Abstract
Adenovirus E1B 55K protein cooperates with E1A gene products to induce cell transformation. E1B 55K mediates its effects by binding to and inhibiting the transcriptional activation and growth-suppression functions of the tumor suppressor p53. Previous studies in vivo have suggested that E1B 55K has an active role in repressing p53 transcriptional activation and that this repression function is directed to specific promoters through E1B 55K’s interaction with DNA-bound p53. Flag-tagged E1B 55K (e55K) was expressed with the baculovirus expression system and immunopurified. Gel filtration, velocity sedimentation centrifugation, and glutaraldehyde cross-linking indicated that e55K is a dimer with a nonglobular conformation. e55K bound directly to purified p53, causing an ∼10-fold increase in p53 affinity for tandem binding sites. Using in vitro transcription assays reconstituted with purified p53, e55K, and HeLa cell nuclear extracts, we found that e55K specifically repressed p53 activation. These results demonstrate that as postulated from earlier transient expression experiments, E1B 55K is a specific repressor of transcription from a promoter with bound p53. Since HeLa nuclear extracts contain little detectable histone protein, E1B 55K probably represses transcription through direct or indirect interactions with the RNA polymerase II transcription machinery.
The cellular phosphoprotein p53 acts as a tumor suppressor, and inactivation of this function is the most prevalent alteration found in human and animal tumors (32, 35). p53 acts as a G1 checkpoint control ensuring completion of DNA replication and the integrity of the genome prior to cells entering the S phase. In response to DNA damage, p53 levels are increased and p53 induces genes whose products result in either growth arrest at the G1/S interface or apoptosis (reviewed in reference 32). p53-inducible genes include p21/WAF21/Cip1 (cyclin/cdk inhibitor) (10, 66), cyclin G (45), Wip1 (a PP2A protein phosphatase) (12), Bax1 (apoptosis inducer) (43), insulin-like growth factor binding protein 3 (5), GADD45 (28), and Mdm2 (46). Mdm2 could be a natural biological regulator of p53 through an autoregulatory feedback loop with p53, since p53 activates Mdm2 expression and Mdm2 inhibits p53 activation (64). As a consequence of the inhibition of p53’s activation by Mdm2, induction of growth arrest and apoptosis are also inhibited (28, 46, 64, 69).
p53 appears to exert most of its biological effects by acting as a sequence-specific DNA-binding transcription factor (Fig. 1) (11) that binds as a tetramer (24) to four tandem, alternatively inverted copies of a 5-bp consensus sequence (7, 9). The transactivation domain maps to the acidic amino-terminal region of the polypeptide (1 to 73 amino acid residues), and it is within this region that p53 has been demonstrated to directly interact with components of the general transcriptional machinery, TATA box-binding protein (TBP), TAFII31, and TFIIH (p62) (41, 42, 58, 60, 65), as well as its negative regulator, Mdm2 (38).
FIG. 1.
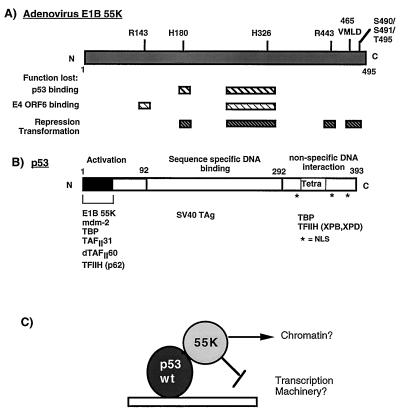
Schematic diagram illustrating the functional domains in Ad E1B 55K (A) and the tumor suppressor p53 (B). (A) Hatched boxes indicate regions in E1B 55K which when mutated result in loss of the described functions. Positions where amino acids have been mutated by linker insertion or site-directed mutagenesis (57, 67) are indicated above the E1B 55K molecule. (B) The activation, DNA binding, and nonspecific DNA binding domains of p53 are indicated. The tetramerization domain (Tetra) is located in the C terminus. Nuclear localization signals (NSL) are indicated by asterisks. Proteins which are known to interact with the defined regions are indicated at the bottom. (C) Model of how E1B 55K may affect the activity of p53. E1B 55K may mediate alterations in chromatin structure or may target the general transcriptional machinery. SV40 TAg, simian virus 40 T antigen.
The inactivation of p53 is critical for the replication of DNA tumor viruses, which require cells to enter the S phase, and which, in the case of adenovirus (Ad), would otherwise induce apoptosis (63). The DNA tumor viruses each encode oncoproteins, such as Ad E1B 55K, human papillomavirus E6, and simian virus 40 T antigen (9, 11, 27, 67), which bind to and inhibit the normal biological functions of p53. Binding of HPV E6 to wild-type p53 results in polyubiquination and subsequent degradation of p53 (53). Simian virus 40 T antigen interacts with the DNA binding domain of p53, thus preventing p53 from binding to DNA and regulating gene expression (11).
The Ad E1B 55K protein also forms a stable complex with p53 in vitro and in vivo (27, 50) and inhibits p53-mediated transcriptional activation (67). E1B 55K binds to the amino-terminal transactivation domain of p53 (27) (Fig. 1). Mutation of p53 at either the proline (amino acid 24) or tryptophan (amino acid 27) residue reduces p53’s affinity for E1B 55K, although the transactivation function remains wild type (38). Previous studies with a panel of E1B 55K linker insertion mutants showed that the ability of E1B 55K to inhibit p53-mediated activation correlated directly with the ability of E1B 55K to transform cells in conjunction with Ad E1A (Fig. 1) (68). Furthermore, the interaction between E1B 55K and p53 was necessary but not sufficient for the repression and transformation functions of E1B 55K, since an E1B 55K insertion mutant at position 443 binds to p53 with wild-type affinity but was defective for both repression of p53 transcriptional activation and transformation (see Fig. 1) (67, 68). These and other studies have indicated that the carboxy-terminal region of E1B 55K and phosphorylation of serine residues 490 and 491 and threonine at 495 within this region are important for the repression functions of E1B 55K (42a, 56, 57, 68). Moreover, these results suggested that E1B 55K has an active role in repression and was not simply preventing p53 from binding to DNA nor sterically blocking the activation domain of p53.
Strong evidence suggesting that E1B 55K has a generalized transcriptional repression activity came from transient expression experiments in which a Gal4-55K fusion was shown to repress expression from several target promoters containing Gal4 binding sites. These promoters did not contain any common activator binding sites, implying that E1B 55K does not inhibit a specific activation mechanism, but rather inhibits a general process required for transcription from most promoters (68). Furthermore, the ability of Gal4-55K to repress transcription was independent of p53, because similar results were obtained from experiments performed with p53-minus cell lines (68). These results suggested the model that E1B 55K acts as a general repressor of RNA polymerase II transcription and that the repression function of E1B 55K is directed toward p53-activated promoters by virtue of its binding to DNA-bound p53. E1B 55K could mediate its repressing effect either by acting directly on the general transcriptional machinery (22, 25) or through modification of chromatin (47) (Fig. 1C).
