Central Illustration
Key Words: contractility, myocyte, myofibril, optical methods, optical tweezers, traction force
Highlights
-
•
Heart failure therapies do not address dysregulated contraction.
-
•
New therapies increasing contractility come from multiscale models of contraction.
-
•
Multiscale models of contraction study disease mechanism from molecules to animals.
-
•
Utilizing multiscale models may create new therapeutic paradigms.
-
•
The authors review how the technologies for these models measure contractility.
Summary
Fundamentally, the heart needs to generate sufficient force and power output to dynamically meet the needs of the body. Cardiomyocytes contain specialized structures referred to as sarcomeres that power and regulate contraction. Disruption of sarcomeric function or regulation impairs contractility and leads to cardiomyopathies and heart failure. Basic, translational, and clinical studies have adapted numerous methods to assess cardiac contraction in a variety of pathophysiological contexts. These tools measure aspects of cardiac contraction at different scales ranging from single molecules to whole organisms. Moreover, these studies have revealed new pathogenic mechanisms of heart disease leading to the development of novel therapies targeting contractility. In this review, the authors explore the breadth of tools available for studying cardiac contractile function across scales, discuss their strengths and limitations, highlight new insights into cardiac physiology and pathophysiology, and describe how these insights can be harnessed for therapeutic candidate development and translational.
A key hallmark of both cardiomyopathies and heart failure (HF) is altered contractility, which is commonly demonstrated by changes in systolic or diastolic function on echocardiography, cardiac magnetic resonance imaging, and/or invasive hemodynamic assessment directly measuring cardiac output.1 Per current American College of Cardiology/American Heart Association guidelines, HF is defined as “a complex clinical syndrome with symptoms and signs that result from any structural or functional impairment of ventricular filling or ejection of blood.”1 HF is subdivided on the basis of left ventricular ejection fraction into heart failure with reduced ejection fraction (HFrEF) (left ventricular ejection fraction <40%), HF with midrange or mildly reduced ejection fraction (left ventricular ejection fraction 41%-49%), and heart failure with preserved ejection fraction (HFpEF) (left ventricular ejection fraction >50%).2 HFrEF, characterized by reduced systolic function, is broadly divided into ischemic and nonischemic forms, but standard guideline-directed medical therapy (GDMT) for HF are agnostic to the etiology of HFrEF. HF with midrange or mildly reduced ejection fraction is a more recently defined category that shares similarities with HFrEF and has some positive clinical benefit from the same GDMT, but research is ongoing to better define this category.2 HFpEF, which is generally characterized by impaired diastolic function, is less well defined clinically and mechanistically, but it represents a growing percentage of HF cases. A subset of nonischemic cardiomyopathies associated with altered contractility are inherited genetic cardiomyopathies, including hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), and restrictive cardiomyopathy. Mutations in sarcomeric and contractile proteins are major causes of inherited cardiomyopathies, with worse outcomes for patients with mutations in sarcomeric genes,3, 4, 5, 6, 7 and therefore, it is unsurprising that many of these mutations affect cardiac contraction. As the burden of HF continues to strain limited resources such as donor hearts, there is an outstanding need to better understand the mechanistic basis of contractile dysfunction and design new therapies.
Although HF and cardiomyopathies are both characterized by altered contractility, current treatments for these diseases do not target cardiac contraction. Current GDMT for HFrEF addresses preventing adverse remodeling of the heart. Although this has improved the prognosis for patients with HFrEF, many patients still progress to end-stage HF, requiring mechanical ventricular assist devices and/or cardiac transplantation.8 In the case of HFpEF, there were no treatments available until recently, with the development of sodium-glucose cotransporter-2 inhibitors.9 With genetic cardiomyopathies, mutation-induced changes in contraction can be observed in genotype-positive patients before the onset of pathologic ventricular remodeling; however, current GDMT focuses on treating patients after the onset of symptomatic disease rather than treating the contractile defects driving the disease pathogenesis before adverse remodeling.10, 11, 12, 13
Given the central role of altered contractility in both HF and cardiomyopathies, there has been a long-standing interest to develop molecules that directly modulate contractility. Inotropes, such as dobutamine and milrinone, increase contractility along with intracellular calcium levels, increasing the risk for malignant arrhythmias and mortality.14 As such, inotropes are typically used in a palliative and critical care setting or as a temporary bridging therapy to ventricular assist devices or cardiac transplantation.1 Recently, there has been a push to develop compounds known as myotropes, which directly target sarcomeric machinery to modulate contractility without affecting intracellular calcium.3,5,7,15, 16, 17 For example, the phase 3 randomized control trial GALACTIC-HF (Registrational Study With Omecamtiv Mecarbil [AMG 423] to Treat Chronic Heart Failure With Reduced Ejection Fraction) demonstrated that omecamtiv mecarbil (OM), a small molecule targeting myosin, increases contractility in patients with HFrEF and improves clinical outcomes.18 Another myosin targeting myotrope, mavacamten, was shown to reduce contractility, and it recently became the first U.S. Food and Drug Administration–approved drug for treating patients with hypertrophic obstructive cardiomyopathy.19 The development of these myotropes was driven by the ability to study and model contractility across scales in preclinical models that span from single molecules to whole organisms. Moreover, these multiscale models have revealed new insights into the pathogenesis of cardiac diseases, and they have been applied to investigate the pathogenicity of variants of unknown significance. It is clear that a multiscale approach to defining contractility has the potential to create new treatment paradigms.
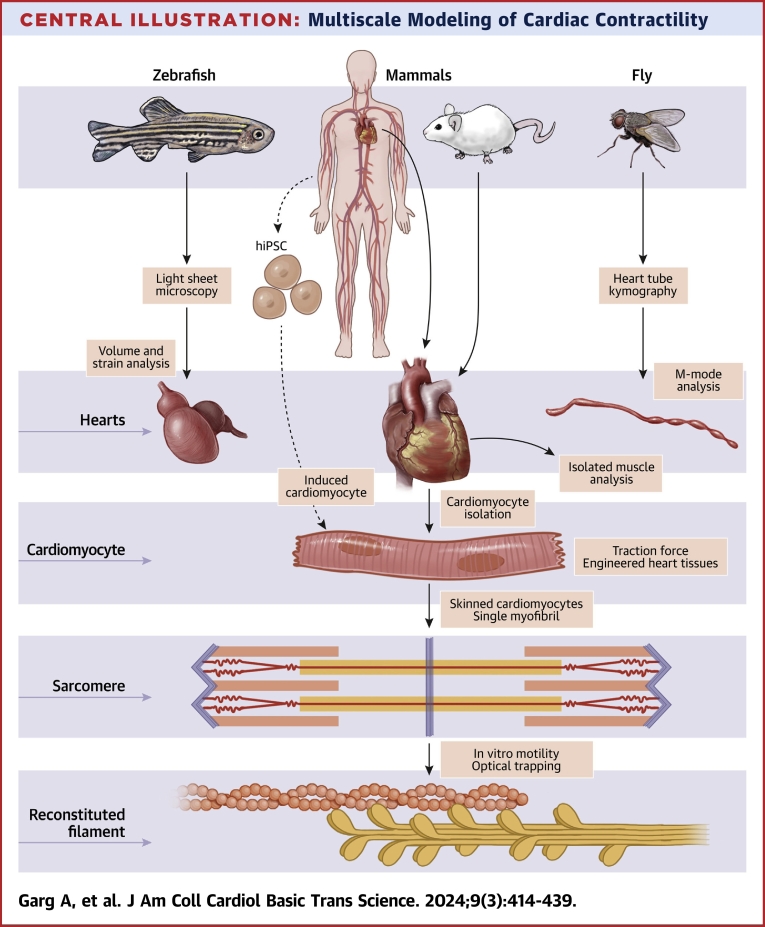
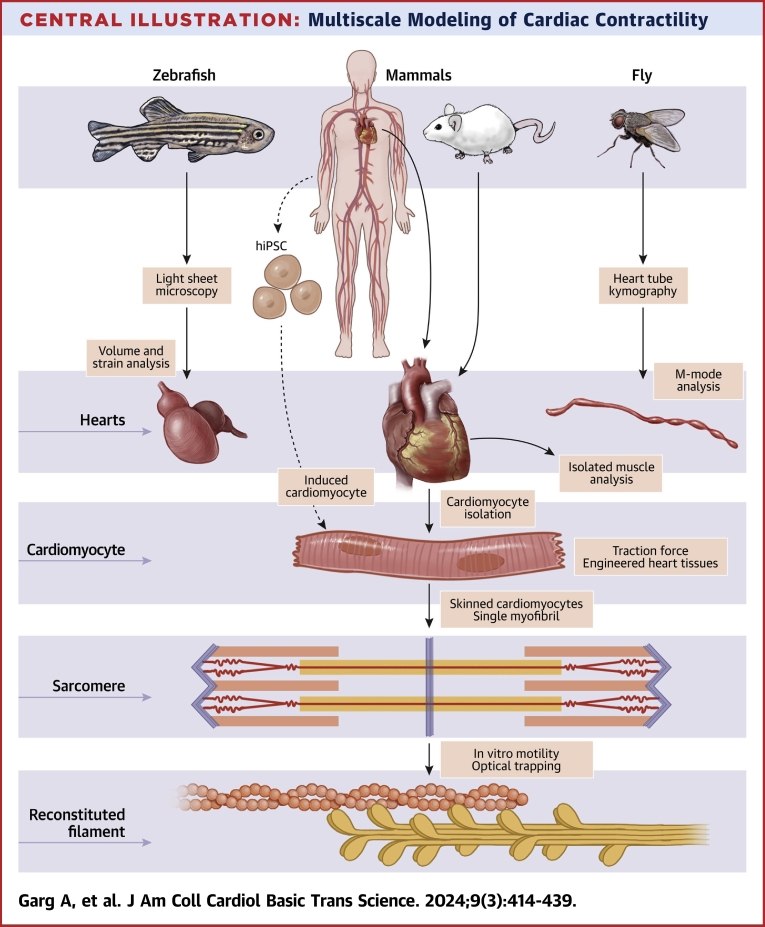
In this review, we describe several multiscale approaches to investigating contractility and provide specific examples of clinically informative insights gleaned from these studies (Central Illustration). A multiscale approach starts with considering the most fundamental interactions between myosin and actin, the principal components of the thick and thin filaments of the sarcomere, respectively. We describe the use of biochemical techniques to reconstitute aspects of contraction at the molecular scale. We follow this with a discussion on contractile studies in skinned cardiomyocytes and single myofibrils that allow biochemical studies while retaining native sarcomeric structures. Then, we explore studies of contractility in human induced pluripotent stem cell–derived cardiomyocytes (iCMs), cardiomyocytes isolated from patients, and engineered heart tissues (EHTs). We close with an overview of selected techniques used to study contractility in model organisms including isolated mouse papillary muscles, Drosophila heart tubes using kymography, and zebrafish hearts using light-sheet microscopy. We discuss the inherent strengths and limitations of each approach and how these tools can complement each other. Finally, we discuss the prospects of these techniques to drive innovative and integrative approaches to studying and treating HF and cardiomyopathies.
Central Illustration.
Multiscale Modeling of Cardiac Contractility
iPSC = induced pluripotent stem cell.
Single-Molecular and Macromolecular Measurements of Contractility
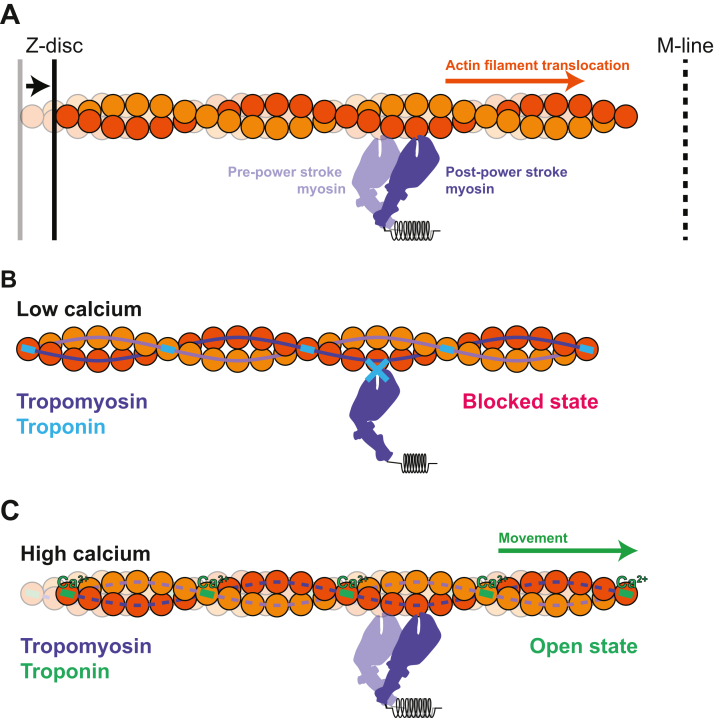
At the molecular scale, force generation occurs in the sarcomere, a macromolecular complex. Force generation in the sarcomere is driven by myosin motors in the thick filament that pull on actin containing thin filaments. This motion draws the Z-discs toward the center of the sarcomere (demarcated by the M-line), resulting in cell shortening and contraction (Figure 1A). Troponin and tropomyosin on the thin filament regulate the calcium-dependent interactions between myosin and actin by sterically blocking myosin binding to the thin filament during diastole (ie, at low calcium levels) (Figure 1B) and enabling these interactions during systole (ie, at high calcium concentrations) (Figure 1C).20,21 In this section, we focus on how in vitro reconstitution techniques can be applied to study the sarcomeric machinery that generates and regulates contraction.
Figure 1.
Calcium-Dependent Actin-Myosin Interactions
(A) Single myosin motor (purple) bound to an actin filament (orange/yellow) in pre– and post–power stroke states that results in movement of the Z-disc (left, solid black line) toward the M-line (right, dashed black line). (B) In the absence of calcium, tropomyosin (dark/light purple line) and troponin (green box) block binding of myosin to the thin filament. This is referred to as the “blocked state.” (C) In the presence of high calcium, calcium (Ca2+) binds to troponin (green), resulting in movement of tropomyosin (dashed light/dark purple lines), allowing myosin binding and thin filament translocation.
Measuring single myosin force generation with optical trapping
A reductionist approach to studying the molecular mechanism of cardiac contractility is the use of reconstituted systems with sarcomere proteins. The simplest contractile unit is a single myosin motor pulling on an actin filament.22 Although several techniques have been developed to study this simplified system, a commonly used tool for studying cardiac myosin is optical trapping.23 Optical trapping uses a tightly focused laser beam to measure nanometer movements and piconewton forces, which are within the range of forces generated by single myosin motors as they interact with actin.24 Importantly, as will be discussed later, optical trapping experiments can directly measure fundamental properties of single myosins related to shortening velocity (eg, myosin’s step size and kinetics), force generation (eg, myosin’s stiffness and unitary force), and power output (eg, load-dependent kinetics). For further information on myosin’s single-molecule properties and their relationships to physiology, the reader is referred to reviews.22,23,25
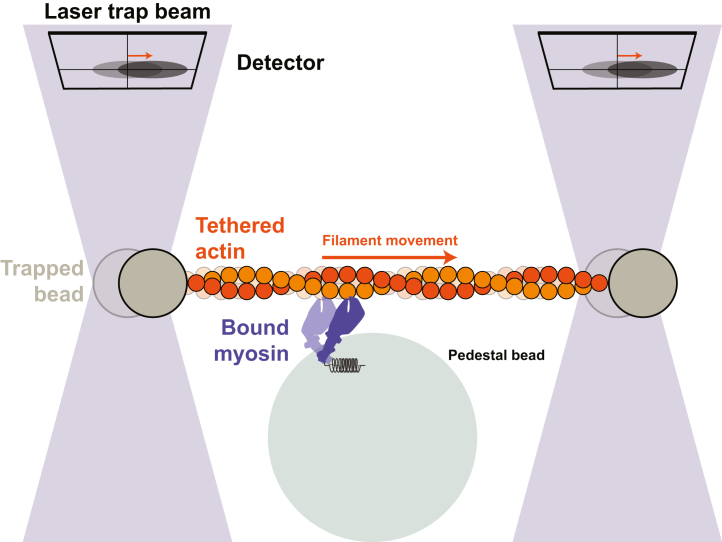
The most common setup for studying single cardiac myosin molecules is a 3-bead arrangement in which a fluorescent actin filament is tethered to 2 optically active beads and then guided to a third surface-bound bead that is sparsely coated with myosin (Figure 2).23,24,26, 27, 28, 29, 30 When the myosin binds to the actin, it displaces the beads, and one can measure both the mechanics (ie, the size of myosin’s displacement) and kinetics (ie, how long myosin remains bound to actin) of this interaction.23 As such, it is an ideal system for studying the coupling between biochemistry and mechanical movements. Optical trapping systems have been used to measure displacements generated by both single27,29, 30, 31, 32 and small ensembles of cardiac myosin motors.33, 34, 35
Figure 2.
3-Bead Optical Trap
Myosin motors are sparsely coated on a fixed pedestal bead (large bead). A single myosin can then bind to and displace an actin filament held between to 2 optically trapped beads (smaller gray beads). When the myosin motor moves the actin filament, a detector senses the displacement of the beads.
