Abstract
A DNA-binding protein (designated DBP) with an apparent molecular mass of 38 kDa was purified to homogeneity from BmN cells (derived from Bombyx mori) infected with the B. mori nucleopolyhedrovirus (BmNPV). Six peptides obtained after digestion of the isolated protein with Achromobacter protease I were partially or completely sequenced. The determined amino acid sequences indicated that DBP was encoded by an open reading frame (ORF16) located at nucleotides (nt) 16189 to 17139 in the BmNPV genome (GenBank accession no. L33180). This ORF (designated dbp) is a homolog of Autographa californica multicapsid NPV ORF25, whose product has not been identified. BmNPV DBP is predicted to contain 317 amino acids (calculated molecular mass of 36.7 kDa) and to have an isoelectric point of 7.8. DBP showed a tendency to multimerization in the course of purification and was found to bind preferentially to single-stranded DNA. When bound to oligonucleotides, DBP protected them from hydrolysis by phage T4 DNA polymerase-associated 3′→5′ exonuclease. The sizes of the protected fragments indicated that a binding site size for DBP is about 30 nt per protein monomer. DBP, but not BmNPV LEF-3, was capable of unwinding partial DNA duplexes in an in vitro system. This helix-destabilizing ability is consistent with the prediction that DBP functions as a single-stranded DNA binding protein in virus replication.
Nucleopolyhedroviruses (NPVs) have large (80- to 180-kb) circular double-stranded DNA (dsDNA) genomes, which replicate in nuclei of infected cells. Despite the widespread use of NPVs for the expression of foreign genes and their potential for pest control, little is known about the mechanism of their replication and the properties of their replication factors. The most widely studied baculovirus, Autographa californica multicapsid NPV (AcMNPV), has the potential to encode about 150 proteins (3), including factors required for virus DNA replication. The products of nine viral genes (ie-1, ie-2, lef-1, lef-2, lef-3, dnahel, dnapol, p35, and lef-7 or pe-38) are necessary and sufficient for efficient replication of transfected plasmid DNAs containing a putative baculovirus replication origin (16, 22). It is likely that DNA polymerase and DNA helicase, which are encoded by the viral genes dnapol and dnahel, respectively (20, 35), form a core of the virus DNA replication machinery. The roles of other factors are less obvious. Single-stranded DNA binding (SSB) protein function was proposed for the protein LEF-3, which binds specifically single-stranded DNA (ssDNA) (10, 14). However, direct proof for the SSB function of LEF-3 in viral DNA replication is lacking. In addition, SSB function was also suggested for LEF-7 on the basis of its predicted amino acid sequence (22). It was recently demonstrated that LEF-1 forms a complex with LEF-2 and may serve as a DNA primase (9). The function of IE-1, IE-2, and PE-38 may result from their ability to activate in trans expression of other genes required for virus replication. The transactivator IE-1 may also participate in the initiation of DNA replication, due to its ability to bind putative replication origins (7, 13, 17, 33). P35 is an inhibitor of apoptosis and may not be involved directly in DNA replication. Its stimulatory effect in the transient-replication assay may result from inhibition of virus-induced apoptosis in cells transfected with the replication genes. Several genes required for DNA replication (six essential and three stimulatory) were also identified in the genome of Orgyia pseudotsugata NPV (1). Homology of these genes to those required for replication of AcMNPV suggests similar replication mechanisms for the two viruses. The genome organization of the Bombyx mori NPV (BmNPV) closely resembles that of AcMNPV. Nineteen homologs of the AcMNPV late expression factor genes (lef genes) were identified in BmNPV (12). At least three of these, ie-2, lef7, and p35, are not essential for virus DNA replication as demonstrated by deletion analysis (12). Because the daughter DNA molecules synthesized under control of the nine essential viral genes appear to be synthesized as concatemers (16, 22, 31, 32), factors required for maturation of nascent DNA and its further processing are still unknown. Although the nine AcMNPV factors were sufficient for efficient DNA replication in Sf cells, an additional viral gene, designated hcf-1, was essential for replication in TN-368 cells (21), indicating dependence of the transient assay on host cell-specific factors. Few proteins involved in NPV DNA replication have been purified from infected cells and characterized in cell-free systems. Among them are AcMNPV DNA polymerase (28, 37), BmNPV DNA polymerase (27), AcMNPV DNA helicase (19), and AcMNPV LEF-3 (10, 14). Isolation of other replication proteins of NPVs is still anticipated.
In this report we describe the purification of a viral DNA-binding protein (designated DBP) from BmNPV-infected cells. DBP binds preferentially to ssDNA and is capable of unwinding duplex DNA. The BmNPV open reading frame (ORF) encoding DBP (dbp gene) is a homolog of AcMNPV ORF25, whose product has not been identified so far.
MATERIALS AND METHODS
Purification of DBP.
Monolayers of BmN (silkworm B. mori) cells were cultured as described earlier (23, 24). The cells (5 × 108) were infected with BmNPV (isolate T3) (23) at a multiplicity of infection of 10 in TC-100 Insect medium (Sigma) supplemented with 10% fetal bovine serum and harvested 26 h postinfection. Infected cells were collected by centrifugation, washed three times with phosphate-buffered saline and once with hypotonic buffer A containing 20 mM HEPES (pH 7.5), 5 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol (DTT), and a set of protease inhibitors (0.2 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 2 μg of E64 per ml, and 2 mM benzamidine), and centrifuged again. The cells were resuspended in an equal volume of buffer A, allowed to swell on ice for 10 min, and then lysed by 15 strokes of a tight-fitting pestle in a Wheaton Dounce homogenizer. The homogenate was centrifuged at 2,000 × g for 10 min to pellet nuclei. The nuclei were resuspended in an equal volume of buffer A containing 3.4 M NaCl, transferred into a 4-ml centrifuge tube, and then incubated on ice for 1 h. The extract was centrifuged at 100,000 × g for 1 h, and the supernatant was dialyzed for 2.5 h against two changes of buffer B (20 mM HEPES [pH 7.5], 10% glycerol, 0.1 M NaCl, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, and the set of protease inhibitors). The extract was clarified by centrifugation at 100,000 × g for 15 min, adjusted to 10 mM EDTA, and loaded onto a 2-ml column of ssDNA cellulose (Sigma) equilibrated with buffer C (10 mM HEPES [pH 7.5], 10% glycerol, 0.1 M NaCl, 1 mM EDTA, 1 mM DTT, and the set of protease inhibitors). The column was washed with 10 ml of buffer C and then successively processed with 10-ml portions of buffer C containing NaCl at final concentrations of 0.2, 0.4, 0.6, 0.7, 0.8, 1.0, 1.4, and 2.0 M. Proteins from each fraction were analyzed by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS–12% PAGE) followed by staining with Coomassie brilliant blue or by Western blotting. After analysis of the protein pattern, the fractions collected were either directly used for experiments or the protein was purified further. In the latter case, the samples were dialyzed against an excess of buffer containing 10 mM Tris-HCl (pH 8.5), 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.4 mM phenylmethylsulfonyl fluoride, and 2 mM benzamidine and were loaded onto a DEAE-Toyopearl 650 (Tosoh) column (0.5 by 2.7 cm) equilibrated with the same buffer. Proteins were eluted from the column by using an NaCl gradient in the buffer, and fractions containing DBP were collected at 30 to 50 mM NaCl. The samples were dialyzed against buffer D (10 mM Tris-HCl [pH 7.5], 50% glycerol, 0.1 mM EDTA, 1 mM DTT, and the set of protease inhibitors) and stored at −20°C. Protein concentrations were determined by SDS-PAGE of portions from the samples followed by optical densitometry of the gel stained with Coomassie brilliant blue. Bovine serum albumin (Bio-Rad) loaded in different amounts on separate lanes of the same gel was used for generation of the calibration curve.
