Abstract
Positive correlations between mates can increase trait variation and prevalence, as well as bias estimates from genetically informed study designs. While past studies of similarity between human mating partners have largely found evidence of positive correlations, to our knowledge, no formal meta-analysis has examined human partner correlations across multiple categories of traits. Thus, we conducted systematic reviews and random-effects meta-analyses of human male-female partner correlations across 22 traits commonly studied by psychologists, economists, sociologists, anthropologists, epidemiologists, and geneticists. Using ScienceDirect, PubMed, and Google Scholar, we incorporated 480 partner correlations from 199 peer-reviewed studies of co-parents, engaged pairs, married pairs, and/or cohabitating pairs that were published on or before August 16, 2022. We also calculated 133 trait correlations using up to 79,074 male-female couples in the UK Biobank (UKB). Estimates of the 22 mean meta-analyzed correlations ranged from rmeta=.08 (adjusted 95% CI=[.03, .13]) for extraversion to rmeta=.58 (adjusted 95% CI=[.50, .64]) for political values, with funnel plots showing little evidence of publication bias across traits. The 133 UKB correlations ranged from rUKB=−.18 (adjusted 95% CI=[−.20, −.16]) for chronotype (being a “morning” or “evening” person) to rUKB=.87 (adjusted 95% CI=[.86, .87]) for birth year. Across analyses, political and religious attitudes, educational attainment, and some substance use traits showed the highest correlations, while psychological (i.e., psychiatric/personality) and anthropometric traits generally yielded lower but positive correlations. We observed high levels of between-sample heterogeneity for most meta-analyzed traits, likely because of both systematic differences between samples and true differences in partner correlations across populations.
Editor summary:
Meta-analyses of 22 traits and UK Biobank analyses of 133 traits find widespread evidence of mate similarity, particularly for social attitudes, education, and substance use traits.
Introduction
Phenotypic resemblance between human mates, spouses, and co-parents (hereafter, “partners”) is a widely studied area of research with applications to an array of disciplines. Quantitative and behavioral geneticists are interested in the magnitudes and mechanisms underlying partner correlations because both can affect genetic and environmental parameters and estimates of interest, informing how model-derived estimates should be interpreted1–4. Sociologists, anthropologists, economists, and other social scientists have studied partner correlations to gain insight into trends in societal values5, the labor market6,7, socioeconomic inequalities5–7, relationship satisfaction, and divorce rates8–13. Understanding patterns of (dis)similarity between mates thus provides useful context for both sociocultural and biological phenomena—as well as their intersection—and can shed light on the forces shaping human mating trends.
Contrary to the maxim “opposites attract,” nonzero phenotypic correlations between human8,14–32 and nonhuman33 mates and partners—a phenomenon referred to as “assortative mating” (AM) when the similarity is present at the beginning of the relationship—are overwhelmingly in the positive direction, with only a handful of examples of significant negative partner correlations (“disassortative mating”) reported in the literature9,15,19,31,33–40. Several potential mechanisms leading to partner resemblance in humans have been described. Phenotypic homogamy (also called “primary phenotypic assortative mating”) occurs when partners match directly on the trait of interest41. While phenotypic homogamy is often conceptualized as partners simply preferring similarity, it can also arise from indirect selection (e.g., in parent-arranged marriages) or when partnerships form within strata that are associated with trait values (e.g., partner correlations for educational attainment arising as an indirect consequence of mate choice occurring within profession). Social homogamy, on the other hand, results when partners assort on non-heritable aspects of a trait29,42. This can occur through several different processes, such as when romantic/marital partnerships form within strata that are heavily determined by social factors (e.g., social networks) that are not genetically correlated with the focal trait. Genetic homogamy describes the opposite phenomenon: when partners assort on heritable aspects of a trait. Genetic homogamy may occur when there is phenotypic homogamy on a trait that is genetically but not environmentally correlated with the trait of interest41,43, resulting in a genetic correlation between partners that is higher than would be expected given the phenotypic correlation for the focal trait. For example, there is evidence of genetic homogamy for educational attainment43 in the UK Biobank, suggesting that partners in this sample are assorting heavily upon a different trait (i.e., intelligence quotient score) that is more genetically than environmentally correlated with education level. Finally, convergence occurs when partners become more similar over time14,19, either due to direct (reciprocal or one-way) phenotypic influences or to the mutual influence of environmental factors shared between partners. Unlike social/phenotypic/genetic homogamy, convergence is not considered a type of AM because it is not a consequence of initial matching (or assortment). Importantly, the processes that lead to partner correlations need not be mutually exclusive, as multiple mechanisms may work in concert to produce an observed correlation between partners for a given trait. For example, it is possible that a mixture of social homogamy and phenotypic homogamy for a given trait is more common than “pure” social homogamy or “pure” phenotypic homogamy. Similarly, it is possible that individuals with a high environmental load for a given trait may preferentially mate with individuals that have a high genetic load for the same trait, forming a gene-environment correlation between partners that would likely appear as phenotypic homogamy.
When occurring on heritable traits, phenotypic and genetic homogamy increase correlations between and within causal genetic variants, which in turn increases both the genetic covariance between relatives and the trait’s genetic and phenotypic variation. Such an increase in variation could manifest as increased prevalence rates of dichotomous traits such as psychiatric disorders29,44, although this effect would only be pronounced under certain conditions (e.g., for rare, highly heritable traits undergoing strong phenotypic AM29). Failing to account for phenotypic and genetic homogamy can also lead to biases in association statistics from genome-wide association studies45, in heritability and genetic correlation estimates based on twin/family designs and single nucleotide polymorphisms46,47, and in the strength of estimated causal associations from Mendelian randomization studies48. Similarly, despite having no genetic consequences, convergence and social homogamy may still increase the environmental and phenotypic variance of a trait and the covariance between relatives to the extent that the trait is shaped by non-genetic parental influences; as a result, these non-genetic mechanisms could still ostensibly affect the prevalence of dichotomous traits. Understanding the magnitude of partner correlations across a wide variety of traits is therefore important for setting expectations of what analyses are at risk of bias due to the influences of homogamy and for guiding efforts to uncover the mechanisms that underlie these correlations.
In addition to contributing to knowledge around the architecture and etiology of traits, research on shared disease/disorder risk factors and health-related behaviors can also have public health implications for screening and intervention practices16,38,49,50 . For example, research on the nature of partner correlations for behavioral similarities could potentially inform how partners can effectively make lifestyle changes together17,51, particularly if and when partner resemblance is due to mutual influences within the relationship. Furthermore, partner correlations can lead to the concentration of certain traits (e.g., education or income) within particular social groups, potentially exacerbating existing social inequalities8. Characterizing patterns of mate similarity—particularly with regard to how they differ across populations—is thus of great sociological relevance.
In the current report, we meta-analyze and compare estimates of 480 male-female partner correlations (taken from 199 independent studies) on 22 traits frequently investigated in this research arena. In addition, we calculate correlations across 133 traits using up to 79,074 male-female partner dyads from the UK Biobank (UKB) sample. These results can help shed light on contemporary human mating and relationship trends, refine the interpretation of heritability estimates, motivate investigation into the various causes of partner correlations across traits, and aid in the choice of design in genetic and non-genetic studies.
Results
Meta-analysis
Our random effects meta-analytic results include a total of 480 partner correlations across 22 (out of an initial 26) traits for which we were able to find results from at least three samples that satisfied our criteria. The total number of partners for each trait ranged from 2,527 (for generalized anxiety) to 2,727,151 (for diabetes). Supplementary Tables 1 and 2 show all studies that we included in our meta-analysis for continuous and dichotomous traits, respectively, as well as the effect sizes for each sample. For comparability across traits, we focus on Pearson’s, Spearman’s, and tetrachoric correlations—some of which were transformed from the original reported statistic—for continuous, ordinal, and dichotomous traits, respectively. Our meta-analyses are based on pooled Fisher Z-transformed estimates from the included studies because this transformation yields a sampling distribution that is approximately normal. We then back-transformed the meta-analyzed estimates and report correlations along with their adjusted confidence intervals (see Table 1).
Table 1:
Results for the random effects meta-analyses of mating partner pairs across 22 traits.
| Trait | r [CI] | k (ks) | N | p-val | I2 total (within-, between-) | τ (within-, between-) | Prediction Interval |
|---|---|---|---|---|---|---|---|
| EA13,15,34,58,82–119 | .55 [.50, .60] | 86 (42) | 1,911,720 | <.0001 | .99 (.40, .59) | .094, .114 | [0.31, 0.72] |
| IQ Score22,103,120–130 | .44 [.23, .61] | 17 (13) | 5,672 | <.0001 | .94 (0,.94) | .000, .223 | [−0.03, 0.74] |
| Political Values13,87,121,131–138 | .58 [.50, .64] | 12 (11) | 11,658 | <.0001 | .80 (.11, .69) | .028, .068 | [0.45, 0.68] |
| Religiosity13,121,131,135,137,139,140 | .56 [.25, .77] | 8 (7) | 6,269 | 0.0022 | .94 (0, .94) | .000, .203 | [0.12, 0.82] |
| PAU28,31,141,142 | .28 [−.21, .66] | 4 | 9,751 | 0.2480 | .62 | .079 | [−0.01, 0.53] |
| Drinking19,49,88,106,143–147 | .42 [.07, .68] | 9 | 7,055 | 0.0187 | .98 | .255 | [−0.17, 0.79] |
| Smk. Cessation16,51,148,149 | .54 [−.28, .91] | 4 | 3,613 | 0.1530 | .84 | .161 | [ 0.02, 0.83] |
| Smk. Initiation16,19,38,49,51,88,143,146,148–152 | .38 [.23, .53] | 16 (13) | 118,856 | <.0001 | .96 (.14, .82) | .054, .130 | [0.09, 0.61] |
| Smk. Quantity15,153–157 | .29 [.07, .68] | 9 (6) | 12,072 | 0.0002 | .82 (.58, .24) | .056, .036 | [0.13, 0.44] |
| Smk. Status16,19,25,38,58,148,149,151,152,155,157–166 | .49 [.38, .59] | 27 (20) | 244,597 | <.0001 | .98 (.24, .74) | .089, .157 | [0.15, 0.72] |
| SUD72,167,168 | .43 [−.73, .95] | 4 (3) | 1,534,416 | >0.999 | 1.00 (0, 1.0) | .000, .223 | [−0.36, 0.86] |
| Agree.34,36,121,132,169–176 | .11 [.03, .20] | 17 (12) | 19,471 | 0.0039 | .82 (.22, .60) | .036, .059 | [−0.04, 0.26] |
| Consc.34,36,121,132,169–176 | .16 [.06, .25] | 18 (12) | 19,930 | <.0001 | .85 (0, .85) | .000, .078 | [−0.02, 0.32] |
| Extrav.13,19,22,34,36,87,120,121,131,132,138,139,154,169–185 | .08 [.03, .13] | 40 (30) | 32,729 | <.0001 | .79 (.50, .29) | .056, .043 | [−0.07, 0.22] |
| Neurot.13,19,22,34,36,121,132,138,139,154,169–178,180–189 | .11 [.08, .15] | 39 (30) | 35,706 | <.0001 | .58 (.52, .06) | .039, .013 | [0.03, 0.19] |
| Open.34,36,121,132,169,171,172,174,176,188,190 | .21 [.10, .31] | 16 (11) | 19,377 | <.0001 | .89 (.06, .83) | .022, .083 | [0.02, 0.38] |
| BMI38,49,50,82,87,88,113,116,118,143,148,152,162,185,191–216 | .16 [.11, .21] | 57 (40) | 123,881 | <.0001 | .94 (.28, .66) | .049, .075 | [−0.02, 0.33] |
| Height19,85,87,113,118,125,147,185,193,194,204,205,211–251 | .24 [.20, .28] | 68 (53) | 293,461 | <.0001 | .97 (.61, .36) | .080, .061 | [0.04, 0.42] |
| WHR49,192,196,203,210,252 | .17 [.03, .29] | 6 | 6,055 | 0.0184 | .59 | .043 | [0.04, 0.28] |
| Depression27,28,31,72,253 | .15 [.03, .28] | 7 (5) | 1,483,486 | 0.0198 | .96 (.01, .95) | .005, .039 | [0.04, 0.26] |
| Diabetes16,27,39,72,152,210,254–257 | .15 [.04, .26] | 12 (10) | 2,727,151 | 0.0046 | .99 (.09, .90) | .023, .087 | [−0.02, 0.31] |
| Gen Anx.28,30,31 | .17 [−.45, .68] | 4 (3) | 2,527 | >0.999 | .54 (.54, 0) | .098, .000 | [−0.20, 0.50] |
Citations after each trait refer to each study that was meta-analyzed for that trait, k = number of samples meta-analyzed, where all samples for a specific trait were determined to be distinct; ks = number of published studies from which the samples were taken; r(df = k−1) = estimate of the mean meta-analyzed random effects partner correlation (Pearson’s r for continuous traits; tetrachoric r for dichotomous traits); Cl = Bonferroni-adjusted 95% confidence intervals (adjusting for 22 tests); N = number of total partner pairs meta-analyzed; p-val = p-value, based on two-sided tests, Bonferroni-adjusted for 22 tests; I2 = Higgins & Thompson’s I2 statistic, a measure of between-/within-study heterogeneity, where overall I2 is the proportion of variance in the meta-analysis that is not due to sampling error; τ = the estimated standard deviation of the true effect size (when applicable, heterogeneity values are given for both the within-study and between-study levels, respectively); Prediction Interval = the interval in which the correlations for future meta-analyses of the trait in question are expected to fall. EA = Educational Attainment; IQ score = Intelligence Quotient Score; PAU = Problematic Alcohol Use; Drinking = Drinking Quantity; Smk. = Smoking; SUD = Substance Use Disorder; Agree. = Agreeableness; Consc. = Conscientiousness; Extrav. = Extraversion, Neurot. = Neuroticism; Open. = Openness to Experience; BMI = Body Mass Index; WHR= Waist-to-Hip Ratio; Gen Anx. = Generalized Anxiety.
Fig. 1 displays the estimates of the mean meta-analyzed correlations for all meta-analyzed traits and, where applicable, the correlations for comparable traits in the UK Biobank (UKB; see Partner Correlations in the UK Biobank), along with the adjusted confidence intervals associated with each trait. The estimates of the mean correlations for the meta-analyzed traits (rmeta) were greater than zero at the two-tailed, Bonferroni-corrected significance level for eighteen traits, with the remaining four estimates being based on only four samples each. rmeta was greater than .35 for nine attitudinal, academic, and substance-related traits, ranging from rmeta =. 38 for smoking initiation to rmeta=.58 for political values. It is important to note that, despite being associated with relatively large point estimates, three of the seven substance use traits we meta-analyzed did not achieve Bonferroni-corrected significance. Estimates of mean meta-analyzed correlations for anthropometric traits and (non-substance related) disorder traits were all low-to-moderate (.15 ≤ rmeta ≤ .24), though the estimate for generalized anxiety (rmeta=.17, padjusted > .999) was not significant. The three lowest estimates we found were for the Big Five personality traits extraversion (rmeta = .08), neuroticism (rmeta = .11), and agreeableness (rmeta = .11). Point estimates for conscientiousness (rmeta = .16) and openness to experience (rmeta = .21) were slightly higher (see Table 1 for Bonferroni-adjusted two-tailed p-values and Bonferroni-adjusted 95% confidence intervals, as well as total sample sizes associated with each meta-analyzed trait).