The optical trapping systems can be modified to measure how external mechanical forces affect myosin’s stepping rate.29,31,36, 37, 38 This is important as myosin in the heart must generate power against external mechanical loads such as peripheral resistance or outflow obstruction seen with aortic valve narrowing or hypertrophic obstructive cardiomyopathy. External force slows the rate of crossbridge cycling, and this gives rise to the force-velocity relationship for muscle.29,31 Importantly, slowing with load influences power production, enabling the myosin to fine-tune its power output in response to physiological and pathologic loads experienced during the cardiac cycle.29, 30, 31 Changes in the force-velocity or force-power relationship are important for contractile function, and they are often observed in failing hearts and hearts with mutations in sarcomeric proteins.39,40
Optical trapping techniques have been applied to study pathologic mutations in β-cardiac myosin, the major human adult ventricular myosin isoform (MYH7, interchangeably referred to as “myosin” in this review).31,37,38,41,42 Some HCM mutations in myosin, such as D239N and H251N, generate greater power output, while some DCM-associated mutations, such as A223T and R237W, decrease power output.43 These findings have contributed to the hypothesis that the HCM and DCM phenotypes due to myosin mutations are caused by hypercontractility and hypocontractility, respectively, at the molecular scale.25 It should be noted that multiple parameters contribute to power output that are not completely captured with the optical trapping assay alone.22,44 For example, the widely studied myosin HCM mutation R403Q, the first mutation associated with HCM,45,46 showed reduced power output when examined at the single-motor level42 but increased power output when examined in the context of more complex systems such as full-length myosin,47 skinned cardiac myocytes and myofibrils,48, 49, 50, 51 intact muscle fibers,52 and targeted mouse models.48,49,51 Thus, the hypercontractility and hypocontractility hypothesis for some MYH7 mutations requires testing at multiple scales.
Optical trapping has also been used to study drugs that bind directly to contractile proteins. Such studies have revealed fundamental insights into the mechanism of OM, a myosin-binding drug that has completed phase 3 clinical trials and has shown some benefits in treating patients with HFrEF.18 Initially, OM was identified in a high-throughput screen for molecules that increase the cardiac myosin adenosine triphosphatase activity, which in isolation would be expected to increase contractile force.3 Consistent with this notion, OM increases the force of contraction in cardiomyocytes and patients at low doses. However, at higher doses, the force of contraction is reduced, and relaxation becomes longer, impairing diastolic function,16,53 suggesting a narrow therapeutic window that may explain the modest benefit in clinical trials. Using optical trapping, it was found that OM-bound myosin heads do not generate force but instead prolong the binding of myosin to the thin filament.31,32 At low concentrations of OM, prolonged binding of myosin to the thin filament leads to cooperative opening of additional myosin binding sites on the thin filament (ie, increased thin filament activation) and enhanced force production.31,32 However, at high OM concentrations, an excessive number of OM-bound myosin heads ineffectually produce force and decrease systolic function. Furthermore, the prolongation of myosin binding to the thin filament keeps the thin filament in an active state as calcium levels drop, consistent with the prolonged time to relaxation and diastolic dysfunction observed in vitro and in animal models.54 In summary, optical trapping can directly measure both the mechanics and kinetics of the actomyosin interaction, revealing mechanistic insights into physiology, pathologic mutations, and drug mechanisms.
Multimolecular systems to understand contractility
The sarcomere is a complex macromolecular environment consisting of many molecules beyond single myosin molecules interacting with a single actin filament. Importantly, the sarcomere displays additional biophysical properties that emerge as complexity is built into the system.
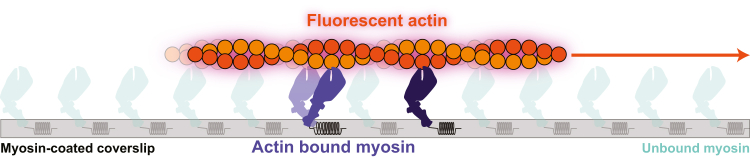
A popular multimolecular assay that can broadly measure actin-myosin–driven contraction is the in vitro actin motility assay.55 In this assay, fluorescently labeled actin filaments are propelled over a bed of myosin, generating movement (Figure 3). Using a standard epifluorescence microscope, one can observe the speed of many filaments moving, which directly correlates with muscle shortening velocity. This assay has the advantages of solution biochemistry (eg, one can completely define the system, including solution conditions that match physiological or pathologic states) and the ability to directly observe changes in contractility. Furthermore, compared with optical trapping, in vitro motility has increased throughput and does not require engineering expertise to setup the system.
Figure 3.
In Vitro Motility Assay
Cartoon showing fluorescently labeled actin gliding on top of a bed of nonfluorescent myosin adsorbed to a glass coverslip. This movement can be measured using a standard fluorescence microscope.
The ability to completely define the system for the in vitro motility assay has enabled studies of actomyosin contractility in the presence of thin-filament regulatory proteins, mechanical load, and intact thick filaments. The thin-filament regulatory proteins troponin and tropomyosin can be added to the assay to recapitulate calcium-dependent regulation of actomyosin interactions.55, 56, 57, 58 With regulated thin filaments, the speed of myosin-driven actin translocation depends on the concentration of calcium, enabling the measurement of thin filament activation (ie, the availability of myosin binding sites on actin).35 In vitro motility assays can also be modified to exert mechanical loads on myosin, enabling the measurement of how load affects the speed of translocation.59, 60, 61 Unlike optical traps that require complex engineering to apply loads to myosin, actin filaments in the in vitro motility assay can be loaded by adding actin-binding proteins (eg, α-actinin or utrophin) that apply a frictional force opposing movement by myosin.60 In contrast to optical trapping which directly quantifies the force produced by single motors, loaded motility assays provide qualitative measurements of force production by many motors referred to as the “ensemble” force. Load can be increased by increasing the concentration of the actin-binding proteins on the surface, which allows generation of qualitative force-velocity curves. Finally, these assays can be conducted using either reconstituted thick filaments from isolated proteins, native thick filaments isolated from tissue, or engineered thick filaments created on DNA scaffolds.62, 63, 64 For example, native thick filaments were used in a motility assay to investigate how myosin-binding protein C affects the speed of thin filament translocation over the thick filament.62,65
The in vitro motility assay has been widely used to characterize the effects of pathogenic mutations in myosin, actin, tropomyosin, troponin, and myosin-binding protein C. Some recent examples include characterizing HCM mutations in the lever arm of MYH7, actin mutations (DCM-associated R312H and HCM-associated R312C), and HCM-associated mutations in tropomyosin (D219V).66, 67, 68 We have used in vitro motility to characterize mutations in troponin T associated with both HCM and DCM.57,58,69 The in vitro motility assay has also been applied to study the effects of small molecules. A recent study showed that OM caused a seemingly paradoxical decrease in motility with increasing concentrations.70 The decreased motility is inconsistent with the initially proposed mechanism of OM as a myosin activator. This key result spurred the optical trapping studies described earlier, which found that OM suppressed myosin’s ability to generate motility.32
Limitations
In vitro reconstitution contractility assays are restricted to a subset of contractile proteins that can be readily purified or expressed in a functional state such as actin, myosin, tropomyosin, the troponin complex, myosin-binding protein C, and small molecules that directly interact with these proteins. Importantly, these specific assays exclude proteins such as titin, which is frequently mutated in inherited DCM. The massive protein titin has an important role in regulating diastolic and systolic function through complex mechanisms.71,72 However, studying the role of integral sarcomere proteins, including titin, can be accomplished in higher order systems such as skinned cardiomyocytes, as discussed in subsequent sections. Although the majority of HCM mutations are found in sarcomeric proteins, which are amenable to these reconstituted contractility assays, the genetic landscape of DCM is much broader, and it includes many proteins not directly involved in sarcomeric contraction.73,74 For example, BAG3 and LMNA mutations are prominent causes of DCM, but these proteins are not directly involved in cardiac contraction, and contractile dysfunction is a secondary, indirect consequence of mutation-induced changes in protein function.75, 76, 77, 78, 79 That being said, it is still possible to study contractile aspects of these diseases in vitro. Native thick and thin filaments can be extracted from cells, tissue from animal models, and human samples to study secondary effects of these mutations on myofilament contractility. Native thick and thin filaments retain post-translational modifications such as phosphorylation and alternative protein isoforms expressed in different pathologic conditions, both of which can regulate myofilament function.65 An alternative approach to studying these mutations is using the higher order systems detailed later. Finally, these reconstituted systems do not recapitulate the complex sarcomere architecture including the spacing between thin and thick filaments.
Skinned Cardiomyocytes and Single Myofibrils
Skinned myocytes or single myofibrils extracted from muscle can be used to study sarcomeric proteins in their native architecture, including full complements of accessory proteins. A skinned cardiomyocyte is a cell that has had its sarcolemma removed by mechanical or chemical means, and their permeabilization allows biochemical manipulation. Permeabilization allows unique simultaneous measurements of force and myosin adenosine triphosphatase rate80, 81, 82 and definition of fine details of muscle physiology using chemically caged compounds that can be activated using light, which is referred to as flash photolysis.83, 84, 85 Permeabilization also has the advantage that it enables studying the direct effects of calcium, drugs, and mutations on contractile function without secondary effects related to altered electrophysiology and calcium handling that are often perturbed in disease.86,87 Moreover, because of their permeabilization, it is possible to exchange certain mutant proteins, as has been done extensively with the troponin complex,88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122 tropomyosin,113,123, 124, 125, 126, 127, 128, 129, 130, 131 myosin light chains,132,133 and even actin.134, 135, 136, 137, 138 Direct protein exchange circumvents the need for patient tissue harboring mutant protein or generation of genetic model organisms. Finally, skinned cardiomyocytes and myofibrils can be processed from unfixed patient biopsy samples and tissue.139,140
Skinned cardiomyocytes
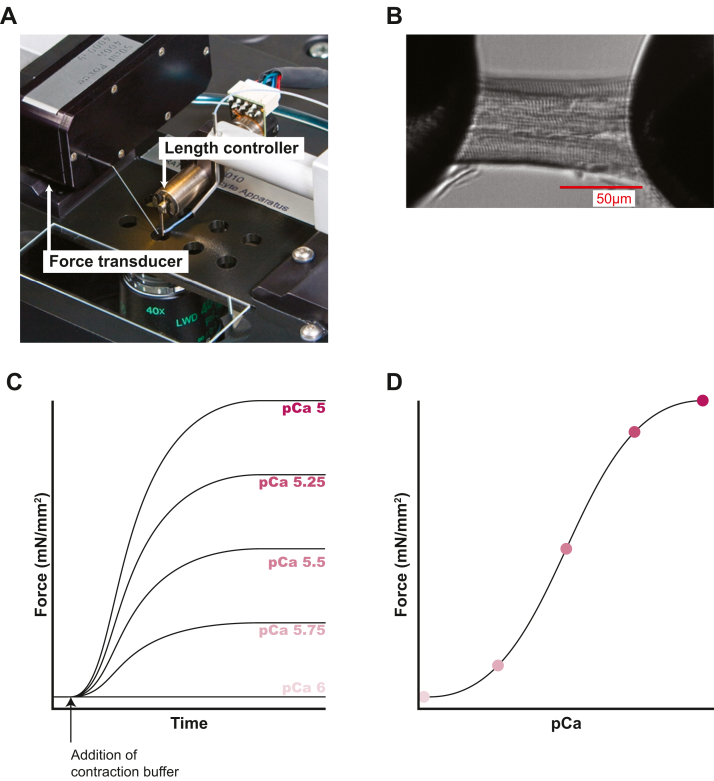
Direct measurement of contractile force in skinned myocytes was first described in skeletal muscle141 and soon after applied to cardiomyocytes.142 In a typical setup for examining skinned myocytes, the myocyte is mounted between a force transducer and a length controller (Figure 4).143 The solution conditions can be controlled by exchanging the bath surrounding the myocyte, and systems have been designed to move samples between solutions (eg, different calcium concentrations). For measurements of length-dependent activation,144 the sarcomere length can be altered by stretching and measured using laser diffraction. More complicated systems have been designed with feedback control of sarcomere length or tension, enabling measurements of additional important parameters such as the force-velocity relationship that can be altered in disease.145,146
Figure 4.
Skinned Cardiomyocyte Contractility
(A) A commercially available system that can be used to measure the force of single myocytes (Aurora Scientific 1600A). The myocyte is mounted between a length mover and a force transducer. Image courtesy of Chris Rand and Aurora Scientific. (B) Magnified view of the attached skinned cardiomyocyte. Reproduced from Greenman et al288 and licensed under a Creative Commons CC BY license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Scale bar in red. (C) Cartoon showing force development in buffers containing differing calcium concentrations. Lower calcium concentrations result in less force generated, with no force generated in the low-calcium pCa 6 (10−6 M calcium) buffer. (D) Cartoon showing the sigmoidal relationship between the maximum force generated and the concentration of calcium (pCa). Colored dots correspond to respectively colored pCa concentrations displayed in C.
The force exerted by the muscle on the force transducer is the sum of active and passive forces. The active force is the force of muscle contraction generated by myosin pulling on thin filaments, and it is related to systolic function. The passive force that resists stretch comes from several sources, most notably the protein titin, though this protein has important roles in both systolic and diastolic function.147, 148, 149 A common measurement in skinned myocyte is the force-calcium relationship, which provides useful information about thin filament activation.150 In this assay, the cardiomyocyte is bathed in solutions containing different amounts of calcium, and the active force generated is measured (Figure 4). This setup has been used to examine multiple pathogenic cardiomyopathy mutations. Studies of mutations in the thin-filament regulatory proteins troponin and tropomyosin have demonstrated that these mutations often affect the calcium sensitivity of activation, where HCM increases calcium sensitivity (which is thought to increase contractility) while DCM decreases calcium sensitivity (which is thought to decrease contractility).88,90,91,94, 95, 96, 97, 98,100,102,104, 105, 106, 107, 108, 109, 110, 111,116, 117, 118, 119, 120, 121, 122,124,126, 127, 128, 129, 130,134, 135, 136, 137, 138 These changes are most consistently seen with mutations in thin-filament regulatory proteins, but this correlation is less consistent for mutations in other protein such as actin and thick-filament proteins.151,152
Similar to the optical trapping and in vitro motility assays, skinned cardiomyocytes can also undergo force-velocity and force-power testing. In these experiments, the skinned muscle is pulled to isometric tension, and then a feedback loop is used to maintain either a constant velocity or constant load as the muscle is allowed to shorten. These techniques were recently applied to study decompensation in a hypertensive animal model.153 Using skinned cardiomyocytes, the investigators found that as hypertensive animals aged, there was a dramatic loss of power output independent of calcium handling. The onset of this power loss was correlated with the onset of HF, reinforcing the importance of looking beyond simple force production and demonstrating the need to consider power production in HF.
Beyond mechanical relationships, skinned cardiomyocytes can also elucidate actin-myosin crossbridge kinetics both in the absence and presence of load. A common technique that is used to measure crossbridge kinetics is the slack-restretch maneuver, in which a myocyte is held at isometric tension, and then the tension is rapidly released and then re-established.154 This causes myosin crossbridges to release from the thin filaments and then reattach, giving a measurement of the rate of tension redevelopment (ktr) which is proportional to the crossbridge cycling rate. This rate has been shown to be altered in some inherited and diabetic cardiomyopathies (discussed later).155, 156, 157 More advanced techniques and modeling are available to measure the rates of key steps of the crossbridge cycle in skinned myocytes, unloaded shortening velocity, and sinusoidal stiffness, all of which can guide downstream biochemical experiments.158, 159, 160, 161, 162, 163, 164, 165, 166
In addition to probing actomyosin interactions, skinned fibers have been instrumental in deciphering the autoregulation of myosin and its perturbation in diseases including cardiomyopathies.22 It has been shown that muscle myosin can form an autoinhibited state known as the “super-relaxed state” (SRX). The presence of the SRX was originally shown in skeletal muscle fibers and later demonstrated in cardiac muscle.167, 168, 169, 170 This state was first observed using skinned fibers, in which the rate of adenosine triphosphate release from relaxed fibers was measured in a pulse-chase experiment using a fluorescently labeled adenosine triphosphate. Autoinhibited myosins in the SRX showed very slow adenosine triphosphate turnover kinetics, while uninhibited myosin in the “disordered relaxed state” had a much faster adenosine triphosphate turnover rate.171,172 Myosins in the disordered relaxed state can generate force in the presence of calcium, whereas myosins in the SRX cannot. Thus, the fraction of myosins in the SRX is a critical determinant of force generation, and this can be dynamically modulated.170 Recent studies of muscle fibers from HCM model organisms have demonstrated that the SRX can be destabilized by many pathogenic mutations, contributing to the hypercontractility seen in patients.133,173, 174, 175, 176 Similarly, skinned cardiomyocytes from a DCM mouse model with a mutation in MYL2 (myosin regulatory light chain), D94A, demonstrated an increase in the population of the SRX, which contributes to hypocontractility.175
Along with genetic cardiomyopathies, skinned cardiomyocytes can be used to study pathologies with complex effects on contractility, which was done in a study of patients with diabetic cardiomyopathy.157 Chronic exposure to elevated glucose causes glycation of key sarcomeric proteins; however, the functional consequences of this modification and its relationship to diabetic cardiomyopathy were not well understood.177 Skinned cardiac muscle fibers were treated with a compound to glycate sarcomeric proteins, and it was shown that glycation reduces the peak tension generated and slows the rate of tension redevelopment. These effects likely contribute the systolic dysfunction in diabetic cardiomyopathy.