Given the central role that aberration of the normal function of p53 has in the development of some cancers, it is important to establish how its activity is regulated and/or inhibited by oncoproteins such as E1B 55K. Furthermore, the mechanism of action by repressors like E1B 55K, which do not simply compete for the DNA binding sites of activating proteins, is not clearly understood for many eukaryotic repressors. Understanding how E1B 55K mediates its effect may yield insights into how other repressors function and how repressors regulate the activity of activators.
Here, we report on the physical and biochemical properties of purified, recombinant E1B 55K expressed from a baculovirus vector and demonstrate through in vitro transcription assays that it is indeed a transcriptional repressor.
MATERIALS AND METHODS
Cells.
Sf9 cells were maintained at 27°C in Grace’s insect medium (Gibco), supplemented with 10% fetal calf serum and 0.4% yeastolate. HeLa S3 suspension cells were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% newborn calf serum.
Plasmid and baculovirus construction.
p5BSE4CAT, which contains five p53 binding sites upstream of the Ad E4 promoter (−38 to +38) driving expression of the chloramphenicol acetyltransferase (CAT) gene, has been described previously (40). pG5E1BCAT contains five Gal4 DNA binding sites upstream of the E1B TATA sequence, which itself lies upstream of the CAT gene (37). pG5E4CAT contains five Gal4 binding sites upstream of the E4 promoter and CAT gene (37).
pAcC12 is a baculovirus transfer vector containing the polyhedron promoter upstream of a polylinker (55). pAc-e55K was constructed by PCR amplification of the Ad E1B 55K sequences with a Flag epitope sequence (MDYKDDDDK) fused to the amino terminus and cloned as an EcoRI-BamHI fragment into the baculovirus transfer vector pAcC12. Most of the PCR-amplified sequences were replaced with wild-type sequences as an NcoI-BamHI fragment from pSRα-55K (67), and the remaining 180-amino-terminal bases and Flag epitope were sequenced to confirm that no errors had been incorporated during PCR amplification.
Recombinant baculovirus Bac-e55K was constructed by cotransfection of BacPAK6 Bsu 361-digested DNA (Clontech) with pAc-e55K into Sf9 insect cells by the calcium phosphate method with modifications (55). Briefly, the DNA-calcium phosphate precipitate was incubated with Sf9 cells plated at 2 × 106 cells/60-mm-diameter plate for 6 h at 27°C. The cells were washed and incubated in Grace’s medium. Twenty-four hours posttransfection, cells were overlayed with 0.7% agarose in Grace’s medium with 0.35% agarose in Grace’s medium 3 and 5 days posttransfection, with the last overlay containing 0.01% neutral red. Plaques of recombinant viruses were picked and assayed for expression of E1B 55K by Western blotting with the anti-55K monoclonal antibody 2A6 (51) and enhanced chemiluminescence (ECL) (Pierce). Recombinant viruses were plaque purified three times prior to amplification of viral stocks for large-scale infections.
Purification of proteins.
Sf9 insect cells were infected with Bac-e55K, the recombinant virus expressing e55K, at a multiplicity of infection of 10 PFU/cell. Cells were harvested 48 h postinfection, and nuclear extracts were prepared (8). Nuclear extracts were incubated with anti-Flag M2 affinity gel (Kodak) for 8 to 12 h at 4°C with rotation. Beads were washed six times with 10 volumes of D buffer (20 mM HEPES, 2 mM EDTA, 20% glycerol, 10 mM β-mercaptoethanol [pH 7.9]) containing 300 mM KCl and 0.1% Nonidet P-40 and twice with 10 column volumes of D buffer containing 100 mM KCl. e55K was specifically eluted with Flag peptide (DYKDDDDK; 1 mg/ml in D buffer containing 100 mM KCl, 60 to 70 μl of peptide, and 100 μl of beads). Purified proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [10% polyacrylamide]) and silver staining. The concentration of e55K was estimated to be 10 ng/μl from silver-stained gels with known bovine serum albumin (BSA) standards.
As a control, Sf9 insect cells were infected with BacPAK6, the viral vector used in the construction of Bac-e55K, and processed in the same manner as e55K extracts. The final fraction from the anti-Flag M2 affinity column would contain any contaminating polypeptides and M2 peptide. This protein fraction served as a negative control and was referred to as BP6.
Influenza virus HA1 epitope-tagged p53 (ep53) was immunopurified from nuclear extracts prepared from vaccinia virus vector-infected HeLa cells as described previously (68). Gal4-AH, a transcriptional activator which consists of an amphipathic α helix fused to the Gal4 DNA binding domain, was purified from Escherichia coli and was a kind gift from Mike Carey (39).
Sucrose density gradient sedimentation.
A mixture (100 μl in D buffer containing 0.3 M KCl) of purified e55K (500 ng) and the molecular mass markers β-amylase (30 μg), alcohol dehydrogenase (ADH) (50 μg), and BSA (25 μg) was layered onto a 5-ml linear 5 to 20% sucrose density gradient in 0.3 M KCl–20 mM HEPES (pH 7.9)–2 mM EDTA–10% glycerol–10 mM β-mercaptoethanol. The gradient was centrifuged for 19 h at 45,000 rpm in a Beckman SW50.1 rotor at 4°C. Fractions (250 μl) were collected and precipitated with 10% trichloroacetic acid in the presence of 20 μg of insulin and 0.1% deoxycholate as carriers and then were analyzed by SDS-PAGE. Fractions containing e55K were detected by Western blotting analysis and ECL with 2A6 antibody. Molecular mass markers were detected by Coomasie blue staining. The molecular mass of e55K was calculated by the equation MA/MB = (dA/dB)3/2, where M is molecular mass and d is the distance from the top of the gradient. The estimated sedimentation coefficient was determined according to the known values for BSA (4.3S [68 kDa]), ADH (7.4S [150 kDa]), and β-amylase (8.9S [200 kDa]).
Gel filtration.
Purified e55K (500 ng) was fractionated by gel filtration through Superose 6 and Superose 12 HR columns (10 by 300 mm; Pharmacia) equilibrated in D buffer containing 0.3 M KCl. The columns were run with a Pharmacia fast-performance liquid chromatograph at 0.5 ml/min at room temperature. Fractions (0.5 ml) were collected, trichloroacetic acid precipitated, and analyzed by SDS-PAGE, followed by Western blot analysis. Molecular mass protein standards were analyzed under the same conditions. These were thyroglobulin (a tetramer of 670 kDa [85Å]), apoferritin (a multimer of 440 kDa [61Å]), β-amylase (a tetramer of 200 kDa), ADH (a tetramer of 150 kDa [46 Å]), BSA (a monomer of 68 kDa [35Å]), and carbonic anhydrase (29 kDa). Columns were checked for reproducibility and varied within 0.3 ml (i.e., <1 fraction). The molecular mass of e55K was determined from the calibration curve of molecular masses of protein standards. The Stokes radius was calculated according to published values for the protein standards and the Porath equation (54).
