Abstract
Human T-cell leukemia virus type 1 (HTLV-1) infects and transforms CD4+ T-lymphocytes both in vivo and in vitro. Although the Tax protein of HTLV-1 has been strongly implicated as a transforming agent, other virally encoded proteins may also play a role in the transformation process. In addition to the rex and tax genes, the pX region of the HTLV-1 genome contains two open reading frames (pX-I and pX-II) which encode the putative viral accessory proteins known as p12I, p30II, and p13II. Mutations in the ACH molecular clone of HTLV-1 that are predicted to abrogate the expression of p12I, p13II and p30II were constructed. These mutations had no effect on viral replication or the immortalization of primary lymphocytes. Although these proteins are dispensable for viral replication and immortalization in vitro, it remains possible that they alter infection in vivo.
Human T-cell leukemia virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia/lymphoma and HTLV-1-associated myelopathy/tropical spastic paraparesis (17, 20, 30, 32). In addition to the gag, pol, and env genes common to most retroviruses, the HTLV-1 genome contains a region between env and the 3′ long terminal repeat (LTR) known as the pX region. The pX region consists of four open reading frames (ORFs), termed pX-I, pX-II, pX-III, and pX-IV, which are alternatively spliced to encode a number of proteins. ORFs pX-IV and pX-III encode the Tax and Rex proteins, which are transcriptional and posttranscriptional regulators of viral gene expression, respectively (3, 13, 21, 22, 34). ORFs pX-I and pX-II are each found in alternatively singly and doubly spliced transcripts which encode a total of three proteins (6, 24, 26). The single- and double-spliced transcripts of pX-I both utilize the same splice acceptor site, and both encode a protein of 12 kDa known as p12I (6, 24, 26). In addition, the double-spliced form of pX-I contains the coding potential for a 27-kDa protein (termed Rof), although expression of this protein in infected cells remains to be demonstrated (6, 26). p12I has distant homology to the bovine papillomavirus (BPV) E5 oncoprotein and has been demonstrated to bind to the 16-kDa subunit of a vacuolar ATPase as well as to the β and γ chains of the interleukin 2 (IL-2) receptor (14, 27, 29). In addition, p12I localizes to cellular endomembranes and perinuclear regions (26) and is capable of cooperating with the BPV E5 protein in the transformation of mouse cells (14). The double-spliced transcript of pX-II encodes a protein of 30 kDa known as p30II (also termed Tof), which localizes to the nucleolus and which has distant homology to the Oct and Pit families of transcription factors (6, 24, 26). The single-spliced pX-II transcript encodes the p13II protein, which consists of the last 87 amino acids (aa) of p30II and which localizes to the nucleus (26). However, despite these initial characterizations of the pX-I- and pX-II-encoded proteins, little is known about the actual role of p12I, p30II, and p13II in viral replication or in the immortalization of primary lymphocytes.
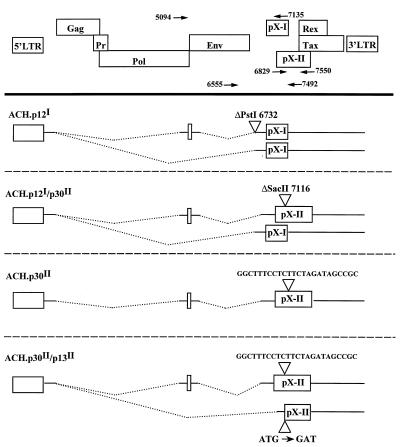
A molecular clone of HTLV-1 known as ACH, which produces infectious virus particles capable of immortalizing primary human lymphocytes (23) and establishing productive infections in rabbits (10), has been previously described. Mutations in the ACH clone that were predicted to result in the loss of expression of the p12I, p30II, and p13II proteins were constructed (Fig. 1). The ACH.p12I mutant contained a deletion of 4 bp of a PstI site as a result of treatment with T4 DNA polymerase, which deletes the adenosine in the conserved AG splice acceptor site utilized by both the single- and double-spliced pX-I transcripts. The ACH.p12I/p30II mutant contained a 2-bp deletion resulting in a frameshift mutation at a SacII site located 291 bp into the 725-bp p30II ORF. In addition to frameshifting the p30II ORF, this mutation also replaced the last 5 aa of p12I with 55 aa derived from a third reading frame. As p12I is predicted to contain two membrane-spanning domains, it is likely that this mutation disrupted the stability and/or processing of p12I. The ACH.p30II