Figure 1: The point estimates of the mean meta-analyzed random effects partner correlations and UK Biobank partner correlations for comparable traits, along with their respective Bonferroni-adjusted 95% confidence intervals.
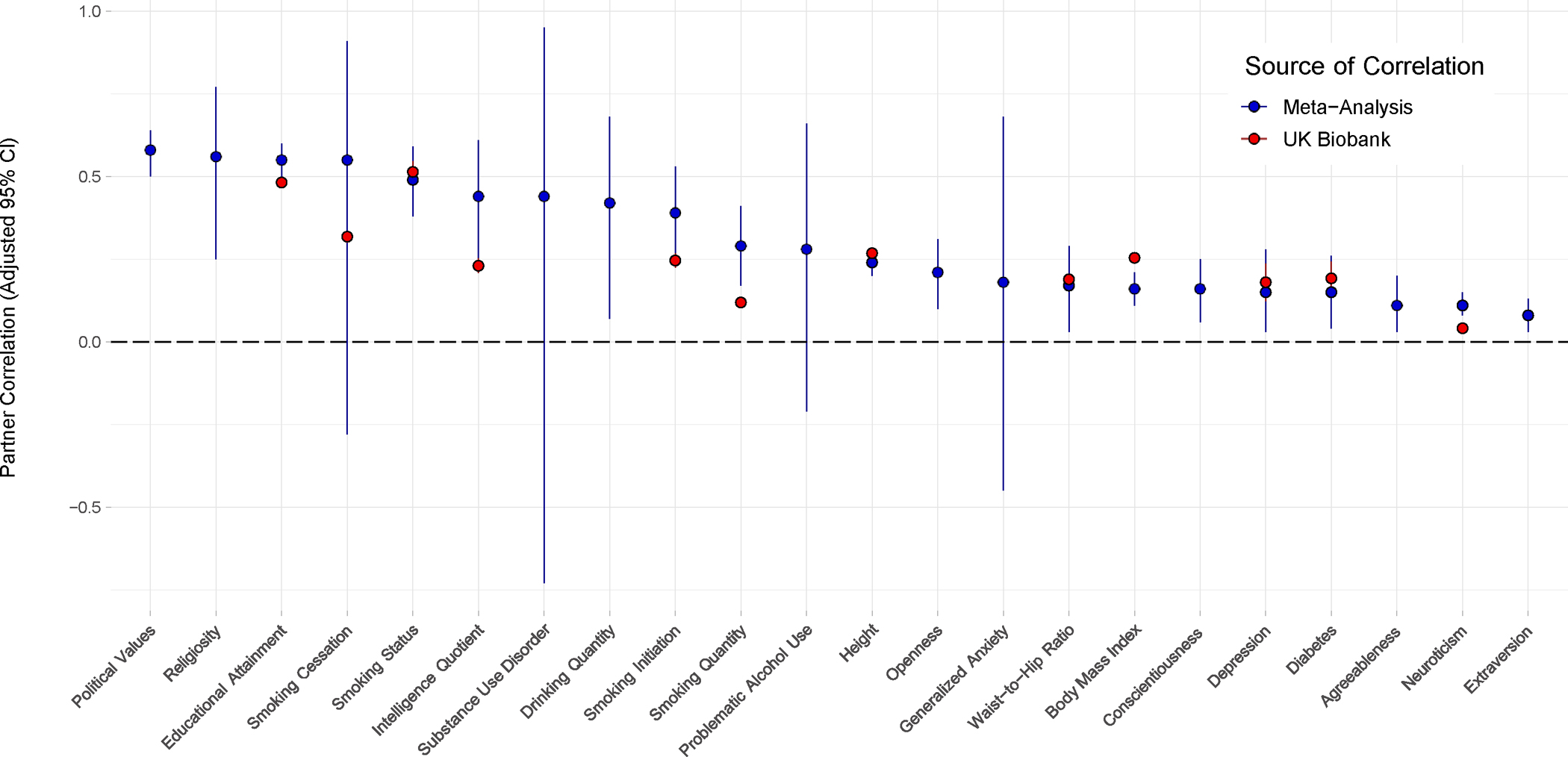
The dark blue points represent the random effects estimates of the mean meta-analyzed correlations for partners, while the red points on the same vertical axis represent the point estimates of the partner correlations for a comparable trait in the UK Biobank, where applicable. To account for multiple testing, meta-analyzed and UK Biobank correlations are shown with Bonferroni-adjusted 95% confidence intervals (adjusting for 22 and 133 traits, respectively). Table 1 and Supplementary Table 4 include the precise sample size and point estimate/adjusted confidence interval/adjusted p-value, etc. for each of these traits in the meta-analysis and UK Biobank, respectively.
Additionally, Table 1 summarizes heterogeneity estimates for each meta-analysis as well as prediction intervals of future studies’ effect sizes. For traits meta-analyzed using random effects models with two levels, we calculated a single Higgins & Thompson’s I2, which estimates the proportion of variation resulting from heterogeneity across a trait’s study-/sample-level effect sizes that is not attributable to sampling error. For traits meta-analyzed using models with three levels (those for which at least one publication reported two or more effect sizes), Table 1 also shows two I2 values: one representing the proportion of variance resulting from heterogeneity within-study and one representing proportion of variance resulting from between-study heterogeneity, which sum to the total I2 (see Meta-analytic Method)52. The median within-study I2 for three-level analyses was .18, while the median between-study I2 was .715. Partitioning within- vs. between-study I2 estimates can yield unstable results, especially for meta-analyses that only involve a small number of effect sizes within a common study, and so should be interpreted with caution. Across our 22 meta-analyses, the median total Higgins & Thompson I2 statistic was .915, reflecting a very high rates of heterogeneity in partner correlations not due to sampling error.
In general, high I2 values may reflect not only high between-sample heterogeneity in estimated effect sizes but also low within-sample estimation error. Thus, the high I2 values we observed may be in part due to the high precision of estimates afforded by the large number of participants comprising many of the samples included in our analysis. To this end, we also report an alternative metric of heterogeneity, τ, that estimates the standard deviation of the true effect size and is unaffected by the precision of individual estimates. The median overall τ was .090 and ranged from .039 (Depression) to .255 (Drinking). τ tended to be positively associated with rmeta, with the average ratio of τ to rmeta (the coefficient of variation) being .373 (SD =.180) across traits.
Overall, our results show that partner correlations are characterized by substantial heterogeneity across samples. Given that traits measured objectively (e.g., height, body mass index, waist-to-hip ratio) were also associated with considerable heterogeneity, our observed I2 and τ estimates suggest that there are substantial differences in true partner correlations for a given trait across populations differentiated by location, time, and/or culture.
Finally, we produced funnel plots for each meta-analyzed trait to assess publication bias. Funnel plots demonstrating a large negative regression slope—particularly those with several studies clustering in the bottom-right but not the bottom-left–are indicative of publication bias. Overall, there were not obvious patterns of asymmetry across traits (Extended Data Figs. 1a–1v). The clearest trend across most of the funnel plots was the large number of points outside the expected triangular region, again reflecting the high heterogeneity in correlations observed across samples.
Partner Correlations in the UK Biobank
We investigated correlations for an initial 140 traits using up to 79,074 pairs of UK Biobank (UKB) participants inferred to be opposite-sex partners (see Analysis of Partner Correlations in the UK Biobank). Seven (5%) of these traits are not included in our primary analyses, in Fig. 2, or in Extended Data Fig. 2 because of low sample size or low endorsement rate. However, results for all 140 traits are presented in Supplementary Table 4, which includes Bonferroni-adjusted two-tailed p-values and Bonferroni-adjusted 95% confidence intervals, as well as summary statistics, cell frequencies and prevalence rates (for dichotomous traits), corresponding UKB field IDs, and trait descriptions. Of the 133 adequately-powered UKB traits presented in Fig. 2 and Extended Data Fig. 2—including 59 continuous, 24 ordinal, and 50 dichotomous traits (estimated using Pearson’s, Spearman’s, and tetrachoric correlations, respectively)—118 partner correlations (88.72%) were significant after correction for multiple testing and fifteen (11.28%) were not. Out of the 118 partner correlations that were significant at the Bonferroni-corrected level, three (2.54%) were negative and 115 traits (97.46%) were positive. The mean partner correlation was .19, and correlations ranged from rUKB= −.18 (chronotype—i.e., being an “evening person” vs. being a “morning person”) to .87 (year of birth).
Figure 2: The UK Biobank partner correlation point estimates for 133 traits, along with their respective Bonferroni-adjusted 95% confidence intervals, grouped by category of the trait.
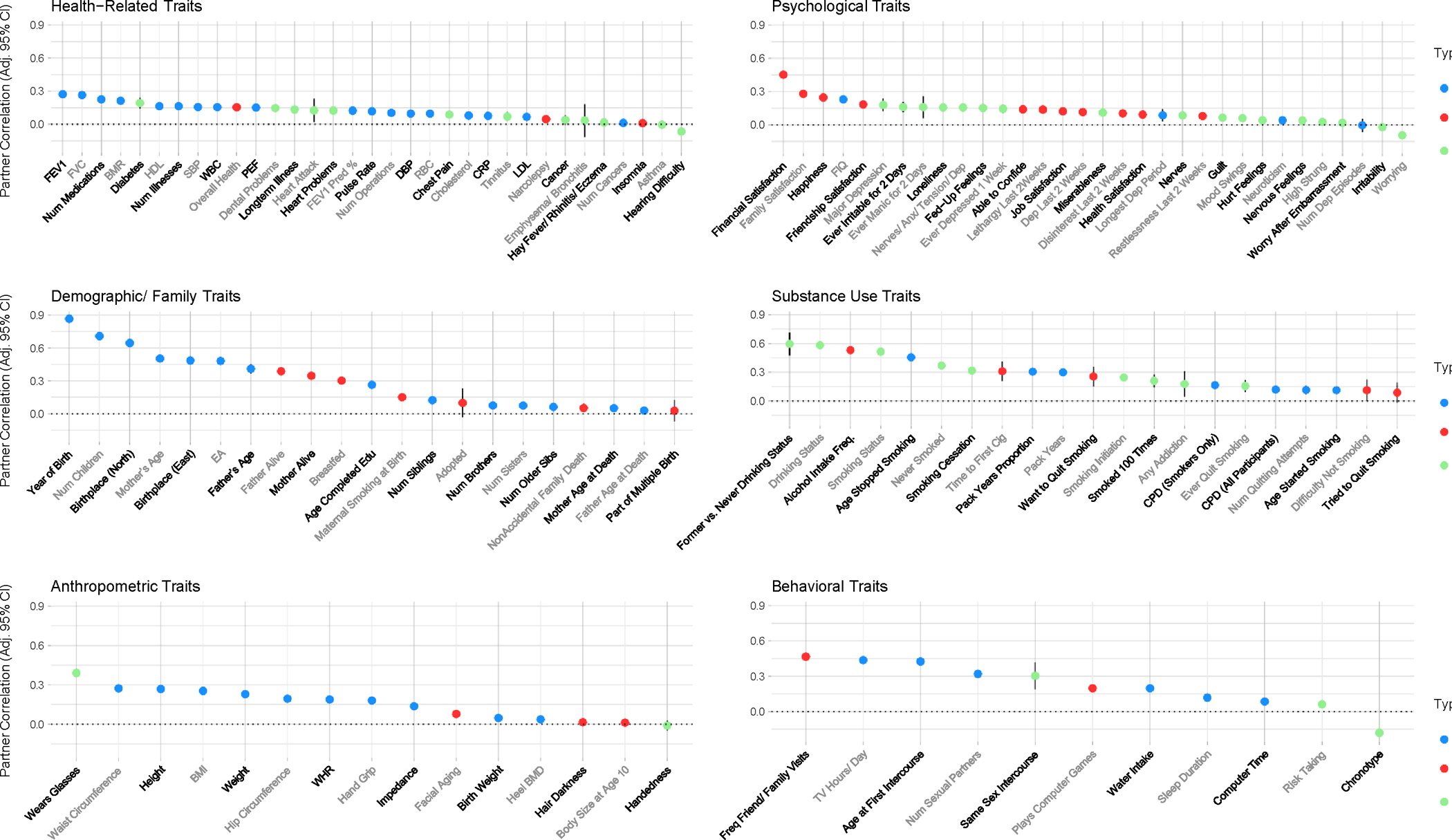
The point estimates on the y-axis represent the estimated partner correlation—along with Bonferroni-adjusted 95% confidence intervals (adjusting for 133 traits)--for the corresponding trait on the x-axis. Estimates are based on up to 79,074 pairs; Supplementary Table 4 includes the precise sample size/point estimate for each trait along with the Bonferroni-adjusted p-values associated with the adjusted 95% confidence intervals depicted in this figure. Traits are grouped according to six categories: health-related, psychological, demographic/family, substance use, anthropometric, and behavioral. Points representing partner correlations for continuous traits (Pearson correlations) are blue; points representing partner correlations for ordinally-coded traits (Spearman correlations) are red; points representing partner correlations for dichotomously-coded traits (Tetrachoric correlations) are light green. Num Dep Episodes = Number of Depressive Episodes; Heel BMD = Heel Bone Mineral Density (in the form of a t-score); LDL = Direct Low-density Lipoprotein Cholesterol, CRP = C-reactive Protein; RBC = Red Blood Cell (Erythrocyte) Count; DBP = Diastolic Blood Pressure; CPD (All Participants) = Cigarettes per Day (Includes Current, Former, and Never Smokers); FEV1 Pred % = Forced Expiratory Volume in 1-Second (FEV1), Predicted Percentage; PEF = Peak Expiratory Flow; WBC = White Blood Cell (Leukocyte) Count; SBP = Systolic Blood Pressure; HDL = High-density Lipoprotein Cholesterol; CPD (Smokers Only) = Cigarettes per Day (Restricted to Current or Former Smokers); WHR = Waist-to-hip Ratio; BMR = Basal Metabolic Rate; FIQ = Fluid Intelligence Quotient; BMI = Body Mass Index; FVC = Forced Vital Capacity; Time to First Cig = Time to First Cigarette; EA = Educational Attainment.
Twelve of the traits that we analyzed in the UKB were also meta-analyzed (see Fig. 1). Pearson and tetrachoric correlations for all but one (cigarettes per day, not restricted to ever-smokers) of these twelve overlapping UKB traits fell within the prediction intervals (distinct from the confidence intervals) of corresponding traits from our meta-analyses; additionally, the Spearman correlation for non-restricted cigarettes per day in the UKB, as well as both correlational effect sizes for cigarettes per day when restricted to ever-smokers, fell in the prediction interval for smoking quantity in the meta-analysis. Across both analyses, there were notable overall findings: for example, estimates of educational attainment and current smoking status ranked amongst the highest correlations while estimates of partner correlations for neuroticism were uniformly low across the two analyses. Additionally, correlational estimates for some traits were notably different across the meta-analysis and UKB, as evidenced by the non-overlapping confidence intervals for a minority of trait pairings in Fig. 1. Because overlapping confidence intervals for the remaining trait pairings do not necessarily indicate a lack of significant difference, we performed a paired test comparing the two groups of correlations across all twelve traits but found no significant difference: t(11) = −1.724, p =.113, mean difference = −.053, 95% CI:[−.121,.015]; V = 23, p = .233.