Molecular mass determination.
The molecular masses and frictional ratio of proteins can be determined by the Stokes radius determined by gel filtration and the sedimentation coefficient obtained from sucrose gradients and the equations (54) M = 6πηNas/(1 − νρ) and f/f0 = a/(3vM/4πN)1/3, where M is molecular weight, a is the Stokes radius, s is the sedimentation coefficient, v is the partial specific volume (set at 0.725 cm3/g), f/f0 is the frictional ratio, η is the viscosity of medium, ρ is the density of medium, and N is Avogadro’s number.
Glutaraldehyde cross-linking.
Purified e55K (100 ng) was incubated for 10 min at 30°C in D buffer containing 0.3 M KCl with increasing concentrations of freshly diluted glutaraldehyde (Sigma) (0, 10−4, 3 × 10−4, 10−3, 3 × 10−3, 10−2, 3 × 10−2, 10−1, 3 × 10−1%) in a final reaction volume of 50 μl. Products were analyzed by standard SDS-PAGE (8% polyacrylamide), except that the stacking gel used had a ratio of acrylamide to bisacrylamide of 80:1 to prevent protein retardation in the stacking gel. Products were detected by Western blot analysis. As a control against nonspecific cross-linking, BSA (100 ng) was treated under the same conditions and SDS-PAGE gels were silver stained.
DNA immunoprecipitation.
The DNA immunoprecipitation assay was performed as described by Yew et al. (68) with modifications. Two DNA probes were 32P labeled by end filling: one containing five Gal4 DNA binding sites isolated as a HindIII-BamHI fragment from pG5E4CAT and one containing five p53 binding sites isolated as a HindIII-Asp-718 fragment from p5BSE4CAT. DNA probe (5 × 105 cpm), purified ep53 (150 ng), and e55K (300 ng) were incubated for 1 h at 30°C in 200 μl of binding buffer (20 mM Tris-Cl [pH 7.4], 100 mM NaCl, 10% glycerol, 1% Nonidet P-40, 5 mM EDTA) containing 50 ng of poly(dI-dC) per μl and 0.5 mg of BSA per ml. Protein A-Sepharose beads (25 μl) were incubated for 2 h at room temperature with 600 μl of hybridoma supernatant of either Ab421 (anti-p53 antibody [19]) or 2A6 (anti-55K antibody [51]) and then washed twice in binding buffer. The antibody-bound protein A-Sepharose beads (25 μl) were then incubated with the protein-DNA mixture for 1 h at room temperature with rotation. The beads were washed twice in binding buffer and incubated with 0.5 mg of proteinase K per ml and 0.1% SDS for 15 min at 37°C. After phenol-chloroform extraction and DNA precipitation, the immunoprecipitated DNA was analyzed by electrophoresis in a 5% polyacrylamide–Tris-borate-EDTA (TBE) gel, followed by autoradiography.
DNase I footprinting.
The DNA probe was prepared from p5BSE4CAT by end filling in of the HindIII site with [32P]dATP and cutting with Asp 718 to generate a 165-bp fragment. The fragment was purified through an 8% nondenaturing polyacrylamide TBE gel and recovered by electroelution. Protein-DNA binding reactions were performed in 25 μl of binding buffer [12 mM HEPES (pH 7.9), 60 mM KCl, 6 mM MgCl2, 0.12 mM EDTA, 12% glycerol, 10 mM β-mercaptoethanol, 0.5 mg of BSA per ml, and 40 μg of poly(dG-dC) per ml] and ∼5 fmol of the DNA probe. After binding for 1 h at 30°C, DNA was digested with 2 μl of DNAse I (1 mg/ml diluted 1:2,000 in 60 mM CaCl2) for 1 min at 30°C. Reactions were stopped by the addition of 25 μl of 2× stop solution (0.2 M NaCl, 0.5% SDS) containing proteinase K (500 μg/ml) and tRNA (20 μg/ml) for 10 min at 37°C. After phenol-chloroform extraction and ethanol precipitation, digestion products were analyzed by electrophoresis through a 6% polyacrylamide–8 M urea–TBE gel, followed by autoradiography.
In vitro transcription.
HeLa nuclear extracts were prepared as described previously (8). Transcription reaction mixtures contained 20 ng of ep53; 10 ng of Gal4-AH; and 4, 8, 12, or 16 μl of e55K (10 ng/μl) or the same volumes of the control BP6 protein fraction as indicated. In vitro transcription reactions were performed essentially as described previously (40). Each transcription reaction mixture contained two DNA templates: p5BSE4CAT (30 ng) and, as an internal control, pG5E1BCAT (30 ng). DNA templates, nuclear extract (60 μg), and other proteins, as indicated, were preincubated for 20 min at 30°C in a 50-μl reaction mix (12 mM HEPES [pH 7.9], 60 mM KCl, 6 mM MgCl2, 0.12 mM EDTA, 12% glycerol, 10 mM β-mercaptoethanol). Transcription was initiated by the addition of ribonucleoside triphosphates (600 μM) and allowed to continue for 1 h at 30°C. Transcription was terminated by the addition of 50 μl of 2× stop solution (0.2 M NaCl, 0.5% SDS) containing proteinase K (500 μg/ml) and tRNA (20 μg/ml) for 10 min at 37°C. Levels of transcription were assayed by primer extension with a γ-32P 5′-end-labeled primer complementary to the coding region of the CAT gene (5′ CTC AAA ATG TTC TTT ACG ATG CCA TTG GGA 3′). Primer extension products were analyzed by electrophoresis through a 10% polyacrylamide–8 M urea–TBE gel, followed by autoradiography.
RESULTS
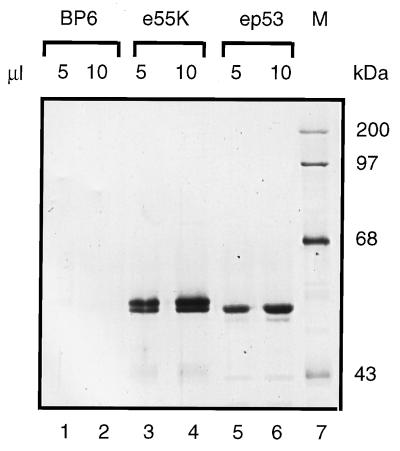
E1B 55K is a phosphoprotein, and previous work has shown that phosphorylation is required for its repression and transformation functions (56, 57). Consequently, it was important to express E1B 55K in a eukaryotic expression system. e55K, tagged at the amino terminus by addition of the eight-residue Flag epitope, was expressed by the baculovirus system and purified in a single step from infected-cell nuclear extracts by immunoaffinity chromatography. The Flag epitope fused to the amino terminus of E1B 55K allowed the specific elution of Flag-tagged protein from the monoclonal antibody M2 with M2 peptide. The affinity purification procedure was valuable, since E1B 55K chromatographed heterogeneously over many ion-exchange resins (unpublished results). e55K was purified to near homogeneity (Fig. 2, lanes 3 and 4) and separated into two closely migrating species, possibly reflecting differences in phosphorylation. BP6, included as a negative control, refers to a protein fraction purified under identical conditions to e55K from nuclear extracts of cells infected with the baculovirus expression vector, BacPAK6, which expresses β-galactosidase instead of e55K (Fig. 2, lanes 1 and 2). No baculovirus or cellular proteins were detected as purifying over the anti-Flag M2 affinity column. ep53 was immunopurified utilizing the HA1 epitope fused to its amino terminus, from the nuclei of HeLa cells dually infected with recombinant vaccinia viruses expressing T7 RNA polymerase and ep53 from a late T7 promoter (68) (Fig. 2, lanes 5 and 6).
