SUMMARY
Human milk oligosaccharides (HMOs) are complex, multi-functional glycans present in human breast milk. They represent an intricate mix of heterogeneous structures which reach the infant intestine in an intact form as they resist gastrointestinal digestion. Therefore, they confer a multitude of benefits, directly and/or indirectly, to the developing neonate. Certain bifidobacterial species, being among the earliest gut colonizers of breast-fed infants, have an adapted functional capacity to metabolize various HMO structures. This ability is typically observed in infant-associated bifidobacteria, as opposed to bifidobacteria associated with a mature microbiota. In recent years, information has been gleaned regarding how these infant-associated bifidobacteria as well as certain other taxa are able to assimilate HMOs, including the mechanistic strategies enabling their acquisition and consumption. Additionally, complex metabolic interactions occur between microbes facilitated by HMOs, including the utilization of breakdown products released from HMO degradation. Interest in HMO-mediated changes in microbial composition and function has been the focal point of numerous studies, in recent times fueled by the availability of individual biosynthetic HMOs, some of which are now commonly included in infant formula. In this review, we outline the main HMO assimilatory and catabolic strategies employed by infant-associated bifidobacteria, discuss other taxa that exhibit breast milk glycan degradation capacity, and cover HMO-supported cross-feeding interactions and related metabolites that have been described thus far.
KEYWORDS: infant microbiome, human milk oligosaccharides, gut microbiota, infant, bifidobacteria
INTRODUCTION
Human breast milk is primarily composed of water, carbohydrates, lipids, and milk proteins to form an early life source of nutrients, while it furthermore contains various minor, yet functional ingredients, such as antibodies (e.g., IgG and IgA), antimicrobial peptides, lactoferrin, and mucins (1–5). A proportion of the human milk-associated carbohydrates, i.e., those larger than lactose and collectively known as human milk oligosaccharides (HMOs), are non-digestible, structurally diverse, multi-functional glycans. Oligosaccharide concentration and diversity in human milk are significantly higher than that of other mammals (1). The availability of HMOs is primarily through feeding of breast milk, which is the universally accepted gold standard for infant nutrition (6). They serve as substrates for select gut microorganisms as they escape host digestion, thereby playing a significant role in shaping the infant microbiota, including the establishment and persistence of a set of health-promoting gut microbes (7). Initial colonization of the infant intestine shortly after birth establishes a life-long mutualistic relationship between the intestinal microbial community and its respective host. This is crucial for the development of pathogen resistance, utilization of dietary components, and interactions with the immune system (8, 9). The formation of the infant gut microbiota is impacted by numerous factors including, but not limited to, mode of delivery, antibiotic administration, geographical and environmental location, and available nutrients (10, 11).
Prevalent taxa in the early infant gut are highly variable and may include Escherichia, Enterococcus, Bifidobacterium, Bacteroides, Streptococcus, and Veillonella (11, 12). Bifidobacterium members are among the earliest colonizers of the infant gut microbiota, in individual infants reaching up to 90% relative abundance (13–15), and certain members within this genus are HMO degraders (11, 13, 16). Specific infant-typical bifidobacterial species, or strains thereof, are equipped with an arsenal of enzymes and transporters to facilitate HMO uptake and metabolism. The first 6–12 months following birth represent a critical stage for the formation and establishment of the human gut microbiota, and infant-associated Bifidobacterium species and HMOs are contributors to the configuration of this microbial community. The microbiota evolves in parallel with the host, which is significant to overall infant health (9) due in part to the metabolites produced from substrate fermentation. As a collection of microbes, complex community dynamics are at play when individual or multiple HMOs are present.
This review will describe HMO structure and composition as well as factors affecting early life microbiota, with particular focus on the intricate interactions between select Bifidobacterium species, and indeed other taxa, and HMOs. Additionally, we provide an overview of the variety and function of metabolites produced from HMO degradation. We also consider HMOs and their similarities to traditional prebiotics applied in infant formula such as galacto-oligosaccharides (GOS) and fructo-oligosaccharides (FOS) as they relate to the infant gut and how they are consumed. Finally, we identify some future considerations and knowledge gaps while illuminating the broad potential of various HMOs in modulating the infant gut microbial composition and the consequent metabolic functions that ensue.
FACTORS AFFECTING EARLY LIFE MICROBIOTA
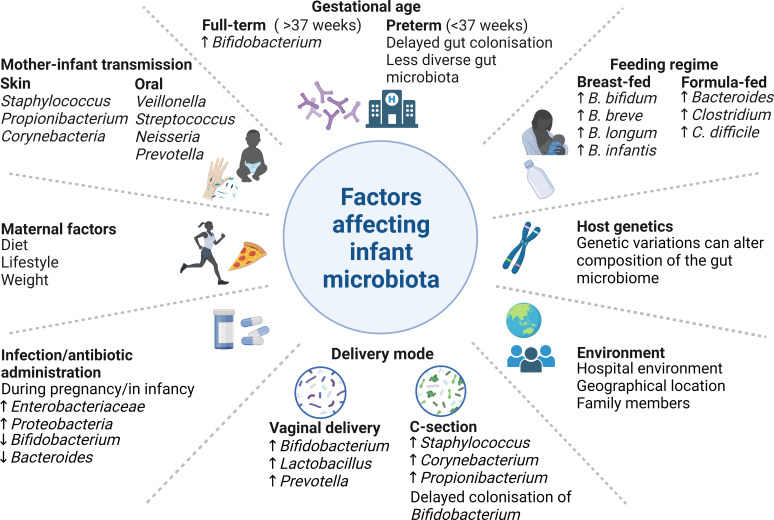
Numerous factors influence the infant gut microbiota establishment and development including maternal diet, antibiotic administration, mode of delivery (i.e., vaginal delivery or delivery by cesarean section), feeding regimen (i.e., breast milk versus formula milk), birth term (full term versus preterm), geographical location, human interaction, mother-infant transfer, and the surrounding environment (Fig. 1). The first year of life is critical for the formation of the infant gut microbiota (6), with bifidobacteria being among the first colonizers (11, 17–19). Bifidobacterial absence in the early life microbial community may expose the infant to undesirable bacteria such as Enterococcus, Enterobacter, Clostridium, and Klebsiella species (12), possibly through a lack of competitive exclusion and antimicrobial metabolite production (16, 20). Antibiotic administration during pregnancy can lead to delayed colonization by bifidobacteria in infants (21) and has been associated with a higher abundance of pathogenic bacteria such as members from Enterobacteriaceae and Proteobacteria (22). Antibiotic usage during prenatal, neonatal, and postnatal periods is thought to retard the appropriate development of the gut microbiota and cause a disruption that may not recover for some 6–12 months after birth (23). Environmental factors such as place of birth, i.e., hospital or home, and geographical location may have an indirect effect on early life microbiota, in part due to cultural habits such as bathing practices, e.g., Japan (24), lifestyle, eating habits and food availability, diversity of human interaction, and climate (23). The various factors influencing the neonatal gut microbial composition have previously been subject to in-depth scrutiny (12, 25, 26) and are summarized in Fig. 1.
Fig 1.
Factors affecting the early infant gut microbiota. Some factors influencing the taxonomic structure, and functionality, of the infant microbiota are displayed here. For further details, see text. Made using BioRender.
The composition of the gut microbiota is highly variable during the early stages of life and is often characterized by low microbial diversity (11). Prevalent taxa in the gut of the neonate may include Escherichia, Enterococcus, Bifidobacterium, Bacteroides, Streptococcus, and Veillonella (9, 11, 12). Bifidobacteria populate the gut, at least in part due to vertical, mother-to-baby transmission and membrane rupture during vaginal delivery (19). C-section can interrupt this natural route of vertical transmission, thus resulting in disruption, delay, and/or reduction of bifidobacterial colonization (19, 27). Immediately following birth, Lactobacillus, Prevotella, or Sneathia spp. dominate the gut in vaginally born infants, whereas Clostridium dominates in infants born via C-section (28). The alpha diversity of the gut microbiome has been shown to be impacted by birth mode, but also age (18, 28, 29). Feeding mode has an impact with breastfeeding increasing certain Bifidobacterium species, such as B. breve and B. bifidum, as well as Ruminococcus in the infant gut, with a lower prevalence of Escherichia coli, Clostridioides difficile, and Bacteroides fragilis (21, 30). In contrast, the gut microbiota of formula-fed infants mainly comprised members from Enterobacteriaceae, and genera such as Streptococcus, Bacteroides, Bifidobacterium, Clostridium, and Atopobium (21, 31, 32). Often, the gut microbiota of a breast-fed infant has a lower microbial diversity, richness, and complexity than that of formula-fed infants (31, 33) but includes a high prevalence of certain bifidobacterial species (13), which will be discussed below. The predominance of bifidobacteria and their intrinsic ability to degrade HMOs is a primary factor in the formation of a less diverse microbial community (29, 34, 35). In contrast to adults, a low microbial diversity, typically dominated by bifidobacteria in breast-fed infants, is favorable due to the health benefits to the infant, e.g., immune system maturation (36), reduction in allergy risk (37), and decreased colonization of potentially undesirable microbes (38). As the gut microbiota develops, bifidobacteria remain present, although to a lesser relative abundance, and compositional shifts are evident at species level (23, 29). The infant microbial composition can be dominated by particular taxa and is much more amenable to change than that of adults, which appears to be crucial, although the true extent remains elusive, in this early stage of microbiota establishment (39). It has been proposed that these taxonomic patterns can be assigned to different infant community state types based on species-level composition (39, 40) and may influence how different substrates, including HMOs, are assimilated and contribute to the corresponding functional benefits.
HUMAN MILK OLIGOSACCHARIDES: STRUCTURE, COMPOSITION, AND VARIATION
One of the most notable aspects of HMOs is their structural diversity. At present, there have been over 200 distinct HMO structures identified in breast milk, with a combined concentration of between 5 and 22 g/L in mature milk (1, 41, 42). Generally, the highest concentration of HMOs is present immediately after birth in colostrum (up to 25 g/L) with a subsequent decrease over the lactation period. HMO concentration and diversity are more abundant in human breast milk when compared to that of other mammals (1). Complex interpersonal variation in HMO composition and structure has been observed due to factors including genetics, lactation stage, and environmental and geographical location. HMOs escape digestion in the small intestine, thus progressing to the colon where they can serve as substrates for select bacteria (1), including certain bifidobacterial populations, thereby facilitating their enhanced abundance.
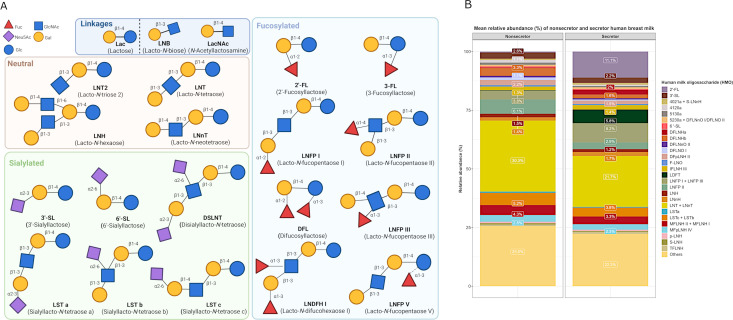
The structural components of HMOs consist of five monosaccharidic building blocks, namely D-glucose (Glc), D-galactose (Gal), N-acetylglucosamine (GlcNAc), L-fucose (Fuc), and N-acetylneuraminic acid (Neu5Ac or sialic acid). Through glycosidic bonds they form complex structures. Generally, all HMOs contain a lactose core (Galβ1-4Glc) at their reducing end, which can be elongated by a β1–3 or β1–6 linkage with one or more units of lacto-N-biose (LNB; Galβ1-3GlcNAc) or its isomer N-acetyllactosamine (LacNAc; Galβ1–4GlcNAc) (43). This provides the basis for the designation of type I and type II structures, respectively (Fig. 2A). This core HMO structure can carry fucose or sialic acid substitutions resulting in three categories: neutral (unsubstituted), fucosylated, or sialylated (Fig. 2A). The most abundant HMO present in milk from secretor mothers (see below in Fig. 2B) is 2′-fucosyllactose (2′-FL), a trisaccharide composed of L-fucose, D-glucose, and D-galactose. Other well-characterized structures include 3-fucosyllactose (3-FL), lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT), 3′-sialyllactose (3′-SL), and 6’-sialyllactose (6′-SL). The biosynthesis of oligosaccharides occurs following the formation of the lactose core when Gal and Glc are coupled by β-galactotransferase in the presence of α-lactalbumin, which is expressed during lactation (44). Lactose can be fucosylated at the terminal Gal by an α1–2 linkage to form 2′-FL or at the reducing end Glc by means of an α1–3 linkage resulting in the formation of 3-FL (45). Lactose can also be sialylated at the terminal Gal in α2–3 or α2–6 linkage to form 3′-SL and 6′-SL. The structural diversity and varying composition of HMOs present in human breast milk are comprehensively reviewed elsewhere, and we therefore refer the interested reader to some excellent recent reviews on this matter (1, 2, 45, 46).
Fig 2.
(A) Schematic representation of select HMO structures. Five basic units form HMOs: Fuc, Neu5Ac, GlcNAc, Gal, and Glc. All HMO structures are composed of a lactose core which can be elongated by LNB (type-1 chain) or N-acetyllactosamine (type-2 chain) via either β1–3 or β1–6 linkages. These structures can be further decorated resulting in three categories: fucosylated, sialylated, and neutral. (B) Mean relative abundance (%) of HMOs in non-secretor and secretor milk profiles. Data adapted from Vinjamuri et al. (47). Percentage labels included for HMOs present >1% relative abundance. Made using BioRender and R (ggplot2 package).
Both inter- and intrapersonal variation influence the type and concentration of HMOs synthesized (45). Specifically, genetic and environmental factors very substantially contribute to the diversity and abundance of (particular) HMOs produced by different women. Such genetic factors, in particular those responsible for the secretor (Se) status and Lewis (Le) blood group, also have a major impact on the composition and concentration of HMOs in breast milk. Approximately 80% of mothers globally, although variations exist among different populations, can express α-1-2-fucosyltransferase (FUT2), enabling them to produce milk containing oligosaccharides such as 2′-FL and lacto-N-fucopentaose (LNFP) I (48). Mothers with the Se gene express FUT2, which is responsible for the synthesis of α-1-2 fucosylated HMOs, whereas those with the Le gene express α-1-3/4-fucosyltransferase, thereby synthesizing α-1-3 or α-1-4 fucosylated HMOs (45). The variations in maternal secretor status are likely to have arisen to confer a host advantage. It has been suggested that the non-secretor phenotype, i.e., those lacking a functional FUT2 enzyme, evolved to better protect the host against diseases such as specific norovirus genotypes (49–51) and Helicobacter pylori (52, 53). Differences in evolutionary pressures, e.g., the risk of particular pathogens or other geographical factors, in countries or areas may have contributed to Se gene polymorphisms. Four milk phenotypes are thus observed; secretor positive and Lewis positive (Se+, Le+), secretor negative and Lewis positive (Se−, Le+), secretor positive and Lewis negative (Se+, Le−), and secretor negative and Lewis negative (Se−, Le−) (45). Se+, Le+ women produce milk containing a higher concentration and a greater variety of HMOs compared to the other three milk phenotypes, with the main distinction being fucosylation (45). Typically, 50%–80% of HMOs are fucosylated (1); however, in Se−, Le− women, a very small proportion of the HMOs (often <10%, although this can vary) (54, 55) carry fucose substitutions such as 3-FL, which could have a significant impact on the functionality resulting from bacterial HMO degradation. Alterations in the HMO concentration occur across the lactation period, with the highest levels found in colostrum with HMO concentration decreasing throughout lactation as milk matures (45). The HMO composition of human milk also changes during lactation, e.g., sialylated HMOs are abundant at the beginning of lactation and subsequently decrease over time (45), whereas 3-FL concentration is reported to increase over the lactation course (41). HMO variation is also evident among lactating mothers of preterm versus mothers of full-term infants, with higher oligosaccharide concentrations being present in milk from the latter (45).
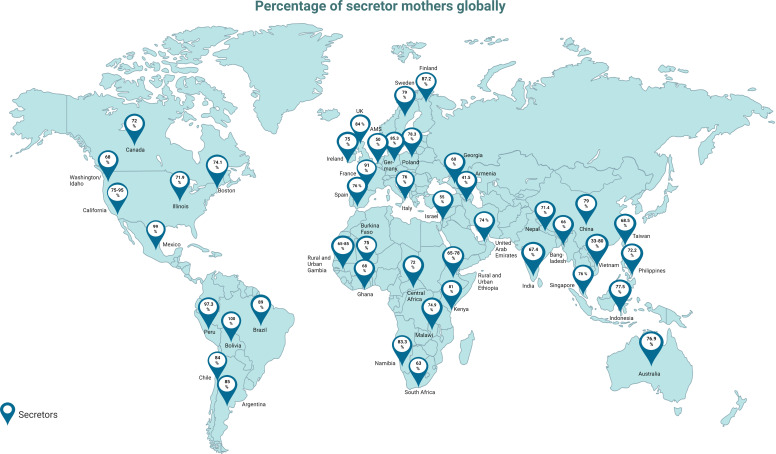
Geographical location, ethnicity, and seasonal changes can be a determining factor in HMO composition and concentration (56, 57). Generally, a relatively high proportion of mothers in the United States [up to 95% (42)] and Europe, e.g., Sweden (79%) (42) and Finland (87.2%) (58), are secretors, whereas many non-secretors inhabit parts of Africa, Central Asia, and Pacific regions (42) (Fig. 3). Secretor status type also varies within Africa, e.g., rural Gambia and South Africa have a greater abundance of non-secretors in contrast to Namibia and Malawi (47). Derrien and colleagues recently observed that 71% of their 105 Kenyan cohort were secretors (40) similar to that of Ireland (74.68% secretors) which has recently been reported (19). In Brazil and Peru, countries with a high proportion of secretors (approximately 98% of the population), 2′-FL is more abundant in mother’s milk compared to that in other countries such as Ghana and rural Gambia, with 68% and 65% being secretors, respectively (42, 59). Additionally, total concentration of HMOs and 2′-FL was shown to be the lowest in Africa, although LNT concentration was shown to be higher compared to other locations such as Germany, China, and Malaysia (60, 61). Interestingly, milk from non-secretor Bangladeshi women contained higher 2′-FL concentrations than that from non-secretors from other locations (57). Variation of HMOs and secretor status across geographical locations has previously been reported in considerable detail (41, 47, 60). Intra-regional variations are also observed, e.g., a recent study by Lahdenperä and colleagues (58), using a Finnish cohort of breastfeeding women, found that, e.g., exposure to residential green areas was positively associated with HMO diversity and concentration. Variation in HMO composition is also evident between those inhabiting rural and urban areas, which has been observed in countries such as Ethiopia and Gambia (42), where, e.g., milk from mothers in urban Gambia had higher levels of LNnT compared to milk from mothers inhabiting rural parts (42). Additionally in Gambia, lower HMO concentrations were found in mothers’ milk during the wet season compared to milk from mothers nursing during the dry season (62). Seasonal differences were also evident in a Canadian study (56), where HMO concentrations varied from winter to spring, e.g., higher LNnT in winter, and higher disialyllacto-N-tetraose (DSLNT) and 6′-SL were observed in spring. Diet has a profound impact on host health, with certain dietary components being associated with variations in HMO composition. A varied diet, including whole grains and vitamins, has been positively correlated to 2′-FL abundance (56, 63). Conversely, a high fat diet has been associated with a decrease in sialylated HMOs in breast milk (63–65). In central Africa, decreased food intake was shown to be associated with lower HMO concentrations (60, 66).
Fig 3.
Percentage of breast milk donors categorized as secretors worldwide. Secretor breast milk contains large amounts of α1-2 fucosylated oligosaccharides in contrast to that of non-secretors. Updated from McGuire et al. (42) with additional data from Cheng and Yeung (67), Vinjamuri et al. (47), Asher et al. (68), Feehily et al. (19), Azad et al. (56), Durham et al. (69), Erney et al. (70), Ferreira et al. (59), Lahdenperä et al. (58), Lewis et al. (71), Liu et al. (72), Liu et al. (73), Menzel et al. (74), Musumeci et al. (75), Moya-Alvarez et al. (60), McJarrow et al. (76), Pell et al. (57), Sudarma et al. (77), and Van Leeuwen et al. (78). Fan et al. (79). (Created with Biorender.com.)
While secretor status remains the predominant factor, other parameters, some of which were discussed above, such as the living environment, geographical location, lactation stage, maternal prepregnancy BMI, age, diet, parity, mode of delivery, infant gestational age, and sex, contribute to inter- and intrapersonal HMO variation (58). Difficulty persists when evaluating independent variables impacting HMO composition and concentration as there are often confounding factors influencing the HMO content simultaneously.
BIFIDOBACTERIA AND HMO UTILIZATION
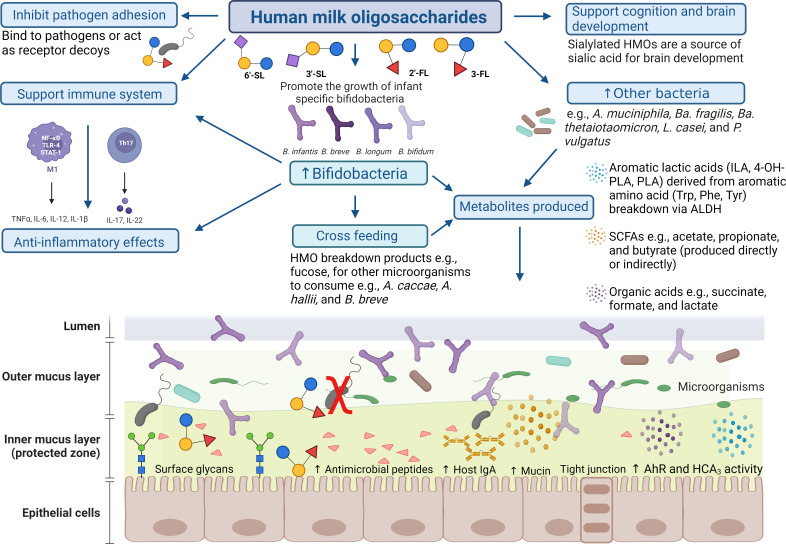
Bifidobacteria, first discovered by Tissier in 1899 from a breast-fed infant fecal sample, are characterized as gram-positive, anaerobic bacteria, often possessing a high G + C genome content. Specific members of Bifidobacterium, a genus which from a taxonomic perspective belongs to the Actinomycetota phylum, can be commonly found in the human gut. The overrepresentation of bifidobacteria in the gut of healthy, breast-fed infants is suggestive of their crucial role in supporting early life health and development of their infant host (11, 80). The beneficial effects of bifidobacteria are at least in part believed to be mediated through the production of carbohydrate-derived metabolites, either directly or indirectly through cross-feeding interactions, examples of which are discussed further below. Short-chain fatty acids (SCFAs) are metabolic end products of (bifido)bacterial fermentation of indigestible carbohydrates, including HMOs. These multi-functional compounds can decrease intestinal pH, impact gut barrier integrity, and prevent pathogen establishment (81–83). Other host interactive molecules produced by bifidobacteria include aromatic lactic acids, mentioned later in this review, exopolysaccharides, conjugated linoleic acid, and pili (35, 84, 85). Beneficial impacts exerted by infant-type bifidobacteria to the host are not solely due to carbohydrates but also nitrogen (86). Urea utilization by B. longum subsp. infantis strains indicates participation in nitrogen homeostasis in early life (87). The higher incidence of pathologies, such as atopy, obesity, and asthma, in infants with reduced bifidobacterial colonization has been reported in several studies (11, 88–90).