Next, we compared correlations for groups of traits within the UKB. We regressed the UKB partner correlation point estimates on the category of trait (a factor with six levels: anthropometric, health-related, psychological, behavioral, demographic/family, and substance use) for the 133 adequately-powered UKB traits, controlling for how the correlations were estimated (Pearson’s, Spearman’s, or Tetrachoric); we used weighted least squares for our estimation because the Levene homogeneity of variance test suggested violation of the equal variances assumption across trait categories when controlling for correlation type (F(5,127) = 9.75, p = 6e-8). There were significant differences between mean correlations depending on the category of trait (F(5,125) = 28.20, p < 2e-16, ηp2 = .106, 95% CI:[.08,.13]). In a follow-up analysis, we found that traits that were anthropometric (rUKB = .15, SE = .031), health-related (rUKB = .11, SE = .013), or psychological (rUKB = .12, SE = .018) yielded significantly lower partner correlations on average than traits related to behavior (rUKB = .22, SE = .059), demography/family (rUKB = .29, SE = .055), or substance use (rUKB = .29, SE = .036) (t(131) = 6.94, p=1.61e-10, Cohen’s d = .88, 95% CI:[.53, 1.20]). Controlling for category of trait, magnitude of partner correlations did not differ based on correlation type (F(2,125) = 2.38, p = .096, ηp2 = .086, 95% CI:[.06,.11]).
Several of the highest positive zero-order UKB partner correlations for demographic variables are unsurprising given that individuals tend to marry and enter into long-term relationships with others who are of similar ages and from nearby geographic locations (e.g., rUKB for north coordinate of birthplace = .65) and because romantic partners often share children with one another (rUKB for number of children = .71). More interestingly, partner correlations for several substance use traits were also very high. For adequately-powered substance use traits, we calculated the highest correlation for former vs. never drinking status (amongst only current non-drinkers, whether or not a person is a previous or never drinker: rUKB = .60), followed by drinking status (whether or not a person currently drinks alcohol: rUKB = .58). We evaluated smoking behavior with multiple measures and, of these, found the highest correlations for smoking status (rUKB = .51) and the age that individuals stopped smoking (restricted to former regular smokers; rUKB = .46) and the lowest correlation for presence and duration of quitting attempts (measured ordinally) in current regular smokers (rUKB = .09, padjusted = .569). While we were underpowered in the UKB to examine concordance for addictions to specific narcotic or prescribed substances, addiction to “any substance or behavior” showed a significant correlation (rUKB = .18, padjusted = .000199), suggesting a modest positive relationship between partners for general addiction.
As expected, educational attainment between partners in the UKB (measured as level of academic qualification/degree achieved) yielded a fairly large correlation (rUKB = .48). Fluid IQ score—evaluated using a brief thirteen-item test–showed a somewhat modest correlation, at rUKB = .23.
Consistent with the results from our meta-analyses, anthropometric, health-related, and psychological (i.e., personality and psychiatric) traits generally showed low-to-moderate correlations, with some of these traits showing no significant correlation between partners. Correlations for height, waist-to-hip ratio, and body mass index were rUKB = .27, rUKB = .19, and rUKB = .25, respectively. Partner correlation for overall neuroticism score was low but significant (rUKB = .04, padjusted < 2.66e-14), and partner correlations for the twelve dichotomous (yes/no) items comprising the neuroticism scale ranged from −.09 to .16 (see Supplementary Table 4). Furthermore, three dichotomous depression traits that we analyzed in the UKB all yielded modest correlations that were similar to one another: rUKB = .15-.18.
We additionally explored multiple traits that did not overlap with those we examined in our meta-analysis. The largest significant negative correlation across all UKB traits was for chronotype, which was dichotomized to indicate whether an individual is a “morning person” or an “evening person” (rUKB = −.18). Furthermore, we found moderate correlations for traits related to sexuality (age of first intercourse: rUKB = .43; number of lifetime sexual partners: rUKB = .32; ever had same-sex intercourse: rUKB = .31). Finally, general happiness (rUKB = .25) and measures of satisfaction with various life aspects (financial situation: rUKB = .45, family relationships: rUKB = .28, friendships: rUKB = .19, job rUKB = .12, and health rUKB = .09) showed small-to-moderate, positive partner correlations.
Discussion
In this study, we collated and synthesized results from a large number of studies on same-trait correlations in opposite-sex/gender couples, spousal pairs, and co-parents (collectively, “partners”) in order to provide a better understanding of the degree to which partners correlate across traits and the degree of between-sample heterogeneity in those correlations. We also analyzed partner correlation for 133 traits in putative partners from the UK Biobank (UKB).
We found widespread evidence of positive correlations across traits, with significant variation across traits with respect to degree of partner similarity. Across both the meta-analyses and UKB raw data analyses, correlations for educational attainment, substance use measures, attitudinal traits, and behavioral traits were often moderate and relatively higher, whereas correlations for anthropometric, psychological (i.e., psychiatric/personality), and health-related traits were typically low to moderate. In our meta-analysis, we calculated the highest mean meta-analyzed partner correlations for political and religious values, educational attainment, IQ score, and some substance use traits, although a few meta-analyses in the latter category produced nonsignificant estimates. Though estimates for other traits were smaller, 18 of 22 traits reached Bonferroni-level significance. Likewise, partner correlations for the large majority (88.72%) of traits we examined in the UKB reached Bonferroni-corrected significance. Overall, educational attainment and certain substance use traits also yielded amongst the highest correlations in the UKB. While point estimates for comparable traits across our two analyses were not significantly different overall, not every UKB estimate was identical to what we observed in the corresponding meta-analysis. This is not surprising given that our meta-analysis revealed strong evidence for between-sample heterogeneity (see below) and given that the UK Biobank likely differed in multiple ways from the typical sample included in our meta-analysis; for example, individuals in the UKB are of a relatively narrow age range, reside in the United Kingdom, and differ as a whole from the general population with respect to several demographic and lifestyle variables (as discussed below in the limitations).
In addition to finding widespread positive associations in mating pairs across traits, our meta-analysis found evidence of substantial within-trait heterogeneity across different samples. Because of this large degree of heterogeneity and because we performed random (rather than fixed) effects meta-analyses, the results presented here should not be interpreted as estimates of a single true partner correlation for a given trait, but rather as estimates of the typical level of partner similarity across many possible levels that might be observed in different populations. Our results suggest that much of the observed between-study and between-sample heterogeneity is due to true differences that exist across populations (cultures, time periods, etc.) from which our samples were drawn. This is sensible given that resemblance between partners can result from personal preferences, social stratification, and/or couple dynamics that are unlikely to be consistent across different cultural contexts for a given trait. However, owing to insufficient diversity in nationality (see limitations) within the samples we included in most of our meta-analyses and to the small number of samples in some of our meta-analyses, we were unable to meaningfully evaluate whether there were significant differences in partner correlations across even broad geographical subdivisions. We were also unable to formally assess changes in partner resemblance over time or control for age in the meta-analyses because publication year served as an inaccurate measure of the year/age at which partners entered into the relationship. Furthermore, although we excluded samples with known biologically related pairs from our meta-analysis, it is possible that varying degrees of unknown or unreported relatedness in couples (i.e., consanguinity) across samples could have contributed to heterogeneity.
In addition to the possible causes described above, the heterogeneity in effect size estimates may be partially due to differences in how constructs were assessed across studies and samples (e.g., differences in the measurement batteries used or differences in measure interpretation). Potentially consistent with this, we observed that the reported prevalence rates of dichotomous traits were highly heterogeneous across supposedly non-ascertained samples, which may have contributed to the heterogeneity we observed in our correlation coefficients (two identical odds ratios translate into different tetrachoric correlations if the prevalence rates in the samples differ). Nevertheless, we observed high levels of heterogeneity in correlations for traits—such as height and body mass index—that are not dichotomous and that are measured in standardized ways, suggesting that measurement invariance and ascertainment are unlikely to be complete explanations. More thoroughly examining sources of heterogeneity in partner correlations across samples remains an important direction for future studies.
There are several implications for the ubiquitous evidence of partner similarity we documented across traits in our meta-analysis. First, partner similarity due to phenotypic or genetic homogamy within a population can bias estimates of genetic correlations53, latent variances from twin/family studies21,47,54, Mendelian randomization48, and SNP-heritability46. To the degree that observed levels of partner similarity are due to these mating mechanisms, our results suggest that, for many traits, some degree of bias due to AM may exist in the aforementioned estimates and designs. Second, genetically informed designs that attempt to model AM often must assume that degree of partner resemblance has been stable across time and place; our observations of high between-sample heterogeneity calls this assumption into question. Such heterogeneity has complex implications with respect to various heritability estimates and dating and marriage trends, as well as long-term implications for genetic parameters such as stabilization of genotypic frequencies (i.e., genetic (dis)equilibrium). Circumventing the caveats resulting from such trends is increasingly possible in genetic designs; for example, the method structural equation modeling-polygenic score (SEM-PGS) can account for various modes of AM even if equilibrium has not been reached for the trait of interest55. Third, partner similarity has the capacity to increase prevalence of disorders and other dichotomous traits through increasing phenotypic variance via increase in environmental/genetic variance, though the precise consequences of partner similarity will depend on which mechanism(s) is/are in play, the magnitude of resemblance, and the heritability of the focal trait. There is evidence demonstrating that, although the resulting increase in prevalence for common traits may be weak, phenotypic AM for a rare, highly heritability trait could substantially increase the trait’s prevalence (i.e., 1.5-fold over one generation and 2.7-fold over ten generations)29. Finally, partner correlations have social and interpersonal implications that are relevant to outcomes of mating partners regardless of whether they procreate. For example, there is longitudinal evidence that smokers with a smoking partner are less likely to quit smoking51, while other findings suggest that being married is associated with reduction in drinking quantity among men but increased drinking quantity among women, with divorce showing the opposite trend56. Thus, knowledge of patterns in partner correlations offers an important understanding of the context shaping complex human traits and refines our interpretations of existing results.
While the current paper included many samples with varying characteristics, there are several further research topics related to human mating relationships that we did not directly assess. For example, past research has found that partner similarity for variables such as academic performance/aspirations, relationship involvement57, political/religious beliefs34, mental distress, exercise, drinking, and smoking58 may be predictive of relationship longevity. In the same vein, several papers have suggested that similarity on personality traits (e.g., happiness) predicts satisfaction and/or relationship longevity59–62, while others have found that partners’ individual characteristics (as opposed to their degree of similarity) are stronger predictors8,9,63,64. Finally, some research comparing partner similarity in male-female, female-female, and male-male couples has pointed to differences across groups for certain demographic, anthropometric, and health-related traits65–67. In light of these potential differences, researchers should further explore how sex/gender make-up interacts with partner resemblance in co-parents to affect rearing environments, as well as how sexual and romantic orientation may be associated with different patterns of similarity within partnerships. These avenues of inquiry have applications that extend beyond the downstream genetic and environmental consequences of AM among procreating pairs and may further help illuminate how social forces shape human relationships.
The current study had limitations that we were not able to address. First, the vast majority of samples in our meta-analyses were drawn from Europe and the United States, with a smaller number coming from East Asia; samples residing in Sub-Saharan Africa, South Asia, Latin America and the Caribbean, Southwest Asia and Northern Africa, and Oceania (the last of which is notably less populous than the other regions) were each present in only a handful of studies. Additionally, participants in the UKB were all between the ages of 40 and 69 when originally recruited, are disproportionately healthy, are less likely to live in socioeconomically deprived areas, and are less likely to smoke and drink on a daily basis. Typically, such restriction of range should diminish the magnitude of correlations towards zero, and the fact that both members of each putative UKB couple (rather than only one) came from this sample may have further exacerbated any effects of such ascertainment on partner similarity. Furthermore, a large majority of those in the UKB are of European ancestry, which is not reflective of the overall human population68; relatedly, we were not able to include cross-ancestry pairs in the UKB because of how relatedness was assessed in this sample (see Analysis of Partner Correlations in the UK Biobank), which may have had an effect on partner correlations for some traits. Additionally, despite comprising a non-negligible share of couples and parents throughout time, partners and co-parents who did not mutually select each other—either because of cultural norms around marriage or because of coercion—are unlikely to be proportionally reflected in recent volunteer samples. For the above reasons, findings from both our meta-analysis and partners in the UKB are unlikely to generalize to all human populations and time periods. Next, there was a broad range of total sample sizes used to calculate partner correlations across traits, suggesting some degree of imbalance in power across trait analyses. Finally, the results of the present study must be interpreted as cross-sectional estimates of resemblance within dyadic partnerships and so are not directly informative with respect to selection mechanisms, longitudinal trends across generations, or trends relating to partner similarity across multiple (concurrent or sequential) partnerships within a single individual. Future studies will be necessary to understand the specific processes underlying partner similarity (i.e., phenotypic/social/genetic homogamy and convergence), including how these processes have changed over time.
In summary, we have conducted a raw data analysis and set of meta-analyses of human partner correlations, with the former drawing from over 79,000 putative partner pairs and the latter being based on estimates from millions of partner pairs and over a century of research. Across both our meta-analyses and our study of partners in the UKB, we found the highest estimates of similarity for traits related to substance use, educational attainment, and attitudes/behaviors, with smaller correlations for personality, psychological, anthropometric, and disorder traits. We also observed high levels of heterogeneity in point estimates across studies for most traits investigated, suggesting that partner resemblance may differ across population. Research into the resemblance between partners and co-parents provides valuable information about the nature of relationships and cultural norms around human mating, as well as information pertinent to the etiological roots of disorders and to the interpretation of results from genetically informed designs.
Methods
Literature Search for the Meta-analysis
For the meta-analytic portion of this paper, we conducted a systematic review of Englishlanguage studies that examined concordance rates in partners for the same—or very similar—complex traits. All included studies were published in peer-reviewed journals on or before August 16, 2022, with no lower limit on when the study could be published, and with studies from books being excluded. The analysis was not preregistered and no protocol was prepared as this was not a hypothesis-driven study.
We selected an initial 26 traits on which to conduct our meta-analysis. While partner concordance has been analyzed for many traits, we focused on those most studied in the literature as well as some less commonly studied dichotomous traits. First, we searched for words pertaining to the traits of interest (with the exception of height; see below and Inclusion and Exclusion Criteria) in conjunction with thirteen terms pertaining to partner correlations and AM (see Supplementary Table 6 for exact search terms used) in the search engines PubMed (using the RISmed package69 in Rstudio Version 1.4.17.17) and ScienceDirect. Additionally, Google Scholar was used for the original submission of this manuscript. Though the latter search engine was not used for the most recent literature review, studies that we recorded in the first iteration of our study were preserved even if they did not appear in the corresponding PubMed or ScienceDirect search. We excluded educational attainment (EA) and body mass index (BMI) from our PubMed search because we had already obtained large sample sizes using only ScienceDirect (1,911,720 partner pairs from 86 samples for EA and 123,881 partner pairs from 57 samples for BMI).
Three authors then worked independently to evaluate whether each paper met the relevant criteria for meta-analysis and then manually collected data from relevant publications. One author then double-checked that each report determined to be applicable was indeed eligible for inclusion; next, all relevant findings from eligible reports were recorded in Supplementary Table 1 or 2 and included for meta-analysis. Supplementary Figs. 1a–1v display forest plots for every trait; each includes every correlation—as presented in Supplementary Table 1 or 2—associated with each included sample, with publications ordered by year and color-coded by geographic region(s) of the sample. Citations that were collated for assessment but that did not meet inclusion criteria are shown in Supplementary Table 3 (with the exception of a small number of studies corresponding to excluded traits, which were included in Supplementary Table 2; see Dichotomous traits), along with the reasons for each exclusion.