FIG. 2.
Silver-stained SDS-PAGE (10% polyacrylamide) gels of purified Flag-tagged 55K (e55K), the control BP6 protein fraction purified under identical conditions to e55K from Sf9 cells infected with a baculovirus vector expressing β-galactosidase, and ep53 proteins. Lanes 1 to 6 contain BP6 (lanes 1 and 2 [5 and 10 μl, respectively]), e55K (lanes 3 and 4 [5 and 10 μl, respectively]), and ep53 (lanes 5 and 6 [5 and 10 μl, respectively]). M, molecular mass markers in kilodaltons (lane 7).
Quaternary structure of E1B 55K.
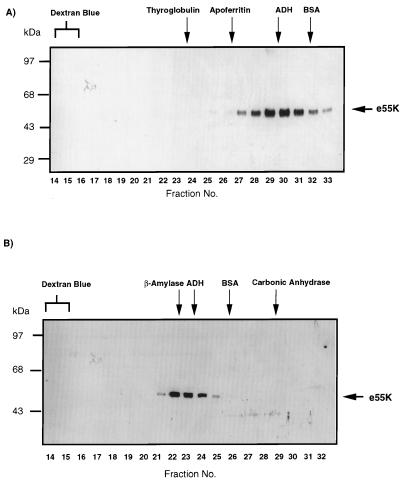
We used gel filtration, first, to determine whether purified e55K behaved as a monodisperse molecular species and, second, to determine the quaternary structure of e55K. It was important to determine the characteristics of purified, soluble e55K, since we had observed that overexpression resulted in high-molecular-mass complexes which were not disrupted by boiling in Laemmli disruption buffer for 30 min. These complexes were minimized by preparing nuclear extracts relatively early after infection with the baculovirus expression vector. We initially used a Superose 6 HR column with an exclusion volume of 40 × 106 Da to characterize immunopurified e55K as described in Materials and Methods. The absence of e55K in the excluded volume (fractions 14 and 15) indicated that the majority of e55K was soluble and was not aggregated (Fig. 3A). The e55K protein eluted as expected for a globular protein with a molecular mass of approximately 150 kDa (fractions 29 to 30) (Fig. 3A). In order to obtain a more accurate estimate of the molecular mass, e55K was resolved over a Superose 12 column which resolves proteins in the molecular mass range of 3 to 300 kDa. Most of the e55K eluted similarly to β-amylase (200 kDa) (Fig. 3B). The elution characteristics of a protein over gel filtration columns correlate better with the protein’s Stokes radius than with its molecular mass (54). By using the elution volumes of the standards of known Stokes radii, the Stokes radius of highly purified e55K was estimated as 52 Å.
FIG. 3.
Gel filtration analysis of e55K through Superose 6 (A) and Superose 12 (B) columns. After chromatography, fractions were trichloroacetic acid precipitated and analyzed by SDS-PAGE and Western blotting with the anti-55K antibody 2A6. e55K was visualized by ECL. Fraction numbers are indicated at the bottom of each panel. The fractions in which the protein standards eluted are indicated at the top of each panel. Protein standards were thyroglobulin (a tetramer of 670 kDa [85Å]), apoferritin (a multimer of 440 kDa [61Å]), β-amylase (200 kDa), ADH (a tetramer of 150 kDa [46Å]), BSA (a monomer of 68 kDa [35Å]), and carbonic anhydrase (29 kDa).
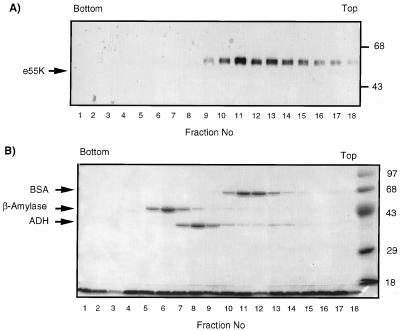
We also examined the velocity of sedimentation of purified e55K by sucrose gradient centrifugation (Fig. 4). If e55K had a nonglobular structure, one would expect it to sediment with a lower apparent molecular mass than that determined by gel filtration analyses. The sedimentation coefficient for e55K was estimated as described in Materials and Methods. The majority of e55K sedimented at ∼4.8S, consistent with a molecular mass of 80 kDa, assuming the protein is globular. This differs significantly from the molecular mass determined by gel filtration, indicating that the protein does not have a globular conformation. Using the values determined for the Stokes radius and sedimentation coefficient of e55K and equations previously described (54) (see Materials and Methods), the estimated molecular mass of purified e55K is ∼103 kDa, suggesting that e55K is a dimer. The frictional ratio was calculated as 1.75 (54). This value indicates that e55K has hydrodynamic properties consistent with an elongated structure, which would explain the apparent discrepancy in molecular mass determinations between gel filtration and sucrose density sedimentation analyses.
FIG. 4.
Sucrose gradient analysis of e55K. e55K, BSA, β-amylase, and ADH were centrifuged together through a 5 to 20% sucrose gradient. Fractions were collected, trichloroacetic acid precipitated, and analyzed by SDS-PAGE, followed by Western blotting with anti-55K antibody 2A6 (A) and Coomassie blue staining (B). The top and bottom of the gradient are indicated. Molecular mass markers are indicated on the right in kilodaltons. Protein molecular mass standards from the bottom of the gradient are β-amylase (8.9S; 200 kDa; tetramer), ADH (7.4S; 150 kDa; tetramer), and BSA (4.3S; 68 kDa; monomer).