mutant contained a linker inserted at the same SacII site, which restored the p12I ORF and introduced a termination codon into the p30II ORF. The ACH.p30II/p13II mutant, constructed by site-specific PCR mutagenesis, contained a deletion of the initiator methionine codon of p13II (changing it to a BglII site), as well as the p30II mutation previously described. The structures of the proviral clones were confirmed by restriction enzyme and nucleotide sequence analyses. These mutations did not affect the ability of the virus to express a functional Tax protein capable of transactivating an HTLV-LTR-luciferase reporter vector when cotransfected with the mutant ACH clones in COS7 or 293T cells (data not shown). In addition, these clones produced comparable amounts of virus particles when transfected in 293T cells with similar Gag and Env protein compositions as determined by Western blotting of concentrated cell culture supernatants with anti-p19MA monoclonal antibody (MAb) 12/1.2 (provided by M. Robert-Guroff) and anti-gp46 SU envelope MAb 0.5α (provided by Y. Matsushita) (data not shown).
FIG. 1.
Mutations constructed in ACH resulting in the loss of expression of p12I, p30II, and p13II. At the top is a schematic drawing of the ORFs in the HTLV-1 genome and the LTRs. The four figures below this represent the transcripts encoding each potential protein product. The triangles indicate the positions of the mutations, and the dotted lines represent the regions of the genome which are removed by splicing. The nucleotide positions are numbered as for the parental ACH clone. The primers used for the confirmation of the mutations (Fig. 2 and 3) are indicated and are designated by the position of the 5′ end of the primer.
These mutants also retained the ability to immortalize primary human peripheral blood mononuclear cells (PBMC). Human PBMC were isolated from normal donors by Ficoll-Paque (Pharmacia, Piscataway, N.J.) centrifugation and activated for 72 h with 10 μg of phytohemagglutinin-P (Gibco, Gaithersburg, Md.) per ml and 50 U of IL-2 per ml, after which they were transfected (107 cells) with 25 μg of the ACH plasmids by electroporation at 250 V and 1,800 μF. The transfected cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (Gibco), 2 mM glutamine, 100 μg of penicillin per ml, 100 U of streptomycin per ml, and 50 U of IL-2 per ml (PBMC growth medium). Cellular proliferation was monitored by MTT conversion assays (19), and cell culture supernatants were collected for p19MA dot blot assays. The wild-type (WT) ACH clone (ACH.wt) immortalized the transfected PBMC on three of four attempts, while the ACH.p12I, ACH.p12I/p30II, ACH.p30II, and ACH.p30II/p13II mutants immortalized the transfected cells on three of four, two of four, four of four, and two of two attempts, respectively. The immortalized cell lines derived from these cultures expressed similar amounts of p19MA antigen in the cell culture supernatants, as determined by dot blot analysis with an anti-p19MA MAb (12/1.2), which suggests the release of similar numbers of viral particles from the infected cells (data not shown). In addition, the cell lines expressed the cell surface markers CD3, CD4, and CD25 but lacked expression of CD8, consistent with an activated T-helper-cell phenotype (data not shown).
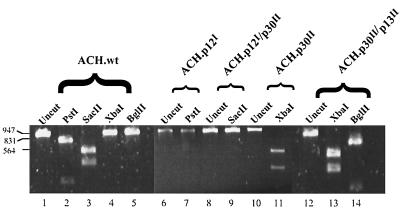
To confirm that the mutations were present in the integrated proviral DNA, a portion of the pX region was amplified by PCR from genomic DNA from the immortalized cell lines. A 937-bp PCR product was amplified from the genomic DNA from the ACH-immortalized cell lines, but not from uninfected PBMC (Fig. 2). The PCR product was then digested with restriction endonucleases (New England Biolabs, Beverly, Mass.) which were diagnostic for the presence or absence of the various mutations (Fig. 2). The ACH.p12I mutation deleted a PstI site, while the ACH.p12I/p30II mutation deleted a SacII site in the fragment. The ACH.p30II mutation added an XbaI site, and the ACH.p30II/p13II mutation added both an XbaI site and a BglII site. According to this analysis, the cell lines examined all contained the original expected mutations in the integrated proviral genome.