Purification of LEF-3.
The fraction collected at 0.6 M NaCl in the course of chromatography of the high-salt extract from BmNPV-infected BmN cells on ssDNA cellulose (see above) was used as a source of BmNPV LEF-3. It was dialyzed against buffer C and loaded onto an ssDNA agarose column (0.5 by 3.8 cm) equilibrated with the same buffer. The column was washed with 8 ml of buffer C containing 0.4 M NaCl and then successively processed with 2.4-ml portions of buffer C containing NaCl in final concentrations of 0.5, 0.6, 0.7, 0.8, and 1.0 M. Each portion was collected into three fractions. Proteins from each fraction were analyzed by SDS–12% PAGE followed by staining with Coomassie brilliant blue. LEF-3 was eluted by 0.6 to 0.8 M NaCl. Fractions enriched in LEF-3 were combined and dialyzed against buffer containing 10 mM Tris-HCl (pH 7.5), 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.4 mM phenylmethylsulfonyl fluoride, and 2 mM benzamidine and loaded onto a DEAE-Toyopearl column (0.5 by 2.7 cm) equilibrated with the same buffer. The column was processed with an NaCl gradient in the buffer, and fractions containing LEF-3 were collected at 0.16 to 0.2 M NaCl. The sample was dialyzed against buffer D and stored at −20°C.
PAGE and Western blotting.
SDS–12% PAGE was performed as described by Laemmli (18). Gels were either fixed and stained with Coomassie brilliant blue or electrophoretically transferred to Clear-blot membrane-p by using a semidry-blot apparatus (Atto) according to the manufacturer’s guidelines. Western blots were probed with a 1:3,000 dilution of rabbit polyclonal antiserum to AcMNPV LEF-3 (10) (a gift from George F. Rohrmann), washed, incubated with a 1:5,000 dilution of goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (ICN Pharmaceuticals), and developed by using an ECL detection system (Amersham) according to the manufacturer’s instructions.
Protein sequencing.
Protein bands corresponding to DBP were identified by staining with Coomassie brilliant blue after SDS–12% PAGE of the purified protein (5.6 μg). Pieces of the gel containing the desired band were excised, transferred to a 1.5-ml Eppendorf tube, and soaked in 0.15 ml of buffer containing 0.1 M Tris-HCl (pH 9.0), 2 mM EDTA, and 0.1% SDS. The protein was digested by treatment with 0.5 μg of Achromobacter protease I for 16 h at 37°C. The sample was clarified by centrifugation in a Suprec-01 filter unit (Takara) and adjusted to pH 2 to 3 by the addition of 70% formic acid. It was then fractionated by high-pressure liquid chromatography on a DEAE column (2 by 30 mm; Senshu Scientific), followed in tandem by a PEGASIL-300 C8 column (2 by 100 mm; Senshu Scientific), with an HP 1100 chromatographic system (Hewlett-Packard). The columns were processed by using a linear gradient of 0 to 60% solvent B (80% acetonitrile, 0.069% trifluoroacetic acid) mixed with solvent A (0.075% trifluoroacetic acid) at a flow rate of 0.2 ml/min for 48 min. The purified peptides were subjected to sequence analysis on a 473A protein sequencer (Applied Biosystems) according to the manufacturer’s instructions. Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectral analysis was performed with a MALDI-TOF REFLEX mass spectrometer (Bruker) with alpha-cyano-4-hydroxycinnamic acid (Sigma) as a matrix.
Mobility shift assay and unwinding assay.
The following oligonucleotides were used as the substrates in binding reactions: 17-mer (TGCCGGGATCATAGAAG), 36-mer (GCAGTGTAGCCACACAGAGTGCCGGGATCATAGAAG), 51-mer(a) (AAGCGGAGTGTATGTGCAGTGTAGCCACACAGAGTGCCGGGATCATAGAAG), 51-mer(b) (CTTCTATGATCCCGGCACTCTGTGTGGCTACACTGCACATACACTCCGCTT), and 64-mer (GATTTCATCTAGCCTTCTATGATCCCGGCACTCTGTGTGGCTACACTGCACATACACTCCGCTT). The oligonucleotides were labeled at the 5′ ends with 32P by using T4 polynucleotide kinase (Takara) and [γ-32P]ATP (ICN Radiochemicals). Unless noted otherwise, binding reactions were carried out in buffer E (10 mM Tris-HCl [pH 8.0], 0.15 M NaCl, 2 mM DTT, 100 μg of bovine serum albumin per ml). Reaction mixtures of 10 μl contained 0.001 to 0.15 pmol of 32P-labeled oligonucleotide probe and 3 μl of protein sample in buffer D. After mixing of the components at 0°C, reaction mixtures were incubated for 15 min at 23°C. Half of each reaction mixture was loaded onto a 6% polyacrylamide (acrylamide-bisacrylamide, 60:1) slab gel (6 by 10 by 0.075 cm) prepared in a buffer containing 20 mM HEPES (pH 8.0) and 0.1 mM EDTA. Electrophoresis was performed and autoradiographs were obtained as previously described (26).
For unwinding assays, DNA duplexes were prepared by annealing of the labeled 17-mer or 36-mer to a 1.2-fold molar excess of the unlabeled 51-mer(b) (*17:51-mer and *36:51-mer duplexes) or by annealing of the labeled 51-mer(a) to a 1.2-fold molar excess of the unlabeled 64-mer (*51:64-mer duplex). Unwinding reactions were carried out in buffer E without NaCl. Reaction mixtures of 10 μl contained 0.005 pmol of the DNA duplex and 3 μl of protein sample in buffer D. After incubation at 23°C, reaction mixtures were treated with 1% SDS and 0.5 mg of proteinase K per ml for 20 min at 23°C and analyzed by electrophoresis in 6% polyacrylamide gels as described above.
Sedimentation in glycerol gradients.
Linear 15 to 30% glycerol gradients were prepared in buffer F (10 mM Tris-HCl [pH 7.5], 0.4 M NaCl, 1 mM EDTA, 1 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride). Protein samples (0.15 ml), after dialysis against buffer F containing 10% glycerol, were layered over 4.0 ml of a glycerol gradient prepared in nitrocellulose tubes for an SW 60.Ti rotor (Beckman). Ovalbumin (45 kDa, 3.6S) and aldolase (158 kDa, 7.4S) (0.2 mg of each) were centrifuged in individual tubes as sedimentation standards. After centrifugation in the SW 60.Ti rotor at 55,000 rpm and 4°C for 24 h, the gradients were fractionated from the bottom with a peristaltic pump.
RESULTS
Purification of DBP.