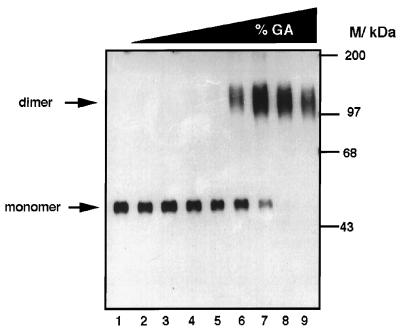
Protein cross-linking was used to further characterize the multimeric structure of purified e55K. Purified e55K was incubated with increasing concentrations of glutaraldehyde (0 to 0.3%) and analyzed by SDS-PAGE, followed by Western blotting (Fig. 5). In the absence of glutaraldehyde, e55K electrophoresed as a monomer of 55 kDa (lane 1). With increasing concentrations of glutaraldehyde, the monomeric form was converted into a dimeric form of approximately 110 kDa. This dimeric form was not cross-linked into higher oligomeric forms, even at the highest glutaraldehyde concentration used, but rather the dimeric form was converted into a more compact (faster) migrating species, characteristic of further intramolecular cross-linking. It is unlikely that higher oligomers were retained in the stacking gel, since a lower than usual ratio of acrylamide to bisacrylamide was used (80:1 compared to 30:1) (62). Furthermore, intermolecular cross-linking did not occur with the protein and glutaraldehyde concentrations used, since no cross-linking of BSA was detected under identical conditions (data not shown). These cross-linking studies confirmed that purified e55K is a dimer.
FIG. 5.
Glutaraldehyde cross-linking analysis of e55K. e55K was incubated with increasing concentrations of glutaraldehyde (%GA): lanes 1 to 9: 0, 10−4, 3 × 10−4, 10−3, 3 × 10−3, 10−2, 3 × 10−2, 10−1, and 3 × 10−1%, respectively. The positions of molecular mass markers (kilodaltons) are indicated. The positions of the monomeric and dimeric forms of e55K are indicated by arrows.
Purified e55K binds purified p53.
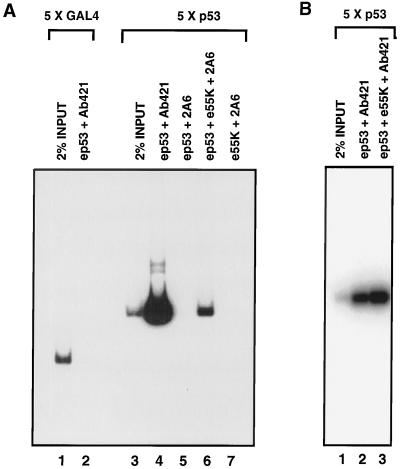
Previous studies have shown that E1B 55K forms a stable complex with p53 in vivo and in vitro (27, 50) and that E1B 55K does not inhibit p53 from binding to its specific DNA sequence (68). We wished to determine, first, whether e55K, purified from insect cells, was functionally active with respect to binding to a p53-DNA complex and, second, whether, by using highly purified proteins, the interaction was direct. These questions were addressed with DNA immunoprecipitation assays. ep53 was purified from nuclear extracts prepared from HeLa cells infected with an ep53-expressing recombinant vaccinia virus (Fig. 2). When ep53 was incubated with a labeled DNA fragment containing five p53 binding sites, this p53-DNA complex was immunoprecipitated with a p53 antibody, Ab421 (Fig. 6A, lane 4), whereas a control DNA fragment containing five Gal4 binding sites was not immunoprecipitated (lane 2), showing that ep53 binds specifically to its binding sites. Furthermore, the p53-DNA complex was not immunoprecipitated by a monoclonal antibody specific for E1B 55K, 2A6 (Fig. 6A, lane 5). When purified e55K was incubated with the p53-DNA complex, an e55K-p53-DNA complex was formed and detected by immunoprecipitation of labeled DNA with the anti-55K monoclonal antibody 2A6 (lane 6). In the absence of p53, e55K did not bind to DNA (lane 7). These results with purified proteins show that e55K binds directly to p53 and that the e55K-p53 complex remains bound to p53 DNA binding sites. Although less labeled DNA was immunoprecipitated with the anti-55K antibody than with the anti-p53 antibody (compare lanes 4 and 6), this was not due to an e55K-induced decrease in p53 binding, as shown in Fig. 6B. When immunoprecipitation was performed with the anti-p53 antibody Ab421, addition of e55K increased the amount of immunoprecipitated DNA (compare lanes 2 and 3).
FIG. 6.
Highly purified e55K interacts with ep53 bound to DNA. (A) Two 32P-labeled DNA fragments were used in the DNA immunoprecipitation assay, one containing five Gal4 binding sites (lanes 1 and 2) and one containing five p53 binding sites (lanes 3 to 7). ep53 (150 ng) was added to binding reaction mixtures in lanes 2, 4, and 6. e55K (150 ng) was added to binding reaction mixtures in lanes 6 and 7. ep53 was immunoprecipitated with an anti-p53 antibody, Ab421, and e55K was immunoprecipitated with the anti-55K antibody 2A6. (B) The 32P-labeled DNA fragment containing five p53 binding sites was incubated with 150 ng of ep53 (lanes 2 and 3) and 150 ng of e55K (lane 3). Immunoprecipitation was with anti-p53 monoclonal antibody 421. For both panels A and B, immunoprecipitated DNA was resolved by PAGE and visualized by autoradiography.
e55K increases the affinity of p53 for its DNA binding sites.
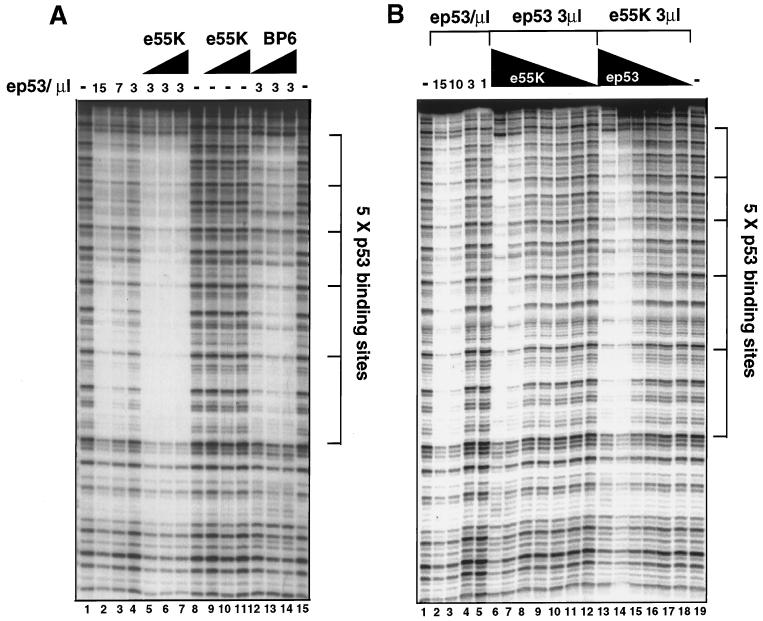
To determine the influence of E1B 55K on p53-DNA binding activity, independently of the influences of the monoclonal antibodies and the immunoprecipitation procedure, we assayed specific p53-DNA binding by DNase I footprinting (Fig. 7). Using a probe with five consecutive p53 binding sites, purified ep53 specifically protected bound DNA in a concentration-dependent manner (Fig. 7A, lanes 2, 3, and 4 with 150, 75, and 30 ng of ep53, respectively). At 30 ng of ep53, protection was only partial (lane 4). However, when increasing amounts of e55K were added to the binding reaction mixture (30, 60, and 90 ng [lanes 5 to 7, respectively]), protection was nearly complete. e55K had no effect on the DNase I pattern, in the absence of ep53 (lanes 9 to 11). These results indicate that E1B 55K increases the affinity of p53 for the tandem binding sites in this probe. This ability of e55K to increase p53 DNA binding activity diminished at lower e55K concentrations (Fig. 7B, lanes 6 to 12).
