SUMMARY
Fecal microbiota transplantation (FMT) is highly effective in preventing recurrent Clostridioides difficile infection (rCDI). However, the mechanisms underpinning its clinical efficacy are incompletely understood. Herein, we provide an overview of rCDI pathogenesis followed by a discussion of potential mechanisms of action focusing on the current understanding of trans-kingdom microbial, metabolic, immunological, and epigenetic mechanisms. We then outline the current research gaps and offer methodological recommendations for future studies to elevate the quality of research and advance knowledge translation. By combining interventional trials with multiomics technology and host and environmental factors, analyzing longitudinally collected biospecimens will generate results that can be validated with animal and other models. Collectively, this will confirm causality and improve translation, ultimately to develop targeted therapies to replace FMT.
INTRODUCTION
Clostridioides difficile is a Gram-positive, spore-forming bacterium that causes the most common nosocomial intestinal infection. The pathology arises from dysbiosis of the intestinal microbiota, usually triggered by antibiotic use, allowing C. difficile to proliferate. The clinical spectrum of C. difficile infection (CDI) ranges from mild diarrhea to toxic megacolon and death.1 The recommended therapy for the primary episode of CDI is either vancomycin, a broad-spectrum Gram-positive antimicrobial, or fidaxomicin, a narrow-spectrum but expensive antibiotic.2 A major clinical challenge is recurrent CDI (rCDI), which occurs in approximately 20% of patients after the primary episode and in 60% of patients following the third episode because no effective standard drug-based therapy exists.3,4
Recently, fecal microbiota transplantation (FMT), also known as intestinal microbiota transfer, has been increasingly adopted into routine clinical practice to treat rCDI because it is the most clinically effective and cost-effective therapy. A successful treatment outcome is usually defined as a lack of CDI recurrence after a follow-up period of at least 8 weeks. Using this criterion, the success rate of FMT has been reported to range from 60% to over 90%, depending on the route of administration (retention enema, nasogastric tube, colonoscopy, or capsules) and study design (randomized placebo-controlled or open-label).5-7 The success rates tend to be lower with randomized placebo-controlled studies than in open-label studies,5 and are also lower in studies using FMT by retention enema than with other routes of delivery.5,7,8 Although highly effective, there are substantial drawbacks with FMT, including infectious risks and sparse long-term safety data.9,10
Although substantial progress has undoubtedly been made in unraveling the “how” of FMT, most human studies are largely associative or correlative, and solely analyze the stool microbiome. Although results from these studies have led to the development of defined microbiota likely to influence FMT efficacy for rCDI, the variable success rate observed across clinical trials demonstrates critical gaps in knowledge about how FMT works. In addition to well-designed clinical trials, a mechanistic understanding of both microbe-microbe and host-microbe interactions that define successful recovery from rCDI is necessary. These include studies using strain-level metagenomic analyses to identify the ecological effects of FMT on the recipient microbiota,11-13 as well as testable hypotheses on mechanisms of the FMT action in human and rodent studies. As such, identifying the key components responsible for the beneficial effect of FMT and the underlying mechanisms should remain a research priority.
In this review, we present an updated overview of C. difficile host-microbe interactions that may additionally influence FMT efficacy for rCDI. We first summarize these concepts in the context of CDI pathogenesis and recurrence. We will then discuss the current understanding of the potential mechanistic actions of FMT for rCDI, focusing on microbial (trans-kingdom), metabolic, immunological, and epigenetic mechanisms potentially underpinning FMT efficacy. We will consider both human observational studies and animal models, as the latter are able to make causal inferences. We will discuss the current research gaps and offer methodological recommendations for future studies to elevate the quality of research and advance translation. We will further explore the challenges and potential mitigation strategies for determining causality in humans.
PATHOGENESIS
CDI
As a complete overview of the pathogenesis of CDI is beyond the scope of this review and has been discussed elsewhere,14 we will highlight C. difficile host-microbial interactions that may influence therapeutic efficacy of FMT (see Figure 1).
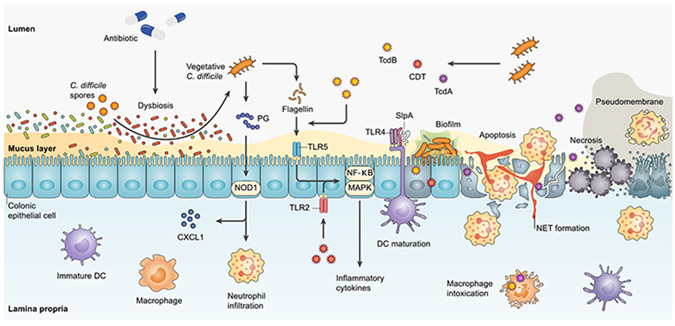
Figure 1. Main mechanisms of action underlying C. difficile pathogenesis.
Antibiotic-induced dysbiosis can establish a favorable condition for the germination of C. difficile spores. The vegetative form of C. difficile, thereafter, can induce pathogenesis mainly by toxin production (TcdA, TcdB, and CDT), SlpA, flagellin, and PG. C. difficile-derived PG interacts with NOD1 to stimulate neutrophil recruitment and CXCL1 production. Neutrophil infiltration and translocation to inflamed tissue result in NET formation. SlpA interaction with TLR4 can accelerate DC maturation and activation. Similar to the stimulation of TLR2 by CDT, flagellin-mediated activation of TLR5, which is promoted by TcdB, triggers transcriptional factors NF-κB and MAPK to provoke the expression of inflammatory cytokines. TcdA, however, mainly triggers programmed cell death pathways and eventually leads to apoptosis, necrosis, and pseudomembrane formation. CDT, C. difficile transferase; CXCL1, CXC chemokine ligand 1; DC, dendritic cell; MAPK, mitogen-activated protein kinase; NET, neutrophil extracellular trap; NF-κB, nuclear factor kappa B; NOD1, nucleotide-binding oligomerization domain 1; PG, peptidoglycan; SlpA, surface layer protein A; TcdA, toxin A; TcdB, toxin B; TLR, Toll-like receptor.
Colonization and germination of C. difficile spores are critical in initiating this toxin-mediated infection, usually occurring in the context of antibiotic-induced dysbiosis. This process is facilitated by adherence to the mucus layer and microbe-microbe interactions with mucin-degrading bacteria. C. difficile uses intestinal mucin as a chemoattractant and energy source with the aid of other gut microbes, including Akkermansia muciniphila, Bacteroides thetaiotaomicron, and Ruminococcus torque.15 C. difficile has been shown to form intestinal biofilms in vivo, with substantial co-colonization with Fusobacterium species.16 These processes result in an acidic intestinal mucus layer, which consists of a higher level of MUC1 and lower MUC2 production.17
Toxins are a major virulence factor, and many C. difficile ribotypes produce up to three distinct toxins: toxin A (TcdA), toxin B (TcdB), and C. difficile transferase or binary toxin (CDT). Almost all clinically significant C. difficile strains produce TcdB, and the epidemic BI/NAP1/027 strain can produce all three toxins. The translocation of C. difficile toxins through receptor-mediated endocytosis leads to pore formation in the endosomal membrane, resulting in actin cytoskeleton disruption and cell rounding, changes in cytokine secretion, and impaired cell proliferation and barrier integrity18; these manifest clinically as diarrhea. Moreover, the induction of cellular apoptosis, especially by TcdA, may contribute to the development of pseudomembranes, crypt damage, and necrotic lesions.19 Activation of inflammatory transcription factors, such as mitogen-activated protein kinase (MAPK) and nuclear factor kappa B (NF-κB), trigger colonic inflammation and the acute influx of host immune cells.20 Also, CDT toxin can trigger the activation of MAPK and NF-κB downstream in a Toll-like receptor 2 (TLR2)/TLR6-dependent manner.21
In addition to secreting toxins, the vegetative cells express other intrinsic immunogenic factors. C. difficile cell wall peptidoglycan (PG) can stimulate CXC chemokine ligand 1 (CXCL1) production and neutrophil infiltration in a nucleotide-binding oligomerization domain 1 (NOD1)-dependent manner. C. difficile surface layer proteins (SLPs) are also involved in the activation of the host innate and adaptive immune response through their effects on the maturation of dendritic cells (DCs).22 C. difficile flagellin stimulation by TLR5 also results in the activation of NF-κB and p38 MAPK in the host epithelial cells. Additionally, TcdB pre-treatment in vitro can potentiate flagellin-induced inflammatory cytokine secretion.23,24
The interaction of C. difficile with other enteric pathogens can also influence its fitness and virulence. Enterococci enriched in the C. difficile-infected gut co-localize with C. difficile in the lumen and in biofilms, markedly enhancing C. difficile colonization and survival.25 Mutually beneficial, C. difficile toxin production is enhanced in the presence of Enterococcus faecalis, and E. faecalis growth is significantly increased in the presence of C. difficile toxin in mouse models.25 Additionally, the core metabolism of C. difficile is significantly altered in the presence of enterococci through the arginine deiminase pathway: E. faecalis depletes arginine and exports high levels of extracellular ornithine, which in turn can be utilized by C. difficile for energy.25
C. difficile recurrence
Approximately 50% of rCDI cases result from reinfection by the original strain.26 The recommended therapy for CDI, vancomycin, contributes to recurrence risk because it is a broad-spectrum antibiotic against Gram-positive bacteria and thus further perpetuates dysbiosis and a loss of colonization resistance. Other risk factors for rCDI include advanced age, concomitant antibiotic use, gastric acid suppression, gastrointestinal surgery, chemotherapy, hematopoietic stem cell transplant, cirrhosis, inflammatory bowel disease (IBD), prior CDI, and infection with a hypervirulent strain such as NAP1/B1/027.27,28 Additionally, recent evidence suggests that the fibronectin-α5β1- and vitronectin-αvβ1-dependent endocytosis of C. difficile spores into gut mucosa significantly contributes to spore persistence and rCDI, as bclA3 gene deletion or pharmacological inhibition of spore internalization reduces recurrence in a mouse model.29 Adaptive host immune responses against TcdA and TcdB may offer some protection against rCDI because high antibody titers are associated with reduced risk30; bezlotoxumab, a monoclonal antibody against TcdB, but not TcdA, has been shown to reduce rCDI risk by 40%.31
FMT
FMT is the process of transferring fecal matter from a carefully screened healthy donor into the gastrointestinal tract of a recipient in order to directly change the recipient’s microbial composition and confer a health benefit.32 Several practice guidelines, including those from the Infectious Diseases of America and the American College of Gastroenterology, have recommended FMT following the second recurrence—or third episode—of CDI.33,34 FMT is regulated in the United States as a biological agent by the Food and Drug Administration (FDA). The use of FMT to treat rCDI is under FDA enforcement discretion,35 and the source of FMT is largely supported by “stool banks” operated by clinical investigators or by OpenBiome. Recently, the FDA approved Rebyota, the first fecal microbiota product for the prevention of rCDI.36 Simultaneous with this decision, the FDA also modified its previous guidance on FMT requiring stool banks that provide FMT products to comply with investigational new drug (IND) requirements.35 Given the high success rate of FMT in preventing C. difficile recurrence,37 there is intense interest in applying this treatment to other chronic conditions associated with intestinal dysbiosis, such as ulcerative colitis (UC).38,39 Therefore, FMT for rCDI has become a paradigm for studying the consequences of host-microbial interactions in relation to pathology and important aspects such as cause and effect relationships.40 In the following sections, we will summarize mechanisms thought to contribute to the prevention of rCDI (see Figure 2).