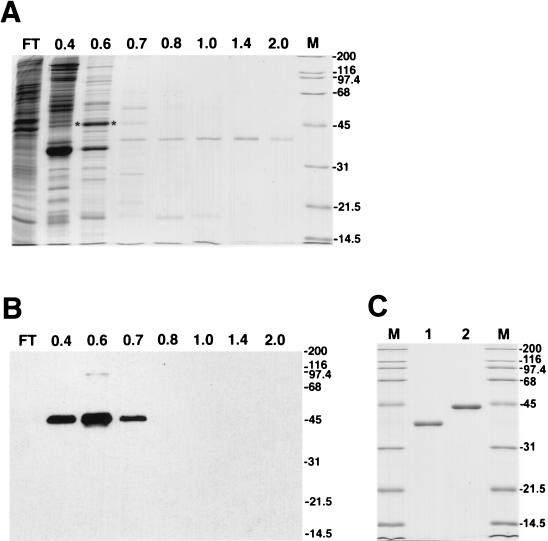
In a search for BmNPV products essential for virus DNA replication, we analyzed for viral proteins possessing a high affinity for ssDNA. The DNA-binding proteins were detected in high-salt extracts of nuclei from BmNPV-infected BmN cells by using chromatography on ssDNA cellulose (Fig. 1). Most of the proteins that bound to ssDNA were eluted from the column at NaCl concentrations lower than 0.6 M. An abundant protein of 44 kDa eluted by 0.6 M NaCl was identified as BmNPV LEF-3 by Western blotting with polyclonal antiserum against AcMNPV LEF-3 (Fig. 1B). The antiserum specifically recognized a protein with an apparent molecular mass of 44 kDa that appeared predominantly in the 0.6 M NaCl fraction (Fig. 1A). One major protein (designated DBP) was present in fractions that eluted at NaCl concentrations higher than 1.0 M (Fig. 1A). It migrated with an apparent molecular mass of 38 kDa. As shown by optical densitometry of the Coomassie blue-stained polyacrylamide gel, ∼97% of the protein in the 1.4 M NaCl fraction and ∼93% of the protein in the 2.0 M NaCl fraction was associated with this single 38-kDa polypeptide band. The total yield of DBP in both fractions from 5 × 108 cells was about 30 μg. For further purification of DBP, the fractions collected after chromatography on ssDNA cellulose were subjected to chromatography on DEAE-Toyopearl. The protein showed a low affinity for the DEAE-resin, but most was retained on the column after loading in the absence of monovalent salt. DBP was eluted by 30 to 50 mM NaCl, and based on SDS-PAGE, it was purified to a homogeneous state (Fig. 1C, lane 1). To compare DBP with a known viral protein also possessing high affinity for ssDNA, we purified LEF-3 from BmNPV-infected BmN cells to a homogeneous state as described in Materials and Methods (Fig. 1C, lane 2). The molecular mass of BmNPV LEF-3 estimated by SDS-PAGE, 44 kDa, corresponds well to the calculated value of 44.8 kDa for the polypeptide predicted to be encoded by BmNPV ORF55, the BmNPV lef-3 gene (12) (GenBank accession no. L33180).
FIG. 1.
Purification of DNA-binding proteins of BmNPV. (A and B) ssDNA cellulose chromatography of nuclear extract from BmNPV-infected BmN cells. The flowthrough (FT) and fractions eluted at the indicated molar concentrations of NaCl were analyzed by SDS–12% PAGE followed by staining with Coomassie brilliant blue (A) or by Western blotting with antiserum to LEF-3 (B). (C) Proteins DBP and LEF-3 after final purification on DEAE-Toyopearl. DBP (0.6 μg) (lane 1) and LEF-3 (0.6 μg) (lane 2) were analyzed by SDS–12% PAGE followed by staining with Coomassie brilliant blue. The migration of molecular size markers (in kilodaltons) is shown in lanes M in panels A and C and on the right side of the blot in panel B. The asterisks show the position of LEF-3 in panel A.
The 38-kDa DNA-binding protein has not been previously identified in insect cells infected with NPVs. The absence of this polypeptide from extracts of uninfected cells suggested that it was of viral origin (data not shown). Therefore, we decided to obtain additional information about this protein presumably encoded by the virus genome.
Identification of DBP as a product of the BmNPV genome.
To identify the source of the 38-kDa DNA-binding protein, we partially or completely sequenced six peptides obtained after digestion with Achromobacter protease I, which cleaves proteins at lysines. All of them were shown to belong to a polypeptide predicted to be encoded by BmNPV ORF16, located at nucleotides (nt) 16189 to 17139 in the BmNPV (isolate T3) genome (GenBank accession no. L33180) (Fig. 2). This ORF was designated the dbp gene. The molecular masses of three other peptides (Fig. 2) were determined by mass spectrometry (MALDI). The values obtained by MALDI were in agreement with molecular masses predicted for digestion products of DBP. The BmNPV dbp gene is predicted to encode a protein of 317 amino acids (36.7 kDa) with an isoelectric point of 7.8. The apparent molecular mass of 38 kDa estimated for DBP by SDS-PAGE corresponds to the mass predicted from the BmNPV dbp gene. The nucleotides surrounding the start codon ATG are consistent with the Kozak rule (AXXATG[A/G]). A putative RNA polymerase II promoter (TATA) is present at nt −69, and a sequence (CAAT) similar to the baculovirus early start site (CAGT) is found 28 nt downstream (Fig. 2). This suggests that dbp may be expressed as an early gene. An ORF homologous to BmNPV dbp in the genome of the closely related virus AcMNPV, ORF25, was described earlier (3). AcMNPV ORF25 potentially encodes a protein of 316 amino acids, which showed 95.9% identity with BmNPV DBP. A similar ORF, ORF43, was also found in the O. pseudotsugata MNPV (2). To date, the mRNA species and protein products encoded by these ORFs have not been identified in NPV-infected cells. BmNPV DBP does not show high homology to any nonbaculovirus protein in the gene bank.
FIG. 2.
DBP is encoded by the BmNPV genome. The nucleotide sequence of BmNPV (isolate T3) dbp (from GenBank; accession no. L33180) is shown. The predicted amino acid sequence of DBP is shown above the nucleotide sequence. Stop codons are indicated by the asterisks. The numbers on the right are amino acid positions. Six peptides were subjected to protein sequencing, and the molecular masses of three peptides were estimated by mass spectrometry (MALDI). The determined amino acid sequences are underlined with thick lines, and those determined by MALDI are underlined with thin lines.
Mobility shift assay of the interaction of DBP with DNA.
Binding of DBP to ssDNA (Fig. 1A) might reflect the biological function of this protein. To study the interaction of DBP with DNA in detail, we used electrophoretic mobility shift analysis. The protein was incubated with 5′-end-labeled 51-mer or 64-mer oligonucleotides, and the products were subjected to nondenaturing PAGE (see Materials and Methods). Before beginning the mobility shift experiments, preparations of DBP were tested for endonucleolytic activity. For this assay, the protein (2.5 μg/ml) was incubated with M13mp7 ssDNA (10 μg/ml) or with pSK+ dsDNA (10 μg/ml) in a reaction mixture containing 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 2 mM DTT for 1 h at 37°C. DNA was then extracted with phenol and phenol-chloroform (1:1), precipitated with ethanol, and analyzed by electrophoresis in a 0.7% agarose gel. No degradation of M13 DNA or conversion of pSK+ DNA from replicative form I (RFI) into RFII (or RFIII) was observed (data not shown).