FIG. 7.
E1B 55K increases the affinity of p53 for its DNA binding sites. DNase I footprinting of DNA-protein complexes. The positions of the p53 binding sites are indicated. (A) Lanes 1, 8, and 15 show the DNase I pattern in the absence of added proteins. Lanes 2 to 4 show decreasing concentrations of purified ep53 (150, 75, and 30 ng, respectively [10 ng/μl]). Lanes 5 to 7 show binding reactions with 3 μl (30 ng) of ep53 plus increasing amounts of e55K (10 ng/μl [3, 6, and 9 μl, respectively]). Lanes 9 to 11 show DNase I footprinting after incubation of the probe with 3, 6, and 9 μl, respectively, of e55K. Lanes 12 to 14 show binding reactions with 3 μl (30 ng) of ep53 plus 3, 6, and 9 μl of the control BP6 fraction. (B) Lanes 1 and 19 show the DNase I digestion pattern in the absence of added protein. Lanes 2 to 5 show footprinting with decreasing amounts of ep53 (10 ng/μl): 15, 10, 3, and 1 μl, respectively. Lanes 6 to 12 show footprinting with 3 μl of ep53 plus decreasing amounts of e55K (100, 30, 10, 3, 1, 0.3, and 0.1 ng, respectively). Lanes 13 to 18 show footprinting in the presence of 30 ng of e55K and decreasing amounts of ep53 (30, 10, 3, 1, 0.3, and 0.1 ng, respectively).
To quantitate the influence of e55K on ep53 DNA binding activity, ep53 binding was assayed over a range of concentrations in the presence and absence of 30 ng of e55K (Fig. 7B). In the presence of e55K, clear protection from DNase I was observed at 10 ng of ep53 (Fig. 7B, lane 14), whereas, in the absence of e55K, an equivalent extent of protection was observed at 100 to 150 ng of ep53 (lanes 2 and 3). These results indicate that the interaction of E1B 55K with p53 increases the affinity of p53 for this probe by a factor of approximately 10. It is worth noting that e55K did not extend the footprint by ep53 (Fig. 7A, lanes 5 to 7, and B, lane 6), thus excluding the possibility that the p53-e55K complex acts like the Drosophila melanogaster repressor Even-skipped (Eve), which can prevent binding of TBP to a TATA box by competitive binding (3).
E1B 55K is a specific repressor of transcription.
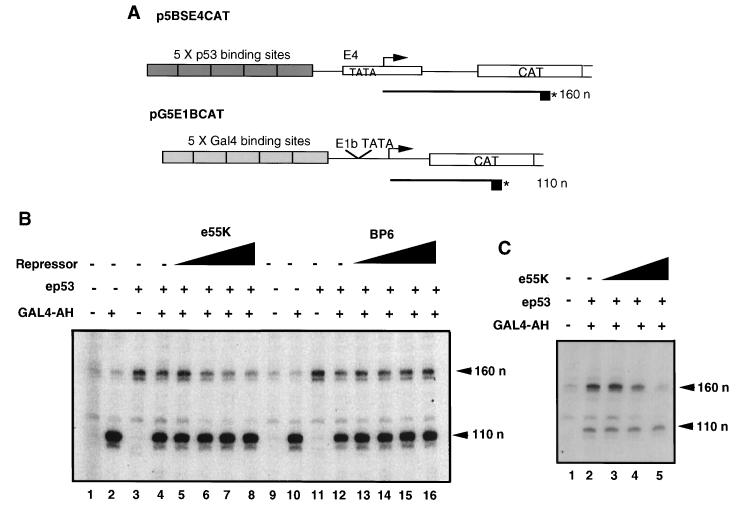
To determine whether E1B 55K can repress the activation of transcription initiation by p53, we performed in vitro transcription reactions by using HeLa cell nuclear extracts (8). Transcription reaction mixtures contained two DNA templates: one containing five p53 binding sites (p5BSE4CAT) and the other containing five Gal4 binding sites (pG5E1BCAT) (Fig. 8A). Transcription from p5BSE4CAT and pG5E1BCAT resulted in primer extension products of 160 and 110 nucleotides, respectively. The latter template was included as an internal control and was responsive to activation by Gal4-AH (Fig. 8B, compare lanes 1 and 2 and 9 and 10). Addition of ep53 (20 ng) specifically activated transcription from the template with p53 binding sites (Fig. 8B, lanes 3 and 11). Addition of both activators ep53 and Gal4-AH resulted in activation from both templates (lanes 4 and 12). Addition of increasing concentrations of e55K (lanes 5 to 8) resulted in a dose-dependent decrease in transcription from the p53-activated promoter, whereas levels of Gal4-AH-activated transcription were unaffected (lanes 5 to 8). Furthermore, reaction mixtures containing the control BP6 protein fraction indicated that neither the M2 peptide nor any contaminating proteins which were not detectable by silver staining were responsible for the inhibition observed with e55K (lanes 13 to 16). In other experiments with another preparation of e55K, specific repression of transcription from the p5BSCAT template was even more dramatic (Fig. 8C). These results show that e55K specifically represses transcription in vitro from a template with p53 binding sites. Since relatively short transcripts were assayed, this suggests that repression is probably occurring during initiation or at an early step in elongation.
FIG. 8.
(A) In vitro transcription templates. p5BSE4CAT contains five p53 binding sites upstream of the Ad E4 TATA box and CAT gene. This template is responsive to activation by p53. The primer extensions of RNAs transcribed from this template are 160 nucleotides (160 n) in length. pG5E1BCAT contains five Gal4 binding sites upstream of the Ad E1B TATA box and CAT gene. This template is responsive to activation by Gal4-AH. Primer extensions of RNAs transcribed from this template are 110 nucleotides (110 n) in length. (B) e55K specifically represses p53-mediated activation at the transcriptional level. In vitro transcription reaction mixtures contained HeLa nuclear extract, template DNAs, and ep53 (20 ng [lanes 3 to 8 and 11 to 16]), Gal4-AH (10 ng [lanes 2, 4 to 8, and 10, and 12 to 16]) and increasing concentrations of e55K (10 ng/μl [lanes 5 to 8, 2, 4, 8, and 16 μl, respectively]) or increasing concentrations of the control BP6 protein fraction (lanes 13 to 16, 2, 4, 8, and 16 μl, respectively). Primer extension products (160 n and 110 n) indicate levels of transcription arising from pB5E4CAT and pG5E1BCAT, respectively and their positions are indicated by arrows. (C) In vitro transcription with templates p5BSCAT and pG5E1BCAT; HeLa nuclear extract; and 40 ng of ep53 (lanes 2 to 5); 10 ng of Gal4-AH; and 30, 70, and 140 ng of e55K (lanes 3, 4, and 5, respectively).