FIG. 2.
Presence of mutations in immortalized-cell genomic DNA. PCR of immortalized-cell genomic DNA with primers 6555 and 7492 results in a fragment of 937 bp. Lanes 1 to 5, expected digestion pattern of the ACH.wt-derived fragment; digestion with PstI results in bands of 760 and 177 bp, while digestion with SacII results in bands of 561 and 376 bp; there are no XbaI or BglII sites in the fragment; lanes 6 and 7, the ACH.p12I mutation deletes the PstI site; lanes 8 and 9, the ACH.p12I/p30II mutation deletes the SacII site; lanes 10 and 11, the ACH.p30II mutation inserts an XbaI site at position 7128, resulting in bands of 573 and 386 bp when the DNA is digested with XbaI; lanes 12 to 14, the ACH.p30II/p13II mutation inserts the same XbaI site plus a BglII site at position 7286, which results in bands of 753 and 206 bp when the DNA is digested with BglII.
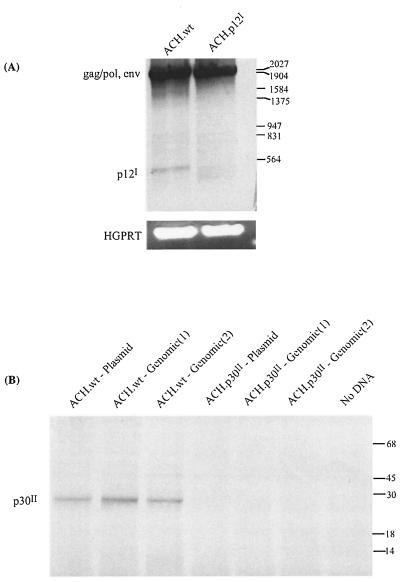
To further confirm the presence of the ACH.p12I and ACH.p30II mutations in the proviruses of the immortalized cell lines, two separate strategies were employed. As the ACH.p12I mutant contains a splice acceptor site deletion, reverse transcription-PCR (RT-PCR) was performed on total RNA isolated from the ACH.wt- and ACH.p12I-immortalized cell lines to confirm the absence of the p12I double-spliced product (Fig. 3A). The primer pair utilized amplifies both a 2,041-bp fragment corresponding to the gag-pol and env transcripts and a 490-bp fragment corresponding to the double-spliced pX-I transcript. The PCR products were separated on a 1% agarose gel, transferred to a nylon membrane, and probed with a digoxigenin (Boehringer Mannheim, Indianapolis, Ind.)-labelled probe derived from the pX-I ORF. Whereas the 2,041-bp fragment was present in both the ACH.wt and ACH.p12I clones, the 490-bp pX-I fragment was only detected in the ACH.wt-immortalized cells. This analysis also confirmed the lack of additional cryptic p12I mRNA splice sites, which could be functioning in the absence of the preferred p12I splice acceptor site, as additional spliced transcripts would have been detected by this probe. This analysis does not, however, exclude the unlikely possibility that the p12I protein could be expressed from a polycistronic message. To assess this possibility, we introduced a polynucleotide linker coding for the influenza hemagglutinin epitope at the 3′ end of the p12I ORF in the ACH.wt and ACH.p12I clones. However, we were unable to detect the expression of the tagged p12I protein with either clone in transiently transfected 293T cells by Western blotting or radioimmunoprecipitation (data not shown). This is most likely due to the low level of expression of the p12I transcript and is consistent with the fact that the translated p12I protein has not yet been detected in HTLV-1-infected cells (6, 24).
FIG. 3.