For each of the traits that we meta-analyzed, Supplementary Fig. 2 presents the number of results for each set of search terms and the number of studies included and excluded for each trait, grouped by reason(s) for exclusion, in accordance with PRISMA guidelines70,71.
Inclusion and Exclusion Criteria
Our analysis was restricted to studies of co-parents, engaged pairs, married pairs, and/or cohabitating pairs, with some studies containing a small number of divorced couples. We refer to the couples or co-parenting dyads as “partners” and refer to the female/woman vs. male/man designations of individuals as “sex/gender” unless referring to a sample that specifically uses one of these terms. Samples of only or predominantly same-sex/gender partners were also excluded (though a small fraction of some samples were likely comprised of such couples by chance); we believe these samples warrant separate analysis because same- and opposite-sex/gender partners have shown different patterns of similarity for some traits66,67 (see Discussion) and because there is less data on the former. For traits such as personality, we required that measures be self-reported, but we permitted case statuses of psychiatric traits reported by family members or partners due to the low numbers of applicable studies for such traits. Except for studies that ascertained partners for the trait of interest, we excluded studies in which partners were ascertained in a way that might have affected the magnitude of concordance for the trait of interest (e.g., concordance for depression in parents of depressed children). We limited our analysis to studies with sample sizes equal to or greater than 100 partner pairs. When sample sizes were reported along with percentages for each cell of a contingency table, we inferred the sample size of each cell by multiplying the percentages by n. When studies reported partner concordance at different waves over time in the same sample, we reported the results from the first wave. For studies conducted in or after 2000, if the n for a sample was not provided or inferable, or if we discovered an inconsistency/point of confusion, we contacted the corresponding author(s) for clarification. In total, we contacted authors from nine different studies and five responded. Although we initially gathered data on rare psychiatric disorders, we did not formally meta-analyze the tetrachoric correlations for these traits because too few studies met our inclusion criteria (see Supplementary Tables 2 and 3).
For consistency, when reported concordance rates were stratified by a variable (i.e., zygosity of a twin sample), we included results for each stratum as a separate entry. We calculated effect sizes from the data reported in primary studies rather than from other meta-analyses or reviews. However, because partner correlations for height have already been meta-analyzed extensively by Stulp et al. (2017)20, we analyzed only samples includes in the Stulp et al. paper for height in place of conducting a literature review for this trait, pooling results the same way that we did for other continuous traits after eliminating studies that met our exclusion criteria.
When we determined that samples for a given trait in multiple studies overlapped or were likely to overlap based on information provided in the publication, we only used the study that had the largest sample size or that we otherwise determined to be more appropriate. We also excluded samples taken from the UK Biobank (UKB) because we calculated partner correlations in this sample ourselves as a separate analysis. Finally, we restricted our meta-analysis to traits for which there were at least three samples that met our criteria.
For continuous and ordinal traits, we only included Pearson/interclass correlations and Spearman rank correlations. When a study presented cross-partner contingency tables for each level of an ordinal trait but a concordance statistic was not reported, we directly calculated the Spearman rank correlation and its corresponding standard error. Because most studies reported them, we analyzed raw correlations when studies reported both the raw correlation and the partial correlation(s) controlling for covariates. However, we included some correlations that controlled for age when the raw correlation was not available because age was a common covariate; additionally, we made occasional exceptions for types of adjustments (i.e. whether the participant was a twin) that attempted to correct for heterogeneity induced by the sampling or measurement scheme used in the study (see Supplementary Tables 1 and 2 for covariates associated with each sample). In every other case, we excluded studies that did not report raw partner correlations.
Dichotomous traits
Within the context of ascertained studies (i.e., those that used probands and controls rather than randomly sampling from a population), we excluded those involving probands taken from a clinical setting (i.e., a hospital or other treatment facility) when the case status of their partners was determined by a different set of criteria (i.e., achieving a certain score on a diagnostic measure but not necessarily having a history of treatment). Furthermore, results from simulation showed that mixing male and female probands with highly discrepant prevalence rates would lead to unacceptable levels of bias. As a result, ascertained studies that did not label discordant partners based on sex/gender and for which such information was not otherwise inferable were eliminated if there was a greater than ~two-fold difference in male and female prevalence rates. Because of possible differences in the strength of calculated concordance based on an all-male proband sample versus that based on an all-female proband sample, we excluded studies that only included single-sex/gender probands. However, when data were available for both a female proband and a male proband (only a single study72), estimates based on both probands (female and male) were included as separate results.
We also restricted our meta-analysis to studies with expected contingency table cell frequencies of five or greater for all cells and observed cell frequencies of greater than zero. Four of the traits in our supplementary tables posed a problem because they are rare (bipolar disorder and schizophrenia) or have not been studied in enough sufficiently large samples (panic disorder and phobia), resulting in under-powered contingency tables in many instances. As a result, there were not enough studies meeting our inclusion criteria to justify formally meta-analyzing these four traits, though we show results from studies on these traits that mostly met our criteria in Supplementary Table 2.
We required that either odds ratios (OR), risk ratios (RR), phi coefficients (Φ), contingency tables (from which an OR/RR and tetrachoric correlation can be calculated), or—if the study was not ascertained (see Effect Size Conversion)–tetrachoric correlations, be reported for dichotomous traits because these effect sizes can be pooled together. No studies that met our inclusion criteria only reported RR and so we do not discuss them further. All effect sizes for dichotomous traits that were not already in the form of a tetrachoric correlation were then converted to a tetrachoric correlation (see Effect Size Conversion).
Effect Size Conversion
To make estimates across continuous and dichotomous traits more comparable, we converted all effect sizes for studies examining dichotomous traits to tetrachoric correlations if the study was not ascertained and did not report one. For non-ascertained studies that reported contingency tables, we simply calculated the corresponding tetrachoric correlation using the polychor() function from the “polycor” package73 in Rstudio Version 1.4.17.17.
If the contingency table was unknown but the OR was reported, we first inferred the contingency table using an R function described in the supplementary methods of Peyrot et al. (2016)29 (which the authors provided to us) and then used the polychor() function to convert it to a tetrachoric correlation. If the values of the contingency table cells were unknown but the Φ was available, we used the phi2tetra() function from the “psych” package74 in Rstudio to convert it to a tetrachoric correlation. Both conversions required inputting male and female prevalence rates, which we based on published male and female prevalence rates for the trait or—if provided or inferable in the study and the sample was not ascertained—the prevalence rates indicated in the study. When gathering prevalence rates from outside sources, we selected those reporting on samples that matched the partner correlation sample as closely as possible with respect to nationality, year, etc. Sample/population prevalence rates for dichotomous traits are reported in Supplementary Table 2 for each applicable meta-analyzed sample.
For ascertained studies, it was necessary to use the aforementioned R function from Peyrot et al. (2016)29—regardless of whether the contingency table was reported–to produce an expected population (non-ascertained) contingency table based on the OR and the population prevalence, prior to converting to a tetrachoric correlation (as described above). This correction is necessary because the case-to-control ratio in an ascertained sample is usually higher than in the general population, and so simply deriving the correlation based on the uncorrected contingency table would lead to a biased estimate.
Meta-analytic Method
We conducted two- or three-level (see below) random effects meta-analyses based on Fisher Z-transformed correlations and their respective variances; we reversed this transformation when reporting final results (e.g., rmeta) for ease of interpretation. Note that evaluating within-sample/study normality was not of primary concern in our meta-analysis because of the large minimum sample size we required (N = 100 pairs; see Inclusion and Exclusion Criteria); additionally, any between-sample/study non-normality was unlikely to drastically affect point estimates75.
We used two-level meta-analysis when none of the publications for a given trait reported multiple effect sizes and used three-level meta-analysis when at least one publication for a given trait reported multiple effect sizes. Two-level random effects meta-analyses allow for the estimation of heterogeneity due to sampling variance versus that due to true effect size differences between populations from which the samples were drawn. Three-level meta-analyses further break down the true effect size differences to capture heterogeneity resulting from variation between studies versus heterogeneity resulting from variation within studies52. Partner correlations estimated in the same study could be more similar due to, for example, measures being taken at the same time or in the same culture or due to the use of common instruments, even if the samples in a study were collected independently.
We utilized restricted maximum likelihood (REML) for both two-level and three-level meta-analyses. For the two-level meta-analyses, we used the rma.uni() function in the “metafor” package76 in Rstudio Version 1.4.17.17 to specify two levels (i.e., standard random effects modeling). For three-level analyses, we used the rma.mv() function, within which we set individual effect sizes (the Z-transformed partner correlations in different samples) to be nested within individual studies for the “random” argument. For both two-level and three-level meta-analyses, we set the test of the coefficient to follow a t-distribution and report two-tailed p-values that were Bonferroni-adjusted for multiple testing, as well as 95% confidence intervals that were Bonferroni-adjusted for multiple testing.
For continuous traits, we specified the effect sizes as Fisher Z-transformed Pearson or r2 Spearman correlations and specified variances as and respectively77. For dichotomous traits, we converted the ascertained contingency table to a tetrachoric correlation corrected for ascertainment, as described above, and used the Fisher Z-transformed tetrachoric correlations for pooling in the meta-analyses. We used polychor() to calculate the untransformed standard error (seut) for non-ascertained studies examining dichotomous traits. For ascertained studies examining dichotomous traits, we estimated seut by creating 1,000 bootstrapped contingency tables, each of size n (the number of partner pairs) and sampling from the study’s (raw, ascertained) contingency table, with replacement, and then estimating the standard deviation of the 1,000 bootstrapped tetrachoric correlations arising from the contingency tables. We then transformed all seut’s for tetrachoric correlations using the delta method78, wherein the standard error of the Z-transformed tetrachoric correlation is equivalent to , and used the square of these transformed standard errors for the variance argument in the rma.uni/rma.mv function. For reporting purposes, all meta-analyzed point estimates were back-transformed to their original format (Pearson’s correlation or tetrachoric correlation), which were recorded alongside their respective Bonferroni-adjusted 95% confidence intervals.
We estimated two sets of heterogeneity parameters, τ and I2, at the between-study level and—when applicable—within-study level. τ estimates the standard deviation of the true effect size at a given level. The total Higgins & Thompson’s I2 represents the percentage/proportion of variance not due to sampling error and is the sum of I2 at the between-study level and I2 at the within-study level. The heterogeneity statistics are detailed in Table 1. We created funnel plots (Extended Data Figs. 1a–1v)—which plot Fisher Z-transformed correlations on the x-axis against their standard errors on the y-axis—to visually inspect whether there was evidence for asymmetry, a potential indicator of publication bias.
Analysis of Partner Correlations in the UK Biobank
In addition to our meta-analyses, we analyzed correlations between inferred male-female partners in the UK Biobank (UKB; Application Number 16651) for an initial 140 ordinal, dichotomous, and continuous traits using R Version 4.2.2 and the University of Colorado Boulder computing cluster resources. We detected putative partners using a similar approach to past studies of partner correlations/AM in the UKB44,53. Beginning with 502,414 individuals, we first restricted our sample to individuals who reported living with a “Husband, wife, or partner” and who did not report living with an unrelated roommate (based on data-field 6141), leaving 359,189 individuals. Using a co-location variable that was previously provided by the UKB, we subsequently limited our sample to pairs of individuals who were living at the same address at the time of recruitment, which narrowed the pool down to 200,707 participants. Though the co-location variable is no longer available from the UKB, the code used for detecting putative mate pairs and conducting the UKB analyses is available at https://github.com/JaredBalbona/UKB-AM-MetaAnalysis79. Afterward, using a series of genomic relationship matrices (GRMs), we removed all pairs of related individuals (p ≥ .05), leaving 175,250 individuals; importantly, because GRMs were calculated within ancestry groups, all cross-ancestry pairs were also removed at this stage. We then kept only pairs who were concordant for the number of people in their household, whether they own or rent their property, and their Townsend Deprivation Index (based on data-fields 709, 680, and 189, respectively), leaving 160,738 participants. Same-sex partners (data-field 31) were then removed, leaving 159,998 participants. Because most discrepancies in self-reported household income (data-field 738) were due either to the pairs choosing two adjacent options (e.g., one individual choosing “18,000 to 30,999” and the other “31,000 to 51,999”) or to non-responses, we removed only pairs whose reported household incomes differed by more than one category, leaving 158,350 individuals. Finally, pairs with age differences of greater than twenty years were removed to minimize the possibility of including stepchild/stepparent pairs, leaving a final sample of 158,148 individuals comprising 79,074 putative mating pairs.
In some cases, we slightly altered the nature of traits by combining information from multiple traits or changing trait coding schemes (see Supplementary Table 4 for traits that were altered and the UKB descriptions of each trait we included). In line with our meta-analytic methods, we used responses from the initial wave when responses were collected at multiple times for the same trait. For each continuous trait, we eliminated partners containing at least one member with an absolute z-score greater than four to reduce the number of invalid responses and then calculated both a Pearson correlation and a Spearman rank correlation to provide two different estimates in the event that unusually-distributed data biased correlations. For zero-inflated data, both Pearson and Spearman rank correlations provide a slight underestimate of the true underlying correlation of two continuous measures where values below some threshold provide an observation of zero, but Spearman correlations tend to be less biased80. For dichotomous traits, we calculated tetrachoric correlations and odds ratios, and for ordinal traits, we calculated only Spearman rank correlations. In addition to the zero-order correlations for UKB partners, we also present partial correlations in Supplementary Table 4 that control for year of birth, whether an individual was born in the British Isles, and each individual’s first ten ancestral principal components. To avoid the re-introduction of covariate effects81, we ranked our ordinal data prior to residualizing. Out of the initial 140 traits, six dichotomous traits did not have expected cell frequencies of five or greater, and one continuous trait was under-powered because of low sample size. These traits were therefore included in Supplementary Table 4 but not in Fig. 2 or Extended Data Fig. 2, where we visualize the point estimates and adjusted confidence intervals for the remaining 133 traits.
Notably, our subsample contains substantially more partners than certain previous research that used inferred partners in the UKB; for example, despite using similar procedures and criteria to detect partners, Yengo et al. (2018)44 detected only 18,984 pairs within the UKB. The discrepancy between these two sample sizes can be attributed to several causes, including our sample containing partners of non-European descent, our approach not requiring perfect matches on household income, and our use of the co-location variable. Additionally, Yengo et al. required that partners be concordant for the number of smokers in their household—a question only asked of non-smokers in the UKB, thus eliminating all pairs that contain one or more current smokers. To evaluate the consequences of our criteria discrepancies, we compared our estimates to those reported in Yengo et al. using an independent samples t-test for all continuous traits that we examined. For each of these traits, we first calculated r1’, the Fisher-transformed partner correlation for n1 available pairs from Yengo et al.’s UKB dataset, and r2’, the Fisher-transformed partner correlation for the same trait in the n2 available pairs unique to our dataset; we then divided the difference between r1’ and r2’ by , the square-root of the sum of their estimated variances. As shown in Supplementary Table 5, the subsamples do not differ substantially for the majority of continuous traits analyzed in both studies, despite Yengo et al.’s more conservative partner detection criteria.
