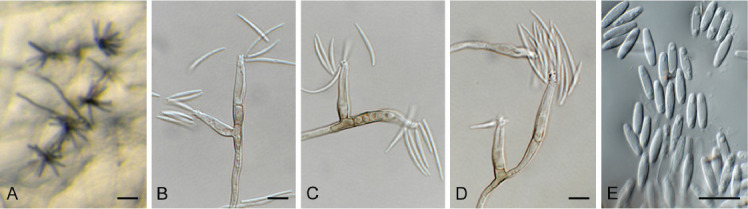
Phomatospora endopteris (CPC 41832). A. Colony on SNA. B–D. Conidiophores and conidiogenous cells giving rise to conidia. E. Conidia. Scale bars = 10 μm.
Description and illustration: Senanayake et al. (2016).
Cultures derived from single ascospores shot from a perithecium onto agar, but material inadequate for description. Ascospores aseptate, fusoid-ellipsoid, pale brown, guttulate, longitudinally striate, ends bluntly rounded, somewhat flattened, 9–10(–11) × 3 μm. Mycelium consisting of hyaline, smooth, branched, septate, 1–1.5 μm diam hyphae. Conidiophores erect, straight to flexuous, arising from superficial hyphae, subcylindrical, 1–2-septate, or reduced to conidiogenous cells, pale brown, smooth, branched or not, 20–50 × 3–3.5 μm. Conidiogenous cells terminal and intercalary, pale brown, smooth, subcylindrical to obclavate, 7–20 × 3–3.5 μm, with terminal rachis of subdenticulate loci, 0.5–1 × 0.5 μm, slightly thickened and darkened. Conidia solitary, aseptate, hyaline, smooth, guttulate, slightly curved, narrowly fusoid, apex subobtuse, base truncate, (11–)13–15(–18) × 1.5 μm.
Culture characteristics: Colonies erumpent, spreading, with sparse to moderate aerial mycelium and smooth, even margin, reaching 18 mm diam after 2 wk at 25 °C. On MEA surface dirty white, reverse ochreous; on PDA and OA surface and reverse dirty white.
Material examined: Netherlands, Utrecht Province, Bilthoven, on stems of Pteridium aquilinum (Dennstaedtiaceae), 24 May 2021, P.W. Crous, HPC 3644 = CBS H-24975, culture CPC 41832 = CBS 149073.
Notes: Phomatospora species are known from marine, aquatic and terrestrial habitats, where they are usually saprobic on submerged wood or decaying twigs (Senanayake et al. 2016). Phomatospora endopteris was isolated from single ascospores that shot onto agar from decaying stems of Pteridium aquilinum. Although the immersed ascomata could not be located, the ascospores were fusoid-ellipsoid and longitudinally striate, which fit well with the circumscription of the genus. Ellis & Ellis (1997) list ascospores of Phomatospora endopteris as 10–11 × 2.5–3 μm, thus fitting with those of the present collection. Phomatospora endopteris is currently not known from sequence data and the blast results below confirm its association with the genus Phomatospora. Although the genus contains more than 100 species names, only a few are currently known from sequence data, namely P. bellaminuta, P. biseriata, P. striatigera, P. uniseriata and P. viticola. The lectotype species, P. berkeleyi, is currently not known from molecular data. See Phukhamsakda et al. (2020) for a recent phylogenetic treatment of the genus.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Phomatospora biseriata (strain MFLUCC 14-0832A, GenBank NR_154640.1; Identities = 478/522 (92 %), 11 gaps (2 %)), Phomatospora sp. (strain UBOCC-A-118152, GenBank MT133745.1; Identities = 523/536 (98 %), two gaps (0 %)), Pyramidospora constricta (strain VG116-5, GenBank OM907745.1; Identities = 537/592 (91 %), 15 gaps (2 %)), Paramicrothyrium chinensis (strain IA20, GenBank KM246198.1; Identities = 531/589 (90 %), 14 gaps (2 %)), and Phomatospora striatigera (strain CBS 133932, GenBank NR_145386.1; Identities = 536/604 (89 %), 24 gaps (3 %)). Closest hits using the LSU sequence were Phomatospora sp. from an oceanic crust in the Indian Ocean (strain UBOCC-A-118152, GenBank MT226561.1; Identities = 723/724 (99 %), no gaps), Phomatospora uniseriata (strain MFLUCC 17-2068, GenBank NG_073857.1; Identities = 806/826 (98 %), one gap (0 %)), and Lanspora cylindrospora (strain NFCCI-4427, GenBank MN168892.1; Identities = 732/753 (97 %), no gaps). Closest hits using the SSU sequence were Phomatospora sp. from an oceanic crust in the Indian Ocean (strain UBOCC-A-118152, GenBank MT226525.1; Identities = 960/963 (99 %), no gaps), Phomatospora biseriata (strain MFLUCC 14-0832A, GenBank NG_065654.1; Identities = 911/914 (99 %), no gaps), and Distoseptispora aquatica (strain MFLUCC 16-1357, GenBank MK828317.1; Identities = 852/858 (99 %), no gaps). Closest hits using the rpb2 sequence had highest similarity to Lanspora coronata (strain AFTOL-ID 736, GenBank DQ470899.1; Identities = 648/840 (77 %), 15 gaps (1 %)), Sporothrix splendens (strain CMW23050, GenBank OM631738.1; Identities = 633/843 (75 %), 31 gaps (3 %)), and Sporothrix fumea (strain CMW26813, GenBank OM631721.1; Identities = 636/849 (75 %), 43 gaps (5 %)). A blast2 comparison with the two Phomatospora rpb2 sequences on GenBank also revealed low similarity: Phomatospora uniseriata (strain MFLUCC 17-2068, GenBank MT394720.1; Identities = 606/817 (74 %), 28 gaps (3 %)), and Phomatospora bellaminuta (strain AFTOL-ID 766, GenBank FJ238345.1; Identities = 612/842 (73 %), 42 gaps (4 %)).
Authors: P.W. Crous & J.Z. Groenewald
Phyllosticta multicorniculata Bissett & M.E. Palm, Canad. J. Bot. 67: 3382. 1989. Fig. 32.
Fig. 32.
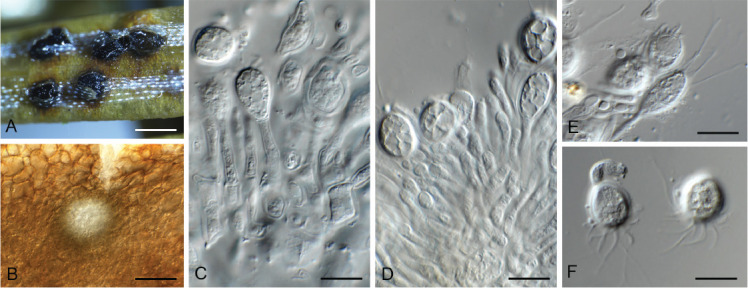
Phyllosticta multicorniculata (CPC 41921). A. Erumpent conidiomata on Abies needle. B. Conidioma with central ostiole. C, D. Conidiogenous cells giving rise to conidia. E, F. Conidia with multiple apical appendages. Scale bars: A = 200 μm, all others = 10 μm.
Description and illustration: Bissett & Palm (1989).
Culture characteristics: Colonies erumpent, spreading, with moderate aerial mycelium and feathery, lobate margin, reaching 30 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface olivaceous grey, reverse iron-grey.
Typus: Canada, Prince Edward Island, Milton Station, on needles of Abies balsamea (Pinaceae), 19 Jul. 1966, C.D. MacCall (holotype DAOM 114552); New Brunswick, Charlotte Co., 1.5 km SW of Little Lepreau, 45.135614° -66.492269°, on buds of A. balsamea, 4 May 2021, D. Malloch, HPC 3633 (epitype designated here CBS H-24979, MBT 10013419, culture ex-epitype CPC 41921 = CBS 149077).
Additional material examined. Canada, New Brunswick, Charlotte Co., 1.5 km SW of Little Lepreau, 45.135614° -66.492269 °, on buds of A. balsamea, 4 May 2021, D. Malloch, HPC 3633, culture CPC 41919 = CBS 149078.
Notes: Conidia of P. multicorniculataare (10–)12–15(–17) × (7–) 10–11(–13) μm, enclosed in a mucilaginous sheath, 1.5 μm thick, with 2–5 conoidal or cylindrical apical appendages, thus closely matching those of the holotype (Bissett & Palm 1989). Within the genus Phyllosticta, P. multicorniculata is peculiar in that it has several apical appendages, whereas most species have only a single, or no apical appendage. In the phylogenetic tree (Fig. 33), P. multicorniculata had a basal position in Phyllostictaceae (76 % / 95 % / 1). Neither the Bayesian nor the RAxML analyses resolve this species as a basal sister lineage; only in the most parsimonious trees does it take up a position as a basal sister lineage but without any support in the parsimony bootstrap consensus tree. Whether this species could represent a novel genus in the future remains to be seen as the phylogenetic trees obtained from the different analyses were inconclusive to clearly separate it from the other species in Phyllosticta. The similarities of the different loci seem to fit within the genetic diversity of the genus (blast results filtered for Phyllostictaceae): ITS (based on the top 2 531 blast hits using GenBank FJ824766) = 82.08 % – 100 %; LSU (based on the top 282 blast hits using GenBank KF766342) = 90.24 % – 100 %; actA (based on the top 345 blast hits using the first 240 nucleotides of GenBank JX025029) = 77.13 % – 100 %; gapdh (based on the top 355 blast hits using GenBank MN556783) = 86.39 % – 100 %; and tef1 (based on the top 249 blast hits using GenBank MN556818) = 79.34 % – 100 %.