Another complex phenotype that has been successfully studied with skinned cardiomyocytes is HFpEF, and this study demonstrated the potential for direct bedside-to-bench studies using samples from patients. As stated previously, HFpEF remains both poorly defined clinically and mechanistically. A recent study used skinned cardiomyocytes from patients with HFpEF to elucidate the contractile effects of this disease.178 In that study, the investigators obtained right ventricular septal biopsies from patients with HFpEF undergoing right heart catheterization, which is a standard diagnostic procedure for cardiologists. Active and passive forces were measured for individual skinned cardiomyocytes from flash-frozen biopsy tissues. The investigators found increased passive force, consistent with diastolic dysfunction but also a striking decrease in calcium-dependent active force generation in myocytes from patients with HFpEF. The defect in active force generation at the skinned myocyte level seemingly differs from the clinical observation that hearts from patients with HFpEF show intact contractility with impaired relaxation. On the basis of their observation, the investigators speculated that applying pro-contractility agents may present a new avenue of treatment for HFpEF; however, further studies are required to test this hypothesis.
Finally, skinned cardiomyocytes can be used to study the effects of direct sarcomeric modulators such as OM and mavacamten, furthering the translational power for this system.53,179, 180, 181, 182, 183, 184, 185, 186 For example, skinned cardiomyocytes from donor and failing hearts were used to explore the effects of OM on force generation, the relaxation rate, and the rate of tension redevelopment.182 Interestingly, although OM increased force generation in the myocytes from failing hearts, relaxation time was increased as was the time to redevelop force and reach maximum force compared with nonfailing donor myocytes. The investigators speculated that high concentrations of OM may compromise diastolic filling, which may blunt the overall inotropic effect of OM, consistent with molecular studies; however, titrating the dose of OM in patients with HF could improve its therapeutic benefit while minimizing the adverse effects and will need to be explored more in the future.
Single-myofibril contractility
In contrast to an array of many parallel sarcomeres, single myofibrils consisting of several sarcomeres linked in series can be isolated from skinned myocytes for mechanical studies.187,188 A single myofibril can give similar information about contractile function as a skinned cardiomyocyte but without contributions of other cellular elements that may confound these measurements such as nonsarcomeric cytoskeletal components. Moreover, the amount of starting material needed for a single myofibril assay is less than that of a skinned cardiomyocyte, which may be beneficial for maximizing replicates when tissue amount can be limiting, such as the case with patient biopsies. Finally, the small size of a single myofibril enables more rapid switching of buffer conditions compared with skinned myocytes.
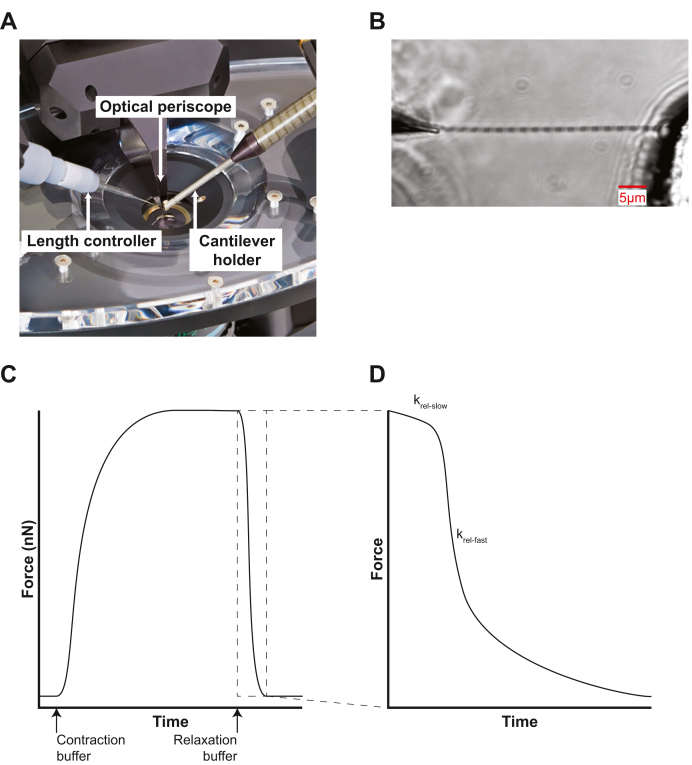
The evolution of single-myofibril techniques was extensively reviewed recently.189 In a typical single myofibril experiment, a myofibril is dissected from a skinned myocyte and then attached between a length mover and a force transducer (eg, pulled glass pipette or cantilever) (Figure 5). A dual-chamber pipette is then brought near the myofibril and different buffers (e.g., high and low calcium buffers) are flowed through each chamber. The dual-chamber pipette can be rapidly moved for quickly switching of buffer conditions. This rapid switching without the need to move the myofibril enables the observation of fast contractile changes that cannot be seen with skinned myocytes.
Figure 5.
Single-Myofibril Contractility
(A) Commercially available system for single myofibril measurements (Aurora Scientific 1700A). The myofibril is mounted between a length mover (left) and cantilever holder (right), which is visualized by an optical periscope (middle). Image courtesy of Chris Rand and Aurora Scientific. (B) Magnified view of attached single myofibril held between a length mover and the stationary force transducer. Approximate scale bar displayed in red. Image used with permission from Aurora Scientific. (C) Cartoon showing the force generated by a single myofibril. The myofibril is initially bathed in relaxing buffer, then bathed in activating buffer, and then relaxing buffer again. Note the difference in the magnitude of forces between a single myofibril shown here and a skinned cardiomyocyte in Figure 4 (nanonewtons vs millinewtons). (D) Cartoon showing the time expanded view of region of interest from C showing transient linear relaxation phase (krel-slow) and the longer exponential relaxation phase (krel-fast).
When a myofibril is rapidly switched from a high-calcium activating buffer to a low-calcium relaxing buffer, the tension relaxes in 2 distinct phases.86 The first phase is short lived, but the rate of relaxation is slow and linear (krel-slow). The rate of this phase is proportional to the rates of thin filament deactivation and crossbridge detachment. This linear phase is not observed in skinned myocytes, as it is not possible to switch buffers fast enough with full myocytes. The linear phase is followed by a fast exponential decay (krel-fast) that relates to the active and passive relaxation forces190 (Figure 5C). This technique was recently used to study the effects of the small molecule danicamtiv that is currently being investigated to increase contractility.191 The investigators found that danicamtiv changes the linear phase of relaxation when calcium levels drop. This suggests that danicamtiv slows the rate of crossbridge release from the thin filament, which would slow the rate of myofibril relaxation and could induce changes in diastolic function. Notably, slowing the rate of myofibril relaxation is thought to attenuate the therapeutic benefit of OM. It is possible that danicamtiv may have similar issues54; however, a recent study showed a more mild effect on diastolic function with danicamtiv compared with OM.16 This mechanistic insight would not have been gleaned from skinned myocytes which do not show a linear phase.
Numerous studies have used single myofibrils to study changes in contractility and kinetics under physiological conditions, in the context of genetic cardiomyopathies, and in the presence of proteins with specific post-translational modifications.189 In one study using a transgenic mouse model of DCM-associated cardiac actin E361G, the investigators showed that single cardiac myofibrils had increased calcium sensitivity but nearly one-half the rate of relaxation for both krel-slow and krel-fast.192 By testing other parameters that change calcium sensitivity, such as the dephosphorylation of troponin I or a small-molecule activator of troponin C, the investigators found that changes in calcium sensitivity correlate with changes in krel-slow and krel-fast. This study and others demonstrate the ability to glean multiple biochemical parameters from a single myofibril experiment.
Limitations
Removal of the sarcolemma influences myofilament organization, passive mechanical properties, calcium responsiveness, and force-length relationships of myofibers compared with intact myofibers.193 Furthermore, probing changes in contractility that rely on electrochemical coupling or cell metabolism are not feasible in demembranated cells. Additionally, though the skinned cardiomyocyte is freely permeable with its bathing solution, measurements are limited by diffusion of buffer into the cells, though this issue is greatly mitigated in single myofibrils. In comparison with biochemical reconstitution systems, contractile changes seen in cardiomyocytes and myofibrils are not as readily relatable to a single molecular defect given the increased molecular complexity of these systems. For example, it is difficult to decipher whether changes in force observed in skinned fibers come from changes in the force generated by individual myosins, the number of active crossbridges, the kinetics of individual crossbridges, and/or the stiffnesses of individual crossbridges. Finally, although the skinned cardiomyocyte and single myofibril experiments are conceptually very similar, they require very different equipment to execute (Figures 4 and 5), with single myofibrils presenting a greater technical challenge.
Measuring Contractility in Living Cells
Cardiac contractility in induced cardiomyocytes
There have been several technical advances that have enabled the study of human cardiomyocytes. One advance is the ability to generate stem cell–derived iCMs.194 These cells can be paired with gene-editing techniques such as CRISPR/Cas9 to generate isogenic cell lines containing mutations or isogenic control cell lines from patients from whom the mutation has been removed.195, 196, 197, 198, 199, 200 The process for creating iCMs from human induced pluripotent stem cells has become substantially more streamlined with multiple protocols and commercially available reagents.201,202 A long-standing challenge with this model is the immaturity of these cells compared with adult cardiomyocytes203,204; however, there are various approaches being used to improve this maturity, which is discussed in a recent review.205 Although these cells cannot recapitulate end-stage HF, they can give powerful insights into the early disease pathogenesis before the onset of maladaptive remodeling.206 Currently, mature cardiomyocytes can be derived only from heart tissue, and protocols have been optimized to obtain these cells from rodents.207 There have been efforts to improve the isolation of human primary cardiomyocytes, with one protocol successfully carrying cells through a cryopreservation and replating cycle.208 However, even with these improvements, experiments taking more than 1 week to complete remain challenging, and there are still major hurdles with obtaining sufficient tissue from appropriately matched patients and with the inherent variability of human samples. Here, we focus on measuring contractility from single cells and microtissues derived from human induced pluripotent stem cells.
Measuring sarcomeric shortening in living cells
Two important contractile parameters of cardiac muscle are fractional shortening (ie, the percentage length change during shortening) and shortening velocity, both of which can be directly measured for individual cardiomyocytes.209 It has recently become possible to measure these parameters for individual sarcomeres within live cardiomyocytes using fluorescently labeled probes. The sarcomere is particularly amenable to these approaches because of its regular, repeating structure. Several approaches have been applied to introduce fluorescent proteins into live sarcomeres, including using small molecules (eg, LifeAct), viral transduction of plasmids for the overexpression of tagged sarcomeric proteins, and gene editing of endogenous proteins to fuse them to fluorescent tags (eg, alpha actinin, myomesin, titin, myosin regulatory light chain).210, 211, 212 A large selection of validated human stem cell lines with genomic expression of tagged sarcomere proteins are available from the Coriell Institute for Medical Research.
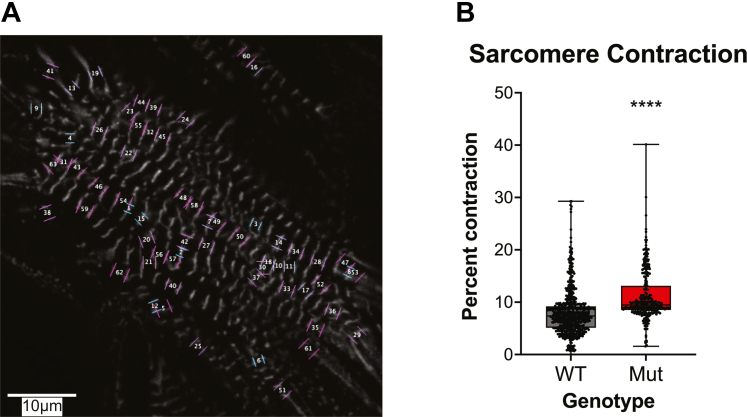
Fluorescently labeled sarcomeric proteins enable the measurement of several important parameters, including sarcomeric length, fractional shortening, and shortening velocity (Figure 6). There are several excellent open-source software packages for the analysis of sarcomeric contraction in living cells, and the reader is referred to one of these packages.213 Sarcomeric tracking was used to study myosin-binding protein C haploinsufficient cells, and that study demonstrated that it is possible to improve hypercontractility in these cells using mavacamten.214 Similarly, this technique has been used to prospectively study several other mutations in TNNI3 (cardiac troponin I), TNNT2 (cardiac troponin T), and MYH7.176,215
Figure 6.
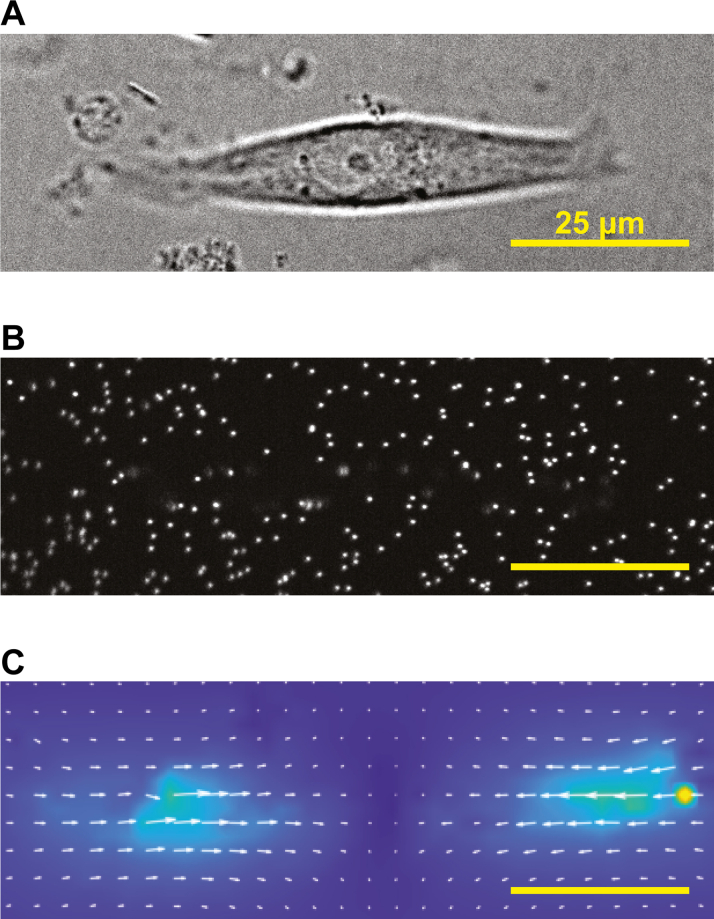
Fluorescent Sarcomere Contractility Tracking in Live iCMs
(A) Stem cell–derived cardiomyocytes expressing ACTN2-mEGFP, which demarcates Z-discs. Sarcomeres were identified and tracked using the SarcTrack software package.213 Note that not all sarcomeres are tracked, on the basis of a filtering algorithm in SarcTrack, which removes sarcomeres that are not reliably tracked for the entirety of the recorded video. (B) Representative data generated using SarcTrack showing a mutant (Mut) cell line that has greater contractility compared with the wild-type (WT) line.
One limitation of this approach is the need for fluorescent proteins to track sarcomeric contractions, which requires either genomic modification or lentiviral overexpression. When generating new lines with fluorescently tagged sarcomeric proteins, it is important to conduct controls to ensure that the fluorescent protein does not affect sarcomeric function.212 Gene editing of isolated cardiomyocytes from patients is currently not possible, because of challenges with long-term culture; however, these cells could be treated with viral overexpression of tagged proteins or peptides. Another limitation is that this technique does not directly measure force. Force-tuned fluorescent tensions sensors have been developed for other cytoskeletal proteins, but these have not yet been applied to measuring forces in the sarcomere.216 Finally, a general theme of single-cell techniques is that it is challenging to apply force to cells to analyze the force-length relationship, although studying force-frequency responses is technically possible.
Measuring cardiomyocyte force generation
Several techniques have been developed for measuring the contractility of single intact cells, but the most frequently used technique is traction force microscopy,217 in which a cardiomyocyte is placed on a deformable hydrogel of known stiffness that contains small, embedded fluorescent beads (Figure 7). When the cardiomyocyte contracts, this force deforms the substrate, displacing the beads. From the known stiffness of the substrate and the displacement of the beads, it is possible to calculate the active force of contraction.211 There are several open-source packages for analyzing traction force experiments, and these programs can calculate important parameters, including peak force, power, and contraction and relaxation rates.211,218,219 Moreover, the stiffness of the substrate can be tuned to several different stiffnesses to mimic physiological and pathologic conditions.211,220,221 Very few cells are needed to perform analysis, which can considerably lessen resource commitment.
Figure 7.