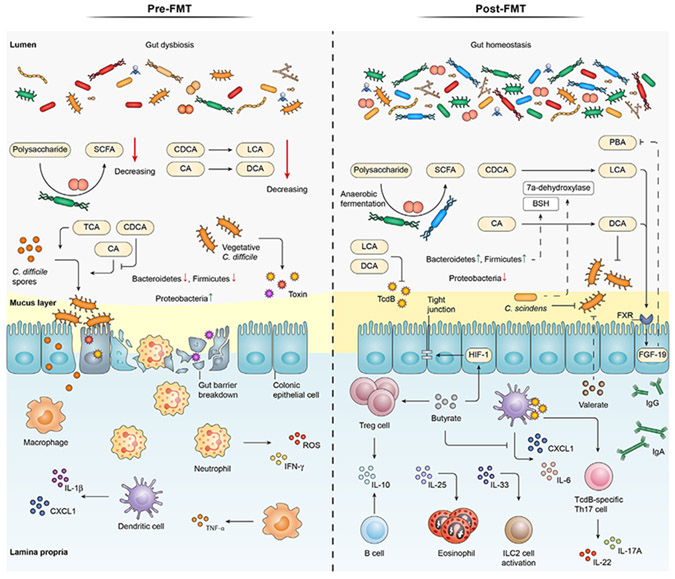
Figure 2. Pre- and post-FMT mechanisms underpinning interactions between C. difficile, intestinal microbiota, and immune system.
During CDI and prior to FMT administration, the gut environment is predominantly accumulated with conjugated bile acids that mostly accelerate C. difficile germination. This is accompanied by the loss of the gut barrier integrity, abundance of inflammation mediators (e.g., ROS, IL-1β, TNF-α, and IFN-γ), impoverishment of Bacteroidetes and Firmicutes phyla, and enrichment of Proteobacteria species. Post-FMT alteration of the gut microbiota composition is characterized by the increased presence of Bacteroidetes and Firmicutes species that promote the production of secondary bile acids (LCA and DCA) from primary bile acids (CDCA, CA). The vegetative growth of C. difficile, the release of TcdB, and the production of PBA are suppressed by LCA and DCA. TcdB interaction with DC leads to the accumulation of TcdB-specific Th17 cells and the subsequent production of IL-17A and IL-22. Furthermore, bacterial production of butyrate can increase the production of tight junction proteins, expand Treg cells, and attenuate the release of pro-inflammatory cytokines. Valerate, however, can interfere with C. difficile colonization. Post-FMT condition also features the abundance of anti-inflammatory cytokines, IgG, and IgA, expansion of eosinophils, and ILC2 cell activation. BSH, bile salt hydrolase; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; FGF, fibroblast growth factor; HIF, hypoxia-inducible factor; IFN-γ, interferon gamma; IgA, immunoglobulin A; IgG, immunoglobulin G; IL, interleukin; ILC2, innate lymphoid type 2 cells; LCA, lithocholic acid; PBA, primary bile acid; ROS, reactive oxygen species; SCFA, short-chain fatty acid; TCA, taurocholic acid; TcdB, toxin B; Th 17, T helper 17; TNF-α, tumour necrosis factor α; Treg, regulatory T cell.
Potential mechanisms contributing to FMT efficacy in rCDI
Restored microbial ecology
The term “microbiome” technically refers to communities composed of all microorganisms, including bacteria, fungi, viruses, protozoa, and parasites, as well as their collective genomes and metabolites in the environment in which they reside. To date, most research has focused on bacterial diversity and community structure before and after FMT.41 Other components of the microbiome, including commensal fungi (the “mycobiome”) and viruses (the “virome”), coexist and interact in ways that may contribute to FMT efficacy,42,43 but these aspects remain understudied.
Effect of FMT on the gut bacteriome.
Prior to FMT, the fecal bacterial community profile of rCDI patients has low diversity and richness, with an over-abundance of potentially pathogenic and putatively inflammatory Proteobacteria, oral bacteria, and oxygen-tolerant bacteria.11,44-47 As early as 7 days after successful FMT, studies have shown a consistent microbial shift, with increased relative abundance of Bacteroidetes and Firmicutes and decreased relative abundance of Proteobacteria; this increased diversity and richness resembles the composition of the donor.7,11,44,46 Using shotgun metagenomics sequencing technology, Aggarwala and colleagues tracked bacterial strains of both donor and recipient and found that 70% of the donor strains, mainly from the orders of Bacteroidales and related Clostridiales, colonized the gut of 13 recipients and persisted up to 5 years post-FMT.48 Bacteroidetes (especially Prevotella species) enrichment after FMT modulates the Bacillis/Clostridia ratio in rCDI patients with concurrent IBD.49 Higher levels of bacterial engraftment, as compared with FMT in other pathologies, were also detected for rCDI patients in three studies using strain-level resolution metagenomics.11-13 Podlesny and colleagues showed that FMT not only resolved taxonomic (i.e., lower α-diversity, altered β-diversity) and functional (i.e., increased relative abundance of oral and oxygen-tolerant species) features of dysbiosis but also resulted in contributions of 60%–90% of donor strains in the recipient microbiota.50 Modeling using meta-analyses of metagenomics data from a wide array of pathologies established that strain engraftment is linked to antibiotic treatment, lower recipient α-diversity, and a higher ratio of species abundance in the donors than in the recipients,11,12 suggesting that high engraftment in rCDI is facilitated by antibiotic-induced dysbiosis and reduced colonization resistance. Given the high success rates of FMT in rCDI, it is likely that engraftment of donor strains plays a key role in re-establishing colonization resistance and preventing CDI recurrence. However, because the recent metagenomics studies compared pre- and post-FMT only in cases with successful treatment outcomes, it is not possible to link engraftment to clinical outcomes. Additional studies are needed to confirm that bacterial engraftment is necessary and essential for FMT to work.
The recent strain-level metagenomic analyses provide an ecological framework for the effects of FMT.11,12 Although the ecological dynamics after FMT are complex (with several ecological processes at play), these studies support the importance of deterministic, niche-based processes for post-FMT microbiome assembly, specifically the competition between and exclusion of closely related recipient and donor strains.11 The outcome of such competition is determined by the fitness of the strains and the relative fitness (adaptation to the gut environment) differences of the incoming and recipient strains. Priority effects, which favor early-arriving strains at an ecological site,51 generally support recipient strains in undisturbed communities11 and provide an explanation for the low levels of strain engraftment in patients with undisrupted microbiota.11 In rCDI patients, depletion of the resident microbiota by antibiotics frees up ecological niches, resulting in increased donor strain engraftment, and effectively overcoming priority effects. In the absence of further perturbation, such as repeated antibiotic exposure or underlying chronic conditions linked to dysbiosis such as IBD, the newly established microbial community appears to remain stable over time.52
Effect of FMT on the gut virome.
A stable and individual-specific viral community exists in a healthy human gut, dominated by temperate bacteriophages, mostly members of crAss-like, Caudovirales, and Microviridae bacteriophages.53-55 While phages act as important modulators of bacterial community structure and metabolism, and their metagenomic composition has been associated with specific diseases, much remains un-known about their actual behavior in the gut.56 Recently, bacteriophages have been shown to modulate both taxonomic composition and functional capacity of the gut microbiome.56-58 For example, Hsu and colleagues showed that bacteriophage transfer in a mouse model nearly altered all Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, including amino acids, peptides, carbohydrates, lipids, nucleotides, cofactors, vitamins, and xenobiotics.56 Campbell and colleagues showed that Bacteroides phage BV01 altered the genome-wide transcriptome profiles of bile acids in vitro.58 However, few studies have examined the gut virome/phageome in the context of FMT for rCDI. They have observed different gut viral abundance and compositions between rCDI patients and stool donors, as well as changes in the recipient virome following FMT. However, our current understanding of the causal role of the virome/phageome on the effects of FMT in rCDI remains vastly incomplete because most studies remain correlative. For example, high levels of donor-derived Caudovirales bacteriophages in the recipients are associated with FMT efficacy in a preliminary study.42 Successful FMT is also positively correlated with the relative abundance of temperate crAss phages, a phage thought to predate on members of the genus Bacteroides, which is decreased in rCDI patients.59-61 Fujumoto et al. found an increased proportion of Microviridae in association with a decreased abundance of Proteobacteria in rCDI patients after FMT, suggesting a potential role of lytic Microviridae in modulating the bacteriome.41 Additional evidence supporting the role of the virome in FMT efficacy comes from a pilot study where five patients did not have further CDI recurrence after receiving sterile fecal filtrate. In that study, remarkably, the viral composition of the recipient, consisting of mostly Caudovirales, resembled that of the donor after treatment, while the bacterial composition did not.62 Further supporting evidence of how the virome may contribute to FMT efficacy stems from the use of C. difficile-targeted phage therapy. For example, Nale and colleagues used a cocktail of four C. difficile myoviruses (CDHM1, 2, 5, and 6) to successfully inhibit C. difficile growth and toxin production in a batch fermentation model.63 Meader et al. showed that specific bacteriophages substantially reduced the C. difficile burden in a human colon model.64 Furthermore, colonization of eukaryotic viruses following FMT may contribute to therapeutic efficacy because their presence is critical for gut homeostasis by modulating both the host immunity and the resident microbiome.65-67
The mutualistic and antagonistic interactions between bacteriophages/eukaryotic viruses, bacteria, and the human host will remain difficult to entangle. Other challenges that need to be overcome include technical limitations of viral enrichment in biological samples, extraction and sequencing library biases toward double-stranded DNA (dsDNA) viruses, removal of single-stranded DNA (ssDNA) and RNA viruses, the limited number of annotated viral genome sequences available in reference databases, and the need to refine and modify methods for viral phylogenetics and taxonomic classifications.