Fig. 33.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE v. 2.1.3 of the Phyllostictaceae and related families LSU nucleotide alignment. Maximum likelihood (> 74 %) and maximum parsimony (> 74 %) bootstrap support values from 1 000 non-parametric bootstrap replicates, and Bayesian posterior probabilities (> 0.84), are shown at the nodes. Thickened lines represent nodes which received full support (100 % / 100 % / 1) from all three analyses. Culture collection or voucher numbers and GenBank accession numbers (superscript) are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. The tree was rooted to Diaporthe perjuncta (voucher BPI 748437; GenBank NG_059064) and the species treated here is highlighted with bold face. The families, orders and class are shown in coloured blocks to the right of the tree. The scale bar indicates the expected number of changes per site.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 41921 had highest similarity to Phyllosticta parthenocissi (deposited in GenBank as Guignardia bidwellii; strain CBS 111645, GenBank FJ824766.1; Identities = 445/536 (83 %), 27 gaps (5 %)), Phyllosticta foliorum (deposited in GenBank as as Guignardia philoprina; strain CBS 447.68, GenBank AF312014.1; Identities = 448/541 (83 %), 32 gaps (5 %)), and Phyllosticta citricarpa (strain PD 010 04421662, GenBank MZ416911.1; Identities = 329/367 (90 %), 15 gaps (4 %)). The ITS sequences of CPC 41921 and 41919 are identical (528/528 nucleotides). Closest hits using the LSU sequence of CPC 41921 were Phyllosticta philoprina (strain CBS 901.69, GenBank KF766342.1; Identities = 765/793 (96 %), no gaps), Phyllosticta styracicola (strain JFRL 03-770, GenBank OQ195355.1; Identities = 759/793 (96 %), no gaps), and Phyllosticta minima (strain CBS 585.84, GenBank KF766382.1; Identities = 759/793 (96 %), no gaps). The LSU sequences of CPC 41921 and 41919 are identical (793/793 nucleotides). Closest hits using the actA sequence of CPC 41921 had highest similarity to Phyllosticta encephalarticola (strain CBS 146014, GenBank MN556783.1; Identities = 462/504 (92 %), nine gaps (1 %)), Phyllosticta austroafricana (strain CBS 144593, GenBank MK442640.1; Identities = 445/485 (92 %), six gaps (1 %)), and Phyllosticta hagahagaensis (strain CBS 144592, GenBank MK442641.1; Identities = 458/504 (91 %), nine gaps (1 %)). The actA sequences of CPC 41921 and 41919 are identical (533/533 nucleotides). Closest hits using the gapdh sequence of CPC 41921 had highest similarity to Phyllosticta hubeiensis (strain LC1654, GenBank JX025029.1; Identities = 304/336 (90 %), no gaps), Phyllosticta rhizophorae (strain NCYUCC 19-0358, GenBank MT363251.1; Identities = 301/336 (90 %), no gaps), and Phyllosticta capitalensis (strain IPN20.1, GenBank KX280619.1; Identities = 300/335 (90 %), no gaps). The gapdh sequences of CPC 41921 and 41919 are identical (545/545 nucleotides). Closest hits using the tef1 (first part) sequence of CPC 41921 had highest similarity to Phyllosticta encephalarticola (strain CBS 146014, GenBank MN556818.1; Identities = 290/332 (87 %), 14 gaps (4 %)), Phyllosticta hagahagaensis (strain CBS 144592, GenBank MK442705.1; Identities = 290/332 (87 %), 14 gaps (4 %)), and Phyllosticta carissicola (strain CPC 25665, GenBank KT950879.1; Identities = 290/332 (87 %), 14 gaps (4 %)). The tef1 sequences of CPC 41921 and 41919 are identical (409/409 nucleotides).
Authors: P.W. Crous, J.Z. Groenewald & D. Malloch
Pleurotheciella aquatica Z.L. Luo et al., Mycol. Progr. 17: 517. 2018. Fig. 34.
Fig. 34.
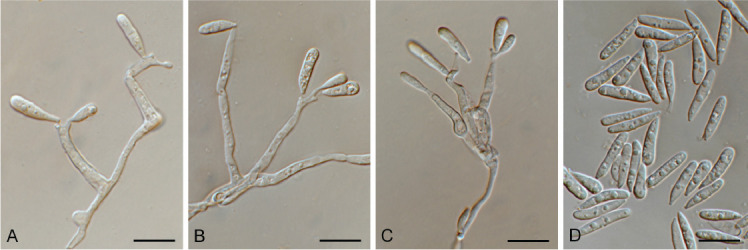
Pleurotheciella aquatica (CPC 44105). A–C. Conidiophores and conidiogenous cells giving rise to conidia. D. Conidia. Scale bars = 10 μm.
Description and illustration: Huang et al. (2022).
Material examined: UK, England, Basingstoke, on Allium schoenoprasum (Alliaceae), May 2022, P.W. Crous, CBS H-25211, culture CPC 44105 = CBS 149694.
Notes: Pleurotheciella aquatica was described from submerged wood collected in China (Huang et al. 2022). This is the first report of this fungus from the UK.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Pleurotheciella aquatica (voucher MFLU 17-0911, GenBank NR_160591.1; Identities = 499/500 (99 %), no gaps), Pleurotheciella fusiformis (strain MFLUCC 16-1356, GenBank MF399235.1; Identities = 494/514 (9 6%), nine gaps (1 %)), and Pleurotheciella uniseptata (strain KAS 4459, GenBank KT278729.1; Identities = 500/534 (94 %), 13 gaps (2 %)). Closest hits using the LSU sequence were Pleurotheciella aquatica (voucher MFLU 17-0911, GenBank NG_066193.1; Identities = 799/800 (99 %), one gap (0 %)), Pleurotheciella fusiformis (strain MFLUCC 17-0115, GenBank MF399249.1; Identities = 755/762 (99 %), one gap (0 %)), and Pleurotheciella lunata (voucher MFLU 17-0913, GenBank NG_066195.1; Identities = 790/800 (99 %), one gap (0 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Pleurotheciella aquatica (strain MFLUCC 17-0464, GenBank MF401405.1; Identities = 655/655 (100 %), no gaps), Pleurotheciella fusiformis (strain MFLUCC 17-0113, GenBank MF401403.1; Identities = 661/699 (95 %), no gaps), and Pleurotheciella uniseptata (strain KUMCC 15-0407, GenBank MF401401.1; Identities = 541/599 (90 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & S. Denman
Ramularia pistaciae Crous, Persoonia 42: 353. 2019. Fig. 35.
Fig. 35.
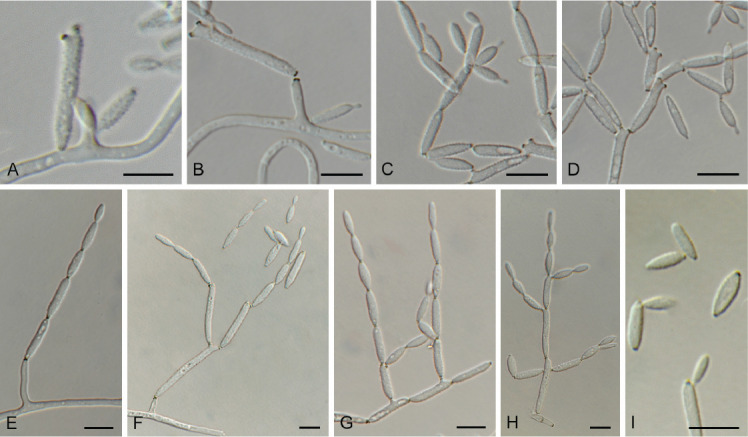
Ramularia pistaciae (CPC 44067). A–H. Conidiophores and conidiogenous cells giving rise to branched conidial chains. I. Conidia. Scale bars = 10 μm.
Description and illustration: Crous et al. (2019a).
Material examined: UK, England, Arundale, on leaf spot on Arbutus unedo (Ericaceae), 14 May 2022, P.W. Crous & S. Denman, HPC 3955, culture CPC 44067 = CBS 149696.
