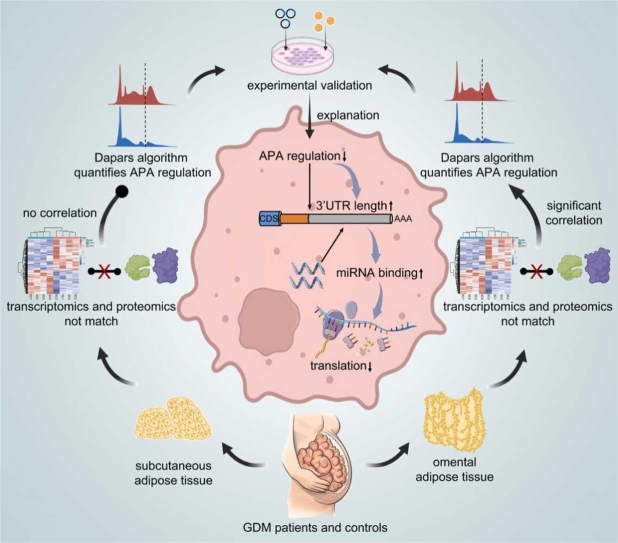
Abstract
In gestational diabetes mellitus (GDM), adipose tissue undergoes metabolic disturbances and chronic low-grade inflammation. Alternative polyadenylation (APA) is a post-transcriptional modification mechanism that generates mRNA with variable lengths of 3' untranslated regions (3'UTR), and it is associated with inflammation and metabolism. However, the role of APA in GDM adipose tissue has not been well characterized. In this study, we conducted transcriptomic and proteomic sequencing on subcutaneous and omental adipose tissues from both control and GDM patients. Using Dapars, a novel APA quantitative algorithm, we delineated the APA landscape of adipose tissue, revealing significant 3'UTR elongation of mRNAs in the GDM group. Omental adipose tissue exhibited a significant correlation between elongated 3'UTRs and reduced translation levels of genes related to metabolism and inflammation. Validation experiments in THP-1 derived macrophages (TDMs) demonstrated the impact of APA on translation levels by overexpressing long and short 3'UTR isoforms of a representative gene LRRC25. Additionally, LRRC25 was validated to suppress proinflammatory polarization in TDMs. Further exploration revealed two underexpressed APA trans-acting factors, CSTF3 and PPP1CB, in GDM omental adipose tissue. In conclusion, this study provides preliminary insights into the APA landscape of GDM adipose tissue. Reduced APA regulation in GDM omental adipose tissue may contribute to metabolic disorders and inflammation by downregulating gene translation levels. These findings advance our understanding of the molecular mechanisms underlying GDM-associated adipose tissue changes.
Keywords: Gestational diabetes mellitus, Alternative polyadenylation, Adipose tissue, Transcriptomics, Proteomics, Metabolic disorders, Inflammation
Graphical Abstract
Highlights
-
•
Alternative polyadenylation landscape of adipose tissue in gestational diabetes mellitus was first depicted through Dapars.
-
•
Alternative polyadenylation influences mRNA structure and translation efficiency by altering the length of the 3'UTR.
-
•
Reduced levels of alternative polyadenylation regulation result in decreased translation of genes.
1. Introduction
The incidence of gestational diabetes mellitus (GDM) and its associated perinatal complications has been increasing, parallel the rise in obesity prevalence [1]. According to the International Diabetes Federation's 2019 estimation, globally, 1 in every 6 live births is affected by GDM [2]. In China, the prevalence is notably high at 14%, with a continuous upward trend [3]. GDM is known to be associated with various adverse pregnancy outcomes, including gestational hypertension and preterm birth. Furthermore, it increases the long-term risk of metabolic syndrome in both mothers and their offspring. Insulin resistance, a fundamental pathophysiological change in GDM, is notably influenced by adipose tissue, which plays a pivotal role in its development [4] Within the context of GDM pathogenesis, adipose tissue experiences metabolic disorders and chronic low-grade inflammation, significantly contributing to insulin resistance [4], [5]. Moreover, post-transcriptional regulation has emerged as a critical factor in the interplay between inflammation and metabolism [6], [7], [8]. The relationship has gained increasing attention, particularly in classical insulin target tissues such as the liver, adipose tissue, and muscles [9], [10], [11].
In recent years, research has unveiled the significant role of alternative polyadenylation (APA) as a widespread post-transcriptional regulation mechanism in eukaryotes [12], [13]. APA, occurring in the 3' untranslated region (3' UTR) of mRNA, can generate transcripts with varying lengths of the 3'UTR. This, in turn, affects the binding of cis-regulatory elements such as microRNAs (miRNAs) and RNA-binding proteins, thereby regulating protein translation efficiency [14]. Studies have demonstrated the profound impact of APA on various biological processes. For instance, SNORD12B, through competitive binding with the APA factor FIP1L1, influences the 3'UTR length of ZBTB4, thereby affecting the expression level of ZBTB4 and glycolipid metabolism in glioblastoma cells [15]. Additionally, FIP1L1 induces 3'UTR shortening of NLRP3 under renal injury and oxidative stress conditions, resulting in the upregulation of NLRP3 expression, exacerbating inflammation, ROS production, and cell apoptosis [16]. Considering the intimate relationship between APA regulation, energy metabolism, and inflammatory response, APA may play an undiscovered role in pathological changes in adipose tissue observed in the context of GDM. Further investigation into the involvement of APA in GDM-related adipose tissue alterations is warranted.
In this study, we analyzed transcriptomic, single-nucleus transcriptomic, and proteomic data of subcutaneous (SC) adipose tissue and omental (OM) adipose tissue from GDM patients and normal pregnant women. An APA event atlas was constructed to compare GDM patients with normal pregnant women, revealing the impact of APA as a post-transcriptional regulatory mechanism on the translation levels of genes related to metabolism and inflammation. These findings underscore the pivotal role of APA regulation in adipose tissue in the pathogenesis of GDM. This study offers new insights for potential therapeutic interventions in GDM, focusing on targeted APA-mediated dysregulation of genes involved in energy metabolism and inflammation.
2. Methods
2.1. Patient inclusion and sample collection
GDM patients (n = 4) and control pregnant women (n = 4) were recruited from the First Affiliated Hospital of Chongqing Medical University. Inclusion criteria comprised maternal age between 18 and 45, gestational age between 37 and 41 weeks, and the necessity for a C-section due to either a scarred uterus or GDM. Exclusion criteria included multiple pregnancies, hypertensive disorders of pregnancy, endocrine diseases (such as hypothyroidism or hyperthyroidism), and immune-mediated diseases. GDM diagnosis was based on the World Health Organization criteria, defined as any degree of glucose intolerance with onset or first recognition during pregnancy[17]. Specifically, considering Chinese population data, cases with oral glucose tolerance tests results during the second trimester of pregnancy exceeding the normal range (blood glucose level: 0 h ≥ 5.1 mmol/L, 1 h ≥ 10 mmol/L, 2 h ≥ 8.5 mmol/L) were diagnosed with GDM. The clinical characteristics of the patients are shown in Additional file 1: Table S1. There were no significant differences in age, BMI, or gestational age between the GDM group and the normal group. Samples were collected from the SC and OM adipose tissue during the C-section and frozen in liquid nitrogen until transcriptomic and proteomic analysis. In total, 16 samples were divided into four groups based on different sources: SC adipose tissue from control cases (NSC group) and GDM patients (GSC group), and OM adipose tissue from control cases (NOM group) and GDM patients (GOM group). The study was approved by the First Affiliated Hospital Ethics Committee of Chongqing Medical University.
2.2. Transcriptomics analysis
High-throughput RNA sequencing was performed for the transcriptomics detection of adipose tissue samples. Briefly, the total amounts and integrity of RNA were assessed using the RNA Nano 6000 Assay Kit with the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Then the library was constructed using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA). After being purified and quantified, the library was sequenced by the Illumina NovaSeq 6000. The image data measured with the high-throughput sequencer were converted into sequence data (reads) by CASAVA base recognition. Clean reads were obtained by removing reads containing adapters, reads containing N bases, and low-quality reads from raw data, then aligned to the reference genome (GRCh38/hg38) using Hisat2 (v2.0.5). FeatureCounts (v1.5.0-p3) were used to count the read numbers mapped to each gene. Finally, the FPKM of each gene was calculated based on the length of the gene and the read count mapped to this gene.
2.3. Single-nucleus RNA sequencing and analysis
Single-nucleus RNA sequencing (snRNA-seq) was carried out using the 10x Genomics Chromium™ platform. Samples from the GOM and NOM groups were mixed separately and then prepared into nuclear suspensions following the 10x Genomics Cell Preparation Guide [18]. The nuclear suspension was loaded into Chromium microfluidic chips with 3' v3 chemistry and barcoded with a 10x Chromium Controller (10x Genomics, CA, USA). RNA from the barcoded nuclei was subsequently reverse-transcribed, and sequencing libraries were constructed with reagents from a Chromium Single Cell 3' v3 reagent kit (10x Genomics, CA, USA) according to the manufacturer's instructions. Sequencing was performed with an Illumina HiSeq 2000 according to the manufacturer's instructions. Raw read sequences produced by the Illumina pipeline in FASTQ format were preprocessed through Trimmomatic software, in which reads with low-quality, adapters, and fewer than 26 bases were discarded. Raw reads were demultiplexed and mapped to the reference genome by the 10x Genomics Cell Ranger pipeline using default parameters [18]. Cellranger and Seurat were applied for data filtration, expression matrix construction, and single-nucleus analyses [19], [20]. In total, 5000 variable genes and 40 principal components (resolution = 0.5) were used for clustering.