Effect of FMT on the gut mycobiome.
Many species of fungi have been detected in the healthy human gut and may play an important role in intestinal homeostasis and disease pathogenesis. Fungi capable of growing in and colonizing the gut are limited to a small number of species, mostly Candida yeast.68 An increase in Candida spp. has been consistently observed to inversely correlate with bacterial diversity across many chronic diseases43,69,70; thus, this can be seen as a marker of dysbiosis. Other commonly detected fungi have dietary or environmental sources (Saccharomyces and Aspergillus) and likely also contribute to microbial ecology.68
Few studies have examined the gut mycobiome in the context of FMT for rCDI. A higher relative abundance of Saccharomyces and Aspergillus has been reported in CDI recipients after successful FMT, whereas non-responders displayed a prominent presence of Candida.43 In addition, the abundance of Candida albicans in donors and Yarrowia spp. in recipients prior to FMT are shown to be negatively correlated with FMT efficacy.43,71 Anti-fungal treatment such as nystatin was associated with the re-establishment of FMT efficacy in a mouse model29 and simultaneously altered the gut mycobiome. However, understanding the effect of the mycobiome on FMT is in its infancy, and further studies are required to characterize the role of the mycobiome after controlling for dietary sources. It is crucial to improve methodological shortcomings in characterizing mycobiota. Specifically, the 18S rRNA gene sequence typically outperforms other markers (e.g., internal transcribed spacer 1 [ITS1], ITS2, and 28S rRNA) because of its ability to amplify and discriminate different species. However, the multicopy nature of rRNA regions in several filamentous fungi results in a strong bias toward those with more copies. In contrast, the ITS region is the standard marker for fungal DNA barcoding. However, primers amplifying ITS1 lead to preferential amplification of on-Dikarya, while ITS2 is biased toward ascomycetes.72 Lastly, the length of the ITS1 and ITS2 markers vary from 50 bp to several kb.73 Incorrect mapping, and thus classification, leads to the inclusion of false positives or the exclusion of valid taxa associated with FMT.
Changes in microbial-derived metabolites
Broadly speaking, the two classes of metabolites best described in FMT studies are bile acids and short-chain fatty acids (SCFAs); these are reviewed below.
Bile acids.
Bile acids are steroids synthesized in the liver from cholesterol, facilitating the absorption of fat and fat-soluble nutrients and also acting as signaling molecules.74 They regulate glucose and energy metabolism as well as bile acid homeostasis through the farnesoid X receptors (FXRs)-fibroblast growth factor (FGF) axis. Primary bile acids (PBAs) produced by the host are exclusively transformed into secondary bile acids through bacteria upon secretion into the intestine, and both forms have been demonstrated to impact C. difficile pathogenesis in addition to host physiology.
Two key gut bacterial enzymes, absent in mammals, are known to facilitate bile acid transformation. First, the taurine and glycine groups can be deconjugated by bile salt hydrolases (BSHs), encoded by genes widely distributed in commensal bacteria, including Bacteroidetes, Firmicutes, and Actinobacteria. Further transformation can occur via a second step, catalyzed by a bile acid inducible gene (baiCD)-encoded 7α-dehydroxylase, which transforms PBAs (cholic acid [CA] and chenodeoxycholic acid [CDCA]) to their respective secondary bile acids: deoxycholic acid (DCA) and lithocholic acid (LCA). Although only a small fraction of bacteria, mainly clusters XIVa and IV Clostridia (e.g., Clostridium scindens), contain 7α-dehydroxylase,75,76 recent studies suggest an underappreciated role for the microbiota in producing other secondary bile acids, including conjugation to other amino acids.77
A large body of work has established the important contribution of bile acids to CDI pathogenesis, based on in vivo and correlative studies. C. difficile spores possess a soluble pseudoprotease receptor, CspC, which is stimulated by cholate-derived bile acids, promoting its germination.78,79 However, this process is competitively inhibited by CDCA.80 In vivo, the inhibitory action of CDCA is probably limited by its low abundance after enterohepatic recirculation.81 Collectively, members of the cholic acid family, including taurocholic acid (TCA) potently induce in vitro C. difficile spore germination, while members of the CDCA family, including LCA and ursodeoxycholic acid (UDCA), inhibit spore germination and growth.79,80,82
Increased levels of PBAs (especially TCA), coupled with diminished secondary bile acids, have been observed in rCDI patients prior to FMT. This altered bile acid composition is restored following successful FMT to resemble that of healthy donors,83-85 which is associated with reversal of intestinal dysbiosis. Furthermore, increased levels of PBAs were also accompanied by reduced BSH levels in rCDI patients prior to FMT.83 Successful FMT has been shown to enrich the gut for BSH-producing microorganisms and restore BSH functionality.85 Similarly, the baiCD operon coding for 7α-dehydroxylase was also lacking in the pre-FMT samples.85 In a landmark study, Buffie and colleagues demonstrated that colonization resistance against C. difficile could be established by the administration of a single bacterium, C. scindens, to antibiotic-treated mice, resulting in the recovery of microbial 7α-dehydroxylase activity that increased DCA levels.86 Furthermore, C. scindens has an inhibitory effect on C. difficile through the secretion of tryptophan-derived antibiotics, 1-acetyl-β-carboline and turbomycin A, which is further augmented in the presence of DCA and LCA.87
Although bile-acid transforming bacteria appear to be involved in C. difficile resistance, the host can also modulate the circulation of PBAs, depending on their presence. Bile acids vary in degrees of affinity for FXR receptors, with CDCA being the most potent endogenous agonist and DCA and LCA being moderate agonists.88 Following FMT, the increased levels of secondary bile acids LCA and DCA, and reduced PBAs CDCA and CA, are associated with upregulation of ileal FXR signaling and a rise in circulating FGF-19 in rCDI patients. The rise in secondary bile acids LCA and DCA and their moderate but collective activation of the FXR receptors may compensate for the decreased level of a more potent ligand CDCA,88 resulting in reduced hepatic PBA synthesis through a negative feedback response,81 creating an unfavorable environment for C. difficile germination. Although these are interesting preliminary human data, it would be challenging to validate the impact of FXR signaling on C. difficile in a mouse model because tauro-beta-muricholic acid (TβMCA), a naturally occurring FXR antagonist in mice, is not found in humans.89
The concept that restored bile acid metabolism plays a key role in establishing colonization and FMT efficacy was recently challenged by Aguirre and colleagues.90 Using a germ-free mouse model with Cyp8b1−/− mutation (cholic acid deficiency), they observed no difference in disease susceptibility between Cyp8b1−/− and Cyp8b1+/− strains mono-associated with C. scindens, despite the absence of cholate-derived secondary bile acids in the Cyp8b1−/− mice. This suggests that 7α-dehydroxylation is dispensable for protection against CDI. The authors demonstrated the ability of C. difficile to use the Stickland pathway to metabolize amino acids to support its growth in the gut, as evidenced by lower proline and glycine (Stickland substrates) and increased 5-aminovalerate (a Stickland metabolic product) in mice mono-colonized with C. scindens, suggesting bile-acid-independent mechanisms for C. difficile to overcome colonization. The importance of Stickland metabolism for C. difficile was also recently observed in another independent study, where mono-colonization of germ-free mice with another amino-acid-fermenting bacterium, Paraclostridium bifermentans, could attenuate CDI.91
SCFAs.
SCFAs are produced by the gut microbiota during the anaerobic fermentation of carbohydrates and amino acids. Once produced, they are absorbed by intestinal epithelial cells (IECs). A multiomics analysis first demonstrated markedly reduced SCFA concentrations in antibiotic-treated murine models.92 The same study noted that a higher concentration of SCFAs provided protection from C. difficile growth. An analysis of the human stool samples following FMT also showed a recovery of the major SCFAs: acetate, propionate, and butyrate.93 Furthermore, this increase was positively correlated with the FMT-induced restoration of unclassified families of Clostridiales and Firmicutes, such as Lachnospiraceae and Ruminococcaceae, which are known SCFA producers.94
The administration of butyrate in an acute CDI mouse model directly promoted the maintenance of the gut epithelial barrier via a hypoxia-inducible factor (HIF)-1-dependent mechanism.95 Butyrate-activated stability in HIF-1 increases epithelial tight junctions of IECs and can potentially resist C. difficile toxin-mediated damage. Although this study found no influence of butyrate on toxin production or C. difficile colonization, it did identify direct protection of IEC integrity and a butyrate-mediated immune-modulatory effect by increased anti-inflammatory cytokine interleukin-10 (IL-10) and decreased pro-inflammatory cytokine IL-6 and chemokine ligand 1.95 Additionally, butyrate attenuated intestinal inflammation by facilitating the extrathymic generation of T regulatory (Treg) cells, a population of T cells able to suppress inflammation, which are discussed further in the adaptive immune section below.96
Another SCFA, valerate, may also play an important role in mediating FMT efficacy. In a chemostat model of CDI, the recovery of valerate was observed only after FMT, unlike other SCFAs that recovered upon antibiotic cessation.97 Although valerate inhibited vegetative growth of several C. difficile ribotypes in a dose-dependent manner, it had no effect on other commensals.97 The 95% reduction of C. difficile total viable counts in mice after oral administration of valerate, and the sustained post-FMT recovery of donor-like valerate concentration in rCDI patients stool samples, further validate the importance of valerate, both in vivo and in vitro.98
Immune-mediated mechanisms of FMT
Although CDI pathogenesis is largely due to the actions of TcdA and TcdB on the IECs,99 most FMT studies have focused on clinical and microbiota-related changes, while immune-related changes remain poorly understood. This section will focus on what is known about the immune effects of FMT in C. difficile patients and how the microbiome influences the immune system. Effects on innate immunity. The innate immune system primarily responds to microbiota in a non-specific manner by cell surface pattern recognition receptors (PRRs) binding to microbe-associated molecular patterns (MAMPs). Activation of PRR signaling results in inflammation and the recruitment of phagocytic cells, such as macrophages and neutrophils. Therefore, some FMT-induced inflammation may be protective in eliminating residual C. difficile via phagocytosis, while an ideal treatment would have minimal induction of pro-inflammatory cytokines, such as IL-23 and IL-6, against novel commensal strains.