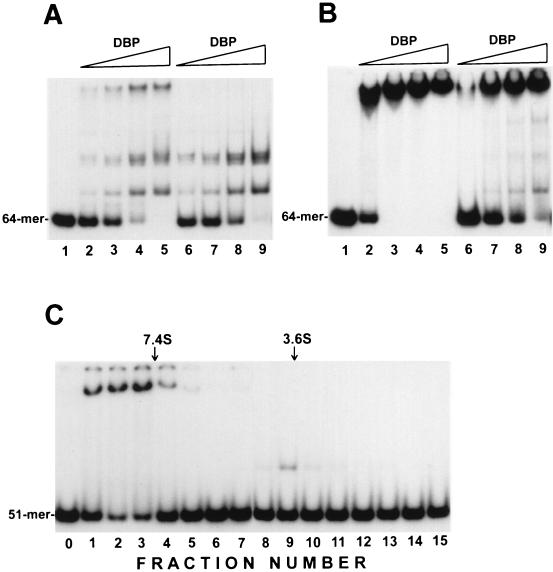
In the first series of binding reactions, we used DBP from the 1.4 M NaCl elution fraction after chromatography on ssDNA cellulose (Fig. 1A). Binding of DBP to the 64-mer in the absence of monovalent salt resulted in the appearance of three major shifted species (Fig. 3A, lanes 2 to 5). Similar retardation patterns were observed after binding reactions with the 51-mer(b) oligonucleotide (data not shown), and these oligonucleotides were used interchangeably. The first species migrating above free oligonucleotide may be a complex of the oligonucleotide with a monomer of DBP, whereas the slowly migrating band in the upper part of the gel may result from binding of the oligonucleotide to a large protein aggregate. Evidence for this interpretation was provided by glycerol gradient fractionation of the DBP sample before the mobility shift assay (Fig. 3C). The protein fraction that cosedimented with ovalbumin (45 kDa, 3.6S) interacted to form the first band above the free oligonucleotide in the mobility shift assay (Fig. 3C, lane 9), suggesting that the respective complexes contain monomers of DBP. The DBP fraction that sedimented ahead of aldolase (158 kDa, 7.4 S) provided the slowly migrating band in the gel (Fig. 3C, lanes 1 to 4), indicating that the respective complexes are formed by the oligonucleotide binding to DBP multimers. The fractionation procedure altered the proportions of different oligomeric forms of DBP, because after centrifugation, the second band above the free oligonucleotides was not visible, whereas the relative amount of slowly migrating complexes was increased. The DBP multimers were presumably due to protein-protein interactions, because pretreatment of the DBP sample prior to centrifugation with an equimolar amount of DNase I did not change the protein sedimentation pattern in glycerol gradients (data not shown). The proportion of DBP oligomers in the DBP preparations was also influenced by the purification procedures. The DBP sample collected after chromatography on DEAE-Toyopearl contained predominantly protein multimers. Only one, slowly migrating band was formed in the absence of salt (Fig. 3B, lanes 1 to 5). Addition of NaCl to the reaction decreased the amount of oligonucleotides bound to the protein aggregates and caused the appearance of the faster-migrating species. Thus, after overnight incubation of the DBP sample at 0.5 M NaCl and subsequent binding in 0.15 M NaCl, complexes of oligonucleotides with protein monomers and higher oligomers were detectable (Fig. 3B, lanes 6 to 9). The inhibition effect of NaCl on generation of the slowly migrating complexes was less pronounced with the DBP sample collected after DEAE-Toyopearl chromatography than with that purified by using ssDNA cellulose (compare Fig. 3A and B). In the latter case, addition of 0.15 M NaCl to the binding mixture precluded the appearance of the slowly migrating species (Fig. 3A, lanes 6 to 9). Therefore, multimerization of DBP and stability of DBP multimers appeared to depend on the concentration of monovalent salts, but this question was not investigated further. The presence of DBP multimers hampered the quantitative estimation of the molar concentration of active protein in the mobility shift assay. Saturation of the oligonucleotide substrates with protein was observed in the presence of a molar excess of DBP.
FIG. 3.
Electrophoretic mobility shift assay of DNA-binding activity of DBP. Reaction mixtures (10 μl) containing 5′-32P-labeled oligonucleotide probe and purified DBP (see Materials and Methods) were incubated for 15 min at 23°C. Portions (5 μl) of reaction mixtures were analyzed by electrophoresis in 6% polyacrylamide gels. (A) Binding of ssDNA cellulose-purified DBP to the 64-mer. Reaction mixtures contained 0.004 pmol of 64-mer and the following amounts of DBP: lanes 2 and 6, 1.7 ng; lanes 3 and 7, 5.2 ng; lanes 4 and 8, 10.4 ng, and lanes 5 and 9, 20.7 ng. No protein was added to the reaction shown in lane 1. Reaction mixtures were incubated in the absence of NaCl (lanes 1 to 5) or in the presence of 0.15 M NaCl (lanes 6 to 9). (B) Binding of DBP collected after chromatography on DEAE-Toyopearl to the 64-mer. Reaction mixtures contained 0.01 pmol of 64-mer and the following amounts of DBP: lanes 2 and 6, 3.6 ng; lanes 3 and 7, 10.6 ng; lanes 4 and 8, 21.4 ng; and lanes 5 and 9, 43 ng. No protein was added to the reaction shown in lane 1. Reaction mixtures were incubated in the absence of NaCl (lanes 1 to 5), or the protein was preincubated for 16 h in the presence of 0.5 M NaCl before addition to the reaction mixtures (final concentration, 0.15 M NaCl) (lanes 6 to 9). (C) Glycerol gradient fractionation of DBP oligomers. DBP (1 μg in 150 μl) collected after chromatography on ssDNA cellulose was layered over 4.0 ml of a gradient of 15 to 30% glycerol in buffer containing 10 mM Tris-HCl (pH 7.5), 0.4 M NaCl, 1 mM EDTA, 1 mM DTT, and 0.2 mM phenylmethylsulfonyl fluoride and was centrifuged in an SW 60.Ti rotor at 55,000 rpm and 4°C for 24 h. The gradient was fractionated from the bottom into 16 fractions, and DNA-binding activity in 5-μl portions was determined for each fraction added to the reaction mixtures containing 0.001 pmol of 5′-labeled 51-mer(b). The positions of the standards centrifuged in separate tubes are shown by arrows: aldolase, 158 kDa, 7.4S (fraction 3.5); ovalbumin, 45 kDa, 3.6S (fraction 9.3). No protein was added to the reaction shown in lane 0.