2.4. Proteomics analysis
Tandem mass tags (TMT) technology was used for proteomics detection. Samples were lysed with SDT (containing 100 mM NaCl) and 1/100 vol of DTT. After incubation for 10 min at 95 °C, the lysate was centrifuged at 12,000 g for 15 min at 4 °C. The supernatant was alkylated with iodoacetamide for 1 h, then mixed with cold acetone at − 20 °C for 2 h. The precipitation was collected after centrifuging at 12000 g for 15 min at 4 °C and finally dissolved with dissolution buffer (8 M Urea, 100 mM TEAB, pH 8.5). Protein concentration was measured using the Bradford protein quantitative kit. Trypsin was used for digestion. The supernatant was loaded into the C18 desalting column and then washed with washing buffer (0.1% formic acid, 3% acetonitrile). The eluents of each sample were lyophilized and reconstituted with 100 μL of 0.1 M TEAB buffer. Acetonitrile-dissolved TMT labeling reagent was added, and the reaction was stopped by adding 8% ammonia after 2 h at room temperature. The sample was fractionated using a C18 column (Waters BEH C18,4.6 ×250 mm, 5 µm) on a Rigol L3000 HPLC system. UHPLC-MS/MS analyses were performed using an EASY-nLC™ 1200 UHPLC system (Thermo Fisher, Germany) coupled with an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher, Germany). The resulting spectra from each run were searched separately against the UniProt database (Homo sapiens, 207393 sequences, released on 3/13/2023) by Proteome Discoverer (PD, Thermo, HFX, and 480). To improve the quality of analysis results, the software PD further filtered the retrieval results. Peptide spectrum matches (PSMs) with a confidence level exceeding 99% are considered reliable PSMs. The reliable protein contains at least 1 unique peptide. The reliable PSMs and proteins were retained and performed with an FDR of no more than 1.0%.
2.5. APA prediction
APA events were predicted and quantified using the DaPars algorithm, which can identify and quantify APA events de novo from RNA-seq data. Concisely, DaPars first identifies a distal polyA site based on RNA-seq and then utilizes a regression model to estimate the precise position of the proximal APA site. Subsequently, it identifies dynamic APAs exhibiting statistically significant differences. The percentage of distal polyA site usage index (PDUI) was calculated as a metric to quantify APA events, with a higher PDUI indicating greater utilization of the distal polyA site (PAS). The difference in average inter-group PDUI values (ΔPDUI) was used to assess variations in PAS utilization between groups. Based on previous literature[21], APA events with │ΔPDUI│ ≥ 0.1 and adjusted P-value (adj. P) < 0.05 were considered statistically significant.
2.6. Plasmid construction
To overexpress the 3'UTR lengthening and shortening isoforms of LRRC25 mRNA, two plasmids, pCS2-LRRC25-LUTR and pCS2-LRRC25-SUTR, were constructed. Full-length cDNA was synthesized based on RNA extracted from human OM adipose tissue using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China), while genomic DNA was extracted from THP-1 cell line using Universal Genomic DNA Purification Mini Spin Kit (Beyotime Biotech, Shanghai, China). The CDS region of LRRC25 and the long and short 3'UTR regions, identified by DaPars, were individually amplified from full-length cDNA and genomic DNA, respectively, using Q5 polymerase (New England Biolabs, MA, USA). The pCS2 vector was digested with restriction endonucleases EcoRI-HF and XhoI (New England Biolabs, MA, USA), and the CDS and 3'UTR inserts were cloned and inserted into the pCS2 vector by homologous reorganization using Hieff Clone® Plus Multi One Step Cloning Kit (Yeasen, Shanghai, China). The primers are listed in Additional file 2: Table S2. The plasmid was transformed into DH5α Chemically Competent Cells (Tsingke Biotech, Beijing, China) and colonies were screened by ampicillin resistance. The plasmid sequence was verified by Tsingke Biotech.
2.7. Cell culture and transfection
THP-1 cells (ATCC TIB-202) were purchased from ATCC and cultured in RPMI-1640 medium supplemented with 10% FBS, 0.05 mM β-mercaptoethanol and 1% penicillin/streptomycin (P/S). Human umbilical vein endothelial cells (HUVECs, ATCC PCS-100–010) were purchased from ATCC and cultured in DMEM-F12 medium supplemented with 10% FBS, 0.1 mg/ml heparin, 0.05 mg/ml endothelial cell growth supplement and 1% (P/S). Primary human adipose-derived stem cells (ADSCs) were provided by the Chongqing Cell Resource Library and cultured in DMEM-F12 medium supplemented with 10% FBS and 1% (P/S). THP-1 derived macrophages (TMDs) were obtained by treating THP-1 cells with 100 nM phorbol 12-myristate 13-acetate (PMA) for 24 h.
The constructed plasmid was transfected into TMDs for 6 h using polyethylenimine linear (Yeasen, Shanghai, China). The siRNA of CSTF3 and PPP1CB was synthesized by Genechem (see sequence in Additional file 2: Table S2) and transfected into cells for 6 h using Lipofectamine RNAiMAX (Invitrogen, CA, USA). After transfection, cells were collected at either 24 h or 48 h for RNA or protein extraction.
2.8. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from human OM adipose tissue using RNAex Pro Reagent (AG, Hunan, China). The synthesis of full-length cDNA followed the same procedure as mentioned above. qRT-PCR was performed with SYBR Green Pro Taq HS (AG, Hunan, China) and the CFX96™ Real-Time System (Bio-Rad, CA, USA). β-Actin was used as a reference gene. The related primers are listed in Table S2.
2.9. Western blot
The OM adipose tissues or TMDs were lysed using RIPA Lysis Buffer (Beyotime Biotech, Shanghai, China) containing Protease Inhibitor Cocktail (Bimake, HOU, USA) and centrifuged at 12,000 g for 10 min at 4 °C. The supernatants were collected, and electrophoresis was performed on the 10% or 12.5% SDS-PAGE. Then proteins were transferred to a PVDF membrane and blocked with 5% nonfat milk. After incubation with the primary antibody overnight at 4 °C, the proteins were incubated with the secondary antibody for 1 h at room temperature, and finally, the protein blots were developed with WesternBright™ ECL (Advansta, CA, USA). The primary antibodies used in Western blot included LRRC25 (Bioss, bs-12339R), CSTF3 (Proteintech, 24290–1-AP) and PPP1CB (Proteintech, 10140–2-AP).
2.10. Flow cytometry
To explore the function of LRRC25, LRRC25 overexpression plasmids were transfected into TMDs for 48 h, followed by a 48-hour treatment with 100 ng/ml lipopolysaccharide (LPS). Based on the different treatments, TMDs were divided into three groups: the control group, the LPS-treated group, and the LRRC25 overexpression + LPS-treated group. The phenotype of TMDs was characterized using flow cytometry following the manufacturer's instructions. Briefly, cells were detached with trypsin and then resuspended and washed with Stain Buffer (BD Pharmingen, CA, USA). Cells were stained with Fixable Viability Stain 620 (BD Pharmingen, CA, USA). Subsequently, APC/Cyanine7 anti-human CD45 antibody and PE anti-human CD86 Antibody (Biolegend, CA, USA) were added to the cell suspension and incubated at 4 °C in the dark for 30 min. APC anti-human CD206 antibody was added to the cell suspension and incubated at room temperature in the dark for 30 min after cell permeabilization with permeabilization reagent (Invitrogen, CA, USA). The fluorescence signal intensity of the cells was detected using a FACSAria III flow cytometer, and FlowJo V.10.8.1 was used for flow cytometry result analysis.
2.11. miRNA Prediction
The miRNAs potentially binding to the 3'UTR were predicted using miRanda 3.3a [22]. In essence, the nucleotide sequences between the proximal and distal PAS of mRNA were retrieved in bulk using UCSC. Subsequently, mature human miRNA sequences were downloaded from miRBase [23]. Finally, miRanda 3.3a was utilized to predict miRNA binding (score=150, energy=−7). The Human microRNA Disease Database was used to identify GDM-associated miRNAs [24].