Eosinophils are an important innate immune cell at mucosal surfaces and may have a protective role in CDI, with undetectable eosinophil counts associated with increased in-hospital mortality and severe sepsis.100 Similarly, a mouse study found that the virulence of the NAP1/027 C. difficile strain was enhanced by suppressing the eosinophilic response through binary toxin CDT.101 Interestingly, restoration of the microbiota-regulated cytokine IL-25 drove colonic accumulation of eosinophils in mice and protected against CDI,102 and a higher level of IL-25 was found in colon biopsies after FMT of CDI patients than in the pre-treatment biopsies.103 Similarly, another microbiota-regulated cytokine, IL-33, increased following FMT in mice and could prevent CDI-associated mortality by activating group 2 innate lymphoid cells.104 Understanding which commensals enhance secretion of these “protective” cytokines and attenuate “damaging” cytokines will help to determine the ideal FMT composition.
Effects on adaptive immunity.
The key adaptive immune cells are T and B cells, which mediate long-lived cellular and humoral immunity, respectively. The concept that adaptive immunity may contribute to FMT efficacy is supported by a study showing that, in mice lacking T and B cells, CDI persisted after FMT while immunocompetent mice fully recovered.105 The two main types of T cells are cytotoxic CD8+ T cells and helper CD4+ T cells (Th), with Th cell subsets including type 1 (Th1), type 2 (Th2), type 17 (Th17), and Treg cells, which encompass subsets that express the transcription factor FOXP3 (FOXP3+ Tregs) and those that are FOXP3neg but secrete high levels of IL-10 (Tr1 cells). Tregs can recognize both self and foreign antigens and play crucial roles in maintaining self-tolerance as well as preventing immunopathology through restraining inflammatory responses and mediating tissue protective and restorative effects.106 Strikingly, a study of CDI in mice deficient in Tregs found that they had increased inflammatory mediators, compromised engraftment of donor bacteria, and could not be cured by FMT.105 Various metabolites, such as SCFAs and vitamins A and D, have been shown to increase Treg numbers and/or function,107 and more recently a role has emerged for secondary bile acids in promoting Treg development. Therefore, it is possible that FMT outcome may be influenced by a recipient’s diet, as a diet rich in particular metabolites can activate Tregs that will establish and maintain tolerance to donor microbiota.
Th17 cells, which secrete IL-17A, play an important role in gut homeostasis and anti-fungal responses, while IFNγ-secreting Th1 (and CD8+) cells are involved in important responses to intracellular pathogens and IL-4-producing Th2 cells are involved in responses to parasites. However, in a dysregulated gut, both Th1 and Th17 cells can drive excessive inflammation.108
Recent work has identified robust CD4+ T cell responses to TcdA and TcdB in CDI patients, with these responses largely composed of Th17 cells. Importantly, rCDI patients had significantly reduced levels of circulating TcdB-specific Th17 cells109 compared with healthy controls. In a follow-up study, it was identified that successful FMT results in a considerable increase in TcdB-specific Th17 cells in rCDI patients, with preliminary data showing that, post-FMT, these cells had increased secretion of IL-17A and IL-22 cytokines.110
A simultaneous increase in systemic anti-toxin IgA and IgG levels was also detected after successful FMT,110 which has been previously associated with a reduced risk of CDI recurrence.108 Consistent with the findings of Cook et al., FMT-induced recovery of CDI in immunocompetent mice was associated with successful engraftment, increased Th17 cells, and increased levels of IL-17A and IL-22 in the large intestine lamina propria.105 However, Th1 cell-deficient (Tbet−/−), IL-17A−/−, and IL-22−/− mice all recovered following FMT, suggesting that the FMT-mediated CDI cure in mice is not solely dependent on Th1 or Th17 cells.105 It has also been proposed that immunosenescence (age-associated immune decline) may contribute to FMT failure,111 as an observational study of four patients receiving sequential FMT for antibiotic refractory fulminant CDI found increased circulating immunosenescent cell populations in a non-responder compared with three FMT responders.111 These data suggest that another mechanistic function of FMT is shaping the total and TcdB-specific CD4+ T cell repertoire, and potentially inducing an anti-aging effect. Taken together, these data suggest that an ideal FMT composition will need to activate a precise, and as yet undetermined, balance of Tregs and other Th cells to preserve intestinal homeostasis. Mechanistic insights gained from in vivo studies highlight the significance of colon-specific immune responses, which require further validation in clinical studies.
Although these human studies of FMT-treated CDI are largely preliminary, they suggest that the adaptive immune system is essential for FMT engraftment and that anti-toxin antibodies as well as Treg and Th17 cell functions are associated with increased efficacy.105,109,110 From studies of host-microbe interactions in the gut, we know how important the microbiome is in shaping immune development, tolerance, and long-lived immunity. Therefore, a big question that remains unexplored is how changing the microbiome through FMT may affect long-lived protective immunity, with one study showing reduced T cell responses to a childhood vaccine post-FMT.110 It will be critical for larger studies to assess immune changes, ideally in both peripheral blood and gut tissue, in parallel with microbiome/metabolome changes to understand the complete mechanisms underpinning the FMT efficacy.
Epigenetic-related mechanisms
In CDI, gut dysbiosis and reduced microbial diversity are likely to alter the levels of nutrients and metabolites, impacting epigenetic pathways and altering gene expression. Recently, FMT has emerged as a useful tool to explore the interrelation between microbiota composition and microRNA (miRNA) expression. To this effect, one study has reported the suppression of circulating miRNAs, small non-coding RNAs that post-transcriptionally regulate gene expression, in two independent cohorts of rCDI patients.112 This effect was subsequently reversed following successful FMT and replicated in FMT-treated mouse models and ex vivo human colonoids. Analyses confirmed that TcdB mediated the suppressive effects of CDI on the miRNAs by dysregulating Drosha expression, an enzyme that plays a prominent role in miRNA biogenesis.112
Specific miRNAs that were upregulated in both rCDI patients and mouse models following successful FMT included miR-26b, miR-23a, miR-150, and miR-28-5p. Overexpression of these miRNAs in human blood resulted in reduced mRNA levels of FGF-21, IL-12B, IL-18, and TNF receptor superfamily member 9 (TNFRSF9) inflammatory gene targets, respectively.112 In the same study, the investigators also determined that combined overexpression of miR-23a-3p and miR-150-5p could protect against TcdB-induced damage to the IEC (see Figure 3). There is still limited understanding of the impact of FMT on the human circulating, fecal, and tissue miRome and wider host epigenome, and further mechanistic studies are required to investigate the long-term epigenetic effects of FMT for rCDI and other disease states associated with gut dysbiosis. Future studies will necessitate comprehensively mapping epitranscriptomic changes associated with CDI, FMT, and dietary manipulation strategies. FMT-regulated miRNAs may represent unique therapeutic targets, which alone or combined with live biotherapeutics may augment therapeutic efficacy against C. difficile and help counteract drug resistance.113
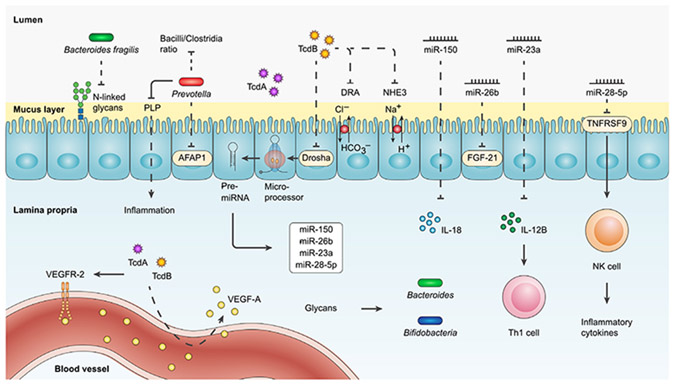
Figure 3. Post-FMT interaction of miRNAs and glycans with the immune system and the gut microbiota.
C. difficile-derived TcdB can reduce the expression of miR-150, miR-26b, miR-23a, and miR-28-5p, which downregulate the expression levels of IL-18, FGF-21, IL-12B, and TNFRSF9 inflammatory gene targets, respectively. IL-12B suppression by miR23a prevents the proliferation of Th1 cell. Likewise, the overexpression of miR-28-5p prevents TNFRSF9 production and subsequently reduces NK cell activation and inflammatory cytokine secretion. TcdB further inhibits the activity of intestinal ion transporter NHE3 and Cl−/HCO3− exchanger protein DRA. TcdB, along with TcdA, can induce vascular permeability and thereby increase the accumulation of VEGF-A and probably the activation of the VEGFR-2. The elevated presence of glycans, in return, accelerates the growth of Bacteroides and Bifidobacteria genera. Bacteroides fragilis can reduce the proportion of N-linked glycans. Prevotella strains decrease the Bacilli/Clostridia ratio, suppress the expression of AFAP1, and catabolize PLP to attenuate inflammation. AFAP1, actin filament-associated protein 1; DRA, down-regulated in adenoma; FGF-21, fibroblast growth factor 21; NHE3, Na+/H+ exchanger 3; NK, natural killer; PLP, pyridoxal-5-phosphate; TcdA, toxin A; TcdB, toxin B; Th 1, T helper 1; TNFRSF9, TNF receptor superfamily member 9; VEGF-A, vascular endothelial growth factor A; VEGFR-2, VEGF-A receptor.
INSIGHTS AND FUTURE DIRECTIONS
As highlighted in this review, our mechanistic understanding of how FMT works in rCDI is still incomplete. Ecological, metabolic, immunological, and epigenetic mechanisms have all been studied at different depths (summarized in Table 1), but their individual contributions remain unclear, and the causal components (bacteria, viruses, fungi, specific metabolites) that contribute to clinical efficacy are still not fully elucidated. The recent meta-analyses11-13 that combined metagenomics with strain-level resolution and predictive modeling clearly established antibiotic treatment, recipient factors (e.g., α-diversity and species distribution), and donor-recipient complementarity as important determinants of engraftment, but these studies were not sufficiently powered to determine whether engraftment is necessary for a successful outcome. Nevertheless, the ecological and statistical approaches established provide frameworks that will inform future studies.