Traction Force Microscopy of Cardiomyocytes
(A) Bright-field image of a patterned stem cell–derived cardiomyocyte on a hydrogel. (B) Fluorescent beads embedded within the hydrogel. (C) Resultant strain map showing forces generated by contraction. This was calculated using the software package Contrax. Note that the strain points toward the center of the cell with maximal strain at opposite poles of the cell.289
A key advantage of a live cell system is that it enables the study of physiology, including signaling pathways, protein isoform switching, and genetic and epigenetic mechanisms. At the molecular scale, point mutations directly affect the structure, function, and/or abundance of the mutated protein, and this molecular dysfunction can lead to the activation of adaptive and maladaptive downstream signaling.44,57,58,69 It is often not possible to examine these changes outside of the context of a live cell, necessitating the pairing of molecular and cellular techniques. For example, we showed that the HCM R92Q mutation in troponin T causes molecular hypercontractility, but this leads to downstream changes in calcium handling, gene expression, and electrophysiology.58 Intact cellular systems have been applied to study nonsarcomeric DCM mutations, including ones in the chaperone protein BAG3, the intermediate filament protein LMNA, and the calcium handling protein PLN.75,76,78,222,223 These studies all revealed reduced contractility, consistent with the known DCM phenotype, but they have very different molecular mechanisms, and the observed changes in contractility are secondary to the direct mutation-induced changes in protein function.44
Limitations
As stated previously, immaturity remains an ongoing issue for studies with iCMs, especially when it comes to electrochemical coupling, metabolism and expressing mature sarcomere protein isoforms.205,224 Mechanistically, deciphering the direct connection between the initial molecular insult and pathogenesis of disease in living cells is more difficult given the inherent complexity of a cell compared with a biochemically reconstituted system or skinned cardiomyocytes. However, this system lacks the complexity of myocardial tissue, which contains additional cell types and structures that may influence the observed contractile phenotype.225,226
Measuring Cardiac Tissue Contractility in Vitro and Ex Vivo
Engineered heart tissues
Multiple platforms have been developed to generate 3-dimensional (3D) human EHTs.219,227, 228, 229, 230, 231, 232, 233, 234, 235, 236 EHTs can recapitulate critical cues experienced in the heart that are missing from most single-cell assays, such as mechanical and electric stimulation, 3D arrangement, and the presence of noncardiomyocyte cell populations.237, 238, 239, 240 EHTs are typically designed in specialized microfabricated or microelectromechanical systems to investigate key parameters of contractile function while limiting the required amount of input material relative to macroscale tissues.228,230,233,234 Recently, it has become possible to generate EHTs on 3D printed scaffolds that geometrically resemble a heart and can recapitulate key aspects of whole-heart contractility.241 The reader is referred to recent reviews on these technologies.239 Currently, it is not possible to make tissues from adult cardiomyocytes, and systems have focused primarily on iCMs or neonatal rat ventricular cardiomyocytes, which readily form tissues.
The most common EHTs used to study contractility consist of iCMs and cardiac fibroblasts embedded in an extracellular matrix (usually collagen or fibrin). Broadly speaking, there are 2 types of EHT systems for measuring contractility: passive systems, which measure forces without allowing the user to actively manipulate the force on the tissue, and active systems, with which the user can manipulate the force.237 One type of passive system is a system in which a linear tissue strip is formed between 2 deformable posts of known stiffness, and then the force of contraction during beating can be calculated from the displacement of the posts (Figure 8).238,242 This system has been used to study several conditions, including cardiomyopathy mutations, doxorubicin-induced cardiotoxicity, and the effects of drugs on tissue function.229,231,243 An advantage of this system is the ability to add in additional cell types, and we recently used this system to investigate how SARS-CoV-2 affects the contractile properties of human heart tissue.244
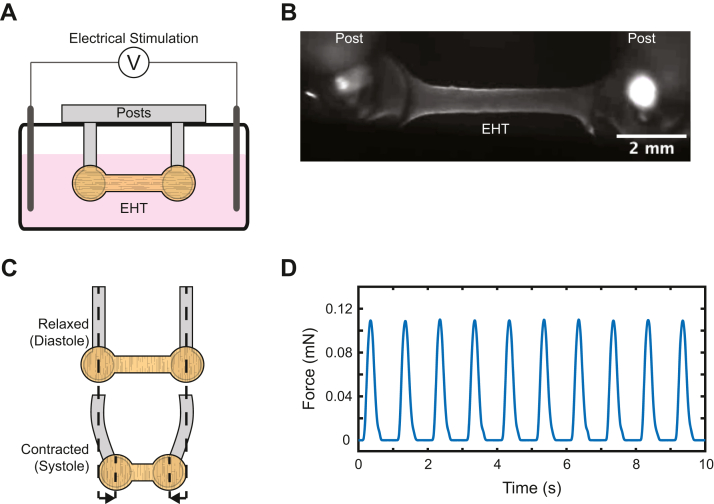
Figure 8.
Force Measurement With Passive Linear EHT System
(A) Side-view schematic of a single engineered heart tissue (EHT) (orange) adhered around posts of known stiffness and immersed in media with electric stimulation. (B) Image of a linear EHT courtesy of Lina Greenberg (Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine). (C) Schematic showing post deformation with contraction. The force can be calculated from the stiffness of the posts and the magnitude of the displacement. (D) Sample force trace showing EHT contraction with electric stimulation.
One example of an active system is a ring-shaped tissue245,246 that can be attached between a force transducer and a length mover, allowing one to measure the active and passive forces of the tissue and establish force-length relationships (ie, Frank-Starling) (Figure 9).247 This ring-tissue configuration was recently applied to study a female patient with severe HF carrying an X-linked Δ45-58 DMD mutation and a heterozygous PLOD3 stop variant.248 iCMs generated with both the Δ45-58 DMD mutation and the PLOD3 mutation showed a decrease in active force and stiffness. Upon correction of the PLOD3 mutation, stiffness, but not active force generation, was restored. Analysis of the EHT showed decreases in collagen synthesis offering a possible explanation to the decrease in tissue stiffness.
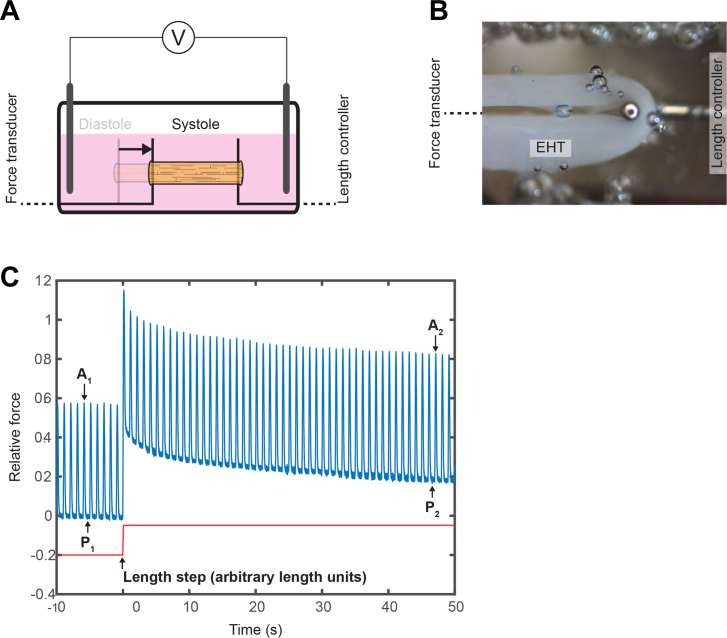
Figure 9.
Force Measurement With Active Ring Shaped EHT System
(A) Side-view schematic of a ring tissue (orange) attached between a force transducer (left) and a length controller (right). The tissue is immersed in media and electrically stimulated. When the tissue contracts, this results in displacement of the force transducer. (B) Partial overhead image of ring tissue attached to force transducer (left, off screen) and length controller (right). (C) Representative force tracing (blue) showing tissue beating before and after a length step (red). The length step results in an increase from the initial passive force (P1) to a new passive force (P2). Active force modestly increases after stretch (A1 vs A2) because of the Frank-Starling mechanism. The tissue shows evidence of viscoelasticity from the increase and subsequent relaxation in total force following the length step. EHT = engineered heart tissue.
Limitations
Although EHTs improve the maturation of iCMs, the iCMs do not reach the same level of maturity as adult cardiomyocytes.239 It should be noted that the required degree of maturity needed to model a disease will vary depending on the disease process being studied. An additional limitation to EHTs is that they require more cells than single-cell techniques. Moreover, although these tissues have a 3D architecture, they lack the organization of cells and extracellular matrix as well as the diversity of cells found in the human heart. Recently, it was demonstrated that it is possible to generate cardiac organoids that contain many of the cell types in the heart, but this technology is still under development.249 There have been advances in 3D printing of heart tissues and the use of decellularized heart scaffolds, but these tools still cannot completely recapitulate this organization.250,251
Intact muscle tissue isolated from model organisms
It is possible to directly study the contractility of isolated intact papillary or trabecular muscles from transgenic animals.252 This technique can be applied to tissue from human patients, but there are major logistical challenges with these studies, as human samples are usually obtained from explanted hearts and require immediate use, unlike skinned cardiomyocytes and myofibrils.253 With that said, there have been successful contractility studies of intact trabeculae muscle obtained from human hearts.254, 255, 256
In contrast to skinned cardiomyocytes and single myofibrils, isolated papillary muscles retain electrochemical coupling with their intact sarcolemma, allowing cardiac conduction. In papillary muscle studies, the muscle tissue is mounted between a length mover and a force transducer. By retaining cell viability and electric coupling, the contractile effects of changes to ion channel conduction, calcium signaling, adrenergic signaling, and additional pathways can be studied. Techniques have also been developed to simultaneously measure force and calcium transients.81,82,257,258 Furthermore, these fibers can be manipulated to measure force-length, force-velocity or force-power, and force-frequency relationships.252,259,260 These tissues retain the native architecture of cells and matrices, enabling the study of complex interactions between cells and the extracellular matrix.
Isolated murine papillary muscle fibers were used to test whether overexpression of SERCA2a could mitigate the impact of diabetic cardiomyopathy.261 In the absence of overexpression, isolated papillary muscles from a diabetic animal model showed a >60% reduction in systolic force and diastolic relaxation; however, these effects were completely reversed with SERCA2a overexpression. In another example the effects of mechanical unloading by heterotopic heart transplantation (ie, connecting a second donor heart to a recipient while maintaining the original heart) were investigated using papillary muscles isolated from a rat DCM model.262 The investigators found that mechanical unloading improved contractility and β-adrenergic sensitivity, which are important clinical targets for preserving overall cardiac function.
Limitations
Unlike skinned cardiomyocytes and single myofibrils, isolated muscles must be used immediately after harvesting. Furthermore, proteins cannot be exchanged into the intact muscle fiber as they can be for skinned cardiomyocytes and single myofibrils. Thus, mutant proteins must already be present in the muscle fiber prior to harvest. Despite these limitations, measuring the contractility of intact muscle remains an important technique given the ability to retain a viable muscle fiber that is capable of being electrically stimulated while retaining other important cell types and matrix organization.
Measuring Contractility in Vivo
In vivo models of cardiac disease provide the most complete model of disease, as they do not require simplifications and they contain the diverse cell types found in the myocardium such as fibroblasts, immune cells, and endothelial cells.240,263 In vivo systems can be used to investigate signaling between organs and organ systems that are critical to normal physiology and disease processes. This is important for understanding complex diseases such as HFpEF, whose pathophysiology is believed to be significantly influenced by cardiac remodeling resulting from endothelial inflammation and oxidative stress.264,265 The mouse is the most popular model organism to use to study cardiac disease, and many tools used to study cardiac function in humans, such as echocardiogram, cardiac magnetic resonance imaging, and invasive catheterization, are also available in mice and have been extensively discussed previously (for a recent review, see Lindsey et al265). Here, we focus on the application of cutting-edge optical imaging techniques to study cardiac function in vivo using fly and zebrafish models.
Drosophila cardiac kymography
Drosophila melanogaster has been an important genetic platform for uncovering the molecular biology underlying development.266,267 Powerful tools have been developed to study Drosophila enabling cost-effective and high-throughput study of genetic perturbations. The Drosophila heart is a linear contractile tube that creates an open circulatory network that transports a mix of blood and interstitial fluids called hemolymph throughout the organism (Figure 10).268 Despite the anatomical differences between Drosophila and humans, Drosophila cardiomyocytes possess a similar sarcomeric architecture with many conserved cardiac regulatory proteins and networks.269
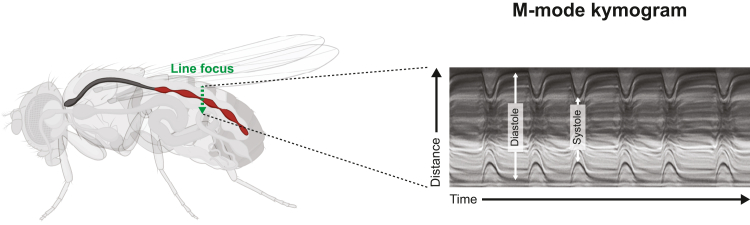
Figure 10.
Drosophila Heart Tube Kymography
Adult heart tube (red) with a line of focus drawn through the walls of the heart (green arrow) is recorded at high frame rate to produce a kymograph (right). Diastole and systole can be observed on the basis of the positioning of the anterior and posterior walls, similar to M-line echocardiography. Fly image created using BioRender.com (left). Kymogram (right) courtesy of Anthony Cammarato (Division of Cardiology, Department of Medicine, Johns Hopkins University School of Medicine).
The Drosophila cardiac tube is routinely visualized by dissection. High–frame rate optical imaging of the cardiac tube enables the determination of end-systolic and end-diastolic diameter, fractional shortening, and shortening and relaxation times and velocities (Figure 10).270 Similar to M-mode echocardiography, a line of focus can be drawn transversely through the cardiac tube, allowing the direct calculation of systolic and diastolic diameters with subsequent derivation of fractional shortening. This method was applied to study the HCM-associated mutation M305L in cardiac actin.271 Mutant hearts showed decreased fractional shortening, prolonged systolic contraction, and prolonged diastolic relaxation. This study demonstrated clear defects in diastolic function with mutation, consistent with what is observed in patients. Similarly, the DCM-associated S532P MYH7 mutation in was studied using this technique.272 The mutant hearts recapitulated a DCM phenotype of reduced systolic function. Taken together, these studies demonstrate the utility of heart tube kymography for studying the in vivo effects of cardiomyopathy-associated mutations.
Zebrafish and light-sheet fluorescence microscopy
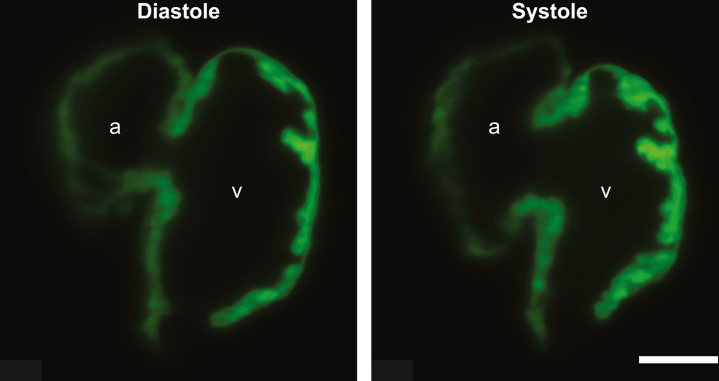
Zebrafish are another important vertebrate model for studying development. Advances in optical technology have made it possible to view zebrafish heart contractility with greater dynamic volumetric resolution similar to human cardiac magnetic resonance imaging. The developing zebrafish heart is visible to optical techniques without dissection. The zebrafish heart is also physiologically different from the human heart, with only a single atrium and ventricle; however, zebrafish models can faithfully recapitulate key aspects of human diseases such as DCM273,274 (Central Illustration).
Several methods have been developed to measure contractility in zebrafish hearts, and recent advances in imaging technologies have enabled studies with dramatically improved spatial and temporal resolution. One such technology is light-sheet fluorescence microscopy (LSFM). LSFM excites a thin section of the sample using a thin sheet of light that is orthogonally projected to the detection pathway. This method requires lower light intensity than other optical methods which reduces phototoxicity. LSFM also has a greater sample penetration than light confocal microscopy.275 Moreover, this technique illuminates only a single sample plane, reducing the background signal. Using this technique, it is possible to generate time synchronized, 3D reconstructions of the heart, allowing accurate volumetric measurements of both diastole and systole (Figure 11). Furthermore, this technique also enables the estimation of global longitudinal strain, which is a common method in clinical echocardiography to ascertain subtle changes in cardiac function that are not captured by 2-dimensional or 3D echocardiography.276 An interesting future application of LSFM may be studying cardiac contractility in Xenopus laevis tadpoles, which are similarly transparent like zebrafish embryos, for laboratories that prefer this model organism. Mutations in sarcomeric proteins such as cardiac troponin that cause DCM in patients have been shown to recapitulate the DCM morphology in genomically engineered X. laevis tadpoles.277 However, the compatibility of LSFM for these applications has yet to be demonstrated.
Figure 11.
Light-Sheet Microscopy of Zebrafish Heart
Single-frame captures of light-sheet fluorescence microscopic videos of fluorescently labeled zebrafish hearts that are 96 hours postfertilization. Scale bar represents 40 μm. Image courtesy of Jamison Leid (Center for Cardiovascular Research, Washington University School of Medicine). a = atrium; v = ventricle.
Limitations
Care needs to be taken when extrapolating from results seen in Drosophila and zebrafish to human studies because of physiological differences in anatomy and protein expression. Moreover, despite the extensive methodological development and validation of LSFM for measuring cardiac contractility, it has yet to be applied to modeling cardiomyopathy. Finally, these techniques require technical expertise, and techniques such as LSFM require sophisticated equipment (reviewed by Reynaud et al278).
Conclusions and Prospects for the Future
The development of multiscale tools for studying cardiac contractility has enabled deeper phenotyping of cardiomyopathies and HF. These studies have revealed new mechanistic categories that lead to contractile defects which have helped drive the development of modulators of cardiac contraction that may eventually complement GDMT. Therapeutics targeting contraction are still new; however, we anticipate that these tools will help fuel the development of the next generation of therapeutics targeting contractility and that these deep phenotyping tools can be used to optimize the characteristics of these compounds.