Human gut-derived bifidobacteria are generally categorized into two groups: “infant type” and “adult type.” Typical infant gut-associated bifidobacteria include species such as B. breve, B. longum subsp. longum (B. longum), B. bifidum, B. pseudocatenulatum, B. longum subsp. infantis (B. infantis), and B. catenulatum subsp. kashiwanohense (B. kashiwanohense) (13, 91, 92). In contrast, B. adolescentis, B. pseudocatenulatum, B. catenulatum subsp. catenulatum, B. longum, and B. dentium are common in the adult gut (93–95). The ubiquitous nature of B. longum has facilitated its identification in the gut microbiota across all age groups (96). This categorization of infant- and adult type is more complex than the presence or absence of one or more species; rather, it is a shift within the bifidobacterial species abundance as the infant develops toward adulthood. One of the defining features between the two categories is the ability of infant-associated bifidobacteria to consume one or more HMOs and/or HMO degradants, which is not typically observed in the adult-type bifidobacterial species. This proven ability to degrade HMOs and derivatives thereof is referred throughout the text in relation to infant-type bifidobacteria. Infant-associated bifidobacteria encode various enzymes and transporters, which is discussed further, to metabolize HMOs, thus offering a competitive advantage over other (bifido)bacteria in the infant gut. Certainly, this ability of infant-associated bifidobacteria to colonize and persist is no doubt due to a variety of factors but includes their ability to metabolize both diet-derived and host glycans such as mucins and HMOs (11).
The underlying mechanisms involved in bifidobacterial, and other microbial, metabolism of HMOs have been the subject of extensive research with the added complexity that not all (infant-associated) Bifidobacterium members can (directly) utilize HMOs. The variation in HMO degradation ability, occurring at both species and strain levels, combined with the substantial number of HMO structures means much remains unanswered. This includes the precise mechanisms involved, the origins of this degradation ability, and the host benefits associated with this metabolic adaptation.
Bifidobacterial HMO utilization in vitro
One of the most prominent, and thus comprehensively studied, HMO degraders in the neonate gastrointestinal system is B. infantis. It has been demonstrated that strains from this species, including B. infantis M-63 and ATCC 15697, can use a broad spectrum of HMO structures, e.g., 2′-FL, 3′-SL, LNnT, and LNT (97–99). There are also strain-specific differences within this species, such as the aforementioned strain M-63 achieving a significantly (P = 0.0004) higher maximum optical density (OD) when 2′-FL was made available when compared to B. infantis Rosell-33 (R0033) (98). Comparable to B. infantis, B. bifidum are in general vigorous consumers of HMOs (100, 101), although there does appear to be some strain-level variation. Gotoh et al. (100) demonstrated that B. bifidum strains JCM 1254, TMC3108, and MC3115 achieved a higher OD (OD >1 at 20 hours) than strain JCM 7004 (OD <1 at 20 hours) in HMO-supplemented media. In a study using 13 B. bifidum strains, most demonstrated a capacity to grow using pooled HMOs from breast milk as well as synthetic HMOs, i.e., LNT, LNnT, 2′-FL, 3-FL, and 6′-SL (102). Four strains, JCM 1255, JCM 1209, JCM 7004, and S28a, struggled to grow using the pooled HMOs (102). Additionally, one of these strains, JCM 1255, was unable to grow using the synthetic HMOs tested, with the exception of LNT (102). Generally, HMO degraders are more common among strains of B. infantis and B. bifidum, in contrast to B. breve and B. longum members where HMO degradation is more sporadic. The ability to consume different HMOs is strain dependent among the latter two species, although LNT metabolism is a common property among strains of either of these species (103). Generally, B. breve can achieve a high level of in vitro growth when either LNT or LNnT is the sole carbohydrate (104, 105). In addition, some B. breve strains (e.g., strains SC95 and SC568) have the capacity to assimilate fucosylated HMOs, e.g., 2′-FL and 3-FL, although this is not widespread across the species (104). Often described as a “scavenger” consumer, B. breve can capitalize on the HMO degradants, such as fucose and sialic acid, released by other avid HMO degraders. This B. breve persistence in the presence of HMOs has been demonstrated in vitro using co-culture growth experiments (98, 106). Pertaining to this, the order in which microbes colonize an environment may be deterministic in the formation of the subsequent community structure, and thus, the functions that occur. Ojima et al. (106) examined the facilitative and inhibitory relationships in vitro between infant-associated bifidobacteria members in response to a pool of nine HMO structures. The authors observed that even with its comparatively low HMO degradation abilities relative to other bifidobacterial species employed in the study, as reflected in both its genotype and phenotype, B. breve was shown to nonetheless achieve a high level of growth in co-culture. Depending on the order in which it was inoculated into the media, B. breve UCC2003 was shown to outcompete B. infantis ATCC 15697, B. bifidum JCM 1254, and B. longum MCC10007 by metabolizing carbohydrates such as fucose, which other species provided from assimilating HMOs (106).
The aforementioned cross-feeding phenomenon has also been observed on a broader scale using in vitro community-based experiments where the initial microbial composition was an important factor when the fermentation of structurally distinct HMOs occurred (Table 1) (107–109). Xu et al. (107) used fecal microbiota groups dominated by either B. longum, B. breve, or Bacteroides to investigate the impact of structurally different HMOs. Taxonomic composition, SCFA production, and gas profiles differed, not solely owing to the various HMOs but more so due to the starting microbial composition. Similarly, a recent in vitro fecal fermentation study (109) using B. longum-dominated microbiotas revealed higher levels of community-produced lactate when neutral HMOs, rather than sialylated structures, were introduced. Taken together, bifidobacterial subpopulations may respond varyingly to distinct HMOs. Concomitantly, HMOs can influence the development of the infant gut microbiota by acting as selective agents for the growth of certain infant-associated (bifido)bacteria.
TABLE 1.
Overview of some recent in vivo- and in vitro-based fermentation investigations of microbial communities as a result of HMO supplementationa
| Type of HMO | Study design | Microbial analysis | Effects on gut microbiota | Metabolite production | Reference |
|---|---|---|---|---|---|
| In vivo | |||||
| 2′-FL and L. reuteri DSM 17938 | Double-blind RCT in infants | 16S rRNA, qPCR | ↑ Bifidobacterium, ↓ C. difficile and K. pneumoniae | ↑ Lactate | Alliet et al. (110) |
| HMO blend (2′-FL, DFL, LNT, 3′-SL, and 6′-SL) | RCT in infants | Shotgun metagenomics | ↑ B. infantis, ↓ C. difficile | ↑ Acetate and lactate | Bosheva et al. (111) |
| Amino acid infant formula with HMOs (2′-FL and LNnT) | Single-arm, multicenter, interventional clinical trial in infants with CMPA. No control group | Shotgun metagenomics | ↑ B. breve, B. bifidum, B. infantis, and B. longum, ↓ Escherichia spp., and Klebsiella spp., | ↑ Acetate | Gold et al. (112) |
| Test infant formula with 2′-FL and LNnT (2:1) | RCT in infants | 16S rRNA | ↑ Bifidobacterium, ↓ Carnobacteriaceae and Escherichia | ↑ Acetate, ↓ succinate, butyrate, propionate, and 5-aminovalerate | Dogra et al. (113) |
| Commercial formula (control) and same formula with 2′-FL (test) | Double-blind RCT in infants | Shotgun metagenomics | 2′-FL = ↑ Bifidobacterium | N/A | Wallingford et al. (114) |
| HMOs (extracted from breast milk) and 2′-FL | Dextran sodium sulphate-induced mouse model 6 weeks old (for prevention of colitis) | 16S rRNA | HMOs = ↑ Muribaculaceae, Bacteroides, and R. torques 2′-FL = ↑ Lactobacillus, Muribaculaceae, and R. torques HMOs and 2′-FL = ↓ Streptococcus, Escherichia-Shigella, and Lactococcus |
HMOs = ↑ acetate, propionate, and butyrate 2′-FL = ↑ acetate and propionate |
Li et al. (115) |
| 2′-FL | Human microbiota-associated mouse model (infant fecal transplant) | 16S rRNA | ↑ Bifidobacteria, Olsenella, and Blautia ↓ Enterorhabdus and Lachnospiraceae_UCG-006 |
↑ Acetate and propionate ↓ butyrate | Qingxue Chen et al. (116) |
| 2′-FL | Healthy sucking rats | 16S rRNA | ↑ Lactobacillus ↓ Enterococcaceae and Streptococcaceae |
↑ Butyric acid | Azagra-Boronat et al. (117) |
| In vitro fermentations | |||||
| HMO6 mix (2′-FL, LNnT, LNT, diFL, 3′-SL, and 6′-SL), 2′-FL, lactose, and 2′-FL/LNnT | baby M-SHIME | 16s rRNA, qPCR | 2′-FL = ↑ Bifidobacteriaceae and Coriobacteriaceae 2′-FL/LNnT = ↑ Brucellaceae HMO6 mix = ↑ B. adolescentis and F. prausnitzii, ↓ Firmicutes Lactose = ↑ Rikenellaceae and Veillonellaceae |
2′-FL = ↑ propionate HMO6 mix = ↑ acetate and butyrate |
Natividad et al. (118) |
| GOS, 2′-FL, or GOS and 2′-FL (1:1 and 3:1) | microMatrix and baby M-SHIME | 16S rRNA | GOS = ↑ Lactobacillus 2′-FL = ↑ Veillonella and Akkermansia GOS and 2′-FL = ↑ Veillonella, Bifidobacterium, Lactobacillus, and Streptococcus GOS and 2′-FL = ↓ Salmonella and Citrobacter GOS or 2′-FL = ↓ Escherichia/Shigella |
GOS and/or 2′-FL = ↑ acetate | Lindner et al. (119) |
| 2′-FL, GOS, and 2′-FL/GOS (1:4 ratio) | In vitro fermentation | 16S rRNA | 2′-FL and 2′-FL/GOS = ↑ Bifidobacterium, Escherichia-Shigella, and Enterococcus GOS = ↑ Bifidobacterium |
↑ Acetate, lactate, succinate, and propionate | Akkerman et al. (120) |
| 2′-FL, 3-FL, 3′-SL, 6′-SL, LNT, LNnT, FOS, and GOS | In vitro fermentation | 16S rRNA |
B. longum-dominated inocula: HMOs and GOS = ↑ B. longum B. breve-dominated inocula: HMOs, GOS, and FOS = ↑ B. breve Bacteroides-dominated inocula: 2′-FL and 3-FL = ↑ B. breve, B. fragilis, and B. vulgatus LNT and LNnT = ↓ B. fragilis and B. vulgatus 3′-SL and 6′-SL = ↓ Ba. vulgatus |
GOS and HMOs (2′-FL, 3-FL, LNT, and LNnT) = ↑ acetate and lactate | Xu et al. (107) |
| 2′-FL, 3-FL, 3′-SL, 6′-SL, LNT, and LNnT, GOS, and FOS | In vitro fermentation | 16S rRNA | HMOs and GOS = ↑ B. longum and E. coli FOS = ↑ Klebsiella pneumoniae |
B. longum-dominant inocula HMOs (2′-FL, 3-FL, LNT, and LNnT) and GOS = ↑ acetate and lactate |
Li et al. (109) |
| 3-FL, LNT2, and GOS/inulin (9:1) | In vitro fermentation (in anaerobic glass fermentation bottles) | 16S rRNA | LNT2 = ↑ Bifidobacterium (B. longum and B. bifidum) and Collinsella 3-FL = ↑ B. longum, Bacteroides (B. dorei), and Enterococcus genus GOS/inulin = ↑ B. longum LNT2, 2′-FL and GOS/inulin= ↓ Escherichia-Shigella |
GOS/inulin = ↑ acetic acid 3-FL = minor amounts of acetic acid (1.1 µmol mg−1) and lactic acid (0.3 µmol mg−1) LNT2 = ↑ acetic acid, butyric acid, succinic acid, and lactic acid |
Kong et al. (121) |
| 3-FL, 3′-SL, 6′-SL, and FOS | In vitro fermentation CoMiniGut model | 16S rRNA | Baby 1: 3-FL, 3′-SL, and 6′-SL = ↑ B. breve, P. distasonis Baby 2: 3-FL, 3′-SL = ↑ B. bifidum, B. adolescentis |
3-FL = ↑ propionic acid FOS = ↑ butyric acid |
Wiese et al. (122) |
| 2′-FL | M-SHIME | 16S rRNA, qPCR | ↑ Bifidobacteriaceae ↓ Veillonellaceae family |
↑ Acetate | Van den Abbeele et al. (123) |
| 2′-FL + L. helveticus Rosell-52 (R0052), 2′-FL + B. bifidum Rosell-71 (R0071), and 2′-FL + B. infantis Rosell-33 (R0033), probiotic combination (80:10:10) +2′-FL, and 2′-FL, | In vitro anaerobic batch culture fermentations | 16S rRNA | FF-fast degrader: 2′-FL, B. bifidum + 2′-FL, B. infantis + 2′-FL = ↑ Lactobacillaceae family FF-slow degrader: 2′-FL + L. helveticus Rosell, probiotic combination + 2′-FL =↓ Streptococcaceae BF-slow and BF-fast degraders: 2′-FL + L. helveticus Rosell, probiotic combination and 2′-FL = ↑ Lactobacillaceae family, ↓ Clostridiaceae |
BF-slow degrader: 2′-FL + L. helveticus Rosell, probiotic combination = ↑ lactic acid BF-fast degrader: 2′-FL, B. bifidum + 2′-FL, and B. infantis + 2′-FL = ↑ acetic acid 2′-FL + L. helveticus Rosell, probiotic combination + 2′-FL = ↑ lactic acid FF- fast degrader: 2′-FL + L. helveticus Rosell, probiotic combination + 2′-FL, 2′-FL, B. bifidum + 2′-FL, and B. infantis + 2′-FL = ↑ acetic acid, succinic acid, and lactic acid FF-slow degrader: 2′-FL + L. helveticus Rosell, probiotic combination + 2′-FL = ↑ lactic acid and succinic acid |
Nogacka et al. (108) |
| 2′-FL | M-SHIME | 16S rRNA, qPCR | Donors 1, 2, and 3 = ↑ Bifidobacterium spp. (B. adolescentis) Donor 1 = ↑ Lachnospiraceae Donor 3 = ↑ Acidaminococcaceae |
Donors 1, 2, and 3 = ↑ acetate, butyrate, and propionate Donor 3 = ↑ lactate |
Van den Abbeele et al. (124) |
| 2′-FL | In vitro fecal batch fermentations | 16S rRNA | FF-fast degrader = ↑ Lactobacillaceae (Lactobacillus) and Enterococcaceae FF-slow degrader = ↑ Enterobacteriaceae and Streptococcaceae BF-fast degrader = ↑ Bifidobacteriaceae (Bifidobacterium) and Enterobacteriaceae BF-slow degrader = ↑ Enterobacteriaceae |
FF-fast and slow degraders = ↑ succinic acid FF- and BF-fast degraders = ↑ acetic acid, ↓ isovaleric acid |
Nogacka et al. (125) |
DFL, 2,3'-di-fucosyllactose; CMPA, cow’s milk protein allergy; RCT, randomized controlled trial; FF, formula fed; BF, breast fed; SHIME, Simulator of the Human Intestinal Microbial Ecosystem; ↑, increase; ↓, decrease.
Knowledge regarding HMO assimilation in other infant-associated bifidobacterial members remains limited, although strains from species such as B. kashiwanohense, B. pseudocatenulatum, and Bifidobacterium longum subsp. suis (B. suis) have demonstrated some degradation abilities. Unlike B. bifidum and B. infantis, where HMO utilization is generalized to the species, HMO utilization in other bifidobacteria tends to be more sporadic. Studies have shown B. kashiwanohense strains DSM 21854 and PV20-2 as well as B. suis BSM11-5 preferred fucosyllactoses over sialylated structures, although this was strain dependent (126, 127). Additionally, it has been suggested that lactodifucotetraose (LDFT) and LNFP I may be acquired and metabolized by B. kashiwanohense JCM 15439T (128), with LNFPI and II degradation also carried out by strains such as B. longum SC596 (129), B. breve SC95 (104), and B. pseudocatenulatum strains SC595 and MP80 (130). Fucose accumulation in the growth medium as a result of fucosylated HMO fermentation has also been reported for B. kashiwanohense DSM 21854 (126), with free fucose unable to support growth (131). However, James et al. (127) deduced that even though growth, as measured using OD, was not substantial with free L-fucose, the metabolite profile altered, indicating some metabolic activity in the presence of this sugar. Additionally, L-fucose accumulation as a by-product from 2′-FL and 3-FL fermentation by B. kashiwanohense APCKJ1 was not observed (127). This, at least partial capacity to consume fucose, is highly intriguing and reinforces the importance of HMO and degradant(s) breakdown on a metabolic level. A recent study has demonstrated that fucose contributes to maintaining the reduction-oxidation balance, and in a medium with low levels of amino acids, fucose will promote bifidobacterial growth (132). Pooled HMOs as the primary energy source have supported select B. pseudocatenulatum strains (130, 133). For example, out of eight infant-derived isolates, four isolates, namely SC585, MP80, MP86, and DSM 20438, were able to achieve a high level of growth (0.94 ≤ maximum OD ≤ 1.17), comparable to B. infantis ATCC 15697 (maximum OD = 1.13), with this ability also evident using purified 2′-FL and 3-FL (130). It was determined that this consumption divergence between infant isolates was driven by the specific ability to metabolize fucosylated HMOs. Furthermore, LNB supplementation has aided growth in a subset of B. pseudocatenulatum tested (33 strains out of 61) (134). For certain strains from both B. kashiwanohense, e.g., APCKJ1 and B. pseudocatenulatum, e.g., MP80 and DSM 20438, putative mechanistic strategies for fucosylated sugar assimilation and catabolism have been described (127, 130). This fucosyllactose degradation specialization may significantly contribute to the formation of the gut metabolome.
Infant-based studies
Culture- and sequencing-based investigations have been instrumental in unraveling the ability of bifidobacteria and other taxa to utilize HMOs, although this has not been investigated extensively in vivo. Many in vivo studies have focused on the ability of breast milk, and/or specific HMOs (either individual structures or mixes), to reduce the development of infant-associated disorders such as necrotizing enterocolitis and evaluate the safety and tolerance of fortified formula milk (135, 136). Moreover, many in vivo investigations focus on 2′-FL and LNnT as these are most frequently added to infant formula. For example, Berger et al. (137) completed a randomized, double-blind study evaluating the gut microbiota of infants (0–14 days old)-fed formula (control) versus the same infant formula with the addition of both 2′-FL and LNnT over a period of 6 months. Following on from this, all infants received the same follow-up formula without HMOs until 12 months of age. Breast-fed infants served as a comparison group with samples taken at 3 and 12 months. At 3 months, the addition of HMOs to infant formula was found to have modulated the microbial composition, bringing it closer to that of the breast-fed infant group, as determined using 16S rRNA sequencing (137). HMO-containing infant formula supported bifidobacterial abundance (median relative abundance at 3 months: 82.81%) compared to the control formula (74.47%) (P < 0.01), although it was still less than breast-fed infants (90.87%) (P < 0.05, compared to control; non-significant compared to the test group) (137). Lower relative abundances of Escherichia and Peptostreptococcaceae were observed in the group fed infant formula containing HMOs compared to the control formula group (137). The supplementation of infant formulas with both 2′-FL and LNnT has been shown to support bifidobacterial abundance in clinical studies elsewhere (112, 113, 137). More specifically, Gold et al. (112), using 16S rRNA sequencing, reported that after 1 month, the microbiota became dominated by HMO-utilizing bifidobacterial species, e.g., B. breve, B. infantis, and B. longum, after supplementation with 2′-FL- and LNnT-containing formula. It must be noted that 16S rRNA sequencing alone for bifidobacteria subspecies identification may be problematic as the 16S gene sequence identity is relatively high (up to 99%) for Bifidobacterium species (138). A reduction in potential pathogens, such as Carnobacteriaceae, Escherichia spp., and Klebsiella spp., was observed in parallel (112, 113). This shift in microbial composition toward that of a breast-fed infant has also been reported for formula fortified with 2′-FL alone (114).
Investigating the associations between the infant microbiota and HMOs consumed can aid in revealing the apparent complex HMO-microbial dynamics. Borewicz and colleagues (139, 140) evaluated breast milk HMO concentrations and infant fecal microbial composition of 24 mother-infant pairs across three time points (2, 6, and 12 weeks old) (139) and of 121 mother-infant pairs at one sampling point (4 weeks old) (140). Redundancy analysis of the operational taxonomy units (OTUs) and estimated consumption of different HMO structures illuminated the potential association between Bifidobacterium OTU 1263 and a high level of HMO consumption. Different HMO structures were shown to be linked with the infant microbiota composition at various time points, such as LNFP III at 2 weeks, 3′-SL at 6 weeks, LNFP I and 2′-FL at 4 weeks (P < 0.05), and LNH at 12 weeks (139, 140). It should be noted that, in some instances, sample size was relatively small, further complicated by additional confounding factors, such as infant age and sex, mode of delivery, etc. Further OTU analysis highlighted numerous associations between bifidobacterial members and various HMO structures. Specific examples include Bifidobacterium OTU 614 associated with 2′-FL, 2,3'-di-fucosyllactose (DFL), LNDFH I, LNFP II, LNFP III, LNH, and LN(n)T, and the presence of Bifidobacterium OTU 418 being significantly correlated (P < 0.05) with high consumption levels of 2′-FL, LN(n)T, LNFP III, LNFP II, LN(n)H, LNDFHI, DFL, LNFP I, LSTa, LSTb, and LSTc (139, 140). In a recent larger-scale study, LNH, a non-fucosylated HMO, was positively associated with higher relative abundances of Bifidobacterium amplicon sequence variants (141). Elsewhere, in a Chinese cohort of 69 mother-infant pairs, it was reported that Bacteroides positively correlated (R1 >0.2, P < 0.05) with 3′-SL present in breast milk, while associations were also observed for certain fecal metabolites, e.g., tryptophan, N-acetyltryptophan, and indolelactic acid (142).
