Abstract
Restriction enzymes are among the best studied examples of DNA binding proteins. In order to find general patterns in DNA recognition sites, which may reflect important properties of protein–DNA interaction, we analyse the binding sites of all known type II restriction endonucleases. We find a significantly enhanced GC content and discuss three explanations for this phenomenon. Moreover, we study patterns of nucleotide order in recognition sites. Our analysis reveals a striking accumulation of adjacent purines (R) or pyrimidines (Y). We discuss three possible reasons: RR/YY dinucleotides are characterized by (i) stronger H-bond donor and acceptor clusters, (ii) specific geometrical properties and (iii) a low stacking energy. These features make RR/YY steps particularly accessible for specific protein–DNA interactions. Finally, we show that the recognition sites of type II restriction enzymes are underrepresented in host genomes and in phage genomes.
INTRODUCTION
Protein–DNA interactions play a fundamental role in cell biology. For instance, the highly specific interactions between transcription factors and DNA are essential for proper gene expression regulation (1). The ‘immune system’ of bacteria and archaea relies on restriction endonucleases (REases) recognizing short sequences in foreign DNA with remarkable specificity and cleaving the target on both strands (2–4). REases are indispensable tools in molecular biology and biotechnology (5–7) and have been studied intensively because of their extraordinary importance for gene analysis and cloning work. In addition, they are important model systems for studying the general question of highly specific protein–nucleic acid interactions (2). REases also serve as examples for investigating structure–function relationships and for understanding the evolution of functionally similar enzymes with dissimilar sequences (3).
Based on subunit composition, cofactor requirements, site specificity and mode of action REases have been classified into four types (8). Enzymes of types I, II and III are parts of restriction–modification (RM) systems, which additionally contain methyltransferases (MTases) adding methyl groups to cytosine or adenine in the host DNA. Type IV REases have no cognate MTases; they recognize and cleave sequences with already modified bases (9) and show only weak specificity (8). RM systems occur ubiquitously among bacteria and archaea (10–12). Their principal biological function is the protection of host DNA against foreign DNA, such as phages and conjugative plasmids (13). Other possible functions are to increase diversity by promoting recombination (13,14) and to act as selfish elements (15,16).
Here we study the recognition sequences of all known type II REases. The main criterion for classifying a restriction enzyme as type II is that it cleaves specifically within or close to its recognition site and does not require ATP hydrolysis. The orthodox type II REase is a homodimer recognizing a palindromic sequence of 4–8 bp. The possible advantage of symmetric recognition sites has already been discussed by the discoverers of restriction enzymes (17). They argued economically that it is ‘much cheaper to specify two identical subunits each capable of recognizing’ the half of the symmetrical sequence than to specify ‘a larger protein capable of recognizing the entire sequence’. This may explain the overwhelming majority of palindromic recognition sequences. However, there are other subtypes too—for instance, type IIA REases that recognize asymmetric sequences (8). Recently, the first example of a type II enzyme (MspI) where a monomer and not a dimer binds to a palindromic DNA sequence (18) has been found.
Much has been written about the evolution of REases. When elaborating on this topic Chinen et al. (19) wondered ‘Why are these recognition sequences so diverse?’ Here we show that these sequences are not as diverse as may appear at first sight. Typical patterns can be identified when focusing on purines and pyrimidines. This is apparent from Table 1, which shows the recognition sequences of all restriction enzymes with known three-dimensional structure.
Table 1.
All type II restriction enzymes with known three-dimensional structure and their cognate DNA recognition sequences [PDB, (20)]
| Enzyme | Source | Recognition sequencea | Purine (1)–pyrimidine (0) pattern |
|---|---|---|---|
| MspI | Moraxella species | CCGG | 0011 |
| FokI | Flavobacterium okeanokoites | GGATG | 11101 |
| EcoRII | Escherichia coli | CCWGG | 00W11 |
| EcoRI | E.coli | GAATTC | 111000 |
| BamHI | Bacillus amyloliquefaciens | GGATCC | 111000 |
| HindIII | Haemophilus influenzae | AAGCTT | 111000 |
| BglII | Bacillus globigii | AGATCT | 111000 |
| BstYI | Bacillus stearothermophilus | RGATCY | 111000 |
| EcoRV | E.coli | GATATC | 110100 |
| Cfr10I | Citrobacter freundii | RCCGGY | 100110 |
| NaeI | Nocardia aerocolonigenes | GCCGGC | 100110 |
| NgoMIV | Neisseria gonorrhoeae | GCCGGC | 100110 |
| HincII | H.influenzae Rc | GTYRAC | 100110 |
| Bse634I | Bacillus species 634 | RCCGGY | 100110 |
| MunI | Mycoplasma species | CAATTG | 011001 |
| PvuII | Proteus vulgaris | CAGCTG | 011001 |
| BsoBI | B.stearothermophilus | CYCGRG | 000111 |
| EcoO109I | E.coli | RGGNCCY | 111N000 |
| BglI | B.globigii | GCCNNNNNGGC | 100NNNNN110 |
The corresponding purine (1)–pyrimidine (0) coding shows that 11/00 is a common pattern in all binding sites.
aRecognition sequence representations use the standard abbreviations (21) to represent ambiguity. R = G or A; K = G or T; S = G or C; B = not A (C or G or T); D = not C (A or G or T); Y = C or T; M = A or C; W = A or T; H = not G (A or C or T); V = not T (A or C or G) and N = A or C or G or T.