Table 1.
Hypothesized mechanisms of the action of FMT in recurrent C. difficile infection (rCDI)
| Mechanisms | Evidence | Knowledge gaps |
|---|---|---|
| Restored colonization resistance through bacterial engraftment and/or modulation of non-bacterial components | animal studies:
|
|
| Direct effect on C. difficile or through modulation of microbial ecology by virome/phageome |
in vitro studies:
|
|
| Inhibition of C. difficile growth and/or germination through bacteria-derived metabolites |
in vitro studies:
|
|
| Modulation of host immune response | animal studies:
|
|
| Modulation of host epigenetic response | human and animal mechanistic studies:112
|
|
Even though FMT is highly effective in preventing rCDI, we still do not understand why a small portion of rCDI patients do not benefit from FMT. Additional ecological factors, such as diet and host genetics, have not been considered in studies of FMT in rCDI and may potentially be the missing links to these “failed” FMT cases. Although diet has been proposed to be relevant because it affects the ecology of microbiome dynamics after FMT, the topic has received virtually no experimental validation. As the relative importance of host genetics on microbiome assembly is rather low (and explains <10% variation),118 diet might influence FMT outcomes in several ways. First, the diet of the recipients would influence the diversity of substrates and resources (in the form of nutrients) that are available for the incoming microbes, therefore directly influencing the niches available for engraftment. As such, pairing donor-recipient combinations based on their dietary patterns and preferences could further optimize efficacy because the donor microbiota would be pre-adapted to the recipient’s diet. Other knowledge gaps to be addressed include whether engraftment or live bacteria are necessary for efficacy, how other non-bacterial components modulate microbial ecology, what relative contributions of adaptive or innate immune response play in outcomes, or whether specific immunological factors such as immune senescence119 or low IgA diversity may affect efficacy.120
Clinical and mechanistic insights provided by FMT in rCDI have extended the potential therapeutic value of FMT to other dysbiosis-associated chronic conditions. A recent search on clinicaltrials.gov yielded 429 studies utilizing FMT in a variety of conditions (accessed on February 7, 2023), highlighting the intense interest surrounding microbiome-based therapeutics. It is worth noting that the pathogenesis of many chronic conditions is complex and multifactorial, where dysbiosis is only a piece of the puzzle and potentially not causal to the pathology.121 For example, IBD is thought to be caused by immune dysregulation, gut dysbiosis, environmental triggers, and genetic susceptibility. Thus, the magnitude of therapeutic benefits and the degree of engraftment following FMT in these chronic conditions would not be expected to be as high or as durable as in rCDI, where dysbiosis is the main pathogenic driver.11,12 The best example is in UC, a form of IBD, the indication for which the strongest evidence from randomized, placebo-controlled trials exists. Irrespective of how FMT was delivered, how frequently FMT was given, or whether the FMT was from a single donor or from pooled multiple donors, the remission rate in mild to moderate UC was only 30%–40%.38,39,122,123 Furthermore, all these studies are relatively short term, with primary outcomes assessed around 7–12 weeks after FMT. Additionally, many responders during the trials ended up with disease flares after they completed the trial.38 FMT for irritable bowel syndrome, on the other hand, has generated conflicting results in randomized, placebo-controlled trials, with some studies demonstrating modest efficacy,124,125 while others showed no benefits.126-128 Promising preliminary results also came from FMT for other indications, such as metabolic syndrome,129,130 hepatic encephalopathy,131 checkpoint-inhibitor-induced colitis,132 graft-versus-host disease,133,134 decolonization of multidrug-resistant organisms,135 to name a few. However, much remains unknown, such as how to select patients most likely to respond favorably to FMT, how to design optimal dosing regimens, or how to improve durability of responses.
Although highly effective, FMT also has several disadvantages. First, there is a risk of transmitting an infectious agent because stool is sourced from a donor, and such risk is highlighted by a death due to extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli which prompted the FDA safety warning.10,136 Emerging pathogens detectable in stool, such as SARS-CoV-2 and monkeypox virus contribute to FMT safety concerns,137,138 making stringent donor testing protocols essential. Second, the composition of each FMT treatment is not known and even varies over time from the same donor. Stool is a complex mixture, and the current regulatory frameworks for drugs do not apply, making regulatory approvals challenging. Third, there is a lack of long-term safety data. As many chronic conditions are associated with intestinal dysbiosis, a donor phenotype could potentially be transferred to a recipient. Fourth, the precise mechanisms of action of FMT remain unknown. Better treatment options for rCDI that are targeted, safe, and donor independent are thus desired.
By recognizing the disadvantages of FMT, a reductionist approach has been taken with the development of more refined live biotherapeutic products. A mixture of 6 phylogenetically diverse intestinal bacteria can resolve relapsing CDI in mice.114 Furthermore, SER-109,115 a product containing only the spore-forming Firmicutes, and MET-2,116 a product which contains 40 strains of rationally selected commensal microbes, have shown promise in treating rCDI patients, with success rates of 88% and 79%, respectively, comparable to those seen with FMT. Interestingly, a mixture of 12 strains of bacteria is not as effective as FMT for rCDI patients in a randomized clinical trial, showing efficacy of 52% when compared with 76% in FMT.139 They highlight that the full microbial spectrum in FMT is not required for clinical efficacy, at least for rCDI. Perhaps there are key strains that can provide “scaffolding” or early functional restoration, and they are essential and permissive for medium and late colonizers. The minimum number of microbes or which microbes required in a consortium to retain efficacy, and whether this is host-factor dependent remains unknown.
To deepen our understanding of FMT mechanisms and to establish causality, human intervention trials using not just stool but stool derivatives with defined compositions and characteristics, or with a defined consortium of bacterial, viral, and metabolic components alone and/or in combinations, will serve as an important experimental platform. These trials should use well-defined outcomes and combine multiomics (metagenomics, transcriptomics, proteomics, metabolomics), host-based (immune phenotyping), and dietary or other environmental factors that analyze samples from both recipients and donors with predictive modeling (e.g., with machine learning or artificial intelligence [AI]) using an ecological framework to determine the relative importance of major determinants of clinical and ecological outcomes. The challenges of integrated multiomics research not only lie with addressing the shortcomings of each “omic” technology but also with how to integrate different molecular datasets. Data libraries need to be further developed, particularly for the virome and metabolome. Additional bioinformatics tools are required to standardize normalization and integration of multi-model experiments. Ideally, clinical assessment and sampling in the recipients should be longitudinal to allow statistical approaches (e.g., mediation analyses) that permit the identification of causal factors. Although animal models and other models (e.g., organoids, organs-on-chips) have limitations in their translatability, they remain important to establish mechanisms, confirm causality, and identify causal components (see Figure 4). This work will provide information to refine FMT approaches (for example, through donor-recipient pairings based on the microbiome and/or diet) while we await the development of refined and targeted biotherapeutics to replace FMT.
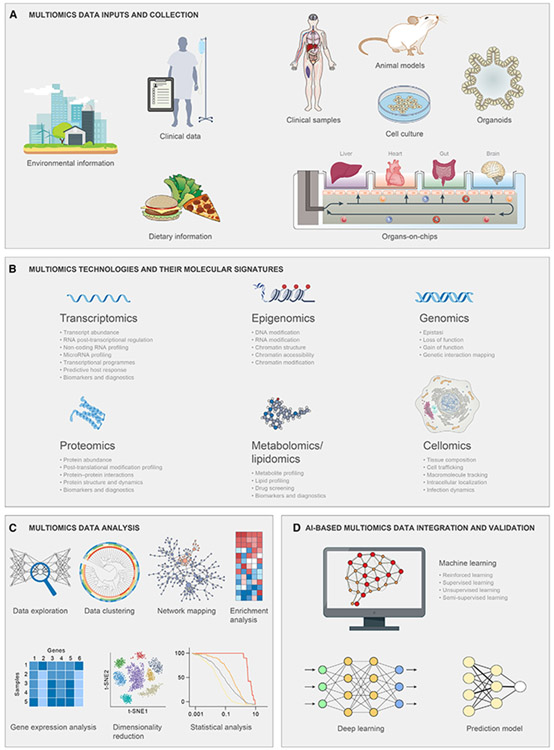
Figure 4. Using integrated multiomics approaches to FMT.
An overview of key omics technologies used in FMT research to capture the relevant molecular signatures. (A) Multiomics data can be obtained from phenomic inputs and a wide spectrum of in vivo and/or in vitro research study designs.
(B) Epigenomics, genomics, transcriptomics, proteomics, and metabolomics are complementary to each other, providing a comprehensive framework for research on FMT. Together, multiple pieces of information from a multiomics method can provide a comprehensive cellular readout that is absent in the outcomes of a single omic approach.
(C) The acquired data from different multiomics technologies should be processed and analyzed accordingly, namely data exploration, clustering, network mapping, enrichment analysis, gene expression analysis, dimensionality reduction techniques, and statistical analysis.
(D) Machine learning, deep learning, and prediction models have paved the way for the integration and validation of separated layers of multiomics data. AI, artificial intelligence.
ACKNOWLEDGMENTS
D.K. acknowledges support by the Canadian Institutes of Health Research and the University of Alberta Hospital Foundation.
Footnotes
DECLARATION OF INTERESTS
D.K. has served on the adjudication board for Finch Therapeutics and has received consulting fees and a speaking honorarium from Rebiotix/Ferring Pharmaceuticals. A.M.S. has received consultation fees from Finch Therapeutics and Rebiotix/Ferring Pharmaceuticals.