Recent in vivo studies focusing on the infant microbiota and HMOs, using either a mix of HMOs (111) or a mix of HMOs in combination with a probiotic (110), have reported the increased abundance of Bifidobacterium such as B. infantis, although this species-level distribution varies between these studies. Both studies demonstrated that the gut microbiota composition of infants-fed formula with added HMO(s) shifted toward that of a breast-fed neonate. Although individual HMOs were not comparatively assessed with the mix of five HMOs (2′-FL, DFL, LNT, 3′-SL, and 6′-SL) in Bosheva et al. (111), the authors postulated that there may be a more pronounced increase of B. infantis relative abundance when compared to a formula containing two HMOs (2′-FL and LNnT) (137), which presents an opportunity for further comparison. Additionally, HMO-fortified formula modulated the gut microbial composition of infants delivered via C-section to become closer to that of vaginally born infants (111, 137). Future in vivo studies evaluating the microbiota response to pooled HMOs, either alone or combined with other prebiotics such as GOS, would be informative, as HMO structures in breast milk are complex and heterogeneous.
In addition to HMOs, clinical trials have also been carried out with other established prebiotics such as GOS and FOS/inulin. In studies where long chain (lc) FOS has been referred to, we will denote this as inulin due to the longer chain structure, whereas the short chain structure will simply be referred to as FOS. The percentage of infants colonized by B. breve and B. adolescentis from enrollment to 4 months of age increased in infants fed GOS more so than the control (143). Furthermore, GOS, when combined with inulin, resulted in a higher abundance of B. longum, B. animalis, B. breve, and B. pseudocatenulatum, with the most notable increase evident for the B. infantis species (144). Administration of a non-hydrolyzed cow’s milk-based formula containing a synbiotic mix of GOS/inulin and B. breve M-16V was shown to shift the infant gut microbiota closer to that of breast-fed infants and compensated for delayed bifidobacterial colonization in infants delivered by C-section compared to the same formula without the synbiotic (145). Moreover, lower Clostridium viable plate counts and higher abundance of Bifidobacterium, as determined using qPCR, were observed in colic infants when supplemented with a GOS-containing formula (146), and similarly, as a result of a GOS-based synbiotic formulation, relative abundance reductions in Ruminococcus gnavus and Blautia were observed (147). Clinical evidence thus far suggests that prebiotics promote restoration in cases of a disrupted gut microbiota and improve diversity while decreasing potentially pathogenic bacteria, for instance following antibiotic treatment or delivery by C-section (110, 111, 137, 145, 148). Despite these insights, at present, there are very few publications related to in vivo studies that compare the effects of HMOs alongside, or in combination with, other prebiotics on the infant gut microbiota.
VARIOUS HMO DEGRADATION STRATEGIES EMPLOYED BY INFANT-ASSOCIATED BIFIDOBACTERIA
Species, subspecies, and, indeed, strain-specific molecular strategies involved in HMO utilization have been the focus of various studies (99, 104, 105, 149). These strain/(sub)species-level differences explain the co-existence of multiple bifidobacterial species and strains in a single niche, as overlapping and distinct metabolic capacities exist within the HMO-degrading bifidobacterial population in the neonatal intestine. Multiple HMO structures can be used by a variety of strains, highlighting their repertoire of human milk carbohydrate utilization mechanisms, many of which are illustrated in Fig. 4, thereby enabling infant-associated bifidobacterial prevalence and abundance. Although HMO metabolism is generally favored by infant-type bifidobacteria, this is not an exclusive ability as bifidobacteria isolated from adults, though probably to a lesser extent, have been shown to metabolize HMOs (150, 151).
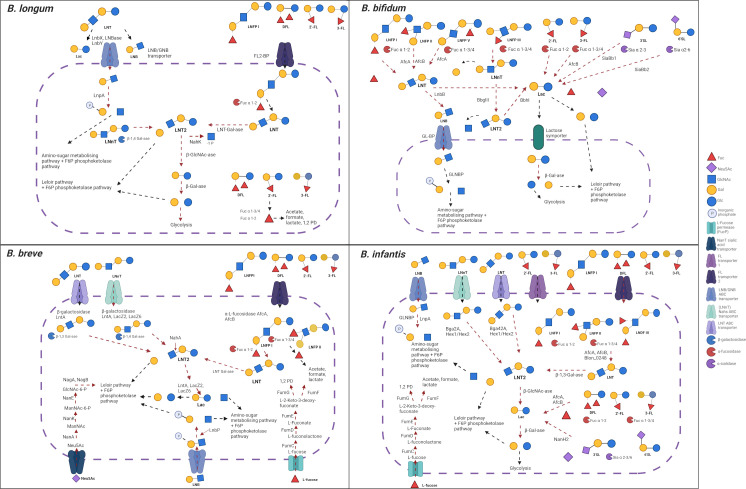
Fig 4.
Overview of HMO metabolism strategies four infant-associated Bifidobacterium species, namely B. longum, B. bifidum, B. breve, and B. infantis, employ. The proteins involved in the breakdown and transportation of some common HMO structure are represented here and discussed in the text. Conserved pathways within a given (sub)species are denoted in blue. Made using BioRender. LN(n)T, lacto-N-(neo)tetraose; LNT 2, lacto-N-triose 2; LNDFH III, lacto-N-difucohexaose III; GlcNac-6-p, N-acetyl-glucosamine-6-phosphate.
The various strategies which infant-related bifidobacteria employ to degrade HMOs can be broadly characterized as intracellular and extracellular approaches (102). The former relies on HMO-specific oligosaccharide transporters, while the latter is based on extracellular HMO-targeting glycosyl hydrolases (GHs). Uptake of intact or partially degraded HMOs is a signature of infant-associated bifidobacteria, although the complete set of molecular mechanisms employed by various bifidobacterial species have yet to be elucidated. B. infantis and B. breve internalize certain HMOs, e.g., LNnT, LNT, and LNB, prior to their intracellular metabolism (104, 105, 134, 152). Additionally, B. infantis has the capacity to internalize and degrade an array of additional fucosylated and sialylated HMOs, e.g., 3-FL, 2′-FL, 3′ SL (98, 153), while also participating in cross-feeding interactions (154), which is discussed in more detail further. Genome sequencing has been instrumental in understanding bifidobacterial-mediated breakdown of HMOs by identifying extensive arsenals of enzymes and transporters. This is particularly true in the case of B. infantis ATCC 15697, in which a 43-kb gene cluster was identified by Sela et al. (149) that harbors genes encoding GHs involved in the stepwise degradation of HMOs, i.e., a sialidase, fucosidase, N-acetyl-β-hexosaminidase, and β-galactosidase, as well as a range of transport-related genes (149). The internalization of HMOs is mediated through solute-binding proteins (SBPs) and associated ATP-binding cassette (ABC) transporters (155). It has been reported that B. infantis ATCC 15697 preferentially consumes small HMOs (degree of polymerization ≤7) over larger structures, which probably reflects the size limitation of associated ABC transporters, although it can still utilize longer-chain HMOs due to extracellular enzymes such as lacto-N-biosidase (101, 156). The ABC transporters and associated SBPs demonstrate exquisite specificity for the imported oligosaccharide structures (128, 157). The high degree of B. infantis adaptation for HMO acquisition is evidenced through the presence of two paralogous fucosyllactose transporters (FL transporter-1 and -2) (128). Sakanaka and colleagues (128) deciphered the overlapping, yet distinct, function of these transporters, which share up to 60% of their SBPs. FL transporter-1 (Blon_0341–0343) can import 2′-FL and 3-FL, in comparison to FL transporter-2 (Blon_2202–2204), which can internalize LDFT and LNFP I, in addition to 2′-FL and 3-FL (128, 155, 158).
Like B. longum, B. breve can assimilate a rather modest number of HMOs and HMO-derived structures. It was previously thought that fucosylated HMO utilization was uncharacteristic of this species; however, in recent years, strategies for HMO assimilation have been identified. In a study conducted by Ruiz-Moyano et al. (104), a GH95 α-fucosidase was identified in all 24 strains examined, whereas the presence of GH29 α-fucosidase was more variable and only detected in five strains. The ability of these strains to metabolize fucosylated HMOs was more prominent in those possessing a GH29 α-fucosidase, e.g., strains SC95, SC568, and SC154, although some possessing this enzyme were not able to degrade 2′-FL in vitro (104), indicating, perhaps, that other activities, such as enzymes or transporters, are necessary. The capacity to utilize acidic HMOs by B. breve was also demonstrated in this study, alongside the identification of an α-sialidase (GH33) (104). Glycoprofiling of a subset of B. breve strains revealed utilization patterns in the presence of total pooled HMOs, including the preferential consumption of specific sialylated HMOs, such as LSTb and monosialyllacto-N-hexaose (S-LNH), over smaller HMOs (104). Interestingly, when individual sialylated HMOs, 3′-SL and 6′-SL, were tested in vitro, growth was not apparent using these smaller molecules (104). Neutral HMOs, e.g., LNT and LNnT, have also been shown to support B. breve growth, both individual structures and when present in pooled HMOs (98, 104). James et al. (105) characterized various metabolic strategies that B. breve UCC2003 employs for LNT (type 1 HMO) and LNnT (type 2 HMO) digestion, determining that overlapping yet distinct metabolic pathways exist for these two HMOs. Reflecting the tactics of B. infantis, B. breve members also internalize LN(n)T using ABC transporters, making the structure(s) available for degradation by intracellular β-galactosidases. One of these enzymes, encoded by lntA, acts on the Galβ-1,3GlcNAc of LNT and Galβ-1,4GlcNAc linkages of LNnT. While the LntA β-galactosidase can act on both LNT and LNnT, the first galactose of LNnT can also be cleaved by other β-galactosidases, LacZ2 and LacZ6 (105). Both lacto-N-triose, which is then hydrolyzed by NahA, and galactose are liberated from the hydrolysis of LN(n)T. The breakdown products from either of these HMOs feed directly or indirectly into the bifid shunt metabolic pathway (105). Once thought of as a “scavenger species,” intensive efforts have revealed that the ability of B. breve strains to acquire and digest HMOs is more extensive than previously recognized.
In contrast to the (predominantly) intracellular approaches described for B. infantis, B. bifidum deploys extracellular hydrolases, e.g., fucosidases, to metabolize HMOs extracellularly and subsequently import the desired degradant carbohydrates (100). Some of these degradants can remain outside the cell for other (bifido)bacteria to acquire (100). Due to the presence of extracellular enzymes, large HMO structures (degree of polymerization >7) can be degraded as there does not seem to be a size limitation in contrast to bifidobacterial species solely depend on ABC transporters to access HMOs (159). B. bifidum provides metabolites, such as fucose and sialic acid, available extracellularly for others to consume as these two HMO-derived carbohydrates are not assimilated by B. bifidum itself (160, 161). To achieve this, a collection of secreted enzymes, including α-L-fucosidases (AfcA and AfcB), lacto-N-biosidase (LnbB), β-1,4-galactosidase III (BbgIII), β-1,3-N-acetylglucosaminidase I (BbhI), and sialidase II, is involved in extracellular HMO breakdown (100, 162). Sialidases from other bifidobacterial strains target α-2,3 and α-2,6 linkages, whereas SiaBb1 and SiaBb2 in B. bifidum target the α-2,3 linkage over the α-2,6 linkage (158, 163). B. bifidum also harbors a greater number of carbohydrate-binding modules in comparison to other species, which may enhance its catabolic ability through the increased GH affinity toward carbohydrates (164). Furthermore, an example of LNB utilization occurs in B. bifidum JCM1254, whereby a galacto-N-biose (GNB)/LNB transporter and GNB/LNB phosphorylase (GLNBP) act on LNB, with the ensuing products (galactose-1-phosphate and GlcNAc) then entering the Leloir and amino-sugar metabolizing pathways, respectively, before being routed to the bifid shunt (100, 101). The extracellularly released LNB can also be utilized by other bifidobacteria that possess a GNB/LNB pathway (158).
In contrast to species such as B. infantis, where HMO catabolism is highly conserved, the ability of B. longum members to assimilate HMOs is variable, and both intra- and extracellular approaches exist (165). This variability was observed by Garrido et al. (129), where B. longum SC596 was shown to vigorously use fucosylated HMOs from pooled breast milk samples, compared to other infant-derived B. longum strains tested due to the presence of a particular gene cluster. A subset of B longum strains, e.g., JCM1217, possess the lnbX gene (LnbX positive), encoding an extracellular lacto-N-biosidase, characterized as part of the GH136 family, which is involved in the degradation of LNT to LNB and lactose (158, 166). Similarly, B. bifidum encodes an extracellular GH20 LnbB (encoded by lnbB) which also hydrolyzes LNT, thereby liberating LNB and lactose (101), although the exact mechanisms differ between the two species. Both LnbB and LnbX are highly specific for activity on LNT, a core structural unit of many HMOs. The B. longum JCM1217encoded lacto-N-biosidase is more flexible in its structural specificity for LNB than its lnbB-encoded counterpart, as it can still release LNB units even if its substrate LNT is decorated with other monosaccharides, e.g., fucose (167). This could provide a route for the persistence of B. longum in the infant gut in that, even though only a limited number of HMOs can be utilized, there could be flexibility within this restricted range. The liberated LNB from LNT can then be transported across the B. longum cell membrane via the GNB/LNB transporter (158, 166). Strain-to-strain variability appears to be more prevalent in HMO consumption by B. longum subspecies members when compared to members of the B. infantis subspecies (103, 153, 168).
OTHER TAXA AND HMO UTILIZATION
HMO utilization in the infant gut is not a phenomenon exclusive to bifidobacteria. Intense research efforts have led to the identification of a broader scope of HMO-degrading microbes residing in the gut, including members belonging to taxa such as Akkermansia, Lactobacillus, Bacteroides, Ruminococcus, and Roseburia, which have developed strategies to consume specific HMO structures. Akkermansia muciniphila has the advantageous ability to consume host mucosal glycans (169). This species, a member of the Verrucomicrobia phylum that colonizes the mucus layer, can in addition to mucin-associated glycans also utilize certain HMOs. Structural similarities exist between mucin-derived glycans and HMOs; therefore, the presence and survival of A. muciniphila in the infant gut may be abetted by its HMO-foraging abilities. Both in silico and in vitro studies have elaborated on the functional capacity of A. muciniphila when HMOs are made available, which followed on from initial studies where positive correlations were observed between the presence of this bacterial species and HMO availability (170). Glycan-degrading enzyme expression has been observed when A. muciniphila is grown using HMOs as a sole carbon/energy substrate (171). Kostopoulos et al. (171) identified key enzymes involved in HMO degradation, including α-L-fucosidases, β-galactosidases, exo-α-sialidases, and β-acetylhexosaminidases and the GH families responsible (e.g., GH2, GH20, GH29, GH33, GH35, and GH95 families), and proposed the mechanistic strategies employed by A. muciniphila. The capacity to consume different HMO structures varied, e.g., a large proportion of 2′-FL and 3′-SL in the growth medium was degraded in contrast to that of 3-FL (171). This led to the proposal of putative degradation pathways for 2′-FL, 3′-SL, LNT, lacto-N-triose II, and lactose by A. muciniphila (171). A. muciniphila is often categorized into four phylogroups (AmI-AmIV), with the type strain, A. muciniphila MucT, assigned to the AmI phylogroup. Luna et al. (172) comparatively assessed the HMO-associated GHs present in 85 A. muciniphila genomes, with representatives from all 4 phylogroups included. GH20 (β-hexosaminidase; lacto-N-biosidase; β-N-acetylglucosaminidase) was consistently the most abundant HMO-associated GH across all phylogroups, with the highest number found in AmII. The lowest number of potential α-fucosidases (e.g., GH29, GH95, and GH141) and N-acetyl β-hexosaminidases (e.g., GH18, GH20, GH84, and GH109) was observed in isolates belonging to AmI (172). The authors then tested the HMO-utilizing ability of four A. muciniphila strains, one representative strain per phylogroup, in vitro. Similar to what has been observed for bifidobacterial members, strain-level variation was observed in terms of the final OD reached when cultivated in the presence of 2′-FL, 6′-SL, and LN(n)T, with strain CSUN-19 (AmIV) consistently achieving the highest OD using the four individual HMOs tested (172). A recent study focusing on the structural characterization of A. muciniphila fucosidases, specifically four assigned to GH29 (AmGH29A-D) and two assigned to GH95 (AmGH95A and AmGH95B), revealed that both AmGH29C and AmGH29D are able to hydrolyze 3-FL yet that only AmGH29C is able to target this glycosidic linkage within the structurally larger LNFP V (173). Broader enzymatic specificity of AmGH95B compared to AmGH95A was also observed as both were active on 2′-FL, while only AmGH95B exhibited activity on 3-FL (173). Relative abundance increase of Akkermansia effected by the inclusion of 2′-FL using in vitro infant fecal fermentations has also been observed by Lindner et al. (119).
Although regularly found at a lower prevalence than bifidobacteria, lactobacilli are frequently identified as members of the infant gut microbiota (28). In contrast to bifidobacteria, the ability to use HMOs is not a widely conserved phenomenon among lactobacilli. Correlation analysis, however, has indicated that degradation of 2′-FL, DFL, LNDFH I, LNT and its isomer LNnT, and LNFP II was significantly associated with a higher relative abundance of lactobacilli (140). In this study, the species level was undetermined, and it is important to consider the impact of other confounding factors. A limited number of strains have the functional capacity to degrade complex HMO structures. One such example is Lactobacillus plantarum ATCC BAA-793, for which pooled HMOs moderately supported growth (OD600nm 0.34 ± 0.1) (174). Many lactobacilli can capitalize on the availability of HMO constituents such as glucose, lactose, and GlcNAc to support growth, thereby acting as “secondary degraders” (175). Lacticaseibacillus rhamnosus strains GG and HN001 have been demonstrated to grow on fucose as the primary energy source (97, 153), although this was not consistent across all L. rhamnosus strains tested (153). Certain HMOs, e.g., LNnT, can be degraded to support growth, albeit moderately in some cases, of particular strains such as Lactobacillus acidophilus strains La-5 and NCFM, Lactiplantibacillus plantarum LP-66, and Limosilactobacillus reuteri DSM 17938 (97). L. plantarum is often identified in a range of environments such as fermented foods, plants, and the human gut (176). This expansive niche colonization is suggestive of the pervasive ability to utilize many substrates, including, to a limited degree, HMOs. Genomic assessment of 54 L. plantarum strains revealed that they all encode GH2 (β-galactosidase), whereas the presence of genes encoding GH20 (β-hexosaminidase/lacto-N-biosidase) was lineage dependent, both of which can be involved in HMO breakdown (177). The authors postulated that the presence of these enzymes is related to lactose degradative abilities and the highly diverse environments where L. plantarum can colonize (177). Some mechanistic strategies have been described, e.g., a novel phosphoenol phosphotransferase system was identified in Lacticaseibacillus casei, which facilitated the importation of LNB (178), and similarly, a system for LacNAc internalization has been described (179); both LNB and LacNAc represent HMO constituents. Additionally, α-L-fucosidase activity as part of the GH29 family (AlfA, AlfB, and AlfC) has been characterized in lactobacilli such as L. casei, Lacticaseibacillus paracasei, and L. rhamnosus, with different strains predicted to carry one, two, or three of such intracellular enzymes (180, 181). Furthermore, an extracellular β-galactosidase described in L. acidophilus NCFM can remove galactose from LNnT resulting in the liberation of LNT2 (97). In a recent study (182), the growth of several Lactobacillus strains (L. acidophilus ATCC 4796, Lactobacillus johnsonii ATCC 33200, L. paracasei ATCC 25302, L. plantarum ATCC 14917, Limosilactobacillus fermentum ATCC 14931, Lactobacillus gasseri ATCC 3323, Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842, and Levilactobacillus brevis ATCC 27303) was examined using preterm human milk, preterm formula, or term formula with added 2′-FL. In comparison to the medium control, all strains were shown to grow using preterm milk and preterm formula, with six strains able to grow using the HMO-supplemented formula (182). Variation occurred between strains when pH was measured after 20 hours growth (182). The authors next scrutinized 452 lactobacilli genomes across 23 species to identify genes encoding putative HMO-degrading GHs (182). Fewer GHs were identified compared to bifidobacterial genomes; nonetheless, most lactobacilli species were predicted to encode members of GH2 and GH42 families, while genes specifying putative GH20 and GH29 family members were less commonly identified (182). Although many lactobacilli do not appear to possess the ability to consume a wide variety of different HMOs, their abundance and functional activity can be stimulated indirectly by extracellularly released carbohydrates and metabolites from HMO degradation by other taxa (183).