MATERIALS AND METHODS
All restriction enzyme binding sites were taken from REBASE [last update March 3, 2005 (10)]. Almost all (98%) known REase recognition sequences belong to type II enzymes. We separated the type II binding sites into symmetric and asymmetric sequences, with just 0.96% belonging to the latter class.
The statistical analysis of sequence patterns is based on counting the frequency of all possible substrings up to a length of 4 bp in the symmetric and asymmetric binding sequences (see Supplementary Table S2). In addition to counting substrings of the actual nucleotide sequence, we also counted substrings according to two different binary coding schemes: purine–pyrimidine coding and ketobase–aminobase coding. For the substring analyses of symmetric sequences we consider only the first half of each sequence (the second half is redundant).
Using a binomial distribution, we calculated P-values that quantify the probability of finding the respective subsequence in a randomized set of binding sites at least as often as in the original binding sites. The P-values take account of the relative abundance of each letter (A, G, R, N etc.) in the binding sites (see Supplementary Table S1).
Analysis of dinucleotide H-bond donor and acceptor clusters
We selected B-DNA crystal structures from PDB (20) with X-ray diffraction resolution ≤1.5 Å. Only structures with Watson–Crick base-pairing, without mismatches and without additional ligands were taken into account. The selected PDB entries are 1D8G, 1D8X, 1D23, 1D49, 1EN3, 1EN8, 1ENN, 232D and 295D. The first and last nucleotides in each sequence were omitted from the analysis.
We calculated the average distance between two canonical (22) H-bond donors (and between two acceptors, respectively), each one belonging to one of two adjacent bases. Donor and acceptor pairs must be oriented towards the major or minor groove; pairs with one partner on the major and one partner on the minor groove were omitted. The DNA backbone was not considered for this analysis. Reported distances are averages for the nine selected crystal structures (see Supplementary Table S3). For each dinucleotide base pair we summed all corresponding reciprocal distance values and thus obtained a quantitative measure for H-bond donor and acceptor clusters of each dinucleotide base pair in the major or minor groove (see Supplementary Table S3). The resulting value integrates the number of acceptors/donors and their distance. Simply counting the number of donor and acceptor pairs gives similar results.
Analysis of DNA geometry and flexibility
We analysed four different datasets for the dinucleotide parameters roll, tilt and twist, and three datasets for shift, slide and rise (see Supplementary Table S4). Olson et al. (23) analysed the flexibility in all these six parameters deduced from protein–DNA and pure DNA crystal complexes (yielding two datasets: OlsDNA and OlsProt-DNA). Scipioni et al. (24) deduced the flexibility in roll, tilt and twist from scanning force microscopy images (dataset Scip). Recently (25), all six parameters were calculated from an extensive analysis of structural databases (dataset Per). These authors also found an excellent agreement between database analysis and corresponding molecular dynamics simulations.
RESULTS
Currently, a total of 3726 different REases from 281 bacterial and 26 archaeal genomes are known (REBASE, last update March 3, 2005). The class type II alone comprises 3654 different REases, recognizing 257 different binding sites (the remainder are isoschizomers). Among these are 176 symmetric sequences (mostly recognized by homodimers) and 81 asymmetric sequences. We statistically analysed all type II binding sites and additionally the small datasets of type I, type III and homing endonucleases.
High GC content in DNA binding sites
Our first observation is the significantly enhanced GC content in all type II binding sites: 68% GC and 32% AT. Ambiguous letters (N, R, Y, K and M) were not taken into account (for the complete statistics of base compositions of type II binding sites, see Supplementary Table S1). In contrast, the mean GC content of the host genomes as well as that of the bacteriophages is on average ∼50%. The GC content of the binding sites thus deviates significantly from this genome-wide average (P < 10−300). We argue that this significantly enhanced GC content reflects biological functionality of the binding sites. Three different facts could play a role in this context. (i) In order to protect themselves, hosts have to methylate the specific binding sites in their own genomes. This happens by methylation of either adenine or cytosine. There are two different methylation sites in cytosine [yielding N4-methylcytosine (m4) and C5-methylcytosine (m5)], but only one methylation site in adenine [yielding N6-methyladenine (m6)] (26). All the known results of methylation sensitivity experiments are collected in REBASE (10). We have counted all m4, m5 and m6 methylations that reliably prevent DNA cutting and found 146, 1350 and 524 methylations, respectively. Evolution may therefore have favoured cytosines (over adenines) in RM binding sites. (ii) GC-rich sequences are more stable than AT-rich sequences because of the better stacking interactions. Furthermore, G and C always form three H-bonds in complementary base-pairing and therefore have a higher binding strength than A and T, which pair with two H-bonds. MTases and endonucleases (like other DNA binding proteins) recognize sequences on a bound double strand better than those on open DNA without H-bonds between the two strands at the ‘open’ site. However, the third fact seems to be the most relevant reason for the high GC content. (iii) One A–T base pair allows for five canonical H-bonds between the bases and the recognizing amino acids, whereas the G–C base pair allows for up to six H-bonds (22), which may be beneficial for protein binding. Generally, type II restriction enzymes exhaust the hydrogen bonding potential of their recognition sequence. In contrast, homing endonucleases do not fully exhaust the hydrogen bonding potential. In support of this notion, the mean GC content in homing enzyme binding sites is only 46% (see Supplementary Table S8).