REFERENCES
- 1.Smits WK, Lyras D, Lacy DB, Wilcox MH, and Kuijper EJ (2016). Clostridium difficile infection. Nat. Rev. Dis. Primers 2, 16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson S, Lavergne V, Skinner AM, Gonzales-Luna AJ, Garey KW, Kelly CP, and Wilcox MH (2021). Clinical practice guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 focused update guidelines on management of Clostridioides difficile infection in adults. Clin. Infect. Dis 73, 755–757. [DOI] [PubMed] [Google Scholar]
- 3.Fekety R, McFarland LV, Surawicz CM, Greenberg RN, Elmer GW, and Mulligan ME (1997). Recurrent Clostridium difficile diarrhea: characteristics of and risk factors for patients enrolled in a prospective, randomized, double-blinded trial. Clin. Infect. Dis 24, 324–333. [DOI] [PubMed] [Google Scholar]
- 4.Keller JJ, and Kuijper EJ (2015). Treatment of recurrent and severe Clostridium difficile infection. Annu. Rev. Med 66, 373–386. [DOI] [PubMed] [Google Scholar]
- 5.Dubberke ER, Lee CH, Orenstein R, Khanna S, Hecht G, and Gerding DN (2018). Results from a randomized, placebo-controlled clinical trial of a RBX2660-A microbiota-based drug for the prevention of recurrent Clostridium difficile infection. Clin. Infect. Dis 67, 1198–1204. [DOI] [PubMed] [Google Scholar]
- 6.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med 368, 407–415. [DOI] [PubMed] [Google Scholar]
- 7.Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, Chang HJ, Coward S, Goodman KJ, Xu H, et al. (2017). Effect of oral capsule- vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: A randomized clinical trial. JAMA 318, 1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly CR, Khoruts A, Staley C, Sadowsky MJ, Abd M, Alani M, Bakow B, Curran P, McKenney J, Tisch A, et al. (2016). Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: A randomized trial. Ann. Intern. Med 165, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perler BK, Chen B, Phelps E, Allegretti JR, Fischer M, Ganapini V, Krajiceck E, Kumar V, Marcus J, Nativ L, et al. (2020). Long-term efficacy and safety of fecal microbiota transplantation for treatment of recurrent Clostridioides difficile infection. J. Clin. Gastroenterol 54, 701–706. [DOI] [PubMed] [Google Scholar]
- 10.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen YB, and Hohmann EL (2019). Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med 381, 2043–2050. [DOI] [PubMed] [Google Scholar]
- 11.Podlesny D, Durdevic M, Paramsothy S, Kaakoush NO, Högenauer C, Gorkiewicz G, Walter J, and Fricke WF (2022). Identification of clinical and ecological determinants of strain engraftment after fecal microbiota transplantation using metagenomics. Cell Rep. Med 3, 100711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt TSB, Li SS, Maistrenko OM, Akanni W, Coelho LP, Dolai S, Fullam A, Glazek AM, Hercog R, Herrema H, et al. (2022). Drivers and determinants of strain dynamics following fecal microbiota transplantation. Nat. Med 28, 1902–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ianiro G, Punčochář M, Karcher N, Porcari S, Armanini F, Asnicar F, Beghini F, Blanco-Míguez A, Cumbo F, Manghi P, et al. (2022). Variability of strain engraftment and predictability of microbiome composition after fecal microbiota transplantation across different diseases. Nat. Med 28, 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abt MC, McKenney PT, and Pamer EG (2016). Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol 14, 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engevik MA, Engevik AC, Engevik KA, Auchtung JM, Chang-Graham AL, Ruan W, Luna RA, Hyser JM, Spinler JK, and Versalovic J (2021). Mucin-degrading microbes release monosaccharides that chemoattract Clostridioides difficile and facilitate colonization of the human intestinal mucus layer. ACS Infect. Dis 7, 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engevik MA, Danhof HA, Auchtung J, Endres BT, Ruan W, Bassères E, Engevik AC, Wu Q, Nicholson M, Luna RA, et al. (2021). Fusobacteriumnucleatum Adheres to Clostridioides difficile via the RadD adhesin to Enhance biofilm Formation in intestinal Mucus. Gastroenterology 160, 1301–1314.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engevik MA, Yacyshyn MB, Engevik KA, Wang J, Darien B, Hassett DJ, Yacyshyn BR, and Worrell RT (2015). Human Clostridium difficile infection: altered mucus production and composition. Am. J. Physiol. Gastrointest. Liver Physiol 308, G510–G524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Liu Z, Perry K, and Jin R (2022). Structure of the glucosyl-transferase domain of TcdA in complex with RhoA provides insights into substrate recognition. Sci. Rep 12, 9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chumbler NM, Farrow MA, Lapierre LA, Franklin JL, and Lacy DB (2016). Clostridium difficile toxins TcdA and TcdB cause colonic tissue damage by distinct mechanisms. Infect. Immun 84, 2871–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Xu S, Xu Q, and Chen Y (2020). Clostridium difficile toxin B induces colonic inflammation through the TRIM46/DUSP1/MAPKs and NF-κB signalling pathway. Artif. Cells Nanomed. Biotechnol 48, 452–462. [DOI] [PubMed] [Google Scholar]
- 21.Simpson M, Frisbee A, Kumar P, Schwan C, Aktories K, and Petri WA (2022). Clostridioides difficile binary toxin is recognized by the toll-like receptor 2/6 heterodimer to induce a nuclear factor-κB response. J. Infect. Dis 225, 1296–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan A, Lynch M, Smith SM, Amu S, Nel HJ, McCoy CE, Dowling JK, Draper E, O’Reilly V, McCarthy C, et al. (2011). A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog. 7, e1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshino Y, Kitazawa T, Ikeda M, Tatsuno K, Yanagimoto S, Okugawa S, Yotsuyanagi H, and Ota Y (2013). Clostridium difficile flagellin stimulates toll-like receptor 5, and toxin B promotes flagellin-induced chemokine production via TLR5. Life Sci. 92, 211–217. [DOI] [PubMed] [Google Scholar]
- 24.Batah J, Denève-Larrazet C, Jolivot PA, Kuehne S, Collignon A, Marvaud JC, and Kansau I (2016). Clostridium difficile flagella predominantly activate TLR5-linked NF-κB pathway in epithelial cells. Anaerobe 38, 116–124. [DOI] [PubMed] [Google Scholar]
- 25.Smith AB, Jenior ML, Keenan O, Hart JL, Specker J, Abbas A, Rangel PC, Di C, Green J, Bustin KA, et al. (2022). Enterococci enhance Clostridioides difficile pathogenesis. Nature 611, 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Czepiel J, Dróżdż M, Pituch H, Kuijper EJ, Perucki W, Mielimonka A, Goldman S, Wultańska D, Garlicki A, and Biesiada G (2019). Clostridium difficile infection: review. Eur. J. Clin. Microbiol. Infect. Dis 38, 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song JH, and Kim YS (2019). Recurrent Clostridium difficile infection: risk factors, treatment, and prevention. Gut Liver 13, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushman FD, Conrad M, Ren Y, Zhao C, Gu C, Petucci C, Kim MS, Abbas A, Downes KJ, Devas N, et al. (2020). Multi-omic analysis of the interaction between Clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe 28, 422–433.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castro-Córdova P, Mora-Uribe P, Reyes-Ramírez R, Cofré-Araneda G, Orozco-Aguilar J, Brito-Silva C, Mendoza-León MJ, Kuehne SA, Minton NP, Pizarro-Guajardo M, et al. (2021). Entry of spores into intestinal epithelial cells contributes to recurrence of Clostridioides difficile infection. Nat. Commun 12, 1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rees WD, and Steiner TS (2018). Adaptive immune response to Clostridium difficile infection: A perspective for prevention and therapy. Eur. J. Immunol 48, 398–406. [DOI] [PubMed] [Google Scholar]
- 31.Wilcox MH, Gerding DN, Poxton IR, Kelly C, Nathan R, Birch T, Cornely OA, Rahav G, Bouza E, Lee C, et al. (2017). Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N. Engl. J. Med 376, 305–317. [DOI] [PubMed] [Google Scholar]
- 32.Gupta S, Allen-Vercoe E, and Petrof EO (2016). Fecal microbiota transplantation: in perspective. Therap. Adv. Gastroenterol 9, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald LC, Gerding DN, Johnson S, et al. (2018). Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis 66, e1–e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly CR, Fischer M, Allegretti JR, LaPlante K, Stewart DB, Limketkai BN, and Stollman NH (2021). ACG clinical guidelines: prevention, diagnosis, and treatment of Clostridioides difficile infections. Am. J. Gastroenterol 116, 1124–1147. [DOI] [PubMed] [Google Scholar]
- 35.Food and Drug Administration (2022). Enforcement policy regarding investigational new drug requirements for use of fecal microbiota for transplantation to treat Clostridioides difficile infection not responsive to standard therapies. https://www.fda.gov/media/86440/download.
- 36.Food and Drug Administration (2022). FDA approves first fecal microbiota product. https://www.fda.gov/news-events/press-announcements/fda-approves-first-fecal-microbiota-product?fbclid=IwAR2We_37cYnGLOcVcU2dY7mTfQ6q5ROmEIZovtVM1ukgNP-pivg0zvgybws.