Bacteroides species are present in the infant gut population to varying extents (28) and commonly possess extensive saccharolytic activities achieved through the presence of polysaccharide utilization loci. Constituent members such as Bacteroides thetaiotaomicron, Bacteroides fragilis, and Bacteroides vulgatus (now Phocaeicola vulgatus) have been shown to exhibit growth in the presence of various HMO structures when used as the primary energy source (153, 184, 185). These include 2′-FL, 3-FL, and 6′-SL, with species differences observed when 3-FL and LDFT were the sole carbohydrate substrates (184). Like A. muciniphila, some Bacteroides members can consume host-derived, mucus-associated glycans, and it has been determined that Ba. thetaiotaomicron and Ba. fragilis induce the same genes for HMO utilization as for such host mucus glycan consumption, although species variation was evident (186, 187). The link between mucin and HMO degradation in the infant gut remains unclear. The overlapping enzyme activities employed in HMO and host mucus glycan degradation include GH33 sialidases, GH2 β-galactosidases, and GH95 and GH29 fucosidases (171, 185). The dual ability to metabolize both mucin-associated glycans and HMOs has been observed in only a limited number of bacteria from genera such as Bacteroides, Akkermansia, and Ruminococcus (171, 186, 188). Furthermore, the characterization of mucin utilization enzymes and pathways for B. bifidum PRL2010 and JCM1254 has been reported (162, 189). B. bifidum, as described above, mainly employs extracellular glycosidases for HMO utilization which could be a significant factor compared to other bifidobacteria which mostly use internal enzymes for HMO hydrolysis, thereby limiting their mucin degradation capacity. As mentioned previously, A. muciniphila is a prominent mucin degrader which uses similar GHs for HMO utilization, e.g., GH2, GH20, GH29, GH33, GH35, and GH95 (171). The ability of these species to degrade both HMOs and mucin may have developed to facilitate their persistence in the gut as the infant develops to adulthood. This opportunistic consumption of HMOs may play a role in the persistence of Bacteroides members in the infant gut. Interestingly, there appeared to be similar growth patterns depending on the nature of the HMO structure for one Bacteroides dorei (now Phocaeicola dorei) isolate, i.e., sialylated (3′-SL and 6′-SL), neutral (LNT and LNnT), and fucosylated structures (2′-FL and DFL) (190). This, however, was not the case for a second P. dorei isolate as variation in the ability to consume either 2′-FL or DFL was observed. The transcriptional response for one P. dorei isolate revealed a broad GH family repertoire facilitating the breakdown of HMOs, including the upregulation of GH2, GH43, and GH97 families (190). However, it appears to be a more generalized response, i.e., no clear association between HMO structure and GH family response when compared to that of bifidobacteria, which apply a more structure-specific approach to HMO degradation (190). For example, and rather unexpectedly, the expression of GH29 and GH95 fucosidases was shown to be upregulated in response to non-fucosylated HMOs, while GH33 sialidases were upregulated in the presence of non-sialylated HMOs (190). Moya-Gonzálvez et al. (191) mined gut metagenomic data from a breast-fed infant to evaluate the presence of GH29 family members. GH29 α-L-fucosidases were annotated in Ba. thetaiotaomicron, Bacteroides caccae, Ruminococcus gnavus, P. vulgatus, P. dorei, and Streptococcus parasanguinis genomes (191). Further characterization of 10 putative α-L-fucosidase-encoding genes revealed substrate specificity. For example, Fuc18 (Ba. thetaiotaomicron), Fuc19A (Ba. caccae), Fuc35B (Ba. caccae), Fuc39 (Ba. thetaiotaomicron), and Fuc1584 (P. vulgatus) were shown to elicit hydrolytic activity on α1,3/4-linked fucose moieties present in 3-FL, while Fuc2358 (S. parasanguinis) was demonstrated to exhibit a broad substrate specificity, releasing fucosyl residues from all tested glycans which included 2′-FL and 3-FL (191). In contrast to many infant-associated bifidobacteria, Bacteroides members have been shown to elicit growth when cultivated on fucose as the sole carbon/energy source (153), indicating that they may profit from HMO degradants released by other taxa.
Disentangling the metabolic pathways employed by certain Roseburia species, e.g., Roseburia inulinivorans and Roseburia hominis, to metabolize particular HMOs was the focus of a recent investigation (192). A combination of genomic analyses and in vitro experiments has highlighted the presence of GH136 homologs involved in HMO degradation as well as an intracellular GH112, an LNB phosphorylase (192), with R. inulinivorans DSM 16841 additionally harboring two fucosidases, GH29 and GH95. The putative SBPs of R. hominis and R. inulinivorans ABC transporters were also described, as upregulation was evident in response to HMOs. Interestingly, R. inulinivorans DSM 16841 and R. hominis DSM 16839 were shown to exhibit limited growth on the most abundant fucosylated and sialylated HMOs, specifically 2′-FL, 3-FL, 3′-SL, and 6′-SL (192). Failure to grow on sialyllactose was consistent with the lack of sialidases present in the corresponding Roseburia genomes. However, growth for both butyrate-producing Roseburia strains was observed when purified HMOs, or the HMO constituent LNB, are available. R. hominis was also able to grow on LNT in contrast to R. inulinivorans (192). In the same study, Eubacterium ramulus DSM 15684 was tested, and growth was observed using LNT (192). Another prominent butyrate producer, Faecalibacterium prausnitzii A2-165 was the focus of a cross-feeding study with B. infantis ATCC 15697, which is discussed in more detail below, where it was proposed that F. prausnitzii promotes the B. infantis sialidase activity when in co-culture with 6′-SL (193). Moderate growth was observed in F. prausnitzii monoculture with 6′-SL (OD600nm 0.5) higher than 2′-FL, 3-FL, and LNT2 (OD600nm 0.2–0.3), although this was less than when in the presence of glucose (final OD600nm of ~1.8) (193). In a clinical trial involving adults with chronic gastrointestinal issues (151), significant increase (P < 0.05) in stool counts of F. prausnitzii was found between the baseline (0 weeks) and the endpoint (6 weeks) when supplemented with 2′-FL-containing nutritional formula. Although this is a preliminary result and the 12 participants were adults, not infants, the ability of F. prausnitzii to consume individual and/or pooled HMOs and the contribution to the microbial metabolic networks that may occur remain largely unexplored and thus require further investigation. Although taxa described here may represent low abundance microbes in the neonatal gut, their presence may have a significant impact on associated microbial functionalities and development of a diverse microbial environment. This could be supported by prominent and efficient HMO users such as bifidobacteria.
Due in part to their structural diversity and specificity, many HMOs remain inaccessible to a range of potentially pathogenic microbes. Salli et al. (153), using in vitro growth experiments, demonstrated very limited utilization of 3-FL by a selection of undesirable microbes, such as certain E. coli strains, Clostridium perfringens E-98861 T, and Cronobacter sakazakii DSM4485. However, 2′-FL and DFL did not support growth of these potential pathogens (153). It should also be noted that single-strain utilization in vitro generally does not necessarily translate to in vivo studies. R. gnavus, a mucin degrader and colonizer of the infant and adult gut (194), possesses various GHs and other enzymes for host glycan degradation. The exact role of R. gnavus in the intestine has not been fully elucidated as it is often referred to as a commensal (195), while a clade purportedly linked to inflammatory bowel disease in adults was also identified in infants (196). Crost et al. (188) reported the growth of both R. gnavus E1 and ATCC 29149 strains when supplemented with 2′-FL and 3-FL but a lack of growth on LN(n)T, and variability between strains on 3′-SL and 6′-SL. However, in Il10−/− mice, supplementation with 2′-FL reduced colitis, and an expansion of R. gnavus abundance was noted (195). Mechanistic strategies have also been proposed for HMO digestion such as the presence of multiple α-L-fucosidases (GH95, and GH29A and 29B subfamilies), which can act on fucosyllactoses (197). Strain-level variation has also been observed. For example, R. gnavus E1 lacks a GH33 sialidase, unlike the GH33 identified in R. gnavus ATCC 29149 and ATCC 35913, which represents an intramolecular trans-sialidase (198, 199). The latter two strains also harbor a nan gene cluster for sialic acid metabolism (188, 199).
Typically, HMOs do not support vigorous growth of potential (opportunistic) pathogens and are more selective for taxa such as infant-associated bifidobacteria, therefore enabling their dominance, both compositionally and functionally, in situ. Concurrently, HMO-mediated reduction of pathogens in vitro is widely recognized, including in cases of enteropathogenic E. coli (200), group B Streptococcus (201–203), Acinetobacter baumannii (204, 205), and certain viral infections [reviewed by Moore et al. (206)]. HMOs have anti-pathogenic capabilities by acting as soluble decoy receptors, thus inhibiting pathogen binding and colonization, potentially resulting in reduced infections (207). Although beyond the scope of this review, the HMO-based host immunological impacts have been reviewed in depth elsewhere (46, 208, 209).
METABOLITES LINKED TO OR DIRECTLY RESULTING FROM HMO METABOLISM
The metabolites produced as a result of HMO metabolism are key variables in microbe-mediated interactions with the host. The beneficial impacts exerted can be direct to the respective host and/or indirect through the enrichment, in both abundance and activity, of other microbes in the gastrointestinal environment (210). One of the most prominent microbial metabolite groups studied resulting from saccharolytic fermentation is represented by the SCFAs, in particular acetate, butyrate, and propionate. Generally, SCFAs are recognized as energy sources for colonocytes, interacting with the immune system, e.g., promoting anti-inflammatory cytokine production and preventing growth of pathogens (211). Some of the metabolites described here, including SCFAs, contribute to the acidification of the gut, thus creating an inhospitable environment for pathogens to thrive, thereby preventing the establishment of enteric disorders. Although the literature predominantly demonstrates the benefits of SCFAs (212), these positive impacts have been more recently questioned (213). Instances where an excessive amount of SCFAs are present and factors such as the host health status (213, 214) may lead to less favorable SCFA-mediated host effects. For instance, higher SCFA concentrations in adults have been associated with obesity and hypertension (214), while propionate has been linked to an increased risk of type 2 diabetes (215). Therefore, their status as beneficial metabolites may be considered controversial and more complex than once previously recognized (216). Many investigations have reported the production of SCFAs and other organic acids, such as lactate and formate, as metabolic end products of microbial fermentation of HMOs, both pooled and individual structures (98, 123, 217). Acetate and lactate are two major end products following HMO metabolism by infant-associated bifidobacteria (98, 217, 218). As bifidobacteria are prominent acetate and lactate producers, they contribute to the beneficial effects exerted by SCFAs, although they appear to lack the metabolic machinery to produce butyrate and propionate (16). However, bifidobacterial metabolites, such as lactate, can participate in cross-feeding interactions, thereby supporting formation of other SCFAs such as butyrate by other gut microbes (219). Lactate itself has beneficial host impacts such as supporting epithelial cell barrier activity (220). It must be considered that lactate may also have negative consequences on host physiology (221), such as acting as a growth substrate for certain pathogens, e.g., Salmonella enterica serovar Typhimurium (222), and possibly promoting biofilm formation and anti-fungal drug resistance, e.g., Candida albicans (223).
HMO structure is a determining factor with respect to microbial utilization, and thus, the metabolic by-products released. For example, variations between fucosylated and sialylated HMOs relating to SCFA production have been noted, with fucosylated HMOs linked to higher amounts of acetate (109). This is particularly true for 2′-FL (123, 124), although it is worth recognizing that it is one of the more well-studied HMOs; therefore, more data are available. Additionally, compared to lactose controls, 2′-FL fermentation by B. infantis Bi-26 resulted in a more diverse metabolite profile, with formate, 1,2-propanediol (1,2-PD), and fucose produced alongside acetic acid and lactate (218). Formate and 1,2-PD can be generated during fucose metabolism as pyruvate is directed away from lactate production (152, 224). This results in an increase in acetyl-CoA, thereby favoring ATP production through a section of the bifid shunt (152, 224). It has been suggested that this shift in metabolic activity, typically resulting in enhanced levels of acetate and formate, enables the restoration of the reduction-oxidation balance and facilitates energy production (98, 218). Fermentation of complex HMOs can liberate core components, e.g., fucose, which subsequently can be utilized and thus result in metabolite production such as 1,2-PD. Therefore, degradation of HMOs, or derivatives thereof, can contribute to propionate production as 1,2-PD can act as a precursor in the formation of this SCFA. Propionate can be involved in the mediation of microbiota and host homeostasis, appetite regulation, and participate in gluconeogenesis in the liver (225, 226). Schwab and colleagues (131) demonstrated that B. infantis can metabolize 2′-FL and 3-FL and, when in co-culture with Eubacterium hallii (now Anaerobutyricum hallii/soehngenii), B. infantis produces acetate, lactate, and 1,2-PD, which can be consumed by A. hallii, resulting in the formation of butyrate, propionate, and formate. This metabolic scheme has also been identified with Limosilactobacillus reuteri ATCC PTA 6475 and B. breve UCC2003, whereby the growth of L. reuteri was enhanced by the 1,2-PD produced from B. breve fucose metabolism (183). This fucose, in turn, was provided by B. bifidum PRL2010 following mucin degradation demonstrating the broader microbiota implications of bifidobacteria in supporting other members of the gut (183). Following a similar pattern, B. infantis co-cultured with F. prausnitzii amassed a significantly (P < 0.0001) higher amount of acetate when compared to a B. infantis monoculture when 6′-SL was the primary energy source, although this was not the case for 2′-FL, 3-FL, and LNT2 (193). HMO fermentation, therefore, may contribute to beneficial metabolite formation either directly or indirectly through sophisticated cross-feeding relationships.
In vitro fermentations using fecal inocula offer a community-level insight into the metabolites produced when HMOs are obtainable by the microbes present (107, 109, 123) (Table 1). In one such study, B. longum-dominant microbial communities were supplemented with individual HMOs (2′-FL, 3-FL, 3′-SL, 6′-SL, LNT, or LNnT), GOS, or FOS (109). The HMOs, varying in structure, and GOS metabolite profiles were comparable, which has also been observed elsewhere (107), with more acetate and lactate produced compared to FOS supplementation. Neutral HMOs, i.e., 2′-FL, 3-FL, LNT, and LNnT, and GOS produced more acetate and lactate compared to sialylated structures. Lactate and succinate, which are intermediate metabolites in SCFA formation, are often positively correlated with microbial HMO degradation, including in the in vitro fecal fermentation situation (107). Both lactate and succinate are microbial metabolic end products, yet they may also act as precursors in the formation of propionate by certain gut microbes (211, 227). Elsewhere, using pooled fecal samples from 12-week-old infants, the fermentation of LNT2 was shown to result in higher total SCFAs and organic acids, such as acetic acid, butyric acid, and lactic acid, than 3-FL or GOS/inulin (9:1) (121). The presence of LNT2 also led to the production of succinic acid, which was absent during 3-FL fermentation (121).
The initial microbial assembly is a predominant contributor to the metabolite profile generated from non-digestible carbohydrate fermentation. A Bacteroides-dominated inoculum, when supplemented with different HMOs, GOS, and FOS, was shown to result in higher succinate yields than two other Bifidobacterium-dominant groups tested, although donor-specific effects were also observed (107). This influence of the starting microbiota on metabolite production has been observed elsewhere, with the designation of “fast degraders” and “slow degraders” by Nogacka et al. (108, 125) contingent on the ability of 2′-FL to be utilized during batch fecal fermentations. Interestingly, both degrader types were evident irrespective of feeding mode, although the low sample size in these two studies means validation is required. The highest levels of acetate and lactate were shown to correspond to fast degraders, which consisted of increased relative abundance of bifidobacteria when compared to that of slow degraders (108, 125). Similarly, Van den Abbeele and colleagues (123, 124), using adapted Simulator of the Human Intestinal Microbial Ecosystem in vitro colon models, demonstrated the rapid fermentation of 2′-FL and enriched bifidobacterial relative abundance, which concomitantly resulted in increased acetate production.
A standout study conducted by Laursen et al. (85) evaluated the breast milk-derived bifidobacterial conversion of aromatic amino acids, e.g., tryptophan, phenylalanine, and tyrosine, to aromatic lactic acids, e.g., indole-3-lactic acid (ILA), phenyllactic acid, and 4-hydroxyphenyllactic acid (85). This is achieved via an aromatic lactate dehydrogenase (ALDH) (85). ILA was shown to activate aryl hydrocarbon receptor (AhR) and hydroxycarboxylic acid receptor 3-dependent pathways, which are involved in the mediation of homeostasis and immune system interaction, e.g., through stimulation of IL-22 (85, 228). These results point to a mechanism by which bifidobacteria-derived aromatic lactic acids can provide protective effects by modulating the infant immune system. Generally, the ability of the Bifidobacterium tested to produce aromatic lactic acids concurred with a capacity to utilize HMOs, suggesting HMOs provide integral support to bifidobacterial growth and production of these beneficial metabolites (85). An important factor to note is that the bifidobacteria in this study are commonly identified in the microbiota of breast-fed infants and that the aromatic lactic acid-producing ability, including the associated gene encoding the ALDH responsible for this conversion, does not appear to be present in bifidobacteria typically associated with a more mature microbiome. This infant-specific phenomenon has been observed elsewhere in relation to ILA production whereby infant-type bifidobacteria strains produced more ILA (1.84 ± 1.07 µg/mL) than those typical of an adult gut (0.4 ± 0.1 µg/mL) (229). Furthermore, B. infantis ATCC 15697-produced ILA has demonstrated anti-inflammatory potential via IL-8 reduction, which was mediated through AhR interaction, resulting in the prevention of IL-8 transcription (230). In-line with these studies was the observation that B. infantis EVC001-produced ILA upregulated immunoregulatory galectin-1 in Th2 and Th17 cells during polarization in vitro (36). Taken together, this suggests that specific metabolic activity, resulting in immunological interactions, is linked to breastfeeding, select infant-associated bifidobacteria, and the ability to consume HMOs. The full implications of aromatic lactic acids and why exactly this conversion occurs by infant-associated bifidobacteria remain to be fully elucidated. This underscores a knowledge gap that exists relating to the adapted functional metabolism of infant-associated microbes and the role of HMOs. Metabolites produced from HMO breakdown can also be directed into other microbial metabolic pathways through intricate cross-feeding interactions, as will be discussed below.
HMO-BASED CROSS-FEEDING INTERACTIONS
Bifidobacteria provide a beneficial effect not only through the degradation of HMOs to the infant directly but also indirectly through close associations between species within the genus and, indeed, with those from other genera inhabiting the infant gut. B. bifidum provides HMO breakdown products, e.g., fucose, for other species, e.g., B. breve, to assimilate (231), thus acting as a driver of community structure through the provision of substrates for others to consume (Fig. 5) (100, 160). The liberation of monosaccharides by B. bifidum from HMOs can play a significant role in the prevalence of other bifidobacterial species, particularly B. breve, in the infant gut (100). The transient release of 3-FL, when part of pooled HMOs is metabolized by B. bifidum R0071, has recently been demonstrated (98). Although B. bifidum can subsequently utilize 3-FL itself, or when co-cultured with B. infantis R0033, an increase in B. infantis R0033 was observed, indicating that (parts of) this HMO is made available for others to consume (98). B. breve can vigorously consume breakdown products from HMO assimilation, thereby facilitating its enrichment in the gut (232). Members of this species can utilize fucose, often made available through the breakdown of HMOs, which is unlike many other infant bifidobacterial species (106). In an in vitro investigation, B. breve M-16V has been shown to exhibit poor growth in mono-culture using 3-FL, 2′-FL, 3′-SL, 6′-SL, and LNnT, although this strain was shown to be able to use the type 1 HMO, LNT (98). However, when in co-culture with B. bifidum R0071 and 3′-SL, growth of B. breve M-16V prevailed (98), highlighting the capacity of B. breve strains to forage on HMO breakdown products for its own prevalence. Cross-feeding between B. breve and B. bifidum involving sialylated HMOs has also been observed elsewhere (161, 233). The extracellular sialidase-mediated release of sialic acid from 6′-SL by B. bifidum ATCC 15696 (161) and 3′-SL by B. bifidum PRL2010 (233) can benefit the growth of B. breve JCM 7019 and UCC2003, respectively.
Fig 5.
Some of the beneficial impacts that occur from human milk oligosaccharide availability to the infant. Select infant-associated bifidobacteria are highly adapted to digest HMOs. Certain members of other taxa also possess metabolic strategies to degrade specific HMO structures. Made using BioRender.
An intricate carbohydrate sharing network among infant-associated bifidobacteria may contribute to their dominance in the neonatal gut. This applies to non-HMO utilizing bifidobacteria, which may be supported by the altruistic behavior of HMO utilizers, thus maximizing nutrient usage. Lawson et al. (217) demonstrated that spent or “conditioned” media from HMO degraders consuming 2′-FL (B. pseudocatenulatum strains LH9, LH11, LH13, and LH14) supported the growth of B. longum LH12, a non-HMO consumer (217). Combinations of strains examined in this study were isolated within the same infant microbial community. However, this mutually beneficial interaction was inconsistent, e.g., B. infantis LH23 2′-FL-derived media did not aid the growth of two B. breve strains, LH21 and LH24 (217). Conditioned nutritional media from HMO utilizer B. longum LH206 grown in the presence of 2′-FL, and to a lesser extent LNnT, were shown to provide modest growth support to B. infantis and B. pseudocatenulatum strains which have been isolated from the same infant (217). Although the exact mechanisms remain elusive, breakdown products were identified, indicating that fucose, galactose, acetate, and GlcNAc are involved in facilitating HMO-based cross-feeding relationships. This phenomenon may contribute to the maturation of the gut microbiota as HMO breakdown products may be exploited by typical adult-associated bifidobacteria such as B. adolescentis, thus assisting metabolic activity and consequent growth of other bifidobacteria (124). There may be particular HMO-degrading strains, rather than species, that are specifically equipped to participate in nutrient sharing with non-HMO-degrading bifidobacteria.
Interactions between bifidobacteria and other SCFA-producing gut microbes, e.g., Anaerostipes caccae (154) and A. hallii/soehngenii (ex-E. hallii) (131), when HMOs are the primary carbohydrate source have been demonstrated in vitro. Trophic interactions can be facilitated by the breakdown of HMOs into simpler carbohydrates, e.g., fucose, and/or providing metabolites, e.g., acetate and 1,2-PD, that encourage the growth of others. As mentioned above, in a study conducted by Cheng et al. (193), the prominent butyrate producer F. prausnitzii was shown to support B. infantis growth when 6′-SL was present as the primary carbohydrate source in co-culture. However, it was observed that F. prausnitzii is not able to substantially consume 6′-SL itself. Additionally, in co-culture, more 6′-SL was consumed, and a significant (P < 0.0001) quantity of acetate produced, compared to B. infantis monoculture with 6′-SL, with the authors postulating that F. prausnitzii promotes B. infantis sialidase activity, although the exact mechanism by which this happens remains unknown (193). In the same study, when in the presence of 2′-FL, the co-culture and B. infantis monoculture reached similar cell density levels, although the growth rate was faster in co-culture conditions, i.e., stationary phase was reached after 32 hours of cultivation compared to 56 hours (193). Mono- and co-culture-based investigations with the butyrate producer A. caccae demonstrated that, in the presence of HMOs, B. infantis (154) and Ba. thetaiotaomicron (185) generate breakdown products e.g., glucose and galactose, available for A. caccae to consume. This, in turn, facilitates the production of butyrate, which, among other beneficial impacts, has been associated with a reduced risk of food allergy development in the infant (234). A recent synbiotic-based study, albeit in adults, showed that a combination of B. infantis and an HMO concentrate from human breast milk during and after antibiotic administration promoted the abundance of B. infantis (235). At the metabolite level, enhanced amounts of lactate and ILA and decreased p-cresol sulfate were measured (235). Additionally, increased levels of Veillonella spp., which are lactate consumers, and the stimulation of Veillonella-produced propionate, both in vitro and in vivo, were observed, indicating a cross-feeding occurrence (235). As discussed above, interest relating to A. muciniphila, a strict anaerobe which has been identified in 1-month-old infants (236), and its ability to degrade HMOs has been accruing in the scientific literature (171, 172). Not only does A. muciniphila have the capacity to assimilate certain complex structures (171, 172), it can also consume constituents such as fucose (171). Although little is known about the dynamics between infant bifidobacterial species and Akkermansia with respect to HMO degradation, there is an opportunity to investigate possible metabolic interactions and whether the degradation mechanisms are comparable and/or complementary.