Notes: Ramularia pistaciae was recently described from leaves of Pistacia lentiscus collected in Rome, Italy (Crous et al. 2019a). This is the first record from the UK, where it is associated with leaf spots of Arbutus unedo.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Ramularia pistaciae (strain CBS 145564, GenBank NR_165576.1; Identities = 491/491 (100 %), no gaps), Ramularia glennii (strain SFI-D21, GenBank MT535837.1; Identities = 480/492 (98 %), two gaps (0 %)), and Ramularia eucalypti (strain SFI-D26, GenBank MT535835.1; Identities = 480/492 (98 %), two gaps (0 %)). Closest hits using the LSU sequence were Ramularia alangiicola (strain CPC 10299, GenBank KX287023.1; Identities = 825/828 (99 %), no gaps), Ramularia eucalypti (strain CBS 120726, GenBank NG_069156.1; Identities = 812/815 (99 %), no gaps), and Ramularia rumicicola (strain CPC 11296, GenBank KX287206.1; Identities = 824/828 (99 %), no gaps (0 %)). Closest hits using the actA sequence had highest similarity to Ramularia pistaciae (strain CBS 145564, GenBank MK876462.1; Identities = 571/575 (99 %), no gaps), Ramularia gaultheriae (strain CBS 299.80, GenBank KX287693.1; Identities = 532/577 (92 %), six gaps (1 %)), and Ramularia unterseheri (strain CBS 124852, GenBank KP894376.1; Identities = 530/576 (92 %), three gaps (0 %)). Closest hits using the gapdh sequence had highest similarity to Ramularia pistaciae (strain CBS 145564, GenBank MK876473.1; Identities = 525/536 (98 %), one gap (0 %)), Ramularia vizellae (strain CPC 25731, GenBank KP894644.1; Identities = 417/457 (91 %), seven gaps (1 %)), and Ramularia lamiigena (strain IRAN 3985C, GenBank MW272941.1; Identities = 385/423 (91 %), seven gaps (1 %)). Closest hits using the his3 sequence had highest similarity to Ramularia tricherae (strain CBS 108973, GenBank KP894799.1; Identities = 346/359 (96 %), no gaps), Ramularia abscondita (strain CBS 114727, GenBank KX288753.1; Identities = 341/356 (96 %), two gaps), and Ramularia variabilis (strain CPC 16866, GenBank KP894829.1; Identities = 345/362 (95 %), no gaps). Closest hits using the rpb2 (first part) sequence had highest similarity to Ramularia gaultheriae (strain CBS 299.80, GenBank KX288569.1; Identities = 620/731 (85 %), no gaps), Ramularia neodeusta (strain CPC 13567, GenBank KX288638.1; Identities = 618/735 (84 %), six gaps (0 %)), and Ramularia cyclaminicola (strain CBS 399.51, GenBank KX288551.1; Identities = 617/736 (84 %), four gaps (0 %)). Closest hits using the tef1 (first part) sequence had highest similarity to Ramularia malicola (strain CBS 119227, GenBank KX288036.1; Identities = 301/308 (98 %), one gap (0 %)), Ramularia rumicicola (strain CPC 11296, GenBank KX288065.1; Identities = 274/330 (83 %), 17 gaps (5 %)), and Ramularia lamii var. lamii (strain CBS 108971, GenBank KX288023.1; Identities = 272/313 (87 %), 13 gaps (4 %)).
Authors: P.W. Crous, J.Z. Groenewald & S. Denman
Ruptoseptoria unedonis (Roberge ex Desm.) Quaedvl. et al., Stud. Mycol. 75: 357. 2013. Fig. 36.
Fig. 36.
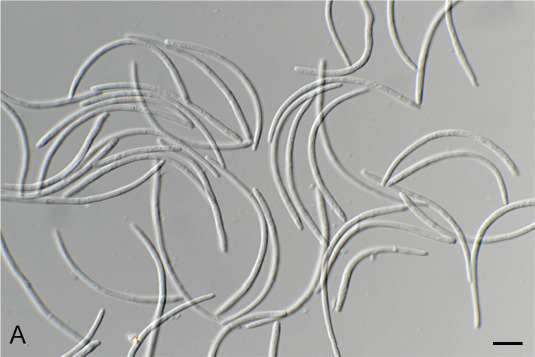
Ruptoseptoria unedonis (CPC 44069). Conidia. Scale bar = 10 μm.
Description and illustration: Quaedvlieg et al. (2013).
Material examined: UK, England, Arundale, on leaf spot on Arbutus unedo (Ericaceae), 14 May 2022, P.W. Crous & S. Denman, HPC 3956, culture CPC 44069 = CBS 149697.
Notes: Ruptoseptoria unedonis is known to cause prominent leaf spotting of Arbutus unedo, and is also known from the UK (Quaedvlieg et al. 2013).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Ruptoseptoria unedonis (strain CBS 355.86, GenBank KF251228.1; Identities = 494/494 (100 %), no gaps), Chuppomyces handelii (strain CBS 113302, GenBank EU167581.1; Identities = 471/496 (95 %), four gaps (0 %)), and Neosonderhenia eucalypti (strain CBS 145081, GenBank NR_165602.1; Identities = 460/499 (92 %), 13 gaps (2 %)). Closest hits using the LSU sequence were Ruptoseptoria unedonis (strain CBS 755.70, GenBank KF251732.1; Identities = 825/826 (99 %), one gap (0 %)), Chuppomyces handelii (strain CBS 113302, GenBank GU214437.1; Identities = 811/828 (98 %), five gaps (0 %)), and Pruniphilomyces circumscissus (strain CPC 36434, GenBank MT223926.1; Identities = 795/827 (96 %), five gaps (0 %)). Closest hits using the rpb2 (first part) sequence had highest similarity to Ruptoseptoria unedonis (strain CBS 755.70, GenBank MF951659.1; Identities = 768/771 (99 %), no gaps), Chuppomyces handelii (strain CBS 113302, GenBank MF951475.1; Identities = 677/773 (88 %), no gaps), and Neopenidiella nectandrae (strain ATCC 200932, GenBank MF951546.1; Identities = 580/740 (78 %), 16 gaps (2 %)).
Authors: P.W. Crous, J.Z. Groenewald & S. Denman
Schizothecium conicum (Fuckel) N. Lundq., Symb. Bot. Upsal. 20(no. 1): 253 (1972) Fig. 37.
Fig. 37.
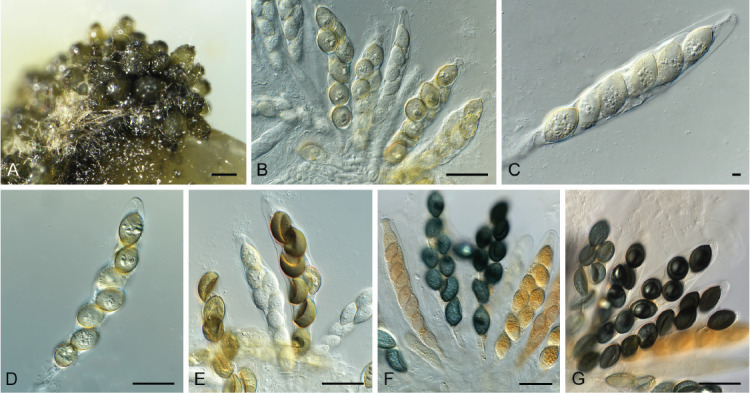
Schizothecium conica (CPC 44110). A. Ascomata on SNA. B–G. Asci with ascospores. Scale bars: A = 180 μm, all others = 10 μm.
Basionym: Cercophora conica Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 245. 1870 [1869–1870].
See MycoBank for synonyms.
Description and illustration: Bell & Mahoney (1995).
Material examined: Netherlands, Overijsel Province, Haaksbergen, Witte Veen, on stems of Juncus effusus (Juncaceae), 28 Apr. 2022, E.R. Osieck, HPC 3962 = WI-55, culture CPC 44110 = CBS 149695.
Notes: Schizothecium conicum is a common species occurring on dung of herbivores, and is here recorded from culms of Juncus effusus in the Netherlands.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Podospora conica (strain CBS 128.94, GenBank AY515356.1; Identities = 519/521 (99 %), one gap (0 %)), Schizothecium carpinicola (strain CBS 228.87, GenBank NR_103589.1; Identities = 515/523 (98 %), two gaps (0 %)), and Podospora vesticola (strain NZ196A5, GenBank AY515365.1; Identities = 514/523 (98 %), two gaps (0 %)). Closest hits using the LSU sequence were Schizothecium vesticola (strain SMH3187, GenBank AY780076.1; Identities = 823/823 (100 %), no gaps), Podospora tetraspora (strain CBS 262.70, GenBank MH871362.1; Identities = 822/823 (99 %), no gaps), and Podospora minicauda (strain CBS 227.87, GenBank MH873757.1; Identities = 818/823 (99 %), no gaps). No LSU sequence of Podospora conica was available for comparison in GenBank.
Authors: P.W. Crous, J.Z. Groenewald & E.R. Osieck
Sporidesmiella pini Crous, Persoonia 47: 259. 2021. Fig. 38.
Fig. 38.