2.12. Bioinformatics Analysis and Statistical Analysis
Gene function enrichment analysis was conducted using Metascape [25]. Principal Component Analysis (PCA) and differential gene analysis were performed in R 4.3.1 using the R packages prcomp and DESeq2, respectively. Visualization of the bioinformatics analysis results, including volcano plots, chord diagrams, heatmaps, and bubble plots, was accomplished using R 4.3.1 with the assistance of R packages ggplot2, circlize, pheatmap, and ggmatrix. IGV 2.16.1 was used for the visualization of RNA-seq results [26]. Venn diagrams, workflow diagrams, and schematics were created in Adobe Illustrator CS6. Statistical graphs were generated using GraphPad Prism 8. Quantification of Western blot image grayscale values was performed using ImageJ 1.54. Continuous variables were assessed using the T test or Mann-Whitney test, while categorical variables were evaluated using the χ2 test or Fisher's exact test. A adj. P less than 0.05 was considered statistically significant. mRNAs with │fold change│≥ 1.2 (│FC│≥1.2) and adj. P < 0.01 were identified as differentially expressed mRNAs (DEmRNAs), and proteins with │FC│≥ 1.2 and adj. P < 0.05 were recognized as differentially expressed proteins (DEPs).
3. Results
3.1. APA is widely present in SC and OM adipose tissue
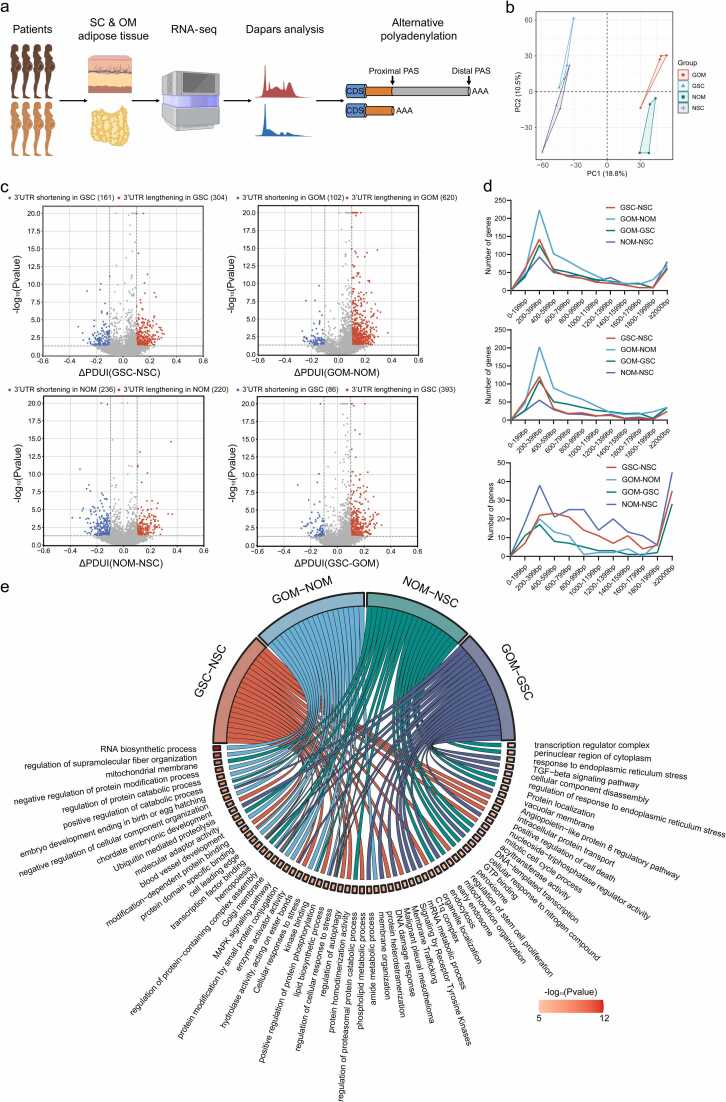
The DaPars algorithm was employed to quantify the dynamics of APA events based on RNA-seq data, presenting results as the Percentage of Distal polyA Site Usage Index (PDUI), to depict the APA landscape of adipose tissue in GDM and normal pregnant women. Considering the significant tissue heterogeneity in APA [27], adipose tissues from different locations (SC and OM) were analyzed (Fig. 1a). A total of 10,834 dynamic APA events were identified in adipose tissue (Additional file 3: Table S3; Additional file 4: Fig. S1a). Principal component analysis (PCA) revealed that PDUI values effectively differentiated samples from different groups (Fig. 1b), indicating significant inter-group differences, particularly between different tissue sources. Separate PCA plots were generated for samples originating from the same tissue source to demonstrate the differences between the GDM and normal groups (Additional file 4: Fig. S1b). The ΔPDUI was calculated to quantify APA differences among groups, enabling the identification of 3'UTR lengthening (ΔPDUI≥0.1) and shortening (ΔPDUI≤−0.1). Four group comparisons (GSC vs NSC, GOM vs NOM, GOM vs GSC, and NOM vs NSC) were conducted to elucidate APA differences across states and locations. As shown in Fig. 1c and Additional file 4: Fig. S1c, the GDM group exhibited significantly more genes with 3'UTR lengthening (ΔPDUI≥0.1, P < 0.05) in both SC and OM adipose tissues than the normal group. This suggests that APA regulation is attenuated in adipose tissues of GDM patients, resulting in increased utilization of distal polyA sites (PAS). However, no significant imbalance in gene expression regarding 3'UTR lengthening or shortening was observed between the normal SC and OM adipose tissues. In contrast, GDM tissues exhibited a higher number of significantly 3'UTR lengthening genes in OM (393 genes) compared to SC (86 genes), indicating a more pronounced attenuation of APA regulation in OM adipose tissue compared to SC adipose tissue. Fig. 1d demonstrates that the distances between the proximal and distal PAS of significantly different APA events across all group comparisons are concentrated mainly in the 200–400 bp range. The functional enrichment analysis of genes with significantly different APA events in each group was conducted using Metascape and revealed associations with stress-related processes (regulation of autophagy, DNA damage response, regulation of cellular response to stress), substance metabolism (lipid biosynthetic process, hydrolase activity, phospholipid metabolic process, amide metabolic process), transcriptional processes (transcription factor binding, mRNA metabolic process, transcription regulator complex), and protein modifications (modification-dependent protein binding, positive regulation of protein phosphorylation, negative regulation of protein modification process) (Fig. 1e; Additional file 4: Fig. S1d).
Fig. 1.
The APA landscape of SC and OM adipose tissues from GDM patients and controls. (a) Flowchart for analyzing adipose tissue APA landscape with the DaPars algorithm. (b) PCA of samples based on PDUI values clearly distinguishes different groups. (c) Volcano plots of 3’UTR lengthening (red) and shortening (blue) genes in different group comparisons. (d) Distances between proximal and distal PAS of significantly different APA events in all group comparisons, including all events (top), lengthening events (middle), and shortening events (bottom). (e) Functional enrichment analysis of genes with significantly different APA events in different group comparisons.
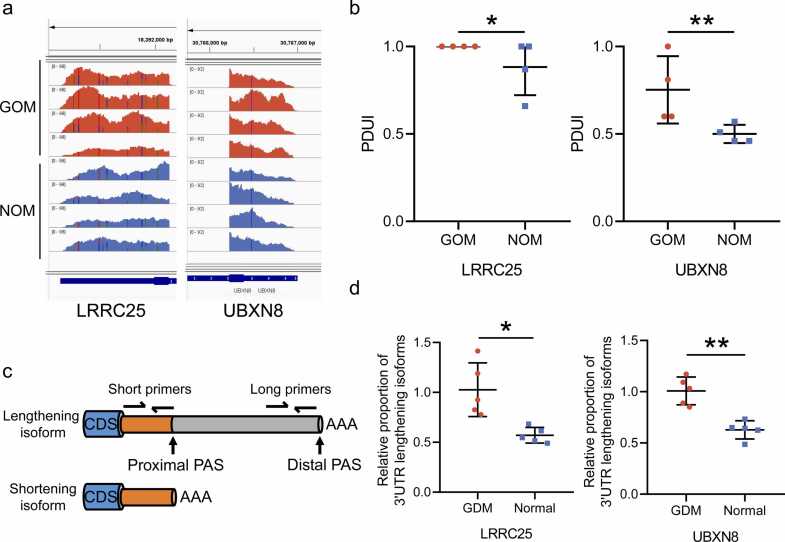
To provide a more intuitive validation of the analysis results, the RNA-seq results of representative genes with significant APA differences were visualized using IGV [26]. Specifically, LRRC25 and UBXN8, identified as significantly 3'UTR lengthening genes in the GOM vs NOM group, exhibited higher utilization of distal PAS in the GOM group (Fig. 2a). These differential 3'UTR profiles between the GOM and NOM groups align with the ΔPDUI calculated by DaPars (Fig. 2b), and similar observations hold in other group comparisons (Additional file 5: Fig. S2). To further validate the expression levels of lengthening and shortening APA isoforms in adipose tissue, two pairs of primers were designed for LRRC25 and UBXN8. The short primers can bind to the common sequence of lengthening and shortening isoforms, while the long primers can only bind to the lengthening isoform (Fig. 2c; Additional file 2: Table S2). Consistent with the RNA-seq and DaPars findings, qRT-PCR results showed that the proportion of 3'UTR lengthening isoform of LRRC25 and UBXN8 was higher in the GOM group than in the NOM group (Fig. 2d). In summary, we have identified the widespread presence of APA regulation in adipose tissue, with an increased number of 3'UTR lengthening genes being a prominent feature of APA regulation in the GDM group. This phenomenon is more pronounced in OM adipose tissue.