INFANT-ASSOCIATED BIFIDOBACTERIA AND PREBIOTIC CARBOHYDRATE METABOLISM
Well-studied prebiotics as additions to infant formula include GOS, FOS/inulin and, more recently, select HMOs such as 2′-FL and LNnT (237–239). Clinical trials in infants have demonstrated that 2′-FL, LNnT, and HMO mixtures when incorporated in standard formula are safe and support growth in infants (240–242). At present, there are seven commercially available HMOs, namely 2′-FL, LNnT, 6′-SL, 3′-SL, 3-FL, LNT, and DFL (243). As noted throughout this review, the heterogeneous mix of HMO glycans is often selective for breast-fed infant-type bifidobacteria (158); conversely, GOS and FOS/inulin are less selective, i.e., bifidobacteria of both infant and adult origin are supported, and more generally, growth and/or activity of a broader range of taxa can be facilitated (153, 243). There are a variety of overlapping, yet distinct, metabolic strategies to utilize these carbohydrates when provided to the microbiota (Table 2).
TABLE 2.
Various mechanisms that members from four infant-associated bifidobacterial species employ for HMO, GOS, and FOS/inulin degradationa
| HMOs | GOS | FOS/inulin | References | |
|---|---|---|---|---|
| B. infantis | ABC transporters, various GHs present in 43 kb “HMO island,” intracellular Bga42A hydrolyze LNT and Bga2A for LNnT. GlcNAc-6-p deacetylase upregulated in the presence of LNT and LNnT to breakdown HMO-bound GlcNAc. GH29 1,3/1,4-α-L-fucosidase hydrolyzes the 3-fucosyl and 4-fucosyl structures of fucosylated HMOs. Predicted intracellular GH33 2,3-2,6-α-sialidases present. | ABC transporter, intracellular GH2 and GH42 β-galactosidases (Bga42A, Bga42B, and Bga42C) for GOS hydrolysis. Bg42A is encoded for 3′-GL hydrolysis. |
ABC transporter, and GH13 (Blon_0128) and GH32 enzymes (Blon_2056) are expressed for FOS and inulin. Blon_0787 is encoded for FOS hydrolysis. An exoinulinase with β-fructofuranosidase and invertase for inulin. | (149, 224, 244–249) |
| B. bifidum | Extracellular LnbB (GH20) liberates LNB from HMOs. Intracellular GLNBP for LNB degradation. Extracellular GH95 1,2-α-L-fucosidase for 2′-FL, GH29 1,3-1,4-α-L-fucosidase for 3-FL, and GH33 exo-α-sialidase for 6′-SL degradation. | GH42 for hydrolysis of GOS GH2 β-1,4-galactosidase and BIF3 for extracellular hydrolysis of GOS. |
Highly variable. GH13 potentially involved. | (7, 159, 189, 244, 247, 250–255) |
| B. breve | ABC transporters, intracellular GH95 and GH29 α-fucosidases, GH33 α-sialidase encoded for fucosylated and sialylated HMO degradation. β-Galactosidases involved in LN(n)T metabolism. | GL-BG transports 6′-GL into the cells for degradation by GH42 β-galactosidase (e.g., BbrY_0422). Endo-β-galactanase (GH53) encoded for 4′-GL degradation. ABC transporter for internalization. |
MFS transporter (lacy), ABC transporter, intracellular β-fructofuranosidase encoded by GH32 family for FOS and inulin hydrolysis. Variable growth observed. | (103, 105, 245, 256, 257) |
| B. longum | GH136 lacto-N-biosidase LnbX-positive, extracellular GH95 and GH29 α-fucosidases. LnbX-negative intracellular degradation of HMOs. GH112 family, GLNBP. GH33 2,3-2,6-α-sialidase. | Extracellular β-galactosidase I (BL0259), endo-β-galactanase (GH53). | Intracellular GH32 β-fructofuranosidase, ABC transporters (BL1164-BL1165), and putative GHs (BL0529 and BL0544) for inulin. | (129, 244, 247, 249, 258–260) |
3′-GL, 3’ galactosyllactose; LN(n)T, lacto-N-(neo)tetraose; LnbX, Lacto-N-biosidase; GH, glycoside hydrolase; GlcNac-6-p, N-acetyl-glucosamine-6-phosphate; SA-C2, sialic acid utilization cluster 2; GLNBP, GNB/LNB phosphorylase.
One of the most intriguing and distinguishing characteristics of HMOs is their structural complexity, diversity, and specificity. It has been observed in fecal fermentations that 2′-FL results in a greater increase in the relative abundance of bifidobacteria when compared to GOS, and when combined, faster fermentation occurred than when GOS was added alone (120). However, the HMO structure should be taken into consideration when comparing against an established prebiotic such as GOS. For example, when 3-FL or GOS/inulin was tested in a fecal fermentation experiment, the relative abundance of B. longum increased but not that of B. bifidum, whereas LNT2 increased both B. bifidum and B. longum relative abundance (121). GOS supports growth of various Bifidobacterium members, such as B. longum and B. breve (107, 109, 120), and infant formula supplemented with GOS has been shown to lead to increased bifidobacterial abundance in infants (143). Structural specificity is also a factor with GOS and FOS, though to a significantly lesser extent than HMOs. Using B. breve UCC2003 as a model strain, it was determined that metabolism of large GOS fractions is associated with the ability to produce an extracellular endogalactanase (261). ABC uptake systems have substrate size limitations meaning larger HMO structures (e.g., greater than a degree of polymerization of 8) cannot be internalized unless first degraded extracellularly (232, 262). Some of the enzymes necessary for GOS metabolism are also involved in LN(n)T catabolism, showing functional overlap between GOS and HMO metabolism in B. breve (105, 261).
As described above, carbohydrate metabolism by B. infantis, LnbX-negative B. longum, and B. breve relies on transport systems to internalize carbohydrates (e.g., ABC-type transporters and phosphoenolpyruvate-dependent phosphotransferase system) and intracellular GHs. Conversely, B. bifidum and LnbX-positive B. longum use extracellular GHs to degrade oligosaccharides, subsequently importing the degradants of choice (100). While the transport systems involved in the internalization of HMOs and GOS are similar, distinct, yet overlapping, GHs encoded for hydrolysis exist. Infant-associated bifidobacteria encode multiple β-galactosidases belonging to GH42 and GH2 families, which participate in the degradation of both HMOs and GOS, in a substrate-specific manner (263). In this study, four bifidobacterial species B. breve, B. bifidum, B. longum, and B. infantis, were shown to possess conserved β-galactosidase activities (Bbr_0529, B216_08266, B8809_0415, and Blon_2016), some of which hydrolyze both HMOs, e.g., LNT, and GOS (263, 264). Enzymes such as GH20 β-hexosaminidases, GH33 sialidase, and GH95 α-fucosidase participate in host-derived oligosaccharide (HMOs/mucin) degradation (244). The glycosidic similarities between HMOs and mucin-derived glycans explain this functional overlap (264, 265).
B. breve members can encode GH95 α-fucosidases for HMO degradation, while GH42 β-galactosidases, e.g., encoded by Bbr_1552, are involved in GOS and particular HMO catabolism (105, 263). Studies focusing on B. breve UCC2003 have established that the β-galactosidase corresponding to locus tag Bbr_0420 exhibits a more narrower spectrum as its expression is upregulated in the presence of GOS but not by certain HMOs, e.g., LNT and LNnT (105, 261). For GOS utilization, two gene clusters encoding β-galactosidase and two SBPs of ABC transporters are present in B. breve involved in 3′-galactosyllactose and 6′-galactosyllactose internalization (256). Galactosyllactoses are components of GOS, but these small oligosaccharides have also been identified in human breast milk (266), whereas FOS/inulin, based on fructose, is not a natural constituent of human milk. Previously, B. breve UCC2003 transcription of the fos operon, encoding a β-fructofuranosidase, has been demonstrated in the presence of FOS (245). Although fructose is generally not provided in early life, frequent colonizers such as B. breve members still have the capacity to utilize fructose (245, 267), demonstrating their broad repertoire for carbohydrate metabolism which contributes to their dominance in the gut microbiota as the infant develops.
B. infantis encodes three GH42 members (Bga42A, Bga42B, and Bga42C) that can act upon GOS and HMOs (246, 258). Although Bga42A in B. infantis ATCC 15697 can prioritize the breakdown of 3-galactosylglucose, present in GOS, over LNT (type I HMO), it still can participate in HMO utilization (246). Interestingly, Bga42A is not part of the HMO cluster in B. infantis ATCC 15697, indicating a high level of adaptation for the assimilation of complex carbohydrates (246, 259). As mentioned above, GLNBP is involved in the degradation of LNB and is structurally similar to GH42 β-galactosidases which can degrade both HMOs and GOS (159, 247). Furthermore, B. infantis has a β-galactosidase encoded by Blon_2016 capable of degrading LNT and LNnT along with GOS (263). Phenotypical differences in GOS degradation exist depending on the structure. Garrido et al. (268) observed that B. infantis isolates (n = 22) grow vigorously on GOS with a DP of up to 3, but only a subset of isolates tested consumed larger structures. B. infantis-encoded β-fructofuranosidases involved in FOS degradation have been characterized, with families such as GH13 (Blon_0128) and GH32 (Blon_2056 and Blon_0787) participating (258, 269). GH29 1,3/1,4-α-L-fucosidase (AfcB) activity is employed for intracellular degradation of fucosylated HMOs such as 3-FL and 2′-FL (270).
B. bifidum DSM 20456 possesses a lacZ-encoded extracellular β-galactosidase potentially involved in the degradation of larger GOS structures, i.e., with a degree of polymerization of 5 and 6 (271). Furthermore, B. bifidum members can encode GH2 and GH42 β-galactosidases for intracellular hydrolysis of GOS including those encoded by BIF1 and BIF2 (258), and extracellular hydrolysis of GOS via BIF3 (250). Liu and colleagues (244) ascertained that 52 B. bifidum strains, both infant- and adult derived, lacked the necessary enzymes for FOS degradation, which translated phenotypically as an absence of growth in the presence of FOS. However, B. bifidum growth in the presence of FOS has been reported, although strain variability was noted (267).
Both intra- and extracellular mechanisms to degrade oligosaccharides are reported in the B. longum species. LnbX-positive B. longum uses extracellular enzymes for HMO degradation, whereas LnbX-negative B. longum uses intracellular enzymes (158). Typically, B. longum members are associated with plant carbohydrate metabolism, which is reflected in the GH families they encode such as GH43, GH10, and GH13 (257, 272). González et al. (260) identified similarities between B. longum LMG 13197 upregulated carbohydrate metabolism genes when grown in infant formula (with added GOS and inulin) and in breast milk in vitro. However, differences were observed for those related to the GNB/LNB operon (specifically BL1638 and BL1639), which were upregulated when the B. longum strain was grown in breast milk.
Although already an established practice (111), there is incomplete understanding with respect to supplementation of formula milk with HMO mixtures. Additionally, HMOs in combination with other prebiotics may elicit more pronounced effects compared to individual HMO supplementation, highlighting an important knowledge gap. Therefore, more clinical studies are needed to determine if a unique oligosaccharide blend can positively modulate the gut microbiota, and the benefits, if any, of using a prebiotic mixture versus individual prebiotics in infant formula.
CONCLUSIONS AND FUTURE CONSIDERATIONS
HMOs can play a major role in shaping the microbial composition, and thus functions that follow, in the neonate intestine. This is a pivotal point in life where the microbiota is established and is highly vulnerable to modulation. Research pertaining to HMOs and microbial utilization, including the broader health implications, has shifted from fundamental to translational in the last several years. Nevertheless, gaps in the scientific understanding remain, particularly at the microbial community and functional levels which, when addressed, would meaningfully expand our knowledge regarding the complex HMO-mediated effects on the gut microbiota.
First, dynamic and multifaceted sharing interactions, both supportive and inhibitory, between microbes in the infant gut can be facilitated by HMOs. It is perceived that extensive nutrient swapping occurs in the extracellular space, and although steps have been taken to establish some of those connections, the exact subtleties remain unknown, including the role of other breast milk bioactive components. Acquisition of nutrients via cross-feeding is a survival strategy for those within bifidobacteria and, indeed, other gut taxa. The proximity of microbes to one another in the gut to facilitate these cross-feeding interactions remains unclear. This is experimentally challenging to quantify, given the complexity of the gut, i.e., the various environments and micro-compartments that exist. Additional models such as organoids or community level metabolic modeling may help determine the importance of microbial spatial distribution for cross-feeding interactions. Although more intricate than the sharing of one substrate from one microbe to another, the true extent of HMO-supported cross-feeding interactions remains unknown. The expansion of HMO consumers beyond bifidobacteria has enabled a broader view of HMO metabolizers and related networks in the infant gut. Further community-based investigations to untangle the details of these networks are warranted, which could be achieved by combining taxonomic analysis with functional-based approaches, such as metabolomics, metatranscriptomics, and metaproteomics, to determine what HMO structure(s) and/or degradant(s) are being used, the subsequent metabolites produced, and how this is being achieved.
HMO structural diversity, composition, and complexity are key factors in the scientific interest in breast milk glycans accrued thus far. Presently, there have been over 200 structures identified, although the known biological relevance in terms of gut modulation remains limited to just several HMO structures. The impact of other less well-characterized HMOs, e.g., 6′-SL, LNH, and DSLNT (273), and indeed those undetected HMOs, remains less studied. Other HMOs present in breast milk, albeit at lower concentrations, may exert important biological impacts. Irrespective of abundance in breast milk, the wide variety of structures means we have only begun to tip the surface of HMO research. Linking to this is the high degree of selective structural utilization by specific bacteria. Investigating HMO metabolizing gene expression in response to structurally varying HMOs, and combinations, would be promising in establishing the strain-level preferential assimilation occurring.
Third, recent efforts have begun to explore how the initial microbial composition impacts the degradation of HMOs. Formation and persistence of the early life microbiota are influenced by numerous factors, including delivery, feeding mode, and surrounding environment. HMO structure depletion, and the subsequent metabolites produced, can vary depending on the taxa present in the gut. Few studies have examined this so far, therefore, presenting an exciting opportunity for further investigation. No study to our knowledge has directly compared what different structures and metabolites can be consumed and produced by microbiota dominated by varying levels of overall bifidobacterial abundance. Few studies have evaluated the impact of the initial microbial assembly when an HMO combination, or in conjunction with established prebiotics, is made available to the microbiota.
Finally, further emphasis on establishing the adapted functional capacity of early life Bifidobacterium, e.g., the production of aromatic lactic acids which have potential to impact early life immune function (85), would be incredibly informative. Certain infant-associated bifidobacteria encode a suite of genes involved in host glycan degradation compared to more plant-derived degradation abilities observed in the adult-type bifidobacteria. This is significant for immune system development and overall infant health. The provision of HMOs via breast milk or fortified infant formula can support the predominance of bifidobacteria in early life and their respective functionalities.
Taken together, it is apparent that HMOs embody a significant element, although they are part of a larger consortium of factors, in the microbiota development of healthy infants. The prevalence of infant-type bifidobacteria, which have genetically evolved to efficiently consume various HMOs, can likely aid in the establishment of the immune system, including a reduction in GI infection risk, through the production of metabolites and interactions with other taxa. Further research to underpin the true extent of HMO degradation patterns among infant bifidobacteria, and indeed other microbes, and the broader health impacts is crucial when moving toward the development of infant formulae to optimally support infant health.
ACKNOWLEDGMENTS
D.v.S. and P.D.C. are members of APC Microbiome Ireland, which is a Centre for Science and Technology (CSET) funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (Grant Numbers SFI/12/RC/2273-412 P1 and SFI/12/RC/2273-P2). Research in the Cotter laboratory is conducted with the financial support of the MASTER project, an Innovation Action funded by the European Commission under the Horizon 2020 Programme under grant number 818368 and by SFI and the Department of Agriculture, Food and Marine under Grant 16/RC/3835 (VistaMilk). This research is financially supported by FrieslandCampina, The Netherlands.
Contributor Information
Paul D. Cotter, Email: paul.cotter@teagasc.ie.
Douwe van Sinderen, Email: d.vansinderen@ucc.ie.
Federico Rey, University of Wisconsin-Madison, Madison, Wisconsin, USA.