As a generalization one might hypothesize that an enhanced GC content may be an important property of protein binding DNA sequences whenever high specificity is needed. It was found that GC-rich DNA sequences have a higher CAP-binding affinity than AT-rich sites (27) (CAP—Escherichia coli catabolite gene activator protein).
Enhanced occurrence of RR/YY dinucleotides in DNA binding sites
We separated the type II enzyme recognition sequences into symmetric and asymmetric sequences. In the case of the former we analysed only the first half of the sequence. For these two subsets we counted the occurrence of subsequences up to size 4 and calculated the corresponding P-values (see Materials and Methods and Supplementary Table S2). The most abundant dinucleotides are GG and CC. However, owing to the high GC content (which affects the P-value) the most significant dinucleotide is GA (P < 10−69 in the symmetric dataset). Other substrings, such as CTG (P < 10−57 in the symmetric dataset) are similarly significant. A much clearer picture is obtained by considering substrings according to the two different binary coding schemes: purine–pyrimidine coding and ketobase–aminobase coding. Table 2 shows that the two dinucleotides RR and YY are the most significant patterns in the large symmetric dataset. In the much smaller asymmetric set, RRR, YYY and YYYY are even more significant, but RR and YY also stand out. In addition, Table 2 shows that there is no comparably significant ketobase–aminobase pattern. Thus, purine–pyrimidine classification seems to be biologically more important than the ketobase–aminobase categorization. This is also underlined by the fact that among all type II recognition sites the number of Rs and Ys (ambiguous binding sites) is about a factor of 26 higher than the number of Ks and Ms (Supplementary Table S1). REases sometimes allow for some degree of ambiguity, as long as the required purine–pyrimidine pattern is ensured.
Table 2.
Purine–pyrimidine and ketobase–aminobase patterns in type II restriction enzyme recognition sequences
| Pattern | Symmetrical recognition sequences | Asymmetrical recognition sequences | ||||||
|---|---|---|---|---|---|---|---|---|
| Purine (1)–pyrimidine (0) | Keto (1)–amino (0) | Purine (1)–pyrimidine (0) | Keto (1)–amino (0) | |||||
| Frequency | P-value | Frequency | P-value | Frequency | P-value | Frequency | P-value | |
| 00 | 1758 | 6.6E−63 | 1097 | 0.61 | 529 | 5.1E−12 | 294 | 1 |
| 01 | 817 | 1 | 1060 | 1 | 214 | 1 | 379 | 0.59 |
| 10 | 903 | 1 | 1278 | 0.01 | 348 | 0.98 | 524 | 2.0E−15 |
| 11 | 1743 | 1.7E−29 | 1389 | 0.01 | 501 | 4.7E−14 | 380 | 0.69 |
| 000 | 348 | 5.5E−08 | 78 | 1 | 288 | 1.5E−24 | 62 | 1 |
| 001 | 328 | 1.8E−08 | 250 | 9.3E−06 | 81 | 1 | 160 | 0.07 |
| 010 | 89 | 1 | 250 | 9.3E−06 | 79 | 1 | 210 | 1.0E−08 |
| 011 | 165 | 0.99 | 302 | 3.3E−10 | 102 | 0.99 | 129 | 0.92 |
| 100 | 269 | 0.04 | 194 | 0.41 | 140 | 0.79 | 142 | 0.52 |
| 101 | 105 | 1 | 117 | 1 | 104 | 0.99 | 156 | 0.16 |
| 110 | 264 | 0.00 | 271 | 1.8E−05 | 193 | 1.0E−05 | 210 | 3.1E−08 |
| 111 | 310 | 8.3E−13 | 132 | 1 | 231 | 1.5E−15 | 128 | 0.95 |
| 0000 | 150 | 3.2E−27 | 14 | 1 | ||||
| 0001 | 3 | 0.59 | 2 | 0.92 | 24 | 0.99 | 31 | 0.99 |
| 0010 | 26 | 0.99 | 91 | 3.4E−08 | ||||
| 0011 | 1 | 0.94 | 3 | 0.42 | 47 | 0.74 | 53 | 0.36 |
| 0100 | 4 | 0.36 | 1 | 0.98 | 32 | 0.99 | 31 | 0.99 |
| 0101 | 9 | 1 | 34 | 0.99 | ||||
| 0110 | 1 | 0.90 | 35 | 0.92 | 81 | 2.4E−05 | ||
| 0111 | 5 | 0.01 | 39 | 0.90 | 27 | 0.99 | ||
| 1000 | 8 | 0.01 | 1 | 0.98 | 78 | 0.00 | 14 | 1 |
| 1001 | 18 | 1 | 83 | 8.2E−06 | ||||
| 1010 | 1 | 0.94 | 2 | 0.68 | 36 | 0.99 | 89 | 2.3E−07 |
| 1011 | 7 | 0.01 | 5 | 0.01 | 45 | 0.73 | 44 | 0.86 |
| 1100 | 3 | 0.54 | 4 | 0.21 | 82 | 2.7E−05 | 24 | 0.99 |
| 1101 | 2 | 0.74 | 2 | 0.41 | 52 | 0.34 | 109 | 2.0E−13 |
| 1110 | 88 | 1.4E−07 | 91 | 1.2E−07 | ||||
| 1111 | 2 | 0.20 | 94 | 2.3E−10 | 20 | 1 | ||
In the pur–pyr coding 1 stands for purine (A, G, R) and 0 for pyrimidine (T, C, S), and in the keto-amino coding 1 stands for a ketobase (G, T, K) and 0 for an aminobase (A, C, M).