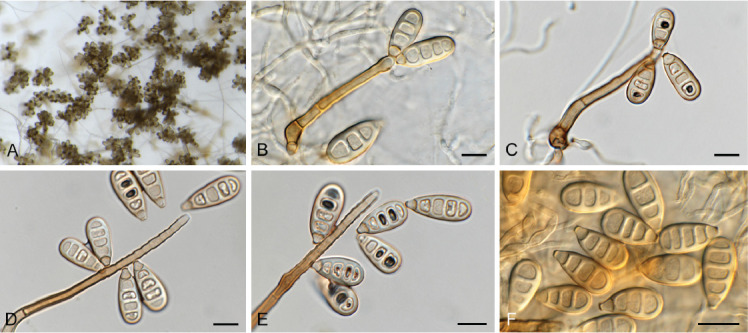
Sporidesmiella pini (CPC 41495). A. Colony on SNA. B–E. Conidiophores and conidiogenous cells giving rise to conidia. F. Conidia. Scale bars = 10 μm.
Description and illustration: Crous et al. (2021b).
Mycelium consisting of hyaline, smooth, branched, septate, 1.5–2 μm diam hyphae. Conidiophores solitary, erect, macronematous, straight to slightly flexuous, brown, smooth- and thick-walled, base swollen, 7–10 μm diam, 3–6-septate, 50–150 × 3.5–4.5 μm. Conidiogenous cells integrated, terminal, subcylindrical, scars inconspicuous, proliferating percurrently with delayed succession, rejuvenating sympodially, pale brown, smooth, 45–65 × 3.5–4 μm. Conidia solitary, dry, obovoid, medium brown, smooth, guttulate, apex obtuse, base slightly darkened, 2.5–4 μm diam, 3–4 distoseptate, with central pore in each septum, (17–)19–21(–24) × (8–)9–10 μm.
Culture characteristics: Colonies erumpent, spreading, with moderate aerial mycelium and smooth, lobate margin, reaching 30 mm diam after 2 wk at 25 °C. On MEA and PDA surface bay with vinaceous pigment, reverse dark brick; on OA surface ochreous.
Material examined: Netherlands, Overijssel Province, Engberts-dijksvenen, near Vriezenveen, 13 m a.s.l., 52°26’53’’N, 06°40’02’’E, on dead culm of Juncus effusus (Juncaceae), 9 Mar. 2021, E.R. Osieck, HPC 3612 = WI-31/#4227 = CBS H-25196, cultures CPC 41494, 41495 = CBS 149045.
Notes: Sporidesmiella pini was recently described from needles of Pinus sylvestris collected in Utrecht Province of the Netherlands (Crous et al. 2021b). This represents a second collection from the Netherlands, namely from Juncus effusus culms collected in Overijssel Province. This represents the 3rd species of Sporidesmiella collected from Juncus in the Netherlands. Their conidial morphology is similar (3–4-distoseptate, obovoid) but Sp. junci and Sp. juncicola have (slightly) longer conidia: viz. (20–) 22–25(–27) × (9–)10(–12) and (20–)28–32(–35) × (8–)9–10 μm (Crous et al. 2021b, 2022a).
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 41494 had highest similarity to Sporidesmiella pini (strain CPC 40067, GenBank OK664747.1; Identities = 559/569 (98 %), five gaps (0 %; four of which involve T-repeats at three positions)), Sporidesmiella hyalosperma (strain CPC 37552, GenBank MT223845.1; Identities = 552/572 (97 %), ten gaps (1 %)), and Sporidesmiella obovoidia (strain MFLUCC 17-2372, GenBank MW286492.1; Identities = 546/570 (96 %), 15 gaps (2 %)). The ITS sequences of CPC 41494 and 41495 are 98 % identical (551/561 nt, including five gaps all involving different numbers of T-repeats at four positions). Closest hits using the LSU sequence of CPC 41494 were Sporidesmiella pini (strain CPC 40067, GenBank NG_081347.1; Identities = 642/642 (100 %), no gaps), Sporidesmiella obovoidia (strain MFLUCC 17-2372, GenBank NG_075412.1; Identities = 639/642 (99 %), no gaps), and Sporidesmiella hyalosperma (strain S-1518, GenBank MK849842.1; Identities = 636/642 (99 %), no gaps). The LSU sequences of CPC 41494 and 41495 are 100 % identical (642/642 nt). Closest hits using the rpb2 (first part) sequence of CPC 41494 had highest similarity to Sporidesmiella pini (strain CPC 40067, GenBank OK651177.1; Identities = 746/747 (99 %), no gaps), Sporidesmiella hyalosperma (strain MFLUCC 18-1013, GenBank MW504070.1; Identities = 695/745 (93 %), no gaps), and Sporidesmiella novae-zelandiae (voucher MFLU 18-2332, GenBank MN124525.1; Identities = 656/747 (88 %), no gaps). Closest hits using the tef1 (second part) sequence of CPC 41494 had highest similarity to Sporidesmiella sp. (strain JAUCC 3436, GenBank OK323223.1; Identities = 338/356 (95 %), no gaps), Sporidesmiella hyalosperma (voucher MFLU 18-2330, GenBank MN194033.1; Identities = 327/342 (96 %), no gaps), and Sporidesmiella aquatica (voucher MFLU 18-1602, GenBank MN194034.1; Identities = 323/342 (94 %), no gaps). No tef1 sequence of Sporidesmiella pini is available for comparison.
Authors: P.W. Crous, J.Z. Groenewald & E.R. Osieck
Thyronectria caraganae Voglmayr et al., Mycol. Progr. 15: 924. 2016. Fig. 39.
Fig. 39.
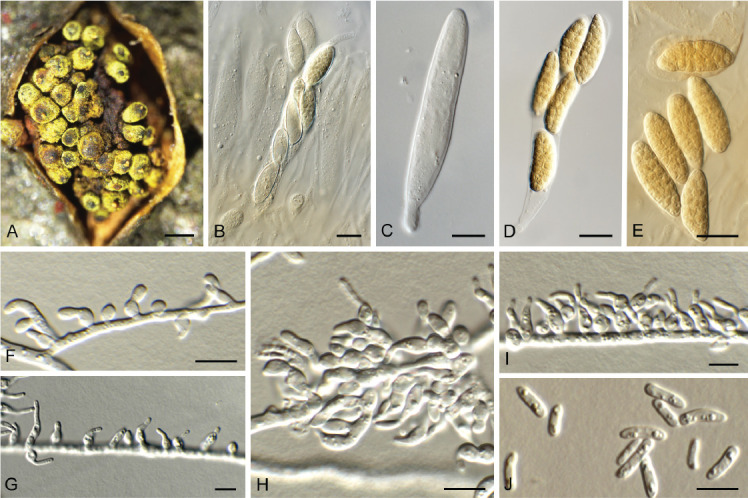
Thyronectria caraganae (CPC 41504). A. Ascomata erumpent from host tissue. B, C. Asci. D, E. Ascospores. F–I. Conidiophores and conidiogenous cells giving rise to conidia. J. Conidia. Scale bars: A = 180 μm, all others = 10 μm.
Description and illustration: Voglmayr et al. (2016).
Mycelium consisting of hyaline, smooth, branched, septate, 1.5–2 μm diam hyphae. Conidiophores reduced to conidiogenous cells, 2–10 × 2–3.5 μm, forming directly on hyphae, varying from hyphal pegs to ampulliform cells, phialidic, solitary or aggregated, giving rise to sporodochia. Conidia allantoid, hyaline, smooth, slightly curved, aseptate, guttulate, ends obtuse, (4–)5–8(–12) × 1.5–2(–3) μm.
Culture characteristics: Colonies flat, spreading, with sparse aerial mycelium, covering dish after 2 wk at 25 °C. On malt extract agar (MEA) surface and reverse sienna; on potato dextrose agar (PDA) surface and reverse pale luteous; on oatmeal agar (OA) surface ochreous.
Typus: Ukraine, Mykolaiv district, Berezansky area, Tashine, on Caragana arborescens (Leguminosae), 16 May 1990, L.V. Smyk (holotype WU 35938); Dvorichna district, Kharkiv region, Krasne Pershe village, National Park Dvorichanskyi, on twigs of C. arborescens, 11 Apr. 2021, A. Akulov, ex CWU (MYC) AS 8120 = HPC 3629 (epitype designated here CBS H-24959, MBT 10013420, culture ex-epitype CPC 41504 = CBS 148949).
Additional material examined: Ukraine, Kharkiv region, Arseniivskyi skyt, Nnat. Park Sviati hory, on dead branch of Caragana arborescens, 1 Aug. 2021, A. Akulov, HPC 3729, culture CPC 42342 = CBS 149509.