One outstanding challenge to the field is the need to integrate data across spatial and temporal scales. New, integrative computational models are being developed that can model aspects of the emergent complexity of these systems.35,279, 280, 281, 282, 283, 284, 285, 286, 287 However, modeling remains difficult, and additional studies that use multiscale techniques are required to establish correlations between results generated from different techniques. This will be best accomplished by collaborative efforts using techniques that span scales of organization. The eventual goal will be to use data from a single experimental system and predict functional outcomes at another scale. For example, one might be able to use biochemical results to predict effects on cardiomyocyte contractility and identify ideal treatment options in silico. With such a platform, we hope it will be eventually possible to rapidly assess functional consequences of genetic variants of unknown significance, identify additional processes for drug targeting, and provide better care for patients overall.
Funding Support and Author Disclosures
This work was supported by the National Institutes of Health (grants R01 HL141086 to Dr Greenberg, R01 HL161185 to Dr Lavine, R35 HL161185 to Dr Lavine, and T32 HL007081 to Dr Garg), the Leducq Foundation Network (grant 20CVD02 to Dr Lavine), the Burroughs Welcome Fund (grant 1014782 to Dr Lavine), the American Heart Association (grant 970198 to Drs Greenberg and Lavine), and the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (grant PM-LI-2019-829 to Drs Lavine and Greenberg). Dr Lavine is the recipient of sponsored research agreements from Amgen, Novartis, Kiniksa, and Implicit Bioscience; and provides consultant services to Medtronic, Kiniksa, and Implicit Biosciences. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank Aurora Scientific and Chris Rand for their images of single-myocyte and single-myofibril systems. The authors thank Lina Greenberg for the images of EHTs and related data traces. The authors thank Anthony Cammarato for sharing images of Drosophila heart tube kymography. The authors thank Jamison Leid for providing light-sheet fluorescence microscopic images of zebrafish hearts.
Footnotes
Lori Walker, MD, served as the Guest Associate Editor for this paper. Michael Bristow, MD, PhD, served as the Guest Editor-in-Chief for this paper.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
References
- 1.Heidenreich P.A., Bozkurt B., Aguilar D., et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2022;79(17):e263–e421. doi: 10.1016/j.jacc.2021.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Savarese G., Stolfo D., Sinagra G., Lund L.H. Heart failure with mid-range or mildly reduced ejection fraction. Nat Rev Cardiol. 2022;19:100–116. doi: 10.1038/s41569-021-00605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik F.I., Hartman J.J., Elias K.A., et al. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 2011;331:1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke M.A., Cook S.A., Seidman J.G., Seidman C.E. Clinical and mechanistic insights into the genetics of cardiomyopathy. J Am Coll Cardiol. 2016;68:2871–2886. doi: 10.1016/j.jacc.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green E.M., Wakimoto H., Anderson R.L., et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNally E.M., Mestroni L. Dilated Cardiomyopathy: genetic determinants and mechanisms. Circ Res. 2017;121:731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He H., Baka T., Balschi J., et al. Novel small-molecule troponin activator increases cardiac contractile function without negative impact on energetics. Circ Heart Fail. 2022;15 doi: 10.1161/CIRCHEARTFAILURE.121.009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsich E.M., Blackstone E.H., Thuita L.W., et al. Heart transplantation: an in-depth survival analysis. J Am Coll Cardiol HF. 2020;8:557–568. doi: 10.1016/j.jchf.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Packer M., Anker S.D., Butler J., et al. Cardiovascular and renal outcomes with empagliflozin in heart failure. N Engl J Med. 2020;383:1413–1424. doi: 10.1056/NEJMoa2022190. [DOI] [PubMed] [Google Scholar]
- 10.Ho C.Y., Sweitzer N.K., McDonough B., et al. Assessment of diastolic function with Doppler tissue imaging to predict genotype in preclinical hypertrophic cardiomyopathy. Circulation. 2002;105:2992–2997. doi: 10.1161/01.cir.0000019070.70491.6d. [DOI] [PubMed] [Google Scholar]
- 11.Mahon N.G., Murphy R.T., MacRae C.A., Caforio A.L., Elliott P.M., McKenna W.J. Echocardiographic evaluation in asymptomatic relatives of patients with dilated cardiomyopathy reveals preclinical disease. Ann Intern Med. 2005;143:108–115. doi: 10.7326/0003-4819-143-2-200507190-00009. [DOI] [PubMed] [Google Scholar]
- 12.Russel I.K., Brouwer W.P., Germans T., et al. Increased left ventricular torsion in hypertrophic cardiomyopathy mutation carriers with normal wall thickness. J Cardiovasc Magn Reson. 2011;13:3. doi: 10.1186/1532-429X-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsey J., Benson L., Rozenblyum E., Friedberg M.K., Mertens L. Early changes in apical rotation in genotype positive children with hypertrophic cardiomyopathy mutations without hypertrophic changes on two-dimensional imaging. J Am Soc Echocardiogr. 2014;27:215–221. doi: 10.1016/j.echo.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Rogers J.G., Butler J., Lansman S.L., et al. Chronic mechanical circulatory support for inotrope-dependent heart failure patients who are not transplant candidates: results of the INTrEPID trial. J Am Coll Cardiol. 2007;50:741–747. doi: 10.1016/j.jacc.2007.03.063. [DOI] [PubMed] [Google Scholar]
- 15.Morgan B.P., Muci A., Lu P.P., et al. Discovery of omecamtiv mecarbil the first, selective, small molecule activator of cardiac myosin. ACS Med Chem Lett. 2010;1:472–477. doi: 10.1021/ml100138q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen S., Sewanan L.R., Jacoby D.L., Campbell S.G. Danicamtiv enhances systolic function and frank-starling behavior at minimal diastolic cost in engineered human myocardium. J Am Heart Assoc. 2021;10 doi: 10.1161/JAHA.121.020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehman S.J., Crocini C., Leinwand L.A. Targeting the sarcomere in inherited cardiomyopathies. Nat Rev Cardiol. 2022;19:353–363. doi: 10.1038/s41569-022-00682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teerlink J.R., Diaz R., Felker G.M., et al. Cardiac myosin activation with omecamtiv mecarbil in systolic heart failure. N Engl J Med. 2021;384:105–116. doi: 10.1056/NEJMoa2025797. [DOI] [PubMed] [Google Scholar]
- 19.Olivotto I., Oreziak A., Barriales-Villa R., et al. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2020;396:759–769. doi: 10.1016/S0140-6736(20)31792-X. [DOI] [PubMed] [Google Scholar]
- 20.McKillop D.F., Geeves M.A. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehman W., Craig R., Vibert P. Ca2+-induced tropomyosin movement in limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 22.Barrick S.K., Greenberg M.J. Cardiac myosin contraction and mechanotransduction in health and disease. J Biol Chem. 2021;297 doi: 10.1016/j.jbc.2021.101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg M.J., Shuman H., Ostap E.M. Measuring the kinetic and mechanical properties of non-processive myosins using optical tweezers. Methods Mol Biol. 2017;1486:483–509. doi: 10.1007/978-1-4939-6421-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finer J.T., Simmons R.M., Spudich J.A. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 25.Spudich J.A. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys J. 2014;106:1236–1249. doi: 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmiter K.A., Tyska M.J., Dupuis D.E., Alpert N.R., Warshaw D.M. Kinetic differences at the single molecule level account for the functional diversity of rabbit cardiac myosin isoforms. J Physiol. 1999;519(Pt 3):669–678. doi: 10.1111/j.1469-7793.1999.0669n.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyska M.J., Hayes E., Giewat M., Seidman C.E., Seidman J.G., Warshaw D.M. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res. 2000;86:737–744. doi: 10.1161/01.res.86.7.737. [DOI] [PubMed] [Google Scholar]
- 28.Sung J., Sivaramakrishnan S., Dunn A.R., Spudich J.A. Single-molecule dual-beam optical trap analysis of protein structure and function. Methods Enzymol. 2010;475:321–375. doi: 10.1016/S0076-6879(10)75014-X. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg M.J., Shuman H., Ostap E.M. Inherent force-dependent properties of β-cardiac myosin contribute to the force-velocity relationship of cardiac muscle. Biophys J. 2014;107:L41–L44. doi: 10.1016/j.bpj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung J., Nag S., Mortensen K.I., et al. Harmonic force spectroscopy measures load-dependent kinetics of individual human beta-cardiac myosin molecules. Nat Commun. 2015;6:7931. doi: 10.1038/ncomms8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C., Kawana M., Song D., Ruppel K.M., Spudich J.A. Controlling load-dependent kinetics of beta-cardiac myosin at the single-molecule level. Nat Struct Mol Biol. 2018;25:505–514. doi: 10.1038/s41594-018-0069-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woody M.S., Greenberg M.J., Barua B., Winkelmann D.A., Goldman Y.E., Ostap E.M. Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat Commun. 2018;9:3838. doi: 10.1038/s41467-018-06193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Debold E.P., Patlak J.B., Warshaw D.M. Slip sliding away: load-dependence of velocity generated by skeletal muscle myosin molecules in the laser trap. Biophys J. 2005;89:L34–L36. doi: 10.1529/biophysj.105.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pant K., Watt J., Greenberg M., Jones M., Szczesna-Cordary D., Moore J.R. Removal of the cardiac myosin regulatory light chain increases isometric force production. FASEB J. 2009;23:3571–3580. doi: 10.1096/fj.08-126672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longyear T., Walcott S., Debold E.P. The molecular basis of thin filament activation: from single molecule to muscle. Sci Rep. 2017;7:1822. doi: 10.1038/s41598-017-01604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woody M.S., Winkelmann D.A., Capitanio M., Ostap E.M., Goldman Y.E. Single molecule mechanics resolves the earliest events in force generation by cardiac myosin. Elife. 2019;8 doi: 10.7554/eLife.49266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vander Roest A.S., Liu C., Morck M.M., et al. Hypertrophic cardiomyopathy beta-cardiac myosin mutation (P710R) leads to hypercontractility by disrupting super relaxed state. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2025030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snoberger A., Barua B., Atherton J.L., et al. Myosin with hypertrophic cardiac mutation R712L has a decreased working stroke which is rescued by omecamtiv mecarbil. Elife. 2021;10 doi: 10.7554/eLife.63691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hugenholtz P.G., Ellison R.C., Urschel C.W., Mirsky I., Sonnenblick E.H. Myocardial force-velocity relationships in clinical heart disease. Circulation. 1970;41:191–202. doi: 10.1161/01.cir.41.2.191. [DOI] [PubMed] [Google Scholar]
- 40.Pouleur H., Rousseau M.F., van Eyll C., Brasseur L.A., Charlier A.A. Force-velocity-length relations in hypertrophic cardiomyopathy: evidence of normal or depressed myocardial contractility. Am J Cardiol. 1983;52:813–817. doi: 10.1016/0002-9149(83)90420-4. [DOI] [PubMed] [Google Scholar]
- 41.Sommese R.F., Sung J., Nag S., et al. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human beta-cardiac myosin motor function. Proc Natl Acad Sci U S A. 2013;110:12607–12612. doi: 10.1073/pnas.1309493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nag S., Sommese R.F., Ujfalusi Z., et al. Contractility parameters of human beta-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spudich J.A. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflugers Arch. 2019;471:701–717. doi: 10.1007/s00424-019-02259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenberg M.J., Tardiff J.C. Complexity in genetic cardiomyopathies and new approaches for mechanism-based precision medicine. J Gen Physiol. 2021;153 doi: 10.1085/jgp.202012662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarcho J.A., McKenna W., Pare J.A., et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 46.Geisterfer-Lowrance A.A., Kass S., Tanigawa G., et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 47.Sarkar S.S., Trivedi D.V., Morck M.M., et al. The hypertrophic cardiomyopathy mutations R403Q and R663H increase the number of myosin heads available to interact with actin. Sci Adv. 2020;6 doi: 10.1126/sciadv.aax0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanchard E., Seidman C., Seidman J.G., LeWinter M., Maughan D. Altered crossbridge kinetics in the alphaMHC403/+ mouse model of familial hypertrophic cardiomyopathy. Circ Res. 1999;84:475–483. doi: 10.1161/01.res.84.4.475. [DOI] [PubMed] [Google Scholar]
- 49.Palmer B.M., Fishbaugher D.E., Schmitt J.P., et al. Differential cross-bridge kinetics of FHC myosin mutations R403Q and R453C in heterozygous mouse myocardium. Am J Physiol Heart Circ Physiol. 2004;287:H91–H99. doi: 10.1152/ajpheart.01015.2003. [DOI] [PubMed] [Google Scholar]
- 50.Belus A., Piroddi N., Scellini B., et al. The familial hypertrophic cardiomyopathy-associated myosin mutation R403Q accelerates tension generation and relaxation of human cardiac myofibrils. J Physiol. 2008;586:3639–3644. doi: 10.1113/jphysiol.2008.155952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer B.M., Wang Y., Teekakirikul P., et al. Myofilament mechanical performance is enhanced by R403Q myosin in mouse myocardium independent of sex. Am J Physiol Heart Circ Physiol. 2008;294:H1939–H1947. doi: 10.1152/ajpheart.00644.2007. [DOI] [PubMed] [Google Scholar]
- 52.Lankford E.B., Epstein N.D., Fananapazir L., Sweeney H.L. Abnormal contractile properties of muscle fibers expressing beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J Clin Invest. 1995;95:1409–1414. doi: 10.1172/JCI117795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagy L., Kovacs A., Bodi B., et al. The novel cardiac myosin activator omecamtiv mecarbil increases the calcium sensitivity of force production in isolated cardiomyocytes and skeletal muscle fibres of the rat. Br J Pharmacol. 2015;172:4506–4518. doi: 10.1111/bph.13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulop G.A., Olah A., Csipo T., et al. Omecamtiv mecarbil evokes diastolic dysfunction and leads to periodic electromechanical alternans. Basic Res Cardiol. 2021;116:24. doi: 10.1007/s00395-021-00866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kron S.J., Spudich J.A. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci U S A. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fraser I.D., Marston S.B. In vitro motility analysis of actin-tropomyosin regulation by troponin and calcium. The thin filament is switched as a single cooperative unit. J Biol Chem. 1995;270:7836–7841. doi: 10.1074/jbc.270.14.7836. [DOI] [PubMed] [Google Scholar]
- 57.Clippinger S.R., Cloonan P.E., Greenberg L., Ernst M., Stump W.T., Greenberg M.J. Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy. Proc Natl Acad Sci U S A. 2019;116:17831–17840. doi: 10.1073/pnas.1910962116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clippinger S.R., Cloonan P.E., Wang W., et al. Mechanical dysfunction of the sarcomere induced by a pathogenic mutation in troponin T drives cellular adaptation. J Gen Physiol. 2021;153 doi: 10.1085/jgp.202012787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bing W., Knott A., Marston S.B. A simple method for measuring the relative force exerted by myosin on actin filaments in the in vitro motility assay: evidence that tropomyosin and troponin increase force in single thin filaments. Biochem J. 2000;350(Pt 3):693–699. [PMC free article] [PubMed] [Google Scholar]
- 60.Greenberg M.J., Moore J.R. The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton (Hoboken) 2010;67:273–285. doi: 10.1002/cm.20441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aksel T., Choe Yu E., Sutton S., Ruppel K.M., Spudich J.A. Ensemble force changes that result from human cardiac myosin mutations and a small-molecule effector. Cell Rep. 2015;11:910–920. doi: 10.1016/j.celrep.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Previs M.J., Beck Previs S., Gulick J., Robbins J., Warshaw D.M. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337:1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mun J.Y., Previs M.J., Yu H.Y., et al. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc Natl Acad Sci U S A. 2014;111:2170–2175. doi: 10.1073/pnas.1316001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Touma A.M., Tang W., Rasicci D.V., et al. Nanosurfer assay dissects beta-cardiac myosin and cardiac myosin-binding protein C interactions. Biophys J. 2022;121:2449–2460. doi: 10.1016/j.bpj.2022.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Previs M.J., Mun J.Y., Michalek A.J., et al. Phosphorylation and calcium antagonistically tune myosin-binding protein C’s structure and function. Proc Natl Acad Sci U S A. 2016;113:3239–3244. doi: 10.1073/pnas.1522236113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones M.R., Tran C., Singh J., Dawson J.F. A gradient of force generation at rest differentiates cardiomyopathy outcomes with variants of actin located at the same residue. J Mol Cell Cardiol Plus. 2022;2 [Google Scholar]