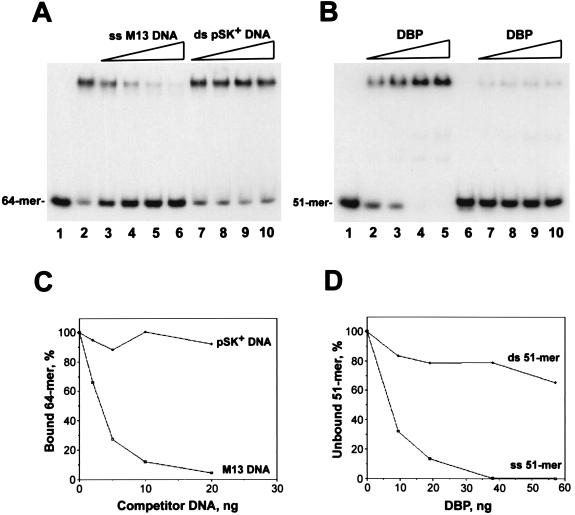
To verify the binding specificity of DBP for ssDNA, we compared the efficiencies of M13 ssDNA and pSK+ dsDNA (RFII) as competitors in binding reactions with the 5′-end-labeled oligonucleotide (Fig. 4A and C). The ssDNA appeared to be at least 1 order of magnitude more efficient a competitor than dsDNA, indicating much higher affinity of DBP for ssDNA than for dsDNA. The higher affinity of DBP for ssDNA was confirmed by a direct comparison of DBP binding to the single-stranded 51-mer oligonucleotide versus the 51-mer duplex (Fig. 4B and D). The preference for binding to ssDNA was demonstrated for preparations of DBP with different proportions of oligomeric forms, including the sample collected after chromatography on ssDNA cellulose, which contained monomers and higher oligomers (data not shown).
FIG. 4.
DBP binds preferentially to ssDNA. Reaction mixtures (10 μl) containing 5′-32P-labeled oligonucleotide probe and DBP collected after chromatography on DEAE-Toyopearl (see Materials and Methods) were incubated in the presence of 0.15 M NaCl for 15 min at 23°C. Portions (5 μl) of reaction mixtures were analyzed by electrophoresis in 6% polyacrylamide gels. (A and C) Competition analysis of DBP with ssDNA and dsDNA. (A) Reaction mixtures contained 0.047 pmol (1 ng) of 64-mer, 30 ng of DBP, and competitor DNA (M13mp9 ssDNA [lanes 3 to 6] or pSK+ dsDNA [RFII] [lanes 7 to 10]) added in the following amounts: lanes 3 and 7, 2 ng; lanes 4 and 8, 5 ng; lanes 5 and 9, 10 ng; and lanes 6 and 10, 20 ng. No competitor was added to the reaction shown in lane 2; no protein was added to the reaction shown in lane 1. (C) Amounts of the bound 64-mers estimated by optical densitometry of an autoradiograph of the gel in panel A. The amount of the bound 64-mers in the absence of competitor (lane 2 in panel A) was taken as 100%. (B and D) Comparison of DBP binding to the single-stranded 51-mer and to the 51-mer duplex. (B) Reaction mixtures contained 0.006 pmol of 51-mer(a) (lanes 1 to 5) or 0.006 pmol of 51-mer duplex obtained by annealing of 51-mer(a) to complementary 51-mer(b) (lanes 6 to 10) and the following amounts of DBP: lanes 2 and 7, 9.5 ng; lanes 3 and 8, 19 ng; lanes 4 and 9, 38 ng; and lanes 5 and 10, 57 ng. No protein was added to the reactions shown in lanes 1 and 6. (D) Amounts of the unbound 51-mers estimated by optical densitometry of an autoradiograph of the gel in panel B.
In similar experiments, we studied binding of BmNPV LEF-3 to 5′-end-labeled oligonucleotides. In agreement with data published previously for AcMNPV LEF-3 (14), BmNPV LEF-3 showed specific binding to ssDNA (data not shown).
Protection by DBP against exonucleolytic digestion of oligonucleotides.
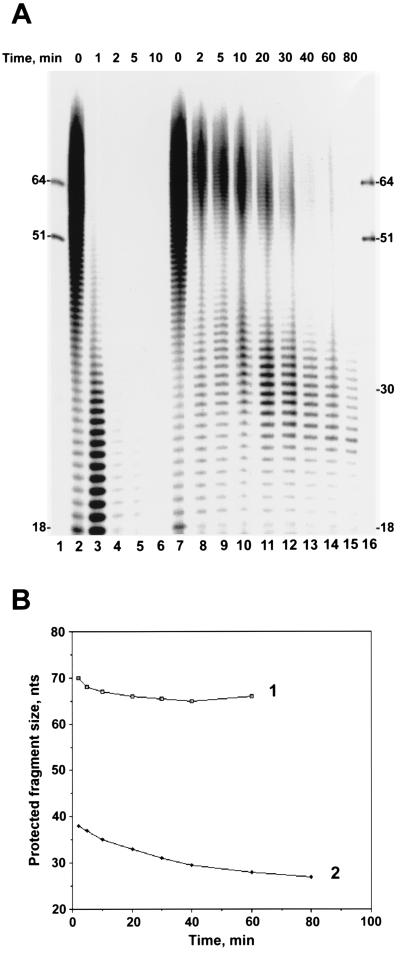
Tightly bound proteins are capable of dramatically changing the capacity of DNA molecules to participate in biological reactions. One of the known effects of SSB proteins is to protect against exonucleolytic digestion of the bound polynucleotide. The size of the DNA fragment protected by the protein monomer provides an estimate of the binding site size for the protein (26). In our study of the biochemical properties of BmNPV DBP, we analyzed the effect of this protein on exonucleolytic digestion of oligonucleotides. Phage T4 DNA polymerase, possessing a potent 3′→5′ exonucleolytic activity specific for ssDNA, was used as the source of exonuclease. To eliminate the effects of oligonucleotide sequence content on the digestion reaction, we used a set of oligo(dT)n with n varying from 12 to ∼100 nt for the DNA substrate. The experimental design was similar to that described previously (26). In the absence of DBP, phage T4 DNA polymerase efficiently digested oligo(dT)n fragments from 3′ ends (Fig. 5A, lanes 2 to 6). No fragments longer than 17 nt persisted in the reaction after a 5-min incubation. In the second reaction, oligo(dT)n was preincubated before the addition of phage T4 DNA polymerase with the preparation of DBP collected after chromatography on ssDNA cellulose and contained monomers and higher oligomers. The retardation pattern for complexes of the 64-mers with the DBP sample used in this experiment is shown in Fig. 3A. Saturation of oligo(dT)n with DBP markedly decreased the digestion rate and revealed two sets of oligonucleotides most efficiently protected. One set of protected fragments was about 66 nt in length. It was visible for about 60 min and disappeared later. Another set of protected fragments was about 30 nt. Although the amount of protected fragments progressively decreased in the course of reaction, the protection pattern was still visible after 80 min. During the last 60 min of the reaction (Fig. 5A, lanes 11 to 15), the mean size of the protected fragments in this set was reduced by only 6 nt, from 33 to 27 nt (Fig. 5B). The data obtained suggests that monomers of DBP are capable of protecting about 30 nt of DNA. This suggests that the binding site size for the DBP monomer is about 30 nt.
FIG. 5.