REFERENCES
- 1. Bode L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22:1147–1162. doi: 10.1093/glycob/cws074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zivkovic AM, German JB, Lebrilla CB, Mills DA. 2011. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A 108 Suppl 1:4653–4658. doi: 10.1073/pnas.1000083107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boquien C-Y. 2018. Human milk: an ideal food for nutrition of preterm newborn. Front Pediatr 6:295. doi: 10.3389/fped.2018.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atyeo C, Alter G. 2021. The multifaceted roles of breast milk antibodies. Cell 184:1486–1499. doi: 10.1016/j.cell.2021.02.031 [DOI] [PubMed] [Google Scholar]
- 5. Ballard O, Morrow AL. 2013. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am 60:49–74. doi: 10.1016/j.pcl.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang S, Li T, Xie J, Zhang D, Pi C, Zhou L, Yang W. 2021. Gold standard for nutrition: a review of human milk oligosaccharide and its effects on infant gut microbiota. Microb Cell Fact 20:108. doi: 10.1186/s12934-021-01599-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Masi AC, Stewart CJ. 2022. Untangling human milk oligosaccharides and infant gut microbiome. iScience 25:103542. doi: 10.1016/j.isci.2021.103542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiciński M, Sawicka E, Gębalski J, Kubiak K, Malinowski B. 2020. Human milk Oligosaccharides: Health benefits, potential applications in infant formulas, and pharmacology. Nutrients 12:266. doi: 10.3390/nu12010266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Turroni F, Milani C, Duranti S, Lugli GA, Bernasconi S, Margolles A, Di Pierro F, van Sinderen D, Ventura M. 2020. The infant gut microbiome as a microbial organ influencing host well-being. Ital J Pediatr 46:16. doi: 10.1186/s13052-020-0781-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dzidic M, Boix-Amorós A, Selma-Royo M, Mira A, Collado MC. 2018. Gut microbiota and mucosal immunity in the neonate. Med Sci (Basel) 6:56. doi: 10.3390/medsci6030056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Linehan K, Dempsey EM, Ryan CA, Ross RP, Stanton C. 2022. First encounters of the microbial kind: perinatal factors direct infant gut microbiome establishment. MRR 1:10. doi: 10.20517/mrr.2021.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 7:e36957. doi: 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan W, Luo B, Zhang X, Ni Y, Tian F. 2021. Association and occurrence of bifidobacterial phylotypes between breast milk and fecal microbiomes in mother-infant dyads during the first 2 years of life. Front Microbiol 12:669442. doi: 10.3389/fmicb.2021.669442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chichlowski M, van Diepen JA, Prodan A, Olga L, Ong KK, Kortman GAM, Dunger DB, Gross G. 2023. Early development of infant gut microbiota in relation to breastfeeding and human milk oligosaccharides. Front Nutr 10:1003032. doi: 10.3389/fnut.2023.1003032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Callaghan A, van Sinderen D. 2016. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol 7:925. doi: 10.3389/fmicb.2016.00925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, Field N, Lawley TD. 2019. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574:117–121. doi: 10.1038/s41586-019-1560-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:852. doi: 10.1016/j.chom.2015.05.012 [DOI] [PubMed] [Google Scholar]
- 19. Feehily C, O’Neill IJ, Walsh CJ, Moore RL, Killeen SL, Geraghty AA, Lawton EM, Byrne D, Sanchez-Gallardo R, Nori SRC, Nielsen IB, Wortmann E, Matthews E, O’Flaherty R, Rudd PM, Groeger D, Shanahan F, Saldova R, McAuliffe FM, Van Sinderen D, Cotter PD. 2023. Detailed mapping of Bifidobacterium strain transmission from mother to infant via a dual culture-based and metagenomic approach. Nat Commun 14:3015. doi: 10.1038/s41467-023-38694-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heiss BE, Ehrlich AM, Maldonado-Gomez MX, Taft DH, Larke JA, Goodson ML, Slupsky CM, Tancredi DJ, Raybould HE, Mills DA. 2021. Bifidobacterium catabolism of human milk oligosaccharides overrides endogenous competitive exclusion driving colonization and protection. Gut Microbes 13:1986666. doi: 10.1080/19490976.2021.1986666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Power SE, O’Toole PW, Stanton C, Ross RP, Fitzgerald GF. 2014. Intestinal microbiota, diet and health. Br J Nutr 111:387–402. doi: 10.1017/S0007114513002560 [DOI] [PubMed] [Google Scholar]
- 22. Vandenplas Y, Carnielli VP, Ksiazyk J, Luna MS, Migacheva N, Mosselmans JM, Picaud JC, Possner M, Singhal A, Wabitsch M. 2020. Factors affecting early-life intestinal microbiota development. Nutrition 78:110812. doi: 10.1016/j.nut.2020.110812 [DOI] [PubMed] [Google Scholar]
- 23. Yao Y, Cai X, Ye Y, Wang F, Chen F, Zheng C. 2021. The role of microbiota in infant health: from early life to adulthood. Front Immunol 12:708472. doi: 10.3389/fimmu.2021.708472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Odamaki T, Bottacini F, Mitsuyama E, Yoshida K, Kato K, Xiao J-Z, van Sinderen D. 2019. Impact of a bathing tradition on shared gut microbe among Japanese families. Sci Rep 9:4380. doi: 10.1038/s41598-019-40938-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chong CYL, Bloomfield FH, O’Sullivan JM. 2018. Factors affecting gastrointestinal microbiome development in neonates. Nutrients 10:274. doi: 10.3390/nu10030274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O’Neill IJ, Sanchez Gallardo R, Saldova R, Murphy EF, Cotter PD, McAuliffe FM, van Sinderen D. 2020. Maternal and infant factors that shape neonatal gut colonization by bacteria. Expert Rev Gastroenterol Hepatol 14:651–664. doi: 10.1080/17474124.2020.1784725 [DOI] [PubMed] [Google Scholar]
- 27. Turroni F, van Sinderen D, Ventura M. 2022. Bifidobacteria: insights into the biology of a key microbial group of early life gut microbiota. MRR 1:2. doi: 10.20517/mrr.2021.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O’Shea CA, Watkins C, Dempsey E, Mattivi F, Tuohy K, Ross RP, Ryan CA, O’Toole PW, Stanton C. 2017. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET cohort. Microbiome 5:21. doi: 10.1186/s40168-017-0240-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Derrien M, Alvarez A-S, de Vos WM. 2019. The gut microbiota in the first decade of life. Trends Microbiol 27:997–1010. doi: 10.1016/j.tim.2019.08.001 [DOI] [PubMed] [Google Scholar]
- 30. Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, et al. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583–588. doi: 10.1038/s41586-018-0617-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, Zhuo N, Wang H, Wang L, Wu D. 2020. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep 10:15792. doi: 10.1038/s41598-020-72635-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Di Guglielmo MD, Franke KR, Robbins A, Crowgey EL. 2022. Impact of early feeding: metagenomics analysis of the infant gut microbiome. Front Cell Infect Microbiol 12:816601. doi: 10.3389/fcimb.2022.816601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC. 2019. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 7:14. doi: 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laursen MF. 2021. Gut microbiota development: influence of diet from infancy to toddlerhood. Ann Nutr Metab 77:1–14. doi: 10.1159/000517912 [DOI] [PubMed] [Google Scholar]
- 35. Saturio S, Nogacka AM, Alvarado-Jasso GM, Salazar N, de Los Reyes-Gavilán CG, Gueimonde M, Arboleya S. 2021. Role of bifidobacteria on infant health. Microorganisms 9:2415. doi: 10.3390/microorganisms9122415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, Olin A, Wang J, Mikes J, Tan Z, et al. 2021. Bifidobacteria-mediated immune system imprinting early in life. Cell 184:3884–3898. doi: 10.1016/j.cell.2021.05.030 [DOI] [PubMed] [Google Scholar]
- 37. Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. 2001. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol 108:516–520. doi: 10.1067/mai.2001.118130 [DOI] [PubMed] [Google Scholar]
- 38. Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, Xu G, Davis JCC, Lebrilla CB, Henrick BM, Freeman SL, Barile D, German JB, Mills DA, Smilowitz JT, Underwood MA. 2017. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere 2:e00501-17. doi: 10.1128/mSphere.00501-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mancabelli L, Tarracchini C, Milani C, Lugli GA, Fontana F, Turroni F, van Sinderen D, Ventura M. 2020. Multi-population cohort meta-analysis of human intestinal microbiota in early life reveals the existence of infant community state types (ICSTs). Comput Struct Biotechnol J 18:2480–2493. doi: 10.1016/j.csbj.2020.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Derrien M, Mikulic N, Uyoga MA, Chenoll E, Climent E, Howard-Varona A, Nyilima S, Stoffel NU, Karanja S, Kottler R, Stahl B, Zimmermann MB, Bourdet-Sicard R. 2023. Gut microbiome function and composition in infants from rural Kenya and association with human milk oligosaccharides. Gut Microbes 15:2178793. doi: 10.1080/19490976.2023.2178793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thum C, Wall CR, Weiss GA, Wang W, Szeto I-Y, Day L. 2021. Changes in HMO concentrations throughout lactation: influencing factors, health effects and opportunities. Nutrients 13:2272. doi: 10.3390/nu13072272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, Kvist LJ, Otoo GE, Brooker SL, Price WJ, Shafii B, Placek C, Lackey KA, Robertson B, Manzano S, Ruíz L, Rodríguez JM, Pareja RG, Bode L. 2017. What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 105:1086–1100. doi: 10.3945/ajcn.116.139980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. doi: 10.1146/annurev.nutr.20.1.699 [DOI] [PubMed] [Google Scholar]
- 44. Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. 2014. Breast milk oligosaccharides: structure-function relationships in the neonate. Annu Rev Nutr 34:143–169. doi: 10.1146/annurev-nutr-071813-105721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bode L, Jantscher-Krenn E. 2012. Structure-function relationships of human milk oligosaccharides. Adv Nutr 3:383S–91S. doi: 10.3945/an.111.001404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bode L. 2015. The functional biology of human milk oligosaccharides. Early Hum Dev 91:619–622. doi: 10.1016/j.earlhumdev.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 47. Vinjamuri A, Davis JCC, Totten SM, Wu LD, Klein LD, Martin M, Quinn EA, Scelza B, Breakey A, Gurven M, Jasienska G, Kaplan H, Valeggia C, Hinde K, Smilowitz JT, Bernstein RM, Zivkovic AM, Barratt MJ, Gordon JI, Underwood MA, Mills DA, German JB, Lebrilla CB. 2022. Human milk oligosaccharide compositions illustrate global variations in early nutrition. J Nutr 152:1239–1253. doi: 10.1093/jn/nxac027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Soyyılmaz B, Mikš MH, Röhrig CH, Matwiejuk M, Meszaros-Matwiejuk A, Vigsnæs LK. 2021. The mean of milk: a review of human milk oligosaccharide concentrations throughout lactation. Nutrients 13:2737. doi: 10.3390/nu13082737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marionneau S, Airaud F, Bovin NV, Le Pendu J, Ruvoën-Clouet N. 2005. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J Infect Dis 192:1071–1077. doi: 10.1086/432546 [DOI] [PubMed] [Google Scholar]
- 50. Larsson MM, Rydell GEP, Grahn A, Rodriguez-Diaz J, Akerlind B, Hutson AM, Estes MK, Larson G, Svensson L. 2006. Antibody prevalence and titer to norovirus (genogroup II) correlate with Secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. J Infect Dis 194:1422–1427. doi: 10.1086/508430 [DOI] [PubMed] [Google Scholar]
- 51. Rydell GE, Kindberg E, Larson G, Svensson L. 2011. Susceptibility to winter vomiting disease: a sweet matter. Rev Med Virol 21:370–382. doi: 10.1002/rmv.704 [DOI] [PubMed] [Google Scholar]
- 52. Lindén S, Mahdavi J, Semino-Mora C, Olsen C, Carlstedt I, Borén T, Dubois A. 2008. Role of ABO secretor status in mucosal innate immunity and H. Pylori infection. PLoS Pathog 4:e2. doi: 10.1371/journal.ppat.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chakrani Z, Robinson K, Taye B. 2018. Association between ABO blood groups and Helicobacter Pylori infection: a meta-analysis. Sci Rep 8:17604. doi: 10.1038/s41598-018-36006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eussen S, Mank M, Kottler R, Hoffmann X-K, Behne A, Rapp E, Stahl B, Mearin ML, Koletzko B. 2021. Presence and levels of galactosyllactoses and other oligosaccharides in human milk and their variation during lactation and according to maternal phenotype. Nutrients 13:2324. doi: 10.3390/nu13072324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu G, Davis JC, Goonatilleke E, Smilowitz JT, German JB, Lebrilla CB. 2017. Absolute quantitation of human milk oligosaccharides reveals phenotypic variations during lactation. J Nutr 147:117–124. doi: 10.3945/jn.116.238279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Azad MB, Robertson B, Atakora F, Becker AB, Subbarao P, Moraes TJ, Mandhane PJ, Turvey SE, Lefebvre DL, Sears MR, Bode L. 2018. Human milk oligosaccharide concentrations are associated with multiple fixed and modifiable maternal characteristics, environmental factors, and feeding practices. J Nutr 148:1733–1742. doi: 10.1093/jn/nxy175 [DOI] [PubMed] [Google Scholar]
- 57. Pell LG, Ohuma EO, Yonemitsu C, Loutet MG, Ahmed T, Mahmud AA, Azad MB, Bode L, Roth DE. 2021. The human-milk oligosaccharide profile of lactating women in Dhaka, Bangladesh. Curr Dev Nutr 5:zab137. doi: 10.1093/cdn/nzab137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lahdenperä M, Galante L, Gonzales-Inca C, Vahtera J, Pentti J, Rautava S, Käyhkö N, Yonemitsu C, Gupta J, Bode L, Lagström H. 2023. Residential green environments are associated with human milk oligosaccharide diversity and composition. Sci Rep 13:216. doi: 10.1038/s41598-022-27317-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ferreira AL, Alves R, Figueiredo A, Alves-Santos N, Freitas-Costa N, Batalha M, Yonemitsu C, Manivong N, Furst A, Bode L, Kac G. 2020. Human milk oligosaccharide profile variation throughout postpartum in healthy women in a Brazilian cohort. Nutrients 12:790. doi: 10.3390/nu12030790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moya-Alvarez V, Eussen S, Mank M, Koyembi J-C, Nyasenu YT, Ngaya G, Mad-Bondo D, Kongoma J-B, Stahl B, Sansonetti PJ, Bourdet-Sicard R. 2022. Human milk nutritional composition across lactational stages in Central Africa. Front Nutr 9:1033005. doi: 10.3389/fnut.2022.1033005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu J, Wu S, Huo J, Ruan H, Xu X, Hao Z, Wei Y. 2020. Systematic characterization and longitudinal study reveal distinguishing features of human milk oligosaccharides in China. Curr Dev Nutr 4:zaa113. doi: 10.1093/cdn/nzaa113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Davis JCC, Lewis ZT, Krishnan S, Bernstein RM, Moore SE, Prentice AM, Mills DA, Lebrilla CB, Zivkovic AM. 2017. Growth and morbidity of Gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep 7:40466. doi: 10.1038/srep40466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li X, Mao Y, Liu S, Wang J, Li X, Zhao Y, Hill DR, Wang S. 2022. Vitamins, vegetables and metal elements are positively associated with breast milk oligosaccharide composition among mothers in Tianjin, China. Nutrients 14:4131. doi: 10.3390/nu14194131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harris JE, Pinckard KM, Wright KR, Baer LA, Arts PJ, Abay E, Shettigar VK, Lehnig AC, Robertson B, Madaris K, Canova TJ, Sims C, Goodyear LJ, Andres A, Ziolo MT, Bode L, Stanford KI. 2020. Exercise-induced 3'-sialyllactose in breast milk is a critical mediator to improve metabolic health and cardiac function in mouse offspring. Nat Metab 2:678–687. doi: 10.1038/s42255-020-0223-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Selma-Royo M, González S, Gueimonde M, Chang M, Fürst A, Martínez-Costa C, Bode L, Collado MC. 2023. Maternal diet is associated with human milk Oligosaccharide profile. Mol Nutr Food Res 66:e2200058. doi: 10.1007/s11130-022-01020-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bottin JH, Eussen S, Igbinijesu AJ, Mank M, Koyembi J-C, Nyasenu YT, Ngaya G, Mad-Bondo D, Kongoma J-B, Stahl B, Sansonetti PJ, Bourdet-Sicard R, Moya-Alvarez V. 2022. Food insecurity and maternal diet influence human milk composition between the infant's birth and 6 months after birth in Central-Africa. Nutrients 14:4015. doi: 10.3390/nu14194015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cheng YJ, Yeung CY. 2021. Recent advance in infant nutrition: human milk oligosaccharides. Pediatr Neonatol 62:347–353. doi: 10.1016/j.pedneo.2020.12.013 [DOI] [PubMed] [Google Scholar]
- 68. Asher AT, Mangel L, Ari JB, Gover O, Ahmad WA, Herzlich J, Mandel D, Schwartz B, Lubetzky R. 2023. Human milk oligosaccharide profile across lactation stages in Israeli women-a prospective observational study. Nutrients 15:2548. doi: 10.3390/nu15112548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Durham SD, Robinson RC, Olga L, Ong KK, Chichlowski M, Dunger DB, Barile D. 2021. A one-year study of human milk oligosaccharide profiles in the milk of healthy UK mothers and their relationship to maternal FUT2 genotype. Glycobiology 31:1254–1267. doi: 10.1093/glycob/cwab057 [DOI] [PubMed] [Google Scholar]
- 70. Erney RM, Malone WT, Skelding MB, Marcon AA, Kleman–Leyer KM, O’Ryan ML, Ruiz–Palacios G, Hilty MD, Pickering LK, Prieto PA. 2000. Variability of human milk neutral oligosaccharides in a diverse population. J Pediatr Gastroenterol Nutr 30:181–192. doi: 10.1097/00005176-200002000-00016 [DOI] [PubMed] [Google Scholar]
- 71. Lewis ZT, Sidamonidze K, Tsaturyan V, Tsereteli D, Khachidze N, Pepoyan A, Zhgenti E, Tevzadze L, Manvelyan A, Balayan M, Imnadze P, Torok T, Lemay DG, Mills DA. 2017. The fecal microbial community of breast-fed infants from Armenia and Georgia. Sci Rep 7:40932. doi: 10.1038/srep40932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu TC, Chang JG, Lin SF, Chang WC, Yang TY, Lin CL, Wang NM, Tsai CH. 2000. Lewis (FUT3) genotypes in Taiwanese, Thai, and Filipino populations. Ann Hematol 79:599–603. [DOI] [PubMed] [Google Scholar]
- 73. Liu S, Mao Y, Wang J, Tian F, Hill DR, Xiong X, Li X, Zhao Y, Wang S. 2023. Lactational and geographical variation in the concentration of six oligosaccharides in Chinese breast milk: a multicenter study over 13 months postpartum. Front Nutr 10:1267287. doi: 10.3389/fnut.2023.1267287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Menzel P, Vogel M, Austin S, Sprenger N, Grafe N, Hilbert C, Jurkutat A, Kiess W, Binia A. 2021. Concentrations of oligosaccharides in human milk and child growth. BMC Pediatr 21:481. doi: 10.1186/s12887-021-02953-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Musumeci M, Simpore J, D’Agata A, Sotgiu S, Musumeci S. 2006. Oligosaccharides in colostrum of Italian and Burkinabe women. J Pediatr Gastroenterol Nutr 43:372–378. doi: 10.1097/01.mpg.0000228125.70971.af [DOI] [PubMed] [Google Scholar]
- 76. McJarrow P, Radwan H, Ma L, MacGibbon AKH, Hashim M, Hasan H, Obaid RS, Naja F, Mohamed HJJ, Al Ghazal H, Fong BY. 2019. Human milk oligosaccharide, phospholipid, and ganglioside concentrations in breast milk from United Arab Emirates mothers: results from the MISC cohort. Nutrients 11:2400. doi: 10.3390/nu11102400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sudarma V, Sunardi D, Marzuki NS, Munasir Z, Asmarinah A, Hidayat A, Hegar B. 2023. Human milk oligosaccharide profiles and the secretor and lewis gene status of Indonesian lactating mothers. Pediatr Gastroenterol Hepatol Nutr 26:266–276. doi: 10.5223/pghn.2023.26.5.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. van Leeuwen SS, Stoutjesdijk E, ten Kate GA, Schaafsma A, Dijck-Brouwer J, Muskiet FAJ, Dijkhuizen L. 2018. Regional variations in human milk oligosaccharides in Vietnam suggest FucTx activity besides FucT2 and FucT3. Sci Rep 8:16790. doi: 10.1038/s41598-018-34882-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fan Y, Vinjamuri A, Tu D, Lebrilla CB, Donovan SM. 2023. Determinants of human milk Oligosaccharides profiles of participants in the STRONG kids 2 cohort. Front Nutr 10:1105668. doi: 10.3389/fnut.2023.1105668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stuivenberg GA, Burton JP, Bron PA, Reid G. 2022. Why are bifidobacteria important for infants? Microorganisms 10:278. doi: 10.3390/microorganisms10020278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leonel AJ, Alvarez-Leite JI. 2012. Butyrate: Implications for intestinal function. Curr Opin Clin Nutr Metab Care 15:474–479. doi: 10.1097/MCO.0b013e32835665fa [DOI] [PubMed] [Google Scholar]
- 82. Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. 2006. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40:235–243. doi: 10.1097/00004836-200603000-00015 [DOI] [PubMed] [Google Scholar]
- 83. Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7:185. doi: 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lin C, Lin Y, Zhang H, Wang G, Zhao J, Zhang H, Chen W. 2022. “Intestinal 'infant-type' bifidobacteria mediate immune system development in the first 1000 days of life”. Nutrients 14:1498. doi: 10.3390/nu14071498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Laursen MF, Sakanaka M, von Burg N, Mörbe U, Andersen D, Moll JM, Pekmez CT, Rivollier A, Michaelsen KF, Mølgaard C, Lind MV, Dragsted LO, Katayama T, Frandsen HL, Vinggaard AM, Bahl MI, Brix S, Agace W, Licht TR, Roager HM. 2021. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol 6:1367–1382. doi: 10.1038/s41564-021-00970-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schimmel P, Stahl B, Knol J, Belzer C. 2023. The infant gut microbiota: in pursuit of non-protein nitrogen. Gut Microbes 15:2211917. doi: 10.1080/19490976.2023.2211917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. You X, Rani A, Özcan E, Lyu Y, Sela DA. 2023. Bifidobacterium Longum subsp. Infantis utilizes human milk urea to recycle nitrogen within the infant gut microbiome. Gut Microbes 15:2192546. doi: 10.1080/19490976.2023.2192546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kalliomäki M, Collado MC, Salminen S, Isolauri E. 2008. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 87:534–538. doi: 10.1093/ajcn/87.3.534 [DOI] [PubMed] [Google Scholar]
- 89. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV. 2016. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22:1187–1191. doi: 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Korpela K, Zijlmans MAC, Kuitunen M, Kukkonen K, Savilahti E, Salonen A, de Weerth C, de Vos WM. 2017. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome 5:26. doi: 10.1186/s40168-017-0245-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fontana F, Alessandri G, Tarracchini C, Bianchi MG, Rizzo SM, Mancabelli L, Lugli GA, Argentini C, Vergna LM, Anzalone R, Longhi G, Viappiani A, Taurino G, Chiu M, Turroni F, Bussolati O, van Sinderen D, Milani C, Ventura M. 2022. Designation of optimal reference strains representing the infant gut bifidobacterial species through a comprehensive multi-omics approach. Environ Microbiol 24:5825–5839. doi: 10.1111/1462-2920.16205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Morita H, Nakano A, Onoda H, Toh H, Oshima K, Takami H, Murakami M, Fukuda S, Takizawa T, Kuwahara T, Ohno H, Tanabe S, Hattori M. 2011. Bifidobacterium kashiwanohense sp. nov., isolated from healthy infant faeces. Int J Syst Evol Microbiol 61:2610–2615. doi: 10.1099/ijs.0.024521-0 [DOI] [PubMed] [Google Scholar]
- 93. Arboleya S, Watkins C, Stanton C, Ross RP. 2016. Gut bifidobacteria populations in human health and aging. Front Microbiol 7:1204. doi: 10.3389/fmicb.2016.01204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Derrien M, Turroni F, Ventura M, van Sinderen D. 2022. Insights into endogenous Bifidobacterium species in the human gut microbiota during adulthood. Trends Microbiol 30:940–947. doi: 10.1016/j.tim.2022.04.004 [DOI] [PubMed] [Google Scholar]