Extended Data
Extended Data Fig. 1. Funnel plots for each meta-analyzed trait.
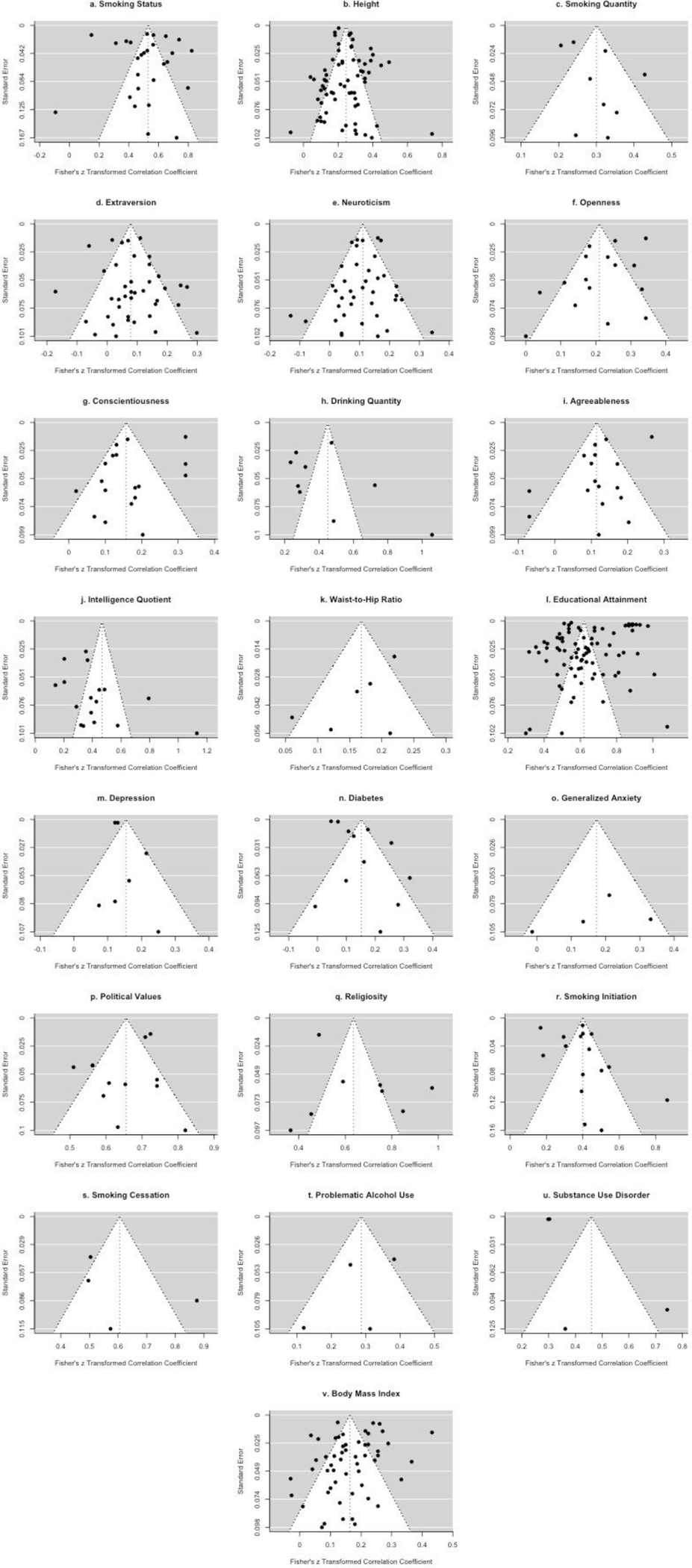
The funnel plots in Extended Data Fig. 1a Smoking Status, Extended Data Fig. 1b Height, Extended Data Fig. 1c Smoking Quantity, Extended Data Fig. 1d Extraversion, Extended Data Fig. 1e Neuroticism, Extended Data Fig. 1f Openness, Extended Data Fig. 1g Conscientiousness, Extended Data Fig. 1h Drinking Quantity, Extended Data Fig. 1i Agreeableness, Extended Data Fig. 1j Intelligence Quotient, Extended Data Fig. 1k Waist-to-Hip Ratio, Extended Data Fig. 1l Educational Attainment, Extended Data Fig. 1m Depression, Extended Data Fig. 1n Diabetes, Extended Data Fig. 1o Generalized Anxiety, Extended Data Fig. 1p Political Values, Extended Data Fig. 1q Religiosity, Extended Data Fig. 1r Smoking Initiation, Extended Data Fig. 1s Smoking Cessation, Extended Data Fig. 1t Problematic Alcohol Use, Extended Data Fig. 1u Substance Use Disorder, and Extended Data Fig. 1v Body Mass Index are designed to assess possible publication bias for each meta-analysis. Here, the Fisher Z-transformed correlations are plotted against their respective standard errors. For dichotomous traits, standard error was estimated using the delta method (see main text).
Extended Data Fig. 2. Partner correlations and Bonferroni-Adjusted 95% confidence intervals for 133 traits in the UK Biobank.
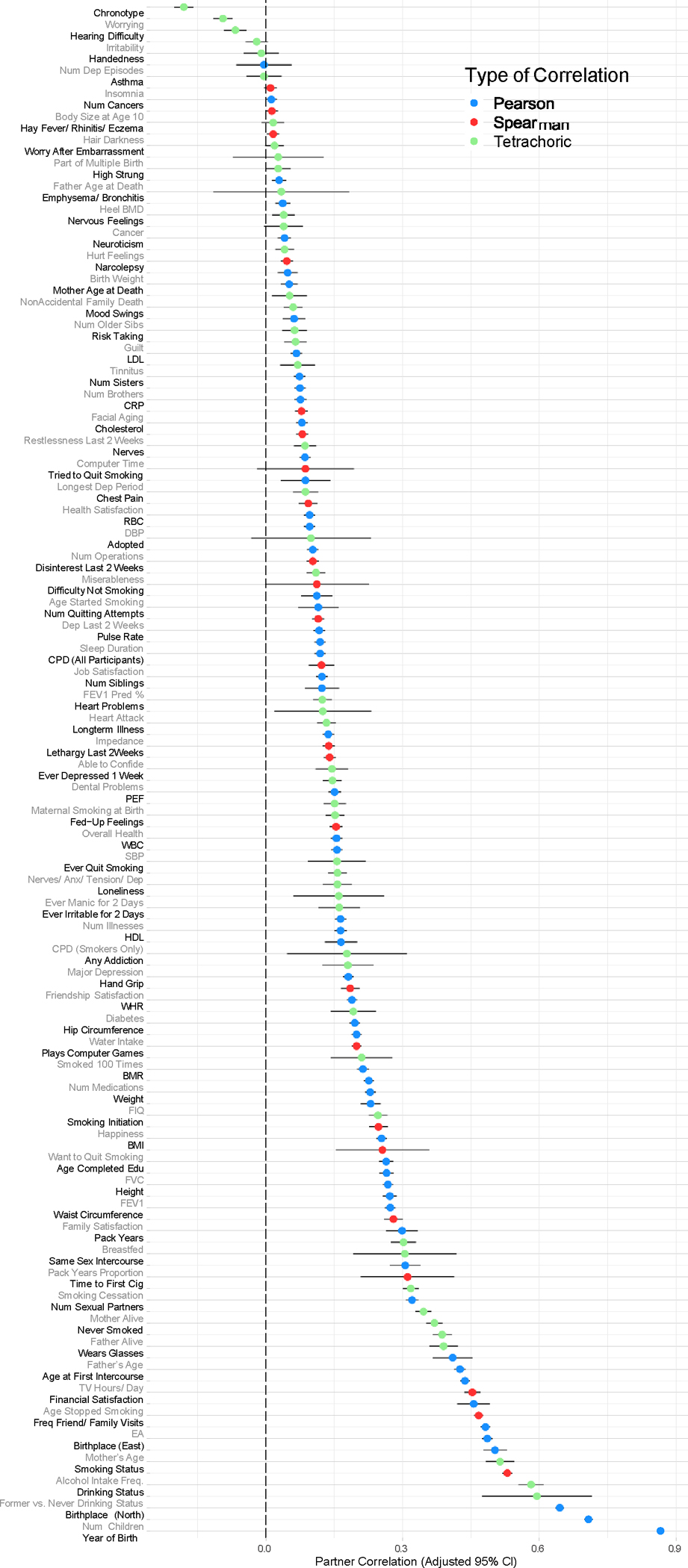
The visualized traits represent partner correlations for all of the adequately-powered UK Biobank traits (out of an original 140). Each estimate is color-coded by the correlation type— Pearson (in blue), Spearman (in red), and Tetrachoric (in green), used for continuous, ordinal, and binary traits, respectively—with the lines depicting the Bonferroni-adjusted 95% confidence interval for each trait. Estimates are based on up to 79,074 pairs; Supplementary Table 4 includes the precise sample size for each trait along with the Bonferroni-adjusted p-values associated with the adjusted 95% confidence intervals depicted in this figure. See main text for description of specific analyses. Num Dep Episodes = Number of Depressive Episodes; Heel BMD = Heel Bone Mineral Density (in the form of a t-score); LDL = Direct Low-density Lipoprotein Cholesterol, CRP = C-reactive Protein; RBC = Red Blood Cell (Erythrocyte) Count; DBP = Diastolic Blood Pressure; CPD (All Participants) = Cigarettes per Day (Includes Current, Former, and Never Smokers); FEV1 Pred % = Forced Expiratory Volume in 1-Second (FEV1), Predicted Percentage; PEF= Peak Expiratory Flow; WBC = White Blood Cell (Leukocyte) Count; SBP = Systolic Blood Pressure; HDL = High-density Lipoprotein Cholesterol; CPD (Smokers Only) = Cigarettes per Day (Restricted to Current or Former Smokers); WHR = Waist-to-hip Ratio; BMR = Basal Metabolic Rate; FIQ = Fluid Intelligence Quotient; BMI = Body Mass Index; FVC = Forced Vital Capacity; Time to First Cig = Time to First Cigarette; EA = Educational Attainment.
Supplementary Material
Acknowledgements
We thank Katerina Zorina-Lichtenwalter for help with ideas and feedback as the project developed. This work was supported by the National Institute of Mental Health R01 Grants MH130448 (MCK; JVB) and MH100141 (MCK), T32 Training Grant MH016880 (KNP), as well as by the National Institute on Drug Abuse T32 Training Grant T32DA017637 (TBH); this work also used resources from the University of Colorado Boulder Research Computing Group, which is supported by the National Science Foundation (Award Nos. ACI-1532235 and ACI-1532236), the University of Colorado Boulder, and Colorado State University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Code Availability
The code for the UKB analyses can be found at: https://github.com/JaredBalbona/UKB-AM-MetaAnalysis79.
Peer review information:
Nature Human Behaviour thanks Laurence Howe, Fartein Torvik, and Yayouk Willems for their contribution to the peer review of this work.
Data Availability
Studies included in the meta-analysis are listed in Supplementary Tables 1 and 2 and cited in Table 1, as well as in the supplementary note; studies excluded from the meta-analysis are listed in Supplementary Table 3. Raw data from the UK Biobank is not publicly available, but summary statistics for most traits are available on the UK Biobank website: https://biobank.ndph.ox.ac.uk/showcase/search.cgi. Note that there is no longer a Field ID corresponding to the co-location variable in the UKB and that the putative partner dataset we created cannot be publicly shared. As such, our UKB partner dataset cannot be directly used/recreated at this time. However, combinations of other variables (e.g., inverse distance to the nearest major road) can potentially be used as proxies for co-location53—in conjunction with the code we have made available—to estimate partner pairs.