Confirmation of the presence of ACH.p12I and ACH.p30II mutations. (A) RT-PCR with total RNA from ACH.wt- and ACH.p12I-immortalized cells and primers 5094 and 7135 amplified a 2,041-bp fragment from the gag-pol and env transcripts, as well as a 490-bp pX-I transcript only in the ACH.wt-immortalized cells. Primers specific for human hypoxanthine-guanine phosphoribosyltransferase (HGPRT) were used as an additional control for RNA quality. (B) In vitro transcription/translation of the p30II ORF cloned from ACH.wt- and ACH.p30II-immortalized cells. The p30II ORF was amplified from both the original plasmid and immortalized-cell DNA with primers 6829 and 7550. An approximately 30-kDa product was made with the WT-derived expression vectors but was absent in the mutant (two separate clones derived from the genomic DNA were utilized).
To confirm the ACH.p30II mutation in the immortalized cell lines, the p30II ORF was amplified by PCR from both the original plasmid and from the genomic DNA from the ACH.wt- and ACH.p30II-immortalized cell lines, cloned into the pTM3 expression plasmid (which places the p30II ORF under the control of the T7 promoter) (16), and expressed in vitro with the TnT-coupled reticulocyte lysate system (Promega, Madison, Wis.). Only the p30II ORF derived from the ACH.wt plasmid or genomic DNA expressed a 30-kDa product, while the product was absent from the ACH.p30II plasmid- and immortalized-cell DNA-derived expression vectors (Fig. 3B).
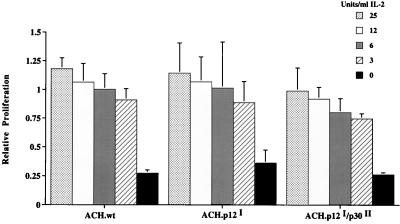
As the p12I protein has been demonstrated to bind to the IL-2 receptor β and γ chains (29) and may therefore influence the IL-2 requirements for cellular proliferation, we examined the IL-2 dependency of the ACH.p12I- and ACH.p12I/p30II-immortalized cell lines. The WT and mutant ACH-immortalized cell lines were all dependent on the addition of exogenous IL-2 for cellular viability and proliferation. Furthermore, over the course of 3 days in culture, the ACH.wt-, ACH.p12I-, and ACH.p12I/p30II-immortalized cell lines all manifested a similar IL-2 dose-response growth relationship (Fig. 4). Finally, long term cultures in gradually decreasing concentrations of IL-2 also revealed no difference in IL-2 dependency, with all cell lines remaining dependent on 3 to 6 U of IL-2 per ml after 6 months in culture (data not shown).
FIG. 4.
IL-2 dependence of mutant-immortalized cell lines. Fifty thousand ACH.wt-, ACH.p12I-, ACH.p12I/p30II-immortalized cells were seeded in triplicate in a 96-well microtiter plate in twofold dilutions of IL-2. Relative proliferation was determined after 72 h in culture by MTT conversion assays. Error bars indicate standard deviations.
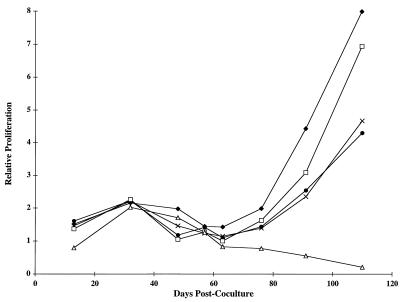
To confirm that the immortalized cell lines were producing virus which was infectious and capable of immortalizing uninfected PBMC, 5 × 105 ACH.wt-, ACH.p12I-, ACH.p12I/p30II-, and ACH.p30II-immortalized cells were exposed to lethal gamma radiation (6,000 rads) and cocultured in PBMC growth medium with 4 × 106 uninfected, activated PBMC and cell viability was monitored by a MTT conversion assay (19). The PBMC alone proliferated for a short period of time, and the irradiated cells alone did not proliferate. In contrast, the cocultured cells continued to proliferate indefinitely, indicating the productive infection and immortalization of the uninfected cells (Fig. 5).
FIG. 5.