- 95. Wong CB, Odamaki T, Xiao J-Z. 2020. Insights into the reason of human-residential bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol Rev 44:369–385. doi: 10.1093/femsre/fuaa010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Odamaki T, Bottacini F, Kato K, Mitsuyama E, Yoshida K, Horigome A, Xiao J-Z, van Sinderen D. 2018. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci Rep 8:85. doi: 10.1038/s41598-017-18391-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Thongaram T, Hoeflinger JL, Chow J, Miller MJ. 2017. Human milk oligosaccharide consumption by probiotic and human-associated bifidobacteria and lactobacilli. J Dairy Sci 100:7825–7833. doi: 10.3168/jds.2017-12753 [DOI] [PubMed] [Google Scholar]
- 98. Walsh C, Lane JA, van Sinderen D, Hickey RM. 2022. Human milk oligosaccharide-sharing by a consortium of infant derived Bifidobacterium species. Sci Rep 12:4143. doi: 10.1038/s41598-022-07904-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zabel BE, Gerdes S, Evans KC, Nedveck D, Singles SK, Volk B, Budinoff C. 2020. Strain-specific strategies of 2'-fucosyllactose, 3-fucosyllactose, and difucosyllactose assimilation by Bifidobacterium longum subsp. infantis Bi-26 and ATCC 15697. Sci Rep 10:15919. doi: 10.1038/s41598-020-72792-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gotoh A, Katoh T, Sakanaka M, Ling Y, Yamada C, Asakuma S, Urashima T, Tomabechi Y, Katayama-Ikegami A, Kurihara S, Yamamoto K, Harata G, He F, Hirose J, Kitaoka M, Okuda S, Katayama T. 2018. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep 8:13958. doi: 10.1038/s41598-018-32080-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. 2008. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol 74:3996–4004. doi: 10.1128/AEM.00149-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, Mills DA. 2015. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep 5:15311. doi: 10.1038/srep15311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Thomson P, Medina DA, Garrido D. 2018. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol 75:37–46. doi: 10.1016/j.fm.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 104. Ruiz-Moyano S, Totten SM, Garrido DA, Smilowitz JT, German JB, Lebrilla CB, Mills DA. 2013. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol 79:6040–6049. doi: 10.1128/AEM.01843-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. James K, Motherway MO, Bottacini F, van Sinderen D. 2016. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep 6:38560. doi: 10.1038/srep38560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ojima MN, Jiang L, Arzamasov AA, Yoshida K, Odamaki T, Xiao J, Nakajima A, Kitaoka M, Hirose J, Urashima T, Katoh T, Gotoh A, van Sinderen D, Rodionov DA, Osterman AL, Sakanaka M, Katayama T. 2022. Priority effects shape the structure of infant-type Bifidobacterium communities on human milk oligosaccharides. ISME J 16:2265–2279. doi: 10.1038/s41396-022-01270-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Xu S, Lane JA, Chen J, Zheng Y, Wang H, Fu X, Huang Q, Dhital S, Liu F, Zhang B. 2022. In vitro infant fecal fermentation characteristics of human milk oligosaccharides were controlled by initial microbiota composition more than chemical structure. Mol Nutr Food Res 66:e2200098. doi: 10.1002/mnfr.202200098 [DOI] [PubMed] [Google Scholar]
- 108. Nogacka AM, Arboleya S, Nikpoor N, Auger J, Salazar N, Cuesta I, Alvarez-Buylla JR, Mantecón L, Solís G, Gueimonde M, Tompkins TA, de Los Reyes-Gavilán CG. 2022. In vitro probiotic modulation of the intestinal microbiota and 2′fucosyllactose consumption in fecal cultures from infants at two months of age. Microorganisms 10:318. doi: 10.3390/microorganisms10020318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li H, Lane JA, Chen J, Lu Z, Wang H, Dhital S, Fu X, Huang Q, Liu F, Zhang B. 2022. In vitro fermentation of human milk oligosaccharides by individual Bifidobacterium longum-dominant infant fecal Inocula. Carbohydr Polym 287:119322. doi: 10.1016/j.carbpol.2022.119322 [DOI] [PubMed] [Google Scholar]
- 110. Alliet P, Vandenplas Y, Roggero P, Jespers SNJ, Peeters S, Stalens J-P, Kortman GAM, Amico M, Berger B, Sprenger N, Cercamondi CI, Corsello G. 2022. Safety and efficacy of a probiotic-containing infant formula supplemented with 2'-fucosyllactose: a double-blind randomized controlled trial. Nutr J 21:11. doi: 10.1186/s12937-022-00764-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bosheva M, Tokodi I, Krasnow A, Pedersen HK, Lukjancenko O, Eklund AC, Grathwohl D, Sprenger N, Berger B, Cercamondi CI, 5 HMO Study Investigator Consortium . 2022. Infant formula with a specific blend of five human milk oligosaccharides drives the gut microbiota development and improves gut maturation markers: a randomized controlled trial. Front Nutr 9:920362. doi: 10.3389/fnut.2022.920362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gold MS, Quinn PJ, Campbell DE, Peake J, Smart J, Robinson M, O’Sullivan M, Vogt JK, Pedersen HK, Liu X, Pazirandeh-Micol E, Heine RG. 2022. Effects of an amino acid-based formula supplemented with two human milk oligosaccharides on growth, tolerability, safety, and gut microbiome in infants with cow's milk protein allergy. Nutrients 14:2297. doi: 10.3390/nu14112297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Dogra SK, Martin F-P, Donnicola D, Julita M, Berger B, Sprenger N. 2021. Human milk oligosaccharide-stimulated Bifidobacterium species contribute to prevent later respiratory tract infections. Microorganisms 9:1939. doi: 10.3390/microorganisms9091939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wallingford JC, Neve Myers P, Barber CM. 2022. Effects of addition of 2-fucosyllactose to infant formula on growth and specific pathways of utilization by Bifidobacterium in healthy term infants. Front Nutr 9:961526. doi: 10.3389/fnut.2022.961526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li A, Ni W, Li Y, Zhang X, Yang J, Ma X, Jia X, Li C, Liu L. 2020. Effect of 2′-fucosyllactose supplementation on intestinal flora in mice with intestinal inflammatory diseases. Int Dairy J 110:104797. doi: 10.1016/j.idairyj.2020.104797 [DOI] [Google Scholar]
- 116. Chen Q, Yin Q, Xie Q, Jiang C, Zhou L, Liu J, Li B, Jiang S. 2022. 2'-fucosyllactose promotes the production of short-chain fatty acids and improves immune function in human-microbiota-associated mice by regulating gut microbiota. J Agric Food Chem 70:13615–13625. doi: 10.1021/acs.jafc.2c04410 [DOI] [PubMed] [Google Scholar]
- 117. Azagra-Boronat I, Massot-Cladera M, Mayneris-Perxachs J, Knipping K, Van’t Land B, Tims S, Stahl B, Garssen J, Franch À, Castell M, Rodríguez-Lagunas MJ, Pérez-Cano FJ. 2019. Immunomodulatory and prebiotic effects of 2'-fucosyllactose in suckling rats. Front Immunol 10:1773. doi: 10.3389/fimmu.2019.01773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Natividad JM, Marsaux B, Rodenas CLG, Rytz A, Vandevijver G, Marzorati M, Van den Abbeele P, Calatayud M, Rochat F. 2022. Human milk oligosaccharides and lactose differentially affect infant gut microbiota and intestinal barrier in vitro. Nutrients 14:2546. doi: 10.3390/nu14122546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lindner C, Looijesteijn E, Dijck H van, Bovee-Oudenhoven I, Heerikhuisen M, Broek T van den, Marzorati M, Triantis V, Nauta A. 2023. Infant fecal fermentations with galacto-oligosaccharides and 2′-fucosyllactose show differential Bifidobacterium longum stimulation at subspecies level. Children (Basel) 10:430. doi: 10.3390/children10030430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Akkerman R, Logtenberg MJ, Beukema M, de Haan BJ, Faas MM, Zoetendal EG, Schols HA, de Vos P. 2022. Combining galacto-oligosaccharides and 2'-fucosyllactose alters their fermentation Kinetics by infant fecal microbiota and influences AhR-receptor dependent cytokine responses in immature dendritic cells. Food Funct 13:6510–6521. doi: 10.1039/d2fo00550f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kong C, Akkerman R, Klostermann CE, Beukema M, Oerlemans MMP, Schols HA, de Vos P. 2021. Distinct fermentation of human milk oligosaccharides 3-FL and LNT2 and GOS/inulin by infant gut microbiota and impact on adhesion of Lactobacillus plantarum WCFS1 to gut epithelial cells. Food Funct 12:12513–12525. doi: 10.1039/d1fo02563e [DOI] [PubMed] [Google Scholar]
- 122. Wiese M, Khakimov B, Nielsen S, Sørensen H, van den Berg F, Nielsen DS. 2018. Cominigut—a small volume in vitro colon model for the screening of gut microbial fermentation processes. PeerJ 6:e4268. doi: 10.7717/peerj.4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Van den Abbeele P, Sprenger N, Ghyselinck J, Marsaux B, Marzorati M, Rochat F. 2021. A comparison of the in vitro effects of 2'fucosyllactose and lactose on the composition and activity of gut microbiota from infants and toddlers. Nutrients 13:726. doi: 10.3390/nu13030726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Van den Abbeele P, Duysburgh C, Vazquez E, Chow J, Buck R, Marzorati M. 2019. 2′-fucosyllactose alters the composition and activity of gut microbiota from formula-fed infants receiving complementary feeding in a validated intestinal model. Journal of Functional Foods 61:103484. doi: 10.1016/j.jff.2019.103484 [DOI] [Google Scholar]
- 125. Nogacka AM, Arboleya S, Nikpoor N, Auger J, Salazar N, Cuesta I, Mantecón L, Solís G, Gueimonde M, Tompkins TA, de Los Reyes-Gavilán CG. 2021. Influence of 2′-fucosyllactose on the microbiota composition and metabolic activity of fecal cultures from breastfed and formula-fed infants at two months of age. Microorganisms 9:1478. doi: 10.3390/microorganisms9071478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bunesova V, Lacroix C, Schwab C. 2016. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol 16:248. doi: 10.1186/s12866-016-0867-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. James K, Bottacini F, Contreras JIS, Vigoureux M, Egan M, Motherway MO, Holmes E, van Sinderen D. 2019. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci Rep 9:15427. doi: 10.1038/s41598-019-51901-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sakanaka M, Hansen ME, Gotoh A, Katoh T, Yoshida K, Odamaki T, Yachi H, Sugiyama Y, Kurihara S, Hirose J, Urashima T, Xiao J-Z, Kitaoka M, Fukiya S, Yokota A, Lo Leggio L, Abou Hachem M, Katayama T. 2019. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci Adv 5:eaaw7696. doi: 10.1126/sciadv.aaw7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, Ugalde JA, German JB, Lebrilla CB, Mills DA. 2016. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep 6:35045. doi: 10.1038/srep35045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shani G, Hoeflinger JL, Heiss BE, Masarweh CF, Larke JA, Jensen NM, Wickramasinghe S, Davis JC, Goonatilleke E, El-Hawiet A, Nguyen L, Klassen JS, Slupsky CM, Lebrilla CB, Mills DA. 2022. Fucosylated human milk oligosaccharide foraging within the species Bifidobacterium pseudocatenulatum is driven by glycosyl hydrolase content and specificity. Appl Environ Microbiol 88:e0170721. doi: 10.1128/AEM.01707-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Schwab C, Ruscheweyh H-J, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C. 2017. Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-Fucose and fucosyllactose degradation. Front Microbiol 8:95. doi: 10.3389/fmicb.2017.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Shiver AL, Sun J, Culver R, Violette A, Wynter C, Nieckarz M, Mattiello SP, Sekhon PK, Friess L, Carlson HK, et al. 2023. A mutant fitness compendium in bifidobacteria reveals molecular determinants of colonization and host-microbe interactions. bioRxiv. doi: 10.1101/2023.08.29.555234 [DOI]
- 133. Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, Matsumoto S, Kurokawa K. 2016. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun 7:11939. doi: 10.1038/ncomms11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Xiao J, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K, Kitaoka M. 2010. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol 76:54–59. doi: 10.1128/AEM.01683-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bode L. 2018. Human milk oligosaccharides in the prevention of necrotizing enterocolitis: a journey from in vitro and in vivo models to mother-infant cohort studies. Front Pediatr 6:385. doi: 10.3389/fped.2018.00385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Masi AC, Embleton ND, Lamb CA, Young G, Granger CL, Najera J, Smith DP, Hoffman KL, Petrosino JF, Bode L, Berrington JE, Stewart CJ. 2021. Human milk oligosaccharide DSLNT and gut microbiome in preterm infants predicts necrotising enterocolitis. Gut 70:2273–2282. doi: 10.1136/gutjnl-2020-322771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Berger B, Porta N, Foata F, Grathwohl D, Delley M, Moine D, Charpagne A, Siegwald L, Descombes P, Alliet P, Puccio G, Steenhout P, Mercenier A, Sprenger N. 2020. Linking human milk oligosaccharides, infant fecal community types, and later risk to require antibiotics. mBio 11:e03196-19. doi: 10.1128/mBio.03196-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ventura M, Canchaya C, Casale AD, Dellaglio F, Neviani E, Fitzgerald GF, van Sinderen D. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int J Syst Evol Microbiol 56:2783–2792. doi: 10.1099/ijs.0.64233-0 [DOI] [PubMed] [Google Scholar]
- 139. Borewicz K, Gu F, Saccenti E, Hechler C, Beijers R, de Weerth C, van Leeuwen SS, Schols HA, Smidt H. 2020. The association between breastmilk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci Rep 10:4270. doi: 10.1038/s41598-020-61024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Borewicz K, Gu F, Saccenti E, Arts ICW, Penders J, Thijs C, van Leeuwen SS, Lindner C, Nauta A, van Leusen E, Schols HA, Smidt H. 2019. Correlating infant fecal microbiota composition and human milk oligosaccharide consumption by microbiota of 1-month-old breastfed infants. Mol Nutr Food Res 63:e1801214. doi: 10.1002/mnfr.201801214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Barnett DJM, Endika MF, Klostermann CE, Gu F, Thijs C, Nauta A, Schols HA, Smidt H, Arts ICW, Penders J. 2023. Human milk oligosaccharides, antimicrobial drugs, and the gut microbiota of term neonates: observations from the KOALA birth cohort study. Gut Microbes 15:2164152. doi: 10.1080/19490976.2022.2164152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ding J, Ouyang R, Zheng S, Wang Y, Huang Y, Ma X, Zou Y, Chen R, Zhuo Z, Li Z, Xin Q, Zhou L, Mei S, Yan J, Lu X, Ren Z, Liu X, Xu G. 2022. Effect of breastmilk microbiota and sialylated oligosaccharides on the colonization of infant gut microbial community and fecal metabolome. Metabolites 12:1136. doi: 10.3390/metabo12111136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sierra C, Bernal M-J, Blasco J, Martínez R, Dalmau J, Ortuño I, Espín B, Vasallo M-I, Gil D, Vidal M-L, Infante D, Leis R, Maldonado J, Moreno J-M, Román E. 2015. Prebiotic effect during the first year of life in healthy infants fed formula containing GOS as the only Prebiotic: a multicentre, randomised, double-blind and placebo-controlled trial. Eur J Nutr 54:89–99. doi: 10.1007/s00394-014-0689-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yu X, Xing Y, Liu H, Chang Y, You Y, Dou Y, Liu B, Wang Q, Ma D, Chen L, Tong X. 2022. Effects of a formula with scGOS/lcFOS (9:1) and Glycomacropeptide (GMP) supplementation on the gut microbiota of very preterm infants. Nutrients 14:1901. doi: 10.3390/nu14091901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Chua MC, Ben-Amor K, Lay C, Neo AGE, Chiang WC, Rao R, Chew C, Chaithongwongwatthana S, Khemapech N, Knol J, Chongsrisawat V. 2017. Effect of synbiotic on the gut microbiota of cesarean delivered infants: a randomized, double-blind, multicenter study. J Pediatr Gastroenterol Nutr 65:102–106. doi: 10.1097/MPG.0000000000001623 [DOI] [PubMed] [Google Scholar]
- 146. Giovannini M, Verduci E, Gregori D, Ballali S, Soldi S, Ghisleni D, Riva E, PLAGOS Trial Study Group . 2014. Prebiotic effect of an infant formula supplemented with galacto-oligosaccharides: randomized multicenter trial. J Am Coll Nutr 33:385–393. doi: 10.1080/07315724.2013.878232 [DOI] [PubMed] [Google Scholar]
- 147. Lagkouvardos I, Intze E, Schaubeck M, Rooney JPK, Hecht C, Piloquet H, Clavel T. 2023. Early life gut microbiota profiles linked to synbiotic formula effects: a randomized clinical trial in European infants. Am J Clin Nutr 117:326–339. doi: 10.1016/j.ajcnut.2022.11.012 [DOI] [PubMed] [Google Scholar]
- 148. Brunser O, Gotteland M, Cruchet S, Figueroa G, Garrido D, Steenhout P. 2006. Effect of a milk formula with prebiotics on the intestinal microbiota of infants after an antibiotic treatment. Pediatr Res 59:451–456. doi: 10.1203/01.pdr.0000198773.40937.61 [DOI] [PubMed] [Google Scholar]
- 149. Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105:18964–18969. doi: 10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Šuligoj T, Vigsnæs LK, Abbeele P den, Apostolou A, Karalis K, Savva GM, McConnell B, Juge N. 2020. Effects of human milk oligosaccharides on the adult gut microbiota and barrier function. Nutrients 12:2808. doi: 10.3390/nu12092808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ryan JJ, Monteagudo-Mera A, Contractor N, Gibson GR. 2021. Impact of 2'-fucosyllactose on gut microbiota composition in adults with chronic gastrointestinal conditions: batch culture fermentation model and pilot clinical trial findings. Nutrients 13:938. doi: 10.3390/nu13030938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Dedon LR, Özcan E, Rani A, Sela DA. 2020. Bifidobacterium infantis metabolizes 2'fucosyllactose-derived and free fucose through a common catabolic pathway resulting in 1,2-propanediol secretion. Front Nutr 7:583397. doi: 10.3389/fnut.2020.583397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Salli K, Hirvonen J, Siitonen J, Ahonen I, Anglenius H, Maukonen J. 2021. Selective utilization of the human milk oligosaccharides 2'-fucosyllactose, 3-fucosyllactose, and difucosyllactose by various probiotic and pathogenic bacteria. J Agric Food Chem 69:170–182. doi: 10.1021/acs.jafc.0c06041 [DOI] [PubMed] [Google Scholar]
- 154. Chia LW, Mank M, Blijenberg B, Bongers RS, van Limpt K, Wopereis H, Tims S, Stahl B, Belzer C, Knol J. 2021. Cross-feeding between Bifidobacterium infantis and Anaerostipes caccae on lactose and human milk oligosaccharides. Benef Microbes 12:69–83. doi: 10.3920/BM2020.0005 [DOI] [PubMed] [Google Scholar]
- 155. Garrido D, Kim JH, German JB, Raybould HE, Mills DA. 2011. oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6:e17315. doi: 10.1371/journal.pone.0017315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. 2007. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem 55:8914–8919. doi: 10.1021/jf0710480 [DOI] [PubMed] [Google Scholar]
- 157. Maqbool A, Horler RSP, Muller A, Wilkinson AJ, Wilson KS, Thomas GH. 2015. The substrate-binding protein in bacterial ABC transporters: dissecting roles in the evolution of substrate specificity. Biochem Soc Trans 43:1011–1017. doi: 10.1042/BST20150135 [DOI] [PubMed] [Google Scholar]
- 158. Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao J-Z, Kitaoka M, Katayama T. 2019. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients 12:71. doi: 10.3390/nu12010071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kitaoka M. 2012. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv Nutr 3:422S–9S. doi: 10.3945/an.111.001420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum Prl2010 in a mucin-based medium. BMC Microbiol 14:282. doi: 10.1186/s12866-014-0282-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Nishiyama K, Nagai A, Uribayashi K, Yamamoto Y, Mukai T, Okada N. 2018. Two extracellular sialidases from Bifidobacterium bifidum promote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe 52:22–28. doi: 10.1016/j.anaerobe.2018.05.007 [DOI] [PubMed] [Google Scholar]
- 162. Turroni F, Bottacini F, Foroni E, Mulder I, Kim J-H, Zomer A, Sánchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107:19514–19519. doi: 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Ashida H, Tanigawa K, Kiyohara M, Katoh T, Katayama T, Yamamoto K. 2018. Bifunctional properties and characterization of a novel sialidase with esterase activity from Bifidobacterium bifidum. Biosci Biotechnol Biochem 82:2030–2039. doi: 10.1080/09168451.2018.1497944 [DOI] [PubMed] [Google Scholar]
- 164. Katoh T, Ojima MN, Sakanaka M, Ashida H, Gotoh A, Katayama T. 2020. Enzymatic adaptation of Bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms 8:481. doi: 10.3390/microorganisms8040481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Odamaki T, Horigome A, Sugahara H, Hashikura N, Minami J, Xiao J-Z, Abe F. 2015. Comparative genomics revealed genetic diversity and species/strain-level differences in carbohydrate metabolism of three probiotic bifidobacterial species. Int J Genomics 2015:567809. doi: 10.1155/2015/567809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Sakurama H, Kiyohara M, Wada J, Honda Y, Yamaguchi M, Fukiya S, Yokota A, Ashida H, Kumagai H, Kitaoka M, Yamamoto K, Katayama T. 2013. Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J Biol Chem 288:25194–25206. doi: 10.1074/jbc.M113.484733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Yamada C, Gotoh A, Sakanaka M, Hattie M, Stubbs KA, Katayama-Ikegami A, Hirose J, Kurihara S, Arakawa T, Kitaoka M, Okuda S, Katayama T, Fushinobu S. 2017. Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome Bifidobacterium longum. Cell Chem Biol 24:515–524. doi: 10.1016/j.chembiol.2017.03.012 [DOI] [PubMed] [Google Scholar]
- 168. Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 286:34583–34592. doi: 10.1074/jbc.M111.248138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- 170. Aakko J, Kumar H, Rautava S, Wise A, Autran C, Bode L, Isolauri E, Salminen S. 2017. Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef Microbes 8:563–567. doi: 10.3920/BM2016.0185 [DOI] [PubMed] [Google Scholar]
- 171. Kostopoulos I, Elzinga J, Ottman N, Klievink JT, Blijenberg B, Aalvink S, Boeren S, Mank M, Knol J, de Vos WM, Belzer C. 2020. Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Sci Rep 10:14330. doi: 10.1038/s41598-020-71113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Luna E, Parkar SG, Kirmiz N, Hartel S, Hearn E, Hossine M, Kurdian A, Mendoza C, Orr K, Padilla L, Ramirez K, Salcedo P, Serrano E, Choudhury B, Paulchakrabarti M, Parker CT, Huynh S, Cooper K, Flores GE, McBain AJ. 2022. Utilization efficiency of human milk oligosaccharides by human-associated akkermansia is strain dependent. Appl Environ Microbiol 88:e0148721. doi: 10.1128/AEM.01487-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Shuoker B, Pichler MJ, Jin C, Sakanaka H, Wu H, Gascueña AM, Liu J, Nielsen TS, Holgersson J, Nordberg Karlsson E, Juge N, Meier S, Morth JP, Karlsson NG, Abou Hachem M. 2023. Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria. Nat Commun 14:1833. doi: 10.1038/s41467-023-37533-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Özcan E, Rozycki MR, Sela DA. 2021. Cranberry proanthocyanidins and dietary oligosaccharides synergistically modulate Lactobacillus plantarum physiology. Microorganisms 9:656. doi: 10.3390/microorganisms9030656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Moya-Gonzálvez EM, Rubio-Del-Campo A, Rodríguez-Díaz J, Yebra MJ. 2021. Infant-gut associated Bifidobacterium dentium strains utilize the galactose moiety and release lacto-N-triose from the human milk oligosaccharides lacto-N-tetraose and lacto-N-neotetraose. Sci Rep 11:23328. doi: 10.1038/s41598-021-02741-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Fidanza M, Panigrahi P, Kollmann TR. 2021. Lactiplantibacillus plantarum-nomad and ideal probiotic. Front Microbiol 12:712236. doi: 10.3389/fmicb.2021.712236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Choi S, Baek M-G, Chung M-J, Lim S, Yi H. 2021. Distribution of bacteriocin genes in the lineages of Lactiplantibacillus plantarum. Sci Rep 11:20063. doi: 10.1038/s41598-021-99683-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Bidart GN, Rodríguez-Díaz J, Monedero V, Yebra MJ. 2014. A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto-N-biose and lacto-N-biose in Lactobacillus casei. Mol Microbiol 93:521–538. doi: 10.1111/mmi.12678 [DOI] [PubMed] [Google Scholar]