References
- 1.Fisher RA The Correlation between Relatives on the Supposition of Mendelian Inheritance. Earth Environ. Sci. Trans. R. Soc. Edinb. 52, 399–433 (1919). [Google Scholar]
- 2.Kong A et al. The nature of nurture: Effects of parental genotypes. Science 359, 424–428 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Eaves L The use of twins in the analysis of assortative mating. Heredity 43, 399–409 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Border R et al. Assortative mating biases marker-based heritability estimators. Nat. Commun. 13, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavryashina MB et al. Interethnic Dynamics among the Indigenous Peoples of Southern Siberia (Demographic Aspect)*. Archaeol. Ethnol. Anthropol. Eurasia 41, 131–142 (2013). [Google Scholar]
- 6.Blundell R, Joyce R, Norris Keiller A & Ziliak JP Income inequality and the labour market in Britain and the US. Honor Sir Tony Atkinson 1944–2017 162, 48–62 (2018). [Google Scholar]
- 7.Schwartz CR Trends and Variation in Assortative Mating: Causes and Consequences. Annu. Rev. Sociol 39, 451–470 (2013). [Google Scholar]
- 8.Luo S Assortative mating and couple similarity: patterns, mechanisms, and consequences. Soc. Personal. Psychol. Compass 11, e12337 (2017). [Google Scholar]
- 9.Watson D et al. Match makers and deal breakers: analyses of assortative mating in newlywed couples. J. Pers. 72, 1029–1068 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Vandermeer MRJ, Kotelnikova Y, Simms LJ & Hayden EP Spousal Agreement on Partner Personality Ratings is Moderated by Relationship Satisfaction. J. Res. Personal. 76, 22–31 (2018). [Google Scholar]
- 11.Glicksohn J & Golan H Personality, cognitive style and assortative mating. Personal. Individ. Differ. 30, 1199–1209 (2001). [Google Scholar]
- 12.Dufouil C & Alpérovitch A Couple similarities for cognitive functions and psychological health. J. Clin. Epidemiol. 53, 589–593 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Feng D & Baker L Spouse similarity in attitudes, personality, and psychological well-being. Behav. Genet. 24, 357–364 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Burgess EW & Wallin P Homogamy in social characteristics. Am. J. Sociol. 49, 109–124 (1943). [Google Scholar]
- 15.Alford JR, Hatemi PK, Hibbing JR, Martin NG & Eaves LJ The politics of mate choice. J. Polit. 73, 362–379 (2011). [Google Scholar]
- 16.Jurj AL et al. Spousal correlations for lifestyle factors and selected diseases in Chinese couples. Ann. Epidemiol. 16, 285–291 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Ask H, Rognmo K, Torvik FA, Røysamb E & Tambs K Non-random mating and convergence over time for alcohol consumption, smoking, and exercise: the Nord-Trøndelag Health Study. Behav. Genet. 42, 354–365 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Rawlik K, Canela-Xandri O & Tenesa A Indirect assortative mating for human disease and longevity. Heredity 123, 106–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price RA & Vandenberg SG Spouse similarity in American and Swedish couples. Behav. Genet. 10, 59–71 (1980). [DOI] [PubMed] [Google Scholar]
- 20.Stulp G, Simons MJP, Grasman S & Pollet TV Assortative mating for human height: a meta-analysis. Am. J. Hum. Biol. 29, e22917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allison DB et al. Assortative mating for relative weight: genetic implications. Behav. Genet. 26, 103–111 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Mascie-Taylor CGN Spouse similarity for IQ and personality and convergence. Behav. Genet. 19, 223–227 (1989). [DOI] [PubMed] [Google Scholar]
- 23.Meyler D, Stimpson JP & Peek MK Health concordance within couples: a systematic review. Soc. Sci. Med. 64, 2297–2310 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Stimpson JP & Peek MK Concordance of chronic conditions in older Mexican American couples. Prev. Chronic. Dis 2, (2005). [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong S & Cho S-I Concordance in the health behaviors of couples by age: a cross-sectional study. J. Prev. Med. Pub. Health 51, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Castelnuovo A, Quacquaruccio G, Donati MB, de Gaetano G & Iacoviello L Spousal concordance for major coronary risk factors: a systematic review and meta-analysis. Am. J. Epidemiol. 169, 1–8 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Hippisley-Cox J Married couples’ risk of same disease: cross sectional study. BMJ 325, 636–636 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galbaud Du Fort G, Bland RC, Newman SC & Boothroyd LJ Spouse similarity for lifetime psychiatric history in the general population. Psychol. Med. 28, 789–802 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Peyrot WJ, Robinson MR, Penninx BWJH & Wray NR Exploring boundaries for the genetic consequences of assortative mating for psychiatric traits. JAMA Psychiatry 73, 1189–1195 (2016). [DOI] [PubMed] [Google Scholar]
- 30.McLeod JD Social and psychological bases of homogamy for common psychiatric disorders. J. Marriage Fam 201–214 (1995). [Google Scholar]
- 31.Maes HH et al. Assortative mating for major psychiatric diagnoses in two population-based samples. Psychol. Med. 28, 1389–1401 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Mathews CA & Reus VI Assortative mating in the affective disorders: a systematic review and meta-analysis. Compr. Psychiatry 42, 257–262 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y, Bolnick DI & Kirkpatrick M Assortative mating in animals. Am. Nat. 181, E125–E138 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Botwin MD, Buss DM & Shackelford TK Personality and mate preferences: five factors in mate selection and marital satisfaction. J. Pers 65, 107–136 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Eysenck HJ & Wakefield JA Psychological factors as predictors of marital satisfaction. Adv. Behav. Res. Ther. 3, 151–192 (1981). [Google Scholar]
- 36.McCrae RR et al. Personality trait similarity between spouses in four cultures. J. Pers. 76, 1137–1164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markey PM & Markey CN Romantic ideals, romantic obtainment, and relationship experiences: the complementarity of interpersonal traits among romantic partners. J. Soc. Pers. Relatsh. 24, 517–533 (2007). [Google Scholar]
- 38.Stimpson JP, Masel MC, Rudkin L & Peek MK Shared health behaviors among older Mexican American spouses. Am. J. Health Behav. 30, 495–502 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Al-Sharbatti SS, Abed YI, Al-Heety LM & Basha SA Spousal concordance of diabetes mellitus among women in Ajman, United Arab Emirates. Sultan Qaboos Univ. Med. J 16, e197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ober C et al. HLA and mate choice in humans. Am. J. Hum. Genet. 61, 497–504 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller MC, Medland SE & Duncan LE Are extended twin family designs worth the trouble? A comparison of the bias, precision, and accuracy of parameters estimated in four twin family models. Behav. Genet. 40, 377–393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cloninger CR Interpretation of intrinsic and extrinsic structural relations by path analysis: theory and applications to assortative mating. Genet. Res. 36, 133–145 (1980). [Google Scholar]
- 43.Robinson MR et al. Genetic evidence of assortative mating in humans. Nat. Hum. Behav. 1, 1–13 (2017). [Google Scholar]
- 44.Yengo L et al. Imprint of assortative mating on the human genome. Nat. Hum. Behav. 2, 948–954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JJ et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Border R et al. Assortative mating biases marker-based heritability estimators. Nat. Commun. 13, 660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coventry WL & Keller MC Estimating the extent of parameter bias in the classical twin design: a comparison of parameter estimates from extended twin-family and classical twin designs. Twin Res. Hum. Genet. 8, 214–223 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Hartwig FP, Davies NM & Davey Smith G Bias in Mendelian randomization due to assortative mating. Genet. Epidemiol. 42, 608–620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Castelnuovo A et al. Cardiovascular risk factors and global risk of fatal cardiovascular disease are positively correlated between partners of 802 married couples from different European countries. Thromb. Haemost. 98, 648–655 (2007). [PubMed] [Google Scholar]
- 50.Knuiman MW, Divitini ML, Bartholomew HC & Welborn TA Spouse correlations in cardiovascular risk factors and the effect of marriage duration. Am. J. Epidemiol. 143, 48–53 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Cobb LK et al. The association of spousal smoking status with the ability to quit smoking: the Atherosclerosis Risk in Communities Study. Am. J. Epidemiol. 179, 1182–1187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrer M, Cuijpers P, Furukawa TA & Ebert DD Doing Meta-Analysis in R. (2019).
- 53.Border R et al. Cross-trait assortative mating is widespread and inflates genetic correlation estimates. Science 378, 754–761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cloninger CR, Rice J & Reich T Multifactorial inheritance with cultural transmission and assortative mating. II. a general model of combined polygenic and cultural inheritance. Am. J. Hum. Genet. 31, 176–198 (1979). [PMC free article] [PubMed] [Google Scholar]
- 55.Balbona JV, Kim Y & Keller MC The estimation of environmental and genetic parental influences. Dev. Psychopathol. 34, 1876–1886 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reczek C, Pudrovska T, Carr D, Thomeer MB & Umberson D Marital histories and heavy alcohol use among older adults. J. Health Soc. Behav. 57, 77–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill CT, Rubin Z & Peplau LA Breakups before marriage: the end of 103 affairs. J. Soc. Issues 32, 147–168 (1976). [Google Scholar]
- 58.Torvik FA, Gustavson K, Røysamb E & Tambs K Health, health behaviors, and health dissimilarities predict divorce: results from the HUNT study. BMC Psychol. 3, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keller MC, Thiessen D & Young RK Mate assortment in dating and married couples. Personal. Individ. Differ. 21, 217–221 (1996). [Google Scholar]
- 60.Olderbak S & Figueredo AJ Shared life history strategy as a strong predictor of romantic relationship satisfaction. J. Soc. Evol. Cult. Psychol. 6, 111 (2012). [Google Scholar]
- 61.Luo S & Klohnen EC Assortative Mating and Marital Quality in Newlyweds: A Couple-Centered Approach. J. Pers. Soc. Psychol 88, 304–326 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Nemechek S & Olson KR Five-factor personality similarity and marital adjustment. Soc. Behav. Personal. Int. J. 27, 309–318 (1999). [Google Scholar]
- 63.Dyrenforth PS, Kashy DA, Donnellan MB & Lucas RE Predicting relationship and life satisfaction from personality in nationally representative samples from three countries: The relative importance of actor, partner, and similarity effects. J. Pers. Soc. Psychol. 99, 690–702 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Arránz Becker O Effects of similarity of life goals, values, and personality on relationship satisfaction and stability: Findings from a two-wave panel study. Pers. Relatsh 20, 443–461 (2013). [Google Scholar]
- 65.Holway GV, Umberson D & Donnelly R Health and health behavior concordance between spouses in same-sex and different-sex marriages. Soc. Curr 5, 319–327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwartz CR & Graf NL Assortative matching among same-sex and different-sex couples in the United States, 1990–2000. Demogr. Res. 21, 843 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Verbakel E & Kalmijn M Assortative mating among Dutch married and cohabiting same-sex and different-sex couples. J. Marriage Fam. 76, 1–12 (2014). [Google Scholar]
- 68.Fry A et al. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am. J. Epidemiol. 186, 1026–1034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kovalchik S RISmed: Download Content from NCBI Databases. (2021).
- 70.Page MJ et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ n71 (2021) doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Page MJ et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ n160 (2021) doi: 10.1136/bmj.n160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nordsletten AE et al. Patterns of nonrandom mating within and across 11 major psychiatric disorders. JAMA Psychiatry 73, 354–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.polycor: Polychoric and Polyserial Correlations version 0.8–1 from R-Forge. https://rdrr.io/rforge/polycor/.
- 74.Revelle W psych: Procedures for Psychological, Psychometric, and Personality Research. (2021).
- 75.Liu Z et al. The normality assumption on between-study random effects was questionable in a considerable number of Cochrane meta-analyses. BMC Med. 21, 112 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viechtbauer W metafor: Meta-Analysis Package for R. (2022).
- 77.Bonett DG & Wright TA Sample size requirements for estimating pearson, kendall and spearman correlations. Psychometrika 65, 23–28 (2000). [Google Scholar]
- 78.Ver Hoef JM Who Invented the Delta Method? Am. Stat 66, 124–127 (2012). [Google Scholar]
- 79.Balbona J JaredBalbona/UKB-AM-MetaAnalysis. (2023).
- 80.Huson LW Performance of Some Correlation Coefficients When Applied to Zero-Clustered Data. J. Mod. Appl. Stat. Methods 6, 530–536 (2007). [Google Scholar]
- 81.Pain O, Dudbridge F & Ronald A Are your covariates under control? How normalization can re-introduce covariate effects. Eur. J. Hum. Genet. 26, 1194–1201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Retnakaran R et al. Spousal concordance of cardiovascular risk factors in newly married couples in china. JAMA Netw. Open 4, e2140578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blackaby DH, Carlin PS & Murphy PD A change in the earnings penalty for British men with working wives: Evidence from the 1980’s and 1990’s. Labour Econ. 14, 119–134 (2007). [Google Scholar]
- 84.Neidhöfer G, Serrano J & Gasparini L Educational inequality and intergenerational mobility in Latin America: A new database. J. Dev. Econ. 134, 329–349 (2018). [Google Scholar]
- 85.Dalmia S & Lawrence PG An empirical analysis of assortative mating in India and the US. Int. Adv. Econ. Res. 7, 443–458 (2001). [Google Scholar]
- 86.Kardum I, Hudek-Knezevic J & Mehic N Similarity indices of the Dark Triad traits based on self and partner-reports: Evidence from variable-centered and couple-centered approaches. Personal. Individ. Differ. 193, 111626 (2022). [Google Scholar]
- 87.Zietsch BP, Verweij KJ, Heath AC & Martin NG Variation in human mate choice: simultaneously investigating heritability, parental influence, sexual imprinting, and assortative mating. Am. Nat. 177, 605–616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dufouil C & Alpérovitch A Couple similarities for cognitive functions and psychological health. J. Clin. Epidemiol. 53, 589–593 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Luo S & Klohnen EC Assortative Mating and Marital Quality in Newlyweds: A Couple-Centered Approach. J. Pers. Soc. Psychol 88, 304–326 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Watkins MP & Meredith W Spouse similarity in newlyweds with respect to specific cognitive abilities, socioeconomic status, and education. Behav. Genet. 11, 1–21 (1981). [DOI] [PubMed] [Google Scholar]
- 91.Hugh-Jones D, Verweij KJH, St. Pourcain B & Abdellaoui A Assortative mating on educational attainment leads to genetic spousal resemblance for polygenic scores. Intelligence 59, 103–108 (2016). [Google Scholar]
- 92.Gayle G-L, Golan L & Soytas MA What is the source of the intergenerational correlation in earnings? J. Monet. Econ 129, 24–45 (2022). [Google Scholar]
- 93.Barban N, De Cao E, Oreffice S & Quintana-Domeque C The effect of education on spousal education: A genetic approach. Labour Econ 71, 102023 (2021). [Google Scholar]