Fig. 2.
APA visualization and experimental validation in the GOM vs NOM group. (a) RNA-seq tracks for two representative APA-related genes (LRRC25, UBXN8). Red tracks represent samples from the GOM group, while blue tracks represent samples from the NOM group. (b) PDUI levels of LRRC25 (left) and UBXN8 (right) between the GOM and NOM groups. (c) Schematic diagram of primer design for lengthening and shortening APA isoforms. (d) qRT-PCR validations for lengthening and shortening APA isoforms of LRRC25 and UBXN8. Results are presented as the relative proportion of 3'UTR lengthening isoform in GDM compared with normal group. Significance levels: * p < 0.05, * * p < 0.01, * ** p < 0.001, the same applies to all figures.
3.2. The combined analysis of transcriptome and proteome revealed inconsistencies between gene transcription and translation levels in GDM adipose tissue
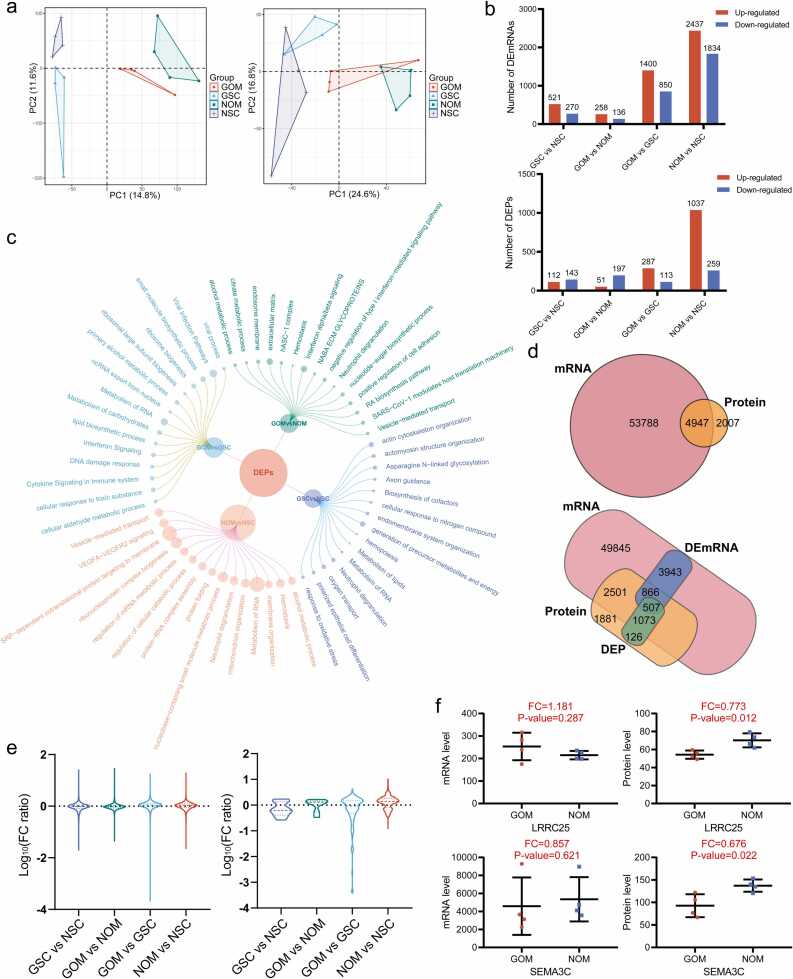
APA, as a post-transcriptional modification, can lead to inconsistencies between transcription and translation levels. Therefore, Proteomic profiling of the adipose tissue was conducted and compared with the transcriptome. The adipose tissue used in both proteomic and transcriptomic profiling was obtained from the same set of clinical samples, thus avoiding biases introduced by the sample source. A total of 58,735 mRNAs and 6954 proteins were identified. The PCA results demonstrate clear clustering of samples in both the transcriptome and proteome, indicating significant inter-group differences, particularly between different tissue sources (Fig. 3a). Additionally, PCA plots of samples from the GDM and normal groups originating from the same tissue source were also generated (Additional file 6: Fig. S3a, b). Differential analysis revealed numerous differentially expressed genes in both the transcriptome and proteome (|FC│≥1.2 and adj. P < 0.01 for DEmRNAs, │FC│≥1.2 and adj. P < 0.05 for DEPs) (Fig. 3b; Additional file 6: Fig. S3c-f; Additional file 7: Table S4). Notably, OM adipose tissue exhibited a higher number of up-regulated genes at both the mRNA and protein levels compared to SC adipose tissue. Moreover, the mRNA level of up-regulated genes was predominant in the GDM group, while down-regulated genes were more prominent at the protein level. Interestingly, there were more differentially expressed genes (DEmRNAs and DEPs) between SC and OM adipose tissues than those between GDM and normal adipose tissues. Subsequent functional enrichment analysis of differentially expressed genes at the protein level revealed enrichment in various functions associated with lipid synthesis, oxidative stress, and metabolism (Fig. 3c; Additional file 8: Table S5). Furthermore, we conducted a comparative analysis of transcriptome and proteome data, identifying 4947 genes that matched at both mRNA and protein levels. These mRNA-protein-matched genes comprised 2446 genes with significant differences at least at one level (mRNA or protein) and 2501 genes with no significant differences (Fig. 3d). Surprisingly, only 5.28% (261 out of 4947 genes) of these genes exhibited a strong positive correlation (r ≥ 0.5) between mRNA and protein expression levels (Additional file 9: Fig. S4a). Among the significantly different genes, only 20.7% (507 out of 2446 genes) were common differentially expressed genes (DEGs), showing significant differences at both the mRNA and protein levels. This finding suggests that a majority (90.5%, 4809 out of 5316 genes) of DEmRNAs did not propagate their significant differences to the protein level.
Fig. 3.
Comparative analyses of the transcriptome and proteome from different adipose groups. (a) PCA of the transcriptome (left) and proteome (right) clearly distinguished different groups. (b) Bar graphs showing the number of DEmRNAs (top) and DEPs (bottom) in different group comparisons. (c) Bubble chart displaying functional enrichment analysis of DEPs in different group comparisons. The size of the points represents the number of related genes. (d) Venn diagrams illustrating gene expression differences between the transcriptome and proteome. (e) Violin plots showing the distribution of FC ratios for non-common DEGs (left) and common DEGs (right) in different group comparisons. The y-axis values are represented as log10(FC ratio) because the FC ratio for some genes is extremely large. (f) mRNA and protein expression levels of two representative genes (LRRC25, SEMA3C) in the GOM and NOM groups indicate large differences between the transcriptome and proteome.
Furthermore, the fold changes of these common differentially expressed genes were examined and the protein-to-mRNA FC ratio for each mRNA-protein-matched gene was calculated and compiled. Interestingly, the non-common differentially expressed genes, namely mRNA-protein matched genes excluding the common differentially expressed genes, exhibited unimodal distributions with peak values close to 0 in all group comparisons (Fig. 3e left; Additional file 10: Table S6). However, for common differentially expressed genes, including 90 genes in the GOM vs GSC group and 315 genes in the NOM vs NSC group, the FC ratios displayed a bimodal distribution, with peaks were not close to 0. For the GSC vs NSC group and the GOM vs NOM group, a typical distribution pattern was not found due to the small number of common differentially expressed genes (10 and 7 genes, respectively) (Fig. 3e right; Additional file 10: Table S6). To illustrate this phenomenon, we present the sequencing results of two GDM-related genes, LRRC25 and SEMA3C, in the GOM and NOM groups (Fig. 3f). These two genes did not show differences at the mRNA level but exhibited significant down-regulation at the protein level in the GOM group. Notably, the APA regulation levels of both genes were significantly decreased. Similar findings were observed in other groups (Additional file 9: Fig. S4b-d). The above results highlight a significant inconsistency between the transcriptome and proteome of adipose tissue, characterized by a low proportion of common differentially expressed genes and substantial differences in expression trends. Furthermore, it is worth mentioning that there were significantly fewer common differentially expressed genes in the GSC vs NSC, and GOM vs NOM groups than in the GOM vs GSC, and NOM vs NSC groups. This suggests that in GDM, the differences between the transcriptome and proteome are more pronounced, and APA regulation may play a more significant role.