DBP protects bound DNA against exonucleolytic hydrolysis. (A) Time course of digestion of dT12–100 with 3′→5′ exonuclease associated with phage T4 DNA polymerase. Two reaction mixtures contained 6 pmol (total nucleotides) of 5′-32P-labeled dT12–100 in a final volume of 50 μl of 10 mM Tris-HCl (pH 7.5)–5 mM MgCl2–0.15 M NaCl–2 mM DTT–100 μg of bovine serum albumin per ml. After incubation for 15 min at 23°C in the absence of DBP (reaction I) or in the presence of 180 ng of DBP (reaction II), 5-μl portions were removed to serve as starting points (lanes 2 and 7, respectively), and then the reaction mixtures were transferred to 30°C and 1 μl (0.8 U) of phage T4 DNA polymerase was added. At the indicated times of incubation, 5-μl portions from the reaction mixtures were transferred onto ice and mixed with 3.5 μl of loading buffer. Portions (4 μl) from each sample were subjected to electrophoresis in a 10% polyacrylamide–8 M urea gel. Migration of the standards, 5′-32P-labeled 51-mer(b) and 64-mer oligonucleotides, is shown in lanes 1 and 16. DBP was collected after chromatography on ssDNA cellulose. (B) Changes in the average sizes of dTn oligonucleotides in two groups of fragments protected by DBP in the course of digestion with 3′→5′ exonuclease associated with phage T4 DNA polymerase. Values calculated for the gel shown in panel A were used for generation of curves 1 and 2.
Destabilization effect of DBP on duplex DNA.
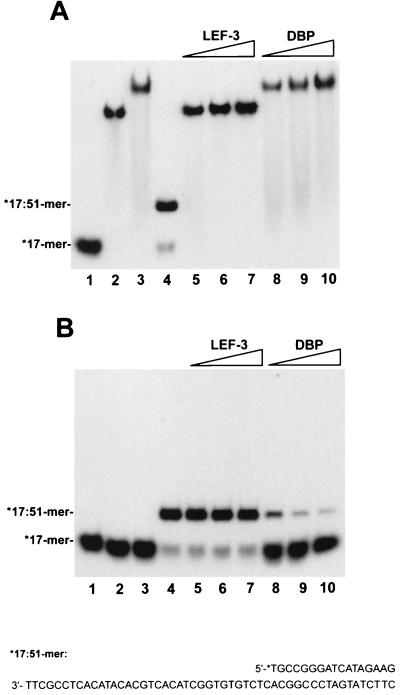
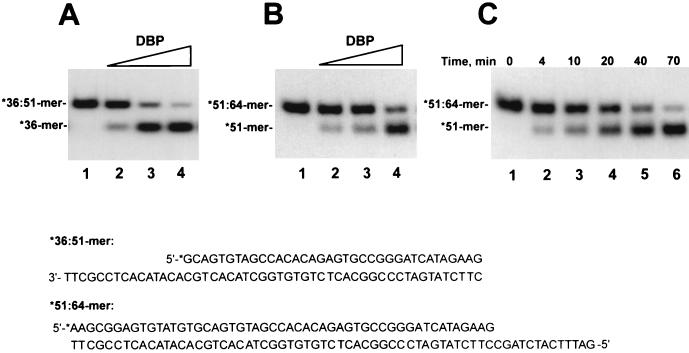
The SSB proteins belong to the group of proteins known as helix-destabilizing proteins. SSB proteins are capable of unwinding DNA duplexes in an ATP-independent manner in in vitro systems (4, 8, 11, 15, 29, 34, 36, 39). To elucidate a possible role for DBP in viral DNA replication, we assayed a destabilization effect of DBP in reaction mixtures containing a partial duplex DNA as the substrate. The first substrate was prepared by annealing a 17-mer oligonucleotide, labeled at 5′ end with 32P, to the unlabeled 51-mer(b) oligonucleotide (see Materials and Methods). In this *17:51-mer, the 34-nt single-stranded region provided a site for binding of proteins. LEF-3, another viral protein specifically binding ssDNA, was used in parallel experiments. Because the melting effect of SSB proteins is extremely sensitive to monovalent salts and MgCl2 (11), the binding reactions were performed in the absence of NaCl. The proteins, DBP and LEF-3, efficiently bound both the free 17-mer oligonucleotide and the 17:51-mer partial duplex, as revealed in the mobility shift assay (Fig. 6A). The electrophoretic mobility of the complexes with free 17-mer did not differ significantly from the mobility of the complexes with 17:51-mer, because both proteins bind DNA as high-molecular-mass oligomers (see above and reference 10). To determine whether the 17-mer was still bound by hydrogen bonds to the 51-mer after binding of the protein to the 17:51-mer, the proteins were inactivated prior to the electrophoresis by treatment with 1% SDS and 0.5 mg of proteinase K per ml (Fig. 6B). Saturation of the 17:51-mer with LEF-3 did not cause detachment of the 17-mer from the complementary 51-mer (Fig. 6B, lanes 5 to 7). The amount of LEF-3 added to the reaction shown in lanes 5 was enough to saturate most 17:51-mers; however, even a fourfold increase in the LEF-3 concentration did not produce any melting (lanes 7). In contrast, binding of DBP to 17:51-mers resulted in detachment of 17-mers (Fig. 6B, lanes 8 to 10). To test for the ability of DBP to melt more stable duplexes, the unwinding reactions were performed with the *36:51-mer and *51:64-mer partial duplexes (Fig. 7). In the *36:51-mer, the single-stranded region for DBP binding was 15 nt in length, and it was adjacent to the 5′ end of the 36-mer. In the *51:64-mer, the single-stranded region was only 13 nt in length, and it was adjacent to the 3′ end of the 51-mer. DBP caused unwinding of both partial duplexes in a dose-dependent manner (Fig. 7A and B). The unwound products were accumulated at a low rate in the reaction carried out at 23°C (Fig. 7C). Therefore, DBP could unwind partial DNA duplexes with either 3′ or 5′ single-stranded tails. This suggests that the helix-destabilizing effect of DBP is nonpolar with respect to the orientation of the single-stranded tail.
FIG. 6.
DBP destabilizes duplex DNA. Reaction mixtures (10 μl) containing free 5′-32P-labeled 17-mer oligonucleotide or this oligonucleotide annealed to a 51-mer(b) oligonucleotide and other components (see Materials and Methods) were incubated with LEF-3 or DBP for 15 min at 23°C. Lanes 1 to 3 contained 0.005 pmol of 17-mer mixed with 0.006 pmol of noncomplementary 51-mer(a); lanes 4 to 10 contained 0.005 pmol of 17-mer annealed to 0.006 pmol of complementary 51-mer(b). No protein was added to the reactions shown in lanes 1 and 4. Other lanes contained LEF-3 at 8 ng (lane 5), 20 ng (lane 6), and 34 ng (lanes 2, 7) and DBP at 14 ng (lane 8), 34 ng (lane 9), and 57 ng (lanes 3, 10). LEF-3 and DBP were collected after chromatography on DEAE-Toyopearl. Portions (5 μl) of reaction mixtures were analyzed by electrophoresis in 6% polyacrylamide gels without additional treatment (A) or after incubation with 1% SDS and 0.5 mg of proteinase K per ml for 20 min at 23°C (B). The sequence of the *17:51-mer is shown at the bottom. The asterisk indicated the radioactive label.
FIG. 7.