- 67.Morck M.M., Bhowmik D., Pathak D., Dawood A., Spudich J., Ruppel K.M. Hypertrophic cardiomyopathy mutations in the pliant and light chain-binding regions of the lever arm of human beta-cardiac myosin have divergent effects on myosin function. Elife. 2022;11 doi: 10.7554/eLife.76805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsaturyan A.K., Zaklyazminskaya E.V., Polyak M.E., et al. De novo Asp219Val mutation in cardiac tropomyosin associated with hypertrophic cardiomyopathy. Int J Mol Sci. 2022;24:18. doi: 10.3390/ijms24010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrick S.K., Greenberg L., Greenberg M.J. A troponin T variant linked with pediatric dilated cardiomyopathy reduces the coupling of thin filament activation to myosin and calcium binding. Mol Biol Cell. 2021;32:1677–1689. doi: 10.1091/mbc.E21-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y., White H.D., Belknap B., Winkelmann D.A., Forgacs E. Omecamtiv mecarbil modulates the kinetic and motile properties of porcine beta-cardiac myosin. Biochemistry. 2015;54:1963–1975. doi: 10.1021/bi5015166. [DOI] [PubMed] [Google Scholar]
- 71.Stohr E.J., Takayama H., Ferrari G. Stretch your heart—but not too far: the role of titin mutations in dilated cardiomyopathy. J Thorac Cardiovasc Surg. 2018;156:209–214. doi: 10.1016/j.jtcvs.2017.10.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tharp C., Mestroni L., Taylor M. Modifications of titin contribute to the progression of cardiomyopathy and represent a therapeutic target for treatment of heart failure. J Clin Med. 2020;9:2770. doi: 10.3390/jcm9092770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puckelwartz M.J., Pesce L.L., Dellefave-Castillo L.M., et al. Genomic context differs between human dilated cardiomyopathy and hypertrophic cardiomyopathy. J Am Heart Assoc. 2021;10 doi: 10.1161/JAHA.120.019944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jordan E., Peterson L., Ai T., et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation. 2021;144:7–19. doi: 10.1161/CIRCULATIONAHA.120.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J., Termglinchan V., Diecke S., et al. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 2019;572:335–340. doi: 10.1038/s41586-019-1406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salvarani N., Crasto S., Miragoli M., et al. The K219T-lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat Commun. 2019;10:2267. doi: 10.1038/s41467-019-09929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhattacharjee P., Dasgupta D., Sengupta K. DCM associated LMNA mutations cause distortions in lamina structure and assembly. Biochim Biophys Acta Gen Subj. 2017;1861:2598–2608. doi: 10.1016/j.bbagen.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 78.Shah P.P., Lv W., Rhoades J.H., et al. Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes. Cell Stem Cell. 2021;28:938–954.e9. doi: 10.1016/j.stem.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin T.G., Myers V.D., Dubey P., et al. Cardiomyocyte contractile impairment in heart failure results from reduced BAG3-mediated sarcomeric protein turnover. Nat Commun. 2021;12:2942. doi: 10.1038/s41467-021-23272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller T., Szczesna D., Housmans P.R., et al. Abnormal contractile function in transgenic mice expressing a familial hypertrophic cardiomyopathy-linked troponin T (I79N) mutation. J Biol Chem. 2001;276:3743–3755. doi: 10.1074/jbc.M006746200. [DOI] [PubMed] [Google Scholar]
- 81.Wen Y., Pinto J.R., Gomes A.V., et al. Functional consequences of the human cardiac troponin I hypertrophic cardiomyopathy mutation R145G in transgenic mice. J Biol Chem. 2008;283:20484–22094. doi: 10.1074/jbc.M801661200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen Y., Xu Y., Wang Y., Pinto J.R., Potter J.D., Kerrick W.G. Functional effects of a restrictive-cardiomyopathy-linked cardiac troponin I mutation (R145W) in transgenic mice. J Mol Biol. 2009;392:1158–1167. doi: 10.1016/j.jmb.2009.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johns E.C., Ryder K.O., Hodson E.A., et al. Investigating the relaxation rate, following diazo-2 photolysis, of a skinned trabecular preparation from guinea-pig hypertrophied left ventricle. Pflugers Arch. 1999;438:771–777. doi: 10.1007/s004249900131. [DOI] [PubMed] [Google Scholar]
- 84.Preston L.C., Lipscomb S., Robinson P., et al. Functional effects of the DCM mutant Gly159Asp troponin C in skinned muscle fibres. Pflugers Arch. 2007;453:771–776. doi: 10.1007/s00424-006-0161-7. [DOI] [PubMed] [Google Scholar]
- 85.Dweck D., Sanchez-Gonzalez M.A., Chang A.N., et al. Long term ablation of protein kinase A (PKA)-mediated cardiac troponin I phosphorylation leads to excitation-contraction uncoupling and diastolic dysfunction in a knock-in mouse model of hypertrophic cardiomyopathy. J Biol Chem. 2014;289:23097–23111. doi: 10.1074/jbc.M114.561472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stehle R., Solzin J., Iorga B., Poggesi C. Insights into the kinetics of Ca2+-regulated contraction and relaxation from myofibril studies. Pflugers Arch. 2009;458:337–357. doi: 10.1007/s00424-008-0630-2. [DOI] [PubMed] [Google Scholar]
- 87.Luo M., Anderson M.E. Mechanisms of altered Ca2+ handling in heart failure. Circ Res. 2013;113:690–708. doi: 10.1161/CIRCRESAHA.113.301651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brenner B., Kraft T., Yu L., Chalovich J. Thin filament activation probed by fluorescence of N-((2-(iodoacetoxy) ethyl)-N-methyl) amino-7-nitrobenz-2-oxa-1, 3-diazole-labeled troponin I incorporated into skinned fibers of rabbit psoas muscle. Biophys J. 1999;77:2677–2691. doi: 10.1016/S0006-3495(99)77102-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chandra M., Montgomery D.E., Kim J.J., Solaro R.J. The N-terminal region of troponin T is essential for the maximal activation of rat cardiac myofilaments. J Mol Cell Cardiol. 1999;31:867–880. doi: 10.1006/jmcc.1999.0928. [DOI] [PubMed] [Google Scholar]
- 90.Ferrara C., Witjas-Paalberends E.R., Piroddi N., et al. A novel HCM mutation in cardiac troponin T primarily alters cross-bridge kinetics and increases the energetic cost of tension generation in human cardiac tissue. https://api.semanticscholar.org/CorpusID:18262940
- 91.López-Dávila A.J., Chalovich J.M., Zittrich S., et al. Cycling cross-bridges contribute to thin filament activation in human slow-twitch fibers. Front Physiol. 2020;11:144. doi: 10.3389/fphys.2020.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szczesna D., Zhang R., Zhao J., Jones M., Guzman G., Potter J.D. Altered regulation of cardiac muscle contraction by troponin T mutations that cause familial hypertrophic cardiomyopathy. J Biol Chem. 2000;275:624–630. doi: 10.1074/jbc.275.1.624. [DOI] [PubMed] [Google Scholar]
- 93.Gomes A.V., Guzman G., Zhao J., Potter J.D. Cardiac troponin T isoforms affect the Ca2+ sensitivity and inhibition of force development. Insights into the role of troponin T isoforms in the heart. J Biol Chem. 2002;277 doi: 10.1074/jbc.M204118200. 35341-3549. [DOI] [PubMed] [Google Scholar]
- 94.Gordon A., Rivera A., Wang C., Regnier M. Cooperative activation of skeletal and cardiac muscle. Adv Exp Med Biol. 2003;538:371–379. doi: 10.1007/978-1-4419-9029-7_34. [DOI] [PubMed] [Google Scholar]
- 95.Kohler J., Chen Y., Brenner B., et al. Familial hypertrophic cardiomyopathy mutations in troponin I (K183Δ, G203S, K206Q) enhance filament sliding. Physiol Genom. 2003;14:117–128. doi: 10.1152/physiolgenomics.00101.2002. [DOI] [PubMed] [Google Scholar]
- 96.Piroddi N., Tesi C., Pellegrino M., Tobacman L., Homsher E., Poggesi C. Contractile effects of the exchange of cardiac troponin for fast skeletal troponin in rabbit psoas single myofibrils. J Physiol. 2003;552:917–931. doi: 10.1113/jphysiol.2003.051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siedner S., Krüger M., Schroeter M., et al. Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. J Physiol. 2003;548:493–505. doi: 10.1113/jphysiol.2002.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sumandea M.P., Pyle W.G., Kobayashi T., de Tombe P.P., Solaro R.J. Identification of a functionally critical protein kinase C phosphorylation residue of cardiac troponin T. J Biol Chem. 2003;278:35135–35144. doi: 10.1074/jbc.M306325200. [DOI] [PubMed] [Google Scholar]
- 99.Burkart E.M., Sumandea M.P., Kobayashi T., et al. Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J Biol Chem. 2003;278:11265–11272. doi: 10.1074/jbc.M210712200. [DOI] [PubMed] [Google Scholar]
- 100.Gafurov B., Fredricksen S., Cai A., Brenner B., Chase P.B., Chalovich J.M. The Δ14 mutation of human cardiac troponin T enhances ATPase activity and alters the cooperative binding of S1-ADP to regulated actin. Biochemistry. 2004;43:15276–15285. doi: 10.1021/bi048646h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vahebi S., Kobayashi T., Warren C.M., de Tombe P.P., Solaro R.J. Functional effects of rho-kinase-dependent phosphorylation of specific sites on cardiac troponin. Circ Res. 2005;96:740–747. doi: 10.1161/01.RES.0000162457.56568.7d. [DOI] [PubMed] [Google Scholar]
- 102.Kruger M., Zittrich S., Redwood C., et al. Effects of the mutation R145G in human cardiac troponin I on the kinetics of the contraction–relaxation cycle in isolated cardiac myofibrils. J Physiol. 2005;564:347–357. doi: 10.1113/jphysiol.2004.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Venkatraman G., Gomes A.V., Kerrick W.G., Potter J.D. Characterization of troponin T dilated cardiomyopathy mutations in the fetal troponin isoform. J Biol Chem. 2005;280:17584–17592. doi: 10.1074/jbc.M409337200. [DOI] [PubMed] [Google Scholar]
- 104.Engel P.L. Dissertation. University of Illinois at Chicago, Health Sciences Center; 2006. Near N-Terminal Region of Troponin I Is Critical in Modifying Feedback Effects of Cross-Bridge Activation. [Google Scholar]
- 105.Narolska N.A., Piroddi N., Belus A., et al. Impaired diastolic function after exchange of endogenous troponin I with C-terminal truncated troponin I in human cardiac muscle. Circ Res. 2006;99:1012–1020. doi: 10.1161/01.RES.0000248753.30340.af. [DOI] [PubMed] [Google Scholar]
- 106.Preston L.C., Watkins H., Redwood C.S. A revised method of troponin exchange in permeabilised cardiac trabeculae using vanadate: functional consequences of a HCM-causing mutation in troponin I. J Muscle Res Cell Motil. 2006;27:585–590. doi: 10.1007/s10974-006-9079-0. [DOI] [PubMed] [Google Scholar]
- 107.Swartz D.R., Yang Z., Sen A., Tikunova S.B., Davis J.P. Myofibrillar troponin exists in three states and there is signal transduction along skeletal myofibrillar thin filaments. J Mol Biol. 2006;361:420–435. doi: 10.1016/j.jmb.2006.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Tombe P.P., Belus A., Piroddi N., et al. Myofilament calcium sensitivity does not affect cross-bridge activation-relaxation kinetics. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1129–R1136. doi: 10.1152/ajpregu.00630.2006. [DOI] [PubMed] [Google Scholar]
- 109.Preston L.C., Ashley C.C., Redwood C.S. DCM troponin C mutant Gly159Asp blunts the response to troponin phosphorylation. Biochem Biophys Res Commun. 2007;360:27–32. doi: 10.1016/j.bbrc.2007.05.221. [DOI] [PubMed] [Google Scholar]
- 110.Robinson P., Lipscomb S., Preston L.C., et al. Mutations in fast skeletal troponin I, troponin T, and β-tropomyosin that cause distal arthrogryposis all increase contractile function. FASEB J. 2007;21(3):896–905. doi: 10.1096/fj.06-6899com. [DOI] [PubMed] [Google Scholar]
- 111.Solzin J., Iorga B., Sierakowski E., et al. Kinetic mechanism of the Ca2+-dependent switch-on and switch-off of cardiac troponin in myofibrils. Biophys J. 2007;93:3917–3931. doi: 10.1529/biophysj.107.111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pinto J.R., Veltri T., Sorenson M.M. Modulation of troponin C affinity for the thin filament by different cross-bridge states in skinned skeletal muscle fibers. Pflugers Arch. 2008;456:1177–1187. doi: 10.1007/s00424-008-0480-y. [DOI] [PubMed] [Google Scholar]
- 113.Tachampa K., Kobayashi T., Wang H., et al. Increased cross-bridge cycling kinetics after exchange of C-terminal truncated troponin I in skinned rat cardiac muscle. J Biol Chem. 2008;283:15114–15121. doi: 10.1074/jbc.M801636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parvatiyar M.S., Pinto J.R., Dweck D., Potter J.D. Cardiac troponin mutations and restrictive cardiomyopathy. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/350706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pinto J.R., Gomes A.V., Jones M.A., et al. The functional properties of human slow skeletal troponin T isoforms in cardiac muscle regulation. J Biol Chem. 2012;287:37362–37370. doi: 10.1074/jbc.M112.364927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lopez-Davila A., Elhamine F., Ruess D., et al. Kinetic mechanism of Ca2+-controlled changes of skeletal troponin I in psoas myofibrils. Biophys J. 2012;103:1254–1264. doi: 10.1016/j.bpj.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wijnker P.J., Foster D.B., Tsao A.L., et al. Impact of site-specific phosphorylation of protein kinase A sites Ser23 and Ser24 of cardiac troponin I in human cardiomyocytes. Am J Physiol Heart Circ Physiol. 2013;304:H260–H268. doi: 10.1152/ajpheart.00498.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Remedios A.M.M., Stienen G.J., van der Velden J., et al. Impact of site-specific phosphorylation of protein kinase. Am J Physiol Heart Circ Physiol. 2013;304:H260–H268. doi: 10.1152/ajpheart.00498.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghashghaee N.B., Awinda P.O., Tanner B.C., Dong W.-J. Functional significance of C-terminal mobile domain of cardiac troponin I. Biophys J. 2017;112:256a–257a. doi: 10.1016/j.abb.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lopez Davila A.J., Zhu L., Fritz L., Kraft T., Chalovich J.M. The positively charged C-terminal region of human skeletal troponin T retards activation and decreases calcium sensitivity. Biochemistry. 2020;59:4189–4201. doi: 10.1021/acs.biochem.0c00499. [DOI] [PubMed] [Google Scholar]
- 121.López-Dávila A.J., Chalovich J.M., Zittrich S., et al. Cycling cross-bridges contribute to thin filament activation in human slow-twitch fibers. Frontiers in Physiology. 2020;11:144. doi: 10.3389/fphys.2020.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Madan A., Viswanathan M.C., Woulfe K.C., et al. TNNT2 mutations in the tropomyosin binding region of TNT1 disrupt its role in contractile inhibition and stimulate cardiac dysfunction. Proc Natl Acad Sci U S A. 2020;117:18822–18831. doi: 10.1073/pnas.2001692117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fujita H., Lu X., Suzuki M., Ishiwata S., Kawai M. The effect of tropomyosin on force and elementary steps of the cross-bridge cycle in reconstituted bovine myocardium. J Physiol. 2004;556:637–649. doi: 10.1113/jphysiol.2003.059956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Siththanandan V.B., Tobacman L.S., Van Gorder N., Homsher E. Mechanical and kinetic effects of shortened tropomyosin reconstituted into myofibrils. Pflugers Arch. 2009;458:761–776. doi: 10.1007/s00424-009-0653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lu X., Heeley D.H., Smillie L.B., Kawai M. The role of tropomyosin isoforms and phosphorylation in force generation in thin-filament reconstituted bovine cardiac muscle fibres. J Muscle Res Cell Motil. 2010;31:93–109. doi: 10.1007/s10974-010-9213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scellini B., Piroddi N., Poggesi C., Tesi C. Extraction and replacement of the tropomyosin–troponin complex in isolated myofibrils. Adv Exp Med Biol. 2010;682:163–174. doi: 10.1007/978-1-4419-6366-6_9. [DOI] [PubMed] [Google Scholar]
- 127.Nixon B.R., Liu B., Scellini B., et al. Tropomyosin Ser-283 pseudo-phosphorylation slows myofibril relaxation. Arch Biochem Biophys. 2013;535:30–38. doi: 10.1016/j.abb.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scellini B., Piroddi N., Flint G., Regnier M., Poggesi C., Tesi C. Impact of tropomyosin isoform composition on fast skeletal muscle thin filament regulation and force development. J Muscle Res Cell Motil. 2015;36:11–23. doi: 10.1007/s10974-014-9394-9. [DOI] [PubMed] [Google Scholar]
- 129.Scellini B., Piroddi N., Matyushenko A.M., et al. The relaxation properties of myofibrils are compromised by amino acids that stabilize α-tropomyosin. Biophys J. 2017;112:376–387. doi: 10.1016/j.bpj.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vitale G., Ferrantini C., Piroddi N., et al. The relation between sarcomere energetics and the rate of isometric tension relaxation in healthy and diseased cardiac muscle. J Muscle Res Cell Motil. 2021;42:47–57. doi: 10.1007/s10974-019-09566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bai F., Groth H.L., Kawai M. DCM-related tropomyosin mutants E40K/E54K over-inhibit the actomyosin interaction and lead to a decrease in the number of cycling cross-bridges. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kazmierczak K., Paulino E.C., Huang W., et al. Discrete effects of A57G-myosin essential light chain mutation associated with familial hypertrophic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2013;305:H575–H589. doi: 10.1152/ajpheart.00107.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kazmierczak K., Liang J., Gomez-Guevara M., Szczesna-Cordary D. Functional comparison of phosphomimetic S15D and T160D mutants of myosin regulatory light chain exchanged in cardiac muscle preparations of HCM and WT mice. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.988066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fujita H., Yasuda K., Niitsu S., Funatsu T., Ishiwata S. Structural and functional reconstitution of thin filaments in the contractile apparatus of cardiac muscle. Biophys J. 1996;71:2307–2318. doi: 10.1016/S0006-3495(96)79465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fujita H., Ishiwata Si. Spontaneous oscillatory contraction without regulatory proteins in actin filament-reconstituted fibers. Biophys J. 1998;75:1439–1445. doi: 10.1016/S0006-3495(98)74062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu X., Bryant M.K., Bryan K.E., Rubenstein P.A., Kawai M. Role of the N-terminal negative charges of actin in force generation and cross-bridge kinetics in reconstituted bovine cardiac muscle fibres. J Physiol. 2005;564:65–82. doi: 10.1113/jphysiol.2004.078055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bai F., Caster H.M., Rubenstein P.A., Dawson J.F., Kawai M. Using baculovirus/insect cell expressed recombinant actin to study the molecular pathogenesis of HCM caused by actin mutation A331P. J Mol Cell Biol. 2014;74:64–75. doi: 10.1016/j.yjmcc.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bai F., Caster H.M., Dawson J.F., Kawai M. The immediate effect of HCM causing actin mutants E99K and A230V on actin–Tm–myosin interaction in thin-filament reconstituted myocardium. J Mol Cell Biol. 2015;79:123–132. doi: 10.1016/j.yjmcc.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 139.Blair C.A., Brundage E.A., Thompson K.L., et al. Heart failure in humans reduces contractile force in myocardium from both ventricles. J Am Coll Cardiol Basic Trans Science. 2020;5:786–798. doi: 10.1016/j.jacbts.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tanner B.C.W., Awinda P.O., Agonias K.B., et al. Sarcomere length affects Ca2+ sensitivity of contraction in ischemic but not non-ischemic myocardium. J Gen Physiol. 2023;155 doi: 10.1085/jgp.202213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hellam D.C., Podolsky R.J. Force measurements in skinned muscle fibres. J Physiol. 1969;200:807–819. doi: 10.1113/jphysiol.1969.sp008723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Meiss R.A. A versatile transducer system for mechanical studies of muscle. J Appl Physiol. 1974;37:459–463. doi: 10.1152/jappl.1974.37.3.459. [DOI] [PubMed] [Google Scholar]