- 94.Colagrossi M, d’Hombres B & Schnepf SV Like (grand)parent, like child? Multigenerational mobility across the EU. Eur. Econ. Rev. 130, 103600 (2020). [Google Scholar]
- 95.Bingley P, Cappellari L & Tatsiramos K Parental assortative mating and the intergenerational transmission of human capital. Eur. Assoc. Labour Econ. World Conf. EALESOLEAASLE Berl. Ger. 25 – 27 June 2020 77, 102047 (2022). [Google Scholar]
- 96.Cave SN, Wright M & von Stumm S Change and stability in the association of parents’ education with children’s intelligence. Intelligence 90, 101597 (2022). [Google Scholar]
- 97.Carmelli D, Swan GE, Hunt SC & Williams RR Cross-spouse correlates of blood pressure in hypertension-prone families in Utah. J. Psychosom. Res. 33, 75–84 (1989). [DOI] [PubMed] [Google Scholar]
- 98.Miller RN Educational assortative mating and time use in the home. Soc. Sci. Res. 90, 102440 (2020). [DOI] [PubMed] [Google Scholar]
- 99.Mishra V & Smyth R Economic returns to schooling for China’s Korean minority. J. Asian Econ. 24, 89–102 (2013). [Google Scholar]
- 100.Anderson G & Leo TW An empirical examination of matching theories: The one child policy, partner choice and matching intensity in urban China. Law Finance 41, 468–489 (2013). [Google Scholar]
- 101.Liu J-T, Hammitt JK & Jeng Lin C Family background and returns to schooling in Taiwan. Econ. Educ. Rev. 19, 113–125 (2000). [Google Scholar]
- 102.Hu A & Qian Z Does higher education expansion promote educational homogamy? Evidence from married couples of the post-80s generation in Shanghai, China. Soc. Sci. Res. 60, 148–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Johnson W, Deary IJ & Iacono WG Genetic and environmental transactions underlying educational attainment. Intelligence 37, 466–478 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xing L, Campbell C, Li X, Noellert M & Lee J Education, class and assortative marriage in rural Shanxi, China in the mid-twentieth century. Res. Soc. Stratif. Mobil 66, 100460 (2020). [Google Scholar]
- 105.Peek MK, Stimpson JP, Townsend AL & Markides KS Well-being in older Mexican American spouses. The Gerontologist 46, 258–265 (2006). [DOI] [PubMed] [Google Scholar]
- 106.Cronkite RC & Moos RH The role of predisposing and moderating factors in the stress-illness relationship. J. Health Soc. Behav 372–393 (1984). [PubMed] [Google Scholar]
- 107.Heath AC et al. No decline in assortative mating for educational level. Behav. Genet. 15, 349–369 (1985). [DOI] [PubMed] [Google Scholar]
- 108.Lykken DT & Tellegen A Is human mating adventitious or the result of lawful choice? A twin study of mate selection. J. Pers. Soc. Psychol. 65, 56 (1993). [DOI] [PubMed] [Google Scholar]
- 109.Kye B & Mare RD Intergenerational effects of shifts in women’s educational distribution in South Korea: Transmission, differential fertility, and assortative mating. Soc. Sci. Res. 41, 1495–1514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ingoglia S, Liga F, Coco AL & Inguglia C Informant discrepancies in perceived parental psychological control, adolescent autonomy, and relatedness psychological needs. J. Appl. Dev. Psychol. 77, 101333 (2021). [Google Scholar]
- 111.Torvik FA et al. Mechanisms linking parental educational attainment with child ADHD, depression, and academic problems: a study of extended families in The Norwegian Mother, Father and Child Cohort Study. J. Child Psychol. Psychiatry 61, 1009–1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shikishima C et al. A simple syllogism-solving test: Empirical findings and implications for g research. Intelligence 39, 89–99 (2011). [Google Scholar]
- 113.Ponzo M & Scoppa V Trading height for education in the marriage market. Am. J. Hum. Biol. 27, 164–174 (2015). [DOI] [PubMed] [Google Scholar]
- 114.Procidano ME & Rogler LH Homogamous assortative mating among Puerto Rican families: Intergenerational processes and the migration experience. Behav. Genet. 19, 343–354 (1989). [DOI] [PubMed] [Google Scholar]
- 115.Chen H, Luo S, Yue G, Xu D & Zhaoyang R Do birds of a feather flock together in China? Pers. Relatsh. 16, 167–186 (2009). [Google Scholar]
- 116.George D et al. Couple similarity on stimulus characteristics and marital satisfaction. Personal. Individ. Differ. 86, 126–131 (2015). [Google Scholar]
- 117.Jacobs JA & Furstenberg FF Jr Changing places: conjugal careers and women’s marital mobility. Soc. Forces 64, 714–732 (1986). [Google Scholar]
- 118.Hur Y-M Assortative mating for personality traits, educational level, religious affiliation, height, weight, and body mass index in parents of a Korean twin sample. Twin Res. 6, 467–470 (2003). [DOI] [PubMed] [Google Scholar]
- 119.Verbakel E & Kalmijn M Assortative Mating Among Dutch Married and Cohabiting Same-Sex and Different-Sex Couples: Homogamy in Same- and Different-Sex Couples. J. Marriage Fam 76, 1–12 (2014). [Google Scholar]
- 120.Phillips K, Fulker DW, Carey G & Nagoshi CT Direct marital assortment for cognitive and personality variables. Behav. Genet. 18, 347–356 (1988). [DOI] [PubMed] [Google Scholar]
- 121.Watson D et al. Match makers and deal breakers: analyses of assortative mating in newlywed couples. J. Pers. 72, 1029–1068 (2004). [DOI] [PubMed] [Google Scholar]
- 122.Tambs K, Sundet JM & Berg K Correlations between identical twins and their spouses suggest social homogamy for intelligence in Norway. Personal. Individ. Differ. 14, 279–281 (1993). [Google Scholar]
- 123.Cattell RB & Willson JL Contributions concerning mental inheritance: I. of intelligence. Br. J. Educ. Psychol. 8, 129–149 (1938). [Google Scholar]
- 124.Johnson W et al. Genetic and environmental influences on the Verbal-Perceptual-Image Rotation (VPR) model of the structure of mental abilities in the Minnesota study of twins reared apart. Intelligence 35, 542–562 (2007). [Google Scholar]
- 125.Keller MC et al. The genetic correlation between height and IQ: shared genes or assortative mating? PLoS Genet. 9, e1003451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nagoshi CT & Johnson RC The ubiquity of g. Personal. Individ. Differ. 7, 201–207 (1986). [Google Scholar]
- 127.Jester JM et al. Intergenerational transmission of neuropsychological executive functioning. Brain Cogn. 70, 145–153 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vinkhuyzen AAE, van der Sluis S, Maes HHM & Posthuma D Reconsidering the Heritability of Intelligence in Adulthood: Taking Assortative Mating and Cultural Transmission into Account. Behav. Genet. 42, 187–198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Loehlin JC, Horn JM & Willerman L Modeling IQ change: evidence from the Texas Adoption Project. Child Dev 993–1004 (1989). [DOI] [PubMed] [Google Scholar]
- 130.Zonderman AB, Vandenberg SG, Spuhler KP & Fain PR Assortative marriage for cognitive abilities. Behav. Genet. 7, 261–271 (1977). [DOI] [PubMed] [Google Scholar]
- 131.Eaves L et al. Comparing the biological and cultural inheritance of personality and social attitudes in the Virginia 30,000 study of twins and their relatives. Twin Res. 2, 62–80 (1999). [DOI] [PubMed] [Google Scholar]
- 132.Leikas S, Ilmarinen V-J, Verkasalo M, Vartiainen H-L & Lönnqvist J-E Relationship satisfaction and similarity of personality traits, personal values, and attitudes. Personal. Individ. Differ. 123, 191–198 (2018). [Google Scholar]
- 133.Martin NG et al. Transmission of social attitudes. Proc. Natl. Acad. Sci. 83, 4364–4368 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Feather NT Family resemblances in conservatism: Are daughters more similar to parents than sons are? J. Pers. 46, 260–278 (1978). [Google Scholar]
- 135.Rozin P Family resemblance in food and other domains: The family paradox and the role of parental congruence. Appetite 16, 93–102 (1991). [DOI] [PubMed] [Google Scholar]
- 136.Bell E, Kandler C & Riemann R Genetic and environmental influences on sociopolitical attitudes: addressing some gaps in the new paradigm. Polit. Life Sci. 37, 236–249 (2018). [DOI] [PubMed] [Google Scholar]
- 137.Abrahamson AC, Baker LA & Caspi A Rebellious teens? Genetic and environmental influences on the social attitudes of adolescents. J. Pers. Soc. Psychol. 83, 1392–1408 (2002). [DOI] [PubMed] [Google Scholar]
- 138.Eysenck HJ & Wakefield JA Jr Psychological factors as predictors of marital satisfaction. Adv. Behav. Res. Ther. 3, 151–192 (1981). [Google Scholar]
- 139.Beer JM, Arnold RD & Loehlin JC Genetic and environmental influences on MMPI factor scales: joint model fitting to twin and adoption data. J. Pers. Soc. Psychol. 74, 818 (1998). [DOI] [PubMed] [Google Scholar]
- 140.Koenig LB, McGue M & Iacono WG Rearing environmental influences on religiousness: An investigation of adolescent adoptees. Personal. Individ. Differ. 47, 652–656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ostermann J, Sloan FA & Taylor DH Heavy alcohol use and marital dissolution in the USA. Soc. Sci. Med. 61, 2304–2316 (2005). [DOI] [PubMed] [Google Scholar]
- 142.Grant JD et al. Spousal concordance for alcohol dependence: evidence for assortative mating or spousal interaction effects? Alcohol. Clin. Exp. Res. 31, 717–728 (2007). [DOI] [PubMed] [Google Scholar]
- 143.Al Rashid K et al. Spousal associations of serum metabolomic profiles by nuclear magnetic resonance spectroscopy. Sci. Rep. 11, 21587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pérusse L, Leblanc C & Bouchard C Familial resemblance in lifestyle components: results from the Canada Fitness Survey. Can. J. Public Health Rev. Can. Sante Publique 79, 201–205 (1988). [PubMed] [Google Scholar]
- 145.Leonard KE & Das Eiden R Husband’s and wife’s drinking: unilateral or bilateral influences among newlyweds in a general population sample. J. Stud. Alcohol. Suppl. 130–138 (1999). [DOI] [PubMed] [Google Scholar]
- 146.Clarke T-K et al. Genetic and shared couple environmental contributions to smoking and alcohol use in the UK population. Mol. Psychiatry 26, 4344–4354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Garn SM, Cole PE & Bailey SM Living together as a factor in family-line resemblances. Hum. Biol. 51, 565–587 (1979). [PubMed] [Google Scholar]
- 148.Pyke SD, Wood DA, Kinmonth AL & Thompson SG Change in coronary risk and coronary risk factor levels in couples following lifestyle intervention. The British Family Heart Study. Arch. Fam. Med. 6, 354–360 (1997). [DOI] [PubMed] [Google Scholar]
- 149.Venters MH, Jacobs DR Jr, Luepker RV, Maimaw LA & Gillum RF Spouse concordance of smoking patterns: the Minnesota Heart Survey. Am. J. Epidemiol. 120, 608–616 (1984). [DOI] [PubMed] [Google Scholar]
- 150.Maes HH et al. Cross-cultural comparison of genetic and cultural transmission of smoking initiation using an extended twin kinship model. Twin Res. Hum. Genet. 21, 179–190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Boomsma DI, Koopmans JR, Van Doornen LJ & Orlebeke JF Genetic and social influences on starting to smoke: a study of Dutch adolescent twins and their parents. Addiction 89, 219–226 (1994). [DOI] [PubMed] [Google Scholar]
- 152.Nakaya N et al. Spousal similarities in cardiometabolic risk factors: A cross-sectional comparison between Dutch and Japanese data from two large biobank studies. Atherosclerosis 334, 85–92 (2021). [DOI] [PubMed] [Google Scholar]
- 153.Wilcox MA, Newton CS & Johnson IR Paternal influences on birthweight. Acta Obstet. Gynecol. Scand. 74, 15–18 (1995). [DOI] [PubMed] [Google Scholar]
- 154.Gleiberman L, Harburg E, DiFranceisco W & Schork A Familial transmission of alcohol use: v. drinking patterns among spouses, tecumseh, michigan. Behav. Genet. 22, 63–79 (1992). [DOI] [PubMed] [Google Scholar]
- 155.Sutton GC Assortative marriage for smoking habits. Ann. Hum. Biol. 7, 449–456 (1980). [DOI] [PubMed] [Google Scholar]
- 156.Cotch MF, Beaty TH & Cohen BH Path analysis of familial resemblance of pulmonary function and cigarette smoking. Am. Rev. Respir. Dis. 142, 1337–1343 (1990). [DOI] [PubMed] [Google Scholar]
- 157.Kalmijn M Intergenerational transmission of health behaviors in a changing demographic context: The case of smoking and alcohol consumption. Soc. Sci. Med. 296, 114736 (2022). [DOI] [PubMed] [Google Scholar]
- 158.Bloch KV, Klein CH, de Souza e Silva NA, Nogueira A. da R. & Salis LHA Socioeconomic aspects of spousal concordance for hypertension, obesity, and smoking in a community of Rio de Janeiro, Brazil. Arq. Bras. Cardiol 80, 179–186, 171–178 (2003). [DOI] [PubMed] [Google Scholar]
- 159.Clark AE & Etilé F Don’t give up on me baby: Spousal correlation in smoking behaviour. J. Health Econ. 25, 958–978 (2006). [DOI] [PubMed] [Google Scholar]
- 160.Ogden MW, Morgan WT, Heavner DL, Davis RA & Steichen TJ National incidence of smoking and misclassification among the U.S. married female population. J. Clin. Epidemiol. 50, 253–263 (1997). [DOI] [PubMed] [Google Scholar]
- 161.Espinosa J & Evans WN Heightened mortality after the death of a spouse: Marriage protection or marriage selection? J. Health Econ. 27, 1326–1342 (2008). [DOI] [PubMed] [Google Scholar]
- 162.Wilson SE The health capital of families: an investigation of the inter-spousal correlation in health status. Soc. Sci. Med. 55, 1157–1172 (2002). [DOI] [PubMed] [Google Scholar]
- 163.Jackson SE, Steptoe A & Wardle J The influence of partner’s behavior on health behavior change: the English Longitudinal Study of Ageing. JAMA Intern. Med. 175, 385–392 (2015). [DOI] [PubMed] [Google Scholar]
- 164.Homish GG & Leonard KE Spousal influence on smoking behaviors in a US community sample of newly married couples. Soc. Sci. Med. 61, 2557–2567 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pai C-W, Godboldo-Brooks A & Edington DW Spousal concordance for overall health risk status and preventive service compliance. Ann. Epidemiol. 20, 539–546 (2010). [DOI] [PubMed] [Google Scholar]
- 166.Machado MPA et al. Alcohol and tobacco consumption concordance and its correlates in older couples in Latin America. Geriatr. Gerontol. Int. 17, 1849–1857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kirillova GP, Vanyukov MM, Kirisci L & Reynolds M Physical maturation, peer environment, and the ontogenesis of substance use disorders. Psychiatry Res. 158, 43–53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sakai JT et al. Mate similarity for substance dependence and antisocial personality disorder symptoms among parents of patients and controls. Drug Alcohol Depend. 75, 165–175 (2004). [DOI] [PubMed] [Google Scholar]
- 169.Rammstedt B & Schupp J Only the congruent survive–personality similarities in couples. Personal. Individ. Differ. 45, 533–535 (2008). [Google Scholar]
- 170.Chopik WJ & Lucas RE Actor, partner, and similarity effects of personality on global and experienced well-being. J. Res. Personal. 78, 249–261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vandermeer MRJ, Kotelnikova Y, Simms LJ & Hayden EP Spousal Agreement on Partner Personality Ratings is Moderated by Relationship Satisfaction. J. Res. Personal. 76, 22–31 (2018). [Google Scholar]
- 172.Watson D, Beer A & McDade-Montez E The role of active assortment in spousal similarity. J. Pers 82, 116–129 (2014). [DOI] [PubMed] [Google Scholar]
- 173.Barelds DP Self and partner personality in intimate relationships. Eur. J. Personal. Publ. Eur. Assoc. Personal. Psychol. 19, 501–518 (2005). [Google Scholar]
- 174.van Scheppingen MA, Chopik WJ, Bleidorn W & Denissen JJA Longitudinal Actor, Partner and Similarity Effects of Personality on Well-Being. J. Pers. Soc. Psychol. 117, e51–e70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dijkstra P & Barelds DPH Self and partner personality and responses to relationship threats. J. Res. Personal. 42, 1500–1511 (2008). [Google Scholar]
- 176.Mosca I & McCrory C Personality and wealth accumulation among older couples: Do dispositional characteristics pay dividends? J. Econ. Psychol. 56, 1–19 (2016). [Google Scholar]
- 177.Dubuis-Stadelmann E, Fenton BT, Ferrero F & Preisig M Spouse similarity for temperament, personality and psychiatric symptomatology. Personal. Individ. Differ. 30, 1095–1112 (2001). [Google Scholar]
- 178.Tambs K, Sundet JM, Eaves L, Solaas MH & Berg K Pedigree analysis of Eysenck Personality Questionnaire (EPQ) scores in monozygotic (MZ) twin families. Behav. Genet. 21, 369–382 (1991). [DOI] [PubMed] [Google Scholar]