DBP unwinds the 36:51-mer and 51:64-mer partial duplexes. (A and B) Dose dependence of the unwinding reaction. Reaction mixtures (10 μl) containing 0.005 pmol of 32P-labeled *36:51-mer (A) or *51:64-mer (B) and other components of the unwinding assay (see Materials and Methods) were incubated with DBP for 30 min at 23°C. No protein was added to the reactions shown in lanes 1. Other reaction mixtures contained the following amounts of DBP collected after chromatography on DEAE-Toyopearl: lanes 2, 2.9 ng, lanes 3, 7.6 ng, and lanes 4, 19 ng. After treatment with 1% SDS and 0.5 mg of proteinase K per ml for 20 min at 23°C, 5-μl portions of reaction mixtures were analyzed by electrophoresis in a 6% polyacrylamide gel. (C) Time course of unwinding of the 51:64-mer partial duplex. A 49-μl reaction mixture containing 0.035 pmol of 32P-labeled *51:64-mer and other components of the unwinding assay was assembled. A 7-μl portion was removed to serve as a starting point (lane 1), and then 18 μl of DBP (114 ng) collected after chromatography on DEAE-Toyopearl was added and the reaction mixture was incubated at 23°C. At the indicated times, 10-μl portions from the reaction mixture were removed and treated with 1% SDS and 0.5 mg of proteinase K per ml for 20 min at 23°C. Portions (5 μl) from the samples were analyzed by electrophoresis in a 6% polyacrylamide gel. The sequences of the *36:51-mer and *51:64-mer are shown at the bottom. The asterisk indicated the radioactive label.
DISCUSSION
We have described a method for the purification of two viral DNA-binding proteins, DBP and LEF-3, from nuclear extracts of BmNPV-infected BmN cells. Purification of AcMNPV LEF-3 was carried out earlier in two laboratories (10, 14). Some modifications were employed in this study. The treatment of the isolated protein with DNase I and the concentration of diluted samples at steps in the purification protocol were not used. At a final stage, we performed chromatography on an anion-exchange resin. Use of the DEAE column permitted us to obtain both proteins in a homogeneous state, as well as to remove possible traces of DNA and to concentrate the samples. The preparation of DBP collected after fractionation of high-salt extracts on ssDNA cellulose contained monomers and higher-order oligomers, whereas after subsequent chromatography on DEAE-Toyopearl, the protein was present only as multimers. The capacity to form multimers in the absence of DNA is not a unique feature among DNA-binding proteins. ICP8, the SSB protein of herpes simplex virus type 1, forms long filaments in solution (30). Phage T4 gene 32 protein forms oligomers due to self-association of monomers (5). The adenovirus DNA-binding protein, DBP, has a C-terminal extension that interacts with another monomer, resulting in the formation of long protein chains (8). Basic and aromatic amino acids of DNA-binding proteins are thought to participate in interaction with DNA substrates. BmNPV DBP contains a motif, K18-R21-Y33-F41-Y53-R64-W71-K80, which presents a perfect match to the consensus sequence, K/RX2–5K/RX4–12F/YX2–14F/YX6–13F/YX1–19K/RX3–26F/Y/WX6–11R/K, found in all SSB proteins from prokaryotic and eukaryotic organisms (38). An additional motif in DBP, K159-K162-F168-Y173-F183-R189-F195-H206, has a single conservative change (R/K→H) at residue 206. The protein does not contain other known motifs such as zinc fingers (C/HX2–5C/HX11–13C/HX2,5C/H), leucine zippers ([LX6] 3L), and nucleoside triphosphate-binding domains (GXXGXGX15–20K; GXGK[S/T] [S/T]).
Isolation of a novel baculovirus DNA-binding protein, BmNPV DBP, poses intriguing question about the possible function of this protein in virus replication. Its preferential binding to ssDNA (Fig. 4), its protection of bound DNA against exonucleases (Fig. 5), and its helix-destabilizing ability (Fig. 6 and 7) are consistent with an SSB function. These properties might be required for initiation and elongation stages in viral DNA replication. Upon initiation in diverse replication systems, SSB proteins accelerate local melting of DNA in an ori region provided by specific ori-binding factors. During elongation, SSB proteins stabilize parental ssDNA chains, thus facilitating the action of DNA helicases, DNA polymerases, and other replication factors. An SSB function was proposed earlier for another viral protein, AcMNPV LEF-3, based on its preferential binding to ssDNA and its abundance in infected cells (14) and its requirement for transient DNA replication (16, 22). The amino acid sequence of AcMNPV LEF-3 has 91.7% identity in a 385-amino-acid overlap to that of BmNPV LEF-3, and both proteins are likely to play the same role in viral DNA replication (12). BmNPV LEF-3 was, in fact, one of the abundant viral proteins in infected BmN cells (Fig. 1) and showed specific binding to ssDNA (data not shown). However, another abundant viral protein, DBP, which also has binding specificity for ssDNA is capable of destabilizing duplex DNA. The binding of SSB proteins favors the production of single-stranded regions resulting from DNA breathing in dsDNA regions. This destabilizes the duplex structure and reduces the temperature required for its melting. For this reason, SSB proteins have also been called helix-destabilizing proteins (6, 25). Although the thermal stability of the short 17-bp duplex in the 17:51-mer is rather low, BmNPV LEF-3 did not cause melting of this duplex upon oversaturation (Fig. 6, lanes 5 to 7). The inability to unwind partial DNA duplexes in an in vitro system does not conform to the SSB function of LEF-3, although it does not eliminate the possibility of this function completely. Because this protein is required for the transient replication of plasmids containing putative viral ori regions in transfection assays (1, 16, 22), other essential roles for LEF-3 might be considered. The copurification of LEF-3 with viral DNA helicase (product of the dnahel gene) observed by other groups (10, 19) corresponds to a role as a possible accessory factor for viral DNA helicase. In contrast to the case for LEF-3, DBP was not detected among viral factors essential for plasmid replication in the transfection assay (1, 16, 22). Moreover, in contrast to most replication factors, DBP was not listed among 18 late gene expression factors (LEFs) identified earlier (22). A host cell nuclear SSB protein, RP-A, might substitute for DBP in the transfection assay, and this protein and/or other factors may permit the initiation of viral DNA replication and may overcome the block to expression of the late viral genes. To clarify the role of DBP, we attempted to isolate BmNPV mutants with a deletion in the dbp gene by using homologous recombination after cotransfection of wild-type BmNPV DNA with a plasmid carrying a deletion in the dbp locus into BmN cells. Our attempts to isolate a BmNPV mutant lacking the dbp gene were unsuccessful, suggesting that dbp is essential for virus replication (data not shown). Further experiments are required for elucidation of the DBP function in the baculovirus infection cycle.
ACKNOWLEDGMENTS
We thank George F. Rohrmann for critical reading of the manuscript and helpful discussion, Jay Evans and George F. Rohrmann for donation of antiserum against AcMNPV LEF-3, Shogo Matsumoto for advice in protein sequencing, and Evgeny Zemskov for help in immunological experiments.
This work was supported, in part, by a Japan Society for Promotion of Science fellowship to V.S.M. and by grants from the CREST (Core Research for Evolutional Science and Technology) and COE (Center of Excellence) programs of the Science and Technology Agency and the Institute of Physical and Chemical Research (RIKEN) to S.M.