Infection of PBMC with virus from immortalized cells. ACH-immortalized cells were exposed to gamma radiation and cocultured with uninfected activated PBMC. Relative cellular proliferation was monitored every 7 to 14 days by MTT conversion assay with 100-μL aliquots of the cultures and was adjusted for the number of culture passages. Whereas the irradiated cells alone do not proliferate (data not shown) and the PBMC alone (Δ) proliferated transiently, the cocultured cells proliferated indefinitely. Cells were immortalized with ACH.wt (⧫), ACH.p12I (•), ACH.p12I/p30II (×), or ACH.p30II (□).
To further quantify the infectivity and immortalizing ability of these mutants, we employed a microtiter infectivity/immortalization assay as described by Persaud et al. (31). In this assay, 104 uninfected activated PBMC were cocultured in PBMC growth medium with 10-fold dilutions (104, 103, 102, and 10) of lethally gamma-irradiated (6,000 rads) ACH mutant-immortalized cells in replicates of 10 to 20 in 96-well microtiter plates. The numbers of cultures which became immortalized were then determined at 9 to 16 weeks after the coculture by microscopic examination of the individual wells. The results of this analysis indicated that the viruses from the ACH.p12I, ACH.p12I/p30II, ACH.p30II, and ACH.p30II/p13II mutant-immortalized cell lines were all capable of infecting and immortalizing the uninfected PBMC with relatively similar efficiencies at the different dilutions of infected cells (Table 1). Furthermore, additional microtiter assays with the ACH.p12I mutant in decreasing concentrations of IL-2 (50, 10, and 2 U/ml) again revealed no difference in immortalizing activity compared to the ACH.wt clone, with 50 U of IL-2 per ml being required for the immortalization of uninfected PBMC for both the WT and p12I mutant (data not shown).
TABLE 1.
Microtiter infectivity/immortalization assay
| Clone | No. of cultures immortalized/total no. of cultures (%) for indicated no. of gamma-irradiated ACH-immortalized cells
|
|||
|---|---|---|---|---|
| 10,000 | 1,000 | 100 | 10 | |
| ACH.wt | 17/20 (85) | 41/50 (82)a | 39/60 (65)a | 9/40 (22)b |
| ACH.p12I | 20/20 (100) | 27/40 (68)b | 17/40 (43)b | 7/20 (35) |
| ACH.p12I/p30II | NDc | 7/10 (70) | 6/20 (30) | 2/20 (10) |
| ACH.p30II | ND | 6/10 (60) | 9/20 (45) | 2/20 (10) |
| ACH.p30II/p13II | 20/20 (100) | 32/40 (80)b | 24/40 (60)b | 8/20 (40) |
Combined results from three separate experiments.
Combined results from two separate experiments.
ND, not done.
In conclusion, the p12I, p30II, and p13II putative accessory proteins of HTLV-1 appear to be dispensable for viral replication and the immortalization of T-lymphocytes in vitro. The role of these proteins in the viral life cycle has remained elusive in part due to their low level of expression in infected cells. Although the pX-I and pX-II transcripts have been detected by RT-PCR in HTLV-1-infected T-cell lines (24), HeLa cells transfected with an HTLV-1 proviral clone (6), freshly isolated cells from HTLV-1-positive individuals (2, 24), and HTLV-1-infected macrophages (25), the translated proteins have not yet been detected. However, it has been reported that a small fraction of HTLV-1-positive individuals may seroconvert to the p30II protein, which would imply its expression in vivo (4). The results reported here are consistent with previous descriptions of naturally occurring HTLV-1 isolates which contain mutations in the pX-I (33) and pX-II (5) ORFs, which may also indicate that these genes are dispensable for certain stages of the viral life cycle or disease pathogenesis. In addition, Derse et al. have recently reported that the p12I and p30II proteins are not required for viral replication or immortalization of T-cells in vitro by using the pCS-HTLV molecular clone of HTLV-1 (12). Our studies extend these results, though, by quantitatively examining immortalization and IL-2-dependent proliferation, as well as by examining a mutation in the p13II ORF. The possibility may exist, however, that the p12I, p30II, and p13II proteins could affect viral replication in a cell-type-specific manner, as has been observed with other retroviral accessory proteins (i.e., human immunodeficiency virus type 1 Vpr and equine infectious anemia virus dUTPase are required for viral replication in quiescent macrophages but not in dividing cells) (1, 11, 36).