Notes: When Voglmayr et al. (2016) described Thyronectria caraganae, no asexual morph was observed, and DNA was extracted directly from the specimen due to the absence of a culture. In this study, we isolated the fungus from a fresh collection, and also observed an asexual morph to develop in vitro, making this an appropriate candidate for epitypification. The two cultures examined here were not genetically identical to each other. For the present, we have chosen not to introduce an additional species pending the collection of more specimens.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence of CPC 41504 had highest similarity to Thyronectria caraganae (voucher WU 35939, GenBank NR_155911.1; Identities = 560/560 (100 %), no gaps), Thyronectria virens (strain NP10, GenBank KM225684.1; Identities = 534/565 (95 %), 12 gaps (2 %)), and Thyronectria abieticola (strain THYA, GenBank OL439231.1; Identities = 532/563 (94 %), 12 gaps (2 %)). The ITS sequences of CPC 41504 and 42342 are 98 % identical (531/542 nt, including three gaps). Closest hits using the LSU sequence of CPC 41504 were Thyronectria caraganae (strain TCA1, GenBank KX514385.1; Identities = 757/757 (100 %), no gaps), Thyronectria aurigera (strain THAU1, GenBank OL439233.1; Identities = 738/757 (97 %), no gaps), and Thyronectria berolinensis (strain CBS 127382, GenBank MH875990.1; Identities = 736/756 (97 %), no gaps). The LSU sequences of CPC 41504 and 42342 are 99 % identical (752/757 nt, no gaps). A manual comparison using the actA sequence of CPC 41504 revealed a similarity of 91 % (224/246 nt) with Thyronectria caraganae (strain TCA1, GenBank KX514382.1). Closest hits using the rpb1 sequence of CPC 41504 had highest similarity to Thyronectria caraganae (strain TCA1, GenBank KX514390.1; Identities = 487/488 (99 %), no gaps), Thyronectria austroamericana (strain GG, GenBank KJ570717.1; Identities = 414/490 (84 %), five gaps (1 %)), and Thyronectria rhodochlora (strain NP1, GenBank KJ570727.1; Identities = 411/490 (84 %), five gaps (1 %)). The rpb1 sequences of CPC 41504 and 42342 are 98 % identical (479/488 nt, no gaps). Closest hits using the rpb2 (first part) sequence of CPC 42342 had highest similarity to Thyronectria rhodochlora (strain NP2, GenBank KJ570751.1; Identities = 722/839 (86 %), six gaps (0 %)), Thyronectria xanthoxyli (strain NP12, GenBank OL440145.1; Identities = 720/837 (86 %), two gaps (0 %)), and Thyronectria berolinensis (strain CBS 127382, GenBank HM534883.1; Identities = 716/835 (86 %), four gaps (0 %)). Closest hits using the tef1 (second part) sequence of CPC 42342 had highest similarity to Thyronectria caraganae (strain TCA1, GenBank KX514396.1; Identities = 720/728 (99 %), no gaps), Metapochonia rubescens (strain CBS 110436, GenBank KJ398795.1; Identities = 800/864 (93 %), no gaps), and Metapochonia bulbillosa (strain P6656, GenBank MT701577.1; Identities = 800/864 (93 %), no gaps). Closest hits using the tub2 sequence of CPC 41504 had highest similarity to Thyronectria caraganae (strain TCA1, GenBank KX514399.1; Identities = 359/359 (100 %), no gaps), Metapochonia bulbillosa (strain CBS 145.70, GenBank KJ398549.1; Identities = 201/224 (90 %), eight gaps (3 %)), and Metapochonia goniodes (strain CBS 891.72, GenBank KJ398551.1; Identities = 202/226 (89 %), seven gaps (3 %)). The tub2 sequences of CPC 41504 and 42342 are 97 % identical (348/359 nt, no gaps).
Authors: P.W. Crous, J.Z. Groenewald & A. Akulov
Trichosphaeria pilosa (Pers.) Fuckel, Jb. nassau. Ver. Naturk. 23–24: 145. 1870. [1869–1870]. Fig. 40.
Fig. 40.
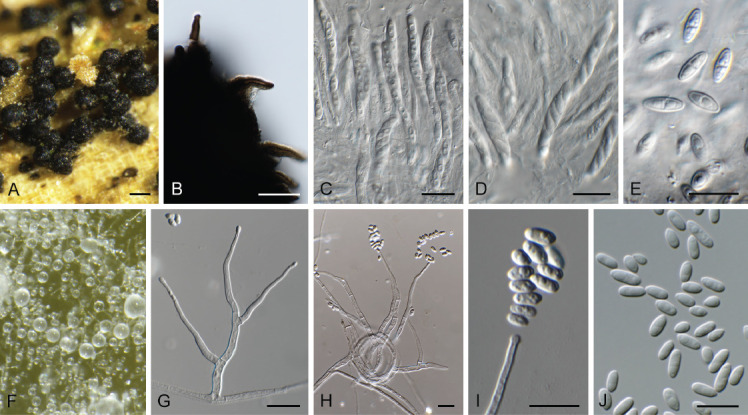
Trichosphaeria pilosa (CPC 42927). A. Ascomata on host tissue. B. Ascomatal wall with setae. C, D. Asci. E. Ascospores. F. Colony on OA. G–I. Conidiophores and conidiogenous cells giving rise to conidia. J. Conidia. Scale bars: A = 180 μm, all others = 10 μm.
Basionym: Sphaeria pilosa Pers., Icon. Desc. Fung. Min. Cognit. 2: 41, tab. 10: 9–10. 1800: Fries, Syst. Mycol. 2(2): 450. 1823.
Description and illustration: Réblová & Gams (2016).
Ascomata in densely aggregated clusters in substrate, superficial, non-stromatic, globose with papillate apex and central ostiole, dark brown to black, 180–250 μm diam, sparsely setose; setae up to 20 μm long, 6–7 μm diam near base, dark brown, 0–1-septate, tapering to subobtuse apex. Ostiole periphysate. Paraphyses hyaline, septate, 2.5–3 μm diam, trabeculae-like. Asci 35–55 × 4–6 μm, cylindrical, short stipitate, apex obtuse, without visible discharge mechanism (in Melzer), 8-spored. Ascospores 7–8(–8.5) × (2.5–)3–3.5 μm, broadly ellipsoid, hyaline, aseptate, becoming medianly septate, guttulate, verruculose, at times with lateral pegs on some ascospores. Mycelium consisting of hyaline, smooth, branched, septate, 1.5–2 μm diam hyphae. Conidiophores erect, solitary, branched below, subverticillately, or unbranched, subcylindrical, hyaline, smooth, 1–3-septate, 60–100 × 3–4 μm. Conidiogenous cells integrated, terminal and lateral, hyaline, smooth, phialidic, subcylindrical to aculeate, apex with cylindrical collarette, 1–2 μm long, 25–50 × 2.5–3 μm. Conidia aggregating in mucoid mass, hyaline, smooth, aseptate, broadly ellipsoid, guttulate, apex subobtuse, tapering to truncate hilum, 0.5 μm diam or base slightly rounded, (4.5–) 5–6(–6.5) ×2.5–3 μm.
Culture characteristics: Colonies flattened, spreading, folded, with moderate aerial mycelium and smooth, lobate margin, reaching 40 mm diam after 2 wk at 25 °C. On malt extract agar (MEA), potato dextrose agar (PDA) and oatmeal agar (OA) surface and reverse olivaceous grey.
Typus: Lectotype designated by Clements & Shear, Gen. fung., Edn 2 (Minneapolis): 261. 1931. Germany, Östricher Wald, on decaying wood of Quercus (Fagaceae), spring, neotype, designated by Réblová & Gams (2016): Fuckel, Fungi Rhen. Exs. no. 946 (M-0280129). Netherlands, Utrecht Province, Nieuw Wulven, near Houten, 1.5 m a.s.l., 52°03’03’’N, 05°09’43’’E, on decayed Salix branch (Salicaceae), 28 Jan. 2022, E.R. Osieck, HPC 3824 = WI-44/#4388 (epitype designated here CBS H-25199, MBT 10013421, culture ex-epitype CPC 42927 = CBS 149698).
Notes: Barr (1990) circumscribed the Trichosphaeriaceae to include Acanthostigma, Eriosphaeria, Rhamphoria and Trichosphaeria, although Barr & Cannon (1994) agreed that the family was heterogeneous. Several taxa were subsequently relocated to Niessliaceae, Lasiosphaeriaceae and Sordariales. Due to these problems, Réblová & Winka (2001) suggested that no additional genera be accepted in the family, until the phylogeny of T. pilosa could be resolved.
Réblová & Gams (2016) examined several collections from Persoon’s fungarium preserved in L under the name S. pilosa. However, these were shown to represent different fungi not corresponding with the protologue. Furthermore, the original illustration [Persoon 1800: tab. 10: 9–10; cited as fig. 2 in Réblová & Gams (2016)] was regarded as insufficient to serve as a nomenclatural type, and therefore a neotype was designated, which correlates well with the epitype we designate in this study.