The high statistical significance of two and more consecutive purines (or pyrimidines) in type II enzyme binding sites points to biological relevance. We present evidence for three mechanisms that are potentially responsible for the observed enrichment of this pattern.
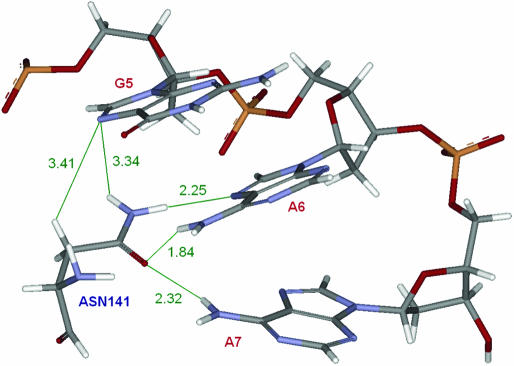
H-bond donor and acceptor clusters. RR/YY steps provide on average stronger H-bond donor (example in Figure 1) and acceptor clusters than other dinucleotides (see Materials and Methods and Supplementary Table S3). Close proximity of acceptor pairs (or donor pairs) on the DNA allows for the establishment of bifurcated H-bonds, which are stronger than canonical single donor–single acceptor interactions. This feature of RR/YY steps potentially facilitates the recognition by and binding of interacting proteins (28). Supplementary Table S3 shows that the average cluster strength of RR/YY steps is higher than that of all other steps. The only (very weak) exception are acceptor clusters in the minor groove, resulting from low strength of the GG/CC step. However, this is counterbalanced by the strong acceptor cluster in the major groove and the donor clusters in the major and minor groove of the GG/CC step. Figure 1 shows an example of a single amino acid (of EcoRI) that potentially interacts with three consecutive purines (GAA) and establishes a bifurcated H-bond.
Figure 1.
Example of an interaction between an H-bond donor cluster (resulting from two adjacent purines AA) and an H-bond acceptor (bifurcated hydrogen bond). The figure shows binding of residue Asn141 from EcoRI to the DNA subsequence 5′-D(GAA)-3′ (only one strand shown). Green lines indicate potential hydrogen donor–acceptor pairs; distances are in angstroms. The structure is according to PDB entry 1CKQ. Note the bending towards the major groove, which reduces the distances between the H-bond donors of the two adenines.
However, there is growing evidence that specific protein–DNA binding is accomplished not only by specific chemical contacts, but also by suitable geometrical arrangement of the DNA and by its propensity to adopt a deformed conformation facilitating the protein binding (29). The following points (ii and iii) show that both properties are better fulfilled by two adjacent purines (or pyrimidines) than by other dinucleotides.
Geometrical arrangement. RR/YY steps allow for a special geometrical arrangement of the DNA (see Materials and Methods and Supplementary Table S4). RR/YY steps are characterized by (a) minimal slide values, without exception; (b) strong tilt in the negative direction [dataset Per deviates somewhat, but ‘tilt is a parameter very sensitive to the choice of calculation method’ (30) and, thus, the consistency of the other three datasets seems remarkable]; and (c) a positive roll in all datasets, which implies positive bending towards the major groove (25). The only exception is the AA/TT step in the Scip dataset. However, AA/TT is by far the least significant dinucleotide of all RR/YY steps (Supplementary Table S2).
Stacking energy. RR/YY steps have a low stacking energy (25) and seem therefore well suited to the often necessary conformational changes during specific protein binding (23,31). Moreover, the stacking energy of all RR/YY steps is anticorrelated with the statistical significance of the RR/YY subsequences (Supplementary Tables S2 and S4). AA/TT has the highest stacking energy and the lowest significance, whereas GA/TC has the lowest stacking energy and the highest significance.
Probably, all three possible reasons for an enhanced frequency of RR/YY steps in type II REase binding sites together play a role in the corresponding specific DNA recognition.
In asymmetric binding sequences longer chains of purines or pyrimidines, such as RRR, YYY and YYYY, are even more significant than RR/YY steps. This could indicate that such substrings are preferred in binding sites. Some dinucleotide parameters, such as stacking energy, more or less add up in longer sequences. On the other hand, a negative correlation between motions at a given base pair step and neighbouring steps was found for most helical coordinates (32).
Binding sites are underrepresented in host and phage genomes
The typical features of type II restriction enzyme binding sites, high GC content and overrepresentation of RR/YY steps, could also be linked to the frequency of these sites in the host and/or phage genomes. To address this question we analysed the genome of E.coli K12 and the known genomes of its phages (33). All four bases are almost equally abundant in both the E.coli genome and the genomes of its phages. Based on this information we can estimate the expected frequency of any given sequence in a randomized genome. Enrichments of sequences are quantified as the ratio of observed versus expected frequency. In addition we calculated weighted ratios, taking into account the number of different enzymes recognizing the same sequence (Supplementary Table S5).
Three findings arise from this analysis: (i) most binding sites are underrepresented in both the host and the phage genomes (possible explanations are that phages try to escape REases and that hosts minimize the methylation effort); (ii) under-(over)representation in host and phage genomes is correlated; and (iii) under(over)representation is correlated with GC content and RR/YY frequency (most underrepresented sequences contain only GC and always contain RR/YY steps). This correlation again underlines the biological importance of these two features.