DISCUSSION
In this report, we described the purification of Ad E1B 55K expressed from a recombinant baculovirus. Using a combination of cross-linking, gel filtration, and velocity sedimentation centrifugation, we found that highly purified e55K exists as a dimer in solution. The estimated frictional coefficient of 1.75 also suggests that E1B 55K has an elongated conformation. The quaternary structure of in vitro-translated Ad12 54K and Ad12 54K isolated from human embryo retinal (HER) cells transformed with Ad12 early region 1 has been studied by velocity sedimentation centrifugation (16). This earlier report suggested that 54K is a tetramer. These studies found that 54K, as either an in vitro-translated protein or the cytoplasmic fraction from transformed cells, sedimented as a single-molecular-mass species of ∼200 kDa. p53 from the transformed cytoplasmic fraction also sedimented as ∼200 kDa. Purified wild-type human p53 has been shown to sediment with an apparent molecular mass of between 44 and 158 kDa (15). The nuclear fraction in HER-transformed cells contained two 54K complexes of 200 and 2,000 kDa, both of which contained p53 (16). Since, however, the in vitro-translated protein was not purified, it is possible that the 200-kDa complex reflects 54K in complex with either p53 and/or other cellular proteins. On the other hand, our studies with highly purified protein clearly indicated that E1B 55K is a dimer.
We have also shown that the purified e55K was functionally active with respect to binding to p53 and that the e55K-p53 protein-protein interactions are direct, since they were observed when highly purified protein preparations were used. As previously demonstrated, e55K did not inhibit p53 from binding to its consensus DNA sequence (68). Rather, we observed that the binding of e55K to p53 significantly increased the affinity of p53 for adjacent binding sites, by a factor of ∼10. There is evidence that conformational changes induced in p53 by either binding of monoclonal antibodies to the carboxy terminus or phosphorylation in the carboxy-terminal domain can stimulate p53 DNA binding activity (32). Also, the interaction of p53 with TFIID increases the affinity of p53 for its specific binding site (6a). E1B 55K binds to the N-terminal activation domain of p53 (27, 38). This interaction might increase p53 DNA-binding activity by inducing a conformational change in the protein. Alternatively, given that e55K is a dimer, each subunit could interact with a tetramer of p53 (15, 62), thereby increasing the cooperativity of p53 binding to adjacent sites. The increased DNA binding activity of a p53-55K complex compared to that of p53 alone probably contributes to E1B 55K repression of p53-activated transcription in vivo. The 10-fold increase in DNA binding affinity (Fig. 7) would be expected to result in increased binding of the complex to low-affinity p53 sites, thereby recruiting the 55K repression domain (68) and inhibiting transcription from neighboring promoters.
We have also shown through direct in vitro transcription assays that e55K is indeed a transcriptional repressor, as postulated from in vivo transfection experiments (67, 68). The in vitro transcription assays utilized HeLa nuclear extracts in which the presence of histones could not be detected when assayed by Western blotting (unpublished result). Furthermore, similar levels of 55K repression have also been observed with general transcription factors purified ∼10-fold from the HeLa nuclear extract, and therefore more extensively purified away from cellular histones (unpublished results). These studies therefore imply that histone deacetylation is not the principal mechanism of the repression by E1B 55K, as has been described for other transcriptional repressors such as Mad-Max or Mxi1-Max (1, 20, 34) and the hormone-inducible nuclear receptors (21, 44).
The interactions of p53 with TAFII31 and TBP have been shown to be important for the activation function of p53 (38, 41, 60). Since E1B 55K also binds to the activation domain of p53, E1B 55K could possibly inhibit p53 activation by blocking these interactions. Consistent with previous results (41, 42), we observed that highly purified p53 coimmunoprecipitated in vitro-translated TBP and TAFII31. However, the addition of highly purified e55K to the binding reaction mixtures, at concentrations at which we observed transcriptional repression in vitro, did not diminish the amount of TBP or TAFII31 that bound to p53 in this assay (unpublished results). Consequently, it seems unlikely that E1B 55K inhibits p53 activation by blocking p53’s interactions with TAFII31 or TBP. Mdm2 also inhibits p53’s ability to activate transcription, induce growth arrest, and induce apoptosis (6, 28, 46, 64, 69). The cocrystal structure of Mdm2 protein and a peptide comprising a region of the activation domain of p53 has been solved (33). These studies showed that an α-helical region of p53 lies in a groove in the Mdm2 protein and that the amino acids in p53, which make important contacts with Mdm2, are identical to those required for activation (Leu-22 and Trp-23). These structural and mutational analyses suggested that Mdm2 and TAFII31 interact with the same surface of p53 (33, 35, 38). Mutation of amino acids 22 and 23 also disrupts the p53-E1B 55K interaction (38). However, mutation of either amino acid 24 or 27 disrupts the p53-E1B 55K interaction without affecting the p53 activation function (38). These amino acids lie on the surface of the p53 α-helix which is not embedded within Mdm2, suggesting that E1B 55K contacts a surface of the p53 helix different from that contacted by Mdm2. This could explain how a tertiary structure of p53-55K-TAFII31 would be possible. Support for TBP being the target for repression by a Drosophila transcriptional repressor Eve has come from studies showing that Eve directly interacts with TBP in vitro (61) and that Eve inhibits TBP and TFIID from binding to the TATA box by cooperative blocking (3). Using DNase I footprinting assays, we have found, however, no evidence to suggest that e55K acts to inhibit TBP from binding to DNA (data not shown).