Because of the uncertain higher-level phylogeny of Trichosphaeria, Réblová & Gams (2016) recommended accepting Trichosphaeria as the only member of the Trichosphaeriaceae, pending further collections of T. pilosa. Currently, Wijayawardene et al. (2022) list 11 genera as belonging to Trichosphaeriaceae (Trichosphaeriales). Based on the phylogenetic tree presented in this study (Fig. 41), T. pilosa is embedded within what was previously recognised as Plectosphaerellaceae (see for example Giraldo & Crous 2019), with Fuscohypha expansa as closest phylogenetic neighbour (97 % / 83 % / 1). Trichosphaeriaceae predates Plectosphaerellaceae (1885 vs 2007), and the latter is therefore reduced to synonym. The family clade is fully supported in all three phylogenetic analyses. On the other hand, the lineage containing Australiascaceae, Glomerellaceae, Malaysiascaceae and Reticulascaceae had low to full support (54 % / 90 % / 1), while the node connecting these families to the Trichosphaeriaceae node was poorly supported in the maximum likelihood analyses (68 % and 38 %). Furthermore, this node was absent in the Bayesian phylogeny as these former families formed a sister lineage to Falcocladiales, Microascales and Torpedosporales with a Bayesian posterior probability value of 0.84 (data not shown). We have therefore retained the use of Glomerellales for those families, while resurrecting Trichosphaeriales for the former family Plectosphaerellaceae. In the phylogenetic analysis of Hyde et al. (2020), “Trichosphaeriaceae” (based on “Brachysporium” groenendalensis; see pages 357 and 364 of Hyde et al. 2020) is genetically distinct from families associated with Glomerellales (see pages 359 and 388 of Hyde et al. 2020). Genera previously considered to belong to Trichosphaeriaceae need to re-evaluated as it is clear that some of these, e.g. Brachysporium, need to be assigned to a different order and family.
Fig. 41.
Consensus phylogram (50 % majority rule) obtained from the maximum likelihood analysis with IQ-TREE v. 2.1.3 of the Sordariomycetes ITS/LSU/tef1 nucleotide alignment. Maximum likelihood (> 74 %) and RAxML (> 74 %) bootstrap support values from 1 000 non-parametric bootstrap replicates, and Bayesian posterior probabilities (> 0.84), are shown at the nodes. Thickened lines represent nodes which received full support (100 % / 100 % / 1) from all three analyses. Culture collection or voucher numbers are indicated for all species. Sequences derived from material with a type status are indicated with a culture or voucher number highlighted with bold face. GenBank accession numbers of the sequences used in the alignment are listed in supplementary Table S2. The tree was rooted to Savoryella lignicola (strain NF00204) and the species treated here is highlighted with bold face. The families, orders and class are shown in coloured blocks to the right of the tree. The scale bar indicates the expected number of changes per site.
With the designation of an epitype in this study, this matter is now resolved, although the recently introduced family Plectosphaerellaceae (2007) is reduced to synonymy. Based on the phylogeny of T. pilosa, the Trichosphaeriaceae presently contains 25 genera.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Verticillium albo-atrum (strain CBS 103.95, GenBank LR026714.1; Identities = 476/496 (96 %), three gaps (0 %)), Plectosphaerella kunmingensis (strain KUMCC 18-0181, GenBank MK993014.1; Identities = 481/511 (94 %), 11 gaps (2 %)), and Plectosphaerella cucumerina (strain ZMXR22 GenBank MT446127.1; Identities = 486/518 (94 %), ten gaps (1 %)). Closest hits using the LSU sequence were Plectosphaerella kunmingensis (voucher KUMCC 18-0181, GenBank MK993015.1; Identities = 838/848 (99 %), no gaps), Gibellulopsis nigrescens (strain CBS 179.40, GenBank MH867573.1; Identities = 836/848 (99 %), no gaps), and Cephalosporium serrae (strain CBS 290.30, GenBank LR025872.1; Identities = 804/816 (99 %), no gaps). No significant hits were obtained using the actA and his3 sequences. Closest hits using the rpb2 (first part) sequence had highest similarity to Verticillium albo-atrum (strain RgVa1, GenBank MZ710737.1; Identities = 641/694 (92 %), no gaps), Plectosphaerella slobbergiarum (strain NL1930002, GenBank MW890074.1; Identities = 510/557 (92 %), no gaps), and Plectosphaerella cucumerina (strain CBS 137.37, GenBank LR026199.1; Identities = 629/691 (91 %), no gaps). Closest hits using the tef1 (first part) sequence had highest similarity to Plectosphaerella ramiseptata (strain CBS 131861, GenBank KY421317.1; Identities = 144/148 (97 %), no gaps), Gliocladium cibotii (strain CBS 299.70H, GenBank EF543807.1; Identities = 147/152 (97 %), no gaps), and Plectosphaerella alismatis (strain CBS 113362, GenBank KY421328.1; Identities = 143/148 (97 %), no gaps). Closest hits using the tef1 (second part) sequence had highest similarity to Plectosphaerella kunmingensis (strain KUMCC 18-0181, GenBank MK993017.1; Identities = 834/894 (93 %), no gaps), Plectosphaerella cucumerina (strain CBS 144925, GenBank LR594767.1; Identities = 837/903 (93 %), no gaps), and Plectosphaerella plurivora (strain GZUIFR-H26.5.1, GenBank MK930456.1; Identities = 834/901 (93 %), no gaps). Closest hits using the tub2 sequence had highest similarity to Plectosphaerella populi (strain CBS 139623, GenBank KY421311.1; Identities = 261/331 (79 %), 15 gaps (4 %)), and Pleurotheciella rivularia (strain CBS 125237, GenBank KT278760.1; Identities = 259/332 (78 %), 17 gaps (5 %)).
Classification: Sordariomycetes, Hypocreomycetidae, Trichosphaeriales, Trichosphaeriaceae.
Order
Trichosphaeriales M.E. Barr, Mycologia 75: 11. 1983.
Family
Trichosphaeriaceae G. Winter [as ‘Trichosphaerieae’], Rabenh. Krypt.-Fl., Edn 2 (Leipzig): 191. 1885.
Synonym: Plectosphaerellaceae W. Gams et al., Nova Hedwigia 85: 476. 2007.
Currently accepted genera
Acremoniisimulans Tibpromma & K.D. Hyde, Fungal Diversity 93: 88. 2018.
Acrostalagmus Corda, Icones fungorum hucusque cognitorum 2: 15. 1838.
Brunneochlamydosporium Giraldo López & Crous, Stud. Mycol. 92: 260. 2018.
Brunneomyces Giraldo et al., Mycol. Progr. 16: 357. 2017.
Chlamydosporiella Giraldo López & Crous, Stud. Mycol. 92: 270. 2018.
Chordomyces Bilanenko et al., Fungal Diversity 76: 55. 2015.
Furcasterigmium Giraldo López, Stud. Mycol. 92: 251. 2018.
Fuscohypha Giraldo López & Crous, Stud. Mycol. 92: 264. 2018.
Gibellulopsis Bat. & H. Maia, Anais Soc. Biol. Pernambuco 16: 153. 1959.
Lectera P.F. Cannon, MycoKeys 3: 28. 2012.
Longitudinalis Tibpromma & K.D. Hyde, Fungal Diversity 87: 155. 2017.
Musicillium Zare & W. Gams, Nova Hedwigia 85: 482. 2007.
Musidium Giraldo López & Crous, Stud. Mycol. 92: 253. 2018.
Nigrocephalum Giraldo López & Crous, Stud. Mycol. 92: 271. 2018.
Paragibellulopsis Giraldo López & Crous, Stud. Mycol. 92: 265. 2018.
Paramusicillium Giraldo López & Crous, Stud. Mycol. 92: 269. 2018.
Phialoparvum Giraldo López & Crous, Stud. Mycol. 92: 265. 2018.
Plectosphaerella Kleb., Phytopathol. Z. 1: 43. 1929.
Sayamraella Giraldo López & Crous, Stud. Mycol. 92: 255. 2018.
Sodiomyces A.A. Grum-Grzhim. et al., Fungal Syst. Evol. 3: 131. 2019.
Stachylidium Link, Mag. Neuesten Entdeck. Gesammten Naturk. Ges. Naturf. Freunde Berlin 3: 15. 1809.
Summerbellia Giraldo López & Crous, Stud. Mycol. 92: 252. 2018.
Theobromium Giraldo López & Crous, Stud. Mycol. 92: 256. 2018.
Trichosphaeria Fuckel, Jahrb. Nassauischen Vereins Naturk. 23–24: 144. 1870.
Verticillium Nees, System der Pilze und Schwämme: 56. 1817.
Authors: P.W. Crous, J.Z. Groenewald & E.R. Osieck
Zaanenomyces versatilis Crous & Osieck, Persoonia 47: 235. 2021. Fig. 42.
Fig. 42.
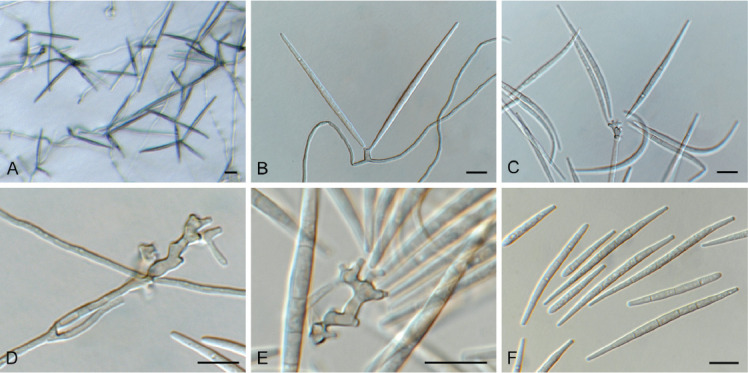
Zaanenomyces versatilis (CPC 42831). A. Colony on SNA. B–E. Conidiophores and conidiogenous cells giving rise to conidia. F. Conidia. Scale bars = 10 μm.