- 37.Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, and Iqbal TH (2017). Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment. Pharmacol. Ther 46, 479–493. [DOI] [PubMed] [Google Scholar]
- 38.Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, et al. (2017). Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228. [DOI] [PubMed] [Google Scholar]
- 39.Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, Katsikeros R, Makanyanga J, Campaniello MA, Mavrangelos C, et al. (2019). Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: A randomized clinical trial. JAMA 321, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrs T, and Walter J (2021). . Pros and cons: is faecal microbiota transplantation a safe and efficient treatment option for gut dysbiosis? Allergy 76, 2312–2317. [DOI] [PubMed] [Google Scholar]
- 41.Fujimoto K, Kimura Y, Allegretti JR, Yamamoto M, Zhang YZ, Katayama K, Tremmel G, Kawaguchi Y, Shimohigoshi M, Hayashi T, et al. (2021). Functional restoration of bacteriomes and viromes by fecal microbiota transplantation. Gastroenterology 160, 2089–2102.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, Ching JYL, Chan PKS, Chan MCW, Wu JCY, et al. (2018). Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut 67, 634–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo T, Wong SH, Cheung CP, Lam K, Lui R, Cheung K, Zhang F, Tang W, Ching JYL, Wu JCY, et al. (2018). Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat. Commun 9, 3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuentes S, van Nood E, Tims S, Heikamp-de Jong I, ter Braak CJ, Keller JJ, Zoetendal EG, and de Vos WM (2014). Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection. ISME J. 8, 1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seekatz AM, Aas J, Gessert CE, Rubin TA, Saman DM, Bakken JS, and Young VB (2014). Recovery of the gut microbiome following fecal microbiota transplantation. mBio 5. e00893–e00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kellingray L, Gall GL, Defernez M, Beales ILP, Franslem-Elumogo N, and Narbad A (2018). Microbial taxonomic and metabolic alterations during faecal microbiota transplantation to treat Clostridium difficile infection. J. Infect 77, 107–118. [DOI] [PubMed] [Google Scholar]
- 47.Brown JR, Flemer B, Joyce SA, Zulquernain A, Sheehan D, Shanahan F, and O’Toole PW (2018). Changes in microbiota composition, bile and fatty acid metabolism, in successful faecal microbiota transplantation for Clostridioides difficile infection. BMC Gastroenterol. 18, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aggarwala V, Mogno I, Li Z, Yang C, Britton GJ, Chen-Liaw A, Mitcham J, Bongers G, Gevers D, Clemente JC, et al. (2021). Precise quantification of bacterial strains after fecal microbiota transplantation delineates long-term engraftment and explains outcomes. Nat. Microbiol 6, 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azimirad M, Jo Y, Kim MS, Jeong M, Shahrokh S, Asadzadeh Aghdaei H, Zali MR, Lee S, Yadegar A, and Shin JH (2022). Alterations and prediction of functional profiles of gut microbiota after fecal microbiota transplantation for Iranian recurrent Clostridioides difficile infection with underlying inflammatory bowel disease: A pilot study. J. Inflamm. Res 15, 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Podlesny D, Arze C, Dörner E, Verma S, Dutta S, Walter J, and Fricke WF (2022). Metagenomic strain detection with SameStr: identification of a persisting core gut microbiota transferable by fecal transplantation. Microbiome 10, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez I, Maldonado-Gomez MX, Gomes-Neto JC, Kittana H, Ding H, Schmaltz R, Joglekar P, Cardona RJ, Marsteller NL, Kembel SW, et al. (2018). Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife 7, e36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ, et al. (2018). Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23, 229–240.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shkoporov AN, and Hill C (2019). Bacteriophages of the human gut: the "known unknown" of the microbiome. Cell Host Microbe 25, 195–209. [DOI] [PubMed] [Google Scholar]
- 54.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, and Gordon JI (2010). Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, and Holtz LR (2015). Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med 21, 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Lyon L, Bry L, Silver PA, and Gerber GK (2019). Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 25, 803–814.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reyes V, Wu M, McNulty NP, Rohwer FL, and Gordon JI (2013). Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. USA 110, 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell DE, Ly LK, Ridlon JM, Hsiao A, Whitaker RJ, and Degnan PH (2020). Infection with Bacteroides phage BV01 alters the Host Transcriptome and Bile Acid Metabolism in a Common Human Gut Microbe. Cell Rep. 32, 108142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, Dinsdale EA, Cinek O, Aziz RK, McNair K, Barr JJ, et al. (2019). Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol 4, 1727–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Draper LA, Ryan FJ, Smith MK, Jalanka J, Mattila E, Arkkila PA, Ross RP, Satokari R, and Hill C (2018). Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome 6, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siranosian BA, Tamburini FB, Sherlock G, and Bhatt AS (2020). Acquisition, transmission and strain diversity of human gut-colonizing crAss-like phages. Nat. Commun 11, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, Cassidy L, Tholey A, Fickenscher H, Seegert D, et al. (2017). Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 152, 799–811.e7. [DOI] [PubMed] [Google Scholar]
- 63.Nale JY, Redgwell TA, Millard A, and Clokie MRJ (2018). Efficacy of an optimised bacteriophage cocktail to clear Clostridium difficile in a batch fermentation model. Antibiotics (Basel) 7, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meader E, Mayer MJ, Steverding D, Carding SR, and Narbad A (2013). Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe 22, 25–30. [DOI] [PubMed] [Google Scholar]
- 65.Dallari S, Heaney T, Rosas-Villegas A, Neil JA, Wong SY, Brown JJ, Urbanek K, Herrmann C, Depledge DP, Dermody TS, et al. (2021). Enteric viruses evoke broad host immune responses resembling those elicited by the bacterial microbiome. Cell Host Microbe 20, 1014–1029.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campbell DE, and Baldridge MT (2021). Enteric viruses seize their immunomodulatory niche. Cell Host Microbe 29, 858–861. [DOI] [PubMed] [Google Scholar]
- 67.Yang JY, Kim MS, Kim E, Cheon JH, Lee YS, Kim Y, Lee SH, Seo SU, Shin SH, Choi SS, et al. (2016). Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-β production. Immunity 44, 889–900. [DOI] [PubMed] [Google Scholar]
- 68.Hallen-Adams HE, and Suhr MJ (2017). Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.García-Gamboa R, Kirchmayr MR, Gradilla-Hernández MS, Pérez-Brocal V, Moya A, and González-Avila M (2021). The intestinal mycobiota and its relationship with overweight, obesity and nutritional aspects. J. Hum. Nutr. Diet 34, 645–655. [DOI] [PubMed] [Google Scholar]
- 71.Kazemian N, Ramezankhani M, Sehgal A, Khalid FM, Kalkhoran AHZ, Narayan A, Wong GK, Kao D, and Pakpour S (2020). The trans-kingdom battle between donor and recipient gut microbiome influences fecal microbiota transplantation outcome. Sci. Rep 10, 18349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, and Kauserud H (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 10, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galloway-Peña JR, and Kontoyiannis DP (2020). The gut mycobiome: the overlooked constituent of clinical outcomes and treatment complications in patients with cancer and other immunosuppressive conditions. PLoS Pathog. 16, e1008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, and Yokota A (2011). Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141, 1773–1781. [DOI] [PubMed] [Google Scholar]
- 75.Kitahara M, Takamine F, Imamura T, and Benno Y (2000). Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol 50, 971–978. [DOI] [PubMed] [Google Scholar]
- 76.Wells JE, and Hylemon PB (2000). Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl. Environ. Microbiol 66, 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, et al. (2020). Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Francis MB, Allen CA, Shrestha R, and Sorg JA (2013). Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLOS Pathog. 9, e1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sorg JA, and Sonenshein AL (2008). Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol 190, 2505–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sorg JA, and Sonenshein AL (2010). Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol 192, 4983–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mullish BH, and Allegretti JR (2021). The contribution of bile acid metabolism to the pathogenesis of Clostridioides difficile infection. Therap. Adv. Gastroenterol 14, 17562848211017725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thanissery R, Winston JA, and Theriot CM (2017). Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45, 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, Bry L, Clish CB, Alm E, and Korzenik JR (2016). Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther 43, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weingarden AR, Dosa PI, DeWinter E, Steer CJ, Shaughnessy MK, Johnson JR, Khoruts A, and Sadowsky MJ (2016). Changes in Colonic Bile Acid Composition following Fecal microbiota Transplantation Are Sufficient to Control Clostridium difficile Germination and Growth. PLoS One 11, e0147210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mullish BH, McDonald JAK, Pechlivanis A, Allegretti JR, Kao D, Barker GF, Kapila D, Petrof EO, Joyce SA, Gahan CGM, et al. (2019). Microbial bile salt hydrolases mediate the efficacy of faecal microbiota transplant in the treatment of recurrent Clostridioides difficile infection. Gut 68, 1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, Liu H, Kinnebrew M, Viale A, et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang JD, Myers CJ, Harris SC, Kakiyama G, Lee IK, Yun BS, Matsuzaki K, Furukawa M, Min HK, Bajaj JS, et al. (2019). Bile acid 7α-Dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem. Biol 26, 27–34.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monaghan T, Mullish BH, Patterson J, Wong GK, Marchesi JR, Xu H, Jilani T, and Kao D (2019). Effective fecal microbiota transplantation for recurrent Clostridioides difficile infection in humans is associated with increased signalling in the bile acid-farnesoid X receptor-fibroblast growth factor pathway. Gut Microbes 10, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, and Bäckhed F (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235. [DOI] [PubMed] [Google Scholar]
- 90.Aguirre AM, Yalcinkaya N, Wu Q, Swennes A, Tessier ME, Roberts P, Miyajima F, Savidge T, and Sorg JA (2021). Bile acid-independent protection against Clostridioides difficile infection. PLoS Pathog. 17, e1010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Girinathan BP, DiBenedetto N, Worley JN, Peltier J, Arrieta-Ortiz ML, Immanuel SRC, Lavin R, Delaney ML, Cummins CK, Hoffman M, et al. (2021). In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29, 1693–1708.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Theriot CM, Koenigsknecht MJ, Carlson PE Jr., Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, and Young VB (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun 5, 3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seekatz AM, Theriot CM, Rao K, Chang YM, Freeman AE, Kao JY, and Young VB (2018). Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe 53, 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martin-Gallausiaux C, Marinelli L, Blottière HM, Larraufie P, and Lapaque N (2021). SCFA: mechanisms and functional importance in the gut. Proc. Nutr. Soc 80, 37–49. [DOI] [PubMed] [Google Scholar]