3.3. The 3'UTR elongation in APA events may lead to a reduction in protein translation
The decrease in APA regulation levels can lead to more mRNAs with 3'UTR lengthening isoform. Lengthening of the 3'UTR enhances the probability of miRNA binding, thereby down-regulating the translation levels of corresponding genes. This mechanism is considered one of the primary ways in which APA contributes to post-transcriptional regulation [28]. To explore the presence of this regulatory mechanism in adipose tissue, comprehensive analyses were performed on the GSC vs NSC and GOM vs NOM groups, revealing more significant differences in both the transcriptome and proteome. The results indicate that ΔPDUI is significantly correlated with both mRNA (OR = 1.994, P = 0.0052) and protein (OR = 0.4223, P = 0.0004) in the GOM vs NOM group (Fig. 4a). Particularly, within the mRNA-protein matched genes of the GOM vs NOM group, 3'UTR lengthening genes demonstrated high mRNA levels but lower protein levels. This aligns with the aforementioned APA regulatory mechanism, indicating that 3'UTR lengthening genes have reduced translation levels due to increased miRNA binding sites. Subsequently, these mRNA-protein matched genes, which conformed to the APA regulatory trends, underwent further screening. Functional enrichment analysis revealed that more than half of these genes (151 out of 269) are involved in the metabolic process (GO:0008152) (Additional file 11: Table S7.1). Further analysis indicated that these metabolism-related genes are primarily associated with three functional categories: energy metabolism (such as lipid biosynthetic process and acetyl-CoA metabolic process), immune regulation (such as inflammatory response and regulation of pattern recognition receptor signaling pathway), and nucleic acid and protein regulation (such as DNA repair and ubiquitin-dependent protein catabolic process) (Fig. 4b; Additional file 11: Table S7.2). These functions are all pivotal in the development of GDM.
Fig. 4.
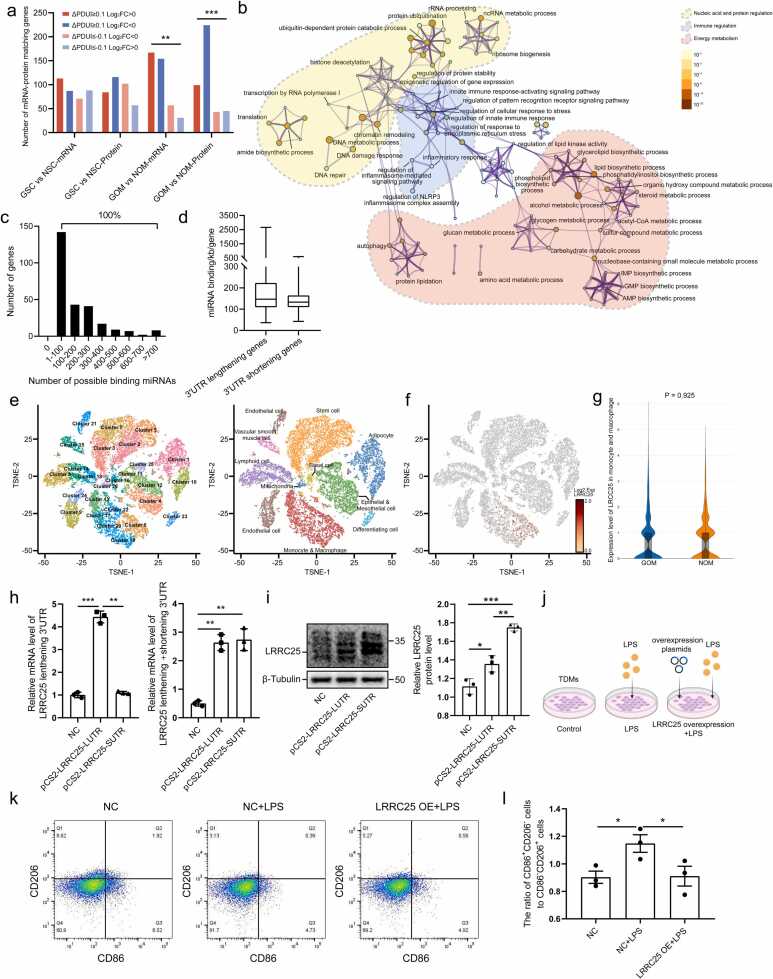
3'UTR elongation leads to reduced protein translation due to increased availability for miRNA binding. (a) Bar chart showing the distribution of mRNA-protein matched genes based on log2(FC) and PDUI values. The chi-squared test results indicate a significant correlation between PDUI and mRNA as well as protein levels in the GOM vs NOM group. (b) APA-regulated genes associated with metabolism further enriched in energy metabolism, immune regulation, and nucleic acids and proteins regulation. (c) The number of genes gained or lost potentially binding miRNAs due to APA regulation in the GOM vs NOM groups. (d) Calculated miRNA binding density per kb for 3’UTR lengthening and shortening genes in the GOM vs NOM groups. (e) tSNE projection of all 28513 sequenced cells in the GOM and NOM groups. Cells were categorized into 26 clusters (left) and further annotated into 11 cell types based on cell markers (right). (f-g) The predominant cell population for LRRC25 (f) and mRNA expression levels in the corresponding populations (g). (h) Expression levels of LRRC25 3'UTR lengthening and total isoforms detected by long and short primers indicate successful transfection of both overexpression plasmids with no significant differences in transfection efficiency. (i) Western blot results showing protein levels of LRRC25 in diferent groups, suggesting that the 3'UTR shortening isoform has higher protein translation efficiency. (j) LRRC25 functional validation schematic. Control group: no treatment; LPS-treated group: treated with 100 ng/ml LPS for 48 h; LRRC25 overexpression + LPS-treated group: transfected with LRRC25 overexpression plasmids for 48 h, then treated with 100 ng/ml LPS for 48 h. (k-l) Flow cytometry results showing that overexpressing LRRC25 reduced the ratio of pro-inflammatory phenotype (CD86+CD206-) to anti-inflammatory phenotype (CD86-CD206+) in response to LPS treatment.
Next, the number of potentially binding miRNAs gained or lost due to 3’UTR lengthening or shortening was calculated using DaPars and miRanda 3.3a [22]. The results showed that each genes gained or lost at least one potentially binding miRNA (Fig. 4c; Additional file 12: Table S8.1). To delve deeper into the significance of these findings, we consulted the Human microRNA Disease Database and discovered that nearly all GDM-related miRNAs documented in the database were encompassed by the heightened potentially binding miRNAs (100 out of 106 miRNAs; Additional file 12: Table S8.2). Furthermore, the Mann-Whitney test was conducted, which indicated that these 3'UTR lengthening genes tended to exhibit a higher miRNA binding density compared to 3'UTR shortening genes (P = 0.066) (Fig. 4d).
In the aforementioned experiments, differences in the APA regulation of LRRC25 between the GOM and NOM groups were observed, as well as inconsistent expression trends between the transcriptome and proteome. To further investigate the impact of APA events on protein translation, LRRC25 was chosen as a representative gene, and experimental validation was conducted. Initially, using snRNA-seq, the key cell types responsible for LRRC25 expression were identified(Fig. 4e; Additional file 13: Fig. S5). LRRC25 was primarily expressed in monocytes and macrophages (Fig. 4f), thus TMDs were selected for further experiments. Additionally, the snRNA-seq results revealed no significant differences in LRRC25 mRNA expression levels between the GOM and NOM groups, supporting the transcriptome results (Fig. 4g). Next, overexpression plasmids for both the 3'UTR lengthening and shortening isoforms were constructed and transfected into TMDs respectively (Additional file 2: Table S2). The LUTR plasmid group exhibited a higher proportion of lengthening 3'UTR compared to the SUTR plasmid group (Fig. 4 h, left), while the total mRNA levels showed no significant difference between the two groups, indicating similar transfection efficiencies (Fig. 4h, right). And the LUTR plasmid group was observed to have lower LRRC25 protein expression levels compared to the SUTR plasmid group (Fig. 4i). Finally, to validate the anti-inflammatory function of LRRC25, TDMs overexpressing LRRC25 were stimulated with LPS to induce their pro-inflammatory transformation(Fig. 4j). The results indicated that overexpression of LRRC25 can reduce the ratio of pro-inflammatory phenotype (CD86+CD206-) to anti-inflammatory phenotype (CD86-CD206+) in response to LPS treatment (Fig. 4k, l). In summary, these results demonstrate that APA regulation can serve as a post-transcriptional regulation mechanism affecting gene translation levels. In OM adipose tissue, genes with 3'UTR lengthening isoform have significantly lower protein levels than genes with 3'UTR shortening isoform.