- 179. Bidart GN, Rodríguez-Díaz J, Pérez-Martínez G, Yebra MJ. 2018. The Lactose operon from Lactobacillus casei is involved in the transport and metabolism of the human milk oligosaccharide core-2 N-acetyllactosamine. Sci Rep 8:7152. doi: 10.1038/s41598-018-25660-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Rodríguez-Díaz J, Monedero V, Yebra MJ. 2011. Utilization of natural fucosylated oligosaccharides by three novel alpha-L-fucosidases from a probiotic Lactobacillus casei strain. Appl Environ Microbiol 77:703–705. doi: 10.1128/AEM.01906-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Curiel JA, Peirotén Á, Langa S, de Vega E, Blasco L, Landete JM. 2022. Characterization and stabilization of the α-L-fucosidase set from Lacticaseibacillus rhamnosus INIA P603. Appl Microbiol Biotechnol 106:8067–8077. doi: 10.1007/s00253-022-12262-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Engevik MA, Stripe LK, Baatz JE, Wagner CL, Chetta KE. 2022. Identifying single-strain growth patterns of human gut microbes in response to preterm human milk and formula. Food Funct 13:5571–5589. doi: 10.1039/d2fo00447j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Cheng CC, Duar RM, Lin X, Perez-Munoz ME, Tollenaar S, Oh J-H, van Pijkeren J-P, Li F, van Sinderen D, Gänzle MG, Walter J. 2020. Ecological importance of cross-feeding of the intermediate metabolite 1,2-propanediol between bacterial gut symbionts. Appl Environ Microbiol 86:e00190-20. doi: 10.1128/AEM.00190-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yu Z-T, Chen C, Newburg DS. 2013. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 23:1281–1292. doi: 10.1093/glycob/cwt065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Chia LW, Mank M, Blijenberg B, Aalvink S, Bongers RS, Stahl B, Knol J, Belzer C. 2020. Bacteroides thetaiotaomicron fosters the growth of butyrate-producing Anaerostipes caccae in the presence of lactose and total human milk carbohydrates. Microorganisms 8:1513. doi: 10.3390/microorganisms8101513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514. doi: 10.1016/j.chom.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. 2010. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem 58:5334–5340. doi: 10.1021/jf9044205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. 2013. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 8:e76341. doi: 10.1371/journal.pone.0076341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. 2011. An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21:437–447. doi: 10.1093/glycob/cwq175 [DOI] [PubMed] [Google Scholar]
- 190. Kijner S, Cher A, Yassour M. 2022. The infant gut commensal Bacteroides dorei presents a generalized transcriptional response to various human milk oligosaccharides. Front Cell Infect Microbiol 12:854122. doi: 10.3389/fcimb.2022.854122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Moya-Gonzálvez EM, Peña-Gil N, Rubio-Del-Campo A, Coll-Marqués JM, Gozalbo-Rovira R, Monedero V, Rodríguez-Díaz J, Yebra MJ. 2022. Infant gut microbial metagenome mining of α-l-fucosidases with activity on fucosylated human milk oligosaccharides and glycoconjugates. Microbiol Spectr 10:e0177522. doi: 10.1128/spectrum.01775-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Pichler MJ, Yamada C, Shuoker B, Alvarez-Silva C, Gotoh A, Leth ML, Schoof E, Katoh T, Sakanaka M, Katayama T, Jin C, Karlsson NG, Arumugam M, Fushinobu S, Abou Hachem M. 2020. Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat Commun 11:3285. doi: 10.1038/s41467-020-17075-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Cheng L, Kiewiet MBG, Logtenberg MJ, Groeneveld A, Nauta A, Schols HA, Walvoort MTC, Harmsen HJM, de Vos P. 2020. Effects of different human milk oligosaccharides on growth of Bifidobacteria in monoculture and co-culture with Faecalibacterium prausnitzii. Front Microbiol 11:569700. doi: 10.3389/fmicb.2020.569700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Sagheddu V, Patrone V, Miragoli F, Puglisi E, Morelli L. 2016. Infant early gut colonization by lachnospiraceae: high frequency of Ruminococcus gnavus. Front Pediatr 4:57. doi: 10.3389/fped.2016.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Grabinger T, Glaus Garzon JF, Hausmann M, Geirnaert A, Lacroix C, Hennet T. 2019. Alleviation of intestinal inflammation by oral supplementation with 2-fucosyllactose in mice. Front Microbiol 10:1385. doi: 10.3389/fmicb.2019.01385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, Bertha M, Cohen M, Garber J, Khalili H, Gevers D, Ananthakrishnan AN, Kugathasan S, Lander ES, Blainey P, Vlamakis H, Xavier RJ, Huttenhower C. 2017. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 9:103. doi: 10.1186/s13073-017-0490-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Wu H, Rebello O, Crost EH, Owen CD, Walpole S, Bennati-Granier C, Ndeh D, Monaco S, Hicks T, Colvile A, Urbanowicz PA, Walsh MA, Angulo J, Spencer DIR, Juge N. 2021. Fucosidases from the human gut symbiont Ruminococcus gnavus. Cell Mol Life Sci 78:675–693. doi: 10.1007/s00018-020-03514-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Le Gall G, de Vos WM, Taylor GL, Juge N. 2015. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun 6:7624. doi: 10.1038/ncomms8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Crost EH, Tailford LE, Monestier M, Swarbreck D, Henrissat B, Crossman LC, Juge N. 2016. The mucin-degradation strategy of Ruminococcus gnavus: the importance of intramolecular trans-sialidases. Gut Microbes 7:302–312. doi: 10.1080/19490976.2016.1186334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Manthey CF, Autran CA, Eckmann L, Bode L. 2014. Human milk oligosaccharides protect against enteropathogenic Escherichia coli attachment in vitro and EPEC colonization in suckling mice. J Pediatr Gastroenterol Nutr 58:165–168. doi: 10.1097/MPG.0000000000000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Ackerman DL, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, Townsend SD. 2017. Human milk oligosaccharides exhibit antimicrobial and antibiofilm properties against group B streptococcus. ACS Infect Dis 3:595–605. doi: 10.1021/acsinfecdis.7b00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Mejia ME, Ottinger S, Vrbanac A, Babu P, Zulk JJ, Moorshead D, Bode L, Nizet V, Patras KA. 2022. Human milk Oligosaccharides reduce murine group B Streptococcus vaginal Colonization with minimal impact on the vaginal Microbiota. mSphere 7:e0088521. doi: 10.1128/msphere.00885-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Lin AE, Autran CA, Szyszka A, Escajadillo T, Huang M, Godula K, Prudden AR, Boons G-J, Lewis AL, Doran KS, Nizet V, Bode L. 2017. Human milk oligosaccharides inhibit growth of group B Streptococcus. J Biol Chem 292:11243–11249. doi: 10.1074/jbc.M117.789974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ackerman DL, Craft KM, Doster RS, Weitkamp J-H, Aronoff DM, Gaddy JA, Townsend SD. 2018. Antimicrobial and antibiofilm activity of human milk oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, and Acinetobacter baumannii. ACS Infect Dis 4:315–324. doi: 10.1021/acsinfecdis.7b00183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Spicer SK, Moore RE, Lu J, Guevara MA, Marshall DR, Manning SD, Damo SM, Townsend SD, Gaddy JA. 2021. Antibiofilm activity of human milk oligosaccharides against multidrug resistant and susceptible isolates of Acinetobacter baumannii. ACS Infect Dis 7:3254–3263. doi: 10.1021/acsinfecdis.1c00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Moore RE, Xu LL, Townsend SD. 2021. Prospecting human milk oligosaccharides as a defense against viral infections. ACS Infect Dis 7:254–263. doi: 10.1021/acsinfecdis.0c00807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Craft KM, Townsend SD. 2018. The human milk glycome as a defense against infectious diseases: rationale, challenges, and opportunities. ACS Infect Dis 4:77–83. doi: 10.1021/acsinfecdis.7b00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Rousseaux A, Brosseau C, Le Gall S, Piloquet H, Barbarot S, Bodinier M. 2021. Human milk oligosaccharides: their effects on the host and their potential as therapeutic agents. Front Immunol 12:680911. doi: 10.3389/fimmu.2021.680911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Triantis V, Bode L, van Neerven RJJ. 2018. Immunological effects of human milk oligosaccharides. Front Pediatr 6:190. doi: 10.3389/fped.2018.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Roager HM, Stanton C, Hall LJ. 2023. Microbial metabolites as modulators of the infant gut microbiome and host-microbial interactions in early life. Gut Microbes 15:2192151. doi: 10.1080/19490976.2023.2192151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. doi: 10.1016/j.cell.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 212. Gill PA, van Zelm MC, Muir JG, Gibson PR. 2018. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther 48:15–34. doi: 10.1111/apt.14689 [DOI] [PubMed] [Google Scholar]
- 213. Xiong R-G, Zhou D-D, Wu S-X, Huang S-Y, Saimaiti A, Yang Z-J, Shang A, Zhao C-N, Gan R-Y, Li H-B. 2022. Health benefits and side effects of short-chain fatty acids. Foods 11:2863. doi: 10.3390/foods11182863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. de la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero R, Velásquez-Mejía EP, Sierra JA, Corrales-Agudelo V, Carmona JA, Abad JM, Escobar JS. 2018. Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients 11:51. doi: 10.3390/nu11010051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, Mujagic Z, Masclee AAM, Jonkers D, Oosting M, Joosten LAB, Netea MG, Franke L, Zhernakova A, Fu J, Wijmenga C, McCarthy MI. 2019. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 51:600–605. doi: 10.1038/s41588-019-0350-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Serino M. 2019. SCFAs - the thin microbial metabolic line between good and bad. Nat Rev Endocrinol 15:318–319. doi: 10.1038/s41574-019-0205-7 [DOI] [PubMed] [Google Scholar]
- 217. Lawson MAE, O’Neill IJ, Kujawska M, Gowrinadh Javvadi S, Wijeyesekera A, Flegg Z, Chalklen L, Hall LJ. 2020. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J 14:635–648. doi: 10.1038/s41396-019-0553-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Zabel B, Yde CC, Roos P, Marcussen J, Jensen HM, Salli K, Hirvonen J, Ouwehand AC, Morovic W. 2019. Novel genes and metabolite trends in Bifidobacterium longum subsp. infantis Bi-26 metabolism of human milk oligosaccharide 2'-fucosyllactose. Sci Rep 9:7983. doi: 10.1038/s41598-019-43780-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Rivière A, Gagnon M, Weckx S, Roy D, De Vuyst L. 2015. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl Environ Microbiol 81:7767–7781. doi: 10.1128/AEM.02089-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Okada T, Fukuda S, Hase K, Nishiumi S, Izumi Y, Yoshida M, Hagiwara T, Kawashima R, Yamazaki M, Oshio T, Otsubo T, Inagaki-Ohara K, Kakimoto K, Higuchi K, Kawamura YI, Ohno H, Dohi T. 2013. Microbiota-derived lactate accelerates colon epithelial cell turnover in starvation-refed mice. Nat Commun 4:1654. doi: 10.1038/ncomms2668 [DOI] [PubMed] [Google Scholar]
- 221. Louis P, Duncan SH, Sheridan PO, Walker AW, Flint HJ. 2022. Microbial lactate utilisation and the stability of the gut microbiome. Gut. Microb 3. doi: 10.1017/gmb.2022.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, Winter SE. 2018. Dysbiosis-associated change in host metabolism generates lactate to support salmonella growth. Cell Host Microbe 23:570. doi: 10.1016/j.chom.2018.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Llibre A, Grudzinska FS, O’Shea MK, Duffy D, Thickett DR, Mauro C, Scott A. 2021. Lactate cross-talk in host-pathogen interactions. Biochem J 478:3157–3178. doi: 10.1042/BCJ20210263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Özcan E, Sela DA. 2018. Inefficient metabolism of the human milk oligosaccharides lacto-N-tetraose and lacto-N-neotetraose shifts Bifidobacterium longum subsp. infantis physiology. Front. Nutr 5:46. doi: 10.3389/fnut.2018.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Hosseini E, Grootaert C, Verstraete W, Van de Wiele T. 2011. Propionate as a health-promoting microbial metabolite in the human gut. Nutr Rev 69:245–258. doi: 10.1111/j.1753-4887.2011.00388.x [DOI] [PubMed] [Google Scholar]
- 226. Chambers ES, Preston T, Frost G, Morrison DJ. 2018. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep 7:198–206. doi: 10.1007/s13668-018-0248-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Lordan C, Thapa D, Ross RP, Cotter PD. 2020. Potential for enriching next-generation health-promoting gut bacteria through prebiotics and other dietary components. Gut Microbes 11:1–20. doi: 10.1080/19490976.2019.1613124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Ehrlich AM, Pacheco AR, Henrick BM, Taft D, Xu G, Huda MN, Mishchuk D, Goodson ML, Slupsky C, Barile D, Lebrilla CB, Stephensen CB, Mills DA, Raybould HE. 2020. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol 20:357. doi: 10.1186/s12866-020-02023-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Sakurai T, Odamaki T, Xiao J-Z. 2019. Production of Indole-3-lactic acid by Bifidobacterium strains isolated from human infants. Microorganisms 7:340. doi: 10.3390/microorganisms7090340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, Zhu W, Walker WA. 2020. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies Infantis is anti-inflammatory in the immature intestine. Pediatr Res 88:209–217. doi: 10.1038/s41390-019-0740-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Centanni M, Ferguson SA, Sims IM, Biswas A, Tannock GW. 2019. Bifidobacterium bifidum ATCC 15696 and Bifidobacterium breve 24B metabolic interaction based on 2'-O-fucosyl-lactose studied in steady-state cultures in a freter-style chemostat. Appl Environ Microbiol 85:e02783-18. doi: 10.1128/AEM.02783-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Sela DA, Mills DA. 2010. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol 18:298–307. doi: 10.1016/j.tim.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. doi: 10.1128/AEM.01114-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Di Costanzo M, De Paulis N, Biasucci G. 2021. Butyrate: a link between early life nutrition and gut microbiome in the development of food allergy. Life (Basel) 11:384. doi: 10.3390/life11050384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Button JE, Cosetta CM, Reens AL, Brooker SL, Rowan-Nash AD, Lavin RC, Saur R, Zheng S, Autran CA, Lee ML, Sun AK, Alousi AM, Peterson CB, Koh AY, Rechtman DJ, Jenq RR, McKenzie GJ. 2023. Precision modulation of dysbiotic adult microbiomes with a human-milk-derived synbiotic reshapes gut microbial composition and metabolites. Cell Host Microbe 31:1523–1538. doi: 10.1016/j.chom.2023.08.004 [DOI] [PubMed] [Google Scholar]
- 236. Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73:7767–7770. doi: 10.1128/AEM.01477-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. 2017. Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:491–502. doi: 10.1038/nrgastro.2017.75 [DOI] [PubMed] [Google Scholar]
- 238. Hegar B, Wibowo Y, Basrowi RW, Ranuh RG, Sudarmo SM, Munasir Z, Atthiyah AF, Widodo AD, Kadim M, Suryawan A, Diana NR, Manoppo C, Vandenplas Y. 2019. The role of two human milk oligosaccharides, 2'-fucosyllactose and lacto-N-neotetraose, in infant nutrition. Pediatr Gastroenterol Hepatol Nutr 22:330–340. doi: 10.5223/pghn.2019.22.4.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Ambrogi V, Bottacini F, Cao L, Kuipers B, Schoterman M, van Sinderen D. 2023. Galacto-oligosaccharides as infant prebiotics: production, application, bioactive activities and future perspectives. Crit Rev Food Sci Nutr 63:753–766. doi: 10.1080/10408398.2021.1953437 [DOI] [PubMed] [Google Scholar]
- 240. Vandenplas Y, Berger B, Carnielli VP, Ksiazyk J, Lagström H, Sanchez Luna M, Migacheva N, Mosselmans JMM, Picaud JC, Possner M, Singhal A, Wabitsch M. 2018. Human milk oligosaccharides: 2’-fucosyllactose (2’-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients 10:1161. doi: 10.3390/nu10091161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Puccio G, Alliet P, Cajozzo C, Janssens E, Corsello G, Sprenger N, Wernimont S, Egli D, Gosoniu L, Steenhout P. 2017. Effects of infant formula with human milk oligosaccharides on growth and morbidity: a randomized multicenter trial. J Pediatr Gastroenterol Nutr 64:624–631. doi: 10.1097/MPG.0000000000001520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Parschat K, Melsaether C, Jäpelt KR, Jennewein S. 2021. Clinical evaluation of 16-week supplementation with 5HMO-mix in healthy-term human infants to determine tolerability, safety, and effect on growth. Nutrients 13:2871. doi: 10.3390/nu13082871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Zhang P, Zhu Y, Li Z, Zhang W, Mu W. 2022. Recent advances on lacto-N-neotetraose, a commercially added human milk oligosaccharide in infant formula. J Agric Food Chem 70:4534–4547. doi: 10.1021/acs.jafc.2c01101 [DOI] [PubMed] [Google Scholar]
- 244. Liu S, Fang Z, Wang H, Zhai Q, Hang F, Zhao J, Zhang H, Lu W, Chen W. 2021. Gene-phenotype associations involving human-residential bifidobacteria (HRB) reveal significant species- and strain-specificity in carbohydrate catabolism. Microorganisms 9:883. doi: 10.3390/microorganisms9050883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Ryan SM, Fitzgerald GF, van Sinderen D. 2005. Transcriptional regulation and characterization of a novel beta-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl Environ Microbiol 71:3475–3482. doi: 10.1128/AEM.71.7.3475-3482.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Viborg AH, Katayama T, Abou Hachem M, Andersen MCF, Nishimoto M, Clausen MH, Urashima T, Svensson B, Kitaoka M. 2014. Distinct substrate specificities of three glycoside hydrolase family 42 β-galactosidases from Bifidobacterium longum subsp. infantis ATCC 15697. Glycobiology 24:208–216. doi: 10.1093/glycob/cwt104 [DOI] [PubMed] [Google Scholar]
- 247. Fushinobu S, Abou Hachem M. 2021. Structure and evolution of the bifidobacterial carbohydrate metabolism proteins and enzymes. Biochem Soc Trans 49:563–578. doi: 10.1042/BST20200163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Warchol M, Perrin S, Grill JP, Schneider F. 2002. Characterization of a purified beta-fructofuranosidase from Bifidobacterium infantis ATCC 15697. Lett Appl Microbiol 35:462–467. doi: 10.1046/j.1472-765X.2002.01224.x [DOI] [PubMed] [Google Scholar]
- 249. Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem 286:11909–11918. doi: 10.1074/jbc.M110.193359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Møller PL, Jørgensen F, Hansen OC, Madsen SM, Stougaard P. 2001. Intra- and extracellular beta-galactosidases from Bifidobacterium bifidum and B. Infantis: molecular cloning, heterologous expression, and comparative characterization. Appl Environ Microbiol 67:2276–2283. doi: 10.1128/AEM.67.5.2276-2283.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Sabater C, Ruiz L, Margolles A. 2021. A machine learning approach to study glycosidase activities from Bifidobacterium. Microorganisms 9:1034. doi: 10.3390/microorganisms9051034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. 2004. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA). J Bacteriol 186:4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017. doi: 10.1093/glycob/cwp082 [DOI] [PubMed] [Google Scholar]
- 254. Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl Environ Microbiol 71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10:1656–1668. doi: 10.1038/ismej.2015.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Sotoya H, Shigehisa A, Hara T, Matsumoto H, Hatano H, Matsuki T. 2017. Identification of genes involved in galactooligosaccharide utilization in Bifidobacterium breve strain YIT 4014T. Microbiology 163:1420–1428. doi: 10.1099/mic.0.000517 [DOI] [PubMed] [Google Scholar]
- 257. Kelly SM, Munoz-Munoz J, van Sinderen D. 2021. Plant glycan metabolism by bifidobacteria. Front Microbiol 12:609418. doi: 10.3389/fmicb.2021.609418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Ioannou A, Knol J, Belzer C. 2021. Microbial glycoside hydrolases in the first year of life: an analysis review on their presence and importance in infant gut. Front Microbiol 12:631282. doi: 10.3389/fmicb.2021.631282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. 2012. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading Type-1 and Type-2 human milk oligosaccharides. Glycobiology 22:361–368. doi: 10.1093/glycob/cwr116 [DOI] [PubMed] [Google Scholar]
- 260. González R, Klaassens ES, Malinen E, de Vos WM, Vaughan EE. 2008. Differential transcriptional response of Bifidobacterium longum to human milk, formula milk, and galactooligosaccharide. Appl Environ Microbiol 74:4686–4694. doi: 10.1128/AEM.00122-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. O’Connell Motherway M, Kinsella M, Fitzgerald GF, van Sinderen D. 2013. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microb Biotechnol 6:67–79. doi: 10.1111/1751-7915.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Alessandri G, van Sinderen D, Ventura M. 2021. The genus Bifidobacterium: from genomics to functionality of an important component of the mammalian gut microbiota. Comput Struct Biotechnol J 19:1472–1487. doi: 10.1016/j.csbj.2021.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Ambrogi V, Bottacini F, O’Sullivan J, O’Connell Motherway M, Linqiu C, Schoemaker B, Schoterman M, van Sinderen D. 2019. Characterization of GH2 and GH42 β-galactosidases derived from bifidobacterial infant isolates. AMB Express 9:9. doi: 10.1186/s13568-019-0735-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Garrido D, Dallas DC, Mills DA. 2013. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology (Reading) 159:649–664. doi: 10.1099/mic.0.064113-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015. Mucin glycan foraging in the human gut microbiome. Front Genet 6:81. doi: 10.3389/fgene.2015.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Samuel TM, Hartweg M, Lebumfacil JD, Buluran KB, Lawenko RB, Estorninos EM, Binia A, Sprenger N. 2022. Dynamics of human milk oligosaccharides in early lactation and relation with growth and appetitive traits of filipino breastfed infants. Sci Rep 12:17304. doi: 10.1038/s41598-022-22244-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Sims IM, Tannock GW. 2020. Galacto- and fructo-oligosaccharides utilized for growth by cocultures of bifidobacterial species characteristic of the infant gut. Appl Environ Microbiol 86:e00214-20. doi: 10.1128/AEM.00214-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Garrido D, Ruiz-Moyano S, Jimenez-Espinoza R, Eom H-J, Block DE, Mills DA. 2013. Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiol 33:262–270. doi: 10.1016/j.fm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Kim J-H, An HJ, Garrido D, German JB, Lebrilla CB, Mills DA, de Crécy-Lagard V. 2013. Proteomic analysis of Bifidobacterium longum subsp. infantis reveals the metabolic insight on consumption of prebiotics and host glycans. PLoS ONE 8:e57535. doi: 10.1371/journal.pone.0057535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. doi: 10.1128/AEM.06762-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Böger M, van Leeuwen SS, Lammerts van Bueren A, Dijkhuizen L. 2019. Structural identity of galactooligosaccharide molecules selectively utilized by single cultures of probiotic bacterial strains. J Agric Food Chem 67:13969–13977. doi: 10.1021/acs.jafc.9b05968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Kujawska M, La Rosa SL, Roger LC, Pope PB, Hoyles L, McCartney AL, Hall LJ. 2020. Succession of Bifidobacterium longum strains in response to a changing early life nutritional environment reveals dietary substrate adaptations. iScience 23:101368. doi: 10.1016/j.isci.2020.101368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Zhu Y, Zhang J, Zhang W, Mu W.. 2022. Recent progress on health effects and biosynthesis of two key sialylated human milk oligosaccharides, 3'-sialyllactose and 6'-sialyllactose. Biotechnol Adv:108058. [DOI] [PubMed] [Google Scholar]