In contrast to our knowledge of activators, relatively little is known regarding the mechanisms utilized by repressors. Repressors can be divided into classes: global and specific. Global repressors, such as Dr1/DRAP (23) and Mot1 (2), repress basal transcription, whereas specific repressors regulate gene expression either by contacting DNA directly through their own sequence-specific DNA binding domains (Kruppel [Kr], Eve, Mad/Max, nuclear hormone receptors) (4, 17, 36) or through protein-protein interactions with a DNA binding protein (E1B 55K via p53, SSN6/TUP1 via α2 or MIG1) (see references 22, 29, and 68 and the references therein). Mechanisms of repression have been reviewed (18, 22, 25, 31). Our interest is focused primarily on repressors which have an active role in repression (i.e., direct repressors). These repressors could either (i) modulate chromatin, (ii) block the assembly of a general transcription factor into the preinitiation complex, or (iii) inhibit the enzymatic function of one of the components of the general transcriptional machinery (phosphorylation of RNA polymerase II carboxy-terminal tail, open complex formation, promoter clearance, elongation, etc.). Furthermore, corepressors could be recruited by the repressor to mediate one of the activities described above. Mad-Max, Mxi1-Max, and the nuclear hormone receptors all recruit a histone deacetylase, HDAC1 or mRPD3, to promoters through their interactions with Sin3A/B, thereby, presumably, resulting in localized histone deacetylation and transcriptional repression (1, 20, 21, 34, 44, 47, 70). This mechanism is also observed in yeast with the DNA-binding repressor protein Ume6 (26). Repressors which directly contact the basal transcription factors include Dr1/DRAP1, Mot1, Eve, Kr, and thyroid hormone receptor (TR), all of which have been shown to repress transcription in vitro (3, 13, 23, 52, 61). Dr1 binds to TBP and thereby prevents entry of TFIIB and TFIIA into the preinitiation complex (23), whereas Mot1 disrupts the TBP-DNA complex in an ATP-dependent manner (2). Eve interacts with TBP (61) and inhibits TBP from binding to DNA (3). Interactions have been observed between the smallest subunit of TFIIE with both Kr (52) and Mdm2 (59), suggesting that these repressors may inhibit TFIIE function. TR inhibits formation of a functional preinitiation complex, possibly through its interaction with TFIIB (13). Other repressors, which have been shown to repress transcription in vitro, include SSN6-TUP1 (49) and KRAB domain repressors (48). Although the mechanisms of repression have not, as yet, been elucidated for these repressors, it is clear that the KRAB repression domain recruits a corepressor to the promoter. Both human KAP-1 and mouse KRIP-1 corepressors have been cloned (14, 30).
In conclusion, we have established a procedure for the purification of E1B 55K, determined the quaternary structure of the e55K molecule, and shown that the purified protein is functionally active for both binding to p53 and for specifically repressing p53 activation at the transcriptional level. It will now be interesting to dissect the mechanism by which E1B 55K represses transcription and ascertain whether mechanisms of repression converge on any particular aspect of transcription.
ACKNOWLEDGMENTS
We gratefully acknowledge Eva Vertelney for the construction of the recombinant baculovirus expressing e55K. We thank Mike Carey for Gal4-AH protein.
This research was supported by grant CA64799 from the NIH. M.E.D.M. was funded by the Jonsson Comprehensive Cancer Center and American Cancer Society (California division) Fellowship J-23-94.
Footnotes
This paper is dedicated to the memory of Carol Newman.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 3.Austin R J, Biggin M D. A domain of the even-skipped protein represses transcription by preventing TFIID binding to a promoter: repression by cooperative blocking. Mol Cell Biol. 1995;15:4683–4693. doi: 10.1128/mcb.15.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 5.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Wu X, Lin J, Levine A J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Chen X, Farmer G, Zhu H, Prywes R, Prives C. Cooperative DNA binding of p53 with TFIID(TBP): a possible mechanism for transcriptional activation. Genes Dev. 1993;7:1837–1849. doi: 10.1101/gad.7.10.1837. [DOI] [PubMed] [Google Scholar]
- 7.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 8.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.el-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 12.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer W E, Vande Woude G F, O’Connor P M, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fondell J D, Roy A L, Roeder R G. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- 14.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X P, Neilson E G, Rauscher F J R. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 15.Friedman P N, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci USA. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grand R J, Mustoe T, Roberts S, Gallimore P H. The quaternary structure of the adenovirus 12 early region 1B 54K protein. Virology. 1995;207:255–259. doi: 10.1006/viro.1995.1074. [DOI] [PubMed] [Google Scholar]
- 17.Han K, Manley J L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 18.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 19.Harlow E, Pim D C, Crawford L V. Complex of simian virus 40 large-T antigen and host 53,000-molecular-weight protein in monkey cells. J Virol. 1981;37:564–573. doi: 10.1128/jvi.37.2.564-573.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 21.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 22.Herschbach B M, Johnson A D. Transcriptional repression in eukaryotes. Annu Rev Cell Biol. 1993;9:479–509. doi: 10.1146/annurev.cb.09.110193.002403. [DOI] [PubMed] [Google Scholar]
- 23.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 24.Jeffrey P D, Gorina S, Pavletich N P. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 25.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 27.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 28.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 29.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 30.Kim S S, Chen Y M, O’Leary E, Witzgall R, Vidal M, Bonventre J V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 32.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 33.Kussie P H, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine A J, Pavletich N P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 34.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 35.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 36.Licht J D, Grossel M J, Figge J, Hansen U M. Drosophila Kruppel protein is a transcriptional repressor. Nature. 1990;346:76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- 37.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 38.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y S, Carey M F, Ptashne M, Green M R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Berk A J. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol Cell Biol. 1995;15:6474–6478. doi: 10.1128/mcb.15.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X, Miller C W, Koeffler P H, Berk A J. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1993;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu H, Levine A J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Martin, M. E. D. Unpublished results.
- 43.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 44.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 47.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 48.Pengue G, Lania L. Kruppel-associated box-mediated repression of RNA polymerase II promoters is influenced by the arrangement of basal promoter elements. Proc Natl Acad Sci USA. 1996;93:1015–1020. doi: 10.1073/pnas.93.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redd M J, Arnaud M B, Johnson A D. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 50.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 51.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 52.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jackle H. Control of transcription by Kruppel through interactions with TFIIB and TFIIEβ. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 53.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 54.Siegel L M, Monty K J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 55.Summers, M. D., and G. E. Smith. 1987. A manual of methods for baculovirus vectors and insect cell culture procedures. Tex. Agric. Exp. Stn. Bull. 1555.
- 56.Teodoro J G, Branton P E. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J Virol. 1997;71:3620–3627. doi: 10.1128/jvi.71.5.3620-3627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teodoro J G, Halliday T, Whalen S G, Takayesu D, Graham F L, Branton P E. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J Virol. 1994;68:776–786. doi: 10.1128/jvi.68.2.776-786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thut C J, Chen J L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 59.Thut C J, Goodrich J A, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Truant R, Xiao H, Ingles C J, Greenblatt J. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J Biol Chem. 1993;268:2284–2287. [PubMed] [Google Scholar]
- 61.Um M, Li C, Manley J L. The transcriptional repressor Even-skipped interacts directly with TATA-binding protein. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang P, Reed M, Wang Y, Mayr G, Stenger J E, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: structure, oligomerization, and transformation. Mol Cell Biol. 1994;14:5182–5191. doi: 10.1128/mcb.14.8.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 64.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 65.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 67.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 68.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 69.Zauberman A, Barak Y, Ragimov N, Levy N, Oren M. Sequence-specific DNA binding by p53: identification of target sites and lack of binding to p53-MDM2 complexes. EMBO J. 1993;12:2799–2808. doi: 10.1002/j.1460-2075.1993.tb05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]