- 143.Gonçalves-Rodrigues P., Almeida-Coelho J., Gonçalves A., et al. In vitro assessment of cardiac function using skinned cardiomyocytes. J Vis Exp. 2020;160 doi: 10.3791/60427. [DOI] [PubMed] [Google Scholar]
- 144.de Tombe P.P., Mateja R.D., Tachampa K., Ait Mou Y., Farman G.P., Irving T.C. Myofilament length dependent activation. J Mol Cell Cardiol. 2010;48:851–858. doi: 10.1016/j.yjmcc.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sonnenblick E.H. Force-velocity relations in mammalian heart muscle. Am J Physiol. 1962;202:931–939. doi: 10.1152/ajplegacy.1962.202.5.931. [DOI] [PubMed] [Google Scholar]
- 146.Chase P.B., Martyn D.A., Hannon J.D. Isometric force redevelopment of skinned muscle fibers from rabbit activated with and without Ca2+ Biophys J. 1994;67:1994–2001. doi: 10.1016/S0006-3495(94)80682-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.LeWinter M.M., Wu Y., Labeit S., Granzier H. Cardiac titin: structure, functions and role in disease. Clin Chim Acta. 2007;375:1–9. doi: 10.1016/j.cca.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 148.Anderson B.R., Granzier H.L. Titin-based tension in the cardiac sarcomere: molecular origin and physiological adaptations. Prog Biophys Mol Biol. 2012;110:204–217. doi: 10.1016/j.pbiomolbio.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Linke W.A. Titin gene and protein functions in passive and active muscle. Annu Rev Physiol. 2018;80:389–411. doi: 10.1146/annurev-physiol-021317-121234. [DOI] [PubMed] [Google Scholar]
- 150.Walker J.S., Li X., Buttrick P.M. Analysing force-pCa curves. J Muscle Res Cell Motil. 2010;31:59–69. doi: 10.1007/s10974-010-9208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Debold E.P., Saber W., Cheema Y., et al. Human actin mutations associated with hypertrophic and dilated cardiomyopathies demonstrate distinct thin filament regulatory properties in vitro. J Mol Cell Cardiol. 2010;48:286–292. doi: 10.1016/j.yjmcc.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chung J.H., Biesiadecki B.J., Ziolo M.T., Davis J.P., Janssen P.M. Myofilament calcium sensitivity: role in regulation of in vivo cardiac contraction and relaxation. Front Physiol. 2016;7:562. doi: 10.3389/fphys.2016.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hanft L.M., Emter C.A., McDonald K.S. Cardiac myofibrillar contractile properties during the progression from hypertension to decompensated heart failure. Am J Physiol Heart Circ Physiol. 2017;313:H103–H113. doi: 10.1152/ajpheart.00069.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wolff M.R., McDonald K.S., Moss R.L. Rate of tension development in cardiac muscle varies with level of activator calcium. Circ Res. 1995;76:154–160. doi: 10.1161/01.res.76.1.154. [DOI] [PubMed] [Google Scholar]
- 155.Brenner B. Effect of Ca2+ on cross-bridge turnover kinetics in skinned single rabbit psoas fibers: implications for regulation of muscle contraction. Proc Natl Acad Sci U S A. 1988;85:3265–3269. doi: 10.1073/pnas.85.9.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McNamara J.W., Singh R.R., Sadayappan S. Cardiac myosin binding protein-C phosphorylation regulates the super-relaxed state of myosin. Proc Natl Acad Sci U S A. 2019;116:11731–11736. doi: 10.1073/pnas.1821660116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Papadaki M., Kampaengsri T., Barrick S.K., et al. Myofilament glycation in diabetes reduces contractility by inhibiting tropomyosin movement, is rescued by cMyBPC domains. J Mol Cell Cardiol. 2022;162:1–9. doi: 10.1016/j.yjmcc.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Metzger J.M., Moss R.L. Effects of tension and stiffness due to reduced pH in mammalian fast- and slow-twitch skinned skeletal muscle fibres. J Physiol. 1990;428:737–750. doi: 10.1113/jphysiol.1990.sp018238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kawai M., Halvorson H. Two step mechanism of phosphate release and the mechanism of force generation in chemically skinned fibers of rabbit psoas muscle. Biophys J. 1991;59:329–342. doi: 10.1016/S0006-3495(91)82227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chase P.B., Martyn D.A., Kushmerick M.J., Gordon A.M. Effects of inorganic phosphate analogues on stiffness and unloaded shortening of skinned muscle fibres from rabbit. J Physiol. 1993;460:231–246. doi: 10.1113/jphysiol.1993.sp019469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhao Y., Kawai M. The effect of the lattice spacing change on cross-bridge kinetics in chemically skinned rabbit psoas muscle fibers. II. Elementary steps affected by the spacing change. Biophys J. 1993;64:197–210. doi: 10.1016/S0006-3495(93)81357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Campbell K.B., Chandra M., Kirkpatrick R.D., Slinker B.K., Hunter W.C. Interpreting cardiac muscle force-length dynamics using a novel functional model. Am J Physiol Heart Circ Physiol. 2004;286:H1535–H1545. doi: 10.1152/ajpheart.01029.2003. [DOI] [PubMed] [Google Scholar]
- 163.Palmer B.M., Suzuki T., Wang Y., Barnes W.D., Miller M.S., Maughan D.W. Two-state model of acto-myosin attachment-detachment predicts C-process of sinusoidal analysis. Biophys J. 2007;93:760–769. doi: 10.1529/biophysj.106.101626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tanner B.C., Breithaupt J.J., Awinda P.O. Myosin MgADP release rate decreases at longer sarcomere length to prolong myosin attachment time in skinned rat myocardium. Am J Physiol Heart Circ Physiol. 2015;309:H2087–H2097. doi: 10.1152/ajpheart.00555.2015. [DOI] [PubMed] [Google Scholar]
- 165.Fenwick A.J., Leighton S.R., Tanner B.C. Myosin MgADP release rate decreases as sarcomere length increases in skinned rat soleus muscle fibers. Biophys J. 2016;111:2011–2023. doi: 10.1016/j.bpj.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Landim-Vieira M., Ma W., Song T., et al. Cardiac troponin T N-domain variant destabilizes the actin interface resulting in disturbed myofilament function. Proc Natl Acad Sci U S A. 2023;120 doi: 10.1073/pnas.2221244120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stewart M., Franks-Skiba K., Cooke R. Myosin regulatory light chain phosphorylation inhibits shortening velocities of skeletal muscle fibers in the presence of the myosin inhibitor blebbistatin. J Muscle Res Cell Motil. 2009;30:17–27. doi: 10.1007/s10974-008-9162-9. [DOI] [PubMed] [Google Scholar]
- 168.Stewart M.A., Franks-Skiba K., Chen S., Cooke R. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc Natl Acad Sci U S A. 2010;107:430–435. doi: 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hooijman P., Stewart M.A., Cooke R. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nag S., Trivedi D.V. To lie or not to lie: super-relaxing with myosins. Elife. 2021;10 doi: 10.7554/eLife.63703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Linari M., Brunello E., Reconditi M., et al. Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature. 2015;528:276–279. doi: 10.1038/nature15727. [DOI] [PubMed] [Google Scholar]
- 172.Reconditi M., Caremani M., Pinzauti F., et al. Myosin filament activation in the heart is tuned to the mechanical task. Proc Natl Acad Sci U S A. 2017;114:3240–3245. doi: 10.1073/pnas.1619484114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Anderson R.L., Trivedi D.V., Sarkar S.S., et al. Deciphering the super relaxed state of human beta-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc Natl Acad Sci U S A. 2018;115:E8143–E8152. doi: 10.1073/pnas.1809540115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mamidi R., Li J., Doh C.Y., Verma S., Stelzer J.E. Impact of the myosin modulator mavacamten on force generation and cross-bridge behavior in a murine model of hypercontractility. J Am Heart Assoc. 2018;7 doi: 10.1161/JAHA.118.009627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yuan C.C., Kazmierczak K., Liang J., Ma W., Irving T.C., Szczesna-Cordary D. Molecular basis of force-pCa relation in MYL2 cardiomyopathy mice: Role of the super-relaxed state of myosin. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2110328119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Toepfer C.N., Garfinkel A.C., Venturini G., et al. Myosin sequestration regulates sarcomere function, cardiomyocyte energetics, and metabolism, informing the pathogenesis of hypertrophic cardiomyopathy. Circulation. 2020;141:828–842. doi: 10.1161/CIRCULATIONAHA.119.042339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Papadaki M., Holewinski R.J., Previs S.B., et al. Diabetes with heart failure increases methylglyoxal modifications in the sarcomere, which inhibit function. JCI Insight. 2018;3(20) doi: 10.1172/jci.insight.121264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Aslam M.I., Hahn V.S., Jani V., Hsu S., Sharma K., Kass D.A. Reduced right ventricular sarcomere contractility in heart failure with preserved ejection fraction and severe obesity. Circulation. 2021;143:965–967. doi: 10.1161/CIRCULATIONAHA.120.052414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mamidi R., Gresham K.S., Li A., dos Remedios C.G., Stelzer J.E. Molecular effects of the myosin activator omecamtiv mecarbil on contractile properties of skinned myocardium lacking cardiac myosin binding protein-C. J Mol Cell Biol. 2015;85:262–272. doi: 10.1016/j.yjmcc.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Utter M.S., Ryba D.M., Li B.H., Wolska B.M., Solaro R.J. Omecamtiv mecarbil, a cardiac myosin activator, increases Ca2+ sensitivity in myofilaments with a dilated cardiomyopathy mutant tropomyosin E54K. J Cardiovasc Pharmacol. 2015;66:347. doi: 10.1097/FJC.0000000000000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gollapudi S.K., Reda S.M., Chandra M. Omecamtiv mecarbil abolishes length-mediated increase in guinea pig cardiac myofiber Ca2+ sensitivity. Biophys J. 2017;113:880–888. doi: 10.1016/j.bpj.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mamidi R., Li J., Gresham K.S., et al. Dose-dependent effects of the myosin activator omecamtiv mecarbil on cross-bridge behavior and force generation in failing human myocardium. Circ Heart Fail. 2017;10 doi: 10.1161/CIRCHEARTFAILURE.117.004257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kieu T.T., Awinda P.O., Tanner B.C. Omecamtiv mecarbil slows myosin kinetics in skinned rat myocardium at physiological temperature. Biophys J. 2019;116:2149–2160. doi: 10.1016/j.bpj.2019.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Governali S., Caremani M., Gallart C., et al. Orthophosphate increases the efficiency of slow muscle-myosin isoform in the presence of omecamtiv mecarbil. Nat Commun. 2020;11:3405. doi: 10.1038/s41467-020-17143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Awinda P.O., Bishaw Y., Watanabe M., Guglin M.A., Campbell K.S., Tanner B.C.W. Effects of mavacamten on Ca. Br J Pharmacol. 2020;177:5609–5621. doi: 10.1111/bph.15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nakanishi T., Oyama K., Tanaka H., et al. Effects of omecamtiv mecarbil on the contractile properties of skinned porcine left atrial and ventricular muscles. Front Physiol. 2022;13 doi: 10.3389/fphys.2022.947206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Iwazumi T. High-speed ultrasensitive instrumentation for myofibril mechanics measurements. Am J Physiol. 1987;252:C253–C262. doi: 10.1152/ajpcell.1987.252.2.C253. [DOI] [PubMed] [Google Scholar]
- 188.Colomo F., Piroddi N., Poggesi C., te Kronnie G., Tesi C. Active and passive forces of isolated myofibrils from cardiac and fast skeletal muscle of the frog. J Physiol. 1997;500(Pt 2):535–548. doi: 10.1113/jphysiol.1997.sp022039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Marston S. Force measurements from myofibril to filament. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.817036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Linke W.A., Ivemeyer M., Labeit S., Hinssen H., Ruegg J.C., Gautel M. Actin-titin interaction in cardiac myofibrils: probing a physiological role. Biophys J. 1997;73:905–919. doi: 10.1016/S0006-3495(97)78123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kooiker KB, Mohran S, Turner KL, et al. Danicamtiv increases myosin recruitment and alters the chemomechanical cross bridge cycle in cardiac muscle. Preprint. bioRxiv. Posted online February 3, 2023. https://doi.org/10.1101/2023.01.31.526380 [DOI] [PMC free article] [PubMed]
- 192.Vikhorev P.G., Song W., Wilkinson R., et al. The dilated cardiomyopathy-causing mutation ACTC E361G in cardiac muscle myofibrils specifically abolishes modulation of Ca2+ regulation by phosphorylation of troponin I. Biophys J. 2014;107:2369–2380. doi: 10.1016/j.bpj.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lewalle A., Campbell K.S., Campbell S.G., Milburn G.N., Niederer S.A. Functional and structural differences between skinned and intact muscle preparations. J Gen Physiol. 2022;154(2) doi: 10.1085/jgp.202112990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kehat I., Kenyagin-Karsenti D., Snir M., et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sun N., Yazawa M., Liu J., et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang L., Kim K., Parikh S., et al. Hypertrophic cardiomyopathy-linked mutation in troponin T causes myofibrillar disarray and pro-arrhythmic action potential changes in human iPSC cardiomyocytes. J Mol Cell Cardiol. 2018;114:320–327. doi: 10.1016/j.yjmcc.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ma N., Zhang J.Z., Itzhaki I., et al. Determining the pathogenicity of a genomic variant of uncertain significance using CRISPR/Cas9 and human-induced pluripotent stem cells. Circulation. 2018;138:2666–2681. doi: 10.1161/CIRCULATIONAHA.117.032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pettinato A.M., Ladha F.A., Mellert D.J., et al. Development of a cardiac sarcomere functional genomics platform to enable scalable interrogation of human TNNT2 variants. Circulation. 2020;142:2262–2275. doi: 10.1161/CIRCULATIONAHA.120.047999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sequeira V., Wang L., Wijnker P.J.M., et al. Low expression of the K280N TNNT2 mutation is sufficient to increase basal myofilament activation in human hypertrophy cardiomyopathy. J Mol Cell Cardiol Plus. 2022;1 doi: 10.1016/j.jmccpl.2022.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lian X., Zhang J., Azarin S.M., et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lyra-Leite D.M., Gutiérrez-Gutiérrez Ó., Wang M., Zhou Y., Cyganek L., Burridge P.W. A review of protocols for human iPSC culture, cardiac differentiation, subtype-specification, maturation, and direct reprogramming. STAR Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.DeLaughter D.M., Bick A.G., Wakimoto H., et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016;39:480–490. doi: 10.1016/j.devcel.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Matsa E., Ahrens J.H., Wu J.C. Human induced pluripotent stem cells as a platform for personalized and precision cardiovascular medicine. Physiol Rev. 2016;96:1093–1126. doi: 10.1152/physrev.00036.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Reilly L., Munawar S., Zhang J., Crone W.C., Eckhardt L.L. Challenges and innovation: Disease modeling using human-induced pluripotent stem cell-derived cardiomyocytes. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.966094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lavine K.J., Greenberg M.J. Beyond genomics-technological advances improving the molecular characterization and precision treatment of heart failure. Heart Fail Rev. 2021;26:405–415. doi: 10.1007/s10741-020-10021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Louch W.E., Sheehan K.A., Wolska B.M. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol. 2011;51:288–298. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Abi Gerges N., Stafford A., Truong K., et al. Measurement of heart contractility in isolated adult human primary cardiomyocytes. J Vis Exp. 2022:186. doi: 10.3791/64394. [DOI] [PubMed] [Google Scholar]
- 209.Kamgoue A., Ohayon J., Usson Y., Riou L., Tracqui P. Quantification of cardiomyocyte contraction based on image correlation analysis. Cytometry A. 2009;75:298–308. doi: 10.1002/cyto.a.20700. [DOI] [PubMed] [Google Scholar]