3.4. CSTF3 and PPP1CB may play key roles in APA regulation in OM adipose tissue
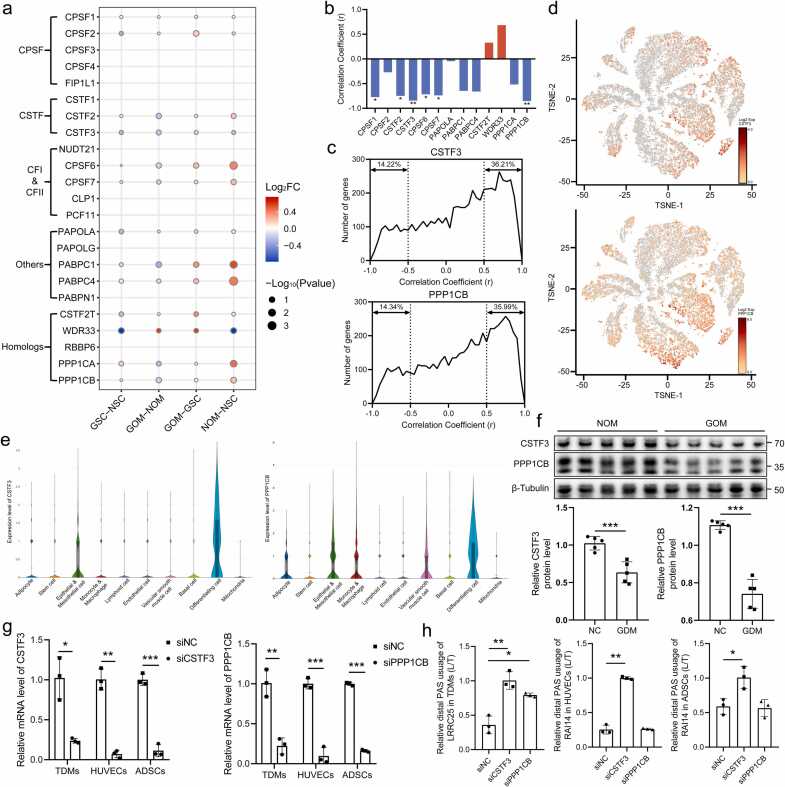
To explore the mechanisms underlying APA dynamics during the development of GDM, various trans-acting factors regulating APA were investigated, including cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CSTF), and cleavage factors I and II (CFI and CFII) [28]. The proteomic results suggest that most trans-acting factors in the GOM group had lower protein levels than those in the NOM group, consistent with the observed increase in 3'UTR lengthening genes in the GOM group (Fig. 5a). The correlation between the protein levels of trans-acting factors and the mean PDUI in OM adipose tissue was calculated to establish the correlation between trans-acting factors and APA. The results showed a significant negative correlation between most trans-acting factors and the mean PDUI values, with CSTF3 (r = −0.843, P = 0.0087) and PPP1CB (r = −0.852, P = 0.0072) showing the strongest correlations with PDUI (Fig. 5b). After establishing the relationship between trans-acting factors and APA, the correlation between trans-acting factors and protein levels was investigated. The correlation of protein levels between trans-acting factors (CSTF3 and PPP1CB) and other genes in OM adipose tissue was calculated. The results showed a skewed distribution of correlation coefficients, with 36.21% and 35.99% of genes displaying strong positive correlations (r > 0.5) with CSTF3 and PPP1CB, respectively (Fig. 5c). The snRNA-seq results demonstrated widespread distribution of CSTF3 and PPP1CB in OM adipose tissue. CSTF3 exhibited prominent expression in epithelial and endothelial cells, as well as differentiating cells. PPP1CB was enriched in adipocytes, epithelial and mesothelial cells, monocytes and macrophages, vascular smooth muscle cells, and differentiating cells (Fig. 5d, e). Western Blot validation confirmed down-regulation of CSTF3 and PPP1CB in the GOM group compared to the NOM group (Fig. 5f). Furthermore, to validate the role of CSTF3 and PPP1CB in APA regulation in different adipose tissue resident cells, CSTF3 and PPP1CB were knocked down in TDMs, HUVECs, and ADSCs (Fig. 5g). Subsequently, the 3'UTR lengthening and shortening isoforms of representative genes were detected (LRRC25 in TDMs and RAI14 in HUVECs and ADSCs). The results showed that knockdown of CSTF3 significantly increased the proportion of 3'UTR lengthening isoform in all three cell types, while knockdown of PPP1CB only functioned in TDMs (Fig. 5h), which suggests that CSTF3 may have a broader role in APA regulation compared to PPP1CB. To sum up, our findings suggest that the decreased level of APA-related regulatory proteins, represented by CSTF3 and PPP1CB, may contribute to reduced APA regulation in OM adipose tissue during the progression of GDM.
Fig. 5.
The decrease in APA-related trans-acting factors causes 3'UTR elongation and reduced protein translation. (a) Differential protein levels of APA-related trans-acting factors in different group comparisons, with 10 out of 23 factors not detected. (b) Pearson correlation coefficients between the protein levels of trans-acting factors and the mean PDUI in OM adipose tissues show a strong correlation between trans-acting factors and APA. (c) Pearson correlation coefficients of protein levels between trans-acting factors (CSTF3 and PPP1CB) and other genes in OM adipose tissue indicate a strong positive correlation between trans-acting factors and protein translation levels. (d) Cell population distribution of CSTF3 (top) and PPP1CB (bottom). (e) Expression levels of CSTF3 (top) and PPP1CB (bottom) in various cell populations. (f) Experimental verification of protein expression levels of CSTF3 and PPP1CB in OM adipose tissue. (g) The knockdown efficiency of CSTF3 and PPP1CB in TDMs, HUVECs and ADSCs. (h) qRT-PCR validations for lengthening and shortening APA isoforms of LRRC25 and RAI14. Results are presented as the relative proportion of 3'UTR lengthening isoforms in different groups.
Fig. S1 Distribution and functional enrichment of APA events across different groups. (a) Heatmap of dynamic APA events in adipose tissue. (b) PCA plots for samples originating from the same tissue source. (c) Scatter plots of mean PDUI values for genes in different group comparisons, with a cutoff value of ∆PDUI = ± 0.1. (d) Functional enrichment networks of significantly different APA events in each group comparison, with clusters of functions connected by edges. The top 20 clusters by P-value are shown.
Fig. S2 RNA-seq tracks and PDUI levels for representative genes in different group comparisons: GSC vs NSC group (a), NOM vs NSC group (b), and GOM vs GSC group (c).
Fig. S3 Distribution of genes in different group comparisons: PCA plots of mRNA (a) and protein (b) from the GDM and normal groups. DEmRNA expression heatmaps (c) and volcano plots (d), DEP expression heatmaps (e), and volcano plots (f). The cut-off values were │FC│≥ 1.2 and adj. P < 0.01 for DEmRNAs, │FC│≥ 1.2 and adj. P < 0.05 for DEPs.
Fig. S4 Gene distribution differences between the transcriptome and proteome. (a) Distribution of Pearson correlation coefficients for protein-mRNA matched genes with the proportion of genes above and below ± 0.5 indicated. (b-d) mRNA and protein expression levels of representative genes in the GSC vs NSC (b), NOM vs NSC (c), and GOM vs GSC (d) groups.
Fig. S5 Partial results from the snRNA-seq analysis: Distribution of cells from GOM and NOM groups in tSNE projection (a) and heatmap of cell markers for different cell populations (b).
Additional material information.
Additional file 1: Table S1 The clinic characteristics of GDM patients and controls.
Additional file 2: Table S2 The primers for qRT-PCR and overexpression vectors and the sequence of siRNA.
Additional file 3: Table S3 The dynamic APA events calculated by Dapars algorithm.
Additional file 4: Fig. S1 Distribution and functional enrichment of APA events across different groups.
Additional file 5: Fig. S2 RNA-seq tracks and PDUI levels for representative genes in different group comparisons.
Additional file 6: Fig. S3 Distribution of DEGs in different group comparisons.
Additional file 7: Table S4 The transcriptomics and proteomics data analysis in different group comparisons.
Additional file 8: Table S5 The functional enrichment analysis of DEPs in different group comparisons.
Additional file 9: Fig. S4 Gene distribution differences between the transcriptome and proteome.
Additional file 10: Table S6 The protein-per-mRNA FC ratio analysis for mRNA-protein matched genes in different group comparisons.
Additional file 11: Table S7 The GO biological process enrichment results for potentially APA-regulated genes.
Additional file 12: Table S8 Predicted binding miRNAs between proximal and distal PAS of mRNA-Protein matched genes and their relation with GDM.
Additional file 13: Fig. S5 Partial results from the snRNA-seq analysis.
4. Discussion
GDM, as one of the most common complications during pregnancy, poses significant risks to the short-term and long-term health of both mothers and fetuses [29], [30]. To advance the scientific comprehension of GDM pathogenesis and enhance its prevention and treatment strategies, it is imperative to delve deeper into its mechanisms. With the progression of sequencing technologies, researchers have endeavored to delineate the GDM landscape through RNA sequencing. However, it is worth noting that numerous genes exhibit conspicuous disparities between mRNA and protein levels. This highlights the insufficiency of relying solely on transcriptomic data for accurately characterizing biological specimens [31]. In light of this, we complemented the transcriptomic analysis with proteomic profiling and unveiled noteworthy differences between these two molecular levels. Specifically, we discovered that APA emerges as a vital post-transcriptional regulation mechanism during the progression of GDM. We observed a decrease in APA regulation within omental adipose tissue, leading to 3'UTR elongation in specific genes and an increased propensity for miRNA binding. Consequently, this alteration results in attenuated translation efficiency of these genes. Importantly, these APA-regulated genes are primarily involved in metabolic processes, exerting discernible effects on energy metabolism and inflammatory response within adipose tissue during the development of GDM.