- 95.Fachi JL, Felipe JS, Pral LP, da Silva BK, Corrêa RO, de Andrade MCP, da Fonseca DM, Basso PJ, Câmara NOS, de Sales E Souza ÉL, et al. (2019). Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-Dependent mechanism. Cell Rep. 27, 750–761.e7. [DOI] [PubMed] [Google Scholar]
- 96.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martinez-Gili L, McDonald JAK, Liu Z, Kao D, Allegretti JR, Monaghan TM, Barker GF, Miguéns Blanco J, Williams HRT, Holmes E, et al. (2020). Understanding the mechanisms of efficacy of fecal microbiota transplant in treating recurrent Clostridioides difficile infection and beyond: the contribution of gut microbial-derived metabolites. Gut Microbes 12, 1810531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McDonald JAK, Mullish BH, Pechlivanis A, Liu Z, Brignardello J, Kao D, Holmes E, Li JV, Clarke TB, Thursz MR, et al. (2018). Inhibiting growth of Clostridioides difficile by restoring valerate, Produced by the Intestinal Microbiota. Gastroenterology 155, 1495–1507.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun X, and Hirota SA (2015). The roles of host and pathogen factors and the innate immune response in the pathogenesis of Clostridium difficile infection. Mol. Immunol 63, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kulaylat AS, Buonomo EL, Scully KW, Hollenbeak CS, Cook H, Petri WA, and Stewart DB (2018). Development and validation of a prediction model for mortality and adverse outcomes among patients with peripheral eosinopenia on admission for Clostridium difficile infection. JAMA Surg. 153, 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, Schwan C, Eichhoff AM, Koch-Nolte F, Lyras D , et al. (2016). The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat. Microbiol 1, 16108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buonomo EL, Cowardin CA, Wilson MG, Saleh MM, Pramoonjago P, and Petri WA (2016). Microbiota-regulated IL-25 increases eosinophil number to provide protection during Clostridium difficile infection. Cell Rep. 16, 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jan N, Hays RA, Oakland DN, Kumar P, Ramakrishnan G, Behm BW, Petri WA, and Marie C (2021). Fecal microbiota transplantation increases colonic IL-25 and dampens tissue inflammation in patients with recurrent Clostridioides difficile. mSphere 6, e0066921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frisbee AL, Saleh MM, Young MK, Leslie JL, Simpson ME, Abhyankar MM, Cowardin CA, Ma JZ, Pramoonjago P, Turner SD, et al. (2019). IL-33 drives group 2 innate lymphoid cell-mediated protection during Clostridium difficile infection. Nat. Commun 10, 2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Littmann ER, Lee JJ, Denny JE, Alam Z, Maslanka JR, Zarin I, Matsuda R, Carter RA, Susac B, Saffern MS, et al. (2021). Host immunity modulates the efficacy of microbiota transplantation for treatment of Clostridioides difficile infection. Nat. Commun 12, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cook L, Stahl M, Han X, Nazli A, MacDonald KN, Wong MQ, Tsai K, Dizzell S, Jacobson K, Bressler B, et al. (2019). Suppressive and gut-reparative functions of human Type 1 T regulatory cells. Gastroenterology 157, 1584–1598. [DOI] [PubMed] [Google Scholar]
- 107.Hoeppli RE, Wu D, Cook L, and Levings MK (2015). The environment of regulatory T cell biology: cytokines, metabolites, and the microbiome. Front. Immunol 6, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hernández Del Pino RE, Barbero AM, Español LÁ, Morro LS, and Pasquinelli V (2021). The adaptive immune response to Clostridioides difficile: A tricky balance between immunoprotection and immunopathogenesis. J. Leukoc. Biol 109, 195–210. [DOI] [PubMed] [Google Scholar]
- 109.Cook L, Rees WD, Wong MQ, Kwok WW, Levings MK, and Steiner TS (2021). Recurrent Clostridioides difficile infection is associated with impaired T helper Type 17 immunity to C difficile toxin B. Gastroenterology 160, 1410–1413.e4. [DOI] [PubMed] [Google Scholar]
- 110.Cook L, Rees WD, Wong MQ, Peters H, Levings MK, and Steiner TS (2021). Fecal microbiota transplantation for recurrent Clostridioides difficile infection enhances adaptive immunity to C difficile toxin B. Gastroenterology 160, 2155–2158.e4. [DOI] [PubMed] [Google Scholar]
- 111.Monaghan TM, Duggal NA, Rosati E, Griffin R, Hughes J, Roach B, Yang DY, Wang C, Wong K, Saxinger L, et al. (2021). A multi-factorial observational study on sequential fecal microbiota transplant in patients with medically refractory Clostridioides difficile infection. Cells 10, 3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Monaghan TM, Seekatz AM, Markham NO, Yau TO, Hatziapostolou M, Jilani T, Christodoulou N, Roach B, Birli E, Pomenya O, et al. (2021). Fecal microbiota transplantation for recurrent Clostridioides difficile infection associates with functional alterations in circulating microRNAs. Gastroenterology 161, 255–270.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Monaghan TM, Polytarchou C, Kao D, Alexander C, and Gurnani P (2022). Therapeutic potential of miRNAs in Clostridioides difficile infection. Future Microbiol. 17, 315–318. [DOI] [PubMed] [Google Scholar]
- 114.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, et al. (2012). Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 8, e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feuerstadt P, Louie TJ, Lashner B, Wang EEL, Diao L, Bryant JA, Sims M, Kraft CS, Cohen SH, Berenson CS, et al. (2022). SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. SER. N. Engl. J. Med 386, 220–229. [DOI] [PubMed] [Google Scholar]
- 116.Kao D, Wong K, Franz R, Cochrane K, Sherriff K, Chui L, Lloyd C, Roach B, Bai AD, Petrof EO, et al. (2021). The effect of a microbial ecosystem therapeutic (MET-2) on recurrent Clostridioides difficile infection: a phase 1, open-label, single-group trial. Lancet Gastroenterol. Hepatol 6, 282–291. [DOI] [PubMed] [Google Scholar]
- 117.Ramesh V, Fralick JA, and Rolfe RD (1999). Prevention of Clostridium difficile -induced ileocecitis with bacteriophage. Anaerobe 5, 69–78. [Google Scholar]
- 118.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. [DOI] [PubMed] [Google Scholar]
- 119.Parker A, Romano S, Ansorge R, Aboelnour A, Le Gall G, Savva GM, Pontifex MG, Telatin A, Baker D, Jones E, et al. (2022). Fecal microbiota transfer between young and aged mice reverses hallmarks of the aging gut, eye, and brain. Microbiome 10, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang C, Mogno I, Contijoch EJ, Borgerding JN, Aggarwala V, Li Z, Siu S, Grasset EK, Helmus DS, Dubinsky MC, et al. (2020). Fecal IgA levels are determined by strain-level differences in Bacteroides ovatus and are modifiable by gut microbiota manipulation. Cell Host Microbe 27, 467–475.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Walter J, Armet AM, Finlay BB, and Shanahan F (2020). Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell 180, 221–232. [DOI] [PubMed] [Google Scholar]
- 122.Rossen NG, Fuentes S, van der Spek MJ, Tijssen JG, Hartman JH, Duflou A, Löwenberg M, van den Brink GR, Mathus-Vliegen EM, de Vos WM, et al. (2015). Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149, 110–118.e4. [DOI] [PubMed] [Google Scholar]
- 123.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi A, Armstrong D, Marshall JK, Kassam Z, Reinisch W, et al. (2015). Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6. [DOI] [PubMed] [Google Scholar]
- 124.Holvoet T, Joossens M, Vázquez-Castellanos JF, Christiaens E, Heyerick L, Boelens J, Verhasselt B, van Vlierberghe H, De Vos M, Raes J, et al. (2021). Fecal microbiota transplantation reduces symptoms in some patients with irritable bowel syndrome with predominant abdominal bloating: short- and long-term results from a placebo-controlled randomized trial. Gastroenterology 160, 145–157.e8. [DOI] [PubMed] [Google Scholar]
- 125.El-Salhy M, Hatlebakk JG, Gilja OH, Bråthen Kristoffersen A, and Hausken T (2020). Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut 69, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aroniadis OC, Brandt LJ, Oneto C, Feuerstadt P, Sherman A, Wolkoff AW, Kassam Z, Sadovsky RG, Elliott RJ, Budree S, et al. (2019). Faecal microbiota transplantation for diarrhoea-predominant irritable bowel syndrome: a double-blind, randomised, placebo-controlled trial. Lancet Gastroenterol. Hepatol 4, 675–685. [DOI] [PubMed] [Google Scholar]
- 127.Halkjær SI, Christensen AH, Lo BZS, Browne PD, Günther S, Hansen LH, and Petersen AM (2018). Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut 67, 2107–2115. [DOI] [PubMed] [Google Scholar]
- 128.Holster S, Lindqvist CM, Repsilber D, Salonen A, de Vos WM, König J, and Brummer RJ (2019). The effect of allogenic versus autologous fecal microbiota transfer on symptoms, visceral perception and fecal and mucosal microbiota in irritable bowel syndrome: A randomized controlled study. Clin. Transl. Gastroenterol 10, e00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143. 913–6.e7. [DOI] [PubMed] [Google Scholar]
- 130.Mocanu V, Zhang Z, Deehan EC, Kao DH, Hotte N, Karmali S, Birch DW, Samarasinghe KK, Walter J, and Madsen KL (2021). Fecal microbial transplantation and fiber supplementation in patients with severe obesity and metabolic syndrome: a randomized double-blind, placebo-controlled phase 2 trial. Nat. Med 27, 1272–1279. [DOI] [PubMed] [Google Scholar]
- 131.Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, et al. (2017). Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology 66, 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, Jiang ZD, Abu-Sbeih H, Sanchez CA, Chang CC, et al. (2018). Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med 24, 1804–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Goeser F, Sifft B, Stein-Thoeringer C, Farowski F, Strassburg CP, Brossart P, Higgins PG, Scheid C, Wolf D, Holderried TAW, et al. (2021). Fecal microbiota transfer for refractory intestinal graft-versus-host disease - Experience from two German tertiary centers. Eur. J. Haematol 107, 229–245. [DOI] [PubMed] [Google Scholar]
- 134.Bilinski J, Lis K, Tomaszewska A, Grzesiowski P, Dzieciatkowski T, Tyszka M, Karakulska-Prystupiuk E, Boguradzki P, Tormanowska M, Halaburda K, et al. (2021). Fecal microbiota transplantation in patients with acute and chronic graft-versus-host disease-spectrum of responses and safety profile. Results from a prospective, multicenter study. Am. J. Hematol 96, E88–E91. [DOI] [PubMed] [Google Scholar]
- 135.Langdon A, Schwartz DJ, Bulow C, Sun X, Hink T, Reske KA, Jones C, Burnham CD, Dubberke ER, Dantas G, et al. (2021). Microbiota restoration reduces antibiotic-resistant bacteria gut colonization in patients with recurrent Clostridioides difficile infection from the open-label PUNCH CD study. Genome Med. 13, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.FDA (2019). Important safety alert regarding use of fecal microbiota for transplantation and risk of serious adverse reactions due to transmission of multi-drug resistant organisms. https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/important-safety-alert-regarding-use-fecal-microbiota-transplantation-and-risk-serious-adverse.
- 137.FDA (2020). Safety Alert Regarding Use of Fecal Microbiota for Transplantation and Additional Safety Protections Pertaining to SARS-CoV-2 and COVID-19. https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/safety-alert-regarding-use-fecal-microbiota-transplantation-and-additional-safety-protections.
- 138.FDA (2022). Safety Alert Regarding Use of Fecal Microbiota for Transplantation and Additional Safety Protections Pertaining to Monkeypox Viru. https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/safety-alert-regarding-use-fecal-microbiota-transplantation-and-additional-safety-protections-0.
- 139.Rode AA, Chehri M, Krogsgaard LR, Heno KK, Svendsen AT, Ribberholt I, Helms M, Engberg J, Schønning K, Tvede M, et al. (2021). Randomised clinical trial: a 12-strain bacterial mixture versus faecal microbiota transplantation versus vancomycin for recurrent Clostridioides difficile infections. Aliment. Pharmacol. Ther 53, 999–1009. [DOI] [PubMed] [Google Scholar]