- 210.Ribeiro A.J., Ang Y.S., Fu J.D., et al. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc Natl Acad Sci U S A. 2015;112:12705–12710. doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ribeiro A.J.S., Schwab O., Mandegar M.A., et al. Multi-imaging method to assay the contractile mechanical output of micropatterned human iPSC-derived cardiac myocytes. Circ Res. 2017;120:1572–1583. doi: 10.1161/CIRCRESAHA.116.310363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sharma A., Toepfer C.N., Ward T., et al. CRISPR/Cas9-mediated fluorescent tagging of endogenous proteins in human pluripotent stem cells. Curr Protoc Hum Genet. 2018;96 doi: 10.1002/cphg.52. 21.11.1-21.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Toepfer C.N., Sharma A., Cicconet M., et al. SarcTrack. Circ Res. 2019;124:1172–1183. doi: 10.1161/CIRCRESAHA.118.314505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Toepfer C.N., Wakimoto H., Garfinkel A.C., et al. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci Transl Med. 2019;11(476) doi: 10.1126/scitranslmed.aat1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pua C.J., Tham N., Chin C.W.L., et al. Genetic studies of hypertrophic cardiomyopathy in Singaporeans identify variants in TNNI3 and TNNT2 that are common in Chinese patients. Circ Genom Precis Med. 2020;13:424–434. doi: 10.1161/CIRCGEN.119.002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Grashoff C., Hoffman B.D., Brenner M.D., et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Harris A.K., Wild P., Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 218.Sala L., van Meer B.J., Tertoolen L.G.J., et al. MUSCLEMOTION: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ Res. 2018;122:e5–e16. doi: 10.1161/CIRCRESAHA.117.312067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Guo J., Simmons D.W., Ramahdita G., et al. Elastomer-grafted iPSC-derived micro heart muscles to investigate effects of mechanical loading on physiology. ACS Biomater Sci Eng. 2021;7:2973–2989. doi: 10.1021/acsbiomaterials.0c00318. [DOI] [PubMed] [Google Scholar]
- 220.Engler A.J., Carag-Krieger C., Johnson C.P., et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.McCain M.L., Yuan H., Pasqualini F.S., Campbell P.H., Parker K.K. Matrix elasticity regulates the optimal cardiac myocyte shape for contractility. Am J Physiol Heart Circ Physiol. 2014;306:H1525–H1539. doi: 10.1152/ajpheart.00799.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Judge L.M., Perez-Bermejo J.A., Truong A., et al. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight. 2017;2 doi: 10.1172/jci.insight.94623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Feyen D.A.M., Perea-Gil I., Maas R.G.C., et al. Unfolded protein response as a compensatory mechanism and potential therapeutic target in PLN R14del cardiomyopathy. Circulation. 2021;144:382–392. doi: 10.1161/CIRCULATIONAHA.120.049844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Cai W., Zhang J., de Lange W.J., et al. An unbiased proteomics method to assess the maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2019;125:936–953. doi: 10.1161/CIRCRESAHA.119.315305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Miragoli M., Gaudesius G., Rohr S. Electrotonic modulation of cardiac impulse conduction by myofibroblasts. Circ Res. 2006;98:801–810. doi: 10.1161/01.RES.0000214537.44195.a3. [DOI] [PubMed] [Google Scholar]
- 226.Hulsmans M., Clauss S., Xiao L., et al. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169:510–522.e20. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Eschenhagen T., Fink C., Remmers U., et al. Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: a new heart muscle model system. FASEB J. 1997;11:683–694. doi: 10.1096/fasebj.11.8.9240969. [DOI] [PubMed] [Google Scholar]
- 228.Legant W.R., Pathak A., Yang M.T., Deshpande V.S., McMeeking R.M., Chen C.S. Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proc Natl Acad Sci U S A. 2009;106:10097–10102. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hinson J.T., Chopra A., Nafissi N., et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Daily N.J., Yin Y., Kemanli P., Wakatsuki T. Improving cardiac action potential measurements: 2D and 3D cell culture. J Bioeng Biomed Sci. 2015;5(3):168. doi: 10.4172/2155-9538.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wijnker P.J., Friedrich F.W., Dutsch A., et al. Comparison of the effects of a truncating and a missense MYBPC3 mutation on contractile parameters of engineered heart tissue. J Mol Cell Cardiol. 2016;97:82–92. doi: 10.1016/j.yjmcc.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 232.Stillitano F., Turnbull I.C., Karakikes I., et al. Genomic correction of familial cardiomyopathy in human engineered cardiac tissues. Eur Heart J. 2016;37:3282–3284. doi: 10.1093/eurheartj/ehw307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Daily N.J., Santos R., Vecchi J., Kemanli P., Wakatsuki T. Calcium transient assays for compound screening with human iPSC-derived cardiomyocytes: evaluating new tools. J Evolv Stem Cell Res. 2017;1:1–11. doi: 10.14302/issn.2574-4372.jesr-16-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Daily N.J., Du Z.-W., Wakatsuki T. High-throughput phenotyping of human induced pluripotent stem cell-derived cardiomyocytes and neurons using electric field stimulation and high-speed fluorescence imaging. Assay Drug Dev Technol. 2017;15:178–188. doi: 10.1089/adt.2017.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mosqueira D., Mannhardt I., Bhagwan J.R., et al. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur Heart J. 2018;39(43):3879–3892. doi: 10.1093/eurheartj/ehy249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Loskill P., Huebsch N. Engineering tissues from induced pluripotent stem cells. Tissue Eng Part A. 2019;25:707–710. doi: 10.1089/ten.TEA.2019.0118. [DOI] [PubMed] [Google Scholar]
- 237.Greenberg M.J., Daily N.J., Wang A., Conway M.K., Wakatsuki T. Genetic and tissue engineering approaches to modeling the mechanics of human heart failure for drug discovery. Front Cardiovasc Med. 2018;5:120. doi: 10.3389/fcvm.2018.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Litvinukova M., Talavera-Lopez C., Maatz H., et al. Cells of the adult human heart. Nature. 2020;588:466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tani H., Tohyama S. Human engineered heart tissue models for disease modeling and drug discovery. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.855763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Koenig A.L., Shchukina I., Amrute J., et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat Cardiovasc Res. 2022;1:263–280. doi: 10.1038/s44161-022-00028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kupfer M.E., Lin W.H., Ravikumar V., et al. In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3D bioprinted, chambered organoid. Circ Res. 2020;127:207–224. doi: 10.1161/CIRCRESAHA.119.316155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hansen A., Eder A., Bonstrup M., et al. Development of a drug screening platform based on engineered heart tissue. Circ Res. 2010;107:35–44. doi: 10.1161/CIRCRESAHA.109.211458. [DOI] [PubMed] [Google Scholar]
- 243.Truitt R., Mu A., Corbin E.A., et al. Increased afterload augments sunitinib-induced cardiotoxicity in an engineered cardiac microtissue model. J Am Coll Cardiol Basic Trans Science. 2018;3:265–276. doi: 10.1016/j.jacbts.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bailey A.L., Dmytrenko O., Greenberg L., et al. SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis. J Am Coll Cardiol Basic Trans Science. 2021;6:331–345. doi: 10.1016/j.jacbts.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zimmermann W.H., Schneiderbanger K., Schubert P., et al. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 2002;90:223–230. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- 246.Tiburcy M., Meyer T., Soong P.L., Zimmermann W.H. Collagen-based engineered heart muscle. Methods Mol Biol. 2014;1181:167–176. doi: 10.1007/978-1-4939-1047-2_15. [DOI] [PubMed] [Google Scholar]
- 247.Goldfracht I., Protze S., Shiti A., et al. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat Commun. 2020;11:75. doi: 10.1038/s41467-019-13868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kameda S., Higo S., Shiba M., et al. Modeling reduced contractility and stiffness using iPSC-derived cardiomyocytes generated from female Becker muscular dystrophy carrier. J Am Coll Cardiol Basic Trans Science. 2023;8(6):599–613. doi: 10.1016/j.jacbts.2022.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hofbauer P., Jahnel S.M., Papai N., et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell. 2021;184:3299–3317.e22. doi: 10.1016/j.cell.2021.04.034. [DOI] [PubMed] [Google Scholar]
- 250.Lu T.Y., Lin B., Kim J., et al. Repopulation of decellularized mouse heart with human induced pluripotent stem cell-derived cardiovascular progenitor cells. Nat Commun. 2013;4:2307. doi: 10.1038/ncomms3307. [DOI] [PubMed] [Google Scholar]
- 251.Ahrens J.H., Uzel S.G.M., Skylar-Scott M., et al. Programming cellular alignment in engineered cardiac tissue via bioprinting anisotropic organ building blocks. Adv Mater. 2022;34 doi: 10.1002/adma.202200217. [DOI] [PubMed] [Google Scholar]
- 252.Uhl S., Freichel M., Mathar I. Contractility measurements on isolated papillary muscles for the investigation of cardiac inotropy in mice. J Vis Exp. 2015;103 doi: 10.3791/53076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pieske B., Schlotthauer K., Schattmann J., et al. Ca2+-dependent and Ca2+-independent regulation of contractility in isolated human myocardium. Basic Res Cardiol. 1997;92(suppl 1):75–86. doi: 10.1007/BF00794071. [DOI] [PubMed] [Google Scholar]
- 254.Chung J.H., Milani-Nejad N., Davis J.P., et al. Impact of heart rate on cross-bridge cycling kinetics in failing and nonfailing human myocardium. Am J Physiol Heart Circ Physiol. 2019;317:H640–H647. doi: 10.1152/ajpheart.00163.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mashali M.A., Saad N.S., Canan B.D., et al. Impact of etiology on force and kinetics of left ventricular end-stage failing human myocardium. J Mol Cell Cardiol. 2021;156:7–19. doi: 10.1016/j.yjmcc.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mashali M.A., Saad N.S., Peczkowski K.K., et al. Mechanical dyssynchrony of isolated left and right ventricular human myocardium in end-stage heart failure. Circ Heart Fail. 2023;16 doi: 10.1161/CIRCHEARTFAILURE.122.009871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Szczesna-Cordary D., Jones M., Moore J.R., et al. Myosin regulatory light chain E22K mutation results in decreased cardiac intracellular calcium and force transients. FASEB J. 2007;21:3974–3985. doi: 10.1096/fj.07-8630com. [DOI] [PubMed] [Google Scholar]
- 258.Kerrick W.G., Kazmierczak K., Xu Y., Wang Y., Szczesna-Cordary D. Malignant familial hypertrophic cardiomyopathy D166V mutation in the ventricular myosin regulatory light chain causes profound effects in skinned and intact papillary muscle fibers from transgenic mice. FASEB J. 2009;23:855–865. doi: 10.1096/fj.08-118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Julian F.J., Sollins M.R. Sarcomere length-tension relations in living rat papillary muscle. Circ Res. 1975;37:299–308. doi: 10.1161/01.res.37.3.299. [DOI] [PubMed] [Google Scholar]
- 260.Paulus W.J., Claes V.A., Brutsaert D.L. Physiological loading of isolated feline cardiac muscle. The interaction between muscle contraction and vascular impedance in the production of pressure and flow waves. Circ Res. 1979;44:491–497. doi: 10.1161/01.res.44.4.491. [DOI] [PubMed] [Google Scholar]
- 261.Lamers J.M. Some characteristics of monocarboxylic acid transfer across the cell membrane of epithelial cells from rat small intestine. Biochim Biophys Acta. 1975;413:465–476. [PubMed] [Google Scholar]
- 262.Muranaka H., Marui A., Tsukashita M., et al. Prolonged mechanical unloading preserves myocardial contractility but impairs relaxation in rat heart of dilated cardiomyopathy accompanied by myocardial stiffness and apoptosis. J Thorac Cardiovasc Surg. 2010;140:916–922. doi: 10.1016/j.jtcvs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 263.Paik D.T., Cho S., Tian L., Chang H.Y., Wu J.C. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol. 2020;17:457–473. doi: 10.1038/s41569-020-0359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Cornuault L., Rouault P., Duplaa C., Couffinhal T., Renault M.A. Endothelial dysfunction in heart failure with preserved ejection fraction: what are the experimental proofs? Front Physiol. 2022;13 doi: 10.3389/fphys.2022.906272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lindsey M.L., Kassiri Z., Virag J.A.I., de Castro Bras L.E., Scherrer-Crosbie M. Guidelines for measuring cardiac physiology in mice. Am J Physiol Heart Circ Physiol. 2018;314:H733–H752. doi: 10.1152/ajpheart.00339.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Bodmer R., Frasch M. In: Heart Development. Rosenthal N., Harvey R., editors. Elsevier; New York: 1999. Genetic determination of Drosophila heart development; pp. 65–90. [Google Scholar]
- 267.Zaffran S., Frasch M. Early signals in cardiac development. Circ Res. 2002;91:457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- 268.Wolf M.J., Rockman H.A. Drosophila, genetic screens, and cardiac function. Circ Res. 2011;109:794–806. doi: 10.1161/CIRCRESAHA.111.244897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Cammarato A., Ahrens C.H., Alayari N.N., et al. A mighty small heart: the cardiac proteome of adult Drosophila melanogaster. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ocorr K., Vogler G., Bodmer R. Methods to assess Drosophila heart development, function and aging. Methods. 2014;68:265–272. doi: 10.1016/j.ymeth.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Viswanathan M.C., Schmidt W., Franz P., et al. A role for actin flexibility in thin filament-mediated contractile regulation and myopathy. Nat Commun. 2020;11:2417. doi: 10.1038/s41467-020-15922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Trujillo A.S., Hsu K.H., Puthawala J., et al. Myosin dilated cardiomyopathy mutation S532P disrupts actomyosin interactions, leading to altered muscle kinetics, reduced locomotion, and cardiac dilation in Drosophila. Mol Biol Cell. 2021;32:1690–1706. doi: 10.1091/mbc.E21-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Bowley G., Kugler E., Wilkinson R., et al. Zebrafish as a tractable model of human cardiovascular disease. Br J Pharmacol. 2022;179:900–917. doi: 10.1111/bph.15473. [DOI] [PubMed] [Google Scholar]
- 274.Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011;91:279–288. doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Huisken J., Swoger J., Del Bene F., Wittbrodt J., Stelzer E.H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 276.Fei P., Lee J., Packard R.R., et al. Cardiac light-sheet fluorescent microscopy for multi-scale and rapid imaging of architecture and function. Sci Rep. 2016;6 doi: 10.1038/srep22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Landim-Vieira M., Johnston J.R., Ji W., et al. Familial dilated cardiomyopathy associated with a novel combination of compound heterozygous TNNC1 variants. Front Physiol. 2019;10:1612. doi: 10.3389/fphys.2019.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Reynaud E.G., Peychl J., Huisken J., Tomancak P. Guide to light-sheet microscopy for adventurous biologists. Nat Methods. 2015;12:30–34. doi: 10.1038/nmeth.3222. [DOI] [PubMed] [Google Scholar]
- 279.Aboelkassem Y., Bonilla J.A., McCabe K.J., Campbell S.G. Contributions of Ca2+-independent thin filament activation to cardiac muscle function. Biophys J. 2015;109:2101–2112. doi: 10.1016/j.bpj.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Sewanan L.R., Moore J.R., Lehman W., Campbell S.G. Predicting effects of tropomyosin mutations on cardiac muscle contraction through myofilament modeling. Front Physiol. 2016;7:473. doi: 10.3389/fphys.2016.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Campbell K.S., Janssen P.M.L., Campbell S.G. Force-dependent recruitment from the myosin off state contributes to length-dependent activation. Biophys J. 2018;115:543–553. doi: 10.1016/j.bpj.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Forsch N., Govil S., Perry J.C., et al. Computational analysis of cardiac structure and function in congenital heart disease: translating discoveries to clinical strategies. J Comput Sci. 2021;52 doi: 10.1016/j.jocs.2020.101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Mijailovich S.M., Prodanovic M., Poggesi C., Geeves M.A., Regnier M. Multiscale modeling of twitch contractions in cardiac trabeculae. J Gen Physiol. 2021;153 doi: 10.1085/jgp.202012604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.McCulloch A.D., Grandi E., Saucerman J.J. Computational models of cardiovascular regulatory mechanisms. J Mol Cell Cardiol. 2021;155:111. doi: 10.1016/j.yjmcc.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 285.Kosta S., Colli D., Ye Q., Campbell K.S. FiberSim: A flexible open-source model of myofilament-level contraction. Biophys J. 2022;121:175–182. doi: 10.1016/j.bpj.2021.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Margara F., Psaras Y., Wang Z.J., et al. Mechanism based therapies enable personalised treatment of hypertrophic cardiomyopathy. Sci Rep. 2022;12 doi: 10.1038/s41598-022-26889-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Longobardi S., Sher A., Niederer S.A. Quantitative mapping of force-pCa curves to whole-heart contraction and relaxation. J Physiol. 2022;600:3497–3516. doi: 10.1113/JP283352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Greenman A.C., Diffee G.M., Power A.S., et al. Treadmill running increases the calcium sensitivity of myofilaments in diabetic rats. J Appl Physiol (1985) 2022;132:1350–1360. doi: 10.1152/japplphysiol.00785.2021. [DOI] [PubMed] [Google Scholar]
- 289.Pardon G, Lewis H, Vander Roest AS, et al. Insights into single hiPSC-derived cardiomyocyte phenotypes and maturation using ConTraX, an efficient pipeline for tracking contractile dynamics. Preprint. biRxiv. Published online March 19, 2021. https://doi.org/10.1101/2021.03.18.436014v1