In this study, we employed the DaPars algorithm to delve into the landscape of APA in adipose tissues from GDM patients and normal pregnant women. Our analysis unveiled a prevalent occurrence of APA across adipose tissues. Through meticulous comparisons across diverse groups, we elucidated two pivotal findings. First, during the progression of GDM, a notable attenuation in APA regulation surfaced, characterized by an heightened number of genes with 3'UTR lengthening in the GDM group compared to the normal group, regardless of the adipose tissue subtype (OM or SC). Second, the diminishing APA regulatory trend in GDM was uneven between the two adipose tissue types. Notably, the reduction in APA levels was more pronounced in OM adipose tissue than in SC adipose tissue. This distinction was starkly evident in the GDM group, where the number of significantly 3'UTR lengthening genes in OM adipose tissue nearly quadrupled that of SC adipose tissue. Conversely, in the normal group, the number of significantly 3'UTR lengthening genes in OM adipose tissue was similar to that in SC adipose tissue. These findings indicate that APA regulation exerts a more substantial influence on OM adipose tissue during GDM. These observations align with previous studies that have also highlighted the tissue-specific and disease-specific aspects of APA regulation. For instance, genes in testes, ovaries, embryonic stem cells, and tumors tend to exhibit short 3'UTRs, whereas neurons predominantly express genes with long 3'UTRs [14], [32]. The alterations in APA regulation levels observed in this study imply that APA plays a crucial role in GDM, constituting the primary focus of our study.
In this study, we investigated the transcriptomic and proteomic results of adipose tissue from different groups. Interestingly, we observed a relatively low concordance in the expression trends and levels of DEmRNAs and DEPs, particularly in the comparisons between the GDM and normal groups. To understand this phenomenon, we analyzed APA regulation levels and found that ΔPDUI was significantly correlated with both mRNA and protein levels. However, the correlation trends exhibited an inverse relationship. Specifically, genes with lengthening 3'UTRs exhibited higher mRNA levels but lower protein translation levels. This phenomenon may be attributed to the elongation of 3'UTRs, which increases the probability of miRNA binding, consequently leading to translational suppression. This regulatory mechanism partially explains the substantial differences observed between the transcriptome and proteome. This mechanism has been corroborated in other studies as well. For instance, APA regulation of ZFR alters miR-579's negative feedback regulation of this gene [33]. miRNA-200a downregulates PTEN expression by binding to its 3'UTR, promoting ovarian cancer cells invasion [34]. In this study, both miRNA binding prediction and experimental validation supported this regulatory mechanism.
The in-depth validation of LRRC25, a gene associated with anti-inflammation, has provided us with valuable insights into the role of APA regulation in the progression of GDM. Inflammation plays a pivotal role in insulin resistance and diabetes, and our multi-omics data suggested that the increased proportion of the 3'UTR lengthening isoform of LRRC25 is responsible for its unchanged mRNA levels but significantly reduced protein levels in GDM. To bolster this hypothesis, we conducted transfections using plasmids containing the lengthening or shortening 3'UTR isoform of LRRC25. Our results confirmed that the increased proportion of the lengthening isoform led to reduced LRRC25 protein levels. LRRC25 belongs to the leucine-rich repeat (LRR)-containing protein family and has been reported to interact with the Rel homology domain of p65/RelA, thereby inhibiting the NF-κB signaling pathway-mediated inflammatory response [35]. Similarly, in this study, TMDs overexpressing LRRC25 displayed a lower pro-inflammatory macrophages (CD86+CD206-) to anti-inflammatory macrophages (CD86-CD206+) ratio after LPS treatment, indicating a dampened inflammatory response. NF-κB is closely associated with the chronic low-grade inflammation induced by obesity, which contributes to the development of insulin resistance [36]. We propose that the decreased LRRC25 protein levels in OM adipose tissue hinder the effective inhibition of the NF-κB signaling pathway, leading to the establishment of an inflammatory environment and promoting the onset of GDM. It's worth noting that, in Feng et al.'s study, TMDs did not exhibit changes in LRRC25 mRNA levels after LPS treatment, but protein levels were altered [35]. This suggests that LRRC25 regulation occurs through post-transcriptional regulation mechanisms, and in line with our findings, this post-transcriptional regulation mechanism is likely related to APA regulation.
Our investigation into LRRC25 unveiled a potential mechanism by which APA may influence inflammation levels. Furthermore, in the functional enrichment analysis of genes regulated by APA, we observed that more than half of these genes were associated with metabolic processes. These metabolism-related genes were further clustered in processes related to energy metabolism, immune responses, and the regulation of nucleic acids and proteins. These findings are particularly relevant considering two prominent characteristics of GDM, namely metabolic dysfunction and low-grade chronic inflammation. In our analysis, we observed enriched functions related to lipid biosynthetic processes, inflammatory responses, and DNA repair. Lipid biosynthesis is crucial for adipocyte development and function. Impaired lipid synthesis, exemplified by the loss of ACLY (ATP citrate lyase) in adipocytes, can lead to reduced fat mass, hepatic lipid accumulation, insulin resistance, and other metabolic disturbances [37]. Similarly, our study identified that ACLY is regulated by APA and decreased in GDM. Another gene regulated by APA that we investigated was ASCC2, a component of the ASCC complex known for its role in DNA damage repair [38]. In this study, ASCC2 was found to be reduced in GDM, indicating a weakened capacity for DNA repair. The chronic inflammation characteristic of GDM contributes to oxidative stress, leading to sustained DNA damage and a concomitant decrease in DNA repair capacity [39]. This phenomenon further exacerbates the compromised equilibrium of energy metabolism observed in GDM [40]. Overall, our findings suggest that APA-regulated genes may wield substantial influence over the onset and progression of GDM.
In addition to investigating the role of APA regulation in GDM adipose tissue, we delved into the factors responsible for changes in APA regulation levels. We identified a widespread decrease in APA-related trans-acting factors in OM adipose tissue of GDM patients. Notably, CSTF3 and PPP1CB displayed robust associations with ΔPDUI and protein levels in GDM and were widely distributed across various cell types in OM adipose tissue. Previous research has reported on these two molecules, which target specific base sequences at the 3'UTR end, thereby influencing the processing of specific poly(A) sites [41]. Studies have shown that elevated levels of trans-acting factors correlate with increased 3'UTR shortening events in various types of tumors [32], [42]. Consistent with this, we found that knocking down CSTF3 and PPP1CB in cells can lead to varying degrees of 3'UTR elongation. Furthermore, it was observed that CSTF3 affects a wider range of cell types compared to PPP1CB, which may be attributed to CSTF3 being the primary component of APA regulation, while PPP1CB is just one of the homologs with substitutability [43].
Our findings suggest that the decline in CSTF3 and PPP1CB levels in GDM is likely a key reason for the increase in 3'UTR lengthening genes and the decrease in protein expression levels. These findings suggest that changes in trans-acting factors such as CSTF3 and PPP1CB may be one of the primary drivers of the altered APA landscape observed in GDM.
This study does have certain limitations that should be acknowledged. First, the sample size of enrolled patients was relatively small. To enhance the statistical power and generalizability of our findings, it is recommended to enroll a larger cohort of GDM patients and normal late-term pregnant individuals in future studies. Second, although our study identified the significance of CSTF3 and PPP1CB in APA regulation in GDM, we did not elucidate the detailed molecular mechanisms underlying their down-regulation. Further investigation is needed to uncover the specific pathways and molecular interactions involved in the regulation of CSTF3 and PPP1CB in GDM adipose tissue.
5. Conclusion
This study represents a pioneering effort in unraveling the dysregulation of APA in GDM adipose tissue, integrating multi-omics analysis with experimental validation. Our findings offer a comprehensive understanding of APA regulation's pivotal role in GDM adipose tissue. Through an in-depth exploration of the transcriptome, proteome, and APA regulation levels in OM and SC adipose tissue, we discovered a systemic decline in APA levels in OM adipose tissue, leading to diminished protein translation. This dysregulation predominantly affects metabolism, consequently influencing energy metabolism and inflammation and contributing to the development of GDM. In summary, our study provides valuable insights into the underlying mechanisms of GDM pathophysiology and paves the way for future investigations aimed at improving clinical care for individuals with GDM.
Ethics approval and consent to participate
The study was approved by the First Affiliated Hospital Ethics Committee of Chongqing Medical University. Informed consent was obtained from all participants included in the study.
Consent for publication
Not applicable.
Funding
This work was supported by the Innovation Project for Doctoral Students at the First Affiliated Hospital of Chongqing Medical University (CYYY-BSYJSCXXM-202210), National Natural Science Foundation of China (81971406, 82271715), the 111 Project (Yuwaizhuan (2016)32), Chongqing Science & Technology Commission (cstc2021jcyj-msxmX0213), Natural Science Foundation of Chongqing, China (CSTB2022NSCQ-MSX1679), Chongqing Municipal Education Commission (KJZD-K202100407), Chongqing Health Commission and Chongqing Science & Technology Commission (2021MSXM121).
CRediT authorship contribution statement
Bingnan Chen: Conceptualization, Data curation, Formal analysis, Writing - original draft. Xuyang Chen: Validation, Methodology. Ruohan Hu: Validation, Methodology, Writing - review & editing. Hongli Li: Validation, Methodology. Min Wang: Validation. Linwei Zhou: Validation. Hao Chen: Validation. JianqiWang: Writing - review & editing. Hanwen Zhang: Writing - review & editing. Xiaobo Zhou: Conceptualization, Project administration, Supervision, Writing - review & editing. Hua Zhang: Conceptualization, Funding acquisition, Project administration, Supervision, Writing - review & editing.
Declaration of Competing Interest
None.