HTLV-1-infected T-cell lines which have become IL-2 independent as well as uncultured lymphocytes from adult T-cell leukemia/lymphoma patients often acquire constitutively activated Jak-STAT signalling pathways (28, 35, 39). This is particularly interesting in light of the fact that p12I has been demonstrated to bind the β and γ chains of the IL-2 receptor (29) and IL-2 signalling is associated with the phosphorylation of STAT proteins (15). Although we do not find an influence of the p12I protein on the IL-2 requirements for proliferation or immortalization in the ACH-immortalized (IL-2 dependent) cell lines, it remains an intriguing possibility that p12I may be necessary for the acquisition of IL-2 independence, either by altering IL-2 receptor signalling or through another unknown mechanism.
The pX regions of the closely related HTLV-2 and bovine leukemia viruses (BLV) also contain a number of spliced ORFs which have varying degrees of homology to the pX-I and pX-II ORFs of HTLV-1 (7, 18, 37). The pX-I ORF of HTLV-2 encodes a nucleolar protein designated p10xI which is 56% similar and 37% identical to the p12I protein (7, 18). The HTLV-2 pX-II ORF encodes a protein known as p28xII which has little or no homology to p30II (7, 18). The HTLV-2 pX-I and pX-II ORFs were previously found to be dispensable for viral replication and immortalization in vitro (18). However, these HTLV-2 ORFs were subsequently shown to be important for viral replication in a rabbit model of HTLV-2 infection (8). Likewise, the R3 and G4 ORFs of BLV, which are also distantly homologous to pX-I and pX-II, were shown to be important for the maintenance of high viral loads in sheep (37, 38). Preliminary results with a rabbit model of HTLV-1 infection have indicated that virus derived from the ACH.p12I mutant is incapable of establishing a productive infection in rabbits, implying a role for p12I in viral replication in vivo (9). Therefore, it remains possible that the HTLV-1 p30II and p13II proteins are also playing a role in viral infectivity or replication in vivo.
Acknowledgments
We thank M. Robert-Guroff for the gift of the 12/1.2 antibody, Y. Matsushita for the gift of the 0.5α antibody, and Cetus for the gift of IL-2. We also thank David Leib for helpful discussions, and Nathaniel Collins and Michael Lairmore for critical reviews of the manuscript and the sharing of unpublished data.
This work was supported by PHS grant CA64317 and training grant 2T32GM07067-22 (M.D.R.).
REFERENCES
- 1.Balotta C, Lusso P, Crowley R, Gallo R C, Franchini G. Antisense phosphorothioate oligodeoxynucleotides targeted to the vpr gene inhibit human immunodeficiency virus type 1 replication in primary human macrophages. J Virol. 1993;67:4409–4414. doi: 10.1128/jvi.67.7.4409-4414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berneman Z N, Gartenhaus R B, Reitz M S, Jr, Blattner W A, Manns A, Hanchard B, Ikehara O, Gallo R C, Klotman M E. Expression of alternatively spliced human T-lymphotropic virus type I pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc Natl Acad Sci USA. 1992;89:3005–3009. doi: 10.1073/pnas.89.7.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cann A J, Rosenblatt J D, Wachsman W, Shah N P, Chen I S Y. Identification of the gene responsible for human T-cell leukemia virus transcriptional regulation. Nature (London) 1985;318:571–574. doi: 10.1038/318571a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y-M A, Chen S-H, Fu C-Y, Chen J Y, Osame M. Antibody reactivities to tumor-suppressor protein p53 and HTLV-1 Tof, Rex, and Tax in HTLV-1 infected people with different clinical status. Int J Cancer. 1997;71:1–7. doi: 10.1002/(sici)1097-0215(19970410)71:2<196::aid-ijc12>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 5.Chou K S, Okayama A, Tachibana N, Lee T H, Essex M. Nucleotide sequence analysis of a full-length human T-cell leukemia virus type I from adult T-cell leukemia cells: a prematurely terminated pX open reading frame II. Int J Cancer. 1995;60:701–706. doi: 10.1002/ijc.2910600522. [DOI] [PubMed] [Google Scholar]
- 6.Ciminale V, Pavlakis G N, Derse D, Cunningham C P, Felber B K. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J Virol. 1992;66:1737–1745. doi: 10.1128/jvi.66.3.1737-1745.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciminale V, D’Agostino D M, Zotti L, Franchini G, Felber B K, Chieco-Bianchi L. Expression and characterization of proteins produced by mRNAs spliced into the X region of the human T-cell leukemia/lymphoma virus type II. Virology. 1995;209:445–456. doi: 10.1006/viro.1995.1277. [DOI] [PubMed] [Google Scholar]
- 8.Cockerell G, Rovnak J, Green P L, Chen I S Y. A deletion in the proximinal untranslated pX region of human T-cell leukemia virus type II decreases viral replication but not infectivity in vivo. Blood. 1996;87:1030–1035. [PubMed] [Google Scholar]
- 9.Collins, N. D., and M. D. Lairmore. 1997. Unpublished data.