DISCUSSION
We presented a statistical analysis of all known DNA recognition sites of type II restriction enzymes. This collection comprises by far the largest group of reliably known specific protein binding sites on DNA. There is hardly any sequence similarity among restriction enzymes (34). REases often use uncommon DNA binding motifs (35), but sometimes also typical structures already known from transcription factors, such as FokI and NaeI, which both use a helix–turn–helix motif. The typical features of type II REase binding sites such as high GC content and many RR/YY steps may also be relevant for other DNA recognition sequences. We have also analysed all known binding sites of type I and type III restriction enzymes and of homing endonucleases (Supplementary Tables S6–S8). However, we found no statistically significant motifs, which is probably due to the small number of sequences of these types. Homing endonucleases are known to bind less specifically (10,36). This lack of specificity could be another explanation for the lack of statistically significant patterns among this class of binding sites. Table 3 shows examples of other DNA binding proteins along with their recognition sequences. Nearly all of them contain RR/YY steps. The average GC content of these sequences is 54%.
Table 3.
Examples of gene regulatory proteins that recognize specific short DNA sequences
| DNA binding protein | Recognition sequence (or consensus motif) | Purine (1)–pyrimidine (0) pattern | References |
|---|---|---|---|
| p53 | RRRCW2GYYYRRRCW2GYYY | 1110W210001110W21000 | (38) |
| MADS box | CCW6GG | 00W611 | (39) |
| ERSE | CCAATN9CCACG | 00110N900101 | (40) |
| Ski oncoprotein | GTCTAGAC | 10001110 | (41) |
| GAL4 | CGGN5TN5CCG | 011N50N5001 | (42) |
| GAL4 in vitro | WGGN10–12CCG | W11N10–12001 | (42) |
| nkx-2.5 | CWTTAATTN | 0W001100N | (43) |
| Bicoid | TCTAATCCC | 000110000 | (44) |
| AP-2 | GCCCCAGGC | 100001110 | (45) |
| Stat5-RE | TTCN3GAA | 000N3111 | (46) |
| GRE | AGAACAN3TGTTCT | 111101N3010000 | (46) |
| SRF | CCW2AW3GG | 00W21W311 | (47) |
| MCM1 | CCYW3N2GG | 000W3N211 | (47) |
| NFκB | GGGACTTTCC | 111100000 | (48) |
| pur repressor | ANGCAANCGNTTNCNT | 1N1011N01N00N0N0 | (49) |
| YY1 | GGCCATCTTG | 1100100001 | (50) |
| NF-1/CTF-1 | TGGN6GCCAA | 011N610011 | (51) |
| PPAR | AGGAAACTGGA | 11111100111 | (52) |
| NFAT | ATTGGAAA | 10011111 | (53) |
| CREA | GCGGAGACCCCAG | 1011111000011 | (54) |
| C/EBP | CCAAT | 00110 | (55) |
| PacC | GCCARG | 100111 | (56) |
| TTK finger1 | GAT | 110 | (57) |
| TTK finger2 | AGG | 111 | (57) |
| Zif finger1 | GCG | 101 | (57) |
| Zif finger2 | TGG | 011 | (57) |
| GLI finger4 | TTGGG | 00111 | (57) |
| GLI finger5 | GACC | 1100 | (57) |
| E.coli sigma factors (binding in −35 region) | (58–60) | ||
| σ70 (primary) | CTTGA | 00011 | |
| σ32 (heat shock) | CTTGAA | 000111 | |
| σ60 (nitr. reg. gene) | CTGGNA | 0011N1 | |
| σ54 (nit. ox. stress) | TTGG CACG | 0011 0101 | |
| σ28 (exter. stress) | CTAAA | 00111 |
We presented three different possible explanations for the amplified occurrence of two neighboured purines (or pyrimidines) in the recognition sites. One argument is that these give stronger H-bond donor and acceptor clusters than any other adjacent base pair and therefore facilitate hydrogen bonds to amino acids. For instance, EcoRV (binding GATATC) establishes multiple contacts to the first 2 bp and the last 2 bp, but none to the middle 2 bp (60).
Evolutionary relatedness of REases recognizing similar sequences would be a completely different explanation for our observed patterns. Although only a few REase crystal structures have been solved so far, it became clear from additional bioinformatics studies that REases belong to at least four unrelated and structurally distinct superfamilies: PD-(D/E)XK, PLD, HNH and GIY-YIG (34). The largest one [PD-(D/E)XK] comprises the two major classes α (EcoRI-like) and β (EcoRV-like) (2). Enzymes belonging to the same superfamily sometimes also have similar recognition sequences. For instance, Eco29kI, NgoMIII and MraI, which are related to the GIY-YIG superfamily, all bind to CCGCGG (61). HpyI (CATG), NlaIII (CATG), SphI (GCATGC), NspHI (RCATGY), NspI (RCATGY), MboII (GAAGA) and KpnI (GGTACC) belong to the HNH superfamily (62), and SsoII (CCNGG), EcoRII (CCWGG), NgoMIV (GCCGGC), PspGI (CCWGG) and Cfr10I (RCCGGY) to the EcoRI branch (63). It has already been argued that these enzymes diverged early in evolution, presumably from a type IIP enzyme that recognized CCxGG or xCCGGx (63). We are not aware of any systematic study of recognition sequence similarity versus membership in superfamilies. However, it is conceivable that sequence similarity (or the corresponding purine–pyrimidine pattern) is evolutionarily conserved. Some positive correlation between amino acid similarity and recognition sequence similarity of restriction enzymes has already been found (64). However, REases are extremely divergent and mostly structurally and evolutionarily unclassified (34). Even related enzymes binding to similar DNA sequences may differ much in the details of protein–DNA interaction. Comparing the cocrystal structures of the related enzymes BamHI and EcoRI, it has been inferred that none of the interactions could have been anticipated from the other structure (65). Lukacs and Aggarwal (66) studied the structures of two related enzyme pairs BglII (AGATCT) versus BamHI (GGATCC) and MunI (CAATTG) versus EcoRI (GAATTC), which both differ in only the outer base of the binding site. For the first pair they found ‘surprising diversity’ in how the common base pairs are recognized, whereas the enzymes of the second pair recognize their common inner and middle base pairs in a nearly identical manner.