Acknowledgments
We would like to express our heartfelt gratitude to all the patients who generously contributed their clinical samples to this research. Thank the Chongqing Cell Resource Library for providing primary human ADSCs.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.03.013.
Contributor Information
Xiaobo Zhou, Email: xiaobo_zhou@cqmu.edu.cn.
Hua Zhang, Email: zh2844@gmail.com.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Data availability
Most of the data generated or analysed during this study are included in this published article and its supplementary information files. The sequencing raw data used and analysed during the current study are available from the corresponding author on reasonable request.
References
- 1.Lende M., Rijhsinghani A. Gestational diabetes: overview with emphasis on medical management. Int J Environ Res Public Health. 2020;17(24):9573. doi: 10.3390/ijerph17249573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sweeting A., Wong J., Murphy H.R., Ross G.P. A clinical update on gestational diabetes mellitus. Endocr Rev. 2022;43(5):763–793. doi: 10.1210/endrev/bnac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Zheng J., Shi W., Du N., Xu X., Zhang Y., et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut. 2018;67(9):1614–1625. doi: 10.1136/gutjnl-2018-315988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed B., Sultana R., Greene M.W. Adipose tissue and insulin resistance in obese. Biomed Pharm. 2021;137 doi: 10.1016/j.biopha.2021.111315. [DOI] [PubMed] [Google Scholar]
- 5.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiol Rev. 2018;98(4):2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agbu P., Carthew R.W. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat Rev Mol Cell Biol. 2021;22(6):425–438. doi: 10.1038/s41580-021-00354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolicka D., Sobolewski C., Correia de Sousa M., Gjorgjieva M., Foti M. mRNA Post-Transcriptional Regulation by AU-Rich element-binding proteins in liver inflammation and cancer. Int J Mol Sci. 2020;21(18):6648. doi: 10.3390/ijms21186648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makita S., Takatori H., Nakajima H. Post-transcriptional regulation of immune responses and inflammatory diseases by RNA-Binding ZFP36 family proteins. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.711633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beilerli A., Kudriashov V., Sufianov A., Kostin A., Begliarzade S., Ilyasova T., et al. Regulation and mechanism of action of miRNAs on insulin resistance in skeletal muscles. Noncoding RNA Res. 2023;8(2):218–223. doi: 10.1016/j.ncrna.2023.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu T., Wu C., Yao X., Han Q., Wang N., Yuan W., et al. AS3MT facilitates NLRP3 inflammasome activation by m(6)A modification during arsenic-induced hepatic insulin resistance. Cell Biol Toxicol. 2023;39(5):2165–2181. doi: 10.1007/s10565-022-09703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X., Lin J., Zhang Y., Kang S., Belkin N., Wara A.K., et al. MicroRNA-181b improves glucose homeostasis and insulin sensitivity by regulating endothelial function in white adipose tissue. Circ Res. 2016;118(5):810–821. doi: 10.1161/CIRCRESAHA.115.308166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang P., Yang Y., Li G., Huang L., Wen M., Ruan W., et al. Alternative polyadenylation by sequential activation of distal and proximal PolyA sites. Nat Struct Mol Biol. 2022;29(1):21–31. doi: 10.1038/s41594-021-00709-z. [DOI] [PubMed] [Google Scholar]
- 13.Tian B., Manley J.L. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18(1):18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber A.J., Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20(10):599–614. doi: 10.1038/s41576-019-0145-z. [DOI] [PubMed] [Google Scholar]
- 15.Dong W., Liu X., Yang C., Wang D., Xue Y., Ruan X., et al. Glioma glycolipid metabolism: MSI2-SNORD12B-FIP1L1-ZBTB4 feedback loop as a potential treatment target. Clin Transl Med. 2021;11(5) doi: 10.1002/ctm2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng T., Tan Y., Qiu J., Xie Z., Hu X., Zhang J., et al. Alternative polyadenylation trans-factor FIP1 exacerbates UUO/IRI-induced kidney injury and contributes to AKI-CKD transition via ROS-NLRP3 axis. Cell Death Dis. 2021;12(6):512. doi: 10.1038/s41419-021-03751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ICD-11 for Mortality and Morbidity Statistics. https://icd.who.int/browse11/l-m/en#/http%3a%2f%2fid.who.int%2ficd%2fentity%2f1320503631. Accessed 2 November 2023.
- 18.10x Genomics. https://www.10xgenomics.com. Accessed 2 November 2023.
- 19.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao T., Zhan D., Qu S., Jiang S., Gan W., Qin W., et al. Transcriptomics-proteomics Integration reveals alternative polyadenylation driving inflammation-related protein translation in patients with diabetic nephropathy. J Transl Med. 2023;21(1):86. doi: 10.1186/s12967-023-03934-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1) doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Z., Shi J., Gao Y., Cui C., Zhang S., Li J., et al. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019;47(D1):D1013–D1017. doi: 10.1093/nar/gky1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafez D., Ni T., Mukherjee S., Zhu J., Ohler U. Genome-wide identification and predictive modeling of tissue-specific alternative polyadenylation. Bioinformatics. 2013;29(13):i108–i116. doi: 10.1093/bioinformatics/btt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Liu L., Qiu Q., Zhou Q., Ding J., Lu Y., et al. Alternative polyadenylation: methods, mechanism, function, and role in cancer. J Exp Clin Cancer Res. 2021;40(1):51. doi: 10.1186/s13046-021-01852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Billionnet C., Mitanchez D., Weill A., Nizard J., Alla F., Hartemann A., et al. Gestational diabetes and adverse perinatal outcomes from 716,152 births in France in 2012. Diabetologia. 2017;60(4):636–644. doi: 10.1007/s00125-017-4206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damm P., Houshmand-Oeregaard A., Kelstrup L., Lauenborg J., Mathiesen E.R., Clausen T.D. Gestational diabetes mellitus and long-term consequences for mother and offspring: a view from Denmark. Diabetologia. 2016;59(7):1396–1399. doi: 10.1007/s00125-016-3985-5. [DOI] [PubMed] [Google Scholar]
- 31.Chua B.A., Van Der Werf I., Jamieson C., Signer R.A.J. Post-transcriptional regulation of homeostatic, stressed, and malignant stem cells. Cell Stem Cell. 2020;26(2):138–159. doi: 10.1016/j.stem.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia Z., Donehower L.A., Cooper T.A., Neilson J.R., Wheeler D.A., Wagner E.J., et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3'-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinske L.C., Galante P.A., Limbeck E., Mohnle P., Parmigiani R.B., Ohno-Machado L., et al. Alternative polyadenylation allows differential negative feedback of human miRNA miR-579 on its host gene ZFR. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0121507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang J.H., Lv Q.Y., Yi Y.X., Liao J., Wang X.W., Zhang W. MicroRNA-200a promotes proliferation and invasion of ovarian cancer cells by targeting PTEN. Eur Rev Med Pharm Sci. 2018;22(19):6260–6267. doi: 10.26355/eurrev_201810_16033. [DOI] [PubMed] [Google Scholar]
- 35.Feng Y., Duan T., Du Y., Jin S., Wang M., Cui J., et al. LRRC25 Functions as an Inhibitor of NF-kappaB Signaling Pathway by Promoting p65/RelA for autophagic degradation. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-12573-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catrysse L., van Loo G. Inflammation and the metabolic syndrome: the tissue-specific functions of NF-kappaB. Trends Cell Biol. 2017;27(6):417–429. doi: 10.1016/j.tcb.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez S., Viola J.M., Torres A., Wallace M., Trefely S., Zhao S., et al. Adipocyte ACLY facilitates dietary carbohydrate handling to maintain metabolic homeostasis in females. Cell Rep. 2019;27(9):2772–2784. doi: 10.1016/j.celrep.2019.04.112. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brickner J.R., Soll J.M., Lombardi P.M., Vagbo C.B., Mudge M.C., Oyeniran C., et al. A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair. Nature. 2017;551(7680):389–393. doi: 10.1038/nature24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pacal L., Varvarovska J., Rusavy Z., Lacigova S., Stetina R., Racek J., et al. Parameters of oxidative stress, DNA damage and DNA repair in type 1 and type 2 diabetes mellitus. Arch Physiol Biochem. 2011;117(4):222–230. doi: 10.3109/13813455.2010.551135. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu I., Yoshida Y., Suda M., Minamino T. DNA damage response and metabolic disease. Cell Metab. 2014;20(6):967–977. doi: 10.1016/j.cmet.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., et al. Systematic profiling of poly(A)+ transcripts modulated by core 3' end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 2015;11(4) doi: 10.1371/journal.pgen.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miles W.O., Lembo A., Volorio A., Brachtel E., Tian B., Sgroi D., et al. Alternative Polyadenylation in Triple-Negative Breast Tumors Allows NRAS and c-JUN to Bypass PUMILIO Posttranscriptional Regulation. Cancer Res. 2016;76(24):7231–7241. doi: 10.1158/0008-5472.CAN-16-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elkon R., Ugalde A.P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
Most of the data generated or analysed during this study are included in this published article and its supplementary information files. The sequencing raw data used and analysed during the current study are available from the corresponding author on reasonable request.