- 10.Collins N D, Newbound G C, Ratner L, Lairmore M D. In vitro CD4+ lymphocyte transformation and infection in a rabbit model with a molecular clone of human T-cell lymphotropic virus type 1. J Virol. 1996;70:7241–7246. doi: 10.1128/jvi.70.10.7241-7246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 12.Derse D, Mikovits J, Ruscetti F. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology. 1997;237:123–128. doi: 10.1006/viro.1997.8781. [DOI] [PubMed] [Google Scholar]
- 13.Felber B K, Paskalis H, Kleinman-Ewing C, Wong-Staal F, Pavlakis G N. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science. 1985;229:675–679. doi: 10.1126/science.2992082. [DOI] [PubMed] [Google Scholar]
- 14.Franchini G, Mulloy J C, Koralnik I J, Monico A L, Sparkowski J J, Andresson T, Goldstein D J, Schlegel R. The human T-cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16-kilodalton subunit of the vacuolar H+ ATPase. J Virol. 1993;67:7701–7704. doi: 10.1128/jvi.67.12.7701-7704.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank D A, Robertson M J, Bonni A, Ritz J, Greenberg M E. Interleukin 2 signalling involves the phosphorylation of STAT proteins. Proc Natl Acad Sci USA. 1995;92:7779–7783. doi: 10.1073/pnas.92.17.7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gessain A, Barin F, Vernant J-C, Gout O, Maurs L, Calendar A, de The G. Antibodies to human T-lymphotropic virus type I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 18.Green P L, Ross T M, Chen I S Y, Pettiford S. Human T-cell leukemia virus type II nucleotide sequences between env and the last exon of tax/rex are not required for viral replication or cellular transformation. J Virol. 1995;69:387–394. doi: 10.1128/jvi.69.1.387-394.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen M B, Nielsen S E, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 20.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita K, Shirakawa S, Miyoshi I. Adult T-cell leukemia antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981;78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue J, Seiki M, Yoshida M. The second pX product p27X-III of HTLV-1 is required for gag gene expression. FEBS Lett. 1986;209:187–190. doi: 10.1016/0014-5793(86)81108-5. [DOI] [PubMed] [Google Scholar]
- 22.Inoue J, Yoshida M, Seiki M. Transcriptional (p40X) and post transcriptional (p27X-III) regulators are required for the expression and replication of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1987;84:3653–3657. doi: 10.1073/pnas.84.11.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimata J T, Wong F H, Wang J J, Ratner L. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology. 1994;204:656–664. doi: 10.1006/viro.1994.1581. [DOI] [PubMed] [Google Scholar]
- 24.Koralnik I J, Gessain A, Klotman M E, Monico A L, Berneman Z N, Franchini G. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphoma virus type I. Proc Natl Acad Sci USA. 1992;89:8813–8817. doi: 10.1073/pnas.89.18.8813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koralnik I J, Lemp J F, Jr, Gallo R C, Franchini G. In vitro infection of human macrophages by human T-cell leukemia/lymphoma virus type I (HTLV-I) AIDS Res Hum Retroviruses. 1992;8:1845–1849. doi: 10.1089/aid.1992.8.1845. [DOI] [PubMed] [Google Scholar]