- 179.Guttman R & Zohar A Spouse similarities in personality items: Changes over years of marriage and implications for mate selection. Behav. Genet. 17, 179–189 (1987). [DOI] [PubMed] [Google Scholar]
- 180.Eysenck HJ Personality, premarital sexual permissiveness, and assortative mating. J. Sex Res. 10, 47–51 (1974). [Google Scholar]
- 181.Patterson CH The relationship of Bernreuter scores to parent behavior, child behavior, urban-rural residence, and other background factors in 100 normal adult parents. J. Soc. Psychol. 24, 3–49 (1946). [DOI] [PubMed] [Google Scholar]
- 182.Terman LM & Buttenwieser P Personality factors in marital compatibility: II. J. Soc. Psychol. 6, 267–289 (1935). [Google Scholar]
- 183.Rushton JP & Bons TA Mate choice and friendship in twins: evidence for genetic similarity. Psychol. Sci. 16, 555–559 (2005). [DOI] [PubMed] [Google Scholar]
- 184.Farley FH & Davis SA Arousal, Personality, and Assortative Mating in Marriage. J. Sex Marital Ther. 3, 122–127 (1977). [DOI] [PubMed] [Google Scholar]
- 185.Sutton GC Do men grow to resemble their wives, or vice versa? J. Biosoc. Sci 25, 25–29 (1993). [DOI] [PubMed] [Google Scholar]
- 186.Abdellaoui A et al. Associations between loneliness and personality are mostly driven by a genetic association with Neuroticism. J. Pers. 87, 386–397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hoffeditz EL Family resemblances in personality traits. J. Soc. Psychol. 5, 214–227 (1934). [Google Scholar]
- 188.Donnellan MB, Conger RD & Bryant CM The Big Five and enduring marriages. J. Res. Personal 38, 481–504 (2004). [Google Scholar]
- 189.Lake RI, Eaves LJ, Maes HH, Heath AC & Martin NG Further evidence against the environmental transmission of individual differences in neuroticism from a collaborative study of 45,850 twins and relatives on two continents. Behav. Genet. 30, 223–233 (2000). [DOI] [PubMed] [Google Scholar]
- 190.Chopik WJ & Johnson DJ Modeling dating decisions in a mock swiping paradigm: An examination of participant and target characteristics. J. Res. Personal. 92, 104076 (2021). [Google Scholar]
- 191.Sebro R, Peloso GM, Dupuis J & Risch NJ Structured mating: patterns and implications. PLoS Genet. 13, e1006655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kim HC et al. Spousal concordance of metabolic syndrome in 3141 Korean couples: a nationwide survey. Ann. Epidemiol. 16, 292–298 (2006). [DOI] [PubMed] [Google Scholar]
- 193.Pennock-Román M Assortative marriage for physical characteristics in newlyweds. Am. J. Phys. Anthropol. 64, 185–190 (1984). [DOI] [PubMed] [Google Scholar]
- 194.Staessen J et al. Familial aggregation of blood pressure, anthropometric characteristics and urinary excretion of sodium and potassium—a population study in two Belgian towns. J. Chronic Dis 38, 397–407 (1985). [DOI] [PubMed] [Google Scholar]
- 195.Katzmarzyk PT, Hebebrand J & Bouchard C Spousal resemblance in the Canadian population: implications for the obesity epidemic. Int. J. Obes. 26, 241–246 (2002). [DOI] [PubMed] [Google Scholar]
- 196.Wu D-M et al. Familial resemblance of adiposity-related parameters: results from a health check-up population in Taiwan. Eur. J. Epidemiol. 18, 221–226 (2003). [DOI] [PubMed] [Google Scholar]
- 197.Zahra J, Jago R & Sebire SJ Associations between parenting partners’ objectively-assessed physical activity and Body Mass Index: A cross-sectional study. Prev. Med. Rep. 2, 473–477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Longini IM Jr, Higgins MW, Hinton PC, Moll PP & Keller JB Genetic and environmental sources of familial aggregation of body mass in Tecumseh, Michigan. Hum. Biol 733–757 (1984). [PubMed] [Google Scholar]
- 199.Tambs K et al. Genetic and environmental contributions to the variance of the body mass index in a Norwegian sample of first- and second-degree relatives. Am. J. Hum. Biol. 3, 257–267 (1991). [DOI] [PubMed] [Google Scholar]
- 200.Jee SH, Suh I, Won SY & Kim MY Familial correlation and heritability for cardiovascular risk factors. Yonsei Med. J. 43, 160–164 (2002). [DOI] [PubMed] [Google Scholar]
- 201.Davillas A & Pudney S Concordance of health states in couples: analysis of self-reported, nurse administered and blood-based biomarker data in the UK Understanding Society panel. J. Health Econ. 56, 87–102 (2017). [DOI] [PubMed] [Google Scholar]
- 202.Moll PP, Burns TL & Lauer RM The genetic and environmental sources of body mass index variability: the Muscatine Ponderosity Family Study. Am. J. Hum. Genet. 49, 1243 (1991). [PMC free article] [PubMed] [Google Scholar]
- 203.van Dongen J, Willemsen G, Chen W-M, de Geus EJC & Boomsma DI Heritability of metabolic syndrome traits in a large population-based sample[S]. J. Lipid Res. 54, 2914–2923 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Prichard I et al. Brides and young couples: partners’ weight, weight change, and perceptions of attractiveness. J. Soc. Pers. Relatsh. 32, 263–278 (2015). [Google Scholar]
- 205.Province MA & Rao DC Path analysis of family resemblance with temporal trends: applications to height, weight, and Quetelet index in northeastern Brazil. Am. J. Hum. Genet. 37, 178 (1985). [PMC free article] [PubMed] [Google Scholar]
- 206.Chien KL et al. Familial aggregation of metabolic syndrome among the Chinese: Report from the Chin-Shan community family study. Diabetes Res. Clin. Pract. 76, 418–424 (2007). [DOI] [PubMed] [Google Scholar]
- 207.Knight B et al. Evidence of genetic regulation of fetal longitudinal growth. Early Hum. Dev. 81, 823–831 (2005). [DOI] [PubMed] [Google Scholar]
- 208.Bouchard C, Perusse L, Leblanc C, Tremblay A & Theriault G Inheritance of the amount and distribution of human body fat. Int. J. Obes 12, 205–215 (1988). [PubMed] [Google Scholar]
- 209.Friedlander Y, Kark JD, Kaufmann NA, Berry EM & Stein Y Familial aggregation of body mass index in ethnically diverse families in Jerusalem. The Jerusalem Lipid Research Clinic. Int. J. Obes. 12, 237–247 (1988). [PubMed] [Google Scholar]
- 210.Silverman-Retana O et al. Spousal concordance in pathophysiological markers and risk factors for type 2 diabetes: a cross-sectional analysis of The Maastricht Study. BMJ Open Diabetes Res. Care 9, e001879 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sear R & Marlowe FW How universal are human mate choices? Size does not matter when Hadza foragers are choosing a mate. Biol. Lett 5, 606–609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Silventoinen K, Kaprio J, Lahelma E, Viken RJ & Rose RJ Assortative mating by body height and BMI: Finnish twins and their spouses. Am. J. Hum. Biol. 15, 620–627 (2003). [DOI] [PubMed] [Google Scholar]
- 213.Mueller WH & Malina RM Differential contribution of stature phenotypes to assortative mating in parents of Philadelphia black and white school children. Am. J. Phys. Anthropol. 45, 269–276 (1976). [DOI] [PubMed] [Google Scholar]
- 214.Heude B et al. Anthropometric relationships between parents and children throughout childhood: the Fleurbaix–Laventie Ville Santé Study. Int. J. Obes. 29, 1222–1229 (2005). [DOI] [PubMed] [Google Scholar]
- 215.Sanchez-Andres A & Mesa M Assortative mating in a Spanish population: effects of social factors and cohabitation time. J. Biosoc. Sci. 26, 441–50 (1994). [DOI] [PubMed] [Google Scholar]
- 216.Salces I, Rebato E & Susanne C Evidence of phenotypic and social assortative mating for anthropometric and physiological traits in couples from the Basque country (Spain). J. Biosoc. Sci. 36, 235–250 (2004). [DOI] [PubMed] [Google Scholar]
- 217.Ahmad M & Gilbert RI Assortative mating for height in Pakistani arranged marriages. J. Biosoc. Sci. 17, 211–214 (1985). [DOI] [PubMed] [Google Scholar]
- 218.Ajala O et al. The relationship of height and body fat to gender-assortative weight gain in children. A longitudinal cohort study (EarlyBird 44). Int. J. Pediatr. Obes. 6, 223–228 (2011). [DOI] [PubMed] [Google Scholar]
- 219.Annest JL, Sing CF, Biron P & Mongeau JG Familial aggregation of blood pressure and weight in adoptive families: III. analysis of the role of shared genes and shared household environment in explaining family resemblance for height, weight and selected weight/height indices. Am. J. Epidemiol. 117, 492–506 (1983). [DOI] [PubMed] [Google Scholar]
- 220.Burgess EW & Wallin P Homogamy in personality characteristics. J. Abnorm. Soc. Psychol. 39, 475 (1944). [Google Scholar]
- 221.Byard PJ, Poosha DVR & Satyanarayana M Genetic and environmental determinants of height and weight in families from Andhra Pradesh, India. Hum. Biol 621–633 (1985). [PubMed] [Google Scholar]
- 222.Byard PJ, Mukherjee BN, Bhattacharya SK, Russell JM & Rao DC Familial aggregation of blood pressure and anthropometric variables in patrilocal households. Am. J. Phys. Anthropol. 79, 305–311 (1989). [DOI] [PubMed] [Google Scholar]
- 223.Dasgupta I, Dasgupta P & Daschaudhuri AB Familial resemblance in height and weight in an endogamous Hahisya caste population of rural West Bengal. Am. J. Hum. Biol. Off. J. Hum. Biol. Assoc. 9, 7–9 (1997). [DOI] [PubMed] [Google Scholar]
- 224.Eckman RE, Williams R & Nagoshi C Marital assortment for genetic similarity. J. Biosoc. Sci. 34, 511–523 (2002). [DOI] [PubMed] [Google Scholar]
- 225.Ellis JA et al. Comprehensive multi-stage linkage analyses identify a locus for adult height on chromosome 3p in a healthy Caucasian population. Hum. Genet. 121, 213–222 (2007). [DOI] [PubMed] [Google Scholar]
- 226.Ginsburg E, Livshits G, Yakovenko K & Kobyliansky E Major gene control of human body height, weight and BMI in five ethnically different populations. Ann. Hum. Genet. 62, 307–322 (1998). [DOI] [PubMed] [Google Scholar]
- 227.Harrison GA, Gibson JB & Hiorns RW Assortative marriage for psychometric, personality and anthropometric variation in a group of Oxfordshire villages. J. Biosoc. Sci. 8, 145–153 (1976). [DOI] [PubMed] [Google Scholar]
- 228.Knuiman MW, Divitini ML & Bartholomew HC Spouse selection and environmental effects on spouse correlation in lung function measures. Ann. Epidemiol. 15, 39–43 (2005). [DOI] [PubMed] [Google Scholar]
- 229.Luo ZC, Albertsson-Wikland K & Karlberg J Target height as predicted by parental heights in a population-based study. Pediatr. Res. 44, 563–571 (1998). [DOI] [PubMed] [Google Scholar]
- 230.Mascie-Taylor CGN Assortative mating in a contemporary British population. Ann. Hum. Biol. 14, 59–68 (1987). [DOI] [PubMed] [Google Scholar]
- 231.McManus IC & Mascie-Taylor CG Human assortative mating for height: non-linearity and heteroscedasticity. Hum. Biol. 56, 617–623 (1984). [PubMed] [Google Scholar]
- 232.Mukhopadhyay N et al. A genome-wide scan for loci affecting normal adult height in the Framingham Heart Study. Hum. Hered. 55, 191–201 (2003). [DOI] [PubMed] [Google Scholar]
- 233.Nagoshi CT & Johnson RC Between- vs. within-family analyses of the correlation of height and intelligence. Soc. Biol. 34, 110–113 (1987). [PubMed] [Google Scholar]
- 234.Nance WE, Corey LA & Eaves LJ A model for the analysis of mate selection in the marriages of twins application to data on stature. Acta Genet. Medicae Gemellol. Twin Res. 29, 91–101 (1980). [DOI] [PubMed] [Google Scholar]
- 235.Pearson K & Lee A On the laws of inheritance in man: I. Inheritance of physical characters. Biometrika 2, 357–462 (1903). [Google Scholar]
- 236.Pieper U Assortative mating in the population of a German and a Cameroon city. J. Hum. Evol. 10, 643–645 (1981). [Google Scholar]
- 237.Pomerat CM Homogamy and infertility. Hum. Biol 8, 19 (1936). [Google Scholar]
- 238.Raychaudhuri A, Ghosh R, Vasulu TS & Bharati P Heritability estimates of height and weight in Mahishya caste population. Int. J. Hum. Genet 3, 151–154 (2003). [Google Scholar]
- 239.Roberts DF, Billewicz WZ & McGregor I Heritability of stature in a West African population. Ann. Hum. Genet. 42, 15–24 (1978). [DOI] [PubMed] [Google Scholar]
- 240.Seki M, Ihara Y & Aoki K Homogamy and imprinting-like effect on mate choice preference for body height in the current Japanese population. Ann. Hum. Biol. 39, 28–35 (2012). [DOI] [PubMed] [Google Scholar]
- 241.Siniarska A Assortative mating of parents and sib-sib similarities in offspring. Stud Hum Ecol 5, 95–112 (1984). [Google Scholar]
- 242.Smith M A research note on homogamy of marriage partners in selected physical characteristics. Am. Sociol. Rev 11, (1946). [Google Scholar]
- 243.Stulp G, Buunk AP & Pollet TV Women want taller men more than men want shorter women. Personal. Individ. Differ. 54, 877–883 (2013). [Google Scholar]
- 244.Stulp G, Buunk AP, Pollet TV, Nettle D & Verhulst S Are human mating preferences with respect to height reflected in actual pairings? PLoS One 8, e54186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Stulp G, Mills M, Pollet TV & Barrett L Non-linear associations between stature and mate choice characteristics for American men and their spouses. Am. J. Hum. Biol. 26, 530–537 (2014). [DOI] [PubMed] [Google Scholar]
- 246.Susanne C Heritability of anthropological characters. Hum. Biol 573–580 (1977). [PubMed] [Google Scholar]
- 247.Tambs K et al. Genetic and environmental contributions to the variance of body height in a sample of first and second degree relatives. Am. J. Phys. Anthropol. 88, 285–294 (1992). [DOI] [PubMed] [Google Scholar]
- 248.Tenesa A, Rawlik K, Navarro P & Canela-Xandri O Genetic determination of height-mediated mate choice. Genome Biol. 16, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.To WWK, Cheung W & Kwok JSY Paternal height and weight as determinants of birth weight in a Chinese population. Am. J. Perinatol. 15, 545–548 (1998). [DOI] [PubMed] [Google Scholar]
- 250.Willoughby RR Somatic homogamy in man. Hum. Biol. 5, 690 (1933). [Google Scholar]
- 251.Xu J et al. Major recessive gene(s) with considerable residual polygenic effect regulating adult height: confirmation of genomewide scan results for chromosomes 6, 9, and 12. Am. J. Hum. Genet. 71, 646–650 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Donahue RP, Prineas RJ, Gomez O & Hong CP Familial resemblance of body fat distribution: the Minneapolis Children’s Blood Pressure Study. Int. J. Obes. Relat. Metab. Disord. J. Int. Assoc. Study Obes 16, 161–167 (1992). [PubMed] [Google Scholar]
- 253.McLeod JD Spouse concordance for depressive disorders in a community sample. J. Affect. Disord. 27, 43–52 (1993). [DOI] [PubMed] [Google Scholar]
- 254.Sun J et al. Prevalence of diabetes and cardiometabolic disorders in spouses of diabetic individuals. Am. J. Epidemiol. 184, 400–409 (2016). [DOI] [PubMed] [Google Scholar]
- 255.Patel SA et al. Chronic disease concordance within Indian households: a cross-sectional study. PLoS Med. 14, e1002395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Jun SY, Kang M, Kang SY, Lee JA & Kim YS Spousal Concordance regarding Lifestyle Factors and Chronic Diseases among Couples Visiting Primary Care Providers in Korea. Korean J. Fam. Med. 41, 183–188 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Watanabe T, Sugiyama T, Takahashi H, Noguchi H & Tamiya N Concordance of hypertension, diabetes and dyslipidaemia in married couples: cross-sectional study using nationwide survey data in Japan. BMJ Open 10, e036281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Studies included in the meta-analysis are listed in Supplementary Tables 1 and 2 and cited in Table 1, as well as in the supplementary note; studies excluded from the meta-analysis are listed in Supplementary Table 3. Raw data from the UK Biobank is not publicly available, but summary statistics for most traits are available on the UK Biobank website: https://biobank.ndph.ox.ac.uk/showcase/search.cgi. Note that there is no longer a Field ID corresponding to the co-location variable in the UKB and that the putative partner dataset we created cannot be publicly shared. As such, our UKB partner dataset cannot be directly used/recreated at this time. However, combinations of other variables (e.g., inverse distance to the nearest major road) can potentially be used as proxies for co-location53—in conjunction with the code we have made available—to estimate partner pairs.