The problem of recognition and binding of a protein to its specific DNA sequence is far from being solved. Heitman and Model (35) substituted amino acids in the binding domain of EcoRI such that some of the original 12 hydrogen bonds contacting the base pairs of the recognition sequence could not be established by the mutant. This change did not affect the binding specificity of EcoRI, but only its enzymatic activity. It was concluded that the hydrogen bonds revealed by the crystal structure are insufficient to fully account for substrate recognition, and additional amino acids must contact the DNA to help discern the substrate (35). The authors argued that protein–DNA interactions can be influenced by sequence-dependent variation of the structure of the DNA backbone [originally suggested by Dickerson (67)], and that the EcoRI enzyme could recognize its cognate sequence because it adopts its unusual bound conformation more readily than other DNA sequences. It was concluded that even with a detailed cocrystal structure it is exceedingly difficult to determine which interactions contribute to sequence-specific DNA recognition (35). Moreover, it has been found that protein binding to DNA is modulated by sequence context outside the recognition site (68) and that different endonucleases have different context preferences (69).
Our work suggests that sometimes only the purine–pyrimidine pattern matters for recognition by a certain biomolecule. Note that R and Y are most frequent among the ambiguous letters in restriction enzyme binding sites. In such cases the exact base would be irrelevant as long as it is a purine (or pyrimidine). Several such examples are already known. For instance, during translation the third base of the codon is nearly always analysed in this binary manner (in the yeast mitochondrial code this is always the case) (70). Another example is the sequential contact model for EcoRI, proposing that during the transition from DNA binding to DNA scission, the contacts to the pyrimidines could either precede or follow the purine contacts observed in the crystal structure (35). It is known that a change in just 1 bp of the cognate site can reduce the ratio kcat/Km for DNA cleavage by a factor of >106 (71). Thus, a transition exchange might generally have a less dramatic effect than a transversion exchange. Such a smaller effect of a transition exchange could also be observed in corresponding pausing experiments (72), which might be important for protein engineering.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank two anonymous referees for valuable comments. This work has been supported by the Bundesministerium für Bildung und Forschung (Grant 0312704E). Funding to pay the Open Access publication charges for this article was provided by the Institute of Molecular Biotechnology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Beyer A., Hollunder J., Nasheuer H.-P., Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics. 2004;3:1083–1092. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujnicki J.M. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the ‘midnight zone’ of homology. Curr. Protein Pept. Sci. 2003;4:327–337. doi: 10.2174/1389203033487072. [DOI] [PubMed] [Google Scholar]
- 4.Pingoud A.M. Restriction endonucleases. In: Gross H.J., editor. Nucleic Acids and Molecular Biology. Vol. 14. Berlin, Heidelberg: Springer-Verlag; 2004. p. 442. [Google Scholar]
- 5.Chandrasegaran S., Smith J. Chimeric restriction enzymes: what is next? Biol. Chem. 1999;380:841–848. doi: 10.1515/BC.1999.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams R.J. Isolation and characterization of an unknown restriction endonuclease. Methods Mol. Biol. 2001;160:431–442. doi: 10.1385/1-59259-233-3:431. [DOI] [PubMed] [Google Scholar]
- 7.Jenkins G.J., Williams G.L., Beynon J., Ye Z., Baxter J.N., Parry J.M. Restriction enzymes in the analysis of genetic alterations responsible for cancer progression. Br. J. Surg. 2002;89:8–20. doi: 10.1046/j.0007-1323.2001.01968.x. [DOI] [PubMed] [Google Scholar]
- 8.Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J., Blumenthal R.M., Degtyarev S.Kh., Dryden D.T., Dybvig K., et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bickle T.A., Kruger D.H. Biology of DNA restriction. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts R.J., Vincze T., Posfai J., Macelis D. REBASE—restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts R.J., Halford S.E. Type II restriction endonucleases. In: Linn S.M., Lioyd R.S., Roberts R.J., editors. Nucleases. Cold Spring Harbor, New York, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 35–88. [Google Scholar]
- 12.Raleigh E.A., Brooks J.E. In: Bacterial Genomes. De Bruijn F.J., Lupski J.R., Weinstock G.M., editors. New York: Chapman and Hall; 1998. pp. 78–92. [Google Scholar]
- 13.Arber W. Promotion and limitation of genetic exchange. Science. 1979;205:361–365. doi: 10.1126/science.377489. [DOI] [PubMed] [Google Scholar]
- 14.Price C., Bickle T.A. A possible role for DNA restriction in bacterial evolution. Microbiol. Sci. 1986;3:296–299. [PubMed] [Google Scholar]