Mycelium consisting of pale brown, smooth, guttulate, branched, septate, 1.5–2 μm diam hyphae, forming hyphal coils. Conidiophores erect, solitary, arising from superficial hyphae, terminal or intercalary, pale brown, smooth, subcylindrical, 15–40 2–3 μm. Conidiogenous cells terminal, pale brown, smooth, 5–25 × 2–3 μm, developing a rachis of denticles, 1–3 × 1–2 μm, not thickened nor darkened, with truncate apices. Conidia solitary, dry, fusoid, widest in middle, hyaline, smooth, guttulate, apices obtuse, hilum truncate, 1–1.5 μm diam, unthickened, not darkened, 3–6(–8)-septate, (35–)50–60(–70) × (2.5–)3 μm.
Culture characteristics: Colonies erumpent, spreading, with moderate aerial mycelium and smooth, lobate margin, reaching 15 mm diam after 2 wk at 25 °C. On MEA, PDA and OA surface and reverse olivaceous grey.
Material examined. Netherlands, Utrecht Province, Nieuw Wulven, on dead stems of Juncus effusus (Juncaceae), Dec. 2021, E. Osieck, HPC 3812 = CBS H-25166, culture CPC 42831 = CBS 149453.
Notes: Zaanenomyces (Tubeufiaceae) was recently introduced for three cercosporoid-like hyphomycetes occurring on culms of Juncus in the Netherlands (Crous et al. 2021b). Zaanenomyces versatilis was described as having conidia that are (3–)7–10(–12)-septate, (16–)43–50(–55) × (2.5–)3(–3.5) μm). The present collection has slightly longer conidia, with less septa, but fits well based on its DNA phylogeny.
Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had highest similarity to Zaanenomyces versatilis (strain CBS 148312, GenBank NR_175227.1; Identities = 556/557 (99 %), no gaps), Zaanenomyces moderatricis-academiae (strain CBS 148315, GenBank NR_175222.1; Identities = 485/505 (96 %), six gaps (1 %)), and Camporesiomyces vaccinii (strain CBS 216.90, GenBank NR_156202.1; Identities = 490/559 (88 %), 15 gaps (2 %)). Closest hits using the LSU sequence were Zaanenomyces versatilis (strain CBS 148312, GenBank NG_081336.1; Identities = 820/820 (100 %), no gaps), Zaanenomyces moderatricis-academiae (strain CBS 148315, GenBank NG_081331.1; Identities = 789/793 (99 %), no gaps), and Helicosporium luteosporum (voucher MFLU 16-2871, GenBank NG_059773.1; Identities = 811/836 (97 %), no gaps). Closest hits using the tef1 (second part) sequence had highest similarity to Helicosporium flavum (strain MFLUCC 16-1230, GenBank KY873285.1; Identities = 765/825 (93 %), two gaps (0 %)), Tubeufia cylindrothecia (strain MFLUCC 17-1792, GenBank MH550968.1; Identities = 755/825 (92 %), two gaps (0 %)), and Helicosporium viridisporum (strain GZCC 22-2008, GenBank OP698087.1; Identities = 754/824 (92 %), no gaps).
Authors: P.W. Crous, J.Z. Groenewald & E.R. Osieck
Fig. 24.
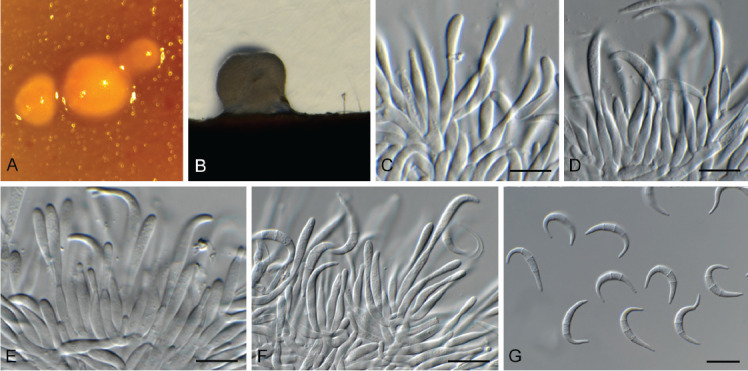
Microcera physciae (CPC 42638). A. Colony sporulating on OA. B. Colony sporulating on PNA. C–F. Conidiophores and conidiogenous cells giving rise to conidia. G. Conidia. Scale bars = 10 μm.
Acknowledgments
We are grateful to the European Union’s Horizon 2020 research and innovation program (RISE) under the Marie Skłodowska-Curie grant agreement No. 101008129, project acronym ‘Mycobiomics’, and the Dutch NWO Roadmap grant agreement No. 2020/ENW/00901156, project ‘Netherlands Infrastructure for Ecosystem and Biodiversity Analysis – Authoritative and Rapid Identification System for Essential biodiversity information’ (acronym NIEBA-ARISE) for funding. We thank Marjan Vermaas for assistance with the photographic plates.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
Supplementary Material: http://fuse-journal.org/
Summary of phylogenetic information for the different analyses in this study
GenBank accession numbers of sequences used to generate the alignment for the placement of Trichosphaeria pilosa.
REFERENCES
- Andersson MA, Salo J, Mikkola R, et al. (2021). Melinacidin-producing Acrostalagmus luteoalbus, a major constituent of mixed mycobiota contaminating insulation material in an outdoor wall. Pathogens 10: 843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmani Z, Abdollahzadeh J, Amini J. et al. (2021). Biscogniauxia rosacearum the charcoal canker agent as a pathogen associated with grapevine trunk diseases in Zagros region of Iran. Scientific Reports 11: 14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ME. (1990). Prodromus to nonlichenized, pyrenomycetous members of class Hymenoascomycetes. Mycotaxon 39: 43–184. [Google Scholar]
- Barr ME, Cannon P. (1994). Discussion 3: Calosphaeriales, Clavicipitales, Coryneliales, Diaporthales, Diatrypales, Halosphaeriales, Hypocreales, Meliolales, Ophiostomatales, Phyllachorales, Sordariales, Trichosphaeriales, and Xylariales. Ascomycete Systematics Problems and Perspectives in the Nineties (Hawksworth DL, ed.) New York & London: Plenum Press: 371–378. [Google Scholar]
- Bell A, Mahoney DP. (1995). Coprophilous fungi from New Zealand. I. Podospora species with swollen agglutinated perithecial hairs. Mycologia 87: 375–396. [Google Scholar]
- Bissett J, Palm ME. (1989). Species of Phyllosticta on conifers. Canadian Journal of Botany 67: 3378–3385. [Google Scholar]
- Boehm EWA, Mugambi GK, Miller AN, et al. (2009). A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Studies in Mycology 64: 49–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U, Kruse J. (2021). Contributions to the knowledge of mycosphaerellaceous genera and species. Schlechtendalia 38: 163–168. [Google Scholar]
- Braun U, Nakashima C, Crous PW, et al. (2018). Phylogeny and taxonomy of the genus Tubakia s. lat. Fungal Systematics and Evolution 1: 41–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral D, Stone JK, Carroll GC. (1993). The internal mycobiota of Juncus spp.: microscopic and cultural observations of infection patterns. Mycological Research 97: 367–376. [Google Scholar]
- Cheek M, Lughadha EC, Kirk P, et al. (2020). New scientific discoveries: Plants and fungi. Plants, People and Planet 2: 371–388. [Google Scholar]
- Crous PW. (1998). Mycosphaerella spp. and their anamorphs associated with leaf spot diseases of Eucalyptus. Mycologia Memoir 21: 1–170. APS Press, MN, USA. [Google Scholar]
- Crous PW, Begoude BAD, Boers J, et al. (2022a). New and Interesting Fungi 5. Fungal Systematics and Evolution 10: 19–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Boers J, Holdom D, et al. (2022b). Fungal Planet description sheets: 1383–1435. Persoonia 48: 261–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, et al. (2019a). Fungal Planet description sheets: 868–950. Persoonia 42: 291–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G, et al. (2020). Fungal Planet description sheets: 1112–1181. Persoonia 45: 251–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, et al. (2021a). New and Interesting Fungi 4. Fungal Systematics and Evolution 7: 255–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Osieck ER, Jurjević Ž, et al. (2021b). Fungal Planet description sheets: 1284–1382. Persoonia 47: 178–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Akulov A, et al. (2019b). New and Interesting Fungi. 2. Fungal Systematics and Evolution 3: 57–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. (eds) (2019c). Fungal Biodiversity. [Westerdijk Laboratory Manual Series No. 1.]. Utrecht: Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L, et al. (2019c). Fungal Planet description sheets: 951–1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Park RF. (1991). Mycosphaerella nubilosa a synonym of M. molleriana. Mycological Research 95: 628–632. [Google Scholar]
- De Vries GA. (1962). Cyphellophora laciniata nov. gen., nov. sp. and Dactylium fusarioides Fragoso et Ciferri. Mycopathologia et Mycologia Applicata 16: 47–54. [Google Scholar]
- Dissanayake LS, Wijayawardene NN, Dayarathne MC, et al. (2021). Paraeutypella guizhouensis gen. et sp. nov. and Diatrypella longiasca sp. nov. (Diatrypaceae) from China. Biodiversity Data Journal 9: e63864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MB, Ellis JP. (1997). Microfungi on land plants: an identification handbook (2nd ed.). Richmond Publishing Co., Slough. [Google Scholar]
- Fan XL, Bezerra JDP, Tian CM, et al. (2018). Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 40: 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gams W. 1971. Cephalosporium-artige Schimmelpilze (Hyphomycetes). G. Fischer Publishing, Stuttgart, Germany. [Google Scholar]
- Giraldo A, Crous PW. (2019). Inside Plectosphaerellaceae. Studies in Mycology 92: 227–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockling SL. 1998. Accessory conidium production in three species of Harposporium and an evaluation of nematophagous members of the genus. Mycological Research 102: 891–896. [Google Scholar]