REFERENCES
- 1.Ahrens C H, Rohrmann G F. Replication of Orgyia pseudotsugata baculovirus DNA: lef-2 and ie-1 are essential and ie-2, p34, and Op-iap are stimulatory genes. Virology. 1995;212:650–662. doi: 10.1006/viro.1995.1523. [DOI] [PubMed] [Google Scholar]
- 2.Ahrens C H, Russell R L, Funk C J, Evans J T, Harwood S H, Rohrmann G F. The sequence of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus genome. Virology. 1997;229:381–399. doi: 10.1006/viro.1997.8448. [DOI] [PubMed] [Google Scholar]
- 3.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 4.Boehmer P E, Lehman I R. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J Virol. 1993;67:711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll R B, Neet K, Goldthwait D A. Studies of the self-association of bacteriophage T4 gene 32 protein by equilibrium sedimentation. J Mol Biol. 1975;91:275–291. doi: 10.1016/0022-2836(75)90380-0. [DOI] [PubMed] [Google Scholar]
- 6.Chase J W, Williams K R. Single-stranded DNA-binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 7.Choi J, Guarino L A. The baculovirus transactivator IE1 binds to viral enhancer elements in the absence of insect cell factors. J Virol. 1995;69:4548–4551. doi: 10.1128/jvi.69.7.4548-4551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker J, Kanellopoulos P N, Loonstra A K, Oosterhout J A W M v, Leonard K, Tucker P A, Vliet P C v d. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 1997;16:1455–1463. doi: 10.1093/emboj/16.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans J T, Leisy D J, Rohrmann G F. Characterization of the interaction between the baculovirus replication factors LEF-1 and LEF-2. J Virol. 1997;71:3114–3119. doi: 10.1128/jvi.71.4.3114-3119.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans J T, Rohrmann G F. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J Virol. 1997;71:3574–3579. doi: 10.1128/jvi.71.5.3574-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgaki A, Strack B, Podust V, Hubscher U. DNA unwinding activity of replication protein A. FEBS Lett. 1992;308:240–244. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- 12.Gomi S, Zhou C E, Yih W, Majima K, Maeda S. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology. 1997;230:35–47. doi: 10.1006/viro.1997.8457. [DOI] [PubMed] [Google Scholar]
- 13.Guarino L A, Dong W. Expression of an enhancer-binding protein in insect cells transfected with the Autographa californica nuclear polyhedrosis virus IE1 gene. J Virol. 1991;65:3676–3680. doi: 10.1128/jvi.65.7.3676-3680.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hang X, Dong W, Guarino L A. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J Virol. 1995;69:3924–3928. doi: 10.1128/jvi.69.6.3924-3928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosoda J, Takacs B, Brack C. Denaturation of T4 DNA by an in vitro processed gene 32 protein. FEBS Lett. 1974;47:338–342. doi: 10.1016/0014-5793(74)81043-4. [DOI] [PubMed] [Google Scholar]
- 16.Kool M, Ahrens C H, Goldbach R W, Rohrmann G F, Vlak J M. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovacs G R, Choi J, Guarino L A, Summers M D. Functional dissection of the Autographa californica nuclear polyhedrosis virus immediate-early 1 transcriptional regulatory protein. J Virol. 1992;66:7429–7437. doi: 10.1128/jvi.66.12.7429-7437.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Laufs S, Lu A, Arrell K, Carstens E B. Autographa californica nuclear polyhedrosis virus p143 gene product is a DNA-binding protein. Virology. 1997;228:98–106. doi: 10.1006/viro.1996.8361. [DOI] [PubMed] [Google Scholar]
- 20.Lu A, Carstens E B. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology. 1991;181:336–347. doi: 10.1016/0042-6822(91)90500-b. [DOI] [PubMed] [Google Scholar]
- 21.Lu A, Miller L K. Differential requirements for baculovirus late expression factor genes in two cell lines. J Virol. 1995;69:6265–6272. doi: 10.1128/jvi.69.10.6265-6272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda S. Gene transfer vectors of a baculovirus, Bombyx mori, and their use for expression of foreign genes in insect cells. In: Mitsuhashi J, editor. Invertebrate cell system applications. I. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 167–181. [Google Scholar]
- 24.Maeda S, Kawai T, Obinata M, Fujiwara H, Horiuchi T, Saeki Y, Sato Y, Furusawa M. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature (London) 1985;315:592–594. doi: 10.1038/315592a0. [DOI] [PubMed] [Google Scholar]
- 25.Meyer R R, Laine P S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikhailov V S, Bogenhagen D F. Effects of Xenopus laevis mitochondrial single-stranded DNA-binding protein on primer-template binding and 3′→5′ exonuclease activity of DNA polymerase γ. J Biol Chem. 1996;271:18939–18946. doi: 10.1074/jbc.271.31.18939. [DOI] [PubMed] [Google Scholar]
- 27.Mikhailov V S, Marlyev K A, Ataeva J O, Kullyev P K, Atrazhev A M. Characterization of 3′→5′ exonuclease associated with DNA polymerase of silkworm nuclear polyhedrosis virus. Nucleic Acids Res. 1986;14:3841–3857. doi: 10.1093/nar/14.9.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller L K, Jewell J E, Browne D. Baculovirus induction of a DNA polymerase. J Virol. 1981;40:305–308. doi: 10.1128/jvi.40.1.305-308.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monaghan A, Webster A, Hay T. Adenovirus DNA binding protein: helix destabilizing properties. Nucleic Acids Res. 1994;22:742–748. doi: 10.1093/nar/22.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Donnell M E, Elias P, Funell B E, Lehman I R. Interaction between the DNA polymerase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus 1. J Biol Chem. 1987;262:4260–4266. [PubMed] [Google Scholar]
- 31.Pearson M, Bjornson R, Pearson G, Rohrmann G. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science. 1992;257:1382–1384. doi: 10.1126/science.1529337. [DOI] [PubMed] [Google Scholar]
- 32.Pearson M N, Bjornson R M, Ahrens C, Rohrmann G F. Identification and characterization of a putative origin of DNA replication in the genome of a baculovirus pathogenic for Orgyia pseudotsugata. Virology. 1993;197:715–725. doi: 10.1006/viro.1993.1647. [DOI] [PubMed] [Google Scholar]
- 33.Rodems S M, Friesen P D. Transcriptional enhancer activity of hr5 requires dual-palindrome half sites that mediate binding of a dimeric form of the baculovirus transregulator IE1. J Virol. 1995;69:5368–5375. doi: 10.1128/jvi.69.9.5368-5375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soengas M S, Gutierrez C, Salas M. Helix-destabilizing activity of phi29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J Mol Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 35.Tomalski M D, Wu J G, Miller L K. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology. 1988;167:591–600. [PubMed] [Google Scholar]
- 36.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J Mol Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Kelly D C. Baculovirus replication: purification and identification of the Trichoplusia ni nuclear polyhedrosis virus-induced DNA polymerase. J Gen Virol. 1983;64:2229–2236. [Google Scholar]
- 38.Wang Y, Hall J D. Characterization of a major DNA-binding domain in the herpes simplex virus type 1 DNA-binding protein (ICP8) J Virol. 1990;64:2082–2089. doi: 10.1128/jvi.64.5.2082-2089.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zijderveld D C, van der Vliet P C. Helix-destabilizing properties of the adenovirus DNA-binding protein. J Virol. 1994;68:1158–1164. doi: 10.1128/jvi.68.2.1158-1164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]