- 15.Naito T., Kusano K., Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–56. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly T.J., Smith H.O. A restriction enzyme from Hemophilus influenzae II. Base sequence of the recognition site. J. Mol. Biol. 1970;51:393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q.S., Kucera R.B., Roberts R.J., Guo H.C. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Chinen A., Naito Y., Handa N., Kobayashi I. Evolution of sequence recognition by restriction-modification enzymes: selective pressure for specificity decrease. Mol. Biol. Evol. 2000;17:1610–1619. doi: 10.1093/oxfordjournals.molbev.a026260. [DOI] [PubMed] [Google Scholar]
- 20.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomenclature Committee of the International Union of Biochemistry. Nomenclature for incompletely specified bases in nucleic acid sequences. Recommendations 1984. Eur. J. Biochem. 1985;150:1–5. doi: 10.1111/j.1432-1033.1985.tb08977.x. [DOI] [PubMed] [Google Scholar]
- 22.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag, NY: 1984. [Google Scholar]
- 23.Olson W.K., Gorin A.A., Lu X.-J., Hock L.M., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scipioni A., Anselmi C., Zuccheri G., Samori B., De Santis P. Sequence-dependent DNA curvature and flexibility from scanning force microscopy images. Biophys. J. 2002;83:2408–2418. doi: 10.1016/S0006-3495(02)75254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pérez A., Noy A., Lanksa F., Luque F.J., Orozco M. The relative flexibility of B-DNA and A-RNA duplexes: database analysis. Nucleic Acids Res. 2004;32:6144–6151. doi: 10.1093/nar/gkh954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bujnicki J.M. Understanding the evolution of restriction-modification systems: clues from the sequence and structure comparisons. Acta Biochim. Pol. 2001;48:935–967. [PubMed] [Google Scholar]
- 27.Gartenberg M.R., Crothers D.M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- 28.Parra R.D., Furukawa M., Gong B., Zeng X.C. Energetics and cooperativity in three-center hydrogen bonding interactions. I. Diacetamide-X dimers (X=HCN, CH3OH) J. Chem. Phys. 2001;115:6030–6035. [Google Scholar]
- 29.Lankaš F. DNA sequence-dependent deformability—insights from computer simulations. Biopolymers. 2004;73:327–339. doi: 10.1002/bip.10542. [DOI] [PubMed] [Google Scholar]
- 30.Lu X.J., Olson W.K. Resolving the discrepancies among nucleic acid conformational analyses. J. Mol. Biol. 1999;285:1563–1575. doi: 10.1006/jmbi.1998.2390. [DOI] [PubMed] [Google Scholar]
- 31.Rozenberg H., Rabinovich D., Frolow F., Hegde R.S., Shakked Z. Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets. Proc. Natl Acad. Sci. USA. 1998;95:15194–15199. doi: 10.1073/pnas.95.26.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zacharias M., Sklenar H. Conformational deformability of RNA: a harmonic mode analysis. Biophys. J. 2000;78:2528–2542. doi: 10.1016/S0006-3495(00)76798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallin P.F., Ussery D. CBS Genome Atlas Database: a dynamic storage for bioinformatic results and sequence data. Bioinformatics. 2004;20:3682–3686. doi: 10.1093/bioinformatics/bth423. [DOI] [PubMed] [Google Scholar]
- 34.Chmiel A.A., Bujnicki J.M., Skowronek K.J. A homology model of restriction endonuclease SfiI in complex with DNA. BMC Struct. Biol. 2005;5:2. doi: 10.1186/1472-6807-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heitman J., Model P. Substrate recognition by the EcoRI endonuclease. Proteins. 1990;7:185–197. doi: 10.1002/prot.340070207. [DOI] [PubMed] [Google Scholar]
- 36.Jurica M.S., Stoddard B.L. Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bian J., Sun Y. p53CP, a putative p53 competing protein that specifically binds to the consensus p53 DNA binding sites: a third member of the p53 family? Proc. Natl Acad. Sci. USA. 1997;94:14753–14758. doi: 10.1073/pnas.94.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parenicova L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida H., Haze K., Yanagi H., Yura T., Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 40.Nicol R., Stavnezer E. Transcriptional repression by v-Ski and c-Ski mediated by a specific DNA binding site. J. Biol. Chem. 1998;273:3588–3597. doi: 10.1074/jbc.273.6.3588. [DOI] [PubMed] [Google Scholar]
- 41.Vashee S., Xu H., Johnston S.A., Kodadek T. How do ‘Zn2 cys6’ proteins distinguish between similar upstream activation sites? Comparison of the DNA-binding specificity of the GAL4 protein in vitro and in vivo. J. Biol. Chem. 1993;268:24699–24706. [PubMed] [Google Scholar]
- 42.Chen C.Y., Schwartz R.J. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J. Biol. Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 43.Burz D.S., Rivera-Pomar R., Jäckle H., Hanes S.D. Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J. 1998;17:5998–6009. doi: 10.1093/emboj/17.20.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayama M., Takahashi K., Kitamuro T., Murakami O., Shirato K., Shibahara S. Transcriptional control of adrenomedullin induction by phorbol ester in human monocytic leukemia cells. Eur. J. Biochem. 2000;267:3559–3566. doi: 10.1046/j.1432-1327.2000.01384.x. [DOI] [PubMed] [Google Scholar]