- Greenhalgh G, Morgan-Jones G. (1964). Some species of Trochila and an undescribed discomycete on leaves of Prunus laurocerasus. Transactions of the British Mycological Society 47: 311–320. [Google Scholar]
- Hansen K, Perry BA, Dranginis AW, et al. (2013). A phylogeny of the highly diverse cup-fungus family Pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traits. Molecular Phylogenetics and Evolution 67: 311–335. [DOI] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, et al. (2017). Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology 86: 53–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Giraldo A, van Doorn R, et al. (2020). The Genera of Fungi - G6: Arthrographis, Kramasamuha, Melnikomyces, Thysanorea, and Verruconis. Fungal Systematics and Evolution 6: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou LW, Giraldo A, Groenewald JZ, et al. (2023). Redisposition of acremonium-like fungi in Hypocreales. Studies in Mycology 105: 23–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-P, Bao D-F, Shen H-W, et al. (2022). Neomonodictys aquatica sp. nov. (Pleurotheciaceae) from a plateau lake in Yunnan Province, China. Biodiversity Data Journal 10: e76842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN, et al. (2020). Refined families of Sordariomycetes. Mycosphere 11: 305–1059. [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, et al. (2018). Taxonomic novelties of hysteriform Dothideomycetes. Mycosphere 9: 803–837. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaubauf S, Tharreau D, Fournier E, et al. (2014). Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Studies in Mycology 79: 85–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat C, Fournier J, Vega M, et al. (2018). Geonectria, a new genus in Bionectriaceae from France. Ascomycete.org 10: 81–85. [Google Scholar]
- Lin CG, Chen Y, McKenzie EHC, et al. (2016). The genus Fusariella. Mycological Progress 15: 1313–1326. [Google Scholar]
- Lücking R, Aime MC, Robbertse B, et al. (2021). Fungal taxonomy and sequence-based nomenclature. Nature Microbiology 6: 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magaña-Dueñas V, Cano-Lira JF, Stchigel AM. (2021). New Dothideomycetes from freshwater habitats in Spain. Journal of Fungi 7: 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchisio VF, Fontana A, Luppi Mosca AM. (1977). Anthopsis deltoidea, a new genus and species of Dematiaceae from soil. Canadian Journal of Botany 55: 115–117. [Google Scholar]
- Morgan-Jones G. (1973). Genera coelomycetarum. VII. Cryptocline Petrak. Canadian Journal of Botany 51: 309–325. [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, et al. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini LE, Petrini O, Leuchtmann A, et al. (1991). Conifer inhabiting species of Phyllosticta. Sydowia 43: 148–169. [Google Scholar]
- Phukhamsakda C, McKenzie EHC, Phillips AJL, et al. (2020). Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102: 1–203. [Google Scholar]
- Punithalingam E, Spooner BM. (2011). Miricatena prunicola (Hyphomycetes), a new genus and species causing leaf spots of Prunus serotina in the UK. Kew Bulletin 66: 637–642. [Google Scholar]
- Quaedvlieg W, Verkley GJM, Shin H-D, et al. (2013). Sizing up Septoria. Studies in Mycology 75: 307–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Y, Muggia L, Moreno LF, et al. (2020). A re-evaluation of the Chaetothyriales using criteria of comparative biology. Fungal Diversity 103: 47–85. [Google Scholar]
- Rao V, De Hoog GS. (1986). New of critical hyphomycetes from India. Studies in Mycology 28: 1–84. [Google Scholar]
- Rayner RW. (1970). A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society. Kew, Surrey, UK. [Google Scholar]
- Réblová M, Gams W. (2016). A revision of Sphaeria pilosa Pers. and re-evaluation of the Trichosphaeriales. Mycological Progress 15: 52. [Google Scholar]
- Réblová M, Hernández-Restrepo M, Fournier J, et al. (2020). New insights into the systematics of Bactrodesmium and its allies and introducing new genera, species and morphological patterns in the Pleurotheciales and Savoryellales (Sordariomycetes). Studies in Mycology 95: 415–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Hernández-Restrepo M, Sklenář F, et al. (2022). Consolidation of Chloridium: new classification into eight sections with 37 species and reinstatement of the genera Gongromeriza and Psilobotrys. Studies in Mycology 103: 87–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réblová M, Winka K. (2001). Generic concepts and correlations in ascomycetes based on morphological and molecular data: Lecythothecium duriligni gen. et sp. nov. with Sporidesmium anamorph and Ascolacicola aquatica sp. nov. Mycologia 93: 478–493. [Google Scholar]
- Rehm H. (1896). Abt. 3. Ascomyceten: hysteriaceen und discomyceten. In: Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz (Rabenhorst L, ed.). Verlag von Eduard Kummer, Leipzig. [Google Scholar]
- Rogers JD, Ju Y-M, Candoussau F. (1996). Biscogniauxia anceps comb. nov. and Vivantia guadalupensis gen. et sp. nov. Mycological Research 100: 669–674. [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkiewicz-Michalska M, Knysak P, Skrobek I, et al. (2017). Cephaliophora tropica: A third European record. Mycotaxon 132: 445–451. [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, et al. 2011. The Genera of Hyphomycetes. CBS Biodiversity Series 9: 1–997. CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands. [Google Scholar]
- Senanayake IC, Al-Sadi AM, Bhat JD, et al. (2016). Phomatosporales ord. nov. and Phomatosporaceae fam. nov., to accommodate Lanspora, Phomatospora and Tenuimurus, gen. nov. Mycosphere 7: 628–641. [Google Scholar]
- Shearer CA, Crane JL, Miller MA. (1976). Illinois fungi VI: Two new species of wood inhabiting hyphomycetes from freshwater. Mycologia 68: 184–189. [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, et al. (1996). Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- Sogonov MV, Castlebury LA, Rossman AY, et al. (2008). Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Studies in Mycology 62: 1–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers AG. (1998). Melampsora and Marssonina pathogens of poplars and willows in New Zealand. European Journal of Forest Pathology 28: 233–240. [Google Scholar]
- Spiers AG, Hopcroft DH. (1998). Morphology of Drepanopeziza species pathogenic to poplars. Mycological Research 102: 1025–1037. [Google Scholar]
- Stamatakis A. (2014). RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1844–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suková M, Chlebicki A. (2004). Fungi on Juncus trifidus in the Czech Republic (II) with taxonomical notes to some species. Czech Mycology 56: 203–221. [Google Scholar]
- Swofford DL. (2003). PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Tsopelas P, Soulioti N, Wingfield MJ, et al. (2021). Ceratocystis ficicola causing a serious disease of Ficus carica in Greece. Phytopathologia Mediterranea 60: 337–349. [Google Scholar]
- Tsuneda A, Hambleton S, Currah RS. (2004). Morphology and phylogenetic placement of Endoconidioma, a new endoconidial genus from trembling aspen. Mycologia 96: 1128–1135. [PubMed] [Google Scholar]
- Voglmayr H, Akulov OY, Jaklitsch WM. (2016). Reassessment of Allantonectria, phylogenetic position of Thyronectroidea, and Thyronectria caraganae sp. nov. Mycological Progress 15: 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira SIR, Groenewald JZ, Nakashima C, et al. (2017). Mycosphaerellaceae – chaos or clarity? Studies in Mycology 87: 257–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arx JA. (1957). Revision der zu Gloeosporium gestellten Pilze. Verhandelingen der Koninklijke Nederlandse Akademie van Wetenschappen, Afd. Natuurkunde, Tweede Reeks 51: 1–153. [Google Scholar]
- Vu D, Groenewald M, de Vries M, et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayawardene NN, Hyde KD, Dai DQ, et al. (2022). Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13: 53–453. [Google Scholar]
- Yang H, Hyde KD, Karunarathna S, et al. (2018). New species of Camptophora and Cyphellophora from China, and first report of sexual morphs for these genera. Phytotaxa 343: 149–159. [Google Scholar]
- Zare R, Gams W, Schroers HJ. (2004). The type species of Verticillium is not congeneric with the plant-pathogenic species placed in Verticillium and it is not the anamorph of ‘Nectria’ inventa. Mycological Research 108: 576–582. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, et al. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology 7: 203–214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of phylogenetic information for the different analyses in this study
GenBank accession numbers of sequences used to generate the alignment for the placement of Trichosphaeria pilosa.