- 26.Koralnik I J, Fullen J, Franchini G. The p12I, p13II, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type I open reading frames I and II are localized in three different cellular compartments. J Virol. 1993;67:2360–2366. doi: 10.1128/jvi.67.4.2360-2366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koralnik I J, Mulloy J C, Andresson T, Fullen J, Franchini G. Mapping of the intermolecular association of the human T cell leukemia/lymphotropic virus type I p12I and the vacuolar H+-ATPase 16 kDa subunit protein. J Gen Virol. 1995;76:1909–1916. doi: 10.1099/0022-1317-76-8-1909. [DOI] [PubMed] [Google Scholar]
- 28.Migone T-S, Lin J-X, Cereseto A, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Constitutively activated Jak-STAT pathway in T-cells transformed with HTLV-1. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 29.Mulloy J C, Crowley R W, Fullen J, Leonard W J, Franchini G. The human T-cell leukemia/lymphotropic virus type 1 p12I protein binds the interleukin-2 receptor β and γc chains and affects their expression on the cell surface. J Virol. 1996;70:3599–3605. doi: 10.1128/jvi.70.6.3599-3605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osame M, Usuku K, Izumo S, Ijichi N, Amitini H, Igata A. HTLV-I associated myelopathy: a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 31.Persaud D, Muñoz J L, Tarsis S L, Parks E S, Parks W P. Time course and cytokine dependence of human T-cell lymphotropic virus type 1 T-lymphocyte transformation as revealed by a microtiter infectivity assay. J Virol. 1995;69:6297–6303. doi: 10.1128/jvi.69.10.6297-6303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratner L, Josephs S F, Starcich B, Hahn B, Shaw G M, Gallo R C, Wong-Staal F. Nucleotide sequence analysis of a variant human T-cell leukemia virus (HTLV-Ib) provirus with a deletion in pX-I. J Virol. 1985;54:781–790. doi: 10.1128/jvi.54.3.781-790.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seiki M, Inoue J, Takeda T, Yoshida M. Direct evidence that the p40x of human T-cell leukemia virus type-1 is a trans-activating transcriptional activator. EMBO J. 1986;5:561–565. doi: 10.1002/j.1460-2075.1986.tb04247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takemoto S, Mulloy J C, Cereseto A, Migone T S, Patel B K R, Matsuoka M, Yamaguchi K, Takatsuki K, Kamihara S, White J D, Leonard W J, Waldmann T, Franchini G. Proliferation of adult T cell leukemia/lymphoma cells is associated with constitutive activation of Jak/STAT proteins. Proc Natl Acad Sci USA. 1997;94:13897–13902. doi: 10.1073/pnas.94.25.13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Threadgill D S, Steagall W K, Flaherty M T, Fuller F J, Perry S T, Rushlow K E, Le Grice S F, Payne S L. Characterization of equine infectious anemia virus dUTPase: growth properties of a dUTPase-deficient mutant J. Virol. 1993;67:2592–2600. doi: 10.1128/jvi.67.5.2592-2600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willems L, Kerkhofs P, Dequiedt F, Portetelle D, Mammerickx M, Burney A, Kettmann R. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc Natl Acad Sci USA. 1994;91:11532–11536. doi: 10.1073/pnas.91.24.11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willems L, Kettmann R, Dequiedt F, Portetelle D, Voneche V, Cornil I, Kerkhofs P, Burney A, Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993;67:4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Kang S-H, Heidenreich O, Okerholm M, O’Shea J J, Nerenberg M I. Constitutive activation of different Jak tyrosine kinases in human T-cell leukemia virus type 1 tax protein or virus-transformed cells. J Clin Invest. 1995;96:1548–1555. doi: 10.1172/JCI118193. [DOI] [PMC free article] [PubMed] [Google Scholar]