- 45.Stoecklin E., Wissler M., Moriggl R., Groner B. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol. Cell. Biol. 1997;17:6708–6716. doi: 10.1128/mcb.17.11.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nurrish S.J., Treisman R. DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 1995;15:4076–4085. doi: 10.1128/mcb.15.8.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karin M., Yamamoto Y., Wang Q.M. The IKK NF-kappa B system: a treasure trove for drug development. Nature Rev. Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 48.Pabo C.O., Sauer R.T. Protein–DNA recognition. Annu. Rev. Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 49.Sakamuro D., Prendergast G.C. New Myc-interacting proteins: a second Myc network emerges. Oncogene. 1999;18:2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 50.Wenzelides S., Altmann H., Wendler W., Winnacker E.L. CTF5—a new transcriptional activator of the NFI/CTF family. Nucleic Acids Res. 1996;24:2416–2421. doi: 10.1093/nar/24.12.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandard S., Muller M., Kersten S. Peroxisome proliferator-activated receptor alpha target genes. Cell. Mol. Life Sci. 2004;61:393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Northrop J.P., Ho S.N., Chen L., Thomas D.J., Timmerman L.A., Nolan G.P., Admon A., Crabtree G.R. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 53.Cubero B., Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lekstrom-Himes J., Xanthopoulos K.G. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 55.Espeso E.A., Penalva M.A. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J. Biol. Chem. 1996;271:28825–28830. doi: 10.1074/jbc.271.46.28825. [DOI] [PubMed] [Google Scholar]
- 56.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular Biology of the Cell, 4th edn. Garland Publishing, NY: 2002. [Google Scholar]
- 57.Harley C.B., Reynolds R.P. Analysis of E.coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jishage M., Iwata A., Ueda S., Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gardner A.M., Gessner C.R., Gardner P.R. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli. Role of NorR and sigma54 in the nitric oxide stress response. J. Biol. Chem. 2003;278:10081–10086. doi: 10.1074/jbc.M212462200. [DOI] [PubMed] [Google Scholar]
- 60.Winkler F.K., Banner D.W., Oefner C., Tsernoglou D., Brown R.S., Heathman S.P., Bryan R.K., Martin P.D., Petratos K., Wilson K.S. The crystal-structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bujnicki J.M., Radlinska M., Rychlewski L. Polyphyletic evolution of type II restriction enzymes revisited: two independent sources of second-hand folds revealed. Trends Biochem. Sci. 2001;26:9–11. doi: 10.1016/s0968-0004(00)01690-x. [DOI] [PubMed] [Google Scholar]
- 62.Saravanan M., Bujnicki J.M., Cymerman I.A., Rao D.N., Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pingoud V., Sudina A., Geyer H., Bujnicki J.M., Lurz R., Luder G., Morgan R., Kubareva E., Pingoud A. Specificity changes in the evolution of type II restriction endonucleases—a biochemical and bioinformatic analysis of restriction enzymes that recognize unrelated sequences. J. Biol. Chem. 2005;280:4289–4298. doi: 10.1074/jbc.M409020200. [DOI] [PubMed] [Google Scholar]
- 64.Jeltsch A., Kröger M., Pingoud A. Evidence for an evolutionary relationship among type-II restriction endonucleases. Gene. 1995;160:7–16. doi: 10.1016/0378-1119(95)00181-5. [DOI] [PubMed] [Google Scholar]
- 65.Newman M., Strzelecka T., Dorner L.F., Schildkraut I., Aggarwal A.K. Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 66.Lukacs C.M., Aggarwal A.K. BglII and MunI: what a difference a base makes. Curr. Opin. Struct. Biol. 2001;11:14–18. doi: 10.1016/s0959-440x(00)00174-3. [DOI] [PubMed] [Google Scholar]
- 67.Dickerson R.E. Base sequence and helix structure variation in B and A DNA. J. Mol. Biol. 1983;166:419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- 68.Beveridge D.L., Barreiro G., Byun K.S., Case D.A., Cheatham T.E., III, Dixit S.B., Giudice E., Lankas F., Lavery R., Maddocks J.H., et al. Molecular dynamics simuletions of the 136 unigue tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(CpG) steps. Biophys. J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engler L.E., Sapienca P., Dorner L.F., Kucera R., Schildkraut I., Jen-Jacobson L. The energetics of the interaction of BamHI endonuclease with its recognition site GGATCC. J. Mol. Biol. 2001;307:619–636. doi: 10.1006/jmbi.2000.4428. [DOI] [PubMed] [Google Scholar]
- 70.Wilhelm T., Nikolajewa S. A new classification scheme of the genetic code. J. Mol. Evol. 2004;59:598–605. doi: 10.1007/s00239-004-2650-7. [DOI] [PubMed] [Google Scholar]
- 71.Taylor J.D., Halford S.E. Discrimination between DNA sequences by the EcoRV restriction endonuclease. Biochemistry. 1989;28:6198–6207. doi: 10.1021/bi00441a011. [DOI] [PubMed] [Google Scholar]
- 72.Jeltsch A., Alves J., Wolfes H., Maass G., Pingoud A. Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry. 1994;33:10215–10219. doi: 10.1021/bi00200a001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.